Abstract
Skeletal muscle is a large organ that accounts for up to half the total mass of the human body. A progressive decline in muscle mass and strength occurs with ageing and in some individuals configures the syndrome of ‘sarcopenia’, a condition that impairs mobility, challenges autonomy, and is a risk factor for mortality. The mechanisms leading to sarcopenia as well as myopathies are still little understood. The Human Skeletal Muscle Proteome Project was initiated with the aim to characterize muscle proteins and how they change with ageing and disease. We conducted an extensive review of the literature and analysed publically available protein databases. A systematic search of peer‐reviewed studies was performed using PubMed. Search terms included ‘human’, ‘skeletal muscle’, ‘proteome’, ‘proteomic(s)’, and ‘mass spectrometry’, ‘liquid chromatography‐mass spectrometry (LC‐MS/MS)’. A catalogue of 5431 non‐redundant muscle proteins identified by mass spectrometry‐based proteomics from 38 peer‐reviewed scientific publications from 2002 to November 2015 was created. We also developed a nosology system for the classification of muscle proteins based on localization and function. Such inventory of proteins should serve as a useful background reference for future research on changes in muscle proteome assessed by quantitative mass spectrometry‐based proteomic approaches that occur with ageing and diseases. This classification and compilation of the human skeletal muscle proteome can be used for the identification and quantification of proteins in skeletal muscle to discover new mechanisms for sarcopenia and specific muscle diseases that can be targeted for the prevention and treatment.
Keywords: Ageing, Skeletal muscle, Proteomics, Mass spectrometry, Post‐translational modifications
Introduction
Skeletal muscle is one of the most abundant tissues in mammals, accounting for up to 40% of the total mass of the human body.1 The human skeletal muscle is a complex organ made up of multinucleated long and cylindrical cells called myofibres that are, in turn, composed of myofibrils. Myofibrils consist of proteins called myofilaments, mainly actins and myosins, that form the sarcomeres, which are the smallest contractile units. The contraction–relaxation cycle in muscle requires energy that is mostly generated aerobically by mitochondria particularly abundant in adult muscle fibres. Contraction is stimulated by motor neurons that are resident in the spinal cord and connect with muscle fibres through a specialized synapsis known as the neuromuscular junction. Acetylcholine released into the neuromuscular junction cleft interacts with specific receptors and generates an action potential across the surface of the muscle cell that rapidly diffuses within the muscle fibres through a network of specialized tubules called T tubules. The action potential triggers the opening of the dihydropyridine receptor that allows influx of calcium (Ca2+) in the cytoplasm that stimulates ryanodine receptors to release more Ca2+ from the sarcoplasmic reticulum. Ca2+, in turn, triggers the interaction between contractile proteins and elicits a shortening of the muscle fibre.2 Such structural and functional complexity is mirrored by the large repertoire of proteins found in skeletal muscle.
Maintenance of muscle mass and effective muscle function throughout life is essential to preserve a healthy and active lifespan and to maintain physical function and autonomy in old age. Conversely, the development of overt sarcopenia has devastating consequences on individuals' life because it is associated with mobility loss, higher risk of disability, nursing home admission, and death.3, 4 Thus, understanding the causal pathways leading to age‐related sarcopenia and develop strategies for prevention and cure of this condition is essential to improve the well‐being and quality of life of a growing number of older individuals in the population. There is strong evidence that diabetes type 2, cancer, chronic kidney diseases, chronic pulmonary disease, and bed rest cause accelerated muscle mass and strength, which is superimposed to the effect of ageing. However, the mechanisms by which ageing and diseases cause sarcopenia are still not fully understood and may be quite different. With the exception of exercise, there is currently no effective treatment for sarcopenia.
Most of current research on sarcopenia is based on clinical characterization, analysis of blood biomarkers, and examination of muscle biopsies.5, 6, 7, 8 Histological examination of muscle specimens is generally considered as the gold standard test for most myopathies but, especially for age‐related sarcopenia, histology lacks diagnostic specificity and has limited value for monitoring the effect of interventions.
We propose that proteomic analysis of muscle tissue, especially using a discovery approach complemented by targeted, quantitative mass spectrometry (MS), may overcome the many limitations of previous approaches to the study of sarcopenia. Specific proteomic signatures, or the overrepresentation or underrepresentation of certain proteins or even the altered stoichiometric interrelationship between proteins, may provide clues on the processes that lead to muscle impairments, perhaps also allowing the diagnosis of the different conditions, including age‐related sarcopenia. For example, changes in mitochondrial proteins may suggest that scarcity of energy is at the root of muscle degeneration. Also, proteomics could be used to monitor at an early stage the potential effectiveness of intervention aimed at normalizing muscle. This approach would add considerable value to the simple monitoring muscle mass and strength. In fact, interventions aimed at expanding muscle mass, such a testosterone or selective androgen receptor modulators, have shown little impact on physical functions such as walking performance; the structural and biochemical reasons for such a discrepancy are not clear.9, 10 On the other hands, modifications in abundance of specific muscle proteins may occur long before any improvement in strength is clinically detectable. Overall, the complementary use of advanced high‐sensitivity discovery‐mode and targeted, quantitative MS/MS proteomic analysis of skeletal muscle specimens may provide new insights into the pathogenesis of skeletal muscle in health and disease through the characterization of protein abundance, quality, and diversity, including their isoforms and post‐translational modifications (PTMs). Application of proteomics may help to identify biomarkers of adaptive or maladaptive biological mechanisms manifested during ageing or diseases.
A critical step for making progress in this field is a comprehensive characterization and classification of the human skeletal proteome. Because it is well known that muscles are different in men and women and change substantially with ageing, such normative data will have to be developed in men and women of different age, possibly free of major conditions or treatment that may affect muscle proteins. Similarly, it is important to have an available annotation system that characterize proteins in terms of their cell localization and function.
We performed a search of the literature of published studies that had reported on skeletal muscle proteomic analyses in humans. The goal of this review is to summarize the state of the art in muscle proteomic research as the first step of a new initiative of the Biology/Disease‐driven Human Proteome Project.11 Then, using data from available databases and an extensive review of the literature, we created a refined annotation system that improved our ability to classify muscle proteins based on their cellular localization and function. Finally, we outline what research should be performed to create a reference atlas of the normal, human skeletal muscle proteome.
Human skeletal muscle proteome
Caveats and challenges in performing and interpreting skeletal muscle proteomics
Human skeletal muscle has mainly been studied in muscle biopsies obtained under local anaesthesia. The percutaneous needle introduced by Bergstrom in 196212 yields useful biopsy material (around 100–300 mg) that has been used successfully for proteomic analysis conducted by MS.13
Previous attempts to characterize the muscle proteomics have been incomplete14 and mostly focused on muscular dystrophies.15, 16 Although there are hundreds of skeletal muscles in the body, nearly all proteomic studies have focused on vastus lateralis, quadriceps, and deltoid muscles, and therefore, most of the current knowledge is skewed towards these muscles. This is problematic because different muscles may have quite distinct protein composition affected by different content in adipose and connective tissue, degree of vascularization and innervation, and fibre type composition. Skeletal muscle fibres can be divided mainly into three main subtypes distinguished by their contractile and metabolic properties: (A) slow, oxidative, type 1 fibres (measured by staining myosin‐7, encoded by MYH7); (B) fast, oxidative, type 2A fibres (measured by myosin 2, encoded by MYH2); (C) fast, glycolytic, type 2x fibres (measured by myosin‐1, encoded by MYH1).17 The relative percentage of fibre types tends to be muscle specific and is also affected by the overall level of physical activity and metabolic diseases such as diabetes.18, 19, 20 All these factors may affect protein synthesis and degradation. The large number of proteins and the wide dynamic range in abundance of different proteins in skeletal muscle are one of main challenges in performing high‐fidelity proteomics. In fact some of the largest (titin) and smallest (phospholamban) proteins in the human proteome are found in this tissue. In addition, sample preparation becomes critically important because the contractile proteins are so abundant that without adequate fractionation, the chance to detect low abundant proteins, such as signalling enzymes, becomes low.
