Zhu et al. mapped architectural changes in genome organization during neutrophil differentiation. They propose that supercontraction and recruitment of heterochromatin to the lamina facilitates the folding of neutrophil genomes into a confined geometry imposed by a multilobular nuclear structure.
Keywords: genome topology, Lamin B receptor, neutrophils, nucleoli, rDNA
Abstract
Neutrophils are responsible for the first line of defense against invading pathogens. Their nuclei are uniquely structured as multiple lobes that establish a highly constrained nuclear environment. Here we found that neutrophil differentiation was not associated with large-scale changes in the number and sizes of topologically associating domains (TADs). However, neutrophil genomes were enriched for long-range genomic interactions that spanned multiple TADs. Population-based simulation of spherical and toroid genomes revealed declining radii of gyration for neutrophil chromosomes. We found that neutrophil genomes were highly enriched for heterochromatic genomic interactions across vast genomic distances, a process named supercontraction. Supercontraction involved genomic regions located in the heterochromatic compartment in both progenitors and neutrophils or genomic regions that switched from the euchromatic to the heterochromatic compartment during neutrophil differentiation. Supercontraction was accompanied by the repositioning of centromeres, pericentromeres, and long interspersed nuclear elements (LINEs) to the neutrophil nuclear lamina. We found that Lamin B receptor expression was required to attach centromeric and pericentromeric repeats but not LINE-1 elements to the lamina. Differentiating neutrophils also repositioned ribosomal DNA and mininucleoli to the lamina—a process that was closely associated with sharply reduced ribosomal RNA expression. We propose that large-scale chromatin reorganization involving supercontraction and recruitment of heterochromatin and nucleoli to the nuclear lamina facilitates the folding of the neutrophil genome into a confined geometry imposed by a multilobed nuclear architecture.
It is now well established that, in interphase nuclei, the chromatin fiber is folded into loops that are anchored by architectural proteins such as CTCF and members of the cohesion complex. Clusters of loops are organized as topologically associating domains (TADs). TADs, in turn, collectively associate with either the euchromatic (A) or heterochromatic (B) compartment. Finally, individual chromosomes fold into territories that only sporadically intermingle (Lieberman-Aiden et al. 2009; Dixon et al. 2012, 2015; Jin et al. 2013; Rao et al. 2014). An additional layer of chromatin architecture involves interactions between the chromatin fiber and nuclear structures (Bickmore and Van Steensel 2013). Particularly well-characterized nuclear structures are the nuclear lamina, Polycomb group (PcG) bodies, transcription factories, and nucleoli. Prominent among interactions involving the chromatin fiber and nuclear structures are the heterochromatin and the nuclear lamina (Melcer and Meshorer 2010; Peric-Hupkes et al. 2010; Meuleman et al. 2013; Towbin et al. 2013), a process mediated by the Lamin B receptor (LBR) and Lamin A/C (Solovei et al. 2013). Likewise, PcG bodies associate with H3K27me3-marked transcriptionally silent chromatin to form an intrachromosomal and interchromosomal network (Denholtz et al. 2013; Vieux-Rochas et al. 2015). Clusters of genes that are coordinately transcribed tend to cluster as transcription factories (Apostolou and Thanos 2008; Li et al. 2012; de Wit et al. 2013). Finally, ribosomal DNA (rDNA) repeat elements are arranged as nucleolar organizer regions that cluster when associated with the RNA polymerase I transcription machinery to form nucleoli, a nuclear body specialized in ribosomal RNA (rRNA) transcription, processing, and assembly of ribosomes (Kalmárová et al. 2007; Németh and Längst 2011).
Neutrophils are terminally differentiated, short-lived circulating cells. Like other leukocytes, they are continuously generated from hematopoietic stem cells and develop through intermediate developmental stages. Neutrophils belong to a special class of immune cells called polymorphonuclear cells that also includes basophils and eosinophils. Polymorphonuclear cells are unique in that their nuclei are ring-shaped or multilobulated, a feature that facilitates rapid migration through the endothelial lining of blood vessels and the interstitial spaces of tissues (Kolaczkowska and Kubes 2013). How the genomes of polymorphonuclear cells are organized into multiple lobes remains largely unknown. A critical component involves the LBR. Homozygous nonsense mutations in the Lbr gene result in an anomaly that is characterized by a failure to generate multilobular nuclei and an accumulation of immature neutrophils (Gaines et al. 2008; Verhagen et al. 2012). Hypolobulated neutrophils are deficient in their ability to migrate through interstitial spaces (Gaines et al. 2008).
Here we mapped architectural changes in genome organization during neutrophil differentiation. We found that the genome of neutrophils was characterized by an extensive loss of local genomic interactions (<3 Mb) and a gain of long-range chromatin interactions (>3 Mb) that spanned vast genomic distances. Population-based simulation of spherical and toroid genomes revealed declining radii of gyration for neutrophil chromosomes. We found that the neutrophil genome underwent widespread chromosome contraction across vast genomic distances, a process termed supercontraction. Supercontraction involved genomic regions located in the B compartment in both progenitors and neutrophils or genomic regions that switched from the A to the B compartment during neutrophil differentiation. The differentiation of neutrophils was also accompanied by the repositioning of centromeres, pericentromeres, long interspersed nuclear element 1 (LINE-1) elements, rDNA, and nucleoli from the nuclear interior to the nuclear lamina. We found that LBR expression was required to attach centromeric and pericentromeric repeats but not LINE-1 elements to the neutrophil nuclear lamina. We propose that supercontraction and recruitment of heterochromatin to the lamina facilitates the folding of neutrophil genomes into a confined geometry imposed by a multilobular nuclear structure. Furthermore, we suggest that rDNA localization regulates protein synthesis and neutrophil life span.
Results
A model system to study multilobed nuclear architecture
To examine how the neutrophil genome folds into a confined environment imposed by a multilobular nuclear structure, we used a previously described cell line that permits the isolation of large numbers of homogeneous myeloid progenitors and multilobular cells. This system uses a promyeloid cell line named ECOMG, which was established by conditional immortalization of myeloid progenitors using an E2A/PBX1–estrogen receptor fusion protein (Sykes et al. 2003). As reported previously, ECOMG cells proliferated indefinitely in the presence of GM-CSF and estrogen, but, upon estrogen withdrawal, readily differentiated into a homogeneous population of multilobular cells (>98%) (Supplemental Fig. S1A). To compare the transcriptomes of in vitro differentiated neutrophils with murine bone marrow-derived neutrophils (BMDNs), RNA was isolated from ECOMG progenitors (progenitors) and in vitro differentiated neutrophils. The transcriptomes of both populations were analyzed using RNA sequencing (RNA-seq) and compared with the transcription signatures of murine BMDNs (Wong et al. 2013). We identified 3192 transcripts that were expressed at higher abundance versus 3378 transcripts that were expressed at lower abundance in differentiated neutrophils versus progenitors (Supplemental Fig. S1B). Notably, we found that the transcription signatures of in vitro differentiated neutrophils closely correlated with primary neutrophils isolated from the bone marrow (Supplemental Fig. S1C). Gene ontology analysis revealed that the transcription signature of in vitro differentiated neutrophils was in agreement with previous microarray assays: Activated genes in neutrophils were enriched for adhesion, cytokine production, migration, phagocytosis, and bacterial killing (Supplemental Fig. S1D,E), whereas repressed genes in neutrophils encoded primarily for proteins associated with metabolism, RNA processing, and translation (Supplemental Fig. S1D). Prominent among the changes in gene expression was the coordinate decline in transcript abundance of ribosomal proteins and RNA polymerase I subunits (Supplemental Fig. S1E, right panel). Taken together, these data indicate that the transcription signature of in vitro differentiated neutrophils resembles that of primary neutrophils and that the differentiation of neutrophils is closely associated with a decline in the expression of genes associated with protein synthesis.
