Abstract
Polymyxin B and E (i.e. colistin) are a family of naturally occurring lipopeptide antibiotics that are our last-line of defense against MDR Gram-negative pathogens. Unfortunately, nephrotoxicity is a dose-limiting factor for polymyxins which limits their clinical utility. Our recent studies demonstrate that polymyxin-induced nephrotoxicity is a result of their extensive accumulation in renal tubular cells. The design and development of safer, novel polymyxin lipopeptides is hampered by our limited understanding of their complex structure-nephrotoxicity relationships. This is the first study to employ a novel targeted chemical biology approach map the polymyxin recognition epitope of a commercially available polymyxin mAb; and demonstrate its utility for mapping the kidney distribution of a novel, less nephrotoxic polymyxin lipopeptide. Eighteen novel polymyxin lipopeptide analogs were synthesized with modifications in the polymyxin core domains, namely the N-terminal fatty acyl region, tripeptide linear segment and the cyclic heptapeptide. Surface Plasmon Resonance epitope mapping revealed that the mAb recognition epitope consisted of the hydrophobic domain (N-terminal fatty acyl and position 6/7) and Dab residues at positions 3, 5, 8 and 9 of the polymyxin molecule. Structural diversity within the hydrophobic domains and Dab 3 position are tolerated. Enlightened with an understating of the structure-binding relationships between the polymyxin mAb and the core polymyxin scaffold, we can now rationally employ the mAb to probe the kidney distribution of novel polymyxin lipopeptides. This information will be vital when designing novel, safer polymyxins through chemical tailoring of the core scaffold and exploring the elusive/complex polymyxin structure-nephrotoxicity relationships.
Keywords: Polymyxin, monoclonal antibody, epitope mapping, chemical biology
Graphical Abstract
Introduction
Antibiotic resistance has evolved into a serious global health concern.1, 2 In the United States over 23,000 people die each year due to infections with antibiotic-resistant bacteria.3, 4 Sadly, the ‘magic bullet’ antibiotics we have used liberally over the past decades are rapidly losing their effectiveness. There has been a steady decline in the number of FDA approved antibiotics, with only 10 antibiotics that are considered “New Molecular Entities” being approved by the FDA from 2004 to 2012.5, 6 Medicine is clearly entering a critical period and there could be catastrophic costs to healthcare and society, if bacteria continue developing resistance to multiple antibiotics at the present rate and at the same time the pipeline continues to dry up.7 Modern healthcare over the last century has been founded on the basis that bacterial infections can be effectively treated using antibiotics. In a world without effective antibiotics, modern surgical and medical procedures that we take for granted will become too dangerous or impossible due to the threat of untreatable bacterial infections. This ‘perfect storm’ has led to the revival of polymyxin B and E (colistin), as the last-line therapy for infections caused by multidrug-resistant (MDR) Gram-negative pathogens.8
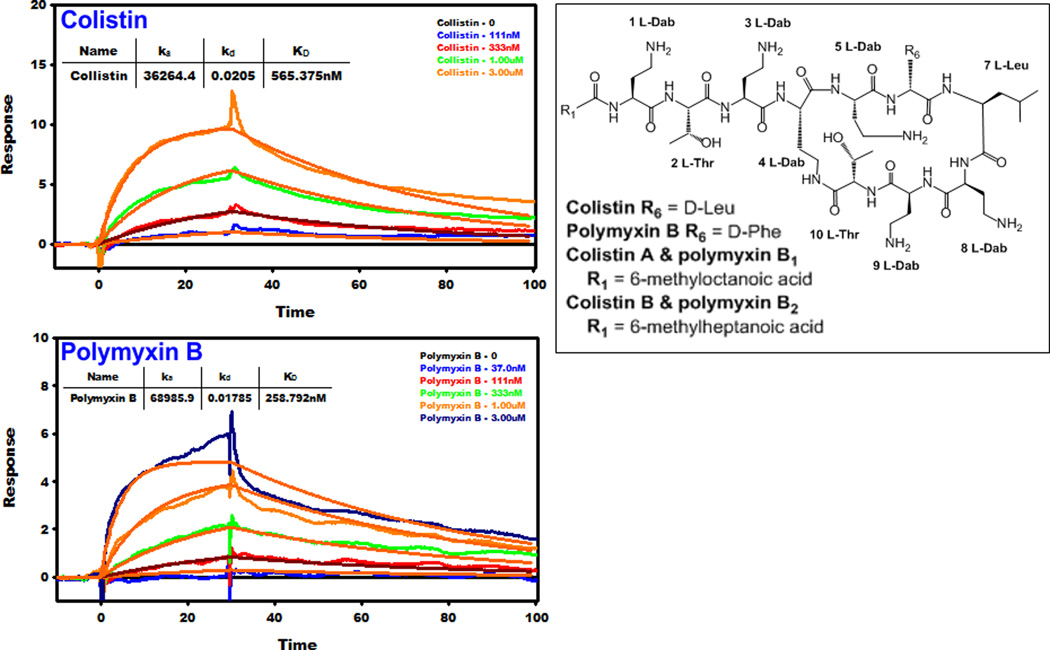
Polymyxins are cationic lipopeptides comprising of hydrophobic and hydrophilic domains (amphipathicity), both of which are pivotal for their antibacterial activity. The core polymyxin scaffold consists of a cyclic heptapeptide linked to a linear tripeptide with an N-terminal fatty acyl tail (Figure 1). Five l-α,γ-diaminobutyric acid (Dab) residues decorate the scaffold, the primary amines of which are positively charged at physiological pH (7.4). Two hydrophobic residues in positions 6 and 7 of the cyclic heptapeptide form an intermediary hydrophobic segment that breaks the succession of cationic Dab residues. Polymyxin B and colistin, are differentiated by a single hydrophobic residue at position 6: d-Phe in polymyxin B and d-Leu in colistin. Both polymyxins are secondary products and are produced by fermentation as mixtures, containing two major components, colistin A and B and polymyxin B1 and B2, which differ by only by a single carbon at the N-terminal fatty acyl tail (i.e C7 vs C8 fatty acyls) (Figure 1). Notwithstanding these differences, cross resistance between colistin and polymyxin B exists.9
Figure 1.
Left panels. SPR sensograms for the binding of mAb clone 45 to colistin and polymyxin B. The inset shows the binding affinity values and the polymyxin concentration injected over the surface. Right panel. The chemical structures of colistin and polymyxin B.
In spite of their excellent antibacterial activity, the polymyxins have an Achilles’ heel, namely dose-limiting nephrotoxicity which can occur in ~60% of patients.10–13 Polymyxin-induced nephrotoxicity is linked to their complex renal handling.14–16 Only a very small fraction of the dose is renally excreted.14–16 The polymyxins are filtered at the glomeruli and then undergo very extensive renal re-absorption leading to accumulation in tubular cells.17–19 Our group has shown that, at concentrations achievable in patients considering the high risk of nephrotoxicity, resistance to the polymyxins can emerge rapidly in vitro in Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae20–22, and resistance rates are increasing in isolates from patients.23 Most worryingly, human infections caused by Escherichia coli that harbour plasmid-mediated colistin resistance have been recently reported in China and Switzerland, potentially as a result of the agricultural use of colistin in animal feed.24–26 Unfortunately, polymyxin resistance often implies a total lack of antibiotic treatment options for infections caused by MDR Gram-negative ‘superbugs’. Clearly, the development of a new generation of polymyxin lipopeptides with a wider therapeutic window is an urgent unmet global medical need.
