Abstract
Human papillomavirus (HPV) is the most common sexually transmitted viral infection. Infection with certain types of HPV pose a major public health risk as these types are associated with multiple human cancers, including cervical cancer, other anogenital malignancies and an increasing number of head and neck cancers. The HPV life cycle is closely tied to host cell differentiation with late viral events such as structural gene expression and viral genome amplification taking place in the upper layers of the stratified epithelium. The DNA damage response (DDR) is an elaborate signaling network of proteins that regulate the fidelity of replication by detecting, signaling and repairing DNA lesions. ATM and ATR are two kinases that are major regulators of DNA damage detection and repair. A multitude of studies indicate that activation of the ATM (Ataxia telangiectasia mutated) and ATR (Ataxia telangiectasia and Rad3-related) pathways are critical for HPV to productively replicate. This review outlines how HPV interfaces with the ATM- and ATR-dependent DNA damage responses throughout the viral life cycle to create an environment supportive of viral replication and how activation of these pathways could impact genomic stability.
Keywords: Papillomavirus, DNA damage, viral replication, life cycle
Introduction
HPVs are small, double-stranded, non-enveloped DNA viruses that infect stratified epithelium at various anatomical sites, including the skin, anogenital tract and oral cavity (1). Of the over 100 hundred types of HPV that have been identified, about 40 of these infect the genital mucosa and are categorized into high-risk and low-risk based on their association with cancer (2, 3). Low-risk types (e.g. HPV 6, HPV11) cause benign genital warts, and rarely progress to malignancy. In contrast, about 15 types are considered high-risk and are the causative agents of 99% of cervical cancers (4). In addition, high-risk HPV types, particularly HPV16, are associated with 25% of head and neck cancers (5).
Many viruses have been shown to manipulate DNA repair pathways in order to efficiently replicate (6). While some viruses have evolved mechanisms to avoid or eliminate DNA repair machinery (e.g. Adenovirus), other viruses (e.g. SV40) activate and utilize the DDR to replicate their genomes (7). Numerous studies support a role for the DDR in facilitating HPV replication, with activation of the ATM and ATR pathways being required for productive viral replication (8–12). Activation of the ATR pathway may also contribute to the establishment of HPV genomes (13), as well as to the stable maintenance of viral genomes in undifferentiated cells (8, 12, 14). Three licensed HPV vaccines offer great promise in reducing the number of deaths due to cervical cancer, however these vaccines are not therapeutic. Understanding how HPV activates and utilizes DNA repair pathways to promote viral DNA synthesis may identify potential therapeutic targets to limit viral replication and block disease progression.
Viral Genome Organization
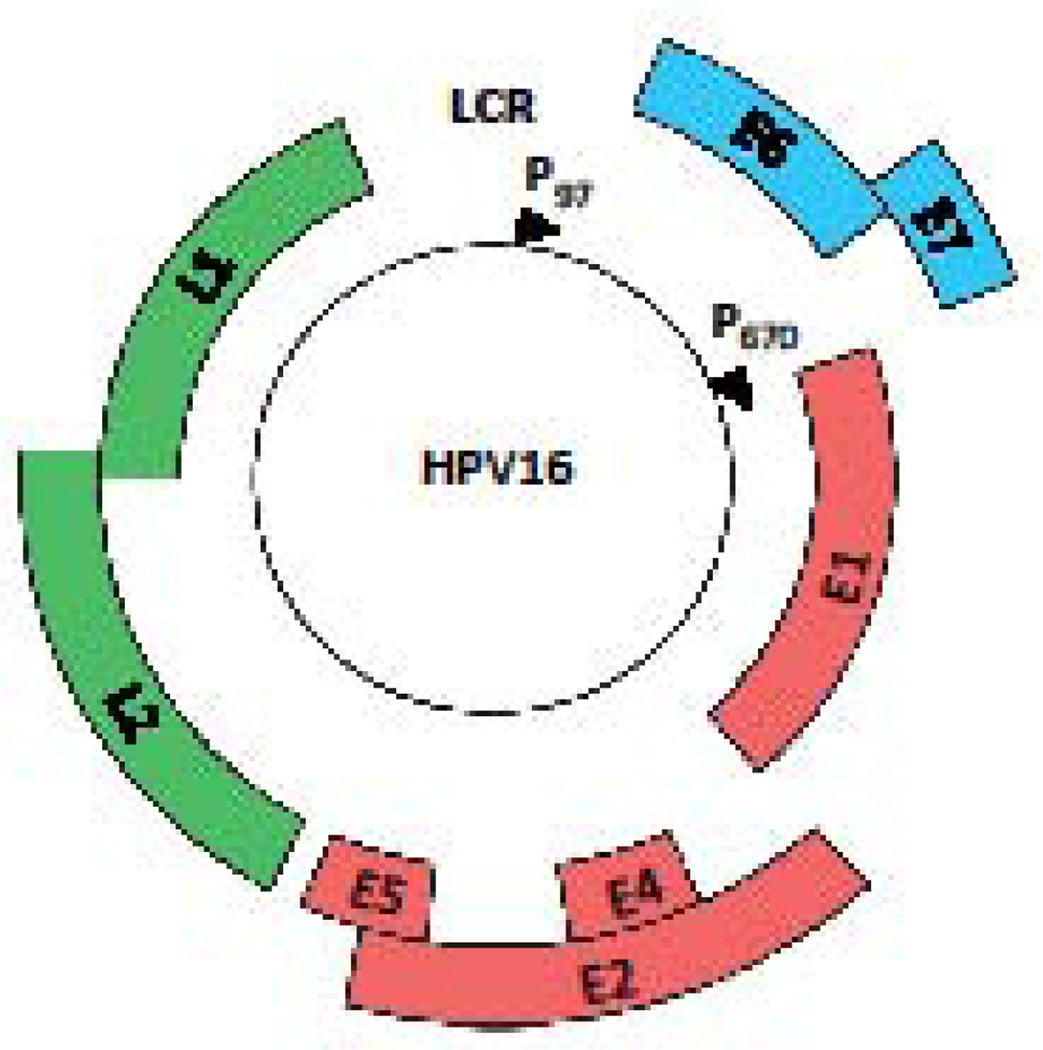
The HPV genome is maintained as a circular extrachromosamal element (episome) of approximately 8kb in length (1). HPV DNA is associated with histones in the virion as well as infected cells, forming a nucleosome pattern similar to that of cellular DNA (15, 16). The viral genome typically contains six to eight open reading frames (ORF) that are designated as early (E) or late (L) (Figure 1). The ORFs are transcribed as polycistronic messages from one strand of the DNA that are alternatively spliced to yield individual gene products (17). The early proteins are non-structural and are involved in a variety of functions ranging from viral genome replication to cell cycle control and immune evasion (18). E1 is an ATP-dependent helicase that also recruits host DNA replication factors to the viral origin of replication (19). E2 cooperates with E1 to facilitate the initiation of viral genome replication, and also regulates viral gene expression from the early promoter, as well as plays a role in viral genome partitioning (20). E6 and E7 are the oncoproteins for the high-risk HPV types and perform a wide variety of functions ranging from inhibiting apoptosis to cell cycle manipulation to immune evasion, all of which contribute to cancer development (2, 21, 22). E4 and E5 play a role in virion release and immune evasion, respectively, and are also required for productive viral replication (23–26). L1 and L2 are structural proteins that form the viral capsid. A regulatory region, designated the LCR (long control region) or URR (upstream regulatory region), is located at the end of L1 and the start of the early region and contains the viral origin of replication, binding sites for E1 and E2, as well as the early promoter and transcription factor binding sites (18).
