Abstract
Significance: “Nitroproteomic” is under active development, as 3-nitrotyrosine in proteins constitutes a footprint left by the reactions of nitric oxide-derived oxidants that are usually associated to oxidative stress conditions. Moreover, protein tyrosine nitration can cause structural and functional changes, which may be of pathophysiological relevance for human disease conditions. Biological protein tyrosine nitration is a free radical process involving the intermediacy of tyrosyl radicals; in spite of being a nonenzymatic process, nitration is selectively directed toward a limited subset of tyrosine residues. Precise identification and quantitation of 3-nitrotyrosine in proteins has represented a “tour de force” for researchers.
Recent Advances: A small number of proteins are preferential targets of nitration (usually less than 100 proteins per proteome), contrasting with the large number of proteins modified by other post-translational modifications such as phosphorylation, acetylation, and, notably, S-nitrosation. Proteomic approaches have revealed key features of tyrosine nitration both in vivo and in vitro, including selectivity, site specificity, and effects in protein structure and function.
Critical Issues: Identification of 3-nitrotyrosine-containing proteins and mapping nitrated residues is challenging, due to low abundance of this oxidative modification in biological samples and its unfriendly behavior in mass spectrometry (MS)-based technologies, that is, MALDI, electrospray ionization, and collision-induced dissociation.
Future Directions: The use of (i) classical two-dimensional electrophoresis with immunochemical detection of nitrated proteins followed by protein ID by regular MS/MS in combination with (ii) immuno-enrichment of tyrosine-nitrated peptides and (iii) identification of nitrated peptides by a MIDAS™ experiment is arising as a potent methodology to unambiguously map and quantitate tyrosine-nitrated proteins in vivo. Antioxid. Redox Signal. 26, 313–328.
Keywords: : nitration, peroxynitrite, free radicals, protein oxidation, proteomics
Introduction
Immediately after the discovery of the beneficial signal-transducing functions of nitric oxide (•NO) in the vasculature and other systems (e.g., vasodilation and neurotransmission), it became evident that, when overproduced, •NO could also participate as a cytotoxic molecule with pathological effects [reviewed in Ignarro (52) and Radi (82)]. Much of •NO-mediated pathogenicity depends on the formation of secondary intermediates such as peroxynitrite anion (ONOO−) and nitrogen dioxide (•NO2) that are typically more reactive and toxic than •NO (82). The formation of reactive nitrogen species from •NO is generally associated with the formation of inflammatory environments and requires the presence of other oxidants such as superoxide anion radical (O2•−), hydrogen peroxide (H2O2), and transition metal centers, the concentration of which is usually increased at inflammatory sites that are characterized by the generation of a nitro-oxidative stress (75, 86). Nitrogen dioxide can also be formed in hydrophobic environments from the reactions of •NO with molecular oxygen, where these species concentrate (32, 62, 82).
One of the molecular footprints left by the reactions of reactive nitrogen species with biomolecules is the nitration (i.e., substitution of a hydrogen atom by a nitro group, -NO2) of protein tyrosine residues in the ortho position to the phenolic hydroxyl group to form 3-nitrotyrosine (Fig. 1). Target proteins can be nitrated by different biochemical pathways that result in the modification of specific tyrosine residues (4). Overall, biological protein tyrosine nitration is an oxidative post-translational modification (PTM) that involves a free radical process with the intermediacy of tyrosyl radicals (82); notably, it has been shown that in spite of being a nonenzymatic process, it is selectively directed under biologically relevant conditions toward a limited subset of tyrosine residues [i.e., typically only one or two tyrosine residues are targets in a nitrated protein under biologically relevant conditions, (84) and vide infra].
FIG. 1.
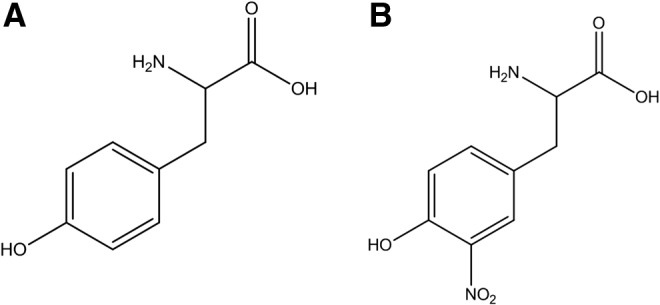
Molecular structures of (A) tyrosine and (B) 3-nitrotyrosine.
In addition to nitration, inflammatory conditions promote other oxidative modifications in tyrosine, such as chlorination, bromination, hydroxylation, and dimerization to 3-chloro-, 3-bromo-, 3-hydroxytyrosine, and 3,3′-dityrosine (82). Moreover, in proteins, another amino acid can be nitrated, namely tryptophan (5, 9, 29, 54, 55, 88), although the existing information and relevance of this modification remains preliminary and scarcely defined yet. The identification of these oxidative modifications is important not only because they may help in identifying preferential nitration pathways (82) but also because of technical difficulties that they may cause in the identification of tyrosine-nitrated proteins (87, 114). For a long time, tyrosine nitration was considered a stable PTM of proteins, although putative evidence on in vivo denitration processes has been provided (4, 43, 106), which requires further confirmation.
Physicochemical Properties and Biological Consequences of Tyrosine Nitration
Tyrosine nitration affects different physicochemical properties of the amino acid residue and of the corresponding protein: (i) causes a decrease in the pKa of the phenolic hydroxyl group, from 10.2 to ∼7.2 for the free amino acids in aqueous solution; this decrease will be affected by the protein environment (i.e., nearby presence of basic vs. acidic amino acid residues) and/or the properties of the milieu (i.e., less polarity will increase the pKa value) (91, 107, 122); (ii) changes the amino acid size by the incorporation of a substituent (30 Å3 larger) (124); (iii) changes light absorption with the appearance of a band centered at 360 nm at acidic pH and 420 nm at alkaline pH (107) (i.e., ionization of the phenol group in 3-nitrotyrosine causes a large red shift in absorptivity); (iv) influences the ionization process of 3-nitrotyrosine under certain mass spectrometry circumstances (4); and (v) increases the hydrophobicity of the nitrated peptide (14, 68). Even when the addition of a nitro group to a tyrosine residue confers particular physicochemical properties to the modified amino acid residue (and the corresponding protein), which may have important functional consequences (4, 82), the small fraction of nitrated protein has questioned its possible biological relevance (82). Protein tyrosine nitration is a process that does not occur randomly but clearly, as a selective process, which means that a relatively limited number of proteins are preferential targets of nitration (less than 110 nitrated proteins, and usually much less, were found in any of all the papers that aimed at identifying nitrated proteins by proteomics methods) (4). This “limited” number of nitrated proteins contrasts with other common PTMs of proteins, such as phosphorylation and acetylation. Enzymatic acetylation of the N-terminus of proteins occurs in 80–90% of higher eukaryotic proteins and acetylation of other lysine residues, most notably in histones and transcription, typically involves several hundreds of different proteins (45, 48, 49). In the same way, it is estimated that about 30% of the ∼30,000 proteins that were identified in the human proteome are substrates for phosphorylation (25). To date, acetylation is a very common and important PTM that regulates many important cell-signaling pathways. Actually, due to its frequency and important role in regulating the metabolism, acetylation has been proposed as a PTM that rivals phosphorylation (58). In the context of •NO-mediated PTMs, the small number of detected and reported tyrosine-nitrated proteins is in sharp contrast with the much larger number of reported S-nitrosated proteins (37, 98); indeed, the “S-nitrosoproteome” has been reported to involve at least 1000 of proteins (90, 98, 117). The, at least, 10-fold larger number of cysteine nitrosated versus tyrosine-nitrated proteins observed is probably due, among other possible factors, to the relatively chemical ease of S-nitrosation reactions. An integrated view of oxidative PTMs in proteins by •NO and •NO-derived oxidants under both homeostatic and stress conditions remains to be established.
Within the small number of nitrated proteins, only one or a few specific tyrosine residues were shown to be nitrated (14, 109, 110). Because the species participating in nitration reactions have short diffusion distances and particular formation sites, nitration reactions can be concentrated on proteins of either intra- or extracellular compartments (82). For example, under both physiological and pathological conditions, mitochondria typically contain larger amounts of nitrated proteins with respect to other cellular compartments, underscoring their role as a continuous source of peroxynitrite (63, 85, 95). On the other hand, in plasma, apolipoprotein A1 (ApoA1) in high-density lipoprotein has a much larger extent of tyrosine nitration than apolipoprotein B in low-density lipoprotein (34), mainly due to the fact that apoA1 binds one of the major extracellular sources of vascular nitrating species myeloperoxidase (MPO) (99, 100); the association of ApoA1-MPO under conditions of excess vascular •NO production promotes the site-specific nitration of Tyr 166 of apoA1 (34, 46, 73, 75, 99, 100).
Thus, although there is reportedly a small fraction of tyrosine-nitrated proteins, nitration can be focused on specific tyrosine/proteins, resulting in loss or gain of important protein functions (4, 82, 110). As mentioned earlier, incorporation of -NO2 group to a tyrosine lowers the pKa of its phenolic -OH by ∼3 pH units and adds a bulky substituent. If placed in a relevant tyrosine residue, nitration can alter protein function and conformation, modify the sensitivity for proteolytic degradation, impose steric restrictions, and inhibit tyrosine phosphorylation. However, to have biological significance, a loss-of-function modification requires a large fraction of protein to become nitrated at specific critical tyrosine residues and it is doubtful that many proteins will undergo such an extent of nitration, with one of the few already well-demonstrated examples being the case of Mn-SOD (16, 63, 64, 84). On the other hand, sometimes, tyrosine nitration results in a gain of function (82). In that case, only a small fraction of a particular nitrated protein can elicit a substantive, “new” biological signal. This initial attractive concept has been already shown in a few proteins such as cytochrome c, which acquires a strong peroxidase activity after nitration (14, 22, 23, 108, 110) and translocates into the cytosol without initiating apoptosis (41). In the same way, nitrated fibrinogen accelerates clot formation (116); protein kinase Cɛ becomes activated and translocates on nitration (8); peroxynitrite transforms nerve growth factor into an apoptotic factor for motor neurons (72); and α-synuclein gains activity due to nitro-oxidative modifications (110).
