Summary
Naïve T cell stimulation activates anabolic metabolism to fuel the transition from quiescence to growth and proliferation. Here we show that naïve CD4+ T cell activation induces a unique program of mitochondrial biogenesis and remodeling. Using mass spectrometry, we quantified protein dynamics during T cell activation. We identified substantial remodeling of the mitochondrial proteome over the first 24 hr of T cell activation to generate mitochondria with a distinct metabolic signature, with one carbon metabolism as the most induced pathway. Salvage pathways and mitochondrial one carbon metabolism, fed by serine, contribute to purine and thymidine synthesis to enable T cell proliferation and survival. Genetic inhibition of the mitochondrial serine catabolic enzyme SHMT2 impaired T cell survival in culture, and antigen-specific T cell abundance in vivo. Thus, during T cell activation, mitochondrial proteome remodeling generates specialized mitochondria with enhanced one carbon metabolism that is critical for T cell activation and survival.
Introduction
To support proliferation, activated T cells, like tumor cells, utilize metabolism to generate precursors required for macromolecular synthesis, energy, stress response and other pro-survival pathways (German and Haigis, 2015; Hirschey et al., 2015; MacIver et al., 2013; Pearce et al., 2013). To initiate an immune response against pathogens, a small number of antigen-specific T cells within the polyclonal repertoire need to proliferate rapidly to generate large numbers of effector cells that can clear pathogens. In T cells, exit from quiescence and entry into the cell cycle is determined by external cues of activation, which also regulate the switch from catabolic to anabolic metabolism. Upon naïve T cell activation, signaling pathways downstream of the T cell receptor through Erk and downstream of the CD28 costimulatory receptor through phosphatidylinositol 3′-kinase (PI3K)/Akt stimulate glucose and glutamine uptake and metabolism (Carr et al., 2010; Frauwirth et al., 2002).
Mitochondria are not inert during metabolic rewiring in T cells. For example, T cell activation induces metabolic flux through the tricarboxylic acid (TCA) cycle to generate citrate for lipid biosynthesis and provide electron donors for the electron transport chain (ETC) (Berod et al., 2014). Activation of the ETC generates energy, and is important for signaling events during T cell activation, mediated in part, through reactive oxygen species (ROS), which stimulate T cell expansion and cytokine production (Sena et al., 2013). Furthermore, effector T cells contain higher numbers of mitochondria than naïve cells (D’Souza et al., 2007; van der Windt et al., 2012). However, the role of mitochondrial proliferation in T cell activation and survival has not been well defined, and highlights a fundamental question: Do cells replicate mitochondria solely to increase their existing metabolic and energetic function, or does mitochondrial proliferation allow development of a new population of organelles with distinct and specialized function?
Here, we describe a synchronized program of mitochondrial biogenesis during activation of naïve T cells. We quantified dynamic changes in mitochondrial and cellular protein composition during the initial activation of naive CD4+ T cells in vitro by conducting mass spectrometry analyses at 4, 9 and 24 hr post-activation. Using this global approach, we discovered mitochondrial proteome remodeling, which resulted in organelles with a new metabolic signature enriched for one carbon metabolism. We demonstrate that this mitochondrial remodeling occurs also in vivo after physiological stimulation of T cells. The resulting enhanced mitochondrial one carbon metabolism is critical for sustained activation and survival of naïve CD4+ T cells in vitro and in vivo.
Results
Naïve CD4+ T cell activation initiates a synchronized program of mitochondrial biogenesis
To systematically define mitochondrial proliferation during early T cell activation, we utilized a well-established system for in vitro activation of purified, sorted naïve CD62LhiCD44loCD25− CD4+ T cells (Figure S1A), using a combination of anti-CD3/anti-CD28 antibodies to mimic the T cell receptor-mediated signal and the CD28 costimulatory signal (Figure 1A). As expected, activation of T cells using these conditions increased cell size (Figure 1B), upregulated early activation markers CD69 and CD25, and downregulated L-selectin (CD62L) (Figure 1C). Proliferation occurred at 48 and 72 hr post-activation (Figure 1D). Standard outputs of metabolic activation, such as glucose and glutamine uptake and lactate secretion were significantly increased (Figure 1E, S1B, and 1F). In addition, intermediates in glycolysis, the pentose phosphate pathway and the TCA cycle, were increased by 4 hr post-activation (Figure 1G). Thus, this reconstituted system of T cell activation recapitulates metabolic reprogramming observed in vivo and identifies a 24-hr window for studying mitochondria prior to cell division.
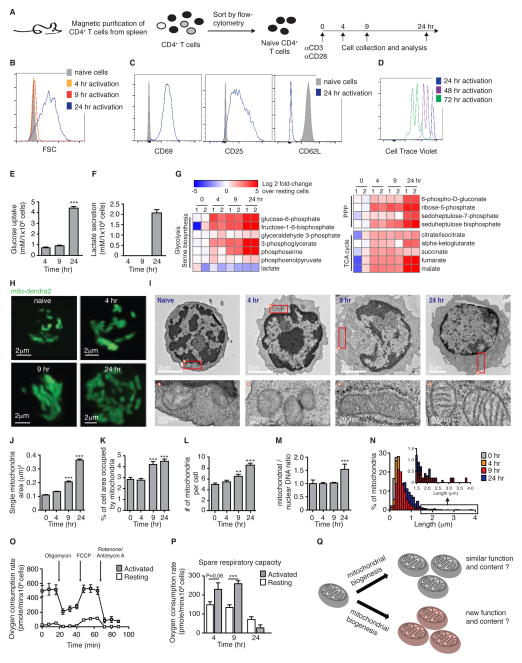
Figure 1. Activation of naïve CD4+ T cell initiates a synchronized program of mitochondrial biogenesis and bioenergetics.
(A) Scheme of experimental design. Purified naïve CD4+ T cells were activated, and harvested at 0, 4, 9 and 24 hr post-activation as indicated. Representative flow-cytometry plots (from >5 individual experiments) demonstrate: (B) activation-induced growth in cell size, (C) up-regulation of CD69 and CD25, and down regulation of the L-selectin CD62L. (D) Representative FACS histograms demonstrating cell proliferation (detected by dilution of Cell Trace Violet) occurring at 48 and 72 hr post-activation. (E) Glucose uptake, and (F) lactate secretion were measured by analyzing the growth media of CD4+ T cells purified and activated as described in Figure 1A (n=4 pools of 4 mice each). (G) Kinetic changes in metabolite levels in activated vs. naïve CD4+ T cells. (H) Representative 3D reconstructions of Z-stacks taken by live-cell imaging of activated CD4+ T cells from PHAMexcised mice. (I) Representative EM micrographs of naive CD4+ T cells at 0, 4, 9 and 24 hr post-activation (scale bar = 2 μm). Rectangle indicates region represented in lower panels, with a scale bar of 200 nm. Asterisk indicates orientation of panel. (J–L) Quantitation of the EM micrographs (n=30 images per sample, 4–5 samples time point) showed changes over time in single mitochondrial area (J), the % of cell area occupied by the mitochondria (K) and the number of mitochondria per cell (L). (M) qPCR analysis of mitochondrial (Cox-I) vs. nuclear (Rp18s) DNA content (n=4 pools of 4 mice). (N) Quantitation of mitochondrial length in EM micrographs. (O) Oxygen consumption rate in resting versus activated (24 hr) T cells. (P) Spare respiratory capacity calculated based on the changes in oxygen consumption rates (n=5 pools of 2 mice each). All results are mean ± SEM of 2–3 individual experiments. **p<0.01, ***p<0.001. (Q) A schematic illustrating potential roles of mitochondrial biogenesis to: 1) increase mitochondria with a similar function or 2) to generate a new population of specialized mitochondria.