Figure 1. Genomic organization of high-risk HPV16.
The open reading frames (ORF) are designated by the colored blocks. The Long Control Region (LCR) contains sequences for transcriptional regulation, as well as the origin of replication. The early promoter (p97 for HPV16) is located upstream of the E6 ORF and the late promoter (p670 for HPV16) is located in the E7 ORF.
The HPV Life Cycle
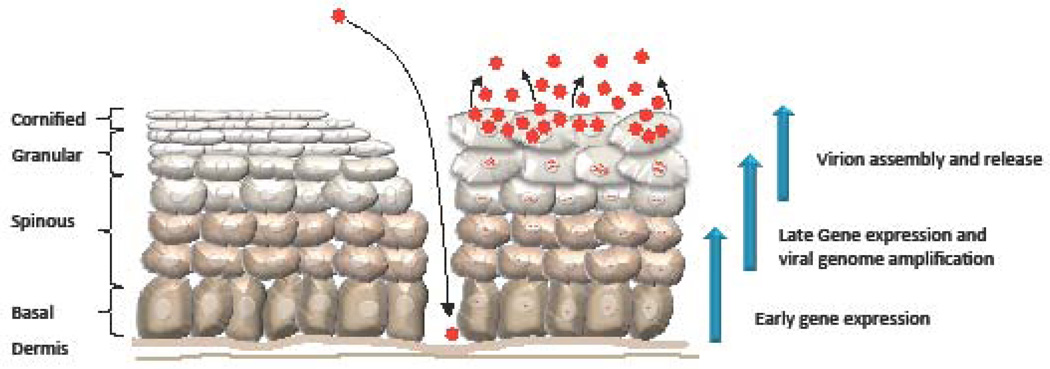
The HPV life cycle is tightly linked to the differentiation of the host cell keratinocyte (Figure 2) (18). Due to the small coding capacity of the virus, HPV is reliant on cellular factors for viral replication. The viral life cycle encompasses three phases of replication; establishment, maintenance and productive, all of which are influenced by the DNA damage response, and these three phases are described briefly below.
Figure 2. The HPV life cycle.
HPV infects basal keratinocytes of the stratified epithelium through a microwound. HPV-infected epithelium is diagrammed on the right, while uninfected epithelium is diagrammed on the left. Upon infection, HPV undergoes establishment replication to 50–100 episomal genome copies per cell and early viral genes are expressed. Viral genomes are stably maintained in the basal layer of the epithelium by replicating in Sphase with cellular DNA. Upon cell division, one daughter cell migrates away from the basal layer and begins to undergo differentiation. Differentiation triggers the productive phase of the viral life cycle, which results in late gene expression and viral genome amplification. Ongoing expression of E6 and E7 deregulates cell cycle control, pushing a subset of post-mitotic differentiating cells back into S-phase, providing cellular DNA synthesis machinery for productive viral replication. Expression of L1 and L2 in the uppermost layers of the epithelium results in the encapsidation of viral genomes and the release of infectious virions as the epithelium is shed.
Establishment Phase
HPVs infect the actively dividing cells of the basal layer of the stratified epithelium to establish infection (27). Virions are thought to gain access to these cells through a microwound (28). Upon host cell entry, viral genomes enter the nucleus, where E1, E2, E6 and E7 are expressed from the early promoter (p97 for HPV16, 31; p105 for HPV18). E2 binds and recruits the E1 helicase to the viral origin of replication (29–31). E1 also binds cellular factors required for viral replication, including polymerase alpha, topoisomerase 1 and Replication Protein A (RPA) (19). Both E1 and E2 are required for the rapid and transient initial amplification of viral genomes to establish a low viral copy number (50–100 copies per cell).
Maintenance phase
Following establishment, the viral genome is maintained is at an approximately stable number in the proliferating basal cells of the epithelium. While E2 is required for stable maintenance of viral episomes, E1 has been shown to be dispensable in some cases (20, 32, 33). Maintenance replication of HPV genomes can occur in an ordered once per S-phase manner, or randomly, whereby some viral genomes may replicate several times, some once and some not at all (34). The mode of replication utilized depends on the type of HPV, the cell line used, as well as the levels of E1, with high levels of E1 driving random choice replication (34). E2 also contributes to episomal maintenance by ensuring equal partitioning of HPV genomes to daughter cells upon cell division (20). E2 accomplishes this by tethering viral genomes to host mitotic chromosomes through chromatin adapter proteins (35). E6 and E7 also contribute to the stable maintenance of viral genomes by providing an environment conducive to viral replication (36). This is accomplished, at least in part, through E6’s ability to bind and degrade p53 (22, 37, 38), which inhibits cell cycle checkpoints and prevents host cell apoptosis. E7 deregulates cell cycle control through its ability to bind and promote the degradation of the tumor suppressor Rb, as well as the related pocket proteins p130 and p107, resulting in the constitutive activation of E2F transcription factors (21, 39, 40). E6 and E7 both contribute to inactivation of innate immune response pathways to block the antiviral response and promote replication (41).
Productive phase
When an infected basal cell divides, one of the daughter cells remains in the actively dividing basal layer, while the other moves upward and begins the process of terminal differentiation. Differentiation activates the productive amplification of viral genomes to hundreds to thousands of copies per cell (2, 42). Differentiation also results in activation of the late promoter, located in the E7 ORF (43), which drives high levels of E1 and E2 to facilitate productive replication (44, 45), as well as E4 and E5 (45), which are also required for efficient viral DNA synthesis (24, 25). How E4 functions in productive replication is not well defined, but may contribute through affects on cell cycle progression (23). E5 helps maintain a proliferative environment upon differentiation (26, 46, 47). The capsid proteins L1 and L2 are also expressed from the late promoter, allowing for encapsidation of newly replicated genomes in the uppermost layers of the epithelium (18).
Normally, epithelial cells committed to differentiation exit the cell cycle, however, HPV must keep differentiating cells active in the cell cycle to provide cellular factors for viral replication. E7’s ability to target Rb for degradation is critical to push post-mitotic cells back into the cell cycle, providing an environment conducive to viral replication in differentiating cells (48–50). As a result, cells supportive of productive replication express markers of differentiation, but also of cell cycle re-entry. Traditionally, E7-induced cell cycle re-entry has been thought to provide an S-phase environment that allows for amplification of viral genomes. However, recent studies indicate that productive replication occurs post-cellular DNA synthesis, as cells transition from S- to G2-phase (51), likely allowing HPV access to cellular factors without competing with cellular DNA synthesis. The mode of replication HPV uses to achieve rapid amplification of viral DNA upon differentiation is not clear. However, there is some evidence to suggest that there is a switch from the bi-directional (theta) replication used to stably maintain viral genomes in undifferentiated cells to a mode that utilizes rolling circle replication and/or recombination (52), which likely involves DNA repair factors, as discussed below. A seminal study from the Laimins lab demonstrated that highrisk HPV31 requires activation of DDR pathways for productive replication (9). Several studies since this initial observation offers further support that HPV productively replicates using DNA repair pathways, specifically through activation of the ATM and ATR DNA damage kinases (8, 10, 11, 53). Below, we first describe the ATM and ATR pathways then discuss how HPV has been shown to influence these DNA damage repair pathways throughout the viral life cycle.
