Abstract
Model microbial systems provide opportunity to understand the genetic bases of ecological traits, their evolution, regulation and fitness contributions. Experimental populations of Pseudomonas fluorescens rapidly diverge in spatially structured microcosms producing a range of surface-colonising forms. Despite divergent molecular routes, wrinkly spreader (WS) niche specialist types overproduce a cellulosic polymer allowing mat formation at the air–liquid interface and access to oxygen. Given the range of ways by which cells can form mats, such phenotypic parallelism is unexpected. We deleted the cellulose-encoding genes from the ancestral genotype and asked whether this mutant could converge on an alternate phenotypic solution. Two new traits were discovered. The first involved an exopolysaccharide encoded by pgaABCD that functions as cell–cell glue similar to cellulose. The second involved an activator of an amidase (nlpD) that when defective causes cell chaining. Both types form mats, but were less fit in competition with cellulose-based WS types. Surprisingly, diguanylate cyclases linked to cellulose overexpression underpinned evolution of poly-beta-1,6-N-acetyl-d-glucosamine (PGA)-based mats. This prompted genetic analyses of the relationships between the diguanylate cyclases WspR, AwsR and MwsR, and both cellulose and PGA. Our results suggest that c-di-GMP regulatory networks may have been shaped by evolution to accommodate loss and gain of exopolysaccharide modules facilitating adaptation to new environments.
Introduction
Numerous instances of convergent and parallel evolution point to the fact that at a certain level evolution is predictable (Martin and Orgogozo, 2013; Orgogozo, 2015). The underlying causes have been the source of extensive study with understanding much accelerated in recent years through application of the tools of genomics and genetics (Jost et al., 2008; Gerstein et al., 2012; Tenaillon et al., 2012; Zhen et al., 2012; Barrick and Lenski, 2013; Herron and Doebeli, 2013). This has shifted focus from selection—often seen as the primary determinant—to genetic details, in particular, the kinds of genes in which mutations are found and their regulatory connections (Gompel and Prud'homme, 2009; Stern, 2013).
Parallel evolution at both phenotypic and genetic levels has been extensively reported in laboratory populations of microbes (Wichman et al., 1999; McDonald et al., 2009; Gerstein et al., 2012; Barrick and Lenski, 2013; Herron and Doebeli, 2013; Lind et al., 2015). Given that replicate populations are typically founded by a single common ancestral type, that selection is often directional (and strong), and population sizes large, such parallelism is not surprising (Wichman et al., 1999). Indeed, it has been commonplace to attribute parallel changes to selection alone. Recent work however shows that even in simple laboratory populations of microbes, the causes are not necessarily so straightforward. For example, studies with experimental populations of Pseudomonas fluorescens have shown that genetic architecture—the differing capacity of loci to translate mutation into phenotypic variation—can generate parallel molecular evolution (McDonald et al., 2009; Lind et al., 2015).
The genome of P. fluorescens encodes 39 putative diguanylate cyclases (DGCs), any one of which, if functional and properly localised in the cell, can be activated by mutation to generate the surface-colonising ‘wrinkly spreader' (WS) niche specialist type. Despite the seemingly large array of mutational routes, evolution takes just three pathways: Wsp, Mws, or Aws (McDonald et al., 2009). If these three commonly trod routes are eliminated, then evolution proceeds via one of thirteen additional pathways, although WS types take twice as long to emerge (despite requiring just a single mutation) (Lind et al., 2015). Interestingly, the fitness of the WS types that arise via mutation in the additional pathways do not differ to those arising from mutation in the commonly used pathways, indicating the role of factors other than selection (Lind et al., 2015). Investigation of underlying genetics has shown that the causes depend on mutational target size (McDonald et al., 2009; Lind et al., 2015). The DGC in each of the commonly trod Wsp, Mws, or Aws pathways is subject to negative regulation and this, combined with the fact that loss of function mutations are more common than gain of function mutations, means that these pathways offer many mutational possibilities to activate their respective DGC and thus produce the WS phenotype (McDonald et al., 2009; Lind et al., 2015). For a DGC-encoding pathway requiring a rare gain-of-function mutation, including promoter mutations, the mutational target size is much reduced and all factors being equal, evolution proceeds via loci with greatest capacity to translate mutation into phenotypic variation (Lind et al., 2015).
A more extreme form of parallelism underpins the phenotypic basis of WS success. Whereas mutations generating WS can activate different DGCs, every WS type ever studied, irrespective of mutational route has, as its basis, the cellulose biosynthetic pathway encoded by the wss operon (Spiers et al., 2002; Lind et al., 2015). At first glance, this seems odd, especially as there are many ways to generate adhesion among cells. Moreover, the genome of SBW25 encodes a number of polysaccharide biosynthetic pathways (Silby et al., 2009)—some of which appear to have been acquired by lateral gene transfer from unrelated bacteria (Silby et al., 2009; Gallie et al., 2015). The expression of a diversity of adhesive components is a key component for biofilm formation on solid surfaces and at air–liquid interfaces and is recognised as a major determinant of the mechanical properties of biofilms and an important ecological trait (Flemming and Wingender, 2010).
Being curious, and aware of the potentially complex interplay between selection and genetic architecture in generating parallel changes in ecological traits, we eliminated the wss operon, encoding the cellulose biosynthetic pathway, from the ancestral type and asked whether this type might find an alternate evolutionary solution to the challenge of colonising the air–liquid interface (Figure 1a). We identified two new ecological strategies (and underlying molecular details): mutants expressing alternate solutions were detected at approximately the same time as cellulose-based mat-forming mutants, but neither delivered the fitness advantage of the later. Thus, for cellulose-based WS types, parallel phenotypic evolution is explained primarily by selection.
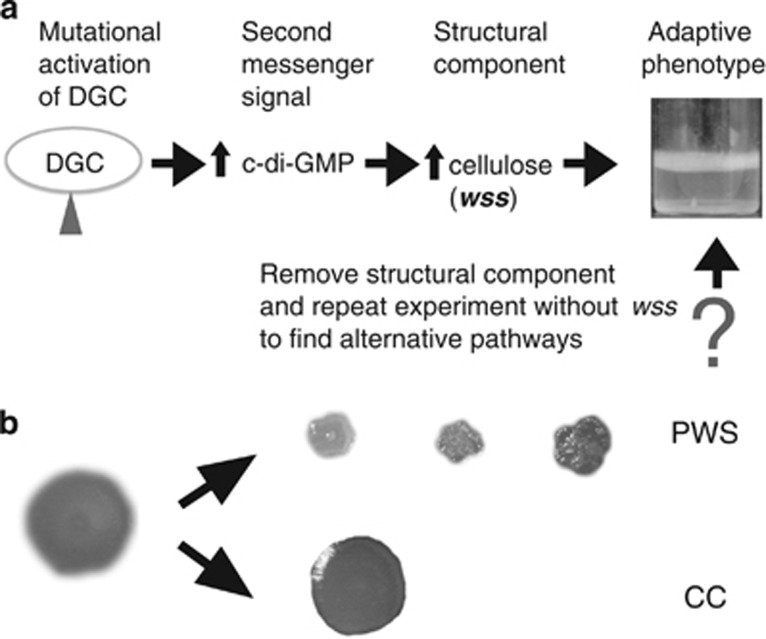
Figure 1.