Neutrophil genomes are enriched for long-range genomic interactions
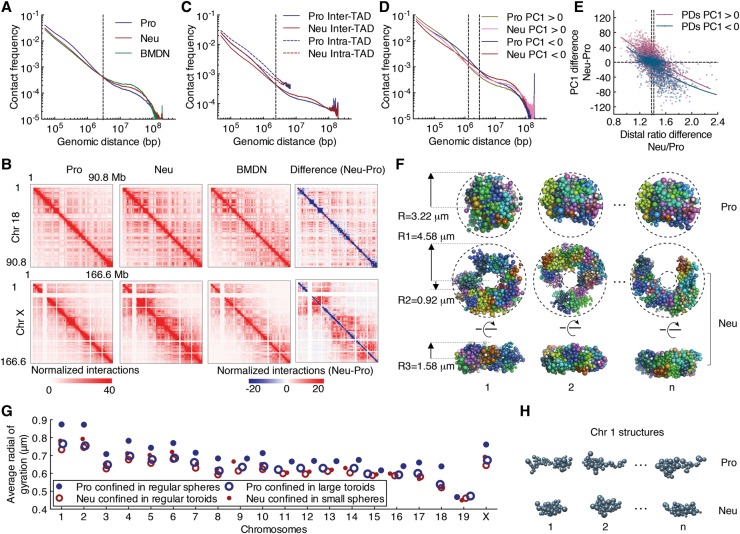
As a first approach to determine how the topology of the neutrophil genome differs from that of mononuclear cells, we used genome-wide chromosome conformation capture analysis followed by high-throughput sequencing (Hi-C) (Rao et al. 2014). We found that neutrophils and BMDN, when compared with progenitors, were depleted of genomic interactions that spanned <3 Mb but were enriched for genomic interactions separated by genomic distances >3 Mb (Fig. 1A). We next constructed chromosome-wide contact maps for autosomes as well as X chromosomes (Fig. 1B). We found that short-range genomic interactions (diagonal) were depleted, whereas interactions across large genomic distances (off-diagonal) were enriched in neutrophils (Fig. 1B). The differences in short-range versus long-range interactions were even more pronounced for X chromosomes (Fig. 1B). The differences were particularly apparent in the differential heat maps constructed by subtracting contact frequencies in progenitors from neutrophils (Fig. 1B, far right panels). The contact matrices revealed a similar distribution for TADs when comparing progenitors versus neutrophils (Supplemental Fig. S2A). The size and numbers of TADs were highly consistent for both cell types (Supplemental Fig. S2B). However, the average intrachromosomal contact probability as a function of genomic distance differed between progenitors and neutrophils. Specifically, we found that intra-TAD interactions and inter-TAD interactions involving adjacent TADs (<3 Mb) were depleted, whereas genomic interactions that spanned multiple TADs (>3 Mb) were enriched for neutrophils (Fig. 1C). We next used principal component analysis to identify the A and B compartments. Genomic regions that displayed a continuum of either positive or negative PC1 values were defined as PC1 domains (PDs) (Lieberman-Aiden et al. 2009). Neutrophils versus progenitors displayed increased numbers of PDs that were smaller in size overall (Supplemental Fig. S2C). Specifically, we identified 488 PDs that switched from the A to the B compartment, whereas 342 PDs switched from the B to the A compartment (Supplemental Fig. S2D; Supplemental Table S1). The size of PDs that switched during the transition from progenitors to neutrophils was significantly smaller than that of average PDs (Supplemental Fig. S2E). We found that genomic interactions involving large genomic distances (>3 Mb) for both compartment A and B were enriched in neutrophils (Fig. 1D). To quantify long-range versus local genomic interactions, we calculated the distal ratio (percentage of contacts that were separated by >3 Mb) for progenitors and neutrophils. We found that the distal ratio was significantly higher in neutrophils when compared with progenitors, and the differences of the distal ratio were inversely correlated with differences in PC1 values (Fig. 1E). Specifically, genomic regions that were associated with declining PC1 values or switched PC1 values often displayed changes in interaction patterns: from short-range interactions in progenitors to long-range genomic interactions in neutrophils (Fig. 1E; Supplemental Table S2). In sum, these data indicate that neutrophil genomes are depleted for local genomic interactions (<3 Mb) but enriched for long-range genomic interactions that span vast genomic distances (>3 Mb).
Figure 1.
The differentiation of neutrophils is closely associated with large-scale changes in intradomain and interdomain interactions. (A) Average contact probabilities as a function of genomic distance for genomic interactions within individual chromosomes at 50-kb resolution for progenitors (Pro), neutrophils (Neu), and BMDN genomes are shown. The dashed line indicates the crossover for contact frequencies at 3 Mb of gneomic separation. (B) Hi-C contact heat maps for chromosome 18 (Chr 18) (top) and the X chromosome (Chr X) (bottom) at 500-kb resolution for progenitors, neutrophils, and BMDNs are shown. The intensity of each pixel represents the normalized number of contacts between a pair of loci. The right panels indicate differential contact heat maps for chromosome 18 and the X chromosome. Subtraction of the progenitor from neutrophil contact matrices shows changes in the normalized number of contacts between the Hi-C samples. Red represents enrichment, whereas blue represents depletion for normalized contact frequencies in neutrophils. (C) Intrachromosomal contact frequency decay curves for intra-TAD and inter-TAD interactions at 50-kb resolution for progenitors and neutrophils. (D) Intrachromosome contact frequency decay curves for the A and B compartments at 25-kb resolution for progenitor and neutrophil genomes are shown. Interactions were classified based on whether both end points are located within the A or B compartment. The crossover in contact frequencies between progenitor and neutrophil genomes is at 1.3 and 3 Mb for the A and B compartments, respectively (dashed lines). (E) Differences in distal ratio (fold change, Neu/Pro) versus the differences in PC1 values for neutrophil and progenitor genomes for each continuous PD is indicated. Color intensity corresponds to the density of the data points. Solid lines indicate nonlinear fit curves. Genome-wide, the distal ratio was significantly higher in neutrophils when compared with progenitors: a median distal ratio fold change of 1.38 for the A compartment versus 1.42 for the B compartment (dashed lines). (F) Three representative structures for progenitor and neutrophil genomes were derived from the structure populations. Modeling was performed for genomes confined as either spherical or toroid nuclear volumes. Shown are top (middle panel) and side (bottom panel) views of modeled neutrophil nuclear structures. The dimensions and shape of the nuclear volumes are indicated and were based on experimental measurements (Supplemental Fig. S2F). (G) The average radius of gyration for each chromosome in neutrophil and progenitor genomes was computed for two different settings. The progenitor and neutrophil genomic structures were modeled using Hi-C reads derived from progenitor and neutrophil genomes folded into spherical or toroid nuclear structures. (H) Representative structures for chromosome 1 are shown for progenitors and neutrophils with a radius of gyration similar to the average value, as indicated in resulting models.
