Abstract
Immune signaling networks must be tunable to alleviate fitness costs associated with immunity and, at the same time, robust against pathogen interferences. How these properties mechanistically emerge in plant immune signaling networks is poorly understood. Here, we discovered a molecular mechanism by which the model plant species Arabidopsis thaliana achieves robust and tunable immunity triggered by the microbe‐associated molecular pattern, flg22. Salicylic acid (SA) is a major plant immune signal molecule. Another signal molecule jasmonate (JA) induced expression of a gene essential for SA accumulation, EDS5. Paradoxically, JA inhibited expression of PAD4, a positive regulator of EDS5 expression. This incoherent type‐4 feed‐forward loop (I4‐FFL) enabled JA to mitigate SA accumulation in the intact network but to support it under perturbation of PAD4, thereby minimizing the negative impact of SA on fitness as well as conferring robust SA‐mediated immunity. We also present evidence for evolutionary conservation of these gene regulations in the family Brassicaceae. Our results highlight an I4‐FFL that simultaneously provides the immune network with robustness and tunability in A. thaliana and possibly in its relatives.
Keywords: incoherent feed‐forward loop, jasmonate, plant immunity, salicylic acid, signaling perturbation
Subject Categories: Evolution, Immunology, Plant Biology
Introduction
Proper processing of signals through signaling networks is central for organisms to respond accordingly to the signals. As such, signaling networks are comprised of recurring regulatory subnetwork structures called network motifs with various information‐processing functions. Feed‐forward loop (FFL), which consists of two regulators and a target, represents a major class of network motifs 1. Each of interactions among the components of a FFL can be either positive (activation) or negative (repression). As a result, there are eight possible structural configurations of FFL. Of these configurations, incoherent type‐4 FFL (I4‐FFL), in which a regulator has a positive effect on the target but a negative effect on the other regulator that positively regulates the target, is rare in biological networks and, therefore, its biological function has rarely been described.
In nature, plants are in constant contact with a wide variety of microbes, which often produce common molecular signatures known as microbe‐associated molecular patterns (MAMPs) 2. Plants sense MAMPs by plasma membrane‐localized pattern recognition receptors and feed this information into signaling networks that finely control the output immune reaction designated as pattern‐triggered immunity (PTI) 2, 3, 4, 5. Since recognized MAMPs are often common to a class of microbes 2, PTI could be triggered by both pathogenic and non‐pathogenic microbes. Therefore, it is vital for plants to avoid unnecessary PTI against non‐pathogenic microbes, as there is a trade‐off between immunity and growth 6, 7, 8, 9. At the same time, it is important to retain PTI that is effective against pathogens that deploy virulence effectors to interfere with immune signaling components 10, 11 and that can function under perturbation due to diverse environmental conditions 12. The molecular mechanisms that allow these properties to emerge from PTI signaling networks are poorly understood.
Plants rely on PTI to resist necrotrophs that actively kill hosts to acquire nutrients as well as to resist biotrophs that require living hosts for multiplication 2, 13. The phytohormone jasmonate (JA) is a major contributor to immunity against necrotrophs 13. JA is produced in response to MAMPs such as flg22 14 and chitin 15, a part of bacterial flagellin and a part of fungal cell walls, respectively. JA biosynthesis requires allene oxide synthase encoded by DELAYED‐DEHISCENCE 2 (DDE2) 16. JA and its derivatives including methyl JA (MeJA) can be converted to JA‐isoleucine (JA‐Ile) 17, 18. Perception of JA‐Ile by the F‐box protein CORONATINE INSENSITIVE 1 (COI1) leads to ubiquitination‐ and proteasome‐dependent degradation of JASMONATE ZIM DOMAIN (JAZ) proteins 19, 20, 21. This liberates downstream transcription factors including MYC2 and its homologues MYC3 and MYC4, which are normally repressed by JAZ proteins in the resting state, thereby activating JA‐mediated transcriptional responses and immunity 22, 23.
Another phytohormone, salicylic acid (SA), is a central regulator of immunity against biotrophs and hemi‐biotrophs such as the bacterial pathogen Pseudomonas syringae 13, 24. Indeed, SA production is activated by the bacterial MAMP flg22 25. Previous studies have identified a number of genes involved in SA biosynthesis and signaling. SALICYLIC ACID‐INDUCTION DEFICIENT 2 (SID2) encodes an isochorismate synthase that is essential for SA biosynthesis through the isochorismate pathway 26. PHYTOALEXIN‐DEFICIENT 4 (PAD4) contributes to MAMP‐induced SA accumulation 25, 27. ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) is essential for pathogen‐induced SA accumulation in Arabidopsis thaliana 28, 29, 30 and encodes a MATE transporter, which was proposed to mediate SA transport from chloroplasts, the site of SID2‐mediated SA biosynthesis, to the cytoplasm 30. SA affects transcriptional regulation of hundreds of genes, including PATHOGENESIS‐RELATED 1 (PR1) 31. SA accumulation and signaling should be tightly controlled, as excessive activation of SA biosynthesis or signaling is associated with growth retardation 6, 32, 33, 34. However, current understanding of the signaling mechanisms regulating SA production is fragmented.
Phytohormone signaling pathways form a complex network, which could confer great regulatory potential to control plant responses to diverse internal and external stimuli 35, 36. For instance, antagonism between JA and SA is thought to be important to activate proper immunity depending on pathogen lifestyles 13, 37. Interestingly, cooperation between JA and SA has been also reported 14, 38. Thus, plants appear to have context‐dependent crosstalk between JA and SA. However, the molecular mechanisms and the biological relevance of the JA–SA crosstalk remain elusive.
Previously, a quantitative model was built to capture signal flows in the network consisting of the JA, SA, PAD4, and ethylene (ET) signaling sectors during PTI 14. The model pointed to JA and PAD4 as the sole determinants of SA signaling activity 14. Here, we report the molecular mechanism by which JA enables robust and tunable SA accumulation during PTI in A. thaliana. Our data demonstrate that JA inhibits expression of PAD4, a positive regulator of EDS5 expression. Paradoxically, JA induces EDS5 expression directly via the transcription factor MYC2. This I4‐FFL explains the negative role of JA on SA accumulation in the intact network and its positive role in the absence of PAD4. We also show that both of these transcriptional effects of JA occur not only in A. thaliana but also in other Brassicaceae species. Taken together, our results highlight the I4‐FFL that allows plants to alleviate the negative impact of SA on fitness as well as to support robust SA accumulation when PAD4 function is compromised.
Results
JA is defined as a repressor or activator of SA accumulation depending on PAD4
To investigate the regulatory relationship between JA and PAD4 in MAMP‐triggered SA accumulation, we measured SA levels in leaves of wild‐type, dde2, pad4, and dde2 pad4 plants after infiltration with flg22. In the wild type, an increase in SA level was observed at 9 h post‐infiltration (Fig 1A). The SA level was elevated in dde2, which is reminiscent of the often described repressive effect of JA on SA. In contrast, in pad4, SA was increased by flg22 treatment, but to a level lower than in wild type, which is consistent with PAD4 being a positive regulator of SA accumulation in response to flg22 25. Strikingly, flg22‐triggered SA accumulation was abolished in dde2 pad4, showing a requirement of JA for SA induction in the absence of PAD4. Similar patterns were observed for expression of the canonical SA marker gene PR1 (Fig 1B), as well as that of At2g26400 and At2g30550 (Fig 1C and D), which was shown to be induced upon challenge with P. syringae pv. tomato DC3000 (Pto) in an SA‐dependent manner 39. In line with the previous study 14, these results demonstrated that JA acts as a repressor or activator of SA accumulation in the presence or absence of PAD4, respectively, during flg22‐triggered PTI.
Figure 1. JA is genetically defined as a repressor or activator of SA accumulation depending on PAD4 .
