Abstract
Sho1p, an integral membrane protein, plays a vital role in the high-osmolarity glycerol (HOG) mitogen-activated protein kinase pathway in the yeast Saccharomyces cerevisiae. Activated under conditions of high osmotic stress, it interacts with other HOG pathway proteins to mediate cell signaling events, ensuring that yeast cells can adapt and remain viable. In an attempt to further understand how the function of Sho1p is regulated through its protein–protein interactions (PPIs), we identified 49 unique Sho1p PPIs through the use of membrane yeast two-hybrid (MYTH), an assay specifically suited to identify PPIs of full-length integral membrane proteins in their native membrane environment. Secondary validation by literature search, or two complementary PPI assays, confirmed 80% of these interactions, resulting in a high-quality Sho1p interactome. This set of putative PPIs included both previously characterized interactors, along with a large subset of interactors that have not been previously identified as binding to Sho1p. The SH3 domain of Sho1p was found to be important for binding to many of these interactors. One particular novel interactor of interest is the glycerol transporter Fps1p, which was shown to require the SH3 domain of Sho1p for binding via its N-terminal soluble regulatory domain. Furthermore, we found that Fps1p is involved in the positive regulation of Sho1p function and plays a role in the phosphorylation of the downstream kinase Hog1p. This study represents the largest membrane interactome analysis of Sho1p to date and complements past studies on the HOG pathway by increasing our understanding of Sho1p regulation.
Keywords: high osmolarity glycerol (HOG) pathway, MAPK pathways, SH3 domains, protein–protein interactions, membrane yeast two-hybrid assay
Introduction
Organisms often face various internal and external stresses and, over time, develop systems to adapt to these challenges. The budding yeast Saccharomyces cerevisiae has long been an excellent model to study many different processes, including those associated with stress, in eukaryotic cells. The high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) stress response pathway ensures that yeast cells adapt and remain viable under conditions of high osmotic stress (reviewed in Ref. [1]). Turgor pressure, and therefore cell viability, is lost during conditions under which the concentration of the solute is higher in the external than in the internal environment of the cell. The HOG pathway consists of two branches mediated by Sln1p and Sho1p (reviewed in Ref. [1]). The signaling cascades mediated by these two branches eventually result in the production of active Hog1p protein kinase, which acts to control the expression of genes necessary for the stress response including those involved in glycerol production, cell cycle arrest, and glycerol uptake [2–7]. Ultimately, the HOG pathway allows for cells to adapt to increased osmolarity through the activation of various cell processes in order to balance the cellular osmotic pressure with that of the external environment.
Sho1p, an activator of one branch of the HOG pathway, is a yeast integral membrane protein that is activated under conditions of severe hyperosmotic stress [2]. Sho1p itself is not thought to be involved in the sensing of extracellular osmolarity, which is performed instead through the mucin-like proteins Msb2p/Hkr1p [8]. Sho1p is composed of four transmembrane domains and a cytoplasmic SH3 domain at its C-terminus. The SH3 domain on Sho1p mediates protein–protein interactions (PPIs) with PXXP-motif-containing protein partners such as the co-scaffold protein Pbs2p [9,10] and Ste20p [11]. Together with Sho1p, Pbs2p binds a large complex of proteins including the G-protein Cdc42p, the kinases Ste20p and Ste11p, and Ste50p, resulting in the downstream activation of Hog1p (reviewed in Ref. [1]). As with many other cellular pathways, the HOG pathway is controlled by various positive and negative regulators. Most notable are protein phosphatases such as Ptp2p, Ptp3p, and Ptc1p (reviewed in Ref. [12]), responsible for the dephosphorylation of various components of the pathway. In addition to the HOG pathway, Sho1p has also been implicated in other cellular processes, such as filamentous growth [13], cell wall integrity [14], and more recently in cytokinesis [15].
As yeast Sho1p has been implicated in various signaling pathways in the cell, it is an excellent model to study the basics of signaling pathways in eukaryotes. Although many components of the HOG pathway have been identified, specific regulators of Sho1p function have largely remained elusive. To date, it has been difficult to study the protein interactors of the full-length form of Sho1p, particularly using proteome-wide interactive proteomic techniques [16–20], which have thus far yielded little information on possible regulators of Sho1p. This is due in part to the considerable hydrophobicity conferred by the multiple transmembrane domains of Sho1p, necessitating the use of technically challenging protein solubilization steps for many traditional approaches. However, recent smaller-scale screens have revealed novel Sho1p interactors such as those involved in cytokinesis [15]. Comprehensive in vivo studies, in which Sho1p is in the context of its natural membrane environment, are also currently lacking. Given its prominent role in the stress response pathway, it is important to identify currently unknown protein partners of Sho1p in order to better understand the signal transduction capabilities of this multifunctional protein, the mechanisms that dictate its signaling specificity, and its modes of regulation.
In this study, we used the membrane yeast two-hybrid (MYTH) assay [21–26] to screen for novel interactors of Sho1p. Using both N- and C-terminally tagged versions of Sho1p as bait, we found 49 unique putative interactors, many of which required the SH3 domain of Sho1p for their interaction. We focused on one of these interactors, the glycerol transporter Fps1p, and confirmed its interaction with Sho1p by two other complementary methods. We also examined protein domains and portions of Sho1p and Fps1p that are important for their interaction. While investigating the cellular role of the Fps1p–Sho1p interaction, using genetic assays and by the examination of downstream Sho1p signaling through the HOG kinase Hog1p, we found that Fps1p is involved in the positive regulation of Sho1p function. These data contribute to our current understanding of the regulation of Sho1p-mediated pathways and implicate Fps1p in the regulation of Sho1p activity.
Results
Construction and verification of Sho1p MYTH bait proteins
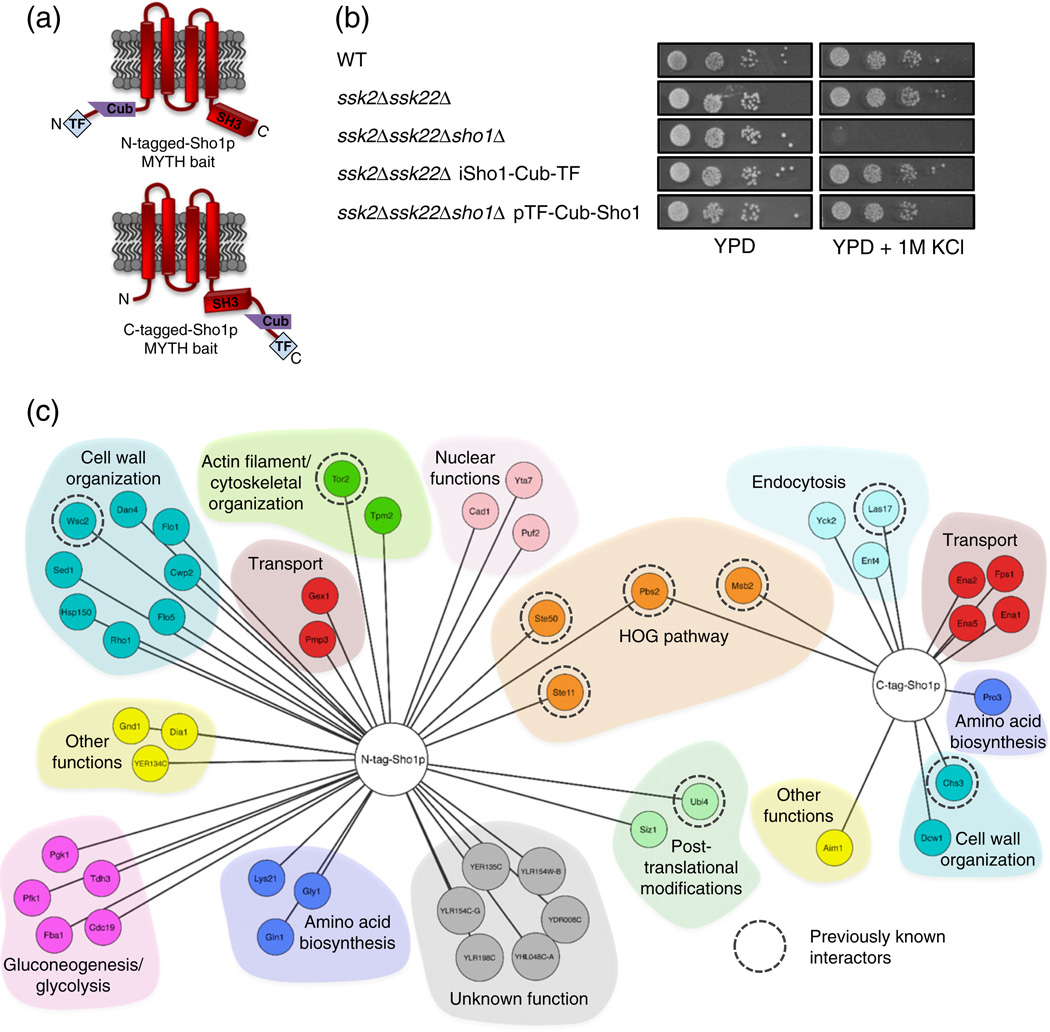
In an attempt to further understand the diverse functions and regulatory processes of Sho1p, we sought to identify its interacting protein partners. Using the Membrane Yeast Two-Hybrid (MYTH) system [21–26], we generated full-length Sho1p baits tagged with the MYTH Cub-transcription factor (TF) fusion cassette at their N- or C-terminus, and expressed them either ectopically from a plasmid (N-tagged Sho1p bait) or under the control of the native SHO1 promoter upon endogenous tagging of the SHO1 locus (C-tagged Sho1p bait) (Fig. 1a). To confirm that MYTH tagging did not disrupt correct localization of Sho1p, we constructed Sho1p baits incorporating YFP into the MYTH tag and examined them by fluorescence microscopy. Localization of N-tagged Sho1p (Supplementary Fig. 1) was varied, as expected, due to its presence in different steps of the vesicle transport pathway. However, a significant fraction of the Sho1p bait localized to the bud neck as previously reported [27] and also co-localized with the plasma-membrane-bound protein Pdr5p (Supplementary Fig. 1). Proper plasma membrane localization of the C-tagged integrated Sho1p bait has been shown previously [22]. Bait expression by detection on a Western blot was assessed for both Sho1p baits. Although the N-tagged Sho1p was not detected by Western blot (data not shown), the integrated C-tagged Sho1p bait showed a band corresponding to the expected size of the tagged protein with antibodies against the LexA portion of the MYTH tag (Supplementary Fig. 2a).
