Abstract
Phosphatidylinositol-3,4,5-trisphosphate (PIP3) mediates signaling pathways as a second messenger in response to extracellular signals. Although primordial functions of phospholipids and RNAs have been hypothesized in the “RNA world”, physiological RNA-phospholipid interactions and their involvement in essential cellular processes has remained a mystery. We explicate the contribution of lipid-binding long non-coding RNAs (lncRNAs) in cancer cells. Among them, Long Intergenic Noncoding RNA for Kinase Activation (LINK-A) directly interacts with AKT pleckstrin homology domain and PIP3 at the single nucleotide level, facilitating AKT-PIP3 interaction and consequent enzymatic activation. LINK-A-dependent AKT hyperactivation leads to tumorigenesis and resistance to AKT inhibitors. Genomic deletions of the LINK-A PIP3-binding motif dramatically sensitized breast cancer cells to AKT inhibitors. Furthermore, meta-analysis showed the correlation between LINK-A expression and incidence of a SNP (rs12095274: A>G), AKT phosphorylation status, and poor outcomes for breast and lung cancer patients. PIP3-binding lncRNA modulates AKT activation with broad clinical implications.
Introduction
Phosphatidylinositol-3,4,5-trisphosphate (PIP3) generated by phosphoinositide 3-kinase (PI3K) mediates the signal transductions that are important for homeostasis and disease, by interacting with protein kinases/phosphatases1,2. PIP3 is recognized by membrane-binding proteins via target-specific binding domains, including the C1 domain3, pleckstrin homology (PH) domain4, and 'Fab1, YOTB, Vac1, EEA1' (FYVE) domains5. The PIP3−PH domain interaction is responsible for signal-dependent membrane recruitment and activation of downstream kinases, such as Protein Kinase B (PKB/AKT), Phosphoinositide-dependent kinase-1 (PDK1) and Bruton's tyrosine kinase (BTK)6–8. Dysregulation of PI3K and downstream AKT activation are involved in many human cancers and diseases9,10. Although AKT is recruited to PIP3 upon ligand stimulation, where AKT is phosphorylated and activated by PDK1 and mTOR complex at Ser473 and Thr308 respectively11, the PH domain of AKT prevents it from being phosphorylated12. The association between the PH domain and PIP3 may cause a conformational change in AKT, making Ser473 accessible to PDK112. Thus, small molecule inhibitors targeting PH domains of AKT e.g. MK2206 are in clinical trials for aggressive cancers alone or in combination with other pathway inhibitors13–15. However, some cancer cells acquire resistance to MK220616,17; therefore, delineation of the mechanisms of resistance is critical for the development of strategies to treat or prevent resistant tumors.
Long non-coding RNAs (lncRNAs) play emerging roles in cell signaling pathways via interactions with protein partners18–22. The observation that RNA molecule association with cellular membranes is involved in formation of the Escherichia coli signal recognition particle23 and regulation of Saccharomyces cerevisiae cell membrane permeability24 support the notion that RNA-lipid interactions might be physiologically important. However, bona fide RNA-phospholipid interactions remain unidentified. The identification of lncRNA-lipid interactions introduces lncRNAs as mediators of signaling pathways relevant to homeostasis and disease.
We show that a lncRNA named Long Intergenic Noncoding RNA for Kinase Activation (LINK-A, also LINC01139) directly and specifically interacts with AKT and PIP3. The LINK-A-AKT and -PIP3 interactions facilitate AKT recruitment and consequent activation. We identified the single nucleotides of LINK-A required for PIP3 and AKT bindings. LINK-A-dependent AKT hyperactivation leads to tumorigenesis and resistance to AKT inhibitors. Genomic deletion of the LINK-A PIP3-binding motif in resistant cells restores MK2206 sensitivity, suggesting that LINK-A confers resistance to targeted therapy in breast cancer. Furthermore, amplification of LINK-A locus in cancer patients substantiates its promise as a clinical biomarker. The meta-analysis uncovered the association between LINK-A expression and high incidence of an SNP (rs12095274:A>G), which further correlated with AKT phosphorylation status, people of African descent, and poor outcomes for breast cancer patients. Our data reveal a PIP3-dependent role of lncRNA in meditating AKT activation and conferring resistance to AKT inhibitors. Clinically, preventing resistance is favorable to treating resistance after it develops; thus, if LINK-A overexpression is observed in patients that develop resistance to AKT inhibitors, this provides a rationale for targeting LINK-A, either alone or in combination with AKT inhibitors, when treating breast tumors.
Results
Identification of lipid-binding lncRNAs
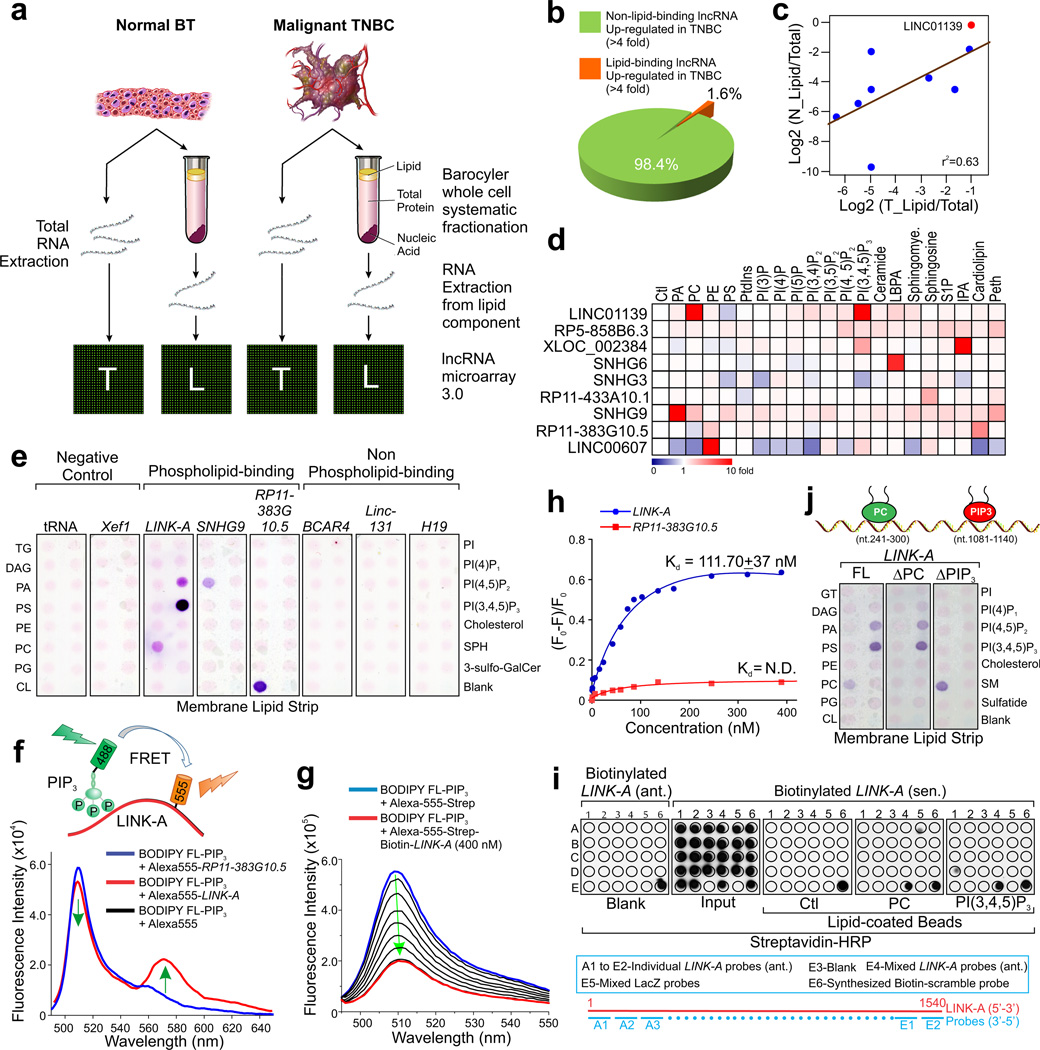
To identify RNAs that naturally associate with cellular lipids, we isolated lipid fractions and bound RNAs from triple-negative breast cancer (TNBC) patient tissues. Fresh-frozen TNBC tissues and their normal counterparts18 were subjected to total RNA extraction or, in parallel, to whole cell fractionation via Hydrostatic Pressure Cycling to form a lipid-containing upper phase, a denatured protein-containing lower phase, and an insoluble fraction containing DNA and RNA25–27. The total RNAs and the RNAs from the lipid fraction were analyzed by LncRNA Array (Fig. 1a and Supplementary Table 1). Using a 4-fold cutoff threshold (tumor vs. normal) and by overlapping the total lncRNA array with the lipid-bound lncRNA array, we identified 9 of 550 lncRNAs (ca. 1.6%) that were associated with the lipid fraction (Fig. 1b). We defined the relative lipid enrichment by comparing the normalized density of the total array with that of the lipid-bound array (Lipid/Total) and found that while all 9 lncRNAs showed consistent lipid enrichment in both normal (N_Lipid/Total) and TNBC tissues (T_Lipid/Total), LINC01139 exhibited the highest lipid enrichment (Fig. 1c and Supplementary Fig. 1a). Furthermore, LINC01139 is upregulated in TNBC compared to its normal counterpart (Supplementary Fig. 1b). Using lipid-coated beads28 pulldown followed by RT-qPCR assay, we confirmed that 7 of the 9 lncRNAs exhibited specificities for various phospholipids (>2 fold enrichment compared to control beads). Among them, LINC01139 (renamed LINK-A) showed enrichment for phosphatidylcholine (PC) and PIP3, with minimal cross reaction with phosphatidylinositol-4,5-biphosphate (PIP2) (Fig. 1d and Supplementary Fig. 1c, d).
Figure 1. Global identification of lipid-interacting lncRNAs and characterization of LINK-A-lipid interaction.
(a) Experimental scheme for identification of lipid-bound lncRNAs in triple-negative breast cancer (TNBC). (b) Pie chart of percentage of TNBC-upregulated lncRNAs that associated with cellular lipids. (c) Scatter blot representing lipid enrichment of top 9 lncRNAs in normal breast tissue (N) or malignant breast cancer (T). X and Y axis represent log2 scale of normalized lncRNA density (Lipid/Total). (d) Heatmap of lipid enrichment of top 9 lncRNAs based on lipid-RNA pulldown followed by RT-qPCR. Red/blue indicates increased/decreased fold change in lipid enrichment over control beads. (e) RNA-lipid overlay assay showing binding of LINK-A to PC and PIP3. In vitro transcribed biotinylated RNA transcripts, as indicated, were applied to membrane lipid strips. (f) Upper panel: graphic illustration of the PIP3-LINK-A interaction detected by FRET assay. Lower panel: fluorescence spectra of BODIPY FL-PIP3 (donor) in the presence of Alexa-555-Strep (blue) or Alexa-555-Strep-biotin-LINK-A (red; λexc = 475 nm). (g) Representative fluorescence spectra of BODIPY FL-PIP3 upon titration of increasing concentrations of LINK-A (0 ~ 400 nM; λexc = 490 nm). (h) Fitting the fluorescence quenching of BODIPY FL-PIP3 induced by LINK-A with one site binding equation. Data fitting yielded a dissociation constant (Kd) of 112 ± 37 nM (mean ± s.e.m. were derived from n=3 independent experiments). (i) In vitro RNA-lipid binding using in vitro transcribed biotinylated LINK-A sense or antisense, and lipid-coated beads followed by dot-blot assays (upper panel). Bottom panel: graphic illustration of oligonucleotides base-pairing LINK-A sequence. (j) Upper panel: graphic illustration of LINK-A-PC/PIP3 binding. Lower panel: RNA-lipid overlay assay showing the binding of full-length LINK-A and PC- or PIP3-binding region deletion transcripts (ΔPC and ΔPIP3, respectively).