(a) Experimental evolution of adaptive divergence in P. fluorescens SBW25. Strong selection for access to oxygen in static microcosms leads to the invasion of mutants that colonise the air–liquid interface. In wild-type P. fluorescens SBW25, wrinkly spreader (WS) mutants repeatedly evolve through mutations activating diguanylate cyclases (DGCs), which in turn activates production of the structural component cellulose, encoded by the wss operon. By removing the genes encoding the cellulose biosynthetic machinery we address the question as to whether there exists alternative structural components for mat formation at the air–liquid interface. (b) Alternative adaptive air–liquid colonising mutants. Two new types of mutants were found after experimental evolution in static microcosms. Colony morphologies on Congo Red agar revealed one type (PWS) that strongly binds Congo Red and has a wrinkled morphology and one type more similar to the ancestral strain (CC).
Materials and methods
Strains and media
P. fluorescens SBW25 (Silby et al., 2009) or derivatives thereof have been used for all experiments except for Escherichia coli genetic tools employed for strain construction and transposon mutagenesis (E. coli DH5-α λpir, E. coli SM10 λpir IS-Ω-kan/hah, E. coli pRK2013). King's medium B (KB) (King et al., 1954) at 28 °C was used to grow P. fluorescens and E. coli was grown in Lysogeny Broth (LB) (Bertani, 1951) at 37 °C. Solid media were KB and LB with 1.5% agar. Colony morphology (Supplementary Figure 1) was photographed after growth for 48 h on KB with 0.8 % agar or KB with 1.5% agar and Congo Red. Swimming motility was assayed in LB plates with 0.25% agar. Antibiotics were used during strain construction at the following concentrations: kanamycin (100 mg l−1), tetracyclin (15 mg l−1), nitrofurantoin (100 mg l−1) and cycloserine (1000 mg l−1). X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at a concentration of 40 mg l−1. For visualisation of extracellular polysaccharides we used Calcofluor (Fluorescent brightener 28) at a concentration of 35 mg l−1 and Congo Red at 10 mg l−1. All strains were stored in glycerol saline at −80 °C.
Strain construction
Strains with scarless deletions spanning wssA-wssJ (PFLU0300-PFLU0309) and/or pgaABCD (PFLU0143-PFLU0146) were constructed using a two-step allelic exchange method (Rainey, 1999; Bantinaki et al., 2007). PCR fragments with ~700 bp homologies to each side of the deleted regions were made using SOE-PCR (Horton et al., 1989) and cloned into suicide plasmid pUIC3 (Rainey, 1999) in E. coli DH5-α λpir. Conjugation was used to transfer the plasmids into SBW25, where the plasmid could not replicate, and transformants with the plasmid integrated though homologous recombination into the desired position in the chromosome could be selected. A cycloserine enrichment protocol was used to select against tetracycline resistant cells, which allowed us to isolate the cells that had lost pUIC3 by plating and screening for loss of the lacZ gene and confirmation of a tetS phenotype. We confirmed the deletions using PCR and Sanger sequencing. Point mutations were reconstructed using a similar protocol as previously described (Rainey, 1999; Bantinaki et al., 2007).
Strains expressing green fluorescent protein (GFP) were constructed using a miniTn7 transposon (miniTn7(Gm)PrrnB P1 gfp-a) (Lambertsen et al., 2004) that allows integration at a defined locus (attTn7) in the chromosome. The transposon was introduced into SBW25 strains in a tri-parental conjugation from E. coli together with the pUX-BF13 plasmid carrying the transposase genes.
Experimental evolution
We inoculated 24 independent glass microcosms (30 ml glass tubes with 6 ml KB) with the Δwss strain and incubated under static condition for 5 days. The cultures were then vortexed twice for 30 s and serial dilutions were spread on KB agar. After 48 h incubation we screened plates for colonies with changed morphology and restreaked them on KB plates for single colonies.
Transposon mutagenesis
Transposon mutagenesis was used to find candidate genes involved in the alternative colony morphology phenotypes (Giddens et al., 2007). Plasmid pCM639 with the IS-Ω-kan/hah transposon was conjugated from E. coli SM10 λpir into strains with changed colony morphology using helper plasmid pRK2013. Transconjugants were selected on KB plates with kanamycin and nitrofurantoin, for counterselection of E. coli, and less than 1000 colonies from each independent conjugation were screened for reversion to the ancestral smooth colony morphology, which were then single colony isolated. The chromosomal locations of the transposons were then determined by an arbitrarily-primed PCR and Sanger sequencing (Manoil, 2000).
Fitness assays
We measured relative fitness using competition assays where one strain was tagged with GFP with a starting ratio of either ~1:1 (referred to as ‘Competition') or 1:100 (referred to as ‘Invasion') as previously described (Lind et al., 2015). Briefly, for the competition assay strains were grown for 16 h shaking before dilution and mixing and allowed to grow for 4 h shaking before the initial ratio was determined using flow cytometry (BD FACS Canto, Beckton, Dickinson and Company, Franklin Lakes, NJ, USA) by counting 100 000 cells. The initial populations were diluted 1000-fold in KB and incubated for 24 h under static conditions and the ratios were again measured and the change in relative frequencies could be determined with high accuracy. Serial dilutions of the initial and final populations were plated on KB plates to determine the number of doublings during the experiment and to examine if adaptive mutants affecting the colony morphology or GFP expression (when possible) had appeared during the experiment and invaded the population. Selection coefficient were calculated as s=[ln(R(t)/R(0))]/[t] where R is the ratio of the two strains and t the number of doublings during the experiment (Dykhuizen, 1990). The fitness cost of the GFP marker was measured using isogenic strains and used to adjust fitness accordingly, so by definition the fitness difference is s=0. Invasion fitness was measured in a similar way except that strains were incubated for 48 h static and initial ratios were determined using serial dilution and plating due to the difficulties of accurately measuring ratios of approximately 1:100 using flow cytometry.
Results
To understand the causes of the repeated evolution of cellulose-based WS forms, the entire wssA-J operon (PFLU0300-PFLU0309; Figure 2a) was deleted from the ancestral SM genotype (PAL26; referred to henceforth as SBW25 Δwss). SBW25 Δwss was inoculated into 24 independent microcosms and propagated without shaking (Figure 2b). After 5 days samples were taken, diluted and plated onto agar. The resulting colonies were screened for changes in colony morphology indicative of alterations to cell surface components characteristic of mat-forming mutants (Figure 2c). Two different distinguishable types were found in all microcosms: one, similar to the previously described WS mutants (Spiers et al., 2002; McDonald et al., 2009; Lind et al., 2015) formed atypical wrinkly colonies (PGA wrinkly spreader (PWS)), whereas the other produced colonies more similar to the ancestral wild type, but with a cell chaining (CC) phenotype observed by microscopy (Figure 1b; Supplementary Figure 2 and below).