Simulating spherical and toroid genomes revealed decreased radii of gyration for neutrophil chromosomes
The unique nuclear shape confers a larger surface to volume ratio in neutrophils compared with progenitors (Supplemental Fig. S2F). To determine how changes in nuclear shape affect genome compaction, we generated three-dimensional (3D) models for mononuclear and multilobular genomes (Kalhor et al. 2012). Specifically, a population of genome structures was generated in which chromatin contacts were statistically consistent with the experimental Hi-C maps that were derived from progenitors and neutrophils (neutrophil: correlation 0.972; progenitor: correlation 0.970). Modeling was performed for genomes confined in either spherical or toroid nuclear shapes (Fig. 1F). We then computed the degree of compaction for each of the chromosomes by measuring the average radii of gyration (Kalhor et al. 2012). We observed substantial differences in the radii of gyration of chromosomes in the two cell types. In neutrophils, the chromosomes were consistently more compacted with substantially lower radii of gyration when compared with progenitors (Fig. 1G,H). To investigate the role of the nuclear shape, we also calculated, using Hi-C reads derived from progenitor cells, the radii of gyration of progenitor genomes confined in toroid nuclear shapes. Notably, progenitor chromosomes displayed smaller radii of gyration even though the nuclear volume remained unchanged (Fig. 1G). When changing neutrophil genomes from toroid to spherically nuclear shapes, their chromosomes showed slightly increased radii of gyration, reflecting a more elongated chromosome shape (Fig. 1G). These observations suggest that differences in nuclear shape can impose distinct folding patterns of genomes: Multilobular toroid-like nuclei tend to favor more compacted chromosome configurations.
Supercontraction of neutrophil genomes
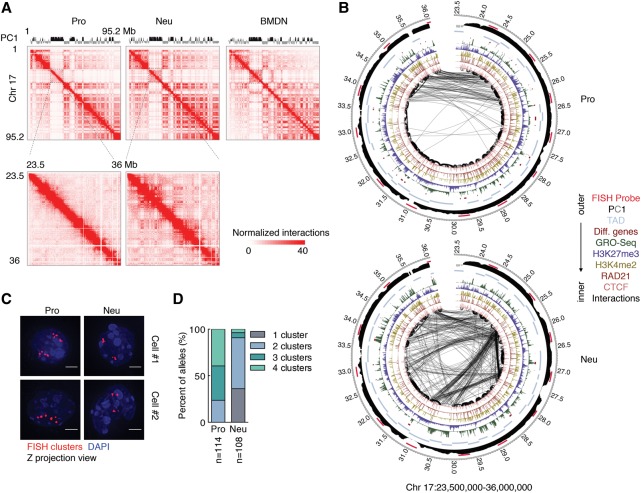
To determine how the increase in long-range genomic interactions as measured by Hi-C relates to spatial contraction, we focused on a 13-Mb genomic region located on chromosome 17. This 13-Mb region includes the top five distal-favored PDs in neutrophil genomes as compared with progenitor genomes identified by distal ratio analysis (Supplemental Table S2). In neutrophils, the 13-Mb region displayed significant enrichment for long-range genomic interactions (Fig. 2A,B). To visualize this region in single cells, we performed 3D-FISH using fluorescently labeled BAC probes that spanned the 13-Mb region (Fig. 2B, red bars). We found that the majority of progenitors displayed three or four fluorescent foci versus one or two foci per allele for neutrophils, indicating that the increase in long-range genomic interactions as observed by Hi-C correlates with spatial contraction (Fig. 2C,D). Here, we refer to spatial contraction across large genomic distances as supercontraction. Taken together, these data indicate that the differentiation of neutrophils is closely associated with spatial contraction across large genomic distances, a process named supercontraction.
Figure 2.
Genomes of differentiating neutrophils undergo large-scale spatial contraction. (A) A Hi-C contact heat map for chromosome 17 at 500-kb resolution (top) and a subgenomic region for chromosome 17 at 50-kb resolution (bottom) for the indicated cell types is shown. The intensity of each pixel represents the normalized number of contacts between a pair of loci. (B) Circos plots show genomic interactions across chromosome 17 derived from progenitor and neutrophil genomes. RAD21, CTCF, H3K4me2, H3K27me3, GRO-seq (global run-on combined with sequencing), cell type-specific gene positions, and PC1 values are shown. Probes used for 3D-FISH are indicated in red. Only significant interactions are shown. P < 0.001, binomial test. The thickness of the connecting lines reflects the significance of genomic interactions (−log P). Bin size, 50 kb. Numbers around the margins indicate genomic positions (in megabases). (C) 3D-FISH in progenitors and neutrophils using BAC probes spanning a 13-Mb region on chromosome 17 (shown in A, bottom). Representative maximum intensity Z-projected images are shown. Original magnification, 100×. (Red) BAC probes; (blue) DAPI staining. Bars, 2 µm. (D) Fractions of the number of clusters per allele were visualized using BAC probes for neutrophils and progenitors. (n) Number of alleles quantified for each sample.
Epigenetic determinants that mark neutrophil differentiation
Previous observations indicated that developmental transitions are associated with the swiching of genes between the A and B compartments (Lin et al. 2012; Dixon et al. 2015). To identify genes that switched compartments during neutrophil differentiation and how these changes relate to the activation of a neutrophil-specific transcription signature, we used GRO-seq (global run-on combined with sequencing). Overall, nascent transcript levels correlated well with those observed for RNA-seq (Supplemental Fig. S3A). Next, we examined how changes in transcript levels during neutrophil differentiation relate to the switching of genes between nuclear compartments. We identified 65 genes that coordinately switched compartments and modulated levels of nascent transcription in differentiating neutrophils (Supplemental Fig. S3B). Out of a total of 1033 genes that were associated with increased nascent transcription in differentiating neutrophils, 39 switched from compartment B to compartment A, whereas 26 out of 2333 genes that displayed decreased nascent transcript levels switched from compartment A to compartment B (Supplemental Fig. S3B,F; Supplemental Table S3).
Given that we found that only a small proportion of genes that switched compartments exhibited coordinate changes in gene expression levels, we reasoned that other epigenetic mechanisms must exist that regulate a neutrophil-specific transcription signature. Hence, we examined progenitors and neutrophils for the genome-wide deposition of 11 epigenetic marks (data not shown). Among these marks, the deposition of H3K27me3 was particularly intriguing. Among the 1033 genes that were activated in neutrophils, we identified 222 genes associated with promoters that were enriched for H3K27me3 in progenitors but depleted of H3K27me3 in neutrophils (Supplemental Fig. S3D–F; Supplemental Table S3). Taken together, these data indicate that the activation of a neutrophil-specific transcription signature is regulated at multiple levels that include changes in nuclear location but primarily involve depletion of H3K27me3 across promoters that are associated with genes that are activated in neutrophils.
