Abstract
In the context of precision medicine, understanding patient-specific variation is an important step in developing targeted and patient-tailored treatment regimens for periodontitis. While several studies have successfully demonstrated the usefulness of molecular expression profiling in conjunction with single classifier systems in discerning distinct disease groups, the majority of these studies do not provide sufficient insights into potential variations within the disease groups.
Aim
The goal of the present study is to discern biological response profiles of periodontitis and non-periodontitis smoking subjects using an informed panel of biomarkers across multiple scales (salivary, oral microbiome, pathogens and other markers).
Materials and Methods
The investigation uses a novel ensemble classification approach (SVA-SVM) to differentiate disease groups and patient-specific biological variation of systemic inflammatory mediators and IgG antibody to oral commensal and pathogenic bacteria within the groups.
Results
Sensitivity of SVA-SVM is shown to be considerably higher than several traditional independent classifier systems. Patient-specific networks generated from SVA-SVM are also shown to reveal cross-talk between biomarkers in discerning the disease groups. High-confidence classifiers in these network abstractions comprised of host responses to microbial infection elucidated their critical role in discerning the disease groups.
Conclusions
Host adaptive immune responses to the oral colonization/infection contribute significantly to creating the profiles specific for periodontitis patients with potential to assist in defining patient-specific risk profiles and tailored interventions.
Keywords: periodontitis, smoking, saliva, serum, oral bacteria
Introduction
Periodontitis is a chronic destructive inflammatory disease that affects a majority of U.S. and global adults (Jin et al., 2011, Eke et al., 2012, Baelum and Lopez, 2013, Eke et al., 2015) and has been associated with significant systemic consequences for general health (Linden et al., 2013, Torumtay et al., 2015, Hajishengallis, 2015, Kumar, 2016). While clinical parameters are used in dental practice to identify disease, several critical limitations in addressing 21st century dentistry exist by constraining diagnostic and prognostic decisions based upon these clinical parameters. A principal issue with the clinical measures is that a significant amount of damage must occur before these diagnostic parameters are able to detect a sufficient level of disease, and are distanced in time from the biologic processes that initiated the disease progression. These clinical parameters, as currently obtained, cannot determine the current status of disease progression, nor effectively predict the rapidity and timing of exacerbation and progression, as well as the likely response to therapeutic intervention. Therefore, identifying signatures of biologic responses that occur early in the kinetics of the disease, presage disease progression, and provide guidance on treatment decisions and more rapid evaluation of treatment success would be of significant value to the field (Armitage, 2013, Slots, 2013, Matthews, 2014).
Disease phenotypes are often preceded by marked changes in the activity of several variables (molecular, microbial and others) across multiple scales. Understanding these changes can be especially helpful in identifying potential biomarkers with the capability to discern disease groups. Recent studies, with complex disease phenotypes, have clearly demonstrated the usefulness of single classifiers in conjunction with molecular profiling in discerning distinct disease groups (Kebschull and Papapanou, 2010, Kebschull et al., 2013). While useful, molecular profiling interrogates the groups of interest only at a single scale and traditional single classifiers do not provide sufficient insights into potential patient-specific variations within the disease groups of interest in their native or “out-of-the box” form. Understanding such variations is especially critical in developing patient-tailored treatment strategies. As importantly, the literature is replete with publications demonstrating differences in host factors (Eren et al., 2015, Winning et al., 2015, Lutfioglu et al., 2016) and oral microbes (Shchipkova et al., 2010, Bizzarro et al., 2013) in smokers compared to non-smokers. However, it is important to recognize disease variation within the smoking population, and thus critical to delineate the unique biological features of health and disease in the smoking population (Haytural et al., 2015, Eren et al., 2015). The present study, applies an ensemble classification framework (SVA: selective voting ensemble classification approach) (Nagarajan et al., 2015b, Nagarajan and Upreti, 2016) for discerning the biology of periodontitis and non-periodontitis populations, while providing insights into potential variations within the periodontitis population. In a recent study (Nagarajan et al., 2015b), we had demonstrated the usefulness of SVA in understanding potential variations between gingivitis and periodontitis populations using four critical salivary biomarkers (IL-1β, IL-6, MIP-1α, MMP-8) corresponding to fundamental biologic processes driving the disease such as inflammation, tissue destruction, and bone remodeling (Ebersole et al., 2013a, Hajishengallis and Sahingur, 2014, Reynolds, 2014, Silva et al., 2015, Hienz et al., 2015). Unlike traditional single classifiers, SVA is an ensemble approach and varies the feature sets in a sample-specific manner using a majority voting strategy revealing potential sample-specific variations within and between the disease groups of interest. In contrast to traditional disease classification based on molecular expression profiling, the present study discerns periodontitis and periodontitis and non-periodontitis subjects using an integrated set of biomarkers. These informed set of biomarkers interrogate the disease groups at multiple scales and comprise of molecular markers, microbial markers (pathogenic, commensal bacteria), and other markers. Such an integrated approach also has the ability to reveal potential cross-talk (Nagarajan et al., 2016b) between the biomarkers across distinct scales in contrast to traditional classification using molecular expression profiles. Performance of SVA (sensitivity, specificity, accuracy) is also compared to those obtained using five different single classifier approaches (Linear Discriminant Analysis (LDA); Quadratic Discriminant Analysis (QDA); Support Vector Machine (SVM); Naïve Bayes (NB) and Classification and Regression Tree (CART)).
Traditional single classifiers, while helpful, use all the biomarkers simultaneously as features in the classification process resulting in a sparse representation of the samples in a high-dimensional space with dimensions of the classifier often comparable to that of the sample size. In contrast, SVA uses pairs of variables for each classifier and pools the results across these ensemble of classifiers using a voting strategy (e.g. majority voting) (Kuncheva, 2004). Normalized vote-counts and differential proclivity estimates from SVA is shown to vary considerably within the periodontitis samples with a subset of borderline cases with equal proclivity to periodontitis and non-periodontitis. The ensemble sets of these borderline cases also exhibited lack of overlap with the rest of the periodontitis samples. Patient-specific network abstractions of the ensemble sets revealed markedly differently topology between samples with varying proclivities. High-confidence classifiers in these networks comprised primarily of microbial biomarkers revealing their usefulness in capturing patient-specific variations within the periodontitis group.
