Microtubules (MTs) are depolymerized within Aspergillus nidulans biofilms in response to gaseous microenvironments regulated by the SrbA hypoxic transcription factor. This mode of MT regulation could contribute to the unique characteristics of fungal biofilms in medical and industrial settings.
Abstract
How microtubules (MTs) are regulated during fungal biofilm formation is unknown. By tracking MT +end–binding proteins (+TIPS) in Aspergillus nidulans, we find that MTs are regulated to depolymerize within forming fungal biofilms. During this process, EB1, dynein, and ClipA form transient fibrous and then bar-like structures, novel configurations for +TIPS. Cells also respond in an autonomous manner, with cells separated by a septum able to maintain different MT dynamics. Surprisingly, all cells with depolymerized MTs rapidly repolymerize their MTs after air exchange above the static culture medium of biofilms. Although the specific gasotransmitter for this biofilm response is not known, we find that addition of hydrogen sulfide gas to growing cells recapitulates all aspects of reversible MT depolymerization and transient formation of +TIPs bars. However, as biofilms mature, physical removal of part of the biofilm is required to promote MT repolymerization, which occurs at the new biofilm edge. We further show MT depolymerization within biofilms is regulated by the SrbA hypoxic transcription factor and that without SrbA, MTs are maintained as biofilms form. This reveals a new mode of MT regulation in response to changing gaseous biofilm microenvironments, which could contribute to the unique characteristics of fungal biofilms in medical and industrial settings.
INTRODUCTION
Microtubules (MTs) are cytoskeletal filaments that act as highways for long-distance intracellular trafficking during interphase and then form the mitotic spindle during mitosis (Kirschner and Mitchison, 1986; Szewczyk and Oakley, 2011). MTs are dynamic polymers of tubulin dimers that are nucleated from MT-organizing centers (MTOCs) containing γ-tubulin (Oakley et al., 1990). MTs also display “dynamic instability,” by which they undergo cycles of growth and shrinkage (Mitchison and Kirschner, 1984). The underlying basis for this behavior is an ongoing race between GTP tubulin addition, resulting in growth, and GTP hydrolysis after its incorporation into the polymer, favoring MT catastrophe (Brouhard, 2015). A variety of plus end–binding/tracking proteins (+TIPS) affect MT dynamic instability. Among the most prominent of these is end-binding protein-1 (EB1; Vaughan, 2005; Honnappa et al., 2006), which promotes MT growth and acts as a landing site for other +TIPS, including the cytoplasmic linker protein of 170 kDa (CLIP-170; Bieling et al., 2008; Dixit et al., 2009) and the cytoplasmic dynein heavy chain (Duellberg et al., 2014).
Much of our understanding of how MTs are regulated comes from studies of actively growing cells. However, many cells exist in nongrowing states over extended periods of time after terminal differentiation and exit from the cell cycle. Similarly, cells will reversibly stop growing in response to stress or when nutrients become limiting, as, for example, can occur when cells form communities within biofilms (Forster et al., 2016). How MTs are regulated in nongrowing cells and the effect of their local growth environment are of interest in microbial as well as cancer biology because any specific type of regulation could be involved in drug resistance. In this regard, during biofilm formation, fungal cells experience microenvironments that modify their physiology, leading to antifungal drug resistance and the generation of dormant persister cells tolerant to antifungals (Taff et al., 2013). In addition, many cancer drugs target MTs (Jordan and Wilson, 2004).
Many fungal species readily form biofilms on a submerged substratum, generating a multilayered community of hyphae (Desai et al., 2014). Within fungal biofilms, inverse gradients of oxygen and nutrients are generated as cell crowding occurs within the biofilm. Cells that initiate biofilm formation are located in the deeper layers of the structure and therefore experience a different microenvironment than cells at the biofilm periphery (Fanning and Mitchell, 2012). Little is known about how the MT cytoskeleton is regulated in cells within fungal biofilms and colonies, although a considerable amount of fungal biomass in nature consists of such nongrowing cells. From a medical perspective, fungi form biofilms on medical implants and in the lungs, sinuses, and urinary tract of immune-compromised patients (Kaur and Singh, 2014; Araujo et al., 2017). Because cells within biofilms are less responsive to antifungal treatments, they contribute to hard-to-treat chronic disease states. Of note, fungal biofilms are sometimes also the preferred mode of growth during industrial production of enzymes, organic acids, and food products because they are more productive than during submerged free-floating growth (Ramage et al., 2011; Gutierrez-Correa et al., 2012).
Understanding how biofilm microenvironments affect the physiology of fungal cells is therefore of medical and industrial interest. Here we report that dissolved gaseous microenvironments within fungal biofilms promote highly reversible MT depolymerization. We further show that hydrogen sulfide mimics these effects and find that the response is regulated via the SrbA hypoxic transcription factor, which is known to control other physiological responses to hypoxia in fungal cells.
RESULTS
EB1 as a marker for microtubule dynamics
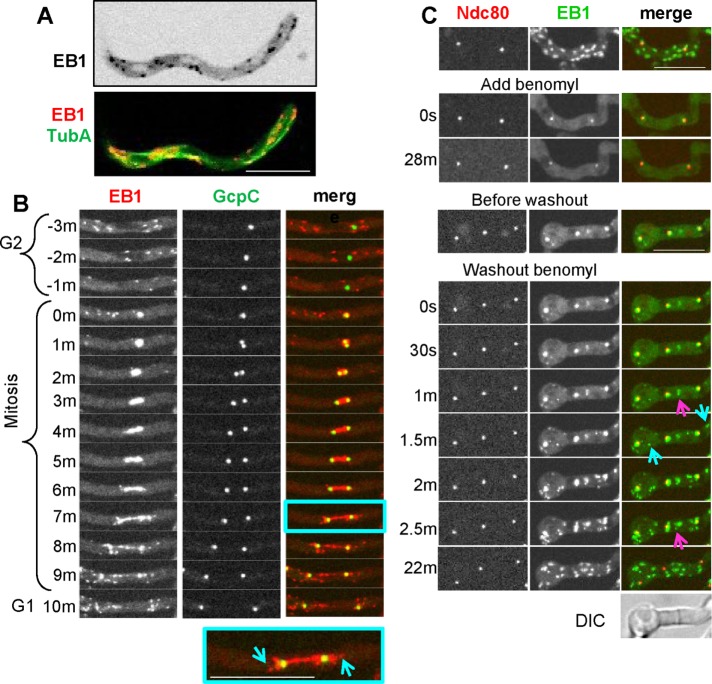
In Aspergillus nidulans, EB1–monomeric red fluorescent protein (mRFP; Campbell et al., 2002)) expressed from its native locus appears as comets at the growing plus ends of MTs as defined by green fluorescent protein (GFP)–TubA (Zeng et al., 2014; Figure 1A and Supplemental Video S1). At the cell tip, EB1-mRFP comets disperse, reflecting the MT depolymerization that occurs in this cellular region (Supplemental Video S1). To track EB1 behavior during mitosis, we used a strain expressing EB1 tagged with mCherry (CR: Shaner et al., 2004) and spindle pole bodies (SPBs) marked with GcpC-GFP (Xiong and Oakley, 2009). During mitosis, cytoplasmic EB1-CR comets are reduced in number as cytoplasmic MTs disassemble, and EB1-CR then locates to forming mitotic spindles (Figure 1B and Supplemental Video S2). During mitotic exit, EB1-CR comets reappear, emanating away from the separating SPBs as astral MTs polymerize and grow into the cytoplasm (Figure 1B and Supplemental Video S2). Benomyl-induced MT depolymerization causes immediate EB1-GFP dispersal from comets (Figure 1C, Add benomyl). Subsequent removal of benomyl allows MTs to repolymerize, and EB1-GFP appears as bursts of comets forming from MTOCs at SPBs (marked by Ndc80-CR; Yang et al., 2004) at septa and within the cytoplasm (Figure 1C, Washout benomyl). EB1 is therefore an excellent marker (Egan et al., 2012; Zeng et al., 2014) for growing MTs in A. nidulans, as it is in other organisms, with EB1 comets reflecting MTs that are growing and its dispersal that MTs are being depolymerized. During MT repolymerization, localized bursts of EB1-GFP comets also functionally define sites of MTOCs.
FIGURE 1:
EB1 is a good indicator of MT dynamics. (A) Localization of EB1-mRFP (grayscale, inverted) and TubA-GFP (green in montage) from Supplemental Video S1 (strain NS133) showing dynamics of EB1 mRFP comets at the plus ends of growing MTs. (B) EB1-CR and the GcpC-GFP SPB marker during mitosis (Supplemental Video S2, strain NS327). EB1-CR localizes to the mitotic spindle and then to astral MTs during mitotic exit (inset, blue arrows). (C) EB1-GFP (strain NS326) disperses from comets when MTs are depolymerized using benomyl and reappears at mobile comets after benomyl washout. EB1-GFP comets reemerge from SPBs (marked by Ndc80), septa (pink arrow), and cytoplasmic foci (blue arrows). Scale bar, 10 μm.
