Abstract
Staphylococcus aureus USA300 represents the dominant community-associated methicillin-resistant S. aureus lineage in the USA, where it is a major cause of skin and soft tissue infections. Previous comparative genomic studies have described the population structure and evolution of USA300 based on geographically restricted isolate collections. Here, we investigated the USA300 population by sequencing genomes of a geographically distributed panel of 191 clinical S. aureus isolates belonging to clonal complex 8 (CC8), derived from the Tigecycline Evaluation and Surveillance Trial program. Isolates were collected at 12 healthcare centres across nine USA states in 2004, 2009 or 2010. Reconstruction of evolutionary relationships revealed that CC8 was dominated by USA300 isolates (154/191, 81 %), which were heterogeneous and demonstrated limited phylogeographic clustering. Analysis of the USA300 core genomes revealed an increase in median pairwise SNP distance from 62 to 98 between 2004 and 2010, with a stable pattern of above average dN/dS ratios. The phylogeny of the USA300 population indicated that early diversification events led to the formation of nested clades, which arose through cumulative acquisition of predominantly non-synonymous SNPs in various coding sequences. The accessory genome of USA300 was largely homogenous and consisted of elements previously associated with this lineage. We observed an emergence of SCCmec negative and ACME negative USA300 isolates amongst more recent samples, and an increase in the prevalence of ϕSa5 prophage. Together, the analysed S. aureus USA300 collection revealed an evolving pan-genome through increased core genome heterogeneity and temporal variation in the frequency of certain accessory elements.
Keywords: MRSA, USA300, Evolution
Data Summary
1. Whole-genome sequence reads and assemblies of Staphylococcus aureus isolates described in this study have been deposited in the European Nucleotide Archive. The accession numbers together with related metadata are provided in Table S1 (available in the online Supplementary Material).
Impact Statement
The expansion of Staphylococcus aureus USA300 and its dominance as a community-associated MRSA clone within the USA has prompted interest in the genomics of this pathogen. The resolving power of whole-genome sequencing has already shed light on the nature of USA300 transmission within defined communities. However, a question of the broader population structure remains. Here, we investigated whole-genome sequences of epidemiologically diverse CC8 isolates, inclusive of 154 USA300 samples. Based on this collection we investigated the wider population structure of USA300 and other CC8 isolates and assessed temporal variation in core and accessory genomes. We report that the USA300 population has become increasingly heterogeneous between 2004 and 2010 at the core genome level, accompanied by subtle changes in the accessory genome involving loss of ACME and SCCmec elements and an increase in the prevalence of ϕSa5 prophage. These findings demonstrate that successful lineages of MRSA are not genetically static and continue to evolve, which may involve loss of molecular markers previously considered to define the lineage. While further surveillance of USA300 genomes is required to confirm wider SCCmec and ACME loss, such genomic changes might impact on the future epidemiology of this important S. aureus clone.
Introduction
Staphylococcus aureus is a major cause of human disease worldwide. One of the most successful lineages of this bacterial species is clonal complex 8 (CC8), from which a number of major methicillin-resistant S. aureus (MRSA) clones have emerged (Deurenberg & Stobberingh, 2008). This includes several pandemic MRSA clones associated with causing healthcare-associated (HA) infections as well as community-associated (CA) infections (Monecke et al., 2011). HA-MRSA CC8 clones include historic Archaic and Iberian clones as well as the more contemporaneous USA500 clone, whereas the current USA300 epidemic clone has emerged as the dominant cause of CA infections in the USA.
The first reports of USA300 date back to the early 2000s, when it was identified in a number of localised outbreaks of CA skin and soft-tissue infections (SSTI) (Centers for Disease Control and Prevention (CDC), 2001, 2003, McDougal et al., 2003; Tenover et al., 2006). Additional reports followed which demonstrated the widespread dissemination of USA300 across the USA (Begier et al., 2004; Kazakova et al., 2005; Miller et al., 2005). Other than SSTI, USA300 has also been reported as a causative agent of CA pneumonia, sepsis, infective endocarditis and osteomyelitis (Francis et al., 2005; Gonzalez et al., 2005; Haque et al., 2007; Seybold et al., 2007). In addition to being responsible for a rising incidence of CA infections, USA300 has also been increasingly reported in association with nasal colonisation (Freitas et al., 2010; Tenover et al., 2008) and HA infections (Gonzalez et al., 2006; Patel et al., 2007; Seybold et al., 2006). In the healthcare setting, USA300 was reported to emerge alongside established HA-MRSA clones (Chua et al., 2008; Limbago et al., 2009; Pasquale et al., 2013; Tattevin et al., 2009) or to replace them (Carey et al., 2010; Liu et al., 2008).
The striking epidemiology of USA300 within the USA has prompted a number of genomic studies aimed at the identification of genetic features that might have promoted its transmission, host colonisation and disease pathogenesis. Early whole-genome sequence analysis of a single USA300 strain revealed that the accessory genome included a Staphylococcal Cassette Chromosome mec (SCCmec) type IVa element, an arginine catabolic mobile element (ACME) type I, a S. aureus pathogenicity island 5 (SaPI5) and a Panton-Valentine leukocidin-carrying ϕSa2 prophage (Diep et al., 2006). These four mobile genetic elements (MGEs) represent common molecular markers of USA300, although analysis of a wider population demonstrated that their carriage is not ubiquitous (Uhlemann et al., 2014). Isolates belonging to USA300 also frequently carry two plasmids: a 27 kb plasmid carrying genes that confer resistance to penicillin (blaZ), erythromycin (msrA and mphBM) bacitracin (bcrA), kanamycin (aphA-3) and streptothricin (sat), and a 3.1 kb cryptic plasmid (Kennedy et al., 2010). The contribution of MGEs to the success of USA300 has been a subject of debate and generation of the hypothesis that the accessory genome has promoted the enhanced transmission of USA300 (Li et al., 2009). In particular, the ACME element has been suggested to promote the survival of USA300 on human skin and persistence within cutaneous abscesses (Thurlow et al., 2013). In contrast, the pathogenesis of USA300 may have been mediated by subtle genetic changes within the core genome (Highlander et al., 2007) together with modulated expression of core virulence determinants such as α-toxin, phenol-soluble modulins (PSMs) and an accessory gene regulator (agr) (Li et al., 2009). Core genome single nucleotide polymorphisms (SNPs) might also result in significantly different virulence phenotypes between USA300 isolates, associated with variable exoprotein expression (Kennedy et al., 2008).
Recently, the genomics of USA300 have been studied on a wider scale, through whole-genome sequencing of larger collections derived from specific geographic regions (Alam et al., 2015; Uhlemann et al., 2014). This led to significant epidemiological findings such as the identification of households as long-term reservoirs of USA300. Comparative genomic analysis has also allowed the diversification of the USA300 lineage to be studied, including evolutionary events surrounding the emergence of the USA300 clade as well as micro-evolution within the population (Uhlemann et al., 2014). This exemplifies the power of whole-genome sequencing for comprehensive analysis of bacterial pathogens, as it allows an in-depth interrogation of phylogenetic relationships, identification of genetic features associated with virulence and antimicrobial resistance, and the reconstruction of significant evolutionary events (Harris et al., 2010; Holden et al., 2013). While previous studies describing the comparative genomics of USA300 focused on geographically restricted isolate collections, here we describe whole-genome sequence analysis of S. aureus CC8 derived from 12 USA healthcare centres located across nine states. The isolates, collected in 2004, 2009 or 2010, were derived from the multi-centre Tigecycline Evaluation and Surveillance Trial (T.E.S.T) study and represent consecutive clinical isolates (methicillin-susceptible and methicillin -resistant) from both the community and hospital settings. As USA300 has become widespread across the USA, an analysis of a collection from geographically distinct locations has allowed us to study a broader population of epidemiologically unrelated USA300 isolates as well as their microevolution over time. For this we interrogated the phylogeny of the CC8 lineage, with particular focus on the USA300 clade, followed by analysis of temporal variation within core and accessory genomes as well as investigation of more subtle polymorphisms that define the phylogenetic structure of the USA300 population.
