Abstract
Studying the embryogenesis of diverse insect species is crucial to understanding insect evolution. Here we review current advances in understanding the development of two emerging model organisms: the wasp Nasonia vitripennis, and the beetle Tribolium castaneum in comparison to the well-studied fruit fly Drosophila melanogaster. Although Nasonia represents the most basally branching order of holometabolous insects, it employs a derived long germband mode of embryogenesis, more like that of Drosophila, while Tribolium undergoes an intermediate germband mode of embryogenesis, which is more similar to the ancestral mechanism. Comparing the embryonic development and genetic regulation of early patterning events in these three insects has given invaluable insights into insect evolution. The similar mode of embryogenesis of Drosophila and Nasonia is reflected in their reliance on maternal morphogenetic gradients. However, they employ different genes as maternal factors, reflecting the evolutionary distance separating them. Tribolium on the other hand relies heavily on self-regulatory mechanisms other than maternal cues, reflecting its sequential nature of segmentation and the need for reiterated patterning.
Introduction
For the past three decades, the fruit fly Drosophila melanogaster has served as the premiere model for understanding the molecular basis of developmental patterning mechanisms. In particular, the array of powerful genetic tools available in this organism has allowed the thorough dissection of the events leading to the establishment of cell fates in the Drosophila embryo, and the regulatory networks underlying this process stand as the mostly thoroughly characterized among multi-cellular organisms. For those interested in the evolution of developmental mechanisms, the highly detailed description of Drosophila embryonic patterning is an appealing starting point for understanding the origin of complex gene regulatory networks, and how these networks can change in the course of evolution.
One of the appealing features of the Drosophila embryo as a developmental model system is that, until gastrulation, it can be thought of as a two dimensional Cartesian coordinate system, with the two orthogonal axes (anterior-posterior (AP) and dorsal-ventral (DV)) being patterned independently, but more or less simultaneously. The ability to rapidly establish cell fates representing the entire future organism at this early stage is enabled by maternally localized mRNAs which provide high levels of positional information that can be interpreted and refined by downstream target genes.
The mode of embryogenesis employed by Drosophila is termed long germ embryogenesis, which for the purposes of this review is defined by the establishment of all segmental fates at the blastoderm stage prior to gastrulation 1, 2. This is a derived mode of embryogenesis that is only found in scattered species among the Holometabola. Insects of more basally branching lineages use a conceptually different mode of embryogenesis, where only the most anterior segments are specified before gastrulation, while the more posterior segments are generated and patterned progressively from a posterior region called the growth zone. This is termed short, or intermediate germ embryogenesis, depending on the number of segments established at the blastoderm stage 1.
Different approaches have been combined to understand how major transitions in embryonic patterning strategies have occurred in the course of insect evolution. One approach has generated a highly detailed understanding of embryogenesis in an insect employing the ancestral, short/intermediate germ mode of embryogenesis. By comparing this to the derived mode found in Drosophila, many insights into how regulatory networks were rewired to pattern a long germ embryo have been revealed. In a complementary strategy, the embryo of an insect that independently derived a long germ mode of embryogenesis has been examined and then compared to Drosophila as well as the short/intermediate germ species. This approach has uncovered common strategies underlying the transition from short to long germ embryogenesis.
Two holometabolous insects, the intermediate germ beetle Tribolium castaneum and the long germ wasp Nasonia vitripennis, have emerged as powerful comparative model organisms with which these topics can be addressed. In this review, we will describe the contributions of these “non-model” models to understanding the evolution of development, and provide perspectives on the bright future such lines of research hold.
A tale of two models: Nasonia and Tribolium
To enable the characterization of developmental regulatory networks at high enough resolution to make evolutionary comparisons meaningful, tools for identifying, cloning and manipulating genes and their regulatory elements must be available and robust in each comparative model system. The availability of such tools is summarized in Table 1 and described in detail below. Access to the fully sequenced genomes of both Nasonia and Tribolium 3, 4 facilitates the identification of Drosophila gene orthologs, including descriptions of complete gene families, and identification of potential regulatory elements. The main method used to manipulate gene function in both of these organisms is parental RNA interference (pRNAi) 5, 6. In this technique, in vitro synthesized dsRNA of the gene of interest is injected into the abdominal hemocoel of female pupae or adults, where it is taken up by the cells, and leads to the degradation of the target mRNA of interest. If developing oocytes take up the dsRNA the effect perdures in the fertilized eggs (unless the gene knock-down causes adult lethality or sterility), and thus the developmental consequences of reducing the expression of any gene of interest can be examined in embryos or early larvae. This technique is so efficient in Tribolium that there is currently an effort underway to knock down and characterize every gene in the genome (Gregor Bucher, personal communication, see http://ibeetle.uni-goettingen.de/).
Table 1.
Techniques and tools available in Drosophila, Tribolium, and Nasonia.
| Drosophila | Tribolium | Nasonia | |
|---|---|---|---|
| in situ hybridization, immunohistochemistry | Y | Y | Y |
| Parental RNAi | N | Y | Y |
| Embryonic RNAi | Y | Y | Y |
| Germline Transformation | Y | Y | y |
| Enhancer Trapping | Y | Y | n |
| Misexpression constructs | Y | Y | n |
| Amenable to Forward genetics | Y | Y | Y |
| Ability to screen in F1 generation | N | N | Y |
| Sequenced Genome | Y | Y | Y |
| Detailed Transcriptome Data | Y | Y | Y |
| Availibility of Custom Transcriptome Microarray | Y | y | Y |
Y = indicates availability is well established, N = technique is not, and is unlikely to become available, y = indicates the availability is preliminary and/or unpublished, n = technique not available, but could conceivably be developed in the near future.
Another critical technique for characterizing developmental networks is the ability to engineer the genome to alter endogenous sequences or express exogenous ones through the use of germ-line transformation. These techniques are well established in Tribolium 7, and have been used to perform a genome-wide insertional mutagenesis and enhancer-trapping screen that has yielded many interesting and useful transgenic lines 8. Furthermore, techniques for tissue and time specific misexpression have been developed by implementing the GAL4-UAS system in Tribolium 9. The methodology in Nasonia is not as advanced, but some promising success has been gained in establishing germ-line transformation in Nasonia (Claude Desplan, personal communication).
Nasonia has an additional attractive feature, which is its haplo-diploid mode of sex determination. Unfertilized eggs develop as haploid males, allowing mutations affecting embryonic patterning to be rapidly identified by screening the progeny of the F1 generation 10. A screen employing this method has already provided valuable insight into the mechanisms patterning the Nasonia embryo 11.
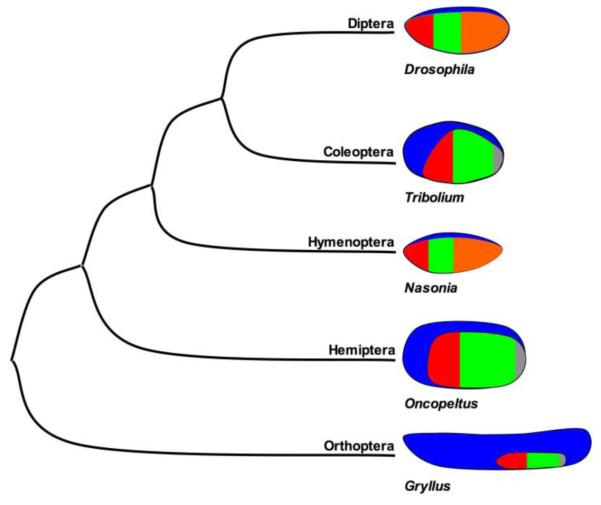
Nasonia and Tribolium represent the two most speciose orders of insects (the Hymenoptera and Coleoptera, respectively). Together with Drosophila, these three comparative models represent most of the diversity of holometabolous insects. Recent molecular phylogenies place the Hymenoptera at the base of the holometabolous radiation (Figure 1), with the Coleoptera and Diptera diverging a relatively short time later. These phylogenetic relationships allow hypotheses concerning the direction of evolutionary change in embryonic patterning as well as trends associated with the different modes of embryogenesis to be tested. For example, characters shared by Tribolium and Drosophila, but not Nasonia, may represent novelties that arose after the divergence of the Hymenoptera from the rest of the Holometabola (alternatively, the character could be ancestral but lost in Nasonia), while characters shared by Nasonia and Drosophila, but not Tribolium, might represent common strategies for dealing with the long germ mode of embryogenesis. Any such hypothesis can then be tested by sampling other insect lineages, including more basally branching hemimetabolous species, for the character of interest. The highly detailed understanding of embryonic patterning now obtainable in Nasonia, Tribolium and Drosophila can be used to generate hypotheses that will both broaden and deepen our understanding of how developmental strategies can evolve. Indeed, such studies have already contributed much to understanding how establishment and patterning of the AP and DV axes of the embryo, as well as the establishment of the germline, have evolved within holometabolous insects.
Figure 1. Phylogenetic relationships of selected insect models, and schematic representations of their early embryonic fate maps.
Blue = extraembryonic tissue, Red = segments of the head, Green = thoracic segments, Orange = abdominal segments, and Gray = growth zone primordium.
AP polarity and patterning
The AP axis of Drosophila is patterned by a hierarchy of regulatory gene functions that progressively increases the resolution and precision of positional information. This process starts with maternal coordinate genes that provide broad, graded information emanating from both ends of the embryo. These gradients are interpreted by the gap genes (the first zygotically activated segmentation genes). The expression domains of these genes subdivide the embryo in large, partially overlapping yet precisely defined blocks, and also provide short-range graded information. The pair-rule genes interpret these short-range gradients and are expressed in a double segment periodicity to give the first hints of the metameric body plan of the fly. Pair-rule gene expression patterns are then interpreted by the segment polarity genes, which function to establish the borders between, and fates within each segment. Contributions to understanding the evolution of patterning mechanisms at each level of this hierarchy have been made by studying Tribolium and Nasonia.
Maternal inputs into AP patterning
In Drosophila, three maternally derived patterning systems provide spatial information that progressively subdivides the embryo along the anterior-posterior axis: the anterior, posterior, and terminal systems. Two of these systems, the anterior and posterior, rely on localized mRNAs responding to the internal polarity of the oocyte (bicoid and nanos, respectively), while the terminal system relies on modifications to the eggshell during oogenesis to provide its patterning function (see Figure 2). Work in Nasonia and Tribolium has provided insights into how strategies for providing patterning information may be correlated with the mode of embryogenesis employed.
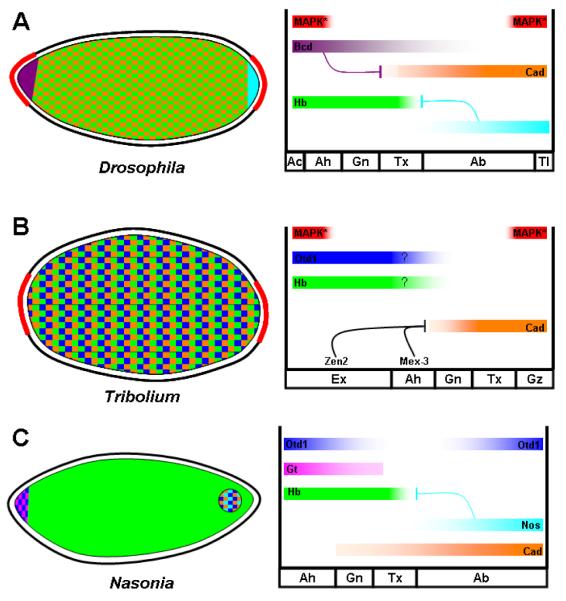
Figure 2. Maternal provision of AP patterning information in Drosophila, Tribolium, and Nasonia.
Left: Schematic representation of Drosophila (A), Tribolium (B), and Nasonia (C) embryos showing distribution of maternally provided mRNAs. Right: representation of resulting protein gradients. Red curves in left panel on A and B represent the distribution of tsl expression, and correspond with the red bars at right representing the resulting activation of MAP kinase in the embryo. Question marks for Otd and Hb in panel B indicate their now doubted role in providing positional information to the early embryo. Ah=anterior head, Gn=gnathal, Tx=thorax, Ab=abdomen, Tl=telson, Gz=growth zone.
The Anterior system
Establishment of cell fates along AP axis of the Drosophila embryo requires a symmetry-breaking event leading to AP polarity and subsequent patterning along the AP axis. In Drosophila both steps are provided by bcd: anterior localization of bcd mRNA provides a symmetry breaking event and its translation into a protein gradient along the AP axis provides positional information to the gap genes for patterning.
