Abstract
Nuclear factor κB (NF-κB) is a master regulator of inflammation and cell death. Whereas most of the activity of NF-κB is regulated through the inhibitor of kB (IκB) kinase (IKK)–dependent degradation of IκB, IKK also phosphorylates subunits of NF-κB. Here, we investigated the contribution of the phosphorylation of the NF-κ B subunit p65 at the IKK phosphorylation site serine 536 (Ser536) in humans, which is thought to be required for the activation and nuclear translocation of NF-κB. Through experiments with knock-in mice (S534A mice) expressing a mutant p65 with an alanine-to-serine substitution at position 534 (the murine homolog of human Ser536), we observed increased expression of NF-κ B–dependent genes after injection of mice with the inflammatory stimulus lipopolysaccharide (LPS) or exposure to gamma irradiation, and the enhanced gene expression was most pronounced at late time points. Compared to wild-type mice, S534A mice displayed increased mortality after injection with LPS. Increased NF-κ B signaling in the S534A mice was at least in part explained by the increased stability of the S534A p65 protein compared to that of the Ser534-phosphorylated wild-type protein. Together, our results suggest that Ser534 phosphorylation of p65 in mice (and, by extension, Ser536 phosphorylation of human p65) is not required for its nuclear translocation, but instead inhibits NF-κ B signaling to prevent deleterious inflammation.
INTRODUCTION
Nuclear factor κB (NF-κB) is a transcription factor that acts as a master regulator of inflammation, controlling the expression of several hundred target genes (1). NF-κB activation is a tightly controlled event, which is largely mediated by the inhibitor of κB (IκB) kinase (IKK)–mediated phosphorylation of IκB, which normally sequestrates NF-κB and prevents it from entering the nucleus (2, 3). After its IKK-mediated phosphorylation, IκB is degraded through proteasome-mediated mechanisms, thereby liberating NF-κB and enabling it to enter the nucleus, where it binds to the promoters of genes with κB sites (2, 3).
Early on, it had been noted that IKK, as well as large number of other kinases, also phosphorylates NF-κB itself (4). Because the cytokine-stimulated phosphorylation of transcription factors, for example, the phosphorylation of c-Jun by c-Jun N-terminal kinase (JNK), is often associated with pathway activation (5), inducible phosphorylation of NF-κB has been considered a key mechanism in the positive regulation of NF-κB activity (6–9). Ser536 (S536) of the p65 subunit represents the site with the most potent inducible phosphorylation in response to inflammatory stimuli (9), and it is highly conserved among different species (10), indicating a potential role in the regulation of inducible NF-κB activity. In contrast to the phosphorylation of Ser276 of p65, which is an essential contributor to NF-κB activation in vitro and in vivo (11, 12), the functional contribution of the IKK-mediated phosphorylation of Ser536 remains poorly understood.
Numerous studies in cells reconstituted with p65 in which Ser536 is substituted by alanine (S536A) showed a requirement for this phosphorylation site for the NF-κB–dependent transcription of target genes in vitro (13–19), consistent with the hypothesis that phosphorylation increases NF-κB transactivation. Moreover, it has been suggested that Ser536 phosphorylation represents a noncanonical pathway through which NF-κB can translocate to the nucleus independently of IκBα degradation (16, 18, 20). Accordingly, cell-permeable peptides that inhibit Ser536 phosphorylation also block the nuclear translocation of NF-κB in cells in vitro (21) and NF-κB–dependent cell survival in vivo (22). Moreover, Ser536 phosphorylation is implicated in multiple inflammatory diseases, including Porphyromonas gingivalis–induced periodontal disease (23), Helicobacter pylori–induced inflammation (24), and the activation of NF-κB by Epstein-Barr virus (EBV) latent infection membrane protein 1 (25, 26). On the other hand, several studies have found either no role or inhibitory effects of Ser536 phosphorylation on nuclear translocation and gene transcription (11, 27). Determination of the contribution of Ser536 phosphorylation by clean genetic methods, such as knock-in approaches, may resolve these controversies and provide insight about the role of Ser536 phosphorylation in regulating inflammation in vivo.
RESULTS
Characterization of S534A knock-in mice
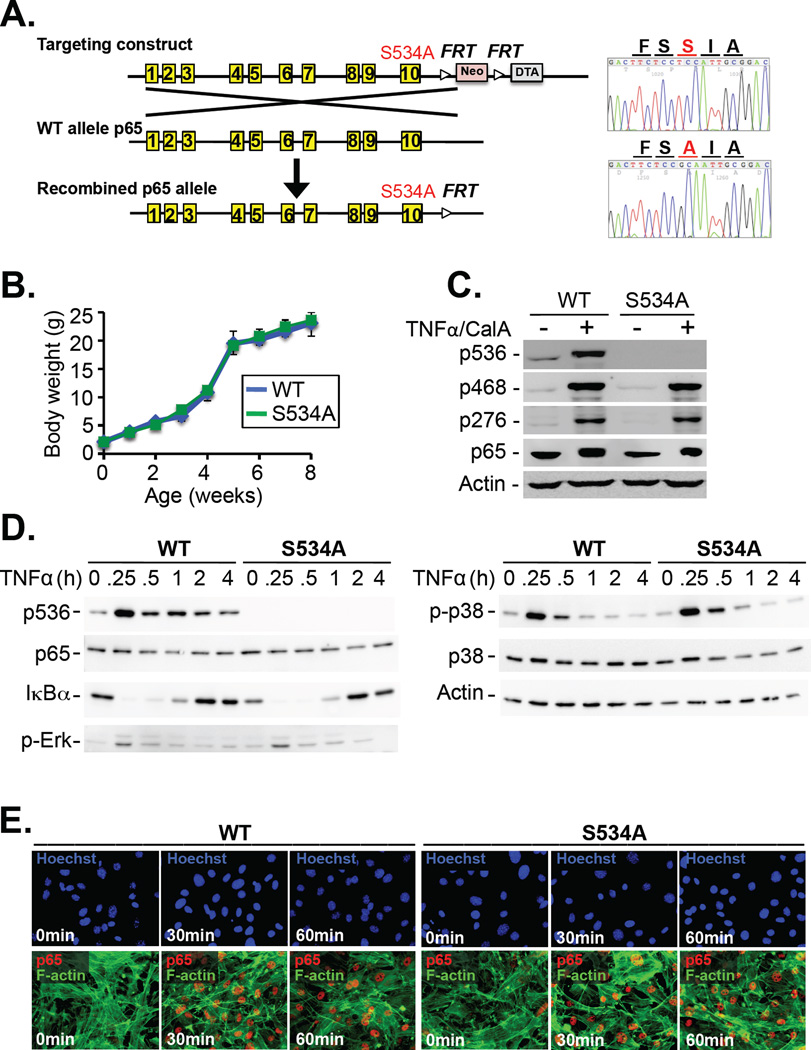
To resolve the conflicting data regarding the role of p65 Ser536 phosphorylation in the regulation of NF-κB activity and understand its function in relevant in vivo settings, we generated knock-in mice (Fig. 1A) expressing a mutant p65 in which Ser534, the murine homolog of Ser536, was substituted by alanine (S534A). In contrast to p65 knockout mice, which die in utero because of massive liver cell death (28), S534A mice were born at a normal Mendelian ratio without any gross abnormalities and displayed normal weight gain, blood counts, and spleen size, as well as the absence of spontaneous liver injury or inflammation at young and advanced ages (Fig. 1B, table S1, fig. S1, A to F). Of note, phosphorylation of Ser534 was completely blunted without affecting p65 phosphorylation at Ser468 or Ser276 (Fig. 1C). Moreover, there was no difference in the tumor necrosis factor (TNF)–stimulated degradation of IκBα or phosphorylation of p38 or extracellular signal–regulated kinase (ERK) in mouse embryonic fibroblasts (MEFs) (Fig. 1D). Microarray analysis did not reveal any substantial differences in baseline gene expression between wild-type mice and S534A mice (table S2).