Characterization of LINK-A as a PIP3-binding lncRNA
LINK-A has been characterized as a long intergenic non-protein coding RNA19,29. We first validated the LINK-A-PIP3/PC interaction using lipid strips30; we used lncRNAs with comparable length as the controls, including BCAR4 (1,309 bp), Linc-131 (1,353 bp), and H19 (2,322 bp) and they did not interact with lipids (Fig. 1e). Consistent with lipid-coated beads pulldown assay (Fig. 1d), SNHG9 and RP11-383G10.5 exhibited specific binding to PA and cardiolipin respectively (Fig. 1e). We then performed a fluorescence resonance energy transfer (FRET) assay31 using soluble, fluorophore-conjugated PIP3 (15 nM) as the donor and Alexa-555-streptavidin-labeled LINK-A or RP11-383G10.5 (0–200 nM) as the acceptors (Fig. 1f, upper panel). When excited at 475 nm, we observed robust FRET signals as indicated by an increase of acceptor emissions at 570 nm with concomitant decreases in the donor emission centered around 513 nm (Fig. 1f, lower panel), signifying specific physical contact between LINK-A and PIP3 in vitro. The addition of LINK-A, but not RP11-383G10.5 (0–400 nM), to PIP3 led to a reduction in the emission intensity (Fig. 1g and Supplementary Fig. 1e). These data allow for calculation of the binding affinity of the LINK-A-PIP3 interaction in solution (Kd=110.7±37 nM) (Fig. 1h), which was stronger than or similar to known protein-PIP3 interactions (200 nM to 2 µM)32,33. The cardiolipin-binding lncRNA RP11-383G10.5 exhibited no detectable binding with PIP3 (Fig. 1h).
Next, we mapped the LINK-A sequences required for PC and PIP3 binding using a dot-blot assay18,19 (Fig. 1i). Pre-incubation of biotinylated LINK-A with PC- or PIP3-coated beads protected the lipid-bound LINK-A sequences from RNase I digestion and hybridization of these RNA sequences to dot-blot showed streptavidin signals on dots A5 (nt. 241–300) and D1 (nt. 1,081–1,140) respectively (Fig. 1i), indicating these regions are responsible for lipid binding. Deletion of PC- and PIP3-binding motifs of LINK-A (referred to as ΔPC and ΔPIP3 LINK-A) abolished its interaction with PC and PIP3 respectively (Fig. 1j). Amplified Luminescent Proximity Homogenous Assay (Alpha)34,35 was applied to calculate the LINK-A-PIP3 binding affinity, finding that PIP3 binds specifically to LINK-A with Kd=142 nM (Supplementary Fig. 1f, g). Competition with short RNA sequences of PIP3 binding motif (nt. 1,081–1,140) but not PC-binding motif revealed a reduced PIP3 binding to LINK-A (Supplementary Fig. 1 g), indicating that nt. 1,081–1,140 of LINK-A is required for PIP3 binding.
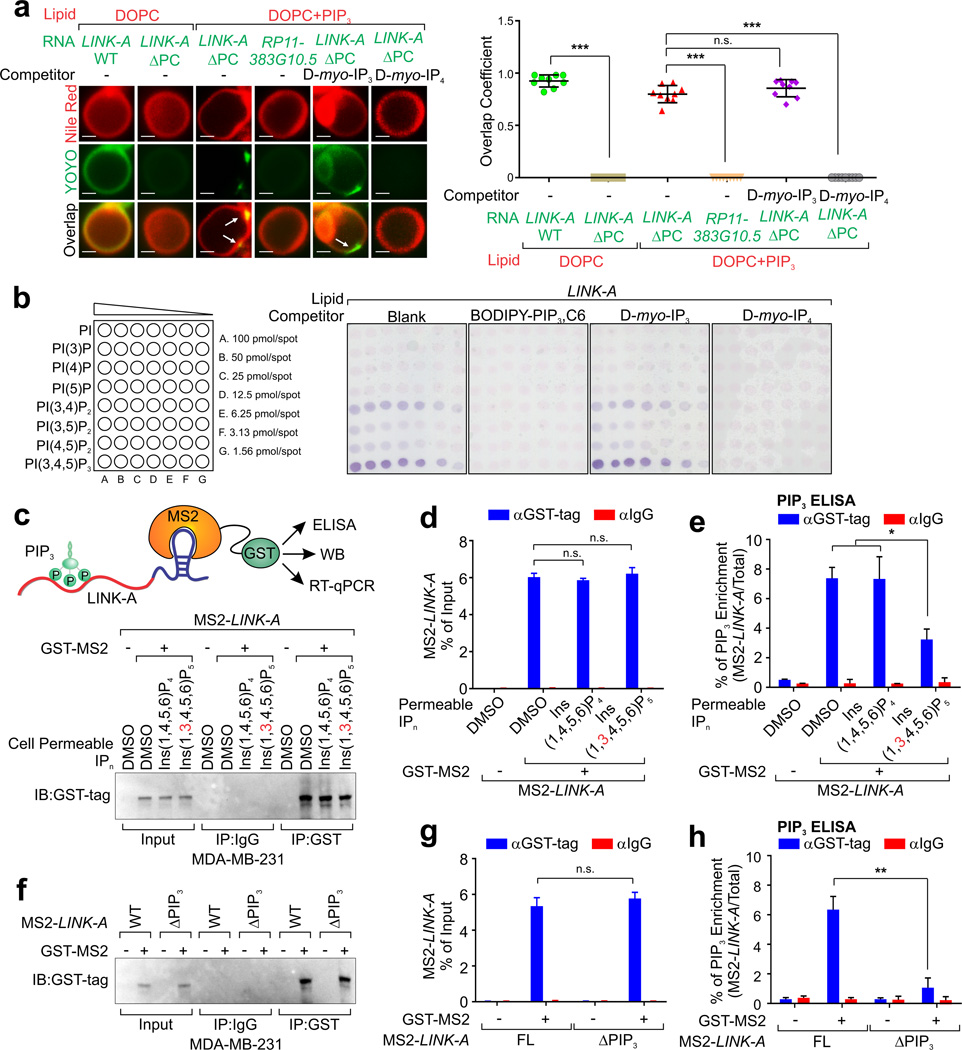
To visualize LINK-A association with phospholipids in the context of a membrane, we generated giant unilamellar vesicles (GUVs) composed of 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) alone or DOPC with PIP3 (1.5% mol)36–38. Full-length but not ΔPC LINK-A or non-lipid-binding lncRNAs H19, Linc-131 and BCAR4 or cardiolipin-binding lncRNA RP11-383G10.5, bound to the surface of the DOPC containing GUVs (Fig. 2a and Supplementary Fig. 2a–c). LINK-A ΔPC was recruited to the surface of the GUVs as clusters, which is consistent with the notion that PIP3 segregates into enriched regions within the plasma membrane39. The PIP3 analog D-myo-Inositol 1,3,4,5-tetrakisphosphate (IP4) but not D-myo- Inositol 1,4,5-trisphosphate (IP3) (100:1 molar ratio to PIP3) abolished this interaction (Fig. 2a). Using PIP arrays, we observed that LINK-A exhibited the association with PIP3, weaker binding to PIP2, and minimal interaction with other PIPs (Fig. 2b). This interaction was abolished in the presence of soluble PIP3 or IP4 (100:1 molar ratio to RNA) but not IP3 (Fig. 2b).
Figure 2. Characterization of LINK-A-PIP3 interaction.
(a) Fluorescence microscopy analysis reveals the interaction between lipid vesicles (visualized by Nile Red) and indicated lncRNA (visualized by YOYO-1), in the absence or presence of the PIP competitor D-myo-Inositol 1,4,5-tetraphosphate (IP3) or D-myo-Inositol 1,3,4,5-tetraphosphate (IP4) at 100:1 molar ratio to PIP3 (IP3/IP4: PIP3). Left panel: representative images. Scale bars, 50 nm. Right panel: the quantification of lncRNA-lipid vesicle interactions is represented by calculations of overlap coefficients between lipid vesicles (channel 1, Nile Red) and lncRNA (channel 2, YOYO-1) (n=9 lipid vesicles; median, one-way ANOVA, n.s. p>0.05 and ***p<0.001). (b) RNA-lipid overlay assay showing the binding of LINK-A to PIPs in the absence or presence of soluble BODIPY®-FL PIP3, IP3, or IP4 at a molar ratio of 100:1 (lipid:RNA). (c–e) Determination of interaction between PIP3 and LINK-A by MS2-TRAP assay. Upper panel: schematic illustration of MS2-TRAP assay (c). MS2-tagged LINK-A and its associated protein/lipid complexes were pulled down by anti-GST antibodies from cells treated with DMSO, cell permeable PI(1,4,5,6)P4 or PI(1,3,4,5,6)P5 (100 µM, 2 hrs) followed by immunoblotting (c, lower panel), RT-qPCR (d), and PIP3 mass ELISA (e). (f–h) MS2-TRAP analysis of full-length LINK-A or ΔPIP3 deletion mutant associated protein/lipid complexes by immunoblotting (f), RT-qPCR (g), and PIP3 mass ELISA (h). For d, e, g and h, mean ± s.e.m. were derived from n=3 independent experiments (n.s. p>0.05, *p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Statistics source data for a are in Supplementary Table 6.
To examine the LINK-A-PIP3 interaction in vivo, we took advantage of the MS2-TRAP (MS2-tagged RNA affinity purification) system40,41 by expressing MS2-tagged FL LINK-A or ΔPIP3 deletion mutant in MDA-MB-231 cells and analyzing the protein-RNA-lipid complex pulled down by GST antibodies (Supplementary Fig. 2d, g and Fig. 2c, upper panel). By pulling down GST-MS2 (Fig. 2c, lower panel), we retrieved similar levels of MS2-LINK-A from cells treated with DMSO, permeable PI(1,4,5,6)P4 or PI(1,3,4,5,6)P5 (Fig. 2d). MS2-LINK-A-associated PIP3 was detected in the presence of DMSO or PI(1,4,5,6)P4 (Fig. 2e) by PIP3 mass ELISA42. Conversely, in the presence of PI(1,3,4,5,6)P5, MS2-LINK-A-PIP3 interaction was impaired (Fig. 2e), suggesting that the 3’-phosphate of PIP3 is important for LINK-A binding. Further, the MS2-tagged LINK-A ΔPIP3 failed to associate with PIP3 (Fig. 2f–h). These observations were not due to aberrant PI3K activity or level as revealed by PI3-Kinase Activity ELISA and PI3K immunoblotting respectively (Supplementary Fig. 2e, f and h, i). Additionally, LINK-A binds to IP4 with an affinity (Kd =210 nM) similar to that for PIP3 (Supplementary Fig. 3a, b), indicating that while the 3’-, 4’-, and 5’-phospho groups of PIP3 are all required, the 3’- phospho group is the most important for LINK-A binding. RNA immunoprecipitation (RIP) assay using a PIP3 antibody showed the LINK-A-PIP3 interaction in vivo, which was abolished by PI3K inhibitor LY294002 (Supplementary Fig. 3c, d).
