Abstract
Cruciferous vegetable intake is associated with reduced risk of bladder cancer, yet mechanisms remain unclear. Cruciferous vegetable isothiocyanates (ITCs), namely sulforaphane (SFN) and erucin (ECN), significantly inhibit histone deacetylase (HDAC) activity in human bladder cancer cells representing superficial to invasive biology (59–83% inhibition with 20μM, 48h treatment), and in bladder cancer xenografts (59±3% ECN inhibition). Individual HDACs inhibited by SFN and ECN include HDACs 1, 2, 4 and 6. Interestingly, global acetylation status of histones H3 or H4 remain unaltered. The interplay between HDAC inhibition and modest modulation of AcH3 and AcH4 status is partially explained by decreased histone acetyl transferase activity (48.8±5.3%). In contrast, a significant decrease in phosphorylation status of all isoforms of histone H1 was observed, concomitant with increased phosphatase PP1β and PP2A activity. Together, these findings suggest that ITCs modulate histone status via HDAC inhibition and phosphatase enhancement. This allows for reduced levels of histone H1 phosphorylation, a marker correlated with human bladder cancer progression. Therefore, ITC-mediated inhibition of histone H1 phosphorylation presents a novel direction of research in elucidating epidemiological relationships and supports future food-based prevention strategies.
Keywords: Cruciferous Vegetables, Broccoli, Bladder Cancer, Isothiocyanates, Sulforaphane, Erucin, Histone Acetylation, Histone Phosphorylation
Graphical Abstract

Introduction
The field of epigenetics, where genomic modifications alter patterns of gene regulation without altering the nucleotide sequence, has emerged as a crucial area for understanding how our genomes function [1–3]. The ability of epigenetic modulators, both pharmacologic and natural compounds from dietary sources, to impact carcinogenesis is beginning to be elucidated. Unlike genetic mutations, which are difficult to reverse, epigenetic modulation is dynamic and may lead to changes in gene regulation that can be manipulated by drugs or phytochemicals found in foods [4–7]. Two principal mechanisms have been identified that contribute to epigenetic modulation of transcription, namely histone modification and DNA methylation [8]. DNA methylation in regions with high GC content (CpG-islands) is the most widely studied modification and is associated with gene silencing [9]. Histones are also key players in transcriptional regulation via binding to DNA, a process modulated by acetylation and phosphorylation. The histones are comprised of four core histones (H2A, H2B, H3 and H4) and the linker histone H1. Two H2A-H2B dimers and one H3-H4 tetramer associate to form the histone octamer, which binds to 147 bp of DNA, wrapping 1.65 turns around the histone octamer to form a nucleosome with about 50 bp of DNA between each nucleosome. Histone H1 is called the “linker histone” because rather than forming part of the nucleosome core, it binds the DNA entry/exit points of the nucleosome [9–11].
Histones are largely globular in structure, except for their N-terminal tails, which are unstructured. It is on these N-terminal tails that histones possess a diverse set of post-translational modifications. These modifications affect chromatin structure and protein-protein interactions and are instrumental in regulating gene transcription. Histone modifications include: acetylation, methylation, phosphorylation, ubiquitination, SUMOlyation, and ADP-ribosylation, among others [9]. A euchromatic gene conformation is usually associated with high levels of acetylation and trimethylated H3K4, H3K36 and H3K79. Conversely, heterochromatin is characterized by low levels of acetylation and high levels of H3K9, H3K27 and H4K20 methylation [9]. Global loss of monoacetylation and trimethylation of histone H4 are common markers detected in human cancer cells [12]. In addition, we have shown a significant increase in phosphorylation of H1 linker histones in the progression from normal bladder epithelial cells to low-grade superficial to high-grade invasive bladder cancer, reflecting bladder carcinogenesis [13–16].
Histone acetylation status is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [17]. An increase in HAT activity and/or an inhibition of HDAC activity, leads to higher histone acetylation status. HDACs are grouped into four classes, based on their sequence homology to their yeast orthologs: class I (HDAC 1, 2, 3 and 8), class IIa (HDAC 4, 5, 7 and 9), class IIb (HDAC 6 and 10), class III (Sirtuins) and class IV (HDAC 11). Class I, II and IV are referred to as “classical” HDACs and have Zn2+-dependent catalytic activity, while class III consists of the Sirtuins and require NAD+ as an essential cofactor [8]. Class I HDACs are found exclusively in the nucleus, while class II shuttle between the nucleus and cytoplasm [12]. The term “HDAC inhibitor” (HDACI) is commonly used for compounds that target classical HDACs [18]. HDACIs have been shown to strongly and selectively induce growth arrest, differentiation and apoptosis in many cancer types [8].
Although the effect of HDACIs on the histone code is increasingly appreciated, this may not be the sole mechanism of action responsible for the anti-cancer effect of HDACIs. Emerging evidence points to HDACI modulation of non-histone targets, including tubulin, p53, Ku70, Hsp9 and STAT3, among many others [8]. These targets can be referred to as the acetylome [19]. In addition, HDACIs have been reported to downregulate Akt phosphorylation by interrupting the interaction between protein phosphatase 1 (PP1) with HDAC 1 or 6 [4]. These emerging targets may more aptly explain the effects of HDACIs on cancer.
We have pursued potential mechanisms whereby cruciferous vegetable phytochemicals may impact bladder carcinogenesis based upon evidence from epidemiologic studies [20, 21]. In the past, we showed the positive correlation of H1 phosphorylation and bladder cancer carcinogenesis and progression. We hypothesize that long-term, low-toxicity dietary modulation of the epigenome may inhibit the carcinogenic process, by preventing silencing of tumor suppressor genes, and reducing H1 phosphorylation, among other mechanisms. We and others have shown the ability of broccoli isothiocyanates, namely sulforaphane (SFN) and erucin (ECN), to inhibit bladder cancer growth in vitro and in vivo by inducing cell cycle modulation in the G2/M phase as well as apoptosis [22–26]. HDACIs have been reported to disrupt the cell cycle in the G2 phase and induce apoptosis which mirrors our findings and led us to suspect epigenetic modulation as a plausible mechanism of action by which these compounds may act. Furthermore, it has been reported that SFN can act as an HDACI [12, 27, 28]. Prior reports by Dashwood and colleagues ([29–31] and reviewed extensively in [27]and [32]) showed that the isothiocyanate, sulforaphane (SFN), behaved as an HDAC inhibitor in colon cells. This was supported by numerous immunoblot experiments measuring acetylated histones including H3 and H4. Immunoblotting of histone modifications is not always reliable due to cross reactivity and interfering neighboring modifications. This evidence led us to further examine the potential of isothiocyanates to impact histone status thereby influencing their potential as epigenetic modulators of bladder carcinogenesis and progression [33]. When we examined this effect using mass spectrometry we did not observe the same extent of HDAC inhibition in bladder cancer cells, and we feel that it is an important observation. However, quite remarkably, we did see a very significant change in the histone phosphorylation, which is an indicator of altered phosphatase / kinase activity.
Here we show cruciferous vegetable isothiocyanates, namely sulforaphane (SFN) and erucin (ECN) significantly inhibited histone deacetylase (HDAC) activity in vitro in human bladder cancer cells representing superficial to invasive biology, as well as in invasive bladder cancer in vivo xenografts. Individual HDAC activity inhibited by SFN and ECN included HDACs 1, 2, 4 and 6 while HDACs 3, 5, 7–10 and SIRT1–2 activity remained unchanged. Interestingly, global acetylation status of histones H3 or H4 as revealed by LC-MS remained unaltered while the interplay between HDAC inhibition and modest modulation of AcH3 and AcH4 status may be partially explained by decreased histone acetyl transferase activity in isothiocyanate treated cells. In contrast to the small changes in acetylated histone status, a significant decrease in phosphorylation status of all isoforms of histone H1 was observed, concomitant with increased phosphatase PP1β and PP2A activity. Taken together, these findings suggest that cruciferous vegetable isothiocyanates SFN and ECN modulate histone status and act via epigenetic mechanisms in human bladder cancer cells via HDAC inhibition and phosphatase enhancement allowing for reduced levels of histone H1 phosphorylation, a mark we previously correlated with human bladder cancer progression [13]. Therefore, SFN- and ECN-mediated targeting of histone H1 phosphorylation via epigenetic modulation presents a novel direction of research to elucidate epidemiological relationships and perhaps support future studies of food based prevention strategies in high risk cohorts.
