Abstract
T-cell CD4+ epitopes are important targets of immunity against infectious diseases and cancer. State-of-the-art methods for MHC class II epitope prediction rely on supervised learning methods in which an implicit or explicit model of sequence specificity is constructed using a training set of peptides with experimentally tested MHC class II binding affinity.
In this paper we present a novel method for CD4+ T-cell eptitope prediction based on modeling antigen-processing constraints. Previous work indicates that dominant CD4+ T-cell epitopes tend to occur adjacent to sites of initial proteolytic cleavage. Given an antigen with known three-dimensional structure, our algorithm first aggregates four types of conformational stability data in order to construct a profile of stability that allows us to identify regions of the protein that are most accessible to proteolysis. Using this profile, we then construct a profile of epitope likelihood based on the pattern of transitions from unstable to stable regions. We validate our method using 35 datasets of experimentally measured CD4+ T cell responses of mice bearing I-Ab or HLA-DR4 alleles as well as of human subjects.
Overall, our results show that antigen processing constraints provide a significant source of predictive power. For epitope prediction in single-allele systems, our approach can be combined with sequence-based methods, or used in instances where little or no training data is available. In multiple-allele systems, sequence-based methods can only be used if the allele distribution of a population is known. In contrast, our approach does not make use of MHC binding prediction, and is thus agnostic to MHC class II genotypes.
Introduction
Epitope-specific CD4+ T cells have been observed to correlate with protection against infections and cancer [1, 2]; in some cases immunization with single epitope peptides were protective [3–5]. However, immunization with CD4+ epitope peptides has also been shown to cause immunopathology and death [6, 7]. These studies emphasize the critical need for further analysis of CD4+ T-cell responses, which would be greatly facilitated by accurate epitope prediction.
Endogenous proteins, such as self proteins and viral proteins, are processed in the cytosol and transported into the ER and loaded onto class I MHC (MHCI) molecules. Exogenous proteins are taken up by endo/phagocytosis and processed into peptides and loaded onto MHC class II (MHCII) molecules. MHCI-peptide complexes bind to specific T-cell receptors on CD8+ T cells, which are cytotoxic, while MHCII-peptide complexes bind T-cell receptors on CD4+ T cells, which are more varied in nature. CD4+ T cells provide numerous protective functions as part of the adaptive immune response, including cytokine-mediated and contact-mediated signals to B cells, CD8+ T cells, and innate-immune cells, as well as direct modes of attack on pathogenic agents. While MHCI and MHCII molecules have scores of alleles, their three-dimensional structures are highly conserved; allele variation occurs primarily in the peptide binding groove and influences antigen peptide specificity. The closed binding grooves of MHCI molecules exhibit a preference for 8- to 11-mers, while the open binding grooves of MHCII molecules are less specific, with bound peptides being between 10 and 30 amino acids long.
Computational methods for predicting MHCII-restricted epitopes are of great interest for understanding immune responses to a variety of pathogens. The most accurate computational approaches to MHC binding prediction are currently based on modeling the sequence preferences for a given (single) MHCI or MHCII allele. While the particular machine learning method can vary, generally sequence-based methods work by using training data obtained from allele-specific MHC binding assays to construct a model that predicts a binding score. For example, the widely used NetMHCII server [8, 9] performs MHCII-restricted epitope prediction and utilizes a position-specific scoring matrix constructed from the training data for a specified allele.
Since MHCI molecules are highly sequence specific, supervised learning methods (e.g., [10]) for epitope prediction have been successful (e.g., [11]). Additionally, antigen processing and loading for the MHCI pathway are more tightly orchestrated, with proteolysis ocurring in one compartment, and binding/loading occurring in another. The MHCII presentation pathway is more challenging to model due to the open binding groove in the class II molecule, but also because antigen processing, loading, and MHC binding happen concurrently. Recently, Wang et al. [12] showed that while a “consensus” approach yields substantially improved MHCII binding predictions, these improvements do not necessarily carry over to the subsequent prediction of CD4+ T-cell immune responses.
Early studies demonstrated that multiple lysosomal endoproteases and exoproteases participate in processing of the antigens and that their activities were partially redundant [13]. Processing steps are thought to occur both before and after peptide binding to the MHCII protein. The elution of nested sets of peptides from naturally loaded MHCII proteins suggested that proteolytic trimming takes place after binding [14]. However, other studies indicated that proteolysis must occur before binding. Watts and coworkers found that presentation of multiple T-cell epitopes in tetanus toxoid depended on an initial proteolytic cleavage by asparagine endoprotease [15]. Presumably, the nicked protein was destabilized enough for unfolding to expose the epitopes for binding to the MHCII protein. Disulffde crosslinks help a protein resist unfolding, and the works of Cresswell and Landry and their coworkers demonstrated that disulfide bonds can block epitope immunogenicity [16], (H.-N. Nguyen et al., unpublished). However, disulfide bonds can also have the opposite effect, to increase T-cell epitope immunogenicity, presumably by stabilizing the antigen against proteolytic destruction [17]. On a more subtle level, dominant epitopes were reported to occur most frequently at sites adjacent to conformationally flexible protein segments, which may serve as entry points for proteolytic processing [18–20]. Several studies have confirmed that these epitopes occur near the ends of peptides generated by limited proteolysis in vitro [21–23].
The intertwined mechanisms of antigen processing and peptide loading are further modulated by the action of HLA-DM and its regulator HLA-DO (or generically DM and DO). DM stimulates peptide exchange in MHCII proteins, and mice that lack DM have altered epitope dominance patterns [24]. DO inhibits DM by blocking the site that interacts with the MHCII protein, and mice lacking DO also have altered epitope-specific responses [25]. We are not aware of any efforts to specifically incorporate mechanisms of antigen processing or DM/DO-regulated peptide exchange for refinement of class II epitope prediction. One indirect effort utilized the SYFPEITHI database [26] of natural MHCII ligands to predict viral peptides that not only bind well to the MHCII protein but also resemble the pools of natural (mostly self) ligands [27].
In this paper, we present an algorithm for MHCII-restricted epitope prediction that utilizes conformational stability data of a given antigen structure. We hypothesize that mechanisms for antigen processing, and thus antigen three-dimensional structure, play an important role in guiding the ultimate presentation of an epitope. Our algorithm uses experimental and predicted conformational stability criteria as input, and computes an epitope likelihood score for any peptide in the antigen sequence.
In single-allele systems, our approach is orthogonal to existing, MHC binding-based prediction while achieving essentially the same accuracy. Thus, in the single-allele setting our method can potentially supplement existing MHCII-binding based prediction schemes. Perhaps the most applicable setting of our approach is in multiple-allele systems. MHC binding-based approaches face the dual challenges of requiring knowledge of the MHC alleles and identifying appropriate weights on each allele. In contrast our method is far more practical: it allows epitope prediction without genotype information while achieving the same or better performance of existing methods in single-allele predictions.
Materials and Methods
Datasets Compiled
We collected 35 datasets of CD4+ T-cell immune response mapping studies. Table 1 provides the details of each antigen and citation to the experimental study. In general each mapping study provides a quantitative profile of immunogenicity for the set of peptides that was tested. In our experiments, we used epitopes characterized in the literature as ground truth (see Table 1, “Antigen” column). For data gathered in the Landry Lab, we used the Wilcoxon signed-rank test to determine epitopes.
Table 1.
