Abstract
The general stress response in Neisseria gonorrhoeae was investigated. Transcriptional analyses of the genes encoding the molecular chaperones DnaK, DnaJ, and GrpE suggested that they are transcribed from σ32 (RpoH)-dependent promoters upon exposure to stress. This was confirmed by mutational analysis of the σ32 promoter of dnaK. The gene encoding the gonococcal RpoH sigma factor appears to be essential, as we could not isolate viable mutants. Deletion of an unusually long rpoH leader sequence resulted in elevated levels of transcription, suggesting that this region is involved in negative regulation of RpoH expression during normal growth. Transcriptional analyses and protein studies determined that regulation of the RpoH-mediated stress response is different from that observed in most other species, in which regulation occurs predominantly at the transcriptional and translational levels. We suggest that an increase in the activity of preformed RpoH is primarily responsible for induction of the stress response in N. gonorrhoeae.
When exposed to environmental stress, bacteria respond by rapidly increasing synthesis of a characteristic set of proteins that allows them to contend with the adverse environment, meanwhile down-regulating the expression of many housekeeping genes. Elevated temperatures have been widely used as a convenient means to study stress responses, and as a result, proteins induced upon increased temperature have been termed heat shock proteins (Hsps). There is strong evidence to suggest that the signal that induces the heat shock response is the accumulation of misfolded and denatured proteins that arise with stress (15).
Many of these stress-induced proteins are chaperones and proteases. Their biological role is to protect cells against the toxic effects generated by exposure to stress, but they also have an important role in protein function during normal growth conditions. The increase in production of these proteins following exposure to stress allows the bacteria to respond to the elevated level of misfolded proteins. The chaperones function to eliminate misfolded proteins in numerous ways, including (i) unfolding these proteins and subsequently promoting proper folding and (ii) targeting unfolded proteins for proteolysis (20). The best known of the chaperones are GroEL/GroES and members of the DnaK chaperone system, including DnaK, DnaJ, and GrpE. The DnaK chaperone system is well characterized in Escherichia coli, where it is the most abundant cytosolic chaperone system and cannot be replaced in vivo (30). These proteins are of particular interest in that in some species they are also involved in regulation of the stress response (14, 47). Induction of specific bacterial genes with stress is most often regulated at the transcriptional level (53), with the mechanisms controlling this induction varying greatly between species.
The alternative sigma factor RpoH is often used to regulate the stress response and has been identified in bacteria from different subdivisions of proteobacteria (36). The majority of species respond to stress by increasing the level of RpoH produced, using positive and/or negative regulatory processes. The positive regulation of rpoH expression in the alpha subdivision seems to occur primarily at the transcriptional and posttranslational levels. The rpoH genes from Caulobacter crescentus (51), Rhodobacter capsulatus (10), and Agrobacterium tumefaciens (33) are positively autoregulated from an RpoH-dependent promoter upon heat shock. In C. crescentus, down-regulation of the stress response is independent of DnaK and RpoH (9). In A. tumefaciens, DnaK-dependent regulation of RpoH has also been shown to occur at the level of activity (34). Some bacteria of this subdivision contain more than one rpoH gene, with each being regulated by different mechanisms (36).
In the majority of the organisms in the gamma subdivision of proteobacteria, regulation of rpoH occurs primarily by translational repression involving a thermosensitive secondary structure in the rpoH mRNA (55). Among the members of this group of bacteria studied, only Haemophilus influenzae (12) and Buchnera aphidicola (40) appear to lack this rpoH mRNA secondary structure. An additional regulatory process is the negative regulation of RpoH levels by the DnaK chaperone system. During nonstress conditions RpoH interacts with DnaK (14), and together with DnaJ and GrpE, RpoH is targeted to proteases for degradation (23). Upon exposure to heat shock DnaK is sequestered by misfolded proteins such that RpoH function is restored (14). Regulation of rpoH in most of these species can also occur, although to a minor extent, at the transcriptional level by way of different promoters which respond to various signals (11).
Other species negatively regulate heat shock genes at the transcriptional level by the controlling inverted repeat of chaperone expression (CIRCE)/HrcA repressor system (57). These include the gram-positive Enterococcus faecium (45) and Streptococcus pyogenes (50). The stress-responsive genes, such as dnaK and groE, are transcribed from conventional σ70 promoters, but their expression is modulated by GroE-dependent binding of the HrcA repressor protein to the CIRCE element (54). This system is also found in some gram-negative bacteria, including Helicobacter pylori, a member of the epsilon subdivision of the proteobacteria (46), and Chlamydia species (49). Only the groE operon of the alpha subdivision of the proteobacteria retains a CIRCE operator sequence (54). These operons are transcribed from a σ32 promoter, and the HrcA/CIRCE control system seems to act to repress groE transcription in nonstress conditions (39). An exception to this is R. capsulatus, which has a CIRCE element upstream of the groE operon but lacks the HrcA protein. It is suggested that the role of the CIRCE element in this species is to stabilize the groE mRNA (22).
Neisseria belongs to the beta subdivision of the proteobacteria. Homologues of the rpoH, dnaK, and groE genes have been identified in members of this subdivision. There have been limited investigations into the nature of the stress response in these species despite the evidence that molecular chaperones induce an immune response (24) and have a role in disease pathogenesis (37). Recent work on the heat shock response of Neisseria meningitidis using microarray technology suggested that the majority of genes are deregulated only at 45°C (16) rather than at 42°C as shown for Neisseria gonorrhoeae (48). Cloning and sequence analysis of the gonococcal groES and groEL homologues revealed that they were organized in a bicistronic operon, an arrangement similar to that found in most bacterial species (48). Transcription of these genes occurs from a σ70 promoter under nonstress conditions, and an elevated level of transcription occurs from an overlapping σ32 promoter following exposure to stress (48).
Here we report the transcriptional analysis of the genes encoding members of the gonococcal DnaK chaperone system and the RpoH sigma factor. We show that transcription of each of the genes encoding the chaperones is induced upon exposure of the gonococci to heat stress, that this transcription is mediated by RpoH, and that it is predominantly regulated at the level of RpoH activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The E. coli strain used in all cloning experiments was DH5α [F− endA1 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80 ΔlacZM15)]. The N. gonorrhoeae strain used was MS11-A (41). JKD484 is a spontaneous rifampin-resistant mutant derivative of MS11-A which contains the conjugative plasmid ptetM25.2 (25). Gonococcal strains JKD487, JKD488, JKD489, JKD491, JKD492, and JKD493 were derived from JKD484. The growth conditions for E. coli and N. gonorrhoeae have been described previously (13).