Methods
Bacterial isolates.
Isolates analysed here derived from the Tigecycline Evaluation and Surveillance Trial (T.E.S.T.), a global multi-centre surveillance study. The trial was initiated in 2004, and since then sites have been added and removed. For the purpose of conducting a longitudinal assessment of S. aureus isolates associated with disease in the USA, we prospectively selected USA sites that were participating between 2004 and 2010 (five sites). A further seven sites active at the time of study were included. A total of 516 S. aureus isolates derived from 2004, 2009 and 2010 were collected and analysed. All isolates belonging to CC8 (n = 191), as determined by multi-locus sequence typing, were selected for this work.
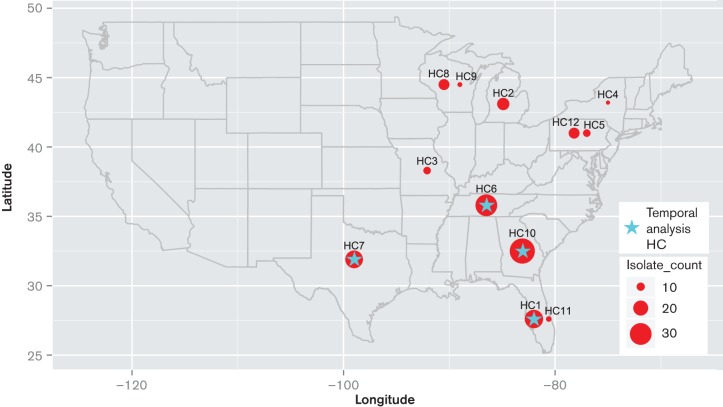
In line with T.E.S.T requirements, the participating sites collected consecutive isolates from patients with a documented infection. Only one isolate per patient was permitted and all body sites were considered an acceptable clinical source. Epidemiological details of the 191 S. aureus CC8 isolates analysed here are presented in Table S1 (available in the online Supplementary Material). Briefly, isolates were derived from the 12 T.E.S.T. sites that were located across nine states of the USA (Fig. 1), with 26 % (50/191) collected in 2004, 31 % (58/191) in 2009 and 43 % (83/191) in 2010. For the purpose of this work the collecting sites were called healthcare centre 1 – 12 (HC1 – HC12). The collection consisted of 46 MSSA and 145 MRSA isolates. Isolates were collected from diverse clinical sources, categorised in this work to represent four main infection types defined as wound, invasive, respiratory, or other. Isolates were collected from patients aged 0 to 95 in various clinical settings, with samples broadly defined in this study as derived from either inpatient or outpatient.
Fig. 1.
Geographic distribution of healthcare centres (HC) that collected isolates analysed in this study. The geographic location on the map represents state coordinates, where the corresponding HC was based. Plots are scaled to represent total number of isolates from corresponding site. HCs that provided isolates used in temporal analyses are marked with a star.
Whole-genome sequencing.
Genomic DNA was isolated using the Qiagen QIAcube system according to manufacturer’s protocol. Tagged DNA libraries were created using a method adapted from a standard Illumina Indexing protocol, as described previously (Hsu et al., 2015). Whole-genome sequencing was performed on the Illumina HiSeq 2000 platform with 100 bp paired-end reads. The Illumina sequence data has been submitted to the European Nucleotide Archive (ENA) and the accession numbers are provided in Table S1.
Whole-genome sequence data analysis.
Single nucleotide polymorphisms (SNPs) were detected by mapping the paired-end reads against the S. aureus USA300_FPR3757 reference genome (GenBank accession NC_007793), using SMALT version 0.5.8. (http://www.sanger.ac.uk/science/tools/smalt-0). The generated whole-genome sequence alignment was curated to exclude accessory regions (mobile genetic elements identified for USA300_FPR3757 reference genome) and variable sites associated with recombination, with the latter detected using Gubbins (Croucher et al., 2014). Based on the curated core genome SNP alignment a maximum-likelihood phylogenetic tree was reconstructed using RAxML version 7.8.6 (Stamatakis, 2014) with the generalised time reversible (GTR) model with GAMMA method of correction for among site rate variation and 100 bootstrap replications (Harris et al., 2010). De novo assembly of whole-genome sequences was performed using Velvet (Zerbino & Birney, 2008) with Velvet Optimiser (http://bioinformatics.net.au/software.velvetoptimiser.shtml) and kmers ranging between 66 % and 90 % of the read-length. Contigs of less than 300 bp were removed. The draft assemblies have been submitted to the ENA with accession numbers provided in Table S1. The assembled contigs were annotated using Prokka (Seemann, 2014). The accessory genome was defined as described previously (Harris et al., 2010).
Isolate genotyping.
Multi-locus sequence typing was performed on assembled genomes using an in-house developed pipeline based on a previously described typing scheme (Enright et al., 2000). Isolates were also spa genotyped by extracting the spa gene variable X region from assembled genomes using previously described primers (Kahl et al., 2005), and determining the spa type with the spaTyper tool (http://spatyper.fortinbras.us). MLST and spa genotypes of all isolates are presented in Table S1. In addition, in silico PCR was used to confirm carriage of prophages (Goerke et al., 2009) and to determine the SCCmec type amongst mecA-positive samples (Milheiriço et al., 2007a, b).
Statistical analyses.
To test the statistical significance of the temporal variation in the prevalence of USA300 isolates as well as selected mobile genetic elements, the likelihood ratio test was applied. The strength of phylogenetic signal of epidemiological trait was measured using phylo.d() function in R package caper based on 1000 permutations. The function computes a D value, which provides a measure of trait dispersion for categorical characters (Fritz & Purvis, 2010). A negative D statistic indicates that a trait is phylogenetically clustered whereas a positive D statistic indicates that a trait is over dispersed across the phylogenetic tree.
Results and Discussion
CC8 population structure overview and temporal variation
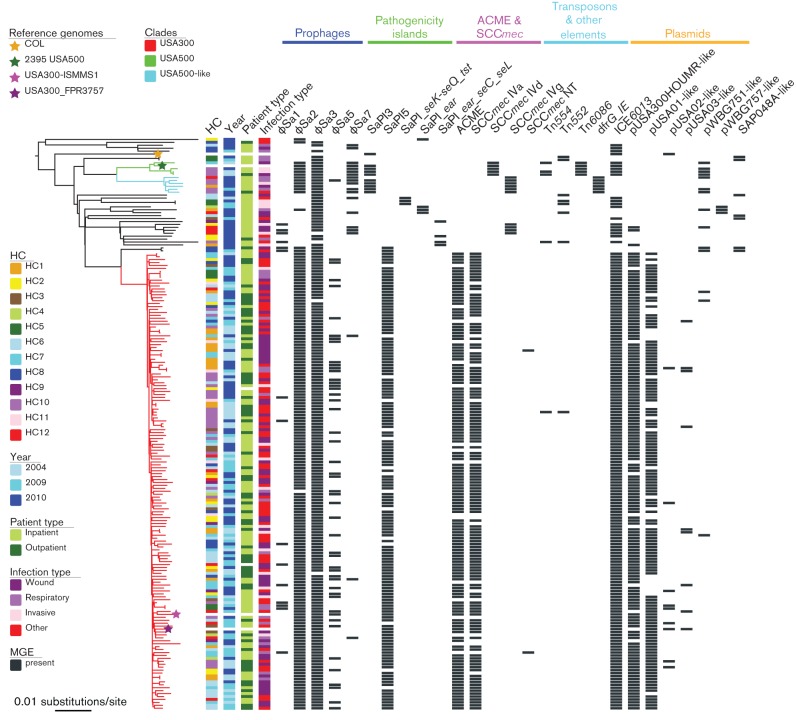
Whole-genome sequences of 191 clinical S. aureus isolates belonging to CC8 were analysed, the majority of which belonged to ST8 (179/191, 94 %; Table S1). A total of ten single-locus variants (SLV) of ST8 were identified, which included six distinct novel STs. The majority of isolates belonged to spa type t008 (146/191, 76 %; Table S1). Alignment of the core genomes (after excluding accessory regions and regions of predicted recombination, resulting in a core genome length of 2 592 236 bp) of all CC8 isolates revealed a total of 10 651 SNP sites. To identify different sub-lineages, a phylogenetic tree of all CC8 samples was reconstructed with the addition of USA300, USA500 and COL reference genomes (Fig. 2). The tree revealed that the majority of isolates belonged to a single dominant clade (154/191, 81 %), which contained the two USA300 reference strains (USA300_FPR3757 and USA300-ISMMS1) and thus was identified to represent the USA300 genotype. The population also contained four isolates that clustered with the USA500 reference strain (2395 USA500) and, as such, represented the USA500 clone. Six further isolates were closely related to the USA500 clade but descended from a distinct node and were labelled here USA500-like. The majority of these USA500 and USA500-like isolates (7/10) originated from a single healthcare centre (HC10, state of Georgia). The reconstructed phylogeny and detection of USA300 and USA500 genotypes in the context of a diverse CC8 population revealed that the two are only distantly related, in contrast to a previous suggestion that USA500 represents a progenitor of the USA300 clone (Li et al., 2009). A number of non-clonal MSSA CC8 isolates were more closely related to the USA300 clade than the USA500 sub-lineage suggesting that the two MRSA clones arose independently. The structure of CC8 phylogeny presented here is in line with a recently published overview of evolutionary relationships between various CC8 genomes (Boyle-Vavra et al., 2015).
