Abstract
Type 1 diabetes (T1D) is a chronic autoimmune disease in which pancreatic β cells are killed by infiltrating immune cells and by cytokines released by these cells. This takes place in the context of a dysregulated dialogue between invading immune cells and target β cells, but the intracellular signals that decide β cell fate remain to be clarified. Alternative splicing (AS) is a complex post-transcriptional regulatory mechanism affecting gene expression. It regulates the inclusion/exclusion of exons into mature mRNAs, allowing individual genes to produce multiple protein isoforms that expand the proteome diversity. Functionally related transcript populations are co-ordinately spliced by master splicing factors, defining regulatory networks that allow cells to rapidly adapt their transcriptome in response to intra and extracellular cues. There is a growing interest in the role of AS in autoimmune diseases, but little is known regarding its role in T1D. In this review, we discuss recent findings suggesting that splicing events occurring in both immune and pancreatic β cells contribute to the pathogenesis of T1D. Splicing switches in Tcells and in lymph node stromal cells are involved in the modulation of the immune response against β cells, while β cells exposed to pro-inflammatory cytokines activate complex splicing networks that modulate β cell viability, expression of neoantigens and susceptibility to immune-induced stress. Unveiling the role of AS in β cell functional loss and death will increase our understanding of T1D pathogenesis and may open new avenues for disease prevention and therapy.
Introduction
Pre-mRNA alternative splicing (AS) is a key post-transcriptional regulatory mechanism that affects gene expression, acting as a major generator of proteomic diversity. It regulates the incorporation of alternative sets of exons into mature mRNA molecules, allowing single genes to produce multiple, structurally distinct mRNA and protein isoforms that may have different biological properties (1). This tightly regulated process provides cells with the ability to rapidly adapt their transcriptome and proteome in response to intra and extracellular cues. Nearly 95% of human genes undergo AS, producing on average six alternative isoforms per gene, thus explaining the discrepancy between the predicted 22 000 protein-coding genes of the human genome and the observed >200 000 protein isoforms (2, 3). The prevalence and extent of AS correlates with organismal complexity, suggesting that AS plays a key role for the development of complex phenotypic traits during evolution (4, 5). AS regulation plays an important role in virtually all biological processes, including cell growth and death, development stage, pluripotency, differentiation, circadian rhythms, response to stimuli and disease (6, 7, 8).
Regulation of AS: the splicing code and the coupling with the transcription machinery
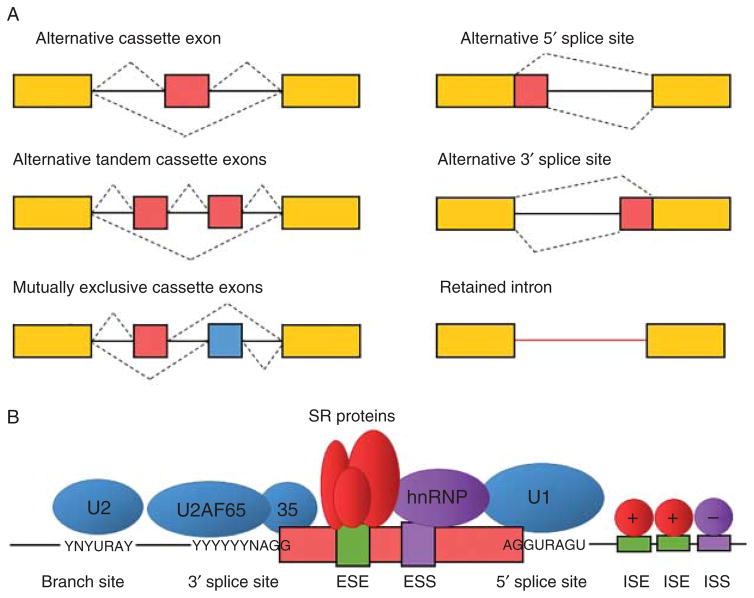
The splicing process involves the retention of exons and removal of introns from the pre-mRNA. This is not a rigid process, showing instead a wide variation by the different usage of alternative exons and other components of the transcribed mRNA that are either included or excluded from the final mRNA isoform (Fig. 1A). This requires the recognition of exons through the identification of a complex code of cis-acting elements within the pre-mRNA molecule (Fig. 1B). The catalytic reactions that occur during the splicing process are mediated by the stepwise assembly of a large and dynamic ribonucleoprotein (RNP) complex called the spliceosome. The spliceo-some is composed by five small nuclear RNP particles (snRNP), U1, U2, U4/U6 and U5, and around 200 proteins (9). The snRNP spliceosomal particles recognize the core splicing signals (5′ splice site, branch site and polypyrimidine tract-3′ splice site) that are essential to carry out the splicing reaction. Core splicing signals are short and degenerate (i.e. not completely conserved) sequences that alone are insufficient to define intron–exon boundaries. Thus, they require the presence of additional cis-acting regulatory sequences to achieve fidelity in the splicing process. AS is accurately regulated by the interplay between these cis-acting regulatory elements and their cognate trans-acting splicing factors, the so-called ‘splicing code’, that either promotes or represses the recognition of core splicing signals (Fig. 1B). Regulatory sequences present in both exons and introns, termed splicing enhancers or silencers, work as binding sites for splicing factors that either enhance or repress splicing depending on their activity and binding position (2, 10). These splicing regulators are RNA-binding proteins (RBPs) of the serine/arginine (SR)-rich proteins and heterogeneous nuclear RNPs (hnRNP) families, as well as other cell-, stage- or tissue-specific proteins such as the NOVA, RBFOX, CELF or MBLN families. These families establish the splicing code and determine in a combinatorial fashion which splice site is selected in specific tissues (for instance, NOVA1 is only expressed in brain and pancreatic β cells) (11, 12, 13, 14). The regulation of AS is accomplished by the relative expression levels of the different RBPs, determining how efficiently different splice sites are used to generate specific mRNA isoforms in different cells and tissues. In addition to the splicing code, defined by RBPs, regulation of AS is also influenced by other mechanisms. For instance, cis-acting RNA–RNA base pairing and RNA secondary structures can control the splice site choice in some genes (15, 16, 17). Splicing occurs mainly co-transcriptionally and is integrated with other regulatory layers controlling gene expression (18). Mechanisms affecting the RNA polymerase transcription rate, such as chromatin structure, histone modifications or DNA methylation, influence the splicing pattern, and recent findings indicate that the splicing, transcription and chromatin organization machineries interact to ensure that AS is properly controlled in time and space (18, 19, 20, 21). Depending on the pattern of exon inclusion/exclusion, AS events can be classified into six major types: cassette exons (an exon that is either retained or skipped), tandem cassette exons (two or more exons that are retained or skipped together), mutually exclusive exons (two exons, where the retention of one involves the skipping of the other and vice versa), alternative 5′ or 3′ splice site (an alternative donor or acceptor site is used changing the exon–exon junction) and retained intron (the intron is not spliced out leading to its inclusion into the coding region) (Fig. 1A).
Figure 1.
Regulation of alternative splicing (AS). (A) Major types of AS events. Exons are represented by boxes and introns by solid lines; constitutive exons are shown in yellow while alternative exons are shown in red or blue; dashed lines represent different splicing events. (B) Schematic representation of snRNP spliceo-somal particles bound to splicing signals (5′ splice site, branch point, polypyrimidine tract-3′ splice site) in the pre-mRNA molecule. Additional cis-acting regulatory sequences that regulate splice site selection in exons and introns are also shown. ESE, exonic splicing enhancer; ESS, exonic splicing silencer; ISE, intronic splicing enhancer; ISS, intronic splicing silencer. Adapted from reference (124).