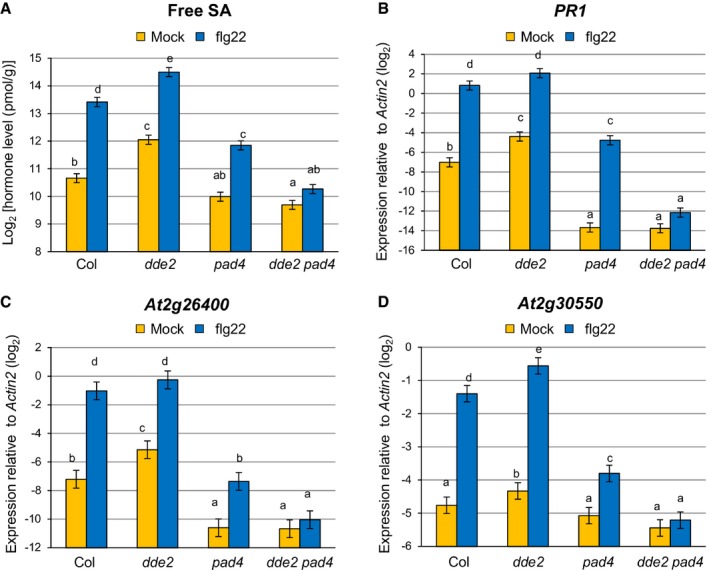
-
AMeasurement of SA levels in leaves infiltrated with water (mock) or 1 μM flg22 at 9 hpi. Bars represent means and standard errors of the SA levels on a log2 scale calculated from two independent experiments using a mixed linear model.
-
B–DRT–qPCR analysis of PR1, At2g26400 and At2g30550 expression in leaves infiltrated with water (mock) or 1 μM flg22 at 9 hpi. Bars represent means and standard errors of the log2 expression level relative to Actin2 (At3g18780) calculated from three independent experiments using a mixed linear model.
JA represses PAD4 expression through the action of MYC transcription factors
Since the enhanced SA accumulation in dde2 was dependent on PAD4 (Fig 1A; compare dde2 and dde2 pad4), we tested whether JA represses PAD4 expression. PAD4 expression was elevated in dde2 as well as in coi1 at 9 h after flg22 treatment (Fig 2A). The transcription factors MYC2 and its homologues MYC3 and MYC4 are important for transcriptional responses to JA, and we found a MYC2‐binding motif (G box; CACATG) in the PAD4 promoter using the online tool Athena (Fig 2B) 40, 41, 42. These observations led us to test whether MYC2 and its homologues MYC3 and MYC4 are responsible for JA‐mediated repression of PAD4 expression. Indeed, increased expression of PAD4 was observed in myc2 myc3 myc4 but not in myc2 (Fig 2A). Thus, these MYCs seem to act redundantly to repress PAD4 expression during flg22‐triggered PTI.
Figure 2. JA represses PAD4 expression through MYC transcription factors.
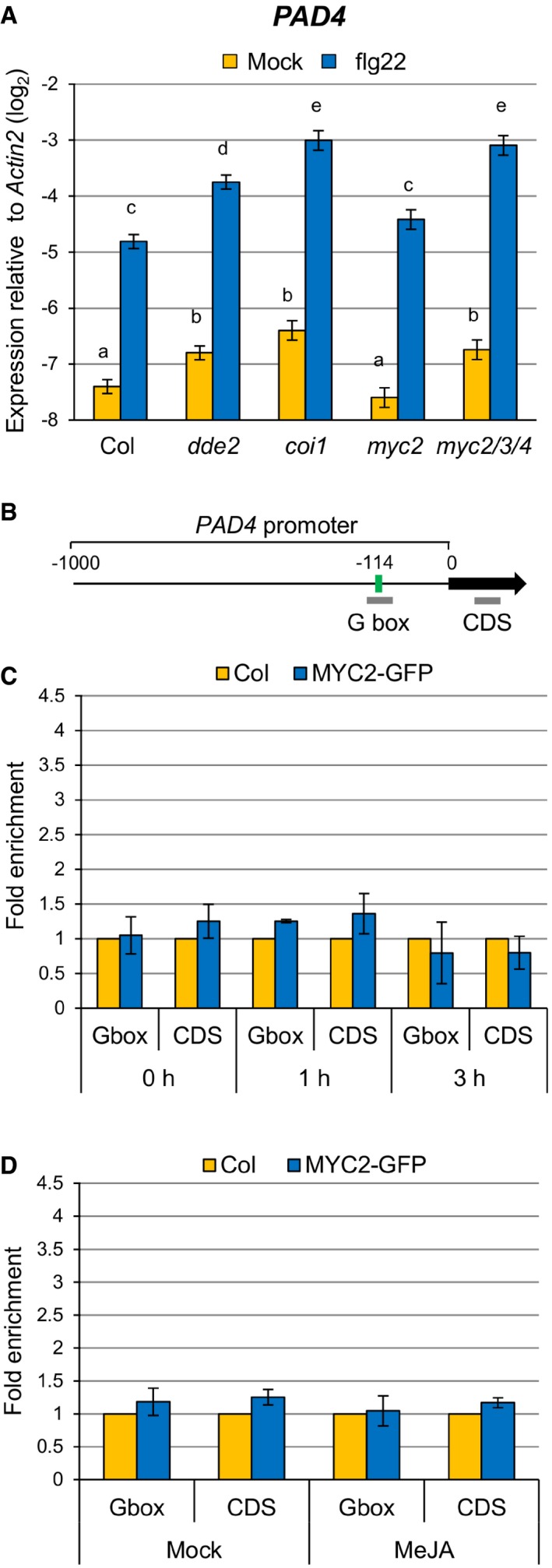
-
ART–qPCR analysis of PAD4 expression in leaves infiltrated with water (mock) or 1 μM flg22 at 9 hpi. Bars represent means and standard errors of the log2 expression level relative to Actin2 calculated from four independent experiments using a mixed linear model.
-
BPAD4 promoter showing the G box motif located 114 bp upstream of the transcription start site. Bold gray horizontal lines show the regions amplified by different qPCR primers.
-
C, DChIP‐qPCR analysis of MYC2 binding to the PAD4 promoter. MYC2‐GFP seedlings were treated with 1 μM flg22 for the indicated time periods (C) or 100 μM MeJA for 3 h (D). Bars represent means and standard errors of the fold enrichment relative to the wild‐type plants set to 1, calculated from two independent experiments.
We then tested whether MYC2 directly binds to the G box motif in the PAD4 promoter in planta by chromatin immunoprecipitation (ChIP) using a transgenic A. thaliana line constitutively expressing the MYC2‐GFP fusion protein (Fig EV1). The enrichment of the G box sequence in immunoprecipitates from MYC2‐GFP plants relative to those from wild‐type plants was determined by qPCR. A DNA segment from the coding sequence (CDS) of PAD4 was used as a negative control. Although these MYC transcription factors contribute to PAD4 repression (Fig 2A), we did not observe direct binding of MYC2 to the PAD4 promoter even after the treatment with flg22 or MeJA (Fig 2C and D). Considering that MYC2, MYC3, and MYC4 are transcriptional activators with shared DNA‐binding specificity 43, it is likely that these MYC transcription factors indirectly repress PAD4 expression through an intermediate factor(s).
Figure EV1. Accumulation of MYC2‐GFP protein in the p35S:MYC2‐GFP transgenic plants.
Total protein was extracted from leaves of 4‐ to 5‐week‐old plants and separated on an SDS–PAGE gel. The MYC2‐GFP protein was detected using an anti‐GFP antibody. Ponceau S staining is shown as a loading control.
JA induces EDS5 expression directly through MYC2
Since JA positively contributes to SA accumulation in the absence of PAD4, we examined expression levels of SID2 and EDS5, both of which are essential for pathogen‐induced SA accumulation 25, 26, 28, 29. At 5 h after flg22 treatment, expression of SID2 was similar in pad4 and dde2 pad4 (Fig 3A). In contrast, expression of EDS5 was significantly lower in dde2 pad4 than in pad4, and EDS5 induction was abolished in dde2 pad4 (Fig 3B), indicating that PAD4 and JA together are responsible for flg22‐triggered EDS5 expression. Importantly, the compromised EDS5 induction in dde2 pad4 was correlated well with the compromised SA induction in dde2 pad4 (Fig 1A), suggesting that EDS5 is the causal gene for the positive role of JA in SA accumulation.
Figure 3. MYC2 directly regulates EDS5 induction by JA.
