Abstract
Chikungunya virus (CHIKV), a reemerging arbovirus, causes a crippling musculoskeletal inflammatory disease in humans characterized by fever, polyarthralgia, myalgia, rash, and headache. CHIKV is transmitted by Aedes species of mosquitoes and is capable of an epidemic, urban transmission cycle with high rates of infection. Since 2004, CHIKV has spread to new areas, causing disease on a global scale, and the potential for CHIKV epidemics remains high. Although CHIKV has caused millions of cases of disease and significant economic burden in affected areas, no licensed vaccines or antiviral therapies are available. In this Review, we describe CHIKV epidemiology, replication cycle, pathogenesis and host immune responses, and prospects for effective vaccines and highlight important questions for future research.
Ecology and epidemiology
Chikungunya virus (CHIKV) is a mosquito-borne virus responsible for periodic and explosive outbreaks of a febrile disease that is characterized by severe and sometimes prolonged polyarthritis. CHIKV was first recognized as a human pathogen after it was isolated from the serum of an infected patient during an outbreak of debilitating arthritic disease in 1952 in present-day Tanzania (1–3). Because of the stooped posture and rigid gait of infected individuals, the disease was given the name chikungunya, a word from the Kimakonde language that translates as “that which bends up” (2). The vast majority of infected individuals develop chikungunya fever, an acute illness notable for rapid onset of fever, incapacitating polyarthralgia and arthritis, rash, myalgia, and headache (Table 1 and ref. 4). Acute CHIKV disease symptomatically resembles dengue fever, and retrospective case reports suggest that CHIKV outbreaks occurred as early as 1779 but were inaccurately attributed to dengue virus (5, 6). However, unlike dengue, a characteristic feature of CHIKV disease is recurring musculoskeletal disease primarily affecting the peripheral joints that can persist for months to years after acute infection (7–10). CHIKV disease is often self-limiting and has a low fatality rate (~0.1%) (11), but manifestations of CHIKV infection that lead to acute and chronic disability have considerable implications, including a substantial impact on quality of life for infected patients as well as considerable economic and community consequences (7, 9, 12).
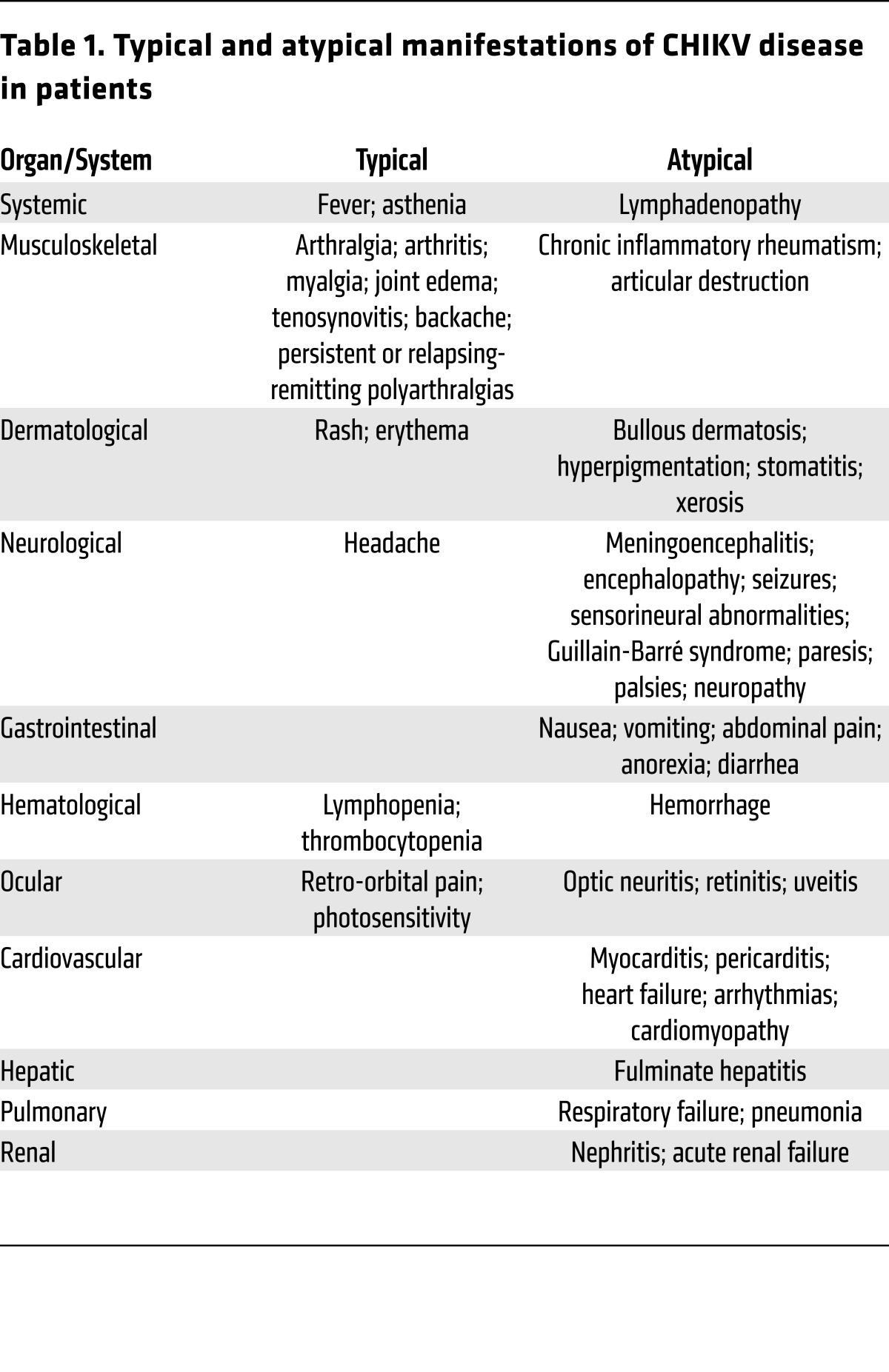
Table 1. Typical and atypical manifestations of CHIKV disease in patients.
Transmission of CHIKV occurs mainly through the bite of an infected Aedes (subgenus Stegomyia) species of mosquito. However, maternal-fetal transmission can occur intrapartum, which results in high rates of infant morbidity (12, 13). Historically, CHIKV has been endemic in tropical and subtropical regions of sub-Saharan Africa and Southeast Asia, where two distinct CHIKV transmission cycles exist. CHIKV is maintained in a rural enzootic transmission cycle, which occurs between various forest or savannah Aedes (Stegomyia) mosquitoes and animal reservoirs (14–17), with nonhuman primates being the presumed major reservoir host (4, 18–21). Occasional introduction of the virus into urban areas is thought to cause periodic outbreaks of CHIKV disease (1, 14). Urban transmission is mediated primarily by Aedes aegypti or Aedes albopictus mosquitoes and occurs in a human-mosquito-human transmission cycle (22). While enzootic sylvan transmission of CHIKV has been well established in Africa, outbreaks in Asia have been mainly attributed to urban human-mosquito-human transmission, although there is limited evidence for enzootic transmission (18, 23–25). Little is known about the factors contributing to the natural maintenance of CHIKV (1, 15, 26, 27), but understanding catalysts that promote CHIKV maintenance and spillover dynamics is essential to combatting emergence and spread of the virus.
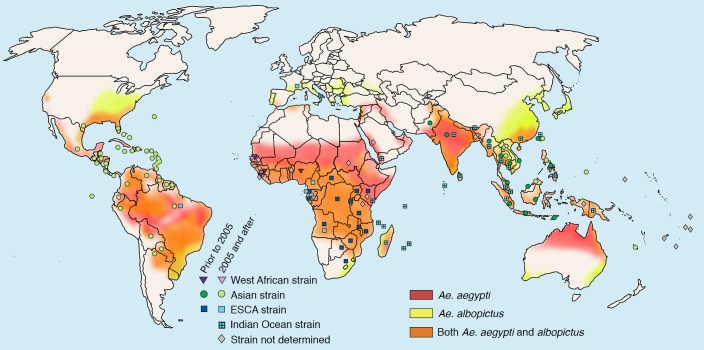
Since the first reports of CHIKV infection in Africa in the 1950s, subsequent epidemics of CHIKV occurred throughout the latter half of the 20th century in countries within Asia and sub-Saharan Africa (Figure 1 and reviewed in ref. 28). Phylogenetic analyses of CHIKV sequences indicate that CHIKV originated in Africa over 500 years ago (29), and a common lineage diverged into two distinct branches, termed West African (WA) and East/Central/South African (ECSA) (28–31). WA strains have been associated mainly with enzoonotic transmission and small focal outbreaks of human disease in countries located in western Africa (Figure 1 and ref. 30). In contrast, strains from the ECSA lineage have repeatedly spread to new regions to cause significant urban epidemics. The first emergence of an ECSA strain outside of Africa is estimated to have occurred between 70 and 150 years ago in Asia (29). The virus continued to circulate in this area, evolving independently of the ECSA lineage into a distinct Asian genotype (29), which has caused numerous outbreaks of CHIKV disease in this region (Figure 1 and ref. 28).
Figure 1. Geographic distribution of endemic CHIKV and its primary vectors, Ae. aegypti and Ae. albopictus.
Countries in which autochthonous cases of CHIKV have been reported are specified with colored symbols representing the distinct viral genotypes detected during outbreaks in that country. West African strains are indicated by purple triangles; Asian strains are indicated by green circles; East/South/Central African (ESCA) strains are indicated with blue squares; strains of the Indian Ocean lineage, a subtype of the ESCA clade, are indicated with blue squares with a cross; and strains whose genotype has not been determined are indicated with gray diamonds. Symbols are shaded to differentiate transmission prior to (darker hue) or after (lighter hue) the reemergence of the virus in the Indian Ocean (ECSA strain in 2005) and Pacific Islands (Asian strain in 2010). Symbols indicate the countries in which natural transmission has occurred and are not meant to indicate precise locations of outbreaks. Overlayed with CHIKV distribution is the geographic range of the two primary vectors responsible for urban mosquito-human-mosquito transmission of the virus. Range of Ae. aegypti is indicated in red; range of Ae. albopictus is indicated in yellow; and areas where both mosquito species are present are indicated in orange. Endemic CHIKV data were obtained from numerous PubMed publications (1, 3, 23, 24, 28, 30–32, 37, 38, 46–62). Range of mosquito data was obtained from (63).
