ABSTRACT
Zika virus (ZIKV) is an emerging human pathogen that is spreading rapidly through the Americas and has been linked to the development of microcephaly and to a dramatically increased number of Guillain-Barré syndrome cases. Currently, no vaccine or therapeutic options for the prevention or treatment of ZIKV infections exist. In the study described in this report, we expressed, purified, and characterized full-length nonstructural protein 5 (NS5) and the NS5 polymerase domain (NS5pol) of ZIKV RNA-dependent RNA polymerase. Using purified NS5, we developed an in vitro nonradioactive primer extension assay employing a fluorescently labeled primer-template pair. Both purified NS5 and NS5pol can carry out in vitro RNA-dependent RNA synthesis in this assay. Our results show that Mn2+ is required for enzymatic activity, while Mg2+ is not. We found that ZIKV NS5 can utilize single-stranded DNA but not double-stranded DNA as a template or a primer to synthesize RNA. The assay was used to compare the efficiency of incorporation of analog 5′-triphosphates by the ZIKV polymerase and to calculate their discrimination versus that of natural ribonucleotide triphosphates (rNTPs). The 50% inhibitory concentrations for analog rNTPs were determined in an alternative nonradioactive coupled-enzyme assay. We determined that, in general, 2′-C-methyl- and 2′-C-ethynyl-substituted analog 5′-triphosphates were efficiently incorporated by the ZIKV polymerase and were also efficient chain terminators. Derivatives of these molecules may serve as potential antiviral compounds to be developed to combat ZIKV infection. This report provides the first characterization of ZIKV polymerase and demonstrates the utility of in vitro polymerase assays in the identification of potential ZIKV inhibitors.
KEYWORDS: RdRp, ZIKV, Zika virus, nonradioactive assay, polymerase assay, polymerase inhibitor
INTRODUCTION
Zika virus (ZIKV) is an emerging arthropod-borne human pathogen in the family Flaviviridae (genus Flavivirus) first isolated in 1947 from a febrile sentinel rhesus monkey in the Zika Forest of Uganda (1). Though mainly transmitted by the Aedes aegypti mosquito, current reports strongly suggest that the virus is being transmitted perinatally, sexually, and via blood transfusion (2–4). ZIKV infections are usually short lasting and self-limiting, with 80% of infected individuals being clinically asymptomatic. Symptoms for patients that become ill are usually mild and non-life threatening. Symptoms include fever, maculopapular rash, joint pain and/or conjunctivitis, muscle pain, headache, and retro-orbital pain. Recently, higher than normal incidences of Guillain-Barré syndrome (GBS), the most frequent cause of non-poliovirus-associated acute flaccid paralysis, and of primary microcephaly cases have been linked to ZIKV outbreaks in French Polynesia and Brazil (5). GBS is a serious disease believed to be initiated by an immune-mediated response to antigenic exposure from certain viruses or bacterial infections. Roughly 20% of the affected patients are left with severe disability, and approximately 5% of the patients die (6). Also of great concern is the apparent correlation of ZIKV infections with a 20-fold increase in the incidence of microcephaly cases reported in Brazil in 2015 (5). The explosive spread of the virus through the Americas and the increasing evidence of teratogenic and neuropathic effects associated with ZIKV infections have caused great concern and prompted the WHO Public Health Emergency Committee to declare ZIKV a global public health emergency (7). Zika is the first mosquito-borne illness to receive an emergency health status, and the WHO is predicting that the spread of the virus will reach global pandemic proportions in 2016 and 2017 with over 4 million infections worldwide. Currently, there are no vaccines or therapeutic options for the prevention or treatment of ZIKV infections. The lessons learned from efforts to develop a vaccine against dengue fever virus (DFV) indicate that development of a prophylactic vaccine to protect against ZIKV may be very challenging and could require more time and effort than the development of antiviral agents.
The mechanism of infection with ZIKV has not been well studied, but the replication cycle of the virus may be similar to that of other Flavivirus members, such as DFV. Presumably, ZIKV genome replication occurs within intracellular compartments in the endoplasmic reticulum and is catalyzed by a membrane-bound viral replication complex consisting of viral nonstructural proteins, viral RNA, and host proteins, the identity of which are unknown. The genome of ZIKV is a single-stranded plus-strand RNA molecule approximately 10.7 kb in length. It serves as a carrier of genomic information and as mRNA for protein translation. Genomic RNA contains two noncoding flanking regions (NCR) known as the 5′-NCR and the 3′-NCR. The ZIKV RNA genome contains a single open reading frame (ORF) encoding a long polypeptide (∼3,400 amino acids), which is cleaved into three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) (8). It is assumed that, like other members of the Flaviviridae family, the ZIKV replication complex first transcribes genomic plus-strand RNA into a complementary minus-strand RNA intermediate, resulting in the formation of a duplex RNA. The minus strand of this duplex serves as a template for multiple rounds of plus-strand RNA synthesis. Viral RNA synthesis occurs through an asymmetric replication cycle in which 10 times more plus-strand RNA than minus-strand RNA is synthesized (9). A key target for the development of a therapeutic antiviral agent is the ZIKV genome-encoded RNA-dependent RNA polymerase (RdRp). By homology with other Flavivirus members, the catalytic region of the enzyme is located within the C-terminal portion of the NS5 protein. Additionally, the NS5 protein also contains methyltransferase activity in the N-terminal portion of the NS5 protein, which is involved in the capping of plus-strand genomic RNA (10–12). The overall architecture of the ZIKV RdRp most likely resembles the canonical right-hand conformation, consisting of fingers, palm, and thumb subdomains, that has been typical for all known polymerase structures and is similar to that reported for the RdRps of other members of the Flaviviridae family, including hepatitis C virus (HCV) and DFV.
Significant nucleoside-based drug discovery and development have been undertaken and are ongoing to combat viral infections caused by Flaviviridae members, such as HCV and dengue virus (13–19). Sofosbuvir, an FDA-approved ribonucleotide analog targeting HCV NS5b polymerase, has been shown to be a very effective and safe drug for the treatment of HCV infection, and these findings provided a proof of concept for the use of ribonucleotide analogs as effective antiviral drugs against Flaviviridae (20, 21). Effective ribonucleoside analogs resemble natural ribonucleosides that can be converted into the corresponding active ribonucleotide 5′-triphosphates, which are then incorporated into nascent viral RNA by viral RdRps. To inhibit viral replication, two properties are desirable for ribonucleoside 5′-triphosphate analogs: (i) efficient incorporation by the viral polymerase and (ii) chain termination. These two criteria are the first to be evaluated for any ribonucleotide analog.
In this report, we demonstrate the expression and purification of both the full-length NS5 (NS5) and the NS5 polymerase domain (NS5pol) of the ZIKV RNA-dependent RNA polymerase. Using the purified NS5 enzyme, we developed an in vitro nonradioactive primer extension assay using a fluorescently labeled primer-template (P/T) pair. The incorporation efficiency and chain termination ability of a series of ribonucleotide 5′-triphosphates of known flavivirus polymerase inhibitors were determined using this assay. Several analog ribonucleotide 5′-triphosphates that were incorporated by the polymerase and caused chain termination were further evaluated in a nonradioactive coupled-enzyme inhibition assay to determine 50% inhibitory concentrations (IC50 values).
RESULTS
RdRp expression and purification.
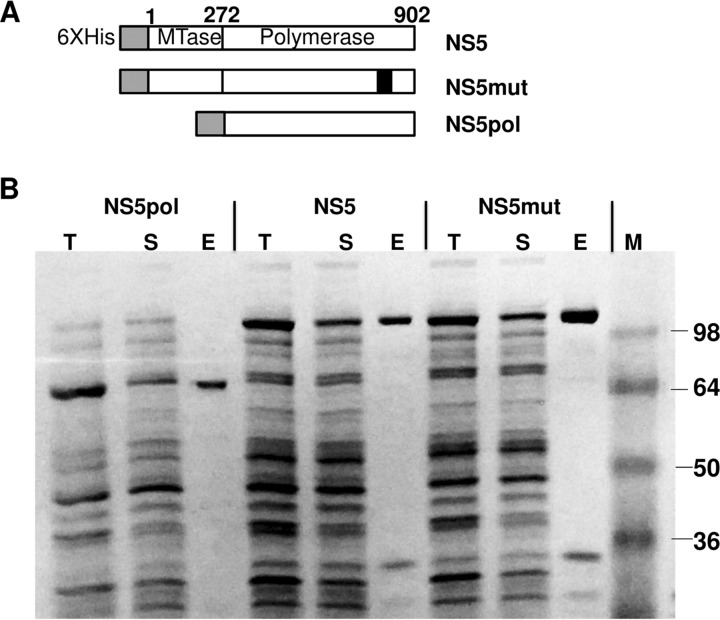
Two versions of ZIKV RNA-dependent RNA polymerase were prepared and tested in this study: full-length NS5 and a truncated NS5 containing the polymerase domain only. The expression vector diagrams are shown in Fig. 1A. A predicted inactive polymerase mutant control (NS5mut) containing an active-site mutation (GDD → GAA) was also prepared. All recombinant proteins contained a 6×His tag at the N terminus to facilitate protein purification. All three variants of ZIKV polymerase were expressed in and purified from Escherichia coli and were found to be partially soluble (Fig. 1B). Interestingly, under the same conditions, both full-length dengue virus NS5 and the RdRp domain of dengue virus NS5 were expressed in E. coli as fully soluble proteins (data not shown). Different strategies were tried to increase ZIKV protein solubility, including the use of different expression E. coli strains (E. coli strain SHuffle T7 Express lysY and Stellar cells), performance of induction at different temperatures (24°C and 16°C), the use of different E. coli cell densities at the time of induction (optical density at 650 nm, 0.1 and 1), and the use of various concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside) for induction (0.01 to 0.1 mM). None of these variations led to a significant increase in ZIKV NS5 protein solubility after expression in E. coli. The highest solubility was achieved by performing induction of ZIKV NS5, NS5pol, and NS5mut expression at 20°C for 2 h. Higher temperatures and/or longer induction/expression times resulted in the majority of the expressed protein being found in the insoluble fraction. After purification, the identity of the purified proteins was determined by mass spectrometry analysis, and the proteins were confirmed to be ZIKV NS5 (data not shown). A few common minor E. coli protein contaminants (<10% of total protein) were also detected in the purified samples by mass spectrometry analysis; those contaminating proteins did not have measurable polymerase activity, as evidenced by the findings for the NS5mut control purified in parallel under the same conditions (see below).