During early biofilm formation, EB1-GFP comets disperse and EB1 locates at bar-like structures
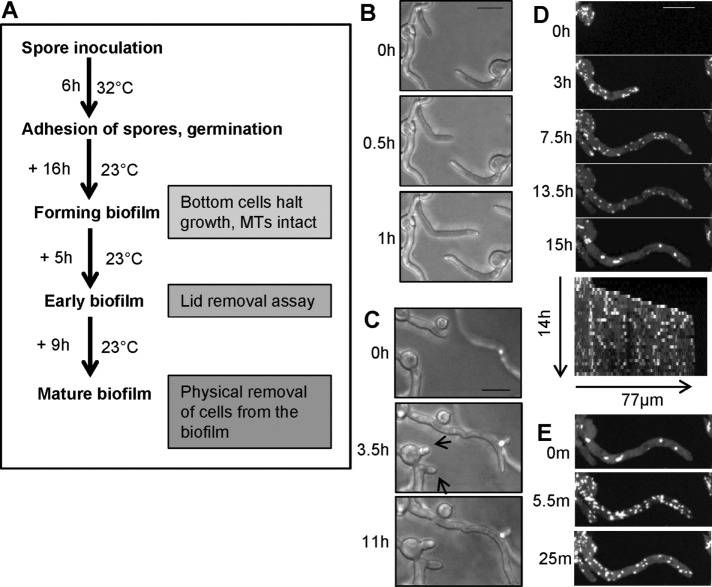
When imaging live cells of A. nidulans, spores are inoculated into the medium, which sink and attach to the bottom of the imaging dish. We then image cells using inverted spinning-disk confocal microscopy. The growth temperature and time of culture during the different stages of biofilm formation imaged are indicated in Figure 2A. All cells initially grow and populate the bottom surface of the culture via polarized tip cell growth. Some tips were seen to apparently detect other cells and change their direction of growth to avoid each other (Figure 2B). As cells grow further and become more crowded, they stop growing, even when there appears to be space for them to grow into (Figure 2C, tip cells, arrows), and begin the early stages of biofilm formation. Although these cells are no longer growing, they maintain active MT dynamics and display EB1-GFP comets. A representative cell is shown in Figure 2D from a time course with images taken at 30-min intervals. Kymographs readily reveal the growing and growth-arrested phases, with EB1-GFP comets being present during both (Figure 2D). The process within a cell population of ∼188 cells is shown in Supplemental Video S3, with the field of view representing a stitched image of 308 × 308 µm. At the early time points, some cells undergo mitosis and have EB1-GFP at mitotic spindles and not at cytoplasmic comets (10 cells at T1 indicated by arrows in Supplemental Video S3). Figure 3 shows the population of cells at the beginning (Figure 3A) and end of the growth period (Figure 3B). During further biofilm maturation, EB1-GFP becomes dispersed in some cells (Figure 3C and Supplemental Video S3), with 68 of ∼188 cells having dispersed EB1-GFP. Of further interest, and unexpectedly, EB1-GFP was also seen to locate at more fibrous and bar-like structures during this period (60 of 188 cells) a configuration unusual for a protein known to locate to the plus ends of growing microtubules (Figure 1A). Imaging at shorter time intervals revealed that EB1-GFP first appeared in a fibrous pattern, which then shrank, leaving more bar-like structures (Figure 4A and Supplemental Video S4). The distribution along fibers was also seen to start in a discontinuous pattern but became more uniform as fibers transitioned into bars. The bars were also seen to break (Figure 4B and Supplemental Video S5) and with time retract and also disappear, as can be seen in the kymographs of Figure 4.
FIGURE 2:
During early stages of A. nidulans biofilm formation, cells stop growing and then undergo distinctive changes in their EB1-GFP behavior reflecting MT depolymerization. (A) Schematic representation of growth conditions used for examining EB1-GFP localization during different stages of biofilm formation. (B) During the initial growth phase of biofilm formation (strain SO1563), cells can change their growth direction when growing toward each other. (C) Cells then stop growing (arrows) as they become further crowded. (D) The montage and kymograph show that after cells stop growing, they initially maintain EB1-GFP comets, but then EB1-GFP starts to disperse and form immobile bars and foci (15 h). (E) Surprisingly, EB1-GFP relocates back to comets within minutes of removal of culture dish lid. Cells were initially grown at 32°C for 6 h, followed by growth for 10, 9, 6, and 21 h at 23 ± 2°C for imaging in B–E, respectively. Scale bar, 10 µm.
FIGURE 3:
EB1-GFP behavior during early stages of A. nidulans biofilm formation as cells reversibly depolymerize their MTs. Stitched images from live-cell imaging during the early stages of biofilm formation in strain SO1563 after growth at 32°C for 6 h followed by growth for the indicated times at 23 ± 2°C (Supplemental Video S3). (A) Initial growth phase when all cells have dynamic MTs with EB1-GFP in comets or at mitotic spindle (arrows). (B) After another 10 h of growth, cells stop growing but maintain dynamic MTs and still have EB1-GFP at comets. (C) Some cells start to depolymerize their MTs, causing EB1-GFP to disperse from comets, and in some cells, EB1-GFP surprisingly also locate at bars. Of note, this response is cell autonomous, and many other cells still display EB1-GFP at comets. (D) After removal of culture dish lid, all cells repolymerize their MTs, and EB1-GFP returns in all as comets (Supplemental Video S7). Scale bar, 50 μm.
FIGURE 4:
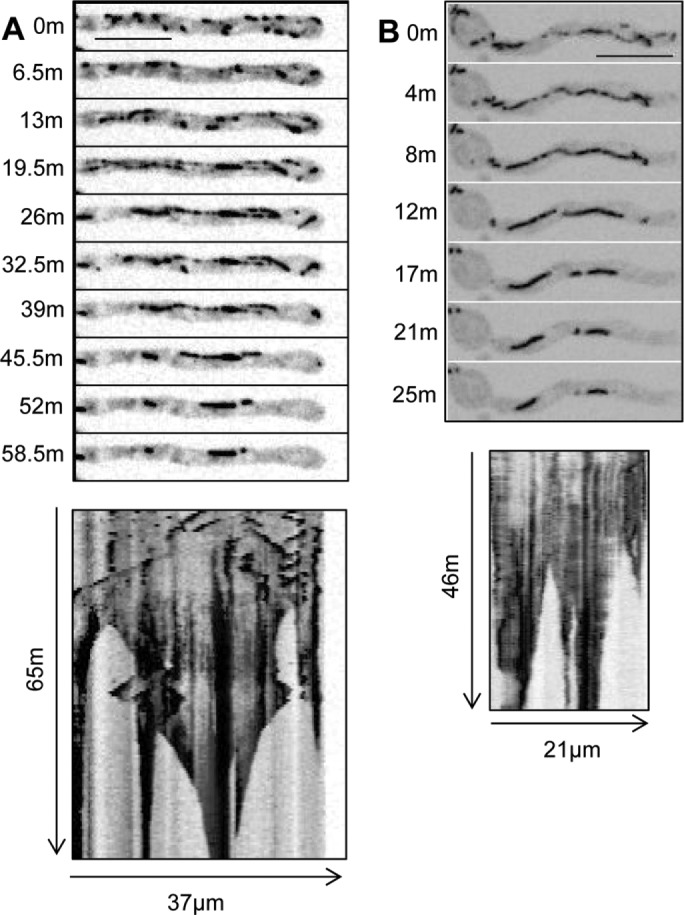
During MT depolymerization within biofilms, EB1-GFP unexpectedly locates to long fibers, which shrink to form bars. (A) Montage and kymograph during the transition of EB1-GFP (strain SO1563) from comets to cables and then to bars (Supplemental Video S4). (B) EB1-GFP distribution on cables is initially uneven but gradually becomes more uniform, followed by splitting and shrinking to form bars (Supplemental Video S5). Scale bar, 10 μm.
Of importance, cells were observed to respond in an autonomous manner, with different cells at the same time displaying EB1-GFP at comets or dispersed and/or at bars (Figure 3C). In fact, even cells separated by a septum were seen to display different locations for EB1-GFP, with the cell on one side of a septum maintaining dynamic EB1-GFP comets, whereas the cell on the other side had EB1-GFP dispersed and/or at bars (Supplemental Video S6). These observations reveal that during the early stages of biofilm formation, cells stop growing and subsequently start to depolymerize their microtubules in a cell-autonomous manner. The data also show that EB1-GFP, in addition to decorating the ends of growing MTs, has the capacity to locate to novel bar-like structures.