Transformation and conjugation of N. gonorrhoeae.
Transformation of N. gonorrhoeae was performed essentially as described previously (5). Erythromycin-resistant transformants from recombination between DNA from the Hermes-2 E. coli and N. gonorrhoeae shuttle plasmid and the ptetM25.2 conjugative plasmid (25) were selected on GC agar plates containing 7 μg of erythromycin/ml. Conjugations were performed by mixing 5 × 108 donor cells and 5 × 109 recipient cells on a small section of a GC agar plate which was incubated overnight at 37°C in a 5% CO2 atmosphere. The growth was transferred into 500 μl of GC broth, and 100-μl aliquots were spread onto GC agar plates containing 7 μg of erythromycin and 10 μg of nalidixic acid/ml for selection of transconjugants.
Recombinant DNA techniques and RNA analysis.
The techniques used were performed as described previously (13). Oligonucleotide primers used are listed in Table 1. Plasmids used are listed in Table 2. Total RNA was prepared from exponentially growing cultures of E. coli and N. gonorrhoeae as described previously (13). The methods used for RNA dot blot and primer extension analysis have been described previously (13). Probes used were 16S rRNA, an 0.83-kb PCR product amplified from N. gonorrhoeae MS11-A using oligonucleotide primers 3260 and 3261 (to confirm equivalent amounts of RNA); dnaK, a 1.37-kb HindIII/ClaI fragment from pJKD1926; dnaJ, a 0.53-kb DraI/ClaI fragment from pJKD2107; grpE, a 0.95-kb PCR product amplified from pJKD2108 using oligonucleotide primers 5610 and 5611; and rpoH, a 0.59-kb HincII/StuI fragment from pJKD2101.
TABLE 1.
Oligonucleotide primers used in this study
| Oligo- nucleotide | Sequence (5′→3′) | Reference or Source |
|---|---|---|
| 3260 | CACACTGGGACTGAGACATG | 6 |
| 3261 | CGGCAGTCTCATTAGAGTGC | 6 |
| 4527 | CATATTGACCCTAGCCGC | This study |
| 5490 | CGCCATTCCCATCATGCG | This study |
| 5492 | CAAGTGCCGATTTATGCG | This study |
| 5493 | TTGGATGGCGGGTAATGC | This study |
| 5494 | TCGGTAGCTGCTCTTGCC | This study |
| 5495 | TTTCCTACTGTCTCGACG | This study |
| 5608 | GTACCCTATTTCCAAACG | This study |
| 5609 | CGGCTTTGAACATGGACG | This study |
| 5610 | TGCCAGAGGTCGAAACCG | This study |
| 5611 | CGGCATACGGGTTGACCG | This study |
| 6034 | TCATCGAGTCTTCACACG | This study |
| 6035 | ACAAGAGTTGGTTGTACC | This study |
| 7070 | CAGGATGAGTTGTTTGGC | This study |
| 7071 | ACCGCCGATACGCAGTTTCAGCC | This study |
| 7078 | CGGCGGGCTGTTTCCCGCTACAGCATGGC | This study |
| 7079 | GCCATGCTGTAGCGGGAAACAGCCCGCCG | This study |
| 7080 | CGATGGGCTGTAAATCTGGCGGGCGGCGGG | This study |
| 7081 | CCCGCCGCCCGCCAGATTTACAGCCCATCG | This study |
| 7082 | TGTAAACCTGATAGCTCAATTCG | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| Hermes-2 | E. coli/N. gonorrhoeae shuttle vector | 25 |
| pKK232-8 | Promoter selection vector | 7 |
| pSU2718 | Chloramphenicol-resistant cloning vector | 29 |
| pUC18 | Ampicillin-resistant cloning vector | 52 |
| pJKD1595 | Hermes-2 containing a promoterless cat gene | 5 |
| pJKD1926 | Clone from a N. gonorrhoeae MS11-A Sau3A1 library in pUC18 (dnaK) | 17 |
| pJKD2101 | 2.37-kb PCR product amplified from N. gonorrhoeae MS11-A genomic DNA with oligonucleotide primers 5490 and 5492, filled in, and inserted into HincII-digested pSU2718 (rpoH) | This study |
| pJKD2107 | 1.5-kb PCR product amplified from N. gonorrhoeae MS11-A genomic DNA with primers 5608 and 5609, filled in, and inserted into HincII-digested pUC18 (dnaJ) | This study |
| pJKD2108 | 0.95-kb PCR product amplified from N. gonorrhoeae MS11-A genomic DNA with primers 5610 and 5611, filled in, and inserted into HincII-digested pSU2718 (grpE) | This study |
| pJKD2122 | 1.1-kb cassette containing recA promoter::cat transcriptional fusion inserted into HincII pJKD2101; same orientation as rpoH | This study |
| pJKD2124 | 1.1-kb cassette containing recA promoter::cat transcriptional fusion inserted into HincII pJKD2101; opposite orientation to rpoH | This study |
| pJKD2238 | 0.33-kb PCR product amplified from N. gonorrhoeae MS11-A genomic DNA with oligonucleotide primers 6034 and 6035 (Fig. 2A), filled in, and inserted into HincII-digested pUC18 (wild-type dnaK upstream region) | This study |
| pJKD2239 | PCR fusion product amplified with oligonucleotide primers 6034 and 6035 using the PCR products amplified from pJKD2238 with oligonucleotide primer combinations 6034-7080 and 6035-7081 (Fig. 2A) as templates; 0.33-kb product was filled in and inserted into HincII-digested pUC18 (mutated dnaK −10 region) | This study |
| pJKD2240 | PCR fusion product amplified with oligonucleotide primers 6034 and 6035 using the PCR products amplified from pJKD2238 with oligonucleotide primer combinations 6034-7078 and 6035-7079 (Fig. 2A) as templates, the 0.33-kb product was filled in and inserted into HincII-digested pUC18 (mutated dnaK −35 region) | This study |
| pJKD2266 | 0.33-kb BamHI-HindIII fragment of pJKD2239 inserted into pKK232-8 | This study |
| pJKD2267 | 0.33-kb BamHI-HindIII fragment of pJKD2240 inserted into pKK232-8 | This study |
| pJKD2268 | 0.48-kb PCR product amplified from N. gonorrhoeae MS11-A genomic DNA with oligonucleotide primers 4527 and 7070 (Fig. 4), filled in, and inserted into HincII-digested pUC18 (wild-type rpoH upstream region) | This study |
| pJKD2270 | PCR fusion product amplified with oligonucleotide primers 4527 and 7070 using the PCR products amplified from pJKD2101 with oligonucleotide primer combinations 4527-7082 and 7070-7071 (Fig. 4) as templates; the 0.36-kb product was filled in and inserted into HincII-digested pSU2718 (deleted rpoH upstream region) | This study |
| pJKD2273 | 0.33-kb BamHI-HindIII fragment of pJKD2238 inserted into pKK232-8 | This study |
| pJKD2282 | Fragment from pJKD2273 containing wild-type dnaK promoter::cat transcriptional fusion in Hermes-2 | This study |
| pJKD2283 | Fragment from pJKD2266 containing mutated dnaK −10 promoter::cat transcriptional fusion in Hermes-2 | This study |
| pJKD2284 | Fragment from pJKD2267 containing mutated dnaK −35 promoter::cat transcriptional fusion in Hermes-2 | This study |
| pJKD2319 | 0.48-kb BamHI-HindIII fragment of pJKD2268 inserted into pKK232-8 | This study |
| pJKD2320 | 0.36-kb BamHI-HindIII fragment of pJKD2270 inserted into pKK232-8 | This study |
| pJKD2325 | Fragment from pJKD2319 containing wild-type rpoH promoter::cat transcriptional fusion in Hermes-2 | This study |
| pJKD2326 | Fragment from pJKD2320 containing deleted rpoH promoter::cat transcriptional fusion in Hermes-2 | This study |
Determination of CAT levels.
