Abstract
Methods to monitor and manipulate the immune system are of enormous clinical interest. For example, the development of vaccines represents one of the earliest and greatest accomplishments of the biomedical research enterprise. More recently, drugs capable of “reawakening” the immune system to cancer have generated enormous excitement. But, much remains to be done. All drugs available today that manipulate the immune system cannot distinguish between “good” and “bad” immune responses and thus drive general and systemic immune suppression or activation. Indeed, with the notable exception of vaccines, our ability to monitor and manipulate antigen-specific immune responses is in its infancy. Achieving this finer level of control would be highly desirable. For example, it might allow the pharmacological editing of pathogenic immune responses without restricting the ability of the immune system to defend against infection. On the diagnostic side, a method to comprehensivel y monitor the circulating, antigen-specific antibody population could provide a treasure trove of clinically useful biomarkers, since many diseases expose the immune system to characteristic molecules that are deemed foreign and elicit the production of antibodies against them. This perspective will discuss the state-of-the-art of this area with a focus on what we consider seminal opportunities for the chemistry community to contribute to this important field.
Graphical Abstract
Introduction
The mammalian adaptive immune system allows us to survive in a world replete with infectious agents. Its two major branches, the cellular system, comprised of T cells, and the humoral system, comprised of B cells and antibodies, provide the means to recognize and neutralize almost any “foreign” molecule (called an antigen). At the core of this remarkable system are diverse receptors of the immunoglobulin family displayed on the surface of B cells (B cell receptors (BCRs) and T cell receptors (TCRs)). To a first approximation, the BCR or TCR from one particular B or T cell is almost identical to that from another cell, with the important exception of the small region of the protein that serves as the antigen-binding pocket of the receptor (Fig. 1)1. Here there is massive diversity resulting from the combinatorial nature with which the DNA encoding these regions is assembled2. Humans have on the order of 109–1010 distinct B cell and T cell receptors in what is called the pre-immune repertoire.
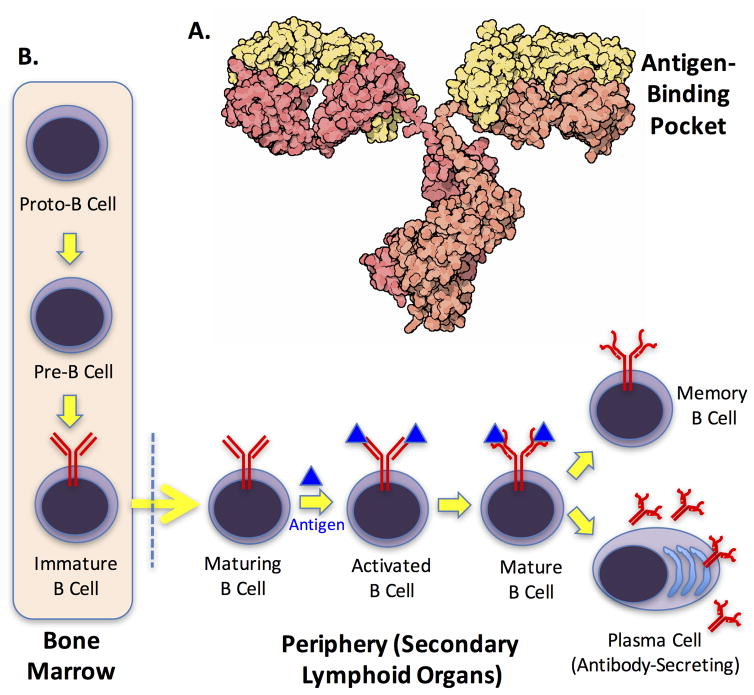
Figure 1. The humoral arm of the adaptive immune system.
A. Structure of an IgG antibody (courtesy of the Protein Data Bank Education Portal (http://pdb101.rcsb.org/motm/21). B. Schematic depiction of the development of B cells. The BCR is shown in red. The yellow arrow crossing the dotted line represents migration of B cells out of the bone marrow into peripheral, secondary lymphoid organs. It is activated upon binding an antigen (blue triangle) that is deemed “foreign”. The transition from an activated B cell to a mature B cell involves affinity maturation of the BCR, indicated by a change in shape. The mature B cell can go on to become a plasmablast, which secretes antibodies, or a memory B cell.
When a receptor engages an antigen that is deemed foreign it triggers intracellular signaling pathways that allow that particular clone to proliferate. Repeated stimulation can trigger a process known as somatic hypermutation1, which can produce receptors with much higher affinity for the foreign antigen (Nature’s version of medicinal chemistry). The B cells (but not T cells) that are stimulated by a foreign antigen can differentiate into plasma cells that pump out large amounts of antibody. The antibody essentially corresponds to a free-floating version of the B cell receptor (Fig. 1). Note that a single antigen can stimulate the proliferation of many different B cell or T cell clones. This is called a polyclonal response, which can occur in two ways. Either BCRs or TCRs with different (but similar) antigen-binding sequences can bind to the same specific region of the antigen (the precise region of the antigen that physically contacts the receptor is called an epitope). Alternatively, different regions of a single antigen could be immunogenic, resulting in the expansion of cells with receptors that are quite different from one another because they bind completely different epitopes displayed by a single antigen. Thus, even a limited number of antigens can produce a relatively complex polyclonal response, a point that will become important as we discuss efforts to monitor adaptive immune responses.
While the adaptive immune system presumably evolved to fight off invading organisms, it is integral to far more disease processes than infection. An obvious example is autoimmunity, a family of diseases in which one or more “self” molecules are mistakenly recognized as foreign and an attack is mounted against the tissues in which these molecules (called autoantigens) are located. For example, the underlying cause of type 1 diabetes (T1D) is an attack of the adaptive immune system on the pancreatic islet cells that produce insulin, resulting in their destruction. In such cases, the haywire immune response is the “bad guy” and the goal is to block the response. This can now be done, at least for B cells, using drugs that either kill off the entire B cell population, such as Rituximab,3 or prevent B cells from being activated in the presence of an antigen4,5, such as Ibrutinib.
On the other end of the spectrum, it is now understood that cancers form with disturbing frequency, but we are saved (most of the time) by a rapid and effective immune response against these “microtumors”. In this case, like infectious disease, the immune system is the “good guy” and it is in our interest to stimulate it. Indeed, it would be difficult to pick a hotter area in the pharmaceutical arena currently than immuno-oncology. Much of the enormous amount of investment flowing into this area aims to capitalize on the discovery of the basic mechanisms by which tumors eventually escape immune surveillance by restraining T cells from destroying them6. It turns out many tumors have “figured out” how to co-opt the natural mechanism by which T cells are “told” to “calm down”. This process, which is critical in preventing rampant autoimmune disease, involves binding of specific ligands to “checkpoint” receptors on the surface of T cells. Tumors present these ligands to T cells and thus restrain what would otherwise be a fatal assault by the T cells that recognize it. Drugs have been developed that block these interactions7, thus “reawakening” the T cells to the presence of the cancer. Remarkable results have been seen in the clinic using these drugs, even in cases of highly metastatic cancers.
While these and other developments are incredibly exciting, our ability to manipulate the adaptive immune system pharmacologically is limited in a fundamental way. The currently available drugs discussed above target proteins that may be B cell- or T cell-restricted, but are present in all B or T cells, regardless of their antigen-binding preference. Thus, when the cellular or humoral immune system is repressed or stimulated pharmacologically, all antigen-specific responses are affected, both good and bad. It is not currently possible to manipulate the adaptive immune system in a way that promotes or suppresses certain antigen-specific responses, but not others, with the major exception of vaccines. This is unfortunate, since one would like to kill off only the antigen-specific B or T cells that are driving an autoimmune reaction and thus not compromise our ability to deal with infections. Conversely, it may be advantageous to stimulate only the T cells that recognize a tumor without the risk of inducing systemic “cytokine storms”8 or triggering autoimmune disease. Clearly, targeting antigen-specific receptors on immune effector cells would have a significant impact on the treatment of many disease classes.
The lack of chemical tools with which to engage B cells, T cells, and antibodies in an antigen-specific fashion is also a roadblock to taking advantage of the enormous opportunity afforded by adaptive immune responses in the area of molecular diagnostics. There is clear evidence in some disease states that a condition-specific immune response predates symptoms by quite some time, often years. For example, autoantibodies against certain pancreatic islet cell antigens can be detected in the serum of patients that go on to become type 1 diabetics long before abnormal blood glucose levels become apparent9,10, reflecting the time it takes for the autoimmune response to wipe out most of the insulin-producing beta cells. It is reasonable to suspect that the adaptive immune system “knows” about many diseases at a pre-symptomatic stage, even if it is not driving the process11. If so, then the circulating antibodies produced against disease-specific antigens would make ideal biomarkers for diagnosis of the disease and for testing the effectiveness of treatment. It would be difficult to overstate the potential utility of simple blood tests that would reveal the development of serious diseases like cancers, neurodegeneration, etc. at an early, pre-symptomatic stage when available therapies are likely to be far more effective. However, to measure the level of a particular antigen-specific antibody, one requires a way to separate it from the large excess of other antibodies in the blood. This is usually accomplished by passing serum over an immobilized antigen, then measuring the amount of antibody retained with a labeled secondary antibody (for example, a rabbit antibody that binds human antibodies regardless of antigen specificity). Unfortunately, we simply do not understand enough about the molecular details of most diseases or how the immune system reacts to them to know exactly what antigen-antibody pairs to use for this purpose. Therefore, the development of unbiased methods to search for these putative immune biomarkers is an extremely high priority.
As is hopefully evident from the above discussion, a significant expansion of our ability to differentiate different antigen-specific antibodies, B cells and T cells could have an enormous impact on clinical medicine, both diagnostically and therapeutically. This Perspective will discuss progress towards the development of chemical tools for this purpose.
Synthetic Epitope Surrogates
Let us first consider the interesting challenge of suppressing a particular antigen-specific immune response without affecting other responses, a process we will call immune editing.
We chose to address this challenge in the context of chronic lymphocytic leukemia (CLL).12 CLL is the second most common form of blood cancer, with approximately 15,000 new patients diagnosed in the U.S. per year. In CLL, which is a hybrid of cancer and autoimmune disease, a single antigen-specific B cell clone is amplified relentlessly, crowding out healthy B cells from lymph nodes and other immune centers, eventually forming a tumor (Fig. 2A). Even after treatment, when patients relapse it is the same B cell clone that grows back12. This monoclonal response means that the pathogenic B cells must be reacting to a particular antigen, though the identities of antigens driving CLL are generally unknown.
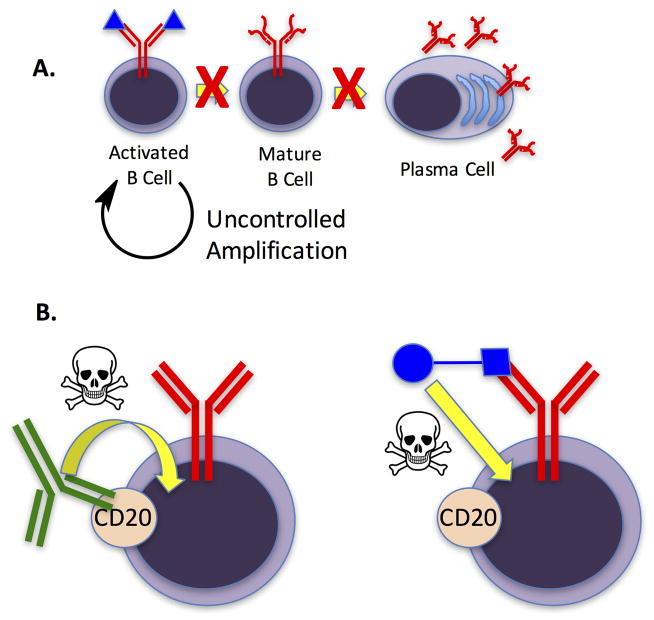
Figure 2. Chronic lymphocytic leukemia (CLL) as a useful test case for immune editing.
A. Cartoon depiction of the defects in B cell maturation observed for CLL B cells. This results in a lack of plasma cells that produce antibodies corresponding to the CLL B cell receptor (BCR). B. Left: Current drugs, such as Rituximab (green antibody) target B cell-restricted receptors such as CD20 and recruit immune effector functions (represented by the skull and crossbones) to kill the cells displaying this receptor at a sufficient density. This strategy eliminates all B cells. Right: An alternative would be to devise a synthetic molecule (blue square) capable of binding selectively to the antigen-binding site of the CLL BCR, which is not found on any other cell. Delivery of a toxin or some molecule that recruits immune effector functions (blue circle) would result in destruction of only the pathogenic B cells without compromising the ability of the immune system to respond to infections.
There exist good drugs for CLL. Antibodies that recognize the B cell-restricted receptor CD20, such as Rituximab, are quite effective in marking B cells for death by binding to CD20 and recruiting immune effector functions to them13 (Fig. 2B). Recently, Ibrutinib, an inhibitor of Bruton’s Tyrosine Kinase (BTK), a key player in the signaling cascade that activates B cells, has come on the market to great acclaim5. As mentioned above, however, these treatments kill or inactivate all B cells. While patients tolerate this remarkably well for some period of time, chronic suppression is undesirable. Thus, the idea of editing the pathogenic CLL B cells without affecting the remainder of the B cell population is attractive in the long run, particularly if this could be done early in the course of the disease prior to the pathogenic cells crowding out the healthy B cells.
A plausible way to do this would be to develop molecules that bind to the antigen-binding pocket of the pathogenic CLL BCR with very high selectivity (Fig. 2B). These could be conjugated to either a suitable toxin14 or a molecule that recruits immune effector functions15–17 (vide infra), hopefully resulting in the selective elimination of only B cells that display the targeted BCR. An extraordinary feature of CLL with respect to this strategy is that soluble antibodies corresponding to the pathogenic BCR will not be present in the circulation because CLL cells are defective in the maturation process that produces plasmablasts (Fig. 2A). Thus, CLL BCR-targeted drugs would not be “distracted” by soluble antibody targets.
