Our findings show that Archaea are a habitual and vital component of human and great ape gut microbiomes but are largely ignored on account of the failure of previous studies to realize their full diversity. Here we report unprecedented levels of archaeal diversity in great ape gut microbiomes, exceeding that detected by conventional 16S rRNA gene surveys. Paralleling what has been reported for bacteria, there is a vast reduction of archaeal diversity in humans. Our study demonstrates that archaeal diversity in the great ape gut microbiome greatly exceeds that reported previously and provides the basis for further studies on the role of archaea in the gut microbiome.
KEYWORDS: archaea, great apes, gut microbiome, microbial diversity, species interactions
ABSTRACT
Archaea are habitual residents of the human gut flora but are detected at substantially lower frequencies than bacteria. Previous studies have indicated that each human harbors very few archaeal species. However, the low diversity of human-associated archaea that has been detected could be due to the preponderance of bacteria in these communities, such that relatively few sequences are classified as Archaea even when microbiomes are sampled deeply. Moreover, the universal prokaryotic primer pair typically used to interrogate microbial diversity has low specificity to the archaeal domain, potentially leaving vast amounts of diversity unobserved. As a result, the prevalence, diversity, and distribution of archaea may be substantially underestimated. Here we evaluate archaeal diversity in gut microbiomes using an approach that targets virtually all known members of this domain. Comparing microbiomes across five great ape species allowed us to examine the dynamics of archaeal lineages over evolutionary time scales. These analyses revealed hundreds of gut-associated archaeal lineages, indicating that upwards of 90% of the archaeal diversity in the human and great ape gut microbiomes has been overlooked. Additionally, these results indicate a progressive reduction in archaeal diversity in the human lineage, paralleling the decline reported for bacteria.
IMPORTANCE Our findings show that Archaea are a habitual and vital component of human and great ape gut microbiomes but are largely ignored on account of the failure of previous studies to realize their full diversity. Here we report unprecedented levels of archaeal diversity in great ape gut microbiomes, exceeding that detected by conventional 16S rRNA gene surveys. Paralleling what has been reported for bacteria, there is a vast reduction of archaeal diversity in humans. Our study demonstrates that archaeal diversity in the great ape gut microbiome greatly exceeds that reported previously and provides the basis for further studies on the role of archaea in the gut microbiome.
INTRODUCTION
Archaea are well-recognized residents of the human gut microbiome (1), but the prevalence, diversity, and evolution of human-associated Archaea are still largely unknown. Previous studies have revealed that Methanobrevibacter is the predominant archaeal genus in the human gut, although a few other genera, such as Methanosphaera and members of the order Methanomassiliicoccales have been recurrently detected in a small percentage of hosts (1). Most human microbiome surveys focus on the bacterial component of gut microbial communities because relatively few sequences classified as Archaea are recovered even when microbiomes are sampled deeply (1–6). Moreover, the universal prokaryotic primers typically used to interrogate microbial communities do not fully capture the breadth of archaeal diversity (1, 7), which has potentially left vast amounts of archaeal diversity unobserved.
Due to these limitations, we hypothesized that the archaeal diversity within the gut microbiomes of great apes is much higher than previously recognized. Using primers designed to target a broad range of archaea, we investigated the diversity of archaeal taxa within the gut microbiomes of great apes. These data revealed that upwards of 90% of the archaeal diversity in human and great ape gut microbiomes has been previously overlooked. Additionally, comparison of archaeal diversity within five great ape species revealed the dynamics of archaeal lineages across evolutionary timescales. Although several archaeal lineages have been maintained throughout hominoid evolution, paralleling what has been reported for bacteria, we observed a vast reduction of archaeal diversity in humans that has been accompanied by the loss of some associations between archaea and bacteria typical of other great apes.
RESULTS
Increased diversity captured with archaeon-targeted primers.
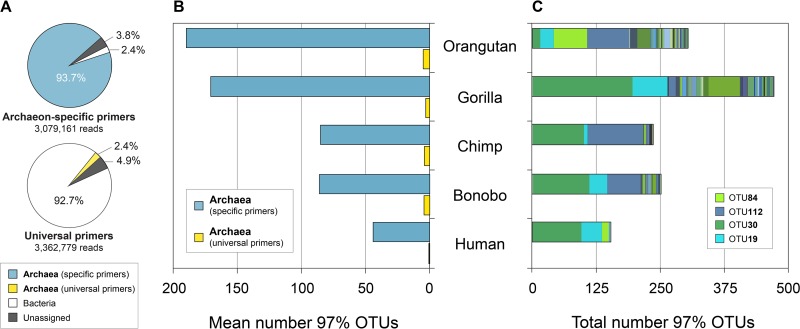
We sequenced the V4-V5 region of the 16S rRNA gene using the archaeon-specific primer pair 516F/915R (see Table S1 in the supplemental material) (8, 9) to evaluate the diversity of Archaea in the gut microbiomes of humans (n = 10) and four other great ape species: Pan troglodytes (chimpanzee [n = 14]), Pan paniscus (bonobo [n = 18]), Gorilla gorilla (gorilla [n = 20]), and Pongo pygmaeus (orangutan [n = 8]). To more accurately compare diversity between species, all individuals from each species were sampled at a single geographic location: humans in the United States, chimpanzees in Tanzania, bonobos in the Democratic Republic of the Congo, gorillas in Cameroon, and orangutans in the United States. Standardization of extraction protocols enabled us to compare the diversity recovered with archaeon-specific primers to that reported for the same samples using 515F/806R primers (Table S1), which are widely used in microbiome studies (6). Of the 3,079,161 reads obtained with the archaeon-specific primers, 3.8% were bacterial in origin, which likely represents a low level of indiscriminate priming since the primers were designed to have complete specificity to Archaea (see Data Set S1 in the supplemental material for primer performance details). Conversely, archaeal reads accounted for only 2.4% of the 3,362,779 reads obtained with the 515F/806R universal prokaryotic primers (Fig. 1A). This low number of archaeal reads obtained with universal primers can be attributed in part to their low specificity to Archaea (Data Set S1). After removing unassigned reads from the archaeal and universal 16S rRNA gene data sets, the diversity within the archaeal data set was analyzed with and without bacterial reads, and all data sets were subsampled to a maximum rarefaction depth of 10,000 reads per sample. Differences in the specificity, binding preferences, and coverage spectrum of the universal prokaryotic and archaeon-specific primers prohibit direct comparisons of the relative abundance of archaea present in the two data sets. Removal of the small number of bacterial reads from the archaeal data set did not significantly change the results, so subsequent analyses are based on the data set excluding bacterial reads (see Data Set S2 for per individual read details).