A remarkable example of loss of enzyme activity linked to nitration in vivo is the mitochondrial enzyme manganese superoxide dismutase (MnSOD). This protein is nitrated by peroxynitrite in Tyr 34 by an Mn-catalyzed process, which leads to enzyme inactivation (63, 64, 82, 119). MnSOD usually circumvents peroxynitrite formation by dismutating superoxide anion radical (O2•−). However, due to kinetic factors, such as the reaction of •NO with O2•− is diffusion controlled, none of the SODs can fully inhibit peroxynitrite formation. Thus, in the presence of sustained fluxes of •NO and O2•−, Mn SOD will be inactivated via peroxynitrite (31), which, in turn, may lead to a vicious circle, where an increase in peroxynitrite formation and/or reactions in mitochondria by any pathophysiological condition may lead to further nitro-oxidative damage (4). This has already been shown in an animal model of renal damage (27). Nitrated and inactivated MnSOD is found in acute and chronic inflammatory processes in both animal models and human diseases (63, 89). Endothelial cells are able to generate reactive oxygen and nitrogen species on exposure to cyclosporine A (CsA), leading to peroxynitrite formation (89). In this model, the authors reported, through the use of proteomic-based methods, that CsA induced nitration of MnSOD-Tyr34 with the concomitant inhibition of the enzyme; the authors were unable to quantitate the amount of nitration of MnSOD, and overexpression of the enzyme was necessary to reach the limit of detection for the Tyr34-containing nitrated peptide (89). It is possible that with the use of a nanoLC plus superior mass spectrometers for quantitation purposes (i.e., triple quadrupole or hybrid triple quadrupole linear ion trap machines), one may be able to quantitate the amount of nitrated MnSOD under the reported conditions. MnSOD nitration via the hemeperoxidase/•NO2 pathway does not lead to significant inactivation of the enzyme (30, 112), and it occurs at tyrosine residues that are more superficial than Tyr 34 such as solvent-exposed Tyr 9 and 11 (112). Interestingly, even in the case of the peroxynitrite-dependent nitration of the metal-depleted form of MnSOD (i.e., apoMnSOD), regio-selectivity at Tyr 34 is lost and overall nitration yield is increased, indicating that the metal center and protein conformation play key roles in defining the target residue and yields of the nitration process (80). Thus, the association of nitration plus inactivation of MnSOD may serve to shed light on the chemical nature of the nitrating species in vivo (82).
Even when tyrosine nitration has been implicated in the loss or gain of function of proteins, it should be noted that not all studies have shown unequivocally that tyrosine nitration is the main/unique cause for the gain or loss of function of the protein, since the nitrating agent and associated oxidants also modify other amino acid residues (i.e., cysteine, methionine, and tryptophan) (4). α-Synuclein is a particular example of the more general situation of an important protein that gains activity due to oxidative modifications (110).
Another important consequence of tyrosine nitration is the modification of the immunogenicity of the modified protein. It is well known that PTM of self-proteins may lead to the generation of new epitopes, triggering an immune response, which may be the cause of an autoimmune disease (4, 70). In apoptotic or inflamed tissues, a variety of PTMs of proteins (including nitrated) have been shown to accumulate (70). The accumulation of nitrotyrosine-containing autologous proteins in inflamed tissues has been reported; these appear as foreign proteins to the immune system and might induce an autoimmune response (4, 17). Elevated levels of anti-nitrotyrosine antibodies have, indeed, been determined in synovial fluid of patients with rheumatoid arthritis and osteoarthritis (57), as well as in the serum of patients with systemic lupus erythematosus (56). Moreover, it has been shown that intratumoral reactive nitrogen species production results in the nitration of the chemokine CCL2, a process associated with the prevention of the infiltration of antigen-specific T-cells to the inner core of the tumor (66).
Mechanisms and Selectivity in Protein Tyrosine Nitration
The nitration of protein tyrosine residues in biology is a free radical process. The best well-known nitration mechanism involves the initial one-electron oxidation of tyrosine to yield tyrosyl radical followed by a diffusion-controlled reaction with nitrogen dioxide (•NO2) to yield 3-nitrotyrosine (10, 82). Several oxidants may promote one-electron tyrosine oxidation and include carbonate radicals (CO3•−), high oxidation state of transition metal centers, hydroxyl radicals, and •NO2, as have been analyzed elsewhere (84). In lipid-rich structures such as biomembranes or lipoproteins, lipid peroxyl (LOO•) and alkoxyl (LO•) radicals can serve as proximal one-electron oxidants (13). In biological systems, •NO2 arises from a variety of sources, most notably the decomposition of peroxynitrite and the hemeperoxidase-dependent oxidation of nitrite (NO2−) (82). Nitrating species are also generated by NO2− under acidic conditions (i.e., HNO2) (93), which accounts for the nitration of gastric proteins, including the formation of nitrated pepsin (92). Regardless of the specific nitration process, protein 3-nitrotyrosine typically represents a fairly stable end product that is generated by the action of •NO-derived oxidants.
Considering the free radical nature of the nitration process, some caveats have to be indicated when correlating proteomic data obtained from exposure of proteins to chemical- versus biologically generated nitrating species. For example, exposure of proteins to a “bolus” of peroxynitrite [half-life of 0.8 s at pH 7,4 and 37°C (83)] can generate large initial concentrations of radical species in the bulk phase, which lead to a high initial level of tyrosyl radicals and, subsequently, 3-nitrotyrosine in solvent-exposed moieties. It is recommended that some of the experiments with peroxynitrite should be performed using a low flux of peroxynitrite and even those with the peroxynitrite donor SIN-1 should do so (14, 96), which better recapitulate biological conditions and provide a way to (i) minimize nonrelevant radical reactions and (ii) allow intramolecular electron transfer processes to modulate sites and extents of tyrosine nitration (e.g., tyrosyl radical repair by adjacent cysteine residue) (11–13, 65).
Tyrosine is usually an abundant amino acid in proteins, and its content typically ranges from about 3% to 4% (10). However, under biologically relevant levels of nitrating species, very few tyrosine residues become nitrated, making nitration a selective free radical process. Factors influencing regio-selectivity include the protein structure, the nature of the proximal nitrating species or mechanisms, the redox environment, and the physicochemical properties of the reaction milieu, all of which will impact the nitrated protein residue (4, 14) (Table 1). The influence of the protein structure on the regio-selectivity of Tyr nitration and of some predictive models on protein nitration sites have been communicated recently elsewhere (4, 12, 15, 84). The overall conclusion is that although there are some factors that favor effective and selective tyrosine nitration, the complexity of the process requires, from a proteomic perspective, a case-by-case analysis, which also assists in revealing the nature of the proximal nitrating species.
Table 1.
Reported Factors Influencing Protein Tyrosine Nitration Regio-Selectivity and Yields
| Factor | Elements | Remarks | Examples | Reference |
|---|---|---|---|---|
| Protein structure | ||||
| Loops | Nearby turn-inducing amino acids (Pro, Gly) favor nitration | Y115, Y76 in RNAse A Y20, Y23 in lysozyme |
(109) | |
| Presence of charged amino acids | Usually, the presence of acid or basic amino acids favors nitration with the participation of hydrogen bond bridges. | Y20, Y23 in lysozyme | (15, 109) | |
| Nearby cysteine residues | Inhibit nitration reactions via tyrosyl radical repair or consumption of nitrogen dioxide | Y25, Y92, and Y97, which are not nitrated in RNAse A | (109) | |
| Tyrosine nitration can be inhibited due to intramolecular electron transfer reactions between Cys and Tyr residues. | Y35 in Fe-SODB (T. cruzi) Cys-Tyr peptides | (18, 38, 65) | ||
| Nearby methionine residues | Tyrosine nitration can be enhanced due to intramolecular electron transfer reactions. | Met-Tyr peptides | (44) | |
| Location of tyrosine residues | Nitration in buried tyrosine residues is hindered if they cannot accommodate the nitro group. | (15) | ||
| Exposure of the aromatic ring to the protein surface | Y76 in RNAse A | (109) | ||
| Electrostatic forces | The presence of nearby positively charged amino acids such as arginine may inhibit nitration due to electrostatic forces. | Y20 in lysozyme has an electrostatic interference with R21. | (109) | |
| Transition metal centers (Fe, Mn, Cu) | Promote peroxynitrite-dependent nitration | Y34 in MnSOD | (64, 80, 89, 119) | |
| Y430 in prostacyclin synthase | (24) | |||
| Hemeperoxidase-binding sites | Promote hemoperoxidase-dependent nitration | Y18, Y166, and Y192 in apoA1 | (34, 99) | |
| Y115 and RNAse A | (109) | |||
| Heme properties and microenvironment | Some hemes promote peroxynitrite-mediated nitration via intermediate formation of oxidizing oxo-heme(IV) species. | Y430 in prostacyclin synthase Y99, Y347, and Y430 in CytC P450 |
(24, 123) | |
| Other hemes inhibit peroxynitrite-depedent nitration via its isomerization to nitrate; thus, nitration of these proteins likely reflects a peroxynitrite-independent mechanism. | Oxyhemoglobin plant leghemoglobin | (67, 96) | ||
| Consensus sequence (lack of) | The existence of a consensus sequence for nitration has not been demonstrated, with the secondary and tertiary structures being the most important factors to determine selectivity rather than a sequence homology. | (15, 109) | ||
| Nitration mechanism | ||||
| Peroxynitrite dependent | Regio-specific nitration by transition metals | Y34 MnSOD | (64, 80, 89, 119) | |
| Nitration of solvent-exposed tyrosines in the presence of CO2 | Y48, Y74, and Y97 in CitC | (14) | ||
| Promotes nitration of tyrosines that are associated to hydrophobic biostructures | Transmembrane KALP spanning peptides Y294, Y295 in SERCA | (13, 50, 118) | ||
| Hemeperoxidase dependent | Mainly directed toward solvent-exposed residues | Y9, Y11 in MnSOD Y18 in apoA1 | (112) | |
| Redox environment | ||||
| Endogenous antioxidants | Glutathione inhibits nitration by a combination of mechanisms, with the most relevant being nitrogen dioxide consumption. | (20, 21, 36) | ||
| Ascorbate inhibits nitration by interactions with oxidizing/nitrating intermediates and also by repair of the tyrosyl radical back to tyrosine. | (40, 51) | |||
| Uric acid is a strong inhibitor of nitration by consuming oxidizing/nitrating intermediates and by interfering in the catalytic cycle of heme peroxidases. | (20, 105) | |||
| Lipid peroxidation processes | Fuel nitration reactions in proteins associated to hydrophobic biostructures by promoting the one-electron oxidation of tyrosine | (10, 12, 13) | ||
| Physicochemical properties of the milieu | ||||
| Hydrophobicity | Limits the diffusion of charged reactive species such as carbonate radicals | (94) | ||
| Excludes hydrophilic anti-nitrating compounds; reactive species such as nitrogen dioxide can concentrate and live longer. | (10, 104) | |||
| pH | pH changes influence peroxynitrite-dependent nitration yields. | (12) | ||
| Acidic conditions favor nitrite-dependent nitration. | Pepsin | (92, 93) | ||
Analytical Methods for the Detection of 3-Nitrotyrosine
The chemical structure of the analyzed species is very important for the development of any analytical method. In that sense, as stated earlier (114), 3-nitrotyrosine is very unusual, because even though it is a small molecule (molecular mass 226.19 g/mol), it has four functional groups: (i) an alpha acid carboxylic group; (ii) an alpha amino group; (iii) a phenolic group; and (iv) a nitro group at the ortho position of the aromatic group. These structural features have been used to develop different methods for the qualitative and quantitative analysis of 3-nitrotyrosine (114). Some approaches are based on chemical modification of one or more of these functionalities to improve analytical performance (e.g., detector response, volatility, chromatographic properties), whereas others target the unmodified form (114). 3-nitrotyrosine in different forms (i.e., native/unlabeled or stable-isotope labeled) has been extensively analyzed by liquid chromatography–mass spectrometry (LC-MS) and liquid chromatography–tandem mass spectrometry (LC-MS/MS) in both the positive and negative electrospray ionization (ESI) modes, and it has been recently reviewed elsewhere (114) (Table 2). In ESI+, protonated native 3-nitrotyrosine ([M+H]+ = m/z 227) produces few significant product ions when fragmented by collision-induced dissociation (CID). Some of these product ions have been used for quantitative analyses, and the use of the immonium ion of nitroTyr (m/z 181.06) is remarkable as it requires a relatively low collision energy to be produced and gives a very good signal (114). In ESI- mode, CID of the deprotonated unlabeled 3-nitrotyrosine ([M-H]− = m/z 225) gives a product ion at m/z 163 and this has been used in quantitative analyses (81). Protein-bound nitro-tyrosine has been related with the risk of coronary artery disease (CAD) (101–103). The authors reported that protein-bound nitrotyrosine levels in plasma were significantly higher among patients with CAD (median 9.1 μmol nitro-tyrosine/mol tyrosine [interquartile range, 4.8–13.8 μmol/mol] vs. 5.2 μmol nitro-tyrosine/mol tyrosine [interquartile range, 2.2–8.4 μmol/mol]; p < 0.001) (101) (Table 2).