Impact of AS on protein function
Splicing changes in protein-coding mRNAs can have profound and diverse effects on protein function, producing splice variants with related, distinct or even opposite functions (1, 22). For instance, several genes of the BCL2 family of apoptotic regulators, such as Bcl2–l1, Mcl1 and BID, produce both pro- and anti-apoptotic isoforms through AS, and changes in the relative ratios of these splice variants may lead to cancer or neurodegenerative diseases by modulating cell death (23). AS can lead to changes on protein localisation, enzymatic activity and interaction with ligands (24). It can also modulate protein-binding properties, modifying interactions with other proteins, nucleic acids or membranes. In some cases, specific protein domains are regulated by AS, modifying protein structure and functions. For instance, AS changes in transcriptions factors can alter the transactivation or DNA-binding domains inducing negative or positive effects on transcription; changes on channel proteins can modify their electrophysiological properties; and gene function may change from dominant negative to constitutively active through AS (24). By regulating the inclusion/exclusion of exons harbouring a stop codon or introducing a frameshift change, AS is often coupled with the nonsense-mediated mRNA decay (NMD), a quality control process that eliminates transcripts containing premature termination codons, indirectly regulating mRNA expression (mRNA expression results from the balance between mRNA transcription and degradation) (25, 26).
The key role of AS in the brain
AS has been extensively studied in the brain, revealing its pivotal role in neural development and the establishment and function of neuronal networks (27). AS is particularly widespread and more highly conserved in the brain than in any other tissue, suggesting that it has contributed to the functional complexity of the CNS during evolution (27). It is thus not surprising that splicing defects lead to neurologic and neuropsychiatric disorders. Mutations or polymorphisms affecting cis-regulatory elements have been identified in several brain-related diseases, including frontotemporal dementia, schizophrenia, bipolar disorder and autism spectrum disorder, among others (28, 29). For instance, alterations on the developmental and tissue-specific AS pattern of the microtubule-associated protein tau (MAPT) give rise to frontotemporal dementia with Parkinsonism (30). Mutations that affect AS of exon 10, encoding a microtubule-binding motif, disturb the normal ratio between isoforms of low and high affinity, increasing microtubule assembly and leading to the formation of neurofibrillary tangles and consequent neurodegeneration (31). On the other hand, other neurological diseases arise from defects in splicing regulatory RBPs, leading to large mRNA splicing alterations (28, 32). For instance, mutations in RBFOX1 have been associated with autism, mental retardation and epilepsy (33). Microsatellite expansions (also called dynamic mutations) can induce aberrant nuclear sequestration of RBPs like MBNL1 and CELF1, leading to neurodegenerative disorders such as myotonic dystrophy, spinocerebellar ataxia or Huntington’s disease (34).
AS is coordinated into regulatory networks
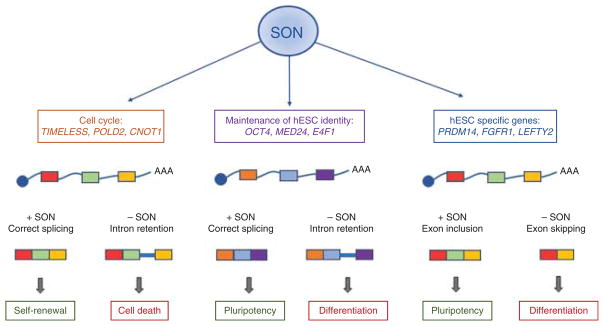
Although AS may seem a chaotic process of independent events, recent evidence indicate that, on the contrary, cells typically coordinate splicing changes into regulatory networks. Splicing networks are regulatory hubs of functionally coherent transcript populations (also known as RNA regulons) co-regulated simultaneously by key splicing factor RBPs. These master splicing factors coordinate the splicing of alternative exons of functionally related genes to promote particular biological outcomes in response to specific signals (35, 36). To date, several splicing networks have been reported, including networks regulating neural development (27), T cell activation (37), stem cell pluripotency (38) (Fig. 2), cell cycle and apoptosis (39) or myogenic differentiation (40).
Figure 2.
Example of a splicing regulatory network controlling pluripotency in human embryonic stem cells (hESC). The spliceosome-associated factor SON regulates the splicing of a network of genes in hESC that are essential to maintain the pluripotency phenotype. Using RNA sequencing, Lu and colleagues (38) showed that SON acts mainly as an intron splicing activator; depletion of SON leads to an increased intron inclusion in genes regulating cell cycle and hESC identity. Transcripts with retained introns are potentially targeted for degradation by the NMD pathway, leading to differentiation, decreased cell survival and loss of hESC phenotype. In addition, SON regulates the inclusion/exclusion of alternative exons in several pluripotency regulatory genes. Adapted from references (38, 125).
Genome-wide methods to analyse AS
The development of different high-throughput technologies during the last decade has fostered a significant increase in the understanding of AS, its regulatory mechanisms, dynamics, evolution and organization into complex networks. The initial genome-wide studies of AS were performed using splicing-sensitive microarrays that typically used short oligonucleotides probes recognizing exon junctions (41). These platforms provided relative quantitation of splicing changes across different tissues, development stages or upon perturbation of specific splicing factors (42, 43, 44, 45, 46). More recently, next generation RNA sequencing (RNA-seq) has emerged as the method of choice to analyse AS at a genomic scale (2, 47). In RNA-seq, cDNA fragments derived from polyadenylated RNAs are amplified and massively sequenced to generate millions of short sequence reads. These reads are later aligned back to the reference transcriptome and analysed using extensive bioinformatics processing, allowing detection and quantification of virtually all RNA transcripts, including splice variants (48). RNA-seq presents several advantages when compared to microarrays, such as low background, increased sensitivity, high reproducibility, lack of cross-hybridization artefacts and unbiased detection of novel transcripts (49, 50, 51). On the other hand, RNA-seq data analysis is computationally intensive, and pipelines typically include several steps, including quality assessment, filtering, alignment, transcript assembly, normalisation, estimation of transcript abundance and statistical analysis to identify differentially expressed genes/transcripts (52, 53). The choice of software to analyse RNA-seq data is not trivial, and different tools can give slightly different results (54, 55, 56). A key issue when analysing AS by RNA-seq is the coverage depth, i.e. the average number of times a nucleotide is read during the sequencing process. Accurate quantification of splicing levels relies on the number of reads that specifically map to exon junctions, and it has been estimated that a depth of ~200 million reads is required to quantify the splicing levels of 80% of all genes (47). Complementary to the methods described above, several methods have been developed to obtain RNA–protein interaction maps on a genome-wide scale (57). Cross-linking and immunoprecipitation followed by high-throughput sequencing (HITS–CLIP) is a technique in which RNA transcripts bound to a given RBP are sequenced after specific purification of RNP complexes (58, 59, 60). These techniques, combined with RNA-seq, allow identification of direct, functional RNA targets and uncover splicing networks regulated by specific RBPs (61).