Air exchange above the biofilm culture medium promotes MT repolymerization
Another entirely unexpected effect on EB1-GFP dynamics was caused after removal of the lid of the culture dish. In cells that had EB1-GFP dispersed and/or at bars, EB1-GFP spontaneously reformed comets after the lid of the culture dish was removed (Figures 2E, 5.5 min, and 3D). This effect was seen even though the lids are triple-vented to allow air exchange during incubation. Live-cell imaging after lid removal revealed the synchronous nature of this response in a population of cells (Supplemental Video S7). In this experiment, all cells imaged (188 of ∼188) repolymerized their MTs regardless of their MT status before lid removal (compare Figure 3, C with D).
These findings indicate that, upon culture lid removal, cells at the early stages of biofilm formation rapidly repolymerize MTs. No media mixing was required to promote this effect, indicating MT repolymerization could involve exchange of dissolved gaseous component(s).
To investigate whether EB1-GFP dispersal during static biofilm culture is specific to the static mode of growth, we asked whether similar behavior would occur within biofilm cells formed under agitation. We used identical growth conditions, except for mixing of the culture dish at 100 rpm on a rotating shaker after an initial static incubation of 8 h at 32°C. We then imaged the dishes immediately after removal from the shaker and observed that EB1-GFP became dispersed within cells of the biofilm formed under rotating conditions (Supplemental Figure S1). This result indicates that biofilm-driven EB1-GFP dispersal is not specific to statically grown biofilms.
As biofilms mature, partial biofilm removal is required to promote MT repolymerization
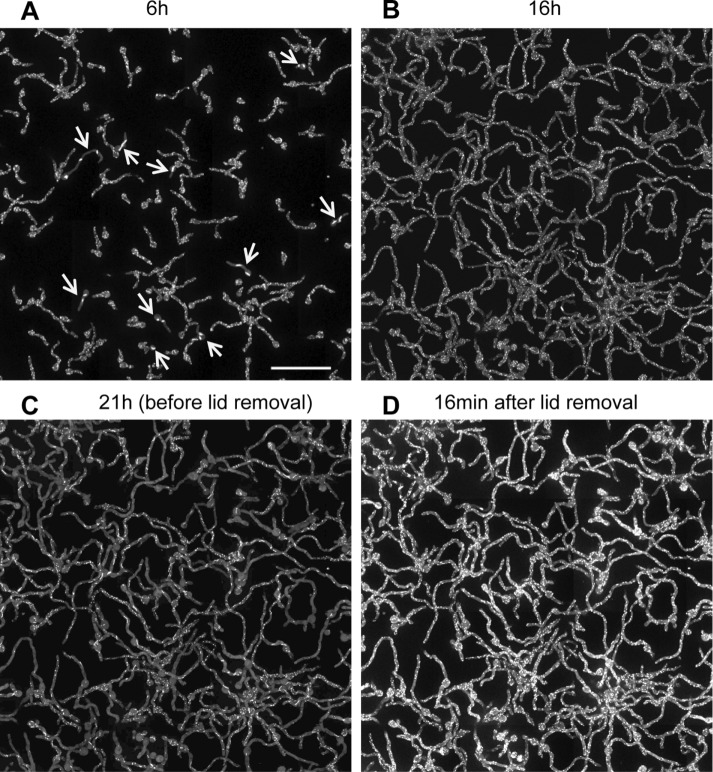
As biofilm cultures matured, EB1-GFP bars also disassembled. In these more mature biofilms, lid removal, or lid removal followed by media mixing above the biofilm, failed to trigger MT repolymerization. We therefore asked whether physically breaking the biofilm might affect MT dynamics. A central region of the biofilm was removed, as depicted in Figure 5A and shown in Supplemental Figure S2. The localization of EB1-GFP was examined over multiple adjacent fields before and after biofilm removal. Image stitching produced a single image covering an 1191 × 218 µm area. Analysis of the ∼400 cells from the undisturbed biofilm shown in Supplemental PowerPoint S1 (play as slide show) revealed that the large majority had EB1-GFP dispersed. A much lower number of cells had EB1-GFP at small foci or bars, and a few were highly vacuolated, with an atypically high nonspecific signal. However, no cells had EB1-GFP at comets, indicating that all had disassembled MTs before removal of part of the biofilm (Figure 5B). After partial biofilm removal, three zones of cells with different patterns of EB1-GFP were apparent (Supplemental PowerPoint 2, play as slide show). Cells in the biofilm nearest the scrape edge repolymerize their MTs, and EB1-GFP appeared as comets (Figure 5C, zone 1). In cells further from the edge (Figure 5C, zone 2), EB1-GFP appeared at bar-like structures, whereas the zone furthest from the edge (Figure 5C, zone 3) contained cells in which EB1-GFP remained dispersed. These experiments indicate that an intact biofilm is required to maintain MTs in a depolymerized state after the biofilm has matured but that cells retain the ability to repolymerize MTs when their microenvironment is physically changed.
FIGURE 5:
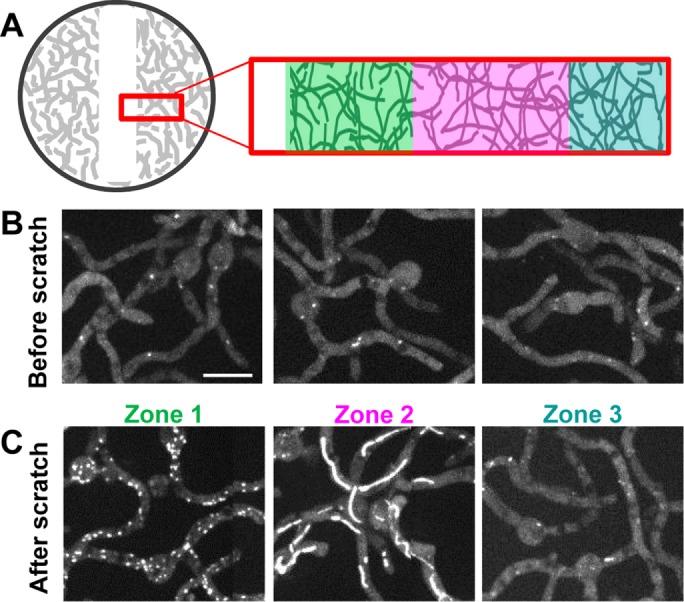
As biofilms mature, partial biofilm removal is required to promote MT repolymerization. (A) Experimental setup in which the local cell density is decreased by scraping off some cells using a micropipette tip (Supplemental Figure S1). (B) Before local cell removal, 40 imaged fields (20 × 2) were stitched together to show that EB1-GFP (strain SO1563) was dispersed. Representative examples of cells; see Supplemental PowerPoint S1 for entire imaged region. (C) Representative examples of cells 30 min after partial biofilm removal, showing EB1-GFP at comets (zone 1), at bars (zone 2), or remaining dispersed (zone 3). See Supplemental PowerPoint S2 for the entire imaged region. To generate the mature biofilm, cells were grown at 32°C for 6 h, followed by 30 h at 23 ± 2°C. Scale bar, 10 μm.
Fungal colony–type growth promotes MT disassembly toward the center of the colony
During fungal colony development, cells grow in a radial fashion at the colony edge. The central region contains a higher density of older, nongrowing cells, whereas the cells at the growing edge are younger and less crowded. We considered the possibility that MTs might be regulated in the different zones of the colony in a manner analogous to that seen during biofilm development. In this scenario, the growing leading edge of the colony would represent stages before biofilm formation and the central zone a more biofilm-like state. To address this, cells were point inoculated at the edge of the microscope imaging dish and allowed to grow across the observation field as depicted in Figure 6A. EB1-GFP was imaged over multiple linked fields from the growing tip cells back into the older, nongrowing cells. Cells closest to the growing edge had EB1-GFP comets reflecting the dynamic MTs in these cells (Figure 6B, zone 1). In the more central region, EB1-GFP was dispersed, indicating that these nongrowing cells had likely disassembled MTs (Figure 6B, zone 3). Between zones 1 and 3, zone 2 also contained nongrowing cells, and many had EB1-GFP located to bar-like structures (Figure 6B, zone 2). Therefore cells within a growing colony undergo changes in their EB1 dynamics in a manner similar to what occurs during biofilm maturation and likely reflect the different microenvironment that cells in the different zones generate and experience that causes their MTs to disassemble.
