Abstract
Deletions on chromosome 22q11.2 are a strong genetic risk factor for development of schizophrenia and cognitive dysfunction. We employed shotgun liquid chromatography–mass spectrometry (LC-MS) proteomic and metabonomic profiling approaches on prefrontal cortex (PFC) and hippocampal (HPC) tissue from Df(16)A+/− mice, a model of the 22q11.2 deletion syndrome. Proteomic results were compared with previous transcriptomic profiling studies of the same brain regions. The aim was to investigate how the combined effect of the 22q11.2 deletion and the corresponding miRNA dysregulation affects the cell biology at the systems level. The proteomic brain profiling analysis revealed PFC and HPC changes in various molecular pathways associated with chromatin remodelling and RNA transcription, indicative of an epigenetic component of the 22q11.2DS. Further, alterations in glycolysis/gluconeogenesis, mitochondrial function and lipid biosynthesis were identified. Metabonomic profiling substantiated the proteomic findings by identifying changes in 22q11.2 deletion syndrome (22q11.2DS)-related pathways, such as changes in ceramide phosphoethanolamines, sphingomyelin, carnitines, tyrosine derivates and panthothenic acid. The proteomic findings were confirmed using selected reaction monitoring mass spectrometry, validating decreased levels of several proteins encoded on 22q11.2, increased levels of the computationally predicted putative miR-185 targets UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110 kDa subunit (OGT1) and kinesin heavy chain isoform 5A and alterations in the non-miR-185 targets serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform, neurofilament light chain and vesicular glutamate transporter 1. Furthermore, alterations in the proteins associated with mammalian target of rapamycin signalling were detected in the PFC and with glutamatergic signalling in the hippocampus. Based on the proteomic and metabonomic findings, we were able to develop a schematic model summarizing the most prominent molecular network findings in the Df(16)A+/− mouse. Interestingly, the implicated pathways can be linked to one of the most consistent and strongest proteomic candidates, (OGT1), which is a predicted miR-185 target. Our results provide novel insights into system-biological mechanisms associated with the 22q11DS, which may be linked to cognitive dysfunction and an increased risk to develop schizophrenia. Further investigation of these pathways could help to identify novel drug targets for the treatment of schizophrenia.
Introduction
Hemizygous deletions in chromosome 22q11.2 occur predominately de novo and cause a deletion syndrome (22q11.2DS), characterized by a broad spectrum of physical and mental manifestations with a variable phenotype. The syndrome has an incidence of 1 in 2000–4000 live births. The physical phenotype is characterized by variably penetrant craniofacial and cardiovascular anomalies, immunodeficiency, short stature and hypocalcaemia. Furthermore, deletion carriers typically present with a range of behavioural and cognitive deficits and 25–30% develop schizophrenia during adolescence or early adulthood.1 22q11.2 deletions account for 1–2% of sporadic schizophrenia cases, exemplifying the role of rare mutations in disease susceptibility. Importantly, there are no major clinical differences in the core schizophrenia phenotype between individuals with schizophrenia who are 22q11.2 microdeletion carriers and those with an idiopathic disease onset.2 A 1.5-Mb critical region, which confers haplo-insufficiency of 27 genes,3, 4 including the candidate schizophrenia susceptibility genes (proline dehydrogenase (PRODH), catechol-O-methyltransferase (COMT) and DHHC palmitoyltransferase (ZDHCC8)), is largely conserved on mouse chromosome 16. This has facilitated generation of mouse models of the human 22q11DS, carrying hemizygous deletions in the syntenic region of chromosome 16. One of the best characterized models is the Df(16)A+/− mouse, which carries a hemizygous 1.3-Mb chromosomal deficiency on chromosome 16 (Df(16)A), encompassing 27 genes, which is syntenic to the minimal 1.5-Mb human 22q11.2 deletion.5 These mice develop deficits in spatial working memory, sensorimotor gating and fear conditioning as well as presenting with hyperactive behaviour.5, 6 In addition, they show abnormalities in synaptic connectivity of hippocampal and cortical neurons, including a lower density of dendritic spines and glutamatergic synapses and altered synaptic plasticity.7
Studies that have examined the effect of Df(16)A on transcriptional networks revealed downregulation of microRNA (miRNA) transcripts,5, 8 resulting from the combined effect of two disrupted genes (Dgcr8 and miR-185) in the critical region. Dgcr8 hemizygosity leads to dysregulation in the production of up to 20% of all miRNAs, while miR-185 levels are reduced by 70–80% owing to a combined effect with the Dgcr8 hemizygosity,9 which leads to an impaired maturation of the pri-miR-185 transcript. Based on the transcriptional profiling, the most robust miRNA target was a novel neuronal regulatory gene 2310044H10Rik/Mirta22, which was confirmed to be specific for miR-185 and has been followed up in functional studies. It was shown that this gene encodes a protein, which is located in the Golgi apparatus and in vesicles and tubular-like extensions of dendrites, mediating at least some of the effects of the 22q11.2 deletions on dendrite and spine formation.10
To date, no other direct miRNA targets have been reported for the 22q11.2DS and it is still not known how the reported transcriptomic5, 11 alterations affect cell biology at the proteomic and metabonomic level. In the disease pathology, proteomic and metabonomic networks are likely to be regulated through the combined interactions of the 22q11.2 deletion gene products, which involve the hemizygosity of four transcription factors and disruption of miRNA-regulated targets. This may involve dysregulation of other transcription factors, chromosome remodelling elements or changes in the expression of signalling proteins. Also, secondary compensation effects might have a role. Here we have carried out a combined shotgun proteomic and metabonomic profiling analysis of brain tissue from Df(16)A+/− mice to investigate the combined effects of miRNA dysregulation and disruption of genes residing in the 22q11.2 region. Our results provide novel insights towards a better understanding of the molecular pathophysiology of psychiatric disorders and cognitive dysfunction.
Materials and methods
For more detailed information, please refer to Supplementary Methods. A flowchart of the molecular profiling methods can be found in Supplementary Figure S1.
Animals
The Df(16)A+/− mice were produced and bred at Columbia University (New York, NY, USA) as described previously.5 Eight-week-old male mice were used for the analyses. Mutant mice carry a hemizygous 1.3-Mb chromosomal deficiency (Df(16)A+/−), which ranges from Dgcr2 to the Hira gene and encompasses a segment syntenic to the 1.5-Mb human 22q11.2 deletion, including 27 protein-coding genes. All animal procedures were performed according to protocols approved by the Columbia University Institutional Animal Care and Use Committee under federal and state regulations.
Multiplex immunoassay profiling
Serum samples from 11 Df(16)A+/− and 10 wild-type mice (cohort 1) were analysed randomized and blinded using a multianalyte profiling platform, comprising multiplexed immunoassays for 58 analytes (Supplementary Table S1) in a Clinical Laboratory Improved Amendments (CLIA)-certified laboratory at Myriad-RBM (Austin, TX, USA) as described previously.12, 13 Data quality was assessed via principal component analysis (PCA), data were checked for normality and significance analysis of mircroarray14 was performed.
Metabonomics
Metabonomic sample preparation and profiling of prefrontal cortical (PFC) and hippocampal (HPC) tissues from 11 Df(16)A+/− and 10 wild-type mice (cohort 1) was performed as described previously.15 In brief, tissue samples were prepared using a two-step process based on extraction into ‘aqueous' and ‘organic' phases for polar and nonpolar metabolites. Quality-control samples were prepared by combining an aliquot from each study sample to produce a representative sample, which was used for column conditioning and data quality assessment as described by Want et al.15 Aqueous extracts were analysed via nano-ultra-performance liquid chromatography–mass spectrometry (UPLC-MS) analysis using a Waters XEVO G2 Q-TOF mass spectrometer coupled online to an Acquity UPLC-MS system (Waters, Milford, MA, USA). Metabolites were separated using a 2.1 × 100 mm2 (1.7 μm) HSS T3 Acquity column, and acquisition was performed in both positive ion mode and negative ion mode. Quality-control samples were injected 10 times at the start of the analytical batch in order to condition the column, and then after every 10 samples throughout the run to assess instrument stability. Samples were run randomized and blinded. Data were processed using the freeware XCMS16, 17 using standard parameters. The output consisted of a matrix of metabolite feature m/z, retention time and intensity values, which was imported into SIMCA-P for multivariate analysis to check data quality and sample outliers. Data were normalized, filtered and checked for normality prior statistical analysis. P-values were determined using Wilcoxon's signed-rank test and corrected to control for multiple hypothesis testing (Benjamini–Hochberg).18 Ratios were calculated for each analyte as the mean intensity values of Df(16)A+/− mice divided by those of wild-type littermates.
Proteomics
For more detailed information, please refer to Supplementary Methods.
Sample preparation and study design
PFC and HPC brain tissues from Df(16)A+/− and wild-type mice were prepared using total lysis protein extraction19, 20 combined with either in-solution (two independent cohorts; cohort 1 (PFC and HPC): 11 Df(16)A+/− vs 10 wild-type mice, cohort 2 (PFC): 13 Df(16)A+/− vs 13 wild-type mice) or in-gel tryptic digestion ((PFC): 20 Df(16)A+/− and 21 wild-type mice, combined from both cohorts to increase statistical power, proteome coverage and avoid sample extraction biases). Samples were subsequently analysed using label-free LC-MSE mass spectrometry, enabling unbiased protein identification and quantification. Mass spectrometrical sample preparation, label-free LC-MSE analysis and selected reaction monitoring (SRM) analysis were performed blinded and randomized. Sample size was chosen based on previous experiments using FC and HPC brain tissues and proteomic guidelines. In all, 8–10 animals per group are sufficient for the proteomic shotgun discovery phase. Power calculations using MSstats showed that to achieve a fold change of 15% with 0.8 power 10 sample are needed. The validity of the results was further proven in 20 Df(16)A+/− and 20 wild-type mice using highly sensitive and robust label-based SRM mass spectrometry.
