Abstract
The electrically conductive pili (e-pili) of Geobactersulfurreducens have environmental and practical significance because they can facilitate electron transfer to insoluble Fe(III) oxides; to other microbial species; and through electrically conductive biofilms. E-pili conductivity has been attributed to the truncated PilA monomer, which permits tight packing of aromatic amino acids to form a conductive path along the length of e-pili. In order to better understand the evolution and distribution of e-pili in the microbial world, type IVa PilA proteins from various Gram-negative and Gram-positive bacteria were examined with a particular emphasis on Fe(III)-respiring bacteria. E-pilin genes are primarily restricted to a tight phylogenetic group in the order Desulfuromonadales. The downstream gene in all but one of the Desulfuromonadales that possess an e-pilin gene is a gene previously annotated as ‘pilA–C’ that has characteristics suggesting that it may encode an outer-membrane protein. Other genes associated with pilin function are clustered with e-pilin and ‘pilA–C’ genes in the Desulfuromonadales. In contrast, in the few bacteria outside the Desulfuromonadales that contain e-pilin genes, the other genes required for pilin function may have been acquired through horizontal gene transfer. Of the 95 known Fe(III)-reducing micro-organisms for which genomes are available, 80 % lack e-pilin genes, suggesting that e-pili are just one of several mechanisms involved in extracellular electron transport. These studies provide insight into where and when e-pili are likely to contribute to extracellular electron transport processes that are biogeochemically important and involved in bioenergy conversions.
Keywords: Geobacter, extracellular electron transfer, e-pilin, type IVa PilA, positive selection, evolution
Data Summary
Supplementary data (Tables S1-–S2 and Figs S1–S4) and phylogenetic trees (Bayesian and maximum-likelihood) have been deposited in FigShare; https://figshare.com/s/87e875d0c5c97c2e5498.
Accession numbers from pilin gene sequences analyzed for this study are provided in Supplementary Table S1.
Impact Statement
It is becoming increasingly apparent that micro-organisms can make electrical connections with other cells as well as abiological materials, such as minerals and electrodes. This form of extracellular electron transfer plays an important role in natural biogeochemical cycles and is being adapted for practical applications in bioremediation, bioenergy and biomaterials. The electrically conductive pili (e-pili) of Geobacter sulfurreducens are an example of one type of electrical connection that has been studied intensively. However, the extent to which e-pili are distributed throughout the microbial world has been uncertain. The results presented here demonstrate that e-pili have arisen as a relatively recent evolutionary event and appear to be primarily restricted to a tight phylogenetic group within the deltaproteobacteria. This finding suggests that most bacteria other than species of the genus Geobacter have alternative strategies for making extracellular electrical connections and that electrically conductive filaments that have been observed in other organisms are likely to have evolved independently of e-pili and may have different mechanisms for conductivity.
Introduction
The concept that electrically conductive pili (e-pili) can enable long-range electron transfer to insoluble minerals (Reguera et al., 2005), to other cells (Rotaru et al., 2014; Shrestha et al., 2013; Summers et al., 2010) and through electrically conductive biofilms (Malvankar et al., 2011; Reguera et al., 2006) is a paradigm shift in microbial electron transport. The distance over which e-pili can conduct electrons and the apparent competitive advantage of this capability in a number of anaerobic environments distinguish electron transfer via e-pili from more typical forms of short-range biological electron transfer (Lovley & Malvankar, 2015; Malvankar & Lovley, 2014; Malvankar et al., 2015; Vargas et al., 2013).
To date, e-pili have only been documented in two species of the genus Geobacter, Geobacter sulfurreducens (Chun, 2014; Malvankar et al., 2011; Reguera et al., 2005) and Gobacter metallireducens (Y. Tan et al., unpublished). The pili of the only other species of the genus Geobacter examined, Geobacter uraniireducens, are poorly conductive (Tan et al., 2016). G. uraniireducens functions without e-pili because it utilizes a soluble electron shuttle to reduce Fe(III) oxides, does not form thick electrically conductive biofilms and does not participate in direct interspecies electron transfer (Rotaru et al. 2015; Tan et al., 2016).
It has been suggested that electrically conductive filaments may facilitate extracellular electron transfer in other organisms (Castro et al., 2014; Eaktasang et al., 2016; Gorby et al., 2006; Venkidusamy et al., 2015; Wanger et al., 2013), but in no instance has it been definitively demonstrated that the filaments are required for this process, and in many instances the composition of the filaments has yet to be determined. In contrast, many lines of evidence support the concept of long-range electron transport along the e-pili of Geobacter sulfurreducens. This includes the findings that: (1) deleting the gene for PilA, the pilus monomer, inhibited electron transport to Fe(III) oxide, interspecies electron exchange, and the development of thick electrically conductive biofilms (Malvankar et al., 2011; Nevin et al., 2009; Reguera et al., 2005; Summers et al., 2010); (2) genetically modifying pilA to yield pili with poor conductivity inhibited Fe(III) oxide reduction and reduced biofilm conductivity (Vargas et al., 2013); (3) a strain of Geobacter sulfurreducens expressing the poorly conductive pili of Pseudomonas aeruginosa was ineffective in Fe(III) oxide reduction and current production (Liu et al., 2014); (4) the individual pilin filaments are electrically conductive (Adhikari et al., 2016; Malvankar et al., 2011; Reguera et al., 2005); and (5) the pili propagate charge similarly to carbon nanotubes (Malvankar et al., 2014).
In the initial study on Geobacter sulfurreducens e-pili, it was noted that the PilA monomer of Geobacter sulfurreducens was much shorter than PilA from most micro-organisms for which a PilA sequence was available (Reguera et al., 2005). It has been speculated that the shorter pilus monomer permits tighter packing, positioning aromatic amino acids (Lovley & Malvankar, 2015) and charged amino acids (Feliciano et al., 2015), in a manner that promotes electron conduction along the length of the pilin. This hypothesis is supported by the finding that G. metallireducens, which also has a short PilA, produces e-pili (Y. Tan, unpublished), whereas the pili of Geobacter uraniireducens, which has a long PilA typical of that found in many micro-organisms, produces pili of low conductivity (Tan et al., 2016).
Many more genome sequences have become available in the decade since the original phylogenetic analysis of the Geobacter sulfurreducens PilA protein (Reguera et al., 2005). The purpose of this study was to further evaluate the phylogenetic distribution of the short pilA gene, (i.e. e-pilin) that appears to yield e-pili, to determine the distribution of e-pilin sequences in the microbial world and to obtain insight into the evolution of this unique electron transfer mechanism.
Methods
Genome data analysis.
Sequence data for all of the bacterial genomes was acquired from the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) or from Genbank at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). Initial analyses were done with analysis tools available on the Integrated Microbial Genomes (IMG) website (img.jgi.doe.gov). The Find Function search tool on IMG was used to identify proteins that carried the type IV pilin N-terminal methylation site GFxxxE (pfam13544), the N_methyl_3 domain (pfam13633), the PilA domain (COG4969), the pilin domain (Pfam001114)and/or the prepilin-type N-terminal cleavage/methylation domain (TIGR02532). These domains were also identified in various proteins with NCBI conserved domain search (Marchler-Bauer et al., 2015) and Pfam search (Finn et al., 2016) tools. These proteins were then individually screened to ensure that they were in fact type IV PilA proteins by comparison to previously characterized PilA proteins available in the NCBI Genbank database with the BLASTp and PSI-BLAST algorithms (Altschul et al., 1997).
Open reading frames in genomes were also screened for the presence of type IV pilin-like motifs with PilFind (Imam et al., 2011), FlaFind (Szabó et al., 2007), Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and Motif Search (http://www.genome.jp/tools/motif/). A hidden Markov model was built from an alignment of 100 different Type IVa PilA protein sequences using the hmmbuild function in HMMER 3 (Eddy, 2008, 2011). Candidate genes in these genomes were identified by comparison to these alignments with the hmmsearch function and then further screened by comparison to sequences in the Genbank database with BLASTp and PSI-BLAST algorithms.
The same domain search tools used to identify type IVa pilA genes were also used to find genes coding for pilin accessory proteins. Open reading frames were scanned for the following motifs: pilW (pfam16074), pilX (pfam14341), pilY1 (pfam05567), pilD (COG1989, pfam01478, and pfam06750), pilS (COG0642, pfam00512, pfam00672, and pfam02518), pilR (COG2204, pfam00072, pfam00158, and pfam02954), pilQ (COG4796, pfam00263, pfam07660, and pfam11741), pilP (COG3168 and pfam04351), pilO (COG3167 and pfam04350), pilN (COG3166 and pfam05137) and pilM (COG4972 and pfam11104). Genes from the xap operon were identified with the following motifs: xapA (TPR_8; pfam13181), xapB (ABC2_membrane_2 pfam;12679), xapD (ABC-type multidrug transport system, ATPase component; COG1131 and pfam00005), xapE (ubiA; COG0382 and pfam01040), xapF (Glycos_transf_2; pfam00535); xapG (ABC2_membrane; COG1682 and pfam01061), xapH (COG1134, pfam00005 and pfam14524), xapI (Methyltransf_21; pfam05050) and xapJ (Glyco_transf_9; COG0859 and pfam01075).
Alpha helices and beta strands were predicted with the Jnet algorithm (Cuff & Barton, 2000) on the JPred4 server (Drozdetskiy et al., 2015) and transmembrane helices were predicted with TMpred (Hofmann & Stoffel, 1993), TMHMM (Krogh et al., 2001), and HMMTOP (Tusnády & Simon, 2001). Signal peptides were identified with PSORTb v. 3.0.2 (Yu et al., 2010) and Signal P v. 4.1 (Petersen et al., 2011).
Phylogenetic analyses.