Conditions and factors affecting the composition of the human skeletal muscle proteome
Skeletal muscle not only contains proteins that pertain to muscle cells but also is unavoidably contaminated by proteins that pertain to adipose, connective, vascular, and neural tissue. To better understand the difference between specimens, proteins should be classified based on the subcellular localization (i.e. contractile apparatus, nuclei, mitochondria, sarcoplasmic reticulum, transverse tubules, Golgi apparatus, sarcolemma, extracellular matrix, ribosomes, cytoskeleton, and the cytosolic fraction)21 function and metabolic and signalling pathways (i.e. glucose metabolism, electron transport chain and OXPHOS system, and calcium homeostasis).22, 23, 24, 25 Early proteomic studies identified that most of the proteins of skeletal muscle are localized in the sarcomere (i.e. myosins, actins, troponins, tropomyosins, and auxiliary proteins of sarcomeric units), representing about 55–60% of the total proteins.18, 22, 26, 27, 28 Mitochondrial proteins constitute around 28% of the transcripts found in skeletal muscle confirming the importance muscle in the energy metabolism.29 Of note, most of these proteins, especially sarcomeric, glycolytic, and mitochondrial proteins exist in different isoforms (i.e. myosin has more than 20 isoforms), therefore highly affecting the functionality, the integrity, and the protein synthesis in skeletal muscle, especially during ageing and some other pathological conditions.
During physical activity or movement, skeletal muscle is highly metabolically demanding, therefore, a very large number of proteins and enzymes related with all the metabolic pathways as well as proteins involved in the cellular stress response such as heat shock proteins (HSPs) will be prone to change in abundance during these states.18, 30
Also limited literature suggests that contractile, structural, and mitochondrial proteins, together with metabolic enzymes, can all be affected by conditions that affect muscle health (i.e. diabetes type 2, obesity, ageing, age‐related diseases, myopathies, and dystrophies) and behavioural stress such as extreme exercise.31 Factors in liquid chromatography‐mass spectrometry (LC‐MS/MS) proteomic studies that have been found to affect muscle proteome composition are summarized in the next sections and in Table 1.
Table 1.
Changes in skeletal muscle protein composition caused by aging, age‐related diseases and skeletal muscle disease. Black arrow: downregulation; Red arrow: upregulation

|
Ageing
Age‐related changes in the contractile apparatus are mainly the result of transition from fast‐twitching fibres (mainly IIx) to slow‐twitching muscle fibres.32, 33, 34 Proteomic studies have been shown that myofibrillar and cytoskeletal proteins are less abundant in vastus lateralis in older compared with middle‐aged women.27, 35 Ageing also appeared to change the abundance of enzymes involved in oxidative metabolism, anaerobic metabolism, and the glycolysis pathway, such as adenosine triphosphate (ATP) syntase β‐chain, Acyl‐CoA dehydrogenase, aconitase, 2‐oxoglutarate dehydrogenase, malate dehydrogenase, creatine kinase, glycogen phosphorylase, glyceraldehyde‐3‐phosphate dehydrogenase, among others.33 Other proteins that are similar to the HSP are involved in detoxification and cytoprotection functions (i.e. carbonic anhydrase 3 or elongation factor 2, and ubiquitin‐40S ribosomal protein S27a) decrease during ageing, with reduced scavenging of reactive oxygen species accumulation of aggregated proteins.30, 35, 36
Exercise and physical inactivity
Although it is well known that skeletal muscle adaptations to exercise depend on duration, intensity, and frequency,37 changes in muscle proteins associated with different types of exercise have not been well characterized. Myofibrillar protein isoforms are most affected, mainly in the distribution of myosin heavy chain isoforms, thin filament isoforms, sarcoplasmic reticulum calcium proteins, mitochondrial proteins, and some chaperones involved in cell stress response. Studies also found (i) changes in fibre type composition; (i) change in enzymes implicated in energetic metabolism, such as the E3 subunit of the pyruvate dehydrogenase complex, fumarate hydratase, malate dehydrogenase, and aspartate aminotransferase; and (iii) expression of some of the components of the electron transport chain such as complex I, IV, and ATP synthase subunits, although there is wide heterogeneity between studies on specific results.38, 39, 40, 41
Long bed rest is known to induce considerable atrophy in lower limb muscles accompanied by a reduction in cross‐sectional area and fascicle length.42 Bed rest proteomic studies with or without exercise have shown muscle fibre atrophy accompanied by a deregulation in myosin heavy chain distribution, downregulation of thin filament proteins, and changes in fibre type composition (increased type I and decreased type IIA fibres) in both, vastus lateralis and soleus muscles. Moreover, these changes are usually accompanied by (i) down‐regulation of proteins involved in aerobic metabolism (i.e. dihydrolipoil dehydrogenase, succinyl‐CoA ligase, and aconitate hydratase between others); (ii) up‐regulation of proteins involved in anaerobic glycolysis such as some isoforms of glycogen phosphorylase and α‐enolase; and (iii) down‐regulation of the antioxidant defence system proteins.43, 44
Hypoxia and extreme environmental stress
The effects of hypoxia in skeletal muscle remain controversial. Early reports suggested that chronic hypoxia might positively affect muscle oxidative capacity and capillarization through up‐regulation of hypoxia inducible factor‐1 alpha, the primary transcriptional response factor for hypoxic adaptation.45, 46 However, continued exposure to hypoxia has negative effects on muscle structure, producing muscle atrophy.47, 48, 49 Using a proteomic approach, it was found that under hypoxic conditions, vastus lateralis contractile proteins such as the γ‐actin isoform were decreased together with other contractile proteins such as D repeat protein 1, vinculin, tubulin, Kelch repeat, and desmin. Furthermore, proteins involved in tissue injury hypoxia (mainly α‐enolase), in oxidative metabolism (i.e. 2‐oxoglutarate dehydrogenase, malate dehydrogenase, aconitate hydratase, and electron transport chain mitochondrial proteins), in fatty acid β oxidation, creatine biosynthesis, lipid peroxidation, and proteins involved in oxidative stress protection were also less abundant under hypoxic conditions.50 A proteomic study that aimed to elucidate the mechanisms by which Tibetans born and living at high altitude could adapt to hypoxia found that the most differentially overexpressed proteins in vastus lateralis compared with subjects living a low altitude were those related with oxidative and glycolytic pathways such as glutathione‐S‐transferase P1‐1, Δ2‐enoyl‐CoA‐hydratase, Nicotinamide adenine dinucleotide (NADH)‐ubiquinone oxidoreductase, myoglobin, glyceraldehyde‐3‐phosphate dehydrogenase, and lactate dehydrogenase, suggesting a metabolic switch to cope with this hypoxic conditions and reinforcing protection against reactive oxygen species tissue damage.51
Conversely, changes in diet and environment, such as cold or hot conditions, can negatively impact muscle physiology.52, 53 The increase of energy expenditure upon cold exposure or dietary changes is called adaptive thermogenesis.54 The exact mechanisms by which skeletal muscle adapts to thermogenesis still remains unknown and may be related to changes in the mitochondrial uncoupling.55 A study evaluating the differences of the human skeletal muscle proteome under cold and overfeeding conditions found that extreme conditions produced higher levels of contractile proteins (including fast and slow isoforms) and metabolic enzymes, mainly related with glycolysis and fatty acid metabolism, suggesting that all these modifications were related to alterations in energy expenditure after cold exposure.56
Metabolic diseases and other pathological conditions
Obesity
Accumulation of adipose tissue that occurs with ageing and the metabolic syndrome, especially ectopic lipid deposition such as enlarged visceral fat depots, the liver, skeletal muscle, and other non‐adipose tissues, tends to be associated with loss of muscle mass as well as decline in muscle quality. Obesity may negatively impact human skeletal muscle through accumulation of lipids such as long‐chain diacyl‐glycerols or ceramides, production of inflammatory mediators, interference with maintenance, and repair mechanisms and other still unknown mechanisms.57 Studies conducted on digitonin extracted cytosolic proteins in rectus abdominal muscle from obese women have shown increased expression of metabolic pathway proteins such as adenylate kinase 1, glyceraldehyde‐3‐phosphate dehydrogenase, aldolase A, fatty acid‐binding protein 3, and pyruvate kinase, and a decrease in carbonic anhydrase 3 compared with lean control subjects, suggesting that these differences represent a compensatory mechanism to counteract the muscle mitochondrial dysfunction associated with obesity already described in other studies23, 58
Type 2 diabetes
There is strong evidence from epidemiological studies that people with insulin resistance or type 2 diabetes experience accelerated decline of muscle strength and muscle mass, although the reason for this association is not clear.59 Defects in the activity of the metabolic enzymes and in the insulin signalling pathways associated with decreased muscle glycogen synthesis have been described in the muscle of these patients.60, 61 Proteomic studies of human skeletal muscle have shown that proteins involved in the tricarboxylic acid cycle (TCA cycle), mitochondrial respiration, and other mitochondrial functions such as ATP synthase β and α isoform and creatine kinase brain isoform, among others, were less abundant in vastus lateralis from insulin‐resistant subjects, suggesting an increased glycolytic and decreased mitochondrial protein abundance together with a shift in muscle properties towards a fast‐twitch pattern in the absence of marked changes in fibre‐type distribution.58, 62 Proteins involved in cytoskeletal structure and function like the actin capping and Z‐disc component and the four isoforms of α‐1 chain of type VI collagen, isoenzymes of myosin regulatory light chain 2A and B, myosin 15, and troponin isoforms were significantly altered together with some chaperone and co‐chaperone proteins.62, 63
Exercise is beneficial in patients with type 2 diabetes and plays a major role in the prevention and control of insulin resistance.64 Proteomic studies have shown that a 4 week training programme of endurance exercise in patients with type 2 diabetes increased proteins in vastus lateralis involved in energy metabolism via glycolysis, TCA cycle, electron transport chain, and β‐oxidation, mainly lactate dehydrogenase B chain, mitochondrial creatine kinase, aspartate aminotransferase acetyl‐coA‐acetyltransferase, cytochrome b‐c1 complex subunit 2, isoform 1 of succinyl‐CoA ligase (ADP‐forming) subunit beta, isoform cytoplasmic of fumarate hydratase, and isocitrate dehydrogenase. There was also a reduction in the levels of glycolytic proteins, including isoform 1 of 6‐phosphofructokinase, fructose‐biophosphate aldolase A, and lactoylglutathione lyase, or glyoxalase 1.65
Specific muscle diseases and neuromuscular disorders
Proteomic studies performed in skeletal muscle from patients with skeletal muscle diseases such as hereditary inclusion body myopathy, myofibrillar myopathies, desminopathies, and some muscle disorders caused by collagen VI mutations have shown that the most abundant changes are found in proteins related to cytoskeleton and sarcomere functions, basically caused by mutations in genes encoding sarcomeric and extra‐sarcomeric proteins.66, 67, 68, 69, 70 This will affect not only the skeletal muscle but also their connectives tissues producing contractures and muscle weakness.