An ECSA strain emerged again during an outbreak in Kenya in late 2004, initiating one of the largest CHIKV epidemics on record, with expansion to areas well beyond the historical range of the virus (4, 32). During this devastating epidemic, the virus spread to a number of islands in the Indian Ocean, India, and parts of Southeast Asia, leading to over 6 million estimated cases of CHIKV disease (Figure 1 and refs. 4, 30, 32, 33). International air travel greatly facilitated the geographic expansion of CHIKV during this epidemic (34–36), with viremic travelers importing an unprecedented number of CHIKV cases into naive countries, including more temperate regions in Europe and the United States (35, 37, 38). Infected travelers often served as sentinels, seeding autochthonous transmission of the virus in naive areas, including in Italy in 2007 (37) and France in 2010 (38).
Historically, urban transmission of CHIKV was vectored mainly by A. aegypti mosquitoes. However, many of the recent outbreaks in the Indian Ocean basin and Southeast Asia have been attributed to circulating strains from the Indian Ocean lineage (IOL), a newly emerged subgroup within the ECSA clade (29). Some strains within this subgroup contain an adaptive mutation (E1-A226V) that increases viral fitness in A. albopictus without compromising replication in A. aegypti (39–45). This mutation has been selected convergently in multiple ECSA strains in different geographic regions during the past decade (31, 34, 46–48). Importantly, the E1-A226V substitution requires epistatic interactions (E1-98A and E2-211T) to allow penetrance in A. albopictus (reviewed in ref. 49). These epistatic variants are lineage-specific, as they have been observed in other ECSA and WA strains but not in strains from the Asian lineage, rendering viruses from the Asian clade genetically constrained in the capacity to adapt to A. albopictus (41). Additional substitutions in viral glycoproteins also augment replicative capacity in A. albopictus (reviewed in ref. 49).
Like that of the ECSA lineage, the geographical range of the Asian lineage has also recently expanded. Since 2011, when the virus was first detected in the Pacific Island region of New Caledonia (50), CHIKV has caused outbreaks on 10 of the 22 countries and territories of the Pacific Islands, with strains from the Asian lineage being responsible for the majority of these outbreaks (51, 52). Beginning in 2012, islands in Oceania were bombarded with not only CHIKV outbreaks, but also epidemics of Dengue virus and Zika virus (50, 53, 54). As with CHIKV, infected travelers are thought to be the main source of Dengue and Zika virus outbreaks in the Pacific. Strains from the Asian and ECSA lineages, including the IOL subgroup, continue to cocirculate and spread within the Indian subcontinent, Southeast Asia, and Oceania (Figure 1 and refs. 41, 47, 55–60).
With such high rates of virus importation by infected travelers, the introduction of CHIKV into the Western Hemisphere seemed inevitable. In December 2013, the first local transmission of CHIKV in the Western Hemisphere in modern history was reported, with autochthonous cases identified in the Caribbean island of French St. Martin (61). The emergence of CHIKV in the Western Hemisphere was remarkable in that the virus spread rapidly to immunologically naive populations in the Caribbean as well as Central, South, and North America. Since the 2013 introduction of CHIKV in the Americas, over 2 million suspected cases caused by endemic transmission have occurred in almost 50 countries, including 12 cases of autochthonous transmission in the United States (Figure 1 and ref. 51). The initial outbreak of CHIKV in the Americas was first attributed to a variant from the Asian lineage. However, the introduction of an ECSA strain into Brazil in 2014 (62) raises a concern for vector adaptation in the Americas and spread of CHIKV to more temperate regions, such as the United States, where A. albopictus has a greater range (Figure 1 and ref. 63). Because of globalization and the expansion and year-round presence of relevant vectors, especially in densely populated urban centers (63), the risk that the virus will become endemic in tropical regions of the Americas remains high. Establishment of CHIKV in the Americas, as well as repeated introduction events, suggests that the virus will continue to spread and that the sporadic and explosive outbreaks of CHIKV observed in Africa and Asia also will likely occur in the Western Hemisphere.
CHIKV structure, genome, and replication cycle
CHIKV is a small (~70 nm in diameter), enveloped virus that is a member of the Old World Semliki Forest virus group of arthritogenic alphaviruses within the Togaviridae family (reviewed in ref. 64). The CHIKV genome is approximately 11,800 nucleotides, constituting a single-stranded, message-sense RNA with a 5′ 7-methylguanosine cap and a 3′ poly-A tail (65). The genome contains two open reading frames (ORFs) separated by a noncoding junction as well as 5′- and 3′-untranslated regions (65, 66). Four essential nonstructural proteins (nsP1, 2, 3, and 4) constitute the RNA replicase (Table 2) and are encoded by the 5′ two-thirds of the genome (65, 66). The 3′ ORF is translated from a subgenomic positive-strand mRNA and encodes six proteins (Table 2), including three major structural proteins (capsid, E1, and E2) (64, 66–68). The CHIKV virion is formed by a lipid bilayer envelope tightly associated with an icosahedral nucleocapsid shell (240 capsid copies) that encapsidates genomic RNA (64, 69). Embedded within the viral envelope are heterodimers of the E1 and E2 glycoproteins arranged in trimers forming an icosahedral lattice (69).
Table 2. CHIKV proteins and functions.
Although the general events of CHIKV replication are comparable to those of other alphaviruses (Figure 2), much more remains to be discovered about the specific biology of CHIKV replication. However, CHIKV displays broad tropism, replicating in many vertebrate and invertebrate cells (70–73); bona fide proteinaceous receptors have not been identified for CHIKV. Glycosaminoglycans, which are expressed on many susceptible cell types, appear to serve as attachment factors to enhance infectivity (64, 74–77). After E2-mediated attachment to cells, receptor-bound particles are internalized mainly by clathrin-mediated endocytosis (Figure 2 and refs. 70, 77, 78). Endosomal acidification triggers conformational changes in the viral glycoproteins, leading to exposure and insertion of the buried E1 fusion loop into the host membrane, which results in fusion of the viral envelope and endosomal membrane (77). Following release into the cytoplasm, the nucleocapsid disassembles to deliver genomic viral RNA (vRNA) into the cytosol for translation (77). The incoming CHIKV genome is directly translated to produce the nonstructural precursor polyprotein P1234, which is cleaved by the virus-encoded protease located in nsP2 into P123 and nsP4 (64, 79). Some strains encode an opal stop codon following nsP3, and low-frequency read-through yields both P123 and P1234 (79). Together, P123 and nsP4, presumably with host proteins, form an early, short-lived viral replicase that synthesizes a full-length negative-sense vRNA (64).
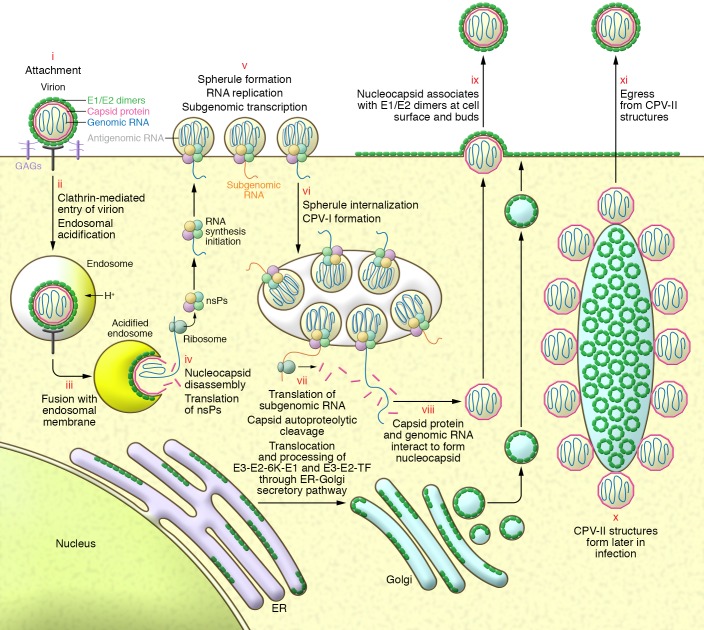
Figure 2. CHIKV replication cycle in mammalian cells.
(i) E2 binds to the cell surface via an unknown receptor and possibly glycosaminoglycans as attachment factors. (ii) CHIKV enters the cell through clathrin-mediated endocytosis. Acidification of endosomes leads to insertion of the fusion peptide in E1 into the endosomal membrane. (iii) Fusion of the viral envelope and endosomal membrane releases nucleocapsid into the cytosol. (iv) Disassembly of the nucleocapsid liberates positive-sense genomic RNA and nonstructural protein (nsP) translation occurs. (v) Four nsPs, together with genomic RNA and presumably host proteins, assemble at the plasma membrane (PM) and modify it to form viral replication compartments (spherules) containing viral dsRNA. nsP1–4 function as a replicase and localize to the spherule neck to generate genomic, antigenomic, and subgenomic vRNAs. (vi) Spherule internalization allows formation of large cytopathic vacuoles (CPV-1) that house multiple spherules. Spherules at the PM or within CPV-I are fully functional. (vii) Translation of subgenomic RNA produces the structural polyprotein, and capsid autoproteolysis releases free capsid into the cytoplasm. Translocation of E3-E2-6K-E1/E2-E2-TK polyproteins into the ER. E2/E1 are posttranslationally modified, transit the secretory system, and are deposited at the PM. (viii) Interaction of capsid and genomic RNA leads to formation of icosahedral nucleocapsids. (ix) Nucleocapsids assemble with E2/E1 at the PM, resulting in budding of mature progeny virions. (x) Later in infection, CPV-IIs form, containing hexagonal lattices of E2/E1 and are studded with nucleocapsids. (xi) CPV-IIs likely serve as transport vehicles and assembly sites for structural proteins, allowing formation of mature virions and egress.
Negative-strand synthesis is linked to formation of viral replication compartments termed spherules, which are small, vesicular structures that form at the plasma membrane (PM) and serve as the site of vRNA replication (Figure 2). The nsPs are thought to localize at the neck of the spherules, which house dsRNA intermediates, protecting them from degradation and recognition by cellular pattern-recognition receptors (64, 80–84). As infection proceeds, spherules are internalized to form large cytopathic vacuoles (CPV-I), which contain markers from endosomal and lysosomal membranes (Figure 2 and refs. 80–82). P123 accumulation leads to complete proteolytic processing of the polyprotein, resulting in a switch of the replicase to an abundant conformer that uses negative-sense vRNA as a template for amplification of genomic positive-sense vRNA as well as transcription of a subgenomic, positive-sense vRNA (81) that encodes the structural polyproteins (capsid, E3, E2, 6K, TF, and E1). Following translation of capsid protein, autoproteolysis releases it from the structural polyprotein, allowing interaction with newly synthesized genomes to catalyze oligomerization and formation of intact nucleocapsids containing a single molecule of the RNA genome (Figure 2 and ref. 64). Translation of the structural polyprotein continues, generating a majority product, E3-E2-6K-E1, and a minor product, E3-E2-TF, due to ribosomal frameshifting (67, 68). A signal sequence present at the N-terminus of E3 traffics the major and minor structural polyproteins through the host secretory pathway, where cleavage by host proteases produces pE2 (E3-E2); 6K or transframe (TF); and E1. 6K and TF are viral accessory proteins that share an N-terminus but have disparate C-termini, resulting from ribosomal frameshifting, and are hypothesized to form ion channels (67). Although 6K and TF are found at low levels in virion particles (68, 85, 86) and appear to contribute to viral budding (68, 87) and pathogenesis (88), their precise roles in glycoprotein processing, assembly, budding, and particle stability remain to be clarified (67, 68, 88).