Fig. 1. S534A mice display normal body weight, IκBα degradation, MAPK activation, and p65 nuclear translocation.
(A) Left: Generation of S534A knock-in mice by BAC recombineering. Right: Base pair substitutions were confirmed by sequencing. (B) Weight gain in wild-type (WT) and S534A mice was monitored at the indicated times. Data are means ± SD of six mice per genotype. (C) WT and S534A MEFs were left unstimulated or were stimulated with TNF-α (30 ng/ml) and 10 nM calyculin A for 15 min. The cells were then analyzed by Western blotting with antibodies specific for p65 phosphorylated at the indicated residues. The antibody against human pSer536-p65 detects mouse pSer534-p65. Actin was used as a loading control. Western blots are representative of three experiments. (D) WT and S534A MEFs were stimulated with TNF-α (30 ng/ml) for the indicated times before being analyzed by Western blotting with antibodies specific for the indicated targets. Data representative of three experiments. (E) WT and S534A MEFs treated with TNF-α (30 ng/ml) for the indicated times were analyzed by immunofluorescence microscopy to detect the nuclear translocation of p65 (red). F-actin was stained by phalloidin (green), whereas nuclei were detected with Hoechst (blue). Images are representative of three experiments.
Ser534 phosphorylation is not required for the nuclear translocation of p65
Because previous studies had suggested a role for Ser536 phosphorylation in the nuclear translocation of p65 or NF-κB–dependent gene transcription (13–16, 20), we next assessed these parameters in S534A MEFs in response to TNF-α. However, S534A MEFs did not show any alterations in p65 nuclear translocation (Fig.1E), activation of an NF-κB–dependent luciferase reporter, or induction of NF-κB–dependent gene expression (fig. S2, A and B).
S534A knock-in mice display increased NF-κB–dependent gene transcription and increased lipopolysaccharide (LPS)-induced mortality
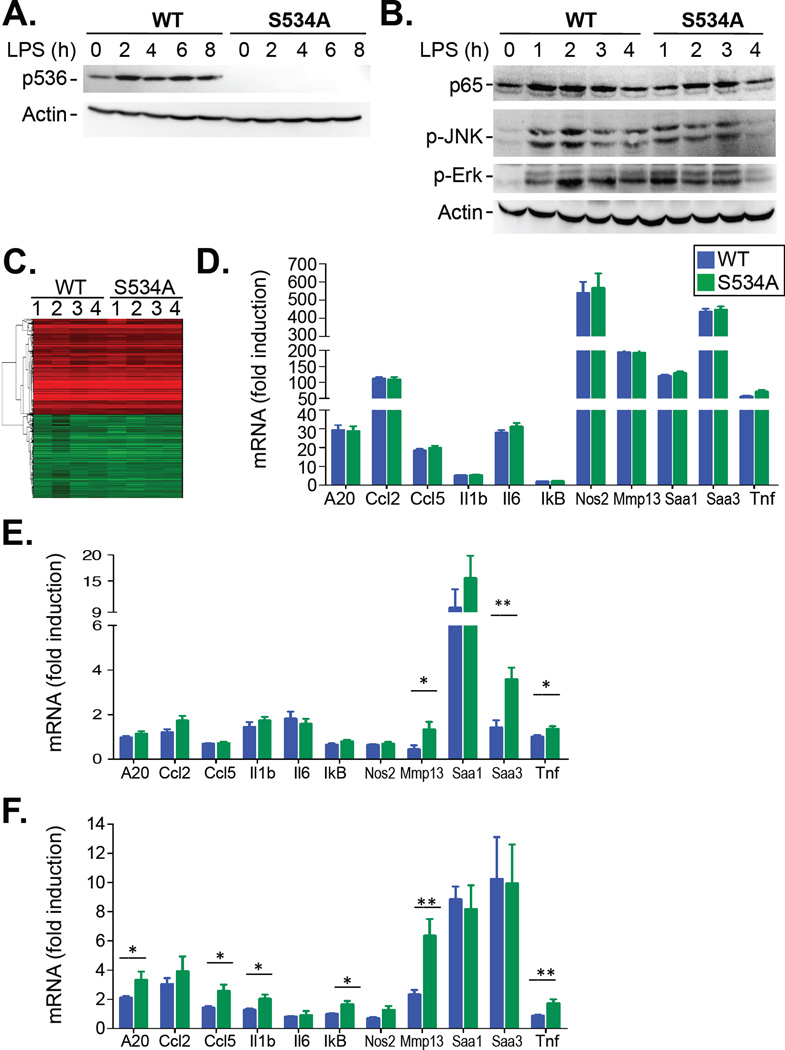
To study the contribution of Ser534 phosphorylation under relevant conditions in vivo, we treated mice with the Toll-like receptor 4 (TLR4) agonist lipopolysaccharide (LPS), a powerful activator of NF-κB and a key contributor to pathogen-induced inflammation and septic shock. Similar to our in vitro data, Ser534 phosphorylation was completely blunted in S534A mice after injection with LPS (Fig. 2A). Moreover, there were no differences in LPS-induced ERK or JNK phosphorylation in vivo between wild-type and S534A mice (Fig. 2B). We did not observe statistically significant differences in overall gene expression (as determined by microarray analysis) or in the expression of 11 well-characterized NF-κB–dependent genes [as determined by quantitative polymerase chain reaction (qPCR) analysis] between S534A and WT mice in the liver and spleen after injection with high-dose (1 mg/kg) LPS (Fig. 2, C and D, fig. S2C, table S3). In contrast, we found increased expression of some NF-κB–dependent genes in the spleens, but not livers, of TNF-treated S534A mice (Fig. 2E and fig. S2D). Similarly, whole-body irradiation induced the increased expression of 6 of 11 NF-κB–responsive genes in the livers of S534A mice compared to those in wild-type mice (Fig. 2F). Surprisingly, in all of these experiments, the expression of NF-κB–dependent genes was not decreased but was increased in LPS-treated S534A mice in comparison to LPS-treated wild-type mice.