FIG 1.
Construction, expression, and purification of ZIKV NS5, NS5pol, and NS5mut. (A) Schematic diagram of the NS5, NS5pol, and NS5mut expression vectors. NS5mut contains an active-site mutation (GDD → GAA) in amino acids 663 to 665. MTase, methyltransferase domain. (B) SDS-PAGE analysis of protein expression and purification. Lanes T, total cell lysate; lanes S, soluble fraction; lanes E, eluate from the HisPur Ni-NTA agarose resin; lane M, protein standard marker. The sizes of protein markers (in kilodaltons) are indicated on the right.
Purified enzyme polymerase reaction conditions.
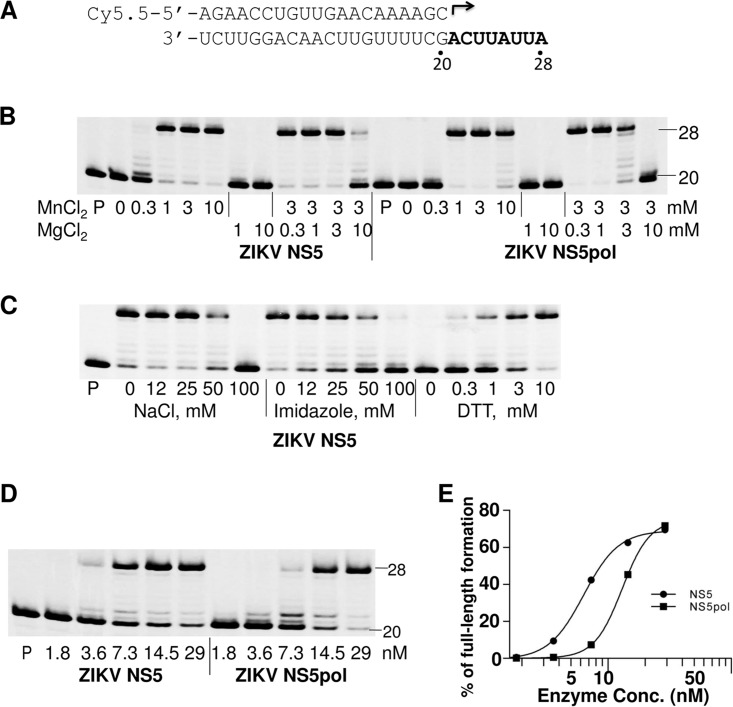
ZIKV NS5 and NS5pol polymerase activity was measured in a primer extension assay using a fluorescently labeled RNA primer (20-mer) annealed to an unlabeled RNA template in the presence of 100 μM natural ribonucleotide triphosphates (rNTPs). The product of the primer extension, a full-length 28-mer RNA, was analyzed by denaturing polyacrylamide gel electrophoresis (urea-PAGE) (Fig. 2A). The primer was fully extended by both enzyme forms in the presence of 1 to 10 mM MnCl2 but not in the presence of 1 to 10 mM MgCl2 (Fig. 2B). Furthermore, the polymerization reaction was inhibited with increasing concentrations of MgCl2 when MgCl2 was added to the standard concentration of MnCl2.
FIG 2.
Analysis of reaction conditions for purified enzymes. Polymerase activity was evaluated in a primer extension assay. (A) P/T duplex composition used in the assay. The 20-mer primer (top) contains a fluorescent label (Cy5.5) at the 5′ end. Arrow, location and direction of primer extension to form a 28-mer product. (B) Analysis of the effect of MnCl2 and MgCl2 on polymerase activity. ZIKV NS5 or NS5pol (15 nM) was incubated with 10 nM P/T complex in a reaction buffer in the presence of the indicated concentrations of MgCl2 or MnCl2, or both. The reactions were initiated by addition of 100 μM NTPs, and the products were separated by denaturing PAGE. Lanes P, primer only. (C) Effect of different NaCl, imidazole, and DTT concentrations on NS5 polymerase activity. The concentrations of the components in the reaction mixtures are indicated under each lane. In the reaction mixtures measuring the effect of NaCl or imidazole, the DTT concentration was 10 mM. No NaCl or imidazole was added to the reaction mixtures for measuring the effect of DTT. (D) Analysis of the effect of the NS5 or NS5pol concentration on polymerase activity. Serial dilutions of NS5 or NS5pol (1.8 to 29 nM; indicated under each lane) and a constant concentration of P/T (10 nM) were used in this assay. The number 20 on the right of the gels in panels B and D indicates the location of the 20-mer unextended primer, and the number 28 indicates the location of the 28-mer full-length product. (E) The percentages of the full-length products in the reactions in panel D were plotted against the enzyme concentrations. The results were fitted to sigmoidal dose-response curves using the GraphPad Prism program.
The effect of the reaction buffer ionic strength on the polymerase activity was tested by adding increasing concentrations (0 to 100 mM) of NaCl or imidazole to the reaction mixture. We found that both NaCl and imidazole were tolerable at low concentrations but inhibited NS5 polymerase activity at concentrations higher than 25 mM (Fig. 2C). The high concentration of NaCl or imidazole may negatively affect the binding of NS5 to RNA through electrostatic interactions, thereby inhibiting overall activity. We also found that ZIKV NS5 requires a relatively high concentration of dithiothreitol (DTT; 10 mM) to achieve maximal polymerase activity (Fig. 2C), suggesting that the cysteine residues in ZIKV NS5 may readily form intra- or intermolecular disulfide bonds, resulting in decreased polymerase activity.
To confirm that the polymerase activity tested in the primer extension reaction is an inherent property of purified NS5, a mutant NS5 (NS5mut, which contains an active-site mutation, GDD → GAA, in amino acid positions 663 to 665 of NS5) was also expressed and purified in parallel with wild-type NS5 (Fig. 1). This mutation caused the complete loss of NS5 polymerase activity (Fig. 3A, lanes 3 and 6), confirming that the primer extension activity in the reaction with nonmutated enzyme could be attributed to purified active ZIKV NS5. To further compare the enzymatic activity of ZIKV NS5 and NS5pol, an enzyme dilution experiment was carried out. In this experiment, different concentrations of NS5 or NS5pol (1.8 nM to 29 nM) were incubated with 10 nM P/T complex, and the reactions were initiated by adding rNTPs to a final concentration of 100 μM and continued for 1 h at 35°C (Fig. 2D). The percentage of full-length product in the total reaction mixture was plotted against the log of the enzyme concentrations (Fig. 2E). The resulting curves were fitted to a hyperbolic S shape. The curves were slightly different for NS5 and NS5pol, suggesting that the polymerase activity of the purified NS5 and NS5pol may have different enzymatic properties. The time course of the primer extension reaction was also evaluated (see Fig. S1 in the supplemental material). The results suggest that the initiation of the primer extension reaction is a slow process and that preincubation in the presence of ATP (the first nucleotide to be incorporated) did not significantly accelerate the formation of the full-length products. It is possible that only a small fraction of NS5 in the reaction mixture is capable of synthesizing RNA and that multiple rounds of enzyme turnover lead to the accumulation of the full-length products. Due to the slow kinetics of the primer extension reaction, a 1-h incubation time was used in the following primer extension assays.