As E1 and E3-E2 transit the secretory pathway (Figure 2), they remain associated as a noncovalent, hetero-oligomeric complex that undergoes conformational changes and posttranslational modifications, including palmitoylation and N-linked glycosylation as well as release of E3 by furin, to form mature spikes at the PM (Figure 2 and refs. 89, 90). Recruitment of intact nucleocapsids to membrane-associated envelope glycoproteins leads to PM budding of assembled particles (64). Late in infection, a second type of virally induced cytopathic vacuole, CPV-II, is formed (Figure 2). These structures contain helical tubular arrays of viral glycoproteins within the vesicles, which are studded with nucleocapsids on their cytoplasmic face (91–93). Their proximity to the PM suggests that CPV-IIs may be an assembly intermediate (92), but it is not clear whether they are necessary for efficient infection or contribute to pathogenesis. CHIKV CPV-I and CPV-II structures also have been observed in mosquito cells (91).
Infection of mammalian cells with CHIKV leads to massive changes, including antiviral responses (e.g., apoptosis, IFN, stress granule formation, and unfolded protein response) and proviral responses (e.g., host cell shutoff and authophagy) (94). CHIKV nsP2 and nsP3 display activities that counteract some of these antiviral responses (94). Authophagy is proposed to play a global pro-CHIKV function in human cells, possibly by limiting apoptosis, and may be a pathogenesis determinant (95–97). Although some additional proviral host factors have been identified (70, 74, 78, 98–100), specific host pathways and mechanisms that promote replication of CHIKV are areas of active research. A comprehensive understanding of the CHIKV replication cycle in both mammalian and mosquito cells is essential for development of effective antivirals.
Disease mechanisms and host immune responses
Studies of CHIKV-infected humans and animals have defined symptoms and immune responses of acute and chronic CHIVK disease, but much of the molecular interplay between virus and host remains to be established. CHIKV-induced disease shares many similarities with illnesses caused by other arthritogenic alphaviruses, with some distinctions observed (reviewed in refs. 101–103). After deposition into the bloodstream or skin through a bite of an infected mosquito, CHIKV replicates at the site of inoculation in fibroblasts and possibly macrophages (Figure 3 and refs. 8, 70, 104, 105). Despite triggering innate immune responses, the virus spreads via lymphatics into the bloodstream, allowing dissemination to several sites of replication, most commonly lymphoid organs (lymph nodes and spleen), skin, and especially tissues where prominent disease symptoms occur (muscle, peripheral joints, and tendons) but also in brain and liver in more severe cases (72, 104–109). Replication of CHIKV in peripheral tissues results in remarkably high serum viral loads (>109 virus particles/ml; ref. 110). Such high-level viremia in humans is rare for most alphaviruses and allows CHIKV to be easily transmitted to mosquitoes via a bloodmeal.
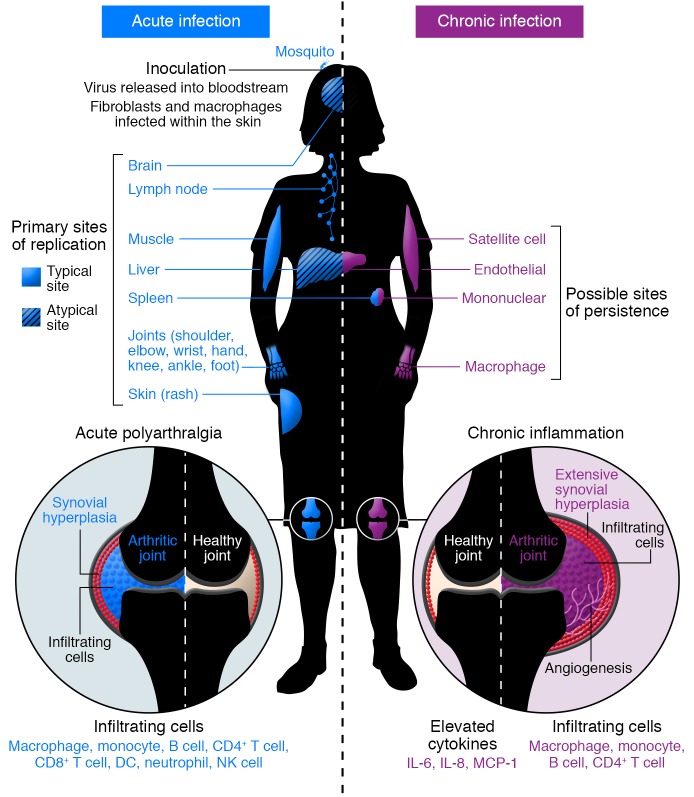
Figure 3. Model of acute and chronic CHIKV pathogenesis.
Acute CHIKV infection begins with transmission of the virus via a bite of an infected mosquito to the skin, where it replicates in susceptible cells, including fibroblasts and macrophages. The virus disseminates through lymphatics and bloodstream to typical (solid) and atypical (hatched) sites of primary replication (indicated in blue). Acute infection elicits an inflammatory response in infected tissues, characterized by an extensive infiltration of mainly macrophages and monocytes, but also neutrophils, NK cells, and lymphocytes in target tissues (indicated in blue), and by elaboration of a number of proinflammatory chemokines and cytokines. Within arthroskeletal tissues, synovial hyperplasia begins. Viral replication and host immune responses cause myalgia and polyarthralgia in distal joints. Chronic CHIKV disease can persist for months or years after acute infection but is often limited to more distal joints. Chronic disease is likely mediated by persistent virus and inflammation. Possible sites of CHIKV persistence include endothelial cells in the liver and other organs, mononuclear cells in the spleen, macrophages within the synovial fluid and surrounding tissues, and satellite cells within the muscle (indicated in purple). Within the chronically infected joint, the continued presence of a subset of infiltrating cells (mainly macrophages, monocytes, and lymphocytes) and specific proinflammatory mediators (IL-6, IL-8, and MCP-1) within the synovial fluid likely contribute to prolonged inflammatory disease. Chronic joint pathology resembles that in rheumatoid arthritis, with significant hyperplasia and angiogenesis. This model is based on human and animal studies.
Acute CHIKV infection elicits robust innate immune responses, leading to elevation of type I IFNs and numerous proinflammatory chemokines, cytokines, and growth factors (Table 2 and refs. 70, 111–115). Type I IFN signaling controls viral replication and pathogenesis during acute infection (104, 114–116). In humans, IFN-α appears early in infection and correlates with viral load (111, 114, 117). Coincident with rising viral loads and IFN-α responses, the vast majority of infected patients experience sudden onset of clinical illness (Table 1), with a small proportion of infected individuals (5%–28%) remaining asymptomatic (118, 119). Acute CHIKV infection is predominated by high fever (39°C–40°C), which can last up to a week and may occur in a biphasic manner (2, 120). After fever onset, most patients develop severe and often debilitating polyarthralgia that is usually bilateral and symmetric, most commonly in ankles, wrists, and phalanges. Other symptoms include arthritis, asthenia, conjunctivitis, gastrointestinal distress, headache, myalgia, and rash, which is usually maculopapular and pruritic (Table 1). In recent epidemics, more atypical and severe symptoms have been observed, including multiple dermatological manifestations, hemorrhage, hepatitis, myocarditis, neurological disorders, and ocular disease (Table 1 and refs. 110, 121–124). Atypical symptoms are most prevalent among vulnerable groups, including neonates, the elderly, and those with underlying comorbidities.
In animal models (reviewed in refs. 20 and 125), innate immune responses triggered by viral replication recruit inflammatory cells to infected musculoskeletal tissues, which are thought to contribute to muscular and articular damage resulting in pain and discomfort in muscles, joints, and tendons (Figure 3). Virus titers decline with the development of CHIKV-specific adaptive immunity. Although viremia is usually cleared 5–7 days after infection and most acute symptoms resolve within 2 weeks, rheumatic manifestations can persist for months to years in a subset of individuals (7–10, 37, 103, 126, 127). The percentage of CHIKV-infected patients reported to develop protracted illness varies greatly (~14% to ~87%), but an average prevalence of approximately 48% among infected patients has been estimated (128). Risk factors for chronic CHIKV disease include age (>45 years), preexisting chronic inflammatory arthropathy, and severity of symptoms during the acute phase (129, 130). Two types of persistent CHIKV-induced rheumatic disorders have been described (131). The vast majority of patients experience chronic musculoskeletal disorders and usually respond to some extent to analgesics, antiinflammatory treatments, and physiotherapy. However, approximately 5% of patients meet the criteria for chronic inflammatory rheumatism, which may be destructive and deforming (131). Thus, the underlying mechanisms of CHIKV disease may involve multiple distinct immunopathological processes. Early differentiation of chronic CHIKV infection would inform disease management, which may vary according to type.