Materials and Methods
Cell Culture
RT4, J82 and UMUC3 human bladder cancer cells were purchased from American Type Culture Collection (Manassas, VA) and cultured in Gibco RPMI-1640 medium (Invitrogen Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum, L-glutamine, penicillin and streptomycin. All cells were grown as monolayer cultures at 37 °C in a 95% air/5% CO2 humidified atmosphere.
In Vivo Mouse Xenografts
Mouse xenograft studies were carried out with strict adherence to protocols approved by the Institutional Animal Care and Use Committee of The Ohio State University as previously described (Animal Protocol 2007A0167-R1; PI: A. Mortazavi) [22]. Briefly, female athymic nude mice (FOXN1nu 4–5 weeks of age), were obtained from Harlan Laboratories (Indianapolis, IN) and acclimated on AIN93G pelleted diet for one week. Mice where then subcutaneously inoculated with 0.05 × 106 UMUC3 cells, in 0.1 mL of Matrigel (BD Biosciences, San Jose, CA; 50% [v/v] in serum-free medium), in the left and right dorsal flank. Three days after injection, mice were randomized into one of three treatment groups (12 mice/group): 1) vehicle control soybean oil oral gavage once daily 2) 295 μmol/kg sulforaphane oral gavage once daily 3) 295 μmol/kg erucin oral gavage once daily. Mice were anesthetized with isofluorane prior to gavage treatments. Mice were sacrificed after 2 weeks, when tumors reached approximately 1.2 cm in diameter. Tumors and bladders were collected at sacrifice and flash frozen in liquid nitrogen. Tumor and bladder tissue protein extraction was performed using the T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Rockford, IL) per the manufacturer’s protocol. The amount of protein was quantified and normalized using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL) and subjected to the HDAC Activity Assay.
HDAC Activity Assay
RT4 cells (1 x 106) and J82 or UMUC3 cells (2 x 106) were seeded overnight in culture dishes in 20 mL of media. Cells were then treated with 5, 10 or 20 μM concentrations of R,S, sulforaphane (LKT Laboratories, Inc., St. Paul, MN) or erucin (LKT Laboratories, Inc., St. Paul, MN) diluted in 10% FBS supplemented RPMI 1640 or DMSO vehicle at a concentration equal to that of drug-treated cells (final DMSO concentration, ≤0.1% by volume) for 48 hours. Cells were collected and lysed in non-denaturing Cell Lysis Buffer according to manufacturer’s protocol (Cell Signaling Technology, Inc., Danvers, MA). Protein concentrations of the cell lysates were determined and normalized by using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). Relative HDAC activity was measured using the HDAC Fluorimetric Assay/Drug Discovery Kit (Enzo Life Sciences, Inc., Farmingdale, NY) per manufacturer’s protocol.
Individual HDAC Activity
UMUC3 cells (2 x 106) were seeded overnight in culture dishes in 20 mL of media. Cells were then treated with 20 μM concentrations of R,S, sulforaphane, erucin or DMSO vehicle control for 48 hours. Cells were collected and lysed in non-denaturing Cell Lysis Buffer as described above followed by protein concentration determination using the BCA Protein Assay as described above. Individual HDACs or SIRTs were immunoprecipitated from normalized protein lysates using the Seize X Protein A/G Immunoprecipitation Kit (Thermo Fisher Scientific, Rockford, IL). The washed beads with bound HDAC/Anti-HDAC complexes were incubated with 150 μL of 100 μM Fluor de Lys® Substrate from the HDAC Fluorimetric Assay/Drug Discovery Kit (Enzo Life Sciences, Inc., Farmingdale, NY) for 10 minutes with rocking (37 °C). Aliquots (30 μL) were withdrawn, mixed with 20 μL Assay buffer and 50 μL Fluor de Lys® Developer, and fluorescence measured according to manufacturer’s protocol. Primary antibodies used were HDAC 1, 2, 3, 4, 5 and 7 (Cell Signaling Technology, Inc., Danvers, MA) and HDAC 6, 8, 9, 10, SIRT1, 2 (Abcam, Cambridge, MA). Western blots were run to confirm that the correct enzymes were immunoprecipitated. Secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
HAT Activity
Cells were seeded overnight in culture dishes at 6 x 106 in 20 mL of media for RT4 and UMUC3 cells and treated with 5, 10 or 20 μM concentrations of erucin diluted in 10% FBS supplemented RPMI 1640 or DMSO vehicle for 48 hours. Nuclear extracts were obtained using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to protocol. Protein concentrations of the cell lysates were determined and normalized by using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). Relative HAT activity was measured using the HAT Assay Kit (Active Motif, Carlsbad, CA) according to protocol except 50 μg of nuclear extracts was substituted for p300.
Western Blotting
Cells were seeded overnight in culture dishes at 1 x 106 in 20 mL of media for RT4 and 2 x 106 in 20 mL of media for UMUC3 cells and treated with 5, 10 or 20 μM concentrations of sulforaphane or erucin for 48 hours. Cells were collected and lysed in SDS Lysis buffer (EMD Millipore, Billerica, MA). Lysate protein concentration was determined and normalized by the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). SDS-PAGE was performed according to manufacturer’s protocols provided with NuPAGE pre-cast gels (Invitrogen Life Technologies, Grand Island, NY) and then transferred onto PVDF membranes (Invitrogen Life Technologies, Grand Island, NY). Proteins were visualized by chemiluminescence (Western Blotting Luminol Reagent, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Primary antibodies used were acetylated histone H3, histone H3, thymidylate synthase (Cell Signaling Technology, Inc., Danvers, MA), p21 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), GAPDH, acetylated tubulin (Sigma-Aldrich, St. Louis, MO), pan-phospho histone H1 and histone H1 (Abcam, Cambridge, MA). Secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. Densitometry using the ImageJ (NIH, Bethesda, MD) program was utilized to quantify relative protein expression.
Histone Extraction
Cells were seeded overnight in culture dishes at 1 x 106 in 20 mL of media for RT4 and 2 x 106 in 20 mL of media for UMUC3 cells and treated with 20 μM concentrations of R,S-sulforaphane (LKT Laboratories, Inc, St. Paul, MN), or erucin (LKT Laboratories, Inc, St. Paul, MN) diluted in 10% FBS supplemented RPMI 1640 or DMSO vehicle (final DMSO concentration, ≤0.1% by volume) for 3 hours. After treatment, the cells were harvested by scraping and then snap frozen. Histones were extracted as described previously [34]. Reagents for histone extraction were obtained from Sigma-Aldrich (St. Louis, MO). Briefly, cell pellets were resuspended in 1 mL of NP-40 lysis buffer [10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine, 1 mM PMSF, protease inhibitor cocktail (1:1000)] and incubated on ice for 5 min. The nuclei were pelleted at 1,500 rpm for 15 min at 4 °C and the pellet washed with 1 mL TBS [10 mM Tris-HCl (pH 7.4), 150 mM NaCl]. 0.2 M sulfuric acid (H2SO4) was added to the washed pellet to extract the histones and it was vortexed and incubated on ice for 30 min. The solution was centrifuged at 12,045 x g for 15 min at 4 °C to remove the cellular debris. 80% acetone was added to the supernatant and precipitated at −20 °C overnight. The precipitated histones were centrifuged at 12,045 x g for 15 min at 4 °C, allowed to air-dry for 10 min and resuspended in HPLC water.