Similar results have been found in patients with amyotrophic lateral sclerosis a severe and fatal neurodegenerative disease characterized by the selective death of motor neurons in the motor cortex, brainstem, and spinal cord.71, 72 The pathogenesis of this disease is still unknown, but it has been suggested that skeletal muscle could play an important pathogenic role.71, 73
Post‐translational modifications in skeletal muscle proteome
Post‐translational modifications are chemical alterations to protein structure, typically catalysed by substrate‐specific enzymes that may change the properties of a protein by proteolytic cleavage or by addition of a modifying group to one or more amino acids.74 More than 300 different types of PTMs are known, and many are implicated in diseases such as cancer, ageing, and age‐related diseases.75, 76 Protein phosphorylation and acetylation play key roles in signal transduction pathways regulating energy metabolism, contractile function, and muscle mass in human skeletal muscle, but the role of protein phosphorylation and acetylation in normal skeletal muscle and in skeletal muscle disorders remains still poorly understood because of technical limitations related to both the stability of PTMs in collected biopsies and limits of proteomic technology (see succeeding text).
In the first global analysis of the in vivo phosphoproteome of human skeletal muscle from vastus lateralis in healthy subjects, 367 phosphorylation sites in 144 phosphoproteins/phosphoprotein groups were described. More than one‐quarter were sarcomeric proteins from the contractile apparatus, such as thin actin and thick myosin‐containing filaments, and M‐line and Z‐disc‐associated proteins. Other phosphorylation sites were identified in some enzymes of glycogen metabolism and in some kinase and phosphates subunits that regulate the phosphorylation of glycogen synthase and phosphorylase.22, 77 In mitochondria isolated from vastus lateralis, other authors identified 155 distinct phosphorylation sites in 77 mitochondrial phosphoproteins mainly involved in apoptosis, oxidative phosphorylation (the most abundant), TCA cycle, fatty transporters and β‐oxidation, amino acid degradation, import machinery and transporters, and calcium homeostasis. The authors suggested that reversible phosphorylation of mitochondrial proteins may play a role in the mitochondrial dysfunction that is found in different pathologies such as muscle disorders, type 2 diabetes, and age‐related diseases.78
In another study, the same investigators examined the effects of 4 h stimulation with physiological levels of insulin on the phosphorylation of mitochondrial proteins in human skeletal muscle in vivo in healthy individuals.79 They identified 207 phosphorylation sites, of which 45% were identified both in basal and insulin‐stimulated samples, whereas almost half of them were identified exclusively in the insulin‐stimulated samples. The most abundant phosphoproteins were those involved in oxidative phosphorylation and the TCA cycle. Furthermore, they found multiple phosphorylation sites in components of the mitochondrial inner membrane organizing system. The authors concluded that these results suggest that insulin might regulate the phosphorylation of mitochondrial proteins in skeletal muscle. Also, a recent phosphoproteomic study in human skeletal muscle showed that some 5' adenosine monophosphate activated protein kinase (AMPK) substrates regulate mitochondrial respiration in response to exercise via phosphorylation.80
Finally, another proteomic study comparing skeletal muscle from obese with or without type 2 diabetes found significant differences in phosphorylation and ubiquitinylations in some of the most important glycolytic enzymes such as glycogen phosphorylase muscle isoform, β‐enolase, and pyruvate kinase 2 together with other contractile and cytoskeleton proteins such as glyceraldehyde 3‐phosphate dehydrogenase, myosin regulatory light chain 2, ventricular/cardiac muscle, and myosin regulatory light chain 2 skeletal muscle.81
Compilation of the current human skeletal muscle proteome
In order to compile a comprehensive atlas of the skeletal muscle proteome, we searched PubMed using the terms ‘skeletal muscle’ combined with either ‘proteome’, ‘proteomics’, ‘MS’, and ‘LC‐MS/MS’, using a filter for ‘human’ and published before December 2015. Papers were excluded if they involved proteomic investigations of human cell lines or studies in which low abundance of proteins were detected using immunological methods rather than proteomic and MS approaches. Thirty‐eight publications fit the aforementioned criteria and were used for compiling the human skeletal muscle proteome (Figure 1). The Uniprot database was used as the authoritative reference for human proteins. Lists of proteins were compiled for each paper based upon protein data in the published tables and/or supplementary tables. In order to combine all protein identifications into one list, International protein index accessions and National Center for Biological Information protein accessions were converted to Uniprot accessions by Uniprot ID conversion.82 Several classes of proteins were removed from the resulting set. Proteins that have been merged into other accession numbers or removed from their respective databases were considered obsolete and removed. Proteins that were duplicated between identifications were also reduced to the single identification. Finally, obvious contaminants, which were decoy proteins, trypsin, and non‐human proteins, were removed. The final filtered list that contained 5431 proteins were identified for the human skeletal muscle proteome. We also identified the top 20 most reported skeletal muscle proteins from the 38 publications (Figure 2).
Figure 1.
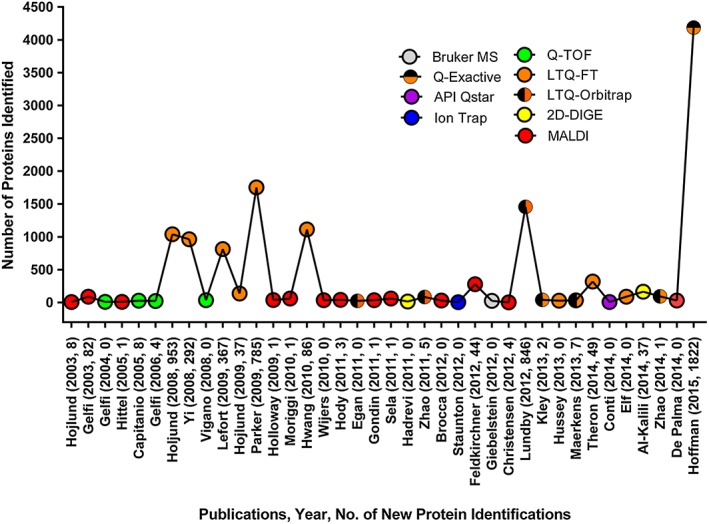
Mass spectrometry‐based human skeletal muscle publications and proteins. We compiled mass spectrometry‐based human skeletal muscle proteome papers since 2003. The X axis contains the first author, year of publication, and how many new proteins were discovered in skeletal muscle in the study. Colour‐coded circles show what mass spectrometer was primarily used by each study. The Y axis shows the total number of proteins identified by each study.
