ABSTRACT
Tobacco smoking is a preventable environmental factor that contributes to a wide spectrum of age-related health outcomes; however, its association with the development of frailty is not yet well established. We examined the associations of self-reported smoking indicators, serum cotinine levels and smoking-related DNA methylation biomarkers with a quantitative frailty index (FI) in 2 independent subsets of older adults (age 50–75) recruited in Saarland, Germany in 2000 – 2002 (discovery set: n = 978, validation set: n = 531). We obtained DNA methylation profiles in whole blood samples by Illumina HumanMethylation450 BeadChip and calculated the FI according to the method of Mitnitski and Rockwood. Mixed linear regression models were implemented to assess the associations between smoking indicators and the FI. After controlling for potential covariates, current smoking, cumulative smoking exposure (pack-years), and time after smoking cessation (years) were significantly associated with the FI (P-value < 0.05). In the discovery panel, 17 out of 151 previously identified smoking-related CpG sites were associated with the FI after correction for multiple testing (FDR < 0.05). Nine of them survived in the validation phase and were designated as frailty-associated loci. A smoking index (SI) based on the 9 loci manifested a monotonic association with the FI. In conclusion, this study suggested that epigenetic alterations could play a role in smoking-associated development of frailty. The identified CpG sites have the potential to be prognostic biomarkers of frailty and frailty-related health outcomes. Our findings and the underlying mechanisms should be followed up in further, preferably longitudinal studies.
KEYWORDS: Aging, epigenetic epidemiology, frailty, smoking methylation, whole blood sample
Introduction
Frailty is an emerging gerontological concept describing a multidimensional syndrome characterized by loss of physiologic reserves, which leads to increased vulnerability to age-related diseases and functional impairment.1,2 As a possible reversible syndrome that has pronounced associations with longevity and other age-related phenotypes, frailty is of particular interest for aging research and has received rapidly growing attention in recent years.3, 4 Previous publications reported that frail people are at higher risk of age-related diseases and higher late-life mortality.4-6 Over the past decade, several approaches have been proposed to define and evaluate frailty.1,7,8 A widely accepted approach proposed by Mitnitski et al. uses a continuous frailty index (FI) to define frailty as the proportion of accumulated health deficits, such as specific diseases, symptoms, signs, or disabilities presented at the time of investigation.1
Cigarette smoking has been shown to be one of the major causes of a wide spectrum of age-related health outcomes.9 However, its association with the development of frailty is not yet well-established, and reported findings were inconsistent: several cross-sectional studies reported that smoking was associated with being less frail,10-12 while most prospective cohort studies asserted that baseline smoking status was predictive of significantly accelerated progression of frailty at follow-up.4,13,14 In addition, the mechanisms underlying the potential linkage between smoking and frailty yet remain unclear. The so far most commonly suggested explanation is chronic inflammation induced by various toxic chemicals produced by tobacco smoking, which is supported by findings of positive associations between increased levels of inflammatory markers,15 such as C-reactive protein and interleukin-6, and higher prevalence and incidence of frailty.16,17
Recent advances in epigenetic research have shown the regulating role of DNA methylation, one of the main forms of epigenetic modification, in the pathways of smoking and smoking-induced diseases.18,19 An increasing number of smoking-related CpG sites in various genes, such as AHRR and F2RL3, have been discovered by epigenome-wide association studies (EWAS) based on whole blood samples and have been shown to be useful as quantitative biomarkers of current and past smoking exposure and predictors of smoking-associated health risks.20-24 Teschendorff et al. constructed a smoking index based on 1,501 smoking-related CpG sites and illustrated that such DNA methylation signatures of smoking could be useful indicators of smoking-induced health disorders.25 Hence, we hypothesized that smoking-induced DNA methylation might also be correlated with frailty. We therefore performed a comprehensive analysis of the associations of self-reported smoking indicators, serum cotinine levels and smoking-related DNA methylation biomarkers with frailty in a large population-based study of older adults in Germany and evaluated a smoking-related methylation-based predictor of frailty.
Results
Participant characteristics
Characteristics of the study population in the discovery and the validation panel were comparable with respect to smoking behaviors and lifestyle factors, as well as frailty categories, as summarized in Table 1. Average age in the 2 subsets was about 62 y. More than half of the participants in each subset were ever smokers (current or former smokers), and around 18% still smoked at the time of recruitment. Current smokers included a larger proportion of younger participants than former and never smokers (Figure S1). The proportion of men was much higher in current smokers than in never smokers: 60.8% vs. 29.4% in the discovery panel and 48.0% vs. 21.1% in the validation panel. Average cumulative smoking exposure (pack-years) of current smokers was considerably higher than that of former smokers in both subsets (discovery set: 36.8 vs. 23.3; validation set: 33.9 vs. 19.9). Average time after smoking cessation (years) of former smokers in both subsets was also similar, approximately 17 y. The majority of participants in both subsets of the study population were overweight or obese, reported no or only low physical activity, and no or low amounts of alcohol drinking. Mean values of the FI were slightly higher among women than among men, increased with age and were higher in ever smokers compared with never smokers in both panels (Table S2).
Table 1.
