ABSTRACT
The metallo-β-lactamase NDM-1 is among the most worrisome resistance determinants and is spreading worldwide among Gram-negative bacilli. A bleomycin resistance gene, bleMBL, downstream of the blaNDM-1 gene has been associated with resistance almost systematically. Here, we characterized the corresponding protein, BRPMBL, conferring resistance to bleomycin, an antitumoral glycopeptide molecule. We have determined whether the expression of the blaNDM-1-bleMBL operon is inducible in the presence of carbapenems and/or bleomycin-like molecules using quantitative reverse transcription-PCR (qRT-PCR), determination of imipenem and zeocin MICs, and carbapenemase-specific activity assays. We showed that the blaNDM-1-bleMBL operon is constitutively expressed. Using electrophoretic mobility shift and DNA protection assays performed with purified glutathione S-transferase (GST)-BRPMBL, we demonstrated that BRPMBL is able to bind and sequester bleomycin-like molecules, thus preventing bleomycin-dependent DNA degradation. In silico modeling confirmed that the mechanism of action required the dimerization of the BRPMBL protein in order to sequester bleomycin and prevent DNA damage. BRPMBL acts specifically on bleomycin-like molecules since cloning and expression of bleMBL in Staphyloccoccus aureus did not confer cross-resistance to any other antimicrobial glycopeptides such as vancomycin and teicoplanin.
KEYWORDS: antimicrobial resistance, cancer treatment, mechanism of action, structure
INTRODUCTION
Bleomycin, initially discovered from Streptomyces verticillus, is an antimitotic-antibiotic compound of the glycopeptide family (1), causing cell death as a result of multiple-strand scissions by direct interaction with DNAs through a mechanism that is still not completely understood (2). It is a medication used to treat several cancers. It is on the World Health Organization's List of Essential Medicines, the most important medications needed in a basic health system (http://www.who.int/medicines/publications/essentialmedicines/en/).
Resistance to bleomycin can be mediated by three different types of proteins or mechanisms: (i) bleomycin hydrolases; (ii) bleomycin N-acetylating enzymes that inactivate bleomycin-like molecules; and (iii) bleomycin-binding proteins (BMLA and BMLT) that act by trapping the bleomycin-like molecules (2, 3). Bleomycin hydrolase from rabbit lungs is an aminopeptidase B-like enzyme that is inhibited by N-ethylmaleimide, leupeptin, puromycin, and divalent cations but is unaffected by chelating agents (4). The bleomycin hydrolase from Saccharomyces cerevisiae (encoded by an open reading frame [ORF] consisting of 483 amino acids [AA]) is a thiol protease with an aminopeptidase activity (5). In S. verticillus, bleomycin resistance is related to the expression of blmA and blmB genes encoding two proteins, BLMA (122 AA) and BAT (301 AA). BLMA is a binding protein with a high affinity for bleomycin, whereas BAT is an acetyltransferase that catalyzes the acetylation of bleomycin, leading to subsequent loss of antibacterial activity, i.e., of the DNA-cleaving activities of bleomycin (6, 7). In methicillin-resistant Staphylococcus aureus (MRSA), acquired bleomycin resistance has been reported to be mediated by another bleomycin-binding protein, named BLMS (8). In Gram-negative bacteria, transposon Tn5 carries a bleomycin resistance gene, named ble. The ble gene product, also called BMLT, is a 126-amino-acid-long bleomycin-binding protein of 126 AA sharing low identity with BLMA (from S. verticillus) and BLMS (from S. aureus).
Recently, a new bleomycin resistance gene, named bleMBL, has been characterized from carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii (9). The bleMBL gene is located downstream of the blaNDM-1 gene, as part of the same operon, and the deduced protein, named BRPMBL, showed 54% amino acid identity with the most closely related Tn5-encoded bleomycin resistance protein, BMLT (9, 10). In addition, it was recently demonstrated that BRPMBL is functional and yields decreased susceptibility to bleomycin and bleomycin-like molecules, such as zeocin, when expressed in Enterobacteriaceae and A. baumannii (9).
The aims of the study were (i) to monitor the expression of the blaNDM-1-bleMBL operon under conditions of carbapenem and bleomycin induction, (ii) to elucidate the bleomycin resistance mechanism related to BRPMBL and its resulting role in DNA protection against bleomycin DNA degradation, and (iii) to identify any possible cross-resistance to other antimicrobial glycopeptides such as vancomycin and teicoplanin that is related to bleMBL expression.