Table 2.
Concentration of Free and Protein-Bound 3-Nitrotyrosine in Human Biological Samples Measured by Validated Liquid Chromatography–Tandem Mass Spectrometry Methods
| Matrix | Source of 3NT | Concentration | LLOD/LLOQ | Reference |
|---|---|---|---|---|
| Plasma | Free | 4.4 nM | 1 fmol/4.4 nM | (121) |
| Plasma | Free | ND | 3.2 fmol/1.6 nM | (39) |
| Urine | Free | <1.6 nmol/mmol creatinine | 25 nM/NR | (71) |
| EBC | Free | 40–1000 pM | LLOQ 40 pM | (42) |
| Urine | Free | 0.3–3 nM | 30 pM/NR | (81) |
| Plasma | Free | NR | 30 pM/NR | (81) |
| Plasma | Free | 1.5 nM | 34 pM/625 pM | (47) |
| Urine | Free | 0.17 ± 0.16 nM | 8.8 pM/44 pM | (59, 60) |
| Plasma | Free | 14 ± 0.7 ng/ml | (38) | |
| Urine | Protein-bound | 60–800 pg/ml | 30 pM/NR | (81) |
| Plasma | Protein-bound | 200–1000 pg/ml | 30 pM/NR | (81) |
| Plasma | Protein-bound | 9.1 μmol nitro-tyrosine/mol tyrosine in patients with CAD | 100 fmol/NR | (69, 101) |
| Lung Tissue | Protein-bound | 79 ± 8 μmol nitro-tyrosine/mol tyrosine in proteins recovered from lung tissue after aeroallergen challenge | 100 fmol/NR | (18, 69) |
Adapted and updated from Tsikas and Duncan (114).
CAD, coronary artery disease; ND, not detectable; NR, not reported.
In our group, 3-nitrotyrosine determinations have been regularly performed according to the method described (65, 69). Basically, the quantification of nitroTyr in biological matrices was performed using an LC-MS approach (triple quad-ion trap MS/high-performance liquid chromatography) with the use of stable isotopically labeled precursors as synthetic internal standards, thus considering the intra-preparative sample losses and monitoring potential artifactual generation of nitroTyr during sample preparation (69). The described method allows rapid and reproducible quantification of nitroTyr in biological and clinical samples at the 100 fmol on column detection limit (69).
Proteomic Methods to Study Tyrosine-Nitrated Proteins
There are at least two main situations where researchers may need to identify tyrosine-nitrated proteins and, moreover, the tyrosine residue(s) that is (are) being nitrated; we will discuss the general strategies we have used to resolve both of them.
In the first case, probably the “simplest” situation, one can imagine the case of a pure protein that is being treated “in vitro” by one or more different nitrating agents. That was the case when we first analyzed the nitration of cytochrome c by tetranitromethane (TNM) and peroxynitrite in the presence or absence of bicarbonate (14). Cytochrome c contains four highly conserved tyrosine residues, three of which are highly exposed to the solvent (Tyr 48, Tyr 74, and Tyr 97). To analyze the feasibility of each tyrosine to become nitrated, we analyzed the time course and site(s) of tyrosine nitration in horse cytochrome c by fluxes of peroxynitrite or TNM (14). In that case, we developed a cation-exchange high-performance liquid chromatography (HPLC) method for purifying each nitrated cytochrome c product. Then, each purified nitrated protein was digested and a peptide map was performed by reverse-phase HPLC with double detection of peptides at 220 and 360 nm. It should be noted that 3-nitrotyrosine could form an internal hydrogen bond between the nitro and the phenolic OH-group, causing its absorption properties to be strongly pH dependent (4, 28). At acidic pH (pH <6), as is usually the case for reverse-phase chromatography of peptides, 3-nitrotyrosine has an absorption maximum at 360 nm wavelength (25, 91), which has been used for the detection of 3-nitrotyrosine-containing peptides by HPLC with ultraviolet-visible spectroscopy under acidic conditions (4, 26, 53, 109).
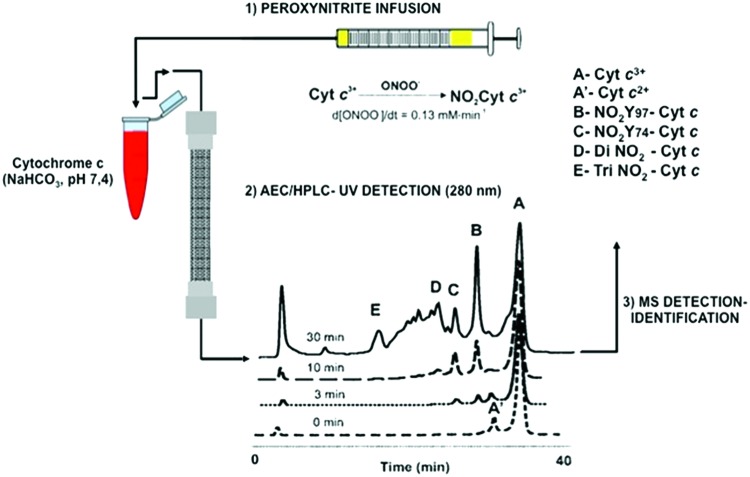
In our study, a flux of peroxynitrite caused a time-dependent formation of different nitrated species (Fig. 2). At low doses of peroxynitrite, the main products were two mono-nitrated cytochrome c at Tyr 97 and Tyr 74, as shown by peptide mapping and matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (MS) analysis (Fig. 2; peak B and C). At higher doses, almost all tyrosine residues in cytochrome c were nitrated, including dinitrated (i.e., Tyr 97 and Tyr 67 or Tyr 74 and Tyr 67) and trinitrated (i.e., Tyr 97, Tyr 74, and Tyr 67) forms of the protein. Interestingly, all mono-, di-, and trinitrated cytochrome c species displayed an increased peroxidase activity and were unable to restore the respiratory function of cytochrome c-depleted mitochondria (14). The nitration pattern of cytochrome c in the presence of TNM was comparable to that obtained with peroxynitrite, but with a relatively increased nitration yield at the buried Tyr 67. Interestingly, cytochrome c nitration by a “peroxidatic” mechanism involving the participation of its heme center (i.e., the initial reaction of H2O2 with the ferric heme) in the presence of either NO2− or •NO (23) likely leads to the predominant nitration of heme-adjacent and solvent-buried Tyr 67, further underscoring the influence of the nitration mechanism in regio-selectivity (Fig. 3). The yields and sites of cytochrome c nitration by the peroxidatic mechanism may be modulated by the presence of cardiolipin, a process that is under investigation in our laboratory.
FIG. 2.
Time course of peroxynitrite-mediated cytochrome c modification. Cytochrome c (1 mM) in 200 mM potassium phosphate buffer and 100 μM DTPA (pH 7.0) was treated with an infusion of peroxynitrite (0.13 mM/min). The reaction mixture (600 μg of protein) was analyzed at different time points by cation-exchange HPLC. HPLC-UV, high-performance liquid chromatography with ultraviolet-visible spectroscopy. Adapted from Batthyany et al. (14) and Hannibal et al. (44). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
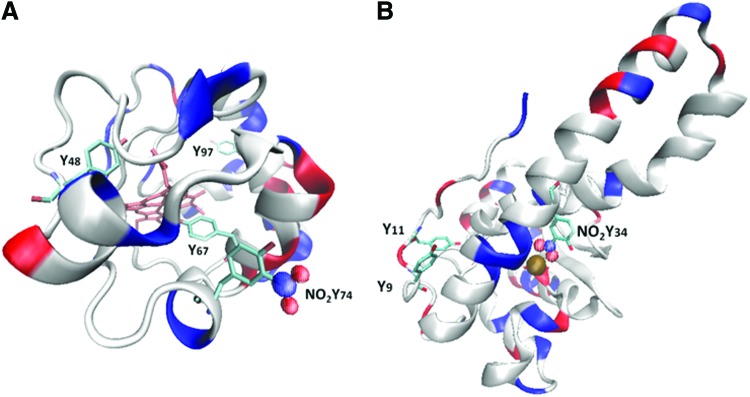
FIG. 3.
Three-dimensional structures of selected regio-specific tyrosine-nitrated proteins. (A) Structure of horse cytochrome c showing nitrated Tyr 74 (modified from pdb 1HRC). Both Tyr 74 and Tyr 97 are prone to nitration by peroxynitrite (14). The solvent buried Tyr 67 adjacent to the heme moiety is prone to nitration by peroxidase-dependent mechanisms (23, 24). Tyr 48, although not readily nitrated, has been found to be phosphorylated (123). The four described tyrosine residues in horse cytochrome c are highly conserved, whereas another tyrosine (Tyr 46) is present in human cytochrome c. (B) Structure of human manganese superoxide dismutase (hMnSOD) showing nitrated Tyr34 (pdb 2ADP). Tyr 34 is part of the superoxide radical entrance channel and active site, and its nitration leads to enzyme inactivation (67, 119). Also shown are surface-exposed Tyr 9 and Tyr 11, which undergo nitration by peroxidase-dependent mechanisms (112). hMnSOD also has Tyr at positions 45, 165, 166, 169, 176, and 193 that are not shown for the sake of clarity in the diagram. The structures were drawn using VMD Software (50). VMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Thus, the purification of mono-, di-, and tri-nitrated species by cation-exchange HPLC coupled with reverse-phase HPLC with peptide detection at 220/360 nm allowed us to better define the sensitivity of each Tyr residue to become nitrated (14, 22). The MALDI-TOF machine we used was equipped with a laser light that emits at 337 nm. Nitrotyrosine-containing peptides are sensitive to this wavelength light and undergo a decomposition process, yielding a unique triplet signal consisting of the expected mass increase of 45 amu for the addition of a nitro group (NO2-Tyr; [M + H+ + 45]) as well as other major peaks corresponding to nitrosotyrosine (NO-Tyr; [M + H+ + 29]) and aminotyrosine (NH2-Tyr; [M + H+ + 15]) (14, 97). In our study, the expected fragmentation pattern of nitrated tyrosine-containing peptides on the ionizations induced by the nitrogen laser of the MALDI-TOF mass spectrometer was also observed and allows us to further identify the nitrated peptides (14). This is due to the fact that the absorbance maximum of 3-nitrotyrosine under acidic conditions is ∼360 nm, which is in the vicinity of the emission wavelength of the nitrogen ion laser (337 nm) (4, 97).