Study population characteristics in the discovery and the validation panela.
| Characteristics | Discovery Panel | Validation Panel |
|---|---|---|
| n | 978 | 531 |
| Age (years) | 62.1 (6.5) | 62.0 (6.6) |
| Sex (male) | 495 (50.6%) | 207 (39.0%) |
| Smoking status | ||
| Current smoker | 181 (18.5%) | 98 (18.4%) |
| Former smoker | 328 (33.5%) | 182 (34.3%) |
| Never smoker | 469 (48.0%) | 251 (47.3%) |
| Pack-years of smoking | ||
| Current smokers | 36.8 (19.3) | 33.9 (17.5) |
| Former smokers | 23.3 (16.3) | 19.9 (15.1) |
| Smoking cessation time (years)b | 17.3 (11.3) | 17.6 (10.6) |
| Body mass indexc | ||
| Underweight or normal weight (< 25.0) | 245 (25.1%) | 162 (30.5%) |
| Overweight (25.0 to <30.0) | 472 (48.4%) | 228 (42.9%) |
| Obese (≥ 30.0) | 258 (26.5%) | 141 (26.6%) |
| Alcohol consumptiond | ||
| Abstainer | 311 (34.1%) | 169 (34.4%) |
| Low | 531 (58.2%) | 290 (59.1%) |
| Intermediate | 53 (5.8%) | 27 (5.5%) |
| High | 17 (1.9%) | 5 (1.0%) |
| Physical activitye | ||
| Inactive | 189 (19.3%) | 109 (20.5%) |
| Low | 433 (44.3%) | 261 (49.2%) |
| Medium or high | 356 (36.4%) | 161 (30.3%) |
| Frailty categories (cut-off points) | ||
| Non-frail (≤ 0.20) | 435 (44.5%) | 263 (49.5%) |
| Pre-frail (0.20 ∼0.45) | 443 (45.3%) | 220 (41.4%) |
| Frail (≥ 0.45) | 100 (10.2%) | 48 (9.0%) |
Mean values (SD) for continuous variables and n (%) for categorical variables.
Former smokers only, data missing for 9 and 3 participants, respectively, in the discovery and the validation panel; cessation time equals age at recruitment minus age at cessation.
Data missing for 3 participants in the discovery panel.
Data missing for 66 and 40 participants, respectively, in the discovery and the validation panel. Categories defined as follows: abstainer, low [women: 0 to <20 g/d, men: 0 to <40 g/d], intermediate [20 to <40 g/d and 40 to <60 g/d, respectively], high [≥ 40 g/d and ≥ 60 g/d, respectively].
Categories defined as follows: inactive [< 1 h of physical activity/week], low [other], medium or high [≥ 2 h of vigorous and ≥ 2 h of light physical activity/week].
Associations between smoking indicators and frailty
In the analyses of associations of self-reported smoking indicators and serum cotinine levels with the FI, 2 linear regression models were used (details are presented in Methods) in the discovery panel, controlling for potential confounders. After adjustment for age and sex, current smoking, cumulative smoking exposure (pack-years), and time after smoking cessation (years) were significantly associated with the FI (P-value <0.05; for time after smoking cessation, an inverse association was observed), while former smoking or serum cotinine levels was not (Table 2). Additional adjustment for alcohol consumption did not alter the results in any relevant manner.
Table 2.
Associations of self-reported smoking indicators and cotinine levels with frailty index in the discovery panela.
| Model 1 |
Model 2 |
||||||
|---|---|---|---|---|---|---|---|
| Self-reported smoking indicators | Estimate | SE d | P-value | Estimate | SE d | P-value | |
| Smoking status | Never smoker | Ref | Ref | ||||
| Former smoker | 0.010 | 0.013 | 0.415 | 1.3 e-3 | 0.014 | 0.948 | |
| Current smoker | 0.021 | 0.011 | 0.034 | 0.027 | 0.012 | 0.036 | |
| Cumulative smoking (pack-years) b | 1.6 e-3 | 4.0 e-4 | < 0.0001 | 1.5 e-3 | 4.3 e-4 | 0.0009 | |
| Smoking cessation time (years) c | −8.5 e-4 | 7.7 e-4 | 0.029 | −3.7 e-4 | 8.0 e-4 | 0.043 | |
| Serum cotinine levels (ng/ml) | 1.0 e-4 | 2.5 e-4 | 0.678 | 1.1 e-4 | 2.5 e-4 | 0.652 | |
Model 1: Adjusted for age (years) and sex (male/female); Model 2: Adjusted for age, sex, alcohol consumption (abstainer/ low/ intermediate/ high).
A pack-year was defined as having smoked 20 cigarettes per day for 1 year, including current and former smokers from the discovery panel.
Cessation time defined as age at the time of recruitment minus age at cessation, only including former smokers from the discovery panel.
SE = standard error.
Of the 151 smoking-related CpG sites, which had been identified ≥ 2 times in previous smoking EWAS as biomarkers of smoking exposure,26 17 passed the threshold of FDR < 0.05 in regression analysis with the FI and thus demonstrated significant associations with the FI in the discovery phase (Table S3). Nine of them were confirmed to be significantly related to FI in the validation panel (Table 3, FDR < 0.05). A sensitivity analysis with additionally adjusting for education background did not lead to any relevant changes (details not shown). Effect sizes (average methylation difference between never and current smokers) of the 9 hypomethylated loci ranged from 1.7 to 9.2%. Six of them were mapped in known genome regions, including cg02657160 (CPOX), cg05673882 (POLK), cg07826859 (MYO1G), cg19859270 (GPR15), cg23667432 (ALPP), and cg25189904 (GNG12). Methylation intensity of each locus in the validation panel was significantly lower in the frail population than that in the non-frail population (P-value <0.05 by Kruskal-Wallis test), and intermediate levels of methylation intensity were observed for the pre-frail population (Figure S2).
Table 3.