Methods and Materials
Patient Population and Clinical Parameters
The study population consisted of (N=117) non-periodontitis subjects (58 periodontally healthy subjects; 59 gingivitis patients; 77.2% female) from ages 21–65 and (N=117) periodontitis patients with ages 22–59 years (55.2% female) who were all smokers. The protocol for this study was approved by the University of Kentucky Institutional Review Board and all participants signed an appropriate consent form. A comprehensive oral and periodontal examination was completed to assess the periodontal health. Inclusion/exclusion criteria for participating in the study: must be smokers, able to complete a questionnaire and sign a consent form, have a minimum of 20 teeth, willing to have blood drawn, whole saliva collected, and have a full periodontal evaluation. The clinical evaluation of the periodontium included mean probing pocket depth (PPD), clinical attachment level, and bleeding on probing (BOP) as we have described previously (Novak et al., 2008, Ebersole et al., 2009, Miller et al., 2014, Nagarajan et al., 2015a). Measures of BOP and PPD were used to categorize the patients: mean PPD ≤2.5 mm for non-periodontitis (ie. Health and Gingivitis) and ≥2.5 mm for periodontitis. Variables such as age (Age), pack years of smoking (Yrs), and salivary cotinine (Cot) levels were also included in the analysis.
Serum Analyses
Serum from a venipuncture blood sample was evaluated from 234 subjects (non-periodontitis smokers, NP, N=117; periodontitis smokers, PD, N=117) groups. An analysis determined antibody levels to a group of oral bacteria: Aggregatibacter actinomycetemcomitans (Aa) strain JP2, Porphyromonas gingivalis (Pg) ATCC 33277, Treponema denticola (Td) ATCC 35405, Streptococcus sanguinis (Ss) ATCC 10556, Actinomyces naeslundii (An) ATCC 49340, Veillonella parvula (Vp) ATCC 10790, Capnocytophaga ochracea (Co) ATCC 33596. An ELISA was used to determine the level of IgG antibody to the bacteria (Ebersole et al., 2008). Serum inflammatory markers, included interleukin-1β (IL-1β), plasminogen activator inhibitor-1 (PAI-1), myeloperoxidase (MPO), interleukin (IL)-10, prostaglandin E2 (PGE2) and cotinine were evaluated as described previously (Ebersole et al., 2002, Hayman et al., 2011, Ebersole et al., 2014, Nagarajan et al., 2016a).
Statistical Analyses
The samples (N = 234) from periodontitis (N = 117) and non-periodontitis (N = 117) smoking population were divided into training and test sets. The classifier was trained on the training set (N = 200) comprising of 100 periodontitis and 100 non-periodontitis samples. Subsequently, its performance was tested on an independent validation cohort (N = 34) or test set comprising of 17 periodontitis and 17 non-periodontitis samples. Biomarkers (N = 13) used in the classification comprised of (a) molecular markers (4 markers, PGE2, IL1β, MPO, PAI), (b) periodontal pathogens (3 pathogens, Aa, Pg, Td), (c) oral commensal bacteria (4 bacteria, Ss, An, Vp, Co) and other variables (2 additional variables, Cotinine levels, Years of Smoking). SVA using Support Vector Machines (SVM) as base classifier using pairs of features for each of the base classifiers was used for classification and shall be referred to as SVA-SVM in the following sections. Performance of SVA-SVM was also compared to those obtained using traditional single classifier systems (LDA, QDA, NB, CART, SVM).
Selective-Voting Ensemble Classification Approach (SVA)
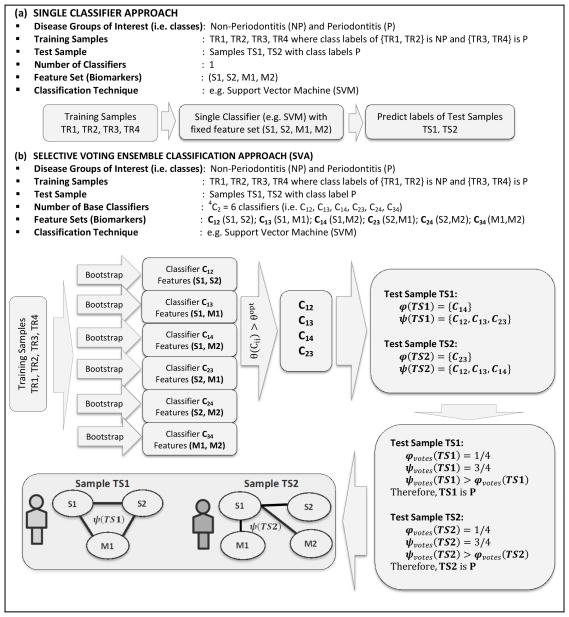
The SVA implementation (Nagarajan and Upreti, 2016) consisted of two main parts working in tandem. The first part determined the optimal sensitivity threshold from the training samples and base classifiers that maximize the overall sensitivity of the ensemble. Subsequently, this optimal sensitivity threshold was used in predicting class membership of an independent validation cohort in the second part. Working principle of SVA and comparison to traditional single classifier systems in discerning the profiles of periodontitis and non-periodontitis groups is elucidated with a simple example in Figure 1 for convenience. A more detailed explanation can be found elsewhere (Nagarajan and Upreti, 2016). The example considers four training samples (TR1, TR2, TR3, TR4). While (TR1, TR2) were chosen to be non-periodontitis samples, (TR3, TR4) were chosen to be periodontitis samples. The two test samples (TS1, TS2) considered were chosen as periodontitis samples. As noted earlier, SVA uses pairs of biomarkers (Figure 1b) as features generating an ensemble of classifiers as opposed to traditional single classifiers that uses all the biomarkers simultaneously in the classification process (Figure 1a). The four biomarkers considered in the example, Figure 1, comprised of salivary markers (S1, S2) and microbial markers (M1, M2), resulting in 4C2 = 6 potential classifiers {C12,C13,C14,C23,C24,C34} with feature sets {S1S2, S1M1, S1M2, S2M1, S2M2, M1M2} respectively. Imposing the optimal sensitivity threshold resulted in a subset of classifiers {C12, C13, C14, C23}. The ensemble sets of the test sample TS1 are given by φ (TS1) = {C14} and ψ (TS1) = {C12, C13, C23} whereas those of TS2 are given by φ (TS2) = {C23} and ψ (TS2) = {C12, C13, C14}. It is important to note that for each of the test samples 3 out of 4 classifiers voted them as periodontitis whereas 1 out of 4 voted them as non-periodontitis. Therefore, their normalized vote counts based on majority voting is identical and given by , indicating equal proclivity of these test samples to the periodontitis as opposed to the non-periodontitis group with (i. e. Δvotes=ψvotes − φvotes = 1/2 > 0).. While the results based on normalized vote-counts from SVA correctly revealed equal proclivity of the test samples TS1 and TS2 to periodontitis, network abstractions of their ensemble sets ψ (TS1)and ψ (TS2) were markedly different, Figure 1. For instance, since ψ (TS1) = {C12, C13, C23}, the corresponding features of the base classifiers are (M1M2, M1M3, M2M3) resulting in a network with three nodes (M1, M2, M3) and three edges corresponding (M1-M2, M1-M3, M2-M3). More importantly, these patient-specific networks not only reveal potential variations within the periodontitis group but also cross-talk between salivary and microbial biomarkers in this example.