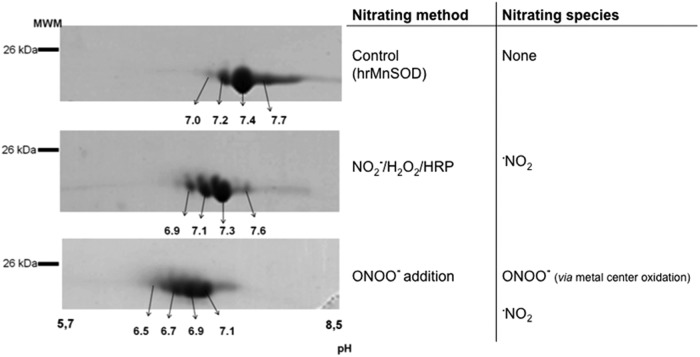
On the other hand, the analysis of the nitroproteome of tyrosine-nitrated proteins in more complex samples requires more complex analytical strategies. Up to now, the study of the “nitroproteome” by two dimensional-electrophoresis (2DE) has resulted in most of the identifications of known endogenously nitrated proteins (4). In our lab, the general strategy based on 2DE fractionation, followed by immunoblotting, in-gel digestion, and MS identification, has been widely used for nitroproteome studies of both single nitrated proteins (e.g., human MnSOD [hMnSOD], Fig. 4) (30) and more complex samples (19, 61, 65). Mn-SOD, an essential mitochondrial antioxidant enzyme, is nitrated and inactivated in vivo under a variety of conditions, leading to mitochondrial dysfunction and/or inflammation (30, 31, 63, 80, 119). The nitration of MnSOD in a critical tyrosine residue (Tyr 34) represents a prime example of an oxidative PTM in vivo that is significant and directly associated with a loss of function (63, 82, 119). hMnSOD contains a total of nine residues and depending on the nitrating agent and mechanism of nitration, one or more of them could be nitrated with a different probability (30, 112). In this regard, observed changes in isoelectric point of tyrosine-nitrated samples (Fig. 4) reflect the decrease in the pKa of each nitrated tyrosine and one should expect the generation of a new spot, more acidic, for each nitrated tyrosine in the protein. In MnSOD, four tyrosine residues were found to be nitrated (Tyr 2, Tyr 9, Tyr 11, and Tyr 34), and the distribution of the nitrated residues depends on the nitrating agent used. If peroxynitrite is the main nitrating agent, Tyr 34 is the primarily nitrated residue at low concentrations of peroxyntrite, with a prevalence of more than 80%, with functional consequences in the enzyme (30, 112). On the other hand, when nitrogen dioxide (•NO2) that is generated by the simultaneous presence of nitrite, H2O2, and a peroxidase is the nitrating agent, the main modified residues are Tyr 2, Tyr 9, and Tyr 11 in a process that has a rather modest impact on the enzyme activity (30, 112). This change in the profile of the nitrated residues affects the pI of the enzyme, where a more acidic pI are found in the enzyme nitrated with peroxynitrite (Fig. 4) (30). Actually, we had an early hint of this phenomenon in a 1D-native gel, when first studying the nitration of cytochrome c by peroxynitrite (22). Native gel electrophoresis showed that peroxynitrite caused a dose-dependent appearance of up to three different species, displaying decreased migration toward the cathode, indicating that each new species was more negatively charged, as expected for the various nitrated tyrosine-containing protein species (22).
FIG. 4.
Two dimensional-polyacrilamide gel electrophoresis (2D-PAGE) analysis of the effect of different nitrating agents on human recombinant MnSOD (hrMnSOD). hrMnSOD was exposed to nitrite and hydrogen peroxide in the presence of horseradish peroxidase (HRP) or peroxynitrite (ONOO−). Changes in isoelectric point are indicated with arrows. Reproduced and adapted from Demicheli et al. (30).
Even when in our experience this has been a very powerful strategy, we are aware that 2DE has several limitations that explain why the identified nitrated proteins are generally abundant and soluble ones. Moreover, sometimes, the major problem with this strategy is the failure to actually detect the nitrated tyrosine residue and this may be more likely due to: (i) limited protein load capability of the technique combined with the low levels of nitro-tyrosine on a given protein and the low abundance of the tyrosine-nitrated protein in a given sample, (ii) the recovery of nitrotyrosine-containing peptides from the gels and/or HPLC nanocolumns during subsequent nLC-MS/MS analysis (4, 19, 61, 65). It is also important to note that another important limitation of this strategy is its difficulty to analyze hydrophobic membrane proteins; this limitation becomes of particular relevance when studying tyrosine nitration in biological samples, since this PTM can be present in protein transmembrane domains and apolipoproteins (4, 10–13, 118, 120, 126, 127). Finally, since a spot may contain more than one protein (depending on the quality of the 2D-gel and the MS being used for the analysis, this number may vary between one and more than 10 proteins/spot), it is sometimes tricky to assign an immuno-positive spot on the blotting membrane to the identified protein. It is, therefore, mandatory to confirm the nitration site of the protein by MS. As an example of this concept, recently, it was found that in 57 immuno-positive anti-nitrotyrosine spots, more than 800 proteins were identified, but only 18 were actually nitrated (76). Possible explanations regarding this unexpected observation in which an excess of “nonnitrated” proteins were detected on immune-positive spots for protein 3-nitrotyrosine will be analyzed in the next section in the context of possible technical limitations.
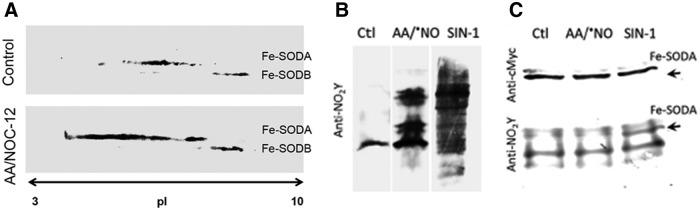
By using the 2DE-MS general strategy, using both an MALDI-TOF/TOF (4800 MALDI TOF/TOF Analyzer, Abi Sciex) and a nano-LC/nano spray ionization with a linear ion trap mass spectrometer (Easy nLC, Thermo; LTQ Velos, Thermo), we have also analyzed the nitration of Trypanosoma cruzi iron superoxide dismutases (Fe-SODs). The reason for studying this process is that, as was previously observed for other T. cruzi antioxidant enzymes (2), T. cruzi Fe-SODs may function as virulence factors in Chagas Disease (American trypanosomiasis), a hypothesis currently under investigation in our laboratory. It is important to note that during T. cruzi invasion to immunostimulated macrophages, T. cruzi proteins become nitrated inside the phagosome by cytotoxic levels of host cell-derived peroxynitrite (6). In this context, the nitration of parasite Fe-SOD in key tyrosine residues during host cell-T. cruzi interactions by •NO-derived oxidants (6, 74) may cause its inactivation and contribute to parasite cell death. In particular, nitration and inactivation of the mitochondrial isoform, Fe-SODA (65), would cause an increase in the intramitochondrial steady-state levels of superoxide radical anion and promote T. cruzi programmed cell death (78). The MALDI-TOF/TOF data allowed the identification of the nitration site of the protein (Fig. 5) (19, 61, 65), but we were unable to explain unequivocally each of the different pIs detected (Fig. 5), indicating how difficult it can be to identify tyrosine nitrated residues with the MS strategies currently available. The 2D-LC-MS analysis of the major spot (pI = 7.5) of Fe-SODB (cytosolic isoform) generated after peroxynitrite treatment (150 μM) revealed the presence of only one nitrated peptide (m/z = 790.6) corresponding to the triple charged molecular ion, assigned to the sequence 31HHQG35YVTKLNAAAQTNSALATK52, which contained the Tyr 35 (Fig. 5B; equivalent to human Tyr 34), thus revealing the preferential nitration of this residue by peroxynitrite under these experimental conditions (65). At higher concentrations of peroxynitrite (600–1000 μM), other peptides containing solvent-exposed Tyr residues were also found to be nitrated (65). In the case of Fe-SODA, the critical Tyr 35 was also preferentially nitrated after exposure to peroxynitrite (100 μM), but in this case, that residue was detected on a dinitrated peptide that also contains Tyr 29 (65). The proteomic data contributed toward demonstrating that the disparate susceptibilities of Tyr 35 in Fe-SOD A and B for nitration are due to the presence of Cys 83 in Fe-SOD B (absent in Fe-SOD A) that inhibits Tyr 35 nitration through an intramolecular electron transfer repair process of the tyrosyl radical intermediate (65). Actually, T. cruzi overexpressers (78, 113) were used to search for modifications of Fe-SOD occurring during nitroxidative stress conditions in living parasites (epimastigote stage). After the induction of Fe-SODA expression (4–6-fold increase with respect to wild type), parasites were incubated in the presence of antimycin A (complex III electron chain inhibitor) plus an •NO donor, to specifically generate peroxynitrite at the mitochondrial cell compartment (79). Afterward, parasite protein extracts were separated by Two Dimensional-Polyacrilamide Gel Electrophoresis and probed with anti-Fe-SODs antibodies. The pI changes observed in Fe-SODA obtained from living parasites during exposure to either endogenous (AA/•NO) or exogenous peroxynitrite were similar to those observed for the recombinant Fe-SODA after in vitro peroxynitrite treatment (65). Notably, Fe-SODB was not significantly altered under these cellular nitroxidative conditions, in agreement with its high resistance to peroxynitrite (65) (Fig. 6).
FIG. 5.
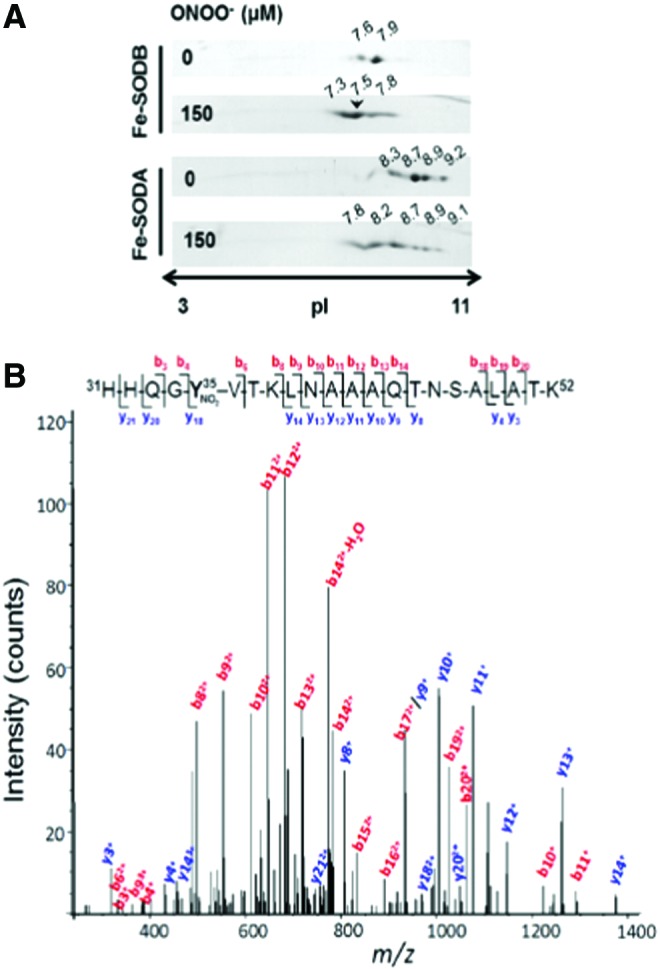
Peptide mapping of Trypanosoma cruzi iron superoxide dismutases (Fe-SODs) after peroxynitrite treatment. (A) T. cruzi Fe-SODs were treated with an excess of peroxynitrite in sodium phosphate buffer (0.2 M) at pH 7.4 and 25°C. Two-dimensional gel electrophoresis was performed as previously described (19, 61, 65). The arrowhead shows the selected Fe-SODB spot (pI 7.5) that was further analyzed by mass spectrometry (MS). (B) MS/MS spectrum of the triply charged ion at m/z 790.6 (M + H+ = 2369.7; retention time 30.3 min) indicated with an arrow in (A). The major N-terminal (b-labeled) and C-terminal (y-labeled) fragment ions that allowed the sequence 31–52 assignment that includes a nitrated tyrosine residue (mascot ion score = 61; p < 0.05) are shown. Inset: amino acid sequence of peptide 31–52, indicating major b and y ions detected by full-scan MS/MS taken from Martinez et al. (65). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 6.