Our research group is currently conducting a program to understand the mechanism of polymyxin-induced nephrotoxicity and discover new polymyxin lipopeptides with improved efficacy and safety over the currently used polymyxin B and colistin. To this end we utilized a commercially available monoclonal antibody (mAb) raised to recognize polymyxin B to map the distribution of polymyxins in the kidneys. Immunohistochemical kidney disposition studies in animal models provide very valuable information on the kidney distribution of polymyxin B and colistin as well as our novel polymyxin lipopeptides. Because modifications to the polymyxin scaffold may have a significant influence on antibody recognition, it is important to understand the potential influence of structural differences on the structure-binding relationship with the polymyxin-mAb. Currently the amino acid residues in polymyxin B scaffold responsible for antibody binding are not known. Our objective was to undertake epitope mapping via Surface Plasmon Resonance (SPR) screening of novel polymyxin lipopeptides from our extensive library that contains a diverse array of modifications to the core domains of the polymyxin scaffold. The ability to map the kidney distribution and structure-nephrotoxicity relationships (SNR) of novel polymyxin lipopeptides will facilitate the chemical tailoring of their core scaffold to develop novel, safer polymyxins against MDR Gram-negative ‘superbugs’.
Results and Discussion
Nephrotoxicity is the dose-limiting factor of the last-line antibiotics polymyxin B and colistin.27–29 We previously reported that polymyxin-induced nephrotoxicity results from the extensive accumulation of polymyxins in renal tubular cells with intracellular concentrations reaching 1,930- to 4,760-fold greater than the extracellular levels.17 The structure-nephrotoxicity relationships (SNR) are a major gap in the knowledge base for developing novel safer polymyxin-like lipopeptides. Towards this end, the present study employs a novel polymyxin lipopeptide library to perform SPR mapping of the polymyxin recognition epitope of a commercially available mAb and demonstrate its utility for mapping the kidney distribution of the novel polymyxin lipopeptide (FADDI-019) that we have previously shown to be less nephrotoxic than polymyxin B and colistin.30, 31 The novel polymxin lipopeptides screened for mAb binding contained a diverse array of structural modifications to the key structural domains of the polymyxin scaffold (Figure 1 and Table 1): 1) N-terminal fatty acyl group; 2) position 6/7 hydrophobic segment and 3) hydrophilic (positively charged) Dab residues. The antimicrobial activity of the lipopeptides against a panel of polymyxin-susceptible and polymyxin-resistant clinical isolates of P. aeruginosa, K. pneumoniae, A. baumannii, and Enterobacter cloacae are documented in Table 2 and discussed for each lipopeptide class below.
Table 1.
Monoclonal antibody binding affinity values for polymyxin lipopeptides.
| Polymyxin lipopeptide |
Polymyxin lipopeptide sequence | KD (µM) |
|---|---|---|
| Colistin | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Leu-Leu-Dab-Dab-Thr* | 0.56 |
| Polymyxin B | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 0.26 |
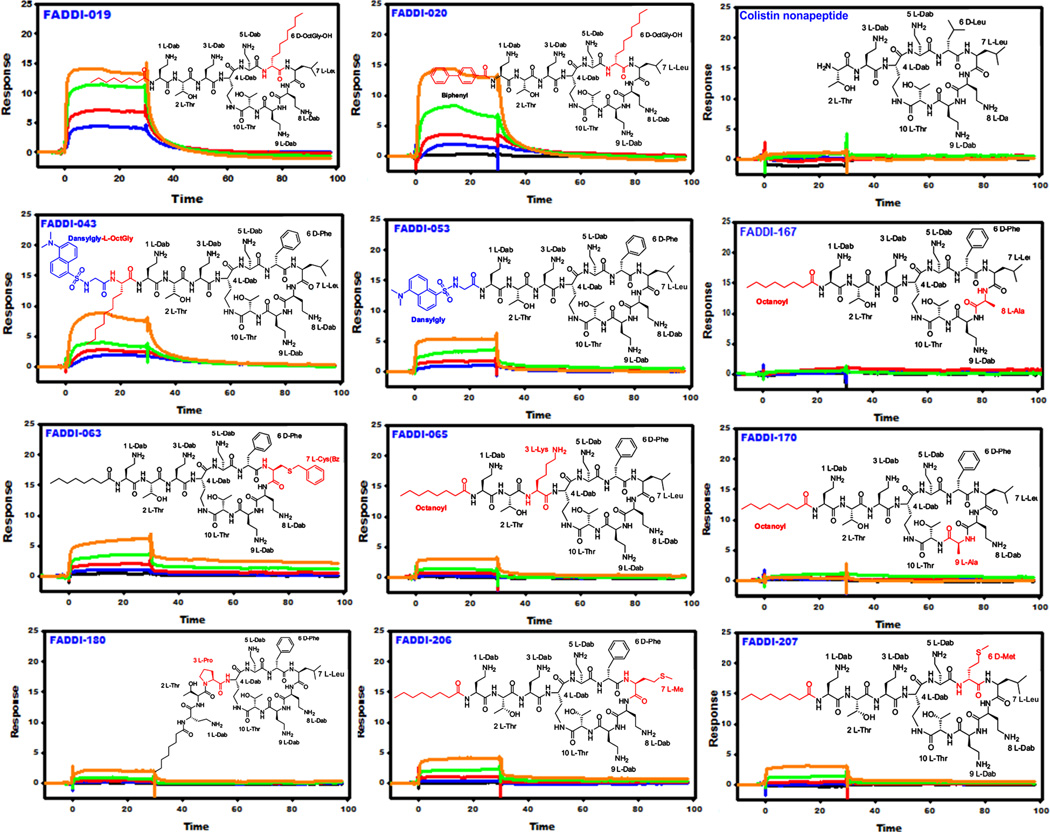
| FADDI-019 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-OctGly-Leu-Dab-Dab-Thr* | 0.43 |
| FADDI-020 | 4-Biphenylcarboxyl-Dab-Thr-Dab-Dab*-Dab-D-OctGly-Leu-Dab-Dab-Thr* | 0.94 |
| FADDI-043 | Dansylgly-OctGly-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 2.71 |
| FADDI-063 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Cys(Bz)-Dab-Dab-Thr* | 5.15 |
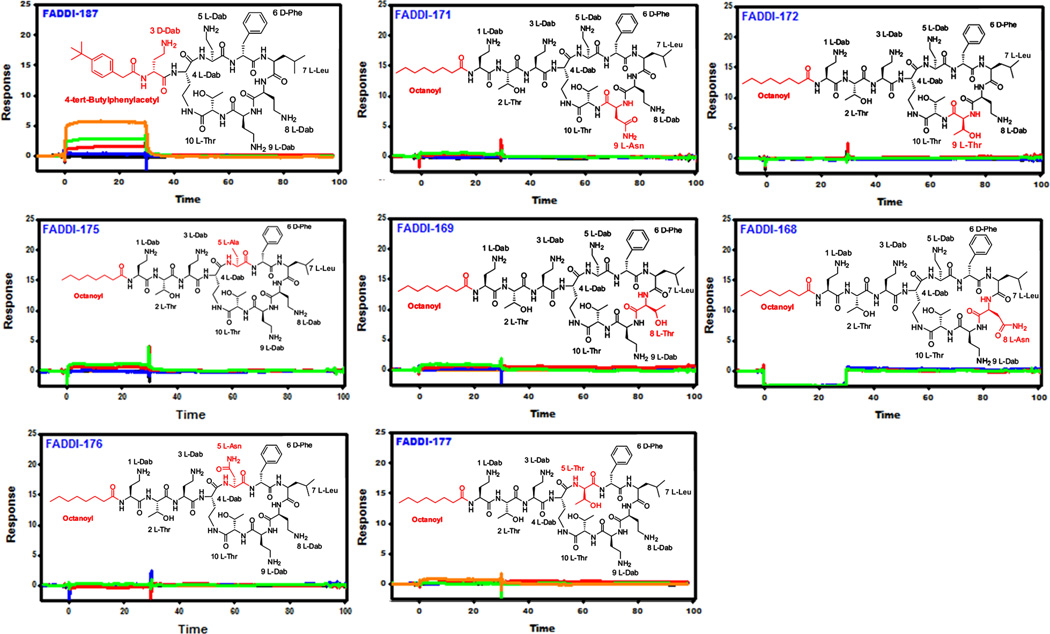
| FADDI-187 | 4-tert-Butylphenylacetyl-D-Dab-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 5.39 |
| FADDI-053 | Dansylgly-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 5.50 |
| FADDI-206 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Met-Dab-Dab-Thr* | 8.80 |
| FADDI-065 | Octanoyl-Dab-Thr-Lys-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 13.1 |
| FADDI-207 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Met-Leu-Dab-Dab-Thr* | 13.4 |
| FADDI-180 | Octanoyl-Dab-Thr-Pro-Dab*-Dab-D-Phe-Leu-Dab-Dab-Thr* | 20.3 |
| FADDI-167 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Ala-Dab-Thr* | NSB |
| FADDI-168 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Asn-Dab-Thr* | NSB |
| FADDI-169 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Thr-Dab-Thr* | NSB |
| FADDI-170 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Ala-Thr* | NSB |
| FADDI-171 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Asn-Thr* | NSB |
| FADDI-172 | Octanoyl-Dab-Thr-Dab-Dab*-Dab-D-Phe-Leu-Dab-Thr-Thr* | NSB |
| FADDI-175 | Octanoyl-Dab-Thr-Dab-Dab*-Ala-D-Phe-Leu-Dab-Dab-Thr* | NSB |
| FADDI-176 | Octanoyl-Dab-Thr-Dab-Dab*-Asn-D-Phe-Leu-Dab-Dab-Thr* | NSB |
| FADDI-177 | Octanoyl-Dab-Thr-Dab-Dab*-Thr-D-Phe-Leu-Dab-Dab-Thr* | NSB |
| Colistin nonapeptide |
Thr-Dab-Dab*-Dab-D-Leu-Leu-Dab-Dab-Thr* | NSB |
NSB. No significant binding (KD>25 µM)
Cyclisation points
Table 2.
