Abstract
Aim of the study
The ability of immune cells in peripheral blood to produce certain cytokines affects tumour-elicited inflammation. The aim of this study was to investigate the gene expression of interleukin 12A (IL-12A), IL-12B, IL-23A, IL-10, IL-6, transforming growth factor β (TGF-β), HDAC3, and iNOS in peripheral blood mononuclear cells (PBMC) from colorectal cancer (CRC) patients.
Material and methods
The venous blood for PBMC isolation was collected preoperatively and 10 days after surgery, from CRC patients. After isolation of total RNA and synthesis of cDNA, quantitative real-time PCR assays were performed.
Results
Our results demonstrated that among investigated cytokine genes IL-10 and TGF-β were significantly upregulated in patients with CRC compared to the control group, while the expression of IL-23 mRNA was significantly decreased in CRC patients. We observed significantly increased mRNA levels in CRC patients’ PBMC before surgery for IL-10 and TGF-β compared to both postoperative and control groups. We also found a significant upregulation of iNOS in early compared to advanced CRC.
Conclusions
Based on the results we can assume that PBMC gene expression programming in CRC patients drives local differentiation of Th cells towards Treg instead of the Th1 anti-tumour subpopulation.
Keywords: mRNA, cytokine, qPCR, HDAC3, iNOS, Treg, CRC
Introduction
Colorectal cancer follows definitive genetic changes including, at least, the adenomatous polyposis coli (APC) tumour suppressor gene and mutations in K-Ras oncogene, providing the reproduction of tumour cells. Genome instability and epigenetic alteration ensure on certain mutant genotypes a selective advantage for tumour growth and present the first enabling hallmark of cancer [1, 2]. The second enabling characteristic involves the inflammatory state of premalignant and frankly malignant lesions, which is driven by cells of the immune system, some of which serve to promote tumour progression through various means [3–5]. Inflammation is likely to be involved with both forms of sporadic as well as heritable colon cancer. The “cancer-related inflammation” as characteristic of tumour illustrates the remarkable progress made in the last decade to describe the role of inflammation in promoting cancer development. The inflammation promotes cancer progression and accomplishes the full malignant phenotype, such as tumour tissue remodelling, angiogenesis, metastasis, and the suppression of the innate anticancer immune response [6, 7]. A specific cause-effect relationship between colorectal cancer angiogenesis and the upregulation of the inducible NOS (iNOS) gene in the tumour specimens has been recently demonstrated [8]. Moreover, the iNOS upregulation requires induction in response to proinflammatory cytokines. Several studies support the concept that histone deacetylase (HDAC) is responsible for transcriptional induction of proinflammatory cytokines and modulation of antigen-presenting cell activity [9, 10].
The presence of distinct cell types of the innate and adaptive immune system in neoplastic lesion is a marker for local antitumoral responses and tumour-elicited inflammation. Additionally, innate and specific immune cells can release different cytokines, responsible for pro- and anti-tumoural effect [11]. Tumour-infiltrating immune cells and cytokine-related signalling pathways are critical components in colorectal cancer initiation and progression. The mixture of cytokines that is locally and systemically produced can either block or facilitate tumour growth [7, 11, 12]. Most intratumoural immune cells are recruited from peripheral blood. Whether they are already programmed or can change their functions after falling into tumour microenvironment remains elusive. The ability of immune cells in peripheral blood to produce certain cytokines and possibility for their reprograming after entering into a tumour is a key feature for tumour growth or immune destruction. Previously, altered gene expression profiles of cytokines at mRNA level in CRC monocytes after in vitro stimulation were demonstrated, and this was associated with tumour development [13].
In view of the above facts, the aim of the present study was to investigate the gene expression of interleukin (IL)-12A, IL-12B, IL-23A, IL-10, IL-6, and TGF-β in peripheral blood mononuclear cells from CRC patients. We also studied the mRNA expression of inducible NO synthase (iNOS) and hystone deacetylase-3 (HDAC3) in CRC patient’s blood cells.