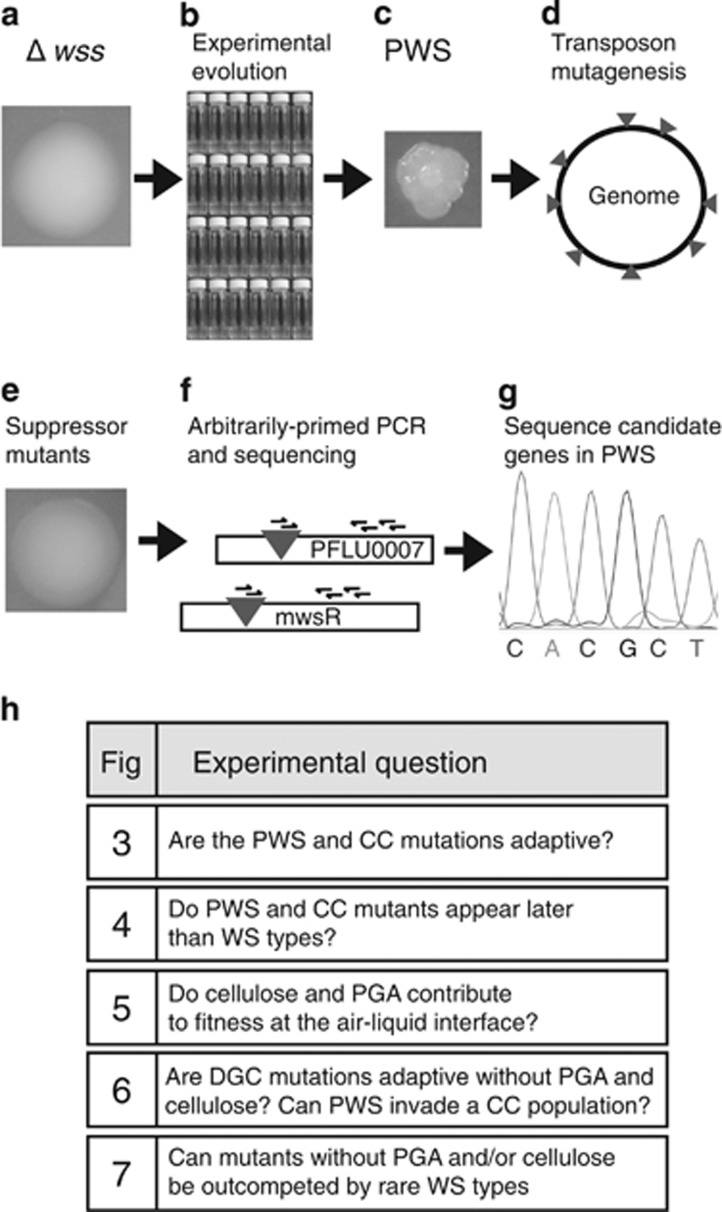
Figure 2.
Experimental flow chart for finding genes underpinning the PWS phenotype and rationale for fitness assays. (a) The wss operon, encoding the cellulose biosynthetic machinery, was genetically deleted from the wild type. (b) Twenty-four independent populations of the cellulose-deficient mutant were subjected to experimental evolution by static incubation, which selects for colonisation of the air–liquid interface. (c) Adaptive mutants were identified by differences in colony morphology on agar plates. (d) PWS mutants were subjected to random transposon mutagenesis. (e) Transposon mutants supressing the PWS colony morphology was isolated on agar plates. (f) PCR and sequencing was used to identify the position of the suppressing transposons in the genome and showed that the PWS phenotype can be reversed by insertions in pgaABCD, PFLU0005, PFLU0007 and a diguanylate cyclase (awsR or mwsR). (g) The mutations causing the PWS phenotype were identified by sequencing of the awsXRO and mwsR genes in PWS mutants. (h) Rationale for the fitness assays and diversification experiments presented in Figures 3, 4, 5, 6, 7.
Genetic basis of mat-forming mutants
To identify genes underpinning expression of the PWS morphotype, transposon mutagenesis was used to identify suppressor mutants (Figure 2d). Nine independent PWS types were subjected to mutagenesis using Tn5-Ω-kan/hah (Giddens et al., 2007) and transconjugants screened for loss of the PWS morphotype (Figure 2e). A total of 72 suppressor mutants were identified. None were capable of colonising the air–liquid interface demonstrating a direct connection between colony morphology and colonisation of the meniscus. Localisation of the transposon by arbitrary-primed PCR (Manoil, 2000) showed that 35 carried insertions in PFLU0143-PFLU0146, six in PFLU0005 and 19 in PFLU0007 (Figure 2f).
PFLU0143-PFLU0146 marks a predicted operon encoding four genes with similarity to the pgaABCD gene cluster found in E. coli and Acinetobacter baumannii that together encode enzymes necessary to produce poly-beta-1,6-N-acetyl-D-glucosamine (PGA or PNAG), an exopolysaccharide involved in surface adhesion (Wang et al., 2004; Gehrig, 2005; Choi et al., 2009). PFLU0005 and PFLU0007 encode a putative two-component response regulator system that is orthologous to PA4032 and PA4036 found in P. aeruginosa, for which no function has been attributed. PFLU0005 encodes an OmpR family DNA binding response regulator transcribed divergently from a predicted operon comprised of PFLU0006 and PFLU0007. PFLU0007 encodes a predicted signal transduction histidine kinase and PFLU0006, a predicted periplasmic protein, with LysM and FecR domains.
In suppressor mutants of three PWS types, transposon insertions were also found in genes encoding the DGCs PFLU5210 (awsR) and PFLU5329 (mwsR) (Figure 2f). Such suppressor mutants were unexpected given the central role of these genes in expression of cellulose-based WS types (McDonald et al., 2009). However, given that spontaneous mutations generating WS arise within these genes, combined with the possibility of regulatory overlap between DGCs affecting cellulose and PGA expression, awsR and mwsR were sequenced in all 24 PWS genotypes (Figure 2g). Six PWS types carried a mutation in awsR and three carried a mutation in mwsR (Supplementary Table 1). In several cases these mutations were identical to those previously shown to cause cellulose-based WS types (McDonald et al., 2009). In light of these findings we considered it possible that mutations in other known mutational routes to cellulose-based WS might also cause PGA-based PWS types. Accordingly, PFLU1224 (wspF, a negative regulator of the DGC WspR) and PFLU5211 (awsX, a negative regulator of the DGC AwsR) were sequenced in the remaining PWS types (McDonald et al., 2009; Figure 2g). Five PWS types were found to harbour mutations in awsX (Supplementary Table 1), again identical to some previously found (McDonald et al., 2009), but no mutations were identified in wspF. The mutational origins of 10 PWS types remain unknown, but at least four of them are dependent on PGA, based on the results from the transposon mutagenesis screen. The PFLU0005 and PFLU0007 genes and upstream regions were sequenced in all 24 PWS strains, but no mutations were found (Figure 2g).
It was not possible to investigate the genetic basis of the CC types by screening for suppressor mutants, because the minor difference in colony morphology compared with the SBW25 Δwss ancestral type made identification of mutants impossible. Nonetheless, microscopy revealed a CC morphology and a motility defect (evident on semi solid agar)—two phenotypes that had previously been associated with mutants in a previous experimental evolution study (Beaumont et al., 2009). Genome-sequencing data available from that study (of line 6B4) suggested candidate genes and Sanger sequencing was used to find the mutational causes. In 23 out of 24 strains a mutation was found in PFLU1301 (nlpD), which encodes a lipoprotein that in E. coli (Uehara et al., 2010), Vibrio cholerae (Moll et al., 2014) and Yersinia pestis (Tidhar et al., 2009) is an activator of AmiC (N-acetylmuramoyl-l-alanine amidase) hydrolase activity required for proper cell division: loss-of-function mutations in nlpD in these species exhibit a similar cellular morphology as observed for the CC types and can be complemented by expression in trans. In all 23 cases the mutation in nlpD was identical—a single Q189* (CAG->TAG) nonsense mutation. Such striking molecular parallelism is difficult to explain without further investigation, but may reflect constraints due to effects on the expression of rpoS (which encodes the stationary phase sigma factor σs) located immediately downstream of nlpD.