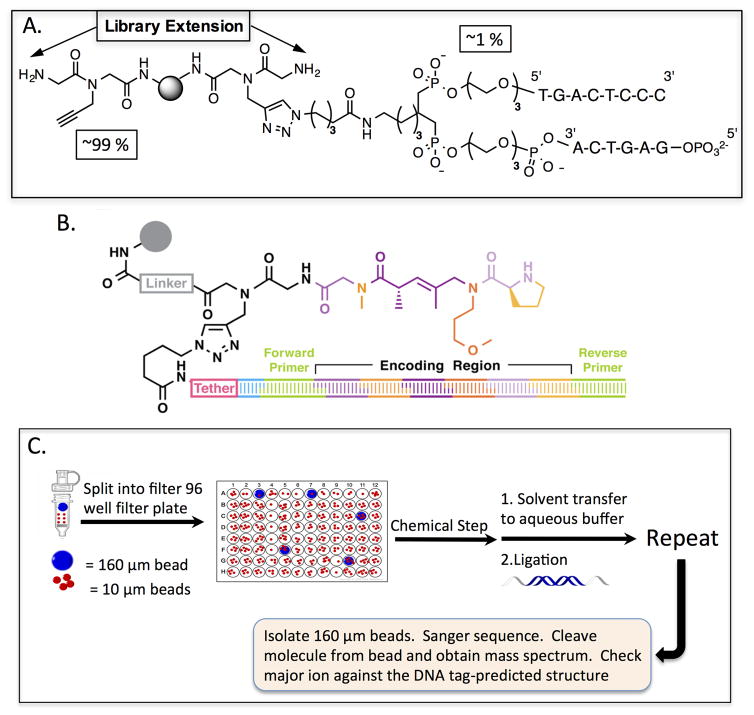
To attempt to identify ligands that bind selectively to these pathogenic BCRs, we developed a high-throughput screening protocol that employed a large library of bead-displayed oligomeric molecules. These libraries are made via split and pool synthesis18,19, resulting in each bead displaying many copies of a single molecule (a so-called one bead one compound (OBOC) library). This is depicted in Fig. 3. To screen these libraries, they are first denuded of ligands for conserved regions of the antibody and ligands for antigen-binding sites of antibodies that are not disease-related (Fig. 4). To do this, the collection of beads is first incubated with human serum and, after washing away unbound proteins, a fluorescently labeled secondary antibody that recognizes any human antibody is added. After another wash, beads that display a strong florescence, indicating antibody binding, are discarded. Presumably, promiscuous compounds that stick to many different proteins are removed in this step as well as uninteresting antibody ligands. To identify ligands to the CLL BCR antigen-binding sites, the remainder of the library is incubated with a soluble version of a patient-derived CLL BCR in the presence of a large excess of competitor proteins and again the antibody-binding beads are identified by fluorescence and isolated. These compounds are released from the bead by cleavage of a common linker and characterized by mass spectrometry20 (Fig. 4).
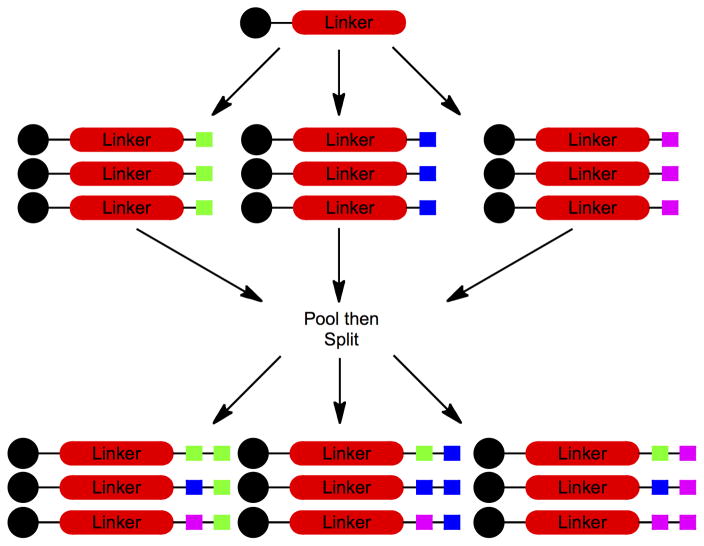
Figure 3. Graphic depiction of the solid phase split and pool synthesis scheme.
A simple example is illustrated of the creation of the nine possible dimers using three different building blocks at each position. The black circles represent the synthesis resin. The colored squares represent the synthetic building blocks (for example amino acids in the construction of a peptide library).
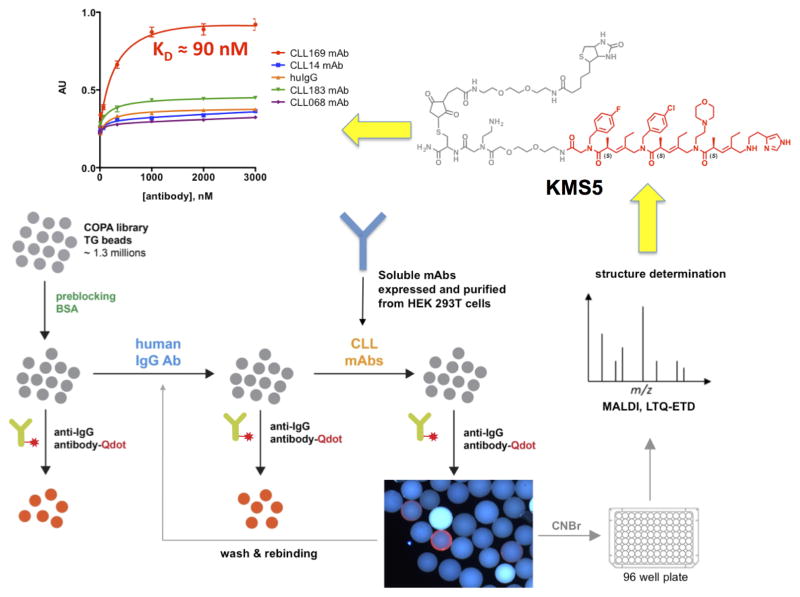
Figure 4. Workflow for the discovery of “epitope surrogates”.
An OBOC library (gray spheres) of over one million compounds comprised of a peptoid unit followed by three COPA21 (chiral oligomer of pentenoic amide) units was first cleared of ligands to uninteresting antibodies. This was done by incubation with a mixture of human IgGs from healthy donors followed, after washing, by addition of a quantum dot-conjugated secondary antibody. After another wash, beads that displayed a high level of fluorescence under a low power fluorescence microscope were removed from the library. The remainder of the bead population was then mixed with soluble versions of the CLL BCR targets. Beads displaying ligands for these species were picked using the same fluorescence-guided method. 16 strongly fluorescent beads were picked. The compounds were released from the beads and sequenced by mass spectrometry. After resynthesis with a new linker including a biotin tag (grey atoms in the chemical structure), the molecules were immobilized on an ELISA plate and binding of the CLL antibodies or control molecules was evaluated (top left graph). The best ligand for the CLL BCR 169 was KMS5 (shown in red), which bound to CLL 169 with a KD of 90 nM. Modified from ref. 22.
In a typical screen of this type against a particular CLL patient’s pathogenic BCR, 16 ligands were identified from a library of about one million compounds22. After re-synthesis and biophysical analysis, we found that the best of these ligands, oligomer KMS5 (Fig. 4), bound to the target antibody with a KD of 90 nM, as determined using an ELISA-like experiment in which immobilized ligand was titrated with increasing antibody22. A fluorescence polarization experiment using a fluorescein-labeled ligand free in solution indicated a KD of 400 nM. The difference almost certainly reflects avidity effects in binding of the bivalent antibody to immobilized KMS5 in the ELISA assay.
We were pleased with the relatively high affinity of this primary screening hit for the antigen-binding site of the antibody. Our first efforts in screening against antibodies employed libraries of peptoids23 (N-substituted oligoglycines). Large libraries of peptoids are easy to construct via iterative addition of the activated ester of bromoacetic acid to an amine followed by displacement of the bromide with a primary amine.24 But these compounds proved to be too “floppy” to provide high affinity ligands routinely. KMS5 is a member of a class of compounds that we call peptoid-inspired, conformationally constrained oligomers (PICCOs)25 in which bromoacetic acid is replaced with a bis-electrophile that contains some type of functional group that enforces conformational constraint. As will be discussed in more detail below, libraries of PICCOs have proven to be far superior sources of protein ligands.
When the fluorescently labeled compound was mixed with patient-derived CLL B cells, no specific binding was observed by flow cytometry, likely due to the low residence time of the ligand-BCR complex during the protocol. However, if the ligand was oligomerized by appending ≈ 20 copies to a biotinylated dextran oligomer26, binding to cells could easily be detected by using flow cytometry22. Gratifyingly, this association was highly selective for B cells displaying only the antigen-specific BCR that was employed in the screen. The ligand failed to interact with several other B cell clones tested. Not surprisingly, KMS5 proved to be highly serum stable 22 since it does not contain natural peptide bonds.
While KMS5 represents a non-optimized primary screening hit, these results show quite clearly that it is indeed possible to obtain highly selective, non-peptidic ligands for the antigen-binding sites of BCRs and antibodies.
Can a single epitope surrogate bind to a polyclonal family of related antibodies?
We have termed synthetic ligands for the antigen-binding sites of immune receptors “epitope surrogates”27. We prefer this term to “epitope mimics” because the latter suggests that the synthetic ligand will bind the immune receptor in the same way as the native epitope. This seems unlikely given that the native epitope and the synthetic ligand will have quite different structures, particularly if the building blocks employed to make the combinatorial libraries from which the surrogates are mined have side chains different from those in proteins. Of course, this remains speculation until structures of the same antibody bound to a native antigen as well as an epitope surrogate are available, which is not the case currently.
An important issue underlies these semantics. Will a synthetic ligand selected to bind a particular monoclonal receptor, let’s call it X1, bind to the other monoclonal receptors (X2 – Xn) that make up the polyclonal response to a particular epitope? The knee-jerk answer is of course it would, since the native epitope does so. Yet, these different monoclonal receptors, while having homologous variable regions are not identical. It could be the case that the ligand makes a critical contact with a residue in the binding site of X1 that does not exist in the sites of X2-Xn because that residue is not critical for binding the native epitope. This is a complication that is rarely encountered with probes that target non-immunoglobulin proteins. While a given protein or RNA target might differ from patient to patient due to single nucleotide polymorphisms (SNPs) in the gene, this level of variability is nowhere near that brought about by the combinatorial process that assembles the variable region of immune receptors combined with somatic hypermutation.
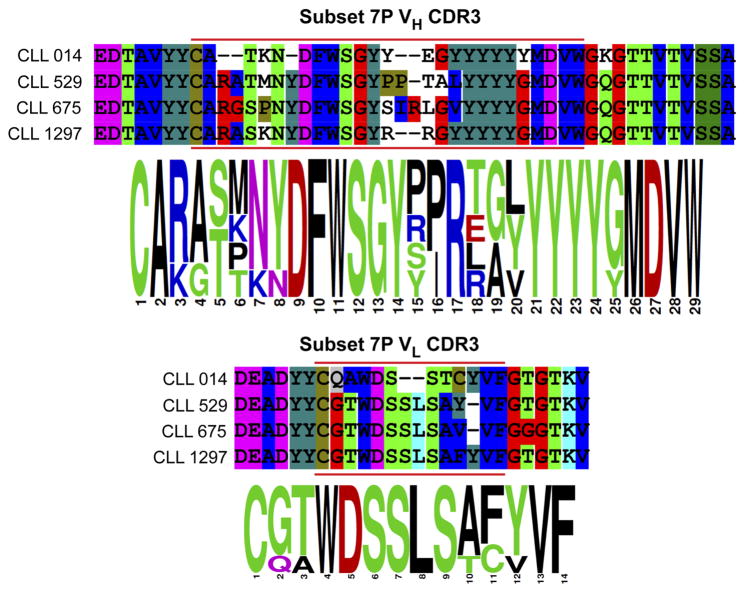
This issue can also be probed in a relatively straightforward way in the context of CLL BCRs. Not all CLL patients have the same or closely related pathogenic BCRs. DNA sequencing of the BCR heavy chain variable region-encoding genes from thousands of CLL patients revealed that only about a third of patients have BCRs that show sequence similarity to the BCRs from other CLL patients (Fig. 4)28. These are called stereotyped receptors. The other two-thirds are apparently unique. Moreover, even in the third of cases that are stereotyped, there are at least 19 different families of receptors, each one presumably reflecting a group of patients reacting to the same antigen. While any one patient is essentially monoclonal for the CLL BCR, different patients in that stereotyped group have related, but non-identical BCRs, as illustrated in Fig. 5. This constitutes a model for the polyclonal response to a given epitope in a single individual. Therefore, the ability of an epitope surrogate to recognize most or all of the different BCRs in a “polyclonal” group can be addressed by screening against a particular member of a stereotyped CLL group, then testing for binding to BCRs obtained from other CLL patients of the same stereotyped subset. This is a very important point with respect to developing drugs that would edit the CLL BCR. If an epitope surrogate does not bind to most or all of the BCRs in a stereotyped subset, then it would be necessary to develop a unique compound for each patient, which is clearly impractical.
Figure 5.
Sequence of the CDR3 region of the heavy and light chains of the BCRs from four CLL patients of the subset 7p stereotype.