MICs (mg/L) of polymyxin lipopeptides against Gram-negative bacteria (polymyxin-resistant isolates in red).
| Bacterial isolate | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymyxin lipopeptide |
P. aeruginosa ATCC 27853 | P. aeruginosaFADDIPA001 | P. aeruginosa FADDI-PA025 | P. aeruginosa FADDI-PA070 | P.aeruginosa FADDI-PA060 | P. aeruginosa FADDI-PA090 | A. baumanniiATCC 19606 | A. baumanniiFADDI-AB110 | A. baumanniiATCC 17978 | A. baumanniiFADDI-AB065 | A. baumanniiFADDI-AB156 | A. baumanniiFADDI-AB167 | K. pneumoniae ATCC 13883 | K. pneumoniae FADDI-KP032 | K. pneumoniae FADDI-KP027 | K. pneumoniae FADDI-KP003 | K. pneumoniae FADDI-KP012 | E. cloacae FADDI-EC006 | E. cloacae FADDI-EC001 | E. cloacae FADDI-EC003 |
| Colistin | 1 | 1 | 2 | >128 | >128 | 8 | 1 | 0.5 | 0.5 | 128 | 16 | 8 | 1 | 1 | >128 | 128 | 32 | <0.125 | 0.25 | <0.125 |
| Polymyxin B | 1 | 1 | 1 | 32 | >32 | 4 | 1 | 0.5 | 1 | 128 | 8 | 16 | 1 | <0.125 | 128 | >32 | 16 | 0.5 | 0.25 | 0.5 |
| FADDI-019 | 4 | 4 | 8 | 4 | 2 | 8 | 1 | 4 | / | 32 | 16 | 8 | 4 | 4 | >32 | 32 | 16 | 2 | 1 | 1 |
| FADDI-020 | 2 | 4 | 4 | 2 | 2 | 8 | 4 | 2 | / | 32 | 4 | 4 | 4 | 2 | >32 | >32 | 8 | 4 | 4 | 2 |
| FADDI-043 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 4 | 4 | 8 | 4 | 8 | 16 | / | / | / | / | / | / |
| FADDI-053 | 2 | 2 | 2 | >32 | / | 16 | 4 | 2 | 1 | >32 | 32 | 8 | 2 | / | / | / | / | / | / | / |
| FADDI-063 | 1 | / | / | >32 | / | / | 1 | / | / | >32 | 2 | / | 16 | / | / | / | / | / | / | / |
| FADDI-065 | 2 | 2 | 8 | >32 | >32 | 32 | 2 | 1 | / | >32 | 32 | 32 | 0.25 | 0.5 | >32 | >32 | >32 | 1 | 0.5 | 0.5 |
| FADDI-167 | 2 | 2 | 32 | >32 | >32 | >32 | 16 | 16 | / | >32 | >32 | >32 | >32 | 1 | >32 | >32 | >32 | 8 | 1 | 2 |
| FADDI-168 | 2 | 4 | 14 | >32 | >32 | 4 | 4 | 8 | / | >32 | >32 | >32 | >32 | 0.5 | >32 | >32 | >32 | 0.5 | 0.5 | 0.5 |
| FADDI-169 | 2 | 4 | 16 | >32 | >32 | >32 | 16 | 32 | / | >32 | >32 | >32 | >32 | 1 | >32 | >32 | 8 | 8 | 1 | 2 |
| FADDI-170 | 2 | 4 | 32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | 4 | >32 | >32 | >32 | 8 | 8 | 8 |
| FADDI-171 | 2 | 4 | 8 | 8 | 8 | 8 | >32 | >32 | / | >32 | >32 | >32 | >32 | 8 | >32 | >32 | >32 | 8 | 16 | 8 |
| FADDI-172 | 4 | 4 | >32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | 4 | >32 | >32 | >32 | >32 | 8 | 16 |
| FADDI-175 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 32 | 32 |
| FADDI-176 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| FADDI-177 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| FADDI-180 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| FADDI-187 | 1 | 2 | 2 | >32 | >32 | >32 | >32 | >32 | / | >32 | >32 | >32 | >32 | 4 | >32 | >32 | >32 | 16 | 26 | 8 |
| FADDI-206 | 1 | 1 | 4 | >32 | >32 | >32 | 1 | 1 | / | >32 | 8 | 2 | 2 | 0.25 | >32 | >32 | >32 | 0.5 | 0.25 | 0.25 |
| FADDI-207 | 1 | 2 | 4 | >32 | >32 | >32 | 8 | 8 | / | >32 | >32 | 16 | 16 | 0.25 | >32 | >32 | >32 | 1 | 0.25 | 0.5 |
/=Not determined
Role of the N-terminal fatty acyl group in antibody binding
The available SAR and SNR data suggest that the N-terminal fatty acyl domain of the polymyxins is essential for antibacterial activity and is partly responsible for the nephrotoxicity of polymyxins.32–40 Not surprisingly, most discovery programs have concentrated on generating semi-synthetic N-terminal analogs in an effort to discover safer polymyxins.36–38, 41 The SPR epitope mapping data reveal that the mAb strongly recognized the octanoyl fatty acyl chain of the native polymyxin scaffold, as removal of the fatty acyl chain (colistin nonapeptide) abolished the binding. Replacement of the octanoyl fatty acyl chain (polymyxin B1) with bulky hydrophobic substituents such as the 4-biphenylcarboxyl moiety (FADDI-020), dansylglycine-octanoyl-glycine (FADDI-043), dansylglycine (FADDI-053) or a 4-tert-butylphenylacetyl (FADDI-187) group decreased mAb binding affinity up to 22-fold for FADDI-053 (Figure 2 and Table 1). It is important to note that FADDI-020 also contained a modification to another structural domain which had an influence on antibody binding. Likewise for FADDI-187 in which the Dab1-Thr2 segment was removed. It appears that while the N-terminal fatty acyl group was critical for antibody binding, the mAb was tolerant to some modification of this structural domain. In summary, the mAb preferentially recognized the linear C8 fatty acyl chain and bulky or aromatic substituents were poorly tolerated.
Figure 2.
SPR sensograms for the binding of mAb clone 45 to novel polymyxin lipopeptides (Black 0.0 µM, Blue 0.037 µM, Red 0.11 µM, Green 0.33 µM, Orange 1.0 µM, Dark Blue 3.0 µM). The insets show the chemical structure of each lipopeptide and the modified regions that differ from the polymyxin B core scaffold are colored red.