Figure 2.
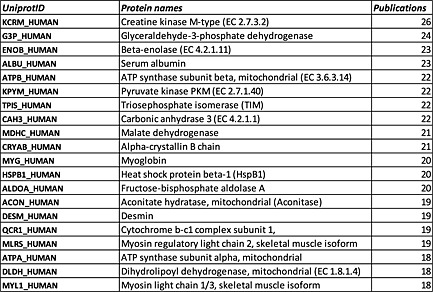
Top 20 proteins in human skeletal muscle proteome. Most reported skeletal muscle proteome from all 38 publications.
The neXtProt database is a recently developed database, which incorporates only high‐quality, well‐studied proteins as the human proteome.83 Therefore, we used this neXtProt to validate our final human skeletal muscle proteome. In our list of 5431, 4947 had neXtProt accessions (Table S1). There were 484 proteins without neXtProt accessions (Table S1, sheet 2) as of December 2015. The high rate of concordance between neXtProt and our dataset shows that we have a proper filtering method that provided a quite comprehensive collection of the proteome.
Many of the earlier studies of the human skeletal muscle proteome involved two‐dimensional gel electrophoresis and Matrix‐assisted laser desorption/ionization‐ time of flight (MALDI‐TOF), a workflow that is mainly useful to identify proteins that are differentially expressed between groups.25, 33, 36, 51, 58, 62 In compiling this list of human skeletal muscle proteins from that manuscripts, we encountered several problems: (i) papers based on earlier technology may have been limited by ambiguous identification of proteins; (ii) most studies did not state the precise criteria used to identify proteins, including false discovery rates (FDR) for peptides and proteins; (iii) some reported isoforms have no accompanying peptide data, thus, it is not possible to determine whether a peptide amino acid sequence that is unique to the specific isoform was actually observed; (iv) most studies did not provide spectra and did not have raw MS data available in a public repository for further inspection and corroboration; and (v) a few studies did not even specify which skeletal muscle was biopsied for study. Some studies used an FDR for proteins >1%, which by current standards is not considered to be sufficiently rigorous. For example, the Plasma Peptide Atlas uses a stringent 1% FDR for proteins (which corresponds to a 0.2% FDR at the peptide level). If such stringent criteria were applied to the human skeletal muscle proteome, as compiled in the present paper, the number of identifications would be anticipated to be less than 5431 non‐redundant proteins in Table S1. However, in certain cases, an investigator may be justified in allowing an FDR up to 5%, in particular for older mass spectrometers. Furthermore, we also note that the question of the correct method of calculating FDR is not completely settled.84, 85 Therefore, for our list, we accept identifications reported at the thresholds from published datasets. In the future, the redundancy of protein identifications and quality of reporting of peptide data from the human skeletal muscle could be improved with standards established by the human skeletal muscle proteome initiative.
Finally, we compared our proteome to three other datasets relevant to the skeletal muscle proteome: a non‐MS, antibody‐based, global human proteome, called Human Protein Atlas28, 86, 87; a mouse proteomics dataset from skeletal muscle88, 89; and an RNA‐seq dataset produced from human skeletal muscle from 361 individuals, from the Genotype‐tissue Expression (GTEx) project.90 While we are interested in the MS‐based human proteome, transcript data and antibody‐based protein data can still inform the analysis of proteomics data, and mouse skeletal muscle is also a useful comparison. In order to translate between transcript and protein, and between mouse and human, we compared all three skeletal muscle studies at the genome level. In mouse skeletal muscle, there were 9887 genes reported from 10 218 proteins, while Human Protein Atlas retrieved 5389 skeletal muscle coding genes, and GTEx contained 7077 skeletal muscle‐specific genes. To reduce the complexity of analysis, we combined the transcriptome data from Protein Atlas and GTEx to 10 962 genes. Our compiled skeletal muscle shows 5024 genes for 5431 proteins. The comparison of these datasets shows that there is likely more of the proteome to be mapped in human skeletal muscle (Figure 3).
Figure 3.
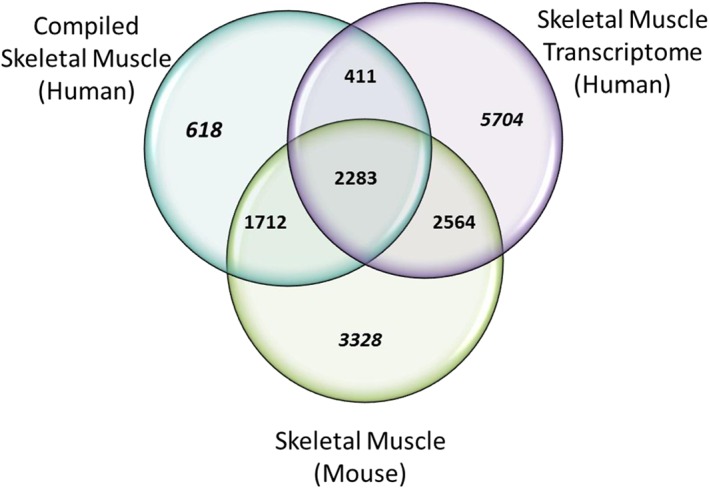
Comparison of approaches to human skeletal muscle proteome. The 5024 genes identified from 38 human skeletal muscle publications (complied skeletal muscle human) were compared with Genotype‐tissue Expression V6 and Protein Atlas skeletal muscle transcriptomics data (10 962) and mouse skeletal muscle proteome data (9887). Mouse skeletal muscle shared 6559 genes (skeletal muscle mouse) and transcriptome skeletal muscle from Protein Atlas and Genotype‐tissue Expression shared 5258 genes (skeletal muscle transcriptome human). Two thousand, two hundred and eighty‐three genes were common between all three skeletal muscle studies. Text in italics are genes unique for each study. Eighty‐eight per cent of the compiled skeletal muscle proteins were present in the transcriptome and mouse skeletal muscle datasets.
Functional annotation of skeletal muscle proteome
To describe the skeletal muscle proteome, we characterized the 5431 compiled proteins based on gene ontology (GO), Uniprot keywords, and manual annotation. In particular, we annotated proteins by Uniprot keywords when they were available. When the Uniprot keyword was not available, we used the GO annotations and then manually searched the literature for an exact annotation. In all proteins with multiple annotations, the literature was consulted, and the protein was manually annotated towards its localization and function in skeletal muscle. Protein numbers were normalized to the total number of ontology annotation terms identified for all the proteins. The justification for this annotation process is as follows: GO provides a set of hierarchical controlled vocabularies split into three categories: cellular component, molecular function, and biological process. However, GO is designed to be species neutral, an approach that tends to describe very broad functional domains.91, 92 UniProt contains a controlled vocabulary that is specific to species. Therefore, to make the annotation more relevant to human biology, we combined the GO annotations with UniProt keyword annotation and direct literature search.
By this process, we were able to annotate 5031 proteins over 7934 terms for the cellular component, 4316 over 5795 for molecular function and, finally, 4395 proteins over 6501 terms for biological function. One hundred and three serum proteins were excluded from both molecular function and biological process. Some proteins are multiple counted because they are in several compartments, or have different molecular functions, or are involved in multiple different biological processes.
Classification of the human skeletal muscle proteome based on the cellular components
Gene Ontology Consortium (GOC) defines the cellular component as ‘locations, at the levels of subcellular structures and macromolecular complexes’. Based on this, we classified 5031 proteins. We found that the majority of the skeletal muscle proteins are compartmentalized in these major compartments: cytoplasm (~30%), cell membrane (~24%), nucleus (~19%), and mitochondria (~11%) (Figure 4A). Membrane proteins include nuclear membrane, cell membrane, inner and outer mitochondrial membrane, matrix membrane, and Golgi membrane. Only a small percentage (7%) of the proteins were not assigned to any compartment (no information available), and we called them as ‘undetermined’. Of note, a high percentage of proteins can be located in multiple compartments (95%) (data not shown).
Figure 4.