Cell extracts required for enzyme assays were prepared from exponentially growing cultures, the protein concentration was determined, and chloramphenicol acetyltransferase (CAT) assays were performed as described previously (13).
Western blotting.
Western blotting was performed as described previously (27). The dilution of the polyclonal antibody raised against E. coli RpoH antiserum was 1/4,000. The first antibody was detected using peroxidase-conjugated anti-rabbit immunoglobulin at a 1/3,000 dilution.
RESULTS
Identification of the dnak, dnaJ, and grpE genes of N. gonorrhoeae.
A BLAST analysis (1) of the N. gonorrhoeae strain FA1090 (GenBank accession number AE004969) genome database revealed regions where the derived amino acid sequence displayed significant similarity with that of the E. coli DnaK/Hsp70 (3), DnaJ/Hsp40 (4), and GrpE (28) proteins (72.4, 59.1, and 34.3% amino acid sequence identity, respectively [data not shown]). Each of these predicted gonococcal proteins has the domains and amino acid residues shown to be important for its function. Each gene appears to form an independent transcriptional unit, unlike the equivalent genes in other species (43). Each gene was amplified or cloned from N. gonorrhoeae MS11-A (Table 2).
Transcription of the dnak, dnaJ, and grpE genes of N. gonorrhoeae is induced upon exposure to heat shock.
RNA dot blot hybridization was employed to determine whether transcription of the gonococcal dnaK, dnaJ, and grpE genes was induced when cells were exposed to heat shock. Probes for each of the genes are described in Materials and Methods. Total RNA was extracted from cultures of N. gonorrhoeae MS11-A following their exposure to the higher temperature of 42°C for various times. A substantial increase in the level of dnaK-, dnaJ-, and grpE-specific transcripts was observed following exposure to heat shock at 42°C for 10 min (Fig. 1A), indicating that transcription of these genes is induced upon exposure to stress. The amount of transcripts continued to increase slightly with longer exposure to a 42°C environment. The detection of transcripts at 37°C indicates that the proposed σ32-dependent promoters for dnaK, dnaJ, and grpE are functional under nonstress conditions.
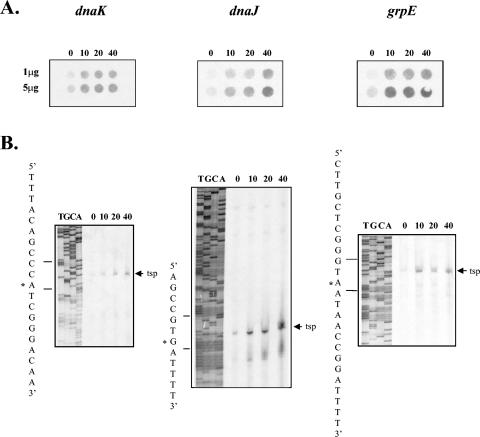
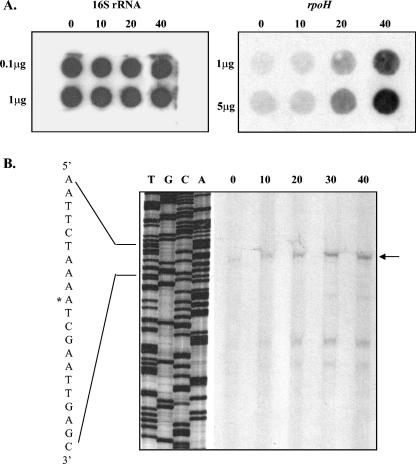
FIG. 1.
Induction of the gonococcal dnaK, dnaJ, and grpE genes upon exposure to heat shock. (A) RNA dot blot hybridizations of RNA extracted from cells exposed to heat shock at 42°C for the number of minutes indicated above each panel. The amount of RNA (μg) transferred to the membranes is indicated to the left. Filters were probed with the gonococcal dnaK, dnaJ, and grpE genes as indicated. (B) Primer extension analysis of the promoter regions of these genes in N. gonorrhoeae MS11-A. Total RNA (50 μg per lane) was extracted from cells that had been heat shocked at 42°C for the number of minutes indicated above each lane. Primer extension products obtained are indicated by arrows. Sequencing ladders adjacent to the primer extension reactions are marked TGCA. The TSPs are indicated by the asterisks on the sequences at the left of each panel and shown in Fig. 2.
To confirm induction of transcription of these genes, and to locate the promoters responsible for their transcription, the transcriptional start points (TSPs) were mapped by primer extension using the same RNA preparations. Oligonucleotide primers 6035 (Fig. 2A; Table 1), 5494 (Fig. 2B; Table 1), and 5495 (Fig. 2C; Table 1) were used to identify the TSPs of dnaK, dnaJ, and grpE, respectively, and were also used in sequencing reactions. Primer extension products were obtained for all the genes at 37°C; however, the signals were greatly intensified with a shift to 42°C (Fig. 1B). In each case, the primer extension product obtained increased in intensity with continued exposure to heat shock. The TSP for dnaK under physiological and stress conditions was mapped to an A residue situated 61 bp upstream of the putative initiation codon (Fig. 1B and 2A). The TSP for dnaJ under stress and nonstress conditions mapped to a G residue and increased with a thermal upshift (Fig. 1B and 2B). Transcription of grpE was initiated from an A residue 42 bp upstream of the putative start codon (Fig. 1B and 2C). The sequence upstream of each stress-induced TSP displayed similarity to the −10 and −35 regions of σ32-dependent promoters (Fig. 2) (8).
