Summary
By interacting with hundreds of protein partners, 14-3-3 proteins coordinate vital cellular processes. Phosphorylation of the small heat shock protein HSPB6 within its intrinsically disordered N-terminal domain activates its interaction with 14-3-3, ultimately triggering smooth muscle relaxation. After analyzing the binding of an HSPB6-derived phosphopeptide to 14-3-3 using isothermal calorimetry and X-ray crystallography, we have determined the crystal structure of the complete assembly consisting of the 14-3-3 dimer and full-length HSPB6 dimer and further characterized this complex in solution using fluorescence spectroscopy, small-angle X-ray scattering and limited proteolysis. We show that selected intrinsically disordered regions of HSPB6 are transformed into well-defined conformations upon the interaction, whereby an unexpectedly asymmetric structure is formed. This structure provides the first-ever atomic resolution snapshot of a human small HSP in functional state, explains how 14-3-3 proteins sequester their regulatory partners, and can inform the design of small-molecule interaction modifiers to be used as myorelaxants.
Keywords: 14-3-3 proteins, small heat shock proteins, intrinsically disordered regions, protein-protein interaction, smooth muscle relaxation, crystal structure, small angle X-ray scattering
Introduction
Although the ability of proteins to fold into a well-defined three-dimensional structure had been traditionally considered a prerequisite of their physiological activity, it was realized recently that functionally vital proteins often include regions lacking a defined three-dimensional structure. Rapidly growing evidence indicates that such intrinsically disordered regions (IDRs) play central roles in protein-protein interactions (Tompa et al., 2015), such as those mediated by the 14-3-3 proteins, key nodes of the eukaryotic signaling network (Pozuelo Rubio et al., 2004; Uhart and Bustos, 2013). These abundant proteins, amounting up to 1.3 % of the proteome mass in some tissues, are widely recognized as coordinators of multiple processes such as cell proliferation, transcription, hormonal production and apoptosis (Aitken, 2006; Pozuelo Rubio et al., 2004). Eukaryotes typically carry several 14-3-3 isoforms (seven in humans), which form homo- and heterodimers (Aitken, 2006). In cellulo, the 14-3-3 proteins interact with over 700 partners, including transcription factors, receptors, protein kinases, phosphatases, proapoptotic proteins and certain enzymes (Pozuelo Rubio et al., 2004; Uhart and Bustos, 2013).
An important partner of the 14-3-3 proteins is HSPB6 (also known as HSP20) (Chernik et al., 2007; Dreiza et al., 2005) belonging to the small heat shock proteins (sHSPs), a large group of ATP-independent cellular chaperones (Mymrikov et al., 2011). HSPB6 consists of a rigid α-crystallin domain (ACD) flanked by N- and C-terminal domains (NTD and CTD) that have a high degree of intrinsic disorder (Mymrikov et al., 2011; Weeks et al., 2014) (Fig.1a). Unlike most members of the sHSP family forming high-order oligomers, HSPB6 is stable in a dimeric state (Bukach et al., 2004; Sluchanko et al., 2011; Weeks et al., 2014). Interaction between the 14-3-3 proteins and HSPB6 (Chernik et al., 2007; Dreiza et al., 2005) is dependent on the phosphorylation of the latter by cyclic nucleotide-dependent protein kinases (Furnish et al., 2010; Komalavilas et al., 2008) at a characteristic motif centered at residue Ser16 (Dreiza et al., 2005; Yaffe et al., 1997). This motif, RRApSAP, follows the ‘canonical’ pattern for 14-3-3 binding, (R|K)X2-3(pS|pT)X(P|G), meaning that a positive charge is required 3-4 residues upstream, and a Pro or Gly residue two residues downstream of the phosphorylated Ser or Thr, while the X positions are not strictly defined (Yaffe et al., 1997). As a result, a stable 90 kDa heterotetramer consisting of the 14-3-3 and phosphorylated HSPB6 (pHSPB6) dimers is formed (Chernik et al., 2007; Sluchanko et al., 2011). The ζ isoform of human 14-3-3 was previously reported to bind pHSPB6 with micromolar affinity (Sluchanko et al., 2011). Being a major substrate for several protein kinases, HSPB6 was hypothesized to act as a phosphoswitchable displacer of other regulatory proteins from 14-3-3 complexes, which should affect the balance of the whole 14-3-3 interactome (Pozuelo Rubio et al., 2004; Uhart and Bustos, 2013). Interestingly, another abundant cellular 14-3-3 partner, vimentin, has also been proposed to sequester the 14-3-3 proteins and displace their other targets in vivo (Tzivion et al., 2000). In line with this model, controlling the 14-3-3/pHSPB6 interaction was shown to be crucial in a number of pathological conditions requiring smooth muscle relaxation, such as subarachnoid hemorrhage, asthma and hypertension (Furnish et al., 2010; Komalavilas et al., 2008). Indeed, an HSPB6-derived phosphopeptide AZX100® (Fig.1a) was able to partially reproduce the relaxation-promoting effect of the full-sized phosphorylated protein (Arno et al., 2014; Furnish et al., 2010; Lopes et al., 2009). It has been suggested that this effect may be due to HSPB6-induced displacement of phosphocofilin from the 14-3-3 complex, followed by cofilin activation by dephosphorylation and actin cytoskeleton remodeling (Dreiza et al., 2005). However, direct 14-3-3/phosphocofilin interaction could not be confirmed in vitro (Sudnitsyna et al., 2012). It is therefore likely that there are other binding partners of 14-3-3, such as phosphatases acting on cofilin in particular, which could be released from the 14-3-3 complex by pHSPB6, providing for an efficient control of cofilin activation (Sudnitsyna et al., 2012).
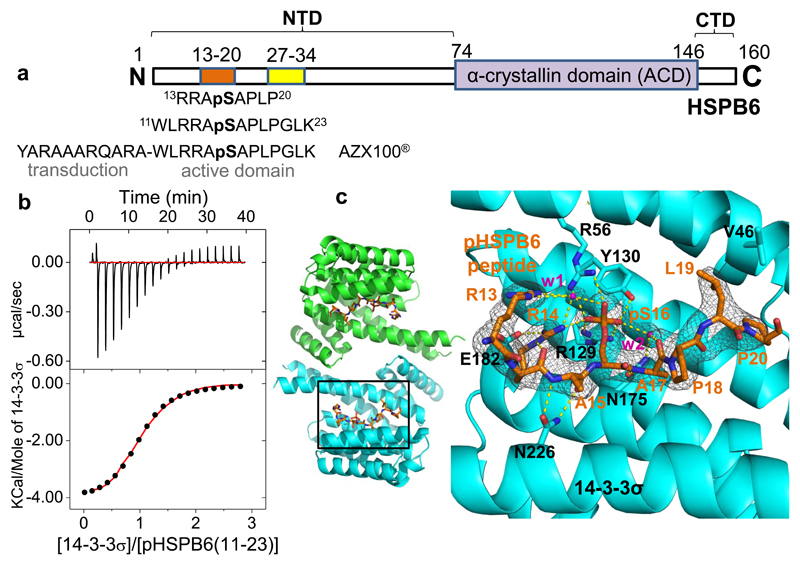
Figure 1. Interaction of 14-3-3σ with HSPB6 phosphopeptides.
a. A schematic diagram of HSPB6. Within its NTD, the 14-3-3 binding motif and the conserved motif 27RLFDQxFG34 are highlighted in orange and yellow, respectively. Below are the sequences of two phosphopeptides used in this study and of the therapeutic phosphopeptide AZX100. b. ITC data for the interaction of 14-3-3σ with the pHSPB6(11-23) phosphopeptide. c. Crystal structure of the 14-3-3σ dimer with the bound pHSPB6(11-23) (orange). A close-up to the right shows the 2Fo-Fc electron density map for the phosphopeptide (grey mesh, contoured at 1σ), ordered waters (w1, w2, magenta) and hydrogen bonds/salt bridges (dashed lines).
Here we have sought a detailed structural and biophysical insight into the interaction between phosphorylated HSPB6 and 14-3-3 proteins. These efforts have culminated in the determination of the atomic structure of the complete regulatory complex of these proteins, both in crystals and in solution. This resolves a long-standing challenge in the field, related to the difficulty of crystallizing proteins with prominent, functional IDRs. Indeed, no atomic structures of full-length mammalian sHSPs in either free or complex form could be determined until now. At the same time, atomic resolution data on the interaction of 14-3-3 with partners have been limited to a number of 14-3-3 complexes with short phosphopeptides as well as a single larger complex involving truncated arylalkylamine N-acetyltransferase in a non-natural stoichiometry (PDB entry 1IB1). The structural data obtained here shed light on the molecular mechanism of action of phosphorylated HSPB6 in the context of its competition with other regulatory partners of 14-3-3 proteins.