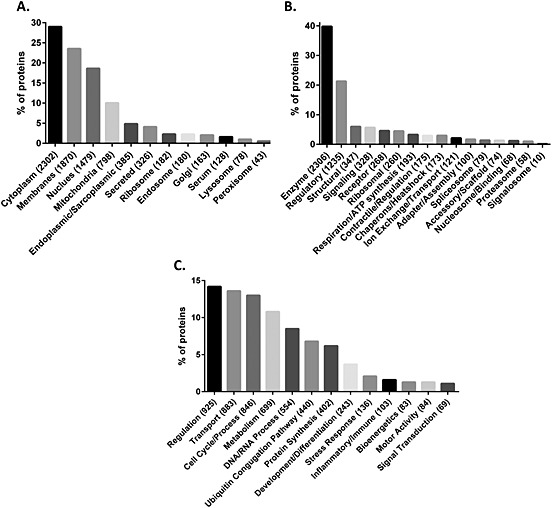
Classification of the human skeletal muscle proteome. Based on gene ontology, Uniprot keywords and manual curation skeletal muscle proteome were classified. (A) Classification of the skeletal muscle proteome based on cellular components, (B) classification of the skeletal muscle proteome based on molecular function, and (C) classification of the skeletal muscle proteome based on the biological process.
Classification of the human skeletal muscle proteome based on the molecular function
GOC defines molecular function as ‘Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level’. Based on this definition, we were able to define the molecular function of 4316 proteins over 5795 annotated terms. Specifically, we identified 18 different major molecular functions (Figure 4B). In brief, 40% of the proteins have an enzymatic function and almost 56% (~1300 proteins) do not have any molecular functional details. Interestingly, about one‐half of the proteins identified in the muscle have regulatory, structural and signalling function underlying the complexity of the muscle as a metabolic and secretory organ, which is not limited to energetic metabolism. In fact, the TCA cycle, electron transport respiratory chain, and ATP synthesis only constitute the 3.1% of the skeletal muscle proteome (Figure 5A).
Figure 5.
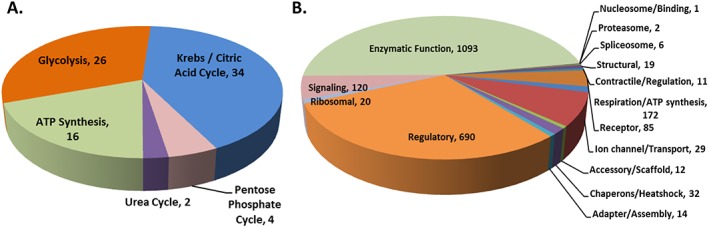
(A) Bioenergetics process from skeletal muscle proteome. The major energy production chains and the number of proteins compiled for each process as shown next to process. (B) Molecular function of enzymes. Enzymes predominate skeletal muscle protein molecular function. Text shows molecular function of enzymes and number of proteins compiled next.
The highest percentage of molecular function annotations is for enzymes (Figure 5B). Changes in the muscle enzyme levels might be a signal of muscle damage or may underlie some myopathies or another defective process during ageing process, but there is no enough information in the literature on this topic.
Classification of the human skeletal muscle proteome based on biological processes
Finally, we classified the skeletal muscle proteome based on biological processes. GOC defines the biological process as ‘a recognized series of events or molecular functions with a defined beginning and end’. Because we aimed for a parsimonious approach, and because we found around 80 different biological processes described, we summarized them into 15 major biological processes, namely, (i) ‘transport process’ that includes protein transport, lipid transport, ion transport, electron transport, endocytosis, and exocytosis; (ii) ‘cell cycle/process’ that includes cell shape, cell adhesion, cell growth, chemotaxis, chemoattraction, cell proliferation, apoptosis, angiogenesis, cell killing, autophagy, cell division, mitosis, and meiosis; (iii) ‘metabolism’ that combines amino acid, carbohydrate, ketone body, fatty acid, glycogen, glycan, lipid, protein, nitrogen compound, and aldehyde metabolism; (iv) ‘bioenergetics process’ that includes glycolysis, Krebs cycle, pentose shunt, urea cycle, and ATP synthesis (Figures 4C and 5A). Based on this, we were able to annotate 4395 proteins over 6501 annotated terms.
Future directions and gaps in knowledge
By performing a comprehensive review of the literature, we have shown that there are at least 5431 different proteins in human skeletal muscle. In spite of the extensive work done so far, our knowledge is still too limited to translate skeletal muscle proteomics into a powerful clinical tool. First of all, there are >200 muscles in the human body, and most of the data available are based on biopsies collected almost exclusively in the quadriceps and deltoid muscle. It is known that there are considerable differences between muscles in protein and enzymatic composition based on the muscle fibre type composition; therefore, some of them will be mainly glycolytic and involved in fast contractions, and others will be primarily aerobic, and with a higher oxidative capacity. Thus, the list of proteins reported here does not apply to other specialized muscle types, such as laryngeal muscles and extra‐ocular muscles in humans. Also, skeletal muscle is highly abundant in different proteoforms that may play an important role in muscle function and could affect the composition of the skeletal muscle proteome. In addition, normative proteomic data on muscle proteomics in disease‐free men and women dispersed over a wide age range and assessed by rigorous proteomic, and MS approaches and state of the art technology are not available. The absence of a solid reference atlas of skeletal muscle proteins makes the identification of changes in the abundances of proteins specifically associated with different disease or conditions a formidable task.
Most of these obstacles are not insurmountable. For example, vastus lateralis biopsies are being performed in the Baltimore Longitudinal Study of Aging, the America's longest‐running scientific study of human ageing, in participants dispersed over a wide age range who are selected to be healthy according to very strict inclusion and exclusion criteria that will increase the likelihood of producing reliable and reproducible proteomic results among others.
Significant technological progress that was made in MS over the last decade substantially improved quality of proteomics data in fundamental biological and pathophysiological research. Proteomic studies became a valuable asset for the global tissue or cell‐specific protein profiling, biomarker discovery, and quantitative proteome studies. Enhanced mass accuracy, acquisition rate, and sensitivity on some of the newly released instruments in combination with advanced liquid chromatography methods allow to record large amounts of information and accurately identify peptides at a very high confidence level. Thousands of proteins can be identified just in a single shotgun experiment in a robust and highly reproducible manner using simplified, accurately controlled sample preparation protocols. Extremely complex biological samples with challenging dynamic ranges, such as skeletal muscle tissues or human plasma, can be analysed in depth by employing advanced offline high performance liquid chromatography/ultra light pressure liquid chromatography (HPLC/UPLC) protein or peptide fractionation methods including ion exchange, basic reverse phase, hydrophilic interaction (HILIC), in solution isoelectric focusing (IEF), or traditional 1D, 2D gel electrophoresis techniques with dramatically increased yield in protein discovery allowing detection of functionally important low‐abundant proteins (the ‘deep proteome’ analysis).93, 94, 95, 96 Furthermore, improved sample preparation and fractionation techniques including isolated organelles such as mitochondrial, nuclear, or membrane fractions as well as IP of Tyr‐phos proteins, pull down of, for example, 14‐3‐3 proteins, pull down of interaction partners of specific signalling enzymes, for example, IRS‐1, and so on can be also used to target highly specialized organelle proteomes and unique protein complexes in various cell models at different biological states.
Quantitative proteomic methods are also evolving rapidly alongside with the improving instrument quality merging both discovery and quantitative proteomic studies in one unified analysis. Recently proposed SWATHTM technology,97 a unique open window acquisition technique available on some of triple TOFs or DIA protocols specific to some of the Orbitraps are used today as a powerful alternative to some of the traditional peptide‐labelling techniques such as isobaric tag labelling (iTRAQ or TMT).98, 99 Peptide or protein‐labelling techniques are used broadly in proteomic studies for the relative and absolute protein quantification in the complex cell or tissue lysate analysis. Targeted methods in proteomics such as selected/multiple reaction monitoring are widely used as accurate quantitative assays but also as additional validation techniques in the discovery studies,100, 101
Methods and instruments are also improving for the detection of PTMs such as phosphorylation, glycosylation, acetylation, ubiquitination, sumoylation, and so on. The observation that acetylation and phosphorylation are often identified on the same peptides and proteins, and probably interact, is a new interesting area of research. PTMs are important to study because they reflect the diversity of protein function.73, 102 Many of the PTMs are difficult to study because they are labile to sample processing and LC‐MS artefacts. For example, O‐GlcNAcylation, an important PTM that rivals phosphorylation in abundance and distribution and has been implicated in chronic diseases, has been especially challenging to detect and measure.103 At least in part, advancements in this field are due to steep improvement of bioinformatics tools and better search algorithms and proteins reference databases. Many proteins have functions that are unknown or not well understood. By studying the proteins with which a particular protein interacts, it is possible to deduce biological functions and pathways using in silico structure‐based approach.104 Protocols have recently been developed for proteomic analysis of formalin‐fixed and paraffin‐embedded tissues and equivalent to proteome inventories obtained from frozen tissue specimens. Thus, using also these tissues could be a valuable resource for retrospective biomarker discovery studies.105
A single technique or method alone is not enough to define or characterize a proteome. Because one of the most challenging issues for the scientific community nowadays is the identification of biological markers, all these advances in the proteomic field together with high‐throughput technologies such as genomics, metabolomics, and other ‘omics’ will allow us to identify molecular pathways and novel protein–protein interactions that are involved not only in pathological conditions but also in normal human biology.
