Abstract
Background and Objective
More than 100 salivary constituents have been found to show levels significantly different in patients with oral squamous cell carcinoma (OSCC) from those found in healthy controls, and therefore have been suggested to be potential salivary biomarkers for OSCC detection. However, many of these potential OSCC salivary biomarkers are also involved in chronic inflammation, and whether the levels of these biomarkers could be affected by the presence of chronic periodontitis (CP) was not known. The objective of this pilot study was therefore to measure the levels of 7 previously-reported potential OSCC salivary mRNA biomarkers in patients with CP and compare them to levels found in OSCC patients and Healthy Controls. The 7 salivary mRNAs were: IL-8, IL-1β, dual specificity phosphatase 1 (DUSP1), H3 histone family 3A (H3F3A), ornithin decarboxylase antizyme 1 (OAZ1), S100 calcium-binding protein P (S100P), and spermidine/spermine N1-acetyltransferase 1 (SAT1).
Material and Methods
Unstimulated whole saliva samples were collected from a total of 105 human subjects from the following 4 study groups: OSCC; CPNS (periodontitis, moderate to severe degree, non-smokers); CPS (periodontitis, moderate to severe degree, smokers); and Healthy Controls. Levels of each mRNA in patient groups (OSCC or CP) relative to the Healthy Controls was determined by a pre-amplification RT-qPCR approach with nested gene-specific primers. Results were recorded and analyzed by Bio-Rad CFX96 Real-Time System. Mean fold changes between each pair of patient vs. control group were analyzed by the Mann-Whitney U test with Bonferroni corrections.
Result
Only S100P showed significantly higher levels in OSCC patients compared to both CPNS patients (p= 0.003) and CPS patients (p=0.007). The difference in S100P levels between OSCC patients and Healthy Controls was also marginally significant (p=0.009). There was no significant difference in the levels of salivary IL-8, IL-1β, and DUSP1 mRNAs between the OSCC patients and the CPNS patients (p=0.510, 0.058, and 0.078, respectively); no significant difference in levels of salivary OAZ1 and SAT mRNAs between the OSCC patients and the CPS patients (p=0.318 and 0.764, respectively); and no significant difference in levels of the H3F3A mRNA between the OSCC patients and either CPS (p=0.449) or Health Controls (p=0.107).
Conclusion
Salivary S100P mRNA could be a reliable biomarker for OSCC detection, regardless of the presence of CP. The presence of CP could significantly affect the levels of the other 6 mRNAs, and negatively influence reliability for using them as biomarkers for oral cancer detection.
Keywords: Saliva, squamous cell carcinoma, chronic periodontitis, S100P protein
Introduction
Saliva constituents have been found to reflect a diseased or physiological state of the human body, and hence could be utilized for diagnostic purposes (1-4) Potential applications not only include detection of infectious diseases, cancers, autoimmune diseases, and cardiovascular diseases (such as acute myocardial infarction), but also monitoring of hormone levels and testing for substance abuse (5-9). Using salivary biomarkers for early detection of oral cancer is a promising non-invasive approach. However, the search for valid biomarkers is complicated by the fact that local inflammation is also commonly found in the oral cavity--due to trauma, dental plaque, infection or certain chronic mucocutaneous inflammatory diseases. Whether such oral inflammation (non-neoplastic conditions) affects the levels of the more-than-100 salivary constituents previously reported as potential OSCC biomarkers is mostly unknown, because in previous studies levels of the potential biomarkers measured in OSCC patients have been compared only with non-OSCC controls (2). In addition, many of those reported potential OSCC salivary biomarkers, such as IL-6 (10-12), IL-8 (10, 13, 14), IL-1β (14, 15), basic FGF (16), and antioxidant enzymes (17), are also important factors involved in inflammation and/or wound healing (18, 19). The levels of some of them indeed have been reported to be significantly higher or lower in periodontitis patients who had no OSCC (20-23). If chronic periodontitis (CP), which affects 48% of the US population age 30 or older (24-26) and 5-15% global population (27), is found to affect levels of a potential biomarker to a degree that there is no significant difference from the levels found in OSCC patients, then use of that biomarker for clinical detection of OSCC could result in significant false positive rates—thereby greatly reducing its value as a non-invasive diagnostic adjunct. Therefore, further validation is urgently needed, to determine the reliability of reported potential salivary biomarkers for OSCC, or for any cancer detection.
As part of a long-term effort to validate potential salivary biomarkers for clinical detection of OSCC, we selected a group of seven promising salivary mRNAs that had been found to show significantly higher levels in some groups of OSCC patients, compared to the levels found in normal controls (15, 28, 29). These 7 mRNAs are: IL-8, IL-1β, dual specificity phosphatase 1 (DUSP1), H3 histone family 3A (H3F3A), ornithin decarboxylase antizyme 1 (OAZ1), S100 calcium-binding protein P (S100P), and spermidine/spermine N1-acetyltransferase 1 (SAT1). The objective of this pilot study was to measure the levels of these salivary mRNAs in patients with CP and compare them to levels found in OSCC patients, which had not been done before.
Material and Methods
1. Patient groups and recruitment
Human subjects were recruited for each of the following 4 groups, during the period from September 1, 2011 to May 15, 2014, from the Stomatology Center, Department of Periodontics and Oral and Maxillofacial Surgery at Texas A&M University - Baylor College of Dentistry (TAMU-BCD), in Dallas and from referrals by local dentists and surgeons who had recently diagnosed OSCC patients. Healthy controls were recruited from the Undergraduate Dental Clinic and the Dental Hygiene Clinic, TAMU-BCD. Groups were defined as follows:
Group A, OSCC: Patients with newly-diagnosed OSCC, tested before any treatment was started
Group B, CPNS: Adult periodontitis patients, moderate to severe degree, who are non-smokers
Group C, CPS: Adult periodontitis patients, moderate to severe degree, who are smokers
Group D, Healthy Controls: Individuals who had no moderate or severe periodontitis, and no oral mucosal disease or lesions (such as oral lichen planus, geographic tongue, candidiasis, or aphthous ulcers).
The inclusion and exclusion criteria for each group of participants are listed in Table 1 (30-33). The rationale for these criteria has been delineated in our previous work (34, 35). The Healthy Controls were recruited by matching the age range of the other three study groups (aged 35 and above), and including approximately equal numbers of males and females.
Table 1.
