Abstract
Mitochondrial BKCa channel, mitoBKCa, regulates mitochondria function in the heart but information on its protein partnerships in cardiac mitochondria is missing. A directed proteomic approach discovered the novel interaction of BKCa with Tom22, a component of the mitochondrion outer membrane import system, and the adenine nucleotide translocator (ANT). The expressed protein partners co-immunoprecipitated and co-segregated into mitochondrial fractions in HEK293T cells. The BKCa 50 amino acid splice insert, DEC, facilitated BKCa interaction with ANT. Further, BKCa transmembrane domain was required for the association with both Tom22 and ANT. The results serve as a working framework to understand mitoBKCa import and functional relationships.
Keywords: Proteomics, BK channel, MaxiK channel, potassium ion channel, mitochondria, heart, Tom22, adenine nucleotide translocator, macromolecular complex
1. Introduction
Mitochondrial BKCa channels (mitoBKCa) are safeguards of cardiac function as their activation protects the heart from ischemic insult, a property that is absent in the knockout animal (Xu et al., 2002; Singh et al., 2013; Soltysinska et al., 2014). One of the proposed mechanisms by which mitoBKCa activation favors cardiac health after ischemic insult is via improved mitochondrial Ca2+ handling and regulation of the mitochondrial permeability transition pore (mPTP) (Singh et al., 2013). Consistent with this idea, in other systems, mitoBKCa pharmacological inhibition produces cytochrome c release, an event associated with the opening of the mPTP and cell death; while Bax (proapoptotic Bcl-2 associated protein X), a mPTP activator, inhibits channel activity (Cheng et al., 2011). In agreement with its protective role, the activation of cardiac mitoBKCa improves basal mitochondrial energetic performance (Aon et al., 2010), whereas silencing BKCa expression reduces cardiac oxidative phosphorylation (Soltysinska et al., 2014).
To exert its function in mitochondria, mitoBKCa must be first transported into this organelle and likely associate with partner proteins, as it does in other regions of the cell (Toro et al., 2013). Supporting this view, a two-hybrid system approach discovered that the regulatory β1 subunit of BKCa channels directly binds to a mitochondrial protein, cytochrome c oxidase subunit I (Ohya et al., 2005).
We have previously demonstrated that the mitoBKCa pore-forming α subunit is encoded by the same gene that encodes the plasma membrane BKCa channel, Kcnma1, and that a 50 amino acid C-terminal splice insert (named DEC) favors BKCa channel targeting into mitochondria of adult cardiomyocytes (Singh et al., 2013). Thus, the overall structure of the mitoBKCa channel is equivalent to that of the well-studied plasma membrane tetrameric channel, with each α-subunit composed of 7 transmembrane segments (S0-S6), a long intracellular C-terminus and an extracellular N-terminus (Meera et al., 1997). However, the protein partners of mitoBKCa are unknown.
Efforts to identify BKCa channel partners at a large scale have been few and restricted to cochlea and brain preparations (Kathiresan et al., 2009; Gorini et al., 2010; Sokolowski et al., 2011; Singh et al., 2016). In this work, we have examined the interactome of mitoBKCa in adult heart (isolated cardiomyocytes and whole ventricle), using a directed proteomic approach aided by co-immunoprecipitation with BKCa antibodies and pull-down with recombinant DEC sequence. Two putative protein partners were selected to validate and further examine the regions in BKCa involved in the associations: i) Tom22 from the mitochondrial import system, and ii) the adenine nucleotide translocator (ANT), which is linked to oxidative phosphorylation and the regulation of mPTP.
2. Materials and methods
2.1. Animals
Sprague-Dawley male rats (3 months old) were used. Protocols received institutional approval.
2.2. Antibodies
The following antibodies were used: Anti-BKCa monoclonal antibody (mAb) (75-022, UC Davis/ NIH NeuroMab facility), Anti-BKCa polyclonal (p) Ab (APC-021, Alomone Labs), Anti-c-Myc mAb (M4439, Sigma), Anti-c-Myc polyclonal pAb (C3956, Sigma), Anti-HA mAb (H3663, Sigma), Anti-HA pAb (H6908, Sigma), anti-DDK mAb (TA50011, OriGene), Goat Anti-Rabbit IgG Alexa Fluor® 680 conjugate (A21109, Invitrogen), and IRDye® 800CW Goat Anti-Mouse IgG (926-32210, Odyssey).
2.3. Clones
Full length human BKCa α-subunit starting from Met1 (BKCa) with or without DEC splice insert (EKKWFTDEPD NAYPRNIQIK PMSTHMANQI NQYKSTSSLI PPIREVEDEC) at the C-terminus were used in all experiments except for Figs. 4, 5 and 7 (see below). Compared to NCBI Accession No. U11058.2 (Wallner et al., 1995), which starts from Met3, constructs starting from Met1 have additional 96 amino acids at the N-terminus that includes a 3xHemagglutinin (HA)-tag upstream Met3 (MANGGGGGGG SSGGGGGGGG SSLRMSSNIH ANHLSLDASS SSSSSSSSSS SSSSSSSSSS VHEPKMYPYD VPDYAGYPYD VPDYAGSYPY DVPDYA; 3xHA tag sequences are in italics). BKCa-DEC has the C-terminal DEC sequence inserted downstream amino acids RDKQN as found in the heart (Singh et al., 2013). Clones starting from Met3 (Accession No. U11058.2) were either untagged or tagged at the N-terminus with c-Myc epitope (Meera et al., 1997) (Figs. 4 and 7) or with HA epitope (Fig. 5). Clones used in deletion constructs experiments (Figs. 4 and 7) were: BKCa (1-1113); C-terminal deletion constructs 1-343, 1-441 and 1-711; and N-terminal deletion constructs 322-1113 and 679-1113; numbers correspond to NCBI Accession No. U11058.2. All BKCa constructs were in pcDNA3. Tom22 construct was the human translocase of outer mitochondrial membrane 22 homolog (Accession No. NM_020243.4) with C-terminal c-Myc and DDK tags in pCMV6. ANT construct was the human mitochondrial adenine nucleotide translocator (Accession No. NM_001151.2) with a C-terminal c-Myc-DDK tag in pCMV6. Tom 22 and ANT clones were purchased from OriGene Technologies (Rockville, MD).
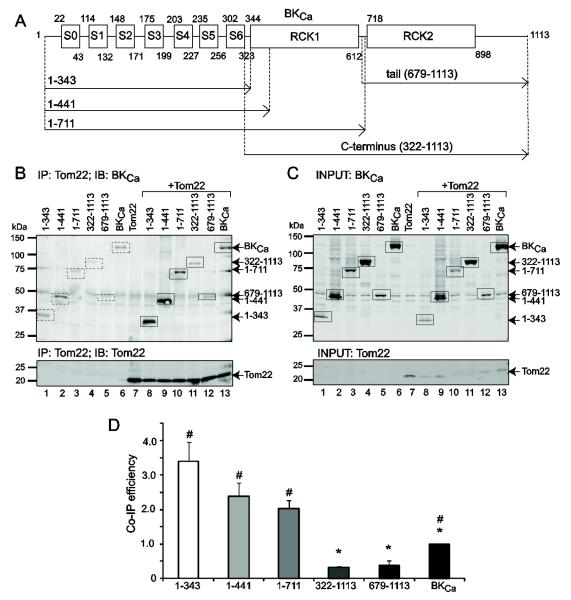
Figure 4. BKCa channel transmembrane region interacts with Tom22.
(A) Scheme of wild type BKCa (1-1113) pore-forming α-subunit and different truncated proteins that were either expressed alone or co-expressed with Tom22. Numbers correspond to amino acid numbers in NCBI Accession No. U11058.2. S0-S6, transmembrane regions form the channel voltage sensor. Linker between S5-S6 forms the tetrameric channel pore region. Regulator of potassium conductance (RCK) 1 and 2 are Ca2+ sensors. (B) Dually labeled immunoblot for BKCa (top) and Tom22 (bottom) showing the ability of Tom22 to co-immunoprecipitate different BKCa channel regions (lanes 8-13, top). BKCa and Tom22 images were separated for clarity. Lanes 1-7 are controls expressing BKCa constructs or Tom22 alone. Squares and arrows mark the expected band sizes of each construct. Background signals (dashed squares, lanes 1-7, top) were much lower than the signals of the co-immunoprecipitated BKCa products (squares, lanes 8-13, top) and were subtracted to calculate Co-IP efficiency in (D). As control, the lower panel shows Tom22 successful immunoprecipitation. Tom22 was immunoprecipitated with anti-DDK mAb. Co-immunoprecipitated BKCa proteins were detected with anti-c-Myc pAb, and Tom22 was detected with anti-DDK mAb. (C) Control immunoblot of input lysates of cells expressing different BKCa regions (upper panel) and Tom22 (lower panel). BKCa constructs were detected with anti-c-Myc pAb and Tom22 was detected with anti-DDK mAb. Squares and arrows mark the expected band sizes. (D) Tom22 efficiency in pulling down different BKCa regions indicates that BKCa channel interacts with Tom22 mainly through the transmembrane domain (construct 1-343). Co-IP efficiency was calculated after subtracting background signals and calculating the ratio of the band intensities of co-immunoprecipitated BKCa constructs (i.e. B, top, lanes 8-13, squares) to corresponding immunoprecipitated Tom22 (i.e. B, lower panel, lanes 8-13), and normalized to BKCa construct intensities in the corresponding input lysates (i.e.. C, top, lanes 8-13). *, p<0.05 with respect to construct 1-343; #, p<0.05 with respect to the C-terminus (322-1113).
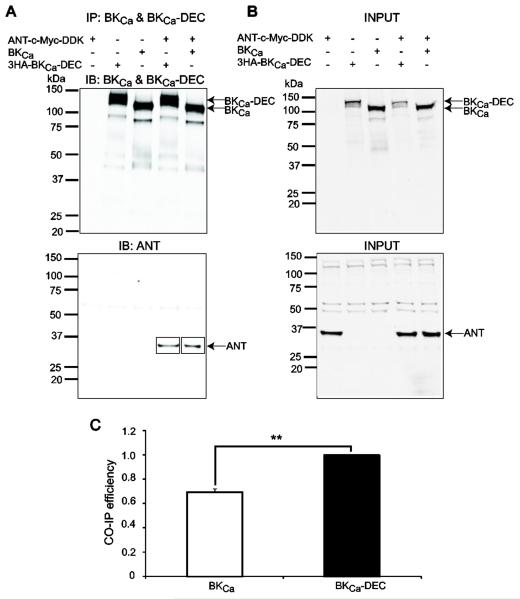
Figure 5. BKCa interacts with ANT in HEK293T cells.
Cells were co-transfected with BKCa, BKCa-DEC and ANT in different combinations (+, present; -, absent). (A) BKCa and BKCa-DEC were immunoprecipitated with anti-BKCa mAb and detected with anti-BKCa pAb (top panel); coimmunoprecipitated ANT was detected with anti-Flag pAb that recognizes the DDK epitope (lower panel, squares). Top and bottom panels are different blots using the same immunoprecipitation products. (B) Input lysates showing proper expression of BKCa (top panel) and ANT (lower panel). BKCa and BKCa-DEC were detected with anti-BKCa pAb and ANT was detected with anti-Flag pAb. (C) Mean values of BKCa and BKCa-DEC efficiency in pulling down ANT show that BKCa-DEC associates better with ANT by about 30%. Co-immunoprecipitation efficiency was calculated as described in the legend to Figure 2 for Tom22. **, p<0.01 , n=3 independent experiments.
Figure 7. Molecular determinant of BKCa channel interaction with ANT.
Different regions of BKCa and wild-type BKCa (see scheme in Fig. 4A) were either expressed alone or co-expressed with ANT. (A) Dually labeled immunoblot shows that immunoprecipitated ANT pulled down wild-type and deletion BKCa constructs (lanes 8-13) to different degrees. Squares and arrows mark the bands of the expected sizes. Dashed squares mark background signals, which were subtracted for the analysis in (C). ANT was immunoprecipitated with anti-DDK mAb. Coimmunoprecipitated BKCa proteins and ANT were detected with anti-c-Myc pAb. (B) Control immunoblot of input lysates of cells expressing different BKCa regions and ANT. (C) ANT efficiency in pulling different BKCa constructs indicates that BKCa channel interacts with ANT mainly through the transmembrane domain (construct 1-343). *, p<0.05 with respect to construct 1-343; #, p<0.05 with respect to construct 322-1113. n=3 independent experiments.
2.4. Expression of recombinant fusion proteins in E. Coli
Glutathione S-Transferase (GST)-DEC and GST in pGEX3 vector were transformed into E. coli BL21DE3 (Invitrogen) and cultured at 37 °C using Luria-Bertani (LB) broth (Thermo Fisher Scientific) supplemented with 100 ng/mL ampicillin until OD600= 0.5. The expression of GST-DEC and GST was then induced by adding IPTG to a final concentration of 0.2 mM. Cultures were continued in a rotary shaker for 4 h at 37°C. Bacteria were centrifuged at 1000 g for 5 min to collect the pellet. The pellet was further lysed using sonication (15 s) at power level 2 in lysis buffer (mM): 50 Tris, pH 7.4, 100 NaCl, 5 MgCl2, 1% Triton X-100, 10% glycerol, supplemented with fresh 2 mM DTT, 200 mM PMSF and 1:500 dilution of protease inhibitor cocktail (11697498001, Roche).
