Significance
RNA polymerases (RNAPs) from all organisms share a common form of regulation: Regulator proteins bind in a conserved “secondary channel” pore on RNAP and alter RNAP activity. In bacteria, multiple such regulators are present in the same cell, but how these bind without mutual interference is unclear. We directly observed binding of single molecules of secondary channel protein GreB to RNAP transcription complexes. Unexpectedly, GreB was not selectively recruited to RNAPs requiring its transcript proofreading function. Instead, GreB transiently bound to multiple types of complexes, eventually finding via random search RNAPs that require its activity. The observations suggest a paradigm by which a regulator can act while minimizing obstruction of a binding site that must be shared with other proteins.
Keywords: secondary channel, total internal reflection fluorescence, transcription elongation, backtracking
Abstract
The secondary channel (SC) of multisubunit RNA polymerases (RNAPs) allows access to the active site and is a nexus for the regulation of transcription. Multiple regulatory proteins bind in the SC and reprogram the catalytic activity of RNAP, but the dynamics of these factors’ interactions with RNAP and how they function without cross-interference are unclear. In Escherichia coli, GreB is an SC protein that promotes proofreading by transcript cleavage in elongation complexes backtracked by nucleotide misincorporation. Using multiwavelength single-molecule fluorescence microscopy, we observed the dynamics of GreB interactions with elongation complexes. GreB binds to actively elongating complexes at nearly diffusion-limited rates but remains bound for only 0.3–0.5 s, longer than the duration of the nucleotide addition cycle but far shorter than the time needed to synthesize a complete mRNA. Bound GreB inhibits transcript elongation only partially. To test whether GreB preferentially binds backtracked complexes, we reconstituted complexes stabilized in backtracked and nonbacktracked configurations. By verifying the functional state of each molecular complex studied, we could exclude models in which GreB is selectively recruited to backtracked complexes or is ejected from RNAP by catalytic turnover. Instead, GreB binds rapidly and randomly to elongation complexes, patrolling for those requiring nucleolytic rescue, and its short residence time minimizes RNAP inhibition. The results suggest a general mechanism by which SC factors may cooperate to regulate RNAP while minimizing mutual interference.
The multisubunit RNA polymerases (RNAPs) that synthesize mRNAs have catalytic subunits that are conserved across all domains of life but incorporate different regulatory structures and mechanisms (1, 2). Despite this divergence, all RNAPs have in common certain key target sites by which they interact with regulatory proteins. One of the most important of these is the secondary channel (SC), a pore extending from the outside surface of RNAP to the enzyme active site deeply buried in the center of the transcription elongation complex (EC) (3).
Even within an individual bacterial species, multiple regulatory factors may interact with the SC. Escherichia coli has at least five proteins that are thought to bind in the RNAP SC to regulate transcription (4). These proteins have similar overall structures (5) but have qualitatively different effects on transcription complexes and thus have distinct roles in regulation.
A well-characterized role of some SC proteins is the endonucleolytic rescue of backtracked ECs (6–8). In response to particular protein factors, template DNA sequences, transcript sequences, or nucleotide deprivation, the EC may undergo “backtracking” in which it reverse threads the nascent RNA and the DNA template by one or more base pair (9–12). In a backtracked conformation the EC is still intact, but the active site is misaligned for nucleotide addition because the RNA 3′ end extends into the SC (13, 14). The E. coli proteins GreA and GreB stimulate endonucleolytic cleavage of the RNA backbone by the RNAP active site to create a new 3′-OH that is correctly positioned at the active site for resumed polymerization. This cleavage activity may function in living cells to reduce the duration of transcriptional pauses that involve backtracking and to rescue ECs stalled by misincorporation of a noncomplementary base at the nascent transcript 3′ end (15, 16).
In contrast to its positive role in promoting transcript synthesis through rescue of backtracked complexes, GreB binding also is predicted to inhibit the process of transcript elongation. In particular, a combination of crystallographic data and structural modeling suggests that when GreB binds in the SC, it is likely to insert into the RNAP active site in a manner that inhibits nucleotide addition to the nascent transcript (13, 17–20). However, functional studies have not demonstrated inhibition by wild-type Gre proteins in vitro (17, 20), raising the question of how the proteins can stimulate transcript cleavage but not interfere with transcript elongation. One possibility is that GreB is specifically recruited only by the backtracked ECs that require rescue (18, 20).
A parallel question relates to the sharing of the SC by multiple SC-binding proteins. Based on their structures and other data, the proteins are widely assumed to compete for the SC (5, 21). However, there is conflicting evidence that competition is limited or nonexistent. For example, DksA is an SC protein with multiple roles during initial polymerase binding and subsequent transcript elongation and termination (18–20). Nevertheless, competition experiments between GreB and DksA show that each protein’s activity is unimpeded in the excess (10- to 300-fold) presence of the other protein, raising the possibility that the proteins are recruited to distinct subpopulations of ECs (18, 20). In contrast, other observations have been taken to suggest that multiple SC factors can bind simultaneously to RNAP; such simultaneous binding is inconsistent with simple binding competition (19, 20, 22).
To resolve the apparent inconsistencies in data on SC protein function, it is essential to define the kinetics of interaction of SC proteins with transcription complexes and how these kinetics change as the complexes proceed through the both active and backtracked intermediates of transcript elongation. Here we use a multiwavelength single-molecule fluorescence microscopy technique, colocalization single-molecule spectroscopy (CoSMoS) (23), to observe the dynamic interaction of GreB molecules with individual ECs that either are undergoing active transcript elongation or are statically locked in defined backtracked or nonbacktracked configurations. Our data contradict the recruitment hypothesis and define a mechanism by which GreB can function in transcript cleavage without substantially interfering with the activities of other SC proteins or with transcript elongation.
Results
GreB Partially Inhibits Transcription Elongation.
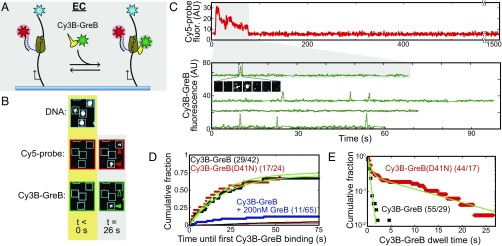
To examine the effects of GreB on EC activity, we used a single-molecule elongation rate assay that assesses elongation rates independently of initiation kinetics and measures the distribution of elongation rates across the molecular population (24). We tethered Alexa Fluor 488 (AF488)-labeled DNA molecules containing the λPR′ promoter (Fig. 1A) to the surface of a passivated flow chamber, observed them by single-molecule total internal reflection fluorescence (TIRF) microscopy, and recorded their locations (Fig. 1 B and C, Left). After incubation with E. coli σ70 RNAP to form open complexes, we removed free RNAP and introduced a solution containing NTPs and a Cy5-labeled probe oligonucleotide complementary to sequences near the 5′ end of the transcript.
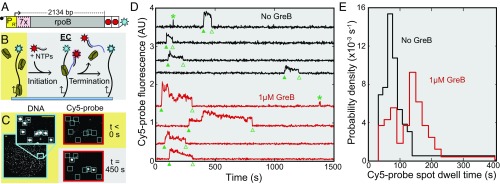
Fig. 1.
Single-molecule measurement of transcript elongation inhibition by GreB. (A) The DNA template (24) contains the PR′ promoter (yellow) driving transcription of seven tandem repeats of a 20-nt hybridization target (pink) followed by the E. coli rpoB gene (dark gray) and ending with the terminators TR′ and Te (red). The upstream end of the DNA was labeled with biotin (black circle), and the downstream end was labeled with an AF488 dye (blue star). (B) Design of the single-round, single-molecule transcription experiment (see text). Production of the nascent transcript (blue line) is detected by TIRF microscopy as the appearance of a colocalized spot of fluorescence from the dye-labeled Cy5-probe oligonucleotide (red star). Termination of transcription releases the RNA from the surface and results in the disappearance of the spot. (C) Images showing examples of DNA (blue) and Cy5-probe (red) fluorescent spots. DNA images show the full-microscope field of view and a magnified inset with individual DNA molecules marked (squares). (Scale bar in Inset: 3 µm.) Cy5-probe images show the surface region corresponding to the magnified inset before and after the start of the experiment; locations of the DNA molecules are marked. (D) Examples of records (offset for clarity) of Cy5-probe fluorescence intensity recorded from 3- × 3-pixel squares centered at the positions of individual DNA molecules in experiments without (black traces) or with (red traces) 1 µM GreB. Filled and empty triangles indicate spot appearance and disappearance events that were scored as the initial detection of and release of a transcript, respectively. Asterisks indicate spots present for durations <30 s, which were likely caused by transient nonspecific Cy5-probe binding and therefore not scored. AU, arbitrary units. (E) Probability density distributions of elongation Cy5-probe spot lifetimes with [red trace; median = 146 ± 6 s (SE); n = 81; seven events >400 s; maximum = 1,140 s] and without (black trace; median = 81 ± 5 s; n = 84; three events >400 s; maximum = 831 s) 1 µM GreB.
Spots of Cy5-probe fluorescence appeared at locations that colocalized with DNA molecules (Fig. 1 C and D). Of the observed DNA molecules, 32 ± 3% (SE; 84/263) exhibited a single colocalization with a duration >30 s that was scored as production of a transcript. Control experiments in which no NTPs were added showed essentially no transcript production (0.9 ± 0.9%; 1/117). The difference between the times at which the Cy5-probe spot appeared and disappeared (Fig. 1D) was used to estimate the time required to elongate the transcript from +172 until transcript release, which is rapid at intrinsic terminators (25, 26). The distribution of these Cy5-probe dwell times (Fig. 1E, black traces) displayed a peak with a long tail, as expected from the previously measured heterogeneity in RNAP elongation rates (27) and approximately consistent with the rates measured in previous single-molecule assays (24). Decreasing the laser exposure of the probe by a factor of ∼15 did not significantly alter the median Cy5-probe dwell time (Fig. S1), indicating that the measured dwell times were not significantly limited by photobleaching.
