Abstract
Cytomotive filaments are essential for the spatial organization in cells, showing a dynamic behavior based on nucleotide hydrolysis. TubZ is a tubulin-like protein that functions in extrachromosomal DNA movement within bacteria. TubZ filaments grow in a helical fashion following treadmilling or dynamic instability, although the underlying mechanism is unclear. We have unraveled the molecular basis for filament assembly and dynamics combining electron and atomic force microscopy and biochemical analyses. Our findings suggest that GTP caps retain the filament helical structure and hydrolysis triggers filament stiffening upon disassembly. We show that the TubZ C-terminal tail is an unstructured domain that fulfills multiple functions contributing to the filament helical arrangement, the polymer remodeling into tubulin-like rings and the full disassembly process. This C-terminal tail displays the binding site for partner proteins and we report how it modulates the interaction of the regulator protein TubY.
Tubulin-like GTPases play a central role in a wide range of physiological processes in cells, plasmids and viruses through their assembly into dynamic cytomotive filaments. In the presence of GTP, these proteins assemble in a head-to-tail fashion and the addition of new filament molecules involves the formation of the complete GTPase active site1. During filament assembly, GTP hydrolysis induces a conformational change that makes filaments prone to depolymerization. Further, there are structural differences between unassembled and assembled monomers unrelated to the nucleotide chemical state, known as structural plasticity, that are key in the modulation of the polymer dynamics2.
TubZ is a GTPase of the tubulin superfamily that functions as the motor component of the DNA positioning system by forming a spindle-like apparatus3. These segregation systems are the survival kits of Bacillus and Clostridium virulence plasmids that ensure their faithful inheritance by daughter cells during division. The plasmid partitioning requires a cis-acting centromere-like DNA sequence (tubC) and a centromere binding protein (TubR) that collectively form the partitioning complex. TubZ interacts with this complex and elicits plasmid trafficking along the cytoplasm4. Furthermore, there is another protein, TubY, that can bind to and modify the dynamics of TubZ filaments5. TubY can also bind to the operon promoter and activate transcription6. The understanding of plasmid segregation is key in order to design new strategies to prevent the spread of virulence genes in growing bacterial populations. Both TubZ and the regulator protein TubY may be ideal targets to block DNA movement within growing bacteria. However, in order to develop novel and specific drugs to target plasmid segregation we need a better understanding of their mechanism of action.
In microtubules, tubulin dimers associate into a tubular-like structure where the continuous addition of GTP-containing tubulin subunits forms caps that stabilize the structure7. Loss of these caps results in rapid depolymerization known as dynamic instability8. The consensus view agrees that during depolymerization a straight to curved structural change in microtubule protofilaments occurs9. However, in the presence of these caps microtubules can undergo treadmilling dynamics10. Like tubulin, TubZ forms polar assemblies that can follow dynamic instability11 and treadmilling3,12. However, TubZ polymerizes in a helical fashion, suggesting that the conformational changes associated with nucleotide hydrolysis and the structural plasticity may be different. Recent cryo-EM structures of 2- and 4-stranded TubZ filaments showed an opening in the longitudinal interface due to GTPase activity13. Further, these structures displayed contacts between the C-terminal tail and upper TubZ monomers within the protofilament, however a lack in continuous electron density made it impossible to describe the interaction mechanism13. Herein, we have analyzed TubZ filament structure with and without GTP caps when considering the full-length protein and C-tail truncations to gain insight into the molecular mechanism of filament dynamics. Our structural and biochemical data reveal that GTP caps retain the helical pattern and that GTP hydrolysis produces an unexpected filament stiffening upon disassembly. Further, the flexible C-tail participates as a multifunctional domain on filament assembly and disassembly.
Results
CbTubZ primarily assembles as a 4-stranded helical filament
According to cryo-EM reconstructions of Bacillus thuringiensis TubZ (BtTubZ) polymers, Montabana and Agard13 proposed that structural changes in TubZ filaments during nucleotide hydrolysis involve an increase from 2 (GTP- γ-S-bound) to 4 (GDP/GTP-bound) protofilaments. We have measured the nucleotide content of BtTubZ filaments assembled with GTP- γ-S and GTP and have found 20% and 80% of GDP-bound molecules, respectively (Methods). Therefore, we wanted to investigate whether the GDP content was necessary to induce such conformational changes.