Liquid Chromatography / Mass Spectrometry
Protein concentration was determined by Bradford analysis (BioRad, Hercules, CA). 30 μg of purified histones were characterized by LC-MS analysis. LC-MS analysis was performed with a Dionex U3000 HPLC (Dionex; Sunnyvale, CA) coupled to a MicroMass Q-TOF (Micromass; Whythenshawe, UK). Reversed-phase separation was carried out on a Discovery Bio Wide Pore C18 column (1.0 mm x 150 mm, 5 μm, 300 Å; Supelco, USA). Mobile phases A and B consisted of water and acetonitrile with 0.05% trifluoroacetic acid, respectively. The flow rate was 25 μL/min and the gradient started at 20% B, increased linearly to 30% B in 2 min, to 35% B in 8 min, 50% B in 20 min, 60% B in 5 min and 95% B in 1 min. After washing at 95% B for 4 min, the column was equilibrated at 20% B for 30 min and a blank was run between each sample injection. The cone voltage on the Q-Tof was 25 V. LC-MS data was deconvoluted using MassLynx 4.1.
Mass Spectrometry Data Analysis
The LC-MS experiment was repeated four times from different starting cell cultures. The raw LC-MS data was summed across each eluting peak in the chromatogram and the resulting summed mass was deconvoluted to produce a zero-charge mass spectrum. Peak heights were used to determine the extent of histone modification for linker histone H1 variants and core histone H3 and H4 variants. The weighted average mass for the distributions of the histone variants H3.1 and H3.2 was calculated to determine the extent of acetylation after SFN and ECN treatment as described previously [35]. The mass*height product for each H3 isoform peak was summed. This summed value was divided by the total height of all of the peaks in the distribution. This calculation was repeated for each replicate. The average extent of acetylation and the standard error were then determined. A similar approach was used to determine the significance of H4 acetylation and H1 phosphorylation changes. In the case of H4, the summed peak heights for the isoforms with 2–4 acetylations were compared to the N-terminally acetylated isoform. For H1, the summed peak heights for all modified phosphorylated isoforms of a particular H1 variant were compared to the unphosphorylated isoform.
Phosphatase Activity Assay
UMUC3 cells were seeded overnight in culture dishes at 2 x 106 in 20 mL of media and treated with 20 μM concentrations of R,S-sulforaphane, or erucin (LKT Laboratories, Inc., St. Paul, MN) diluted in 10% FBS supplemented RPMI 1640 or DMSO vehicle (final DMSO concentration, ≤0.1% by volume) for 3 hours. Cells were collected and lysed in non-denaturing Cell Lysis Buffer according to manufacturer’s protocol (Cell Signaling Technology, Danvers, MA). Protein concentrations of the cell lysates were determined and normalized by using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). PP1β and PP2A were immunoprecipitated and phosphatase activity was assayed using the Malachite Green Phosphatase Assay (EMD Millipore, Billerica, MA).
Statistics
Statistical significance for activity assays and western blot densitometry analysis was tested by two-tailed Student’s t-test, using Excel or one way analysis of variance (ANOVA) followed by either the Student-Newman-Keuls multiple comparisons test or Bonferroni post hoc analysis, using Instat software (GraphPad, La Jolla, CA). Significance testing for mass spec based analyses were performed using ANOVA via Microsoft Excel. An F-test was first performed to determine if the variances were equal. A two-tailed Student’s T-test was then performed to ascertain if the changes in extent of acetylation or phosphorylation were significant between treatments. p values of ≤ 0.05 indicated statistically significant results with levels of significance reported as *, p ≤ 0.05; **, p < 0.01; ***, p < 0.001.
Results
Sulforaphane and erucin cause inhibition of HDAC activity
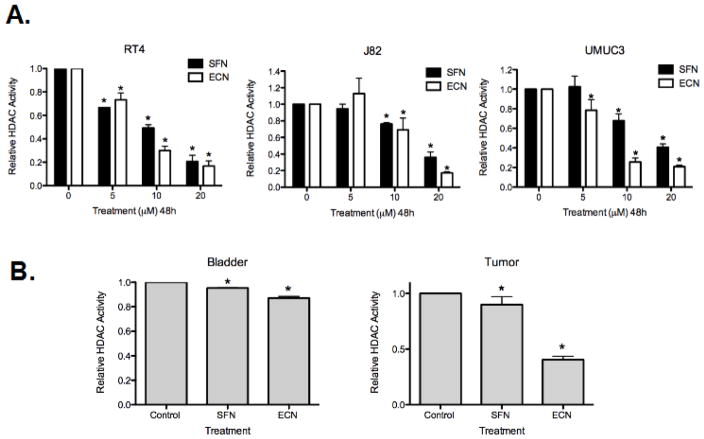
We have recently shown that broccoli isothiocyanates (ITCs), particularly sulforaphane (SFN) and erucin (ECN) have the ability to significantly inhibit bladder cancer in vitro and in vivo. There is some evidence that SFN can act as an HDACI in prostate and colon cancers [12, 27]. We tested if the ability of broccoli ITCs to inhibit bladder cancer is in association with epigenetic modulation. To do this, we first examined the ability of the broccoli ITCs SFN and ECN to inhibit HDAC activity in a panel of human bladder cancer cells ranging from superficial (RT4) to invasive (J82 and UMUC3). We found that SFN and ECN cause robust inhibition of HDAC activity in these cell lines (p<0.05 for all values reported below). After 48h treatment at 20 μM, SFN caused 79.3 ± 5% inhibition of HDAC activity and ECN caused 83.2 ± 4.2% inhibition, in RT4 cells. With similar treatment, SFN caused 64% ± 6.5 inhibition and ECN caused 82.6 ± 1.4% inhibition in J82 cells; and SFN caused 59.38% ± 3.4 inhibition and ECN caused 79.2 ± 1.7% inhibition in UMUC3 cells (Figure 1A). Overall, ECN exhibited a greater HDACI effect than SFN. Since we observed a greater SFN/ECN-mediated effect in the ability to inhibit HDAC activity in UMUC3 cells when compared to J82 cells, both invasive human bladder cancer cell lines, we chose to focus on the UMUC3 cell line to model invasive tumor cells throughout the remainder of the study. We next determined the relevance of our findings in an in vivo setting, utilizing a UMUC3 subcutaneous xenograft model treated with either control vehicle gavage (soybean oil), 295 μmol/kg bodyweight (bw) SFN gavaged daily or 295 μmol/kg bw ECN gavaged daily for 2 weeks. We found a mild inhibition of HDAC activity in normal bladder tissue treated with SFN (4.8 ± 0.2% inhibition) and ECN (13 ± 1.5% inhibition) and greater inhibition in UMUC3 xenograft tumor tissue, which was much more striking with ECN (59.6 ± 0.3% inhibition) treatment compared to SFN (10 ± 0.7% inhibition), mimicking our in vitro data (Figure 1B).
Figure 1. Sulforaphane and erucin inhibits HDACs in bladder cancer in vitro and in vivo.
A. Human bladder cancer cells ranging from superficial (RT4) to invasive (J82 and UMUC3) were treated with DMSO vehicle control or 5, 10 and 20 μM sulforaphane (SFN) and erucin (ECN) for 48h. Protein lysates were obtained under non-denaturing conditions followed by concentration determination and normalization via the BCA Assay. HDAC activity was assessed by Fluor de Lys fluorometric HDAC activity assay. The data was collected, normalized to the DMSO “0” control and is expressed as the mean Relative HDAC Activity ± standard deviation (SD) and represents three independent experiments. Statistical significance was set at *, p<0.05 relative to DMSO controls. B. HDAC activity was also assessed using tissue from a mouse xenograft study where athymic nude mice were subcutaneously injected with 0.05 × 106 UMUC3 cells and gavaged daily with either vehicle control (soybean oil), 295 μmol/kg body weight SFN or 295 μmol/kg body weight ECN (n = 12 mice/group) for 2 weeks until the tumors reached approximately 1.2 cm in diameter and were sacrificed. Protein extraction was performed from normal bladder tissue (Bladder) and UMUC3 tumor xenografts (Tumor) from each treatment group; the amount of protein was quantified/normalized and subjected to the HDAC Activity Assay. HDAC activity assay data was collected and normalized to DMSO treated mice and is expressed as the mean Relative HDAC Activity ± SD and represents data from four bladder tissue samples (normal bladder tissue, labeled “Bladder”) and four UMUC3 tumor samples (UMUC3 tumor xenograft, labeled “Tumor”) from each treatment group. Statistical significance is set at *, p<0.05 relative to the DMSO Control.