The MIC data revealed FADDI-020 displays an improved activity against the polymyxin-resistant strains of P. aeruginosa, A. baumannii and K. pneumoniae (Table 2). The MICs of FADDI-020 against the polymyxin-susceptible strains were generally higher compared to polymyxin B and colistin.31 The MICs of FADDI-043 were fairly consistent ranging from 4 to16 mg/L across both the polymyxin-susceptible and -resistant strains examined.42 The MICs of FADDI-053 were largely comparable to those of polymyxin B and colistin, with an approximately 2-fold variation across most of the MICs. FADDI-187 was largely inactive against all of the test strains, only showing activity (MICs 1–2 mg/L) against three of the polymyxin-susceptible strains tested.
Role of the position 6/7 hydrophobic segment in antibody binding
In addition to the N-terminal fatty acyl chain, polymyxins feature a second hydrophobic domain, the position 6/7 segment in the heptapeptide ring that has been implicated in the plasma protein binding of polymyxins (Figure 1).43 More importantly, we have previously reported that the substitution of the segment with D-Octylglycine fatty acyl moieties significantly improved activity against polymyxin-resistant bacteria and lowers nephrotoxicity.31 The D-Phe6 acts as a turn inducing residue that is critical for attaining the active β-turn conformation of the polymyxin molecule.44–46 The D-Phe6-L-Leu7 segment of polymyxin B appears to be important for mAb recognition. This is particularly evident with colistin where a small reduction in the hydrophobicity within this segment as per the D-Leu6 substitution resulted in a 2-fold reduction in mAb binding affinity (Figures 1 and 2; Table 1). The hydrophobicity of the amino acids at this position was critical as evident from the substitution of either positions 6 or 7 with methionine (FADDI-206 and FADDI-207), an amino acid structurally similar to leucine but less hydrophobic, which resulted in an up to 50-fold reduction in mAb binding affinity when compared to polymyxin B and colistin. Incorporation of a residue at position 6 (D-OctGly6 FADDI-019) and 7 (Cystine-Benzoyl7 FADDI-063) with increased hydrophobicity resulted in a ~2-fold and 19-fold reduction in mAb binding affinity, respectively. Together these results demonstrated that the hydrophobic position 6/7 segment is important for mAb recognition.
In terms of the antibacterial activity, the hydrophobic side chains of the position 6/7 segment are believed to insert into the bacterial outer membrane (OM) and help stabilize complex formation with LPS; through hydrophobic interactions with the lipid A fatty acyl chains.45, 46 The MIC data revealed that FADDI-019 is similarly antibacterial as FADDI-020 with an improved activity against the polymyxin-resistant strains of P. aeruginosa, A. baumannii and K. pneumoniae (Table 2).31 The MICs of FADDI-019 were generally lower against the polymyxin-susceptible strains compared to polymyxin B and colistin. FADDI-206 and FADDI-207 showed generally comparable MICs to those of polymyxin B and colistin.
Role of the hydrophilic positively charged Dab residues in antibody binding
In addition to the two aforementioned hydrophobic regions, the positive charges of the five Dab residues were also identified as major nephrotoxic ‘hot spots’ in the polymyxin scaffold.8, 19, 39, 47–52 The epitope mapping data revealed that the Dab residues at positions 5 (FADDI 175–177), 8 (FADDI 167–169) and 9 (FADDI 170–172) are critical for mAb recognition as substitution at any these positions led to a complete loss of mAb binding (Figures 1&2; Table 1). The mAb also appeared to recognize the Dab3 position as the exchange of Dab3 with a lysine residue (FADDI-065) resulted in a 44-fold decrease in binding affinity while a change to a Pro (FADDI-180) resulted in a 67-fold decrease in binding affinity. In summary, the mAb preferentially recognized the cationic Dab residues at positions 3, 5, 8 and 9 of the polymyxin molecule.
The exact order of the Dab residues within the primary sequence of the polymyxin scaffold ensures the correct placement of the positive charges for electrostatic interactions with the negative phosphates of lipid A, which is critical for antimicrobial activity.51 The MIC data revealed FADDI-175, FADDI-176, FADDI-177 and FADDI-180 were inactive against all of the strains tested with MICs >32 mg/L. The MIC profiles of the lipopeptides FADDI-065, FADDI-167, FADDI-168, FADDI-169, FADDI-170, FADDI-171 and FADDI-172 were very similar; all of these lipopeptides were largely inactive, except against a few of the polymyxin-susceptible strains of P. aeruginosa (MICs 2–8 mg/L), K. pneumoniae FADDI-KP032 (MICs 0.5–8 mg/L) and against the three E. cloacae strains tested (MICs 0.5–16 mg/L).
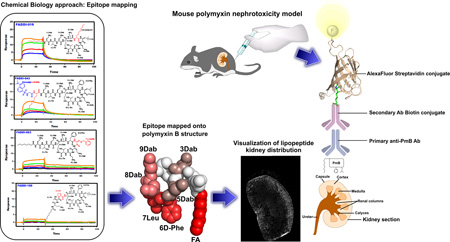
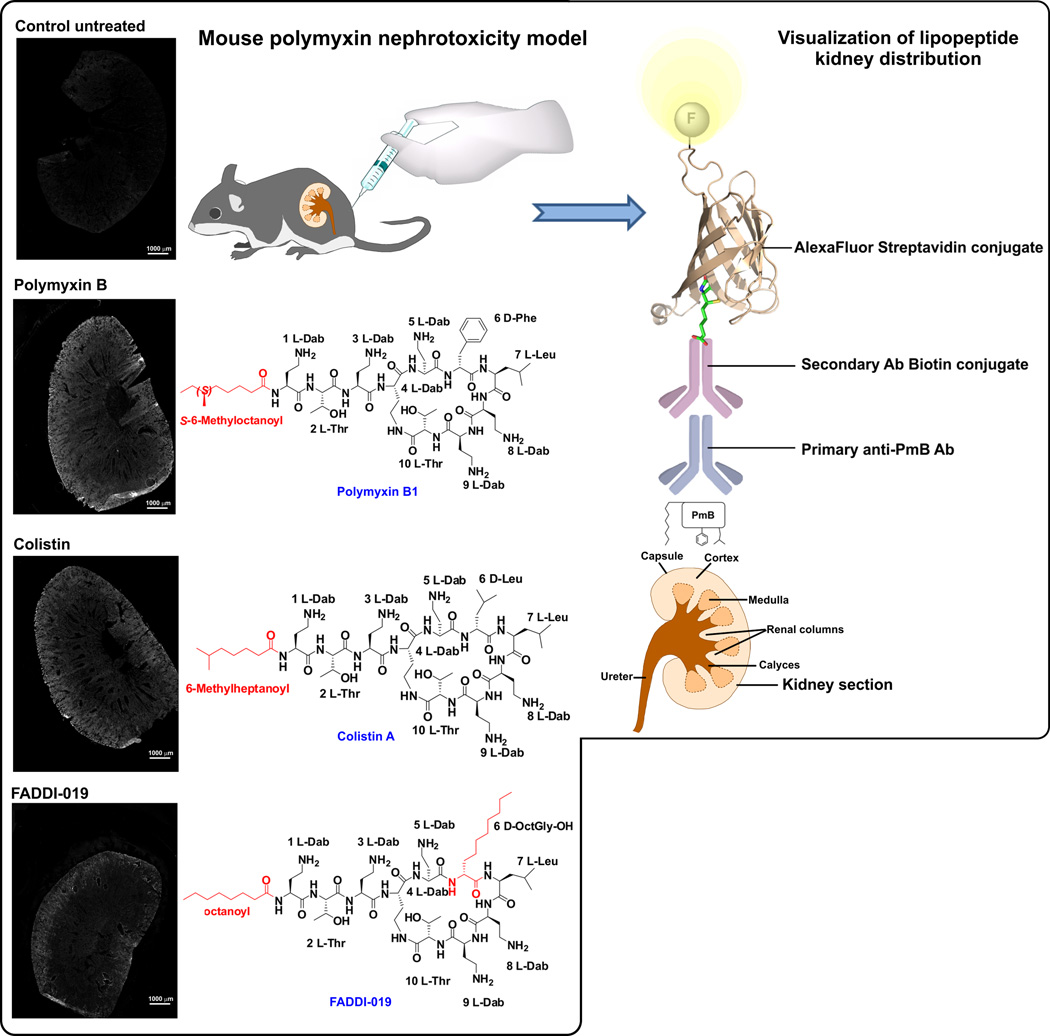
Imaging the localization of polymyxins in bacterial cells and mouse kidneys using the polymyxin B mAb
As a proof-of-concept, we employed in situ immunohistochemical staining of the kidney sections from a mouse treated with polymyxin B (d-Phe6-l-Leu7), colistin (d-Leu6-l-Leu7) and the novel FADDI-019 (d-Octylglycine6-l-Leu7), which we have previously shown to be less nephrotoxic than polymyxin B and colistin.31 The imaging data revealed that polymyxin B and colistin were predominantly distributed within the renal cortex and to a lesser extent within the medulla of the kidney (Figure 3). These findings are in line with our recent immunohistochemical and correlative microscopy studies which demonstrated that polymyxins accumulate at extremely high concentrations within renal tubular cells of the renal cortex.17, 19 In contrast, FADDI-019 in which position 6 is substituted with a more hydrophobic d-Octylglycine amino acid was distributed largely within the outer layer of the renal cortex. Our recent data demonstrate that increasing the hydrophobicity of position 7 can substantially increase plasma protein binding of polymyxin molecules.31 Differences in pharmacokinetics may cause the different localization of polymyxins in the mouse kidney as shown using the polymyxin B mAb and further pharmacological studies are warranted (Figure 3).