2.5. Isolation of cardiomyocytes from left ventricle
Sprague-Dawley rats were anesthetized and injected with heparin (200 IU/kg, i.v.). After 15 min hearts were harvested in ice-cold Tyrode’s solution (mM): 130 NaCl, 5.4 KCl, 1 MgCl2, 0.6 Na2HPO4, 10 glucose, 5 taurine, 10 2,3-butanedione monoxime, and 10 HEPES, pH 7.4, oxygenated with 95% O2-5% CO2 (v/v), and mounted on a modified Langendorff apparatus with an 80 cm H2O constant pressure. After 5-10 min of perfusion with Tyrode’s solution at 37°C, the hearts were next perfused for 15 min with Tyrode’s solution containing 372 U/mL Collagenase Type-2 and 1.0 U/mL Protease Type-XIV, and washed for 10-15 min with KB solution (mM): 25 KCl, 10 KH2PO4, 5 creatine, 2 MgSO4, 20 glucose, 20 taurine, 100 K-glutamate, 10 aspartic acid, 5 HEPES, 0.5 EGTA, and 1% (wt/v) BSA, pH 7.2 oxygenated with 95% O2-5% CO2 (v/v). After washing, the left ventricles were tweezed into pieces in KB solution to release cells. Isolated cardiomyocytes were filtered through a 100 μm strainer, and centrifuged at 1,000 g for 2 min.
2.6. Isolation of “crude” mitochondria and Percoll-purification
i) “Crude” mitochondria. Isolated cardiomyocytes or left ventricles were homogenized with a Potter-Elvehjem homogenizer (20 rapid strokes) on ice and using isolation buffer A (mM): 70 sucrose, 210 mannitol, 50 Tris-HCl, and 1 Na2-EDTA, pH 7.4. The homogenate was centrifuged at 2,400 g for 5 min at 4°C. The supernatant was then centrifuged at 17,000 g for 10 min at 4°C. The pellet containing “crude” mitochondria was either further purified (ventricle samples) using Percoll gradient or directly resuspended in lysis buffer (isolated cardiomyocyte samples) (section 2.7). ii) Percoll purification. “Crude” mitochondria was carefully added onto 3 mL of 30% (v/v) Percoll (Graham , 2001) in buffer B (mM): 250 sucrose, 10 Na-HEPES, 1 Na2-EDTA, pH 7.4. Samples were centrifuged in a fixed angle rotor at 50,000 g for 45 min. After ultracentrifugation, three clear layers were observed, and labeled as M1, M2 and M3 (Singh et al., 2012). The M3 fraction, corresponding to the purified mitochondria, was carefully isolated and resuspended in 1 mL of isolation buffer A and centrifuged at 17,000 g for 10 min. The pellet was washed twice by resuspension with the same buffer and centrifuged again at 17,000 g for 5 min each. The purified mitochondria were lysed within 2 h after isolation.
2.7. Mitochondria and cardiomyocyte lysates
Mitochondria or isolated cardiomyocyte preparations were incubated at 4°C (on a rotatory shaker for 1 h) in cell lysis buffer (1 and 3 mL, respectively) (mM): 50 Tris, 150 NaCl, 5 EDTA, 0.1% Nonylphenyl Polyethylene Glycol (NP-40 alternative, Calbiochem), and 0.25% Na-deoxycholate, pH 7.4 supplemented with protease inhibitor cocktail (one tablet/50 mL) and 200 mM PMSF. After lysis, samples were centrifuged at 17,000 g for 10 min at 4 °C and the supernatant was saved at −80°C for future use. Protein concentration was measured with Bio-Rad Protein Assay method.
2.8. Pull down assay and SDS-PAGE
Recombinant fusion protein GST-DEC and GST (control) were used. Glutathione Sepharose 4B beads (GE healthcare) were first prewashed with bacteria lysis buffer and then incubated with 3 mg of GST or GST-DEC on a rotatory shaker at 4 °C for 3 h. After binding of GST-DEC and GST onto Glutathione Sepharose 4B beads, each sample was further washed with bacteria lysis buffer 3 times, following incubation with 2 mg protein of cardiomyocyte cell lysates or mitochondrial lysates on a rotatory shaker at 4 °C for 2 h. Each sample was then incubated with SDS loading buffer for 1 h at 37 °C and centrifuged at 10,000 g for 3 min. The supernatant was separated on a 4-20% SDS-PAGE at room temperature. The protein bands were stained with SYPRO-RUBY stain according to the manufacturer’s instructions (Bio-Rad).
2.9. Immunoprecipitation for proteomics
Mitochondrial lysates (see section 2.7) were precleared with 10 μL protein A/G resin/mg protein (Pierce Biotechnology Inc.) for 1 h at 4 °C. Next, samples were centrifuged at 2,400 g for 5 min at 4 °C to remove protein A/G resin. The supernatant was collected, and the protein concentration was calculated according to the Bio-Rad method. Anti-BKCa antibodies (2 μg) or IgGs (2 μg, control) were incubated for 2 h at 4 °C with 10 μL protein A/G resin on a rotatory shaker. Unbound antibodies were then removed by centrifugation 5 times at 2,400 g for 1 min at 4 °C. Mitochondria lysates (2 mg protein each) were added to immobilized anti-BKCa and IgG antibodies, and incubated on a rotatory shaker overnight at 4 °C. Samples were washed 5 times with cell lysis buffer at 2,400 g for 1 min each at 4 °C. Laemmli sample buffer (30 μL of 3X buffer) was added, and samples were incubated at 37 °C for 1 h. Eluted proteins were centrifuged at 17,000 g for 10 min at 4 °C, and separated on a 4-20% SDS-PAGE at room temperature. Protein bands were then stained with SYPRO-RUBY.
2.10. Mass spectrometry analysis of proteins
Plugs of SYPRO-RUBY stained 1-D gels containing GST- and GST-DEC bound proteins or anti-BKCa and IgG bound proteins were excised using a sterile scalpel blade. Sections (~2 mm) ranging from 10 kDa to 250 kDa were collected and further digested with trypsin. Protein digestion and elution were carried as described earlier (Singh et al., 2009; Singh et al., 2013). Liquid chromatography, mass spectrometry in tandem (LC/MS/MS) was performed using a Thermo LTQ-Orbitrap XL mass spectrometer equipped with an Eksigent Nano Liquid chromatography-1D plus system and an Eksigent auto sampler at the University of California Los Angeles W. M. Keck Proteomic Center. Sequences were assigned using the Swiss-Prot Rat database and the MASCOT Daemon search engine. The output data included MASCOT score, molecular weight, number of peptides matched, peptide sequences, sequence coverage, rank, and p-value. To select candidate proteins, the score value of individual spectra was set at a 95% confidence level and a MASCOT score > or =21. Peptides were selected as specified by the Peptide Prophet algorithm. GST-DEC or BKCa specific interacting proteins were defined as those not found in the samples using GST alone or IgG, respectively.
2.11. Cell culture
HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% (v/v) FBS, 2 units/mL streptomycin and penicillin at 37 °C in a humidity-controlled CO2 [5% (v/v)] incubator.
2.12. Transient transfection
HEK293T cells were transfected at 70% confluence using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Plasmid DNAs were incubated with Lipofectamine 2000 in OPTI-MEM (Invitrogen) for 20 min at room temperature (20-25 °C). Plasmid-Lipofectamine mixtures were then incubated with cells in OPTI-MEM at 37 °C in a CO2 incubator for 5-7 h. Afterwards, DMEM containing 20% FBS was added to reach a final FBS concentration of 10%. Because cotransfection of Tom22 or ANT with BKCa significantly decreased the expression of BKCa, we first transfected BKCa constructs to allow their expression followed by a second transfection of Tom22 or ANT 24 h later. All constructs were transfected with 10 μg plasmid/100 mm dish with the exception of Tom22 that was transfected at 1 μg plasmid/100 mm dish in experiments involving truncated BKCa channels. After 48 h from the second transfection, cells were collected and lysed for further analysis.
2.13. Western blot
HEK293T cells or cellular fractions were treated with cell lysis buffer (see section 2.7) containing protease inhibitor cocktail (one tablet/50 mL), and incubated for 30 min at 4 °C with shaking. Samples were then centrifuged at 17,000 g for 10 min at 4 °C, and the supernatant was collected as lysates. Lysate proteins (20 μg/lane) were separated on 4–20% SDS/PAGE and transferred to nitrocellulose membranes. Nitrocellulose membranes were blocked with 5% (wt/v) milk in TBS (150 mM NaCl, 20 mM Tris-HCl, pH 7.4) at room temperature for 60 min and incubated with antibodies (2 μg/mL) overnight. Next day, membranes were washed 3 times with TBS and incubated with 0.01 μg/mL secondary Abs for 1 h at room temperature. After washing 3 times, membranes were visualized and scanned with the Odyssey Imaging System (Li-Cor Biosciences).
2.14. Co-immunoprecipitation of heterologously expressed proteins
HEK293T cells expressing different combinations of BKCa constructs, Tom22 or ANT were cultured on 100 mm dish and lysed with cell lysis buffer containing protease inhibitor cocktail. Lysates were centrifuged at 17,000 g for 10 min at 4 °C and the supernatant was cleared with 10 μL protein A/G resin/mg protein for 1 h at 4 °C on a rotatory shaker. The precleared lysates (1 mg protein) were incubated with 10 μL protein A/G resin preconjugated with 2 μg antibodies (Anti-HA mAb or anti-BKCa mAb for BKCa and BKCa-DEC; anti-c-Myc mAb for Tom22, ANT, and BKCa constructs; anti-DDK mAb for Tom22 and ANT; and Flag pAb for ANT) in a total volume of 500 μL overnight at 4 °C. After incubation, samples were centrifuged at 2,400 g for 1 min and washed for 5 times with cell lysis buffer. The proteins were eluted from the beads with 30 μL 3X Laemmli sample buffer at 37 °C for 1 h. After elution, samples were centrifuged at 17,000 g for 10 min at 4°C and separated on 4-20% SDS-PAGE along with whole cell lysates. Separated proteins were transferred to nitrocellulose membranes and immunoblotted with respective antibodies (1 μg/mL). Single- or double-labeled immunoblots were imaged as described above.
2.15. Cell fractionation
HEK293T cells were collected and homogenized in isolation buffer A (mM): 70 sucrose, 210 mannitol, 1 Na2-EDTA, 50 Tris-HCl, pH 7.4 using a Potter-Elvehjem homogenizer (3 rapid strokes). The homogenate was centrifuged at 2,400 g for 5 min in an Eppendorf tube. The supernatant was collected and centrifuged at 17,000 g for 10 min. The pellet containing “crude” mitochondria was resuspended in 50 μL of isolation buffer A, and the supernatant was kept. The “crude” mitochondria preparation was further purified by Percoll gradient centrifugation (see section 2.6) and lysed within 2 h after isolation. The supernatant after isolating the “crude” mitochondria was used to isolate the “crude” membrane fraction and cytosolic fraction. The supernatant was centrifuged in the same fixed angle rotor at 100,000 g for 30 min. The pellet was collected as “crude” membrane fraction. The supernatant containing ribosomes and cytosol was named here cytosolic fraction. All fractions were lysed in cell lysis buffer (see section 2.7) for Western blot analysis together with whole-cell lysate.
2.16. Statistics
Data are presented as means ± S.E. Statistical comparisons between groups were made using Student’s t test. Multiple comparisons were done with ANOVA followed by the Tukey test. A p value<0.05 was considered statistically significant. All experiments were performed using at least three independent preparations.
3. Results
3.1. Identification of mitoBKCa-associated cardiac mitochondrial proteins
mitoBKCa-associated proteins were identified using a directed proteomic approach using recombinant GST-DEC and specific anti-BKCa mAb (Misonou et al., 2006). The BKCa C-terminal DEC insert is known to support BKCa mitochondrial expression in adult cardiomyocytes (Singh et al., 2013), and thus, was expected to associate with mitochondrial partners. Three protocols were used to maximize the isolation of BKCa protein partners: 1) pull down from “crude” mitochondria of isolated cardiomyocytes using GST-DEC and GST (control); 2) pull down from Percoll-purified mitochondria of left ventricle using anti-BKCa mAb and IgG (control); and 3) pull down from whole cardiomyocytes with GST-DEC and GST (control).
The “crude” mitochondria preparation had the advantage of using solely rat cardiomyocytes. However, due to material limitations, further Percoll purification of this preparation was impractical; instead, we used the GST-DEC fusion protein that, we expected, would favor specific recognition of mitochondrial proteins. To identify proteins interacting with BKCa regions other than DEC, we used a specific BKCa mAb to immunoprecipitate proteins from the Percoll-purified mitochondria. We also investigated whether GST-DEC could pick up mitochondrial interacting proteins in whole ventriculocyte lysates. Indeed, with this approach using the whole ventriculocyte lysates with GST-DEC as bait yielded 49 mitochondrial partners out of a total of 341 partners (~14% mitochondrial proteins; in 3 independent experiments). This result is consistent with experiments using: i) GST-DEC to pull down proteins in “crude” mitochondria from isolated cardiomyocytes that revealed 68 mitochondrial partners out of a total of 561 partners (~12% mitochondrial proteins; in 3 independent experiments), and ii) anti-BKCa mAb to immunoprecipitate proteins from Percoll-purified mitochondria from left ventricle that detected 77 mitochondrial partners out of a total of 428 partners (~17%; in 3 independent experiments). Overall, a total of 1079 different proteins were identified as partners of BKCa in cardiomyocytes and left ventricle using the above mentioned protocols. The locations were catalogued into intracellular organelles i.e. mitochondrion (151 proteins), endoplasmic reticulum (44 proteins), Golgi (17 proteins), and nucleus (151 proteins) (Tables 1 and 2); cytoplasm (322 proteins); and plasma membrane (223 proteins). The remaining 171 proteins were catalogued as “others” by the UniProt Knowledgebase.
Table 1.