Sulforaphane and erucin inhibit HDACs 1, 2, 4 and 6
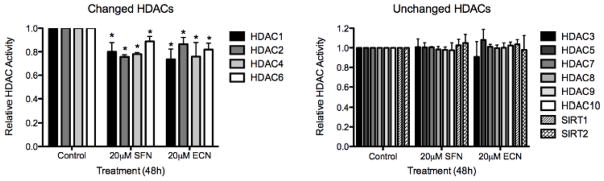
Because we observed strong HDAC inhibition with SFN and ECN treatment in the in vitro and in vivo models of bladder cancer, we next determined which HDACs demonstrated altered activity in response to these compounds. We quantified each individual HDAC activity through immunoprecipitation combined with HDAC activity assay. After testing HDAC 1–10 and Sirtuin 1 and 2, significant (p<0.05) inhibition was exhibited by both SFN and ECN (48h treatment at 20 μM) in HDACs 1, 2, 4 and 6, but no significant changes in HDACs 3, 5, 7, 8, 9 or 10 or the Sirtuins were observed. SFN inhibited HDAC 1 activity by 20 ± 7.8% and ECN inhibited it by 26.4 ± 8.7%. HDAC 2 was inhibited 24.4 ± 1.7% by SFN and 13.56 ± 5.8% by ECN. HDAC 4 was inhibited 22 ± 1.3% by SFN and 24.1± 12% by ECN and HDAC 6 was inhibited 11.2 ± 4.3% by SFN and 18.1± 5.4% by ECN (Figure 2).
Figure 2. Sulforaphane and erucin inhibit HDAC1, 2, 4 and 6 with no effect on other HDACs or Sirtuins.
UMUC3 (invasive human bladder cancer cells) were treated with either DMSO vehicle control, 20 μM sulforaphane (SFN) or erucin (ECN) for 48h and protein lysates were obtained under non-denaturing conditions. Input protein concentration was determined and normalized via the BCA Assay. Subsequently, each HDAC/Sirtuin was immunoprecipitated (IP) sequentially and protein beads were utilized to assess HDAC activity of each individual IP HDAC/SIRT. The data was collected and normalized to the DMSO vehicle condition and is expressed as the mean Relative HDAC or SIRT Activity ± SD and represents three independent experiments. Statistical significance was set at *, p<0.05 relative to the DMSO (Control).
Histone acetylation status is not robustly modulated by SFN or ECN treatment
After observing significant inhibition of HDAC activity upon treatment of human bladder cancer cells with SFN and ECN, our goal was to determine the acetylation status of histones in these cells. HDAC inhibition typically causes increased acetylation of histones, common targets of HDACIs. We treated RT4 and UMUC3 human bladder cancer cells with SFN and ECN, extracted histones and used a global LC-MS proteomic approach to determine changes in histone acetylation. We examined the histone acetylation profiles at the following treatment times and doses: 4 μM SFN or ECN for 24h and 20 μM SFN or ECN for 3h, 6h, 12h, 24h and 48h. The LC-MS profiles showed no statistically significant change in global histone acetylation (Supplemental Figures 1–6, 13–21). While this result is unexpected, the lack of a global increase does not rule out increases in acetylation on the core histones. Because LC-MS is a read-out of the summation of all acetylations, small increases in a few sites may not significantly alter the global profile.
To determine changes in acetylation with greater site specificity, we utilized western blotting with pan anti-Ac and specific anti-Ac antibodies [36]. We used a panel of dosages ranging from 5–20 μM which includes the previously reported dose of 15 μM. Small, yet significant increases in AcH3 status (1.4 ± 0.2 fold increase with 20 μM ECN and 1.76 ± 0.2 with 20 μM SFN, treatment for 48h, p<0.05) in the non-invasive cell line (RT4) was observed (Figure 3A). However, the invasive cell line (UMUC3) displayed the opposite trend as 20 μM SFN and 20 μM ECN significantly decreased the relative expression of AcH3. We did not observe any changes in histone H4 acetylation in these cell lines, by western blot analysis (data not shown). Interestingly, p21, a commonly modulated tumor suppressor gene by HDACI showed a strong increase in expression in RT4 cells (2.6 ± 0.7 fold increase with 20 μM ECN and 2.3 ± 0.6 with 20 μM SFN, treatment for 48h, p<0.05), however was not appreciably modulated in UMUC3 cells (Figure 3A). In addition, thymidylate synthase, which has been shown to be significantly down-regulated by HDACIs in bladder cancer cell lines through microarray analysis [37], also showed significant down-regulation by SFN and ECN, in both RT4 and UMUC3 cell lines (44.5 ± 0.8% decrease with 20 μM ECN and 65.4 ± 0.5% with 20 μM SFN, treatment for 48h) (Figure 3A).The lack of observed increase in acetylation status of histones by both LC-MS and western blotting, in the context of robust HDACI, led us to speculate that HDACI may be targeting cytoplasmic substrate to a greater extent than nuclear histones. We quantified the effects of 48h treatment of 20 μM SFN and ECN on protein levels of acetylated tubulin, a common acetylated cytoplasmic protein [38], and found a similar trend to our histone data. SFN treatment led to a modest (21%) but non-significant increase in acetylated tubulin and ECN treatment led to an 87% decrease in acetylated tubulin (p<0.0001) in UMUC3 cells (Figure 3B).
Figure 3. Histone acetylation status is not robustly modulated by sulforaphane and erucin treatment.
A. RT4 and UMUC3 cells were treated with 5, 10 and 20 μM SFN and ECN for 48h, protein lysates were obtained and analyzed by western blot analysis utilizing a site-specific (K9 and K13) acetylated histone H3 (AcH3) antibody. The antibody was raised against amino acids 1–20 of histone H3 (ARTKQTAR[K*]STGG[K*]APRKQLC, where K* is acetylated). P21 and thymidylate synthase (TS) protein expression was also analyzed and protein expression was quantified by densitometry relative to GAPDH. The data was normalized to the DMSO control “C” and is expressed as the mean (AcH3, p21, and Thymidylate Synthase Protein) Relative Densitometry (ratio to control) ± SD and represents three independent experiments. Statistical significance was set at *, p<0.05 relative to the “C” control. B. UMUC3 cells were treated with 20 μM SFN or ECN for 48h, protein lysates were obtained and analyzed by western blot analysis utilizing acetylated tubulin antibody and quantified by densitometry relative to GAPDH. The data was normalized to the DMSO treated control and is expressed as the mean Ac-Tubulin Protein (Fold Change) ± standard error of the mean (SEM) and represents 3 independent experiments. Statistical significance was set as *, p<0.05 relative to the DMSO control. Representative gels in (A–B) are shown from three independent experiments.