Results for Epitope Prediction.
| Allele | Antigen | # epitopes | # peptides | Stability1 | MHC binding2 |
|---|---|---|---|---|---|
| I-Ab | HIV gp120 (JRFL)3 | 5 | 46 | 2 | 2 |
| Y. pestis LcrV4 | 7 | 53 | 3 | 4 | |
| Tuberculosis antigen 85A [40] | 5 | 28 | 2 | 0 | |
| Tuberculosis antigen 88 [41] (4LVQ) | 2 | 30 | 1 | 1 | |
| Cholera toxin B [42] (1FGB) | 1 | 23 | 0 | 1 | |
| Y. pestis CAF14 | 8 | 26 | 3 | 3 | |
| Friend virus env [43] | 6 | 20 | 4 | 3 | |
| GFP [44] (2QLE) | 1 | 17 | 1 | 1 | |
| HIV gp120 (89.6) [45] | 8 | 47 | 2 | 2 | |
| SIV gp1203 | 11 | 80 | 3 | 2 | |
| Bacteriophage T4 HSP105 | 2 | 20 | 1 | 1 | |
| Hepatitis B virus [46] (3J2V) | 1 | 13 | 0 | 1 | |
| HRSV M2-1 [47] (4C3B) | 1 | 23 | 0 | 1 | |
| HSP 16 [48] (3W1Z) | 1 | 13 | 0 | 0 | |
| Listerialysin O [49] (4CDB) | 1 | 48 | 0 | 1 | |
| Tuberculosis MPT51 [50] | 1 | 25 | 0 | 0 | |
| Tuberculosis PstS [51] (1PC3) | 3 | 32 | 1 | 1 | |
| VSV glycoprotein [52] (2CMZ) | 2 | 44 | 1 | 1 | |
| West Nile virus env [53] (2I69) | 1 | 78 | 0 | 1 | |
| West Nile virus ns3 [53] (2WV9) | 2 | 116 | 0 | 1 | |
| Yellow Fever env [54] | 1 | 96 | 0 | 1 | |
| HLA-DR4 | Chlamydia CPAF [55] | 5 | 58 | 1 | 1 |
| Anthrax protective antigen [38] | 15 | 74 | 3 | 6 | |
| Anthrax lethal factor [38] | 16 | 89 | 3 | 7 | |
| Dengue env [56] | 14 | 26 | 4 | 2 | |
| Burkholderia agellin [38] | 2 | 37 | 1 | 0 | |
| Human | Adenovirus serotype 5 hexon [57] | 16 | 133 | 7 | - |
| Tuberculosis antigen 85A [58] | 14 | 28 | 4 | - | |
| Birch pollen Bet v 1 [59] | 5 | 50 | 2 | - | |
| Hepatitis C ns3 [60] | 7 | 45 | 2 | - | |
| M. leprae HSP70 [61] | 6 | 60 | 3 | - | |
| Polio vp1 [62] | 5 | 25 | 1 | - | |
| Tick-borne encephalitis protein E [63] | 12 | 121 | 4 | - | |
| Tetanus toxoid [64] | 10 | 51 | 4 | - | |
| Wasp Ves v 1 [65] | 7 | 65 | 2 | - | |
Number of epitopes recovered at the 80th percentile scoring threshold for stability-based epitope prediction.
Number of epitopes recovered at the 80th percentile scoring threshold for MHC binding-based epitope prediction.
H. Nguyen et al., Vaccine, In press.
H. Nguyen et al., Vaccine, In press.
Landry and coworkers, unpublished data.
To apply our stability-based prediction method, we utilize crystallographic B-factors, solvent-accessible surface area, an estimate of local instability in the antigen structure (COREX [28, 29] residue stabilities) and a statistic of evolutionary sequence divergence (Shannon sequence entropy). Each parameter provides a quantitative measure of the local conformational flexibility and therefore the likelihood of proteolysis at any particular position in the protein [30]. All antigen structures considered in our experiments were solved using X-ray crystallography with the exception of that for Bet v 1, which was solved by NMR. B-factors are a measure of local conformational disorder, which is an indication of how easily the structure may be deformed for binding in a protease active site [31]. Solvent accessible area quantifies accessibility to proteases as well as local disorder. COREX provides a score of the probability of unfolding at each amino acid, and has been validated by correlation with hydrogen-deuterium exchange protection NMR experiments [28]. Hydrogen exchange occurs on much longer timescales than the conformational fluctuations that are captured by B-factors, and therefore it provides complementary information about the likelihood of proteolysis [32]. Sequence entropy is correlated with solvent-accessible surface area [33], and it provides information on protein segments that were not present in the crystallographic/NMR structure. For this analysis, backbone amide-nitrogen B-factors were extracted from the PDB entries given in Table 1. Likewise, average root-mean-square deviations for the backbone amide nitrogens were extracted from the PDB entry for Bet v 1. Solvent-accessible surface area was calculated with the crystallographic or NMR structures using Molmol [34]. We computed the COREX/BEST [28, 35] score using the provided web interface [29]. For analysis of sequence entropy, all protein sequence entries having 50%–95% identity to the target were collected from the UniProtKB/Swiss-Prot or UniProtKB database using Blastp. The UniProtKB database was used when Bastp returned fewer than 25 sequences from the UniProtKB/Swiss-Prot database. The sequences were aligned using ClustalW [36], and then the Shannon sequence entropy was calculated using BioEdit [37].
Algorithm1
The input to our algorithm is the antigen sequence and the four sources of conformational stability data mentioned above: crystallographic B-factors, solvent-accessible surface area, COREX residue stabilities and sequence conservation. Our algorithm proceeds in three main steps. In the first step, we preprocess the input structural data into a suitably smooth representation. For the second step, we first compute a z-score statistic that characterizes the aggregate conformational stability at each residue relative to the input structure. Then we segment the structure into regions of conformational stability (and instability) by thresholding the computed z-score statistic. In the final step, we construct the output epitope likelihood by applying a chosen weighting scheme that emphasizes the C- or N-terminal flanks of unstable regions.
We first preprocess the data to register the antigen sequence to the structure. Since a PDB file corresponding to the antigen sequence can have missing residues, we address gaps by assigning default values for each data type that indicate a minimum level of stability. For example, a missing B-factor would be set to be the maximum B-factor value observed in the PDB file. Then, as a preprocessing step to smooth the data, we take a windowed average across each dataset using a window size of 15 residues.
After smoothing the data, we compute z-scores for every residue with respect to each dataset. We view the z-scores for each type of data as independent observations of antigen conformational stability. Using the analog of Fisher’s method for combining test statistics, we combine z-scores to obtain an aggregate z-score for each residue of the antigen. This aggregate z-score characterizes the extent to which our sources of structural data “agree” that the antigen is conformationally unstable at a given position, and thus attempts to model likely sites of proteolysis. Then we segment the protein using this aggregate z-score by classifying any residue with an aggregate z-score greater than zero as stable and all other residues unstable. In the next step, we will refer to any continguous stretch of stable amino acids as a stable region, and any other contiguous stretch of amino acids as unstable regions. We note that by definition, this definition segments the protein into alternating stable and unstable regions.