The Bcd gradient of Drosophila acts as a morphogen with both permissive and instructive functions that provides positional information along most of the length of the embryo. The permissive function of the Bcd gradient arises from the ability of this protein to bind and repress translation of caudal mRNA into protein where it would interfere with anterior development 12, 13. The instructive function arises from the differential sensitivity of AP target genes to Bcd concentration, such that the expression of genes that respond to low levels of Bcd extends further towards the posterior, while the expression of those that are less sensitive is restricted to the anterior 14, 15. Although essential to AP patterning in Drosophila, bcd is an evolutionary novelty of higher Diptera 16. It is derived from the Hox cluster, but unlike most Hox proteins, possesses a Lysine (K) at position 50 of its homeodomain, giving it a distinct DNA binding affinity in comparison to typical Hox homeodomains which possess a glutamine (Q) at this position 17.
Localized mRNAs, in combination with permissive and instructive protein functions, are also critical in patterning the anterior half of the Nasonia embryo 18. Nv-orthodenticle1 (Nv-otd1) mRNA is tightly localized to the anterior (and posterior, see below) pole of the embryo 19, where a gradient of translated protein performs the instructive function (Figure 2). pRNAi against Nv-otd1 significantly shifts the anterior fatemap, resulting in the loss of all head segments, including some thoracic segments in the most extreme cases. Nv-otd1 does not act alone, but rather cooperates with Nv-hunchback (Nv-hb) in activating target genes. This is similar to the cooperation of Bcd with Hb in activating anterior target genes in Drosophila, and indicates that cooperation between K50 homeodomain proteins and Hb is an ancient strategy for anterior patterning in holometabolous insects 19 (Figure 2).
No clear analog of the permissive function of Bcd has been identified for Nv-Otd1, which is not surprising, given the posterior localization of Nv-cad mRNA (see below). However, an anteriorly localized, permissive patterning factor with novel function has been found in the wasp. Nv-giant (gt) mRNA is localized to the anterior pole of the oocyte (Figure 2). After egg activation it disperses, forming a cap of Nv-gt mRNA at the anterior pole at the early blastoderm stage. Once translated into protein, this early source of Nv-Gt is critical to prevent the expansion of Nv-Kr and concomitant loss of Nv-hb expression in the anterior half of the embryo 20. When Nv-gt and Nv-Kr are knocked down simultaneously, most of the anterior patterning defects seen in Nv-gt single pRNAi, are rescued, demonstrating that the primary function of maternal Nv-gt is to prevent the expression of Nv-Kr in the anterior half of the embryo, either by repressing it directly or indirectly through activation of Nv-hb.
The situation in Tribolium appears to differ significantly from what has been observed in Nasonia and Drosophila. This is likely due, at least in part, to differences in the blastodermal fate map of short/intermediate type embryos. Instead of many head segments that require high levels of patterning information to set their borders, most of the anterior end of the Tribolium embryo is devoted to extraembryonic cell fates, fates that are restricted to the dorsal side of the embryo in Drosophila and Nasonia. Thus, it is not clear whether Tribolium needs the high levels of patterning information provided by localized mRNAs as in Nasonia and Drosophila.
However, positional information is needed in Tribolium at least to specify the border between embryonic and extraembryonic tissues. It was proposed that Tribolium orthodenticle1 (Tc-otd1), which is provided maternally and appeared to result in an anterior to posterior protein gradient due to translational repression 21 (Figure 2), along with hunchback act as bicoid-like morphogens in Tribolium. Recently, doubt has been cast on the putative role of Tc-otd1 in providing positional information along the AP axis 22, 23. However, anteriorly localized mRNAs have been found in Tribolium (Bucher, Farzana et al. 2005), and seem to have effects on early fatemap (S. J. Brown, manuscript under preparation).
The Posterior System
In Drosophila, posterior patterning relies on two major maternal factors: caudal (cad) and nanos (nos) (Figure 2). Together these two genes, acting in distinct ways, ensure proper specification of abdominal fates: cad encodes a transcription factor that activates posterior target genes, while nos encodes a translational factor that acts permissively for abdominal specification. Both of these genes are conserved in Tribolium and Nasonia, and analyses of their functions have revealed both conserved and divergent strategies for patterning the posterior segments in insects.
1. caudal function and regulation
In the early Drosophila embryo, ubiquitous maternal cad mRNA, translationally repressed by the Bicoid gradient, produces a reciprocal Cad gradient 12, 13. The posterior patterning function of Cad provides activating input to pair-rule genes to form posterior stripes but only weakly activates posterior gap genes. It does not appear to act as a morphogen, and it seems that the repressive function of Bcd on cad mRNA is mainly to prevent ectopic Cad from disrupting head patterning 24.
Nv-cad mRNA is localized to the posterior pole of the oocyte, and upon egg laying it diffuses, forming a posterior to anterior gradient. This posterior localization of maternal Nv-cad obviates the need for translational repression at the anterior. Nv-cad has a more fundamental role in patterning the wasp embryo, as the strongest mutant and pRNAi phenotypes delete all abdominal and thoracic segments, while the strongest Drosophila cad mutants have variable defects restricted to the abdomen. 25 In addition, Nv-cad functions higher in the patterning hierarchy than does its fly counterpart, as it is absolutely required for the proper expression of posterior gap genes 25.
caudal also plays a critical role in patterning most of the segmented embryo in Tribolium. At the embryonic stage just prior to the onset of gastrulation (termed differentiated blastoderm stage), there are three developmentally distinct regions: the extraembryonic membrane primordia, the anterior head (pregnathal), and segmented germband (that part of the embryo patterned by pair-rule genes, including gnathal and trunk segments). Tc-cad is responsible for specifying this last region (Figure 2); its knock down leads to the complete loss of posterior head and all trunk segments. The phenotype, even more severe than that of Nv-cad RNAi, is also observed in most non-Drosophila insects 26, 27, indicating a larger ancestral role for Caudal in general body patterning. An important downstream target of Tc-Cad is likely to be Tc-even-skipped (Tc-eve), a gene which is critical for the maintenance of the progressive segmentation process of Tribolium (see below the section on pair-rule genes).
As is the case for its Drosophila and Nasonia orthologs, Tc-Cad function must be repressed in the anterior regions of the embryo. However, Tribolium does not contain a bcd ortholog and Tc-cad mRNA is not posteriorly localized as it is in Nasonia. The beetle uses another strategy: two zygotically activated genes, Tc-mex3, expressed in the anterior head primordia, and Tc-zen2, expressed in the serosal anlage (the anteriormost extraembryonic region), are required to repress Tc-Cad protein production in these regions of the differentiated blastoderm embryo. Interestingly, the C. elegans ortholog of Tc-mex3 is also involved in translational regulation of the worm cad ortholog, indicating that this mechanism for regulating cad expression along the AP axis was in place in at least the common ancestor of the Ecdysozoa 28, 29.
2. nanos regulation and function
In the fly, localization of nanos (nos) mRNA to the posterior pole of the embryo is critical for proper patterning of the abdominal segments. Only localized nos mRNA (which represents just 4% of total nos mRNA present in the embryo) is translated 30, giving rise to a gradient of Nos protein in the posterior half of the embryo that represses translation of maternal hb mRNA, which would, if translated, interfere with posterior gap gene regulation, and result in the disruption of posterior abdominal segments 31. Localization of nos mRNA occurs late in oogenesis and depends on properly assembled germ plasm 32. Thus, osk, vas, and tudor mutants have abdominal defects identical to that of nos.
So far no expression or functional data have been presented for nos in Tribolium, so it is not clear what, if any, role this factor plays in patterning the beetle embryo. Intriugingly, Nanos Response Elements (NREs), the sequence motifs through which Nanos exerts its repressive influence, have been detected in the 3′ UTRs of both Tc-otd1 and Tc-hb, and both of these genes show evidence of translational repression at the posterior pole 21, 33. However, the functional significance of these NREs requires further validation.
In Nasonia, Nv-otd1 and Nv-hb mRNAs both also possess NREs in their 3′ UTRs, and both also show posterior translational repression 19, 34. Nasonia nanos (Nv-nos) mRNA is localized to the posterior pole, in the oosome, a structure that is the wasp equivalent of polar granules in the fruitfly. When Nv-nos is knocked down via pRNAi, the domain of maternal Nv-Hb protein expression expands to the posterior pole, revealing that the role of Nv-nos in repressing Nv-hb translation is conserved with its Drosophila ortholog. Nv-nos pRNAi also leads to the misregulation of posterior gap genes, and the eventual loss of posterior segments in the larva 35.
Regulation of the localization and function of Nv-nos shows some conservation with, as well as significant divergence from, the strategies employed in Drosophila. Like their Drosophila counterparts, Nv-vas and Nv-osk are required for the proper translation of Nv-nos mRNA at the posterior. Also as in the fly, this function of germ plasm genes seems to be in opposition to the activity of smaug, which represses translation of unlocalized nos mRNA 36. A major difference between the wasp and fly is that while nos localization in Drosophila is completely abolished in germ plasm deficient mutants, Nv-nos shows significant posterior localization after Nv-vasa or Nv-osk pRNAi, despite the absence of the oosome. Another major difference in the function of Nv-nos is that its action is delayed due to maternal provision of Nv-Hb protein, which is absent in Drosophila, but which is also found in the holometabolous locust Schistocerca 37. The lack of maternal Hb protein may have been important to the evolution of the rapid early syncytial cleavage divisions prior to blastoderm formation in Drosophila, which are considerably more rapid than in Nasonia.
The terminal system
In addition to the Bcd gradient, the terminal system is involved in patterning the termini of Drosophila embryo 38. torso, encoding a receptor tyrosine kinase 39, and its ligand trunk are provided maternally and ubiquitously to the embryo 40. Spatial specificity for the system is first provided by the expression of torso-like (tsl) in the anterior and posterior follicle cells during oogenesis 41, and later by incorporation of Tsl protein into the vitelline membrane 42. tsl is required for production of the active form of trunk, by a still unknown mechanism, which defines the regions of terminal system activity at both embryonic poles 43, 44. Targets of Torso signaling include the terminal gap genes tailless (tll) and huckebein (hkb) 45, which are initially expressed in caps covering both anterior and posterior poles. Both tll and hkb are important in patterning the unsegmented posterior region, the posterior abdominal segments, and the head 46 (however, it was shown that in case of increased bcd expression, the terminal system is dispensable for head development 47).
In Tribolium, Tc-tsl is activated in the follicle cells located at both ends of the oocyte. Both Tc-torso and Tc-tsl knockdown result in the same defects at both embryo poles (no serosa is formed anteriorly, and no post-blastodermal segments emanate from posterior), indicating that Torso signaling plays similar roles in beetles and flies 48. However, Tc-tll is activated only at the posterior terminus in the early blastoderm. Only later, in the germ rudiment, is it activated in the head 49 (note that the Tribolium head anlage is located mid-embryo and not at the anterior pole, as in Drosophila). This indicates that the terminal system must signal through a gene other than Tc-tll. Torso signaling at the posterior pole of the Tribolium embryo is required for both Wnt signaling and zygotic activation of Tc-cad (with the possibility that Torso signaling is regulating zygotic Tc-cad through Wnt signaling), and thereby affecting post-blastodermal segmentation 48. However, it is not known whether this posterior action of Torso signaling is carried out through Tc-tll. Anterior Torso signaling is required for Tc-zen1 activation, and hence its importance in serosa formation.
An ortholog of tailless is expressed at the posterior pole and in the head anlage at the anterior during the blastoderm stages of Nasonia 50. However, these expression domains are not regulated by the torso signaling cascade, as no ortholog of trunk could be found in the Nasonia genome, and activated MAP kinase is not observed at the poles of the embryo (JAL, personal observation). Rather, both the anterior and posterior domains of Nv-tll depend on Nv-otd1 for their activation 50. The terminal system is also lacking in the honey bee Apis mellifera 51, indicating that either the terminal system originated after the divergence of Coleoptera and Diptera from the Hymenoptera, or that the terminal system was lost specifically in the Hymenoptera. Sampling from more basally branching hemimetabolous lineages will be required to resolve this question.
Gap genes
In the fly, the gap genes function at the second level of the embryonic AP patterning hierarchy, and are generally the first zygotic genes to interpret the maternal gradients. The transcription factors encoded by the gap genes form short-range gradients that are generally mutually repressive, leading to precise overlap of opposed gradients. Loss of gap gene function leads to the misregulation of downstream pair-rule genes and the loss of one or more contiguous blocks of segment primordia. In addition to their role in regulating pair-rule genes, gap genes also regulate the domains of Hox gene expression 52, 53.
Although limited, the data on the function of gap genes in Nasonia indicate that their functions and interactions are generally similar to their fly counterparts. In the first functional study of Nasonia embryogenesis, a number of potential gap gene mutations were identified by their phenotypic similarity to fly mutants, 10. One of these has been identified as a mutation in the Nasonia hunchback (Nv-hb) ortholog 34. This mutation is of particular interest, since it affects only zygotic expression of the protein, yet results in a loss of segments (all of the head and most thorax) that is greater than the combined loss of maternal and zygotic contributions of hb in the fly. pRNAi against Nv-Kr and Nv-gt also showed that these genes possess canonical gap gene functions, and act at similar positions along the AP axis as their fly homologs 20, 35.
