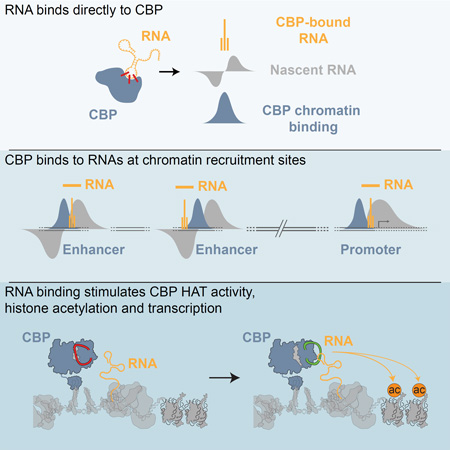
Summary
CBP/p300 are transcription co-activators whose binding is a signature of enhancers, cis-regulatory elements that control patterns of gene expression in multicellular organisms. Active enhancers produce bi-directional enhancer RNAs (eRNAs) and display CBP/p300 dependent histone acetylation. Here, we demonstrate that CBP binds directly to RNAs in vivo and in vitro. RNAs bound to CBP in vivo include a large number of eRNAs. Using steady-state histone acetyltransferase (HAT) assays we show that an RNA binding region in the HAT domain of CBP— a regulatory motif unique to CBP/p300—allows RNA to stimulate CBP’s HAT activity. At enhancers where CBP interacts with eRNAs, stimulation manifests in RNA-dependent changes in the histone acetylation mediated by CBP, such as H3K27ac, and by corresponding changes in gene expression. By interacting directly with CBP, eRNAs contribute to the unique chromatin structure at active enhancers, which in turn is required for regulation of target genes.
Graphical abstract
Introduction
Enhancers confer spatiotemporal specificity to gene expression and orchestrate gene expression patterns in response to environmental and/or developmental stimuli (Lam et al., 2014). Enhancers account for the majority of transcription factor binding sites in the genome and have a characteristic chromatin structure. (Heinz et al., 2015). The number of potential enhancer elements greatly exceeds the number of coding regions, however most are maintained in a silent or poised state until an activating signal is received (Rada-Iglesias et al., 2011). Upon activation, binding of cell-type or temporal-specific transcription factors (TFs) to cognate DNA motifs recruits transcription co-activators, such as histone acetyltransferases CBP and p300 and RNA polymerase II (PolII) (Creyghton et al., 2010; Li et al., 2013). This results in two hallmarks of active enhancers: the production of bi-directional non-coding RNA transcripts known as enhancer RNAs (eRNAs) (Hah et al., 2011; Kim et al., 2010; Wang et al., 2011; Ørom et al., 2010) and elevated histone 3 lysine 27 acetylation (H3K27ac) (Creyghton et al., 2010).
Increasing evidence points to a direct role for eRNAs in enhancer function (Kim and Shiekhattar, 2015). eRNAs regulate looping between enhancers and promoters through recruitment of cohesin (Hsieh et al., 2014; Li et al., 2013), increase chromatin accessibility (Mousavi et al., 2013), and aid the recruitment of mediator to promoters (Lai et al., 2013). Consistently, depletion of eRNAs affects transcription from enhancer-associated genes (Kim et al., 2010; Lai et al., 2013; Li et al., 2013; Melo et al., 2013; Mousavi et al., 2013). At poised genes, eRNAs promote release of paused PolII into the gene body (Schaukowitch et al., 2014).
Several non-coding RNAs (ncRNAs) interact with chromatin-modifying enzymes and modulate enzymatic function (Cifuentes-Rojas et al., 2014; Di Ruscio et al., 2013; Kaneko et al., 2014a; 2014b; 2013; Wongtrakoongate et al., 2015; Yang et al., 2014). Binding of RNA to Polycomb repressive complex 2 (PRC2) decreases its methyltransferase activity and recruitment to promoters (Beltran et al., 2016; Cifuentes-Rojas et al., 2014; Kaneko et al., 2013; 2014a; 2014b). ncRNA transcribed proximal to the C/EBPA locus interacts with DNA methyltransferase 1 (DNMT1) to block DNA methylation at the promoter (Di Ruscio et al., 2013). A key question is whether eRNAs play a similar role in modulating chromatin at enhancers. Indeed, eRNA transcripts do not direct methylation changes at enhancers upon activation, which result from active transcription by PolII (Kaikkonen et al., 2013).
Elevated H3K27ac is another hallmark of active enhancers, produced by CBP and p300 (Jin et al., 2011; Tie et al., 2009). CBP/p300 bind a vast array of TFs through transactivation domains (TADs), acting as a transcription network ‘hub’ (Bedford et al., 2010; Wang et al., 2013). The broad interactome results in nearly-universal recruitment of CBP and p300 to enhancers, such that CBP/p300 occupancy is a key feature (Creyghton et al., 2010) and identifies enhancers genome-wide (May et al., 2012). The enzymatic activity of CBP and p300 is localized in their catalytic histone acetyltransferase (HAT) domain and targets both histones and numerous TF substrates, including p53 and PolII (Barlev et al., 2001; Jin et al., 2011; Schröder et al., 2013; Tie et al., 2009; Wang et al., 2008a). Acetylation of PolII by CBP promotes PolII release into the gene body (Schröder et al., 2013). H3K27ac correlates with regions of active transcription genome-wide (Tie et al., 2009; Wang et al., 2008b), and CBP-mediated acetylation can directly stimulate transcription by improving TF recruitment efficiency and promoter escape of PolII (Stasevich et al., 2014). Targeting the p300-HAT domain alone to enhancers using de-activated Cas9 nuclease (dCas9) and enhancer-specific guide RNAs (gRNAs) increases expression of enhancer-associated genes (Hilton et al., 2015). Thus, p300-mediated acetylation is sufficient for gene activation at endogenous enhancers and loci.
RNA could potentially impact the function of CBP and p300. Transcription of the antisense ncRNA khps1 from the promoter of the proto-oncogene SPHK1 helps to recruit p300 to the SPHK1 promoter (Postepska-Igielska et al., 2015). Moreover, stable depletion of eRNA transcribed from an enhancer for chorionic gonadotropin alpha (Cga) decreased H3K27ac (Pnueli et al., 2015). A key open question is whether there is a direct mechanistic connection between eRNA and CBP function at regulatory elements.
Here we demonstrate a direct interaction between CBP and RNA, including a large population of eRNAs. In vivo crosslinking and sequencing identified a subset of CBP-bound eRNAs associated with genes requiring CBP for their transcription. eRNA binding directly stimulates acetyltransferase activity of CBP in vitro, modulating H3K27ac levels. In cells, eRNA transcripts modulate their local chromatin environment, regulating H3K27ac to promote gene expression.
Results
CBP interacts with RNA in vivo
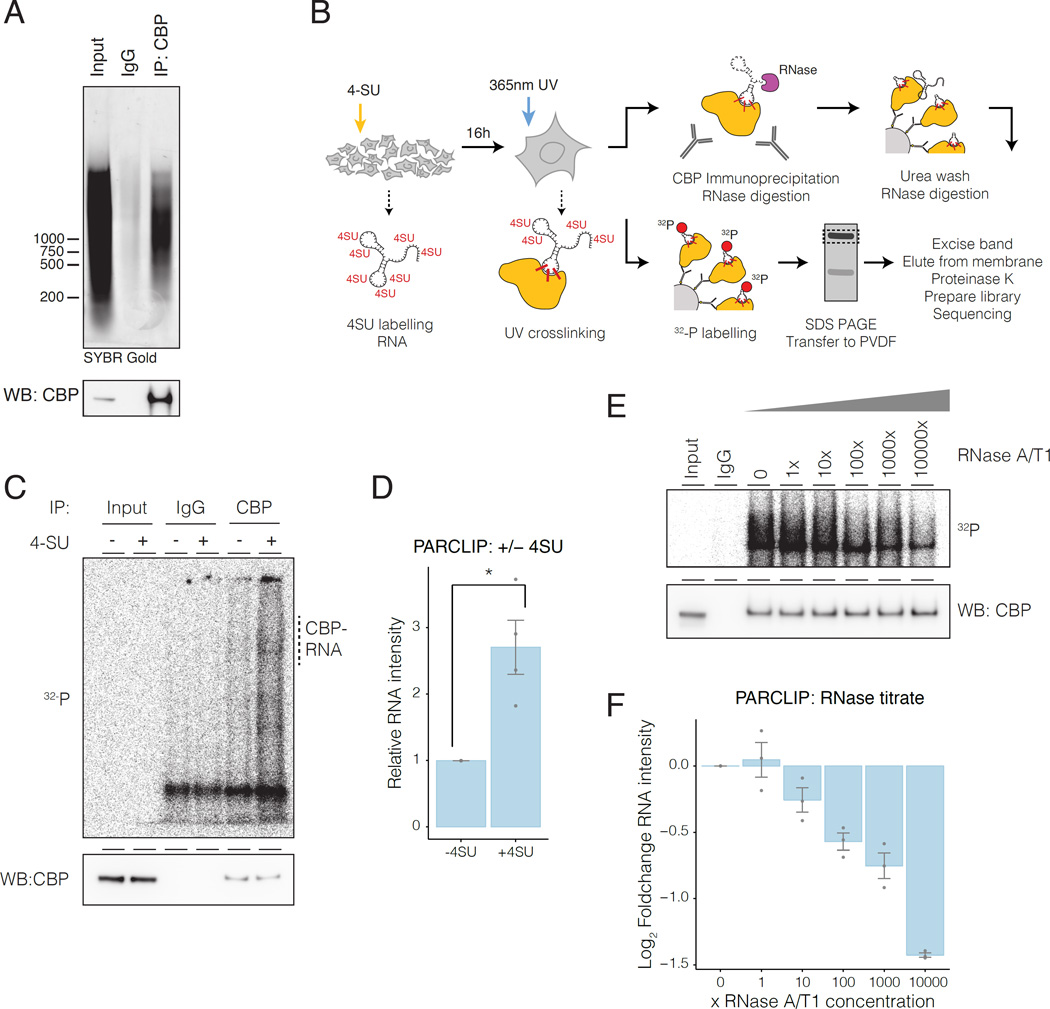
The ability of CBP to interact directly with RNA in cells is untested, although some evidence suggests interactions (Castello et al., 2012; G Hendrickson et al., 2016; Postepska-Igielska et al., 2015). To validate the association of CBP with RNA without crosslinking, we carried out an RNA immunoprecipitation (RIP) experiment under native conditions from mouse embryonic fibroblasts (MEFs), and found that CBP IP co-purifies a population of bound RNA (Figure 1A, Figure S1A). To determine whether immunoprecipitated RNAs were interacting directly with CBP rather than indirectly through another CBP-bound protein, we used Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) (Figure 1B) (Hafner et al., 2010; Huppertz et al., 2014). We observed PAR-CLIP signal corresponding to CBP that was dependent on labeling of the bound RNA with 4-thiouridine (4-SU) (Figure 1C, Figure S1B-E). We note that, similar to other PAR-CLIP results (Hafner et al., 2010; Kaneko et al., 2013; 2014a), cross-linked CBP-RNA complexes migrated in a less defined band due to crosslinking with heterogeneous RNAs. Labeling was sensitive to RNase treatment, confirming that the material crosslinked to CBP was RNA (Figure 1E-F; S1F-H).
Figure 1. CBP interacts with RNA in vivo.
A) Native RNA-IP of CBP. Top, RNA immunprecipitated with CBP. Bottom, CBP western blot.
B) PAR-CLIP protocol. 4-Thiouridine (4-SU).
C) CBP PAR-CLIP required 4-SU: top, autoradiography; bottom, CBP western blot..
D) Quantification of CBP PAR-CLIP. Error bars represent mean +/− s.e.m; n=4.
E) CBP PAR-CLIP signal was sensitive to RNAse. 1× RNAse cocktail contained: RNAse A (0.01mU/ul) + RNase T1 (0.4mU/ul).
F) Quantification of RNase titration. Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test: *P< 0.05; **P< 0.01; ***P< 0.001.
CBP-bound RNAs arise from sites of CBP binding
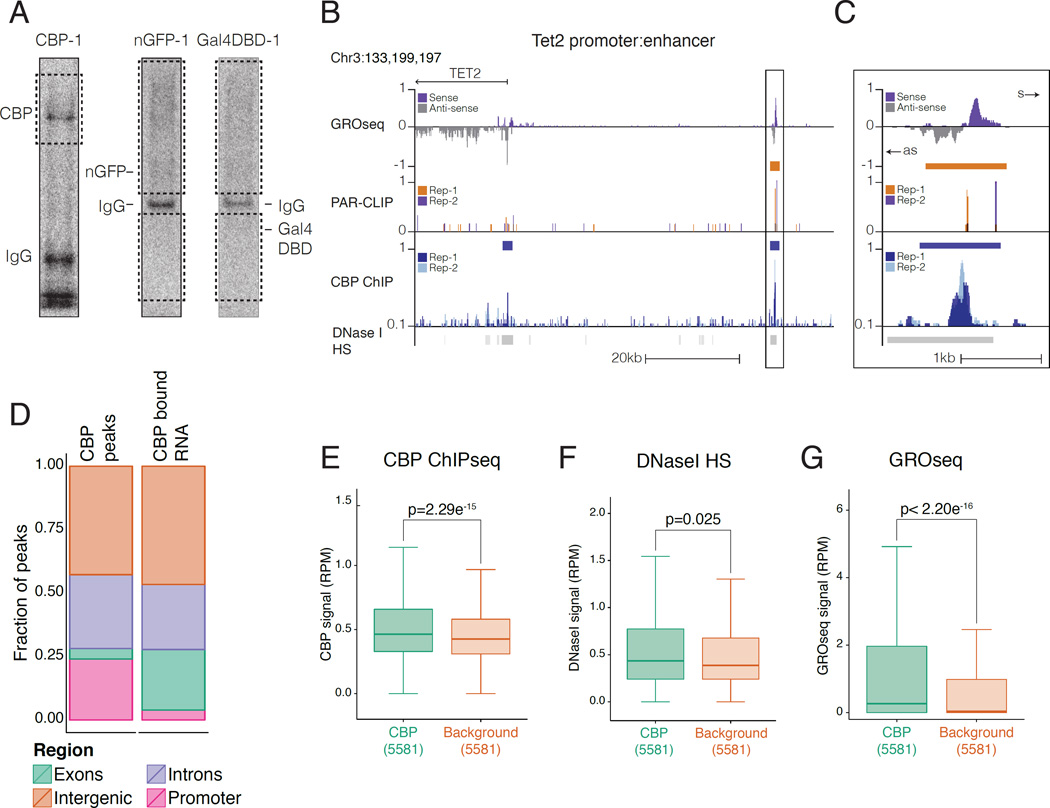
To investigate how CBP interacts with RNA across the genome, we sequenced CBP PAR-CLIP libraries from two biological replicates. To control for background RNA binding events (Friedersdorf and Keene, 2014), we also prepared libraries for nuclear-localized and Flag epitope-tagged GFP (nGFP) and for the DNA binding domain of the yeast TF Gal4p (Gal4-DBD) (two replicates each) (Figure 2A, Figure S2A). Statistically enriched RNA regions were identified on individual and pooled replicate datasets using PARalyzer v1.1, which takes advantage of the T-to-C transitions caused by photo-crosslinking to identify interaction sites (Corcoran et al., 2011). CBP PAR-CLIP read densities from each replicate were well correlated in the RNA regions called by PARalyzer on the separate replicates (Pearson's coefficient = 0.93; Pearson's coefficient = 0.95 for common regions between replicates) (Figure S2B). Having confirmed the reproducibility of our CBP PAR-CLIP sequencing experiments, we pooled the replicates to identify a consensus set of CBP-bound RNA regions for downstream analyses. This resulted in 11,041 CBP-bound RNAs (CBP-RNAs) with a median size of 19bp (Figure S2C). There were 2367 bound RNAs in common between Gal4-DBD and nGFP controls, 2922 between CBP and Gal4-DBD, and 2020 between CBP and nGFP (Figure S2D–F); in total, ~71% (7,835) of CBP-RNAs did not appear in the background controls, suggesting that these represent specific signal from CBP-associated RNAs (Figure S2G, Table S1). We did not detect RNA-dependent interactions between CBP and other eRNA interacting proteins, such as Mediator (Lai et al., 2013) or NELF (Schaukowitch et al., 2014) under native (Figure S2H) or PAR-CLIP (Figure S2I) conditions. Using published ChIPseq data (Kagey et al., 2010; Sun et al., 2011), we found no difference in Med-1 signal at CBP-RNAs compared to background RNAs (Figure S2J), although NELF signal was enriched at CBP-RNAs, suggesting active transcription at these regions (Figure S2J). Thus, we were unable to detect co-binding of CBP and other eRNA interacting proteins to the same RNAs.
Figure 2. CBP-RNAs arise at regions of CBP enrichment.
A) Autoradiography of CBP PAR-CLIP and nls-GFP (nGFP) and yeast Gal4 DNA binding domain (Gal4-DBD). Membrane excised for sequencing (Dashed box).
B) UCSC genome browser view of CBP-RNA upstream of the Tet2 promoter. Top; GROseq signal (purple=sense, grey=antisense); CBP-RNA (PARalyzer) orange bars; 2 replicates PAR-CLIP reads (orange/purple); CBP peaks (blue bars); 2 replicates CBP reads (light/dark blue).
C) Close-up of box from G). Sense (s) and antisense (as).
D) Distribution of CBP and CBP-RNA by genome region.
E-G) Read density at CBP-RNAs and control RNAs. CBP peaks were randomly downsampled to match the size of the background dataset (5581); F) CBP ChIPseq signal (GSE54453);
G) DNaseI hypersensitivity (ENCODE, GSE37074); H) GROseq (GSM1524922). P-values from Mann-Whitney U-test.