β cell death in type 1 diabetes and the role of AS
Type 1 diabetes (T1D) is a chronic autoimmune disease where putative environmental factors (viral infections, dietary components, etc.) interact with predisposing genes to trigger an autoimmune assault against pancreatic β cells (62). The disease is characterized by pancreatic islet inflammation (insulitis) and progressive β cell loss by apoptosis (63, 64, 65). The incidence of T1D is increasing, and it is expected that new cases of T1D in European children will double between 2005 and 2020 (66). This, and the fact that there are presently no adequate approaches to prevent or cure the disease (67), makes T1D one of the great health challenges of the 21st century.
Pancreatic β cells and the immune system in T1D: dialogue and misunderstanding
Inflammation contributes to both the early induction and secondary amplification of the immune assault against the β cells. Inflammatory mediators contribute to β cell functional suppression and subsequent apoptosis, inhibit or stimulate β cell regeneration and may cause peripheral insulin resistance (63). These different roles of inflammation take place in the context of a ‘dialogue’ between invading immune cells and the target β cells. This dialogue is mediated by cytokines/chemokines released by β cells and immune cells and by putative immunogenic signals delivered by dying or modified β cells (63) and is modulated by candidate genes for the disease, acting at both the immune system and pancreatic β cell levels (11, 62, 68, 69). Progressive loss of β cell mass is a central feature of T1D, and immune cells contribute to β cell apoptosis by cell-to-cell interactions, via the Fas–FasL and perforin–granzyme systems, and by releasing pro-inflammatory cytokines such as interleukin 1β (IL1β), tumor necrosis factor α (TNFα), interferon γ (IFNγ) and IL17 (63, 70, 71). Cytokine-induced β cell apoptosis depends on the activation of complex gene networks regulated by transcription factors such as NFκB (72, 73) and STAT-1 (74). However, the mechanisms by which autoimmunity is triggered and aggravated and the nature of the intracellular signals that decide the β cell fate between survival and death remain to be clarified.
Destruction of β cells in T1D is mediated by CD8+T cells that recognise target epitopes presented by HLA Class I molecules (75). However, most dominant epitopes are not recognised in many patients, suggesting that more universal immunodominant epitopes, critical for amplifying the autoimmune assault and amenable to biomarker and therapeutic development, remain to be identified. The emergence of selected antigen sequences in pancreatic β cells as T cell targets might be favoured by defects in central and/or peripheral tolerance. Several β cell-restricted antigens are expressed by medullary thymic epithelial cells (mTECs) and presented to developing T cells (76) during a quality control process that deletes most autoreactive T cells. Not all self-antigens, however, are expressed in mTECs, and some are expressed as incomplete isoforms that lack key epitopes (77). Thus, the ‘immune self-image’ presented in the thymus is incomplete, and inflammation-induced modifications in pancreatic β cell AS (11) may lead to generation of neoantigens that contribute to amplify and accelerate β cell destruction.
The emerging role of AS in T1D and other autoimmune diseases
Our understanding of the importance of AS in the development of autoimmune diseases is just emerging. Triggering of autoimmune diseases depends on a complex interaction between multiple genetic and environmental factors interconnected thought regulatory mechanisms that intervene at different stages of disease evolution. AS alterations caused by defects in both cis-acting regulatory elements (i.e. mutations or single nucleotide polymorphisms affecting splicing enhancers or silencers) and transacting factors (i.e. defects in splicing factors) have been suggested to play a role in the development of auto-immune diseases, such as multiple sclerosis, myasthenia gravis and systemic lupus erythematosus (78, 79, 80, 81). For instance, a polymorphism associated with high risk of multiple sclerosis alters the splicing of the interleukin-7 receptor (IL7R) in T cells, decreasing the ratio of transmembrane to soluble isoforms and causing immune dysfunction (78).
In the context of T1D, recent data suggest that AS alterations acting at both immune and pancreatic β cells, and in some cases affecting disease susceptibility genes, may contribute to the pathogenesis of the disease. AS switches have been reported to modulate the immune response against β cells and contribute to the progression of T1D. CTLA-4 is a T lymphocyte regulatory gene associated with risk of T1D and other autoimmune diseases (82, 83). CTLA-4 regulates the ‘choice’ of T cells between proliferative responsiveness and tolerance. Studies in NOD mice and other models have shown that differential expression of CTLA-4 splice variants impacts on T cell function and the overall immune response. Thus, while splice variants lacking the transmembrane domain exacerbate the autoimmune pathology, variants lacking the ligand-binding domain are protective (83, 84, 85). Importantly, AS is more frequent in the critical MHC region (a region that accumulates more than 40% of the genetic risk associated with T1D (86)) than it is genome wide (87).
AS defects in pancreatic lymph nodes may underlie the breakdown in peripheral self-tolerance that contributes to insulitis (88). Deformed epidermal autoregulatory factor 1 (Deaf1) is a transcriptional regulator that controls the expression of peripheral tissue antigens in lymph node stromal cells; these peripheral tissue antigens are important to ‘educate’ T regulatory cells (Tregs) and maintain tolerance (89). Deaf1 is spliced into a dominant negative variant (Deaf1–Var1) in pancreatic lymph nodes of T1D patients (89), and Deaf1–Var1 expression is reduced in the pancreatic lymph nodes of diabetes-prone NOD mice that escape diabetes development (90). Inflammation and hyperglycaemia drive Deaf1 splicing independently through activation of two different splicing factors, Srsf10 and Ptbp2 respectively (90). These data suggest a complementary role for AS in the regulation of auto-immunity: since deletional tolerance and induction of Tregs is at least in part mediated by expression of peripheral tissue antigens, Deaf1 splicing and consequent decrease in function may allow the persistence of an increased number of autoreactive T cells, thus aggravating the autoimmune attack against β cells (90).
AS changes regulate β cell responses to immune-induced stress
Modifications of AS within pancreatic β cells may contribute to β cell dysfunction and death through modulation of the expression of pro-apoptotic proteins, generation of neoantigens that lead to presentation of novel β cell epitopes and subsequent amplification of the autoimmune response (see above), modification of the surface location of antigens and/or introduction of changes in the post-translational configuration of proteins (78, 91, 92, 93). Several apoptotic regulators, including members of the BCL2 family, undergo AS (94), leading to the generation of different protein isoforms with distinct functions, locations and/or pro-apoptotic activity (95). Thus, caspase 2 (96), caspase 9 (97), BCLX (98, 99), MCL1 (99, 100) and the pro-apoptotic BH3-only protein Bim (101, 102) undergo AS in other cell types. These proteins play an important role in the regulation of cytokine-induced pancreatic β cell apoptosis (103, 104), with a central role for the pro-apoptotic BH3-only protein Bim (104, 105, 106, 107).