The Inclusion and Exclusion Criteria of the Study Groups (OSCC= Oral Squamous Cell Carcinoma).
| Groups | Inclusion Criteria- Patient who.... | Exclusion Criteria |
|---|---|---|
| A: OSCC | 1. Had pathology reports of OSCC and had oral lesions of OSCC at the time of saliva collection. | Subjects who had... 1. Used any corticosteroids or immunosuppressants, for any reason, less than 1 week prior to saliva collection 2. Oral lichen planus, bone marrow transplants, hepatitis C, lupus erythematosus, or Sjögren's syndrome. 3. Had previous radiation therapy to the head and neck area |
| B: Chronic Periodontitis–Moderate to Severe Degree, Non-smokers | 1. Had at least two interproximal sites on different teeth showing clinical attachment level ≥ 4mm, or at least two interproximal sites on different teeth showing pocket depth ≥ 5mm*, and 2. Had had no periodontal treatment within 6 months prior to participation in the study; and 3. Had no previous diagnosis of oral epithelial dysplasia or OSCC; and 4. Stated they are non-smokers† or ex-smokers‡, and consume fewer than 14 alcoholic drinks§ per week. |
|
| C: Chronic Periodontitis-Moderate to Severe Degree, Smokers | 1. Had at least two interproximal sites on different teeth showing clinical attachment level ≥ 4mm, or at least two interproximal sites on different teeth showing pocket depth ≥ 5mm*, and 2. Had had no periodontal treatment within 6 months prior to participation in the study; and 3. Had no previous diagnosis of oral epithelial dysplasia or OSCC; and 4. Stated they are smokers. |
|
| E: Normal Controls (non-OSCC, non-periodontitis) | Subjects who... 1. Had no history of OLP, epithelial dysplasia or OSCC; and 2. Had fewer than 2 interproximal sites with clinical attachment level ≥ 4mm or fewer than 2 interproximal pocket depths ≥ 5mm,* and 3. Stated that they are non-smokers† or ex-smokers‡ and consume fewer than 14 alcoholic drinks§ per week. |
The clinical attachment level refers to the distance between the cemento-enamel junction of the tooth and the base of the gingival sulcus. The pocket depth refers to the distance from the free gingival margin to the bottom of the gingival sulcus. This definition of moderate to severe periodontitis is based on Eke et al. (25)
“Non-smoker” was defined as a person who ha d smoked fewer than 100 cigarettes in his/her lifetime and had not smoked at all within one calendar year prior to beginning participation in the study, and who had not smoked pipe or cigar or used smokeless tobacco, for any more than a total of 6 months in their lifetime, and not at all within the calendar year prior to beginning participation in the study (22,23)
“Ex-smoker” was defined as a person whose last smoking or other use of tobacco products was at least 20 years prior to beginning participation in the study (24)
“One alcoholic drink” was defined as approximately 150 ml of wine, 330 ml of beer or 30 ml of hard liquor (23)
** “Smoker” is defined as a person who has smoked more than 100 cigarettes in his/her lifetime and/or has smoked within one calendar year prior to beginning participation in the study, and who has smoked pipe or cigar or used smokeless tobacco, for more than a total of 6 months in their lifetime, and/or within the calendar year prior to beginning participation in the study.
Convenience sampling was used, in which any OSCC and CP patients who were interested in participating and fit the criteria were included in the study. Clinical information about the participants, including the OSCC clinical stage, pocket depth and/or clinical attachment levels of the CP patients, was retrieved from the database at TAMU-Baylor College of Dentistry, or provided by referring doctors. The recruitment protocol used was approved by the Institutional Review Board of TAMU-BCD, and informed consent was obtained from each participant prior to saliva collection.
2. Saliva collection
Unstimulated whole saliva samples were collected between 6am and 12pm, using previously described methods (35, 36). Briefly, subjects were asked to not eat, drink or perform any kind of oral hygiene procedures prior to saliva collection. Just before saliva collection, a cup of water was given to participants for rinsing. Five minutes after rinsing, participants were asked to spit into a 50ml sterile plastic tube kept in ice. A maximum of 8ml of saliva was collected within 30 minutes.
3. Sample processing
Saliva samples were processed immediately after collection according to a previously published method (35). Briefly, the saliva samples were centrifuged at 2,600g for 15 minutes, at 4°C. The supernatant was separated from the pellet, and the RNase inhibitor (Superase-In, Ambion Inc., Austin, TX) was added to the supernatant-- 5ul Superase-In/ml of supernatant. All samples were stored at −80 °C in aliquots until further use.
4. RT- pre-amplification and qPCR
A pre-amplification RT-qPCR approach with nested gene-specific primers was used, according to the previously described methods (28, 35, 37). After thawing, the total RNA of the saliva sample was extracted, using the RNeasy mini kit (Qiagen, Valencia, CA), and all samples were then treated with RNase-free DNase (Qiagen, Valencia, CA) to remove genomic DNA.
Nested PCR assay, a RT-PCR pre-amplification followed by qPCR, was used, according to Hu et al. (37). The outer and inner primer sequences for the seven mRNA biomarkers (IL-8, IL-1β, DUSP1, H3F3A, OAZ1, S100P, and SAT1), along with two reference genes (RPS9 and β-actin), were adopted from previous studies (28, 35). The qPCR was performed with Bio-Rad ITAQ Fast SYBR ROX kit (Bio-Rad, Hercules, CA) on Bio-Rad CFX96 Real-Time System (Bio-Rad, Hercules, CA). Each saliva sample was tested in triplicate. An inter-run calibrator of 0.125 ug/ul human RNA (Strategene qPCR Human Reference Total RNA, Agilent Technologies, Santa Clara, CA) was added in each experiment along with the saliva samples. The amount of each of the 7 mRNAs was normalized by β–actin and RPS9 (28). The results were recorded by the Bio-Rad CFX Manager Software version 2.0. Descriptive tests (minimum, maximum, range, sum, count, mean, median, and standard deviation) were used to describe and summarize study results. Analysis of variance (ANOVA) and the Chi square test were used to evaluate the mean difference of age and gender distribution, respectively, among the study groups. The Kruskal-Wallace test was used to analyze the differences in the relative quantity (ΔCq) of each salivary mRNA biomarker among the study groups. The Mann-Whitney U test was applied as a post hoc test for pairwise comparisons of normalized relative quantity (ΔΔCq) between each pair of study groups. Bonferroni corrections were applied in order to control inflated type I error due to multiple comparisons. Since there were 6 pairwise tests, p values needed to be less than 0.0083 (0.05/6 = 0.0083) to be able to reject the null hypotheses. The mean fold changes between each pair of patients vs. the control group were calculated by the Pfaffl method (38).
Results
After excluding saliva samples that had blood contamination or were insufficient in amount, 105 valid samples were analyzed. Demographic information and OSCC stages are listed in Table 2. In the OSCC group, 40% of participants were diagnosed as being in clinical Stage I, and 32% in Stage IV. There was no significant difference in the mean age among the four study groups (p=0.073). However, there was a significant difference in gender distribution among the four groups (p=0.001). In order to investigate whether this difference in gender distribution might be a confounding factor to the salivary biomarker levels, we then used the independent T test to analyze the mean differences in the relative quantity (ΔCq) of each salivary mRNA biomarker between males and females in the Healthy Control group. There was no significant difference in the relative quantity of each of the 7 salivary mRNAs between males and females (Table 3).
Table 2.