We compared the distribution of CBP PAR-CLIP signal with published CBP ChIPseq (Kasper et al., 2014) and GROseq (Meng et al., 2014) datasets in MEFs. Genome browser views of bound RNA regions and corresponding reads, such as at a target enhancer upstream of Tet2 (Figure 2B-C), revealed increased PAR-CLIP read density. In many locations such as the Tet2 enhancer, CBP PAR-CLIP reads mapped to regions of chromatin showing nascent transcription (by GROseq) and CBP occupancy (by ChIPseq; Figure 2B-C). The majority of CBP-RNAs (73%) were found in intergenic (47%) or intronic regions (26%) of the genome similar to CBP itself (72%; 43% intergenic, 29% intronic) (Figure 2D). The remaining CBP-RNAs arise from exons (23%) and very few (4%) from promoters (Figure 2D). As only 4% of CBP chromatin binding sites are at exons, we examined the distribution at exons and promoters in greater detail. The high level of exonic CBP-RNAs relative to CBP binding appears to result from increased false-positives due to the high transcription and stability of exons. First, actual PAR-CLIP reads are enriched at promoters and distal sites, compared to exons (Figure S2K). Second, the enrichment for CBP-RNAs at exons was relatively low, especially when compared to high transcription at exons (Figure S2K-L). Third, the TSS-1kb window used to define promoters limited the number of CBP-RNAs assigned to promoters. Therefore, the distribution of CBP-RNAs reflects patterns of CBP chromatin binding genome-wide.
Compared to RNAs bound in the negative control libraries, CBP-RNAs were enriched for CBP ChIPseq signal (Figure 2E) and were slightly more likely to comprise accessible chromatin (Figure 2F). CBP-RNAs had more nascent transcription by GROseq (Figure 2G), and were enriched for more stable RNA transcripts in total RNAseq data (Figure S2M). We then compared levels of nascent transcription (GROseq) to stable RNA transcripts (total RNAseq) at CBP-RNAs. The CBP-RNAs most enriched for GROseq reads also displayed high-read density in total RNAseq data (Figure S2N) and vice versa (Figure S2O). Nevertheless, CBP-RNAs with low enrichment in total RNAseq still demonstrated transcription by GROseq (Figure S2O, bottom deciles). Consistent with this, CBP-RNAs arising from stable transcripts at exons were strongly enriched for total RNAseq reads, but GROseq reads were strongly enriched at promoters and distal regions (Figure S2P). This reflects the ability of GROseq to detect the often-unstable, nascent RNA transcripts at these locations, where most CBP-RNAs arise (Figure 2D), and suggests that CBP-RNAs contain an abundant population of nascent RNAs. There was an inverse correlation between peak-to-peak distance of CBP ChIPseq and PAR-CLIP signal when considering intersecting CBP and CBP-RNAs (Figure S2Q), with a higher Spearman co-efficient at short distances. Importantly, there was a consistent and more robust correlation with CBP ChIP signal for CBP-RNAs than for Gal4-DBD or nGFP control RNAs (Figure S2Q). Taken together these data demonstrate that CBP-RNAs localize at sites with nascent RNA transcription and within regions of chromatin bound by CBP.
CBP binds to eRNAs
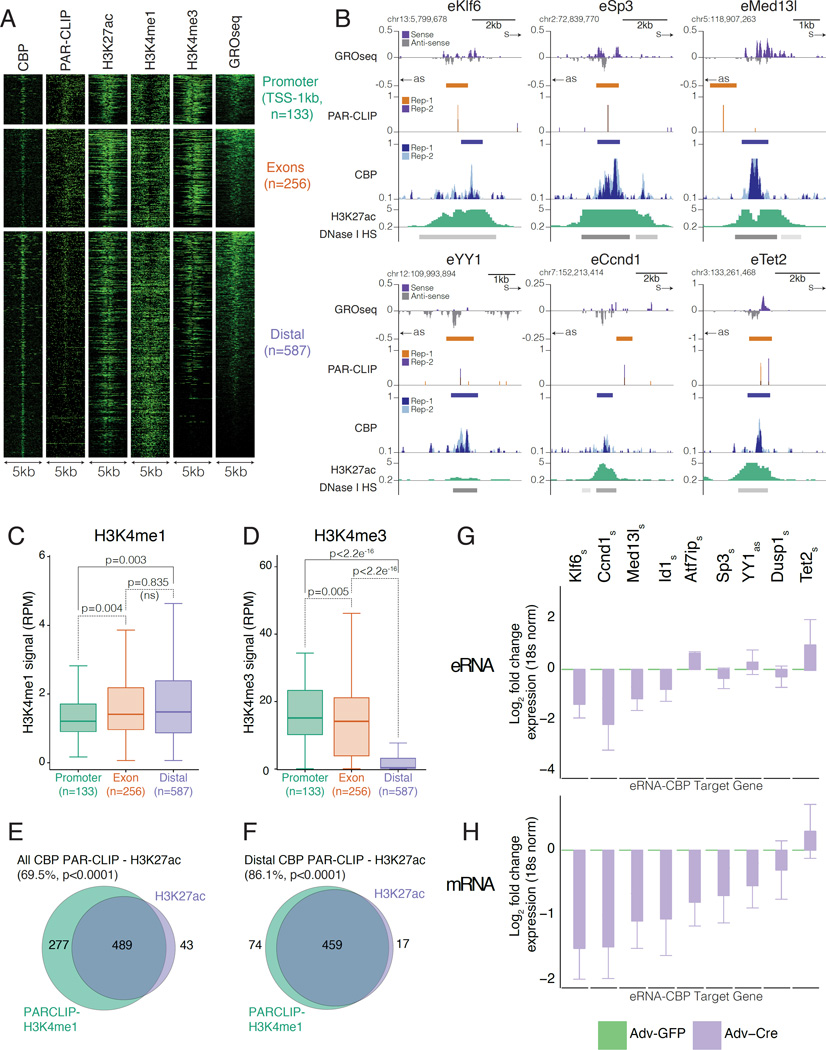
As CBP-RNAs originated from transcribed chromatin regions occupied by CBP, two hallmarks of enhancers, we sought to determine whether CBP-RNAs corresponded to eRNAs. To account for the typical length of eRNAs, we focused on 1,138 locations where the center of the CBP-RNAs was less than 3.5kb from the nearest CBP peak and where CBP-ChIPseq and PAR-CLIP signal were strongly correlated (Figure S2Q) (De Santa et al., 2010; Kim et al., 2010). These CBP-RNAs close to CBP-ChIPseq peaks showed strong enrichment for PAR-CLIP reads (Figure S3A) compared to CBP-RNAs arising further from a CBP peak. They were also enriched for CBP ChIPseq and GROseq reads (Figure S3B-C), suggesting a positive relationship between PAR-CLIP signal and levels of CBP recruitment and transcription. To account for differences in CBP chromatin binding genome-wide (Figure 2D), we divided CBP peaks proximal to CBP-RNAs into promoter (TSS-1kb, 133 peaks, 13.5%), exonic (256 peaks, 26.2%) and distal (intergenic/intronic, 587 peaks 60.1%) sub groups (Figure 3A, Figure S3D). Sites with the highest enrichment for PAR-CLIP reads corresponded to regions most enriched for CBP in all three groups; the highest levels of CBP are found at the most enriched PAR-CLIP sites in promoter and distal regions - classical regions of CBP binding (Figure S3E). Locations with highest CBP binding (Figure S2F) and GROseq signal (Figure S3G) displayed the greatest enrichment for PAR-CLIP reads across all three groups. The distribution of reads across all three groups demonstrated slight enrichment for CBP binding at promoters and distal locations (Figure S3H-I), lower levels of transcription at distal sites (Figure S3J-K) but enrichment for PAR-CLIP reads at promoters and distal sites compared to exons (Figure S3L-M). The abundance of CBP-RNAs corresponded to the level of CBP chromatin binding at actively transcribed regions. Therefore, RNA binding to CBP appears to be locus-specific: CBP-RNAs arise from transcribed regions at locations where CBP binds to chromatin.
Figure 3. CBP binds to eRNAs and directs gene expression from CBP-eRNA enhancers.
A) Heatmaps of RNA bound CBP peaks (<3.5kb) show +/− 2.5kb window from centre of CBP peak, reads were binned over 50bp.
B) UCSC genome browser views of CBP-eRNAs proximal to Klf6, Sp3, Med13l, YY1, Ccnd1 and Tet2. Colours as in Figure 2B and H3K27ac (green). Sense (s) and antisense (as).
C-D) Reads at CBP-RNAs: C) H3K4me3; D) H3K4me1. P-values from Mann-Whitney U-test.
E-F) Venn diagrams show CBP ChIPseq peaks with H3K4me1 intersecting H3K27ac for: E) CBP-RNAs; F) CBP-eRNAs. P-values from permutation test with random regions restricted to TSS-40kb.
G-H) RT-qPCR of G) CBP-eRNAs (s=sense, as=antisense); and H) Nearest gene. Data show log2 fold change between control adenoviral GFP (Adv-GFP, green) or knockdown adenoviral Cre (Adv-Cre, purple); n=3.
The large number of distal CBP-RNAs (Figure 2D) reflects the prevalence of CBP binding to enhancers in distal regions of the genome (Kasper et al., 2014). Distal CBP-RNA sites display bi-directional RNA transcription, enriched CBP occupancy, more accessible chromatin (increased DNAse I hypersensitivity) and elevated levels of H3K27ac (Figure 3B). Furthermore, distal sites have higher H3K27ac (Figure S3N) and nascent transcription by GROseq (Figure S3O) compared to non-RNA interacting control CBP binding sites. Moreover, distal CBP-RNAs are enriched for H3K4me1 and depleted for H3K4me3. CBP therefore appears to interact with eRNAs at distal enhancers (referred to hereafter as CBP-eRNA). We note that within CBP-eRNA loci, distal regions of strong GROseq signal are associated with elevated H3K4me3 (Figure 3A) reflecting more active transcription at these sites. We asked whether CBP-eRNAs had a chromatin signature of active enhancers, characterized by elevated H3K27ac. Of 766 CBP-RNAs intersecting H3K4me1, 532 (69.5%) also overlapped H3K27ac peaks (Figure 3E, Figure S3Q-S). However, of 533 distal CBP-eRNAs that intersect H3K4me1, 459 (86.1%) overlapped H3K27ac (Figure 3F, Figure S3P-R) and thus displayed an active enhancer signature.
Enhancers often regulate expression of adjacent genes (Kim et al., 2010; Lai et al., 2013; Li et al., 2013; Melo et al., 2013; Mousavi et al., 2013). Gene ontology (GO) analysis revealed that the genes nearest to CBP-eRNAs are significantly enriched for transcription regulators, including TFs, mediator subunits and epigenetic regulators (Figure S3S and Table S2). Furthermore, motif analysis identified binding sites for known CBP-interacting TFs (Table S3) (Bedford et al., 2010). The amount of CBP and p300 bound at promoters and enhancers is a poor predictor of the CBP/p300 requirement for expression of proximal genes (Kasper et al. 2014). Genes associated with CBP-eRNA (Table S4) tend to be downregulated upon CBP knockdown relative to genes not associated with CBP-eRNA (Figure S3T) (data from published microarray (Kasper et al., 2014)). We used RT-qPCR to examine the effect of CBP knockdown (control in Figure S3U) on newly identified eRNA transcripts and associated mRNA expression at a subset of genes (Figure 3G-H). The majority of tested mRNAs (8/9; 89%) were downregulated upon CBP knockdown (Figure 3H), and 4/8 (50%) of the corresponding eRNAs were also decreased (Figure 3G), indicating that these selected genes are direct targets of CBP. Overall, our data support the conclusion that distal CBP-eRNA regions represent a subset of active enhancers where eRNAs bind to CBP, and whose associated genes require CBP activity for expression.
In vitro
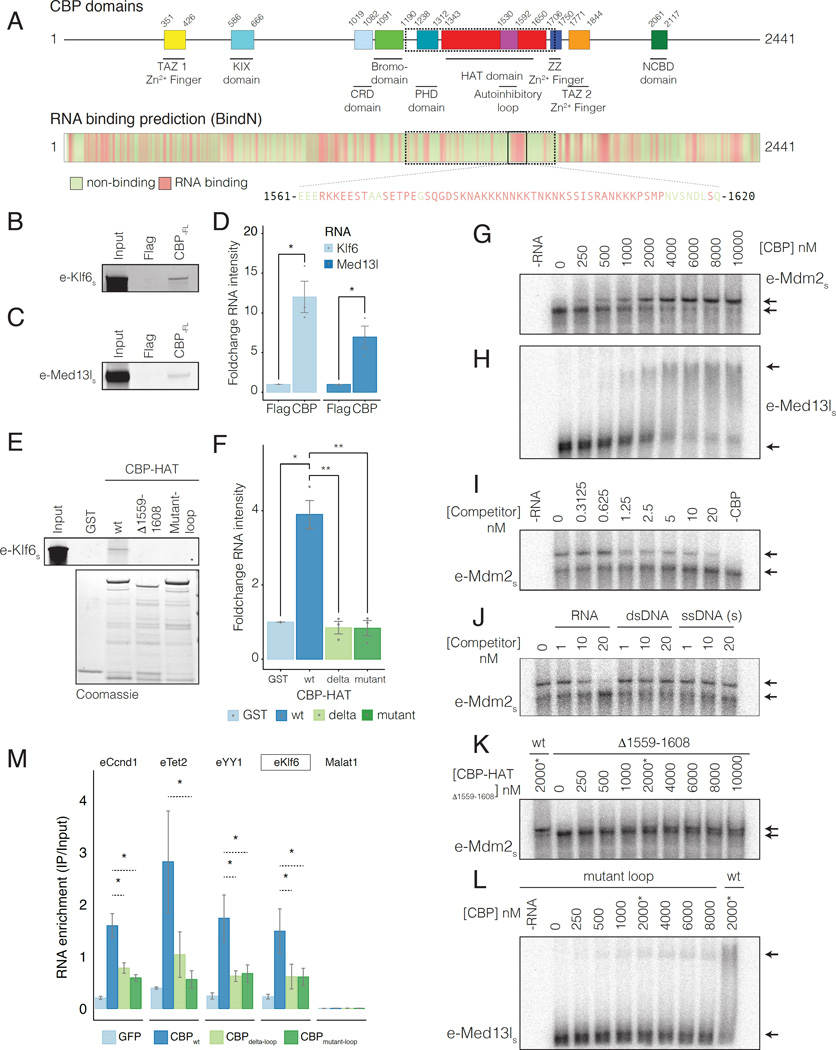
To investigate RNA binding to predicted RNA binding regions (RBRs) within CBP (BindN (Wang and Brown, 2006), Figure 4A, Figure S4A), we purified full length CBP (CBP-FL) from Sf9 cells (Figure S4B, left panel). Using an RNA pull-down (Bonasio et al., 2014) with in vitro transcribed RNA probes corresponding to eRNA-Klf6 (Figure 4B) and eRNA-Med13l (Figure 4C), we observed RNA interactions with CBP-FL (but not in control experiments), suggesting that CBP directly binds to RNA (Figure 4C-D, S4C-D).
Figure 4. In vitro reconstitution of CBP RNA binding.
A) CBP domains and RNA binding prediction (BindN). Non-binding (green) and binding residues (red). Magnified sequence shows predicted RBR in CBP-HATwt.
B-C) In vitro pull down of B) eRNA-Klf6s and C) eRNA-Med13ls. s=sense, as=antisense strand. Replicate images in Figure S4C-D.
D) Quantification of RNA-pulldown data in B-C. n=3.
E) In vitro pull down of eRNA-Klf6s. RNA Input and protein fractions in Figure S4E.
F) Quantification of RNA-pulldown data in B-C. n=3.
G-H) RNA EMSA of eRNAs using CBP-HATwt. G) eRNA-Mdm2s; H) eRNA-Med13ls.
I-J) Competition binding RNA EMSAs. Binding of 2nM 32-P radiolabelled eRNA-Mdm2 to CBP-HATwt (2000nM) was competed with: I) 0–20nM unlabelled eRNA-Mdm2; J) 1nM, 10nM and 20nM un-labeled eRNA-Mdm2 (RNA), dsDNA or ssDNA with the same sequence.
K) RNA EMSA using: K) CBP-HATdelta-loop and eRNA-Mdm2; L) CBP-HATmutant-loop and eRNA-Med13l. RNA was titrated with 0–8000nM CBP-HAT. (*) CBP-HATwt (2000nM).
M) RBR mediates RNA binding to FL-CBP in vivo. PAR-CLIP for GFP-tagged CBPwt, CBPdelta-loop or CBPmutant-loop in MEFs was followed by RT-qPCR. Control lncRNA Malat-1 was not identified by PARalyzer v1.1. P-values from two-tailed Student’s t-test: *P< 0.05; **P< 0.01; ***P< 0.001.
RNA binding predictions for CBP (Figure 4A) suggested a highly basic region (residues 1561–1620) within the core HAT domain might contain the observed RNA binding activity. This region shares >90% identity with p300 and comprises basic residues that are evolutionarily conserved and disordered (Delvecchio et al., 2013; Liu et al., 2008; Wang et al., 2008a). To investigate the predicted RBR in the HAT domain, CBP1196–1718 (CBP-HATwt), which contains all functional domains necessary for HAT activity (Delvecchio et al., 2013), was purified from bacteria (Figure S4B, right panel). RNA pull-down using sequences from nascent eRNAs (e-Klf6, e-Tet2, e-Klf4), processed lncRNAs (HOTAIR1–300, Gas5, Meg3) and exonic RNAs (Id-1 (exon-1), Bbs2 (exon-1), Klf2 (exon-3)) (Figure 4E-F, Figure S4E-G), demonstrated that CBP-HATwt in isolation could bind to multiple different RNA sequences. However, we noted that binding to eRNA sequences was more consistent than binding to lncRNAs and exons (Figure S4G). Thus, CBP-HATwt recapitulated the interaction of CBP-FL with RNA (Figure 4B-D, Figure S4C-D), and displayed a general rather than sequence-specific RNA binding activity in vitro. As CBP-RNAs were enriched at sites of CBP chromatin binding (Figure 2), this supports a genomic locus-specific binding model, where RNAs transcribed proximal to CBP can bind to the RBR independent of RNA sequence.