Cellular detection of Fe-SODA nitroxidative modifications. (A) 2D-PAGE. T. cruzi Fe-SODA overexpressers were treated with AA (5 μM) plus NOC-12 (5 mM; t1/2 100 min at pH 7.4), SIN-1 (5 mM), or ONOO− (300 μM) for 3 h at room temperature. After treatment, samples were processed and subjected to 2D-PAGE. Membranes were probed with the specific Fe-SODA and Fe-SODB antibodies. (B) 3-nitrotyrosine detection in parasites. After treatment in the conditions described in A, parasite extracts were separated in 15% SDS PAGE gels and transferred to nitrocellulose membranes. Membranes were probed with the specific anti-3-nitrotyrosine antibody. (C) Immunoprecipitation of nitrated Fe-SODA. Parasite extracts as described earlier were incubated overnight at 4°C in the presence of the monoclonal c-Myc antibody that recognized the 9E10 epitope of Fe-SODA in the presence of protein A/G-agarose. Immunoprecipitated proteins were run in 15% SDS-PAGE gel, electro-transferred to nitrocellulose, and revealed using anti-c-Myc antibody and anti-3-nitrotyrosine antibody [taken from Martinez et al. (65)].
Proposal of an Alternative Method to Improve the Sensitivity and Selectivity for the MS-Analysis of Tyrosine-Nitrated Proteins
Precise quantitation and identification of nitrated tyrosine residues in proteins can be a “tour de force” for researchers working in the redox biology and chemistry area: The difficulties probably arise from the relatively small number of nitrated tyrosine versus total tyrosine residues (meaning the presence of one modified peptide per a large number of native ones), hydrophobicity of tyrosine-nitrated peptides, instability of the modification when analyzed by MALDI ionization, and other still unknown factors. To partially solve these problems, we consider that a good analytical approach should be the combination of the best orthogonal methods to separate proteins and peptides in tandem, with the more sensitive MS-based method to detect nitrated tyrosine peptides. In that sense, the “ideal” method should separate total proteins by 2D electrophoresis followed by immunodetection of Tyr-nitrated spots. Regarding this approach, one should carefully analyze the specificity of the anti-nitrotyrosine antibody and also remember that the identification of the spot is carried out on a membrane after transferring the protein from the gel, and the in-gel digestion is performed on the gel. Thus, there are at least two technical issues (i.e., antibody specificity and mistake when selecting the superimpose place to cut) that may affect the quality and robustness of the experimental result (76).
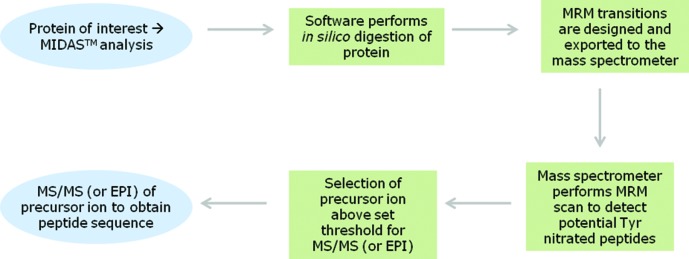
Proteins present in those spots (it is always important to remember that one single, well-resolved spot in a 2D gel typically contains more than one protein) should be identified by traditional MS methods (61, 76, 114). Once all the proteins present in that spot have been identified, one should be able to predict, in silico (software assisted) (114), all the possible Tyr-containing tryptic peptides (114) and apply a targeted mass spectrometric workflow to the sensitive identification of nitrated Tyr residues (115). The MIDAS™ (multiple reaction monitoring [MRM] initiated detection and sequencing or MRM-triggered MS/MS method) MS strategy employs MRM to search for all putative peptides that are specifically modified in a target protein. Positive MRMs above a preset threshold trigger an MS/MS (or an enhanced product information [EPI]) experiment to confirm the nature and site of the modification (2, 115). The selection of which fragment ions are the best option to use for both Q1 and Q3 for the MRM experiment is either based on previously acquired MS/MS identifications or predicted in silico based on the peptide sequence (1, 3). With these theoretical data, researchers may use a triple quadrupole MS or a hybrid triple quadrupole-ion trap MS to run the MIDAS experiment method to detect any possibly Tyr-nitrated peptide in that spot (2), obtaining the best sensitivity for both ID and quantitation of the peptide (Fig. 7). To further confirm the nitration of the peptide/protein, it may be wise to perform an EPI of the detected peptide to obtain all the MS/MS data to unequivocally confirm the nitration site. In this sense, the use of an ESI source-based MS method to identify nitrotyrosine-containing peptides has several advantages compared with an MALDI source: (i) ESI does not induce the characteristic photochemical decomposition of nitrotyrosine produced under MALDI conditions; (ii) under ESI conditions, nitrotyrosine fragmentation produces an immonium ion (m/z 181.06) that is a very useful footprint of a nitroTyr-containing peptide. Thus, the presence of this footprint in the EPI experiment will increase the confidence of the identification.
FIG. 7.
MIDAS™ MS strategy for the detection of Tyr-nitrated residues and peptides. EPI, enhanced product information; MRM, multiple reaction monitoring. Adapted from Unwin et al. (115). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Also, the formation of this immonium ion allows the design of another alternative method to increase the sensitivity of the identification/quantitation of nitrated tyrosine-containing peptides. In this case, researchers may use the precursor ion scanning mode to search for the immonium ion for nitrotyrosine, which will specifically identify a 3-nitrotyrosine-containing peptide/protein (77). Unfortunately, as stated elsewhere (125), a huge amount of non-nitrated tryptic peptides will negatively affect the characterization and identification of nitropeptides. For that reason, it is highly recommended to associate an immunoaffinity enrichment method for nitropeptides before the MS analysis.
Conclusions and Perspectives
The area of “nitroproteomics” continues to be under active development and has revealed to be quite challenging in technical terms. Proteomic approaches have revealed key features of protein tyrosine nitration both in vivo and in vitro, including its selectivity in terms of nitrated proteins and regio-specificity of nitrated residues. Of note is the relatively limited number of nitrated proteins detected under both normal and oxidative stress conditions, in comparison to other PTMs, including the NO-dependent process of S-nitrosylation, even when nitration is a more stable PTM than S-nitrosylation. However, it is challenging to identify, map, and quantitate nitrotyrosine-containing proteins due to the low abundance of this oxidative modification in biological samples and its unfriendly behavior in MS-based technologies, that is, MALDI, ESI, and CID (125).
Tyrosine-nitrated proteins are detected in cells and tissues under normal physiological conditions, revealing a basal continuous flux of nitrating species; the levels of nitrated proteins are strongly increased under conditions of enhanced formation of •NO and/or oxidants, and they are typically strongly associated with disease onset and progression. The differential nitration of proteins under basal and pathological conditions remains to be clearly established, although it is important to consider that nitrated proteins are typically contained in the same subcellular or extracellular compartment where nitrating species are formed due to the short diffusion distances and biological half-life of species such as peroxynitrite and •NO2. Although most of the current available proteomic information on nitrated proteins is concentrated in mammalian systems, nitrated proteins are also found in plants that typically have an active redox nitrogen metabolism and some proteomic analysis with identification of regio-specific tyrosine nitrated residues have been recently published (96). Proteomic analysis of nitrated proteins in intracellular pathogens such as Escherichia coli (35), Mycobacterium tuberculosis (111), and T. cruzi (6, 7, 33) secondary to the release of reactive nitrogen intermediates to neutrophils and macrophage phagosomes as part of the cytotoxic cellular immune response awaits analysis.
The combined use of 2D gel electrophoresis with immunochemical detection of 3-nitrotyrosine followed by identification of proteins by regular MS/MS approaches plus the immuno enrichment of Tyr-nitrated peptides and the ID of those nitrated peptides by a MIDAS experiment (see before) is arising as a potent methodology to undoubtedly characterize and quantitate tyrosine-nitrated proteins in vivo. For this approach, the use of a highly specific and sensitive antibody, well characterized, is a key element in the quality and confidence of the results (19). Finally, MS-based proteomics, independently of all the other factors that may affect the quality of the results (i.e., sample preparation, mass spectrometer use, general methodological strategy used, etc.), always depends on the software that will be used to analyze and interpret the data; this is also of particular relevance when analyzing tyrosine-nitrated peptides/proteins.
Proteomic approaches to study protein tyrosine nitration alone or in combination with immunochemical and/or bioanalytical methods represent potent tools for (i) identification of protein targets and molecular footprints in a variety of pathological conditions associated to oxidative stress, (ii) identification of the proximal nitrating species in vivo via the peptide mapping data, (iii) revealing structural and functional effects of nitration of specific protein residues in proteins, and (iv) obtaining pure modified protein species nitrated on specific tyrosine residues only to be used in further studies. In parallel to tyrosine nitration, proteomic studies can also assist in assessing parallel oxidative and related PTMs (e.g., addition of electrophiles) that may simultaneously contribute to changes in protein structure and function and have an impact in biological outcome. New methodological developments and research in proteomics of nitrated proteins open opportunities for identification of specific nitrated proteins and peptides as biomarkers and predictors of specific disease conditions, their possible participation in signaling cascades, and their role in inflammatory, immune, and degenerative diseases (66, 70, 76).
