Graphical abstract
Ryanodine receptors (RyR) and inositol 1,4,5-trisphosphate receptors (InsP3R) are the predominant intracellular Ca2+ release channels in cells, localized predominately in the sarco- and endoplasmic reticulum (ER), the major intracellular Ca2+ reservoirs, and regulating diverse cellular processes. The two channel families are each comprised of three distinct genes with wide-spread expression throughout the body. RyR1 plays an important role in skeletal muscle where, through an interaction with voltage-gated Ca2+ channels (VGCC) in the transverse tubule membrane, it mediates excitation-contraction coupling. RyR2 is expressed in the heart, where it is activated during each heartbeat by Ca2+ influx through plasma membrane VGCC by the process of Ca2+-induced Ca2+ release. RyR3 is expressed in muscle in some species, as well as in the brain. The type 1 InsP3R (InsP3R-1) is highly expressed in cerebellar Purkinje neurons, although it and the other two InsP3R isoforms have widespread expression patterns. All three InsP3Rs are ligand gated ion channels, activated by binding to InsP3 generated by engagement of plasma membrane receptors coupled to the activation of phospholipases C [1]. These families of ion channels are among the largest known. As homo-tetramers, RyRs have a molecular mass ~ 2.3 MDa. InsP3R homo- and hetero-tetramers are smaller, with a molecular mass ~1.2 MDa.
Amino acid sequence analyses and secondary structural predictions indicated that the RyR and InsP3R families are evolutionary homologs. The channels are tetramers; most of their molecular mass resides in the cytoplasm with only limited exposure to the ER lumen; and they exhibit similar domain organizations in their amino termini, the region of InsP3 binding in the InsP3Rs, and in a structurally conserved core of six transmembrane segments in each molecule with striking sequence homology in the ion conduction pore region, near the carboxyl termini in both families. This homology is reflected in their similar permeation properties: both families are relatively non-selective Ca2+ channels with significant monovalent cation permeabilities [1]. Besides shared structural homology, the channels also share physiological regulation by various intracellular messengers. The most important of these is Ca2+ and ATP. In addition, the channels have unique regulation by other ligands, for example InsP3 for the InsP3R and caffeine and ryanodine for the RyR. Both families of ion channels interact with many other proteins, which confer regulation of the channel activities, often by allosteric modulation of the sensitivities to other ligands such as InsP3 and Ca2+.
Although X-ray crystallographic structures have been obtained for the InsP3R binding domain [2-4] and for various cytoplasmic domains of the RyR [5-7], no crystal structure of a full-length member of either Ca2+ release channel family have been obtained. However, the RyR has been a favorite macromolecule for structural biologists employing cryo-electron microscopy (cryo-EM) of isolated solubilized proteins [8]. All three mammalian RyR isoforms and the InsP3R-1 have been characterized by cryo-EM, but until recently the three-dimensional reconstructions were limited to about 10Å resolution [8, 9]. This resolution was sufficient to establish that all three RyR isoforms shared a similar structure, to distinguish between closed and open states, and to determine the general location of the binding sites of interacting proteins [8]. But higher resolutions are necessary to reveal what we really want to see, including the structural basis for ion permeation, the ligand binding sites and the detailed inter-and intermolecular interactions that couple ligand binding to conformational changes associated with gating.
The field has now entered a new era, and these are exciting times for researchers interested in the structural biology of intracellular Ca2+-release channels. With advances in single-particle cryo-EM technologies and structure analysis and modeling software, quaternary structures of the massive RyR with ever-improving resolutions have become available at unprecedented rates over the past two years [10-16]. The 2016 studies provide structures with sufficient resolution (~ 3.6–5Å) to reveal not only the backbones, but also side chains of > 90% of the amino residues in the types 1 and 2 RyR channels for both open and closed conformations. Although cryo-EM acquisition of quaternary structures of the InsP3R has lagged behind that of RyR, a structure with near-atomic resolution (~4.7 Å) was published for the apo-state of the InsP3R-1 channel (in the absence of InsP3 and Ca2+) in 2015 [17] showing the backbones of most residues of the channel and the residue side chains in the transmembrane helices. Notably, however, none of these structures resolved the major ligand binding sites that regulate the activities of the channels. Now, des Georges et al. [13] have described open and closed RyR1 channel structures observed under a multitude of ligand conditions (with and without Ca2+, ATP, caffeine and ryanodine), with the binding sites of these ligands located by comparing the structures obtained in the presence and absence of various combinations of ligands. Because of the homology of the two channel families and their analogous ligand regulation by Ca2+ and ATP, we have here compared the quaternary structures of the two channels to ask whether insights gained from these new structures might provide insights into the structural bases of ligand binding in InsP3Rs.
