Abstract
The life cycle of human papillomaviruses (HPV) is tightly regulated by the differentiation state of mucosal and cutaneous keratinocytes. To counteract viral infection, constitutively expressed cellular factors, which are defined herein as restriction factors, directly mitigate viral gene expression and replication. In turn, some HPV gene products target these restriction factors and abrogate their anti-viral effects to establish efficient gene expression and replication programs. Ironically, in certain circumstances, this delicate counterbalance between viral gene products and restriction factors facilitates persistent infection by HPVs. This review serves to recapitulate the current knowledge of nuclear restriction factors that directly affect the HPV infectious cycle.
Keywords: HPV, Sp100, IFI16, FIT1, Restriction factor, Intrinsic immunity
Graphical Abstract
1. Introduction
The human immune system consists of both adaptive and non-adaptive responses that function to restrict and clear viral pathogens. Adaptive immune responses are tailored specifically to individual pathogens, while non-adaptive immunity (comprised of “intrinsic” and “innate” responses) provides an initial barrier to invading pathogens (Iwasaki and Medzhitov, 2015). All three types of immunity (intrinsic, innate, and adaptive) contribute to the restriction and clearance of viral pathogens in humans. The innate immune response is not specific for individual pathogens but is rapidly induced when an invading pathogen is detected. Restriction factors that mediate intrinsic immune responses to pathogens are pre-existing in cells (though they may also be induced) and function to quickly shut down viral infections (Yan and Chen, 2012).
Intrinsic anti-viral immunity is mediated through cellular restriction factors that directly, and immediately, inhibit infection at various steps in the viral life cycle. Viral infection can be restricted upon cell entry, trafficking through the endosome and trans-golgi network, uncoating, or nuclear entry. Restriction factors can also inhibit transcription and translation of viral genes, replication of the viral genome, and assembly/release of viral particles). As described below, the HPV infectious cycle has three main phases: viral entry and establishment; persistent infection in undifferentiated cells; and, productive infection in differentiated cells. Host factors could restrict any of these stages of infection to prevent, or reduce, production of progeny virions. However, one characteristic of intrinsic restriction factors, is that viruses have almost always evolved strategies to evade or counteract the anti-viral function (Yan and Chen, 2012).
Other chapters in this Special Issue will focus on related topics of innate immunity to HPV (Westrich et al., 2016), entry and trafficking of papillomaviruses (DiGiuseppe et al., 2016a), and interferon and NFκB induced pathways (Hong and Laimins, 2016). In this article, we will focus on intrinsic nuclear factors that directly restrict papillomavirus replication and transcription.
2. HPV Infectious Cycle
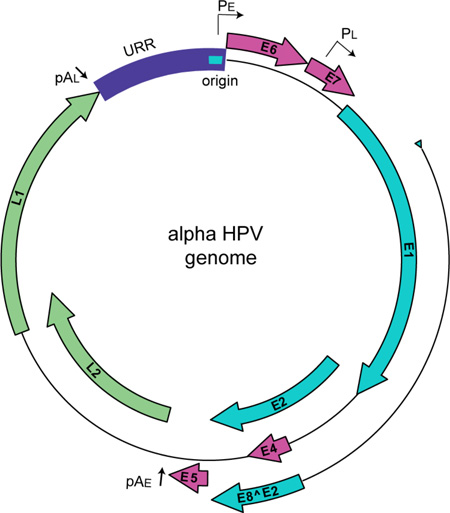
Papillomaviridae are a family of non-enveloped, DNA viruses that infect a wide spectrum of vertebrate hosts and are trophic for human mucosal and cutaneous keratinocytes. HPVs are associated with a multitude of human pathologies, including benign and malignant tumors (Cubie, 2013). Additionally, a subset of viruses within the alpha genus are deemed “high-risk” HPVs; they are the etiological agents of nearly all cases of human cervical cancer and are highly associated with other cancers such as oropharyngeal carcinoma (Gillison et al., 2000). HPVs have a small, dsDNA genome that is approximately 8,000 base pairs in length. All papillomavirus genomes have four core viral open reading frames (ORFs): E1, the viral helicase important for viral genome replication (Bergvall et al., 2013); E2, the helicase loader and major transcriptional regulatory protein (McBride, 2013); E8^E2, a transcriptional repressor (Dreer et al., 2016b); and the major and minor capsid proteins, L1 and L2 (Buck et al., 2013; Wang and Roden, 2013). Moreover, many PVs encode additional ORFs: E4, E5, E6, E7 and E10 (DiMaio and Petti, 2013; Doorbar, 2013; Roman and Munger, 2013; Van Doorslaer and McBride, 2016; Van Doorslaer et al., 2013; Vande Pol and Klingelhutz, 2013). These accessory genes encode products that are involved with cell cycle deregulation, immune evasion and recruitment of host factors for replication. Figure 1 demonstrates a prototypical HPV genome from the alpha-PV genus, which encodes many of these gene products.
Figure 1. HPV genome.
Alpha-HPVs have a circular dsDNA genome of approximately 8,000 base pairs. Viral ORFs are shown as colored block arrows. Viral early and late promoters are shown as, PE and PL, respectively, and polyadenylation sites as pAE and pAL. The origin of replication is shown as turquoise bar in the Upstream Regulatory Region (URR).
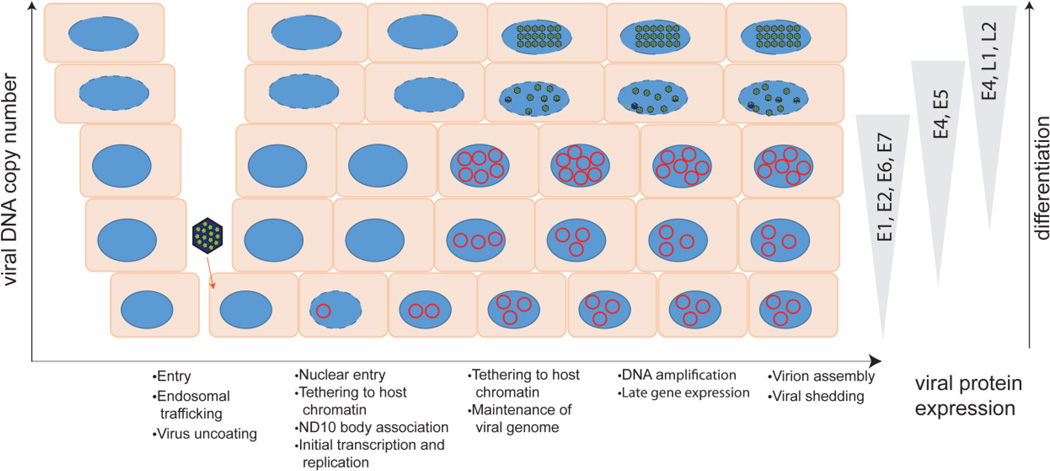
The HPV life cycle is entirely coincident with, and codependent on, the differentiation process of the host epithelium. The virus infects, and establishes a persistent infection in, the basal cells of the epithelium that it accesses through micro-abrasions. As the infected cells progress through differentiation, viral DNA is amplified, late genes are expressed, and the virus assembles in the most superficial layers of the epithelium (See Figure 2).
Figure 2. Overview of the viral infectious cycle.
The virus accesses the basal keratinocytes through a microabrasion. After entering the cell, the virus traffics through the endosome. Breakdown of the nuclear envelope during mitosis allows the virus to enter the nucleus and viral DNA is observed on mitotic chromosomes, in complex with the L2 protein. The L2 genome complex then localizes to ND10 bodies and early gene expression occurs. After a short burst of replication, the genome is maintained at a low copy number in the dividing cells in the lower levels of the epithelium. As the infected keratinocytes differentiate, the genome is amplified to high levels and late genes are expressed. The viral genome is assembled in capsids in the superficial layers of the epithelium, and viruses are shed from the surface in viral-laden squames. The different steps in the viral life cycle are listed below the diagram; host restriction factors can interfere at many stages of infection.
The HPV capsid consists of the two viral proteins, L1 and L2, which are both necessary for infection (Holmgren et al., 2005; Richards et al., 2006). L1 is the major structural capsid protein, and initially interacts with heparin sulfate proteoglycans (HSPGs) on the cell surface (Giroglou et al., 2001; Joyce et al., 1999) and extracellular matrix of the basement membrane of the epithelium (Kines et al., 2009; Selinka et al., 2007). This interaction induces a conformational shift within the capsid that exposes an epitope on the minor capsid protein, L2 (Richards et al., 2006). The virus binds a secondary (as yet uncharacterized) receptor, enters the cell by endocytosis and is trafficked through the endosomal pathways into the trans-golgi network. Many viruses protect their genomes in capsids until viral DNA is delivered to the nucleus. However, HPV is uncoated as it traffics through the endosomal compartment, and L2 is the only capsid protein that remains associated with the viral DNA as it moves to the Golgi network (Day et al., 2013). Finally, the virus arrives at the nuclear membrane, where it requires cell division to occur (resulting in nuclear membrane breakdown) before it gains access to the nucleus (Aydin et al., 2014; Pyeon et al., 2009). Recent evidence shows that the L2-genome complex is delivered to the nucleus in a membranous vesicle that traffics along microtubules and binds host chromosomes, remaining there until the end of mitosis (DiGiuseppe et al., 2016b). The viral genome is protected within these vesicles until about four hours after the completion of mitosis.