RESULTS
Expression of blaNDM-1-bleMBL is not inducible.
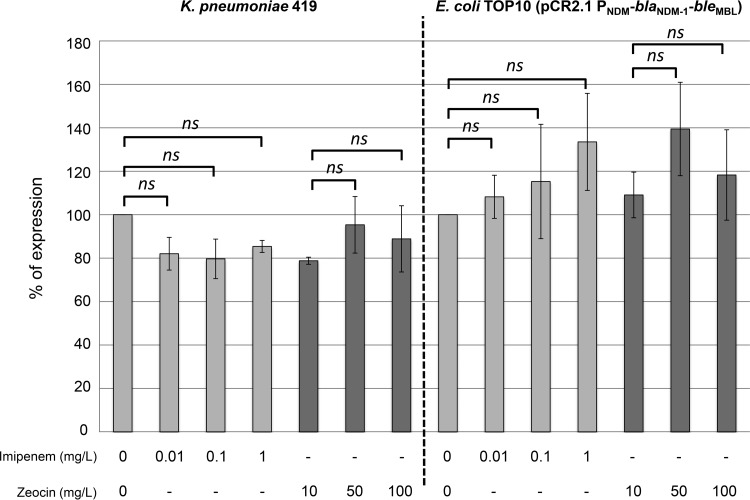
Both the blaNDM-1 and bleMBL genes have been shown to be expressed from the same promoter located upstream of the operon (9). Induction of this promoter in the presence of imipenem or bleomycin-like molecule subinhibitory concentrations was assessed using quantitative reverse transcription-PCR (qRT-PCR) with the NDM-1-producing Klebsiella pneumoniae 419 clinical isolate (11) and with Escherichia coli TOP10 harboring the recombinant plasmid pCR2.1 PNDM-blaNDM-1-bleMBL (9). As shown in Fig. 1, no significant modification of the expression of the blaNDM-1-bleMBL operon was observed in the presence of subinhibitory concentrations of imipenem or zeocin (a bleomycin analogue). Similarly, addition of zeocin to the culture medium of NDM-1-producing enterobacterial clinical isolates did not induce higher levels of resistance to imipenem (Table 1). Furthermore, the absence of NDM-1 induction in the presence of zeocin was confirmed by measuring the specific activity of the carbapenemase on crude protein extracts of an E. coli TOP10 (pCR2.1 PNDM-blaNDM-1-bleMBL) clone and of three NDM-1-producing clinical isolates (E. coli 471, K. pneumoniae 419, and K. pneumoniae DIN) (Table 1). Moreover, the addition of imipenem at different concentrations (0.01, 0.1, and 1 mg/liter) in the culture medium of E. coli TOP10 (pCR2.1 PNDM-blaNDM-1-bleMBL) did not induce zeocin resistance (see Fig. S1 in the supplemental material). However, an additive effect of the two molecules was observed (Fig. S1).
FIG 1.
Relative expression levels of the operon blaNDM-1-bleMBL under conditions of imipenem or zeocin induction. Statistical analysis was performed on the results of 3 independent experiments using Student's t test. P values of <0.05 were considered to represent statistically significant differences. ns, not significant.
TABLE 1.
MIC of and specific enzymatic activity in response to imipenem after induction with increasing concentrations of zeocin
| Strain | Genotypea |
MIC of imipenem (mg/liter) on Mueller-Hinton agar supplemented with zeocin at the indicated concn (mg/liter)b |
Specific activities (mU/mg of protein) using imipenem as the substrate after induction with zeocin at the indicated concn (mg/liter)c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| blaNDM−1 | bleMBL | 0 | 10 | 50 | 100 | 500 | 0 | 10 | 50 | |
| E. coli TOP10 (pCR2.1) | − | − | 0.12 | − | − | − | − | ND | ND | ND |
| E. coli TOP10 (pCR2.1 PNDM-blaNDM-1-bleMBL) | + | + | >32 | >32 | >32 | >32 | 16 | 2.92 ± 0.26 | 3.07 ± 0.14ns | 2.84 ± 0.21ns |
| E. coli 271 | + | + | 6 | 6 | 6 | 4 | 2 | 4.67 ± 1.33 | 7.17 ± 1.15ns | 5.24 ± 0.65ns |
| K. pneumoniae 419 | + | + | 1.5 | 1 | 1 | 1 | 0.5 | 1.31 ± 0.73 | 2.09 ± 0.52ns | 3.27 ± 0.88ns |
| K. pneumoniae DIN | + | + | 4 | 2 | 2 | 1.5 | 1.5 | 6.21 ± 1.08 | 3.88 ± 1.26ns | 7.91 ± 1.45ns |
−, absence of the corresponding gene; +, presence and expression of the corresponding gene.