Fig. 1.
Application of Membrane Yeast Two-Hybrid (MYTH) to the Sho1p interactome. (a) Sho1p MYTH baits used in this study. The location of MYTH tag fusions (at the N- or the C-terminus of the protein) is indicated. (b) Sho1p baits are functional in the HOG pathway. Wildtype cells (WT) and deletion strains (as indicated) in the presence or absence of MYTH-tagged Sho1p bait were 10-fold serially diluted (from a starting OD0.05), spotted onto solid media, grown for 2 days at 30 °C, and imaged. All images are from the same plate. (c) Proteins identified as interacting protein partners of N-tagged (TF-Cub-Sho1p) and C-tagged (Sho1p-Cub-TF) Sho1p by the MYTH method. Proteins are grouped by colour according to function, and those found in previously reported physical or genetic interaction screens are indicated by a broken circle.
To test whether the fused Sho1p constructs were functional in the HOG pathway, we examined the growth of cells containing targeted disruptions of the two functional branches of the HOG pathway, on salt-containing media, in the presence and absence of MYTH-tagged Sho1p (Fig. 1b). As expected, the deletion of SHO1, SSK2, and SSK22 together rendered both branches non-functional, and the cells were incapable of growing on media containing 1 M KCl. When this triple deletion strain was transformed with the N-terminally tagged Sho1p MYTH bait, however, the growth of these cells was similar to that of ssk2Δssk22Δ cells, which are non-functional in only the Sln1-mediated branch of the HOG pathway. Likewise, the C-terminal tagging of Sho1p in the ssk2Δssk22Δ background did not affect the growth of these cells on media containing 1 M KCl when compared to ssk2Δssk22Δ cells with untagged Sho1p present. Therefore, these data confirm that both N- and C-terminally tagged versions of the Sho1p MYTH bait were functional in the context of the HOG pathway.
We next tested for the self-activation and proper expression of our Sho1p MYTH baits. To this end, we transformed control prey constructs, composed of Ost1p (an unrelated protein not predicted to physically interact with Sho1p) fused to our MYTH prey tag (Nub), into strains containing the N-terminally tagged, plasmid-based Sho1p bait, as well as into a strain containing the C-terminally tagged, genomically integrated bait. Two versions of Nub were used, NubG and NubI. As previously shown [23], NubI (and consequently NubI-tagged proteins) spontaneously interacts with Cub on the bait protein (independent of a bait–prey interaction), leading to a positive read-out on MYTH screen selection plates if the Cub-containing bait is properly expressed. Conversely, NubG contains a mutation that prevents spontaneous interaction, and thus, NubG-tagged proteins will only interact if a physical interaction occurs between bait and prey. Bait self-activation and expression was tested as performed previously [23], and our MYTH screens were subsequently performed with 50 mM and 10 mM3-AT, respectively, for the plasmid-based and the integrated Sho1p tags.
Sho1p physically interacts with a variety of proteins
Using these bait constructs, we initially tested for physical interactions between Sho1p and a small panel of previously known interactors, followed by large-scale MYTH screens against NubG-tagged cDNA and gDNA libraries. Putative interactors were tested for bait dependency as previously described [23] by retransforming plasmids expressing interactors identified from the screen into a strain containing the Sho1p bait, or an unrelated protein (the ABC transporter Snq2p) as bait, in order to confirm that putative interactors were specific to Sho1p. The Sho1p MYTH analysis, in total, identified 51 putative Sho1p interactions, across 49 unique proteins (Fig. 1c). Of these, 9 have been previously shown to be associated with Sho1p, as determined by physical and/or genetic screens, with 5 of these having been identified in two or more separate studies as annotated in the BioGRID protein interaction database [28]. The remaining 40 represent novel Sho1p interactors, 6 of which are of unknown biological function. These results indicate that screening for PPIs through the MYTH method enables the identification of many novel interactors for a membrane-bound protein such as Sho1p.
Many previously known interactors bind Sho1p through its SH3 protein interaction domain. To determine which of our candidate interactors are dependent on the SH3 domain of Sho1p for interaction, we generated a MYTH-tagged version of Sho1p in which the SH3 domain was removed (Sho1ΔSH3-Cub-TF), and tested it for an interaction against a small panel of putative Sho1p interactors found in our screens. Sho1ΔSH3-Cub-TF has a similar expression level as full-length Sho1-Cub-TF bait as detected by Western blot (Supplementary Fig. 2a). We found that a substantial subset of the identified candidates in the MYTH screen required the SH3 domain of Sho1p to fully interact (Supplementary Fig. 2b), including Fps1p, Las17p, Dcw1p, and Chs3p. On the other hand, many other candidates (Yck2p, Ent4p, Aim1, and Pro3p, as well as the tandem array Ena1p/Ena2p/Ena5p) retained their interaction with Sho1p in the absence of the SH3 domain, implying that they bind Sho1p through another portion of the protein.
Confirmation of Sho1p interactors through BiFC and co-IPs
To further validate our Sho1p interactome, we used two different complementary assays, co-immunoprecipitations (co-IPs) and bimolecular fluorescence complementation (BiFC). For co-IP validation, 33 out of the 49 unique Sho1p interactors, available as tandem affinity purification (TAP)-tagged [29] strains tagged at their C-termini, were selected (Supplementary Table 1). Those interactors that were not available in the TAP collection or that have been previously reported in the literature as interacting with Sho1p were not included in the confirmation set. Into these TAP-tagged strains, we transformed a plasmid expressing Sho1p tagged at the C-terminus with an HA tag. Cells were lysed, and the TAP-tagged protein, along with any interacting proteins, was purified with calmodulin beads. Interacting HA-tagged Sho1p was then identified with an anti-HA antibody via Western blot. Of the 33 strains, 28 exhibited Sho1p-HA expression in whole cell lysates (Supplementary Fig. 3 and Supplementary Table 1). Of these 28 putative Sho1p interactors tested by co-IP, 20 (71%) tested were positive for an interaction (Fig. 2a, Supplementary Fig. 3, and Supplementary Table 1).
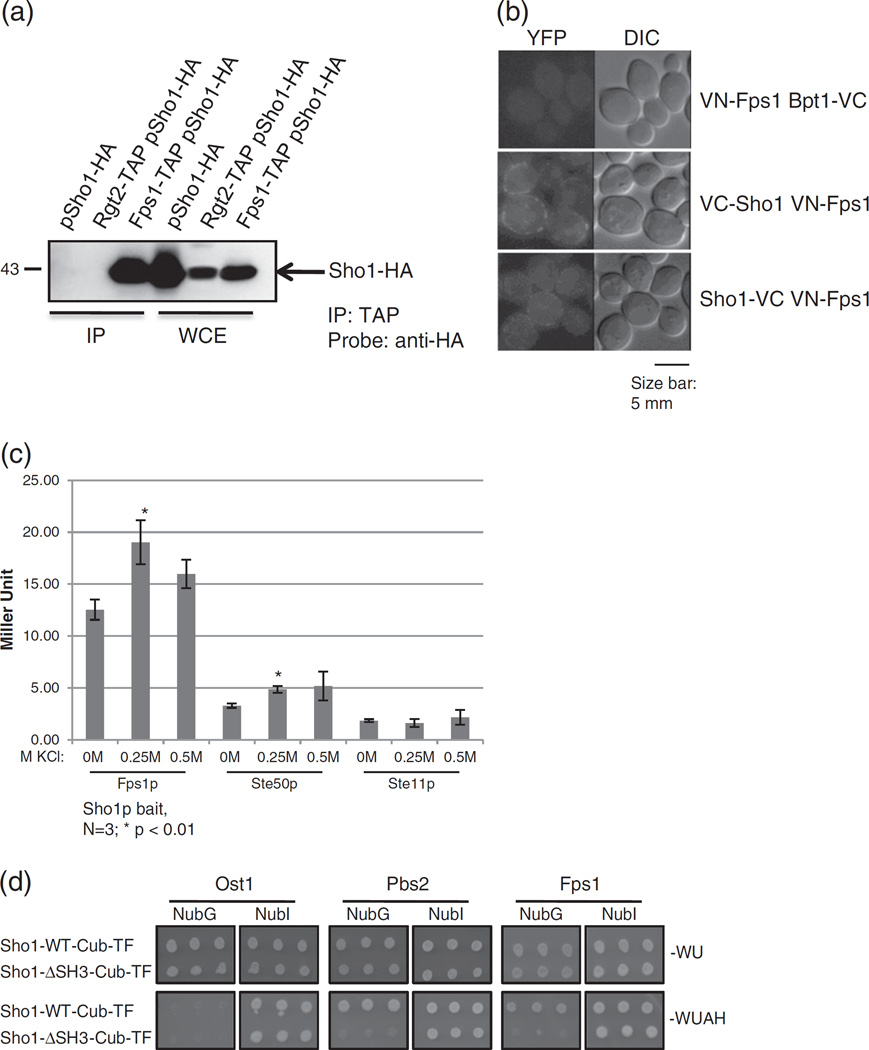
Fig. 2.