Differentiating neutrophils are enriched for long-range genomic interactions involving transcriptionally silent regions
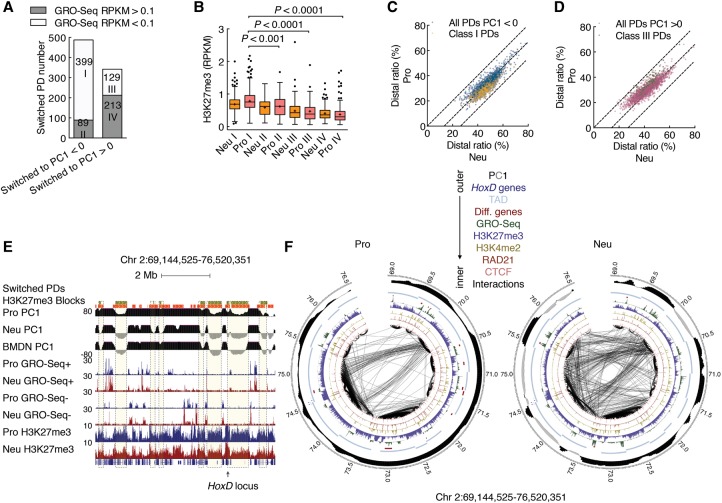
To further evaluate the switching of genomic regions between compartments during neutrophil differentiation, we segregated switched PDs as either transcriptionally active or transcriptionally silent domains as defined by the abundance of nascent transcription. We found that 82% of PDs that switched from compartment A to B were transcriptionally silent in both progenitors and neutrophils (class I) (Fig. 3A). Notably, class I PDs were highly enriched for H3K27me3 and chromosomal contacts that spanned large genomic distances in neutrophils (Fig. 3B–D). Prominent among class I PDs were the Hox loci. Multiple class I PDs located in genomic regions that flank the HoxD locus displayed a concerted decrease in intradomain interactions and a concurrent increase in long-range genomic interactions in neutrophils (Fig. 3E,F). Previous studies have revealed that, in embryonic stem cells, genomic regions associated with the HoxD locus interact with other regions that are characterized by a high abundance of H3K27me3 to generate a network of intrachromosomal and interchromosomal interactions across the A compartment (Denholtz et al. 2013; Vieux-Rochas et al. 2015). In contrast, we found that, in neutrophils, a network of genomic interactions marked by H3K27me3 was associated with the B compartment. We next inspected progenitors and neutrophils for CTCF and RAD21 binding. CTCF occupancy remained unchanged during the transition from progenitors to neutrophils (Supplemental Fig. S4A). However, we found that, in differentiated neutrophils, RAD21 occupancy was depleted across 4142 bound sites (Supplemental Fig. S4B). Inspection of significant interactions with endpoints in 5-kb regions surrounding CTCF peaks revealed similar interaction frequencies for both cell types (Supplemental Fig. S4C). When focused on interactions with endpoints across a 5-kb region surrounding differential RAD21-bound sites, we found that neutrophils displayed significantly lower interaction frequencies (Supplemental Fig. S4C). For example, within a 13-Mb region on chromosome 11, two domains that were depleted for RAD21 occupancy and maintained CTCF binding were depleted of intradomain interactions but enriched for intrachromosomal genomic interactions that spanned vast genomic distances (Supplemental Fig. S5, top panel, dotted purple boxes). Notably, both domains associated with this region were localized in the B compartment (Supplemental Fig. S5, top panel, dotted purple boxes). Among the four classes of switched domains, class I PDs were most severely depleted of RAD21 occupancy (Supplemental Fig. S4D). Taken together, these data indicate that the differentiation of neutrophils is closely associated with de novo long-range chromosomal interactions involving genomic regions located in the B compartment in both progenitors and neutrophils or genomic regions that switched from the A to the B compartment that were enriched for H3K27me3 and depleted of RAD21 occupancy.
Figure 3.
Differentiating neutrophils establish a network of genomic interactions involving transcriptionally silent regions. (A) Bar plot showing the numbers of four classes of PDs that switched compartments during neutrophil differentiation. The switched PDs were classified into two subcategories according to GRO-seq transcript abundance: transcriptionally active (reads per kilobase per million mapped read [RPKM] > 0.1) in either progenitors or neutrophils or transcriptionally silent (RPKM < 0.1) in both progenitors and neutrophils. Numbers in bars indicate totals of PDs. Note that 82% of PDs that switched from the A to the B compartment in neutrophils (class I) were transcriptionally silent in progenitors. (B) Box and whisker plot showing the abundance of H3K27me3 in four classes of PDs that switched compartments in progenitors and neutrophils. P-value, Kruskal-Wallis test. (C) Scatter plot comparing the distal ratio of class I PDs and inactive PDs for neutrophil genomes. Color intensity corresponds to the density of the data points. (D) Scatter plot comparing the distal ratio of class III PDs and active PDs for neutrophil genomes. Color intensity corresponds to the density of the data points. (E) Genome browser snapshot of the Hoxd locus and flanking genomic regions. The deposition of H3K27me3-marked regions is shown for progenitors and neutrophils. Green filled boxes represent class I PDs. Orange filled boxes represent H3K27me3-marked regions. PC1 values as well as read densities for nascent RNA (GRO-seq) and H3K27me3 are shown. (F) Circos plot showing genomic interactions within the same genomic region as in E for progenitor and neutrophil genomes as indicated. RAD21, CTCF, deposition of H3K4me2 and H3K27me3, nascent transcript abundance (GRO-seq), cell type-specific gene positions, and PC1 values are shown. Only significant interactions are shown. P < 0.001, binomial test. The thickness of the connecting lines reflects the significance of the interaction (−log P). Bin size, 50 kb. Numbers at the margins indicate genomic position (in megabases). Colors indicate CTCF, RAD21, H3K4me2, H3K27me3, GRO-seq, cell type-specific genes, TADs, HoxD family, PC1 tracks, and FISH probes (color key above).