The ErbB receptor tyrosine kinases activate PI3K and downstream AKT in a ligand-dependent manner43,44. We reasoned that LINK-A-PIP3 interaction is triggered by EGF stimulation. We observed that LINK-A was enriched in the membrane fraction upon EGF treatment (Supplementary Fig. 4a–c). Rescue experiments after LINK-A was knocked down by Locked Nucleic Acids (LNA) and followed by reintroduction of LNA-resistant FL LINK-A or LINK-A deletion mutants (ΔPC and ΔPIP3 respectively) (Supplementary Fig. 4d, e), indicated that depletion of LINK-A diminished LINK-A-PIP3 interaction, which was rescued by FL or ΔPC LINK-A but not ΔPIP3 LINK-A (Supplementary Fig. 4f). These results validate that LINK-A bind PIP3 in a specific manner.
LINK-A facilitates AKT recruitment to PIP3 and enzymatic activation of AKT
Hyperactivation of PI3K and AKT-PIP3 interaction directs tumor growth and metastasis45,46. We examined the role of LINK-A in AKT-PIP3 interaction using lipid strips, finding that although recombinant AKT associated with PIP3 in vitro, the presence of FL LINK-A enhanced the AKT-PIP3 interaction in a PIP3 binding-dependent manner (Fig. 3a). We further measured the Kd of the AKT-PIP3 interaction in the presence or absence of LINK-A. Using Alpha-based saturation and competition assays, we calculated that Kd for the AKT-PIP3 interaction without LINK-A is between 304–349 nM, which is consistent with literature32 (Fig. 3b–d). AKT exhibited minor binding with PIP2 (Kd=1,243 nM) and no binding with PI(4)P (Kd=N.D.) (Fig. 3c, middle and right panels). In the presence of FL but not ΔPIP3 LINK-A, AKT-PIP3 binding was enhanced 16 fold (Kd =19 nM) (Fig. 3d). These data suggest that AKT, PIP3, and LINK-A form a complex upon ligand stimulation, which was confirmed by RIP assay (Fig. 3e). Further, knockdown of LINK-A impaired AKT phosphorylation at Thr308 and Ser473, under both basal and EGF stimulation conditions, leading to impaired AKT activation as revealed by diminished phospho-GSK-3β (Fig. 3f, g). LINK-A depletion also abolished AKT-PIP3 interaction in vivo (Fig. 3h). Next, we examined whether LINK-A modulates AKT kinase activity, finding that LINK-A knockdown impaired AKT kinase activity in EGF-treated cells and overexpression of FL but not ΔPIP3 LINK-A rescued AKT activation (Fig. 3i). The presence of FL but not ΔPIP3 LINK-A enhanced AKT enzymatic activity in a polyPIPosomes-mediated in vitro assay6 (Supplementary Fig. 4g). These data suggest that LINK-A enhances AKT-PIP3 interaction to facilitate the AKT enzymatic activation.
Figure 3. LINK-A enhances AKT-PIP3 interaction and AKT kinase activation.
(a) Lipid-RNA-protein overlay assay revealing AKT-PIP3 interaction in the absence or presence of FL LINK-A or ΔPIP3 deletion transcript. Cardiolipin-binding lncRNA RP11-383G10.5 serves as a negative control. (b) Schematic illustration of Alpha-based AKT-PIP3 binding assay. (c) Saturation curve used to determine Kd of the interactions between AKT and biotinylated-PIP3 (left panel), -PIP2 (middle panel), and -PI(4)P (right panel) in Alpha format (mean ± s.e.m. were derived from n=3 independent experiments). (d) Left panel: competition binding assay to determine Kd for a biotinylated PIP3 (0.4 µM)-His6-AKT (0.4 µM) interaction in the presence of indicated RNA transcripts. Unlabeled PIP3 was titrated from 33 µM to 0.25 nM. Right panel: summary of Kd values (mean ± s.e.m. were derived from n=3 independent experiments). (e) Association between LINK-A and AKT in vivo detected by RIP assay using AKT antibody in MDA-MB-231 cells treated with EGF. (f and g) Immunoblotting of AKT pathway components in MDA-MB-231 cells transfected with control or LINK-A siRNAs followed by EGF treatment (f), or transduced with control or LINK-A shRNA (g). (h) Immunoblotting of AKT retrieved by immunoprecipitates using indicated PIP-coated beads from MDA-MB-231 cell transfected with indicated siRNAs followed by EGF treatment. (i) Quantification of AKT kinase activity in cell lysates extracted from MDA-MB-231 cells transfected with LNA against LINK-A followed by overexpression of indicated rescue vectors and EGF treatment. For e and i, mean ± s.e.m. were derived from n=3 independent experiments (*p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
Molecular mechanisms of LINK-A-PIP3-AKT interactions
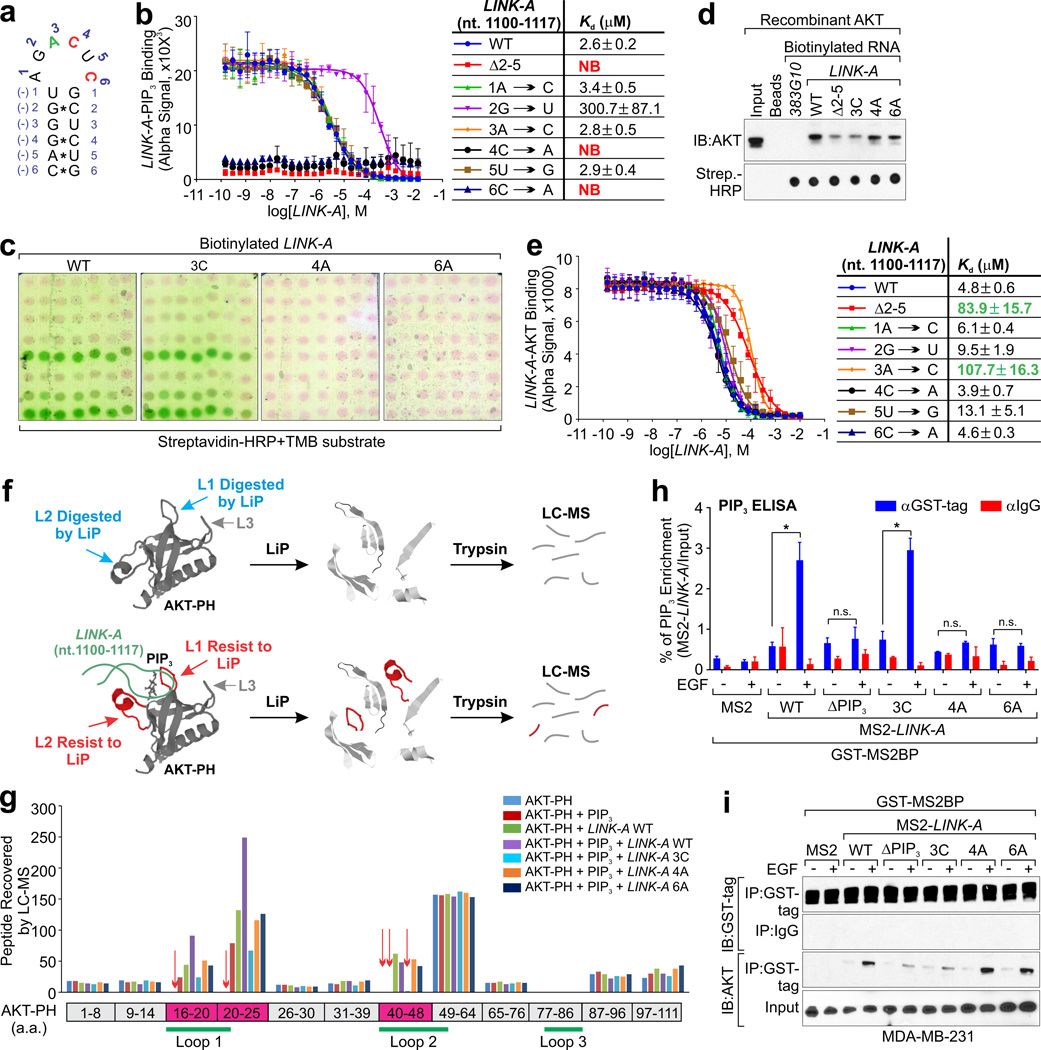
Structural analysis has shown that RNA loops may be important for protein associations47,48. The secondary structure of LINK-A is involved in its interaction with BRK and LRRK219 (Supplementary Fig 5a, black circle) and possibly in PIP3 interaction (Supplementary Fig. 5a, red circle). The 60-nt RNA oligonucleotides of the PIP3 binding motif (nt. 1,081–1,140) harbors an identical stem-loop structure based on computational calculation (Supplementary Fig. 5b, red circle). We synthesized Digoxigenin (DIG)-labeled 18-nt RNA oligonucleotides harboring wild-type LINK-A sequence (nt.1,100–1,117, 5’-CAGGGUAGACUCGCUCUG-3’, loop underlined) (Fig. 4a), RNA oligonucleotides with the 4 central nucleotides of the loop removed or single nucleotides of the loop region mutated (Supplementary Fig. 5c). Alpha assay indicated an adequate interaction between wild-type LINK-A and PIP3 in vitro, with Kd of 2.6 µM (Fig. 4b). Deletion of the loop region (Δ2–5) abolished RNA-PIP3 interaction (Fig. 4b). Interestingly, RNA oligonucleotides with C to A mutations at either position 4 or 6 showed undetectable interaction with PIP3 (Fig. 4b), suggesting the importance of nucleotides C1109 and C1111 of LINK-A in mediating PIP3 binding. RNA-PIPs overlay assay also exhibited undetectable association of PIP3 with either LINK-AC1109A (referred to as 4A) or LINK-AC1111A (referred to as 6A) (Fig. 4c and Supplementary Fig. 5d). Using a similar strategy, the 18-nt RNA oligonucleotide representing LINK-A (nt.1100–1117) directly associated with recombinant AKT in vitro (Fig. 4e). Nucleotide A1108 at position 3 of the stem-loop was required to mediate LINK-A-AKT interaction (Fig. 4e). LINK-A A1108 to C mutant (referred to as 3C) exhibited similar binding to PIP3 as wild-type LINK-A (Fig. 4b), but impaired binding to AKT in vitro (Fig. 4d).
Figure 4. Molecular mechanisms of LINK-A-PIP3-AKT interaction.