Material and methods
Subject
A group of 19 Bulgarian patients with CRC, who underwent surgical resection of a tumour, were included in the study. Cases with new diagnosis of CRC attending the University Hospital and St. Ivan Rilsky Hospital in Stara Zagora, Bulgaria between October 2009 and November 2011 were selected. The histopathological examination confirmed the diagnosis of cancer. The mean age of the total group of CRC patients was 63.75 ±7.68 years. Tumour grading and staging was performed according to the tumour–node–metastasis (TNM) classification. Ten patients were with early, non-metastatic (stage I and II), and 9 were advanced, metastatic (stage III and IV) CRC.
With the consent of the Local Ethics Board, peripheral venous blood (3 ml) was collected one day before surgery (preoperative) and 10 days after surgery (postoperative) from CRC patients and from 22 healthy donors, in sterile tubes with EDTA, and 1 ml was used for PBMC isolation immediately.
Criteria for eligible patients included no history of prior surgery for colon or rectal tumours, and no known hereditary cancer, ulcerative colitis, or Crohn’s disease. All patients and healthy volunteers gave informed consent for this research.
Peripheral blood mononuclear cells isolation
Peripheral blood mononuclear cells (PBMC) were isolated by Histopaque-1077 density gradient centrifugation. The interface containing PBMC was harvested and washed twice with cold RPMI-1640 medium.
RNA extraction
Total RNA was isolated from PBMC using innuPREP blood RNA isolation kit AJ Roboscreen (Leipzig; Germany) with the additional step of treatment with DNase I to remove traces of genomic DNA. The total RNA was quantified by spectrophotometric analysis. To remove traces of genomic DNA, total RNAs (1 mg) were treated with RNase-free DNase I following the manufacture’s instruction.
Reverse transcription
Synthesis of cDNA was performed manually according to the manufacturer’s instructions with a High-Capacity cDNA Archive kit (Applied Biosystems, USA) that used random primers and MutliScribe TM MuLV reverse transcriptase enzyme. Incubation conditions for reverse transcription was 10 minutes at 25°C followed by 2 hours at 37°C and was performed on a GeneAmp PCR System 9700 (Applied Biosystems, USA).
Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The following validated PCR primers and TaqMan MGB probes (6FAM-labeled) were used: IL-12B (assay ID: Hs00233688_m1), IL-12A (Hs00168405_m1), IL-23A (Hs00372324_m1), IL-10 (Hs00174086_m1), IL-6 (NM_000600), TGFβ1 (NM_000660), iNOS (Hs00167248_m1), and HDAC3 (Hs00187320_m1), Primerdesign, UK. Eukaryotic 18S ribosomal RNA (Hs99999903_m1) was used as an endogenous control.
An aliquot of 5 µl of the RT reaction was amplified in duplicate in a final volume of 20 µl using a TaqMan Universal PCR Master Mix and Gene Expression Assay mix, containing specific forward and reverse primers and labelled probes for target genes and endogenous control (Applied Biosystems, USA). The thermocycling conditions were: initial 10-minute incubation at 95°C followed by 40 cycles of denaturation for 15 seconds at 95°C and annealing/extension for 1 minute at 60°C. PCR data were collected with Sequence Detection System (SDS) software, version 1.3.1.
Statistical analysis
Relative quantitative evaluation of cytokine mRNAs was performed by the comparative ∆∆Ct method. The mean ∆Ct obtained in PBMC of healthy donors for each cytokine mRNA was used as a calibrator after normalisation to endogenous control-18S rRNA. The results are presented as an n-fold difference relative to the calibrator (RQ = 2-∆∆Ct).
The data were expressed as mean and standard deviation (SD). Student’s t-test was used to determine the statistical differences between mean values. Differences were considered significant when the p value was less than 0.05.