Next, we used RNA electrophoretic mobility shift assays (EMSA) to evaluate RNA binding of CBP-HATwt to radiolabeled eRNA target sequences (Table S5). All sequences displayed a robust mobility shift when titrated with CBP-HATwt (Figure 4G-H, S4H-I). Moreover, binding to labeled eRNA-Mdm2 (Figure 4I) was outcompeted by identical unlabeled eRNA. Importantly, single strand (ss) and double strand (ds) DNA with the same sequence could not compete with the CBP-eRNA interactions (Figure 4J), demonstrating that CBP-HATwt binds specifically to RNA in vitro.
The predicted RBR in the HAT domain (Q1559-K1608) (Figure S4A) forms a highly basic disordered loop which was proteolytically cleaved in x-ray crystallographic structures of the p300 HAT domain (Delvecchio et al., 2013; Liu et al., 2008). Binding of eRNA-Klf6 to CBP in pull-downs (Figure 4E-F, Figure S4E) and eRNA-Mdm2 in EMSA (Figure 4K) was severely decreased when Q1559-K1608 was deleted (CBP-HATdelta-loop) (Figure S4J). To test whether the charge of the loop was important for RNA binding, we mutated basic residues within the loop to acidic residues not predicted to interact with RNA (CBP-HATmutant-loop) (Figure S4J). We maintained lysine residues that are known targets for CBP mediated auto-acetylation regulating CBP-HAT activity and positively-charged residues important for lysine targeting by CBP (Liu et al., 2008; Thompson et al., 2001; 2004). Notably, the acetylation state of the RBR loop was important for RNA binding, as CBP-HATwt only exhibited detectable RNA binding when co-expressed with lysine deacetylase ySir2 (Figure S4K), suggesting interplay between auto-acetylation of the RBR loop and RNA binding. In contrast, CBP-HATmutant-loop displayed severe defects in RNA binding when co-expressed with ySir2, both in RNA pull-downs with eRNA-Klf6 (Figure 4E-F, Figure S4E) and in RNA EMSAs using radiolabelled eRNA probes (Figure 4L), confirming the importance of basic residues within the RBR for RNA binding to the CBP-HAT domain.
We next asked whether CBP-eRNAs could interact with the identified RBR in CBP using PAR-CLIP in cells. We quantified binding of CBP-eRNA to full-length CBPwt, CBPdelta-loop or CBPmutant-loop in cells by PAR-CLIP followed by RT-qPCR (Figure 4M, control in Figure S4L). CBPdelta-loop and CBPmutant-loop showed drastically reduced binding to CBP-eRNAs compared to CBPwt (Figure 4M), suggesting that the identified RBR within the regulatory loop is the predominant but not the only site of interaction for CBP with eRNAs. Overall, the in vitro RNA binding experiments demonstrate that CBP can directly associate with RNA. Moreover, a specific RNA-binding sequence located within a basic loop that is part of the HAT domain, constitutes a previously unrecognized RBR within CBP. The binding data suggest that CBP can bind to RNA in a locus-specific manner, independent of RNA sequence; thus a bias towards eRNAs is observed due to the high recruitment of CBP to enhancer regions where it interacts with nascent eRNA transcripts.
CBP acetyltransferase activity is stimulated by RNA binding
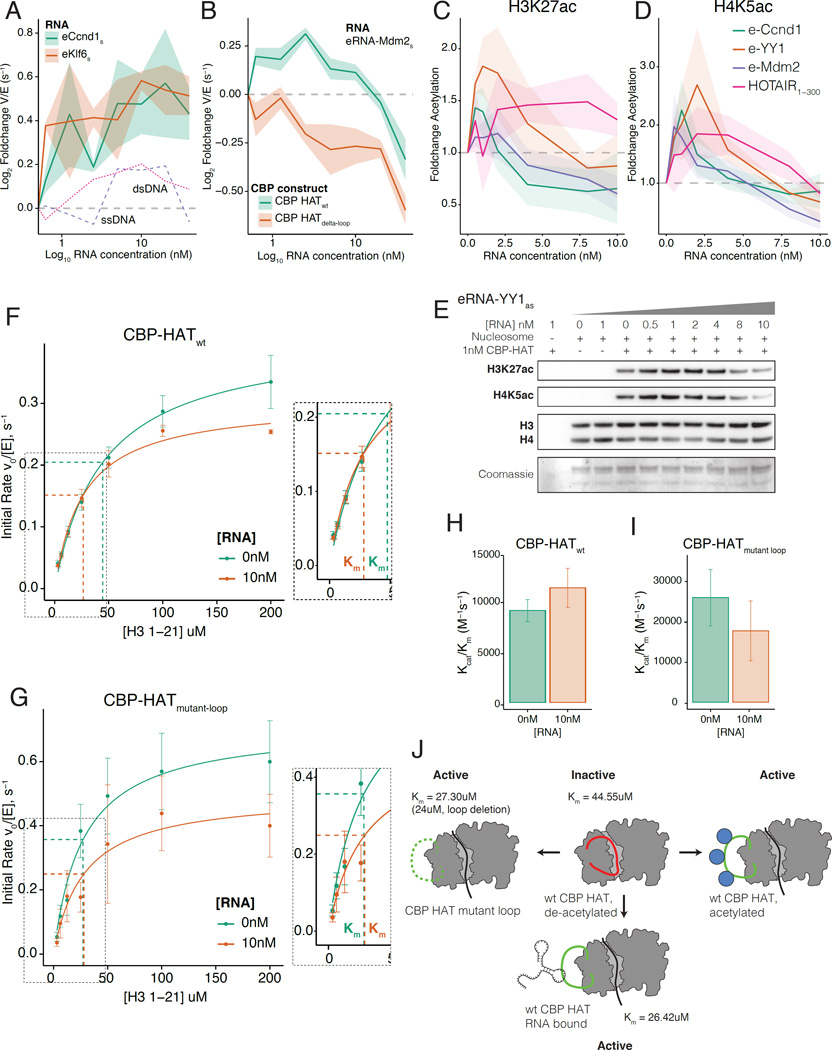
Having defined an RBR within the HAT domain of CBP, we next investigated whether RNA binding affects the HAT activity of CBP, using a radioactive filter-binding assay (Figure S5A). Increasing concentrations of two different eRNAs, eRNA-Ccnd1 and eRNA-Klf6, stimulated CBP HAT activity (Figure 5A). Both eRNAs stimulated activity in a dose dependent manner, but the magnitude and pattern of activity differed slightly between eRNA species. There was no equivalent stimulation when titrating control Ccnd1 ssDNA or dsDNA (Figure 5A). Stimulation was independent of RNA sequence, but required RNA binding to the RBR: HOTAIR1–300 and Id-1 (exon-1), which bound robustly to CBP-HATwt (Figure S5B) stimulated activity, but Meg3, which bound poorly, did not (Figure S5B-C). Importantly, HAT activity did not increase in CBP deleted for the RBR (CBP-HATdelta-loop, Figure 5B). Thus, RNA binding to the RBR within the HAT domain of CBP stimulates HAT activity.
Figure 5. CBP acetyltransferase activity is stimulated by RNA binding.
A) RNA stimulated CBP HAT activity in filter binding assays. 5nM CBP-HATwt was titrated with 0–40nM eRNA-Ccnd1 and eRNA-Klf6 or control (dsDNA/ssDNA with same sequence). Data shows fold change in rate (Vmax/[E], (s−1)) from 0nM RNA. Shaded regions show mean +/− s.e.m (RNA n=3, control n=1). s=sense, as=antisense strand RNA.
B) Stimulation of CBP HAT activity required the RBR. 1nM CBP-HATwt or CBP-HATdelta-loop was titrated with 0–40nM eRNA-Mdm2. Shaded regions show mean +/− s.e.m (n=4).
C-E) Western blot HAT assay using recombinant nucleosome substrate. 1nM CBP-HATwt was titrated with 0–10nM eRNA-YY1as. RNA stimulated: C) H3K27ac (H3 normalized) and; D) H4K5ac (H4 normalized). E) Western blot for H3K27ac, H4K5ac and H3/H4. Coomassie (bottom panel) shows individual histones.
F-G) Steady state filter binding assay. Michaelis-Menten plots for: F) CBP-HATwt; G) CBP-HATmutant-loop. Reactions contained 0nM (green) or 10nM (orange) eRNA-Mdm2. Concentrations of CBP-HAT domain and acetyl-CoA were 10nM and 100uM respectively. H3-1-21 peptide from 0–200uM (n=4). Derived kinetic parameters for Km and Kcat in Figure S5K.
H-I) Specificity constant (Kcat/Km(H3-1-21)) for reaction with 0nM or 10nM eRNA-Mdm2. H) CBP-HATwt; I) CBP-HATmutant-loop.
J) Mechanism for stimulation of CBP-HAT activity by RNA binding.
Next, we determined whether RNA binding increases CBP-dependent histone modifications. CBP acetylates distinct lysine residues, including H3K27ac and H3K18ac on histone H3 and H4K5ac on histone H4 in vitro (Henry et al., 2013). We repeated the HAT assay using reconstituted recombinant nucleosomes (Figure S5D) and assayed by western blot. The level of both H3K27ac and H4K5ac increased as eRNA concentration was titrated (Figure 5C-E, Figure S5E-G), confirming that increased activity with RNA translates to higher H3K27ac (Figure 5C) and H4K5ac (Figure 5D) in vitro. Stimulation was independent of RNA sequence; eRNA-YY1 (Figure 5E), eRNA-Mdm2, eRNA-Ccnd1 and lncRNA-HOTAIR1–300 all stimulated both H3K27ac and H4K5ac (Figure S5E-G), although the different sequences generated different profiles of acetylation. The effect was also observed for recombinant H3 in isolation (Figure S5H, quantified in Figure S5I). The dose response is U-shaped; at high molar ratios of RNA to CBP-HAT there was a drop in H3K27ac (Figure 5C, Figure S5I) and H4K5ac (Figure 5D), which could result from CBP interactions with RNA outside of the primary CBP-RBR at high RNA concentrations. Consistent with this hypothesis, the activity of CBP-HATdelta-loop also decreased at high RNA concentrations (Figure 5B); we note that most eRNAs have low abundance in different systems (Kim et al., 2010; Lam et al., 2013; Li et al., 2013). When the experiment was repeated with recombinant human MOF (hMOF) there was no stimulation of H4K16ac, the major acetylation site of hMOF (Figure S5J). MYST-type acetyltransferases such as hMOF, although also regulated by acetylation, do not contain a CBP/p300-like RBR (Wang et al., 2008a).
RNA binding to the HAT domain of CBP through the identified RBR, therefore stimulates acetyltransferase activity in an RNA concentration-dependent manner, generating increased levels of H3K27ac and H4K5ac in vitro.
Mechanism of RNA stimulation of acetyltransferase activity
CBP/p300 are regulated by a basic activation loop that blocks substrate binding to the active site (Liu et al., 2008; Thompson et al., 2004; Wang et al., 2008a). The loop is displaced by acetylation, which permits substrate binding. In steady state HAT assays, this causes a decrease in the concentration of substrate needed to reach half maximum acetylation rate (Km) (Thompson et al., 2004). Remarkably, the location of the CBP/p300 activation loop largely overlaps the position of the RBR within the HAT domain (Figure 4A, Figure S4A). We therefore tested whether eRNA binding to the RBR could stimulate the HAT activity of CBP through a similar reduction of auto-inhibition.
We adapted our radioactive filter binding assay to evaluate the steady-state kinetic mechanism of stimulation of CBP activity by eRNA (Bowers et al., 2010) (Figure 5F-G; kinetic parameters are summarized in Figure S5K and a model for activation in Figure 5J). Our calculated value of Km(H3-1-21) for CBP-HATwt (Km(H3-1-21)= 44.55 +/− 3.99 uM) (Figure 5F) closely matches previous values for recombinant p300 HAT domain (Km(H4-15) = 40 +/− 8 uM) (Thompson et al., 2004). Consistent with our model, addition of 10nM eRNA-Mdm2 decreased the Km(H3-1-21) of CBP-HATwt from 44.55 +/− 3.99 to 26.42 +/− 3.55uM (Figure 5F). We note that while 10nM eRNA also slightly decreased the Kcat (Figure 5F), the specificity constant Kcat/Km was increased, reflecting an increase in catalysis (Figure 5H). Hence, in common with acetylation of the activation loop, the stimulation of CBP HAT activity by eRNA binding is due to its increased affinity for histone substrate rather than to an alteration of the catalytic mechanism.
To confirm this mechanism, CBP-HATmutant-loop, which does not bind to eRNA (Figure 4N), displayed no change in the Km with 10nM eRNA (Figure 5G). Indeed, the Km of CBP-HATmutant-loop in the absence of eRNA was already decreased (Km(H3-1-21)= 27.30 +/−5.65 uM) compared to CBP-HATwt (Km(H3-1-21)= 44.55 +/− 3.99 uM) (Figure S5K). This indicates that mutation of basic residues to acidic residues within the loop is sufficient to displace the loop from the active site and thus mimic the effect of eRNA binding. Inclusion of eRNA, which does not interact with the mutant loop, has no further effect on the Km and consequently there is no increase in Kcat/Km(H3-1-21) (Figure 5I, Figure S5K). Importantly, Kcat with 10nM RNA was similar for both CBP-HATmutant-loop and CBP-HATwt, which suggested that the decrease in Kcat observed in both cases was independent of eRNA binding to the RBR (compare Figures 5F and 5G, values in Figure S5K). However, as 10nM RNA did not change the Km of CBP-HATmutant-loop, the specificity constant Kcat/Km was slightly decreased (Figure 5I, Figure S5K). These data indicate that eRNA binding to the RBR within the activation-loop results in its displacement from the active site of CBP. This allows improved substrate binding, decreases the Km in the presence of eRNA, and thus enhances the HAT activity of CBP (see model in Figure 5J).
eRNAs modulate the acetyltransferase activity of CBP in vivo
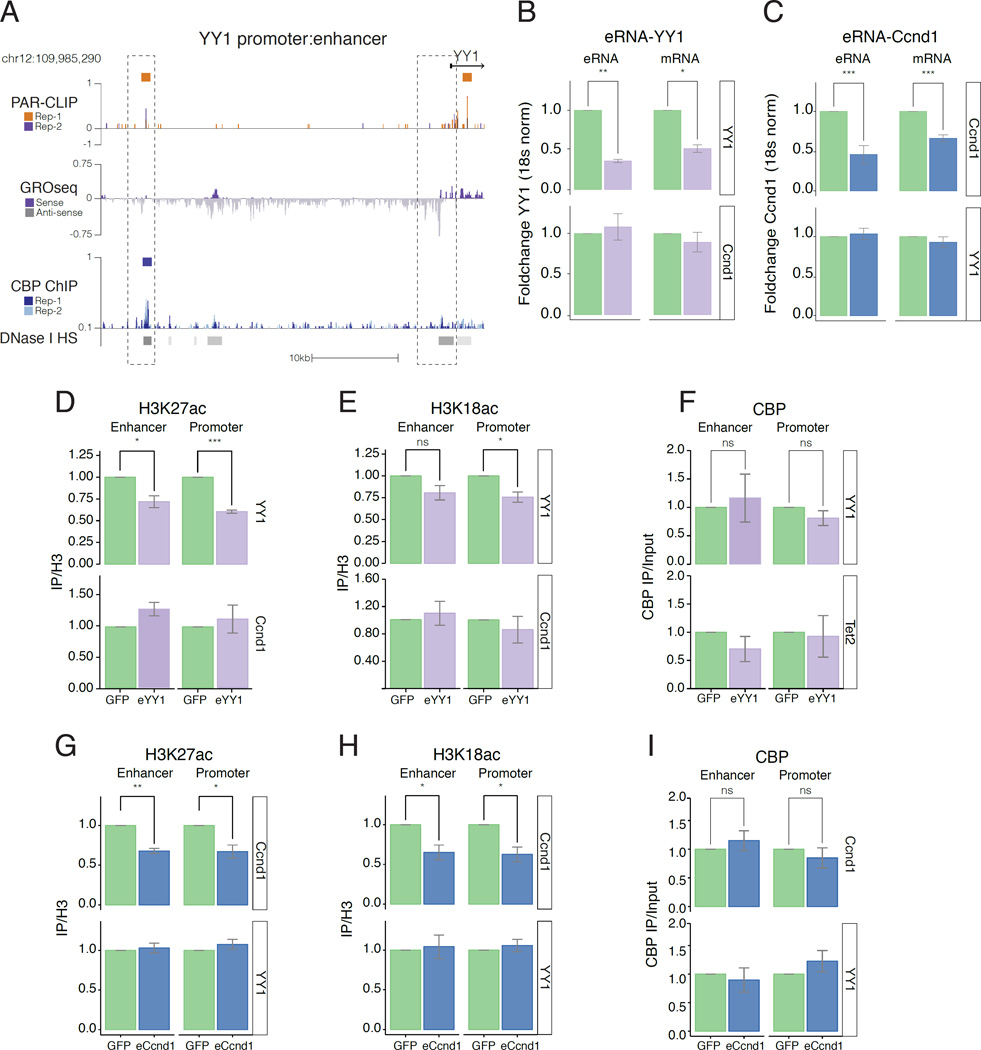
Our data demonstrate that CBP binds to RNAs transcribed close to CBP binding sites such as at the YY1 enhancer (Figure 6A); recruitment of CBP to active enhancers results in binding to eRNAs, and that eRNA binding to the HAT domain of CBP stimulates its acetyltransferase activity.
Figure 6. Localized eRNA binding can stimulate acetyltransferase activity of CBP in vivo.
A) UCSC genome browser of YY1 enhancer and promoter. Colours as in Figure 2B.
B-C) Depletion of PAR-CLIP eRNA using antisense olignucleotide (ASO) targeting: B) eRNA-YY1as (purple); and C) eRNA-Ccnd1s (blue); GFP-control (green). RT-qPCR shows fold-change in eRNA and associated mRNA at target (top) or control gene (bottom). Error bars represent mean +/− s.e.m; n=4.
D-F) ChIP-qPCR following depletion of eRNA-YY1 (purple) or GFP-control (green). Foldchange (IP/H3) for: D) H3K27ac; E) H3K18ac and F) CBP (IP/Input) at YY1 and control gene Ccnd1 (bottom). Error bars represent mean +/− s.e.m; n=4.