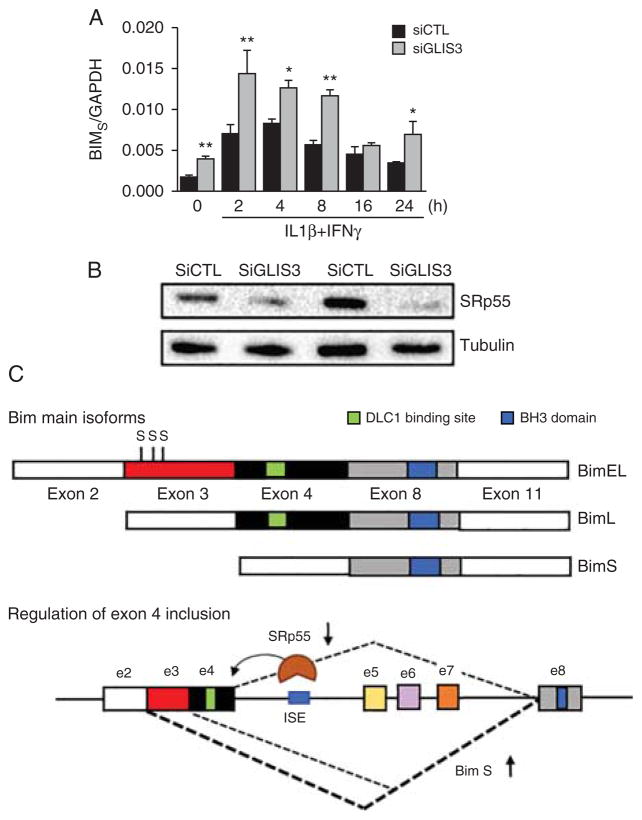
Our group was the first to show that inflammation induces extensive changes in β cell AS (46). Using microarray analysis of rat primary β cells exposed to pro-inflammatory cytokines (IL1β plus IFNγ or TNFα plus IFNγ), we found that cytokines modify the expression of nearly 50 splicing factors and other RBPs involved in the splicing machinery. Cytokines were also found to modify the splicing of nearly 20% of all genes expressed in β cells. Pathway enrichment analysis indicated that these splicing changes affect many genes involved in cell death signalling (46). Of particular relevance, in a subsequent study we observed that GLIS3, a candidate gene for diabetes, contributes to β cell death by indirectly regulating the AS of the pro-apoptotic gene BIM (105). GLIS3 is a transcription factor that plays a critical role in pancreatic development and in the maintenance of the β cell differentiated phenotype, and severe inactivating mutations in GLIS3 cause neonatal diabetes (108). Genome-wide association study indicates that GLIS3 is one of the rare genes showing association with both T1D and T2D (109, 110, 111). We found that decreased expression of GLIS3, besides having a negative impact on β cell function and phenotype, increases β cell apoptosis both basally and after cytokine exposure (105). The observed increase of apoptosis was due to modulation of AS of the pro-apoptotic BH3-only protein Bim, favouring the expression of the most pro-apoptotic isoform, Bim Small (Bim S). The activity of Bim is controlled by AS, generating three main isoforms, namely Bim extra large (EL), Bim large (L) and Bim S (101). Bim EL and Bim L contain exon 4, encoding a dynein light chain 1 (DLC1) binding site. This domain maintains these isoforms in a relatively inactive form through their binding to the dynein motor complex and consequent sequestration to the cytoskeleton (112). On the other side, Bim S is not subject to post-translational regulation, remaining free to exert its potent pro-apoptotic activity (101). We found that decreased levels of GLIS3 reduces the expression of the splicing factor SRp55, which in turn regulates the inclusion of Bim exon 4, leading to an increase of Bim S expression and consequent higher β cell apoptosis (105) (Fig. 3).
Figure 3.
The T1D and T2D candidate gene GLIS3 regulates splicing of the pro-apoptotic BH3-protein Bim through modulating expression of the splicing factor SRp55. Insulin-producing INS-1E cells were transfected with control or GLIS3 siRNA. After 48 h, cells were incubated with cytokines and collected at different time points for real-time PCR analyses. (A) mRNA expression of Bim S after GLIS3 KD. Results are means±S.E.M. (n-4). *P<0.05, **P<0.01 and vs siCTL. Paired t test. (B) Representative blot of two independent experiments showing SRp55 protein expression after GLIS3 silencing. (C) Schematic representation of BIM isoforms and its regulation by SRp55. The BIM gene contains 11 exons that are alternatively spliced to produce isoforms with different pro-apoptotic properties. Bim EL and Bim L, but not Bim S, contain exon 4 coding for a DLC1 binding site that allows sequestration to the cytoskeleton and consequent decrease in pro-apoptotic effect. In addition, Bim EL contains exon 3, which encodes for three serine residues (shown with S) that are subject to phosphorylation, thus targeting the isoform for proteosomal degradation. Bim S is not subject to any known post-translational regulation, being the most potent apoptosis inducer among the three isoforms. SRp55 promotes the inclusion of exon 4 through binding to a predicted ISE located in intron 4 (126). Thus, decreased expression of SRp55 leads to increased Bim S expression and consequent augmented apoptosis. Data adapted from reference (105).
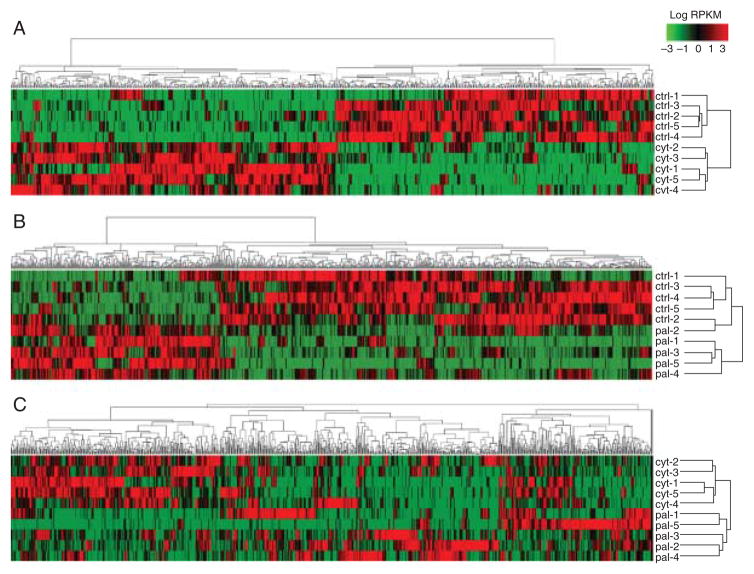
As mentioned above, RNA-seq allows a robust analysis of AS events at a genomic scale. Transcriptome analysis of human islets using this technique provided further insights into the β cell transcriptome and its modulation by pro-inflammatory cytokines (11). We found that many splicing factors are significantly enriched in human islets when compared with other tissues, and that, surprisingly, several so-called ‘neuron-specific’ splicing factors such as NOVA, RBFOX or CELF RBPs are also expressed in human β cells. In line with the previous microarray data (46), we found that pro-inflammatory cytokines modify the expression of >30 splicing-regulating RBPs and induce AS changes in >3000 genes. Comparison of RNA-seq datasets of human islets exposed to cytokines or to palmitate, a saturated fatty acid that contributes to β cell failure in models of T2D, indicate that inflammation induces a specific AS signature that is different from that induced by the metabolic stressor (Fig. 4) (11, 113) (unpublished data), suggesting that different β cell stressors induce different and stress-specific ‘splicing signatures’. Enrichment analysis using IPA, DAVID or GO databases suggest that cytokine-induced AS changes affect key functions/pathways in β cells, such as cell death and apoptosis, cellular growth and proliferation, antigen presentation, mitochondria dysfunction, unfolded protein response and several immune cell-related pathways, among others. Taken together, these data support the idea that early islet inflammation, through modulation of the expression of key RBPs in β cells, leads to the activation of specific AS regulatory networks that modulate β cell viability and/or susceptibility to immune-induced stress. Some of these networks may regulate the cross-talk between β cells and immune cells during insulitis, giving rise to the generation of neoepitopes recognized by CD8+T cells and consequent amplification of the immune assault. Other networks may contribute to β cell death by modifying apoptosis-related proteins or key pathways for β cell survival.