DNA Damage Repair Pathways
Reliable maintenance of genomic integrity is an essential function of every cell. In order to ensure faithful replication and maintenance of chromosomes, several mechanisms exist to detect and repair damaged DNA. Depending on the type of damage that occurs, the cell has a variety of repair pathways at its disposal. As a group, these pathways are collectively known as the DNA damage response (DDR). The three major regulators of the DDR are the ATM, ATR and DNA-PK (DNA-dependent protein kinase) kinases, which belong to the PI 3-Kinase (PIK) family of kinases (54). Double strand DNA breaks (DSBs) can be repaired by either high fidelity, homologous recombination (HR) that requires ATM activity, or by the more error prone non-homologous end joining (NHEJ) response, which is regulated by DNA-PK (54). Conversely, ATR is required for repair of single-strand DNA that can result from stalled replication forks or processing of DSBs. (55, 56). Numerous studies have demonstrated that HPV exploits both the ATM and ATR DDR pathways in order to successfully replicate its genome (8, 12, 57). In contrast, little is known regarding the importance of HPV’s interaction with the DNA-PK pathway for the viral life cycle. Through activation of the ATM and ATR pathways, HPV is able to recruit DNA repair factors necessary for viral genome replication, as well as provide cellular factors for viral replication, which appears to be especially important upon differentiation.
ATM DNA damage repair pathway
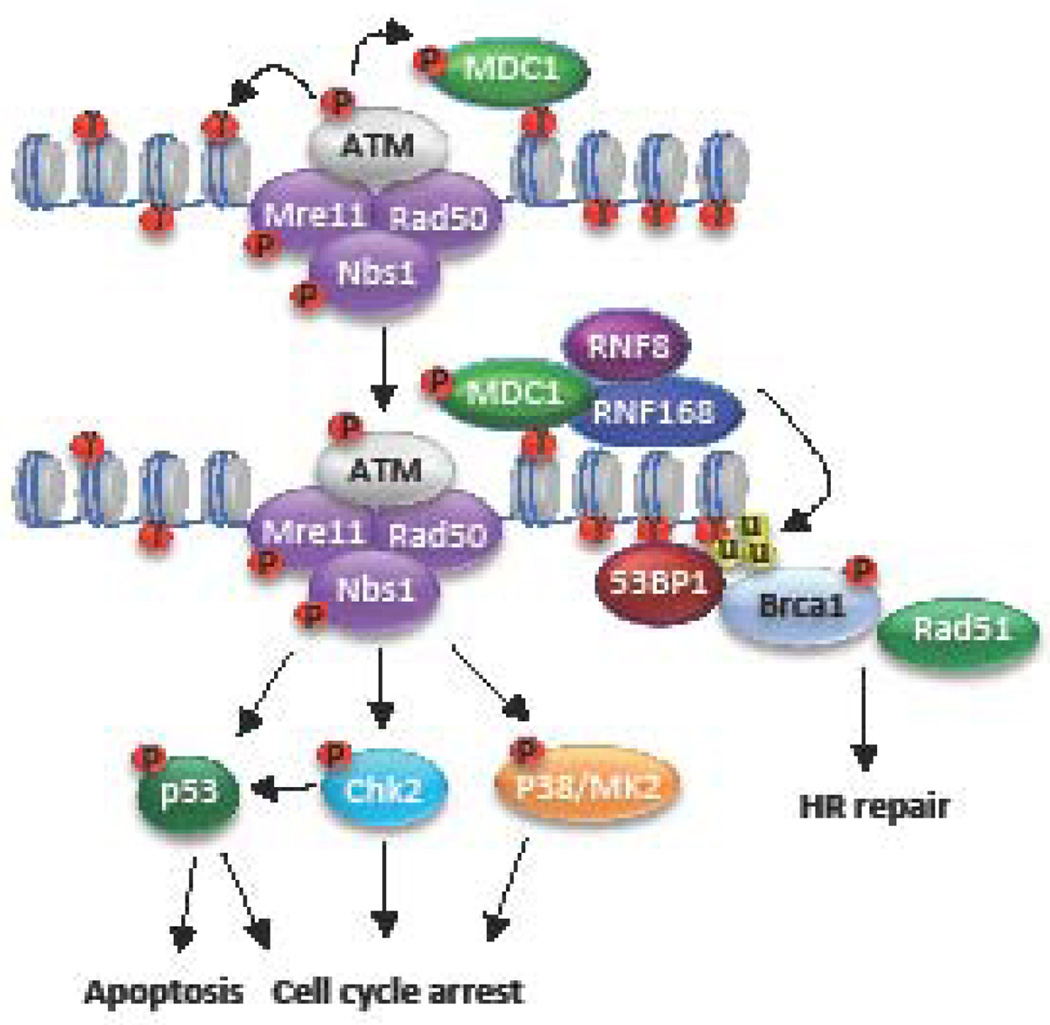
The ATM DDR pathway is typically activated in response to DNA DSBs and primarily repairs DNA through HR (Figure 3). In response to ionizing radiation, DSBs are recognized by the MRN complex, which is composed of Mre11, Rad50, and NBS1 (54). Mre11 has endonuclease and exonuclease activities important for preparing broken DNA ends for HR repair (58, 59). Rad50 is a member of the structural maintenance of chromosomes (SMC) family of proteins and interacts with the broken ends of DNA in the DSB (54). NBS1 directly binds and localizes ATM to sites of DNA damage (60, 61). ATM is activated by its recruitment to DSBs by NBS1, as well as acetylation by the TIP60 acetyltransferase (62–65). Activated ATM coordinates repair of DSBs by phosphorylating numerous downstream targets, including proteins involved in HR repair, such as NBS1, BRCA1, and the endonuclease CtIP (66). Mre11, along with CtIP and BRCA1, mediate resection of the broken ends, resulting in ssDNA that is rapidly coated by the ssDNA binding protein complex RPA. RPA is then replaced by the Rad51 recombinase, which is recruited by the BRCA2/PALB2 complex (54, 67). Rad51 forms nucleofilaments on ssDNA that invade into a homologous template, most often the sister chromatid, resulting in repair through recombination. HR is restricted to the S and G2 phases due to the availability of the sister chromatid, as well as cell cycle-dependent regulation of DNA end resection, which generates the 3’ single-stranded DNA necessary for homology search (68). Extensive formation of ssDNA prevents the binding of the KU70/KU80 heterodimer, which recruits DNAPK for NHEJ, resulting in the inhibition of NHEJ and the commitment of DNA repair towards HR (68).
Figure 3. Schematic of the ATM DNA Damage Pathway.
ATM is activated in response to DSBs and repairs damaged DNA through homologous recombination (HR). DSBs breaks are sensed by the MRN (Mre11, Rad50, and Nbs1) complex, which recruits and activates ATM through direct interaction with NBS1. Activated ATM in turn phosphorylates numerous downstream targets (p), including H2AX (γ), in turn providing a docking site for the binding of MDC1. MDC1 recruits the ubiquitin ligases RNF8/RNF168, which ubiquitylate (u) H2A/H2AX and promotes the recruitment of other DNA repair factors, including 53BP1 and BRCA1. ATM signaling to Chk2, p53 and the p38/MK2 kinase complex facilitates cell cycle arrest and DNA repair, or apoptosis of the damage is too extensive.