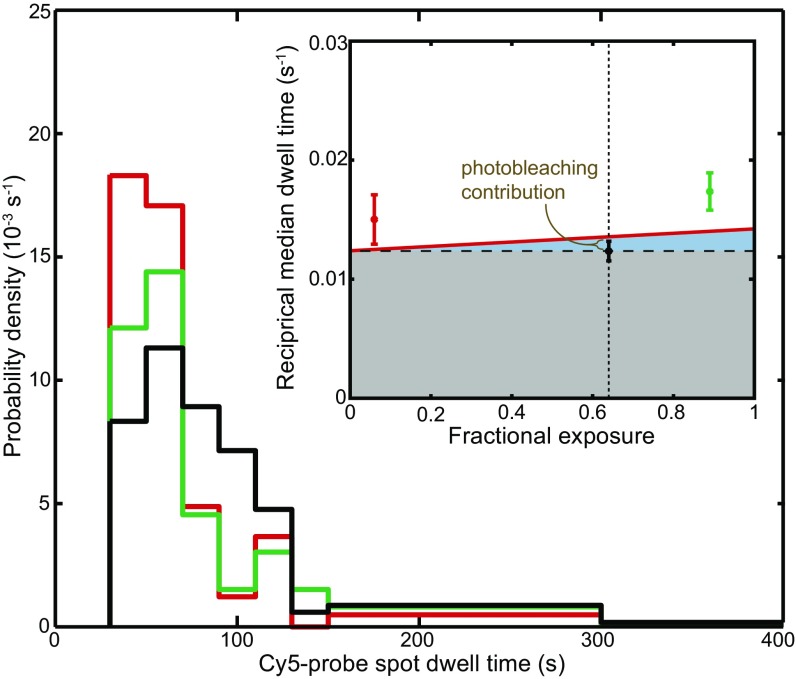
Fig. S1.
Measurement of the Cy5-probe photobleaching rate. Probe lifetimes in the absence of GreB measured at a 633-nm laser fractional exposure of 0.64 are replotted from Fig. 1E (black trace) together with analogous data collected at fractional exposures of 0.06 (red trace) and 0.89 (green trace). For the 0.06, 0.64, and 0.89 fractional exposure experiments, n = 67–84 measurements and 8, 3, and 1 event(s), respectively, are not shown because their values exceeded 400 s. (Inset) Reciprocal of the median dwell time (±SE) for each of the three factional exposures. The weighted linear fit (solid red line) yields a slope of 0.002 ± 0.003 (SE) and intercept 0.012 ± 0.002/s. Photobleaching is assumed to be proportional to exposure, so that the intercept (gray region) corresponds to the effective dissociation rate, and the height of the line above the intercept (blue region) corresponds to the photobleaching rate at a particular exposure. The photobleaching rate at the 0.64 exposure used in Fig. 1E is calculated to be 0.001 ± 0.002/s (tan bracket), implying that photobleaching does not appreciably shorten the dwell time distributions shown there.
GreB binds to backtracked ECs with an apparent Kd variously estimated to be ∼10–360 nM (18, 28). When a concentration of wild-type GreB >> Kd (1 µM) was added, the Cy5-probe median dwell times increased significantly (Fig. 1 D and E, red traces). This increase is consistent with a model in which EC elongation is partially inhibited during time intervals when GreB is bound (Discussion and Fig. S2).
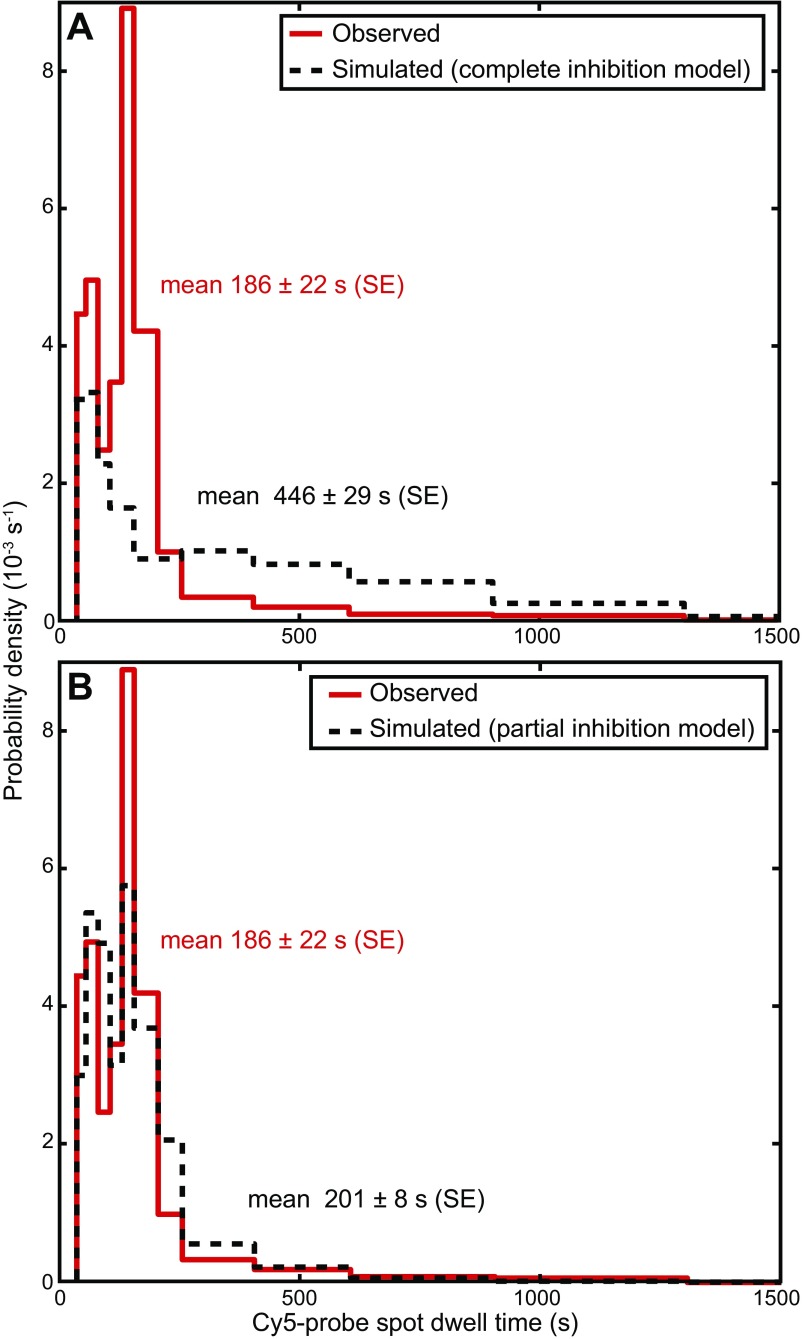
Fig. S2.
Monte Carlo simulations of GreB inhibition of transcript elongation. Distribution of Cy5-probe spot dwell times in the presence of 1 µM GreB (red trace; from Fig. 1E) compared with predictions (black dashed line) from models in which GreB bound to the EC completely (A) or partially (B) inhibits transcript elongation. Each model prediction was calculated from 840 Monte Carlo simulations (1-ms step time) of GreB binding and dissociation trajectories using the probe dwell times measured in the absence of GreB (Fig. 1E, black trace) and the Cy3B-GreB kon and τ values measured on steady-state elongating complexes (Table 1). To account for the subpopulation of complexes that do not bind GreB (∼20%; see Fig. 2D, black trace), that subpopulation was randomly selected from the starting Cy5-probe spot dwell distribution and was left unaltered. In A, each dwell time was assumed to be lengthened by the summed duration of GreB-bound intervals in the simulations, corresponding to a model in which the ECs do not elongate when bound to GreB. In this simulation there were no free parameters. Because the simulation in A (black dashed line) agreed poorly (Kolmogorov–Smirnov test; P = 6.8 × 10−5) with the measurements (red trace), we conducted additional simulations in which GreB-bound ECs were assumed to elongate at a rate that was a fixed fraction, x, of the elongation rate in the absence of bound GreB. The black dashed line in B shows the simulation results for x = 0.40 (P = 0.91), a value that was chosen to produce the best agreement with the measurements (red trace) by minimizing the least-squares deviations between simulation and measurements. The effects of photobleaching were included in all simulations using the photobleaching rate measured in Fig. S1.
GreB Interaction with Steady-State ECs Is Highly Dynamic.
Since GreB partially inhibits elongation, how does it avoid interfering with EC function? To address this question, we examined the dynamics of GreB interaction with individual ECs during steady-state elongation. For these experiments, we first prepared a GreB construct with a relocated, surface-accessible cysteine (rcGreB), and fluorescently labeled it with a Cy3B-monomaleimide dye (Cy3B-GreB) (Materials and Methods). A structural model (Fig. S3A) suggests that the G82C mutation and labeling of the cysteine are not expected to interfere with the GreB–EC interaction. Consistent with this expectation, Cy3B-GreB produces the same cleavage fragments as wild-type GreB and displays similar cleavage kinetics (Fig. S3 B and C). We then repeated the experiment in Fig. 1 in the presence of 2 nM Cy3B-GreB and simultaneously recorded the appearance of Cy5-probe spots and Cy3B-GreB spots at the locations of template DNA molecules (Fig. 2 A and B). During time intervals in which Cy5-probe was present at a DNA location for >30 s, indicating the presence of an EC, we detected transient appearance of Cy3B-GreB fluorescent spots (Fig. 2C).