G-I) ChIP-qPCR following depletion of eRNA-Ccnd1 (blue) or GFP-control (green). Foldchange (IP/H3) for: G) H3K27ac; H) H3K18ac and I) CBP (IP/Input) at Ccnd1 and control gene YY1 (bottom). Error bars represent mean +/− s.e.m; n=4. P-values from two-tailed Student’s t-test: *P< 0.05; **P< 0.01; ***P< 0.001.
We next asked whether depletion of CBP-eRNAs in cells reduces histone acetylation in a locus-specific manner. RT-qPCR following transfection of antisense-oligonucleotides (ASO) targeting eRNA-YY1 (Figure 6A, Figure S6A), compared to GFP ASO control, revealed specific depletion of the targeted eRNA-YY1 and reduction of the adjacent coding mRNA (Figure 6B, Figure S6E). Importantly, eRNA and mRNA at non-targeted Ccnd1 loci (Figure 6B, bottom panel; Figure S6E) was not significantly reduced upon depletion of eRNA-YY1. We note that eRNA-YY1 depletion also slightly affects expression of Tet2 mRNA, likely indirectly through depletion of YY1 protein, as the Tet2 promoter contains a YY1 motif (TRANSFAC, (Matys et al., 2006)). Similarly, ASO that depleted eRNA-Ccnd1 (Figure S6B) reduced levels of eRNA and adjacent coding mRNA at Ccnd1, but not at non-targeted YY1 and Tet2 loci (Figure 6C, Figure S6E).
We depleted eRNA-YY1 and found a similar reduction of H3K27ac (Figure 6D) and H3K18ac (Figure 6E), whereas there was no significant change in acetylation at the Ccnd1 control enhancer and promoter loci (Figure 6D-E, bottom panel) or at Tet2 control loci (Figure S6F-G, bottom panel). There was also no change in CBP occupancy at enhancer or promoter regions (Figure 6F). Similarly, depletion of eRNA-Ccnd1 decreased H3K27ac (Figure 6G) and H3K18ac (Figure 6H) at the Ccnd1 enhancer and promoter, but produced no change in acetylation at the YY1 enhancer and promoter (Figure 6G-H, bottom panels) and no change in CBP occupancy (Figure 6I). Acetylation was also reduced at the Tet2 enhancer and promoter upon depletion of eRNA-Tet2 (Figure S6H-I), with no alteration in CBP binding (Figure S6J) or acetylation at YY1 control loci. (we note that this did not lead to decreased Tet2 mRNA, consistent with our previous observation that loss of CBP did not reduce Tet2 mRNA levels in Figure 3H). Importantly, for all three examples, eRNA depletion caused decreased acetylation specifically at the target enhancer and promoter, but not at control enhancer and promoter loci.
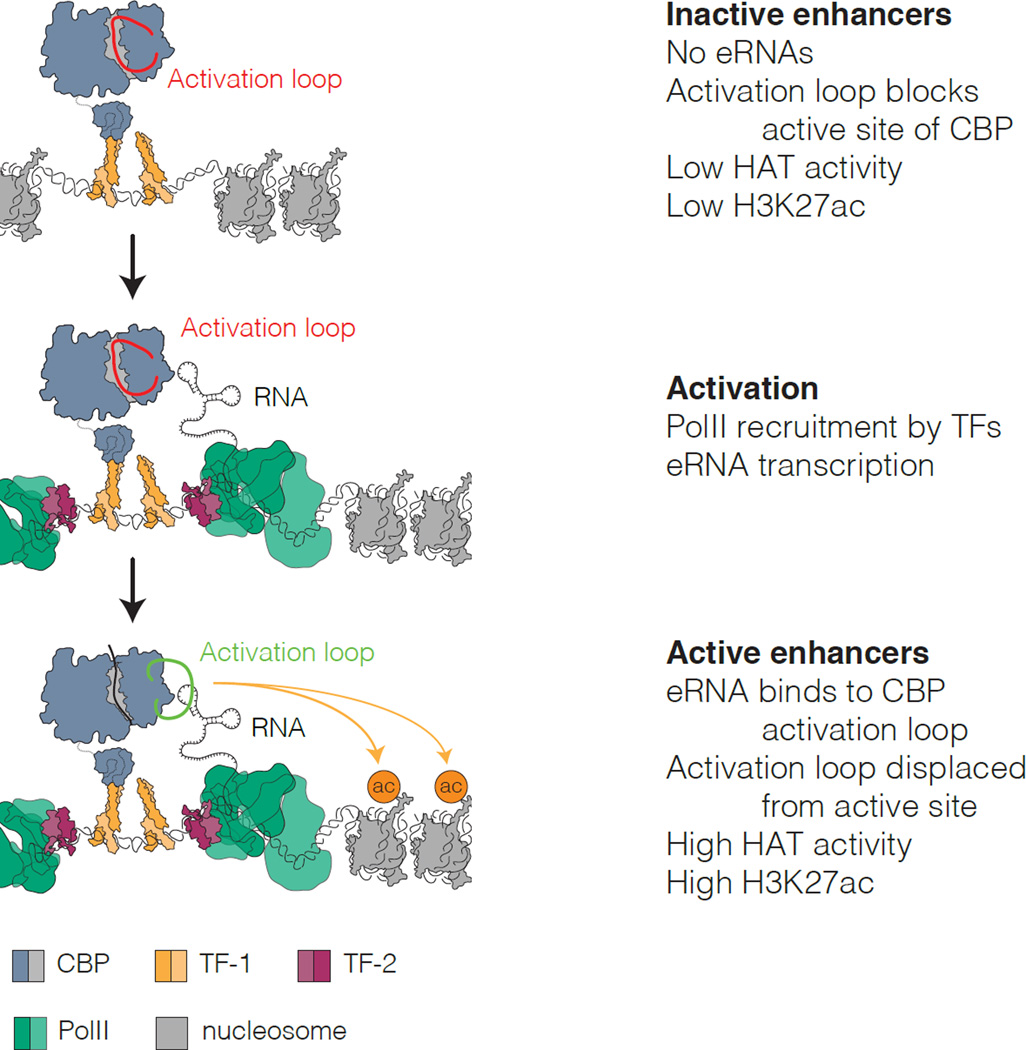
Our findings lead to a model (Figure 7) where CBP binding to specific eRNA transcripts regulates CBP acetyltransferase activity in cis. This results in a local increase in CBP-dependent histone acetylation at the same enhancer and target promoter regions. eRNA binding therefore contributes to enhancer-specific patterns of histone acetylation at regulatory regions.
Figure 7. Model for RNA stimulation of CBP HAT activity during enhancer activation.
i) At inactive enhancers, CBP activity is limited by the activation loop, which occupies the active site.
ii) During activation, recruitment of PolII by bound TFs results in eRNA transcription.
iii) eRNAs bind to the CBP HAT domain RBR, displacing the activation loop, stimulating the HAT activity of CBP and increasing H3K27ac at the enhancer and associated promoter.
Discussion
Our results provide direct evidence of RNA binding to CBP in cells. We show that CBP interacts in a locus-specific manner with RNAs transcribed proximal to sites of CBP chromatin binding. By interacting with a CBP/p300-specific RBR located within its catalytic HAT domain, RNA stimulates the HAT activity of CBP. This observation has particular importance at enhancers, where CBP binding, histone acetylation, and eRNA production are interconnected. Localizing acetyltransferase activity to enhancers is sufficient to promote transcription of associated genes (Hilton et al., 2015). Our findings link these events in a mechanistic manner: eRNA binding to CBP in cis stimulates the localized acetyltransferase activity at enhancers, increasing H3K27ac and H3K18ac at the enhancer and, importantly, at the target promoter, thereby promoting gene expression. Nevertheless, further work is needed to fully elucidate the role of CBP dependent acetylation at enhancers in regulating gene expression.
The mechanism of regulation by activation loop is unique to CBP/p300 among acetyltransferases (Wang et al., 2008a). Remarkably, we show that this same motif renders the HAT activity of CBP sensitive to RNA (Figure 5F-G). Our observation that acetylation was important for RNA binding (Figure S4K) suggests a possible interplay between RNA binding and auto-acetylation of the loop region. CBP is therefore a further example of a chromatin-modifying enzyme whose activity can be regulated by direct binding of ncRNAs, similar to PRC2 (Cifuentes-Rojas et al., 2014; Kaneko et al., 2014b) and DNMT1 (Di Ruscio et al., 2013). However, for CBP, RNA binding contributes to an active chromatin profile through stimulation of CBP HAT activity.
Our data suggests that CBP could bind to many RNA sequences with broadly similar affinities (Figure 4). Binding could therefore be promiscuous, similar to PRC2 (Cifuentes-Rojas et al., 2014; Davidovich et al., 2015; Kaneko et al., 2013; 2014a). RNA binding to CBP also appears to be locus-specific: CBP binds to RNAs where it is recruited to chromatin. CBP activity at promoters could also be regulated by RNA, although the prevalence of enhancer bound CBP (Kasper et al., 2014) results in a strong bias towards eRNA binding. Promiscuous, locus-specific RNA binding has obvious benefits for a transcriptional co-activator such as CBP with a broad genome-wide binding profile, allowing RNAs to stimulate local CBP activity regardless of their sequence. Moreover, RNAs could potentially generate locus-specific activity and acetylation profiles, thus enabling fine-tuning of target gene expression. This model enables a de-coupling of CBP/p300 recruitment from histone acetylation and transcriptional output, helping to explain why CBP/p300 recruitment is a poor predictor of gene activation (Bedford et al., 2010; Kasper et al., 2014). Consistently, depletion of CBP-eRNAs decreased H3K27ac and H3K18ac specifically at associated promoters and modulated gene expression (Figure 6F-G, I-J).
In conclusion, we show that RNAs transcribed proximal to CBP binding sites directly interact with CBP in cis. Binding to the RBR within the catalytic HAT domain of CBP – a region critical for regulating HAT activity - allows substrate to bind more easily and thereby stimulates the HAT activity of CBP. At active enhancers, CBP binds to eRNAs leading to elevated histone acetylation and increased transcription of target genes (Figure 7). In this model, CBP-mediated histone acetylation can be regulated independently from CBP recruitment, and allows enhancer- and gene-specific tuning of acetylation. By stimulating CBP activity, RNA binding can generate a tailored chromatin environment at target genes to fine-tune transcriptional output.
STAR Methods
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Please direct any requests for further information or reagents to the lead contact: Professor Shelley L. Berger (bergers@mail.med.upenn.edu), Cell and Developmental Biology and Epigenetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
wt Mouse Embryonic Fibroblasts (MEFs) (ATCC, CRL-2991) were cultured in complete DMEM (10% FBS, 1% Penn-strep) at 37°C in 5% CO2. CBP/p300 flox/flox MEFs (kindly provided by Paul Brindle (Kasper et al., 2014)) were cultured in complete DMEM (10% FBS, 0% Penn-strep) at 37°C in 5% CO2. GP2-293 cells were cultured in complete DMEM (10% FBS, 1% Penn-strep) at 37°C in 5% CO2.
Bacterial cultures
For expression of recombinant proteins in bacteria, Rosetta 2 DE3 cells (EMDMillipore) were grown in Luria Broth (LB) of 2YT broth. Cells were grown at 37°C with shaking at 225rpm until OD600nm = 0.4–0.6. Expression was induced with 1mM IPTG. Expression time and temperature were optimized for individual constructs (details in Protein expression and purification). Cells were harvested by centrifugation at 4800rpm for 15 minutes.
Sf9 cultures
For expression of full-length CBP, Sf9 insect cells (ThermoFisher) were grown in serum-free Sf900-III medium (ThermoFisher). Suspension cultures (1–2e6 cells/ml) were infected with high-titer passage 2 (P2) virus stock at a multiplicity of infection (MOI) of 1. Cells were harvested after 24h.
METHOD DETAILS
RNA immunoprecipitation
Native RNA immunoprecipitation (Native RIP)
Native RIP experiments were carried out from nuclear extracts. Cells were harvested and washed with PBS + 1 mM EDTA, then with Buffer A (10 mM Tris pH 7.9 4°C, 1.5 mM MgCl2, 10 mM KCl, supplemented with Complete EDTA free protease inhibitor (Roche)). Cells were lysed by incubating with Lysis Buffer (Buffer A + 0.2% IGEPAL CA-630) for 10 minutes on ice. Nuclei were isolated by centrifugation at 2,500×g for 5 min and lysed in Buffer C (20 mM Tris pH 7.9 4°C, 25% glycerol, 400 mM NaCl, 1.5 mM MgCl2, 10 mM EDTA, 0.4 u/μl murine RNAse inhibitor (NEB), Complete EDTA free protease inhibitor (Roche) for 30 min at 4°C. Lysates were cleared by centrifugation at 20,000×g for 30 min. Protein-RNA complexes were immunoprecipitated by incubating with a depleting amount of antibody for 3h at 4°C in RIP buffer (20 mM Tris pH 7.9 4°C, 200 mM KCl, 0.05% IGEPAL CA-630, 10 mM EDTA). Immunocomplexes were recovered with Protein A dynabeads (ThermoFisher) blocked with BSA for 30 minutes. Beads were washed twice with RIP Wash Buffer (RIP Buffer + 0.05% NP40, 1mM MgCl2) and contaminating DNA was removed by incubating with 2.5U Turbo DNase I (ThermoFisher) for 10 minutes at 21°C. 20% of resuspended beads were retained for western blot. RNA was purified using Trizol (ThermoFisher) and isopropanol precipitation. A second incubation with 2.5U Turbo DNase I at 37°C for 30 minutes ensured no DNA contamination. Immunoprecipitated RNA was resolved on a 6% denaturing TBE Urea gel and imaged with SYBR gold.
PAR-CLIP
For Photoactivatable ribonucleoside–enhanced CLIP (PAR-CLIP), Cells were incubated for 16h with 150uM 4-thiouridine (4-SU). Plates were washed with PBS before cells were cross-linked using 150mJ/cm2 365nM UV light with a Stratalinker UV crosslinker (Stratagene). All subsequent stages of PAR-CLIP were carried out immediately to prevent degradation of cross-linked CBP. Cells were harvested by scraping in PBS supplemented with 1mM EDTA and lysed by incubating for 10 minutes on ice with CLIP lysis buffer A (20mM Hepes KOH pH7.9, 150mM KCl, 5mM EDTA, 2% EMPIGEN, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 0.1% SDS, 2mM DTT) supplemented with Complete EDTA free protease inhibitor (Roche), Murine RNase inhibitor (NEB), Turbo DNase I (ThermoFisher) and sonicated for 2× 30s cycles in a bioruptor (Diagenode). Cell lysate was cleared by centrifugation at 20000×g for 30 minutes. The cleared cell lysates were subjected to an initial partial RNAse digestion step using RNase/T1 cocktail (4000×) (ThermoFisher) for 5 min at 21°C to improve immunoprecipitation efficiency (Friedersdorf and Keene, 2014; Hafner et al., 2010; Huppertz et al., 2014). Immunopreciptations were carried out using a depleting amount of anti-CBP antibody (Santa Cruz, sc-583) for 2 hours at 4°C. Lysates were then subjected to a second, more stringent RNAse treatment using RNase A/T1 cocktail (10×) for 15 minutes at 37°C before recovering immunocomplexes for 30 minutes with BSA blocked Protein A dynabeads (ThermoFisher). The digestion with higher RNase concentration was important for optimizing transfer efficiency of the large (>250KDa) CBP-RNA complexes. Beads were collected and washed twice with CLIP wash buffer (20mM Tris HCl pH 7.9 (4°C), 400mM KCl, 5mM EDTA, 0.5mM EGTA, 2% EMPIGEN, 0.7% Na-deoxycholate, 0.5% N-lauroylsarcosine, 2M Urea, 2mM DTT, protease inhibitors) then IP50 buffer (20mM Tris HCl pH 7.9 (4°C), 50mM KCl, 0.2mM EDTA, 10% Glycerol, 2mM DTT), before resuspending in 1× Turbo DNase buffer. CLIP wash buffer contains 2M Urea, which was important for removing non-specific RNA binding whilst retaining immunoprecipitated protein-RNA complexes. Contaminating DNA was removed by incubation with 2.5U Turbo DNase I for 30 minutes at 37°C. Beads were washed with CLIP wash buffer and IP50 buffer. Cross-linked RNA complexes were dephosphorylated by incubating with Recombinant Shrimp Alkaline Phosphatase (rSAP, NEB) for 15 minutes at 37°C, washing with Phosphatase wash buffer (20mM Tris HCl pH 7.9 (4°C), 400mM KCl, 10mM EDTA, 10mM EGTA, 0.05% IGEPAL CA-630, 2mM DTT) then T4-PNK buffer (50mM Tris HCl pH 7.5 (at room temperature (RT)), 50mM NaCl, 10mM MgCl2, 5mM DTT). Protein-RNA complexes were then labelled by incubating with 5U T4 PNK (NEB) with 10uCi γ-32P-ATP for 15 minutes at 37°C, then for 5 minutes with 100uM unlabelled ATP. Labelled material was washed again with CLIP wash buffer and IP50 before denaturing with 4× LDS loading buffer (ThermoFisher) and resolving on a 3–8% Tris-Acetate gel. For RNase titration experiments, following labelling, samples were incubated with different RNAse concentrations for 15 minutes at 37°C. Gels were transferred to 0.2uM PVDF membrane for 12h at 40V, then exposed to autoradiography screens or x-ray film for 12–24h to image labelled RNA complexes. Autoradiography screens were imaged using a Typhoon Phosphorimager (GE Healthcare). For western blot analysis, PAR-CLIP membranes were probed with anti-CBP antibody (Abcam, ab2832) for 16h).
Sequencing libraries for PAR-CLIP seq
For sequencing experiments, labelled CBP-RNA bands (or control bands) were excised from the membrane and eluted by incubating with RNA elution buffer (50mM Hepes pH7.0, 50mM NaCl, 1mM EDTA, 2mM CaCl2, 1% SDS) and Proteinase K (4mg/ml, NEB) for 30 minutes at 37°C, then for 30 minutes at 55°C with RNA elution buffer + 3.5M Urea and Proteinase K. Proteinase K was pre-incubated for 15 minutes at 37°C before use to remove contaminating RNAse. RNA was then purified using acidic phenol:chloroform (ThermoFisher) and ethanol precipitation.