HAT activity is also inhibited by broccoli ITCs
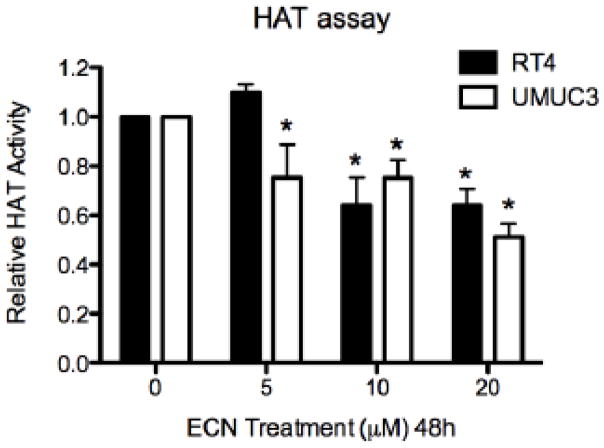
Because we observed inhibition of HDAC activity, but not appreciable increases in the status of acetylated histones, we quantified histone acetyltransferase (HAT) activity as modulated by SFN and ECN treatment. If HAT activity was also inhibited, in addition to inhibition of HDAC activity, this would explain the lack of large increases in acetylated status of the histones. We found that in RT4 cells 20 μM of ECN decreased HAT activity by 35.9% ± 6.6% and in UMUC3 cells by 48.9 ± 5.4%, with 48h treatment (Figure 4).
Figure 4. HAT activity is also inhibited by broccoli ITCs.
UMUC3 cells were treated with DMSO vehicle control or 5, 10, 20 μM SFN or ECN for 48h, nuclear extract was obtained under non-denaturing conditions and HAT activity was assessed. The data was normalized to the DMSO vehicle control and is expressed as the mean Relative HAT Activity ± SD and represents three independent experiments. Statistical significance was set at *, p<0.05 relative to the DMSO control.
Modulation of histone H1 phosphorylation by SFN and ECN treatment
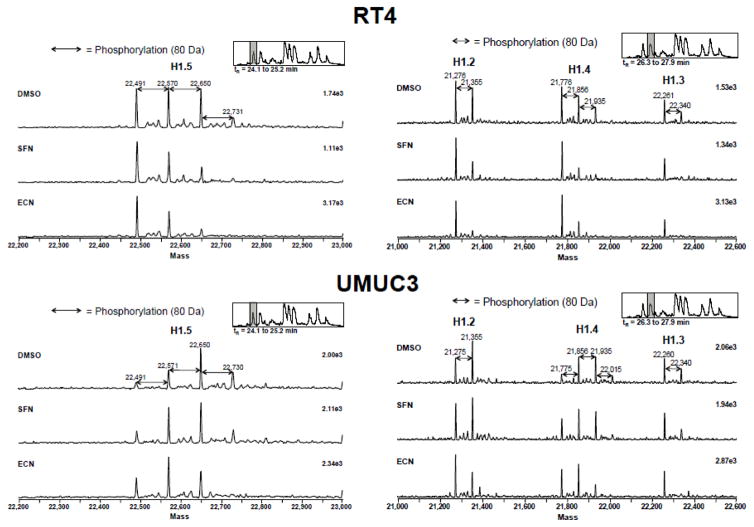
Although we did not see large increases in the acetylation status of histones, in spite of large decreases in HDAC activity, the LC-MS analysis revealed a novel histone modification change in response to SFN and ECN treatment. Histone H1 phosphorylation decreased upon treatment with SFN and ECN (20 μM, 3h). We did not observe global phosphorylation changes in the LC-MS results of the core histones. The deconvoluted mass spectra of the four histone H1 variants (H1.5, H1.2, H1.4 and H1.3) from RT4 and UMUC3 with DMSO vehicle control, SFN, and ECN treatment are shown in Figure 5 and Supplementary Figures 7–12. For RT4 cells, there was a dramatic decrease in phosphorylation for all linker histone variants upon ECN treatment. In addition, there was a decrease in phosphorylation of the linker histone variants H1.2, H1.4 and H1.3 upon SFN treatment. For UMUC3, there was a decrease in phosphorylation status upon ECN treatment.
Figure 5. Modulation of histone H1 phosphorylation occurs with SFN and ECN treatment.
RT4 and UMUC3 cells were treated with DMSO vehicle control or 20 μM SFN or ECN for 3h, cells were flash frozen, histones extracted and analyzed by LC-MS. The deconvoluted mass spectra of all four variants of histone H1 (H1.2, H1.3, H1.4 and H1.5) of RT4 and UMUC3 is shown. An 80 Da shift to the right indicates an increase in phosphorylation of histone H1. A representative spectra is shown from three independent experiments.
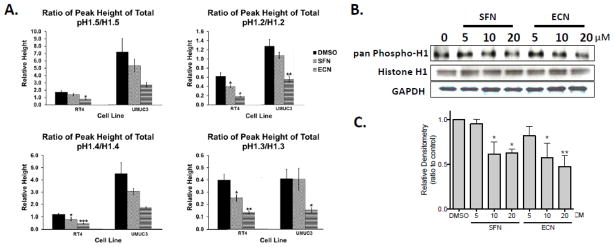
Quantification of the ratio of summed peak heights for the phosphorylated isoforms to unphosphorylated isoforms of the individual histones is presented in Figure 6A. There was a significant decrease in phosphorylation status of all of the linker histone variants (H1.5, H1.2, H1.4 and H1.3) upon ECN treatment for the RT4 cell line. In addition, there was a significant decrease in phosphorylation status of the linker histone variants H1.2, H1.4 and H1.3 upon SFN treatment for the RT4 cell line. For the UMUC3 cells, while trends are observed for H1.4 and H1.5, only significant decreases in relative phosphorylation were observed in H1.2 and 1.3 with ECN and non-significant trends observed with SFN. The decrease in phosphorylation status found by mass spectrometry with SFN and ECN treatment was validated by western blot analysis utilizing a pan-phospho histone H1 antibody in UMUC3 cells. Treatment with 20 μM SFN caused a 37 ± 6% decrease in pan-phospho histone H1 status and 20 μM ECN caused a 53 ± 11% decrease (3h treatment) (Figure 6B–C).
Figure 6. Modulation of histone H1 phosphorylation occurs earlier and appears to be more robust than histone acetylation changes with SFN and ECN treatment.
Data from the treatment of RT4 and UMUC3 cells with DMSO, SFN or ECN treatment (20 μM, 3h) is shown. A. The ratio of the summed peak heights from the LC-MS data for the phosphorylated isoforms to the unphosphorylated isoform for all four variants of histone H1 (H1.2, H1.3, H1.4 and H1.5) is shown. Data represents four independent experiments, error bars represent ± standard error and statistically significant differences are indicated (*, p<0.05; **, p<0.01; ***, p<0.001). B. Histone H1 phosphorylation changes were also analyzed by western blot analysis where representative gels are shown from at least two independent experiments. C. Densitometry from the western blot analysis from B. is shown. The data was normalized to the DMSO control and is expressed as the mean Relative Densitometry (ratio to control) ± SEM and represents two independent experiments. Statistical significance was set at *, p<0.05 and **, p<0.01 relative to the DMSO control.
The decrease in phosphorylation occurs earlier and appears to be more robust than histone acetylation changes with SFN and ECN treatment. The raw LC-MS data for all of the core histones was analyzed as above. No apparent difference in acetylation status was observed for any of the cores histones (Supplementary Figures 1–6, 13–21). For each H3 variant in each cell line the weighted average mass was calculated (data not shown). Similar analysis was carried out for histone H4 (data not shown). No increase in acetylation was observed with SFN or ECN treatment (20 μM, 3h) except for a decrease (p<0.05) in acetylation in the RT4 cell line with the SFN treatment.
Increased PP1β and PP2A activity occurs with SFN and ECN treatment
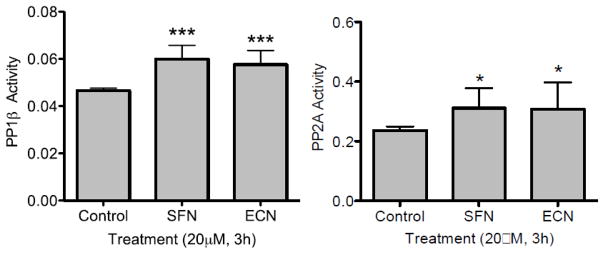
One reasonable explanation for a decrease in phosphorylation is that cells are arrested and/or undergoing apoptosis. The time point used (3h) in this study was carefully chosen to evaluate the early effects of SFN and ECN prior to cell death. Another explanation for decreased H1 phosphorylation is alteration of phosphatase activity. HDACIs have been reported to disrupt the interaction between protein phosphatase 1 (PP1) and HDAC 1 or 6 [4] leading to higher phosphatase activity. Therefore, a phosphatase activity assay was performed on the UMUC3 cells treated with DMSO or 20 μM concentrations of SFN or ECN at the early 3h time point. PP1β and PP2A phosphatase activity was assayed and the results are summarized in Figure 7. Significant increases in PP1β and PP2A activity (***, p<0.001 and *, p<0.05) occurred for all treatments (SFN and ECN) relative to the control.