FIGURE 6:
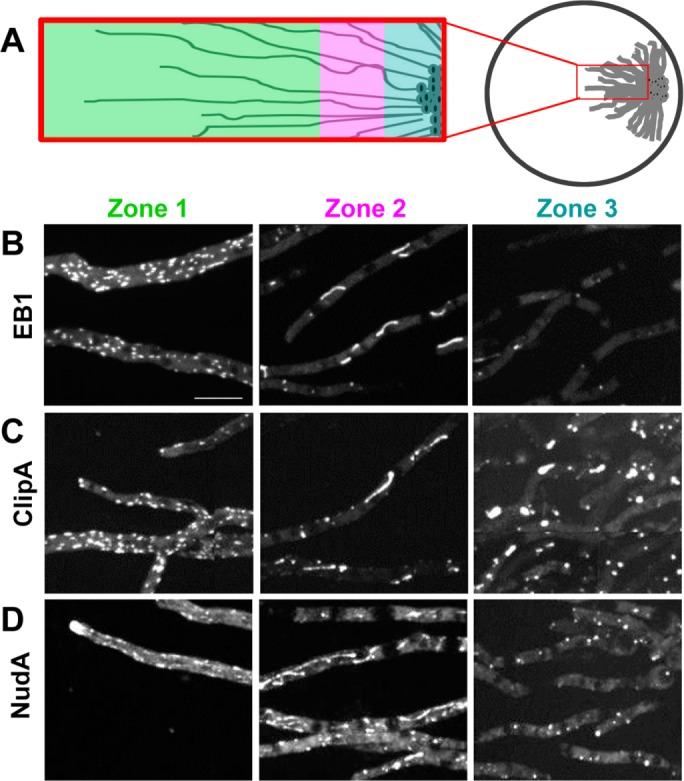
Cells within colonies display different locations for +TIPs proteins. Colony growth generates distinct zones of cells with polymerized and depolymerized MTs. (A) Growth pattern and zones having different states of MT polymerization as reflected in the locations of (B) EB1-GFP (strain SO1563), (C) ClipA-GFP (strain NS141), and (D) dynein heavy chain NudA-3XGFP (strain NS176). Scale bar, 10 μm.
Dynein and CLIP-170 (ClipA) display similar dynamics to EB1 during colony formation
To investigate whether EB1-GFP was unique in its behavior, particularly with regard to its location to bar-like structures, we investigated two additional +TIPS proteins: ClipA (orthologue of Clip-170) and NudA (cytoplasmic dynein heavy chain). Like EB1, ClipA and NudA interact with MTs and locate to their plus ends (Xiang et al., 2000; Efimov et al., 2006; Egan et al., 2012). Both ClipA-GFP and NudA-3XGFP displayed similar distributions in the three zones of the fungal colony as observed for EB1-GFP. In zone 1, populated by growing tip cells and nongrowing subapical cells, ClipA-GFP appeared as comets representing the ends of growing MTs (Figure 6C, zone 1) as previously reported (Efimov et al., 2006). NudA-3XGFP similarly located to comets of cells in zone 1, as well as more distinctly at the growing cell tips (Figure 6D, zone 1), also as previously reported (Xiang et al., 2000; Egan et al., 2012). However, like EB1-GFP, in intermediate zone 2, ClipA-GFP and NudA-3XGFP appeared dispersed and in bar-like structures, with neither of them locating as comets in such cells (Figure 6, C and D, zone 2). Finally, in central zone 3, ClipA-GFP and NudA-3XGFP were largely dispersed but also located to immobile foci (Figure 6, C and D, zone 3). Similar immobile ClipA-GFP and NudA-3XGFP foci were seen after MT disassembly using benomyl, indicating that the immobile foci in the central zone cells represent effects caused by MT disassembly (Supplemental Figure S3).
Hydrogen sulfide promotes reversible microtubule disassembly
MT repolymerization caused by culture dish lid removal (Figure 3D and Supplemental Video S7) suggests that soluble gases might be involved in MT depolymerization and perhaps generation of the novel EB1-GFP bar-like structures. As part of a separate study using dithiothreitol (DTT) to activate the unfolded protein response in A. nidulans (Saloheimo et al., 2003), we observed that DTT together with sodium thiosulfate (which was in the medium as a sulfur supplement) promoted rapid MT disassembly, although addition of each alone did not elicit this response. Prior heating of DTT and thiosulfate together before adding them to cells greatly enhanced this effect and generated the distinctive smell of hydrogen sulfide. This suggested that hydrogen sulfide, known to be formed from DTT and thiosulfate (Olson et al., 2013), might promote MT disassembly. This was further tested by using sodium hydrosulfide (NaHS), a commonly used H2S-releasing chemical, which promoted rapid MT disassembly. We therefore studied the effects of NaHS addition on MT dynamics of growing cells in more detail.
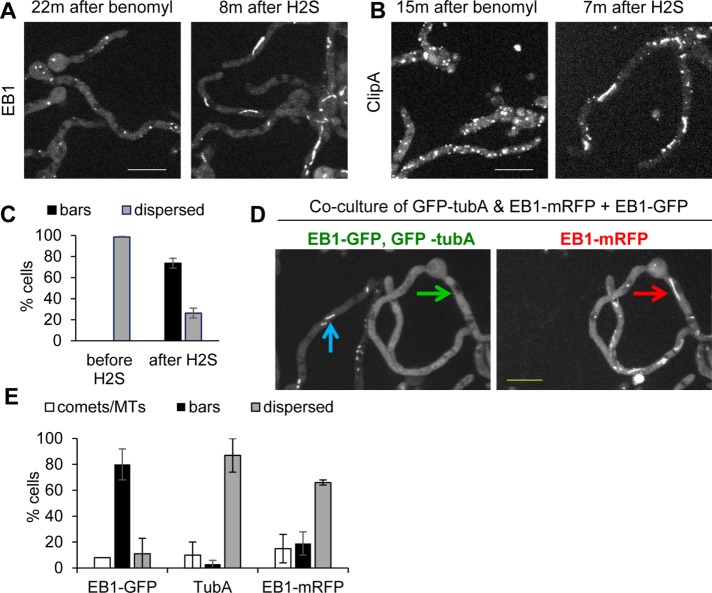
After addition of 50 µM NaHS, MTs were disassembled and EB1-GFP dispersed throughout cells. However, in many cells, MTs then repolymerized and EB1-GFP reappeared as comets (unpublished data). This reversible effect is presumably due to transient release and then volatilization of H2S (Olson et al., 2013). Increasing NaHS levels to 100 µM or higher promoted longer-lasting MT disassembly, and EB1-GFP remained dispersed for an extended period (>1 h). The effect of NaHS on EB1-GFP was rapid (Figure 7A and Supplemental Video S8). Of importance, EB1-GFP did not just remain dispersed after NaHS addition but, over the course of minutes, appeared as thin fibers and bar-like structures (Figure 7, A and B, and Supplemental Video S8), which are reminiscent of the EB1-GFP structures observed in cells within biofilms and colonies undergoing MT depolymerization (Figures 2C, 4, A and B, and 6). Although somewhat variable, a pattern emerged for the way in which EB1-GFP transitioned from being at comets to filaments and bars after NaHS addition. Initially, EB1-GFP quickly dispersed throughout cells, with some foci of EB1-GFP present near shadows (Figure 7A). The shadows represent soluble EB1-GFP excluded from nuclei, indicating that EB1-GFP foci are at SPBs. Similar foci are generated after MT depolymerization using benomyl (compare Figures 7A and 1C). The foci might therefore represent remnant MTs at MTOCs that are resistant to NaHS and benomyl treatment. In the second phase, EB1-GFP appeared at long filaments that were initially thin but thickened with time, generating more bar-like structures (Supplemental Video S8).