Figure 1.
Working principle of traditional single classifier system C using salivary (S1, S2) and microbial (M1, M2) biomarkers simultaneously as features in the classification process is shown in (a) whereas those of SVA using pairs of biomarkers as features and an ensemble of classifiers {C12,C13,C14,C23,C24,C34} is shown in (b). Imposing the optimal sensitivity threshold in SVA returns a subset of classifiers {C12,C13,C14,C23}. The training samples (TR1,TR2,TR3,TR4) were used to learn the classifiers and used subsequently to predict the labels of the test samples (TS1,TS2). The normalized vote-counts of the periodontitis test samples TS1 and TS2 using majority voting is identical , indicating equal proclivity of TS1 and TS2 to periodontitis group. However, network abstractions of their ensemble sets exhibit markedly different topologies revealing patient-specific variations within the periodontitis subjects and cross-talk between the salivary and microbial biomarkers.
Results
Optimal sensitivity threshold of SVA-SVM from the training sample
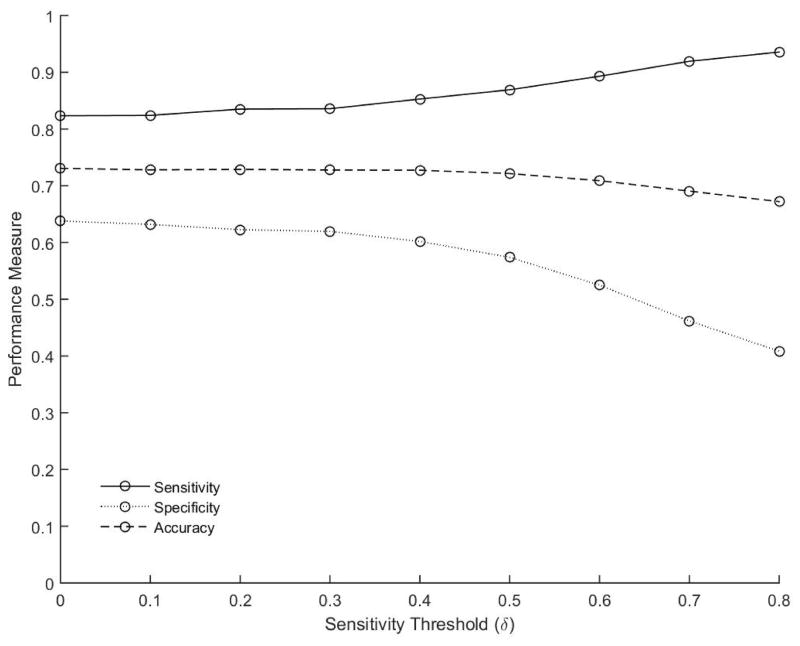
The optimal sensitivity threshold estimated on the training samples (N=200) using SVA-SVM is shown in Figure 2. Of interest is to note that the sensitivity of the ensemble (Figure 2, solid line) exhibits an increasing trend with the sensitivity threshold of the base classifiers δ. The increasing trend of the ensemble sensitivity is also accompanied by a decreasing trend in the ensemble specificity (Figure 2, dotted line) with the ensemble accuracy remaining more or less constant around 0.7 (Figure 2, dashed line). The increase in the overall sensitivity of the ensemble is especially pronounced when δ ≥ 0.5 with δ = 0.5 representing the case where only base classifiers that classify at least 50% of the periodontitis samples correctly are members of the ensemble. The sensitivity of the ensemble peaked around δ = 0.8. Any further increase in δ resulted in very few base classifiers preventing reliable estimation of the ensemble performance. Since the primary objective of the present study and biomarker development in general is increased sensitivity, the optimal sensitivity threshold for selection of the base classifiers was set at δ = 0.8 for SVA-SVM.
Figure 2.
Performance measures sensitivity (solid line), specificity (dotted line) and accuracy (dashed line) estimated on the training sample as a function of the sensitivity threshold δ of the base classifiers using SVA-SVM is shown. Each circle represents the average estimate over (N = 100) independent realizations.
Performance of SVA-SVM on an independent validation cohort (test sample)
Performance of traditional single classifiers (LDA, QDA, NB, CART, SVM) and SVA-SVM with δ = 0.5 and δ = 0.8 on the independent validation cohort comprising of (N = 34 samples; 17 Periodontitis, 17 Non-Periodontitis) is shown in Table 1. Mean performance measures (sensitivity, specificity, accuracy) along with their standard deviation estimated from (N = 100) realizations for each of the classification techniques is shown in Table 1. Among the single classifiers SVM had the highest average sensitivity (~60%) whereas Naïve Bayes had the highest average specificity (~70%). The average accuracies of the single classifiers were comparable with no apparent differences. However, SVA-SVM had markedly higher average sensitivity ~70% for δ = 0.5 and ~85% for δ = 0.8 higher than all the single classifiers. The average accuracy of SVA-SVM was also comparable to that of the single classifiers.
Table 1.