Results
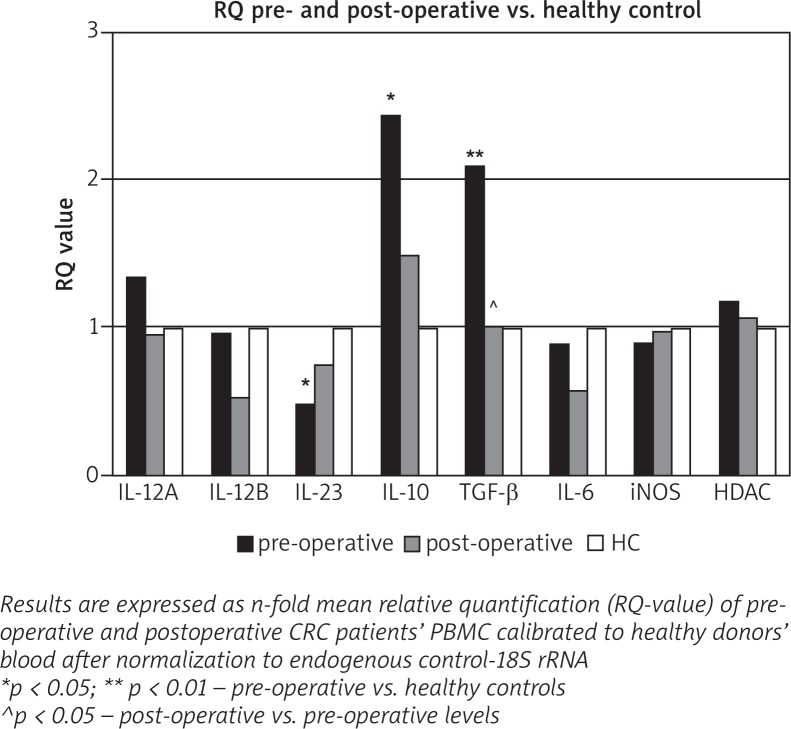
We investigated the expression of cytokine genes IL-12A, IL-12B, IL-23A, IL-17, IL-10, IL-6, and TGF-β in PBMC from patients before surgery (preoperative) and 10 days after surgery (postoperative) in the same patients, and the results were compared to the data taken from normal healthy volunteers. The results are presented in Fig. 1.
Fig. 1.
Relative quantity of cytokine gene expression in PBMC before and 10 days after surgery of CRC patients and healthy controls
We observed significantly increased mRNA in CRC patients’ blood before surgery for IL-10 and TGF-β (p = 0.034 and p = 0.0048, respectively) in comparison with healthy donors. When compared data from pre- and post-operative blood, significantly decreased mRNA quantity postoperatively was detected for TGF-β alone.
Our results demonstrated that among investigated cytokine genes IL-10 and TGF-β were significantly upregulated in patients with CRC compared to the control group, while the expression of IL23 mRNA was significantly decreased in CRC patients. Although the expression of IL-23A was downregulated, the other gene of IL-23 cytokine for subunit IL-12p40, IL-12B was not suppressed in preoperative blood compared to healthy donors.
iNOS and HDAC3 mRNA levels were similar in preoperative and postoperative CRC patients compared to healthy donors’ blood.