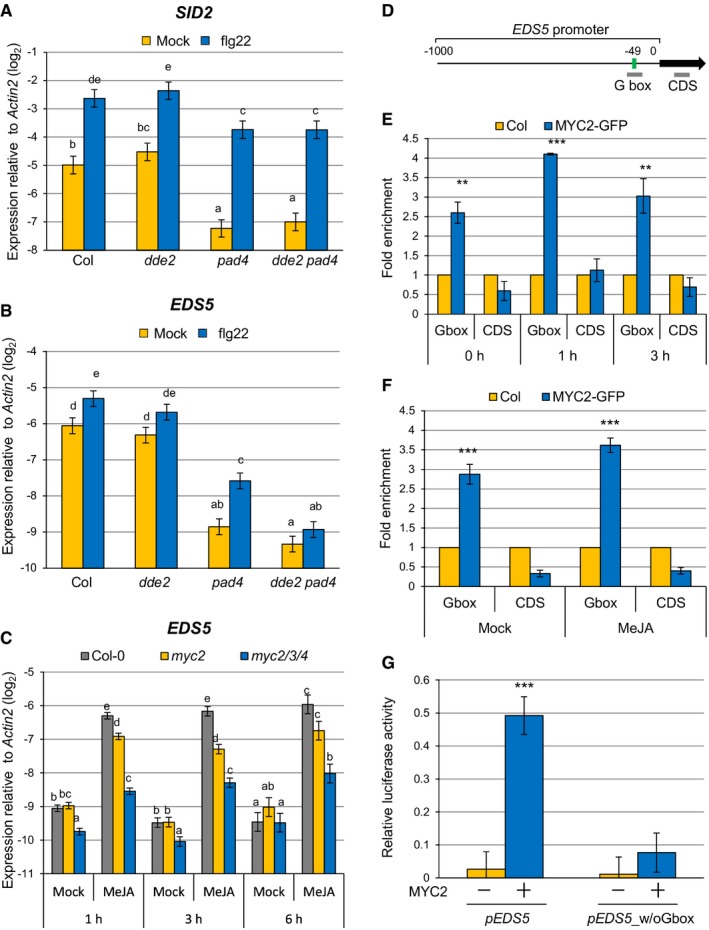
-
A, BRT–qPCR analysis of SID2 (A) and EDS5 (B) expression in leaves infiltrated with water (mock) or 1 μM flg22 at 5 hpi. Bars represent means and standard errors of the log2 expression levels relative to Actin2 calculated from four independent experiments using a mixed linear model.
-
CRT–qPCR analysis of EDS5 expression in seedlings treated with water (mock) or 100 μM MeJA for the indicated time periods. Bars represent means and standard errors of the log2 expression level relative to Actin2 calculated from two independent experiments using a mixed linear model.
-
DEDS5 promoter showing the G box motif located 49 bp upstream of the transcription start site. Bold gray horizontal lines show the regions amplified by different qPCR primers.
-
E, FChIP‐qPCR analysis of MYC2 binding to the EDS5 promoter. MYC2‐GFP seedlings were treated with 1 μM flg22 for the indicated time periods (E) or 100 μM MeJA for 3 h (F). Bars represent means and standard errors of the fold enrichment relative to the wild‐type plants set to 1, calculated from two independent experiments.
-
GLuciferase reporter assays using EDS5 promoters with or without G box. Luc reporter construct driven by the wild‐type EDS5 promoter (pEDS5) or the EDS5 promoter without G box (pEDS5_w/oGbox) was transfected with or without 35S‐MYC2 plasmid to Arabidopsis protoplasts. Bars represent means and standard errors of the Luc activity relative to the internal control (Luc derived from Renilla spp. driven by 35S promoter) calculated from three independent experiments each with three biological replicates.
To explore the mechanism by which JA regulates EDS5 expression, the promoter sequence of EDS5 was searched for cis elements using the Athena analysis tool. We found a canonical G box (CACGTG), the binding site for MYC transcription factors, in close proximity to the transcription start site of EDS5 (Fig 3D). This prompted us to test whether MYC2 and its homologues MYC3 and MYC4 are responsible for EDS5 induction by JA. In the wild‐type plants, MeJA treatment induced EDS5 expression at the three time points tested, while EDS5 expression was significantly reduced in myc2 and myc2 myc3 myc4 (Fig 3C), demonstrating that these MYCs are required for JA‐mediated EDS5 induction. We then performed ChIP experiments using MYC2‐GFP plants treated with or without flg22 or MeJA to test whether MYC2 directly binds to the EDS5 promoter. We found a significant enrichment of the promoter sequence containing the G box motif in all the conditions tested, but no enrichment was observed for a DNA segment in the CDS of EDS5 used as a negative control (Fig 3E and F). To test whether the G box in the EDS5 promoter is required for MYC2‐mediated transcriptional activation of EDS5, we carried out luciferase (Luc) reporter assays using Arabidopsis protoplasts. Expression of MYC2 significantly induced the wild‐type EDS5 promoter‐driven Luc activity, whereas deletion of the G box abolished this MYC2‐mediated transcriptional activation (Fig 3G). Taken together, these results indicate that MYC2 directly binds to the EDS5 promoter and controls EDS5 induction by JA.
Reconstitution of EDS5 expression restores flg22‐triggered SA accumulation and immunity in dde2 pad4
To test for a causal link between JA‐mediated EDS5 expression and SA accumulation, we generated transgenic lines expressing EDS5 under two different promoters in dde2 pad4. In two independent lines expressing EDS5 from the constitutive 35S promoter, EDS5 expression was higher than in the wild type and was not altered after flg22 treatment (Fig 4A). The expression level of EDS5 was more than eightfold higher in p35S:EDS5 line #1 than in line #2 (Fig 4A). Another transgenic line expressing EDS5 from the SID2 promoter showed the wild‐type level of EDS5 expression after mock treatment and slightly higher expression of EDS5 compared to the wild type after flg22 treatment (Fig 4A). This is in accordance with our finding that SID2 was responsive to flg22 in dde2 pad4 (Fig 3A). Induction of SA accumulation and PR1 expression by flg22 was detected in p35S:EDS5 line #1 but not in line #2 (Fig 4B and C). The pSID2:EDS5 line also showed restored SA accumulation and PR1 expression after flg22 treatment (Fig 4B and C) although the expression level of EDS5 was lower than in p35S:EDS5 line #2. Thus, a minimal level of EDS5 expression, which is not achieved in dde2 pad4, is required for flg22‐triggered SA accumulation. These results also suggest that transcriptional induction of EDS5 in response to flg22 can overcome the need to constitutively express EDS5 at a very high level for flg22‐triggered SA accumulation. As EDS5 is inducible by flg22, this transcriptional induction might be a critical part of flg22‐triggered SA accumulation. Overall, our data clearly established a causal connection between compromised EDS5 expression or induction and the compromised SA accumulation in dde2 pad4 in response to flg22.
Figure 4. Reconstitution of EDS5 expression restores flg22‐triggered SA accumulation and flg22‐PTI in dde2 pad4 .
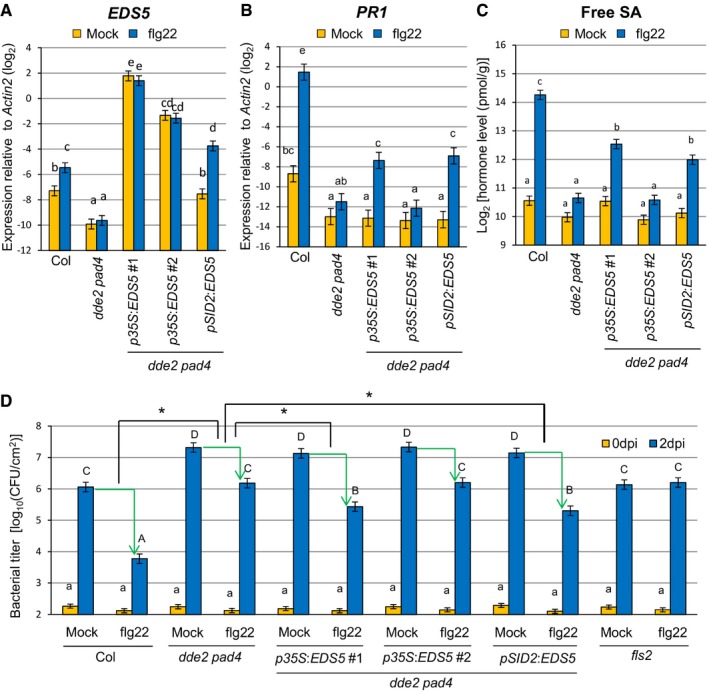
-
A, BRT–qPCR analysis of EDS5 (A) and PR1 (B) expression in leaves of Col, dde2 pad4, p35S::EDS5 lines and a pSID2::EDS5 line infiltrated with water (mock) or 1 μM flg22. The expression levels of EDS5 and PR1 were measured at 5 hpi and 9 hpi, respectively. Bars represent means and standard errors of the log2 expression levels relative to Actin2 calculated from two independent experiments using mixed linear models.