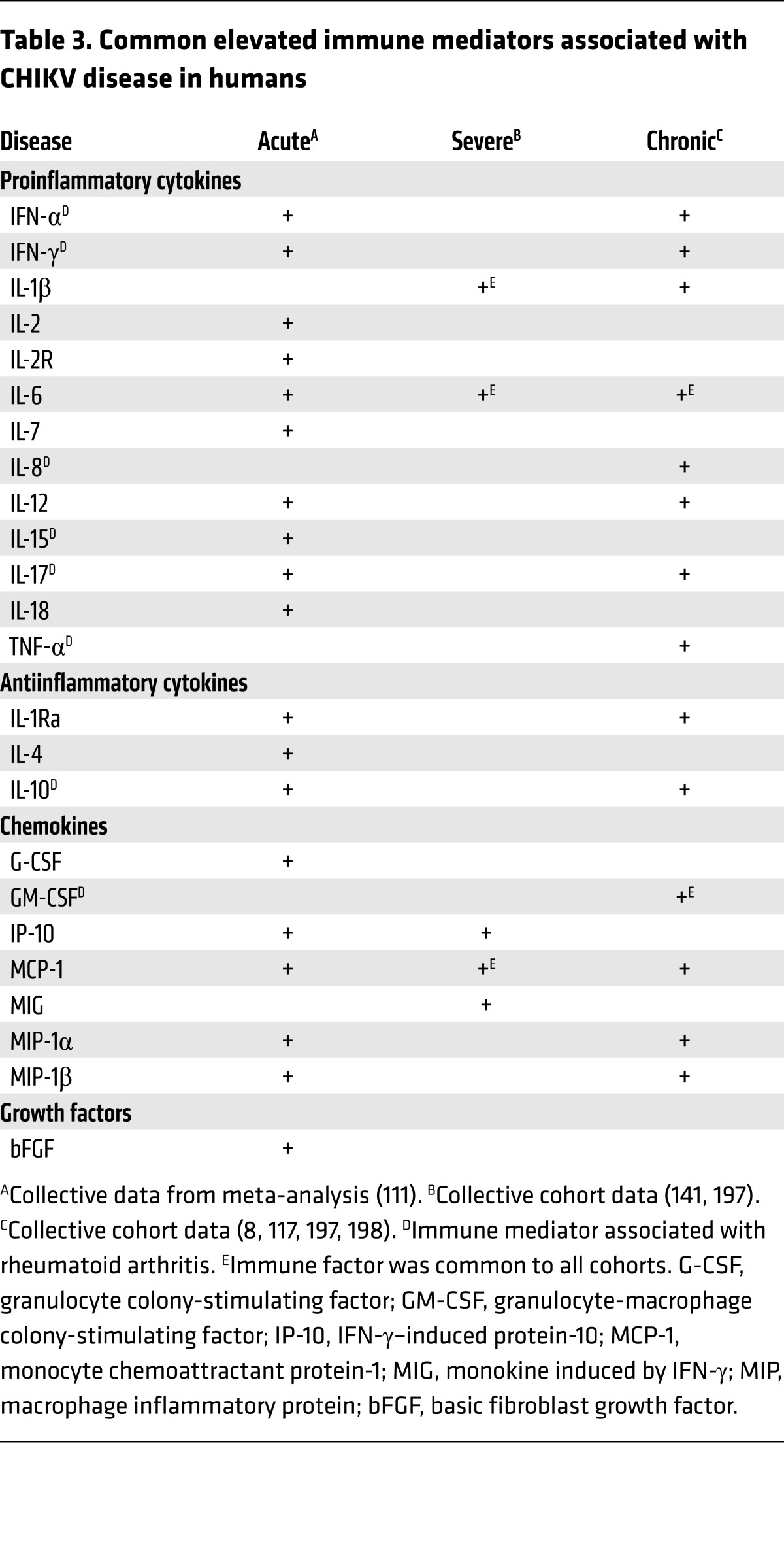
Symptoms of acute CHIKV disease are caused by direct cellular damage and local inflammation, but the specific contributions of viral replication and the host immune response to CHIKV infection are yet to be completely unraveled. CHIKV infection is cytopathic and induces apoptosis, resulting in direct tissue injury (70, 132, 133). Numerous cell types, many of which are located at sites of disease, are susceptible to CHIKV, including chondrocytes, endothelial cells, fibroblasts, hepatocytes, macrophages, monocytes, muscle satellite cells, myocytes, and osteoblasts (8, 72, 75, 104–107, 113, 116, 134–139). A meta-analysis of immune mediators from geographically distinct cohorts (111) revealed a common signature profile during acute CHIKV disease in humans (Table 3). Several of these soluble factors are associated with the progression of rheumatoid arthritis (140). In multiple cohorts, elevated levels of IL-1β, IL-6, and monocyte chemoattractant protein-1 (MCP-1) correlate with disease severity (117, 141), and high levels of IL-6 and GM-CSF correlate with persistent arthralgia (refs. 104, 117, and Table 3). Thus, an imbalance of immune mediators required for effective antiviral defense also likely contributes to CHIKV pathogenesis. Although CHIKV infection is controlled by the innate immune response in both hematopoietic and nonhematopoietic cells (113, 114), the specific cellular targets of infection that contribute to the orchestration of these responses in vivo remain largely undefined.
Table 3. Common elevated immune mediators associated with CHIKV disease in humans.
Although many hematopoietic cells appear refractory to CHIKV infection (66, 70, 114), monocytes and macrophages are targeted by CHIKV and contribute to virus-induced pathogenesis in both humans and animals (reviewed in ref. 142). Activated macrophages are the primary infiltrating cell in infected tissues (Figure 3 and refs. 8, 72, 105–107), and elevated levels of MCP-1, the major chemoattractant for monocytes and macrophages, correlate with high viral loads in persons with acute CHIKV infection (8, 106, 111–113, 117, 143). Depletion of macrophages in mice results in reduced CHIKV-associated musculoskeletal disease but significantly prolonged viremia (106), highlighting independent functions of macrophages in CHIKV disease. Monocytes and macrophages have been detected in synovial fluid from chronic CHIKV patients (8) and animals (105), and CHIKV RNA and protein are detectable in synovial macrophages from humans (8) and nonhuman primates (NHPs) (105) during the chronic phase of infection. These data suggest that macrophages are a source of persistent virus and contribute to CHIKV-induced arthropathy. Plasmacytoid dendritic cells, NK cells, and neutrophils also infiltrate infected tissues during acute CHIKV infection (8, 143–146), but their role in CHIKV control remains to be clarified.
Acute infection in humans leads to activation and proliferation of CD8+ T cells, while a CD4+ T cell response is dominant during the chronic phase of CHIKV disease (8, 115). Although activated, CD8+ T cells do not appear to mediate CHIKV clearance or disease in animals (125). In contrast, studies using mice deficient in various types of lymphocytes implicate CD4+ T cells as inflammatory mediators in infected tissues (125, 147). However, these cells also may contribute to viral clearance (147). Tregs are involved in CHIKV pathology, as expansion of Tregs reduces CHIKV disease by selectively inhibiting CHIKV-specific CD4+ effector T cells (148). In addition, γδ T cells, which are abundant in skin, protect against CHIKV disease, as γδ T cell–deficient mice display exacerbated CHIKV infection (149).
Development of CHIKV-neutralizing antibodies is essential to control CHIKV viremia (125, 137, 147, 150, 151). CHIKV-infected humans, mice, and NHPs develop potent and neutralizing IgM and IgG antibodies that control viremia and confer cross-protection against secondary CHIKV infections in animal models (125, 137, 143, 147, 150–152). In humans, IgM levels are detected within 5–7 days after the onset of symptoms (153), peak several weeks after infection (154), and begin to wane over the next several months (154). An IgG response can be detected approximately 7–10 days after onset of illness, often after viremia has been cleared (37, 154–156). Many mouse and human IgM and IgG antibodies broadly and potently neutralize CHIKV (157–162). The most potently neutralizing antibodies target domains A and B of the E2 glycoprotein, with those targeting domain B often displaying broad neutralization against multiple strains of CHIKV and other related alphaviruses (157, 159–161). In B cell–deficient (μMT) mice, persistent, steady-state viremia occurs following CHIKV infection (147, 150), further highlighting the importance of neutralizing antibodies in mitigating CHIKV disease.
Although antibodies control viral load, CHIKV antigen and RNA have been detected in muscle and synovial biopsies from patients with chronic CHIKV disease, and muscle satellite cells and synovial macrophages have been proposed to be reservoirs for persistent CHIKV in humans (refs. 8, 72, and Figure 3). Persistent CHIKV infection also has been observed for weeks to months in experimentally infected mice (137, 147) and NHPs (105, 163), with vRNA detected in joint tissue (150), muscle, and spleen (105). In addition, persistent IgM levels have been detected in patients and animals experiencing chronic arthritis (8, 105, 126, 153, 164, 165). These data suggest that persistent CHIKV antigen, or perhaps infection, in protected cellular reservoirs triggers inflammation that culminates in chronic CHIKV rheumatic disease (Figure 3). Further investigation to identify specific CHIKV reservoirs and mechanisms of persistence will greatly inform illness management and development of therapeutics for chronic CHIKV pathologies.
Prospective CHIKV treatments and vaccines
The recent reemergence and worldwide spread of CHIKV render this virus an important public health threat. Although the mortality rate of CHIKV disease is modest, the debilitating and chronic nature of CHIKV disease and its associated economic burden are important considerations for the development of specific treatments. To date, no licensed anti-CHIKV therapeutics or vaccines are available, and only palliative care using analgesics, antipyretics, and NSAIDs is recommended to alleviate symptoms. As traditional vector control has met with only limited success in CHIKV containment, there is a significant need for safe, efficacious, and economical CHIKV treatments or vaccines to mitigate viral spread and limit disease burden.
A number of therapeutic strategies to combat CHIKV have been investigated (reviewed in ref. 166). To date, ribavirin is the only FDA-licensed drug tested in humans that effected a positive outcome in CHIKV-infected patients (167). High-throughput screening of chemical libraries (168–171), as well as synthesis of designer drugs (172, 173), has identified promising candidate CHIKV antivirals. Anti-CHIKV therapeutics also have been discovered or developed to directly target the viral replication cycle, including stages of entry, protein synthesis, genome replication, or enzymatic functions. Drugs indirectly targeting host factors required for efficient replication also have been identified (reviewed in ref. 166). Although compound discovery has unearthed a number of putative CHIKV antivirals, further testing in animal models and humans is required for clinical advancement of these drugs.