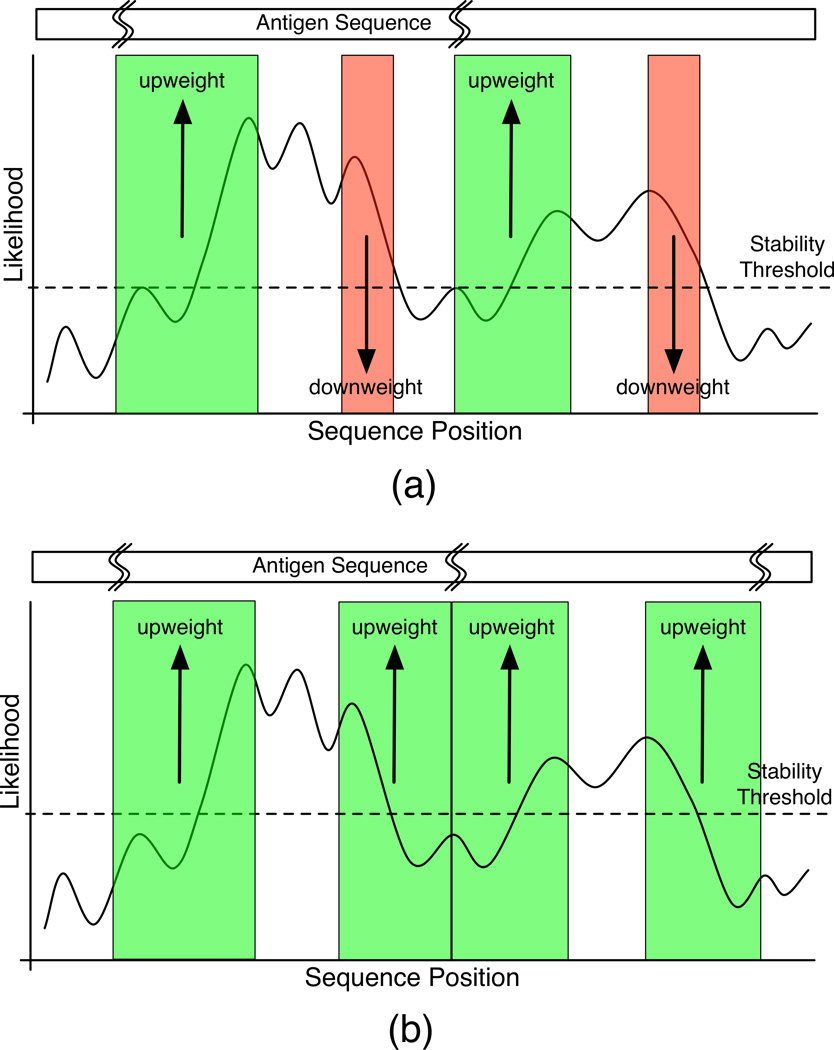
To construct our epitope likelihood score, we first set the epitope likelihood of any residue in an unstable region to be zero. Stable residues adopt their z-score as their epitope likelihood (Figure 1, black curve above dotted line). Then, we selectively upweight likelihoods in regions of the protein that transition from unstable to stable or vice versa (Figure 1). For exposition, consider an unstable region and the C-terminally adjacent stable region. First, we magnify the epitope likelihoods for first third of the stable region by a factor of three. Then, we assign epitope likelihoods by linearly interpolating from the midpoint of the unstable region to the midpoint of the upweighted portion of the C-terminal flanking stable region. This same upweighting method can be applied to the N-terminal flank or to both the C-terminal and N-terminal flanks of an unstable region. We allow the user to choose which particular scheme will be applied for a given input antigen. In the results presented here, single-allele datasets made use of C-terminal weighting only (Figure 1(a)), whereas human datasets made use of weighting on both flanks of unstable regions (Figure 1(b)).
Figure 1. Epitope Likelihood construction.
Epitope likelihood is constructed by reweighting the aggregate stability score in relation to an assumed proteolytic site at the midpoint of an unstable region. Depending on the context we (a) upweight regions downstream of proteolytic sites, or (b) regions both upstream and downstream of proteolytic sites (upweighted regions are shaded in green and downweighted regions are shaded in red).
MHC Binding Score Prediction
For each of the antigens above, we computed MHC binding affinity to use as an alternate analysis of predicted immunogenicity. We used NetMHCII [8, 9] to perform prediction for each antigen sequence. The resulting prediction score (the 1-log50 K(aff) entry in the output) was used as an epitope likelihood score. We note that IEDB [38] also has a set of tools, among which are the NetMHCII server itself. We chose to use NetMHCII for flexibility; NetMHCII allows the selection of individual peptide lengths used in the peptide mapping assays for each antigen, which varied in length from 15 amino acids to 20 amino acids.
Performance Criteria and Evaluation
In our work, we use two metrics for evaluating predictive performance with respect to a given a set of epitopes determined experimentally (e.g., with an assay for T-cell proliferation or IFN-γ ELISPOT). First, we consider how many epitopes are recovered by the 90th and 80th percentile scores for a given method of prediction (as shown in Table 1). These two thresholds were choisen on the basis of rates of experimental epitope discovery, which ranged from 8% to 19% of tested peptides corresponding to proteins that were mapped (see below). The same threshold has been used in recent work evaluating the 5 correlation between MHC-binding and T-cell activation assays [39]. We note that in general it is difficult to choose the “correct” threshold that best balances true and false positive rates; it is for this reason that we also use the AUC metric to evaluate performance on single-allele and human datasets.
For each threshold, we classify the performance of a method based on how many epitopes are correctly identified. These results provide an evaluation of how effective a particular method is in a real-world setting where a set of peptides must be chosen for testing. To establish baseline performance, we compare both MHC binding-based and stability-based prediction results against the expected number of epitopes at a given threshold. In practice, this baseline approach would correspond to choosing peptides at random with a probability based on an estimated epitope frequency.
In prior work, evaluation of MHC binding performance (e.g., [12]) has typically been characterized by the area under the receiver-operator characteristic (ROC) curve. As above, for these evaluations peptides have an underlying classification as a binder or nonbinder (e.g. determined experimentally) and any method for binding prediction is rated as to its effectiveness in predicting the underlying classification. In our approach, we compute epitope likelihood on a per antigen basis, and to compare predictions across antigens we normalize scores to be in the range [0, 1]. Then, predictions for a collection of peptides spanning multiple antigens can be evaluated in the same manner as MHC binding. We computed ROC curves in this way for epitope prediction in both single-allele systems (Figure 2 (c)) and humans (Figure 3 (b)).
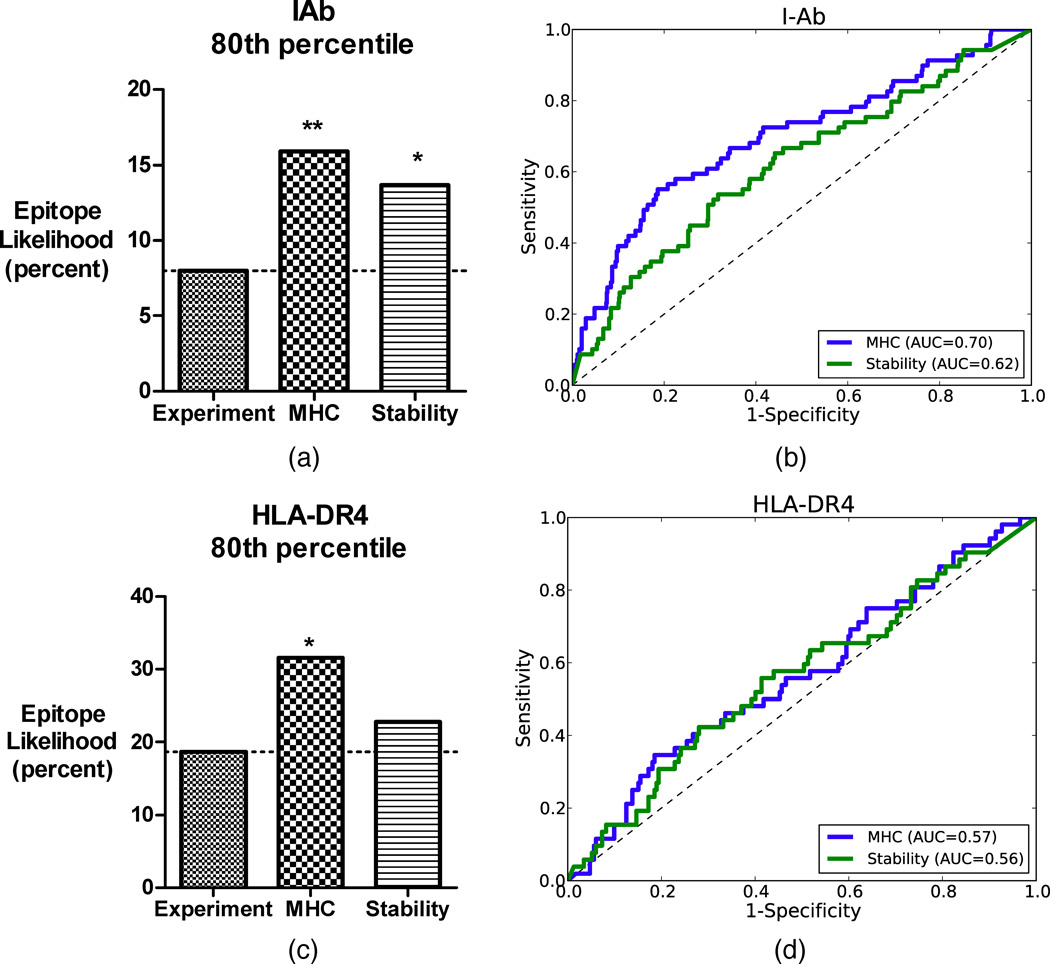
Figure 2. Prediction accuracy and ROC curves for single-allele datasets.