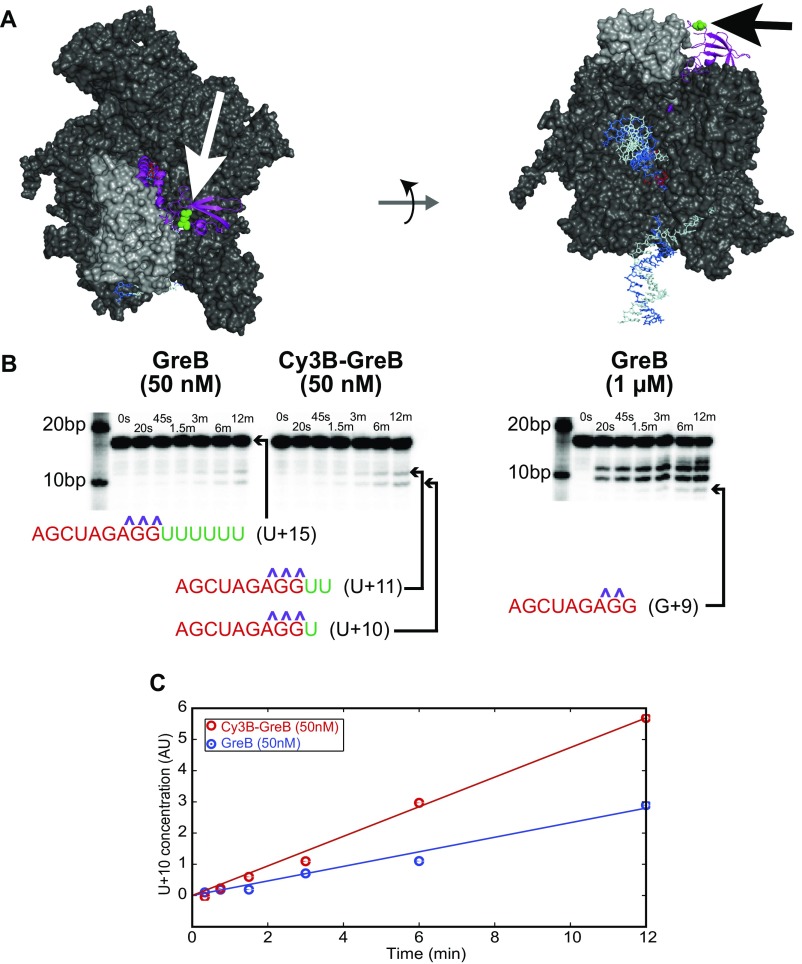
Fig. S3.
Cy3B-GreB is as effective as GreB in rescuing backtracked ECs. (A) Structural model of GreB (magenta) bound to an E. coli EC. RNAP is shown in dark gray, and the lineage-specific trigger loop sequence insertion [variously called “β′GNCD” (45), “β′i6” (46), “β′In6” (47), or “SI3” (48)] is shown in light gray. RNA (red) and template (light blue) and nontemplate (dark blue) DNA strands are visible on the periphery of the EC. GreB E82, the residue that was mutated to cysteine in the Cy3B-GreB construct, is highlighted (green; arrows). The EC structure was modified from Opalka et al. (46) to correct the positioning of SI2 and ω according to more recently published structures [e.g., Protein Data Bank (PDB) ID code 4FYK in ref. 49]. To position GreB in this model, we first aligned the structure derived from the Gre-C1 chimera/Thermus RNAP cocrystal (PDB ID code 4WQT; ref. 13) with the EC model using the PyMOL align function. We then aligned the E. coli GreB structure from PDB ID code 2P4V (35) to Gre-C1. (B) GreB and Cy3B-GreB show similar transcript cleavage kinetics. Transcript cleavage was assayed by treating reconstituted 98-bp EC−6 (prepared as in Fig. 3E but without the AF488 and biotin labels) with GreB or Cy3B-GreB for the indicated times followed by separation on a denaturing 25% polyacrylamide gel and phosphorimaging of the 5′-end–labeled [32P]-RNA. Labels indicate the identity of the starting RNA (U+15) and the cleavage products inferred from their lengths; phosphorothioate linkages are indicated (purple). The greater cleavage seen at 1 µM GreB demonstrates that GreB is subsaturating in reactions at 50 nM. (C) Kinetics of the appearance of the U+10 cleavage product at 50 nM GreB. Linear fits (lines) reveal that the cleavage rate with Cy3B-GreB is approximately twofold larger than with GreB.
Fig. 2.
GreB binding to and dissociation from individual ECs during steady-state elongation. (A) Experimental design. GreB (2 nM) labeled with Cy3B (green star) is detected by TIRF microscopy at the locations of AF488-labeled DNA molecules (cyan star) during intervals in which the presence of Cy5-labeled probe (red star) fluorescence indicates the existence of an EC. (B) Examples of TIRF images recorded at three wavelengths of a region (3.9 × 4.6 µm) of the flow chamber taken before (gold background) and after (gray background) the start of the experiment. Filled and empty triangles indicate the presence or absence of a DNA colocalized fluorescent spot, respectively. (C, Upper) An example of a record of Cy5-probe fluorescence recorded from a 3- × 3-pixel square centered at the position of one DNA molecule. (Lower) Examples of Cy3B-GreB fluorescence recorded from the same region and from three other DNA molecules. In all cases, the recording shows only the time interval during which a Cy5-probe fluorescent spot was present. (Inset) Images (1.1 × 1.1 µm) of the Cy3B-GreB spot corresponding to the indicated florescence peak (∼0.24 s per frame). (D) Cumulative distribution of times to the first Cy3B-GreB binding. The plot displays the fraction of ECs that first bound Cy3B-GreB at a time less than or equal to the indicated time after Cy5-probe binding. Separate experiments were conducted with 2 nM Cy3B-GreB (black circles), 2 nM Cy3B-GreB(D41N) (red circles), and 2 nM Cy3B-GreB plus 200 nM GreB (blue circles). Background data (black and red lines) were collected from the corresponding recordings at randomly selected locations that did not contain visible DNA molecules. Fits (green curves) to a background-corrected exponential model (29) yielded second-order association rate constant (kon) and active fraction (Af) values (Table 1). (E) Cumulative dwell-time distributions for Cy3B-GreB (black) and Cy3B-GreB(D41N) (red) fluorescent spots on ECs. Exponential or biexponential background-corrected fits (green) (29) yielded characteristic dwell times (Table 1). In D and E, numbers in parentheses give the number of observed Cy3B-GreB–binding events followed by the number of ECs analyzed. The number of observed Cy3B-GreB–binding events at background areas in D were 16/307 for Cy3B-GreB and 11/357 for Cy3B-GreB(D41N).
To measure the rate at which GreB binds to ECs, we determined the time that elapsed before the first observed binding of Cy3B-GreB to each EC (29). Nearly all these binding events were specific, because little binding was seen at randomly chosen background locations not containing a DNA molecule or in a control experiment in which a saturating concentration of unlabeled GreB was also present (Fig. 2D). Similarly, in a control in which RNAP was omitted, the frequency of GreB-binding events was reduced 100-fold. Fits to the distributions of binding times (Materials and Methods and Fig. 2D) revealed that a majority of ECs (80 ± 6%) were capable of binding Cy3B-GreB and did so with a near-diffusion-limited rate constant on the order of 107 M−1 s−1 (Table 1). Interestingly, a minority of the ECs (∼20%) appeared incapable of binding Cy3B-GreB even though they were active and had similar elongation rates (the median Cy5-probe spot durations were 53 ± 6 s and 61 ± 9 s, respectively, for ECs observed to bind or to not bind GreB). The origin of the binding-inactive EC subpopulation is unclear. Nevertheless, it is apparent that most ECs in the experiment are actively elongating at normal velocities and bind GreB at a fast, near-diffusion-limited rate. Based on these observations, it is likely that in steady-state elongation these complexes spend all or most of the time in states capable of binding GreB.
Table 1.
Kinetics of Cy3B-GreB interactions with ECs
| GreB construct | Type of EC | kon,107 M−1 s−1 | Binding-competent fraction, Af | Dwell time distribution fit parameters* | KD, nM† |
| Cy3B-GreB | Steady-state elongating (Fig. 2 D and E) | 2.7 ± 1.0 | 0.80 ± 0.06 | τ: 0.4 ± 0.1 s | 93 ± 36 |
| Static EC0 (Fig. 4 E and F) | 3.2 ± 0.1 | 1‡ | τ: 0.29 ± 0.01 s | 106 ± 4 | |
| Static EC−6 (Fig. 4 E and F) | 1.9 ± 0.1 | 1‡ | τ: 0.29 ± 0.02 s | 184 ± 8 | |
| Cy3B-GreB (D41N) | Steady-state elongating (Fig. 2 D and E) | 3.2 ± 1.4 | 0.81 ± 0.08 | τ1: 0.6 ± 0.1 s | N.D. |
| a: 73 ± 8% | |||||
| τ2: 11 ± 2 s | |||||
| Static EC0 (Fig. S5) | N.D. | N.D. | τ1: 0.99 ± 0.04 s | N.D. | |
| a: 99% [98, 99.3] | |||||
| τ2: 10.4 [6, 13] s | |||||
| Static EC−6 (Fig. S5) | N.D. | N.D. | τ1: 0.83 ± 0.08 s | N.D. | |
| a: 92 ± 4% | |||||
| τ2: 4.9 ± 1.6 s |
Indicated uncertainties are ±SE, except where bootstrap analysis indicated an asymmetrical confidence interval; in those cases the 68% confidence interval is given in brackets. For each fit the number of observations is reported in the referenced figure. N.D., not determined.
From single or biexponential fit.
KD is calculated as 1/(τ kon).
All complexes exhibited GreB binding.
We also examined the kinetic stability of the GreB-EC complexes. Under our experimental conditions, Cy3B-GreB fluorescence at ECs disappeared rapidly (Fig. 2E), and its dwell time, τ = 0.4 ± 0.1 s (Table 1), was not appreciably affected by photobleaching (Fig. S4). These data are inconsistent with proposals that ECs maintain a kinetically stable association with GreB during elongation (19, 22). Instead, the data suggest that bound GreB typically dissociates from elongating complexes on a time scale (∼0.4 s) not much longer than that required for a cycle of nucleotide addition to the transcript (∼0.1 s at 20 °C).
Fig. S4.