Demographics, Mean Age (Years), Age Range (Years) and Clinical Information of the Study Groups.
| Groups | N | Mean Age (Range) | Gender | OSCC Stage | Smoking History | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | I | II | III | IV | S | Ex | NS | |||
| A: OSCC | 25 | 57.96 (27-81) | 18 | 7 | 10 | 4 | 3 | 8 | 11 | 4 | 10 |
| B: Chronic Periodontitis, Non-smokers | 26 | 60.65 (40-78) | 7 | 19 | - | 26 | |||||
| C: Chronic periodontitis, Smokers | 25 | 50.92 (29-82) | 19 | 6 | - | 25 | |||||
| D: Normal Controls | 29 | 55.93 (29-77) | 14 | 15 | - | 29 | |||||
Table 3.
Comparison of the Relative Quantity (ΔCq) of Salivary mRNA Biomarkers Between Males and Females in the Healthy Controls.
| Salivary mRNA | Gender (N) | Relative Quantity (ΔCq) Mean ± Standard Deviation |
T test result (p value) |
|---|---|---|---|
| IL-8 | M (14) | 0.76 ± 2.03 | 0.841 |
| F (15) | 0.60 ± 2.10 | ||
| OAZ1 | M (14) | 0.05 ± 1.09 | 0.988 |
| F (15) | 0.04 ± 0.87 | ||
| S100P | M (14) | 1.80 ± 2.44 | 0.660 |
| F (15) | 1.45 ±1.68 | ||
| IL-1β | M (14) | −0.26 ± 1.07 | 0.400 |
| F (15) | 0.10 ±1.15 | ||
| H3F3A | M (14) | 0.65 ± 1.75 | 0.437 |
| F (15) | 0.18 ± 1.47 | ||
| SAT1 | M (14) | −1.57 ± 1.15 | 0.938 |
| F (15) | −1.54 ± 1.03 | ||
| DUSP1 | M (14) | 1.57 ±0.88 | 0.730 |
| F (15) | 1.69 ± 1.00 |
The mean and standard deviation of the threshold temperature (Cq), and the normalized mean and median Cqs (ΔCqs) of each mRNA in each study group are listed in Table 4. The median fold changes of the 7 salivary mRNAs in each patient group, compared to the levels found in the Healthy Control group, and the statistical results for each pair of the study groups are shown in Figure 1. Extremely low levels of salivary OAZ1 and SAT1 mRNAs were found in the CPNS patient group when compared to the Healthy Controls (0.03 fold for both mRNAs).
Table 4.
Mean and Standard Deviation of the Threshold Temperature (Cq), and Normalized Mean and Median Cqs (ΔCq) of Each mRNA in Each Study Group.
| Salivary mRNAs | Group A (OSCC) | Group B (Chronic Periodontitis, Non-smokers) | Group C (Chronic periodontitis, Smokers) | Group D (Healthy Control) | ||||
|---|---|---|---|---|---|---|---|---|
| Cq | ΔCq (Median) | Cq | ΔCq (Median) | Cq | ΔCq (Median) | Cq | ΔCq (Median) | |
| IL-8 | 16.83 ± 2.50 | −2.63 ± 1.23 (−2.51) | 13.59 ± 1.22 | −2.68 ± 0.77 (−2.67) | 18.57±3.11 | −1.29 ± 1.55 (−0.99) | 22.94 ± 4.07 | 0.68 ± 2.03 (0.97) |
| OAZ1 | 18.76 ± 2.01 | −0.70 ± 0.59 (−0.64) | 21.93 ± 3.70 | 5.66 ± 3.37 (5.49) | 18.96 ± 2.49 | −0.90 ± 0.60 (−0.92) | 21.85 ± 3.36 | 0.05 ± 0.97 (0.23) |
| S100P | 19.80 ± 2.58 | 0.34 ± 1.44 (0.19) | 17.86 ± 1.84 | 1.59 ± 1.30 (1.20) | 21.58 ± 3.46 | 1.72 ± 1.79 (1.01) | 23.42 ± 4.08 | 1.62 ± 2.05 (1.16) |
| IL-1β | 16.72 ± 2.24 | −2.74 ± 0.71 (−2.79) | 13.20 ± 1.09 | −3.07 ± 0.38 (−3.00) | 17.52 ± 4.48 | −2.34 ± 3.89 (−1.57) | 21.73 ± 3.28 | −0.07 ± 1.11 (−0.29) |
| H3F3A | 19.23 ± 2.26 | −0.23 ± 0.99 (−0.1) | 14.40 ± 1.07 | −1.87 ± 0.39 (−1.88) | 19.89 ± 3.57 | 0.03 ± 1.29 (−0.01) | 22.21 ± 4.02 | 0.41 ± 1.60 (0.44) |
| SAT1 | 17.19 ± 2.16 | −2.27 ± 0.92 (−2.46) | 19.76 ± 2.87 | 3.49 ± 2.67 (3.44) | 17.82 ± 2.29 | −2.04 ± 1.33 (−2.07) | 20.24 ± 2.73 | −1.56 ± 1.07 (−1.42) |
| DUSP1 | 19.25 ± 2.31 | −0.21 ± 0.69 (−0.32) | 16.41 ± 1.16 | 0.14 ± 0.59 (0.17) | 20.67 ± 2.80 | 0.81 ± 0.90 (0.49) | 23.43 ± 3.29 | 1.63 ± 0.93 (1.41) |
Figure 1, A-G.