Results
Binding of HSPB6 phosphopeptides to the 14-3-3 proteins
To better understand the primary interaction between 14-3-3 and pHSPB6 at molecular level, we have employed two synthetic phosphopeptides which correspond to HSPB6 residues 13-20 and 11-23 respectively (Fig.1a). Importantly, the latter is equivalent to the active C-terminal moiety of the AZX100 peptide (Lopes et al., 2009). Isothermal titration calorimetry (ITC) experiments have shown that phosphopeptide 11-23 binds to 14-3-3γ the predominant isoform in muscle tissue (Horie et al., 1999), with an apparent KD of 2.3 ± 0.3 μM. A similar value was obtained with the human 14-3-3σ isoform (KD = 6.3 ± 0.5 μM; Fig.1b), and both complexes showed the expected stoichiometry of one peptide per 14-3-3 subunit. It should be noted that these interactions are stronger than, for instance, binding of phosphopeptides of C-RAF (KD=16.7 ± 2.0 μM with 14-3-3ζ (Molzan and Ottmann, 2012)) and p53 (KD =16.3 ± 0.7 μM with 14-3-3σ (Schumacher et al., 2010)). The strength of the primary interaction hints towards a highly stable complex between the full-length phosphorylated HSPB6 and 14-3-3, as indeed could be shown below.
We have also co-crystallized human 14-3-3σ with both phosphopeptides 13-20 and 11-23 and determined the atomic structures of the resulting complexes at 2.4 and 2.5Å resolution, respectively (Tables 1 and S1). The peptides bind in the characteristic groove of the 14-3-3 molecule in identical conformations (Fig.1c). Also for the longer peptide, only residues 13-20 were found ordered in the electron density map, suggesting that the upstream and downstream residues are mobile and not critically involved in the binding. The phosphate moiety forms salt bridges with the conserved 14-3-3σ residues Arg56 and Arg129 and is H-bonded to Tyr130 (Fig.1c). The main-chain atoms of the residues immediately adjacent to the phosphoserine, i.e. Ala15 and Ala17, are H-bonded to 14-3-3 residues Asn226 and Asn175, respectively. In addition, stabilizing interactions are observed within the phosphopeptide itself, as Arg14 directly coordinates the phosphate group of pS16 (and also forms a salt bridge with Glu182 of 14-3-3), while the residues Arg13 and Ala17 are each H-bonded to the phosphate via ordered water molecules. Finally, residues Leu19 and Pro20 of the peptide pack against a rather apolar side of the α3 helix of 14-3-3 near residue Val46 (Fig.1c).
Table 1. X-ray data collection and refinement statistics (See also Table S1).
| 14-3-3σ Clu3 (1-231) / pHSPB6 (13-20) peptide | 14-3-3σ WT (1-231) / pHSPB6 (11-23) peptide | 14-3-3σ Clu1 (1-231) / pHSPB6 (1-149) # | HSPB6 ACD / N-peptide | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | C2 | C2221 | P3122 |
| Cell dimensions: a, b, c (Å) | 55.6, 104.0, 123.1 | 90.1, 79.0, 75.4 | 125.1, 341.3,144.6 | 105.7, 105.7,112.0 |
| α, β, γ (°) | 90, 93.9, 90 | 90, 102.5, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution range (Å) 1 |
48 - 2.4 [48-11] (2.46-2.4) |
43 - 2.5 [43-11] (2.56-2.5) |
48 - 4.5 [48-20] (4.62-4.5) |
48 – 2.6 [48-12] (2.67-2.6) |
| Rmerge 2 | 0.099 [0.032] (1.53) |
0.150 [0.040] (1.82) |
0.314 [0.066] (2.16) |
0.088 [0.029] (2.68) |
| Rmeas | 0.116 [0.038] (1.79) |
0.175 [0.051] (2.12) |
0.337 [0.072] (2.23) |
0.093 [0.031] (2.81) |
| <I / σ> | 8.6 [25.9] (0.9) |
7.1 [22.4] (0.9) |
5.8 [22.2] (1.1) |
18.2 [63.5] (0.9) |
| CC1/2 | 1.00 (0.62) | 1.00 (0.60) | 0.99 (0.34) | 1.00 (0.51) |
| Completeness (%) | 99.7 (100) | 99.6 (100) | 99.6 (99.9) | 100 (100) |
| Redundancy | 3.7 (3.8) | 3.7 (3.8) | 7.9 (8.1) | 11.3 (11.3) |
| Refinement | ||||
| No. of reflections: total | 54511 | 17876 | 18722 | 22684 |
| ‘free’ set | 2718 | 1040 | 934 | 1097 |
| Rwork (%) 3 | 18.5 | 20.9 | 23.6 | 20.7 |
| Rfree (%) 3 | 21.4 | 25.0 | 25.6 | 22.7 |
| No. of 2:2 complexes /asu | 2 | 1 | 3 | 2.5 |
| No. of non-H atoms: protein/ion/water | 7409 /32 /361 | 3733 /0 /127 | 15845 /48 /0 | 3375 /20 /14 |
| Solvent content (%) | 62 | 48 | 60 | 67 |
| R.m.s.d. bond lengths (Å)/angles (°) | 0.010 / 1.6 | 0.010 / 1.2 | 0.008 / 1.1 | 0.010 / 1.3 |
| Ramachandran favoured/outlier (%) | 97.7 / 0.1 | 97.4 / 0.2 | 93.9 / 0.8 | 95.4 / 0.5 |
| Molprobity score / percentile | 1.32 / 99th | 1.55 / 99th | 2.23 / 100th | 2.12 / 95th |
| PDB code | 5LU1 | 5LU2 | 5LTW | 5LUM |
Statistics for the lowest and highest resolution shells are indicated in square brackets and parentheses, respectively
All merging statistics as defined in XSCALE (Kabsch, 2010)
As defined in Buster (Blanc et al., 2004).
Isomorphous crystals could be obtained for the 14-3-3σ Clu1 (1-231) complexes with either pHSPB6(1-149) truncation, full-length pHSPB6 (1-160), or the 7-153 truncation but in the two latter cases the diffraction never exceeded 7Å.
Interaction between the full-length pHSPB6 and 14-3-3
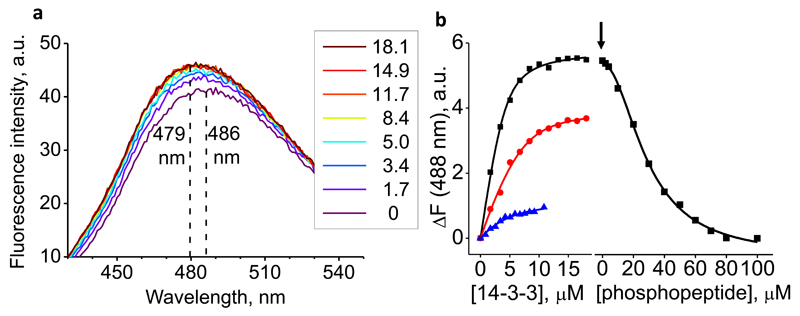
Next, we have analyzed the binding of phosphorylated HSPB6 to 14-3-3 in physiological solution, in which both proteins are predominantly present as dimers (Bukach et al., 2004; Chernik et al., 2007; Sluchanko et al., 2011). ITC measurements were done but revealed a complex thermal pattern which was difficult to interpret, supporting a multistage process of complex assembly (see Discussion). Instead, fluorescence spectroscopy has been used. To this end, phosphorylated full-length HSPB6 was stoichiometrically labeled at its unique Cys46 residue with an environmentallysensitive 1,5-I-AEDANS moiety. Upon titration with 14-3-3σ, the fluorescence peak of pHSPB6AEDANS shifted from 486 to 479 nm and its amplitude increased up to a saturation point (Fig.2a). Analysis of the resulting binding curve (Fig.S1) yielded an apparent KD of the 14-3-3σ/pHSPB6AEDANS complex at 0.56 ± 0.2 µM. This value suggests that the interaction of 14-3-3σ with intact pHSPB6 is significantly stronger than the interaction with the isolated HSPB6 phosphopeptide described above. In line with that, the following titration of the saturated 14-3-3σ/pHSPB6AEDANS complex with increasing amounts of the 11-23 phosphopeptide revealed a sigmoidal decay in the fluorescence signal with an IC50 of 28.9 ± 0.9 µM (Fig.2b), indicating that only excessive concentrations of the phosphopeptide could expel pHSPB6 from the complex. Moreover, titration of the AEDANS-labeled pHSPB6 by 14-3-3σ in the presence of equimolar concentration of the phosphopeptide and even a 7-fold excess of the latter also resulted in a saturable curve (Fig.2b), suggestive of a retained 14-3-3σ/pHSPB6AEDANS interaction. These competition experiments have also allowed an independent estimate of an apparent dissociation constant for the 14-3-3σ/phosphopeptide interaction at 5.2 ± 0.5 µM, in good agreement with our ITC data (Fig.1b).