Fig. 2. S534A mice show increased expression of NF-κB–dependent genes in specific settings.
(A) WT (n=4) and S534A (n=4) mice were injected i.v. with LPS (20 mg/kg) and sacrificed at the indicated times. Liver extracts were then analyzed by Western blotting with antibodies specific for the indicated targets. Western blots are representative of four experiments. (B) WT and S534A mice were injected i.v. with LPS (1 µg/kg) for the indicated times. Liver extracts were then analyzed by Western blotting with antibodies specific for the indicated proteins. Western blots are representative of three experiments. (C and D) WT (n = 9) and S534A (n = 9) mice were injected i.v. with LPS (1 mg/kg) and sacrificed 4 hours later. (C) Liver tissue was subjected to microarray analysis as described in Materials and Methods, and the data are presented as a heatmap showing genes with greater than a 2 log-fold change in expression and FDR < 0.05 in comparison to untreated mice. Red indicates genes that were increased in expression; green indicates genes that were decreased in expression. (D) Liver tissue from the indicated mice was subjected to qPCR analysis of the expression of the indicated NF-κB–responsive genes. Data are means ± SD of nine mice per genotype and show the fold-increase in mRNA abundance relative to that in untreated liver samples (E) WT (n = 8) and S534A (n = 10) mice were injected i.v with TNF-α (5 µg/kg) and were sacrificed 4 hours later. Splenic RNA was extracted and subjected to qPCR analysis of the expression of the indicated NF-κB–dependent genes. Data are means ± SD of at least eight mice per genotype (F) WT (n = 14) and S534A (n = 14) mice received whole-body irradiation (12 Gy) and were sacrificed 4 hours later. Liver RNA was extracted and subjected to qPCR analysis of the expression of the indicated NF-κB–dependent genes. Data are means ± SD of fourteen mice per genotye. *P < 0.05, **P < 0.01.
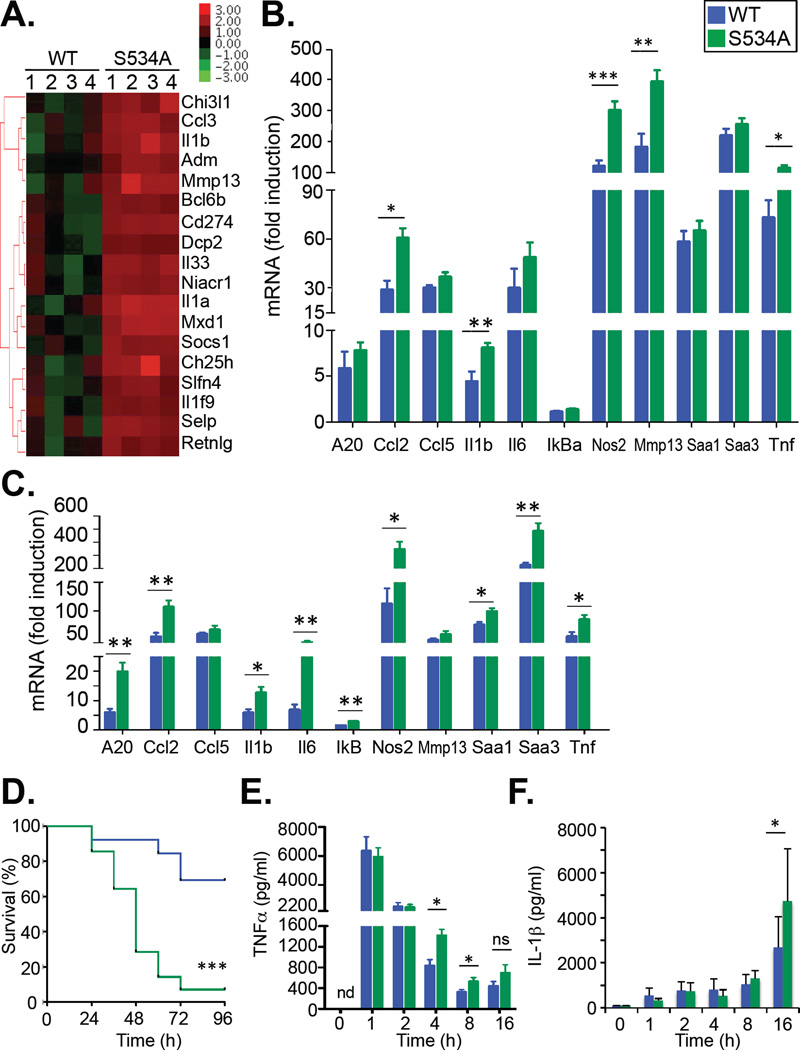
One important difference between high-dose LPS injection and the gamma irradiation and TNF models was the degree by which the expression of NF-κB–dependent genes was induced, with much lower induction of most genes in the latter two models. Therefore, we reasoned that the injection of high-dose LPS may have induced maximal expression of inflammatory genes and masked the rather moderate effects of the S534A mutation that were observed in other models in our readouts. Based on this hypothesis, we investigated the effects of lower doses of LPS in wild-type and S534A mice. Microarray analysis revealed that the expression of 18 genes was statistically significantly different [false discovery rate (FDR) < 0.05] between LPS-treated wild-type and S534A mice (Fig. 3A and table S4). Of these 18 genes, five (Cd274, Chi3l1, Il1a, Il1b, and Selp) are listed as NF-κB target genes (http://www.bu.edu/nf-kb/gene-resources/targetgenes/), demonstrating a statistically significant enrichment (χ2 probability < 2.2 × 10-16) of NF-κB–dependent genes in the pool of genes with significantly (FDR < 0.05) altered expression. Three additional genes have been described as being NF-κB–dependent: Ccl3 (30), Ch25h (31), and Mmp13 (32). Hence, a large percentage (that is, 8 of 18 genes) of the genes with statistically significantly altered expression was NF-κB–dependent. Again, all of these NF-κB–dependent genes were not decreased, but were increased in expression in LPS-treated S534A mice in comparison to LPS-treated wild-type mice. These data were confirmed by the qPCR analysis of several genes from this array (fig. S3), as well as of the set of 11 NF-κB–dependent genes described earlier, of which approximately half were statistically significantly increased in S534A mice in comparison to wild-type mice at this lower concentration of LPS (Fig. 3B).
Fig. 3. S534A mice display increased expression of NF-κB–dependent genes and mortality.