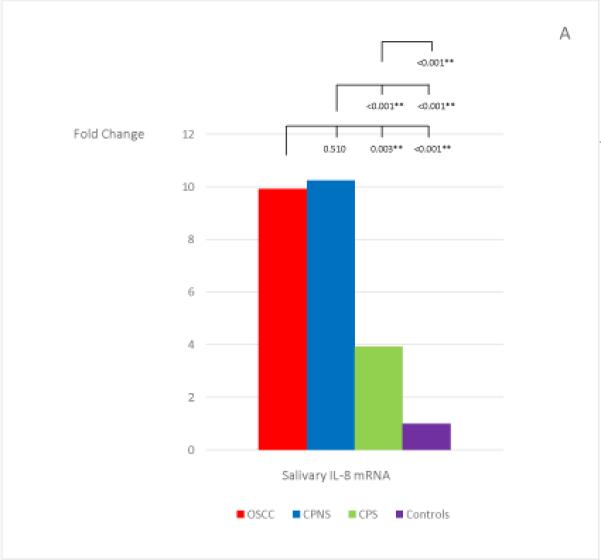
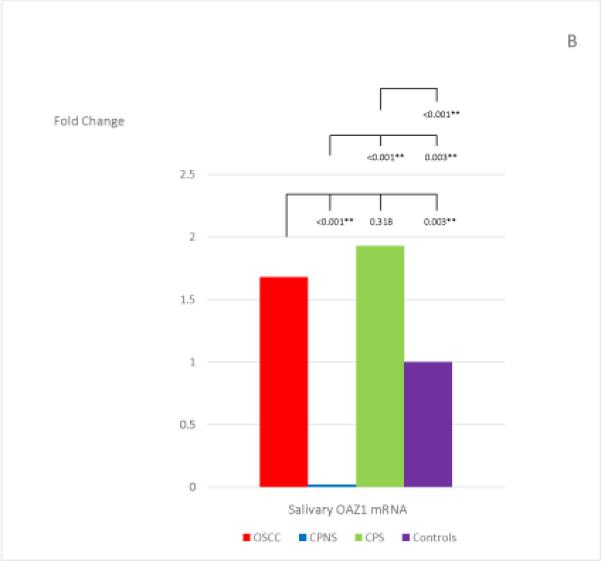
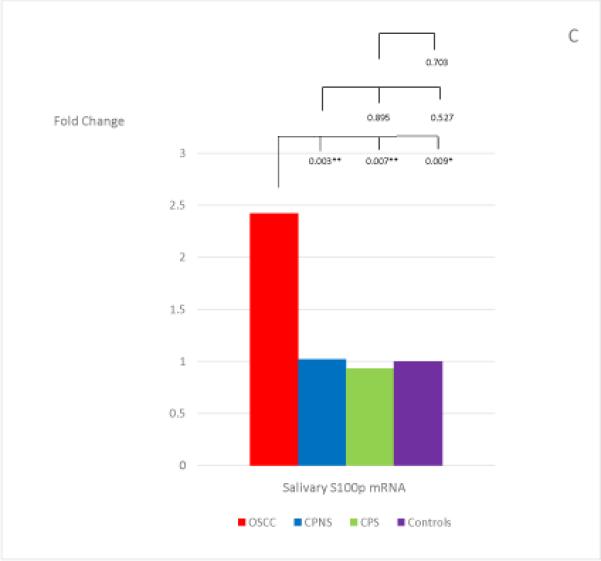
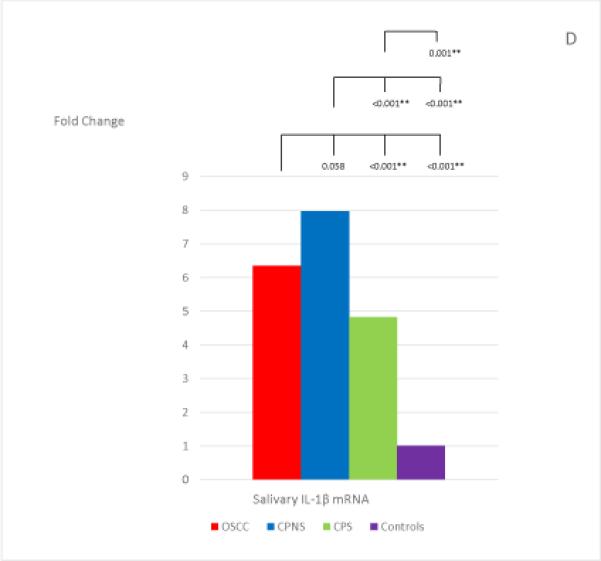
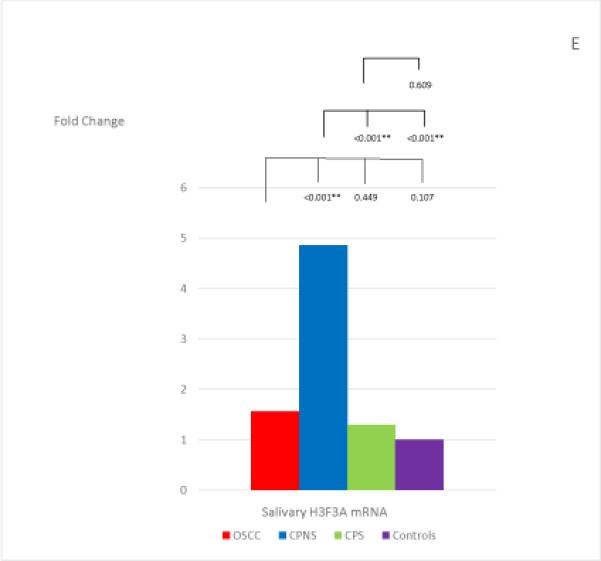
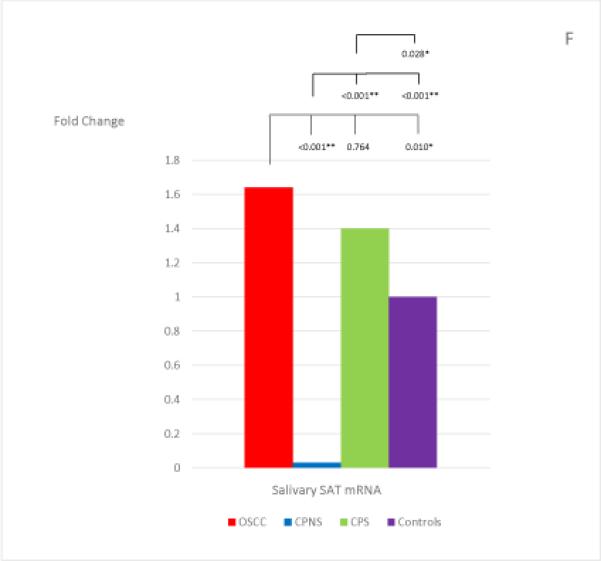
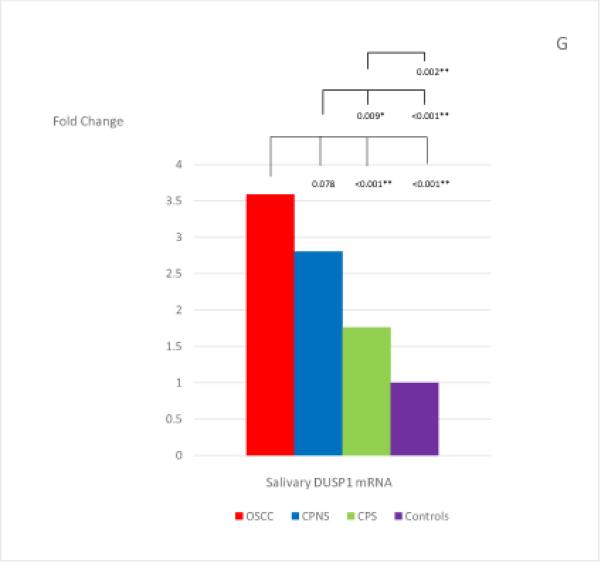
The mean fold changes of the 7 salivary mRNAs in each patient group when compared to the levels found in the Healthy Control group, and the statistical results for each pair of the study groups. Bracket with the p-value indicates the statistical result for each pair of study groups (“*” indicates marginal significance and “**” indicates statistical significance).