Significant associations between methylation of smoking-related CpG sites and the frailty index in the validation panela.
| Mean β value (Standard deviation) |
Associations with the FI |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG site | Chrb | Gene | Never smoker | Former smoker | Current smoker | Effect sizec (Smoking) | Correlation coefficients d | Changes of the FI per SD (SE) e | P-value | FDR |
| cg01127300 | 22 | Unassigned | 0.490 (0.051) | 0.471 (0.055) | 0.459 (0.058) | −0.031 | −0.130 | −0.020 (0.0049) | 5.70 e-5 | 4.82 e-4 |
| cg02657160 | 3 | CPOX | 0.824 (0.020) | 0.814 (0.029) | 0.799 (0.029) | −0.025 | −0.091 | −0.012 (0.0048) | 1.19 e-2 | 2.54 e-2 |
| cg05673882 | 5 | POLK | 0.303 (0.049) | 0.282 (0.047) | 0.269 (0.050) | −0.034 | −0.068 | −0.022 (0.0051) | 2.30 e-5 | 3.86 e-4 |
| cg07826859 | 7 | MYO1G | 0.556 (0.044) | 0.537 (0.043) | 0.526 (0.044) | −0.030 | −0.024 | −0.025 (0.0076) | 9.00 e-4 | 3.83 e-3 |
| cg14753356 | 6 | Unassigned | 0.425 (0.052) | 0.402 (0.057) | 0.381 (0.064) | −0.044 | −0.220 | −0.022 (0.0086) | 1.03 e-2 | 2.54 e-2 |
| cg19589396 | 8 | Unassigned | 0.641 (0.050) | 0.623 (0.047) | 0.619 (0.053) | −0.022 | −0.104 | −0.021 (0.0058) | 2.96 e-4 | 1.68 e-3 |
| cg19859270 | 3 | GPR15 | 0.869 (0.016) | 0.854 (0.023) | 0.837 (0.027) | −0.032 | −0.117 | −0.012 (0.0049) | 1.50 e-2 | 2.84 e-2 |
| cg23667432 | 2 | ALPP | 0.629 (0.038) | 0.613 (0.039) | 0.611 (0.035) | −0.017 | −0.040 | −0.017 (0.0068) | 1.20 e-2 | 2.54 e-2 |
| cg25189904 | 1 | GNG12 | 0.461 (0.061) | 0.409 (0.070) | 0.369 (0.080) | −0.092 | −0.066 | −0.014 (0.0050) | 5.43 e-3 | 1.85 e-2 |
Adjusted for age (years), sex (male/female), alcohol consumption (abstainer/ low/ intermediate/ high), leukocyte distribution (Houseman algorithm) and random batch effects of methylation measurement.
Nine of the 17 loci identified by the discovery panel were confirmed in the validation set.
Chromosomal position, according to GRCh37/hg19.
Effect size = Mean βcurrent smoker – Mean βnever smoker.
Spearman's rank-order correlation coefficients, all loci showed significant correlations with the FI (P-value <0.0001).
SD = the overall standard deviation for each CpG site, SE = standard error.
Smoking index (SI) and frailty
We constructed a SI based on the 9 identified smoking-related loci. The SI was significantly associated with the FI in the fully-adjusted mixed linear model (P-value < 0.0001). Furthermore, the SI showed a monotonic linear dose-response relationship with the FI (Fig. 1). An increase in the SI by one standard deviation was roughly associated with a 0.04-unit increase of the FI.
Figure 1.
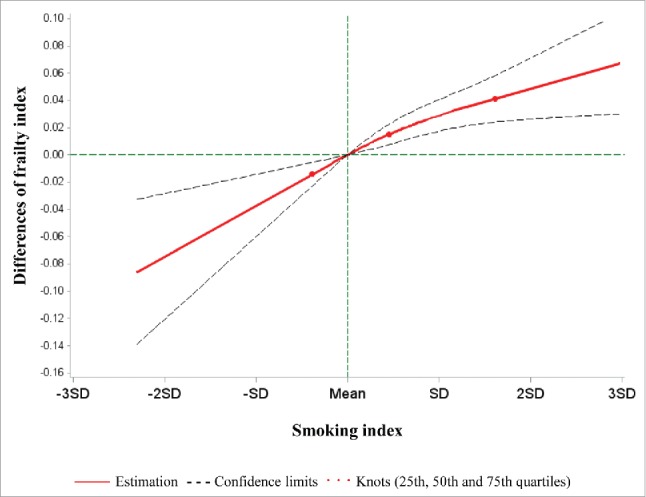
Best-fitting model for the association of the smoking index with the frailty index in the validation panel. Red lines: Estimation; dashed lines: confidence limits; red dots: knots (25th, 50th, and 75th quartiles); green lines: reference lines.
As shown in Table 4, current smoking was associated with an increase in the FI by 3.7 percent units compared with never smoking, whereas an increase of the FI by 3.3, 6.2 and 7.5 percent units was observed in the participants in the 2nd, 3rd, and 4th quartiles of the SI compared with those in the first quartile after adjustment for potential covariates (all P-values < 0.05). In the model considering both smoking indicators simultaneously, the effect estimate for current smoking was strongly reduced and no longer statistically significant, while the estimates for the SI quartiles remained essentially unchanged, suggesting that the impact of smoking on FI may be partly mediated by smoking-associated methylation changes.
Table 4.
Associations of smoking status and the methylation-based smoking index with the frailty index in the validation panel, with and without mutual adjustment for the 2 smoking indicatorsa.
| Model including smoking status only |
Model including smoking index only |
Model including both smoking indicators |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Smoking indicators | Estimate | SEb | P-value | Estimate | SE b | P-value | Estimate | SE b | P-value | ||
| Smoking status | Never smoker | Ref | Ref | ||||||||
| Former smoker | 0.019 | 0.018 | 0.293 | −0.010 | 0.019 | 0.610 | |||||
| Current smoker | 0.037 | 0.015 | 0.013 | 0.023 | 0.015 | 0.116 | |||||
| Smoking index | 1st Quartile | Ref | Ref | ||||||||
| 2nd Quartile | 0.033 | 0.017 | 0.048 | 0.034 | 0.017 | 0.043 | |||||
| 3rd Quartile | 0.062 | 0.018 | 0.0008 | 0.060 | 0.019 | 0.0016 | |||||
| 4th Quartile | 0.075 | 0.019 | < 0.0001 | 0.078 | 0.020 | 0.0001 | |||||
All model controlling for age (years), sex (male/female), and alcohol consumption (abstainer/ low/ intermediate/ high). The models including the smoking index were additionally adjusted for the leucocyte distribution estimated by Houseman algorithm.