Abbreviations Used
- 2DE
two dimensional-electrophoresis
- 2D-PAGE
two dimensional-polyacrilamide gel electrophoresis
- ApoA1
apolipoprotein A1
- CAD
coronary artery disease
- CID
collision-induced dissociation
- CO3•−
carbonate radical
- CsA
cyclosporine A
- EPI
enhanced product information
- ESI
electrospray ionization
- Fe-SOD
iron superoxide dismutase
- H2O2
hydrogen peroxide
- hMnSOD
human MnSOD
- HNO2
nitrous acid
- HPLC
high-performance liquid chromatography
- LC-MS
liquid chromatography–mass spectrometry
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- MALDI-TOF
matrix-assisted laser desorption/ionization–time of flight
- MIDAS™
multiple reaction monitoring-initiated detection and sequencing
- Mn-SOD
manganese superoxide dismutase
- MPO
myeloperoxidase
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- nLC
nano liquid chromatography
- •NO
nitric oxide
- •NO2
nitrogen dioxide
- NO2−
nitrite
- -NO2
nitro group
- O2•−
superoxide anion radical
- -OH
phenolic group
- ONOO−
peroxynitrite anion
- PTM
post-translational modification
- TNM
tetranitromethane
- Tyr; Y
tyrosine
- UV
ultraviolet-visible spectroscopy
Acknowledgments
This work was supported by grants of Agencia Nacional de Investigación e Innovación (FCE_2014_104233), Universidad de la Republica (CSIC), and the National Institutes of Health (RO1 AI095173) to R.R. and Universidad de la República (CSIC) and Institut Pasteur de Montevideo to CB. Additional support to the investigators was obtained from Programa de Desarrollo de Ciencias Básicas (PEDECIBA), Universidad de la República (CEINBIO, Espacio Interdisciplinario), Centro de Biología Estructural del Mercosur (CeBEM), and Ridaline and Tecniplast through Fundación Manuel Perez.
References
- 1.MIDAS™ Workflow Designer: Software for the Optimization of Targeted Peptide Experiments. AB/Sciex Tech Note, 2007
- 2.Targeted, Hypothesis-Driven Mass Spectrometry: MRM Initiated Detection and Sequencing using the MIDAS™ Workflow for Faster, More Intelligent and Sensitive Protein Discovery and Characterization. AB/Sciex Tech Note 114TN38-01, 2007
- 3.MRMPilot™ Software: Developing MRM Assays for Peptide Quantitation. AB/Sciex Tech Note 115PB07-02, 2008
- 4.Abello N, Kerstjens HA, Postma DS, and Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res 8: 3222–3238, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Alvarez B, Rubbo H, Kirk M, Barnes S, Freeman BA, and Radi R. Peroxynitrite-dependent tryptophan nitration. Chem Res Toxicol 9: 390–396, 1996 [DOI] [PubMed] [Google Scholar]
- 6.Alvarez MN, Peluffo G, Piacenza L, and Radi R. Intraphagosomal peroxynitrite as a macrophage-derived cytotoxin against internalized Trypanosoma cruzi: consequences for oxidative killing and role of microbial peroxiredoxins in infectivity. J Biol Chem 286: 6627–6640, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez MN, Piacenza L, Irigoin F, Peluffo G, and Radi R. Macrophage-derived peroxynitrite diffusion and toxicity to Trypanosoma cruzi. Arch Biochem Biophys 432: 222–232, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Balafanova Z, Bolli R, Zhang J, Zheng Y, Pass JM, Bhatnagar A, Tang XL, Wang O, Cardwell E, and Ping P. Nitric oxide (NO) induces nitration of protein kinase Cepsilon (PKCepsilon), facilitating PKCepsilon translocation via enhanced PKCepsilon -RACK2 interactions: a novel mechanism of no-triggered activation of PKCepsilon. J Biol Chem 277: 15021–15027, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, and Challis GL. Cytochrome P450-catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat Chem Biol 8: 814–816, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartesaghi S, Ferrer-Sueta G, Peluffo G, Valez V, Zhang H, Kalyanaraman B, and Radi R. Protein tyrosine nitration in hydrophilic and hydrophobic environments. Amino Acids 32: 501–515, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Bartesaghi S, Peluffo G, Zhang H, Joseph J, Kalyanaraman B, and Radi R. Tyrosine nitration, dimerization, and hydroxylation by peroxynitrite in membranes as studied by the hydrophobic probe N-t-BOC-l-tyrosine tert-butyl ester. Methods Enzymol 441: 217–236, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Bartesaghi S, Valez V, Trujillo M, Peluffo G, Romero N, Zhang H, Kalyanaraman B, and Radi R. Mechanistic studies of peroxynitrite-mediated tyrosine nitration in membranes using the hydrophobic probe N-t-BOC-L-tyrosine tert-butyl ester. Biochemistry 45: 6813–6825, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Bartesaghi S, Wenzel J, Trujillo M, Lopez M, Joseph J, Kalyanaraman B, and Radi R. Lipid peroxyl radicals mediate tyrosine dimerization and nitration in membranes. Chem Res Toxicol 23: 821–835, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batthyany C, Souza JM, Duran R, Cassina A, Cervenansky C, and Radi R. Time course and site(s) of cytochrome c tyrosine nitration by peroxynitrite. Biochemistry 44: 8038–8046, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Bayden AS, Yakovlev VA, Graves PR, Mikkelsen RB, and Kellogg GE. Factors influencing protein tyrosine nitration—structure-based predictive models. Free Radic Biol Med 50: 749–762, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayir H, Kagan VE, Clark RS, Janesko-Feldman K, Rafikov R, Huang Z, Zhang X, Vagni V, Billiar TR, and Kochanek PM. Neuronal NOS-mediated nitration and inactivation of manganese superoxide dismutase in brain after experimental and human brain injury. J Neurochem 101: 168–181, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Birnboim HC, Lemay AM, Lam DK, Goldstein R, and Webb JR. Cutting edge: MHC class II-restricted peptides containing the inflammation-associated marker 3-nitrotyrosine evade central tolerance and elicit a robust cell-mediated immune response. J Immunol 171: 528–532, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Brennan ML, Wu W, Fu X, Shen Z, Song W, Frost H, Vadseth C, Narine L, Lenkiewicz E, Borchers MT, Lusis AJ, Lee JJ, Lee NA, Abu-Soud HM, Ischiropoulos H, and Hazen SL. A tale of two controversies: defining both the role of peroxidases in nitrotyrosine formation in vivo using eosinophil peroxidase and myeloperoxidase-deficient mice, and the nature of peroxidase-generated reactive nitrogen species. J Biol Chem 277: 17415–17427, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Brito C, Naviliat M, Tiscornia AC, Vuillier F, Gualco G, Dighiero G, Radi R, and Cayota AM. Peroxynitrite inhibits T lymphocyte activation and proliferation by promoting impairment of tyrosine phosphorylation and peroxynitrite-driven apoptotic death. J Immunol 162: 3356–3366, 1999 [PubMed] [Google Scholar]
- 20.Carballal S, Bartesaghi S, and Radi R. Kinetic and mechanistic considerations to assess the biological fate of peroxynitrite. Biochim Biophys Acta 1840: 768–780, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carballal S, Trujillo M, Cuevasanta E, Bartesaghi S, Moller MN, Folkes LK, Garcia-Bereguiain MA, Gutierrez-Merino C, Wardman P, Denicola A, Radi R, and Alvarez B. Reactivity of hydrogen sulfide with peroxynitrite and other oxidants of biological interest. Free Radic Biol Med 50: 196–205, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Cassina AM, Hodara R, Souza JM, Thomson L, Castro L, Ischiropoulos H, Freeman BA, and Radi R. Cytochrome c nitration by peroxynitrite. J Biol Chem 275: 21409–21415, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Castro L, Eiserich JP, Sweeney S, Radi R, and Freeman BA. Cytochrome c: a catalyst and target of nitrite-hydrogen peroxide-dependent protein nitration. Arch Biochem Biophys 421: 99–107, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Chen YR, Chen CL, Chen W, Zweier JL, Augusto O, Radi R, and Mason RP. Formation of protein tyrosine ortho-semiquinone radical and nitrotyrosine from cytochrome c-derived tyrosyl radical. J Biol Chem 279: 18054–18062, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci 25: 596–601, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Crow JP. and Ischiropoulos H. Detection and quantitation of nitrotyrosine residues in proteins: in vivo marker of peroxynitrite. Methods Enzymol 269: 185–194, 1996 [DOI] [PubMed] [Google Scholar]
- 27.Cruthirds DL, Novak L, Akhi KM, Sanders PW, Thompson JA, and MacMillan-Crow LA. Mitochondrial targets of oxidative stress during renal ischemia/reperfusion. Arch Biochem Biophys 412: 27–33, 2003 [DOI] [PubMed] [Google Scholar]
- 28.De Filippis V, Frasson R, and Fontana A. 3-Nitrotyrosine as a spectroscopic probe for investigating protein protein interactions. Protein Sci 15: 976–986, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degendorfer G, Chuang CY, Hammer A, Malle E, and Davies MJ. Peroxynitrous acid induces structural and functional modifications to basement membranes and its key component, laminin. Free Radic Biol Med 89: 721–733, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Demicheli V, Moreno DM, Jara G, Lima A, Carbajal S, Rios N, Batthyany C, Ferrer-Sueta G, Quijano C, Estrin D, Martí M, and Radi R. Mechanism of the reaction of human Mn-superoxide dismutase with peroxynitrite: nitration of critical tyrosine-34. Biochemistry 55: 3403–3417, 2016 [DOI] [PubMed] [Google Scholar]
- 31.Demicheli V, Quijano C, Alvarez B, and Radi R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic Biol Med 42: 1359–1368, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Denicola A, Batthyany C, Lissi E, Freeman BA, Rubbo H, and Radi R. Diffusion of nitric oxide into low density lipoprotein. J Biol Chem 277: 932–936, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Dhiman M, Nakayasu ES, Madaiah YH, Reynolds BK, Wen JJ, Almeida IC, and Garg NJ. Enhanced nitrosative stress during Trypanosoma cruzi infection causes nitrotyrosine modification of host proteins: implications in Chagas’ disease. Am J Pathol 173: 728–740, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiDonato JA, Aulak K, Huang Y, Wagner M, Gerstenecker G, Topbas C, Gogonea V, DiDonato AJ, Tang WH, Mehl RA, Fox PL, Plow EF, Smith JD, Fisher EA, and Hazen SL. Site-specific nitration of apolipoprotein A-I at tyrosine 166 is both abundant within human atherosclerotic plaque and dysfunctional. J Biol Chem 289: 10276–10292, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans TJ, Buttery LD, Carpenter A, Springall DR, Polak JM, and Cohen J. Cytokine-treated human neutrophils contain inducible nitric oxide synthase that produces nitration of ingested bacteria. Proc Natl Acad Sci U S A 93: 9553–9558, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folkes LK, Trujillo M, Bartesaghi S, Radi R, and Wardman P. Kinetics of reduction of tyrosine phenoxyl radicals by glutathione. Arch Biochem Biophys 506: 242–249, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Forrester MT, Foster MW, Benhar M, and Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med 46: 119–126, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frost MT, Halliwell B, and Moore KP. Analysis of free and protein-bound nitrotyrosine in human plasma by a gas chromatography/mass spectrometry method that avoids nitration artifacts. Biochem J 345 Pt 3: 453–458, 2000 [PMC free article] [PubMed] [Google Scholar]