Phylogenetic trees were generated with the maximum likelihood method using MEGA v. 6.0 software (Tamura et al., 2013). Before trees were constructed, the Find Best DNA/Protein Models program was run on sequences previously aligned in GUIDANCE2. The PilA–C phylogeny was generated with the Dayhoff model (Dayhoff et al., 1978) with Gamma Distributed rates among sites. The PilA phylogeny was generated with the LG model (Le & Gascuel, 2008) with Gamma Distributed with invariant sites set as the rate among sites. All tests of phylogeny were determined with the Bootstrap Method using 500 replicates. Relative tree node ages were determined with MEGA v. 6.0 and FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Amino acid and nucleotide sequence alignments were generated with MAFFT (Katoh & Standley, 2013) and PRANK (Löytynoja & Goldman, 2005) algorithms. GUIDANCE2 (Sela et al., 2015) was used to identify and eliminate unreliable residues, columns and sequences in all alignments.
Tajima’s Neutrality Test (Tajima, 1989) was conducted on amino acid and nucleotide sequence alignments in MEGA6 (Tamura et al., 2013). Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated.
Results and Discussion
Distribution of e-pilin genes in microbial enomes
More than 250 different genomes from a diversity of Gram-negative and Gram-positive bacteria (95 of which were Fe(III)-respiring bacteria) were scanned for the presence of type IVa pilin genes using previously characterized type IVa PilA sequences from such species as Neisseria gonorrhoeae (Craig et al., 2006), Geobacter sulfurreducens (Reguera et al., 2005), Pseudomonas aeruginosa (Craig et al., 2003), Myxococcus fulvus (Sun et al., 2000), Escherichia coli (Bieber et al., 1998), and Shewanella oneidensis (Gorgel et al., 2015) as queries. This analysis revealed that truncated PilA proteins, like those in Geobacter sulfurreducens and Geobacter metallireducens are primarily found in micro-organisms in the order Desulfuromondales (Table 1, Fig. 1). These truncated pilA genes are predicted to encode mature pilin structural proteins with only 60–90 amino acids, compared with the >120 amino acid residues typically found in PilA subunits of most other bacteria (Table 1). In the few instances in which pili conductivity has been directly measured, truncated pilA genes code for proteins that give rise to conductive pili (Geobacter sulfurreducens and Geobacter metallireducens), whereas longer pilA genes yield pili with poor conductivity (Geobacter uraniireducens and Pseudomonas aeruginosa) (Liu et al., 2014; Tan et al., 2016). Therefore, the truncated pilA genes were designated e-pilin to distinguish them from longer type IVa pilA genes that are more commonly found in bacteria.
Table 1. Predicted size and phylogenetic classification of type IVa pili detected in various Fe(III)-reducing bacteria.
| Fe(III)-reducing bacterium | Type IVa pilA type | Immature pilin size (aa) | Mature pilin size (aa) | Leader sequence length | Accession number | Number of beta strands at C terminus* |
|---|---|---|---|---|---|---|
| Geobacter bemidjiensis | e-pilin | 76 | 66 | 10 | Gbem_2590 | 1 |
| Geobacter bremensis | e-pilin | 74 | 64 | 10 | K419DRAFT_00801 | 0 |
| Pelobacter seleniigenes | e-pilin | 70 | 59 | 11 | N909DRAFT_0006 | 0 |
| Geobacter sp. OR-1 | e-pilin | 74 | 64 | 10 | WP_041974243 | 0 |
| Geobacter sp. M18 | e-pilin | 74 | 64 | 10 | GM18_2492 | 0 |
| Geobacter sp. M21 | e-pilin | 74 | 64 | 10 | GM21_1636 | 0 |
| Desulfuromonas sp. TF | e-pilin | 75 | 64 | 11 | DTFDRAFT_03630 | 0 |
| Geoalkalibacter ferrihydriticus | e-pilin | 72 | 61 | 11 | Ga0056053_00657 | 0 |
| Geoalkalibacter subterraneus | e-pilin | 79 | 66 | 13 | WP_040199521 | 0 |
| Desulfuromonas thiophila | e-pilin | 70 | 59 | 11 | Ga0056074_12312 | 0 |
| Geobacter metallireducens | e-pilin | 69 | 59 | 10 | Gmet_1399 | 0 |
| Geobacter lovleyi | e-pilin | 70 | 60 | 10 | Glov_2096 | 0 |
| Geobacter sulfurreducens | e-pilin | 90 | 61 | 29 | GSU1496 | 0 |
| Geobacter pickeringii | e-pilin | 70 | 60 | 10 | Ga0069007_111762 | 0 |
| Desulfuromusa kysingii | e-pilin | 72 | 60 | 12 | Ga0056096_02700 | 0 |
| Geobacter argillaceus | e-pilin | 75, 75 | 65, 65 | 10, 10 |
Ga0052872_01800 Ga0052872_01802 |
0, 0 |
| Geobacter soli | e-pilin | 75 | 65 | 10 | WP_039645155 | 0 |
| Geopsychrobacter electrodiphilus | e-pilin | 75 | 64 | 11 | D888DRAFT_2042 | 0 |
| Pelobacter propionicus | e-pilin | 74 | 64 | 10 | Ppro_1656 | 0 |
| ‘Desulfuromonas subbituminosa' | Long type IVa pilA | 132 | 125 | 7 | Ga0064601_106193 | 3 |
| Geobacter uraniireducens | Long type IVa pilA | 203 | 193 | 10 | Gura_2677 | 8 |
| Geobacter daltonii | Long type IVa pilA | 218 | 208 | 10 | Geob_3369 | 10 |
| ‘Desulfuromonas soudanensis'WTL | Long type IVa pilA | 204 | 193 | 11 | Ga0069009_112157 | 8 |
| Pelobacter carbinolicus | Long type IVa pilA | 138, 196 | 131, 185 | 7, 11 | Pcar_2154, Pcar_2144 | 3, 7 |
| Desulfobacter postgatei | Long type IVa pilA | 216 | 205 | 11 | DespoDRAFT_1114 | 8 |
| Desulfuromonas acetoxidans | No type IVa pilA detected | |||||
| Desulfobulbus propionicus | No type IVa pilA detected | |||||
| Desulfobacterium autotrophicum | e-pilin | 73 | 59 | 14 | HRM2_27700 | 0 |
| Thermincola ferriacetica | Long type IVa pilA | 113 | 100 | 13 | TferDRAFT_00608 | 2 |
| ‘Candidatus Acidianus copahuensis' | No type IVa pilA detected | |||||
| Acidicaldus organivorans | No type IVa pilA detected | |||||
| Carboxydothermus ferrireducens | Long type IVa pilA | 150 | 137 | 13 | CarfeDRAFT_00001450 | 8 |
| Deferrisoma camini | Long type IVa pilA | 144, 131 | 130, 125 | 14, 5 | DefcaDRAFT_3089, DefcaDRAFT_3087 | 2, 3 |
| Desulfovibrio frigidus | No type IVa pilA detected | |||||
| Fervidicella metallireducens | Long type IVa pilA | 126 | 114 | 12 | Q428_01340 | 4 |
| Desulfovibrio ferrireducens | No type IVa pilA detected | |||||
| Desulfovibrio vulgaris | Long type IVa pilA | 141 | 128 | 13 | Ga0076800_111227 | 6 |
| ‘Thermincola potens' | Long type IVa pilA | 146 | 138 | 8 | WP_049771692 | 8 |
| Carboxydothermus hydrogenoformans | Long type IVa pilA | 150 | 137 | 13 | CHY_0635 | 8 |
| Shewanella oneidensis | Long type IVa pilA | 138 | 126 | 12 | SO0417 | 3 |
| Anaeromyxobacter dehalogenans | Long type IVa pilA | 195 | 188 | 7 | A2cp1_0669 | 7 |
| Ardenticatena maritima | No type IVa pilA detected | |||||
| Thermoanaerobacter ethanolicus | Long type IVa pilA | 121 | 107 | 14 | TeCCSD1DRAFT_1919 | 3 |
| ‘Thermoanaerobacter cellulolyticus' | Long type IVa pilA | 138 | 133 | 5 | N907DRAFT_0879 | 6 |
| Bacillus infernus | No type IVa pilA detected | |||||
