Abstract
The biochemical networks found in living organisms include a huge variety of control mechanisms at multiple levels of organization. While the mechanistic and molecular details of many of these control mechanisms are understood, their exact role in driving cellular behaviour is not. For example, yeast glycolysis has been studied for almost 80 years but it is only recently that we have come to understand the systemic role of the multitude of feedback and feed-forward controls that exist in this pathway. In this article, control theory is discussed as an approach to dissect the control logic of complex pathways. One of the key issues is distinguishing between the terms control and regulation and how these concepts are applied to regulated enzymes such as phosphofructokinase. In doing so, one of the paradoxes in metabolic regulation can be resolved where enzymes such as phosphofructokinase have little control but, nevertheless, possess significant regulatory influence.
Keywords: control, negative feedback, metabolism
1. Introduction
‘It is apparent that feedback systems theory is becoming of increasing significance to most life scientists, … ’ This was written in 1973 by Richard Jones in his unique book Principles of biological regulation [1]. Similar sentiments can be found in other texts of the period. In reality, feedback systems theory was of little significance to most life scientists at that time, particularly in the cell and molecular communities. So it was for the next 40 years. The reason for the disinterest is difficult to understand but it is likely to be a blend of factors including the lack of training, cultural resistance to change and the perceived difficulty in making quantitative measurements. More recently, there has been a significant groundswell in interest in applying more quantitative approaches to problems in cell biology, particularly with the influx of expertise from quantitative disciplines such as computer science, electrical and control engineering. For example, yeast glycolysis was one of the first pathways to be studied but it is only in recent years that we have come to understand the systemic role of the multitude of feedback and feed-forward control that exist in this pathway [2–5].
This article covers some aspects of control of cellular pathways with particular emphasis on negative feedback and the phosphofructokinase paradox.1
2. Historical context
Historically, we can trace interest in regulated systems back to Ctesibius (250 BC), a Greek scientist thought to have explored the use of a simple float system to maintain a constant supply of water to a water clock. This is the first recorded instance of the use of negative feedback in a man-made device. Some of this early work was taken up during the golden age of Medieval Islam between the ninth and twelfth centuries where a great variety of sophisticated clocks and other automata were invented [6]. However, it was not until the seventeenth century when industrialization began to put pressure on the need for regulated devices that progress in the field quickened. A series of smaller developments culminated in the invention by James Watt in 1788 AD of the governor. Watt repurposed the way fantails orientated windmills into the wind to regulate the speed of the newly developed steam engine. It was this event that ushered in a sustained interest and a clearer understanding of systems that could self regulate. In the 1930s, Harold Black, while travelling to work on a ferry, realized how he could use negative feedback to build distortion free amplifiers for the burgeoning telephone industry. This stimulated further theoretical analysis and the rise of modern control theory. These and other historical insights can be found in the review by Bennett [7] or the book by Mayr [8].
In 1940, Dische [9,10] discovered the inhibition of hexokinase-mediated glucose phosphorylation in red blood cell haemolysates by phosphoglyceric acid. However, this discovery went largely unnoted until Umbarger [11] and Yates & Pardee [12] published their discovery in 1956 of feedback inhibition in the isoleucine biosynthesis pathway and the inhibition of aspartate transcarbamylase in Escherichia coli. This ultimately led to the celebrated work on allosteric enzymes by Monod et al. in 1965 [13]. It is from this background that various strands of control theory applied to cellular pathways emerged.
Joseph Higgins in 1959 completed his PhD thesis on ‘A theoretical study of the kinetic properties of sequential enzyme reactions’ [14], where he introduced the notion of the reflection coefficient (later to be called the control coefficient [15]) [16,17]. This measured how much influence a given input had on a variable in a reaction pathway. This work influenced later developments particularly the work by Heinrich [18] in Berlin and Kacser [19] in Edinburgh. Each group derived additional inspiration from other disciplines. In the case of Heinrich, it was dynamical systems theory [20–23] and for Kacser it was the role of genetic mutations in influencing the phenotype and later on the work by Sewall Wright [24] on the proposed physiological explanation for genetic dominance. This meant that each group had a particular flavour to their approach but mathematically they were identical. A third, and important strand culminated in the work by Savageau at Michigan, who developed biochemical systems theory [25–28]. This had a more direct route from classical control theory. All three are extremely noteworthy and had a profound influence on many research groups both experimental and theoretical, especially in Europe [29–36]. More recently, researchers such as Ingalls [37] and Rao [38] have reconciled these approaches with classical control theory and it is now recognized that metabolic control analysis, biochemical systems analysis and classical control theory are one and the same thing but with different emphases.
Much of this work has taken a great deal of time to enter mainstream cell and molecular biology and only in recent years have undergraduate textbooks begun to discuss some of the main results [39] that have emerged from this work. Still, much remains to be done to infuse even the most basic aspects of control theory into both our undergraduate programmes and even our seasoned researchers. It is surprising that in metabolic engineering circles, particularly in the USA, intuition is still an important tool to direct the engineering of metabolic systems.
2.1. Side topic: elasticities
Elasticities (also called kinetic orders in biochemical systems theory) describe how sensitive a reaction rate is to changes in reactant, product and effector concentrations. They are defined by
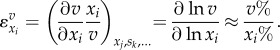 |
2.1 |
They represent the degree to which changes are transmitted by factors that directly affect the reaction rate.