Figure 2. Interaction of AEDANS-labeled phosphorylated HSPB6 with 14-3-3σ studied by fluorescence spectroscopy.
a. Changes in the AEDANS fluorescence spectrum (excitation at 336 nm) for 4.2 µM pHSPB6AEDANS solution upon titration with increasing concentrations of 14-3-3σ as indicated (in µM, colored lines). b. Changes in fluorescence intensity (ΔF) at 488 nm upon titration of 4.2 µM pHSPB6AEDANS solution by 14-3-3σ in the absence of the HSPB6 phosphopeptide 11-23, followed (at the time point indicated by an arrow) by titration with phosphopeptide. Alternatively, ΔF was measured for 4.2 µM (red line, circles) or 2.8 µM (blue line, triangles) pHSPB6AEDANS solution supplemented with 4.2 µM or 20 µM phosphopeptide, respectively, and titrated with 14-3-3σ. See also Fig. S1.
Crystal structure of the complete 14-3-3/pHSPB6 complex
The main challenge of this study was to determine the atomic structure of the regulatory complex formed by the full-length 14-3-3 and pHSPB6. To this end we have systematically attempted the crystallization of highly pure complexes formed by recombinant in vivo phosphorylated HSPB6 with each of the seven human 14-3-3 isoforms. This yielded reproducible crystals for the 14-3-3σ isoform complex only (Table S1), where the initial diffraction was limited to ~8Å resolution. At this point, we have designed two mutants of 14-3-3σ using the surface entropy reduction approach (Goldschmidt et al., 2007) (designated Clu1 and Clu3, each carrying a replacement of three consecutive residues to alanines, see Experimental Procedures) as well as HSPB6 variants with short terminal truncations. Using these mutants as well as scanning >100 crystals at a microfocus synchrotron radiation beamline eventually allowed us to improve the resolution to 4.5Å (Table 1) and determine the crystal structure of the full complex.
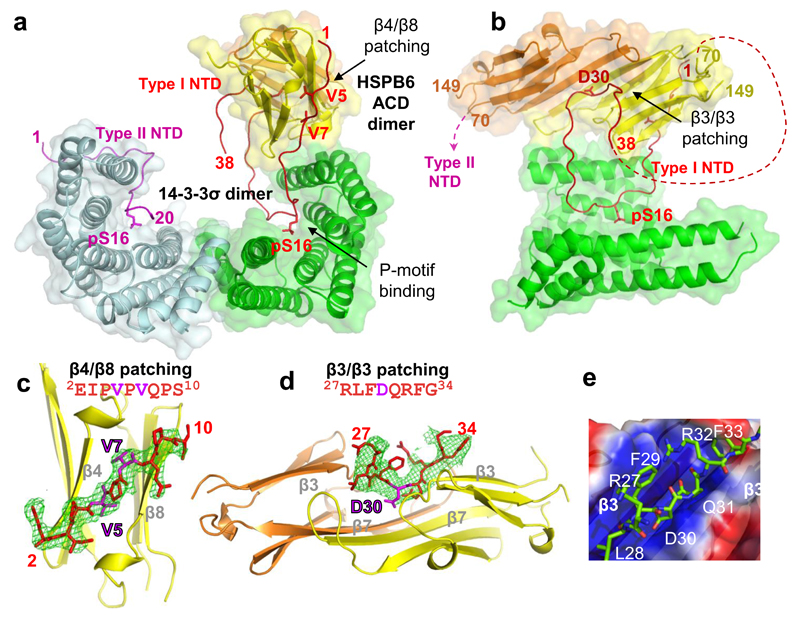
There are three 14-3-3σ/pHSPB6 complexes per asymmetric unit of the crystals. Despite differences in the crystal lattice contacts made by each of the complexes, the overall structure of the three is very similar (Fig.S2a). The binding of pHSPB6 does not cause any major conformational changes in the 14-3-3 dimer (Cα r.m.s.d. 0.90Å compared to the phosphopeptide-bound structure). Remarkably, while both phosphorylated N-terminal IDRs of HSPB6 interact with the binding grooves of the 14-3-3 dimer in the same way, the ACD dimer of HSPB6 attaches to only one 14-3-3 subunit resulting in an unexpected asymmetric structure (Figs.3a, b). The binding involves two C-terminal α-helices of the 14-3-3 molecule forming an antiparallel motif, and three surface loops (connecting the strands β3-β4, β5-β7 and β8-β9 respectively) of the nearest ACD. The binding interface features a salt bridge between Arg224 (14-3-3) and Glu86 (HSPB6) and masks about 400Å2, corresponding to 4.3% of the total accessible surface of the ACD dimer, suggesting that this interface is specific (see Fig.S2b for additional detail).
Figure 3. Crystal structure of the 14-3-3σ/pHSPB6 regulatory complex.
a, b. Ribbon diagram of the heterotetramer shown in two perpendicular views (second 14-3-3 molecule omitted in panel b for clarity). The ordered parts of the HSPB6 NTDs of type I and II are colored red and magenta, respectively. Important residues are labeled. The semi-transparent surfaces of 14-3-3 dimer and ACD dimer are also shown. Due to disordering of residues 39 to 69 there is ambiguity in the interconnection of the N-terminal parts and the ACDs: the NTD of type I could connect to the yellow-colored ACD and the NTD of type II to the orange-colored ACD (as indicated by dashed lines in panel b), or vice versa. c. β4/β8 groove patching observed in the co-crystals of the HSPB6 ACD with the N-terminal peptide 2EIPVPVQPS10. For clarity, only one ACD chain is shown. An Fo-Fc difference (omit) map for the peptide is shown at 2.6Å resolution, contoured at 2.5σ (green mesh). d. Patching of the shared β3/β3 groove of the ACD dimer by the conserved region of the NTD, as observed in the 14-3-3σ/pHSPB6 complex. A corresponding 4.5Å resolution difference (omit) map is shown at 2.5σ contour level (green mesh). e. A close-up of the shared groove patching. A semitransparent surface of the ACD dimer is colored by electrostatic potential from -1 (red) to +1 kT/e (blue). See also Fig. S2.
The structure of the 14-3-3σ/pHSPB6 complex reveals that the β-sandwich ACD of HSPB6 dimerizes via the APII type interface of the aligned β7 strands (Laganowsky et al., 2010), confirming a hypothesis that this is the preferred dimerization mode for mammalian ACDs (Baranova et al., 2011; Hochberg et al., 2014; Mymrikov et al., 2010). The ACD dimer features a relatively flat common β-sheet resulting from the β7-β7 alignment (Fig.3d), just like that found in the crystal structure of HSPB6 ΔN56 truncation (Weeks et al., 2014) (Cα r.m.s.d. 0.79Å after superimposing dimers), rather than a curved shape that had been suggested from solid-state NMR data for αB-crystallin (Jehle et al., 2010). In addition, our new structure shows that, in the full-length HSPB6, the ACD fold starts with strand β3 (residues 74-79), in contrast to HSPB1 and HSPB5 which seem to contain an additional short strand β2 (Hochberg et al., 2014).
Ordering of functional IDRs upon the complex formation
Importantly, the obtained 4.5Å electron density map of the 14-3-3σ/pHSPB6 complex reveals several functionally relevant regions of the HSPB6 NTD (residues 1-71). In each of three complexes found in the crystallographic asymmetric unit, residues 1 to 20 of both NTDs as well as residues 27 to 38 of one of the NTDs could be traced. In addition, in one of the six NTDs, residues 21 to 26 were additionally traced, so that the structure of the entire region encompassing residues 1 to 38 could be established.
First, both phosphoserine-containing regions of the HSPB6 dimer were found to bind in the corresponding 14-3-3 grooves (Fig.3a), in a conformation virtually identical to that observed in the 14-3-3/phosphopeptide 13-20 complex (Cα r.m.s.d. of 0.15Å for residues 14-19). The observed structure thus confirms the hypothesis that HSPB6 fully sequesters the 14-3-3 dimer making it unavailable for interaction with other phosphorylated partners.