Fig. 2.
Midpoint-rooted phylogenetic tree of the 191 analysed S. aureus CC8 isolates. Four reference genomes were included (corresponding nodes marked with a star). Branches of USA300, USA500 and USA500-like clades have been highlighted. The tree has been annotated to show the association between phylogenetic distribution and healthcare centre (HC) of origin (excluding reference genomes), year of collection (excluding reference genomes), patient type (excluding reference genomes), infection type (excluding reference genomes) and carriage of mobile genetic elements (MGE). The latter shows the distribution of identified prophages, pathogenicity islands (where novel sequence was identified, name has been assigned based on virulence gene content), transposons and other elements such as ICE6013 and dfrG insertion element, and plasmids (name assigned based on close similarity to previously identified plasmid). Further details on variable carriage of virulence genes within ϕSa2 and ϕSa3 prophages, as well as variable distribution of antimicrobial resistance genes within pUSA300HOUMR-like and pUSA03-like plasmids have been provided in Fig. S1. The tree was annotated using EvolView (Zhang et al., 2012).
Isolates belonging to the USA300 clade were detected amongst samples from all healthcare centres. Some genotypic heterogeneity was observed within the USA300 clade, which contained six SLVs of ST8. We also observed a level of spa type diversity amongst USA300 isolates. While the majority belonged to spa type t008 (132/154, 86 %), 10 other spa types were identified, including t024 (8/154, 5 %) and t681 (3/154, 2 %). The detected genotype diversity of USA300 isolates based on multi- and single-locus genotyping underscores the benefit of applying core genome sequence phylogeny for comprehensive identification of clinically significant sub-lineages such as S. aureus USA300.
Focusing on the USA300 clade only, we investigated the strength of phylogenetic signal in the distribution of isolates based on the healthcare centre of origin, year of isolation, patient type and infection type. For this we calculated the D statistic, which provides a measure of trait dispersion. A random distribution of isolates based on geographic origin was observed for the majority of healthcare centres (D range 0.17–1.22), with exception of HC1 (D = −0.68). Phylogenetic clustering was also observed for isolates collected in 2004 (D = −0.08) with random distribution of isolates from 2009 and 2010 (D = 0.86 and D = 0.85, respectively). No phylogenetic signal could be observed based on patient type (outpatient, D = 0.73; inpatient, D = 0.91) or infection type (wound, D = 0.48; respiratory, D = 0.67; invasive, D = 1.1).
We next investigated whether the CC8 population structure and the distribution of USA300 genotype varied at different sampling time points. For this, isolates collected in 2004 together with 2009 and 2010 collections derived from matched healthcare centres were used (Fig. 1). We found that USA300 was the dominant clone in all years. No significant variation in the distribution of USA300 was observed between 2004 (46/50, 92 %) and 2009 (23/24, 96 %) collections (p = 0.52). However, the 2010 collection (29/40, 73 %) revealed a significantly lower frequency of USA300 genotype in comparison with both 2004 (p < 0.05) and 2009 (p < 0.05). This suggests that the significant drop in the prevalence of USA300 observed for 2010 was likely specific to that year rather than a time trend. Outside of the USA300 clade, the USA500-like clone was observed only amongst more recent samples with a single isolate identified in 2009 (1/24, 4 %) and a comparatively higher prevalence in 2010 (4/40, 10 %).
Accessory genome of CC8
The genomes of 191 S. aureus CC8 isolates were analysed for the distribution of variable regions, followed by annotation to define the accessory component. Overall, a diverse mobile genetic repertoire was identified across the lineage, although isolates belonging to USA300, USA500 and USA500-like clones revealed the presence of clade-associated MGEs (Fig. 2).
In total, five different prophage types were identified. The most common was ϕSa3, found in 96 % of isolates, although variation in carriage of prophage-encoded virulence factors was observed (Fig. S1). Nearly all ϕSa3-positive USA300 isolates carried the immune evasion cluster (IEC) composed of scn, chp and sak genes, which was also detected amongst some non-USA300 isolates. The second most common prophage, due to the dominance of USA300 within this population, was ϕSa2. All but one USA300 isolate carried the Panton-Valentine leukocidin-positive ϕSa2USA300, which was also detected in three non-USA300 isolates. Three further prophages were identified within the analysed CC8 population: ϕSa1, ϕSa5 and ϕSa7. All three were detected at considerably lower frequency than ϕSa3 or ϕSa2. Prophage ϕSa1 was detected sporadically but across the entire CC8 population, whereas ϕSa7 was prevalent mostly amongst non-USA300 MRSA isolates and found in all USA500 and the majority of USA500-like (5/6) samples. In contrast, prophage ϕSa5 was identified predominantly within the USA300 clade (47/154, 30 %), and was randomly distributed (D = 1.66) across the USA300 phylogeny suggesting multiple acquisition events rather than prevalence due to clonal propagation of ϕSa5-positive isolates. No putative virulence genes were identified within the sequence of any of the three prophages. We investigated whether acquisition of these prophages might affect the bacterial host thorough gene inactivation at the insertion site. Analysis of the chromosomal environment surrounding the site of prophage insertion revealed that ϕSa1 and ϕSa7 inserted within intergenic locations downstream of the iron-sulphur cluster assembly protein gene sufB (between coding sequences corresponding to USA300_FPR3757 SAUSA300_0822 and SAUSA300_0823), and the heme transport protein gene isdB (between coding sequences corresponding to USA300_FPR3757 SAUSA300_1027 and SAUSA300_1028), respectively. In contrast, prophage ϕSa5 were found inserted within a coding sequence responsible for a putative metal-binding protein with iron-sulphur oxidoreductase domain (SAUSA300_1858), which would likely interfere with its expression.
Other accessory virulence genes amongst analysed CC8 isolates were associated with various pathogenicity islands, with SaPI5 identified in majority of USA300 isolates (139/154, 90 %). Nearly all USA500 (3/4) and USA500-like (5/6) isolates carried the SaPI3, which similarly to SaPI5, encodes the seK and seQ enterotoxin genes.
