Abstract
For efficient delivery of required genetic elements we utilized high-capacity adenoviral vectors in the past allowing high transgene capacities of up to 36 kb. Previously we explored the hyperactive Sleeping Beauty (SB) transposase (HSB5) for somatic integration from the high-capacity adenoviral vectors genome. To further improve this hybrid vector system we hypothesized that the previously described hyperactive SB transposase SB100X will result in significantly improved efficacies after transduction of target cells. Plasmid based delivery of the SB100X system revealed significantly increased integration efficiencies compared with the previously published hyperactive SB transposase HSB5. After optimizing experimental setups for high-capacity adenoviral vectors-based delivery of the SB100X system we observed up to eightfold and 100-fold increased integration efficiencies compared with the previously published hyperactive SB transposase HSB5 and the inactive transposase mSB, respectively. Furthermore, transposon copy numbers per cell were doubled with SB100X compared with HSB5 when using the identical multiplicity of infection. We believe that this improved hybrid vector system represents a valuable tool for achieving stabilized transgene expression in cycling cells and for treatment of numerous genetic disorders. Especially for in vivo approaches this improved adenoviral hybrid vector system will be advantageous because it may potentially allow reduction of the applied viral dose.
Introduction
Gene therapy is an effort to cure diseases on a genomic level by utilizing nucleic acid as a drug. The development of this emerging medical field is ongoing for more than 20 years and has successfully reached the bedside which was demonstrated by recent pioneering studies for diseases either curable by gene transfer into hematopoietic stem cells or human liver.1,2,3,4 However, challenging problems remain which have to be addressed in the future and further improvements of the used vectors are desirable to continue this process.
Currently used vectors in the clinic are still lacking efficiency and safety for systemic in vivo delivery of large transgenes, and therefore high-capacity adenoviral vectors (HCAdV) represent attractive alternatives. They lack all sequences coding for viral proteins, leading to a transgene capacity of up to 36 kb5 which is remarkably higher compared with retroviruses and lentiviruses (~10 kb)6 or Adeno-associated virus (AAV) (~5 kb).7 By using HCAdV instead of early generation adenoviral vectors lacking only one or more of the early adenoviral genes, the toxicity problems associated with these vectors due to leaky expression of adenoviral genes in the transduced target cell have been significantly improved.8,9,10,11 The newest generation of adenoviral vectors offers the lowest immunogenicity and toxicity currently available. The HCAdV can be used for ex vivo treatment (cells are explanted from the patient, treated, and reimplanted)12 or for systemic in vivo administration.10 Since the production of the HCAdV using a helper virus to provide all viral proteins in trans5,13,14 is highly modular, it is possible to exchange the viral proteins in an easy way leading to a different tropism and escape from neutralizing antibodies of the vector. A novel cloning system was recently introduced by Mueck-Haeusl and colleagues using a platform based on bacterial artificial chromosomes and recombineering.15
An attribute which sets back the use of adenoviral vectors for a broad range of diseases has been shown that HCAdVs are replication deficient due to the lack of a retention and replication mechanisms leading to loss of the therapeutic DNA during mitosis. This is especially of interest for the treatment of cells having a short generation time such as blood cells16 because copy numbers of the vector rapidly decline and the therapeutic effect is lost. Attempts have been made in the past to engineer mitotically stable adenoviral vectors either by somatic integration into the genome or by episomal persistence. Examples of genetic elements for integration of the therapeutic transgene into the host genome included PhiC31,17 retrovirus,18 adeno-associated virus,19,20,21,22 and the Sleeping Beauty (SB) transposase system.10,23,24 Two different attempts have been made to construct episomally persisting adenovirus hybrid vectors. One system was based on Epstein-Barr virus replication and retention25,26 and the other a nonviral plasmid replicon containing a scaffold matrix attachment region.27
Here we evaluated a significantly improved version of an HCAdV hybrid system combining high transduction efficiencies of adenoviral vectors with the hyperactive SB transposase system based on SB100X for enhanced somatic integration. The SB transposase system, derived from the TC1/mariner family of transposable elements, integrates the therapeutic transgene, cloned between two inverted repeat (IR) sequences, close to randomly at TA dinucleotides within the human genome in a copy and paste manner.24,28 Currently the SB transposase system is reaching the clinic in a trail using ex vivo engineered T-cells.29 Since the efficacy of the wild type transposase was relatively low, several attempts have been made to engineer hyperactive forms of the SB transposase molecule. This improvement of the SB transposase system after the awakening from its domestic sleep in the salmonid genome in 1997 was based on a rational approach leading to the hyperactive SB transposase versions SB11, HSB5, and SB100X.30,31,32 The latest version of SB transposase shows a 100-fold increased enzymatic activity compared with its original version. Since the systemic plasmid based delivery of SB transposase in large mammals remains a major challenge, several HCAdV-SB transposase hybrid vectors were constructed in the past.10,23 In this study, we are introducing the latest and most active version of a HCAdV-SB transposase hybrid vector based on hyperactive SB100X and show that this system is significantly enhanced.
Results
Principle and vectors used to establish the improved adenoviral hybrid vector system utilizing hyperactive transposase SB100X
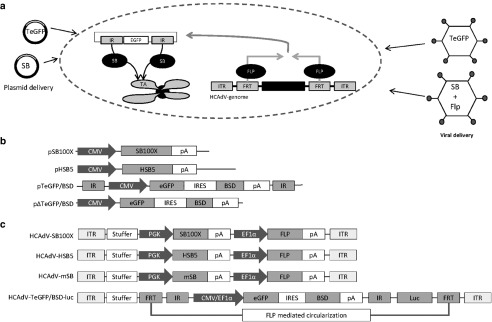
In this study, we aimed at developing a significantly improved generation of a mitotically stable high-capacity adenoviral vector (HCAdV) hybrid system by taking advantage of enhanced SB-mediated integration. The hyperactive SB system is based on a two vector system which can either be delivered by plasmids or viral vectors. After viral or nonviral delivery into target cells and transport to the nucleus, the whole transposon cassette is excised and circularized by Flp recombinase and subsequently mobilized and integrated by SB transposase into the host cell genome between TA dinucleotides (Figure 1a). The detailed mechanism was shown and discussed elsewhere.10,23,33 Since efficient SB transposase mediated integration was demonstrated to work most efficiently from circularized constructs as shown in Figure 1a, the transposon needs to be excised from the HCAdV donor vector by Flp recombinase mediated recombination.10,23,33
Figure 1.