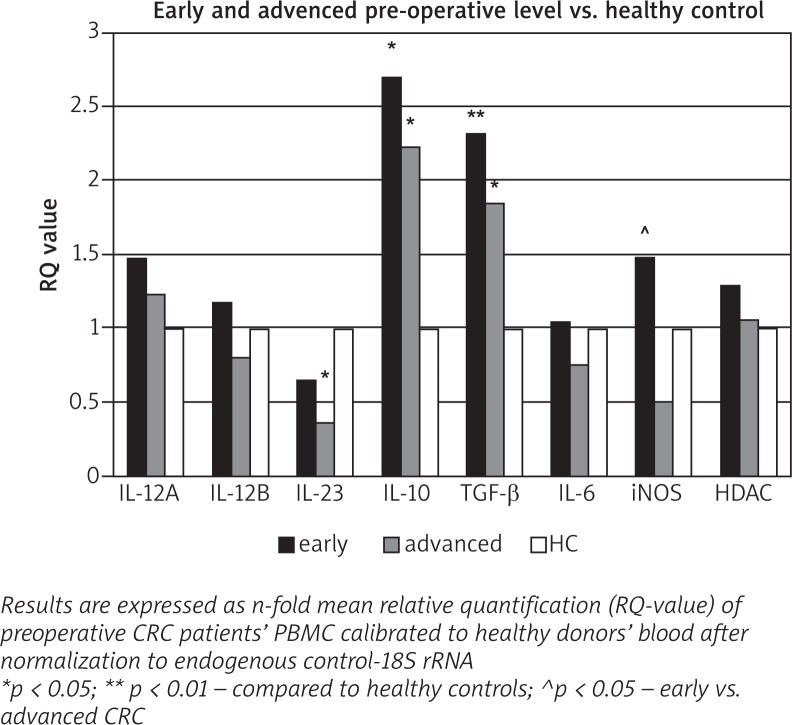
The results for the expression of the investigated genes in CRC preoperative blood after stratification of patients in early and advanced stages are presented as dCt in Table 1 and as RQ in Fig. 2. We showed that IL-10 and TGF-β genes were significantly upregulated in early cases as well as in advanced cases of CRC, compared to the control group. The expression of IL23A gene was significantly decreased in advanced CRC alone. Slightly enhanced IL-12A and IL-12B mRNA quantities in CRC early stages without statistical significance were found.
Table 1.
Gene expression levels in pre-operative PBMC of patients with early and advanced CRC and healthy controls
| Target gene | Average dCt ± SD | RQ | ||||
|---|---|---|---|---|---|---|
| Early CRC | Advanced CRC | Healthy controls | Early vs. advanced | Early vs. HC | Advanced vs. HC | |
| IL-12A | 16.25 ±1.7 | 16.5 ±1.2 | 16.8 ±0.93 | 1.27 p = 0.76 |
1.46 p = 0.26 |
1.15 p = 0.48 |
| IL-12B | 21.2 ±2.2 | 21.75 ±1.5 | 21.43 ±1.06 | 1.69 p = 0.59 |
1.17 p = 0.69 |
0.69 p = 0.52 |
| IL-23 | 15.17 ±1.73 | 16.01 ±1.4 | 14.55 ±0.86 | 2.01 p = 0.34 |
0.65 p = 0.19 |
0.32 p = 0.00163 |
| IL-10 | 15.44 ±2.1 | 15.72 ±1.98 | 16.87 ±1.65 | 1.13 p = 0.8 |
2.69 p = 0.064 |
2.38 p = 0.127 |
| TGF-β | 15.57 ±0.93 | 15.9 ±1 | 16.78 ±0.74 | 1.26 p = 0.56 |
2.31
p = 0.0059 |
1.84
p = 0.049 |
| IL6 | 23.33 ±1.67 | 23.8 ±2.18 | 23.39 ±0.97 | 1.43 p = 0.67 |
1.04 p = 0.93 |
0.73 p = 0.58 |
| iNOS | 27.61 ±2.36 | 29.17 ±1.55 | 28.18 ±0.96 |
2.95
p = 0.053 |
1.48 p = 0.43 |
0.503
p = 0.064 |
| HDAC3 | 17.02 ±0.78 | 17.3 ±0.95 | 17.39 ±0.45 | 1.214 p = 0.95 |
1.292; p = 0.202 |
1.06; p = 0.776 |
Fig. 2.
Relative quantity of cytokine gene expression in pre-operative PBMC of patients with early and advanced of CRC and healthy controls
Overall analysis showed that the expression at mRNA level of investigated IL-12A, IL-12B, and IL-6 genes in patients’ blood did not alter depending on development of colorectal cancer.
Although the expression of both iNOS and HDAC3 in early CRC blood was enhanced, statistically significant differences were found only for iNOS between patients with early and advanced stages. It should be noted that our study is limited by a relatively small number of early and advanced cases, and these results must be interpreted with caution and need to be confirmed in larger studies.