The majority of USA300 isolates carried ACME (137/154, 89 %) and SCCmec type IVa elements (130/154, 84 %), which were not found in any other isolates. Where the elements were absent this was predicted to have resulted from a deletion event, with corresponding isolates dispersed across the phylogenetic tree. In line with a previous report (Uhlemann et al., 2014), an association between the absence of SCCmec and ACME was observed, with majority of ACME-negative USA300 isolates also lacking the SCCmec (13/17, 76 %). This is not surprising considering the proximal location of these elements in the USA300 chromosome. Nearly all SCCmec-positive USA300 isolates carried a complete type IVa element although two isolates carried unusual recombinant variants of SCCmec IVa, which arose through putative recombination of SCCmec with ACME or another SCCmec element (Fig. S2). We also identified one isolate carrying a remnant SCC element lacking the mecA gene that exhibited similarity to SCCpbp4 from Staphylococcus epidermidis ATCC 12228. Finally, non-USA300 mecA-positive isolates carried either SCCmec IVd or SCCmec IVg. The carriage of SCCmec correlated with the methicillin susceptibility phenotype (Table S1) except for a single SCCmec-positive isolate, which was identified as MSSA (PFESA2067). Further investigation revealed the presence of a single base insertion in mecA gene resulting in a frame-shift mutation and premature stop codon, which would likely lead to a loss of mecA expression.
The analysed CC8 population revealed a high prevalence of an accessory region recognised as integrative conjugative element 6013 (ICE6013) (Smyth & Robinson, 2009). The element was detected in all USA300 and USA500 isolates but was absent in all USA500-like samples. The ICE6013 does not appear to carry any specific virulence or resistance genes and lacks insertion specificity (Smyth & Robinson, 2009), integrating at a number of sites amongst the analysed CC8 isolates, including both inter- and intragenic positions (Fig. S3). In the USA300 isolates however, ICE6013 was only found inserted within a putative membrane protein gene (SAUSA300_0606), and thus an insertional inactivation of this particular coding sequence might provide selective advantage to USA300. Interestingly, this particular ICE6013 insertion site differs from the USA300_FPR3757 reference genome (Diep et al., 2006), which also contains this element, but is identical to ICE6013 location within the USA300-ISMMS1 genome (Altman et al., 2014).
There was only a sporadic carriage of resistance-associated transposon elements such as tetM-positive Tn6086, ermA- and spc-positive Tn554, and bla operon-carrying Tn552. All USA500-like isolates carried a 3.3 kb insertion element containing the trimethoprim resistance gene dfrG, not found elsewhere in this collection. Amongst USA300 isolates, the antimicrobial resistance genes were carried predominantly on plasmids, with the majority of samples (146/154, 95 %) carrying a large plasmid resembling a previously reported 27 kb pUSA300HOUMR (Highlander et al., 2007). The plasmid contains genes that confer resistance to various antimicrobials including penicillin (blaZ), kanamycin (aphA-3), streptothricin (sat), bacitracin (bcrA) and erythromycin (msrA, mphBM), and homologous plasmids have been previously reported in other USA300 isolates (Kennedy et al., 2010; Uhlemann et al., 2014). However, amongst the 146 USA300 isolates carrying the pUSA300HOUMR-like plasmid, 12 carried a variant that lacked some of the resistance genes described above, with at least five different variants detected (Fig. S1). The pUSA300HOUMR-like plasmid was not confined to the USA300 clade within this collection, and was detected in several other isolates. A few USA300 isolates (8/154, 5 %) carried a circa 34 kb plasmid, which partly resembled the 37 kb pUSA03 that contains genes conferring resistance to erythromycin (ermC) and mupirocin (ileS-2) (Diep et al., 2006). At least three variants of this pUSA03-like plasmid were observed (Fig. S1). All lacked the ermC gene, and only some carried the ileS-2 resistance determinant. In addition some of the ileS-2-positive variants also carried the kanamycin resistance gene addD. Also detected amongst USA300 isolates (3/154, 2 %) was a 2.5 kb ermC-positive pWBG751-like plasmid and a small tetracycline resistance (tetK) plasmid (8/154, 5 %), resembling the 4.4 kb pUSA02 (Diep et al., 2006).
Temporal variation in the accessory genome
We further analysed whether the variable distribution of selected MGEs was associated with the year of sampling. For this, we compared the distribution of SCCmec, ACME and ϕSa5 amongst USA300 isolates collected in 2004 with the 2009 and 2010 collections from matched healthcare centres (Table 1). All USA300 isolates carried SCCmec in 2004, with a single negative isolate in 2009. The frequency of SCCmec carriage was lowest amongst 2010 isolates, which represented a significant drop in comparison with both 2004 (p < 0.05) and 2009 (p < 0.05) groups. Emergence of SCCmec-negative USA300 isolates resulting from a sporadic loss of the element has been previously reported, however at lower frequency than observed here (Uhlemann et al., 2014). As such the reduced frequency of SCCmec amongst USA300 isolates from 2010 might be specific to that year. Nevertheless a reported shift in antimicrobial prescription practices in the US between 2004 and 2008, away from β-lactams in favour of MRSA-active drugs when treating SSTI might have influenced the epidemiology of CA-MRSA infections (Talan et al., 2011). Lower frequency of beta-lactam usage could result in a lower selective pressure to maintain SCCmec leading to gradual loss of this element. While USA300 has been recognised predominantly as an MRSA lineage, infections caused by MSSA USA300 have also been reported suggesting that transmission of USA300 may not be entirely dependent on SCCmec carriage (McCaskill et al., 2007; Talan et al., 2011). In support of the hypothesis that a shift in epidemiology of MRSA might have occurred around 2010, we also observed an increase in the prevalence of non-USA300 SCCmec-negative isolates at this sampling time point.
Table 1. Temporal variation in the distribution of selected mobile genetic elements among USA300 isolates. Based on isolates derived from healthcare centres selected for temporal analysis (HC1, HC6, HC7, HC10) .
| 2004 | 2009 | 2010 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N* | SCCmec | ACME | ΦSa5 | N* | SCCmec | ACME | ΦSa5 | N* | SCCmec | ACME | ΦSa5 | |
| HC1 | 14 | 14 | 14 | 2 | na | na | na | na | 8 | 6 | 7 | 5 |
| HC6 | 6 | 6 | 6 | 1 | 12 | 12 | 12 | 4 | 8 | 7 | 7 | 3 |
| HC7 | 8 | 8 | 8 | 3 | 10 | 9 | 8 | 2 | 5 | 2 | 4 | 3 |
| HC10 | 18 | 18 | 18 | 2 | 1 | 1 | 1 | 0 | 8 | 6 | 6 | 3 |
| Total | 46 | 46 | 46 | 8 | 23 | 22 | 21 | 6 | 29 | 21 | 24 | 14 |
| p† | 0.19 | <0.05 | 0.7 | |||||||||
| p‡ | <0.05 | <0.05 | <0.05 | |||||||||
| p§ | <0.05 | 0.39 | 0.07 | |||||||||
Total number of USA300 isolates identified for corresponding centre and year.
p value after controlling for each HC, for the distribution of corresponding MGE among USA300 isolates in 2004 and 2009.
p value after controlling for each HC, for the distribution of corresponding MGE among USA300 isolates in 2004 and 2010.
p value after controlling for each HC, for the distribution of corresponding MGE among USA300 isolates in 2009 and 2010.