Design of the vectors and plasmids used for this study. (a) Basic principle of the Sleeping Beauty (SB) transposase hybrid vector system. Transposon enhanced green fluourescent protein (TeGFP) and SB transposase are codelivered into one cell using plasmid-based (left) or adenoviral delivery (right). The gene of interest (eGFP) is cloned between SB transposon derived 5′ and 3′inverted repeats (IR) resulting into the transposon TeGFP. When the transposase and the transposon, which in case of adenoviral delivery was excised by Flp recombination in the target cell, cotransduce one cell, two SB transposase proteins bind at each IR sequence. This results in excision of the transposon which in a further step is integrated at TA dinucleotides present in the host chromosomes of the cell. (b) DNA sequences contained in plasmids pSB100X, pHSB5, pTeGFP/BSD, and p▵TeGFP/BSD used for transposition assays. (c) Design of the high capacity adenoviral vectors (HCAdV) used in this study. HCAdV-SB100X describes the novel SB transposase vector based on the latest version of Sleeping Beauty transposase (SB100X). HCAdV-HSB5 represents the precursor version of HCAdV-SB100X containing the hyperactive SB transposase HSB5 instead of SB100X. HCAdV-mSB is a vector with an inactive form of SB transposase (mSB). HCAdV-TeGFP/BSD matches a newly constructed reporter vector to evaluate different transposase efficiencies. It contains a biscistronic construct expressing blasticidin for selection of stably transduced cells and eGFP to evaluate transposon derived transgene expression. CMV, cytomegalovirus promoter; SB100X, hyperactive SB transposase; HSB5, hyperactive SB transposase; mSB, inactive version of SB transposase; pA, polyadenoylation signal; IR, inverted repeat; IRES, internal ribosomal entry site; eGFP, enhanced green fluorescent protein; BSD, blasticidin; PGK, phosphoglycerate kinase promoter; EF1α, elongation factor alpha promoter; FRT, Flp recombinase recognition sites; Luc, luciferase.
To establish and optimize the system, we constructed various plasmids and HCAdVs which are displayed in Figure 1b and Figure 1c. We aimed to explore the hyperactive SB transposase SB100X in the context of an adenoviral hybrid vector and to compare this setup with the previously published adenoviral hybrid vector system utilizing the hyperactive SB transposase HSB5.10,24 As negative control an inactive version of the transposase (mSB) carrying a point mutation in the catalytic domain was used. To quantify integration events and to visualize transgene expression, we generated a transposon containing an internal ribosomal entry site based bicistronic construct expressing enhanced green fluorescent protein (eGFP) and the selection marker blasticidin (BSD) under the control of the CMV promoter. This expression cassette was either delivered as a plasmid (pTeGFP/BSD) or as a viral vector (HCAdV-TeGFP/BSD-luc).
Vectors HCAdV-SB100X and HCAdV-TeGFP/BSD-luc were produced using an established protocol.5,14 In brief, after release of the viral genome from the plasmid (see Supplementary Figure S1) vectors were amplified by serial passaging. After large-scale amplification vectors were purified by cesium chloride gradient ultracentrifugation and titrated (see Supplementary Figure S1). Note that the viral amplification production protocol was slightly modified and optimized for the HCAdV-SB100X vector (see also Materials and Methods section). Before performing further experiments the vector HCAdV-TeGFP/BSD-luc was evaluated for eGFP expression (see Supplementary Figure S1).
Functionality of DNA sequences contained in adenoviral vectors for achieving mitotic stability
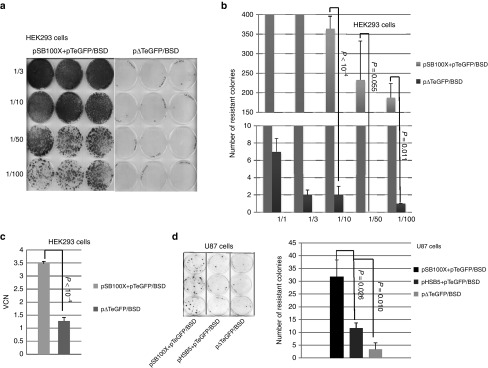
To characterize and to show functionality of DNA sequences contained in the adenoviral hybrid vector system described in Figure 1, we performed plasmid-based colony-forming assays in human embryonic kidney cells (HEK293). We cotransfected the hyperactive SB100X encoding plasmid (pSB100X) in which transposase expression is driven by the cytomegalovirus promoter with either the transposon containing plasmid pTeGFP/BSD or the control plasmid p▵TeGFP/BSD lacking IRs at a molar ratio of 1:3 of transposase to transposon. In case of the control, plasmid stuffer DNA was added to assure that equal molar ratios were transfected into both groups, respectively. Before splitting the cells at different ratios (1:3, 1:10, 1:50, and 1:100), equal transfection efficiencies were evaluated by eGFP expression 48 hours after transfection (data not shown). Experiments were performed in triplicates and after blasticidin selection resistant colonies were stained with methylene blue (Figure 2a and Figure 2b). We observed more than 60-fold increased transposition efficiencies for SB100X compared with the control groups (Figure 2a and Figure 2b). Note that for groups that received SB100X, which were split 1:1 and 1:3, colonies were not countable anymore because they grew to complete confluency. As shown in Figure 2c vector genome copy numbers (VCN) per cell were increased twofold compared with surviving colonies in the control group.
Figure 2.