BKCa channel-interacting mitochondrial proteins identified by LC/MS/MS
| Proteins | Uniprot ID | Mass, Da | Score | Peptides |
|---|---|---|---|---|
| Oxidative Phosphorylation | ||||
| NADH-ubiquinone oxidoreductase 75 kDa subunit (C,B) | NDUS1_RAT | 80331 | 785 | 18 |
| ATP synthase protein 8 (B) | ATP8_RAT | 7637 | 514 | 4 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit (C) | DHSA_RAT | 72596 | 501 | 10 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 (C) | NDUS2_RAT | 52927 | 268 | 8 |
| Cytochrome c oxidase subunit 4 isoform 1 (C) | COX41_RAT | 19559 | 266 | 10 |
| ATP synthase-coupling factor 6 (B) | ATP5J_RAT | 12487 | 236 | 3 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2 (B) | NDUV2_RAT | 27703 | 140 | 3 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 4 (A,B) | NDUS4_RAT | 19785 | 135 | 4 |
| Cytochrome b-c1 complex subunit Rieske (B) | UCRI_RAT | 29712 | 128 | 7 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10 (C) |
NDUAA_RAT | 40753 | 127 | 5 |
| ATP synthase subunit e (B) | ATP5I_RAT | 8249 | 121 | 2 |
| Cytochrome c oxidase subunit 7A2 (A,B,C) | CX7A2_RAT | 9347 | 95 | 1 |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit(A) | DHSB_RAT | 32607 | 95 | 3 |
| Cytochrome c oxidase subunit 6C-2 (B) | CX6C2_RAT | 8449 | 93 | 4 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 (C) |
NDUAB_RAT | 15129 | 73 | 2 |
| NADH dehydrogenase [ubiquinone] flavoprotein 3 (B,C) | NDUV3_RAT | 11934 | 72 | 3 |
| ATP synthase subunit epsilon (B) | ATP5E_RAT | 5820 | 46 | 1 |
| ATP synthase subunit d, mitochondrial (A, C) | ATP5H_RAT | 18809 | 44 | 2 |
| Cytochrome c oxidase subunit 1 (B) | COX1_RAT | 56956 | 40 | 3 |
| ATP synthase subunit a ((B), (C)) | ATP6_RAT | 25034 | 38 | 1 |
| Cytochrome c oxidase subunit 6A2 (B) | CX6A2_RAT | 10594 | 30 | 1 |
| Cytochrome b-c1 complex subunit 8 (B) | QCR8_RAT | 9843 | 29 | 3 |
| ATP synthase lipid-binding protein (C) | AT5G1_RAT | 14463 | 29 | 2 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 6 (B) | NDUS6_RAT | 12946 | 27 | 3 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 4 (C) |
NDUF4_RAT | 20146 | 27 | 4 |
| Cytochrome c oxidase subunit 6C-1 (C) | CX6C1_RAT | 8547 | 26 | 1 |
| ATP synthase subunit s (C) | ATP5S_RAT | 23765 | 25 | 1 |
| NADH-ubiquinone oxidoreductase chain 4 (C) | NU4M_RAT | 51937 | 21 | 2 |
|
| ||||
| Valine, Propionate and Butanoate Metabolism | ||||
| Trifunctional enzyme subunit alpha (A) | ECHA_RAT | 83297 | 796 | 20 |
| 3-hydroxyacyl-CoA dehydrogenase type-2 (A) | HCD2_RAT | 27343 | 125 | 4 |
| Short-chain specific acyl-CoA dehydrogenase (A) | ACADS_RAT | 45022 | 87 | 1 |
| Malonyl-CoA decarboxylase (C) | DCMC_RAT | 55298 | 81 | 5 |
| Isovaleryl-CoA dehydrogenase (C) | IVD_RAT | 46862 | 64 | 2 |
| 2-Oxoisovalerate dehydrogenase subunit alpha (B, C) | ODBA_RAT | 50418 | 50 | 2 |
| Methylcrotonoyl-CoA carboxylase beta chain (A, C) | MCCB_RAT | 61992 | 43 | 2 |
| Acyl-coenzyme A synthetase ACSM2 (A) | ACSM2_RAT | 64617 | 41 | 3 |
| Propionyl-CoA carboxylase beta chain (C) | PCCB_RAT | 59216 | 38 | 2 |
| Methylcrotonoyl-CoA carboxylase subunit alpha (A, B, C) | MCCA_RAT | 79564 | 25 | 2 |
| 3-Hydroxyisobutyrate dehydrogenase (A) | 3HIDH_RAT | 35679 | 24 | 2 |
| 3-Hydroxyisobutyrate dehydrogenase (A) | 3HIDH_RAT | 35679 | 24 | 2 |
| Succinate-semialdehyde dehydrogenase (B) | SSDH_RAT | 56723 | 23 | 1 |
| Propionyl-CoA carboxylase alpha chain (A, C) | PCCA_RAT | 82198 | 23 | 5 |
| Dihydrolipoyl dehydrogenase (B) | DLDH_RAT | 54574 | 22 | 3 |
|
| ||||
| TCA Cycle | ||||
| Pyruvate dehydrogenase E1 component subunit alpha, somatic form (A) |
ODPA_RAT | 43883 | 448 | 9 |
| Malate dehydrogenase (B) | MDHM_RAT | 36117 | 161 | 7 |
| Succinyl-CoA ligase [GDP-forming] subunit alpha (B) | SUCA_RAT | 36524 | 134 | 5 |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (C) |
ODP2_RAT | 67637 | 127 | 6 |
| Fumarate hydratase (A) | FUMH_RAT | 54714 | 126 | 8 |
| Isocitrate dehydrogenase [NAD] subunit beta(A) | IDH3B_RAT | 42612 | 125 | 10 |
| [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2 (B) | PDK2_RAT | 46304 | 105 | 7 |
| [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 (A) | PDK1_RAT | 49392 | 88 | 6 |
| Isocitrate dehydrogenase [NAD] subunit gamma 1 (A) | IDHG1_RAT | 43223 | 34 | 3 |
|
| ||||
| Fatty Acid Metabolism | ||||
| Very long-chain specific acyl-CoA dehydrogenase (C) | ACADV_RAT | 71047 | 801 | 16 |
| Carnitine O-palmitoyltransferase 1 (A, B) | CPT1B_RAT | 89129 | 701 | 13 |
| Carnitine O-palmitoyltransferase 2 (A, C) | CPT2_RAT | 74634 | 210 | 7 |
| Long-chain-fatty-acid--CoA ligase 6 (A, B) | ACSL6_RAT | 79156 | 52 | 4 |
| Peroxisomal 3,2-trans-enoyl-CoA isomerase (A) | PECI_RAT | 43336 | 50 | 3 |
|
| ||||
| Arginine and Alanine Metabolism | ||||
| Glutamate dehydrogenase 1 (C) | DHE3_RAT | 61719 | 190 | 12 |
| Amine oxidase [flavin-containing] A (A, C) | AOFA_RAT | 60097 | 111 | 4 |
| Delta-1-pyrroline-5-carboxylate dehydrogenase (B) | AL4A1_RAT | 62286 | 86 | 4 |
| Carbamoyl-phosphate synthase (C) | CPSM_RAT | 165673 | 25 | 7 |
| Amine oxidase [flavin-containing] B (B) | AOFB_RAT | 59049 | 24 | 2 |
|
| ||||
| Glycine, Serine and Threonine Metabolism | ||||
| 5-Aminolevulinate synthase, nonspecific (B) | HEM1_RAT | 71830 | 28 | 5 |
| Sarcosine dehydrogenase (A) | SARDH_RAT | 102573 | 21 | 2 |
|
| ||||
| PPAR Signaling Pathway | ||||
| Glycerol kinase (A) | GLPK_RAT | 58238 | 37 | 2 |
| Sterol 26-hydroxylase (C) | CP27A_RAT | 60980 | 30 | 3 |
|
| ||||
| Drug Metabolism | ||||
| Glutathione S-transferase P (A) | GSTP1_RAT | 23652 | 80 | 2 |
| Glutathione S-transferase kappa 1 (A) | GSTK1_RAT | 25590 | 27 | 2 |
|
| ||||
| Import Mechanism | ||||
| Sorting and assembly machinery component 50 homolog (C) | SAM50_RAT | 52384 | 372 | 12 |
| GrpE protein homolog 1 (B, D) | GRPE1_RAT | 24510 | 303 | 9 |
| Mitochondrial-processing peptidase subunit alpha (C) | MPPA_RAT | 59083 | 165 | 7 |
| Mitochondrial import inner membrane translocase subunit TIM16 (C) |
TIM16_RAT | 13722 | 103 | 1 |
| Mitochondrial import receptor subunit TOM22 (B) | TOM22_RAT | 15481 | 94 | 1 |
| Mitochondrial import inner membrane translocase subunit Tim23 (C) |
TIM23_RAT | 22064 | 45 | 2 |
| Mitochondrial import receptor subunit TOM40 (A) | TOM40_RAT | 38294 | 35 | 2 |
| Mitochondrial import receptor subunit TOM70 (A) | TOM70_RAT | 68143 | 24 | 2 |
|
| ||||
| Transport (Channels and Carriers)* | ||||
| Voltage-dependent anion-selective channel protein 1 (A, C) | VDAC1_RAT | 30851 | 173 | 4 |
| Phosphate carrier protein (C) | MPCP_RAT | 39876 | 50 | 2 |
| ADP/ATP translocase 2 ((B),E) | ADT2_RAT | 33108 | 43 | 2 |
| ATP-binding cassette sub-family B member 7 (C) | ABCB7_RAT | 82848 | 40 | 6 |
| Voltage-dependent anion-selective channel protein 3 (C) | VDAC3_RAT | 31178 | 32 | 2 |
| Tricarboxylate transport protein (B) | TXTP_RAT | 34156 | 24 | 4 |
| Calcium-binding mitochondrial carrier protein SCaMC-2 (C) | SCMC2_RAT | 53060 | 24 | 1 |
|
| ||||
| Pyrimidine Metabolism | ||||
| GTP:AMP phosphotransferase (C) | KAD3_RAT | 25479 | 98 | 3 |
| Thymidylate synthase (A) | TYSY_RAT | 35280 | 30 | 2 |
| Thioredoxin reductase 2 (A) | TRXR2_RAT | 57167 | 23 | 4 |
|
| ||||
| Programmed Cell Death | ||||
| Apoptosis-inducing factor 1 (C) | AIFM1_RAT | 66966 | 133 | 6 |
| Dynamin-like 120 kDa protein (A) | OPA1_RAT | 111751 | 115 | 5 |
| Mitochondrial fission 1 protein (C) | FIS1_RAT | 17041 | 43 | 1 |
| Dynamin-1-like protein (A) | DNM1L_RAT | 84369 | 37 | 4 |
| Chaperone activity of bc1 complex-like (C) | ADCK3_RAT | 72750 | 24 | 2 |
| Receptor-interacting serine/threonine-protein kinase 3 (A) | RIPK3_RAT | 52715 | 24 | 3 |
| Superoxide dismutase (A, B, C) | SODM_RAT | 24887 | 24 | 1 |
| Protein TBRG4 (A) | TBRG4_RAT | 71592 | 24 | 4 |
|
| ||||
| Regulation | ||||
| LETM1 and EF-hand domain-containing protein 1 (C) | LETM1_RAT | 83635 | 203 | 10 |
| Elongation factor G (C) | EFGM_RAT | 84089 | 96 | 5 |
| Mitofusin-2 (C) | MFN2_RAT | 86809 | 37 | 4 |
| Thioredoxin-dependent peroxide reductase (A, B) | PRDX3_RAT | 28563 | 30 | 1 |
| Lon protease homolog, mitochondrial (B,C) | LONM_RAT | 106296 | 27 | 9 |
| Single-stranded DNA-binding protein (A) | SSBP_RAT | 17444 | 25 | 1 |
| DNA polymerase subunit gamma-1 (A) | DPOG1_RAT | 137909 | 23 | 3 |
|
| ||||
| Others# | ||||
| Stress-70 protein (C) | GRP75_RAT | 74097 | 1023 | 22 |
| ATP-dependent Clp protease ATP-binding subunit clpX-like (C) | CLPX_RAT | 69963 | 288 | 9 |
| Acyl-coenzyme A thioesterase 2 (B) | ACOT2_RAT | 49955 | 178 | 5 |
| Electron transfer flavoprotein subunit beta (B) | ETFB_RAT | 27898 | 145 | 8 |
| A-kinase anchor protein 1 (C) | AKAP1_RAT | 92660 | 95 | 2 |
| Up-regulated during skeletal muscle growth protein 5 (A, B) | USMG5_RAT | 6460 | 92 | 2 |
| Cytochrome b5 type B (A, B) | CYB5B_RAT | 16312 | 89 | 3 |
| [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1 (C) | PDP1_RAT | 61739 | 68 | 1 |
| Nucleoside diphosphate-linked moiety X motif 19 (A) | NUD19_RAT | 40426 | 68 | 1 |
| Growth hormone-inducible transmembrane protein (A) | GHITM_RAT | 37324 | 66 | 2 |
| [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase (C) | BCKD_RAT | 46673 | 63 | 1 |
| Iron-sulfur cluster assembly 1 homolog (C) | ISCA1_RAT | 14311 | 57 | 2 |
| ATPase family AAA domain-containing protein 3 (A, B) | ATAD3_RAT | 66889 | 53 | 8 |
| Peroxiredoxin-5 (B) | PRDX5_RAT | 22507 | 53 | 1 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase (A) | ECH1_RAT | 36491 | 52 | 2 |
| Tyrosyl-tRNA synthetase, mitochondrial (C) | SYYM_RAT | 52936 | 51 | 1 |
| Quinone oxidoreductase-like protein 2 (C) | QORL2_RAT | 38095 | 47 | 2 |
| Uncharacterized protein C2orf47 homolog (A, C) | CB047_RAT | 33411 | 39 | 1 |
| Transmembrane protein 126A (A, C) | T126A_RAT | 21758 | 39 | 3 |
| CDGSH iron-sulfur domain-containing protein 1 (A) | CISD1_RAT | 12260 | 37 | 1 |
| Brain protein 44 (B) | BR44_RAT | 14306 | 36 | 2 |
| Coiled-coil domain-containing protein 90B (A) | CC90B_RAT | 29895 | 34 | 3 |
| ES1 protein homolog (A, B) | ES1_RAT | 28497 | 34 | 2 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase (C) | ETFD_RAT | 69010 | 32 | 5 |
| Glycerol-3-phosphate acyltransferase 1 (C) | GPAT1_RAT | 94568 | 32 | 2 |
| 39S ribosomal protein L14 (C) | RM14_RAT | 16017 | 32 | 2 |
| Fumarylacetoacetate hydrolase domain-containing protein 1 (C) | FAHD1_RAT | 24750 | 31 | 1 |
| Kynurenine/alpha-aminoadipate aminotransferase (A, B, C) | AADAT_RAT | 48096 | 29 | 2 |
| GTP-binding protein Rhes (B) | RHES_RAT | 30576 | 28 | 2 |
| Oxidation resistance protein 1 (C) | OXR1_RAT | 93209 | 27 | 3 |
| Enoyl-CoA hydratase domain-containing protein 3 (B) | ECHD3_RAT | 32650 | 27 | 4 |
| Protein Mpv17 (C) | MPV17_RAT | 19842 | 27 | 1 |
| Surfeit locus protein 1 (C) | SURF1_RAT | 35060 | 26 | 3 |
| RRP15-like protein (A) | RRP15_RAT | 31078 | 26 | 3 |
| Serine/threonine-protein phosphatase PGAM5 (A) | PGAM5_RAT | 32269 | 25 | 1 |
| UPF0629 protein C17orf42 homolog (C) | CQ042_RAT | 41911 | 25 | 1 |
| Leucine-rich PPR motif-containing protein (C) | LPPRC_RAT | 157808 | 24 | 9 |
| Stomatin-like protein 2 (A, B) | STML2_RAT | 38504 | 24 | 1 |
| Growth arrest and DNA damage-inducible proteins-interacting protein 1 (A) |
G45IP_RAT | 26565 | 23 | 7 |
| Probable Xaa-Pro aminopeptidase 3 (A) | XPP3_RAT | 57103 | 23 | 2 |
| NLR family member X1 (A) | NLRX1_RAT | 108548 | 22 | 4 |
| Beta-lactamase-like protein 2 (B) | LACB2_RAT | 32749 | 22 | 1 |
| Translational activator of cytochrome c oxidase 1 (A) | TACO1_RAT | 33190 | 22 | 5 |
| Pyruvate carboxylase (A, C) | PYC_RAT | 130436 | 22 | 4 |
| Kinesin-like protein KIF1B (A) | KIF1B_RAT | 205411 | 21 | 7 |
| Protein FAM54B (A) | FA54B_RAT | 32166 | 21 | 2 |
| 39S ribosomal protein L38 (C) | RM38_RAT | 45095 | 21 | 1 |
| Aldehyde dehydrogenase X (B) | AL1B1_RAT | 58102 | 21 | 1 |
| Patatin-like phospholipase domain-containing protein 7 (A) | PLPL7_RAT | 151240 | 21 | 1 |
GST-DEC specific interacting protein (see Methods) detected in pulldown samples using mitochondrial lysates from isolated cardiomyocytes;
same as in A but using whole-cell cardiomyocyte lysates;
BKCa-specific interacting protein detected in immunoprecipitation samples using monoclonal anti-BKCa antibody and lysates of Percoll purified mitochondria from left ventricle (see Methods).