The mutually repressive interactions between gap genes also appear to be conserved in Nasonia, at least those between Nv-Kr and Nv-hb. In cases where the domain of either of these genes is altered by pRNAi or mutant backgrounds, the other expands or contracts to maintain a sharp border between the posterior edge of the Nv-hb domain and the anterior edge of the Nv-Kr domain20.
Tribolium orthologs of hunchback, giant and Krüppel are required for proper segmentation in the beetle 22, 54, 55. Surprisingly, Tc-knirps is important for head development but has only a minor effect on trunk segmentation 56. In addition, a newly identified gap gene, milles-pattes (mlpt) 57 is unusual in that it encodes a series of small peptides, rather than a transcription factor.
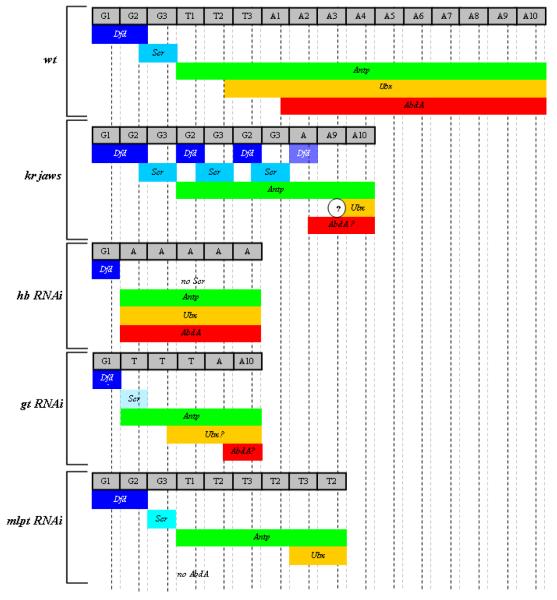
In wildtype beetle embryos, the expression of Tc-hb, Tc-gt, Tc-Kr and Tc-mlpt changes dynamically between their initiation in the blastoderm stage and their resolution into domains covering one or more segments in the head and germband. Tc-hb, provided maternally, is found ubiquitously and then clears from the posterior end. This early expression domain resolves to the serosa, while a new posterior blastoderm domain arises and resolves to the anterior head through first thoracic segment. Finally, a domain arises in abdominal segments A7 through the posterior end of the germband as those segments are added 33. Tc-gt is also expressed in a broad blastoderm domain that resolves to cover the anterior head as well as the mandibular and maxillary segments. An additional Tc-gt domain arises at the posterior pole of the embryo in the growth zone, the region from which the abdominal segments arise. As the germband elongates this domain broadens and splits into two stripes, one in third thoracic segment (T3) the other in the second abdominal segment (A2) 54. Tc-Kr expression initiates in the posterior blastoderm, eventually resolving to cover the thorax 55. Tc-mlpt expression initates in the blastoderm and resolves into domains covering the anterior head and mandibular segment, T2 – A4, and a small domain in A7 57.
Although on the surface, gap gene functions in Tribolium appear similar to those of their Drosophila and Nasonia counterparts, in the end they are quite different. As in Nasonia and Drosophila, Tribolium gap genes are expressed in domains that overlap several segment primordia, show some evidence of cross-regulation (see Figure 3 for a comparison between expression domains and regulatory relationships between gap genes in both Drosophila and Tribolium), and regulate pair-rule and homeotic genes to some extent. Their loss of function through RNAi or mutation leads to large patterning gaps in the resulting cuticles. However, careful analysis of the segmentation process during embryogenesis revealed that these apparent deletion phenotypes are actually combinations of germband truncation (halted elongation) and homeotic transformations 22, 23, 55, 58 (see Figure 4 for a comparison between gap genes phenotypes of Drosophila and Tribolium and Figure 5 for Hox genes expression patterns for Tribolium in wildtype as well as gap genes mutants or RNAi embryos). Moreover, only very few pair-rule stripes appear to be specifically regulated by particular gap genes (S. J. Brown and M. Klingler, unpublished data), and most pair-rule pattern changes can be explained by halted elongation.
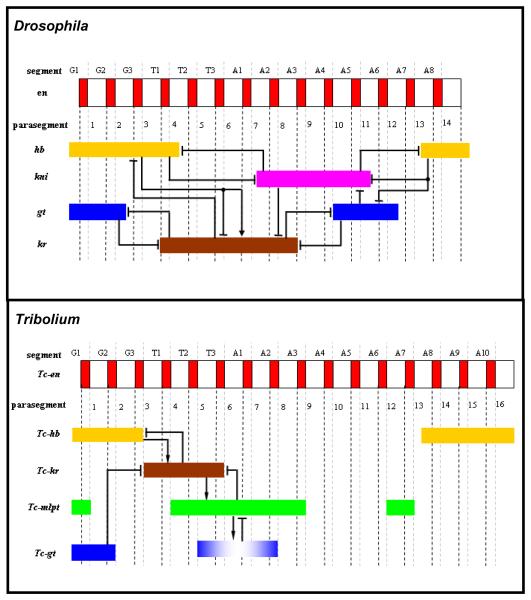
Figure 3. Comparison of gap gene expression patterns and their regulatory relationships in Drosophila and Tribolium.
Top panel: Late blastodermal expression patterns of gap genes (non-terminal, non-head) in Drosophila and their regulatory relationships. Bottom panel: post-blastodermal expression patterns of gap genes (non-terminal, non-head) in Tribolium after they emanate from the growth zone and before they fade, and a parsimonious interpretation of their regulatory relationships. An arrow represents positive regulation and a blunt line represents negative regulation. G= gnathal segment, T= thoracic segment, A= abdominal segment.
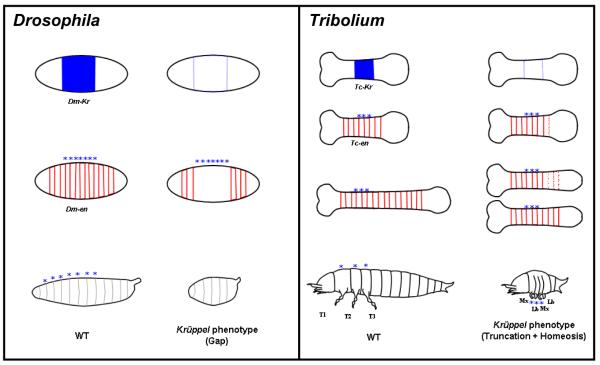
Figure 4. Comparison of gap genes phenotypes in Drosophila and Tribolium.
Expression of Krüppel (in blue) and engrailed (in red) and the resulting larval cuticles (bottom of each panel) in WT (left of each panels) and Krüppel mutants (right of each panel). Left panel (Drosophila): The strongest Krüppel phenotype in Drosophila results in loss of seven segments (marked by stars) within the expression domain of the gene Krüppel, which is a typical “gap” phenotype. Right panel (Tribolium): In the Krüppel mutant (jaws), the segments within the Tc-Krüppel expression domain (marked by stars) form properly, and only a few more posterior segments develop, which are disorganized at first but later regulated to form intact segments. In addition to this “truncation” phenotype, a homeotic transformation is observed in the larval cuticle, in which the three thoracic segments and the first abdominal segment are transformed to gnathal identity (maxillary-labial-maxillary-labial). This phenotype is best described as “truncation plus homeosis”, rather than “gap”. The yet unexplained “truncation after expression domain” of Tribolium gap genes in contrast to “gap within expression domain” of Drosophila is proved valuable here to show clearly the homeotic effect of gap genes phenotypes in Tribolium, whereas it is precluded by the loss of segments in Drosophila (except for some hypomorphic mutants).
Figure 5. Hox gene expression in wt and gap gene RNAi and mutant embryos.
The identity of segments that are formed is described in the gray boxes at the top of each panel. Diluted color in an expression domain indicates reduced expression. A question mark inside an expression domain indicates that this expression domain is not reported in literature. A question mark in a circle at the border of an expression domain indicates that the exact location of this border is not reported precisely in literature. G= gnathal segments, T= thoracic segments, A= abdominal segments.
Pair-rule genes
Seven stripes of primary pair-rule gene expression (even-skipped, runt and hairy) are positioned in the Drosophila blastoderm by input from specific combinations of maternal and gap genes 59. These primary pair-rule genes interact to regulate one another and the secondary pair-rule genes (ftz, odd, odd-paired, paired and slp) 60-62. A complex network of interactions between primary and secondary pair-rule genes has been described 63 that positions stripes of the segment polarity genes wingless (wg) and engrailed (en) to define the borders between segments. Although the use of wg and en to define segmental boundaries is highly conserved, the expression patterns and functions of pair-rule gene orthologs differ greatly among insects and arthropods 64.
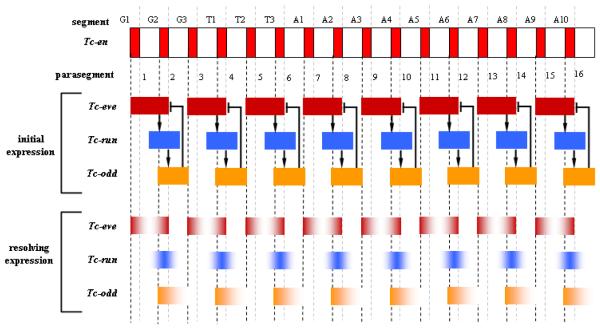
Similar to their Drosophila counterparts, Tribolium pair-rule genes are expressed in patterns of double segment periodicity 65-68. However, in contrast to the simultaneous formation of pair-rule gene stripes in the Drosophila blastoderm, the Tribolium orthologs of Drosophila pair-rule genes are expressed in stripes sequentially from anterior to posterior, starting in the blastoderm and continuing during germband elongation. RNAi studies revealed that certain genes, Tc-ftz, Tc-hairy and Tc-odd-paired, are not required for proper segmentation, despite their striped expression patterns 67, 69. Classical pair-rule phenotypes are observed in Tc-prd and Tc-slp RNAi or mutant embryos 68, 70, but depletion of Tc-even-skipped (Tc-eve), Tc-runt (Tc-run) or Tc-odd-skipped (Tc-odd) leads to loss of all (in the case of Tc-eve) or almost all (in the case of Tc-run and Tc-odd) gnathal and trunk segments 67. Epistatis analysis revealed a negative feedback loop in which Tc-eve activates Tc-run, which activates Tc-odd, which in turn represses Tc-eve 67 (Figure 6) (however, this analysis was done using pupal injection; so while this verifies the circuit in the blastoderm phase, it is yet to be verified in the germband phase using embryonic injection). This negative feedback loop could serve as a self-regulatory segmentation mechanism, employing no or little cues from upstream maternal or gap genes. This is in contrast to the fly case, where pair-rule genes are under strict control of upstream maternal and gap genes.
Figure 6. Initial and resolving expressions of primary pair-rule genes in Tribolium.
A negative feedback loop between the three primary pair-rule genes Tc-eve, Tc-run, and Tc-odd are responsible for their initial double-segmental periodic expression. Later, they resolve into segmental periodic expression through a yet to be identified genetic mechanism. This later segmental primary pair-rule expression regulates downstream genes to ultimately position segment polarity genes. An arrow represents positive regulation and a blunt line represents negative regulation. G= gnathal segment, T= thoracic segment, A= abdominal segment. It is wothnoting that these regulatory relationships were verified for the anteriormost segments. Further expriments are needed to verify it for the following segments (using embryonic RNAi).
Components of this negative feedback loop in Tribolium are categorized as primary pair-rule genes, since the depletion of any one of the three affects the expression of the other two, as well as the expression of Tc-prd and Tc-slp, which, hence, are categorized as secondary pair-rule genes. Although the functions of prd and slp are conserved in that they regulate segment polarity genes in both beetles and flies, the evolutionary flexibility of these modules is reflected in the register of slp function; it is necessary for odd-numbered en stripes in flies, but for even-numbered stripes in beetles 68. In Drosophila, stripes of en expression are precisely positioned by eve in odd-numbered segments and by ftz in even-numbered segments through complex interactions with several secondary pair-rule genes 63. As noted above, the beetle orthologs of many of these genes are not required for proper segmentation. However, Tc-eve may regulate segment polarity genes in every segment since it can, in combination with Tc-prd or Tc-runt, activate Tc-en 67, 68. It is not known if other, as yet unidentified, genes function as secondary pair-rule genes in Tribolium. However, the different interactions between pair-rule genes that have evolved in beetles and flies still produce the same outcome; juxtaposed stripes of wg and en that define the segmental boundaries. This underscores the evolutionary modularity of the segmentation gene network in insects.