Quantification of transposition events based on plasmid transfection in HEK293 and U87 cells. We evaluated functionality of plasmids in HEK293 cells using colony forming assays allowing for selection of cells in which transposition events occurred. After plasmid transfection cells were split at different ratios and blasticidin selection was started. (a) HEK293 cells were transfected using a molar ratio of 3:1 (transgene: transposase). The negative control received the plasmid pΔTeGFP/BSD and stuffer DNA, respectively. Two days post-transfection cells were split at different ratios (1/3, 1/10, 1/50, and 1/100) and selection was started using blasticidin. Methylene blue staining was performed between 2 and 6 weeks after initial splitting depending in the number of surviving colonies and colonies counted. (b) Quantification of transposition events. The diagram shows the experimental group which received plasmids pSB100X and pTeGFP/BSD or the control group which was transfected with the plasmid pΔTeGFP and stuffer DNA to reach equal molar ratios. For the splitting ratios 1/1 and 1/3, colonies could not be counted manually in case of the group which received plasmids pSB100X and pTeGFP/BSD. Groups which received the control plasmid without IRs (pΔTeGFP) revealed a significantly reduced number of colonies. (c) Transgene copy numbers (VCN) in groups which received the plasmids pSB100X and pTeGFP/BSD or the control plasmid pΔTeGFP and stuffer DNA. To measure VCN genomic DNA was isolated and a quantitative PCR performed. (d) Colony forming assay performed in U87 glioblastoma cells comparing the previous SB transposase version HSB5 and the improved version SB100X. The control group received the plasmid pΔTeGFP/BSD. Selection was started 24 hours post-transfection. After blasticidin selection colonies were stained (left panel). Quantification of colonies (right panel) revealed that pSB100X was superior compared with all other groups including the precursor pHSB5 and pΔTeGFP/BSD. Error bars indicate mean ± SD. The P-values are provided and a significant difference between the two groups was confirmed if the P-value was < 0.05. *P < 0.05, ** P < 0.01, ***P < 0.001, ****P < 10–4, n.s.: not significant. SB100X, hyperactive SB transposase; HSB5, hyperactive SB transposase; HEK293, human embryonic kidney cells; TeGFP, transposon enhanced green fluourescent protein; SB, Sleeping Beauty; BSD, blasticidin; VCN, vector genome copy numbers; PCR, polymerase chain reaction.
To show that our results are cell line independent and to use a cell line which was also used for HCAdV based colony forming assays in our previous study,27 we aimed at confirming our findings in human glioblastoma cells line U87. Thus, U87 cells were cotransfected with the SB transposase encoding plasmids (pSB100X and pHSB5) and the transposon containing plasmid pTeGFP/BSD. The negative control group received the plasmid p▵TeGFP/BSD and stuffer DNA. As shown in Figure 2d SB100X resulted in the highest number of colonies also compared with the precursor SB transposase version HSB5.
Characterization and efficiencies of the high-capacity adenoviral vectors system utilizing SB100X for somatic integration
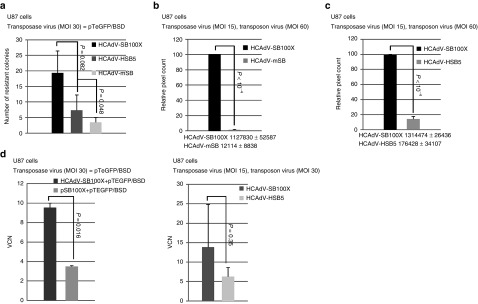
Consequently we performed experiments using the HCAdV hybrid-vector system as delivery platform for the transposase integration machinery in U87 cells. Initially we used plasmid delivery of the transposon (plasmid pTeGFP/BSD) in concert with HCAdV delivery of SB100X. We transfected the transposon DNA followed by infection with the respective transposase encoding adenoviral vector (HCAdV-SB100X, HCAdV-HSB5, and HCAdV-mSB) using a multiplicity of infection (MOI) of 30. After the selection process was completed, cell colonies were stained. HCAdV-SB100X resulted in a higher number of colonies as compared to HCAdV-HSB5 (2.7-fold) or the negative control group which received mSB (fivefold) (Figure 3a and Supplementary Figure S2a). Later we tested different experimental settings for the adenovirus hybrid vector system for which the transposon donor and the transposase were delivered by the HCAdV. For the first experiment U87 cells were infected at MOIs 15 for the vectors HCAdV-SB100X and HCAdV-mSB, and the transposon encoding vector was applied at MOI 60 (Figure 3b). After the selection process was completed the analyses were performed based on the software CellProfiler enabling pixel based analysis. The software was used for colony forming assays performed with U87 cells, since efficiency was high and resulting colonies too close to be counted manually (see Supplementary Figure S2b). Between HCAdV-SB100X and HCAdV-mSB there was a nearly 100-fold difference based on pixel surface coverage (Figure 3b and Supplementary Figure S2b). Figure 3c shows an experiment in which we compared HCAdV-SB100X and HCAdV-HSB5 in U87 cells at MOI 15 for the transposase and MOI 30 for the transposon-donor vector HCAdV-TeGFP/BSD-FRT/IR. The efficiency of HCAdV-SB100 was > sevenfold higher compared with HCAdV-HSB5.
Figure 3.
Quantification of colony forming numbers based on viral transduction of U87 glioblastoma-derived cells. (a) This figure shows the results of a colony forming assay performed in U87 cells after cotransduction of the transposon containing plasmid pTeGFP/BSD and the transposase encoding viral vectors HCAdV-SB100X, HCAdV-HSB5, and HCAdV-mSB. (b) Results of a colony forming assay comparing HCAdV-SB100X and HCAdV-mSB at MOI 30 after coinfection with the transposon vector HCAdV-pTeGFP/BSD at MOI 60. After the selection process was completed remaining colonies were stained. Colored pixels were counted in a pixel based assay. There was ~ 100-fold difference between the HCAdV-SB100X and HCAdV-mSB. The number of absolute pixels is shown below the diagram as mean ± SD. (c) U87 cells were infected with the transposase vectors HCAdV-SB100X and HCAdV-HSB5 at MOI 15 and the transposon donor vector HCAdV-TeGFP/BSD-luc at MOI 30. There was a sevenfold difference in transposition efficiencies for HCAdV-SB100X compared with HCAdV-HSB5. The number of absolute pixel numbers is shown as mean ± SD below the diagram. (d) Transgene copy numbers from an experiment using plasmid delivery for the transposon and viral delivery for the transposase (left panel) and an experiment using solely HCAdV for delivery of transposon and transposase (right panel). Error bars indicate mean ± SD. The P-values are provided and a significant difference between the two groups was confirmed if the P-value was < 0.05. * P < 0.05, **P < 0.01, ***P < 0.001, ****P < 10–4, n.s.: not significant. MOI, multiplicity of infection; SB100X, hyperactive SB transposase; HSB5, hyperactive SB transposase; mSB, inactive version of SB transposase; TeGFP, transposon enhanced green fluorescent protein; BSD, blasticidin; Luc, luciferase; HCAdV, high-capacity adenoviral vectors.
