ABSTRACT
The airway mucosa expresses protective interferon (IFN) and inflammatory cytokines in response to respiratory syncytial virus (RSV) infection. In this study, we examine the role of bromodomain containing 4 (BRD4) in mediating this innate immune response in human small airway epithelial cells. We observe that RSV induces BRD4 to complex with NF-κB/RelA. BRD4 is functionally required for expression of the NF-κB-dependent inflammatory gene regulatory network (GRN), including the IFN response factor 1 (IRF1) and IRF7, which mediate a cross talk pathway for RIG-I upregulation. Mechanistically, BRD4 is required for cyclin-dependent kinase 9 (CDK9) recruitment and phospho-Ser 2 carboxy-terminal domain (CTD) RNA polymerase (Pol) II formation on the promoters of IRF1, IRF7, and RIG-I, producing their enhanced expression by transcriptional elongation. We also find that BRD4 independently regulates CDK9/phospho-Ser 2 CTD RNA Pol II recruitment to the IRF3-dependent IFN-stimulated genes (ISGs). In vivo, poly(I·C)-induced neutrophilia and mucosal chemokine production are blocked by a small-molecule BRD4 bromodomain inhibitor. Similarly, BRD4 inhibition reduces RSV-induced neutrophilia, mucosal CXC chemokine expression, activation of the IRF7-RIG-I autoamplification loop, mucosal IFN expression, and airway obstruction. RSV infection activates BRD4 acetyltransferase activity on histone H3 Lys (K) 122, demonstrating that RSV infection activates BRD4 in vivo. These data validate BRD4 as a major effector of RSV-induced inflammation and disease. BRD4 is required for coupling NF-κB to expression of inflammatory genes and the IRF-RIG-I autoamplification pathway and independently facilitates antiviral ISG expression. BRD4 inhibition may be a strategy to reduce exuberant virus-induced mucosal airway inflammation.
IMPORTANCE In the United States, 2.1 million children annually require medical attention for RSV infections. A first line of defense is the expression of the innate gene network by infected epithelial cells. Expression of the innate response requires the recruitment of transcriptional elongation factors to rapidly induce innate response genes through an unknown mechanism. We discovered that RSV infection induces a complex of bromodomain containing 4 (BRD4) with NF-κB and cyclin-dependent kinase 9 (CDK9). BRD4 is required for stable CDK9 binding, phospho-Ser 2 RNA Pol II formation, and histone acetyltransferase activity. Inhibition of BRD4 blocks Toll-like receptor 3 (TLR3)-dependent neutrophilia and RSV-induced inflammation, demonstrating its importance in the mucosal innate response in vivo. Our study shows that BRD4 plays a central role in inflammation and activation of the IRF7-RIG-I amplification loop vital for mucosal interferon expression. BRD4 inhibition may be a strategy for modulating exuberant mucosal airway inflammation.
KEYWORDS: BRD4, CDK9, transcriptional elongation, RSV, Pol II CTD, phosphorylation, acetylation
INTRODUCTION
The airway epithelium is a highly dynamic barrier that initiates innate and inflammatory responses to aerobiological exposures. Epidemiological studies have shown that RNA viral infections are linked to exacerbations of airway dysfunction (1, 2). In particular, the paramyxovirus respiratory syncytial virus (RSV) is a major cause of acute pulmonary disease in children, adults with chronic airway disease, and the hospitalized elderly. Without an effective vaccine, an estimated 2.1 million children under 5 years of age require medical attention in the United States annually (3). In predisposed and immunologically naive children, RSV can produce lower respiratory tract infection (LRTI), a disease complicated by postinfectious sequelae, including recurrent episodic wheezing and allergic sensitization (4, 5). Along with viral replication, the host signaling response to RSV infection is thought to play a significant role in disease pathogenesis.
RSV targets and productively replicates in the airway epithelium. Upon production of RSV-associated molecular patterns, including double-stranded RNA (dsRNA) and 5′ phosphorylated RNA, cytoplasmic germ line-encoded pattern recognition receptors (PRRs) trigger the innate response (6, 7). This response integrates reactive oxygen stress generation with a ubiquitin ligase-IκB kinase (IKK) pathway, with the two cooperating to produce posttranslational activation of NF-κB and interferon (IFN) regulatory factor 3 (IRF3) transcription factors (8, 9). Computational studies of the integrated innate response to viral patterns have identified that NF-κB-induced IRF signaling forms a positive cross talk pathway converging on the retinoic acid-inducible gene (RIG-I) PRR (10, 11). Mechanistically, activated NF-κB- and inducible IRF isoforms (IRF1 and IRF7) bind and synergistically activate RIG-I mRNA expression at the transcriptional level. RIG-I upregulation further promotes IRF1 and IRF7 expression, enhancing type I IFN production (12), resulting in a positive autoamplification loop. NF-κB also directly controls the expression of cytokine networks in the epithelium, including CC chemokines (Exodus-1, RANTES, and MIP-1α and -1β), CXC chemokines (I-TAC; GRO-α, -β, and -γ; and interleukin-8 [IL-8]), CX3C chemokine, defensins, thymic stromal lymphopoietin, and others (13, 14). In cooperation with IRF, NF-κB regulates type I and III IFNs. These signals cooperate with IRF-dependent IFN-stimulated genes (ISGs) to activate airway dendritic cells (DCs) to express the CD-40, -80, and -86 costimulatory molecules, activating their migration to draining pulmonary lymph nodes (DLNs). In the DLNs, activated DCs stimulate cytotoxic T lymphocytes, including NK and CD4/8 memory T cells that serve as second-line protective innate immunity. In this way, the epithelium plays a central role in the induction of virus-induced inflammation and the downstream adaptive immune response (15). Importantly, although the innate response is critical for limiting pathogen spread, an exuberant innate response is linked to disease pathogenesis. In this regard, our earlier studies showed that inhibition of the NF-κB arm of the mucosal innate response reduces weight loss, chemokine production, and airway inflammation in a mouse model of RSV infection (16). Understanding the molecular control of inflammatory and antiviral genes therefore has therapeutic significance.
Because rapid activation of NF-κB and IRF signaling is vital for host protection from viral pathogens, innate response genes are highly inducible, immediate early genes maintained in an “open” chromatin configuration engaged by a “paused” RNA polymerase. Upon innate pathway activation, the positive transcription elongation factor (P-TEFb) complex is rapidly recruited to the promoters of both NF-κB- and IRF-dependent innate immune response (IIR) genes (17–19). Kinases within the P-TEFb complex, including cyclin-dependent kinase 9 (CDK9), catalyze phosphorylation at Ser 2 of the heptad repeats of the carboxy-terminal domain (CTD) of the large subunit of RNA polymerase (Pol) II, RPB1, as well as negative elongation factors. Phospho-Ser 2 CTD RNA Pol II then enters a processive mode, producing full-length, fully spliced mRNAs (18–20). Through this mechanism, P-TEFb plays a central role in activating mRNA expression in both the NF-κB and IRF3 arms of the innate pathway (18, 19, 21).
We previously observed that innate pathway activation dynamically transitions P-TEFb into an activated state by dissociating it from inhibitory 7SK small nuclear RNA (19) and forming a distinct protein interaction network involving over 407 proteins (22). These studies showed that innate activation reprogrammed the CDK9 complex to associate with RNA/DNA helicases, ribosomal/translation elongation subunits, and unfolded protein response proteins. The innate response-inducible CDK9-interacting proteins produce a functional complex capable of coordinated mRNA expression, splicing, export, and translation, enabling rapid transcriptional responses in the innate response (22, 23). Analyses integrating the inducible protein network with gene expression patterns indicated that the activated P-TEFb complex is reprogrammed from the control of housekeeping gene regulatory networks (GRNs) to those controlling apoptosis, cell cycle processes, DNA repair, cytoskeleton organization, and stress and immune responses (22).
The mechanism by which CDK9 is recruited to innate genes is not fully understood. Activated CDK9 binds to sequence-specific DNA binding factors, including NF-κB/RelA (21, 24) and STAT3 (25), responsible for recruiting CDK9 to inducible gene promoters. Specifically, we found that CDK9 targets to inflammatory GRNs in a RelA-dependent manner (19, 21, 24). In this study, we further extend our study of the CDK9-interacting complex. Here, we observe that RSV infection induces BRD4 expression, interaction with CDK9 and NF-κB/RelA, and its recruitment to immediate early innate genes. Using short interfering RNA (siRNA)-mediated knockdown, we observe that BRD4 is functionally required for the recruitment and stabilization of CDK9 and phospho-Ser 2 CTD RNA Pol II to NF-κB-dependent inflammatory genes, including the IRF1/7-RIG-I amplification loop and downstream IRF-dependent IFN-stimulated genes (ISGs). A small-molecule inhibitor of histone acetyllysine (Lys) binding pocket blocks virus-induced BRD4·CDK9 binding and phospho-Ser 2 CTD Pol II formation on the IRF-RIG-I cross talk pathway and its downstream innate GRN. We observe that disruption of BRD4 bromodomain (BD) interactions blocks both Toll-like receptor 3 (TLR3)-initiated and RIG-I-driven airway inflammation in vivo. We conclude that the action of the BRD4 epigenetic reader is essential for activity of CDK9, formation of phospho-Ser 2 CTD Pol II, and histone acetyltransferase activity mediating transcriptional elongation, inflammation, and antiviral activity.
RESULTS
RSV induces changes in the BRD4 abundance associated with NF-κB activation.
We have established a model of RSV infection using human small airway epithelial cells (hSAECs) relevant for the study of innate responses in RSV-induced LRTIs. hSAECs are immortalized with human telomerase and CDK4 expression; these maintain a stable epithelial morphology, express differentiated cytokeratin (CK) isoforms, form pseudostratified columnar epithelial structures, undergo cell cycle arrest, and transition into mesenchymal cells like primary bronchiolar cells (26–28). Moreover, hSAECs have similar genomic signatures and exhibit similar RSV-induced innate responses as those of primary SAECs (22, 27, 29). Upon RSV adsorption, the initiation of RSV transcription triggers the innate response. Our previous work has shown that this response is mediated by RSV genomic RNA binding to the cytoplasmic RIG-I PRR, resulting in coupled NF-κB translocation and transcriptional activation by Ser 276 phosphorylation (19, 30).
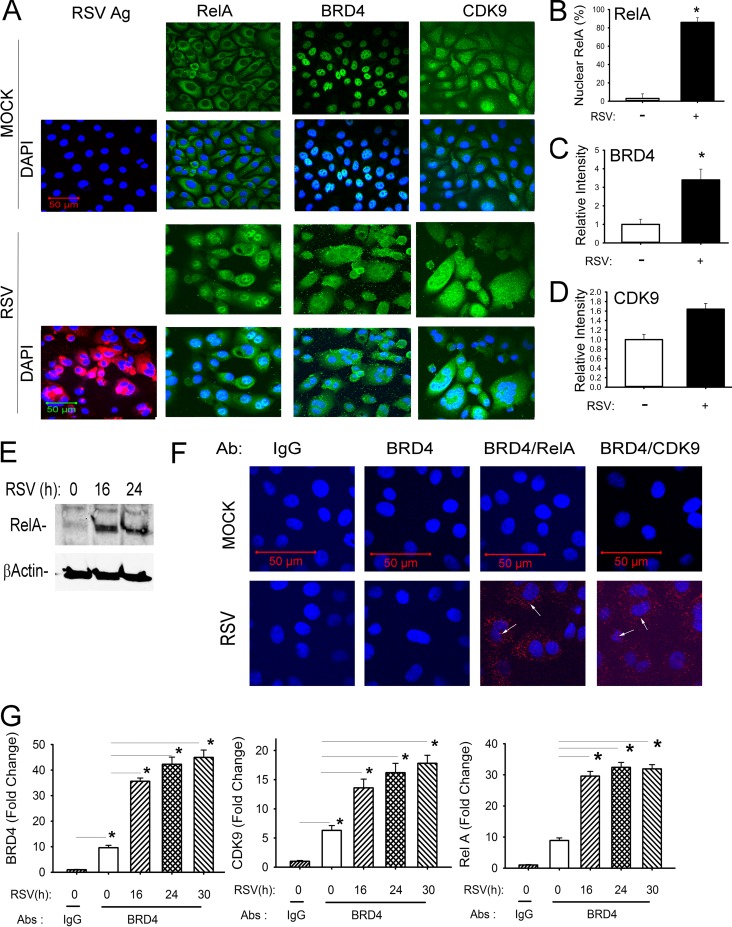
Using this model, we examined the effect of RSV replication on the abundance and subcellular distribution of CDK9 and BRD4 by confocal immunofluorescence microscopy. We observed that under these conditions, RSV infection was uniform throughout the cell population (Fig. 1A). In uninfected cells, BRD4 and CDK9 are primarily nuclear proteins, whereas NF-κB/RelA is cytoplasmic. In response to RSV infection, RelA translocates into the nucleus (Fig. 1A and B). Additionally, RSV infection induces a 3.5-fold increase in BRD4 abundance (Fig. 1C), associated with an increase in BRD4 signal in both the nuclear and cytoplasmic compartments (Fig. 1A). A less dramatic 1.6-fold increase in CDK9 expression is seen upon RSV infection. In RSV-infected cells, CDK9 is also in both cytoplasmic and nuclear compartments (Fig. 1A and D).
FIG 1.
RSV induces activation of the NF-κB/RelA-BRD4 complex. (A) Indirect immunofluorescence assays of RSV-infected hSAECs. hSAECs were exposed to sucrose (mock) or RSV (MOI of 1.0) for 24 h. After fixation, cells were stained with anti-RSV, -RelA, -BRD4, or -CDK9 Abs as indicated. Secondary detection was performed with Alexa 568 (red)-conjugated goat anti-rabbit IgG. Nuclei were counterstained with DAPI (blue). For each antigen (Ag), Alexa staining is shown in the top row; the merged DAPI image is shown in the bottom row. Images were acquired by confocal microscopy at ×63 magnification. (B) Quantification of RelA. Nuclear RelA was scored in 5 independent fields. Shown is percentage of RelA-positive cells (n = 100 for each). (C) Quantification of BRD4. Relative fluorescence intensity was quantified in 5 separate fields. (D) Quantification of CDK9. Quantification of CDK9 relative fluorescence intensity is presented as in panel C. (E) Activation of NF-κB/RelA in response to RSV infection. Nuclear extracts from uninfected (time zero) or RSV-infected (16 and 24 h p.i.) cells were prepared and analyzed by Western immunoblotting. (Top panel) Staining with anti-RelA Ab. (Bottom panel) β-Actin staining was used as an internal control. These data confirm nuclear translocation of RelA (A). (F) In situ proximal ligation assay of mock- or RSV-infected hSAECs (MOI of 1.0) for 24 h. Cells were fixed and subjected to PLA using IgG, anti-mouse BRD4 (anti-mBRD4) Ab alone, anti-mBRD4 plus anti-rabbit RelA, or anti-mBRD4 plus anti-CDK9 as indicated. Nuclei are counterstained in DAPI (blue); interacting proteins appear as red foci in both cytoplasm and nucleus (arrows). Images were acquired by confocal microscopy at ×100 magnification. (G) IP-SID-SRM-MS of BRD4 complex from control and RSV-infected hSAECs. Whole-cell lysates were immunoprecipitated using IgG or anti-BRD4 Abs. (Left panel) BRD4 abundance was detected by monitoring precursor-product transitions of 3 proteotypic peptides. Shown is the fold change in BRD4 signal abundance. (Middle panel) CDK9 abundance in BRD4 complexes was measured by SID-SRM-MS. (Right panel) RelA abundance. *, P < 0.05.