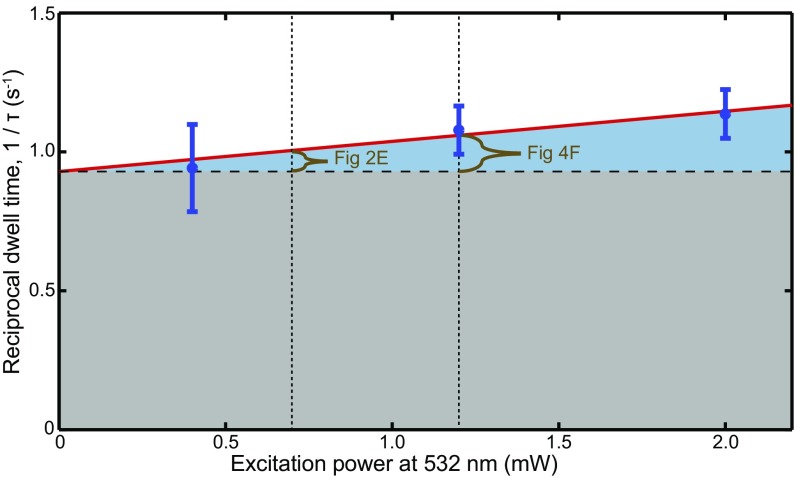
Photobleaching has a negligible effect on observed Cy3B-GreB dwell times. Reciprocal dwell times (±SE) of Cy3B-GreB(D41N) from EC−6 complexes in the absence of NTPs were measured at three different 532-nm laser excitation powers (blue). Cy3B-GreB(D41N) has longer initial dwell times than Cy3B-GreB in all ECs studied and therefore is a more stringent test of photobleaching. The weighted linear fit (red line) yields a power-specific photobleaching rate of 0.11 ± 0.10 mW−1 s−1. At the excitation powers used in the experiments to examine Cy3B-GreB dwell times (0.7 mW in Fig. 2E and 1.2 mW in Fig. 4F), the reciprocal dwell times (brown) are 0.08 ± 0.07 and 0.13 ± 0.13 s−1, corresponding to 3.2 ± 2.8% and 3.7 ± 3.7%, respectively, of the koff = 1/τ values calculated from the uncorrected dwell times reported in Table 1. The negligible photobleaching supports the use of GreB-absent intervals (29) to measure the association rate constants in Fig. 4 D and E.
To test the functional significance of the observed binding of Cy3B-GreB to ECs, we also examined the binding and dissociation of Cy3B-GreB(D41N), a mutant that lacks one of the coiled-coil domain tip acidic residues that are required for transcript cleavage activity (6). The mutant association rate is identical to of wild type (Fig. 2D). However, once bound, the dissociation of the mutant is multiexponential and on average is much slower than in the wild type (Fig. 2E and Table 1). Thus, a point mutation that abrogates cleavage activity also dramatically alters the observed binding, suggesting that observed binding events are relevant to the cleavage activity of wild-type GreB. From the results presented thus far, it is unclear whether the faster dissociation of wild-type GreB is a consequence of transcript cleavage or whether it is caused by different binding interactions between ECs and Cy3B-GreB vs. Cy3B-GreB(D41N).
Reconstitution of ECs Stabilized in Backtracked and Nonbacktracked Configurations.
The foregoing results show that GreB binds rapidly and transiently to complexes undergoing steady-state elongation. However, in the steady state RNAP-DNA complexes can be in multiple different on-pathway and paused states (30, 31). The experiments described above do not address which state or states of the EC are capable of binding GreB. In particular, we wanted to ask whether GreB is recruited preferentially to backtracked complexes, as has been proposed (19, 20, 22, 32).
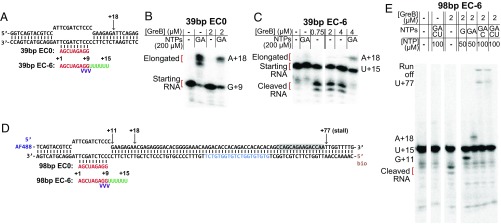
To prepare ECs in defined states, we reconstituted complexes by mixing RNAP in the absence of NTPs with preformed scaffolds consisting of a short RNA hybridized to a duplex DNA containing a noncomplementary bubble region (33). Complexes designed to be nonbacktracked incorporated a 9-nt RNA complementary to the template strand bubble sequence (39-bp EC0) (Fig. 3A). Consistent with the nonbacktracked state, the RNA in these complexes was not cleaved when GreB was added, and a significant fraction of RNAs (52–77%) were elongated when NTPs were added either in the absence or presence of GreB (Fig. 3B). We also prepared complexes designed to be stably backtracked by 6 nt using an RNA that contained a noncomplementary six-uridine 3′ extension (39-bp EC−6) (Fig. 3A) (18, 34). To reduce cleavage of the RNA by the intrinsic endonuclease activity of RNAP, this RNA contained phosphorothioate linkages bridging positions +7 through +10. As expected for backtracked complexes, untreated EC−6 complexes did not elongate the RNA when challenged with NTPs (Fig. 3C). However, a portion of the RNA in the EC−6 samples was cleaved upon GreB addition, and 30–50% of these cleaved RNAs were elongated upon the subsequent addition of NTPs. Thus, essentially all RNA synthesis observed on the EC−6 complexes was GreB dependent.
Fig. 3.
Reconstituted nonbacktracked and backtracked ECs. (A) Diagram of the DNA (black)/RNA (red/green) scaffolds used to prepare 39-bp EC0 and EC−6. Phosphorothioate linkages in the EC−6 RNA are indicated (purple). (B and C) Cleavage of and transcript elongation by 39-bp EC0 (B) and EC−6 (C) complexes after 10–15 min incubation at the indicated concentrations of GreB followed by chase with the indicated subset of NTPs. The 5′-end [32P]-labeled RNA cleavage and elongation products were visualized by phosphorimaging in a denaturing 23% polyacrylamide gel; bands are marked with the RNA length and the identity of the 3′ terminal nucleotide. (D) Diagram of the nucleic acid scaffolds used to prepare 98-bp template EC0 and EC−6 for single-molecule assays. A downstream sequence (blue) is identical to the fluorescent DNA oligonucleotide used to detect nascent RNA. Gray shading shows the template sequence corresponding to the portion of the RNA 3′ end anticipated to be inside RNAP when these ECs are stalled at +77 by elongation in the absence of UTP. (E) GreB-stimulated cleavage of and transcript elongation by EC−6 complexes on the 98-bp template.
To adapt the reconstituted complexes for single-molecule analysis, we prepared versions of the EC0 and EC−6 complexes with a longer downstream DNA segment (Fig. 3D). These 98-bp templates incorporate biotin and dye for surface tethering and visualization. The noncomplementary transcription bubble in the reconstituted complexes (Fig. 3 A and D) cannot reanneal during transcript elongation and thus in principal could interfere with the movement of RNAP out of the bubble and therefore with transcript elongation. However, ECs formed on the 98-bp scaffolds behaved similarly to the corresponding short complexes upon exposure to GreB and could produce (albeit with low efficiency) RNAs tens of nucleotides in length when provided with the appropriate combinations of NTPs (Fig. 3 D and E). Thus, when exposed to NTPs, a fraction of the 98-bp template complexes produce nascent RNAs of sufficient length that their 5′ ends are expected to extend out of the RNAP exit channel, allowing detection of transcription by oligonucleotide probe hybridization.
Identifying Individual Elongation-Competent Reconstituted ECs.
To perform single-molecule experiments to examine the dynamics of GreB binding to individual reconstituted EC0 and EC−6 complexes, we immobilized the preformed dye- and biotin-labeled complexes (Fig. 3D) on the surface of a microscope flow chamber in the absence of NTPs. The two types of complexes were added sequentially, and their locations were recorded after each addition (Materials and Methods). Consequently, individually identified EC0 and EC−6 complexes were present simultaneously in the same chamber so that they could be observed under identical conditions.
In each chamber we initially performed an internal control in which we measured the binding of a Cy5 probe complementary to a downstream target sequence (Fig. 3D, green) in the absence of NTPs. Because this sequence is not present in the initial scaffold RNA, this control measurement served to confirm that nonspecific Cy5-probe binding was minimal (Fig. 4 A and B, interval i). A small number of complexes that bound the Cy5 probe at this stage (typically 2–9%) were excluded from further analysis.
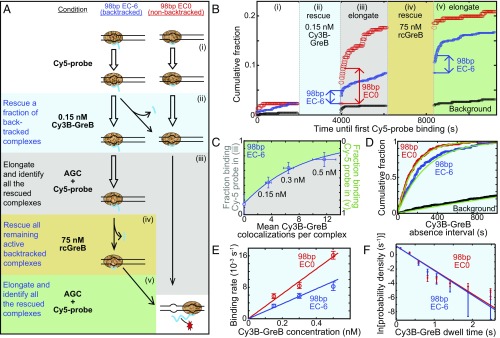
Fig. 4.