SE = standard error.
Discussion
In the present study, we systematically investigated the association of tobacco smoking with frailty in a population-based cohort of older adults. We observed that current smoking, cumulative smoking exposure (pack-years), and time after smoking cessation (years) were significantly associated with the frailty index (FI). Furthermore, we identified 9 previously confirmed smoking-related CpG sites to be also associated with the FI, and the smoking index (SI) based on them manifested a monotonic dose-response relationship with the FI. The association between the SI and the FI remained essentially unchanged in a model including both self-reported smoking and the SI, whereas the association between self-reported smoking and the FI was strongly alternated in such a model.
Findings based on both self-reported and epigenetic indicators of smoking showed that active smoking is associated with frailty. In the present study, we found that current smoking, as well as cumulative smoking exposure (pack-years), was significantly associated with worsening frailty status, which is consistent with previous studies.14,27 Serum cotinine levels, an established biomarker of short-term smoking exposure,28 showed a positive correlation with the FI in the same direction as current smoking, albeit without statistical significance. Furthermore, former smoking showed a much weaker association with frailty compared with current smoking, suggesting that smoking cessation could reduce the accumulation of deficits, which would be in accordance with the significant inverse association between time after smoking cessation (years) and the FI. In line with these findings, clear patterns of the lowest and intermediate methylation levels, respectively, among current and former smokers, compared with never smokers were consistently observed among the 9 hypomethylated smoking-related loci. The observed decreasing trend of the FI after smoking cessation was also in line with the previous findings that the methylation levels of smoking-related CpG sites might “recover” after quitting smoking.21,29
Prior to the present study, epigenetic changes of several promoter CpG islands and global DNA methylation were proposed to explain the development of frailty syndrome.30,31 To the best of our knowledge, this is the first study identifying specific CpG sites that were not only associated with frailty but also are well established as smoking-related loci. Among them, 3 loci were located in the genome regions related to inflammation that could promote the progression of frailty. In particular, loci cg07826859 (MYO1G) and cg19859270 (GPR15) have been reported to play a role in the formation of chronic inflammation via regulating the activity of T cells.32,33 Locus cg25189904 is located on the south shore of a CpG island spanning the promoter of GNG12 that has been suggested to be an important factor in the overall inflammatory signaling cascade.34 Moreover, another 3 loci are mapped to genes linked to the risks of various age-related cancers. Locus cg02657160 is located in a predicted poised promoter region in CPOX, which is 60 kb far from cg19859270 (GPR15), and both loci have been recently found to be associated with increased risk of lung cancer mortality.35 The corresponding genes POLK for cg05673882 and ALPP for cg23667432 are oncogenes that play roles in the formation of several age-related cancers, including breast, prostate, lung, and ovarian cancers.36-39 The biologic functions of the remaining 3 loci within unassigned genome regions are not yet known, only locus cg01127300 has been disclosed as a potential epigenetic regulator of obesity,40 one of the deficits of the frailty syndrome.41 Interestingly, neither of the most strongly smoking-related CpG sites, cg05575921 (AHRR) or cg03636183 (F2RL3),26 was identified to be related to the FI in the present study. The 2 corresponding genes may simply have no biologic actions that contribute to the development or progression of frailty.
The FI has been proposed as one of the noteworthy aging parameters by defining the accumulation of deficit in epidemiological and clinical settings.42 Previous studies have evaluated its relationships with 2 other aging estimators, telomere length,43,44 and age-related DNA methylation changes (“epigenetic clock”).45,46 Telomere length was not associated with frailty, neither in Asian nor in Caucasian populations.43,44 In contrast, the epigenetic clock was found to be significantly associated with frailty-related phenotypes and the FI. Marioni et al. reported significant correlations between the epigenetic clock and cognitive functioning, grip strength and lung function.46 In addition, Breitling et al. showed that the epigenetic clock was correlated with the FI and further confirmed the robustness of this association across diverse domains of health deficits.45 Our previous study on the relationship between smoking-induced DNA methylation and the epigenetic clock intriguingly found 7 out of the 9 now identified loci to be associated with the epigenetic clock as well,47 including cg01127300, cg02657160 (CPOX), cg05673882 (POLK), cg07826859 (MYO1G), cg14753356, cg19589396 and cg23667432 (ALPP). The associations of smoking-related methylation with the FI and the epigenetic clock consequently share some pronounced commonalities, though the underlying causality certainly requires further investigation. These overlapping CpG sites have the potential to be investigated by pertinent research to discover the common mechanisms of aging reflected by the epigenetic clock and frailty.
Major strengths of this study include the relatively large sample size with detailed information on a broad range of covariates and validation in an independent group. Several limitations should be acknowledged as well. First, the associations of DNA methylation with smoking exposure and FI in whole blood may be affected by smoking-related shifts in leukocyte distribution.48 Hence, we adjusted for leukocyte distribution by the Houseman algorithm,49 which limits the confounding from differential blood counts to the greatest possible extent. Furthermore, all deficits for the FI construction were self-reported and possibly affected by reporting bias. Confounding from inter-individual heritable genetic variations might also impair the interpretability of the observed DNA methylation patterns.50 Finally, our study was cross-sectional, limiting our capability for drawing conclusions on causality and direction of effects.