Fitness effects
We performed a range of fitness assays and studied the dynamics of diversification in order to determine why WS types dominate model radiations established from the ancestral SM type and to investigate the contribution of cellulose and PGA to fitness at the air–liquid interface (Figure 2h). PWS strains with representative DGC-activating mutations in awsR (T27P), awsX (ΔY77-Q87), mwsR (M926I) and one of the nlpD (Q189*) mutants that defined the CC type, were selected for further characterisation. Given that loss-of-function mutations in wspF are the most common mutation underpinning evolution of cellulose-based WS types (McDonald et al., 2009), a wspF (ΔT226-G275) mutation was introduced into SBW25 Δwss to see whether there might be phenotypic effects.
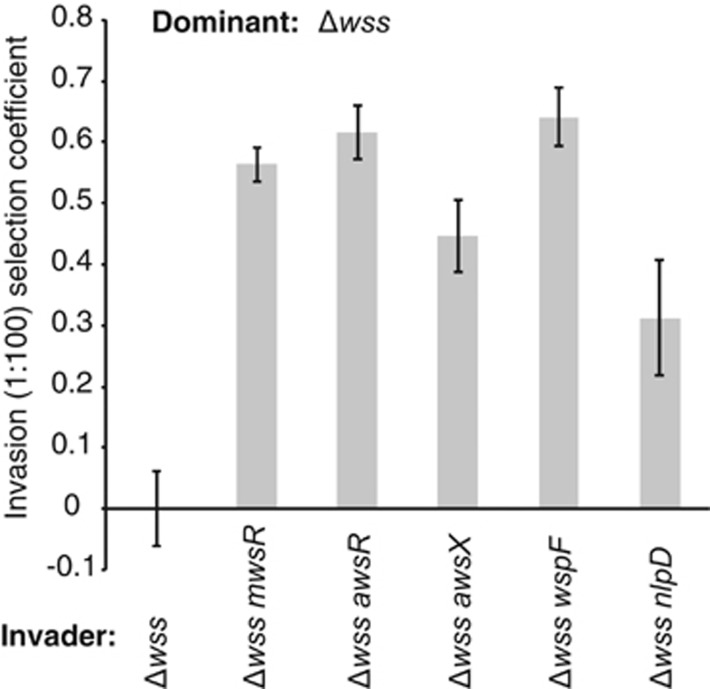
All PWS mutants colonised the air–liquid interface of static broth microcosms and were found to rapidly invade ancestral SBW25 Δwss populations, demonstrating a fitness advantage thus explaining their appearance during experimental evolution (Figure 3). Surprisingly, despite no obvious effects on colony morphology, the Δwss wspF mutant had a frequency dependent fitness advantage that was similar to the PWS types. This suggests that wspF mutants (in the Δwss background) are expected to arise at similar frequencies as the other PWS mutants (mutational target size is known to be similar), but their smooth colony morphology on agar plates would preclude detection. The Δwss nlpD mutant showed a lower fitness than the PWS mutants (two-tailed t-tests P<0.01) and invaded at approximately half the rate of genotypes with DGC-activating mutations in mwsR, awsR, awsX and wspF (Figure 3).
Figure 3.
Fitness contribution of non-cellulosic traits to competitive performance at the air–liquid interface. Both the PWS types, with mutations in mwsR, awsR, awsX and wspF, and the CC type with a mutation in nlpD show a strong frequency dependent fitness advantage in a 1:100 invasion assay against the ancestral SM population. Data are average values from four independent replicates, error bars represent ±1 s.d.
Dynamics of diversification
Given that both PWS and CC types increased in frequency from rare, we considered it of value to examine the adaptive radiation of ancestral (SM) SBW25 and SBW25 Δwss through the course of five days (as per Rainey and Travisano, 1998). Examining the evolutionary dynamics of the radiation stands to contribute understanding of the timing of mutant origin and the interplay with fitness effects. For example, were PWS and CC types to arise after WS types then this would suggest the involvement of priority effects to explain the dominance of WS types (Fukami et al., 2007). Conversely, should PWS and CC types arise before WS types then this would suggested that the dominance of WS is a consequence of superior fitness. Accordingly, 24 replicate microcosms for each ancestral type were founded on day 0 and four microcosms (for each type) were harvested on successive days. The phenotype of the resulting colonies was scored as previously, but with SM-type colonies subject to an additional test to check for loss of motility (CC types).
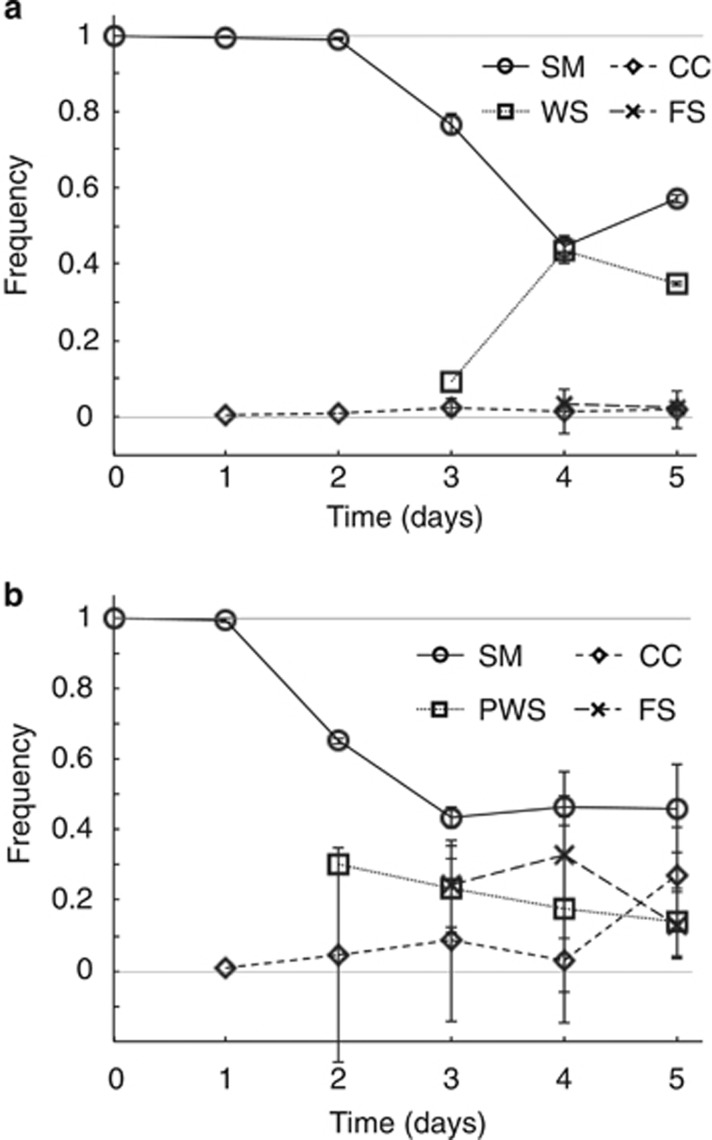
The dynamics of diversification are shown in Figure 4. Diversification of SM SBW25 (Figure 4a) showed a typical pattern with WS arising on day 3 and Fuzzy Spreader (FS) types at day 4 (Ferguson et al., 2013). Interestingly, CC types were detected on day 1 but never increased beyond a small fraction. No PWS types were observed. In diversifying populations of SBW25 Δwss (Figure 4b), CC types were also detected on the first day and increased in frequency through the five-day period. PWS types were detected from day 2 and FS types from day 3.
Figure 4.
Adaptive radiation in static microcosms over the course of five days for (a) ancestral SBW25 and (b) SBW25 Δwss. Data are average values from four independent replicates, error bars represent ±1 s.d.