- 39.Gaut JP, Byun J, Tran HD, and Heinecke JW. Artifact-free quantification of free 3-chlorotyrosine, 3-bromotyrosine, and 3-nitrotyrosine in human plasma by electron capture-negative chemical ionization gas chromatography mass spectrometry and liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Biochem 300: 252–259, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Gebicki JM, Nauser T, Domazou A, Steinmann D, Bounds PL, and Koppenol WH. Reduction of protein radicals by GSH and ascorbate: potential biological significance. Amino Acids 39: 1131–1137, 2010 [DOI] [PubMed] [Google Scholar]
- 41.Godoy LC, Munoz-Pinedo C, Castro L, Cardaci S, Schonhoff CM, King M, Tortora V, Marin M, Miao Q, Jiang JF, Kapralov A, Jemmerson R, Silkstone GG, Patel JN, Evans JE, Wilson MT, Green DR, Kagan VE, Radi R, and Mannick JB. Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc Natl Acad Sci U S A 106: 2653–2658, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goen T, Muller-Lux A, Dewes P, Musiol A, and Kraus T. Sensitive and accurate analyses of free 3-nitrotyrosine in exhaled breath condensate by LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci 826: 261–266, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Grune T, Blasig IE, Sitte N, Roloff B, Haseloff R, and Davies KJ. Peroxynitrite increases the degradation of aconitase and other cellular proteins by proteasome. J Biol Chem 273: 10857–10862, 1998 [DOI] [PubMed] [Google Scholar]
- 44.Hannibal L, Tomasina F, Capdevila DA, Demicheli V, Tortora V, Alvarez-Paggi D, Jemmerson R, Murgida DH, and Radi R. Alternative conformations of cytochrome c: structure, function, and detection. Biochemistry 55: 407–428, 2016 [DOI] [PubMed] [Google Scholar]
- 45.Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, Stockli J, Burchfield JG, Jensen TE, Jothi R, Kiens B, Wojtaszewski JF, Richter EA, and James DE. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab 22: 922–935, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, DiDonato JA, Levison BS, Schmitt D, Li L, Wu Y, Buffa J, Kim T, Gerstenecker GS, Gu X, Kadiyala CS, Wang Z, Culley MK, Hazen JE, Didonato AJ, Fu X, Berisha SZ, Peng D, Nguyen TT, Liang S, Chuang CC, Cho L, Plow EF, Fox PL, Gogonea V, Tang WH, Parks JS, Fisher EA, Smith JD, and Hazen SL. An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat Med 20: 193–203, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hui Y, Wong M, Zhao SS, Love JA, Ansley DM, and Chen DD. A simple and robust LC-MS/MS method for quantification of free 3-nitrotyrosine in human plasma from patients receiving on-pump CABG surgery. Electrophoresis 33: 697–704, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Humphrey SJ, Azimifar SB, and Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol 33: 990–995, 2015 [DOI] [PubMed] [Google Scholar]
- 49.Humphrey SJ, James DE, and Mann M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol Metab 26: 676–687, 2015 [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W, Dalke A, and Schulten K. VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 1996 [DOI] [PubMed] [Google Scholar]
- 51.Hunter EP, Desrosiers MF, and Simic MG. The effect of oxygen, antioxidants, and superoxide radical on tyrosine phenoxyl radical dimerization. Free Radic Biol Med 6: 581–585, 1989 [DOI] [PubMed] [Google Scholar]
- 52.Ignarro LJ. Biosynthesis and metabolism of endothelium-derived nitric oxide. Annu Rev Pharmacol Toxicol 30: 535–560, 1990 [DOI] [PubMed] [Google Scholar]
- 53.Jiao K, Mandapati S, Skipper PL, Tannenbaum SR, and Wishnok JS. Site-selective nitration of tyrosine in human serum albumin by peroxynitrite. Anal Biochem 293: 43–52, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki H, Ikeda K, Shigenaga A, Baba T, Takamori K, Ogawa H, and Yamakura F. Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells. Free Radic Biol Med 50: 419–427, 2011 [DOI] [PubMed] [Google Scholar]
- 55.Kawasaki H, Shigenaga A, Uda M, Baba T, Ogawa H, Takamori K, and Yamakura F. Nitration of tryptophan in ribosomal proteins is a novel post-translational modification of differentiated and naive PC12 cells. Nitric Oxide 25: 176–182, 2011 [DOI] [PubMed] [Google Scholar]
- 56.Khan F. and Ali R. Antibodies against nitric oxide damaged poly L-tyrosine and 3-nitrotyrosine levels in systemic lupus erythematosus. J Biochem Mol Biol 39: 189–196, 2006 [DOI] [PubMed] [Google Scholar]
- 57.Khan F. and Siddiqui AA. Prevalence of anti-3-nitrotyrosine antibodies in the joint synovial fluid of patients with rheumatoid arthritis, osteoarthritis and systemic lupus erythematosus. Clin Chim Acta 370: 100–107, 2006 [DOI] [PubMed] [Google Scholar]
- 58.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J 19: 1176–1179, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li XS, Li S, and Kellermann G. A novel mixed-mode solid phase extraction coupled with LC-MS/MS for the re-evaluation of free 3-nitrotyrosine in human plasma as an oxidative stress biomarker. Talanta 140: 45–51, 2015 [DOI] [PubMed] [Google Scholar]
- 60.Li XS, Li S, and Kellermann G. Tailored 96-well muElution solid-phase extraction combined with UFLC-MS/MS: a significantly improved approach for determination of free 3-nitrotyrosine in human urine. Anal Bioanal Chem 407: 7703–7712, 2015 [DOI] [PubMed] [Google Scholar]
- 61.Lima A, Duran R, Schujman GE, Marchissio MJ, Portela MM, Obal G, Pritsch O, de Mendoza D, and Cervenansky C. Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J Proteomics 74: 1720–1734, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Miller MJ, Joshi MS, Thomas DD, and Lancaster JR., Jr. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc Natl Acad Sci U S A 95: 2175–2179, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacMillan-Crow LA, Crow JP, Kerby JD, Beckman JS, and Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc Natl Acad Sci U S A 93: 11853–11858, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacMillan-Crow LA. and Thompson JA. Tyrosine modifications and inactivation of active site manganese superoxide dismutase mutant (Y34F) by peroxynitrite. Arch Biochem Biophys 366: 82–88, 1999 [DOI] [PubMed] [Google Scholar]
- 65.Martinez A, Peluffo G, Petruk AA, Hugo M, Pineyro D, Demicheli V, Moreno DM, Lima A, Batthyany C, Duran R, Robello C, Marti MA, Larrieux N, Buschiazzo A, Trujillo M, Radi R, and Piacenza L. Structural and molecular basis of the peroxynitrite-mediated nitration and inactivation of Trypanosoma cruzi iron-superoxide dismutases (Fe-SODs) A and B: disparate susceptibilities due to the repair of Tyr35 radical by Cys83 in Fe-SODB through intramolecular electron transfer. J Biol Chem 289: 12760–12778, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molon B, Ugel S, Del Pozzo F, Soldani C, Zilio S, Avella D, De Palma A, Mauri P, Monegal A, Rescigno M, Savino B, Colombo P, Jonjic N, Pecanic S, Lazzarato L, Fruttero R, Gasco A, Bronte V, and Viola A. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J Exp Med 208: 1949–1962, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreno DM, Marti MA, De Biase PM, Estrin DA, Demicheli V, Radi R, and Boechi L. Exploring the molecular basis of human manganese superoxide dismutase inactivation mediated by tyrosine 34 nitration. Arch Biochem Biophys 507: 304–309, 2011 [DOI] [PubMed] [Google Scholar]
- 68.Nakagawa H, Komai N, Takusagawa M, Miura Y, Toda T, Miyata N, Ozawa T, and Ikota N. Nitration of specific tyrosine residues of cytochrome C is associated with caspase-cascade inactivation. Biol Pharm Bull 30: 15–20, 2007 [DOI] [PubMed] [Google Scholar]
- 69.Nicholls SJ, Shen Z, Fu X, Levison BS, and Hazen SL. Quantification of 3-nitrotyrosine levels using a benchtop ion trap mass spectrometry method. Methods Enzymol 396: 245–266, 2005 [DOI] [PubMed] [Google Scholar]
- 70.Ohmori H. and Kanayama N. Immunogenicity of an inflammation-associated product, tyrosine nitrated self-proteins. Autoimmun Rev 4: 224–229, 2005 [DOI] [PubMed] [Google Scholar]
- 71.Orhan H, Vermeulen NP, Tump C, Zappey H, and Meerman JH. Simultaneous determination of tyrosine, phenylalanine and deoxyguanosine oxidation products by liquid chromatography-tandem mass spectrometry as non-invasive biomarkers for oxidative damage. J Chromatogr B Analyt Technol Biomed Life Sci 799: 245–254, 2004 [DOI] [PubMed] [Google Scholar]
- 72.Pehar M, Vargas MR, Robinson KM, Cassina P, England P, Beckman JS, Alzari PM, and Barbeito L. Peroxynitrite transforms nerve growth factor into an apoptotic factor for motor neurons. Free Radic Biol Med 41: 1632–1644, 2006 [DOI] [PubMed] [Google Scholar]
- 73.Peluffo G, Calcerrada P, Piacenza L, Pizzano N, and Radi R. Superoxide-mediated inactivation of nitric oxide and peroxynitrite formation by tobacco smoke in vascular endothelium: studies in cultured cells and smokers. Am J Physiol Heart Circ Physiol 296: H1781–H1792, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peluffo G, Piacenza L, Irigoin F, Alvarez MN, and Radi R. L-arginine metabolism during interaction of Trypanosoma cruzi with host cells. Trends Parasitol 20: 363–369, 2004 [DOI] [PubMed] [Google Scholar]
- 75.Peluffo G. and Radi R. Biochemistry of protein tyrosine nitration in cardiovascular pathology. Cardiovasc Res 75: 291–302, 2007 [DOI] [PubMed] [Google Scholar]
- 76.Peng F, Li J, Guo T, Yang H, Li M, Sang S, Li X, Desiderio DM, and Zhan X. Nitroproteins in human astrocytomas discovered by gel electrophoresis and tandem mass spectrometry. J Am Soc Mass Spectrom 26: 2062–2076, 2015 [DOI] [PubMed] [Google Scholar]
- 77.Petersson AS, Steen H, Kalume DE, Caidahl K, and Roepstorff P. Investigation of tyrosine nitration in proteins by mass spectrometry. J Mass Spectrom 36: 616–625, 2001 [DOI] [PubMed] [Google Scholar]
- 78.Piacenza L, Irigoin F, Alvarez MN, Peluffo G, Taylor MC, Kelly JM, Wilkinson SR, and Radi R. Mitochondrial superoxide radicals mediate programmed cell death in Trypanosoma cruzi: cytoprotective action of mitochondrial iron superoxide dismutase overexpression. Biochem J 403: 323–334, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piacenza L, Peluffo G, Alvarez MN, Kelly JM, Wilkinson SR, and Radi R. Peroxiredoxins play a major role in protecting Trypanosoma cruzi against macrophage- and endogenously-derived peroxynitrite. Biochem J 410: 359–368, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, and Radi R. Reaction of peroxynitrite with Mn-superoxide dismutase. Role of the metal center in decomposition kinetics and nitration. J Biol Chem 276: 11631–11638, 2001 [DOI] [PubMed] [Google Scholar]