To determine VCN in virus based experiments we isolated genomic DNA from pools of selected cells and performed quantitative polymerase chain reaction (PCR). The left panel of Figure 3d shows copy numbers of the transgene within the colonies of the experiment using the HCAdV for transposase delivery in combination with plasmid delivery for the transposon donor. We measured up to 10 VCN per cell for this group and up to four VCN for a group which received the SB100X system using plasmid-based delivery. As shown in the right panel of Figure 3d, VCNs were also increased (> twofold) in U87 cells which were infected with the SB100X based hybrid vector system if directly compared with the previous version of the adenoviral hybrid vector system utilizing HSB5 for somatic integration.
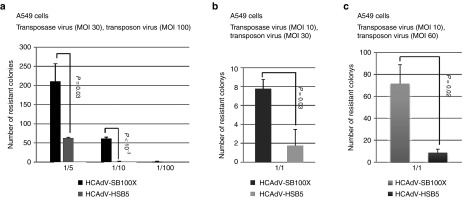
To rule out that this is not a cell line dependent phenomenon, we performed experiments in adenocarcinoma human alveolar basal epithelial (A549) cells (Figure 4). We infected cells at MOI 100, for the transposon donor vector and MOI 30, for the SB100X encoding HCAdV vector. Two days later, infection cells were split at different ratios (1/5, 1/10, and 1/100) and blasticidin selection was started. In concordance to our results obtained in U87 cells, HCAdV-SB100X showed significantly increased numbers of colonies compared with HCAdV-HSB5 (up to 10-fold for splitting ratio 1:10) (Figure 4a). We confirmed this trend in the second experiment performed in A549 cells when using a lower total MOI (MOI 30, for the transposon donor vector and MOI 10, for the SB100X encoding vector) as shown in Figure 4b. Representative single colonies stably expressing eGFP in A549 cells after selection are displayed in Supplementary Figure S2c. Finally, we doubled the MOI of the transposon vector (MOI 60) while keeping the MOI for transposase stable (MOI 10). This resulted in a significant increase in colony forming numbers (> eightfold) (Figure 4c). To show true transposition in surviving colonies we performed PCR analyses to prove that the selection process was sufficient to lose all episomal adenoviral vector genome copies of the HCAdV vector carrying the transgene. We conducted a vector-specific PCR which amplifies the transgene blasticidin which should be detectable in all resistant colonies and a second PCR which amplifies the left arm of the adenoviral genome including inverted terminal repeat (ITR) and packaging signal (see Supplementary Figure S3a). The latter PCR should only be positive if the adenoviral DNA molecule itself was maintained during selection pressure. We analyzed six individual clones derived from single colonies which were either transduced with HCAdV-HSB5 or HCAdV-SB100X. As expected we detected the transgene blasticidin in all analyzed colonies. However, we could show absence of the adenoviral DNA in five of six analyzed samples indicating true transposition in most of our colonies. One of our colonies showed a positive signal for the adenoviral ITR and the packing signal (see Supplementary Figure S3b).
Figure 4.
Performance of the viral hybrid vector system in human A549 adenocarcinoma cells. A549 cells were infected a two different total MOIs for transposon (HCAdV-TeGFP/BSD-luc) and transposase vectors (HCAdV-SB100X and HCAdV-HSB5). Two days postinfection, cells were split at different ratios (1/5, 1/10, and 1/100), maintained under blasticidin selection pressure and stained with methylene blue. (a) A549 cells were infected at an MOI of 30 for the transposase vector and MOI 100 for the transposon vector. (b) Colony forming assay performed at lower MOI using MOI of 10 of the transposase vector and MOI 30 for the transposon vector. (c) Colony forming assay performed at an MOI of 10 for the transposase vector and an elevated MOI for the transposon vector (MOI 60). Error bars indicate mean ± SD. The P-values are provided and a significant difference between the two groups was confirmed if the P-value was < 0.05. *P < 0.05, ** P < 0.01, ***P < 0.001, ****P < 10–4, n.s.: not significant. MOI, multiplicity of infection; SB100X, hyperactive SB transposase; HSB5, hyperactive SB transposase; mSB, inactive version of SB transposase; TeGFP, transposon enhanced green fluorescent protein; BSD, blasticidin; Luc, luciferase; HCAdV, high-capacity adenoviral vectors.
Discussion
The aim of this study was to construct and evaluate a new third generation high capacity adenoviral hybrid vector for somatic persistence in rapidly dividing cells. The integration machinery was based on SB transposase mediated integration. SB transposase based adenoviral hybrid vectors have been explored in previous studies.10,23 However, increased efficacy may be desirable to further improve the system and to potentially decrease the required viral dose to achieve stabilized transgene expression at therapeutic levels. In this study, we tested the latest optimized form of SB transposase SB100X32 offering significantly increased efficiencies compared with precursor versions. Note that SB100X was also used in other gene transfer studies utilizing other viral vector systems for efficient delivery of the genetic cargo. This included lentiviral vectors,34 baculovirus for long-term gene expression in the eye35 and Adeno-associated virus (AAV).36 This study complements these previous studies showing that the hybrid HCAdV system is efficient in delivering SB100X leading to remarkable integration efficiencies. Nevertheless it remains to be evaluated which viral vector system may be the most efficient technology for a specific application in gene therapy. It is likely that the molecular design and the viral vector type used for delivery of the SB transposase system needs to be adapted to the specific application.