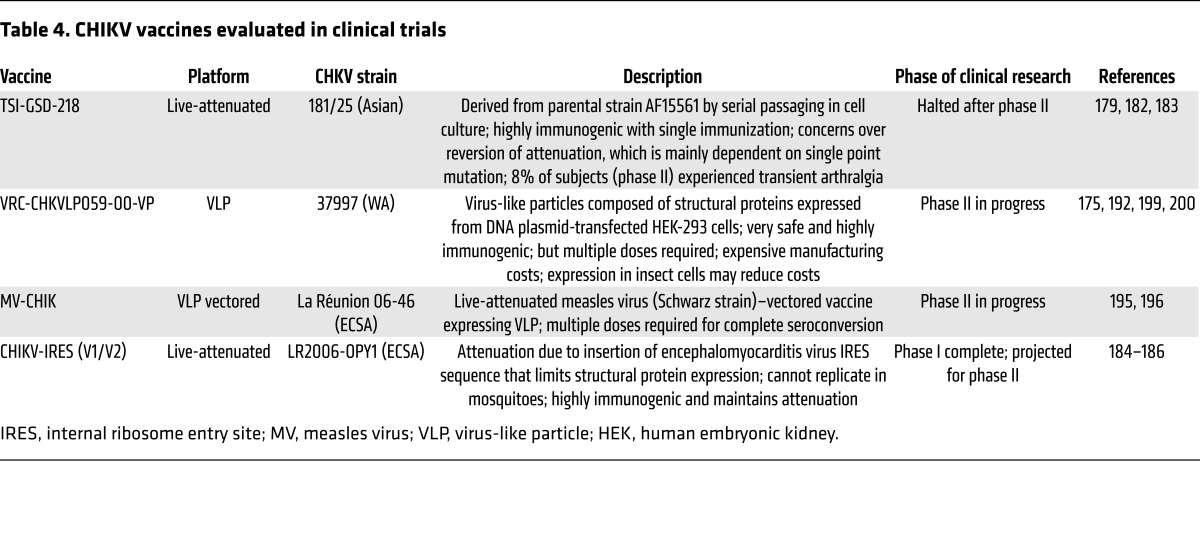
Since humoral immune responses control CHIKV infection and persistence (104, 125, 137, 147, 150, 151) and neutralizing CHIKV antibodies are often protective across CHIKV clades as well as other alphaviruses (152, 161), the use of monoclonal antibodies as prophylactics or therapeutics against CHIKV may offer another avenue for CHIKV control (reviewed in ref. 174). Passive transfer of antisera (104, 106, 175) or isolated monoclonal antibodies protects against CHIKV disease in mice (160, 161, 176) or NHPs (177). Furthermore, in postexposure therapeutic trials, monoclonal antibodies protect against CHIKV disease in mice, even when administered at late times of infection (160, 161), suggesting that immunotherapy would be effective for treatment of CHIKV infection. However, the best strategy to prevent further spread of CHIKV is likely to be an effective vaccine. As evidence suggests that a single vaccine against CHIKV should protect against all CHIKV strains and provide lifelong immunity, and humans are the primary amplifying host during urban epidemics, rapidly deployed vaccination of at-risk populations could curtail mounting epidemics. Furthermore, immunization of tourists and military personnel traveling to endemic or epidemic areas would protect travelers and decrease importation of the virus to naive populations. Various CHIKV vaccine strategies have been investigated, with many in preclinical and clinical studies (reviewed in ref. 178). Here, we summarize various vaccine candidates, focusing on those that have advanced to human studies (Table 4).
Table 4. CHIKV vaccines evaluated in clinical trials.
Live-attenuated vaccines offer effective and lasting immunity, often with a single dose and low cost of production. The first reported live-attenuated CHIKV vaccine is strain 181/25 (also called TSI-GSD-218), which was developed by passaging of clinical isolate AF15561 extensively in cell culture (Table 4 and ref. 179). Strain 181/25 is attenuated but immunogenic in both mice and NHPs (138, 179, 180), and vaccination with 181/25 protects NHPs against CHIKV challenge (179). Strain 181/25 showed no adverse effects in a phase I clinical trial (181), and although it was highly immunogenic in phase II clinical trials, approximately 8% of subjects developed mild, transient arthralgia (182). Attenuation of 181/25 is attributable to only two point mutations in E2 (T12I and G82R) (180); the mutation at residue 82, which enhances glycosaminoglycan binding (74, 75), is the dominant attenuating factor (180). However, residue 82 is also a major determinant of CHIKV clearance, as it influences neutralization by CHIKV-specific antibodies (150). Indeed, reversion at residue 82 in 181/25 was observed in mice (75, 180) and humans (183), indicating instability of attenuation and raising concerns about safety. Therefore, other strategies for development of live-attenuated vaccines for CHIKV have been investigated.
One such live-attenuated candidate, CHIKV/IRES, is a vaccine that contains an internal ribosomal entry site (IRES) in place of the subgenomic promoter (Table 4). This modification decreases the expression of viral structural proteins (184, 185), which attenuates replication in mammalian cells and prevents replication in mosquito cells, as the IRES is not functional in insect cells (184). The CHIKV/IRES vaccine is safe, highly immunogenic, and efficacious in mice and NHPs (184, 185), while remaining attenuated (186). Other live-attenuated CHIKV vaccine strategies include deletions in viral genes (187, 188), modification of glycosaminoglycan binding (138), alphavirus chimeras (189, 190), and codon alteration (191). As reversion is a major safety concern with live-attenuated vaccines, a CHIKV vaccine containing a combination of attenuation strategies could diminish reversion and pathogenicity.
Virus-like particles (VLPs) represent another promising vaccine strategy (Table 4). VLPs are generated by expression of the CHIKV structural cassette from a DNA expression plasmid transfected into human cells (175). The expressed structural proteins form particles that are indistinguishable from intact virions but are replication-incompetent because of the lack of genomic vRNA (175). In both mice and NHPs, the VLP vaccine elicits high-titer neutralizing antibodies that are protective against heterologous strains (175). In a three-dose escalation phase I trial, the VLP vaccine was safe, well tolerated, highly immunogenic, with a 100% seroconversion rate in all dose cohorts after booster immunizations (192), and cross-protective against multiple CHIKV strains (193). Although the VLP vaccine is currently in phase II clinical trials (NCT02562482, ClinicalTrials.gov), concerns over cost and manufacturing efficiency have prompted other strategies to generate VLPs. One uses a recombinant baculovirus-insect cell expression system that yields VLPs eliciting strong neutralizing antibodies, which are protective in mice (194). Another VLP vaccine strategy uses a recombinant, live-attenuated measles virus (MV) vector (MV-CHIKV) (Table 4 and ref. 195). In preclinical studies using Ifnar–/– mice, MV-CHIKV elicited robust and cross-neutralizing immune responses that protect against lethal challenge (195). In a phase I clinical trial of MV-CHIKV, neutralizing antibodies were detected in a dose-dependent manner with 100% seroconversion after booster immunization in all three dose cohorts, despite the presence of preexisting antivector immunity (196). Although the vaccine was reasonably tolerated at the two lower doses, 58% of the high-dose cohort exhibited adverse events related to vaccination (196). This vaccine is the third CHIKV vaccine to enter phase II clinical trials (NCT02861586).
In addition to these vaccine candidates, other recent CHIKV vaccine approaches investigated in preclinical studies include inactivated particles, subunit, and DNA- and recombinant virus–vectored vaccines (reviewed in ref. 178). Although substantial progress has been made in the development of CHIKV vaccines, a number of difficulties will need to be overcome. The sporadic nature of CHIKV epidemics makes it difficult to identify at-risk populations for phase III clinical trials. As CHIKV epidemics have mostly occurred in developing nations, the most utilitarian vaccine should induce a robust and durable immune response with a single dose at a low cost. However, even though vaccination is the most promising means to protect people in regions with endemic CHIKV, the challenging balance between vaccine safety, immunogenicity, and economical constraints of production may impede CHIKV vaccine development and licensing.
Conclusion
As CHIKV continues to emerge and disseminate, millions of people will experience the significant burden of chronic, incapacitating CHIKV disease. Our current understanding of CHIKV pathobiology indicates that CHIKV-elicited immune responses serve both protective and pathogenic functions, with several types of immune cells contributing to CHIKV-induced rheumatic disease. The duplicity and varied manifestations of CHIKV immunopathology pose challenges to the development of effective treatments. Although recent studies have contributed to a better understanding of the basic biology of CHIKV replication and disease, future work on virus-vector interactions, molecular mechanisms of viral replication, careful deconstruction of the multifaceted CHIKV-induced immune responses, and development of therapeutic interventions will be required to combat CHIKV transmission and illness.
Acknowledgments
This work was supported by Public Health Service awards F32 AI096833 (to LAS), R01 AI123348 (to LAS and TSD), and U54 AI057157 for the Southeast Regional Center of Excellence for Emerging Infections and Biodefense (to LAS and TSD). We thank Anthony Lentscher, Nicole McAllister, and Danica Sutherland for critical review of the manuscript. We apologize to all whose work could not be referenced because of formatting and length restrictions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(3):737–749.
https://doi.org/10.1172/JCI84417.