In conclusion, this study raises the possibility that active smoking could affect frailty development through DNA methylation changes at specific smoking-related CpG sites. Nevertheless, prospective longitudinal studies with repeated measurements of smoking status, DNA methylation profiles, the FI and causality analyses possibly along the lines of Mendelian randomization51 will be needed to conclusively clarify the causality in the observed associations among smoking exposure, DNA methylation and frailty.
Methods
Study design and population
Study subjects were selected from the ESTHER study, an ongoing statewide population-based cohort study conducted in Saarland, a state located in southwestern Germany. Details of the study design have been reported previously.52,53 Briefly, 9,949 older adults (aged 50–75 years) were enrolled by their general practitioners during a routine health check-up between July 2000 and December 2002, and followed up thereafter. The current cross-sectional analysis is based on data and biospecimen collected at baseline. Two independent subgroups were selected as the discovery and the validation panel for DNA methylation analyses as described previously.47 Briefly, the discovery panel included 1,000 participants recruited consecutively at the start of the ESTHER study between July and October 2000. The validation panel included 548 participants randomly selected from participants recruited between October 2,000 and March 2001. The study was approved by the ethics committees of the University of Heidelberg and the state medical board of Saarland, Germany. Written informed consent was obtained from all participants.
Data collection
Information on socio-demographic characteristics, lifestyle factors, and health status at baseline was obtained by standardized self-administered questionnaires. Participants were asked about past and present cigarette, cigar, and pipe smoking behaviors and were then categorized into current, former and never smokers. Detailed information on smoking history was also obtained from questionnaires, including age at initiation and smoking intensities at various ages, as well as the age of quitting smoking for former smokers.54 Twenty-two and 17 participants with missing information on smoking status were excluded from the discovery and the validation panel, respectively. Additional information on body mass index (BMI) was extracted from a standardized form filled by the general practitioners during the health check-ups.
Laboratory analyses
Blood samples were taken during the health check-up and stored at −80°C until further processing. Serum cotinine levels of participants in the discovery panel were measured using the customized version of an enzyme-linked immunosorbent assay (Inspec II-Cotinine-EIA; Mahsan Diagnostika) as described previously.28 DNA from whole blood samples was collected using a salting out procedure.55 DNA methylation profiles of 151 smoking-related loci which had been identified ≥ 2 times in previous smoking EWAS26 were extracted by the Illumina HumanMethylation450 BeadChip (Illumina, San Diego, CA, USA). As described previously,,56 samples were analyzed following the manufacturer's instruction at the Genomics and Proteomics Core Facility of the German Cancer Research Center, Heidelberg, Germany. Illumina's GenomeStudio® (version 2,011.1; Illumina Inc.) was used to extract DNA methylation signals from the scanned arrays (Module version 1.9.0; Illumina Inc.). The methylation level of a specific CpG site was quantified as a β value ranging from 0 (no methylation) to 1 (full methylation). According to the manufacturer's protocol, no background correction was done and data were normalized to internal controls provided by the manufacturer. All controls were checked for inconsistencies in each measured plate. Signals of probes with a detection P-value > 0.05 were excluded from analysis. We used the Illumina normalization and preprocessing method implemented in Illumina's GenomeStudio®.
Frailty index (FI)
As described previously,4 a continuous FI was derived for all participants to define frailty by an accumulation of deficits approach.7 The FI counts individuals' health deficits, which can be symptoms, signs, blood markers, disabilities, and diseases. The index is the ratio of the deficits present divided by the total number of deficits considered and ranges from 0 (the absence of any deficit) to 1 (the presence of all deficits). According to pre-defined selection criteria,7 we selected 34 out of 50 potential deficits assessed in the ESTHER study (Table S1), that were associated with the general health status, accumulated with age, did not saturate too early, had more than 1% prevalence, and did not have a high prevalence (>50%) at younger ages (50–60 years). Missing values in the variables needed for the FI calculation were dealt with by multiple imputations (20 data sets imputed). According to previously determined cut-off points for the FI, study participants were assigned to the groups of non-frail (FI ≤ 0.20), pre-frail (0.20 < FI < 0.45) and frail (FI ≥ 0.45).4
Statistical analyses
Major socio-demographic characteristics, lifestyle factors, and smoking behaviors of both the discovery and the validation panel were summarized by descriptive statistics.
First, we investigated the associations of self-reported smoking indicators [smoking status (current/ former/ never smoker), cumulative smoking exposure (pack-years, in current and former smokers) and smoking cessation time (years, in former smokers only), as independent variables] and serum cotinine levels (ng/ml, as independent variable) with the FI (as dependent variable) in the discovery panel. Two linear regression models were used, controlling for potential confounding factors. Model 1 was adjusted for age (years) and sex (male/ female), and Model 2 was additionally adjusted for alcohol consumption [abstainer, low (women: 0 to <20 g/d, men: 0 to <40 g/d), intermediate (20 to <40 g/d and 40 to <60 g/d, respectively), high (≥ 40 g/d and ≥ 60 g/d, respectively)]. Indicators with a P-value < 0.05 were considered as frailty-associated factors.
Furthermore, we selected the 151 smoking-related loci that had been identified ≥ 2 times in previous smoking EWAS as potential biomarkers of smoking exposure.26 Associations of their methylation levels (as independent variables) with the FI (as dependent variable) were analyzed by a mixed linear regression model with methylation assay batch as a random effect, controlling for age, sex, alcohol consumption, and the leukocyte distribution estimated by the Houseman algorithm.49 After correction for multiple testing by the false discovery rate (FDR, Benjamini-Hochberg method),57 CpG sites with a corrected P-value < 0.05 were selected from the discovery panel and then replicated in the validation panel. Loci with a FDR < 0.05 in the validation panel were eventually identified as frailty-associated CpG sites.