Grpe1 is equivalent to Mge1 (nucleotide exchange factor of Hsp70).
Peptides found (TAVAPIE, YFPTQALNFAFK) are identical in ADP/ATP translocase 1.
Some proteins were re-assigned manually to known functions according to UniProt.
Proteins in “Transport” group were assigned manually. The number of peptides correspond to the highest score of each identified protein.
Table 2.
BKCa channel-interacting proteins from identified by LC/MS/MS.
| Proteins | Uniprot ID | Mass, Da | Score | Peptides |
|---|---|---|---|---|
| Endoplasmic Reticulum | ||||
| Extended synaptotagmin-1 (B,C) | ESYT1_RAT | 121369 | 139 | 6 |
| 78 kDa glucose-regulated protein (B,C) | GRP78_RAT | 72473 | 136 | 4 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (A, B,C) | AT2A1_RAT | 110707 | 65 | 2 |
| Carnitine O-acetyltransferase (A, C) | CACP_RAT | 71211 | 65 | 4 |
| Erlin-2 (B) | ERLN2_RAT | 37915 | 62 | 4 |
| Elongation factor 1-gamma (A,B) | EF1G_RAT | 50371 | 57 | 3 |
| Serpin H1 (B) | SERPH_RAT | 46602 | 51 | 3 |
| Cytochrome P450 2C11 (A) | CP2CB_RAT | 57658 | 39 | 3 |
| Calreticulin (A) | CALR_RAT | 48137 | 38 | 1 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 (B) |
RPN2_RAT | 69149 | 37 | 2 |
| Sterol regulatory element-binding protein 2 (A, C) | SRBP2_RAT | 124096 | 37 | 2 |
| Calnexin (A, B) | CALX_RAT | 67612 | 36 | 2 |
| ORM1-like protein 3 (C) | ORML3_RAT | 17451 | 34 | 1 |
| Reticulon-4 (C) | RTN4_RAT | 126766 | 33 | 3 |
| Antigen peptide transporter 2 (A) | TAP2_RAT | 78063 | 32 | 3 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 (A) | PLOD3_RAT | 85577 | 31 | 2 |
| Cytochrome P450 2A2 (A) | CP2A2_RAT | 56480 | 30 | 3 |
| 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4- isomerase type 2 (A) |
3BHS2_RAT | 42592 | 29 | 1 |
| HCLS1-associated protein X-1 (C) | HAX1_RAT | 31429 | 29 | 2 |
| Inositol 1,4,5-trisphosphate receptor type 1 (A, C) | ITPR1_RAT | 316486 | 29 | 3 |
| Transmembrane protein 214 (A) | TM214_RAT | 77486 | 29 | 2 |
| Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (A) |
NSDHL_RAT | 40671 | 27 | 1 |
| Squalene synthase (A) | FDFT_RAT | 48703 | 26 | 1 |
| Protein ERGIC-53-like (A) | LMA1L_RAT | 56571 | 26 | 2 |
| BET1 homolog (A) | BET1_RAT | 13336 | 26 | 2 |
| Protein ERGIC-53 (B) | LMAN1_RAT | 58206 | 26 | 1 |
| Cytochrome P450 3A1 (B) | CP3A1_RAT | 58222 | 26 | 1 |
| Carboxylesterase 3 (B) | CES3_RAT | 62393 | 25 | 2 |
| Choline-phosphate cytidylyltransferase B (B) | PCY1B_RAT | 42159 | 25 | 1 |
| Reticulocalbin-2 (A) | RCN2_RAT | 37410 | 25 | 2 |
| Cytochrome P450 2C12, female-specific (C) | CP2CC_RAT | 56453 | 24 | 1 |
| Cytochrome P450 2E1 (C) | CP2E1_RAT | 56990 | 24 | 2 |
| Multiple coagulation factor deficiency protein 2 homolog (A) | MCFD2_RAT | 16252 | 24 | 1 |
| Thrombospondin-4 (A) | TSP4_RAT | 110882 | 24 | 1 |
| DnaJ homolog subfamily C member 3 (A,C) | DNJC3_RAT | 57981 | 24 | 3 |
| Lipase maturation factor 2 (A) | LMF2_RAT | 80431 | 23 | 1 |
| Phosphatidylinositol-glycan biosynthesis class W protein (A,C) | PIGW_RAT | 57205 | 23 | 1 |
| LDLR chaperone MESD (A) | MESD_RAT | 25314 | 23 | 1 |
| Uncharacterized glycosyltransferase AER61 (C) | AER61_RAT | 62340 | 23 | 2 |
| Inhibitor of nuclear factor kappa-B kinase-interacting protein (B) |
IKIP_RAT | 42391 | 22 | 3 |
| Sphingosine-1-phosphate lyase 1 (A) | SGPL1_RAT | 64287 | 22 | 4 |
| Dehydrogenase/reductase SDR family member 7B (C) | DRS7B_RAT | 35662 | 22 | 2 |
| UDP-glucose:glycoprotein glucosyltransferase 1 (C) | UGGG1_RAT | 177061 | 22 | 3 |
| Poly [ADP-ribose] polymerase 16 (C) | PAR16_RAT | 37088 | 21 | 2 |
|
| ||||
| Golgi | ||||
| Ras-related protein Rab-10 (C) | RAB10_RAT | 23072 | 102 | 2 |
| Ras-related protein Rab-1A (C) | RAB1A_RAT | 22891 | 67 | 2 |
| Calcium-transporting ATPase type 2C member 1 (B) | AT2C1_RAT | 101519 | 39 | 2 |
| Nucleobindin-2 (A) | NUCB2_RAT | 50173 | 39 | 1 |
| Phosphofurin acidic cluster sorting protein 1 (A, B) | PACS1_RAT | 105034 | 32 | 2 |
| Golgin subfamily A member 2 (A) | GOGA2_RAT | 113232 | 31 | 7 |
| Caveolin-3 (A), B) | CAV3_RAT | 17904 | 31 | 1 |
| CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3- sialyltransferase (A) |
SIAT6_RAT | 42340 | 29 | 3 |
| Protein O-linked-mannose beta-1,2-N- acetylglucosaminyltransferase 1 (A) |
PMGT1_RAT | 75670 | 28 | 1 |
| Alpha-1,3-mannosyl-glycoprotein 2-beta-N- acetylglucosaminyltransferase (A) |
MGAT1_RAT | 51839 | 27 | 1 |
| Myomegalin (A, B,C) | MYOME_RAT | 263193 | 26 | 7 |
| Vacuolar protein sorting-associated protein 54 (B) | VPS54_RAT | 109913 | 24 | 3 |
| Acid phosphatase-like protein 2 (C) | ACPL2_RAT | 55664 | 24 | 1 |
| Golgi phosphoprotein 3-like (A) | GLP3L_RAT | 33072 | 23 | 3 |
| Carbohydrate sulfotransferase 11 (B) | CHSTB_RAT | 42112 | 23 | 3 |
| Beta-1,4-mannosyl-glycoprotein 4-beta-N- acetylglucosaminyltransferase (A) |
MGAT3_RAT | 62552 | 22 | 1 |
| Golgi SNAP receptor complex member 2 (B) | GOSR2_RAT | 24649 | 21 | 1 |
|
| ||||
| Nucleus | ||||
| Vimentin (A, B,C) | VIME_RAT | 53757 | 942 | 16 |
| Alpha-actinin-4 (A, B,C) | ACTN4_RAT | 105306 | 319 | 18 |
| Keratin, type I cytoskeletal 13 (A, C) | K1C13_RAT | 48098 | 208 | 9 |
| Lamin-B1 (A) | LMNB1_RAT | 66794 | 126 | 7 |
| Caveolin-2 (B) | CAV2_RAT | 18482 | 124 | 1 |
| Histone H2A type 1-C (A) | H2A1C_RAT | 14097 | 123 | 2 |
| Cullin-associated NEDD8-dissociated protein 2 (A) | CAND2_RAT | 141238 | 117 | 4 |
| 60S ribosomal protein L23a (A, C) | RL23A_RAT | 17684 | 104 | 1 |
| Homeobox protein Nkx-6.1 (A, B,C) | NKX61_RAT | 37723 | 89 | 1 |
| Cyclin-dependent kinase 7 (Fragment) (A) | CDK7_RAT | 37402 | 59 | 2 |
| DNA-directed RNA polymerase I subunit RPA1 (A, B,C) | RPA1_RAT | 196235 | 54 | 3 |
| Centrosomal protein of 57 kDa (A, C) | CEP57_RAT | 57404 | 49 | 4 |
| Heterogeneous nuclear ribonucleoproteins A2/B1 (A) | ROA2_RAT | 37512 | 49 | 1 |
| Serine/threonine-protein kinase TNNI3K (A, B) | TNI3K_RAT | 93985 | 44 | 1 |
| Mitotic spindle assembly checkpoint protein MAD2B (A, B) | MD2L2_RAT | 24615 | 39 | 1 |
| Speckle targeted PIP5K1A-regulated polyA polymerase (A) | STPAP_RAT | 95516 | 38 | 2 |
| Myc-induced nuclear antigen (A) | MINA_RAT | 53638 | 37 | 2 |
| DNA mismatch repair protein Msh2 (A) | MSH2_RAT | 104704 | 37 | 5 |
| Snurportin-1 (A) | SPN1_RAT | 41337 | 37 | 1 |
| Transcription initiation factor TFIID subunit 6 (A) | TAF6_RAT | 73295 | 36 | 1 |
| Ras association domain-containing protein 2 (B) | RASF2_RAT | 38168 | 36 | 1 |
| 60S ribosomal protein L11 (A) | RL11_RAT | 20468 | 36 | 1 |
| Serine/threonine-protein kinase MAK (A, C) | MAK_RAT | 70280 | 35 | 1 |
| Tyrosine-protein kinase Fer (Fragment) (A) | FER_RAT | 37479 | 35 | 2 |
| Coiled-coil and C2 domain-containing protein 1B (A) | C2D1B_RAT | 93818 | 34 | 3 |
| Splicing regulatory glutamine/lysine-rich protein 1 (C) | SREK1_RAT | 56930 | 34 | 3 |
| Zinc finger protein 57 (A) | ZFP57_RAT | 47584 | 34 | 2 |
| UPF0027 protein C22orf28 homolog (A) | CV028_RAT | 55727 | 33 | 1 |
| Scaffold attachment factor B1 (A, B,C) | SAFB1_RAT | 104960 | 33 | 4 |
| Transmembrane protein 109 (A,B) | TM109_RAT | 26282 | 33 | 2 |
| DNA topoisomerase 1 (A, C) | TOP1_RAT | 91159 | 32 | 3 |
| Hypoxia-inducible factor 3-alpha (A) | HIF3A_RAT | 73470 | 32 | 1 |
| Nucleoporin GLE1 (A) | GLE1_RAT | 79955 | 31 | 4 |
| Double-stranded RNA-specific editase 1 (A) | RED1_RAT | 78218 | 31 | 2 |
| Doublesex- and mab-3-related transcription factor C1 (A) | DMRTC_RAT | 24055 | 31 | 2 |
| Origin recognition complex subunit 1 (C) | ORC1_RAT | 96840 | 30 | 2 |
| 5′-AMP-activated protein kinase subunit gamma-1 (A) | AAKG1_RAT | 37534 | 30 | 1 |
| Transforming growth factor beta regulator 1 (A, C) | TBRG1_RAT | 45384 | 30 | 2 |
| High mobility group nucleosome-binding domain-containing protein 3 (C) |
HMGN3_RAT | 10176 | 30 | 1 |
| Histone acetyltransferase MYST2 (C) | MYST2_RAT | 71154 | 30 | 2 |
| Zinc finger and BTB domain-containing protein 44 (A, B) | ZBT44_RAT | 50987 | 30 | 3 |
| DNA (cytosine-5)-methyltransferase 1 (A, B) | DNMT1_RAT | 185054 | 30 | 5 |
| Nasal embryonic luteinizing hormone-releasing hormone factor (A) |
NELF_RAT | 60701 | 30 | 4 |
| Zinc finger and BTB domain-containing protein 24 (A) | ZBT24_RAT | 79355 | 29 | 3 |