Performance of traditional single classifiers and SVA-SVM on an independent validation cohort
| Classification Technique | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Traditional Single Classifiers | |||
| LDA | 0.50 ± 0.13 | 0.67 ± 0.13 | 0.58 ± 0.08 |
| QDA | 0.51 ± 0.10 | 0.56 ± 0.11 | 0.54 ± 0.07 |
| Naïve Bayes | 0.32 ± 0.06 | 0.70 ± 0.09 | 0.51 ± 0.05 |
| CART | 0.59 ± 0.18 | 0.53 ± 0.17 | 0.56 ± 0.10 |
| SVM | 0.60 ± 0.15 | 0.56 ± 0.15 | 0.58 ± 0.09 |
| Selective Voting Ensemble Classification Approach | |||
| SVA-SVM with δ = 0.5 | 0.70 ± 0.07 | 0.45 ± 0.03 | 0.58 ± 0.04 |
| SVA-SVM with δ = 0.8 | 0.85 ± 0.08 | 0.27 ± 0.09 | 0.56 ± 0.05 |
Patient-specific variations in the periodontitis test samples
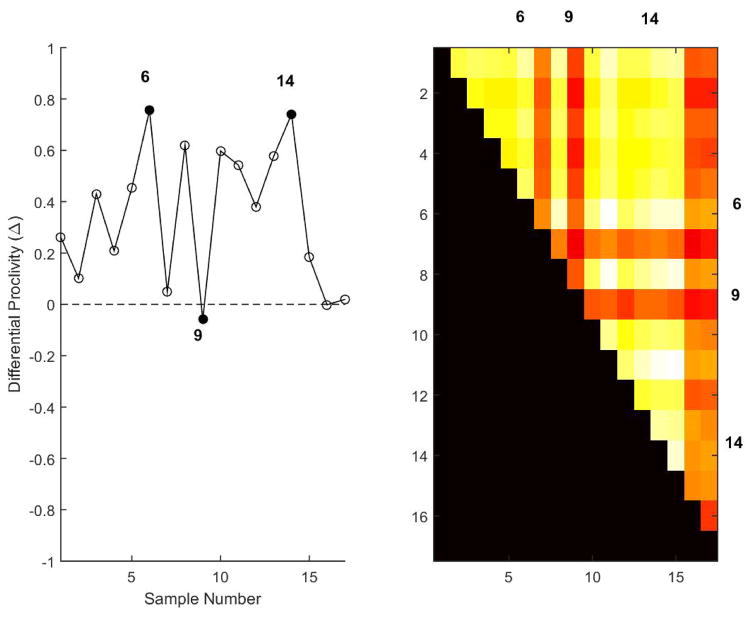
As noted earlier, unlike traditional single classifiers SVA-SVM can reveal patient-specific variations within disease groups while discerning the disease groups by adapting the feature sets in a patient-specific manner. Such variations are reflected in the normalized vote-counts, hence the differential proclivity estimates of the samples. Differential proclivity estimates, Figure 3 revealed considerable variation within the periodontitis test samples (N = 17) with a subset of borderline samples exhibiting equal proclivity (Δ ~ 0) to periodontitis and non-periodontitis groups. Three representative samples (6, 14, 9) with markedly proclivities were identified from Figure 3. Samples 6 and 14 had similar proclivities (Δ ~ 0.75) markedly different from that of the borderline sample 9(Δ ~ 0). Ensemble sets of the borderline samples also failed to exhibit considerable overlap with the rest of the periodontitis test samples and accompanied by characteristic dark streaks in the heatmap representation of the consensus map (Nagarajan and Upreti, 2016), Figure 3.
Figure 3.
Average differential proclivity estimates of the periodontitis test samples (N = 17) is shown on the left averaged over (N = 100) independent realizations. Three representative samples (6, 14, 9) with varying differential proclivities are shown in dark circles (left). Dashed line represents samples with equal proclivity to periodontitis and non-periodontitis groups with dark and white arrows representing increasing proclivity to periodontitis and non-periodontitis groups. Samples 6 and 14 have similar differential proclivities (Δ ~ 0.75) in contrast to that of the borderline sample 9 (Δ ~ 0).
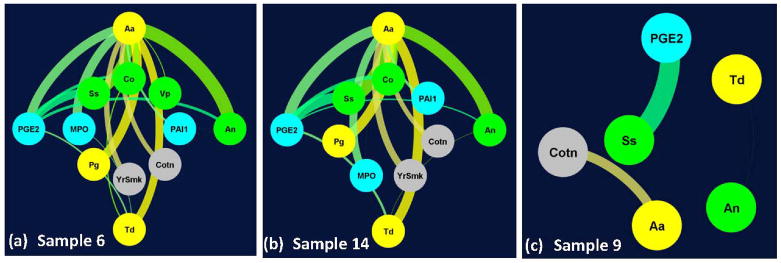
Patient-specific network (PSN) abstraction of the ensemble sets were subsequently generated with the nodes representing the features or biomarkers and edges representing the classifiers in the ensemble sets. The thickness of the edges correspond to the classifier confidence across (N = 100) independent realizations. PSN of the three representative periodontitis test samples (6, 14, 9) is shown in Figure 4. Unlike differential proclivity, Figure 3, PSN revealed critical nodes, their interactions and variation in the overall topology with considerable similarity between (6, 14) in contrast to that of sample 9. From Figure 4, 11 out of the 13 variables were retained in the PSNs of samples 6 and 14 whereas only 6 were retained in the PSN of sample 9. Unlike PSNs of samples 6 and 14, those of sample 9 were fragmented. High-confidence edges across samples 6 and 14 primarily comprised of periodontal pathogens (Aa, Pg, Td) and their concerted working with oral commensal bacteria (An, Ss) and molecular marker (PGE2), Figures 4a–4b. High-confidence edges representing base classifiers with >40% confidence common to samples (6, 14) were composed of pathogenic bacteria (Aa-Pg; Aa-Td), commensal bacteria (Aa-Ss; Aa-Co; An-Co) and their interaction (Aa-An; Td-Co; Td-An). The other prominent edges common to samples 6 and 14 also consisted of interaction between the microbiome and molecular entities with PGE2 playing a critical role (PGE2-Aa; PGE2-Td; PGE2-Ss; PGE2-An; PGE2-Co). In contrast, many of these high-confidence edges were absent in the PSN corresponding to sample 9, Figure 4c. However, a subset of the high-confidence classifiers (PGE2-Ss; Aa-Cotn) was present consistently across these three representative samples. While the edge Td-An was also present in sample 9, its confidence barely passed the cut-off (40%). Ranking the classifier confidence across the 17 periodontitis test samples also revealed that biomarkers (Aa-An and Aa-Ss) to be highly ranked classifiers (top two) across a majority of these samples. These results essentially elucidate the critical role of pathogenic and commensal bacteria and their concerted effort with other biomarkers in discerning periodontitis and non-periodontitis smoking subjects while revealing patient-specific variations within the periodontitis group. Interaction between the microbiome and molecular markers may also provide novel insights into variations in the underlying molecular mechanisms due to microbial challenge across these subjects.
Figure 4.