−, no growth.
ND, not determined; ns, not significant. Statistical analysis was performed on the results of 3 independent experiments using Student's t test. P values of <0.05 were considered significantly different.
BRPMBL binds to bleomycin-like molecules.
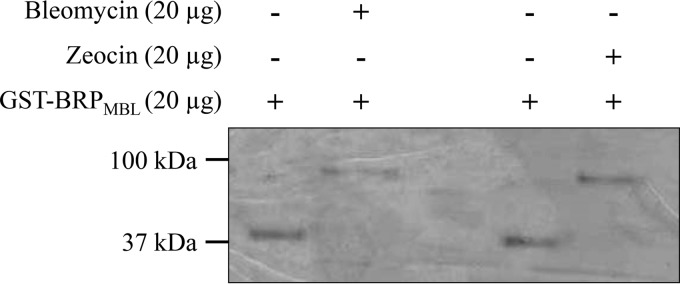
The purified soluble glutathione S-transferase (GST)-BRPMBL fusion protein had a molecular mass corresponding to the expected size of 40 kDa, as assessed by 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2). An electrophoretic mobility shift assay (EMSA) using purified GST-BRPMBL (20 μg) and 20 μg of bleomycin or zeocin showed that the GST-BRPMBL protein migration was delayed in the presence of bleomycin-like molecules (Fig. 2). This result indicates that BRPMBL could bind the bleomycin-like molecules. In addition, the observed shift of migration was approximately 50 to 60 kDa, corresponding to the molecular mass of the GST-BRPMBL (40 kDa) plus that of bleomycin or zeocin (∼14 kDa), suggesting a dimerization of the BRPMBL in the presence of bleomycin-like molecules.
FIG 2.
SDS-PAGE profiles of GST-BRPMBL, preincubated with bleomycin or zeocin. Lanes 1 and 3, SDS-PAGE profiles in the presence of GST-BRPMBL (20 μg); lane 2, SDS-PAGE profile in the presence of bleomycin (20 μg); lane 4, SDS-PAGE profile in the presence of zeocin (20 μg).
BRPMBL inhibits bleomycin-induced DNA damages.
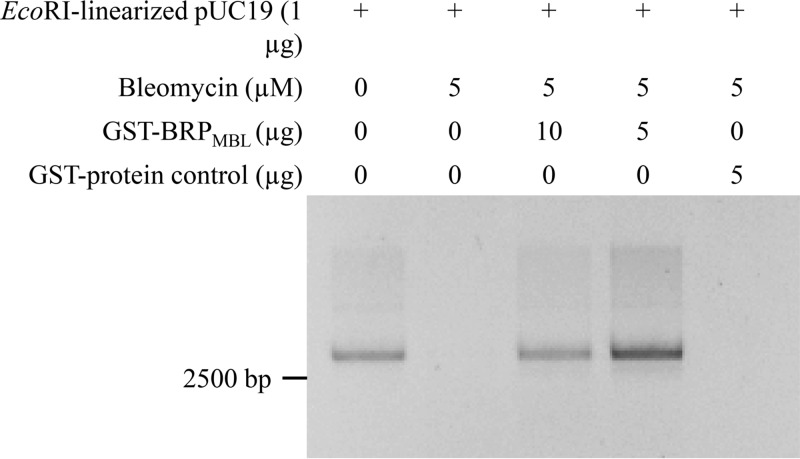
Resistance to DNA-damaging agents was assayed using EcoRI-linearized pUC19 (1 μg), used as probe DNA, in the presence of a mixture of bleomycin and the GST-BRPMBL used for the EMSA. The DNA protection assay demonstrated that bleomycin degraded the linearized plasmid DNA (as the DNA band disappeared) and that the excess of BRPMBL prevented the DNA cleavage induced by bleomycin from occurring (Fig. 3). The specificity was assessed with 5 μg of GST-MVP used as a control protein, and no protection of the DNA damage was observed (12).
FIG 3.
Electrophoretic profile of EcoRI-linearized pUC19 after incubation with bleomycin alone or with bleomycin and GST-BRPMBL.
In silico three-dimensional model of the BRPMBL-bleomycin complex.