The total number of colonies in plasmid based experiments was lower for all experimental settings in U87 cells when compared with experiments performed in HEK293 cells. We speculate that this may have two reasons. The protocol for the colony forming assay performed in U87 cells was different since selection was performed without splitting cells after transfection. Furthermore, initial transfection efficiencies vary between different cell lines. The other reason could be that the cell line and the origin of the tissue may have an influence on transposition efficiencies which was also shown by another study by Kolacsek and colleagues.37
For our experiments using the HCAdV for delivery of the transposase we observed that SB100X was more efficient compared with the precursor HSB5 and the inactive SB transposase mSB. When compared with HCAdV-HSB5 the gain of efficiency was > sevenfold for the SB100X based system, demonstrating that SB100X is superior compared with HSB5. This might be of special interest for in vivo experiments for which transposon and transposase are codelivered via two HCAdVs. Higher efficiency may allow injection of decreased vectors doses which may improve the toxicity profile related to incoming viral particles and expression of recombinases such as SB100X and Flp recombinase. Overall, improved efficiency of the SB transposase hybrid vector system may also allow decreasing the required viral load for achieving therapeutic levels to treat a genetic disease, which may correspond to reduced vector-related toxicity.
Moreover, the VCN was increased for the SB100X based hybrid vector system in A549- and U87 cells. On first sight, this observation may be advantageous because it may lead to increased expression levels, but genotoxicity risks may be increased if more integration sites at different loci in the host genome are detected. Notably, in U87 cells adenoviral vector delivery of the transposon system was more efficient when performing colony forming assays. Plasmid delivery of the identical transposon led to decreased numbers of resistant colonies. This could be explained by limitations of the transfection protocols for the respective cell line. Overall experiments in both cell lines clearly indicated that the MOI seems to play a crucial role for transposition.
It was shown that recombinant adenovirus genomes can persist extrachromosomally for a prolonged time especially in quiescent cells.8,9 Furthermore, there is evidence that adenovirus DNA molecules are more stable compared with nonviral DNA38 and that HCAdV genomes can somatically integrate at low frequencies.39 Therefore, it can be hypothesized that in our experiments these features of HCAdV genomes may contribute to background transgene expression. PCR analyses revealed that one isolated cell clone maintained HCAdV DNA and it can be speculated that in this particular cell clone the HCAdV genome was integrated into the host genome.
Next steps should involve experiments which utilize the high-capacity adenoviral hybrid vector system utilizing SB100X in primary cells. In our previous studies we successfully applied the precursor version of the hybrid vector system utilizing hyperactive SB transposase HSB5 instead of SB100X in primary cells in vivo in small and large animals.10,24 The only differences of both systems are the point mutations in the SB100X coding sequence and therefore, we also expect the SB100X-based system to work sufficiently in primary cells.
In this study, we renounced from performing integration site analysis for SB100X and HSB5-mediatd transposition since this was already performed in depth in other studies.24,36,40 It can be concluded from these studies that no significant differences can be detected regarding the integration profile between the hyperactive SB transposase SB100X and its precursor versions. Moreover the transposition machinery itself is independent from the viral or nonviral delivery method and therefore, we expect a comparable integration profile as shown for the previous studies. Although SB100X transposase clearly improves efficacy of the adenovirus SB transposase hybrid-vector system, some challenges still remain to overcome. This includes the necessity of using Flp recombinase for circularization from the adenoviral vector genome and the requirement of using two vectors coinfecting a single cell simultaneously. Latter is necessary since coexistence of transposon and transposase in the production cell line would inhibit vector production. One strategy to overcome this limitation could be micro-RNA mediated suppression of SB-transposase in the HCAdV producer cell lines. Such an approach was previously shown for expression of cytotoxic zinc fingers from adenoviral vectors.41 Another strategy could be the usage of an inducible system driving expression of the SB transposase. In summary, in this study we produced, established and evaluated a novel, significantly improved high capacity adenoviral vector system for somatic integration and stabilized transgene expression. In a further step, this hybrid vector system can be evaluated in preclinical studies. Especially for direct in vivo gene therapy, we believe that this system will play an important role in developing novel concepts for therapeutic approaches.
Materials and methods
Plasmids used in this study. The plasmids pZac-SB100,36 pHM5-PmeI,10 pHM5-Pepito-FRT,27 pHM5-Pepito-▵S/MAR-FRT/pΔTeGFP,27 p cytomegalovirus -HSB5,23 pEPito,42 and p phosphoglycerate kinase(PGK)-ΔTP23 were published previously and the plasmid pZAC2.1 was obtained from Jim Wilson (University of Pennsylvania, Philadelphia, PA).
To generate the HCAdV production plasmid pHCAdV-TeGFP/BSD-IR-luc, the following cloning steps were performed. pHM5-TeGFP/BSD-IR/FRT-luc was generated in this study and is based on pHM5-FRT/IR.10 A PCR using pEPito as template was used to amplify the transgene including cytomegalovirus promoter, eGFP cDNA, an IRES sequence and the blasticidin encoding DNA. To amplify the specific PCR product, primers GFP Forw (5′- GAT ACT CGA GTC GCC ACC ATG GTG AGC AAG-3′) and Blast rev (5′- CTA GCG GCC GCT ATT TAG ATC CTT AGC CCT C-3′) were used. The PCR product was digested using NotI and XhoI and the resulting DNA fragment was inserted into the vector pZac2.1, which was opened using the same enzymes. The resulting plasmid was digested with SmaI and the transgene cloned into the pHM5-FRT/IR resulting into the plasmid pHM5-TeGFP/BSD-IR/FRT. A luciferase expression cassette was added by digesting pHM5-FRT-IR using I-CeuI and cloning of a PCR product derived from pGL3-control vector (Promega, Madison, WI) using primers I-CEU-luc-forw (5′-CTA AGG TAG CGA AGC TCG AGA TCT GCG ATC TGC-3′) and PmeI-luc-rev (5′-CGC CGT TTA AAC CGA TTT TAC CAC ATT TGT-3′). This resulted in the plasmid pHM5-TeGFP/BSD-IR/FRT-luc. Finally the complete transposon and the luciferase expression cassette were released by I-CeuI and PI-SceI restriction enzyme digest and cloned into the HCAdV cloning vector pAdFTC8 using the same restriction enzyme sites. This resulted in the final HCAdV production plasmid pHCAdV-TeGFP/BSD-IR-luc.