Patient-specific networks for representative periodontitis test samples (6, 14) shown in (a, b) had markedly different topology from that of sample 9 (c). For clarity, variables corresponding to (i) pathogenic bacteria Aggregatibacter actinomycetemcomitans (Aa), Porphyromonas gingivalis (Pg), Treponema denticola (Td), (ii) commensal bacteria Streptococcus sanguinis (Ss), Actinomyces naeslundii (An), Veillonella parvula (Vp), Capnocytophaga ochracea (Co), (iii) molecular markers interleukin 1β (IL1b), prostaglandin E2 (PGE2), plasminogen activator inhibitor-1 (PAI-1), myeloperoxidase (MPO) and (iv) other markers corresponding to years of smoking (Yrs), salivary cotinine level (Cot) and age (Age) are shown in yellow, green, cyan and gray colors in (a, b, c) respectively. Each edge corresponds to a classifier with its thickness proportional to the classifier confidence in each of the subplots. For clarity, only edges corresponding to classifiers whose confidence > 40% is shown in (a, b, c). In (a) and (b) the top four highly connected nodes (Aa, PGE2, An, Td) are shown in the outer circle.
Discussion
Since periodontitis represents a persistent inflammatory response to chronic biofilms inhabiting the subgingival crevice (Hajishengallis, 2014, Nibali et al., 2014) the current disease paradigm engages a model of variations in the quantity and quality of the oral microbiome with disease. These microbial changes generate a dysregulated inflammatory response or are in response to this dysregulated response, resulting in a microbial dysbiosis that exacerbates the tissue destructive processes (Hajishengallis et al., 2012, Lamont and Hajishengallis, 2015). Thus, measures of this disruption of biologic homeostasis can be detected in the oral cavity and systemically, and biomolecules representing the various stages of progression of the infection and destructive inflammatory response evaluated (Ebersole et al., 2013b, Lappin et al., 2013, Salazar et al., 2013, Salminen et al., 2014, Gumus et al., 2014, Saraiva et al., 2014, Longo et al., 2014, Ebersole et al., 2015, Torumtay et al., 2015). Also to be noted in examining host responses in periodontitis is the clear role of adaptive immunity, and particularly humoral immune responses in the development of, or protection from, periodontitis (Ebersole et al., 2001, Ebersole et al., 2013a). It is well documented that the host responds to oral bacteria with generally elevated levels of antibody to putative oral pathogens in both serum and gingival crevicular fluid in disease; however, some reports have suggested that severe generalized disease may show some decreased antibody levels (Califano et al., 1999, Wang et al., 2005, Takeuchi et al., 2006, Hwang et al., 2014). Treatment of periodontal disease is typically associated with early increases, but longer-term decreases with successful therapy in antibody specific for oral bacteria (Ebersole et al., 1985, Mooney et al., 1995, Beikler et al., 1999, Sakai et al., 2001, Yamazaki et al., 2004). While many of these biomolecules have been detected in saliva and correlate with periodontal disease, a wide array of them have also been detected in serum associated with chronic inflammation related to the oral disease (Fain, 2006, Zakynthinos and Pappa, 2009, Cierny et al., 2014, Wu et al., 2010). Nevertheless, a clinical history needs to be a portion of the patient characterization to minimize false-positive responses in periodontally healthy subjects.
Consequently, the use of a panel of potential biomarkers, whether in serum, gingival crevicular fluid, or saliva could help dentistry move towards the era of precision medicine (Flores et al., 2013, Schmidt, 2014, Cesario et al., 2014). This concept is consistent with the programmatic emphasis of the National Institute for Dental and Craniofacial Research described as “Ongoing analyses of … information in many fields of biomedicine are uncovering new approaches for diagnosing and managing disease based on molecular signatures, rather than relying mainly on symptoms and clinical assessment”. One of the objectives of this initiative is to discern distinct disease groups from an integrated panel of biomarkers across multiple scales using sophisticated biomedical informatics approaches while providing insights into patient-specific variations, as well as potential interactions between these biomarkers. Classical approaches using traditional single classifier systems have been useful but often investigate differences between the disease groups at a single scale (e.g. molecular expression profiling). Single classifier systems also use all the biomarkers simultaneously in classification process and do not provide insights into potential cross-talk between these biomarkers. The dimensionality of feature space of single classifier systems using all features is often comparable to the sample size affecting their overall performance and generalizability. Ensemble classification approaches such as SVA used in the present study overcomes these limitations by projecting the samples in a two-dimensional space. More importantly, SVA is shown to reveal potential cross-talk and interactions between the biomarkers across multiple scales as well as variations within the disease groups. These interactions are modeled as weighted undirected graphs from the ensemble sets returned by SVA. Also of interest was targeting a group of smokers, since there is conclusive evidence for the negative impact of this environmental challenge on the expression, severity, altered therapeutic response, and response to regenerative procedures (Reynolds, 2014, Michalowicz et al., 2014, Johannsen et al., 2014, Eke et al., 2016). Of critical consequence in developing these biologic models is to discern how they can be utilized to improve professional clinical decisions and patient care. We believe that this transformation will need to occur in at least three steps. First, to delineate the appropriate panel of biomarkers in specific diagnostic fluids; second, develop improved methods for more rapid assessment of actionable biomarker profiles; and, finally, to implement these 21st century approaches towards modifying the current clinical and insurance based decision paradigm for a procedure-based doctrine of treating existing disease versus a preemptive, preventive approach to this oral infection and disease.
The results presented demonstrate the sensitivity of SVA to be considerably higher than that of traditional single classifier systems. Proclivity estimates from SVA also revealed considerable variation within the periodontitis group. Ensemble sets of three representative periodontitis test samples and their proclivities and patient-specific networks were investigated. The patient-specific network abstraction of the borderline sample with equal proclivity to non-periodontitis and periodontitis was markedly different from the other two samples. Based on these results, the responses to the oral bacteria, reflecting the oral colonization/infection challenge to the host of both pathogens and commensal bacteria were highly informative in creating the profiles delineating the periodontitis patients. Thus, this adaptive immune response, which appears to be underappreciated within the context of the recent emphasis on innate immune and inflammatory response cells and molecules, needs to be re-examined in more detail as a fundamental interactive component that differentiates destructive periodontitis from health. These types of findings are generally consistent with existing microbiome data (Shchipkova et al., 2010, Bizzarro et al., 2013, Camelo-Castillo et al., 2015) and individual analyte host response data (Barbour et al., 1997, Apatzidou et al., 2005, Al-Ghamdi and Anil, 2007, Hayman et al., 2011, Lutfioglu et al., 2016) supporting differences in colonization and local and systemic reactivity of smokers compared to nonsmokers. As importantly, the subset of periodontitis patients that were determined to present distinct profiles, including non-standard and unlinked responses, will be of particular interest relative to the details of their disease and treatment response experiences. This concept also incorporates an important limitation of this cross-sectional study design. Some of the greatest value of modeling disease initiation and progression would be for the biomarker complex to be capable of predicting the disease process in an individual patients. Thus, this innovative approach to evaluating the biomarker profiles will require a prospective longitudinal study design to address this important gap in the field.