FIGURE 7:
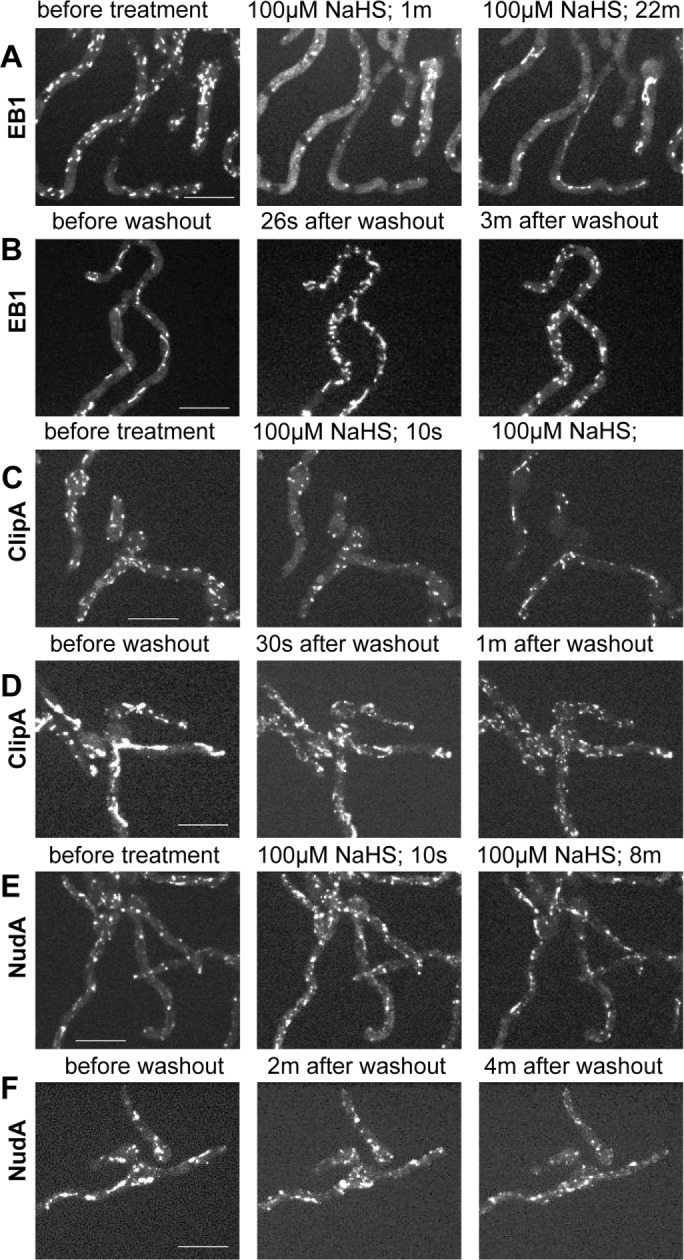
Treatment with the H2S donor NaHS reversibly depolymerizes MTs. (A) After treatment with 100 μM NaHS, EB1-GFP (strain NS326) initially disperses from comets but then locates to fibrous and bar-like structures (Supplemental Video S8). (B) After washout, EB1-GFP rapidly returned to comets, indicating immediate MT repolymerization (Supplemental Video S9). (C) ClipA-GFP (strain NS141) also dispersed and then located to bar-like structures upon NaHS addition (Supplemental Video S10). (D) ClipA-GFP also relocated to mobile comets immediately after washout (Supplemental Video S11). (E) On NaHS treatment, NudA-3XGFP (strain NS329) rapidly transitioned into fibrous and bar-like structures (Supplemental Video S12) and it also (F) resumed its dynamic behavior after NaHS washout (Supplemental Video S13). Scale bar, 10 μm.
To investigate whether the effects of NaHS are toxically irreversible, we removed it by media exchange. After media exchange, EB1-GFP bars and filaments dissolved, and starburst-like clusters of EB1-GFP comets appeared at multiple sites within seconds (Figure 7B and Supplemental Video S9). It is therefore possible to promote the transition of EB1-GFP from comets to filaments/bars and back to comets by addition and removal of H2S (Figure 7, A and B), closely mimicking how EB1-GFP behaves naturally within biofilms but on a much faster time scale.
Experiments tracking ClipA-GFP or NudA-3XGFP after addition of 100 µM NaHS and its subsequent removal revealed similar effects as seen for EB1-GFP (Figure 7, C–F, and Supplemental Videos S10–S13), with some subtle differences. ClipA-GFP, like EB1-GFP, largely dispersed immediately after NaHS addition and also located to multiple foci. The time taken between addition of NaHS to taking the first image (<10 s) was often sufficient to trigger this effect. ClipA-GFP was less apparent at fibrous structures, however, before appearing at bar-like structures when compared to EB1-GFP (Figure 7C and Supplemental Video S10). On washout, ClipA-GFP in bars and foci immediately converted into the normal comet-like ClipA-GFP behavior as seen before NaHS addition (Figure 7D and Supplemental Video S11). NudA-3XGFP, on the other hand, first appeared more directly at fibrous-like structures before appearing at bars in response to NaHS (Figure 7E and Supplemental Video S12). It also rapidly resumed its normal dynamic behavior after NaHS washout (Figure 7F and Supplemental Video S13).
Effects of MT depolymerization on EB1-GFP filaments and bars
To determine whether the bar-like structures to which EB1-GFP locates are potentially MT based, we asked whether they could be disassembled by benomyl treatment. Addition of benomyl failed to disrupt the bars generated after NaHS treatment (unpublished data). We therefore asked whether prior MT disassembly using benomyl would prevent the transition of dispersed EB1-GFP into bar-like structures upon NaHS treatment. Surprisingly, although EB1-GFP was effectively dispersed by benomyl (Figure 8A), subsequent cotreatment with NaHS still promoted the formation of EB1-GFP bar-like structures (Figure 8, A and C, after H2S; and Supplemental Video S14). The effect of MT depolymerization on ClipA-GFP bar formation was also tested, and its bar location abilities did not change dramatically after H2S treatment (Figure 8B and Supplemental Video S15).
FIGURE 8:
EB1-GFP bars may not represent MT-based structures. (A) Benomyl treatment causes EB1-GFP (strain NS326) dispersal from comets, and subsequent NaHS addition then promotes EB1-GFP bar formation (Supplemental Video S14). (B) As also shown in Supplemental Figure S2, benomyl causes ClipA-GFP (strain NS141) to locate at immobile foci, but ClipA still relocates to bars upon treatment with NaHS (Supplemental Video S15). (C) Quantitation (mean ± SE) for EB1-GFP in benomyl-treated cells before and after NaHS treatment. The experiment was performed three times with 65 cells. (D) Coculture of strains expressing EB1-GFP (strain SO1563) and GFP-TubA and EB1-mRFP (strain NS133) during biofilm formation shows that when the majority of EB1-GFP cells display bars (blue arrow), none of the GFP-TubA locates to bars (green arrow). Within the same cell, at the time EB1-mRFP locates to bars (red arrow), GFP-tubA does not (green arrow). (E) Quantitation (mean ± SE) of EB1-GFP, GFP-TubA, and EB1-mRFP location to comets/MTs or bars or dispersed during biofilm formation. The experiment was performed twice with 122 cells. Scale bar, 10 µm.
To investigate further whether MTs or MT remnants could be involved in EB1 bar formation within biofilms, we cocultured a strain expressing both GFP-TubA and EB1-mRFP with a strain expressing only EB1-GFP until the biofilm stage at which EB1 locates to bars. EB1-GFP located clearly at bars (80% of cells), but virtually no cells had GFP-TubA localized at bar-like structures, including those in which EB1-mRFP located at bars (Figure 8, D and E). This result further indicates that MTs might not be involved in the formation of EB1-GFP bars. In these experiments, we also determined that EB1-mRFP was less likely to appear at bars than EB1-GFP even when grown under identical conditions (Figure 8E).
Microtubule depolymerization during biofilm formation is regulated by SrbA, the A. nidulans sterol regulatory element–binding protein 1 hypoxic transcription factor
To gain insight into the potential regulatory systems involved in controlling microtubule depolymerization in biofilms, we considered whether hypoxia might be involved. Similar to other fungi, including Schizosaccharomyces pombe, in which the regulatory pathway was discovered (Hughes et al., 2005), adaptation to hypoxia in A. nidulans depends on the homologue of the hypoxic transcription factor sterol regulatory element–binding protein 1 (Sre1). In A. nidulans and Aspergillus fumigatus, Sre1 is called SrbA (Willger et al., 2008; Bat-Ochir et al., 2016). As in other fungi, deletion of srbA in A. nidulans is known not to affect growth on agar plates under normoxic conditions (21% O2, ambient air) but impinges growth when conditions are hypoxic (1% O2 and 99% N2; Bat-Ochir et al., 2016). To test whether SrbA is similarly required for growth in liquid medium, srbA was deleted, and strains without SrbA were tested for growth in closed-tube cultures. This was done by using untreated medium or medium purged with N2 and by replacing air in the space above the liquid medium with N2. After growth, cells were washed in water and their dry weights determined. Compared to the wild-type (WT) control, srbA-deleted cells grew to a dry weight of 40% in closed, nonpurged tube cultures. The N2 purging limited WT growth to 70% of the control; however, growth of the srbA-deleted cells was far more limited and reached only 1.3% of the control. This indicates that SrbA is required for normal liquid medium growth under O2-limiting conditions, as previously shown for agar plate colony culture (Bat-Ochir et al., 2016).
We next compared the response of EB1-GFP during biofilm formation in a WT strain to one in which srbA was deleted (ΔsrbA). In WT cells, as described earlier, EB1-GFP became dispersed and formed bars during biofilm formation. Under identical growth conditions and time of culture, however, ΔsrbA cells failed to disperse EB1-GFP, which instead remained as mobile comets (Figure 9A). The effects were quantitated by counting the number of cells with EB1-GFP comets over binned time windows during biofilm formation and maturation (Figure 9B). This analysis revealed that whereas WT cells increasingly lost their EB1-GFP comets, the srbA-null continued to display EB1-GFP at comets for hours. The lack of EB1-GFP dispersal in the absence of SrbA indicates that during biofilm formation, the microenvironment starts to become hypoxic and that cells adapt through the SrbA regulatory system to promote MT depolymerization.