Neutrophil genomes are enriched for interchromosomal interactions
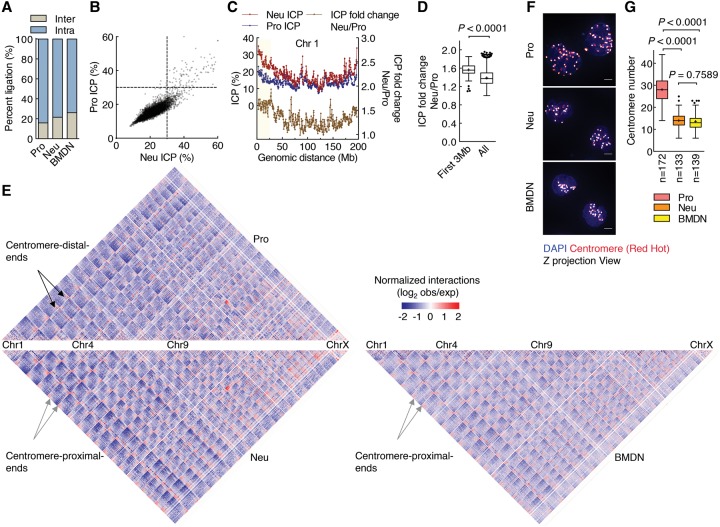
To determine whether interchromosomal interactions were modulated during neutrophil differentiation, we analyzed progenitors and neutrophils for interchromosomal contact frequencies (Kalhor et al. 2012). We found that interchromosomal contacts were highly enriched in neutrophil genomes when compared with progenitors (Fig. 4A,B). For individual chromosomes, we found that centromere-proximal ends (genomic regions located within ∼20 Mb from chromosome ends) were most enriched for interchromosomal interactions in differentiated neutrophils (Fig. 4C,D). We next constructed genome-wide contact maps. We found that, although the chromosomal interaction pattern appeared largely unchanged, all pairs of centromere-proximal ends displayed enrichment for genomic contacts in neutrophils when compared with progenitors (Fig. 4E). A similar trend was observed for primary BMDNs (Fig. 4E). To validate these findings in single cells, we performed 3D-FISH using fluorescently labeled centromeric probes (Fig. 4F). Consistent with the Hi-C contact map, we found that neutrophils displayed decreased numbers of centromeric fluorescent foci compared with progenitors, a pattern indicative of spatial clustering (Fig. 4G). Taken together, these observations indicate that neutrophil differentiation is associated with enrichment for interchromosomal interactions that involve centromere-proximal ends.
Figure 4.
Differentiating neutrophils are enriched for interchromosomal interactions. (A) Neutrophil and BMDN genomes exhibit higher percentages of interchromosomal interactions than progenitor genomes. The fractions of intrachromosomal and interchromosomal contacts from progenitors, neutrophils, and BMDNs are indicated. (B) Scatter plot comparing ICP values derived from neutrophil and progenitor genomes. ICP values were calculated as the ratio of interchromosomal interaction frequency versus the frequencies of all genomic interactions for each 500-kb interval. Color intensity corresponds to the density of data points. (C) Plots showing ICP values derived from progenitors and neutrophils (red and blue lines, left Y-axis) and ICP differences (Neu/Pro) between the two cell types (yellow line, right Y-axis) for chromosome 1 at 500-kb resolution. (D) Box and whisker plot showing ICP fold change (Neu/Pro) comparison for centromeric-proximal regions (<3 Mb) with genome-wide average fold change values. P-value, Mann-Whitney test. (E) Hi-C contact heat map comparison between neutrophils (bottom left), BMDN (bottom right), and progenitors (top) at 500-kb resolution. Colors indicate the log ratio of the observed interaction frequency to expected interaction frequency (obs/exp). (Blue) Lower than expected; (red) higher than expected; (black arrows) centromere-distal end zone; (gray arrows) centromere-proximal end zone. (F) 3D-FISH analysis of progenitors and neutrophils using a fluorescently labeled centromeric probe. Representative maximum intensity Z-projections of image stacks are shown. Original magnification, 100×. (Red hot) Intensity of centromere staining; (blue) DAPI staining. Bars, 2 µm. (G) Box and whisker plot showing centromere numbers revealed using Volocity software. (n) Number of cells quantified for each sample. P-value, one-way ANOVA test.
LBR expression is required to attach centromeric and pericentromeric but not LINE-1 elements to the neutrophil lamina
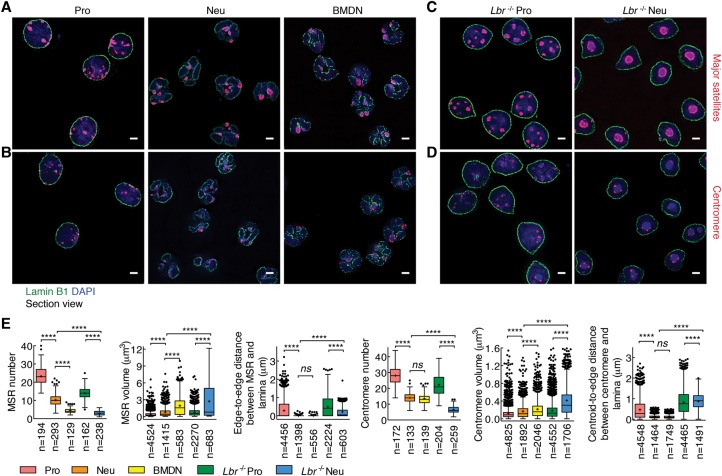
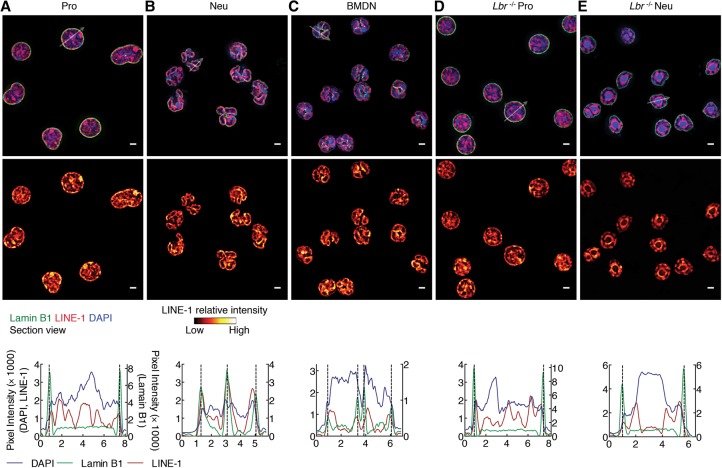
To determine how centromeric and pericentromeric heterochromatin is positioned in progenitors and neutrophils, we used immuno-3D-FISH. In progenitors, centromeres and major satellite repeats were localized at both the lamina and the nuclear interior, whereas, in neutrophils, they localized primarily at the lamina (Fig. 5A,B,E). To determine how heterochromatic repeat elements were tethered at the neutrophil lamina, we generated LBR-deficient progenitors using CRISPR–cas9-mediated deletion (Supplemental Fig. S6A–C; Cong et al. 2013). Lbr−/− progenitors were differentiated and examined for localization of pericentromeric and centromeric repeat elements (Fig. 5C–E). We found that centromeric regions in undifferentiated Lbr−/− progenitors were numerous and primarily organized as small spherical compact bodies (Fig. 5C–E). In contrast, centromeres in Lbr−/− neutrophils were fewer in number and localized as large clusters that were irregular in shape and located away from the lamina (Fig. 5C–E). Despite the striking repositioning of centromeric and pericentromeric DNA away from the lamina, we detected a rim of heterochromatin, reflected by DAPI staining, that remained associated with the lamina in differentiated Lbr−/− neutrophils. To explore the possibility that LINEs were retained at the lamina of LBR-deficient neutrophils, we performed 3D-FISH using a truncated LINE-1 element as probe. We found that, in progenitors, LINE-1 elements were dispersed throughout the nucleus but that, in differentiated neutrophils, they repositioned to the lamina (Fig. 6A–C). Notably and distinct from what was observed for other heterochromatic repeat elements, a fraction of LINE-1 elements remained associated with the nuclear lamina, whereas another fraction wrapped around chromocenters in LBR-deficient neutrophils (Fig. 6E). Thus, LBR expression is required to anchor centromeric and pericentromeric but not LINE-1 elements to the neutrophil nuclear lamina.