| Bacillus subterraneus | No type IVa pilA detected | |||||
| Caloramator australicus | Long type IVa pilA | 134 | 126 | 8 | WP_008908368 | 5 |
| ‘Desulfotomaculum reducens' | Long type IVa pilA | 120 | 107 | 13 | Dred_1042 | 2 |
| Anaeromyxobacter sp. strain FW109-5 | Long type IVa pilA | 198 | 191 | 7 | Anae109_0680 | 4 |
| Desulfosporosinus orientis | Long type IVa pilA | 141 | 129 | 12 | Desor_0988 | 6 |
| Acidithiobacillus ferrooxidans | Long type IVa pilA | 178 | 163 | 15 | AFE_0416 | 6 |
| Acidithiobacillus ferrivorans | Long type IVa pilA | 169 | 158 | 11 | Ga0058672_17875 | 6 |
| Desulfosporosinus meridei | Long type IVa pilA | 130, 133 | 114, 117 | 15, 16 | Desmer_0976, Desmer_0977 | 4, 6 |
| Aeromonas hydrophila | Long type IVa pilA | 128 | 121 | 7 | AHA_0692 | 3 |
| Shewanella algae | Long type IVa pilA | 138 | 126 | 12 | BryDRAFT_00594 | 4 |
| Rhodoferax ferrireducens | Long type IVa pilA | 160 | 152 | 8 | Rfer_1265 | 4 |
| Acidicaldus organivorans | No type IVa pilA detected | |||||
| Acidimicrobium ferroxidans | No type IVa pilA detected | |||||
| Acidiphilum cryptum | No type IVa pilA detected | |||||
| ‘Acidithrix ferrooxidans' | Long type IVa pilA | 177 | 144 | 33 | AFO_01365 | 5 |
| Acidobacterium capsulatum | No type IVa pilA detected | |||||
| Acidocella facilis | No type IVa pilA detected | |||||
| Alicyclobacillus contaminans | No type IVa pilA detected | |||||
| ‘Alkaliphilus metalliredigens' | Long type IVa pilA | 112 | 98 | 14 | Amet_3479 | 3 |
| Clostridium beijerinckii | Long type IVa pilA | 189 | 182 | 7 | Cbei_4216 | 5 |
| Desulfitobacterium hafniese | Long type IVa pilA | 171 | 156 | 15 | Dhaf_3553 | 8 |
| Desulfitobacterium metallireducens | Long type IVa pilA | 172 | 163 | 9 | Desme_2113 | 9 |
| Ferrimicrobium acidiphilum | No type IVa pilA detected | |||||
| Ferrimonas baelerica | Long type IVa pilA | 170 | 165 | 5 | Fbal_0401 | 7 |
| Geothrix fermentans | No type IVa pilA detected | |||||
| Geovibrio thiophilus | Long type IVa pilA | 147, 142 | 140, 134 | 7, 8 | K300DRAFT_1049, K300DRAFT_1050 | 4, 3 |
| Leptospirillum ferriphilum | No type IVa pilA detected | |||||
| Pantoea agglomerans | Long type IVa pilA | 151 | 145 | 6 | Ga0004745_2907 | 5 |
| Rhodobacter capsulatus | No type IVa pilA detected | |||||
| Rhodopseudomonas palustris | No type IVa pilA detected | |||||
| Rhodopseudomonas sphaeroides | No type IVa pilA detected | |||||
| Sulfobacillus acidophilus | No type IVa pilA detected | |||||
| Sulfobacillus thermosulfidooxidans | No type IVa pilA detected | |||||
| Sulfurospirillum barnesii | No type IVa pilA detected | |||||
| Thermoanaerobacter siderophilus | No type IVa pilA detected | |||||
| Thermus scotoductus | Long type IVa pilA | 117 | 111 | 6 | WP_038068616 | 4 |
| Serratia fonticola | No type IVa pilA detected | |||||
| Tepidimicrobium xylanilyticum | Long type IVa pilA | 142 | 127 | 15 | Ga0056071_02505 | 2 |
| Clostridium celerecrescens | No type IVa pilA detected | |||||
| Enterococcus gallinarum | No type IVa pilA detected | |||||
| Pelosinus fermentans | No type IVa pilA detected | |||||
| Shewanella amazonensis | Long type IVa pilA | 157 | 145 | 12 | Sama_0370 | 4 |
| Shewanella putrefaciens | Long type IVa pilA | 152 | 136 | 16 | Sput200_3560 | 4 |
| Shewanella baltica | Long type IVa pilA | 187 | 171 | 16 | Sbal175DRAFT_2072 | 7 |
| Shewanella peizotolerans | Long type IVa pilA | 110 | 95 | 15 | swp_4760 | 3 |
| Shewanella decolorationis | Long type IVa pilA | 136 | 124 | 12 | SdecDRAFT01_04314 | 4 |
| Shewanella frigidimarina | Long type IVa pilA | 148 | 136 | 12 | Sfri_3782 | 4 |
| Shewanella loihica | Long type IVa pilA | 145 | 134 | 11 | Ga0069557_1249 | 4 |
| Shewanella pealeana | Long type IVa pilA | 185 | 172 | 13 | Spea_3315 | |
*Secondary structure predictions were made with the Jnet algorithm (Cuff & Barton, 2000) on the Jpred4 server ((Drozdetskiy et al., 2015)
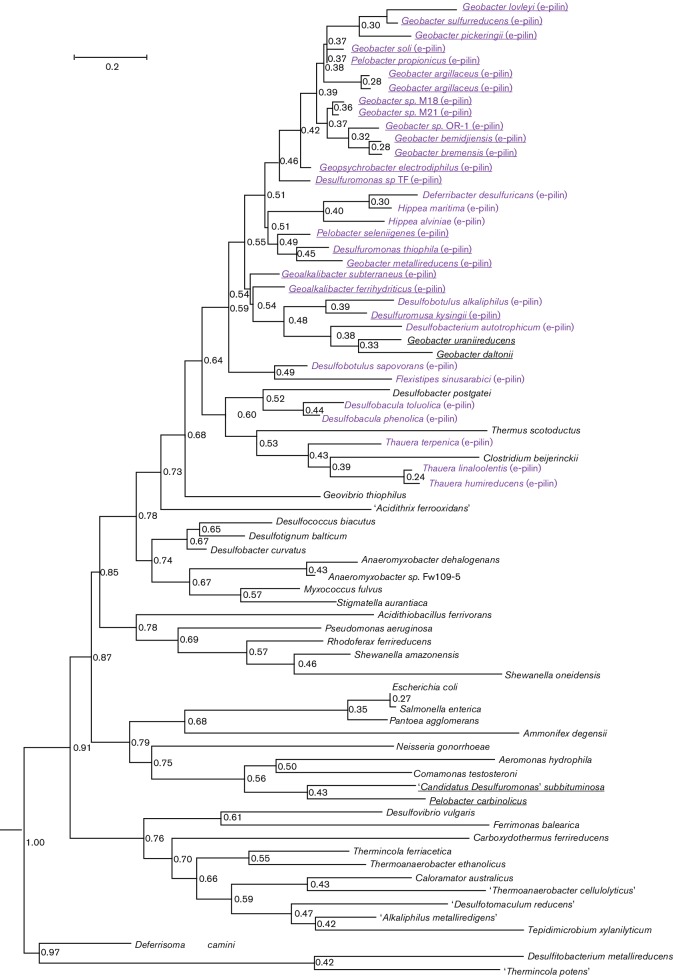
Fig. 1.
Phylogenetic tree generated with the maximum-likelihood algorithm comparing type IVa PilA proteins from various Fe(III)-reducing species and species with well-characterized type IVa PilA proteins. Desulfitobacterium metallireducens and ‘Thermincola potens' were used as outgroups. Organisms with e-pilin proteins are identified with purple font, and all species from the order Desulfuromondales are underlined. Relative divergence times are shown for each node and the root age was set at 1.0.
All of the e-pilin sequences lack the C-terminal portion of the sequence found in longer, more typical, PilA proteins, which code for beta sheet structures that form a large globular head (Craig et al., 2004; Giltner et al., 2012). Analysis of the structure of the Geobacter sulfurreducens e-pilus suggested that the lack of this C-terminus sequence permits the pilin monomers to pack more tightly than the larger pilin monomers found in most bacteria, yielding a thinner pilus diameter and positioning amino acids in patterns that confer conductivity (Malvankar et al., 2015).
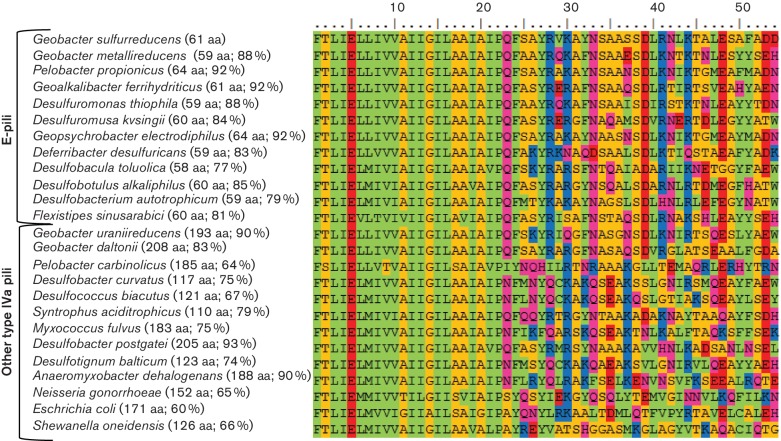
As previously noted in the analysis of the Geobacter sulfurreducens PilA sequence (Malvankar et al., 2015; Reardon & Mueller, 2013; Reguera et al., 2005), the N-termini of the e-pilin proteins share many similarities with previously described type IVa PilA found in members of genera such as Neisseria, Pseudomonas, Escherichia and Myxococcus (Giltner et al., 2012; Mattick, 2002). It is highly conserved and consists of one or two alpha helices and a transmembrane motif and aligns closely with other previously described type IVa pilin proteins (Figs 2 and S1, available in the online Supplementary Material).
Fig. 2.
Alignment of the N-terminus (first 54 amino acids) of mature type IVa PilA proteins from various taxonomic groups. The numbers in parentheses represent the size of the mature pilin protein and the percentage similarity to PilA from Geobacter sulfurreducens. The Lesk color scheme was used to shade the amino acids. All small nonpolar residues (G, A, S, T) are orange; all hydrophobic residues (C, V, I, L, P, F, Y, M, W) are green; all polar residues (N, Q, H) are magenta; all negatively charged (D, E) are red; and all positively charged residues (K, R) are blue.
Consistent with other type IVa pilins, all of the e-pilin proteins have a phenylalanine at the N terminus (Fig. 2), and the majority have leader peptides with less than 12 amino acids (Table 1). The exception is the leader peptide of Geobacter sulfurreducens, which is 29 amino acids long and more characteristic of a type IVb PilA.