To visualize mitochondrial biogenesis during early T cell activation, we utilized live-cell microscopy in T cells isolated from PhAMexcised mice, which ubiquitously express a mitochondrial-localized version of the fluorescent protein dendra-2 (Pham et al., 2012). Images at 4, 9 and 24 hr post-activation revealed a dramatic increase in mitochondrial mass (Figure 1H). To quantify these prominent changes, we examined mitochondria by electron microscopy (EM) (Figure 1I). Mitochondrial area increased by 2- and 4-fold at 9 and 24 hr post-activation, respectively (Figure 1J). As expected, activation induced cell growth (Figures S1C), but the relative area occupied by mitochondria in the cytosol increased significantly (Figure 1K). Mitochondrial number (Figure 1L) and mitochondrial DNA (mtDNA; Figure 1M) also doubled. These studies reveal a unique model of robust and synchronized mitochondrial biogenesis in early T cell activation.
Evidence of synchronicity was apparent from distinct stages of mitochondrial morphology. Naïve CD4+ T cells contained fragmented, rounded mitochondria. After 9 hr, mitochondria appeared hyperfused, and this elongated intermediate resolved back to a fragmented morphology by 24 hr (Figure 1H, 1I and 1N). We connected these morphological changes to bioenergetic capacity (Figure S1D–E). Strikingly, after mitochondria proliferated (24 hr post-activation), basal respiration reached full capacity, 10-fold increased compared to resting cells (Figures 1O and S1F–G), leaving no spare respiratory capacity (Figure 1P). Thus, T cells induce a program of synchronized mitochondrial activation and biogenesis, which occurs within 24 hr, resulting in mitochondria which function at their maximal respiratory rate.
Quantitative proteomics identifies mitochondrial remodeling
This synchronized system of mitochondrial biogenesis provided us with a unique opportunity to address the fundamental question of whether biogenesis merely replicates existing mitochondria, or generates a distinct population of mitochondria with specialized function (Figure 1Q). We quantified changes in the cellular proteome during T cell activation using tandem mass spectrometry after isobaric peptide tagging, and analyzed the dynamics of the global proteome, as well as proteins within the mitochondria (Figure 2A). We quantified over 5500 proteins and their expression kinetics during the first 24 hr of T cell activation, prior to cell division (Table S1). Reproducibility was high between biological replicates (Figure 2B, S2A). By 24 hr post-activation, the majority of cellular proteins were increased by at least 2-fold (Figure 2C), as anticipated given the massive cell growth (Figure 1B). Proteins were grouped into clusters according to kinetics and magnitude of induction, with some proteins induced more than 500-fold (Figure 2D–2E, S2B). Cluster 1 (induced 6–100 fold at 4 hr), represented the earliest-induced proteins, which we speculate could be ‘drivers’ of T cell activation. Consistent with this idea, cluster 1 included known early activation markers (CD69 and CD25) and transcriptional regulators of T cell activation and function (MYC, NR4A, JUNB, NFKBID; Figure 2F) (Arnold et al., 2012; Sekiya et al., 2013; Wang et al., 2011). Thus, this dataset may serve as a hypothesis-generating tool for identifying novel immune regulators and for identifying drivers of exit from quiescence and/or entry into cell cycle.
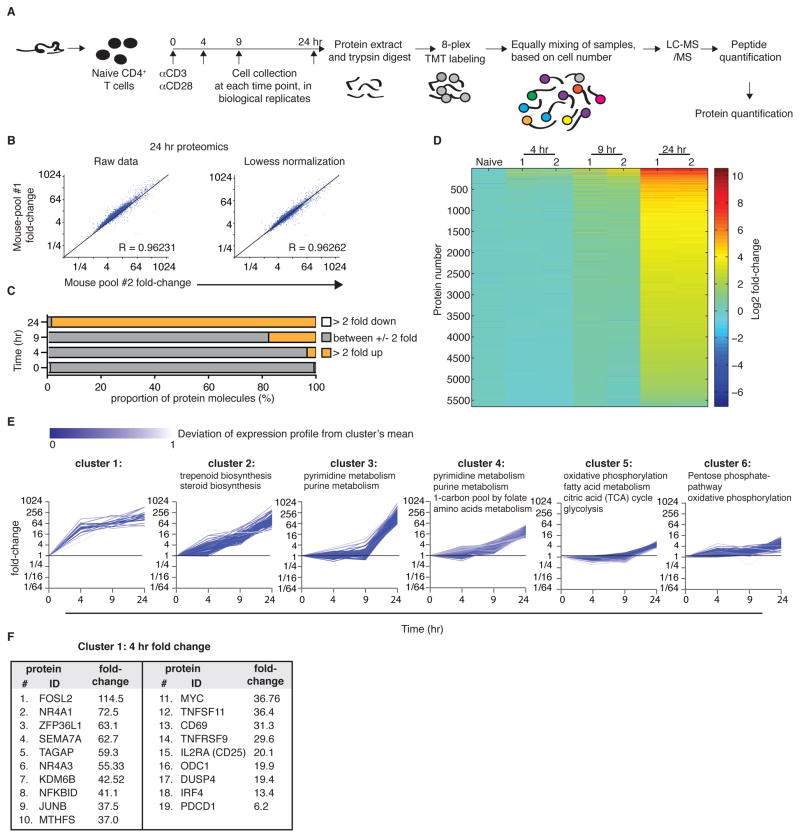
Figure 2. Quantitative proteomics identifies differential induction of metabolic pathways during activation of naïve CD4+ T cells.
(A) Experimental scheme. Naïve CD4+ T cells were purified from two separate pools of mice, activated using plate-bound anti-CD3/anti-CD28, collected, and processed by protein extraction and digestion. The peptide pool from each of the 8 samples was labeled with a specific TMT label. Pools were equally mixed, based on cell numbers and analyzed by LC-MS/MS, to quantify proteins. (B) Scatter plots showing the biological replicates at 24 hr are well correlated. (C) Graph demonstrating the overall induction in protein content during T cell activation. Values are the average of the two biological replicates at each time. (D) Heatmap showing the kinetics of changes in CD4+ T cell proteomics following activation. (E) 6 Representative clusters (see Figure S2 for clusters 7–12) of proteins that share similar expression kinetics during T cell activation. KEGG pathway analysis identified the specific metabolic pathways enriched within each cluster. (F) A list of the proteins in cluster 1, representing the proteins with the greatest induction at 4 hr.