The adaptive contributions of cellulose and PGA
Transposon insertions in pgaABCD responsible for reverting the PWS morphology to ancestral SM suggest that the encoded exopolysaccharide—a probable PGA—is likely to serve as the primary structural determinant of mat formation. Like cellulose, overproduction of PGA is presumed to prevent cells from separating fully after cell division thus facilitating colonisation of the air–liquid interface.
To elucidate the roles of PGA and the possible overlap/interaction with cellulose—the latter suggested by the fact that mutations in several DGC regulators affect both PGA and cellulose production—a set of genotypes were constructed combining DGC-activating mutations in mwsR, awsR, awsX and wspF with deletions of the wss operon, the pga operon, and both wss and pga operons. All strains with mutations in mwsR, awsR, awsX and wspF had reduced motility on soft agar (0.25%), regardless of the absence of cellulose and/or PGA, which shows that reduced motility is not sufficient for mat formation. These genotypes were competed against an isogenic set of strains carrying an intact wss operon and a GFP reporter. The GFP marker allowed flow cytometry to be used to determine the initial and final ratios. The starting ratio for these experiments was 1:1 and assays were performed in static broth microcosms where growth was primarily at the air–liquid interface.
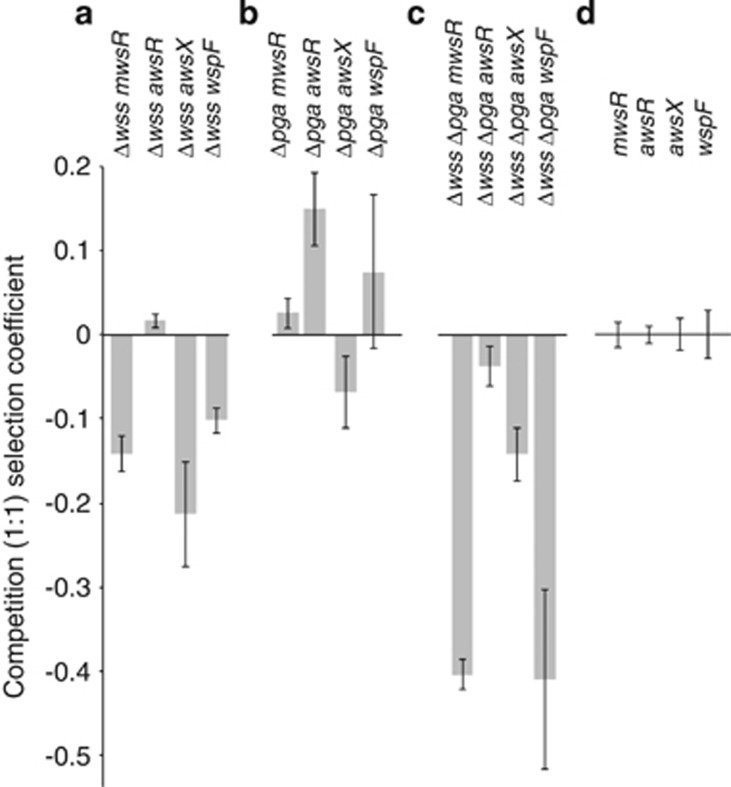
First, we focused on the adaptive contribution of cellulose. This involved competing genotypes lacking the wss operon (but harbouring the PGA operon) and carrying DGC-activating mutations in mwsR, awsR, awsX and wspF, with an isogenic set of strains that carried both PGA and cellulose biosynthetic loci. Because PWS types have not previously been detected when evolution in static broth microcosms proceeds from ancestral SM (cellulose-encoding) types, we expected to see cellulose overexpressing WS types outcompete cellulose negative, PGA overexpressing types. This expectation was realised for mwsR, awsX and wspF generated WS genotypes (two-sided t-tests, P<0.01), but not for awsR (P=0.74), where the PGA overexpressing PWS type was equivalent in fitness to the cellulose overexpressing WS type (Figure 5a). The result for the awsR genotype raises the possibility that AwsR affects expression of some additional and unidentified adhesive factor (this is further considered below). Overall, these data demonstrate significant contribution of cellulose to colonisation of the air–liquid interface as previously recognized (Spiers et al., 2002; Spiers et al., 2003).
Figure 5.
Competition of WS mutants against isogenic strains without wss and/or pga. Reconstructed mwsR, awsR, awsX and wspF mutants with GFP tags are competed 1:1 with strains carrying the same mutation but with (a) wss deleted, (b) pga deleted or (c) both wss and pga deleted to reveal the adaptive contribution of each exopolysaccharide. (d) Control competitions against unlabelled WS strains, defined as a selection coeffiecient s=0. Data are average values from four independent replicates, error bars represent ±1 s.d.
Next, the contribution of PGA was examined by competing genotypes lacking PGA against genotypes overexpressing cellulose and carrying PGA. Given the significant contribution of cellulose to fitness at the air–liquid interface, a first order expectation was that deletion of PGA would have little impact on competitive performance in this niche. The expectation was largely realised (Figure 5b), although the Δpga genotype carrying the awsR DGC-activating mutation showed evidence of enhanced fitness (two-tailed t-test P=0.03).
zFinally we explored the combined contribution of PGA and cellulose to colonisation of the meniscus. In monoculture all Δwss Δpga mutants were deficient in colonisation of the air–liquid interface, whereas this was not the case for mutants lacking just the cellulose biosynthetic locus wss (Supplementary Figure 1). mwsR and wspF mutants lacking PGA and cellulose were significantly less fit (two-sided t-tests P<10−5) than the PGA and cellulose proficient competitor, indicating that both contribute toward competitive performance at the air–liquid interface (Figure 5c) . The two aws mutants (awsX and awsR) were less fit (two-sided t-tests P<0.05), but not nearly as compromised, further supporting the view that aws affects the expression of some additional factor that contributes a component of fitness.
There is reason to treat this conclusion with caution: the above experiments were founded with equal ratios of competitors, one of which overproduces both PGA and cellulose. This could result in the establishment of physical interactions that might allow a genotype with minimal capacity to grow at the air–liquid interface to hitchhike with the common competitor. We therefore additionally performed competition assays where genotypes carrying DGC-activating mutations were mixed at a ratio of 1:100 with isogenic SM (non-adhesive producing) strains tagged with GFP and lacking either pga, or wss and pga. The choice of competitor ensures that there is little possibility for the rare strain to hitchhike with the common genotype.