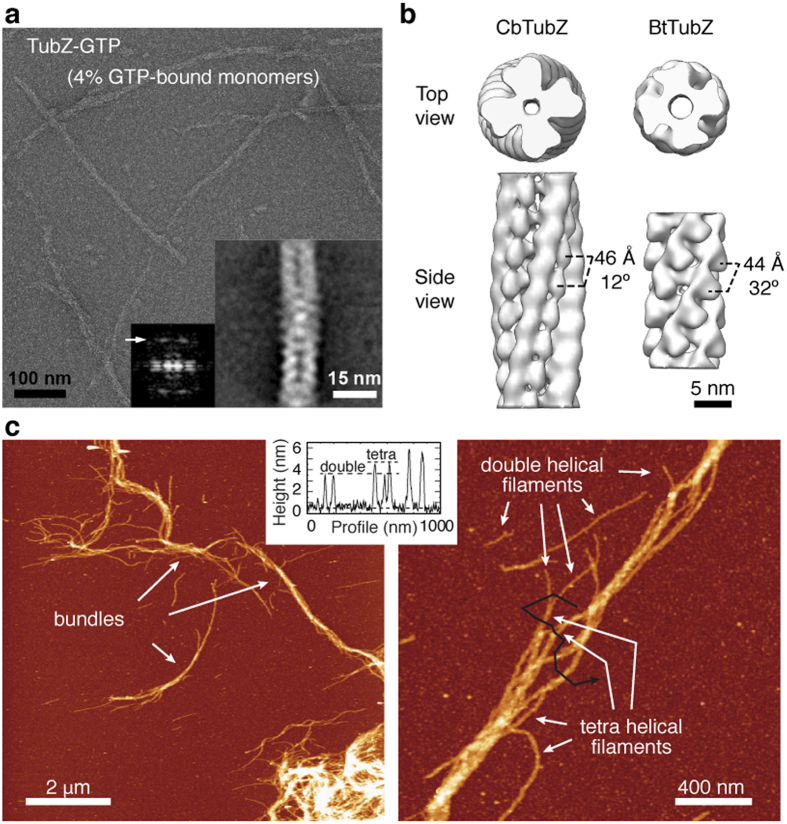
To better understand the architecture of TubZ filaments and how its nucleotide-bound state affects its structure, we analyzed wild type Clostridium botulinum TubZ (CbTubZ) polymers and mutated versions at the nucleotide-binding site (CbTubZT100A) and the catalytic loop (CbTubZE200A). These proteins assembled in the presence of GTP and GTP- γ-S, but the resulting filaments showed different hydrolysis ratios5. Further, the nucleotide content and the size of the defined polymer’s caps14 differed considerably. For the wild type protein, 4% of GTP-bound molecules were observed while up to 20% and 80% GTP-bound molecules were observed for the CbTubZT100A and CbTubZE200A mutants, respectively5. We combined electron microscopy (EM) and atomic force microscopy (AFM) to get an average projection of the filaments and an accurate measurement of their heights that provided useful information on polymer 3D structure. Surprisingly, the filaments analyzed yielded similar averaged structures including those grown in the presence of GTP- γ-S (Fig. 1a and Fig. S1a). The filaments shared a 4-stranded helical arrangement with a rise of ~46 Å and an azimuthal angle of ~12° (Fig. 1b and Fig. S1b). These filaments had mean heights of 3.2 ± 0.4 nm (measured from a mean basal line), and mean widths of 27 ± 4 nm (measured as the full-width at half-maximum height) (N > 50, Fig. 1c). However, mean widths of 13.6 nm were deduced after the correction of the tip-convolution effect when considering the tip size of 10 nm (as provided by Nanosensor, Switzerland). This value is slightly lower than that obtained in previous cryo-EM reconstructions13, probably due to a dehydration effect on AFM samples when imaged in air15. By contrast, BtTubZ 4-stranded filaments have a rise of ~44 Å and an azimuthal angle of ~32°13, indicating that despite sharing a common core5,16, CbTubZ and BtTubZ display structural differences that underlie distinct helical patterns. In our images we also identified thinner filaments with mean heights of 2.7 ± 0.3 nm and mean widths of 16 ± 3 nm (5.6 nm after correction) that likely correspond to 2-stranded forms (Fig. 1c and Fig. S2a)5, however we were unable to obtain acceptable EM averages due to their low frequency.
Figure 1. Characterization of TubZ filaments.
(a) Negative stain EM showing wild type CbTubZ filaments. Inset corresponds to the averaged filament and its Fourier transform. Arrow indicates a 4.8-nm longitudinal spacing between molecules. (b) Top and side views of our CbTubZ low-resolution filament versus BtTubZ filament13 filtered at a similar low-resolution level. Both filaments are 4-stranded but show different longitudinal distances between monomers and azimuthal angles. (c) AFM images of CbTubZT100A filaments, showing 2 – and 4-stranded filaments and bundles. The black line in the second image corresponds to the height profile shown in the graph. Color scale from dark to bright corresponds to 0–11 nm.
It may be argued that in vitro filament assembly could differ from cell-based filament assembly. One explanation for this could be differences within the cellular environment such as macromolecular crowding. Moreover, higher viscosity and the excluded volume effect would increase the likelihood of intermolecular interactions17. Differences in the filament assembly of tubulin-like FtsZ polymers have been described in diluted versus crowded conditions18. To simulate the crowded cytoplasm of a bacterium we studied TubZ polymerization using Ficoll-70, a chemically inert sucrose-based polymer that occupies space in order to mimic molecular crowding but does not interact with the protein19. In these conditions, TubZ filaments retained their helical structure displaying widths corresponding to 2-stranded (7.7 ± 0.7 nm) and 4-stranded (13.7 ± 2.2 nm) polymers (N > 50, Fig. S2a,b), in agreement within the error of AFM measurements. However, filament dynamics decreased significantly in the presence of Ficoll (Fig. S2c).
Our findings strongly suggest that the functional CbTubZ polymer is a 4-stranded filament and that the 2-stranded filament may constitute an intermediate stage during polymerization that rapidly moves toward a 4-stranded form, regardless of nucleotide content. By contrast, BtTubZ is 2-stranded filament when assembled with GTP- γ-S13,20. Hence, GTP- γ-S might blocks BtTubZ filaments on the double helical state, which could be a non-functional filament during DNA trafficking.