Early in infection (presumably after the completion of mitosis), the L2:DNA complex becomes localized adjacent to nuclear structures known as Nuclear Domain 10 (ND10) bodies (Day et al., 2004). The L2 protein reorganizes proteins within the ND10 structure, making it conducive as a site for the establishment of transcription and replication of the viral DNA (Becker et al., 2003; Florin et al., 2002). Early viral transcripts are synthesized shortly after infection (Ozbun, 2002), and this is initiated by cellular factors since the HPV virion does not contain any transcriptional regulatory proteins. These early transcripts encode the E1 and E2 viral replication proteins, which are required to initiate viral DNA replication. The HPV genome then undergoes a minor burst of viral DNA amplification (the first of the three stages of viral DNA replication). E1 and E2 cooperatively bind to the replication origin, to unwind the strands of the viral genome (McBride, 2008). This process promotes initiation of replication at the origin, which proceeds using cellular factors to synthesize viral DNA. This unscheduled DNA synthesis engages the cellular DNA damage response (DDR) (McKinney and McBride, 2016; Reinson et al., 2013), which likely alerts cell restriction pathways. In fact, expression of the E1 protein is sufficient to induce the ATM/ATR DNA damage response pathways (Fradet-Turcotte et al., 2011; Reinson et al., 2013; Sakakibara et al., 2011).
The E2 protein also serves as the master regulator of viral transcription (McBride, 2013). The binding of E2 to E2 binding sites (E2BS) located in the Upstream Regulatory Region (URR) of the viral genome can positively or negatively regulate the transcriptional activity of the early promoter, depending on the levels of E2 and the ratio of E2 isoforms that function as competitors of E2BS (Ammermann et al., 2008; Fertey et al., 2010). These isoforms are known by various names, but most often E8^E2 (this issue (Dreer et al., 2016b)). E8^E2 represses early viral transcription as well as viral DNA replication (Ammermann et al., 2008) by interacting with the cellular NCoR/SMRT corepressor complexes (Dreer et al., 2016a). Thus, the viral E8^E2 protein restricts the viral life cycle, which helps avoid immune detection and promotes persistent/latent infection.
During the persistent, maintenance phase of the viral life cycle, the virus goes to great lengths to limit DDR signaling by keeping levels of viral DNA at low levels, and limiting the presence of the replication proteins in the nucleus. This is carried out shuttling the E1 protein to the cytoplasm when it is not required for replication (Deng et al., 2004). There is also evidence that the E1 protein might not be necessary for this mode of replication (Egawa et al., 2012; Kim and Lambert, 2002). Cellular factors that can further inhibit viral replication (and will be discussed in detail below) are p53 and cellular miRNAs that target the E1 and E2 coding regions.
In addition to its role in initiation of DNA replication, the E2 protein is responsible for ensuring that newly replicated viral genomes are efficiently transferred from parent to daughter cells as they undergo mitosis (Bastien and McBride, 2000; Ilves et al., 1999; Skiadopoulos and McBride, 1998). This process is essential for stable genome maintenance in infected, undifferentiated basal keratinocytes and is critical to the long-lived, persistent nature of HPV infections.
The E6 and E7 oncoproteins of the high-risk alpha-PVs contribute to many different processes in the early stages of viral life cycle (Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013)) but, most notably, they disable major cell cycle checkpoints to provide the virus with an environment conducive to replication in stratified epithelial tissue. E6 and E7 also interfere with interferon and NFκB signaling pathways (Beglin et al., 2009; Chang and Laimins, 2000; Tummers and Burg, 2015). There are many similarities between tumor suppressor pathways and immune signaling pathways and it has been proposed that the primary reason that oncogenic viruses target these pathways is, in fact, for immune evasion (Moore and Chang, 2010)
Following loss of keratinocyte attachment to the basement membrane during the process of differentiation, HPVs switch from a low-copy, maintenance mode of viral replication to a productive replication phase. The mode of replication is again dependent on E1 and E2 (Egawa et al., 2012; Ozbun and Meyers, 1998), and amplifies viral DNA to high copy number. E1 and E7 independently promote a DNA damage response (DDR) that recruits factors that the virus can hijack to replicate its DNA outwith S-phase (Gautam and Moody, 2016; McKinney et al., 2015). During this phase, there is sustained synthesis of intermediate (E1, E2 and E4) and late (L1 and L2) transcripts. The E4 protein is the most abundant protein present in the upper layers of infected epithelium and interacts with keratins to reorganize the dense, rigid network of dead or dying cells in the stratum corneum, to facilitate viral transmission (Doorbar, 2013). Production of high levels of capsid proteins L1 and L2 results in encapsidation of viral DNA within the nucleus, concomitant with assembly of viral capsids.
3. Papillomaviruses and the intrinsic immune response
The epithelium itself provides the first line of defense against viral pathogens; this physical barrier is further protected by a mucus layer containing anti-microbial peptides or defensins (Buck et al., 2006). But occasionally the virus breaches this barrier through microabrasions (Roberts et al., 2007), which allows it access to the progenitor basal cells. The ensuing persistent, differentiation-dependent life cycle is also a strategy to escape from immune detection, as high level gene expression, and cytopathic effects, occur only in short-lived, terminally differentiated cells (Stanley, 2012). HPVs enter cells via endocytic pathways (Hindmarsh and Laimins, 2007; Popa et al., 2012; Smith et al., 2007); however, HPVs do not strongly activate endosomal immune sensors during entry (Tummers and Burg, 2015). In agreement, typical antiviral response factors (IFN-β, TNF-α, IFI16, IL-6 and RIG-I) are not induced during the initial stages of infection by canine papillomavirus (Luff et al., 2013). The viral capsid is not disassembled until it reaches the late endosome and the viral genome remains in complex with host histones and the minor capsid protein, L2 as it escapes the endosomal compartment (Bergant Marusic et al., 2012; Kamper et al., 2006). Furthermore, the L2-minichromosome complex remains in membrane vesicles that track to host chromosomes when the nuclear membrane breaks down during cell division (DiGiuseppe et al., 2016b), further protecting it from immune recognition.
Upon entry into the nucleus, the virus encounters additional intrinsic defenses that restrict viral replication and transcription, and this will be the focus of this review. The viral genome-L2 complex is observed localized to ND10 bodies early after infection (Day et al., 2004), presumably after the cell has progressed through mitosis (Pyeon et al., 2009). It has been shown that the viral DNA initially localizes with mitotic chromatin (Cerqueira and Schelhaas, 2015; DiGiuseppe et al., 2016b), but it is unclear how the viral DNA is trafficked from mitotic chromatin to ND10. ND10 bodies are thought to play a role in anti-viral defense and many DNA viruses localize here early in infection, but disrupt the bodies or some of their components (Maul, 1998). The PML protein is necessary for both BPV1-pseudovirus and wart-derived BPV1 virions to efficiently transcribe their packaged genomes upon infection (Day et al., 2004). HPV18 infection of primary human keratinocytes is also slightly impaired when PML is reduced by siRNA treatment (Stepp et al., 2013). Therefore, while ND10 bodies are considered to be anti-viral, they provide a favorable nuclear location for the initial stages of HPV infection. Once the genome has arrived at ND10, viral transcription is required to express the E1 and E2 proteins in order to replicate the viral DNA. This restricted gene amplification likely involves the DDR machinery, and many DDR factors localize, at least transiently, to ND10 bodies (Barr et al., 2003; Lombard and Guarente, 2000; Xue et al., 2003; Zhong et al., 1999). Therefore, this site might be advantageous for the virus to initiate the early stages of viral replication and transcription. Conversely, the ND10 bodies contain repressive factors, such as Sp100 and Daxx; however, viruses have evolved various ways to interfere with their functions.
During the maintenance phase of infection, the viral genome replicates in synchrony with the host genome and is likely tolerated by the cell better than the multiple rounds of unscheduled DNA amplification that occur at the early and late stages of infection. However, the virus is still under immunosurveillance, as immunosuppression or immunodeficiency results in a great increase in productive infection (Leiding and Holland, 2012). Viral proteins expressed at this stage (E5, E6, and E7) counteract many pathways such as the interferon response, DNA damage response and antigen presentation (DiMaio and Petti, 2013; Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013) and the virus-host interaction reaches a balance that allows the virus to be tolerated at low levels.