FIG 1 .
Archaeal diversity in great ape gut microbiomes. (A) Proportion of total reads assigned to archaea when gut microbiomes are surveyed with archaeon-specific primers and with the 515F/806R universal prokaryotic primers, along with percentages of bacterial and taxonomically unassigned reads. Percentages are based on the complete set of fecal samples from five great ape host species: Homo sapiens (n = 10), Pan troglodytes (n = 14), Pan paniscus (n = 18), Gorilla gorilla (n = 20), and Pongo pygmaeus (n = 8). (B) Mean numbers of archaeal OTUs when gut microbiomes are surveyed with archaeon-specific primers and with universal prokaryotic primers. Sequences were rarefied to 10,000 reads per sample, and OTUs clustered at 97% sequence identity. (C) Total numbers of archaeal OTUs when gut microbiomes are surveyed with archaeon-specific primers. Color coding of four of the most prevalent archaeal OTUs detected in great apes is indicated. Sequences were rarefied to 10,000 reads per sample, and OTUs clustered at 97% sequence identity.
Performance of the primers used in this study. Primer performance was evaluated using TestPrime 1.0 implemented on the SILVA database (http://www.arb-silva.de/search/testprime/). For both primer pairs, the performance was calculated allowing either 0 or 1 mismatch. Coverage = (matches/eligible) × 100, specificity = 100 − (outgroup matches/outgroup matchable) × 100, Accessions = number of sequences in the taxonomic path, Eligible = number of regions with sequence data at the position of the primer pair, Mismatch = number of mismatched regions with the primer pair in the taxonomic path, and No Data = number of regions with no sequence data at the position of the primer. Download DATA SET S1, XLSX file, 0.1 MB (75.7KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Archaeal read counts pre- and postfiltering for each individual. Total reads for each individual using archaeon-specific primer pair Arch516F/Arch915R after quality filtering (Phred score above Q20). Shown are the percentages of chimeric sequences, singletons, unassigned, and bacterial reads detected per individual. Download DATA SET S2, XLSX file, 0.1 MB (52.5KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Numbers of OTUs detected with (A) VSEARCH and (B) UCLUST. Download TABLE S1, PDF file, 0.1 MB (31.8KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The mean number of archaeal operational taxonomic units (OTUs) detected in each great ape species, based on a 97% sequence identity threshold, was substantially larger when interrogating the samples with the archaeon-specific primers (orangutans, 161 OTUs for archaeon-targeted primers and 7 for universal primers; gorillas, 135 OTUs for archaeon-targeted primers and 7 for universal primers; bonobos, 71 OTUs for archaeon-targeted primers and 6 for universal primers; chimpanzees, 69 OTUs for archaeon-targeted primers and 7 for universal primers; humans, 37 OTUs for archaeon-targeted primers and 1 for universal primers) (Fig. 1B). In terms of overall counts, we identified a total of 646 archaeal 97% OTUs, with over 100 in each species (orangutans, 302; gorillas, 470; bonobos, 247; chimpanzees, 235; and humans, 120) (Fig. 1C). Even at sequencing depths of 200,000 reads, the universal primers capture less than 20% of archaeal diversity that is detected at 10,000 reads with the archaeon-specific primers (see Data Set S3 in the supplemental material). Samples were also interrogated with the universal primer pair that is commonly used to assay bacterial diversity in microbial communities (10). The mean numbers of 97% OTUs detected in each great ape species were n = 533 for orangutans, n = 330 for gorillas, n = 383 for bonobos, n = 467 for chimpanzees, and n = 204 for humans, consistent with bacterial diversity reported in reference 6.
Numbers of archaeal reads and OTUs detected with the universal and archaeon-specific primer pairs. Total numbers of archaeal reads and 97% OTUs in each sample detected with universal prokaryotic primer pair 515F/806R were counted at original read depths (i.e., before rarefraction). Read depths obtained with archaeon-specific primers were rarefied to the number of archaeal reads obtained with universal prokaryotic primers (column G) prior to computing total numbers of OTUs (column J) and the percentage detected with universal versus archaeal primers (column K). Download DATA SET S3, XLSX file, 0.1 MB (47.9KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomic assignment of 16S rRNA gene sequences are typically performed with the SILVA (11), RDP (12, 13), or Greengenes (14) databases, but these classifiers have rather limited training sets for Archaea. In the entire set of 646 archaeal 97% OTUs, 200 (31%) could not be classified as any genus, and 39 (6%) were classified as the incorrect phylum (e.g., assigned to phylum Crenarchaeota but exhibited best BLAST hits to members of the phylum Thaumarchaeota). Because assignment of archaeal OTUs to known taxa is problematic, we analyzed the phylotypic diversity without relying on a reference-based taxonomy and instead base our analyses on 94% OTUs and 97% OTUs, which have traditionally been considered sequence thresholds that delineate genera and species, respectively (15).
At 97% clustering, 74 OTUs were common to all five great ape species (see Fig. S1A in the supplemental material). When considering only OTUs constituting the core archaeal microbiome (defined as those OTUs present in at least 80% of the individuals in each host species), nine 97% OTUs are shared by all species. The core archaeal microbiome comprises only 9% of the total number of 97% OTUs detected in gorillas but 26% of the archaeal diversity in orangutans (Fig. S1A and B), indicating that there is no simple relationship between the total diversity harbored by a species and the scope of its core microbiome.
Number of archaeal 97% OTUs shared among great ape species. (A) Venn diagram showing the number of archaeal 97% OTUs shared between species, with the full number of archaeal 97% OTUs detected in each host species shown under the species name. (B) Venn diagram showing the number of core archaeal 97% OTUs shared between species, with the full number of core OTUs detected in each host species shown under the species name. Download FIG S1, PDF file, 0.1 MB (75.9KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the core archaeal OTUs from each species (incorporating one representative sequence per OTU) revealed two lineages—OTU30 and OTU84—that could be unequivocally assigned to the known human gut associates Methanobrevibacter smithii and Methanosphaera stadtmanae (see Fig. S2 in the supplemental material). The 97% OTU corresponding to Methanobrevibacter smithii (OTU30) is present in all species sampled and occurs at the highest relative abundance in a majority of the humans (Fig. 1C; see Data Set S4 in the supplemental material). In addition to Methanobrevibacter smithii, Methanomassiliicoccales sp. (OTU112) and Methanobrevibacter sp. (OTU19) were present in all humans sampled, and Methanosphaera stadtmanae was present in 90% of humans sampled. Despite the extensive OTU diversity detected with archaeon-specific primers, none of the OTUs classified as the order Methanomassiliicoccales matched the reference species typically detected in the human microbiome (Fig. S2). Although many of the 14 core archaeal OTUs in the human gut microbiome are present in at least one other ape species (Fig. S1B), it is not possible to infer their gain or loss during great ape diversification because several are shared exclusively by paraphyletic taxa or are only sporadically present in other species.