(a) Schematic illustration of the stem-loop structure of LINK-A nt. 1,100–1,117. (b) Competition Alpha binding assay to determine Kd for a biotinylated PIP3: DIG-LINK-A oligonucleotides interaction in the presence of full-length LINK-A. Unlabeled LINK-A was titrated from 10 mM to 0.1 nM (mean ± s.e.m. were derived from n=3 independent experiments). (c) RNA-lipid overlay assay showing the binding of wild-type LINK-A or mutants to PIP array using TMB substrates. (d) In vitro RNA pull-down using biotinylated RNAs as indicated and recombinant AKT. The immunoprecipitates were subjected to IB detection using anti-AKT antibody. The presence of RNA transcripts were detected by streptavidin-HRP using dot-blot assay. (e) Competition binding assay to determine Kd for a DIG-LINK-A: His-AKT interaction in the presence of FL LINK-A titrated from 10 mM to 0.1 nM (mean ± s.e.m. were derived from n=3 independent experiments). (f) Schematic illustration of LiP followed by LC-MS. (g) Recombinant AKT PH domain, and/or PIP3, wild-type LINK-A, 3C, 4A or 6A mutant were subjected to LiP followed by LC-MS. Recovered peptide number by LC-MS for each fragment of AKT PH domain were shown. (h and i) MS2-tagged LINK-A and its associated protein/lipid complexes were pulled down by anti-GST antibodies from cells transfected with indicated expression vectors followed by PIP3 mass ELISA (h) or IB detection using indicated antibodies (i). For h, mean ± s.e.m. was derived from n=3 independent experiments (n.s. p>0.05 and *p<0.05, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
Crystallographic analysis indicated that AKT PH domain harbors three variable loops (referred as L1: aa. 16–21; L2: aa. 40–52; L3: aa. 80–81) (Fig. 4f)49,50. The phospho-groups of IP4 forms hydrogen bonds with Lys14 and Arg23, which are flanking L149,50. We hypothesize that the formation of LINK-A-PIP3-AKT complex may cause potential conformational change of AKT PH domain, which can be studied by limited proteolysis51. We applied Limited Proteolysis (LiP) followed by liquid chromatography–mass spectrometry (LC-MS) analysis (LiP-LC-MS)52,53 to analyze the AKT PH domain (Fig. 4f). The loop regions of AKT PH domain (aa. 16–24 referred to as L1 and aa. 40–48 referred to as L2) were digested by LiP and were not detected by LC-MS (Fig. 4g and Supplementary Table 2). For the PIP3-bound AKT PH domain, L1 resisted partial digestion and was detected by LC-MS (Fig. 4g and Supplementary Table 2), which is consistent with the previous studies49,50. Under the same conditions, no peptides were recovered from the L2 region, suggesting that L2 of the AKT PH domain is not involved in interacting with PIP354. In the presence of LINK-A, both L1 and L2 resisted LiP, probably due to LINK-A-AKT association and/or potential conformational change of the PH domain (Fig. 4g and Supplementary Table 2). Peptides recovered from the L1 region were increased when both PIP3 and LINK-A associated with the PH domain, suggesting a synergistic effect of LINK-A in stabilizing the PIP3-AKT-LINK-A complex. In the presence of LINK-A 3C mutant, fewer peptides were detected, which suggests that the synergistic effect was lost when LINK-A-AKT interaction was abolished. Furthermore, LINK-A 4A and 6A mutants failed to protect L1 from LiP, further supporting the notion that LINK-A-PIP3 interaction enhances PIP3-AKT interaction (Fig. 4g and Supplementary Table 2). Two factors that likely contribute to the synergistic effect are: 1) direct LINK-A-AKT interaction and 2) AKT conformational change in response to this interaction.
We further generated MS2-tagged LINK-A 3C, 4A and 6A mutants for an MS2-TRAP assay (Fig. 4h). Compared to WT LINK-A, 3C exhibited similar interaction with PIP3; however, ΔPIP3, 4A and 6A mutants failed to interact with cellular PIP3 as revealed by PIP3 mass ELISA (Fig. 4h), although similar amounts of MS2-tagged LINK-A were retrieved (Supplementary Fig. 5e). We also determined that LINK-A ΔPIP3 and 3C mutants exhibited impaired association with AKT, while 4A and 6A were as competent as WT LINK-A (Fig. 4i). Therefore, we identified the single nucleotides of LINK-A that are required for PIP3 and AKT binding respectively and provide structural evidence underlying LINK-A facilitated AKT-PIP3 interaction.
Functional relevance of LINK-A-PIP3 interaction in AKT activation
We determined that there are roughly 150 copies of the LINK-A per MDA-MB-231 cell (Supplementary Fig. 6a, b) and the copy number was reduced to 10–15 per cell by LNA, which was restored by exogenously expressed LINK-A or the ΔPIP3 deletion mutant (Supplementary Fig. 6b). LINK-A knockdown abolished EGF-induced AKT phosphorylation and cell proliferation (Supplementary Fig. 6c, d). Reintroduction of FL LINK-A, but not the ΔPIP3 mutant, restored AKT/GSK-3β phosphorylation and tumor cell proliferation (Supplementary Fig. 6c, d). Less than 10 copies of LINK-A are found in normal mammary gland epithelial cells (MCF-10A). Stably expressing FL LINK-A in MCF-10A cells, to about 150 copies per cell, amplified AKT/GSK-3β phosphorylation and cell proliferation (Supplementary Fig. 6e–g).
We used a pair of colorectal cancer cell lines (DLD-1 PIK3CA+/+ and DLD-1 PIK3CA+/−) in which PIP3 levels were genetically altered. Parental DLD-1 PIK3CA+/+ cells harbors about 0.6 pmol PIP3 per 5×106 cells before EGF stimulation, and about 2.8 pmol PIP3 after EGF stimulation (Fig. 5a). In DLD-1 PIK3CA+/− cells, the PIP3 level per 5×106 cells was about 0.2 pmol (Fig. 5a). Consistently, the EGF-induced AKT phosphorylation at Thr308 and Ser473 were abolished in DLD-1 PIK3CA+/− cells (Fig. 5b). We also measured the copy number of LINK-A in DLD-1 PIK3CA+/+ and PIK3CA+/− cells, finding that both cell lines harbored about 10 copies of LINK-A (Fig. 5d). We then chose to deliver the in vitro transcribed and capped wild-type LINK-A or mutant intracellularly into DLD-1 cells (Fig. 5c). We attempted to deliver 150 copies of LINK-A, and RT-qPCR indicated that single DLD-1 cells contained about 120 copies of LINK-A after delivery (Fig. 5d), of which wild-type LINK-A, but not ΔPIP3 mutant, dramatically enhanced the EGF-induced AKT phosphorylation (Fig. 5e).
Figure 5. Functional involvement of LINK-A-PIP3 interaction in mediating AKT activation.
(a and b) Detection of PIP3 (a) and phospho-AKT (b) level in DLD-1 PIK3CA+/+ or PIK3CA−/− cells with or without EGF stimulation. (c) Schematic illustration of intracellular delivery of PIP3 and/or LINK-A to DLD-1 cells. (d and e) RT-qPCR determination of LINK-A copy number (d) or IB detection of indicated proteins (e) in DLD-1 PIK3CA+/+ cells delivered with indicated RNA transcripts with or without EGF stimulation. (f–h) PIP3 mass ELISA (f), RT-qPCR determination of LINK-A copy number (g) or IB detection of indicated proteins (h) in DLD-1 PIK3CA+/− cells delivered with PIP3 and/or LINK-A transcripts with or without EGF stimulation. (i–k) PIP3 mass ELISA (i), RT-qPCR determination of LINK-A copy number (j) or IB detection of indicated proteins (k) in DLD-1 PIK3CA+/− cells delivered with PIP3 and indicated LINK-A deletion transcripts with or without EGF stimulation. (l) IB detection of indicated proteins in DLD-1 PIK3CA+/− cells delivered with PIP3 and indicated LINK-A single nucleotide mutated transcripts with or without EGF stimulation. (m and n) IB detection of immunoprecipitated AKT (m) and quantification of AKT-associated PIP3 (n) in DLD-1 PIK3CA+/+ cells delivered with indicated LINK-A single nucleotide mutated transcripts with or without EGF stimulation. For a, d, f, g, i, j and n, mean ± s.e.m. were derived from n=3 independent experiments (n.s. p>0.05, *p<0.05, and ***p<0.001, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
Further, in DLD-1 PIK3CA+/− cells, delivery of PIP3 (3 pmol per 5×106 cells) increased cellular PIP3 to about 2.2 pmol per 5×106 cells (Fig. 5c, f). However, restored cellular PIP3 levels marginally mediated EGF-induced AKT phosphorylation (Fig. 5h). Delivery of LINK-A (150 copies per cell) alone minimally affected AKT phosphorylation (Fig. 5g, h), while combined delivery of PIP3 and LINK-A robustly enhanced AKT phosphorylation (Fig. 5f–h). To determine if LINK-A-PIP3 interaction but not either alone, is important in enhancing the activation of the EGF-AKT pathway, we delivered WT LINK-A, ΔPIP3, or ΔPC mutants to DLD-1 PIK3CA+/− cells in the presence of PIP3 (Fig. 5i–k). Despite comparable delivery efficiencies, FL LINK-A but not ΔPIP3 mutants enhanced EGF-triggered, PIP3-mediatd AKT phosphorylation (Fig. 5k). Consistently, co-delivery of PIP3 with WT LINK-A, but not 3C, 4A, or 6A mutants in DLD-1 PIK3CA+/− cells led to robust AKT phosphorylation (Fig. 5l and Supplementary Fig. 6h, i). Furthermore, delivery of ~120 copies of WT LINK-A, but not 3C, 4A or 6A mutant, enhanced recruitment of AKT to PIP3 in vivo upon EGF stimulation in DLD-1 PIK3CA+/+ cells (Fig. 5m, n and Supplementary Fig. 6j). Taken together, our data indicate that about 150 copies of LINK-A is functionally sufficient to facilitate EGF-triggered AKT-PIP3 interaction and subsequent AKT activation.