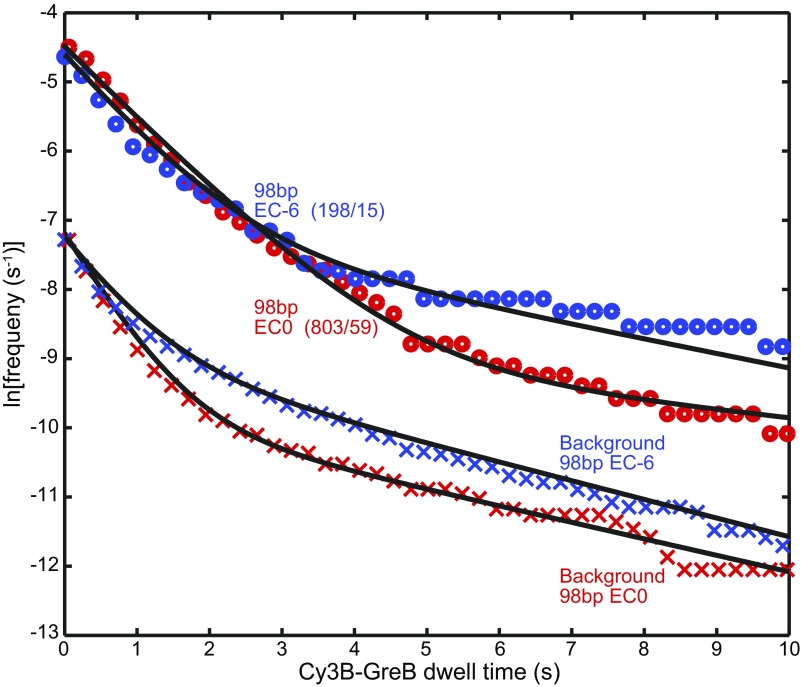
Single-molecule analysis of GreB binding to backtracked and nonbacktracked ECs. (A) The five-step experimental protocol used to identify functional reconstituted ECs and observe their GreB binding. In each subsequent step of the protocol, the reagents from the preceding step were removed by flushing the reaction chamber before the indicated new reagents were added. ECs contain RNA polymerase core (tan), RNA (blue), and dsDNA (black). Closed arrows indicate changes in inferred complex structure; open arrows indicate no change. (B) An example of a record of transcript production detected on individual ECs using the protocol in A. The graph shows the cumulative fraction of complexes that exhibited a colocalized Cy5-probe spot by the indicated time for the EC0 preparation (red; n = 215 total complexes), EC−6 preparation (blue; n = 706), and randomly selected sites without DNA (black; n = 640). The Cy5-probe was not present in solution during intervals ii and iv. Abrupt increases in Cy5-probe binding upon incubation with NTPs during the first 100 s of intervals iii and v demonstrate the elongation of nonbacktracked ECs present initially (red arrow) or produced from backtracked complexes by transcript cleavage that occurred during intervals ii and iv (blue arrows). (C) The fraction of EC−6 complexes that were rescued (i.e., did not remain backtracked) during interval ii increased as the concentration of Cy3B-GreB used in interval ii was increased. Points show how the fraction (±SE) of functional EC−6 complexes that did (Right Axis) or did not (Left Axis) survive interval ii varies with the mean number of Cy3B-GreB–binding events observed on the surviving complexes. Data are taken from the experiment in B and from additional experiments using the indicated concentrations of Cy3B-GreB during interval ii. The exponential fit (curve) yields a cleavage probability of 0.08 per GreB binding. (D) Cumulative distributions (points) of time intervals separating successive Cy3B-GreB–binding events recorded from functional (see text) EC0 (red; n = 132 intervals) and EC−6 (blue; n = 137 intervals) complexes and from control sites without DNA (black; n = 186 intervals) during interval ii of B (0.15 nM Cy3B-GreB). Lines are fits to a background-corrected exponential model (29). (E) Background-corrected apparent first-order Cy3B-GreB–binding rate constants (open circles) determined as in D at Cy3B-GreB concentrations of 0.15, 0.3, and 0.5 nM; n = 73–416. Weighted linear fits (lines) yielded second-order binding-rate constants, kon (Table 1). (F) Background-corrected distributions of Cy3B-GreB dwell times (circles) on functional (see text) EC0 (red; 666 dwells) or EC−6 (blue; 254 dwells) complexes during interval ii pooled from experiments at 0.15, 0.3, and 0.5 nM Cy3B-GreB. Lines indicate exponential fits yielding characteristic dwell time τ values (Table 1).
After the control, we followed an experimental protocol designed (i) to identify the subpopulations of ECs competent for transcript cleavage and elongation (for EC−6) or competent for elongation (for EC0) and (ii) to measure the binding of Cy3B-GreB to only this competent subpopulation, thus excluding from our analysis any RNAP that had not formed an authentic EC. First, the complexes were incubated for 1,400 s with ≥0.15 nM Cy3B-GreB without NTPs (Fig. 4 A and B, interval ii). During this interval, we recorded colocalized individual Cy3B-GreB–binding events as in Fig. 2C, Lower. Typically only 1–10 GreB-binding events were observed on each complex, consistent with the low Cy3B-GreB concentration used. The complexes then were tested by removing the GreB and adding the Cy5 probe (5 nM) together with ATP, CTP, and GTP (0.5 mM each) to determine which were capable of transcript elongation (Fig. 4 A and B, interval iii). Next, to detect all complexes competent for transcript cleavage, we performed an incubation with unlabeled GreB (rcGreB) (Materials and Methods) at a much higher concentration (75 nM) in the absence of NTPs (Fig. 4 A and B, interval iv), again followed by removing the GreB and performing an NTPs/Cy5-probe incubation (Fig. 4 A and B, interval v).
During the initial NTP incubation (Fig. 4 A and B, interval iii), we observed that a fraction of the nonbacktracked EC0 complexes acquire a Cy5-probe spot in the first 100 s (Fig. 4B, red arrow), consistent with rapid production of the +77 stalled complex (Fig. 3D). This fraction was small [7.5 ± 1% (S.E.) of n = 670 EC0 complexes], consistent with the possible presence of inactive complexes, the difficulty of quickly elongating out of the noncomplementary bubble as observed in the ensemble experiments, or both (Fig. 3E). However, the EC0 rescue detected by Cy5-probe binding was reproducible and was much larger than Cy5-probe binding to randomly chosen areas that did not contain visible complexes (0.7 ± 0.2%; n = 2,011). As expected, few additional complexes (1.0 ± 0.4%) bound the Cy5 probe during a subsequent NTPs incubation (Fig. 4 A and B, interval v), showing that nearly all complexes capable of quickly producing the full transcript had already done so during the first NTPs incubation.
The behavior of backtracked EC−6 complexes was qualitatively different from that of EC0. Less than half of the total active EC−6 complexes showed prompt elongation after the first incubation with NTPs (Fig. 4B, blue arrow in interval iii; 3.0 ± 0.6% of n = 706 complexes). More than half elongated only after the second NTPs incubation (Fig. 4B, blue arrow in interval v; 3.8 ± 0.7%), the condition in which EC0 complexes displayed essentially no elongation over background. This finding is consistent with the expectation that, unlike EC0, functional EC−6 complexes require GreB-triggered transcript cleavage to activate their elongation activity. The data also confirm that the subnanomolar concentration of Cy3B-GreB used in rescue (Fig. 4 A and B, interval ii) was low enough that it activated only a fraction of the potentially activated complexes.
To verify our interpretation that the Cy3B-GreB binding during interval ii caused the jump in Cy5-probe hybridization upon NTPs challenge (interval iii), we further analyzed data obtained using a range of subnanomolar Cy3B-GreB concentrations in interval ii. As the concentration used was increased, the fraction of rapid Cy5-probe–binding events observed in interval iii increased. Furthermore, the fraction of functional complexes that survived interval ii and thus were rescued in interval iv decreased exponentially with the number of Cy3B-GreB–binding events to the functional complexes seen during interval ii (Fig. 4C, green). The observations are consistent with each GreB–binding event having a fixed, low probability, P = 0.08 (Materials and Methods), of rescuing the complex from the backtracked state by cleaving the RNA. This probability presumably would be higher for unmodified RNA than we measured for the phosphorothioate-modified RNA used in these experiments to reduce spontaneous cleavage.
Kinetics of GreB Interaction with Stabilized Backtracked and Nonbacktracked ECs.
Having identified the DNA molecules that contained correctly reconstituted, functional ECs, we examined the kinetics of GreB binding and dissociation during interval ii only in these complexes. In particular, for EC0 complexes we analyzed only the subpopulation that rapidly bound the Cy5 probe when challenged by NTPs in interval iii, ensuring that the complexes were functional. For EC−6 complexes we analyzed only complexes that displayed rapid Cy5-probe binding in interval v, ensuring that the complexes were functional and that they had remained in the backtracked state throughout the entirety of interval ii.
The distribution of time intervals separating successive Cy3B-GreB colocalizations to active complexes was exponential (Fig. 4D), consistent with a simple, single-step binding process. Measurements at multiple Cy3B-GreB concentrations revealed that the second-order association rate constants for the EC0 and EC−6 complexes differed by less than a factor of two (Fig. 4E and Table 1). Notably, the data show that GreB does not bind to backtracked complexes more quickly than to nonbacktracked; association with EC0 is actually faster than with EC−6 by a factor of 1.7. We also measured the dissociation rates of Cy3B-GreB from the nonbacktracked and backtracked complexes (Fig. 4F) and observed nearly identical distributions. Photobleaching was negligible under these experimental conditions (Fig. S4). Taken together, these data indicate that backtracked complexes neither preferentially bind nor preferentially stabilize bound GreB relative to nonbacktracked complexes (Table 1).
The ability to study binding to functional EC0 and EC−6 complexes also allowed us to examine whether the differences between binding of Cy3B-GreB and Cy3B-GreB(D41N) to ECs in steady-state elongation (Fig. 2E and Table 1) resulted from the inability of the mutant to stimulate transcript cleavage. Cy3B-GreB(D41N) binding frequencies and dissociation kinetics were essentially indistinguishable between EC0 and EC−6 (Fig. S5 and Table 1). Nevertheless, dissociation kinetics were slower (and more complex) for Cy3B-GreB(D41N) than for Cy3B-GreB, as was apparent in the steady-state EC experiments (Fig. 2E). Thus, the slower dissociation of the mutant is caused by the altered interactions of the mutant with ECs and not by the mutant’s inability to cleave backtracked transcript: Dissociation on EC−6 was equivalently slow to that on EC0 in which backtracked transcript was absent and therefore cleavage presumably was not possible.
Fig. S5.
Cumulative frequency distributions of Cy3B-GreB(D41N) dwell times on EC0 and EC−6. The plot shows the observed frequencies of dwell times of the indicated duration or longer in experiments on active EC0 (red) and EC−6 (blue) complexes. The distributions of dwell times at locations of complexes (○) and background locations (×) are indicated together with fits (lines) yielding the background-corrected fit parameters shown in Table 1. Data are from experiments using the protocol in Fig. 4A with 0.3 nM Cy3B-GreB(D41N) during interval ii and 75 nM rcGreB during interval v. The values in parentheses give the number of observed Cy3B-GreB–binding events followed by the number of ECs or background areas analyzed.