So far the only pair-rule ortholog described from Nasonia is Nv-paired (prd). Unlike its fly ortholog, Nv-prd expression initiates progressively from anterior to posterior, and shows no classical pair-rule expression, rather it appears in segmental stripes that alternate between strong and weak expression 71. However, it is likely that genes with pair-rule functions exist in Nasonia, as several mutants with classic pair-rule phenotypes have been identified 10. Preliminary evidence also indicates that the pattern of pair-rule expression is similar to that of the honey bee, where pair-rule stripes are initiated progressively from anterior to posterior 72. In addition, the most posterior abdominal segments may not be patterned by the pair-rule paradigm, as only six pair-rule stripes are observed at the blastoderm stage; the later segmental stripes appear to progressively split from the most posterior pair-rule stripe 72 (Claude Desplan, personal communication). This may indicate that elements of the progressive growth and patterning that typify short germ embryogenesis are maintained in many “long-germ” embryos.
Segment polarity genes
Once segment polarity genes have been activated by pair-rule genes in Drosophila, their expression is maintained through a gene circuit including the transcription factor En, and two signaling pathways, Wnt and Hedgehog 73. En and the components of the signaling pathways are highly conserved and have been identified in the both Nasonia and Tribolium. In Tribolium, RNAi analysis indicates both the Wnt and Hedgehog pathways play conserved roles in the segment polarity gene module 74-76.
In Tribolium, loss of canonical Wnt signaling via depletion of Tc-arrow (which encodes a co-receptor for canonical Wnt pathway) or Tc-porcupine (which encodes a Wnt ligand processing component), leads to germband truncation in the growth zone 77. Detailed RNAi analysis revealed that multiple Wnts, specifically Wnt1 and WntD/8, are involved in posterior growth and patterning 78. Additional studies will be required to determine the molecular mechanisms sensitive to Wnt signaling in the growth zone, but the need for a posterior source of Wnt signaling is conserved in other short germ insects 79, 80, other arthropods 81 and indeed most other metazoans 82. Interestingly, although wg is expressed at the posterior pole of Drosophila eggs, it is not required for germband elongation or posterior patterning; mutations in genes encoding Wnt pathway components lead to segment polarity defects 83.
Homeotic genes
Homeotic genes were the basis of the first “evo-devo” studies in many organisms 84, and like most insects examined to date, Drosophila, Nasonia and Tribolium contain a single complement of homeotic genes, including labial, maxillopedia, Deformed, Sex combs reduced, Antennapedia, Ubx, abd-A and Abd-B. As in other organisms, they direct regional development along the AP axis. Their expression domains are similar in beetles and flies, and loss of function phenotypes affect similar body regions. In homeotic mutants, one developmental fate is replaced by another; thus, legs may appear where mouth parts should be 85, depending on the resulting combination of homeotic genes expressed there (one exception to this is Tc-labial in Tribolium, which upon RNAi leads to deletion of the respective segment: intercalary 86). In addition to specifying a specific regional identity along the AP axis, homeotic genes also repress anterior development; complete loss of homeotic gene function in a particular segment or region results in antennae developing on that segment or the segments in that region 87, and in the most extreme case, antennae develop on every segment 88.
DV patterning
A key player in patterning the DV axis in Drosophila is the NFκB transcription factor Dorsal. Dorsal protein is expressed in all cells of the early Drosophila embryo. However, its nuclear uptake occurs in the form of a gradient with maximal levels on the ventral side of the embryo 89. The differential expression of Dorsal target genes define various cell fates (broadly, the mesoderm, ectoderm and extraembryonic fates) along the DV axis (Figure 7) 90, 91. Cross-regulatory relationships between these target genes further refine DV gene expression domains, in a manner reminiscent of Bicoid and gap gene action in AP axis patterning. For example, while twist (twi) and snail (sna) respond to high levels of nuclear Dorsal ventrally to specify the mesoderm, they also repress expression of many genes that are activated by intermediate and low levels of nuclear Dorsal (e.g., single-minded (sim), rhomboid (rho) and short gastrulation (sog)) 92, 93.
Figure 7. DV fate map and expression domains of genes involved in DV patterning in Drosophila.
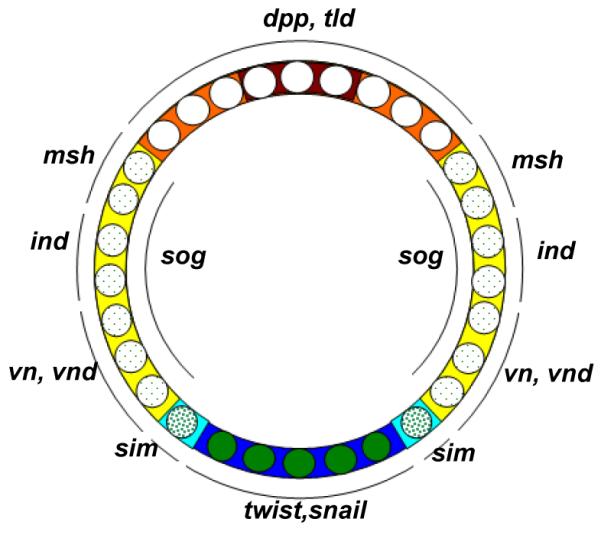
Different concentrations of nuclear Dorsal drive different sets of downstream genes and ultimately (with the help BMP and EGF signaling) define DV fates. Blue: mesoderm, cyan: mesectoderm, yellow: neurogenic ectoderm, orange: dorsal ectoderm, red: amnioserosa, opaque green circles: high nuclear Dorsal concentration, heavily dotted green circles: intermediate nuclear Dorsal concentration, lightly dotted green circles: low nuclear Dorsal concentration, white nuclei: no nuclear Dorsal.
Dorsal target genes of particular interest are those that encode components of other signaling pathways. For example, the BMP ligand dpp and the protease tolloid (tld) are both strongly repressed by Dorsal, and are thus restricted to the dorsal side of the embryo 94, 95, while sog (an extracellular BMP binding protein), activated by low Dorsal levels 96(and repressed by snail), is expressed in broad lateral stripes. The arrangement of these factors inhibits BMP signaling in the ectoderm, and produces peak levels of BMP signaling at the dorsal midline that are critical to specifying the amnioserosa. This latter phenomenon results from the binding of Sog to Dpp, which prevents signaling in ventral regions and enhances diffusion. The following cleavage of Sog by Tld on the dorsal side of the embryo releases transported Dpp from Sog mediated inhibition 97. In addition, a local gradient of the rho protease in mirror image ventrolateral stripes, regulated by intermediate levels of Dorsal activation and Snail repression, provides EFG signaling activity 98, 99. The nuclear Dorsal gradient, in combination with BMP and EGF signaling further refines patterning of neuroectoderm into three domains defined by the columnar genes: vnd, ind, and msh 100.
There are similarities as well as differences in DV patterning between Tribolium and Drosophila. As in Drosophila, the ventral-most expression of Tc-twist and Tc-snail defines the mesoderm, and adjacent stripes of Tc-sim demarcate the mesectoderm 101, 102. As in Drosophila, a ventral domain of Tc-sog (somewhat wider than the ventrolateral domain of sog in Drosophila) is needed to transport Tc-Dpp to the dorsal side of the embryo where it is released from Tc-Sog by Tc-Tld 103, 104. Active EGF signaling in the ventral neuroectoderm in Tribolium presumably regulates columnar gene expression 105. Major blastoderm fate map differences between Tribolium and Drosophila are visible in the regulation of dpp. While dpp is restricted to the dorsal side of the embryo of Drosophila, Tc-dpp is initially expressed in a weak AP gradient, and then in an oblique stripe along the AP axis that defines the boundary between embryonic and extraembryonic cell fates. Despite these differences, the gradient of activated BMP signaling (as detected by phosphorylated-MAD) centered on the dorsal midline, is conceptually similar in the fly and beetle 103.
While nuclearized Dorsal is essential to activate most DV patterning genes in Drosophila, it plays a less prominent role in Tribolium. The Tc-Dorsal gradient fades at the end of the blastoderm stage (see below), completely disappearing prior to gastrulation 106. This transient Tc-Dorsal expression initiates (but does not maintain) the most ventral fates. Tc-twist expression is activated at the ventral midline of the blastoderm and continues to be expressed at the posterior end of the germband after the Tc-Dorsal gradient disappears. As a result, abdominal segments emanating from the caudal end of the embyro continue to be correctly patterned with DV fates. Tc-twist expression is never initiated after Tc-dorsal depletion by RNAi, producing embryos that are symmetrical along the DV axis 107. However, additional sources of patterning capacity exist, as demonstrated by the self-regulatory features of BMP signaling. The initial AP-asymmetric, DV-symmetric expression of Tc-dpp during early blastoderm phase is transformed into both AP and DV asymmetric BMP signalling activity in the late blastoderm by Tc-Sog mediated transportation towards the dorsal side103. In Tc-dorsal RNAi, probably due to the depletion of Tc-sog, the late blastodermal BMP signalling loses its DV asymmetry and becomes purly AP asymmetric. As a result, during germband elongation, additional DV symmetric stripes of BMP form along the AP axis in cells distant from the initial peak of BMP activity, indicating that a BMP-mediated self-regulatory mechanism is at work 107 (Figure 8). Interestingly, this same phenotype is not observed in Tc-sog RNAi embryos where DV fate specification is mostly normal, but with an additional peak of BMP activity replacing Tc-sog expression at the ventral midline 103. This indicates that another factor downstream of Tc-Dorsal is capable of orienting BMP activity, and emphasizes the self-regulatory properties of BMP signaling in Tribolium.
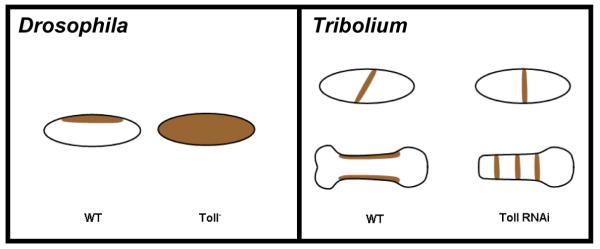
Figure 8. Comparison between Toll phenotype in Drosophila and Tribolium.
Left panel (Drosophila): dpp (brown) is expressed dorsally in WT (left) and along the entire DV axis in a Toll mutant (right). Right panel (Tribolium): Early blastodermal Tc-dpp is expressed as an oblique stripe in WT (left upper embryo), and loses this obliqueness in Toll RNAi (right upper embryo). In Tribolium, the initial orientation of balstodermal Tc-dpp dictates the orientation of later reiterated germband expression of Tc-dpp. Consequently, Tc-dpp reiterates only twice in the DV direction in WT probably due to space constraints (left lower embryo), and many times in the AP direction in Toll RNAi (right lower embryo).
The source of nuclear Dorsal gradient
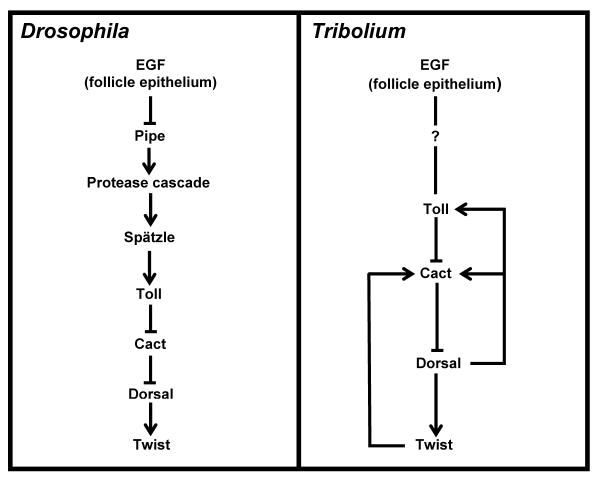
In contrast to the Bcd protein gradient that forms by diffusion of newly translated protein from a localized mRNA source at the anterior pole, there is no localized point-like source from which to initiate a Dl nuclear uptake gradient 108. Formation of the gradient can be traced back to the dorsal localization of gurken (grk) mRNA in the late oocyte, which results in dorsal activation of EGF signaling in the adjacent follicle epithelium. EGF signaling represses pipe, restricting its expression to the ventral side of the follicle epithelium 109. Pipe, a sulfotransferase, modifies vitelline membrane proteins 110 to define the region in which a protease cascade is initiated that ultimately activates the Toll ligand Spätzle (Spz). In the embryo, binding of the Spz to the Toll receptor leads to the phosphorylation and degradation of the Dorsal inhibitor Cactus (Cact), permitting translocation of Dorsal to the nucleus 111. A still unanswered question is how the homogenous, sharp-bordered pipe domain is translated into the graded distribution of Toll activation through Spz. Some clues come from mutants in which Pipe expression is wider than in wildtype, leading to two ventrolateral peaks of Dorsal nuclear uptake instead of one ventral peak 112. This suggests that behind the Spätzle gradient is a self-regulatory mechanism with a domain of action that is restricted ventrally by the maternal factor Pipe. Extending the Pipe domain makes it possible to adopt more than one peak of nuclear Dorsal 113.