All signals are transmitted via elasticities.
Elasticities are central to understanding control and regulation in a biochemical network.
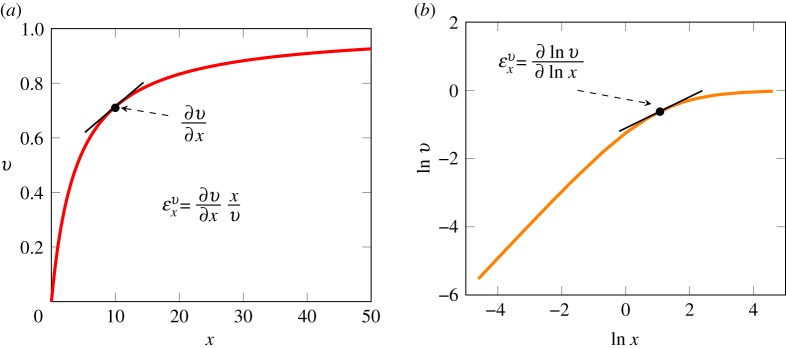
Figure 1.
(a) The slope of the reaction rate, v, versus the reactant concentration, x, scaled by both the reactant concentration and reaction rate yields the elasticity,  . (b) If the log of the reaction rate and log of the reactant concentration are plotted, the elasticity can be read directly from the slope of the curve. Curves were generated by assuming
. (b) If the log of the reaction rate and log of the reactant concentration are plotted, the elasticity can be read directly from the slope of the curve. Curves were generated by assuming 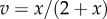 . Note that we use the partial derivative notation for the slope, because the reaction rate could potentially be changed by other factors such as product which we keep constant when measuring the gradient of the response to x. (Online version in colour.)
. Note that we use the partial derivative notation for the slope, because the reaction rate could potentially be changed by other factors such as product which we keep constant when measuring the gradient of the response to x. (Online version in colour.)
For a kinetic rate law such as v = kx, where v is the reaction rate, k is the rate constant and x the concentration of reactant, the elasticity, 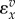 can be derived by taking the derivative with respect to x and scaling the result:
can be derived by taking the derivative with respect to x and scaling the result:
For a generalized irreversible mass-action law such as
the elasticity for species xi is ni. For a simple reversible mass-action reaction rate law such as
| 2.2 |
where we have used s to represent the concentration of reactant and p the product, the elasticities for the substrate and product are given by
| 2.3 |
and
| 2.4 |
where vf is the forward rate, vr is the reverse rate and v is the net rate. Note that 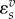 is positive and
is positive and 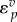 negative. In general, the elasticity for an effector that results in an increase in reaction rate will be positive and negative if the effector results in a decrease in the reaction rate.
negative. In general, the elasticity for an effector that results in an increase in reaction rate will be positive and negative if the effector results in a decrease in the reaction rate.
If we divide equations (2.3) and (2.4) top and bottom by 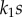 , and noting that the equilibrium constant Keq is given by the ratio
, and noting that the equilibrium constant Keq is given by the ratio 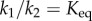 , the mass–action ratio by
, the mass–action ratio by 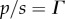 and
and 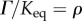 , we can express the elasticities in the form
, we can express the elasticities in the form
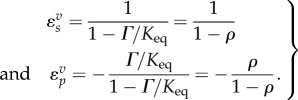 |
2.5 |
These expressions can vary over a wide range of values. Far from equilibrium ( )
) 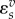 will lie close to 1.0, whereas
will lie close to 1.0, whereas 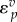 will be close to −0.0. When operating close to equilibrium however (
will be close to −0.0. When operating close to equilibrium however (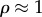 ), the same elasticities will tend to
), the same elasticities will tend to 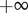 and
and 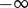 , respectively.
, respectively.
For a simple enzyme mechanism governed by the Briggs–Haldane relationship, 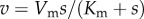 , where Vm is the maximal velocity, Km is the substrate concentration at half-maximal rate and s is the substrate concentration, the elasticity with respect to substrate it given by
, where Vm is the maximal velocity, Km is the substrate concentration at half-maximal rate and s is the substrate concentration, the elasticity with respect to substrate it given by 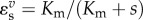 . The substrate elasticity shows a range of values from zero at high substrate concentrations to one at low substrate concentrations. When the enzyme is near saturation, it is naturally unresponsive to further changes in substrate concentration, hence the elasticity is near zero. A summary of behaviours and the corresponding value for the elasticity is shown in table 1.
. The substrate elasticity shows a range of values from zero at high substrate concentrations to one at low substrate concentrations. When the enzyme is near saturation, it is naturally unresponsive to further changes in substrate concentration, hence the elasticity is near zero. A summary of behaviours and the corresponding value for the elasticity is shown in table 1.
Table 1.