Sequencing libraries for PAR-CLIP experiments were prepared using NEB next small RNA library set for Illumina sequencing (New England Biolabs) according to manufacturers instructions. Libraries were checked for quality by bioanalyzer before and after size selection by Pippin prep (Sage science) and were 50bp single-end sequenced on an Illumina NextSeq 500.
Bioinformatic analyses
PAR-CLIP analysis
Following de-multiplexing, sequences corresponding to illumina adaptors were removed using Cutadapt v1.7.1 (Martin, 2011) and reads greater than 15nt in length were retained (cutadapt -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -e 0.1 -m 15). Samples were aligned to Mouse genome assembly mm10 using Bowtie 1.1 (-p 3 -v 2 -m 10 --best —strata) (Langmead et al., 2009). Following alignment, aligned reads were converted to bed format using Samtools (Li et al., 2009) and filtered to remove any non T>C mismatches according to original protocols (Corcoran et al., 2011). Filtered reads were then used for peak calling using PARalyzer v1.1, requiring a minimum count of 5 reads for generating groups, clusters or kernel density estimation (KDE); a minimum cluster size of 15 nucleotides and a minimum conversion count of 1 T>C conversion (Corcoran et al., 2011). All replicates and control datasets were treated identically when processing with PARalyzer v1.1.
General Bioinformatic analysis
All downstream processing following alignment to mm10 and Peak calling was done using the mm9 reference genome. Genome co-ordinates were converted from mm10 to mm9 using the UCSC Genome Browser LiftOver tool.
All genome wide read counts were normalised per million reads mapped (RPM) and visualized with UCSC Genome Browser (http://genome.ucsc.edu/). BigWig UCSC Genome browser tracks were generated by normalising (RPM) and converting reads to bedgraph format (Bedtools v2.15 - genomecov -bg -g -scale) (Quinlan and Hall, 2010), then to BigWig format using UCSC Genome Browser bedGraphToBigWig v4.0.
Intersection of CBP-PAR-CLIP enriched RNAs with control enriched RNAs, CBP peaks and chromatin features (H3K27ac (GSM1000139), H3K4me1 (GSM769028), H3K4me3 (GSM769029) and DNaseI HS (GSM1014199)) was done using Bedtools v2.15 (Quinlan and Hall, 2010) (intersect -wa -u). For comparison of read density enrichment at CBP bound RNAs and background RNAs, loci in either dataset intersecting repetitive regions were removed with repeatmasker and CBP bound RNAs intersecting background RNAs were removed using Bedtools v2.15 (-v). The remaining CBP bound RNAs were randomly downsampled to match the size of the background dataset (5581). For intersection with genome regions, intersection was made in the order Promoter (1kb window upstream of TSS)> Exons > Introns > Intergenic. All gene co-ordinates were obtained from Refseq. CBP ChIPseq peaks were called independently on each replicate CBP ChIPseq dataset (GSE54453) using MACs (v1.4, default parameters) (Zhang et al., 2008). Only peaks called independently in each dataset that were present in both datasets were retained. The correlation of CBP ChIPseq read density at CBP ChIPseq peaks intersecting CBP RNAs or control RNAs with increasing centre to centre distance (100bp increments) between peaks was calculated using custom in-house scripts.
Read density histograms and read density enrichment over defined peak regions were calculated using custom in-house scripts. All genome wide read counts were normalised per million reads mapped (RPM). MEF GROseq data was obtained from GSM1524922 (Meng et al., 2014). All downstream analysis was done using R in Rstudio; significance P-values for tag enrichment were calculated using a two-sided Mann-Whitney U-test in R. Heat maps were generated using custom in-house scripts. Heat maps were centred on CBP peaks intersecting CBP PAR-CLIP enriched RNA <3.5kb away. All distances were measured centre-centre (c-c).
Gene expression microarray data from CBP/p300 flox/flox MEFS (GSE54452) was analysed initially using Partek Genome Analysis software. Refseq annotated genes proximal to eRNA-CBP sites or Distal CBP bound enhancers were identified using custom in-house scripts that compare the distance from the centre of called peaks to the nearest Refseq annotated TSS to locate the nearest genes. Gene sets were randomly down-sampled using R. All subsequent numerical analysis was carried out using R in R studio. GO analysis was carried out using DAVID (Huang et al., 2009). Motif analysis of CBP-eRNA sites was carried out using the MEME suite (Bailey and Elkan, 1994) and TOMTOM (Gupta et al., 2007) and using SeqPos (He et al., 2010) with the Jaspar database in the Galaxy/Cistrome suite.
Publicly available datasets
We used a number of publicly available datasets to aid analysis of our PAR-CLIP data:
CBP ChIPseq (GSE54453) and microarray data (GSE54452) from CBP/p300 flox/flox MEFs both from (Kasper et al., 2014)).
Mouse Embryonic Fibroblast GROseq data GSM1524922 (Meng et al., 2014)).
Mouse Embryonic Fibroblast Total RNAseq data: GSE45284 (GSM1100746, GSM1100747, GSM1100748) (Bezzi et al., 2013) and GSE27843 (GSM687308,GSM687309) (Lienert et al., 2011)
NELF ChIPseq in Mouse Embryonic Fibroblasts GSE24113: NELFb (GSM593398), Input (GSM593399) (Sun et al., 2011).
Med-1 ChIPseq in Mouse Embryonic Fibroblasts (GSM560353) (Kagey et al., 2010).
From ENCODE:
H3K27ac (GSM1000139), H3K4me1 (GSM769028), H3K4me3 (GSM769029) Ren Lab (Yue et al., 2014). (GSE31039, LICR histone (Shen et al., 2012)).
DNaseI HS (GSM1014199) Stam Lab (GSE37074 UWash DNaseI) (Yue et al., 2014).
Molecular biology and cloning
All PCR reactions were carried out using a standard 50ul reaction mix (0.5ul PfuUltra II Fusion HS DNA Polymerase (Agilent), 1× PFu Ultra II buffer, 5% DMSO, 0.2uM Fwd/Rvs primer, 200uM dNTP mix, template DNA). DNA was denatured at 95°C, and extension was carried out at 72°C for targets <10kb and 68°C for templates >10kb. See Table S6 for all Primer sequences used for cloning experiments.
Cloning of CBP (full-length)
For expression of recombinant proteins, full-length CBP was cloned into pFastBAC HTA (ThermoFisher), modified with an in-frame N-terminal 1× Flag tag and 6×HisTag. All PCR reactions were carried out using PfuUltra II Fusion HS DNA Polymerase (Agilent). CBP was cloned in two parts using single copy Xba1 site at position 3294–3299; CBP1–3299 was cloned into BamHI/XbaI sites in pFastBac HTA using primers (Fwd: DB_PCR_1; Rvs: DB_PCR_2); CBP3294–7326 was then cloned into XbaI/HindIII sites in the pFastBAC HTA-CBP1–3299 construct using primers (Fwd: DB_PCR_3; Rvs: DB_PCR_4). Cloning re-forms the native XbaI site in full length CBP (Plasmid: pFB:CBP-FLwt); all constructs were checked for correct frame by sequencing through restriction sites.
For expression of full-length CBP in mammalian cell lines, CBP was initially cloned into pAcGFP-C1 (Clontech) modified to remove the SacI restriction site. CBP was cloned in two separate reactions: Firstly, a silent SalI restriction site was introduced into FL-CBP at position 3464 using primers (Fwd: DB_PCR_5; Rvs: DB_PCR_6). CBP1–3464 was cloned in frame with an N-terminal GFP tag using HindIII/SalI sites in the pAcGFP-C1 vector using primers (Fwd: DB_PCR_7; Rvs: DB_PCR_8). CBP3465–7326 was cloned into pAcGFP-C1 using SalI/BamHI sites in the vector with primers (Fwd: DB_PCR_9; Rvs: DB_PCR_10). CBP3465–7326 was sub-cloned into SalI/BamHI sites in pAcGFP-C1-CBP1–3464 to reconstruct FL-CBP in frame with the N-terminal GFP-tag. Cloning reforms the silent SalI site in full length CBP; all constructs were checked for correct sequence and frame by sequencing through restriction sites. N-terminal GFP-tagged FL-CBP was sub-cloned with an additional in-frame N-terminal Flag- epitope tag into BamHI/NotI sites in the pTRE3G vector using primers (Fwd: DB_PCR_11; Rvs: DB_PCR_12). All work with viral vectors was carried out using One Shot Stbl3 E. coli (ThermoFisher) to reduce recombination of repeat sequences.
Cloning of CBP-HATwt constructs
CBP1196–1718 (CBP-HATwt) was cloned into BamHI/NotI sites in pGEX-4T-1 (Novagen) encoding an in-frame GST tag (plasmid: pGEX4T1:CBP-HATwt). To generate CBP-HATdelta-loop mutants, primers (Fwd: DB_PCR_13; Rvs: DB_PCR_14) were used to amplify CBP-HATwt in pGEX-4T-1 in reverse directions to delete residues (aa1559–1608, 4677–4732bp) using PFu Ultra II high fidelity DNA polymerase (Agilent) (plasmid: pGEX4T1:CBP-HATdelta-loop). For CBP-HATmutant loop constructs, we inserted two SacI sites using silent mutations at positions 4612 and 5089 into CBP-HATwt in pGEX-4T-1 using primers (Fwd: DB_PCR_15, Rvs: DB_PCR_16). The insert sequence coding for wild-type CBP-HAT with mutant loop residues (aa1559–1608, 4677–4732bp, lower case in sequence below) was ordered as a gBlock gene fragment (IDT) with flanking SacI sites (see Table S6 for sequence). The insert sequence was amplified by PCR, digested with SacI and ligated into SacI digested CBP-HATwt. Inserts were checked for correct frame and orientation by sequencing (plasmid: pGEX4T1:CBPmutant-loop).
Cloning of CBP mutant constructs
To clone full-length CBPdelta-loop and CBPmutant loop into pTRE3G, constructs were generated using the same method as for CBP-HAT domain constructs (see above) in pAcGFP-C1-CBP3465–7326. Mutant CBP3465–7326 was then sub-cloned into SalI/BamHI sites in pAcGFP-C1-CBP1–3464 to reconstruct FL-CBP in frame with the N-terminal GFP-tag. Cloning re-forms the silent SalI site in full-length CBP; all constructs were checked for correct frame by sequencing through restriction sites (plasmid: pAcGFPC1:CBP-FLwt; pAcGFPC1:CBP-FLdelta-loop; pAcGFPC1:CBP-FLmutant-loop). N-terminal GFP-tagged FL-CBP was sub-cloned with an additional in-frame N-terminal Flag-epitope tag into BamHI/NotI sites in the pTRE3G vector using primers (Fwd: DB_PCR_17; Rvs: DB_PCR_18) (plasmid: pTRE3G:CBP-FLwt; pTRE3G:CBP-FLdelta-loop; pTRE3G:CBP-FLmutant-loop). All work with viral vectors was carried out using One Shot Stbl3 E. coli (ThermoFisher) to reduce recombination of repeat sequences.
Cloning of ySir2
ySir2 was cloned into pET28-b+ (EMDMillipore) at EcoRI/HindIII sites using primers (Fwd: DB_PCR_19, Rvs: DB_PCR_20) (plasmid: pET28b+:ySir2).
Protein expression and purification
CBP (full length)
Flag tagged full-length CBP in pFastBAC HTA was packaged into baculovirus and transfected into Sf9 cells for expression. Sf9 insect cells (ThermoFisher) were grown in serum-free Sf900-III medium (ThermoFisher). Suspension cultures (1–2e6 cells/ml) were infected with high-titer passage 2 (P2) virus stock at a multiplicity of infection (MOI) of 1. Cells were harvested after 24h to achieve optimal expression and purity. Sf9 expression was courtesy of David Schultz and the Wistar protein expression facility. All subsequent purification stages were completed on a single day to limit degradation of purified CBP fractions. For purification of full length CBP, Sf9 cell pellets from 2L culture were re-suspended in Buffer A (20mM TrisHCl pH7.3 (RT), 300mM NaCl, 2mM MgCl2, 5% Glycerol, 2mM DTT) supplemented with 0.1% NP40, 0.1mM PMSF, Complete EDTA free protease inhibitor (Roche), 1mM Benzamidine, Benzonase (5U/ml final), 1mM Na-Butyrate and lysed by sonication. Cell lysates were cleared by centrifugation (18000rpm, 45 minutes, 4°C) and filtered through a 0.45uM syringe filter. Cleared lysates were loaded on to a 5ml Ni-Affinity HisTrap HP column (GE healthcare) using an Akta purifier FPLC. The column was washed with Buffer A and eluted with Buffer B (Buffer A + 500mM Imidazole). Fractions corresponding to eluted protein were collected and loaded on to a gravity flow column (Biorad) packed with 3 ml Flag-M2 agarose (Sigma) and equilibrated with Buffer A. Flow through was collected and re-loaded on to packed column two more times to ensure maximum yield of purified CBP. The loaded column was washed three times with buffer A, and bound protein was eluted with 3× Flag elution buffer (Buffer A + 100ug/ml 3× Flag-peptide (Sigma)). Elution was repeated 3× to ensure maximum yield. Fractions with CBP were pooled and concentrated using a Vivaspin centrifugal concentrator (GE Healthcare) and loaded on a Superdex 200 16/60 size exclusion column equilibrated with Buffer A. Fractions containing CBP were collected and concentrated using a Vivaspin centrifugal concentrator (GE Healthcare). Protein concentration was determined using a nanodrop with extinction co-efficient and molecular weight parameters and verified by Bradford assay (Biorad). Purified protein was checked for RNase contamination using an RNase Alert lab test kit (Ambion) and dialysed into Buffer D (Buffer A + 50% glycerol + 0.2mM EDTA). Fractions corresponding to eluted protein were collected at each purification stage and checked for purity by SDS PAGE on a 3–8% Tris acetate gel. Purified protein was divided into single use aliquots, flash frozen in liquid nitrogen and stored at −80°C.
CBP-HAT domain constructs
CBP-HAT domain constructs (CBP-HATwt, CBP-HATmutant-loop, CBP-HATdelta1559–1608) in pGEX-4T-1 (Ampicillin) were expressed in Rosetta 2 DE3 cells (EMDMillipore) in Luria Broth (LB). Constructs were co-expressed with ySir2 in pET-28b+ (Kanamycin); co-expression improved yield by limiting indiscriminate acetylation of host proteins and autoacetylation of expressed CBP-HATwt domain (Thompson et al., 2004). Co-expression with ySir2 was required for RNA binding to CBP (Figure S4K). Cells were grown at 37°C with shaking at 225rpm until OD600nm = 0.4–0.6, then shifted to 22°C and induced with 1mM IPTG for 4 hours. Cells were harvested by centrifugation at 4800rpm for 15 minutes.
Cell pellets were lysed in GST Buffer A (20mM TrisHCl pH7.3 (RT), 300mM NaCl, 0.2mM EDTA, 1mM DTT, 5% Glycerol, 10mM Na-Butyrate and supplemented with Complete protease inhibitor cocktail (Roche). Cells were lysed by sonication. Cell lysates were cleared by centrifugation at 18000 rpm for 45 minutes at 4°C. Cell lysates were filtered with a 0.45uM syringe filter and loaded on to an equilibrated 5ml GSTrap HP column (GE Healthcare) using an Akta Purifier FPLC at 4°C (GE Healthcare). The column was washed with GST Buffer A; bound protein was eluted using a linear gradient of 0–100% GST Buffer B (GST Buffer A + 10mM Glutathione (reduced)). Purity was determine by SDS PAGE using a 4–12% Bis-Tris gel run in 1× MOPS SDS buffer. Eluted fractions containing CBP-HAT constructs were pooled and concentrated using a Vivaspin centrifugal concentrator (GE healthcare). Concentrated protein was dialysed overnight into GST-Buffer D (GST Buffer A + 20% glycerol). Protein concentration was determined using a nanodrop with calculated extinction co-efficient and molecular weight parameters. Purified protein was checked for RNase contamination using an RNase Alert lab test kit (ThermoFisher). Purified protein was divided into single use aliquots, flash frozen in liquid nitrogen and stored at −80°C.
Nucleosomes
Recombinant human nucleosomes were reconstituted using established protocols (Lowary and Widom, 1998; Luger et al., 1999; Tanaka et al., 2004) described in detail below. Human histones in pET15-b+ (EMDMillipore) were expressed in Rosetta 2 DE3 cells (EMDMillipore) in 2YT broth. Cells were resuspended in Histone buffer A (50mM TrisHCl pH8.0 (RT), 500mM NaCl, 1mM PMSF, 5% Glycerol) and lysed by sonication. Cell lysates were cleared by centrifugation at 18000 rpm for 45 minutes at 4°C. Cell pellets were retained and resuspended in Histone buffer B (50mM TrisHCl pH8.0 (RT), 500mM NaCl, 1mM PMSF, 5% Glycerol, 6M Urea). Supernatents were loaded on to a gravity flow column (Biorad) packed with 3ml Ni-NTA agarose (Qiagen) and equilibrated with Histone buffer B, washed three times with Histone buffer B and eluted with Histone Elution buffer (Histone buffer B + 500mM imidazole) and checked for purity by SDS PAGE. Purified histones were dialysed into MilliQ water + 2mM B-mercaptoethanol and lyophilized. To reconstitute histone octamers, lyophilized histones were dissolved in Unfolding buffer (7M guanidinium HCL, 20mM Tris-HCl pH7.5 (RT), 10mM DTT). Equimolar ratios of H2A, H2B, H3 and H4 were mixed and dialysed against Re-folding buffer (2M NaCl, 10mM Tris-HCl pH7.5 (RT), 1mM EDTA, 5mM B-mercaptoethanol). Trace precipitate was removed by centrifugation and the re-folded octamer was purified on a Superdex 200 16/60 size exclusion column equilibrated with Re-folding buffer. Purity and stoichiometry of the reconstituted octamer was checked by SDS-PAGE. Nucleosome core particles were then re-constituted using a 207bp Widom-601 DNA sequence (Lowary and Widom, 1998) via stepwise dialysis from 2M to 250mM KCl over a 48h period (Luger et al., 1999; Peterson, 2008). Purity and stoichiometry was confirmed by SDS-PAGE and DNA binding to the reconstituted nucleosome was visualized by 6% Native PAGE stained with SYBR gold (Figure S5D).