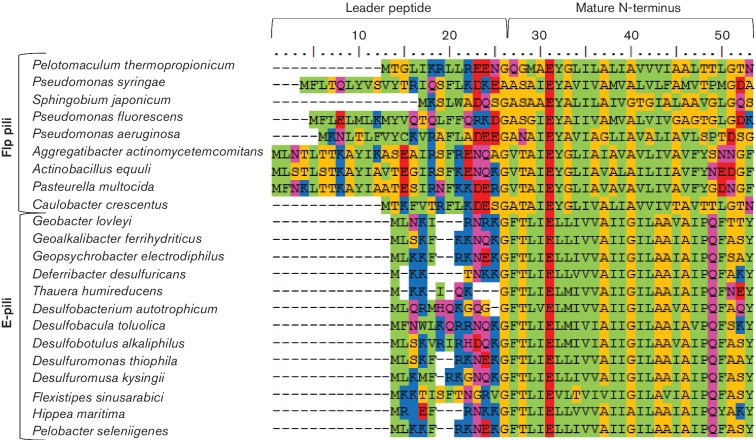
The size of these type IVa e-pilins is similar to that of the major subunits of Tad pili (Flp) found in Aggregatibacter actinomycetemcomitans (Kachlany et al., 2001; Tomich et al., 2007) and Peudomonas aeruginosa (Bernard et al., 2009; Burrows, 2012), which range in size from 50 to 80 amino acid residues. However, Flp pili are type IVb pili with longer leader peptides and a different nonpolar hydrophobic residue (alanine or valine) at the N-terminus following removal of the signal sequence (Fig. 3).
Fig. 3.
Alignment of the N-terminus of immature Flp pili and E-pili from various taxonomic groups. The Lesk color scheme was used to shade the amino acids. All small nonpolar residues (G, A, S, T) are orange; all hydrophobic residues (C, V, I, L, P, F, Y, M, W) are green; all polar residues (N, Q, H) are magenta; all negatively charged (D, E) are red; and all positively charged residues (K, R) are blue.
Positive selection of the e-pilin gene in the Desulfuromondales
With a few exceptions, most of the e-pilin genes cluster in the same phylogenetic clade, suggesting that they evolved from a common ancestral pilA gene (Fig. 1). This clade branches off from other type IVa PilA proteins at a relative time of 0.55 when the root node divergence age is set at 1.0. The majority of nodes for longer PilA proteins found in various Fe(III)-reducing bacteria and other well-studied bacteria such as members of the genera Neisseria, Shewanella, Pseudomonas and Myxococcus, on the other hand, have relative times that are > 0.7.
Tajima’s test of neutrality (Tajima, 1989) indicated that the Desulfuromondales e-pilin amino acid and nucleotide sequences are undergoing positive selection; Tajima's D=6.03 for the protein alignment and Tajima's D=3.5 for the nucleotide alignment. These results suggest that there is a strong selective pressure on the conservation of e-pilin genes within the order Desulfuromondales and that these organisms have been exposed to environmental conditions under which e-pili provide a competitive advantage.
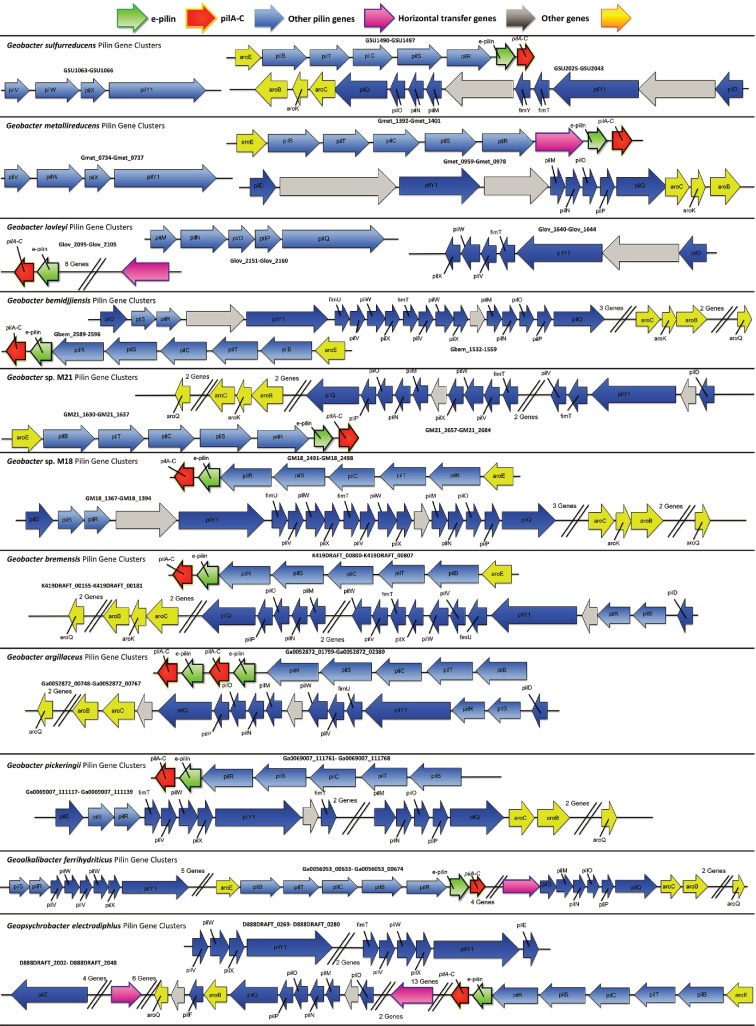
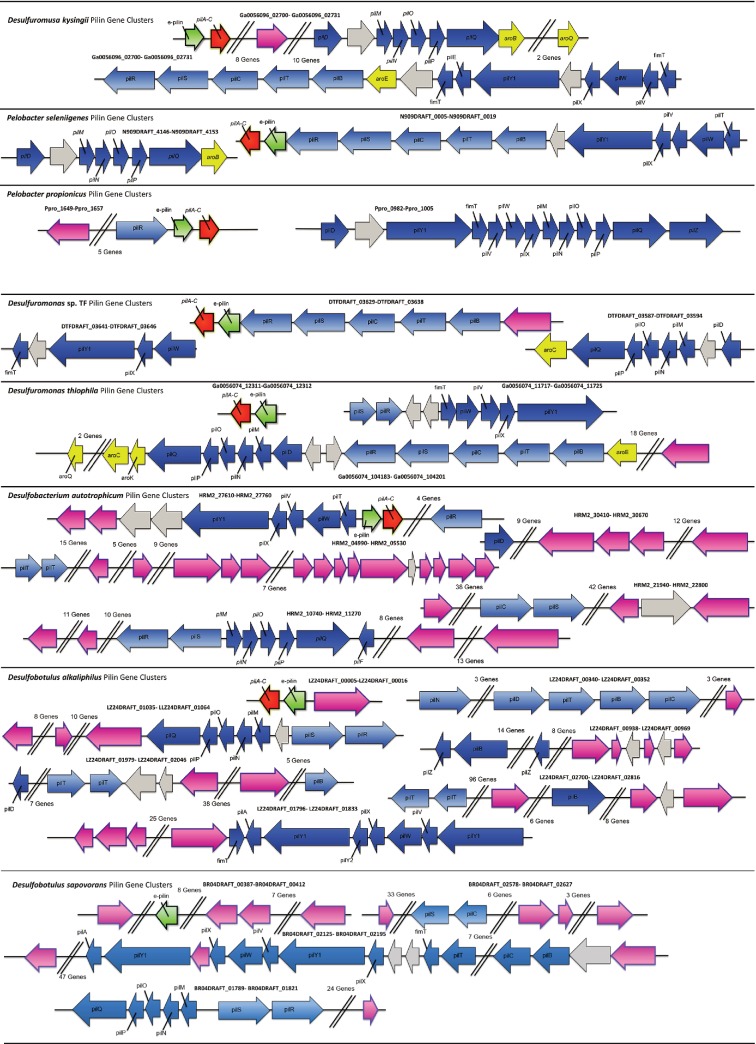
Most (77 %) of the bacteria in the order Desulfuromondales possess e-pilin genes. The exceptions are Gobacter uraniireducens, Geobacter daltoni, ‘Desulfuromonas soudanensis', ‘Desulfuromonas subbituminosa', Desulfuromonas acetoxidans and Pelobacter carbinolicus. With the exception of a gene coding for an IS4 family transposase located directly upstream from the Geobacter uraniireducens pilA gene (Gura_2677), there is little evidence of horizontal gene transfer within pilin gene clusters in these six species (Fig. 4, Table S1, available in the online Supplementary Material). Desulfuromonas. acetoxidans does not possess a type IVa pilA gene, and the pilA gene cluster found in the ‘Desulfuromonas subbituminosa' genome is more similar to those of other deltaproteobacteria (Fig. S2). Pilus gene clusters associated with ‘Desulfuromonas soudanensis', Pelobacter carbinolicus, Geobacter daltonii and Geobacter uraniireducens are characteristic of species of the order Desulfuromondales with e-pilin genes (Fig. S2).
Fig. 4.
Distribution of pilin genes within the genomes of bacteria that are predicted to possess e-pili.
Hypothetical protein gene associated with e-pilin genes
In the genomes of all members of the order Desulfuromondales with e-pilin genes, except the genome of Geobacter soli, the adjacent gene downstream from e-pilin, is a gene that codes for a small (86–136 amino acids) hypothetical protein with multiple beta strands and a signal peptide (Table 2). This gene was annotated as ‘pilA–C’ in the Geobacter sulfurreducens genome, with the expectation that its protein was a component of the Geobacter sulfurreducens pilin protein. However, this annotation is incorrect because denatured pili do not contain a protein of the predicted size (Reguera et al., 2005; Tan et al., 2016; Veazey et al., 2011).