Sho1p physically interacts with Fps1p. (a) Confirmation of the Sho1p–Fps1p interaction by co-immunoprecipitation (co-IP). An HA-tagged Sho1p plasmid was transformed into an untagged strain, a strain encoding genomically TAP-tagged Rgt2p (negative control), or genomically TAP-tagged Fps1p. Cells were lysed, the TAP-tagged protein immunoprecipitated with calmodulin beads (“IP” lanes), and electrophoresed on a SDS-PAGE gel along with whole cell extract (“WCE” lanes) to determine protein expression. Proteins were transferred onto nitrocellulose membrane and the interacting HA-tagged protein was visualized with an anti-HA antibody. Molecular weight marker is shown at the left. (b) Confirmation of the Sho1p–Fps1p interaction by Bimolecular Fluorescence Complementation (BiFC). Fps1p, and Sho1p, as well as the unrelated protein Bpt1p, was tagged genomically with the N-terminal fragment of YFP (VN) or the C-terminal fragment of YFP (VC) as indicated. Cells expressing tagged Fps1p alongside either Bpt1p or Sho1p were imaged to detect YFP fluorescence, indicative of a physical interaction. A 5-µm-size bar has been added as a size reference. (c) The Sho1p–Fps1p interaction is increased under hyperosmotic conditions. MYTH reporter strains containing the Sho1p bait, along with Fps1p, Ste50p, or Ste11p preys were assessed for β-galactosidase activity by monitoring for the hydrolyzation of ONPG after growth in media with the indicated KCl concentrations. Miller units were calculated and plotted, and error bars are shown. Asterisk (*) denotes a statistically significant difference compared to 0M KCl. (d) The full-length Fps1p requires the SH3 domain of Sho1p for interaction. A plasmid expressing the full-length Fps1 prey, tagged at its N-terminus with NubG or NubI, or other N-tagged prey control constructs were transformed into a yeast reporter strain lacking the endogenous SHO1 gene alongside plasmid expressing C-terminally MYTH-tagged, full-length Sho1p (Sho1-WT-Cub-TF) or Sho1p lacking its SH3 domain (Sho1ΔSH3-Cub-TF). Cells were spotted in triplicate onto solid media that selected for the plasmids only (−WU) or selected for an interaction between the bait and prey proteins (−WUAH) and were incubated for 4 days before imaging.
We also used the BiFC method [30] as a secondary method to confirm the putative Sho1p interactions. Of the set of 49 unique Sho1p interactors, we attempted to confirm 36 (available in a haploid BiFC-tagged collection) by BiFC (Supplementary Table 1). Haploid strains, expressing the putative interactors at their endogenous loci tagged with the N-terminal fragment of YFP (VN) at their C-terminus [31], were mated to haploid strains expressing Sho1p endogenously tagged at either terminus with the C-terminal fragment of YFP (VC) and the diploid strains examined to detect the presence of YFP fluorescence, which is indicative of a physical interaction. Of the 36 putative Sho1p interactors tested by BiFC, 24 (67%) tested positive for an interaction (Supplementary Fig. 4 and Supplementary Table 1), and almost all showed fluorescence at the plasma membrane.
In summary, 28 putative Sho1p interactors were tested via co-IP, 36 were tested by BiFC, and 28 were tested by both methods. Of these 28, 27 (96%) were confirmed by either BiFC or co-IP, and 12 (43%) were confirmed by both BiFC and co-IP. Altogether, of the 49 unique Sho1p interactors we identified through MYTH, 39 (80%) were either confirmed by at least one of our other testing methods (co-IP or BiFC) or previously reported in the literature, indicating the high quality of the Sho1p interactome mapped by the MYTH assay.
Sho1p interacts physically with the glycerol transporter Fps1p
One of the interesting candidate interactors requiring the Sho1p SH3 domain for its interaction is the glycerol transporter Fps1p (Supplementary Fig. 2b). In both co-IP and BiFC confirmation assays, Fps1p was found to bind Sho1p, but not the unrelated proteins Rgt2 (co-IP; Fig. 2a) and Bpt1 (BiFC; Fig. 2b). Specifically, in the co-IP experiments (Fig. 2a), lanes from cells expressing only Sho1p-HA or cells expressing Rgt2-TAP and Sho1p-HA did not have a band size corresponding to Sho1p-HA after the immunoprecipitation of the TAP tag. On the other hand, lanes corresponding to cells containing both Fps1p-TAP and Sho1p-HA showed a band corresponding to the expected size of Sho1p-HA, illustrating that these two proteins interact. In the BiFC experiments (Fig. 2b), cells containing N-tagged Fps1p and N- or C-tagged Sho1p showed intense fluorescence at the plasma membrane, while a strain tagged with VC at the C-terminus of an unrelated protein, Bpt1p, together with Fps1p tagged at its N-terminus with VN, showed no fluorescence (Fig. 2b).
As both Sho1p and Fps1p have been implicated in the HOG pathway, we next tested for an effect of salt on the interaction between the two proteins. To assess protein binding quantitatively, we employed a liquid-based assay using ortho-nitrophenyl-β-galactoside (ONPG) as a substrate to assess β-galactosidase activity. Binding between Sho1p and Fps1p was increased modestly at higher salt concentrations (0.25 M and 0.5 M KCl) compared to the Sho1p interacting proteins (and HOG pathway components) Ste50p and Ste11p (Fig. 2c). This difference in interactions shows that it is likely Fps1p specific and is not due to a general increase in expression of HOG pathway components under hyperosmotic conditions.
All together, these data show that the interaction found in the MYTH screen between Sho1p and Fps1p occurs in vivo at the plasma membrane, can be robustly detected using a number of distinct methodologies, and the interaction is modestly increased with hyperosmotic stress. Although previously implicated as a component of the cellular HOG response by closing in response to stress to prevent escape of glycerol [32], Fps1p has not been reported to physically interact with Sho1p at the plasma membrane. Therefore, we decided to focus on the characterization of the biological consequences of the Sho1p–Fps1p interaction.
Sho1p and Fps1p interact through the SH3 domain of Sho1p and the N-terminus of Fps1p
We next decided to thoroughly investigate the involvement of the SH3 domain of Sho1p in its interaction with Fps1p. As a control, a previously known interactor of Sho1p, Pbs2p, was also tested and interacted with full-length Sho1p, but not with Sho1p missing its SH3 domain (Sho1ΔSH3) in the MYTH system. As expected, the full-length NubG-tagged version of Fps1p interacted with the Sho1p bait, while the interaction was abolished with Sho1ΔSH3 (Fig. 2d). To try to determine which residues of the Sho1p SH3 domain were responsible for this interaction, we tested three SH3 point mutants, ΔAY, I49A, and Y54A. These SH3 mutants were chosen for a variety of reasons. ΔAY (at residues 17–18) is part of a binding surface on the Sho1p SH3 domain and renders Sho1p deficient in activating the HOG pathway even though the protein exhibits wildtype binding to Pbs2p [10]. Position 49 is a key residue on surface II of SH3 domains and is often involved in binding the extended regions of PXXP-containing peptides or non-PXXP-containing peptides [33]. The Y54A mutant was chosen as it abrogates binding in vitro to the PXXP-containing site from Pbs2p [10]. We found that all of the Sho1p SH3 mutants tested retained their interaction with full-length Fps1p in the MYTH assay (Supplementary Fig. 5).
As other HOG pathway proteins such as Ste20p and Pbs2p have been reported to bind the SH3 domain of Sho1p [9–11], we wanted to determine whether the absence of these proteins, including Fps1p, have an effect on their binding. MYTH reporter strains (wildtype or lacking STE20, PBS2, or FPS1) harbouring a Sho1p bait, as well as Ste20p, Pbs2p, or Fps1p prey, were spotted on selective media. In all strains, Sho1p was equally able to bind Fps1p and Pbs2p (Supplementary Fig. 6). Ste20p, on the other hand, appeared to not be able to bind Sho1p in the MYTH system (Supplementary Fig. 6).
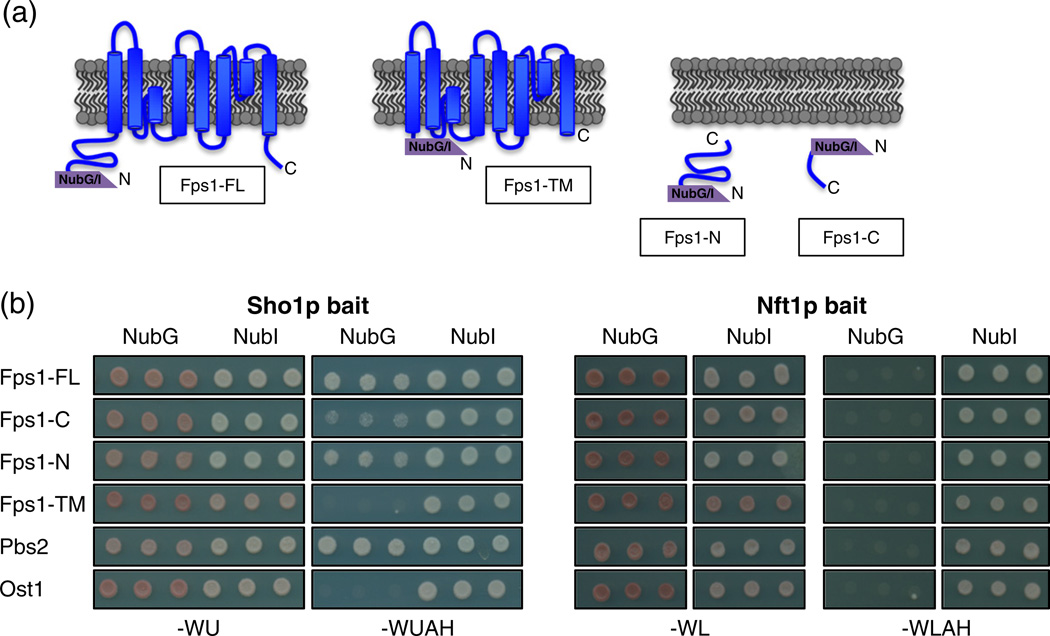
In order to determine which region of Fps1p was important for the interaction with Sho1p, we constructed truncation mutants of Fps1p and tested for portions of the protein that can interact with full-length Sho1p in the MYTH system (Fig. 3a). As a control, we also tested for the binding of these Fps1p fragments to the unrelated transporter Nft1p. Both the N-terminal (Fps1-N) and the C-terminal (Fps1-C) soluble domains of Fps1p were able to bind Sho1p, with Fps1-N fragment binding much more strongly than the weakly binding Fps1-C (Fig. 3b, left panel). In contrast, the middle portion Fps1p, containing the eight transmembrane segments (Fps1-TM), was unable to bind Sho1p (Fig. 3b, left panel). When tagged with NubI, Fps1-TM showed an interaction with NubI, demonstrating that the non-binding of the NubG version is not due to a lack of expression of the MYTH-tagged protein. All of the NubG-tagged Fps1p fragments were unable to bind to the unrelated Nft1p bait (Fig. 3b, right panel), showing that the binding observed was Sho1p specific.