In contrast to the stable gradient of nuclear Dorsal in Drosophila, Tribolium nuclear Dorsal is highly dynamic. Starting as a very broad domain of nuclear localization along the DV axis, Tc-Dorsal clears from the dorsal side in a gradually shrinking gradient with its peak at the ventral side, and finally vanishes at the differentiated blastoderm stage 106. While the formation of the Drosophila Dorsal is to a great extent set by maternal factors, the dynamics of Tc-Dorsal gradient is generated mainly through its positive regulation of zygotic Tc-Toll as well as its own negative regulator, Tc-cact 107. The negative component of this self-regulatory system is stabilized by the inclusion of the autoactivation of Tc-Twist as a late positive regulator of Tc-cact transcription, which is required to keep Tc-Dorsal out of the nuclei during the germband extension stages.
The exact molecular interactions among Tc-Dorsal, Tc-Toll, and Tc-Cact that give rise to the dynamic nature of the Tc-Dorsal gradient are as yet unknown. The regulatory relationships between these factors indicate both negative and positive feedback loops, which in principle could generate self-regulated patterns by local activation and long-range inhibition in the mode proposed by Meinhardt and Gierer 114. The positive feedback loop between Tc-Toll and Tc-Dorsal satisfies the local activation requirement, while, in principle, Tc-Cact could satisfy the lateral inhibition requirement; however, it not yet know whether Tc-Cact has a long range effect, which is an important requirement of the model 107.
Self-regulatory mechanisms of the type found for Tc-Dorsal gradient formation are capable in principle of generating patterns by intensifying random early fluctuations without any maternal bias. However, in order to be useful in establishing axial polarity, such a system needs a mechanism to consistently orient itself perpendicula to the AP axis. This function is carried out by asymmetric activation of EGF signaling in the follicular epithelium overlying the oocyte nucleus during oogenesis. When EGF signaling is disrupted by pRNAi, the affects seen in the resulting embryos are highly variable, some show chaotically-positioned, duplicate peaks of nuclear Tc-Dorsal, while others produce Tc-Dorsal gradients oriented perpendicular to the DV axis 115. Yet to be discovered is the sequence of events connecting EGF signaling in the follicle epithelium of the Tribolium oocyte to the point at which the symmetry of early nuclear Tc-Dorsal expression is lost.
Nasonia is an attractive model in which to further explore the evolution of DV patterning mechanisms, for much the same reasons it is attractive for studying the AP axis: at gastrulation, DV genes homologs are expressed in very similar patterns in Nasonia and Drosophila 115. What is known so far is that, as in Tribolium and Drosophila, proper establishment of the DV axis depends on maternal EGF signaling 115. In an interesting parallel, mRNA for the Nasonia EGF ligand is localized to the dorsal cortex, much like grk is localized in Drosophila, which is in sharp contrast to the lack of localized factors in Tribolium. Thus, heavier reliance on localized mRNAs also extends to patterning the DV axis in long germ insects.
Head development
The embryonic head anlage of Drosophila is partitioned by means of segment-polarity genes into six segments (listed in a posterior to anterior order): the labium, maxillary and mandible (composing the gnathocephalon), and intercalary, antenna, and ocular (composing the procephalon). Molecular arguments for the existence of a labral segment have been found in Drosophila 116 but not in other insects 86, 117. Segmentation in the gnathocephalon is under the control of the segmentation hierarchy that defines the trunk segments, while procephalon segmentation is controlled by a different set of genes, the so-called “head gap-like genes” (orthodenticle (otd), buttonhead (btd), empty-spiracles (ems) and sloppy-paired) 118, 119. These genes are expressed in overlapping domains, and mutations in them result in segmental deletions similar to the effects of gap gene mutations in the trunk (hence the name “gap-like”). Therefore, it was thought that they regulate the patterning of segment-polarity genes in the procephalon in a fashion similar to that of the trunk gap genes (directly or with the help of second order regulators instead of pair rule genes 120). However, later studies indicate that head gap-like genes play only a permissive role for segment polarity genes expression, and do not regulate their metameric postions 121, leaving the problem of head segmentation unresolved 122.
The situation is similar in Tribolium, where the head gap-like genes (Tc-otd, Tc-btd, and Tc-ems) are not even extensively overlapping 123. Tc-otd is the only one of these genes to have a gap-like phenotype (antennal and intercalary segments are missing in late embryonic knockdown). Tc-ems knockdown results in a partial deletion of the antennal segment, while Tc-btd knockdown has no effect at all, ruling out possible gap-like functions in Tribolium 123. However, RNAi analysis of the knirps homolog in Tribolium produces a gap in the head pattern 56, which is in contrast to the normal early metameric patterning and late head defects of knirps mutants in Drosophila 124. Thus, Tc-knirps joins Tc-otd as a head gap-like gene in Tribolium 122.
In Drosophila, head gap-like genes receive input from three maternal systems: the anterior, terminal and DV systems 46. In Tribolium, the terminal and DV systems are likely to be responsible for the high-level partitioning of the embryo into extraembryonic, head and trunk, mediated by the cross-regulation of Tc-zen1, Tc-mex3, and Tc-cad (see AP section). So far, Tc-ems, Tc-knirps, Tc-collier and Tc-labial have been shown to regulate segment polarity expression in the head 125. Their exact mode of action and more genetic factors regulating their fine-tuning and ultimately positioning the metameric expression of segment-polarity genes are yet to be discovered.
Mapping embryonic head segments to their larval counterparts in Drosophila and insects in general was and still is subject to much debate 117. This is mainly because the larval head of the best-studied insect, Drosophila, undergoes a complex process of involution that introduces technical difficulties to mutant screens and expression assays. In addition, the derived nature of the Drosophila is not representative of insect head development in general. In Tribolium, head segments are initially collinear with the AP axis during embryonic development (the later bend and zipper morphogenetic movements, however, change the situation such that the eye becomes located posterior to the antenna – see below), making this beetle, with its available genetic tools, a good model system for studying typical insect head development 122. To support this line of inquiry, many larval morphological markers as well as many embryonic genetic markers have been characterized in the Tribolium head. Comparison of gene expression domains in Tribolium and Drosophila has helped reassess the assignment of morphological structures to individual segments 126. These markers have also been used to compare embryonic and larval phenotypes in a variety of mutants, providing new insight into the fate map of the insect head 127. Recently the “bend and zipper” model was formulated to describe the morphogenetic movements the insect head undergoes during development 127. In addition, evidence supporting the hypothesis of the non-segmental, appendage-bearing nature of labrum was presented 86. However, more technical advances in Tribolium are required to give unequivocal answers to the problem of Tribolium and insect head development in general.
In contrast to the previously mentioned processes, head patterning is not well understood in Drosophila. Hence, it will be the task of future Tribolium research to establish the paradigm in this species while Drosophila and Nasonia provide interesting cases of parallel evolutionary adaptations to long germ embryogenesis. Additionally, in Drosophila the head involution is an intriguing evolutionary novelty the evolution of which remains to be understood.
Germline specification
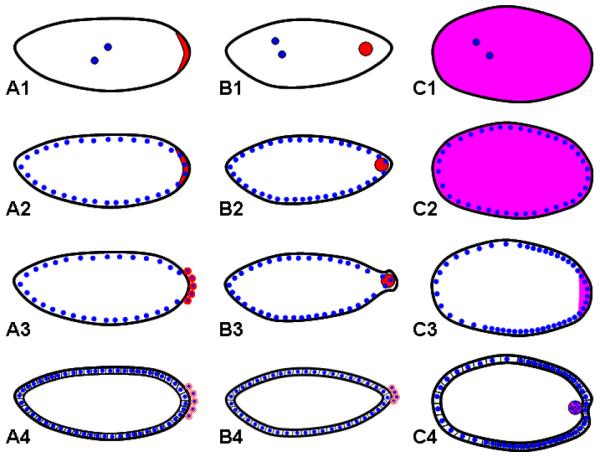
In Drosophila, the germline is specified by cytoplasmic inclusions called polar granules, which are produced specifically at the posterior pole of the oocyte and the embryo (Figure 10, A1-A2). When the syncytial nuclei arriving at the plasma membrane during blastoderm formation become associated with these structures, they take on the germ line fate and become pole cells (Figure 10, A3-A4). The gene oskar (osk) is both necessary and sufficient to induce the polar granules 128, and thus the activity of this gene must be tightly regulated.
Figure 10. Germ cell formation in Drosophila, Nasonia, and Tribolium.
Red=osk mRNA, magenta=Tc-vasa mRNA, blue=nuclei.
A1-C1: In the early cleavage stages of embryogenesis, osk mRNA is localized in the posterior pole plasm of Drosophila (A1) and oosome of Nasonia (B1), while Tc-vasa is ubiquitous in Tribolium (C1).
A2-C2: As the syncytial blastoderm forms, posterior nuclei interact with the pole plasm (A2), or the oosome (B2) in fly and wasp, respectively, while nuclei enter an apparently homogenous environment in Tribolium (C2).
A3-C3: Individual nuclei that enter the posterior pole plasm bud singly, forming the pole cells in Drosophila (A3). In contrast, the oosome along with multiple nuclei bud simultaneously from the posterior in Nasonia (B3). In the later blastoderm stages of Tribolium, Tc-vasa mRNA is cleared from the embryo, save for the most posterior pole.
A4-C4: Production of pole cells is completed, and osk mRNA begins to be degraded in the cellular blastoderm stage in both Drosophila (A4) and Nasonia (B4). Posterior cells that retained Tc-vasa expression delaminate from the blastoderm into the interior of the embryo (C4) and will become the primordial germ cells of the beetle.
Neither maternally synthesized polar granules, nor early specification of the primordial germ cells in the form of pole cells, are ancestral features of insect development. These features are missing from all described hemimetabolous species, as well as from some holoemetabolous taxa, such as Tribolium and Apis 4, 129. These species use zygotic induction mechanisms to specify germline fate at varying times later in embryonic development 130. On the other hand, Drosophila-like, maternal inheritance modes of germline determination are found in most hymenopterans (including Nasonia) and dipterans, and in many beetles 131. osk had, until recently, only been found in the genomes of Drosophilids and other Diptera, leading to the idea that osk was a novelty of the Diptera 132.
Recently, an osk ortholog was discovered in the genome of Nasonia. It is a component of the oosome (the functional equivalent of the posterior pole plasm in Drosophila), and is taken up into the pole cells as they form, until it is degraded during the cellular blastoderm stage in a pattern quite similar to Drosophila osk (Fig 10, B1-B4). pRNAi against Nv-osk leads to the loss of the oosome and pole cells, as well as posterior patterning defects 131.
The discovery of an oskar ortholog performing a conserved function in generating maternal germ plasm in Nasonia has shed light on the evolutionary history of germline determination among insects. First, the latest time for the origin of the oskar gene is pushed back to the last common ancestor of the Holmetabola, indicating that it was lost independently multiple times (at least in the lineages leading Apis, Tribolium and Bombyx). Second, the general conservation of the regulatory networks both upstream and downstream of Nv-osk in the formation of germ plasm and pole cells, indicates that there was a single origin of the maternal inheritance mode of germline determination that coincided with oskar, and that the strong correlation of losses of oskar with losses of maternal inheritance are not coincidental 131.
The fact that the lack of pole cells is likely secondarily derived in Tribolium makes understanding how the germline is established in this species very interesting. So far little data exist. Tc-Vasa protein is not maternally localized, unlike the Vasa orthologs in Drosophila and Nasonia 131. However, its initially ubiquitous mRNA (Figure 10, C1-C2) appears to be selectively degraded, so that it is only found at the posterior pole at the onset of gastrulation (Figure 10, C3), after which it marks the primordial germ cells (Figure 10, C4) 133. It will be interesting to see how this degradation is regulated and what factors are responsible for the induction of the rest of the germline program.
Morphogenesis
During the blastoderm stages when cell fates are being determined, the Drosophila embryo is a relatively simple two dimensional structure. Once cellularization has been completed and cell fates have been determined, numerous cell movements and rearrangements including gastrulation, dorsal closure, and germ band extension occur which transform the embryo into a dynamic, three-dimensional entity 134. Equivalent movements also occur in Tribolium and Nasonia, however the means by which they are carried out vary significantly due to the combination of the different embryonic architectures, and independent evolution of strategies to solve morphological problems in each species’ respective lineage.
Gastrulation
In Drosophila gastrulation begins with the formation of the ventral furrow, which forms more or less simultaneously along the entire AP axis of the embryo. In this process, cells on the ventral surface of the embryo expressing twist and snail contract at their apical sides, which leads to the internalization of the presumptive mesoderm, and formation of the ventral midline when the formerly lateral, single-minded (sim) expressing cells of the mesectoderm meet and fuse over the internalized mesoderm (Figure 11) 134.