LINK-A confers resistance to AKT inhibitors
There are four types of inhibitors targeting the AKT PH domain that are being developed: 1) phosphatidyl-inositol ether lipid analogs, alkyl-phospho-lipids or other inositol phosphate derivatives, such as perifosine55; 2) sulfonamides56; 3) purine/pyrimidine analogues, such as triciribine57; and 4) allosteric compounds, including MK220658. Since LINK-A enhances AKT activation by interacting with AKT’s PH domain and PIP3, high LINK-A expression may antagonize the effect of AKT inhibitors including perifosine and MK2206. We confirmed that perifosine and MK2206 inhibited PIP3-AKT interaction with an IC50 value of 8.1 nM and 28.7 nM respectively, while AKT kinase domain inhibitors exhibited minimal effects (Fig. 6a). In vitro, the presence of FL LINK-A, but not LINK-A ΔPIP3, antagonized the inhibitory effect of MK2206 and perifosine by 753- and 140- fold respectively (IC50 = 6.4 µM and 4.8 µM) (Fig. 6b, c). Then, we genetically deleted the PIP3 binding motif (PIP3-BM) of LINK-A using CRISPR/Cas9 technology in MDA-MB-231 cells (Fig. 6d and Supplementary Fig. 7a–d). Two independent single-cell clones of LINK-A PIP3-BM−/− cells showed that AKT failed to interact with PIP3 (Fig. 6e–f), leading to diminished AKT phosphorylation (Fig. 6g). We performed an Alpha assay to quantify the EGF-induced AKT phosphorylation in PIP3-BM−/− cells by measuring the amounts of total AKT and phospho-AKT simultaneously, in response to a series of 3-fold dilutions of EGF (Fig. 6h). The EGF concentration that induced 50% phosphorylation of AKT over total AKT was defined as EGF EC50 (Fig. 6i). In LINK-A PIP3-BM+/+ cells, about 11.0 nM of EGF was required to induce 50% AKT phosphorylation, while in PIP3-BM−/− cells 94.5 nM of EGF was needed (Fig. 6i). In rescue experiments, intracellularly delivered WT LINK-A restored sensitivity to EGF stimulation; however, LINK-A 3C, 4A, and 6A mutants failed to rescue this phenotype (Fig. 6i). Pharmacologic validation was performed using 1:2 dilution of MK2206 under the EGF EC80 (200 nM) condition (Fig. 6j). In LINK-A PIP3-BM+/+ cells, 16.2 µM MK2206 was needed to half the AKT phosphorylation (Fig. 6j). However, in LINK-A PIP3-BM−/− cells, only 65.7 nM of MK2206 was sufficient to inhibit AKT activation (Fig. 6j). Intracellular delivery of WT LINK-A, but not 3C, 4A or 6A mutant, into LINK-A PIP3-BM−/− cells restored resistance to MK2206 (Fig. 6j).
Figure 6. LINK-A confers resistance to AKT inhibitors.
(a) Competition binding assay to determine IC50 of indicated AKT inhibitors for inhibiting AKT-PIP3 binding (mean ± s.e.m. were derived from n=3 independent experiments). (b and c) Competition binding assays were used to determine IC50 of MK2206 (b) or perifosine (c) for inhibiting AKT-PIP3 binding in the presence of indicated RNA transcripts (mean ± s.e.m. were derived from n=3 independent experiments). (d) Schematic illustration of genetic depletion of LINK-A PIP3 binding sequence. (e) Upper panel: schematic illustration of detection of AKT-bound PIP3 in LINK-A genetic editing cells. Lower panel: IP and IB detection using anti-AKT antibody in LINK-A PIP3-BM+/+ and PIP3-BM−/− cells. (f and g) PIP3 mass ELISA detection of AKT-bound PIP3 (f) or IB detection of indicated proteins (g) in LINK-A PIP3-BM+/+ and PIP3-BM−/− cells with or without EGF stimulation. (h) Schematic illustration of quantitative measurement of AKT phosphorylation using Alpha assay. (i and j) Percentage of Alpha signal of p-AKT over total-AKT in LINK-A PIP3-BM+/+ and PIP3-BM−/− cells intracellularly delivered with indicated RNA transcript upon titration of EGF stimulation (i) or in the presence of EGF (200 nM) and titration of MK2206 (j). The EC50 of EGF (i, right panel) and IC50 of MK2206 (j, right panel) were shown (mean ± s.e.m. were derived from n=3 independent experiments). For f, mean ± s.e.m. were derived from n=3 independent experiments (n.s. p>0.05 and *p<0.05, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
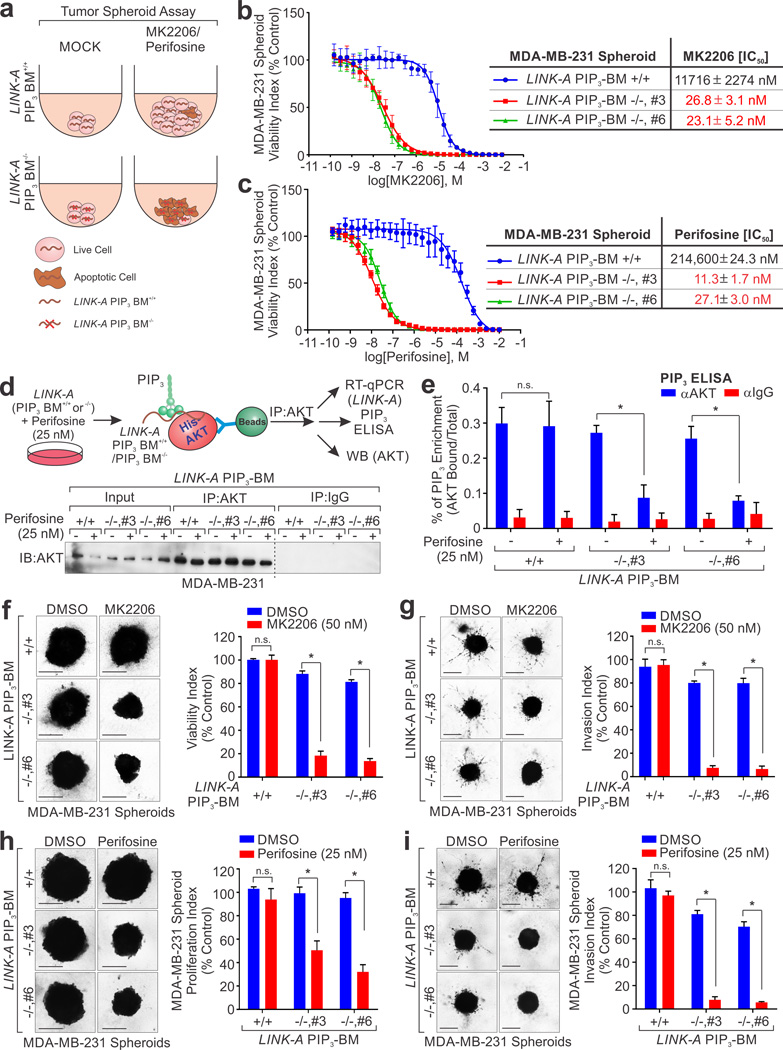
Next, we used 3-dimentional tumor spheroid assay59 to evaluate the impact of LINK-A–PIP3 interaction on AKT inhibitor efficacy (Fig. 7a). About 11.7 µM MK2206 was needed to inhibit LINK-A PIP3-BM+/+ tumor spheroid growth (Fig. 7b). However, only about 50 nM MK2206 was sufficient to inhibit LINK-A PIP3-BM−/− spheroid growth by half (Fig. 7b). The cell viability index indicated that 214 µM perifosine was needed to induce 50% cell death of LINK-A PIP3-BM+/+ spheroid. LINK-A PIP3-BM−/− spheroids were sensitized to perifosine by about 20,000 fold, with an IC50 value of 11.3 nM (Fig. 7c). For LINK-A PIP3BM−/− cells, only 25 nM perifosine was sufficient to inhibit AKT-PIP3 interaction in vivo (Fig. 7d, e). Furthermore, 50 nM MK2206 or 25 nM perifosine efficiently inhibited proliferation and invasion of LINK-A PIP3-BM−/− spheroids (Fig. 7f–i), while the same dose elicited an undetectable effect on LINK-A PIP3BM+/+ spheroids (Fig. 7f–i). These data indicate that LINK-A-PIP3 interaction confers resistance to AKT inhibitors.
Figure 7. Depletion of LINK-A sensitize cancer cells to treatment of AKT inhibitors.
(a) Schematic illustration of tumor spheroid assay using LINK-A PIP3-BM+/+ and PIP3-BM−/− cells. (b and c) 3-D spheroid fluorometric proliferation/viability assay conducted using LINK-A PIP3-BM+/+ or LINK-A PIP3-BM−/− spheroids to determine IC50 of MK2206 (b) or perifosine (c) (mean ± s.e.m. were derived from n=3 independent experiments). (d) Upper panel: schematic illustration of detection of AKT-bound PIP3 in LINK-A genetic editing cells treated with perifosine. Lower panel: IB detection using anti-AKT antibody in LINK-A PIP3-BM+/+ or LINK-A PIP3-BM−/− cells treated with 25 nM perifosine for 1 hour. The dotted line indicates the boundary of two separate blots whose uncropped images are shown in Supplementary Fig. 9. (e) PIP3 mass ELISA determination of AKT-associated PIP3 in LINK-A PIP3-BM+/+ or LINK-A PIP3-BM−/− cells treated with 25 nM perifosine for 1 hour. (f and g) Growth (f) and invasion (g) of LINK-A PIP3-BM+/+ or LINK-A PIP3-BM−/− spheroids after 4 days treatment with 50 nM MK2206. Left panels: phase contrast images of spheroids. Scale bars, 300 µm; Right panels: quantitative analysis of surface area for spheroids growth and invasion treated with DMSO or MK2206 (n=3 independent experiments based on 4 spheroids per group), presented as proliferation and invasion index respectively. (h and i) Growth (h) and invasion (i) of LINK-A PIP3-BM+/+ or LINK-A PIP3-BM−/− spheroids after 4 days treatment with 25 nM perifosine. Left panels: phase contrast images of spheroids. Scale bars, 300 µm; Right panels: quantitative analysis of surface area for spheroids growth and invasion treated with DMSO or perifosine (n=3 independent experiments based on 4 spheroids per group), presented as proliferation and invasion index respectively. For e–i, mean ± s.e.m. were derived from n=3 independent experiments (n.s. p>0.05 and *p<0.05, two-tailed paired Student’s t-test). Statistics source data for f, g, h and i are in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
Genetic mutation and amplification of LINK-A correlate with AKT phosphorylation in human cancer
By analyzing somatic copy number variants amongst clinical samples, we found that the LINK-A gene locus is amplified in multiple cancer types including breast cancer (Fig. 8a). We also identified genetic mutations within or adjacent to the LINK-A gene locus in breast and lung cancer patient samples (Supplementary Fig. 8a–d). Intriguingly, mutation of a single nucleotide polymorphism (SNP) downstream of the LINK-A transcription unit correlated with LINK-A expression and breast cancer patient outcomes (Fig. 8b, c and Supplementary Fig. 8a). Likewise, NCBI dbSNP Reference SNP (refSNP) Cluster Report showed that African women with advanced breast cancer exhibit the highest incidence of double allele mutations of this SNP compared to Europeans and Asians (Fig. 8d).
Figure 8. Genetic variation of LINK-A correlates with breast cancer risk.