Label-free LC-MSE analysis of PFC and HPC tissue
Brain tissue samples were analysed individually in technical duplicates using a splitless UPLC (10 kpsi nanoAcquity; Waters) coupled online to a Waters Q-TOF Premier mass spectrometer. Data were acquired in expression mode (MSE). The procedure, quality assessment and data processing were performed as described previously.21 LC-MSE data were processed using the ProteinLynx Global Server (PLGS) v.2.4. (Waters) and Rosetta Elucidator v.3.3 (Rosetta Biosoftware, Seattle, WA, USA) was used for time and mass/charge alignment of mass spectrometric data. The Swiss-Prot mouse reference (March, 2013) proteome was used for protein identification searches. Only peptides detected in both replicates and in >80% of samples were included in further analysis. Protein abundance changes were determined using the MSstats package22 based on linear mixed-effects models, which views peptides mapped to the same protein as replicate measurements of protein abundance, following log2 transformation and exclusion of peptide intensity values that were >3 s.d. from the mean of each group. The MSstats package included quality assessment and PCA on the raw data was carried out to detect data outliers. The P-values were adjusted to control the false-discovery rate at a cutoff of 0.05 following the Benjamini–Hochberg procedure.23
Label-based SRM mass spectrometry
Digested FC and HPC proteomes (20 Df(16)A+/− and 20 wild-type mice), prepared using in-solution digestion from total lysates, were analysed using targeted SRM mass spectrometry on a Xevo TQ-S mass spectrometer (Waters) coupled to a nanoAcquity UPLC system (Waters) as described previously.13, 20, 24 Multiplex SRM assays were developed using a high-throughput strategy.25 Physiochemical criteria for selecting tryptic peptides were based on peptide count, uniqueness and quality of transitions. Transitions were selected based on software internal predictions, discovery proteomics data and spectral data (NIST spectral libraries26) and calculated using Skyline version 1.2.0.3425.27 Quantitative SRM measurements comparing abundance levels of 40 proteins between Df(A)16+/− mice and wild-type littermates were performed in scheduled SRM acquisition mode. Heavy isotope labelled peptide versions (JPT Peptide Technologies, Berlin, Germany) were spiked in the peptide mixture for accurate quantification and identification. For each peptide, at least three transitions were monitored for the heavy and light versions. The final transitions can be found in Supplementary Table S2. Samples were run blinded, randomized and blocked28 in triplicates, and blanks and quality-control peptide injections (yeast alcohol dehydrogenase)24 were run alternating after each biological replicate. Resulting SRM data were analysed using Skyline and statistical analysis, testing for differential abundance among Df(A)16+/− and wild-type mice, was conducted using SRMstats23 with the ‘expanded technical replication' setting. Data quality was checked using quality plots in the MSstats package and by employing PCA on the raw data. P-values were corrected to control for multiple hypothesis testing (Benjamini–Hochberg).18
Results
Serum characterization—quantitative serum immunoassay profiling
Immunodeficiency is one of the key features of the 22q11.2DS, but the degree and spectrum of severity varies widely. A large number of studies have provided evidence suggestive of an immunological component of schizophrenia29 and other psychiatric disorders.30, 31 We hypothesized that immune dysfunction, as reflected in the levels of immune and metabolic circulation markers, may contribute to the elevated risk of schizophrenia in the 22q11.2DS.32 Analysis of 58 such markers (Supplementary Table S1) in the serum of the Df(16)A+/− mouse using a multiplex immunoassay platform resulted in the identification of a significant decrease in matrix metalloproteinase 9 (MMP-9; ratio=0.74, P=0.039, P*=0.20) in the Df(16)A+/− (n=11) compared with wild-type (n=10) mice.
Brain characterization—quantitative proteomics
In the proteomic discovery phase, shotgun profiling of PFC and HPC tissue was carried out to identify proteins differentially expressed in the Df(16)A+/− mouse model compared with wild-type mice. The main aim was to identify affected protein networks that might contribute to the development of psychiatric and cognitive phenotypes in the 22q11.2DS, as well as novel miR-185 targets. We focused on the PFC and HPC because these brain regions have previously been characterized at the transcriptomic level5, 11 and are implicated in the pathology of various psychiatric disorders. Special emphasis was placed on the PFC as the most extensively characterized brain region in the Df(16)A+/− mouse.5, 11 Total lysis extracts from PFC tissues of two independent cohorts of Df(16)A+/− mice (11 Df(16)A+/− vs 10 wild-type / 13 Df(16)A+/− vs 13 wild-type) were analysed, as well as from the HPC of one of the cohorts (11 Df(16)A+/− vs 10 wild-type). Additionally, PFC tissue of both cohorts were combined (n=40) and prepared with a different gel-based procedure to increase proteome coverage and statistical power. Shotgun profiling analysis resulted in the identification of 578 distinct proteins (9605 peptides) in the PFC (cohort 1) of which 59 proteins showed significant genotypic differences (P*<0.05). Likewise, 715 distinct proteins (14 334 peptides) (cohort 2) were identified in cohort 2, with 186 proteins showing abundance differences between genotypes. In total, 552 distinct proteins (11 193 peptides) were identified in the combined cohorts and 104 of these had significantly changed protein levels. In the HPC, we identified 570 distinct proteins (8934 peptides) of which 63 were significantly different in the mutant mice (Supplementary Table S3).
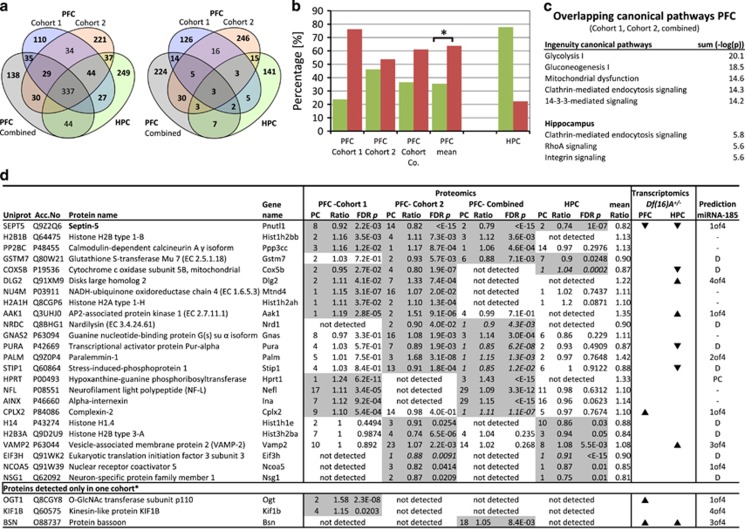
In order to identify robust alterations of Df(16)A+/− hemizygosity, we cross-compared all proteomic profiling studies among each other and to previous transcriptomic profiling studies5, 11 of the Df(16)A+/− mice (Figure 1a). Most consistently, we found an average decrease of 20% in septin 5 levels (Figure 1d), a protein encoded in the 22q11.2DS critical region. In the case of other 22q11.2DS encoded proteins, we detected a reduction in the levels of protein DGCR14 (ES2EL) (ratio=0.85, P=8.2 × 10−6, 1 peptide) in one of the proteomic cohorts. We further detected a 30% increase in the levels of the hypoxanthine-guanine phosphoribosyltransferase (HPRT) in the Df(16)A+/− mice. Two mini-cassettes of the Hprt gene are part of the viral transfection vector where they serve as a positive selection marker for successful recombination and deletion of the 22q11.2 critical region. The identified changes in these proteins substantiate the technical robustness of our proteomic platform. All other 22q11.2DS-encoded proteins, which could serve as positive controls, were below the detection limit, probably owing to low or absent expression in the brain.33
Figure 1.
(a) Venn diagrams of the overlap of all identified proteins (left) and of all significantly altered proteins (right) identified in label-free LC-MSE (liquid chromatography–mass spectrometry) proteomic profiling studies of Df(16)A+/− mice compared with wild-type mice (Supplementary Table S4 for full list). (b) Percentages of significantly increased (red) and decreased (green) proteins in Df(16)A+/− mice across all proteomic studies. Mean percentages show significantly increased protein levels in the prefrontal cortex (PFC; p<0.05, t-test). PFC combined are samples from both cohorts combined and in-gel digested (see Material and methods). As the hippocampal (HPC) was only investigated from one cohort, significant levels could not be established. (c) Top ingenuity pathway analysis: altered pathways in the PFC and HPC. (d) Detailed information of the overlapping proteins identified as significantly changing across all proteomic screens. Proteins were sorted by the number of proteomic studies in which they have been detected as significantly altered. Only proteins with a mean ratio of >1.1 or <0.9 across all identified studies are displayed. P-values were determined using MSstats and corrected to control for multiple hypothesis testing after Benjamini–Hochberg.114 (Supplementary Table S3 for full information). Proteins identified by one peptide (italic) were included if they were overlapping between the proteomic screens. Results were compared with the transcriptomic results using Affymetrix chips published by Stark et al.5 and Feneleon et al.11 Stark reported 716 transcripts altered in the FC and 85 transcripts in the HPC (false-discovery rate=0.01). Significantly changed transcripts are marked with an ▴ and ▾ according to their reported fold change direction in comparison to wild-type mice. Four different bioinformatic miRNA target prediction tools were used to assess if the upregulated proteins are potential Mir-185 targets. Xof4, predicted in X of the four predictions; D, downregulation; PC, positive control.
Other identified proteins, which showed alterations in abundance, are either primary or secondary effects of abnormal miRNA metabolism or hemizygosity of 22q11.2 genes. For example, we identified a decrease in glutathione S-transferase Mu7 (GSTM7) in three of the proteomic studies, which could be a secondary effect of the Txnrd2 (thioredoxin reductase 2) hemizygosity. Both proteins have been implicated in the regulation of the intracellular redox environment.34, 35, 36, 37 Furthermore, we discovered robust decreases of mitochondrial proteins (cytochrome c oxidase subunit 5B, mitochondrial (COX5B), NADH-ubiquinone oxidoreductase chain 4 (NU4M)). A possible reason for these alterations is the involvement of several 22q11.2 genes (Prodh, Comt, Txtp, Slc25a1) in mitochondrial function. Indeed, using the IPA software, we confirmed that ‘mitochondrial dysfunction' was a common pathway affected across all profiling studies (Figure 1c).