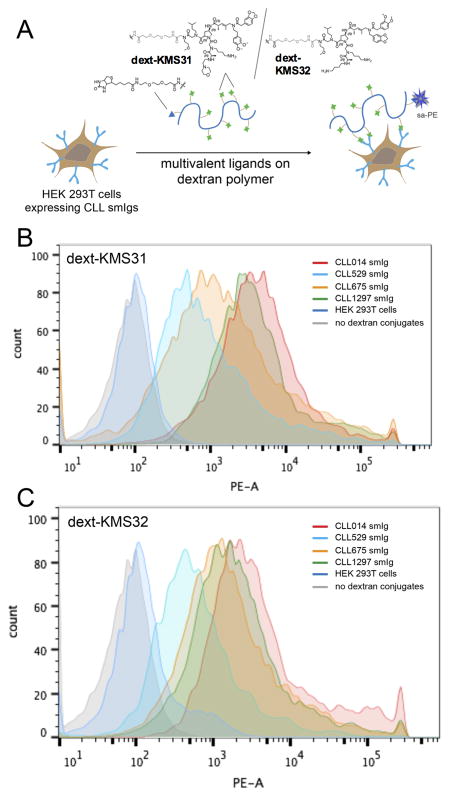
We recently completed a study addressing this question29. A CLL BCR was cloned from a patient (CLL014) belonging to the stereotyped subset 7p, which represents a highly aggressive CLL. The sequences of the heavy and light chain CDR3 regions of CLL014 and three other BCRs from patients in the subset 7p stereotype are shown in Fig. 5, illustrating the high level of homology, but also several differences. All of these BCRs were expressed as soluble IgGs, and the CLL014 IgG was used in a screen against more than a million oligomers using the same general protocol shown in Fig. 4. The two best hits obtained from this screen (called KMS31 and KMS32; Fig. 6) bound to the CLL014 IgG with KDs of 50 nM and 140 nM, respectively, as determined using ELISA assays, and are highly selective for CLL014 over BCRs from other CLL patients outside the subset 7p stereotype. They were then tested for binding to the soluble IgG versions of the other CLL BCRs from the stereotyped 7p subset. The data show clearly that the KMS31 and KMS32 indeed bind to the three other subset 7p BCRs. The lowest affinity interaction was with CLL1297, which KMS31 and KMS32 bound with 7-fold and 3.5-fold lower affinities, respectively29. These modest affinity differences were reflected in the binding of dextran conjugates of KMS31 and KMS32 to cells expressing these BCRs (Fig. 6). The highest affinity binding was to cells expressing membrane-anchored CLL014, but binding to cells expressing the other subset 7p receptors was observed at a level well above that of binding to control cells or cells expressing a CLL BCR outside of subset 7p (Fig. 6). These data show that epitope surrogates are capable of engaging different members of a polyclonal antibody population though, as one might have expected, there will be quantitative differences in the affinity for different clones in the population.
Figure 6. Flow cytometry analysis of ligand binding to cells displaying stereotyped CLL smIgs on HEK 293T cells.
(A) The cell binding assay with dextran conjugated multimeric ligands. CLL mAbs from subset 7p and other control CLL IgG were transiently expressed on the surface of HEK 293T cells. They were incubated with a biotinylated dextran polymer displaying 20–30 copies of the KMS31 or KMS32 ligands or controls followed by staining with phycoerythrin-conjugated streptavidin (sa-PE). (B) Histograms showing the binding of dext-KMS31 on the cells expressing stereotyped smIgs from subset 7P and controls, as analyzed by flow cytometry. (C) Same as B, but using dextran-conjugated KMS32. Reprinted with permission from ref. 25.
Synthetic analogues of antibody-drug conjugates
In order to edit an epitope-specific humoral immune response, whether in CLL or any other disease, the BCR-binding molecule must deliver some sort of “payload” that kills cells displaying ligand-binding BCRs (Fig. 2B). This work is in progress. The following two sections discuss strategies to achieve this.
An obvious possibility is to tether the epitope surrogate to a highly toxic molecule, such as monomethyl aurestatin (MMA) or an enediyne, in direct analogy with antibody-drug conjugates (ADCs).14,30 This would likely be the payload of choice if binding of the epitope surrogate triggers endocytosis of the BCR, which is likely, but has yet to be determined. In some ADCs, the linker connecting the antibody and the payload contains a protease site that results in its cleavage upon endocytosis. A similar strategy would likely be best for an epitope surrogate-drug conjugate. An important point is that it should be far simpler to construct conjugates with simple, synthetically manipulable epitope surrogates than is the case with antibodies. Indeed, the generation of homogeneous ADCs is a significant challenge31. Another point is that it should be straightforward to create conjugates that carry many equivalents of the payload, perhaps allowing the use of molecules that need not be quite as hyper-toxic as enediynes or MMA. For example, we have shown that conjugation of epitope surrogates that target BCRs or antibodies on a dextran oligomer reliably provides molecules able to bind with good avidity26. In other words, two of the displayed surrogates can occupy the two arms of the IgG simultaneously26, providing high affinity. There remain many sites on the dextran for further modification, which could be used to attach the payload. However, this strategy would make it difficult to prepare a single, defined species. Thus, an alternative would be to create a single, defined oligomer in which a large, but precise, number of payload molecules are attached to the epitope surrogate. For example, one could make a primary amine derivative of the payload molecule and employ the highly efficient peptoid sub-monomer synthesis32 to incorporate at least 10–20 equivalents of payload in the conjugate. An advantage of either approach is that different payloads could easily be included in the conjugate. This would discourage resistance arising to the conjugate resulting from mutations that reduce the efficacy of a single payload.
In summary, while much work remains to be done, it seems likely that completely synthetic molecules capable of editing epitope-specific immune responses will be available in the near future, at least for B cells.
While this discussion has focused on the use of epitope surrogates targeted to antigen-specific BCRs, it should not be terribly difficult to develop highly selective synthetic ligands for many different cancer-restricted receptors (CD19, HER2, etc.) using methods similar to those employed to obtain the BCR ligands (Fig. 4). Thus, it is quite reasonable to imagine that synthetic analogues of ADCs could be developed for a variety of cancers. From a commercial point of view, antibodies are attractive as targeting molecules due to their favorable pharmacological properties, including a long circulating half-life. However, it seems silly to employ a 150 kD carrier to deliver approximately two equivalents of a 600 Dalton toxin, particularly since the conjugation chemistry is challenging31.
Chemical dimerizers that target immune effector functions to blood cancers
As mentioned above, we do not yet know if it will generally be the case that engagement of a CLL BCR with an epitope surrogate will trigger internalization. If not, then an intriguing alternative to chemical toxins as the payload would be to tether the surrogate to a molecule that recruits immune effector functions to the target cell. In other words, develop a chemical dimerizer to recruit the killing power of the immune system to the pathogenic target cell. Chemical dimerizers were the brainchild of Stuart Schreiber, Gerald Crabtree and colleagues33–36, who were inspired by their discovery that the natural products cyclosporine and FK-506 work through a mechanism that involves the formation of a ternary complex in which the drug is sandwiched between two different proteins37. In other words, it heterodimerizes them. They went on to demonstrate that synthetic molecules comprised of ligands for two different signaling proteins connected by appropriate linkers could be used to manipulate many signaling pathways by forcing the proteins recognized into close proximity.
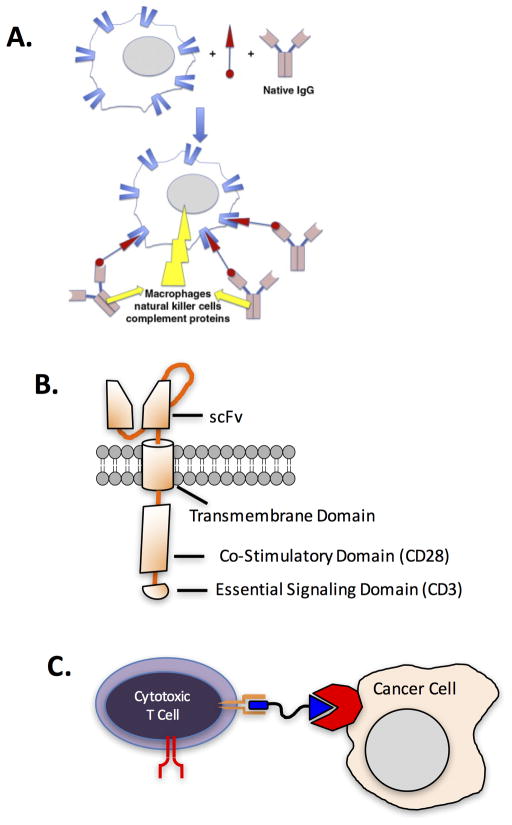
David Spiegel and colleagues have applied the chemical dimerizer concept to the goal of recruiting immune effector function to target cells that they wish to eliminate17,38,39, marking the first experimental realization of the concept shown in Fig. 7A. Specifically, they constructed dimerizers they call ARMs (antibody recruiting molecules) that are comprised of a receptor-binding molecule tethered to an antigen for antibodies common in almost all people, such as rhamnose or dinitrophenols (DNPs)40. The ARM thus attracts antibodies to a target cell displaying the receptor recognized by the small molecule. If there is a sufficiently high concentration of the target receptor on the cell surface, the antibody can recruit effector functions, such as the complement cascade or cytotoxic T cells, to kill the target cell. These workers have demonstrated the efficacy of this approach in vitro38 and in a mouse xenograft model for prostate cancer39. So in order to edit epitope-specific immune responses in general, and develop highly selective drugs for CLL in particular, one possibility is to create DNP- or rhamnose-epitope surrogate conjugates.
Figure 7. Chemical dimerizers to attract immune effector functions to a target cell.
A. Chemical dimerizers (red) that attract antibodies to a target cell. Reprinted with permission from ref. 41. B. Schematic illustration of a chimeric antigen receptor (CAR). The scFv (single chain fragment variable) determines the binding selectivity of the CAR T cell. C. A hypothetical chemical dimerizer (blue) that would mediate the co-localization of a target cell and a cytotoxic T cell. In this particular case, the dimerizer would bind to a cancer-restricted receptor (red) and a checkpoint receptor such as CTLA4 or PD1 (orange). The red forked shape represents the TCR, which would not be essential for cancer cell recognition in this scheme.
Another way to achieve the goal of bringing an antibody to the CLL cell is to create pre-formed epitope surrogate-antibodies chimeras. This can be done using the novel catalytic antibody developed by Barbas and co-workers42. This species, which was originally identified in a screen of antibodies capable of catalyzing aldol condensations43, is able to link covalently to any eta-lactam or 1,3-diketone (the former linkage is irreversible and thus preferred). An epitope surrogate linked to a eta-lactam unit could be mixed with the Barbas antibody to create a stable epitope-surrogate-antibody conjugate.
Alternatively, a cue could be taken from the exploding interest in chimeric antigen receptor T cell (CAR-T cell) technology44. In this approach, a patient’s own effector T cells are modified to express an artificial receptor comprised of an extracellular single chain variant (scFv) of an antibody, which includes the antigen-binding site, fused to an intracellular signaling domain (usually from CD3)(Fig. 7B). The latter results in activation of the T cell when the former binds to clustered antigens on the target cell. The CAR is essentially a kind of artificial TCR that reprograms the binding selectivity of a T cell. Whereas native T cells only recognize MHC-peptide complexes via their TCR, the CAR-T technology theoretically allows T cells to be directed towards any target of interest for which there exists a good antibody. Another way to look at it is to consider the CAR a dimerizer that attracts a cytotoxic T cell to a target cell. To date, CAR-T cells displaying an anti-CD19 (another B cell-restricted receptor) have shown great promise clinically in the treatment of B cell cancers44. There is enormous interest in expanding CAR-T cell therapy to other cancers, though this will likely require the discovery of target receptors that are truly unique to other cancers, since even modest expression of a CAR-T cell target on healthy tissue runs the risk of life-threatening toxicity. Another issue with the widespread use of CAR-T cells is that they will be expensive and tricky to make on an industrial scale.
It is quite interesting therefore to imagine directing native T cells to pathogenic CLL cells with a chemical dimerizer (Fig. 7C). Ideally, the T cell-binding unit in such a dimerizer would activate the T cell in addition to attracting it to the cancer target. There are several T cell-restricted receptors that might fit the bill as the target for this ligand. For example, two receptors displayed on the surface of T cells, called CTLA-4 and PD-1, are involved in quieting T cell responses. As mentioned above, these “immune checkpoints”7,45 are exploited by some tumors that display ligands to these receptors, thus blunting the immune response against them greatly. Antibody drugs have recently come into clinical use that antagonize these ligand-receptor interactions and thus “reawaken” the T cells to the presence of the cancer6. It seems reasonable that one could discover synthetic CTLA-4 and PD-1 antagonists that would serve as the T cell recruiting arm of the type of chemical dimerizer shown in Fig. 7C, since this might favor activation of the T cells recruited to the cancer cell by the other arm of the dimerizer.
The downside of systemic checkpoint inhibition is that it results in activation not of simply the tumor-associated lymphocytes, but many other T cells throughput the body. This can cause dangerous “cytokine storms” and even trigger the development of autoimmune problems8. Thus, it might be preferable to construct dimerizers comprised of a high affinity, perhaps even irreversible, ligand for the target cell, but only a moderate affinity antagonist for CTLA-4 or PD-1. In this way, one might achieve preferential activation of the T cells associated with the tumor target since this would result in the presentation of the T cell ligands in a clustered fashion on the tumor target, perhaps driving higher affinity through avidity effects.
Targeting TCRs
The above discussion, which has focused on CLL, involves editing antigen-specific B cells. But what about editing T cell responses? There are reasons to believe that this would be even more useful. Many autoimmune diseases are thought to be driven primarily by autoreactive T cells. If one wishes to target these diseases at their core and block the action of the autoimmune T cells, selective targeting of the pathogenic cells will be critical. There is no analogue of Rituximab for T cells. While patients tolerate complete B cell depletion remarkably well, at least for limited periods of time, T cell depletion is lethal.
The key reagents will be molecules that target the TCR with high affinity and selectivity. This is almost completely virgin territory. The native ligand of a TCR is a peptide bound to an MHC. Peptides alone will not bind to a TCR. The absence of any natural, low molecular mass ligands for a TCR may have discouraged any serious effort to find drug-like TCR ligands. Nonetheless, several years ago we decided to evaluate the feasibility of targeting an antigen-specific TCR46. The study design employed a mouse model for multiple sclerosis (MS), called experimental autoimmune encephalomyelitis (EAE)47. Mice are immunized with a myelin-derived peptide under conditions designed to break tolerance. The resultant anti-myelin autoimmune response results in demyelination and ultimately a neurological deficit. It is known that myelin-reactive CD4+ T cells play a prominent role in this MS model48.