(A and B) WT (n = 7) and S534A (n = 7) mice were injected i.v. with LPS (1 µg/kg) and sacrificed 4 hours later. (A) Liver tissue was then subjected to microarray analysis. The heatmap shows those genes that were differentially regulated in expression in the livers of LPS-treated S534A mice compared to those in the livers of LPS-treated WT mice (FDR <0.05). (B) Liver tissue from the indicated LPS-treated mice was subjected to qPCR analysis of the expression of the indicated NF-κB–dependent genes. Data are means ± SD of seven mice per genotype. (C) WT and S534A mice were injected i.v. with LPS (1 µg/kg) and then were sacrificed eight hours later. Liver RNA was extracted and subjected to qPCR analysis of the expression of the indicated NF-κB–dependent genes. Data are means ± SD of at least six mice per genotype. (D to F) WT (n = 13) and S534A mice (n = 14) were injected i.v. with LPS (20 mg/kg). (D) Survival was monitored for 96 hours. (E and F) Serum concentrations of TNF-α (E) and IL-1β (F) were determined by ELISA. Data are means ± SD of at least 12 mice per genotype and time point). *P < 0.05, **P < 0.01, ***P < 0.001.
Note that LPS doses as low as 1 µ g/kg still led to a several hundred-fold induction in the expression of some NF-κB target genes (Fig. 3B). Furthermore, LPS-induced inflammation and morbidity in humans typically occur at very low concentrations (in the nanogram range) (29). Differences in the expression of NF-κB–dependent genes induced by lower dose LPS were even more pronounced at later time points in the liver (Fig. 3C) and could only be detected at late, but not early, time points in the spleen (fig. S4 A and B). To further investigate the functional consequences of this altered expression of NF-κB–dependent genes, we treated mice with a sublethal dose of LPS (20 mg/kg). Consistent with our findings that S534A mice displayed increased inflammatory gene expression, we observed statistically significantly increased LPS-induced mortality in S534A mice in comparison to wild-type mice (Fig. 3D, 69.2% vs. 7.1%, P < 0.001). Accordingly, the serum concentrations of the proinflammatory cytokines interleukin-1β (IL-1β) and TNF-α, two key contributors to LPS-induced lethality, were increased in S534A mice compared to those of wild-type controls (Fig. 3, E and F). Collectively, these data demonstrate that Ser534 phosphorylation acts as a negative regulator of NF-κB–dependent gene expression and inflammation, and that its effect is moderate and therefore best revealed at submaximal stimulation or borderline lethal doses.
Ser534 phosphorylation decreases the stability of NF-κB p65
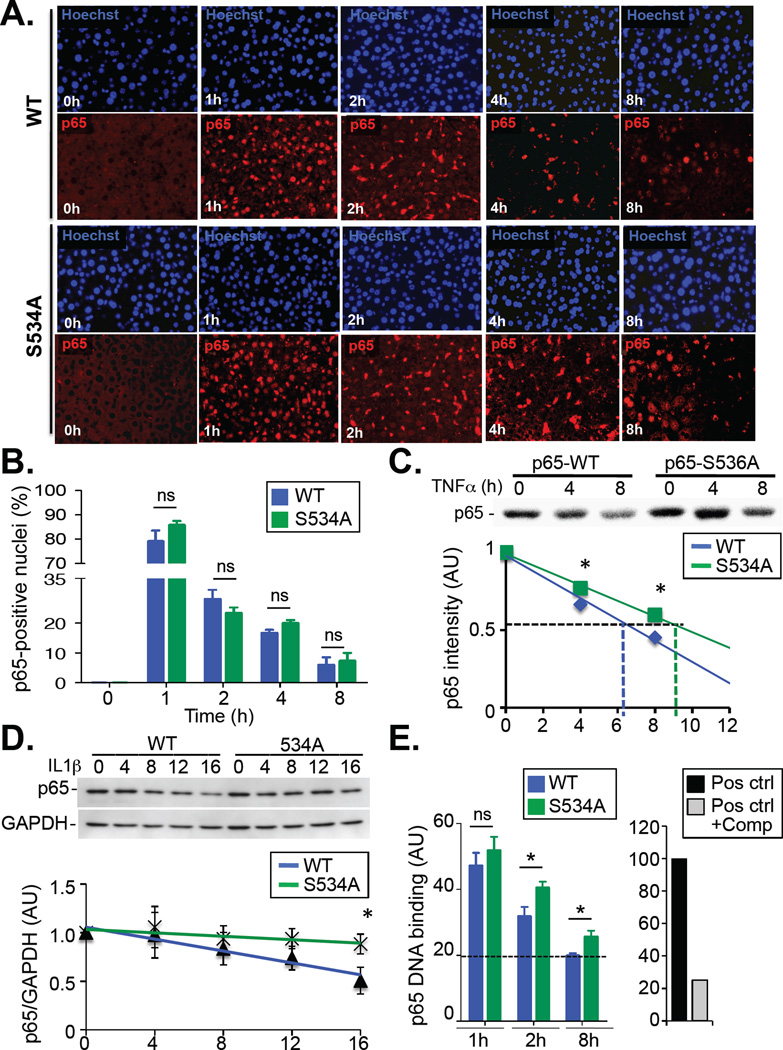
To understand the mechanism by which Ser534 phosphorylation inhibits NF-κB activity, we next investigated the nuclear translocation of NF-κB in liver from wild-type and S534A mice. Similar to our in vitro data from experiments with MEFs, we did not observe any difference in the initial nuclear translocation of p65 in hepatocytes after injection of mice with a sublethal dose of LPS; moreover, there were not significant differences in the abundance of nuclear p65 in hepatocytes from S534A and wild-type mice at late time points (Fig. 4, A and B). Previous studies suggested a role for Ser536 phosphorylation in regulating the half-life of the p65 protein (39, 40), which would be consistent with our findings that differences in NF-κB–dependent gene expression and cytokine production were more pronounced at later time points, that is, at a time when a proportion of p65 may have undergone degradation. Consistent with these previous studies, we observed a small, but statistically significant, increase in the half-life of 35S-labeled S536A p65 protein compared to that of labeled wild-type p65 (Fig. 4C, 9 hours 44 min vs. 7 hours 03 min; P < 0.05) in transfected human embryonic kidney (HEK) 293 cells (which enabled clean immunoprecipitation of p65). This finding was further confirmed by a cycloheximide pulse-chase experiment in primary MEFs (Fig. 4D). Moreover, we observed a decrease in the abundance of p65 in primary macrophages from wild-type mice after treatment with LPS, which was blunted in primary macrophages from LPS-treated S534A mice (fig. S5A). Note that S534A macrophages also displayed a more substantial increase in the expression of several NF-κB–dependent genes than did macrophages from wild-type mice (fig. S5B). Consistent with these findings, nuclear extracts from the livers of LPS-treated S534A mice showed identical NF-κB DNA-binding activity to that of extracts from LPS-treated wild-type mice at early time points, but showed a substantial increase at later time points (Fig. 4E). Together, these observations support the hypothesis that increased LPS-induced inflammation and lethality in S534A mice are a result of the prolonged action of NF-κB. The moderate increase in the half-life of the S536A p65 protein is consistent with the moderate phenotype of S534A mice. Because (i) IKK phosphorylates p65 at multiple sites, including Ser468 (33) and Ser547 (fig. S6, A and B) and (ii) Ser468 phosphorylation also reduces the expression of NF-κB–dependent genes (33, 34), it is likely that Ser534/Ser536 acts in concert with additional p65 phosphorylation sites, such Ser468 and Ser547, and that a more profound phenotype would be observed in double or triple mutant mice.