In order to identify proteins that might be upregulated owing to miR-185 hemizygosity, we analysed all upregulated detected proteins using bioinformatic miRNA target prediction tools (MirDB, Targetscan, MirBase, DIANA), which were based on thermodynamics (DIANA), seed complementarity (Targetscan) or support vector machines (MirDB). Potential miR-185 targets are disk large homolog 2 (DLG2), AP2-associated protein kinase 1 (AAK1), sodium/potassium-transporting ATPase subunit alpha-3 (AT1A3), O-GlcNAc transferase subunit p110 (OGT1) and protein bassoon (BSN), which were found to be upregulated at the proteomic and transcriptomic levels.5
We were not able to identify a significant increase in sarcoplasmatic/endoplasmic reticulum calcium ATPase 2 (SERCA2—Atp2a2), another recently proposed target of 22q11.2DS-associated miRNA dysregulation,38 in any of our proteomic screens. This is consistent with Fenelon et al.,11 who failed to confirm a change in the transcript and protein levels of Atp2a2.
Remarkably, the differentially expressed proteins showed an overall increase in abundance levels across all FC proteomic studies (Figure 1b), which is likely owing to a decrease in about 20% of the miRNAs5 (including miR-185) and subsequent upregulation of targets. This finding is consistent with the transcriptomic profiling study by Fenelon et al.11 On the other hand, the HPC proteome showed a trend towards downregulation of differentially expressed proteins. Because of the lack of replication studies, further confirmation of the opposing changes in the HPC is needed. Nevertheless, the results are consistent with reported miRNA changes in PFC and HPC identified through transcriptomic profiling, as nearly twice the number of miRNAs were found to be decreased in the PFC (59 miRNAs) than in the HPC (30 miRNAs).5
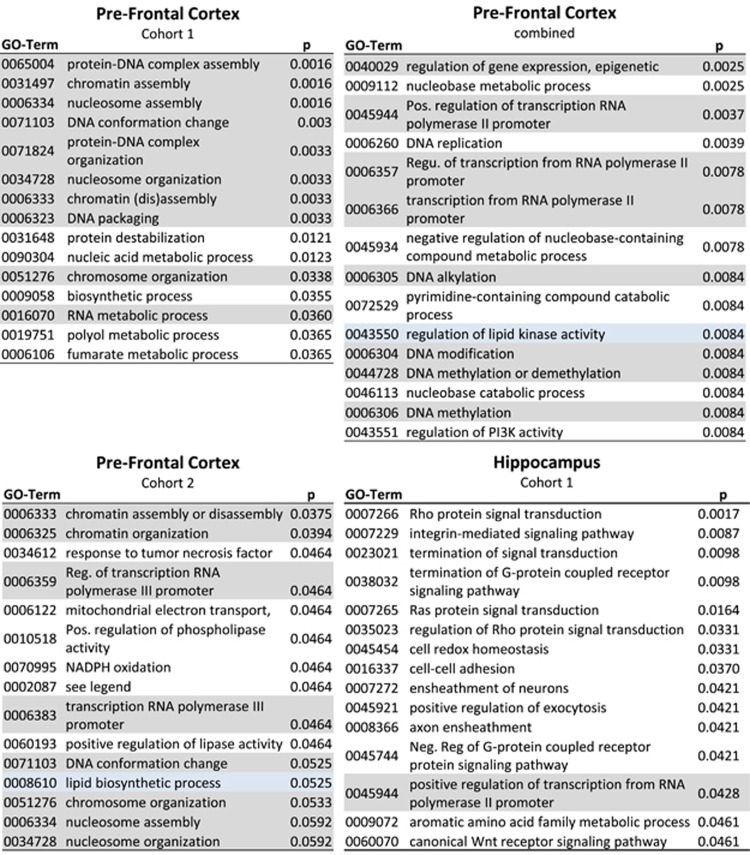
In silico analysis revealed an overrepresentation of developmentally regulated pathways related to nucleosome and chromatin assembly and chromatin-level regulation across all PFC proteomic studies (Figure 2). Further pathway analysis using the IPA software allowed us to identify overlaps between canonical pathways. The proteomic alterations were associated with the canonical pathways ‘glycolysis I', ‘gluconeogenesis' and ‘mitochondrial dysfunction' (Figure 1c) in the PFC and ‘Rho signalling' in the HPC, which was also reflected in the enrichment analysis.
Figure 2.
Significantly enriched pathways identified by gene set enrichment analysis using GOstats (proteins per pathway >3). The top 15 significant Gene Ontotlogy (GO) terms are displayed. GO terms falling into the category of chromosomal regulation are highlighted in grey. A full colour version of this figure is available at the Molecular Psychiatry journal online.
Brain characterization—quantitative metabonomics
We next carried out metabonomic profiling of PFC and HPC tissues from the Df(16)A+/− mouse in order to determine whether the above changes in proteins could be linked to alterations in metabolites. Therefore, after the initial identification of a metabolite candidate, the sample was re-run and the peaks were subjected to fragmentation in an MS/MS experiment. Identification of a specific metabolite was accomplished though comparison of the fragmentation pattern of a candidate metabolite with the known MS/MS fragmentation pattern in public and in-house databases. This analysis showed significant changes in 21 m/z-retention time pairs in the PFC, and we were able to identify 7 of these metabolites via MS/MS fragmentation. In the HPC, we detected 27 significantly changed m/z-retention time pairs, which we could match to 12 metabolites using metabolome databases and identified 4 via MS/MS fragmentation (Table 1, Supplementary Table S5). The most significant change in the PFC was a decrease in ceramide phosphoethanolamine (SP0302), which is a sphingomyelin analog. SP0302 may be linked to the hemizygosity of the ZDHHC8 palmitoyltransferase in the Df(16)A+/− mouse, as palmitoyl-CoA is a precursor in the ceramide synthesis. Alongside, further ceramides were found to be decreased in the PFC and S-palmitoyl-L-cysteine was decreased in the HPC. In the HPC, we also found that sphingomyelin (SM(d17:1/24:1) was the most significantly increased metabolite (ratio=1.72, P=0.031). We were able to validate sphingomyelin's databank identification using tandem MS/MS. We detected changes in further lipids such as phosphatidylcholines and glycerolipids, which reflect the changes in lipid metabolism found in the proteomic pathway analysis of Df(16)A+/− mice (Figure 2). In addition, several carnitines, which have a role in fatty acid metabolism, were found to be decreased (Table 1). Furthermore, proteomic abnormalities in aromatic amino acids could be confirmed via metabonomic profiling, detecting changes in different tyrosine derivates in both regions (N-stearoyl tyrosine and metyrosine). Although we were not able to validate the identification of these metabolites using MS/MS, their changes were reflected in the protein set enrichment analysis of the HPC ‘aromatic amino-acid family metabolic process'. A decrease in panthothenic acid—essential in the synthesis of coenzyme-A (CoA)—was also reflected in the proteomic pathway analysis with CoA biosynthesis being affected in cohort 1, where we found increased levels of two of the key enzymes in this pathway.
Table 1. Differentially altered metabolites identified by metabolic profiling of PFC and HPC brain tissue from Df(16)A +/− mice (11 vs 10).