Clinical Relevance.
Scientific rationale for the study: Various studies have suggested a relationship between
Scientific rationale for the study
Varied inflammatory and immune responses to the oral microbiomes lead to considerable heterogeneity in smokers with periodontitis. Understanding patient-specific variation using an array of biomarkers can assist in developing patient-tailored interventions.
Principal findings
The results identified a subset of smokers with periodontitis with markedly different host response profiles. Patient-specific network abstractions also revealed the interplay between the various biomarkers across multiple scales including oral microbiome and their critical role in discerning smokers with periodontitis and non-periodontitis.
Practical implications
Biomarkers of the oral microbiome can be useful in discerning biological underpinnings of periodontitis and non-periodontitis in smokers, as well as revealing potential variations within this periodontitis population
Acknowledgments
This work was supported by USPHS grants P20RR020145, P20GM103538, and UL1TR000117 from the National Institutes of Health. We thank M.J. Steffen and J. Stevens for expert technical assistance in the evaluation of the biomarkers in the serum samples. The authors have stated explicitly that there are no conflicts of interest in connection with this article
References
- Al-Ghamdi HS, Anil S. Serum antibody levels in smoker and non-smoker saudi subjects with chronic periodontitis. J Periodontol. 2007;78:1043–1050. doi: 10.1902/jop.2007.060431. [DOI] [PubMed] [Google Scholar]
- Apatzidou DA, Riggio MP, Kinane DF. Impact of smoking on the clinical, microbiological and immunological parameters of adult patients with periodontitis. J Clin Periodontol. 2005;32:973–983. doi: 10.1111/j.1600-051X.2005.00788.x. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Learned and unlearned concepts in periodontal diagnostics: a 50-year perspective. Periodontol 2000. 2013;62:20–36. doi: 10.1111/prd.12006. [DOI] [PubMed] [Google Scholar]
- Baelum V, Lopez R. Periodontal disease epidemiology - learned and unlearned? Periodontol 2000. 2013;62:37–58. doi: 10.1111/j.1600-0757.2012.00449.x. [DOI] [PubMed] [Google Scholar]
- Barbour SE, Nakashima K, Zhang JB, Tangada S, Hahn CL, Schenkein HA, Tew JG. Tobacco and smoking: environmental factors that modify the host response (immune system) and have an impact on periodontal health. Crit Rev Oral Biol Med. 1997;8:437–460. doi: 10.1177/10454411970080040501. [DOI] [PubMed] [Google Scholar]
- Beikler T, Karch H, Ehmke B, Klaiber B, Flemmig TF. Protective effect of serum antibodies against a 110-kilodalton protein of Actinobacillus actinomycetemcomitans following periodontal therapy. Oral Microbiol Immunol. 1999;14:281–287. doi: 10.1034/j.1399-302x.1999.140503.x. [DOI] [PubMed] [Google Scholar]
- Bizzarro S, Loos BG, Laine ML, Crielaard W, Zaura E. Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. J Clin Periodontol. 2013;40:483–492. doi: 10.1111/jcpe.12087. [DOI] [PubMed] [Google Scholar]
- Califano JV, Schifferle RE, Gunsolley JC, Best AM, Schenkein HA, Tew JG. Antibody reactive with Porphyromonas gingivalis serotypes K1–6 in adult and generalized early-onset periodontitis. J Periodontol. 1999;70:730–735. doi: 10.1902/jop.1999.70.7.730. [DOI] [PubMed] [Google Scholar]
- Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomas I. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. 2015;6:119. doi: 10.3389/fmicb.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario A, Auffray C, Russo P, Hood L. P4 medicine needs P4 education. Curr Pharm Des. 2014;20:6071–6072. doi: 10.2174/1381612820666140314145445. [DOI] [PubMed] [Google Scholar]
- Cierny JT, Unal ER, Flood P, Rhee KY, Praktish A, Olson TH, Goetzl L. Maternal inflammatory markers and term labor performance. Am J Obstet Gynecol. 2014;210:447 e441–446. doi: 10.1016/j.ajog.2013.11.038. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Cappelli D, Holt SC. Periodontal diseases: to protect or not to protect is the question? Acta Odontol Scand. 2001;59:161–166. doi: 10.1080/000163501750266756. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Cappelli D, Mathys EC, Steffen MJ, Singer RE, Montgomery M, Mott GE, Novak MJ. Periodontitis in humans and non-human primates: oral-systemic linkage inducing acute phase proteins. Ann Periodontol. 2002;7:102–111. doi: 10.1902/annals.2002.7.1.102. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Dawson DR, 3rd, Morford LA, Peyyala R, Miller CS, Gonzalez OA. Periodontal disease immunology: 'double indemnity' in protecting the host. Periodontol 2000. 2013a;62:163–202. doi: 10.1111/prd.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Holt SC, Hansard R, Novak MJ. Microbiologic and immunologic characteristics of periodontal disease in Hispanic americans with type 2 diabetes. J Periodontol. 2008;79:637–646. doi: 10.1902/jop.2008.070455. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Nagarajan R, Akers D, Miller CS. Targeted salivary biomarkers for discrimination of periodontal health and disease(s) Front Cell Infect Microbiol. 2015;5:62. doi: 10.3389/fcimb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Novak MJ, Michalowicz BS, Hodges JS, Steffen MJ, Ferguson JE, Diangelis A, Buchanan W, Mitchell DA, Papapanou PN. Systemic immune responses in pregnancy and periodontitis: relationship to pregnancy outcomes in the Obstetrics and Periodontal Therapy (OPT) study. J Periodontol. 2009;80:953–960. doi: 10.1902/jop.2009.080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Schuster JL, Stevens J, Dawson D, 3rd, Kryscio RJ, Lin Y, Thomas MV, Miller CS. Patterns of salivary analytes provide diagnostic capacity for distinguishing chronic adult periodontitis from health. J Clin Immunol. 2013b;33:271–279. doi: 10.1007/s10875-012-9771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Steffen MJ, Thomas MV, Al-Sabbagh M. Smoking-related cotinine levels and host responses in chronic periodontitis. J Periodontal Res. 2014;49:642–651. doi: 10.1111/jre.