Prevalence of archaeal OTUs in the great ape gut microbiome species. The nine core 97% OTUs present in all great ape host species are shown in gray. The core 97% OTUs of each species (i.e., those present in >80% of sampled individuals) are in black boldface (related to Fig. S2). Download DATA SET S4, XLSX file, 0.1 MB (77.3KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic diversity of archaeal 97% OTUs. Unrooted maximum likelihood phylogeny of the 110 archaeal core 97% OTUs is shown. The phylogeny was inferred by PhyML (GTR+Γ4) and is based on a representative sequence for each OTU. OTUs included in the analysis were present in >80% of individuals from each species, and the nine OTUs present in all great ape host species are indicated in red. The scale bar represents average number of substitutions per site. The reliability for the internal branches of the ML tree was assessed by the bootstrapping method (100 bootstrap replicates) and the approximate likelihood ratio test (aLRT–SH-Like) (51). Dots at nodes denote aLRT values: black, >0.8; gray, >0.5; white, <0.5. Download FIG S2, PDF file, 0.1 MB (75.7KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Declining archaeal diversity in the gut microbiome during great ape diversification.
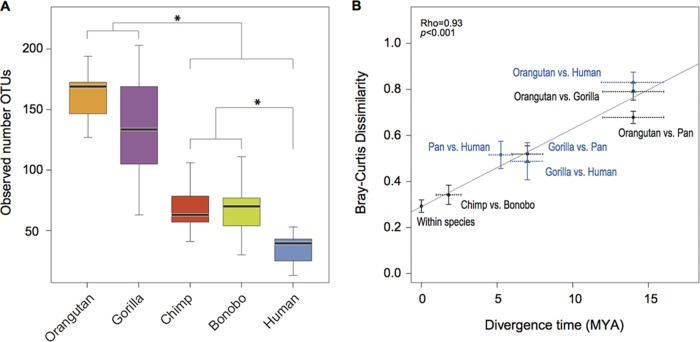
We compared the numbers of 97% OTUs recovered from each of the five host species to determine if archaeal diversity within the gut microbiome has changed over the evolutionary history of great apes: phylogenetic relationship of great apes = ({[human (chimp, bonobo)], gorilla}, orangutan). Paralleling what has been reported for bacteria (6), archaeal diversity is lower in humans than in other great apes (Fig. 2A; see Fig. S3A in the supplemental material). More than 90% of the archaeal 97% OTUs detected in humans, chimpanzees, and bonobos were detected in one or more of the other great ape species, whereas gorillas and orangutans harbor many unique OTUs (Fig. S1A). Principal component analysis of both unweighted and unweighted UniFrac distances differentiated host genera, but the two species of Pan broadly overlapped in archaeal diversity (Fig. S3B and C). Because the two Pan species are from geographically separated locations—bonobos reside in the Democratic Republic of the Congo and chimpanzees in Tanzania—the similarities in their microbiome compositions are a not consequence of shared habitats.
FIG 2 .
Changes in archaeal diversity during great ape diversification. (A) Alpha diversity of archaeal OTUs in great ape gut microbiomes. Box-and-whiskers plots show high, low, and median values, with the lower and upper edges of each box denoting the first and third quartiles, respectively. Species comparisons yielding statistically significant differences are indicated by asterisks (*, P < 0.05, Wilcoxon rank-sum test after Bonferroni correction). (B) Association between microbiome divergence, indexed by Bray-Curtis dissimilarity of 97% OTUs, and host divergence time (Spearman correlation; rho = 0.93, P < 0.001). Black crosses denote comparisons within all species and between species, excluding humans, and blue crosses denote comparisons between humans and other great ape species. Capped verticals of each cross denote the 99% confidence interval of pairwise Bray-Curtis dissimilarities, and the dotted horizontal lines show the range of divergence times estimated for the species pair considered. A significant correlation between Bray-Curtis dissimilarities and divergence times was also ascertained by a Mantel test (P < 0.01, 10,000 replicates).
Diversity of archaeal 97% OTUs in the gut microbiomes of great apes. (A) Diversity measured by Shannon’s index, Simpson’s index, Fisher’s alpha, and Chao1. Box-and-whiskers plots show high, low, and median values, with the lower and upper edges of each box denoting the first and third quartiles, respectively. Species comparisons yielding statistically significant differences are indicated by asterisks (*, P < 0.05, Wilcoxon rank-sum test after Bonferroni correction). (B) Principal coordinate analysis of unweighted UniFrac distances of the archaeal component of the gut microbiomes of five great ape species. (C) Principal coordinate analysis of weighted UniFrac distances of the archaeal component of the gut microbiomes of five great ape species. Download FIG S3, PDF file, 0.1 MB (80.4KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Despite differences among species in diet, habitat, and lifestyles, it has been shown that the bacterial composition of the gut microbiome has changed at a relatively clock-like rate during the diversification of great apes (6). The same is true for Archaea: for 97% OTUs, the Bray-Curtis dissimilarity between the archaeal communities of each pair of species increases linearly with the divergence times between species (r2 = 0.93, P = 0.0001) (Fig. 2B).