References
- 1.Lumsden WH. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. II. General description and epidemiology. Trans R Soc Trop Med Hyg. 1955;49(1):33–57. doi: 10.1016/0035-9203(55)90081-X. [DOI] [PubMed] [Google Scholar]
- 2.Robinson MC. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955;49(1):28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 3.Ross RW. The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 1956;54(2):177–191. doi: 10.1017/S0022172400044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiberville SD, et al. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013;99(3):345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey DE. Chikungunya and dengue: a case of mistaken identity? J Hist Med Allied Sci. 1971;26(3):243–262. doi: 10.1093/jhmas/xxvi.3.243. [DOI] [PubMed] [Google Scholar]
- 6.Halstead SB. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerging Infect Dis. 2015;21(4):557–561. doi: 10.3201/eid2104.141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couturier E, et al. Impaired quality of life after chikungunya virus infection: a 2-year follow-up study. Rheumatology (Oxford) 2012;51(7):1315–1322. doi: 10.1093/rheumatology/kes015. [DOI] [PubMed] [Google Scholar]
- 8.Hoarau JJ, et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010;184(10):5914–5927. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- 9.Schilte C, et al. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis. 2013;7(3):e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sissoko D, et al. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis. 2009;3(3):e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renault P, Josseran L, Pierre V. Chikungunya-related fatality rates, Mauritius, India, and Reunion Island. Emerging Infect Dis. 2008;14(8):1327. doi: 10.3201/eid1408.080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gérardin P, et al. Multidisciplinary prospective study of mother-to-child chikungunya virus infections on the island of La Réunion. PLoS Med. 2008;5(3):e60. doi: 10.1371/journal.pmed.0050060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gérardin P, et al. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: the CHIMERE cohort study on Reunion Island. PLoS Negl Trop Dis. 2014;8(7):e2996. doi: 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diallo D, et al. Landscape ecology of sylvatic chikungunya virus and mosquito vectors in southeastern Senegal. PLoS Negl Trop Dis. 2012;6(6):e1649. doi: 10.1371/journal.pntd.0001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg. 1999;60(2):281–286. doi: 10.4269/ajtmh.1999.60.281. [DOI] [PubMed] [Google Scholar]
- 16.Jupp PG, McIntosh BM. Aedes furcifer and other mosquitoes as vectors of chikungunya virus at Mica, northeastern Transvaal, South Africa. J Am Mosq Control Assoc. 1990;6(3):415–420. [PubMed] [Google Scholar]
- 17.Jupp PG, McIntosh BM, Dos Santos I, DeMoor P. Laboratory vector studies on six mosquito and one tick species with chikungunya virus. Trans R Soc Trop Med Hyg. 1981;75(1):15–19. doi: 10.1016/0035-9203(81)90005-5. [DOI] [PubMed] [Google Scholar]
- 18.Inoue S, et al. Distribution of three arbovirus antibodies among monkeys (Macaca fascicularis) in the Philippines. J Med Primatol. 2003;32(2):89–94. doi: 10.1034/j.1600-0684.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 19.Paul SD, Singh KR. Experimental infection of Macaca radiata with Chikungunya virus and transmission of virus by mosquitoes. Indian J Med Res. 1968;56(6):802–811. [PubMed] [Google Scholar]
- 20.Broeckel R, Haese N, Messaoudi I, Streblow DN. Nonhuman Primate Models of Chikungunya Virus Infection and Disease (CHIKV NHP Model) Pathogens. 2015;4(3):662–681. doi: 10.3390/pathogens4030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh BM, et al. Further studies on the Chikungunya outbreak in Southern Rhodesia in 1962. I. Mosquitoes, wild primates and birds in relation to the epidemic. Ann Trop Med Parasitol. 1964;58:45–51. doi: 10.1080/00034983.1964.11686213. [DOI] [PubMed] [Google Scholar]
- 22.Weaver SC. Evolutionary influences in arboviral disease. Curr Top Microbiol Immunol. 2006;299:285–314. doi: 10.1007/3-540-26397-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchette NJ, Rudnick A, Garcia R. Alphaviruses in Peninsular Malaysia: II. Serological evidence of human infection. Southeast Asian J Trop Med Public Health. 1980;11(1):14–23. [PubMed] [Google Scholar]
- 24.Sam IC, et al. Chikungunya Virus in Macaques, Malaysia. Emerging Infect Dis. 2015;21(9):1683–1685. doi: 10.3201/eid2109.150439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfe ND, et al. Sylvatic transmission of arboviruses among Bornean orangutans. Am J Trop Med Hyg. 2001;64(5-6):310–316. doi: 10.4269/ajtmh.2001.64.310. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal A, et al. Evidence of experimental vertical transmission of emerging novel ECSA genotype of Chikungunya Virus in Aedes aegypti. PLoS Negl Trop Dis. 2014;8(7):e2990. doi: 10.1371/journal.pntd.0002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavale M, et al. Venereal transmission of chikungunya virus by Aedes aegypti mosquitoes (Diptera: Culicidae) Am J Trop Med Hyg. 2010;83(6):1242–1244. doi: 10.4269/ajtmh.2010.09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81(pt 2):471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 29.Volk SM, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010;84(13):6497–6504. doi: 10.1128/JVI.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88(pt 9):2363–2377. doi: 10.1099/vir.0.82858-0. [DOI] [PubMed] [Google Scholar]
- 31.Schuffenecker I, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3(7):e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49(6):942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8(7):491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 34.de Lamballerie X, Leroy E, Charrel RN, Ttsetsarkin K, Higgs S, Gould EA. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virol J. 2008;5:33. doi: 10.1186/1743-422X-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibney KB, et al. Chikungunya fever in the United States: a fifteen year review of cases. Clin Infect Dis. 2011;52(5):e121–e126. doi: 10.1093/cid/ciq214. [DOI] [PubMed] [Google Scholar]
- 36.Hochedez P, et al. Chikungunya infection in travelers. Emerg Infect Dis. 2006;12(10):1565–1567. doi: 10.3201/eid1210.060495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezza G, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370(9602):1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 38.Grandadam M, et al. Chikungunya virus, southeastern France. Emerging Infect Dis. 2011;17(5):910–913. doi: 10.3201/eid1705.101873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3(12):e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One. 2009;4(8):e6835. doi: 10.1371/journal.pone.0006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsetsarkin KA, et al. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci U S A. 2011;108(19):7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1(4):310–317. doi: 10.1016/j.coviro.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsetsarkin KA, et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. doi: 10.1038/ncomms5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazeille M, et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2(11):e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arias-Goeta C, Mousson L, Rougeon F, Failloux AB. Dissemination and transmission of the E1-226V variant of chikungunya virus in Aedes albopictus are controlled at the midgut barrier level. PLoS One. 2013;8(2):e57548. doi: 10.1371/journal.pone.0057548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hapuarachchi HC, et al. Re-emergence of Chikungunya virus in South-east Asia: virological evidence from Sri Lanka and Singapore. J Gen Virol. 2010;91(pt 4):1067–1076. doi: 10.1099/vir.0.015743-0. [DOI] [PubMed] [Google Scholar]
- 47.Sam IC, et al. Chikungunya virus of Asian and Central/East African genotypes in Malaysia. J Clin Virol. 2009;46(2):180–183. doi: 10.1016/j.jcv.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Kumar NP, Joseph R, Kamaraj T, Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol. 2008;89(pt 8):1945–1948. doi: 10.1099/vir.0.83628-0. [DOI] [PubMed] [Google Scholar]
- 49.Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao-Lormeau VM, Musso D. Emerging arboviruses in the Pacific. Lancet. 2014;384(9954):1571–1572. doi: 10.1016/S0140-6736(14)61977-2. [DOI] [PubMed] [Google Scholar]
- 51. Centers for Disease Control and Prevention. Chikungunya Geographic Distribution. CDC Web site. http://www.cdc.gov/chikungunya/geo/index.html Updated May 12, 2016. Accessed January 9, 2017.
- 52.Nhan TX, Musso D. The burden of chikungunya in the Pacific. Clin Microbiol Infect. 2015;21(6):e47–e48. doi: 10.1016/j.cmi.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Musso D, Cao-Lormeau VM, Gubler DJ. Zika virus: following the path of dengue and chikungunya? Lancet. 2015;386(9990):243–244. doi: 10.1016/S0140-6736(15)61273-9. [DOI] [PubMed] [Google Scholar]
- 54.Roth A, et al. Concurrent outbreaks of dengue, chikungunya and Zika virus infections — an unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012-2014. Euro Surveill. 2014;19(41):pii:20929. doi: 10.2807/1560-7917.es2014.19.41.20929. [DOI] [PubMed] [Google Scholar]
- 55.Arankalle VA, et al. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol. 2007;88(Pt 7):1967–1976. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- 56.Dupont-Rouzeyrol M, et al. Chikungunya virus and the mosquito vector Aedes aegypti in New Caledonia (South Pacific Region) Vector Borne Zoonotic Dis. 2012;12(12):1036–1041. doi: 10.1089/vbz.2011.0937. [DOI] [PubMed] [Google Scholar]
- 57.Horwood PF, et al. Outbreak of chikungunya virus infection, Vanimo, Papua New Guinea. Emerging Infect Dis. 2013;19(9):1535–1538. doi: 10.3201/eid1909.130130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mombouli JV, et al. Chikungunya virus infection, Brazzaville, Republic of Congo, 2011. Emerging Infect Dis. 2013;19(9):1542–1543. doi: 10.3201/eid1909.130451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wangchuk S, et al. Chikungunya fever outbreak, Bhutan, 2012. Emerging Infect Dis. 2013;19(10):1681–1684. doi: 10.3201/eid1910.130453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu D, et al. Chikungunya virus with E1-A226V mutation causing two outbreaks in 2010, Guangdong, China. Virol J. 2013;10:174. doi: 10.1186/1743-422X-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cassadou S, Boucau S, Petit-Sinturel M, Huc P, Leparc-Goffart I, Ledrans M. Emergence of chikungunya fever on the French side of Saint Martin island, October to December 2013. Euro Surveill. 2014;19(13):pii:20752. doi: 10.2807/1560-7917.es2014.19.13.20752. [DOI] [PubMed] [Google Scholar]
- 62.Nunes MR, et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kraemer MU, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kuhn RJ. Togaviridae. In: Knipe DM, Howley PM, eds. Fields Virology. Philadelphia, Pennsylvania, USA: Lippincott, Williams, and Wilkins; 2013:629–650. [Google Scholar]
- 65.Khan AH, Morita K, Parquet mDel C, Hasebe F, Mathenge EG, Igarashi A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol. 2002;83(pt 12):3075–3084. doi: 10.1099/0022-1317-83-12-3075. [DOI] [PubMed] [Google Scholar]