Other important targets of ATM include the histone variant H2AX, as well as Chk2, and p53 (69–71). In response to DNA damage, ATM rapidly phosphorylates H2AX (referred to as γH2AX), which nucleates the assembly of DNA repair factors at sites of DNA damage in a highly ordered fashion (69). γH2AX recruits the scaffolding protein MDC1, which is also a substrate of ATM. pMDC1 recruits the ubiquitin ligases RNF8 and RNF168, which ubiquitylate H2A/H2AX, facilitating the recruitment of BRCA1, as well as 53BP1 (54, 72). Phosphorylation of Chk2 results in arrest in G2 phase through phosphorylation and inactivation of the CDC25 phosphatase and its downstream effector CDK1 (73). A temporary halt in the cell cycle allows time for the DNA to be repaired before continuation of the cell cycle. Activation of p53 also leads to cell cycle arrest, and in the case of unrepaired damage, cell death through apoptosis (71).
ATR DNA damage pathway
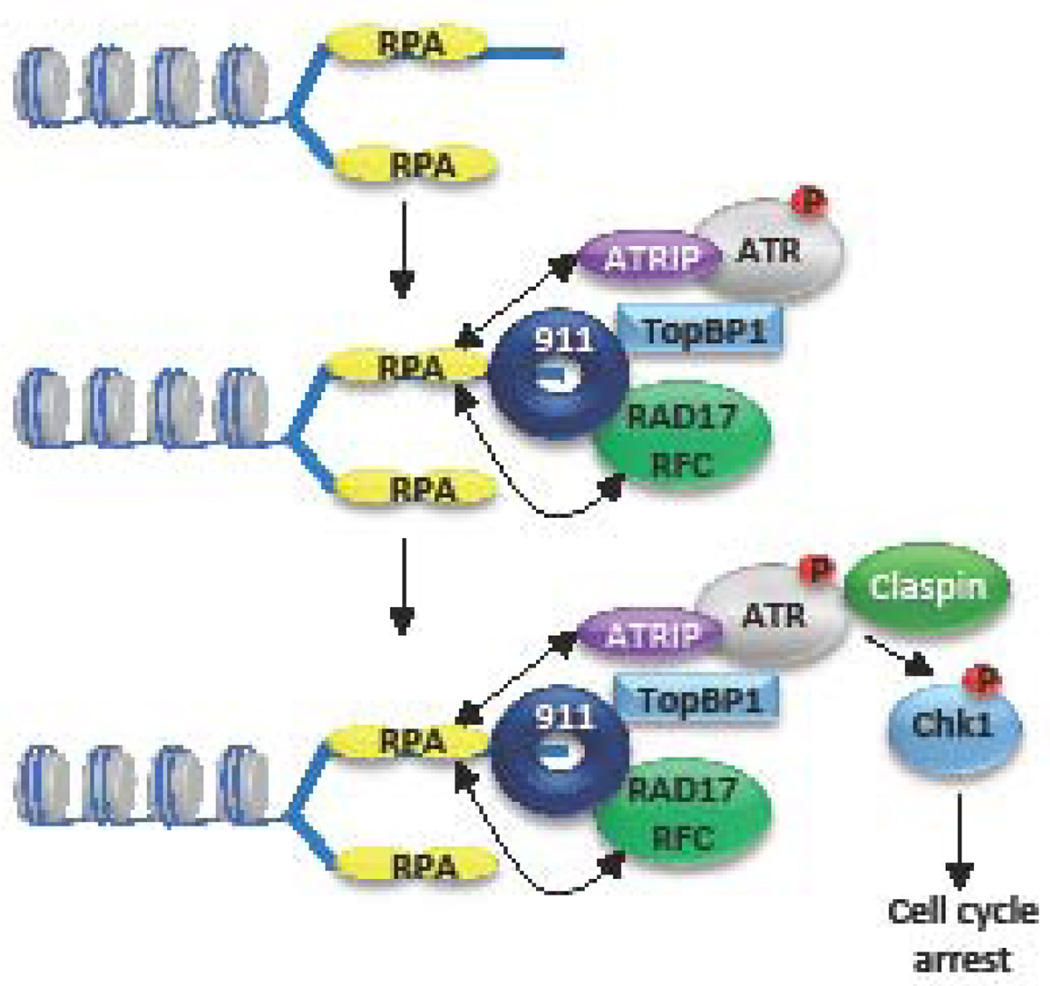
ATR is the central coordinator of the DDR relating to replication stress and replication fork stalling (55, 56). Replication stress occurs due to the slowing down or stalling of replication fork progression and/or DNA synthesis and results in large regions of RPA-coated ssDNA (Figure 4). ATRIP, the regulatory partner of ATR, directly binds RPA, allowing ATR/ATRIP to recognize ssDNA at sites of DNA damage or stalled replication forks (74). In addition, RPA-ssDNA recruits the Rad17/RFC complex, which loads the 9-1-1 complex (RAD9-HUS1-RAD1) on to DNA. Rad9 interacts with the topoisomerase IIbinding protein 1 (TopBP1), which binds and stimulates the kinase activity of ATR. Activated ATR then phosphorylates its downstream effector Chk1 through the mediator protein claspin (54, 55, 75–78). ATR/Chk1 activity is critical for cells to cope with intrinsic cellular stresses. The ATR/Chk1 pathway protects the genome against DNA damage and replication stress by regulating and coordinating multiple cellular processes including fork stabilization and restart, inhibition of replication origin firing, while preventing progression through the cell cycle while replication is completed (56). Recent studies have shown that the activity of ATR/Chk1 is particularly important for the survival of cancer cells, which typically exhibit high levels of replication stress (79–81).
Figure 4. Schematic representation of the ATR DNA damage response pathway.
ATR is activated in response to single-stranded DNA generated by replication stress (stalled replication forks) or resected DSBs. Single-stranded DNA (ssDNA) is coated by the tripartite replication protein A (RPA) complex, which recruits the ATR regulator ATRIP. ssDNA-RPA also recruits the Rad17/RFC complex to the replication fork, which loads the 9-1-1 complex (RAD9-HUS1-RAD1). Rad9 then interacts with TopBP1, which plays a critical role in activating ATR. Claspin recruitment to the complex allows ATR to phosphorylate Chk1 (p) and other downstream effectors, which mediate cell cycle arrest to allow for DNA damage repair.