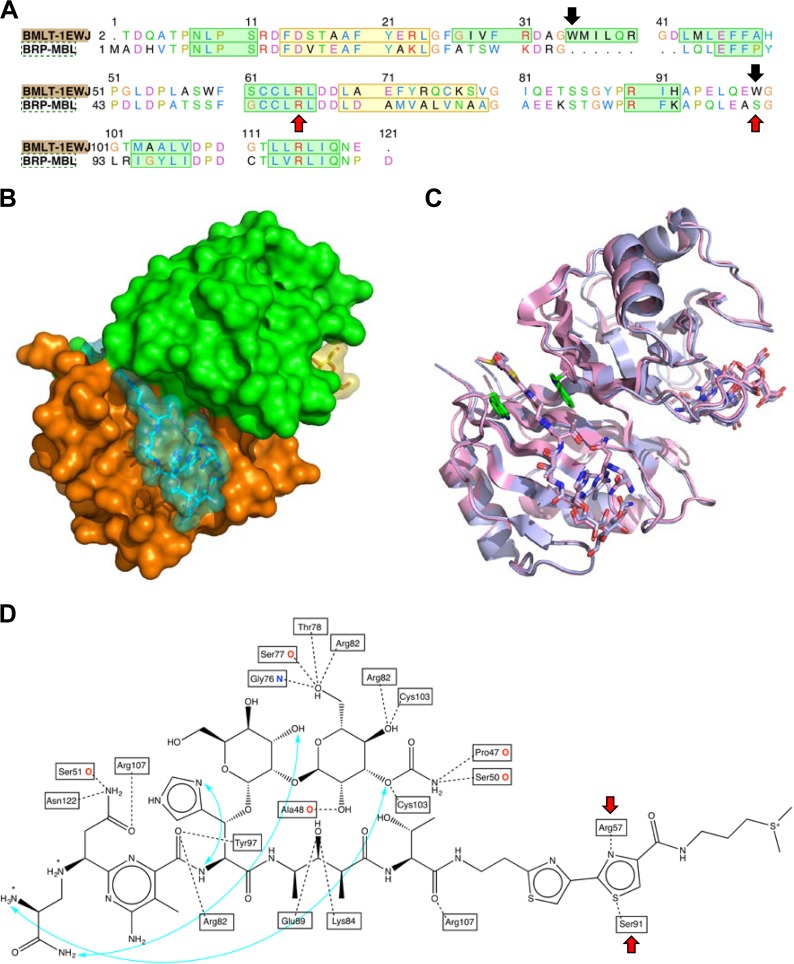
The three-dimensional model of BRPMBL in complex with bleomycin was built by homology with the BLMT structure (PDB code 1EWJ) (13) as the template (see Fig. 4A for the alignment used in this process). The model needed to be constructed directly as a dimer, in the presence of the bleomycin ligand, in order to avoid important clashes in the resulting structure (Fig. 4B; see also Fig. S2). A comparison of the BLMT template to the BRPMBL model (Fig. 4C) showed very similar conformations for the proteins and for the bleomycin ligands, with the exception of the region consisting of AA 34 to 42, which represents an 8-amino-acid deletion in the BRPMBL protein sequence. That region contains one of the two tryptophan (Trp) residues that interact with the bithiazole moiety of bleomycin in the bleomycin-BLMT complex (Fig. 4C).
FIG 4.
BRPMBL interactions with a bleomycin substrate. (A) Sequence alignment between BRPMBL and BMLT that was used for building the three-dimensional model of BRPMBL. Black arrows indicate Trp35 and Trp99 from BLMT (which are lost in the BRPMBL) involved in the π-π interactions with the bithiazole moiety of bleomycin. Red arrows indicate Arg57 and Ser91 of BRPMBL involved in the bleomycin bithiazole fragment interaction. (B) Surface representation of the BRPMBL model (chains A and B colored in orange and green, respectively) in complex with two bleomycin molecules (colored in cyan and yellow). (C) Superposition of BRPMBL (blue) and BMLT (magenta) dimers in complex with the corresponding bleomycin molecules. Two Trp residues from BMLT that interact with the bithiazole moiety from bleomycin are colored in green. (D) Chemical structure of bleomycin. Hydrogen bond interactions with the BRPMBL binding site are represented as dashed lines. Most of these interactions are established with the side chains of binding site residues, but where the backbone is involved, an “O” or “N” follows the residue name. Intramolecular hydrogen bonds in bleomycin are represented as arrows colored in cyan. Red arrows indicate Arg57 and Ser91 of BRPMBL involved in the bleomycin bithiazole fragment interaction.