To identify ligands for the TCR of anti-myelin CD4+ T cells, the entire population of spleen-derived CD4+ T cells was collected from EAE mice as well as control mice not immunized with the peptide antigen. These cells were labeled with red and green dyes respectively. A library of about 300,000 bead-displayed peptoids49–51 was then incubated with a mixture of the red- and green-labeled T cells. Beads that bound only to the red (EAE) T cells and not the green (healthy) T cells were visualized under a low power fluorescence microscope and manually removed from the population (Fig. 6). The peptoids they displayed were released from the beads and characterized by tandem mass spectrometry. The thinking behind this screening strategy is that the major difference between the EAE and control T cell populations would be a preponderance of anti-myelin T cells in the former. Thus, if a bead uniquely bound only the red-labeled cells, there was an excellent chance that the peptoid it displays is a target for the TCR of an anti-myelin T cell. This was indeed shown to be the case by a variety of follow-up validation experiments using one of these peptoids, called AG12A46. To estimate the affinity of this species for the antigen-specific TCR it recognized, a titration was done in which increasing amounts of a biotinylated DOPA-conjugate of the peptoid was added to T cells bearing the target TCR or to control T cells displaying a different antigen-specific TCR. Sodium periodate was added to trigger cross-linking of the DOPA moiety to a nucelophilic residue in the TCR52,53 and the amount of covalent product was assessed by flow cytometry after staining with labeled streptavidin. These experiments indicated that the KD of the peptoid-EAE TCR complex is on the order of 40–50 μM46. As mentioned previously, floppy peptoids generally do not exhibit high affinity for protein targets, so this weak binding was not surprising. However, binding did appear to be selective as no binding of the peptoid to the control TCR was observed. As another measure of binding selectivity, a conjugate of the EAE TCR-binding peptoid with Ru(bpy)32+ was synthesized. When photolyzed with visible light, this Ru(II) complex generates singlet oxygen efficiently. This results in the oxidative modification and inactivation of the protein to which the peptoid is bound54. It was demonstrated that incubation of the Ru(bpy)32+-AG12A conjugate with EAE T cells followed by irradiation ablated the ability of these T cells to proliferate when they were subsequently exposed to the myelin peptide antigen and antigen-presenting cells. In contrast, when the Ru(bpy)32+-AG12A conjugate was irradiated in the presence of T cells that recognized a different peptide antigen, it had no effect on their ability to proliferate.
These experiments remain, to best of our knowledge, the only published demonstration of selective binding of an antigen-specific TCR by synthetic ligands. While the affinity of the TCR-binding peptoids was unimpressive, this at least constitutes proof of principle that it is feasible to identify ligands able to distinguish between antigen-specific TCRs. Of course, to even begin to think about editing antigen-specific T cell responses much better ligands for TCRs will be required. As is discussed briefly below, we are hopeful that libraries of far more conformationally constrained molecules will serve as a rich source of much higher affinity ligands. If such molecules were in hand, then one could imagine deleting clonal T cell populations in the same way as was discussed above for B cells.
Can antibody profiling provide a general route to pre-symptomatic diagnostic tests?
Most of the discussion above has focused on manipulating antigen-specific immune responses through chemical editing. But what about using similar technology to simply monitor immune responses? We believe that this provides enormous opportunities in the diagnostic realm27. There has long been speculation that the adaptive immune response could serve as a treasure trove of biomarkers, promising a revolution in medical diagnostics11. The idea is that the pathophysiological chemistry of a particular disease will expose the immune system to molecules that are deemed foreign and thus trigger the production of antigen-specific B cells, T cells and antibodies against them. Antibodies are perhaps the most interesting candidate biomarkers in this regard since they are so readily available from blood. If we could learn to associate a particular set of antibodies with a disease state, then monitoring the levels of these antibodies in the blood would constitute a simple diagnostic test. An attractive feature of this idea is that it seems highly likely that the immune system would “know” about a disease long before symptoms were apparent. Thus, if a panel of such tests were routinely available and could be administered at an annual physical, for example, then many diseases could be caught at an early stage, leading to far more effective treatment in many cases. Another advantage of this approach is that it would be relatively straightforward to create highly multiplexed tests to monitor dozens of IgG antibodies in the blood simultaneously, as will be described in more detail below. This is important if the goal is pre-symptomatic diagnosis, since it would require only a single, small, sample of blood from the patient and could presumably be done for a relatively low cost.
The rate-limiting step in realizing this dream is, of course, the discovery of the right antibodies to measure as disease-specific biomarkers. The most common approach is to test candidate antigens as capture agents for antibody biomarkers. If one knows enough about the mechanism of the disease, it may be possible to make an educated guess about what proteins or other biomolecules might trigger an adaptive immune response. This candidate antigen can be immobilized on a surface, such as an ELISA plate, and incubated with patient serum. After washing, the level of captured antibody is then measured easily by subsequent hybridization with a secondary anti-human IgG antibody labeled with a suitable probe, such as a fluorescent dye. In other words, the disease-specific antigen acts as a “capture agent” for the antibodies of interest, separating them from the millions of other antibodies present in the blood so that their levels can be measured.
Unfortunately, it is rarely the case that we know enough about the disease state and the way in which the immune system responds to it that the candidate approach proves fruitful. Thus, some sort of search for interesting antibodies has to be done. One popular approach is to conduct a differential screen in which some collection of candidate antigens is exposed to case and control serum with the intent of identifying a molecule that retains far more antibodies from the diseased samples than the controls. This approach has been tried many times with protein55,56, lipid57,58, glycan59 or peptide arrays60–63, but it has a poor success rate. Obviously, this will only work if the native antigen is in the collection of molecules tested against the serum samples. If, for example, the native autoantigen is a protein that is post-translationally modified in some unusual way, then it will not be present on an array of recombinant proteins. Perhaps this is why the success rate is low. The fact that many of the major rheumatoid arthritis autoantibodies recognize citrullinated proteins64 is a case in point.
A better, though more laborious, approach is to use case and control serum samples to stain patient-derived tissue. If one observes a higher level of antibody staining from the case rather than the control samples, then it provides a starting point. In favorable cases, staining different tissues with case serum can provide a clue as to what the candidate antigen might be 65 given that much is known about gene expression profiles in many different cell types. A good example of this was the discovery by Lennon and co-workers that the water channel protein Aquaporin 4 (AQP4) is the antigen recognized by pathogenic antibodies that drive the autoimmune disease neuromyelitis optica (NMO)65–67. Serum autoreactivity to AQP4 now serves as a reliable blood test for this disease68,69. If tissue distribution does not provide sufficient clues, biochemical fractionation of other approaches can be used to separate the many different molecules in a tissue extract and these can be tested for antibody binding until the antigen is discovered. The major advantage over probing recombinant protein arrays and the like is that the tissue should have the antigen present in its native form.
A very promising method to search for disease-specific immunoglobulins is to deep sequence the variable regions of tens of thousands of circulating B cells70. For some time, this technique was limited by the fact that it is impossible to maintain native pairings of heavy chain- and light chain-encoding RNAs when a pool of cells is lysed. But methods have been published recently that allow the separation of 10,000–50,000 individual plasmablasts into different wells of a microtiter plate. Well-specific barcoding of the PCR primers employed to amplify the CDR regions of the genes allow the paired heavy and light chain sequences to be recognized even after all of the PCR amplicons are mixed together prior to deep sequencing71–73. A similar strategy could be employed for T cell receptor (TCR) characterization. A significant limitation of sequencing plasmablasts is that under normal circumstances the number of circulating plasmablasts is low. Many antibodies are produced by non-circulating B cells, for example those residing in the bone marrow. Thus, it is not possible to use plasmablast sequencing as a surrogate for the circulating antibody population without resorting to painful, invasive biopsies. There are some exceptions, such as soon after vaccination when large numbers of plasmablasts are found in the circulation, and this has been the main application of this technology to date70,71,73, though a paper on characterizing autoimmune B cell populations has appeared74. Only recently has direct characterization of circulating antibody populations been attempted using mass spectrometry-based proteomics.72,75 This is a daunting technical challenge. Moreover, identification of the peaks representing peptides from the antigen-binding site is currently dependent on guidance from deep sequencing information, so it does not surmount the limitations of inaccessibility of many plasmablasts.
In favorable cases such as post-vaccination, the sequencing data provide a beautiful overview of the different clones that arise during an autoimmune response, allowing one to follow the evolution of an immune response as never before70. While it is impossible to glean anything from these data regarding the antigen recognized by disease-specific antibodies identified by deep sequencing, monoclonal antibodies can be cloned and expressed, then used to “fish” for the native antigen in an appropriate tissue extract (vide infra). Thus, for some conditions, the discovery of disease-specific antibodies through deep sequencing is likely to be a productive endeavor.
High-throughput screening for epitope surrogates
While the approaches described immediately above are exciting, we felt there remained room for the development of new antibody profiling technologies. Therefore, about eight years ago we began to explore a different approach in which we conducted differential serum screening experiments to identify synthetic ligands that bind antibodies rich in the serum of patients or animals with a given disease, but they were absent or present at much lower levels in the control sera. We were not searching for the native antigen at this stage, but rather an epitope surrogate27. A rough analogy to this idea would be drug development. Many drugs occupy the same surface of an enzyme as a native substrate, yet they do not obviously resemble that substrate structurally. We hope that by using libraries with a large diversity of chemical descriptor space, molecular surrogates for a variety of post-translationally modified or otherwise unusual antigens could be discovered.
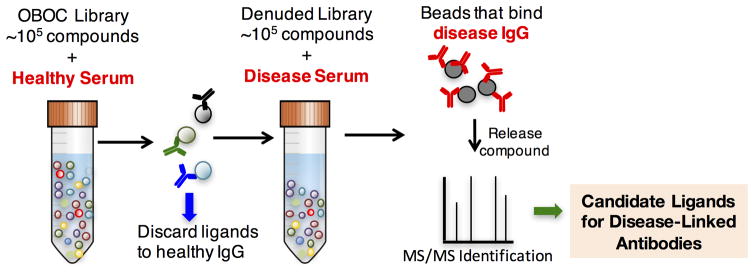
The workflow that we have developed is shown in Fig. 8. A pool of control serum samples is incubated with an OBOC library of 105–106 different molecules. The beads are TentaGel, comprised of an amine-functionalized polystyrene core onto which is grafted a thick coating of amine-terminated polyethylene glycol that resists non-specific protein binding51. Beads that retain significant amounts of uninteresting antibodies at this stage are visualized by subsequent addition of a secondary antibody attached to a quantum dot76 or magnetic microspheres77. They are then removed from the population manually under a low power fluorescence microscope76 or by using a powerful magnet77,78. The remainder of the library is then incubated with a pool of serum sample obtained from patients or animals with the disease of interest. Again, beads that retain significant amounts of antibody are visualized with a labeled secondary antibody and collected. These are the epitope surrogate candidates. In some cases, these beads are stripped of antibody and re-subjected to the entire process using different control and case serum samples to ensure that they are likely to be bona fide ligands for interesting antibodies79. Finally, the compounds are released from the resin and their structure is determined by tandem mass spectrometry20,22.
Figure 8.
Workflow for the discovery of synthetic epitope surrogates. See text for details.
Optimization of the system
This process works and we reported its successful application to two autoimmune diseases where the nature of the offending autoantibodies was already known: an animal model for multiple sclerosis47 and the human disease neuromyelitis optica (NMO)65. In each case, we obtained synthetic molecules that were indeed ligands for the antigen-binding site of disease-linked antibodies80,81. We also reported promising preliminary results that indicated there may be autoantibodies associated with human Alzheimer’s disease 80, a contention supported soon thereafter by other studies using different techniques 63,82. However, our progress was quite slow due to a number of technical limitations27, so before attempting to apply the technique widely, we instead focused on addressing these issues.
First, screening combinatorial libraries on beads has a notoriously high false positive rate83,84. False positives are beads that “light up” as though they display high affinity ligands for the protein target, but when the compound is re-synthesized and tested for binding in solution or even when immobilized on a different surface, it binds the target poorly or not at all. A great deal of time and effort was wasted on the re-synthesis and characterization of these ultimately useless compounds. This problem is not unique to serum antibody screening.83 Eventually, we discovered that a major reason false positives are so common is that the density with which compounds are displayed on the surface of TentaGel bead can vary enormously from bead to bead84, perhaps an unavoidable consequence of the grafting process by which they are made. A small percentage of the beads display the compound at a very high density. Thus, even if a bead-displayed molecule is a terrible ligand, when the target protein enters this “molecular kelp forest”, it never escapes due to the enormously high local concentration of the displayed compound and the bead is scored as a hit. Thankfully, these very high-density beads are rare in the population. Thus, a simple way to avoid being fooled by them is to use a redundant library in which several beads display the same compound (i.e., the number of beads used in the library synthesis is several times larger than the theoretical diversity of the library). This is because it is highly unlikely that, in a large library, a given compound would be found more than once on these relatively rare, ultra-dense beads. Indeed, we have found that redundant hits are almost always bona fide ligands, while “singleton” hits have a much higher chance of being false positives.84 While this sounds simple, it represented an enormous technical advance in the bead screening field.