HPV manipulation of the DNA damage response pathways
Utilization of the ATM pathway during the viral life cycle
The observation by the Laimins lab that high-risk HPV31 positive cells exhibit constitutive activation of an ATM-dependent DDR was the first indication that HPVs may manipulate DNA repair pathways for viral replication (9). ATM signaling is observed in undifferentiated and differentiated high-risk HPV31 positive cells, and is characterized by phosphorylation of the downstream effector Chk2, as well as NBS1 and BRCA1. Activation of the ATM-dependent DDR has since been extended to the high-risk types HPV16 and HPV18 (82). Despite markers of ATM activation throughout the viral life cycle, inhibition of ATM kinase activity has no detectable effect on episomal maintenance, but completely blocks productive replication (9). Although these results suggest that ATM activity is only required for replication in differentiating cells, it is important to note that knocking down ATM expression using siRNAs results in a 70% decrease in HPV16 copy number (14). The ATM effector Chk2 is also required for productive replication (9). ATM-driven Chk2 activity is necessary for activation of caspase-7, whose activity is required for cleavage of the E1 helicase and productive replication (9, 83). Inhibition of Chk2 activity also results in decreased phosphorylated Cdc25c levels in HPV31 positive cells (9), suggesting that HPV may utilize ATM signaling, at least in part, to provide the G2 arrested environment required for amplification of viral genomes. In addition, HPV utilizes ATM to activate the p38/MK2 pathway, which is also required for productive viral replication and acts in parallel to Chk2 and Chk1 to elicit DNA repair (53, 84).
Understanding how, as well as why HPV activates the ATM response for viral replication has been an area of intense interest. Interestingly, ATM activation seems to occur in an MRN-independent manner in HPV positive keratinocytes (10), suggesting a non-canonical mechanism of ATM DDR activation (85), which has also recently been suggested for Adenovirus infection (86). Both E1 and E7 proteins are sufficient when expressed alone to induce ATM activity. In undifferentiated cells, expression of E1 induces ATM activation in a manner likely dependent on its ability to non-specifically bind and unwind cellular DNA, leading to DSB formation (82, 87). When co-expressed with E2 however, E1 and E2 form nuclear viral replication foci that recruit numerous DNA repair factors, including phosphorylated ATM (pATM), pChk2, pNBS1, BRCA1 and Rad51, as well as pChk1 and TopBP1, two components of the ATR response (13, 82, 88). In organotypic raft cultures derived from either HPV18 positive or HPV18 E7-expressing keratinocytes, Banerjee et al demonstrated that activation of ATM, Chk2 and Chk1 occurs in differentiating cells of the stratified epithelium (89), indicating a role for E7 in eliciting the ATM activity required for productive viral replication. Similar results were observed in HPV31 organotypic raft cultures (9). Interestingly, HPV31 E7 binds to the phosphorylated form of ATM (9), but whether this interaction influences the activation of downstream effectors, or directs ATM to distinct targets is currently unclear.
Recent studies have shown that the Rb binding domain of HPV31 E7 is required to maintain high levels of total and phosphorylated ATM, Chk2 and Chk1, the MRN complex, as well as the HR factors Rad51 and BRCA1 in infected cells (90). Interestingly, E7 does not increase levels of DNA repair factors strictly through transcription, as one would expect given that targeted degradation of Rb results in the constitutive activation of E2F factors. Rather, the Rb binding domain of E7 allows for HPV to broadly increase the half-life of DNA repair factors (90). Although the mechanism is currently unclear, E7 may influence protein stability through binding to DNA repair factors and has been shown to interact with NBS1 (10), BRCA1 (91) and Rad50 (10), in addition to pATM (9). Alternatively, E7-dependent activation of ATM may lead to increased protein stability of its downstream targets. E7 has also been shown to regulate the activation of ATM through the immune regulator STAT-5, whose activity is also required for productive viral replication (92). STAT-5 may regulate ATM activation in HPV positive cells by increasing the activity of the TIP60 acetyltransferase, which is required for productive replication and must acetylate ATM to allow for its full activation (65, 93). These studies provide a link between the innate immune response and DNA damage signaling in HPV positive cells. How E7 leads to STAT-5 activation is currently unclear, but is postulated to require E7’s Rb binding domain (92).
Multiple ATM effectors localize to sites of replication in HPV positive cells, including pATM, γH2AX, pChk2, γH2AX, 53BP1, RPA, the MRN complex, and the HR proteins Rad51 and BRCA1 (10, 82, 94, 95). Gillespie at al demonstrated that γH2AX is bound to HPV genomes (94), which may be important in facilitating the recruitment of downstream DDR factors to HPV genomes (96). The finding that Rad51 and BRCA1 localize to sites of viral DNA synthesis suggests that HPV may utilize ATM activity to drive productive replication in a recombination-dependent manner. Indeed, productively replicating viral genomes exhibit branched structures consistent with strand invasion and unidirectional or rolling circle replication, which are not observed during maintenance replication (52). Importantly, Rad51 and BRCA1, as well as the MRN complex are required for productive viral replication, but not for episomal maintenance (10, 11). Disruption of the MRN complex through knockdown of NBS1 expression prevents the localization of Mre11 and Rad50, as well as Rad51 to viral genomes upon differentiation (10), suggesting that MRN-dependent resection may be required for Rad51 loading onto viral DNA. In support of this, inhibition of Mre11’s nuclease activity blocks productive viral replication (10). The binding of Rad51, as well as NBS1 to HPV DNA is dependent on the deacetylase Sirtuin 1 (SIRT1), which is upregulated in HPV positive cells and required for productive viral replication (97, 98). SIRT1, as well as TIP60 both influence the DSB repair pathway of choice to HR through modification of cellular chromatin (99, 100), and may provide a similar role in HPV positive cells through effects on viral chromatin (96). As mentioned previously, BRCA1 also influences repair pathway choice to HR by promoting end resection (68). Given the requirement of Rad51, BRCA1, the MRN complex, SIRT1 and TIP60 in productive replication, HPV may utilize ATM activity to direct the recruitment of HR factors to viral genomes rather than NHEJ factors, as recently been shown for SV40 (101). How HR plays a role in the productive replication of HPV is currently unclear. Productive replication may result in DSBs and/or stalled replication forks that require HR for repair or fork restart, respectively. Additionally, if HPV switches to a rolling circle replication mechanism upon differentiation, HR may be used to resolve concatemers into individual viral genomes suitable for packaging. As HR is considered a relatively error-free process, HPV may preferentially use this method of repair to achieve high fidelity amplification of viral genomes. Furthermore, productive replication is thought to take place in a G2/M arrested environment, when HR is most active.
Utilization of the ATR pathway during the HPV life cycle
While manipulation of the ATM DDR pathway by HPV for viral replication has become well established, less is known about HPV’s engagement of the ATR DDR. However, recent studies have shown that high-risk HPVs activate the ATR/Chk1 pathway and may require its activity for all three phases of replication (8, 9, 13, 82). Examining the impact of the DDR on establishment replication has largely been achieved through transient transfection of viral DNA, as producing large amounts of virus for de novo infection is cumbersome. Using a U20S-based model system, Reinson et al. demonstrated that transient transfection of HPV18 genomes induces the activation of ATR and the accumulation of ATR components (γH2AX, ATRIP, and TopBP1) at viral replication centers in an E1-dependent manner (13). These results suggest that initial amplification of viral genomes may result in replication stress that activates the ATR pathway. In addition, 2D gel analysis of transiently replicating viral genomes revealed structures consistent with bi-directional theta replication, as well as unidirectional replication, which may involve recombination and suggests that the initial round of viral replication may occur through two distinct mechanisms (13). Upon initial infection, ATR may engage recombination/repair factors at viral replication sites that are important for the establishment of viral genomes. Though specific DNA repair factors have not been identified that are necessary for establishment replication, these studies indicate that recombination may be important for both the initial amplification of viral genomes, as well as for productive replication upon differentiation.