Among the 7 salivary mRNAs, only the S100P mRNA showed significantly higher levels in OSCC patients when compared to levels found in both periodontitis patient groups: CPNS (p=0.003) and CPS (p=0.007). The difference in levels between OSCC patients and Healthy Controls was also marginally significant (p=0.009). Three salivary mRNAs, IL-8, IL-1β, and DUSP1, showed the same tendency in that significantly higher levels were found in OSCC patients when compared to the levels found in CPS (p=0.003; p<0.001; and p<0.001, respectively) and Healthy Controls (p<0.001; p<0.001; and p<0.001, respectively); however, no significant difference were noted in the levels between OSCC and CPNS (p=0.510; p=0.058; and p=0.078, respectively). In addition, those 3 mRNAs also showed marginally significant or significantly higher levels in CPNS compared to the levels both in CPS (p<0.001; p<0.001; and p=0.009, respectively), and in Healthy Controls (p<0.001; p<0.001; and p<0.001, respectively). On the other hand, OAZ1 and SAT1 mRNAs both showed marginally significant or significantly higher levels in OSCC patients compared to levels in both CPNS (p<0.001 and p<0.001, respectively) and Healthy Controls (p=0.003 and p=0.010, respectively) but not CPS (p=0.318 and p=0.764, respectively). Both mRNAs also showed significantly lower levels in CPNS compared to levels in both CPS (p<0.001 and p<0.001, respectively) and Healthy Controls (p=0.003 and p<0.001, respectively).
Salivary H3F3A mRNA showed a significantly higher level in CPNS patients when compared to all other study groups: OSCC (p<0.001), CPS (p<0.001), and Healthy Controls (p<0.001). However, it did not show significantly different levels in OSCC patients compared to levels found in either CPS (p=0.449) or Health Controls (p=0.107).
Discussion
The 7 salivary mRNAs investigated in the present study had all been found previously to show significantly higher levels in OSCC patients compared to Healthy Controls, in 5 different cohorts of patients; and therefore, had been suggested as good salivary biomarker candidates for OSCC detection (15, 29). Our results, in comparing the levels found in OSCC patients and Healthy Controls, are mostly consistent with the 2 previously published studies, except that we did not find significantly different levels in the H3F3A mRNA between the OSCC patients and our Healthy Controls. However, in our study the presence of advanced periodontitis appears to significantly affect the levels of 5 out of these 7 salivary mRNAs which would affect their reliability as biomarkers for OSCC detection. The CPNS patients showed no significantly different levels of salivary IL-8, IL-1β, or DUSP1 mRNAs, compared with their levels found in OSCC patients; and the CPS patients showed no significantly different levels of salivary OAZ1 and SAT1 mRNAs. Although our findings seem disappointing, in light of the large research study that previously included these 5 mRNAs in a panel of the most promising salivary biomarkers for OSCC detection (29), we believe it is best to raise awareness among researchers that there is an urgent need to validate any promising salivary biomarkers for cancer detection by at least testing them against the common oral inflammatory disease, periodontitis. With this additional step of validation, a new panel or a modified panel from the study by Elashoff et al. (29) may be identified eventually—not only avoiding false positives but perhaps providing even higher sensitivity and specificity, which would be more useful for clinicians and their patients.
Among the 7 salivary mRNAs, only S100P showed significantly higher levels in OSCC patients compared to levels in both CP patient groups (regardless of smoking history), and the differences were also marginally significant between OSCC patients and Healthy Controls after Bonferroni corrections. In our previous work, we also found that salivary S100P mRNA showed significantly higher levels in OSCC patients when compared to the levels found both in OSCC patients-in-remission (who were at least two years post-treatment without any recurrence) and in patients with oral lichen planus (OLP)--a chronic, T-cell mediated inflammatory disease (regardless of OLP disease activity) (35). Taken together, these results make salivary S100P a very promising candidate as an OSCC-specific biomarker, whether or not the patient might also have a common chronic inflammatory condition in the oral cavity.
Our findings also suggest that the expression of S100P may be a distinguishing event in the development of OSCC from chronic inflammation. Indeed, these findings are consistent with current knowledge regarding the role of S100P in carcinogenesis, in which it contributes mainly to the process of invasion and metastasis (39, 40), a phenomenon that does not occur in the epithelial cells in chronic oral inflammatory diseases such as periodontitis and OLP. In addition, overexpression of S100P mRNA has been found specifically in the OSCC cells which were resistant to detachment-induced apoptosis, indicating its involvement in the metastatic process in OSCC (41). The S100P expression in OSCC appeared to be regulated by B-cell lymphoma/leukemia 10 (BCL 10), an apoptotic regulatory protein, through the STAT1/ATF4 signaling pathway (42). At this time, whether the increased salivary level of S100P in the OSCC patients found in our study reflects an increased S100P mRNA expression in the OSCC tissue is still unclear. There has been only one study so far investigating the mRNA expression of S100P in OSCC. Sapkota et al. (43) investigated the profile of 16 S100 gene-family members, including S100P, in OSCC patients in Sudan, and the authors did not find significantly increased levels of S100P mRNAs in the OSCC tissue specimens when compared to healthy controls. However, there is evidence to suggest that changes in gene expression of the S100 gene family members may vary among OSCCs that develop via different carcinogenesis mechanisms. Sapkota et al. (43) has pointed out that the high incidence of OSCC in Sudan is not associated with tobacco products as it is in Western countries, as social and cultural constraints make smoking and smokeless tobacco habits relatively uncommon there. OSCC in that country is strongly attributed to extensive consumption of toombak, an oral snuff containing a high amount of N-nitrosamine. They noted that while the S100A7 mRNA levels in OSCC patients showed no significant difference from the healthy controls from the Sudan population, its levels were significantly up-regulated in the OSCC patients from Norway when compared to paired healthy controls (43). In fact, up-regulated S100A7 mRNA expression has also been found in OSCC patients in other Western countries such as Germany and Poland (44-46). A study in Canada also found that over-expression of S100A7 protein in OSCC patients appeared to be associated with a reduced cancer-free survival period (47).
We found that the levels of salivary mRNAs of IL-8, IL-1β, and DUSP1 appeared to be significantly elevated by the presence of either OSCC or periodontal inflammation (in both smokers and non-smokers), when compared to the Health Controls. Levels of these 3 salivary mRNAs were elevated in CPNS patients to a degree that there was no significant difference from the levels found in OSCC patients, rendering them unreliable as salivary biomarkers for OSCC detection. In addition, when comparing the levels of these 3 salivary mRNAs between the smokers and non-smokers in CP patients, smoking appeared to reduce the levels of all 3 of them. Both IL-8 and IL-1β are proinflammatory cytokines and have been found to show significantly increased levels in periodontal inflammation as well as in OSCC (15, 18, 35, 48-50). Although the research on smoking effect on the various inflammatory cytokines in periodontal disease is still inconclusive, our findings are consistent with the conclusion of most previous studies that in periodontitis patients, smokers appeared to have decreased levels of several proinflammatory cytokines, including IL-8 and IL-1β (51-53).