| mz_Rt pair | Ion mode | HMDB/Chempub ID | Name | Class/function | Exact mass | Delta (mz−exact mass) | MS/MS | Ratio | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Frontal cortex | |||||||||
| 645.5_1323 | + | LMSP03020039 | PE-Cer(d14:2(4E,6E)/19:0) PE-Cer(d15:2(4E,6E)/18:0) | Ceramide phosphoethanolamines (SP0302) | 644.4893 | 6.5E-05 | ID | 0.64 | 0.0001 |
| 426.4_681 | + | HMDB06464 HMDB05065 | Elaidic carnitine Oleoylcarnitine | Acyl carnitine | 425.3505 | 4.5E-03 | ID | 0.59 | 0.0101 |
| 232.2_270 | + | HMDB02013 | Butyrylcarnitine | Acyl carnitine | 231.147 | 4.4E-03 | ID | 0.33 | 0.0133 |
| 448.3_667 | + | 74380333 123060515 | N-docosanoyl taurine N-stearoyl tyrosine | N-acyl amine N-acyl amine | 447.3382 447.3349 | 4.0E-04 3.7E-03 | NP | 0.70 | 0.0133 |
| 765.6_1179 | + | 123064881 | PG(O-20:0/16:0)/PG(O-16:0/20:0) | Glycerophosphoglycerol | 764.5931 | 1.6E-03 | ID | 0.69 | 0.0220 |
| 367.1_609 | + | 8439 | Salicin 6-phosphate | Glycoside phosphate | 366.0716 | 3.0E-03 | NP | 1.44 | 0.0350 |
| 650.4_1019 | + | 135642574 | PC(16:0/9:0(CHO)) | Phophatidylcholine | 649.4319 | 4.5E-03 | NP | 0.71 | 0.0350 |
| 530.5_1146 | + | - | Water loss from #16 (548.5, 1146sec) | — | — | — | ID | 0.58 | 0.0350 |
| 130_25 | + | HMDB33561 | 2-Acetyl-4,5-dihydrothiazole | — | 129.0248 | 1.4E-03 | NP | 0.70 | 0.0435 |
| 184.1_1095 | + | HMDB33141 | Fragment of a phosphatidylcholine | — | 183.0796 | 5.0E-04 | ID | 1.33 | 0.0435 |
| 550.6_1035 | + | LMSP00000005 | Cer(m18:1(4E)/18:0) | Ceramide/sphingolipid | 549.5485 | 0.00013 | ID | 0.55 | 0.0435 |
| 631.6_1302 | + | LMSP03020003 | PE-Cer(d14:1(4E)/18:1(9Z))/PE-Cer(d14:2(4E,6E)/18:0)/PE-Cer(d16:2(4E,6E)/16:0) | Ceramide phosphoethanolamines | 630.4737 | 0.00022 | ID | 0.68 | 0.0435 |
| 252.1_526 | + | HMDB00101 | Deoxyadenosine | Purine nucleosides and analogues | 251.101 | 0.00412 | NP | 2.62 | 0.0435 |
| 643.5_1269 | + | HMDB07312 | DG(18:3(9Z,12Z,15Z)/20:2(11Z,14Z)/0:0)(iso2) (several) | Diacylglycerol | 642.5223 | 9.8E-03 | NP | 0.60 | 0.0435 |
| Hippocampus | |||||||||
| 382.3_619 | + | 4266008 24701448 | Prostaglandin F2 alpha dimethyl amide 5,6-DiHETrE-EA (several) | Prostaglandin Endocannabinoid | 381.2879 | 5.8E-03 | NP | 0.39 | 0.0033 |
| 771.6_1472 | + | 123068775 123067209 | SM(d18:2/21:0) PA(O-20:0/22:2(13Z,16Z)) | Ceramide posphocholine Glycerophosphate | 770.6302 770.6189 | 4.5E-03 6.8E-03 | NP NP | 0.57 | 0.0229 |
| 359.2_661 | + | HMDB12983 | Kinetensin 1-3 | Peptide | 358.2329 | 5.9E-03 | NP | 0.56 | 0.031 |
| 799.7_1372 | + | 123068785 | SM(d17:1/24:1) or SM(d18:2/23:0) | Sphingomyelin | 798.6615 | 7.8E-03 | ID | 1.72 | 0.031 |
| 360.3_662 | + | — | S-palmitoyl-L-cysteine | Palmitoylated residue | 359.2494 | 5.7E-03 | NP | 0.59 | 0.031 |
| 140.1_1645 | + | 207572 5035 | 4-Amino-5-hydroxymethyl-2-methylpyrimidine L-Histidinal | Thiamin metabolism Histidin metabolism | 139.0746 | 1.7E-04 | NP | 0.35 | 0.042 |
| 545.3_725 | + | — | PC (no database match, Isotope of 544.3 (PC 20:4)) | Phophatidylcholine | — | — | T | 0.67 | 0.042 |
| 882.6_1302 | + | 123061384 | PC(22:5(4Z,7Z,10Z,13Z,16Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Phophatidylcholine | 881.5935 | 7.7E-03 | NP | 0.69 | 0.042 |
| 479.3_760 | — | — | Phospholipid—ID cannot be confirmed | — | — | — | T | 0.55 | 3E-05 |
| 218.1_300 | — | 149588 | Pantothenic acid | Vitamine | 219.1107 | 4.6E-05 | NP | 0.80 | 0.019 |
| 194.1_632 | — | HMDB14903 HMDB29217 | Metyrosine Tyrosine methylester | Tyrosine | 195.0895 | 2.3E-04 | NP | 0.61 | 0.023 |
| 566.3_758 | — | LMGP01050056 | PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0) | Glycerophosphocholine | 567.3325 | 3.2E-05 | ID | 1.30 | 0.023 |
| 567.3_758 | — | LMGP01050056 | Isotope of 566.3_758 | ||||||
| 480.3_746 | — | LMGP01060010 | PC(O-16:0/0:0) | Phosphatidylcholine | 481.3532 | −7.8E-05 | ID | 1.43 | 0.043 |
| 532.3_815 | — | LMSP02010055 | Cer(d14:2(4E,6E)/20:1(11Z)) | N-acylsphingosines (ceramides) | 533.4808 | -0.0003 | ID | 0.66 | 0.043 |
Abbreviations: HFC, hippocampus; ID, identified; MS/MS, tandem mass spectrometry; NP, cannot (dis)prove ID; PFC, prefrontal cortex; T, tentative, class indicated but not database match. Databank searches in METLIN, Lipidmaps, HMDB and MS/MS fragmentation were employed for metabolite identification.
Proteomic validation
We developed highly specific and sensitive targeted SRM assays in order to follow-up and validate alterations identified in the proteomic discovery phase. We assayed for potential miRNA targets (OGT1, ADA10) as well as altered proteins identified in the LC-MSE profiling approach (PP2BC, NRDC). The inclusion of the miR-185 targets was based on LC-MSE analysis, bioinformatic predictions and the shotgun proteomics analysis-derived in silico pathway analysis. We further included 6 of the 27 22q11.2 gene products to determine whether or not the mRNA changes are reflected at the protein level and could serve as potential positive controls. The vesicular glutamate transporter 1 (VGLU1) was included based on evidence obtained from the transcriptomic screen where VGLU1 was one of the three transcripts found to be significantly upregulated in both PFC and HPC.5 Unfortunately, we were not able to develop assays for DLG2 and BSN owing to assay design issues. We further extended the validation screen to include proteins of pathways that have been implicated in some of the major symptoms observed in 22q11.2DS. This included the mammalian target of rapamycin (mTOR) signalling pathway, which has been linked to autism-like behaviour, cognitive function and glutamatergic signalling. Details of the composition of the multiplex assay can be found in Table 2.
Table 2. Significantly changed proteins identified using label-based LC-SRM (targeted proteomics) in the PFC and HPC of the Df(16)A +/− mouse compared with wild-type mice (PFC=20 vs 20, HPC=11 vs 10).
|
Prefrontal cortex |
Hippocampus |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Proteomics |
Proteomics |
|||||||||||||||||
|
Discovery (D) |
Targeted proteomics (SRM) |
D |
Targeted proteomics (SRM) |
|||||||||||||||
| Transcriptomics | 10 vs 10 | 12 vs 10 | 20 vs 20 | 20 vs 21 | Transcriptomics | 10 vs 10 | 11 vs 10 | |||||||||||
| Uniprot | Acc. no. | Protein | Gene | Label | Ratio | P | P* | Ratio | P | P* | ||||||||
| 22q11.2 critical gene products+vector construct | ||||||||||||||||||
| Sep-05 | Q9Z2Q6 | Septin-5 | Sep-05 | 22q11.2 | ⇓ | ▾ | ▾ | ▾ | ▾ | 0.78 | 5.2E-11 | 1.1E-09 | ⇓ | ▾ | ▾ | 0.85 | 1.8E-04 | 4.9E-04 |
| RANG | P34022 | Ran-specific GTPase-activating protein | Ranbp1 | 22q11.2 | ⇓ | — | — | — | ▾ | 0.55 | <E-15 | <E-15 | ⇓ | — | ▾ | 0.47 | <E-15 | <E-15 |
| Q8JZU2 | Q8JZU2 | Protein Slc25a1 | Slc25a1 | 22q11.2 | ⇓ | — | — | — | ▾ | 0.79 | 4.8E-08 | 7.5E-07 | ⇓ | — | ▾ | 0.64 | 6.7E-10 | 5.9E-09 |
| PROD | Q9WU79 | Proline dehydrogenase 1, mitochondrial | Prodh | 22q11.2 | ⇓ | — | — | — | ▾ | 0.62 | 3.5E-07 | 4.4E-06 | ⇓ | — | ▾ | 0.90 | 0.26 | 0.30 |
| COMT | O88587 | Catechol O-methyltransferase | Comt | 22q11.2 | ⇓ | — | — | — | ▾ | 0.76 | 1.4E-06 | 1.4E-05 | ⇓ | — | ▾ | 0.71 | 1.6E-03 | 3.6E-03 |
| HPRT | P00493 | Hypoxanthine-guanine phosphoribosyltransferase | Hprt1 | Vector construct | * | ▴ | — | — | ▴ | 1.56 | <E-15 | <E-15 | — | — | ▾ | 1.77 | <E-15 | <E-15 |