The interactions between bleomycin and the binding site residues of BRPMBL are shown in Fig. 4D. All the fragments present in the structure of bleomycin formed a large number of hydrogen bonds with the surrounding residues, with the exception of the terminal γ-aminopropyldimethylsulfonium, located at the end of the long groove, which showed no particular interaction and was probably highly flexible, in agreement with the crystallographic data previously obtained for the complex bleomycin-BLMA (3). It is also noteworthy that the three intramolecular hydrogen bonds maintained the relative rigidity of the core of the bleomycin structure (Fig. 4D). The π-π interactions observed between the bithiazole moiety of bleomycin and Trp35 and Trp99 from BLMT are lost in the BRPMBL-bleomycin complex. Indeed, Trp35 is part of the 8-AA deletion in the region AA 35 to 42 and Trp99 is replaced by a Ser residue in BRPMBL (Ser91) (Fig. 4A). However, the bithiazole fragment is solidly anchored in the BRPMBL binding site groove through hydrogen bond interactions with the side chains of Arg57 and Ser91 (Fig. 4D).
BRPMBL does not confer resistance to other antimicrobial glycoproteins.
In order to identify any cross-resistance to other antimicrobial glycopeptides such as vancomycin and teicoplanin that is related to bleMBL expression, the bleMBL gene was cloned into a shuttle vector (pPRT plasmid) that allows replication and expression in Gram-negative (E. coli TOP10) and Gram-positive (S. aureus CIP658) species. Although the MICs of bleomycin and zeocin were >256 mg/liter for the recombinant S. aureus CIP658 strain (pPRT-bleMBL) (versus 8 mg/liter and 2 mg/liter, respectively, for the wild-type S. aureus CIP658 strain), no decreased susceptibility could be observed for glycopeptides (vancomycin and teicoplanin) compared to the susceptibility of the parental strain (data not shown).
DISCUSSION
NDM is a recently described carbapenemase that has rapidly disseminated worldwide, with the Indian subcontinent being an important reservoir. NDM is currently among the most threatening antimicrobial resistance traits in Enterobacteriaceae (14). The blaNDM genes have been almost systematically found in association with the bleMBL gene encoding BRPMBL, a putative bleomycin resistance protein. Despite its low (54%) amino acid sequence identity with the other known BRPs, BRPMBL was shown to be a functional protein conferring resistance to bleomycin-like molecules. The genes encoding BRPMBL and NDM are coexpressed from the same promoter, located upstream of the blaNDM gene and in the right extremity of insertion sequence ISAba125 (9). This coexpression suggested coselection by either of the two molecules with carbapenems and bleomycin. Thus, not only the usage of carbapenem but also that of bleomycin-like molecules might be responsible for the selection of NDM producers. We have demonstrated here that neither carbapenems nor bleomycin-like molecules have any impact on the expression of the blaNDM-bleMBL operon. This result is in agreement with the fact that, most often, promoter elements brought by insertion sequences lead to constitutive expression of genes located downstream, as observed for other carbapenemases (15, 16).
As observed for other BRPs (17), the role of BRPMBL, once produced in a hypermutable E. coli strain (9), in DNA stabilization has been demonstrated. However, the exact mechanism of action of BRPBML has still not been elucidated. Here, we have shown that BRPMBL prevents the occurrence of bleomycin-mediated DNA damage by directly binding bleomycin-like molecules. Several three-dimensional structures of bleomycin-binding proteins are available in the Protein Data Bank (18): bleomycin resistance protein from Streptoalloteichus hindustanus ble (Shble) (PDB codes 1BYL, 1XRK, and 2ZHO), transposon Tn5-carried bleomycin resistance determinant BLMT (PDB codes 1ECS, 1EWJ, 1MH6, and 1NIQ), and bleomycin-binding protein from Streptomyces verticillus BLMA (PDB codes 1JIE, 1JIF, and 1QTO). Those proteins, even though sharing, respectively, 18%, 52%, and 16% amino acid identity with BRPMBL only, have similar three-dimensional (3D) structures, suggesting that they may be used as bases for BRPMBL modeling. The crystal structures are dimers, and a few of them are complexed with two bleomycin molecules, which are located symmetrically at the monomer-monomer interfaces (13). In silico modeling suggested that the three-dimensional model of the BRPMBL needed to be constructed directly as a dimer, in the presence of the bleomycin ligand. These results were confirmed by EMSA, which showed that the BRPMBL protein dimerized in the presence of bleomycin-like molecules.