Table 2. Predicted properties of ‘PilA–C’ proteins from members of the genus Geobacter and other bacteria with e-pilin genes .
| Organism | GeneID | beta-strands* | alpha helices* | Size (aa) | Signal peptide?† |
|---|---|---|---|---|---|
| Desulfuromusa kysingii | Ga0056096_02701 | 7 | 2 | 119 | + |
| Geopsychrobacter electrodiphilus | D888DRAFT_2041 | 4 | 3 | 125 | + |
| Pelobacter seleniigenes | N909DRAFT_0005 | 5 | 2 | 128 | + |
| Geobacter bemidjiensis | Gbem_2589 | 8 | 3 | 136 | + |
| Geobacter bremensis | K419DRAFT_00800 | 5 | 3 | 129 | + |
| Geobacter sp. OR-1 | WP_041974245 | 5 | 1 | 122 | + |
| Geobacter sp. M18 | GM18_2491 | 5 | 3 | 120 | + |
| Geobacter sp. M21 | GM21_1637 | 4 | 2 | 117 | + |
| Geobacter soli | Not present | ||||
| Pelobacter propionicus | Ppro_1657 | 3 | 2 | 86 | − |
| Geobacter argillaceus | Ga0052872_01799 | 6 | 2 | 121 | + |
| Geobacter argillaceus | Ga0052872_01801 | 5 | 2 | 121 | + |
| Desulfuromonas sp. TF | DTFDRAFT_03629 | 4 | 2 | 124 | + |
| Geoalkalibacter ferrihydriticus | Ga0056053_00658 | 5 | 1 | 106 | + |
| Geoalkalibacter subterraneus | WP_040199522 | 6 | 2 | 121 | + |
| Desulfuromonas thiophila | Ga0056074_12311 | 6 | 1 | 109 | + |
| Geobacter metallireducens | Gmet_1400 | 6 | 1 | 113 | + |
| Geobacter lovleyi | Glov_2095 | 4 | 3 | 119 | + |
| Geobacter sulfurreducens | GSU1497 | 6 | 2 | 124 | + |
| Geobacter pickeringii | Ga0069007_111761 | 5 | 1 | 109 | + |
| Desulfobotulus alkaliphilus | LZ24DRAFT_00005 | 7 | 1 | 138 | + |
| Flexistipes sinusarabici | Flexsi_2289 | 5 | 4 | 128 | − |
| Desulfobotulus sapovorans | Not present | ||||
| Desulfobacterium autotrophicum | HRM2_27710 | 6 | 2 | 120 | + |
| Desulfobacula phenolica | Not present | ||||
| Desulfobacula toluolica | Not present | ||||
| Thauera linaloolentis | Not present | ||||
| Hippea maritima | Hipma_0736 | 6 | 2 | 107 | + |
| Hippea alviniae | G415DRAFT_0873 | 4 | 2 | 109 | + |
| Deferribacter desulfuricans | DEFDS_1271 | 4 | 1 | 107 | + |
| Thauera humireducens | Not present | ||||
| Thauera terpenica | Not present |
*Alpha helices and beta strands were identified with the Jnet algorithm (Cuff & Barton, 2000) on the Jpred 4 server (Drozdetskiy, 2015).
†Signal peptides were predicted with PSORTb v. 3.0 (Wagner, 2010) and SignalP 4.1 (Nielsen,1997).
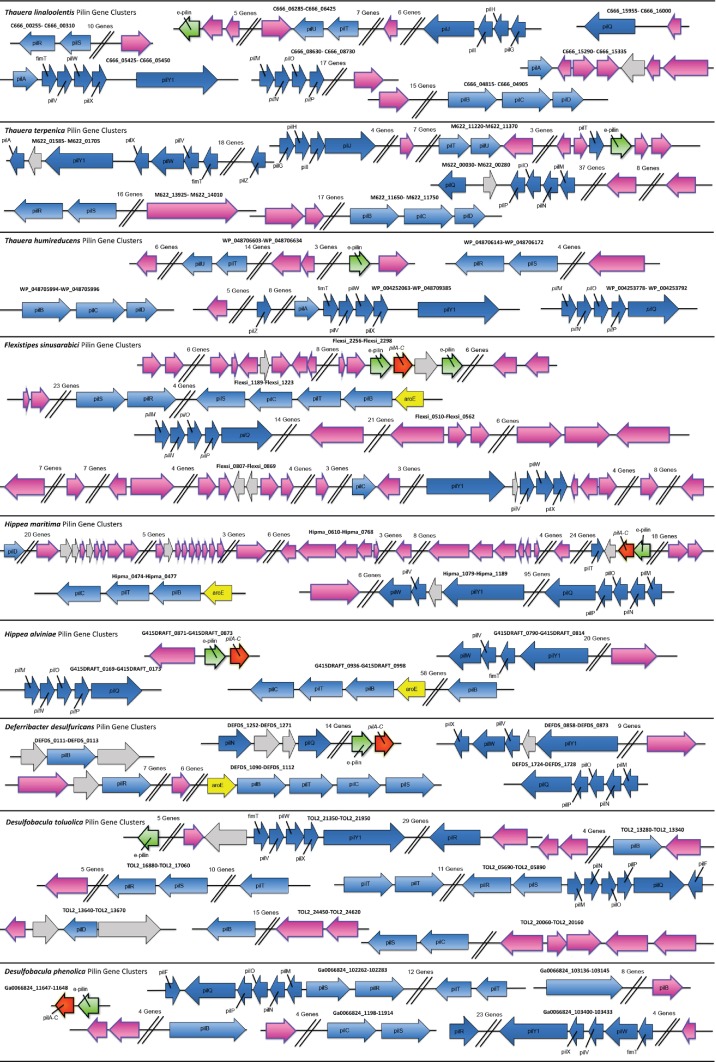
Although both the C-terminus of long type IVa PilA and ‘PilA–C’ proteins have similar topology (beta strands and alpha helices), it is unlikely that they evolved from the same protein. If the ‘pilA–C’ gene was derived from the C-terminus of a typical type IV PilA gene it would only be composed of beta strands and the proteins would be more homologous (Fig. 5). However, all of the ‘PilA–C’ proteins have one to three alpha helices (Fig. S3) and the proteins form two distinct phylogenetic clades. Because of the great divergence between ‘pilA–C’ and the C-terminus of typical long pilA genes, it was difficult to reconstruct an accurate phylogenetic tree comparing all of the taxonomic groups discussed thus far in this paper. Therefore, a relative time tree comparing ‘PilA–C’ and the C-terminus of long PilA proteins was constructed with only sequences from members of the genus Geobacter. It is apparent that even within the same genus these two proteins are highly divergent and that they evolved separately around the same time forming two distinct phylogenetic clades (Fig. 5).
Fig. 5.
Phylogenetic tree generated by the maximum-likelihood algorithim comparing amino acid sequences from the C-terminus of Geobacter PilA to Geobacter ‘PilA–C’. Values represent relative times when each branch node evolved compared with a root node age set at 1.0. Geobacter uraniireducens and Geobacter daltonii were used as outgroups.
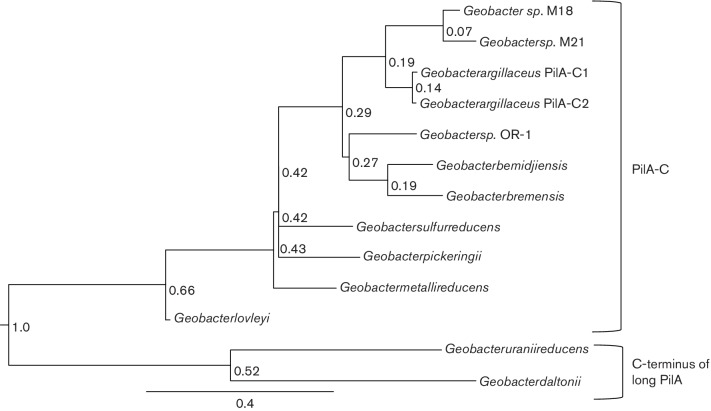
There is little homology between ‘PilA–C’ and other characterized proteins, however, all of the Desulfuromondales ‘PilA–C’ proteins except P. propionicus have signal peptides, suggesting that they are either extracellular or localized to the outer membrane, and their N-termini are similar to the porin-like protein OmpX from Klebsiella pneumonia and Escherichia coli (Fig. 6). The ‘PilA–C’ protein shares many other features with porin proteins (Galdiero et al., 2012); multiple beta strands, a signal sequence of approximately 21 amino acids at its N-terminus, low hydrophobicity and few to no cysteine residues. The only porin-related feature that is lacking in ‘PilA–C’ is a C-terminal phenylalanine, which is important for membrane insertion. A porin-like protein (usher protein; PapC) is associated with type I pili and P-pili in E. coli (Ford et al., 2012; Waksman & Hultgren, 2009). PapC facilitates translocation of the major pilin subunit across the outer membrane and acts as an assembly platform for the pilus. Although no sequence homology between PilA–C and PapC is apparent, it is possible that they play a similar role in pilus assembly.
Fig. 6.
Alignment of N-termini from OmpX and PilA-C proteins. The numbers in parentheses represent the percentage similarity to OmpX from Klebsiella pneumoniae. The Lesk color scheme was used to shade the amino acids. All small nonpolar residues (G, A,S, T) are orange; all hydrophobic residues (C, V, I, L, P, F, Y, M, W) are green; all polar residues (N, Q, H) are magenta; all negatively charged (D, E) are red; and all positively charged residues (K, R) are blue.
Another difference between type I pilin and type IV e-pilin systems is the absence of a chaperone protein in the e-pilin gene cluster. The type I chaperone (PapD) has two immunoglobulin-like domains and facilitates pilin subunit folding and delivery to PapC. Only two e-pilin-harboring taxa, Geobacter bemidjiensis (Gbem_1552) and strain M18 (GM18_1387), have PapD-like domain proteins. These PapD-like proteins are not located in the e-pilin gene cluster, rather they are found in the vicinity of pilMNOPQ and only have a single immunoglobulin-like domain. The pilVWXY1 gene cluster in many other e-pilin-harboring members of the order Desulfuromondales, on the other hand, contains a gene coding for another chaperone, thiol:disulfide interchange protein (DsbC), which helps fold secreted proteins, such as pili, by enabling the formation of disulfide bonds (Zapun et al., 1995). While most type IV pilin proteins have disulfide bonds (Giltner et al., 2012), including all of the long type IVa pili discussed here, e-pili lack the cysteine residues that would be required for the formation of disulfide bonds. Therefore, it seems that this protein is not likely to be involved in posttranslational modification of e-pili.