Surprisingly, metabolic pathways associated with activated T cells, like glycolysis, pentose phosphate pathway, and oxidative phosphorylation were enriched within clusters 5 and 6, in the group of least-induced proteins (Figure 2E). Instead, clusters associated with highest induction were enriched for enzymes of lipid biosynthesis (cluster 2) and nucleotide metabolism (clusters 3 and 4; Figure 2E). Thus, our proteomic data point to major nodes of metabolic reprogramming yet to be examined in T cell activation.
To examine specifically the mitochondrial proteome, we used the MitoCarta database (Pagliarini et al., 2008) as a filter to generate a list of 552 mitochondrial proteins detected in our database. Most mitochondrial proteins were induced upon CD4+ T cell activation (Figure 3A). Yet, we observed a high degree of variability in the magnitude of induction, ranging from 2-fold to greater than 200-fold induction (Figure 3A), indicating that mitochondrial biogenesis is accompanied by substantial mitochondrial proteome remodeling. To identify the pathways enriched or underrepresented, we performed gene set enrichment analysis (GSEA) on mitochondrial proteins, based on the proteins’ individual induction Z score at each time point (Figure 3B). One carbon metabolism was the only significantly induced pathway, while fatty acid catabolism, TCA cycle and oxidative phosphorylation were among the suppressed pathways (Figure 3B).
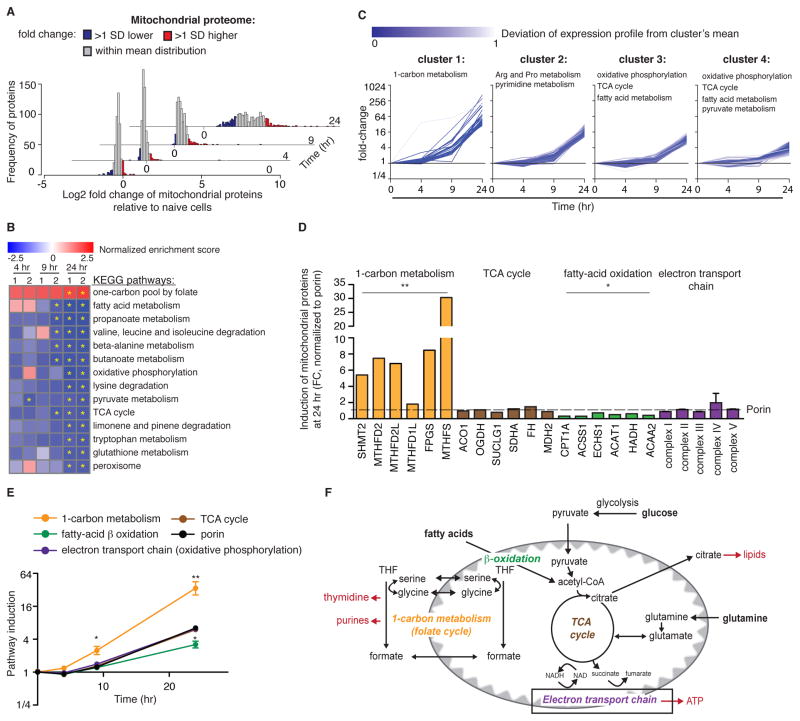
Figure 3. Mitochondrial protein composition is changed with T cell activation.
(A) Kinetic distribution of mitochondrial proteome induction following CD4+ T cell activation. Color codes show in blue: proteins induced more than one standard deviation below mean distribution, in red: proteins induced more than one standard deviation above mean distribution, and in grey: proteins induced within mean distribution. Values are the average of the two biological replicates at each time. (B) GSEA analysis of the mitochondrial proteome indicating in blue, pathways that were significantly downregulated by 24 hr post-activation, and in red: pathways that were significantly upregulated by 24 hr post-activation. Analysis was performed on the full list of mitochondrial proteins, pre-ranked based on their fold-change induction compared to naïve T cells. Yellow asterisk indicates P<0.05. (C) The mitochondrial proteome was segregated into 4 clusters based on protein level kinetics following T cell activation. KEGG pathway analysis identified the specific metabolic pathways enriched within each cluster. (D) Fold-change induction of individual proteins in pathways of one carbon metabolism, TCA cycle, fatty acid oxidation 24 hr post-activation. Electron transport chain complexes are shown as the average level of individual subunits within each complex. Values are the average of two biological replicates, normalized to porin (E) Average induction of mitochondrial metabolic pathways from panel D compared to porin. (F) Schematic of central metabolic pathways in the mitochondria. Results are mean ± SEM. *p<0.05, **p<0.01 (Student’s t-test comparing each of the metabolic pathways to porin).
We next classified mitochondrial proteins into four clusters (clusters 1–4), based on the kinetics and magnitude of induction (Figure 3C). KEGG pathway analysis highlighted one carbon metabolism (folate cycle) as the only significantly enriched pathway in cluster 1 that represents the earliest and most highly induced proteins (Figure 3C). Interestingly, this analysis also identified significant enrichment in enzymes involved in oxidative phosphorylation and the TCA cycle in the least-induced clusters, whose proteome fraction decreased during T cell activation (clusters 3 and 4). The selective elevation of enzymes in the mitochondrial one carbon pathway compared to the TCA cycle, electron transport chain, and fatty acid oxidation was apparent when we compared the individual proteins within these pathways (Figure 3D,E). While enzymes of the TCA cycle and electron transport chain were induced similarly to porin, enzymes in fatty acid oxidation were under-represented. In contrast, enzymes in the one carbon metabolic pathway were induced 6–30 fold greater than porin (Figure 3D,E). Taken together, our analysis of the mitochondrial proteome demonstrated that T cell activation induces mitochondrial proliferation and proteome remodeling, leading to enrichment in enzymes of one carbon metabolism (Figures 3D–F).
Enzymes of the one carbon metabolism are induced in vivo during T cell activation
To validate the unique proteomic signature in early T cell activation, we optimized a set of antibodies to probe proteins by Western blotting or FACS analysis. First, we compared the expression of enzymes in the one carbon metabolic pathway in naive and activated cells by Western blotting. We observed a massive induction of these enzymes (SHMT1, SHMT2, MTHFD1, MTHFD2), as well as in enzymes of pyrimidine biosynthesis (DHODH, UMPS), consistent with our proteomic dataset. This observation contrasted with metabolic proteins that were not elevated (MDH2, OGDH, ASL), or proteins that were significantly under-expressed (CPT1A, OTC) in our proteomic dataset (Figures 4A–B). These results are striking, as enzymes within the TCA cycle, ETC, or fatty acid oxidation are the typical markers of mitochondrial biogenesis.
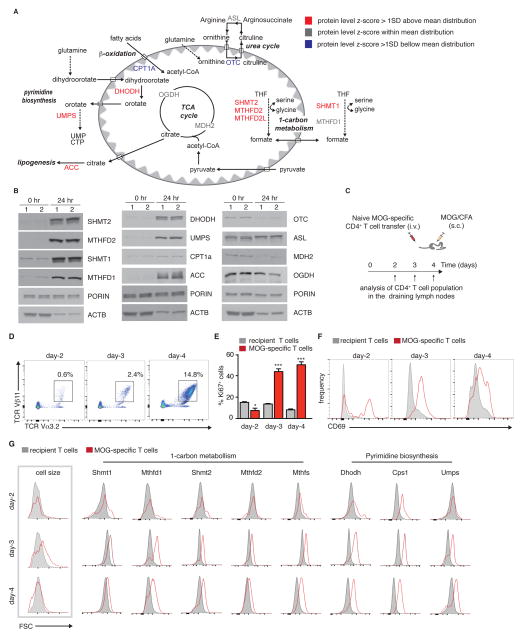
Figure 4. Enzymes involved in one carbon metabolism and pyrimidine biosynthesis are induced in vivo in antigen specific T cells.