Figure 4.
Alternative splicing signature in human islets exposed to pro-inflammatory cytokines. Heat maps representing the expression of differentially expressed mRNA isoforms (DEIs) in human islet samples exposed to: (A) control (ctrl) vs cytokines (cyt; IL1β+IFNγ); (B) control (ctrl) vs the free fatty acid palmitate (pal); and (C) cytokines vs palmitate. DEIs were obtained from analysis of previous RNA-seq datasets generated by our group (11, 113) (unpublished data), filtered according to their log2 fold change ratio (only DEIs with values higher than four are represented) and hierarchically clustered using GenePattern modules (reads per kilobase of transcript per million values log transformed, not centred and not normal normalised). Green colour indicates low expressed isoforms while red colour indicates high expressed isoforms.
NOVA1, a ‘neuron-specific’ splicing factor, controls β cell function and survival
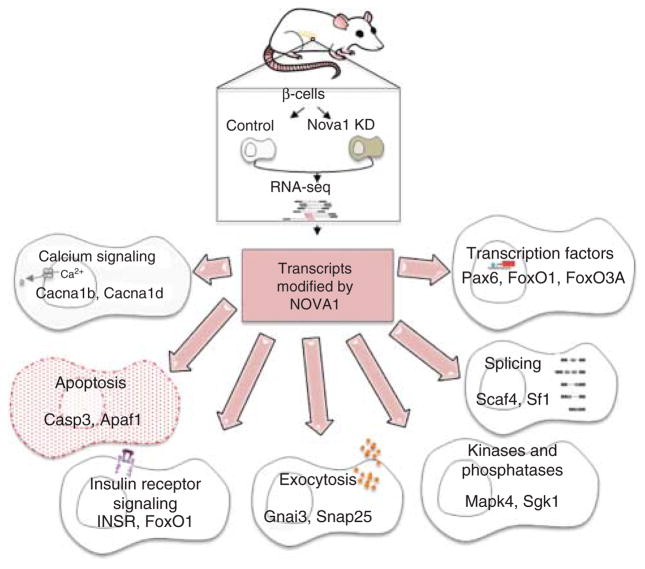
To further investigate the role of individual splicing factors in the regulation of β cell function and survival, we studied the ‘neuron-specific’ RBP NOVA1 using siRNA silencing coupled with RNA-seq (12) (Fig. 5). NOVA1 silencing modified the splicing of nearly 5000 transcripts (11% of the total isoforms) in fluorescence-activated cell sorting (FACS)-purified primary rat β cells. Pathway analysis indicated that many of these genes are involved in exocytosis, apoptosis, insulin receptor signalling, splicing and transcription. In line with these findings, NOVA1 silencing-impaired insulin release by inducing splicing changes in genes regulating exocytosis, such as Snap25 and PLCβ1, and decreased voltage-dependent Ca2+ currents by modifying splicing of voltage-gated ion channels (12). Interestingly, pro-inflammatory cytokines down-regulate NOVA1 expression, and silencing NOVA1 using specific siRNAs increases apoptosis basally and after cytokine treatment in rodent and human β cells, indicating a key role for NOVA1 in the maintenance of β cell viability. Apoptosis induced by NOVA1 silencing is mediated by the intrinsic pathway of apoptosis due to up-regulation of the transcription factor FoxO3A and consequent increase of Bim expression (12).
Figure 5.
Impact of NOVA1 silencing on the rat pancreatic β cell transcriptome. FACS-purified rat β cells were transfected with control or NOVA1 siRNA and then RNA-seq to detect changes in gene and transcript expression, as well as in splicing patterns. Enrichment pathway analysis using IPA software indicates that NOVA1-mediated splicing regulates several key gene networks essential for β cell function and survival. Adapted from reference (12).
Estimating the disease progression in T1D – an unmet need
Destruction of β cells in T1D is mediated by autoreactive T cells and macrophages as a result of a loss of immune tolerance (63, 64, 65). However, we do not know whether β cell loss in T1D is slow and progressive, relapsing and remitting, or late and rapid. The limited success of drugs that block autoimmunity suggests that therapies should be introduced early, prior to clinical diagnosis of T1D (67, 114, 115). In order to decide on the best therapy, and to detect eventual protective effects on β cell survival and mass once treatment starts, it is crucial to identify novel biomarkers that reflect the state of β cell health and the activation of the immune system.
As discussed above, transcriptome analysis suggest that stressed β cells of pre-diabetic individuals exhibit unique gene expression signatures, including AS of specific mRNA species and consequent production of new proteins. Thus, differentially expressed and alternatively spliced transcripts have potential as specific serum/plasma biomarkers of β cell stress and as biomarkers for β cell imaging (116). Bioinformatics comparison of the β cell transcriptome under control and pro-inflammatory conditions, or from β cells isolated from T1D patients, is thus a rational approach to discover β cell neoantigens that contribute for T cell activation. Potential neoantigens can be selected from a pool of splice variants that are β cell-specific, up-regulated by inflammation and not expressed in the thymus and other tissues. The identification of key epitopes of T cell activation against pancreatic β cells can be then used as biomarkers to monitor disease progression. Furthermore, immunodominant epitopes are potential therapeutic targets that can be used for tolerogenic vaccination in prevention strategies or be modulated in T1D patients to reduce the autoimmune assault against β cells.
Conclusions and future perspectives
Accumulating evidence indicates that splicing networks and master splicing factors have a key role in maintaining cell identity and phenotype, as well as in regulating cell adaptation to extracellular cues (117, 118). Indeed, splicing networks enable the precise coordination and cross-talk between different signalling pathways. A major challenge in the RNA splicing field is to determine the combinations of cis-elements that discriminate splice sites and govern splicing patterns, known as the ‘splicing code’. This knowledge would allow us to predict RBP splice targets and infer splicing regulatory networks. Although this code is not yet completely understood, significant advances have been made in recent years with the development of computational tools that integrate features on the pre-mRNA sequence (RBP-binding motifs) with RNA-seq transcriptomic data and HITS–CLIP RNA–protein interaction maps (119, 120, 121). Furthermore, important progress has been made in the development of methods to manipulate splicing for therapy (122). Different molecular tools allow modulation of splicing patterns, enabling to increase or decrease specific isoforms through targeting of its regulatory cis-elements (123). These tools include antisense oligonucleotides (AON), modified small nuclear RNAs, trans-splicing and small molecule compounds.
As described above, an emerging picture in the context of T1D suggests that AS events in both immune and pancreatic β cells regulate autoimmunity and the β cell responses to immune-induced stress (Fig. 6). Thus, identifying critical AS switches, master splicing factors and key splicing networks regulating β cell phenotype, survival and susceptibility to stress will shed light on the mechanisms underlying T1D pathogenesis and may open novel strategies for disease prevention or treatment. The identification of splice variants acting as β cell autoantigens, modulators of immune response or β cell survival, may lead to the development of novel therapeutic strategies for T1D based on splicing modulation. In line with this possibility, the use of an AON-targeted splice-switching approach against the CTLA-4 gene, which modulates T cell activation and proliferation, reduced the incidence of insulitis and diabetes in diabetes-prone NOD mice (84). Another potential therapeutic target to enhance β cell survival is the short isoform of the pro-apoptotic BH3-protein Bim (Bim S). AON targeting against splicing motifs regulating the inclusion of the exon encoding the dynein-binding site could allow decreasing the expression of Bim S, thus reducing immune-induced β cell apoptosis (Fig. 3).