Summary of elasticity ranges corresponding to different reaction orders and behaviour.
| situation | elasticity |
|---|---|
| first order | 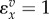 |
| saturated enzyme | 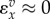 |
| close to equilibrium | 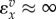 |
| cooperative enzyme | 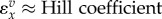 |
The fourth row of table 1 suggests that the elasticity for an enzyme that shows cooperativity with respect to one of its reactants will roughly be of the order of the Hill coefficient of the reactant. This result can be easily show for a reaction that obeys the Hill equation:
| 2.6 |
where h is called the Hill coefficient, x is the substrate concentration, Vm the maximal rate and Kd the dissociation constant. If we differentiate this equation with respect to the reactant concentration x and apply the appropriate scaling, we obtain the following elasticity coefficient (see electronic supplementary material for derivation [40]):
| 2.7 |
The maximum value the elasticity can reach is h when 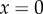 . Another way to look at this is to examine the relationship between the elasticity and the degree of saturation by reactant. Note that for many enzymes that show cooperativity there will be multiple binding sites for reactant molecules. The degree of saturation is therefore the average number of binding site occupied by reactant. Expression (2.8) is derived in the electronic supplementary material:
. Another way to look at this is to examine the relationship between the elasticity and the degree of saturation by reactant. Note that for many enzymes that show cooperativity there will be multiple binding sites for reactant molecules. The degree of saturation is therefore the average number of binding site occupied by reactant. Expression (2.8) is derived in the electronic supplementary material:
| 2.8 |
where Y is the fraction of bindings sites occupied by reactant. The less saturated the enzyme (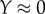 ), the closer the elasticity is to the Hill coefficient. Moreover, at half saturation (
), the closer the elasticity is to the Hill coefficient. Moreover, at half saturation (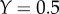 ), the elasticity is given by
), the elasticity is given by 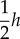 . For other mechanisms that can generate cooperativity, for example, the Adair model [41], the Monod et al. model [13] or the reversible Hill equation [42], the overall elasticity response will be different but in each case the elasticity will approach a maximum of h where h is the equivalent Hill coefficient or number of binding sites [40].
. For other mechanisms that can generate cooperativity, for example, the Adair model [41], the Monod et al. model [13] or the reversible Hill equation [42], the overall elasticity response will be different but in each case the elasticity will approach a maximum of h where h is the equivalent Hill coefficient or number of binding sites [40].
3. Control and regulation
Two words that are used frequently in the biological literature are ‘control’ and ‘regulation’ [43]. One of the first things to clarify is the meaning these words will have in this article (figure 2). In the vernacular, the word control usually means the ability to influence, command or to restrain a situation or process; therefore, we define control as the ability to direct or command behaviour. Given this definition, quantifying the degree of control is straightforward and can be achieved by measuring the effectiveness a given input has on a particular output of the system.
Figure 2.
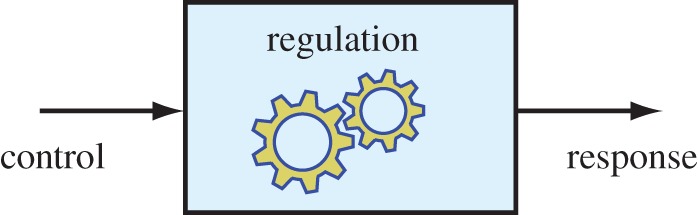
Control and regulation can be differentiated by their mode of action on and within a system. Control can be considered the ability of a system to respond to perturbations from outside the system boundary, whereas regulation is the mechanism by which that response is achieved. (Online version in colour.)
Regulation is more difficult to pin down but can be thought of as the mechanism or process that allows control to be achieved. We therefore define regulation as the mechanism that responds to unforseen disturbances or control actions that result in homeostasis or specific state changes to a system. Quantifying regulation is however difficult and various attempts have been made to do so with varying degrees of effectiveness [44–49].
To put these definitions in context, consider a room temperature thermostat. Control is what allows a user to set the temperature to a specific level. The sensors, feedback and actuators that make up the regulatory system ensure that the temperature setting is achieved.
3.1. Quantifying control
In the control theory of cellular reaction pathways, control is quantified by measuring the influence a parameter has on a system variable. Common parameters include enzyme concentrations, pathway sources and sinks, and various interventions such as drugs, nutrients such as glucose in glycolysis or EGF for the MAPK pathway. Common variables include the fluxes through metabolic pathways and molecular concentrations in metabolic, signalling and gene regulatory pathways. Given that cellular systems are highly nonlinear, we are forced mathematically to consider the effect of small changes to parameters on variables. Of course it is experimentally possible to make large changes, but the theory to help guide researches in these cases is not yet well developed [50–54].
We describe the control an enzyme has over a steady state flux, J or concentration, x, by using the control coefficients [18, 55–57] where 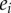 is the concentration of enzyme. These are defined as follows:
is the concentration of enzyme. These are defined as follows:
There are some subtleties with respect to the definition of these coefficients, and we refer readers to the text by Schuster & Heinrich [58] for more details. The coefficients are dimensionless values that are roughly equal to the ratio of percentage changes in the variable and parameter. In metabolic research, these are frequently measured as a means to gauge the influence particular steps have on fluxes and metabolite concentrations.
More important is the relationship between the control coefficients and elasticities. Because the elasticities describe the behaviour of individual reaction steps, describing control coefficients in terms of elasticities allows us to understand how particular steps have more control than others in terms of enzyme kinetic properties. This can give considerable insights into the workings of a pathway. We see examples of this in the remainder of the article.
4. Unregulated pathways
Let us first consider the properties of an unregulated pathway, this will allow us to contrast its behaviour to one that includes a negative feedback loop. Consider the pathway shown in figure 3 that includes four reaction steps and three metabolite species, 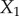 –
–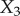 .
.
Figure 3.
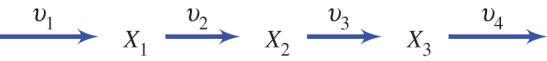
Simple four-step unbranched pathway. (Online version in colour.)