(A) Schematic showing central metabolic pathways in the mitochondria, listing representative enzymes, color-coded based on their fold-change at 24 hr post-activation. In blue: proteins induced more than one standard deviation below mean distribution, in red: proteins induced more than one standard deviation above mean distribution, and in grey: proteins induced within mean distribution. (B) Protein quantitation by western blot of the enzymes listed in (A) using porin and β-actin as loading controls. (C) Experimental strategy. Naïve, MOG-specific CD4+ T cells were transferred into C57Bl/6 wild-type recipients, which were immunized with MOG35-55 peptide in CFA. Mice (n=4 per time point) were sacrificed at days 2, 3 and 4 following immunization for analysis of the T cells in the draining lymph nodes by flow cytometry. (D) Representative plots showing percentages of MOG-specific T cells TCRβ11/TCRα3.2 in the draining lymph nodes following immunization, and (E) percentages of proliferating 2D2 MOG-specific cells (Ki67+), compared to host CD4+ T cell population. *p<0.05, ***p<0.001 (Student’s t-test comparing wild-type and 2D2-specific T cells at each time point). (F) Representative plots showing the kinetics of CD69 expression on MOG-specific 2D2 T cells following immunization. (G) Representative plots showing changes in cell size (FSC), and expression of metabolic enzymes in MOG-specific 2D2 T cells following immunization by flow cytometry. N=4 mice per time point in each experiment. Experiments were performed 2–3 times. Results are mean ± SEM.
To test if this mitochondrial and metabolic rewiring recapitulated in a physiological setting, we examined expression of enzymes after in vivo T cell activation. We transferred myelin oligodendrocyte (MOG) antigen-specific 2D2 TCR transgenic CD4+ T cells into C57Bl/6 wild-type mice, immunized the mice with MOG 35–55 peptide, and monitored antigen-specific T cell activation in the draining lymph nodes by flow cytometry on days 2, 3 and 4 after immunization (Figure 4C). As anticipated, 2D2 TCR transgenic T cells (TCR Vα3.2+/Vβ11+) accumulated in the draining lymph nodes (Figure 4D) and proliferated (Figure 4E). CD69, an early T cell activation marker, was induced only on MOG-specific T cells following immunization (Figure 4F).
Next, we assessed metabolic reprograming in activated MOG-specific CD4+ T cells by measuring the expression of enzymes involved in one carbon metabolism and pyrimidine biosynthesis by flow cytometry. Strikingly, many of these enzymes were induced in antigen-specific T cells as early as 2 days post-immunization (Figure 4G), in parallel to induction of CD69 (Figure 4F) and prior to cell division (Figure 4E). Thus, this proteomic dataset defines a distinct metabolic signature, which provides a novel tool for probing early metabolic activation of T cells in single cells by FACS and in vivo.
T cells activate mitochondrial one carbon metabolism
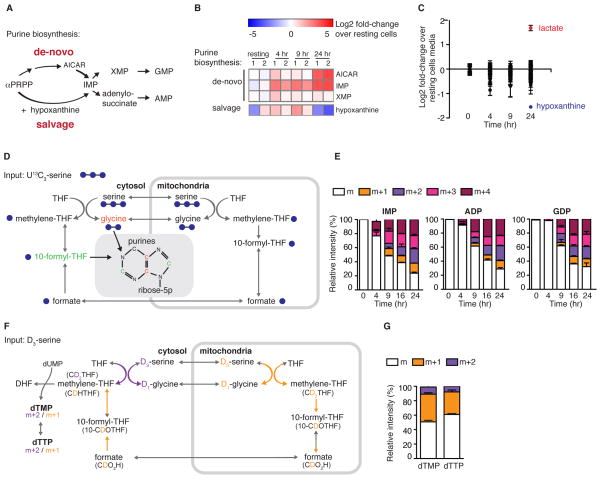
Carbon units derived from the folate-dependent one carbon pathway are used for thymidylate and de-novo purine biosynthesis. In addition, existing purine derivatives, like hypoxanthine, can be recycled in salvage pathways to generate new purines (Figure 5A). Thus, we examined the levels of these metabolites during T cell activation. Consistent with proteomic data, IMP (inosine monophosphate), generated by purine salvage and de novo biosynthesis, was induced 4–6 fold as early as 4 hr post-activation (Figure 5B). Interestingly, by 24 hr, the intracellular level of AICAR, a substrate that precedes IMP in de novo purine biosynthesis was highly induced (10–13 fold), while hypoxanthine, a substrate used for IMP synthesis in the purine salvage pathway was depleted (Figure 5B). Furthermore, hypoxanthine was the most depleted metabolite in the culture media by 24 hr post-activation (Figure 5C), suggesting that T cells engage purine salvage and de novo biosynthesis early upon activation.
Figure 5. Mitochondrial one carbon metabolism is induced in CD4+ T cells upon activation, and contributes to de novo purine biosynthesis.
(A) Schematic showing major metabolic pathways contributing to purine biosynthesis. αPRPP – phosphoribosyl pyrophosphate; AICAR-5-Aminoimidazole carboxamide ribonucleotide; IMP – inosine monophosphate; XMP-xanthosine monophosphate; GMP – guanosine monophosphate; AMP-adenosine monophosphate. (B) Heatmap showing Log2 fold-change of intermediates in purine biosynthetic pathways in activated compared to naïve T cells, measured by LC-MS. (C) Changes in culture media composition in cultures of naïve T cells and of T cells at 4, 9 and 24 hr post-activation, highlighting lactate secretion (red) and hypoxanthine consumption (blue). (D) Metabolic tracing strategy of the one carbon metabolic pathway using uniformly labeled 13C-serine, highlighting the incorporation of its products: glycine (13C2) and two molecules of 10-formyl-THF (13C1) into the purine ring, thru de-novo purine biosynthesis. Incorporation of one molecule of 13C 10-formyl-THF will give rise to a mass of m+1. m+2 is the result of incorporation of two molecules of 13C 10-formyl-THF or one labeled 13C2-glycine. m+3 indicates addition of one molecule of 13C 10-formyl-THF and one labeled 13C2-glycine, and m+4 is the result of addition of two 13C 10-formyl-THF molecules and one labeled 13C2-glycine. THF: tetrahydrofolate. (E) 13C labeling of representative purine molecules. (F) Metabolic tracing strategy using D3-serine, to differentiate flux thru the mitochondrial versus the cytosolic arm of one carbon metabolism by monitoring the labeling pattern of thymidylate. DHF: dihydrofolate. (G) Activated T cells produce predominantly the m+1 isotopomer of dTMP and dTTP, indicative of mitochondrial rather than cytosolic flux. All results are mean ± SEM.