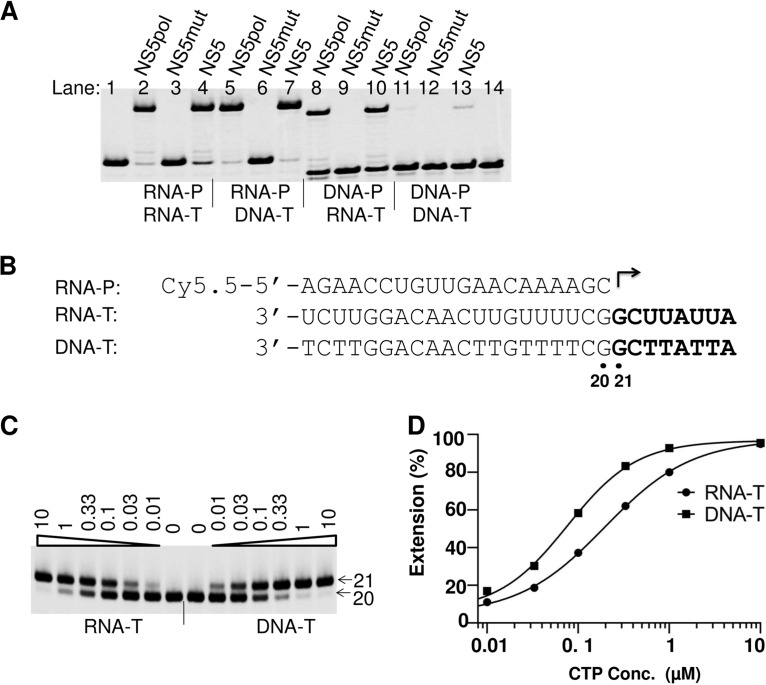
FIG 3.
ZIKV RdRp template specificity. (A) Effect of using different P/T duplexes on efficiency of primer extension. NS5 (30 nM), NS5pol (30 nM), or NS5mut (60 nM) was incubated with 10 nM P/T complex in the polymerase reaction buffer; the reactions were initiated by addition of 100 μM (each) rNTP. The products were analyzed by denaturing PAGE. The primers (RNA-P and DNA-P) and templates (RNA-T and DNA-T) in the P/T duplexes used in the reaction mixtures are indicated under each lane. The RdRp enzymes used in the reaction mixtures are indicated above the lanes. (B) Sequences of the primer and the templates used in the assays whose results are shown in panel C. (C) Analysis of CTP incorporation using RNA or single-stranded DNA as the template. The templates used in the reaction mixtures are indicated at the bottom. ZIKV NS5 and RNA-P/RNA-T or RNA-P/DNA-T duplexes were mixed in the reaction buffer, and the reactions were initiated by addition of the indicated concentrations of CTP (in micromolar; indicated above each lane). The products of the reactions were analyzed by denaturing PAGE. Arrow labeled 20, location of the 20-mer primer; arrow labeled 21, location of the 21-mer extended product. (D) The percentages of the extended products in panel C were plotted against the CTP concentration, and the results were fitted to a sigmoidal dose-response curve to generate K1/2. For the RNA template, K1/2, CTP = 0.20 μM; and for the DNA template, K1/2, CTP = 0.076 μM.
ZIKV NS5 enzyme specificity for primer and template composition.
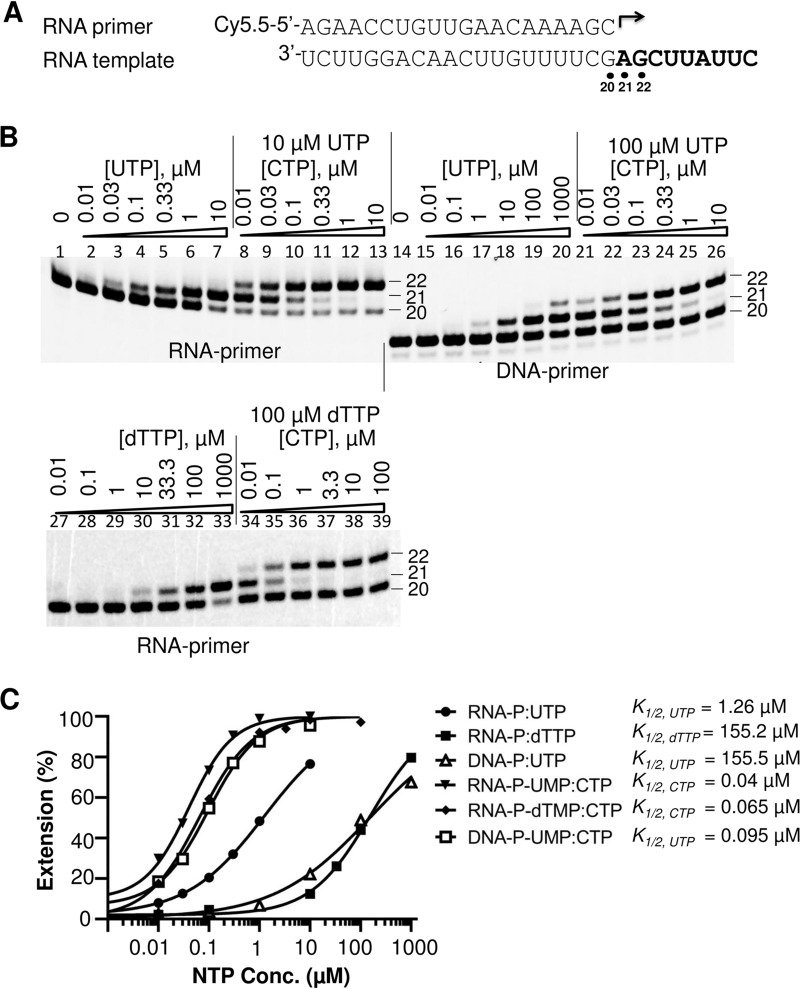
The natural activity of ZIKV NS5 is presumably to serve as an RdRp. To test whether ZIKV NS5 can utilize other templates, four different primer-template duplexes (RNA primer-RNA template, RNA primer-DNA template, DNA primer-RNA template, and DNA primer-DNA template) were used as P/T pairs in the primer extension assay. Both ZIKV NS5 and NS5pol could efficiently extend an RNA primer that was annealed to an RNA or a DNA template (Fig. 3A, lanes 2 and 4 and lanes 5 and 7, respectively). The efficiency of RNA elongation using an RNA or DNA template was further evaluated in a single ribonucleotide incorporation assay (Fig. 3C). In this assay, the efficiency of ribonucleotide incorporation was tested by measuring the ribonucleotide concentration at which half of a 20-mer RNA primer is extended into the 21-mer RNA product (K1/2). When RNA was used as a template in this assay, the K1/2 value for CTP was measured to be 0.20 μM; when DNA was used as a template under conditions that were otherwise the same, K1/2 was measured to be 0.076 μM (Fig. 3D). These results suggest that DNA can serve as a template for ZIKV NS5 with an efficiency similar to that of an RNA template. We also found that DNA could be extended when it was used as a primer for ZIKV NS5 and NS5pol to synthesize RNA in cases when the DNA was paired with an RNA template (Fig. 3A, lanes 8 and 10). In contrast, a DNA primer paired with a DNA template was poorly extended by NS5 and NS5pol (Fig. 3A, lanes 11 and 13). In the DNA primer-RNA template case, the efficiency of full-length product formation was dramatically decreased compared to that when an RNA primer-RNA template was used (Fig. 3A, lanes 2 and 4 versus lanes 8 and 10). To further investigate the use of DNA as a primer, a single nucleotide incorporation assay was performed to measure the K1/2 values of ribonucleotides following either DNA or RNA primers under reaction conditions that were otherwise the same. When DNA was used as a primer, the K1/2 value for the first ribonucleotide, UTP (K1/2, UTP), was 155.5 μM, which was 120-fold higher than that when RNA served as a primer (Fig. 4B and C). In contrast, the K1/2 values of CTP (K1/2, CTP), the second ribonucleotide following the first U, were not substantially different (K1/2, CTP = 0.095 μM for the DNA primer and 0.040 μM for the RNA primer; Fig. 4C). These results suggest that the efficiency of incorporation of the first ribonucleotide was significantly impacted when DNA was used as a primer, but the efficiency of incorporation of the following ribonucleotide approached normal values, i.e., the values obtained when RNA was used as a primer. It should be noted that the effect of a long DNA primer on the efficiency of incorporation of the next ribonucleotide is different from a misincorporation of a single deoxynucleotide following an RNA primer. In the last case, the incorporation efficiency of the next ribonucleotide was minimally affected (Fig. 4B, bottom, and C): the K1/2 values of CTP incorporation into the RNA-UMP and RNA-dTMP primers were 0.040 and 0.065 μM, respectively.
FIG 4.