Fig. 3.
The N- and C-terminus of Fps1p are important for Sho1p interaction. (a) Schematic diagram of NubG and NubI MYTH prey constructs expressing full-length Fps1p (Fps1-FL), the Fps1p C-terminus (Fps1-C), the Fps1p N-terminus (Fps1-N), and the Fps1p transmembrane domain (amino acids 256–530; Fps1-TM). (b) Constructs from (a) were transformed into NMY51 reporter cells along with full-length Sho1p bait or Nft1p bait as an unrelated control and were spotted in triplicate onto solid media that selected for the plasmids only (−WU or −WL) or selected for an interaction between the bait and prey proteins (−WUAH or −WLAH). Note that, for a given media, all cells were spotted onto the same plate. Plates were incubated at 30 °C for 4 days and then imaged.
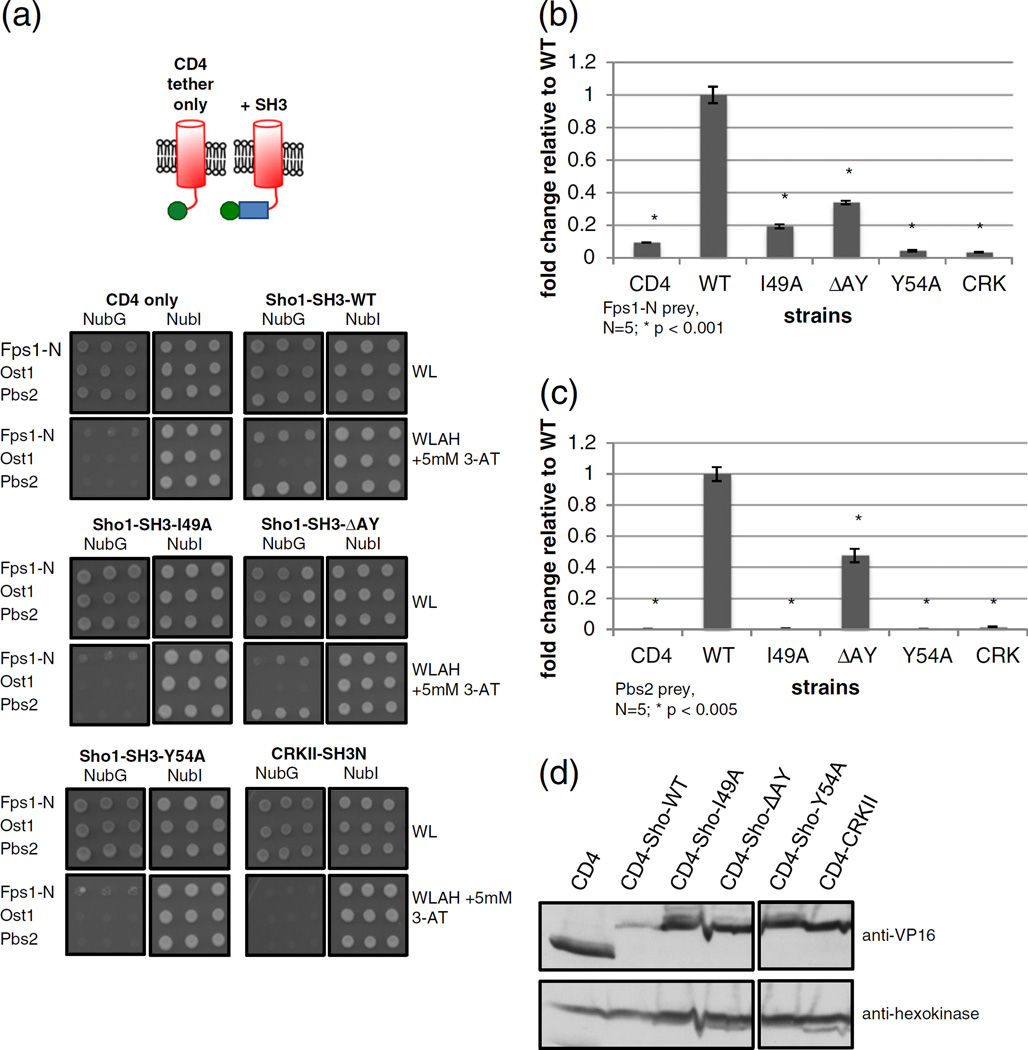
As Sho1p-Fps1p binding appears to occur largely in the N-terminal soluble domain of Fps1p, we next tested this domain for binding to SH3 domainmutants of Sho1p. As the MYTH assay requires the bait to be membrane bound, we used a single-pass transmembrane domain, from the human T-cell surface glycoprotein CD4, to tether the soluble Sho1p SH3 domain to the plasma membrane, and tested for binding to Fps1-N, the unrelated prey Ost1p, and Pbs2p (Fig. 4a). Compared to the CD4 tether only, or the unrelated N-terminal SH3 domain from the human Crk-II protein, wildtype Sho1p SH3 was able to bind to Fps1-N strongly, while there was weak binding to Fps1-N by Sho1-SH3-I49A and Sho1-SH3-ΔAY. Sho1-SH3-Y54A, on the other hand, showed no binding to Fps1-N. To assess protein binding quantitatively, we employed the liquid-based ONPG assay and found that the ΔAY, I49A, and Y54A mutants bound 0.3-, 0.2-, and 0.04-fold less than wildtype, respectively (Fig. 4b). The above mentioned experiments were also repeated with the full-length Pbs2p protein (Fig. 4a and c). ΔAY bound 0.5-fold less than wildtype, and the I49A and Y54A mutants showed no binding. Our Pbs2p binding data for ΔAY and Y54 correspond to those of previously reported in vitro binding assays [10], while the binding affinities of Pbs2p to the I49A mutant have not yet been reported in the literature. All of the above CD4-Sho1p SH3mutantswere found to be expressed as well, if not slightly better than wildtype Sho1p-SH3 when monitored by Western blot (Fig. 4d).
Fig. 4.
Specific residues in Sho1p SH3 are involved in binding to Fps1-N. Plasmids expressing the CD4 tether only, CD4-Sho1-SH3 (wildtype or various point mutants as indicated), or an unrelated SH3 domain (CRKII-SH3N) were co-transformed into reporter cells along with prey plasmids expressing a NubG- or NubI-tagged N-terminal fragment of Fps1p (Fps1-N) or full-length Ost1p or Pbs2p. Cells were (a) spotted in triplicate onto solid media that selected for the plasmids only (−WL) or selected for an interaction between the bait and prey proteins (−WLAH + 5 mM 3-AT). Note that, for a given media, all cells were spotted onto the same plate. Plates were incubated at 30 °C for 5 days and then imaged. The same strains containing (b) Fps1-N prey or (c) Pbs2p prey were assessed for β-galactosidase activity by monitoring for the hydrolyzation of ONPG. Miller units were calculated and the fold change over wildtype is plotted and error bars are shown. Asterisk (*) denotes a statistically significant difference compared to wildtype. (d) Whole cell lysates of log phase cells from (b) were electrophoresed and transferred onto nitrocellulose membrane, and the expression of bait proteins or an unrelated protein (hexokinase as a loading control) was determined using anti-VP16 (for MYTH-tagged baits) or anti-hexokinase antibodies.
The N-terminal domain of Fps1p has recently been implicated in the binding of Hog1p, leading to the phosphorylation and dissociation of the Fps1p regulator Rgc2p [34]. Dissociation of Rgc2p leads to the closing of the Fps1p under hyperosmotic conditions. We next tested for the interaction of Sho1p-Fps1 in hog1Δ, rgc1Δ, and rgc2Δ mutants to try to determine if this interaction occurs preferentially with the closed or open states of Fps1p. Our results indicate that the Sho1p–Fps1p interaction is not dependent on the presence of these genes, as the interaction was unaffected in all of the mutant strains tested (Supplementary Fig. 7).
Taken together, these data show that residues in both N- and C-termini of Fps1p are likely involved in its interaction with Sho1p, with Fps1-N playing a more prominent role. In addition, Fps1-N is able to bind to the wildtype Sho1p SH3 domain, while various point mutations in this domain reduced its binding affinity to or rendered it incapable of binding to Fps1-N.