To test directly whether metabolic flux through the one carbon pathway was induced by T cell activation, we stimulated naïve CD4+ T cells in media containing uniformly labeled 13C3-serine (U13C-serine). Metabolism of U13C-labeled serine by the one carbon metabolic pathway generates uniformly labeled 13C2-glycine and 13C-labeled 10-formyl-THF, which are consumed by the de novo purine biosynthetic pathway, resulting in 13C labeling of purines (Figure 5D). IMP labeling was evident after 4 hr of activation, followed by the labeling of down-stream purine species at 9 hr (Figure 5E, S3A–E). Serine can be synthesized from 3-phosphoglycerate, an intermediate in glycolysis. To test whether T cells engage in de-novo serine biosynthesis, we monitored 13C incorporation into phosphoserine and serine following stimulation in media containing uniformly labeled 13C6-glucose (U13C6-glucose; Figure S3F). Indeed, in T cells activated for 24 hr, a significant fraction of phosphoserine and serine was labeled with 13C (Figure S3G–H). Taken together, our metabolic tracing analysis confirmed that CD4+ T cells utilized one carbon metabolism upon activation, and highlighted serine as an important donor of one carbon units.
Mammalian cells contain parallel pathways in the mitochondria and cytosol for generating one carbon units from serine, and enzymes of both cellular compartments were highly induced with T cell activation (Figures 3 and 4). To test whether both compartments contributed to production of one carbon units in activated T cells (Figure 5E), we utilized a strategy based on 2,2,3-2H3-serine (D3-serine) tracing (Figure 5F; (Herbig et al., 2002)). Metabolism of D3-serine via the mitochondrial arm ultimately generates cytosolic methylene-THF (after export from mitochondria as formate) labeled with one deuterium (CDHTHF), whereas direct D3-serine metabolism by the cytosolic arm generates methylene-THF labeled with two atoms of deuterium (CD2THF). These two species give rise to dTMP and dTTP labeled with either one or two deuteriums (m+1 and m+2; Figure 5F). Analysis of dTMP and dTTP labeling patterns showed a higher fraction of the m+1 compared to m+2 (Figure 5G). Thus, in activated T cells, the majority of one carbon units are generated in the mitochondria.
SHMT2 is critical for mitochondrial one carbon metabolism and T cell survival
We probed the physiological relevance of mitochondrial one carbon metabolism by studying the effect of the knockdown (KD) of SHMT2, the first enzyme in this pathway (Figure 6A). To genetically manipulate primary CD4+ T cells, we developed a protocol in which naïve CD4+ T cells were activated with anti-CD3/anti-CD28, transduced 24 hours later with a retroviral vector, rested in IL-2/IL-7 and restimulated by anti-CD3/anti-CD28 (Figure S4A; modified from (Kurachi et al., 2014)). Upon restimulation, these ‘resting’ T cells increased in size and proliferated (Figure S4B, S4C), upregulated the early activation markers CD69 and CD25, and downregulated CD62L (Figure S4D). Reactivation also induced a 2-fold increase in mtDNA (Figure S4E). Importantly, this induction of mitochondrial mass was accompanied by a large increase in SHMT2 expression (Figure S4F). Thus, restimulated CD4+ T cells showed similar immune activation markers, cell proliferation and mitochondrial biogenesis as naïve T cells during their initial activation.
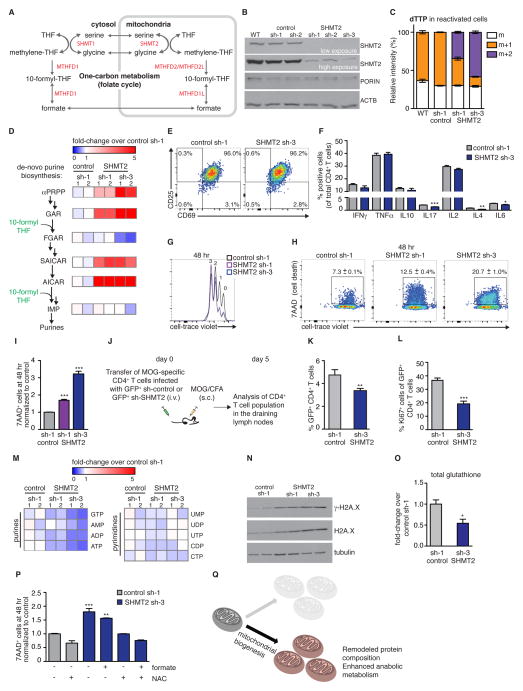
Figure 6. Genetic inhibition of mitochondrial one carbon metabolism impairs T cell survival in vitro and in vivo.
(A) Schematic of enzymes in mitochondrial and cytosolic one carbon metabolism. SHMT: Serine hydroxymethyltransferase; MTHFD: methylenetetrahydrofolate dehydrogenase; MTHFD1L: methylenetetrahydrofolate dehydrogenase 1-like. (B) Protein quantitation of SHMT2 in non-infected cells (WT) or cells infected with retrovirus containing individual shRNA sequences to target LacZ (control) or SHMT2. Resting CD4+ T cells infected with either sh-LacZ (control, sh-1) or sh-SHMT2 (sh-1, sh-3) were used for the following experiments (C–O): (C) Cells were reactivated in media containing D3-serine for 48 hr, and m+1 and m+2 dTTP isotopomer were analyzed by LC-MS. (D) Pathway on left indicates metabolites in de novo purine synthesis, 10-formyl THF (green) indicates addition of one carbon units. Double arrows indicate multiple enzyme steps. Heat map shows fold-change in metabolite in SHMT KD versus control LacZ sh cells (48 hr after reactivation). (E–I) Representative plots from control and SHMT2 knockdown T cells 48 hr after reactivation showing (E) activation markers (CD69 and CD25) (F) intracellular cytokine levels, (G) cell proliferation, and (H–I) survival (measured by 7-AAD incorporation). (J) Experimental strategy. Resting CD4+, MOG-specific 2D2 TCR transgenic T cells infected with retrovirus expressing GFP+ sh-control or GFP+ sh-SHMT2 were adoptively transferred into wild type C57BL/6 recipient mice that were immunized with MOG/CFA. 5 days post-immunization, the mice were sacrificed and the draining lymph nodes analyzed by FACS. (K) % GFP+ cells of total CD4+ T cells, in the draining lymph node. (L) % of proliferating cells (Ki67+) of infected (GFP+) CD4+ T cells. Experiment was performed two times with n=7 mice per group for each experiment. (M) Cells were reactivated for 48 hr as for panel D. Heatmap summarizes the levels of purines (left panel) and pyrimidines (right panel). (N) Protein quantitation of γ-H2A.X at 48 hr post reactivation. (O) Total glutathione levels at 48 hr post-reactivation, measured by LC-MS. (P) Cells were reactivated +/− formate (1 mM) and +/− NAC (7.5 mM) and analyzed for cell death (7-AAD+) by flow cytometry. (Q) Model: mitochondrial biogenesis in T cells gives rise to remodeled mitochondria with enhanced anabolic functions. **p<0.01, ***p<0.001 (Student’s t-test (F,K,L); One-Way ANOVA followed by Tukey’s multiple comparisons test (I,O), showing significant changes over sh-control cell (G). Results are mean ± SEM.