| miRNA targets | ||||||||||||||||||
| GSK3B | Q9WV60 | Glycogen synthase kinase-3 beta | Gsk3b | miRNA target (2/4) | ⇑ | NS | — | ▾ | ▴ | 1.14 | 4E-06 | 3.2E-05 | NS | — | ▴ | 1.16 | 6.1E-04 | 1.5E-03 |
| OGT1 | Q8CGY8 | O-GlcNAc transferase subunit p110 | Ogt | miRNA target (1/4) + Figure 1d | ⇑ | ▴ | — | — | ▴ | 1.22 | 1.3E-02 | 3.5E-02 | NS | — | ▴ | 1.20 | 8.9E-03 | 1.6E-02 |
| KIF5A | P33175 | Kinesin heavy chain isoform 5A | Kif5a | miRNA target (1/4) | NS | — | — | ▴ | ▴ | 1.22 | 3.6E-02 | 0.09 | ⇑ | — | NS | 1.15 | 0.18 | 0.22 |
| A4 | P12023 | Amyloid beta A4 protein | App | miRNA target (1/4) | NS | — | — | — | NS | 1.13 | 6.5E-02 | 0.13 | NS | — | NS | 1.11 | 0.16 | 0.21 |
| KDM2A | P59997 | Lysine-specific demethylase 2A | Kdm2a | miRNA target (2/4) | NS | — | — | — | NS | 1.02 | 0.77 | 0.86 | NS | — | NS | 1.06 | 0.71 | 0.74 |
| RHOA | Q9QUI0 | Transforming protein RhoA | Rhoa | miRNA target, Liu et al.,115 | NS | NS | NS | NS | NS | 1.05 | 0.37 | 0.48 | NS | ▾ | NS | 1.08 | 0.05 | 0.08 |
| MBOA5 | Q91V01 | Lysophospholipid acyltransferase 5 | Lpcat3 | miRNA target (1/4) + Figure 1d | NS | — | — | — | NS | 1.09 | 0.13 | 0.21 | NS | — | NS | — | — | — |
| ADA10 | O35598 | Disintegrin metalloproteinase domain-containing protein 10 | Adam10 | miRNA target (3/4) | NS | (▴) | — | — | NS | 1.03 | 0.59 | 0.73 | NS | NS | NS | 1.04 | 0.696 | 0.7443 |
| ANFY1 | Q810B6 | Ankyrin repeat and FYVE domain-containing protein 1 | Ankfy1 | miRNA target (1/4) | NS | — | — | — | NS | 1.02 | 0.85 | 0.91 | NS | NS | NS | 1.22 | 0.05 | 0.07 |
| Discovery | ||||||||||||||||||
| PP2BC | P48455 | Serine/threonine-protein phosphatase 2B catalytic γ isoform | Ppp3cc | Figure 1d | NS | ▴ | ▴ | ▴ | ▴ | 1.06 | 1.1E-02 | 3.1E-02 | NS | NS | ▴ | 1.21 | 4.7E-10 | 4.8E-09 |
| PP2BB | P48453 | Serine/threonine-protein phosphatase 2B catalytic β isoform | Ppp3cb | NS | NS | NS | NS | ▴ | 1.16 | 5.8E-04 | 2.4E-03 | NS | NS | ▴ | 1.16 | 8.9E-03 | 1.6E-02 | |
| NFL | P08551 | Neurofilament light polypeptide (NF-L) | Nefl | Figure 1d | NS | ▴ | - | ▴ | ▴ | 1.09 | 1.8E-03 | 6.3E-03 | NS | NS | ▴ | 1.15 | 1.6E-04 | 4.9E-04 |
| AINX | P46660 | Alpha-internexin (Alpha-Inx) | Ina | Figure 1d | NS | ▴ | — | ▴ | NS | 0.96 | 0.31 | 0.42 | NS | NS | ▴ | 1.21 | 3.0E-08 | 2.3E-07 |
| NRDC | Q8BHG1 | Nardilysin (EC 3.4.24.61) | Nrd1 | Figure 1d | NS | — | ▾ | ▾ | NS | 1.02 | 0.60 | 0.73 | NS | NS | NS | 0.99 | 0.89 | 0.90 |
| VGLU1 | Q3TXX4 | Vesicular glutamate transporter 1 (VGluT1) | Slc17a7 | Transcriptomics5 | ⇑ | NS | — | — | ▴ | 1.16 | 1.6E-06 | 1.4E-05 | ⇑ | NS | ▴ | 1.19 | 1.5E-04 | 4.8E-04 |
| Implicated pathways | ||||||||||||||||||
| mTOR–autism spectrum disorders and mental retardation | ||||||||||||||||||
| MTOR | Q9JLN9 | Serine/threonine-protein kinase mTOR | Mtor | NS | — | — | — | ▴ | 1.28 | 4.8E-04 | 2.1E-03 | NS | NS | NS | 1.10 | 0.30 | 0.34 | |
| MK03 | Q63844 | Mitogen-activated protein kinase 3 (ERK-1) | Mapk3 | NS | ▴ | — | — | ▴ | 1.16 | 2.3E-05 | 1.5E-04 | NS | NS | ▴ | 1.23 | 3.7E-05 | 1.3E-04 | |
| MK01 | P63085 | Mitogen-activated protein kinase 1 (ERK-2) | Mapk1 | NS | ▴ | — | — | ▴ | 1.11 | 5.9E-05 | 3.0E-04 | NS | NS | ▴ | 1.07 | 6.3E-03 | 1.2E-02 | |
| PEA15 | Q62048 | Astrocytic phosphoprotein PEA-15 | Pea15 | NS | ▴ | — | — | ▴ | 1.15 | 5.0E-05 | 2.8E-04 | NS | — | ▾ | 0.91 | 2.8E-02 | 4.4E-02 | |
| KPCG | P63318 | Protein kinase C gamma type | Prkcg | ⇑ | (▴) | NS | NS | ▴ | 1.11 | 1.2E-03 | 4.6E-03 | NS | NS | ▴ | 1.21 | 6.2E-07 | 2.9E-06 | |
| Glutamatergic signalling—schizophrenia and affective disorders | ||||||||||||||||||
| NMDZ1 | P35438 | Glutamate receptor ionotropic, NMDA 1 (NMD-R1) | Grin1 | Hippocampus network5 | NS | — | — | — | ▴ | 1.15 | 0.015 | 3.8E-02 | ⇑ | NS | ▴ | 1.86 | 1.1E-12 | 2.3E-11 |
| GRIA1 | P23818 | Glutamate receptor 1 (GluR-1) | Gria1 | Hippocampus network5 | ⇑ | NS | — | — | NS | 1.08 | 0.18 | 0.26 | ⇑ | NS | ▴ | 1.43 | 2.7E-06 | 1.0E-05 |
| GRIA2 | P23819 | Glutamate receptor 2 (GluR-2) | Gria2 | NS | — | NS | (▴) | ▴ | 1.12 | 4E-04 | 2E-03 | NS | ▾ | ▴ | 1.25 | 1.9E-12 | 2.9E-11 | |
| GRIA3 | Q9Z2W9 | Glutamate receptor 3 (GluR-3) | Gria3 | Hippocampus network5 | NS | — | NS | — | NS | 1.03 | 0.64 | 0.76 | ⇑ | — | ▴ | 1.30 | 1.1E-03 | 2.6E-03 |
| KCC2A | P11798 | Calcium/calmodulin-dependent protein kinase type II α | Camk2a | Hippocampus network5 | ⇑ | NS | ▴ | — | NS | 1.05 | 0.20 | 0.28 | ⇑ | NS | ▴ | 1.27 | 1.6E-07 | 1.1E-06 |
| KCC2G | Q923T9 | Calcium/calmodulin-dependent protein kinase type II γ | Camk2g | NS | NS | NS | NS | NS | 1.05 | 0.11 | 0.20 | NS | NS | ▴ | 1.25 | 1.7E-04 | 4.9E-04 | |
| SYT1 | P46096 | Synaptotagmin-1 | Syt1 | Hippocampus network5 | NS | NS | ▾ | ▴ | NS | 1.05 | 0.07 | 0.13 | ⇑ | NS | NS | 1.08 | 0.05 | 0.08 |
| DLG4 | Q62108 | Disks large homolog 4 | Dlg4 | Hippocampus network5 | ⇑ | — | — | NS | NS | 1.00 | 0.81 | 0.88 | ⇑ | NS | ▴ | 1.17 | 7.6E-05 | 2.6E-04 |
| NCDN | Q9Z0E0 | Neurochondrin (Norbin) | Ncdn | ⇑ | NS | NS | ▴ | (▴) | 1.09 | 0.05 | 0.10 | NS | NS | ▴ | 1.27 | 3.0E-03 | 6.2E-03 | |
| Housekeeping | ||||||||||||||||||
| RL8 | P62918 | 60S ribosomal protein L8 | Rpl8 | ⇓ | — | — | — | NS | 1.05 | 0.35 | 0.46 | NS | NS | — | — | — | — | |
| MAP2 | O08663 | Methionine aminopeptidase 2 | Metap2 | NS | NS | NS | NS | NS | 1.04 | 0.21 | 0.29 | NS | NS | NS | 1.06 | 0.04 | 0.06 | |
| RS3A | P97351 | 40S ribosomal protein S3a | Rps3a | NS | — | — | — | NS | 1.04 | 0.11 | 0.20 | NS | NS | (▴) | 1.12 | 0.04 | 0.06 | |
Abbreviations: HFC, hippocampus; NS, not significant. Significantly (P*<0.05) downregulated and upregulated in proteomic profiling (▾▴) or transcriptomic profiling5, 8 (⇑⇓), in brackets if only P<0.05; PFC, prefrontal cortex; —, not detected.
Findings are compared with label-free LC-MSE proteomic discovery profiling and transcriptomic profiling results. Italicized cells indicates consistency across the studies. Column ‘Label' indicates implication of protein (for example, predicted miRNA target by Xof4 bioinformatic prediction tools). P-values were determined using SRMstats and corrected for multiple hypothesis testing (P*).18
The SRM analysis (Table 2) validated the expected decrease in all the five tested 22q11.2 gene products and an increase of the HPRT peptides from the vector cassette, showing the technical robustness of the developed method. Importantly, SRM confirmed an increase in the miR-185-target OGT1 in PFC and HPC tissue. Alterations of OGT1 have been identified in the transcriptomics5 and in proteomics (Figure 1) high-throughput screens of the PFC. The OGT1 enzyme exhibits protein N-acetylglucosaminyltransferase activity, which leads to attachment of O-GlcNAc onto intracellular proteins.39 Furthermore, we were able to validate an increase in levels of the glycogen synthase kinase-3 beta (GSK3b) and kinesin heavy chain isoform 5A (KIF5A), a microtubule-dependent molecular motor that is important for neuronal functions especially for GABAA receptor transport.40 Similar to the transcriptomic and PROTEOMIC profiling analyses, we did not find altered levels of the miR-185 target transforming protein RhoA (RHOA). This might be due to secondary regulation caused by the hemizygous genes or downregulated miRNA that might involve alterations in transcriptional levels.