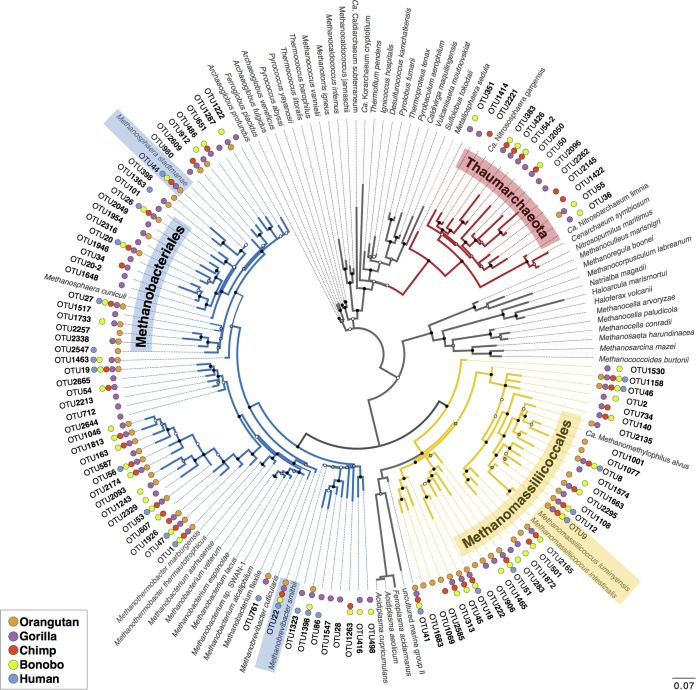
When examined at the level of 94% OTUs, the archaeal diversity within the great ape microbiomes shows patterns consistent with those detected with 97% OTUs—most notably, there is a decrease in diversity in humans (see Fig. S4 in the supplemental material). Overall, the great apes harbored a total of 104 archaeal 94% OTUs at a rarefaction depth of 10,000 reads. Phylogenetic analysis of one representative of each of these 104 OTUs with 58 reference archaeal 16S rRNA gene sequences (i.e., well-classified representatives from the major archaeal lineages with complete genomes) placed most of the 94% OTUs in the orders Methanobacteriales, Methanomassiliicoccales, and Nitrososphaerales (Fig. 3). Members of these orders have previously been detected in the human gut microbiome (1), but as noted above, only Methanobrevibacter is reported to be a habitual resident.
FIG 3 .
Phylogenetic diversity of archaeal lineages in the great ape gut microbiome. Shown is an unrooted maximum likelihood tree of 102 archaeal 94% OTUs detected in great apes (numbered OTUs) and 58 archaeal 16S reference sequences (Latin binomes). Phylogeny was inferred by PhyML (GTR+Γ4). Colored branches denote the major three archaeal groups to which all identified OTUs belong: Methanobacteriales (blue), Methanomassiliicoccales (yellow), and Thaumarchaeota (red). OTUs identical or with very high similarity to archaeal strains previously identified in the human gut microbiome are highlighted: M. smithii and M. stadtmanae in blue and M. luminyensis and “Ca. Methanomassiliicoccales alvus” in yellow. Solid circles at branch termini show the OTU distribution among host species, with OTUs considered present if found in any individual of a species. The scale bar represents the average number of substitutions per site. Reliability of internal branches was assessed by bootstrapping (100 replicates) and the approximate likelihood ratio test (aLRT–SH-Like). Dots at nodes denote aLRT values: black, >0.8; gray, >0.5; white, < 0.5.
Diversity of archaeal 94% OTUs in the gut microbiomes of great apes. Diversity was measured by the following indices: observed OTUs, Shannon’s index, Simpson’s index, Fisher’s alpha, and Chao1. Box-and-whiskers plots show high, low, and median values, with the lower and upper edges of each box denoting the first and third quartiles, respectively. Species comparisons yielding statistically significant differences are indicated by asterisks (*, P < 0.05, Wilcoxon rank-sum test after Bonferroni correction). Download FIG S4, PDF file, 0.1 MB (58KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mapping the phylogenetic distribution of 94% OTUs in each host species revealed that there is much more diversity of Methanobacteriales- and Methanomassiliicoccales-related OTUs in humans than has been previously recognized. The 94% OTUs clustering within the phylum Thaumarchaeota (order Nitrososphaerales) were recovered exclusively from the nonhuman apes (Fig. 3). Moreover, not one 94% OTU is uniquely present in humans, whereas the nonhuman great ape species, particularly gorillas and orangutans, each possess many unique 94% OTUs. Most 94% OTUs could not be assigned to a prescribed archaeal genus, and the phylogenetic distance between the OTUs that we recovered and the reference genomes demonstrates that there are vast amounts of archaeal diversity in the great ape microbiome that have not previously been observed (Fig. 3).
Microbial associations maintained throughout hominoid evolution.
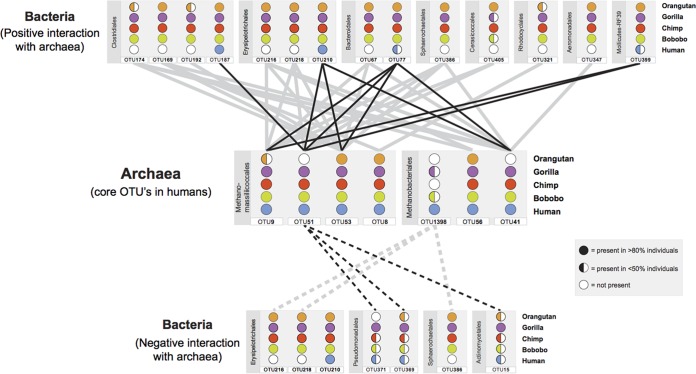
Because certain archaea in the gut are known to be metabolically dependent on bacteria, we searched for associations between 94% archaeal OTUs and the bacterial genera present in the gut microbiomes of humans and great apes. We considered only those significant negative and positive cooccurrence relationships (P < 0.001 after Bonferroni correction) involving the core archaeal 94% OTUs of humans having Spearman’s rho values less than −0.50 or greater than 0.50 with a human- or great ape-associated bacterial genus (see Data Set S5 in the supplemental material). We identified 17 bacterial genera that are significantly positively or negatively correlated with seven of these archaeal 94% OTUs (Fig. 4). Notably, there were no significant associations between the archaeal OTUs assigned as Methanobrevibacter smithii (OTU22) or Methanosphaera stadtmanae (OTU44), both of which are common constituents in the gut microbiome, and any of the bacterial genera. Overall, we identified more bacterial associations with Methanomassiliicoccales-related OTUs than with Methanobacteriales-related OTUs: Prevotella (OTU77) was found to be positively associated with three Methanomassiliicoccales-related OTUs (OTU9, OTU51, and OTU53) and one Methanobacteriales-related OTU (OTU41). Most of these significant associations occur between archaeal and bacterial genera that were not present in the human gut microbiome. There were positive associations between Methanomassiliicoccales-related OTU53 and Methanobacteriales-related OTU41 with Sphaerochaeta, Butyrivibrio, Oribacterium, and unclassified members of the family Erysipelotrichaceae and the order Bacteroidales, none of which were detected in our human samples (Fig. 4). However, 14 bacterium-archaeon associations that occur in humans are present in multiple great ape species, such as the positive associations among Methanomassiliicoccales-related OTUs (OTU9, OTU51, and OTU53) and Clostridiales (OTU187), Erysipelotrichales (OTU210), Bacteroidales (OTU77) and Mollicutes-RF39 (OTU399), and between Methanobacteriales-related OTU41 and Bacteroidales (OTU77) and Erysipelotrichales (OTU210) (Fig. 4), and the negative associations between Methanomassiliicoccales-related OTU51 and Pseudomonadales (OTU369, OTU371) and Actinomycetales (OTU15).