Discussion
Understanding the mechanisms of EC regulation has been hindered by lack of information about the dynamics of protein interactions with the SC, a key regulatory target. We now have directly observed and quantified the kinetics of GreB interaction with ECs. Methods of preparing ECs often produce mixtures of correctly assembled and inactive complexes; the multi-wavelength single-molecule fluorescence methods allowed us to restrict analysis to the subset of complexes confirmed to be correctly assembled and elongation competent. We observed the binding and dissociation kinetics for steady-state elongation (Fig. 2) (during which the EC cycles rapidly through the different intermediates of the nucleotide-addition cycle) and for static complexes designed to be stably locked into either the nonbacktracked configuration or into the 6-nt backtracked configuration that is a good substrate for GreB-stimulated cleavage (Fig. 4). GreB was observed to bind rapidly to ECs with a kon ∼107 M−1 s−1, at or near the diffusion limit. However, binding was transient and short-lived, with a GreB residence time on the RNAP of <1 s. This time is orders of magnitude less that the time required to synthesize a long messenger RNA under the experimental conditions and is not much longer than the time required for a single cycle of nucleotide addition under the same conditions.
We observed almost identical kinetic values for nonbacktracked complexes, backtracked complexes, and complexes in steady-state elongation reactions (Table 1), indicating that GreB is neither preferentially recruited to nor preferentially stabilized by backtracked ECs. This result suggests a model in which GreB functions by nonselectively binding to all configurations of ECs to locate by random sampling the small minority of complexes that are, at a given time, arrested in a backtracked state and thus require rescue via endonucleolytic cleavage. The fast binding rate and short residence time likely serve to accelerate this random search.
The two-helix bundle of GreB has a patch of positive surface charge that has been proposed to interact with the backtracked RNA 3′ end in the SC (35, 36). The observation that the GreB binding and release kinetics are the same on backtracked and nonbacktracked complexes suggests that the RNA–GreB interaction does not make a substantial contribution to the energy barrier to GreB binding or dissociation.
There are alternative proposals about how the multiple SC-binding proteins present in a bacterial cell work together to regulate ECs. Based on data from experiments on Thermus thermophilus Gre, it was suggested that SC proteins remain stably bound to an individual EC throughout the production of a given transcript molecule (19, 22). Our results show this kinetically stable binding is not true for SC proteins in general; we observe that GreB binding ECs is short-lived (Figs. 2E and 4F) but that GreB nevertheless can productively rescue backtracked complexes (Fig. 4 A–C). Conversely, data from Fe+2-mediated protein cleavage studies (18) have been taken to indicate that GreB binds to reconstituted 2-nt backtracked complexes and not to nonbacktracked complexes (18, 32). However, the cleavage assay does not measure GreB binding directly: The extent of cleavage depends on both binding and cleavage efficiency, and cleavage efficiency may be different in backtracked and nonbacktracked complexes because of increased metal ion binding by the former (37). Our results based on the direct observation of binding show essentially identical association rates and binding constants for nonbacktracked and 6-nt backtracked complexes (Table 1).
Just as GreB and other proteins compete for binding to the SC in ECs, the same proteins may compete for binding to the RNAP holoenzyme SC during transcription initiation. However, there is little reason to suppose that interactions with holoenzyme are identical to those with ECs. Thus, measuring the kinetics of GreB and other SC-binding proteins interactions with holoenzyme will be required to determine whether the competition mechanism proposed here for ECs also applies to initiation complexes.
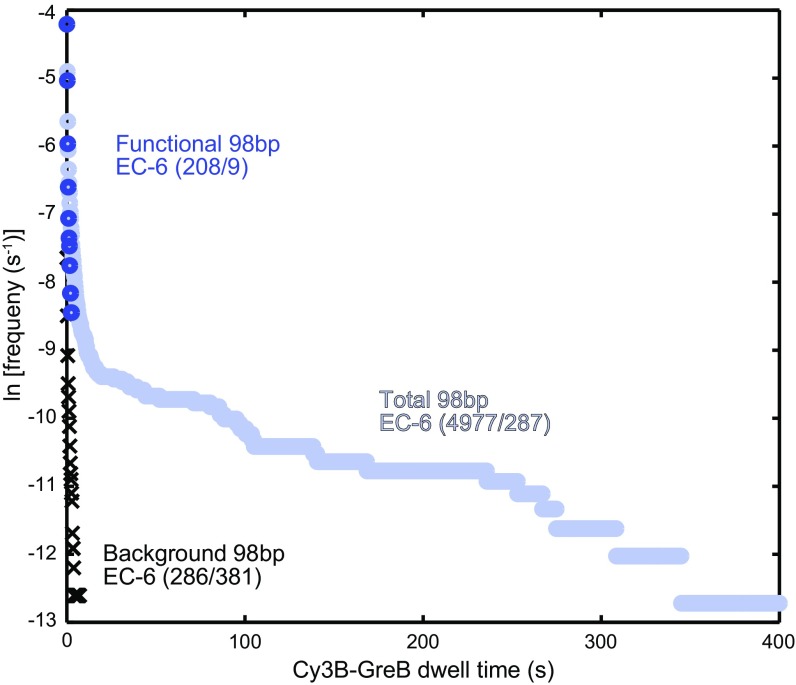
In our experiments using reconstituted ECs, we tested each individual complex to determine if it was elongation competent. In particular, we required that EC0 complexes promptly exhibit transcript probe hybridization upon incubation with NTPs and that EC−6 complexes exhibit hybridization only after GreB-induced transcript cleavage and incubation with NTPs (Fig. 4A). These conservative criteria may underestimate the number of functional complexes (e.g., because phosphorothioate RNA cleavage is inefficient, because escape from the noncomplementary bubble in the template was sometimes slow, or because the GreB concentration used was not sufficient to rescue all complexes properly). Nevertheless, as has been observed previously (e.g., ref. 33), it is clear that only a fraction of the DNA molecules were incorporated into correctly assembled ECs in our experiments. Our single-molecule functional tests reinforce the view that bulk biochemical experiments on reconstituted complex preparations can produce misleading results because the preparations are mixtures of functional and non- or poorly functioning complexes. When we selected only functional complexes, all our experiments with wild-type GreB produced the single-exponential lifetime distributions (Fig. 4 D and F) characteristic of a one-step reaction by a kinetically homogeneous population. In contrast, the same analysis on the complete population of reconstituted complexes revealed more complex, multicomponent dynamics (Fig. S6). These observations suggest that caution is necessary in interpreting the results of bulk experiments on reconstituted transcription complexes, because both active and inactive complexes may be present.
Fig. S6.
Multicomponent Cy3B-GreB dissociation kinetics from the full population of reconstituted EC−6. The plot shows the observed frequencies of Cy3B-GreB dwell times of the indicated duration or longer on all complexes in the EC−6 preparation (light blue) or on only the functional subpopulation (blue). The distribution of dwell times at control background locations (black) is shown also. Data are from experiments using the protocol in Fig. 4A with 0.5 nM Cy3B-GreB during interval ii and 50 nM rcGreB during interval v. Values in parentheses give the number of observed Cy3B-GreB–binding events followed by the number of ECs or background areas analyzed.
Structural models suggest that EC-bound GreB is likely to inhibit transcript elongation by interfering with the operation of the RNAP trigger loop, by stabilizing a catalytically inactive conformation of the polymerase, or both (13, 38). Consistent with this hypothesis, we observe the inhibition of transcript elongation in the presence of 1 µM GreB (Fig. 1E), a concentration that is expected to be nearly saturating based on our measurements (Table 1). However, our data are inconsistent with a model in which GreB-bound ECs do not elongate at all (Fig. S2A). Instead, they suggest that EC–GreB complexes are capable of transcript elongation but that the average elongation velocity is only 40% of that in the absence of GreB (Fig. S2B). Therefore, we hypothesize that, even in the GreB-bound state, the polymerase–GreB complex has the ability to adopt conformations that allow it to catalyze transcript elongation.
We find that the inactivating D41N mutation substantially alters the interaction of GreB with the EC: We observed only transient EC–GreB interactions, whereas a significant fraction of EC–GreB(D41N) complexes persist at least 25-fold longer (Fig. 2E, red trace). Significantly, we observe nearly identical kinetics of GreB(D41N) interacting with backtracked and nonbacktracked ECs (Table 1). Thus, the inability of the catalytically inactive mutant to cleave and release backtracked RNA is not responsible for this long-lived population. Additionally, these data suggest that the absence of any long-lived population in experiments with wild-type GreB is similarly independent of transcript cleavage (i.e., the departure of GreB from ECs is not coupled to RNA cleavage). One possible explanation for these data is that GreB(D41N) is able to sequester ECs into a kinetically stable alternative complex not accessible with wild-type GreB. This interpretation is consistent with the strong suppression of RNAP activity previously seen with GreA and GreB tip-residue mutants (17).
To function effectively in a cell, GreB must avoid wholesale inhibition of transcript elongation and also not prevent access to the SC by other proteins (e.g., GreA and DksA) that have different activities with respect to ECs (32, 34, 39). On the other hand GreB must be able to locate and repair backtracked ECs quickly in the presence of an enormous excess of active, nonbacktracked ECs. The observed short GreB dwell times, only slightly longer than the nucleotide addition cycle, minimize its occupation of the SC, and the fast, diffusion-limited association rate should help GreB interrogate large numbers of ECs quickly to rescue the subset of complexes that are backtracked. These kinetic properties may serve to allow efficient GreB function while still leaving the SC mostly unoccupied to allow interaction with competing SC factors, an idea which can be tested in future experiments in which multiple SC factors are present simultaneously.
Materials and Methods
DNA and Plasmids.
A pET-15b expression plasmid encoding N-terminal His6-tagged GreB from E. coli (generously provided by S. Darst, Rockefeller University, New York) (39) was modified using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) using the primer sequences 5′-CGATTACTCC CCTCACATGCGGCAAAGTCTTTTTTGGCG-3′ to create the E82C mutation, 5′-CGTGTGTATCTCACTAAAAGTCTGGAAAATCTCAAAATCGTC-3′ for C68S, and 5′-GGTGACCTGGGCCGCAAGTCTGGGCAACCGCAGCG-3′ for D41N (IDT DNA); mutation sequences are underlined. Constructs were verified by sequencing.