The HCAdV production plasmid pHCAdV-SB100 was generated as follows: First the Flp recombinase expression cassette consisting of the Ef1a promotor, the Flp recombinase cDNA and the polyA signal was amplified using the primers EF1afw (5′-GTA AGT GCC GTG TGT GGT TCC C-3′) and PolyA rev (5′-AAT TCG CCC TTT GGG AAA ATC AAA AGA -3′). As template for this PCR reaction the plasmid pFTC-HSB5-Flp was used. The PCR product was cloned into the plasmid pHM5-PmeI using the restriction enzyme PmeI resulting in the plasmid pHM5-Flp. Subsequently the cDNA of SB100X was PCR amplified from pZac-SB100 using Primers SB100 FW (5′-GCC CTT CTA GTA TTT GGT AGC ATT GCC-3′) and SB100 REV(5′- CTA GAG GTA CCA CGC GTG AAT-3′) and the fragment was then cloned into the PmeI site of pPGK-ΔTP. The SB100X expression cassette expressed under control of the PGK promoter was then PCR amplified using primers PGKFW (5′-AAT TCT ACC GGG TAG GGG AGG C-3′) and SB100REV2 (5′ CCA GCT GGT TCT TTC CGC CT-3′). This fragment was then cloned into pHM5-FLP,23 which was digested with KpnI and blunted with T4 DNA polymerase resulting in the plasmid pHM5-Flp-SB100. After ClaI digest of this plasmid, homologous arms flanking the Flp-SB100 cassette for subsequent recombinieering into pBHCA-galK-kan harboring the adenoviral 5′ ITR, the adenoviral packing signal Ψ, and the 3′ ITR43 were generated by PCR using primers BHCA-Pepi-5′ (5′-CGC TTA ATG CGC CGC TAC AGG GCG CGT GGG GTG AGC GCG TCA ATT AAC CC-3′) and BHCA-Pepi-3′ (5′-CCC GAA CAT GAC GGT TAG TTG GAT GGC TCC GGG CGC GCC ATA AGA GCT CG-3′). This PCR fragment was then inserted via galK based counter-selection recombineering into pBHCA-galK-kan43 replacing galK-Kan with the Flp-SB100 cassette. This resulted in the HCAdV production plasmid pHCAdV-SB100X.
Vector production. Viruses HCAdV-HSB510 and HCAdV-mSB23 were described previously and recombinant vectors HCAdV-SB100X and HCAdV-TeGFP/BSD-IR were produced in this study. For virus production of HCAdV-SB100X and HCAdV-TeGFP/BSD-IR, a previously established protocol was applied.5,14 In brief, the HCAdV vector genomes were released by PmeI (HCAdV-SB100X) and NotI (HCAdV-TeGFP/BSD-IR) (see Supplementary Figure S1) and the linearized HCAdV genome was transfected into 116 cells producer cells.14 Pre-amplification steps were performed in tissue culture dishes using the previously published helper virus introduced by Palmer and Ng.14 For large scale amplification a spinner flask system was used and the vectors were purified using cesium chloride gradient ultracentrifugation as described previously.5,14 As first attempts to amplify HCAdV-SB100X failed until we infected with the helper virus 6 hours prior to the HCAdV infection during the serial amplification steps.
For titration and measurement of HCAdV transducing units, A549 cells were infected using different volumes of the final vector preparation. After 3 hours, cells were washed to eliminate noninfectious particles. Then DNA isolation was conducted and a quantitative PCR was performed using the primers TQ-eGFP-RH-fw (5′-GAA GCG CGA TCA CAT GGT-3′) and TQ-eGFP-RH-rv (5′-CCA TGC CGA GAG TGA TCC-3′) for EGFP containing vector and primers SBnf1 (5′-GGT GGC AGC ATC ATG TTG TG-3′) and SBnr2 (5'-CCT TCC TCA TGA TGC CAT CTA TT-3') for SB transposase containing vectors. A quantitative PCR was performed using the following protocol: a 5 minute denaturation step at 95°C was followed by 39 rounds of 15 seconds denaturation at 95°C, annealing and extension at 60° for 60 seconds (SYBR mix Biorad, Thermocylcer C1000 Touch (Biorad, Hummelgasse, Wien Austria) ).
Cell culture. All experiments were performed in HEK293, human adenocarcinoma alveolar basal epithelial-derived A549 cells, and U87 glioblastoma cells. For production of the HCAdV vectors 116 cells14 were used. HEK293, U87, and A549 cells were cultured in Dulbecco's modified eagle medium (DMEM, PAN-Biotech, Aidenbach, Germany). For culture of U87 cells dishes were collagenized with Collagen A (Biochrom, Berlin, Germany) according to the manufacturer's protocol. For 116 cells we used minimal essential medium (MEM Eagle, Pan-Biotech) additionally supplemented with 100 µg/ml hygromycin B for sustained selection for CRE expression during virus production. CRE expression is crucial to remove the packing signal from the helper virus used to supply essential viral proteins. In contrast to the helper virus the HCAdV genome contains a normal packaging not flanked by loxP sites. This assures that predominantly HCAdV genomes are packed into assembled virus particles. Fetal bovine serum (FBS, PAA, Pasching, Austria) was added to 10% together with 0,1 mg/ml penicillin-streptomycin (PAA).
Colony forming assay. For the plasmid based colony forming assays in HEK293, cells were grown to a density of ~70% in 6-well plates. Plasmid DNA was purified using a commercially available plasmid DNA isolation kit (Peqlab, Erlangen, Germany and Qiagen, Hilden, Germany). Transfection was performed using Fugene 6 (Promega). Before transfection was performed copy numbers were adjusted based on DNA concentration measured using a spectrophotometer (Eppendorf, Hamburg, Germany). After 2 days, transfection cells were split at different ratios and selection with blasticidin (Carl Roth, Karlsruhe, Germany) was started with a concentration of 4 µg/ml. Also medium was changed at average 3 times per week. For the viral and nonviral experiments using U87 cells, selection was started after 2 days by performing media exchange. For viral colony forming assays using A549 cells, cells cultured in 6-well plates were infected and 2 days later split at different ratios. Subsequently selection was started. After several weeks of successful selection, shown by 100% eGFP expressing colonies, cells were stained using methylene blue (Sigma-Aldrich, Munich, Germany) or DNA was isolated using a commercial DNA extraction kit (Peqlab).
Surface analysis for the percentage of covered dishes with cells was performed (in case of U87 cells) using the open source software CellProfiler available at www.cellprofiler.org from the Broad Institute44 and the analysis module “Measure Image Area Occupied”.