Second, we could trace the very N-terminal residues 1-12 of both HSPB6 chains, which accommodate distinct conformations (designated as type I and II, see Figs.3a,b). Interestingly, we have found that the type I N-terminus ‘patches’ the β4/β8 groove on the side of the nearest ACD within one heterotetramer, whereas the type II chain engages in a similar patching across a crystal lattice contact (Figs.3a and S2). In each case, the residues V5 and V7 insert into the hydrophobic core of the ACD (Fig.3c). Moreover, we could obtain an independent insight into this phenomenon by co-crystallizing the isolated ACD domain of HSPB6 (residues 72 to 149) with an N-terminal peptide of HSPB6 which corresponds to residues 2 to 10 (EIPVPVQPS, N-peptide). The resulting crystal structure at 2.6Å resolution readily reveals the patching of the β4/β8 grooves at either side of the ACD dimer by well-ordered peptides (Fig.3c), fully confirming the patching mechanism observed in the 14-3-3σ/pHSPB6 complex (Fig.S2c). Previously, such patching of the β4/β8 grooves by hydrophobic residues has been observed mainly for the C-terminal IPI/V motif (Delbecq et al., 2012; Hochberg et al., 2014; Weeks et al., 2014), which is present in most human sHSPs but missing in HSPB6. Here we show that the unusual N-terminal 3IPVPV7 motif of HSPB6 (Weeks et al., 2014) is also capable of such patching, which could be the case in the free HSPB6 dimer as well.
Most interestingly, the crystal structure of the 14-3-3σ/pHSPB6 complex also reveals the RLFDQxFG motif (HSPB6 residues 27-34) which is the sole highly conserved region within the NTD of vertebrate sHSPs (Heirbaut et al., 2014). We have found that in the type I chain this motif becomes ordered upon its insertion into the distinct shared groove of the ACD dimer formed by the two β3 strands (Bagneris et al, 2009) (Figs.3b,d). Such an insertion is favored by the interaction of the negatively charged Asp30 residue and the positive potential at the floor of the shared groove (Fig.3e). The groove has room for only a single copy of the motif. Correspondingly, per heterotetramer, only one conserved NTD motif (in type I chain) was found in the bound state, while the second motif (in type II chain) remains disordered. The proximal half of the NTD (residues 39-69) is disordered in all chains, leaving an ambiguity in the interconnection of the N-terminal parts and the ACDs (see Fig.3b for details).
Solution data on the 14-3-3/pHSPB6 complex
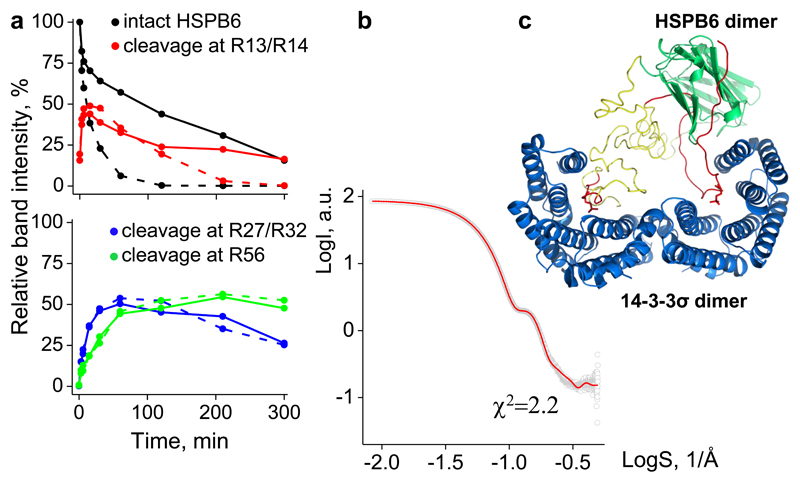
The tracing of the NTD within the 14-3-3/pHSPB6 complex could also be verified in solution using limited proteolysis. Free HSPB6 is most efficiently cleaved by trypsin at residues R13 and R14, but in the complex with 14-3-3γ this cleavage is considerably slower (Fig.4a and Fig.S3). This is in line with our structural data, as the residues are clearly protected once bound within the phosphomotif-binding groove (see Fig.3a). In contrast, the cleavage at residues R27 and R32 proceeds with nearly the same efficiency in both free and complexed pHSPB6, although at a yet slower rate than the cleavage at R13/R14 within the complex (Fig.4a). This implies that residues R27 and R32 are quite protected both in the free HSPB6 dimer and the complex.
Figure 4. Analysis of the 14-3-3/pHSPB6 complex in solution.
a. Time course of trypsinolysis of pHSPB6 alone (dashed lines) and in complex with 14-3-3γ (solid lines), showing the evolution of band intensities for intact pHSPB6 and its cleavage products at indicated sites. All band intensities are expressed as a percentage of the intact pHSPB6 band at zero time. See also Fig. S3. b. Solution SAXS curve for the complex (grey circles) overlaid by the calculated scattering from the full atomic model (red line). c. A cartoon of the full model, including the crystal structure (14-3-3σ dimer in blue, HSPB6 ACD dimer in green and the ordered parts of the NTD in red) supplemented with the flexible parts of the NTD (yellow). See also Figs. S4, S5.
Finally, we have used small angle X-ray scattering (SAXS) data from the 14-3-3σ/pHSPB6 complex in solution to model its complete molecular structure. A dimensionless Kratky plot of the scattering data (Fig.S4a) revealed a pronounced peak characteristic for globular proteins and no significant rise towards higher s values, suggesting that the overall 14-3-3σ/pHSPB6 complex is rather rigid. To model the complex, the obtained crystal structure has been supplemented with the missing parts of the two NTDs (11% of total residues in the complex) towards maximizing the fit between the experimental data and the SAXS curve calculated from the full-atom model. A model producing good fit could be achieved, with χ2 value of 2.2 for the full range of scattering data (Fig.4b, S4). Alternative models (see Fig. S5 and Experimental Procedures) failed to yield a satisfactory fit. These results indicate that the crystallographic structure is principally preserved in solution, whereby both NTDs, while being inherently dynamic, accommodate relatively compact conformations and reside within the open cavity formed by the interacting 14-3-3 and ACD dimers (Fig.4c).
Druggable pockets within the 14-3-3/pHSPB6 interface
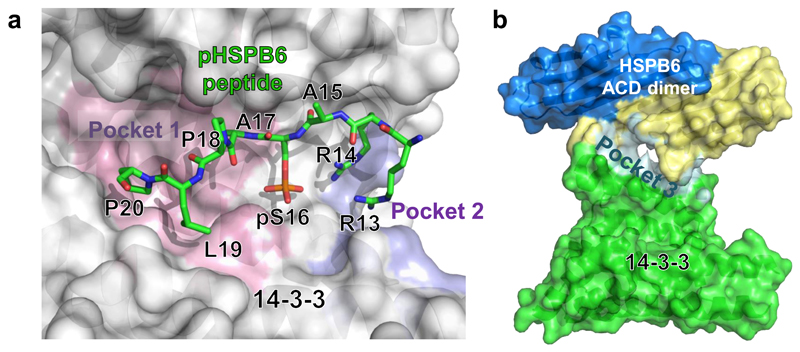
Computational analysis of the 14-3-3σ/pHSPB6 complex using the PockDrug method (Hussein et al., 2015) reveals two ‘druggable’ pockets within the 14-3-3 binding groove located in the vicinity of HSPB6 residues 13-14 and 17-20 respectively, as well as a third pocket within the relatively loose 14-3-3/ACD interface (Fig.5 and Table S2). These findings open a perspective of rational design of novel myorelaxant drugs. Firstly, our structural data enable the optimization of the small molecules that directly mimic the pHSPB6 binding to 14-3-3 (An et al., 2011) but are free of the limitations of the peptide-based drug candidates such as AZX100®. Secondly, it appears promising to develop compounds which would modulate the interaction of 14-3-3 with the naturally present pHSPB6, rather than completely block the primary 14-3-3 binding groove. The strength of this approach is its selectivity towards the specific binding partner, i.e. pHSPB6. Supporting such a possibility, a recently developed small-molecule stabilizer of the interaction between 14-3-3 and the Gab2 protein (Bier et al., 2016) was found to bind to our highest-scoring ‘pocket 1’ (Fig.5).
Figure 5. ‘Druggable’ pockets in the 14-3-3σ/pHSPB6 complex.
a. View of the 14-3-3σ groove (semi-transparent surface) which accommodates the pHSPB6(13-20) peptide (green sticks). Analysis using the PockDrug method (Hussein et al., 2015) reveals two druggable pockets at either end of the bound phosphopeptide (pockets 1 and 2 coloured pink and violet, respectively). b. Another prominent druggable pocket (pocket 3, cyan) is found at the 14-3-3σ/ACD interface. See also Table S2.