Using this optimized strategy, naïve CD4+ T cells were infected with retrovirus targeting multiple sequences for either GFP+ sh-LacZ (control sh-1-2) or GFP+ sh-SHMT2 (SHMT2 sh-1-3). After 5 days of ‘resting’, the cells were sorted for GFP+ cells, reactivated, and SHMT2 depletion was validated by western blotting (Figure 6B). Sequences for SHMT2 sh-1 and SHMT2 sh-3 demonstrated efficient knockdown and were used for further experiments. To test if reduction of SHMT2 blocked mitochondrial one carbon metabolism, we performed metabolic flux analysis with D3-serine in the SHMT2 KD and control cells (as in Figure 5F). Knockdown of SHMT2 decreased the mitochondrial contribution of one carbon units to the total cellular pool (m+1 dTTP), while increasing the share of one carbon units derived from the cytosolic pathway (m+2 dTTP; Figure 6C).
We next used, LC-MS to measure levels of intermediates in de novo purine biosynthesis (Figure 6D). SHMT2 knockdown resulted in an accumulation of α–PRPP, GAR, SAICAR, and AICAR metabolites upstream of 10-formyl THF incorporation. Notably, FGAR, a product of 10-formyl-THF reaction with GAR transformylase, was reduced in SHMT2 sh-3 cells, which had the lowest levels of SHMT2 (Figure 6D). Levels of IMP, which is also downstream of the salvage pathway (Figure 5A), were not affected (Figure 6D). These data suggest that the mitochondrial pathway is critical for supplying one carbon units for de novo purine synthesis.
We next asked if mitochondrial one carbon metabolism was critical for T cell activation. We measured early activation markers CD69 and CD25, and found that levels of these surface molecules were comparable in control and SHMT2 KD T cells (Figure 6E), indicating that expression of these immune activation markers was not dependent on mitochondrial one carbon metabolism. In addition, intracellular levels of key cytokines were not strongly affected by SHMT2 deletion (Figure 6F). Likewise, SHMT2 KD cells displayed comparable cell proliferation to control cells (Figures 6G and S5A). Notably, T cells with low SHMT2 showed a 2–3-fold increase in cell death compared with control cells, at 48 and 72 hr post-reactivation (Figures 6H,I and S5B,C), demonstrating that mitochondrial one carbon metabolism is essential for T cell survival and cannot be fully compensated by increased purine scavenging or flux through cytosolic one carbon pathway.
To assess the physiological significance of SHMT2 in T cells in vivo and at later time points, we adoptively transferred MOG-specific 2D2 CD4+ T cells that had been infected with either GFP+ SHMT2 sh-3 or GFP+ control sh-1 vector into wild-type recipient mice. We immunized the recipient mice with MOG 35–55 peptide, and assessed GFP+ antigen-specific T cells in the draining lymph nodes at day 5 post-immunization (Figure 6J). There were significantly fewer GFP+ SHMT2 KD T cells, compared to GFP+ sh-control T cells (Figure 6K). Moreover, expression of the cell proliferation marker Ki67 was reduced 50% in GFP+ SHMT2 KD T cells that persisted in vivo compared to GFP+ sh-control T cells (Figure 6L), suggesting that inhibition of mitochondrial one carbon metabolism reduced antigen-specific T cell abundance in vivo, via decreased proliferation and increased cell death.
To investigate the mechanism by which downregulation of SHMT2 impairs cell survival, we tested whether inhibition of de-novo purine biosynthesis in SHMT2 KD T cells caused nucleotide imbalance, which we hypothesized would contribute to an accumulation of DNA damage and increased cell death. Compared to control cells, SHMT2 KD cells had up to a 50% reduction in purine levels. By contrast, cellular levels of pyrimidines were intact (Figure 6M and S5D,E). To assess DNA damage we measured levels of γH2A.X, a marker of double-stranded DNA breaks, and found a significant increase in DNA damage in SHMT2 KD T cells (Figure 6N).
In addition to generating one carbon units for purine biosynthesis, SHMT2 has a critical role in maintaining redox state in cancer cells via glutathione synthesis (to which it can contribute the essential substrate glycine) and mitochondrial NADPH production (Ducker et al., 2016; Fan et al., 2014; Mehrmohamadi et al., 2014; Tedeschi et al., 2013; Ye et al., 2014). Therefore, we compared glutathione levels in control and SHMT2 KD T cells. Total glutathione levels were significantly reduced in SHMT2 KD T cells (Figure 6O). We next tested whether SHMT2 KD T cells could be rescued from death by the addition of formate (product of mitochondrial one carbon metabolism). Formate alone did not rescue T cell death. Cancer cells defective in the mitochondrial one-carbon pathway depend on extracellular glycine for glutathione synthesis, and this dependency can be rescued by N-acetylcysteine. Accordingly, we tested whether the combination of formate and N-acetyl cysteine (NAC), a commonly used antioxidant and a glutathione precursor, could rescue SHMT2 KD T cells from death. This combination completely rescued death in SHMT2 knockdown T cells (Figure 6P). In sum, our study elucidates a novel program of mitochondrial biogenesis that results in mitochondria with altered proteomic composition distinguished by an enrichment of the one carbon metabolism pathway, which we found to be critical for T cell survival and proliferation (Figure 6Q).
Discussion
Our findings highlight an active role for mitochondria in supporting T cell proliferation and survival. While early studies of tumor and immune cells suggested a predominant role for glycolysis (DeBerardinis et al., 2008; Hirschey et al., 2015), more recent work has identified a role for mitochondria in metabolic reprogramming, emphasizing functions for TCA cycle intermediates (such as α-ketoglutarate and citrate), respiration and amino acid metabolism to support cellular proliferation and energetics (Berod et al., 2014; Metallo et al., 2012; Sullivan et al., 2015). However, we still have an incomplete molecular understanding of how mitochondria contribute to cellular metabolic adaptation, proliferation and survival. In this study, we discovered naïve T cell activation induces a synchronized program of mitochondrial biogenesis and remodeling specialized to support the anabolic state of rapidly dividing T cells.
Mitochondrial biogenesis occurs through growth and division of the pre-existing organelles and involves orchestrated transcription and translation of both mitochondrial-encoded and nuclear-encoded genes (Dominy and Puigserver, 2013). A long-standing question has been whether cells induce mitochondria proliferation to: 1) increase the existing pools of mitochondria, or 2) generate a new pool of specialized mitochondria (Figure 1Q). Mitochondrial biogenesis has been observed in a number of models, including brown fat activation, exercise, and calorie restriction (Finley et al., 2012; Freyssenet et al., 1996; Martin-Montalvo and de Cabo, 2013; Nisoli et al., 2005). However, to our knowledge, T cell activation provides one of the few robust and synchronized models of biogenesis that can be recapitulated ex vivo, allowing us to address these fundamental questions.