Before proceeding, we should alert the reader that in the many papers published on RyR and InsP3R structures, multiple domain nomenclatures have been used. For clarity here, we use the domain names and residue numbers from [13, 14] for rabbit RyR1 and those from [17] for rat InsP3R-1. Furthermore, the primary sequence comparisons we mention refer to comparisons among all rabbit and human RyR isoforms and among all rat and human InsP3R isoforms unless stated otherwise.
One of the major revelations in [13] is that each of the three newly discovered ligand-binding sites for Ca2+, ATP and caffeine is at the interface(s) of multiple domains, with contributions to binding by residues from different domains. Furthermore, all residues in each site come from the same RyR molecule, so there are four of each kind of binding site in a RyR channel. This is also true for the InsP3-binding site in the InsP3R [3]. Unlike the InsP3-binding site that is constituted of residues from sequentially adjacent domains (β-TF2 and ARM1), the domains involved in each of the newly discovered binding sites in the RyR are not sequentially adjacent. In fact, some of the residues in the sole Ca2+-binding site identified are over 1000 residues apart. That explains why these sites were not previously identified by analysis of the RyR primary sequence. All of the residues involved in ligand interactions as well as many residues in their sequential vicinity are conserved in all RyR, suggesting the functional importance of the identified binding site. Notably, this level of conservation extends to the corresponding domains in InsP3R (See Extended Data figure 8 of [16]), thus allowing accurate residue alignments between corresponding RyR and InsP3R domains. From these alignments, it is possible to locate residues in InsP3R that correspond to those in the Ca2+, ATP and caffeine-binding sites in RyR. A second major revelation is that the three activating ligand-binding sites are in close spatial proximity to each other, clustered around the CTD and close, by comparison with the prodigious height of the channel, to the cytoplasmic face of the membrane.
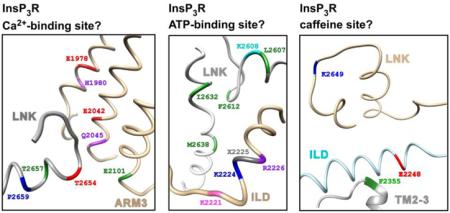
The RyR Ca2+-binding site coordinates a Ca2+ ion with the carboxylate side chains of E3893 and E3967 in the Csol and the backbone carbonyl of T5001 in the CTD; and H3895 and Q3970 in the Csol contribute to the second coordination sphere of the Ca2+ [13]. All of these resides are conserved in all InsP3R (Fig. 1A), corresponding to E1978, E2024 in the ARM3, T2654 in the LNK (red residues), and H1980, Q2045 in the ARM3 (purple), respectively [16]. Importantly, these residues are in close spatial proximity in the InsP3R-1 cryo-EM structure. These clues strongly suggest that these residues can also form a Ca2+-binding site in the InsP3R channel, although resolution of the available cryo-EM InsP3R structure [17] is not high enough to locate residue side-chains in that region, so currently it is impossible to determine if those InsP3R residues can properly coordinate Ca2+. Furthermore, the RyR structure reveals that hydrogen bonds formed between E4032 in the Csol and T5004 and Q5006 in the CTD help to stabilize the interface between the Csol and CTD, and therefore can be of functional importance to the spatially-nearby Ca2+ binding site. According to sequence alignment, E4032 and T5004 correspond to E2101in ARM3 and T2657 in LNK of InsP3R-1, respectively (green residues in Fig. 1A). Q5006 corresponds to P2659 in the LNK of InsP3R (blue). These three residues are conserved among InsP3R. However, in the InsP3R structure, E2101 is too distant from T2657 and P2659 to interact with them. So the functional importance of these residues to the stabilization of the Ca2+-binding site cannot be confirmed from the available InsP3R structure.
Fig.