Molecular analyses of surviving cell clones after selection. After completion of the selection procedure genomic DNA was isolated from resistant cell pools derived from transduced HEK293- and U87 cells. To determine VCN, a transgene EGFP specific quantitative PCR was performed using the primers TQ-eGFP-RH-fw (5′-GAA GCG CGA TCA CAT GGT-3′) and TQ-eGFP-RH-rv (5′- CCA TGC CGA GAG TGA TCC-3′). A quantitative PCR was performed using the following protocol: a 5 minute denaturation step at 95°C was followed by 39 rounds of 15 seconds denaturation at 95°C, annealing and extension at 60° for 60 seconds (SYBR mix Biorad, Thermocylcer C1000 Touch (Biorad)).
To analyze surviving colonies derived from A549 cells on a molecular level, we isolated and amplified single cell colonies after the initial selection procedure was completed. When the colonies had grown to confluency in a 6 cm tissue culture dish, we prepared genomic DNA using the Qiagen Blood and Tissue kit which were then analyzed by two PCR reactions. The first PCR amplifies the transgene blasticidin which should be detectable in all resistant colonies and the second PCR amplifies the left arm of the adenoviral genome including ITR and packaging signal which results in a PCR product specific for the adenoviral DNA molecule which was maintained during selection pressure. Subsequently we performed a standard PCR using Phusion High-fidelity Polymerase (NEB) according to the NEB protocol using ITR and packaging signal-specific primers ADITR5′ (5′-GGC GGG TGA CGT AGT AGT GT-3′) and ADITR3′ (5′–GCG GAA AAC ACC TGA GAA AA-3′), and transgene-specific primers BSD5′ (5′-CCG CAT CTT CAC TGG TGT-3′) and BSD3′ (5′-GCT CAA GAT GCC CCT GTT CT-3′).
Statistical analysis. All experiments were performed using at least triplicates. All data are reported as mean ± SD unless otherwise noted. Statistical comparison was made using the two-tailed student's t-test, and a value of P < 0.05 was considered to be relevant compared to the respective control group.
Ethics statement. In the context of this study no research involving human subjects (including human material or human data) was performed.
SUPPLEMENTARY MATERIAL Figure S1. Production of the high-capacity vectors HCAdV-SB100X and HCAdV-TeGFP/BSD-luc. Figure S2. Sample colonies of the performed experiments. Figure S3. Molecular analyses of single colonies derived from A549 cells.
Author contributions
P.B. performed the majority of shown experiments, participated in the design of the study and contributed to drafting the manuscript. W.Z. made substantial contributions to conception and design. M.S. helped in drafting the manuscript and E.S. was involved in producing viral vectors analyzed in this study. A.E. conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript. The authors declare that there are no competing interests.
Acknowledgments
This work was supported by the PhD programme of the University Witten/Herdecke (P.B.) and the E-Rare project Transposmart (A.E.). We thank Zsuzsanna Izsvak (Max Delbrück Center for Molecular Medicine, Berlin, Germany) and Zoltan Ivics (Paul-Ehrlich-Institut, Langen, Germany) for providing the plasmid encoding SB100X. We would like to acknowledge that Dr. Martin Mück-Häusl (current address: Institute of Virology, Helmholtz Zentrum München, Munich, Germany) was involved in cloning the high-capacity vector HCAdV-SB100X. We would like to thank Phil Ng (Baylor College, Houston, USA) for providing 116 cells and the helper virus for HCAdV production.
Supplementary Material
References
- Aiuti, A, Biasco, L, Scaramuzza, S, Ferrua, F, Cicalese, MP, Baricordi, C et al. (2013). Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi, A, Montini, E, Lorioli, L, Cesani, M, Fumagalli, F, Plati, T et al. (2013). Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341: 864. [DOI] [PubMed] [Google Scholar]
- Nathwani, AC, Reiss, UM, Tuddenham, EG, Rosales, C, Chowdary, P, McIntosh, J et al. (2014). Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 371: 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina, S, Pai, SY, Gaspar, HB, Armant, M, Berry, CC, Blanche, S et al. (2014). A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med 371: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, L, Hausl, MA, Rauschhuber, C, Wolf, NM, Kay, MA and Ehrhardt, A (2009). A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc 4: 547–564. [DOI] [PubMed] [Google Scholar]
- Kumar, M, Keller, B, Makalou, N and Sutton, RE (2001). Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther 12: 1893–1905. [DOI] [PubMed] [Google Scholar]
- Grieger, JC and Samulski, RJ (2005). Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol 79: 9933–9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt, A and Kay, MA (2002). A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood 99: 3923–3930. [DOI] [PubMed] [Google Scholar]
- Schiedner, G, Morral, N, Parks, RJ, Wu, Y, Koopmans, SC, Langston, C et al. (1998). Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet 18: 180–183. [DOI] [PubMed] [Google Scholar]
- Hausl, MA, Zhang, W, Müther, N, Rauschhuber, C, Franck, HG, Merricks, EP et al. (2010). Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther 18: 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral, N, O'Neal, W, Rice, K, Leland, M, Kaplan, J, Piedra, PA et al. (1999). Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc Natl Acad Sci USA 96: 12816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H, Cao, H, Wohlfahrt, M, Kiem, HP and Lieber, A (2008). Tightly regulated gene expression in human hematopoietic stem cells after transduction with helper-dependent Ad5/35 vectors. Exp Hematol 36: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, RJ, Chen, L, Anton, M, Sankar, U, Rudnicki, MA and Graham, FL (1996). A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA 93: 13565–13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D and Ng, P (2003). Improved system for helper-dependent adenoviral vector production. Mol Ther 8: 846–852. [DOI] [PubMed] [Google Scholar]
- Mück-Häusl, M, Solanki, M, Zhang, W, Ruzsics, Z and Ehrhardt, A (2015). Ad 2.0: a novel recombineering platform for high-throughput generation of tailored adenoviruses. Nucleic Acids Res 43: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov, DM, Papayannopoulou, T, Stamatoyannopoulos, G and Lieber, A (2000). Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J Virol 74: 2567–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt, A, Yant, SR, Giering, JC, Xu, H, Engler, JA and Kay, MA (2007). Somatic integration from an adenoviral hybrid vector into a hot spot in mouse liver results in persistent transgene expression levels in vivo. Mol Ther 15: 146–156. [DOI] [PubMed] [Google Scholar]