In the last phase of infection, viral DNA replication and transcription are induced to high levels by keratinocyte differentiation (Bedell et al., 1991). There is less immune surveillance in these terminally differentiated cells, which permits the persistently infected cells in the lower layers to coexist in a lesion with overlying virus producing cells (Stanley and Sterling, 2014). However, cellular restriction factors can still modulate viral activity in the differentiated epithelium, suggesting that the viral-host arms race continues in the latest stages of viral infection (Stanley and Sterling, 2014).
4. Host Factors that restrict transcription and Replication of Papillomaviruses
In this section we describe specific host factors that restrict HPV transcription and replication. We also describe how factors that are restriction factors for other viruses, modulate the HPV life cycle.
4.1 ND10 factor: PML
ND10 bodies (also known as PML nuclear bodies) are spherical, sub-nuclear structures that are integral to many different functions within the cell (Lallemand-Breitenbach and de The, 2010). As described above, HPV genomes traffic here during the early stages of infection. The PML protein is the critical nucleating factor, or scaffolding protein, upon which the ND10 body is organized (Ishov et al., 1999). Interaction with Ubc9, a SUMO-E2 ligase, deposits SUMO moieties on PML (Duprez et al., 1999), and promotes oligomerization of PML dimers, ultimately resulting in the assembly of the ND10 scaffold (Shen et al., 2006). Proteins interact with ND10 through one of two primary mechanisms: either direct protein-protein interactions with the PML backbone, or, more commonly, through SUMO-mediated interactions (Van Damme et al., 2010). The ND10 structure is highly dynamic, and undergoes changes in composition depending on the needs of the cell and the stage of the cell cycle. During interphase, ND10 bodies are observed as punctate nuclear foci, and many ND10 interacting proteins localize here. However, as cells progress towards mitosis, the punctate nuclear staining typical of PML is lost and the PML protein is observed in globular structures, clearly distinct from mitotic chromosomes and without SUMO (Everett et al., 1999b).
DNA viruses both interact with and manipulate these structures (Maul, 1998). Upon entering the nucleus many DNA viruses initiate their transcription and replication programs at, or adjacent to, ND10 bodies. At the same time, viral capsid, tegument or immediate-early proteins simultaneously reorganize or degrade ND10 associated factors (Scherer and Stamminger, 2016). The presence of the PML protein itself plays a positive role in initial papillomavirus infection (Day et al., 2004; Stepp et al., 2013). So, while PML does not suppress HPV infection, the ND10 bodies are an important location for the virus and contain other factors that restrict viral transcription and replication. PML is a depot for transcription factors, replication factors, and DNA damage repair proteins, and so association of the virus with PML may be a strategy for the virus to focus cellular and viral factors adjacent to replicating viral DNA.
4.2 ND10 factor: Daxx
Daxx is a component of ND10 bodies and contributes to its role in intrinsic immunity (Everett et al., 1999a). Daxx serves as a chaperone that assists in formation of nucleosomes and promotes specific deposition of H3.3 onto histone-free DNA (Lewis et al., 2010). Daxx associates with the chromatin-remodeling component, ATRX (α-thalassemia X-linked mental retardation protein) at ND10 (Xue et al., 2003), recruiting histone deacetylases, mobilizing ATRX to heterochromatin, and facilitating nucleosome deposition (Lewis et al., 2010; Schreiner and Wodrich, 2013). Daxx is also implicated in the repression of viral transcription for many DNA viruses (such as HCMV and adenovirus) and, like PML, is antagonized by viral proteins from several different virus types during the early stages of infection (Scherer and Stamminger, 2016). During HCMV infection, Daxx inhibits viral IE gene expression and is targeted for proteasomal degradation by the viral tegument protein pp71 (Saffert and Kalejta, 2006). Similar processes have been observed in HSV-1 infection, where ICP0 antagonizes the anti-viral effects of the Daxx-ATRX complex (Lukashchuk and Everett, 2010), and for adenovirus where E1B-55K protein inhibits Daxx and targets ATRX for proteasomal degradation (Schreiner et al., 2013).
However, contrary to its role in the life cycle of other DNA viruses, Daxx has a positive role in HPV transcription. In U2OS cells in which Daxx has been downregulated, both HPV early transcription and viral DNA replication are reduced (Kivipold et al., 2015). Similarly, Daxx depletion in primary human keratinocytes modestly impairs early viral gene expression and initial amplification of the HPV genome (Stepp et al., 2013). Expression of the minor capsid protein, L2, strongly recruits nuclear Daxx to ND10 bodies (Florin et al., 2002), while displacing Sp100 (Florin et al., 2002; Stepp, 2015) and L2 interacts with Daxx, even in the absence of PML (Becker et al., 2003). Thus, while L2 appears to behave like other viral factors that antagonize ND10 associated restriction factors, it seems to recruit Daxx. Further studies are required to determine the mechanistic role of L2 on Daxx protein function, and to determine whether ND10 localization of L2 is necessary for the early (upon infection), or late (during packaging) stages of infection.
4.3 ND10 factor: Sp100
There are four distinct isoforms of the Sp100 protein (Sp100A, Sp100B, Sp100C, and Sp100 HMG), some of which constitutively associate with the ND10 bodies (Negorev et al., 2006). Each isoform contains a dimerization domain and a domain that mediates binding of Sp100 to HP1 (Sternsdorf et al., 1999). Sp100B, −C, and −HMG have an additional SAND (Sp100, AIRE-1, NucP41/45, and DEAF-1) DNA binding domain that mediates direct binding to unmethylated CpG sequences (Bottomley et al., 2001; Isaac et al., 2006). Additionally, Sp100C and HMG isoforms contain either additional HMG box DNA binding motifs (Sp100HMG), or bromodomain and plant homeodomain (PHD) finger chromatin interaction motifs (Sp100C). The presence of these chromatin association domains and DNA binding motifs in the Sp100 isoforms strongly implicate these proteins in the regulation of gene expression. Sp100 has been linked with cellular transcriptional regulation due to its colocalization and interaction with nuclear regulatory factors like heterochromatin protein-1 alpha (HP-1α) (Seeler et al., 1998), and transcriptional intermediary factor (TIF) (Seeler et al., 2001), as well as coregulation of several cellular genes (Seeler et al., 1998 ; Wasylyk et al., 2002; Zong et al., 2000). Yet, surprisingly little is known about the overarching role of Sp100 in cellular gene regulation.
In contrast, well-characterized studies of adenoviruses and herpesviruses underscore the importance of Sp100 in the context of viral infection and implicates these proteins as potential effectors in the response to viral infection. Early studies indicated that Sp100, like other core ND10 proteins was a repressor of viral infection. However, more detailed analyses indicate that the shortest and most abundant isoform, Sp100A activates transcription, whereas the longer isoforms repress transcription (Berscheminski et al., 2014; Negorev et al., 2006; Newhart et al., 2013; Wilcox et al., 2005). Sp100 proteins are upregulated by interferon and viral proteins alter, displace or degrade Sp100 (Guldner et al., 1992; Lavau et al., 1995). In cells infected with adenovirus, the immediate early proteins expressed from the E1 and E4 ORFs mediate a dramatic morphological change in the ND10 bodies, expelling the repressive Sp100 isoforms −B, −C, and −HMG before forming replication factories within PML-containing tracks; however, the activating isoform Sp100A is retained and hijacked by the virus (Berscheminski et al., 2014; Doucas et al., 1996). Adenovirus infection abrogates binding of Sp100A to HP-1β, which results in decondensation of chromatin and enhanced viral gene expression (Berscheminski et al., 2014).
HSV-1 DNA also localizes adjacent to ND10 structures (Maul et al., 1996) and it is thought that viral transcription and replication initiate here. Nucleoplasmic Sp100B, −C, and −HMG isoforms repress early HSV transcription, outside PML bodies, while Sp100A accumulates at ND10 and does not significantly impact HSV-1 ICP0 or ICP4 production (Negorev et al., 2006). In order to mitigate the anti-viral functions of ND10 bodies, HSV-1 ICP0 targets Sp100 and PML for proteasomal degradation (Gu and Roizman, 2003).
Incoming HPV18 genomes (introduced by transfection, or by quasivirus infection) are similarly subjected to Sp100-mediated transcriptional repression with a concomitant inhibition of initial amplification of viral DNA (Stepp et al., 2013). These effects on initial infection also abrogate the establishment of persistent HPV18 infection, as measured by a quantitative colony forming assay. However, Sp100 has either minimal (Habiger et al., 2016), or no (Stepp et al., 2013) significant effects on viral transcription or replication in cells maintaining HPV31 or HPV18 genomes. During the late stages of HPV31 infection, PML and Sp100 localize next to replication foci and, in some instances, infiltrate the replication factories (Stepp, 2015). Additionally, Sp100 represses late viral transcription and vegetative viral DNA amplification (Stamos and McBride, unpublished; Stepp, 2015). These results suggest that Sp100 represses transcription and DNA amplification during both early and late stages of the HPV life cycle.