GTP caps are essential for maintenance of 4-stranded filament morphology
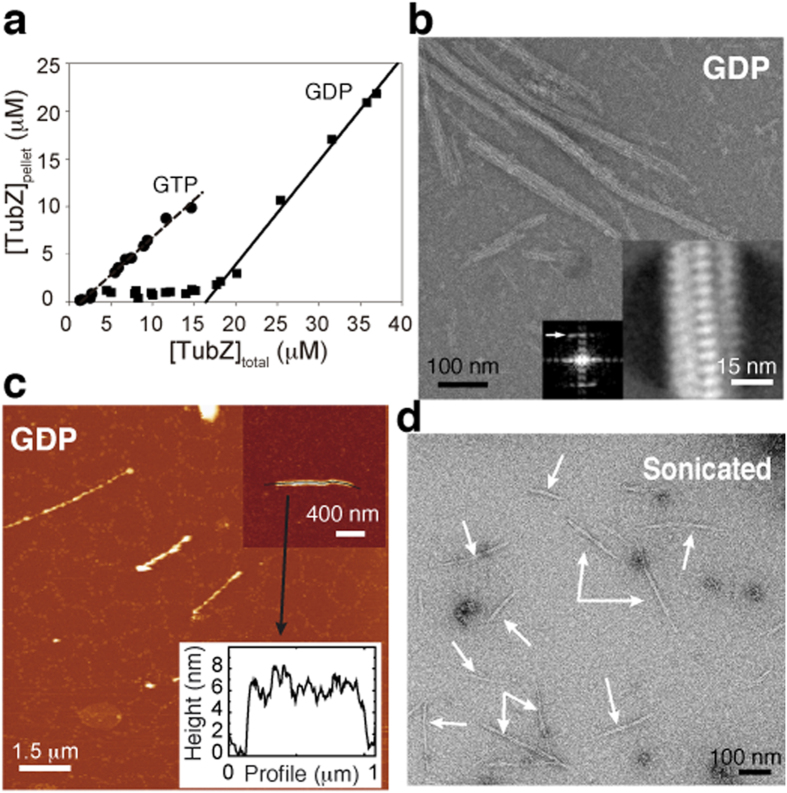
To unravel the molecular mechanism underlying filament dynamics, we analyzed the effect of GTP caps release. First, we forced the formation of pure GDP-bound filaments lacking GTP caps by supplying only GDP during the assembly reaction. Despite a lower affinity between monomers (higher critical concentration), we found that CbTubZ assembled cooperatively onto stable filaments (Fig. 2a). However, the polymer structure was different than the 4-stranded filament described above, showing both wider (20–40 nm) and thicker (5–8 nm) dimensions (Fig. 2b,c). Furthermore, the resulting filament was straight and appeared rigid, but retained the intermonomer spacing (measured from the averaged image Fourier transform) and some degree of helical trend (Fig. 2b).
Figure 2. Effect of GTP hydrolysis on filament structure.
(a) Critical concentration measurements of CbTubZ in buffer containing Ficoll (200 g/L) and GTP/Mg2+ or GDP/Mg2+ (b). Negative stain EM showing CbTubZ filaments polymerized in the presence of GDP/Mg2+. Inset shows averaged filaments and the corresponding Fourier transform. Arrow indicates 4.8-nm longitudinal spacing between molecules. (c) AFM images of CbTubZ filaments polymerized in the presence of GDP/Mg2+. The black line corresponds to the height profile shown in the graph. Color scale from dark to bright correspond to 0–11 nm. (d) Negative stain EM of CbTubZ filaments assembled with GTP/Mg2+ and sonicated for 10 min, showing the stiffening effect.
Next, we analyzed the effect of removing 4% of GTP-bound molecules that stabilize CbTubZ filaments (GTP caps). To this end, we polymerized TubZ in a solution containing an excess of GTP and Mg2+ and examined the resulting filaments after disrupting them by mechanical force (sonication). We identified multiple straight and short filaments as breakdown products of GTP-formed TubZ filaments (Fig. 2d). Since CbTubZ polymer is mainly GDP-bound, it is reasonable to assign these short products as the GDP-bound state.
We hypothesize that in the presence of GTP caps, lattice contacts may control and maintain the 4-stranded helical arrangement, similar to what has been described for microtubules21. Unlike tubulin, however, there is no protofilament curving, but rather filament stiffening, thereby promoting its disassembly. In TubZ, protofilaments are helical and the straightening induced by GTP hydrolysis may generate the necessary forces to break longitudinal contacts. Here, fewer contacts would make a weaker interface favoring monomer detaching.
Role of the C-tail during the assembly process
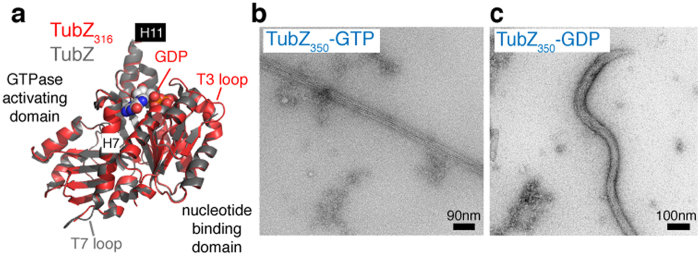
The C-tail of TubZ is flexible and ranges in length and amino acid composition from short (~20 residues) and acidic to long (more than 60 residues) and basic16,22. Since the C-tail makes contacts with upper subunits within the filament during polymerization13,23, we further investigated its functional implication. The C-tail of CbTubZ is long (50 residues) and acidic, so we generated three truncated mutants: CbTubZ350, lacking the last 8 residues that are not charged; CbTubZ331, lacking the same residues as for CbTubZ350 plus an acidic patch and CbTubZ316, lacking the entire flexible tail (Fig. S3a). All three constructs were expressed and purified as soluble proteins at similar levels as the wild type protein. Similarly, all showed equivalent circular dichroism spectra, suggesting that the truncations did not affect the core structure (Fig. S3b). Further, we solved the crystal structure of CbTubZ316 at 2.4 Å resolution (PDB 4XCQ, Table S1), which closely superposed with the apo full-length protein (PDB 3V3T) with an r.m.s.d of 0.37 Å (Fig. 3a). Moreover, the nucleotide-binding affinities of these mutants were similar to those of the wild type protein (5–6 × 106 M−1, Fig. S3c), suggesting that the nucleotide-binding step is not limiting during the polymerization process.