(a) Somatic copy number variants (CNV) of LINK-A gene locus in multiple cancer types (n=376, 1,096, 36, 57, 573, 518, 470, 302, 504, 134, 185, 87, 179, 547, 593, 185, 412, 626, 1,090, 514, 80, 521, 443, 498, 532, 124, 166, 505, 616, 458, 883, 261, 48, 290, 92 and 66 tumor samples respectively). The boxes show the median and the interquartile range. The boxes show the median±1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile ranges from the box boundaries The red dot lines showing threshold of 0.2 in segment mean values was considered an amplification while −0.2 was considered a deletion. (b) SNP rs12095274 is significantly associated with survival of breast cancer in TCGA samples (n=765, 137, and 16 patients, log rank test). (c) SNP rs12095274 correlates with LINK-A expression in TCGA samples (n=765, 137, and 16 patients; ***p<0.001two-tailed Wilcoxon test). The boxes show the median±2 quartile, with whiskers extending to the most extreme data point within 2 interquartile ranges from the box boundaries. (d) Population distribution of SNP rs12095274. (e and f) Pearson’s correlation analysis comparing staining density between LINK-A and phospho-AKT (Thr308) (e) or phospho-AKT (Ser473) (f) status (n=40 breast tumors; ***p<0.001, Fisher’s exact test). (g) SNP rs12095274 correlates with phospho-AKT staining density in breast cancer tissues (n=79, 16 and 2 breast tumors; *p<0.05, median±1 quartile, one-way ANOVA). (h) SNP rs12095274 correlates with LINK-A expression detected by RT-qPCR in breast cancer tissues (n=102, 17 and 4 breast tumors; *p<0.05, median±1 quartile, one-way ANOVA). (i) Schematic illustration of LNA injection and PET scan. (j) Left panel: Representative PET/CT images of nude mice bearing breast tumors injected with scrambled or LINK-A LNA. White dotted lines indicate area of the breast tumors and black dotted lines indicate area of muscle tissues that were analyzed for in vivo glucose uptake. Right panel: Statistical analysis of in vivo glucose uptake on day 3 and day 6 (n=3 independent experiments based on 3 mice per group). (k) Effects of LINK-A LNA on growth of MDA-MB-231 xenografts in nude mice (n=3 mice per group). The arrow indicates the tumor burden. For j and k, mean ± s.e.m. were derived from n=3 independent experiments (*p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Statistics source data for j and k are in Supplementary Table 6.
We assessed LINK-A expression in breast cancer tissue samples (clinical information listed in Supplementary Table 3) by RNAscope®, finding that high LINK-A expression correlated with breast cancer malignancy and unfavorable outcomes (Supplementary Fig. 8e, f). Our data suggest strong correlation between LINK-A expression and AKT phosphorylation at Thr308 and Ser473 (r2=0.7392 and 0.6659, p<0.0001 respectively) (Fig. 8e, f and Supplementary Fig. 8g). We confirmed that tissue samples with single or double allele mutations (A–G) of the aforementioned LINK-A SNP exhibited elevated LINK-A expression and increased AKT phosphorylation at Thr308 (Fig. 8g, h). These data indicates the importance of LINK-A in regulating the PIP3-AKT signaling pathway in human cancer.
Finally, we determined that LINK-A is required for cell proliferation and anti-apoptosis (Supplementary Fig. 8h–k). We next examined the therapeutic value of LNAs targeting LINK-A in an orthotopic mouse models of breast cancer. LNA administration inhibited in vivo tumor glucose uptake revealed by positron emission tomography (PET) scanning (Fig. 8i, j). Tumor burden was considerably diminished with LINK-A LNA treatment compared to the control group (Fig. 8k), suggesting that by using LNAs to block LINK-A in vivo, glycolysis and tumorigenesis can be repressed in breast cancer cells.
Discussion
We have identified a cohort of lncRNAs that associates with various membrane phospholipids, including PC and PIP3. Our data show that LINK-A-PIP3 interaction facilitates AKT activation upon EGF stimulation, indicating that this interaction is important in both homeostasis and cancer. The association between the AKT PH domain and PIP3 may cause a conformational change in AKT, making the phosphorylation sites accessible to its activating kinases. Our data demonstrate that LINK-A promoted the recruitment of AKT to PIP3, which enhances AKT phosphorylation.
Many key regulators exhibit high binding affinities for PIP3, with lower binding affinities for other PIPs33. Our data indicate that LINK-A exhibits strong PIP3 binding affinity with a Kd around 111.7 nM in the liquid phase. This binding affinity is similar to or stronger than that of many known PIP3 binding proteins. For example, PLC-δ1 and AKT binds PI(1,4,5)P3 with Kd 210 nM and 400 nM respectively32,60. Therefore, the interaction between LINK-A and PIP3 is probably biologically relevant.
The consequence of LINK-A-PIP3 interaction is intriguing. As a proof of concept, we examined the AKT kinase activity in the presence of both LINK-A and PIP3. Our data demonstrate that in vitro, LINK-A promoted AKT recruitment to PIP3; in vivo, LINK-A-PIP3 and LINK-A-AKT interactions are required for AKT phosphorylation. LiP-LC-MS analysis provides further support that LINK-A association with the L1 and L2 loop of the AKT PH domain, which may elicit a conformational change, facilitating L1loop interaction with PIP3, given that the 3’-phosphate of IP4 associates with L1 loop50. These effects further stabilize the AKT-PIP3 interaction, leading to a more “open” structure and enhanced AKT enzymatic activation. It is cautionary that LiP-LC-MS only provides initial evidence for potential conformational change of target protein under different conditions, which will pave the way for ultimate structural resolution of RNA-lipid by nuclear magnetic resonance (NMR) and/or X-ray crystallography.
LINK-A is highly expressed in breast cancer and correlated with outcomes of breast cancer patient. In human cancer, LINK-A is likely upregulated by two mechanisms: genetic amplification and disease-related SNPs that modulate LINK-A expression. Our study suggests that cancer with high LINK-A expression may exhibit resistance to AKT inhibitors, with implications for clinical trials and the development of inhibitors. LINK-A could likely serve as a biomarker to predict resistance to AKT PH domain inhibitors. Breast cancer patients could be stratified based on LINK-A expression levels to help determine an individual’s sensitivity to AKT inhibitor treatment. These findings demonstrate the importance of lncRNAs in cancer biology and signify the physiological relevance of RNA-PIP3 interaction.
Methods
Tissue samples
Fresh-frozen triple-negative breast cancers and adjacent normal tissues were purchased from Asterand Bioscience and clinical parameters were list in a previous study18. Two sets of fresh-frozen breast cancer tissues (Nanjing Cohort and Duke Cohort) were collected from Yixing People’s Hospital (Yixing, Jiangsu Province, China) and Duke University respectively. The study protocol was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China) and Duke University Health System. All tissue samples were collected in compliance with informed consent policy. Detailed clinical information is listed in Supplementary Table 3.
Cell lines, transfection, treatments and cellular assays
Human breast cancer cell lines MDA-MB-231, MDA-MB-468 and human mammary gland epithelial cell line MCF-10A were purchased from American Type Culture Collection (ATCC). DLD-1 PIK3CA+/+ and PIK3CA+/− cells were purchased from Sigma-Aldrich. All cell lines were authenticated by autosomal STR profiles provided by Characterized Cell Line core (MD Anderson Cancer Center). All cell lines were free of mycoplasma contamination tested by vendors using MycoAlert kit from Lonza. siRNA and plasmid transfections were performed using DharmaFECT4 (Dharmacon) and Lipofectamine® 3000 (Life Technologies) respectively. None of the cell lines used was found in the database of commonly misidentified cell lines that are maintained by ICLAC and NCBI Biosample. For growth factor treatment, cells were serum starved for 24 hrs followed by treatment with indicated concentration of EGF (Peprotech) for 20 mins. For competition binding assay, cells were pretreated with 100 µM cell permeable PI(1,4,5,6)P4 or PI(1,3,4,5,6)P5 for 2 hrs. For pharmacologic inhibition, cells or tumor spheroids were treated with MK-2206 or Perifosine (SelleckChem) with the indicated time and concentrations. Membrane and cytoplasmic fractionation were prepared using ReadyPrep™ Protein Extraction Kit (Membrane I) (Bio-Rad). Immunoblotting, immunoprecipitation, cell proliferation and apoptosis assays were performed as previously described19. LINK-A PIP3-binding motif knockout cell lines were generated using the CRISPR/Cas9 genome editing system by Gene Editing/Cellular Model Core Facility (MD Anderson Cancer Center).
RNA biology assays
All lncRNAs were in vitro transcribed and purified as previously described18,19. To isolate lipid-bound RNAs, total lipids from the fresh-frozen triple-negative breast cancers and their adjacent normal tissues were extracted by Barocycler® NEP3229 using Pressure Enhanced Systems Biology Kit (Pressure BioScience). The lipid-bound RNAs were further purified by Trizol®. LncRNA array hybridization and data analysis were performed as previously described18. RNAScope®, RIP assay, in vitro RNA-lipid binding coupled with dot-blot assay, immunohistochemistry and image quantification was performed as previously described18,19. Intracellular delivery of 3´-0-Me-m7G(5')ppp(5')G capped LINK-A was performed using TransIT®-mRNA Transfection Kit (Mirus Bio). The MS2-TRAP assay was performed as described previously40,41.
Lipid reagents, antibodies and siRNA, shRNA and LNA™
All reagents for lipid experiments were purchased from Echelon Biosciences. PIP3 were quantified using PIP3 mass ELISA kit (Echelon Biosciences). PI3K activity was measured using PI3-Kinase Activity ELISA (Echelon Biosciences). Intracellular PIP3 delivery was performed using PI(3,4,5)P3 Shuttle PIP™ Kit (Echelon Biosciences).
The antibodies used in this study are summarized in Supplementary Table 4. Commercially available Lincode SMARTpool siRNA targeting LINK-A (R-027622) was used in this study. The oligonucleotides for shRNA targeting LINK-A were designed based on Lincode SMARTpool siRNA sequence and cloned into pLKO.1-Puro vector. LNAs targeting LINK-A or a scrambled sequence were designed and synthesized by Exiqon. Detailed sequences were listed in the Supplementary Table 5.
Cloning Procedures
Mammalian expression vectors for full-length LINK-A and various deletion mutants were constructed by subcloning the gene sequences into pCDNA3.1 (+) backbone (Life Technologies), pBabe retroviral expression vector, or MS2-24x-pCNDA vector41. To generate LNA#5-resistant LINK-A expression vectors used in the rescue experiments, LNA#5 targeting sequence ACA GCT CAT TTA TCC A was mutated to ACA GGC GAT TTA TCC A. All single-point and deletion mutations were generated using QuikChange™ Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Detailed oligonucleotide sequences are listed in the Supplementary Table 5.
RNA/protein-lipid overlay assay
Membrane lipid strip or PIP array was blocked with RNA-lipid binding buffer (50 mM HEPES pH 7.0, 50 mM NaCl, 5 mM MgCl2, and 2 mM CaCl2) supplemented with 3% BSA and 10 µg/ml yeast tRNA and then incubated with in vitro transcribed RNAs (1 µg/ml) in RNA-lipid binding buffer supplemented with 50 U/ml RNase inhibitor. The strip/array was washed three times with RNA-lipid binding buffer supplemented with 0.05% NP-40 and further incubated with Streptavidin-HRP. The strip/array was washed three times as described above and the bound RNA was detected by TMP Precipitating reagent. For PIP competition experiments, D-myo-Inositol 1,3,4,5-tetraphosphate (IP4), D-myo-Inositol 1,4,5-tetraphosphate (IP3) or BODIPY®FL-PIP3 was added in RNA-lipid overlay assay at 100:1 molar ratio (lipid: RNA). For lipid-RNA-protein overlay assay, after blocking once and washing three times, the strip was further incubated with indicated proteins followed by washes as described above. Strips were probed with the specific primary antibody followed by peroxidase-conjugated secondary antibodies.