- 81.Radabaugh MR, Nemirovskiy OV, Misko TP, Aggarwal P, and Mathews WR. Immunoaffinity liquid chromatography-tandem mass spectrometry detection of nitrotyrosine in biological fluids: development of a clinically translatable biomarker. Anal Biochem 380: 68–76, 2008 [DOI] [PubMed] [Google Scholar]
- 82.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A 101: 4003–4008, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Radi R. Peroxynitrite, a stealthy biological oxidant. J Biol Chem 288: 26464–26472, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radi R. Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc Chem Res 46: 550–559, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Radi R, Cassina A, Hodara R, Quijano C, and Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med 33: 1451–1464, 2002 [DOI] [PubMed] [Google Scholar]
- 86.Radi R, Peluffo G, Alvarez MN, Naviliat M, and Cayota A. Unraveling peroxynitrite formation in biological systems. Free Radic Biol Med 30: 463–488, 2001 [DOI] [PubMed] [Google Scholar]
- 87.Randall LM, Manta B, Hugo M, Gil M, Batthyany C, Trujillo M, Poole LB, and Denicola A. Nitration transforms a sensitive peroxiredoxin 2 into a more active and robust peroxidase. J Biol Chem 289: 15536–15543, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rebrin I, Bregere C, Gallaher TK, and Sohal RS. Detection and characterization of peroxynitrite-induced modifications of tyrosine, tryptophan, and methionine residues by tandem mass spectrometry. Methods Enzymol 441: 283–294, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Redondo-Horcajo M, Romero N, Martinez-Acedo P, Martinez-Ruiz A, Quijano C, Lourenco CF, Movilla N, Enriquez JA, Rodriguez-Pascual F, Rial E, Radi R, Vazquez J, and Lamas S. Cyclosporine A-induced nitration of tyrosine 34 MnSOD in endothelial cells: role of mitochondrial superoxide. Cardiovasc Res 87: 356–365, 2010 [DOI] [PubMed] [Google Scholar]
- 90.Rhee KY, Erdjument-Bromage H, Tempst P, and Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A 102: 467–472, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riordan JF, Sokolovsky M, and Vallee BL. Environmentally sensitive tyrosyl residues. Nitration with tetranitromethane. Biochemistry 6: 358–361, 1967 [DOI] [PubMed] [Google Scholar]
- 92.Rocha BS, Gago B, Barbosa RM, Lundberg JO, Mann GE, Radi R, and Laranjinha J. Pepsin is nitrated in the rat stomach, acquiring antiulcerogenic activity: a novel interaction between dietary nitrate and gut proteins. Free Radic Biol Med 58: 26–34, 2013 [DOI] [PubMed] [Google Scholar]
- 93.Rocha BS, Gago B, Barbosa RM, Lundberg JO, Radi R, and Laranjinha J. Intragastric nitration by dietary nitrite: implications for modulation of protein and lipid signaling. Free Radic Biol Med 52: 693–698, 2012 [DOI] [PubMed] [Google Scholar]
- 94.Romero N, Peluffo G, Bartesaghi S, Zhang H, Joseph J, Kalyanaraman B, and Radi R. Incorporation of the hydrophobic probe N-t-BOC-L-tyrosine tert-butyl ester to red blood cell membranes to study peroxynitrite-dependent reactions. Chem Res Toxicol 20: 1638–1648, 2007 [DOI] [PubMed] [Google Scholar]
- 95.Sacksteder CA, Qian WJ, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, 2nd, Smith RD, Smith DJ, Squier TC, and Bigelow DJ. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry 45: 8009–8022, 2006 [DOI] [PubMed] [Google Scholar]
- 96.Sainz M, Calvo-Begueria L, Perez-Rontome C, Wienkoop S, Abian J, Staudinger C, Bartesaghi S, Radi R, and Becana M. Leghemoglobin is nitrated in functional legume nodules in a tyrosine residue within the heme cavity by a nitrite/peroxide-dependent mechanism. Plant J 81: 723–735, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sarver A, Scheffler NK, Shetlar MD, and Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom 12: 439–448, 2001 [DOI] [PubMed] [Google Scholar]
- 98.Seth D. and Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol 15: 129–136, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shao B, Pennathur S, and Heinecke JW. Myeloperoxidase targets apolipoprotein A-I, the major high density lipoprotein protein, for site-specific oxidation in human atherosclerotic lesions. J Biol Chem 287: 6375–6386, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shao B, Tang C, Heinecke JW, and Oram JF. Oxidation of apolipoprotein A-I by myeloperoxidase impairs the initial interactions with ABCA1 required for signaling and cholesterol export. J Lipid Res 51: 1849–1858, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shishehbor MH, Aviles RJ, Brennan ML, Fu X, Goormastic M, Pearce GL, Gokce N, Keaney JF, Jr., Penn MS, Sprecher DL, Vita JA, and Hazen SL. Association of nitrotyrosine levels with cardiovascular disease and modulation by statin therapy. JAMA 289: 1675–1680, 2003 [DOI] [PubMed] [Google Scholar]
- 102.Shishehbor MH, Brennan ML, Aviles RJ, Fu X, Penn MS, Sprecher DL, and Hazen SL. Statins promote potent systemic antioxidant effects through specific inflammatory pathways. Circulation 108: 426–431, 2003 [DOI] [PubMed] [Google Scholar]
- 103.Shishehbor MH. and Hazen SL. Inflammatory and oxidative markers in atherosclerosis: relationship to outcome. Curr Atheroscler Rep 6: 243–250, 2004 [DOI] [PubMed] [Google Scholar]
- 104.Signorelli S, Moller MN, Coitino EL, and Denicola A. Nitrogen dioxide solubility and permeation in lipid membranes. Arch Biochem Biophys 512: 190–196, 2011 [DOI] [PubMed] [Google Scholar]
- 105.Simic MG. and Jovanovic SV. Antioxidation mechanisms of uric acid. J Am Chem Soc 111: 5778–5782, 1989 [Google Scholar]
- 106.Smallwood HS, Lourette NM, Boschek CB, Bigelow DJ, Smith RD, Pasa-Tolic L, and Squier TC. Identification of a denitrase activity against calmodulin in activated macrophages using high-field liquid chromatography—FTICR mass spectrometry. Biochemistry 46: 10498–10505, 2007 [DOI] [PubMed] [Google Scholar]
- 107.Sokolovsky M, Riordan JF, and Vallee BL. Conversion of 3-nitrotyrosine to 3-aminotyrosine in peptides and proteins. Biochem Biophys Res Commun 27: 20–25, 1967 [DOI] [PubMed] [Google Scholar]
- 108.Souza JM, Castro L, Cassina AM, Batthyany C, and Radi R. Nitrocytochrome c: synthesis, purification, and functional studies. Methods Enzymol 441: 197–215, 2008 [DOI] [PubMed] [Google Scholar]
- 109.Souza JM, Daikhin E, Yudkoff M, Raman CS, and Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Arch Biochem Biophys 371: 169–178, 1999 [DOI] [PubMed] [Google Scholar]
- 110.Souza JM, Peluffo G, and Radi R. Protein tyrosine nitration—functional alteration or just a biomarker? Free Radic Biol Med 45: 357–366, 2008 [DOI] [PubMed] [Google Scholar]
- 111.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, and Nathan C. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A 98: 9901–9906, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Surmeli NB, Litterman NK, Miller AF, and Groves JT. Peroxynitrite mediates active site tyrosine nitration in manganese superoxide dismutase. Evidence of a role for the carbonate radical anion. J Am Chem Soc 132: 17174–17185, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor MC. and Kelly JM. pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol 6: 32, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsikas D. and Duncan MW. Mass spectrometry and 3-nitrotyrosine: strategies, controversies, and our current perspective. Mass Spectrom Rev 33: 237–276, 2014 [DOI] [PubMed] [Google Scholar]
- 115.Unwin RD, Griffiths JR, and Whetton AD. A sensitive mass spectrometric method for hypothesis-driven detection of peptide post-translational modifications: multiple reaction monitoring-initiated detection and sequencing (MIDAS). Nat Protoc 4: 870–877, 2009 [DOI] [PubMed] [Google Scholar]
- 116.Vadseth C, Souza JM, Thomson L, Seagraves A, Nagaswami C, Scheiner T, Torbet J, Vilaire G, Bennett JS, Murciano JC, Muzykantov V, Penn MS, Hazen SL, Weisel JW, and Ischiropoulos H. Pro-thrombotic state induced by post-translational modification of fibrinogen by reactive nitrogen species. J Biol Chem 279: 8820–8826, 2004 [DOI] [PubMed] [Google Scholar]
- 117.Vanzo E, Ghirardo A, Merl-Pham J, Lindermayr C, Heller W, Hauck SM, Durner J, and Schnitzler JP. S-nitroso-proteome in poplar leaves in response to acute ozone stress. PLoS One 9: e106886, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Viner RI, Ferrington DA, Williams TD, Bigelow DJ, and Schoneich C. Protein modification during biological aging: selective tyrosine nitration of the SERCA2a isoform of the sarcoplasmic reticulum Ca2+-ATPase in skeletal muscle. Biochem J 340 (Pt 3): 657–669, 1999 [PMC free article] [PubMed] [Google Scholar]
- 119.Yamakura F, Taka H, Fujimura T, and Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J Biol Chem 273: 14085–14089, 1998 [DOI] [PubMed] [Google Scholar]
- 120.Yamamoto T, Maruyama W, Kato Y, Yi H, Shamoto-Nagai M, Tanaka M, Sato Y, and Naoi M. Selective nitration of mitochondrial complex I by peroxynitrite: involvement in mitochondria dysfunction and cell death of dopaminergic SH-SY5Y cells. J Neural Transm (Vienna) 109: 1–13, 2002 [DOI] [PubMed] [Google Scholar]
- 121.Yi D, Ingelse BA, Duncan MW, and Smythe GA. Quantification of 3-nitrotyrosine in biological tissues and fluids: generating valid results by eliminating artifactual formation. J Am Soc Mass Spectrom 11: 578–586, 2000 [DOI] [PubMed] [Google Scholar]
- 122.Yokoyama K, Uhlin U, and Stubbe J. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J Am Chem Soc 132: 8385–8397, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu H, Lee I, Salomon AR, Yu K, and Huttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta 1777: 1066–1071, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zamyatnin AA. Protein volume in solution. Prog Biophys Mol Biol 24: 107–123, 1972 [DOI] [PubMed] [Google Scholar]
- 125.Zhan X, Wang X, and Desiderio DM. Mass spectrometry analysis of nitrotyrosine-containing proteins. Mass Spectrom Rev 34: 423–448, 2015 [DOI] [PubMed] [Google Scholar]
- 126.Zhang H, Joseph J, Feix J, Hogg N, and Kalyanaraman B. Nitration and oxidation of a hydrophobic tyrosine probe by peroxynitrite in membranes: comparison with nitration and oxidation of tyrosine by peroxynitrite in aqueous solution. Biochemistry 40: 7675–7686, 2001 [DOI] [PubMed] [Google Scholar]
- 127.Zhang H, Joseph J, and Kalyanaraman B. Hydrophobic tyrosyl probes for monitoring nitration reactions in membranes. Methods Enzymol 396: 182–204, 2005 [DOI] [PubMed] [Google Scholar]