In terms of flux control, the pathway shows some interesting behaviour. The connectivity theorem [18,55] allows us to express the ratio of adjacent flux control coefficients in terms of corresponding adjacent elasticity coefficients. For a species, X, flanked by two steps, i and 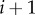 , the flux connectivity theorem can be written as
, the flux connectivity theorem can be written as
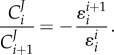 |
By taking the ratio of all elasticities along a unbranched unregulated pathway, it is possible to show that the following statement is true:
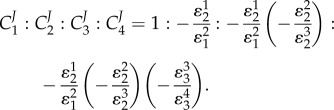 |
Note that an elasticity term such as 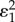 refers to the elasticity of reaction two with respect to species one. For a pathway of arbitrary length, the nth term, corresponding to
refers to the elasticity of reaction two with respect to species one. For a pathway of arbitrary length, the nth term, corresponding to 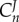 , will equal
, will equal
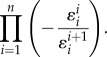 |
If we assume that the enzymes are operating below saturation, so that their behaviour can be approximated by the rate law, 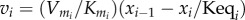 , then we can replace the substrate elasticities by
, then we can replace the substrate elasticities by 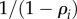 and the product elasticities by
and the product elasticities by 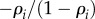 (see (2.5)). Using these substitutions, the ratios of flux control coefficients become [55]
(see (2.5)). Using these substitutions, the ratios of flux control coefficients become [55]
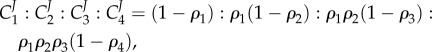 |
4.1 |
or for an arbitrary length pathway, the nth term, where 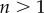 , is equal to
, is equal to
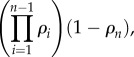 |
4.2 |
and 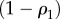 when
when 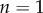 . This is an important result, because by just knowing the equilibrium constants and the concentrations of the intermediate pools, it is possible to obtain an idea of the relative strengths of the flux control coefficients across the pathway. Better still, if we invoke the flux summation theorem [57], we can obtain the absolute values of the flux control coefficients in terms of the mass-action ratio terms, ρ (equations (4.3)). In equation (4.3), k refers to the kth step and n the number of steps in the pathway:
. This is an important result, because by just knowing the equilibrium constants and the concentrations of the intermediate pools, it is possible to obtain an idea of the relative strengths of the flux control coefficients across the pathway. Better still, if we invoke the flux summation theorem [57], we can obtain the absolute values of the flux control coefficients in terms of the mass-action ratio terms, ρ (equations (4.3)). In equation (4.3), k refers to the kth step and n the number of steps in the pathway:
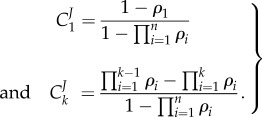 |
4.3 |
This is potentially a very useful expression for metabolic engineers. Given an unregulated segment of metabolism, and knowing the equilibrium constants for each reaction together with the measured metabolites, it is possible to get an approximate value for the control at each reaction step. Those steps with the largest control are targets for engineering. Note that once engineered control will shift to other steps and by remeasuring the new metabolite levels new targets can be identified.
4.1. Front-loading
Equation (4.3) also suggests a pattern in the distribution of flux control. Consider the flux control in the first and last step of a four step pathway:
The disequilibrium ratio, ρ, can be shown [40] to equal the ratio of the forward and reverse rates of a reaction, 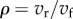 . Because the forward rate will always be greater than the reverse rate for a pathway showing a positive net rate, the disequilibrium ratio will always be less than 1,
. Because the forward rate will always be greater than the reverse rate for a pathway showing a positive net rate, the disequilibrium ratio will always be less than 1, 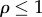 . This means that given two product expressions such as
. This means that given two product expressions such as 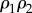 and
and 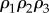 , the second term will be smaller than the first. By comparing
, the second term will be smaller than the first. By comparing 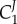 and
and 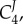 we can show that
we can show that 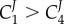 . In general, given the assumptions that have been made, namely enzymes working below saturation, we can state that
. In general, given the assumptions that have been made, namely enzymes working below saturation, we can state that
This means that in a unbranched pathway control will tend to be concentrated upstream, a behaviour that can be called front-loading. To understand why this should be the case, we must consider how the control coefficients are expressed in terms of elasticities.
For a pathway with n steps where n is even (if n is odd there are sign changes but the general form stays the same, see electronic supplementary material), the flux control coefficient equations are given by the following general equations [59]:
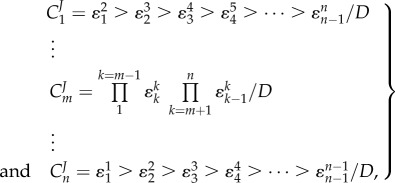 |
4.4 |
where D is the common denominator in the expressions (see electronic supplementary material for more details). If we look carefully at 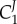 , then we see that the numerator is the product of all the reactant elasticities. This implies that a perturbation in
, then we see that the numerator is the product of all the reactant elasticities. This implies that a perturbation in 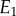 ‘hops’ from one enzyme to the next until it reaches the end of the pathway. Conversely, the control coefficient of the last enzyme,
‘hops’ from one enzyme to the next until it reaches the end of the pathway. Conversely, the control coefficient of the last enzyme, 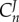 , includes all the product elasticities, that is the perturbation ‘hops’ from one enzyme to the next until it reaches the beginning of the pathway. Hofmeyr called this a ‘chain of local effects’ [60].