To confirm NF-κB/RelA cytoplasmic-nuclear translation, Western blot analysis shows enhanced RelA abundance in the nuclear extracts, indicating cytoplasmic-to-nuclear translocation (Fig. 1E), in a manner consistent with earlier reports (18, 30–32). To examine whether RSV-activated NF-κB/RelA associated with BRD4 or CDK9, we performed in situ proximity ligation assays (PLAs). In this assay, control or RSV-infected hSAECs are fixed and stained with anti-mouse BRD4 (anti-mBRD4) antibody (Ab) alone or with anti-rabbit RelA (anti-rRelA) or rCDK9 Abs. The slides are then stained with species-specific anti-mouse or -rabbit IgG coupled to complementary oligonucleotides. Molecular interactions between the two primary antibodies are detected after enzymatic ligation of coupled oligonucleotides and PCR amplification and appear as fluorescent foci in confocal microscopy. Controls performed in the absence of primary antibody or the presence of anti-mBRD4 Ab show only that no signal is generated (Fig. 1F). Similarly, in uninfected hSAECs, few BRD4-CDK9 interactions (2 ± 3 foci/cell) are detected (Fig. 1F, top). In contrast, upon RSV infection, numerous BRD4-CDK9-interacting foci (40 ± 15 foci/cell) are observed in both the cytoplasmic and nuclear compartments, indicating that RSV induces association of BRD4-CDK9 (Fig. 1F, bottom). A similar pattern of inducible association is observed for BRD4-RelA interactions (Fig. 1F, bottom). In uninfected cells, 2 ± 3 foci/cell of BRD4-RelA complexes are seen versus the 100 ± 20 foci/cell in RSV-infected cells. These data indicated to us that RSV replication induces NF-κB activation and formation of BRD4-RelA and BRD4-CDK9 complexes in situ.
To quantify changes in BRD4 abundance and to confirm that RSV induces BRD4 complex formation with NF-κB and CDK9, we performed quantitative selected reaction monitoring (SRM) mass spectrometry (MS) measurements for transcription factor enrichment after BRD4 immunoprecipitation (IP). SRM-MS assays specifically monitor the abundance of proteotypic peptides specific for a target protein using stable isotope dilution (SID) enabled by high-resolution mass spectrometers and are superior to Western blot assays in quantification and analyte specificity (33). SID-SRM-MS assays developed for BRD4, CDK9, and RelA were applied to BRD4 complexes immunoprecipitated from a time course of RSV-infected cells (22). In control hSAECs, we observed significant BRD4 signals in IPs produced by anti-BRD4 Ab versus that of IgG, indicating IP specificity (Fig. 1G, left). We observed a significant 3.5-fold increase in BRD4 abundance in a time-dependent manner in RSV-infected cells relative to that of uninfected hSAECs, peaking at 4.5-fold 30 h postinfection (Fig. 1G, left). Similarly, CDK9 was enriched 7-fold relative to IgG in the BRD4 IPs from control cells (Fig. 1G, middle). A similar 4-fold time-dependent increase in CDK9 in the BRD4 complex was observed during the time course of RSV infection (Fig. 1G, center). A parallel analysis was performed for RelA association. In a pattern similar to that of CDK9, RelA was detectable in the BRD4 complex in unstimulated cells (Fig. 1G, right). RSV infection induced an ∼4-fold increase in RelA abundance in the BRD4 complex. Together, we interpret these data as showing that the abundance of BRD4-CDK9 and BRD4-RelA complexes was increased in response to RSV infection and that this increase was independent of changes in subunit stoichiometry.
BRD4 is functionally required for RSV-mediated induction of NF-κB-dependent cytokines.
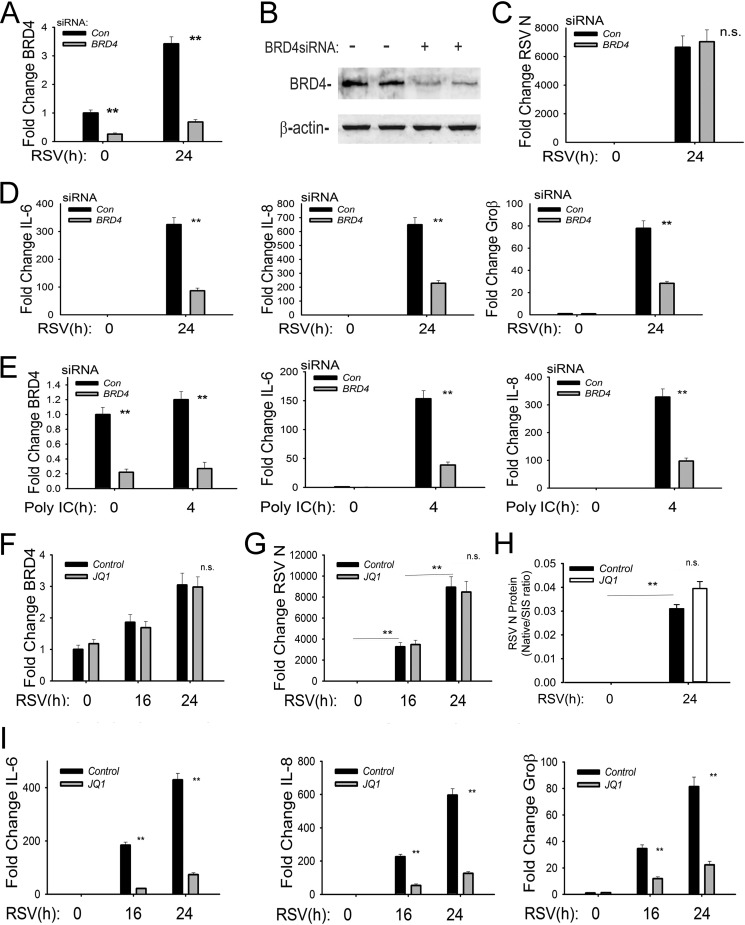
To determine if BRD4 is functionally required for NF-κB-dependent inflammatory cytokine expression, we examined the effect of RSV infection in the absence or presence of BRD4 silencing using siRNA-mediated transfection. Relative to control cells, cells transfected with BRD4-selective siRNA showed an 80% inhibition of steady-state BRD4 mRNA levels (Fig. 2A). RSV induced a 3.4-fold increase in BRD4 mRNA in RSV-infected cells, consistent with its enhanced protein expression (compare with Fig. 1A), further confirming the observation that BRD4 expression is activated by the innate pathway. Reduction in BRD4 protein expression by siRNA-mediated silencing was further confirmed by Western blotting. In untransfected cells, BRD4 is an abundant nuclear protein that migrates at an apparent molecular mass of 150 kDa, corresponding to the long isoform of BRD4 (BRD4L) (Fig. 2B). In BRD4 siRNA transfectants, BRD4L steady-state levels were reduced by 80% (Fig. 2B). To examine whether BRD4 knockdown affected RSV transcription, we separately measured expression of RSV N mRNA; here, the 6,500-fold inductions of RSV N mRNA were similar in control and BRD4 knockdowns (Fig. 2C). Interpreting this finding together with our previous findings that CDK9 inhibition has no effect on RSV replication (19), we conclude that SAECs express BRD4L and that RSV transcription is independent of the transcriptional elongation complex.
FIG 2.
Functional role of BRD4 in RSV-induced NF-κB-dependent gene expression. (A) siRNA-mediated silencing of BRD4. BRD4 mRNA abundance was measured in hSAECs transfected with 100 nM scrambled siRNA (control) or BRD4-specific siRNA. Forty-eight hours later, cells were infected with RSV (MOI of 1.0). BRD4 mRNA abundance was measured by Q-RT-PCR. Shown is BRD4 abundance normalized to cyclophilin mRNA in each sample expressed as fold change relative to control hSAECs. **, P < 0.01 in comparing silenced versus control siRNA-treated cells. (B) BRD4 protein depletion. Whole-cell extracts were prepared from control or BRD4 siRNA-transfected cells as described above. Two replicates of each treatment condition were analyzed. BRD4 abundance was determined using anti-BRD4 Ab in a Western blotting assay. β-Actin was stained as a loading control. The 150-kDa BRD4 isoform was reduced to less than 50% of that of control cells. (C) Effect of BRD4 silencing on RSV transcription. Total cellular RNA from the experiment shown in panel A was analyzed for RSV N transcript by Q-RT-PCR. Shown are changes in RSV N normalized to cyclophilin mRNA in each sample expressed as fold change relative to control hSAECs. n.s., not significant. (D) Q-RT-PCR for NF-κB-dependent IL-6, IL-8, and Groβ genes normalized to cyclophilin and expressed as fold change over uninfected control siRNA transfectants. **, P < 0.01 compared to control siRNA treatment. (E) Q-RT-PCR for BRD4, IL-6, and IL-8 mRNA in response to the TLR3 agonist poly(I·C). Control or BRD4 siRNA-transfected cells were stimulated with poly(I·C) for 4 h. Fold change in each mRNA species is shown. **, P < 0.01 compared to control siRNA treatment. (F) Effect of BRD4 inhibitor (JQ1) on BRD4 expression. Control or JQ1-treated cells were mock or RSV infected (MOI of 1). Shown are changes in BRD4 normalized to cyclophilin mRNA relative to control hSAECs. n.s., not significant. (G) Effect of JQ1 on RSV transcription. Samples from panel F were analyzed for changes in RSV N mRNA transcripts shown as fold change. **, P < 0.01 compared to control siRNA treatment. (H) Effect of JQ1 on RSV translation/secretion. Cell supernatants were measured for RSV N protein using SID-SRM. (I) Effect of BRD4 inhibitor (JQ1) on RSV-induced NF-κB-dependent genes. Shown are results of Q-RT-PCR for IL-6, IL-8, and Groβ gene expression in response to RSV infection in the absence or presence of JQ1. **, P < 0.01 compared with vehicle treatment.
We next analyzed the role of BRD4 in RSV-induced, NF-κB-dependent immediate early genes. In RSV-infected hSAECs, RSV replication induced a 320-fold change in IL-6 mRNA, a 650-fold induction of IL-8 mRNA, and a 78-fold induction of Groβ mRNA relative to mock-infected cells (Fig. 2E). In contrast, the RSV-induced IL-6 expression in BRD4-silenced cells was reduced to 25% of that seen in control siRNA-transfected cells (P < 0.01) (Fig. 2D, left panel). Statistically significant inhibition of RSV-inducible IL-8 and Groβ expression was also observed in the BRD4-silenced cells (Fig. 2D, middle and right panels). We conclude that BRD4 is functionally important for these NF-κB-dependent immediate early inflammatory GRNs downstream of the RIG-I pathway.
To investigate whether BRD4 was required for TLR3-induced GRNs, control transfected or BRD4-silenced hSAECs were stimulated with the TLR3 ligand extracellular poly(I·C) for 4 h. BRD4 knockdown was also confirmed by reduction of BRD4 mRNA; over this shorter time course, BRD4 mRNA was less strongly induced (1.2-fold) than that observed in RSV infection (Fig. 2E) and the levels of BRD4 mRNA were less than 25% of those in control siRNA-transfected cells in both control and poly(I·C)-treated cells (Fig. 2E). In response to poly(I·C) stimulation, a similar robust IL-6 induction was observed, peaking 170-fold over that of unstimulated cells. In contrast, in BRD4-silenced cells, IL-6 levels were 25% of those of control cells (P < 0.01) (Fig. 2E, middle panel). Similarly, the 300-fold induction of IL-8 was inhibited significantly by BRD4 silencing (P < 0.01) (Fig. 2E, right panel). We interpret these results to indicate that BRD4 is functionally required for expression of NF-κB-dependent immediate early genes in response to both the RIG-I and TLR3 signaling pathways.
BRD4 BD interactions are essential for inducible inflammatory gene expression.
The bromodomain (BD) of the BET family of chromatin readers is the major domain for acetyl-Lys binding to modified histones and transcription factors (34, 35). We therefore examined the effect of disrupting BD interactions in RSV- and poly(I·C)-induced NF-κB-dependent inflammatory gene expression. hSAECs were treated with a highly selective small-molecule inhibitor (JQ1) of the BET BDs, a molecule that competitively binds the acetyl-Lys recognition pockets of BRD4 BD-1 and -2 at Kd (dissociation constant) values of ∼50 and 90 nM, respectively, displacing BRD4 from chromatin (36). In contrast to the siRNA knockdown experiments, JQ1 treatment did not affect the 3-fold increase in BRD4 mRNA expression produced by RSV infection (Fig. 2F). Similarly, JQ1 treatment had no effect on the dramatic increase in RSV N transcription (Fig. 2G). We also examined the presence of RSV virions in cellular supernatants using selective SRM-MS assays developed by us previously (29). All RSV proteins were detected in the supernatant in patterns identical to that shown for RSV N protein, where cells treated with JQ1 had slightly higher levels of RSV proteins (Fig. 2H). Together, these data indicate that RSV transcription, translation, and secretion occur independently of BRD4-BD interactions.
The effect of JQ1 on NF-κB-dependent gene expression was next measured in response to RSV infection. In vehicle-treated control cells, 24 h of RSV infection induced an ∼400-fold increase of IL-6 expression; this induction was markedly blunted to less than 20% of untreated cells by JQ1 treatment (Fig. 2I, left panel). Similar patterns of induction and inhibition by JQ1 were found for IL-8 and Groβ genes (Fig. 2I, middle and right panels). Based on these data, we conclude that BD interaction is functionally important for virus-inducible expression of NF-κB-dependent GRNs.
BRD4 is required for NF-κB-mediated induction of the IRF-RIG-I amplification loop.
Activation of viral PRR signaling produces a dramatic amplification of RIG-I mRNA expression, an autoamplification loop essential for the sustained antiviral ISG expression (10, 30). Our recent computational model of the integrated innate response predicted—and experimentally verified—that RIG-I amplification is downstream of an NF-κB-IRF cross talk pathway. Here, the reactive oxygen species (ROS)-induced NF-κB phosphorylation is required for upregulation of IRF1 and IRF7 transcription factors (10, 11). In addition, we previously observed that RSV-activated RelA binds to the IRF7 promoter and is required for IRF7 expression (11); that both RelA and IRF7 bind to the RIG-I promoter, potentiating RIG-I expression (10); and that RIG-I is required for sustained type I IFN expression using siRNA-mediated silencing (30). With IFN′s secondary effect on IRF1/7 upregulation, a positive regulatory loop is established, producing an antiviral response (10, 11, 37). The functional role of NF-κB in IRF1-RIG-I amplification has not been systematically examined.