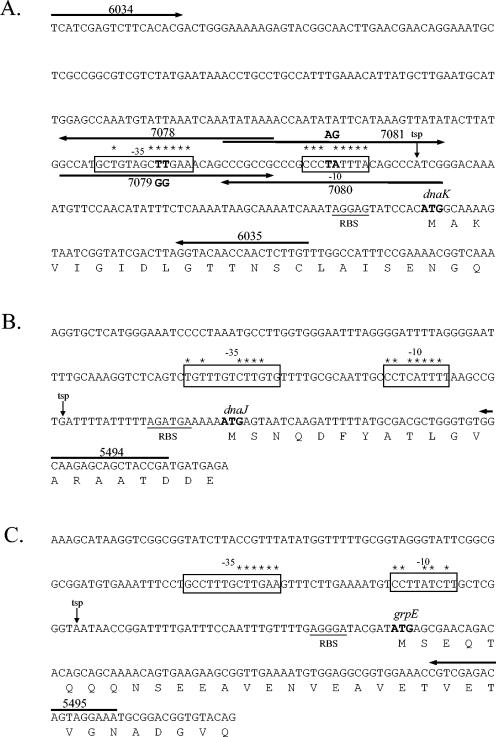
FIG. 2.
Nucleotide sequences of the promoter regions of the dnaK (A), dnaJ (B), and grpE (C) genes from N. gonorrhoeae. The initiation codon for each gene is shown in boldface. The deduced amino acid sequences of the proteins are shown beneath the corresponding nucleotide sequences. Putative ribosome binding sites (RBS) are underlined. Oligonucleotide primers are indicated by numbered arrows. The σ32 promoter sequences are boxed (−35 and −10), and asterisks indicate nucleotides identical to those in the consensus sequence (8). In panel A, boldface letters in the −35 and −10 boxes indicate those nucleotides changed in site-directed mutagenesis experiments. The introduced changes are indicated above or below the boxes. Downward-pointing arrows represent TSPs determined by primer extension analysis (Fig. 1).
dnaK is transcribed from a σ32 promoter.
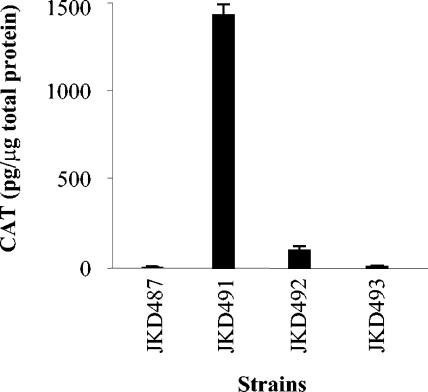
To verify that the putative σ32 promoters were responsible for the increased transcription observed under stress conditions, site-directed mutagenesis of the promoter upstream of dnaK was performed. The nucleotide changes made to bases within the −10 and −35 regions (Fig. 2) were designed to render RpoH incapable of recognizing the promoter. The wild-type, mutated −10, and mutated −35 promoter regions were fused to a promoterless cat gene and introduced into N. gonorrhoeae JKD484 using plasmids pJKD2282, pJKD2283, and pJKD2284 (Table 2), generating strains JKD491, JKD492, and JKD493, respectively. As a negative control, a promoterless cat gene was also introduced into this background using plasmid pJKD1595 (Table 2) to give strain JKD487.
Exponentially growing cultures of the gonococcal strains containing the promoter::cat transcriptional fusions were subjected to heat shock at 42°C for 20 min. Cell extracts of the cultures were prepared, and CAT levels were determined (Fig. 3). Basal levels of CAT were obtained for the negative control strain JKD487. As expected, much higher levels of CAT were obtained for strain JKD491, which contains the wild-type dnaK promoter region and establishes the activity of the σ32-dependent promoter in this background. The CAT levels obtained for strains JKD492 and JKD493, which contain the mutated −10 and −35 boxes, respectively, were greatly reduced, approximately 17- and 80-fold, respectively. These results indicate that the putative σ32-dependent promoter is responsible for transcription of the dnaK gene and the bases mutated in the −35 region have a more pronounced effect on sigma binding than those mutated in the −10 region (Fig. 2A).
FIG. 3.
Transcriptional analysis of the gonococcal dnaK upstream region in N. gonorrhoeae strains JKD491, JKD492, and JKD493, which contain promoter::cat transcriptional fusions integrated into the conjugative plasmid ptetM25.2. These strains carry the wild-type dnaK promoter region, the mutated −10 region, and the mutated −35 region, respectively. Strain JKD487 was the negative control containing a promoterless cat gene. Cell extracts were prepared from strains exposed to heat shock at 42°C for 20 min. The CAT levels shown are the means of results of four separate assays. The error bars represent standard deviations.
Identification and nucleotide sequence analysis of the rpoH gene from N. gonorrhoeae.
A BLAST search (1) of the N. gonorrhoeae strain FA1090 (GenBank accession number AE004969) genome database with the E. coli RpoH amino acid sequence (26) indicated the presence of an rpoH homologue in N. gonorrhoeae (50% amino acid similarity [data not shown]). Based on flanking FA1090 sequences, oligonucleotide primer pair 5490-5492 (Table 1) was designed to amplify rpoH and flanking regions from N. gonorrhoeae MS11-A genomic DNA by PCR. Flanking regions were included to enable subsequent construction of an rpoH mutant (see below). Analysis of the gonococcal RpoH sequence indicates that proposed functional domains and residues have been conserved. These sequences include regions required for promoter recognition and binding to RNA polymerase (32, 35). rpoH was cloned from N. gonorrhoeae MS11-A (Table 2).
The rpoH gene from N. gonorrhoeae appears to be essential for growth.