Figure 7. An increase in PP1β and PP2A activity occurs with SFN and ECN treatment.
UMUC3 cells were treated with DMSO vehicle control or 20 μM SFN or ECN for 3h, cells were lysed in non-denaturing cell lysis buffer, PP1β and PP2A were immunoprecipitated and phosphatase activity was assayed. The data was collected in triplicate and is expressed as the mean Phosphatase Activity (O.D.) ± SD and represents four independent experiments. A significant increase in PP1β and PP2A activity resulted for all of the treatments (SFN and ECN) relative to the control where statistically significant differences are indicated (*, p<0.05, ***, p<0.001).
Discussion
The potential of dietary components, such as broccoli isothiocyanates (ITCs), to prevent bladder cancer carcinogenesis and progression through modification of the cancer epigenome, is an attractive hypothesis. We have shown that broccoli ITCs have the ability to significantly inhibit bladder cancer cell growth in vitro in diverse human bladder cancer cell lines (RT4, J82, UMUC3) and in vivo, through an UMUC3 xenograft model [22]. This inhibition was associated with decreases in cell viability, induction of apoptosis and cell cycle accumulation in the G2/M phases [22, 39]. However, the molecular mechanisms responsible remained unclear. Therefore, the goal of the present study was to examine mechanisms that may underlie these observations. Since SFN has been reported to decrease HDAC activity, we first examined HDAC inhibition as a potential mechanism of action in multiple bladder cancer model systems [29, 30, 36, 40]. We report that broccoli ITCs, sulforaphane (SFN) and erucin (ECN) have strong HDACI activity (Figure 1). We saw stronger HDAC inhibition by ECN than SFN, which is the most commonly studied of cruciferous vegetable phytochemical metabolites. Perhaps a stronger focus on ECN as a potential epigenetic modulator would be fruitful as it can be found at high concentrations in other cruciferous vegetables such as arugula [41, 42]. To the best of our knowledge, this is the first report of HDAC inhibition by erucin.
It is important to note however that although the modification of the epigenetic histone code has been shown to regulate the transcription of a defined set of genes through chromatin remodeling, there is increasing evidence suggesting that this may not be the only mechanism for HDAC inhibitor-mediated growth inhibition and apoptosis in cancer cells [43].While histones are an important target of HDACs, the antitumor effects of HDAC inhibitors might also be attributed to transcription-independent mechanisms by modulating the acetylation status of a series of non-histone targets, such as Nrf2 which has been shown to be a target of SFN’s bioactivity. It is also important to consider the action of HDACIs on Akt downregulation via alteration of HDAC / protein phosphatase complex formation [8].
We found that the specific HDACs inhibited are HDAC 1, 2, 4 and 6 which belong to classes I, IIa and IIb (Figure 2). HDAC 1 and 2 overexpression has been associated with increased proliferation, HDAC 4 with increased angiogenesis and HDAC 6 with increased fibroblast migration [18]. Interestingly, HDAC 6 does not affect acetylation status of histones, but rather targets cytoplasmic proteins, such as Ku70 and Hsp90. Since we saw robust HDAC inhibition, we assessed the status of acetylated histones and found small and insignificant changes in acetylation status of histones. However, resulting markers of this inhibition, such as p21 up-regulation and thymidylate synthesis inhibition was observed (Figure 3A).
SFN has been reported to increase acetylation of the core histone variants H3 and H4 both globally and locally in human embryonic kidney cells [29], human colorectal cancer cells [29], prostate cancer epithelial cells [36], and in the tissues of a mouse model of colon cancer (Apcmin)[30]. However, it has also been reported that no change in histone acetylation was observed in human breast cancer cell lines, even with HDAC inhibition [40]. In addition, it has also been shown that SFN’s HDAC inhibitory effect depends on the invasiveness of the in vitro experimental cell line studied [39]. Our results show that although HDACs are strongly inhibited by both SFN and ECN, the resulting effect on the acetylation status of histones is modest at best, as seen in the RT4 cell line, or can actually lead to a decreased acetylation status as seen in the UMUC3 cells (Figure 3A). We found no changes in the global acetylation of H3 or H4 by LC-MS. However, by western blot, we found significant changes in the AcH3 status (increase in the non-invasive cell line (RT4) and decrease in the invasive cell line (UMUC3)). The lack of global changes in the LC-MS analysis could be due to a lack of sensitivity of the assay and not necessarily the lack of global changes. A targeted LC-MS/MS technique could resolve this uncertainty and perhaps shed light on the difference in the western blot results. Recently Ben Garcia [44] and other groups have developed robust protocols for some histone modifications, which still do not provide complete coverage. Development of these targeted assays were beyond the scope of this project, but would be an excellent extension of this work as these targeted techniques evolve. However, the unbiased nature of the LC-MS strongly suggests the HDACI activity is not as profound as the western blot data suggests and thus alternate mechanisms should be considered. Motivated by our observations that HDAC6, an important modifier of cytoplasmic proteins rather than histones, was inhibited in our IP panel (Figure 2), we looked at the acetylation status of tubulin, a commonly acetylated cytoplasmic marker. However, this too showed similar results to our histone data. This data may be partly explained by the dual inhibition of both HDAC and HAT activity by SFN and ECN (Figure 4).
In contrast to a modest impact on histone acetylation, we observed some striking changes in histone H1 phosphorylation (Figures 5 and 6, Supplementary Figures 7–12). We have recently shown that when comparing normal human bladder tissue with superficial and invasive bladder cancers, there is a robust increase in phosphorylation of histone H1 status, making phosphorylated histone H1 a potential biomarker of bladder carcinogenesis and progression [13]. Interestingly, we found that SFN, and to a much greater extent, ECN has the ability to reverse this phosphorylation. Phosphatase PP1β and PP2A activity increased upon treatment with SFN and ECN (Figure 7). A prevailing hypothesis suggested by Ching Shi Chen [45] is that the HDACIs can interrupt PP1 binding to the HDAC complex. When we looked at PP1β and PP2A activity we observed a similar phenotype as they observed for the HDACI AR42. This disruption of the interaction between PP1 and HDAC 1 or 6 frees PP1 to dephosphorylate proteins. These data indicate that by observing the phosphorylation changes of histone H1, we are likely monitoring downstream effects of increased phosphatase activity. Although SFN and ECN have not been previously reported to affect phosphatase activity, it has been reported that treatment with SFN results in a significant decrease in phosphorylated Akt in a human ovarian cancer cell line [46], as Akt is dephosphorylated by PP1 and PP2A [47]. The decrease of phosphorylated Akt previously reported and the observed decrease in linker histone phosphorylation are likely, at least partly, a downstream effect of the increase in phosphatase activity upon treatment with SFN and ECN. Thus we assert that SFN and ECN phosphatase activation may play a greater mechanistic role than HDAC inhibition. This novel assertion is supported by our observation that phosphorylation changes occur earlier and more robustly than acetylation changes (Figure 6). These results support a novel mechanism of action compared to previous work. A more detailed examination of the intracellular signaling pathways affected by the presence of dietary isothiocyanates is an excellent subject of future research in this area.