Similar to other viruses, Sp100 B, C, and HMG isoforms repress HPV transcription, while Sp100A has no effect (Habiger et al., 2016; Stepp, 2015). Notably, Sp100A and Sp100C isoforms are specifically downregulated at the mRNA level in a variety of cell lines harboring HPV16 and HPV31 genomes (Habiger et al., 2016). All Sp100 isoforms are induced by interferon, but E6 downregulates constitutive IFN-κ expression in keratinocytes by hypermethylation of its promoter (Habiger et al., 2016; Reiser et al., 2011; Rincon-Orozco et al., 2009). Thus, E6 may inhibit IFN-κ expression to downregulate the abundance of the Sp100 isoforms to subvert specifically innate immune functions mediated by these proteins. Therefore, the Sp100 family of proteins are important restriction factors for HPV.
4.4 p53, FANCD2 and the DNA Damage Response
The “high risk” HPV E6 and E7 proteins are considered oncogenes, and manipulate many steps in DDR and cell cycle regulatory pathways. While this promotes an environment conducive to viral DNA replication (and inadvertently promotes oncogenesis), it has been proposed that the primary reason that these pathways are targeted is to disable anti-viral immune surveillance pathways (Moore and Chang, 2010).
Oncogenic HPV infection induces DNA damage signaling and p53 induction, which must be tempered to allow persistent replication of the viral genome. The viral E7 protein promotes cell cycle progression by inactivating pRb and p21, and deregulating E2F (Roman and Munger, 2013) to provide an environment conducive to viral DNA replication. However, the constant drive to promote proliferation causes nucleotide deficiency and promotes replication stress and genomic instability (Bester et al., 2011). This results in activation of p53, which should induce growth arrest, but E6 binds and degrades p53 to allow continued replication of viral DNA (Vande Pol and Klingelhutz, 2013). This ability of E6 to inactivate p53 is essential for persistent DNA replication (Flores et al., 2000; Park and Androphy, 2002; Thomas et al., 1999) and HPV genomes mutated in E6 can only replicate in cells in which p53 has been inactivated (Brimer and Vande Pol, 2014; Lorenz et al., 2013).
Although DNA damage signaling can be detrimental to persistent HPV infection, it is beneficial for the productive phase of replication. Therefore, HPVs must inactivate some components of the DDR that can restrict the viral infectious cycle, but they also take advantage of other facets that are conducive for viral replication. The E1 and E2 proteins are induced to high levels in differentiating cells (Klumpp and Laimins, 1999) to amplify viral DNA. Both E1 and E7 activate the DNA damage response in these cells and this is required for genome amplification (Fradet-Turcotte et al., 2011; Gauson et al., 2015; Moody and Laimins, 2009; Reinson et al., 2013; Sakakibara et al., 2011). It is thought that engagement of the DDR machinery results in an influx of DNA damage and repair factors to replication foci to facilitate replication in differentiated cells (Gautam and Moody, 2016; Sakakibara et al., 2013).
Inactivation of p53 by E6 is also necessary for vegetative amplification of viral DNA. Viruses that cannot disarm p53 are also unable to amplify viral DNA in differentiated cells (Kho et al., 2013; Wang et al., 2009). Although much of the p53-mediated inhibition of HPV replication can be attributed to its role in checkpoint control, p53 can also directly inhibit viral DNA synthesis (Akgul et al., 2003; Ilves et al., 2003; Lepik et al., 1998). This inhibition can be mediated through direct interaction of p53 with the E2 protein (Brown et al., 2008; Massimi et al., 1999), which results in direct inhibition of viral DNA replication. Thus, p53 can be classified as a factor that restricts the viral life cycle.
The Fanconi anemia (FA) DNA repair pathway contains 15 genes involved in the error-free repair of intra-strand DNA cross-links and double stranded breaks. Individuals with a defect in any one of these genes are highly susceptible to HPV infection and have increased incidence of cancer development. These individuals are extremely vulnerable to squamous cell carcinomas in sites susceptible to HPV associated cancers, yet many of the resulting cancers are HPV negative (Park et al., 2016; van Zeeburg et al., 2008). E7 activates the FA pathway, and induces increased genetic instability in cells defective in the FA pathway (Spardy et al., 2007). The resulting genetic instability can render the FA-deficient cells no longer dependent on E7 (Park et al., 2016). Furthermore, in FA-deficient cells, productive HPV genome replication in differentiated cells is significantly higher, suggesting that the functional FA pathway restricts the HPV life cycle (Hoskins et al., 2012). The details of this restriction have yet to be elucidated, but there is precedent that other viruses manipulate the FA pathway for replication. For example FA-mediated suppression of NHEJ promotes productive HSV-1 infection (Karttunen et al., 2014). More research is required to determine how the FA pathway suppresses HPV productive replication.
4.5 IFIT1 (p56)
Type I interferon (IFN) treatment of HPV infected cells causes the rapid loss of extrachromosomal HPV genomes (Chang et al., 2002; Herdman et al., 2006; Lace et al., 2015; Turek et al., 1982). This is, at least in part, due to induction of the protein p56 (also known as IFIT1 or ISG56) (Saikia et al., 2010; Terenzi et al., 2008). The IFIT1 family of proteins are encoded by a group of interferon-stimulated genes (ISG) that are induced by viral infection (Fensterl and Sen, 2011). They share common structural elements called multiple tetratricopeptide motifs that mediate protein-protein interactions with effector proteins resulting in inhibition of many viral and cellular processes (Fensterl and Sen, 2011). The N-terminal region of IFIT1 binds directly to the C-terminal region of HPV18 E1 (Terenzi et al., 2008). This interaction inhibits E1’s sequence-specific binding to the viral replication origin and blocks E1’s helicase function, thus inhibiting replication of HPV DNA (Saikia et al., 2010; Terenzi et al., 2008). Additionally, IFIT1 substantially inhibits the interaction of E1 with E2 (Saikia et al., 2010). Combined, these actions by IFIT1 act to inhibit significantly replication of the HPV genome.
4.6 IFI16
Many restriction factors sense, and respond to, incoming or foreign DNA. Most of these sensors are located in the cytosol, but more recently a nuclear DNA sensing factor, IFI16, has been described (Dempsey and Bowie, 2015). IFI16 is a member of the PHYIN family of proteins, and it recognizes invading DNA (and distinguishes from host DNA) in a length-dependent, sequence-independent manner (Morrone et al., 2014). IFI16 binds to foreign DNA in a cooperative manner, and the resulting oligomeric, filamentous complexes recruit STING. This induces the expression of IFN-β in a TBK-1 dependent manner (Unterholzner et al., 2010). IFI16s requirement for a minimal sequence length of naked DNA ensures that host DNA exposed in the linker region between nucleosomes, or in transcription bubbles, are not recognized (Morrone et al., 2014).
IFI16 is a restriction factor for several other DNA viruses, including HSV-1 and HCMV (Gariano et al., 2012; Orzalli et al., 2013), and it induces anti-viral inflammasome activity against KSHV and EBV (Ansari et al., 2013; Roy et al., 2016). However, while IFI16 restricts incoming HSV-1 DNA, it does not restrict SV40 DNA packaged in nucleosomes and delivered to the nucleus by transfection (Orzalli et al., 2013).
IFI16 is constitutively expressed in keratinocytes (Gariglio et al., 2002). Downregulation of IFI16 greatly increases HPV18 genome amplification in differentiated keratinocytes (Lo Cigno et al., 2015) and overexpression of IFI16 restricts both HPV18 replication and transcription in U2OS cells. This is mediated by modification of the chromatin state of the viral genome (Lo Cigno et al., 2015). IFI16 induces an increase of repressive heterochromatic marks and a decrease in activating euchromatic marks on both the early and late promoters (Lo Cigno et al., 2015). (Lo Cigno et al., 2015). Interestingly, IFI16 is also able to reduce transcription from integrated genomes, possibly involving the Sp1 binding site in the LCR (Lo Cigno et al., 2015).(De Andrea et al., 2015)
It is not known how exactly IFI16 mediates this change in chromatin state in HPV; in its epigenetic-based restriction of HCMV, it is speculated that IFI16 binding to the viral genome induces a conformational change that promotes the recruitment of histone modification enzymes (Orzalli et al., 2013). These epigenetic modifications function to considerably reduce replication and viral transcription by making the viral genome less accessible to transcription and replication factors. However, the antiviral effect of IFI16 observed in HPV positive cells is independent of its role as a sensor of foreign DNA or activation of interferon.