Activation of the ATR/Chk1 pathway is also required for the stable maintenance of viral episomes (8, 12, 14), and recent studies have found that ATR/Chk1 activation is also required for productive viral replication (8, 12). Both E7 and E1 can independently activate the ATR/Chk1 pathway (8, 82, 87). E7 induces ATR/Chk1 activation through a STAT5-mediated increase in TopBP1 expression (8). The mechanism by which E1 induces ATR/Chk1 activity is currently unclear, though may occur through replication stress or DSBs at viral replication centers, or from non-specific binding and unwinding of cellular DNA. The ATR pathway is constitutively active in HPV positive cells (8, 9), suggesting that virally infected cells are constantly under replication stress. The expression of high-risk E6 and E7 proteins induces replication stress through unscheduled entry into the cell cycle, promoting cellular DNA synthesis in the absence of a sufficient supply of nucleotides (102, 103). Activation of the ATR/Chk1 pathway in response to replication stress induces a re-wiring of E2F signaling, resulting in increased expression of genes involved in DNA repair and replication to prevent DNA damage and allow for survival (104). Recent studies from our lab have shown that HPV31 uses the ATR/Chk1 pathway to increase expression of the small subunit of the ribonucleotide reductase complex, RRM2, in an E7- and E2F1-dependent manner (12). RRM2 is required for the de novo synthesis of dNTPs and is necessary for the maintenance of nucleotide pools for productive replication in differentiating HPV31 positive cells (12, 105). These results suggest that E7-induced cell cycle re-entry upon differentiation results in replication stress and ATR/Chk1/E2F1 activation that HPV hijacks to maintain E2F signaling, in turn providing an environment supportive of productive viral replication.
HPV, DNA Damage and Genomic Instability
Although HPV must activate DNA repair pathways for replication, HPV-immortalized cells and HPV-associated cancers exhibit genomic instability (2, 106), which is critical for carcinogenesis and suggests that DNA repair is attenuated during progression to malignancy. High-risk E6 and E7 proteins can independently induce DNA damage, resulting in numerical and chromosomal structural instability (107). Furthermore, high-risk E7 drives proliferation in the presence of DNA damage by inhibiting the DNA damage checkpoint response through proteolytic degradation of claspin (108). As shown by Bester et al, high-risk E6 and E7 expression results in replication stress and DNA damage that results in genomic instability (102). In HPV-associated cancers, the viral genome is frequently found integrated into the host cell genome, resulting in deregulated expression of E6 and E7 that can further fuel genomic instability (109, 110). Integration is random, but often occurs near common fragile sites (111), which are particularly prone to replication stress and DNA damage (112). E2, along with cellular Brd4 are thought to nucleate viral replication foci near fragile sites, providing a convenient supply of DNA repair factors for viral DNA synthesis (113). As such, replication of high-risk HPV types near cellular areas prone to DNA breakage may in turn facilitate viral integration, which subsequently drives cancer development (113). Although E6 and E7 are considered the main contributors to genomic instability (114), expression of E1 and E2 from viral episomes can lead to replication from integrated HPV origins, resulting in the amplification of viral DNA sequences and structural alterations in the flanking cellular sequences (115). DNA structures resulting from this “onion skin” type of replication results in the activation of ATM and the recruitment of both HR and NHEJ machinery (115), the latter of which can promote mutations and translocations through error-prone repair (54). These results suggest that the co-existence of viral episomes with integrated copies of HPV can result in genomic instability that contributes to carcinogenesis.
Conclusions
In this review, we have described how HPV manipulates the ATM and ATR DNA repair pathways for viral replication (Figure 5). HPV activates ATM mainly through E7 and E1 and its activity seems to be primarily required upon differentiation to drive productive replication in a recombination-dependent manner. Indeed, several other viruses also exhibit a dependence on the DDR and replication-associated recombination for replication, including SV40 (116), HSV (117–119), KSHV (120), and EBV (121, 122). The activation of the ATR/Chk1 pathway in response to replication stress in HPV positive cells may ensure the presence of cellular factors required for viral DNA synthesis through maintenance of E2F signaling. This may be particularly important upon differentiation and re-entry into the cell cycle. Numerous repair factors localize to sites of HPV replication and are likely important for viral replication during the establishment, maintenance, as well as productive phases of the viral life cycle. Understanding how these DNA repair factors contribute to HPV replication is an important area of future research. The development of more selective inhibitors of DDR components (e.g. ATM, ATR, Chk1, Chk2) has generated intense interest in targeting the DNA repair pathways for cancer treatment (123, 124). Further understanding of how HPV induces DNA damage and activation of repair pathways for viral replication, and in turn how HPV negatively impacts the repair of cellular DNA, may identify DDR targets that can be exploited therapeutically to treat HPV-associated diseases (124).
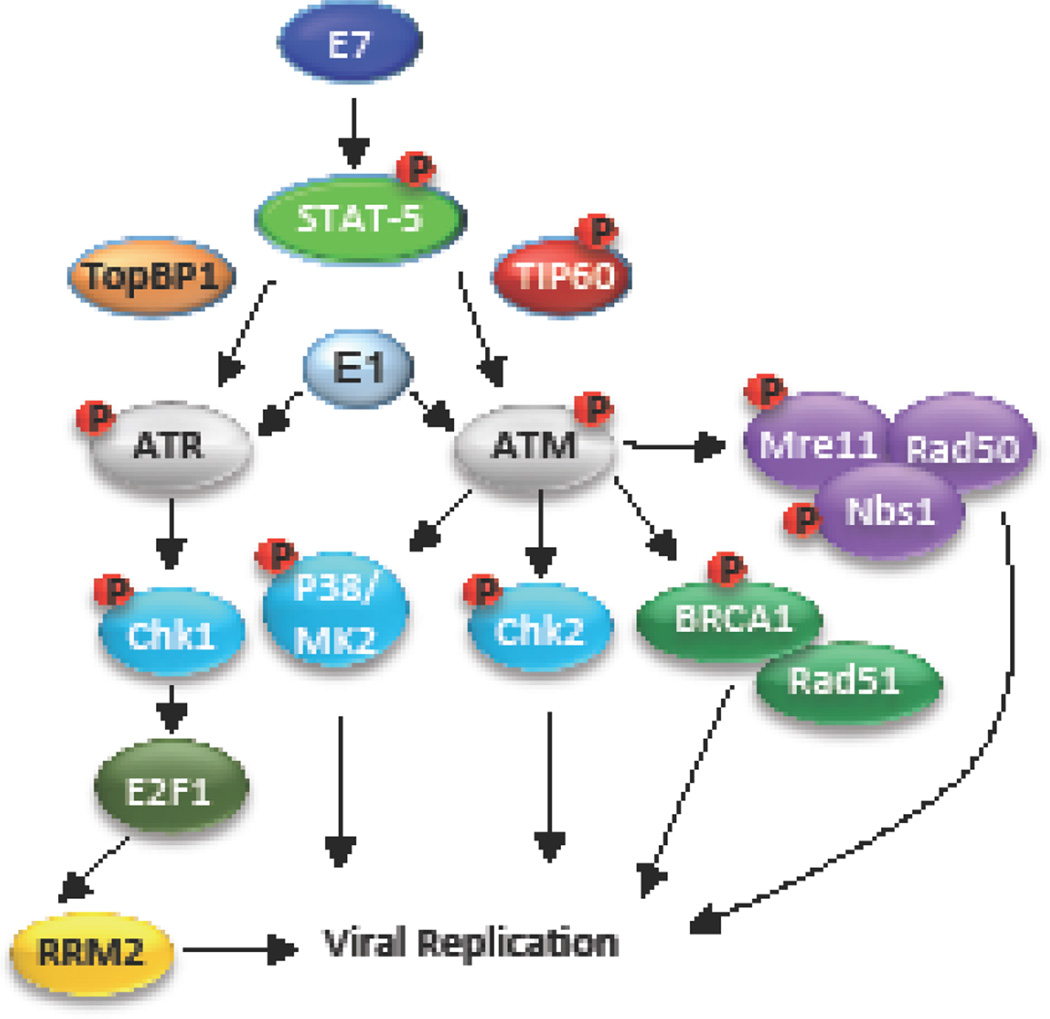
Figure 5. Model for HPV regulation of the ATM and ATR DNA repair pathways.