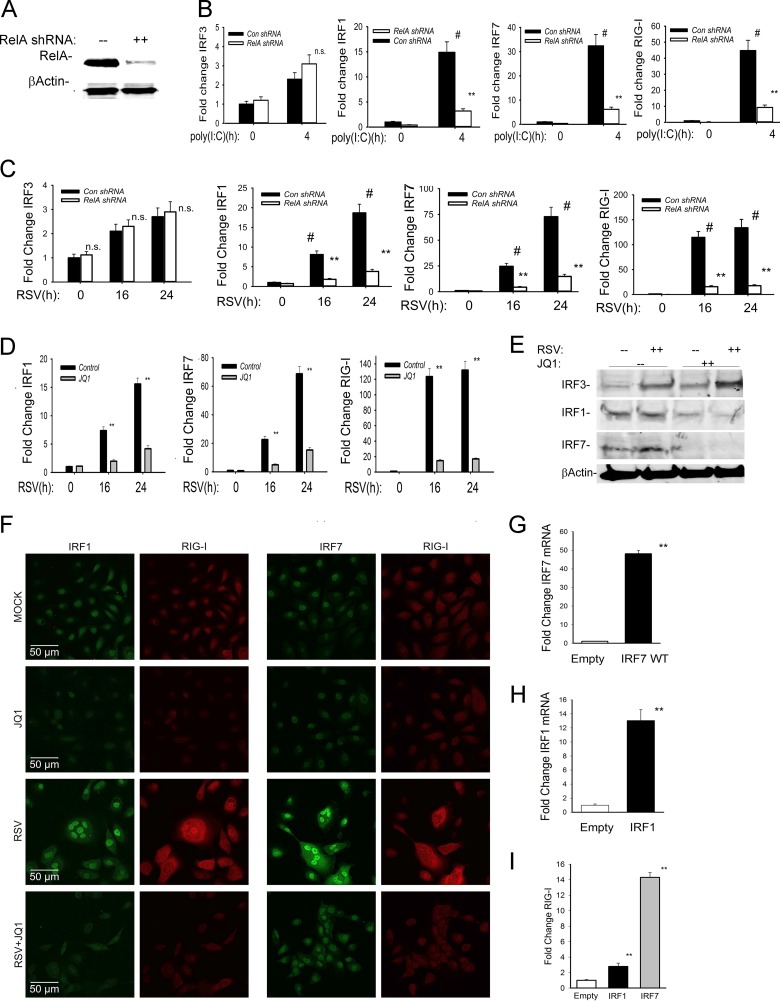
To determine whether NF-κB is required for IRF1/7 expression, we constructed hSAECs stably expressing a doxycycline (Dox)-inducible RelA short hairpin RNA (shRNA). Dox treatment produced greater than 90% inhibition of total cellular RelA protein in Western immunoblot assays (Fig. 3A). We next stimulated cells with poly(I·C) to examine the effect on IRF isoform expression. In both wild-type (WT) and RelA-silenced cells, IRF3 mRNA expression was weakly induced 2.5-fold in response to 4 h of poly(I·C) treatment (Fig. 3B, leftmost panel). In contrast, the 15-fold induction of IRF1 expression and 32-fold induction of IRF7 expression were significantly reduced in RelA-silenced cells (Fig. 3B, two middle panels). We noted that the 42-fold induction of RIG-I was similarly reduced to less than 8-fold after RelA silencing (Fig. 3B, rightmost panel), indicating that RelA was functionally required for TLR3-mediated expression of IRF1, IRF7, and RIG-I.
FIG 3.
BRD4 is required for the NF-κB-IRF-RIG-I amplification loop. (A) RelA knockdown. hSAECs were stably transfected with doxycycline-induced control (Con) or RelA-directed shRNA. Western immunoblot analysis was performed on Dox-treated control or RelA shRNA-expressing cells. (Top panel) Anti-RelA. (Bottom panel) β-Actin staining as loading control. (B) RelA dependence for IRF-RIG-I expression in response to poly(I·C). Wild-type (control shRNA) or RelA-silenced hSAECs were stimulated in the absence or presence of poly(I·C). Shown are fold changes in IRF3, IRF1, IRF7, and RIG-I gene expression by Q-RT-PCR. **, P < 0.01 compared with control shRNA-expressing cells; n.s., not significant; #, P < 0.01 versus untreated cells. (C) RelA dependence for IRF-RIG-I expression in response to RSV. WT or RelA-silenced hSAECs were mock or RSV infected for 16 or 24 h. Shown are fold changes for IRF3, IRF1, IRF7, and RIG-I gene expression by Q-RT-PCR. **, P < 0.01 compared with controls. #, P < 0.01 versus uninfected cells. (D) The BRD4 BD is required for activation of the RSV-induced IRF-RIG amplification loop. Q-RT-PCR for IRF1, IRF7, and RIG-I mRNAs in hSAECs treated with JQ1 prior to mock or RSV infection (MOI of 1) for indicated times (hours). **, P < 0.01 compared to vehicle. (E) Effect of JQ1 on IRF3 and RelA activation. hSAECs were treated with JQ1 prior to mock or RSV infection (MOI of 1) for 24 h. Shown are results of Western immunoblot analysis of nuclear extracts probed with anti-IRF3, anti-IRF1, or anti-IRF7 RelA Abs. β-Actin staining is the loading control. (F) Microscopic confocal visualization of IRF1, IRF7, and RIG-I expression in control or JQ1-treated cells subjected to mock or RSV infection (MOI of 1.0, 24 h). IgG, IRF1, and IRF7 were detected by Alexa Fluor 488 (green), and RIG-I was detected by anti-rabbit or Alexa Fluor 568 (red)-conjugated anti-goat IgG. Confocal images are shown at ×63 magnification. (G) Ectopic IRF7 expression. Q-RT-PCR for IRF7 in hSAECs transfected with empty vector or IRF7-expressing lentivirus. **, P < 0.01 compared to empty-vector-transfected cells. (H) Ectopic IRF1 expression. Q-RT-PCR for IRF1 in hSAECs transfected with empty vector or IRF1-expressing lentivirus. **, P < 0.01 compared to empty-vector-transfected cells. (I) Effect of ectopic IRF1 or IRF7 expression on RIG-I expression. hSAECs transfected with empty vector or IRF1- or IRF7-expressing lentivirus were analyzed for RIG-I expression by Q-RT-PCR. Shown is fold change in RIG-I expression normalized to cyclophilin. **, P < 0.01 compared to empty-vector transfectants.
The role of RelA in activation of the IRF1/7 and RIG-I pathway was also examined in response to RSV. In WT and RelA-silenced cells, RSV induced similar time-dependent increases in IRF3 mRNA (Fig. 3C). In contrast, 24 h of RSV infection produced an 18- and 75-fold induction of IRF1 and IRF7 mRNAs, respectively, inductions that were significantly abrogated to 4- and 15-fold, respectively, in the RelA-silenced cells (Fig. 3C, middle panels). Furthermore, in response to RSV, RIG-I was induced more highly and at an earlier time than peak IRF1/7 expression, increasing by 110-fold within 16 h of RSV infection (Fig. 3C, rightmost panel). These data further support the functional requirement of RelA for IRF1/7-RIG-I amplification mediated by the RIG-like helicase (RLH) class of PRRs.
To test whether BRD4 was functionally required for this NF-κB-IRF-RIG-I cross talk pathway, we examined the effect of JQ1 treatment on RSV-inducible IRF isoform expression. Here, replication produced a time-dependent 15-fold activation of IRF1 and 70-fold activation of IRF7 mRNAs 24 h after RSV infection (Fig. 3D). Both inductions were significantly inhibited by JQ1 treatment. Consistent with the synergistic interactions, the 120-fold induction of RIG-I expression achieved an expression plateau 16 h after RSV infection, at times preceding the peaks in IRF1 or IRF7 expression. Importantly, RIG-I amplification was significantly reduced by JQ1 treatment (Fig. 3D, right panel). To extend these studies, we examined the effect of JQ1 on RSV-induced nuclear translocation of IRF isoforms. Nuclear extracts from control or RSV-infected hSAECs in the absence or presence of JQ1 were prepared and analyzed by Western immunoblot assay. We found that RSV induced IRF3 nuclear translocation independently of BRD4 inhibition (Fig. 3E). In contrast, IRF1 and IRF7 expression was significantly reduced by JQ1 treatment, in a manner consistent with the inhibitory effects of JQ1 on IRF1 and IRF7 mRNA expression (compare Fig. 3D and E). Collectively, these data indicate that BRD4 is required for expression of the inducible IRF1/7 isoforms, isoforms which are primarily RelA dependent.
Confocal immunofluorescence assays were conducted to further explore the role of BRD4 in RSV-induced IRF and RIG-I expression. In uninfected cells, IRF1 and RIG-I expression was very low, but both proteins were markedly upregulated by RSV infection (Fig. 3F, left columns). Here, IRF1 staining was intense in the RSV-infected cells, including multinucleated forms. Importantly, in the cells with high levels of IRF1 expression, RIG-I was also highly expressed. In three independent fields counted, 100% of the IRF1-positive cells (n = 50) were also RIG-I positive. In contrast, in JQ1-treated cells, RSV infection did not induce IRF1 or RIG-I expression. Interestingly, we also noted that the amount of syncytia was significantly reduced by JQ1 treatment (see Discussion). Similar findings were observed for RSV-induced IRF7 expression and colocalization with RIG-I-expressing cells (Fig. 3F, right columns). In this setting, 100% of the IRF7-expressing cells were also RIG-I positive. The finding that RIG-I expression is upregulated only in those cells expressing IRF1 or IRF7 further suggests the functional requirement of IRF1 and IRF7 isoforms in RIG-I amplification.
To mechanistically extend the role of IRF1/7 on RIG-I amplification, we investigated whether IRF1 and IRF7 expression was sufficient for RIG-I production. Separately, IRF7 or IRF1 was stably expressed in hSAECs (Fig. 3G and H, respectively). Under these conditions, either IRF1 or IRF7 expression was sufficient to transactivate the endogenous RIG-I promoter (Fig. 3I), establishing its functional redundancy and sufficiency in RIG-I expression. Interpreting these functional data together with our previous work demonstrating RelA binding to IRF1, IRF7, and RIG-I (11) indicates that BRD4 is functionally important in mediating NF-κB/RelA activation of the IRF cross talk pathway converging on the RIG-I PRR.
Functional role of BRD4 in mucosal antiviral signaling.
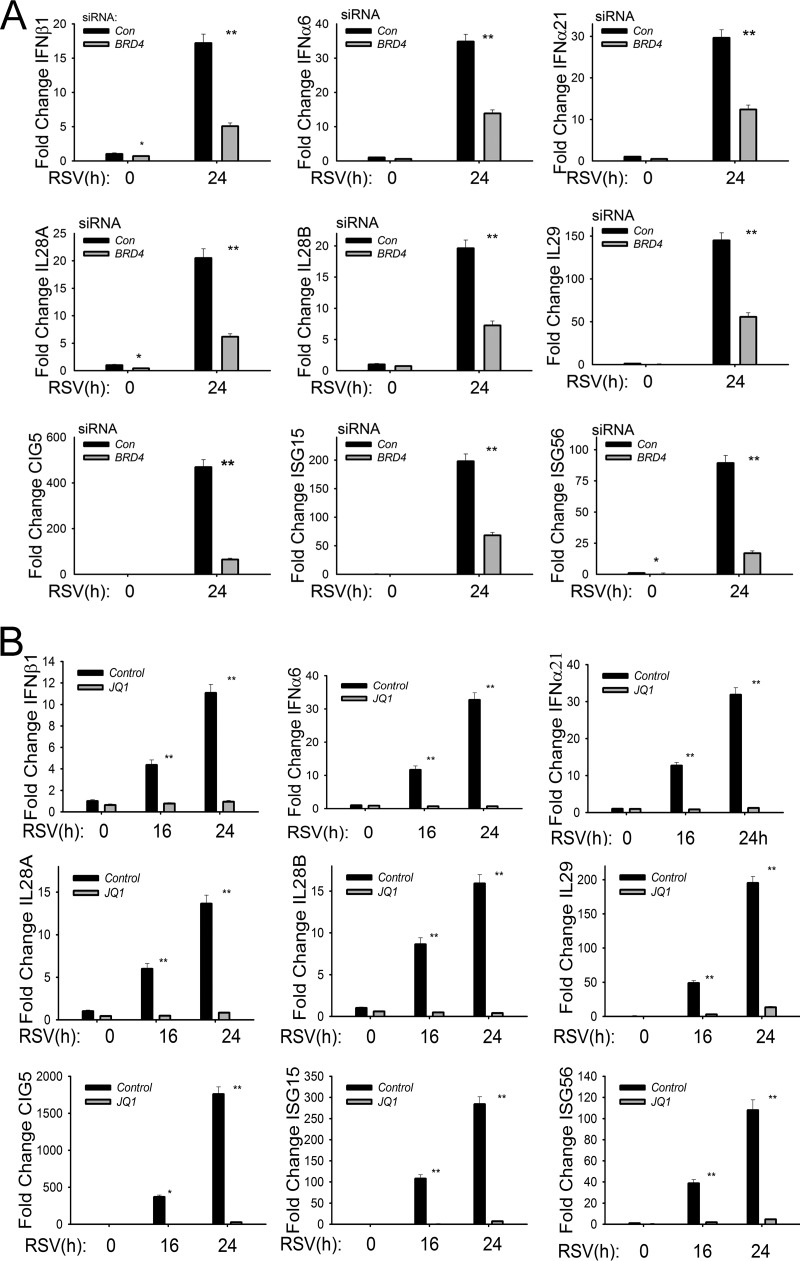
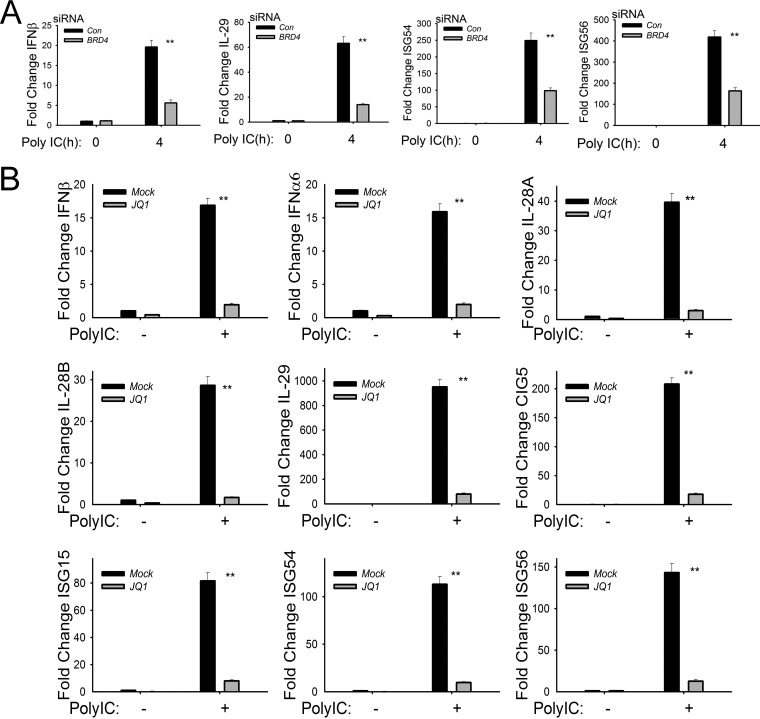
The coordinate activity of NF-κB and IRF signaling leads to the expression of 23 cytokines, chemokines, and type I and III interferons (IFNs) in RSV-infected epithelial cells (13, 14, 19, 38). Type I and III IFNs are central mediators for the mucosal antiviral response by inducing transcription of the ISG network to reduce viral replication (39, 40). Of the mucosal IFNs, type I (IFN-α/β) and type III (IL-28A, IL-28B, and IL-29) are functionally the most potent antiviral cytokines expressed by epithelial cells (38, 41, 42). To examine whether BRD4 was required for IFN production, we examined the effect of BRD4 silencing on RSV-induced IFN and ISG expression. In control siRNA-transfected cells, RSV induced IFNβ1 expression by 17-fold, which was reduced to only ∼5-fold induction in BRD4-silenced cells (P < 0.01) (Fig. 4A, top panels). Similar results were observed for the 35-fold induction of IFNα6 and 29-fold induction of IFNα21, whose expression was reduced to 12- and 11-fold, respectively, by BRD4 silencing (P < 0.01) (Fig. 4A, top panels). We also observed that BRD4 silencing reduced the highly RSV-inducible expression of the type III IFNs IL-28A and -B and IL-29, whose expression was reduced to less than 30% of that of control transfectants (Fig. 4A, middle panels). These data indicate that BRD4 is functionally required for RSV-induced type I and III IFN responses in epithelial cells.