4.7 APOBEC
An important host restriction factor, best studied in the context of HIV infection, is the APOBEC family of nucleic acid editing factors. APOBECs convert cytidines into uridine in RNA, but there is also evidence that certain APOBECs can induce mutations in genomic DNA (Shinohara et al., 2012). In fact, APOBEC proteins are upregulated in many cancers and the resulting APOBEC generated mutagenesis is widespread (Roberts et al., 2013). APOBEC3A and 3B are upregulated in HPV-associated cancers and this seems to be mediated by E7 functions (Warren et al., 2015a). Most HPV-related studies have focused on the ability of some members of the APOBEC family to edit HPV genomes in cancer cells, but APOBEC’s role as an intrinsic restriction factor of HPV infection remains less clear.
APOBECs edit single stranded DNA, so any action on HPV genomes must occur during viral transcription or replication. APOBEC editing has been detected in a small proportion of HPV containing lesions; genomes derived from some HPV1 infected cutaneous warts and HPV16 associated pre-malignant lesions show mutations consistent with APOBEC editing (Vartanian et al., 2008). Additionally, interferon treatment of W12 cells (cervical neoplasia derived cells containing extrachromosomal HPV16 genomes) induces endogenous APOBEC enzymes and results in mutation of viral DNA (Wang et al., 2014). Quasiviruses and pseudoviruses packaged in the presence of exogenous overexpressed hA3A show decreased early transcription (Ahasan et al., 2015; Warren et al., 2015b) and there is some evidence that this might interfere with packaging through interaction with the L1 protein (Ahasan et al., 2015). However, the precise role of APOBECs in the productive HPV infectious cycle is still not clear. HPVs are genetically very stable, and evolve at a very slow rate (Rector et al., 2007), indicating that putative APOBEC-induced mutations are very rarely, if ever, fixed. However, alpha-HPVs, which mostly infect the anogenital mucosa, are relatively depleted in TC dinucleotides, the preferred target of APOBEC. This led Warren et al. to propose that, over a very long evolutionary period, alpha-HPVs have evolved to escape APOBEC editing (Warren et al., 2015a). Clearly, further studies are required to assess whether the APOBEC proteins restrict the HPV life cycle in natural infection.
4.8 miRNA
Micro RNAs are small, non-coding regulatory RNAs that serve to bind to mRNA and silence gene expression. The opportunity to silence viral gene expression in a sequence specific manner, at the mRNA level, represents a powerful tool that host cells use as restriction factors against viral pathogens. Host miRNAs restrict many aspects of DNA virus (HBV, HCMV, and HSV-1) life cycles (Russo and Potenza, 2011).
HPV regulates the expression of many cellular miRNAs (Zheng and Wang, 2011). Of these, only a few are known to directly affect the viral infectious cycle. However, miR-145, has a pronounced, differentiation-dependent decrease in abundance in HPV31 infected cells, apparently mediated by E7 (Gunasekharan and Laimins, 2013; Gunasekharan et al., 2016). It is hypothesized that a sequence in the E1 open reading frame (ORF) is the primary target of miR-145, but there is also limited binding to the E2 ORF (Gunasekharan and Laimins, 2013). Overexpression of miR-145 results in sharply reduced genome amplification and results in diminished late viral transcription, indicating an inhibitory role in the viral life cycle (Gunasekharan and Laimins, 2013). It is speculated that HPV relies on the inhibitory effects of miR-145 early in the life cycle to keep the copy number low during low level maintenance replication in undifferentiated cells (Gunasekharan and Laimins, 2013). Other viruses, such as KSHV and HSV-1, rely on virally-encoded miRNAs to maintain a latent state (Lu et al., 2010; Umbach et al., 2008). However, HPV does not express its own miRNAs (Cai et al., 2006) and thus must rely on manipulating or overcoming the host cells’ miRNA to maintain immune evasion.
4.9 C/EBPβ
CCAAT/enhancer binding proteins (C/EBPs) play a role in the regulation of differentiation program of many different cell types. C/EBPβ is present in the nuclei of keratinocytes and is capable of binding the URRs of various HPV types (Bauknecht and Shi, 1998; Wang et al., 1996). Additionally, HPV E2 directly binds both C/EBPα and C/EBPβ (Bauknecht and Shi, 1998; Hadaschik et al., 2003). C/EBPβ inhibits both the replication and transcription of HPV (Wang et al., 1996). Further studies have revealed that C/EBPβ has three isoforms, I (LAP), II (LAP*), and III (LIP), transcribed from a single, intron-less gene with alternative start codons for each of the three isoforms (Baer and Johnson, 2000; Descombes and Schibler, 1991). LAP activates HPV late transcription by binding the HPV late promoter in a differentiation-dependent manner (Gunasekharan et al., 2012). Conversely, LIP has an opposite effect on the HPV late transcription. The mechanism for this repression is the formation of LIP-LAP dimers which bind to HPV late promoter sequences and prevent activation by the LAP transactivation domain. Upon differentiation, HPV positive cells show a significant decrease in LIP abundance, increasing the ratio of LAP-LAP homodimers and allowing high rates of transcription of late genes (Gunasekharan et al., 2012). Although not considered traditional restriction factors, the LIP C/EBPβ isoforms directly restrict the HPV life cycle.
4.10 EVERs
Most individuals with the rare genetic disease epidermodysplasia verruciformis have loss of function mutations in one of two genes, EVER1/TMC6 or EVER2/TMC8 (Ramoz et al., 2002). These individuals have extreme susceptibility to infection by beta HPVs (but not other HPVs or other pathogens) and suffer life-long persistent HPV infections of the skin (Orth, 2006). While there is evidence that the EVER deficiency might result in defects in the adaptive immune system (Lazarczyk et al., 2012), the EVER1 and EVER 2 proteins also complex with zinc transporter-1 (ZnT-1) in keratinocytes to control intracellular zinc concentration (Lazarczyk et al., 2008). When functional, this complex serves to limit intracellular zinc levels in the nucleus, resulting in downregulation of zinc-stimulated transcription factors (such as MTF-1) and AP-1 family members such as c-jun. It is proposed that in alpha-HPV infected cells, the E5 protein binds to the EVER/ZnT-1 complex and blocks this negative modulation (Lazarczyk et al., 2008), whereas beta-HPVs do not encode E5 proteins. However, the exact mechanism by which EVER serves to restrict HPV infection is unclear. Clearly, more studies are needed to further elucidate the mechanism of β-HPV restriction by EVER1/2.
5. Viral Factors that Counteract Host Intrinsic Defenses
A common feature of intrinsic restriction factors is that viruses have evolved mechanisms, or encode proteins, that enable them to avoid or antagonize the anti-viral function of restriction factors (Yan and Chen, 2012). In fact, viruses often takes advantage of host restriction factors to promote latency or persistence (Lieberman, 2016).
One of the earliest defenses that HPV encounters during infection is the ND10 bodies. Many DNA viruses localize to these structures during the initial stages of infection and encode gene products that directly affect the anti-viral activities of these structures. The HPV minor capsid protein L2 is responsible for trafficking the viral genome to the nucleus (Day et al., 2004). On arrival at ND10, however, L2 reorganizes the ND10 body by recruiting Daxx and displacing Sp100 (Becker et al., 2003; Florin et al., 2002; Stepp, 2015). This displacement results in the degradation of the Sp100 protein, an event blocked in the presence of the proteasome inhibitor, MG132 (Florin et al., 2002). This suggests that L2 not only localizes the viral genome to ND10, but also might condition the ND10 environment to be more conducive to the establishment of viral genomes.
Papillomaviruses have a very limited coding capacity; thus it is more cost effective to disrupt cellular signaling pathways than to directly interact with, and inactivate, host restriction factors. The E6 and E7 proteins manipulate many pathways such as cell cycle regulation, the DNA damage response and differentiation, and as discussed above there is significant overlap with these pathways and those of immune surveillance (Moore and Chang, 2010). However, E6 and E7 also directly counteract interferon and NFκB signaling pathways (Beglin et al., 2009; Chang and Laimins, 2000; Reiser et al., 2011; Tummers and Burg, 2015), and E2 can also downmodulate interferon and STING (Sunthamala et al., 2014).
The persistent life cycle of papillomaviruses requires escape from host restriction factors at different temporal stages of the infectious cycle, during many diverse cellular processes, and in multiple differentiation states of the host cell. It is remarkable that they not only manage to evade intrinsic immune surveillance mechanisms but also take advantage of these processes to establish long-term infection of the host.
Table 1.
HPV Restriction Factors and their viral targets
| Restriction Factor |
Viral Target | Key Reference |
|---|---|---|
| Sp100 | Viral transcription | (Habiger et al., 2016; Stepp, 2015; Stepp et al., 2013) |
| miR-145 | E1 RNA | (Gunasekharan and Laimins, 2013) |
| p56 | E1 Protein | (Saikia et al., 2010) (Terenzi et al., 2008) |
| IFI16 | Early and Late Promoters | (Lo Cigno et al., 2015) |
| C/EBPβ | URR, Late Promoter | (Gunasekharan et al., 2012) |
| p53 | E2 Protein | (Brown et al., 2008; Massimi et al., 1999) |
| EVERs | Viral transcription | (Lazarczyk et al., 2008) |
| APOBEC | unknown | (Warren et al., 2015b) |
| FANCD2 | unknown | (Hoskins et al., 2012) |
Highlights.