-
CMeasurement of SA levels in leaves of Col, dde2 pad4, p35S::EDS5 lines and a pSID2::EDS5 line infiltrated with water (mock) or 1 μM flg22 at 9 hpi. The means and standard errors calculated from two independent experiments using a mixed linear model are shown on a log2 scale.
-
DBacterial growth assay in leaves of Col, dde2 pad4, p35S::EDS5 lines and a pSID2::EDS5 line infiltrated with Pto (OD600 = 0.0002) together with water (mock) or 1 μM flg22. The bacterial titers at 0 or 2 dpi were measured. Bars represent means and standard errors of two independent experiments with at least 4 or 12 biological replicates for 0 dpi or 2 dpi in each experiment, respectively.
To test whether the restored SA accumulation in the transgenic lines is relevant for immunity, we measured Pto growth. Leaves were co‐infiltrated with Pto and flg22 and sampled at 2 days after infiltration. Co‐infiltration of flg22 inhibited Pto growth in the wild type but not in fls2, a mutant lacking the receptor for flg22 (Fig 4D). This reduction in bacterial growth, termed flg22‐triggered PTI, was calculated by subtracting the log10‐transformed bacterial titer in flg22‐treated leaves from that in mock‐treated leaves. Flg22‐triggered PTI was much less in dde2 pad4 than in the wild type. Importantly, flg22‐triggered PTI was significantly higher in the transgenic lines with restored SA accumulation than in dde2 pad4 plants (Fig 4D). Given the genetic requirement for JA in flg22‐triggered EDS5 expression and SA accumulation in pad4 (Figs 1A and 3B), we conclude that JA enables robust flg22‐triggered PTI by supporting SA accumulation through MYC2‐activated EDS5 expression.
Distinct effects of JA on bacterial resistance depending on PAD4
Our genetic perturbation and reconstitution approach illustrates an I4‐FFL consisting JA (MYC transcription factors), PAD4, and EDS5 (Fig 5A). To further investigate the roles of the I4‐FFL in plant immunity, we assessed effects of exogenous MeJA application on flg22‐triggered PTI against Pto in the wild type, dde2 pad4, and the transgenic p35S:EDS5 #1 and pSID2:EDS5 lines with restored flg22‐triggered SA accumulation. MeJA reduced flg22‐triggered PTI in the wild type but enhanced it in dde2 pad4 (Fig 5B), demonstrating that the negative effect of JA is dominant in the presence of PAD4, whereas the positive effect of JA is evident in the absence of PAD4. MeJA had no effect on flg22‐triggered immunity in the transgenic lines, suggesting that the positive role of JA in the absence of PAD4 is to support SA accumulation via EDS5 expression. These results are consistent with our I4‐FFL model, in which JA negatively or positively regulates SA‐mediated bacterial resistance in the presence or absence of PAD4, respectively.
Figure 5. Distinct effects of JA on bacterial resistance depending on PAD4 .
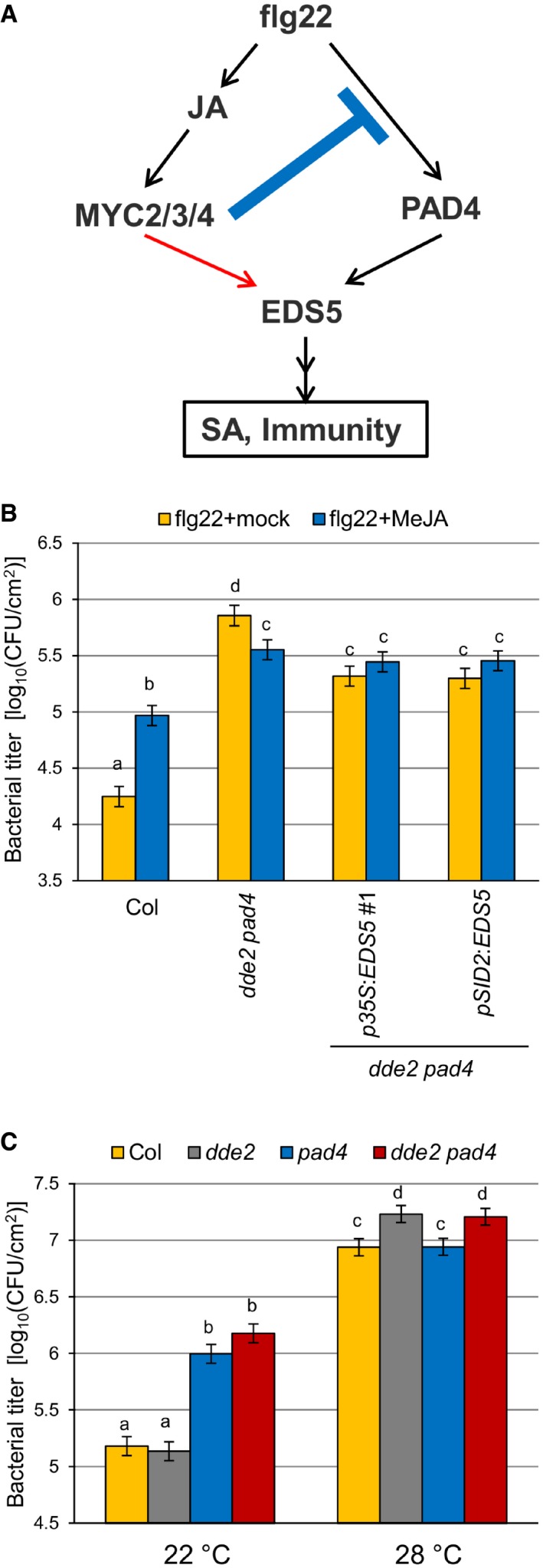
- A model of the incoherent type‐4 feed‐forward loop consisting of JA, PAD4, and EDS5. The blue line and the red arrow indicate negative and positive effects of JA on the network output, respectively.
- Bacterial growth assay in leaves of Col, dde2 pad4, p35S::EDS5 line #1, and pSID2::EDS5 line infiltrated with Pto (OD600 = 0.0002) and 1 μM flg22 with or without treatment of 1 mM MeJA. The bacterial titers at 2 dpi were measured. Bars represent means and standard errors of three independent experiments each with at least 10 biological replicates.
- Bacterial growth assay in leaves of Col, dde2, pad4, and dde2 pad4 infiltrated with Pto (OD600 = 0.0002) at 22 or 28°C. The bacterial titers at 2 dpi were measured. Bars represent means and standard errors of two (22°C) or three (28°C) independent experiments each with at least 10 biological replicates.
PAD4‐regulated signaling to SA activation is perturbed at high temperature such as 28°C 44. To investigate the biological importance of the I4‐FFL in a more natural context, we measured Pto growth in the wild type, dde2, pad4, and dde2 pad4 at 22°C and 28°C. As shown in Fig 5C, pad4 was more susceptible to Pto than the wild type at 22°C. Such enhanced susceptibility of pad4 was not observed at 28°C, indicating that PAD4 function in Pto resistance is compromised at this temperature. Interestingly, dde2 and dde2 pad4 supported more Pto growth than the wild type and pad4, respectively, at 28°C. No significant differences in Pto growth between Col and dde2 and between pad4 and dde2 pad4 were observed at 22°C. The effects of dde2 mutation at 22°C might be masked by coronatine produced by Pto, which activates JA signaling by acting as a molecular mimic of JA‐Ile 20, 45. Overall, these results support a biological significance of the I4‐FFL for conferring JA‐mediated bacterial resistance under perturbation of PAD4 at high temperature, which can naturally occur.