, includes all the product elasticities, that is the perturbation ‘hops’ from one enzyme to the next until it reaches the beginning of the pathway. Hofmeyr called this a ‘chain of local effects’ [60].
If we look at any intermediate enzyme step we find two groups of elasticities, one group representing the perturbation travelling downstream via the substrate elasticities and the other representing the perturbation travelling upstream via product elasticities. The numerators in the flux control equations therefore indicate the routes taken by a disturbance [60,61].
Given a reversible mass-action rate law, such as  , the elasticities are given by
, the elasticities are given by
 |
From these equations and assuming that 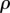 , we can state that
, we can state that
That is, the absolute value of the substrate elasticity is always greater than the product elasticity. Given that the control coefficient of an upstream enzyme will have more numerator substrate elasticities than product elasticities, it follows that the numerator will be larger when compared with an enzyme further downstream which will have more of the smaller value product elasticities. What this means is that perturbations at a downstream enzyme will be attenuated compared with a similar perturbation at an upstream step. Hence the control coefficients upstream will on average be larger.
The origins of the asymmetry between the substrate and product elasticities is a thermodynamic one. If the thermodynamic gradient were to be reversed, so that the pathway flux travelled ‘upstream’, the elasticity values exchange so that now the front loading occurs downstream, although ‘downstream’ is now ‘upstream’ because the flux has reversed.
4.2. Rate-limiting steps
How do the control coefficients relate to the concept of the rate-limiting step? Although the definition of a rate-limiting step varies in the literature, one common interpretation is that the rate-limiting—also called the rate-determining step—in a metabolic pathway is the slowest step and thereby determines the overall rate of the pathway [62]. The term ‘slowest’ is not always defined but in chemistry it is often the step with the smallest rate constant. In a metabolic pathway, one could imagine it to be the step with the smallest maximal velocity, Vm. This implies that increasing the amount of enzyme that catalyses the rate-limiting step will increase the overall rate through the pathway. This gives a direct connection between the flux control coefficient and the rate-limitingness of a step. A high flux control coefficient means that the step is rate-limiting.
4.2.1. Why is not there just one rate-limiting step?
The classic view of metabolic regulation is that there is one rate-limiting step in a pathway. This is sometimes even stated as a definition, because there can only be one slowest step in a pathway and hence one rate-limiting step. What is meant by the slowest step is not often specified but is usually considered the step with the smallest forward maximal rate (Vm). Other terms that are widely used in the literature to describe pathway control include pacemaker, choke point, rate-determining and probably the least meaningful, the key step.
The idea of a slowest step dictating the overall rate is appealing and there are many everyday precedents that support this view. Examples include traffic flow or customer lines at cashpoint tills in a supermarket. In both cases, the rate at which cars or customers are ‘processed’ is independent of the number of customers in the line or the length of the backup in the traffic. This behaviour means that both the road congestion and the limited number of cashiers makes these steps limit the overall rate of processing. In a biochemical pathway, the rate of a process is a function of the concentration of substrate and product. In the analogy with traffic congestion, it is as if the flow of cars is a function of how many cars are backed up. Because biochemical pathways do not work the same way as traffic flow, the analogy breaks down. The degree to which a particular reaction step limits flow is a function of many factors and not just the step in question.
Traffic congestion and the customer line are rate-limiting because the only way to increase the flow is to either widen the road or increase the number of cash tills. This means there is a single factor that determines the rate of flow. This is not the case in a biochemical pathway.
5. A simple feedback model
Now that we have looked at the distribution of flux control in a unbranched pathway we can now turn our attention to pathways that include negative feedback loops. Negative feedback is widespread in biochemical and physiological systems. On the face of it, negative feedback is a simple process that involves subtracting a portion of the output from the input (figure 4).
Figure 4.
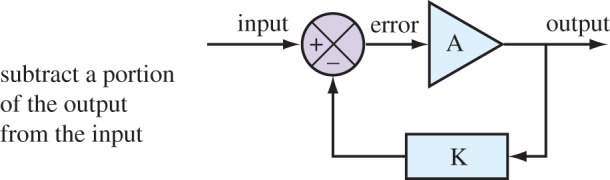
In the engineering community, negative feedback systems are often depicted in a way that attempts to separate the problem away from the specific implementation details. This diagram illustrates a typical example used by engineers and strips the problem down to its essentials, so that a common analysis can be applied no matter what the actual device implementation might be. Although perhaps not obvious at this stage, it is possible to map each of the indicated components in the figure to equivalent parts in a biological control system. This mapping is described in more detail in a later section. (Online version in colour.)
To understand the behaviour of a system with negative feedback, let us first consider a graphical method. Consider a very simple two step pathway with a negative feedback loop (figure 5). This is an idealized diagram that is not meant to imply a specific mechanism.
Figure 5.
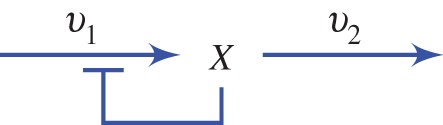
Simplest pathway with negative feedback. Species X inhibits the reaction rate  . (Online version in colour.)
. (Online version in colour.)