Figure 3. Effect of C-tail on TubZ assembly.
(a) Comparison of full length and CbTubZ316 structures, highlighting the main domain arrangements and the most important secondary structure elements. TubZ316 displays GDP at the nucleotide-binding pocket, an ordered T3 loop and a disordered T7 loop (which is ordered in the full-length protein). In both structures the C-terminal helix H11 is in the same position, but neither contain an ordered C-tail. (b,c) Negative stain EM of CbTubZ350 stiff filament assembled in the presence of GTP/Mg2+ (b) and flexible filament polymerized with GDP/Mg2+ (c).
Similar to other C-tail truncated TubZs, our CbTubZ mutants showed no assembly in diluted solutions13,24. However, based on the differing kinetics observed in crowded versus diluted conditions, we examined filament assembly with the mutated proteins in an in vitro crowded environment using Ficoll-70. While all three mutants polymerized cooperatively, the critical concentration increased progressively in parallel with the absence of longer sequences from the tail region (Fig. S3d), indicating reduced polymer elongation affinity as compared with the wild type protein. Although these mutants retained the ability to polymerize, none showed significant levels of GTPase activity (Fig. S3e), which may be related to a defect on the formation of the GTPase active site (i.e. the insertion of the catalytic loop in the GTP binding pocket of the lower subunit). Indeed, the assembled polymers no longer displayed a helical arrangement (Fig. 3b, Fig. S3f), instead they resembled the wild type GDP-bound filament (Fig. 2b). This suggests that the modulation of the C-tail tip interactions may contribute to the stiffening of the filament after hydrolysis. Further, the C-tail movement during GTP hydrolysis could provide a simple explanation for the clashes observed for TubZ-GDP C-tail, as described during its docking into the GTP-like (GMPCPP) cryoEM filament structure23.
In the head-to-tail assembly process the addition of a new monomer to the tip of the filament implies that the C-tail is probably free, because there are no upper subunits available to form additional contacts with the protein. Here, we show that by removing the very end of the C-tail (8 residues) the filament morphology changes dramatically. Therefore, during assembly those amino acids may make the initial contacts (with the most distant monomer in the row), after which the charged residues can participate in the stabilization of the tail (as previously described23) through interaction with more proximal molecules. Our results suggest that the C-tail domain is directly involved in modulating filament assembly as well as facilitating the formation of a functional helical filament. Differences in tail length may affect the degree of monomer rotation within the filament and hence its twist. In fact, BtTubZ (with more than 60 residues) and CbTubZ (with 50 residues) assemble into 4-stranded filaments with distinct helical pitches (above). By contrast, phages 201 ϕ2-1 and KZ TubZs (both with 20 residues) polymerize into 3-stranded filaments23 and have longer helical pitches (~51 nm)25.
We further wanted to understand how nucleotide hydrolysis induces depolymerization. In order to answer this question, we assembled truncated C-tail CbTubZs in the presence of GDP. The resulting filaments were curly rather than straight, denoting increased flexibility (Fig. 3c and Fig. S3g). This is probably due to a weakening of the longitudinal interactions between monomers as mention previously13. We hypothesize that this softening of the longitudinal interactions produced by GTP hydrolysis is the last step preceding subunit disassembly from the filament (see the proposed model below, Fig. 5).
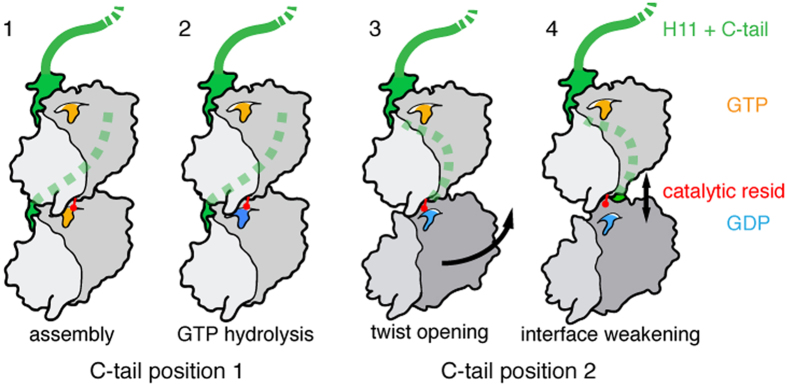
Figure 5. Model of TubZ filament dynamics.
For simplicity, we show only two consecutive molecules in the filament: (1) GTP-bound state induces the assembly, whereas the C-tail (green and dashed line when behind a protein subunit) establishes specific contacts with upper subunits leading to the generation of a twist and allowing the formation of the canonical longitudinal interface; (2) the catalytic residue (red) is correctly positioned and reaches GTP (yellow), which is hydrolyzed into GDP (blue); (3) the enzymatic reaction induces changes in the bottom molecule, changing the C-tail interacting surface with the upper subunits and opening the twist between both molecules; (4) the GDP-bound state results in weakening of the longitudinal interface, likely due to loss of contacts. C-tail truncated proteins do not establish the specific contacts with upper subunits within the filament, blocking the formation of the canonical longitudinal interface. Accordingly, GTP assembly leads to state 3 filaments, while GDP-bound polymers are in state 4.