FIG 4 .
Archaeon-bacterium associations in great ape gut microbiomes. Networks of positive and negative associations between archaeal 94% OTUs and bacterial genera are shown. Associations are based on relative abundances and include only those involving the seven archaeal 94% OTUs comprising the core archaeal microbiome of humans. Distributions of archaeal OTUs and bacterial genera among great ape species are indicated by solid and open circles, respectively. Lines denote statistically significant associations (P = 0.001) with a Spearman’s rho value of less than −0.5 or greater than 0.5. Black lines show positive and negative associations between archaeal OTUs and bacterial genera present in humans, whereas gray lines show associations of the same archaeal OTUs with bacterial genera not detected in humans. (See Data Set S5 for additional taxonomic information.)
Cooccurrence relationships between archaea and bacteria present in the gut microbiomes of great apes. Only statistically significant associations (P = 0.001) with a Spearman’s rho value less than −0.5 or greater than 0.5 are shown. Associations between the nine core 97% OTUs (i.e., those present in all host species) and bacterial genera not detected in human samples are shown in gray. Associations involving these core OTUs and bacterial genera present in humans (and other great apes) are shown in black. Download DATA SET S5, XLSX file, 0.1 MB (13.8KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Surveys of the microbial diversity within thousands of individuals (2–6, 16) have led to the view that relatively few archaeal lineages are regular constituents of the human gut microbiome. Until recently, only two methanogenic species, Methanobrevibacter smithii and Methanosphaera stadtmanae, both in the order Methanobacteriales, had been detected in the human gut (1). But the isolation of additional strains representing a new archaeal order, Methanomassiliicoccales, demonstrated that other methanogens are occasionally present (17–21). Using primers designed to target archaeal rRNA genes—as opposed to the “universal” prokaryotic primers employed in the majority of studies—we show that previous surveys and strain isolation procedures have vastly underestimated the extent of archaeal diversity in the gut microbiome, even when interrogated at very high sequencing depths.
Methanobrevibacter smithii has been reported to occur in 64% to 95% of individuals sampled (22, 23), and several studies suggest that this species is the most abundant methanogen in the human gut and constitutes up to 10% of the anaerobes found in the colon (16, 24, 25). In contrast, Methanosphaera stadtmanae and other members of the order Methanomassiliicoccales occur in a minority of humans and are present in low abundance (16, 17, 22, 26, 27). Consistent with previous studies, we recovered OTUs representing M. smithii, M. stadtmanae, and Methanomassiliicoccales species in the majority of the humans sampled. Although we sampled a relatively small number of humans from a single geographic location, we observed M. stadtmanae and Methanomassiliicoccales at much higher frequencies than previously reported, suggesting that these taxa are much more prevalent than currently recognized.
Archaea belonging to nonmethanogenic lineages such as halophiles, Nitrososphaera, and Sulfolobus have also been detected sporadically in the human gut microbiome (2–6, 28–31). Despite the high sequencing depth at which we surveyed Archaea (averaging over 40,000 reads per sample), we detected no OTUs belonging to nonmethanogenic lineages: all of the archaeal OTUs we identified in humans fell within the methanogenic orders Methanobacteriales and Methanomassiliicoccales. Given the limitations of our sampling scheme, it is possible that nonmethanogenic archaea are occasionally present in the human gut microbiome. However, we recovered 120 97% OTUs in humans, over 100 times the number detected in the same samples with universal prokaryotic primers. In addition, 14 of these OTUs were detected in at least 80% of humans, indicating that Archaea are prevalent in the human gut.
The numbers of archaeal OTUs in the gut microbiomes of orangutans, gorillas, bonobos, and chimpanzees are high, with each species containing at least twice the 97% OTUs of humans and having representatives of the phylum Thaumarchaeota, which were not detected in our human samples. Other studies have identified Nitrososphaera, an ammonium oxidizer in the phylum Thaumarchaeota, in gut microbiome studies of United States and Malawian populations (1, 5, 29), as well as on human skin (32); therefore, the lack of detection of Thaumarchaeota in our human samples could be due to sampling bias. Due to their sporadic occurrence and relatively low frequencies in gut microbiomes, Thaumarchaeota could be transitory and not habitual gut constituents. The fact that 74 of the 97% OTUs from the orders Methanobacteriales and Methanomassiliicoccales were detected in all great ape species, and nine of these shared OTUs are present in the majority of individuals in each species, suggests that these lineages occurred in the ancestor of all great apes and have possibly coevolved with their hosts for millions of years, as reported for some bacterial lineages (33).
By studying the gut microbiome great apes in a phylogenetic context, we can begin to gain insight into how the composition of the gut microbiome has changed over time. Analysis of humans, chimpanzees, bonobos, gorillas, and orangutans reveals that there has been a loss of archaeal diversity during the diversification of great apes, particularly in the human lineage. Because methanogens help degrade plant polysaccharides by using the end products of bacterial fermentation (17–21, 34), and some methanogens are capable of reducing methanol, which is generated through bacterial degradation of pectin, mainly from fruits (22, 23, 35), the higher archaeal diversity in nonhuman apes could reflect their plant-based diets: gorillas and orangutans, which are principally herbivorous, have a higher level of archaeal diversity than do omnivorous chimpanzees and humans. Although recent studies have shown that captivity can cause reductions in bacterial diversity of gut microbiomes (36, 37), the orangutans, which were all sampled from a zoo colony, averaged the highest level of archaeal diversity of all great apes—even higher than that of gorillas, chimpanzees, and bonobos, which were sampled from wild populations. Moreover, our surveys of bacterial diversity using universal prokaryotic primers revealed that orangutans averaged the highest level of bacterial diversity as well. Since microbiome reductions are often ascribed to dietary restrictions of plant material, the maintenance of high levels of diversity in captive orangutans likely reflects their diverse plant-based diet, which includes more than a dozen families of fruits, grains, roots, and leafy vegetables. Nutrients from fruits, stems, and leaves are fermented in the colon, which is much larger in nonhuman apes—the human colon occupies 17 to 23% of the digestive tract, whereas in chimpanzees, orangutans, and gorillas it is 52 to 54% (38). This reduction in fermentation-assisting archaea, like colon size, could be a response to higher-quality foods, which can be digested in the small intestine (38).