Papillomavirus transcription and replication can be restricted throughout the lifecycle
Papillomaviruses subvert host restriction factors to complete their life cycle
Restriction of viral infection can facilitate persistent HPV infection
Acknowledgments
We thank all members of the McBride laboratory for helpful discussions. We apologize to colleagues whose primary work we could not cite, because of space limitations. A portion of this review is derived from the Introduction chapter in the thesis of WHS (Stepp, 2015). This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahasan MM, Wakae K, Wang Z, Kitamura K, Liu G, Koura M, Imayasu M, Sakamoto N, Hanaoka K, Nakamura M, Kyo S, Kondo S, Fujiwara H, Yoshizaki T, Mori S, Kukimoto I, Muramatsu M. APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity. Biochem Biophys Res Commun. 2015;457(3):295–299. doi: 10.1016/j.bbrc.2014.12.103. [DOI] [PubMed] [Google Scholar]
- Akgul B, Karle P, Adam M, Fuchs PG, Pfister HJ. Dual role of tumor suppressor p53 in regulation of DNA replication and oncogene E6-promoter activity of epidermodysplasia verruciformis-associated human papillomavirus type 8. Virology. 2003;308(2):279–290. doi: 10.1016/s0042-6822(02)00133-2. [DOI] [PubMed] [Google Scholar]
- Ammermann I, Bruckner M, Matthes F, Iftner T, Stubenrauch F. Inhibition of transcription and DNA replication by the papillomavirus E8-E2C protein is mediated by interaction with corepressor molecules. J.Virol. 2008;82(11):5127–5136. doi: 10.1128/JVI.02647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, Chikoti L, Lu J, Everly D, Chandran B. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. Journal of virology. 2013;87(15):8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog. 2014;10(5):e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer M, Johnson PF. Generation of Truncated C/EBPβ Isoforms by in VitroProteolysis. Journal of Biological Chemistry. 2000;275(34):26582–26590. doi: 10.1074/jbc.M004268200. [DOI] [PubMed] [Google Scholar]
- Barr SM, Leung CG, Chang EE, Cimprich KA. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Current biology : CB. 2003;13(12):1047–1051. doi: 10.1016/s0960-9822(03)00376-2. [DOI] [PubMed] [Google Scholar]
- Bastien N, McBride AA. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology. 2000;270(1):124–134. doi: 10.1006/viro.2000.0265. [DOI] [PubMed] [Google Scholar]
- Bauknecht T, Shi Y. Overexpression of C/EBPβ represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein. Journal of virology. 1998;72(3):2113–2124. doi: 10.1128/jvi.72.3.2113-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Florin L, Sapp C, Sapp M. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology. 2003;314(1):161–167. doi: 10.1016/s0042-6822(03)00447-1. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J.Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglin M, Melar-New M, Laimins L. Human papillomaviruses and the interferon response. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29(9):629–635. doi: 10.1089/jir.2009.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergant Marusic M, Ozbun MA, Campos SK, Myers MP, Banks L. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic. 2012;13(3):455–467. doi: 10.1111/j.1600-0854.2011.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology. 2013;445(1–2):35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berscheminski J, Wimmer P, Brun J, Ip WH, Groitl P, Horlacher T, Jaffray E, Hay RT, Dobner T, Schreiner S. Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J Virol. 2014;88(11):6076–6092. doi: 10.1128/JVI.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nature structural biology. 2001;8(7):626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- Brimer N, Vande Pol SB. Papillomavirus E6 PDZ interactions can be replaced by repression of p53 to promote episomal human papillomavirus genome maintenance. J Virol. 2014;88(5):3027–3030. doi: 10.1128/JVI.02360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C, Kowalczyk AM, Taylor ER, Morgan IM, Gaston K. P53 represses human papillomavirus type 16 DNA replication via the viral E2 protein. Virol J. 2008;5:5. doi: 10.1186/1743-422X-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Day PM, Thompson CD, Lubkowski J, Lu W, Lowy DR, Schiller JT. Human alpha-defensins block papillomavirus infection. Proc.Natl.Acad.Sci.U.S.A. 2006;103(5):1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Day PM, Trus BL. The papillomavirus major capsid protein L1. Virology. 2013;445(1–2):169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. Journal of virology. 2006;80(21):10890–10893. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira C, Schelhaas M. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog. 2015;11(2):e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJE, Laimins LA. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. Journal of Virology. 2000;74(9):4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJE, Pena L, Sen GC, Park JK, Laimins LA. Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. Journal of Virology. 2002;76(17):8864–8874. doi: 10.1128/JVI.76.17.8864-8874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubie HA. Diseases associated with human papillomavirus infection. Virology. 2013;445(1–2):21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Day PM, Baker CC, Lowy DR, Schiller JT. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(39):14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol. 2013;87(7):3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Andrea M, Lo Cigno I, Albertini S, Borgogna C, Peretti A, Johnson K, Chandran B, Landolfo S, Gariglio M. Cytokine. 2015;76:104. doi: 10.1128/JVI.00013-15. Id: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey A, Bowie AG. Innate immune recognition of DNA: A recent history. Virology. 2015;479:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin BY, Jin G, Wheeler CG, Ma T, Harper JW, Broker TR, Chow LT. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J.Virol. 2004;78(24):13954–13965. doi: 10.1128/JVI.78.24.13954-13965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the sam mRNA. Cell. 1991;67(3):569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe S, Bienkowska-Haba M, Guion LG, Sapp M. Cruising the cellular highways: How human papillomavirus travels from the surface to the nucleus. Virus Research. 2016a doi: 10.1016/j.virusres.2016.10.015. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe S, Luszczek W, Keiffer TR, Bienkowska-Haba M, Guion LG, Sapp MJ. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc Natl Acad Sci U S A. 2016b;113(22):6289–6294. doi: 10.1073/pnas.1600638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445(1–2):99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445(1–2):80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10(2):196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- Dreer M, Fertey J, van de Poel S, Straub E, Madlung J, Macek B, Iftner T, Stubenrauch F. Interaction of NCOR/SMRT Repressor Complexes with Papillomavirus E8^ E2C Proteins Inhibits Viral Replication. PLoS Pathog. 2016a;12(4):e1005556. doi: 10.1371/journal.ppat.1005556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreer M, Van de Poel S, Stubenrauch F. Control of viral replication and transcription by the papillomavirus E8^E2 protein. Virus Research. 2016b doi: 10.1016/j.virusres.2016.11.005. In Press. [DOI] [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de The H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112(Pt 3):381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Egawa N, Nakahara T, Ohno S, Narisawa-Saito M, Yugawa T, Fujita M, Yamato K, Natori Y, Kiyono T. The E1 protein of human papillomavirus type 16 is dispensable for maintenance replication of the viral genome. J Virol. 2012;86(6):3276–3283. doi: 10.1128/JVI.06450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Earnshaw WC, Pluta AF, Sternsdorf T, Ainsztein AM, Carmena M, Ruchaud S, Hsu WL, Orr A. A dynamic connection between centromeres and ND10 proteins. J Cell Sci. 1999a;112(Pt 20):3443–3454. doi: 10.1242/jcs.112.20.3443. [DOI] [PubMed] [Google Scholar]
- Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. J Cell Sci. 1999b;112(Pt 24):4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. The ISG56/IFIT1 gene family. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31(1):71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertey J, Ammermann I, Winkler M, Stoger R, Iftner T, Stubenrauch F. Interaction of the papillomavirus E8--E2C protein with the cellular CHD6 protein contributes to transcriptional repression. J Virol. 2010;84(18):9505–9515. doi: 10.1128/JVI.00678-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J Virol. 2000;74(14):6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Schafer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology. 2002;295(1):97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J Virol. 2011;85(17):8996–9012. doi: 10.1128/JVI.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariano GR, Dell’Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. The Intracellular DNA Sensor IFI16 Gene Acts as Restriction Factor for Human Cytomegalovirus Replication. Plos Pathogens. 2012;(1):8. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio M, Azzimonti B, Pagano M, Palestro G, de Andrea M, Valente G, Voglino G, Navino L, Landolfo S. Immunohistochemical expression analysis of the human interferon-inducible gene IFI16, a member of the HIN200 family, not restricted to hematopoietic cells. Journal of interferon & cytokine research. 2002;22(7):815–821. doi: 10.1089/107999002320271413. [DOI] [PubMed] [Google Scholar]
- Gauson EJ, Donaldson MM, Dornan ES, Wang X, Bristol M, Bodily JM, Morgan IM. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2 mediated DNA replication. J Virol. 2015 doi: 10.1128/JVI.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam D, Moody CA. Impact of the DNA Damage Response on Human Papillomavirus Chromatin. PLoS Pathog. 2016;12(6):e1005613. doi: 10.1371/journal.ppat.1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. Journal of the National Cancer Institute. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J.Virol. 2001;75(3):1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci U S A. 2003;100(15):8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldner HH, Szostecki C, Grotzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. Journal of immunology. 1992;149(12):4067–4073. [PubMed] [Google Scholar]