The C-tail modulates interactions with TubY and the formation of tubulin-like rings
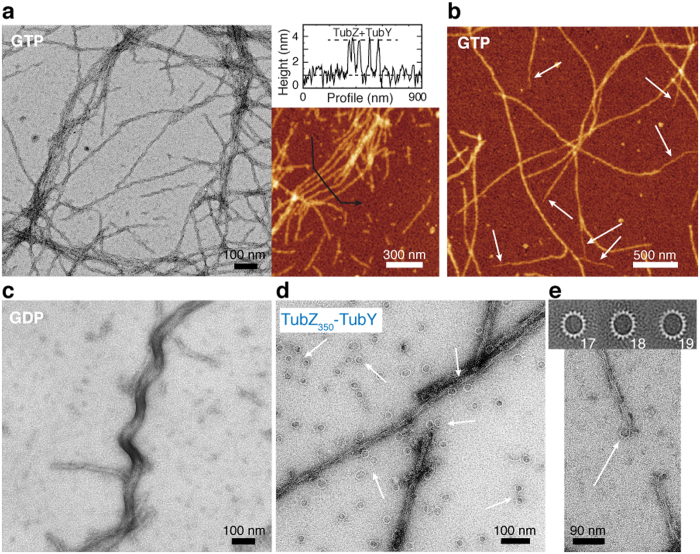
We previously demonstrated that TubY binding to TubZ affected filament dynamics, however we could not identify the underlying molecular mechanism5. Now we have analyzed the effect of filament nucleotide content on TubY binding. Sedimentation experiments revealed that TubY interacts similarly with wild type, CbTubZT100A, and CbTubZE200A filaments (Fig. S4a). Furthermore, the resulting polymers were also similar according to EM and AFM images showing mean heights of 2.7 ± 0.4 nm (Fig. 4a). The height values measured were slightly lower than expected due to an increase in the basal plane caused by the presence of a greater quantity of protein absorbed on the surface (see Methods). Indeed, the amplitude of noise in the basal plane was 1.4 ± 0.4 nm as compared with 0.9 ± 0.2 nm when measured in the absence of TubY. Interestingly, we observed thinner extensions at the ends of some filaments (white arrows in Fig. 4b), with mean heights of 2.0 ± 0.2 nm, suggesting coating of TubZ filaments by TubY. Hence, the binding region is likely accessible along the whole filament, regardless of the nucleotide content. Surprisingly, TubY clearly modified the wild type GDP-bound polymer structure (Fig. 2b), which became more flexible and in turn resembled the GDP-bound filaments of C-tail truncated proteins (Figs 3c and 4c). This result supports a regulatory function of TubY in modulating the dynamics of TubZ filaments by modifying subunit interactions.
Figure 4. TubY - TubZ interaction.
(a) Negative stain EM (left) and AFM (right) of CbTubZ-TubY filaments. The height profile shown in the graph was taken in the direction of the black arrow. (b) AFM images of CbTubZE200A-TubY filaments. White arrows indicate thinner extensions at the ends of some filaments. Color scale from dark to bright is 0–11 nm in AFM images. (c–e) EM images of TubY binding to GDP-bound CbTubZ filaments (c), TubZ350 filaments and rings formed in the presence of TubY (d) and detail of ring formation upon disassembly from filaments tips (e, bottom). (e) Different types of averaged structures obtained after ring processing, including the number of monomers in each ring (top).
Considering our findings regarding the role of the C-tail in TubZ filament assembly and its interaction with TubR16, we examined the role of the C-tail in TubY binding. Light scattering experiments revealed interactions with all the truncated proteins used in this study (Fig. S4b). Interestingly, TubY binding induced the rearrangement of CbTubZ350 filaments into tubulin-like rings (Fig. 4d), suggesting that the C-tail domain may modulate this interaction. We previously described the formation of similar rings during TubY binding to TubZ filaments in the presence of the partition complex TubRC5. Thus, TubY and the segrosome may compete for the C-tail tip, and TubY may promote the filament re-arrangement into rings only when the TubRC complex blocks the TubZ C-terminal tip.
The formation of rings is a recurring subject in tubulins26,27,28. These rings are equivalent to coiled protofilaments with a certain degree of curvature. Their formation requires a significant remodeling of the subunit-subunit longitudinal and lateral contacts. In some images, we could observe ring-like structures “peeling off” at the ends of filaments (Fig. 4e and Fig. S4c), resembling typical images that can be observed for depolymerizing microtubules. This supports the view that contacts within the rings are related to the longitudinal contacts within the filament. Image processing of ~1200 independent rings revealed an inherent flexibility with ring sizes ranging from 17 to 19 monomers (rings diameter, 24–29 nm; Fig. 4e), as also described for tubulin and FtsZ rings26, suggesting a similar degree of inter-monomer bending. Taken together, our results show that despite marked differences (helical versus straight filaments), TubZ conserves inter-subunit interfaces and the bending ability of tubulin-like proteins.