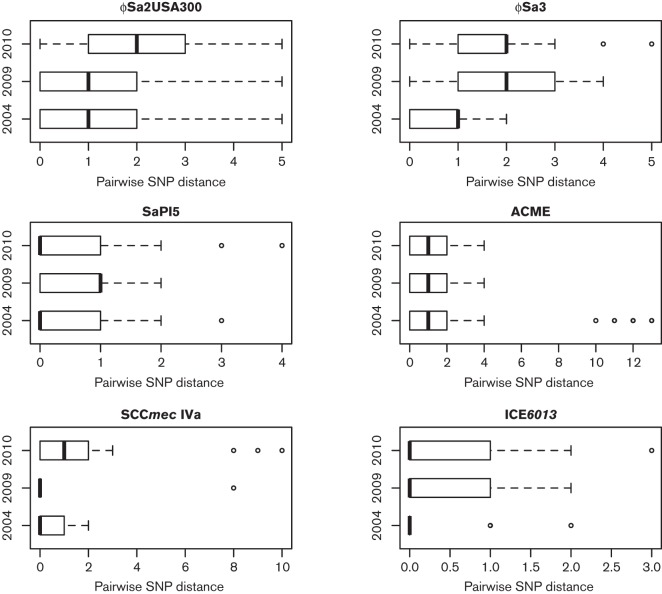
As described above, carriage of ACME was associated with the distribution of SCCmec amongst USA300 isolates. Therefore in line with SCCmec prevalence, loss of ACME was only observed in 2009 and 2010 samples, both groups showing a significantly lower number of ACME-positive USA300 isolates when contrasted with the 2004 group (p < 0.05). Although the prevalence of ACME-negative USA300 isolates was more notable in 2010, the difference between 2009 and 2010 was not significant (p = 0.39).
The sample size of our collection was relatively low and thus the observed temporal variation in the distribution of SCCmec and ACME among USA300 isolates, and the apparent loss of these elements in more recent samples, would require further surveillance. However, the occurrence of SCCmec-negative and ACME-negative USA300 isolates was not limited to a particular healthcare centre (Table 1). Furthermore, the absence of phylogenetic association between USA300 isolates lacking either SCCmec or ACME suggests that forces driving the loss of these important MGEs are acting upon the wider USA300 population.
In addition to changes in prevalence of SCCmec and ACME, we observed a trend of increasing prophage ϕSa5 frequency, although only the difference between 2004 and 2010 isolates was statistically significant (p < 0.05). The overall frequency of ϕSa5 carriage by USA300 isolates collected in 2009 and 2010 was higher than the prevalence reported for New York City isolates (16 %) (Uhlemann et al., 2014). However, the presence of ϕSa5 prophage within the latter collection was associated with a particular USA300 sub-clade, suggesting that carriage of this prophage might contribute to expansion of more successful sub-populations (Uhlemann et al., 2014).
The USA300 population analysed in this study demonstrated a stable temporal prevalence of antimicrobial resistance-associated MGEs. All isolates collected in 2004 carried the multiple drug resistance pUSA300HOUMR-like plasmid, which was also found in the majority of 2009 (20/23, 87 %) and 2010 isolates (28/29, 97 %) from matched healthcare centres. A dip in pUSA300HOUMR-like plasmid prevalence amongst 2009 isolates was specific to a single centre and thus unlikely to represent a wider trend. It has been previously observed that carriage of aminoglycoside- and bacitracin-resistance genes associated with the pUSA300HOUMR-like plasmid might mediate resistance to certain topical antimicrobial agents available over-the-counter in the USA (Suzuki et al., 2011). It is likely, therefore, that a continuous selective pressure has been maintaining this element within the USA300 population.
We also investigated whether the chromosomally-inserted MGEs carried by USA300 (ϕSa2, ϕSa3, SaPI5, ACME, SCCmec IVa and ICE6013) diversified over time leading to temporal variation in the distribution of pairwise SNP distances. To discount recombination-mediated variation, alignments of each MGE sequence were first queried for the presence of recombination regions and if detected, the corresponding isolate was removed from the analysis (up to two isolates for ICE6013, ϕSa3 and SCCmec IVa were removed). We found that the analysed MGEs were well conserved between the isolates, with a median pairwise SNP distance for all elements equal to 1. When comparing the median of pairwise SNP distances for individual MGEs between the 2004, 2009 and 2010 isolates, temporal variation was observed for all except ACME and ICE6013 elements (Fig. 3). For all, an increase of median pairwise SNP distance by 1 was observed at either or both later sampling time points.
Fig. 3.
Temporal variation in pairwise SNP distance distributions between chromosomally-inserted MGEs carried by USA300 isolates. Based on isolates derived from healthcare centres selected for temporal analysis (HC1, HC6, HC7, HC10).
Outside the USA300 clade, it was noteworthy that the temporally distributed USA500-like sub-population differed markedly in its accessory genome from the closely related USA500 clone and could be distinguished based on SCCmec type, carriage of dfrG insertion element and absence of the ICE6013.
Core genome microevolution of USA300 clade
We investigated further the core genomes of USA300 isolates and found that the population formed a heterogeneous clade with a median pairwise SNP distance of 88 and a range of 0–165 SNPs. We then investigated if USA300 population has become more heterogeneous over time by comparing pairwise SNP distances of isolates collected in 2004 with the panels from 2009 and 2010 (isolates from matched healthcare centres only). We observed an increase in median pairwise SNP distance values between 2004 [62, median absolute deviation (MAD) = 13] and 2009 (103, MAD = 13) as well as 2010 (98, MAD = 15) groups. As previously reported (Kennedy et al., 2008), the USA300 isolates demonstrate an elevated ratio of non-synonymous-to-synonymous SNPs (dN/dS), associated with the recent clonal expansion of this lineage and accumulation of SNPs that have not yet been eliminated by purifying selection (Rocha et al., 2006). We observed a dN/dS median of 0.71 (MAD = 0.19) across USA300 core genomes, in comparison to the median 0.22 (MAD = 0.01) dN/dS ratio for non-USA300 isolates. In order to determine the dN/dS ratio of mutations that were acquired independently by the USA300 isolates after each diverged from the last common ancestor, we determined the dN/dS ratios on terminal branches of the USA300 clade. For clonal or very closely related isolates (up to 2 pairwise SNPs distance) a single representative was selected, and isolates that accumulated less than a total of 10 SNPs since divergence from the last common ancestor, or showed either no synonymous or no non-synonymous mutations were excluded. A total of 139 USA300 isolates were analysed and revealed a median dN/dS ratio of 2.6 (MAD = 1.1), which corresponded to a previously reported dN/dS ratio for another USA300 collection (Kennedy et al., 2008). Analysis of temporal changes in dN/dS ratio revealed equally high median values for 2004 (2.33, MAD = 1.23), 2009 (2.5, MAD = 0.4) and 2010 (2.8, MAD = 1.5) altogether indicating that purifying selection has not yet acted upon the USA300 population (Rocha et al., 2006).
To interrogate more closely the phylogenetic structure of the USA300 clade, we repeated the phylogeny reconstruction for USA300 isolates only (Fig. 4). Genomic divergence within the USA300 clade, based on branch lengths, was most pronounced on terminal branches and thus was strain-specific. In line with previous studies describing the population structure of USA300 isolates, the generated phylogeny revealed low resolution of inter-strain evolutionary relationships due to short internal branches. We therefore investigated further the clustering patterns of the innermost branches descending immediately from the root of USA300 clade, by focusing on branches that after each evolutionary split gave rise to a majority of the descending population. This revealed a subtle and sequential diversification process involving the formation of nested clades (Fig. 4). Based on the collection analysed in this study, at least six fine-scale divergence events descending from the root of the clade could be identified, each involving acquisition of up to five SNPs (Table 2). Where branches emerged due to multiple SNPs, in each case the mutations were independent in relation to their genomic position and thus did not represent a recombination. Nearly all mutations occurring in protein-coding sequence were non-synonymous (11/12) and involved genes of various functions. Four of the non-synonymous SNPs occurring across three distinct branches were identified in genes associated with antimicrobial resistance. This included a gene encoding a putative drug resistance transporter ErmB/QacA (SAUSA300_2126), a gene encoding an efflux pump (SAUSA300_2489) recently associated with resistance to linoleic and arachidonic acids that are present on the skin and contribute to the low pH of this environment (Alnaseri et al., 2015), and the genes gyrA (SAUSA300_006) and parC/grlA (SAUSA300_1251) associated with fluoroquinolone resistance. The emergence of fluoroquinolone resistance in the USA300 population was previously described and estimated to have occurred around 1995 (Uhlemann et al., 2014). Amongst other genes affected by non-synonymous SNPs was a putative virulence determinant (SAUSA300_1322), which belongs to the cvfC operon and contributes to control of haemolysin production and detergent resistance through positive regulation of thyA gene (Ikuo et al., 2010).