Effect of using DNA or RNA as a primer by ZIKV NS5 RdRp. (A) Composition of the primer and template in the RNA-RNA P/T duplex used in the assay whose results are shown in panel B. The corresponding DNA primer is listed in Table 1 and was used in the DNA-RNA P/T. (B) Single nucleotide incorporation assay using RNA or DNA as a primer. The primers used in the P/T duplexes are indicated under the panels. The serial dilution of the first (UTP or dTTP) or second (CTP) nucleotide that was used in the assays is indicated above each lane. For assay of incorporation of the second nucleotide, 10 μM UTP was used as a first nucleotide following the RNA primer (lanes 8 to 13), 100 μM UTP was used as a first nucleotide following the DNA primer (lanes 21 to 26), and 100 μM dTTP was used as the first nucleotide following the RNA primer (lanes 34 to 39). The locations of the primers (both are 20-mers) as well as of the first nucleotide extension products (21-mers) and the second nucleotide extension products (22-mers) are indicated by the corresponding numbers. Note that DNA-containing products migrate faster than the corresponding RNA products. (C) Quantitative analysis of UTP, dTTP, and CTP incorporation efficiency from panel B. UTP and dTTP incorporation efficiency was evaluated on the basis of the extension of the 20-mer to 21-mer products and the incorporation efficiency of CTP was evaluated on the basis of the extension of 21-mer to 22-mer products, as described in the Fig. 3 legend.
Chain termination.
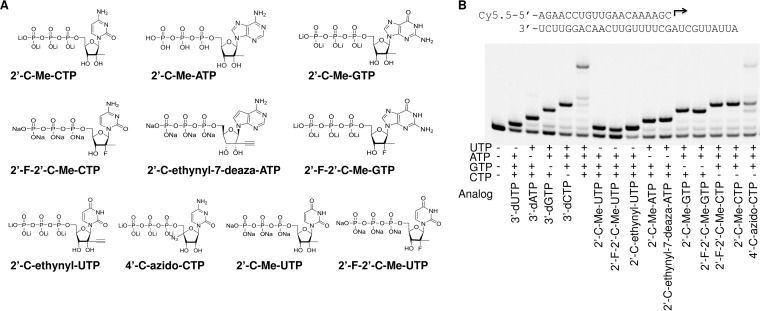
Chain termination is one of the major mechanisms of inhibition of viral RNA polymerases by ribonucleotide analogs. Several ribonucleotide 5′-triphosphate inhibitors that have activities against other Flaviviridae members (Fig. 5A) and that are currently on the market or in various stages of preclinical or clinical development were tested for their ability to cause chain termination during RNA synthesis by ZIKV NS5. Primer extension reactions were performed in the presence of different rNTP analogs complemented with three natural rNTPs (Fig. 5B). 3′-dUTP, 3′-dATP, 3′-dGTP, and 3′-dCTP are obligate chain terminators and served as positive controls for chain termination. As expected, incorporation of each 3′-deoxynucleotide triphosphate (3′-dNTP) fully prevented extension of a product in the presence of the next correct ribonucleotide(s). Similarly, β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine-5′-triphosphate (2′-F-2′-C-Me-UTP), β-d-2′-β-C-methyluridine-5′-triphosphate (2′-C-Me-UTP), β-d-2′-β-C-ethynyluridine-5′-triphosphate (2′-C-ethynyl-UTP), β-d-2′-β-C-methyladenosine-5′-triphosphate (2′-C-Me-ATP), β-d-2′-β-C-ethynyl-7-deaza-adenosine-5′-triphosphate (2′-C-ethynyl-7-deaza-ATP), β-d-2′-β-C-methylguanosine-5′-triphosphate (2′-C-Me-GTP), β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-triphosphate (2′-F-2′-C-Me-GTP), β-d-2′-β-C-methylcytidine-5′-triphosphate (2′-C-Me-CTP), and β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylcytidine-5′-triphosphate (2′-F-2′-C-Me-CTP) caused chain termination after incorporation into nascent RNA. Interestingly, the 2′-C-Me- or 2′-C-ethynyl-modified ribonucleotide analog 5′-triphosphates caused immediate chain termination after incorporation, while incorporation of 4′-C-azidocytidine-5′-triphosphate (4′-C-azido-CTP) caused only partial chain termination. Further analysis showed that the incorporation of 4′-C-azidocytidine in the nascent RNA chain resulted in a decreased efficiency of incorporation of the next correct ribonucleotide by as much as 25-fold (Fig. S2). It has also been reported that the incorporation of 4′-C-azidocytidine into HCV RNA could block the synthesis of the complementary strand (14, 22). This mechanism of inhibition still needs to be investigated.
FIG 5.
(A) Chemical structures of the ribonucleotide 5′-triphosphate analogs used in the assay. (B) Analysis of chain termination ability of ribonucleotide analogs in the reaction with ZIKV NS5 RdRp. The P/T duplex used in the assay is indicated at the top. The polymerase reactions were initiated by addition of the ribonucleotide analogs at 100 μM and of the three complementing natural ribonucleotides at 10 μM each, as indicated under each lane, and the products of the reactions were resolved by denaturing PAGE.
Incorporation efficiency of ribonucleotide analogs.
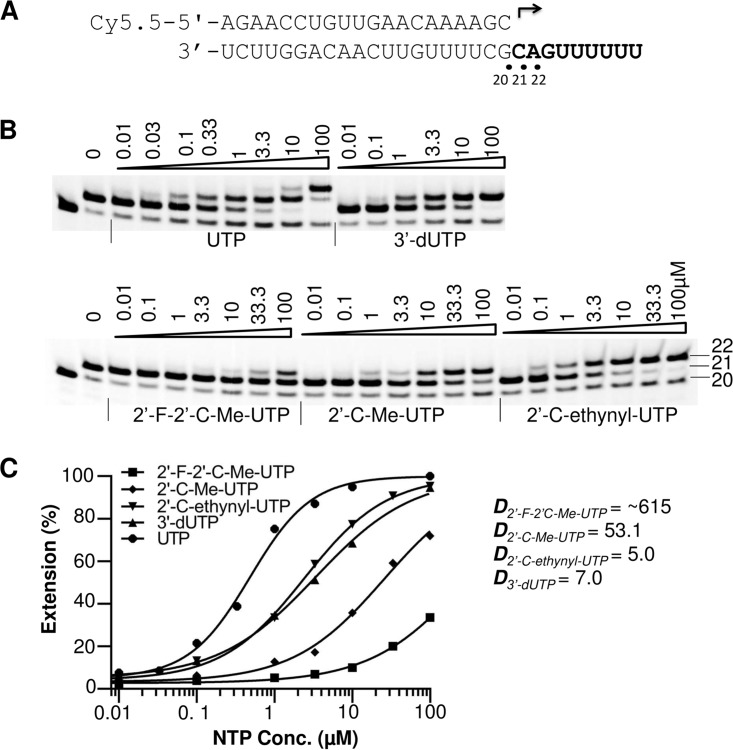
Since cells contain high levels of natural rNTPs (23), ribonucleotide analogs have to compete with the natural rNTPs for incorporation in order to inhibit NS5 polymerase activity. To act as a competitive, alternative substrate inhibitor of an RdRp, a ribonucleotide analog must be incorporated into the growing RNA chain. The relative incorporation efficiency of ribonucleotide analogs versus natural ribonucleotides plays a major role in the ability of analogs to inhibit polymerase activity. In this study, the relative incorporation efficiencies of ribonucleotide analogs were evaluated by measuring the K1/2 value of RNA synthesis in a single ribonucleotide incorporation assay and by comparing that value to the K1/2 value of natural rNTPs determined under the same reaction conditions. Discrimination values were calculated by dividing the K1/2 of the rNTP analogs by the K1/2 of the natural rNTPs and are referred to as Danalog values. To avoid the effect of the first nucleotide incorporation, which can be affected by the processes of P/T binding and by variable rates of first nucleotide incorporation, different templates were designed such that the first incorporated ribonucleotide would be a natural ribonucleotide used at a high concentration to promote the initiation, and the tested analog would be incorporated as the second nucleotide in the product (Table 1). An example of the P/T design and testing of UTP analogs is shown in Fig. 6. In this assay the measured K1/2 values of the UTP analogs 2′-F-2′-C-Me-UTP, 2′-C-Me-UTP, 2′-C-ethynyl-UTP, and 3′-dUTP were ∼297.6 μM, 25.72 μM, 2.421 μM, and 3.376 μM, respectively; these K1/2 values were compared with the K1/2 of UTP (0.484 μM) measured under the same conditions. The calculated Danalog values were ∼615, 53.1, 5.0, and 7.0, respectively (Fig. 6C). These results suggest that the 2′-C-ethynyl modification of UTP was better tolerated by ZIKV NS5 than the 2′-C-Me modification. The 2′-F and 2′-C-Me double modification further decreased the incorporation efficiency of the ribonucleotide analog by ZIKV NS5. Since this assay is designed to measure the incorporation of the second nucleotide (elongation only), we have confirmed that the results are not affected by the absence of Mg2+ ions in the assay. Analogous assays in the presence of 3 mM MnCl2 plus 3 mM MgCl2 in the reaction mixture showed the same analog's discrimination trends (Fig. S3). The discrimination values of all tested ribonucleotide analogs are summarized in Table 2. The results show that the discrimination values of 2′-C-Me-ATP, 2′-C-ethynyl-7-deaza-ATP, and 2′-C-ethynyl-UTP were the lowest, suggesting the high efficiency of incorporation of these compounds by ZIKV NS5. Derivatives of these ribonucleotide analogs may serve as potential antiviral molecules to combat ZIKV infection. It has been shown before that 2′-C-ethynyl-7-deaza-ATP is also a potent inhibitor of HCV and dengue virus RdRps, and its antiviral activity has been extensively studied (16, 18, 24).