Discussion
In this prospective study, we aimed to show that CRC is causally involved in induction of aberrant systemic cytokine expression, and to clarify which cytokine genes from PBMC are involved. mRNA levels of expression of following cytokines were investigated: IL-6, IL-12A, IL-12B, IL-23A, IL-10, and TGF-β. We selected this panel of cytokines considering the available data that demonstrated their involvement in differentiation of Th1, Th17, and Treg subpopulations. Th17 and Treg cells are known to promote tumour development, while Th1 is mainly responsible for anti-tumour immune response.
Colorectal cancer-affected tissue is invaded by immune cells from the host, suggesting that the activity of these cells may have a potential impact upon cancer development. Within the tumour mass the huge infiltration of white blood cells, as pivotal players in the tumour microenvironment, is well documented [14, 15].
A critical role of inflammation in colon tumorigenesis, including initiation, promotion, progression, and metastasis has been shown [7, 11]. Colorectal cancer mucosa-invaded immune cells synthesised large numbers of cytokines, which drive inflammation and have a huge impact upon both cancer development and anti-tumour immune response. The molecular signature of tumour-infiltrated immune cells includes the gene expression pattern for cytokines that they produced. Thus far, a strong association has been shown between CRC development and changes of cytokine gene expression in tumour mucosa [16–18]. Recently, data have showed that colorectal cancer progression was closely associated with infiltration of Th17 inflammatory cells, and the main cytokines related with their differentiation and function are IL-6, IL-23, TGF-β, and IL-17 [19, 20]. Moreover, tumour-infiltrating inflammatory mononuclear cells are the main source of iNOS in tumour specimens, which upregulate gene expression correlated with tumour progression and mediate blood supply in early CRC [8, 21]. Although several transcription factors are implicated in cytokine gene transcription, HDACs have been described as key regulators of cytokine expression [9, 22].
Apart from local tumour-elicited inflammation, cancer patients frequently present with changes in numerous systemic parameters, used as cancer biomarkers. These include changes in peripheral immune cell number and alternations in the level of serum inflammatory cytokines [23, 24]. Here, we demonstrated a well expressed upregulation of IL-10 and TGF-β cytokine in PBMC of CRC patients (preoperative blood) compared to healthy individuals. In addition, eradication of tumour (post-operative blood) leads to significantly decreased expression of IL-10 and TGF-β. These findings suggest that the tumour induces aberrant changes in gene expression, and indicate that levels of cytokine gene expression are associated with the presence of CRC. However, it is unclear whether these changes in gene expression reflect the anti-cancer immune response or the cancer itself.
The TGF-b signalling pathway plays an important role in controlling cell proliferation and differentiation involved in colorectal carcinogenesis. In addition, there is evidence that excess production and/or activation of TGF-b by cancer cells can contributed to the tumour progression by paracrine mechanisms involving neoangiogenesis, production of stroma and proteases, and subversion of immune surveillance mechanisms in tumour hosts [25]. Moreover, the activity of TGF-β on stromal cells increases the efficiency of organ colonisation by CRC [26]. Within the tumour microenvironment TGF-β also exerts a predominantly immunosuppressive effect on CD8+ cytotoxic T-lymphocytes and has been shown as an active player in tumour immune evasion. In light of these findings upregulation of TGF-β1 expression in CRC patients’ blood sustains our hypothesis that changes in gene expression reflect the cancer itself. In the same direction are data for participation of TGF-β1 in differentiation of Treg and Th17 subpopulations, which are involved in tumour-promoting inflammation [27]. It has been demonstrated that TGF-β and IL-10 maintain differentiation of Treg cells, while TGF-β in combination with IL-6 and IL-23 are closely connected to differentiation and maintenance of Th17 cells [28, 29].