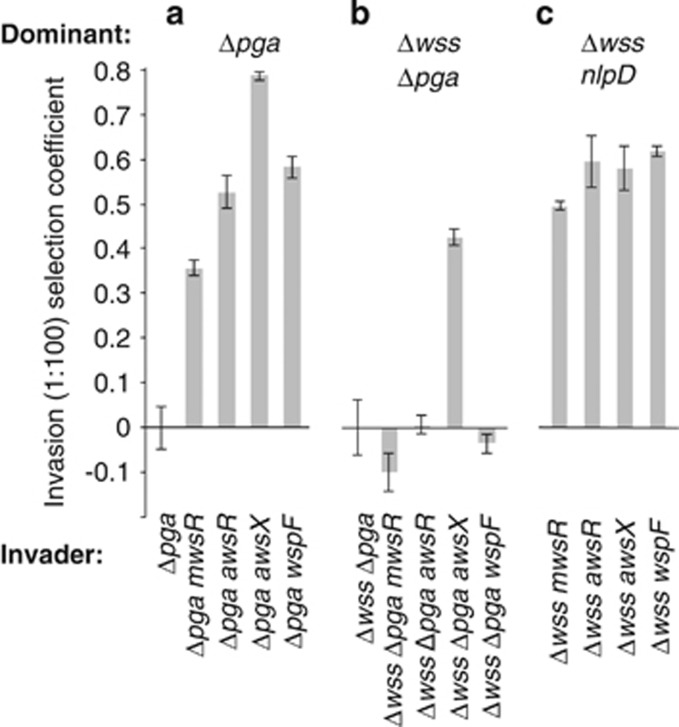
The results show that genotypes carrying the cellulose biosynthetic operon and DGC-activating mutations, but lacking pga can readily invade from rare against numerically dominant population of SM devoid of PGA (SM Δpga; Figure 6a; one-sided t-tests, P<10−3). However, loss of both pga and wss abolishes the adaptive advantage of these mutants (one-sided t-tests, P>0.1), except in the case where the DGC-activating mutation is in awsX (Figure 6b; one-sided t-tests, P<10−3). Interestingly this mutant also produces a slightly wrinkled, Congo Red positive, colony on agar plate culture (Supplementary Figure 2). It thus seems that the aws operon affects the expression of some yet to be determined adhesive factor. Interestingly, the effect is not seen in the awsR mutant, perhaps reflecting the fact that when AwsX (the negative regulator of the AwsR DGC) is inactive, AwsR achieves a level of activation beyond that possible through mutations in awsR alone. Figure 6 also shows the ability of PWS types to invade from rare populations dominated by CC types (Figure 6c).
Figure 6.
Ability of genotypes with WS-generating DGC mutations, but lacking wss and/or pga to invade from rare against isogenic non-adhesive producing SM types. (a) Strains with mutations activating DGCs that lack pga, but express cellulose, readily invade an isogenic SM population without the use of PGA. (b) Strains with DGC-activating mutations that lack both wss and pga cannot invade an isogenic SM population with the exception of awsX. (c) All Δwss PWS mutants can invade a Δwss nlpD population. Data are average values from four independent replicates, error bars represent ±1 s.d.
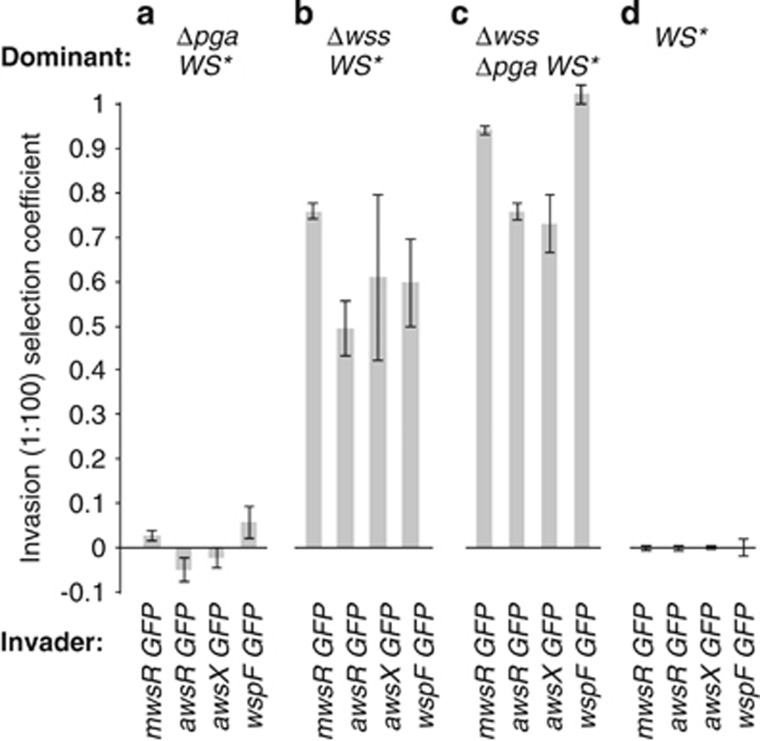
Last we asked if WS mutants (caused by mutations in mwsR, awsR, awsX or wspF) could invade from rare, isogenic populations lacking either pga, wss, or pga and wss. The results confirm the competitive superiority of cellulose-based mats. Absence of pga alone did not allow invasion of any of the otherwise isogenic DGC-activated WS types (Figure 7a), which suggests a limited role for pga when wss is present. However, WS types could rapidly invade a WS genotype lacking wss (Figure 7b). A similar, but even more pronounced effect was apparent when the common type lacked both wss and pga operons (Figure 7c).
Figure 7.
Ability of WS types carrying DGC-activating mutations in Mws, Aws and Wsp to invade from rare isogenic DGC-activated mutant types lacking wss and/or pga. (a) WS mutants cannot invade isogenic populations with a deletion of pga, but rapidly invade populations without wss (b). (c) Invasion rate is greatest against WS mutants lacking both wss and pga. (d) Control experiment for invasion of isogenic WS*-GFP versus WS*, both with wss and pga intact. WS* for the dominant population represent strains with identical WS mutations in mwsR, awsR, awsX or wspF as those of the invading strain. Data are average values from four independent replicates, error bars represent ±1 s.d.
Discussion
The number of mutational routes to solve a selective problem is of importance for understanding the causes of ecological success, parallel evolution and for evolutionary forecasting. If only one solution is possible, then there is no surprise to find only one pathway used. However, if there are multiple pathways to an adaptive phenotype, and yet only one is ever observed, then the causes of bias are of interest (Orgogozo, 2015). We have previously shown that the large diversity of mutational pathways underpinning generation of the wrinkly spreader phenotype all involve activation of DGCs and overproduction of cellulose (Lind et al., 2015). Despite the similar fitness effects due to activation of the different mutational pathways, only three of 39 potential pathways are routinely used. This bias stems from the fact that the three commonly trod pathways afford greatest target size (McDonald et al., 2009; Lind et al., 2015).
In this study our focus has been parallel phenotypic evolution of the WS type—a distinctive class of phenotype that uniquely evolves from constitutive activation of genes required for production of a cellulosic polymer. Whereas in previous studies of mutational routes we sequentially removed pathways encoding DGCs and observed recruitment of previously unused DGC-encoding pathways (Lind et al., 2015), here we eliminated ability of the ancestral genotype to produce cellulose and asked whether this type could converge on new non-cellulose-based solutions for mat formation (Figure 1). Two alternate ecological strategies were identified: one involved the genes pgaABCD predicted to encode the polysaccharide PGA and the other, a curious CC phenotype. Both alternate strategies were of reduced fitness compared to the cellulose-based WS types indicating that the selective advantage of mats, whose fabric is cellulose, explains phenotypic parallelism of WS types arising during the course of adaptive radiation.
Additional support for this conclusion comes from time-course experiments in which adaptive radiation was allowed to proceed from an ancestral SM type devoid of the cellulose-encoding wss operon. Both CC and PWS types were detected within the first two days of the radiation indicating that a low frequency of mutation does not limit the possibility that these types are detected when the radiation proceeds from the ancestral SM type (with the wss operon intact). Rather, the low fitness of CC and PWS types relative to WS types precludes the possibility that they increase to detectable levels. Of note is the presence of CC types after 24 h (in radiations from both SM and SM Δwss). This suggests the possibility that the CC phenotype is generated at high frequency sufficient to ensure that despite its low fitness it gains an early foothold in a vacant niche, only to be eliminated by WS types once they rise to prominence. This draws attention to the interplay between the timing at which a particular niche specialist mutant arises and ecological opportunity (Fukami et al., 2007).