Further validated proteins were serine/threonine protein phosphatase 2B catalytic subunit gamma isoform (PP2BC), the neurofilament light polypeptide (NFL) and VGLU1. The latter was one of the only three proteins found in the transcriptomic screen in both PFC and HPC. The extracellular signal–regulated kinase (ERK) pathway was most significantly affected in the PFC, while abnormal glutamatergic signalling was predominately found in the HPC. The latter finding is consistent with published transcriptomic profiling results from the Df(16)A+/− mice.5
Discussion
To the best of our knowledge, this study represents the first comprehensive quantitative proteomic and metabonomic characterization of a 22q11.2DS mouse model. We assume that the phenotype of this disease model is due to cumulative effects of abnormal miRNA regulation and hemizygosity of genes residing within the 22q11.2 chromosome region, which requires analysis on the systems level rather than by targeting the effects of individual genes.41, 42, 43
Multiplex immunoassay profiling analysis identified altered serum levels of only one protein, the endopeptidase MMP-9, which has been implicated in many pathological conditions such as cancer, cardiovascular disease,44 amyotrophic lateral sclerosis45 and stroke.46, 47 Recently, MMP-9 has been reported to have a role in the plasticity of the central nervous system48 by regulating the activity of PFC.49 A functional polymorphism of the MMP-9 gene in schizophrenia50 and bipolar disorder type II51 has also been reported. The identified alterations might serve as surrogate readouts for the psychiatric comorbidities in the 22q11.2DS and help to elucidate the molecular mechanisms of the immunodeficiency in 22q11.2DS patients.
The proteomic brain profiling analysis revealed PFC and HPC changes in various molecular pathways associated with chromatin remodelling and RNA transcription, indicative of an epigenetic component of 22q11.2DS. Recently, it has been shown that developmentally regulated genes involved in chromatin remodelling are overrepresented among targets of de novo mutations in schizophrenia.52, 53 Therefore, this study highlights a possible contribution of chromatin remodelling in the manifestation of the psychiatric and cognitive phenotype in 22q11.2DS. Other associated pathways were glycolysis, gluconeogenesis, lipid biosynthesis and the mitochondrial transport chain. Effects on the latter two pathways were also reflected in the transcriptomic enrichment analysis of the Df(16)A+/− mouse5 and together with lipid metabolism have frequently been linked to psychiatric conditions.54, 55 Altered levels of sphingomyelin and ceramide have been reported in erythrocytes and postmortem brain tissues of schizophrenic patients.56
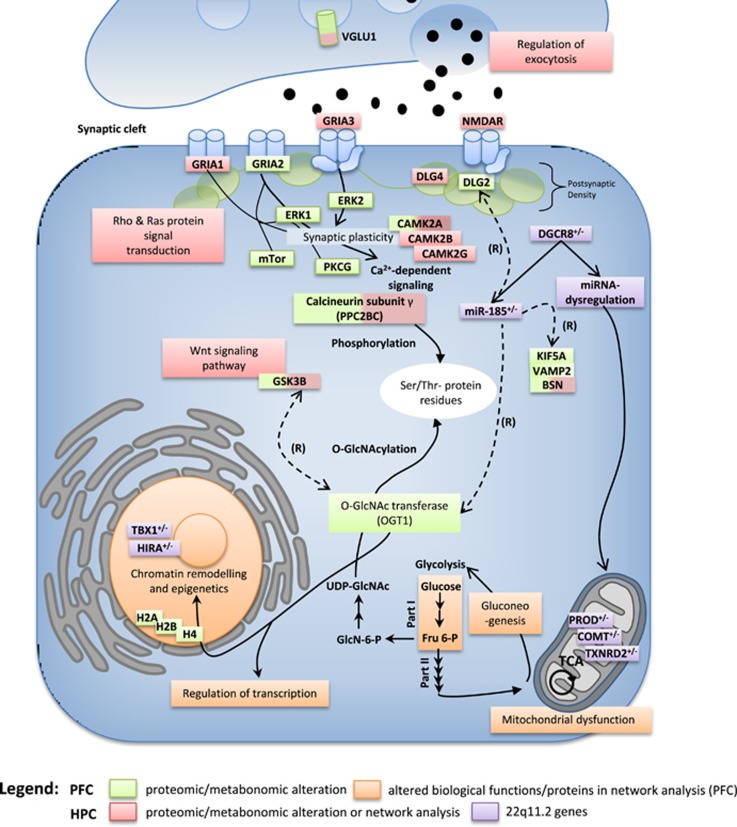
Based on the proteomic and metabonomic findings, we created a schematic model summarizing the most prominent molecular network findings in the Df(16)A+/− mouse. Interestingly, all implicated pathways can be linked through one of the most consistent and strongest proteomic candidates, (OGT1), which is a predicted miR-185 target (Figure 3).
Figure 3.
Schematic model summarizing the findings of the proteomic and metabonomic profiling analyses. Green boxes: altered levels of molecules determined by proteomic or metabonomic profiling. Orange boxes: altered pathways determined by in silico pathway analysis using IPKB or gene set enrichment analysis. Increased levels of O-GlcNAc transferase subunit p110 (OGT1) lead to abnormal O-GlcNAcylation, which affects chromatin remodelling and transcriptional regulation. Levels of the OGT1 substrate UDP-GlcNac are generated by glycolysis and gluconeogenesis (both found to be enriched). Both glutamatergic and Ca2+ signalling are mainly affected in the hippocampus (HPC). (R)=regulates. IPKB, Ingenuity Protein Knowledge Base; PFC, prefrontal cortex.
OGT1 adds single terminal N-actylglucosamine (O-GlcNAc) residues to serine/threonine residues of specific target proteins.39, 57 This represents a crucial signalling modification relevant for cellular functions, such as epigenetic regulation, transcription activation processes,58, 59 chromatin remodelling,60 translation, proteasomal degradation, signal transduction and stress responses.61 Cellular substrates recognized by OGT1 are mostly present in macromolecular complexes, such as nuclear pores,62, 63, 64, 65 the transcriptional machinery66, 67, 68 and vesicle docking components.69, 70, 71, 72 Consistently, we identified abnormalities in biological functions associated with these complexes in the Df(16)A+/− brain and most of these were found in the PFC. Interestingly, we identified robust alterations of several histone proteins in the Df(16)A+/− mouse model (Figure 1d). O-GlcNAcylation occurs on all four core-nucleosomal histone proteins (H2A, H2B, H3 and H4).73 However, such chromatin and transcriptional-level abnormalities could also be due to the hemizygosity for four transcription factors (for example, Tbx1, Hira) in the Df(16)A+/− mouse. Abnormalities in glycosylation have already been postulated to occur in the Df(16)A+/− mouse by a bioinformatic enrichment analysis, which found coordinated dysregulation of Golgi-related putative miR-185 targets in the Df(16)A+/− mouse.9 Furthermore, the miR-185 target Mirta22 has been localized to the Golgi apparatus. This is the first study to suggest that changes in OGT activity may be linked to the above glycosylation alterations. This is also supported by the fact that in silico analysis of the 25 identified human OGT interacting proteins showed an enrichment of intra Golgi-vesicle-mediated transport and vitamin biosynthetic processes.74 Also, bioinformatic analysis of an OGT-interactome substantiated the role of OGT in transcriptional regulation.74 A potential role for O-GlcNAcylation in modifying synaptic efficacy and cognition has previously been suggested.75 O-GlcNAcylation of AMPA receptors GluA2 has been linked to long-term depression in HPC synapses.76 OGT1 is also involved in mitochondrial function,77 consistent with the finding of alterations in the levels of mitochondrial proteins in this study. Overexpression of OGT1 leads to decreases in proteins involved in the respiratory chain and the TCA cycle,77 as well as altered mitochondrial morphology. The majority of O-GlcNAc-modified proteins are soluble nuclear or cytoplasmic proteins that are modified in response to cellular or environmental cues, such as growth factors, signalling molecules, glucose and other nutrient fluxes and stressors. O-GlcNAcylation has also been implicated in the aetiology of human disorders, including type II-diabetes,78 Alzheimer's disease79 and cancer.80 Notably, O-GlcNAcylation has also been shown to be a negative regulator of insulin signalling.81 Transgenic mice overexpressing OGT in muscle and fat show elevated insulin levels and insulin resistance.81, 82 Interestingly, schizophrenia patients show an increased prevalence of diabetes, impaired glucose tolerance83 and metabolic syndrome.84 Similarly, molecular analyses of postmortem brain tissues and blood cells have substantiated these findings.85, 86, 87 We also confirmed changes in GSK3beta and KIF5A, a microtubule-dependent molecular motor that is important for neuronal function, especially for GABAA receptor transport.40 In contrast, we did not find changes in the known miRNA target RHOA in any of the omics experiments. This might be due to the secondary effects of 22q11.2-encoded proteins. Further novel candidate miRNA targets implicated in both this and previous transcriptomic studies5, 9 include BSN, VAMP2, AAK1 and DLG2.
We were also able to validate the increased levels of PP2BC and NFL in the model. PP2BC is involved in synaptic plasticity and has already been shown to be associated with schizophrenia in genome-wide association studies,88, 89 via genome-wide DNA methylation analysis90 and transcriptomic studies.91 As PP2BC phosphorylates proteins on serine or threonine residues, it may have a role in the extensive crosstalk between O-GlcNAcylation and phosphorylation involved in cellular signalling.92 Interestingly, VGLU1 was increased significantly in our proteomics and also the transcriptomic (PFC and HPC) screens. This protein has already been shown to be increased in bipolar disorder and major depressive disorder93 and decreased in schizophrenia.94
We also tested proteins that have already been implicated at the pathway level in major psychiatric symptoms observed in 22q11.2DS. The first of these was the mTOR pathway, which has been linked to autism spectrum disorder,95, 96, 97 cognition98, 99 and glutamate signalling.100, 101, 102 The SRM analysis showed that mTOR was specifically affected in the PFC together with changes in the ERK1/2, PKCG and CAMK2 isoforms, which are essential components of NMDAR-related signal transduction. ERK is regulated by the activity of dopamine, serotonin and glutamate receptors.103 ERK signalling has also been implicated in the mechanism of action of mood stabilizers104 and antipsychotics105 and has been shown to be affected in social behaviour106 and autism spectrum disorder.107, 108, 109
In contrast to the above findings, we found a robust increase in proteins relating to glutamatergic signalling in the HPC. This is consistent with the findings of the transcriptomic profiling study of the Df(16)A+/− mouse, which found upregulation of a multimodal gene interaction network in the HPC associated with DLG4 and glutamatergic synapses.5 The HPC has been implicated in the cognitive deficits in 22q11 as alterations in the gross morphology of this brain region have been observed in 22q11.2DS patients,110, 111, 112 and this positively correlates with cognitive impairment.112 Interestingly, PKCG was also found to be increased in the HPC. Furthermore, O-GlcNAcylation of AMPA receptor GluA2 is associated with NMDA receptor and PKC-independent long-term depression in Ca3-Ca1 synapses.113 Therefore, our findings of alterations in the levels of OGT1 support the possibility that disruption of O-GlcNAcylation pathways could be involved in cognitive dysfunction processes.