The role of DUSP1 in OSCC is currently under investigation, and recent evidence suggests that it acts as a tumor supressor gene which regulates cancer-associated inflammation, and is significantly silenced in OSCC (54, 55). Interestingly, despite that salivary DUSP1 mRNA level has been found to be significantly elevated in OSCC patients when compared to healthy controls (15, 29, 35), DUSP1 mRNA and protein expression was found to be decreased in OSCC tissue when compared to the adjacent non-tumor control tissue (55). Although nicotine has recently been found to induce degradation of DUSP1 (56), the specific role of DUSP1 in periodontitis has not been studied so far. However, given the fact that DUSP1 is a key regulator of innate immune responses to mico-organisms (57), it would not be surprising if DUSP1 is eventually found to be involved in periodontitis.
An interesting finding in our study is that the salivary biomarkers that showed significantly different levels between the OSCC and the two periodontitis patient groups (CPNS and CPS) are mutually exclusive, i.e., mRNAs of IL-8, IL-1β, and DUSP1 levels were significantly higher in OSCC patients compared to those found in CPS, while mRNAs of OAZ1 and SAT (but not IL-8, IL-1β, and DUSP1) levels were significantly higher in OSCC patients compared to those found in CPNS. For the findings regarding IL-8, IL-1βand DUSP1 mRNAs, both CPS and CPNS indeed showed marginally significant or significantly higher levels of IL-8, IL-1β and DUSP1 when compared to the levels found in Healthy Controls (Fig. 1). The significantly higher levels found in OSCC patients when compared to the CPS patients were due to a reduction in the levels of these 3 mRNAs in the CPS group, and we speculated that this was due to the smoking habit which has been reported to decrease inflammatory cytokines such as IL-8 and IL-1β (51-53), as discussed above. In the same way, the significantly higher levels of OAZ1 and SAT1 found in OSCC patients when compared to the CPNS patients were also mostly attributed to the significantly lower levels of these two mRNAs in the CPNS group. In fact, the levels of these two mRNAs were significantly lower in CPNS when compared to the levels found in all other groups including Healthy Controls (Fig. 1). When the smoking factor is added to the periodontitis patients, as in the CPS group, it appears to increase the levels of both OAZ1 and SAT1 mRNAs, and that effect is indeed the same effect of OSCC to these two salivary mRNAs, as both OAZ1 and SAT1 showed marginally significant or significantly higher levels in OSCC patients when compared to those found in Healthy Controls (see Fig. 1). These findings support and reinforce the well-known notion that smoking is a risk factor for OSCC. In addition, the increase in both OAZ1 and SAT1 levels in CPS patients has reached a degree that results in no significant difference in their levels when compared to the levels found in the OSCC patients. We unfortunately do not have a Healthy Control group who are smokers for comparison. However, all previous published studies regarding these 7 salivary mRNAs did include smokers in their Healthy Controls (15, 28, 29). Therefore, this finding combined with the findings in previously published studies suggest that while both OAZ1 and SAT1 mRNAs could be potential OSCC salivary biomarkers in individuals who are non-smokers, regardless of periodontal status, they would not be reliable OSCC salivary biomarkers in periodontitis patients who have a smoking habit, a known risk factor for OSCC.
The dramatically low levels of salivary OAZ1 and SAT1 mRNAs in CPNS patients are indeed a novel finding in our study and worth further discussion. OAZ1 is a member of ornithine decarboxylase antizymes (OAZ) which inhibit ornithine decarboxylase (ODC). Both ODC and SAT1 are key regulatory enzymes in the polyamine biosynthesis and interconversion pathway (58-60). As increased ODC activity has been found in most human cancers (58, 61), and it has been suggested that OAZ functions as a tumor suppressor (62-64). Increased ODC mRNA expression due to smoking also has been reported in the ventricular myocardium of hypertensive rats (65), and in epithelium and smooth muscles of the airway in asthma patients (66). However, smoking effect to OAZ1 and SAT1 in periodontitis patients is unknown, except for our finding in the CPS group in this study. Since all our Healthy Control group patients were non-smokers, our finding in CPNS indicate that periodontal inflammation alone could significantly reduce salivary OAZ1 and SAT1 mRNAs. This finding indicates dysregulation of polyamine in periodontal disease, and significantly lower salivary levels of OAZ1 suggest a possible higher level of ODC in CP patients. This also appears to be consistent with a previously reported finding that salivary ODC levels were significantly higher in CP patients compared to healthy controls (67). Although salivary mRNAs of OAZ1 and SAT1 have been found to show significantly higher levels in OSCC patients (15, 28, 29), their levels did not appear to be significantly higher in patients who have oral lichen planus (OLP), another chronic oral inflammatory disease with a different pathogenesis from periodontitis, when compared to healthy controls (35). The dramatically low levels of salivary OAZ1 and SAT1 mRNAs, which is the opposite trend from OSCC and is a different trend from OLP, also suggest the possibility that these two salivary mRNAs may be unique salivary biomarkers for periodontitis. This leads to a new direction for further research in the relationship between periodontitis and polyamine biosynthesis.
In conclusion, we found that only salivary S100P mRNA could be a reliable biomarker for OSCC detection, regardless of the presence of CP; and that the presence of CP would affect reliability for using 6 of the 7 promising salivary mRNA biomarkers for oral cancer detection.
Acknowledgements
This study was funded by the National Institute of Dental and Craniofacial Research (1R21DE018757-01A2), by the Texas A&M Health Science Center Faculty Bridge Grant, by the Baylor Oral Health Foundation, and by the Office of the Associate Dean for Research, and the Department of Diagnostic Sciences, Texas A&M University-Baylor College of Dentistry. All authors declare no conflicts of interest. We wish to thank Dr. Ole Brinkmann for technical advices on qPCR with saliva samples, and Dr. David Wong, Associate Dean of Research at UCLA, for his invaluable advice and support, and all doctors who have assisted us in the process of patient recruitment for this study.