The involvement of PGA solves a long-standing gap in knowledge evident from earlier exploration of WS types (Spiers et al., 2002; Spiers et al., 2003). In these previous studies transposon insertions in the Wsp pathway rendered the WS type indistinguishable from the ancestral SM phenotype, whereas mutations in the cellulose biosynthetic cluster, while reverting the wrinkled morphology to smooth, did not completely abolish the production of a Congo Red-binding factor responsible for adhesion to glass (Spiers et al., 2003). Salmonella typhimurium and E. coli rdar morphs, which are phenotypically similar WS and also caused by DGC-activating mutations, produce both cellulose and thin aggregative fimbriae (Romling et al., 2000; Zogaj et al., 2001). Microscopy of WS types revealed no evidence for the involvement of extracellular appendages (Spiers et al., 2003). The results of this work indicate that the previously unknown Congo Red-binding factor is PGA. Moreover, this work also shows that the PGA phenotype requires a two-component system encoded by PFLU0005 and PFLU0007. This constitutes a plausible system for transcriptional control, thus it appears that PGA is subject to both transcriptional control and post-translational control. In E. coli, c-di-GMP functions primarily as an allosteric activator that binds directly to PgaC and PgaD to increase glycosyltransferase activity (Steiner et al., 2013). The presence of a homologous two-component system (PA4032 and PA4036) in P. aeruginosa, which lacks the pgaABCD genes, suggests that the PFLU0005-PFLU0007 system might not exclusively regulate PGA expression or that its regulatory role is indirect.
PGA production delivers a clear fitness advantage to ancestral SM types that lack capacity to produce cellulose, but PGA seems to make little contribution to fitness when the cellulose biosynthetic pathway is intact. This is evident in Figures 5 and 7 and most apparent in Figure 7a where cellulose and PGA proficient genotypes carrying DGC-activating mutations are of equivalent fitness to the same genotype lacking the genes necessary to produce PGA (in the sensitive invasion from rare assay). Similarly, mutants expressing PGA, but not cellulose, are at a significant fitness disadvantage relative to the same genotypes that also produce cellulose (Figure 7b).
That both cellulose and PGA are effected by the same DGC-activating mutations is consistent with studies of adhesive overproducing mutants in other bacteria (Malone et al., 2010; Romling et al., 2013). Here, matters seem even more complex given that activation of AwsR (the DGC from the Aws pathway) appears to elicit production of a third, as yet unknown, Congo Red-binding polymer. A number of on-going and unresolved questions arise surrounding the mapping between DGCs and their targets. The first is the extent to which the connections revealed here—based on constitutive activation of DGCs—are indicative of those that hold in the ancestral genotype where DGCs are activated on receipt of specific signals, and possibly to levels never achieved by mutational activation. There is thus need for caution in reaching firm conclusions. Nonetheless, the connections revealed here provide a working hypothesis and prompt questions concerning specificity, hierarchy and magnitude of effects.
On the basis of colony phenotype alone, Aws and Mws appear to exert the greatest effect on PGA activation, with Wsp activation inducing no detectable change in colony morphology. Nonetheless, as fitness and Congo Red-binding assays show, Wsp activation does have an effect on PGA production (Figure 3; Supplementary Figure 2). Such differences raise the issue of whether the effects reflect genuine physiological differences in magnitude and specificity, or whether they are unintended consequences of c-di-GMP overactivation. Two pieces of data suggest the former. Firstly, data from fitness assays demonstrate a contribution of PGA toward fitness at the air–liquid interface: capacity of a PGA and cellulose proficient WS type to invade from rare is marginally higher when both PGA and Wss are intact (Figure 7b versus Figure 7c). This suggests that both are activated at the same time, as opposed to PGA activation being a spill-over effect of c-di-GMP that is targeted specifically to cellulose with effects on PGA evident only when Wss is absent. Secondly, the Aws pathway, when activated, elicits production of a third Congo Red-binding adhesive factor that is unaffected by activation of either Mws or Wsp. Interestingly the degree of activation of this third adhesive factor appears to depend on whether the DGC-activating mutation is in awsX or awsR. This is most readily understood from the perspective of the activating mutation: AwsX is a negative regulator of AwsR (Malone et al., 2010; McDonald et al., 2009) and our speculation is that when AwsX is defective, the level of AwsR activity is greater than achieved by a mutation directly within awsR.
The CC types were defined by mutations in nlpD. Strikingly, 23/24 mutants had identical base pair substitutions C565T (the mutation in the remaining mutant remains unknown), which cause a truncated protein (Q189*). This mutation was not present in the ancestral genotype and reversion of the mutation reversed the CC phenotype. Loss of nlpD has been shown to lead to a lack of activation of AmiC in E. coli, an amidase required for murein cleavage and proper cell division. The E. coli mutants also display the segmented cell morphology phenotype observed in CC types. Precisely how the chaining and segmented cell phenotype contributes to colonisation of the air–liquid interface is not known. Clearly the mutants have some capacity to adhere to the meniscus, but this may stem from adhesion to glass, with the chaining of cells allowing entanglement sufficient to bring about the formation of a weak mat. However, given the extreme molecular parallelism observed combined with the lack of mutations in amiC (which causes a similar phenotype (Uehara et al., 2010)), it is conceivable that the adaptive contribution is not directly related to loss of nlpD activity. Instead, effects maybe attributable to changes in expression of rpoS, which in Pseudomonas putida reportedly harbours a promoter in the nlpD open reading frame, with a transcriptional start site six base pairs downstream of the nlpD mutation (Kojic et al., 2002). Effects on rpoS expression may thus constrain the mutational target and explain the parallelism. In support of this hypothesis, σS, encoded by rpoS, is a positive transcriptional activator of the exopolysaccharide Psl in Pseudomonas aeruginosa (Irie et al., 2010).
Homologues of the pgaABCD operon have a limited presence among Pseudomonas species and are found in P. fluorescens and P. protegens, but not in P. aeruginosa, P. syringae, P. stutzeri or P. putida (Winsor et al., 2011; Whiteside et al., 2013). However, homologues are present in a variety of more distantly related gamma-proteobacteria including E. coli, Citrobacter, Serratia, Klebsiella and Yersinia, but not in Salmonella, Proteus or Vibrio (Whiteside et al., 2013). This patchy phylogenetic distribution suggests that lateral gene transfer, possibly in combination with gene loss, has played a role in the evolutionary history of pgaABCD. A similar patchy distribution is also evident for other exopolysaccharide biosynthetic clusters, such as colanic acid and those encoded by wss, pel and psl—all important structural components of bacterial biofilms and all subject to regulation by the c-di-GMP network (Hengge, 2009; Mann and Wozniak, 2012; Romling et al., 2013; Gallie et al., 2015; Martin et al., 2016). This begs the question as to how such ecological traits can be accommodated upon acquisition (Lind et al., 2010; Knöppel et al., 2014). One possibility is that the c-di-GMP network has evolved in such a way as to allow rapid regulatory integration of newly acquired structural loci. Such modular organisation, combined with a flexible regulatory network, might confer a degree of evolvability on Pseudomonas that underpins its capacity to adapt to novel conditions and invade new ecological niches (Spiers et al., 2000; Rainey and Cooper, 2004). In this context, this work adds to the increasing number of studies that show how model microbial populations can contribute understanding of the mechanistic bases of biofilm formation and biofilm community evolution with particular relevance to the establishment of chronic infections (Smith et al., 2006; Malone et al., 2012) and more generally to the challenge of surface colonisation (Martin et al., 2016).