Discussion
Recent evidence indicates that IDRs play critical roles in protein-protein interactions, frequently acting via a phosphorylation-dependent mechanism (Bah et al., 2015; Lee et al., 2010; Tompa et al., 2015). The interaction between 14-3-3 protein and HSPB6, which is involved in the regulation of smooth muscle relaxation, is a prime example of this functional paradigm. We have shown here that the pHSPB6 dimer has a submicromolar apparent affinity to the 14-3-3 dimer, which exceeds the affinity of an isolated HSPB6 phophopeptide by roughly a factor of 10. This pronounced avidity is due to both simultaneous binding of two phosphomotifs and a direct interaction between one 14-3-3 subunit and the ACD dimer which we could observe in the full complex structure. Given high intracellular concentrations of HSPB6 (~1% of soluble protein in muscle (Kato et al., 1994)), its strong affinity for 14-3-3 demonstrated here implies that this sHSP, once phosphorylated, can indeed act a potent competitor for 14-3-3 binding in the cellular context (Dreiza et al., 2005; Sudnitsyna et al., 2012).
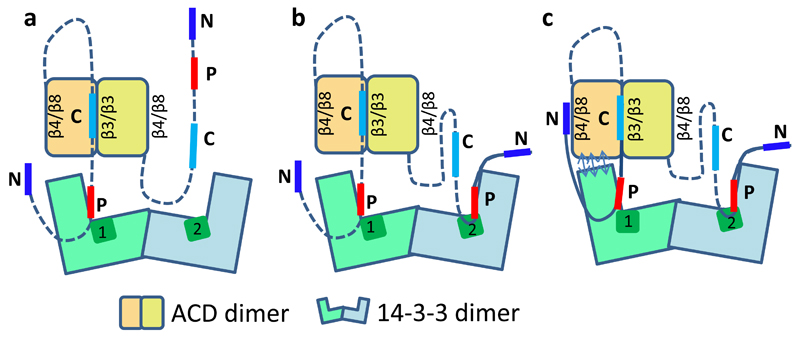
The data presented here provide for the understanding of the 14-3-3/pHSPB6 complex formation at molecular level. We predict that this process starts with the interaction of one phosphoserine-containing motif of the HSPB6 dimer with one of the binding grooves of the 14-3-3 dimer (Fig.6). This greatly facilitates the attachment of the second motif, resulting in a synergistic, doubly-anchored binding. As the result, the 14-3-3 dimer becomes fully engaged, making it unavailable for other phosphopeptide-dependent interactions, while the flexible NTDs are likely to still allow for a considerable freedom of the ACD dimer position. Thereafter the ACD dimer docks onto the two C-terminal α-helices of one 14-3-3 subunit, breaking the two-fold symmetry. Notably, simultaneous attachment of both ACDs to both 14-3-3 chains via the mentioned interface is not possible stereochemically (see Fig.3a). The newly formed tight complex is relatively rigid and facilitates further ordering of the NTDs of HSPB6, which are confined in a cavity formed by the folded domains of both proteins.
Figure 6. Proposed mechanism of the 14-3-3/pHSPB6 regulatory complex formation.
Key regions of the HSPB6 NTD are labeled as follows: N, the N-terminal region (residues 1-10); P, the 14-3-3-binding region (residues 13-20) including the phosphoserine 16; C, the ‘central’ conserved region (residues 27-34). One of the two central regions may occupy the shared β3/β3 groove of the ACD dimer already in the free state. a. The first phosphomotif of dimeric pHSPB6 binds in the groove of one 14-3-3 subunit. b. The second phosphomotif binds to the other 14-3-3 subunit. c.The ACD dimer docks onto the C-terminal lobe of the proximal 14-3-3 subunit (double-sided arrows), which induces ordering of additional N-terminal residues of the engaged HSPB6 chain (solid lines), resulting in a compact assembly. The second NTD is accommodated in the cavity formed and is likely to undergo partial ordering as well.
Moreover, the structural characterization of the 14-3-3/pHSPB6 complex gives an unprecedented snapshot of a mammalian sHSP. Indeed, within the complex we were able to trace 38 amino-acid residues at the N-terminus of human HSPB6, corresponding to more than half of the NTD. The latter domain, predicted to be largely disordered in this and other sHSPs while in the free form (Weeks et al., 2014), plays an essential role in both chaperoning function and the large oligomer formation observed for most sHSPs (Heirbaut et al., 2014; Mymrikov et al., 2011). Ordering of the NTDs within the 14-3-3/pHSPB6 complex follows the principle of induced folding, which appears to be a general feature observed upon the interaction of IDR-containing proteins with their partners (Wright and Dyson, 2009). As the result, the IDRs can act as ‘molecular rheostats’ adjusting their structure depending on a particular partner (Flock et al., 2014; Lee et al., 2010), a capacity that is likely to be relevant towards the chaperoning function of sHSPs as well. Along the same lines, the fact that the 14-3-3/HSPB6 complex deviates from the ‘natural’ two-fold symmetry implies that the NTDs of HSPB6 are able to adopt different conformations even upon the interaction with the same partner (the two equivalent subunits of 14-3-3), revealing an interesting case of the IDRs’ versatility. Such behavior appears to be a common theme in sHSPs, as the large oligomers of human αB-crystallin were recently shown to include two distinct conformations of its NTD (Mainz et al., 2015), while similar nonequivalence of the NTDs has also been observed in yeast (Hanazono et al., 2013) and plant sHSPs (van Montfort et al., 2001).
Of particular importance is the observed binding of the highly conserved N-terminal motif RLFDQxFG in the shared groove of the ACD dimer. We hypothesize that this interaction is an inherent feature contributing to the dimer stability in HSPB6 and most likely other mammalian sHSPs. This correlates with our findings on the impact of this conserved motif on the functional properties of HSPB6 (Heirbaut et al., 2014). Most interestingly, a class of drug-like molecules was recently shown to bind to the same shared groove in α-crystallins (Makley et al., 2015). Such binding results in α-crystallin stabilization and ultimately helps to prevent its aggregation, opening a perspective of eye cataract treatment.
Last but not least, the presence of IDRs in all mammalian sHSPs (Weeks et al., 2014) is the likely reason why none of them could ever be crystallized in their entirety. Fortuitously, the formation of the 14-3-3/pHSPB6 complex leads to partial ordering of the NTDs within the cavity made by the 14-3-3 dimer and the asymmetrically bound ACD dimer. This ordering is apparently just sufficient to enable the crystal formation, provided the 14-3-3σ isoform is being used, allowing us to resolve this long-standing enigma.
Experimental Procedures
Synthetic peptides
HSPB6 phosphopeptides and the N-terminal peptide (residues 2 to 10), all without terminal modifications and at >98% purity, were obtained from Severn Biotech (UK) and Proteogenix (France) respectively. Fmoc chemistry and C18 reverse phase HPLC were used.
Cloning, expression and purification of pHSPB6 constructs
The pET23 vector-based expression construct for the full-length (residues 1-160) wild-type untagged HSPB6 was described before (Bukach et al., 2004). In addition, we have used the pETHSUL vector (Weeks et al., 2007) to overexpress the ACD of HSPB6 (residues 72 to 149) as well as its truncated variant (residues 1-149) lacking the CTD. Previously the CTD was found largely disordered in the crystal structure of N-terminally truncated HSPB6 (ΔN56) (Weeks et al., 2014). Finally, another HSPB6 construct including small truncations at both N- and C-termini (residues 7-153) was cloned into the pET23 vector.
Phosphorylated HSPB6 constructs were obtained in E. coli by co-expression with protein kinase A (PKA). To this end, His-tagged catalytically active subunit of mouse PKA was cloned into pACYC vector (chloramphenicol resistance) using NcoI and BamHI restriction sites. E.coli C41(DE3) competent cells were simultaneously transformed with the HSPB6 plasmid (ampicillin resistance) and the latter PKA plasmid under selection on both antibiotics. Selected clones were used for expression in 2 l of 3-fold Luria-Bertani media supplied with the two antibiotics. After 8 h shaking at 37 ºC, 0.3 mM isopropyl-β-D-thiogalactoside was added to enhance protein expression for another 20-24 h at 30 ºC.