| Nuclear pore complex protein Nup155 (A, B) | NU155_RAT | 156387 | 29 | 2 |
| Period circadian protein homolog 2 (B) | PER2_RAT | 137539 | 28 | 2 |
| Far upstream element-binding protein 2 (A, B) | FUBP2_RAT | 74466 | 28 | 1 |
| Paired box protein Pax-8 (A) | PAX8_RAT | 49118 | 28 | 1 |
| Coiled-coil domain-containing protein 55 (C) | CCD55_RAT | 64370 | 28 | 2 |
| PIN2/TERF1-interacting telomerase inhibitor 1 (A) | PINX1_RAT | 36916 | 28 | 2 |
| Coiled-coil domain-containing protein KIAA1826 homolog (A) | K1826_RAT | 41264 | 28 | 1 |
| Probable ATP-dependent RNA helicase DDX46 (A, C) | DDX46_RAT | 117826 | 28 | 6 |
| Lethal(3)malignant brain tumor-like protein 2 (A) | LMBL2_RAT | 80057 | 28 | 1 |
| DNA-directed RNA polymerase III subunit RPC3 (A) | RPC3_RAT | 60903 | 27 | 2 |
| DNA polymerase delta catalytic subunit (A) | DPOD1_RAT | 124948 | 27 | 5 |
| Nuclear pore complex protein Nup54 (A, C) | NUP54_RAT | 55825 | 27 | 1 |
| ATP-dependent RNA helicase DDX39 (A) | DDX39_RAT | 49591 | 27 | 4 |
| Histone deacetylase 4 (A, C) | HDAC4_RAT | 119377 | 27 | 3 |
| DNA topoisomerase 2-alpha (A) | TOP2A_RAT | 173853 | 27 | 6 |
| PDZ domain-containing protein 2 (A, C) | PDZD2_RAT | 296389 | 27 | 4 |
| Coiled-coil domain-containing protein 104 (A) | CC104_RAT | 39736 | 27 | 2 |
| Neurofibromin (C) | NF1_RAT | 320473 | 27 | 1 |
| Forkhead box protein J1 (C) | FOXJ1_RAT | 45994 | 27 | 2 |
| Origin recognition complex subunit 2 (A) | ORC2_RAT | 66094 | 27 | 3 |
| Transcriptional adapter 2-alpha (A, B) | TAD2A_RAT | 52107 | 27 | 2 |
| Telomerase protein component 1 (A, B,C) | TEP1_RAT | 295173 | 26 | 5 |
| Transcription factor EC (A, C) | TFEC_RAT | 35495 | 26 | 2 |
| SAP domain-containing ribonucleoprotein (A) | SARNP_RAT | 23647 | 26 | 2 |
| Transcription elongation factor A protein-like 8 (A) | TCAL8_RAT | 13553 | 26 | 1 |
| Protein AATF (A) | AATF_RAT | 59461 | 26 | 2 |
| DNA-directed RNA polymerase I subunit RPA2 (A) | RPA2_RAT | 129462 | 26 | 2 |
| Zinc finger CCCH domain-containing protein 18 (C) | ZCH18_RAT | 105702 | 26 | 2 |
| Transcription factor AP-2-alpha (A, C) | AP2A_RAT | 48316 | 26 | 3 |
| Neuronal PAS domain-containing protein 4 (C) | NPAS4_RAT | 88004 | 26 | 1 |
| Clusterin-associated protein 1 (B) | CLUA1_RAT | 46543 | 26 | 2 |
| Apolipoprotein A-IV (A) | APOA4_RAT | 44429 | 25 | 1 |
| 60S ribosomal protein L3 (A), B) | RL3_RAT | 46392 | 25 | 4 |
| 40S ribosomal protein S13 (A) | RS13_RAT | 17212 | 25 | 2 |
| Phosphatase and actin regulator 3 (A, B) | PHAR3_RAT | 58596 | 25 | 2 |
| Histone H3.3 (A) | H33_RAT | 15376 | 25 | 2 |
| SKI family transcriptional corepressor 1 (A) | SKOR1_RAT | 101312 | 25 | 3 |
| Protein GRINL1A (A, B) | GRL1A_RAT | 41305 | 25 | 1 |
| Cell growth-regulating nucleolar protein (A, C) | LYAR_RAT | 44110 | 25 | 2 |
| Pancreas/duodenum homeobox protein 1 (C) | PDX1_RAT | 30983 | 25 | 1 |
| DNA repair protein RAD50 (A, C) | RAD50_RAT | 154772 | 25 | 15 |
| RAC-gamma serine/threonine-protein kinase (B) | AKT3_RAT | 56217 | 25 | 2 |
| Leucine-rich repeat flightless-interacting protein 1 (C) | LRRF1_RAT | 80484 | 25 | 2 |
| Something about silencing protein 10 (C) | SAS10_RAT | 53949 | 24 | 1 |
| Structural maintenance of chromosomes protein 3 (A, B,C) | SMC3_RAT | 138761 | 24 | 4 |
| MACRO domain-containing protein 1 (Fragment) (C) | MACD1_RAT | 29081 | 24 | 2 |
| ZW10 interactor (C) | ZWINT_RAT | 30295 | 24 | 3 |
| Protein Dom3Z (A, C) | DOM3Z_RAT | 45784 | 24 | 2 |
| Kazrin (C) | KAZRN_RAT | 87433 | 24 | 3 |
| General transcription factor 3C polypeptide 1 (A, B,C) | TF3C1_RAT | 245004 | 24 | 3 |
| Suppression of tumorigenicity 18 protein (A) | ST18_RAT | 115074 | 24 | 2 |
| Myocyte-specific enhancer factor 2A (A) | MEF2A_RAT | 53391 | 24 | 2 |
| Homeobox protein MOX-2 (A) | MEOX2_RAT | 33869 | 24 | 1 |
| Ubiquitin carboxyl-terminal hydrolase 16 (A) | UBP16_RAT | 95242 | 24 | 3 |
| Sentrin-specific protease 2 (A) | SENP2_RAT | 67837 | 24 | 1 |
| Far upstream element-binding protein 1 (A, B) | FUBP1_RAT | 67326 | 24 | 3 |
| Hepatoma-derived growth factor-related protein 2 (A) | HDGR2_RAT | 74088 | 24 | 1 |
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-1 (A) |
PLCB1_RAT | 139113 | 24 | 1 |
| Selenocysteine insertion sequence-binding protein 2 (A,B,C) | SEBP2_RAT | 94154 | 24 | 5 |
| Protein salvador homolog 1 (B) | SAV1_RAT | 45133 | 24 | 3 |
| Structural maintenance of chromosomes protein 1A (A) | SMC1A_RAT | 143743 | 23 | 5 |
| DNA-binding protein SMUBP-2 (A) | SMBP2_RAT | 109400 | 23 | 3 |
| Myocyte-specific enhancer factor 2D (A, B) | MEF2D_RAT | 54563 | 23 | 3 |
| Nuclear pore membrane glycoprotein 210 (A, B) | PO210_RAT | 204943 | 23 | 2 |
| Myb-binding protein 1A (A) | MBB1A_RAT | 153046 | 23 | 4 |
| Fidgetin-like protein 1 (A) | FIGL1_RAT | 74891 | 23 | 1 |
| Smad nuclear interacting protein 1 (A) | SNIP1_RAT | 44953 | 23 | 1 |
| Aryl hydrocarbon receptor nuclear translocator 2 (A) | ARNT2_RAT | 78536 | 23 | 1 |
| H/ACA ribonucleoprotein complex subunit 1 (A, B) | GAR1_RAT | 23167 | 23 | 1 |
| Putative rRNA methyltransferase 3 (A, B,C) | RRMJ3_RAT | 95165 | 23 | 2 |
| REST corepressor 2 (C) | RCOR2_RAT | 58116 | 23 | 1 |
| PR domain zinc finger protein 2 (C) | PRDM2_RAT | 189890 | 23 | 2 |
| MutS protein homolog 5 (B,C) | MSH5_RAT | 93362 | 23 | 2 |
| E3 SUMO-protein ligase PIAS3 (C) | PIAS3_RAT | 69289 | 23 | 1 |
| Transcription factor IIIB 50 kDa subunit (C) | BRF2_RAT | 47421 | 22 | 2 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (C) | F263_RAT | 64491 | 22 | 1 |
| Vitamin D3 receptor (B) | VDR_RAT | 48467 | 22 | 2 |
| Meiosis-specific nuclear structural protein 1 (A, B) | MNS1_RAT | 61316 | 22 | 5 |
| Cell division cycle-associated protein 7 (A) | CDCA7_RAT | 43909 | 22 | 2 |
| Kinesin-like protein KIF22 (A, C) | KIF22_RAT | 73239 | 22 | 5 |
| Protein timeless homolog (A) | TIM_RAT | 139562 | 22 | 4 |
| U11/U12 small nuclear ribonucleoprotein 35 kDa protein (A) | U1SBP_RAT | 29186 | 22 | 2 |
| Nuclear pore complex protein Nup85 (A, B) | NUP85_RAT | 29186 | 22 | 2 |
| Interferon-stimulated 20 kDa exonuclease-like 2 (A) | I20L2_RAT | 41684 | 22 | 3 |
| ATPase WRNIP1 (A) | WRIP1_RAT | 72687 | 22 | 2 |
| Nucleolar RNA helicase 2 (A) | DDX21_RAT | 86540 | 22 | 4 |
| Period circadian protein homolog 1 (A, B) | PER1_RAT | 137562 | 22 | 2 |
| U4/U6.U5 tri-snRNP-associated protein 1 (A, B,C) | SNUT1_RAT | 91127 | 22 | 2 |
| Intraflagellar transport protein 172 homolog (C) | IF172_RAT | 199245 | 22 | 2 |
| Serine/threonine-protein kinase PRP4 homolog (A) | PRP4B_RAT | 117335 | 22 | 3 |
| CLK4-associating serine/arginine rich protein (C) | CLASR_RAT | 76932 | 22 | 3 |
| Nuclear pore complex protein Nup98-Nup96 (A) | NUP98_RAT | 198358 | 21 | 1 |
| Nucleolar GTP-binding protein 1 (A) | NOG1_RAT | 74640 | 21 | 3 |
| Protein SCAF8 (A) | SCAF8_RAT | 139874 | 21 | 2 |
| Myogenin (A, B,C) | MYOG_RAT | 33224 | 21 | 2 |
| Ester hydrolase C11orf54 homolog (C) | CK054_RAT | 35427 | 21 | 2 |
| Uncharacterized protein C1orf103 homolog (C) | CA103_RAT | 83074 | 21 | 4 |
| Poly(ADP-ribose) glycohydrolase (A, B) | PARG_RAT | 110417 | 21 | 1 |
| Dual specificity protein kinase CLK3 (C) | CLK3_RAT | 59247 | 21 | 2 |
| Scm-like with four MBT domains protein 1 (A, B,C) | SMBT1_RAT | 98984 | 21 | 1 |
| Chromodomain-helicase-DNA-binding protein 8 (A, B,C) | CHD8_RAT | 292449 | 21 | 1 |
| Digestive organ expansion factor homolog (C) | DEF_RAT | 88169 | 21 | 2 |
| Uncharacterized protein C12orf32 homolog (C) | CL032_RAT | 27281 | 21 | 1 |
| Regulator of G-protein signaling 14 (C) | RGS14_RAT | 59968 | 21 | 1 |
| Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (C) |
ZCRB1_RAT | 24758 | 21 | 2 |
| Spliceosome RNA helicase Bat1 (B) | UAP56_RAT | 49460 | 21 | 1 |
| E3 SUMO-protein ligase PIAS2 (B) | PIAS2_RAT | 64303 | 21 | 1 |
GST-DEC specific interacting protein (see Methods) detected in pulldown samples using mitochondrial lysates from isolated cardiomyocytes.;
same as in A but using whole-cell cardiomyocyte lysates;
BKCa specific interacting protein (see Methods) detected in immunoprecipitation samples using monoclonal anti-BKCa antibody and lysates of Percoll purified mitochondria from left ventricle.