- Zheng, C, Baum, BJ, Iadarola, MJ and O'Connell, BC (2000). Genomic integration and gene expression by a modified adenoviral vector. Nat Biotechnol 18: 176–180. [DOI] [PubMed] [Google Scholar]
- Gonçalves, MA, Holkers, M, Cudré-Mauroux, C, van Nierop, GP, Knaän-Shanzer, S, van der Velde, I et al. (2006). Transduction of myogenic cells by retargeted dual high-capacity hybrid viral vectors: robust dystrophin synthesis in duchenne muscular dystrophy muscle cells. Mol Ther 13: 976–986. [DOI] [PubMed] [Google Scholar]
- Recchia A, Parks RJ, Lamartina S, Toniatti C, Pieroni L, Palombo F, et al. (1999) Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc Nat Acad Sci USA 96:2615–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H and Lieber, A (2006). A helper-dependent capsid-modified adenovirus vector expressing adeno-associated virus rep78 mediates site-specific integration of a 27-kilobase transgene cassette. J Virol 80: 11699–11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H, Shayakhmetov, DM, Leege, T, Harkey, M, Li, Q, Papayannopoulou, T et al. (2005). A capsid-modified helper-dependent adenovirus vector containing the beta-globin locus control region displays a nonrandom integration pattern and allows stable, erythroid-specific gene expression. J Virol 79: 10999–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant, SR, Ehrhardt, A, Mikkelsen, JG, Meuse, L, Pham, T and Kay, MA (2002). Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol 20: 999–1005. [DOI] [PubMed] [Google Scholar]
- Zhang, W, Muck-Hausl, M, Wang, J, Sun, C, Gebbing, M, Miskey, C et al. (2013). Integration profile and safety of an adenovirus hybrid-vector utilizing hyperactive sleeping beauty transposase for somatic integration. PLoS One 8: e75344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel, F and Kochanek, S (2004). Long-term transgene expression in proliferating cells mediated by episomally maintained high-capacity adenovirus vectors. J Virol 78: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, JS, Gallaher, SD and Berk, AJ (2010). Delivery of an EBV episome by a self-circularizing helper-dependent adenovirus: long-term transgene expression in immunocompetent mice. Gene Ther 17: 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigtlander, R, Haase, R, Mück-Hausl, M, Zhang, W, Boehme, P, Lipps, HJ et al. (2013). A novel adenoviral hybrid-vector system carrying a plasmid replicon for safe and efficient cell and gene therapeutic applications. Mol Ther Nucleic Acids 2: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics, Z, Hackett, PB, Plasterk, RH and Izsvák, Z (1997). Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510. [DOI] [PubMed] [Google Scholar]
- Kebriaei, P, Huls, H, Jena, B, Munsell, M, Jackson, R, Lee, DA et al. (2012). Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum Gene Ther 23: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed, H, Izsvák, Z, Walisko, O and Ivics, Z (2004). Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther 9: 292–304. [DOI] [PubMed] [Google Scholar]
- Yant, SR, Park, J, Huang, Y, Mikkelsen, JG and Kay, MA (2004). Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol 24: 9239–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés, L, Chuah, MK, Belay, E, Jerchow, B, Manoj, N, Acosta-Sanchez, A et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41: 753–761. [DOI] [PubMed] [Google Scholar]
- Boehme, P, Doerner, J, Solanki, M, Jing, L, Zhang, W and Ehrhardt, A (2015). The sleeping beauty transposon vector system for treatment of rare genetic diseases: an unrealized hope? Curr Gene Ther 15: 255–265. [DOI] [PubMed] [Google Scholar]
- Staunstrup, NH, Moldt, B, Mátés, L, Villesen, P, Jakobsen, M, Ivics, Z et al. (2009). Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther 17: 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen, TA, Laakkonen, JP, Alasaarela, L, Airenne, KJ and Ylä-Herttuala, S (2014). Sleeping Beauty-baculovirus hybrid vectors for long-term gene expression in the eye. J Gene Med 16: 40–53. [DOI] [PubMed] [Google Scholar]
- Zhang, W, Solanki, M, Müther, N, Ebel, M, Wang, J, Sun, C et al. (2013). Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells. PLoS One 8: e76771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolacsek, O, Erdei, Z, Apáti, A, Sándor, S, Izsvák, Z, Ivics, Z et al. (2014). Excision efficiency is not strongly coupled to transgenic rate: cell type-dependent transposition efficiency of sleeping beauty and piggyBac DNA transposons. Hum Gene Ther Methods 25: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, L and Ehrhardt, A (2009). Persistence of high-capacity adenoviral vectors as replication-defective monomeric genomes in vitro and in murine liver. Hum Gene Ther 20: 883–896. [DOI] [PubMed] [Google Scholar]
- Harui, A, Suzuki, S, Kochanek, S and Mitani, K (1999). Frequency and stability of chromosomal integration of adenovirus vectors. J Virol 73: 6141–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt, B, Miskey, C, Staunstrup, NH, Gogol-Döring, A, Bak, RO, Sharma, N et al. (2011). Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol Ther 19: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydaminova, K, Ye, X, Wang, H, Richter, M, Ho, M, Chen, H et al. (2015). Efficient genome editing in hematopoietic stem cells with helper-dependent Ad5/35 vectors expressing site-specific endonucleases under microRNA regulation. Mol Ther Methods Clin Dev 1: 14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, R, Argyros, O, Wong, SP, Harbottle, RP, Lipps, HJ, Ogris, M et al. (2010). pEPito: a significantly improved non-viral episomal expression vector for mammalian cells. BMC Biotechnol 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mück-Häusl, M, Solanki, M, Zhang, W, Ruzsics, Z and Ehrhardt, A (2015). Ad 2.0: a novel recombineering platform for high-throughput generation of tailored adenoviruses. Nucleic Acids Res 43: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, TR, Kang, IH, Wheeler, DB, Lindquist, RA, Papallo, A, Sabatini, DM et al. (2008). CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.