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Taubman MA, Smith DJ, Haffajee AD. Effect of subgingival scaling on systemic antibody responses to oral microorganisms. Infect Immun. 1985;48:534–539. doi: 10.1128/iai.48.2.534-539.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, Genco RJ. Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 – 2012. J Periodontol. 2015:1–18. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ Cdc Periodontal Disease Surveillance workgroup: James Beck GDRP. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- Eke PI, Wei L, Thornton-Evans GO, Borrell LN, Borgnakke WS, Dye B, Genco RJ. Risk Indicators for Periodontitis in US Adults: National Health and Nutrition Examination Survey (NHANES) 2009 – 2012. J Periodontol. 2016:1–18. doi: 10.1902/jop.2016.160013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren G, Turkoglu HO, Atmaca H, Atilla FG. Evaluation of GCF MMP-1, MMP-8, TGF-beta1, PDGF-AB, and VEGF levels in periodontally healthy smokers. Turk J Med Sci. 2015;45:850–856. [PubMed] [Google Scholar]
- Fain JN. Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam Horm. 2006;74:443–477. doi: 10.1016/S0083-6729(06)74018-3. [DOI] [PubMed] [Google Scholar]
- Flores M, Glusman G, Brogaard K, Price ND, Hood L. P4 medicine: how systems medicine will transform the healthcare sector and society. Per Med. 2013;10:565–576. doi: 10.2217/PME.13.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumus P, Nizam N, Nalbantsoy A, Ozcaka O, Buduneli N. Saliva and serum levels of pentraxin-3 and interleukin-1beta in generalized aggressive or chronic periodontitis. J Periodontol. 2014;85:e40–46. doi: 10.1902/jop.2013.130281. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Sahingur SE. Novel inflammatory pathways in periodontitis. Adv Dent Res. 2014;26:23–29. doi: 10.1177/0022034514526240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman L, Steffen MJ, Stevens J, Badger E, Tempro P, Fuller B, McGuire A, Al-Sabbagh M, Thomas MV, Ebersole JL. Smoking and periodontal disease: discrimination of antibody responses to pathogenic and commensal oral bacteria. Clin Exp Immunol. 2011;164:118–126. doi: 10.1111/j.1365-2249.2010.04314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haytural O, Yaman D, Ural EC, Kantarci A, Demirel K. Impact of periodontitis on chemokines in smokers. Clinical oral investigations. 2015;19:979–986. doi: 10.1007/s00784-014-1314-2. [DOI] [PubMed] [Google Scholar]
- Hienz SA, Paliwal S, Ivanovski S. Mechanisms of Bone Resorption in Periodontitis. J Immunol Res. 2015;2015:615486. doi: 10.1155/2015/615486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang AM, Stoupel J, Celenti R, Demmer RT, Papapanou PN. Serum antibody responses to periodontal microbiota in chronic and aggressive periodontitis: a postulate revisited. J Periodontol. 2014;85:592–600. doi: 10.1902/jop.2013.130172. [DOI] [PubMed] [Google Scholar]
- Jin LJ, Armitage GC, Klinge B, Lang NP, Tonetti M, Williams RC. Global oral health inequalities: task group--periodontal disease. Adv Dent Res. 2011;23:221–226. doi: 10.1177/0022034511402080. [DOI] [PubMed] [Google Scholar]
- Johannsen A, Susin C, Gustafsson A. Smoking and inflammation: evidence for a synergistic role in chronic disease. Periodontol 2000. 2014;64:111–126. doi: 10.1111/j.1600-0757.2012.00456.x. [DOI] [PubMed] [Google Scholar]
- Kebschull M, Guarnieri P, Demmer RT, Boulesteix AL, Pavlidis P, Papapanou PN. Molecular differences between chronic and aggressive periodontitis. J Dent Res. 2013;92:1081–1088. doi: 10.1177/0022034513506011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull M, Papapanou PN. The use of gene arrays in deciphering the pathobiology of periodontal diseases. Methods Mol Biol. 2010;666:385–393. doi: 10.1007/978-1-60761-820-1_24. [DOI] [PubMed] [Google Scholar]
- Kumar PS. From focal sepsis to periodontal medicine: A century of exploring the role of the oral microbiome in systemic disease. J Physiol. 2016 doi: 10.1113/JP272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuncheva LI. Combining pattern classifiers: methods and algorithms. John Wiley & Sons; 2004. [Google Scholar]
- Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin DF, Apatzidou D, Quirke AM, Oliver-Bell J, Butcher JP, Kinane DF, Riggio MP, Venables P, McInnes IB, Culshaw S. Influence of periodontal disease, Porphyromonas gingivalis and cigarette smoking on systemic anti-citrullinated peptide antibody titres. J Clin Periodontol. 2013;40:907–915. doi: 10.1111/jcpe.12138. [DOI] [PubMed] [Google Scholar]
- Linden GJ, Lyons A, Scannapieco FA. Periodontal systemic associations: review of the evidence. J Periodontol. 2013;84:S8–S19. doi: 10.1902/jop.2013.1340010. [DOI] [PubMed] [Google Scholar]
- Longo PL, Artese HP, Rabelo MS, Kawamoto D, Foz AM, Romito GA, Dib SA, Mayer MP. Serum levels of inflammatory markers in type 2 diabetes patients with chronic periodontitis. J Appl Oral Sci. 2014;22:103–108. doi: 10.1590/1678-775720130540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfioglu M, Aydogdu A, Sakallioglu EE, Alacam H, Pamuk F. Gingival crevicular fluid interleukin-8 and lipoxin A4 levels of smokers and nonsmokers with different periodontal status: a cross-sectional study. J Periodontal Res. 2016;51:471–480. doi: 10.1111/jre.12324. [DOI] [PubMed] [Google Scholar]
- Matthews DC. Prevention and treatment of periodontal diseases in primary care. Evid Based Dent. 2014;15:68–69. doi: 10.1038/sj.ebd.6401036. [DOI] [PubMed] [Google Scholar]
- Michalowicz BS, Hyman L, Hou W, Oates TW, Jr, Reddy M, Paquette DW, Katancik JA, Engebretson SP Diabetes & Periodontal Therapy Trial Study T. Factors associated with the clinical response to nonsurgical periodontal therapy in people with type 2 diabetes mellitus. J Am Dent Assoc. 2014;145:1227–1239. doi: 10.14219/jada.2014.92. [DOI] [PubMed] [Google Scholar]