In many environments, including the human gut, archaea live in syntrophy with bacteria (39), and there are numerous archaeal OTUs whose presence or absence coincided with particular bacterial genera in the microbiomes of great apes. We identified positive associations of the bacterial genus Prevotella (OTU77 in Fig. 4) with Methanobrevibacter (OTU41) and Methanomassiliicoccales (OTU9, OTU51, and OTU53). OTUs in Methanobrevibacter and Prevotella were previously assigned as members of distinct gut enterotypes (40), so their cooccurrence in all great ape species was not anticipated, although later studies have reported positive associations between these taxa (29, 41). Most of the bacterium-archaeon associations that we identify have never previously been observed, but that is because most involve bacterial taxa not present in human samples. This suggests that along with the decrease in archaeal and bacterial diversity in the human gut microbiome, many associations between bacteria and archaea have been lost. However, several bacterium-archaeon associations are maintained in multiple great ape species, suggesting evolutionary conservation and providing a starting point for future investigations on the role of archaea in the gut microbiome.
Here, we show that much of the archaeal diversity in the gut microbiome has been overlooked due to the reliance on prescribed sets of universal prokaryotic primers to characterize microbiomes. High numbers of OTUs were detected despite the limited numbers of sites and samples that were examined for each species, suggesting that all great apes possess extensive species-level diversity of Archaea in their gut microbiomes. Nearly all Archaea we identified in great apes belong to the orders Methanobacteriales and Methanomassiliicoccales; therefore, they are all likely hydrogen-utilizing methanogens. The few hydrogen-utilizing methanogens (M. smithii, M. stadtmanae, Methanomassiliicoccus luminyensis, “Candidatus Methanomassiliicoccus intestinalis,” and “Candidatus Methanomethylophilus alvus”) already known to occur in the human gut are capable of using different or multiple substrates for methanogenesis, including carbon dioxide, methanol, or the methyl compounds monomethylamine, dimethylamine, and trimethylamine (1). Because methanogens compete for hydrogen in the gut, it is possible that some taxa are capable of performing metabolic capabilities that allow them to interact with different hydrogen-producing organisms and maintain distinct niches. Although we observed a decrease in archaeal diversity in the human lineage, we were able to identify several archaeal OTUs that are conserved across all great ape species, and their persistence implies a continual role in the breakdown of dietary compounds throughout hominoid evolution.
MATERIALS AND METHODS
Sample sources and DNA extraction procedures.
Samples representing five great ape species were obtained from the following sources. (i) For humans, DNAs were purified from fecal samples of 10 individuals following the method of Shannon et al. (42). (Sample sources and designations are presented in Data Set S3.) (ii) For chimpanzees, DNAs, purified from field-collected fecal samples from 15 individuals (Pan troglodytes), were selected from those reported in reference 6. DNA extraction procedures and bacterial contents of samples, as surveyed with 16S rRNA gene universal prokaryotic primer pair 515F/806R, are also presented in reference 6. (Sample sources and designations are presented in Data Set S3.) (iii) For gorillas, DNA samples, purified from field-collected fecal samples from 20 individuals (Gorilla gorilla), were selected from those reported in reference 6. DNA extraction procedures and bacterial contents of samples, as surveyed with 16S rRNA universal prokaryotic primer pair 515F/806R, are also presented in reference 6. (Sample sources and designations are presented in Data Set S3.) (iv) For bonobos, DNA samples, purified from the field-collected fecal samples from 18 individuals (Pan paniscus) were obtained from Moeller et al. (6). (Sample sources and designations are presented in Data Set S3.) (v) For orangutans, fecal samples were obtained from 10 individuals (Pongo pygmaeus) from a colony at the Atlanta Zoo. (Sample sources and designations are presented in Data Set S3.) Upon collection, each sample was mixed with an equal volume of RNAlater (Ambion) and stored frozen at −80°C. Total DNA from each fecal sample was extracted from 400-mg aliquots following the bead-beating procedure described in reference 43.
Sample preparation and DNA sequencing.
DNAs were subjected to PCR amplification targeting the V4-V5 variable region of archaeal 16S rRNA genes using the primer set Arch516F/Arch915R (8, 9). Three replicate PCRs were performed on each sample, and the amplicons generated from each set of three reactions were pooled and quantified on a Qubit 2.0 fluorometer (Invitrogen). Replicate samples were pooled and individually bar coded according to procedures outlined in reference 10. Amplicons were purified with AMPure XP beads (Beckman Coulter, Inc.). Bar-coded samples were pooled and sequenced on the Illumina MiSeq at the Genome Sequencing and Analysis Facility at the University of Texas at Austin.
Sequencing data corresponding the V4 region, as amplified with universal prokaryotic primers 515F/806R, were obtained from published sources: data for gorillas, bonobos, and chimpanzees were from reference 6. Because samples from humans and orangutans had not previously been characterized for microbial diversity with the universal prokaryotic primer set, PCR amplifications of the corresponding V4 region of 16S rRNA were performed on these samples with the 515F/806R primer pair using the protocol described above.
Sequence filtering and analysis.
Sequence reads from the universal prokaryotic primer data sets and the archaeon-specific primer data set were processed in QIIME (44). For the Archaea-specific primer data set, FASTQ files were quality filtered with split_libraries_fastq.py, allowing a minimum Phred score of Q20. Forward and reverse Illumina reads were joined with join_paired_ends.py. Chimeric sequences, identified using USEARCH6.1 implemented in the identify_chimeric_seqs.py script in QIIME, were removed (3% of the total reads). OTUs were clustered at both 94% sequence identity and 97% sequence identity by UCLUST, as implemented in pick_open_reference_otus.py. Additionally, OTUs were clustered using VSEARCH (43). On average, the VSEARCH clustering method detected slightly higher numbers of OTUs than UCLUST (Tables S1A and B), so to be conservative, all subsequent analyses were performed on OTUs clustered by UCLUST. Sequence reads were initially searched against the July 2015 release of the SILVA 94% and 97% reference database (http://www.arb-silva.de/download/arb-files) (11). Sequences that did not find matches in the SILVA database at 94% or 97% were subsequently clustered into de novo OTUs with UCLUST. All OTUs represented by <10 reads were removed, as were any reads that matched mitochondrial or chloroplast sequences and those that could not taxonomically assigned to Bacteria or Archaea (“unassigned reads”). A reference sequence from each archaeal OTU (clustered at 97% and at 94% sequence identity) was selected and aligned using PyNAST (10). This alignment was used to construct a phylogenetic tree using FastTree (45) within QIIME. To compare the extent of archaeal diversity captured with universal prokaryotic primers to that detected with archaeon-specific primers, the data set was analyzed both with and without bacterial reads. Downstream analyses, including the taxonomic assignment of OTUs, and estimates of alpha and beta diversity were conducted using the QIIME workflow core_diversity_analysis.py, applying a sampling depth of 10,000 reads per sample and default parameters. Rarefaction depths were chosen manually to exclude samples with low total sequences (see Data Set S2 for sample details). The final sample sizes for each species after rarefaction were as follows: Homo sapiens, n = 10; Pan troglodytes, n = 14; Pan paniscus, n = 18; Gorilla gorilla, n = 20; and Pongo pygmaeus, n = 8.