Our results demonstrate significant upregulation of the expression levels of IL-10 and TGF-β in CRC blood cells, which decreased after tumour eradication, almost paralleling those of healthy donors’ levels. We hypothesise that the observed gene expression alteration in PBMC could be associated with epigenetic modification, such as histone deacetylation and DNA methylation. Thus, epigenetic modification of these genes will maintain the same pattern of expression in lymphocytes after entry into the tumour microenvironment. Although we did not find changes in HDAC and iNOS expression in CRC blood, it does not exclude an active role of these enzymes in epigenetic alterations. In a mouse model of inflammation-promoting intestinal cancer the inflammation is associated with increased global aberrant DNA methylation [30, 31]. Another argument for the role of methylation is that it showed overexpression of DNA methyl transferases (Dnmts), which are implicated in methylation in various human cancers [32, 33]. Moreover, methylation modulated gene expression present in the inflammation-prone tissue prior to formation of colon cancers [33, 34]. These assumptions are in accordance with our previous findings that monocytes isolated from colorectal cancer patients exhibit a hyporesponsiveness to LPS stimulation compared with healthy people, and that hyporesponsiveness was strongly expressed in monocytes from advanced, and then early stages of CRC [14]. Further studies are needed to clarify whether gene expression patterns in blood lymphocytes and monocytes are related to epigenetic alteration.
Although the upregulated expression of HDAC3 in colon neoplasia has been well documented [34] data for transcriptional activation of the HDAC3 gene in mononuclear cells of CRC patients are not available. Our results showed that the HDAC3 gene in PBMC of CRC patients does not change its transcriptional activity pre- or post-operatively compared to healthy donors.
Obviously, CRC generates not only the local inflammatory microenvironment, named as tumour-elicited inflammation, but also promotes systemic changes that are favourable for cancer progression. A part of these systemic effects, which cancer development induces, includes reprogramming of gene expression in blood immune cells. Here we demonstrated upregulation of TGF-β and IL-10 mRNA expression in peripheral immune cell, which can serve as blood biomarkers for CRC development.
Bearing in mind the above, we could assume that PBMC gene expression programming in CRC cancer patients triggers local differentiation of Th cells towards Treg instead of the Th1 anti-tumour subpopulation. At the same time, TGF-β produced by peripheral immune cells of CRC patients additionally depresses anti-tumour immune response at the level of Th cells, cytotoxic T lymphocytes, and natural killer cells, while increasing the numbers of Tr cells.
In conclusion, we suppose that the established tumour-elicited inflammation can contribute to multiple hallmark capabilities by supplying bioactive molecules, not only to the benefit of tumour growth, but also affecting epigenetic alternation in immune blood cells, resulting in gene expression reprogramming and leading to ineffective antitumor immune response.
This work was supported by Grants: no. 4/2009 and 1/2016 from the Fund for Scientific and Mobile Project from the Faculty of Medicine at Trakia University, Stara Zagora, Bulgaria.
The authors declare no conflict of interest.
References
- 1.Cruz-Bustillo CD. Molecular genetics of colorectal cancer. Rev Esp Enferm Dig. 2004;96:48–9. doi: 10.4321/s1130-01082004000100007. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg R. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 4.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–81. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 5.Demaria S, Pikarsky E, Karin M, et al. Cancer and inflammation: promise for biologic therapy. J Immunother. 2010;33:335–51. doi: 10.1097/CJI.0b013e3181d32e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 7.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and Colon Cancer. Gastroenterology. 2010;138:2101–14. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 8.Cianchi F, Cortesini C, Fantappie O, et al. Inducible nitric oxide synthase expression in human colorectal cancer. Am J Pathol. 2003;162:793–801. doi: 10.1016/S0002-9440(10)63876-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leoni F, Fossati G, Lewis E, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–12. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woan K, Sahakian E, Sotomayor E, Seto E, Villagra A. Modulation of antigen presenting cells by HDAC inhibitors: implications in autoimmunity and cancer. Immunol Cell Biol. 2012;90:55–65. doi: 10.1038/icb.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klampfer L. Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets. 2011;11:451–64. doi: 10.2174/156800911795538066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrone C, Dranoff G. Dual roles for immunity in gastrointestinal cancers. J Clin Oncol. 2010;28:4045–51. doi: 10.1200/JCO.2010.27.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanilov N, Miteva L, Dobreva Z, Jovchev J, Cirovski G, Stanilova S. Monocytes expression of IL-12 related and IL-10 genes in association with development of colorectal cancer. Mol Biol Rep. 2012;39:10895–902. doi: 10.1007/s11033-012-1987-z. [DOI] [PubMed] [Google Scholar]
- 14.DeNardo DG, Johansson M, Coussens LM. Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev. 2008;27:11–8. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 15.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol. 2009;86:1065–73. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 16.Csiszar A, Szentes T, Haraszti B, Balazs A, Petranyi G, Pocsik E. The pattern of cytokine gene expression in human colorectal carcinoma. Pathology Oncology Research. 2004;10:109–11. doi: 10.1007/BF02893465. [DOI] [PubMed] [Google Scholar]
- 17.Baier PK, Wolff-Vorbeck G, Eggstein S, Baumgartner U, Hopt UT. Cytokine expression in colon carcinoma. Anticancer Res. 2005;25:2135–40. [PubMed] [Google Scholar]
- 18.Stanilov N, Miteva L, Mintchev N, Stanilova S. High expression of Foxp3, IL-23p19 and surviving mRNA in colorectal carcinoma. Int J Colorectal Dis. 2009;24:151–7. doi: 10.1007/s00384-008-0588-8. [DOI] [PubMed] [Google Scholar]
- 19.Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. The phenotype of human Th17 cells and their precursors, the cytokines that mediate their differentiation and the role of Th17 cells in inflammation. Int Immunol. 2008;20:1361–8. doi: 10.1093/intimm/dxn106. [DOI] [PubMed] [Google Scholar]
- 20.Zou W, Restifo N. TH17 cells in tumour immunity and immunotherapy. Nat Rev Immunol. 2010;10:248–56. doi: 10.1038/nri2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy HK, Wali RK, Kim Y, et al. Inducible nitric oxide synthase (iNOS) mediates the early increase of blood supply (EIBS) in colon carcinogenesis. FEBS Lett. 2007;581:3857–62. doi: 10.1016/j.febslet.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, Dalpke AH. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606. doi: 10.1111/j.1365-2567.2007.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chechlinska M, Kowalewska M, Nowak R. Systemic inflammation as a confounding factor in cancer biomarker discovery and validation. Nat Rev Cancer. 2010;10:2–3. doi: 10.1038/nrc2782. [DOI] [PubMed] [Google Scholar]
- 24.Stanilov N, Miteva L, Deliyski T, Jovchev J, Stanilova S. Advanced colorectal cancer is assosiated with enhanced Interleukin-23 and Interleukin-10 serum level. LabMedicine. 2010;41:159–63. [Google Scholar]
- 25.Prud’homme GJ. Pathobiology of transforming growth factor β in cancer, fibrosis and immunologic disease, and therapeutic considerations. Lab Invest. 2007;87:1077–91. doi: 10.1038/labinvest.3700669. [DOI] [PubMed] [Google Scholar]
- 26.Calon A, Espinet E, Palomo-Ponce S, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Xu K, Wu J, et al. The changes of Th17 cells and the related cytokines in the progression of human colorectal cancers. BMC Cancer. 2012;12:418–27. doi: 10.1186/1471-2407-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veldhoen M, Hocking RJ, Atkins CL, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Kleinewietfeld M, Hafler D. The plasticity of human Treg and Th17 cells and its role in autoimmunity. Semin Immunol. 2013;25:305–12. doi: 10.1016/j.smim.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borinstein S, Conerly M, Dzieciatkowski S, Biswas S, Trobridge P, Washington M. Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model. Mol Carcinog. 2010;49:94–103. doi: 10.1002/mc.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niwa T, Tsukamoto T, Toyoda T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 32.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–42. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 33.Hahn MA, Hahn T, Lee DH, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–9. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–58. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]