Fig. 4. S534 phosphorylation affects DNA binding and gene expression by NF-κB at late time points through regulation of p65 stability.
(A and B) WT and S534A mice were injected i.v. with LPS (20 mg/kg) and sacrificed after the indicated times. (A) Liver tissue was analyzed by immunohistochemistry to monitor the translocation of p65 (red) to the nucleus (blue). (B) The percentages of cells with p65-positive nuclei were quantified. Data are means ± SD of five mice per genotype and time point. (C) Top: HEK 293 cells overexpressing human M2-p65 or M2-S536A-p65 were treated with TNF-α and then subjected to pulse-chase analysis for the indicated times to determine the half-life of p65 protein. Bottom: Data are means ± SD of three independent experiments, each performed in triplicate. (D) Top: WT and S534A MEFs were treated with cycloheximide (30 µg/ml) and then were left unstimulated or were stimulated with IL-1β for the indicated times. Samples were analyzed by Western blotting with antibodies against the indicated proteins. Bottom: The relative abundance of p65 protein, normalized to that of GAPDH, was determined for the indicated times by densitometric analysis. Data are means ± SD of three experiments. (E) Left: NF-κB DNA binding activity was determined in nuclear extracts from the livers of LPS-treated WT and S534A mice (n = 6 mice per genotype and time point). A value of 100 represents the positive control (recombinant human p65). The dashed line indicates NF-κB binding activity at baseline. Right: The competitive oligonucleotide suppressed DNA binding by the positive control. *P < 0.05; n.s., not significant. Data are means ± SD of six mice per genotype.
DISCUSSION
Phosphorylation of transcription factors serves as rapid and powerful mechanism to regulate transcription in a positive or negative fashion in many signaling pathways, including those mediated by signal transducer and activator of transcription (STAT) proteins, Smads, c-Jun, and NF-κB (35, 36). In the case of NF-κB, the IKK-mediated phosphorylation of IκB was identified as the key mechanism for regulating the nuclear translocation of NF-κB (3, 9). In contrast, the role of inducible phosphorylation of NF-κB is much less understood. Phosphorylation of Ser536 of the p65 subunit is the most common phosphorylation event associated with NF-κB activation that is described in the literature (10, 13–21, 23–26, 37, 38), but its role remains controversial. Our data from experiments with newly generated knock-in mice now suggest an inhibitory effect of Ser534/Ser536 phosphorylation on the regulation of NF-κB activity. Although this effect was moderate and did not result in any spontaneous phenotype, it was observed in different disease models, such as LPS-induced shock, TNF-induced inflammation, and gamma irradiation. This is in contrast to transcription factors that have a key role in the regulation of inflammation, such as c-Jun, for which inducible phosphorylation by dedicated kinases, such as JNK, is essential in promoting pathway activation (5). Our data suggest that Ser534 phosphorylation inhibits NF-κB activity by decreasing the half-life of the p65 protein without affecting the kinetics of its nuclear import or export.
Because the effects of the S534A mutation in the knock-in mice were moderate, they could only be revealed at submaximal doses of LPS, that is, at lower doses of LPS when investigating NF-κB–dependent gene expression and at a sublethal dose of LPS when investigating LPS-induced mortality. Our finding that Ser534 phosphorylation enhanced p65 half-life in multiple cell types is consistent with two previous studies that also showed the increased half-life of S536A mutants (39, 40). Note that the effects of the S534A mutation were most substantial at later time points of stimulation (for example, 8 hours after LPS injection) in both the liver and spleen, which is consistent with the finding that Ser536 phosphorylation reduces p65 half-life. Previous studies demonstrated a role for the E3 ubiquitin ligases Cullin-based ligase 2 (CUL2) and Copper metabolism MURR1 domain–containing 1 (COMMD1) in the degradation of p65 in cultured cells in response to both Ser468 and Ser536 phosphorylation (40, 41). Because of the mild effect of S534A phenotype in the liver, the absence of effects at some time points, and the much lower effects in tissues such as the spleen, we could not determine to what degree these cofactors regulate p65 stability in vivo. It is likely that IKK-mediated Ser536 phosphorylation reduces the stability of p65 protein through the same mechanism in vitro and in vivo. Given that both Ser468 (33, 34) and Ser536 negatively regulate p65 stability and the transcription of NF-κB–dependent genes, these two residues may functionally synergize to negatively regulate p65 stability and the activity of the NF-κB pathway in vivo. Moreover, in the current study we also detected a third IKK phosphorylation site at Ser547 (fig. S6). Although previous reports (10) and our current study found that Ser534/Ser536 represents the main phosphorylation site of p65, which accounts for about 90% of IKK-mediated phosphorylation, it is likely that double or triple knock-in mice harboring mutations at Ser468, Ser536, and Ser547would present a stronger phenotype.
Future studies, involving ChIP-seq, for example, are needed to determine whether Ser534/Ser536-phosphorylated p65 binds to specific promoters or enhances the expression of a specific set of NF-κB target genes. These studies would also enable us to determine the half-life of Ser534/Ser536-phosphorylated p65 at specific promoters or enhancers and discriminate between direct and indirect effects on gene regulation. We can therefore not exclude the possibility that the negative effect of Ser534 phosphorylation on NF-κB–dependent gene transcription that we observed is mediated by additional mechanisms. Nonetheless, the physiological relevance of Ser534 phosphorylation was demonstrated by our finding of a substantial increase in LPS-induced mortality in the S534A mice. In summary, our data suggest that Ser534/Ser536 phosphorylation of p65 is a negative regulator of the NF-κB pathway in vivo and that IKK-mediated signals may therefore regulate inflammatory gene expression both in a positive (through the phosphorylation and degradation of IκBα) and negative manner (through the phosphorylation of p65). These opposing functions may provide the basis for the fine-tuning of NF-κB–targeted therapies in chronic inflammatory diseases.
MATERIALS AND METHODS
Mice
Mice were maintained on a 12-hour dark-light cycle with free access to food and water. For all experiments, 8- to 10-week-old mice were used unless otherwise indicated in the figure legends. All animal procedures were approved by the Columbia University Institutional Animal Care and Use Committee and were in accordance with the Guide for the Care and Use of Laboratory Animals issued by the National Institutes of Health.