In conclusion, this is the first system-based study to identify proteomic and metabonomic abnormalities in a 22q11.2DS mouse model. The main effects were brain region specific and involved proteins associated with chromatin modulation pathways, along with alterations in lipid and energy metabolism pathways. As one of the most robust candidates, we were able to identify OGT1, a potential miR-185 target, and postulated that O-GlcNAc might have effects on many of the affected pathways. These results provide further insights into the molecular basis of the synaptic, circuitry and behavioural deficits of the 22q11.1 mouse model and potentially of the human syndrome. A shortcoming of this study is that the employed approaches focus on global changes of FC and HPC tissue, although these brain regions are comprised of different subpopulations of brain cells, which might lead to a dilution of the magnitude of the changes. Further analysis into these brain regions and of the role of OGT1 in these effects might help to elucidate its neuronal functions and increase our understanding of the development and pathogenesis of schizophrenia and other psychiatric disorders in 22q11.2DS and in the wider population.
Acknowledgments
This research was kindly supported by the Stanley Medical Research Institute (SMRI) and the Dutch Fund for Economic Structure Reinforcement (No. 0908), the NeuroBasic PharmaPhenomics project and the National Institute of Mental Health (Grants MH67068 to MK and JAG and MH077235 to JAG). BX has been supported in part by a National Alliance for Research on Schizophrenia and Depression Young Investigator Award. We thank Kim Stark and Yan Sun for help with the generation of mouse cohorts. EJW acknowledges Waters Corporation for funding.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
SB is a director of Psynova Neurotech. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci 2010; 11: 402–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Hodgkinson K, Chow EW, Correia S, Scutt LE, Weksberg R. 22q11 deletion syndrome in adults with schizophrenia. Am J Med Genet 1998; 81: 328–337. [PMC free article] [PubMed] [Google Scholar]
- Burn J, Takao A, Wilson D, Cross I, Momma K, Wadey R et al. Conotruncal anomaly face syndrome is associated with a deletion within chromosome 22q11. J Med Genet 1993; 30: 822–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet 2000; 9: 2421–2426. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 2008; 40: 751–760. [DOI] [PubMed] [Google Scholar]
- Drew LJ, Crabtree GW, Markx S, Stark KL, Chaverneff F, Xu B et al. The 22q11.2 microdeletion: fifteen years of insights into the genetic and neural complexity of psychiatric disorders. Int J Dev Neurosci 2010; 29: 259–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB et al. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci 2008; 11: 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res 2010; 1338: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 2013; 152: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci 2005; 8: 34–42. [DOI] [PubMed] [Google Scholar]
- Fenelon K, Xu B, Lai CS, Mukai J, Markx S, Stark KL et al. The pattern of cortical dysfunction in a mouse model of a schizophrenia-related microdeletion. J Neurosci 2013; 33: 14825–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Rahmoune H, Tricklebank M, Guest PC, Bahn S. A targeted multiplexed proteomic investigation identifies ketamine-induced changes in immune markers in rat serum and expression changes in protein kinases/phosphatases in rat brain. J Proteome Res 2015; 14: 411–421. [DOI] [PubMed] [Google Scholar]
- Wesseling H, Guest PC, Lee CM, Wong EHF, Rahmoune H, Bahn S. Integrative proteomic analysis of the NMDA NR1 knockdown mouse model reveals effects on central and peripheral pathways associated with schizophrenia and autism spectrum disorders. Mol Autism 2014; 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc 2013; 8: 17–32. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal Chem 2006; 78: 779–787. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 2008; 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57: 289–300. [Google Scholar]
- Martins-de-Souza D, Menezes de Oliveira B, dos Santos Farias A, Horiuchi RS, Crepaldi Domingues C, de Paula E et al. The use of ASB-14 in combination with CHAPS is the best for solubilization of human brain proteins for two-dimensional gel electrophoresis. Brief Funct Genomic Proteomic 2007; 6: 70–75. [DOI] [PubMed] [Google Scholar]
- Wesseling H, Gottschalk MG, Bahn S. Targeted multiplexed selected reaction monitoring analysis evaluates protein expression changes of molecular risk factors for major psychiatric disorders. Int J Neuropsychopharmacol 2014; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Ma D, Garcia-Perez I, Tsang TM, Kluge W, Schwarz E et al. Molecular validation of the acute phencyclidine rat model for schizophrenia: identification of translational changes in energy metabolism and neurotransmission. J Proteome Res 2012; 11: 3704–3714. [DOI] [PubMed] [Google Scholar]
- Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics 2012; 13 (Suppl 16): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Picotti P, Huttenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R et al. Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol Cell Proteomics 2012; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Rahmoune H, Tricklebank M, Guest PC, Bahn S. A targeted multiplexed proteomic investigation identifies ketamine-induced changes in immune markers in rat serum and expression changes in protein kinases/phosphatases in rat brain. J Proteome Res 2015; 14: 411–421. [DOI] [PubMed] [Google Scholar]
- Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods 2010; 7: 43–U45. [DOI] [PubMed] [Google Scholar]
- Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA et al. A high-confidence human plasma proteome reference set with estimated concentrations in peptideatlas. Mol Cell Proteomics 2011; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010; 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg AL, Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res 2009; 8: 2144–2156. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Santos A, Lucio Teixeira A, Salgado JV. Evidence for an immune role on cognition in schizophrenia: a systematic review. Curr Neuropharmacol 2014; 12: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M, Schmidt MJ, Mirnics K. Immune system gene dysregulation in autism and schizophrenia. Dev Neurobiol 2012; 72: 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller N. Immunology of major depression. Neuroimmunomodulation 2014; 21: 123–130. [DOI] [PubMed] [Google Scholar]
- Ross HE, Guo Y, Coleman K, Ousley O, Miller AH. Association of IL-12p70 and IL-6:IL-10 ratio with autism-related behaviors in 22q11.2 deletion syndrome: a preliminary report. Brain Behav Immun 2013; 31: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Haskell GT, Peters AZ, Sikich L, Lieberman JA, LaMantia AS. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc Natl Acad Sci USA 2003; 100: 14433–14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim SM, Lee RT. Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance. Antioxid Redox Signal 2013; 18: 1165–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 2005; 45: 51–88. [DOI] [PubMed] [Google Scholar]
- Raza H, Robin MA, Fang JK, Avadhani NG. Multiple isoforms of mitochondrial glutathione S-transferases and their differential induction under oxidative stress. Biochem J 2002; 366(Pt 1): 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude A, Ven Murthy MR, Mirault ME. Mitochondrial thioredoxin system: effects of TrxR2 overexpression on redox balance, cell growth, and apoptosis. J Biol Chem 2004; 279: 27302–27314. [DOI] [PubMed] [Google Scholar]
- Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci 2012; 32: 14132–14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Sawaguchi S, Kamemura K, Okajima T. Intracellular and extracellular O-linked N-acetylglucosamine in the nervous system. Exp Neurol 2015; 274: 166–174. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yin X, Takei Y, Seog DH, Homma N, Hirokawa N. Molecular motor KIF5A is essential for GABA(A) receptor transport, and KIF5A deletion causes epilepsy. Neuron 2012; 76: 945–961. [DOI] [PubMed] [Google Scholar]
- Moya-Alvarado G, Gershoni-Emek N, Perlson E, Bronfman FC. Neurodegeneration and Alzheimer's disease. What can proteomics tell us about the Alzheimer's brain? Mol Cell Proteomics 2015; 15: 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Kreutz MR. Proteomics of the synapse - a quantitative approach to neuronal plasticity. Mol Cell Proteomics 2015; 15: 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Guest PC, Lago SG, Bahn S. Technological advances for deciphering the complexity of psychiatric disorders: merging proteomics with cell biology. Int J Neuropsychopharmacol 2014; 17: 1327–1341. [DOI] [PubMed] [Google Scholar]
- Yabluchanskiy A, Ma YG, Iyer RP, Hall ME, Lindsey ML. Matrix metalloproteinase-9: many shades of function in cardiovascular disease. Physiology 2013; 28: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz-Zajac M, Mroczko B, Slowik A. Matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMPs) in amyotrophic lateral sclerosis (ALS). J Neural Transm (Vienna) 2014; 121: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzepa J, Kurzepa J, Golab P, Czerska S, Bielewicz J. The significance of matrix metalloproteinase (MMP)-2 and MMP-9 in the ischemic stroke. Int J Neurosci 2014; 124: 707–716. [DOI] [PubMed] [Google Scholar]
- Chaturvedi M, Kaczmarek L. MMP-9 inhibition: a therapeutic strategy in ischemic stroke. Mol Neurobiol 2014; 49: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek L, Lapinska-Dzwonek J, Szymczak S. Matrix metalloproteinases in the adult brain physiology: a link between c-Fos, AP-1 and remodeling of neuronal connections? EMBO J 2002; 21: 6643–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okulski P, Jay TM, Jaworski J, Duniec K, Dzwonek J, Konopacki FA et al. TIMP-1 abolishes MMP-9-dependent long-lasting long-term potentiation in the prefrontal cortex. Biol Psychiatry 2007; 62: 359–362. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK, Skibinska M, Kapelski P, Kaczmarek L, Hauser J. Functional polymorphism of the matrix metalloproteinase-9 (MMP-9) gene in schizophrenia. Schizophr Res 2009; 109: 90–93. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK, Skibinska M, Leszczynska-Rodziewicz A, Kaczmarek L, Hauser J. Matrix metalloproteinase-9 gene and bipolar mood disorder. Neuromolecular Med 2009; 11: 128–132. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci 2012; 15: 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 2012; 44: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M et al. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci 2012; 13: 293–307. [DOI] [PubMed] [Google Scholar]