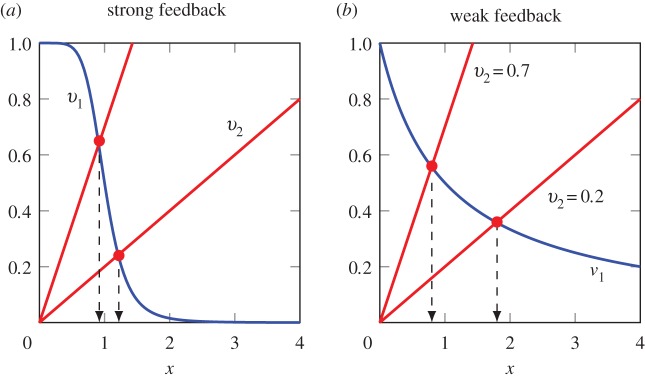
Figure 6 shows two plots, one with strong and the other with weak feedback. The plots show the reaction rates  and
and 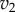 as a function of the intermediate species, X. We assume that the second step follows first-order kinetics, so that the
as a function of the intermediate species, X. We assume that the second step follows first-order kinetics, so that the 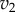 is a straight line. The feedback–response curve shows a decline from high to low as X increases. For strong feedback, the decline is steep (left plot). If we change the rate constant for the second step, this changes the slope of
is a straight line. The feedback–response curve shows a decline from high to low as X increases. For strong feedback, the decline is steep (left plot). If we change the rate constant for the second step, this changes the slope of 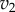 ; this is equivalent to applying a perturbation in the system. In the case of weak feedback, changes in
; this is equivalent to applying a perturbation in the system. In the case of weak feedback, changes in 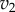 result in significant changes to X, this is because the feedback response is shallow. In contrast, when we have strong feedback (left panel), where the slope is very steep, any changes in
result in significant changes to X, this is because the feedback response is shallow. In contrast, when we have strong feedback (left panel), where the slope is very steep, any changes in 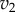 results in only small changes in X. The effect of negative feedback is to buffer the concentration of X in the face of changes to
results in only small changes in X. The effect of negative feedback is to buffer the concentration of X in the face of changes to 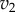 .
.
Figure 6.
Plot of 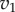 and
and 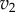 versus the concentration of X for a simple two step pathway with negative feedback. Two perturbations in
versus the concentration of X for a simple two step pathway with negative feedback. Two perturbations in 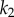 that determines
that determines 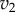 are shown. In panel (a), where the feedback is strong, changes in
are shown. In panel (a), where the feedback is strong, changes in 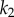 have hardly any effect on X. In the panel (b), the same change in
have hardly any effect on X. In the panel (b), the same change in 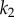 results in a much larger change in X. This illustrates the homeostatic effect of negative feedback. (a)
results in a much larger change in X. This illustrates the homeostatic effect of negative feedback. (a) 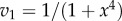 and (b)
and (b) 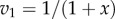 . (Online version in colour.)
. (Online version in colour.)
The graphical explanation is very instructive but we can get a better appreciation of how effective negative feedback is by taking an algebraic approach via a thought experiment. Consider that reaction rates 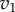 and v2 are determined by two enzymes E1 and
and v2 are determined by two enzymes E1 and 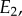 respectively. Changing either enzyme will result in a change to the steady-state level of X and the steady-state reaction rates v. Consider a small change in E1 of magnitude
respectively. Changing either enzyme will result in a change to the steady-state level of X and the steady-state reaction rates v. Consider a small change in E1 of magnitude 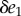 . This will have a number of effects, it will increase v1 which in turn will increase X which in turn will increase v2. We can represent this using a perturbation path:
. This will have a number of effects, it will increase v1 which in turn will increase X which in turn will increase v2. We can represent this using a perturbation path: 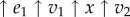 . Eventually, the system will settle to a new steady state. We can describe these changes by focusing on the change in v1 and v2. The change in v2, which we designate
. Eventually, the system will settle to a new steady state. We can describe these changes by focusing on the change in v1 and v2. The change in v2, which we designate 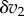 , came about as a result of the change
, came about as a result of the change 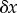 . Because we are only considering small changes, we can express the change
. Because we are only considering small changes, we can express the change 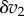 in terms of
in terms of 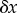 using the relation
using the relation
| 5.1 |
where the derivative 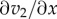 measures how responsive v2 is to changes in X. The relationship at this point is approximate, because we are considering finite though small changes to X. The derivative can be computed if we know the rate law for v2. For example, if we assume that the rate law is
measures how responsive v2 is to changes in X. The relationship at this point is approximate, because we are considering finite though small changes to X. The derivative can be computed if we know the rate law for v2. For example, if we assume that the rate law is 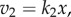 then the derivative is k2. We can also use a similar strategy to compute the change in v1 as a result of the change
then the derivative is k2. We can also use a similar strategy to compute the change in v1 as a result of the change 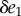 . This time the change in v1 is a result of two changes, the change in E1 itself and the change in X. We can express these changes by summing the two individual contributions:
. This time the change in v1 is a result of two changes, the change in E1 itself and the change in X. We can express these changes by summing the two individual contributions:
| 5.2 |
We have two equations, one describing the change in v1 (5.2) and the other in v2 (5.1). Because we allowed the system to settle to a new steady state, we can also state that the change in reaction rates must be the same (otherwise it would not be at steady state). That is we can assert that 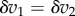 . With this in mind, we equate the two equations and write
. With this in mind, we equate the two equations and write
Solving for the ratio 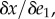 we obtain
we obtain
In the limit, as we make the change 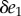 smaller and smaller, the left-hand side converges to the derivative
smaller and smaller, the left-hand side converges to the derivative 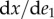 and we can replace the approximation by an exact equality:
and we can replace the approximation by an exact equality:
We can go one step further and scale the derivatives to eliminate units. Multiplying both sides by e1 and dividing both sides by x yields the scaled derivatives:
The scaled derivatives on the right-hand side are the elasticities, 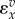 and the scaled left-hand term is the scaled sensitivity coefficient or concentration control coefficient,