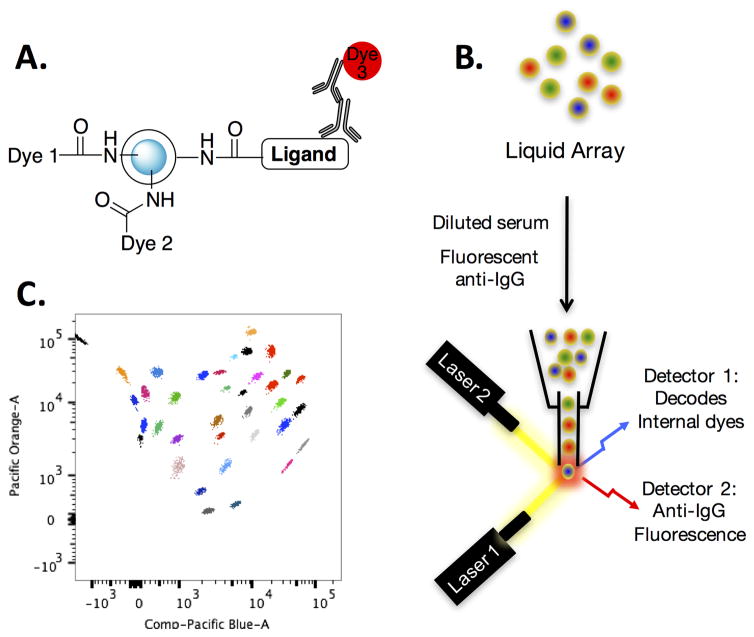
Another bottleneck was how to test screening hits rapidly against large numbers of serum samples in order to evaluate whether they are useful leads or not. Early attempts to use peptoids affixed covalently to maleimide-coated ELISA plates did not work well, due to poor signal to noise. This was likely a combination of the low affinity of the peptoids for serum antibodies and the propensity of antibodies to bind non-specifically to the plastic surface of ELISA plates. Moreover, ELISA assays are relatively consumptive of serum and many samples are available only in small amounts. Thus, we developed a multiplexed assay that uses color-coded TentaGel microspheres as the scaffold (Fig. 9). The 10 μm TentaGel beads were color coded by linking a particular amount of the dyes Pacific Blue (PB) and Pacific Orange (PO) covalently to the hydrophobic interior of the bead that is inaccessible to protein. The amine groups on the hydrophilic exterior region of the bead were bromoacetylated and then the appropriate ligand was attached via thioether formation. The level and ratio of the PB and PO dyes encodes the identity of the ligand on the surface. After washing, the amount of antibody retained on each is marked with a secondary antibody carrying a third color. After another wash, the beads are analyzed using a flow cytometer, since the 10 μm TentaGel beads are about the size of a small mammalian cell. As each bead passes single file through the analyzer, the PB/PO intensity and ratio identify the ligand displayed on the surface and the third channel measuring the dye affixed to the secondary antibody quantifies the amount of primary antibodies captured on the bead (Fig. 9). Up to 50 independent ligand-antibody measurements can be made using only 1 μl of serum, making it ideal for the analysis of precious samples. Our system is inspired by one marketed by the company Luminex, which invented the bead coding technology85, but differs in a few important details that make it more effective for monitoring small molecule-antibody interactions. For one, TentaGel microspheres are coated with 50–80% w/w with polyethylene glycol (PEG), shielding the “sticky” polystyrene core from deposition of IgG nonspecifically, reducing background noise inherent in serological assays. Additionally, the encoding dyes are linked covalently to internal amines in our system, but only absorbed physically into the latex beads employed by Luminex. Covalent attachment precludes the dyes from “leaking out” during coupling of the synthetic ligand to the surface, which is done in organic solvents.
Figure 9. Multiplexed analysis of small molecule-antibody interactions in serum samples.
A. Depiction of the beads that display a ligand on the surface and contain a mixture of two dyes in the protein-inaccessible interior. B. The beads are incubated with a small amount of serum (≈ 1 μl) followed by a fluorescently labeled secondary antibody. They are then analyzed using a flow cytometer with two lasers. One “reads” the code and the other quantifies the amount of secondary antibody on each bead. C. A sample of the type of data that are obtained in this experiment. See ref. 86 for details.
Discovery of epitope surrogates and native antigens in the mouse model for type 1 diabetes (T1D)
With these important technical improvements established, we proceeded to attempt to apply this technology to the discovery of novel epitope surrogates and native autoantigens in T1D. T1D is, in our opinion, an excellent system in which to further develop this approach. As mentioned above, the cause of T1D is an autoimmune assault on the pancreas, which destroys the insulin-producing beta cells. Without insulin, it is impossible to control blood sugar appropriately. Autoreactive antibodies and T cells appear years prior to the onset of insulin deficiency87. This means that it takes the autoimmune reaction quite some time to destroy enough of the insulin-producing beta cells to finally push the patient into crisis. Unfortunately, about a quarter of T1D goes undiagnosed until this crisis point, which involves the patient going into ketoacidosis. This is especially true in infants and several die each year from this condition88. This highlights the desirability of having a blood test for T1D that is capable of picking up oncoming disease at a pre-symptomatic phase so that insulin treatment can be started prior to the “crash”. Moreover, exciting advances in making pancreatic beta cells on a large scale from stem cells89 holds out the prospect of Type 1 diabetics receiving routine islet transplants, once a suitable container is devised for these cells that would shield them from immune attack but allow insulin and glucose to diffuse freely. Again, early detection of the disease would be highly desirable to allow such a transplant to be done far in advance of the complete loss of native insulin production.
The good news is that pre-symptomatic detection of the disease is possible. Several T1D-specific autoantigen/autoantibody pairs have been discovered painstakingly over the last few decades using the candidate approach90,91. These include insulin, GAD6592, IA-293 and ZnT894. The bad news is that many of these T1D autoantigens perform poorly in ELISA assays, apparently because fixing them to a surface disrupts interaction with the autoantibodies. This has forced investigators to employ radioimmunoassays (RIAs) using soluble antigens to measure T1D autoantibody levels10,95–97. RIAs are tedious and notoriously challenging to reproduce. This is a major problem. Since less than 5% of children tested randomly will have T1D-related autoantibodies, a blood test must be extremely convenient and economical in order to screen the general population periodically, for example at an annual physical. The RIA does not remotely qualify. Recently, measurement of anti-insulin and anti-GAD65 autoantibodies by electrochemiluminescence using the MesoScale Discovery (MSD) platform has been achieved,98,99 which employs soluble, labeled autoantigens and thus avoids their immobilization. More esoteric platforms are also being considered.100 Whether any of these efforts will result in a blood test that could be used clinically in a routine fashion remains to be determined. Finally, there remains a need for the discovery of additional T1D biomarkers, because about 20% of T1D patients escape detection using the known autoantigens. Clearly, there remains a need for more and better pre-symptomatic T1D antibody biomarkers and improved methods for monitoring them in a multiplexed fashion.
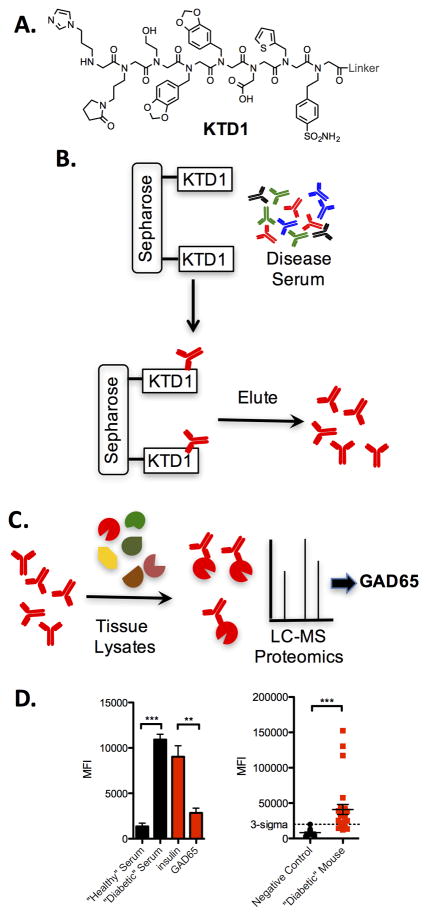
Our initial investigations used a mouse model of T1D called the non-obese diabetic (NOD) mouse101 to test the second-generation antigen surrogate screening approach. Like human patients, NOD mice develop hyperglycemia after a long, asymptomatic period. A screen using the basic protocol shown schematically in Fig. 8 was carried out using an OBOC peptoid library of octamers (this was done prior to the availability of PICCO libraries). Of the four compounds that were identified as hits after this rigorous screen, only one, peptoid KTD1 (Fig. 10A), was isolated on two different beads, so we focused on characterization of this putative autoantibody ligand. KTD1 validated nicely. In the Luminex-like flow assay (Fig. 9), KTD1 retained 4–5 times more antibodies from the serum of a particular NOD mouse than a control mouse79. Importantly, the binding of antibodies from NOD mouse serum was competed almost entirely by an excess of soluble KTD1. We have found it critical to perform these competition experiments, as well as other controls, as some serum samples have high levels of “sticky”, promiscuous antibodies that can bind non-selectively to many different immobilized molecules. When dozens of serum samples obtained from different mice were analyzed, we found that KTD1 was able to pick up T1D autoantibodies from 40% of diabetic mice. Unfortunately, there was also a 10% false positive rate (2/20 control mice evinced above-background levels of antibody binding to KTD1). This represents off-target binding to antibodies not relevant to T1D.
Figure 10. Discovery of a synthetic surrogate for a GAD65 epitope.
A. Structure of the compound identified in the screen (KTD1). B. Schematic representation of the protocol employed to enrich serum antibodies that recognize KTD1. C. Cartoon of the immunoprecipitation protocol employed to characterize the protein recognized by KTD1-binding antibodies in a pancreatic tissue lysate. The major candidate was GAD65. D. Left: Levels of IgG antibodies bound by immobilized KTD1 from serum obtained from a healthy mouse or a diabetic mouse (black bars). Addition of excess soluble GAD65, but not soluble insulin, competed binding of antibodies from the T1D serum sample to immobilized KTD1 (red bars). Right: Level of IgG antibody binding to immobilized GST-GAD65 from healthy control mice (black) and T1D mice (red). The “3-sigma” line represents the mean of the control sample measurements plus three standard deviations.
As a diagnostic agent, KTD1 would be of modest utility due to the significant false positive rate. Nonetheless, we hoped that it would be good enough to act as an affinity reagent to enrich the antibodies it binds (Fig. 10B). This proved to be true, allowing us to use the enriched antibody preparation to probe tissue extracts by Western blotting and immunoprecipitation for molecules recognized by the KTD1-binidng autoantibodies79. We found that a 65 kD protein was recognized by these antibodies and not control antibodies. Mass spectrometry revealed the identity of this protein to be GAD65 (Fig. 10C), which is known to be expressed on the surface of pancreatic beta cells102. A number of other pieces of evidence supported the idea that GAD65 is indeed a bona fide T1D autoantigen, including the fact that soluble GAD65, but not other proteins, competed binding of autoantibodies to KTD179 (Fig. 10D) Moreover, when a GST-mouse GAD65 fusion protein was immobilized on glutathione-modified TentaGel beads, it captured above background levels of antibodies from 80% of the NOD mice and none of the controls (Fig. 10D)79. Importantly, the entire signal was competed by soluble GST-mouse GAD65, but not by GST or other control proteins. The discovery of GAD65 as a novel T1D autoantigen in NOD mice79 demonstrates the power of this system.
Interestingly, GAD65 has a history as a T1D autoantigen. It is known to be a T cell antigen in the NOD mouse and a humoral antigen in human T1D patients95,97,98. This was initially also assumed to be the case in the NOD mouse, since GAD65 immobilized on an ELISA plate did capture antibodies from NOD mouse serum, albeit somewhat weakly.103 However, this was judged to be a non-specific interaction because the binding could not be competed by excess soluble GAD65.96,104 As mentioned above, this competition experiment is crucial in evaluating the specificity of any raw signal one sees in an ELISA or ELISA-like assay. However, in these experiments, the investigators employed rat, porcine or human GAD65 as the soluble competitor. We found that NOD mouse antibodies that exhibit a high affinity for mouse GAD65 do not bind to human GAD65, despite a high level of sequence identity between the two proteins79. Thus, the rejection of GAD65 as a NOD mouse humoral antigen is apparently due to a false assumption that the mouse antibodies would bind to GAD65 protein produced by other organisms.105 This highlights the importance of taking into account potential species-specific antibody-antigen interactions.
Beyond peptoids: Development of libraries of conformationally constrained oligomers as a source of higher affinity antibody ligands
The identification of GAD65 as a mouse NOD autoantigen was an encouraging validation of the power of the combinatorial technology for biomarker discovery. Moreover, GST-GAD65 appears to be a relatively convenient protein to produce in large quantities and employ as a capture agent in clinically relevant assays106. However, as will be discussed in more detail below, this will not always be the case. Sometimes proteins are just too difficult to produce in quantity or are not well enough behaved to be used as capture agents in large-scale assay development. In many cases therefore, the development of high sensitivity and specificity diagnostic tests based solely on synthetic epitope surrogates would be desirable. The low affinity that peptoids have for protein targets and the observation of a significant false positive rate for KTD179, reflecting off-target binding, are highly problematic in this regard. Thus, over the last few years, even as we have tried to improve the screening technology using peptoid libraries as a convenient source of compounds, we have been engaged in a parallel effort to develop libraries that are likely to be a source of much higher affinity and more selective protein ligands than peptoids21,107–111. Our guiding hypothesis, as mentioned above, is that the structure of peptoids imposes few conformational constraints on the main chain and that this inherent floppiness is detrimental to high affinity binding. Indeed, even the amide bond exists as a mixture of cis and trans isomers112. As a simple first step towards higher affinity ligands, we strove to develop libraries of compounds that would retain the many favorable properties of peptoids, including ease of synthesis32, characterization by MS113, etc., but would replace the methylene unit of the basic peptoid structure with more elaborate groups that would impose significant conformational constraints.