TABLE 1.
Sequences of labeled and unlabeled oligonucleotides used in the study
| Oligonucleotide | Sequencea |
|---|---|
| Labeled RNA primer | Cy5.5-5′-AGAACCUGUUGAACAAAAGC-3′ |
| Labeled DNA primer | Cy5.5-5′-AGAACCTGTTGAACAAAAGC-3′ |
| RNA template 1 | 3′-UCUUGGACAACUUGUUUUCGGCUUAUUA-5′ |
| DNA template | 3′-TCTTGGACAACTTGTTTTCGGCTTATTA-5′ |
| RNA template 2 | 3′-UCUUGGACAACUUGUUUUCGAGCUUAUUC-5′ |
| Template for ATP analog K1/2 | 3′-UCUUGGACAACUUGUUUUCGAUCGUUAUUA-5′ |
| Template for UTP analog K1/2 | 3′-UCUUGGACAACUUGUUUUCGCAGUUUUUU-5′ |
| Template for GTP analog K1/2 | 3′-UCUUGGACAACUUGUUUUCGACUUAUUA-5′ |
| Template for CTP analog K1/2 | 3′-UCUUGGACAACUUGUUUUCGUGCAUUAUUA-5′ |
| Template for UTP analog IC50 | 5′-A30AACAGGUUCUAGAACCU GUU-3′ |
Boldface type indicates a single-stranded template part after P/T duplex formation.
FIG 6.
Efficiency of UTP analog incorporation by ZIKV NS5. (A) P/T sequence used to assay UTP analogs. (B) Polymerase reactions were performed in the presence of 10 μM GTP as the first nucleotide and increasing concentrations (in micromolar) of UTP or UTP analogs, as indicated above each lane. The identity of the tested ribonucleotide is indicated at the bottom. The locations of the 20-mer primer and 21- and 22-mer first and second nucleotide extension products are indicated on the right. (C) Quantitative analysis of UTP and UTP analog incorporation. The incorporation efficiency was evaluated on the basis of the extension of 21-mer to 22-mer products. The measured K1/2 values for UTP, 3′-dUTP, 2′-F-2′-C-Me-UTP, 2′-C-Me-UTP, and 2′-C-ethynyl-UTP were 0.484 μM, 3.376 μM, ~297.6 μM, 25.72 μM, and 2.421 μM, respectively. Danalog values were calculated as K1/2, analog/K1/2, UTP and are shown to the right of the graph.
TABLE 2.
Discrimination values of ribonucleotide analogs
| Ribonucleotide analog | Danaloga (mean ± SD) |
|---|---|
| ATP analogs | |
| 3′-dATP | 1.4 ± 0.5 |
| 2′-C-Me-ATP | 3.7 ± 0.4 |
| 2′-C-ethynyl-7-deaza-ATP | 5.7 ± 1.2 |
| GTP analogs | |
| 3′-dGTP | 3.7 ± 1.3 |
| 2′-C-Me-GTP | 12.9 ± 0.1 |
| 2′-F-2′-C-Me-GTP | 19.3 ± 4.0 |
| CTP analogs | |
| 3′-dCTP | 2.2 ± 0.5 |
| 2′-F-2′-C-Me-CTP | 9.8 ± 1.1 |
| 2′-C-Me-CTP | 6.7 ± 0.7 |
| 4′-C-azido-CTP | 437.5 ± 79.9 |
| UTP analogs | |
| 3′-dUTP | 5.7 ± 1.2 |
| 2′-F-2′-C-Me-UTP | 660.5 ± 64.3 |
| 2′-C-Me-UTP | 53.2 ± 0.1 |
| 2′-C-ethynyl-UTP | 4.9 ± 0.1 |
Danalog value = K1/2, analog/K1/2, natural. Data are from 2 or 3 independent experiments.
Inhibition of ZIKV NS5 by UTP analogs (IC50 measurement).
A nonradioactive assay based on chemiluminescent detection of pyrophosphate (PPi) was developed to measure the inhibition of ZIKV NS5 activity by analog rNTPs. In this assay, the polymerase activity is measured by quantifying the PPi released during the RNA synthesis reaction. After the polymerase reaction, the released PPi is converted to ATP in a reaction catalyzed by ATP sulfurylase using adenosine 5′-phosphosulfate sodium salt (APS) as a substrate, and the generated ATP is quantified in the same reaction tube using a sensitive chemiluminescent luciferase assay (25–28). An RNA template that contains a polyadenine sequence at the 5′ end and that can form a hairpin structure at the 3′ end (Table 1) was used as a primer-template in this assay. UTP analogs that can be incorporated by ZIKV NS5 and cause chain termination (see above) were tested in this assay (Table 3). The results showed that the IC50s for 2′-C-Me-UTP, 2′-F-2′-C-Me-UTP, 2′-C-ethynyl-UTP, and 3′-dUTP were 5.78, 90.76, 0.46, and 0.67 μM, respectively, when measured in the presence of 1 μM competing UTP. The IC50s of these analogs against dengue virus type 2 NS5 (prepared using the same methods) were also determined and showed a rank of activities similar to those measured for ZIKV NS5 (Table 3). Our data suggest that 2′-C-ethynyl-UTP is the most potent inhibitor of ZIKV NS5 and dengue virus NS5 polymerases, while 2′-C-Me-UTP is less active and 2′-F-2′-C-Me-UTP is the least potent in the enzyme inhibition assay.
TABLE 3.
UTP analog discrimination values and IC50s determined in ZIKV and dengue virus polymerase assays
| Analog ribonucleotide | ZIKV NS5 |
DFV NS5 |
||
|---|---|---|---|---|
| Danaloga (mean ± SD) | IC50 (μM) | Danaloga (mean ± SD) | IC50 (μM) | |
| 3′-dUTP | 5.7 ± 1.2 | 0.67 | 3.8 ± 1.7 | 0.68 |
| 2′-F-2′-C-Me-UTP | 660.5 ± 64.3 | 90.76 | 678 ± 258 | 55.13 |
| 2′-C-Me-UTP | 53.2 ± 0.1 | 5.78 | 55.6 ± 38.5 | 4.94 |
| 2′-C-ethynyl-UTP | 4.9 ± 0.1 | 0.46 | 6 | 0.33 |
Danalog value = K1/2, analog/K1/2, natural. Data are from 2 or 3 independent experiments.
DISCUSSION
In this study, recombinant full-length ZIKV NS5 RdRp and its polymerase domain were both successfully constructed, expressed, and purified. To our knowledge, this is the first report of the enzymatic characterization of ZIKV NS5. Using purified NS5, we have developed an in vitro nonradioactive primer extension assay and have demonstrated that it can be used as a tool to identify ribonucleotide analog substrates and inhibitors of ZIKV RdRp.
The most common problems with the expression of a viral protein in an E. coli system are protein solubility and stability. In our study, different ZIKV NS5 variants expressed in E. coli were only partially soluble. As described in the Results section, different protein expression strategies were tested, but none of the strategies led to significant improvements in ZIKV NS5 protein solubility. Under the same expression and purification conditions, both full-length NS5 and the RdRp subdomain of DFV could be expressed as fully soluble proteins (data not shown). Both ZIKV and dengue virus belong to the Flavivirus genus, and the NS5 protein sequences show 60% amino acid sequence identity, but the expressed proteins have noticeably different solubilities. Perhaps some sequences or domains in ZIKV NS5 promote protein aggregation when ZIKV NS5 is expressed in E. coli. Further optimization of ZIKV NS5 expression conditions could be employed to increase the solubility of ZIKV NS5 in E. coli.
Purified ZIKV NS5 can extend an RNA primer annealed to an RNA template in a Mn2+-dependent fashion. The dependence of polymerase activity upon divalent metal ions has been demonstrated for other viral RNA polymerases (29–31). Interestingly, Mg2+, the most common divalent metal ion used by many RNA polymerases, cannot support ZIKV NS5 primer extension activity. The effect of Mn2+ preference is not unique to the ZIKV polymerase and has been observed with other viral polymerases, including the NS5 protein of Japanese encephalitis virus (JEV), also a Flavivirus member (32, 33). In our study, a high concentration of Mg2+ inhibited ZIKV NS5 polymerase activity even in the presence of Mn2+, indicating perhaps that Mg2+ can compete with Mn2+ for binding to the active site of the ZIKV NS5.