Discussion
Unlike tubulin, TubZ molecules must rotate during polymerization to generate a helical filament. The correct formation of the longitudinal interface is paramount in order for hydrolysis to yield a dynamic filament. The relative rotation between the N- and C-terminal domains gives rise to a twist in the TubZ protofilament20. However, when 2, 3 or 4 protofilaments wrap around each other, the rotation must change to conserve the same longitudinal interface. Our findings suggest that the C-tail is a flexible domain that drives this change and enables the correct positioning of monomers; being responsible for the helical pitch and the number of protofilaments of the resulting assembly. Further, this domain is also involved in the filament stiffening upon disassembly. Based on our results and previous findings, we propose a model to explain TubZ filament dynamics (Fig. 5). TubZ in solution is prone to assemble upon GTP binding, as also described for tubulin29. The first interaction of the newly added TubZ molecule may result in a certain degree of rotation in the monomer20, but its positioning is inappropriate for GTP hydrolysis. Depending on the length of the C-tail, the addition of one or several molecules is necessary so the C-tail tip can make an initial contact, which is followed by the stabilization of the entire C-tail via the charged residues (Step 1). Once the molecule is correctly positioned and the catalytic loop reaches the terminal gamma phosphate, GTP hydrolysis can occurs (Step 2). GTPase activity triggers the opening of the twist in the longitudinal subunit-subunit interface13 and a rearrangement of the C-tail binding interactions with upper molecules happens (Step 3). At this point filament stiffening does not occur due to the stability of the filament lattice and the presence of GTP caps. However, once the GTP caps are lost a conformational change occurs that triggers depolymerization. Moreover, the longitudinal subunit-subunit interfaces become too weak to hold the molecules together (Step 4), leading to the release of those molecules during filament disassembly. In fact, the crystal structure of phages 201 ϕ2-1 and KZ TubZ (both in the presence of GDP) show a protofilament-like arrangement that may constitute two stages of the weakening process after GTP hydrolysis. In both structures, the C-tail is already in a post-hydrolysis state. However, KZ TubZ may represent an initial stage with a larger (999.6 Å2, versus 205 Å2 in 201ϕ2-1 TubZ) and more proximal interface area25 (i.e. the distance between the catalytic loop and GDP is 6.9 Å, versus 12.2 Å in 201ϕ2-1 TubZ). The proposed model can fit both treadmilling and dynamic instability. In the presence of a GTP cap, depolymerization occurs preferentially from the filament minus end, where the subunit-subunit interface may be weaker due to the inherent filament polarity. However, if the cap is lost, the entire filament becomes stiffer, thus triggering a complete disassembly.
The C-tail tip plays a dual role, on one hand, it modulates monomer binding with upper subunits within the filament and, on the other hand, it interacts with the segrosome (TubRC) and TubY, probably once the tail is stabilized through charged residues. Based on results showing that the segrosome only binds at the TubZ filament end12, we propose that although TubY binds to the whole filament, it only facilitates filament disassembly at the segrosome-attached end. Thereby, contributing to DNA movement along the bacteria cytoplasm.
Recent studies have proposed that tubulin and tubulin-like FtsZ rings display opposite curvatures when protofilaments bend to form a ring. This has been explained in terms of the different localization of the C-terminus within the ring30. Further studies will be required to elucidate the biological function of TubZ rings and their structural arrangement with regards to their bending orientation, the positioning of the C-tail and the presence of TubY during this filament rearrangement.
A key finding of our study is the importance of the TubZ C-tail on protein assembly and filament disassembly modulation. Since TubZ is the motor protein in plasmid encoded segregation systems, inhibiting its assembly or filament dynamics would in turn seriously affects DNA trafficking during bacterial cell division. Considering that high virulence Bacillus and Clostridium strains carry plasmids encoding this kind of partition system, perturbing the plasmid maintenance would lead to a completely lost of the bacteria toxicity. According to our results, the design of such new drugs would involve peptides mimicking different TubZ C-tails. Further studies are warranted that would answer the effectiveness of these kinds of compounds and their applicability in a population of growing bacteria.
Methods
Cloning, protein expression and purification
Wild type TubZ and the mutants TubZT100A, and TubZE200A, and TubY226 were cloned as previously described5. TubZ C-terminal truncated proteins were obtained by PCR mutagenesis using our wild type construct vector (a modified pET28 vector that includes an N-terminal His tag followed by a 3C protease site) as a template. All proteins were expressed and purified as previously described5 and stored at −80 °C. New constructs were analyzed by mass spectrometry.
TubZ assembly
The time course of polymerization was monitored by right angle light scattering analysis of 0.5-mL samples at 350 nm (0.5 nm excitation and emission slits widths) at 30 °C, except for controls and AFM experiments (25 °C). Proteins were diluted in PKE assembly buffer (50 mM Pipes/KOH [pH 6.5], 100 mM potassium acetate, 1 mM EDTA) with or without 200 g/L Ficoll-70 (prepared in PKE buffer), and polymerization was initiated by the addition of nucleotide (1 mM GTP, 0.1 mM GTP- γ-S or 2 mM GDP) and 6 mM magnesium acetate. Polymer formation was quantified by sedimentation assays. Samples were prepared as described above and centrifuged for 30 min at 60,000 rpm in a TLA100 rotor. The supernatants were carefully withdrawn and the pellets were resuspended in the same volume of buffer and both samples were loaded in consecutive lanes of 12% polyacrylamide gels. Gels were stained with Coomassie Blue, scanned with a Bio-Rad CS-800 calibrated densitometer and analyzed with Quantity One software (Bio-Rad). Alternatively, protein concentration was measured with the Bio-Rad protein assay kit using spectrophotometrically calibrated TubZ standards for the calibration curve. The critical concentration was determined based on the relation between the amounts of protein in the pellet relative to the total amount of protein at different protein concentrations.
Electron microscopy
Samples from polymerization experiments were adsorbed onto carbon-coated grids and negatively stained with 2% uranyl acetate. Images were taken at various magnifications with a JEOL 1230 electron microscope equipped with a 16 Mpixel CMOS TVIPS Temcam-F416 camera and operated at 100 kV. For image processing micrographs were captured at 40,000x nominal magnification (2.84 Å/pixel). For filament structural analysis, straight segments were manually selected and boxed at 200 × 200 and 128 × 128 pixels using EMAN31. For each sample, the extracted particles (CbTubZ GTP ~ 8000, CbTubZ GTP- γ-S ~ 4600, CbTubZ GDP ~ 7500, CbTubZE200A GTP ~ 20,000 and CbTubZT100A GTP ~ 12,500) were classified and averaged using the CL2D algorithm32. Helical reconstruction was performed with CbTubZ GTP data (128 × 128 pixels per particles) using the Iterative Helical Real Space Reconstruction Protocol (IHRSR33). An initial non-symmetrized three-dimensional reconstruction was performed using EMAN to obtain an initial estimation of the helical parameters, rise, and angle between monomers. A featureless cylinder of the appropriate diameter was used as starting model. Different values around the estimated ones were used as starting points in the IHRSR scripts to analyze parameter convergences. All runs converged on a similar final reconstruction. Rings were obtained from assembled TubZ350 in the presence of GTP and magnesium acetate after the addition of TubY226 (in a 1:1 ratio) to the solution. Images were selected manually and subsequently classified and averaged using XMIPP34.