Finally, although the terminal γ-aminopropyldimethylsulfonium, located at the end of the long groove of the BRPMBL protein, was found to be highly flexible, we demonstrated that BRPMBL acts specifically on bleomycin-like molecules and thus that the results explain why other antimicrobial glycopeptides, such as vancomycin and teicoplanin, could not be sequestered by BRPMBL.
MATERIALS AND METHODS
Bacterial strains.
The previously described pCR2.1 PNDM-blaNDM-1-bleMBL E. coli TOP10 clone (9) and clinical isolates K. pneumoniae 419 (11), E. coli 271 (19), and K. pneumoniae DIN (9) expressing the blaNDM-1-bleMBL operon were used for the monitoring of blaNDM-1-bleMBL operon expression upon induction with imipenem and bleomycin-like molecules. The BL21(DE3) E. coli strain (Life Technologies, Cergy-Pontoise, France) was used as the host for BRPMBL expression and purification. The S. aureus CIP658 strain was used as the host for cloning experiments with pAT28-bleMBL plasmid.
Susceptibility testing.
MICs of imipenem were determined on Mueller-Hinton agar (bioMérieux, La Balme-les-Grottes, France) using the Etest technique (bioMérieux) following the manufacturer's instructions.
Zeocin susceptibility testing was performed using the disc diffusion method following the European Society of Clinical Microbiology and Infectious Diseases (EUCAST) recommendations (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/Manual_v_5.0_EUCAST_Disk_Test.pdf) on Mueller-Hinton agar supplemented with imipenem (Sigma-Aldrich, Saint Quentin Fallavier, France) at various concentrations (0, 0.01, 0.1, and 1 mg/liter). Discs impregnated with various amounts of zeocin (20 μg, 50 μg, 100 μg, 200 μg, 500 μg, and 1,000 μg) were extemporaneously prepared by adding 10 μl of diluted zeocin solutions (ThermoFisher, Villebon, France) to sterile dried discs.
Carbapenemase-specific activity.
The specific activities with 100 μM imipenem as the substrate of the crude protein extracts obtained from overnight cultures of blaNDM-1-bleMBL-expressing isolates, namely, E. coli TOP10 (pCR2.1 PNDM-blaNDM-1-bleMBL), K. pneumoniae 419, E. coli 271, and K. pneumoniae DIN, were determined with an Ultrospec 2000 UV spectrophotometer (Amersham Pharmacia Biotech, Orsay, France) as previously described (20). One unit of enzyme activity was defined as the amount of enzyme that hydrolyzes 1 μmol of substrate per min. Bacterial isolates were grown overnight in brain heart infusion medium supplemented with zeocin at 0, 2, or 200 mg/liter.
qRT-PCR.
The pCR2.1 PNDM-blaNDM-1-bleMBL E. coli TOP10 clone and K. pneumoniae 419 isolate were grown overnight in BHI medium supplemented with 0, 0.01, 0.1, and 1 mg/liter of imipenem or 0, 10, 50, and 100 mg/liter of zeocin. RNAs were recovered using an RNeasy minikit (Qiagen, Courtaboeuf, France). In order to quantify expression of the bleMBL gene, one-step qRT-PCR (SYBR green Rotor gene; Qiagen, France) was used as previously described (21) with primers rtBleoFw (5′-TATCCTGACCTCGACCCAGC-3′) and rtBleoRv (5′-ATCAGGTAGCCGATCCTCAGG-3′). The levels of expression of the bleMBL gene and the 16S rRNA gene were quantified with the same RNA extracts as described previously (21). Expression levels were standardized relative to the transcription level of the constitutively expressed 16S rRNAs with primers rt16SFw (5′-GGACGGGTGAGTAATGTCTG-3′) and rt16SRv (5′-TCTCAGACCAGCTAGGGATCG-3′). The transcriptional levels, determined by qRT-PCR, were interpreted using the threshold cycle (2ΔΔCT) method as described previously (22).
BRPMBL purification.