By establishing the Skeletal Muscle Proteome Project Initiative, which recently (October 2015) was presented at the Human Proteome Organization World Conference in Vancouver, Canada, we will be able to characterize the proteome of the normal human skeletal muscle during the entire life cycle, including protein variants and PTMs. For this purpose, we aim at promoting progress in this area by establishing a collaborative network of scientists that can exchange experiences, understand failure, start creating some initial standard guidelines about the performance of muscle biopsies, sample preparation and fractionation, quality control, statistical processing, and archival of muscle proteins so that we can start pulling data created by different laboratory and create the critical mass of information that is needed to address the substantial challenges posed by this field.
Funding
National Institutes of Health (NIH) (grants R01 AG027012); Intramural Research Program (IRP) of the National Institute on Aging (NIA).
Conflict of interest
C.D. is a full‐time employee of Thermo Scientific. The rest of the authors have declared no conflict of interest.
Supporting information
Table S1. Legend X: Present Compiled Skeletal Muscle Proteome.
Legend X: Present Compiled Skeletal Muscle Proteome.
Acknowledgements
This work was support by NIH (grants R01 AG027012) and the National Institutes of Health (NIH), Intramural Research Program (IRP) of the National Institute on Aging (NIA). The authors certify that they comply with the ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2015.106
Gonzalez‐Freire, M. , Semba, R. D. , Ubaida‐Mohien, C. , Fabbri, E. , Scalzo, P. , Højlund, K. , Dufresne, C. , Lyashkov, A. , and Ferrucci, L. (2017) The Human Skeletal Muscle Proteome Project: a reappraisal of the current literature. Journal of Cachexia, Sarcopenia and Muscle, 8: 5–18. doi: 10.1002/jcsm.12121.
References
- 1. Janssen I, Heymsfield SB, Wang ZM, Ross R. Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J Appl Physiol (1985) 2000;89:81–8. [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez‐Freire M, de Cabo R, Studenski SA, Ferrucci L. The neuromuscular junction: aging at the crossroad between nerves and muscle. Front Aging Neurosci Frontiers 2014;6:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dam T‐T, Peters KW, Fragala M, Cawthon PM, Harris TB, McLean R, et al. An evidence‐based comparison of operational criteria for the presence of sarcopenia. J Gerontol A Biol Sci Med Sci 2014;69:584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McLean RR, Shardell MD, Alley DE, Cawthon PM, Fragala MS, Harris TB, et al. Criteria for clinically relevant weakness and low lean mass and their longitudinal association with incident mobility impairment and mortality: the foundation for the National Institutes of Health (FNIH) sarcopenia project. J Gerontol A Biol Sci Med Sci 2014;69:576–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anton SD, Woods AJ, Ashizawa T, Barb D, Buford TW, Carter CS, et al. Successful aging: advancing the science of physical independence in older adults. Ageing Res Rev 2015;24:304–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cesari M, Rolland Y, Abellan Van Kan G, Bandinelli S, Vellas B, Ferrucci L. Sarcopenia‐related parameters and incident disability in older persons: results from the “invecchiare in Chianti” study. J Gerontol A Biol Sci Med Sci 2015;70:457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meng Y, Wu H, Yang Y, Du H, Xia Y, Guo X, et al. Relationship of anabolic and catabolic biomarkers with muscle strength and physical performance in older adults: a population‐based cross‐sectional study. BMC Musculoskelet Disord 2015;16:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Su J, Ekman C, Oskolkov N, Lahti L, Strom K, Brazma A, et al. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle 2015;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi SM, Lee B‐M. Comparative safety evaluation of selective androgen receptor modulators and anabolic androgenic steroids. Expert Opin Drug Saf 2015;14:1773–1785. [DOI] [PubMed] [Google Scholar]
- 10. Velders M, Diel P. How sex hormones promote skeletal muscle regeneration. Sports Med 2013;43:1089–1100. [DOI] [PubMed] [Google Scholar]
- 11. Aebersold R, Bader GD, Edwards AM, van Eyk JE, Kussmann M, Qin J, et al. The biology/disease‐driven human proteome project (B/D‐HPP): enabling protein research for the life sciences community. J Proteome Res 2013;12:23–7. [DOI] [PubMed] [Google Scholar]
- 12. Bergstrom J. Muscle electrolytes in man. Determined by neutron activation analysis on needle biopsy specimens. Scand J Clin Lab Invest 1962;14:511–3. [Google Scholar]
- 13. Ohlendieck K. Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Rev Proteomics 2010;7:283–96. [DOI] [PubMed] [Google Scholar]
- 14. Parker KC, Walsh RJ, Salajegheh M, Amato AA, Krastins B, Sarracino DA, et al. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J Proteome Res 2009;8:3265–3277. [DOI] [PubMed] [Google Scholar]
- 15. Brinkmeier H, Ohlendieck K. Chaperoning heat shock proteins: proteomic analysis and relevance for normal and dystrophin‐deficient muscle. Proteomics Clin Appl 2014;8:875–95. [DOI] [PubMed] [Google Scholar]
- 16. Ohlendieck K. Proteomic identification of biomarkers of skeletal muscle disorders. Biomark Med 2013;7:169–86. [DOI] [PubMed] [Google Scholar]
- 17. Schiaffino S. Fibre types in skeletal muscle: a personal account. Acta Physiol (Oxf) 2010;199:451–63. [DOI] [PubMed] [Google Scholar]
- 18. Capitanio D, Viganò A, Ricci E, Cerretelli P, Wait R, Gelfi C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two‐dimensional gel electrophoresis. Proteomics 2005;5:2577–86. [DOI] [PubMed] [Google Scholar]
- 19. Burniston JG, Hoffman EP. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev Proteomics 2011;8:361–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalyani RR, Corriere M, Ferrucci L. Age‐related and disease‐related muscle loss: the effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol 2014;2:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohlendieck K. Proteomic profiling of skeletal muscle plasticity. Muscles Ligaments Tendons J 2012;1:119–26. [PMC free article] [PubMed] [Google Scholar]
- 22. Højlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, et al. Characterization of the human skeletal muscle proteome by one‐dimensional gel electrophoresis and HPLC‐ESI‐MS/MS. Mol Cell Proteomics 2008;7:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.23. Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, et al. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 2010;59:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol 2014;49:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 2013;280:4294–314. [DOI] [PubMed] [Google Scholar]
- 26. Gelfi C, De Palma S, Cerretelli P, Begum S, Wait R, Begum S. Two‐dimensional protein map of human vastus lateralis muscle. Electrophoresis WILEY‐VCH Verlag 2003;24:286–95. [DOI] [PubMed] [Google Scholar]
- 27. Staunton L, Zweyer M, Swandulla D, Ohlendieck K. Mass spectrometry‐based proteomic analysis of middle‐aged vs. aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int J Mol Med 2012;30:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baraibar MA, Gueugneau M, Duguez S, Butler‐Browne G, Bechet D, Friguet B. Expression and modification proteomics during skeletal muscle ageing. Biogerontology Springer Netherlands 2013;14:339–52. [DOI] [PubMed] [Google Scholar]
- 29. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue‐based map of the human proteome. Science American Association for the Advancement of Science 2015;347 1260419–9:. [DOI] [PubMed] [Google Scholar]
- 30. Morton JP, Kayani AC, McArdle A, Drust B. The exercise‐induced stress response of skeletal muscle, with specific emphasis on humans. Sports Med 2009;39:643–62. [DOI] [PubMed] [Google Scholar]
- 31. Gelfi C, Vasso M, Cerretelli P. Diversity of human skeletal muscle in health and disease: contribution of proteomics. J Proteomics 2011;74:774–95. [DOI] [PubMed] [Google Scholar]