Our analysis of the mitochondrial proteome demonstrates substantial proteome remodeling: mitochondrial proliferation in activated T cells gives rise to distinct mitochondria with a specialized function. The most striking change in activated T cell mitochondria was the massive induction of enzymes involved in folate-mediated one carbon metabolism, a pathway that generates one carbon units for purine and thymidylate synthesis, as well as NAD(P)H (Fan et al., 2014; Fox and Stover, 2008; Ye et al., 2014) (Figures 3, 4). The mitochondria and cytosol contain parallel pathways for generating and processing one carbon units. Yet, our data show that these two compartments are not redundant, as intact cytosolic one carbon metabolism was insufficient to support T cell proliferation and survival in vivo, when mitochondrial one carbon metabolism was genetically impaired (Figure 6). SHMT2 KD T cells accumulated DNA damage, partially due to reduced purine levels. Yet, only the combination of one carbon units (formate) and anti-oxidants (NAC) could rescue cell death, consistent with previous studies linking mitochondrial one carbon flux with NADPH production and glutathione biosynthesis (Fan et al., 2014; Mehrmohamadi et al., 2014; Tedeschi et al., 2013; Ye et al., 2014). A previous study further linked SHMT2 to mitochondrial dTTP synthesis and mtDNA integrity (Anderson et al., 2011). In tumors, knockdown of SHMT2 causes mitochondrial dysfunction, and increased ROS production (Ye et al., 2014). Thus, mitochondrial biogenesis during T cell activation produced specialized mitochondria, which may be critical for anabolic metabolism, bioenergetics and/or controlling oxidative stress.
This study led to surprising insights into T cell metabolism. When analyzing proteomic data, we expected metabolic pathways previously linked to T cell activation, such as glycolysis, pentose phosphate pathway, glutaminolysis and oxidative phosphorylation to be enriched in activated T cells. Remarkably, all of these pathways were among the least-induced proteins (i.e., those proteins whose fractional abundance in mitochondria decreases with T cell activation; Figure 2), suggesting that increased central carbon metabolism during T cell activation is mediated by post-translational events, such as enzyme localization or covalent modification, rather than metabolic enzyme abundance (Deprez et al., 1997; Wieman et al., 2007). Moreover, our list of early-induced proteins (Cluster 1, Figure 2) contains many master regulators of T cell activation, like MYC and TNF-related immunoregulatory proteins. Intriguingly, this group also includes proteins little studied in immunology, which we speculate may be critical drivers of T cell biology. To illustrate this point, at 4 hr post-activation, 5,10-methenyltetrahydrofolate synthetase (MTHFS) protein level was elevated by >35 fold. This enzyme converts 5-formyl-THF, a stable folate species utilized by cells for one carbon storage into 5,10-methenyl-THF, which can be used in one carbon metabolism (Fox and Stover, 2008). Intriguingly, accumulation of 5-formyl-THF has been observed in dormant cells, including seeds and spores (Kruschwitz et al., 1994). In naïve T cells, 5-formyl THF could serve as store for one carbon units that can be rapidly mobilized upon activation.
Our findings reveal similarities in mitochondrial metabolism in cancer cells and immune cells. Expression levels of mitochondrial enzymes of the one carbon pathway (SHMT2, MTHFD2 and MTHFD1L) are strongly correlated with rates of proliferation across a large array of cancer cells, and associated with increased mortality among breast cancer patients (Jain et al., 2012). In some cancers, the mitochondrial one carbon pathway is a significant contributor to the cellular pool of NADPH (Fan et al., 2014; Piskounova et al., 2015), and SHMT2 KD increases oxidative stress and promotes cell death under hypoxia (Ye et al., 2014). Drugs that target folate-related enzymes, like methotrexate, a DHFR inhibitor, have long been used in the clinic to treat inflammatory diseases including both rheumatoid arthritis and cancer. Our studies suggest that specifically targeting the mitochondrial arm of one carbon metabolism could provide a more focused approach for treating immune-mediated diseases. Moreover, a diet free of serine and glycine reduced tumor growth in cancer xenograft mouse models (Maddocks et al., 2013). A similar approach might potentially be used to ameliorate T cell mediated pathologies such as autoimmunity.
In sum, our study defines a program of mitochondrial proteome remodeling and biogenesis during early T cell activation, which identified a novel component of T cell activation -increased mitochondrial one carbon metabolism. In addition, our proteomic dataset enabled us to define novel biomarkers for monitoring metabolism in small numbers of cells under physiological conditions in vivo. This approach provides a powerful means for identifying biomarkers for monitoring the activation and metabolic status of multiple immune cell types, as well as other cell types, in physiological settings, including cancer.
Experimental procedures
Mice
7–10 week old mice were used for all experiments. Wild-type C57BL/6 and myelin oligodendrocyte glycoprotein 2D2 T cell receptor transgenic mice (Tcra2D2,Tcrb2D21Kuch) were purchased from the Jackson Laboratory (Bar Harbor, ME). PhAMexcised mice were the generous gift of Dr. David Chan (Pham et al., 2012). Experimental mice were housed in specific pathogen-free conditions at Harvard Medical School and used in accordance with animal care guidelines from the Harvard Medical School Standing Committee on Animals and the National Institutes of Health.
Culture and stimulation of naïve CD4+ T cells
Naïve CD4+CD62LhiCD44loCD25− cells were sorted by flow cytometry (purity >99%), cultured at 37°C and 5% CO2 in complete RPMI media, and activated in vitro with 4 μg/mL plate-bound anti-CD3 (clone 1H5-2C11; BioXCell) and anti-CD28 (clone 37.51; BioXCell). In some experiments, IL-7 (5 ng/mL; R&D Systems), formate (1mM; Sigma) and n-acetyl cysteine (7.5mM; Sigma) were added to culture media. See Supplemental Experimental Procedures for culture conditions, retroviral transduction and metabolic tracing experiments.
Adoptive transfer & Immunization
Naïve (CD62LhiCD44loCD25−) CD4+ T cells were sorted from spleens of TCR-transgenic 2D2 (MOG-specific) mice, and transferred by tail vein injection into WT recipients (1–2 × 106 cells per mouse). In some experiments, the cells were retrovirally transduced prior to adoptive transfer. For MOG 35–55 immunizations (referred to as “MOG/CFA”), mice were injected subcutaneously with 100μg of MOG 35–55 (UCLA Biopolymers Facility) in a 1:1 emulsion with H37RA CFA (Sigma) on the mouse flanks. At different time points (as indicated), the mice were euthanized and inguinal lymph nodes (dLN) were harvested for flow cytometric analyses.
Flow cytometry
In vitro cultured cells, or cells isolated from lymphoid organs of immunized mice were collected, resuspended in staining buffer (PBS containing 1% fetal bovine serum and 2 mM EDTA), and stained. Antibodies are listed in the Supplemental text. For intracellular staining, the FoxP3 fix/perm kit (eBioscience) was used after surface staining. All flow cytometry was analyzed with an LSR II (BD biosciences) using standard filter sets, and FlowJo software (TreeStar).