(a, c) Overall accuracy of prediction at the 80th percentile threshold of the random baseline, stability-based prediction and MHC binding-based prediction for IAb and HLA-DR4 data, respectively. (b, d) ROC curves for stability-based prediction (in green) and MHC binding-based prediction (blue).
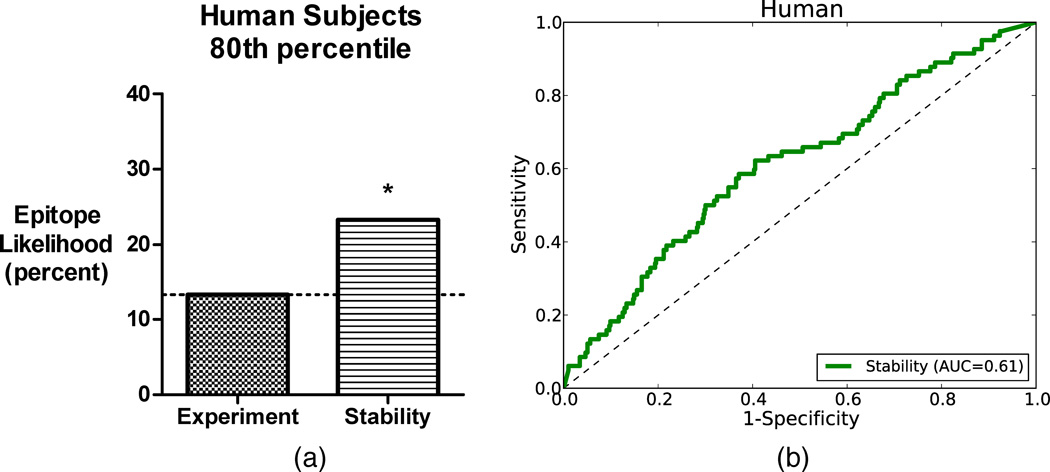
Figure 3. Prediction accuracy and ROC for human data.
(a) Overall accuracy of prediction at the 80th percentile threshold of the random baseline and stability-based prediction. (b) ROC curve for stability-based prediction.
Results and Discussion
We considered a total of 35 datasets in which epitopes were experimentally determined. For each single-allele MHC dataset, we compared the performance of our algorithm against a popular sequence-based method, NetMHCII [8, 9]. For multiple allele (i.e., humans) datasets, MHC binding methods can only be used when the alleles (or allele distribution) of the population is known. This information was not available for the datasets we considered and thus no MHC binding-based prediction was possible.
Table 1 shows the number of epitopes identified when we consider the top-scoring peptides at the 80th percentile threshold for each method. When evaluating predictive power, we compare both methods against the naive random approach, in which a peptide is predicted to be an epitope with a probability equal to the epitope frequency for that antigen.
Results for single-allele systems
Epitope-predictions based on stability and based on MHC-peptide binding were evaluated for accuracy in epitope-mapping results obtained in immunized mice. Each experimental map was generated with a scan for T-cell responses using an overlapping series of peptides that spanned the complete antigen. The input requirements for the epitope predictions limit the number of epitope-mapping studies that may be used for the evaluation. For predictions based on conformational stability, the high-resolution structure of the antigen must have been solved by X-ray crystallography or nuclear magnetic resonance. For predictions based on MHC binding, the T-helper epitopes must have been mapped in mice that have a single well-characterized MHCII protein. The evaluation also excluded studies on mammalian antigens, for which some epitopes may have been suppressed by negative selection. Epitope maps of influenza antigens were excluded because epitope placement is exceptional, in that epitopes consistently appear on the N-terminal flanks of flexible sites, possibly due to viral modifications to antigen processing mechanisms [66]. The usefulness of excluding influenza epitopes highlights that fact that antigen-processing steps play an important role in shaping CD4+ epitope dominance. It is likely that additional organisms modulate epitope dominance through their influence on antigen processing. The present approach creates a formalism for handling these influences and ultimately incorporating them into the algorithm.
These limitations reduced the available experimental systems to C57BL/6 mice (70 epitopes in 21 antigens) and HLA-DR4-transgenic mice (52 epitopes in 5 antigens). Several epitope-mapping studies in BALB/c mice have been reported, but the number of epitopes discovered is low. The BALB/c strain also has two MHCII proteins (I-Ad and I-Ed), which potentially complicate the MHCII binding prediction and the interpretation of mapping data. Thus, results from BALB/c mice were not included. The several mapping studies in HLA-DR1- and HLA-DQ8-transgenic mice yielded only a small number of epitopes, and thus they were not included. For comparison, the IEDB lists 377 I-Ab-restricted epitopes in 146 non-mammalian antigens. Most of these epitopes were not included in the present study because the high-resolution structure of the antigen was not available (e.g., a membrane protein or intrinsically disordered protein) or because a limited set of peptides (usually prescreened for I-Ab binding) were tested for a T-cell response.
In C57BL/6 and HLA-DR4 mice, the various antigens primed as few as one and as many as eleven epitopes, which were discovered by testing with peptide sets of 20-89 peptides. In general the density of epitopes is similar to that previously reported for a collection of nine antigens and allergens [66]. For the collection of 21 antigens in C57BL/6 mice, the rate of epitope discovery is the number of epitopes divided by the number of test peptides (aggregated over all antigens, to account for antigen size). Expressed as a percentage, the rate was 8%. This rate imposes a lower limit on the accuracy of epitope prediction because 8% of peptides randomly selected from the test set are expected to contain an epitope.
Accuracy of epitope prediction in C57BL/6 mice was evaluated by comparing the 80th percentile of predicted epitopes to the experimentally discovered epitopes (Figure 2(a)). Using our stability-based method for epitope prediction, 13% of peptides scoring in the 80th percentile of predicted immunogenicity were actually observed as epitopes (see Supplementary Information Figures S1–S21 for full details of predictions for all antigens). This is a significant improvement over random peptide selection (p = 0.02). Prediction based on peptide-binding to the I-Ab MHCII achieved a success rate of 15%, and was also significant (p = 0.002). In HLA-DR4 transgenic mice, epitopes were discovered at a rate of 19% of the test peptides. Epitope prediction at the 80th percentile using stability-based prediction and HLA-DR4 binding-based prediction yielded accuracies of 23% and 32%, respectively (Figure 2 (c), see Supplementary Information Figures S22–S226 for full details of predictions for all antigens). HLA-DR4 binding-based prediction was significantly better than random (p = 0.03) at this threshold.