Conservation and diversification of JA‐mediated regulation of PAD4 and EDS5 in Brassicaceae
The importance of the I4‐FFL identified in this study could be reflected by evolutionary conservation in plants. To address this point, we used the A. thaliana EDS5 protein sequence to identify related proteins in some Brassicaceae species, tomato and rice whose genome sequences and gene annotations are available. Construction of a phylogenetic tree using the related proteins suggests that the EDS5 clade is conserved in the family Brassicaceae but not in other plants (Fig EV2). Since our results suggest that MYC2 controls JA‐mediated EDS5 induction through binding to the CACGTG G box motif (Fig 3E–G), we surveyed 500 bp upstream of the transcription start sites (hereafter referred to as “promoters”) and 5′‐UTRs of these EDS5 orthologues for this motif. Interestingly, the G box motif was found in the EDS5 promoters of A. thaliana, Arabidopsis lyrata, Capsella grandiflora, and Eutrema salsugineum, whereas it was located in the 5′‐UTRs in Capsella rubella and Brassica rapa (Fig EV3). MeJA treatment induced EDS5 expression in A. thaliana, A. lyrata, and E. salsugineum, but not in C. rubella (Fig 6A). This is in line with the presence or absence of the G box motif in the promoters. Capsella rubella was responsive to MeJA in other ways, as exemplified by induction of a homologue of the A. thaliana VSP2, a JA responsive gene (Fig EV4). The inducibility of EDS5 by JA is not correlated to the phylogenetic distance within Brassicaceae 46. Thus, these results may suggest that the JA‐mediated EDS5 regulation emerged in the ancestor of Brassicaceae and C. rubella has lost it.
Figure EV2. Phylogenetic analysis of putative EDS5 orthologues.
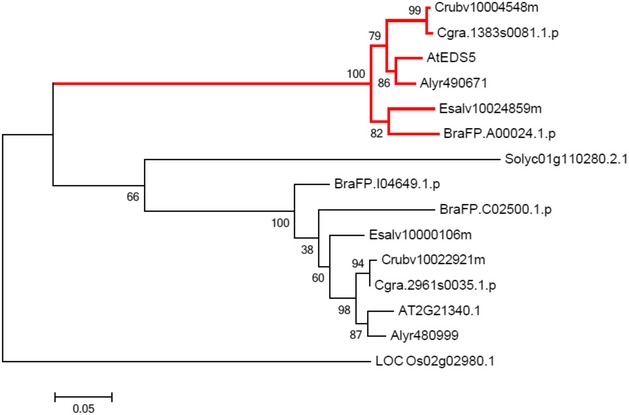
The proteins belonging to the same group as Arabidopsis thaliana EDS5 were identified by OrthoMCL. A maximum likelihood phylogenetic tree was constructed based on the amino acid sequences using the MEGA6 software. The EDS5 clade is highlighted by red lines.
Figure EV3. Conservation of G boxes in EDS5 promoters of Brassicaceae species.
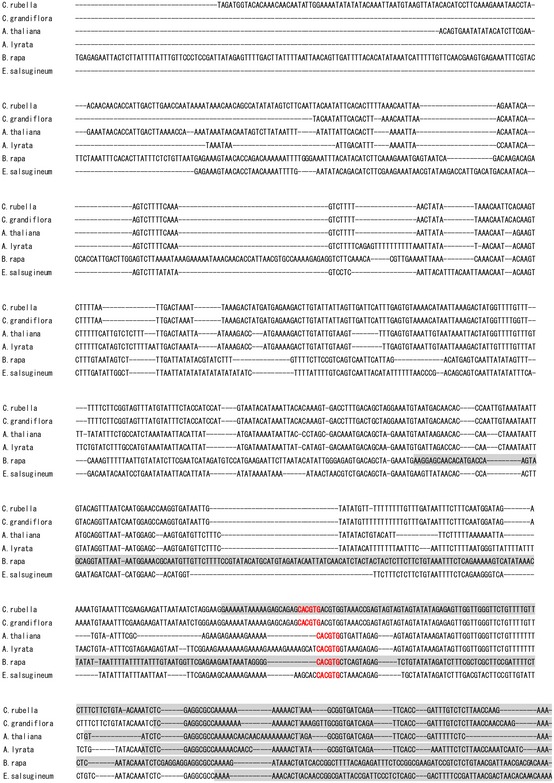
The 500 bp upstream of the transcription start sites of EDS5 and the 5′‐UTRs was aligned using MUSCLE. The 5′‐UTRs were highlighted by gray boxes. The CACGTG G box motif was shown in bold red letters.
Figure 6. Conservation and diversification of the transcriptional regulation of EDS5 and PAD4 by JA in Brassicaceae .
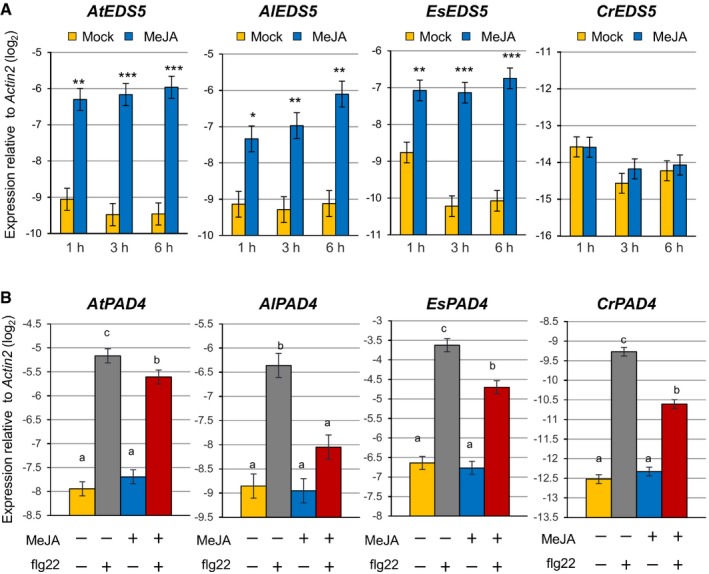
-
A, BRT–qPCR analysis of EDS5 (A) and PAD4 (B) expression in seedlings of Arabidopsis thaliana, Arabidopsis lyrata, Capsella rubella, and Eutrema salsugineum. In (A), seedlings were treated with mock (water) or MeJA (100 μM) for the indicated time periods. Asterisks indicate statistically significant differences from the mock controls at each time point (*P < 0.05, **P < 0.01, ***P < 0.001; two‐tailed t‐tests). In (B), seedlings were treated with mock (water) or MeJA (100 μM) for 3 h, followed by treatment with mock (water) or flg22 (1 μM) for 30 min. The Benjamini–Hochberg method was used to adjust P‐values (two‐tailed t‐tests) for correcting multiple hypothesis testing, and statistically significant differences were indicated by different letters (adjusted P‐value < 0.05). Bars represent means and standard errors of the log2 expression levels relative to Actin2 calculated from two independent experiments using mixed linear models.
Figure EV4. Capsella rubella is responsive to JA.
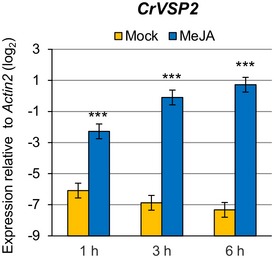
RT–qPCR analysis of VSP2 expression in C. rubella seedlings after treatment with water (mock) or 100 μM MeJA for the indicated time periods. Bars represent means and standard errors of the log2 expression levels relative to Actin2 calculated from two independent experiments using a mixed linear model. Asterisks indicate statistically significant differences compared to the mock controls at each time point (***P < 0.001; two‐tailed t‐tests).
PAD4 is conserved among flowering plants 47. We therefore tested whether JA‐mediated repression of PAD4 expression is conserved among Brassicaceae. A. thaliana, A. lyrata, C. rubella, and E. salsugineum plants were treated with mock or MeJA, followed by flg22 treatment. In A. thaliana, MeJA treatment had no effect on PAD4 expression but inhibited PAD4 induction by flg22 (Fig 6B). As in A. thaliana, MeJA had an inhibitory effect on PAD4 induction by flg22 in the other three species (Fig 6B). Thus, the repressive effect of JA on PAD4 expression during flg22‐PTI appears to be conserved in Brassicaceae.