FPS1 positively regulates SHO1
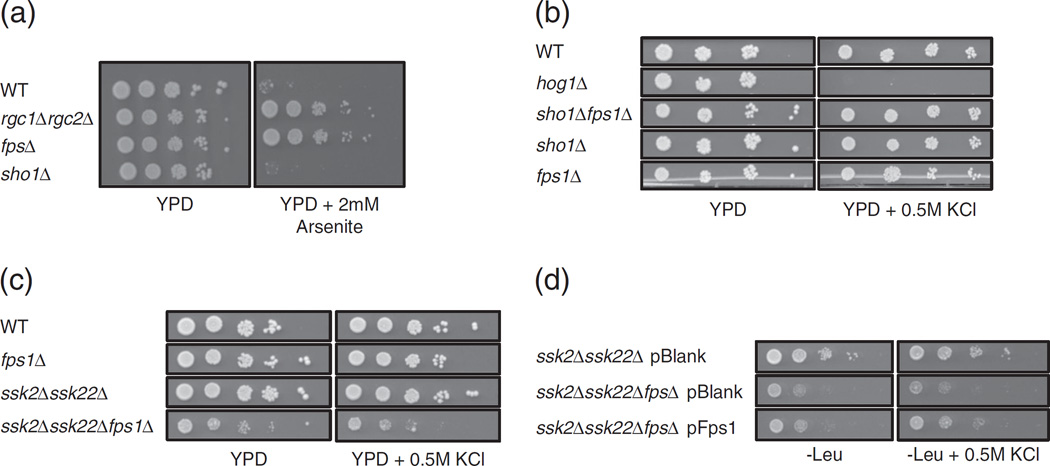
The data described above establish a physical interaction between Fps1p and Sho1p. Next, we wanted to determine how these proteins function in the regulation of each other and in the context of the HOG pathway. First, we tested the function of Fps1p in the presence and absence of SHO1. Along with the transport of glycerol, Fps1p is involved in the import of various compounds such as arsenite [35]. We proceeded to assess the sensitivity of various mutant strains to arsenite (Fig. 5a). As expected, wildtype cells (containing Fps1p) had a natural growth defect in media containing arsenite as previously reported [35]. Conversely, a strain with a deletion of FPS1, or a deletion in the known positive regulators of FPS1, rgc1Δrgc2Δ, was resistant to arsenite as previously reported [36]. Notably, we found that cells lacking functional SHO1 had a sensitivity to arsenite similar to that of wildtype strains, showing that Fps1p is still functional in strains deleted for SHO1.
Fig. 5.
Fps1p positively regulates Sho1p. (a and b) Wildtype cells (WT) and deletion strains (as indicated) were 10-fold serially diluted (from a starting OD600 of 0.5) and were spotted onto solid rich media (YPD) with or without 2 mM arsenite or 0.5 M KCl. Plates were grown at 30 °C for 4 days and then imaged. (c) Cells with the indicated gene deletions were diluted and spotted onto solid rich media with or without 0.5 M KCl as described in (a). Plates were grown at 30 °C for 3 days and then imaged. (d) A blank plasmid or a plasmid expressing Fps1p was transformed into strains from (c). Cells were diluted and spotted as in (a) onto plasmid selective plates (−Leu) with or without 0.5 M KCl, grown at 30 °C for 2 days, and then imaged. Note that images in each panel were taken from the same agar plate.
We tested the function of the HOG pathway in the absence of FPS1 by examining the phenotype of fps1Δ cells on media containing salt (Fig. 5b). As expected, cells lacking Hog1p have a strong growth defect on media containing 0.5 M KCl. On the other hand, wildtype cells (in which Fps1p closes in response to stress) or cells deleted for FPS1 (in which glycerol is unable to efficiently be exported from the cell) do not have a growth defect. sho1Δ cells grow similarly to wildtype cells in the presence of 0.5 M KCl, as the compensating Sln1p-mediated branch of the HOG pathway is still functional. When both SHO1 and FPS1 are deleted, these cells are not growth deficient. Together, these results reveal that the absence or presence of FPS1 does not affect the growth of cells that are functional only in the Sln1p-mediated branch of the HOG pathway.
In contrast, FPS1 does appear to be important for the growth of cells functional only in the Sho1pmediated branch of the HOG pathway, as a triple deletion strain in which both the Sln1p-mediated branch and FPS1 are absent (ssk2Δssk22Δfps1Δ) displayed a growth deficiency on standard rich media that increased in severity in the presence of salt, an effect that was notably absent in both ssk2Δssk22Δ double deletion and fps1Δ single deletion strains (Fig. 5c). This growth defect was partially rescued when a plasmid expressing FPS1 was added back to this triple deletion strain, as compared to cells containing a blank plasmid (Fig. 5d). Therefore, these data suggest that Sho1p malfunctions in the context of the HOG pathway in the absence of FPS1, implicating Fps1p as a positive regulator of Sho1p function.
FPS1 regulates Hog1p signaling through the Sho1p pathway
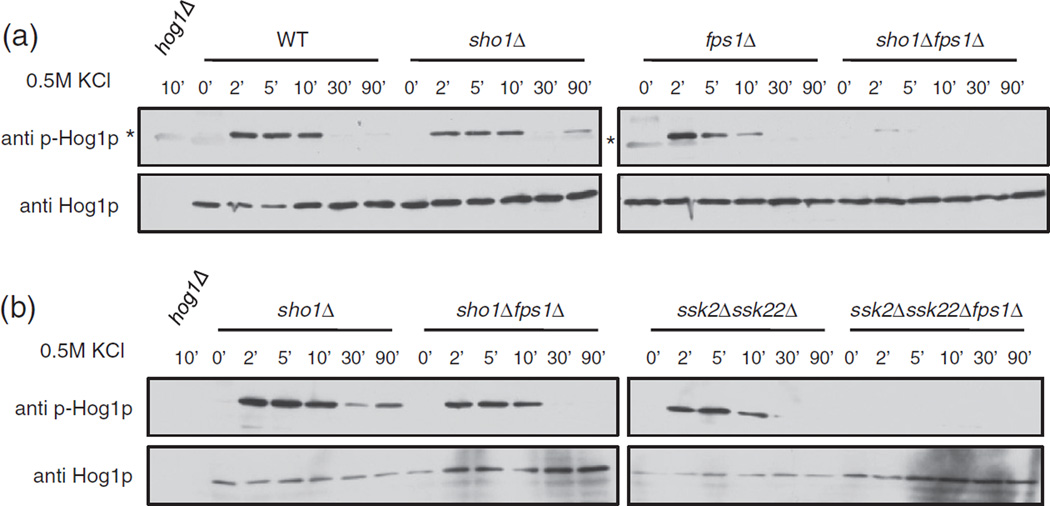
To investigate the possible role of FPS1 as a positive regulator of Sho1p, we decided to examine downstream signaling events. Specifically, we monitored the phosphorylation of Hog1p, a kinase involved in the HOG signaling cascade downstream of Sho1p. We examined whole cell extracts from wildtype, sho1Δ, fps1Δ, and sho1Δfps1Δ cells at various time points after the addition of 0.5 M KCl, and detected total Hog1p and phosphorylated Hog1p (p-Hog1p) with appropriate primary antibodies on a Western blot (Fig. 6a). In all strains, p-Hog1p was not detectable at time 0 (before salt addition). In the wildtype strain, the p-Hog1p signal increased when salt was added, peaking at 2–10 min, and then decreased to a constant low/undetectable level at 30–90 min after the addition of salt. Cells lacking the Sho1p-mediated upstream branch of the HOG pathway (sho1Δ strain) had overall lower Hog1p phosphorylation levels as expected, which decreased to a very low level by 30 min and was slightly increased by 90 min. Interestingly, in fps1Δ cells, p-Hog1p levels peaked at 2 min and gradually decreased at 5–10 min until there was no detectable p-Hog1p at 30 min. p-Hog1p levels in the double mutant strain, sho1Δfps1Δ, were lowered, with a slightly detectable peak at 2 min after salt addition, and no p-Hog1p detected after 2 min.
Fig. 6.
Hog1p phosphorylation is affected by FPS1. Cells, wildtype (WT) or with the indicated gene deletions, were grown and 0.5 M KCl was added for the indicated times in minutes. Growth was stopped by centrifugation and freezing of the cell pellets. Cells were then lysed, and proteins were electrophoresed onto an SDS-PAGE gel and transferred onto a nitrocellulose membrane. Phosphorylated Hog1p (p-Hog1p) and total Hog1p were visualized with appropriate antibodies. The exposure time in (b) is higher than that of (a). Asterisk (*) denotes a faint background band present in some anti-p-Hog1p blots.
To monitor the contribution of FPS1 to the regulation of signaling in the two branches of the HOG pathway, we tested for the Hog1p phosphorylation pattern in cells deleted in each of the Sho1p- and Sln1p-mediated branches (Fig. 6b). In cells in which only the Sln1p-mediated pathway is functional (sho1Δ), the absence of FPS1 (sho1Δ fps1Δ) resulted in a generally lowered amount of phosphorylated Hog1p, peaking at 10 min after the addition of salt. On the other hand, in cells in which only the Sho1p branch is functional (ssk2Δssk22Δ), the phenotype was much more severe when FPS1 was not present (ssk2Δssk22Δfps1Δ), as phosphorylated Hog1p could not be detected in the triple mutant. Taken together, our results support the model that Fps1p functions to positively regulate Sho1p, and a lack of FPS1 in the cell results in a change in the downstream phosphorylation of the HOG pathway kinase Hog1p.