Figure 3.
Immunohistochemical staining with mAb clone 45 of kidney sections from mice treated with polymyxin lipopeptides. Right panel. Schematic diagram of the mouse nephrotoxicity model and in situ image development procedure employed for the visualization of the kidney distribution of the polymyxin lipopeptides.
Polymyxins exert their antimicrobial activity through an electrostatic interaction of the cationic Dab residues with the negatively charged phosphate groups of the lipid A of LPS. This complex formation displaces divalent cations (Ca2+ and Mg2+) that bridge adjacent LPS molecules.8, 51, 53 Subsequently, hydrophobic interactions occur between the N-terminal fatty acyl tail and the position 6 and 7 hydrophobic segment of the polymyxin molecule and the lipid A fatty acyl chains, thereby becoming imbedded in the bacterial OM. It is hypothesized that this ‘self-promoted’ uptake mechanism leads to disruption of the cell envelope resulting in bacterial killing.51, 54–56 To further validate the epitope mapping data, we attempted to employ the mAb to detect polymyxin B on A. baumannii ATCC19606 cells. Interestingly, the mAb was unable to detect polymyxin B in the OM of A. baumannii cells (data not shown). Presumably, once the polymyxin B has complexed with lipid A,51 its hydrophobic domains (that form most of the mAb recognition epitope) become imbedded in the OM and are therefore shielded from mAb binding.
Conclusions
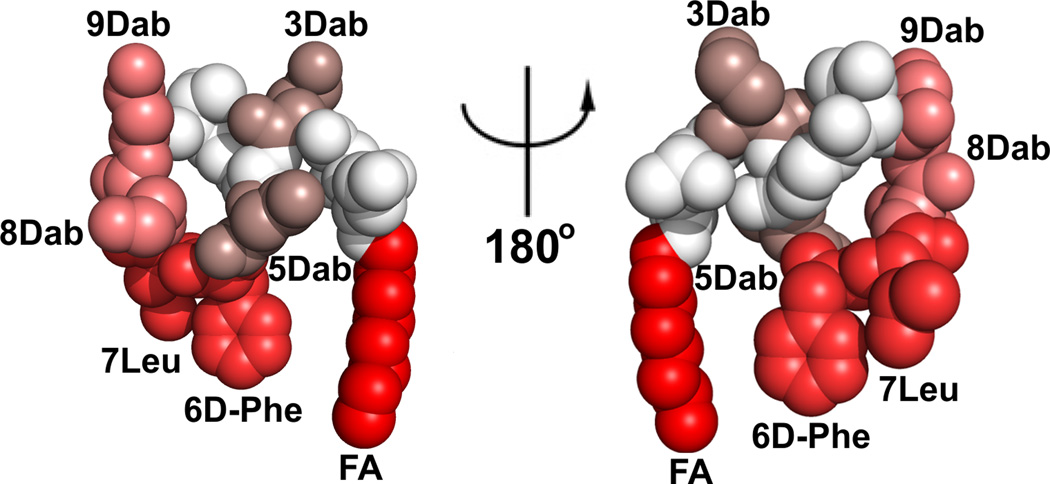
In summary, this is the first chemical biology study to map the polymyxin recognition epitope of a commercially available polymyxin mAb. Both the hydrophobic domains (N-terminal fatty acyl and position 6/7) and Dab residues, in particular Dabs 3, 5, 8 and 9, of the polymyxin are components of the polymyxin mAb recognition epitope (Figure 4). Notwithstanding, the lipopeptide library SPR screening and FADDI-019 imaging data indicate that structural diversity within the hydrophobic domains and Dab 3 position are tolerated. This means that the mAb may be of considerable utility for probing the kidney distribution of novel polymyxin lipopeptides, as we have demonstrated using FADDI-019. The ability to effectively map the kidney distribution of novel polymyxin lipopeptides using a polymyxin-specific mAb with a defined epitope map will facilitate the elucidation of the ever-elusive structure-nephrotoxicity relationships of the polymyxins. This in turn may open up opportunities for the development of novel, safer polymyxins through the careful chemical tailoring of the core scaffold.
Figure 4.
Monoclonal antibiody clone 45 epitope ‘HOTSPOTS’ on the polymyxin B structure. The SPR KD values were mapped onto the NMR solution structure of polymyxin B1 (higher affinity binding is represented by a brighter red coloring). The polymyxin B1 structure is shown in CPK representation and the two views are rotated by 180° about the y-axis.
Experimental Section
Chemical reagents
Anti-polymyxin B mouse IgM antibody (clone 45) was obtained from Thermo Fisher Scientific (Rockford, Illinois, USA). Polymyxin B (sulfate), colistin (sulfate), triisopropylsilane (TIPS), trifluoroacetic acid, (TFA) and diphenylphosphorylazide (DPPA), Streptavidin were purchased from Sigma-Aldrich (Sydney, New South Wales, Australia). Piperidine and diisopropylethylamine (DIPEA) were obtained from Auspep (Melbourne, Victoria, Australia). Fmoc-Dab(Boc)-OH was purchased from Try-lead Chem (Hangzhou, Zhejiang,China). Fmoc-Thr(tBu)-OH and Fmoc-Leu-OH were from Mimotopes (Melbourne, Australia). Fmoc-Dab(ivDde)-OH, Fmoc-D-Phe-OH, Fmoc-Dap-OH, Fmoc-D-Dap-OH and 1H-Benzotriazolium-1-[bis(dimethylamino)methylene]-5-chloro hexafluoro-phosphate-(1-),3-oxide (HCTU) were purchased from Chem-Impex International (Wood Dale, Illinois, USA). Fmoc-Thr(tBu)-TCP-Resin was obtained from Intavis Bioanyltical Instruments (Köln, Cologne,Germany). Dimethylformamide (DMF), methanol (MeOH), diethyl ether, dichloromethane (DCM), hydrochloric acid and acetonitrile were purchased from Merck (Melbourne, Victoria, Australia). Polymyxin lipopeptide stock solutions were prepared with Milli-Q water (Millipore, North Ryde, New South Wales, Australia) and filtered using 0.22-µm syringe filters (Sartorius, Melbourne, Victoria, Australia). Solutions were stored at 4°C for up to one month.57
Lipopeptide synthesis
Synthesis procedures for specific lipopeptides were previously described in detail by our group.31,58 Briefly, the protected linear peptides were synthesised on a CEM Liberty Microwave automated peptide synthesizer using Fmoc solid-phase peptide chemistry. Synthesis was carried out using TCP-Resin, pre-loaded with Fmoc-Thr(tBu)-OH (loading 1.0 mmol/g), 0.25 mmol scale (260 mg of resin). Fmoc-amino acid coupling and the N-terminal octanoyl group was performed as follows: 5 molar equivalents of Fmoc amino acid and HCTU with activation using DIPEA in DMF over 2 min at room temperature then for 4 min at 50°C (25W microwave power). Fmoc deprotection was performed as follows: 20% piperidine in dimethylformamide (1 × 30s, 1 × 3 min) at 75°C (35W microwave power). Resin was removed from the synthesiser, transferred to a synthesis syringe and treated with 2% hydrazine in DMF (4 × 15 min), removing the ivDde group. The resin was then washed with MeOH (2 × 2 min) and diethyl ether (1 × 2 min), air dried under vacuum. The protected linear peptide was cleaved from the resin by washing with 1% TFA in DCM (1 × 5 min, 3 × 10 min). The residue was dissolved in 50% acetonitrile/water and freeze dried. The protected linear peptide was dissolved in DMF (10 mL) to which DPPA (3 eq. relative to the loading of the resin) and DIPEA (6 eq. relative to the loading of the resin) were added. The solution was stirred overnight at room temperature and then concentrated under vacuum. The residue was taken up in a solution of 5% TIPS in TFA and stirred at room temperature for 2 h. TFA was removed under a nitrogen stream and the crude cyclic peptide was precipitated with cold diethyl ether, collected by centrifugation and air-dried. The residue was then taken up in Milli-Q water and desalted with a Vari-Pure IPE SAX column. The crude cyclic lipopeptide was subjected to RP-HPLC purification and LC-MS analysis as described below.