We used the identified frailty-associated loci to construct a smoking index (SI) according to Teschendorff et al.'s algorithm,25 to measure the deviation of DNA methylation in a given sample from a normal reference, with the mean taken over from the identified loci. In more detail, we computed the mean β value () and standard deviation () across never smokers of the given data set, and then defined the SI as
where is −1(+1) if the smoking-associated CpG, c, is hypomethylated (hypermethylated) in smokers and where is the β value of this CpG in samples s.25 We calculated the SI for each participant and assessed its association with the FI by mixed linear regression models with adjustment for potential covariates in the validation panel. Subsequently, we used restricted cubic spline functions using the SAS macro from Desquilbet et al. to evaluate the dose-response relationship of the SI with the FI, controlling for age, sex, alcohol consumption, and the leukocyte distribution. The 25th, 50th, and 75th percentiles of the SI were selected as knots.
Finally, we intended to assess whether and to what extent the association between smoking and FI might be mediated by smoking-associated methylation changes. We assessed and compared the associations of self-reported smoking status (current/ former/ never smoker) and the methylation-based SI (in quartiles) with the FI (as dependent variable) when both types of smoking indicators were simultaneously included as categorical independent variables in mixed linear regression models, adjusting for age, sex, and alcohol consumption, in the validation panel. The models including the SI were additionally adjusted for the leukocyte distribution estimated by the Houseman algorithm.49
Data cleaning and all aforementioned analyses were performed by SAS version 9.3 (SAS Institute Inc., Cary, NC, USA). Analyses on the FI were performed in the 20 imputed data sets and results were combined by the SAS procedure PROC MIANALYZE, taking into account the variation between the results of the 20 data sets.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work of Xu Gao is supported by the grant from the China Scholarship Council (CSC). The authors gratefully acknowledge contributions of DKFZ Genomics and Proteomics Core Facility, especially Melanie Bewerunge-Hudler and Matthias Schick, in the processing of DNA samples and performing the laboratory work, and Mr. Jonathan Heiss for providing the estimation of leukocyte distribution.
Funding
The ESTHER study was supported in part by the Baden-Württemberg state Ministry of Science, Research and Arts (Stuttgart, Germany) and from the German Federal Ministry of Education and Research (Berlin, Germany).
References
- 1.Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. Scientific World J 2001; 1:323-36; PMID:12806071; http://dx.doi.org/ 10.1100/tsw.2001.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman H, Ferrucci L, Guralnik J, Hogan DB, Hummel S, Karunananthan S, Wolfson C. Frailty: an emerging research and clinical paradigm—issues and controversies. J Gerontol A Biol Sci Med Sci 2007; 62:731-7; PMID:17634320; http://dx.doi.org/24671603 10.1093/gerona/62.7.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Jazwinski SM. Quantitative measures of healthy aging and biological age. Healthy aging research 2015; 4:26; PMID:26005669; http://dx.doi.org/24671603 10.12715/har.2015.4.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saum KU, Dieffenbach AK, Muller H, Holleczek B, Hauer K, Brenner H. Frailty prevalence and 10-year survival in community-dwelling older adults: results from the ESTHER cohort study. Eur J Epidemiol 2014; 29:171-9; PMID:24671603; http://dx.doi.org/ 10.1007/s10654-014-9891-6 [DOI] [PubMed] [Google Scholar]
- 5.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. The Lancet 2013; 381:752-62; PMID:23395245; http://dx.doi.org/24666801 10.1016/S0140-6736(12)62167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulmala J, Nykanen I, Hartikainen S. Frailty as a predictor of all-cause mortality in older men and women. Geriatr Gerontol Int 2014; 14:899-905; PMID:24666801; http://dx.doi.org/ 10.1111/ggi.12190 [DOI] [PubMed] [Google Scholar]
- 7.Searle SD, Mitnitski A, Gahbauer EA, Gill TM, Rockwood K. A standard procedure for creating a frailty index. BMC Geriatr 2008; 8:24; PMID:18826625; http://dx.doi.org/ 10.1186/1471-2318-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, Seeman T, Tracy R, Kop WJ, Burke G. Frailty in older adults evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001; 56:M146-M57; PMID:11253156; http://dx.doi.org/23879634 10.1093/gerona/56.3.M146 [DOI] [PubMed] [Google Scholar]
- 9.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 2006; 3:e442; PMID:17132052; http://dx.doi.org/23879634 10.1371/journal.pmed.0030442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard R, Searle S, Mitnitski A, Rockwood K. Effect of smoking on the accumulation of deficits, frailty and survival in older adults: a secondary analysis from the Canadian Study of Health and Aging. J Nutr Health Aging 2009; 13:468-72; http://dx.doi.org/ 10.1007/s12603-009-0085-y [DOI] [PubMed] [Google Scholar]