- Gunasekharan V, Hache G, Laimins L. Differentiation-dependent changes in levels of C/EBPbeta repressors and activators regulate human papillomavirus type 31 late gene expression. J Virol. 2012;86(9):5393–5398. doi: 10.1128/JVI.07239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan V, Laimins LA. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. Journal of virology. 2013;87(10):6037–6043. doi: 10.1128/JVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan VK, Li Y, Andrade J, Laimins LA. Post-Transcriptional Regulation of KLF4 by High-Risk Human Papillomaviruses Is Necessary for the Differentiation-Dependent Viral Life Cycle. PLoS Pathog. 2016;12(7):e1005747. doi: 10.1371/journal.ppat.1005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiger C, Jager G, Walter M, Iftner T, Stubenrauch F. Interferon Kappa Inhibits Human Papillomavirus 31 Transcription by Inducing Sp100 Proteins. J Virol. 2016;90(2):694–704. doi: 10.1128/JVI.02137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaschik D, Hinterkeuser K, Oldak M, Pfister HJ, Smola-Hess S. The Papillomavirus E2 protein binds to and synergizes with C/EBP factors involved in keratinocyte differentiation. J.Virol. 2003;77(9):5253–5265. doi: 10.1128/JVI.77.9.5253-5265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman MT, Pett MR, Roberts I, Alazawi WO, Teschendorff AE, Zhang XY, Stanley MA, Coleman N. Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis. 2006;27(11):2341–2353. doi: 10.1093/carcin/bgl172. [DOI] [PubMed] [Google Scholar]
- Hindmarsh PL, Laimins LA. Mechanisms regulating expression of the HPV 31 L1 and L2 capsid proteins and pseudovirion entry. Virol.J. 2007;4:19. doi: 10.1186/1743-422X-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol. 2005;79(7):3938–3948. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Laimins L. Manipulation of the innate immune response by human papillomaviruses. Virus Research. 2016 doi: 10.1016/j.virusres.2016.11.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins EE, Morreale RJ, Werner SP, Higginbotham JM, Laimins LA, Lambert PF, Brown DR, Gillison ML, Nuovo GJ, Witte DP, Kim MO, Davies SM, Mehta PA, Butsch Kovacic M, Wikenheiser-Brokamp KA, Wells SI. The fanconi anemia pathway limits human papillomavirus replication. J Virol. 2012;86(15):8131–8138. doi: 10.1128/JVI.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Kadaja M, Ustav M. Two separate replication modes of the bovine papillomavirus BPV1 origin of replication that have different sensitivity to p53. Virus Res. 2003;96(1–2):75–84. doi: 10.1016/s0168-1702(03)00174-6. [DOI] [PubMed] [Google Scholar]
- Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. Journal of Virology. 1999;73(5):4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac A, Wilcox KW, Taylor JL. SP100B, a repressor of gene expression preferentially binds to DNA with unmethylated CpGs. J Cell Biochem. 2006;98(5):1106–1122. doi: 10.1002/jcb.20841. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147(2):221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nature immunology. 2015;16(4):343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J.Biol.Chem. 1999;274(9):5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- Kamper N, Day PM, Nowak T, Selinka HC, Florin L, Bolscher J, Hilbig L, Schiller JT, Sapp M. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J.Virol. 2006;80(2):759–768. doi: 10.1128/JVI.80.2.759-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karttunen H, Savas JN, McKinney C, Chen YH, Yates JR, 3rd, Hukkanen V, Huang TT, Mohr I. Co-opting the Fanconi anemia genomic stability pathway enables herpesvirus DNA synthesis and productive growth. Molecular cell. 2014;55(1):111–122. doi: 10.1016/j.molcel.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho EY, Wang HK, Banerjee NS, Broker TR, Chow LT. HPV-18 E6 mutants reveal p53 modulation of viral DNA amplification in organotypic cultures. Proc Natl Acad Sci U S A. 2013;110(19):7542–7549. doi: 10.1073/pnas.1304855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lambert PF. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology. 2002;293(1):10–14. doi: 10.1006/viro.2001.1305. [DOI] [PubMed] [Google Scholar]
- Kines RC, Thompson CD, Lowy DR, Schiller JT, Day PM. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc.Natl.Acad.Sci.U.S.A. 2009;106(48):20458–20463. doi: 10.1073/pnas.0908502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivipold P, Vosa L, Ustav M, Kurg R. DAXX modulates human papillomavirus early gene expression and genome replication in U2OS cells. Virol J. 2015;12:104. doi: 10.1186/s12985-015-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp DJ, Laimins LA. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology. 1999;257(1):239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- Lace MJ, Anson JR, Haugen TH, Dierdorff JM, Turek LP. Interferon treatment of human keratinocytes harboring extrachromosomal, persistent HPV-16 plasmid genomes induces de novo viral integration. Carcinogenesis. 2015;36(1):151–159. doi: 10.1093/carcin/bgu236. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11(5):871–876. [PubMed] [Google Scholar]
- Lazarczyk M, Dalard C, Hayder M, Dupre L, Pignolet B, Majewski S, Vuillier F, Favre M, Liblau RS. EVER proteins, key elements of the natural anti-human papillomavirus barrier, are regulated upon T-cell activation. PloS one. 2012;7(6):e39995. doi: 10.1371/journal.pone.0039995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarczyk M, Pons C, Mendoza JA, Cassonnet P, Jacob Y, Favre M. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J.Exp.Med. 2008;205(1):35–42. doi: 10.1084/jem.20071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiding JW, Holland SM. Warts and all: Human papillomavirus in primary immunodeficiencies. J Allergy Clin Immun. 2012;130(5):1030–1048. doi: 10.1016/j.jaci.2012.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepik D, Ilves I, Kristjuhan A, Maimets T, Ustav M. p53 protein is a suppressor of papillomavirus DNA amplificational replication. J Virol. 1998;72(8):6822–6831. doi: 10.1128/jvi.72.8.6822-6831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman PM. Epigenetics and Genetics of Viral Latency. Cell host & microbe. 2016;19(5):619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Cigno I, De Andrea M, Borgogna C, Albertini S, Landini MM, Peretti A, Johnson KE, Chandran B, Landolfo S, Gariglio M. The Nuclear DNA Sensor IFI16 Acts as a Restriction Factor for Human Papillomavirus Replication through Epigenetic Modifications of the Viral Promoters. J Virol. 2015;89(15):7506–7520. doi: 10.1128/JVI.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Guarente L. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 2000;60(9):2331–2334. [PubMed] [Google Scholar]
- Lorenz LD, Rivera Cardona J, Lambert PF. Inactivation of p53 rescues the maintenance of high risk HPV DNA genomes deficient in expression of E6. PLoS Pathog. 2013;9(10):e1003717. doi: 10.1371/journal.ppat.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. Journal of virology. 2010;84(6):2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff JA, Yuan H, Suter MM, Muller EJ, Schlegel R, Moore PF. Canine keratinocytes upregulate type I interferons and proinflammatory cytokines in response to poly(dA:dT) but not to canine papillomavirus. Vet Immunol Immunopathol. 2013;153(3–4):177–186. doi: 10.1016/j.vetimm.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84(8):4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimi P, Pim D, Bertoli C, Bouvard V, Banks L. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene. 1999;18(54):7748–7754. doi: 10.1038/sj.onc.1203208. [DOI] [PubMed] [Google Scholar]
- Maul GG. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20(8):660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217(1):67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- McBride AA. Replication and partitioning of papillomavirus genomes. Advances in virus research. 2008;72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AA. The papillomavirus E2 proteins. Virology. 2013;445(1–2):57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, McBride AA. 2016 [Google Scholar]
- McKinney CC, Hussmann KL, McBride AA. The Role of the DNA Damage Response throughout the Papillomavirus Life Cycle. Viruses. 2015;7(5):2450–2469. doi: 10.3390/v7052450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human Papillomaviruses Activate the ATM DNA Damage Pathway for Viral Genome Amplification upon Differentiation. Plos Pathogens. 2009;5(10):e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10(12):878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proceedings of the National Academy of Sciences. 2014;111(1):E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev DG, Vladimirova OV, Ivanov A, Rauscher F, 3rd, Maul GG. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J Virol. 2006;80(16):8019–8029. doi: 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Negorev DG, Rafalska-Metcalf IU, Yang T, Maul GG, Janicki SM. Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site. Mol Biol Cell. 2013;24(9):1454–1468. doi: 10.1091/mbc.E12-09-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth G. Genetics of epidermodysplasia verruciformis: Insights into host defense against papillomaviruses. Semin.Immunol. 2006;18(6):362–374. doi: 10.1016/j.smim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proceedings of the National Academy of Sciences. 2013;110(47):E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J Virol. 2002;76(22):11291–11300. doi: 10.1128/JVI.76.22.11291-11300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology. 1998;248(2):218–230. doi: 10.1006/viro.1998.