FIG 4.
Functional requirement for BRD4 in RSV-induced type I and type III IFN and ISG expression. (A) Q-RT-PCR for type I IFNs (IFNβ, IFNα6, and IFNα21), type III IFNs (IL-28A and -B and -29), and ISGs (CIG5 and ISG-15 and -56). hSAECs were transfected with scrambled siRNA (control) or BRD4-specific siRNA. Forty-eight hours later, cells were RSV infected (MOI of 1.0). **, P < 0.01 compared with control siRNA. (B) Q-RT-PCR of control or JQ1-treated hSAECs mock or RSV infected (MOI of 1) for indicated times (hours). *, P < 0.05 compared with control siRNA; **, P < 0.01 compared with vehicle.
Previous work has shown that the ISGs are transcriptionally activated by RSV-induced translocation of cytoplasmic IRF3 that binds to highly conserved IFN-stimulated response element (ISRE) sites in the proximal ISG promoters (19). Knockdown experiments have shown that the ISGs are purely IRF3 dependent and are independent of NF-κB/RelA (10, 19). We observed that the highly RSV-inducible ISGs CIG5 and ISG-15 and -56 were also significantly reduced by BRD4 silencing (Fig. 4A, bottom panels). Together, these data indicate that BRD4 is a common mediator of both NF-κB and IRF3 ISG regulatory networks constituting the innate immune response.
BRD4 BD interactions are essential for RSV-inducible mucosal IFN responses.
In a similar approach as that used in the investigation of the NF-κB-inducible genes, we examined whether the BRD4 BD was required for the RSV-induced increase in type I and III IFNs and ISGs; all were substantially inhibited by JQ1 treatment (Fig. 4B). In this experiment, we noted that the effect of the JQ1 inhibitor was greater than that produced by the siRNA-mediated knockdown. For example, the 12-fold induction of IFNβ1 24 h after RSV infection was less than 1.5-fold in JQ1-treated cells versus a 5-fold induction observed in BRD4-silenced cells (compare Fig. 4A and B). RSV induction of the type III IFNs IL-28A/B and -29 was dramatically reduced by JQ1 treatment, as was the expression of ISGs CIG5 and ISG-15 and -56 (Fig. 4B). These differences may be due to incomplete inhibition of BRD4 expression by siRNA silencing or a possible contribution of other BET proteins, such as BRD2, in RSV-induced innate signaling.
To examine the role of BRD4 in TLR3-activated expression of the IFN-ISG response network, we analyzed IFN and ISG expression in BRD4-silenced hSAECs stimulated with poly(I·C). The 20-fold induction of IFNβ and 60-fold induction of IL-29 induced by 4 h of poly(I·C) treatment were significantly reduced to <20% of that of control siRNA-transfected cells 4 h after poly(I·C) treatment (P < 0.01) (Fig. 5A). ISG-54/ISG-56 expression was also significantly reduced in cells with BRD4 silencing (Fig. 5A). Similar results were obtained in poly(I·C)-stimulated cells in the absence or presence of JQ1, where significant reductions of type I IFNs (IFNβ and IFNα6), type III IFNs (IL-28-A/B and IL-29), and ISG-15/-54/-56 were observed by JQ1 treatment (Fig. 5B). From these experiments, we conclude that BRD4 mediates both the IRF- and NF-κB-dependent innate response network through BD interactions in the RIG-I and TLR3 signaling pathways.
FIG 5.
Functional requirement of BRD4 for poly(I·C)-induced IFN-I and -III and ISG expression. (A) Q-RT-PCR of IFNβ, IL-29, and ISG-54 and -56 in BRD4-silenced hSAECs in response to poly(I·C). **, P < 0.01 compared with control siRNA. (B) Q-RT-PCR of IFNβ, IL-29, and ISG-54 and -56 in JQ1-treated cells in response to poly(I·C). **, P < 0.01 compared with mock treatment (vehicle).
BRD4 is required for the formation of active transcriptional elongation complexes on NF-κB-dependent immediate early inflammatory genes.
Our previous work has shown that the NF-κB- and IRF3-dependent immediate early genes controlling inflammation and antiviral response in the innate response are mediated by transcriptional elongation (18, 19). This process occurs in genes in open chromatin configuration engaged with hypophosphorylated RNA Pol II. Upon cellular activation, P-TEFb recruitment results in phosphorylation of Ser 2 in the heptad repeats of the carboxy-terminal domain (CTD) of RNA polymerase II. Phospho-Ser 2 CTD RNA Pol II is able to productively elongate fully spliced innate defense genes. The functional role of BRD4 in this process has not been investigated. We hypothesized that the acetyl-Lys binding function of BRD4 would be important in CDK9 recruitment and phospho-Ser 2 CTD RNA Pol II formation.
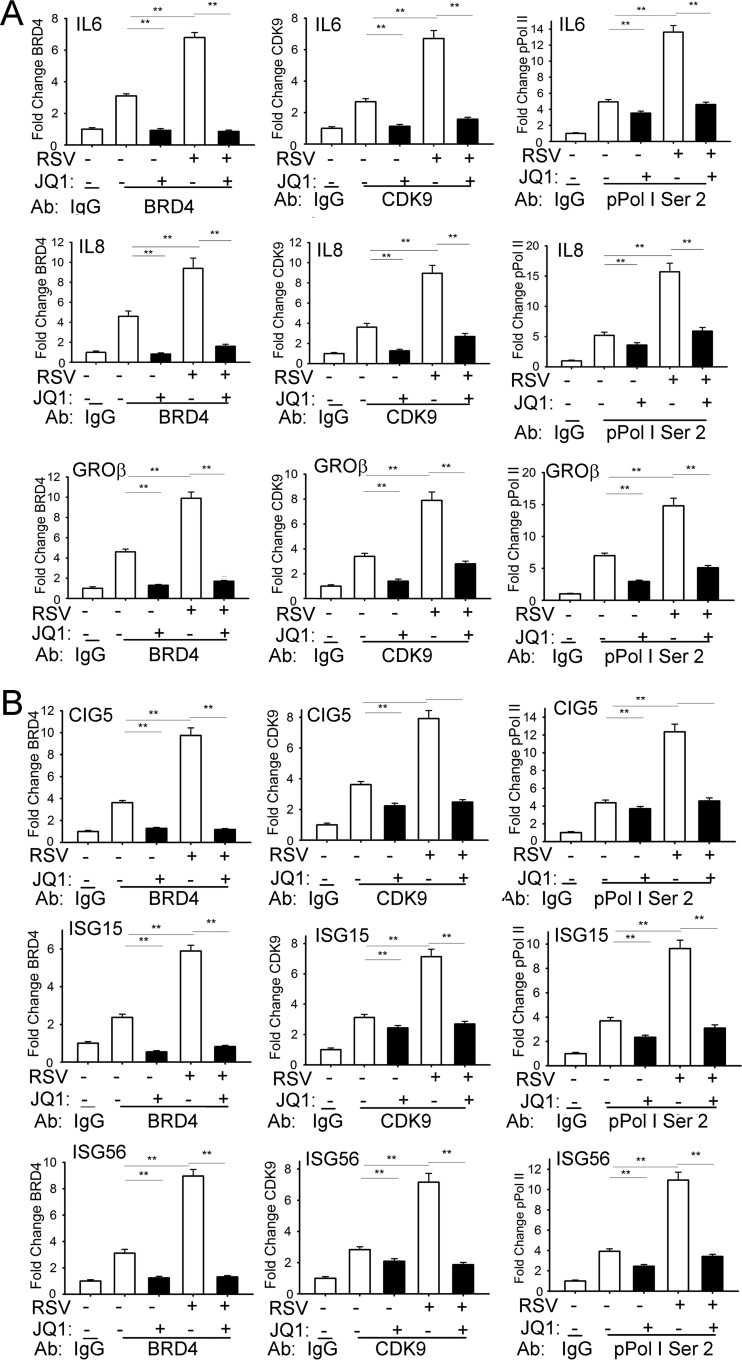
We applied a highly sensitive two-step cross-linking chromatin immunoprecipitation (XChIP) technique that we developed earlier to examine the role of BRD4 on CDK9 and phospho-Ser 2 Pol II recruitment to innate inflammatory and antiviral genes (19, 43, 44). In unstimulated cells, BRD4 is associated with the IL-6 promoter (3-fold over nonspecific IgG) and its binding is further induced 7-fold 16 h after RSV infection (Fig. 6A, left panels). JQ1 treatment completely inhibited both basal and RSV-induced BRD4 binding to IL-6, consistent with the requirement for BD acetyl-Lys binding for chromatin association (45). Examination of CDK9 association shows basal CDK9 binding that is increased ∼3-fold after RSV infection. Importantly, CDK9 binding is completely disrupted by JQ1 treatment (Fig. 6A, middle panels). Finally, the 14-fold induction of phospho-Ser 2 CTD RNA Pol II binding by RSV is reduced to basal levels by JQ1 treatment (Fig. 6A, right panels). The binding of BRD4, CDK9, and phospho-Ser 2 RNA Pol II for both the IL-8 and Groβ promoters showed similar patterns of RSV induction and JQ1 inhibition (Fig. 6A, middle and bottom rows). These data indicate that RSV-induced recruitment of CDK9 and formation of phospho-Ser 2 CTD RNA Pol II are dependent on acetyl-Lys binding activity of BRD4.
FIG 6.
BRD4 is required for CDK9 recruitment and p-Pol II activation. (A) Chromatin immunoprecipitation (XChIP) assays of NF-κB-dependent genes. hSAECs were treated with JQ1 (10 μM) prior to mock (−) or RSV (+; MOI of 1) infection for 16 h. For each gene, fold change is shown relative to nonspecific control (IgG) using Q-gPCR. **, P < 0.01 compared to JQ1 treatment or infection. (B) XChIP of ISGs from hSAECs treated with JQ1 (10 μM) prior to mock (-) or RSV (+; MOI of 1) infection for 16 h. **, P < 0.01 compared to JQ1 treatment or infection.
BRD4 forms active transcriptional elongation complexes on RSV-induced ISGs.
Finally, we used XChIP to examine the effect of disrupting the BRD4-acetyl-Lys interactions on CDK9 recruitment and phospho-Ser 2 CTD RNA Pol II formation on the CIG5 and ISG-15 and -56 promoters. Our previous work has shown that these genes are absolutely dependent on IRF3 and independent of direct NF-κB/RelA binding (10, 19). Like the binding patterns on NF-κB-dependent genes, RSV induced BRD4 recruitment to the CIG5 promoter, an effect that was blocked by JQ1 treatment (Fig. 6B). RSV-inducible CDK9 binding and phospho-Ser 2 CTD RNA Pol II formation were also significantly inhibited by JQ1 (Fig. 6B, top row). Similar patterns were observed for the ISG-15 and -56 promoters (Fig. 6B, middle and bottom rows). Together, these data suggest that the mechanism for JQ1 inhibition of the viral pattern-induced innate immune response is mediated by its inhibition of active transcriptional elongation complex formation in both NF-κB-dependent and IRF3-dependent ISGs.
BRD4 mediates poly(I·C)-induced inflammation in vivo.
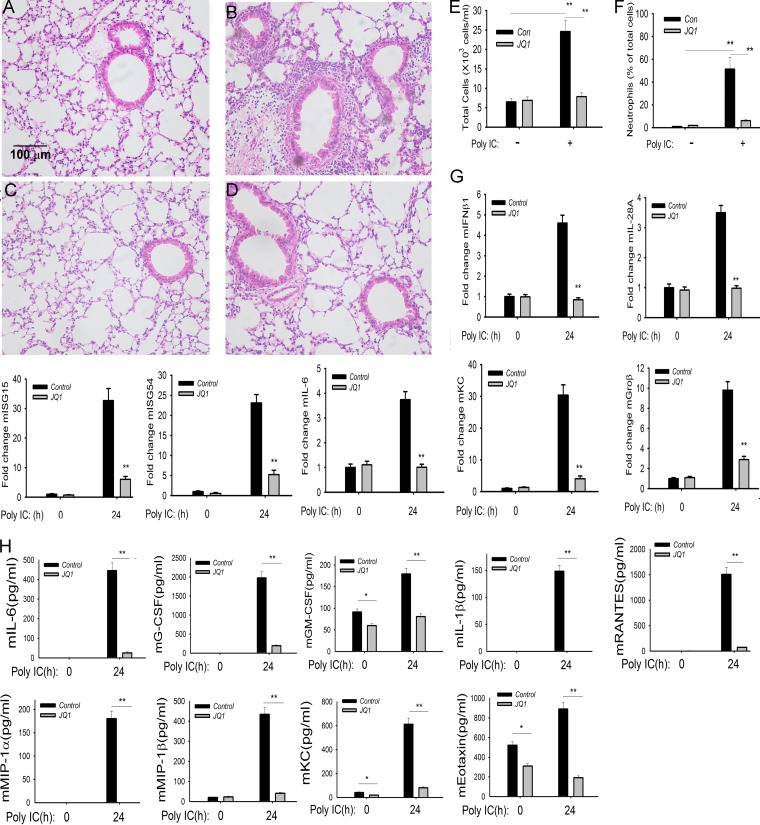
To further explore the role of BRD4 in mediating innate inflammation, we examined its role in mediating TLR3-induced inflammation in vivo. Previous work has shown that intranasal (i.n.) administration of poly(I·C) induces TLR3-dependent activation of the mucosal innate response (46), linked to activation of protective IFN type I expression (47) and induction of airway hyperreactivity (48). Mice were administered poly(I·C) in the absence or presence of JQ1 pretreatment. We observed that poly(I·C) induced a profound polymorphonuclear leukocytic (neutrophilic) inflammation around the small and medium-sized airways 24 h after introduction (compare Fig. 7B with A). This neutrophilic inflammation was completely inhibited by JQ1 administration (Fig. 7D). Concordantly, the number of bronchoalveolar lavage fluid (BALF) leukocytes in poly(I·C)-treated mice increased nearly 4 times over that of phosphate-buffered saline (PBS)-treated mice, an induction completely inhibited by JQ1 treatment (Fig. 7E). Also, the percentage of BALF neutrophils increased from 1% in PBS-treated mice to 51.4% in RSV-infected mice and was reduced to 17% in the JQ1-treated mice (Fig. 7F).