Fig. 4.
Mid-point rooted phylogenetic tree of S. aureus USA300 isolates. Two most closely related non-USA300 isolates were included as an out-group (isolates PFESA2332 and PFESA1492, red branches). Also, two reference genomes were included and corresponding nodes are marked with a star. Branches were highlighted to show consecutive clades of the nested clade structure, and labelled 1 to 6. SNPs that define each labelled clade are described in Table 2.
Table 2. Descriptive summary of SNPs identified on branches of early divergence nested clades across the phylogenetic tree of analysed USA300 isolates. The phylogeny is presented in Fig. 4.
| Clade | FPR3757 Position | SNP Type* | CDS ID | Product | Base change | AA change |
|---|---|---|---|---|---|---|
| 1 | 2300219 | NS | SAUSA300_2126 | drug resistance transporter, EmrB/QacA subfamily | A -> G | V156A |
| 2 | 150941 | NS | SAUSA300_0132 | glycosyl transferase | T -> A | D271E |
| 714167 | I | − | − | A -> G | − | |
| 1453525 | NS | SAUSA300_1322 | virulence factor C, CvfC3 | G -> A | A281V | |
| 1877205 | NS | SAUSA300_1701 | NAD(FAD)-utilizing dehydrogenase | G -> A | R228H | |
| 1915352 | I | − | − | A -> G | − | |
| 3 | 386867 | I | − | − | A -> T | − |
| 4 | 906507 | I | − | − | C -> A | − |
| 1693739 | NS | SAUSA300_1543 | oxygen-independent coproporphyrinogen III oxidase | C -> T | A145T | |
| 1908169 | NS | SAUSA300_1725 | transaldolase | C -> T | V6I | |
| 2230768 | S | SAUSA300_2068 | hypothetical protein | T -> C | − | |
| 2690087 | NS | SAUSA300_2489 | antibiotic transport-associated protein | G -> T | Q795K | |
| 5 | 7282 | NS | SAUSA300_0006 | DNA gyrase GyrA | C -> T | S84L |
| 1374405 | NS | SAUSA300_1251 | DNA topoisomerase ParC/GrlA | C -> A | S80Y | |
| 6 | 837108 | NS | SAUSA300_0750 | hypothetical protein | T -> G | D17E |
| 1074815 | NS | SAUSA300_0980 | putative membrane protein | A -> G | N71D |
* SNP type: NS – non-synonymous; I – intergenic; S – synonymou.
To further resolve the observed USA300 tree topology and formation of nested clades, we have reconstructed the phylogeny of the studied USA300 isolates together with 329 genomes of previously sequenced USA300 isolates derived from New York city (Uhlemann et al., 2014). The tree topology of this expanded USA300 collection also revealed a nested clade population structure (Fig. S4). The clade -defining mutations corresponded to the polymorphisms described above (Table 2) demonstrating that the early diversification changes identified in this work can be detected across wider USA300 population.
Conclusions
This study provides an overview of the S. aureus CC8 population structure based on whole-genome sequence analysis of clinical isolates derived from nine USA states, collected in 2004, 2009 and 2010. Unsurprisingly, it was dominated by the USA300 clone. The non-USA300 isolates represented a fifth of the CC8 population and included the USA500 clone as well as a closely related USA500-like genotype with a distinct accessory genome composition, which emerged at the later sampling time point. The USA500 clone emerged prior to the appearance and dissemination of the USA300 genotype but has been diminishing in prevalence since (David et al., 2015; Nelson et al., 2015). This trend was also observed in our collection, with the majority of USA500 isolates identified amongst the 2004 samples. Furthermore, the distribution of both USA500 and USA500-like isolates was geographically restricted as most originated from a single site (HC10). This is in contrast to geographically dispersed USA300 isolates. There may also be additional regional differences associated with USA500 that we did not detect owing to the majority of USA500 isolates being detected from just one site. For example, Benson et al. (2014) identified that most of the USA500 isolates obtained from patients in New York area harboured the IS256 element located in close proximity to the rot gene. In our analysis we identified the presence of this IS element in the genomes of all USA500 isolates and a single USA500-like strain. However, insertion of IS256 next to rot was observed for only one of the USA500 strains.
The analysis of temporal variation in the population structure of CC8 isolates indicated a significant drop in the prevalence of USA300 around 2010, in comparison with both 2004 and 2009. However, due to the limitations of our study such as narrow sampling time points, small sample size and lack of isolates representing the Western USA we could not verify if this dip in frequency of USA300 clone represented a broader event or a time trend. A recent review reported that there has been an overall increase in spatio-temporal prevalence of USA300 across the USA between 2000 and 2013 in the context of MRSA epidemiology (Carrel et al., 2015). However, regional differences as well as year-on-year fluctuations were also observed, demonstrating a need for a national surveillance program that would allow more accurate monitoring of the epidemiology of clinically significant S. aureus clones such as USA300.
Based on this geographically diverse collection of CC8 isolates, we investigated the USA300 clade for evidence of microevolution. The observed limited phylogeographic clustering suggests a general lack of geographical expansion events resulting from emergence of locally adapted clones. This is in agreement with findings that at a local community level, multiple introductions of USA300 occur (Uhlemann et al., 2014). Analysis of core genomes revealed that USA300 isolates are becoming more heterogeneous as defined by a temporal increase in pairwise SNP distances, which is underlined by a stable pattern of elevated dN/dS ratios. However, an analysis of the wider phylogenetic relationships defined by branches descending immediately from the root of the clade revealed the presence of subtle polymorphisms shared across the population. The pattern of accumulative SNP acquisition as defined by the formation of nested clades suggests that the USA300 ancestral isolates continued to diversify shortly after the emergence of the clade but prior to its clonal expansion. Although determining the phenotypic significance of the acquired SNPs is beyond the scope of this work, we speculate that at least some might represent adaptive evolution such as development of antimicrobial resistance.
Our data also suggests temporal changes in the composition of the USA300 accessory genome involving small-scale loss of SCCmec and ACME elements and increased frequency of ϕSa5 carriage. These findings are subject to the previously described limitations of this study and thus might not be indicative of a continuous temporal trend of USA300 accessory genome re-shuffle. Still, the collection analysed in this study represents an unbiased sample of S. aureus CC8 population derived from diverse clinical sources, providing a unique insight into the broader epidemiology and evolution of USA300 in the context of its parental lineage. This allowed us to capture the apparent instability of SCCmec and ACME carriage amongst more recent USA300 isolates. This is significant considering some strategies for identification of USA300 isolates based on defined genetic markers such as carriage of SCCmec or ACME (Alam et al., 2015; David et al., 2013), which might hinder identification of emerging USA300 variants that have lost these MGEs leading to their under-reporting. Also, taking into consideration the putative role of SCCmec and ACME in the successful dissemination of S. aureus USA300, it will be important to understand what leads to the loss of these elements. We speculate that the loss of SCCmec may have been driven by changes in antimicrobial prescription practices in the USA. It is more challenging to hypothesize what factors could have contributed to the drop in ACME prevalence. However, the observation that absence of SCCmec and ACME coincided in majority of isolates, suggests that the elements are frequently lost in a single deletion event. This scenario is supported by previous findings that the CcrAB recombinases of SCCmec can mobilise ACME as well as catalyse an excision of the entire ACME– SCCmec composite island (Diep et al., 2008). While further work is required to investigate how this might impact on the epidemiology of USA300, the analysis presented here of spatially diverse USA300 isolates gives a new insight into the wider evolutionary changes occurring within this highly successful sub -lineage, and provides ground for further surveillance of this clone.