The untagged full-length pHSPB6 and the 7-153 truncation were purified using ammonium sulfate fractionation and ion-exchange and hydrophobic interaction chromatography as described earlier (Bukach et al., 2004; Mymrikov et al., 2010; Sluchanko et al., 2011). The His-tagged SUMO fusion for pHSPB6(1-149) was purified using subtractive immobilized metal-affinity chromatography (IMAC) and gel-filtration essentially as described (Weeks et al., 2014). During this procedure, the fusion was cleaved with SUMO hydrolase resulting in a target protein without exogenous residues. The obtained samples were fully phosphorylated at a single residue Ser16 as verified by fragmentation of 1429.7 Da phosphopeptide using tandem mass-spectrometry on a MALDI TOF/TOF ultrafleXtreme mass-spectrometer (Bruker, Germany). Purification of the (unphosphorylated) HSPB6 ACD construct followed the same procedure as for pHSPB6(1-149).
Isolation of 14-3-3 constructs
Cloning, overexpression and purification of the full-length human 14-3-3γ and 14-3-3ζ were described previously (Chernik et al., 2007; Sluchanko et al., 2011). In addition, expression plasmids for human 14-3-3 isoforms with truncation of a short disordered region at the C-terminus were kindly provided by the laboratory of Christian Ottmann (Eindhoven University). These plasmids were used to express the following fragments: 14-3-3σ (1-231), 14-3-3τ (1-230), 14-3-3ε (1-232), 14-3-3β (1-232) and 14-3-3η (1-235). The expression constructs contained an N-terminal His-tag that was cleaved off at a tobacco etch virus protease site during IMAC-based purification, leaving exogenous residues GAMGS at the N-terminus of the final product.
In an attempt to improve crystallization, mutations in 14-3-3σ protein were designed following the surface entropy reduction (SER) approach (Goldschmidt et al., 2007) using an online server at http://www.doe-mbi.ucla.edu/Services/SER. The following clusters of triple mutations were used: 159KKE161 → AAA, designated Clu1 (top SERp score = 6.17) and 75EEK77 → AAA, designated Clu3 (SERp score = 4.73). The mutants were prepared by site-directed mutagenesis of the C-terminally truncated 14-3-3σ protein (1-231), and cloned into a modified pET28 vector. The vector contains the N-terminal His-tag followed by a 3C protease cleavage site. After subtractive IMAC purification and cleavage, the final products retained three exogenous residues GPH at their N-terminus. The triple mutants Clu1 and Clu3 have indeed helped to crystallize the complexes of 14-3-3σ(1-231) with pHSPB6(1-149) and the pHSPB6(13-20) peptide, respectively (see Fig. S2b).
All final protein samples appeared as a single band on a Coomassie-stained SDS-PAGE. The pure proteins were dialyzed against 20 mM Tris-HCl buffer (pH 7.6) containing 150 mM NaCl, 0.1 mM EDTA, 15 mM ME or 2 mM DTT, 0.05 mM PMSF, concentrated on 10 kDa cutoff Amicon concentrators (Millipore) and stored frozen at -20 ºC. Protein concentration was determined spectrophotometrically at 280 nm.
Isothermal titration calorimetry
The binding of the pHSPB6(11-23) phosphopeptide to either 14-3-3γ or 14-3-3σ was analyzed using a MicroCal iTC200 System (GE Healthcare Life Sciences, USA) at 30 °C in a 20 mM Tris-HCl buffer (pH 7.6) containing 150 mM NaCl, 0.1 mM EDTA and 2 mM β-mercaptoethanol. 2 μl aliquots of the 14-3-3 protein were injected into the 0.2 ml cell containing the phosphopeptide to obtain a complete binding isotherm. Concentration of pHSPB6(11-23) in the cell was either 44 μM (titrated by 0.5 mM 14-3-3γ) or 100 μM (titrated by 1 mM 14-3-3σ). Measurements have been repeated at least three times with a variation of protein and peptide concentrations. The obtained data were corrected for heat of dilution measured by injecting 14-3-3 into a buffer solution. Titration curves were fitted using MicroCal Origin 9.0 software.
Fluorescence spectroscopy
Full-length HSPB6 was phosphorylated by PKA in vitro (Sluchanko et al., 2011) and stoichiometric phosphorylation at the Ser16 residue (Beall et al., 1999) was confirmed by native gel electrophoresis. Thereafter the pHSPB6 sample was buffer-exchanged into 20 mM HEPES buffer (pH 7.5) containing 100 mM NaCl. To reduce the SH groups, 20 mM DTT was added for 20 min at room temperature and then its excess was removed by using NAP-5 (Illustra, GE Healthcare) desalting columns. Immediately afterwards, a 6-fold molar excess of fresh aqueous solution of 5-[2-(Iodoacetamido)ethylamino]naphthalene-1-sulfonic acid (1,5-I-AEDANS) was added and incubated for 1h at room temperature and then overnight at 4 °C. After removal of the unbound label using an ultraconcentration device (Millipore) and subsequently a NAP-5 column, the extent of labeling was estimated spectrophotometrically at 336 nm using extinction coefficient of AEDANS of 5700 M-1 cm-1, and reached 90%. AEDANS fluorescence was followed using a Cary Eclipse spectrofluorometer (Varian Inc.) equipped with a thermocontrolled multicell holder. Titration of AEDANS-labeled phosphorylated HSPB6 by human 14-3-3σ (1-231), added in 1-2 µl portions, mixed by pipetting and allowed to equilibrate for 5 min, was performed in quartz cuvettes (600 µl) at 30°C. The experiments were also done in the presence of the HSPB6 11-23 phosphopeptide. The peptide contains one native tryptophan residue (Trp11), enabling an accurate determination of its concentration by absorbance at 280 nm. Titration curves were buffer-corrected and then fitted using MicroCal Origin 9.0 software by either a classical quadratic equation (in the absence of phosphopeptide) or, in a competition assay, by a cubic equation according to the method by Wang (Wang, 1995), to estimate the apparent binding constant.
Crystallization
Initial search for crystallization conditions was done using commercial screens including PACT, Procomplex (Qiagen), Wizard I+II (Emerald BioStructures), Index (Hampton Research) and JCSG+ (Molecular Dimensions). Sitting drops containing 0.2 μl protein at above 10 mg/ml concentration (See Table S1) and 0.2 μl precipitant solution were set up in 96-well plates using a Mosquito robot (TTL Labtec). The crystallizations were incubated at either 4 or 20 ºC and monitored using a RockImager 1000 (Formulatrix). Crystallization optimization was performed manually in 1-2 μl sitting drops.
To form the 14-3-3/phosphopeptide complexes prior to crystallization, an 14-3-3 isoform (in 50 mM Tris buffer, pH 7.5, containing 150 mM NaCl, 0.1 mM EDTA, 2 mM MgCl2 and 2 mM DTT) was supplemented with 2 to 6-fold molar excess of the pHSPB6 (13-20) or (11-23) phosphopeptide.
The complexes of 14-3-3 proteins and phosphorylated HSPB6 variants were prepared and purified using size-exclusion chromatography (SEC) immediately before crystallization essentially as described in (Chernik et al., 2007). Briefly, the individual proteins were mixed at 1.2-1.5 molar excess of pHSPB6 and incubated for 15-20 min at 30-37 °C. The sample was then concentrated to above 10 mg/ml and loaded on a Superdex 200 column (120 ml) equilibrated with 50 mM HEPES buffer (pH 7-7.5), 150 mM NaCl, 0.1 mM EDTA and 2 mM DTT. The main peak corresponding to the stable 2:2 complex was concentrated and used for crystallization. Different combinations of 14-3-3 isoforms and truncation or deletion constructs of phosphorylated HSPB6 were tested. All 7 isoforms of 14-3-3 (β, γ, ζ, ε, σ, η, τ) formed stable complexes with all pHSPB6 constructs. However, no crystals could be obtained unless the 14-3-3σ isoform was used. The presence of both partners in the crystals was confirmed by SDS-PAGE. After optimization, macrocrystals could be reproducibly grown within several days for the complex of 14-3-3σ(1-231) with each of the following pHSPB6 variants: full-length (1-160), 1-149 and 7-153. The exact pHSPB6 construct was not significant. Two crystal forms both diffracting to 6-8Å resolution were obtained: prisms in the space group P6x22 and plates in the space group C2221. Using the above-described Clu1 and Clu3 triple SER mutants of 14-3-3σ(1-231) for complex formation did not result in any radical changes in the crystals’ appearance, but however somewhat improved their diffraction quality.
Finally, the N-terminal peptide of HSPB6 in powder form was added to the HSPB6 ACD solution in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl to achieve about ~3x molar excess of the peptide prior to crystallization.
Optimized crystallization conditions for all complexes are given in Table S1.