In order to overexpress and purify BRPMBL, the bleMBL gene from E. coli 271 was amplified (19) with primers pET41b-ble-SpeI-F (5′-GAGTCACTAGTGCTGACCACGTCACCCC-3′) and pET41b-ble-XhoI-R (5′-GAGTCCTCGAGTCAGTCGGGGTTCTGGATC-3′). The resulting product was introduced into PCRblunt pTOPO (Life Technologies) and expressed in E. coli TOP10 (Life Technologies) for amplification and subcloning into SpeI- and XhoI-restricted pET41b+ (Novagen, Darmstadt, Germany). BRPMBL was expressed at a high level as a fusion protein with 220-AA glutathione S-transferase (GST). GST-BRPMBL fusion constructs were expressed in E. coli BL21(DE3) upon induction with IPTG (isopropyl-β-d-thiogalactopyranoside) at 0.1 mM during 16 h at 25°C, and the crude protein extract was prepared using NaCl (100 mM)- and lysozyme (50 mg/liter)-containing phosphate buffer (50 mM, pH 7), sonication, DNase (5 mg/liter), RNase (10 mg/liter), and high-speed centrifugation as recommended by the manufacturer (Qiagen, les Ulis, France). The resulting GST-BRPMBL was purified by glutathione chelate affinity (Qiagen) on an Akta purifier (GE Healthcare, Les Ulis, France) and dialyzed in 50 mM Tris-HCl (pH 8).
BRPMBL-bleomycin-like molecule binding assay.
The capacity of binding of the GST-BRPMBL to bleomycin was assayed by electrophoretic mobility shift assay (EMSA) by using purified GST-BRPMBL (20 μg) and 20 μg bleomycin or zeocin in 30 μl of 10 mM Tris-HCl (pH 7.5) at 16°C. After 3 h of incubation, 30 μl of the reaction mixture was loaded onto a 10% nondenaturing polyacrylamide gel, as previously described (13, 23, 24).
DNA protection assay.
The DNA protection assay was performed as previously described (24, 25). Briefly, GST-BRPMBL (0, 5, and 10 μg) or a GST-protein control, named GST-major vault protein (GST-MVP) (12) (5 μg), was mixed with 5 μM bleomycin–10 mM Tris-HCl (pH 7.5). After incubation at 25°C for 5 min, 0.1 μg of EcoRI-linearized pUC19, 0.1 mM FeSO4, and 1 mM dithiothreitol were added to achieve a total volume of 20 μl. The mixture was incubated at 25°C; after 10 min, 40 mM EDTA was added. The solution (10 μl) was separated by agarose (1%) gel electrophoresis.
Generation of BRPMBL-overexpressing S. aureus.
To express BRPMBL in S. aureus, the pPRT-bleMBL derivative plasmid was constructed. The bleMBL gene was amplified from E. coli 271 with primers pAT18-BamHI-bleF (5′-GAGTCGGATCCATGGCTGACCACGTCA-3′) and pAT18-XbaI-bleR (5′-GAGTCTCTAGATCAGTCGGGGTTCTGG-3′) and introduced into the BamHI- and XbaI-restricted pPRT plasmid, a pAT18 derivative plasmid (26). In the pPRT-bleMBL plasmid, BRPMBL was expressed under the control of the promoter region of the protease gene from Lactococcus lactis subsp. cremoris, which is active in Gram-positive bacterial species (26). The pPRT-bleMBL plasmid was introduced into S. aureus CIP658 by electroporation using erythromycin (5 mg/liter) as the selective agent as previously described (27).
Molecular modeling.
The three-dimensional model of BRPMBL was built using Modeler v9.16 (28), with the structure of BMLT (PDB code 1EWJ) as the template (13). The structure was constructed as a dimer, with two bleomycin molecules positioned at the interface of the monomers. The protein-ligand interactions were analyzed with UCSF Chimera (29), and the images for sequence alignment and protein-ligand complexes were generated using UCSF Chimera and Pymol, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
This work was partially funded by a grant from the University Paris-Sud and INSERM (U914), France. L.D., D.G., B.I.I., and T.N. are members of the Laboratory of Excellence LERMIT supported by a grant from ANR (ANR-10-LABX-33).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02413-16.