HPV engages both the ATM and ATR DDR pathways in order to recruit host factors and promote viral genome replication. E7 and E1 can independently induce ATM and ATR activation. Activation of ATM is crucial for DNA repair through HR and may serve to direct HR repair machinery to viral genomes (MRN, BRCA1, Rad51), which is required for productive replication. ATM-mediated Chk2 activity is required for productive replication and may contribute through activation of a G2/M arrest. Replication stress in HPV positive cells likely leads to activation of the ATR response, leading to the phosphorylation of Chk1, which results in elevated E2F1 protein levels. In turn, E2F1 upregulates RRM2 expression, allowing for the increased production of dNTPs required for productive viral replication. See text for additional detail.
Highlights.
The life cycle of HPV is dependent on the differentiation of stratified epithelium
HPV requires the ATM and ATR DNA damage response pathways for viral replication
HPV replication requires the recruitment of DNA repair factors to viral genomes
HPV uses ATM activity to productively replicate in a recombination-dependent manner
ATR activity provides cellular factors for viral replication upon differentiation
Acknowledgments
Funding: This work was supported by the National Institutes of Health (1R01CA181581; to C.A.M.) and the American Cancer Society (A14-0113; to C.A.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egawa N, Egawa K, Griffin H, Doorbar J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses. 2015;7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 3.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 7.Luftig MA. Viruses and the DNA Damage Response: Activation and Antagonism. Annu Rev Virol. 2014;1:605–625. doi: 10.1146/annurev-virology-031413-085548. [DOI] [PubMed] [Google Scholar]
- 8.Hong S, Cheng S, Iovane A, Laimins LA. STAT-5 Regulates Transcription of the Topoisomerase IIbeta-Binding Protein 1 (TopBP1) Gene To Activate the ATR Pathway and Promote Human Papillomavirus Replication. MBio. 2015;6:e02006–e02015. doi: 10.1128/mBio.02006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anacker DC, Gautam D, Gillespie KA, Chappell WH, Moody CA. Productive replication of human papillomavirus 31 requires DNA repair factor Nbs1. J Virol. 2014;88:8528–8544. doi: 10.1128/JVI.00517-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chappell WH, Gautam D, Ok ST, Johnson BA, Anacker DC, Moody CA. Homologous Recombination Repair Factors Rad51 and BRCA1 Are Necessary for Productive Replication of Human Papillomavirus 31. J Virol. 2015;90:2639–2652. doi: 10.1128/JVI.02495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anacker DC, Aloor HL, Shepard CN, Lenzi GM, Johnson BA, Kim B, Moody CA. HPV31 utilizes the ATR-Chk1 pathway to maintain elevated RRM2 levels and a replication-competent environment in differentiating Keratinocytes. Virology. 2016;499:383–396. doi: 10.1016/j.virol.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinson T, Toots M, Kadaja M, Pipitch R, Allik M, Ustav E, Ustav M. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J Virol. 2013;87:951–964. doi: 10.1128/JVI.01943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards TG, Helmus MJ, Koeller K, Bashkin JK, Fisher C. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J Virol. 2013;87:3979–3989. doi: 10.1128/JVI.03473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favre M, Breitburd F, Croissant O, Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J Virol. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stunkel W, Bernard HU. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol. 1999;73:1918–1930. doi: 10.1128/jvi.73.3.1918-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson C, Schwartz S. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat Rev Microbiol. 2013;11:239–251. doi: 10.1038/nrmicro2984. [DOI] [PubMed] [Google Scholar]
- 18.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology. 2013;445:35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride AA. The papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Wilson R, Fehrmann F, Laimins LA. Role of the E1--E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J Virol. 2005;79:6732–6740. doi: 10.1128/JVI.79.11.6732-6740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehrmann F, Klumpp DJ, Laimins LA. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J Virol. 2003;77:2819–2831. doi: 10.1128/JVI.77.5.2819-2831.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day PM, Schelhaas M. Concepts of papillomavirus entry into host cells. Curr Opin Virol. 2014;4:24–31. doi: 10.1016/j.coviro.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohr IJ, Clark R, Sun S, Androphy EJ, MacPherson P, Botchan MR. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 30.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egawa N, Nakahara T, Ohno S, Narisawa-Saito M, Yugawa T, Fujita M, Yamato K, Natori Y, Kiyono T. The E1 protein of human papillomavirus type 16 is dispensable for maintenance replication of the viral genome. J Virol. 2012;86:3276–3283. doi: 10.1128/JVI.06450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K, Lambert PF. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology. 2002;293:10–14. doi: 10.1006/viro.2001.1305. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann R, Hirt B, Bechtold V, Beard P, Raj K. Different modes of human papillomavirus DNA replication during maintenance. J Virol. 2006;80:4431–4439. doi: 10.1128/JVI.80.9.4431-4439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride AA. Replication and partitioning of papillomavirus genomes. Adv Virus Res. 2008;72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci U S A. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 39.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 40.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 41.Beglin M, Melar-New M, Laimins L. Human papillomaviruses and the interferon response. J Interferon Cytokine Res. 2009;29:629–635. doi: 10.1089/jir.2009.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maglennon GA, McIntosh P, Doorbar J. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology. 2011;414:153–163. doi: 10.1016/j.virol.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klumpp DJ, Laimins LA. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology. 1999;257:239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- 45.Hummel M, Hudson JB, Laimins LA. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kajitani N, Satsuka A, Kawate A, Sakai H. Productive Lifecycle of Human Papillomaviruses that Depends Upon Squamous Epithelial Differentiation. Frontiers in Microbiology. 2012;3 doi: 10.3389/fmicb.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regan JA, Laimins LA. Bap31 is a novel target of the human papillomavirus E5 protein. J Virol. 2008;82:10042–10051. doi: 10.1128/JVI.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 49.Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J Virol. 2000;74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garner-Hamrick PA, Fostel JM, Chien WM, Banerjee NS, Chow LT, Broker TR, Fisher C. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J Virol. 2004;78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow LT, Duffy AA, Wang HK, Broker TR. A highly efficient system to produce infectious human papillomavirus: Elucidation of natural virus-host interactions. Cell Cycle. 2009;8:1319–1323. doi: 10.4161/cc.8.9.8242. [DOI] [PubMed] [Google Scholar]
- 52.Flores ER, Lambert PF. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satsuka A, Mehta K, Laimins L. p38MAPK and MK2 pathways are important for the differentiation-dependent human papillomavirus life cycle. J Virol. 2015;89:1919–1924. doi: 10.1128/JVI.02712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinney CC, Hussmann KL, McBride AA. The Role of the DNA Damage Response throughout the Papillomavirus Life Cycle. Viruses. 2015;7:2450–2469. doi: 10.3390/v7052450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Czornak K, Chughtai S, Chrzanowska KH. Mystery of DNA repair: the role of the MRN complex and ATM kinase in DNA damage repair. J Appl Genet. 2008;49:383–396. doi: 10.1007/BF03195638. [DOI] [PubMed] [Google Scholar]