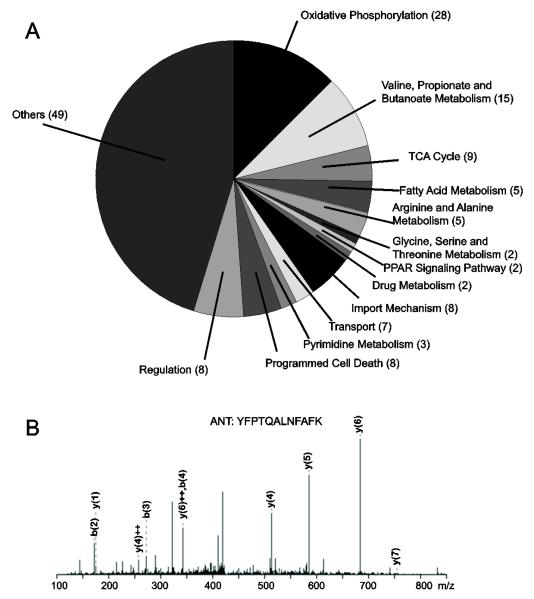
In this work, we focused our analysis on the 151 mitochondrial proteins (Table 1). A functional annotation analysis of these proteins using the bioinformatic tool Database for Annotation, Visualization and Integrated Discovery (DAVID) revealed that BKCa channel is associated with 13 distinctive biological processes in the mitochondria (Fig. 1A). Among those, of special interest were import mechanisms (8 proteins), oxidative phosphorylation (28 proteins), transport (7 proteins), TCA cycle (9 proteins), fatty acid metabolism (5 proteins), and programmed cell death (8 proteins). As an example, a mass spectrum obtained for the adenine nucleotide translocator, ANT is given in Fig. 1B. The uncovered relationship between BKCa channel and the predicted biological activities underscores the potential role of BKCa channel in contributing to the functional integrity of mitochondria and consequently of cardiac cells.
Figure 1. Mitochondrial partners of cardiac mitoBKCa predict new channel functions.
(A) Gene ontology analysis using DAVID of mitochondrial BKCa proteome revealed mitoBKCa potential involvement with 13 mitochondrial functions in the heart. Numbers in parenthesis correspond to the number of proteins allocated to each group. “Transport” group was entered manually. Isolation of BKCa partners was performed using pull-down and immunoprecipitation with: 1) Recombinant GST-DEC and mitochondrial lysates of isolated cardiomyocytes, 2) Recombinant GST-DEC and whole-cardiomyocyte lysates, and 3) Monoclonal anti-BKCa antibody and lysates of Percoll-purified mitochondria from left ventricle. Controls included GST pull-down and IgG immunoprecipitation (see Methods). A total of 151 mitochondrial proteins with a score>21 were identified. (B) Example of mass spectrum of a peptide matching ANT.
3.2. mitoBKCa interacts with Tom22 in HEK293T cells
One of the partners found in the interactome associated with the DEC sequence was Tom22 (matching peptide LQMEQQQQLQQR; Table 1), a protein located on the outer mitochondrial membrane. Tom22 is known to serve as an initial recognition site for protein import into mitochondria (Mayer et al., 1995). Thus, we deemed it important to corroborate its association with BKCa as this would permit to start setting the building blocks of mitoBKCa import pathway. Thus, we co-expressed BKCa with or without DEC (3HA-tagged N-terminal of Met3) with Tom22 (C-terminal c-Myc tagged) in HEK293T cells for co-immunoprecipitation analysis by immunoprecipitating BKCa or BKCa-DEC and examining the immunoprecipitation products for Tom22. As controls, we also expressed each protein alone.
Figure 2A shows in the same blot the immunoprecipitation of BKCa and BKCa-DEC together with the successful co-immunoprecipitation of Tom 22 by both BKCa variants. In separate experiments, immunoprecipitation of Tom22 yielded bands of similar sizes as the ones observed in the co-immunoprecipitation products confirming Tom22 molecular size (not shown). The strong signal near 100 kDa likely corresponds to IgG composite or non-specific labeling of the secondary antibody. The specificity of the co-immunoprecipitation was confirmed by reverse coimmunoprecipitation with BKCa (see section 3.4). Fig. 2B demonstrates in the same blot the proper expression of each clone in the input lysates. The co-immunoprecipitation (Co-IP) efficiency of the two BKCa variants was calculated as previously described (Li et al., 2013) by the ratio of the band intensities of co-immunoprecipitated Tom22 (as in Fig. 2A, squares) to those of immunoprecipitated BKCa variant (as in Fig. 2A, arrows), and normalized to the corresponding Tom22 signals in the input lysates (as in Fig. 2B, arrowhead). Figure 2C shows the mean values demonstrating that BKCa and BKCa-DEC had a similar ability to interact with Tom22 (0.89±0.18 vs. 0.91±0.15; n=3 each). These results are consistent with the proteomic data and demonstrate that Tom22 association with BKCa in HEK293T cells does not depend on the presence of DEC sequence in BKCa.
Figure 2. BKCa interacts with Tom22 in HEK293T cells.
Cells were co-transfected with BKCa, BKCa-DEC and Tom22 in different combinations (+, present; -, absent). Co-immunoprecipitated products are highlighted with squares. (A) Immunoblot demonstrating that BKCa and BKCa-DEC pull down Tom22. BKCa constructs were immunoprecipitated with anti-HA pAb and detected with anti-HA mAb (arrows) and co-immunoprecipitated Tom22 was detected with anti-c-Myc mAb (arrowhead, squares). Primary antibodies were incubated at the same time. (B) Immunoblot showing proper expression of BKCa constructs (arrows) and Tom22 (arrowhead) in the input lysates. (C) BKCa and BKCa-DEC efficiency in pulling down Tom22 is similar (n=3 each). Co-IP efficiency was determined by first subtracting background signals, which in this case were negligible, and calculating the ratio of the band intensities of co-immunoprecipitated Tom22 (i.e. A, squares) to corresponding immunoprecipitated BKCa constructs (i.e.. A, arrows), and normalized to Tom22 intensity in the corresponding input lysates (i.e. B, arrowhead). In this and following figures, image analysis was performed with MetaMorph Image Analysis Software (Molecular Devices).
3.3. Both mitoBKCa and Tom22 are imported into mitochondria of HEK293T cells
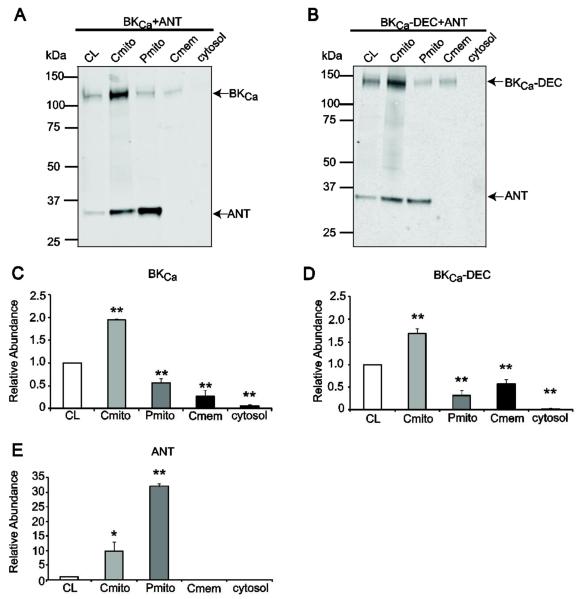
We next examined the subcellular localization of co-expressed Tom22 and BKCa through cell fractionation and immunoblotting. Figure 3A,B (upper panels) and Fig. 3C,D show that both BKCa and BKCa-DEC were significantly enriched in the “crude” mitochondria fraction (Cmito). Importantly, BKCa isoforms were also detected in the purified mitochondria fraction (Pmito), indicating that BKCa was imported into mitochondria in HEK293T cells. Interestingly, Tom22 was found in all of the fractions examined (A,B, bottom panels; E) including the cytosol and crude membranes. This finding cannot be explained by inadequate separation since, as shown later in section 3.6, a typical mitochondrial marker, ANT, was only present in “crude” and purified mitochondrial fractions. The mean relative abundance values of BKCa and BKCa-DEC in Fig. 3C,D show a very similar pattern of expression in the different subcellular fractions. Both variants gave the strongest signal in “crude” mitochondria (BKCa=1.62±0.13 and BKCa-DEC=1.26±0.11, n=3 each) suggesting that the majority of BKCa channels were localized to mitochondria or in mitochondria-associated membranes. In agreement, the purified mitochondria fraction displayed a considerable amount of BKCa channel signal (BKCa=0.73±0.09, BKCa-DEC=0.45±0.17, n=3, each). As expected from previous studies (Ma et al., 2007) both variants were also present in “crude” membranes (BKCa=0.36±0.07, BKCa-DEC=0.65±0.20, n=3 each), whereas their presence in the cytosolic fraction was minimal (BKCa=0.02±0.03, BKCa-DEC=0.02±0.001, n=3 each).
Figure 3. BKCa and Tom22 are imported into mitochondria in HEK293T cells.
(A and B) Dually labeled immunoblots of BKCa (A, top) or BKCa-DEC (B, top) co-expressed with Tom22 (bottom) in different cellular fractions: CL, whole cell lysate; Cmito, “crude” mitochondria; Pmito, Percoll-purified mitochondria; Cmem, “crude” membranes; cytosol, cytosolic fraction. BKCa and Tom22 images were separated for clarity. In panel (A), BKCa and Tom22 blots are from different experiments. BKCa constructs were recognized with anti-HA mAb (upper panels); Tom22 was recognized with anti-c-Myc pAb (lower panels). (C-E) Mean values of BKCa, BKCa-DEC and Tom22 relative abundance in different cellular fractions show that BKCa, BKCa-DEC and Tom22 were imported into “crude” and purified mitochondrial fractions, though not enriched. Values for Tom22 were pooled from both BKCa and BKCa-DEC experiments. *, p<0.05 compared to CL. **, p<0.01 compared to CL. n=3 independent experiments.
The mean relative quantification of Tom22 in different fractions is shown in Fig. 3E. As stated earlier, Tom22 distribution spanned all fractions examined. Nevertheless, the expression and distribution of Tom22 in the “crude” mitochondria (0.94±0.20, n=6 each) and the purified mitochondria (0.59±0.25, n=6 each) was robust. Thus, the results indicate that both BKCa variants and Tom22 are targeted into mitochondria, though not exclusively, in HEK293T cells.
3.4. The BKCa channel interacts with Tom22 through its transmembrane region
To identify the regions in BKCa channel relevant for its association with Tom22, we performed coimmunoprecipitations with BKCa N- and C-terminal deletions. Figure 4A shows a scheme of wild-type BKCa (1-1113 amino acids) where the arrows delineate the regions spanned by each construct (the numbers correspond to the amino acids numbering). All BKCa constructs were tagged with c-Myc epitope at the N-terminus. C-terminal truncates were: 1-343, 1-441 and 1-711; N-terminal deletion constructs were: 322-1117 covering the whole C-terminus and 679-1113 that includes the regulator for potassium conductance 2 (RCK2) domain, also called the “tail” region. Tom22 contained a DDK tag at the C-terminus. HEK293T cells were transfected with BKCa constructs alone (control) or in conjunction with Tom22, and with Tom22 alone (control). All samples were subjected to immunoprecipitation using anti-DDK mAb targeting Tom22, and different truncated BKCa proteins were detected with anti-c-Myc pAb (Fig. 4B, top). The co-immunoprecipitated signals of 1-343, 1-441, and 1-711 constructs were strong (lanes 8-10, solid squares) compared to the background signals of BKCa proteins expressed alone (lanes 1-3, dashed squares), whereas co-immunoprecipitated C-terminus (322-1113) and “tail” (679-1113) were barely detected (lanes 11,12, solid squares) and practically absent in the control (lanes 4 and 5, dashed squares). As reference, lane 13 shows the co-immunoprecipitated wild type BKCa. Note that this coimmunoprecipitation corresponds to the reverse coimmunoprecipitation of Fig. 2A. Figure 4B (bottom panel) shows the successful immunoprecipitation of Tom22 for the same experiment, and Fig. 4C displays the expression of all clones in the input lysates. The co-immunoprecipitation (co-IP) efficiency of different BKCa regions was calculated by first subtracting the corresponding background signals from the coimmunoprecipitation signals of BKCa constructs (as in Fig. 4B, top) and normalizing to the corresponding immunoprecipitated Tom22 (as in Fig. 4B, bottom) and BKCa input signals (as in Fig. 4C, top, lanes 8-13). The co-IP efficiency value of wild-type BKCa was set to 1. Constructs 1-343, 1-441, and 1-711 showed a similar ability to interact with Tom22 (1-343=3.43±0.90, 1-441= 2.42±0.60, 1-711=2.04±0.42; n=3 each, p>0.05), which is much higher than that observed for wild-type BKCa. In contrast, both the C-terminus and tail lost their ability to efficiently coimmunoprecipitate Tom22 (C-terminus=0.33±0.05, tail=0.42±0.21; n=3 each). Note that although construct 1-711 contains part of the C-terminus (up to the first amino acids of the “tail” segment) it maintained a significant interaction with Tom22. Taken together, the results suggest that the interaction between BKCa channel and Tom22 is mainly through the transmembrane domain of BKCa channel and that the “tail” region exerts an inhibitory action on the association of both proteins.