X-ray data collection
Diffraction data (Table 1) were collected at 100K using 0.98Å radiation at Proxima 2a beamline of Synchrotron Soleil (France) by means of ADSC Quantum 315 and Eiger 9M dectors. Crystals were mounted on nylon loops and flash-cooled in liquid nitrogen. For all crystals, the precipitant solution was supplemented with 15-20% ethylene glycol for cryoprotection. For the 14-3-3/pHSPB6 complexes, multiple data collection sessions were alternated by further attempts to improve crystal quality, including additives, growth temperature variation, seeding, dehydration, alternative cryoprotectants and crosslinking, with limited success. The best data were eventually obtained using 14-3-3σ Clu1 (1-231)/pHSPB6(1-149) plates with a size of about 150x100x30 μm3. Screening many similar crystals, as well as the use of the helical data collection protocol at the microfocus beamline were necessary.
Crystal structure solution and refinement
Diffraction data integration and scaling were performed using XDS/Xscale (Kabsch, 2010) and Xdsme (Legrand, 2009). Phasing of the 14-3-3σ/peptide complexes was done by molecular replacement in MolRep (Vagin and Teplyakov, 2010) using a 14-3-3σ dimer from the PDB entry 3IQJ as a search model. The HSPB6 ACD / N-peptide complex was also phased by molecular replacement using the ACD extracted from the crystal structure of HSPB6 ΔN56 fragment (PDB entry 4JUT). Automated refinement was carried out with Buster (Blanc et al., 2004). Manual model correction was done in Coot (Emsley and Cowtan, 2004). The quality statistics of the final models is given in Table 1. Phasing of the 14-3-3σ Clu1 (1-231)/pHSPB6 (1-149) complex was done in MolRep using a dimeric 14-3-3σ/pHSPB6(13-20) peptide complex as a search model, initially positioning three of them. Thereafter a MolRep real-space search (i.e. in a map calculated with the available phases) using an HSPB6 ACD dimer from PDB entry 4JUT could position three such dimers. The missing N-terminal regions of HSPB6 were added in Coot on the basis of difference electron density maps. Subsequent refinement in Buster included a rigid-body positional refinement of all chains, followed by an all-atom refinement with additional restraints provided by the non-crystallographic symmetry as well as by the ‘target’ high-resolution structures. To this end, the refined 14-3-3σ/pHSPB6(13-20) complex (2.4Å resolution) and the HSPB6 ACD / N-peptide complex (2.6Å resolution) were used. Only the overall temperature factors, together with the TLS parameters per each domain, were refined. After the structure of the 14-3-3σ Clu1 (1-231) / pHSPB6 (1-149) complex had been established, using rigid-body refinement against the data for both the 14-3-3σ Clu1 (1-231) / full-length pHSPB6 and the 14-3-3σ Clu1 (1-231) / pHSPB6 (7-153) complexes we could confirm the same overall structure, but no further refinement was carried out due to limited resolution (<7Å) of the data. All structural figures were prepared using Pymol 1.6.9 (Schrödinger), which included calculations of the electrostatic potential using adaptive Poisson-Boltzmann solver plug-in for Pymol (Baker et al., 2001). Atomic coordinates and structure factors have been deposited to PDB under the assession codes indicated in Table 1.
Limited proteolysis
Purified pHSPB6 (1 mg/ml), its mixture with 1.2 molar excess of untagged 14-3-3γ, or the same amount of 14-3-3γ alone were pre-incubated for 30 min at 37 °C. Thereafter tosyl phenylalanyl chloromethyl ketone (TPCK) treated trypsin was added at protease:pHSPB6 ratio of 1:1500. The cleavage was at 37 °C in 20 mM Tris-HCl buffer, pH 7.6, containing 150 mM NaCl, 0.1 mM EDTA and 15 mM ME. At various time intervals the reaction was stopped by addition of the SDS-PAGE sample buffer containing 2.5 mM PMSF as quencher. The samples were boiled and analyzed on SDS-PAGE (12-20% polyacrylamide), and additionally desalted and analyzed using MALDI TOF/TOF ultrafleXtreme mass-spectrometer (Bruker, Germany). Quantitative densitometric analysis of the bands was performed using ImageJ 1.48. The bands resulting from the cleavage at residues R27 and R32, although discernible on the gels (Fig. S3), were integrated together in Fig.4a.
SAXS data collection and processing
SAXS data for the SEC-purified 14-3-3σ (1-231)/pHSPB6 (1-149) complex at different concentrations (1-9 mg/ml) in 50 mM TRIS buffer (pH 7.5) with 150 mM NaCl, 0.1 mM EDTA and 2 mM DTT were recorded for the momentum transfer range 0 < s < 0.5 Å-1, where s = 4π sin Θ/λ, Θ being the scattering angle. The data were collected at 283 K using radiation with λ = 1.2Å at the BM29 beamline (ESRF, Grenoble) equipped with a Pilatus 1M detector (Dectris, Baden, Switzerland). No significant radiation damage was detected. Even though the dependence of the concentration-normalized scattering profiles on sample concentration was only minor, the data were taken to produce the infinite dilution-extrapolated curve using the program Almerge from the ATSAS 2.6.0 package (Petoukhov et al., 2012). To create a complete atomic model of the complex, the crystal structure was first supplemented by a Cα-trace of the missing parts of the NTD using multiple runs of the program Coral (Petoukhov et al., 2012) towards an optimal fit to the SAXS data in a range of 0 < s < 0.3 Å-1. This involved the residues 39-69 in the type I chain and residues 1-12 and 21-69 in the type II chain, totalling to ~11% of the complex mass. The resulting models were validated against the SAXS curve for the full range of scattering data using Crysol (Petoukhov et al., 2012). This procedure showed a good convergence, as a cluster of obtained models yielded a good match to the experimental data, with the χ2 ranging between 2.3 and 3. 8 (Figs.S4b,c), and the model radius of gyration Rg between 30.6 and 30.7Å. Two ways of interconnecting the ordered N-terminal parts and the two ACDs of the crystallographic complex have been tried and yielded comparably good-fitting models. Thereafter the BBQ script of Bioshell package (Gront and Kolinski, 2006) and the program Scwrl 4.0 (Krivov et al., 2009) were used to convert the Cα-trace of the best-fitting Coral-derived model to a full-atom model, resulting in final χ2 = 2.2 (Fig. 4b). In addition, we have noticed that the size of the ACD dimer matches the cavity formed by the 14-3-3 dimer, and used a rigid-body docking to construct an alternative symmetric model of the complex (Fig. S5). However, even after modeling of the missing parts of the NTD using Coral this alternative model failed to produce any reasonable fit to the experimental SAXS data (χ2 = 169).
Supplementary Material
Highlights.
X-ray structure of the 14-3-3/pHSPB6 heterotetrameric complex was obtained
N-terminal domain of HSPB6 becomes partially ordered within the complex
Synergistic binding explains effective sequestering of 14-3-3 by pHSPB6
First-ever atomic resolution snapshot of a human small HSP in a functional state
Acknowledgments
We are grateful to Seppe Leysen and Christian Ottmann (Eindhoven University of Technology, The Netherlands) for expression plasmids and initial protein samples of some 14-3-3 isoforms, to Alexander Shkumatov for help with protein purification and SAXS measurements, and to Kristina V. Tugaeva for help with fluorescence labeling. We also thank William Shepard and colleagues at Synchrotron Soleil, France, and the staff of the BM29 beamline, European Synchrotron Radiation Facility, France, for assistance with X-ray data collection, and Sandra Greive for critical reading of the manuscript. N.N.S. acknowledges the short-term fellowships from FEBS (in 2012) and EMBO (ASTF#637-2014). This research was further supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement N°283570), by grants from Russian Foundation for Basic Research 14-04-00146 to N.N.S. and 16-04-00016 to N.B.G., by the Program “Molecular and cell biology” of the Russian Academy of Sciences to N.N.S., the Wellcome Trust grant 098230 to A.A.A., the KULeuven grant OT13/097 to S.V.S., and the Research Foundation Flanders (FWO) grants G069708N, G093615N and WO03315N to S.V.S.
Footnotes
Author Contributions. N.N.S., N.B.G. and S.V.S. conceived the project. N.N.S., S.B. and S.V.S. prepared and crystallized protein complexes. N.N.S., S.D.W. and S.V.S. collected and processed diffraction data including SAXS. A.A.K. performed ITC experiments. N.N.S. performed fluorescence assays. N.N.S., S.D.W., A.A.A., N.B.G. and S.V.S. analyzed the structures. All authors discussed the results. N.N.S. and S.V.S. wrote the manuscript with input from S.D.W., A.A.A. and N.B.G.
Authors declare no conflict of interest.