Selection is a potent evolutionary force. P. fluorescens readily adapts to the challenge of growth in spatially structured microcosms via mutations that cause constitutive expression of DGCs, overproduction of c-di-GMP and adhesive polymers (Martin et al., 2016). It has long been known that cellulose confers a significant selective advantage to WS types (Spiers et al., 2003)—types that are also found in P. aeruginosa (Smith et al., 2006; Marvig et al., 2015) and Burkholderia cepacia (Traverse et al., 2013) from the lungs of cystic fibrosis patients. Here we have shown that in absence of cellulose, alternate ecological solutions to growth at the air–liquid interface are achievable; that these also involve recruitment of adhesive polymers—most notable PGA—but that the fitness conferred by PGA-based WS types is less than that available via cellulose. Selection thus explains parallel evolution cellulose-based WS types. Of particular significance is the fact that PGA and other adhesive factors are integrated within what appears to be a flexible c-di-GMP regulon.
There are numerous unanswered questions concerning the nature of the c-di-GMP regulatory system and the ecological factors shaping its evolution (Hengge, 2009; Srivastava and Waters, 2012; Kulasekara et al., 2013; Romling et al., 2013; Lori et al., 2015). A particularly pressing issue concerns the ecological significance of adhesive traits in environments where expression of these traits is subject to gene regulation (Gal et al., 2003; Giddens et al., 2007), as opposed to overexpression upon mutational activation, which is the norm in laboratory models of evolution. This requires that the mechanistic insight stemming from use of microbial model systems be applied to natural communities (Rainey et al., 2014), where, despite their often daunting complexity, there exists opportunity to explore the contribution of ecological differences among species to community structure, function and evolution (Jessup et al., 2004).
Acknowledgments
This work was supported by the Marsden Fund Council from New Zealand Government funding, administered by the Royal Society of New Zealand.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Bantinaki E, Kassen R, Knight CG, Robinson Z, Spiers AJ, Rainey PB. (2007). Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics 176: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. (2013). Genome dynamics during experimental evolution. Nat Rev Genet 14: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. (2009). Experimental evolution of bet hedging. Nature 462: 90–93. [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. (2009). The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191: 5953–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE. (1990). Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst 21: 373–398. [Google Scholar]
- Ferguson GC, Bertels F, Rainey PB. (2013). Adaptive divergence in experimental populations of Pseudomonas fluorescens. V. insight into the niche specialist ‘Fuzzy Spreader' compels revision of the model Pseudomonas radiation. Genetics 195: 1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. (2010). The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Fukami T, Beaumont HJ, Zhang XX, Rainey PB. (2007). Immigration history controls diversification in experimental adaptive radiation. Nature 446: 436–439. [DOI] [PubMed] [Google Scholar]
- Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. (2003). Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol Ecol 12: 3109–3121. [DOI] [PubMed] [Google Scholar]
- Gallie J, Libby E, Bertels F, Remigi P, Jendresen CB, Ferguson GC et al. (2015). Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens. PLoS Biol 13: e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig SM. (2005). Adaptation of pseudomonas fluorescens SBW25 to the air-liquid interface: a study in evolutionary genetics. University of Oxford: Oxford, UK.
- Gerstein AC, Lo DS, Otto SP. (2012). Parallel genetic changes and nonparallel gene-environment interactions characterize the evolution of drug resistance in yeast. Genetics 192: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM et al. (2007). Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci USA 104: 18247–18252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B. (2009). The causes of repeated genetic evolution. Dev Biol 332: 36–47. [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273. [DOI] [PubMed] [Google Scholar]
- Herron MD, Doebeli M. (2013). Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol 11: e1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB et al. (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol Evol 19: 189–197. [DOI] [PubMed] [Google Scholar]
- Jost MC, Hillis DM, Lu Y, Kyle JW, Fozzard HA, Zakon HH. (2008). Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol Biol Evol 25: 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- Knöppel A, Lind PA, Lustig U, Näsvall J, Andersson DI. (2014). Minor fitness costs in an experimental model of horizontal gene transfer in bacteria. Mol Biol Evol 31: 1220–1227. [DOI] [PubMed] [Google Scholar]
- Kojic M, Aguilar C, Venturi V. (2002). TetR family member PsrA directly binds the Pseudomonas rpoS and psrA promoters. J Bacteriol 184: 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. (2013). c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife 2: e01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. (2004). Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol 6: 726–732. [DOI] [PubMed] [Google Scholar]
- Lind PA, Tobin C, Berg OG, Kurland CG, Andersson DI. (2010). Compensatory gene amplification restores fitness after inter-species gene replacements. Mol Microbiol 75: 1078–1089. [DOI] [PubMed] [Google Scholar]
- Lind PA, Farr AD, Rainey PB. (2015). Experimental evolution reveals hidden diversity in evolutionary pathways. eLife 4: 07074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN et al. (2015). Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523: 236–239. [DOI] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C et al. (2010). YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog 6: e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Manfredi P, Dotsch A, Blanka A, Bos R et al. (2012). The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog 8: e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Wozniak DJ. (2012). Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36: 893–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C. (2000). Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol 326: 35–47. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. (2013). The Loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- Martin M, Holscher T, Dragos A, Cooper VS, Kovacs AT. (2016). Laboratory evolution of microbial interactions in bacterial biofilms. J Bacteriol 198: 2564–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig RL, Sommer LM, Molin S, Johansen HK. (2015). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47: 57–64. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Gehrig SM, Meintjes PL, Zhang XX, Rainey PB. (2009). Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll A, Dorr T, Alvarez L, Chao MC, Davis BM, Cava F et al. (2014). Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. J Bacteriol 196: 3937–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgogozo V. (2015). Replaying the tape of life in the twenty-first century. Interface Focus 5: 20150057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. (1998). Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Rainey PB. (1999). Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol 1: 243–257. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Cooper TF. (2004). Evolution of bacterial diversity and the origins of modularity. Res Microbiol 155: 370–375. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Desprat N, Driscoll WW, Zhang XX. (2014). Microbes are not bound by sociobiology: response to Kummerli and Ross-Gillespie (2013). Evolution 68: 3344–3355. [DOI] [PubMed] [Google Scholar]
- Römling U, Rohde M, Olsen A, Normark S, Reinkoster J. (2000). AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol 36: 10–23. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM et al. (2009). Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 103: 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AJ, Buckling A, Rainey PB. (2000). The causes of Pseudomonas diversity. Microbiology 146: 2345–2350. [DOI] [PubMed] [Google Scholar]
- Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. (2002). Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. (2003). Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol 50: 15–27. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Waters CM. (2012). A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J Bacteriol 194: 4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Lori C, Boehm A, Jenal U. (2013). Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32: 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. (2013). The genetic causes of convergent evolution. Nat Rev Genet 14: 751–764. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD et al. (2012). The molecular diversity of adaptive convergence. Science 335: 457–461. [DOI] [PubMed] [Google Scholar]
- Tidhar A, Flashner Y, Cohen S, Levi Y, Zauberman A, Gur D et al. (2009). The NlpD lipoprotein is a novel Yersinia pestis virulence factor essential for the development of plague. Plos One 4: e7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. (2013). Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA 110: E250–E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. (2010). Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J 29: 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Preston JF3rd, Romeo T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186: 2724–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside MD, Winsor GL, Laird MR, Brinkman FS. (2013). OrtholugeDB: a bacterial and archaeal orthology resource for improved comparative genomic analysis. Nucleic Acids Res 41: D366–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. (1999). Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY et al. (2011). Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39: D596–D600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Aardema ML, Medina EM, Schumer M, Andolfatto P. (2012). Parallel molecular evolution in an herbivore community. Science 337: 1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. (2001). The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39: 1452–1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.