The PR′ transcription template was prepared by PCR from plasmid pCDW115 (24). Nascent transcripts were detected by hybridization of 5′-GTGTGTGGTCTG TGGTGTCT/3Cy5Sp/-3′ (IDT DNA).
SNAP-Tagged E. coli Core RNAP.
To generate RNAPSNAP, core RNAP with the β′ subunit fused to a C-terminal SNAP-tag, the SNAP-tag from pSNAP (New England Biolabs) was PCR-amplified using primers 5′-CACAACGTCGACATGGACAAAGATTGCGAAATGAAAC-3′ and 5′-CACAACAAGCTTACTCGAGTCCTGGCGCGCCTATACCTGCAGG-3', digested with SalI and HindIII (New England Biolabs), and then ligated into XhoI-, HindIII-cut pRM756 (40) to give pRM775, a plasmid that directs the expression of E. coli rpoA, rpoZ, rpoB, and rpoC::SNAP-His10. E. coli strain BLR λDE3 was transformed with pRM775 and grown in Luria broth supplemented with kanamycin (25 μg/mL) in Fernbach flasks shaken at 200 rpm at 37 °C to an apparent OD600 of 0.5–0.6. RNAP overexpression then was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the cells were shifted to 30 °C for 5 h with continued shaking at 200 rpm. The cells then were harvested by centrifugation and lysed by sonication. RNAPSNAP was purified from the lysate as described previously (41) by polyethylenimine precipitation and affinity chromatography using nickel-agarose and heparin-Sepharose columns (5 mL HiTrap; GE Life Sciences). Purified RNAPSNAP was dialyzed into storage buffer [20 mM Tris⋅Cl (pH 8), 250 mM NaCl, 20 μM ZnCl2, 1 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 20% (vol/vol) glycerol] and stored in small aliquots at −80 °C until use.
Reconstitution of Backtracked and Nonbacktracked ECs.
Scaffolds for 98-bp EC0 and EC−6 complexes contained DNA from Eurofins and partially phosphorothioate-modified RNA from GE Healthcare Dharmacon or IDT DNA. The two DNA strands (Fig. 4D) were combined in reconstitution buffer [10 mM Tris⋅Cl (pH 8), 40 mM KCl, 5 mM MgCl2] to final concentrations of 1.8 μM each and then were annealed by raising the temperature to 95 °C followed by cooling (1 °C/min) to 60 °C. The solution then was mixed with an equal volume of RNA (1.8 μM in reconstitution buffer) and annealed by raising the temperature of the solution to 50 °C followed by cooling (1 °C/min) to 30 °C. The annealed scaffold solution then was mixed at 1:2 (vol/vol) with RNAPSNAP [7.5 μM in 10 mM Tris (pH 8), 100 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 25% glycerol] and was incubated at 33 °C for 20 min. ECs typically were used within 1 h.
To verify polymerase incorporation into reconstituted complexes, RNAPSNAP was labeled with SNAP-Surface 549 (New England Biolabs) in solution using a 3:1 dye/core ratio to yield RNAPSNAP549. RNAPSNAP549 EC−6 tethered to a slide surface was observed to have 92% of the AF488-labeled DNA fluorescence spots colocalized with a RNAPSNAP549 spot.
GreB Preparation.
GreB proteins used in this study were wild type (GreB) or contained a relocated cysteine (rcGreB; E82C/C68S mutant) or contained both the relocated cysteine and a mutation that suppresses transcript cleavage [GreB(D41N); E82C/C68S/D41N mutant]. Plasmids coding for these proteins (pET15b-GB, pET15b-GBdm, and pET15b-GreBdm-D41N, respectively) were transformed into BL21(DE3) cells for protein expression. Cells were grown in LB or 2xYT medium at 37 °C with 100 mg/mL ampicillin or carbenicillin to O.D. ∼0.4 and then were induced with 1–2 mM IPTG. Cells were harvested after 3 h of induction at 37 °C (wild type) or after 6 h of induction at 25 °C (mutants). In some cases, cells were treated to remove periplasmic material as described (42) and then were suspended in lysis buffer [40 mM Tris⋅Cl (pH 8.0), 1 M NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), and 0.1 mM PMSF], frozen in liquid nitrogen, and stored at −80 °C for 1–3 d. Cells were thawed on ice and further lysed using lysozyme (300 μg/mL) or by sonication. Universal nuclease (1 μL; 88702; Pierce) was sometimes added, and the supernatant was clarified by centrifugation at 14,000 × g and was passed twice through a 0.45-μm filter. The protein was isolated by affinity chromatography in column (HisTrap HP 17-5248-02; GE Healthcare) or batch (Ni-NTA agarose; 30210; Qiagen) format. Protein was eluted in binding buffer [40 mM Tris⋅Cl (pH 8.0), 0.6 M NaCl, 0.1 mM TCEP, and 5 mM imidazole] either through a gradient from 5–500 mM imidazole or in steps of 50 mM and 250 mM imidazole. Some GreB preparations were further purified on Sephadex G-75 eluted with elution buffer [40 mM Tris⋅Cl (pH 8.0), 1 M NaCl, 5% glycerol, and 0.1 mM TCEP]. All preparations were dialyzed into storage buffer [40 mM Tris (pH 8.0), 1 M NaCl, 40% glycerol, and 0.1 mM TCEP], frozen in liquid nitrogen, and stored at −80 °C.
Fluorescent Labeling of GreB and Purification of Labeled Protein.
To prepare Cy3B-GreB or Cy3B-GreB(D41N), rcGreB or GreB(D41N), respectively, was labeled with Cy3B-maleimide monoester (GE Healthcare) using a 10:1 molar ratio of dye to protein in labeling buffer [40 mM Tris⋅HCl (pH 8.0), 1 M NaCl, 0.3 mM TCEP] at a total volume of 100–200 μL. Reactions were incubated at 4 °C for 2–4 h. Labeled GreB was separated from free dye on Sephadex G-25 Superfine gel filtration medium eluted with labeling buffer. Protein concentrations and dye-labeling stoichiometries were calculated using ε280,protein = 35,560 M−1 cm−1, ε550,Cy3B = 130,000 M−1 cm−1, and ε280,Cy3B = 10,400 M−1 cm−1. Labeling stoichiometries ranged from 74 to 92%. Labeled GreB was stored at 4 °C in labeling buffer and was used within 2 wk or was frozen in liquid nitrogen in labeling buffer containing 50% (vol/vol) glycerol and 1.5 mg/mL BSA (126615; EMD Millipore) and was stored at −80 °C.
GreB-Stimulated Cleavage and Subsequent Elongation of Reconstituted Transcription Complexes.
Bulk GreB cleavage and elongation assays were performed at 37 °C on ECs reconstituted with RNA that was 5′-end labeled using T4 polynucleotide kinase (New England Biolabs) and γ-[32P]-ATP (Perkin-Elmer). The indicated concentration of GreB was incubated with reconstituted complexes (diluted to ∼100 nM in reconstitution buffer). In some experiments, complexes then were tested for elongation activity by the addition of nucleoside triphosphates (Roche Applied Science) and further incubation for another 10–15 min. Reactions were quenched by the addition of an equal volume of stop buffer [89 mM Tris⋅HCl (pH 8.0), 25 mM EDTA, 10 M urea, and 0.5% (wt/vol) each bromophenol blue and xylene cyanol]. RNA products were separated on 23% or 25% denaturing polyacrylamide gels, and band intensities were visualized via phosphorimaging (Typhoon 9410; GE Life Sciences). Radiolabeled RNA size markers were prepared from Decade Marker (AM7778; Thermo Fisher).
Microscopy.
Single-molecule experiments were conducted using a multiwavelength micromirror TIRF microscope (23) equipped with lasers at wavelengths 488, 532, and 633 nm for fluorescence excitation and at 785 nm for autofocus (43). Experiments were conducted in AB buffer [50 mM Tris⋅Cl (pH 8.0), 130 mM KCl, 5 mM MgCl2, 1 mM DTT, and 1 mg/mL BSA] for EC0 and EC−6 complexes and in KOP buffer [50 mM Tris⋅OAc (pH 8.0), 100 mM KOAc, 27 mM NH4OAc, 8 mM MgOAc, 2 mM DTT, 1 mg/mL BSA, and 3.5% (wt/vol) polyethylene glycol 8000 (EMB Chemicals)] for steady-state elongating complexes. Where noted, an oxygen scavenging system (OSS) was used as described previously (23). Only small differences (2.5-fold in kon and less than fourfold in τ) were detected in control experiments in which the kinetic constants for EC−6 complexes in the two buffers were compared.
TIRF experiments were conducted in flow chambers constructed (23) using glass slides passivated with a (mPEG-SG2000)/(biotin-PEG-SVA5000) (Laysan Bio) mixture (44). Streptavidin-coated fluorescent beads (fiducial markers for drift correction) (29) from the stock solution (T-10711; Molecular Probes) then were added to the chamber at a dilution of ∼1:420,000, typically achieving a surface density of one to three beads in each 65-μm diameter field of view. The chamber was incubated with 0.013 mg/mL streptavidin or NeutrAvidin (31000; Thermo Fisher) for about 45 s (44) and then was flushed clear twice with five chamber-volumes of buffer.
Steady-state elongation experiments were done largely as described previously (24). In brief, KO buffer (KOP without PEG) with OSS containing 50–200 pM template DNA derivatized with AF488 (Molecular Probes) and biotin was added to the NeutrAvidin-coated chamber. DNA density was monitored via 488-nm laser excitation, and the chamber was flushed with buffer after a density of ∼300 or more spots within the field was achieved. E. coli σ70 RNAP holoenzyme (Epicenter) was diluted to 0.2 nM in KO buffer and incubated in the chamber for 15 min to allow the formation of open complexes. Free RNAP was flushed out with KO buffer, and then transcription was initiated by the introduction of KOP buffer with OSS containing 10 nM Cy5 probe, 1 mM NTPs, and GreB as noted.