References
- Aitken A. 14-3-3 proteins: a historic overview. Semin Canc Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- An SS, Askovich PS, Zarembinski TI, Ahn K, Peltier JM, von Rechenberg M, Sahasrabudhe S, Fredberg JJ. A novel small molecule target in human airway smooth muscle for potential treatment of obstructive lung diseases: a staged high-throughput biophysical screening. Respir Res. 2011;12:8. doi: 10.1186/1465-9921-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arno AI, Gauglitz GG, Barret JP, Jeschke MG. New molecular medicine-based scar management strategies. Burns : journal of the International Society for Burn Injuries. 2014;40:539–551. doi: 10.1016/j.burns.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, Slingsby C. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392:1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-dimensional structure of alpha-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol. 2011;411:110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Beall A, Bagwell D, Woodrum D, Stoming TA, Kato K, Suzuki A, Rasmussen H, Brophy CM. The small heat shock-related protein, HSP20, is phosphorylated on serine 16 during cyclic nucleotide-dependent relaxation. J Biol Chem. 1999;274:11344–11351. doi: 10.1074/jbc.274.16.11344. [DOI] [PubMed] [Google Scholar]
- Bier D, Bartel M, Sies K, Halbach S, Higuchi Y, Haranosono Y, Brummer T, Kato N, Ottmann C. Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface. ChemMedChem. 2016;11:911–918. doi: 10.1002/cmdc.201500484. [DOI] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Bukach OV, Seit-Nebi AS, Marston SB, Gusev NB. Some properties of human small heat shock protein Hsp20 (HspB6) Eur J Biochem. 2004;271:291–302. doi: 10.1046/j.1432-1033.2003.03928.x. [DOI] [PubMed] [Google Scholar]
- Chernik I, Seit-Nebi A, Marston S, Gusev N. Small heat shock protein Hsp20 (HspB6) as a partner of 14-3-3gamma. Mol Cell Biochem. 2007;295:9–17. doi: 10.1007/s11010-006-9266-8. [DOI] [PubMed] [Google Scholar]
- Delbecq SP, Jehle S, Klevit R. Binding determinants of the small heat shock protein, alphaB-crystallin: recognition of the 'IxI' motif. EMBO J. 2012;31:4587–4594. doi: 10.1038/emboj.2012.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiza CM, Brophy CM, Komalavilas P, Furnish EJ, Joshi L, Pallero MA, Murphy-Ullrich JE, von Rechenberg M, Ho YS, Richardson B, et al. Transducible heat shock protein 20 (HSP20) phosphopeptide alters cytoskeletal dynamics. FASEB J. 2005;19:261–263. doi: 10.1096/fj.04-2911fje. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Furnish EJ, Brophy CM, Harris VA, Macomson S, Winger J, Head GA, Shaver EG. Treatment with transducible phosphopeptide analogues of the small heat shock-related protein, HSP20, after experimental subarachnoid hemorrhage: prevention and reversal of delayed decreases in cerebral perfusion. J Neurosurg. 2010;112:631–639. doi: 10.3171/2009.7.JNS09730. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gront D, Kolinski A. BioShell--a package of tools for structural biology computations. Bioinformatics. 2006;22:621–622. doi: 10.1093/bioinformatics/btk037. [DOI] [PubMed] [Google Scholar]
- Hanazono Y, Takeda K, Oka T, Abe T, Tomonari T, Akiyama N, Aikawa Y, Yohda M, Miki K. Nonequivalence observed for the 16-meric structure of a small heat shock protein, SpHsp16.0, from Schizosaccharomyces pombe. Structure. 2013;21:220–228. doi: 10.1016/j.str.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Heirbaut M, Beelen S, Strelkov SV, Weeks SD. Dissecting the functional role of the N-terminal domain of the human small heat shock protein HSPB6. PLoS One. 2014;9:e105892. doi: 10.1371/journal.pone.0105892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg GK, Ecroyd H, Liu C, Cox D, Cascio D, Sawaya MR, Collier MP, Stroud J, Carver JA, Baldwin AJ, et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci U S A. 2014;111:E1562–1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, Suzuki M, Takahashi E, Tanigami A. Cloning, expression, and chromosomal mapping of the human 14-3-3gamma gene (YWHAG) to 7q11.23. Genomics. 1999;60:241–243. doi: 10.1006/geno.1999.5887. [DOI] [PubMed] [Google Scholar]
- Hussein HA, Borrel A, Geneix C, Petitjean M, Regad L, Camproux AC. PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins. Nucleic Acids Res. 2015;43:W436–442. doi: 10.1093/nar/gkv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, Rajagopal P, Bardiaux B, Markovic S, Kuhne R, Stout JR, Higman VA, Klevit RE, van Rossum BJ, Oschkinat H. Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers. Nat Struct Mol Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Goto S, Inaguma Y, Hasegawa K, Morishita R, Asano T. Purification and characterization of a 20-kDa protein that is highly homologous to alpha B crystallin. J Biol Chem. 1994;269:15302–15309. [PubMed] [Google Scholar]
- Komalavilas P, Penn RB, Flynn CR, Thresher J, Lopes LB, Furnish EJ, Guo M, Pallero MA, Murphy-Ullrich JE, Brophy CM. The small heat shock-related protein, HSP20, is a cAMP-dependent protein kinase substrate that is involved in airway smooth muscle relaxation. Am J Physiol Lung Cell Mol Physiol. 2008;294:L69–78. doi: 10.1152/ajplung.00235.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Benesch JL, Landau M, Ding L, Sawaya MR, Cascio D, Huang Q, Robinson CV, Horwitz J, Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci U S A. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand P. xdsme. 2009 https://code.google.com/archive/p/xdsme/ed.
- Lopes LB, Furnish EJ, Komalavilas P, Flynn CR, Ashby P, Hansen A, Ly DP, Yang GP, Longaker MT, Panitch A, et al. Cell permeant peptide analogues of the small heat shock protein, HSP20, reduce TGF-beta1-induced CTGF expression in keloid fibroblasts. J Invest Dermatol. 2009;129:590–598. doi: 10.1038/jid.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainz A, Peschek J, Stavropoulou M, Back KC, Bardiaux B, Asami S, Prade E, Peters C, Weinkauf S, Buchner J, et al. The chaperone alphaB-crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat Struct Mol Biol. 2015;22:898–905. doi: 10.1038/nsmb.3108. [DOI] [PubMed] [Google Scholar]
- Makley LN, McMenimen KA, DeVree BT, Goldman JW, McGlasson BN, Rajagopal P, Dunyak BM, McQuade TJ, Thompson AD, Sunahara R, et al. Pharmacological chaperone for alpha-crystallin partially restores transparency in cataract models. Science. 2015;350:674–677. doi: 10.1126/science.aac9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzan M, Ottmann C. Synergistic binding of the phosphorylated S233- and S259-binding sites of C-RAF to one 14-3-3zeta dimer. J Mol Biol. 2012;423:486–495. doi: 10.1016/j.jmb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Mymrikov EV, Bukach OV, Seit-Nebi AS, Gusev NB. The pivotal role of the beta 7 strand in the intersubunit contacts of different human small heat shock proteins. Cell Stress Chaperones. 2010;15:365–377. doi: 10.1007/s12192-009-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Mondry J, Thiel P, Weyand M, Ottmann C. Structure of the p53 C-terminus bound to 14-3-3: implications for stabilization of the p53 tetramer. FEBS Lett. 2010;584:1443–1448. doi: 10.1016/j.febslet.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Sluchanko NN, Sudnitsyna MV, Chernik IS, Seit-Nebi AS, Gusev NB. Phosphomimicking mutations of human 14-3-3zeta affect its interaction with tau protein and small heat shock protein HspB6. Arch Biochem Biophys. 2011;506:24–34. doi: 10.1016/j.abb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Sudnitsyna MV, Seit-Nebi AS, Gusev NB. Cofilin weakly interacts with 14-3-3 and therefore can only indirectly participate in regulation of cell motility by small heat shock protein HspB6 (Hsp20) Arch Biochem Biophys. 2012;521:62–70. doi: 10.1016/j.abb.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Tompa P, Schad E, Tantos A, Kalmar L. Intrinsically disordered proteins: emerging interaction specialists. Curr Opin Struct Biol. 2015;35:49–59. doi: 10.1016/j.sbi.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- Uhart M, Bustos DM. Human 14-3-3 paralogs differences uncovered by cross-talk of phosphorylation and lysine acetylation. PLoS One. 2013;8:e55703. doi: 10.1371/journal.pone.0055703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- Weeks SD, Baranova EV, Heirbaut M, Beelen S, Shkumatov AV, Gusev NB, Strelkov SV. Molecular structure and dynamics of the dimeric human small heat shock protein HSPB6. J Struct Biol. 2014;185:342–354. doi: 10.1016/j.jsb.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.