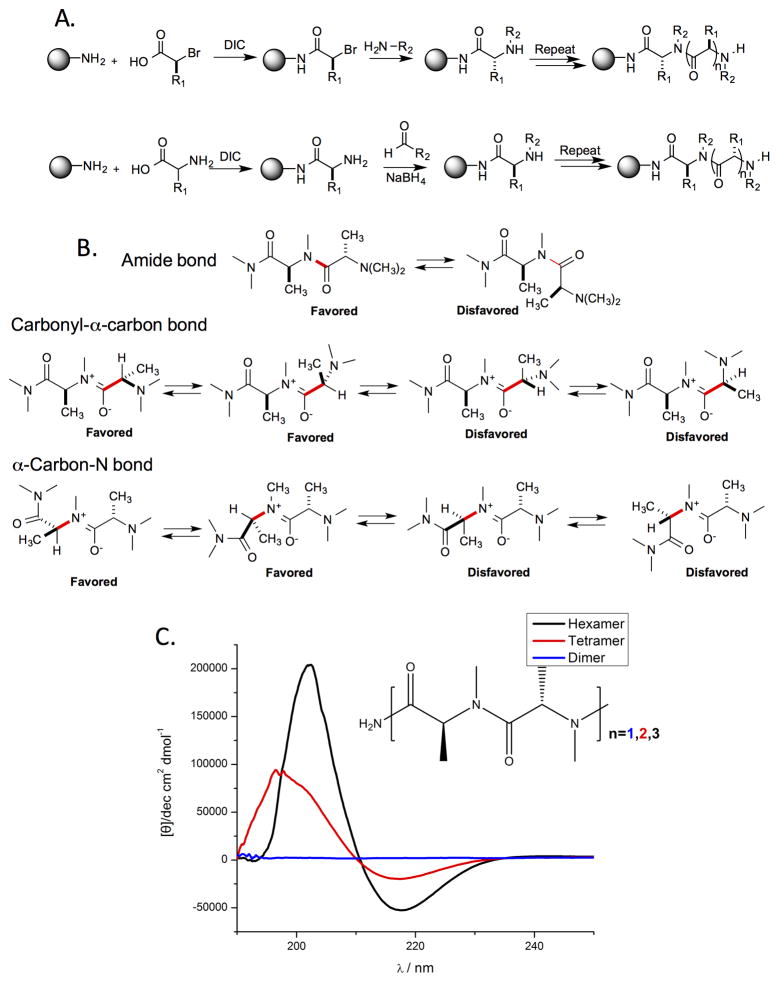
One of the simplest examples of this approach is the construction of libraries of peptide tertiary amides (PTAs), in other words, oligomers of N-alkylated amino acids110,111,114 (Fig. 11). The juxtaposition of the chiral center of an amino acid and an N-substituted amide introduces conformational constraints in the main chain due to strong allylic 1,3-strain considerations (Fig. 11B), as was first demonstrated in the study of natural products that contain N-methylated amino acids.115,116 Unlike either peptides or peptoids, even short oligomers of PTAs are highly structured, as demonstrated by the striking CD spectra of even tetra- and hexa-N-methyl alanine (Fig. 11C).110,115
Figure 11. Peptide tertiary amides (PTAs).
A. Two methods for the synthesis of PTAs. B. Illustration of the non-bonded steric interactions that provide PTAs with significant conformational constraints. C. Circular dichroism spectra of di-, tetra, and hexa-N-methylalanine. Reprinted with permission from ref. 110.
PTAs with diverse N-alkylation can be made in two ways. Some □-amino acids can be converted to chiral 2-bromoacids with retention of stereochemistry117 and these bromides can be employed as sub-monomers in a peptoid-type synthetic scheme110 (Fig. 11A, top). Alternatively, an N-terminal amino acid can be treated with a variety of aldehydes and sodium cyanoborohydride to effect N-alkylation via reductive amination114 (Fig. 11A, bottom).
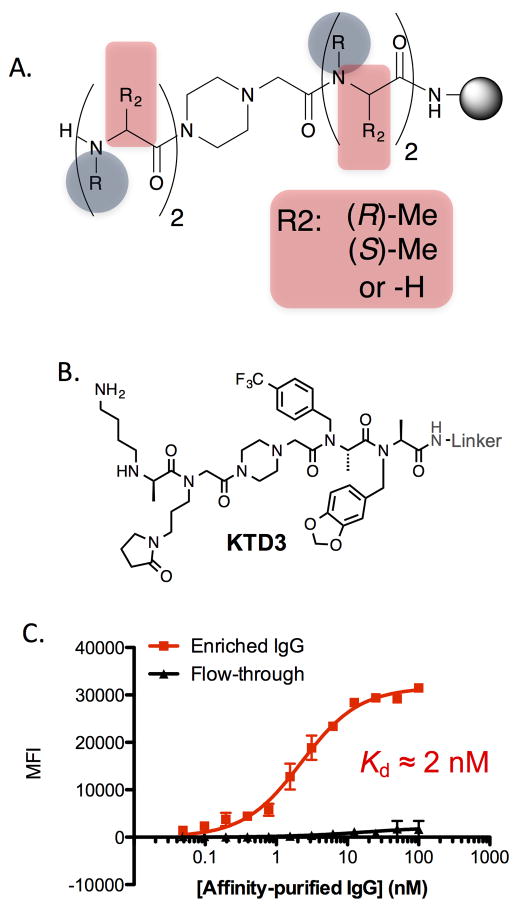
The major obstacle to making PTA oligomers is the difficulty of the amide bond coupling reactions to create the hindered tertiary amides, an issue well known from the N-methyl peptide literature118–120. Indeed, if neither the substituent on the chiral center or the amide nitrogen (R1 and R2 in Fig. 11A) is methyl, then the next amide bond is almost impossible to form in high yield with anything other than a very small, highly reactive acylating reagent such as chloroacetyl chloride111. Therefore, we restricted our initial library synthesis efforts to oligomers of N-alkylated alanines. Even so, stringing more than 2–3 such residues together without some type of break in the steric congestion is difficult and so the libraries that we have created avoid trying to do so. For example, a library of mixed PTA and peptoid molecules was created in which the two C- and N-terminal units were separated by a piperazine spacer (Fig. 12A, R2 = H, (R)-Me or (S)-Me).
Figure 12. Screening a PTA library provides high affinity ligands for antibodies linked to type 1 diabetes in the NOD mouse.
A. General structure of the library employed in screening. B. Structure of KTD3 and KTD4, hits that resulted from screening this library using the protocol shown in Fig. 8. C. Titration of immobilized KTD3 with IgG antibodies from NOD mice. Black: Antibodies that flow through a KTD3 affinity column. Red: Antibodies that were retained by a KTD3 affinity column (and then eluted from the column).
KTD3 (Fig. 12B) was isolated from this library using a screening protocol identical to that which yielded the peptoid KTD1. KTD3 thus was thus considered a potential surrogate for a NOD mouse autoantigen. In contrast to the peptoid KTD1 described above, KTD3 was a high affinity antibody ligand, displaying a “KD”121 of about 2 nM when immobilized on TentaGel beads (Fig. 12C)122. Not surprisingly, this high affinity resulted in a much better signal to noise in a serology assay with the level of antibodies captured by KTD3 being approximately 60-fold higher from NOD mouse serum than control serum. This superior performance was clearly due to the conformational constraints provided by the juxtaposed chiral center and N-alkylated nitrogen. The des-methyl (i.e. peptoid) analogue of KTD3 or any of the diastereomers that result from switching the chirality of any single stereocenter failed to bind T1D-related antibodies122.
Another conformationally constrained molecule, KTD4 (Fig. 12B), isolated from a different library using the same screening strategy, also displayed improved characteristics106. Neither KTD3 nor KTD4 bound significant levels of antibodies from control mouse sera, demonstrating excellent selectivity. When tested against a larger number of mouse serum samples, KTD3 and KTD4 bound above background levels of antibodies from about 40% of the mice. They did not compete with the peptoid KTD1 or with GST-GAD65, indicating that they are surrogates for an epitope present on some other antigen.122,123
Efforts to identify the native antigens recognized by the antibodies that bind KTD3 and KTD4 are in progress. Nonetheless, the fact that they do not bind anti-GAD65 antibodies suggested that when used in combination with GST-GAD65, these epitope surrogates could form part of a highly effective multiplexed test for mouse T1D. We recently demonstrated that this is indeed the case106. A blinded analysis of 40 mouse serum samples (20 controls and 20 diabetic) using a mixture of color-coded 10 μm TentaGel displaying KTD3, KTD4 or GST-GAD65 provided perfect results. All of the NOD mice were identified as such and there were no false positive calls106. This multiplexed analysis, which also included several control ligands, required only 1 μl of serum from each mouse.
Discovery of phosphoperipherin as a novel human T1D autoantigen
These results strongly suggested that a similar screening campaign might reveal new autoantigens for human T1D. As mentioned above, this is an important goal, since the currently known autoantibodies provide about 80% diagnostic sensitivity for pre-symptomatic diagnosis of T1D and only through the use of tedious RIA assays.
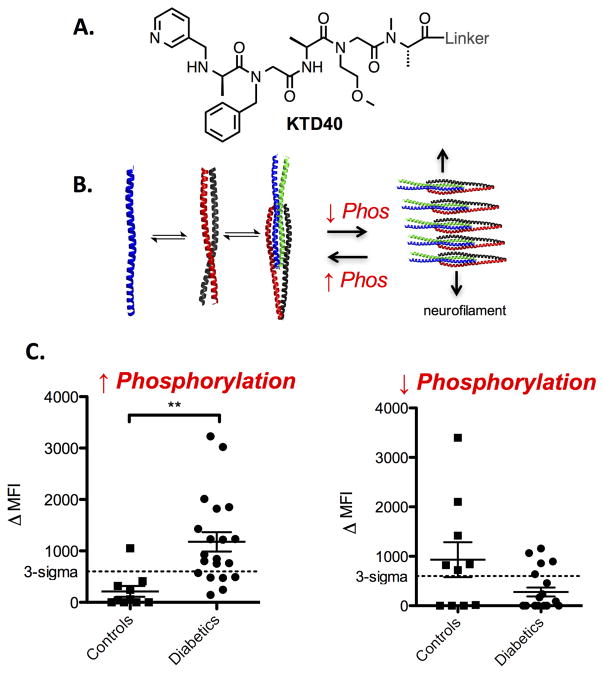
We recently completed the first stage of this effort successfully124. A library was screened in a fashion similar to that described for the NOD mouse model (Fig. 8), but using samples obtained from patients. A compound called KTD40 (Fig. 13) was identified as a ligand for antibodies that appeared to be present in a single T1D patient but not in healthy controls. This was worrisome. It was entirely possible that the antibodies bound by KTD40 had nothing to do with T1D. Perhaps this individual had picked up an infection a couple of weeks before being sampled. Nonetheless, we decided to press on to determine if this lead would be productive.
Figure 13. Discovery of phospho-peripherin as a major humoral autoantigen for type 1 diabetes.
A. Structure of KTD40, a small molecule that binds antibodies from a T1D patient. B. Cartoon of the self-association of peripherin and the effect of phosphorylation on this interaction (PDB: 4YV3). C. Binding of serum antibodies to immobilized human peripherin. The graph on the left depicts the results using natively phosphorylated peripherin. The graph on the right shows the results with the same protein treated with a phosphatase.
Mass spectrometric analysis of the proteins retained by the KTD40-binding antibodies identified a neurofilament protein called peripherin as the candidate autoantigen. We expressed peripherin in mammalian cells, immobilized it on 10 μm TentaGel beads and used the Luminex-like assay86 to evaluate its ability to distinguish between control and T1D serum samples. As shown in Fig. 13C, immobilized peripherin retained antibodies at above background levels from two-thirds of T1D patients, with a single false positive out of 10 control patients124. It is important to note that anti-peripherin antibodies were present in patients with pre-symptomatic T1D. That is they have autoantibodies against at least one of the previously described T1D autoantigens, but are not yet hyperglycemic.
An interesting feature of the peripherin-antibody interaction that we discovered is that it is utterly dependent on phosphorylation of the protein. When peripherin is dephosphorylated enzymatically, it fails to distinguish T1D serum samples from controls (Fig. 13C). Phosphorylation supports dimerization of the protein, but discourages filament formation (Fig. 13B) and we found that the dimer is the species “seen” by the T1D-related autoantibodies124. This highlights the fact that it would have been impossible to identify peripherin as a native autoantigen for T1D using an array of recombinant proteins in which native post-translational modifications are not likely to be present on the proteins. Another lesson to be learned from this study is that the small molecules that come out of the serum screening process are perhaps most useful as affinity reagents with which to enrich the antibodies of interest, thus setting the stage for isolation of the native antigen. As a diagnostic reagent, KTD40 would not itself be very useful as it only picked up one diabetic patient, but it provided a pathway to the discovery of a new autoantigen that reacts with antibodies present in two thirds of diabetics. Presumably, KTD40 is a surrogate for a particular epitope on phosphoperipherin that is targeted by a minority of the anti-peripherin polyclonal antibody population.
Towards a clinically relevant test for pre-symptomatic T1D
It is possible that the discovery of anti-phosphoperipherin antibodies in most type 1 diabetics may close the gap between the current 80% sensitivity of autoantibody-based T1D diagnostics and the desired 100% sensitivity. This is being addressed currently. But even if this is the case, the deployment of a routine clinical test would be facilitated tremendously by moving beyond the use of the autoantigenic proteins themselves as capture agents. As stated above, most of the known T1D autoantigens fail to bind antibodies when immobilized on a surface. While this is not the case for phosphoperipherin, it, unfortunately, is not a great candidate for a clinical reagent either. The protein is easily dephosphorylated during purification and tends to aggregate and precipitate. So our goal will be to assemble a suite of synthetic epitope surrogates that bind a sufficient fraction of the polyclonal antibody populations to provide an inexpensive, high sensitivity, high specificity multiplexed test when mounted on the color-coded TentaGel beads. However, assembling a suite of such molecules using the methods that we have employed to date, (differential screening of libraries with case and control serum pools) would be a long road indeed. From libraries of 100,000 or so molecules we tend to isolate only 1–3 bona fide epitope surrogates. Clearly, we are not digging deeply into the “immunoproteome” of autoantibodies with our current technology.
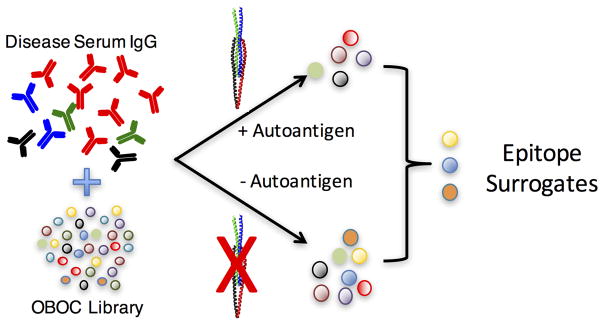
A stratagem that may be helpful would to carry out differential screens in which the serum pool consists entirely of samples obtained from T1D patients, but include in one tube a saturating amount of a known, soluble autoantigen (insulin, GAD65, phosphoperipherin, etc.). The presence of the autoantigen should prevent binding of cognate autoantibodies to bead-displayed ligands. Therefore, compounds identified as antibody-binding molecules in the absence of the soluble autoantigen, but not in its presence, are excellent candidates for surrogates of epitopes found in that autoantigen (Fig. 14). But we almost certainly need to screen far larger libraries as well. Given the low hit rate in these screens it will be important to screen larger numbers of structurally diverse compounds. It is likely that we will need to sort through tens of millions of molecules, not hundreds of thousands, to obtain a full suite of epitope surrogates.