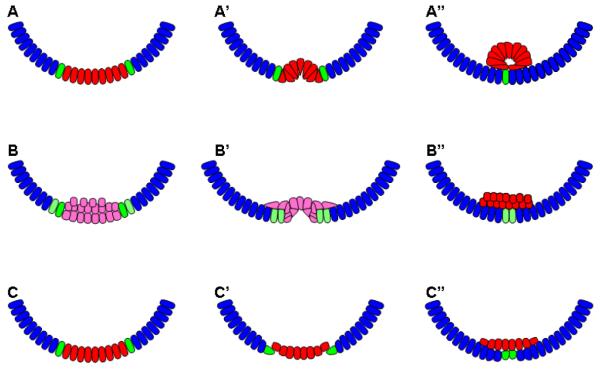
Figure 11. Mechanisms of gastrulation in Drosophila, Tribolium, and Nasonia.
A-A”: Gastrulation by furrow formation employed by Drosophila, and Tribolium in the anterior segments. B-B”: Gastrulation mode found in the segments deriving from the growth zone of Tribolium. A multilayered mass of cells loses its epithelial character and is internalized by the migration of the lateral ectodermal cells. C-C”: Hymenopteran mode of gastrulation, where the mesoderm forms a stiff plate, the ectoderm breaks contact with mesoderm and migrates ventrally until the free edges meet and fuse at the ventral midline. Blue= lateral ectoderm, Green=mesectoderm, Red= mesoderm precursors expressing twist, Pink= mesoderm precursors not expressing twist. Other cell fates have been omitted for clarity.
The mesoderm is also ventrally located in Tribolium, it internalizes after cellularization of the blastoderm, which, during early stages, resembles invagination of the ventral furrow in Drosophila 101. In the fly, gastrulation is among the first morphogenetic movements, and is completed before the major movements of germ band extension and dorsal closure have been initiated. In contrast, gastrulation in Tribolium occurs first in the context of germ rudiment condensation, and continues during convergent extension of the germ band. In addition, extraembryonic membranes are enveloping the embryo 135. Furthermore, both specification of new mesodermal cell populations and ventral closure leading to the formation of the ectodermal ventral midline occur progressively from anterior to posterior 101, contrasting sharply to the rapid, coordinated invagination along the AP axis in the fly. Finally, while a ventral furrow similar to that of Drosophila is observed at certain positions along the axis, the morphogenetic movements involved in internalizing the mesoderm and closing the ventral ectoderm vary along the axis during Tribolium gastrulation. In segments derived from the growth zone, the presumptive mesodermal cells, which unlike Drosophila and more anterior Tribolium segments, do not express twist, lose their epithelial character to form a mass of multilayered tissue, and appear to be internalized chaotically as the ectoderm closes over them 101 (Figure 11 B-B”). Interestingly, after internalization at least a subset of the presumptive mesoderm cells reinitiate Tc-twi expression (Figure 11 B”). In addition, the mesoderm remains relatively flat in the head region, with ectodermal cells crawling over this substrate to meet at the midline (similar to what is seen in hymenopteran gastrulation, see below, Figure 11 C-C”). It will be interesting to see if these different morphogenetic movements are the result of different cell signaling interactions among the different cell types involved, or whether they result simply from different forces impinging on the mesoderm at different AP axis positions.
The cellular and molecular underpinnings of gastrulation have been relatively unexplored in Nasonia. Observations on living embryos have shown, however, that the general features of Nasonia 136 are similar to the unique strategies that are used in many other Hymenoptera 137. Here, no ventral furrow forms, rather the prospective mesoderm remains as a stiff sheet of cells while the ectodermal cells at the border of mesoderm break their lateral contact with their mesodermal neighbors (Figure 11 C-C’). The resulting free edges move across the rigid mesoderm cells until they meet and fuse, forming the ventral midline (Figure C”). This process also occurs in an anterior to posterior progression, again in contrast to Drosophila 137(JAL personal observation). These observations indicate that the molecular and cellular basis of Nasonia gastrulation also differ from that of the fly.
Extraembryonic membranes
In Drosophila, the vast majority of cells present at the end of the blastoderm stage will give rise to the future tissues of the embryo. However there is a small population of cells restricted to the dorsal midline fated to become the amnioserosa. This single extraembryonic membrane in the fly is important for proper germband retraction 138 and dorsal closure of the embryo 139, and disappears in the course of the latter process. While the amnioserosa is critical for successful completion of embryogenesis, it is extremely reduced in comparison to the extraembryonic membranes found in most other insects 140.
Tribolium possesses a more typical insect complement of and investment in extraembryonic membranes 141. There are distinct serosal and amnion tissues, the former occupying a large anterior-dorsal cap, while the latter is a relatively restricted domain located at the anterior and dorsal margins of the embryonic anlage 29. At the onset of gastrulation, these membranes undergo radical rearrangements. The serosal cells flatten and expand, and eventually move to envelope the embryo proper, causing the latter to sink slightly into the yolk. The amnion remains closely associated with the embryo during these movements. After the completion of germband extension and retraction, it is thought that the amnion and serosa fuse by cell intercalation of the two tissues into one, which eventually weakens and ruptures, thus uncovering the embryo. This membrane contracts to the dorsal side of the embryo, and eventually disappears as the embryonic flanks expand and meet at the dorsal midline 29.
As in Drosophila, specification of extraembryonic membranes in Tribolium depends on transcription factors encoded by zerknuellt (zen) genes 29. There are two zen paralogs in Tribolium (Tc-zen1 and Tc-zen2); both of which are expressed in an anterior cap at the blastoderm stage corresponding to the presumptive serosa. Tc-zen2 has an additional later domain in the amnion. Knockdown of Tc-zen1 has dramatic effects on blastodermal patterning. The serosa is completely absent (and thus serosal Tc-zen2 expression is also lacking), the amnion expands dorsally and anteriorly, and the anterior head anlage expands to the anterior pole of the egg. In addition, the embryo never sinks into the yolk, but remains on the surface of the egg throughout development. Despite these major early patterning defects, the majority of larvae look largely normal, indicating that there are mechanisms for compensating for the dramatic expansion of the head, and that the expanded amnion has some capacity to rescue the loss of the serosal fate or that the insect can develop in the absence of the serosa 29.
In contrast to Tc-zen1, the defects seen after Tc-zen2 pRNAi are initially mild. No defects are seen at the blastoderm stage. However, after germband extension, the amnion fails to fuse with the serosa, and does not degenerate on the ventral side, thus preventing ventral eversion of the embryo. The persistent amnion contracts ventrally, dragging the dorsal flanks with it, leading to the inversion of the embryo trapped within the yolk 29.
The complicated movements of covering and uncovering the embryo in the course of extraembryonic development in Tribolium might seem superfluous, or at least unnecessarily baroque, given the ability of apparently wild type embryos to hatch in the absence of these movements and the entire serosal tissue after Tc-zen1 pRNAi. However, the serosa likely has important roles beyond embryogenesis in, for example, providing protection to the embryo from a variety of environmental insults 141. These insults are likely to be more extreme for embryos laid outside of laboratory conditions, thus the proper formation of the extraembryonic membranes is likely to be critical for the success of the organism in the wild. It is interesting to note that reduction in extrambryonic membranes is observed in many long germ species, which may indicate that deposition of the egg in a less harsh environment may be an ecological factor that contributes to the evolution of this type of embryogenesis.
The extraembryonic membranes of the long germ embryo of Nasonia are again comparatively less well described, but the data that do exist indicate that they possess both Drosophila and Tribolium-like features. Observations of living embryos indicate that unlike Drosophila, both a distinct serosa and an amnion are present in the wasp 136. However, like the fly these tissues are defined in a very narrow domain restricted to the dorsal midline of the embryo 34 (JAL, personal observation). At the onset of gastrulation, the presumptive serosa expands from its dorsal domain, and crawls over the embryo proper until it completely envelops the embryo on the ventral side, while the amnion appears to expand from the dorsal flank of the embryo to cover the yolk until the completion of dorsal closure 34, 136. Further experiments are required to determine whether the specification of these two fates bears any similarity to either Drosophila or Tribolium mechanisms.
Conclusions and Perspectives
The tripartite comparative system of Drosophila, Nasonia, and Tribolium has allowed a number of significant insights into how early embryogenesis has evolved among holometabolous insects, especially in relation to their changing embryonic patterning environments. One striking observation was that, although the specific molecules and some of the details differ, both of the long germ species use very similar strategies based on localized mRNAs to rapidly specify cell fates along the entire AP axis of the early embryo. This strategy includes an instructive anterior factor (bcd in Drosophila, Nv-otd1 in Nasonia), factors permissive for anterior patterning (also bcd in Drosophila, maternal anteriorly localized Nv-gt and the posterior localization of Nv-cad in Nasonia), and a permissive posterior factor (nanos orthologs in both species). Nasonia employs an additional posterior instructive factor, the posterior aspect of Nv-otd1 localization. The dependence of long germ species on localized mRNAs extends to the DV axis, as both Nasonia and Drosophila use localized mRNA encoding EGF ligands to establish a stable axis. The nature of the long germ patterning system is likely a major factor driving the acquisition of precisely localized sources of positional information, as embryos of this type must specify a large number of distinct cell fates in a short period of time, and thus require precise and robust patterning information at a very early stage. Although localized factors have been found in Tribolium 142 that are functionally important (S. J. Brown, manuscript under preparation), it seems that they give guidance for self-regulatory mechanisms, other than giving extensive positional information as in the Drosophila case (E. El-Sherif, S. J. Brown, manuscript under preparation).
The reduced dependence of Tribolium on localized mRNAs is likely due in part to its mode of embryogenesis, where significantly fewer cell fates are established at a time accessible to maternally provided mRNAs. In place of maternally provisioned patterning information, Tribolium seems to rely on another powerful strategy for establishing pattern, namely emergent patterning based on self-regulatory systems. Examples of these include the zygotic self regulatory and feed-forward loops of the DV axis, the negative feedback pair-rule loop required for patterning and progressive growth of the germband, and perhaps the spatial and temporal cross regulation of the gap genes in the germband stage.
The ability to break with the Drosophila paradigm is another major development made possible by the establishment of Nasonia and Tribolium as comparative model systems. The discovery of the novel peptide encoding gene mlpt as a gap gene in Tribolium, the discovery that Wnt signaling is required for posterior growth zone and patterning, and the discovery of mex-3 as an anciently conserved regulator of cad among the ecdysozoa are illustrative of this power. These types of discoveries should only increase as more genome wide and unbiased analyses of gene function in both species start bearing fruit.
Additional advances in understanding the developmental systems of Tribolium and Nasonia will come from taking advantage of the rapidly increasing set of transgenic and genomic tools and techniques available in these organisms. For example, germline transformation allows the assessment of putative enhancer regions that can be identified by genomic techniques (such as those discussed below). In addition, the development of techniques for conditionally inducible expression, introduction of fluorescent hybrid proteins, and live imaging will allow for a much more detailed description and genetic analysis of developmental events. This will be especially valuable for dynamic processes, such as the operation of the growth zone or the movements of the extraembryonic membranes, where static observations on fixed material are not sufficient for rigorous interpretation.
The availability of fully sequenced genomes allows for the application of powerful techniques for comprehensively unraveling the regulatory interactions underlying development. These include genome-wide analysis of expression level perturbation (e.g. microarrays) and genome-wide discovery of transcription factor binding sites (e.g. ChIP-seq). The combination of such techniques should provide robust descriptions of gene regulatory networks robust enough to compare meaningfully with those already described for Drosophila.
Finally, the evolutionary significance of the similarities and differences in embryonic patterning among Nasonia, Tribolium, and Drosophila will only be fully clarified by placing them in a more robust phylogenetic and ecological context. For example, sampling of other short germ species both within the beetles, and among hemimetabolous insects will allow the differentiation of characteristics that are typical for short germ embryos, and those which arose uniquely in Tribolium. Reciprocally, additional long germ species, particularly those among the beetles, will be particularly informative in identifying potential constraints or common strategies used in the transition from short germ embryogenesis. Such broad sampling of insect developmental data is increasingly feasible, given the wide applicability of pRNAi, and the increasing power of next generation sequencing techniques to rapidly generate candidate gene sequence data.
Understanding the evolutionary meaning of such expanded developmental data will require a more detailed understanding of the ecological contexts under which these embryos develop and hatch. These conditions could result in selection on the speed of embryogenesis, the relative need for protective extraembryonic membranes, or the relative complexity of the hatching larva. All of these potential selective pressures could have major impacts on the strategies used for early patterning and embryogenesis. For these experiments, the highly developed techniques and resources available in Tribolium and Nasonia will confer on them a status as rich sources of hypothesis generation in relation to the numerous emerging satellite models roughly equivalent to that which Drosophila held in the early days of the wasp and beetle systems. Hopefully the process of reciprocal illumination will continue whereby the results obtained in satellite systems spur further experiments in core model species and vice versa, leading to both a broad and deep understanding of the evolution of developmental mechanisms throughout the insects.
Figure 9. Regulatory relationships between major components in DV axis patterning.