Generation of S534A knock-in mice
Exon 10 of the p65 gene was subcloned into the TOPO plasmid (Life Technologies), and the S534A mutation was introduced through two nucleotide mutations with the QuikChange Mutagenesis kit (Stratagene), thus generating a codon modification from TCC (serine) to GCA (alanine). Sequential rounds of homologous recombination enabled us to introduce the mutated exon 10, a FRT-Neo-FRT-LoxP and DTA cassette on a bacterial artificial chromosome, thus generating the p65 S534A knock-in targeting construct. After transfection, two embryonic stem (ES) cell clones were identified by polymerase chain reaction (PCR)-based screening and injected into C57Bl6/J blastocysts to produce germline chimeras. Chimeras were mated with mice expressing Flipase (FLPe) to excise the Neo cassette after recombination of the FRT sites. The offspring were screened for the targeted allele without the Neo cassette and the FLPe transgene. After five backcrosses to C57Bl6/J mice, the mice were interbred to homozygosity.
In vivo models of NF-κB activation
Age-matched, 8- to 10-week-old wild-type (WT) and S534A male and female mice were injected intravenously (i.v.) with low (1 µg/kg), high (1 mg/kg), or lethal (20 mg/kg) doses of LPS (Sigma-Aldrich, 055:B5) and were sacrificed 1, 2, 4, or 8 hours later. For LPS-induced shock, age-matched, 8- to 10-week-old WT and S534A mice were injected i.v. with LPS (20 mg/kg). Survival was then examined every 8 hours. In a separate experiment, mice were bled 1, 2, 4, 8, and 16 hours after the LPS challenge, and their serum concentrations of TNF-α and IL-1β were measured by enzyme-linked immunosorbent assay (ELISA). For TNF-α–induced NF-κB activation, mice were injected i.v. with TNF-α (5 mg/kg, R&D Systems) and were sacrificed 4 hours later. For irradiation-induced NF-κB activation, mice were exposed to 12 Gy total body gamma irradiation and sacrificed 4 hours later.
Genotyping
Genotyping of S534A knock-in mice was performed through amplification of the genomic region containing the S534A mutation with forward (5’-TCCATGTCTCACTCCACAGC-3’) and reverse (5’-CACTCCCCAGAATGTGTACG-3’) primers coupled with digestion with Mfe I (because an Mfe I restriction site was generated with the S534A mutation). The PCR product digested by Mfe I yielded either one 289-bp band (for the WT allele) or 158- and 131-bp bands (for the S534A allele). Genotyping of the FNFL cassette remnant (one recombined FRT and one loxP site) downstream of the targeted region can also be performed with forward (5’-GCTAAAGGGGGCAGTCTTCT-3’) and reverse (5’-GCCTGGATCTGATTCCAAAA-3’) primers, which yields 366-bp (for the WT allele) and 507-bp (for the S534A allele) fragments.
[35S] methionine pulse-chase and cycloheximide chase assays
Human embryonic kidney (HEK) 293 cells were transfected with lipofectamine (Life Technologies) with plasmids encoding human M2-p65 or M2-S536A-p65. Forty-eight hours later, the transfected cells were starved for 60 min in methionine-deficient in Dulbecco's Modified Eagle Medium DMEM [supplemented with 10% dialyzed fetal calf serum (FCS)] and then underwent metabolic labeling for 12 min with 80 µ Ci/ml of [35S]-Methionine (Perkin Elmer). The pulse-labeled cells were chased after treatment with TNF-α (1 ng/ml, R&D systems) for different times (0, 4, or 8 hours) in complete DMEM supplemented with 10 mM cold methionine, and then the cells were lysed in radioimmunoprecipitation (RIPA) buffer. The radiolabeled p65 protein in the samples was isolated by immunoprecipitation with anti-M2 magnetic beads (Sigma-Aldrich), separated by SDS-PAGE, and visualized with a Typhoon 9400 PhosphorImager (Amersham). To measure the degradation of p65, MEFs were serum-starved for 12 hours in DMEM, 0.1% FCS, which was followed by treatment with cycloheximide (30 µg/ml, Sigma-Aldrich) and stimulation with IL-1β (30 ng/ml, R&D Systems). Total protein lysates were collected at different time points (0, 4, 8, and 16 hours) and were subjected to standard Western blotting analysis of p65 and GAPDH.
Isolation of MEFs
MEFs were isolated from 13.5-day post-coitum embryos. Briefly, fresh embryos were washed in sterile phosphate-buffered saline (PBS) after cutting away the brain and all red organs. The remaining material was minced and digested in 0.05% trypsin-EDTA solution for 30 min at 37°C under agitation. After the addition of warm DMEM, 10% FCS to stop digestion by trypsin, the culture medium was filtered through a 100-µm cell strainer. Cells were then pelleted after a low-speed centrifugation, resuspended in 10 ml of warm DMEM, 10% FCS, and plated out at 1 embryo equivalent per 10-cm dish.
Isolation of bone marrow–derived macrophages
Bone marrow–derived macrophages (BMDMs) were isolated from retired WT and S534A breeders. Briefly, tibias and femurs of donors were flushed with MEM alpha (Life Technologies) medium to obtain the bone marrow. After filtration through a 70-µm cell strainer, the bone marrow cells were washed and plated in MEM alpha medium supplemented with 10% FCS and M-CSF (20 ng/ml, R&D Systems). Subsequently, the BMDMs were treated with LPS and analyzed by Western blotting and qPCR to determine the relative abundances of p65 protein and mRNAs of NF-κB target genes, respectively.
Western blotting analysis
Protein extracts from cells or liver tissue were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and incubated with antibodies specific for human and mouse (human/mouse) p65 (1:1000, SC-372-G, Santa Cruz Biotechnology or ab7970, Abcam), human/mouse pSer536-p65 (1:1000, #3033, Cell Signaling), human/mouse pSer468-p65 (1:1000, #3039, Cell Signaling), human/mouse pSer276-p65 (1:1000, #3037, Cell Signaling,), human/mouse IκBα (1:1000, #4814, Cell Signaling,), pERK (1:1000, #9106, Cell Signaling), pp38 (1:1000, #9215, Cell Signaling), p38 (1:1000, #9212, Cell Signaling), or pJNK (1:1000, #9251, Santa Cruz Biotechnology) before being stripped and incubated with antibodies against actin (1:5000, #691002, MP Biomedicals) or GAPDH (1:20000, G9295, Sigma-Aldrich).
Immunohistochemistry and immunocytofluorescence staining
Immunohistochemical staining was performed on paraffin liver sections after 10% formalin fixation with primary antibody against p65 (1:200, SC-372-G, Santa Cruz) and Alexa Fluor 594–conjugated secondary antibody (Life Technologies). Nuclei were stained with Hoechst 33342 (Sigma Chemical Co.). For immunocytofluorescence, primary MEFs were fixed with 4% paraformaldehyde and stained with primary antibody against p65 (1:100, SC-372-G) and Alexa Fluor 594–conjugated secondary antibody. Counterstaining was performed Alexa Fluor 488–conjugated Phalloidin (Molecular Probes). Nuclei were stained with Hoechst 33342.