Figure 14.
Schematic depiction of a comparative screen designed to identify synthetic surrogates of epitopes present on a known autoantigen.
Discovery of epitope surrogates from DNA-encoded libraries
The only way to access millions of compounds practically is to employ DNA-encoded libraries. The largest such collections are comprised of ribosomally synthesized peptides or cyclic peptides, where it is possible to generate libraries putatively containing 1014 compounds125,126. While these stupendous numbers are attractive, our bias is to avoid libraries of unmodified peptides for this effort because we fear that this is the wrong region of “chemical space” in which to search, much like using an array of recombinant proteins. It is important to point out, however, that this is a bias on our part and it may be interesting to try this approach60, particularly if it could somehow be combined with enzymatic modification of the libraries to introduce post-translational modifications as additional diversity elements.
More interesting to us are the recent advances in creating large, DNA-encoded libraries of synthetic molecules127–129. In the most common version of this technology, split and pool synthesis is conducted in solution, rather than on the solid-phase, with a DNA “headpiece” that also contains a pendant amine group. After first ligating onto the headpiece a primer template site, molecules are built step by step off of the amine. Every time a chemical operation is done, an encoding piece of DNA is ligated onto the end of the DNA chain. At the conclusion of the synthesis, a reverse primer template site is added. The library of small molecule-DNA conjugates are mixed together with a target protein. After a suitable incubation, the target protein is pulled out of solution along with any DNA-encoded small molecules bound stably to it. The encoding DNAs are amplified by PCR and sequenced. One can compare these sequences to some control experiment, for example one in which the entire library is subjected to deep sequencing, to determine what compounds are highly enriched in the target protein pull-down.
While highly attractive, there are significant limitations to DNA-encoded synthetic libraries. Perhaps most significantly, the chemistry that is employed currently is limited, for two major reasons. First, in the current methodology, as the encoding DNA increases in length, the large polyanion is not soluble in most organic solvents, limiting one to reactions that are highly tolerant of water. Second, many conditions used routinely in organic synthesis are incompatible with DNA. Thus, currently available DNA-encoded libraries, while large, tend to be quite simple and scaffold-poor, meaning that they are largely comprised of one, or a few, structural core element and almost all of the diversity is in the “side chains” that extend from this core. This is in stark contrast to typical high-throughput screening collections, which are highly scaffold-diverse. Thus, an important goal in this field will be to explore a much larger swath of organic reactions to assess their utility in the construction of DNA-encoded libraries with much greater scaffold diversity. For example, it was recently demonstrated that Suzuki couplings can be conducted in the presence of DNA with tolerable levels of damage130. Therefore, Buchwald-Hartwig-type couplings may also be feasible. It seems likely that a variety of organocatalytic reactions should be suitable in this setting, though to our knowledge this has not yet been addressed. Hopefully, over the next few years, a much richer palette of reactions for the synthesis of DNA-encoded libraries will become available.
In collaboration with our colleague Brian Paegel and his co-workers, we have been interested in adapting DNA encoding technology to solid-phase synthesis of OBOC libraries. As described in a recent paper131 from the Paegel laboratory, a first generation system has been established. This approach employs the same headpiece that was developed for solution-phase synthesis, but this unit is added to a small percentage of the common linker sites on a TentaGel bead via a copper-catalyzed Huisgen cycloaddition (Fig. 15A). As in the established technology, after addition of a PCR primer template site, the encoded library is created by split and pool synthesis where a chemical step and an oligonucleotide ligation step are conducted during each split (Fig. 15B). The construction of DNA-encoded libraries by solid-phase synthesis should have some advantages over solution phase methods because the solubility of the DNA chain becomes irrelevant, allowing a variety of organic solvents to be used.
Figure 15. DNA-encoded OBOC libraries 131.
A. Depiction of the bead architecture in which a small percentage of the surface sites are modified with a “headpiece” via Click chemistry132. B. Cartoon of a hypothetical oligomer encoded by a DNA strand. C. Quality control by lot sampling. A few thousand 160 μm TentaGel beads are included with millions of 10 μm beads during the synthesis of the library in a 96 well plate format. After completion of the library, the larger beads are easily separated from the smaller beads. The compound on these beads is analyzed by LC-MS for purity and the encoding DNA is Sanger sequenced to determine if the encoding tag correctly predicts the molecular ion in the mass spectrum.
Another advantage of solid-phase synthesis of DNA-encoded libraries is that quality control becomes feasible. While a 10 μm bead has too little compound on it for direct analysis, we dope into the library a few thousand 160 μm beads (Fig. 15C). Control experiments have demonstrated that the chemistry proceeds identically on 10 μm and 160 μm TentaGel beads. Thus, after completion of the library synthesis, the 160 μm beads can be separated and the substantial amount of compound on these large beads can be analyzed for purity by LC-MS. The DNA tag can be Sanger sequenced to determine if it predicts the mass of the molecular ion correctly. This represents a significant advance over solution phase synthesis, since in this mode it is very difficult to assess the quality of a synthetic library.
With DNA encoding, mass spectrometry-based identification of the bead-displayed compound is no longer required. Since PCR amplification of the tag and deep sequencing provides such high sensitivity, much smaller beads with miniscule amounts of material on them can be used. We now routinely employ 10 μm beads, which pass easily through a flow cytometer, allowing screening hits to be identified in a much more convenient, semi-automated fashion (McEnaney, et al., in preparation). There are about 2 billion 10 μm TentaGel beads per gram, as compared to about 3 million 90 μm beads. Thus, many copies of OBOC libraries containing millions of compounds can be made via split and pool synthesis131.
We aim to deploy DNA-encoded OBOC libraries for the discovery of epitope surrogates in the immediate future. Libraries containing tens of millions of conformationally constrained oligomers, as well as the far more efficient FACS-based screening of beads, should be extremely useful in digging much deeper into the immunoproteome. We hope that this will support the facile discovery of a more comprehensive set of epitope surrogates for the development of diagnostic assays and the discovery of new biology.
Summary and Conclusions
The antigen-binding sites of immune receptors (antibodies, BCRs and TCRs) have not generally been considered targets for small molecules. However, our work and that of others have shown that non-peptidic compounds that recognize these surfaces with high affinity and selectivity can be discovered via high-throughput screening. Perhaps not surprisingly, oligomeric compounds with significant conformational constraints tend to be far superior ligands relative to “floppy” compounds such as peptides or peptoids. While this epitope surrogate technology is still very young, the results to date suggest that this approach may have a significant impact in chemical biology and medicine. Perhaps most importantly, differential screening of large libraries of conformationally constrained oligomers against case and control serum samples is capable of identifying diagnostically useful antibody-small molecule complexes. Enrichment of these antibodies using the oligomer as an affinity reagent provides a powerful route to the discovery of new antigens that the adaptive immune system “sees” in the course of the disease. Our work to date has validated the utility of this novel approach to biomarker discovery in autoimmune disease79,81,106,123,124 and, in the process, we have solved a variety of nagging technical problems that grossly limited the pace of this research.84,86 The stage is now set to attempt to discover antibody biomarkers that would have enormous diagnostic utility, such as for early stage cancers or neurodegenerative diseases.
Of potential therapeutic utility, incorporating BCR or TCR ligands into chemical dimerizers may allow the selective killing of pathogenic cells whilst sparing healthy cells. In these efforts, B or T cell could be the target cell or the effector. In the former case, the goal would be to chemically edit antigen-specific immune responses. In the latter one would attempt to attract immune effector functions to non-immunological target cells, such as solid tumors. We hope that this article will stimulate interest in this nascent area of chemical biology and spur innovative research on the use of chemical tools to monitor and manipulate the adaptive immune system.
Acknowledgments
Work in the Kodadek laboratory described in this article was supported by the National Institutes of Health (DP3 DK094309-01), the Fidelity Foundation, the Goodman Family Foundation, the DARPA Fold F(X) program (N66001-14-2-4057) and Opko Inc. We are grateful to Prof. M. Disney (TSRI) for a critical reading of this manuscript.
Footnotes
Conflict of Interest Statement
T.K. is a significant shareholder in Opko, Inc., which is attempting to commercialize some of the diagnostic technology described in this Perspective.
Literature Cited
- 1.Murphy KL. Janeway’s Immunobiology. 8. Garland Science; 2014. [Google Scholar]
- 2.Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, Chen J. Immunol Today. 1992;13:306. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 3.Edwards JC, Cambridge G. Nat Rev Immunol. 2006;6:394. doi: 10.1038/nri1838. [DOI] [PubMed] [Google Scholar]
- 4.Buggy JJ, Elias L. Int Rev Immunol. 2012;31:119. doi: 10.3109/08830185.2012.664797. [DOI] [PubMed] [Google Scholar]
- 5.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, Johnson AJ, Sukbuntherng J, Chang BY, Clow F, Hedrick E, Buggy JJ, James DF, O’Brien S. N Engl J Med. 2013;369:32. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Drake CG, Pardoll DM. Cancer Cell. 2015;27:450. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pico de Coana Y, Choudhury A, Kiessling R. Trends Mol Med. 2015;21:482. doi: 10.1016/j.molmed.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Bakacs T, Mehrishi JN, Moss RW. Immunobiology. 2012;217:583. doi: 10.1016/j.imbio.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, Winkler C, Ilonen J, Veijola R, Knip M, Bonifacio E, Eisenbarth GS. J Am Med Assoc. 2013;309:2473. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verge CF, Gianani R, Kawasaki E, Yu L, Pietropaolo M, Jackson RA, Chase HP, Eisenbarth GS. Diabetes. 1996;45:926. doi: 10.2337/diab.45.7.926. [DOI] [PubMed] [Google Scholar]
- 11.Anderson KS, LaBaer J. J Proteome Res. 2005;4:1123. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiorazzi N, Rai KR, Ferrarini M. N Engl J Med. 2005;352:804. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 13.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, Lamanna N, Coiffier B, Pettitt AR, Ma S, Stilgenbauer S, Cramer P, Aiello M, Johnson DM, Miller LL, Li D, Jahn TM, Dansey RD, Hallek M, O’Brien SM. N Engl J Med. 2014;370:997. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderl J, Faulstich H, Hechler T, Kulke M. Methods Mol Biol. 2013;1045:51. doi: 10.1007/978-1-62703-541-5_4. [DOI] [PubMed] [Google Scholar]
- 15.Murelli RP, Zhang AX, Michel J, Jorgensen WL, Spiegel DA. J Am Chem Soc. 2009;131:17090. doi: 10.1021/ja906844e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodadek T. Curr Opin Chem Biol. 2010;14:721. doi: 10.1016/j.cbpa.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEnaney PJ, Parker CG, Zhang AX, Spiegel DA. ACS Chem Biol. 2012;7:1139. doi: 10.1021/cb300119g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 19.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Nature. 1991;354:84. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar M, Pascal BD, Steckler C, Micalizio GC, Kodadek T, Chalmers MJ. J Amer Soc Mass Spec. 2013;24:1026. doi: 10.1007/s13361-013-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aquino C, Sarkar M, Chalmers MJ, Mendes K, Kodadek T, Micalizio G. Nat Chem. 2011;4:99. doi: 10.1038/nchem.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar M, Liu Y, Morimoto J, Peng H, Aquino C, Rader C, Chiorazzi N, Kodadek T. Chem Biol. 2014;21:1670. doi: 10.1016/j.chembiol.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Proc Natl Acad Sci U S A. 1992;89:9367. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J Amer Chem Soc. 1992;114:10646. [Google Scholar]
- 25.Kodadek T, McEnaney PJ. Chemical communications (Cambridge, England) 2016;52:6038. doi: 10.1039/c6cc00617e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morimoto J, Sarkar M, Kenrick S, Kodadek T. Bioconjugate Chem. 2014;25:1479. doi: 10.1021/bc500226j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodadek T. Chem Biol. 2014;21:1066. doi: 10.1016/j.chembiol.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agathangelidis A, Darzentas N, Hadzidimitriou A, Brochet X, Murray F, Yan XJ, Davis Z, van Gastel-Mol EJ, Tresoldi C, Chu CC, Cahill N, Giudicelli V, Tichy B, Pedersen LB, Foroni L, Bonello L, Janus A, Smedby K, Anagnostopoulos A, Merle-Beral H, Laoutaris N, Juliusson G, di Celle PF, Pospisilova S, Jurlander J, Geisler C, Tsaftaris A, Lefranc MP, Langerak AW, Oscier DG, Chiorazzi N, Belessi C, Davi F, Rosenquist R, Ghia P, Stamatopoulos K. Blood. 2012;119:4467. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar M, Liu YJQ, Peng H, Morimoto J, Rader C, Chiorazzi N, Kodadek T. J Biol Chem. 2016;291:7558. doi: 10.1074/jbc.M115.701656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bander NH, Czuczman MS, Younes A. Clin Adv Hematol Oncol. 2012;10:1. [PubMed] [Google Scholar]