Discussion
The maintenance of the internal environment in a cell, despite different and often extreme conditions in the external environment, is important for the maintenance of various functions essential for an organism's survival. Therefore, the study of the PPIs involved in the HOG pathway is important for determining the methods by which proteins come together to mediate a proper cellular reaction to extracellular stress. In this study, we took advantage of the MYTH method in budding yeast, already proven to be useful in the elucidation of protein interactors of several yeast and human integral membrane proteins [22,24,26], to search for physical interactors of Sho1p, and therefore find additional regulators of the Sho1p branch of the HOG pathway. Using the MYTH method, along with two other complementary PPI assays, we generated for the first time a high-quality Sho1p interactome, which includes 40 novel Sho1p interactors. The MYTH screen coverage was maximized by using both N- and C-tagged baits, expressed either ectopically from a plasmid or endogenously under native promoter control. In addition to previously known interactors, confirmation of this interaction set using two different assays showed that a large percentage of our interactors (80%) were able to interact under different assay conditions, whether stably by immunoprecipitation as shown by our co-IP data or under in vivo conditions (similar to the MYTH assay, but with a different tag and in diploid cells) as in our BiFC dataset. Our BiFC data also revealed that almost all of our confirmed interactions occurred at the plasma membrane, the cell compartment in which putative Sho1p regulators are likely to reside. Therefore, the interactors identified are unlikely to be spurious interactors but rather are physiologically relevant to the maintenance and regulation of the Sho1p branch of the HOG pathway. We identified proteins in a variety of cellular pathways, including 6 interactors with currently unknown functions. Follow-up experiments will allow us to determine what role our validated interactors may play in the HOG pathway or other Sho1p-mediated processes, perhaps as direct regulators of Sho1p itself.
Notably, we identified Fps1p as an interacting protein. This interaction was confirmed with a full-length Fps1p prey by MYTH, in addition to co-IP and BiFC methods, and was found to slightly increase in response to hyperosmotic conditions. It is important to note that Fps1p is the first integral membrane protein identified that can interact with Sho1p through the SH3 domain, a domain that has been shown to be important for binding to cytosolic Sho1p interactors [9–11,15]. Although the full-length Sho1p SH3 mutants that we tested did not seem to affect binding to full-length Fps1p in the MYTH assay, testing of binding of the SH3 domain itself, with the N-terminal fragment of Fps1p, revealed that the Sho1p SH3 residues I49, AY at residues 17–18, and Y54 are involved in binding to Fps1p. As Y54 is a key residue in the PXXP binding groove (surface I), I49 is the central residue on surface II [33], and Y18 is beside I49 in the Sho1p SH3 domain structure (PDB ID: 2VKN), it is likely that both surface I and surface II of the SH3 domain are involved in Fps1p binding.
Consistent with the involvement of the cytosolic Sho1p SH3 domain, we found that the membrane-localized portion of Fps1p is not able to bind Sho1p and that binding occurs mostly at the N-terminal cytosolic domain of Fps1p, with some contribution by the C-terminal cytosolic domain. Neither the N-terminal domain nor the C-terminal domain of Fps1p contains a PXXP motif; therefore, it is likely that the SH3 domain of Sho1p, through both surface I and surface II, binds via a non-canonical motif as has been reported for other SH3-domain-containing proteins [33,37,38]. In addition, it is possible that the previously reported reduced activity of the ΔAY mutant in HOG pathway activation [10] is due to its decreased binding to Fps1p. As both N- and C-termini of Fps1p contain residues important for its regulation [39,40], it would be interesting to narrow down the residues in the Fps1p cytosolic domains that participate in Sho1p binding and to determine the link, if any, to Fps1p regulation.
Fps1p is already a known component of the HOG response, as it closes rapidly to prevent the escape of glycerol in response to stress [32]. In the case of acetic acid stress, Fps1p behaves as a downstream component of the HOG pathway, as it has been reported to be phosphorylated by Hog1p resulting in its downregulation [41]. This also appears to occur with arsenite stress [42]. In the case of salt stress, it has been reported recently that Hog1p is involved in the phosphorylation of the Fps1p regulator, Rgc2p, by binding to the cytosolic N-terminus of Fps1p. Phosphorylation of the Fps1p-bound Rgc2p leads to its uncoupling from Fps1p and closing of the channel [34]. As we have shown that Sho1p appears to interact with Fps1p in the absence or presence of Hog1p or the Rgc regulators, we cannot conclude at this time if the Sho1p–Fps1p interaction occurs with the opened or closed state of Fps1p. Therefore, our finding that physical binding between Sho1p and Fps1p occurs at the plasma membrane implicates Fps1p in a novel role as an upstream component of the salt-induced HOG pathway, in mediating the HOG response at the level of Sho1p.
With the physical interaction of Sho1p and Fps1p at the plasma membrane confirmed and characterized, we next investigated the functional relevance of this interaction. Through monitoring the growth of various single, double, and triple deletion strains, we found that Fps1p plays an important role in the positive regulation of Sho1p-mediated pathway function. The regulation of Sho1p extends downstream through this branch of the HOG pathway, as an absence of FPS1 appears to result in a decrease in the amount of phosphorylated Hog1p. In order to conclusively determine whether this phenotype was linked to the Sln1- or Sho1-mediated pathway and not solely due to the absence of a glycerol channel, we tested the requirement for FPS1 under conditions where only one HOG pathway was functional. Although the downregulation of Hog1p phosphorylation was seen in both cases, the phosphorylation defect in the absence of FPS1 was more severe when only the Sho1p-mediated pathway was functional, and it correlated with the reduced growth of this mutant under salt stress. The smaller decrease in Hog1p phosphorylation levels between the sho1Δfps1Δ double mutant and the sho1Δ single mutant, in contrast, may explain the lack of a growth defect of the double mutant on 0.5 M KCl media, as sho1Δfps1Δ cells likely have enough Hog1p signaling in order to survive under these conditions. The fact that phosphorylation changes occur in this mutant, however, does suggest a potential association of FPS1 with the Sln1p-mediated pathway and warrants future investigation, perhaps under different salt shock conditions. It is also important to note that, in both the ssk2Δssk22Δfps1Δ and sho1Δfps1Δ mutants, the Hog1p phosphorylation defect was much worse than in fps1Δ cells, showing that the phenotype seen was likely not due to a decrease in glycerol escape in these cells.
The mechanism for the role of Fps1p in the Sho1p-mediated branch is currently unknown. Our preliminary analysis shows that it is likely not due to the disturbance of physical interactions between Sho1p and the osmosensor Msb2 or Sho1p's binding to the kinase Ste11p (data not shown). One possibility is that the physical binding between Sho1p and Fps1p may induce a conformational change in Sho1p that prevents binding to other untested HOG pathway proteins, resulting in faulty downstream signaling. A thorough investigation of other components of the large Sho1p protein complex, such as the recently identified Sho1p SH3 binder Ste11p [11], may uncover a defect in the recruitment or phosphorylation of one or more of these upstream components of the pathway when Fps1p is absent.
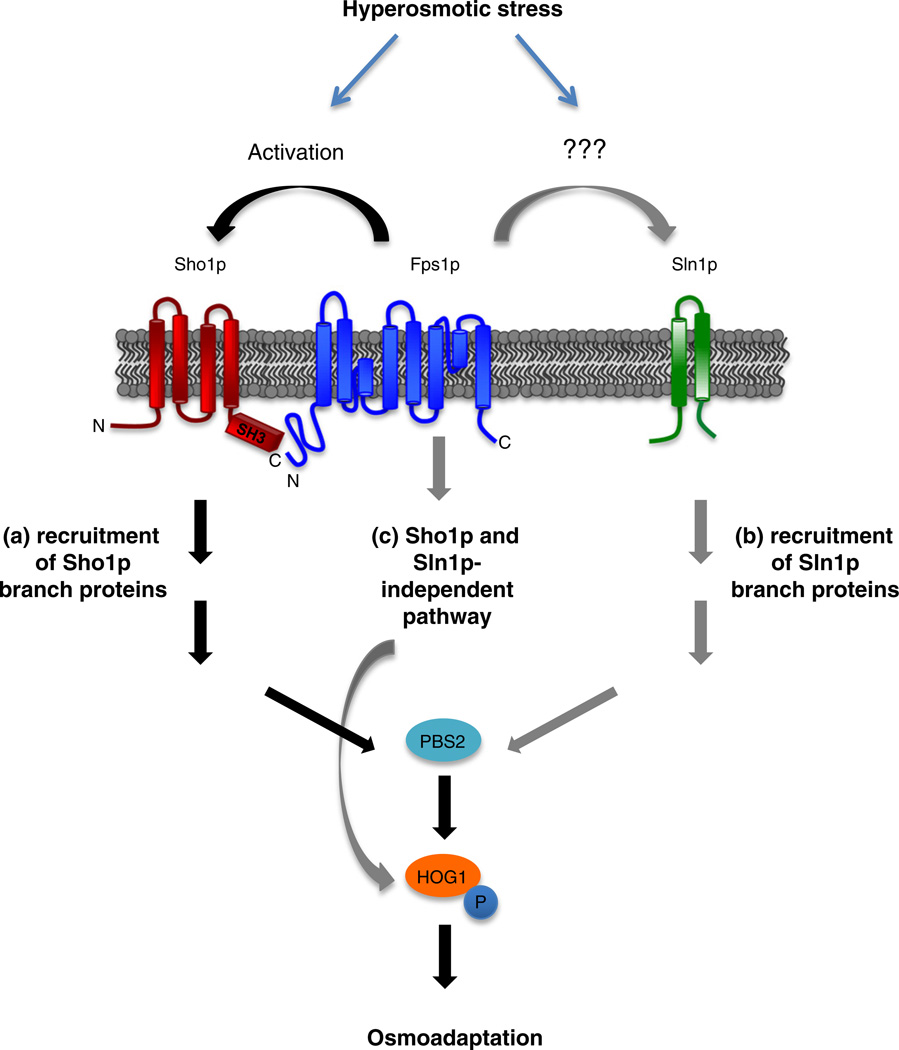
In a study looking at acetate stress, the activation of Hog1p was shown to be dependent on FPS1, although the phenotype seen in this case was quite severe, with almost no Hog1p phosphorylation present [43]. It was also reported that Hog1p phosphorylation levels were very mildly affected in fps1Δ cells under acidic, low-salt conditions (0.1 M NaCl), with slightly more sustained but overall lower p-Hog1p levels when compared to cells with functional Fps1p [43]. When stressed with acetate, regulation occurs through a feedback-based mechanism in which acetate internalized by Fps1 leads to the activation of the HOG pathway, Hog1p phosphorylation, and eventual phosphorylation and downregulation of Fps1p itself [43]. Our data, as summarized in Fig. 7, clearly demonstrate that p-Hog1p levels are moderately affected in fps1Δ cells at 0.5 M KCl and that this effect is dependent on Sho1p: Sho1p and Fps1p are present together at the membrane and are able to physically bind through the SH3 domain of Sho1p and the N-terminus of Fps1p, and cell growth and Hog1p phosphorylation are severely compromised without FPS1 and this effect was dependent on the Sho1p-mediated branch of the HOG pathway. An effect of Fps1p on the Sln1p-mediated pathway or on a Sho1p- or Sln1p-independent pathway cannot be excluded and warrants future study.