Acknowledgements
We thank the core sequencing and informatics teams at the Sanger Institute for their assistance as well as the following employees of Pfizer for their helpful suggestions: Li Hao, Robert Donald and Elizabeth Bieger. S. R. H. and J. P. were supported by Wellcome Trust grant 098051. This work was funded by Pfizer Vaccine Research and Development. N. M., C. Y. T. and A. S. A. are employees of Pfizer and may own stock in the company.
Supplementary Data
Supplementary File 1
Abbreviations:
- ACME
arginine catabolic mobile element
- CA-MRSA
community-associated methicillin-resistant Staphylococcus aureus
- CC
clonal complex
- CDS
coding DNA sequence
- HA-MRSA
hospital-associated methicillin-resistant Staphylococcus aureus
- HC
healthcare centre
- ICE6013
integrative conjugative element 6013
- MAD
median absolute deviation
- MGE
mobile genetic element
- MRSA
methicillin-resistant Staphylococcus aureus
- MSSA
methicillin-susceptible Staphylococcus aureus
- SaPI
staphylococcal pathogenicity island
- SCCmec
Staphylococcal Cassette Chromosome mec
- SNP
single nucleotide polymorphism
- SLV
single locus variant
- ST
sequence type
- T.E.S.T.
Tigecycline Evaluation and Surveillance Trial
References
- Alam M. T., Read T. D., Petit R. A., Boyle-Vavra S., Miller L. G., Eells S. J., Daum R. S., David M. Z.(2015). Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio 6e00054. 10.1128/mBio.00054-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnaseri H., Arsic B., Schneider J. E., Kaiser J. C., Scinocca Z. C., Heinrichs D. E., McGavin M. J.(2015). Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J Bacteriol 1971893–1905. 10.1128/JB.02607-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D. R., Sebra R., Hand J., Attie O., Deikus G., Carpini K. W., Patel G., Rana M., Arvelakis A.(2014). Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am J Transplant 142640–2644. 10.1111/ajt.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begier E. M., Frenette K., Barrett N. L., Mshar P., Petit S., Boxrud D. J., Watkins-Colwell K., Wheeler S., Cebelinski E. A., Connecticut Bioterrorism Field Epidemiology Response Team (2004). A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin Infect Dis 391446–1453. 10.1086/425313 [DOI] [PubMed] [Google Scholar]
- Benson M. A., Ohneck E. A., Ryan C., Alonzo F., Smith H., Narechania A., Kolokotronis S. O., Satola S. W., Uhlemann A. C.(2014). Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93664–681. 10.1111/mmi.12682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Vavra S., Li X., Alam M. T., Read T. D., Sieth J., Cywes-Bentley C., Dobbins G., David M. Z., Kumar N.(2015). USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus. MBio 6e02585–e02514. 10.1128/mBio.02585-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A. J., Della-Latta P., Huard R., Wu F., Graham P. L., Carp D., Saiman L.(2010). Changes in the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol 31613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel M., Perencevich E. N., David M. Z.(2015). USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg Infect Dis 211973. 10.3201/eid2111.150452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2001). Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison-mississippi, 2000. MMWR Morb Mortal Wkly Rep 50919. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2003). Methicillin-resistant Staphylococcus aureus infections among competitive sports participants-Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-–2003. MMWR Morb Mortal Wkly Rep 52793. [PubMed] [Google Scholar]
- Chua T., Moore C. L., Perri M. B., Donabedian S. M., Masch W., Vager D., Davis S. L., Lulek K., Zimnicki B.(2008). Molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates in urban Detroit. J Clin Microbiol 462345–2352. 10.1128/JCM.00154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Page A. J., Connor T. R., Delaney A. J., Keane J. A., Bentley S. D., Parkhill J., Harris S. R.(2014). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. Z., Acree M. E., Sieth J. J., Boxrud D. J., Dobbins G., Lynfield R., Boyle-Vavra S., Daum R. S.(2015). Pediatric Staphylococcus aureus isolate genotypes and infections from the dawn of the community-associated methicillin-resistant S. aureus epidemic era in Chicago, 1994 to 1997. J Clin Microbiol 532486–2491. 10.1128/JCM.00096-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. Z., Taylor A., Lynfield R., Boxrud D. J., Short G., Zychowski D., Boyle-Vavra S., Daum R. S.(2013). Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 51814–819. 10.1128/JCM.02429-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E.(2008). The evolution of Staphylococcus aureus. Infect Genet Evol 8747–763. 10.1016/j.meegid.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A.(2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. The Lancet 367731–739. [DOI] [PubMed] [Google Scholar]
- Diep B. A., Stone G. G., Basuino L., Graber C. J., Miller A., des Etages S. A., Jones A., Palazzolo-Ballance A. M., Perdreau-Remington F.(2008). The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 1971523–1530. 10.1086/587907 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G.(2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 381008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. S., Doherty M. C., Lopatin U., Johnston C. P., Sinha G., Ross T., Cai M., Hansel N. N., Perl T.(2005). Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis 40100–107. 10.1086/427148 [DOI] [PubMed] [Google Scholar]
- Freitas E. A., Harris R. M., Blake R. K., Salgado C. D.(2010). Prevalence of USA300 strain type of methicillin-resistant Staphylococcus aureus among patients with nasal colonization identified with active surveillance. Infect Control 31469–475. [DOI] [PubMed] [Google Scholar]
- Fritz S. A., Purvis A.(2010). Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv Biol 241042–1051. 10.1111/j.1523-1739.2010.01455.x [DOI] [PubMed] [Google Scholar]
- Goerke C., Pantucek R., Holtfreter S., Schulte B., Zink M., Grumann D., Bröker B. M., Doskar J.(2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 1913462–3468. 10.1128/JB.01804-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B. E., Martinez-Aguilar G., Hulten K. G., Hammerman W. A., Coss-Bu J., Avalos-Mishaan A., Mason E. O., Kaplan S. L.(2005). Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics 115642–648. 10.1542/peds.2004-2300 [DOI] [PubMed] [Google Scholar]
- Gonzalez B. E., Rueda A. M., Shelburne S. A., Musher D. M., Hamill R. J., Hultén K. G.(2006). Community-associated strains of methicillin-resistant Staphylococcus aureus as the cause of healthcare-associated infection. Infect Control 271051–1056. [DOI] [PubMed] [Google Scholar]
- Haque N. Z., Davis S. L., Manierski C. L., Vager D., Donabedian S. M., Perri M. B., Sabbagh R., Cheema F.(2007). Infective endocarditis caused by USA300 methicillin-resistant Staphylococcus aureus (MRSA). Int J Antimicrob Agents 3072–77. 10.1016/j.ijantimicag.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T., Quail M. A., Nickerson E. K., Chantratita N., Gardete S., Tavares A., Day N.(2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander S. K., Hultén K. G., Qin X., Jiang H., Yerrapragada S., Mason E. O., Shang Y., Williams T. M., Fortunov R. M.(2007). Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7 10.1186/1471-2180-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Hsu L. Y., Kurt K., Weinert L. A., Mather A. E., Harris S. R., Strommenger B., Layer F., Witte W.(2013). A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23653–664. 10.1101/gr.147710.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. Y., Harris S. R., Chlebowicz M. A., Lindsay J. A., Koh T. H., Krishnan P., Tan T. Y., Hon P. Y., Grubb W. B.(2015). Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol 1681. 10.1186/s13059-015-0643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuo M., Kaito C., Sekimizu K.(2010). The cvfC operon of Staphylococcus aureus contributes to virulence via expression of the thyA gene. Microb Pathog 491–7. 10.1016/j.micpath.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Kahl B. C., Mellmann A., Deiwick S., Peters G., Harmsen D.(2005). Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J Clin Microbiol 43502–505. 10.1128/JCM.43.1.502-505.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakova S. V., Hageman J. C., Matava M., Srinivasan A., Phelan L., Garfinkel B., Boo T., McAllister S., Anderson J.(2005). A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 352468–475. 10.1056/NEJMoa042859 [DOI] [PubMed] [Google Scholar]