- 66.Solignat M, Gay B, Higgs S, Briant L, Devaux C. Replication cycle of chikungunya: a re-emerging arbovirus. Virology. 2009;393(2):183–197. doi: 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Firth AE, Chung BY, Fleeton MN, Atkins JF. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snyder JE, et al. Functional characterization of the alphavirus TF protein. J Virol. 2013;87(15):8511–8523. doi: 10.1128/JVI.00449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voss JE, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468(7324):709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 70.Sourisseau M, et al. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007;3(6):e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gay B, Bernard E, Solignat M, Chazal N, Devaux C, Briant L. pH-dependent entry of chikungunya virus into Aedes albopictus cells. Infect Genet Evol. 2012;12(6):1275–1281. doi: 10.1016/j.meegid.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 72.Ozden S, et al. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2(6):e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salvador B, Zhou Y, Michault A, Muench MO, Simmons G. Characterization of Chikungunya pseudotyped viruses: Identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology. 2009;393(1):33–41. doi: 10.1016/j.virol.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silva LA, et al. A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol. 2014;88(5):2385–2397. doi: 10.1128/JVI.03116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashbrook AW, et al. Residue 82 of the Chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J Virol. 2014;88(21):12180–12192. doi: 10.1128/JVI.01672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Acharya D, Paul AM, Anderson JF, Huang F, Bai F. Loss of glycosaminoglycan receptor binding after mosquito cell passage reduces Chikungunya virus infectivity. PLoS Negl Trop Dis. 2015;9(10):e0004139. doi: 10.1371/journal.pntd.0004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Duijl-Richter MK, Hoornweg TE, Rodenhuis-Zybert IA, Smit JM. Early events in Chikungunya virus infection-from virus cell binding to membrane fusion. Viruses. 2015;7(7):3647–3674. doi: 10.3390/v7072792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernard E, et al. Endocytosis of Chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS One. 2010;5(7):e11479. doi: 10.1371/journal.pone.0011479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol. 2010;84(22):11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spuul P, Balistreri G, Kääriäinen L, Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. J Virol. 2010;84(15):7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol. 2001;75(8):3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thaa B, et al. Differential PI3K-Akt-mTOR activation by Semliki Forest and chikungunya virus, dependent on nsP3 and connected to replication complex internalisation. J Virol. 2015;89(22):11420–11437. doi: 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One. 2016;11(3):e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaedigk-Nitschko K, Schlesinger MJ. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology. 1990;175(1):274–281. doi: 10.1016/0042-6822(90)90209-A. [DOI] [PubMed] [Google Scholar]
- 86.Lusa S, Garoff H, Liljeström P. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology. 1991;185(2):843–846. doi: 10.1016/0042-6822(91)90556-Q. [DOI] [PubMed] [Google Scholar]
- 87.Loewy A, Smyth J, von Bonsdorff CH, Liljeström P, Schlesinger MJ. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69(1):469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taylor A, et al. Effects of an in-frame deletion of the 6k gene locus from the genome of Ross River virus. J Virol. 2016;90(8):4150–4159. doi: 10.1128/JVI.03192-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryan C, Ivanova L, Schlesinger MJ. Effects of site-directed mutations of transmembrane cysteines in sindbis virus E1 and E2 glycoproteins on palmitylation and virus replication. Virology. 1998;249(1):62–67. doi: 10.1006/viro.1998.9281. [DOI] [PubMed] [Google Scholar]
- 90.Ivanova L, Schlesinger MJ. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67(5):2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen KC, Kam YW, Lin RT, Ng MM, Ng LF, Chu JJ. Comparative analysis of the genome sequences and replication profiles of chikungunya virus isolates within the East, Central and South African (ECSA) lineage. Virol J. 2013;10:169. doi: 10.1186/1743-422X-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soonsawad P, et al. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to Alphavirus budding. J Virol. 2010;84(21):11145–11151. doi: 10.1128/JVI.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sjöberg M, Garoff H. Interactions between the transmembrane segments of the alphavirus E1 and E2 proteins play a role in virus budding and fusion. J Virol. 2003;77(6):3441–3450. doi: 10.1128/JVI.77.6.3441-3450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fros JJ, Pijlman GP. Alphavirus infection: host cell shut-off and inhibition of antiviral responses. Viruses. 2016;8(6):166. doi: 10.3390/v8060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joubert PE, et al. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J Exp Med. 2012;209(5):1029–1047. doi: 10.1084/jem.20110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Judith D, et al. Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 2013;14(6):534–544. doi: 10.1038/embor.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krejbich-Trotot P, et al. Chikungunya triggers an autophagic process which promotes viral replication. Virol J. 2011;8:432. doi: 10.1186/1743-422X-8-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog. 2013;9(12):e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Das I, et al. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS One. 2014;9(6):e100531. doi: 10.1371/journal.pone.0100531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scholte FE, et al. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J Virol. 2015;89(8):4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Assunção-Miranda I, Cruz-Oliveira C, Da Poian AT. Molecular mechanisms involved in the pathogenesis of alphavirus-induced arthritis. Biomed Res Int. 2013;2013:973516. doi: 10.1155/2013/973516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hawman DW, et al. Mechanisms underlying the pathogenesis of arthritogenic alphaviruses: host immune responses and virus persistence. Future Virol. 2014;9(5):441–444. doi: 10.2217/fvl.14.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suhrbier A, Jaffar-Bandjee MC, Gasque P. Arthritogenic alphaviruses — an overview. Nat Rev Rheumatol. 2012;8(7):420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- 104.Couderc T, et al. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008;4(2):e29. doi: 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Labadie K, et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010;120(3):894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gardner J, et al. Chikungunya virus arthritis in adult wild-type mice. J Virol. 2010;84(16):8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morrison TE, et al. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. Am J Pathol. 2011;178(1):32–40. doi: 10.1016/j.ajpath.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chua HH, et al. A fatal case of chikungunya virus infection with liver involvement. Med J Malaysia. 2010;65(1):83–84. [PubMed] [Google Scholar]
- 109.Chusri S, Siripaitoon P, Hirunpat S, Silpapojakul K. Case reports of neuro-Chikungunya in southern Thailand. Am J Trop Med Hyg. 2011;85(2):386–389. doi: 10.4269/ajtmh.2011.10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Das T, et al. Chikungunya fever: CNS infection and pathologies of a re-emerging arbovirus. Prog Neurobiol. 2010;91(2):121–129. doi: 10.1016/j.pneurobio.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 111.Teng TS, et al. A systematic meta-analysis of immune signatures in patients with acute Chikungunya virus infection. J Infect Dis. 2015;211(12):1925–1935. doi: 10.1093/infdis/jiv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruiz Silva M, van der Ende-Metselaar H, Mulder HL, Smit JM, Rodenhuis-Zybert IA. Mechanism and role of MCP-1 upregulation upon chikungunya virus infection in human peripheral blood mononuclear cells. Sci Rep. 2016;6:32288. doi: 10.1038/srep32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Her Z, et al. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J Immunol. 2010;184(10):5903–5913. doi: 10.4049/jimmunol.0904181. [DOI] [PubMed] [Google Scholar]
- 114.Schilte C, et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med. 2010;207(2):429–442. doi: 10.1084/jem.20090851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wauquier N, Becquart P, Nkoghe D, Padilla C, Ndjoyi-Mbiguino A, Leroy EM. The acute phase of Chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J Infect Dis. 2011;204(1):115–123. doi: 10.1093/infdis/jiq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gardner CL, Burke CW, Higgs ST, Klimstra WB, Ryman KD. Interferon-alpha/beta deficiency greatly exacerbates arthritogenic disease in mice infected with wild-type Chikungunya virus but not with the cell culture-adapted live-attenuated 181/25 vaccine candidate. Virology. 2012;425(2):103–112. doi: 10.1016/j.virol.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chow A, et al. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J Infect Dis. 2011;203(2):149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ayu SM, et al. Seroprevalence survey of Chikungunya virus in Bagan Panchor, Malaysia. Am J Trop Med Hyg. 2010;83(6):1245–1248. doi: 10.4269/ajtmh.2010.10-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dupuis-Maguiraga L, Noret M, Brun S, Le Grand R, Gras G, Roques P. Chikungunya disease: infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Negl Trop Dis. 2012;6(3):e1446. doi: 10.1371/journal.pntd.0001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ng KW, et al. Clinical features and epidemiology of chikungunya infection in Singapore. Singapore Med J. 2009;50(8):785–790. [PubMed] [Google Scholar]
- 121.Economopoulou A, et al. Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Réunion. Epidemiol Infect. 2009;137(4):534–541. doi: 10.1017/S0950268808001167. [DOI] [PubMed] [Google Scholar]
- 122.Borgherini G, et al. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis. 2007;44(11):1401–1407. doi: 10.1086/517537. [DOI] [PubMed] [Google Scholar]
- 123.Tandale BV, et al. Systemic involvements and fatalities during Chikungunya epidemic in India, 2006. J Clin Virol. 2009;46(2):145–149. doi: 10.1016/j.jcv.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 124.Rajapakse S, Rodrigo C, Rajapakse A. Atypical manifestations of chikungunya infection. Trans R Soc Trop Med Hyg. 2010;104(2):89–96. doi: 10.1016/j.trstmh.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 125.Teo TH, Lum FM, Lee WW, Ng LF. Mouse models for Chikungunya virus: deciphering immune mechanisms responsible for disease and pathology. Immunol Res. 2012;53(1–3):136–147. doi: 10.1007/s12026-012-8266-x. [DOI] [PubMed] [Google Scholar]
- 126.Borgherini G, et al. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin Infect Dis. 2008;47(4):469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- 127.Gérardin P, et al. Perceived morbidity and community burden after a Chikungunya outbreak: the TELECHIK survey, a population-based cohort study. BMC Med. 2011;9:5. doi: 10.1186/1741-7015-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rodriguez-Morales AJ, Cardona-Ospina JA, Villamil-Gómez W, Paniz-Mondolfi AE. How many patients with post-chikungunya chronic inflammatory rheumatism can we expect in the new endemic areas of Latin America? Rheumatol Int. 2015;35(12):2091–2094. doi: 10.1007/s00296-015-3302-5. [DOI] [PubMed] [Google Scholar]
- 129.Gérardin P, et al. Predictors of Chikungunya rheumatism: a prognostic survey ancillary to the TELECHIK cohort study. Arthritis Res Ther. 2013;15(1):R9. doi: 10.1186/ar4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thiberville SD, Boisson V, Gaudart J, Simon F, Flahault A, de Lamballerie X. Chikungunya fever: a clinical and virological investigation of outpatients on Reunion Island, South-West Indian Ocean. PLoS Negl Trop Dis. 2013;7(1):e2004. doi: 10.1371/journal.pntd.0002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Javelle E, Ribera A, Degasne I, Gaüzère BA, Marimoutou C, Simon F. Specific management of post-chikungunya rheumatic disorders: a retrospective study of 159 cases in Reunion Island from 2006–2012. PLoS Negl Trop Dis. 2015;9(3):e0003603. doi: 10.1371/journal.pntd.0003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Krejbich-Trotot P, Denizot M, Hoarau JJ, Jaffar-Bandjee MC, Das T, Gasque P. Chikungunya virus mobilizes the apoptotic machinery to invade host cell defenses. FASEB J. 2011;25(1):314–325. doi: 10.1096/fj.10-164178. [DOI] [PubMed] [Google Scholar]
- 133.Dhanwani R, Khan M, Alam SI, Rao PV, Parida M. Differential proteome analysis of Chikungunya virus-infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics. 2011;11(10):1936–1951. doi: 10.1002/pmic.201000500. [DOI] [PubMed] [Google Scholar]
- 134.Ziegler SA, Lu L, da Rosa AP, Xiao SY, Tesh RB. An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg. 2008;79(1):133–139. [PubMed] [Google Scholar]