- 32. Miljkovic N, Lim J‐Y, Miljkovic I, Frontera WR. Aging of skeletal muscle fibers. Ann Rehabil Med 2015;39:155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gelfi C, Viganò A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, et al. The human muscle proteome in aging. J Proteome Res 2006;5:1344–53. [DOI] [PubMed] [Google Scholar]
- 34. Berger MJ, Doherty TJ. Sarcopenia: prevalence, mechanisms, and functional consequences. Interdiscip Top Gerontol 2010;37:94–114, doi:10.1159/000319997. [DOI] [PubMed] [Google Scholar]
- 35. Theron L, Gueugneau M, Coudy C, Viala D, Bijlsma A, Butler‐Browne G, et al. Label‐free quantitative protein profiling of vastus lateralis muscle during human aging. Mol Cell Proteomics 2014;13:283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hittel DS, Hathout Y, Hoffman EP, Houmard JA. Proteome analysis of skeletal muscle from obese and morbidly obese women. Diabetes 2005;54:1283–8. [DOI] [PubMed] [Google Scholar]
- 37. Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 2013;17:162–84. [DOI] [PubMed] [Google Scholar]
- 38. Holloway KV, O'Gorman M, Woods P, Morton JP, Evans L, Cable NT, et al. Proteomic investigation of changes in human vastus lateralis muscle in response to interval‐exercise training. Proteomics 2009;9:5155–5174. [DOI] [PubMed] [Google Scholar]
- 39. Egan B, Dowling P, O'Connor PL, Henry M, Meleady P, Zierath JR, et al. 2‐D DIGE analysis of the mitochondrial proteome from human skeletal muscle reveals time course‐dependent remodeling in response to 14 consecutive days of endurance exercise training. Proteomics 2011;11:1413–1428. [DOI] [PubMed] [Google Scholar]
- 40. Gondin J, Brocca L, Bellinzona E, D'Antona G, Muffiuletti NA, Miotti D, et al. Neuromuscular electrical stimulation training induces atypical adaptations of the human skeletal muscle phenotype: a functional and proteomic analysis. J Appl Physiol 2011;110:433–450. [DOI] [PubMed] [Google Scholar]
- 41. Hody S, Leprince P, Sergeant K, Renaut J, Croisier JL, Wang F, et al. Human muscle proteome modifications after acute or repeated eccentric exercises. Med Sci Sports Exerc 2011;43:2281–2296. [DOI] [PubMed] [Google Scholar]
- 42. LeBlanc AD, Schneider VS, Evans HJ, Pientok C, Rowe R, Spector E. Regional changes in muscle mass following 17 weeks of bed rest. J Appl Physiol (1985) 1992;73:2172–8. [DOI] [PubMed] [Google Scholar]
- 43. Moriggi M, Vasso M, Fania C, Capitanio D, Bonifacio G, Salanova M, et al. Long term bed rest with and without vibration exercise countermeasures: effects on human muscle protein dysregulation. Proteomics 2010;10:3756–74. [DOI] [PubMed] [Google Scholar]
- 44. Brocca L, Cannavino J, Coletto L, Biolo G, Sandri M, Bottinelli R, et al. The time course of the adaptations of human muscle proteome to bed rest and the underlying mechanisms. J Physiol 2012;590:5211–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynafarje B. Myoglobin content and enzymatic activity of human skeletal muscle – their relation with the process of adaptation to high altitude. Tech Doc Rep SAMTDR USAF Sch Aerosp Med. 1962;SAM‐TDR‐62‐89:8p [DOI] [PubMed]
- 46. Mason S, Johnson RS. The role of HIF‐1 in hypoxic response in the skeletal muscle. Adv Exp Med Biol 2007;618:229–44. [DOI] [PubMed] [Google Scholar]
- 47. Hoppeler H, Vogt M. Muscle tissue adaptations to hypoxia. J Exp Biol 2001;204:3133–9. [DOI] [PubMed] [Google Scholar]
- 48. Lundby C, Calbet JAL, Robach P. The response of human skeletal muscle tissue to hypoxia. Cell Mol Life Sci 2009;66:3615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murray AJ. Metabolic adaptation of skeletal muscle to high altitude hypoxia: how new technologies could resolve the controversies. Genome Med 2009;1:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Viganò A, Ripamonti M, De Palma S, Capitanio D, Vasso M, Wait R, et al. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics 2008;8:4668–79. [DOI] [PubMed] [Google Scholar]
- 51. Gelfi C, De Palma S, Ripamonti M, Wait R, Eberini I, Bajracharya A, et al. New aspects of altitude adaptation in Tibetans: a proteomic approach. FASEB J 2004;18:612–614. [DOI] [PubMed] [Google Scholar]
- 52. Mora‐Rodriguez R, Del Coso J, Estevez E. Thermoregulatory responses to constant versus variable‐intensity exercise in the heat. Med Sci Sports Exerc 2008;40:1945–52. [DOI] [PubMed] [Google Scholar]
- 53. Fernandez‐Elias VE, Hamouti N, Ortega JF, Mora‐Rodriguez R. Hyperthermia, but not muscle water deficit, increases glycogen use during intense exercise. Scand J Med Sci Sports 2015;25:126–34. [DOI] [PubMed] [Google Scholar]
- 54. Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature 2000;404:652–60. [DOI] [PubMed] [Google Scholar]
- 55.1. Wijers SLJ, Schrauwen P, Saris WHM, van Marken Lichtenbelt WD. Human skeletal muscle mitochondrial uncoupling is associated with cold induced adaptive thermogenesis. PLoS One 2008;3:e1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wijers SLJ, Smit E, Saris WHM, Mariman ECM, van Marken Lichtenbelt WD. Cold‐ and overfeeding‐induced changes in the human skeletal muscle proteome. J Proteome Res 2010;9:2226–35. [DOI] [PubMed] [Google Scholar]
- 57. Akhmedov D, Berdeaux R. The effects of obesity on skeletal muscle regeneration. Front Physiol 2013;4:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Giebelstein J, Poschmann G, Højlund K, Schechinger W, Dietrich JW, Levin K, et al. The proteomic signature of insulin‐resistant human skeletal muscle reveals increased glycolytic and decreased mitochondrial enzymes. Diabetologia 2012;55:1114–1117. [DOI] [PubMed] [Google Scholar]
- 59. Kalyani RR, Corriere M, Ferrucci L. Age‐related and disease‐related muscle loss: the effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol 2014;2:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petersen KF, Dufour S, Savage DB, Bilz S, Solomon G, Yonemitsu S, et al. The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proc Natl Acad Sci U S A 2007;104:12587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Højlund K. Metabolism and insulin signaling in common metabolic disorders and inherited insulin resistance. Dan Med J 2014;61:B4890. [PubMed] [Google Scholar]
- 62. Højlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, et al. Proteome analysis reveals phosphorylation of ATP synthase β‐subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J Biol Chem 2003;278:10436–42. [DOI] [PubMed] [Google Scholar]
- 63. Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Højlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 2010;59:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Colberg SR, Sigal RJ, Fernhall B, Regensteiner JG, Blissmer BJ, Rubin RR, et al. Exercise and type 2 diabetes: the American College of Sports Medicine and the American Diabetes Association: joint position statement. Diabetes Care 2010;33:e147–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hussey SE, Sharoff CG, Garnham A, Zhengping Y, Bowen BP, Mandarino LJ, Hargreaves M. Effect of exercise on the skeletal muscle proteome in patients with type 2 diabetes. Med Sci Sports Exerc 2013;45:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sela I, Krentsis IM, Shlomai Z, Sadeh M, Dabby R, Argov Z, Ben‐Basset H, Mitrani‐Rosenbaum S, The proteomic profile of hereditary inclusion body myopathy . PLoS One 2011;6:e16334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Feldkirchner S, Schessl J, Müller S, Schoser B, Hanisch FG. Patient‐specific protein aggregates in myofibrillar myopathies: laser microdissection and differential proteomics for identification of plaque components. Proteomics 2012;12:3598–3609. [DOI] [PubMed] [Google Scholar]