FIG 7.
BRD4 mediates poly(I·C)-induced inflammation and neutrophilia in vivo. C57BL/6 mice were pretreated with or without JQ1 (50 mg/kg) by the intraperitoneal (i.p.) route. Twenty-four hours later, mice were given another administration of JQ1 via the i.p. route and stimulated intranasally with either 50 μl of PBS or 50 μl PBS plus 100 μg poly(I·C) for 24 h. (A) Hematoxylin and eosin (H&E) staining of paraffin-embedded lung from PBS-treated C57BL/6 mouse. A representative section is shown. Magnification, ×10. The bar is the same for all images. (B) H&E staining of paraffin-embedded lung from poly(I·C)-treated mouse. (C) H&E staining of paraffin-embedded lung from JQ1-treated mouse. (D) H&E staining of paraffin-embedded lung from RSV-infected and JQ1-treated mouse. Note that the marked poly(I·C) induction of peribronchiolar neutrophils is blocked by JQ1. (E) Cell counts in the BALF expressed as total number of cells × 103/ml. (F) Percentage of neutrophils in BALF. **, P < 0.01 compared to untreated samples. (G) Q-RT-PCR of IFN, ISG, and NF-κB-dependent gene expression in total lung RNA. **, P < 0.01 compared to untreated control. (H) Cytokine concentration in BALF analyzed by multiplex enzyme-linked immunosorbent assay. **, P < 0.01.
Analysis of innate signaling gene expression program in the lungs demonstrated that poly(I·C) induced coordinate expression of type I and III IFNs (mIFNβ and mIL-28A/B) and ISG-15 and -54 mRNAs by quantitative real-time PCR (Q-RT-PCR) (Fig. 7G). Similarly, JQ1 also inhibited the highly poly(I·C)-inducible NF-κB-dependent inflammatory genes mIL-6, mKC, and mGroβ (Fig. 7G).
Correspondingly, poly(I·C) induced dramatic inductions of cytokine levels measured in the bronchoalveolar lavage fluid (BALF). Here, mIL-6 protein was increased from below the limit of detection in control cells to 400 pg/ml in the BALF in response to poly(I·C) (Fig. 7H). Similar dramatic inductions of mouse granulocyte colony-stimulating factor (mG-CSF), mouse granulocyte-macrophage colony-stimulating factor (mGM-CSF), mIL-1β, mRANTES, and others were observed in the BALF in response to poly(I·C) (Fig. 7H). Of these, mGM-CSF and mRANTES were the most highly upregulated cytokines, increasing to 2,000 pg/ml and 1,500 pg/ml, respectively. In the BALF of JQ1-treated animals, these values were dramatically reduced >95%, nearly to those of untreated mice. Together, these data indicate that BRD4 is required for TLR3-induced neutrophilia and innate response in vivo.
BRD4 mediates RSV-induced inflammation in a mouse model.
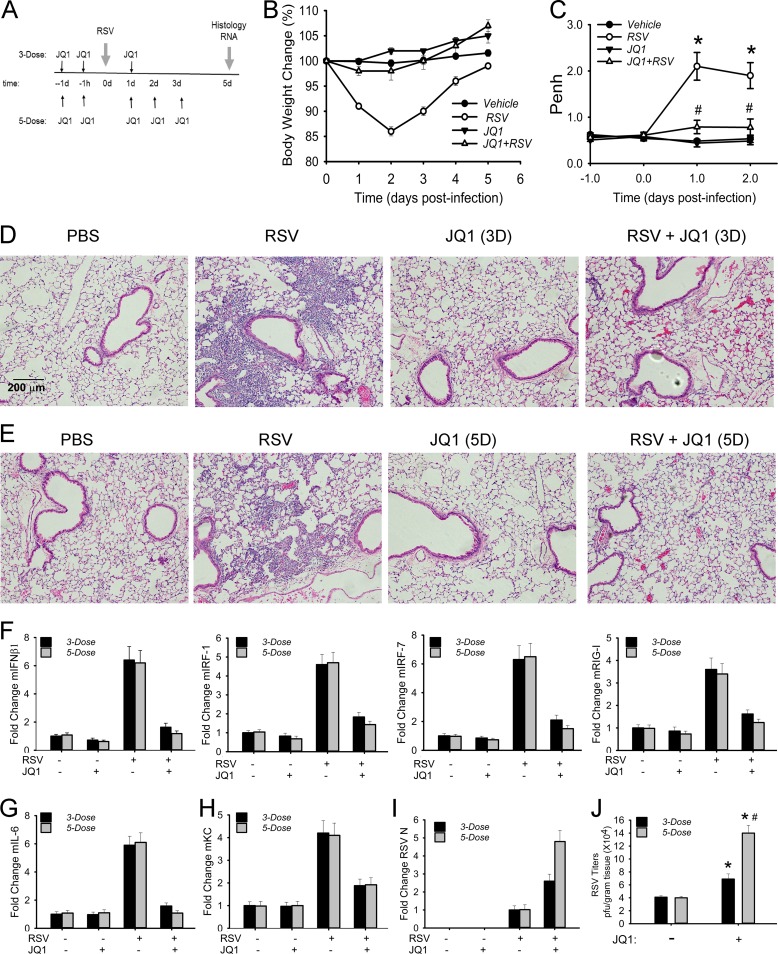
To further explore the role of BRD4 in mediating RSV-induced innate inflammation in vivo, we examined the effect of JQ1 on airway inflammation in a standardized C57BL/6 mouse model of RSV infection. In these series of experiments, we conducted two regimens: (i) a short time course to examine the role of BRD4 in the initial inflammatory response and (ii) a long time course to examine the effect of BRD4 on disease and airway obstruction.
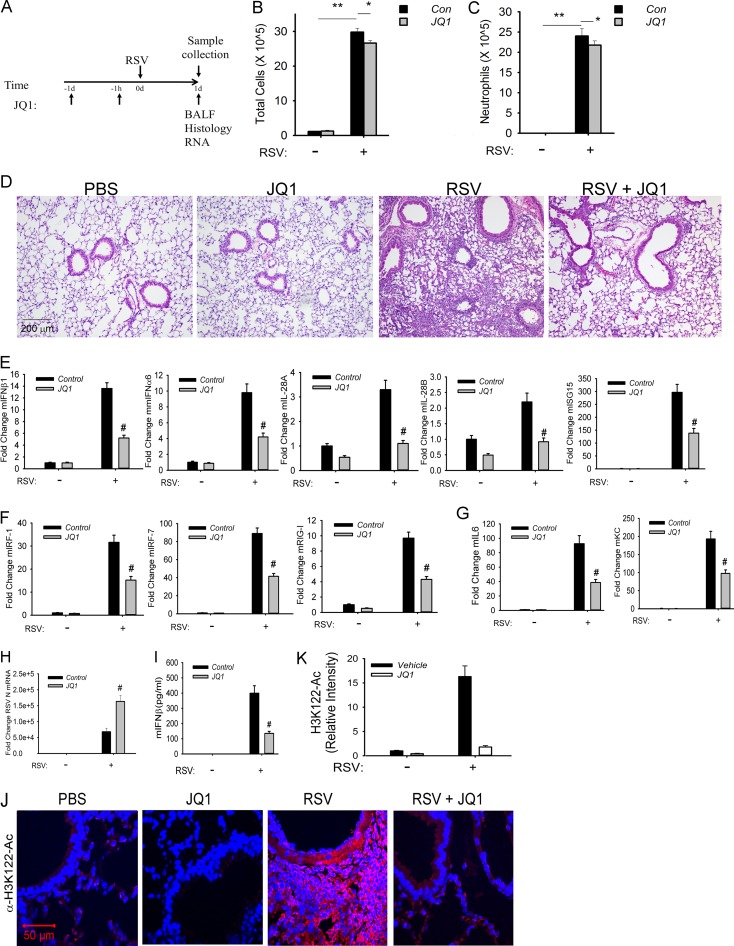
In the short time course, vehicle-only- or JQ1-pretreated mice were RSV infected via the intranasal route for 24 h prior to the assessment of inflammation (Fig. 8A). In this model, the total cell counts in the BALF from RSV-infected mice were increased by 10-fold relative to PBS-treated controls (P < 0.001) (Fig. 8B). In contrast, the cell count in the BALF from JQ1-treated mice was significantly reduced relative to vehicle controls (P < 0.05) (Fig. 8C). The major cell population affected by JQ1 treatment appeared to be the neutrophil population (P < 0.05) (Fig. 8C); no significant differences in macrophages were observed (data not shown).
FIG 8.
BRD4 mediates RSV-induced airway inflammation. (A) Experimental strategy for 1-day time course of RSV infection. (B) Total cell counts in the BALF. Cell numbers are presented as 105/ml from n = 5 mice/treatment group. **, P < 0.001 for RSV versus mock groups; *, P < 0.05 for RSV versus RSV plus JQ1. (C) BALF neutrophils. Neutrophils are shown as the total number. **, P < 0.001 for RSV versus mock groups; *, P < 0.05 for RSV versus RSV plus JQ1. (D) H&E staining of paraffin-embedded mouse lung. Shown are representative sections at ×10 magnification. (E) Q-RT-PCR of type I and III IFN expression. Data are shown as fold change relative to cyclophilin control. #, P < 0.01 compared to vehicle. (F) Q-RT-PCR of IRF-RIG-I amplification loop. #, P < 0.01 compared to vehicle. (G) Q-RT-PCR of NF-κB-dependent genes. #, P < 0.01 compared to vehicle. (H) Q-RT-PCR of RSV N expression. #, P < 0.01 compared to vehicle. (I) IFN-β concentration in BALF analyzed by multiplex enzyme-linked immunosorbent assay. #, P < 0.01 compared to vehicle. (J) Immunofluorescence staining of H3K122 acetylation (H3K122-Ac) in paraffin-embedded sections for each treatment group. Red, H3K122-Ac staining; blue, nuclei counterstained with DAPI. Confocal images are shown at ×63 magnification. (K) Quantification of H3K122-Ac. Relative changes in fluorescence intensity are shown.
Histological assessment indicated that RSV-infected mice exhibited a robust peribronchial and alveolar leukocytic infiltration compared to that of mice treated with PBS or JQ1 alone (Fig. 8D). In contrast, the JQ1-pretreated mice had markedly less alveolar leukocytic infiltrate with small residual peribronchial inflammation in response to RSV (Fig. 8D).
Analysis of the innate response indicated that RSV infection induced coordinate expression of type I IFNs (mIFNα6 and mIFNβ1), type III IFNs (IL-28A/B), and the ISG regulatory network (Fig. 8E). Expression of these IFN genes/ISGs was significantly reduced by JQ1 treatment. Similarly, the IRF1/7 and RIG-I amplification loop was activated by RSV and inhibited by JQ1 (Fig. 8F). The NF-κB-dependent mIL-6 and KC genes were also sensitive to JQ1 treatment (Fig. 8G). This inhibitory pattern of the innate pathway occurred despite the presence of increased RSV transcription in response to JQ1, consistent with the role of the innate pathway in the initial restriction of RSV replication (Fig. 8H). Cytokine expression in the BALF was significantly reduced for IFN-β, mGM-CSF, and mKC (Fig. 8I). Interestingly, expression of Th2 cytokines IL-10 and IL-13 was increased in the BALFs from JQ1-treated mice (data not shown).
To provide direct evidence that RSV activates BRD4 in vivo, we took advantage of the finding that BRD4 is an atypical histone acetyltransferase that uniquely acetylates histone H3 on Lys (K) 122, destabilizing the nucleosome and promoting more efficient transcriptional elongation (49). In vehicle-treated and JQ1-treated mice, H3K122-Ac staining was weak (Fig. 8J). In contrast, RSV infection induced a 15-fold upregulation of H3K122-Ac staining in the mucosa in vehicle-treated mice that was significantly abrogated by JQ1 treatment (Fig. 8K). Together, these data indicate that RSV induces mucosal BRD4 activity that mediates mucosal inflammation in vivo.
BRD4 mediates RSV disease and airway obstruction.
We next examined the effect of BRD4 inhibition in a longer (5-day) time course of RSV infection. Mice were treated with vehicle or JQ1 in two different regimens: (i) a 3-dose JQ1 regimen involving JQ1 administration 24 h and 1 h immediately before infection, followed by a single administration of JQ1 24 h after RSV infection; and (ii) a 5-dose JQ1 regimen that included two additional JQ1 administrations 2 and 3 days after RSV infection (Fig. 9A). For both regimens, histological assessment and gene expression assays were conducted 5 days after RSV infection. Mice receiving either regimen maintained normal body weight throughout the time course of infection; the 3-dose treatment is shown for simplicity (Fig. 9B). In RSV-infected mice not receiving JQ1 treatment, we observed a significant induction of baseline enhanced pause (Penh) 1 and 2 days postinfection (p.i.), indicating airway obstruction; this obstructive physiology was prevented in the JQ1 treatment group (Fig. 9C). In histological analysis, a dense peribronchial and alveolar leukocytic infiltration persisted for the 5-day treatment. This infiltration was significantly reduced in response to either the 3-dose (Fig. 9D) or 5-dose (Fig. 9E) regimen.
FIG 9.