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park RB, Androphy EJ. Genetic Analysis of High-Risk E6 in Episomal Maintenance of Human Papillomavirus Genomes in Primary Human Keratinocytes. Journal of Virology. 2002;76(22):11359–11364. doi: 10.1128/JVI.76.22.11359-11364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JW, Pitot HC, Lambert PF. Loss of Dependence on Continued Expression of the Human Papillomavirus 16 E7 Oncogene in Cervical Cancers and Precancerous Lesions Arising in Fanconi Anemia Pathway-Deficient Mice. mBio. 2016;7(3) doi: 10.1128/mBio.00628-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8(4):e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5(2):e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat.Genet. 2002;32(4):579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- Rector A, Lemey P, Tachezy R, Mostmans S, Ghim S-J, Van Doorslaer K, Roelke M, Bush M, Montali RJ, Joslin J. Ancient papillomavirus-host co-speciation in Felidae. Genome biology. 2007;8(4):1. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinson T, Toots M, Kadaja M, Pipitch R, Allik M, Ustav E, Ustav M. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J Virol. 2013;87(2):951–964. doi: 10.1128/JVI.01943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J, Hurst J, Voges M, Krauss P, Munch P, Iftner T, Stubenrauch F. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85(21):11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc.Natl.Acad.Sci.U.S.A. 2006;103(5):1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, Bachmann A, Ritter TM, Dondog B, Ly R, Bosch FX, Zawatzky R, Rosl F. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 2009;69(22):8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- Roberts JN, Buck CB, Thompson CD, Kines R, Bernardo M, Choyke PL, Lowy DR, Schiller JT. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nature medicine. 2007;13(7):857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature genetics. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445(1–2):138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Dutta D, Iqbal J, Pisano G, Gjyshi O, Ansari MA, Kumar B, Chandran B. Nuclear Innate Immune DNA Sensor IFI16 is Degraded During Lytic Reactivation of Kaposi’s Sarcoma-Associated Herpesvirus (KSHV): Role of IFI16 in Maintenance of KSHV Latency. Journal of Virology, JVI. 01003-01016. 2016 doi: 10.1128/JVI.01003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Potenza N. Antiviral effects of human microRNAs and conservation of their target sites. FEBS letters. 2011;585(16):2551–2555. doi: 10.1016/j.febslet.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80(8):3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia P, Fensterl V, Sen GC. The inhibitory action of P56 on select functions of E1 mediates interferon’s effect on human papillomavirus DNA replication. J Virol. 2010;84(24):13036–13039. doi: 10.1128/JVI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara N, Chen D, McBride AA. Papillomaviruses use recombination-dependent replication to vegetatively amplify their genomes in differentiated cells. PLoS Pathog. 2013;9(7):e1003321. doi: 10.1371/journal.ppat.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara N, Mitra R, McBride AA. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J Virol. 2011;85(17):8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Stamminger T. Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J Virol. 2016;90(13):5850–5854. doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S, Burck C, Glass M, Groitl P, Wimmer P, Kinkley S, Mund A, Everett RD, Dobner T. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013;41(6):3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S, Wodrich H. Virion factors that target Daxx to overcome intrinsic immunity. J Virol. 2013;87(19):10412–10422. doi: 10.1128/JVI.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Losson R, Desterro JM, Hay RT, Chambon P, Dejean A. Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol Cell Biol. 2001;21(10):3314–3324. doi: 10.1128/MCB.21.10.3314-3324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci U S A. 1998;95(13):7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinka HC, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J.Virol. 2007;81(20):10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Molecular cell. 2006;24(3):331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Io K, Shindo K, Matsui M, Sakamoto T, Tada K, Kobayashi M, Kadowaki N, Takaori-Kondo A. APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells. Sci Rep. 2012;2:806. doi: 10.1038/srep00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos MH, McBride AA. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72(3):2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Campos SK, Ozbun MA. Human papillomavirus type 31 uses a caveolin 1-and dynamin 2-mediated entry pathway for infection of human keratinocytes. J.Virol. 2007;81(18):9922–9931. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, Duensing S. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol. 2007;81(23):13265–13270. doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos J, McBride AA. unpublished. [Google Scholar]
- Stanley MA. Epithelial cell responses to infection with human papillomavirus. Clinical microbiology reviews. 2012;25(2):215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley MA, Sterling JC. Host responses to infection with human papillomavirus. Current problems in dermatology. 2014;45:58–74. doi: 10.1159/000355964. [DOI] [PubMed] [Google Scholar]
- Stepp WH. The Role of the Nuclear Domain 10 Proteins in the Human Papillomavirus Lifecycle. Georgetown University; 2015. [Google Scholar]
- Stepp WH, Meyers JM, McBride AA. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio. 2013;4(6):e00845-00813. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem. 1999;274(18):12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Sunthamala N, Thierry F, Teissier S, Pientong C, Kongyingyoes B, Tangsiriwatthana T, Sangkomkamhang U, Ekalaksananan T. E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-kappa transcription in keratinocytes. PloS one. 2014;9(3):e91473. doi: 10.1371/journal.pone.0091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. The EMBO journal. 2008;27(24):3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc.Natl.Acad.Sci.U.S.A. 1999;96(15):8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers B, Burg SH. High-risk human papillomavirus targets crossroads in immune signaling. Viruses. 2015;7(5):2485–2506. doi: 10.3390/v7052485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek LP, Byrne JC, Lowy DR, Dvoretzky I, Friedman RM, Howley PM. Interferon induces morphologic reversion with elimination of extrachromosomal viral genomes in bovine papillomavirus-transformed mouse cells. Proc Natl Acad Sci U S A. 1982;79(24):7914–7918. doi: 10.1073/pnas.79.24.7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. 2010;6(1):51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, McBride AA. Molecular archeological evidence in support of the repeated loss of a papillomavirus gene. Sci Rep. 2016;6:33028. doi: 10.1038/srep33028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2013;41(Database issue):D571–D578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeeburg HJ, Snijders PJ, Wu T, Gluckman E, Soulier J, Surralles J, Castella M, van der Wal JE, Wennerberg J, Califano J, Velleuer E, Dietrich R, Ebell W, Bloemena E, Joenje H, Leemans CR, Brakenhoff RH. Clinical and molecular characteristics of squamous cell carcinomas from Fanconi anemia patients. Journal of the National Cancer Institute. 2008;100(22):1649–1653. doi: 10.1093/jnci/djn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445(1–2):115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320(5873):230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu K, Yuan F, Berdichevsky L, Taichman LB, Auborn K. C/EBPbeta is a negative regulator of human papillomavirus type 11 in keratinocytes. Journal of virology. 1996;70(7):4839–4844. doi: 10.1128/jvi.70.7.4839-4844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23(2):181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Roden RB. L2, the minor capsid protein of papillomavirus. Virology. 2013;445(1–2):175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wakae K, Kitamura K, Aoyama S, Liu G, Koura M, Monjurul AM, Kukimoto I, Muramatsu M. APOBEC3 deaminases induce hypermutation in human papillomavirus 16 DNA upon beta interferon stimulation. J Virol. 2014;88(2):1308–1317. doi: 10.1128/JVI.03091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Pyeon D, Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3 in papillomavirus restriction, evolution and cancer progression. Oncotarget. 2015a;6(37):39385–39386. doi: 10.18632/oncotarget.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol. 2015b;89(1):688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Schlumberger SE, Criqui-Filipe P, Wasylyk B. Sp100 Interacts with ETS-1 and Stimulates Its Transcriptional Activity. Molecular and Cellular Biology. 2002;22(8):2687–2702. doi: 10.1128/MCB.22.8.2687-2702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomaviruses. Virus Research. 2016 doi: 10.1016/j.virusres.2016.11.023. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox KW, Sheriff S, Isaac A, Taylor JL. SP100B is a repressor of gene expression. J Cell Biochem. 2005;95(2):352–365. doi: 10.1002/jcb.20434. [DOI] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proceedings of the National Academy of Sciences. 2003;100(19):10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nature immunology. 2012;13(3):214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-M, Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2011;1809(11):668–677. doi: 10.1016/j.bbagrm.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18(56):7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- Zong RT, Das C, Tucker PW. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 2000;19(15):4123–4133. doi: 10.1093/emboj/19.15.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]