Figure 6.
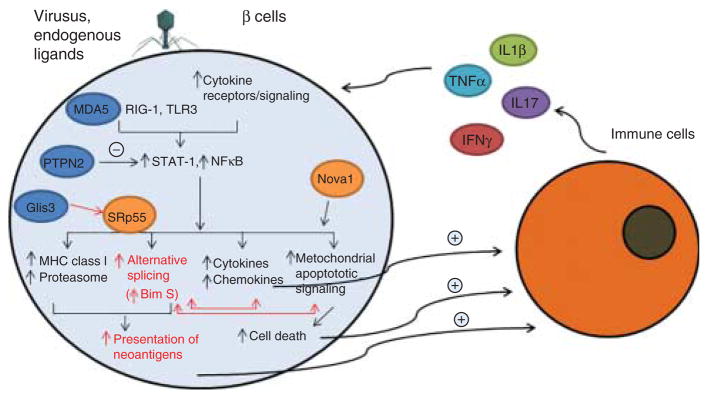
Proposed role for alternative splicing (AS) in the dialogue between pancreatic β cells and the immune system in early T1D. In the early stages of insulitis, β cells are exposed to the locally produced cytokines IL1β, IFNγ, TNFα and IL17 and/or to ‘danger signals’ provided by viruses or endogenous ligands of IFIH1/MDA5 and other innate immune response sensors such as RIG-I and TLR3. These signals activate transcription factors such as STAT-1 and NFκB (46, 63, 71). The T1D candidate gene PTPN2 provides negative feedback for both STAT-1 and mitochondrial pro-apoptotic signals (106, 127), while the candidate gene GLIS3 modulates Bim splicing via inhibition of the splicing factor SRp55 (105) (see also Fig. 3). The splicing factor NOVA1 has a major role in the regulation of AS in β cells, being involved in insulin secretion and apoptosis (12) (see also Fig. 5). Downstream of these and other factors there is modification of AS and up-regulation of the machinery for antigen presentation. This may generate presentation of neoantigens that trigger or augment β cell recognition by the immune system. Additional signals provided by β cells to the immune system include the release of chemokines and cytokines and cell death. AS may both modulate mitochondrial apoptotic signals and the actions of chemokines. Activated immune cells, attracted by the local production of chemokines, will produce more cytokines and chemokines, perpetuating the local inflammatory response and consequent changes in AS. Inflammation is probably modulated by T1D candidate genes (shown in blue circles) such as MDA5, PTPN2 and GLIS3 (11, 63, 105, 106, 127). If this process is not interrupted, genetically predisposed individuals will eventually evolve to clinical diabetes.
Future systems biology approaches that combine transcriptomics, bioinformatics and biochemical analyses should elucidate the critical role of AS in β cell demise and help to develop novel therapies to protect β cells in early T1D, a major unmet need.
Acknowledgments
Funding
This work was supported by the Fonds National de la Recherche Scientifique (FNRS) under grants T.0036.13 and FRFS-Welbio CR-2015A-06, Belgium, the European Union (projects Betaimage, Naimit and BetaBat in the Framework Programme 7 of the European Commission); the Juvenile Diabetes Foundation; the Helmsley Type 1 Diabetes Program; and the NIH–NIDDK–HIRN Consortium. J Juan-Mateu is supported by a MSCA fellowship grant from the Horizons 2020 EU programme (Project reference: 660449).
We thank Dr J V Turatsinze for help and advice on RNA-seq data analysis, and Dr L Mascali for help in the preparation of the NOVA1 figure.
Biography
Invited Author’s profile
Decio L Eizirik, M.D, PhD, is Full Professor and Director of the ULB Center for Diabetes Research, Medical Faculty, Universite Libre de Bruxells (ULB), Belgium. He has published over 300 full papers and has received several prestigious awards. Prof Eizirik’s research focuses on the molecular mechanisms regulating insulitis and β cell apoptosis in type 1 diabetes and on the search for novel approaches to prevent the progressive loss of β cell mass in diabetes.

Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
References
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Reviews Molecular Cell Biology. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annual Review of Biochemistry. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H, Klein R, Majewski J, Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nature Genetics. 2004;36:915–916. doi: 10.1038/ng0904-915. author reply 916–7. [DOI] [PubMed] [Google Scholar]
- 5.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Research. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature Reviews Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 9.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 11.Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, Ortis F, Santin I, Colli ML, Barthson J, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genetics. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villate O, Turatsinze JV, Mascali LG, Grieco FA, Nogueira TC, Cunha DA, Nardelli TR, Sammeth M, Salunkhe VA, Esguerra JL, et al. Nova1 is a master regulator of alternative splicing in pancreatic beta cells. Nucleic Acids Research. 2015;42:11818–11830. doi: 10.1093/nar/gku861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends in Biochemical Science. 2000;25:381–388. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 14.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nature Reviews Molecular Cell Biology. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 15.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Current Opinion in Genetics & Development. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May GE, Olson S, McManus CJ, Graveley BR. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA. 2011;17:222–229. doi: 10.1261/rna.2521311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annual Review of Biochemistry. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 19.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Molecular Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gomez Acuna LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Munoz MJ, Allo M, et al. Transcriptional elongation and alternative splicing. Biochimica et Biophysica Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cellular and Molecular Life Sciences: CMLS. 2004;61:2189–2199. doi: 10.1007/s00018-004-4001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yap K, Makeyev EV. Regulation of gene expression in mammalian nervous system through alternative pre-mRNA splicing coupled with RNA quality control mechanisms. Molecular and Cellular Neurosciences. 2013;56:420–428. doi: 10.1016/j.mcn.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. PNAS. 2015;112:3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. American Journal of Medical Genetics. 2011;156B:382–392. doi: 10.1002/ajmg.b.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niblock M, Gallo JM. Tau alternative splicing in familial and sporadic tauopathies. Biochemical Society Transactions. 2012;40:677–680. doi: 10.1042/BST20120091. [DOI] [PubMed] [Google Scholar]
- 31.D’Souza I, Schellenberg GD. Regulation of tau isoform expression and dementia. Biochimica et Biophysica Acta. 2005;1739:104–115. doi: 10.1016/j.bbadis.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Advances in Experimental Medicine and Biology. 2014;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bill BR, Lowe JK, Dybuncio CT, Fogel BL. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. International Review of Neurobiology. 2013;113:251–267. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan R, Sharma S, Xia Q, Garber K, Jin P. Towards understanding RNA-mediated neurological disorders. Journal of Genetics and Genomics. 2014;41:473–484. doi: 10.1016/j.jgg.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Keene JD. RNA regulons: coordination of post-transcriptional events. Nature Reviews Genetics. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 36.Papasaikas P, Tejedor JR, Vigevani L, Valcarcel J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Molecular Cell. 2015;57:7–22. doi: 10.1016/j.molcel.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 37.Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, Zhu W, Ajith S, Blencowe BJ, Lynch KW. Alternative splicing networks regulated by signaling in human T cells. RNA. 2012;18:1029–1040. doi: 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Goke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nature Cell Biology. 2013;15:1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Research. 2010;38:7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA. 2008;14:197–203. doi: 10.1261/rna.868008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes and Development. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biology. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann B, Castelo R, Blanchette M, Boue S, Rio DC, Valcarcel J. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-1-r11. R11-2009-10-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortis F, Naamane N, Flamez D, Ladriere L, Moore F, Cunha DA, Colli ML, Thykjaer T, Thorsen K, Orntoft TF, et al. Cytokines interleukin-1β and tumor necrosis factor-α regulate different transcriptional and alternative splicing networks in primary β-cells. Diabetes. 2010;59:358–374. doi: 10.2337/db09-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes and Development. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 48.Martin JA, Wang Z. Next-generation transcriptome assembly. Nature Reviews Genetics. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 49.Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Research. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biology. 2011;9 doi: 10.1186/1741-7007-9-34. 34-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindner R, Friedel CC. A comprehensive evaluation of alignment algorithms in the context of RNA-seq. PLoS ONE. 2012;7:e52403. doi: 10.1371/journal.pone.0052403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robles JA, Qureshi SE, Stephen SJ, Wilson SR, Burden CJ, Taylor JM. Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. BMC Genomics. 2012;1 doi: 10.1186/1471-2164-13-484. 484-2164-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-91. 91-2105-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engstrom PG, Steijger T, Sipos B, Grant GR, Kahles A, Ratsch G, Goldman N, Hubbard TJ, Harrow J, Guigo R, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nature Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonseca NA, Marioni J, Brazma A. RNA-Seq gene profiling – a systematic empirical comparison. PLoS ONE. 2014;9:e107026. doi: 10.1371/journal.pone.0107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature Reviews Genetics. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods in Molecular Biology. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. PAR-CliP – a method to identify transcriptome-wide the binding sites of RNA binding proteins. Journal of Visualized Experiments. 2010;41:2034. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Structural & Molecular Biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santin I, Eizirik DL. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and beta-cell apoptosis. Diabetes, Obesity and Metabolism. 2013;15:71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- 63.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nature Reviews Endocrinology. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 64.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32:468–478. doi: 10.1016/j.immuni.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G EURODIAB Study Group. Incidence trends for childhood type 1 diabetes in Europe during 1989;–2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. 2009;373:2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 67.Skyler JS. Immune intervention for type 1 diabetes mellitus. International Journal of Clinical Practice. 2011;170:61–70. doi: 10.1111/j.1742-1241.2010.02580.x. [DOI] [PubMed] [Google Scholar]
- 68.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nature Reviews Genetics. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 69.Bergholdt R, Brorsson C, Palleja A, Berchtold LA, Floyel T, Bang-Berthelsen CH, Frederiksen KS, Jensen LJ, Storling J, Pociot F. Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein–protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61:954–962. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas HE, McKenzie MD, Angstetra E, Campbell PD, Kay TW. Beta cell apoptosis in diabetes. Apoptosis. 2009;14:1389–1404. doi: 10.1007/s10495-009-0339-5. [DOI] [PubMed] [Google Scholar]
- 71.Arif S, Moore F, Marks K, Bouckenooghe T, Dayan CM, Planas R, Vives-Pi M, Powrie J, Tree T, Marchetti P, et al. Peripheral and islet interleukin-17 pathway activation characterizes human autoimmune diabetes and promotes cytokine-mediated beta-cell death. Diabetes. 2011;60:2112–2119. doi: 10.2337/db10-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardozo AK, Heimberg H, Heremans Y, Leeman R, Kutlu B, Kruhoffer M, Orntoft T, Eizirik DL. A comprehensive analysis of cytokine-induced and nuclear factor-κ B-dependent genes in primary rat pancreatic beta-cells. Journal of Biological Chemistry. 2001;276:48879–48886. doi: 10.1074/jbc.M108658200. [DOI] [PubMed] [Google Scholar]
- 73.Eldor R, Yeffet A, Baum K, Doviner V, Amar D, Ben-Neriah Y, Christofori G, Peled A, Carel JC, Boitard C, et al. Conditional and specific NF-κB blockade protects pancreatic beta cells from diabetogenic agents. PNAS. 2006;103:5072–5077. doi: 10.1073/pnas.0508166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore F, Naamane N, Colli ML, Bouckenooghe T, Ortis F, Gurzov EN, Igoillo-Esteve M, Mathieu C, Bontempi G, Thykjaer T, et al. STAT1 is a master regulator of pancreatic β-cell apoptosis and islet inflammation. Journal of Biological Chemistry. 2011;286:929–941. doi: 10.1074/jbc.M110.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, Zaremba A, Rackham C, Allen JS, Tree TI, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. Journal of Clinical Investigation. 2008;118:3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Derbinski J, Kyewski B. How thymic antigen presenting cells sample the body’s self-antigens. Current Opinion in Immunology. 2010;22:592–600. doi: 10.1016/j.coi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. PNAS. 2008;105:15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evsyukova I, Somarelli JA, Gregory SG, Garcia-Blanco MA. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biology. 2010;7:462–473. doi: 10.4161/rna.7.4.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu M, Kakoulidou M, Giscombe R, Pirskanen R, Lefvert AK, Klareskog L, Wang X. Identification of CTLA-4 isoforms produced by alternative splicing and their association with myasthenia gravis. Clinical Immunology. 2008;128:374–381. doi: 10.1016/j.clim.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 80.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jonsen A, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nature Genetics. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 81.Tian Y, Apperson ML, Ander BP, Liu D, Stomova BS, Jickling GC, Enriquez R, Agius MA, Sharp FR. Differences in exon expression and alternatively spliced genes in blood of multiple sclerosis compared to healthy control subjects. Journal of Neuroimmunology. 2011;230:124–129. doi: 10.1016/j.jneuroim.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 83.AlFadhli S, Nizam R. Differential expression of alternative splice variants of CTLA4 in Kuwaiti autoimmune disease patients. Gene. 2014;534:307–312. doi: 10.1016/j.gene.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 84.Mourich DV, Oda SK, Schnell FJ, Crumley SL, Hauck LL, Moentenich CA, Marshall NB, Hinrichs DJ, Iversen PL. Alternative splice forms of CTLA-4 induced by antisense mediated splice-switching influences autoimmune diabetes susceptibility in NOD mice. Nucleic Acid Therapeutics. 2014;24:114–126. doi: 10.1089/nat.2013.0449. [DOI] [PubMed] [Google Scholar]
- 85.Gerold KD, Zheng P, Rainbow DB, Zernecke A, Wicker LS, Kissler S. The soluble CTLA-4 splice variant protects from type 1 diabetes and potentiates regulatory T-cell function. Diabetes. 2011;60:1955–1963. doi: 10.2337/db11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Current Diabetes Reports. 2011;11:533–542. doi: 10.1007/s11892-011-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vandiedonck C, Taylor MS, Lockstone HE, Plant K, Taylor JM, Durrant C, Broxholme J, Fairfax BP, Knight JC. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histo-compatibility complex. Genome Research. 2011;21:1042–1054. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yip L, Fathman CG. Type 1 diabetes in mice and men: gene expression profiling to investigate disease pathogenesis. Immunologic Research. 2014;58:340–350. doi: 10.1007/s12026-014-8501-8. [DOI] [PubMed] [Google Scholar]