BRD4 mediates RSV-induced disease. (A) Experimental strategy for 5-day time course of RSV infection. (B) Changes in body weights during the course of the experiment (n = 5 mice/treatment group). (C) Enhanced pause (Penh) from whole-body plethysmography at baseline. Penh values are presented as means ± standard errors of the means (n = 4 mice/treatment group). *, P < 0.05 for RSV versus RSV plus 3-dose JQ1 treatment regimen. #, not significant versus vehicle treatment. (D) H&E staining for representative sections from 3-dose JQ1 regimen. Shown are representative sections at ×10 magnification. The bar shown is the same for all images. (E) H&E staining for representative sections from 5-dose JQ1 regimen. Shown are representative sections at ×10 magnification. (F) IFN-β and IRF-RIG-I amplification loop. Fold change for indicated gene by Q-RT-PCR. (G) NF-κB-dependent IL-6 mRNA. Fold change by Q-RT-PCR. (H) NF-κB-dependent KC mRNA expression. #, P < 0.01 for RSV infection versus RSV plus JQ1 treatment. (I) RSV transcription expression. RSV N mRNA by Q-RT-PCR in total lung RNA. (J) RSV titers. *, P < 0.05 for JQ1 treatment versus vehicle. #, P < 0.05 for 3-dose versus 5-dose regimen.
Both treatment regimens reduced expression of type I IFN and the NF-κB-dependent IRF1/7 and RIG-I amplification loop in total lung RNAs (Fig. 9F), along with canonical NF-κB-dependent immediate early cytokines IL-6 and KC (Fig. 9G). In a manner consistent with the reduction of type I IFNs and ISGs, viral transcription was increased 3- and 5-fold relative to RSV infection alone in the 3-dose and 5-dose regimens, respectively (Fig. 9I). These changes in transcription were paralleled by direct measurement of RSV titers (6 × 104 and 14 × 104 PFU/g tissue [Fig. 9J]). Collectively, these data indicate that small-molecule inhibition of BRD4 attenuates RSV-induced inflammation, disease manifestations, and airway hyperreactivity, despite enhanced viral replication.
DISCUSSION
RSV is a major human pathogen responsible for severe lower respiratory tract infections worldwide. Lacking an efficacious vaccine, nearly all children in the United States are infected with RSV; infants with naive immune systems or predisposing chronic lung disease (bronchopulmonary dysplasia or cystic fibrosis) or who live in smoking households are more likely to develop more severe lower respiratory tract infections with RSV. The airway epithelium plays a central role in the initiation and maintenance of the pulmonary innate immune response (50, 51). Upon exposure to viral pathogen-associated molecular patterns, germ line-encoded RIG-I and TLR3 PRRs activate ROS stress and inducible transcription factors. These pathways coordinate expression of highly inducible immediate early genes encoding secreted factors whose actions stimulate neighboring epithelia and components of the immune system to trigger dendritic cell activation and the downstream adaptive immune response (18, 19). In response to nuclear ROS signals, coupled posttranslational modifications of RelA enable complex formation with P-TEFb, an event necessary for rapid transcriptional activation of immediate early genes (19, 21, 37). In this study, we investigate the requirement for and mechanism of BRD4 in controlling innate signaling downstream of the RIG-I and TLR3 PRRs. BRD4 inducibly complexes with RelA and CDK9 and is functionally required for effective activation of NF-κB-dependent immediate early cytokine genes in response to viral patterns (schematically diagrammed in Fig. 10). BRD4 functions as a coactivator in the NF-κB-RIG-I cross talk pathway upstream of ISG expression. Our data show that BRD4 is required for both the NF-κB-inducible expression of IRF1 and IRF7 isoforms and RIG-I activation by recruiting CDK9 forming phospho-Ser 2 CTD RNA Pol II and acetylating histone H3. Our studies are the first to demonstrate the functional role of BRD4 in mediating TLR3-induced airway inflammation and its activation (and functional role) in RSV-induced disease. Importantly, BRD4 inhibition significantly reduces the RSV-induced cytokine response, airway inflammation, and airway obstruction. Collectively, these data indicate that BRD4 is a critical regulator of the innate immune response mediating mucosal inflammation.
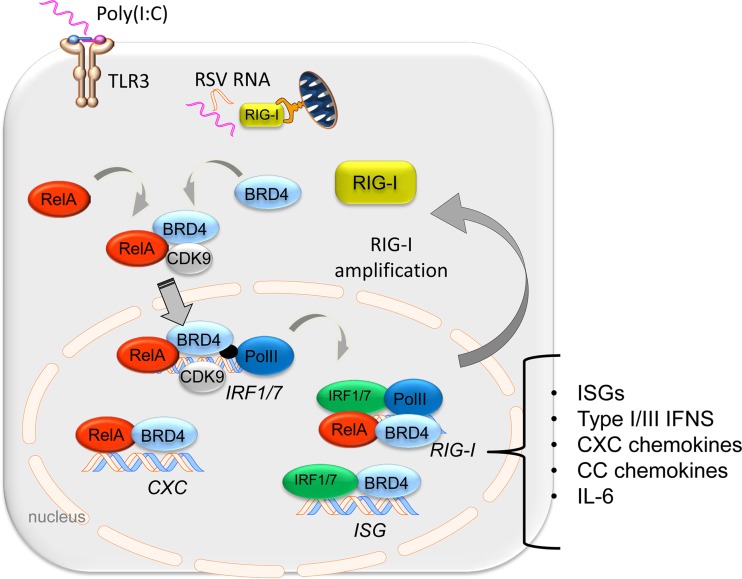
FIG 10.
BRD4 plays a central role in transcriptional elongation in the mucosal innate immune response. Schematic diagram. Pattern recognition receptors RIG-I and TLR3 trigger the acetyl histone reader BRD4 to complex with CDK9 and NF-κB/RelA. The function of BRD4 is required for the mucosal inflammatory response, amplification of IRF1/7-RIG-I amplification loop, and ISG regulatory network bridging CDK9 recruitment and formation of phospho-Ser 2 CTD RNA Pol II. In this manner, BRD4 mediates mucosal inflammation to viral patterns in vivo.
Transcriptional elongation in the IIR.
Although virtually all steps in transcriptional regulation are regulated, our studies have shown that the rapid gene expression mediating the innate immune response (IIR) is primarily mediated by the process of transcriptional elongation (18, 19). In resting epithelial cells, highly inducible innate response genes are maintained in an open chromatin configuration bound by hypophosphorylated RNA Pol II (52) Activation of the innate response transitions the polymerase into its transcriptional elongation mode, producing full-length, fully spliced mRNAs. Previously, we showed that CDK9 is a major mediator of inducible gene expression and RNA Pol II phosphorylation (18, 19). In unstimulated cells, CDK9 is distributed in two states: as an inactive complex with 7SK snRNA and HEXIM (53). We have found that innate activation alters the equilibrium between inactive and active CDK9 complex, shifting the inactive 7SK snRNA-associated fraction to bind BRD4 (19). Our IP-SRM assays support this observation that RSV triggers formation of increased abundance of the RelA-BRD4-CDK9 complex, without apparently affecting the stoichiometry of the complex. We are the first to demonstrate here that activation of the innate immune pathway induces BRD4 mRNA and protein abundance. Moreover, the PLA imaging studies suggest that the RelA-BRD4-CDK9 complex forms in situ in RSV-infected cells and that the complex is undergoing active nucleocytoplasmic shuttling.
Unbiased proteomics studies have shown that innate signaling restructures the CDK9 protein interaction network to acquire new biological functions, including RNA/DNA helicase activity, ribosomal binding, translation elongation components, and unfolded protein response proteins (22). This complex contains proteins involved in multiple steps in RNA processing—transcriptional elongation, mRNA splicing, transport, and initiation of translation, leading to the concept of CDK9's role in “cotranscriptionality” (22, 23). To this point, we found that RNA splicing proteins associated with CDK9 are required for alternative splicing of redox signaling genes (22). Although the full spectrum of regulatory activities of the CDK9 macromolecular complex is still being explored, we do know that CDK9 kinase activity is required for the induction of NF-κB-dependent inflammatory genes as well as IRF-dependent ISGs in RSV infection (18, 19).
Here, we extend our understanding of the role of BRD4 in the integrated innate response to TLR3 and RIG-I PRR signaling. Our XChIP studies indicate that BRD4 inhibition reduces formation of phospho-Ser 2 CTD RNA Pol II. The mechanisms by which BRD4 inhibition reduces phospho-Ser 2 CTD RNA Pol II formation may be complex. Others have shown that BRD4 is itself an RNA CTD Pol II kinase (54). Our data here show that BRD4 is required for CDK9 binding (CDK9 is a kinase with endogenous phospho-Ser 2 RNA CTD activity [55]). Finally, we also note that BRD4 is a kinase that phosphorylates CDK9 as well (56). More studies will be required to understand the role of BRD4 kinase activity in CTD formation in innate signaling, perhaps using BRD4 kinase-dead mutants or selective inhibitors of the BRD4 kinase as they become available.
BRD4 dynamically redistributes in the genome during the IIR.
In unstimulated cells, BRD4's affinity for binding diacetylated histone H4 (on Lys 5, 8, and 12) and H3 (on Lys 9 and 16) results in its primary distribution throughout the genome associated with H3K27-Ac-enriched enhancers of actively expressed genes (57). Although a number of studies have implicated BRD4 in control of housekeeping gene expression, BRD4 interaction is dynamically reconfigured through indirect transcription factor interaction and directly binding acetylated Lys side chains of regulatory histones.
We and others have shown that BRD4 binds activated NF-κB/RelA via Ser 276 phosphorylation-dependent p300/CBP acetylation (18, 21, 34). Detailed mutagenesis studies of the BRD4 BDs indicate that the upstream BD1 preferentially binds acetylated RelA (34). Through its complex formation with NF-κB/RelA, the ternary cytoplasmic RelA-CDK9-BRD4 complex is targeted to immediate early response genes. Consistent with our studies in viral pattern-induced BRD4 reprogramming, genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) studies that have shown an important role of NF-κB/RelA in the formation of dense BRD4 foci (“superenhancers”) in upstream inflammatory gene response networks (58). Our studies clearly demonstrate that NF-κB/RelA targets the CDK9-BRD4 complex to inflammatory genes and the IRF1/7-RIG-I cross talk pathway. Whether viral patterns induce BRD4-enriched superenhancers during the innate response will require further investigation.
We earlier discovered that the nuclear phosphatidylinositol 3-kinase (PI3 kinase) ataxia telangiectasia mutated (ATM) mediates nuclear ROS signaling in response to RSV infection upstream of RelA Ser 276 phosphorylation by the mitogen and stress kinase (11, 59). Additionally, RSV replication induces cellular senescence via ROS and DNA double-strand breaks associated with the ATM-TP53 pathway in a mouse model (60). Interestingly, BRD4 controls double-stranded DNA damage responses through the ATM pathway. In the presence of specific splice isoforms of BRD4, chromatin is compacted, preventing the formation of phosphorylated gH2AX foci, ATM activation, and TP53-mediated cell cycle arrest (61). The role of BRD4 in mediating termination of the RSV-induced DNA damage response and the effect on cellular senescence are intriguing and will be subject to further exploration.
Our immunofluorescence studies demonstrate that RSV induces the abundance of BRD4 as well as its histone acetyltransferase activity, resulting in increased formation of H3K122-Ac marks (49). These studies are the first to our knowledge that demonstrate BRD4 activation in vivo. H3K122 acetylation is a unique posttranslational modification on GC-rich bodies of genes that destabilizes the histone octamer, promoting nucleosome eviction, accelerating the transit of RNA Pol II through densely packed nucleosomes. The individual contributions of BRD4 serine kinase activity (for CDK9 and Pol II) and histone acetyltransferase activity in transcriptional elongation of the innate pathway are currently unknown to us; further studies will be required to dissect these individual functions.
An unanticipated finding in our study was the significant inhibition of RSV-induced multinucleated cell (syncytium) formation by the BRD4 inhibitor. RSV-induced syncytia are produced by furin-like protease processing of the RSV class I fusion glycoprotein (F) and formation of a cell surface complex with RSV G and SH (62). Interestingly, differential display studies have found that syncytium formation also requires cell expression of cytokeratin 17 (CK17) (63). Interestingly, these studies have found that cellular CK17 expression is highly induced by RSV infection via NF-κB (63). Although we have not investigated CK expression in this study, we suspect that JQ1 also interferes with NF-κB-dependent CK17 expression, accounting for the inhibition of syncytia.
BRD4 recruitment to innate genes is mediated by both acetylated transcription factor and histone interactions.
Recruitment of CDK9-BRD4 complex to NF-κB-dependent inflammatory genes is mediated by complex formation with the activated RelA transcription factor (18). These studies indicate that ROS stress triggers a coupled posttranslational modification of RelA by Ser 276 phosphorylation coupled to p300/CBP-mediated acetylation (18, 59). Site-directed mutation to prevent acetylation prevents BRD4 complex formation and interferes with formation of the transcription elongation complex on target genes (18).
A surprising discovery of these studies is that BRD4 is required for expression of ISGs, genes that are primarily driven by IRF3 binding to ISRE sites and lack RelA binding sites (19). Although RIG-I upregulation is important in ISG expression, BRD4 also independently binds to ISG promoters. These findings are consistent with the understanding that BRD4 recruitment to chromatin may be mediated by different mechanisms. We are aware of studies on IFN-induced gene expression, where BRD4 helps recruit CDK9 to hyperacetylated histones (64). However, the transcription start site (TSS) regions of IFN-inducible innate genes in epithelium are frequently nucleosome depleted (65), thus suggesting the presence of an alternative mechanism for BRD4 recruitment in a cell-type-dependent manner. More work will be required to identify the mechanism by which BRD4 is recruited to the ISGs.
BRD4 is required for NF-κB-initiated amplification of the IRF-RIG-I pathway controlling IFN production.
RIG-I is a major primary PRR in the epithelial cell innate response to pathogen patterns elaborated by RSV infection; siRNA silencing of RIG-I shows profound inhibition of type I and III IFNs and downstream ISG expression (30). The RIG-I PRR is one of the most highly upregulated genes in the antiviral response and codependent on de novo IRF1/7 expression (10) and inducible RelA-CDK9 recruitment (59). We provide the direct demonstration that NF-κB is required for activation of the IRF-RIG-I pathway and that ectopic IRF1 and IRF7 are sufficient to induce RIG-I expression. Additionally, we interpret our findings in RelA wild-type (WT) cells that RIG-I is maximally activated before IRF1 or IRF7 mRNA expression reaches its peak to suggest a nonlinear or synergistic interaction between these transcription factors (Fig. 3C). In this study, we demonstrate that RelA controls IRF1 and IRF7 expression and that this mechanism requires BRD4 for transcriptional elongation. Similarly, downstream RIG-I expression is also BRD4 dependent and mediated by active phospho-Ser 2 RNA Pol II CTD formation. The rapid RIG-I expression engendered by synergistic feed-forward activity of RelA and IRF coupled to transcriptional elongation ensures a rapid and robust ISG response to viral infections.
BRD4 mediates TLR3-dependent neutrophilia in vivo.