Regions in the InsP3R structure corresponding to ligand binding sites in the RyR
Three groups of residues participate in ATP binding to the RyR channel. Four non-polar residues M4954 in S6c and F4959, T4979 and L4985 in the CTD provide hydrophobic interactions with the adenine base of the ATP; positive charges in K4211, K4214 and R4215 in the TaF interact with the negatively-charged phosphate tail of the ATP; and the hydroxyl groups in the ribose ring of the ATP can interact with the charged E4955 in the S6c. Sequence alignment between RyR and InsP3R maps the four adenine-interacting residues to L2607, F2612, I2632 and M2638 in the LNK (green residues in Fig. 1B), all of which are non-polar residues that are totally conserved in InsP3R. Among the phosphate-interacting positively-charged residues in RyR, K4211 in RyR is mapped to K2224 (blue) in the ILD of InsP3R, which is conserved in all InsP3R. R4215 in RyR is mapped to non-polar L2225 in the ILD (grey) and this residue is poorly conserved in InsP3R. However, the next residue R2226 is totally conserved in InsP3R (purple), so it may contribute to the interactions with the ATP phosphate group. K4214 in RyR is mapped to position 2221 in InsP3R-1 (magneta). The positive charge is conserved in InsP3R-1 and 2 (K and R, respectively), but InsP3R-3 has a glutamate in that position. Interestingly, this difference may in part account for the difference in ATP regulation of InsP3R single-channel activity observed between InsP3R-1 [18] and InsP3R-3 [19]. The ribose-interacting E4955 in RyR is mapped to K2608 in the LNK (cyan), which is conserved in all InsP3R. Although the charges on these residues are different, the positive charge in K2608 should still be able to interact with the hydroxyl groups in the ATP ribose ring. All of these residues are located in a small spatial region in the InsP3R structure (Fig. 1B). These clues together suggest that, independent of the Walker A type motifs previously identified in the InsP3R sequence [20, 21], the InsP3R channel probably has ATP-binding sites similar to those in the RyR channel.
The caffeine-binding site in the RyR structure consists of at least three residues: I4996 in the CTD and W4716 in the S2S3 that bind caffeine through non-polar interactions; and E4239 in the TaF that interacts with caffeine through hydrogen bonds. Sequence alignment maps I4996 and W4716 in RyR to K2649 in the LNK (magenta residue in Fig. 1C) and F2355 (green) in the TM2-3 of InsP3R. Both residues are well conserved in InsP3R. E4239 in RyR is mapped to E2248 (red) in the ILD of InsP3R, which is only conserved in InsP3R-1 and 2. The charged K2649 is not likely to have strong interaction with the non-polar region of caffeine like its corresponding counterpart I4996 in RyR. Furthermore, K2649 and E2248 are too far apart spatially to form a binding site for the small caffeine molecule (Fig. 1C). Thus, it is unlikely that the InsP3R channel has caffeine binding sites similar to those in the RyR channel. Accordingly, caffeine has not been shown to activate the InsP3R channel, in contrast to its effects on the RyR.
The insights into the structural determinants of ligand binding in RyR1 provided by [13] suggest a structural basis for Ca2+ and ATP regulation of the InsP3R. Of great benefit would be a similarly high-resolution, ligand-bound InsP3R structure to validate the correlations we have described here. Until then, single channel recordings of InsP3R channels with predicted key ligand binding residues mutated will prove informative. The activity of the InsP3R is most significantly regulated by InsP3 and Ca2+, although it is important to note that InsP3 and ATP regulate channel activity mainly by modifying the sensitivity of the channels to Ca2+ regulation [1]. Importantly, the InsP3R is physiologically regulated by cytoplasmic Ca2+ with a biphasic concentration dependence, with Ca2+ at low concentrations activating the channel, and Ca2+ at higher concentrations inhibiting it [22]. This suggests that InsP3R channels should have two distinct Ca2+-binding sites, activating and inhibiting. Which, if either, is the site predicted from the RyR structure? The predominant effect of ATP on the InsP3R is potentiation of channel activity by enhancing the sensitivity of the channel to Ca2+ activation [18]. It is interesting to note that the ATP and Ca2+ binding sites observed in [13] are in close physical proximity, which might provide a structural basis for a functional interaction between the two ligands and therefore suggest that the observed site corresponds to the activating Ca2+ site in the InsP3R. Because InsP3 appears to activate the InsP3R by effectively decreasing the Ca2+ affinity of an inhibitory Ca2+ site [22], it is possible that a second, inhibitory Ca2+ binding site is unique to InsP3Rs since RyRs are not regulated by InsP3. On the other hand, biphasic Ca2+ concentration dependence of RyR gating has been observed [23, 24], so an additional site may exist that was not reported in [13]. Indeed, the RyR has been proposed to have a luminal Ca2+ binding site [24] that was not observed in [13], suggesting that future structures may reveal additional Ca2+ binding sites. Evidence exists for even more Ca2+ binding sites in the InsP3R [1, 25]. Ca2+ modulates channel activity by binding to sites that regulate Ca2+ activation [22], InsP3-dependent Ca2+ inhibition [22], InsP3-independent Ca2+ inhibition [26], channel inactivation and channel recruitment [27]. Another Ca2+ binding site appears to regulate the properties of the InsP3-independent Ca2+ inhibition site [28]. Thus, a picture with one single Ca2+ sensor being responsible for Ca2+ regulation of InsP3R channel activity seems simplistic. A more complex picture with multiple functional Ca2+ sensors and their molecular identities remains to be revealed. Perhaps the resolution revolution in cryo-EM will provide the light.