FIGURE 9:
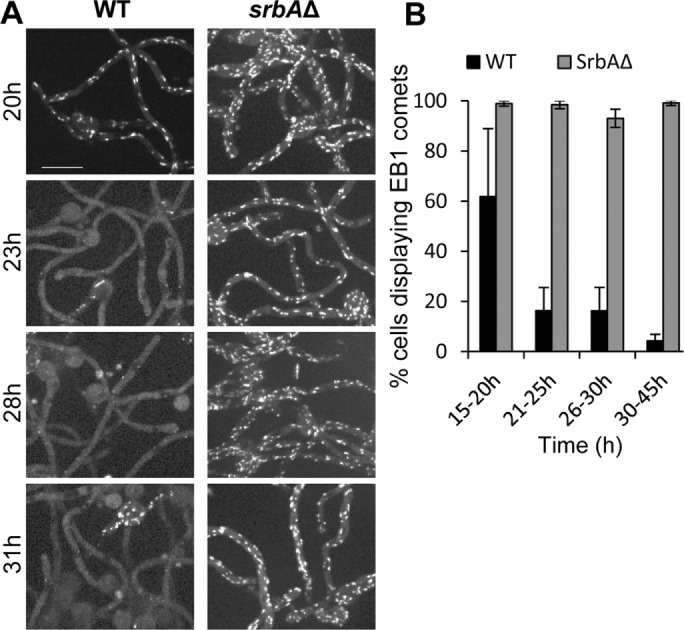
MT disassembly during biofilm formation requires the SrbA hypoxic transcription factor. (A) WT (strain SO1563) and SrbAΔ (strain SO1567) strains were imaged during early biofilm formation as the WT cells stop growing and depolymerize MTs and so have dispersed EB1-GFP. (B) Quantitation (mean ± SE) of EB1-GFP comets during this transition stage of the two strains. Cells were grown at 37°C for 7 h, followed by growth at 18°C for the indicated times. The experiment was performed three times with 2445 cells. Scale bar, 10 μm.
DISCUSSION
This study found that MTs are disassembled within forming A. nidulans biofilms. The process likely involves soluble gases because MTs rapidly reassemble after simple air exchange at the biofilm culture surface. This level of MT regulation is a controlled response mediated by the srbA hypoxic response transcription factor. We propose that stopping repeated cycles of MT polymerization would help to prevent energy loss within the biofilm. This study therefore uncovers a previously unrealized level of MT regulation involving changes in fungal biofilm gaseous microenvironments.
Three stages of MT behavior during early biofilm formation
Our analysis indicates that after adhesion and initiation of growth, the next stage of biofilm formation involves the cessation of growth of the initiator cells at the biofilm base. We previously noticed that growth is inhibited as colonies grow toward each other, forming a zone of growth inhibition (Supplemental Figure S4), and others reported evidence of self-inhibition in Mucor and Aspergillus (Bottone et al., 1998) that promotes unidirectional outward colony growth. A. nidulans cells therefore have a mechanism by which they detect and grow away from each other (Figure 2B) or, when more crowded, inhibit each other’s growth (Figure 2C). Such effects operate at the base of the biofilm as cells are initially surrounded by other growing cells and with time become increasingly crowded. We therefore propose that cell crowding contributes to cell growth inhibition during the early stages of biofilm formation. The nature of this negative growth regulation is unknown. However, the opposite type of regulation in Neurospora crassa, cell-to-cell attraction, is under oscillatory mitogen-activated protein kinase control via an unknown chemotropic attractant (Glass et al., 2004; Fleissner and Glass, 2007; Fleissner et al., 2009; Jonkers et al., 2014). It is interesting that N. crassa cells are actively attracted to each other and physically join together, whereas A. nidulans cells appear to be under an opposite type of regulation to actively avoid and inhibit each other’s growth . Why cells of these two fungi respond to their own kind in the opposite manner is an intriguing question.
After cells stop growing, they initially maintain active MTs, indicating that their growth inhibition is not due to limited nutrient availability or metabolic shutdown. As the biofilm matures, cells depolymerize their MTs, and EB1-GFP disperses and also locates to novel fibrous and then bar-like structures, as do NudA-3XGFP and ClipA-GFP. It is unclear what structure(s) these +TIPS proteins are locating to or potentially forming. Using benomyl, we showed that the novel +TIPS structures are formed in growing cells in response to H2S even after MTs have been depolymerized, suggesting that the +TIPS fibers and bars might not be MT based. Supporting this possibility, we did not detect similar fibrous or bar-like structures when directly tracking MTs using the GFP-tagged α-tubulin TubA during biofilm formation. Of interest, during carbon source starvation, quiescent budding and fission yeast cells form MT bar-like arrays (Laporte et al., 2013, 2015) and we do not discount the possibility that the +TIPS Aspergillus biofilm bars are potentially MT based. For example, such MTs could be resistant to benomyl and/or not contain TubA tubulin. Other alternative possibilities are that the +TIPS are forming de novo structures as seen for some metabolic proteins that form cytoophidia (Greek for “cellular snakes”; Liu, 2016) or proteins that form higher-order assemblies to generate amyloids, signalosomes, and granules (Wu and Fuxreiter, 2016). Further studies are required to characterize the novel +TIPs structures. However, because of their distinctive and transient appearance, they provide a readily monitored characteristic of a specific physiological state within maturing A. nidulans biofilms.
MT depolymerization during biofilm formation is a regulated process in response to developing hypoxia
In theory, MT disassembly within biofilms could be the result, for example, of cells dying or running out of tubulin proteins. However, MTs rapidly repolymerize after air exchange above the culture medium. This indicates that such cells are unlikely to be dying or have limited tubulin. However, as the biofilm matures further and EB1-GFP is dispersed, physical removal of part of the biofilm is required to promote MTs to repolymerize. During biofilm maturation, an extracellular matrix is formed that encases cells within the biofilm structure (Mitchell et al., 2016). Potentially, this matrix might prevent the gaseous exchange that promotes MT repolymerization, therefore requiring its removal to promote MT polymerization. Consistent with this idea, MTs repolymerize first at the edge of the remaining biofilm after partial biofilm removal.
Our findings further indicate that MT disassembly is regulated in response to changing biofilm gaseous microenvironment under regulatory control mediated by the hypoxic transcription factor SrbA. Without SrbA function, cells become stuck at a stage of biofilm formation after they stop growing but maintain active MT dynamics. The cells remain stuck at this stage and continue to display EB1-GFP at comets, whereas in cells with SrbA function, EB1-GFP is dispersed and forms bars. The SrbA regulatory response is therefore required to enable cells to promote MT depolymerization and also the formation of +TIPS filaments and bars. The response also potentially involves the accumulation of H2S, which, of importance, we showed can, under normoxic conditions, promote both MT disassembly and formation of the characteristic +TIPS filament and bar-like structures. We do not have further data to support the idea that naturally generated H2S plays a role within biofilms, but H2S is known to mimic the effects of hypoxia and has been implicated in oxygen sensing (Olson, 2015). H2S can act as a signaling molecule via modification of protein cysteine residues (Paul and Snyder, 2012; Aroca et al., 2015; Filipovic, 2015), although this level of regulation has not been shown in fungal cells, and also has an inhibitory effect on cytochrome C oxidase and respiration (Matallo et al., 2014; Modis et al., 2014; Szabo et al., 2014). Further analysis will therefore be required to define the relationships between hypoxia and MT depolymerization, and it will be interesting to test for the presence of H2S within biofilms. We suggest that the biological function of this regulation is to prevent wasteful energy loss as hypoxia develops within biofilms.
Prior studies showed that as biofilms form, including those generated by fungi, hypoxic microenvironments can be generated to the extent, for example, that Candida albicans biofilms can support the growth of obligate anaerobic bacteria (Fox et al., 2014). In our experiments, we find that although hypoxia likely starts to develop very early during biofilm formation, hypoxic conditions alone may not suffice to promote MT disassembly. If hypoxia alone promotes MT disassembly, then all cells should respond in the same manner as hypoxia develops. However, we see that cells respond autonomously within biofilms even though they would be experiencing the same hypoxic microenvironment. Even cells separated by septa, and so within micrometers of each other, can maintain different states of EB1-GFP behavior. Therefore MT disassembly likely involves interplay between the microenvironment and differing physiological states between individual cells rather than being solely a direct consequence of the changing microenvironment.
Potential insights into fungal pathogenicity
Our findings provide new insight into how the physiology of cells within a fungal biofilm become modified and so might be informative in terms of the roles played by biofilms during fungal infections and subsequent treatments. Two aspects of fungal biofilm biology believed to help mediate resistance to antifungal treatments are the physical barrier conferred by the biofilm and the residence within the biofilm of persister cells recalcitrant to drug therapies (Taff et al., 2013).