Fig. 7.
Fps1p positively regulates the Sho1p branch of the HOG pathway. Hyperosmotic stress triggers the activation of the Sho1p and Sln1p branches of the HOG pathway. (a) The presence of Fps1p positively regulates the Sho1p branch, possibly by binding to the SH3 domain of Sho1p, resulting in the phosphorylation of downstream Hog1p and osmoadaptation of the cell. (b) Fps1p may also positively regulate the Sln1p branch by an unidentified mechanism. (c) It is also possible that there exists a yet uncharacterized Sho1p- and Sln1p-independent pathway of Hog1p regulation by Fps1p.
In conclusion, the membrane interaction proteomics approach applied to Sho1p presented in this study provides a significant contribution in the understanding of the Sho1p-mediated HOG pathway in yeast. In addition, as budding yeast is an exceptional model for understanding similar pathways in other organisms, the data generated should increase our understanding of the regulation of mitogen-activated protein kinase pathways in general.
Materials and Methods
Strains, plasmids, and transformations
Cells denoted as “wildtype” are BY4741 or the MYTH reporter strain THY.AP4 or NMY51 unless otherwise noted. All yeast transformations were performed using a standard LiOAc/PEG protocol. ssk2Δ::HIS3, ssk22Δ::NAT, sho1Δ::URA3, ste20Δ::KANMX, fps1Δ::KanMX, rgc1Δ::KanMX, rgc2Δ::NAT, and pbs2Δ::KANMX, as well as the double rgc1Δ::KanMX rgc2Δ::NAT genomic deletion strains, were constructed by transformation into BY4741, BY4742 or the MYTH reporter strain NMY51 of a PCR product amplified from a plasmid template containing a drug or nutritional marker. Strains containing double or triple gene deletions were constructed by transformation or the mating of single deletion strains, sporulation, tetrad dissection, and selection of the expected nutritional/drug markers. fps1Δ::KanMX, sho1Δ::KanMX, andhog1Δ::KanMX were obtained from S. Hohmann (University of Gothenburg, Sweden) and C. Boone and B. Andrews (University of Toronto, Canada). rgc1Δrgc2Δ (DL3207) is from Ref. [36].
Sho1p (Sho1-Cub-TF) and Sho1ΔSH3p (Sho1ΔSH3-Cub-TF) baits tagged, C-terminally with the MYTH tag, at the endogenous locus of SHO1 was generated as described in Ref. [23]. Plasmid-based C-terminally tagged full-length Sho1p, Sho1-ΔSH3, and Sho1p SH3 point mutant baits were constructed by the amplification of the Cub-TF tag from pL2 [23] and co-transformation into yeast of this PCR product along with an EcoRI linearized pRS316 plasmid marked with URA3 containing wildtype full-length Sho1p, Sho1-ΔSH3, or Sho1p SH3 point mutants expressed from the endogenous Sho1p promoter. Sho1p SH3 mutants ΔSH3, Y54A, and ΔAY were previously described [10], and the I49A mutant was constructed similarly. The Nft1p MYTH bait was constructed as in Ref. [23] with pAMBV. The pCD4 bait (membrane tether) plasmid was from Ref. [23]. The SH3 domain of Sho1p (wildtype and point mutants) was inserted into pCD4 by PCR amplification of the SH3 domains from the plasmids described above, followed by standard gap repair into restriction digested pCD4. The N-terminal SH3 domain of Crk-II was PCR amplified from a Crk-II-containing plasmid [44] and cloned into pCD4 similarly.
pOst1-NubG and pOst1-NubI plasmids are as described in Ref. [23]. Full-length preys pNubG/I-Pbs2, pNubG/I-Ste20, and pNubG/I-Fps1, as well as Fps1p N-terminal (Fps1-N), C-terminal (Fps1-C), and transmembrane domain (Fps1-TM; amino acids 256–530) preys, were constructed as described in Ref. [23] by PCR amplification of PBS2 or FPS1 ORF (open reading frame) or the residues of interest from yeast genomic DNA, followed by integration by gap repair into restriction digested pPR3-N plasmids encoding NubG or NubI, as appropriate. pFps1 (Fig. 5d), expressing untagged full-length Fps1, was constructed by the PCR amplification of Fps1p from yeast genomic DNA with primers containing stop codons on the 3′ primer, followed by integration into the pCMBV MYTH bait plasmid [23]. All strain manipulations and plasmid constructs were confirmed through a combination of restriction enzyme digestions, PCR confirmations, and/or DNA sequencing.
Sho1p MYTH bait construction, validation, and MYTH screen
MYTH reporter strains THY.AP4 and NMY51, containing reporter genes HIS3, ADE2, and lacZ downstream of lexA operators, were obtained from Dualsystems Biotech. The N-terminally tagged Sho1p bait was constructed in the pBT3-N MYTH bait vector, and the integrated bait strain was constructed from the pL2 plasmid template as described in Ref. [23]. The MYTH screen itself was performed essentially as in Ref. [23], by the high efficiency transformation of 5 µg of an N- or a C-terminally tagged yeast cDNA or gDNA prey library into THY.AP4 containing either N- or C-terminally tagged bait. We obtained a coverage of 2-fold for each library (approximately 1 million transformants obtained, each library containing approximately 500,000 clones). Transformants were plated onto − WAH + 10 mM 3-AT (for C-tagged integrated Sho1p bait) or −WLAH + 50 mM 3-AT (for N-tagged, plasmid-based Sho1p bait) to select for cells containing interacting bait–prey pairs. Cells were also plated onto −W(integrated bait) or −WL (plasmid-based bait) to calculate transformation efficiency. After incubation at 30 °C for 3–5 days, transformants were patched onto the same interaction selecting plates, along with a second set of plates containing 5-bromo-4-chloro-3-indolyl-β,d-galactopyranoside (X-Gal). Prey plasmids containing putative interactors were then recovered (NucleoSpin 96-well miniprep kit), transformed into XL10 Gold Escherichia coli competent cells, purified from E. coli, and sequenced (Quintara Biosciences). From each screen, a maximum of 95 transformants were sequenced. Plasmids containing putative hits were then transformed back into a strain containing Sho1p bait, or an unrelated bait, Snq2p, to select for interactors that interact specifically with Sho1p. The interactome of Sho1p, containing both those interactors found through the screens themselves and preys that were verified individually, was visualized using Cytoscape v3.0.2 [45].
ONPG β-galactosidase assays
Reporter strains, harbouring both bait and prey plasmids, were grown to log phase (in media containing indicated concentrations of KCl as needed), pelleted, and resuspended in 1 ml Buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.35% mercaptoethanol, and 0.005% SDS); chloroform was added and vortexed. Cells were incubated at 30 °C for 15 min, followed by the addition of 150 µl of ONPG solution (13.3 M ortho-nitrophenyl-β-galactoside (ONPG), 78 mM KH2PO4, and 122 mM K2HPO4). The reaction was incubated at 30 °C and quenched by the addition of 1 M Na2CO3. OD420 of the supernatant was measured, and Miller units were calculated [units = 1000 × OD420/(time × volume × OD600)]. The assay was repeated for a number of samples (as indicated), followed by calculation of standard deviations. Statistical significance was calculated using a t test (Fig. 2, compared to 0 MKCl; Fig. 4, compared to CD4-Sho1p SH3 wildtype). In Fig. 4, results are plotted as a fold change relative to wildtype Sho1p SH3.
Confocal microscopy
To verify the proper localization of the MYTH baits, we grew cells containing both pPdr5-RFP and pTF-YFP-Cub-Sho1p overnight in plasmid-selecting media with 2% raffinose, re-grew them for 2 h in 2% galactose to induce expression of Pdr5-RFP, and examined them at a magnification of 630× using a Leica (DMI 6000 B) microscope with YFP, RFP, and DIC (differential interference contrast) filters. Image processing was performed with Volocity software (PerkinElmer). pPdr5-RFP was constructed by gap repair by the co-transformation of SmaI digested pCRGU(containing a galactose-induced promoter, RFP, and URA3 marker) and the PDR5 ORF was PCR amplified from yeast genomic DNA. pTF-YFP-Cub-Sho1p was constructed as in Ref. [24].
Confirmation of the Sho1p–Fps1p interaction by co-IP and BiFC
Cells containing a TAP tag [46], tagged at the genomic locus of the preys of interest, were transformed with a plasmid encoding C-terminally taggedSho1p-HAexpressed from an inducible galactose promoter [47].Cells were grown, lysed, immunoprecipitated, and Western blotted with rabbit anti-HA antibody as previously described [24].