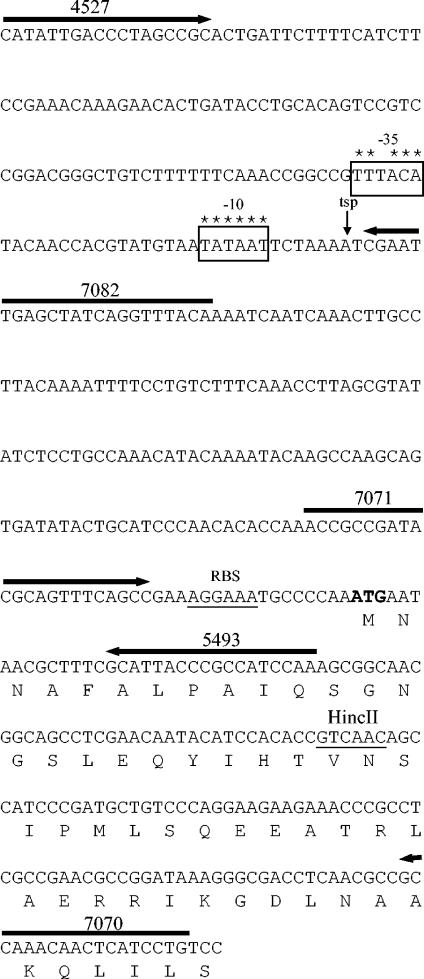
An attempt was made to mutate the rpoH gene from N. gonorrhoeae by insertional activation with an antibiotic resistance cassette. A 1.1-kb cassette containing the gonococcal recA promoter fused to a promoterless cat gene (5) was cloned into a HincII site located 70 bp downstream of the putative translation start site in the rpoH gene in pJKD2101 (Fig. 4). The antibiotic resistance cassette was cloned in both orientations, giving plasmids pJKD2122 and pJKD2124 (Table 2). Each plasmid was transformed into N. gonorrhoeae MS11-A in an attempt to detect integration into the gonococcal chromosome by allelic exchange at the rpoH locus. As an E. coli rpoH mutant was temperature sensitive (56), plates were incubated at 30 and 37°C to determine whether this phenotype applied to the gonococcal mutant. No chloramphenicol-resistant transformants were obtained for cultures incubated at 30°C. Only a few chloramphenicol-resistant transformants were recovered from those cultures incubated at 37°C. Southern hybridization suggested that these transformants resulted from either random integration into sites other than the rpoH gene or a single crossover event, resulting in both an inactivated and an intact copy of the rpoH gene. These results strongly suggest that the rpoH gene of N. gonorrhoeae may be essential for growth at 30 and 37°C.
FIG. 4.
Nucleotide sequence of the promoter region of the rpoH gene from N. gonorrhoeae. The ATG initiation codon is shown in boldface. The deduced amino acid sequence of the gene is shown beneath the corresponding nucleotide sequences. The putative ribosome binding site (RBS) is underlined. Oligonucleotide primers are indicated by the numbered arrows. The σ70 promoter sequences (−35 and −10) are boxed, and asterisks indicate nucleotides identical to those in the consensus sequence (18). The TSP is indicated by the downward-pointing arrow. The HincII site used in attempting to construct the rpoH mutant is indicated.
The rpoH gene from N. gonorrhoeae is transcribed from a σ70 promoter under steady-state and stress conditions.
RNA dot blot hybridization was used to determine the transcriptional levels of rpoH after heat shock. Total RNA was extracted from an exponentially growing culture of N. gonorrhoeae MS11-A following exposure to heat shock at 42°C. The rpoH-specific mRNA increased upon heat shock, but only after 20 min, and became much more intense after 40 min of exposure (Fig. 5A), indicating that transcription of rpoH is induced by stress. The delayed increase in the level of rpoH mRNA compared to dnaK, dnaJ, and grpE mRNA (Fig. 1) suggests that an increase in rpoH transcription is not responsible for the increased transcription of these genes.
FIG. 5.
Induction of the gonococcal rpoH gene upon exposure to heat shock. (A) RNA dot blot hybridizations of RNA extracted from cells exposed to heat shock at 42°C for the number of minutes indicated above each well. The amount of RNA (μg) transferred to the membranes is indicated to the left of each panel. Filters were probed with the gonococcal 16S rRNA and rpoH genes as indicated. (B) Primer extension analysis of the rpoH upstream region of N. gonorrhoeae MS11-A. Total RNA (50 μg per lane) was extracted from cells heat shocked at 42°C for the number of minutes indicated above each lane. Primer extension products obtained using oligonucleotide primer 5493 (Fig. 4) are indicated by the arrow. The sequencing ladder adjacent to the primer extension reaction lanes is marked TGCA. The TSP is indicated by the asterisk on the sequence at the left and is also marked on Fig. 4.
To map the rpoH promoter, primer extension analysis was performed using the same RNA preparations. The oligonucleotide primer 5493 was used for the primer extension and sequencing reactions (Fig. 4). A weak product was obtained for rpoH at 37°C, and this signal intensified following the temperature shift (Fig. 5B). The increase in transcription was slight at 10 min and more prominent at 20 min, consistent with the RNA dot blot hybridization results (Fig. 5A). The TSP mapped to an A residue located 181 bp upstream of the putative start codon (Fig. 4). The sequence upstream of the TSP displayed strong similarity to the −10 and −35 regions of σ70-dependent promoters (18). Several shorter products (Fig. 5B) appear to be artifactual, as they are not associated with consensus promoter sequences. Therefore, N. gonorrhoeae appears to use a single σ70-dependent promoter for transcription in the absence or presence of stress conditions.
RpoH synthesis is induced by heat shock.
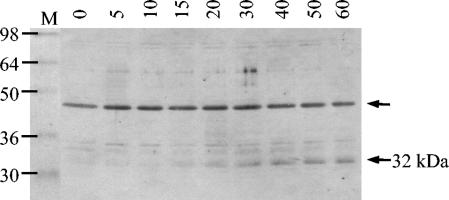
Expression of the gonococcal RpoH protein was investigated to establish whether the rate of synthesis increases with the observed increase in transcription. Exponentially growing cells of N. gonorrhoeae MS11-A were heat shocked at 42°C and subsequently harvested. Cell extracts were prepared, and equivalent amounts of protein were separated on a sodium dodecyl sulfate-polyacrylamide gel. Western blot analysis was performed with a polyclonal antibody raised against E. coli RpoH (Fig. 6). Fortuitously, the antibody cross-reacted with several proteins in the gonococcal extracts, demonstrating an equivalent amount of protein in each sample. These proteins were not recognized by normal rabbit serum. A protein of approximately 32 kDa that was recognized by the antibody was substantially induced following incubation at 42°C. This induction seemed to occur at around 15 min and increased with prolonged exposure to heat shock. It coincided with the gradual increase in rpoH transcription as determined by RNA dot blot analysis and primer extension (Fig. 5). Transcription of dnaK, dnaJ, and grpE is induced as early as 10 min after a thermal upshift. The delayed increase in RpoH protein levels supports the suggestion that neither an increase in transcription nor an increase in translation of rpoH is responsible for induction of the stress genes.
FIG. 6.