Discussion
It is vital for plants to invoke robust immunity against pathogens that interfere with immune signaling and, at the same time, to minimize fitness costs associated with immunity. This is particularly relevant to PTI, since it is activated by MAMPs which do not distinguish pathogens from other beneficial or benign microbes. In this study, we identified an I4‐FFL consisting of JA, PAD4, and EDS5 in the PTI signaling network in A. thaliana. JA induces EDS5 expression directly via the transcription factor MYC2 while repressing expression of PAD4 which positively contributes to EDS5 expression. I4‐FFL is rare in biological networks and, therefore, its biological function has rarely been characterized 48, 49. In the context of PTI, PAD4 repression by JA is functionally dominant in the intact network of wild‐type plants, which explains reduction in SA accumulation in pad4 and increase in dde2. However, in the absence of PAD4, the positive contribution of JA to SA accumulation becomes apparent. Consistently, SA induction in response to flg22 was abolished in dde2 pad4. The JA‐mediated suppression of PAD4 expression is likely important to alleviate the negative impact of SA on plant growth 6, 32, 33, 34. In contrast, the JA‐mediated EDS5 induction provides robust SA accumulation in flg22‐triggered immunity when PAD4 cannot fulfill its function, for example, due to pathogen effectors or environmental factors.
A mechanism by which JA inhibits SA accumulation was uncovered by characterizing the mode of action of the JA‐mimicking bacterial phytotoxin coronatine produced by P. syringae 50. It was demonstrated that MYC2 transcriptionally activates the NAC (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) transcription factors ANAC019, ANAC055, and ANAC072, which repress the SA biosynthesis gene SID2 and induce the SA catabolism gene BSMT1. However, no significant increase in SID2 expression was observed in dde2 during flg22‐triggered PTI (Fig 3A). Thus, the negative effect of JA on SID2 expression is not the cause of antagonistic effects of JA on SA accumulation in the context of flg22‐triggered PTI at least in our hands. In contrast, our genetic evidence indicates that the repressive effect of JA on SA accumulation is dependent on PAD4 in flg22‐triggered PTI, as introducing pad4 mutation into dde2 abolished flg22‐triggered SA accumulation. Consistently, JA represses PAD4 expression in a manner dependent on MYC2, MYC3, and MYC4. The JA‐mediated repression of PAD4 expression could explain the previous observation that expression of a marker gene of PAD4 signaling activity (At5g46960) was elevated in dde2 14. Overall, our genetic evidence suggests a novel mechanism for JA‐mediated suppression of SA accumulation through MYC transcription factors. However, our ChIP experiment did not support direct binding of MYC2 to the PAD4 promoter. It is also unlikely that PAD4 repression by JA is directly mediated by the NACs downstream of the MYCs because there is no NAC‐binding site present in the PAD4 promoter 50. Further studies will be required to unravel the mechanism of the negative regulation of PAD4 expression by JA in PTI.
Although most studies of JA–SA crosstalk have reported antagonistic interactions, cooperative interactions between the two phytohormones have been observed under some conditions 14, 38. However, the underlying mechanism is unknown. In the present study, we show that JA transcriptionally activates EDS5 directly through MYC2. This transcriptional regulation is causally linked to JA‐mediated SA accumulation and immunity in pad4, as reconstitution of EDS5 expression or induction restored flg22‐triggered SA accumulation and immunity in dde2 pad4. In addition, exogenous MeJA application enhanced flg22‐triggered immunity in dde2 pad4 but not in the transgenic p35S:EDS5 #1 and pSID2:EDS5 lines with restored flg22‐triggered SA accumulation. By making use of the fact that PAD4‐regulated signaling to SA activation is highly influenced by temperature 44, we showed that JA confers bacterial resistance under perturbation of PAD4 at 28°C. Thus, we propose that the robust SA accumulation and immunity enabled by JA have a substantial role, when plants face situations in which PAD4 function is perturbed by environmental factors such as high temperature and likely by pathogen effectors. With respect to the latter situation, it is noteworthy that some bacterial effectors target EDS1, which is required for PAD4 function 51, 52.
It would be interesting to discuss effects of coronatine in the framework of the I4‐FFL identified in this study. Coronatine is a JA‐mimicking virulence factor that suppresses SA‐mediated immunity to promote bacterial growth 20, 45, 50. Consistently, we observed that MeJA treatment after flg22 infiltration promotes Pto growth in the wild type. However, in dde2 pad4, MeJA treatment reduced Pto growth. Thus, coronatine may have a negative impact on bacterial virulence when combined with other effectors that interfere with PAD4 activity as well as under environmental conditions in which PAD4 cannot fulfill its function.
Although A. thaliana is an excellent model system to study molecular and genetic aspects of plant biology, it is becoming increasingly important to expand our knowledge to other plant species 46. In this study, we took advantage of the family Brassicaceae, to which A. thaliana belongs, for studying evolutionary conservation of the gene regulation that we identified in A. thaliana. Our results indicate that the repressive effect of JA on PAD4 expression during PTI is conserved not only in A. lyrata and C. rubella, close relatives of A. thaliana, but also in E. salsugineum, a relatively phylogenetically distant species from A. thaliana. Thus, the repression of PAD4 by JA may be a common regulatory mechanism for tunable SA accumulation during PTI in Brassicaceae. Since PAD4 is conserved in flowering plants 47, it would be interesting to test whether JA represses PAD4 expression during PTI in plant species outside Brassicaceae.
In contrast to PAD4, our phylogenetic analysis highlighted a Brassicaceae‐specific clade to which A. thaliana EDS5 belongs, suggesting that the role of EDS5 in SA accumulation might be restricted to this family. Interestingly, our gene expression data together with promoter analysis pointed to a good correlation between the presence or absence of the CACGTG G box motif in the promoters and the inducibility of EDS5 by JA in Brassicaceae. We note that in C. rubella, in which JA does not induce EDS5, the CACGTG sequence is present downstream of the transcription start site and transcribed as a part of the 5′‐UTR 53. Thus, C. rubella might have lost JA‐mediated EDS5 induction by changing the transcription start site. This might also hold true for B. rapa, as the G box motif is located in the 5′‐UTR (Brassica rapa FPsc v1.3, DOE‐JGI, http://phytozome.jgi.doe.gov/). Overall, our comparative analysis suggests that EDS5 and its transcriptional regulation by JA are an innovation of the family Brassicaceae.
In conclusion, our results highlight an I4‐FFL that simultaneously provides robust and tunable regulation of SA response during PTI in A. thaliana. The transcriptional effects of JA on EDS5 and PAD4 appear to be highly conserved in the family Brassicaceae. Whether or not this reflects evolutionary conservation of the I4‐FFL deserves further study.
Materials and Methods
Plant materials and growth conditions
Arabidopsis plants were grown in a chamber at 22°C with a 10‐h light period and 60% relative humidity for 3 weeks and then in another chamber at 22°C with a 12‐h light period and 60% relative humidity. The A. thaliana accession Col‐0 was the background of all Arabidopsis mutants used in this study. Arabidopsis dde2‐2 16, pad4‐1 27, dde2‐2 pad4‐1 54, coi1‐1 19, jin1‐9/myc2 (SALK_017005) 55, myc2 myc3 myc4 43, and fls2 (SAIL_691C4) 56 were described previously. The MYC2‐GFP overexpression plants were obtained from Dr. Hironaka Tsukagoshi (Meijo University, Japan). Seedlings of A. thaliana, A. lyrata (MN47), C. rubella (N22697), and E. salsugineum (Shandong) were grown on solidified half‐strength Murashige and Skoog (MS) medium supplemented with 1% sucrose under a 10‐h light period at 22°C.
Chemicals
MeJA (392705) and flg22 were purchased from Sigma (Munich, Germany) and EZBiolab Inc. (Westfield, IN, USA), respectively.