As with the e-pilin gene, Tajima’s test of neutrality (Tajima, 1989) suggests that the ‘pilA–C’ gene is undergoing positive selection within the order Desulfuromondales; Tajima's D=5.71 for the protein alignment and Tajima's D=3.88 for the nucleotide alignment. The frequent occurrence of ‘pilA–C’ genes in species of the order Desulfuromondales with genomes that encode e-pili suggests that ‘pilA–C’ is likely to play an important role in proper e-pilin expression or function. Further study of the location and function of the protein encoded by the ‘pilA–C’ gene is warranted.
Additional conserved gene organization associated with e-pilin genes
The genome region around the e-pilin genes in species of the order Desulfuromondales species is highly conserved (Fig. 4, Table S1). The e-pilin gene is found within a cluster of genes including pilB and pilT (both ATPase proteins involved in pilin assembly and retraction), pilC (platform protein), pilS and pilR (two-component regulator proteins involved in pilin expression) and genes involved in synthesis and export of surface polysaccharides and biofilm formation (xap genes) (Rollefson et al., 2011). A gene coding for shikimate dehydrogenase (aroE) is also frequently found in this gene cluster. This gene codes for a protein involved in the synthesis of aromatic amino acids, which are required for e-pili conductivity (Adhikari et al., 2016; Vargas et al., 2013).
Additional genes coding for proteins required for proper assembly and function of e-pili are the same as those from other type IVa pili such as Myxococcus, E. coli, Pseudomonas aeruginosa and Neisseria gonorrhoeae (Ayers et al., 2010; Nivaskumar & Francetic, 2014). PilY1 is a pilin tip adhesion protein that helps pili adhere to surfaces. PilV, PilW and PilX are all minor pilin proteins. PilQ is a secretin protein that allows translocation of the pilin subunits across the membrane. PilD is a prepilin peptidase. PilM, PilN, PilO and PilP form a secretin-associated sub complex. Desulfuromondales pilin accessory and minor pilin proteins are homologous to well characterized pilin genes (Table S2, Fig. S4).
In the majority of species of the order Desulfuromondales, genes coding for these pilin assembly proteins are found in one or two gene clusters (Fig. 4). These clusters also possess genes coding for other aromatic amino acid synthesis proteins (aroC, aroK and aroB) and numerous genes coding for glycosyltransferase proteins that may be involved in glycosylation of pilin proteins.
Many of the Desulfuromondales pilin accessory proteins were approximatley 45–80 % similar to proteins from E. coli, Pseudomonas aeruginosa, Nisseria gonorrhoeae and Myxococcus fulvus with the exception of the minor pilin proteins PilX, PilY1, PilW and PilV (Table S2). For the most part, Desulfuromondales pilin accessory proteins form unique phylogenetic clades (Fig. S4). Although e-pilin proteins from species from other orders cluster with species from the order Desulfuromondales, their pilin accessory proteins fall into separate clades. In addition, long type IVa PilA proteins from species of the order Desulfuromondales do not cluster with e-pilin proteins, but most of their pilin accessory proteins cluster with those of e-pilin-harboring Desulfuromondales.
Horizontal transfer of e-pilin outside the Desulfuromondales
Few bacteria outside the Desulfuromondales have e-pilin genes. Those that do include Desulfobacterium autotrophicum, Desulfobotulus alkaliphilus, Desulfobotulus sapovorans, Thauera linaloolentis, Thauera terpenica, Thauera humireducens, Flexistipes sinusarabici, Hippea maritima, Hippea alvinae, Deferribacter desulfuricans, Desulfobacula toluolica, Desulfobacula phenolica and Desulfurobacula sp. TS (Fig. 1, Table 3). With the exception of Desulfobacterium autotrophicum (Lovley et al., 1993), none of these organisms are known to grow via Fe(III) respiration. However, one of the primary functions of e-pili may be interspecies electron exchange (Rotaru et al., 2015), a recently discovered microbial capability (Summers et al., 2010) for which few micro-organisms have been evaluated. Many of the organisms outside the Desulfuromondales that have e-pilin genes were isolated from habitats where species of the order Desulfuromondales are typically found and there is evidence of horizontal gene transfer in regions of their genomes where the e-pilin genes are located.
Table 3. Characteristics of e-pilin genes found in non-Fe(III)-reducing bacterial species.
| Organism | Type IVa pilin type | Immature pilin size (aa) | Mature pilin size (aa) | Leader sequence length | Accession number | #beta strands at C terminus* |
|---|---|---|---|---|---|---|
| Desulfobacterium autotrophicum | epilin | 73 | 59 | 14 | HRM2_27700 | 0 |
| Hippea maritima | epilin | 68 | 59 | 9 | Hipma_0737 | 0 |
| Hippea alviniae | epilin | 70 | 61 | 9 | G415DRAFT_0872 | 0 |
| Deferribacter desulfuricans | epilin | 67 | 59 | 8 | DEFDS_1270 | 0 |
| Flexistipes sinusarabici | epilin | 74, 70 | 67, 60 | 7, 10 |
Flexsi_2291, Flexsi_2288 |
1, 0 |
| Desulfobacula toluolica | epilin | 71 | 58 | 13 | TOL2_21350 | 0 |
| Desulfobacula phenolica | epilin | 72 | 59 | 13 | Ga0066824_11648 | 1 |
| Desulfobacula sp. TS | epilin | Partial sequence | Ga0097800_108051 | 0 | ||
| Desulfobotulus alkaliphilus | epilin | 73 | 60 | 13 | LZ24DRAFT_00006 | 0 |
| Desulfobotulus sapovorans | epilin | 71 | 58 | 13 | BR04DRAFT_00394 | 0 |
| Thauera linaloolentis | epilin and long type 4a pilin | 59, 150, 168 | 52, 144, 161 | 7, 6, 7 |
C666_06285, C666_05425, C666_15290 |
0, 4, 5 |
| Thauera terpenica | epilin and long type 4a pilin | 59, 137 | 52, 128 | 7, 9 |
M622_11345, M622_01585 |
0, 3 |
| Thauera humireducens | epilin and long type 4a pilin | 150, 60 | 144, 52 | 6, 8 |
WP_048709378, WP_048706629 |
4, 1 |
Beta strands were identified with the Jnet algorithm (Cuff & Barton, 2000) on the Jpred 4 server(Drozdetskiy, 2015).
For example, the genome of Flexistipes sinusarabici contains two e-pilin genes, one of which is located next to a putative ‘pilA-C’, that cluster with e-pili from members of the genus Geobacter and closely related organisms. However, there are nine prophage/transposon-related genes in the vicinity of these genes, compared with few, if any, within genomes of members of the genus Desulfuromondales (Fig. 4, Table S1). Furthermore, genes coding for other pilin components are scattered throughout the Flexistipes sinusarabici genome and are flanked by numerous horizontal transfer genes.
The Deferribacter desulfuricans genome contains both the e-pilin and ‘pilA-C’ genes together in a cluster. Other genes required for pilin assembly are scattered throughout the genome in four other clusters and prophage- and transposon-related genes are found in two of these clusters (Fig. 4, Table S1).
In a similar manner, the genomes from Hippea maritima and Hippea alviniae contain both e-pilin and pilA–C genes, but other pilin-related genes are scattered throughout their genomes and flanked by horizontal transfer genes (Fig. 4, Table S1). Genes for E-pilin and ‘PilA–C’ are not present in the two other available genomes of members of the genus Hippea, Hippea jasoniae and '‘‘Hippea medeae'. ‘Hippea medeae' (D891DRAFT_0589), which has not been cultured, has a pilin protein that clusters with non-delta-related long type IVa pili from such genera as Pseudomonas (Fig. 1). No type IVa pili genes were detected in the genome of Hippea jasoniae.
Desulfobacterium autotrophicum is one of the few microbes that appears to have obtained the e-pilin gene via horizontal gene transfer with an adjacent ‘pilA–C ’ gene (Fig. 4, Table S1) that has been verified to reduce Fe(III) (Lovley et al., 1993) and Mn(IV) (Lovley & Phillips, 1994). Other pilin genes are scattered throughout the genome in five different gene clusters flanked with numerous horizontal transfer genes.
Two genomes of members of the genus Desulfobotulus are available. Both the e-pilin and ‘pilA-C ’ genes are found in Desulfobotulus alkaliphilus but only the e-pilin gene is present in the Desulfobotulus sapovorans genome. Similar to other species that are not members of the order Desulfuromondales, other pilin assembly genes are scattered throughout the genome and are flanked with genes indicative of horizontal transfer (Fig. 4, Table S1).
E-pilin genes were also detected in the genomes of three other sulfate reducers from the genus Desulfobacula, Desulfobacula toluolica, Desulfobacula phenolica and strain TS (Kim et al., 2014; Kuever et al., 2001; Rabus et al., 1993). Both Desulfobacula toluolica and Desulfobacula phenolica have an e-pilin gene, but no gene for ‘PilA–C,’ and strain TS only has a partial e-pilin gene (missing three amino acids at the N-terminus) that is located near a gap in the genome’s draft assembly (Tables 2 and 3). While a small hypothetical protein with five beta strands is found directly downstream of the e-pilin gene in Desulfobacula phenolica, it is unlikely to be ‘pilA–C ’ because it lacks a signal peptide. The Desulfobacula phenolica e-pilin gene is also unusual and has a beta strand at its C-terminus. The genome of strain TS is not sufficiently assembled for analysis, however both Desulfobacula toluolica and Desulfobacula phenolica have the full suite of genes required for pilus assembly scattered throughout the genome in seven different gene clusters that are flanked by prophage- and transposon related genes (Fig. 4, Table S1).