Figure 5.
Pericentormeric and centromeric heterochromatin repositions to the lamina in differentiating neutrophils. (A) Pericentromeric heterochromatin in progenitors and neutrophils as well as BMDNs was visualized using immuno-3D-FISH. The lamina was stained using an antibody directed against Lamin B1. Pericentromeric DNA was identified using fluorescently labeled major satellite repeats (MSRs) as a probe. Representative image sections are presented as digitally magnified images. Original magnification, 100×. (Red) FISH foci; (green) Lamin B1; (blue) DAPI staining. Bars, 2 µm. (B) Centromeres in progenitors and neutrophils as well as BMDNs were visualized as described in A but using a centromeric repeat element as a probe. (C) MSR elements in Lbr−/− progenitors and Lbr−/− neutrophils were visualized using immuno-3D-FISH as described in A. (D) Centromeres in Lbr−/− progenitors and Lbr−/− neutrophils were visualized using immuno-3D-FISH as described in A. (E) Box and whisker plot showing the quantification of the number, volume, and distance to the lamina of MSRs and centromeres for progenitors, neutrophils, BMDNs, Lbr−/− progenitors, and Lbr−/− neutrophils using Volocity software. The distance of the MSR to the lamina that separates the edge of the MSR from the edge of the lamina was measured. The distance of the centromere to the lamina was measured using the centroid of centromere dots and the edge of the lamina. (n) Number of cells or FISH foci quantified for each sample. P-value, one-way ANOVA test for numbers, Kruskal-Wallis test for volume or distance. (****) P < 0.0001; (ns) not significant.
Figure 6.
LINE-1 elements reposition during neutrophil differentiation. (A–E) The repositioning of LINE-1 elements to the nuclear lamina during neutrophil differentiation is independent of LBR expression. (Top and middle panels) Immuno-FISH using an antibody directed against Lamin B1 and a LINE-1 probe for progenitors (A), neutrophils (B), BMDNs (C), Lbr−/− progenitors (D), and Lbr−/− neutrophils (E). Representative image sections are presented as digitally magnified images. Original magnification, 100×. (Top) (Red) LINE-1; (green) Lamin B1; (blue) DAPI staining. (Middle) (Red hot) The intensity of FISH signal. Bars, 2 µm. White arrows indicate the diameter across the nucleus. (Bottom) Radial distribution of the fluorescence intensity signal for Lamin B1 (green line, right Y-axis), LINE-1 (red line, left Y-axis), and DAPI (blue line, left Y-axis).
Repositioning of rDNA repeats in neutrophils
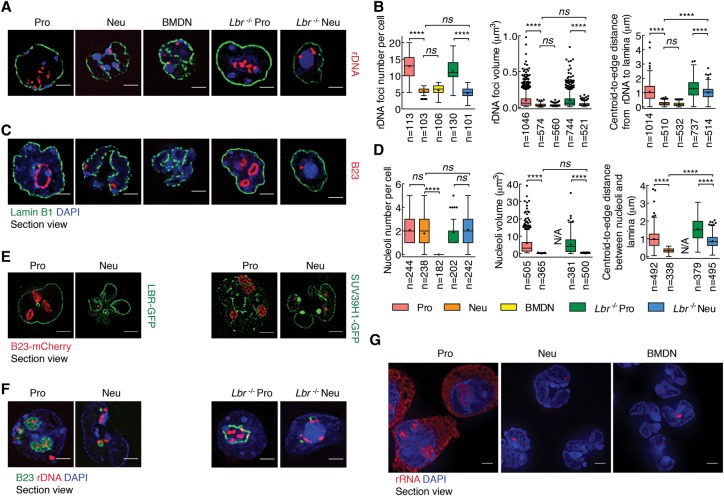
In addition to centromeres, pericentromeres, and LINE-1 elements, rDNA constitutes yet another type of repeat element. To examine whether rDNA also repositions during neutrophil differentiation, we used 3D-FISH. While clusters of rDNA in progenitors were readily detectable in the nuclear interior from progenitors, rDNA foci were far fewer in numbers and predominantly localized near the neutrophil nuclear lamina (Fig. 7A,B). To further characterize the nucleolus in differentiated neutrophils, we examined progenitors and neutrophils for B23, a nucleolar protein that marks nucleoli. We found that, during neutrophil differentiation, nucleoli condensed in size and repositioned to the nuclear lamina in an LBR-dependent manner (Fig. 7C,D). To directly visualize the repositioning and disassembly of nucleoli in live cells, we labeled nucleoli with B23-mCherry, heterochromatin with SUV39H1-GFP, and the nuclear lamina with LBR-GFP. Briefly, progenitor cells were transduced with retroviruses expressing these fluorescent proteins and imaged in live progenitors and differentiated neutrophils. This analysis revealed that, consistent with the 3D-FISH analysis, the nucleolus condensed into small nuclear bodies that localized near the neutrophil lamina (Fig. 7E). To examine how the folding pattern of rDNA relates to the nucleolar structure, we performed immuno-3D-FISH (Fig. 7E). We found that, in progenitors, B23 was wrapped around arrays of rDNA but that this distinct structure disassembled in differentiated neutrophils (Fig. 7F). Since previous studies demonstrated that Lamin B1 expression maintains the functional plasticity of nucleoli, we restored expression of Lamin B1 in neutrophils (Martin et al. 2009). We found that forced Lamin B1 expression was not sufficient to restore a nucleolar structure in differentiated neutrophils (Supplemental Fig. S7). Finally, to determine whether and how a change in nucleolar structure and nuclear location relates to rRNA abundance, we performed RNA-FISH. While ribosomal transcript abundance was high in progenitors, rRNA expression was barely detectable in differentiated neutrophils as well as in neutrophils isolated directly from bone marrow (Fig. 7G). Collectively, these data indicate that, in differentiating neutrophils, nucleoli reorganize and reposition to the nuclear lamina, a process that is closely associated with a halt in rRNA synthesis.
Figure 7.