Cloning and plant transformation
The coding sequence (without introns) of EDS5 (AT4G39030) was amplified by PCR using PrimeSTAR HS DNA polymerase (Takara‐Clontech, Saint‐Germain‐en‐Laye, France) and cloned into the pENTR/D‐TOPO vector following the manufacturer's protocol (Life Technologies, Darmstadt, Germany) to generate pENTR_EDS5. The promoter sequence of SID2 (At1g74710) 57 and the Nos terminator sequence from pER8 58 were amplified by PCR and cloned into the NotI and AscI sites of pENTR_EDS5, respectively, to generate pENTR_pSID2_EDS5_Nos. pENTR_EDS5 and pENTR_pSID2_EDS5_Nos were then recombined into the Gateway‐compatible binary vectors pFAST‐R02 59 and pFAST‐R01 59 , respectively, through the LR reaction (Invitrogen). Primers used are listed in Appendix Table S1. All plasmids constructed in this study were verified by sequencing. Arabidopsis thaliana dde2 pad4 plants were transformed using Agrobacterium tumefaciens stain GV3101 as described 16.
Statistical analysis
Statistical analysis was performed using the mixed linear model function, lmer, implemented in the package lme4 in the R environment. When appropriate, raw data were log‐transformed to meet the assumptions of the mixed linear model. For the t‐tests, the standard errors were calculated using the variance and covariance values obtained from the model fitting. The Benjamini–Hochberg methods were applied to correct for multiple hypothesis testing when all pairwise comparisons of the mean estimates were made in a figure.
RNA extraction, cDNA synthesis, and quantitative PCR
Leaves of 4‐ to 5‐week‐old plants were infiltrated with 1 μM flg22 or mock (water) using a needleless syringe and collected at the indicated time points. Seedlings were submerged into liquid half‐strength MS medium containing 100 μM MeJA or mock (water) for the indicated time period and, if required, transferred to new liquid half‐strength MS medium containing 1 μM flg22 or mock. Total RNAs were isolated using TriFast (peqlab, Erlangen, Germany), followed by cDNA synthesis using superscript II (Life Technologies). Real‐time PCR was performed using EvaGreen (Biotium, Hayward, CA, USA) on the iQ5 Multicolor Real‐Time PCR Detection System (Bio‐Rad, Munich, Germany) or the CFX Connect Real‐Time PCR Detection System (Bio‐Rad). Primers used are listed in Appendix Table S1. The following models were fit to the relative Ct value data compared to Actin2: Ctgyr = GYgy + Rr + egyr, where GY, genotype:treatment interaction and random factors; R, biological replicate; e, residual; Ctytr = YTyt + Rr + eytr, where YT, treatment:time interaction and random factors; R, biological replicate; e, residual. The mean estimates of the fixed effects were used as the modeled relative Ct values, visualized as the relative log2 expression values, and compared by two‐tailed t‐tests.
SA measurement
Leaves of 4‐ to 5‐week‐old plants were infiltrated with mock (water) or 1 μM flg22. Samples were harvested 9 h after the treatment and stored at −80°C. SA measurement was performed as described previously 60. The following model was fit to log2‐transformed SA levels (pmol/g fresh weight): SAgyr = GYgy + Rr + egyr, where GY, genotype:treatment interaction and random factors; R, biological replicate; e, residual. The mean estimates of the fixed effects were compared by two‐tailed t‐tests.
Bacterial growth assay
Bacterial growth assays were performed essentially as described previously 54. For measuring flg22‐triggered immunity, bacterial suspensions were co‐infiltrated with 1 μM flg22 into leaves of 4‐ to 5‐week‐old plants using a needleless syringe. For assessing effects of MeJA, 1 mM MeJA was sprayed onto 4‐ to 5‐week‐old plants shortly after infiltration of bacterial suspensions and 1 μM flg22. For assessing effects of temperature, 4‐ to 5‐week‐old plants were grown, infiltrated with bacterial suspension, and kept at 22 or 28°C throughout the experiments. Log10‐transformed colony‐forming units (cfu) per cm2 leaf surface area were calculated, and the following model was fit to the data: CFUgyr = GYgy + Rr + egyr, where GY, genotype:treatment interaction and random factors; R, biological replicate; e, residual. Flg22‐triggered immunity was calculated by subtracting the modeled bacterial titers in flg22‐treated plants from those in the mock‐treated plants.
Chromatin immunoprecipitation
Tissue fixation and chromatin immunoprecipitation were carried out as described 61 with some modifications. Briefly, 2‐week‐old seedlings grown in liquid half‐strength MS medium supplemented with 1% sucrose were treated with 1 μM flg22 for 1 or 3 h. Untreated seedlings were also harvested. Alternatively, seedlings were treated with mock (water) or 100 μM MeJA for 3 h. After fixation in 1% formaldehyde solution, tissues were frozen in liquid nitrogen and stored at −80°C. Frozen tissues (~1 g) were ground in liquid nitrogen using a mortar and pestle and suspended in 3 ml of lysis buffer [50 mM Tris–HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 1% Triton X‐100, 50 μM MG132 (Sigma), and complete protease inhibitor cocktails (04693132001; Roche, Mannheim, Germany) or proteases inhibitor cocktail (P9599; Sigma)]. The suspension was sonicated twice on the Bioruptor Next Gen UCD‐300 sonication system (Diagenode, Seraing, Belgium) for 10 min at 4°C, followed by centrifugation at 20,000 × g for 10 min at 4°C. The supernatant was used as the starting material for chromatin immunoprecipitation using anti‐GFP antibody (Ab290; Abcam, Cambridge, UK). Aliquots of the supernatant were kept as input samples. The samples were analyzed by quantitative PCR using primers listed in Appendix Table S1. The percentage of input values of the ChIP DNA was further normalized over the value obtained for the Actin7 promoter (AT5G09810). Fold enrichment was then calculated by taking ratios between normalized results from wild‐type plants and from MYC2‐GFP plants. For statistical analysis, the following model was fit to log2‐transformed values of the normalized value data: Ctgyr = GYgy + Rr + egyr, where GY, genotype:treatment interaction and random factors; R, biological replicate; e, residual. The mean estimates of the fixed effects were compared by two‐tailed t‐tests.
Luciferase reporter assay
The WT EDS5 promoter was amplified by PCR (PrimeSTAR HS DNA polymerase; Takara‐Clontech) using pEDS5_F and pEDS5_R (with HindIII and BamHI restriction sites, respectively) listed in Appendix Table S1, designed as recommended by the In‐Fusion HD cloning kit. For the EDS5 promoter without the G box, two fragments were amplified by PCR using two sets of primers, pEDS5_F and pEDS5w/oGbox_R and pEDS5w/oGbox_F and pEDS5_R, respectively (Appendix Table S1) and then fused by PCR using pEDS5_F and pEDS5_R. These promoter sequences were cloned into HindIII/BamHI‐digested pBI221‐LUC using In‐Fusion HD cloning kit (Takara‐Clontech) to generate pBI221_pEDS5::LUC and pBI221_pEDS5w/oGbox::LUC. pENTR_MYC2 used in this study was obtained from Dr. Haitao Cui (Max Planck Institute for Plant Breeding Research, Germany) and recombined into pAM‐PAT vector (35S promoter) with the Gateway LR clonase (Invitrogen) to obtain the pAM‐PAT_MYC2 vector.
EDS5 promoter activity assays were performed by transient expression in Arabidopsis Col‐0 protoplasts as described previously 62. Protoplasts were transfected with pBI221_pEDS5::LUC or pBI221_pEDS5w/oGbox::LUC in the presence or absence of pAM‐PAT_MYC2. The pPTRL plasmid 63 was included for normalization of transformation efficiency, which expresses Renilla luciferase under the 35S promoter. Nineteen hours post‐transfection, protoplasts were harvested and luciferase assay was performed by Dual‐Luciferase reporter assay system (Promega) and Centro LB 960 Microplate Luminometer (Berthold Technologies).
Phylogenetic analysis
The whole protein sequences of A. thaliana, A. lyrata, C. rubella, C. grandiflora, E. salsugineum, B. rapa, tomato, and rice were retrieved from Phytozome 64 and used for identification of putative orthologous groups using the OrthoMCL program 65. The proteins belonging to the same group as A. thaliana EDS5 were aligned using MUSCLE 66. A maximum likelihood phylogenetic tree was constructed using the MEGA6 software 67. To visualize conservation of G boxes, 500 bp upstream of the transcription start sites and 5′‐UTRs of the Brassicaceae EDS5 were retrieved from Phytozome and aligned using MUSCLE.