Reference
- 1.Wong DT. Salivary diagnostics for oral cancer. Journal of the California Dental Association. 2006;34:303–308. [PubMed] [Google Scholar]
- 2.Cheng YS, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clinical and translational medicine. 2014;3:3. doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Xiao H, Wong DT. Salivary biomarkers for clinical applications. Mol Diagn Ther. 2009;13:245–259. doi: 10.1007/BF03256330. [DOI] [PubMed] [Google Scholar]
- 4.Castagnola M, Picciotti PM, Messana I, et al. Potential applications of human saliva as diagnostic fluid. Acta Otorhinolaryngol Ital. 2011;31:347–357. [PMC free article] [PubMed] [Google Scholar]
- 5.Lima DP, Diniz DG, Moimaz SA, Sumida DH, Okamoto AC. Saliva: reflection of the body. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2010;14:e184–188. doi: 10.1016/j.ijid.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Flake C, Arafa J, Hall A, Ence E, Howard K, Kingsley K. Screening and detection of human papillomavirus (HPV) high-risk strains HPV16 and HPV18 in saliva samples from subjects under 18 years old in Nevada: a pilot study. BMC oral health. 2012;12:43. doi: 10.1186/1472-6831-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore C, Crouch D. Oral fluid for the detection of drugs of abuse using immunoassay and LC-MS/MS. Bioanalysis. 2013;5:1555–1569. doi: 10.4155/bio.13.115. [DOI] [PubMed] [Google Scholar]
- 8.Lee D, Huestis MA. Current knowledge on cannabinoids in oral fluid. Drug testing and analysis. 2014;6:88–111. doi: 10.1002/dta.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller CS, Foley JD, 3rd, Floriano PN, et al. Utility of salivary biomarkers for demonstrating acute myocardial infarction. J Dent Res. 2014;93:72S–79S. doi: 10.1177/0022034514537522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-kappaB dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect Prev. 2005;29:42–45. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Katakura A, Kamiyama I, Takano N, et al. Comparison of salivary cytokine levels in oral cancer patients and healthy subjects. The Bulletin of Tokyo Dental College. 2007;48:199–203. doi: 10.2209/tdcpublication.48.199. [DOI] [PubMed] [Google Scholar]
- 12.SahebJamee M, Eslami M, AtarbashiMoghadam F, Sarafnejad A. Salivary concentration of TNFalpha, IL1 alpha, IL6, and IL8 in oral squamous cell carcinoma. Medicina oral, patologia oral y cirugia bucal. 2008;13:E292–295. [PubMed] [Google Scholar]
- 13.St John MA, Li Y, Zhou X, et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 14.Arellano-Garcia ME, Hu S, Wang J, et al. Multiplexed immunobead-based assay for detection of oral cancer protein biomarkers in saliva. Oral diseases. 2008;14:705–712. doi: 10.1111/j.1601-0825.2008.01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, St John MA, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 16.Vucicevic Boras V, Cikes N, Lukac J, Virag M, Cekic-Arambasin A. Salivary and serum interleukin 6 and basic fibroblast growth factor levels in patients with oral squamous cell carcinoma. Minerva Stomatol. 2005;54:569–573. [PubMed] [Google Scholar]
- 17.Bahar G, Feinmesser R, Shpitzer T, Popovtzer A, Nagler RM. Salivary analysis in oral cancer patients: dNA and protein oxidation, reactive nitrogen species, and antioxidant profile. Cancer. 2007;109:54–59. doi: 10.1002/cncr.22386. [DOI] [PubMed] [Google Scholar]
- 18.Mankan AK, Lawless MW, Gray SG, Kelleher D, McManus R. NF-kappaB regulation: the nuclear response. J Cell Mol Med. 2009;13:631–643. doi: 10.1111/j.1582-4934.2009.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hom D. Growth factors in wound healing. Otolaryngol Clin North Am. 1995;28:933–953. [PubMed] [Google Scholar]
- 20.Canakci C, Cicek Y, Yildirim A, Sezer U, Canakci V. Increased levels of 8-hydroxydeoxyguanosine and malondialdehyde and its relationship with antioxidant enzymes in saliva of periodontitis patients. European Journal of Dentistry. 2009;3:100–106. [PMC free article] [PubMed] [Google Scholar]
- 21.Ebersole JL, Schuster JL, Stevens J, et al. Patterns of salivary analytes provide diagnostic capacity for distinguishing chronic adult periodontitis from health. Journal of clinical immunology. 2013;33:271–279. doi: 10.1007/s10875-012-9771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma M, Bairy I, Pai K, et al. Salivary IL-6 levels in oral leukoplakia with dysplasia and its clinical relevance to tobacco habits and periodontitis. Clinical oral investigations. 2011;15:705–714. doi: 10.1007/s00784-010-0435-5. [DOI] [PubMed] [Google Scholar]
- 23.Gursoy UK, Kononen E, Uitto VJ, et al. Salivary interleukin-1beta concentration and the presence of multiple pathogens in periodontitis. Journal of clinical periodontology. 2009;36:922–927. doi: 10.1111/j.1600-051X.2009.01480.x. [DOI] [PubMed] [Google Scholar]
- 24.Albandar JM. Underestimation of periodontitis in NHANES surveys. J Periodontol. 2011;82:337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- 25.Burt B. Position paper: epidemiology of periodontal diseases. J Periodontol. 2005;76:1406–1419. doi: 10.1902/jop.2005.76.8.1406. [DOI] [PubMed] [Google Scholar]
- 26.Data statistics/ periodontal (gum) disease. National Institute of Dental and Craniofacial Research; Anonymous. [Google Scholar]
- 27.Dye BA. Global periodontal disease epidemiology. Periodontology 2000. 2012;58:10–25. doi: 10.1111/j.1600-0757.2011.00413.x. [DOI] [PubMed] [Google Scholar]
- 28.Brinkmann O, Kastratovic DA, Dimitrijevic MV, et al. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47:51–55. doi: 10.1016/j.oraloncology.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elashoff D, Zhou H, Reiss J, et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev. 2012;21:664–672. doi: 10.1158/1055-9965.EPI-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mashberg A, Boffetta P, Winkelman R, Garfinkel L. Tobacco smoking, alcohol drinking, and cancer of the oral cavity and oropharynx among U.S. veterans. Cancer. 1993;72:1369–1375. doi: 10.1002/1097-0142(19930815)72:4<1369::aid-cncr2820720436>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 31.Blot WJ, McLaughlin JK, Winn DM, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 32.Lewin F, Norell SE, Johansson H, et al. Smoking tobacco, oral snuff, and alcohol in the etiology of squamous cell carcinoma of the head and neck: a population-based case- referent study in Sweden. Cancer. 1998;82:1367–1375. doi: 10.1002/(sici)1097-0142(19980401)82:7<1367::aid-cncr21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Eke PI, Page RC, Wei L, Thornton-Evans G, Genco RJ. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol. 2012;83:1449–1454. doi: 10.1902/jop.2012.110664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng YS, Jordan L, Gorugantula L, Schneiderman E, Chen H, Rees T. Salivary interleukins 6 and 8 in oral cancer patients and in patients with chronic oral inflammatory diseases. Journal of Periodontology. 2014;85:956–965. doi: 10.1902/jop.2013.130320. [DOI] [PubMed] [Google Scholar]