Lipid-RNA pull-down assay
The lipid-coated beads were blocked with RNA-lipid binding buffer (see above) supplemented with 3% BSA, 10 µg/ml yeast tRNA, and lipid phosphate phosphatase inhibitors (XY-14, 3-a-aminocholestane, SF1670) followed by incubation with cell lysate. The beads were washed with RNA-lipid binding buffer supplemented with 0.05% NP-40. RNAs were eluted by RIP elution buffer (100 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) and recovered by RNA Clean & Concentrator™-5 (Zymo Research) for RT-qPCR analysis.
RNA-lipid binding Fluorescence Resonance Energy Transfer (FRET) assay
200 nM BODIPY® FL Phosphatidylinositol 3,4,5-trisphosphate (BODIPY -PIP3, λexc=503 nm, λemi=513 nm; Donor) was prepared in RNA-lipid binding buffer (see above). Biotinylated-LINK-A or RP11-383G10.5 was labeled with Alexa Fluor® 555 Streptavidin Conjugates. 400 µl BODIPY-PIP3 (20 nM) (donor) was equally aliquoted into two fractions: one incubated with 200 µl 20 nM Alexa-555-Strep-Biotin-LINK-A or RP11-383G10.5 (acceptor) while the other serves as control by incubation with 200 µl of 20 nM Alexa-555-Strep in the RNA-lipid binding buffer. The spectra were collected from 490 nm to 650 nm with slit widths set at 1 nm and the excitation wavelength set at 475 nm to minimize the contribution from directly excited acceptor emission. The spectrum of acceptor alone with the excitation at 475 nm was also recorded and used as the background to be subtracted from the Donor/Acceptor FRET spectrum.
Fluorescence quenching assay
20 nM BODIPY FL-PIP3 (200 nM stock solutions prepared in RNA-lipid binding buffer) was titrated with a stock solution of LINK-A or RP11-383G10.5 (3 mM prepared in RNA-lipid binding buffer). The spectra were collected from 495 nm to 550 nm with the excitation set at 490 nm and the slit widths set at 1 nm. The final titrated concentration of LINK-A or RP11-383G10.5 was 400 nM. The titration curve was fitted by using the one-site binding equation Y=Bmax * X/(Kd + X), Y = (F0 − F)/F0, in which F0 and F are the fluorescence intensities at 513 nm in the absence or presence of LINK-A and RP11-383G10.5 respectively.
RNA-lipid vesicles binding assay
DOPC giant vesicles were prepared as previously described37,38.The lipid vesicles composed of DOPC mixed with 1.5 mol% Phosphatidylinositol 3,4,5-trisphosphate diC16 [PI(3,4,5)P3 diC16] were generated as previously described36 except the membranes were labeled with Nile red (Life Technologies). RNAs in the RNA-lipid binding buffer (see above) were mixed with the cationic cyanide fluorescence dye YOYO-1 iodide (1 mM) (ThermoFisher). The suspension of lipid vesicles and the solution of RNA/YOYO were gently mixed at a volume ratio of 10:1 and mounted on glass coverslips. For PIP competition experiments, IP4 or IP3 was added in RNA-lipid vesicle binding assay at 100:1 molar ratio (IP4/IP3: PIP3). Images were recorded with a Cooke SensiCam charge-coupled device camera and processed with the use of the SlideBook software package (Intelligent Imaging Innovations).
Determination of Kd for LINK-A-PIP3 and AKT-PIP3 interaction by Alpha assay
Saturation curve was generated to determine the Kd of the interaction between Digoxigenin (DIG)-labeled LINK-A and biotin-labeled lipids in the Alpha format. The Kd was further determined by a competition experiment in which unlabeled LINK-A was titrated from 0.4 mM to 0.05 nM. In details, triplicate samples containing RNA and lipid at indicated concentrations diluted in RNA-lipid binding buffer were transferred, 10 µL to each well, to a ½ area 96-well assay plate then incubated at room temperature for 1 hr. 10 µl of 5xAnti-DIG AlphaLISA® acceptor beads (100 µg/ml) were added to each well. The plate was placed on an orbital shaker for 10 minutes then incubated at room temperature for 1 hr. Following incubation, 10 µl of 5× Streptavidin donor beads (100 µg/ml) were added to each well and incubated 30 mins at room temperature. The plate was read on the EnSpire Multimode Plate Reader (PerkinElmer). The saturation binding curve and competitive inhibition curve were generated based on Alpha signal readings by fitting to a nonlinear regression “saturation binding” model and a “log (inhibitor) vs. response-Variable slope (four parameters)” model respectively (GraphPad Prism 7 software). For IC50 determination, a serial 2-fold dilution of synthetic LINK-A competitors (sequences are listed in Supplementary Table 5) starting at 0.4 mM to 0.05 nM was supplied in RNA-lipid binding buffer and incubated with the reactions. IC50 values were derived by a Log (inhibitor) vs. response-Variable slope (four parameters) model for competitive inhibition curve using GraphPad Prism 7 software.
To determine the Kd value of the AKT-PIP3 interaction with or without lncRNAs, Alpha assays were performed as described above. For saturation curve assay, 0.1 µM, 0.2 µM, and 0.4 µM His-tagged recombinant AKT were incubated with a titration of biotin-PI(3,4,5)P3, -PI(4,5)P2, -PI(4)P as indicated. For the competition binding assay, the unlabeled PI(3,4,5)P3 were used as competitors at a series of 2-fold dilutions ranging from 33 µM to 0.25 nM. The Kd value between AKT and PIP3 were determined using GraphPad Prism 7 software.
Mapping of RNA-protein/lipid single nucleotide binding sites
LINK-A-PIP3 interaction was assessed by AlphaLISA® technology (Perkin Elmer). Synthetic DIG-labeled LINK-A and His6-AKT (ThermoFisher Scientific) and 5 µg/ml of anti-DIG donor beads and anti-6xHis AlphaLISA® acceptor beads, were incubated in RNA-protein binding buffer (50 mM Tris-HCl pH 7.9, 10% Glycerol, 100 mM KCl, 5 mM MgCl2, 10 mM β-ME 0.1% NP- 40) for 1 hr. The Alpha signal was collected with an EnSpire Multimode Plate Reader. To determine the Kd, increasing concentrations of unlabeled LINK-A were added in addition to the labeled RNA and protein or lipid. The Kd values were derived from curve fitting based on a competitive-inhibitor model in GraphPad Prism 7.
Limited proteolysis (LiP) followed by LC-MS
LiP followed by LC-MS was modified based on LiP-SRM analysis52. Briefly, bacterially-expressed AKT PH domain (2 mg/ml), alone or in the presence of PIP3 (2 mM) and/or LINK-A wild-type or mutant oligonucleotides (2 mM), were incubated in buffer (20 mM HEPES, pH7.5, 150 mM KCl and 10 mM MgCl2) and Protease K at room temperature for 5 mins. The digestion was stopped by transferring the reaction mixture to a tube containing guanidine hydrochloride crystals to a final concentration of 7.4 M and by boiling for 3 mins. The digestion mixtures were then subjected to complete tryptic digestion using Immobilized trypsin (Promega). The peptides were subjected to LC-MS analysis at Proteomic and Metabolic core facility of MD Anderson Cancer Center.
Determination of endogenous cellular AKT activation
Simultaneous detection of total and phosphorylated AKT levels in cellular kinase assays was performed using Alpha SureFire® Total AKT1 and AKT1 (p-Ser473) assay kits (PerkinElmer). Data normalization was performed by dividing the raw Alpha signal generated for p-AKT by the raw Alpha signal for total AKT and multiplying by 100, which represents % of AKT activation.
In vitro and in vivo AKT kinase assay
For in vitro AKT kinase activity, 20 ng of recombinant AKT, in vitro transcribed LINK-A wild-type or PIP3 binding domain deletion mutant, and/or PIP3 containing polyPIPosomes (Echelon Biosciences) were incubated in a 1:1:1 molar ratio. For the in vivo AKT kinase assay, the cell lysates were extracted from MDA-MB-231 cells with indicated treatments and the AKT kinase activity was measured by a non-radioactive AKT kinase activity kit (Enzo Life Sciences).
SNP genotyping assay
Genomic DNA from fresh-frozen breast cancer tissues were extracted, purified, and further detected by real-time PCR using TaqMan® Sample-to-SNP™ Kit (ThermoFisher).
3-D spheroid proliferation/viability and invasion assay
Spheroid growth and invasion of parental MDA-MB-231 and its genomically edited derivative cells were conducted using Cultrex® 3-D Spheroid Fluorometric Proliferation/Viability Assay kit and Cultrex® 3D Spheroid BME Cell Invasion Assay kit (Trevigen) respectively according to vendor’s instruction.
In vivo tumorigenesis and glucose uptake study
All animal experiments were performed in accordance with protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. Female athymic Nu/Nu mice (4–6 weeks old) arrived in our facility were randomly put into cages with five mice each. They were injected with respective tumor cells in the unit of cages, which were randomly selected. 5 × 106 of tumor cells (mixed with Matrigel at a 1:1 ratio) were injected into mammary fat pads of age-matched athymic female nude mice (5 mice for each group based on power calculations, which will allow us to detect a ~30% difference in tumor growth and/or glucose uptake between groups at the 95% confidence level). Tumor size was measured weekly using a caliper, and tumor volume was calculated using the standard formula: 0.54×L×W2, where L is the longest diameter and W is the shortest diameter. When tumors reached volumes of 150–200 mm3, mice were randomly divided into groups and intravenously injected with LINK-A or scrambled LNAs (25 mg/kg) every other day for three times. Then mice were imaged and analyzed with [18F]-FDG for in vivo glucose uptake. [18F]-FDG was administered via a single tail-vein injection; further PET/CT images were scanned and collected by using the Inveon CT/PET system (Siemens). During the uptake period, mice were awake and maintained on a heating pad. Images were reconstructed using two dimensional ordered subsets expectation maximization (OSEM) algorithm. PET and CT image fusion and image analysis were performed using software ASIPro 5.2.4.0 (Siemens). The investigators were not blinded to allocation during experiments and outcome assessment.
Somatic copy number variation (CNV)
The CNVs of LINK-A gene loci were analyzed based on previous publications61. Briefly, copy-number data from the TCGA portal were downloaded. The segment mean represents the scope of genomic copy number changes for the LINK-A segmental region. A threshold of 0.2 in segment mean values was considered an amplification while −0.2 was considered a deletion61.