Epigenetic modifications can be largely grouped into two main categories; DNA methylation and histone modifications. It is the interplay and sum of the interactions between these epigenetic factors that leads to the observed outcome [9]. Here our observed outcome is inhibition of bladder cancer cell growth by broccoli ITCs, and we see this outcome associated with epigenetic modulations, including acetylation changes of histone H3 and hypo-phosphorylation of histone H1. Future studies will focus on the persistence of the effect of the epigenetic modification and cell viability [32, 48, 49].
Furthermore, we need to determine if these changes in histone H1 also occur in vivo with treatment of sulforaphane and erucin. Finally, it would be important to determine gene promoter regions exhibiting histone acetylation and phosphorylation changes to determine which genes are targeted by these epigenetic changes. A further study of other histone modification changes such as methylation and ubiquitylation would further enhance our understanding of the epigenetic effects of broccoli ITCs in bladder cancer. Overall, our work highlights the ability of a readily available class of dietary phytochemicals associated with reduced risk of bladder carcinogenesis to modulate the epigenome of a common cancer and supports further work into the ability of dietary interventions to serve in long-term cancer prevention through epigenetic modulation [50].
Conclusions
In conclusion, our findings suggest that cruciferous vegetable isothiocyanates SFN and ECN modulate epigenetic function in human bladder cancer cells via HDAC inhibition and phosphatase enhancement allowing for reduced levels of histone H1 phosphorylation, a potential biomarker we previously correlated with human bladder cancer carcinogenesis and progression [13]. Thus, SFN- and ECN-mediated targeting of histone H1 phosphorylation via epigenetic modulation presents a novel potential preventative and treatment strategy for human bladder cancer. In addition, to the best of our knowledge, this is the first report of modulation of epigenome with ECN, an abundant phytochemical in many cruciferous vegetables, such as broccoli and arugula. Overall, our work supports the hypothesis that relevant dietary phytochemicals may modulate the epigenome of a common cancer and supports further work into the ability of dietary interventions to serve in long-term cancer prevention through epigenetic modulation.
Supplementary Material
Significance.
Collectively, our findings suggest that the cruciferous vegetable isothiocyanates: sulforaphane (SFN) and erucin (ECN), impact histones status in bladder cancer cells by modulating specific HDACs and HATs, and enhancing phosphatase activity, resulting in reduction of histone H1 phosphorylation. These findings are significant due to the fact that our previous work positively correlated histone H1 phosphorylation with bladder cancer carcinogenesis and progression. Therefore, we propose that SFN and ECN may inhibit bladder carcinogenesis via epigenetic modulation of gene expression associated with histone H1 phosphorylation. These efforts may elucidate biomarkers useful in epidemiologic studies related to cruciferous vegetable intake and cancer risk or provide intermediate biomarkers for food-based clinical intervention studies in high-risk cohorts.
Highlights.
Broccoli and other cruciferous vegetables intake has been associated with reduced risk of bladder cancer.
Histone proteomics modifications after exposure to broccoli isothiocyanates, sulforaphane (SFN) and erucin (ECN), were analyzed in bladder cancer.
SFN and ECN reduced histone deacetylase activity in vitro and in vivo.
Interestingly, the global acetylation status of histones H3 or H4 remained largely unchanged, which can be partly explained by concomitant inhibition of histone acetyltransferases.
Histone H1 phosphorylation increases through bladder cancer carcinogenesis and progression; yet SFN and ECN increase phosphatase PP1β and PP2A activity and decrease histone H1 phosphorylation.
Acknowledgments
Funding Sources
NIH/NCCAM F31AT006486, NIH/NIGMS 5T32GM068412-04, NIH/NCI OSU CCC P30 CA16058 and the Molecular Carcinogenesis and Chemoprevention Program, OSU CCC James Cancer Hospital Shoen Cancer Prevention Research Fund and Bladder Cancer Prevention Fund; OSU Center for Functional Foods and Research Entrepreneurship (CAFFRE). C.R.L. is a recipient of a National Institutes of Health T32 Award in Oncology Training Fellowship at The Ohio State University Comprehensive Cancer Center, T32 CA009338.
ABBREVIATIONS
- AcH3
acetylated histone H3
- AcH4
acetylated histone H4
- ECN
erucin
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- HDACI
HDAC inhibitor
- IP
immunoprecipitation
- ITCs
isothiocyanates
- LC-MS
liquid chromatography-mass spectrometry
- SFN
sulforaphane
Footnotes
Author Contributions: The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Willbanks A, Leary M, Greenshields M, Tyminski C, Heerboth S, Lapinska K, Haskins K, Sarkar S. The Evolution of Epigenetics: From Prokaryotes to Humans and Its Biological Consequences. Genetics & epigenetics. 2016;8:25–36. doi: 10.4137/GEG.S31863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues D, Jeronimo C, Henrique R, Belo L, de Lourdes Bastos M, de Pinho PG, Carvalho M. Biomarkers in bladder cancer: A metabolomic approach using in vitro and ex vivo model systems. International journal of cancer. 2016;139(2):256–68. doi: 10.1002/ijc.30016. [DOI] [PubMed] [Google Scholar]
- 3.Harb-de la Rosa A, Acker M, Kumar RA, Manoharan M. Epigenetics application in the diagnosis and treatment of bladder cancer. The Canadian journal of urology. 2015;22(5):7947–51. [PubMed] [Google Scholar]
- 4.Kim WJ, Kim YJ. Epigenetics of bladder cancer. Methods in molecular biology. 2012;863:111–8. doi: 10.1007/978-1-61779-612-8_6. [DOI] [PubMed] [Google Scholar]
- 5.Navarro SL, Li F, Lampe JW. Mechanisms of action of isothiocyanates in cancer chemoprevention: an update. Food & function. 2011;2(10):579–87. doi: 10.1039/c1fo10114e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner AE, Terschluesen AM, Rimbach G. Health promoting effects of brassica-derived phytochemicals: from chemopreventive and anti-inflammatory activities to epigenetic regulation. Oxidative medicine and cellular longevity. 2013;2013:964539. doi: 10.1155/2013/964539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhauser C. Epigenetic impact of dietary isothiocyanates in cancer chemoprevention. Current opinion in clinical nutrition and metabolic care. 2013;16(4):405–10. doi: 10.1097/MCO.0b013e328362014e. [DOI] [PubMed] [Google Scholar]
- 8.Lin HY, Chen CS, Lin SP, Weng JR, Chen CS. Targeting histone deacetylase in cancer therapy. Medicinal research reviews. 2006;26(4):397–413. doi: 10.1002/med.20056. [DOI] [PubMed] [Google Scholar]
- 9.Portela A, Esteller M. Epigenetic modifications and human disease. Nature biotechnology. 2010;28(10):1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Aguilera A, Fernandez-Arroyo S, Cuyas E, Luciano-Mateo F, Cabre N, Camps J, Lopez-Miranda J, Menendez JA, Joven J. Epigenetics and nutrition-related epidemics of metabolic diseases: Current perspectives and challenges. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 2016;96:191–204. doi: 10.1016/j.fct.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Tollefsbol TO. Impact of Epigenetic Dietary Components on Cancer through Histone Modifications. Current medicinal chemistry. 2015;22(17):2051–64. doi: 10.2174/0929867322666150420102641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27(2):344–9. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telu KH, Abbaoui B, Thomas-Ahner JM, Zynger DL, Clinton SK, Freitas MA, Mortazavi A. Alterations of histone H1 phosphorylation during bladder carcinogenesis. Journal of proteome research. 2013;12(7):3317–26. doi: 10.1021/pr400143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harshman SW, Hoover ME, Huang C, Branson OE, Chaney SB, Cheney CM, Rosol TJ, Shapiro CL, Wysocki VH, Huebner K, Freitas MA. Histone H1 phosphorylation in breast cancer. Journal of proteome research. 