Spatial distribution of rDNA and nucleoli in differentiating neutrophils. (A) rDNA repositions during neutrophil differentiation. Immuno-FISH using an antibody directed against Lamin B1 and a fluorescently labeled rDNA probe in progenitors, neutrophils, BMDNs, Lbr−/− progenitors, and Lbr−/− neutrophils. Representative image sections are presented as digitally magnified images. Original magnification, 100×. (Red) rDNA; (green) Lamin B1; (blue) DAPI staining. Bars, 2 µm. (B) Box and whisker plot showing the quantification of the numbers, volumes, and distances to the lamina of rDNA foci using Volocity software. The distance of rDNA foci to the lamina was measured between the centroid of dots and the edge of the lamina. (n) Number of cells or fluorescently labeled foci quantified for each sample. P-value, one-way ANOVA test for numbers, Kruskal-Wallis test for volumes or distances. (****) P < 0.0001; (ns) not significant. (C) Nucleoli reorganize during neutrophil differentiation. Immunofluorescence staining using an antibody directed against Lamin B1 and Nucleophosmin (B23) in progenitors, neutrophils, BMDNs, Lbr−/− progenitors, and Lbr−/− neutrophils. Representative image sections are presented as digitally magnified images. Original magnification, 100×. (Red) B23; (green) Lamin B1; (blue) DAPI staining. Bars, 2 µm. (D) Box and whisker plot showing the quantification of the numbers, volumes, and distances to the lamina of nucleoli using Volocity software. The distance of nucleoli to the lamina was measured between the centroid of foci and the edge of the lamina. (n) Number of cells or nucleoli quantified for each sample. P-value, one-way ANOVA test for number, Kruskal-Wallis test for volume or distance. (****) P < 0.0001; (ns) not significant. (E) Live-cell imaging in progenitors and neutrophils. To visualize changes in nucleolar structure in live cells, nucleoli were marked by B23-mCherry, and the nuclear lamina was labeled using an LBR-GFP fusion protein; SUV39H1-GFP was generated to visualize the heterochromatin. ECOMG cells were transduced with vectors expressing LBR-GFP or SUV39H1-GFP and B23-mCherry. The transduced cells were differentiated into neutrophils. Representative image section shows the relative position of nucleoli to lamina and major satellites. Original magnification, 100×. (Red) B23; (green) LBR-GFP or SUV39H1-GFP; (blue) DAPI staining. Bars, 2 µm. Note that, in progenitors, nucleoli were readily detectable using B23-mCherry as large distinct regions located in the interior of the nucleus. Marked variations in size and shape, occasionally with very large and irregular shapes, were observed. Usually, no more than three nucleoli per cell were detected. (F) Immuno-FISH using an antibody directed against B23 and a fluorescently labeled rDNA probe in progenitors, neutrophils, Lbr−/− progenitors, and Lbr−/− neutrophils. Representative image section shows that rDNA is bridging the mininucleolar structure to major satellites. Original magnification, 100×. (Red) rDNA; (green) B23; (blue) DAPI staining. Bars, 2 µm. (G) RNA-FISH using fluorescently labeled rDNA as a probe in progenitors, neutrophils, and BMDNs. Representative image sections are presented as digitally magnified images. Original magnification, 100×. (Red) rRNA; (blue) DAPI staining. Bars, 2 µm. Note that the decrease in the nucleolar size in neutrophils is associated with a marked decline in nucleolar as well as cytoplasmic rRNA. The neutrophil mininucleolus body still maintains trace amounts of rRNA.
Discussion
Among the wide spectrum of cells that comprise the immune system, polymorphonuclear cells, including neutrophils, eosinophils, and basophils, are unique in that their nuclei are organized into multiple lobes. Their special nuclear structure has raised the question of how their genome architecture differs from that of mononuclear cells. We used a previously developed in vitro model system of neutrophil differentiation and demonstrated that multilobular cells generated by this approach closely resemble neutrophils (in terms of transcription signatures and genome architecture) isolated from the murine bone marrow, permitting a detailed study of how segmentation of nuclei into multiple lobes affects genome topology and how such changes related to neutrophil physiology. We found that the transition from a mononuclear to a multilobular nucleus was not associated with large-scale changes in the number and sizes of TADs. However, neutrophil differentiation was accompanied by a spectrum of alterations in genome topology that included genomic interactions across vast genomic distances involving heterochromatic regions, switching of genomic regions from the A to the B compartments, and large-scale repositioning of repeat elements. Genomic regions that switch compartments and contract during developmental progression have been described previously. Prominent among these are loci encoding for regulators that control B cell fate, including EBF1 and FOXO1 (Lin et al. 2012). Likewise, antigen receptor loci switch nuclear location and undergo locus contraction in developing B cells (Jhunjhunwala et al. 2008; Lin et al. 2012). Supercontraction in neutrophils, however, is distinct, since it involves transcriptionally silent regions and performs a structural rather than a regulatory role. Supercontraction involves enrichment for genomic contacts involving genomic regions located in the B compartment or regions that switch from the A to the B compartment. The most striking epigenetic feature associated with supercontraction was the deposition of H3K27me3, marking pairs of interacting genomic regions that switched from the A to the B compartment. However, the repositioning from the A to the B compartment was not associated with a domain-wise redistribution of H3K27me3. Hence, it seems unlikely that H3K27me3 is the only player that promotes supercontraction. Rather, we propose that, as suggested by the simulation, the remodeling of the genome into a toroid nuclear structure is imposed by physical forces that are facilitated by the deposition of H3K27me3, ultimately leading to large-scale chromosome contraction.
The changes in nuclear topology during neutrophil differentiation also involve the repositioning of genomic repeats, including centromeres, pericentromeres, and LINE-1 elements. LINE-1 elements are scattered across the genome and enriched in the B compartment and reposition at a global scale during neutrophil differentiation. A role for a subset of LINE-1 elements in structuring the genome has been proposed previously for X-chromosome inactivation and chromosome contraction (Chow et al. 2010; Corbel et al. 2013; Hall et al. 2014). Along the same line, we suggest that, during neutrophil differentiation, LINE-1 elements promote chromosome contraction by sequestering large portions of the neutrophil genome to the lamina.
LBR expression and the generation of multilobular nuclei
Finally, as observed previously and confirmed here, the generation of multilobular nuclei requires LBR expression in neutrophils (Gaines et al. 2008; Verhagen et al. 2012). Additinally, we demonstrate here that LBR expression is essential to sequester pericentromeric heterochromain to the lamina but is not essential for heterochromatic clustering in differentiating neutrophils. The mechanism by which LBR promotes a multilobular nuclear shape remains to be determined but can now be addressed experimentally using conditionally induced LBR-deficient ECOMG cells. Previous studies and the observations described here demonstrate that LBR expression in neutrophils is essential for the generation of multilobular nuclear structures and to sequester pericentromeric heterochromatin to the nuclear lamina. We note that neutrophils display substantial differences in nuclear morphology across species. The majority of neutrophils isolated from invertebrates as well as several reptiles (snakes and turtles), named heterophils, is mononuclear in shape. It will be of interest to determine whether differences in LBR expression between heterophil and vertebrate multilobular cells underpin the distinctive features associated with neutrophil genomes across the animal kingdom.