REFERENCES
- 1.Umezawa H, Maeda K, Takeuchi T, Okami Y. 1966. New antibiotics, bleomycin A and B. J Antibiot (Tokyo) 19:200–209. [PubMed] [Google Scholar]
- 2.Sugiyama M, Kumagai T. 2002. Molecular and structural biology of bleomycin and its resistance determinants. J Biosci Bioeng 93:105–116. doi: 10.1016/S1389-1723(02)80001-9. [DOI] [PubMed] [Google Scholar]
- 3.Sugiyama M, Kumagai T, Hayashida M, Maruyama M, Matoba Y. 2002. The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus. J Biol Chem 277:2311–2320. doi: 10.1074/jbc.M103278200. [DOI] [PubMed] [Google Scholar]
- 4.Sebti SM, DeLeon JC, Lazo JS. 1987. Purification, characterization, and amino acid composition of rabbit pulmonary bleomycin hydrolase. Biochemistry 26:4213–4219. doi: 10.1021/bi00388a006. [DOI] [PubMed] [Google Scholar]
- 5.Enenkel C, Wolf DH. 1993. BLH1 codes for a yeast thiol aminopeptidase, the equivalent of mammalian bleomycin hydrolase. J Biol Chem 268:7036–7043. [PubMed] [Google Scholar]
- 6.Sugiyama M, Kumagai T, Shionoya M, Kimura E, Davies JE. 1994. Inactivation of bleomycin by an N-acetyltransferase in the bleomycin-producing strain Streptomyces verticillus. FEMS Microbiol Lett 121:81–85. doi: 10.1111/j.1574-6968.1994.tb07079.x. [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama M, Thompson CJ, Kumagai T, Suzuki K, Deblaere R, Villarroel R, Davies J. 1994. Characterisation by molecular cloning of two genes from Streptomyces verticillus encoding resistance to bleomycin. Gene 151:11–16. doi: 10.1016/0378-1119(94)90626-2. [DOI] [PubMed] [Google Scholar]
- 8.Sugiyama M, Kumagai T, Matsuo H, Bhuiyan MZ, Ueda K, Mochizuki H, Nakamura N, Davies JE. 1995. Overproduction of the bleomycin-binding proteins from bleomycin-producing Streptomyces verticillus and a methicillin-resistant Staphylococcus aureus in Escherichia coli and their immunological characterisation. FEBS Lett 362:80–84. doi: 10.1016/0014-5793(95)00218-X. [DOI] [PubMed] [Google Scholar]
- 9.Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 56:1693–1697. doi: 10.1128/AAC.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blot M, Meyer J, Arber W. 1991. Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage to Escherichia coli K-12. Proc Natl Acad Sci U S A 88:9112–9116. doi: 10.1073/pnas.88.20.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Al Maskari Z, Al Rashdi F, Bernabeu S, Nordmann P. 2011. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J Antimicrob Chemother 66:304–306. doi: 10.1093/jac/dkq428. [DOI] [PubMed] [Google Scholar]
- 12.Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P. 2011. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog 7:e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruyama M, Kumagai T, Matoba Y, Hayashida M, Fujii T, Hata Y, Sugiyama M. 2001. Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin. J Biol Chem 276:9992–9999. doi: 10.1074/jbc.M009874200. [DOI] [PubMed] [Google Scholar]
- 14.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BiomedRes Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu WS, Yao SM, Fung CP, Hsieh YP, Liu CP, Lin JF. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob Agents Chemother 51:3844–3852. doi: 10.1128/AAC.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bostock JM, Miller K, O'Neill AJ, Chopra I. 2003. Zeocin resistance suppresses mutation in hypermutable Escherichia coli. Microbiology 149:815–816. doi: 10.1099/mic.0.C0111-0. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother 54:4914–4916. doi: 10.1128/AAC.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn Microbiol Infect Dis 74:88–90. doi: 10.1016/j.diagmicrobio.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Naas T, Cuzon G, Truong HV, Nordmann P. 2012. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan JS, Reed A, Chen F, Stewart CN Jr. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong MS, Hwang EY, Kim HT, Yoo MA, Jang SB. 2009. Purification of caudal-related homeodomain transcription factor and its binding characterization. J Microbiol Biotechnol 19:1557–1564. doi: 10.4014/jmb.0905.05021. [DOI] [PubMed] [Google Scholar]
- 24.Mori T, Mizuta S, Suenaga H, Miyazaki K. 2008. Metagenomic screening for bleomycin resistance genes. Appl Environ Microbiol 74:6803–6805. doi: 10.1128/AEM.00873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatignol A, Durand H, Tiraby G. 1988. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett 230:171–175. doi: 10.1016/0014-5793(88)80665-3. [DOI] [PubMed] [Google Scholar]
- 26.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99–104. doi: 10.1016/0378-1119(91)90546-N. [DOI] [PubMed] [Google Scholar]
- 27.Trieu-Cuot P, Derlot E, Courvalin P. 1993. Enhanced conjugative transfer of plasmid DNA from Escherichia coli to Staphylococcus aureus and Listeria monocytogenes. FEMS Microbiol Lett 109:19–23. doi: 10.1111/j.1574-6968.1993.tb06137.x. [DOI] [PubMed] [Google Scholar]
- 28.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.