and the scaled left-hand term is the scaled sensitivity coefficient or concentration control coefficient, 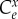 . Replacing the right-hand derivatives with the corresponding elasticities leads to
. Replacing the right-hand derivatives with the corresponding elasticities leads to
We can simplify this expression further. The reaction rate v1 is usually a linear function of e1. For example, in the Briggs–Haldane equation, the reaction rate is given by 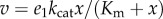 . Differentiating this rate law with respect to e1 and scaling yields:
. Differentiating this rate law with respect to e1 and scaling yields: 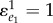 . Using this result gives
. Using this result gives
| 5.3 |
A similar analysis can be done where e2 is perturbed (see electronic supplementary material). In this case, we obtain the sensitivity of X with respect to e2:
| 5.4 |
Expressions (5.3) and (5.4) measure how much enzymes E1 and E2 control the steady state concentration of intermediate X. We can also consider how the steady state reaction rates v1 and v2 are affected by perturbations in E1 and E2. This is often of importance to metabolic engineers who are interested in increasing rates of production. At steady state, the reaction rates are often called the fluxes and abbreviated to J1 and J2. For a unbranched pathway such as this example, both fluxes are equal at steady state, so that the flux through the pathway is simply referred to as J. Expressing the change in flux as a result of a perturbations to the enzymes levels can be derived using a similar approach (see electronic supplementary material) to yield two flux control coefficient equations for E1 and E2:
| 5.5 |
Expressions (5.5) tell us how much enzymes E1 and E2 control the steady state flux. The key point here is that changes in enzyme concentration, or equivalently enzyme activity, must be brought about by an external action.
The approach taken can be applied to pathways of any length and complexity but becomes more tedious for larger systems. In addition to indicating how much control a particular reaction step has over concentrations and fluxes, the expressions tell us one more thing. The right-hand sides are in terms of elasticities and indicate how the control is brought about. In equations (5.5), 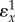 represents the strength of the feedback. The value will be negative, because we are dealing with negative feedback, that is increases in X will decrease
v1. We can investigate how the magnitude of the feedback affects the ability of e1 and e2 to control the concentration and flux through the pathway.
represents the strength of the feedback. The value will be negative, because we are dealing with negative feedback, that is increases in X will decrease
v1. We can investigate how the magnitude of the feedback affects the ability of e1 and e2 to control the concentration and flux through the pathway.
5.1. Engineering control theory
There is an enormous body of knowledge related to control theory [63–65] that arose as a result of the need to include regulating devices in areas ranging from steam engines, space flight and the Internet. In the last section, a simple analysis was made of a feedback system using approaches developed from metabolic control analysis (MCA) and biochemical systems theory (BST). How does this analysis and the broader literature on MCA and BST relate to the existing body of engineering control theory? This has been covered in detail by Ingalls [37] and Rao et al. [38]; here we give a flavour of the connection particularly in relation to negative feedback. Not all effects of negative feedback will be discussed here and the reader is referred to some recent articles for additional details [66,67].
The classic diagram often used to depict negative feedback in control theory is that shown in figure 4. In this diagram, K is the fraction of output that is fed back to the summing junction. The summing junction computes the difference between the input (also called the set-point depending on the application), and the signal fed back from the output. The output is a function of the error, often a simple proportional relationship (hence called proportional control). To put it in more concrete terms, the A block might be a heater in a room and the output the room's temperature. The input is the desired temperature. If the room is hotter than the desired temperature, the error signal will be negative, so that the heater is turned down. The opposite happens if the room is cooler than the desired temperature.
We can derive the relationship between the input and output in figure 4 by noting the following relationships. The error signal can be written as the difference between the input and the return loop: 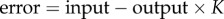 . The output can be written in terms of the gain A and the error:
. The output can be written in terms of the gain A and the error: 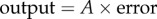 . We can combine both equations and solve for the output in terms of the input (see electronic supplementary material for details):
. We can combine both equations and solve for the output in terms of the input (see electronic supplementary material for details):
In control theory, this is called the closed-loop transfer function. In practice, it is often expressed in the frequency domain but this need not concern us here. What is more interesting is that the control coefficient equations (5.3)–(5.5) are related directly to the closed-loop transfer function [37]. The only difference is that whereas the full transfer function is defined over a range of input frequencies the control coefficients are defined only at a single frequency of zero, the so-called DC response. We therefore state the following equivalence [37,57]:
The input in the close-loop transfer function matches the enzyme elasticity, 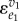 , but could be any input into v1 by substituting
, but could be any input into v1 by substituting 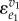 with the elasticity of the input of interest. The other terms need rearranging. If we divide top and bottom by
with the elasticity of the input of interest. The other terms need rearranging. If we divide top and bottom by 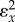 , we obtain
, we obtain
From this we can determine that
This generalizes to any length feedback loop. The feedback term, K will always be equal to the negative of the feedback elasticity. The expression for A, 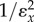 is the control coefficient
is the control coefficient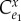 , when there is no feedback. It is the control coefficient for the unregulated system (set
, when there is no feedback. It is the control coefficient for the unregulated system (set 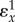 to zero in
to zero in 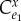 ) and is the gain of the system without feedback assuming everything else being equal. Figure 7 overlays the generalized feedback diagram with the equivalent elasticity terms. The error computed from the set point and feedback is the difference between the two elasticities
) and is the gain of the system without feedback assuming everything else being equal. Figure 7 overlays the generalized feedback diagram with the equivalent elasticity terms. The error computed from the set point and feedback is the difference between the two elasticities 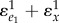 (recall that
(recall that 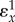 is negative).
is negative).