It is unclear why the polymerase activity of purified ZIKV NS5 is not supported by Mg2+ in this study. In cells, Mg2+ would likely be the divalent metal ion utilized by the viral NS5 protein since the intracellular concentrations of Mg2+ are significantly higher than those of Mn2+. It is possible that cellular cofactors involved in the formation of the ZIKV replicase complex affect the divalent metal ion requirements of the NS5 polymerase. The mechanism of Mn2+ preference needs further characterization, as does the influence of Mn2+ on the ribonucleotide 5′-triphosphate substrate specificity of ZIKV NS5.
It is interesting that DNA can serve as a template for ZIKV NS5 to synthesize RNA. A similar flexibility has also been shown to some degree for other viral RdRps (34, 35). We also showed that ZIKV NS5 can use DNA as a primer to synthesize RNA, although the efficiency of priming is lower than that when RNA is used as a primer. We have also observed that both Zika and dengue virus NS5 proteins can incorporate 2′-deoxynucleotide triphosphates (2′-dNTPs) into RNA with some efficiency (Fig. 4B, bottom, and data not shown). Because 2′-dNTPs are present in cells at relatively high concentrations, it is possible that Flavivirus RdRps may sometimes misincorporate natural 2′-dNTPs into viral RNA and continue to use such mutated RNAs for further replication. It is speculated that being able to use DNA–3′-OH as a primer or to use a template with misincorporated DNA nucleotides enables the NS5 polymerase to continue synthesis of RNA after a deoxynucleotide misincorporation without abortion of the replication process, and to correct such mutations in the next round of RNA synthesis/copying. At the same time, the Flavivirus RdRps do not waste resources copying from DNA since the efficiency of synthesis initiation from double-stranded DNA is very low (Fig. 3A). This property of using DNA as a primer or a template is not unique to the Flavivirus genus. RdRps of other RNA viruses, e.g., brome mosaic virus and vesicular stomatitis virus, have been shown to use DNA, RNA, and chimeric templates to synthesize RNA (34, 36).
During the preparation of the manuscript, a communication by Sacramento et al., posted on the bioRxiv website, suggested that sofosbuvir, a ribonucleotide analog approved for use for the treatment of HCV infection, impairs Zika virus replication (37). Data from that non-peer-reviewed communication also suggest that the active form of sofosbuvir, 2′-F-2′-C-Me-UTP, can inhibit ZIKV RNA-dependent RNA polymerase activity very efficiently, with the IC50 being as low as 0.38 μM. Our data show that 2′-F-2′-C-Me-UTP can serve as a substrate for ZIKV NS5 and can cause chain termination upon incorporation. This result also indicates that it could serve as an inhibitor of ZIKV NS5, but our data showed that the incorporation efficiency was low (discrimination value, ∼660). To verify the effect of 2′-F-2′-C-Me-UTP inhibition of ZIKV NS5 activity, the inhibition potency (IC50) of 2′-F-2′-C-Me-UTP and of several other UTP analogs was measured in a polymerase inhibition assay. In our assay, the competing natural UTP concentration was 1 μM. The results indicate that 2′-F-2′-C-Me-UTP was not a very potent inhibitor (IC50 = 90.76 μM). On the other hand, the IC50 of 2′-C-ethynyl-UTP was 0.46 μM, which shows that this ribonucleotide analog is a potent inhibitor of ZIKV NS5. Using purified dengue virus NS5 RdRps, we obtained similar results: the IC50s for 2′-F-2′-C-Me-UTP and 2′-C-ethynyl-UTP were 55.13 and 0.33 μM, respectively. In the study by Sacramento et al., modified UTPs (biotin-UTP and DIG-UTP) rather than natural UTP were used as competing 5′-triphosphates (37). The low apparent IC50 for 2′-F-2′-C-Me-UTP (0.38 μM) may suggest that 2′-F-2′-C-Me-UTP can efficiently compete with the modified UTPs, but had the natural UTP been used in the assay, the result might have been different. Our in vitro inhibition assay suggests that 2′-F-2′-C-Me-UTP is not a potent inhibitor of ZIKV NS5. Nevertheless, if the intracellular concentration of 2′-F-2′-C-Me-UTP in target cells/organs can reach concentrations that are high enough, it may still inhibit ZIKV replication.
Another recent publication, by Eyer et al., indicated that a 50 μM concentration of sofosbuvir cannot inhibit Zika virus replication in Vero cell culture (38). In the last study, the authors also showed that 2′-C-methylated nucleosides are more potent inhibitors of ZIKV replication in Vero cell culture, which agrees well with our results of relatively low discrimination for 2′-C-methylated ribonucleotides by ZIKV polymerase.
While the polymerase activity of our purified ZIKV NS5 was sufficient enough to measure K1/2 and to determine discrimination values, it was not high enough to measure all IC50s in the inhibition assay. The IC50s of rNTP analogs are usually measured in reaction mixtures containing competing natural ribonucleotides at 1 μM or less. Under these conditions, our ZIKV NS5 was not very efficient at synthesizing RNA using templates containing poly(C), poly(G), or poly(U) sequences. As a result, so far, only the IC50s of different UTP analogs have been determined.
Measurement of a ribonucleotide analog discrimination versus natural ribonucleotide is an efficient way to search for rNTP analogs that can be efficiently recognized and incorporated by viral polymerases. For rNTP analogs that cause chain termination after incorporation, our data also suggest that IC50s correlate well with the discrimination values. Therefore, the discrimination values may serve as an efficient and economical way for evaluating different ribonucleotide analogs as candidates for antiviral drug development.
MATERIALS AND METHODS
Chemicals.
ATP, UTP, CTP, GTP, 3′-dUTP, 3′-dATP, 3′-dGTP, 3′-dCTP, 2′-dATP, 2′-dCTP, 2′-dGTP, and 2′-dTTP were purchased as 100 mM solutions from TriLink Biotechnologies (San Diego, CA). Urea, taurine, DTT, MnCl2, imidazole, adenosine 5′-phosphosulfate sodium salt (APS), and IPTG were purchased from Sigma (St. Louis, MO). LB medium, bovine serum albumin (BSA), NaCl, Triton X-100, protease inhibitor cocktail, ATP sulfurylase, Tris-HCl (pH 7.5), and HisPur Ni-nitrilotriacetic acid (NTA) agarose resin were purchased from Thermo Fisher Scientific (Waltham, MA). An In-Fusion HD cloning kit and Stellar competent cells were purchased from Clontech (Mountain View, CA). The pMal-c5X vector was purchased from New England Biolabs (NEB; Ipswich, MA). Luciferase and d-luciferin were purchased from Promega (Madison, WI). Fluorescently labeled RNA and DNA oligonucleotides (Cy5.5-RNA or Cy5.5-DNA, respectively), as well as unlabeled RNA and DNA oligonucleotides, were chemically synthesized and purified by high-performance liquid chromatography by Integrated DNA Technologies (Coralville, IA). Cy5.5-RNA and Cy5.5-DNA primer sequences are shown in Table 1. β-d-2′-Deoxy-2′-α-fluoro-2′-β-C-methyluridine-5′-triphosphate (2′-F-2′-C-Me-UTP), β-d-2′-β-C-methyluridine-5′-triphosphate (2′-C-Me-UTP), β-d-2′-β-C-ethynyluridine-5′-triphosphate (2′-C-ethynyl-UTP), β-d-2′-β-C-methyladenosine-5′-triphosphate (2′-C-Me-ATP), β-d-2′-β-C-ethynyl-7-deaza-adenosine-5′-triphosphate (2′-C-ethynyl-7-deaza-ATP), β-d-2′-β-C-methylguanosine-5′-triphosphate (2′-C-Me-GTP), β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-triphosphate (2′-F-2′-C-Me-GTP), β-d-2′-β-C-methylcytidine-5′-triphosphate (2′-C-Me-CTP), β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylcytidine-5′-triphosphate (2′-F-2′-C-Me-CTP), and 4′-C-azidocytidine-5′-triphosphate (4′-C-azido-CTP) were synthesized in-house.
Plasmid construction and protein expression.
A codon-optimized cDNA sequence of ZIKV NS5 (amino acids 2522 to 3423; strain H/PF/2013; GenBank accession number KJ776791.1) was synthesized de novo by Blue Heron Biotechnology (Bothell, WA). The ZIKV NS5 polymerase portion was PCR amplified and cloned into a pMal-c5X vector under tac promoter control. Briefly, the PCR-amplified fragment of ZIKV NS5 (amino acids 1 to 902) or NS5pol (amino acids 272 to 902) containing an N-terminal His tag sequence was fused with a PCR-linearized pMal-c5X vector (without a maltose binding protein sequence) using the In-Fusion HD cloning kit (Clontech, Mountain View, CA). A plasmid carrying a ZIKV NS5 mutant (NS5mut) with an active site mutation (GDD → GAA) in amino acid positions 663 to 665 of NS5 was constructed by fusion of a chemically synthesized 60-bp DNA fragment containing a GAA motif mutation with a PCR-linearized ZIKV NS5 expression plasmid. For protein expression, the plasmids were transformed into Stellar competent cells (Clontech). Expression vector pMal-c5X contains a lacI gene, which allows the inducible expression of NS5 in Stellar cells. The transformed cells were grown to an optical density at 600 nm of 1 in LB medium containing 100 μg/ml ampicillin at 37°C. The cells were cooled down in a 4°C refrigerator for 30 min. MgCl2 and ZnCl2 were added to final concentrations of 1 mM and 50 μM, respectively. Protein expression was induced at 20°C for 2 h by the addition of 0.1 mM IPTG. Cells were harvested by centrifugation at 4,000 × g for 20 min at 4°C. The cell pellets were stored at −80°C before further processing.