RNA in vitro transcription and 32-P radio-labeling
eRNA probes were designed according to the presence of nascent RNA transcripts in GROseq data and to incorporate the location of cross-linked RNA in PAR-CLIP data. No cap or poly-A tail was added. Sequences corresponding to eRNA targets (Table S5) were ordered as gBlock gene fragments (IDT) and cloned into pIDT-smart vectors digested with EcoRV (NEB) by blunt end cloning. Templates for in vitro transcription reactions were PCR amplified using PFu Ultra II high fidelity DNA polymerase (Agilent) with primers to include a T7 RNA polymerase promoter site at the 5’ end of the sequence (Table S6) and confirmed to be the expected size by agarose gel electrophoresis. In vitro transcription reactions were carried out using a MAXIscript T7 RNA polymerase in vitro transcription kit (ThermoFisher) for 2 hours at 37°C. DNA templates were then removed by digestion with Turbo DNase I (ThermoFisher) for 30 minutes at 37°C. Transcribed RNA templates were resolved on a 6% denaturing TBE Urea gel. RNA bands were excised from the gel and eluted using RNA elution buffer (50mM BisTris pH6.7, 300mM Na-Acetate, 1% SDS, 10mM EDTA) and ethanol precipitated. Purified RNA probes were re-suspended in BTE (10mM Bis-Tris pH6.7 (RT), 1mM EDTA) and quantified by Qubit (ThermoFisher).
For end-labelling reactions, RNAs were dephosphorylated with calf intestinal alkaline phosphatase (CIP) (In a 20ul Reaction: 1–100pmol RNA, 1× CIP buffer (50mM Tris, pH 8.5, 0.1 mM EDTA, pH 8.0), 0.1U/ul CIP (NEB)) for 1 hour at 37°C followed by purification with acidic Phenol: Chloroform (ThermoFisher) and ethanol precipitation. De-phosphorylated RNA was then end labeled with T4 Polynucleotide Kinase and [γ-32P] ATP (In a 20ul Reaction: 1–100pmol RNA, 25 pmol [γ-32P]ATP (7000 Ci/mmol, 150 mCi/ml), 1× Kinase buffer 50 mM Tris, pH 7.5, 10 mM MgCl2, 5 mM DTT), 0.5 U/ml T4 PNK (NEB) for 1 hour at 37°C. Un-incorporated nucleotides were removed with a Mini Quickspin RNA column (Roche) and the labeled RNAs were gel purified from 6% denaturing TBE Urea gel as above. RNA was ethanol precipitated, re-suspended in BTE and quantified by Cerenkov counting.
Purification of ssDNA and dsDNA probes
To purify ssDNA probes, target sequences were amplified by PCR from template DNA using one biotinylated and one non-biotinylated primer. PCR products were gel purified to remove unbound primers. Purified, biotinylated dsDNA probes were conjugated to streptavidin M-280 magnetic beads (ThermoFisher) in 1× Binding Wash buffer (10mM TrisHCl pH7.5, 0.5mM EDTA, 1M NaCl) then washed with 4× with 1× Binding Wash buffer. dsDNA was then denatured chemically by resuspending beads in 0.2M NaOH and incubating for 6 minutes at 22°C in a thermomixer. Supernatent containing ssDNA was transferred to an equal volumne of 5M Ammonium acetate pH6.8, then purified by isopropanol precipitation. ssDNA probes were then gel purified to ensure correct sizing, and checked on a denaturing 6% TBE Urea gel stained with SYBR gold.
dsDNA probes were amplified by PCR from template DNA using PFu Ultra II high fidelity DNA polymerase (Agilent) and gel purified. All purified DNA probes were quantified by Qubit (ThermoFisher).
In vitro RNA binding assays
In vitro pull-down RNA binding assays
RNA binding assays were carried out using recombinant proteins and re-folded RNA probes. RNA probes were re-folded as above. Flag-tagged full-length CBP or GST-tagged CBP-HAT domain constructs were incubated with 20nM re-folded RNA with rotation at 4°C for 1 hour in RNA Buffer A (20mM Tris HCl pH7.4 (RT), 200mM KCl, 0.2mM EDTA, 2mM DTT, 10% Glycerol, 0.05% NP40, 10mM EDTA). Protein-RNA complexes were recovered with Glutathione agarose beads (ThermoFisher) or Flag-M2 agarose (Sigma) for 30 minutes at 4°C with rotation. Beads were washed three times with RIP wash buffer (RNA Buffer A + 1mM MgCl2). Samples for protein analyses were resuspended in LDS sample loading buffer and resolved by SDS-PAGE. Samples for RNA analyses were purified using Trizol and resolved on a denaturing 6% TBE Urea gel stained with SYBR gold. Replicate experiments were fully independent experiments completed on different days, and included multiple independently purified protein and RNA samples.
RNA electrophoretic mobility shift assay
RNA electrophoretic mobility shift assays (RNA EMSAs) used established protocols. 32-P radiolabelled RNA was diluted to 20,000cpm, 20nM with unlabeled RNA in BTE. RNA was re-folded by incubating for 5 minutes at 95°C before snap-cooling on ice for 5 minutes. RNA re-folding buffer (10mM Bis-Tris pH6.7, 50mM KCl, 10mM MgCl2; 1× final) was added to RNA on ice. For re-folding, RNA was transferred to an ice-cold metal block and allowed to fold by warming to room temperature (~21°C) for 30 minutes. Binding reactions were assembled in RNase free, low adhesion microcentrifuge tubes. Reactions contained 1× RNA binding buffer (20mM Tris HCl pH7.4, 100mM KCl, 1mM EDTA, 1% glycerol, 0.05% NP40, 2mM DTT, 0.5mM ZnCl2, 0.4U/ul murine RNAse inhibitor (NEB)), 0.1mg/ml BSA (NEB), 0.1mg/ml yeast tRNA (Sigma). Unless otherwise stated, 2000cpm (2nM) of re-folded RNA (Or BTE + 1× RNA re-folding buffer) was added to each reaction. For competition binding experiments, the desired concentration of un-labelled RNA, ssDNA or dsDNA was added along with labeled RNA. Binding reactions were started by adding the required concentration of CBP-HAT domain construct and allowed to proceed for 30 minutes at 4°C. Reactions were loaded immediately on a 6% Native polyacrylamide gel (29:1 Acrylamide:Bis-acrylamide) pre-run for 1h at 100V in 0.5X TBE, and run for 2h at 100V. Gels were vacuum dried for 60 minutes at 80°C and exposed to autoradiography screen or X-ray film for 12h before imaging with a Typhoon phosphorimager (GE Healthcare). Replicate experiments were fully independent experiments completed on different days, and included independently purified protein and RNA samples. Densitometric analysis was carried out in FIJI; binding curves were calculated using non-linear regression in R.
Histone acetyltransferase assays
Steady state kinetic characterization and analysis
Steady state HAT assays were carried out using protocols adapted from (Bowers et al., 2010; Henry et al., 2013; Thompson et al., 2004). RNA probes were refolded as above. Reactions were assembled in RNAse Free, low adhesion microcentrifuge tubes (Eppendorf). Reactions contained 1× HAT assay buffer (50mM Tris-HCl pH 7.5 (RT), 5% glycerol, 0.1mM EDTA, 50mM KCl), 1mM DTT, 10mM Na-Butyrate, 1× Complete EDTA protease inhibitor cocktail (Roche), 0.4U/ul murine RNAse inhibitor (NEB), 0.1mg/ml BSA (NEB), 0–200nM H31–21 peptide (Anaspec) and the required concentration of RNA probe. 3H labeled Acetyl Co-enzyme A (3H-acetyl CoA) (specific activity = 3.95 Ci/mmol, 0.1uCi/ul, Perkin Elmer) was diluted in a 1:1 ratio with unlabeled acetyl CoA (Sigma) to give a final concentration of 100uM and 0.3uCi per reaction. 10nM of purified CBP-HAT domain construct was added to the reaction tubes at 20s intervals and incubated at 30°C for 10 minutes to allow binding of RNA and H3 peptide substrate to reach equilibrium. The reaction was started at 20s intervals by adding 3H-acetyl CoA and run for exactly 10 minutes to estimate initial rate (Bowers et al., 2010). After 10 minutes, reactions were quenched at 20s intervals by pipetting half of the reaction mix (15ul) on to p81 Whatman paper (GE Healthcare). Reactions were dried at room temperature for 20 minutes, then paper was washed 3× 15 minutes in 50mM Sodium carbonate/bicarbonate pH9.3 before rinsing for 20s in acetone and drying for 20 minutes at room temperature. The remaining half of the reaction was spotted on to p81 Whatman paper but not washed to determine the total radiation in each reaction, allowing calculation of acetylated peptide formation (uM product/s)(Berndsen and Denu, 2005). Ultima Gold liquid scintillation fluid (Perkin Elmer) was added to the dried filter paper in scintilliation tubes, and incorporated 3H-acetyl CoA (cpm) was counted using a liquid scintillation counter (Perkin Elmer). Background reaction controls were included containing all reaction components excluding the CBP-HAT domain construct and +/− RNA to measure background labeling of peptide. Control reactions were also included with unlabeled acetyl CoA and RNA was purified using Trizol to ensure RNA integrity for the duration of the experiment (Figure S5L). Replicate experiments were fully independent experiments completed on different days. To determine kinetic parameters, the initial rates of reaction were fit to the Michaelis-Menten equation using non-linear regression in R.
RNA dose curve HAT assays
RNA dose curve HAT assays using 3H-acetyl CoA followed the steady state HAT assay protocol with some variations. Following re-folding, RNA was diluted by serial dilution to achieve the desired range of concentrations (0.625–40nM final). Reactions contained 1× HAT assay buffer (50mM Tris-HCl pH 7.5 (RT), 5% glycerol, 0.1mM EDTA, 50mM KCl), 1mM DTT, 10mM Na-Butyrate, 1× Complete EDTA protease inhibitor cocktail (Roche), 0.4U/ul murine RNAse inhibitor (NEB), 0.1mg/ml BSA (NEB), 80nM H31–21 peptide (Anaspec) and the required dilution of RNA probe. 1nM or 5nM of purified CBP-HAT domain construct was added to the reaction tubes at 20s intervals and incubated at 30°C for 10 minutes to allow RNA binding to reach equilibrium. The reaction was started at 20s intervals by adding 3H-acetyl CoA and run for exactly 30 minutes. Replicate experiments were fully independent experiments completed on different days, and included independently purified protein and RNA samples.
Western blot HAT assays followed the RNA dose curve HAT assay for 3H-acetyl CoA with some variations. Reactions contained 1mM recombinant purified human H3.1, 1nM CBP-HAT domain construct and were titrated with 0–10nM re-folded RNA. Reactions were allowed to proceed for 30 minutes and halted by adding LDS loading buffer. Labelled H3 was resolved on a 12% Bis Tris gel run in MES buffer, transferred to 0.2uM pvdf membrane and probed with antibodies against H3K27ac, H3K18ac, H4K16ac (all Active motif), H3 or H4 (Abcam). Western blots were imaged with a Fujifilm gel imager. Densitometric analysis was carried out in FIJI; H3K27ac and H3K18ac band densities were normalized by H3 loading control densities and used to calculate a fold change in acetylation relative to background controls. Replicate experiments were fully independent experiments completed on different days, and included independently purified protein and RNA samples.
Cell culture manipulations
Adenovirus infections
For deletion of endogenous, floxed CBP/p300, cells were infected with Cre-recombinase or GFP-adenovirus (10,000 GC/ml, Penn Vector Core) in the presence of polybrene (8ug/ml). After 24 hours, virus containing media was replaced with complete DMEM (10% FBS, 0% Penn-strep) at 37°C in 5% CO2. Cells were then grown for 5-days following initial infection for efficient CBP/p300 deletion (Kasper et al., 2014).
Retroviral transduction
Retroviral transduction of CBP constructs into MEFs was done using the pRetroX system (Clontech). Full-length CBP constructs were cloned into pRetroX-pTRE3G vector (see molecular biology section for cloning details). pRetroX-Tet3G vector was packaged in GP2-293 cells using either pEco or p10A1 envelope packaging vectors. Packaged retroviral supernatents were used to infect CBP/p300 flox/flox MEFs and selected for infection using 500ug/ml G418 (Corning). Following selection, cells were maintained in 200ug/ml G418. CBP constructs in pRetroX-pTRE3G were packaged in GP2-293 cells using either pEco, p10A1 or VSV-G envelope packaging vectors. pRetroX-Tet3G CBP/p300 flox/flox MEFs were transduced using packaged retroviral supernatents supplemented with 6–8ug/ml Polybrene for up to 3 rounds of transduction; cells were selected using 2.5ug/ml Puromycin and 200ug/ml G418. Expression of transduced constructs was induced using 500–1000ng/ml Doxycycline (Dox) for at least 72h. For CBP deletion/replacement experiments, retrovirally transduced MEFs were treated with 500–1000ng/ml Dox for 24h before infection with Cre- or GFP-adenovirus.
ASO transfection
We designed multiple antisense oligonucleotide (ASO) sequences to target eRNA-CBP sequences identified in our PAR-CLIP experiments for depletion, including ASO’s targeting both sense and antisense eRNA strands at the target enhancers. ASOs were designed using an antisense oligonucleotide design tool (IDT) and screened for efficient knockdown by RT-qPCR. We tested ASOs that produced significant depletion of the targeted eRNA (Figure 6B-C, Figure S6E). ASO targeting eYY1 (as), eCcnd1 (s), e-Tet2 (s) or GFP control (see Table S5 for sequences) were used at 1uM concentration. Sequences were transfected using lipofectamine RNAi-MAX transfection reagent (ThermoFisher). Cells were incubated for 6–12 hours with ASO-lipofectamine complexes in OptiMEM media (GIBCO), then media was replaced with Complete DMEM +10% FBS for a further 24h before harvesting.
Quantitative PCR
RT-qPCR
Cells were harvested using Trizol reagent (ThermoFisher) according to manufacturers instructions. Cells were lysed by resuspending in Trizol reagent. RNA was purified with chloroform and isopropanol precipitated. Purified RNA was resuspended in BTE. Contaminating DNA was removed by incubation with Turbo DNase I (ThermoFisher) for 30 minutes at 37°C. RNA was purified again with acidic Phenol Chloroform (ThermoFisher) followed by ethanol precipitation. Reverse transcription reactions were done using random primers with a High Capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was carried out with Power SYBR (Applied Biosystems) using primers for target eRNA or coding mRNA sequences (Table S6).
Chromatin Immunoprecipitation (ChIP)
For ChIP experiments, cells were crosslinked for 10 minutes at room temperature using formaldehyde (1% final). Cells were harvested by scraping and washed twice in cold PBS. Cells were lysed for by rotating for 10 minutes at 4°C in ChIP lysis buffer 1 (50mM HEPES-KOH, pH7.5, 140mM NaCl, 1mM EDTA, 10% Glycerol, 0.5% NP40, 0.25% Triton X-100, 10mM Na-Butyrate, Complete protease inhibitor cocktail (Roche)). Nuclei were pelleted by centrifugation at 2500rcf for 5 minutes, re-suspended in ChIP Buffer 2 (10mM Tris-HCl pH7.3 (RT), 200mM NaCl, 1mM EDTA, 0.5mM EGTA, 10mM Na-Butyrate, Complete protease inhibitor cocktail) and incubated with rotation for 10 minutes at room temperature. Nuclei were pelleted by centrifugation at 2500rcf for 5 minutes and re-suspended in ChIP lysis buffer 3 (10mM Tris-HCl pH8.0 (7.3@RT) 200mM NaCl 1mM EDTA 0.5mM EGTA 0.1% Na-deoxycholate 0.5% N-lauroylsarcosine, 0.1% SDS) for shearing. Chromatin was sheared to a 250–300bp average size using a Covaris S220, using 1ml millitubes and a high cell number shearing protocol for 15 minutes. Lysates were cleared by centrifugation at 20,000×g for 20 minutes. Concentration of lysates was determined by BCA assay, and IP’s were done using equal concentrations (0.3–1.5mg) of sheared chromatin. Immunoprecipitations were performed by incubating with 2–4ug antibody overnight at 4°C with rotation. Immunocomplexes were recovered by adding blocked Protein A dynabeads for 1–2h and washed 5 times in ChIP wash buffer (50mM HEPES-KOH pH7.5, 500mM LiCl, 1mM EDTA, 1% NP40, 0.7% Na-deoxycholate, 0.1% N-lauroylsarcosine) and 1× in ChIP final wash buffer (10mM Tris-HCl pH8.0 (4°C), 1mM EDTA, 50mM NaCl). Immunocomplexes were eluted by incubating at 65°C for 30 minutes in ChIP elution buffer (50mM Tris-HCl pH8.0 (RT), 10mM EDTA, 1% SDS) and cross-linking was reversed by incubating overnight at 37°C. Immunoprecipitated DNA was treated with RNAse A (0.2mg/ml final) for 2 hours at 37°C, then Proteinase K (0.2mg/ml final) for 2 hours at 55°C. DNA was then purified by phenol:chloroform extraction and ethanol precipitation and resuspended in 1× TE buffer. qPCR was carried out with Power SYBR (Applied Biosystems) using primers for target eRNA or control RNA sequences (Table S6).