Electron Microscopy: sample preparation, data collection and analysis
T cells were fixed with 0.1 M cacodylate buffer, pH 7.4, containing 2% glutaraldehyde, 2% paraformaldehyde, and mixed with 4% low melting-point agarose (details in supplemental text). Imaging was done using a 1400 Transmission Electron Microscope (JEOL) equipped with a side mount Gatan Orius SC1000 digital camera at a nominal magnification of 20K (naïve, 4 and 9 hr) or 12K (24 hr). Micrographs were analyzed using Volocity 3D image analysis software (PerkinElmer).
Live cell imaging
Naïve CD4+ T cells derived from PHAMexcised mice(Pham et al., 2012) were activated in a 6 well plate on 25 mm round glass coverslips (Warner Instruments) pre-coated with anti-CD3/anti-CD28. At the desired time points, the coverslips were placed in an Attofluor cell chamber (Invitrogen) for imaging by confocal microscopy (See Supplemental text).
Real-time PCR
Total DNA was isolated using DNeasy kit (Qiagen). cDNA was synthesized using the iSCRIPT kit (BioRad). Quantitative PCR analysis was performed with SYBR green, using a LightCycler 480 (Roche). Rp18s: F: 5′-ACCTGTCTTGATAACTGCCCGTGT-3′, R: 5′-TAATGGCAGTGATGGCGAAGGCTA-3′; Cox1: F: 5′-ACTTGCAACCCTACACGGAGGTAA-3′, R: 5′-TCGTGAAGCACGATGTCAAGGGAT-3′.
Proteomics sample preparation and data analysis
Total cell proteins were extracted from frozen cell pellets, followed by enzymatic digestion and TMT labeling of peptides (See Supplemental Experimental Procedures). Labeled peptides were fractionated, and analyzed on a Thermo Orbitrap Fusion mass spectrometer. See Supplemental Experimental Procedures for detailed protocols of sample preparation and data extraction. Fold-change data were Lowess normalized and collapsed into unique protein representation to average fold-change of all peptides. The sorted heatmap of the log2 fold-changes was generated using SPIN (Tsafrir et al., 2005). Proteins were clustered using the Agglomerative Clustering function from the SciKit-Learn machine learning library in Python (scikit-learn.org). Clustering analysis compared a protein’s abundance relative to naive T cells at 4, 9, and 24 hr following activation, in each of two experimental replicates (so that six data points per protein were used for clustering). Values from the two replicates were averaged for drawing the curves in Figures 2 and 3. All proteins with at least 2 detected peptides were included in this analysis. The Molecular Signatures Database (MSigDB, Broad Institute; http://software.broadinstitute.org/gsea/msigdb/annotate.jsp) was used for KEGG pathway analysis. For mitochondrial proteins, each sample was z-scored, normalized (average of 0 and standard deviation of 1) for gene set enrichment analysis (Broad Institute; http://software.broadinstitute.org/gsea/index.jsp) (Subramanian et al., 2005).
Metabolomics and metabolite uptake and tracing
Glucose and glutamine uptake, and lactate secretion were measured by analyzing growth media using the BioProfile Flex Analyzer (Nova Biomedical). For carbon and deuterium tracing, and non-labeled metabolomics, samples were processed for analysis by reversed-phase ion-pairing chromatography coupled with negative-mode electrospray-ionization high-resolution MS on a stand-alone orbitrap (Thermo). See Supplemental Expereimental Procedures for details on sample preparation, data collection and analysis.
Statistical analysis
All statistical analyses were performed using Prism (Graphpad Software). We used unpaired Student’s t-test when comparing two groups, and one-way analysis of variance (ANOVA) followed by Tukey-Kramer’s post hoc analysis, when comparing 3 groups.
Supplementary Material
Acknowledgments
The authors thank the Electron Microscopy Core at BIDMC. This study was supported in part by grants from NIH/NIAID HHSN272201100018C to MCH and AHS, F. Hoffmann-La Roche Ltd. to AHS and MCH, the Glenn Foundation for Medical Research to MCH, and the Ludwig center at Harvard to MCH. NRH was supported by The European Molecular Biology Organization long-term postdoctoral fellowship. DS was supported by a PhD Fellowship from Portuguese Foundation for Science and Technology (FCT-MCTES).
Footnotes
Author Contribution Conceptualization, N.R.H., M.C.H., A.H.S.; Methodology, N.R.H., D.S., J.M.G., P.T.S., N.D., S.G., J.D.R., A.H.S., M.C.H.; Formal Analysis, N.R.H., F.K.S, M.S.; Investigation, N.R.H., D.S., J.M.G., P.T.S., A.R., S.B.L., N.D., J.B.S., A.H.S., M.C.H.; Writing-Original Draft, N.R.H. and M.C.H.; Writing-Review & Editing, N.R.H., J.D.R., A.H.S., M.C.H.; Visualization, N.R.H and M.C.H.; Supervision, N.R.H. and M.C.H.; Funding Acquisition, A.H.S. and M.C.H.
References
- Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15163–15168. doi: 10.1073/pnas.1103623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB, Beutler B. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12286–12293. doi: 10.1073/pnas.1209134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature medicine. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. Journal of immunology. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza AD, Parikh N, Kaech SM, Shadel GS. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. The Journal of biological chemistry. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- Dominy JE, Puigserver P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harbor perspectives in biology. 2013:5. doi: 10.1101/cshperspect.a015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metabolism. 2016 doi: 10.1016/j.cmet.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Lee J, Souza A, Desquiret-Dumas V, Bullock K, Rowe GC, Procaccio V, Clish CB, Arany Z, Haigis MC. Skeletal muscle transcriptional coactivator PGC-1alpha mediates mitochondrial, but not metabolic, changes during calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2931–2936. doi: 10.1073/pnas.1115813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Freyssenet D, Berthon P, Denis C. Mitochondrial biogenesis in skeletal muscle in response to endurance exercises. Archives of physiology and biochemistry. 1996;104:129–141. doi: 10.1076/apab.104.2.129.12878. [DOI] [PubMed] [Google Scholar]
- German NJ, Haigis MC. Sirtuins and the Metabolic Hurdles in Cancer. Current biology : CB. 2015;25:R569–583. doi: 10.1016/j.cub.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, DeBerardinis RJ, Diehl AM, Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA, et al. Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol. 2015;35(Suppl):S129–150. doi: 10.1016/j.semcancer.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruschwitz HL, McDonald D, Cossins EA, Schirch V. 5-Formyltetrahydropteroylpolyglutamates are the major folate derivatives in Neurospora crassa conidiospores. J Biol Chem. 1994;269:28757–28763. [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nature immunology. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annual review of immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, de Cabo R. Mitochondrial metabolic reprogramming induced by calorie restriction. Antioxidants & redox signaling. 2013;19:310–320. doi: 10.1089/ars.2012.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrmohamadi M, Liu X, Shestov AA, Locasale JW. Characterization of the usage of the serine metabolic network in human cancer. Cell reports. 2014;9:1507–1519. doi: 10.1016/j.celrep.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H, Metzger D, Chambon P, Yoshimura A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nature immunology. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi PM, Markert EK, Gounder M, Lin H, Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, DiPaola RS, Hirshfield KM, et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell death & disease. 2013;4:e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21:2301–2308. doi: 10.1093/bioinformatics/bti329. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18:1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.