As the biofilm generated in our experiments matured, it appears to form an effective barrier to the gaseous exchange required to allow MTs to repolymerize in cells at the base of the biofilm. Such cells have not lost the ability to repolymerize their MTs but require physical removal of part of the biofilm to promote their MTs to repolymerize. These findings suggest that the A. nidulans biofilm might confer a barrier to very small molecules in solution and, as it matures, could help maintain and perhaps enforce the inactive state of the base cells within the biofilm. In effect, the biofilm barrier might not only provide an effective physical barrier protecting internal cells from antifungal treatments but also help to maintain them in a dormant physiological state, which might help further confer resistance to treatment. The nature of the biofilm barrier remains to be established and is only functionally defined here as a biofilm property that makes MT repolymerization more resistant to gaseous exchanges above the biofilm medium.
Our findings also provide insights into the physiological modifications occurring within biofilms that might play roles in the generation of persister cells and into their resistance to treatment. For example, because cells at the base of the biofilm have stopped growing and are in a dormant state with no MTs, they are unlikely to respond to drugs that target MTs or enzymes involved in growth processes. This reversible semidormant state might therefore contribute to the physiological characteristics that enable persister cells to tolerate antifungal treatments. In a similar vein, cells within mammalian solid tumors experience hypoxia, which induces resistance to MT-based chemotherapies, but in this case, MTs are more stable rather than less stable under hypoxic conditions (Yoon et al., 2005; Peng et al., 2010; Das et al., 2015). It will be interesting to unravel the similarities and differences of the roles of hypoxic microenvironments with MT regulation between biofilms and solid tumors and whether such knowledge might be leveraged for pharmacological interventions.
Results from analysis of SrbA in A. fumigatus support the idea that the physiological changes involving MTs we have defined during biofilm formation are of potential significance to the pathology and treatment of fungal infections. In the absence of SrbA A. nidulans cells fail to depolymerize their MTs normally during biofilm formation. In A. fumigatus, biofilm formation is compromised in the absence of SrbA (Vaknin et al., 2016), and deleted strains are additionally sensitive to azole drugs and have greatly reduced virulence in murine models of invasive pulmonary aspergillosis (Willger et al., 2008). It is possible that some of the effects seen in the absence of SrbA in A. fumigatus could potentially involve the continued presence of active MT dynamics during early stages of biofilm formation and their failure to enter the controlled dormant state that our studies show normally develops in cells at the base of biofilms.
Conclusions
Our analysis provides new insight into the physiological changes that occur in cells during the formation of a fungal biofilm involving the regulated depolymerization of MTs. These changes in cell physiology are likely in response to early-developing hypoxic microenvironments within the biofilm and are under control of the hypoxic transcription factor SrbA. Our findings indicate that one of the responses mediated by the hypoxic SrbA transcription factor is to promote MT depolymerization. This would help preserve energy levels while at the same time leave cells in a state poised to reestablish normal MT dynamics if their microenvironment changed. As part of this study, we additionally found that +TIPS proteins, which normally locate to the plus ends of MTs as comets, can also naturally exist at fibrous and bar-like structures as a characteristic phenotype within cells as biofilms are formed. These structures are immediately converted into comets in response to air exchange above the culture medium of the forming biofilm as MTs are triggered to repolymerize. Identical +TIPS structures can be artificially promoted by H2S, and these too immediately convert into comets on removal of H2S. The work therefore makes new connections between biofilm gaseous microenvironments, MT regulation, SrbA, and, potentially, H2S signaling.
MATERIALS AND METHODS
Strains, media, and drug treatments
Strains (Table 1) were generated by classical genetic methodologies (Pontecorvo et al., 1953; Todd et al., 2007) or using gene-targeting constructs generated via fusion PCR and transformation (Yang et al., 2004; Nayak et al., 2006). All of the proteins examined in this study were tagged at their endogenous locus. The medium for microscopy contained glucose (10 g/l), trace elements, urea (10 mM), MgSO4 (2 mM), KCl (7 mM), and phosphate buffer (6 mM KH2PO4 and 6 mM K2HPO4⋅3H2O). The medium was supplemented with uridine (1.2 g/l) and uracil (1.12 g/l), arginine (6.4 g/l), and sodium thiosulfate (3.2 mM) if required. A 1 M DTT (Sigma-Aldrich) stock was made in sterile deionized water freshly before use. A 3 mg/ml fresh stock of NaHS⋅9H2O (Sigma-Aldrich) was made in ice-cold phosphate buffer (100 mM) before each treatment. Benomyl (0.4 mg/ml stock made in ethanol) was added to a final concentration of 2.4 μg/ml (De Souza and Osmani, 2011).
TABLE 1:
Strains used in this study.
| Strain | Genotype |
|---|---|
| SO1563 | EB1-GFP::pyroAAf; pyrGAf; pyrG89; pyroA4; wA3 (fwA1?; chaA1?) |
| SO1567 | EB1-GFP::pyroAAf; ΔsrbA::pyrGAf; pyrG89; pyroA4; wA3 (fwA1?; chaA1?) |
| SO1529 | EB1-GFP:: pyroAAf(pyroA4); pyrGAf(pyrG89); argB2; nirA14; wA3 |
| NS326 | EB1-GFP::pyroAAf; Ndc80-CR::pyrGAf; ΔnkuA::argB; pyrG89; argB2; nirA14; (pyroA4?) |
| NS133 | EB1-mRFP::pyrGAf; GFP-tubA; pyrG89; argB2; wA3; (fwA1?; chaA1?; nirA14?) |
| NS327 | EB1-CR::pyroAAf; Gcp3-GFP::riboBAf; pyrG89; pyroA4; wA3; sE15; nirA14; wA3; (fwA1?, riboB2?) |
| NS141 | ClipA-GFP::pyrGAf; ΔyA::NLSdsRed; pyrG89; argB2; fwA1; (nirA14?) |
| NS176 | NudA-3XGFP::pyrGAf; AN3906-CR::pyroAAf; ΔnkuA::argB; pyrG89; pyroA4; argB2; sE15; nirA14; wA3; (fwA1?; chaA1?) |
| NS329 | NudA-3XGFP::pyrGAf; Ndc80-CR::pyrGAf; Gcp3-GCP::riboBAf; ΔnkuA::argB; argB2; pyrG89; pyroA4; nirA14; (riboB2?) |
| R153 | wA3; pyroA4 |
A question mark indicates that marker status is unknown due to epistasis to other markers in the strain. Underline indicates auxotrophy. All strains carry veA1.
Cell culture and microscopy
A. nidulans spores were generated by agar overlay or spread plate methods for 36–42 h at 32°C and harvested in 0.2% Tween-80 or spore stock solution (0.02% Tween-80 plus 0.85% NaCl). For biofilm experiments, 3 ml of liquid medium mixed with spores (106 per ml) was added to glass-bottomed microwell dishes (MatTek, Ashland, MA) and grown for 6 h at 32°C followed by 20 h (early biofilm) or 30 h (more mature biofilm) at room temperature (21–25°C) or 7 h at 37°C followed by 20 h at 18°C. These temperatures were used to facilitate the timing of imaging. If all growth is completed at room temperature, with no temperature shifts, the same results are obtained as reported in this study (unpublished data). For imaging biofilm cells formed under mixing conditions, after inoculation, dishes were incubated for 8 h at 32°C with no mixing and then placed on a rotating orbital shaker at 100 rpm at room temperature. Cells were imaged immediately after their removal from the shaker. For growth as submerged colonies, 3- to 5-μl spores in MAG top-agar (0.8% wt/vol) was inoculated on one edge of an empty microwell dish, and 3 ml of minimal medium was secondarily added after the agar solidified. The cells began to grow out from the agar mass into the liquid medium after 24 h and were imaged between 38 and 48 h. For H2S treatments, spores (105 per ml) were grown in 3 ml of medium at 32°C for 3 h followed by growth at 25°C for 12–16 h before imaging. Microscopy was done using either a 60×/1.49 numerical aperture (NA) or 100×/1.4 NA total internal reflection fluorescence objective as described (Govindaraghavan et al., 2014), and image analyses were performed using ImageJ, version 1.47k. Larger images were generated from several fields of view using the automated stitching program in the Ultraview Volocity software (PerkinElmer). All images presented are maximum intensity projections.
Supplementary Material
Acknowledgments
We thank all of the members of the Osmani lab, especially Angela B. Davis and Dale E. Lingo, for assistance with experiments. We thank Samara Reck-Peterson (University of California, San Diego, La Jolla, CA) for providing the NudA3X-GFP–tagged strain (RPA349; Egan et al., 2012). We also thank Herb Arst (Imperial College, London, United Kingdom), Naioki Takaya (University of Tsukuba, Tsukuba, Japan), Sven Krappmann (University Hospital Erlangen and Friedrich-Alexander University, Erlangen, Germany), Jorge Amich (Würzburg University Hospital, Julius-Maximilians University, Würzburg, Germany), and Berl Oakley (University of Kansas, Lawrence, KS) for helpful insights. This research was funded in part by National Institutes of Health Grant GM042564 to S.A.O.