Genes coding for e-pili were also found in the genomes of Thauera linaloolentis, Thauera humireducens and Thauera terpenica, but none of these genes are accompanied by ‘pilA–C.’ The e-pilin gene and other pilin gene clusters in all three of these genomes is flanked by prophage- and transposon-related genes and the GC content of the e-pilin gene from Thauera humireducens, Thauera terpenica and Thauera linaloolentis is 15 %, 10 % and 19 % lower than the GC content of their respective genomes. All three of these genomes also have genes coding for long type IVa PilA proteins (Thauera linaloolentis has two long pilA). The e-pilin gene seems to be isolated within each of the genomes, whereas the long pilA gene clusters with genes required for proper pilin assembly (Fig. 4 and Table S1). Furthermore, the putative e-pilin protein in Thauera humireducens has a beta strand at its C-terminus similar to that found in long PilA proteins, suggesting that this putative e-pilin gene may instead be a truncation of a long pilA gene duplication.
Available genomes from two other denitrifying species of the genus Thauera, Thauera phenylacetica and Thauera aminoaromatica, do not contain genes for e-pilin, but do have genes coding for long type IVa pili.
Most Fe(III)-reducing bacteria lack e-pili
Genes for identifiable e-pilin are not present in the majority of known Fe(III)-reducing micro-organisms with sequenced genomes (Table 1). Only 21 % of the available 95 genomes contain e-pilin genes (Table 1). About half of the genomes without e-pilin genes contain genes for long type IVa PilA (Table 1).
There are a number of instances in which e-pili are not required for extracellular electron transfer. For example, some soluble natural organic compounds, such as humic substances, can act as electron shuttles (Lovley et al., 1996). They are reduced at the outer cell surface (Voordeckers et al., 2010) and then carry electrons to the surface of Fe(III) oxides (Lovley et al., 1996) or other cells (Smith et al., 2015), alleviating the need for long-range electron transport via e-pili. Other environmental components, such as sulfur compounds, may also act as electron shuttles (Nevin & Lovley, 2000). Alternatively, natural organic matter may chelate Fe(III) which can also be reduced at the cell surface (Nevin & Lovley, 2002). Direct contact of the outer cell surface with an insoluble electron acceptor, such as when Geobacter sulfurreducens cells are in direct contact with electrodes (Bond & Lovley, 2003), is another possibility, but the importance of this type of electron transfer in nature is not well understood. These alternative strategies for long-range electron transport may be most favored when rates of microbial metabolism are slow. For example, species of the genus Rhodoferax, which lack e-pili, predominated in a subsurface environment where rates of Fe(III)-reduction were low, but when Fe(III) reduction was artificially stimulated in this same environment as a bioremediation strategy there was a strong selection for species of the genus Geobacter (Mouser et al., 2009; Zhuang et al., 2011).
There is also the possibility that some Fe(III)-reducing micro-organisms that do not have e-pili employ other, independently evolved, types of electrically conductive filaments. For example, Rhodopsuedomonas palustris strain RP2 specifically produced electrically conductive filaments under Fe(III)-reducing conditions (Venkidusamy et al., 2015). It has yet to be determined whether these filaments are required for Fe(III) reduction, and the composition of the filaments is as yet unknown, but given that available Rhodopseudomonas palustris genomes lack e-pilin genes (Table 1), the filaments of Rhodopseudomonas palustris strain RP2 could offer an alternative strategy for extracellular electron exchange. Other examples of electrically conductive filaments putatively involved in extracellular electron exchange are being increasingly documented (Castro et al., 2014; Eaktasang et al., 2016; Li & Li, 2014). Further evaluation of the mechanisms for extracellular electron transfer in these organisms is also warranted.
Conclusions
The results suggest that e-pili of Geobacter sulfurreducens and Geobacter metallireducens, and presumably close relatives, are a relatively recent evolutionary development. The ability to transport electrons over multiple cell distances to minerals or other cells may be one of the key attributes that permits members of the genus Geobacter to outcompete other micro-organisms when there is strong selective pressure for rapid growth on Fe(III) minerals; electron exchange via direct interspecies electron transfer; or under artificial conditions when an electrode is provided as an electron acceptor and the growth of electrically conductive biofilms is favored (Lovley et al., 2011). The acquisition of e-pilin genes in micro-organisms outside the order Desulfuromondales has only been experimentally linked to the capacity for extracellular electron transfer in a few strains. Further investigation of the physiological capabilities of other species with a focus not only on Fe(III) oxide reduction, but also on direct interspecies electron transfer is warranted.
In addition to their significance in anaerobic microbial ecology, e-pili are of interest as a sustainable bioelectronic material. The genomic analysis reported here has identified many new e-pilin genes that may have value for the development of conductive materials. Studies to screen the conductivity of these newly identified e-pili are underway.
Acknowledgements
This research was supported by grants N00014-13-1-0550 and N00014-13-1-0549 from the Office of Naval Research.
Supplementary Data
Supplementary File 1
Abbreviation:
- e-pilin
electrically conductive pilin
References
- Adhikari R. Y., Malvankar N. S., Tuominen M. T., Lovley D. R.(2016). Conductivity of individual Geobacter pili. RSC Adv. 68354–8357. 10.1039/C5RA28092C [DOI] [Google Scholar]
- Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D.(1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 253389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M., Howell P. L., Burrows L. L.(2010). Architecture of the type II secretion and type IV pilus machineries. Future Microbiol 51203–1218. 10.2217/fmb.10.76 [DOI] [PubMed] [Google Scholar]
- Bernard C. S., Bordi C., Termine E., Filloux A., de Bentzmann S.(2009). Organization and PprB-dependent control of the Pseudomonas aeruginosa tad locus, involved in Flp pilus biology. J Bacteriol 1911961–1973. 10.1128/JB.01330-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber D., Ramer S. W., Wu C. Y., Murray W. J., Tobe T., Fernandez R., Schoolnik G. K.(1998). Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 2802114–2118. 10.1126/science.280.5372.2114 [DOI] [PubMed] [Google Scholar]
- Bond D. R., Lovley D. R.(2003). Electricity production by Geobacter sulfurreducens attached to electrodes. Appl Environ Microbiol 691548–1555. 10.1128/AEM.69.3.1548-1555.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows L. L.(2012). Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66493–520. 10.1146/annurev-micro-092611-150055 [DOI] [PubMed] [Google Scholar]
- Castro L., Vera M., Muñoz JÁ., Blázquez M. L., González F., Sand W., Ballester A.(2014). Aeromonas hydrophila produces conductive nanowires. Res Microbiol 165794–802. 10.1016/j.resmic.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Chun A. L.(2014). Bacterial nanowires: an extended membrane. Nat Nanotechnol 9750. 10.1038/nnano.2014.230 [DOI] [PubMed] [Google Scholar]
- Craig L., Pique M. E., Tainer J. A.(2004). Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2363–378. 10.1038/nrmicro885 [DOI] [PubMed] [Google Scholar]
- Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., et al. (2003). Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell 111139–1150. [DOI] [PubMed] [Google Scholar]
- Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A.(2006). Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23651–662. 10.1016/j.molcel.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Cuff J. A., Barton G. J.(2000). Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40502–511. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Schwartz R. M., Orcutt B. C.(1978). A Model of Evolutionary Change in Proteins. Atlas of Protein Sequence and Structure 345–352. Edited by Dayhoff M. O.Washington DC: National Biomedical Research Foundation. [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G. J.(2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43W389–394. 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaktasang N., Kang C. S., Lim H., Kwean O. S., Cho S., Kim Y., Kim H. S.(2016). Production of electrically-conductive nanoscale filaments by sulfate-reducing bacteria in the microbial fuel cell. Bioresour Technol 210. 10.1016/j.biortech.2015.12.090 [DOI] [PubMed] [Google Scholar]
- Eddy S. R.(2008). A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput Biol 4e1000069. 10.1371/journal.pcbi.1000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R.(2011). Accelerated profile HMM searches. PLoS Comput Biol 7e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano G. T., Steidl R. J., Reguera G.(2015). Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys Chem Chem Phys 1722217–22226. 10.1039/C5CP03432A [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44D279–285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford B., Verger D., Dodson K., Volkan E., Kostakioti M., Elam J., Pinkner J., Waksman G., Hultgren S.(2012). The structure of the PapD–PapGII pilin complex reveals an open and flexible P5 pocket. J Bacteriol 1946390–6397. 10.1128/JB.06651-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S., Falanga A., Cantisani M., Tarallo R., Della Pepa M. E., D'Oriano V., Galdiero M.(2012). Microbe–host interactions: structure and role of Gram-negative bacterial porins. Curr Protein Pept Sci 13843–854. 10.2174/138920312804871120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner C. L., Nguyen Y., Burrows L. L.(2012). Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76740–772. 10.1128/MMBR.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorby Y. A., Yanina S., McLean J. S., Rosso K. M., Moyles D., Dohnalkova A., Beveridge T. J., Chang I. S., Kim B. H., et al. (2006). Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci U S A 10311358–11363. 10.1073/pnas.0604517103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgel M., Ulstrup J. J., Bøggild A., Jones N. C., Hoffmann S. V., Nissen P., Boesen T.(2015). High-resolution structure of a type IV pilin from the metal-reducing bacterium Shewanella oneidensis. BMC Struct Biol 154. 10.1186/s12900-015-0031-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W.(1993). TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374166. [Google Scholar]