Finally, we would like to raise a cautionary note concerning the alignment of primary sequences between InsP3R and RyR. An unexpected fact revealed by the high-resolution quaternary structures of InsP3R and RyR is that the corresponding linker sequences between the transmembrane helices in the two molecules can be very different in length: S2S3 in RyR has ~ 120 residues while TM2-3 in InsP3R has only 9; TM3-4 in InsP3R has ~ 35 residues but there is practically no S3S4 in RyR; and S5S6 in RyR has ~ 45 residues while TM5-6 in InsP3R has 103. This, together with weak sequence homology in extensive regions of InsP3R and RyR, can cause misalignment of primary sequences of RyR and InsP3R. For instance, Efremov et al. [16] aligned D4815 in RyR, a conserved residue in S4, with R2466 in InsP3R solely according to the primary sequences of RyR and InsP3R. In fact, according to the subsequently published quaternary structure of InsP3R [17], R2466 is located in the TM5-6 domain. des Georges et al. arrived at the wrong conclusion that E4032 in RyR is not conserved in InsP3R [13] because their sequence alignment was off by over 100 residues. The alignment in [16] correctly shows that E4032 in RyR corresponds to E2101 in InsP3R, the mutation of which affected both activation and inhibition of InsP3R by Ca2+ [29], in a similar manner as mutation of E4032 affected the RyR [30]. Thus, quaternary structures of InsP3R and RyR must be taken into consideration during primary sequence comparisons between the two molecules.
HIGHLIGHTS.
High resolution cryo-EM ryanodine receptor (RyR1) structures have been determined
The structures reveal binding sites for Ca2+, ATP, caffeine and ryanodine
The structures have been compared with an InsP3 receptor structure
RyR structures provide insights into Ca2+ and ATP binding sites in InsP3 receptors
Acknowledgments
Supported by NIH grants and R01-GM114042 (D.-O.D.M) R37-GM056328 (J.K.F.).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foskett JK, White C, Cheung KH, Mak D-OD. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosanac I, Yamazaki H, Matsu-Ura T, Michikawa T, Mikoshiba K, Ikura M. Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 3.Bosanac I, Alattia JR, Mal TK, Chan J, Talarico S, Tong FK, et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- 4.Lin CC, Baek K, Lu Z. Apo and InsP3-bound crystal structures of the ligand-binding domain of an InsP3 receptor. Nat Struct Mol Biol. 2011;18:1172–1174. doi: 10.1038/nsmb.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Petegem F. Ryanodine receptors: allosteric ion channel giants. J Mol Biol. 2015;427:31–53. doi: 10.1016/j.jmb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Lau K, Van Petegem F. Crystal structures of wild type and disease mutant forms of the ryanodine receptor SPRY2 domain. Nat Commun. 2014;5:5397. doi: 10.1038/ncomms6397. [DOI] [PubMed] [Google Scholar]
- 7.Yuchi Z, Yuen SM, Lau K, Underhill AQ, Cornea RL, Fessenden JD, et al. Crystal structures of ryanodine receptor SPRY1 and tandem-repeat domains reveal a critical FKBP12 binding determinant. Nat Commun. 2015;6:7947. doi: 10.1038/ncomms8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagenknecht TC, Liu Z. Electron microscopy of ryanodine receptors. Curr Top Membr. 2010;66:27–47. doi: 10.1016/S1063-5823(10)66002-4. [DOI] [PubMed] [Google Scholar]
- 9.Serysheva II. Toward a high-resolution structure of IP(3)R channel. Cell Calcium. 2014;56:125–132. doi: 10.1016/j.ceca.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng W, Shen H, Wu J, Guo W, Pan X, Wang R, et al. Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2. Science. 2016;354:aah5324. doi: 10.1126/science.aah5324. [DOI] [PubMed] [Google Scholar]