- Miller CS, Foley JD, 3rd, Floriano PN, Christodoulides N, Ebersole JL, Campbell CL, Bailey AL, Rose BG, Kinane DF, Novak MJ, McDevitt JT, Ding X, Kryscio RJ. Utility of salivary biomarkers for demonstrating acute myocardial infarction. J Dent Res. 2014;93:72S–79S. doi: 10.1177/0022034514537522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney J, Adonogianaki E, Riggio MP, Takahashi K, Haerian A, Kinane DF. Initial serum antibody titer to Porphyromonas gingivalis influences development of antibody avidity and success of therapy for chronic periodontitis. Infect Immun. 1995;63:3411–3416. doi: 10.1128/iai.63.9.3411-3416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Miller CS, Dawson D, 3rd, Al-Sabbagh M, Ebersole JL. Patient-Specific Variations in Biomarkers across Gingivitis and Periodontitis. PLoS One. 2015a;10:e0136792. doi: 10.1371/journal.pone.0136792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Miller CS, Dawson D, 3rd, Al-Sabbagh M, Ebersole JL. Crosstalk between clinical and host-response parameters of periodontitis in smokers. J Periodontal Res. 2016a doi: 10.1111/jre.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Miller CS, Dawson D, Al-Sabbagh M, Ebersole JL. Cross-talk between clinical and host-response parameters of periodontitis in smokers. J Periodontal Res. 2016b:n/a–n/a. doi: 10.1111/jre.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Miller CS, Dawson D, III, Al-Sabbagh M, Ebersole JL. Patient-specific variations in biomarkers across gingivitis and periodontitis. PloS one. 2015b;10:e0136792. doi: 10.1371/journal.pone.0136792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Upreti M. An Approach for Deciphering Patient-Specific Variations with application to Breast Cancer Molecular Expression Profiles. J Biomed Inform. 2016 doi: 10.1016/j.jbi.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Nibali L, Henderson B, Sadiq ST, Donos N. Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases. J Oral Microbiol. 2014:6. doi: 10.3402/jom.v6.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak MJ, Potter RM, Blodgett J, Ebersole JL. Periodontal Disease in Hispanic Americans With Type 2 Diabetes. J Periodontol. 2008;79:629–636. doi: 10.1902/jop.2008.070442. [DOI] [PubMed] [Google Scholar]
- Reynolds MA. Modifiable risk factors in periodontitis: at the intersection of aging and disease. Periodontol 2000. 2014;64:7–19. doi: 10.1111/prd.12047. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Shimauchi H, Ito HO, Kitamura M, Okada H. Porphyromonas gingivalis-specific IgG subclass antibody levels as immunological risk indicators of periodontal bone loss. J Clin Periodontol. 2001;28:853–859. doi: 10.1034/j.1600-051x.2001.028009853.x. cpe280907 [pii] [DOI] [PubMed] [Google Scholar]
- Salazar MG, Jehmlich N, Murr A, Dhople VM, Holtfreter B, Hammer E, Volker U, Kocher T. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J Clin Periodontol. 2013;40:825–832. doi: 10.1111/jcpe.12130. [DOI] [PubMed] [Google Scholar]
- Salminen A, Gursoy UK, Paju S, Hyvarinen K, Mantyla P, Buhlin K, Kononen E, Nieminen MS, Sorsa T, Sinisalo J, Pussinen PJ. Salivary biomarkers of bacterial burden, inflammatory response, and tissue destruction in periodontitis. J Clin Periodontol. 2014;41:442–450. doi: 10.1111/jcpe.12234. [DOI] [PubMed] [Google Scholar]
- Saraiva L, Rebeis ES, de Martins ES, Sekiguchi RT, Ando-Suguimoto ES, Mafra CE, Holzhausen M, Romito GA, Mayer MP. IgG sera levels against a subset of periodontopathogens and severity of disease in aggressive periodontitis patients: a cross-sectional study of selected pocket sites. J Clin Periodontol. 2014;41:943–951. doi: 10.1111/jcpe.12296. [DOI] [PubMed] [Google Scholar]
- Schmidt C. Leroy Hood looks forward to P4 medicine: predictive, personalized, preventive, and participatory. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju416. [DOI] [PubMed] [Google Scholar]
- Shchipkova AY, Nagaraja HN, Kumar PS. Subgingival microbial profiles of smokers with periodontitis. J Dent Res. 2010;89:1247–1253. doi: 10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N, Abusleme L, Bravo D, Dutzan N, Garcia-Sesnich J, Vernal R, Hernandez M, Gamonal J. Host response mechanisms in periodontal diseases. J Appl Oral Sci. 2015;23:329–355. doi: 10.1590/1678-775720140259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. Periodontology: past, present, perspectives. Periodontol 2000. 2013;62:7–19. doi: 10.1111/prd.12011. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Aramaki M, Nagasawa T, Umeda M, Oda S, Ishikawa I. Immunoglobulin G subclass antibody profiles in Porphyromonas gingivalis-associated aggressive and chronic periodontitis patients. Oral Microbiol Immunol. 2006;21:314–318. doi: 10.1111/j.1399-302X.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- Torumtay G, Kirzioglu FY, Ozturk Tonguc M, Kale B, Calapoglu M, Orhan H. Effects of periodontal treatment on inflammation and oxidative stress markers in patients with metabolic syndrome. J Periodontal Res. 2015 doi: 10.1111/jre.12328. [DOI] [PubMed] [Google Scholar]
- Wang D, Kawashima Y, Nagasawa T, Takeuchi Y, Kojima T, Umeda M, Oda S, Ishikawa I. Elevated serum IgG titer and avidity to Actinobacillus actinomycetemcomitans serotype c in Japanese periodontitis patients. Oral Microbiol Immunol. 2005;20:172–179. doi: 10.1111/j.1399-302X.2005.00208.x. [DOI] [PubMed] [Google Scholar]
- Winning L, Patterson CC, Cullen KM, Stevenson KA, Lundy FT, Kee F, Linden GJ. The association between subgingival periodontal pathogens and systemic inflammation. J Clin Periodontol. 2015;42:799–806. doi: 10.1111/jcpe.12450. [DOI] [PubMed] [Google Scholar]
- Wu T, Sajitharan D, Mohan C. Biomarkers of rheumatoid arthritis: recent progress. Expert Opin Med Diagn. 2010;4:293–305. doi: 10.1517/17530059.2010.492828. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Ueki-Maruayama K, Honda T, Nakajima T, Seymour GJ. Effect of periodontal treatment on the serum antibody levels to heat shock proteins. Clin Exp Immunol. 2004;135:478–482. doi: 10.1111/j.1365-2249.2003.02375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakynthinos E, Pappa N. Inflammatory biomarkers in coronary artery disease. Journal of cardiology. 2009;53:317–333. doi: 10.1016/j.jjcc.2008.12.007. [DOI] [PubMed] [Google Scholar]