- Imam S., Chen Z., Roos D. S., Pohlschröder M.(2011). Identification of surprisingly diverse type IV pili, across a broad range of Gram-positive bacteria. PLoS One 6e28919. 10.1371/journal.pone.0028919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany S. C., Planet P. J., Desalle R., Fine D. H., Figurski D. H., Kaplan J. B.(2001). flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol 40542–554. 10.1046/j.1365-2958.2001.02422.x [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M.(2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Park S. J., Jung M. Y., Kim J. G., Min U. G., Hong H. J., Rhee S. K.(2014). Draft genome sequence of an aromatic compound-degrading bacterium, Desulfobacula sp. TS, belonging to the Deltaproteobacteria. FEMS Microbiol Lett 3609–12. 10.1111/1574-6968.12591 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L.(2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kuever J., Könneke M., Galushko A., Drzyzga O.(2001). Reclassification of Desulfobacterium phenolicum as Desulfobacula phenolica comb. nov. and description of strain SaxT as Desulfotignum balticum gen. nov., sp. nov. Int J Syst Evol Microbiol 51171–177. 10.1099/00207713-51-1-171 [DOI] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O.(2008). An improved general amino acid replacement matrix. Mol Biol Evol 251307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Li Y., Li H.(2014). Type IV pili of Acidithiobacillus ferrooxidans can transfer electrons from extracellular electron donors. J Basic Microbiol 54226–231. 10.1002/jobm.201200300 [DOI] [PubMed] [Google Scholar]
- Liu X., Tremblay P. L., Malvankar N. S., Nevin K. P., Lovley D. R., Vargas M.(2014). A Geobacter sulfurreducens strain expressing Pseudomonas aeruginosa type IV pili localizes OmcS on pili but is deficient in Fe(III) oxide reduction and current production. Appl Environ Microbiol 801219–1224. 10.1128/AEM.02938-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Malvankar N. S.(2015). Seeing is believing: novel imaging techniques help clarify microbial nanowire structure and function. Environ Microbiol 172209–2215. 10.1111/1462-2920.12708 [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Phillips E. J.(1994). Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl Environ Microbiol 602394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Coates J. D., Blunt-Harris E. L., Phillips E. J. P., Woodward J. C.(1996). Humic substances as electron acceptors for microbial respiration. Nature 382445–448. 10.1038/382445a0 [DOI] [Google Scholar]
- Lovley D. R., Roden E. E., Phillips E. J. P., Woodward J. C.(1993). Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Marine Geology 11341–53. 10.1016/0025-3227(93)90148-O [DOI] [Google Scholar]
- Lovley D. R., Ueki T., Zhang T., Malvankar N. S., Shrestha P. M., Flanangan K. A., Aklujkar M. A., Butler J. E., Giloteaux L., et al. (2011). Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv Micro Physiol 591–100. [DOI] [PubMed] [Google Scholar]
- Löytynoja A., Goldman N.(2005). An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A 10210557–10562. 10.1073/pnas.0409137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvankar N. S., Lovley D. R.(2014). Microbial nanowires for bioenergy applications. Curr Opin Biotechnol 2788–95. 10.1016/j.copbio.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Malvankar N. S., Vargas M., Nevin K. P., Franks A. E., Leang C., Kim B. C., Inoue K., Mester T., Covalla S. F., et al. (2011). Tunable metallic-like conductivity in microbial nanowire networks. Nat Nanotechnol 6573–579. 10.1038/nnano.2011.119 [DOI] [PubMed] [Google Scholar]
- Malvankar N. S., Vargas M., Nevin K., Tremblay P. L., Evans-Lutterodt K., Nykypanchuk D., Martz E., Tuominen M. T., Lovley D. R.(2015). Structural basis for metallic-like conductivity in microbial nanowires. MBio 6e00084. 10.1128/mBio.00084-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvankar N. S., Yalcin S. E., Tuominen M. T., Lovley D. R.(2014). Visualization of charge propagation along individual pili proteins using ambient electrostatic force microscopy. Nat Nanotechnol 91012–1017. 10.1038/nnano.2014.236 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res 43D222–226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S.(2002). Type IV pili and twitching motility. Annu Rev Microbiol 56289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- Mouser P. J., N'Guessan A. L., Elifantz H., Holmes D. E., Williams K. H., Wilkins M. J., Long P. E., Lovley D. R.(2009). Influence of heterogeneous ammonium availability on bacterial community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uranium-contaminated groundwater. Environ Sci Technol 434386–4392. 10.1021/es8031055 [DOI] [PubMed] [Google Scholar]
- Nevin K. P., Lovley D. R.(2000). Potential for nonenzymatic reduction of Fe(III) via electron shuttling in subsurface sediments. Environ. Sci. Technol 342472–2478. 10.1021/es991181b [DOI] [Google Scholar]
- Nevin K. P., Lovley D. R.(2002). Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol J 19141–159. 10.1080/01490450252864253 [DOI] [Google Scholar]
- Nevin K. P., Kim B. C., Glaven R. H., Johnson J. P., Woodard T. L., Methé B. A., Didonato R. J., Covalla S. F., Franks A. E., et al. (2009). Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4. 10.1371/journal.pone.0005628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M., Francetic O.(2014). Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 18431568–1577. 10.1016/j.bbamcr.2013.12.020 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H.(2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Rabus R., Nordhaus R., Ludwig W., Widdel F.(1993). Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol 591444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon P. N., Mueller K. T.(2013). Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem 28829260–29266. 10.1074/jbc.M113.498527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G., McCarthy K. D., Mehta T., Nicoll J. S., Tuominen M. T., Lovley D. R.(2005). Extracellular electron transfer via microbial nanowires. Nature 4351098–1101. 10.1038/nature03661 [DOI] [PubMed] [Google Scholar]
- Reguera G., Nevin K. P., Nicoll J. S., Covalla S. F., Woodard T. L., Lovley D. R.(2006). Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 727345–7348. 10.1128/AEM.01444-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollefson J. B., Stephen C. S., Tien M., Bond D. R.(2011). Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J Bacteriol 1931023–1033. 10.1128/JB.01092-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotaru A.-E., Shrestha P. M., Liu F., Shrestha M., Shrestha D., Embree M., Zengler K., Wardman C., Nevin K. P., Lovley D. R.(2014). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7408–415. 10.1039/C3EE42189A [DOI] [Google Scholar]
- Rotaru A.-E., Woodard T. L., Nevin K. P., Lovley D. R.(2015). Link between capacity for current production and syntrophic growth in Geobacter species. Front Microbiol 6. 10.3389/fmicb.2015.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I., Ashkenazy H., Katoh K., Pupko T.(2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res 43W7–W14. 10.1093/nar/gkv318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha P. M., Rotaru A. E., Aklujkar M., Liu F., Shrestha M., Summers Z. M., Malvankar N., Flores D. C., Lovley D. R.(2013). Syntrophic growth with direct interspecies electron transfer as the primary mechanism for energy exchange. Environ Microbiol Rep 5904–910. 10.1111/1758-2229.12093 [DOI] [PubMed] [Google Scholar]
- Smith J. A., Nevin K. P., Lovley D. R.(2015). Syntrophic growth via quinone-mediated interspecies electron transfer. Front Microbiol 6121. 10.3389/fmicb.2015.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers Z. M., Fogarty H. E., Leang C., Franks A. E., Malvankar N. S., Lovley D. R.(2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 3301413–1415. 10.1126/science.1196526 [DOI] [PubMed] [Google Scholar]
- Sun H., Zusman D. R., Shi W.(2000). Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol 101143–1146. 10.1016/S0960-9822(00)00705-3 [DOI] [PubMed] [Google Scholar]
- Szabó Z., Stahl A. O., Albers S., Kissinger J. C., Driessen A. J., Pohlschröder M.(2007). Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 189772–778. 10.1128/JB.01547-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F.(1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Adhikari R. Y., Malvankar N. S., Ward J. E., Nevin K. P., Woodard T. L., Smith J. A., Snoeyenbos-West O. L., Franks A. E., et al. (2016). The Low Conductivity of Geobacter uraniireducens Pili Suggests a Diversity of Extracellular Electron Transfer Mechanisms in the Genus Geobacter. Front Microbiol 7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich M., Planet P. J., Figurski D. H.(2007). The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 5363–375. 10.1038/nrmicro1636 [DOI] [PubMed] [Google Scholar]
- Tusnády G. E., Simon I.(2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17849–850. 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- Vargas M., Malvankar N. S., Tremblay P. L., Leang C., Smith J. A., Patel P., Snoeyenbos-West O., Synoeyenbos-West O., Nevin K. P., Lovley D. R.(2013). Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. MBio 4e00105–00113. 10.1128/mBio.00210-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey J. P., Reguera G., Tessmer S. H.(2011). Electronic properties of conductive pili of the metal-reducing bacterium Geobacter sulfurreducens probed by scanning tunneling microscopy. Physical Review E 84 10.1103/PhysRevE.84.060901 [DOI] [PubMed] [Google Scholar]
- Venkidusamy K., Megharaj M., Schroder U., Karouta F., Mohan S., V, Naidu R.(2015). Electron transport through electrically conductive nanofilaments in Rhodopseudomonas palustris strain RP2. RSC Adv.5100790–100798. [Google Scholar]
- Voordeckers J. W., Kim B. C., Izallalen M., Lovley D. R.(2010). Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl Environ Microbiol 76 2371–2375. 10.1128/AEM.02250-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G., Hultgren S. J.(2009). Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7765–774. 10.1038/nrmicro2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanger G., Gorby Y., El-Naggar M. Y., Yuzvinsky T. D., Schaudinn C., Gorur A., Sedghizadeh P. P.(2013). Electrically conductive bacterial nanowires in biphosphonate-related osteonecrosis of the jaw biofilms. Oral and Maxillofacial Path 11571–78. [DOI] [PubMed] [Google Scholar]
- Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 261608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A., Missiakas D., Raina S., Creighton T. E.(1995). Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 345075–5089. 10.1021/bi00015a019 [DOI] [PubMed] [Google Scholar]
- Zhuang K., Izallalen M., Mouser P., Richter H., Risso C., Mahadevan R., Lovley D. R.(2011). Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J 5305–316. 10.1038/ismej.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1.Holmes, Dang, Walker, and Lovley, FigShare https://figshare.com/s/87e875d0c5c97c2e5498 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1