Accession numbers
The accession numbers for the genes discussed in this article are as follows: AtActin2 (At3g18780), AtDDE2 (AT5G42650), AtCOI1 (AT2G39940), AtMYC2 (AT1G32640), AtMYC3 (AT5G46760), AtMYC4 (At4G17880), AtEDS5 (AT4G39030), AtPAD4 (AT3G52430), AtSID2 (At1g74710), AtPR1 (At2G14610), AlActin2 (342019), AlEDS5 (490671), AlPAD4 (938122), EsActin2 (Thhalv10020949m), EsEDS5 (Thhalv10024859m), EsPAD4 (Thhalv10011112m), CrActin2 (Carubv10013961m), CrEDS5 (Carubv10004548m), CrPAD4 (Carubv10016970m and Carubv10016967m), and CrVSP2(Carubv10001708 m).
Author contributions
AM and KT conceived and designed the experiments. AM, TN, MCS‐R, TMW, SA, and KT performed the experiments. AM and KT analyzed and discussed the data. DB contributed to generation of the plant materials. AM and KT wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Dr. Kei Hiruma (Nara Institute for Science and Technology) for the myc2/jin‐9 mutant, Dr. Philippe Reymond (University of Lausanne) for the myc2 myc3 myc4 mutant, Dr. Hironaka Tsukagoshi (Meijo University) for the p35S::MYC2‐GFP line, Dr. Haitao Cui (MPIPZ) for pENTR_MYC2, Dr. Nobutaka Mitsuda (AIST) for pPTRL, Dr. Thomas Griebel (MPIPZ) for help in SA measurement and ChIP experiments, Drs. Rainer Birkenbihl (MPIPZ) and Yasuomi Tada (Nagoya University) for help in ChIP experiments, and Drs. Fumiaki Katagiri and Jane Glazebrook (University of Minnesota) for critical reading of the manuscript. This work was supported by the Max Planck Society and Deutsche Forschungsgemeinschaft grant SFB670 (KT), and a Grant‐in‐Aid for the Japanese Society for the Promotion of Science (JSPS) Fellows (AM). AM was a recipient of a Postdoctoral Fellowship for Research Abroad from JSPS.
EMBO Reports (2017) 18: 464–476
References
- 1. Shoval O, Alon U (2010) SnapShot: network motifs. Cell 143: 326.e1 [DOI] [PubMed] [Google Scholar]
- 2. Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- 3. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- 4. Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- 5. Macho Alberto P, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- 6. Heil M, Baldwin IT (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- 7. Alcázar R, Reymond M, Schmitz G, de Meaux J (2011) Genetic and evolutionary perspectives on the interplay between plant immunity and development. Curr Opin Plant Biol 14: 378–384 [DOI] [PubMed] [Google Scholar]
- 8. Huot B, Yao J, Montgomery BL, He SY (2014) Growth‐defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7: 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smakowska E, Kong J, Busch W, Belkhadir Y (2016) Organ‐specific regulation of growth‐defense tradeoffs by plants. Curr Opin Plant Biol 29: 129–137 [DOI] [PubMed] [Google Scholar]
- 10. Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dou D, Zhou J‐M (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- 12. Hua J (2013) Modulation of plant immunity by light, circadian rhythm, and temperature. Curr Opin Plant Biol 16: 406–413 [DOI] [PubMed] [Google Scholar]
- 13. Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- 14. Kim Y, Tsuda K, Igarashi D, Hillmer Rachel A, Sakakibara H, Myers Chad L, Katagiri F (2014) Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 15: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doares SH, Syrovets T, Weiler EW, Ryan CA (1995) Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc Natl Acad Sci USA 92: 4095–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Malek B, van der Graaff E, Schneitz K, Keller B (2002) The Arabidopsis male‐sterile mutant dde2‐2 is defective in the allene oxide synthase gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- 17. Staswick PE (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole‐3‐acetic acids in an assay for adenylation. Plant Cell Online 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Staswick PE (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis . Plant Cell Online 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie DX, Feys BF, James S, Nieto‐Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate‐regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- 20. Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheard LB, Tan X, Mao H, Withers J, Ben‐Nissan G, Hinds TR, Kobayashi Y, Hsu F‐F, Sharon M, Browse J et al (2010) Jasmonate perception by inositol‐phosphate‐potentiated COI1–JAZ co‐receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song S, Qi T, Wasternack C, Xie D (2014) Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol 21: 112–119 [DOI] [PubMed] [Google Scholar]
- 23. Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seyfferth C, Tsuda K (2014) Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front Plant Sci 5: 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F (2008) Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- 26. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- 27. Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase‐like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nawrath C (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in Arabidopsis, is a member of the mate transporter family. Plant Cell Online 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nawrath C, Metraux JP (1999) Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serrano M, Wang B, Aryal B, Garcion C, Abou‐Mansour E, Heck S, Geisler M, Mauch F, Nawrath C, Metraux JP (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion‐like transporter EDS5. Plant Physiol 162: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vlot AC, Dempsey DMA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- 32. Zhang YL, Goritschnig S, Dong XN, Li X (2003) A gain‐of‐function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1‐1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mauch F, Mauch‐Mani B, Gaille C, Kull B, Haas D, Reimmann C (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25: 67–77 [DOI] [PubMed] [Google Scholar]
- 34. Saleh A, Withers J, Mohan R, Marques J, Gu YN, Yan SP, Zavaliev R, Nomoto M, Tada Y, Dong XN (2015) Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pieterse CMJ, Van der Does D, Zamioudis C, Leon‐Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- 36. Mine A, Sato M, Tsuda K (2014) Toward a systems understanding of plant‐microbe interactions. Front Plant Sci 5: 423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mur LAJ (2005) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana . PLoS Genet 9: e1004015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- 41. Abe H, YamaguchiShinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought‐ and abscisic acid‐regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- 43. Fernández‐Calvo P, Chini A, Fernández‐Barbero G, Chico J‐M, Gimenez‐Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco‐Zorrilla JM et al (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alcázar R, Parker JE (2011) The impact of temperature on balancing immune responsiveness and growth in Arabidopsis . Trends Plant Sci 16: 666–675 [DOI] [PubMed] [Google Scholar]
- 45. Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv tomato. Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- 46. Koenig D, Weigel D (2015) Beyond the thale: comparative genomics and genetics of Arabidopsis relatives. Nat Rev Genet 16: 285–298 [DOI] [PubMed] [Google Scholar]
- 47. Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker Jane E (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630 [DOI] [PubMed] [Google Scholar]
- 48. Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- 49. Mangan S, Alon U (2003) Structure and function of the feed‐forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng XY, Spivey NW, Zeng WQ, Liu PP, Fu ZQ, Klessig DF, He SY, Dong XN (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J, Shine MB, Gao QM, Navarre D, Jiang W, Liu C, Chen Q, Hu G, Kachroo A (2014) Enhanced disease susceptibility 1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol 165: 1269–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhattacharjee S, Halane MK, Kim SH, Gassmann W (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- 53. Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo Y‐L, Steige K, Platts AE, Escobar JS, Newman LK et al (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45: 831–835 [DOI] [PubMed] [Google Scholar]
- 54. Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson JP (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis . Plant Cell Online 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- 57. Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X et al (2009) Ethylene insensitive3 and ethylene insensitive3‐like1 repress salicylic acid induction deficient2 expression to negatively regulate plant innate immunity in Arabidopsis . Plant Cell Online 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guo HS, Fei JF, Xie Q, Chua NH (2003) A chemical‐regulated inducible RNAi system in plants. Plant J 34: 383–392 [DOI] [PubMed] [Google Scholar]
- 59. Shimada TL, Shimada T, Hara‐Nishimura I (2010) A rapid and non‐destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana . Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- 60. Villajuana‐Bonequi M, Elrouby N, Nordström K, Griebel T, Bachmair A, Coupland G (2014) Elevated salicylic acid levels conferred by increased expression of isochorismate synthase 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J 79: 206–219 [DOI] [PubMed] [Google Scholar]
- 61. Yamaguchi N, Winter CM, Wu M‐F, Kwon CS, William DA, Wagner D (2014) Protocol: chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoo S‐D, Cho Y‐H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- 63. Ohta M, Ohme‐Takagi M, Shinshi H (2000) Three ethylene‐responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22: 29–38 [DOI] [PubMed] [Google Scholar]
- 64. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N et al (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