Finally, we also consider the area under the ROC curve for a threshold-independent comparison of MHC binding-based prediction and stability-based prediction (Figure 2(b, d)). For I-Ab data, stability-based prediction achieves an AUC of 0.62 (p = 0.0007), while MHC-based prediction achieves an AUC of 0.70 (p < 0.0001). For HLA-DR4 data, while MHC-based prediction achieved significance at the 80% threshold, neither method achieves an AUC that is significantly better than random epitope selection. Interestingly a simple combination of the methods, in which we compute an epitope likelihood score that is the product of the normalized individual scores, achieves an AUC of 0.61 and is significant (p = 0.01). The poor performance of both predictions may be related to the heterologous expression of HLA-DR4. Peptide loading or peptide exchange may be abnormal in the transgenic mice due, for example, to the lack of HLA-DM. The mouse homolog DM radically alters the CD4+ epitope dominance pattern for Leishmania LACK [24], and presentation of an HLA-DR4-restricted arthritogenic epitope was essentially eliminated by co-expression of HLA-DM in APC [67].
Results for human subjects
Antigen processing-based epitope prediction was evaluated for the aggregated results of mapping studies performed in human subjects (see Supplementary Information Figures S27–S35 for full details of predictions for all antigens). As noted above for the single-allele studies, the analysis was limited to non-self antigens for which a crystal structure was available and where the epitope-mapping employed a complete series of overlapping peptides. The identification of a discrete set of all epitopes for a given antigen is difficult for systems with multiple MHC alleles because the immunogenicity of some epitopes could be strongly allelle-specific. If the investigators identified a discrete set of epitopes, then these were included as the immunodominant set. If the investigators reported a frequency of response for all peptides, then the peptides of the 90th percentile were included as the immunodominant set. In all, 82 epitopes were identified as immunodominant in 9 antigens. Since the antigens were scanned with a total of 578 peptides, this represents a rate of discovery for immunodominant epitopes of 14%. We take this to be the threshold for useful accuracy of epitope prediction in this set of antigens.
At the 80th percentile, the accuracy of 23% achieved significance (p = 0.01), Figure 3(a)). To gain additional perspective, we also consider the receiver-operator characteristic (ROC) taken over all predictions for human alleles (Figure 3(b)). Our algorithm achieves an area under the curve of 0.61 (p = 0.001). Taken together, the accuracy and AUC results for human data demonstrates that our method is about twice as effective the random baseline. In practice this would mean screening about half as many peptides to identify the desired number of epitopes.
We now discuss two specific antigens, Bet v 1 and adenovirus type 5 (Ad5), and rationalize our predictions with what was observed in epitope mapping studies.
Bet v 1 in birch-allergic human subjects
T-helper epitope maps from human populations are difficult to interpret due to genetic heterogeneity of class II MHC proteins. Scores of different class II alleles are represented in some populations, and individuals may express six different alleles. For most epitopes, the restricting MHC allele is not known or multiple alleles contribute to presentation. In spite of multiple sources of variability, strong epitope dominance is still observed in the human immune responses.
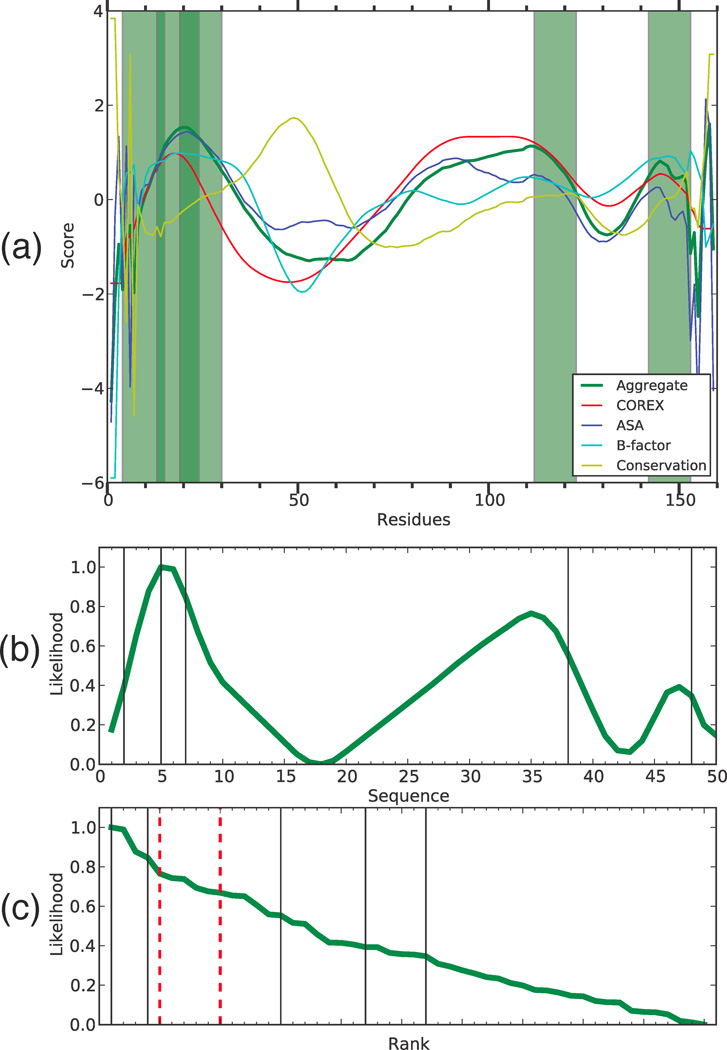
Bet v 1 of birch pollen is one of the most thoroughly studied allergens. Bohle and coworkers mapped the T-helper epitopes of the 159-residue Bet v 1 in a group of 57 birch-allergic subjects using a set of 50 overlapping 12-mer peptides [59]. As expected for a heterogeneous population, the T-helper epitopes were distributed over most of the protein. Forty-three peptides obtained a response from at least one subject, and 35 peptides obtained a response from at least two subjects. Here, we define the immunodominant epitopes as the five most frequently immunogenic peptides (90th percentile), each of which obtained a response from at least 11 subjects (peptides 2, 5, 7, 38, and 48). The single most frequently immunogenic epitope (peptide 48) obtained a response in 32 subjects.
The analysis of conformational stability in Bet v 1 found two major dips in stability and predicted epitopes in the transitions from low-to-high stability on both sides of the stability minima, resulting in three peaks of predicted immunogenicity (Figure 4(a, b)). This equal weighting of N- and C-terminal flanks was adopted on the basis of a previous analysis of immune responses in outbred populations to nine different antigens/allergens [66]. Four Bet v 1 peptides in the 90th percentile of predicted immunogenicity were located in the first major peak, and they include the observed epitopes 5 and 7. Although having generally lower predicted immunogenicity, the second and third peaks each included one of the observed epitopes.
Figure 4. Birch Bet v 1 predictions.
(a) Conformational stability data is shown by residue, epitopes are shaded green bars. (b) The computed epitope likelihood score for sequential 12-mers (epitopes shown as black lines) is shown along with (c) the rank-order view of the scores with 90th and 80th percentile thresholds (red dashed lines).
The fact that the highly dominant peptide 48 coincided with the smallest of the three peaks of predicted immunogenicity in Bet v 1 suggests that that the prediction failed to capture an important aspect of the mechanism for dominance. We posit that the missing element involves the ease with which a proteolytic fragment dissociates from the otherwise intact antigen. The current algorithm assigns high immunogenicity to peptides that are located within stable segments adjacent to highly flexible segments. This weighting recognizes the probability of initial cleavage in the flexible site and assigns immunogenicity to the adjacent stable segment. It does not account for the requirement that the MHCII protein gain access to the stable segment, which is likely to involve the dissociation of the epitope-containing segment from other stable segments. Clearly, these considerations will demand a more sophisticated model than is currently implemented. Nevertheless, our method performs well across all scoring thresholds, achieving an AUC of 0.76.