- Kennedy A. D., Otto M., Braughton K. R., Whitney A. R., Chen L., Mathema B., Mediavilla J. R., Byrne K. A., Parkins L. D.(2008). Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci U S A 1051327–1332. 10.1073/pnas.0710217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. D., Porcella S. F., Martens C., Whitney A. R., Braughton K. R., Chen L., Craig C. T., Tenover F. C., Kreiswirth B. N.(2010). Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol 484504–4511. 10.1128/JCM.01050-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Diep B. A., Villaruz A. E., Braughton K. R., Jiang X., DeLeo F. R., Chambers H. F., Lu Y.(2009). Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 1065883–5888. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbago B., Fosheim G. E., Schoonover V., Crane C. E., Nadle J., Petit S., Heltzel D., Ray S. M., Harrison L. H., Active Bacterial Core surveillance MRSA Investigators (2009). Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 471344–1351. 10.1128/JCM.02264-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Graber C. J., Karr M., Diep B. A., Basuino L., Schwartz B. S., Enright M. C., O'Hanlon S. J., Thomas J. C.(2008). A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in Sansan francisco 2004–2005. Clin Infect Dis 461637–1646. [DOI] [PubMed] [Google Scholar]
- McCaskill M. L., Mason E. O., Kaplan S. L., Hammerman W., Lamberth L. B., Hultén K. G.(2007). Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. Pediatr Infect Dis J 261122–1127. 10.1097/INF.0b013e31814536e0 [DOI] [PubMed] [Google Scholar]
- McDougal L. K., Steward C. D., Killgore G. E., Chaitram J. M., McAllister S. K., Tenover F. C.(2003). Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 415113–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheiriço C., Oliveira D. C., de Lencastre H.(2007a). Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother 6042–48. 10.1093/jac/dkm112 [DOI] [PubMed] [Google Scholar]
- Milheiriço C., Oliveira D. C., de Lencastre H.(2007b). Update to the multiplex PCR695 strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother 513374–3377. 10.1128/AAC.00275-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. G., Perdreau-Remington F., Rieg G., Mehdi S., Perlroth J., Bayer A. S., Tang A. W., Phung T. O.(2005). Necrotizing fasciitis caused by community associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med 3521445–1453. 10.1056/NEJMoa042683 [DOI] [PubMed] [Google Scholar]
- Monecke S., Coombs G., Shore A. C., Coleman D. C., Akpaka P., Borg M., Chow H., Ip M., Jatzwauk L.(2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6e17936. 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. U., Bizzarro M. J., Baltimore R. S., Dembry L. M., Gallagher P. G.(2015). Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in the decade following implementation of an active detection and isolation program. J Clin Microbiol 532492–2501. 10.1128/JCM.00470-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale T. R., Jabrocki B., Salstrom S. J., Wiemken T. L., Peyrani P., Haque N. Z., Scerpella E. G., Ford K. D., Zervos M. J., Ramirez J. A.(2013). Emergence of methicillin resistant Staphylococcus aureus USA300 genotype as a major cause of late-onset nosocomial pneumonia in intensive care patients in the USA. Int J Infect Dis 17e398–e403. 10.1016/j.ijid.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Patel M., Kumar R. A., Stamm A. M., Hoesley C. J., Moser S. A., Waites K. B.(2007). USA300 genotype community-associated methicillin-resistant Staphylococcus aureus as a cause of surgical site infections. J Clin Microbiol 453431–3433. 10.1128/JCM.00902-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P., Smith J. M., Hurst L. D., Holden M. T., Cooper J. E., Smith N. H., Feil E. J.(2006). Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol 239226–235. 10.1016/j.jtbi.2005.08.037 [DOI] [PubMed] [Google Scholar]
- Seemann T.(2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics, btu153. [DOI] [PubMed] [Google Scholar]
- Seybold U., Kourbatova E. V., Johnson J. G., Halvosa S. J., Wang Y. F., King M. D., Ray S. M., Blumberg H. M.(2006). Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 42647–656. 10.1086/499815 [DOI] [PubMed] [Google Scholar]
- Seybold U., Talati N. J., Kizilbash Q., Shah M., Blumberg H. M., Franco-Paredes C.(2007). Hematogenous osteomyelitis mimicking osteosarcoma due to community associated methicillin-resistant Staphylococcus aureus. Infection 35190–193. 10.1007/s15010-007-6058-3 [DOI] [PubMed] [Google Scholar]
- Smyth D. S., Robinson D. A.(2009). Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J Bacteriol 1915964–5975. 10.1128/JB.00352-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.(2014). Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 301312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Yamada K., Nagao M., Aoki E., Matsumoto M., Hirayama T., Yamamoto H., Hiramatsu R., Ichiyama S.(2011). Antimicrobial ointments and methicillin-resistant Staphylococcus aureus USA300. Emerg Infect Dis 171917–1920. 10.3201/eid1710.101365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talan D. A., Krishnadasan A., Gorwitz R. J., Fosheim G. E., Limbago B., Albrecht V., Moran G. J., for The EMERGEncy ID Net Study Group (2011). Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis 53144–149. 10.1093/cid/cir308 [DOI] [PubMed] [Google Scholar]
- Tattevin P., Diep B. A., Jula M., Perdreau-Remington F.(2009). Methicillin-resistant Staphylococcus aureus USA300 clone in long-term care facility. Emerg Infect Dis 15953. 10.3201/eid1506.080195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., McAllister S., Fosheim G., McDougal L. K., Carey R. B., Limbago B., Lonsway D., Patel J. B., Kuehnert M. J.(2008). Characterization of Staphylococcus aureus isolates from nasal cultures collected from individuals in the United States in 2001 to 2004. J Clin Microbiol 462837–2841. 10.1128/JCM.00480-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., McDougal L. K., Goering R. V., Killgore G., Projan S. J., Patel J. B., Dunman P. M.(2006). Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 44108–118. 10.1128/JCM.44.1.108-118.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow L. R., Joshi G. S., Clark J. R., Spontak J. S., Neely C. J., Maile R., Richardson A. R.(2013). Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13100–107. 10.1016/j.chom.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann A.-C., Dordel J., Knox J. R., Raven K. E., Parkhill J., Holden M. T., Peacock S. J., Lowy F. D.(2014). Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community.Pediatr Infect Dis J 1116738–6743. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E.(2008). Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res 18821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gao S., Lercher M. J., Hu S., Chen W. H.(2012). Evolview, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40 W569–W572. 10.1093/nar/gks576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1. Altman, D.R., Sebra, R., Hand, J., Attie, O., Deikus, G., Carpini, K.W.D., Patel, G., Rana, M., Arvelakis, A., Grewal, P. & Dutta, J. GenBank accession NZ_CP007176 (2014). [DOI] [PMC free article] [PubMed]
- 2. Benson, M.A., Ohneck, E.A., Ryan, C., Alonzo, F., Smith, H., Narechania, A., Kolokotronis, S.O., Satola, S.W., Uhlemann, A.C., Sebra, R. & Deikus, G. GenBank accession NZ_CP007499 (2014). [DOI] [PMC free article] [PubMed]
- 3. Diep, B.A., Gill, S.R., Chang, R.F., Phan, T.H., Chen, J.H., Davidson, M.G., Lin, F., Lin, J., Carleton, H.A., Mongodin, E.F. & Sensabaugh, G.F. GenBank accession NC_007793 (2006).
- 4. Gill, S.R., Fouts, D.E., Archer, G.L., Mongodin, E.F., DeBoy, R.T., Ravel, J., Paulsen, I.T., Kolonay, J.F., Brinkac, L., Beanan, M. & Dodson, R.J. GenBank accession NC_002951 (2005).
- 5. Uhlemann, A.C., Dordel, J., Knox, J.R., Raven, K.E., Parkhill, J., Holden, M.T., Peacock, S.J. & Lowy, F.D. ENA accession PRJEB2870 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1