- 31.Chudasama V, Maruani A, Caddick S. Nat Chem. 2016;8:114. doi: 10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- 32.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J Am Chem Soc. 1992;114:10646. [Google Scholar]
- 33.Diver ST, Schreiber SL. J Am Chem Soc. 1997;119:5106. [Google Scholar]
- 34.Crabtree GR, Schreiber SL. Trends in Biochem Sci. 1996;21:418. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 35.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 36.Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Proc Natl Acad Sci USA. 1996;93:4604. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. Clin Immunol Immunopathol. 1996;80:S40. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 38.McEnaney PJ, Fitzgerald KJ, Zhang AX, Douglass EF, Jr, Shan W, Balog A, Kolesnikova MD, Spiegel DA. J Am Chem Soc. 2014;136:18034. doi: 10.1021/ja509513c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubrovska A, Kim C, Elliott J, Shen W, Kuo TH, Koo DI, Li C, Tuntland T, Chang J, Groessl T, Wu X, Gorney V, Ramirez-Montagut T, Spiegel DA, Cho CY, Schultz PG. ACS Chem Biol. 2011;6:1223. doi: 10.1021/cb200222s. [DOI] [PubMed] [Google Scholar]
- 40.Jakobsche CE, Parker CG, Tao RN, Kolesnikova MD, Douglass EF, Jr, Spiegel DA. ACS Chem Biol. 2013;8:2404. doi: 10.1021/cb4004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kodadek T. Curr Opin Chem Biol. 2011;14:721. doi: 10.1016/j.cbpa.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rader C, Sinha SC, Popkov M, Lerner RA, Barbas CF., 3rd Proc Natl Acad Sci USA. 2003;100:5396. doi: 10.1073/pnas.0931308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rader C, Turner JM, Heine A, Shabat D, Sinha SC, Wilson IA, Lerner RA, Barbas CF. J Mol Biol. 2003;332:889. doi: 10.1016/s0022-2836(03)00992-6. [DOI] [PubMed] [Google Scholar]
- 44.Barrett DM, Singh N, Porter DL, Grupp SA, June CH. Annu Rev Med. 2014;65:333. doi: 10.1146/annurev-med-060512-150254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allison JP. J Am Med Assoc. 2015;314:1113. [Google Scholar]
- 46.Gocke AR, Udugamasooriya DG, Archer CT, Lee J, Kodadek T. Chem Biol. 2009;16:1133. doi: 10.1016/j.chembiol.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Racke MK. Curr Protoc Neurosci. 2001;Chapter 9(Unit 9):7. doi: 10.1002/0471142301.ns0907s14. [DOI] [PubMed] [Google Scholar]
- 48.Zamvil SS, Steinman L. Annu Rev Immunol. 1990;8:579. doi: 10.1146/annurev.iy.08.040190.003051. [DOI] [PubMed] [Google Scholar]
- 49.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Proc Natl Acad Sci USA. 1992;89:9367. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Methods Enzymol. 1996;267:437. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 51.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. J Am Chem Soc. 2003;125:13995. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 52.Burdine L, Gillette TG, Lin HJ, Kodadek T. J Am Chem Soc. 2004;126:11442. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- 53.Liu B, Burdine L, Kodadek T. J Am Chem Soc. 2006;128:15228. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J, Udugamasooriya DG, Lim HS, Kodadek T. Nat Chem Biol. 2010;6:258. doi: 10.1038/nchembio.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lueking A, Possling A, Huber O, Beveridge A, Horn M, Eickoff H, Schuchardt J, Lehrach H, Cahill DJ. Mol Cell Proteomics. 2003;2:1342. doi: 10.1074/mcp.T300001-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Hultschig C, Kreutzberger J, Seitz H, Konthur Z, Bussow K, Lehrach H. Curr Opin Chem Biol. 2006;10:4. doi: 10.1016/j.cbpa.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quintana FJ, Farez MF, Viglietta V, Iglesias AH, Merbl Y, Izquierdo G, Lucas M, Basso AS, Khoury SJ, Lucchinetti CF, Cohen IR, Weiner HL. Proc Natl Acad Sci USA. 2008;105:18889. doi: 10.1073/pnas.0806310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, Steinman L, Robinson WH. Nat Med. 2006;12:138. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Bhat R, Sobel RA, Huang W, Wang LX, Olsson T, Steinman L. Drug Develop Res. 2014;75:172. doi: 10.1002/ddr.21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Yu JQ, Sreekumar A, Varambally S, Shen R, Giachero D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiyan AM. N Engl J Med. 2005;355:16. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 61.Robinson WH, Steinman L, Utz PJ. Proteomics. 2003;3:2077. doi: 10.1002/pmic.200300583. [DOI] [PubMed] [Google Scholar]
- 62.Robinson WH, Steinman L, Utz PJ. Biotechniques. 2002;(Dec. Suppl):66. [PubMed] [Google Scholar]
- 63.Restrepo L, Stafford P, Magee DM, Johnston SA. Ann Neurol. 2011;70:286. doi: 10.1002/ana.22405. [DOI] [PubMed] [Google Scholar]
- 64.Avouac J, Gossec L, Dougados M. Ann Rheum Dis. 2006;65:845. doi: 10.1136/ard.2006.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lennon VA, Kryzer TJ, Pittock SJ, Verkman AS, Hinson SR. J Exp Med. 2005;202:473. doi: 10.1084/jem.20050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinson SR, Pittock SJ, Lucchinetti CF, Roemer SF, Fryer JP, Kryzer TJ, Lennon VA. Neurology. 2007;69:2221. doi: 10.1212/01.WNL.0000289761.64862.ce. [DOI] [PubMed] [Google Scholar]
- 67.Saikali P, Cayrol R, Vincent T. Autoimmun Rev. 2009;9:132. doi: 10.1016/j.autrev.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Neurology. 2006;66:1485. doi: 10.1212/01.wnl.0000216139.44259.74. [DOI] [PubMed] [Google Scholar]
- 69.Waters P, Vincent A. Int MS J. 2008;15:99. [PubMed] [Google Scholar]
- 70.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. Nat Biotechnol. 2014;32:158. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM, Sokolove J, Robinson WH. Clin Immunol. 2014;151:55. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, Ellington AD, Dorner T, Marcotte EM, Boutz DR, Georgiou G. Proc Natl Acad Sci USA. 2014;111:2259. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho SL, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A. Nature. 2014;509:55. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, Lu DR, Cai X, Wagner CA, Lindstrom TM, Sokolove J, Robinson WH. Arthritis Rheumatol. 2014;66:2706. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boutz DR, Horton AP, Wine Y, Lavinder JJ, Georgiou G, Marcotte EM. Anal Chem. 2014;86:4758. doi: 10.1021/ac4037679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olivos HJ, Baccawat-Sikder K, Kodadek T. ChemBiochem. 2003;4:1242. doi: 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- 77.Mendes K, Ndungu JM, Clark L, Kodadek T. ACS Comb Sci. 2015;17:506. doi: 10.1021/acscombsci.5b00090. [DOI] [PubMed] [Google Scholar]
- 78.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Chem Biol. 2010;17:38. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doran TM, Simanski S, Kodadek T. ACS Chem Biol. 2015;10:401. doi: 10.1021/cb5007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Cell. 2011;144:132. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raveendra B, Hao W, Baccala R, Reddy MM, Schilke J, Bennett JL, Theofiliopolous AN, Kodadek T. Chem Biol. 2013;20:350. doi: 10.1016/j.chembiol.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagele E, Han M, Demarshall C, Belinka B, Nagele R. PLoS One. 2011;6:e23112. doi: 10.1371/journal.pone.0023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lian W, Upadhyaya P, Rhodes CA, Liu Y, Pei D. J Am Chem Soc. 2013;135:11990. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Doran TM, Gao Y, Mendes K, Dean S, Simanski S, Kodadek T. ACS Comb Sci. 2014;16:259. doi: 10.1021/co500030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vignali DA. J of Immunol Methods. 2000;243:243. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 86.Doran TM, Kodadek T. ACS Chem Biol. 2013;9:339. doi: 10.1021/cb400806r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eisenbarth GS. N Engl J Med. 1986;314:1360. doi: 10.1056/NEJM198605223142106. [DOI] [PubMed] [Google Scholar]
- 88.Ali K, Harnden A, Edge JA. BMJ. 2011;342:d294. doi: 10.1136/bmj.d294. [DOI] [PubMed] [Google Scholar]
- 89.Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Cell. 2014;159:428. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lieberman SM, DiLorenzo TP. Tissue Antigens. 2003;62:359. doi: 10.1034/j.1399-0039.2003.00152.x. [DOI] [PubMed] [Google Scholar]
- 91.Pihoker C, Gilliam LK, Hampe CS, Lernmark A. Diabetes. 2005;54(Suppl 2):S52. doi: 10.2337/diabetes.54.suppl_2.s52. [DOI] [PubMed] [Google Scholar]
- 92.Baekkeskov S, Aanstoot HJ, Christgau S, Reetz A, Solimena M, Cascalho M, Folli F, Richter-Olesen H, De Camilli P. Nature. 1990;347:151. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- 93.Atkinson MA, Maclaren NK. J Clin Invest. 1993;92:1608. doi: 10.1172/JCI116745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC. Proc Natl Acad Sci USA. 2007;104:17040. doi: 10.1073/pnas.0705894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petersen JS, Hejnaes KR, Moody A, Karlsen AE, Marshall MO, Hoier-Madsen M, Boel E, Michelsen BK, Dyrberg T. Diabetes. 1994;43:459. doi: 10.2337/diab.43.3.459. [DOI] [PubMed] [Google Scholar]
- 96.Bonifacio E, Atkinson M, Eisenbarth G, Serreze D, Kay TW, Lee-Chan E, Singh B. Diabetes. 2001;50:2451. doi: 10.2337/diabetes.50.11.2451. [DOI] [PubMed] [Google Scholar]
- 97.Bonifacio E, Lampasona V, Bernasconi L, Ziegler AG. Diabetes. 2000;49:202. doi: 10.2337/diabetes.49.2.202. [DOI] [PubMed] [Google Scholar]
- 98.Miao D, Guyer MS, Dong F, Jiang L, Steck AK, Rewers M, Eisenbarth GS, Yu L. Diabetes. 2013;62:4174. doi: 10.2337/db13-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu L, Dong F, Miao D, Fouts AR, Wenzlau JM, Steck AK. Diabetes care. 2013;36:2266. doi: 10.2337/dc12-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang B, Kumar RB, Dai H, Feldman BJ. Nat Med. 2014;20:948. doi: 10.1038/nm.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anderson MS, Bluestone JA. Annu Rev Immunol. 2005;23:447. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 102.Kim J, Richter W, Aanstoot HJ, Shi Y, Fu Q, Rajotte R, Warnock G, Baekkeskov S. Diabetes. 1993;42:1799. doi: 10.2337/diab.42.12.1799. [DOI] [PubMed] [Google Scholar]
- 103.Tisch R, Yang XD, Singer SM, Liblau RS, Fugger L, McDevitt HO. Nature. 1993;366:72. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 104.Yu L, Robles DT, Abiru N, Kaur P, Rewers M, Kelemen K, Eisenbarth GS. Proc Natl Acad Sci USA. 2000;97:1701. doi: 10.1073/pnas.040556697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Velloso LA, Kämpe O, Eizirik DL, Hallberg A, Andersson A, Karlsson FA. Diabetologia. 1993;36:39. doi: 10.1007/BF00399091. [DOI] [PubMed] [Google Scholar]
- 106.Doran TM, Morimoto J, Simanski S, Kodadek T. Mol BioSyst. 2015;11:3156. doi: 10.1039/c5mb00521c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suwal S, Kodadek T. Org Biomol Chem. 2013;11:2088. doi: 10.1039/c3ob27476d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suwal S, Kodadek T. Org Biomol Chem. 2014;12:5831. doi: 10.1039/c4ob00829d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aditya A, Kodadek T. ACS Comb Sci. 2012;14:164. doi: 10.1021/co200195t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao Y, Kodadek T. Chem Biol. 2013;20:360. doi: 10.1016/j.chembiol.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morimoto J, Kodadek T. Mol BioSyst. 2015;11:2770. doi: 10.1039/c5mb00308c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoo B, Kirshenbaum K. Curr Opin Chem Biol. 2008;12:714. doi: 10.1016/j.cbpa.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 113.Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. J Comb Chem. 2006;8:417. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- 114.Pels K, Kodadek T. ACS Comb Sci. 2015;17:152. doi: 10.1021/acscombsci.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang S, Prabpai S, Kongsaeree P, Arvidsson PI. Chem Commun (Cambridge, U K) 2006:497. doi: 10.1039/b513277k. [DOI] [PubMed] [Google Scholar]
- 116.Kessler H, Chatterjee J, Doedens L, Operrer F, Biron E, Hoyer D, Schmid H, Gilon C, Hruby VJ, Mierkes DF. Adv Exp Med Biol. 2009;611:229. doi: 10.1007/978-0-387-73657-0_106. [DOI] [PubMed] [Google Scholar]
- 117.Izumiya N, Nagamatsu A. Bull Chem Soc Japan. 1952;25:265. [Google Scholar]
- 118.Falb E, Yechezkel T, Salitra Y, Gilon C. J Pept Res. 1999;53:507. doi: 10.1034/j.1399-3011.1999.00049.x. [DOI] [PubMed] [Google Scholar]
- 119.Thern B, Rudolph J, Jung G. Angew Chem, Int Ed. 2002;41:2307. doi: 10.1002/1521-3773(20020703)41:13<2307::AID-ANIE2307>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 120.Videnov GI, Kaiser D, Kempter MC, Jung G. Angew Chem, Intl Ed. 1996;35:1503. [Google Scholar]
- 121.The term “KD” is used loosely here as the molecule is binding to a oligoclonal population of antibodies and thus the measured binding constant represent a weighted average of the various true equilibrium dissociation constants of the monoclonal antibodies that make up the oligoclonal population.
- 122.Doran TM, Gao Y, Simanski S, McEnaney P, Kodadek T. Bioorg Med Chem Lett. 2015;25:4910. doi: 10.1016/j.bmcl.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doran TM, Morimoto J, Simanski S, McEnaney PJ, Kodadek T. Mol Biosyst. 2015;11:3156. doi: 10.1039/c5mb00521c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Doran TM, Morimoto J, Simanski S, Koesema EJ, Clark LF, Pels K, Stoops SL, Pugliese A, Skuyler JS, Kodadek T. Cell Chemical Biology. 2016;23 doi: 10.1016/j.chembiol.2016.04.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roberts RW, Szostak JW. Proc Natl Acad Sci USA. 1997;94:12297. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ohuchi M, Murakami H, Suga H. Curr Opin Chem Biol. 2007;11:537. doi: 10.1016/j.cbpa.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 127.Clark MA. Curr Opin Chem Biol. 2010;14:396. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 128.Scheuermann J, Neri D. Chembiochem. 2010;11:931. doi: 10.1002/cbic.201000066. [DOI] [PubMed] [Google Scholar]
- 129.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. J Am Chem Soc. 2007;129:13137. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ding Y, Clark MA. ACS Comb Sci. 2015;17:1. doi: 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]
- 131.MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM. ACS Comb Sci. 2015;17:518. doi: 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]