The exceptional immunodominance of peptide 48 was studied by Bohle and co-workers [23], and has been attributed to its early and abundant processing and presentation. Peptides presented by dendritic cells were compared to the proteolytic fragments generated in a time-course of Bet v 1 digestion in lysosomal extracts. The most abundantly presented peptides corresponded to fragments that were generated only at early time-points (0.5–3 hr). It remains unclear why peptides generated at later time-points (5–24 hr) are poorly presented. The late peptides appear to be in equal or greater abundance compared to the early peptides at the respective time points, and thus proteolytic destruction of late peptides seems to be ruled out. Mechanisms of intracellular trafic may be responsible. During the maturation of dendritic cells, ubiquitination in the C-terminal tails redirects MHCII from the lysosome to the cell surface [68], and thus the MHCII may not be available to bind peptides that emerge from late stages of antigen processing.
Adenovirus type 5 hexon in HIV-vaccine trial participants
Pre-existing immunity to Ad5 has been linked to weaker responses to Ad5-based vaccines. Initial attention focused on the ability of the antibodies to neutralize the vaccine, and this spurred the development of adenoviruses having low prevalence in humans and little antibody crossreactivity. However, the highly conserved T-cell responses to the hexon subunit of Ad5 recently have also been implicated in the weak responses to Ad5 vaccines [57]. Thus, we sought to examine the relationship of structure and T-cell epitope dominance in hexon, a 947-residue capsid protein.
McElrath and coworkers [57] mapped T-helper epitopes for the Ad5 hexon using 133 overlapping 15-mer peptides with the PBMC of 32 subjects participating in an HIV-vaccine trial. Although approximately half of the subjects were sero-negative for Ad5, most reacted to at least one T-cell epitope, probably because the T-cell epitopes are conserved in other adenoviruses to which the subjects had been exposed (e.g., Ad1 or Ad2). As expected for a heterogeneous population, epitopes were distributed over a large portion (40%) of the hexon sequence. For the present comparison to epitope prediction, we designated as immunodominant the 16 peptides that stimulated a T-cell response from two or more subjects (corresponding to the 88th percentile of the tested peptides).
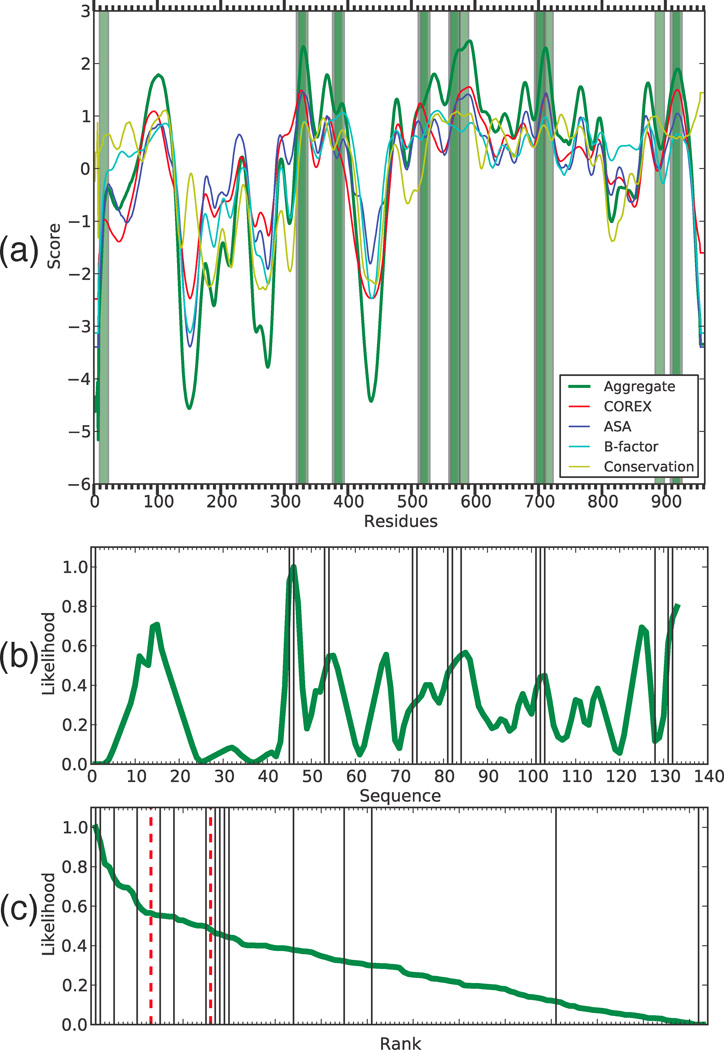
The analysis of hexon conformational stability found at least eight major dips in stability, which gave rise to adjacent peaks of epitope likelihood (Figure 5(a, b)). Peptides in the 90th percentile of predicted epitopes touched five peaks of epitope likelihood. Two peaks contained four peptides that match observed epitopes (peptides 45, 46, 131, and 132). Although the epitopes occured as two pairs of overlapping peptides, it is not clear that each pair should be considered a single epitope because the restricting MHC alleles are unknown. An alternative explanation is that the pairs of epitopes arose from the same abundantly processed antigen fragments, as suggested by their locations at the transitions from low-to-high stability. If we reach down to the 80th percentile, our stability-based approach accurately predicts three more epitopes (peptides 54, 82, and 84).
Figure 5. Adenovirus type 5 hexon predictions.
(a) Conformational stability data is shown by residue, epitopes are shaded green bars. (b) The computed epitope likelihood score for sequential 15-mers (epitopes shown as black lines) is shown along with (c) the rank-order view of the scores with 90th and 80th percentile thresholds (red dashed lines).
In summary for Ad5 hexon, 4 of 13 peptides in the 90th percentile and 7 of 26 peptides in the 80th percentile were observed as epitopes. Of the eight transitions from low-to-high stability (peaks of epitope likelihood), four contained epitopes in the vaccine trial participants. Overall, our method achieves an AUC of 0.76 for this dataset.
Conclusions
In this paper we have developed a method for epitope prediction based solely on antigen processing constraints. Our method achieves significant predictive power, despite the fact that it does not consider the sequence specificity of MHC class II binding. Importantly, a conformational stability-based approach to epitope prediction is orthogonal to existing methods that rely solely on sequence preferences for MHC binding to predict epitopes.
Given this dichotomy, a natural question is whether it is possible to combine the two methods to make improved predictions in single-allele systems. In prior work, researchers have taken a consensus approach [12] in which the results of multiple MHC binding-based predictors are aggregated to obtain improved results. The resulting “consensus” approach from this work is currently implemented in the IEDB [38]. In the course of developing the current antigen processing-based prediction algorithm, we also tested a simple hybrid approach in which stability-based prediction and MHC binding-based prediction were assumed to be independent and were combined accordingly. We found a mixed set of results, wherein any improvement in epitope identification was offset by an increase in false positive rates.
We believe it is important to further refine the construction of epitope likelihood, by conditioning the computation of the score both on antigen processing and MHC binding affinity, in a manner analogous to the class I-restricted epitope prediction tool, NetCTL. However, class II prediction must overcome a considerably larger pool of alleles in the human gene pool, a potentially smaller influence of peptide-binding affinity, and a complex interaction between antigen processing and MHC binding. The present studies take an important first step toward addressing the influence of antigen processing. We are exploring supervised learning methods analogous to those used for training MHC binding affinity predictors, which use training sets of peptides with experimentally determined MHC binding affinity. We seek to generalize these approaches with the inclusion of conformational stability criteria. That is, rather than applying a predetermined upweighting to the flanks of an unstable domain (and adjacent stable domains), we would include conformational stability data in the training data, and rely on the training regime to identify appropriate joint weightings of both conformational stability and MHC binding affinity.