Atomic force microscopy
Samples of TubZ filaments were prepared as described above using a protein concentration of 10 μM in a Thermostat Plus (Eppendorf). For TubZ filaments coated with TubY, an additional 10 μM of TubY was added to the sample. Filaments were deposited onto freshly cleaved mica surface pre-treated with 20 μl MgCl2 (100 mM). After one minute of adsorption, the sample was gently washed with 3 ml Milli-Q water supplemented with 1 mM GTP and 6 mM magnesium acetate. GDP bound filaments were assembled as above, but with the addition of 20 μM wild type CbTubZ. Assembly time was set to 1 h and the mica was functionalized with 0.5% glutaraldehyde for the duration of the incubation time. GDP-bound filaments were adsorbed on the functionalized mica for 1 min and the sample was subsequently washed gently with 2 mL Milli-Q water supplemented with 2 mM GDP and 6 mM magnesium acetate. All AFM samples were dried by simple blotting; without using nitrogen gas. Samples were imaged in air at room temperature and low humidity using tapping mode with amplitude of 5 nm and a scan rate of 1 line·s−1 using an AFM from Nanotec (Nanotec Electrónica, Madrid, Spain). PointProbePlus were used (PPP-NCH, Nanosensors, Switwerland). Image processing consisted of a plane subtraction and flattening using the WSxM freeware35.
Circular dichroism
Spectra were acquired at 25 °C using a 0.1 mm cell in a thermostated cell holder. Sample scanning conditions were as follows: 1 nm bandwidth; 1 nm measurement interval; 20 nm min−1 scan speed; 4 s time constant. Scans of each sample or buffer were repeated 4 times and the results averaged. CD data (millidegrees) were reduced to mean residue ellipticity values (degrees cm2 dmol−1) with the Jasco software and plotted.
Crystallization and structure determination
TubZ316 crystals were grown at 22 °C in reservoir solution containing 0.1 M Na-citrate [pH 5.5], 0.1 M NaCl, 12% PEG4000 (Molecular Dimensions/MemStart-MemSys screen) in the presence of a peptide containing the last 8 residues of TubZ (NDFFSKYM) using the sitting-drop vapor-diffusion technique. Crystals were cryo-protected with 22% glycerol before being mounted in a nylon loop and flash-cooled in liquid nitrogen. X-ray diffraction data were collected at beamline PROXIMA 1 beamline (SOLEIL Synchrotron, France) using a wavelength 0.98011 Å and a DECTRIS PILATUS 6 M detector. Datasets were indexed, integrated, and scaled using XDS36 and the HKL reflection file was converted to a MTZ file using XDSCONV and the CCP4 suite supported program CAD37. Initial coordinates were determined by molecular replacement using PHASER38 and the coordinates for Apo-TubZ (Protein Data Bank (PDB) ID 3V3T). Ligand GDP coordinates and restrains were generated using ELBOW39 and fitted into the electron density maps using LigandFit40. No extra density was observed for the peptide. This was followed by several cycles of refinement using PHENIX41, which also was used for water picking, and iterative model building with COOT42. The final model was validated using MolProbidity43. Figures were generated using Pymol (Schrödinger).
GTPase activity and polymer-bound nucleotide
GTP hydrolysis was monitored by taking 10 μL aliquots from protein polymerizing reactions at different times and measuring the release of inorganic phosphate in the malachite green assay at 650 nm44. For GTP-γ-S, hydrolysis and polymer-bound nucleotide samples were analyzed by HPLC45.
mant-GTP binding affinity
Fluorescence anisotropy of free and protein-bound mant-GTP was measured with a spectrofluorimeter, with excitation and emission wavelengths of 357 nm (2 nm band pass) and 445 nm (5 nm band pass), respectively, using 2 × 10 mm cells at 25 °C. The binding of mant-GTP to TubZ causes a significant increase in its fluorescence that could lead to the calculation of erroneous apparent Kb. To avoid this error, the ratios between the fluorescence intensities of TubZ-bound and free mant-GTP were determined from the different constructs (RTubZ = 2.2; RTubZ350 = 2.7; R TubZ331 = 2.7; RTubZ316 = 2.4) and used to correct all anisotropy-based binding calculations. To measure the Kb, mant-GTP (500 nM) was titrated with TubZ proteins at different concentrations (0.075–10 μM) in PKE buffer. The converged data were used to calculate the binding constant46.
Additional Information
Accession codes (Protein Data Bank 4XCQ).
How to cite this article: Fuentes-Pérez, M. E. et al. TubZ filament assembly dynamics requires the flexible C-terminal tail. Sci. Rep. 7, 43342; doi: 10.1038/srep43342 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Jan Löwe (MRC-LMB) and Eva Nogales (University of California, Berkeley) and Hailey Gahlon (Imperial Colledge) for discussion and advice; the staff at the PROXIMA 1 beamline (SOLEIL, Paris, France) for assistance. This work was supported by Ministerio de Ciencia e Innovación grant RYC-2011-07900 (MAO), Ministerio de Economía y Competitividad and European Regional Development Fund co-funding grants BFU2013-47014-P (MAO) and SAF2014-52301-R (OL), Ministerio de Economía y Competitividad grant FIS2014-58328-P (FM-H), and BES-2015-071244 PhD associated fellowship (A.M.-G) European Community 7FP (FP7/2007–2013) under BioStruct-X (grant agreement N° 283570).