- 68. Maerkens A, Kley RA, Olivé M, Theis V, van der Ven PF, Reimann J, Milting H, Schreiner A, Uszkoreit J, Eisenacher M, Barkovits K, Güttsches AK, Tonillo J, Kuhlmann K, Meyer HE, Schröder R, Tegenthoff M, Fürst DO, Müller T, Goldfarb LG, Vorgerd M, Marcus K. Differential proteomic analysis of abnormal intramyoplasmic aggregates in desminopathy. J Proteomics 2013;90:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kley RA, Maerkens A, Leber Y, Theis V, Schreiner A, van der Ven PFM, Uszkoreit J, Stephan C, Eulitz S, Euler N, Kirschner J, Müller K, Meyer HE, Tegenthoff M, Fürst DO, Vorgerd M, Müller T, Marcus K. A combined laser microdissection and mass spectrometry approach reveals new disease relevant proteins accumulating in aggregates of filaminopathy patients. Mol Cell Proteomics 2013;12:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Palma S, Capitanio D, Vasso M, Braghetta P, Scotton C, Bonaldo P, Lochmüller H, Muntoni F, Ferlini A, Gelfi C. Muscle proteomics reveals novel insights into the pathophysiological mechanisms of collagen VI myopathies. J Proteome Res 2014;13:5022–5030. [DOI] [PubMed] [Google Scholar]
- 71. Conti A, Riva N, Presca M, Iannaccone S, Cannistraci CV, Corbo M, Previtali SC, Quattrini A, Alessio M. Increased expression of myosin binding protein H in the skeletal muscle of amyotrophic lateral sclerosis patients. Biochim Biophys Acta 1842;2014:99–106. [DOI] [PubMed] [Google Scholar]
- 72. Elf K, Shevchenko G, Nygren I, Larsson L, Bergquist J, Askmark H, Artemenko K. Alterations in muscle proteome of patients diagnosed with amytrophic lateral sclerosis. J Proteomics 2014;108:55–64. [DOI] [PubMed] [Google Scholar]
- 73. Poppe L, Rue L, Robberecht W, Van Den Bosch L. Translating biological findings into new treatment strategies for amyotrophic lateral sclerosis (ALS). Exp Neurol 2014;262:138–51. [DOI] [PubMed] [Google Scholar]
- 74. Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol 2006;7:391–403. [DOI] [PubMed] [Google Scholar]
- 75. Mann M, Jensen ON. Proteomic analysis of post‐translational modifications. Nat Biotechnol 2003;21:255–61. [DOI] [PubMed] [Google Scholar]
- 76. Jensen ON. Modification‐specific proteomics: characterization of post‐translational modifications by mass spectrometry. Curr Opin Chem Biol 2004;8:33–41. [DOI] [PubMed] [Google Scholar]
- 77. Lundby A, Secher A, Lage K, Nordsborg NB, Dmytriyev A, Lundby C, et al. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat Commun 2012;3:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao X, León IR, Bak S, Mogensen M, Wrzesinski K, Højlund K, Jensen ON. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics 2011;10 M110.000299:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhao X, Bak S, Pedersen AJT, Jensen ON, Højlund K. Insulin increases phosphorylation of mitochondrial proteins in human skeletal muscle in vivo . J Proteome Res 2014;13:2359–2369. [DOI] [PubMed] [Google Scholar]
- 80. Hoffman NJ, Parker BL, Chaudhuri R, Fisher‐Wellman KH, Kleinert M, Humphrey SJ, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise‐regulated kinases and AMPK substrates. Cell Metab 2015;22:922–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Giebelstein J, Poschmann G, Højlund K, Schechinger W, Dietrich JW, Levin K, Beck‐Nielsen H, Podwojski K, Stühler K, Meyer HE, Klein HH. The proteomic signature of insulin‐resistant human skeletal muscle reveals increased glycolytic and decreased mitochondrial enzymes. Diabetologia 2012;55:1114–1117. [DOI] [PubMed] [Google Scholar]
- 82. Anon . UniProt: a hub for protein information. Nucleic Acids Res 2015;43:D204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lane L, Argoud‐Puy G, Britan A, Cusin I, Duek PD, Evalet O, et al. neXtProt: a knowledge platform for human proteins. Nucleic Acids Res 2012;40:D76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics 2015;14:2394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tyanova S, Temu T, Carlson A, Sinitcyn P, Mann M, Cox J. Visualization of LC‐MS/MS proteomics data in MaxQuant. Proteomics 2015;15:1453–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pontén F, Jirström K, Uhlen M. The human protein atlas – a tool for pathology. J Pathol 2008;216:387–93. [DOI] [PubMed] [Google Scholar]
- 87.87. Lindskog C, Linné J, Fagerberg L, Hallström BM, Sundberg CJ, Lindholm M, et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody‐based profiling. BMC Genomics BioMed Central Ltd 2015;16:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Deshmukh AS, Murgia M, Nagaraj N, Treebak JT, Cox J, Mann M. Deep proteomicsof mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics 2015;14:841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Murgia M, Nagaraj N, Deshmukh AS, Zeiler M, Cancellara P, Moretti I, et al. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep 2015;16:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. GTEx Consortium. Human genomics . The genotype‐tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.91. Primmer CR, Papakostas S, Leder EH, Davis MJ, Ragan MA. Annotated genes and nonannotated genomes: cross‐species use of gene ontology in ecology and evolution research. Mol Ecol 2013;22:3216–41. [DOI] [PubMed] [Google Scholar]
- 92. The Gene Ontology Consortium . The gene ontology project in 2008. Nucleic Acids Res 2008;36:D440–4, doi:10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shi T, Zhou J‐Y, Gritsenko MA, Hossain M, Camp DG , Smith RD, et al. IgY14 and SuperMix immunoaffinity separations coupled with liquid chromatography‐mass spectrometry for human plasma proteomics biomarker discovery. Methods 2012;56:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Megger DA, Bracht T, Meyer HE, Sitek B. Label‐free quantification in clinical proteomics. Biochim Biophys Acta 1834;2013:1581–1590. [DOI] [PubMed] [Google Scholar]
- 95. Evans C, Noirel J, Ow SY, Salim M, Pereira‐Medrano AG, Couto N, Pandhal J, Smith D, Pham TK, Karunakaran E, Zou X, Biggs CA, Wright PC. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 2012;404:1011–1127. [DOI] [PubMed] [Google Scholar]
- 96. Hoedt E, Zhang G, Neubert TA. Stable isotope labeling by amino acids in cell culture (SILAC) for quantitative proteomics. Adv Exp Med Biol 2014;806:93–106. [DOI] [PubMed] [Google Scholar]
- 97. Michalski A, Damoc E, Hauschild J‐P, Lange O, Wieghaus A, Makarov A, et al. Mass spectrometry‐based proteomics using Q exactive, a high‐performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics 2011;10 M111.011015:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics 2012;11 O111.013698:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data‐independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11 O111.016717:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sun H, Qiu J, Chen Y, Yu M, Ding F, Gu X. Proteomic and bioinformatic analysis of differentially expressed proteins in denervated skeletal muscle. Int J Mol Med 2014;33:1586–1596. [DOI] [PubMed] [Google Scholar]
- 101. Narumi R, Tomonaga T. Quantitative analysis of tissue samples by combining iTRAQ isobaric labeling with selected/multiple reaction monitoring (SRM/MRM). Methods Mol Biol 2016;1355:85–101. [DOI] [PubMed] [Google Scholar]
- 102. Zhao Y, Jensen ON. Modification‐specific proteomics: strategies for characterization of post‐translational modifications using enrichment techniques. Proteomics 2009;9:4632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ma J, Hart GW. O‐GlcNAc profiling: from proteins to proteomes. Clin Proteomics 2014;11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Martiny VY, Carbonell P, Lagorce D, Villoutreix BO, Moroy G, Miteva MA. In silico mechanistic profiling to probe small molecule binding to sulfotransferases. PLoS One 2013;8:e73587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sprung RW Jr, Brock JW, Tanksley JP, Li M, Washington MK, Slebos RJ, Liebler DC. Equivalence of protein inventories obtained from formalin‐fixed paraffin‐embedded and frozen tissue in multidimensional liquid chromatography‐tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics 2009;8:1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2015. J Cachexia Sarcopenia Muscle 2015;6:315–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Legend X: Present Compiled Skeletal Muscle Proteome.
Legend X: Present Compiled Skeletal Muscle Proteome.