ELISAs
Serum concentrations of TNF-α and IL-1β were determined by a conventional sandwich ELISA with 96-well plates (R&D Systems) according to the manufacturer’s instructions.
RNA extraction and qPCR analysis
Total RNA was extracted from mouse livers or spleens with Trizol reagent (Life Technologies), column-purified with a high pure RNA Tissue kit (Roche), and reverse-transcribed with a High capacity cDNA reverse transcription kit (Life Technologies) to generate complementary DNA (cDNA). The abundance of cDNA was then determined by quantitative PCR (qPCR) analysis with an Applied Biosystems 7300 PCR cycler using ABI Taqman primers and probes specific for A20, Ccl2, Ccl3, Ccl5, Ch25h, Il1a, Il1b, Il6, Nos2, Mmp13, Saa1, Saa3, Selp, Socs1, and Tnfa (Life Technologies). All PCR products were quantified with relative standard curves, and the abundance of each mRNA of interest was normalized to that of 18S ribosomal RNA.
NF-κB DNA-binding assay
The DNA-binding activity of NF-κB p65 was assessed with an NF-κB Transcription Factor Assay Kit (Cayman Chemical) according to the manufacturer’s instructions. Briefly, nuclear protein extracts (10 µg) were incubated in 96-well plates coated with immobilized, double-stranded oligonucleotides containing an NF-κB response element, and the absorbance in each well was read at 450 nm with a Benchmark Plus microplate spectrophotometer (Bio-Rad).
NF-κB reporter assay
MEFs were transduced with an adenovirus expressing NF-κB–driven luciferase and an adenovirus expressing β-galactosidase at a multiplicity of infection (MOI) of 30 for both. Twenty-four hours later, the MEFs were serum-starved overnight and then were left untreated or were treated with TNF-α (30 ng/ml) or IL-1β (5 ng/ml) for 6 hours. The luciferase activity of the cell lysates was assessed with a luciferase assay kit (Promega).
Microarray analysis
Total RNA was isolated from the livers of WT or S534A mice with a high pure RNA Tissue Kit (Roche). RNA was analyzed on Affymetrix 1.0ST chips (Affymetrix) according to the manufacturer’s instructions. Briefly, 150 ng of RNA was used for cDNA synthesis and terminal labeling with the Ambion WT expression and terminal labeling kit. Normalization by Robust Multichip Algorithm and determination of differential expression by Limma in the R/Bioconductor statistical computing environment was performed as previously described (42). A significance cutoff of the Benjamini-Hochberg false discovery rate (FDR) of < 0.05 was used. Complete linkage hierarchical clustering was performed on statistically significant genes with |log2FC| >1 with Cluster 3.0 and JavaTreeview software. The list of NF-κB target genes was based on the list of NF-κB target genes in humans compiled by Thomas Gilmore and co-workers (http://www.bu.edu/nf-kb/gene-resources/target-genes/). Only experimentally confirmed direct target genes were used, which resulted in a total of 360 distinct genes. We identified 333 mouse homologs with HomoloGene (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/homologene), of which 295 were found to be on the Mouse 430.2 chip, which contains 20,674 unique genes in all. The overrepresentation of NF-κB genes was calculated by the c2 method. Microarray results were submitted to the Gene Expression Omnibus (GEO) database with the accession numbers GSE67072 and GSE67178.
Mutagenesis and in vitro kinase assays
Mutagenesis of glutathioneS-transferase (GST)-p65 to generate single, double, and triple mutants was performed with the Quikchange mutagenesis kit (Stratagene) as described previously (33). In vitro kinase assays were performed with recombinant human IKKβ in combination with GST-p65 fusion proteins as previously described (33). Kinase assays were performed in 20 mM Hepes (pH 7.7), 2 mM MgCl2, 2 mM MnCl2, 1 mM DTT, and 5 µM ATP for 30 min at 30°C in the presence of 1 µCi of 32P-ATP. The kinase reactions were then loaded onto 10% acrylamide gels, and the gels were dried and exposed to film.
Statistical analysis
All data are expressed as means ± SD. For the comparison of two groups with a normal distribution, a two-sided, unpaired t test was used. For the comparison of two groups without a normal distribution, the Mann-Whitney test was used. P <0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank C.-S. Lin from the Irving Cancer Center Transgenic mouse facility for help in generating the S534A knock-in mice.
Funding: This study was supported by NIH grant 5U54CA163111 (to R.F.S.) and a grant from the Fondation pour la Recherche Médicale (to J.P.).
Footnotes
Fig. S1. Phenotypic characterization of S534A knock-in mice.
Fig. S2. Determination of NF-κB activation in MEFs, spleens, and livers from wild-type and S534A mice.
Fig. S3. Analysis of gene expression in the livers of wild-type and S534A mice injected with low-dose LPS.
Fig. S4. Expression of NF-κB–dependent genes in the spleens of wild-type and S534A mice at early and late time points after injection with low-dose LPS.
Fig. S5. Analysis of p65 protein abundance and NF-κB target gene expression in macrophages from WT and S534A mice.
Fig. S6. IKK phosphorylates p65 at multiple sites.
Table S1. Complete blood counts in wild-type and S534A mice.
Table S2. Hepatic gene expression profiles in untreated wild-type and S534A mice.
Table S3. Hepatic gene expression profiles in wild-type and S534A mice treated with high-dose LPS.
Table S4. Hepatic gene expression profiles in wild-type and S534A mice treated with low-dose LPS.