Supplementary Material
Acknowledgments
RRM was supported by NSF CAREER IIS-0643768. TC and SJL were supported by NIH R01-AI080367 (to SJL).
Footnotes
The source code for our current implementation along with the datasets is available upon request.
References
- 1.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512:324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 3.McNeal MM, Basu M, Bean JA, Clements JD, Choi AH, et al. Identification of an immunodominant CD4+ T cell epitope in the VP6 protein of rotavirus following intranasal immunization of BALB/c mice. Virology. 2007;363:410–418. doi: 10.1016/j.virol.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 4.Miyazawa M, Fujisawa R. Restriction of Friend virus-induced erythroid cell proliferation by CD4+ T-lymphocytes that recognize a single gp70 epitope. Leukemia. 1997;11(Suppl 3):227–229. [PubMed] [Google Scholar]
- 5.Kurtz JR, Petersen HE, Frederick DR, Morici LA, McLachlan JB. Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect Immun. 2014;82:2424–2433. doi: 10.1128/IAI.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold IC, Hitzler I, Engler D, Oertli M, Agger EM, et al. The C-terminally encoded, MHC class II-restricted T cell antigenicity of the Helicobacter pylori virulence factor CagA promotes gastric preneoplasia. J Immunol. 2011;186:6165–6172. doi: 10.4049/jimmunol.1003472. [DOI] [PubMed] [Google Scholar]
- 7.Penaloza-MacMaster P, Barber DL, Wherry EJ, Provine NM, Teigler JE, et al. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science. 2015;347:278–282. doi: 10.1126/science.aaa2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M, Lund O. NN-align: An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007:8. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, et al. Reliable prediction of t-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moutaftsi M. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nature Biotechnology. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Sidney J, Dow C, Mothe B, Sette A, et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lennon-Dumenil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol. 2002;14:15–21. doi: 10.1016/s0952-7915(01)00293-x. [DOI] [PubMed] [Google Scholar]
- 14.Nelson CA, Vidavsky I, Viner NJ, Gross ML, Unanue ER. Amino-terminal trimming of peptides for presentation on major histocompatibility complex class II molecules. Proc Natl Acad Sci USA. 1997;94:628–633. doi: 10.1073/pnas.94.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoniou AN, Blackwood SL, Mazzeo D, Watts C. Control of antigen presentation by a single protease cleavage site. Immunity. 2000;12:391–398. doi: 10.1016/s1074-7613(00)80191-0. [DOI] [PubMed] [Google Scholar]
- 16.Maric M, Arunachalam B, Phan UT, Dong C, Garrett WS, et al. Defective antigen processing in GILT-free mice. Science. 2001;294:1361–1365. doi: 10.1126/science.1065500. [DOI] [PubMed] [Google Scholar]
- 17.Mirano-Bascos D, Steede NK, Robinson JE, Landry SJ. Influence of disulfide-stabilized structure on the specificity of helper T-cell and antibody responses to HIV envelope glycoprotein gp120. J Virol. 2010;84:3303–3311. doi: 10.1128/JVI.02242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry SJ. Local protein instability predictive of helper t-cell epitopes. Immunology Today. 1997;18:527–532. doi: 10.1016/s0167-5699(97)01152-3. [DOI] [PubMed] [Google Scholar]
- 19.Dai G, Steede NK, Landry SJ. Allocation of helper T-cell epitope immunodominance according to three-dimensional structure in the human immunodeficiency virus type I envelope glycoprotein gp120. J Biol Chem. 2001;276:41913–41920. doi: 10.1074/jbc.M106018200. [DOI] [PubMed] [Google Scholar]
- 20.Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, et al. Localization of cd4+ t cell epitope hotspots to exposed strands of hiv envelope glycoprotein suggests structural inuences on antigen processing. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown SA, Lockey TD, Slaughter C, Slobod KS, Surman S, et al. T cell epitope “hotspots” on the HIV Type 1 gp120 envelope protein overlap with tryptic fragments displayed by mass spectrometry. AIDS Res Hum Retroviruses. 21:165–170. doi: 10.1089/aid.2005.21.165. [DOI] [PubMed] [Google Scholar]
- 22.Carmicle S, Steede NK, Landry SJ. Antigen three-dimensional structure guides the processing and presentation of helper t-cell epitopes. Mol Immunol. 2006 doi: 10.1016/j.molimm.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Mutschlechner S, Egger M, Briza P, Wallner M, Lackner P, et al. Naturally processed t cell-activating peptides of the major birch pollen allergen. J Allergy Clin Immunol. 125:711–718. doi: 10.1016/j.jaci.2009.10.052. 718 e1-718 e2. [DOI] [PubMed] [Google Scholar]
- 24.Nanda NK, Bikoff EK. DM peptide-editing function leads to immunodominance in CD4 T cell responses in vivo. J Immunol. 2005;175:6473–6480. doi: 10.4049/jimmunol.175.10.6473. [DOI] [PubMed] [Google Scholar]
- 25.Liljedahl M, Winqvist O, Surh CD, Wong P, Ngo K, et al. Altered antigen presentation in mice lacking H2-O. Immunity. 1998;8:233–243. doi: 10.1016/s1074-7613(00)80475-6. [DOI] [PubMed] [Google Scholar]
- 26.Rammensee H1 ENBOSS Bachmann J. SYFPEITHI: database for mhc ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 27.Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human cd4+ t cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilser VJ, Freire E. Structure based calculation of the equilibrium folding pathway of proteins: Correlation with hydrogen exchange protection factors. J Mol Biol. 1996;262:756–772. doi: 10.1006/jmbi.1996.0550. [DOI] [PubMed] [Google Scholar]
- 29.Vertrees J, Barritt P, Whitten S, Hilser VJ. COREX/BEST server: a web browser-based program that calculates regional stability variations within protein structures. Bioinformatics. 2005;21:3318–3319. doi: 10.1093/bioinformatics/bti520. [DOI] [PubMed] [Google Scholar]
- 30.Hubbard SJ, Beynon RJ, Thornton JM. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures. Protein Eng. 1998;11:349–359. doi: 10.1093/protein/11.5.349. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard SJ, Eisenmenger F, Thornton JM. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 1994;3:757–768. doi: 10.1002/pro.5560030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck M, Boyd J, Redfield C, MacKenzie DA, Jeenes DJ, et al. Structural determinants of protein dynamics: analysis of 15N NMR relaxation measurements for main-chain and side-chain nuclei of hen egg white lysozyme. Biochemistry. 1995;34:4041–4055. doi: 10.1021/bi00012a023. [DOI] [PubMed] [Google Scholar]
- 33.Mirny LA, Shakhnovich EI. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 1999;291:177–196. doi: 10.1006/jmbi.1999.2911. [DOI] [PubMed] [Google Scholar]
- 34.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 35.Hilser VJ, B GME, Oas TG, Kapp G, Whitten ST. A statistical thermodynamic model of the protein ensemble. Chem Rev. 2006;106:1545–1558. doi: 10.1021/cr040423+. [DOI] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 38.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazor R, Tai CH, Lee B, Pastan I. Poor correlation between T-cell activation assays and HLA-DR binding prediction algorithms in an immunogenic fragment of Pseudomonas exotoxin A. J Immunol Methods. 2015 doi: 10.1016/j.jim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huygen K, Lozes E, Gilles B, Drowart A, Paliet K, et al. Mapping of TH1 helper T-cell epitopes on major secreted mycobacterial antigen 85A in mice infected with live Mycobacterium bovis BCG. Infect Immun. 1994;62:363–370. doi: 10.1128/iai.62.2.363-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romano M, Denis O, D'Souza S, Wang XM, Ottenhoff TH, et al. Induction of in vivo functional Db-restricted cytolytic T cell activity against a putative phosphate transport receptor of Mycobacterium tuberculosis. J Immunol. 2004;172:6913–6921. doi: 10.4049/jimmunol.172.11.6913. [DOI] [PubMed] [Google Scholar]