Left panel (Drosophila): the DV axis patterning network in Drosophila is mostly a linear cascade with the exception of multiple positive and negative feedback loops within the protease cascade components. Right panel (Tribolium): nothing is known about the involvement of a protease cascade in Tribolium similar to that involved in Drosophila. However, Tribolium posses multiple feedback loops at levels more downstream.
Contributor Information
Ezzat El-Sherif, Program of Genetics, Kansas State University, Manhattan, Kansas..
Jeremy A. Lynch, Institute for Developmental Biology, University of Cologne, Cologne, Germany.
Susan J. Brown, Division of Biology, Kansas State University, Manhattan, Kansas sjbrown@ksu.edu
References
- 1.Davis GK, Patel NH. Short, long, and beyond: Molecular and embryological approaches to insect segmentation. Annual Review of Entomology. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- 2.Tautz D, Friedrich M, Schröder R. Insect embryogenesis - what is ancestral and what is derived? Development. 1994;(Supplement):193–199. [Google Scholar]
- 3.Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJP, et al. Functional and Evolutionary Insights from the Genomes of Three Parasitoid Nasonia Species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJP, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 5.Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12:R85–86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 6.Lynch JA, Desplan C. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat Protoc. 2006;1:486–494. doi: 10.1038/nprot.2006.70. [DOI] [PubMed] [Google Scholar]
- 7.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 8.Trauner J, Schinko J, Lorenzen MD, Shippy TD, Wimmer EA, Beeman RW, Klingler M, Bucher G, Brown SJ. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. Bmc Biology. 2009;7 doi: 10.1186/1741-7007-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schinko JB, Weber M, Viktorinova I, Kiupakis A, Averof M, Klingler M, Wimmer EA, Bucher G. Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. Bmc Developmental Biology. 2010;10 doi: 10.1186/1471-213X-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pultz MA, Zimmerman KK, Alto NM, Kaeberlein M, Lange SK, Pitt JN, Reeves NL, Zehrung DL. A genetic screen for zygotic embryonic lethal mutations affecting cuticular morphology in the wasp Nasonia vitripennis. Genetics. 2000;154:1213–1229. doi: 10.1093/genetics/154.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pultz MA, Pitt JN, Alto NM. Extensive zygotic control of the anteroposterior axis in the wasp Nasonia vitripennis. Development. 1999;126:701–710. doi: 10.1242/dev.126.4.701. [DOI] [PubMed] [Google Scholar]
- 12.RiveraPomar R, Niessing D, SchmidtOtt U, Gehring WJ, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 13.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 14.Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 15.Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 16.Stauber M, Jackle H, Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 18.da Fonseca RN, Lynch JA, Roth S. Evolution of axis formation: mRNA localization, regulatory circuits and posterior specification in non-model arthropods. Current Opinion in Genetics & Development. 2009;19:404–411. doi: 10.1016/j.gde.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature. 2006;439:728–732. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- 20.Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science. 2007;315:1841–1843. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- 21.Schröder R. The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature. 2003;422:621–625. doi: 10.1038/nature01536. [DOI] [PubMed] [Google Scholar]
- 22.Marques-Souza H, Aranda M, Tautz D. Delimiting the conserved features of hunchback function for the trunk organization of insects. Development. 2008;135:881–888. doi: 10.1242/dev.018317. [DOI] [PubMed] [Google Scholar]
- 23.Kotkamp K, Klingler M, Schoppmeier M. Apparent role of Tribolium orthodenticle in anteroposterior blastoderm patterning largely reflects novel functions in dorsoventral axis formation and cell survival. Development. 2010;137:1853–1862. doi: 10.1242/dev.047043. [DOI] [PubMed] [Google Scholar]
- 24.Mlodzik M, Gibson G, Gehring WJ. Effects of Ectopic Expression of Caudal during Drosophila Development. Development. 1990;109:271–277. doi: 10.1242/dev.109.2.271. [DOI] [PubMed] [Google Scholar]
- 25.Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, Desplan C. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development. 2006;133:3973–3982. doi: 10.1242/dev.02576. [DOI] [PubMed] [Google Scholar]
- 26.Shinmyo Y, Mito T, Matsushita T, Sarashina I, Miyawaki K, Ohuchi H, Noji S. caudal is required for gnathal and thoracic patterning and for posterior elongation in the intermediate-germband cricket Gryllus bimaculatus. Mech Dev. 2005;122:231–239. doi: 10.1016/j.mod.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Wilson MJ, Havler M, Dearden PK. Giant, Kruppel, and caudal act as gap genes with extensive roles in patterning the honeybee embryo. Dev Biol. 339:200–211. doi: 10.1016/j.ydbio.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Schoppmeier M, Fischer S, Schmitt-Engel C, Lohr U, Klingler M. An Ancient Anterior Patterning System Promotes Caudal Repression and Head Formation in Ecdysozoa. Current Biology. 2009;19:1811–1815. doi: 10.1016/j.cub.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 29.van der Zee M, Berns N, Roth S. Distinct functions of the Tribolium zerknullt genes in serosa specification and dorsal closure. Current Biology. 2005;15:624–636. doi: 10.1016/j.cub.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 30.Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126:659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 32.Ephrussi A, Dickinson LK, Lehmann R. oskar Organizes the Germ Plasm and Directs Localization of the Posterior Determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 33.Wolff C, Sommer R, Schroder R, Glaser G, Tautz D. Conserved and divergent expression aspects of the Drosophila segmentation gene hunchback in the short germ band embryo of the flour beetle Tribolium. Development. 1995;121:4227–4236. doi: 10.1242/dev.121.12.4227. [DOI] [PubMed] [Google Scholar]
- 34.Pultz MA, Westendorf L, Gale SD, Hawkins K, Lynch J, Pitt JN, Reeves NL, Yao JCY, Small S, Desplan C, et al. A major role for zygotic hunchback in patterning the Nasonia embryo. Development. 2005;132:3705–3715. doi: 10.1242/dev.01939. [DOI] [PubMed] [Google Scholar]
- 35.Lynch JA, Desplan C. Novel modes of localization and function of nanos in the wasp Nasonia. Development. 2010;137:3813–3821. doi: 10.1242/dev.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smibert CA, Wilson J, Kerr K, Macdonald P. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 37.Patel NH, Hayward DC, Lall S, Pirkl NR, DiPietro D, Ball EE. Grasshopper hunchback expression reveals conserved and novel aspects of axis formation and segmentation. Development. 2001;128:3459–3472. doi: 10.1242/dev.128.18.3459. [DOI] [PubMed] [Google Scholar]
- 38.Sprenger FN-V C. The terminal system of axis determination in the drosophila embryo. In: Bate MA A, editor. The development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor Laboratory; 1993. pp. 365–386. [Google Scholar]
- 39.Sprenger F, Trosclair MM, Morrison DK. Biochemical analysis of torso and D-raf during Drosophila embryogenesis: implications for terminal signal transduction. Mol Cell Biol. 1993;13:1163–1172. doi: 10.1128/mcb.13.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casanova J, Furriols M, McCormick CA, Struhl G. Similarities between trunk and spatzle, putative extracellular ligands specifying body pattern in Drosophila. Genes Dev. 1995;9:2539–2544. doi: 10.1101/gad.9.20.2539. [DOI] [PubMed] [Google Scholar]
- 41.Stevens LM, Frohnhofer HG, Klingler M, Nusslein-Volhard C. Localized requirement for torso-like expression in follicle cells for development of terminal anlagen of the Drosophila embryo. Nature. 1990;346:660–663. doi: 10.1038/346660a0. [DOI] [PubMed] [Google Scholar]
- 42.Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. The Drosophila embryonic patterning determinant torsolike is a component of the eggshell. Curr Biol. 2003;13:1058–1063. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- 43.Furriols M, Casanova J. In and out of Torso RTK signalling. Embo J. 2003;22:1947–1952. doi: 10.1093/emboj/cdg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WX. Functions and mechanisms of receptor tyrosine kinase Torso signaling: lessons from Drosophila embryonic terminal development. Dev Dyn. 2005;232:656–672. doi: 10.1002/dvdy.20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigel D, Jurgens G, Klingler M, Jackle H. Two gap genes mediate maternal terminal pattern information in Drosophila. Science. 1990;248:495–498. doi: 10.1126/science.2158673. [DOI] [PubMed] [Google Scholar]
- 46.Jurgens GH V. The terminal regions of the body pattern. In: Bate MA A, editor. The development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor Laboratory; 1993. pp. 687–746. [Google Scholar]
- 47.Schaeffer V, Killian D, Desplan C, Wimmer EA. High bicoid levels render the terminal system dispensable for Drosophila head development. Development. 2000;127:3993–3999. doi: 10.1242/dev.127.18.3993. [DOI] [PubMed] [Google Scholar]
- 48.Schoppmeier M, Schröder R. Maternal Torso Signaling Controls Body Axis Elongation in a Short Germ Insect. 2005;15:2131–2136. doi: 10.1016/j.cub.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 49.Schröder R, Eckert C, Wolff C, Tautz D. Conserved and divergent aspects of terminal patterning in the beetle Tribolium castaneum. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6591–6596. doi: 10.1073/pnas.100005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch JA, Olesnicky EC, Desplan C. Regulation and function of tailless in the long germ wasp Nasonia vitripennis. Dev Genes Evol. 2006 doi: 10.1007/s00427-006-0076-5. [DOI] [PubMed] [Google Scholar]
- 51.Wilson MJ, Dearden PK. Tailless patterning functions are conserved in the honeybee even in the absence of Torso signaling. Developmental Biology. 2009;335:276–287. doi: 10.1016/j.ydbio.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Akam M. The Molecular-Basis for Metameric Pattern in the Drosophila Embryo. Development. 1987;101:1. [PubMed] [Google Scholar]
- 53.RiveraPomar R, Jackle H. From gradients to stripes in Drosophila embryogenesis: Filling in the gaps. Trends in Genetics. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 54.Bucher G, Klingler M. Divergent segmentation mechanism in the short germ insect Tribolium revealed by giant expression and function. Development. 2004;131:1729–1740. doi: 10.1242/dev.01073. [DOI] [PubMed] [Google Scholar]
- 55.Cerny AC, Bucher G, Schroder R, Klingler M. Breakdown of abdominal patterning in the Tribolium Kruppel mutant jaws. Development. 2005;132:5353–5363. doi: 10.1242/dev.02154. [DOI] [PubMed] [Google Scholar]
- 56.Cerny AC, Grossmann D, Bucher G, Klingler M. The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Developmental biology. 2008;321:284–294. doi: 10.1016/j.ydbio.2008.05.527. [DOI] [PubMed] [Google Scholar]
- 57.Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 58.Marques-Souza H. Phd thesis. 2007. Evolution of the gene regulatory network controlling trunk segmentation in insects. [Google Scholar]
- 59.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 60.Ingham P, Gergen P. Interactions between the pair-rule genes runt, hairy, even-skipped and fushi tarazu and the establishment of periodic pattern in the Drosophila embryo. Development. 1988;104:51–60. [Google Scholar]
- 61.Gutjahr T, Vanario-Alonso CE, Pick L, Noll M. Multiple regulatory elements direct the complex expression pattern of the Drosophila segmentation gene paired. Mech Dev. 1994;48:119–128. doi: 10.1016/0925-4773(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 62.Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez L, Chaouiya C, Thieffry D. Segmenting the fly embryo: logical analysis of the role of the segment polarity cross-regulatory module. Int J Dev Biol. 2008;52:1059–1075. doi: 10.1387/ijdb.072439ls. [DOI] [PubMed] [Google Scholar]
- 64.Damen WG. Evolutionary conservation and divergence of the segmentation process in arthropods. Dev Dyn. 2007;236:1379–1391. doi: 10.1002/dvdy.21157. [DOI] [PubMed] [Google Scholar]
- 65.Sommer RJ, Tautz D. Involvement of an orthologue of the Drosophila pair-rule gene hairy in segment formation of the short germ-band embryo of Tribolium (Coleoptera) Nature. 1993;361:448–450. doi: 10.1038/361448a0. [DOI] [PubMed] [Google Scholar]
- 66.Patel NH, Condron BG, Zinn K. Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- 67.Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Developmental biology. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aranda M, Marques-Souza H, Bayer T, Tautz D. The role of the segmentation gene hairy in Tribolium. Development genes and evolution. 2008;218:465–477. doi: 10.1007/s00427-008-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maderspacher F, Bucher G, Klingler M. Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Development genes and evolution. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- 71.Keller RG, Desplan C, Rosenberg MI. Identification and characterization of Nasonia Pax genes. Insect Molecular Biology. 2010;19:109–120. doi: 10.1111/j.1365-2583.2009.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Binner P, Sander K. Pair-rule patterning in the honeybee Apis mellifera: Expression of even-skipped combines traits known from beetles and fruitfly. Development Genes and Evolution. 1997;206:447–454. doi: 10.1007/s004270050074. [DOI] [PubMed] [Google Scholar]