Western blot analysis of N. gonorrhoeae MS11-A exposed to heat shock at 42°C for the number of minutes indicated above each lane and whole cell extracts subsequently prepared. Equal volumes of extract from each sample were fractionated on a 12% polyacrylamide gel electrophoresis gel. The membrane was incubated with the polyclonal antibody raised against E. coli RpoH. Prestained standards (in kDa) served as the markers (M). Proteins recognized by the antiserum are indicated by arrows.
RpoH synthesis does not seem to be subject to translational regulation.
A search for elements involved in expression of rpoH in other species was performed. In the majority of gamma proteobacteria, rpoH regulation seems to occur primarily at the translational level. The 5′ end of the mRNA contains a sequence that forms an internal secondary structure involving the ribosome binding site, initiation codon, and downstream 20 nucleotides, termed the downstream box, which represses rpoH translation at physiological temperatures due to the inaccessibility of the ribosome binding site (55). Following a temperature increase, the structure partially melts, permitting an enhanced level of translation due to the complementarity of the downstream box with 16S rRNA (31). Sequence analysis indicates that the gonococcal rpoH transcript lacks the ability to form such a structure, in part due to the absence of the downstream box (data not shown). This was confirmed via a prediction of the secondary structure of the gonococcal rpoH mRNA (data not shown) using the MULFOLD program (21) and implies that regulation of rpoH in the gonococcus is unlike the regulation in most gamma proteobacteria.
The leader sequence may regulate transcription of the gonococcal rpoH gene.
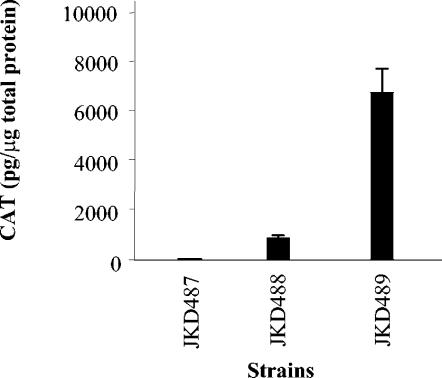
The discovery that the rpoH gene appears to be regulated in part at the transcriptional level led to closer examination of the upstream region and a search for potential regulatory elements. An unusual finding was the presence of the relatively long leader sequence of 172 bp. To determine the transcriptional significance, if any, of the leader sequence, the sequence was deleted, and transcriptional activity from the rpoH promoter region in N. gonorrhoeae was evaluated. The wild-type and deleted promoter regions were fused to a promoterless cat gene and introduced into N. gonorrhoeae JKD484 using plasmids pJKD2325 and pJKD2326 (Table 2), generating strains JKD488 and JKD489, respectively. As a negative control, a promoterless cat gene was also introduced into this background by using plasmid pJKD1595 (Table 2) to give strain JKD487.
Exponentially growing cultures of the gonococcal strains containing the promoter::cat transcriptional fusions were subjected to heat shock at 42°C for 20 min, since rpoH transcription increases substantially by this time (Fig. 5). Cell extracts of the cultures were prepared, and CAT levels were determined (Fig. 7). Basal levels of CAT were obtained for the negative control strain JKD487. Deletion of the leader sequence in strain JKD489 resulted in an approximate sevenfold increase in CAT compared to that in JKD488 containing the wild-type rpoH upstream region, suggesting an important role for the leader sequence in the transcription of rpoH. Analyses of E. coli indicated that the same TSP is used in the deleted construct and that transcript levels from this construct are higher than those in the nondeleted construct (data not shown).
FIG. 7.
Transcriptional analysis of the gonococcal rpoH upstream region in N. gonorrhoeae strains JKD488 and JKD489, which contain promoter::cat transcriptional fusions integrated into the conjugative plasmid ptetM25.2. These strains carry the wild-type rpoH promoter region and that with the deleted leader sequence, respectively. Strain JKD487, containing a promoterless cat gene, was the negative control. Cell extracts were prepared from strains exposed to heat shock at 42°C for 20 min. The CAT levels shown are the means of results from four separate assays. The error bars represent standard deviations.
DISCUSSION
The stress responses of a number of species have been studied, and expression of the chaperones DnaK, DnaJ, and GrpE has generally been found to be regulated at the transcriptional level upon exposure to heat shock (54). We investigated the heat shock response in N. gonorrhoeae and characterized the genes encoding the molecular chaperones DnaK, DnaJ, and GrpE from N. gonorrhoeae FA1090 and MS11-A. Analysis of flanking regions indicated that in the gonococci, these genes were not arranged in an operon, an organization that is common in many species (43). It is possible that this is because different levels of dnaK and dnaJ expression are required in N. gonorrhoeae. However, this end result has been achieved with E. coli because of the infrequent translation initiation of dnaJ mRNA (4, 44). To confirm induction of these genes upon heat shock, RNA dot blot hybridization and primer extension analyses were employed (Fig. 1). All three genes were significantly induced upon exposure to heat stress. A similar result had previously been obtained for the gonococcal groE operon (48).
There appear to be two primary groups of transcriptional regulatory strategies employed by bacteria to regulate the dnaK chaperone system. The majority of gram-positive bacteria and several gram-negative bacteria utilize a negative control mechanism in which stress exposure displaces a repressor protein bound to a CIRCE element located near the promoter. This element is highly conserved among these species but absent from N. gonorrhoeae. Several species of gram-negative bacteria have adopted a positive mode of regulation of the dnaK operon where the alternative sigma factor, RpoH, directs transcription under stress (12, 32, 38). Tauschek et al. (48) demonstrated that transcription of the gonococcal groE operon during stress is initiated from a σ32 promoter.
Primer extension analysis of dnaK, dnaJ, and grpE revealed that transcription under nonstress and heat shock conditions emanated from promoters with similarity to the −10 and −35 regions of σ32 promoters (Fig. 2). Alignment of the consensus sequences for heat shock promoters of E. coli (8) and alpha purple proteobacteria (42) with those identified in this study and that from the gonococcal groE operon (48) revealed that the gonococcal σ32 promoters shared similarity to both of the consensus sequences. In the −10 box of the gonococcal genes there is some variation among the stretch of C residues found in the E. coli consensus. Based upon the above alignment, site-directed mutagenesis was performed on the potential dnaK promoter to assess its activity in N. gonorrhoeae (Fig. 3). This confirmed that the σ32 promoter identified by primer extension analysis is in fact responsible for dnaK transcription.
We were unable to create a gonococcal rpoH mutant by insertional inactivation, suggesting that this gene may be essential for viability as is the case in E. coli (56). This finding was not surprising since a basal level of the proteins encoded by the heat shock genes it transcribes would be required for normal growth and the maintenance of cellular functions.