- 60.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo j. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 62.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 63.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 64.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 65.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 67.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceccaldi R, Rondinelli B, D'Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakkenist CJ, Kastan MB. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair (Amst) 2015;36:8–12. doi: 10.1016/j.dnarep.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 72.Luijsterburg MS, van Attikum H. Close encounters of the RNF8th kind: when chromatin meets DNA repair. Curr Opin Cell Biol. 2012;24:439–447. doi: 10.1016/j.ceb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 74.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 78.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, D'Artista L, Schleker T, Guerra C, Garcia E, Barbacid M, Hidalgo M, Amati B, Fernandez-Capetillo O. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakakibara N, Mitra R, McBride AA. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J Virol. 2011;85:8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc Natl Acad Sci U S A. 2007;104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burgess RC, Misteli T. Not All DDRs Are Created Equal: Non-Canonical DNA Damage Responses. Cell. 2015;162:944–947. doi: 10.1016/j.cell.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shah GA, O'Shea CC. Viral and Cellular Genomes Activate Distinct DNA Damage Responses. Cell. 2015;162:987–1002. doi: 10.1016/j.cell.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. Nuclear accumulation of the papillomavirus E1 helicase blocks Sphase progression and triggers an ATM-dependent DNA damage response. J Virol. 2011;85:8996–9012. doi: 10.1128/JVI.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gauson EJ, Donaldson MM, Dornan ES, Wang X, Bristol M, Bodily JM, Morgan IM. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication. J Virol. 2015;89:4980–4991. doi: 10.1128/JVI.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Banerjee NS, Wang HK, Broker TR, Chow LT. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J Biol Chem. 2011;286:15473–15482. doi: 10.1074/jbc.M110.197574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson BA, Aloor HL, Moody CA. The Rb binding domain of HPV31 E7 is required to maintain high levels of DNA repair factors in infected cells. Virology. 2016;500:22–34. doi: 10.1016/j.virol.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, Fan S, Meng Q, Ma Y, Katiyar P, Schlegel R, Rosen EM. BRCA1 interaction with human papillomavirus oncoproteins. J Biol Chem. 2005;280:33165–33177. doi: 10.1074/jbc.M505124200. [DOI] [PubMed] [Google Scholar]
- 92.Hong S, Laimins LA. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog. 2013;9:e1003295. doi: 10.1371/journal.ppat.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong S, Dutta A, Laimins LA. The acetyltransferase Tip60 is a critical regulator of the differentiation-dependent amplification of human papillomaviruses. J Virol. 2015;89:4668–4675. doi: 10.1128/JVI.03455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gillespie KA, Mehta KP, Laimins LA, Moody CA. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J Virol. 2012;86:9520–9526. doi: 10.1128/JVI.00247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakakibara N, Chen D, Jang MK, Kang DW, Luecke HF, Wu SY, Chiang CM, McBride AA. Brd4 is displaced from HPV replication factories as they expand and amplify viral DNA. PLoS Pathog. 2013;9:e1003777. doi: 10.1371/journal.ppat.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gautam D, Moody CA. Impact of the DNA Damage Response on Human Papillomavirus Chromatin. PLoS Pathog. 2016;12:e1005613. doi: 10.1371/journal.ppat.1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langsfeld ES, Bodily JM, Laimins LA. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog. 2015;11:e1005181. doi: 10.1371/journal.ppat.1005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Allison SJ, Jiang M, Milner J. Oncogenic viral protein HPV E7 up-regulates the SIRT1 longevity protein in human cervical cancer cells. Aging (Albany NY) 2009;1:316–327. doi: 10.18632/aging.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sowd GA, Mody D, Eggold J, Cortez D, Friedman KL, Fanning E. SV40 utilizes ATM kinase activity to prevent non-homologous end joining of broken viral DNA replication products. PLoS Pathog. 2014;10:e1004536. doi: 10.1371/journal.ppat.1004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spardy N, Duensing A, Hoskins EE, Wells SI, Duensing S. HPV-16 E7 reveals a link between DNA replication stress, FANCD2 and alternative lengthening of telomeres (ALT)-associated PML bodies (APBs) Cancer Res. 2008;68:9954–9963. doi: 10.1158/0008-5472.CAN-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bertoli C, Klier S, McGowan C, Wittenberg C, de Bruin RA. Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell-cycle transcription. Curr Biol. 2013;23:1629–1637. doi: 10.1016/j.cub.2013.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 106.Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer. 2004;109:157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- 107.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- 108.Spardy N, Covella K, Cha E, Hoskins EE, Wells SI, Duensing A, Duensing S. Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin. Cancer Res. 2009;69:7022–7029. doi: 10.1158/0008-5472.CAN-09-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu T, Ferber MJ, Cheung TH, Chung TK, Wong YF, Smith DI. The role of viral integration in the development of cervical cancer. Cancer Genet Cytogenet. 2005;158:27–34. doi: 10.1016/j.cancergencyto.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 110.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- 112.Sarni D, Kerem B. The complex nature of fragile site plasticity and its importance in cancer. Curr Opin Cell Biol. 2016;40:131–136. doi: 10.1016/j.ceb.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 113.Jang MK, Shen K, McBride AA. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014;10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009;5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boichuk S, Hu L, Hein J, Gjoerup OV. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J Virol. 2010;84:8007–8020. doi: 10.1128/JVI.00334-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schumacher AJ, Mohni KN, Kan Y, Hendrickson EA, Stark JM, Weller SK. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012;8:e1002862. doi: 10.1371/journal.ppat.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78:4783–4796. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 120.Dheekollu J, Chen HS, Kaye KM, Lieberman PM. Timeless-dependent DNA replication-coupled recombination promotes Kaposi's Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability. J Virol. 2013;87:3699–3709. doi: 10.1128/JVI.02211-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J Virol. 2009;83:6641–6651. doi: 10.1128/JVI.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dheekollu J, Deng Z, Wiedmer A, Weitzman MD, Lieberman PM. A role for MRE11, NBS1, and recombination junctions in replication and stable maintenance of EBV episomes. PLoS One. 2007;2:e1257. doi: 10.1371/journal.pone.0001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 124.Wieringa HW, van der Zee AG, de Vries EG, van Vugt MA. Breaking the DNA damage response to improve cervical cancer treatment. Cancer Treat Rev. 2016;42:30–40. doi: 10.1016/j.ctrv.2015.11.008. [DOI] [PubMed] [Google Scholar]