- 11.Romero‐Ortuno R. Frailty Index in Europeans: association with determinants of health. Geriatr Gerontol Int 2014; 14:420-9; PMID:23879634; http://dx.doi.org/ 10.1111/ggi.12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale CR, Cooper C, Deary IJ, Sayer AA. Psychological well-being and incident frailty in men and women: the English Longitudinal Study of Ageing. Psychol Med 2014; 44:697-706; PMID:23822897; http://dx.doi.org/ 10.1017/S0033291713001384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JS, Auyeung T-W, Leung J, Kwok T, Woo J. Transitions in frailty states among community-living older adults and their associated factors. J Am Med Dir Assoc 2014; 15:281-6; PMID:24534517; http://dx.doi.org/ 10.1016/j.jamda.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 14.Etman A, Kamphuis CB, Van der Cammen TJ, Burdorf A, Van Lenthe FJ. Do lifestyle, health and social participation mediate educational inequalities in frailty worsening? Eur J Public Health 2015; 25:345-50; PMID:25061232; http://dx.doi.org/ 10.1093/eurpub/cku093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima G, Iliffe S, Walters K. Smoking as a predictor of frailty: a systematic review. BMC Geriatr 2015; 15:131; PMID:26489757; http://dx.doi.org/ 10.1186/s12877-015-0134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale CR, Baylis D, Cooper C, Sayer AA. Inflammatory markers and incident frailty in men and women: the English Longitudinal Study of Ageing. Age 2013; 35:2493-501; PMID:23543263; http://dx.doi.org/ 10.1007/s11357-013-9528-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puts MT, Visser M, Twisk JW, Deeg DJ, Lips P. Endocrine and inflammatory markers as predictors of frailty. Clin Endocrinol 2005; 63:403-11; PMID:16181232; http://dx.doi.org/ 10.1111/j.1365-2265.2005.02355.x [DOI] [PubMed] [Google Scholar]
- 18.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet 2013; 4:132; PMID:23882278; http://dx.doi.org/ 10.3389/fgene.2013.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philibert RA, Beach SR, Brody GH. The DNA methylation signature of smoking: an archetype for the identification of biomarkers for behavioral illness. Nebr Symp Motiv 2014; 61:109-27; PMID:25306781; http://dx.doi.org/ 10.1007/978-1-4939-0653-6_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shenker NS, Ueland PM, Polidoro S, van Veldhoven K, Ricceri F, Brown R, Flanagan JM, Vineis P. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology 2013; 24:712-6; PMID:23867811; http://dx.doi.org/ 10.1097/EDE.0b013e31829d5cb3 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Yang R, Burwinkel B, Breitling LP, Brenner H. F2RL3 methylation as a biomarker of current and lifetime smoking exposures. Environ Health Perspect 2014; 122:131-7; PMID:24273234; http://dx.doi.org/ 10.1289/ehp.1306937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philibert R, Hollenbeck N, Andersen E, McElroy S, Wilson S, Vercande K, Beach SR, Osborn T, Gerrard M, Gibbons FX, et al.. Reversion of AHRR Demethylation Is a Quantitative Biomarker of Smoking Cessation. Front Psychiatry 2016; 7:55; PMID:27092088; http://dx.doi.org/ 10.3389/fpsyt.2016.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J 2012; 33:2841-8; PMID:22511653; http://dx.doi.org/ 10.1093/eurheartj/ehs091 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Schöttker B, Ordonez-Mena J, Holleczek B, Yang R, Burwinkel B, Butterbach K, Brenner H. F2RL3 methylation, lung cancer incidence and mortality. Int J Cancer 2015; 137:1739-48; PMID:25821117; http://dx.doi.org/ 10.1002/ijc.29537 [DOI] [PubMed] [Google Scholar]
- 25.Teschendorff AE, Yang Z, Wong A, Pipinikas CP, Jiao Y, Jones A, Anjum S, Hardy R, Salvesen HB, Thirlwell C, et al.. Correlation of Smoking-Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA Oncol 2015; 1:476-85; PMID:26181258; http://dx.doi.org/ 10.1001/jamaoncol.2015.1053 [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics 2015; 7:113; PMID:26478754; http://dx.doi.org/ 10.1186/s13148-015-0148-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fugate Woods N, LaCroix AZ, Gray SL, Aragaki A, Cochrane BB, Brunner RL, Masaki K, Murray A, Newman AB. Frailty: emergence and consequences in women aged 65 and older in the Women's Health Initiative Observational Study. J Am Geriatr Soc 2005; 53:1321-30; PMID:16078957; http://dx.doi.org/ 10.1111/j.1532-5415.2005.53405.x [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Florath I, Saum KU, Brenner H. Self-reported smoking, serum cotinine, and blood DNA methylation. Environ Res 2016; 146:395-403; PMID:26826776; http://dx.doi.org/ 10.1016/j.envres.2016.01.026 [DOI] [PubMed] [Google Scholar]
- 29.Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, Vinuela A, Grundberg E, Nelson CP, Meduri E, et al.. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 2014; 9:1382-96; PMID:25424692; http://dx.doi.org/ 10.4161/15592294.2014.969637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collerton J, Gautrey HE, van Otterdijk SD, Davies K, Martin-Ruiz C, von Zglinicki T, Kirkwood TB, Jagger C, Mathers JC, Strathdee G. Acquisition of aberrant DNA methylation is associated with frailty in the very old: findings from the Newcastle 85+ Study. Biogerontology 2014; 15:317-28; PMID:24770842; http://dx.doi.org/ 10.1007/s10522-014-9500-9 [DOI] [PubMed] [Google Scholar]