HPLC purification and LC-MS analysis of lipopeptides
Crude cyclic lipopeptides were purified by RP-HPLC on a Waters Prep LC system with a Waters 486 tuneable absorbance detector (214 nm). A Phenomenex Axia column (Luna C8(2), 250 × 50.0 mm ID, 100 Å, 10 micron) was employed with a gradient of 0–60% Buffer B over 60 min at a flow rate of 40 mL/min; Buffer A was 0.1% TFA/Water and Buffer B was 0.1% TFA/Acetonitrile. Collected fractions were analysed using a Shimadzu 2020 LCMS system, with a photodiode array detector (214 nm) coupled to an electrospray ionization source and single quadrupole mass analyzer. RP-HPLC was performed with a Phenomenex column (Luna C8(2), 100×2.0 mm ID) with an eluting gradient of 80% acetonitrile in 0.05% aqueous TFA over 10 min at 0.2 mL/min (Buffer A: 0.05%TFA/water; Buffer B: 0.05%TFA/acetonitrile). Mass spectra were acquired in the positive ion mode in the scan range of 200 – 2,000 m/z.
Bacterial strains and MIC determination
P. aeruginosa, K. pneumoniae, A. baumannii, and E. cloacae strains were obtained from the American Type Culture Collection (Rockville, Maryland, USA) or were clinical isolates. Bacterial isolates were stored at −80°C in tryptone soy broth (Oxoid, Thermo Fisher Scientific, Adelaide, South Australia, Australia) with 20% glycerol (Ajax Finechem, Seven Hills, New South Wales, Australia). The MICs of the polymyxin lipopeptides were determined by broth microdilution.59 Experiments were performed with Cation-adjusted Mueller-Hinton Broth (CaMHB) in 96-well polypropylene microtitre plates. Wells were inoculated with 100 µL of bacterial suspension prepared in CaMHB (containing ~106 colony forming units (cfu) per mL) and 100 µL of CaMHB containing increasing concentrations of the lipopeptide (0 to 128 mg/L). The MIC was defined as the lowest concentration at which visible growth was inhibited following 18 h incubation at 37°C.
Surface Plasmon Resonance (SPR)
Immobilization of the anti-polymyxin B mouse IgM monoclonal antibody onto a CM5 sensor chip surface was achieved using a double capture method that utilized a streptavidin-coated chip to load biotin anti-IgM conjugate (CaptureSelect™, Life Technologies, Melbourne, Victoria, Australia) followed by a capture of the IgM antibody. Streptavidin was simultaneously immobilized in all four channels of a CM5 sensor chip docked in a Biacore T200 instrument equilibrated with 1× HBS-P+ buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05% [v/v] Tween-20®). Immobilization was performed at 37°C at a constant flow rate of 10 µL/min, using standard amide coupling chemistry recommended by the chip manufacturer. Streptavidin was diluted in 10 mM sodium acetate buffer pH 4.5 to a final concentration of 100 µg/mL. This approach resulted in immobilization of 12,500 response units (1 RU = 1 pg protein/mm2) with minimal variation between the four channels on the same chip. Freshly immobilized streptavidin surfaces were further conditioned with three consecutive 30 sec injections of conditioning solution consisting of 50 mM NaOH and 1 M NaCl. All subsequent capture and binding experiments were performed at 25°C with 1× HBS-EP+ as instrumentrunning buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% [v/v] Tween-20). CaptureSelect™ biotin anti-IgM conjugate was diluted to 10 µg/mL in 1× HBS-EP+ and injected onto a streptavidin chip surface in channels 3 and 4 at 10 µL/min for 420 sec resulting in a capture of approximately 7,500 RU of protein. Anti-polymyxin B mouse IgM was diluted 1:100 in 1× 1× HBS-EP+ and injected in channel 4 only at 5 µL/min for 2× 120 sec. After allowing the captured anti-polymyxin B mouse IgM baseline to stabilize for 3 minutes, final captured levels were estimated to be ~1,450 RU. To evaluate binding interactions, serial dilutions of polymyxin lipopeptides (cf. Table 1) were prepared in 1× HBS-EP+ buffer and sequentially injected over immobilized proteins in channels 3 and 4 with the association and dissociation phases monitored for 30 sec and 180 sec, respectively. Control injections of the instrument running buffer (‘‘zero-buffer’’ blank solution) were also included for double referencing purposes. No regeneration of anti-polymyxin B mouse IgM surface was required between binding cycles as bound polymyxin lipopeptides fully dissociated within 60 to 180 sec in the running buffer. All SPR sensorgrams were processed using Scrubber software (www.biologic.com.au). Sensorgrams were first zeroed on the y axis and then x-aligned at the beginning of each injection. Bulk refractive index changes were removed by subtracting the response of the reference flow cell (channel 3) responses. The average response of all blank injections was subtracted from all analyte injections and blank sensorgrams to remove systematic artefacts in the experimental and reference flow cells. Scrubber software was then used to extract ka and kd rate parameters from the processed data sets by globally fitting to a 1:1 binding model. The affinity (KD) was calculated from the quotient kd/ka. Alternatively, for rapidly dissociating interactions, KDestimates were derived using steady-state affinity algorithm.
Epitope mapping onto the polymyxin B structure
The Ab binding epitope was mapped onto the three-dimensional structure of polymyxin B using the coordinates from the NMR solution structure of polymyxin B1 when bound to E. coli LPS.45, 46 Molecular visualization was performed using the software package PYMOL (Schrödinger, Cambridge, Massachusetts, USA).
Immunostaining of polymyxins in mouse kidneys and in bacterial cells
The animal study was approved by the Monash Institute of Pharmaceutical Sciences Animal Ethics Committee and performed in accordance with the Australian National Health and Medical Research Council (NHMRC) guidelines for the care and use of animals for scientific purposes. Female Swiss mice (6–8 weeks, 20–25 g) had free access to food and water during the experiment. Polymyxin B, colistin, FADDI-019 or sterile saline was administered subcutaneously to mice (n = 3) over three days (accumulated dose 175 mg/kg, with 35, 10, 10, 20 mg/kg administered at 2 h intervals on day one; 35, 10, 10, 15 at 2 h intervals on day two; and a final dose of 30 mg/kg on day three). Mice were euthanized on day 3 via isoflurane overdose 2 h after the last dose and kidneys were collected, frozen and sectioned at 12 µm. Sections were fixed in cold acetone, dried and treated with 1% hydrogen peroxide (5 min). Endogenous mouse IgG was blocked using the Vector M.O.M kit (Vector Labs, Burlingame, California, USA) as per manufacturer’s instructions. Sections were incubated with anti-polymyxin B mouse IgM mAb diluted to 1:500 overnight at 4°C, washed and incubated with M.O.M biotinylated anti-mouse secondary link (Vector Labs, Burlingame, California, USA) for 10 min. Following incubation with AlexaFluor647 streptavidin conjugate (1:500) (Life Technologies, Melbourne, Victoria, Australia), sections were mounted with Dako fluorescence mounting medium (Dako, Sydney, New South Wales, Australia) and imaged using a MetasystemsVSlide Scanner.