EC0 and EC−6 were examined in the same sample by tethering both types of complexes to a streptavidin-coated flow chamber through the biotin tag on the template DNA molecules. The two types of complexes were distinguished by attaching them sequentially to the chamber surface and recording the locations of the fluorescence spots from the AF488-labeled DNAs. To minimize photobleaching of DNA in the experimental field of view used for data acquisition, a different accumulation field was used to monitor the increasing surface density of complexes during initial attachment. Specifically, AB buffer with OSS containing 20–50 pM EC0 was added to the flow cell, and spot density in the accumulation field was observed as described above. The lane then was flushed, and images of the experimental field were recorded to establish locations for the nonbacktracked complexes. The process was repeated with EC−6. Approximately 40% of the complexes were eliminated from further analysis because of the proximity of neighbors (29). For each successive step in the five-step protocol outlined in Fig. 4A, the chamber was first flushed with two or more chamber-volumes of AB supplemented with OSS to remove the reagents from the preceding step, followed by the introduction of the reagents for the following step in two or more chamber-volumes of AB plus OSS.
Data Analysis.
Image and kinetics analysis was performed using custom software and algorithms for automatic spot detection, spatial drift correction, colocalization, plotting and analyzing kinetic data, and determining association and dissociation rates, active fractions, and amplitudes (https://github.com/gelles-brandeis/CoSMoS_Analysis) (29).
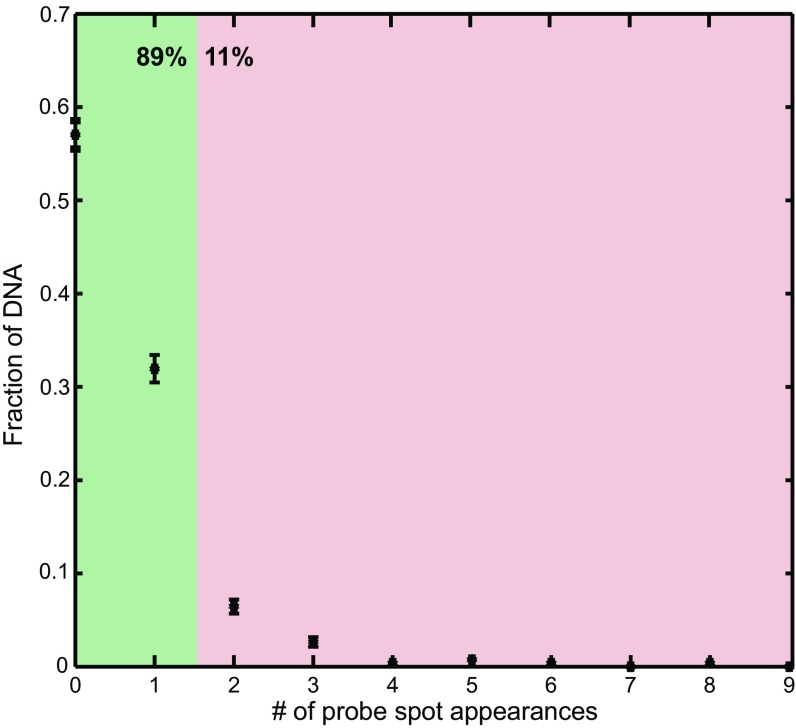
In the experiments in Fig. 1, elongation times were estimated by analyzing the dwell times of fluorescent Cy5-probe spots that colocalized to AF488 DNA. Cy5-probe spots lasting less than 30 s were assumed to represent nonspecific binding rather than authentic hybridization to transcript and were excluded from analysis. When only events with a duration >30 s were considered, most (89%; 234/263) AF488 DNA locations exhibited either zero or one Cy5 probe-binding event (Fig. S7). A substantial fraction (57%) of the 263 DNAs showed zero events, presumably because of inefficient open complex formation at the subnanomolar concentration of holoenzyme used to make open complexes. A small fraction (11%) showed more than one event, possibly because of probe dissociation or blinking. Those that contained one event were judged likely to reflect a single-round transcript elongation event and were used in analysis. In Fig. 2, analysis of fluorescence intensity from Cy3B-GreB was restricted to the times and locations of these elongation events and additionally excluded a minority (11%; 5/47) that had a probe duration >300 s.
Fig. S7.
Distribution of the number of Cy5-probe spots recorded on individual DNA molecules. Points show the fraction of DNA molecules that had the indicated number of separate Cy5-probe spot colocalizations with a duration >30 s detected in the experiment shown by the black traces in Fig. 1 D and E.
In the experiments in Fig. 4, active reconstituted complexes were identified using the colocalization of fluorescent spots arising from the AF488-labeled complex DNA and Cy5-labeled oligonucleotide complementary to transcript RNA. Dwell times >10 s were scored as Cy5–oligonucleotide colocalization events.
The data in Fig. 4C were fit with a single exponential yielding 8.39 GreB-binding events per rescue. With the measured labeling efficiency of 0.74 mol dye/mol protein for the Cy3B-GreB preparation used in this experiment, we infer there are on average 8.39/0.74 = 11.34 binding events per rescue, corresponding to a rescue probability of P = 0.08. This calculation may underestimate the probability because the RNA consists of a mixture of phosphorothioate diastereomers, and it is possible that only some of these are cleavable.
Acknowledgments
We thank David Harbage, Benjamin Morehouse, and Marc Hoemberger for help with protein purification and labeling. This work was supported by NIH Grants R01GM043369 and R01GM38660 and by the G. Harold & Leila Y. Mathers Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616525114/-/DCSupplemental.
References
- 1.Cramer P. Common structural features of nucleic acid polymerases. BioEssays. 2002;24(8):724–729. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- 2.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9(2):85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 3.Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118(3):281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Zenkin N, Yuzenkova Y. New insights into the functions of transcription factors that bind the RNA polymerase secondary channel. Biomolecules. 2015;5(3):1195–1209. doi: 10.3390/biom5031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stepanova EV, Shevelev AB, Borukhov SI, Severinov KV. [Mechanisms of action of RNA polymerase-binding transcription factors that do not bind to DNA]. Russian. Biofizika. 2009;54(5):773–790. [PubMed] [Google Scholar]
- 6.Sosunova E, et al. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100(26):15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaway RC, Kong SE, Conaway JW. TFIIS and GreB: Two like-minded transcription elongation factors with sticky fingers. Cell. 2003;114(3):272–274. doi: 10.1016/s0092-8674(03)00607-x. [DOI] [PubMed] [Google Scholar]
- 8.Koulich D, et al. Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 1997;272(11):7201–7210. doi: 10.1074/jbc.272.11.7201. [DOI] [PubMed] [Google Scholar]
- 9.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272(24):15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 10.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34(Pt 6):1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 11.Nudler E. RNA polymerase active center: The molecular engine of transcription. Annu Rev Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446(7137):820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 13.Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57(3):408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Cheung ACM, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 15.Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6(6):1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 16.Imashimizu M, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16(1):98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22(23):6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol. 2013;425(1):82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenkin N. Multiple personalities of the RNA polymerase active centre. Microbiology. 2014;160(Pt 7):1316–1320. doi: 10.1099/mic.0.079020-0. [DOI] [PubMed] [Google Scholar]
- 20.Roghanian M, Yuzenkova Y, Zenkin N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active centre. Nucleic Acids Res. 2011;39(10):4352–4359. doi: 10.1093/nar/gkq1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinella D, Potrykus K, Murphy H, Cashel M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J Bacteriol. 2012;194(2):261–273. doi: 10.1128/JB.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuzenkova Y, Roghanian M, Zenkin N. Multiple active centers of multi-subunit RNA polymerases. Transcription. 2012;3(3):115–118. doi: 10.4161/trns.19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91(3):1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harden TT, et al. Bacterial RNA polymerase can retain σ70 throughout transcription. Proc Natl Acad Sci USA. 2016;113(3):602–607. doi: 10.1073/pnas.1513899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H, Artsimovitch I, Landick R, Gelles J. Nonequilibrium mechanism of transcription termination from observations of single RNA polymerase molecules. Proc Natl Acad Sci USA. 1999;96(23):13124–13129. doi: 10.1073/pnas.96.23.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132(6):971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolić-Nørrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J Biol Chem. 2004;279(5):3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17(1):99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman LJ, Gelles J. Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods. 2015;86:27–36. doi: 10.1016/j.ymeth.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adelman K, et al. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci USA. 2002;99(21):13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami KS. Structural biology of bacterial RNA polymerase. Biomolecules. 2015;5(2):848–864. doi: 10.3390/biom5020848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furman R, Sevostyanova A, Artsimovitch I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40(8):3392–3402. doi: 10.1093/nar/gkr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daube SS, von Hippel PH. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992;258(5086):1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 34.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72(3):459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 35.Vassylyeva MN, et al. The carboxy-terminal coiled-coil of the RNA polymerase β′-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8(11):1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulish D, et al. The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 2000;275(17):12789–12798. doi: 10.1074/jbc.275.17.12789. [DOI] [PubMed] [Google Scholar]
- 37.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313(5786):518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 38.Opalka N, et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114(3):335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014;53(5):766–778. doi: 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Windgassen TA, et al. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42(20):12707–12721. doi: 10.1093/nar/gku997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50(6):882–893. doi: 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnusdottir A, Johansson I, Dahlgren L-G, Nordlund P, Berglund H. Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates. Nat Methods. 2009;6(7):477–478. doi: 10.1038/nmeth0709-477. [DOI] [PubMed] [Google Scholar]
- 43.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14(1):170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman LJ, Mumm JP, Gelles J. RNA polymerase approaches its promoter without long-range sliding along DNA. Proc Natl Acad Sci USA. 2013;110(24):9740–9745. doi: 10.1073/pnas.1300221110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chlenov M, et al. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase β′ subunit. J Mol Biol. 2005;353(1):138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 46.Opalka N, et al. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8(9):e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: Sequence analysis. J Mol Biol. 2010;395(4):671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278(14):12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 49.Molodtsov V, et al. X-ray crystal structures of Escherichia coli RNA polymerase with switch region binding inhibitors enable rational design of squaramides with an improved fraction unbound to human plasma protein. J Med Chem. 2015;58(7):3156–3171. doi: 10.1021/acs.jmedchem.5b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]