Neutrophils rapidly recruit to the airways upon poly(I·C) stimulation and represent a hallmark of the early innate immune response. A body of work has shown that extracellular poly(I·C) is a potent activator of TLR3 signaling mediating NF-κB activation via MyD88, a pathway independent of the RIG-I PRR. Because neutrophils lack TLR3 expression (66), neutrophil recruitment is an indirect process, primarily mediated by epithelial secretion of chemotactic chemokines such as IL-8/Groα/β in humans and mKC/mGroα/β in mice. Based on the sensitivity of mKC/mGroα/β to JQ1 treatment, we think how JQ1 blocks acute poly(I·C)-induced neutrophilia is via its inhibitory action on transcriptional elongation of NF-κB-dependent chemotactic chemokines. These data validate BRD4 as a target for treatment of acute lung inflammation mediated by viral patterns triggering the TLR3 pathway.
BRD4 mediates immunopathogenesis of RSV infection in the mouse.
RSV infections are associated with ∼120,000 hospitalizations annually in the United States (67); these severely RSV-infected children are subsequently at increased risk for the development of recurrent wheezing and asthma in later childhood (3, 4). In humans with fatal RSV infections, epithelial sloughing, accumulation of mucus plugs, and neutrophil and mononuclear interstitial infiltrates are observed (68, 69). The mouse model of acute RSV infection reproduces the initial events in RSV-induced pneumonia with rapid defects in alveolar fluid clearance, activation of innate cytokine networks, acute neutrophilic inflammation, and small airway obstruction (70–72). Earlier studies have shown that RSV induces mucosal NF-κB activation (72) and expression of epithelial cytokine networks, including CC chemokines (RANTES and MIP-1α and -1β), CXC chemokines (I-TAC and GRO-α, -β, and -γ), CX3C chemokines, type I and III IFNs, defensins, and thymic stromal lymphopoietin (71, 73). Our previous studies showing that blockade of the mucosal NF-κB interferes with RSV-induced pathology implicate the epithelial innate response in disease manifestations (16).
Our findings that the highly inducible expression of epithelium-derived chemokines and type I/III IFNs is significantly reduced by JQ1 treatment implicate the transcriptional elongation pathway in acute RSV pneumonia in vivo. These conclusions are consistent with our earlier experiments, where RSV-induced chemokine expression and airway inflammation were also inhibited by an IκB kinase (IKK) inhibitor (16). In both of these studies, interference with the innate pathway was predictably associated with enhanced RSV replication. Despite enhanced viral replication, blockade of the innate response was associated with reduced weight loss and bronchial obstruction. This result suggests to us that the innate pathway plays an important initiating role in RSV disease. It will be of interest in future studies to examine how BRD4 inhibitors affect innate response-driven adaptive immune responses and viral clearance.
Our demonstration that BRD4 is a common mediator of the TLR3- and RIG-I-induced host response identifies this component of the transcriptional elongation complex as a potential target for modulating the mucosal inflammatory response and/or neutrophil-mediated inflammatory responses to viral patterns.
MATERIALS AND METHODS
Ethics statement.
Male 12-week-old C57BL/6J mice were purchased from Harlan Inc. (Houston, TX) and housed under pathogen-free conditions in the animal research facility of the University of Texas Medical Branch, Galveston, TX, under protocol numbers 1312058 and 9001002 in accordance with the National Institutes of Health and University of Texas Medical Branch institutional guidelines for animal care.
Cell culture, treatment, and inducible RelA silencing.
Human small airway epithelial cells (hSAECs) were immortalized using human telomerase/CDK4 as previously described (26, 27). hSAECs were grown in small airway epithelial cell growth medium (SAGM; Lonza, Walkersville, MD) in a humidified atmosphere of 5% CO2. Poly(I·C) sodium salt was obtained from Sigma (catalog no. P1530; St. Louis, MO) and used at 10 μg/ml in cell culture. JQ1 was purchased from Cayman Chemical (Ann Arbor, MI) and used at 10 μM (36).
hSAECs expressing doxycycline (Dox)-regulated scrambled (control) or RelA shRNAs were established using puromycin selection. Briefly, Tripz Tet-on inducible lentiviral scrambled or RelA shRNA plasmids (Dharmacon, Thermo Fisher Scientific, Lafayette, CO) were reverse transfected with packing construct into BOS23 cell lines, and supernatants were harvested. Naive hSAECs were infected and selected for puromycin resistance (4 μg/ml). Puromycin-resistant hSAECs were pooled and characterized. shRNA expression was produced by addition of doxycycline (Dox; Sigma-Aldrich, St. Louis, MO) to the culture medium for 5 days (2 μg/ml).
Virus preparation and infection.
The human RSV Long strain was grown in HEp-2 cells and prepared as described previously (41). The viral titer of purified RSV pools was varied from 8 to 9 log PFU/ml, determined by a methylcellulose plaque assay. Viral pools were aliquoted, quick-frozen on dry ice-ethanol, and stored at −70°C until they were used.
Q-RT-PCR.
For gene expression analyses, 1 μg of RNA was reverse transcribed using SuperScript III in a 20-μl reaction mixture with gene-specific primers (19, 30, 59). PCRs included an initial denaturation for 90 s at 95°C, followed by 40 cycles of 15 s at 94°C, 60 s at 60°C, and 1 min at 72°C in an iCycler (Bio-Rad). PCR products were subjected to melting curve analysis to ensure that a single amplification product was produced. Quantification of relative changes in gene expression was calculated using the threshold cycle (ΔΔCT) method (19, 26), and expression as the fold change between experimental and control samples was normalized to internal control cyclophilin.
siRNA-mediated BRD4 silencing.
Commercially obtained, control or BRD4 short interfering RNAs (siRNAs) (Dharmacon, Thermo Fisher Scientific, Lafayette, CO) were reverse transfected at a 100 nM concentration using TransIT-siQUEST transfection reagent (Mirus Bio Corp.). Forty-eight hours later, cells were infected with RSV or not (multiplicity of infection [MOI], 1.0) for 24 h or treated with poly(I·C) (10 μg/ml) for 4 h as indicated. Cells were washed with PBS twice and lysed in TRI reagent (Sigma-Aldrich) for isolation of total RNA.
Lentivirus-mediated gene overexpression.
Human IRF1 and IRF7 cDNAs (plasmid templates were kindly provided by Charles M. Rice [74] and Fanxiu Zhu [75], respectively) were cloned into the lentiviral vector pLV-tetO-CMV-SV40-Puro-LoxP (kindly provided by George R. Stark [76]). To produce infectious lentiviruses, each construct was transfected into 293FT packaging cells using Lipofectamine 2000 (Invitrogen). The supernatant media were collected 48 h after transfection and frozen in aliquots at −80°C. hSAECs were infected in the presence of 4 μg/ml Polybrene and selected 48 h later with 2 μg/ml of puromycin for 3 days.
Two-step XChIP.
XChIP was performed as described previously (77). hSAECs (4 × 106 to 6 × 106) in 100-mm dishes were washed twice with PBS. Protein-protein cross-linking was first performed with disuccinimidyl glutarate (Pierce) followed by protein-DNA cross-linking with formaldehyde. Equal amounts of sheared chromatin were immunoprecipitated overnight at 4°C with 4 μg indicated Ab in ChIP dilution buffer (77). Immunoprecipitates were collected with 40 μl protein A magnetic beads (Dynal Inc.), washed, and eluted in 250 μl elution buffer for 15 min at room temperature. Samples were de-cross-linked in 0.2 M NaCl at 65°C for 2 h. The precipitated DNA was phenol-chloroform extracted, precipitated by 100% ethanol, and dried.
Q-gPCR.
Gene enrichment in ChIP was determined by quantitative real-time genomic PCR (Q-gPCR) using region-specific PCR primers (43). Standard curves were generated using a dilution series of genomic DNA (from 40 ng to 25 μg) for each primer pair. The fold change of DNA in each immunoprecipitate was determined by normalizing the absolute amount to input DNA reference and calculating the fold change relative to that amount in unstimulated cells.
Immunofluorescence microscopy.
hSAECs were plated on cover glasses pretreated with rat tail collagen (Roche Applied Sciences). After indicated infection times and treatments, the cells were fixed with 4% paraformaldehyde in PBS, permeabilized by 0.1% Triton X-100, blocked, and incubated with indicated primary rabbit polyclonal Ab as described previously (18, 78). After incubation with Alexa goat anti-rabbit Ab, cells were washed and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were visualized with a Zeiss fluorescence LSM510 confocal microscope at ×63 magnification (43, 76). Fluorescence intensity was quantified in 5 independent fields using ImageJ.
In situ PLA.
hSAECs were grown overnight in 6-well chamber slides, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and incubated with IgG or primary Abs to RelA, CDK9, or BRD4. Slides were then subjected to proximity ligation assay (PLA) using the Duolink PLA kit from OLink Bioscience (Uppsala, Sweden) according to the manufacturer's instructions. The nuclei were counterstained with DAPI, and the PLA signals were visualized in a fluorescence microscope (Olympus) at ×20 magnification.
IP.
Nuclei were suspended in radioimmunoprecipitation assay buffer (RIPA; 150 mM NaCl, 1 mM Na2-EDTA, 1% IGEPAL CA-630, 1% sodium deoxycholate, 20 mM Tris-HCl [pH 7.5]) with Complete protease inhibitor cocktail (Sigma-Aldrich) and 0.1% IGEPAL CA-630 (MP Biomedicals), sonicated 4 times, and centrifuged at 12,000 rpm for 10 min. The supernatants were collected and were quantified for protein concentrations. Equal volumes of nuclear lysates were incubated overnight at 4°C with 4 μg of IgG or CDK9 Ab in ChIP dilution buffer (11). Immune complexes were collected with 40 μl protein A magnetic beads (Dynal Inc.). The beads were washed with PBS for three times and then resuspended in 30 μl of 50 mM ammonium hydrogen carbonate (pH 7.8). The samples were then subjected to on-bead tryptic digestion.
SRM.
Immune complexes on beads were reduced with dithiothreitol (10 mM, 30 min at room temperature), alkylated with iodoacetamide (30 mM, 2 h at 37°C), and digested with trypsin (2 μg, 24 h at 37°C) as described previously (77, 79). After digestion, the supernatant was collected. The beads were washed with 50 μl of 50% acetonitrile (ACN) three times, and the supernatant was pooled and dried. The tryptic digests were then reconstituted in 30 μl of 5% formic acid-0.01% trifluoroacetic acid (TFA). An aliquot of 10 μl of diluted stable-isotope-labeled signature peptides was added to each tryptic digest. These samples were desalted with a ZipTip C18 cartridge. The peptides were eluted with 80% ACN and dried with a SpeedVac system.
SID-SRM-MS assays of BRD4, CDK9, and RelA used the qualified proteotypic peptides DAQEFGADVR (BRD4), DPYALDLIDK (CDK9), and TPPYADPSLQAPVR (RelA) as previously described (77). Stable-isotope-labeled standard (SIS) peptides were chemically synthesized incorporating isotopically labeled [13C615N4]arginine or [13C615N2]lysine to a 99% isotopic enrichment (Thermo Scientific) and quantified by amino acid analysis. BRD4 protein complexes were immunoprecipitated with anti-BRD4 Ab and captured by protein A magnetic beads (Dynal Inc.). The proteins were trypsin digested on the beads as described above. The tryptic digests were then reconstituted in 30 μl of 5% formic acid-0.01% trifluoroacetic acid (TFA). An aliquot of 10 μl of 50-fmol/μl diluted stable-isotope-labeled standard peptides was added to each tryptic digest. These samples were desalted with a ZipTip C18 cartridge, eluted with 80% ACN, and dried. The peptides were reconstituted in 30 μl of 5% formic acid-0.01% TFA and were directly analyzed by liquid chromatography (LC)-SRM-MS. LC-SRM-MS analysis was performed with a TSQ Vantage triple quadrupole mass spectrometer equipped with a nanospray source (Thermo Scientific, San Jose, CA). Eight to 10 targeted proteins were analyzed in a single LC-SRM run. The online chromatography was performed using an Eksigent NanoLC-2D high-performance liquid chromatography (HPLC) system (AB Sciex, Dublin, CA). An aliquot of 10 μl of each of the tryptic digests was injected on a C18 reverse-phase nano-HPLC column (PicoFrit; 75 μm by 10 cm; tip inside diameter [i.d.], 15 μm) at a flow rate of 500 nl/min with 20 min of 98% A, followed by a 15-min linear gradient from 2 to 30% mobile phase B (0.1% formic acid-90% acetonitrile) in mobile phase A (0.1% formic acid). The TSQ Vantage was operated in high-resolution SRM mode with Q1 and Q3 set to 0.2 and 0.7-Da full-width half maximum (FWHM). All acquisition methods used the following parameters: 2,100-V ion spray voltage, a 275°C ion transferring tube temperature, and a collision-activated dissociation pressure at 1.5 mtorr. The S-lens voltage used the values in the S-lens table generated during MS calibration.
All SRM data were manually inspected to ensure peak detection and accurate integration. The chromatographic retention time and the relative product ion intensities of the analyte peptides were compared to those of the stable-isotope-labeled standard peptides. The acceptable variation of the retention time between the analyte peptides and their standard counterparts was within 0.05 min, and the difference in the relative product ion intensities of the analyte peptides and stable-isotope-labeled standard (SIS) peptides was below 20%. The peak areas in the extract ion chromatography of the native and stable-isotope-labeled standard peptides were integrated using Xcalibur 2.1. Three to 5 SRM transitions were monitored for each proteotypic peptide. The default values for noise percentage and baseline subtraction window were used. The ratio between the peak areas of native and SIS versions of each peptide was calculated.
Mouse models of poly(I·C) and RSV infection.
For poly(I·C) stimulation, mice were pretreated in the absence or presence of JQ1 (50 mg/kg of body weight, via the intraperitoneal [i.p.] route) 1 day prior to poly(I·C) stimulation. The next day, animals were given another JQ1 injection immediately followed by intranasal (i.n.) administration of phosphate-buffered saline (PBS; 50 μl) or poly(I·C) (100 μg dissolved in 50 μl PBS). One day later, the mice were euthanized. For bronchoalveolar lavage (BAL), the trachea was cannulated and 1 ml of PBS was introduced by syringe. The BALF was analyzed for total and differential cell counts and quantified for secreted cytokines/chemokines using multiplex immunoassays (BioPlex). Half of each lung was fixed with 10% (vol/vol) neutral buffered formalin for 3 days, processed into paraffin blocks, and cut into 5-μm sections for hematoxylin and eosin (H&E) staining. The other half of each lung was immediately frozen in liquid N2 and pulverized. One milliliter of TRI reagent was added, followed by the extraction of total RNA per the manufacturer's directions. The total RNA was reverse transcribed, and gene expression was quantified using Q-RT-PCR.
For RSV challenge, mice were treated with JQ1 (50 mg/kg body weight, i.p.) on the day prior to RSV infection and 1 h prior to RSV infection. For RSV infection, 107 PFU of purified RSV was administered in 50 μl PBS i.n. Uninfected mice were sham treated with PBS alone. Animals were weighed and examined daily for disease. For the short time course experiment, 1 day after inoculation, mice were euthanized, BALF was collected, RNA was extracted, and lungs were fixed for histopathology. For the 5-day time course, mice were given either JQ1 1 day after infection (3-dose treatment) or three daily doses of JQ1 (on days 1, 2, and 3, constituting the 5-dose treatment). The experiment was terminated 5 days postinfection (p.i.). Unrestrained, whole-body plethysmography (Buxco Electronics, Inc., Sharon, CT) was used to measure the enhanced pause (Penh) to evaluate baseline airway obstruction.