3.5. mitoBKCa interacts with ANT in HEK293T cells
From the potential partners of BKCa in Table 1, we next selected ANT (matching peptides TAVAPIE, YFPTQALNFAFK) to substantiate the association of the two proteins by biochemical means. Although the analysis software reported ANT2 as the matching protein, the identified peptides are identical in ANT1, which is the isoform that predominates in heart tissue (Graham et al., 1997; Dorner et al., 1999). ANT is of special interest: in addition to its role in exchanging ADP/ATP across the inner mitochondrial membrane, ANT serves as regulator of the mPTP (Morciano et al., 2015; Halestrap and Richardson, 2015). Moreover, its potential association with mitoBKCa offers a molecular link between regulatory components of the mPTP and the role of mitoBKCa in the regulation of mPTP (Cheng et al., 2011; Singh et al., 2013).
Figure 5A shows co-immunoprecipitation experiments performed using HEK293T cells co-expressing ANT (c-Myc-DDK tagged) with BKCa (untagged) and BKCa-DEC (HA tagged) (lanes 4 and 5) or expressing ANT, BKCa-DEC or BKCa alone (lanes 1-3). In these experiments, we immunoprecipitated BKCa and tested the blots for BKCa as control (Fig. 5A, top panel) and for co-immunoprecipitated ANT (Fig. 5A, lower panel). Signals of co-immunoprecipitated ANT (squares, lanes 4 and 5) were only present when both ANT and BKCa (or BKCa-DEC) were co-expressed. Confirming the molecular mass of ANT, its immunoprecipitation yielded products with similar molecular mass (not shown). For reverse co-immunoprecipitation see Fig. 7 (lane 13). Fig. 5B demonstrates proper expression of all clones in the input lysates. Note that although expression of BKCa-DEC was lower in the input lysate (Fig. 5B, upper panel, lanes 2 and 4), its immunoprecipitation was quite efficient yielding bands as intense as those for BKCa (Fig. 5A, top panel). The co-immunoprecipitation efficiency, calculated as in Fig. 2E, demonstrates that the DEC sequence in BKCa-DEC enhances the ability of ANT to associate with BKCa by ~30% (from 0.69±0.02 to 1, n=3). This result is in contrast to Tom22, which interacted equally well with both BKCa and BKCa-DEC.
3.6. mitoBKCa and ANT are imported into mitochondria of HEK293T cells
Next, we investigated the subcellular localization of BKCa (or BKCa-DEC) and ANT co-expressed in HEK293T cells using cell fractionation and double-labeled immunoblots. Figure 6 illustrates the distribution of BKCa (A) and BKCa-DEC (B) when co-expressed with ANT. Bands around ~135 kDa correspond to BKCa (A), at ~140 kDa correspond to BKCa-DEC (B) and bands at ~32 kDa correspond to ANT (A,B). In contrast to Tom22 that distributed along all fractions (Fig. 3), ANT showed a clear-cut enrichment in the “crude” and purified mitochondrial fractions indicating that at the time of cell harvesting the vast majority, if not all, synthesized ANT had reached its mitochondrial destination. In line with this view, no signals were detected in the “crude” membrane or cytosolic fractions (within the resolution of our immunoblots). On the other hand, BKCa and BKCa-DEC followed the same expression trend as when co-expressed with Tom22 (Fig. 3) with both BKCa and BKCa-DEC concentrated in the “crude” mitochondrial fraction but still detected in the purified mitochondrial fraction.
Figure 6. BKCa and ANT are imported into mitochondria of HEK293T cells.
A and B, Dually labeled immunoblots of BKCa (A) or BKCa-DEC (B) co-expressed with ANT show that the three constructs were imported into “crude” and purified mitochondria fractions. Note the enrichment of ANT in the purified mitochondria fraction. Labels are as in Fig. 3. BKCa was recognized with anti-HA mAb. ANT was recognized with anti-c-Myc pAb. C, D, and E, Quantification of BKCa, BKCa-DEC and ANT relative abundance in different cellular fractions. *, p<0.05 compared to CL; **, p<0.01compared to CL. n=3 independent experiments.
The mean relative abundance of each protein in the different fractions with respect to cell lysates (set to 1) is shown in Fig. 6C-E. Mean values for BKCa (C) and BKCa-DEC (D) in “crude” mitochondria fractions were: BKCa=1.96±0.002 and BKCa-DEC=1.68±0.10 (n=3 each), and in the purified mitochondria fraction were: BKCa=0.56±0.10 and BKCa-DEC=0.31±0.10 (n=3 each). In contrast, the distribution in the “crude” membrane and cytosolic fractions were minimal with the exception of BKCa-DEC in “crude” membranes (“crude” membranes: BKCa=0.23±0.10, BKCa-DEC=0.57±0.10; cytosol: BKCa=0.06±0.03, BKCa-DEC=0.02±0.03; n=3 each). ANT mean relative abundance values (E) demonstrated a vast enrichment (32 times) in the purified mitochondrial fraction and a robust segregation in the “crude” mitochondria fraction (10 times) with respect to cell lysates. Mean values were: 32.05±0.99 and 9.70±3.20 (n=3 each), respectively. These results are consistent with the fact that ANT is the most abundant protein in the inner mitochondrial membrane (Liu and Chen, 2013) and validate the cell fractionation protocol used here. Moreover, they indicate that the association between ANT and BKCa must occur in mitochondria.
3.7. ANT interacts with BKCa through the channel transmembrane domain
Figure 5C showed that the presence of the DEC sequence can facilitate, but does not determine, the interaction between BKCa and ANT. Thus, other regions in BKCa must be of relevance for the association between the two molecules. To address this point, we performed coimmunoprecipitation experiments to define the major region(s) in BKCa involved in its association with ANT. The same deletion constructs as in Fig. 4C that lack DEC insert were co-expressed or not (control) with ANT in HEK293T cells. Figure 7A shows an example of a coimmunoprecipitation experiment using anti-DDK mAb to immunoprecipitate ANT and anti-c-Myc pAb to detect immunoprecipitated ANT and co-immunoprecipitated BKCa proteins in the same blot. Lanes 8-13 show the co-immunoprecipitated BKCa constructs together with the immunoprecipitated ANT; lanes 1-6 show the corresponding background signals in the absence of ANT expression and lane 7 shows expression of ANT alone.
Signals of 1-343, 1-441, and 1-711 were strong (lanes 8-10, squares), whereas, the C-terminus (322-1113) and “tail” (679-1113) signals were low (lanes 11-12, squares). As reference, lane 13 shows the co-immunoprecipitation of wild type BKCa. Note that the coimmunoprecipitated BKCa signals (squares) are much stronger than the corresponding background signals (dashed squares). The immunoblot of corresponding input lysates is shown in Fig. 7B. After subtraction of background signals, the co-immunoprecipitation signals of BKCa constructs were normalized to the corresponding ANT signals in the immunoprecipitates and to the BKCa signals in the input lysates to obtain the Co-IP efficiency compared to wild-type BKCa. Compared to wild-type BKCa, the Co-IP efficiency was higher for constructs 1-343 and 1-441 (1-343=2.05±0.69; 1-441=1.65±0.34; n=3 each). Protein 1-711 showed very similar ability to interact with ANT (0.91±0.20, n=3 each) compared to the full length BKCa; whereas, the C-terminus 322-1113 (0.17±0.05, n=3 each) and “tail” 679-1113 (0.33±0.03, n=3 each) had a marked decrease in their ability to associate with ANT. Overall, the results indicate that in HEK293T cells the interaction between BKCa and ANT is mainly through the transmembrane domain of BKCa (1-343) and that the whole C-terminus and not only the “tail” as for Tom22 exerts an inhibitory effect in the interaction.
4. Discussion
4.1. Cardiac BKCa mitochondrial interactome: physiological relevance
This study is the first to address the potential interactome of BKCa channels specifically in mitochondria, in particular of cardiac mitochondria. In total, we identified 151 mitochondrial proteins that can interact with BKCa. These partner proteins are involved in 13 physiological mitochondrial functions, including oxidative phosphorylation, TCA cycle, import, and metabolism (Fig. 1 and Table 1). We focused on the heart because mitoBKCa has a proven physiological role in this organ, acting as a shield from ischemia and reperfusion injury (Xu et al., 2002; Singh et al., 2013; Soltysinska et al., 2014; Balderas et al., 2015), while its activity improves basal mitochondrial respiratory performance (Aon et al., 2010). Moreover, in astrocytoma cells mitoBKCa electrical activity is influenced by substrates of oxidative phosphorylation (Bednarczyk et al., 2013). Consistent with these functional studies, we found mitochondrial BKCa partner proteins belonging to four complexes (complex I, III, IV, and V) of the respiratory chain including ATP synthase, cytochrome c oxidase and NADH dehydrogenase. Interestingly, in our previous whole brain proteomic studies, we also found these respiratory chain proteins forming part of the BKCa interactome (Singh et al., 2016). It is worth mentioning that cytochrome c oxidase subunit 1 directly binds to the regulatory β1 subunit of BKCa channel in cardiac cells (Ohya et al., 2005). Because BKCa was found here to also be a potential partner of this subunit of cytochrome c oxidase (Table 1), it is possible that mitoBKCa can form a tripartite complex with these subunits in the heart.
mitoBKCa also participates in the regulation of the mPTP. Inhibition of its activity either by gene silencing in ischemia-reperfusion experiments (Singh et al., 2013) or by pharmacological inhibition under basal conditions (Cheng et al., 2011) promotes the opening of mPTP. The current view on mPTP molecular composition supports ATP synthase dimers or ATP subunit c as the core of mPTP surrounded by several regulatory subunits that include ANT (Giorgio et al., 2013; Halestrap and Richardson, 2015; Morciano et al., 2015; Izzo et al., 2016). That ATP synthase and ANT form a supercomplex (ATP synthasome) together with the inorganic phosphate carrier (PiC) is well established (Chen et al., 2004), although in rat cardiac mitochondria ANT can also be observed independently of ATP synthase (Nuskova et al., 2015). The proteomic data yielded several ATP synthase subunits as candidate partners of BKCa as well as ANT and PiC. This novel information sets the background for future studies to learn whether BKCa forms part of a megacomplex with the ATP synthase. As a first step, in this work, we chose to study the association of BKCa with ANT (Figs. 5-7) (see below).
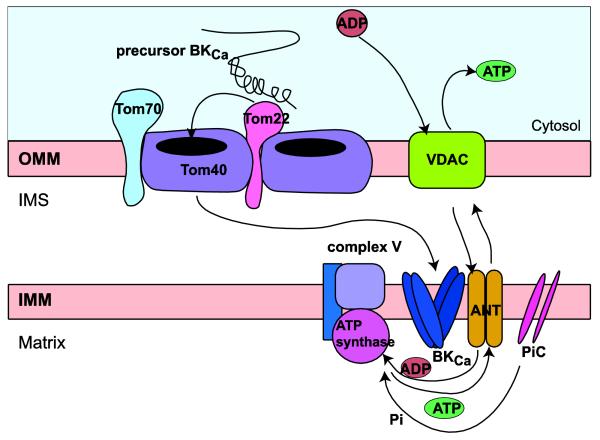
mitoBKCa is a nuclear-encoded protein and thus, must be imported into mitochondria to fulfill its multiple functions. As will be discussed below, our data also gives initial clues about the mechanisms that could support mitoBKCa import.
4.2. Tom22, a potential mitochondrial import mechanism for BKCa channel
A total of 8 proteins of the mitochondrial import system were found as potential partners of BKCa (Table 1). Specifically, we found the receptor proteins Tom22 and Tom70, and the protein-conducting channel Tom40 of the outer membrane TOM complex (Wenz et al., 2015); and Tim23, Tim16, GrpE1, and MPPA of the inner membrane Tim23-PAM classical presequence import pathway (Schulz et al., 2015). Although BKCa lacks a presequence, there is precedence in the literature for atypical proteins (e.g. phosphate carrier) that only contain C-terminal or internal targeting sequences, like BKCa, that utilize the Tim23-PAM pathway (Schulz et al., 2015). Thus, BKCa may be another example of this type of unconventional precursor proteins.
In this work, we scrutinized the association of the receptor protein Tom22 with BKCa and BKCa-DEC. As mentioned before, the C-terminal DEC splice insert favors BKCa expression in mitochondria of adult cardiomyocytes making it a candidate for an import signal. Interestingly, both BKCa isoforms (with or without DEC) associated equally well with Tom22 (Fig. 2). Several explanations are possible: i) that the DEC insert is not essential for the association with Tom22 and that other regions in BKCa are relevant, ii) that the DEC insert is substrate of another component of the import pathway, or iii) that BKCa utilizes different import mechanisms depending on the cell type.
Our results also show that both BKCa isoforms coexist with Tom22 in the “crude” and purified mitochondrial fractions (Fig. 3) demonstrating that both isoforms are able to be imported into mitochondria, and supporting the idea that their association can occur in this organelle. In co-expressing HEK293T cells, BKCa isoforms were also present in the “crude” membrane fraction; this is expected as BKCa can have multiple subcellular locations including the plasma membrane. Tom22 abundance in the cytosolic fraction (likely containing ribosomes) may be an indication of its synthesis at the time of cell harvesting in these cells, but the detection in “crude” membranes is at present enigmatic (Fig. 3E). Previous studies in expressing HeLa cells have shown faint signals in these fractions (Nakamura et al., 2004). The distribution of Tom22 is in clear contrast with the sharp enrichment of ANT in mitochondrial fractions (Fig. 6) arguing against a deficiency in the fractionation protocol or a misstargeting effect due to overexpression. In any event, the results demonstrate beyond doubt that BKCa can associate with Tom22 via its transmembrane segment and in doing so does not require the presence of the C-terminal DEC insert (Fig. 4). The association could be indirect via a yet to be discovered linking partner or direct protein-protein interaction between the two molecules. A simple picture in BKCa import route based on these results and proteomic data can include the recognition of BKCa precursor protein by Tom22 and translocation via Tom40 to reach the inner mitochondrial membrane (Fig. 8) perhaps driven by the Tim23-PAM complex. At present, we can not exclude the possibility that BKCa association with Tom22 could serve another function independent of its import. In fact, new non-assembled intermediate complexes have been recently discovered by proteomics between components of the Tom system (Tom40 and Tom22) and Porin 1 (the voltage dependent anion channel homolog in yeast) (Muller et al., 2016).