2014;13(5):2453–67. doi: 10.1021/pr401248f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Hoover ME, Dang X, Shomo AA, Guan X, Marshall AG, Freitas MA, Young NL. Quantitative Mass Spectrometry Reveals that Intact Histone H1 Phosphorylations are Variant Specific and Exhibit Single Molecule Hierarchical Dependence. Molecular & cellular proteomics : MCP. 2016;15(3):818–33. doi: 10.1074/mcp.M114.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic acids research. 2013;41(21):9593–609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Wei W, Zhou DX. Histone Acetylation Enzymes Coordinate Metabolism and Gene Expression. Trends in plant science. 2015;20(10):614–21. doi: 10.1016/j.tplants.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends in plant science. 2002;7(6):263–70. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- 19.Aksnes H, Hole K, Arnesen T. Molecular, cellular, and physiological significance of N-terminal acetylation. International review of cell and molecular biology. 2015;316:267–305. doi: 10.1016/bs.ircmb.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Michaud DS, Spiegelman D, Clinton SK, Rimm EB, Willett WC, Giovannucci EL. Fruit and vegetable intake and incidence of bladder cancer in a male prospective cohort. Journal of the National Cancer Institute. 1999;91(7):605–13. doi: 10.1093/jnci/91.7.605. [DOI] [PubMed] [Google Scholar]
- 21.Tang L, Zirpoli GR, Guru K, Moysich KB, Zhang Y, Ambrosone CB, McCann SE. Consumption of raw cruciferous vegetables is inversely associated with bladder cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17(4):938–44. doi: 10.1158/1055-9965.EPI-07-2502. [DOI] [PubMed] [Google Scholar]
- 22.Abbaoui B, Riedl KM, Ralston RA, Thomas-Ahner JM, Schwartz SJ, Clinton SK, Mortazavi A. Inhibition of bladder cancer by broccoli isothiocyanates sulforaphane and erucin: characterization, metabolism, and interconversion. Molecular nutrition & food research. 2012;56(11):1675–87. doi: 10.1002/mnfr.201200276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Shan Y. Sulforaphane retards the growth of UM-UC-3 xenographs, induces apoptosis, and reduces survivin in athymic mice. Nutrition research (New York, NY) 2012;32(5):374–80. doi: 10.1016/j.nutres.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Tang L, Zhang Y, Jobson HE, Li J, Stephenson KK, Wade KL, Fahey JW. Potent activation of mitochondria-mediated apoptosis and arrest in S and M phases of cancer cells by a broccoli sprout extract. Mol Cancer Ther. 2006;5(4):935–44. doi: 10.1158/1535-7163.MCT-05-0476. [DOI] [PubMed] [Google Scholar]
- 25.Shan Y, Sun C, Zhao X, Wu K, Cassidy A, Bao Y. Effect of sulforaphane on cell growth, G(0)/G(1) phase cell progression and apoptosis in human bladder cancer T24 cells. International journal of oncology. 2006;29(4):883–8. [PubMed] [Google Scholar]
- 26.Ding Y, Paonessa JD, Randall KL, Argoti D, Chen L, Vouros P, Zhang Y. Sulforaphane inhibits 4-aminobiphenyl-induced DNA damage in bladder cells and tissues. Carcinogenesis. 2010;31(11):1999–2003. doi: 10.1093/carcin/bgq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Seminars in cancer biology. 2007;17(5):363–9. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bricker GV, Riedl KM, Ralston RA, Tober KL, Oberyszyn TM, Schwartz SJ. Isothiocyanate metabolism, distribution, and interconversion in mice following consumption of thermally processed broccoli sprouts or purified sulforaphane. Molecular nutrition & food research. 2014;58(10):1991–2000. doi: 10.1002/mnfr.201400104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer research. 2004;64(16):5767–74. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 30.Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20(3):506–8. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Experimental biology and medicine. 2007;232(2):227–34. [PMC free article] [PubMed] [Google Scholar]
- 32.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. The Journal of nutrition. 2009;139(12):2393–6. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tortorella SM, Royce SG, Licciardi PV, Karagiannis TC. Dietary Sulforaphane in Cancer Chemoprevention: The Role of Epigenetic Regulation and HDAC Inhibition. Antioxidants & redox signaling. 2015;22(16):1382–424. doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Harshman SW, Liu S, Ren C, Xu H, Sallans L, Grever M, Byrd JC, Marcucci G, Freitas MA. Assaying pharmacodynamic endpoints with targeted therapy: flavopiridol and 17AAG induced dephosphorylation of histone H1.5 in acute myeloid leukemia. Proteomics. 2010;10(23):4281–92. doi: 10.1002/pmic.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, Freitas MA, Tsai MD. Identification of histone demethylases in Saccharomyces cerevisiae. The Journal of biological chemistry. 2007;282(19):14262–71. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27(4):811–9. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2(2):151–63. [PubMed] [Google Scholar]
- 38.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society reviews. 2011;40(1):387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 39.Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Molecular nutrition & food research. 2011;55(7):999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pledgie-Tracy A, Sobolewski MD, Davidson NE. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Mol Cancer Ther. 2007;6(3):1013–21. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- 41.Herz C, Hertrampf A, Zimmermann S, Stetter N, Wagner M, Kleinhans C, Erlacher M, Schuler J, Platz S, Rohn S, Mersch-Sundermann V, Lamy E. The isothiocyanate erucin abrogates telomerase in hepatocellular carcinoma cells in vitro and in an orthotopic xenograft tumour model of HCC. Journal of cellular and molecular medicine. 2014;18(12):2393–403. doi: 10.1111/jcmm.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azarenko O, Jordan MA, Wilson L. Erucin, the major isothiocyanate in arugula (Eruca sativa), inhibits proliferation of MCF7 tumor cells by suppressing microtubule dynamics. PloS one. 2014;9(6):e100599. doi: 10.1371/journal.pone.0100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braszewska-Zalewska A, Bernas T, Maluszynska J. Epigenetic chromatin modifications in Brassica genomes. Genome / National Research Council Canada = Genome / Conseil national de recherches Canada. 2010;53(3):203–10. doi: 10.1139/g09-088. [DOI] [PubMed] [Google Scholar]
- 44.Arnaudo AM, Link AJ, Garcia BA. Bioorthogonal Chemistry for the Isolation and Study of Newly Synthesized Histones and Their Modifications. ACS Chem Biol. 2016;11(3):782–91. doi: 10.1021/acschembio.5b00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CS, Weng SC, Tseng PH, Lin HP, Chen CS. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. The Journal of biological chemistry. 2005;280(46):38879–87. doi: 10.1074/jbc.M505733200. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhuri D, Orsulic S, Ashok BT. Antiproliferative activity of sulforaphane in Akt-overexpressing ovarian cancer cells. Mol Cancer Ther. 2007;6(1):334–45. doi: 10.1158/1535-7163.MCT-06-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao L, Gong LL, Yuan D, Deng M, Zeng XM, Chen LL, Zhang L, Yan Q, Liu JP, Hu XH, Sun SM, Liu J, Ma HL, Zheng CB, Fu H, Chen PC, Zhao JQ, Xie SS, Zou LJ, Xiao YM, Liu WB, Zhang J, Liu Y, Li DW. Protein phosphatase-1 regulates Akt1 signal transduction pathway to control gene expression, cell survival and differentiation. Cell death and differentiation. 2010;17(9):1448–62. doi: 10.1038/cdd.2010.16. [DOI] [PubMed] [Google Scholar]
- 48.Royston KJ, Tollefsbol TO. The Epigenetic Impact of Cruciferous Vegetables on Cancer Prevention. Current pharmacology reports. 2015;1(1):46–51. doi: 10.1007/s40495-014-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atwell LL, Beaver LM, Shannon J, Williams DE, Dashwood RH, Ho E. Epigenetic Regulation by Sulforaphane: Opportunities for Breast and Prostate Cancer Chemoprevention. Current pharmacology reports. 2015;1(2):102–111. doi: 10.1007/s40495-014-0002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veeranki OL, Bhattacharya A, Tang L, Marshall JR, Zhang Y. Cruciferous vegetables, isothiocyanates, and prevention of bladder cancer. Current pharmacology reports. 2015;1(4):272–282. doi: 10.1007/s40495-015-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.