- 89.Yip L, Creusot RJ, Pager CT, Sarnow P, Fathman CG. Reduced DEAF1 function during type 1 diabetes inhibits translation in lymph node stromal cells by suppressing Eif4g3. Journal of Molecular Cell Biology. 2013;5:99–110. doi: 10.1093/jmcb/mjs052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yip L, Fuhlbrigge R, Taylor C, Creusot RJ, Nishikawa-Matsumura T, Whiting CC, Schartner JM, Akter R, von Herrath M, Fathman CG. Inflammation and hyperglycemia mediate Deaf1 splicing in the pancreatic lymph nodes via distinct pathways during type 1 diabetes. Diabetes. 2015;64:604–617. doi: 10.2337/db14-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diez J, Park Y, Zeller M, Brown D, Garza D, Ricordi C, Hutton J, Eisenbarth GS, Pugliese A. Differential splicing of the IA-2 mRNA in pancreas and lymphoid organs as a permissive genetic mechanism for autoimmunity against the IA-2 type 1 diabetes autoantigen. Diabetes. 2001;50:895–900. doi: 10.2337/diabetes.50.4.895. [DOI] [PubMed] [Google Scholar]
- 92.Ng B, Yang F, Huston DP, Yan Y, Yang Y, Xiong Z, Peterson LE, Wang H, Yang XF. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. Journal of Allergy and Clinical Immunology. 2004;114:1463–1470. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang F, Chen IH, Xiong Z, Yan Y, Wang H, Yang XF. Model of stimulation-responsive splicing and strategies in identification of immunogenic isoforms of tumor antigens and autoantigens. Clinical Immunology. 2006;121:121–133. doi: 10.1016/j.clim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 94.Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Molecular and Cellular Biology. 2012;32:954–967. doi: 10.1128/MCB.06130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Molecular Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/S0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 97.Seol DW, Billiar TR. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. Journal of Biological Chemistry. 1999;274:2072–2076. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 98.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. Bcl-X, a Bcl-2-Related Gene that Functions as a Dominant Regulator of Apoptotic Cell Death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 99.Montes M, Cloutier A, Sanchez-Hernandez N, Michelle L, Lemieux B, Blanchette M, Hernandez-Munain C, Chabot B, Sune C. TCERG1 regulates alternative splicing of the Bcl-x gene by modulating the rate of RNA polymerase II transcription. Molecular and Cellular Biology. 2012;3:751–762. doi: 10.1128/MCB.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. Journal of Biological Chemistry. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- 101.Weber A, Paschen SA, Heger K, Wilfling F, Frankenberg T, Bauerschmitt H, Seiffert BM, Kirschnek S, Wagner H, Hacker G. BimS-induced apoptosis requires mitochondrial localization but not interaction with anti-apoptotic Bcl-2 proteins. Journal of Cell Biology. 2007;177:625–636. doi: 10.1083/jcb.200610148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clybouw C, Merino D, Nebl T, Masson F, Robati M, O’Reilly L, Hubner A, Davis RJ, Strasser A, Bouillet P. Alternative splicing of Bim and Erk-mediated Bim(EL) phosphorylation are dispensable for hematopoietic homeostasis in vivo. Cell Death and Differentiation. 2012;1:1060–1068. doi: 10.1038/cdd.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Allagnat F, Cunha D, Moore F, Vanderwinden JM, Eizirik DL, Cardozo AK. Mcl-1 downregulation by pro-inflammatory cytokines and palmitate is an early event contributing to beta-cell apoptosis. Cell Death and Differentiation. 2011;18:328–337. doi: 10.1038/cdd.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gurzov EN, Eizirik DL. Bcl-2 proteins in diabetes: mitochondrial pathways of beta-cell death and dysfunction. Trends in Cell Biology. 2011;21:424–431. doi: 10.1016/j.tcb.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 105.Nogueira TC, Paula FM, Villate O, Colli ML, Moura RF, Cunha DA, Marselli L, Marchetti P, Cnop M, Julier C, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genetics. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Santin I, Moore F, Colli ML, Gurzov EN, Marselli L, Marchetti P, Eizirik DL. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic beta-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes. 2011;60:3279–3288. doi: 10.2337/db11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barthson J, Germano CM, Moore F, Maida A, Drucker DJ, Marchetti P, Gysemans C, Mathieu C, Nunez G, Jurisicova A, et al. Cytokines tumor necrosis factor-α and interferon-γ induce pancreatic beta-cell apoptosis through STAT1-mediated Bim protein activation. Journal of Biological Chemistry. 2011;286:39632–39643. doi: 10.1074/jbc.M111.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, Sosa-Pineda B, Jensen J, Pierreux CE, Lemaigre FP, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Molecular and Cellular Biology. 2009;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Senee V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nature Genetics. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 110.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genetics. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Day CL, Puthalakath H, Skea G, Strasser A, Barsukov I, Lian LY, Huang DC, Hinds MG. Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands. Biochemical Journal. 2004;377:597–605. doi: 10.1042/bj20031251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cnop M, Abdulkarim B, Bottu G, Cunha DA, Igoillo-Esteve M, Masini M, Turatsinze JV, Griebel T, Villate O, Santin I, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63:1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 114.Skyler JS. Primary and secondary prevention of Type 1 diabetes. Diabetic Medicine. 2013;30:161–169. doi: 10.1111/dme.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.von Herrath M, Peakman M, Roep B. Progress in immune-based therapies for type 1 diabetes. Clinical and Experimental Immunology. 2013;172:186–202. doi: 10.1111/cei.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bouckenooghe T, Flamez D, Ortis F, Goldman S, Eizirik DL. Identification of new pancreatic beta cell targets for in vivo imaging by a systems biology approach. Current Pharmaceutical Design. 2010;16:1609–1618. doi: 10.2174/138161210791164117. [DOI] [PubMed] [Google Scholar]
- 117.Jangi M, Sharp PA. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiong HY, Barash Y, Frey BJ. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics. 2011;2:2554–2562. doi: 10.1093/bioinformatics/btr023. [DOI] [PubMed] [Google Scholar]
- 120.Barash Y, Vaquero-Garcia J. Splicing code modeling. Advances in Experimental Medicine and Biology. 2014;825:451–466. doi: 10.1007/978-1-4939-1221-6_13. [DOI] [PubMed] [Google Scholar]
- 121.Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. PNAS. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spitali P, Aartsma-Rus A. Splice modulating therapies for human disease. Cell. 2012;148:1085–1088. doi: 10.1016/j.cell.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 123.Havens MA, Duelli DM, Hastings ML. Targeting RNA splicing for disease therapy. Wiley Interdisciplinary Reviews RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Current Opinion in Cell Biology. 2012;24:323–332. doi: 10.1016;/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 125.Lu X, Ng HH, Bubulya PA. The role of SON in splicing, development, and disease. Wiley Interdisciplinary Reviews RNA. 2014;5:637–646. doi: 10.1002/wrna. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hara H, Takeda T, Yamamoto N, Furuya K, Hirose K, Kamiya T, Adachi T. Zinc-induced modulation of SRSF6 activity alters Bim splicing to promote generation of the most potent apoptotic isoform BimS. FEBS Journal. 2013;280:3313–3327. doi: 10.1111/febs.12318. [DOI] [PubMed] [Google Scholar]
- 127.Moore F, Colli ML, Cnop M, Esteve MI, Cardozo AK, Cunha DA, Bugliani M, Marchetti P, Eizirik DL. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-γ-induced pancreatic beta-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]