For detection of polymyxin B bound to bacterial cells, exponential phase A. baumannii ATCC19606 cells (OD 600nm 0.1) were harvested by centrifugation and washed twice with PBS. The cells were then treated with polymyxin B at ×0.5 MIC (0.5 mg/L) for 1 hr and washed with PBS. The cells were then incubated for 5 hrs with anti-polymyxin B mouse IgM mAb diluted to 1:500 at 4°C, washed twice with PBS and imaged as described above.
Acknowledgments
J.L., T.V., R.L.N., P.E.T. and K.D.R. are supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI098771) and J.L. and T.V. are also supported by R01 AI111965. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. J.L. is an Australian NHMRC Senior Research Fellow. T.V. is an Australian NHMRC Industry Career Development Research Fellow.
Abbreviations
- Bz
benzyl
- CaMHB
Cation-Adjusted Mueller-Hinton broth
- Dab
diaminobutyric acid
- KD
binding affinity constant
- LPS
lipopolysaccharide
- MeOH
methanol
- MIC
minimum inhibitory concentration
- MDR
multi-drug resistant
- OM
outer membrane
- SNR
structure-nephrotoxicity relationships
- TFA
trifluoroacetic acid
Footnotes
The authors declare no competing financial interest.
References
- 1.Wernli D, Haustein T, Conly J, Carmeli Y, Kickbusch I, Harbarth S. A Call for Action: The Application of the International Health Regulations to the Global Threat of Antimicrobial Resistance. Plos Medicine. 2011;8(4) doi: 10.1371/journal.pmed.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lodato EMaK. World Health organization; 2013. W Background Paper 6.1 Antimicrobial resistance. [Google Scholar]
- 3.Ventola CL. The antibiotic resistance crisis: part 2: management strategies and new agents. P T. 2015;40(5):344–352. [PMC free article] [PubMed] [Google Scholar]
- 4.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 5.The bacterial challenge - time to react. Ejhp Practice. 2009;15(5):15–15. [Google Scholar]
- 6.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 7.Cars O, Hogberg LD, Murray M, Nordberg O, Sivaraman S, Lundborg CS, So AD, Tomson G. Meeting the challenge of antibiotic resistance. BMJ. 2008;337:a1438. doi: 10.1136/bmj.a1438. [DOI] [PubMed] [Google Scholar]
- 8.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. Pharmacology of polymyxins: new insights into an 'old' class of antibiotics. Future microbiol. 2013;8(6):711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 10.Landman D, Georgescu C, Martin DA, Quale J. Polymyxins revisited. Clin Microbiol Rev. 2008;21(3):449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akajagbor DS, Wilson SL, Shere-Wolfe KD, Dakum P, Charurat ME, Gilliam BL. Higher incidence of acute kidney injury with intravenous colistimethate sodium compared with polymyxin B in critically ill patients at a tertiary care medical center. Clin. Infect. Dis. 2013;57:1300–1303. doi: 10.1093/cid/cit453. [DOI] [PubMed] [Google Scholar]
- 12.Hartzell JD, Neff R, Ake J, Howard R, Olson S, Paolino K, Vishnepolsky M, Weintrob A, Wortmann G. Nephrotoxicity associated with intravenous colistin (colistimethate sodium) treatment at a tertiary care medical center. Clin. Infect. Dis. 2009;48:1724–1728. doi: 10.1086/599225. [DOI] [PubMed] [Google Scholar]
- 13.Kubin CJ, Ellman TM, Phadke V, Haynes LJ, Calfee DP, Yin MT. Incidence and predictors of acute kidney injury associated with intravenous polymyxin B therapy. J. Infect. 2012;65(1):80–87. doi: 10.1016/j.jinf.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Coulthard K, Milne R, Nation RL, Conway S, Peckham D, Etherington C, Turnidge J. Steady-state pharmacokinetics of intravenous colistin methanesulphonate in patients with cystic fibrosis. J Antimicrob Chemother. 2003;52(6):987–992. doi: 10.1093/jac/dkg468. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Milne RW, Nation RL, Turnidge JD, Smeaton TC, Coulthard K. Pharmacokinetics of colistin methanesulphonate and colistin in rats following an intravenous dose of colistin methanesulphonate. J Antimicrob Chemother. 2004;53(5):837–840. doi: 10.1093/jac/dkh167. [DOI] [PubMed] [Google Scholar]
- 16.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin. Infect. Dis. 2013;57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 17.Azad MA, Roberts KD, Yu HH, Liu B, Schofield AV, James SA, Howard DL, Nation RL, Rogers K, de Jonge MD, Thompson PE, Fu J, Velkov T, Li J. Significant accumulation of polymyxin in single renal tubular cells: a medicinal chemistry and triple correlative microscopy approach. Anal Chem. 2015;87(3):1590–1595. doi: 10.1021/ac504516k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun B, Azad MA, Nowell CJ, Nation RL, Thompson PE, Roberts KD, Velkov T, Li J. Cellular Uptake and Localization of Polymyxins in Renal Tubular Cells Using Rationally Designed Fluorescent Probes. Antimicrob Agents Chemother. 2015;59(12):7489–7496. doi: 10.1128/AAC.01216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun B, Azad MA, Wang J, Nation RL, Thompson PE, Roberts KD, Velkov T, Li J. Imaging the distribution of polymyxins in the kidney. J Antimicrob Chemother. 2014;70(8):827–829. doi: 10.1093/jac/dku441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poudyal A, Howden BP, Bell JM, Gao W, Owen RJ, Turnidge JD, Nation RL, Li J. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother. 2008;62(6):1311–1318. doi: 10.1093/jac/dkn425. [DOI] [PubMed] [Google Scholar]
- 21.Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Jacob J, Sidjabat HE, Paterson DL, Nation RL, Li J. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother. 2011;55(12):5685–5695. doi: 10.1128/AAC.05298-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother. 2012;56(10):5103–5112. doi: 10.1128/AAC.01064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Y, Chai D, Wang R, Liang B, Bai N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother. 2012;67(7):1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases. 2015;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 25.Paterson DL, Harris PNA. Colistin resistance: a major breach in our last line of defence. The Lancet Infectious Diseases. 2015 doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 26.Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. The Lancet Infectious Diseases. 2016:S1473–S3099. doi: 10.1016/S1473-3099(16)00006-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Velkov T, Thompson PE, Nation RL, Li J. Structure--activity relationships of polymyxin antibiotics. J Med Chem. 2010;53(5):1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azad MA, Finnin BA, Poudyal A, Davis K, Li J, Hill PA, Nation RL, Velkov T. Polymyxin B induces apoptosis in kidney proximal tubular cells. Antimicrob Agents Chemother. 2013;57(9):4329–4335. doi: 10.1128/AAC.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azad MA, Akter J, Rogers KL, Nation RL, Velkov T, Li J. Major pathways of polymyxin-induced apoptosis in rat kidney proximal tubular cells. Antimicrob Agents Chemother. 2015;59(4):2136–2143. doi: 10.1128/AAC.04869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appelmelk BJ, Su D, Verweij-van Vught AM, Thijs BG, MacLaren DM. Polymyxin B-horseradish peroxidase conjugates as tools in endotoxin research. Anal Biochem. 1992;207(2):311–316. doi: 10.1016/0003-2697(92)90017-2. [DOI] [PubMed] [Google Scholar]