For the data set generated with universal prokaryotic primers, the FASTQ files for the 10 humans, 14 chimpanzees, 18 bonobos, 20 gorillas, and 8 orangutans were filtered for quality with split_libraries_fastq.py, allowing a minimum Phred score of Q20. Forward and reverse Illumina reads were joined with join_paired_ends.py, and the joined paired reads were trimmed to equal lengths. Chimeric sequences, identified using USEARCH6.1 implemented in the identify_chimeric_seqs.py script in QIIME, were removed (amounting to 1.5% of total reads). OTUs clustered at 97% sequence identity were chosen by UCLUST, as implemented in pick_open_reference_otus.py. Sequence reads were initially searched against the July 2015 release of the SILVA 97% reference database (http://www.arb-silva.de/download/arb-files) (11). Sequences that did not find matches in the SILVA data set were clustered into de novo OTUs with UCLUST. All OTUs represented by less than 10 reads were removed, as were any reads that matched mitochondrial or chloroplast sequences and those that could not be taxonomically assigned to Bacteria or Archaea. Downstream analyses, including taxonomic classification of OTUs and estimates of alpha and beta diversity, were conducted in the QIIME workflow core_diversity_analysis.py, applying a sampling depth of 10,000 reads per sample and default parameters. Statistical tests and boxplots were completed in R, and Venn diagrams using the tool from http://bioinformatics.psb.ugent.be/webtools/Venn/. Great ape divergence times follow those reported in reference 46. The coverage and specificity of the primers used in this study were evaluated with TestPrime 1.0 implemented on the SILVA database (Data Set S1) (47).
Phylogenetic analysis.
The 16S rRNA sequences from 58 complete archaeal genomes were obtained from the RDP (13), combined with reference sequences from each archaeal OTU, and aligned with MUSCLE v3.8.31 (48). Sequences were trimmed manually in SeaView (49), and a maximum likelihood tree was inferred with PhyML v3.1 using the GTR+Γ4 model (50). The reliability for the internal branches of the maximum likelihood trees was assessed by bootstrapping and the approximate likelihood ratio test (aLRT–SH-Like) (51).
Association networks and correlation analysis.
We tested the associations between 102 archaeal OTUs (clustered at 94%) and the 415 bacterial genera (clustered at 97% and classified to genera using the RDP classifier) using Spearman’s rank correlation (rho) implemented in R. We extracted statistically significant (P < 0.001, after Bonferroni correction) negative and positive cooccurrence relationships with Spearman’s rho values less than −0.50 or greater than 0.50 that involved archaeal OTUs present in >80% of all human samples. Association networks were visualized with Cytoscape (42).
Ethics statement.
The study protocol was approved by the Yale University Human Investigation Committee. Informed consent was obtained from all human subjects as described in our approved protocol no. 1106008725.
Accession number(s).
All sequence data have been deposited into the NCBI Sequence Read Archive under accession no. PRJNA371799 and PRJNA338781.
ACKNOWLEDGMENTS
This work was supported by NIH awards R01GM101209 and R3GM118038 to H.O. and R35GM118159 to A.L.G.
We thank Emily Putnam and Travis Middleton for technical assistance and Kim Hammond for help with the preparation of the figures. We thank Joseph Mendelson, Martine Peters, Beatrice Hahn, Anne Goodall, Jane Goodall, and Anne Pusey for providing the great ape samples.
REFERENCES
- 1.Gaci N, Borrel G, Tottey W, O’Toole PW, Brugère JF. 2014. Archaea and the human gut: new beginning of an old story. World J Gastroenterol 20:16062–16078. doi: 10.3748/wjg.v20.i43.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, Ochman H. 2014. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A 111:16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eloe-Fadrosh EA, Ivanova NN, Woyke T, Kyrpides NC. 2016. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat Microbiol 1:15032. doi: 10.1038/nmicrobiol.2015.32. [DOI] [PubMed] [Google Scholar]
- 8.Stahl DA. 1991. Development and application of nucleic acid probes, p 205–248. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 9.Takai K, Horikoshi K. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. [Google Scholar]
- 16.Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. 2007. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A 104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, O’Toole PW, Brugère JF. 2014. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrel G, Harris HM, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O’Toole PW, Brugère JF. 2013. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi: 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrel G, Harris HM, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E, Peyret P, Gribaldo S, O’Toole PW, Brugère JF. 2012. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol 194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borrel G, O’Toole PW, Harris HM, Peyret P, Brugère JF, Gribaldo S. 2013. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol 5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlas A, Robert C, Gimenez G, Drancourt M, Raoult D. 2012. Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated archaea species. J Bacteriol 194:4745–4745. doi: 10.1128/JB.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M. 2009. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Million M, Angelakis E, Maraninchi M, Henry M, Giorgi R, Valero R, Vialettes B, Raoult D. 2013. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int J Obes 37:1460–1466. doi: 10.1038/ijo.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller TL, Wolin MJ, Conway de Macario E, Macario AJ. 1982. Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol 43:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihajlovski A, Alric M, Brugère JF. 2008. A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res Microbiol 159:516–521. doi: 10.1016/j.resmic.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Dridi B, Henry M, Richet H, Raoult D, Drancourt M. 2012. Age-related prevalence of Methanomassiliicoccus luminyensis in the human gut microbiome. APMIS 120:773–777. doi: 10.1111/j.1600-0463.2012.02899.x. [DOI] [PubMed] [Google Scholar]
- 28.Rieu-Lesme F, Delbès C, Sollelis L. 2005. Recovery of partial 16S rDNA sequences suggests the presence of Crenarchaeota in the human digestive ecosystem. Curr Microbiol 51:317–321. doi: 10.1007/s00284-005-0036-8. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam YD, Chang HW, Kim KH, Roh SW, Kim MS, Jung MJ, Lee SW, Kim JY, Yoon JH, Bae JW. 2008. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol 46:491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- 31.Oxley APA, Lanfranconi MP, Würdemann D, Ott S, Schreiber S, McGenity TJ, Timmis KN, Nogales B. 2010. Halophilic archaea in the human intestinal mucosa. Environ Microbiol 12:2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- 32.Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhary PP, Gaci N, Borrel G, O’Toole PW, Brugère JF. 2015. Molecular methods for studying methanogens of the human gastrointestinal tract: current status and future directions. Appl Microbiol Biotechnol 99:5801–5815. doi: 10.1007/s00253-015-6739-2. [DOI] [PubMed] [Google Scholar]