Footnotes
The authors declare no competing financial interests.
Author Contributions M.A.O., O.L. and F.M.-H. designed research, M.A.O., M.E.F.-P., R.N.-R., A.M.-G. and D.J.-R. performed research, M.A.O., M.E.F.-P., R.N.-R., A.M.-G., O.L. and F.M.-H. analyzed data, and M.A.O., O.L. and F.M.-H. wrote the paper.
References
- Nogales E., Downing K. H., Amos L. A. & Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5, 451–458 (1998). [DOI] [PubMed] [Google Scholar]
- Oliva M. A. & Andreu J. M. In eLS Citable Reviews doi: 10.1002//9780470015902.a0025586 (2014). [DOI]
- Larsen R. A. et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev 21, 1340–1352 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J. C. & Funnell B. E. Plasmid Partition Mechanisms. Microbiol Spectr 2, doi: 10.1128/microbiolspec.PLAS-0023-2014 (2014). [DOI] [PubMed] [Google Scholar]
- Oliva M. A., Martin-Galiano A. J., Sakaguchi Y. & Andreu J. M. Tubulin homolog TubZ in a phage-encoded partition system. Proc Natl Acad Sci USA 109, 7711–7716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y. et al. A novel transcriptional activator, tubX, is required for the stability of Bacillus sphaericus mosquitocidal plasmid pBsph. J Bacteriol 196, 4304–4314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellberg C., Cade N. I., Holmes D. & Surrey T. The size of the EB cap determines instantaneous microtubule stability. Elife 5, doi: 10.7554/eLife.13470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. & Kirschner M. Dynamic instability of microtubule growth. Nature 312, 237–242 (1984). [DOI] [PubMed] [Google Scholar]
- Nogales E. & Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr Opin Struct Biol 37, 90–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. A. et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol 107, 1437–1448 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M. L. et al. A bacteriophage tubulin harnesses dynamic instability to center DNA in infected cells. Elife 3, doi: 10.7554/eLife.03197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. & Lowe J. Reconstitution of a prokaryotic minus end-tracking system using TubRC centromeric complexes and tubulin-like protein TubZ filaments. Proc Natl Acad Sci USA 112, E1845–1850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montabana E. A. & Agard D. A. Bacterial tubulin TubZ-Bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis. Proc Natl Acad Sci USA 111, 3407–3412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. & Erickson H. P. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids: evidence for a capping mechanism. J Biol Chem 283, 8102–8109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F., Colchero J. & Baro A. M. DNA height in scanning force microscopy. Ultramicroscopy 96, 167–174 (2003). [DOI] [PubMed] [Google Scholar]
- Ni L., Xu W., Kumaraswami M. & Schumacher M. A. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA 107, 11763–11768, doi: 1003817107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. X., Rivas G. & Minton A. P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys 37, 375–397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J. M. et al. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J Biol Chem 278, 37664–37671 (2003). [DOI] [PubMed] [Google Scholar]
- Venturoli D. & Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am J Physiol Renal Physiol 288, F605–613 (2005). [DOI] [PubMed] [Google Scholar]
- Aylett C. H., Wang Q., Michie K. A., Amos L. A. & Lowe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci USA 107, 19766–19771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G. M. et al. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell 157, 1117–1129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer J. A. et al. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149, 1488–1499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr E. A. et al. The Structure and Assembly Mechanism of a Novel Three-Stranded Tubulin Filament that Centers Phage DNA. Structure 22, 539–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S. & Hayashi I. Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid. J Biol Chem 287, 32103–32112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett C. H., Izore T., Amos L. A. & Lowe J. Structure of the Tubulin/FtsZ-Like Protein TubZ from Pseudomonas Bacteriophage PhiKZ. J Mol Biol 425, 2164–2173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P. & Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J Cell Biol 135, 5–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wang H. W. & Niederstrasser H. Tubulin rings: which way do they curve? Curr Opin Struct Biol 13, 256–261 (2003). [DOI] [PubMed] [Google Scholar]
- Erickson H. P., Anderson D. E. & Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74, 504–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey R. M., Diaz J. F. & Andreu J. M. The nucleotide switch of tubulin and microtubule assembly: a polymerization-driven structural change. Biochemistry 45, 5933–5938 (2006). [DOI] [PubMed] [Google Scholar]
- Housman M., Milam S. L., Moore D. A., Osawa M. & Erickson H. P. FtsZ Protofilament Curvature Is the Opposite of Tubulin Rings. Biochemistry 55(29), 4085–4091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S. J., Baldwin P. R. & Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- Sorzano C. O. et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171, 197–206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman E. H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- de la Rosa-Trevin J. M. et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- Horcas I. et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum 78, 013705 (2007). [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta cryst D, Biol crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta cryst D, Biol crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty N. W., Grosse-Kunstleve R. W. & Adams P. D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta cryst D, Biol crystallogr 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Klei H., Adams P. D., Moriarty N. W. & Cohn J. D. Automated ligand fitting by core-fragment fitting and extension into density. Acta cryst D, Biol crystallogr 62, 915–922 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta cryst D, Biol crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta cryst D, Biol crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta cryst D, Biol crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Fukui K. & Kometani K. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem 99, 1465–1472 (1986). [DOI] [PubMed] [Google Scholar]
- Oliva M. A. et al. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J Biol Chem 278, 33562–33570 (2003). [DOI] [PubMed] [Google Scholar]
- Ruiz-Avila L. B. et al. Synthetic inhibitors of bacterial cell division targeting the GTP-binding site of FtsZ. ACS Chem Biol 8, 2072–2083 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.