- 31.Velkov T, Roberts KD, Nation RL, Wang J, Thompson PE, Li J. Teaching 'old' polymyxins new tricks: new-generation lipopeptides targeting Gram-negative 'superbugs'. ACS Chem. Biol. 2014;16(9):1172–1117. doi: 10.1021/cb500080r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Visser PC, Kriek NM, van Hooft PA, Van Schepdael A, Filippov DV, van der Marel GA, Overkleeft HS, van Boom JH, Noort D. Solid-phase synthesis of polymyxin B1 and analogues via a safety-catch approach. J Pept Res. 2003;61(6):298–306. doi: 10.1034/j.1399-3011.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 33.O'Dowd H, Kim B, Margolis P, Wang W, Wu C, Lopez SL, Blais J. Preparation of tetra-Boc-protected polymyxin B nonapeptide. Tetrahedron Letters. 2007;48(11):2003–2005. [Google Scholar]
- 34.Okimura K, Ohki K, Sato Y, Ohnishi K, Sakura N. Semi-synthesis of polymyxin B (2–10) and colistin (2–10) analogs employing the Trichloroethoxycarbonyl (Troc) group for side chain protection of alpha,gamma-diaminobutyric acid residues. Chem Pharm Bull (Tokyo) 2007;55(12):1724–1730. doi: 10.1248/cpb.55.1724. [DOI] [PubMed] [Google Scholar]
- 35.Okimura K, Ohki K, Sato Y, Ohnishi K, Uchida Y, Sakura N. Chemical Conversion of Natural Polymyxin B and Colistin to Their N-Terminal Derivatives. Bulletin of the Chemical Society of Japan. 2007;80(3):543–552. [Google Scholar]
- 36.Sakura N, Itoh T, Uchida Y, Ohki K, Okimura K, Chiba K, Sato Y, Sawanishi H. The Contribution of the N-Terminal Structure of Polymyxin B Peptides to Antimicrobial and Lipopolysaccharide Binding Activity. Bulletin of the Chemical Society of Japan. 2004;77(10):1915–1924. [Google Scholar]
- 37.Chihara S, Yahata M, Tobita T, Koyama Y. Chemical Synthesis and Characterization of n-Fattyacyl Mono-Aminoacyl Derivatives of Colistin Nonapeptide. Ag Biol Chem. 1974;38:1767–1777. [Google Scholar]
- 38.Chihara S, Yahata M, Tobita T, Koyama Y. Chemical Synthesis, Isolation and Characterization of α-N-Fattyacyl Colistin Nonapeptide with Special Reference to the Correlation between Antimicrobial Activity and Carbon Number of Fattyacyl Moiety. Ag Biol Chem. 1974;38:521–529. [Google Scholar]
- 39.Tsubery H, Ofek I, Cohen S, Fridkin M. N-terminal modifications of Polymyxin B nonapeptide and their effect on antibacterial activity. Peptides. 2001;22(10):1675–1681. doi: 10.1016/s0196-9781(01)00503-4. [DOI] [PubMed] [Google Scholar]
- 40.Vaara M. The outer membrane permeability-increasing action of linear analogues of polymyxin B nonapeptide. Drugs Exp Clin Res. 1991;17(9):437–443. [PubMed] [Google Scholar]
- 41.Magee TV, Brown MF, Starr JT, Ackley DC, Abramite JA, Aubrecht J, Butler A, Crandon JL, Dib-Hajj F, Flanagan ME, Granskog K, Hardink JR, Huband MD, Irvine R, Kuhn M, Leach KL, Li B, Lin J, Luke DR, MacVane SH, Miller AA, McCurdy S, McKim JM, Jr, Nicolau DP, Nguyen TT, Noe MC, O'Donnell JP, Seibel SB, Shen Y, Stepan AF, Tomaras AP, Wilga PC, Zhang L, Xu J, Chen JM. Discovery of Dap-3 polymyxin analogues for the treatment of multidrug-resistant Gram negative nosocomial infections. J Med Chem. 2013;56(12):5079–5093. doi: 10.1021/jm400416u. [DOI] [PubMed] [Google Scholar]
- 42.Deris ZZ, Swarbrick JD, Roberts KD, Azad MA, Akter J, Horne AS, Nation RL, Rogers KL, Thompson PE, Velkov T, Li J. Probing the penetration of antimicrobial polymyxin lipopeptides into Gram-negative bacteria. Bioconjug. Chem. 2014;25(4):750–760. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azad MA, Huang JX, Cooper MA, Roberts KD, Thompson PE, Nation RL, Li J, Velkov T. Structure-activity relationships for the binding of polymyxins with human alpha-1-acid glycoprotein. Biochem Pharmacol. 2012;84(3):278–291. doi: 10.1016/j.bcp.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meredith JJ, Dufour A, Bruch MD. Comparison of the structure and dynamics of the antibiotic peptide polymyxin B and the inactive nonapeptide in aqueous trifluoroethanol by NMR spectroscopy. J Phys Chem B. 2009;113(2):544–551. doi: 10.1021/jp808379x. [DOI] [PubMed] [Google Scholar]
- 45.Pristovsek P, Kidric J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J Med Chem. 1999;42(22):4604–4613. doi: 10.1021/jm991031b. [DOI] [PubMed] [Google Scholar]
- 46.Pristovsek P, Kidric J. The search for molecular determinants of LPS inhibition by proteins and peptides. Curr Top Med Chem. 2004;4(11):1185–1201. doi: 10.2174/1568026043388105. [DOI] [PubMed] [Google Scholar]
- 47.Tsubery H, Ofek I, Cohen S, Fridkin M. Structure-activity relationship study of polymyxin B nonapeptide. Adv. Exp. Med. Biol. 2000;479:219–222. doi: 10.1007/0-306-46831-X_18. [DOI] [PubMed] [Google Scholar]
- 48.Vaara M, Siikanen O, Apajalahti J, Fox J, Frimodt-Moller N, He H, Poudyal A, Li J, Nation RL, Vaara T. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob Agents Chemother. 2010;54(8):3341–3346. doi: 10.1128/AAC.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaara M, Vaara T. The novel polymyxin derivative NAB739 is remarkably less cytotoxic than polymyxin B and colistin to human kidney proximal tubular cells. Int J Antimicrob Agents. 2013;41(3):292–293. doi: 10.1016/j.ijantimicag.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Vaara M, Fox J, Loidl G, Siikanen O, Apajalahti J, Hansen F, Frimodt-Moller N, Nagai J, Takano M, Vaara T. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob. Agents Chemother. 2008;52(9):3229–3236. doi: 10.1128/AAC.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velkov T, Thompson PE, Nation RL, Li J. Structure-activity relationships of polymyxin antibiotics. J. Med. Microbiol. 2010;53(5):1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voitenko VG, Bayramashvili DI, Zebrev AI, Zinchenko AA. Relationship between structure and histamine releasing action of polymyxin B and its analogues. Agents Actions. 1990;30(1–2):153–156. doi: 10.1007/BF01969025. [DOI] [PubMed] [Google Scholar]
- 53.Pristovsek P, Kidric J. The search for molecular determinants of LPS inhibition by proteins and peptides. Curr. Top. Med. Chem. 2004;4(11):1185–1201. doi: 10.2174/1568026043388105. [DOI] [PubMed] [Google Scholar]
- 54.Hancock RE. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5(1):37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 55.Hancock RE. Peptide antibiotics. Lancet. 1997;349(9049):418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 56.Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16(2):82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 57.Li J, Milne RW, Nation RL, Turnidge JD, Coulthard K. Stability of colistin and colistin methanesulfonate in aqueous media and plasma as determined by high-performance liquid chromatography. Antimicrob. Agents Chemother. 2003;47(4):1364–1370. doi: 10.1128/AAC.47.4.1364-1370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts KD, Azad MAK, Wang J, Horne AS, Thompson PE, Nation RL, Velkov T, Li J. Antimicrobial Activity and Toxicity of the Major Lipopeptide Components of Polymyxin B and Colistin: Last-Line Antibiotics against Multidrug-Resistant Gram-Negative Bacteria. ACS Infectious Diseases. 2015;1(11):568–575. doi: 10.1021/acsinfecdis.5b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wayne PA. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Twenty-third informational Supplement M100-S23. 2013 [Google Scholar]