- 135.Chen CI, et al. Comparative pathogenesis of epidemic and enzootic Chikungunya viruses in a pregnant Rhesus macaque model. Am J Trop Med Hyg. 2010;83(6):1249–1258. doi: 10.4269/ajtmh.2010.10-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rudd PA, et al. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J Virol. 2012;86(18):9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hawman DW, et al. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J Virol. 2013;87(24):13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gardner CL, et al. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis. 2014;8(2):e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rohatgi A, et al. Infection of myofibers contributes to increased pathogenicity during infection with an epidemic strain of chikungunya virus. J Virol. 2014;88(5):2414–2425. doi: 10.1128/JVI.02716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kinne RW, et al. Macrophages in rheumatoid arthritis. Arthritis Res Ther. 2000;2(3):189–202. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ng LF, et al. IL-1β, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS One. 2009;4(1):e4261. doi: 10.1371/journal.pone.0004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Long KM, Heise MT. Protective and pathogenic responses to Chikungunya virus infection. Curr Trop Med Rep. 2015;2(1):13–21. doi: 10.1007/s40475-015-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Poo YS, et al. CCR2 deficiency promotes exacerbated chronic erosive neutrophil-dominated Chikungunya virus arthritis. J Virol. 2014;88(12):6862–6872. doi: 10.1128/JVI.03364-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stoermer KA, et al. Genetic ablation of arginase 1 in macrophages and neutrophils enhances clearance of an arthritogenic alphavirus. J Immunol. 2012;189(8):4047–4059. doi: 10.4049/jimmunol.1201240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Petitdemange C, et al. Unconventional repertoire profile is imprinted during acute Chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011;7(9):e1002268. doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Teo TH, et al. Caribbean and La Réunion Chikungunya virus isolates differ in their capacity to induce proinflammatory Th1 and NK cell responses and acute joint pathology. J Virol. 2015;89(15):7955–7969. doi: 10.1128/JVI.00909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poo YS, et al. Multiple immune factors are involved in controlling acute and chronic Chikungunya virus infection. PLoS Negl Trop Dis. 2014;8(12):e3354. doi: 10.1371/journal.pntd.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee WW, et al. Expanding regulatory T cells alleviates chikungunya virus-induced pathology in mice. J Virol. 2015;89(15):7893–7904. doi: 10.1128/JVI.00998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Long KM, et al. γδ T cells play a protective role in Chikungunya virus-induced disease. J Virol. 2015;90(1):433–443. doi: 10.1128/JVI.02159-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hawman DW, et al. Pathogenic Chikungunya virus evades B cell responses to establish persistence. Cell Rep. 2016;16(5):1326–1338. doi: 10.1016/j.celrep.2016.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lum FM, Teo TH, Lee WW, Kam YW, Rénia L, Ng LF. An essential role of antibodies in the control of Chikungunya virus infection. J Immunol. 2013;190(12):6295–6302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kam YW, et al. Sero-prevalence and cross-reactivity of chikungunya virus specific anti-E2EP3 antibodies in arbovirus-infected patients. PLoS Negl Trop Dis. 2015;9(1):e3445. doi: 10.1371/journal.pntd.0003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Grivard P, et al. Molecular and serological diagnosis of Chikungunya virus infection. Path Biol (Paris) 2007;55(10):490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 154.Kam YW, et al. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med. 2012;4(4):330–343. doi: 10.1002/emmm.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kam YW, et al. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. J Virol. 2012;86(23):13005–13015. doi: 10.1128/JVI.01780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Panning M, Grywna K, van Esbroeck M, Emmerich P, Drosten C. Chikungunya fever in travelers returning to Europe from the Indian Ocean region, 2006. Emerging Infect Dis. 2008;14(3):416–422. doi: 10.3201/eid1403.070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fox JM, et al. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell. 2015;163(5):1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jin J, et al. Neutralizing monoclonal antibodies block Chikungunya virus entry and release by targeting an epitope critical to viral pathogenesis. Cell Rep. 2015;13(11):2553–2564. doi: 10.1016/j.celrep.2015.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lam S, Nyo M, Phuektes P, Yew CW, Tan YJ, Chu JJ. A potent neutralizing IgM mAb targeting the N218 epitope on E2 protein protects against Chikungunya virus pathogenesis. MAbs. 2015;7(6):1178–1194. doi: 10.1080/19420862.2015.1083664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pal P, et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013;9(4):e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Smith SA, et al. Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against Chikungunya virus. Cell Host Microbe. 2015;18(1):86–95. doi: 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Warter L, et al. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol. 2011;186(5):3258–3264. doi: 10.4049/jimmunol.1003139. [DOI] [PubMed] [Google Scholar]
- 163.Messaoudi I, et al. Chikungunya virus infection results in higher and persistent viral replication in aged rhesus macaques due to defects in anti-viral immunity. PLoS Negl Trop Dis. 2013;7(7):e2343. doi: 10.1371/journal.pntd.0002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chopra A, Anuradha V, Lagoo-Joshi V, Kunjir V, Salvi S, Saluja M. Chikungunya virus aches and pains: an emerging challenge. Arthritis Rheum. 2008;58(9):2921–2922. doi: 10.1002/art.23753. [DOI] [PubMed] [Google Scholar]
- 165.Malvy D, et al. Destructive arthritis in a patient with chikungunya virus infection with persistent specific IgM antibodies. BMC Infect Dis. 2009;9:200. doi: 10.1186/1471-2334-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Abdelnabi R, Neyts J, Delang L. Towards antivirals against chikungunya virus. Antiviral Res. 2015;121:59–68. doi: 10.1016/j.antiviral.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ravichandran R, Manian M. Ribavirin therapy for Chikungunya arthritis. J Infect Dev Ctries. 2008;2(2):140–142. doi: 10.3855/T2.2.140. [DOI] [PubMed] [Google Scholar]
- 168.Ashbrook AW, et al. Antagonism of the Sodium-Potassium ATPase Impairs Chikungunya Virus Infection. MBio. 2016;7(3):e00693-16. doi: 10.1128/mBio.00693-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gigante A, et al. Identification of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones as novel inhibitors of Chikungunya virus replication. J Med Chem. 2014;57(10):4000–4008. doi: 10.1021/jm401844c. [DOI] [PubMed] [Google Scholar]
- 170.Kaur P, et al. Inhibition of chikungunya virus replication by harringtonine, a novel antiviral that suppresses viral protein expression. Antimicrob Agents Chemother. 2013;57(1):155–167. doi: 10.1128/AAC.01467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lucas-Hourani M, et al. A phenotypic assay to identify Chikungunya virus inhibitors targeting the nonstructural protein nsP2. J Biomol Screen. 2013;18(2):172–179. doi: 10.1177/1087057112460091. [DOI] [PubMed] [Google Scholar]
- 172.Bassetto M, et al. Computer-aided identification, design and synthesis of a novel series of compounds with selective antiviral activity against chikungunya virus. Antiviral Res. 2013;98(1):12–18. doi: 10.1016/j.antiviral.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 173.Das PK, et al. Design and validation of novel chikungunya virus protease inhibitors. Antimicrob Agents Chemother. 2016;60(12):7382–7395. doi: 10.1128/AAC.01421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Clayton AM. Monoclonal antibodies as prophylactic and therapeutic agents against Chikungunya virus. J Infect Dis. 2016;214(suppl 5):S506–S509. doi: 10.1093/infdis/jiw324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Akahata W, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 2010;16(3):334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Selvarajah S, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in Chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. 2013;7(9):e2423. doi: 10.1371/journal.pntd.0002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pal P, et al. Chikungunya viruses that escape monoclonal antibody therapy are clinically attenuated, stable, and not purified in mosquitoes. J Virol. 2014;88(15):8213–8226. doi: 10.1128/JVI.01032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schwameis M, et al. Chikungunya vaccines in development. Hum Vaccin Immunother. 2016;12(3):716–731. doi: 10.1080/21645515.2015.1101197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE, Lupton HW. Development of an attenuated strain of Chikungunya virus for use in vaccine production. Vaccine. 1986;4(3):157–162. doi: 10.1016/0264-410X(86)90003-4. [DOI] [PubMed] [Google Scholar]
- 180.Gorchakov R, et al. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol. 2012;86(11):6084–6096. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McClain DJ, et al. Immunologic interference from sequential administration of live attenuated alphavirus vaccines. J Infect Dis. 1998;177(3):634–641. doi: 10.1086/514240. [DOI] [PubMed] [Google Scholar]
- 182.Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. Phase II safety and immunogenicity study of live Chikungunya virus vaccine TSI-GSD-218. Am J Trop Med Hyg. 2000;62(6):681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- 183.Hoke CH, Jr, et al. US Military contributions to the global response to pandemic Chikungunya. Vaccine. 2012;30(47):6713–6720. doi: 10.1016/j.vaccine.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 184.Plante K, et al. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 2011;7(7):e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Roy CJ, et al. Chikungunya vaccine candidate is highly attenuated and protects nonhuman primates against telemetrically monitored disease following a single dose. J Infect Dis. 2014;209(12):1891–1899. doi: 10.1093/infdis/jiu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Plante KS, Rossi SL, Bergren NA, Seymour RL, Weaver SC. Extended preclinical safety, efficacy and stability testing of a live-attenuated Chikungunya vaccine candidate. PLoS Negl Trop Dis. 2015;9(9):e0004007. doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hallengärd D, et al. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol. 2014;88(5):2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Piper A, et al. Chikungunya virus host range E2 transmembrane deletion mutants induce protective immunity against challenge in C57BL/6J mice. J Virol. 2013;87(12):6748–6757. doi: 10.1128/JVI.03357-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang E, et al. Chimeric alphavirus vaccine candidates for chikungunya. Vaccine. 2008;26(39):5030–5039. doi: 10.1016/j.vaccine.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang E, Kim DY, Weaver SC, Frolov I. Chimeric Chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. J Virol. 2011;85(17):9249–9252. doi: 10.1128/JVI.00844-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nougairede A, De Fabritus L, Aubry F, Gould EA, Holmes EC, de Lamballerie X. Random codon re-encoding induces stable reduction of replicative fitness of Chikungunya virus in primate and mosquito cells. PLoS Pathog. 2013;9(2):e1003172. doi: 10.1371/journal.ppat.1003172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chang LJ, et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet. 2014;384(9959):2046–2052. doi: 10.1016/S0140-6736(14)61185-5. [DOI] [PubMed] [Google Scholar]
- 193.Goo L, et al. A virus-like particle vaccine elicits broad neutralizing antibody responses in humans to all Chikungunya virus genotypes. J Infect Dis. 2016;214(10):1487–1491. doi: 10.1093/infdis/jiw431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Metz SW, et al. Effective Chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl Trop Dis. 2013;7(3):e2124. doi: 10.1371/journal.pntd.0002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Brandler S, et al. A recombinant measles vaccine expressing Chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine. 2013;31(36):3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 196.Ramsauer K, et al. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based Chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis. 2015;15(5):519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 197.Kelvin AA, et al. Inflammatory cytokine expression is associated with chikungunya virus resolution and symptom severity. PLoS Negl Trop Dis. 2011;5(8):e1279. doi: 10.1371/journal.pntd.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chaaitanya IK, et al. Role of proinflammatory cytokines and chemokines in chronic arthropathy in CHIKV infection. Viral Immunol. 2011;24(4):265–271. doi: 10.1089/vim.2010.0123. [DOI] [PubMed] [Google Scholar]
- 199.Akahata W, Nabel GJ. A specific domain of the Chikungunya virus E2 protein regulates particle formation in human cells: implications for alphavirus vaccine design. J Virol. 2012;86(16):8879–8883. doi: 10.1128/JVI.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Metz SW, et al. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine. 2013;31(51):6092–6096. doi: 10.1016/j.vaccine.2013.09.045. [DOI] [PubMed] [Google Scholar]