Abbreviations used:
- Clip-170
cytoplasmic linker protein of 170 kD
- DTT
dithiothreitol
- EB1
end-binding protein-1
- GFP
green fluorescent protein
- mRFP
monomeric red fluorescent protein
- MT
microtubule
- MTOC
microtubule-organizing center
- NA
numerical aperture
- SPB
spindle pole body
- Sre1
sterol regulatory element-binding protein 1
- +TIP
plus-end binding/tracking protein
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0750) on January 5, 2017.
REFERENCES
- Araujo D, Henriques M, Silva S. Portrait of candida species biofilm regulatory network genes. Trends Microbiol. 2017;25:62–75. doi: 10.1016/j.tim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Aroca A, Serna A, Gotor C, Romero LC. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 2015;168:334–342. doi: 10.1104/pp.15.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bat-Ochir C, Kwak JY, Koh SK, Jeon MH, Chung D, Lee YW, Chae SK. The signal peptide peptidase SppA is involved in sterol regulatory element-binding protein cleavage and hypoxia adaptation in Aspergillus nidulans. Mol Microbiol. 2016;100:635–655. doi: 10.1111/mmi.13341. [DOI] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone EJ, Nagarsheth N, Chiu K. Evidence of self-inhibition by filamentous fungi accounts for unidirectional hyphal growth in colonies. Can J Microbiol. 1998;44:390–393. doi: 10.1139/w98-009. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Mol Biol Cell. 2015;26:1207–1210. doi: 10.1091/mbc.E13-10-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das V, Stepankova J, Hajduch M, Miller JH. Role of tumor hypoxia in acquisition of resistance to microtubule-stabilizing drugs. Biochim Biophys Acta. 2015;1855:172–182. doi: 10.1016/j.bbcan.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Desai JV, Mitchell AP, Andes DR. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med. 2014;4:a019729. doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani SA. A new level of spindle assembly checkpoint inactivation that functions without mitotic spindles. Cell Cycle. 2011;10:3805–3806. doi: 10.4161/cc.10.22.18187. [DOI] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci USA. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellberg C, Trokter M, Jha R, Sen I, Steinmetz MO, Surrey T. Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat Cell Biol. 2014;16:804–811. doi: 10.1038/ncb2999. [DOI] [PubMed] [Google Scholar]
- Efimov VP, Zhang J, Xiang X. CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol Biol Cell. 2006;17:2021–2034. doi: 10.1091/mbc.E05-11-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Mitchell AP. Fungal biofilms. PLoS Pathog. 2012;8:e1002585. doi: 10.1371/journal.ppat.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic MR. Persulfidation (S-sulfhydration) and H2S. Handb Exp Pharmacol. 2015;230:29–59. doi: 10.1007/978-3-319-18144-8_2. [DOI] [PubMed] [Google Scholar]
- Fleissner A, Glass NL. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryotic Cell. 2007;6:84–94. doi: 10.1128/EC.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A, Leeder AC, Roca MG, Read ND, Glass NL. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci USA. 2009;106:19387–19392. doi: 10.1073/pnas.0907039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster TM, Mogavero S, Drager A, Graf K, Polke M, Jacobsen ID, Hube B. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell Microbiol. 2016;18:1709–1715. doi: 10.1111/cmi.12657. [DOI] [PubMed] [Google Scholar]
- Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24:2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Rasmussen C, Roca MG, Read ND. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 2004;12:135–141. doi: 10.1016/j.tim.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Govindaraghavan M, Anglin SL, Osmani AH, Osmani SA. The Set1/COMPASS histone H3 methyltransferase helps regulate mitosis with the CDK1 and NIMA mitotic kinases in Aspergillus nidulans. Genetics. 2014;197:1225–1236. doi: 10.1534/genetics.114.165647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Correa M, Ludena Y, Ramage G, Villena GK. Recent advances on filamentous fungal biofilms for industrial uses. Appl Biochem Biotechnol. 2012;167:1235–1253. doi: 10.1007/s12010-012-9555-5. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic plus TIP networks. Mol Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Leeder AC, Ansong C, Wang Y, Yang F, Starr TL, Camp DG, 2nd, Smith RD, Glass NL. HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 2014;10:e1004783. doi: 10.1371/journal.pgen.1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- Kaur S, Singh S. Biofilm formation by Aspergillus fumigatus. Med Mycol. 2014;52:2–9. doi: 10.3109/13693786.2013.819592. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Laporte D, Courtout F, Pinson B, Dompierre J, Salin B, Brocard L, Sagot I. A stable microtubule array drives fission yeast polarity reestablishment upon quiescence exit. J Cell Biol. 2015;210:99–113. doi: 10.1083/jcb.201502025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Courtout F, Salin B, Ceschin J, Sagot I. An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence. J Cell Biol. 2013;203:585–594. doi: 10.1083/jcb.201306075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL. The cytoophidium and its kind: filamentation and compartmentation of metabolic enzymes. Annu Rev Cell Dev Biol. 2016;32:349–372. doi: 10.1146/annurev-cellbio-111315-124907. [DOI] [PubMed] [Google Scholar]
- Matallo J, Vogt J, McCook O, Wachter U, Tillmans F, Groeger M, Szabo C, Georgieff M, Radermacher P, Calzia E. Sulfide-inhibition of mitochondrial respiration at very low oxygen concentrations. Nitric Oxide. 2014;41:79–84. doi: 10.1016/j.niox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KF, Zarnowski R, Andes DR. Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 2016;12:e1005828. doi: 10.1371/journal.ppat.1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Modis K, Bos EM, Calzia E, van Goor H, Coletta C, Papapetropoulos A, Hellmich MR, Radermacher P, Bouillaud F, Szabo C. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part II. Pathophysiological and therapeutic aspects. Br J Pharmacol. 2014;171:2123–2146. doi: 10.1111/bph.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Olson KR. Hydrogen sulfide as an oxygen sensor. Antioxid Redox Signal. 2015;22:377–397. doi: 10.1089/ars.2014.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR, Deleon ER, Gao Y, Hurley K, Sadauskas V, Batz C, Stoy GF. Thiosulfate: a readily accessible source of hydrogen sulfide in oxygen sensing. Am J Physiol Regul Integr Comp Physiol. 2013;305:R592–R603. doi: 10.1152/ajpregu.00421.2012. [DOI] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. H(2)S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- Peng WX, Pan FY, Liu XJ, Ning S, Xu N, Meng FL, Wang YQ, Li CJ. Hypoxia stabilizes microtubule networks and decreases tumor cell chemosensitivity to anticancer drugs through Egr-1. Anat Rec (Hoboken) 2010;293:414–420. doi: 10.1002/ar.21086. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Ramage G, Rajendran R, Gutierrez-Correa M, Jones B, Williams C. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol Lett. 2011;324:89–97. doi: 10.1111/j.1574-6968.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Valkonen M, Penttila M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol. 2003;47:1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Szabo C, Ransy C, Modis K, Andriamihaja M, Murghes B, Coletta C, Olah G, Yanagi K, Bouillaud F. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br J Pharmacol. 2014;171:2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Oakley BR. Microtubule dynamics in mitosis in Aspergillus nidulans. Fungal Genet Biol. 2011;48:998–999. doi: 10.1016/j.fgb.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff HT, Mitchell KF, Edward JA, Andes DR. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013;8:1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RB, Davis MA, Hynes MJ. Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Nat Protoc. 2007;2:811–821. doi: 10.1038/nprot.2007.112. [DOI] [PubMed] [Google Scholar]
- Vaknin Y, Hillmann F, Iannitti R, Ben Baruch N, Sandovsky-Losica H, Shadkchan Y, Romani L, Brakhage A, Kniemeyer O, Osherov N. Identification and characterization of a novel Aspergillus fumigatus rhomboid family putative protease, RbdA, involved in hypoxia sensing and virulence. Infect Immun. 2016;84:1866–1878. doi: 10.1128/IAI.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Fuxreiter M. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell. 2016;165:1055–1066. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Oakley BR. In vivo analysis of the functions of gamma-tubulin-complex proteins. J Cell Sci. 2009;122:4218–4227. doi: 10.1242/jcs.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryotic Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res. 2005;65:2761–2769. doi: 10.1158/0008-5472.CAN-04-4122. [DOI] [PubMed] [Google Scholar]
- Zeng CJ, Kim HR, Vargas Arispuro I, Kim JM, Huang AC, Liu B. Microtubule plus end-tracking proteins play critical roles in directional growth of hyphae by regulating the dynamics of cytoplasmic microtubules in Aspergillus nidulans. Mol Microbiol. 2014;94:506–521. doi: 10.1111/mmi.12792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.