- 31.Bellizzi D, D'Aquila P, Montesanto A, Corsonello A, Mari V, Mazzei B, Lattanzio F, Passarino G. Global DNA methylation in old subjects is correlated with frailty. Age 2012; 34:169-79; PMID:21336567; http://dx.doi.org/ 10.1007/s11357-011-9216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerard A, Patino-Lopez G, Beemiller P, Nambiar R, Ben-Aissa K, Liu Y, Totah FJ, Tyska MJ, Shaw S, Krummel MF. Detection of rare antigen-presenting cells through T cell-intrinsic meandering motility, mediated by Myo1g. Cell 2014; 158:492-505; PMID:25083865; http://dx.doi.org/ 10.1016/j.cell.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koks G, Uudelepp ML, Limbach M, Peterson P, Reimann E, Koks S. Smoking-Induced Expression of the GPR15 Gene Indicates Its Potential Role in the Chronic Inflammatory Pathologies. Am J Pathol 2015; 185:2898-2906; PMID:26348578; http://dx.doi.org/ 10.1016/j.ajpath.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 34.Larson KC, Lipko M, Dabrowski M, Draper MP. Gng12 is a novel negative regulator of LPS-induced inflammation in the microglial cell line BV-2. Inflamm Res 2010; 59:15-22; PMID:19568691; http://dx.doi.org/ 10.1007/s00011-009-0062-2 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Breitling LP, Balavarca Y, Holleczek B, Schöttker B, Brenner H. Comparison and combination of blood DNA methylation at smoking-associated genes and at lung cancer-related genes in prediction of lung cancer mortality. Int J Cancer 2016; 139:2482-92; PMID:27503000; http://dx.doi.org/ 10.1002/ijc.30374 [DOI] [PubMed] [Google Scholar]
- 36.Dai ZJ, Liu XH, Ma YF, Kang HF, Jin TB, Dai ZM, Guan HT, Wang M, Liu K, Dai C, et al.. Association Between Single Nucleotide Polymorphisms in DNA Polymerase Kappa Gene and Breast Cancer Risk in Chinese Han Population: A STROBE-Compliant Observational Study. Medicine (Baltimore) 2016; 95:e2466; PMID:26765445; http://dx.doi.org/ 10.1097/MD.0000000000002466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav S, Mukhopadhyay S, Anbalagan M, Makridakis N. Somatic Mutations in Catalytic Core of POLK Reported in Prostate Cancer Alter Translesion DNA Synthesis. Hum Mutat 2015; 36:873-80; PMID:26046662; http://dx.doi.org/ 10.1002/humu.22820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michiels S, Danoy P, Dessen P, Bera A, Boulet T, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis 2007; 28:1731-9; PMID:17494052; http://dx.doi.org/ 10.1093/carcin/bgm111 [DOI] [PubMed] [Google Scholar]
- 39.Ravenni N, Weber M, Neri D. A human monoclonal antibody specific to placental alkaline phosphatase, a marker of ovarian cancer. MAbs 2014; 6:86-94; PMID:24247025; http://dx.doi.org/ 10.4161/mabs.27230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol 2015; 16:1; PMID:25583448; http://dx.doi.org/ 10.1186/s13059-014-0569-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard RE, Lang IA, Llewellyn DJ, Rockwood K. Frailty, body mass index, and abdominal obesity in older people. J Gerontol A Biol Sci Med Sci 2010; 65:377-81; PMID:19942592; http://dx.doi.org/20559726 10.1093/gerona/glp186 [DOI] [PubMed] [Google Scholar]
- 42.Fulop T, Larbi A, Witkowski JM, McElhaney J, Loeb M, Mitnitski A, Pawelec G. Aging, frailty and age-related diseases. Biogerontology 2010; 11:547-63; PMID:20559726; http://dx.doi.org/ 10.1007/s10522-010-9287-2 [DOI] [PubMed] [Google Scholar]
- 43.Saum KU, Dieffenbach AK, Muezzinler A, Muller H, Holleczek B, Stegmaier C, Butterbach K, Schick M, Canzian F, Stammer H, et al.. Frailty and telomere length: cross-sectional analysis in 3537 older adults from the ESTHER cohort. Exp Gerontol 2014; 58:250-5; PMID:25150678; http://dx.doi.org/ 10.1016/j.exger.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 44.Woo J, Tang NL, Suen E, Leung JC, Leung PC. Telomeres and frailty. Mech Ageing Dev 2008; 129:642-8; PMID:18809425; http://dx.doi.org/ 10.1016/j.mad.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 45.Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 2016; 8:21; PMID:26925173; http://dx.doi.org/ 10.1186/s13148-016-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, et al.. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 2015; 44:1388-96; PMID:25617346; http://dx.doi.org/8055125 10.1093/ije/dyu277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Zhang Y, Breitling LP, Brenner H. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget 2016; PMID:27276709; http://dx.doi.org/8055125 10.18632/oncotarget.9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz J, Weiss ST. Cigarette smoking and peripheral blood leukocyte differentials. Ann Epidemiol 1994; 4:236-42; PMID:8055125; http://dx.doi.org/ 10.1016/1047-2797(94)90102-3 [DOI] [PubMed] [Google Scholar]
- 49.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86; PMID:22568884; http://dx.doi.org/ 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2013; 2:e00523; PMID:23755361; http://dx.doi.org/ 10.7554/eLife.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23:R89-98; PMID:25064373; http://dx.doi.org/ 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schöttker B, Haug U, Schomburg L, Kohrle J, Perna L, Muller H, Holleczek B, Brenner H. Strong associations of 25-hydroxyvitamin D concentrations with all-cause, cardiovascular, cancer, and respiratory disease mortality in a large cohort study. Am J Clin Nutr 2013; 97:782-93; PMID:23446902; http://dx.doi.org/ 10.3945/ajcn.112.047712 [DOI] [PubMed] [Google Scholar]
- 53.Gao X, Mons U, Zhang Y, Breitling LP, Brenner H. DNA methylation changes in response to active smoking exposure are associated with leukocyte telomere length among older adults. Eur J Epidemiol 2016; 31: 1231-1241; PMID:27832427; http://dx.doi.org/3344216 10.1007/s10654-016-0210-2 [DOI] [PubMed] [Google Scholar]
- 54.Gao X, Zhang Y, Breitling LP, Brenner H. Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget 2016; 7:59017; PMID:27323854; http://dx.doi.org/3344216 10.18632/oncotarget.10007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16:1215; PMID:3344216; http://dx.doi.org/ 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schöttker B, Brenner H. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia 2015; 59:130-8; PMID:26433941; http://dx.doi.org/ 10.1007/s00125-015-3773-7 [DOI] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol 1995; 57:289-300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