- 35.Cheng YS, Jordan L, Rees T, et al. Levels of potential oral cancer salivary mRNA biomarkers in oral cancer patients in remission and oral lichen planus patients. Clinical oral investigations. 2014;18:985–993. doi: 10.1007/s00784-013-1041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navazesh M. Methods for collecting saliva. Ann NY Acad Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu Z, Zimmermann BG, Zhou H, et al. Exon-level expression profiling: a comprehensive transcriptome analysis of oral fluids. Clin Chem. 2008;54:824–832. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nuclei Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011;41:885–892. doi: 10.1007/s00726-010-0495-5. [DOI] [PubMed] [Google Scholar]
- 40.Tothova V, Gibadulinova A. S100P, a peculiar member of S100 family of calcium- binding proteins implicated in cancer. Acta virologica. 2013;57:238–246. doi: 10.4149/av_2013_02_238. [DOI] [PubMed] [Google Scholar]
- 41.Kupferman M, Patel V, Sriuranpong V, et al. Molecular analysis of anoikis resistance in oral cavity squamous cell carcinoma. Oral Oncol. 2007;43:440–454. doi: 10.1016/j.oraloncology.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Wu TS, Tan CT, Chang CC, et al. B-cell lymphoma/leukemia 10 promotes oral cancer progression through STAT1/ATF4/S100P signaling pathway. Oncogene. 2015;34:1207–1219. doi: 10.1038/onc.2014.43. [DOI] [PubMed] [Google Scholar]
- 43.Sapkota D, Bruland O, Bue O, et al. Expression profile of the S100 gene family members in oral squamous cell carcinomas. J Oral Pathol Med. 2008;37:607–615. doi: 10.1111/j.1600-0714.2008.00683.x. [DOI] [PubMed] [Google Scholar]
- 44.Kesting MR, Sudhoff H, Hasler RJ, et al. Psoriasin (S100A7) up-regulation in oral squamous cell carcinoma and its relation to clinicopathologic features. Oral Oncol. 2009;45:731–736. doi: 10.1016/j.oraloncology.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Winter J, Pantelis A, Reich R, et al. Risk estimation for a malignant transformation of oral lesions by S100A7 and Doc-1 gene expression. Cancer investigation. 2011;29:478–484. doi: 10.3109/07357907.2011.597813. [DOI] [PubMed] [Google Scholar]
- 46.Tyszkiewicz T, Jarzab M, Szymczyk C, et al. Epidermal differentiation complex (locus 1.q21) gene expression in head and neck cancer and normal mucosa. Folia histochemica et cytobiologica / Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2014;52:79–89. doi: 10.5603/FHC.2014.0018. [DOI] [PubMed] [Google Scholar]
- 47.Kaur J, Matta A, Kak I, et al. S100A7 overexpression is a predictive marker for high risk of malignant transformation in oral dysplasia. Int J Cancer. 2014;134:1379–1388. doi: 10.1002/ijc.28473. [DOI] [PubMed] [Google Scholar]
- 48.Milward M, Chapple I, Wright H, Millard J, Matthews J, Cooper P. Differential activation of NF-κB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol. 2007;148:307–324. doi: 10.1111/j.1365-2249.2007.03342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duarte P, de Oliveira M, Tambeli C, Parada C, Casati M, Nociti FJ. Overexpression of interleukin-1β and interleukin-6 may play an important role in periodontal breakdown in type 2 diabetic patients. J Periodont Res. 2007;42:377–381. doi: 10.1111/j.1600-0765.2006.00961.x. [DOI] [PubMed] [Google Scholar]
- 50.Moutsopoulos N, Madianos P. Low-grade inflammation in chronic infectious diseases, Paradigm of periodontal infections. Annals of the New York Academy of Sciences. 2006;1088:251–264. doi: 10.1196/annals.1366.032. [DOI] [PubMed] [Google Scholar]
- 51.Johannsen A, Susin C, Gustafsson A. Smoking and inflammation: evidence for a synergistic role in chronic disease. Periodontology 2000. 2014;64:111–126. doi: 10.1111/j.1600-0757.2012.00456.x. [DOI] [PubMed] [Google Scholar]
- 52.Tymkiw KD, Thunell DH, Johnson GK, et al. Influence of smoking on gingival crevicular fluid cytokines in severe chronic periodontitis. Journal of clinical periodontology. 2011;38:219–228. doi: 10.1111/j.1600-051X.2010.01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawlinson A, Grummitt JM, Walsh TF, Ian Douglas CW. Interleukin 1 and receptor antagonist levels in gingival crevicular fluid in heavy smokers versus non-smokers. Journal of clinical periodontology. 2003;30:42–48. doi: 10.1034/j.1600-051x.2003.300107.x. [DOI] [PubMed] [Google Scholar]
- 54.Khor GH, Froemming GR, Zain RB, et al. DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma. International journal of medical sciences. 2013;10:1727–1739. doi: 10.7150/ijms.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Hyer JM, Yu H, D'Silva NJ, Kirkwood KL. DUSP1 phosphatase regulates the proinflammatory milieu in head and neck squamous cell carcinoma. Cancer Res. 2014;74:7191–7197. doi: 10.1158/0008-5472.CAN-14-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y, Song P, Zhang W, et al. Activation of AMPKalpha2 in adipocytes is essential for nicotine-induced insulin resistance in vivo. Nature medicine. 2015;21:373–382. doi: 10.1038/nm.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham SM, Clark AR. Dual-specificity phosphatase 1: a critical regulator of innate immune responses. Biochemical Society transactions. 2006;34:1018–1023. doi: 10.1042/BST0341018. [DOI] [PubMed] [Google Scholar]
- 58.Pegg A. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- 59.Pegg A. Spermidine/spermine-N′-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrino Metab. 2008;294:E995–E1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 60.Heller J, Fong W, Canellakis E. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci USA. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shantz L, Levin V. Regulation of ornithine decarboxylase during oncogenic transformation: mechanisms and therapeutic potential. Amino Acids. 2007;33:213–223. doi: 10.1007/s00726-007-0531-2. [DOI] [PubMed] [Google Scholar]
- 62.Koike C, Chan D, Zetter B. Sensitivity to polyamine-induced growth arrest correlates with antizyme induction in prostate carcinoma cells. Cancer Res. 1999;59:6109–6112. [PubMed] [Google Scholar]
- 63.Iwata S, Sato Y, Asada M, et al. Anti-tumor activity of antizyme which targets the ornithine carboxylase (ODC) required for cell growth and transformation. Oncogene. 1999;18:165–172. doi: 10.1038/sj.onc.1202275. [DOI] [PubMed] [Google Scholar]
- 64.Tsuji T, Usui S, Aida T, et al. Induction of epithelial differentiation and DNA demethylation in hamster malignant oral keratinocyte by ornithine decarboxylase antizyme. Oncogene. 2001;20:24–33. doi: 10.1038/sj.onc.1204051. [DOI] [PubMed] [Google Scholar]
- 65.Meurrens K, Ruf S, Ross G, Schleef R, von Holt K, Schluter KD. Smoking accelerates the progression of hypertension-induced myocardial hypertrophy to heart failure in spontaneously hypertensive rats. Cardiovascular research. 2007;76:311–322. doi: 10.1016/j.cardiores.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 66.Bergeron C, Boulet LP, Page N, et al. Influence of cigarette smoke on the arginine pathway in asthmatic airways: increased expression of arginase I. The Journal of allergy and clinical immunology. 2007;119:391–397. doi: 10.1016/j.jaci.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 67.Ozer L, Elgun S, Ozdemir B, Pervane B, Ozmeric N. Arginine-nitric oxide-polyamine metabolism in periodontal disease. J Periodontol. 2011;82:320–328. doi: 10.1902/jop.2010.100199. [DOI] [PubMed] [Google Scholar]