Author contributions: J.-P.P. generated knock-in mice, designed experiments, performed analysis, and drafted the manuscript; C.H. performed experiments and drafted the manuscript; C.K. and T.L. performed experiments; R.A.F. performed microarray analysis; and R.F.S. conceptualized and oversaw the study, designed experiments, and drafted the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES and NOTES
- 1.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 2.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annual Review of Immunology. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunological reviews. 2012;246:239–253. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercurio F, Murray BW, Shevchenko A, Bennett BL, Young DB, Li JW, Pascual G, Motiwala A, Zhu H, Mann M, Manning AM. IkappaB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Molecular and Cellular Biology. 1999;19:1526–1538. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes & Development. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz ML, Mattioli I, Buss H, Kracht M. NF-kappaB: a multifaceted transcription factor regulated at several levels. Chembiochem. 2004;5:1348–1358. doi: 10.1002/cbic.200400144. [DOI] [PubMed] [Google Scholar]
- 7.Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cellular Signalling. 2010;22:1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes & Development. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. The Journal of Biological Chemistry. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki T, Sakon S, Sasazuki T, Sakurai H, Doi T, Yagita H, Okumura K, Nakano H. Phosphorylation of serine 276 is essential for p65 NF-kappaB subunit-dependent cellular responses. Biochemical and Biophysical Research Communications. 2003;300:807–812. doi: 10.1016/s0006-291x(02)02932-7. [DOI] [PubMed] [Google Scholar]
- 12.Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S. Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes & development. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappa B through utilization of the Ikappa B kinase and activation of the mitogen-activated protein kinase p38. The Journal of Biological Chemistry. 2001;276:18934–18940. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 14.O'Mahony AM, Montano M, Van Beneden K, Chen LF, Greene WC. Human T-cell lymphotropic virus type 1 tax induction of biologically Active NF-kappaB requires IkappaB kinase-1-mediated phosphorylation of RelA/p65. The Journal of biological chemistry. 2004;279:18137–18145. doi: 10.1074/jbc.M401397200. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Tang E, Guan K, Wang CY. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. Journal of Immunology. 2003;170:5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki CY, Barberi TJ, Ghosh P, Longo DL. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. The Journal of Biological Chemistry. 2005;280:34538–34547. doi: 10.1074/jbc.M504943200. [DOI] [PubMed] [Google Scholar]
- 17.Adli M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. The Journal of Biological Chemistry. 2006;281:26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 18.Douillette A, Bibeau-Poirier A, Gravel SP, Clement JF, Chenard V, Moreau P, Servant MJ. The proinflammatory actions of angiotensin II are dependent on p65 phosphorylation by the IkappaB kinase complex. The Journal of Biological Chemistry. 2006;281:13275–13284. doi: 10.1074/jbc.M512815200. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed AU, Sarvestani ST, Gantier MP, Williams BR, Hannigan GE. Integrin-linked kinase modulates lipopolysaccharide- and Helicobacter pylori-induced nuclear factor kappaB-activated tumor necrosis factor-alpha production via regulation of p65 serine 536 phosphorylation. The Journal of Biological Chemistry. 2014;289:27776–27793. doi: 10.1074/jbc.M114.574541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohuslav J, Chen LF, Kwon H, Mu Y, Greene WC. p53 induces NF-kappaB activation by an IkappaB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. The Journal of Biological Chemistry. 2004;279:26115–26125. doi: 10.1074/jbc.M313509200. [DOI] [PubMed] [Google Scholar]
- 21.Oakley F, Teoh V, Ching ASG, Bataller R, Colmenero J, Jonsson JR, Eliopoulos AG, Watson MR, Manas D, Mann DA. Angiotensin II activates I kappaB kinase phosphorylation of RelA at Ser 536 to promote myofibroblast survival and liver fibrosis. Gastroenterology. 2009;136:2334–2344. e2331. doi: 10.1053/j.gastro.2009.02.081. [DOI] [PubMed] [Google Scholar]
- 22.Moles A, Sanchez AM, Banks PS, Murphy LB, Luli S, Borthwick L, Fisher A, O'Reilly S, van Laar JM, White SA, Perkins ND, Burt AD, Mann DA, Oakley F. Inhibition of RelA-Ser536 phosphorylation by a competing peptide reduces mouse liver fibrosis without blocking the innate immune response. Hepatology. 2013;57:817–828. doi: 10.1002/hep.26068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi H, Hirano T, Whitmore SE, Morisaki I, Amano A, Lamont RJ. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLoS Pathogens. 2013;9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb A, Yang XD, Tsang YH, Li JD, Higashi H, Hatakeyama M, Peek RM, Blanke SR, Chen LF. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO reports. 2009;10:1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JE, Kim SY, Lim SY, Kieff E, Song YJ. Role of Ca2+/calmodulin-dependent kinase II-IRAK1 interaction in LMP1-induced NF-kappaB activation. Molecular and Cellular Biology. 2014;34:325–334. doi: 10.1128/MCB.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song YJ, Jen KY, Soni V, Kieff E, Cahir-McFarland E. IL-1 receptor-associated kinase 1 is critical for latent membrane protein 1-induced p65/RelA serine 536 phosphorylation and NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2689–2694. doi: 10.1073/pnas.0511096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattioli I, Sebald A, Bucher C, Charles RP, Nakano H, Doi T, Kracht M, Schmitz ML. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. Journal of Immunology. 2004;172:6336–6344. doi: 10.4049/jimmunol.172.10.6336. [DOI] [PubMed] [Google Scholar]
- 28.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 29.Sauter C, Wolfensberger C. Interferon in human serum after injection of endotoxin. Lancet. 1980;2:852–853. doi: 10.1016/s0140-6736(80)90189-0. [DOI] [PubMed] [Google Scholar]
- 30.Grove M, Plumb M. C/EBP, NF-kappa B, and c-Ets family members and transcriptional regulation of the cell-specific and inducible macrophage inflammatory protein 1 alpha immediate-early gene. Molecular and Cellular Biology. 1993;13:5276–5289. doi: 10.1128/mcb.13.9.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradere JP, Kluwe J, De Minicis S, Jiao JJ, Gwak GY, Dapito DH, Jang MK, Guenther ND, Mederacke I, Friedman R, Dragomir AC, Aloman C, Schwabe RF. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology. 2013;58:1461–1473. doi: 10.1002/hep.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohno S, Im HJ, Knudson CB, Knudson W. Hyaluronan oligosaccharides induce matrix metalloproteinase 13 via transcriptional activation of NFkappaB and p38 MAP kinase in articular chondrocytes. The Journal of Biological Chemistry. 2006;281:17952–17960. doi: 10.1074/jbc.M602750200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwabe RF, Sakurai H. IKKbeta phosphorylates p65 at S468 in transactivaton domain 2. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2005;19:1758–1760. doi: 10.1096/fj.05-3736fje. [DOI] [PubMed] [Google Scholar]
- 34.Buss H, Dorrie A, Schmitz ML, Frank R, Livingstone M, Resch K, Kracht M. Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. The Journal of Biological Chemistry. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- 35.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 36.Whitmarsh AJ, Davis RJ. Regulation of transcription factor function by phosphorylation. Cellular and Molecular Life Sciences : CMLS. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Guan PP, Wang T, Yu X, Guo JJ, Wang ZY. Aggravation of Alzheimer's disease due to the COX-2-mediated reciprocal regulation of IL-1beta and Abeta between glial and neuron cells. Aging Cell. 2014;13:605–615. doi: 10.1111/acel.12209. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhao Y, Banerjee S, Dey N, LeJeune WS, Sarkar PS, Brobey R, Rosenblatt KP, Tilton RG, Choudhary S. Klotho depletion contributes to increased inflammation in kidney of the db/db mouse model of diabetes via RelA (serine)536 phosphorylation. Diabetes. 2011;60:1907–1916. doi: 10.2337/db10-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Hu L, Tong X, Ye X. Casein kinase 1gamma1 inhibits the RIG-I/TLR signaling pathway through phosphorylating p65 and promoting its degradation. Journal of Immunology. 2014;192:1855–1861. doi: 10.4049/jimmunol.1302552. [DOI] [PubMed] [Google Scholar]
- 41.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO reports. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.