BiFC analysis was performed as previously described [24] by genomic integration of the VC or VN tag (N- or C-terminal fragment of YFP Venus) at the N- or C-terminus of the ORFs of interest in BY4741 or BY4742, as indicated, followed by cell mating, diploid selection, and analysis of YFP fluorescence by microscopy. The location of the VC tag (at the N- or C-terminus of Sho1p) was selected such that the tag terminus corresponded accordingly between the MYTH and BiFC assays. VN-tagged bait strains were as described in Ref. [31]. Image acquisition and processing for Fig. 2b were performed using an epifluorescence microscope (E800; Nikon) and SlideBook software, and images in Supplementary Fig. 4 were captured using a spinning-disk confocal microscope (Ultra-VIEW VoX CSU-X1 system; PerkinElmer) and Volocity software, as previously described in Ref. [24].
Western blots
Cells were lysed by one of two methods. The first method was essentially as in Ref. [48], by the addition of 0.2 M NaOH, followed by a 10-min incubation on ice, and proteins precipitated with a further 10-min incubation with trichloroacetic acid. The sample was centrifuged at 12,000g for 5 min, and the pellet was resuspended in a mixture of 3× sample buffer and 1 M Tris base. In the second method, cells were pelleted, washed extensively with 10% trichloroacetic acid followed by 1 M Tris buffer or 1 M Hepes buffer, pelleted, and quickly frozen. Thawed pellets were resuspended in 5× sample buffer, bead beated with glass beads, and boiled. Proteins were electrophoresed on a 10% SDS-PAGE gel and transferred onto a nitrocellulose or PVDF membrane. The membrane was blocked with 5% bovine serum albumin or 10% milk in TBST. MYTH baits were detected with rabbit anti-VP16 antibody (Sigma) or anti-LexA antibody (Santa Cruz Biotechnology) and with equal loading monitored with anti-hexokinase antibody (Rockland Immunochemicals) or anti-RVS167 antibody.
Hog1p Western blots
Overnight cultures of a wildtype strain or strains containing a deletion in the genomic locus of the indicated genes were grown in YPAD liquid media. The saturated culture was diluted and re-grown for a further 3 h, salt-shocked with the addition of 0.5 M KCl, and pelleted at the indicated time points, and the pellet was quickly flash frozen. Cells were lysed, electrophoresed, and transferred onto nitrocellulose as described above. The membrane was blocked with 5% bovine serum albumin in TBST and probed with a rabbit polyclonal anti-Hog1p antibody to detect totalHog1p (Santa Cruz Biotechnology) or a rabbit polyclonal anti-p-p38 antibody to detect phosphorylated Hog1p (Santa Cruz Biotechnology) as indicated. A donkey anti-rabbit-HRP secondary antibody (GE Healthcare) was used, followed by detection by ECL (Pierce SuperSignal West Pico).
Supplementary Material
Acknowledgments
We would like to thank the Huh, Boone, Andrews, Hohmann, Levin, and Emili laboratories for strains and reagents; S. Stumpf, S. Ivantsi, and A. Hanif for assistance with experiments; B. Garcia and A. Zhu for plasmids and reagents; and all members of the Stagljar laboratory for comments and suggestions. This work was supported by grants from the Canadian Institutes of Health Research to M.B. and I.S.; grants from the Canadian Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, Ontario Genomics Institute, Canadian Cystic Fibrosis Foundation, Canadian Cancer Society, Pancreatic Cancer Canada, and University Health Network to I.S.; and a grant from the National Institutes of Health (R01-GM76375) to H.-O.P.
Abbreviations used
- HOG
high-osmolarity glycerol
- PPI
protein–protein interaction
- MYTH
membrane yeast two-hybrid
- TF
transcription factor
- BiFC
bimolecular fluorescence complementation
- co-IP
co-immunoprecipitation
- TAP
tandem affinity purification
- ONPG
ortho-nitrophenyl-β-galactoside
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2015.01.016.
References
- 1.Hohmann S. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett. 2009;583:4025–4029. doi: 10.1016/j.febslet.2009.10.069. [DOI] [PubMed] [Google Scholar]
- 2.O'Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 4.Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Ariño J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- 5.Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 6.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, et al. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatebayashi K, Tanaka K, Yang H-Y, Yamamoto K, Matsushita Y, Tomida T, et al. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 10.Marles JA, Dahesh S, Haynes J, Andrews BJ, Davidson AR. Protein–protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol Cell. 2004;14:813–823. doi: 10.1016/j.molcel.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka K, Tatebayashi K, Nishimura A, Yamamoto K, Yang H-Y, Saito H. Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci Signal. 2014;7:ra21. doi: 10.1126/scisignal.2004780. [DOI] [PubMed] [Google Scholar]
- 12.Saito H, Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- 13.O'Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bermejo C, Rodríguez E, García R, Rodríguez-Peña JM, Rodríguez de la Concepción ML, Rivas C, et al. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labedzka K, Tian C, Nussbaumer U, Timmermann S, Walther P, Müller J, et al. Sho1p connects the plasma membrane with proteins of the cytokinesis network through multiple isomeric interaction states. J Cell Sci. 2012;125:4103–4113. doi: 10.1242/jcs.105320. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, et al. Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci U S A. 2005;102:12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlecht U, Miranda M, Suresh S, Davis RW, St Onge RP. Multiplex assay for condition-dependent changes in protein–protein interactions. Proc Natl Acad Sci U S A. 2012;109:9213–9218. doi: 10.1073/pnas.1204952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, et al. An in vivo map of the yeast protein interactome. Science. 2008;80:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 20.Babu M, Vlasblom J, Pu S, Guo X, Graham C, Vizeacoumar FJ, et al. Interaction landscape of membrane–protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 21.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paumi CM, Menendez J, Arnoldo A, Engels K, Iyer KR, Thaminy S, et al. Mapping protein–protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Snider J, Kittanakom S, Damjanovic D, Curak J, Wong V, Stagljar I. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5:1281–1293. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- 24.Snider J, Hanif A, Lee ME, Jin K, Yu AR, Graham C, et al. Mapping the functional yeast ABC transporter interactome. Nat Chem Biol. 2013;9:565–572. doi: 10.1038/nchembio.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paumi CM, Chuk M, Chevelev I, Stagljar I, Michaelis S. Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J Biol Chem. 2008;283:27079–27088. doi: 10.1074/jbc.M802569200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MH, Kalaidzidis Y, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal. 2009;2:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 27.Reiser V, Salah SM, Ammerer G. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat Cell Biol. 2000;2:620–627. doi: 10.1038/35023568. [DOI] [PubMed] [Google Scholar]
- 28.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 30.Sung M-K, Huh W-K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein–protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 31.Sung M-K, Lim G, Yi D-G, Chang YJ, Bin YE, Lee K, et al. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res. 2013;23:736–746. doi: 10.1101/gr.148346.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamás MJ, Luyten K, Sutherland FC, Hernandez A, Albertyn J, Valadi H, et al. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Lee CD, Rath A, Davidson AR. Recognition of noncanonical peptides by the yeast Fus1p SH3 domain: elucidation of a common mechanism for diverse SH3 domain specificities. J Mol Biol. 2008;377:889–901. doi: 10.1016/j.jmb.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Reiter W, Dohnal I, Gregori C, Beese-Sims S, Kuchler K, et al. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 2013;27:2590–2601. doi: 10.1101/gad.229310.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wysocki R, Chéry CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, et al. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol Microbiol. 2001;40:1391–1401. doi: 10.1046/j.1365-2958.2001.02485.x. [DOI] [PubMed] [Google Scholar]
- 36.Beese SE, Negishi T, Levin DE. Identification of positive regulators of the yeast fps1 glycerol channel. PLoS Genet. 2009;5:e1000738. doi: 10.1371/journal.pgen.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kami K, Takeya R, Sumimoto H, Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J. 2002;21:4268–4276. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottger G, Barnett P, Klein AT, Kragt A, Tabak HF, Distel B. Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol Biol Cell. 2000;11:3963–3976. doi: 10.1091/mbc.11.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedfalk K, Bill RM, Mullins JGL, Karlgren S, Filipsson C, Bergstrom J, et al. A regulatory domain in the C-terminal extension of the yeast glycerol channel Fps1p. J Biol Chem. 2004;279:14954–14960. doi: 10.1074/jbc.M313126200. [DOI] [PubMed] [Google Scholar]
- 40.Karlgren S, Filipsson C, Mullins JGL, Bill RM, Tamás MJ, Hohmann S. Identification of residues controlling transport through the yeast aquaglyceroporin Fps1 using a genetic screen. Eur J Biochem. 2004;271:771–779. doi: 10.1111/j.1432-1033.2004.03980.x. [DOI] [PubMed] [Google Scholar]
- 41.Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorsen M, Di Y, Tängemo C, Morillas M, Ahmadpour D, Van der Does C, et al. The MAPK Hog1p modulates Fps1pdependent arsenite uptake and tolerance in yeast. Mol Biol Cell. 2006;17:4400–4410. doi: 10.1091/mbc.E06-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollapour M, Shepherd A, Piper PW. Presence of the Fps1p aquaglyceroporin channel is essential for Hog1p activation, but suppresses Slt2(Mpk1)p activation, with acetic acid stress of yeast. Microbiology. 2009;155:3304–3311. doi: 10.1099/mic.0.030502-0. [DOI] [PubMed] [Google Scholar]
- 44.Petschnigg J, Groisman B, Kotlyar M, Taipale M, Zheng Y, Kurat CF, et al. The mammalian-membrane two-hybrid assay (MaMTH) for probing membrane–protein interactions in human cells. Nat Methods. 2014;11:585–592. doi: 10.1038/nmeth.2895. [DOI] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 47.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam MH, Urban-Grimal D, Bugnicourt A, Greenblatt JF, Haguenauer-Tsapis R, Emili A. Interaction of the deubiquitinating enzyme Ubp2 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One. 2009;4:e4259. doi: 10.1371/journal.pone.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.