- Oresic M, Seppanen-Laakso T, Sun D, Tang J, Therman S, Viehman R et al. Phospholipids and insulin resistance in psychosis: a lipidomics study of twin pairs discordant for schizophrenia. Genome Med 2012; 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhle C, Reichel M, Gulbins E, Kornhuber J. Sphingolipids in psychiatric disorders and pain syndromes. Handb Exp Pharmacol 2013; 216: 431–456. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan K, Durning S, Wells L. Functional O-GlcNAc modifications: implications in molecular regulation and pathophysiology. Crit Rev Biochem Mol Biol 2014; 49: 140–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta 2009; 1800: 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem 2012; 287: 23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci USA 2013; 110: 5169–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus BD, Love DC, Hanover JA. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol 2009; 41: 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Hart GW. O-GlcNAc and the control of gene expression. Biochim Biophys Acta 1999; 1473: 161–171. [DOI] [PubMed] [Google Scholar]
- Hanover JA. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J 2001; 15: 1865–1876. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Berlin WK, Starr CM, Hanover JA. Characterization of nuclear pore protein p62 produced using baculovirus. Protein Expr Purif 1995; 6: 546–554. [DOI] [PubMed] [Google Scholar]
- Hanover JA. The nuclear pore: at the crossroads. FASEB J 1992; 6: 2288–2295. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 2002; 110: 69–80. [DOI] [PubMed] [Google Scholar]
- Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem 2001; 276: 10570–10575. [DOI] [PubMed] [Google Scholar]
- Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem 1995; 270: 18961–18965. [DOI] [PubMed] [Google Scholar]
- Cole RN, Hart GW. Cytosolic O-glycosylation is abundant in nerve terminals. J Neurochem 2001; 79: 1080–1089. [DOI] [PubMed] [Google Scholar]
- Cole RN, Hart GW. Glycosylation sites flank phosphorylation sites on synapsin I: O-linked N-acetylglucosamine residues are localized within domains mediating synapsin I interactions. J Neurochem 1999; 73: 418–428. [DOI] [PubMed] [Google Scholar]
- Ding M, Vandre DD. High molecular weight microtubule-associated proteins contain O-linked-N-acetylglucosamine. J Biol Chem 1996; 271: 12555–12561. [DOI] [PubMed] [Google Scholar]
- Griffith LS, Schmitz B. O-linked N-acetylglucosamine is upregulated in Alzheimer brains. Biochem Biophys Res Commun 1995; 213: 424–431. [DOI] [PubMed] [Google Scholar]
- Xu Q, Yang C, Du Y, Chen Y, Liu H, Deng M et al. AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res 2014; 42: 5594–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng RP, He X, Guo SJ, Liu WF, Tao Y, Tao SC. Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics 2014; 14: 1020–1030. [DOI] [PubMed] [Google Scholar]
- Kanno T, Yaguchi T, Nagata T, Mukasa T, Nishizaki T. Regulation of AMPA receptor trafficking by O-glycosylation. Neurochem Res 2010; 35: 782–788. [DOI] [PubMed] [Google Scholar]
- Taylor EW, Wang K, Nelson AR, Bredemann TM, Fraser KB, Clinton SM et al. O-GlcNAcylation of AMPA receptor GluA2 is associated with a novel form of long-term depression at hippocampal synapses. J Neurosci 2014; 34: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EP, Villar MT, E L, Lu J, Selfridge JE, Artigues A et al. Altering O-linked beta-N-Acetylglucosamine cycling disrupts mitochondrial function. J Biol Chem 2014; 289: 14719–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics 2013; 10: 365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster S, Welleford AS, Triplett JC, Sultana R, Schmitz B, Butterfield DA. Increased O-GlcNAc levels correlate with decreased O-GlcNAcase levels in Alzheimer disease brain. Biochim Biophys Acta 2014; 1842: 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell 2014; 54: 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008; 451: 964–969. [DOI] [PubMed] [Google Scholar]
- McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC et al. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA 2002; 99: 10695–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelman LM, Walsh PI, Sharifi N, Collins P, Thakore JH. Impaired glucose tolerance in first-episode drug-naive patients with schizophrenia. Diabet Med 2007; 24: 481–485. [DOI] [PubMed] [Google Scholar]
- Newcomer JW. Metabolic syndrome and mental illness. Am J Manag Care 2007; 13(7 Suppl): S170–S177. [PubMed] [Google Scholar]
- Guest PC, Wang L, Harris LW, Burling K, Levin Y, Ernst A et al. Increased levels of circulating insulin-related peptides in first-onset, antipsychotic naive schizophrenia patients. Mol Psychiatry 2010; 15: 118–119. [DOI] [PubMed] [Google Scholar]
- Harris LW, Guest PC, Wayland MT, Umrania Y, Krishnamurthy D, Rahmoune H et al. Schizophrenia: metabolic aspects of aetiology, diagnosis and future treatment strategies. Psychoneuroendocrinology 2012; 38: 752–766. [DOI] [PubMed] [Google Scholar]
- Herberth M, Koethe D, Cheng TM, Krzyszton ND, Schoeffmann S, Guest PC et al. Impaired glycolytic response in peripheral blood mononuclear cells of first-onset antipsychotic-naive schizophrenia patients. Mol Psychiatry 2010; 16: 848–859. [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M et al. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci USA 2003; 100: 8993–8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti E, Scassellati C, Minelli A, Valsecchi P, Bonvicini C, Pasqualetti P et al. Schizophrenia susceptibility and NMDA-receptor mediated signalling: an association study involving 32 tagSNPs of DAO, DAOA, PPP3CC, and DTNBP1 genes. BMC Med Genet 2013; 14: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wockner LF, Noble EP, Lawford BR, Young RM, Morris CP, Whitehall VL et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry 2014; 4: e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood SL, Burnet PW, Harrison PJ. Decreased hippocampal expression of the susceptibility gene PPP3CC and other calcineurin subunits in schizophrenia. Biol Psychiatry 2005; 57: 702–710. [DOI] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal 2010; 3: ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezato A, Meador-Woodruff JH, McCullumsmith RE. Vesicular glutamate transporter mRNA expression in the medial temporal lobe in major depressive disorder, bipolar disorder, and schizophrenia. Bipolar Disord 2009; 11: 711–725. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Harrison PJ. Decreased expression of vesicular glutamate transporter 1 and complexin II mRNAs in schizophrenia: further evidence for a synaptic pathology affecting glutamate neurons. Schizophr Res 2005; 73: 159–172. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron 2014; 84: 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci 2013; 16: 1537–1543. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013; 493: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J, Marin P. mTOR in brain physiology and pathologies. Physiol Rev 2015; 95: 1157–1187. [DOI] [PubMed] [Google Scholar]
- Troca-Marin JA, Alves-Sampaio A, Montesinos ML. Deregulated mTOR-mediated translation in intellectual disability. Prog Neurobiol 2012; 96: 268–282. [DOI] [PubMed] [Google Scholar]
- Burket JA, Benson AD, Tang AH, Deutsch SI. NMDA receptor activation regulates sociability by its effect on mTOR signaling activity. Prog Neuropsychopharmacol Biol Psychiatry 2015; 60: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 2011; 480: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci 2006; 26: 2167–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Pascoli V, Svenningsson P, Paul S, Enslen H, Corvol JC et al. Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proc Natl Acad Sci USA 2005; 102: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Creson T, Zhang L, Li P, Du F, Yuan P et al. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. J Neurosci 2004; 24: 6590–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Mol Psychiatry 2005; 10: 79–104. [DOI] [PubMed] [Google Scholar]
- Satoh Y, Endo S, Nakata T, Kobayashi Y, Yamada K, Ikeda T et al. ERK2 contributes to the control of social behaviors in mice. J Neurosci 2011; 31: 11953–11967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz TA, Guilmette E, Gosink MM, Fischer JE, Fitzgerald LW, Stephenson DT et al. Transcriptomic analysis of genetically defined autism candidate genes reveals common mechanisms of action. Mol Autism 2013; 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E, Fennoy I, Laraque D, Kanem N, Brown G, Mitchell J. Autism and developmental abnormalities in children with perinatal cocaine exposure. J Natl Med Assoc 1992; 84: 315–319. [PMC free article] [PubMed] [Google Scholar]
- Yang K, Sheikh AM, Malik M, Wen G, Zou H, Brown WT et al. Upregulation of Ras/Raf/ERK1/2 signaling and ERK5 in the brain of autistic subjects. Genes Brain Behav 2011; 10: 834–843. [DOI] [PubMed] [Google Scholar]
- Chow EW, Zipursky RB, Mikulis DJ, Bassett AS. Structural brain abnormalities in patients with schizophrenia and 22q11 deletion syndrome. Biol Psychiatry 2002; 51: 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbane M, Schaer M, Farhoumand R, Glaser B, Eliez S. Hippocampal volume reduction in 22q11.2 deletion syndrome. Neuropsychologia 2006; 44: 2360–2365. [DOI] [PubMed] [Google Scholar]
- Deboer T, Wu Z, Lee A, Simon TJ. Hippocampal volume reduction in children with chromosome 22q11.2 deletion syndrome is associated with cognitive impairment. Behav Brain Funct 2007; 3: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EW, Wang K, Nelson AR, Bredemann TM, Fraser KB, Clinton SM et al. O-GlcNAcylation of AMPA receptor GluA2 is associated with a novel form of long-term depression at hippocampal synapses. J Neurosci 2014; 34: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Picotti P, Huttenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R et al. Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol Cell Proteomics 2011; 11: M111 014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J et al. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett 2011; 301: 151–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.