To determine the mechanisms responsible for regulation of the gonococcal rpoH gene, a search for the regulatory elements operative in other organisms was performed. The lack of a σ32 consensus sequence in the rpoH upstream region suggested that this gene is not autoregulated (Fig. 4). The positive regulatory element, the downstream box, was lacking from the gonococcal transcript, and the 5′ region did not form the characteristic secondary structure (data not shown) responsible for thermal regulation in E. coli and several other gamma subdivision proteobacteria (31, 32, 55). RNA dot blot hybridization and primer extension analyses were performed (Fig. 5). As for dnaK, dnaJ, and grpE, transcription of rpoH increased substantially following temperature stress. The primer extension experiments showed that this increase was evident at 20 min. Transcription emanated from a σ70 promoter (Fig. 4), and no other promoters were identified under the conditions tested. This is in contrast to the situation in many other species, in which transcription occurs from multiple promoters (11). The level of RpoH protein was investigated following heat shock and appeared to increase at around 15 min (Fig. 6). The concomitant increase in transcription and translation suggests that rpoH from N. gonorrhoeae is controlled, at least partially, at the level of transcription following heat shock. However, the levels of dnaK, dnaJ, and grpE increased prior to the increase observed for rpoH, suggesting that activation of preformed RpoH was sufficient to induce the heat shock response. Such an energy-efficient mode of regulation would be particularly advantageous to N. gonorrhoeae since it could respond rapidly to stress stimuli in vivo. Therefore, the increased amount of RpoH obtained by transcription late in the heat shock response is probably a subsidiary mechanism for sustaining the elevated expression of Hsps. The immediate increase in heat shock protein synthesis prior to an increase in the RpoH level has also been observed for A. tumefaciens (34).
In E. coli, the DnaK chaperone system modulates the cellular level of RpoH by regulating its synthesis, stability, and activity (14, 47). The control of activity and stability is exerted by preferential binding of the DnaK system to RpoH under normal conditions, rendering the DnaK system sensitive to proteolysis and sequestering it away from core RNA polymerase (19). This form of activity and stability control of RpoH may be occurring in N. gonorrhoeae. With stress, the DnaK chaperone system could preferentially bind to misfolded or denatured proteins so that RpoH was free to bind to core RNA polymerase and induce transcription of the heat shock genes.
To further elucidate the regulatory mechanism responsible for the transcriptional regulation of rpoH from N. gonorrhoeae, the uncharacteristically long leader sequence was deleted, and transcriptional activity in N. gonorrhoeae was assessed. As determined by measuring CAT protein levels (Fig. 7), deletion of the leader sequence led to a significantly higher level of transcription. A similar result was found for E. coli. This may be a result of the different secondary structures adopted by each of the individual transcripts which may effect transcription or translation. The secondary structure of the intact rpoH promoter region would be more extensive and complex than the deleted one and may therefore decrease mRNA stability or increase the frequency of premature transcription termination (2). Alternatively, these results could suggest the binding of a repressor to the rpoH leader sequence. This is to be investigated.
Overall, the data presented here suggest that the gonococcal stress response is regulated predominantly at the level of activity immediately following a thermal upshift. An increase in rpoH transcription and a subsequent increase in RpoH levels are then observed following ongoing exposure to heat shock. The stability of RpoH, the mechanism involved in regulation of rpoH transcription, and the role of the leader sequence have yet to be determined.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia.
We thank Bernd Bukau for the polyclonal antibodies to the E. coli RpoH protein. We acknowledge the Gonococcal Genome Sequencing Project, supported by USPHS/NIH grant number AI38399, and B. A. Roe, L. Song, S. P. Lin, X. Yuan, S. Clifton, Tom Ducey, Lisa Lewis, and D. W. Dyer at the University of Oklahoma.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Avedissian, M., D. Lessing, J. W. Gober, L. Shapiro, and S. L. Gomes. 1995. Regulation of the Caulobacter crescentus dnaKJ operon. J. Bacteriol. 177:3479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., and E. A. Craig. 1984. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc. Natl. Acad. Sci. USA 81:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell, J. C., K. Tilly, E. Craig, J. King, M. Zylicz, and C. Georgopoulos. 1986. The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene. A gene that encodes a heat shock protein. J. Biol. Chem. 261:1782-1785. [PubMed] [Google Scholar]
- 5.Black, C. G., J. A. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, C. G., J. A. Fyfe, and J. K. Davies. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208:61-66. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J. 1984. Plasmid vectors for the selection of promoters. Gene 27:151-160. [DOI] [PubMed] [Google Scholar]
- 8.Cowing, D. W., J. C. Bardwell, E. A. Craig, C. Woolford, R. W. Hendrix, and C. A. Gross. 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. USA 82:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva, A. C., R. C. Simao, M. F. Susin, R. L. Baldini, M. Avedissian, and S. L. Gomes. 2003. Downregulation of the heat shock response is independent of DnaK and σ32 levels in Caulobacter crescentus. Mol. Microbiol. 49:541-553. [DOI] [PubMed] [Google Scholar]
- 10.Emetz, D., and G. Klug. 1998. Cloning and characterization of the rpoH gene of Rhodobacter capsulatus. Mol. Gen. Genet. 260:212-217. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, J. W., V. Vaughn, W. A. Walter, F. C. Neidhardt, and C. A. Gross. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419-432. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 13.Fyfe, J. A., C. S Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ70 promoter during growth in vitro. J. Bacteriol. 177:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamer, J., G. Multhaup, T. Tomoyasu, J. S. McCarty, S. Rudiger, H. J. Schonfeld, C. Schirra, H. Bujard, and B. Bukau. 1996. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor σ32. EMBO J. 15:607-617. [PMC free article] [PubMed] [Google Scholar]
- 15.Goff, S. A., and A. L. Goldberg. 1985. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock proteins. Cell 41:587-595. [DOI] [PubMed] [Google Scholar]
- 16.Guckenberger, M., S. Kurz, C. Aepinus, S. Theiss, S. Haller, T. Leimbach, U. Panzner, J. Weber, H. Paul, A. Unkmeir, M. Frosch, and G. Dietrich. 2002. Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol. 184:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton, C. W. 1999. Analysis of repeated sequence elements in pathogenic Neisseria. Ph.D. thesis. Monash University, Victoria, Australia.