Protein purification and identification.
Cell pellets were resuspended in sonication buffer (20 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl, 0.5% Triton X-100, 10 mM DTT, 2 mM MgCl2, 50 μM ZnCl2, 30 mM imidazole, 1× protease inhibitor cocktail). Cell disruption was performed on ice for 10 min using an ultrasound probe sonicator. The cell extract was clarified by centrifugation at 16,000 × g for 20 min at 4°C. The supernatant was incubated with HisPur Ni-NTA agarose resin with gentle rocking for 1 h at 4°C. The resin was then washed 5 times with 10 volumes of wash buffer (20 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 2 mM MgCl2) containing 30 mM imidazole and then one time with wash buffer containing 80 mM imidazole. The protein was eluted from the resin with 1 volume of elution buffer (20 mM Tris-HCl, pH 7.5, 10% glycerol, 50 mM NaCl, 0.5% Triton X-100, 10 mM DTT, 300 mM imidazole). The eluted enzyme was adjusted to 50% glycerol and stored at −80°C before use. Protein identification by mass spectrometry was performed by MS Bioworks (Ann Arbor, MI). The concentration of a targeted protein was measured by SDS-PAGE using BSA (Sigma, St. Louis, MO) as a standard.
Primer and template annealing.
To generate RNA or DNA primer-template complexes, 1 μM fluorescently labeled oligonucleotide primer (Cy5.5-RNA or Cy5.5-DNA) and 4 μM unlabeled DNA or RNA template were mixed in 50 mM NaCl in deionized RNase- and DNase-free water, incubated at 95°C for 10 min, and then slowly cooled to room temperature. The annealed primer and template (P/T) complex was stored at −20°C in the dark before use in primer extension assays.
Primer extension polymerase activity assay.
ZIKV NS5 polymerase activity was determined in a primer extension reaction using a fluorescently labeled RNA primer-RNA template complex. A typical primer extension reaction was performed in a 20-μl reaction mixture containing reaction buffer (5 mM Tris-HCl, pH 7.5, 10 mM DTT, 5 mM MnCl2, 0.5% Triton X-100, 0.01 U RNasin, 10% glycerol), 10 nM P/T complex, and 15 nM NS5 protein. The reaction was initiated by the addition of rNTPs at a final concentration of 100 μM, unless otherwise specified, followed by incubation for 1 h at 35°C. The reactions were quenched by the addition of 40 μl quenching buffer (8 M urea, 90 mM Tris base, 29 mM taurine, 10 mM EDTA, 0.02% SDS, 0.1% bromophenol blue). The quenched samples were denatured at 95°C for 15 min, and the primer extension products were separated using 15% denaturing polyacrylamide gel electrophoresis (urea-PAGE) in 1× TTE buffer (90 mM Tris base, 29 mM taurine, 0.5 mM EDTA). After electrophoresis, the gels were scanned using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The images were analyzed, and the proper RNA bands were quantified using Image Studio software Lite (version 4.0; LI-COR Biosciences, Lincoln, NE).
Analysis of chain termination ability of ribonucleotide analogs.
The primer extension reactions were performed as described above, and each reaction mixture included three natural rNTPs and an analog rNTP as a replacement for the fourth natural rNTP. Briefly, 10 nM RNA primer-RNA template was incubated with 15 nM NS5 in a reaction buffer (5 mM Tris-HCl, pH 7.5, 10 mM DTT, 5 mM MnCl2, 0.5% X-100, 0.01 U RNasin, 10% glycerol), and the reaction was initiated by adding both rNTPs and an analog rNTP. The final concentrations of natural rNTPs and analog 5′-triphosphates were 10 μM and 100 μM, respectively. The reaction mixtures were incubated at 35°C for 1 h, and the reaction was quenched with quenching buffer. The quenched samples were heated at 95°C for 15 min and analyzed by denaturing PAGE as described above.
Measurement of ribonucleotide analog incorporation efficiency.
Different templates were designed to test individual analog rNTPs. The particular primer and template sequences are shown in Table 1. The templates were designed such that the tested analog rNTP was the second ribonucleotide to be incorporated. The first natural ribonucleotide was added at a high concentration to promote the formation of polymerase-P/T and of the elongation complexes. Different concentrations of the ribonucleotide analogs tested were added to reaction mixtures containing 10 nM P/T complex and 15 nM NS5 in a reaction buffer (5 mM Tris-HCl, pH 7.5, 10 mM DTT, 5 mM MnCl2, 0.5% X-100, 0.01 U RNasin, 10% glycerol) to initiate the reactions. After 1 h of incubation at 35°C, the reactions were quenched with quenching buffer and analyzed by denaturing PAGE as described above.
Quantitation of extension products and data analysis.
After electrophoresis, the gels were scanned using an Odyssey infrared imaging system. The intensity of the different RNA bands was quantified using Image Studio software Lite (version 4.0).
The incorporation efficiencies of the different rNTP analogs were evaluated by measurement of the K1/2 and the corresponding discrimination values (39, 40).
K1/2 calculations.
In a single-nucleotide incorporation assay, the percent ribonucleotide incorporation was calculated as follows: amount of product band/(amount of product band + amount of unextended substrate band). The percent incorporation was plotted against the tested ribonucleotide concentration, and the data were fitted to a hyperbolic equation to generate the K1/2 value, i.e., the ribonucleotide concentration at which 50% of product formation was reached. The data were analyzed with GraphPad Prism software using the following equation: Y = bottom + (top − bottom)/{1 + 10^[(log K1/2 − S) × Hill slope]}, where Y is the percent incorporation observed, top is the maximal incorporation percent, bottom is the percent of incorporation under a condition in which no ribonucleotide analog is added (background signal), S is the log of the concentration of the analog rNTP tested, and K1/2 is the ribonucleotide concentration at which 50% of the substrate was converted to the product.
Analog rNTP discrimination (Danalog) values.
Discrimination (Danalog) was defined as the incorporation efficiency of an rNTP analog relative to the incorporation efficiency of the corresponding natural rNTP by the polymerase. The Danalog value is calculated as K1/2, analog/K1/2, natural, where K1/2, analog is the K1/2 measured for an analog rNTP and K1/2, natural is the K1/2 of the corresponding natural rNTP measured under the same reaction conditions. A lower discrimination value indicates a better efficiency of incorporation of a tested rNTP analog by the polymerase.
Inhibition of ZIKV NS5 by UTP analogs (IC50 measurement).
A 1 μM solution of RNA template (5′-A30AACAGGUUCUAGAACCU GUU-3′) in 100 mM NaCl in water was incubated at 98°C for 5 min and slowly cooled to room temperature to allow the formation of the intramolecular hairpin (41). The RNA polymerase inhibition assays were performed in a 15-μl reaction mixture containing reaction buffer (5 mM Tris-HCl, pH 7.5, 10 mM DTT, 5 mM MnCl2, 0.5% Triton X-100, 0.01 U RNasin, 10% glycerol), 10 nM RNA template, and 30 nM NS5 protein. The reaction was initiated by the addition of pyrophosphate (PPi)-free UTP (1 μM) mixed with various concentrations (0.01 to 100 μM) of PPi-free ribonucleotide analogs (all rNTPs were pretreated with pyrophosphatase to remove contaminating PPi), followed by incubation for 1 h at 35°C. The reaction mixture without the RNA template served as a control for nonspecific PPi generation. After incubation, the reaction mixtures (15 μl) were transferred into a 96-well white microtiter plate. To quantify the PPi generated in the polymerase reaction, 85 μl of the luciferase reaction mixture containing 13 nM luciferase, 33 μU/ml ATP sulfurylase, 160 μM d-luciferin, 6 μM APS, 5 mM MgCl2, 10 mM KCl, and 50 mM Tris-HCl, pH 7.5, was added to each well containing 15 μl the polymerase reaction mixtures, and the plate was incubated at room temperature for 5 min. The luciferase activity was measured using a luminometer (Veritas; Promega, Madison, WI). Sodium pyrophosphate standards (3.125, 12.5, 50, and 200 nM) were used to generate a calibration curve for calculation of the PPi concentration in the polymerase reactions.
Data analysis and IC50 calculations.