PAR-CLIP RT-qPCR
Cells were incubated for 16h with 150uM 4-thiouridine (4-SU). Plates were washed with PBS before cells were cross-linked using 400mJ/cm2 365nM UV light with a Stratalinker UV crosslinker (Stratagene). Cleared cell lysates were prepared as for regular PAR-CLIP (see above). Immunopreciptations used equal concentrations (1–2mg per IP) of cleared cell lysate with a depleting amount of anti-GFP antibody (Abcam, ab290) for 2 hours at 4°C. 10% of cleared lysate was retained for input samples. Immunocomplexes were recovered for 30 minutes with BSA blocked Protein G dynabeads (ThermoFisher). Beads were collected and washed three times with CLIP wash buffer (as above) then IP50 buffer (as above), before resuspending in 1× Turbo DNase buffer. Contaminating DNA was removed by incubation with 2.5U Turbo DNase I for 30 minutes at 37°C. Beads were washed twice with CLIP wash buffer and IP50 buffer. Beads were resuspended in 1ml IP50 and transferred to a clean microcentrifuge tube for RNA purification. Proteinase K (20mg/ml, Roche) was diluted to 2mg/ml in Proteinase K (PK) buffer (50mM Hepes pH7.0, 50mM NaCl, 1mM EDTA, 1% SDS) and pre-incubated for 15 minutes at 37°C before use to remove contaminating RNase. Beads were collected and resupended in PK buffer mix (2mg/ml PK final), then incubated for 1h at 37°C to digest cross-linked protein. Immunopreciptated RNA and Input RNA was purified using acidic phenol:chloroform (ThermoFisher) and ethanol precipitation. RNA pellets were resuspended in 1× BTE buffer and used for Reverse transcription reactions using random primers with a High Capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was carried out with Power SYBR (Applied Biosystems) using primers for target eRNA or control RNA sequences (Table S6).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical details of experiments are included in the Figures or Figure legends.
DATA AND SOFTWARE AVAILABILITY
Data resources
The GEO repository series accession number for the CBP PAR-CLIP and background control (yeast Gal4-DBD and nls-GFP) PAR-CLIP datasets, as well as read density profiles for CBP-PAR-CLIP is GSE75684 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75684).
ADDITIONAL RESOURCES
Supplementary Material
A) RNA binding prediction (BindN, (Wang and Brown, 2006)) for CBP-HAT domain. Predictions for non-binding residues (green) and binding residues (red) are indicated. Residues forming the activation loop and residues 1559–1608 deleted or mutated to prevent RNA binding are highlighted.
B) Purification of flag-tagged recombinant CBP from Sf9 cells (Left panel) and GST tagged CBP-HAT domain from E.coli (Right panel). Both gels were stained with Coomassie.
C-D) Replicate gel images of Figure 4B-C; In vitro pull down of C) eRNA-Klf6s and D) eRNA-Med13ls using flag-tagged CBP. s=sense, as=antisense strand RNA.
E) In vitro pull down of eRNA-Klf6 using GST-tagged CBP HAT domain constructs (see also Figure 4C). RNA fraction stained with SYBR gold is shown in top panel; protein fractions stained with coomassie are shown in bottom panel.
F) In vitro pull down of diverse RNA sequences using CBP-HATwt. eRNA sequences eRNA-Tet2s, eRNA-Klf4s, eRNA-Klf6s (left); lncRNA sequences HOTAIR1-300, Gas5, Meg3 (middle); exonic RNA sequences (ID-1 (exon1), Bbs2 (exon-1), Klf2 (exon-2) (right). All RNAs were resolved using 6% TBE Urea gels, apart from Meg3 (resolved using denaturing agarose formaldehyde gel) and stained using SYBR Gold.
G) Quantification of RNA-pulldown data in Figure S4F. n=3.
H) Binding curves showing eRNA binding to CBP-HATwt.
I) RNA EMSA of eRNA probes using CBP-HATwt (0–10000nM); Top) eRNA-YY1s; Middle) eRNA-Klf6s; Bottom) eRNA-Ccnd1s.
J) GST CBP-HAT domain constructs used in EMSAs showing CBP-HATwt, CBP-HATdelta-loop and CBP-HATmutant loop constructs. Constructs were designed with reference to predicted RNA binding residues (Figure S4A) and residues deleted in available X-ray crystallographic structures of the p300 HAT domain. In the mutant loop sequence, mutated residues are highlighted in lower case.
K) RNA EMSA showing requirement for de-acetylation for RNA binding. GST-CBP-HATwt was expressed without co-expressing ySir2 and purified (CBP-HATac). 2nM eRNA-YY1 was titrated with 0–10000nM CBP-HATac.
L) Western blot for immunoprecipitated GFP-CBP (control for Figure 4M).
Alignment and pre-processing of PAR-CLIP reads.
A) Read counts in PAR-CLIP datasets (2× replicates of CBP PAR-CLIP and control nGFP and Gal4-DBD PAR-CLIP) following sequencing, pre-processing and alignment.
B) Output from PARalyzer v1.1 (Corcoran et al., 2011) showing identified CBP bound RNAs and control bound RNAs.
C) Intersection between CBP bound RNAs and control bound RNAs.
Enriched Gene ontology (GO) terms (DAVID) for nearest genes to 587 Distal CBP-eRNA sites (FDR<10).
A) Enriched GO terms.
B-E) Gene lists for nearest genes to 587 Distal CBP-eRNA sites corresponding to the top 4 enriched GO categories.
Analysis of underlying motifs at distal eRNA bound CBP peaks revealed binding sites for known CBP-interacting transcription factors.
A) Motif discovery with MEME (Bailey and Elkan, 1994), motif comparison with TOMTOM (Gupta et al., 2007).
B) Motif analysis using SeqPos (Galaxy/Cistrome) (He et al., 2010).
Gene expression changes following deletion of CBP/p300 in CBP/p300 flox/flox MEFs using adenoviral Cre. Data from GSE54452, (Kasper et al., 2014). Genes validated by RT-qPCR are shown in bold.
A) All genes.
B) Genes proximal to CBP-eRNAs.
C) Genes associated with CBP-eRNAs in Transcription factor related GO categories (see Table S3).
A) RNA sequences used for in vitro assays.
B) Primers used for in vitro transcription of RNA probes.
C) ASO sequences used for eRNA depletion experiments in Figure 6.
A) RT-qPCR primers.
B) ChIP-qPCR primers.
C) Cloning PCR primers.
D) Mutant loop DNA sequence. Lower case residues indicate mutated sequence compared to CBP-HATwt used to generate mutated loop.
A) Whole western blot view of Figure 1A
B) Whole autoradiography and western blot view of Figure 1C. CBP PAR-CLIP signal required 4-SU.
C) Western blot for CBP. Input was not 32-P labeled.
D-E) Example replicates of Figure 1C quantified in Figure 1D, CBP PAR-CLIP required 4-SU: Top, Autoradiography; Bottom, western blot for CBP on PAR-CLIP membrane. Input was not 32-P labeled, but resolved and transferred to the same membrane.
F-G) Whole autoradiography (F) and western blot (G) view of Figure 1E, CBP PAR-CLIP signal was sensitive to increasing RNAse concentrations. 1× RNAse cocktail contained: RNAse A (0.01mU/ul) + RNase T1 (0.4mU/ul).
H) Example replicate autoradiography of Figure 1E, quantified in Figure 1F.
A) Autoradiography of 2nd biological replicate of CBP PAR-CLIP and nls-GFP (nGFP) and yeast Gal4 DNA binding domain (Gal4-DBD) background control membranes. Dashed box shows regions of membrane excised for library preparation.
B) Correlation of PAR-CLIP reads from individual replicates 1 and 2 over CBP enriched RNAs called from replicate 1 (5926) and 2 (5322). Correlation of reads from replicate 1 and 2 over common peaks that were called independently in both replicates (3229) are highlighted in red.
C) Distribution of sizes of CBP-RNA regions following assignment of statistically enriched RNAs with PARalyzer v1.1.
D-G) Intersection of CBP-RNAs with background control RNAs: D) Gal4-DBD control intersecting nGFP control; E) CBP enriched RNA intersecting Gal4-DBD control; F) CBP enriched RNA intersecting nGFP control; G) CBP enriched RNA intersecting Gal4-DBD and nGFP control RNAs.
H) Immunoprecipitation of CBP under Native RNA-IP conditions +/− treatment with 1mg/ml RNase A; WB for Mediator (Med-1 subunit) or NELF (NELF-e subunit).
I) Immunoprecipitation of CBP under PAR-CLIP conditions; WB for Mediator (Med-1 subunit) or NELF (NELF-e subunit).
J) Comparison of Mediator (GSM560353 (Kagey et al., 2010), left panel) and NELFb (GSE24113 (Sun et al., 2011), right panel) read density over CBP-RNAs and enriched control RNAs. CBP peaks were randomly downsampled to match the size of the background dataset (5581).
K-L) Enrichment of reads at CBP-RNAs sorted by genome regions from Figure 2D; L) CBP-RNA (PAR-CLIP) signal; M) Nascent RNA transcription (GROseq) signal (Promoter = 1kb upstream of TSS).
M) Read density of total RNAseq datasets (GSM687308, right panel; GSM1100747, left panel) at CBP-RNAs.
N-O) Comparison of nascent transcription (GROseq) and total RNAseq at CBP-RNAs; N) Enrichment of total RNAseq reads at CBP-RNAs. CBP-RNAs were grouped into deciles based on GROseq signal (10th decile = most enriched for GROseq reads). Boxplot shows total RNAseq reads (GSM1100747); O) Enrichment of GROseq signal at CBP-RNAs. CBP-RNAs were grouped into deciles based total RNAseq signal (10th decile = most enriched for total RNAseq reads). Boxplot shows total GROseq reads.
P) Comparison of GROseq and total RNAseq (GSM1100747) enrichment at CBP-RNAs sorted by genome regions from Figure 2D. P-values from Mann-Whitney U-test.
Q) Spearman correlation co-efficient between PAR-CLIP and CBP ChIPseq reads over 1kb intersecting bound RNA regions at increasing peak-to-peak distances. Plot shows correlation for CBP bound RNAs (purple), Gal4-DBD (orange) and nGFP (green) background control bound RNAs. Grey dashed line highlights a 3.5kb peak-peak distance.
A-C) Comparison of read density (RPM) over enriched RNA regions <3.5kb from nearest CBP peak and >3.5kb from nearest CBP peak; A) CBP PAR-CLIP; B) CBP ChIPseq; C) GROseq. P-values from Mann-Whitney U-test.
D) Histogram showing distance from RNA bound CBP peaks to nearest TSS. Peaks were divided into Promoter (TSS-1kb), Exon and Distal (intronic/intergenic) sub groups.
E) Comparison of CBP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on PAR-CLIP enrichment. Boxplots show enrichment for CBP ChIPseq reads at each decile (10th decile = most enriched for total PAR-CLIP reads).
F) Comparison of PAR-CLIP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on CBP enrichment. Boxplots show enrichment for PAR-CLIP reads at the top five deciles for CBP signal (10th decile = most enriched for total CBP ChIPseq reads).
G) Comparison of PAR-CLIP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on GROseq enrichment. Boxplots show enrichment for PAR-CLIP reads at the top five deciles for GROseq signal (10th decile = most enriched for total GROseq reads).
H-M) Comparison of RNA-CBP peaks in Promoter, Exon and Distal subgroups. Profiles show 50bp bins over 5kb windows centered on the CBP peak and were normalized by number of peaks in each genome region subgroup; Promoter (green), Exon (orange), Distal (purple). Grey boxes show the 1kb window used for quantification in box-plots; H) Read density profiles of CBP reads; I) Quantification of CBP read density; J) Read density profiles of GROseq reads; K) Quantification of GROseq read density; L) Read density profiles of PAR-CLIP reads; M) Quantification of PAR-CLIP read density. All reads were normalized per million mapped reads (RPM). P-values from Mann-Whitney U-test.
N) Boxplot shows H3K27ac reads over RNA bound CBP peaks (green) or non-RNA bound CBP peaks (orange). P-values from Mann-Whitney U-test. non-RNA bound CBP peaks were randomly downsampled to match CBP-eRNA peaks (976).
O) GROseq read density profiles (RPM) comparing RNA bound CBP peaks (green) or non-RNA bound CBP peaks (orange). non-RNA bound CBP peaks were randomly downsampled to match CBP-eRNA peaks (976).
P-R) Venn diagrams showing overlap between: P) RNA bound CBP peaks and H3K4me1; Q) RNA bound CBP peaks and H3K27ac; R) All CBP peaks with H3K4me1 peaks and H3K27ac peaks.
S) GO analysis (DAVID, (Huang et al., 2009)) of nearest genes to PAR-CLIP CBP peaks. (FDR <1, sorted by FDR).
T) Comparison of CBP-eRNA associated genes with genes not associated with CBP-eRNA. Control population was downsized to match the 1146 genes proximal to CBP-eRNA (Gene expression microarray data from GSE54452, (Kasper et al., 2014)). P-values from Mann-Whitney U-test.
U) RT-qPCR data showing fold change in CBP mRNA following infection with control adenoviral GFP (Adv-GFP, green) or knockdown with Adenoviral Cre (Adv-Cre, purple). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test.
A) Schematic of filter binding HAT assay.
B) In vitro pull-down of RNA sequences using CBP-HATwt. lncRNA HOTAIR1–300, exonic RNA (ID-1 (exon1) and lncRNA Meg3. All RNAs were resolved using 6% TBE Urea gels, apart from Meg3 (resolved using denaturing agarose formaldehyde gel) and stained using SYBR Gold (same experiment as Figure S4F).
C) RNA dose curve filter binding assays: RNA dependent stimulation of CBP acetyltransferase activity. 5nM CBP-HATwt was titrated with 0–40nM RNA (lncRNA HOTAIR1–300, exonic RNA (ID-1 (exon1) and lncRNA Meg3). Data shows fold change in rate of Acetyl-CoA incorporation (Vmax)/Concentration enzyme ([E], (s−1)) from 0nM RNA. Shaded regions represent mean +/− s.e.m (RNA n=3).
D) Native PAGE showing DNA binding to reconstituted nucleosomes (top panel). Coomassie staining showing individual histones (bottom panel).
E-G) Western blot HAT assay using recombinant nucleosome substrate. 1nM CBP-HATwt was titrated with 0–10nM E) eRNA-Mdm2s; F) eRNA-Ccnd1s; G) lncRNA HOTAIR1–300. The reaction was resolved by SDS-PAGE and probed for H3K27ac, H4K5ac and H3/H4 by western blot. Coomassie staining (bottom panel) shows individual histones.
H) Western blot HAT assay. 1nM CBP-HATwt was titrated with 0–10nM eRNA-Mdm2 (top), eRNA-PAPPA (middle) (Melo et al., 2013) or long noncoding RNA HOTAIR (bottom) (Kaneko et al., 2013). Reaction was resolved by SDS-PAGE and probed for H3K27ac by western blot. Reactions contained 20uM purified recombinant H3.1 and 200uM Acetyl CoA.
I) Quantification of Western blot HAT assays in Figure 5C and Figure S5B. Densitometry was determined in FIJI. For quantified Western blots, n=4).
J) Western blot HAT assay: human MOF (hMOF). 1nM hMOF HAT domain was titrated with 0–10nM eRNA-Mdm2. Reaction was resolved by SDS-PAGE and probed for H3K27ac by western blot. (hMOF was a kind gift of Ronen Marmorstein). Reactions contained 20uM purified recombinant H4 and 200uM Acetyl CoA.
K) Kinetic parameters derived from steady state HAT assays. All analysis was carried out using nls regression in R.
L) RNA integrity was checked before and after each HAT assay (Example control gel of eRNA-Mdm2 from steady state HAT assays). Control reactions contained 200uM unlabeled Acetyl-CoA; RNA was extracted with Trizol and resolved on a denaturing 6% TBE urea gel stained with SYBR gold.
A-C) UCSC genome browser view of GROseq signal (+ strand purple, - strand grey) and CBP-RNAs (orange bars) at enhancer and promoter regions for: A) YY1; B) Ccnd1; C) Tet2.
D) ChIP-qPCR for H3K27ac (left) and H3K18ac (right) at enhancer and promoter regions following knockdown of CBP and p300. CBP/p300 flox/flox MEFs were infected with control adenoviral GFP (Adv-GFP, Green) or adenoviral Cre (Adv-Cre, purple/blue/orange). Data shows foldchange in IP/H3. Error bars represent mean +/− s.e.m; n=3.
E) ASO mediated depletion of PAR-CLIP eRNA. RT-qPCR data showing foldchange in expression of eRNA and associated mRNA following transfection of ASO targeting eRNA-YY1antisense (purple), eRNA-Ccnd1sense (blue), eRNA-Tet2sense (orange) or GFP-control (green). Depletion efficiency was determined from expression of eRNA and mRNA at YY1 (top), Ccnd1 (middle) or Tet2 (bottom). Error bars represent mean +/− s.e.m; n=4.
F-G) ChIP-qPCR at enhancer and promoter regions following transfection of ASO targeting eRNA-YY1 (purple) or GFP-control (green) showing foldchange in IP/H3 for: F) H3K27ac and G) H3K18ac at targeted gene and non-targeted control gene Tet2 (control, bottom). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test. (Data is from the same experiment as Figure 6F-H)
H-I) ChIP-qPCR at enhancer and promoter regions following transfection of ASO targeting eRNA-Tet2 (orange) or GFP-control (green) showing foldchange in IP/H3 for: H) H3K27ac and I) H3K18ac at targeted gene Tet2 and non-targeted control gene YY1 (bottom). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test.
J) CBP ChIP-qPCR at targeted gene Tet2 and non-targeted control gene YY1. Data shows foldchange in CBP IP/Input. Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test: *P< 0.05; **P< 0.01; ***P< 0.001.
Acknowledgments
We thank P. Brindle and L. Kasper for CBP/p300 flox/flox MEFs. R. Marmorstein for hMOF; C. He for Meg3 RNA; H. Goodarzi for advice. J. Chen for experiments not included in the manuscript. H. Fan lab for advice and equipment. R. Marmorstein, D. Schultz and J. Dorsey for advice on experiments. P. Sen, P. Shah, M. Sammons and A. Twelvetrees provided considerable support, advice and critical reading of the manuscript. SLB is supported by NIH grant R01 CA078831. RB was supported by an NIH Innovator Award (DP2MH107055), the Searle Scholars Program (15-SSP-102), the March of Dimes Foundation (1-FY-15-344), and the W.W. Smith Charitable Trust (C1404).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The GEO repository series accession number for the CBP PAR-CLIP and background control (yeast Gal4-DBD and nls-GFP) PAR-CLIP datasets, as well as read density profiles for CBP-PAR-CLIP is GSE75684.