Cellular inflammation.
Total cell counts were determined by staining 50 μl of BALF with trypan blue and counting viable cells using a hemocytometer. For differentials, 100 μl of BALF was used to generate cytospin preparations. Slides were dried, fixed, and stained with Protocol Hema3 (Fisher Diagnostics, Middletown, VA). A total of 300 cells were counted per sample using light microscopy.
Statistical analysis.
One-way analysis of variance (ANOVA) was performed when looking for time differences followed by Tukey's post hoc test to determine significance. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
Core laboratory support was through the UTMB Optical Imaging Laboratory and the SCMM Selected Reaction Monitoring facility.
This work was supported, in part, by funding from NIH grants NIAID AI062885 (A.R.B.), UL1TR001439 (A.R.B.), and NIEHS ES006676 and NSF grant DMS-1361411/DMS-1361318 (A.R.B.).
REFERENCES
- 1.Johnston NW, Sears MR. 2006. Asthma exacerbations. 1. Epidemiology. Thorax 61:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston SL. 2005. Overview of virus-induced airway disease. Proc Am Thorac Soc 2:150–156. doi: 10.1513/pats.200502-018AW. [DOI] [PubMed] [Google Scholar]
- 3.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohapatra SS, Boyapalle S. 2008. Epidemiologic, experimental, and clinical links between respiratory syncytial virus infection and asthma. Clin Microbiol Rev 21:495–504. doi: 10.1128/CMR.00054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigurs N, Aljassim F, Kjellman B, Robinson PD, Sigurbergsson F, Bjarnason R, Gustafsson PM. 2010. Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax 65:1045–1052. doi: 10.1136/thx.2009.121582. [DOI] [PubMed] [Google Scholar]
- 6.Loo Y-M, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol Rev 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins PA, Sriskandan S. 2005. Mammalian Toll-like receptors: to immunity and beyond. Clin Exp Immunol 140:395–407. doi: 10.1111/j.1365-2249.2005.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Bertolusso R, Tian B, Zhao Y, Vergara LA, Sabree A, Iwanaszko M, Lipniacki T, Brasier A, Kimmel M. 2014. Dynamic cross talk model of the epithelial innate immune response to double-stranded RNA stimulation: coordinated dynamics emerging from cell-level noise. PLoS One 9:e93396. doi: 10.1371/journal.pone.0093396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang L, Choudhary S, Tian B, Boldogh I, Yang C, Ivanciuc T, Ma Y, Garofalo RP, Brasier AR. 2015. Ataxia telangiectasia mutated kinase mediates NF-kappaB serine 276 phosphorylation and interferon expression via the IRF7-RIG-I amplification loop in paramyxovirus infection. J Virol 89:2628–2642. doi: 10.1128/JVI.02458-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian B, Zhang Y, Luxon BA, Garofalo RP, Casola A, Sinha M, Brasier AR. 2002. Identification of NF-kappaB-dependent gene networks in respiratory syncytial virus-infected cells. J Virol 76:6800–6814. doi: 10.1128/JVI.76.13.6800-6814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Luxon BA, Casola A, Garofalo RP, Jamaluddin M, Brasier AR. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J Virol 75:9044–9058. doi: 10.1128/JVI.75.19.9044-9058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitsett JA, Alenghat T. 2015. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haeberle HA, Casola A, Gatalica Z, Petronella S, Dieterich HJ, Ernst PB, Brasier AR, Garofalo RP. 2004. IkappaB kinase is a critical regulator of chemokine expression and lung inflammation in respiratory syncytial virus infection. J Virol 78:2232–2241. doi: 10.1128/JVI.78.5.2232-2241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall NF, Price DH. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 18.Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. 2011. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J Virol 85:11752–11769. doi: 10.1128/JVI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian B, Zhao Y, Kalita M, Edeh CB, Paessler S, Casola A, Teng MN, Garofalo RP, Brasier AR. 2013. CDK9-dependent transcriptional elongation in the innate interferon-stimulated gene response to respiratory syncytial virus infection in airway epithelial cells. J Virol 87:7075–7092. doi: 10.1128/JVI.03399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price DH. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20:2629–2634. doi: 10.1128/MCB.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. 2008. RelA Ser276 phosphorylation is required for activation of a subset of NF-κB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol Cell Biol 28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Zhao Y, Kalita M, Li X, Jamaluddin M, Tian B, Edeh CB, Wiktorowicz JE, Kudlicki A, Brasier AR. 2015. Systematic determination of human cyclin dependent kinase (CDK)-9 interactome identifies novel functions in RNA splicing mediated by the DDX5/17 RNA helicases. Mol Cell Proteomics 14:2701–2721. doi: 10.1074/mcp.M115.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perales R, Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8:327–337. [DOI] [PubMed] [Google Scholar]
- 25.Hou T, Ray S, Brasier AR. 2007. The functional role of an interleukin 6-inducible CDK9-STAT3 complex in human gamma-fibrinogen gene expression. J Biol Chem 282:37091–37102. doi: 10.1074/jbc.M706458200. [DOI] [PubMed] [Google Scholar]
- 26.Tian B, Li X, Kalita M, Widen SG, Yang J, Bhavnani SK, Dang B, Kudlicki A, Sinha M, Kong F, Wood TG, Luxon BA, Brasier AR. 2015. Analysis of the TGFbeta-induced program in primary airway epithelial cells shows essential role of NF-kappaB/RelA signaling network in type II epithelial mesenchymal transition. BMC Genomics 16:529. doi: 10.1186/s12864-015-1707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. 2004. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 28.Kalita M, Tian B, Gao B, Choudhary S, Wood TG, Carmical JR, Boldogh I, Mitra S, Minna JD, Brasier AR. 2013. Systems approaches to modeling chronic mucosal inflammation. Biomed Res Int 2013:505864. doi: 10.1155/2013/505864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Jamaluddin M, Zhang Y, Sun H, Ivanciuc T, Garofalo RP, Brasier AR. Systematic analysis of cell-type differences in the epithelial secretome reveals insights into pathogenesis of RSV-induced lower respiratory tract infections. J Immunol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol 81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garofalo R, Sabry M, Jamaluddin M, Yu RK, Casola A, Ogra PL, Brasier AR. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J Virol 70:8773–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamaluddin M, Casola A, Garofalo RP, Han Y, Elliott T, Ogra PL, Brasier AR. 1998. The major component of IkappaBalpha proteolysis occurs independently of the proteasome pathway in respiratory syncytial virus-infected pulmonary epithelial cells. J Virol 72:4849–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Brasier AR. 2013. Applications of selected reaction monitoring (SRM)-mass spectrometry (MS) for quantitative measurement of signaling pathways. Methods 61:313–322. doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. 2009. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol 29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhary S, Boldogh I, Brasier AR. 2016. Inside-out signaling pathways from nuclear reactive oxygen species control pulmonary innate immunity. J Innate Immun 8:143–155. doi: 10.1159/000442254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Lu M, Tian B, Li K, Garofalo RP, Prusak D, Wood TG, Brasier AR. 2009. Expression of an IKKgamma splice variant determines IRF3 and canonical NF-kappaB pathway utilization in ssRNA virus infection. PLoS One 4:e8079. doi: 10.1371/journal.pone.0008079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamaluddin M, Wang S, Garofalo RP, Elliott T, Casola A, Baron S, Brasier AR. 2001. IFN-beta mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am J Physiol Lung Cell Mol Physiol 280:L248–L257. [DOI] [PubMed] [Google Scholar]
- 43.Tian B, Yang J, Brasier AR. 2012. Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol Biol 809:105–120. doi: 10.1007/978-1-61779-376-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowak DE, Tian B, Brasier AR. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 45.Nishiyama A, Mochizuki K, Mueller F, Karpova T, McNally JG, Ozato K. 2008. Intracellular delivery of acetyl-histone peptides inhibits native bromodomain-chromatin interactions and impairs mitotic progression. FEBS Lett 582:1501–1507. doi: 10.1016/j.febslet.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stowell NC, Seideman J, Raymond HA, Smalley KA, Lamb RJ, Egenolf DD, Bugelski PJ, Murray LA, Marsters PA, Bunting RA, Flavell RA, Alexopoulou L, San Mateo LR, Griswold DE, Sarisky RT, Mbow ML, Das AM. 2009. Long-term activation of TLR3 by poly(I:C) induces inflammation and impairs lung function in mice. Respir Res 10:43. doi: 10.1186/1465-9921-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerrero-Plata A, Baron S, Poast JS, Adegboyega PA, Casola A, Garofalo RP. 2005. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J Virol 79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aeffner F, Traylor ZP, Yu EN, Davis IC. 2011. Double-stranded RNA induces similar pulmonary dysfunction to respiratory syncytial virus in BALB/c mice. Am J Physiol Lung Cell Mol Physiol 301:L99–L109. doi: 10.1152/ajplung.00398.2010. [DOI] [PubMed] [Google Scholar]
- 49.Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS. 2016. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 23:540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holt PG, Strickland DH, Wikstrom ME, Jahnsen FL. 2008. Regulation of immunological homeostasis in the respiratory tract. Nat Rev Immunol 8:142–152. doi: 10.1038/nri2236. [DOI] [PubMed] [Google Scholar]
- 51.Knight DA, Holgate ST. 2003. The airway epithelium: structural and functional properties in health and disease. Respirology 8:432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 52.Hsin J-P, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ III, Singer DS. 2012. BRD4 is an atypical kinase that phosphorylates serine 2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. 2002. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou M, Huang K, Jung K-J, Cho W-K, Klase Z, Kashanchi F, Pise-Masison CA, Brady JN. 2009. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J Virol 83:1036–1044. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, Siebenlist U, O'Shea JJ, Ozato K. 2014. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, Yang T, Wang H, Luscinskas FW, Croce KJ, Bradner JE, Plutzky J. 2014. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell 56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang L, Choudhary S, Zhao Y, Edeh CB, Yang C, Boldogh I, Brasier AR. 2014. ATM regulates NF-kappaB-dependent immediate-early genes via RelA Ser 276 phosphorylation coupled to CDK9 promoter recruitment. Nucleic Acids Res 42:8416–8432. doi: 10.1093/nar/gku529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martínez I, García-Carpizo V, Guijarro T, García-Gomez A, Navarro D, Aranda A, Zambrano A. 2016. Induction of DNA double-strand breaks and cellular senescence by human respiratory syncytial virus. Virulence 7:427–442. doi: 10.1080/21505594.2016.1144001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. 2013. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heminway BR, Yu Y, Tanaka Y, Perrine KG, Gustafson E, Bernstein JM, Galinski MS. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801–805. doi: 10.1006/viro.1994.1245. [DOI] [PubMed] [Google Scholar]
- 63.Domachowske JB, Bonville CA, Rosenberg HF. 2000. Cytokeratin 17 is expressed in cells infected with respiratory syncytial virus via NF-kappaB activation and is associated with the formation of cytopathic syncytia. J Infect Dis 182:1022–1028. doi: 10.1086/315841. [DOI] [PubMed] [Google Scholar]
- 64.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. 2011. Determinants of nucleosome organization in primary human cells. Nature 474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayashi F, Means TK, Luster AD. 2003. Toll-like receptors stimulate human neutrophil function. Blood 102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 67.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. 1999. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA 282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 68.Welliver TP, Garofalo RP, Hosakote Y, Hintz KH, Avendano L, Sanchez K, Velozo L, Jafri H, Chavez-Bueno S, Ogra PL, McKinney L, Reed JL, Welliver RC Sr. 2007. Severe human lower respiratory tract illness caused by respiratory syncytial virus and influenza virus is characterized by the absence of pulmonary cytotoxic lymphocyte responses. J Infect Dis 195:1126–1136. doi: 10.1086/512615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aherne WT, Bird T, Court SDB, Gardner PS, McQuillin J. 1970. Pathological changes in virus infections of the lower respiratory tract in children. J Clin Pathol 23:7–18. doi: 10.1136/jcp.23.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peebles RS, Graham BS. 2005. Pathogenesis of respiratory syncytial virus infection in the murine model. Proc Am Thorac Soc 2:110–115. doi: 10.1513/pats.200501-002AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haeberle HA, Kuziel WA, Dieterich HJ, Casola A, Gatalica Z, Garofalo RP. 2001. Inducible expression of inflammatory chemokines in respiratory syncytial virus-infected mice: role of MIP-1alpha in lung pathology. J Virol 75:878–890. doi: 10.1128/JVI.75.2.878-890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haeberle HA, Takizawa R, Casola A, Brasier AR, Dieterich HJ, Van Rooijen N, Gatalica Z, Garofalo RP. 2002. Respiratory syncytial virus-induced activation of nuclear factor-kappaB in the lung involves alveolar macrophages and Toll-like receptor 4-dependent pathways. J Infect Dis 186:1199–1206. doi: 10.1086/344644. [DOI] [PubMed] [Google Scholar]
- 73.Lee HC, Headley MB, Loo YM, Berlin A, Gale M Jr, Debley JS, Lukacs NW, Ziegler SF. 2012. Thymic stromal lymphopoietin is induced by respiratory syncytial virus-infected airway epithelial cells and promotes a type 2 response to infection. J Allergy Clin Immunol 130:1187–1196. doi: 10.1016/j.jaci.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang Q, Fu B, Wu F, Li X, Yuan Y, Zhu F. 2012. ORF45 of Kaposi's sarcoma-associated herpesvirus inhibits phosphorylation of interferon regulatory factor 7 by IKKepsilon and TBK1 as an alternative substrate. J Virol 86:10162–10172. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. 2013. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J 32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao YX, Widen SG, Jamaluddin M, Tian B, Wood TG, Edeh CB, Brasier AR. 2011. Quantification of activated NF-kappa B/RelA complexes using ssDNA aptamer affinity-stable isotope dilution-selected reaction monitoring-mass spectrometry. Mol Cell Proteomics 10:M111.008771. doi: 10.1074/mcp.M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalita MK, Sargsyan K, Tian B, Paulucci-Holthauzen A, Najm HN, Debusschere BJ, Brasier AR. 2011. Sources of cell-to-cell variability in canonical nuclear factor-kappaB (NF-kappaB) signaling pathway inferred from single cell dynamic images. J Biol Chem 286:37741–37757. doi: 10.1074/jbc.M111.280925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Y, Tian B, Edeh CB, Brasier AR. 2013. Quantitation of the dynamic profiles of the innate immune response using multiplex selected reaction monitoring-mass spectrometry. Mol Cell Proteomics 12:1513–1529. doi: 10.1074/mcp.M112.023465. [DOI] [PMC free article] [PubMed] [Google Scholar]