- 18.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman, C., and R. D'Ari. 1998. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr. Opin. Microbiol. 1:204-209. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger, J. A., D. H. Turner, and M. Zuker. 1990. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 183:281-306. [DOI] [PubMed] [Google Scholar]
- 22.Jager, S., A. Jager, and G. Klug. 2004. CIRCE is not involved in heat-dependent transcription of groESL but in stabilization of the mRNA 5′-end in Rhodobacter capsulatus. Nucleic Acids Res. 32:386-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanemori, M., K. Nishihara, H. Yanagi, and T. Yura. 1997. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 179:7219-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann, S. H. 1990. Heat shock proteins and the immune response. Immunol. Today 11:129-136. [DOI] [PubMed] [Google Scholar]
- 25.Kupsch, E. M., D. Aubel, C. P. Gibbs, A. F. Kahrs, T. Rudel, and T. F. Meyer. 1996. Construction of Hermes shuttle vectors: a versatile system useful for genetic complementation of transformable and non-transformable Neisseria mutants. Mol. Gen. Genet. 250:558-569. [DOI] [PubMed] [Google Scholar]
- 26.Landick, R., V. Vaughn, E. T. Lau, R. A. VanBogelen, J. W. Erickson, and F. C Neidhardt. 1984. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell 38:175-182. [DOI] [PubMed] [Google Scholar]
- 27.Laskos, L., J. P. Dillard, H. S. Seifert, J. A. Fyfe, and J. K. Davies. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilise the pilE σ54 promoter. Gene 208:95-102. [DOI] [PubMed] [Google Scholar]
- 28.Lipinska, B., J. King, D. Ang, and C. Georgopoulos. 1988. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucleic Acids Res. 16:7545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez, E., B. Bartolome, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 30.Mogk, A., C. Schlieker, C. Strub, W. Rist, J. Weibezahn, and B. Bukau. 1999. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278:17615-17624. [DOI] [PubMed] [Google Scholar]
- 31.Morita, M. T., Y. Tanaka, T. S. Kodama, Y. Kyogoku, H. Yanagi, and T. Yura. 1999. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev. 13:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakahigashi, K., H. Yanagi, and T. Yura. 1995. Isolation and sequence analysis of rpoH genes encoding σ32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 23:4383-4390. [PMC free article] [PubMed] [Google Scholar]
- 33.Nakahigashi, K., H. Yanagi, and T. Yura. 1998. Regulatory conservation and divergence of σ32 homologs from gram-negative bacteria: Serratia marcescens, Proteus mirabilis, Pseudomonas aeruginosa, and Agrobacterium tumefaciens. J. Bacteriol. 180:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahigashi, K., H. Yanagi, and T. Yura. 2001. DnaK chaperone-mediated control of activity of a σ32 homolog (RpoH) plays a major role in the heat shock response of Agrobacterium tumefaciens. J. Bacteriol. 183:5302-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narberhaus, F., and S. Balsiger. 2003. Structure-function studies of Escherichia coli RpoH (σ32) by in vitro linker insertion mutagenesis. J. Bacteriol. 185:2731-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, Y., H. Mitsui, T. Sato, and K. Minamisawa. 2001. Two RpoH homologs responsible for the expression of heat shock protein genes in Sinorhizobium meliloti. Mol. Gen. Genet. 264:902-912. [DOI] [PubMed] [Google Scholar]
- 37.Pannekoek, Y., J. Dankert, and J. P. van Putten. 1992. Identification and characterization of a cross-reactive and a unique B-cell epitope on the hsp60 homologue from Neisseria gonorrhoeae. FEMS Microbiol. Lett. 78:23-29. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Santos, J., J. Collado-Vides, M. Garcia-Varela, and M. C. Gomez-Eichelmann. 2001. Conserved regulatory elements of the promoter sequence of the gene rpoH of enteric bacteria. Nucleic Acids Res. 29:380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen, R., K. Buttner, D. Becher, K. Nakahigashi, T. Yura, M. Hecker, and E. Z. Ron. 2002. Heat shock proteome of Agrobacterium tumefaciens: evidence for new control systems. J. Bacteriol. 184:1772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, S., and H. Ishikawa. 1997. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J. Bacteriol. 179:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal, E., E. Billyard, M. So, S. Storzbach, and T. F. Meyer. 1985. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell 40:293-300. [DOI] [PubMed] [Google Scholar]
- 42.Segal, R., and E. Z. Ron. 1995. The dnaKJ operon of Agrobacterium tumefaciens: transcriptional analysis and evidence for a new heat shock promoter. J. Bacteriol. 177:5952-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal, R., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 44.Sell, S. M., C. Eisen, D. Ang, M. Zylicz, and C. Georgopoulos. 1990. Isolation and characterization of dnaJ null mutants of Escherichia coli. J. Bacteriol. 172:4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva Laport, M., M. R. da Silva, C. Costa Silva, M. do Carmo de Freire Bastos, and M. Giambiagi-deMarval. 2003. Heat-resistance and heat-shock response in the nosocomial pathogen Enterococcus faecium. Curr. Microbiol. 46:313-317. [DOI] [PubMed] [Google Scholar]
- 46.Spohn, G., A. Danielli, D. Roncarati, I. Delany, R. Rappuoli, and V. Scarlato. 2004. Dual control of Helicobacter pylori heat shock gene transcription by HspR and HrcA. J. Bacteriol. 186:2956-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straus, D., W. Walter, and C. A. Gross. 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of σ32. Genes Dev. 4:2202-2209. [DOI] [PubMed] [Google Scholar]
- 48.Tauschek, M., C. W. Hamilton, L. A. Hall, C. Chomvarin, J. A. Fyfe, and J. K. Davies. 1997. Transcriptional analysis of the groESL operon of Neisseria gonorrhoeae. Gene 189:107-112. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, A. C., and M. Tan. 2004. Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J. Bacteriol. 186:3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodbury, R., and W. G. Haldenwang. 2003. HrcA is a negative regulator of the dnaK and groESL operons of Streptococcus pyogenes. Biochem. Biophys. Res. Commun. 302:722-727. [DOI] [PubMed] [Google Scholar]
- 51.Wu, J., and A. Newton. 1997. The Caulobacter heat shock sigma factor gene rpoH is positively autoregulated from a σ32-dependent promoter. J. Bacteriol. 179:514-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 53.Yura, T., H. Nagai, and H. Mori. 1993. Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 47:321-350. [DOI] [PubMed] [Google Scholar]
- 54.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:153-158. [DOI] [PubMed] [Google Scholar]
- 55.Yuzawa, H., H. Nagai, H. Mori, and T. Yura. 1993. Heat induction of σ32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 21:5449-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, Y. N., N. Kusukawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor σ32. J. Bacteriol. 170:3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]