- 34.Probst AJ, Auerbach AK, Moissl-Eichinger C. 2013. Archaea on human skin. PLoS One 8:e65388. doi: 10.1371/journal.pone.0065388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, Ochman H. 2016. Cospeciation of gut microbiota with hominids. Science 353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, Travis DA, Long HT, Tuan BV, Minh VV, Cabana F, Nadler T, Toddes B, Murphy T, Glander KE, Johnson TJ, Knights D. 2016. Captivity humanizes the primate microbiome. Proc Natl Acad Sci U S A 113:10376–10381. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan Y, Zhang H, Gao X, Shang S, Wu X, Chen J, Zhang W, Zhang W, Jiang M, Zhang B, Chen P. 2016. Comparison of the bacterial communities in feces from wild versus housed sables (Martes zibellina) by high-throughput sequence analysis of the bacterial 16S rRNA gene. AMB Express 6:98. doi: 10.1186/s13568-016-0254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milton K. 1999. A hypothesis to explain the role of meat-eating in human evolution. Evol Anthropol 8:11–21. [Google Scholar]
- 39.Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. 2013. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 40.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Weissenbach J, Ehrlich SD, Bork P, MetaHIT Consortium . 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanderhaeghen S, Lacroix C, Schwab C. 2015. Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol Lett 362:fnv092. doi: 10.1093/femsle/fnv092. [DOI] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignment. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, et al. . 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Performance of the primers used in this study. Primer performance was evaluated using TestPrime 1.0 implemented on the SILVA database (http://www.arb-silva.de/search/testprime/). For both primer pairs, the performance was calculated allowing either 0 or 1 mismatch. Coverage = (matches/eligible) × 100, specificity = 100 − (outgroup matches/outgroup matchable) × 100, Accessions = number of sequences in the taxonomic path, Eligible = number of regions with sequence data at the position of the primer pair, Mismatch = number of mismatched regions with the primer pair in the taxonomic path, and No Data = number of regions with no sequence data at the position of the primer. Download DATA SET S1, XLSX file, 0.1 MB (75.7KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Archaeal read counts pre- and postfiltering for each individual. Total reads for each individual using archaeon-specific primer pair Arch516F/Arch915R after quality filtering (Phred score above Q20). Shown are the percentages of chimeric sequences, singletons, unassigned, and bacterial reads detected per individual. Download DATA SET S2, XLSX file, 0.1 MB (52.5KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Numbers of OTUs detected with (A) VSEARCH and (B) UCLUST. Download TABLE S1, PDF file, 0.1 MB (31.8KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Numbers of archaeal reads and OTUs detected with the universal and archaeon-specific primer pairs. Total numbers of archaeal reads and 97% OTUs in each sample detected with universal prokaryotic primer pair 515F/806R were counted at original read depths (i.e., before rarefraction). Read depths obtained with archaeon-specific primers were rarefied to the number of archaeal reads obtained with universal prokaryotic primers (column G) prior to computing total numbers of OTUs (column J) and the percentage detected with universal versus archaeal primers (column K). Download DATA SET S3, XLSX file, 0.1 MB (47.9KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of archaeal 97% OTUs shared among great ape species. (A) Venn diagram showing the number of archaeal 97% OTUs shared between species, with the full number of archaeal 97% OTUs detected in each host species shown under the species name. (B) Venn diagram showing the number of core archaeal 97% OTUs shared between species, with the full number of core OTUs detected in each host species shown under the species name. Download FIG S1, PDF file, 0.1 MB (75.9KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of archaeal OTUs in the great ape gut microbiome species. The nine core 97% OTUs present in all great ape host species are shown in gray. The core 97% OTUs of each species (i.e., those present in >80% of sampled individuals) are in black boldface (related to Fig. S2). Download DATA SET S4, XLSX file, 0.1 MB (77.3KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic diversity of archaeal 97% OTUs. Unrooted maximum likelihood phylogeny of the 110 archaeal core 97% OTUs is shown. The phylogeny was inferred by PhyML (GTR+Γ4) and is based on a representative sequence for each OTU. OTUs included in the analysis were present in >80% of individuals from each species, and the nine OTUs present in all great ape host species are indicated in red. The scale bar represents average number of substitutions per site. The reliability for the internal branches of the ML tree was assessed by the bootstrapping method (100 bootstrap replicates) and the approximate likelihood ratio test (aLRT–SH-Like) (51). Dots at nodes denote aLRT values: black, >0.8; gray, >0.5; white, <0.5. Download FIG S2, PDF file, 0.1 MB (75.7KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diversity of archaeal 97% OTUs in the gut microbiomes of great apes. (A) Diversity measured by Shannon’s index, Simpson’s index, Fisher’s alpha, and Chao1. Box-and-whiskers plots show high, low, and median values, with the lower and upper edges of each box denoting the first and third quartiles, respectively. Species comparisons yielding statistically significant differences are indicated by asterisks (*, P < 0.05, Wilcoxon rank-sum test after Bonferroni correction). (B) Principal coordinate analysis of unweighted UniFrac distances of the archaeal component of the gut microbiomes of five great ape species. (C) Principal coordinate analysis of weighted UniFrac distances of the archaeal component of the gut microbiomes of five great ape species. Download FIG S3, PDF file, 0.1 MB (80.4KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diversity of archaeal 94% OTUs in the gut microbiomes of great apes. Diversity was measured by the following indices: observed OTUs, Shannon’s index, Simpson’s index, Fisher’s alpha, and Chao1. Box-and-whiskers plots show high, low, and median values, with the lower and upper edges of each box denoting the first and third quartiles, respectively. Species comparisons yielding statistically significant differences are indicated by asterisks (*, P < 0.05, Wilcoxon rank-sum test after Bonferroni correction). Download FIG S4, PDF file, 0.1 MB (58KB, pdf) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cooccurrence relationships between archaea and bacteria present in the gut microbiomes of great apes. Only statistically significant associations (P = 0.001) with a Spearman’s rho value less than −0.5 or greater than 0.5 are shown. Associations between the nine core 97% OTUs (i.e., those present in all host species) and bacterial genera not detected in human samples are shown in gray. Associations involving these core OTUs and bacterial genera present in humans (and other great apes) are shown in black. Download DATA SET S5, XLSX file, 0.1 MB (13.8KB, xlsx) .
Copyright © 2017 Raymann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.