- 11.Bai XC, Yan Z, Wu J, Li Z, Yan N. The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening. Cell Res. 2016;26:995–1006. doi: 10.1038/cr.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei R, Wang X, Zhang Y, Mukherjee S, Zhang L, Chen Q, et al. Structural insights into Ca(2+)-activated long-range allosteric channel gating of RyR1. Cell Res. 2016;26:977–994. doi: 10.1038/cr.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, et al. Structural Basis for Gating and Activation of RyR1. Cell. 2016;167:145–157. doi: 10.1016/j.cell.2016.08.075. e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, et al. Structure of a mammalian ryanodine receptor. Nature. 2015;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Z, Bai XC, Yan C, Wu J, Li Z, Xie T, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015;517:39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- 17.Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, et al. Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 2015;527:336–341. doi: 10.1038/nature15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mak D-OD, McBride S, Foskett JK. ATP regulation of type 1 inositol 1,4,5-trisphosphate receptor channel gating by allosteric tuning of Ca2+ activation. J Biol Chem. 1999;274:22231–22237. doi: 10.1074/jbc.274.32.22231. [DOI] [PubMed] [Google Scholar]
- 19.Mak D-OD, McBride S, Foskett JK. Regulation by Ca2+ and inositol 1,4,5-trisphosphate (InsP3) of single recombinant type 3 InsP3 receptor channels. Ca2+ activation uniquely distinguishes types 1 and 3 InsP3 receptors. J Gen Physiol. 2001;117:435–446. doi: 10.1085/jgp.117.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betzenhauser MJ, Wagner LE, 2nd, Park HS, Yule DI. ATP regulation of type-1 inositol 1,4,5-trisphosphate receptor activity does not require walker A-type ATP-binding motifs. J Biol Chem. 2009;284:16156–16163. doi: 10.1074/jbc.M109.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betzenhauser MJ, Wagner LE, 2nd, Iwai M, Michikawa T, Mikoshiba K, Yule DI. ATP modulation of Ca2+ release by type-2 and type-3 inositol (1, 4, 5)-triphosphate receptors. Differing ATP sensitivities and molecular determinants of action. J Biol Chem. 2008;283:21579–21587. doi: 10.1074/jbc.M801680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mak D-OD, McBride S, Foskett JK. Inositol 1,4,5-trisphosphate activation of inositol trisphosphate receptor Ca2+ channel by ligand tuning of Ca2+ inhibition. Proc Natl Acad Sci USA. 1998;95:15821–15825. doi: 10.1073/pnas.95.26.15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissner G. Ryanodine receptor/Ca2+ release channels and their regulation by endogenous effectors. Annu Rev Physiol. 1994;56:485–508. doi: 10.1146/annurev.ph.56.030194.002413. [DOI] [PubMed] [Google Scholar]
- 24.Laver DR. Regulation of ryanodine receptors from skeletal and cardiac muscle during rest and excitation. Clin Exp Pharmacol Physiol. 2006;33:1107–1113. doi: 10.1111/j.1440-1681.2006.04500.x. [DOI] [PubMed] [Google Scholar]
- 25.Foskett JK, Mak D-OD. Regulation of IP3R channel gating by Ca2+ and Ca2+ binding proteins. In: Serysheva II, editor. Curr Top Membr. Vol. 66. Elsevier; Amsterdam; 2010. pp. 235–272. Structure-Function of Ca2+ Release Channels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak D-OD, McBride SM, Foskett JK. Spontaneous channel activity of the inositol 1,4,5-trisphosphate (InsP3) receptor (InsP3R). Application of allosteric modeling to calcium and InsP3 regulation of InsP3R single-channel gating. J Gen Physiol. 2003;122:583–603. doi: 10.1085/jgp.200308809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ionescu L, Cheung KH, Vais H, Mak D-OD, White C, Foskett JK. Graded recruitment and inactivation of single InsP3 receptor Ca2+-release channels: implications for quantal Ca2+ release. J Physiol. 2006;573:645–662. doi: 10.1113/jphysiol.2006.109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mak D-OD, McBride SM, Petrenko NB, Foskett JK. Novel regulation of calcium inhibition of the inositol 1,4,5-trisphosphate receptor calcium-release channel. J Gen Physiol. 2003;122:569–581. doi: 10.1085/jgp.200308808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu H, Nosyreva E, Miyakawa T, Wang Z, Mizushima A, Iino M, et al. Functional and biochemical analysis of the type 1 inositol (1,4,5)-trisphosphate receptor calcium sensor. Biophys J. 2003;85:290–299. doi: 10.1016/S0006-3495(03)74474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fessenden JD, Chen L, Wang Y, Paolini C, Franzini-Armstrong C, Allen PD, et al. Ryanodine receptor point mutant E4032A reveals an allosteric interaction with ryanodine. Proc Natl Acad Sci U S A. 2001;98:2865–2870. doi: 10.1073/pnas.041608898. [DOI] [PMC free article] [PubMed] [Google Scholar]