- 42.Cong Y, Bowdon HR, Elson CO. Identification of an immunodominant T cell epitope on cholera toxin. Eur J Immunol. 1996;26:2587–2594. doi: 10.1002/eji.1830261108. [DOI] [PubMed] [Google Scholar]
- 43.Messer RJ, Lavender KJ, Hasenkrug KJ. Mice of the resistant H-2(b) haplotype mount broad CD4(+) T cell responses against 9 distinct Friend virus epitopes. Virology. 2014;456-457:139–144. doi: 10.1016/j.virol.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King C, Garza EN, Mazor R, Linehan JL, Pastan I, et al. Removing T-cell epitopes with computational protein design. Proc Natl Acad Sci USA. 2014;111:8577–8582. doi: 10.1073/pnas.1321126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T, Steede NK, Nguyen HN, Freytag LC, McLachlan JB, et al. A comprehensive analysis of contributions from protein conformational stability and mhcii-peptide binding affinity to cd4+ epitope immunogenicity in hiv-1 envelope glycoprotein. J Virol. 2014 doi: 10.1128/JVI.00789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milich DR, McLachlan A, Moriarty A, Thornton GB. Immune response to hepatitis B virus core antigen (HBcAg): localization of T cell recognition sites within HBcAg/HBeAg. J Immunol. 1987;139:1223–1231. [PubMed] [Google Scholar]
- 47.Liu J, Ruckwardt TJ, Chen M, Johnson TR, Graham BS. Characterization of respiratory syncytial virus M- and M2-specific CD4 T cells in a murine model. J Virol. 2009;83:4934–4941. doi: 10.1128/JVI.02140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vordermeier HM, Harris DP, Lathigra R, Roman E, Moreno C, et al. Recognition of peptide epitopes of the 16,000 MW antigen of Mycobacterium tuberculosis by murine T cells. Immunology. 1993;80:6–12. [PMC free article] [PubMed] [Google Scholar]
- 49.Geginat G, Schenk S, Skoberne M, Goebel W, Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 2001;166:1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki M, Aoshi T, Nagata T, Koide Y. Identification of murine H2-Dd- and H2-Ab-restricted T-cell epitopes on a novel protective antigen, MPT51, of Mycobacterium tuberculosis. Infect Immun. 2004;72:3829–3837. doi: 10.1128/IAI.72.7.3829-3837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vordermeier HM, Harris DP, Moreno C, Singh M, Ivanyi J. The nature of the immunogen determines the specificity of antibodies and T cells to selected peptides of the 38 kDa mycobacterial antigen. Int Immunol. 1995;7:559–566. doi: 10.1093/intimm/7.4.559. [DOI] [PubMed] [Google Scholar]
- 52.Burkhart C, Freer G, Castro R, Adorini L, Wiesmuller KH, et al. Characterization of T-helper epitopes of the glycoprotein of vesicular stomatitis virus. J Virol. 1994;68:1573–1580. doi: 10.1128/jvi.68.3.1573-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brien JD, Uhrlaub JL, Nikolich-Zugich J. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol. 2008;181:8568–8575. doi: 10.4049/jimmunol.181.12.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology. 2002;296:117–124. doi: 10.1006/viro.2002.1432. [DOI] [PubMed] [Google Scholar]
- 55.Li W, Murthy AK, Lanka GK, Chetty SL, Yu JJ, et al. A T cell epitope-based vaccine protects against chlamydial infection in HLA-DR4 transgenic mice. Vaccine. 2013;31:5722–5728. doi: 10.1016/j.vaccine.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nascimento EJ, Mailliard RB, Khan AM, Sidney J, Sette A, et al. Identification of conserved and HLA promiscuous DENV3 T-cell epitopes. PLoS Negl Trop Dis. 2013;7:e2497. doi: 10.1371/journal.pntd.0002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frahm N, DeCamp AC, Friedrich DP, Carter DK, Defawe OD, et al. Human adenovirus-specific T cells modulate HIV-specific T cell responses to an Ad5-vectored HIV-1 vaccine. J Clin Invest. 2012;122:359–367. doi: 10.1172/JCI60202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roche PW, Peake PW, Billman-Jacobe H, Doran T, Britton WJ. T-cell determinants and antibody binding sites on the major mycobacterial secretory protein MPB59 of Mycobacterium bovis. Infect Immun. 1994;62:5319–5326. doi: 10.1128/iai.62.12.5319-5326.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jahn-Schmid B, Radakovics A, Luttkopf D, Scheurer S, Vieths S, et al. Bet v 1142-156 is the dominant T-cell epitope of the major birch pollen allergen and important for cross-reactivity with Bet v 1-related food allergens. J Allergy Clin Immunol. 2005;116:213–219. doi: 10.1016/j.jaci.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 60.Gerlach JT, Ulsenheimer A, Gruner NH, Jung MC, Schraut W, et al. Minimal T-cell-stimulatory sequences and spectrum of HLA restriction of immunodominant CD4+ T-cell epitopes within hepatitis C virus NS3 and NS4 proteins. J Virol. 2005;79:12425–12433. doi: 10.1128/JVI.79.19.12425-12433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams E, Britton W, Morgan A, Sergeantson S, Basten A. Individuals from different populations identify multiple and diverse T-cell determinants on mycobacterial HSP70. Scand J Immunol. 1994;39:588–596. doi: 10.1111/j.1365-3083.1994.tb03417.x. [DOI] [PubMed] [Google Scholar]
- 62.Graham S, Wang EC, Jenkins O, Borysiewicz LK. Analysis of the human T-cell response to picornaviruses: identification of T-cell epitopes close to B-cell epitopes in poliovirus. J Virol. 1993;67:1627–1637. doi: 10.1128/jvi.67.3.1627-1637.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwaiger J, Aberle JH, Stiasny K, Knapp B, Schreiner W, et al. Specificities of human CD4+ T cell responses to an inactivated avivirus vaccine and infection: correlation with structure and epitope prediction. J Virol. 2014;88:7828–7842. doi: 10.1128/JVI.00196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.James EA, Bui J, Berger D, Huston L, Roti M, et al. Tetramer-guided epitope mapping reveals broad, individualized repertoires of tetanus toxin-specific CD4+ T cells and suggests HLA-based differences in epitope recognition. Int Immunol. 2007;19:1291–1301. doi: 10.1093/intimm/dxm099. [DOI] [PubMed] [Google Scholar]
- 65.Bohle B, Zwolfer B, Fischer GF, Seppala U, Kinaciyan T, et al. Characterization of the human T cell response to antigen 5 from Vespula vulgaris (Ves v 5) Clin Exp Allergy. 2005;35:367–373. doi: 10.1111/j.1365-2222.2005.02180.x. [DOI] [PubMed] [Google Scholar]
- 66.Landry SJ. Three-dimensional structure determines the pattern of CD4+ T-cell epitope dominance in influenza virus hemagglutinin. J Virol. 2008;82:1238–1248. doi: 10.1128/JVI.02026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amria S, Hajiaghamohseni LM, Harbeson C, Zhao D, Goldstein O, et al. HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur J Immunol. 2008;38:1961–1970. doi: 10.1002/eji.200738100. [DOI] [PubMed] [Google Scholar]
- 68.Ma JK, Platt MY, Eastham-Anderson J, Shin JS, Mellman I. MHC class II distribution in dendritic cells and B cells is determined by ubiquitin chain length. Proc Natl Acad Sci USA. 2012;109:8820–8827. doi: 10.1073/pnas.1202977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.