The PPi concentration was calculated on the basis of the PPi calibration curve. The PPi generated not as a result of the polymerase reaction (nonspecific signal) was subtracted by using a no-RNA control. The percent polymerase activity was calculated as follows: ([PPi]analog/[PPi]UTP) × 100, where [PPi]analog is the PPi concentration measured in the polymerase reaction with various concentrations of analog and 1 μM UTP added, and [PPi]UTP is the PPi concentration measured in the polymerase reaction with only 1 μM UTP added.
IC50, the analog concentration at which the polymerase activity was reduced by 50%, was calculated by fitting the data to the equation Y = bottom + (top − bottom)/{1 + 10^[(log IC50 − S) × Hill slope]}, where Y is the percent polymerase activity, top is the maximal polymerase activity with no ribonucleotide analog added (no inhibition), bottom is the percent minimal polymerase activity (high inhibition), S is the log10 concentration of the rNTP analog tested, and IC50 is the ribonucleotide concentration at which 50% of the inhibition of polymerase activity is achieved. The reported IC50s are the means of three independent measurements.
Supplementary Material
ACKNOWLEDGMENT
Partial funding for this project was provided by Georgia Research Alliance (GRA) grant GRA.VL16.C3.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01967-16.
REFERENCES
- 1.Dick G, Kitchen S, Haddow A. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Besnard M, Lastère S, Teissier A, Cao-Lormeau V, Musso D. 2014. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill 19:pii=20751. doi: 10.2807/1560-7917.ES2014.19.13.20751. [DOI] [PubMed] [Google Scholar]
- 3.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau V-M. 2015. Potential sexual transmission of Zika virus. Emerg Infect Dis 21:359. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musso D, Gubler DJ. 2016. Zika virus. Clin Microbiol Rev 29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 6.Yuki N, Hartung H-P. 2012. Guillain-Barré syndrome. N Engl J Med 366:2294–2304. doi: 10.1056/NEJMra1114525. [DOI] [PubMed] [Google Scholar]
- 7.Gulland A. 2016. Zika virus is a global public health emergency, declares WHO. BMJ 352:i657. doi: 10.1136/bmj.i657. [DOI] [PubMed] [Google Scholar]
- 8.Kuno G, Chang G-J. 2007. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol 152:687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach B, Murray C, Thiel H, Rice C. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Coloma J, Jain R, Rajashankar KR, García-Sastre A, Aggarwal AK. 2016. Structures of NS5 methyltransferase from Zika virus. Cell Rep 16:3097–3102. doi: 10.1016/j.celrep.2016.08.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA. 2010. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res 87:125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi P-Y, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentile I, Borgia F, Buonomo RA, Castaldo G, Borgia G. 2013. A novel promising therapeutic option against hepatitis C virus: an oral nucleotide NS5B polymerase inhibitor sofosbuvir. Curr Med Chem 20:3733–3742. doi: 10.2174/09298673113209990178. [DOI] [PubMed] [Google Scholar]
- 14.Powdrill MH, Bernatchez JA, Götte M. 2010. Inhibitors of the hepatitis C virus RNA-dependent RNA polymerase NS5B. Viruses 2:2169–2195. doi: 10.3390/v2102169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malet H, Massé N, Selisko B, Romette J-L, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan SG, Lescar J. 2008. The flavivirus polymerase as a target for drug discovery. Antiviral Res 80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y-L, Yin Z, Duraiswamy J, Schul W, Lim CC, Liu B, Xu HY, Qing M, Yip A, Wang G, Chan WL, Tan HP, Lo M, Liung S, Kondreddi RR, Rao R, Gu H, He H, Keller TH, Shi PY. 2010. Inhibition of dengue virus RNA synthesis by an adenosine nucleoside. Antimicrob Agents Chemother 54:2932–2939. doi: 10.1128/AAC.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sofia MJ, Chang W, Furman PA, Mosley RT, Ross BS. 2012. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J Med Chem 55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 18.Latour DR, Jekle A, Javanbakht H, Henningsen R, Gee P, Lee I, Tran P, Ren S, Kutach AK, Harris SF, Wang SM, Lok SJ, Shaw D, Li J, Heilek G, Klumpp K, Swinney DC, Deval J. 2010. Biochemical characterization of the inhibition of the dengue virus RNA polymerase by beta-d-2′-ethynyl-7-deaza-adenosine triphosphate. Antiviral Res 87:213–222. doi: 10.1016/j.antiviral.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y-L, Yokokawa F, Shi P-Y. 2015. The search for nucleoside/nucleotide analog inhibitors of dengue virus. Antiviral Res 122:12–19. doi: 10.1016/j.antiviral.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Degasperi E, Aghemo A. 2014. Sofosbuvir for the treatment of chronic hepatitis C: between current evidence and future perspectives. Hepat Med 6:25–33. doi: 10.2147/HMER.S44375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cholongitas E, Papatheodoridis GV. 2014. Sofosbuvir: a novel oral agent for chronic hepatitis C. Ann Gastroenterol 27:331–337. [PMC free article] [PubMed] [Google Scholar]
- 22.Waheed Y, Bhatti A, Ashraf M. 2013. RNA dependent RNA polymerase of HCV: a potential target for the development of antiviral drugs. Infect Genet Evol 14:247–257. doi: 10.1016/j.meegid.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Traut TW. 1994. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 24.Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL, Grobler JA, LaFemina RL, Markel EJ, Migliaccio G, Prhavc M, Stahlhut MW, Tomassini JE, MacCoss M, Hazuda DJ, Carroll SS. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother 48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyrén P, Lundin A. 1985. Enzymatic method for continuous monitoring of inorganic pyrophosphate synthesis. Anal Biochem 151:504–509. doi: 10.1016/0003-2697(85)90211-8. [DOI] [PubMed] [Google Scholar]
- 26.Nyrén P. 1987. Enzymatic method for continuous monitoring of DNA polymerase activity. Anal Biochem 167:235–238. doi: 10.1016/0003-2697(87)90158-8. [DOI] [PubMed] [Google Scholar]
- 27.Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem 242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 28.Lahser FC, Malcolm BA. 2004. A continuous nonradioactive assay for RNA-dependent RNA polymerase activity. Anal Biochem 325:247–254. doi: 10.1016/j.ab.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Ferrari E, Wright-Minogue J, Fang JW, Baroudy BM, Lau JY, Hong Z. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol 73:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417–1426. doi: 10.1016/S0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 31.Ng KK-S, Arnold JJ, Cameron CE. 2008. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol 320:137–156. https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/18268843/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold JJ, Ghosh SKB, Cameron CE. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem 274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 33.Yu F, Hasebe F, Inoue S, Mathenge E, Morita K. 2007. Identification and characterization of RNA-dependent RNA polymerase activity in recombinant Japanese encephalitis virus NS5 protein. Arch Virol 152:1859–1869. doi: 10.1007/s00705-007-1007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel RW, Bellon L, Beigelman L, Kao CC. 1999. Use of DNA, RNA, and chimeric templates by a viral RNA-dependent RNA polymerase: evolutionary implications for the transition from the RNA to the DNA world. J Virol 73:6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deval J, D'Abramo CM, Zhao Z, McCormick S, Coutsinos D, Hess S, Kvaratskhelia M, Götte M. 2007. High resolution footprinting of the hepatitis C virus polymerase NS5B in complex with RNA. J Biol Chem 282:16907–16916. doi: 10.1074/jbc.M701973200. [DOI] [PubMed] [Google Scholar]
- 36.Morin B, Whelan SP.. 2014. Sensitivity of the polymerase of vesicular stomatitis virus to 2′ substitutions in the template and nucleotide triphosphate during initiation and elongation. J Biol Chem 289:9961–9969. doi: 10.1074/jbc.M113.542761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sacramento CQ, de Melo GR, Rocha N, Hoelz LVB, Mesquita M, de Freitas CS, Fintelman-Rodrigues N, Marttorelli A, Ferreira AC, Barbosa-Lima G, Bastos MM, de Mello Volotao E, Tschoeke DA, Leomil L, Bozza FA, Bozza PT, Boechat N, Thompson FL, de Filippis AM, Bruning K, Souza T. 2016. July 6. The clinically approved antiviral drug sofosbuvir impairs Brazilian Zika virus replication. bioRxiv doi: 10.1101/061671. [DOI] [Google Scholar]
- 38.Eyer L, Nencka R, Huvarová I, Palus M, Alves MJ, Gould EA, De Clercq E, Růžek D. 2016. Nucleoside inhibitors of Zika virus. J Infect Dis 214:707–711. doi: 10.1093/infdis/jiw226. [DOI] [PubMed] [Google Scholar]
- 39.Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. 2013. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fung A, Jin Z, Dyatkina N, Wang G, Beigelman L, Deval J. 2014. Efficiency of incorporation and chain termination determines the inhibition potency of 2′-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob Agents Chemother 58:3636–3645. doi: 10.1128/AAC.02666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niyomrattanakit P, Abas SN, Lim CC, Beer D, Shi P-Y, Chen Y-L. 2011. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J Biomol Screen 16:201–210. doi: 10.1177/1087057110389323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.