Figure 8. Scheme of potential mitoBKCa mport route and interactions in mitochondria.
Overall the results suggest that the precursor BKCa could be recognized by Tom22 in the outer mitochondrial membrane (OMM) and imported by Tom 40 to reach its final destination in the inner mitochondrial membrane (IMM). At the IMM, mitoBKCa interacts with ADP/ATP translocase (ANT). The interaction could be independent or in combination with known partners of ANT, e.g. the synthasome (ATP synthase and the phosphate carrier, PiC), and implies a new role of mitoBKCa in mitochondrial metabolism. Of note, proteomic analysis (Table 1) points to the possibility that BKCa may also interact with the voltage-dependent anion channel VDAC, the ATP synthase and PiC. VDAC, allows ATP/ADP exchange with the intracellular milieu. IMS, intermembrane space.
4.3. BKCa interaction with ANT
We have demonstrated the novel interaction between ANT and BKCa in co-expressing HEK293T cells (Figs. 5 and 7). As for Tom22, both BKCa isoforms (with and without DEC) could associate with ANT; however, the presence of DEC sequence improved their association. The trend in subcellular distribution of BKCa (with or without DEC) co-expressed with ANT including the relative abundance in the purified mitochondria fraction was similar to that observed with Tom22 (Figs. 3 and 6) indicating that its distribution to mitochondria was not biased by the co-expressed partner. At the time of harvesting of the cells, ANT was dramatically enriched in the purified mitochondria fraction and BKCa though concentrated in the “crude” mitochondria fraction was also present in purified mitochondria (Fig. 6). Because ANT was so enriched in the purified mitochondria fraction, the likelihood that the interaction between the two proteins happens in the mitochondria is high. The fact that ANT resides in the inner mitochondrial membrane gives further support to the notion that BKCa resides in this location as well (Fig. 8) (Xu et al., 2002). The association of BKCa with ANT in the inner mitochondrial membrane could be in conjunction with the ATP synthasome as mentioned before (Fig. 8) or independent of it. In any case, the interaction of BKCa and ANT discovered in this work takes place via the transmembrane domain of BKCa (Fig. 7) either directly with ANT or via an unknown intermediary partner. Together our results open a new window to understand how BKCa channel regulates the activity of mPTP inasmuch as ANT is a regulator of mPTP (Halestrap and Richardson, 2015). A functional coupling between BKCa and ANT could be an additional factor in the complex regulation of mPTP.
5. Conclusion
The results from this work demonstrate the novel interactions of BKCa with Tom22 and ANT, and provide a molecular framework to better understand the intricacies of BKCa mitochondrial import mechanism and function.
Highlights.
151 mitochondrial BKCa channel protein partners are revealed by proteomics
A molecular framework defining BKCa mitochondrial protein relationships is provided
Novel partners of BKCa are Tom22 import protein and ANT nucleotide translocator
BKCa interaction with ANT is enhanced by the 50 amino acid BKCa splice insert, DEC
The BKCa transmembrane domain is required for the interaction with Tom 22 and ANT
Acknowledgements
This work was supported by the National Institutes of Health [grant HL107418 to LT, ES, RO].
Abbreviations
- ANT
adenine nucleotide translocator
- BKCa
large conductance voltage- and Ca2+-dependent K+ channel
- BKCa-DEC
BKCa containing DEC splice insert at the C-terminus
- co-IP
co-immunoprecipitation
- DEC
a 50 amino acid C-terminal BKCa splice insert
- GST
glutathione S-transferase
- LC/MS/MS
liquid chromatography, mass spectrometry in tandem
- mAb
monoclonal antibody
- mitoBKCa
mitochondrial BKCa
- mPTP
mitochondrial permeability transition pore
- pAb
polyclonal antibody
- Tom22
mitochondrial import receptor subunit Tom22
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aon MA, Cortassa S, Wei AC, Grunnet M, O'Rourke B. Energetic performance is improved by specific activation of K+ fluxes through K(Ca) channels in heart mitochondria. Biochim.Biophys.Acta. 2010;1797:71–80. doi: 10.1016/j.bbabio.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderas E, Zhang J, Stefani E, Toro L. Mitochondrial BKCa channel. Front Physiol. 2015;6:104. doi: 10.3389/fphys.2015.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarczyk P, Wieckowski MR, Broszkiewicz M, Skowronek K, Siemen D, Szewczyk A. Putative Structural and Functional Coupling of the Mitochondrial BK Channel to the Respiratory Chain. PLoS.One. 2013;8:e68125. doi: 10.1371/journal.pone.0068125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ko Y, Delannoy M, Ludtke SJ, Chiu W, Pedersen PL. Mitochondrial ATP synthasome: three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J.Biol.Chem. 2004;279:31761–31768. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Gulbins E, Siemen D. Activation of the permeability transition pore by Bax via inhibition of the mitochondrial BK channel. Cell Physiol Biochem. 2011;27:191–200. doi: 10.1159/000327944. [DOI] [PubMed] [Google Scholar]
- Dorner A, Olesch M, Giessen S, Pauschinger M, Schultheiss HP. Transcription of the adenine nucleotide translocase isoforms in various types of tissues in the rat. Biochim.Biophys.Acta. 1999;1417:16–24. doi: 10.1016/s0005-2736(98)00245-4. [DOI] [PubMed] [Google Scholar]
- Giorgio V, von SS, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabo I, Lippe G, Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci U S A. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini G, Ponomareva O, Shores KS, Person MD, Harris RA, Mayfield RD. Dynamin-1 co-associates with native mouse brain BKCa channels: proteomics analysis of synaptic protein complexes. FEBS Lett. 2010;584:845–851. doi: 10.1016/j.febslet.2009.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat.Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- Graham JM. Purification of a crude mitochondrial fraction by density-gradient centrifugation. Curr.Protoc.Cell Biol. 2001 doi: 10.1002/0471143030.cb0304s04. Chapter 3, Unit. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Richardson AP. The mitochondrial permeability transition: a current perspective on its identity and role in ischaemia/reperfusion injury. J.Mol.Cell Cardiol. 2015;78:129–141. doi: 10.1016/j.yjmcc.2014.08.018. [DOI] [PubMed] [Google Scholar]
- Izzo V, Bravo-San Pedro JM, Sica V, Kroemer G, Galluzzi L. Mitochondrial Permeability Transition: New Findings and Persisting Uncertainties. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Kathiresan T, Harvey M, Orchard S, Sakai Y, Sokolowski B. A protein interaction network for the large conductance Ca(2+)-activated K(+) channel in the mouse cochlea. Mol.Cell Proteomics. 2009;8:1972–1987. doi: 10.1074/mcp.M800495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang Z, Koh H, Lu R, Jiang Z, Alioua A, Garcia-Valdes J, Stefani E, Toro L. The beta1-Subunit of the MaxiK Channel Associates with the Thromboxane A2 Receptor and Reduces Thromboxane A2 Functional Effects. J.Biol.Chem. 2013;288:3668–3677. doi: 10.1074/jbc.M112.426585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen XJ. Adenine nucleotide translocase, mitochondrial stress, and degenerative cell death. Oxid.Med.Cell Longev. 2013;2013:146860. doi: 10.1155/2013/146860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Nakata T, Zhang G, Hoshi T, Li M, Shikano S. Differential trafficking of carboxyl isoforms of Ca2+-gated (Slo1) potassium channels. FEBS Lett. 2007;581:1000–1008. doi: 10.1016/j.febslet.2007.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Nargang FE, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meera P, Wallner M, Song M, Toro L. Large conductance voltage- and calcium-dependent K+ channel, a distinct member of voltage-dependent ion channels with seven N-terminal transmembrane segments (S0-S6), an extracellular N terminus, and an intracellular(S9-S10) C terminus. Proc.Natl.Acad.Sci.U.S.A. 1997;94:14066–14071. doi: 10.1073/pnas.94.25.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Menegola M, Buchwalder L, Park EW, Meredith A, Rhodes KJ, Aldrich RW, Trimmer JS. Immunolocalization of the Ca2+-activated K+ channel Slo1 in axons and nerve terminals of mammalian brain and cultured neurons. J Comp Neurol. 2006;496:289–302. doi: 10.1002/cne.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morciano G, Giorgi C, Bonora M, Punzetti S, Pavasini R, Wieckowski MR, Campo G, Pinton P. Molecular identity of the mitochondrial permeability transition pore and its role in ischemia-reperfusion injury. J.Mol.Cell Cardiol. 2015;78:142–153. doi: 10.1016/j.yjmcc.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Muller CS, Bildl W, Haupt A, Ellenrieder L, Becker T, Hunte C, Fakler B, Schulte U. Cryo-slicing Blue Native-Mass Spectrometry (csBN-MS), a Novel Technology for High Resolution Complexome Profiling. Mol.Cell Proteomics. 2016;15:669–681. doi: 10.1074/mcp.M115.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Suzuki H, Sakaguchi M, Mihara K. Targeting and assembly of rat mitochondrial translocase of outer membrane 22 (TOM22) into the TOM complex. J.Biol.Chem. 2004;279:21223–21232. doi: 10.1074/jbc.M314156200. [DOI] [PubMed] [Google Scholar]
- Nuskova H, Mracek T, Mikulova T, Vrbacky M, Kovarova N, Kovalcikova J, Pecina P, Houstek J. Mitochondrial ATP synthasome: Expression and structural interaction of its components. Biochem.Biophys.Res.Commun. 2015;464:787–793. doi: 10.1016/j.bbrc.2015.07.034. [DOI] [PubMed] [Google Scholar]
- Ohya S, Kuwata Y, Sakamoto K, Muraki K, Imaizumi Y. Cardioprotective effects of estradiol include the activation of large-conductance Ca(2+)-activated K(+) channels in cardiac mitochondria. Am.J.Physiol Heart Circ.Physiol. 2005;289:H1635–H1642. doi: 10.1152/ajpheart.00016.2005. [DOI] [PubMed] [Google Scholar]
- Schulz C, Schendzielorz A, Rehling P. Unlocking the presequence import pathway. Trends Cell Biol. 2015;25:265–275. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Singh H, Li M, Hall L, Chen S, Sukur S, Lu R, Caputo A, Meredith AL, Stefani E, Toro L. MaxiK channel interactome reveals its interaction with GABA transporter 3 and heat shock protein 60 in the mammalian brain. Neuroscience. 2016;317:76–107. doi: 10.1016/j.neuroscience.2015.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Lu R, Bopassa JC, Meredith AL, Stefani E, Toro L. mitoBKCa is encoded by the Kcnma1 gene, and a splicing sequence defines its mitochondrial location. Proc Natl Acad Sci U S A. 2013;110:10836–10841. doi: 10.1073/pnas.1302028110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Lu R, Rodriguez PF, Wu Y, Bopassa JC, Stefani E, Toro L. Visualization and quantification of cardiac mitochondrial protein clusters with STED microscopy. Mitochondrion. 2012;12:230–236. doi: 10.1016/j.mito.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Warburton S, Vondriska TM, Khakh BS. Proteomics to identify proteins interacting with P2X2 ligand-gated cation channels. J.Vis.Exp. 2009 doi: 10.3791/1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski B, Orchard S, Harvey M, Sridhar S, Sakai Y. Conserved BK channel-protein interactions reveal signals relevant to cell death and survival. PLoS.One. 2011;6:e28532. doi: 10.1371/journal.pone.0028532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysinska E, Bentzen BH, Barthmes M, Hattel H, Thrush AB, Harper ME, Qvortrup K, Larsen FJ, Schiffer TA, Losa-Reyna J, Straubinger J, Kniess A, Thomsen MB, Bruggemann A, Fenske S, Biel M, Ruth P, Wahl-Schott C, Boushel RC, Olesen SP, Lukowski R. KCNMA1 encoded cardiac BK channels afford protection against ischemia reperfusion injury. PLoS.One. 2014;9:e103402. doi: 10.1371/journal.pone.0103402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro L, Li M, Zhang Z, Singh H, Wu Y, Stefani E. MaxiK channel and cell signalling. Pflugers Arch. 2013 doi: 10.1007/s00424-013-1359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner M, Meera P, Ottolia M, Kaczorowski GJ, Latorre R, Garcia ML, Stefani E, Toro L. Characterization of and modulation by a beta-subunit of a human maxi KCa channel cloned from myometrium. Receptors.Channels. 1995;3:185–199. [PubMed] [Google Scholar]
- Wenz LS, Opalinski L, Wiedemann N, Becker T. Cooperation of protein machineries in mitochondrial protein sorting. Biochim.Biophys.Acta. 2015;1853:1119–1129. doi: 10.1016/j.bbamcr.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Xu W, Liu Y, Wang S, McDonald T, Van Eyk JE, Sidor A, O'Rourke B. Cytoprotective role of Ca2+- activated K+ channels in the cardiac inner mitochondrial membrane. Science. 2002;298:1029–1033. doi: 10.1126/science.1074360. [DOI] [PubMed] [Google Scholar]