Author Contributions
Study was conceived by SLB and DB. SLB and DB initiated and led the study with input from RB, RS and DR. DB, SLB, RB and RS designed experiments. DB carried out all experiments. DB and GD analyzed high-throughput sequencing data. DB, SLB and RB wrote the manuscript. All authors reviewed and commented on the manuscript.
References
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods. 2005;36:321–331. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. Elife. 2014;3:e02637. doi: 10.7554/eLife.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Mol Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Pnas. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Biol. 2013 doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedersdorf MB, Keene JD. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G Hendrickson D, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RA, Kuo Y-M, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry. 2013;52:5746–5759. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee G-SM, Chen S, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proceedings of the National Academy of Sciences. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. Embo J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Mol Cell. 2014a;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014b;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Qu C, Obenauer JC, McGoldrick DJ, Brindle PK. Genome-wide and single-cell analyses reveal a context dependent relationship between CBP recruitment and gene expression. Nar. 2014;42:11363–11382. doi: 10.1093/nar/gku827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013 doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013 doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014 doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schübeler D. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 2011;7:e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nar. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit W, Oude Vrielink JAF, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Meng F-L, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon K-R, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’Orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs Promote Transcription by Establishing Chromatin Accessibility at Defined Genomic Loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL. Salt gradient dialysis reconstitution of nucleosomes. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5113. pdb.prot5113. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin α-subunit gene. Proceedings of the National Academy of Sciences. 2015;112:4369–4374. doi: 10.1073/pnas.1414841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo J-Y, Liu X, Watts JK, Martinez C, Kim T-K. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, et al. Acetylation of RNA polymerase II regulates growth-factor induced gene transcription in mammalian cells. Mol Cell. 2013;52:314–324. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014 doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- Sun J, Pan H, Lei C, Yuan B, Nair SJ, April C, Parameswaran B, Klotzle B, Fan J-B, Ruan J, et al. Genetic and genomic analyses of RNA polymerase II-pausing factor in regulation of mammalian transcription and cell growth. Journal of Biological Chemistry. 2011;286:36248–36257. doi: 10.1074/jbc.M111.269167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, Yokoyama S. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013;70:3989–4008. doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nar. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008a;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008b;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtrakoongate P, Riddick G, Fucharoen S, Felsenfeld G. Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes. PLoS Genet. 2015;11:e1005615. doi: 10.1371/journal.pgen.1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) RNA binding prediction (BindN, (Wang and Brown, 2006)) for CBP-HAT domain. Predictions for non-binding residues (green) and binding residues (red) are indicated. Residues forming the activation loop and residues 1559–1608 deleted or mutated to prevent RNA binding are highlighted.
B) Purification of flag-tagged recombinant CBP from Sf9 cells (Left panel) and GST tagged CBP-HAT domain from E.coli (Right panel). Both gels were stained with Coomassie.
C-D) Replicate gel images of Figure 4B-C; In vitro pull down of C) eRNA-Klf6s and D) eRNA-Med13ls using flag-tagged CBP. s=sense, as=antisense strand RNA.
E) In vitro pull down of eRNA-Klf6 using GST-tagged CBP HAT domain constructs (see also Figure 4C). RNA fraction stained with SYBR gold is shown in top panel; protein fractions stained with coomassie are shown in bottom panel.
F) In vitro pull down of diverse RNA sequences using CBP-HATwt. eRNA sequences eRNA-Tet2s, eRNA-Klf4s, eRNA-Klf6s (left); lncRNA sequences HOTAIR1-300, Gas5, Meg3 (middle); exonic RNA sequences (ID-1 (exon1), Bbs2 (exon-1), Klf2 (exon-2) (right). All RNAs were resolved using 6% TBE Urea gels, apart from Meg3 (resolved using denaturing agarose formaldehyde gel) and stained using SYBR Gold.
G) Quantification of RNA-pulldown data in Figure S4F. n=3.
H) Binding curves showing eRNA binding to CBP-HATwt.
I) RNA EMSA of eRNA probes using CBP-HATwt (0–10000nM); Top) eRNA-YY1s; Middle) eRNA-Klf6s; Bottom) eRNA-Ccnd1s.
J) GST CBP-HAT domain constructs used in EMSAs showing CBP-HATwt, CBP-HATdelta-loop and CBP-HATmutant loop constructs. Constructs were designed with reference to predicted RNA binding residues (Figure S4A) and residues deleted in available X-ray crystallographic structures of the p300 HAT domain. In the mutant loop sequence, mutated residues are highlighted in lower case.
K) RNA EMSA showing requirement for de-acetylation for RNA binding. GST-CBP-HATwt was expressed without co-expressing ySir2 and purified (CBP-HATac). 2nM eRNA-YY1 was titrated with 0–10000nM CBP-HATac.
L) Western blot for immunoprecipitated GFP-CBP (control for Figure 4M).
Alignment and pre-processing of PAR-CLIP reads.
A) Read counts in PAR-CLIP datasets (2× replicates of CBP PAR-CLIP and control nGFP and Gal4-DBD PAR-CLIP) following sequencing, pre-processing and alignment.
B) Output from PARalyzer v1.1 (Corcoran et al., 2011) showing identified CBP bound RNAs and control bound RNAs.
C) Intersection between CBP bound RNAs and control bound RNAs.
Enriched Gene ontology (GO) terms (DAVID) for nearest genes to 587 Distal CBP-eRNA sites (FDR<10).
A) Enriched GO terms.
B-E) Gene lists for nearest genes to 587 Distal CBP-eRNA sites corresponding to the top 4 enriched GO categories.
Analysis of underlying motifs at distal eRNA bound CBP peaks revealed binding sites for known CBP-interacting transcription factors.
A) Motif discovery with MEME (Bailey and Elkan, 1994), motif comparison with TOMTOM (Gupta et al., 2007).
B) Motif analysis using SeqPos (Galaxy/Cistrome) (He et al., 2010).
Gene expression changes following deletion of CBP/p300 in CBP/p300 flox/flox MEFs using adenoviral Cre. Data from GSE54452, (Kasper et al., 2014). Genes validated by RT-qPCR are shown in bold.
A) All genes.
B) Genes proximal to CBP-eRNAs.
C) Genes associated with CBP-eRNAs in Transcription factor related GO categories (see Table S3).
A) RNA sequences used for in vitro assays.
B) Primers used for in vitro transcription of RNA probes.
C) ASO sequences used for eRNA depletion experiments in Figure 6.
A) RT-qPCR primers.
B) ChIP-qPCR primers.
C) Cloning PCR primers.
D) Mutant loop DNA sequence. Lower case residues indicate mutated sequence compared to CBP-HATwt used to generate mutated loop.
A) Whole western blot view of Figure 1A
B) Whole autoradiography and western blot view of Figure 1C. CBP PAR-CLIP signal required 4-SU.
C) Western blot for CBP. Input was not 32-P labeled.
D-E) Example replicates of Figure 1C quantified in Figure 1D, CBP PAR-CLIP required 4-SU: Top, Autoradiography; Bottom, western blot for CBP on PAR-CLIP membrane. Input was not 32-P labeled, but resolved and transferred to the same membrane.
F-G) Whole autoradiography (F) and western blot (G) view of Figure 1E, CBP PAR-CLIP signal was sensitive to increasing RNAse concentrations. 1× RNAse cocktail contained: RNAse A (0.01mU/ul) + RNase T1 (0.4mU/ul).
H) Example replicate autoradiography of Figure 1E, quantified in Figure 1F.
A) Autoradiography of 2nd biological replicate of CBP PAR-CLIP and nls-GFP (nGFP) and yeast Gal4 DNA binding domain (Gal4-DBD) background control membranes. Dashed box shows regions of membrane excised for library preparation.
B) Correlation of PAR-CLIP reads from individual replicates 1 and 2 over CBP enriched RNAs called from replicate 1 (5926) and 2 (5322). Correlation of reads from replicate 1 and 2 over common peaks that were called independently in both replicates (3229) are highlighted in red.
C) Distribution of sizes of CBP-RNA regions following assignment of statistically enriched RNAs with PARalyzer v1.1.
D-G) Intersection of CBP-RNAs with background control RNAs: D) Gal4-DBD control intersecting nGFP control; E) CBP enriched RNA intersecting Gal4-DBD control; F) CBP enriched RNA intersecting nGFP control; G) CBP enriched RNA intersecting Gal4-DBD and nGFP control RNAs.
H) Immunoprecipitation of CBP under Native RNA-IP conditions +/− treatment with 1mg/ml RNase A; WB for Mediator (Med-1 subunit) or NELF (NELF-e subunit).
I) Immunoprecipitation of CBP under PAR-CLIP conditions; WB for Mediator (Med-1 subunit) or NELF (NELF-e subunit).
J) Comparison of Mediator (GSM560353 (Kagey et al., 2010), left panel) and NELFb (GSE24113 (Sun et al., 2011), right panel) read density over CBP-RNAs and enriched control RNAs. CBP peaks were randomly downsampled to match the size of the background dataset (5581).
K-L) Enrichment of reads at CBP-RNAs sorted by genome regions from Figure 2D; L) CBP-RNA (PAR-CLIP) signal; M) Nascent RNA transcription (GROseq) signal (Promoter = 1kb upstream of TSS).
M) Read density of total RNAseq datasets (GSM687308, right panel; GSM1100747, left panel) at CBP-RNAs.
N-O) Comparison of nascent transcription (GROseq) and total RNAseq at CBP-RNAs; N) Enrichment of total RNAseq reads at CBP-RNAs. CBP-RNAs were grouped into deciles based on GROseq signal (10th decile = most enriched for GROseq reads). Boxplot shows total RNAseq reads (GSM1100747); O) Enrichment of GROseq signal at CBP-RNAs. CBP-RNAs were grouped into deciles based total RNAseq signal (10th decile = most enriched for total RNAseq reads). Boxplot shows total GROseq reads.
P) Comparison of GROseq and total RNAseq (GSM1100747) enrichment at CBP-RNAs sorted by genome regions from Figure 2D. P-values from Mann-Whitney U-test.
Q) Spearman correlation co-efficient between PAR-CLIP and CBP ChIPseq reads over 1kb intersecting bound RNA regions at increasing peak-to-peak distances. Plot shows correlation for CBP bound RNAs (purple), Gal4-DBD (orange) and nGFP (green) background control bound RNAs. Grey dashed line highlights a 3.5kb peak-peak distance.
A-C) Comparison of read density (RPM) over enriched RNA regions <3.5kb from nearest CBP peak and >3.5kb from nearest CBP peak; A) CBP PAR-CLIP; B) CBP ChIPseq; C) GROseq. P-values from Mann-Whitney U-test.
D) Histogram showing distance from RNA bound CBP peaks to nearest TSS. Peaks were divided into Promoter (TSS-1kb), Exon and Distal (intronic/intergenic) sub groups.
E) Comparison of CBP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on PAR-CLIP enrichment. Boxplots show enrichment for CBP ChIPseq reads at each decile (10th decile = most enriched for total PAR-CLIP reads).
F) Comparison of PAR-CLIP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on CBP enrichment. Boxplots show enrichment for PAR-CLIP reads at the top five deciles for CBP signal (10th decile = most enriched for total CBP ChIPseq reads).
G) Comparison of PAR-CLIP signal at RNA-bound CBP peaks (<3.5kb) in Promoter, Exon and Distal groups. CBP peaks were sorted into deciles based on GROseq enrichment. Boxplots show enrichment for PAR-CLIP reads at the top five deciles for GROseq signal (10th decile = most enriched for total GROseq reads).
H-M) Comparison of RNA-CBP peaks in Promoter, Exon and Distal subgroups. Profiles show 50bp bins over 5kb windows centered on the CBP peak and were normalized by number of peaks in each genome region subgroup; Promoter (green), Exon (orange), Distal (purple). Grey boxes show the 1kb window used for quantification in box-plots; H) Read density profiles of CBP reads; I) Quantification of CBP read density; J) Read density profiles of GROseq reads; K) Quantification of GROseq read density; L) Read density profiles of PAR-CLIP reads; M) Quantification of PAR-CLIP read density. All reads were normalized per million mapped reads (RPM). P-values from Mann-Whitney U-test.
N) Boxplot shows H3K27ac reads over RNA bound CBP peaks (green) or non-RNA bound CBP peaks (orange). P-values from Mann-Whitney U-test. non-RNA bound CBP peaks were randomly downsampled to match CBP-eRNA peaks (976).
O) GROseq read density profiles (RPM) comparing RNA bound CBP peaks (green) or non-RNA bound CBP peaks (orange). non-RNA bound CBP peaks were randomly downsampled to match CBP-eRNA peaks (976).
P-R) Venn diagrams showing overlap between: P) RNA bound CBP peaks and H3K4me1; Q) RNA bound CBP peaks and H3K27ac; R) All CBP peaks with H3K4me1 peaks and H3K27ac peaks.
S) GO analysis (DAVID, (Huang et al., 2009)) of nearest genes to PAR-CLIP CBP peaks. (FDR <1, sorted by FDR).
T) Comparison of CBP-eRNA associated genes with genes not associated with CBP-eRNA. Control population was downsized to match the 1146 genes proximal to CBP-eRNA (Gene expression microarray data from GSE54452, (Kasper et al., 2014)). P-values from Mann-Whitney U-test.
U) RT-qPCR data showing fold change in CBP mRNA following infection with control adenoviral GFP (Adv-GFP, green) or knockdown with Adenoviral Cre (Adv-Cre, purple). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test.
A) Schematic of filter binding HAT assay.
B) In vitro pull-down of RNA sequences using CBP-HATwt. lncRNA HOTAIR1–300, exonic RNA (ID-1 (exon1) and lncRNA Meg3. All RNAs were resolved using 6% TBE Urea gels, apart from Meg3 (resolved using denaturing agarose formaldehyde gel) and stained using SYBR Gold (same experiment as Figure S4F).
C) RNA dose curve filter binding assays: RNA dependent stimulation of CBP acetyltransferase activity. 5nM CBP-HATwt was titrated with 0–40nM RNA (lncRNA HOTAIR1–300, exonic RNA (ID-1 (exon1) and lncRNA Meg3). Data shows fold change in rate of Acetyl-CoA incorporation (Vmax)/Concentration enzyme ([E], (s−1)) from 0nM RNA. Shaded regions represent mean +/− s.e.m (RNA n=3).
D) Native PAGE showing DNA binding to reconstituted nucleosomes (top panel). Coomassie staining showing individual histones (bottom panel).
E-G) Western blot HAT assay using recombinant nucleosome substrate. 1nM CBP-HATwt was titrated with 0–10nM E) eRNA-Mdm2s; F) eRNA-Ccnd1s; G) lncRNA HOTAIR1–300. The reaction was resolved by SDS-PAGE and probed for H3K27ac, H4K5ac and H3/H4 by western blot. Coomassie staining (bottom panel) shows individual histones.
H) Western blot HAT assay. 1nM CBP-HATwt was titrated with 0–10nM eRNA-Mdm2 (top), eRNA-PAPPA (middle) (Melo et al., 2013) or long noncoding RNA HOTAIR (bottom) (Kaneko et al., 2013). Reaction was resolved by SDS-PAGE and probed for H3K27ac by western blot. Reactions contained 20uM purified recombinant H3.1 and 200uM Acetyl CoA.
I) Quantification of Western blot HAT assays in Figure 5C and Figure S5B. Densitometry was determined in FIJI. For quantified Western blots, n=4).
J) Western blot HAT assay: human MOF (hMOF). 1nM hMOF HAT domain was titrated with 0–10nM eRNA-Mdm2. Reaction was resolved by SDS-PAGE and probed for H3K27ac by western blot. (hMOF was a kind gift of Ronen Marmorstein). Reactions contained 20uM purified recombinant H4 and 200uM Acetyl CoA.
K) Kinetic parameters derived from steady state HAT assays. All analysis was carried out using nls regression in R.
L) RNA integrity was checked before and after each HAT assay (Example control gel of eRNA-Mdm2 from steady state HAT assays). Control reactions contained 200uM unlabeled Acetyl-CoA; RNA was extracted with Trizol and resolved on a denaturing 6% TBE urea gel stained with SYBR gold.
A-C) UCSC genome browser view of GROseq signal (+ strand purple, - strand grey) and CBP-RNAs (orange bars) at enhancer and promoter regions for: A) YY1; B) Ccnd1; C) Tet2.
D) ChIP-qPCR for H3K27ac (left) and H3K18ac (right) at enhancer and promoter regions following knockdown of CBP and p300. CBP/p300 flox/flox MEFs were infected with control adenoviral GFP (Adv-GFP, Green) or adenoviral Cre (Adv-Cre, purple/blue/orange). Data shows foldchange in IP/H3. Error bars represent mean +/− s.e.m; n=3.
E) ASO mediated depletion of PAR-CLIP eRNA. RT-qPCR data showing foldchange in expression of eRNA and associated mRNA following transfection of ASO targeting eRNA-YY1antisense (purple), eRNA-Ccnd1sense (blue), eRNA-Tet2sense (orange) or GFP-control (green). Depletion efficiency was determined from expression of eRNA and mRNA at YY1 (top), Ccnd1 (middle) or Tet2 (bottom). Error bars represent mean +/− s.e.m; n=4.
F-G) ChIP-qPCR at enhancer and promoter regions following transfection of ASO targeting eRNA-YY1 (purple) or GFP-control (green) showing foldchange in IP/H3 for: F) H3K27ac and G) H3K18ac at targeted gene and non-targeted control gene Tet2 (control, bottom). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test. (Data is from the same experiment as Figure 6F-H)
H-I) ChIP-qPCR at enhancer and promoter regions following transfection of ASO targeting eRNA-Tet2 (orange) or GFP-control (green) showing foldchange in IP/H3 for: H) H3K27ac and I) H3K18ac at targeted gene Tet2 and non-targeted control gene YY1 (bottom). Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test.
J) CBP ChIP-qPCR at targeted gene Tet2 and non-targeted control gene YY1. Data shows foldchange in CBP IP/Input. Error bars represent mean +/− s.e.m; n=4; P-values from two-tailed Student’s t-test: *P< 0.05; **P< 0.01; ***P< 0.001.