Detection and statistical analysis of somatic mutations
The TCGA database provides somatic mutations detected from whole genome and whole-exome sequencing from matched tumor and normal samples based on methodologies published previously62. Data were imputed for over 20 million SNPs using data from the 1000 Genomes Project (the Phase III integrated variant set release, across 2,504 samples) as a reference. We phased the haplotype with Shapeit v2 and performed imputation with IMPUTE2. Poorly imputed SNPs defined by an information measure Is <0.80 with IMPUTE2 were excluded from the analyses. Tests of associations between imputed SNPs and lung or breast cancer were performed under a probabilistic dosage model in SNPTEST v2.5 after adjusting by age, gender, pack-year of smoking, and the top PCA. The expression of LINK-A was log transformed with formula log2(x+1). Linear regression was used to evaluate the association between SNPs and gene expression. TCGA RNA-seq level 3 data for all cancers, tumor and normal, were processed and normalized; we used the RSEM normalized values for gene expression. Fold changes in LINK-A gene expression between tumor and normal tissues were calculated using median expression of tumors and normal tissues. The significance of differential gene expression change in LINK-A were calculated using a two-tailed Wilcoxon test for multiple hypothesis testing. For the SNP incidence in different populations, the data was retrieved from NCBI dbSNP Reference SNP (refSNP) Cluster Report: rs12095274 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=12095274).
Statistics & reproducibility
The experiment was set up to use 3–5 samples/repeats per experiment/group/condition to detect a 2-fold difference with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies. All experiments including IP/IB and lipid strips were carried out with three biological replicates. Panels in Figs 2a, 7f–i, 8j–k and Supplementary Figs 2a, 8e, f, k shows a representative image of three independent experiments. Relative quantities of gene expression level were normalized to B2M. The relative quantities of RIP samples were normalized by individual inputs, respectively. Results are reported as mean ± standard error of the mean (s.e.m) of three independent experiments. Each exact n value is indicated in the corresponding figure legend or in the figure. Comparisons were performed using two tailed paired Student’s t-test, one-way ANOVA and Wilcoxon test (n.s., p>0.05, *p<0.05, **p<0.01 and ***p<0.001), as indicated in individual figures. Fisher’s exact test was implemented for statistical analyses of the correlation between markers and clinical parameters. For survival analysis, the expression of LINK-A or phosphorylation density of indicated proteins was treated as a binary variant and divided into ‘high’ and ‘low’ level. Kaplan-Meier survival curves were compared using the log rank test. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Data availability
Lipid-binding lncRNA array data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE92414. Previously published microarray data that were re-analyzed here are available under accession code GSE6068918. Previously published structural data that were used to support the findings of this study were obtained from the Protein Data Bank under codes 1H1049. Mass spectrometry data that support the findings of this study have been deposited in ProteomeXchange with the primary accession code PXD005636. The data supporting the somatic mutation detection and analysis in this study were derived from the TCGA Research Network: http://cancergenome.nih.gov/. Source data for 2a, 7f–i, 8j–k and Supplementary Figs 2a, 8e, f, k have been provided as Supplementary Table 6. All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We are grateful to Dr. P. Marshall in Southern University at New Orleans and J. King in University of North Texas, Center for Human Identification for Barocycler usage, Drs. E. Lauwers and P. Verstreken from VIB Center for the Biology of Disease, Belgium for discussing the procedures of PIP3 vesicles generation. We thank Dr. Guohui Wan, Pennsylvania State University for providing MS2-24x-pCDNA construct. We thank Drs. Junjie Chen and Lei Li from Gene Editing/Cellular Model Core Facility of MD Anderson Cancer Center, for assistance with CRISPR-cas9 mediated gene editing. We thank Bih-Fang Pan from Proteomics and Metabolomics Core Facility of MD Anderson Cancer Center, for assistance with mass spectrometry analysis. We thank Dr. Lei Zheng, Department of Biochemistry & Molecular Biology, University of Texas, Health science Center at Houston, for consulting of structural analysis. We thank D. Aten for assistance with figure presentation. This work was supported by NIH grant (R01GM112003) to Y.Z., NIH R00 award (R00DK094981), UT Startup and UT STARS grants to C.R.L., and the NIH R00 award (R00CA166527), CPRIT award (R1218), UT Startup and UT STARS grants to L.Q.Y.
Footnotes
Author Contributions A.F.L., Q.S.H., C.L.L. devised and performed most experiments. Z.X., K.L., S.Y.W. helped with biochemistry and lipids studies. G.L. M. and Y.B. performed FRET assays. Y.Y generated CRISPR-cas9 KO cell line. D.H.H. performed mass spectrometry analysis for LiP assay. J.W.Z., Y.Z and J.R.M. provided clinical specimens assisted with pathological analyses. The histological staining were performed by K.L. TCGA and bioinformatics data analysis were performed by C.W., Z.B.H, H.B.S., J.Y., J.L., L.H., and H.L. P.K.P. helped with manuscript preparation. M.C.H. contributed to discussion and data interpretation. L.Q.Y. and C.R.L. initiated and supervised the project and wrote the paper with input from all authors.
References
- 1.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annual review of medicine. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 3.Ono Y, et al. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park WS, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaullier JM, et al. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 6.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 7.Le Good JA, et al. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 8.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton's tyrosine kinase in B cell malignancies. Nat Rev Cancer. 2014;14:219–232. doi: 10.1038/nrc3702. [DOI] [PubMed] [Google Scholar]
- 9.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 11.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokoe D, et al. Dual Role of Phosphatidylinositol-3,4,5-trisphosphate in the Activation of Protein Kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 13.Yap TA, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol. 2011;29:4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- 14.Wisinski KB, et al. Phase I Study of an AKT Inhibitor (MK-2206) Combined with Lapatinib in Adult Solid Tumors Followed by Dose Expansion in Advanced HER2+ Breast Cancer. Clin Cancer Res. 2016;22:2659–2667. doi: 10.1158/1078-0432.CCR-15-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudis C, et al. A phase 1 study evaluating the combination of an allosteric AKT inhibitor (MK-2206) and trastuzumab in patients with HER2-positive solid tumors. Breast cancer research : BCR. 2013;15:R110. doi: 10.1186/bcr3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi L, et al. PDK1-mTOR signaling pathway inhibitors reduce cell proliferation in MK2206 resistant neuroblastoma cells. Cancer cell international. 2015;15:91. doi: 10.1186/s12935-015-0239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stottrup C, Tsang T, Chin YR. Upregulation of AKT3 Confers Resistance to the AKT Inhibitor MK2206 in Breast Cancer. Molecular cancer therapeutics. 2016;15:1964–1974. doi: 10.1158/1535-7163.MCT-15-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing Z, et al. lncRNA Directs Cooperative Epigenetic Regulation Downstream of Chemokine Signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin A, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Arun G, Akhade VS, Donakonda S, Rao MR. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol. 2012;32:3140–3152. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 24.MacIntosh GC, Bariola PA, Newbigin E, Green PJ. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes? Proc Natl Acad Sci U S A. 2001;98:1018–1023. doi: 10.1073/pnas.98.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mindaye ST, Ra M, Lo Surdo J, Bauer SR, Alterman MA. Improved proteomic profiling of the cell surface of culture-expanded human bone marrow multipotent stromal cells. J Proteomics. 2013;78:1–14. doi: 10.1016/j.jprot.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Gross V, et al. Tissue fractionation by hydrostatic pressure cycling technology: the unified sample preparation technique for systems biology studies. Journal of biomolecular techniques : JBT. 2008;19:189–199. [PMC free article] [PubMed] [Google Scholar]
- 27.Olszowy PP, Burns A, Ciborowski PS. Pressure-assisted sample preparation for proteomic analysis. Anal Biochem. 2013;438:67–72. doi: 10.1016/j.ab.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Lichtenbergova L, Snitko Y, Cho W. A phospholipase A2 kinetic and binding assay using phospholipid-coated hydrophobic beads. Anal Biochem. 1997;250:109–116. doi: 10.1006/abio.1997.2200. [DOI] [PubMed] [Google Scholar]
- 29.Bubb KL, et al. Scan of human genome reveals no new Loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez Y, et al. Lipid binding by the Unique and SH3 domains of c-Src suggests a new regulatory mechanism. Scientific reports. 2013;3:1295. doi: 10.1038/srep01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huranova M, et al. In vivo detection of RNA-binding protein interactions with cognate RNA sequences by fluorescence resonance energy transfer. RNA. 2009;15:2063–2071. doi: 10.1261/rna.1678209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frech M, et al. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 34.Ke J, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20:1377–1382. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 35.Pedram Fatemi R, et al. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. Journal of biomolecular screening. 2015;20:1132–1141. doi: 10.1177/1087057115594187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khuong TM, et al. Synaptic PI(3,4,5)P3 is required for Syntaxin1A clustering and neurotransmitter release. Neuron. 2013;77:1097–1108. doi: 10.1016/j.neuron.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 37.Janas T, Yarus M. Visualization of membrane RNAs. RNA. 2003;9:1353–1361. doi: 10.1261/rna.5129803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janas T, Janas T, Yarus M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 2006;34:2128–2136. doi: 10.1093/nar/gkl220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Richards DA. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biology open. 2012;1:857–862. doi: 10.1242/bio.20122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon JH, Srikantan S, Gorospe M. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58:81–87. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han C, et al. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell reports. 2014;8:1447–1460. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa C, et al. Measurement of PIP3 levels reveals an unexpected role for p110beta in early adaptive responses to p110alpha-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27:97–108. doi: 10.1016/j.ccell.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarden Y, Shilo BZ. SnapShot: EGFR signaling pathway. Cell. 2007;131:1018. doi: 10.1016/j.cell.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 45.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nature reviews. Drug discovery. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 47.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gosai SJ, et al. Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Current biology : CB. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 50.Milburn CC, et al. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panchenko AR, Luthey-Schulten Z, Wolynes PG. Foldons, protein structural modules, and exons. Proc Natl Acad Sci U S A. 1996;93:2008–2013. doi: 10.1073/pnas.93.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Y, et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 53.Liu F, Fitzgerald MC. Large-Scale Analysis of Breast Cancer-Related Conformational Changes in Proteins Using Limited Proteolysis. J Proteome Res. 2016;15:4666–4674. doi: 10.1021/acs.jproteome.6b00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gambhir A, et al. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophysical journal. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gills JJ, Dennis PA. Perifosine: update on a novel Akt inhibitor. Current oncology reports. 2009;11:102–110. doi: 10.1007/s11912-009-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahad AM, et al. Development of sulfonamide AKT PH domain inhibitors. Bioorganic & medicinal chemistry. 2011;19:2046–2054. doi: 10.1016/j.bmc.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 58.Hirai H, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Molecular cancer therapeutics. 2010;9:1956–1967. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 59.Zanoni M, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Scientific reports. 2016;6:19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemmon MA, Ferguson KM, O'Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 61.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Molecular cancer research : MCR. 2014;12:485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Lipid-binding lncRNA array data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE92414. Previously published microarray data that were re-analyzed here are available under accession code GSE6068918. Previously published structural data that were used to support the findings of this study were obtained from the Protein Data Bank under codes 1H1049. Mass spectrometry data that support the findings of this study have been deposited in ProteomeXchange with the primary accession code PXD005636. The data supporting the somatic mutation detection and analysis in this study were derived from the TCGA Research Network: http://cancergenome.nih.gov/. Source data for 2a, 7f–i, 8j–k and Supplementary Figs 2a, 8e, f, k have been provided as Supplementary Table 6. All other data supporting the findings of this study are available from the corresponding author on request.