- 73.DiNardo S, Heemskerk J, Dougan S, O'Farrell PH. The making of a maggot: patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev. 1994;4:529–534. doi: 10.1016/0959-437x(94)90068-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choe CP, Brown SJ. Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Developmental biology. 2009;325:482–491. doi: 10.1016/j.ydbio.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 75.Farzana L, Brown SJ. Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol. 2008;218:181–192. doi: 10.1007/s00427-008-0207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oppenheimer DI, MacNicol AM, Patel NH. Functional conservation of the wingless-engrailed interaction as shown by a widely applicable baculovirus misexpression system. Curr Biol. 1999;9:1288–1296. doi: 10.1016/s0960-9822(00)80050-0. [DOI] [PubMed] [Google Scholar]
- 77.Bolognesi R, Fischer TD, Brown SJ. Loss of Tc-arrow and canonical Wnt signaling alters posterior morphology and pair-rule gene expression in the short-germ insect, Tribolium castaneum. Development genes and evolution. 2009;219:369–375. doi: 10.1007/s00427-009-0299-3. [DOI] [PubMed] [Google Scholar]
- 78.Bolognesi R, Farzana L, Fischer TD, Brown SJ. Multiple Wnt Genes Are Required for Segmentation in the Short-Germ Embryo of Tribolium castaneum. Current Biology. 2008;18:1624–1629. doi: 10.1016/j.cub.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, Ohuchi H, Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Dev. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 80.Angelini DR, Kaufman TC. Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev Biol. 2005;283:409–423. doi: 10.1016/j.ydbio.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 81.McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WG. Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr Biol. 2008;18:1619–1623. doi: 10.1016/j.cub.2008.08.045. [DOI] [PubMed] [Google Scholar]
- 82.Martin BL, Kimelman D. Wnt signaling and the evolution of embryonic posterior development. Curr Biol. 2009;19:R215–219. doi: 10.1016/j.cub.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 84.Angelini DR, Kaufman TC. Comparative developmental genetics and the evolution of arthropod body plans. Annu Rev Genet. 2005;39:95–119. doi: 10.1146/annurev.genet.39.073003.112310. [DOI] [PubMed] [Google Scholar]
- 85.Shippy TD, Brown SJ, Denell RE. Maxillopedia is the Tribolium ortholog of proboscipedia. Evol Dev. 2000;2:145–151. doi: 10.1046/j.1525-142x.2000.00055.x. [DOI] [PubMed] [Google Scholar]
- 86.Posnien N, Bashasab F, Bucher G. The insect upper lip (labrum) is a nonsegmental appendage-like structure. Evol Dev. 2009;11:480–488. doi: 10.1111/j.1525-142X.2009.00356.x. [DOI] [PubMed] [Google Scholar]
- 87.Brown SJ, Shippy TD, Beeman RW, Denell RE. Tribolium Hox genes repress antennal development in the gnathos and trunk. Mol Phylogenet Evol. 2002;24:384–387. doi: 10.1016/s1055-7903(02)00205-1. [DOI] [PubMed] [Google Scholar]
- 88.Stuart JJ, Brown SJ, Beeman RW, Denell RE. A deficiency of the homeotic complex of the beetle Tribolium. Nature. 1991;350:72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- 89.Steward R, Zusman SB, Huang LH, Schedl P. The dorsal protein is distributed in a gradient in early Drosophila embryos. Cell. 1988;55:487–495. doi: 10.1016/0092-8674(88)90035-9. [DOI] [PubMed] [Google Scholar]
- 90.Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- 91.Reeves GT, Stathopoulos A. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stathopoulos A, Levine M. Dorsal gradient networks in the Drosophila embryo. Dev Biol. 2002;246:57–67. doi: 10.1006/dbio.2002.0652. [DOI] [PubMed] [Google Scholar]
- 93.Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: Insights from postgenome technologies. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20072–20076. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ray RP, Arora K, Nusslein-Volhard C, Gelbart WM. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development. 1991;113:35–54. doi: 10.1242/dev.113.1.35. [DOI] [PubMed] [Google Scholar]
- 95.Kirov N, Childs S, O'Connor M, Rushlow C. The Drosophila dorsal morphogen represses the tolloid gene by interacting with a silencer element. Mol Cell Biol. 1994;14:713–722. doi: 10.1128/mcb.14.1.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Francois V, Solloway M, O'Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 1994;8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 97.O'Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown SJ, Parrish JK, Beeman RW, Denell RE. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mechanisms of development. 1997;61:165–173. doi: 10.1016/s0925-4773(96)00642-9. [DOI] [PubMed] [Google Scholar]
- 99.Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 100.Von Ohlen T, Doe CQ. Convergence of dorsal, Dpp, and Egfr signaling pathways subdivides the Drosophila neuroectoderm into three dorsal-ventral columns. Developmental Biology. 2000;224:362–372. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- 101.Handel K, Basal A, Fan X, Roth S. Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Development Genes and Evolution. 2005;215:13–31. doi: 10.1007/s00427-004-0446-9. [DOI] [PubMed] [Google Scholar]
- 102.Cande J, Goltsev Y, Levine MS. Conservation of enhancer location in divergent insects. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14414–14419. doi: 10.1073/pnas.0905754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van der Zee M, Stockhammer O, von Levetzow C, da Fonseca RN, Roth S. Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16307–16312. doi: 10.1073/pnas.0605154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.da Fonseca RN, van der Zee M, Roth S. Evolution of extracellular Dpp modulators in insects: The roles of tolloid and twisted-gastrulation in dorsoventral patterning of the Tribolium embryo. Developmental Biology. 2010;345:80–93. doi: 10.1016/j.ydbio.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 105.Wheeler SR, Carrico ML, Wilson BA, Skeath JB. The Tribolium columnar genes reveal conservation and plasticity in neural precursor patterning along the embryonic dorsal-ventral axis. Developmental Biology. 2005;279:491–500. doi: 10.1016/j.ydbio.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 106.Chen G, Handel K, Roth S. The maternal NF-kappa B/Dorsal gradient of Tribolium castaneum: dynamics of early dorsoventral patterning in a short-germ beetle. Development. 2000;127:5145–5156. doi: 10.1242/dev.127.23.5145. [DOI] [PubMed] [Google Scholar]
- 107.da Fonseca RN, von Levetzow C, Kaischeuer P, Basal A, van der Zee M, Roth S. Self-regulatory circuits in dorsoventral axis formation of the short-germ beetle Tribolium castaneum. Developmental Cell. 2008;14:605–615. doi: 10.1016/j.devcel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 108.Roth S. The origin of dorsoventral polarity in Drosophila. Philos Trans R Soc Lond B Biol Sci. 2003;358:1317–1329. doi: 10.1098/rstb.2003.1325. discussion 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peri F, Technau M, Roth S. Mechanisms of Gurken-dependent pipe regulation and the robustness of dorsoventral patterning in Drosophila. Development. 2002;129:2965–2975. doi: 10.1242/dev.129.12.2965. [DOI] [PubMed] [Google Scholar]
- 110.Zhang ZY, Stevens LM, Stein D. Sulfation of Eggshell Components by Pipe Defines Dorsal-Ventral Polarity in the Drosophila Embryo. Current Biology. 2009;19:1200–1205. doi: 10.1016/j.cub.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rushlow CA, Han K, Manley JL, Levine M. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell. 1989;59:1165–1177. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 112.Roth S, Schupbach T. The relationship between ovarian and embryonic dorsoventral patterning in Drosophila. Development. 1994;120:2245–2257. doi: 10.1242/dev.120.8.2245. [DOI] [PubMed] [Google Scholar]
- 113.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 114.Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 115.Lynch JA, Peel AD, Drechsler A, Averof M, Roth S. EGF Signaling and the Origin of Axial Polarity among the Insects. Current Biology. 2010;20:1042–1047. doi: 10.1016/j.cub.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schmidt-Ott U, Technau GM. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development. 1992;116:111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- 117.Scholtz G, Edgecombe GD. The evolution of arthropod heads: reconciling morphological, developmental and palaeontological evidence. Dev Genes Evol. 2006;216:395–415. doi: 10.1007/s00427-006-0085-4. [DOI] [PubMed] [Google Scholar]
- 118.Cohen SM, Jurgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 119.Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- 120.Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- 121.Wimmer EA, Cohen SM, Jackle H, Desplan C. buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development. 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- 122.Posnien N, Schinko JB, Kittelmann S, Bucher G. Genetics, development and composition of the insect head--a beetle's view. Arthropod Struct Dev. 2010;39:399–410. doi: 10.1016/j.asd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 123.Schinko JB, Kreuzer N, Offen N, Posnien N, Wimmer EA, Bucher G. Divergent functions of orthodenticle, empty spiracles and buttonhead in early head patterning of the beetle Tribolium castaneum (Coleoptera) Developmental biology. 2008;317:600–613. doi: 10.1016/j.ydbio.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 124.Gonzalez-Gaitan M, Rothe M, Wimmer EA, Taubert H, Jackle H. Redundant functions of the genes knirps and knirps-related for the establishment of anterior Drosophila head structures. Proc Natl Acad Sci U S A. 1994;91:8567–8571. doi: 10.1073/pnas.91.18.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schaeper ND, Pechmann M, Damen WG, Prpic NM, Wimmer EA. Evolutionary plasticity of collier function in head development of diverse arthropods. Dev Biol. 344:363–376. doi: 10.1016/j.ydbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 126.Economou AD, Telford MJ. Comparative gene expression in the heads of Drosophila melanogaster and Tribolium castaneum and the segmental affinity of the Drosophila hypopharyngeal lobes. Evol Dev. 2009;11:88–96. doi: 10.1111/j.1525-142X.2008.00305.x. [DOI] [PubMed] [Google Scholar]
- 127.Posnien N, Bucher G. Formation of the insect head involves lateral contribution of the intercalary segment, which depends on Tc-labial function. Dev Biol. 2010;338:107–116. doi: 10.1016/j.ydbio.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 128.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 129.Weinstock GM, Robinson GE, Gibbs RA, Worley KC, Evans JD, Maleszka R, Robertson HM, Weaver DB, Beye M, Bork P, et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dearden PK. Germ cell development in the Honeybee (Apis mellifera); Vasa and Nanos expression. Bmc Developmental Biology. 2006;6 doi: 10.1186/1471-213X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lynch JA, Ozuak O, Khila A, Abouheif E, Desplan C, Roth S. The Phylogenetic Origin of oskar Coincided with the Origin of Maternally Provisioned Germ Plasm and Pole Cells at the Base of the Holometabola. PLoS Genet. 2011;7:e1002029. doi: 10.1371/journal.pgen.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, Kimura K, Milshina NV, Hasselmann M, Gempe T, et al. Patterns of conservation and change in honey bee developmental genes. Genome Research. 2006;16:1376–1384. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schröder R. vasa mRNA accumulates at the posterior pole during blastoderm formation in the flour beetle Tribolium castaneum. Development Genes and Evolution. 2006;216:277–283. doi: 10.1007/s00427-005-0054-3. [DOI] [PubMed] [Google Scholar]
- 134.Leptin M. Gastrulation in Drosophila: The logic and the cellular mechanisms. Embo Journal. 1999;18:3187–3192. doi: 10.1093/emboj/18.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Handel K, Grunfelder CG, Roth S, Sander K. Tribolium embryogenesis: a SEM study of cell shapes and movements from blastoderm to serosal closure. Development Genes and Evolution. 2000;210:167–179. doi: 10.1007/s004270050301. [DOI] [PubMed] [Google Scholar]
- 136.Bull AL. Stages of Living Embryos in the Jewel Wasp Mormoniella-(Nasonia)-Vitripennis-(Walker)(Hymenoptera, Pteromalidae) International Journal of Insect Morphology & Embryology. 1982;11:1–23. [Google Scholar]
- 137.Fleig R, Sander K. Embryogenesis of the Honeybee Apis-Mellifera L (Hymenoptera, Apidae) - an Sem Study. International Journal of Insect Morphology & Embryology. 1986;15:449–462. [Google Scholar]
- 138.Yip MLR, Lamka ML, Lipshitz HD. Control of germ-band retraction in Drosophila by the zinc-finger protein HINDSIGHT. Development. 1997;124:2129–2141. doi: 10.1242/dev.124.11.2129. [DOI] [PubMed] [Google Scholar]
- 139.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. Journal of Cell Biology. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rafiqi AM, Lemke S, Ferguson S, Stauber M, Schmidt-Ott U. Evolutionary origin of the amnioserosa in cyclorrhaphan flies correlates with spatial and temporal expression changes of zen. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:234–239. doi: 10.1073/pnas.0709145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Panfilio KA. Extraembryonic development in insects and the acrobatics of blastokinesis. Developmental Biology. 2008;313:471–491. doi: 10.1016/j.ydbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 142.Bucher G, Farzana L, Brown SJ, Klingler M. Anterior localization of maternal mRNAs in a short germ insect lacking bicoid. Evolution & Development. 2005;7:142–149. doi: 10.1111/j.1525-142X.2005.05016.x. [DOI] [PubMed] [Google Scholar]