Repositioning of rDNA during neutrophil differentiation
During neutrophil differentiation, nucleoli mediate some of the most striking changes in nuclear architecture. We found that, during neutrophil differentiation, rDNA was sequestered at the lamina in a heterochromatic environment and was associated with a virtual absence of rRNA expression. The decline in rRNA abundance may have implications for neutrophil physiology. It is well established that at least a subset of neutrophils is short-lived cells with a half-life in peripheral blood that is less than several hours. Although still to be proven, it may very well be that the life span of neutrophils is controlled at least in part by rRNA abundance. We note that, likewise, other terminally differentiated cells, including erythrocytes, platelets, keratinocytes, and lens fiber cells, are associated with low ribosome numbers, and it will be of interest to determine whether the repositioning of rDNA is also associated with decreased protein synthesis and life span in these cell types.
Conclusion
In sum, we propose that the large-scale changes in nuclear topology established during neutrophil differentiation are orchestrated by both physical and molecular mechanisms that involve changes in geometric confinement imposed by a multilobular nuclear structure as well as enrichment for intrachromosomal interactions across vast genomic distances and the repositioning of repeat elements from the nuclear interior to the lamina.
Materials and methods
Cell culture and plasmids
Cells were maintained in a 37°C humidified incubator in the presence of 5% CO2. HEK293T cells were cultured in DMEM (Invitrogen, 11995-073) supplemented with 10% fetal bovine serum and penicillin–streptomycin–glutamine (Invitrogen, 10378-016). GM-CSF-dependent ECOMG cells were grown in RPMI 1640 (Invitrogen, 11875-093) with 10% FBS and penicillin–streptomycin–glutamine by the addition of 1:100 conditioned medium (∼10 ng/mL GM-CSF) isolated from a B16 melanoma cell line stably transfected with a murine GM-CSF construct. β-Estradiol (Sigma, E2758), where applicable, was added to the medium at a final concentration of 1 µM from a 10,000× stock in 100% ethanol. Differentiation of ECOMG cells into granulocytes was induced by withdrawal of β-estradiol as described previously (Sykes et al. 2003). CD11b+Ly6G+ was used as a marker to validate granulocytic differentiation at day 5 (data not shown).
In order to ectopically express Lamin B1-GFP, LBR-GFP, SUV39H1-GFP, and B23-mCherry, their coding sequences were amplified from ECOMG cell cDNA. Via BamHI and EcoRI sites, the PCR products were ligated into the multiple cloning site of a pMys-IRES-TAC vector. Retroviral supernatant was obtained through HEK293T transfection using calcium phosphate precipitation and vectors encoding for Lamin B1-GFP, LBR-GFP, SUV39H1-GFP, and B23-mCherry in conjunction with the packaging plasmid pCL-Eco. Two days after infection, samples underwent enrichment for infected cells through the use of anti-human CD25 magnetic beads (Miltenyi Biotech) and an AutoMACS separator (Miltenyi Biotech). Plasmids and maps will be provided on request.
Population-based structure modeling
We generated models of 3D genome structures using our recently introduced population-based modeling approach, which constructs a population of 3D genome structures derived from and fully consistent with the Hi-C data (Kalhor et al. 2012; Tjong et al. 2016). By embedding the genome structural model in 3D space and applying additional spatial constraints (e.g., all chromosomes must lie within the nuclear volume, and no two chromosome domains can overlap), it is possible to deconvolute the ensemble-based Hi-C data into a set of plausible structural states. The data preprocessing, normalization, and modeling were described previously (Kalhor et al. 2012; Tjong et al. 2016). Chromatin is represented at the level of macrodomains at ∼3-Mb resolution, which was inferred from the Hi-C data. Analysis of Hi-C and other methods are detailed in the Supplemental Material.
Accession number
The Hi-C, GRO-seq, RNA-seq, and chromatin immunoprecipitation combined with high-throughput sequencing and methylated DNA immunprecipitation sequencing data were deposited at the NCBI Gene Expression Omnibus (GEO) database and are accessible through GSE93127.
Supplementary Material
Acknowledgments
Sequencing was performed at the Institute for Genomics Medicine Genomics Center at the University of California at San Diego, supported by grant P30CA023100. Imaging was performed at the microscopy core of the School of Medicine at the University of California at San Diego, supported by grants CA23100 and NS047101. Y.Z. was supported by a Cancer Research Institute post-doctoral fellowship. F.A. was supported by the National Science Foundation (CAREER 1150287) and the Arnold and Mabel Beckman Foundation. C.M. was supported by the National Insitutes of Health (DK107977, AI00880, AI09599, and AI102853).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.293910.116.
References
- Apostolou E, Thanos D. 2008. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-γ gene expression. Cell 134: 85–96. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas system. Science 339: 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel C, Diabangouaya P, Gendrel A-V, Chow JC, Heard E. 2013. Unusual chromatin status and organization of the inactive X chromosome in murine trophoblast giant cells. Development 140: 861–872. [DOI] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. 2013. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13: 602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PHL, Festuccia N, Nora EP, Welling M, et al. 2013. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature 501: 227–231. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, Berliner N. 2008. Mouse neutrophils lacking lamin B-receptor expression exhibit aberrant development and lack critical functional responses. Exp Hematol 36: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. 2014. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 156: 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJM, Grosveld FG, Knoch TA, Murre C. 2008. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell 133: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen C-A, Schmitt AD, Espinoza CA, Ren B. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmárová M, Smirnov E, Mašata M, Koberna K, Ligasová A, Popov A, Raška I. 2007. Positioning of NORs and NOR-bearing chromosomes in relation to nucleoli. J Struct Biol 160: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, Kubes P. 2013. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 13: 159–175. [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Chen S, Maya-Mendoza A, Lovric J, Sims PFG, Jackson DA. 2009. Lamin B1 maintains the functional plasticity of nucleoli. J Cell Sci 122: 1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Meshorer E. 2010. The silence of the LADs: dynamic genome-lamina interactions during ESC differentiation. Cell Stem Cell 6: 495–497. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-hupkes D, Kind J, Beaudry J, Pagie L, Kellis M, Reinders M, Wessels L, Van Steensel B. 2013. Constitutive nuclear lamina—genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Längst G. 2011. Genome organization in and around the nucleolus. Trends Genet 27: 149–156. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. [DOI] [PubMed] [Google Scholar]
- Sykes DB, Scheele J, Pasillas M, Kamps MP. 2003. Transcriptional profiling during the early differentiation of granulocyte and monocyte progenitors controlled by conditional versions of the E2a-Pbx1 oncoprotein. Leuk Lymphoma 44: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, Zhou Y, Li H, Zhou XJ, Le Gros MA, et al. 2016. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci 113: E1663–E1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Sandoval A, Gasser SM. 2013. Mechanisms of heterochromatin subnuclear localization. Trends Biochem Sci 38: 356–363. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, de Graaf CA, Baldwin TM, Goradia A, Collinge JE, Kile BT, Metcalf D, Starr R, Hilton DJ. 2012. Reduced lymphocyte longevity and homeostatic proliferation in lamin B receptor-deficient mice results in profound and progressive lymphopenia. J Immunol 188: 122–134. [DOI] [PubMed] [Google Scholar]
- Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. 2015. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci 112: 4672–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJL, Ritchie W, Ebner OA, Selbach M, Wong JWH, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.