Figure 7.
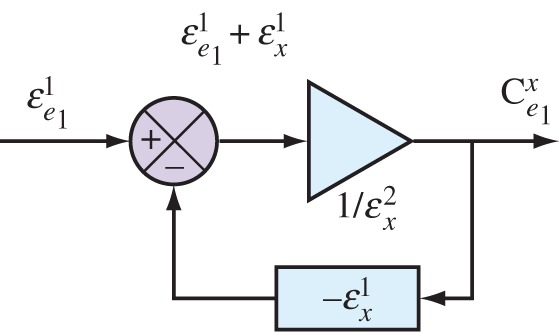
Mapping the standard engineering feedback control diagram to equivalent biochemical components. (Online version in colour.)
The product 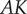 has a special meaning in control theory and is called the loop gain. It is one of the factors that determines the stability of a system with negative feedback and general performance of the system. In the reaction pathway,
has a special meaning in control theory and is called the loop gain. It is one of the factors that determines the stability of a system with negative feedback and general performance of the system. In the reaction pathway, 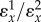 is the loop gain. For the unregulated pathway,
is the loop gain. For the unregulated pathway, 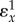 will be absent and under these conditions the control coefficient equals
will be absent and under these conditions the control coefficient equals 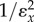 . We can therefore rewrite the loop gain as
. We can therefore rewrite the loop gain as
where 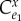 is computed for the unregulated pathway. The minus signs comes from the observation that K is equal to the negative of the feedback elasticity. This generalizes to any length pathway with feedback. For example, for the four step pathway where X3 is the return signal (figure 9), the loop gain equals
is computed for the unregulated pathway. The minus signs comes from the observation that K is equal to the negative of the feedback elasticity. This generalizes to any length pathway with feedback. For example, for the four step pathway where X3 is the return signal (figure 9), the loop gain equals
| 5.6 |
where 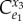 is the control coefficient for the unregulated but equivalent system and
is the control coefficient for the unregulated but equivalent system and 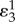 is the elasticity of the feedback loop. The control coefficient for the unregulated equivalent system can be easily determined by simply setting the negative feedback elasticity to zero in the control coefficient for the regulated pathway.
is the elasticity of the feedback loop. The control coefficient for the unregulated equivalent system can be easily determined by simply setting the negative feedback elasticity to zero in the control coefficient for the regulated pathway.
Figure 9.
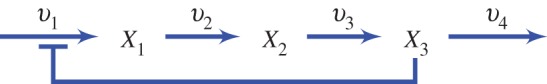
Four-step unbranched pathway with negative feedback. (Online version in colour.)
5.2. Control and negative feedback
Let us now consider what factors influence the degree of control that enzymes have in a pathway with negative feedback. Using the simple negative feedback pathway in figure 5, we can construct a table (table 2) that shows the different combinations for the elasticities and the corresponding values for the control coefficients. Of particular interest are the first two rows.
Table 2.
Behaviour of the simple feedback pathway under different regimes.
 (feedback) (feedback) |
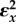 |
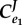 |
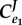 |
|---|---|---|---|
strong, 
|
low, 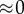
|
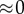 |
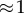 |
weak, 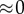
|
high, 
|
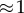 |
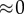 |
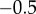 |
0.5 | 0.5 | 0.5 |
When the feedback is strong, 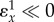 , most influence is found on the second step, the first step, v1 has virtually no influence. This is very typical of a pathway with negative feedback.
, most influence is found on the second step, the first step, v1 has virtually no influence. This is very typical of a pathway with negative feedback.
The switch over as the strength of the feedback increases can be seen in figure 10.
Figure 10.
Switch-over of flux control from the first step to the second step as a function of the feedback strength. (Online version in colour.)
The flux control summation theorem states that the sum of all the flux control coefficients in a pathway must sum to one [55,56]. This means that control is conserved so that if control disappears from one part of a pathway it must reappear elsewhere. This is a fundamental constraint on the control of flux in biochemical pathways. When there is a lack of control on the regulated step, v1, control will move to the second step, v2. This behaviour is the basis for the supply/demand metabolic architecture put forward by Hofmeyr and co-workers [68–70]. This control pattern ensures that the pathway flux is determined by demand (which has the higher flux control coefficient) rather than by supply (figure 8).
Figure 8.
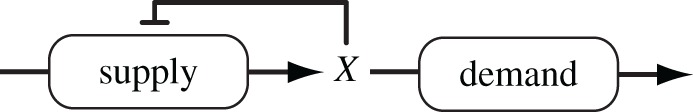
Supply/demand architecture based on negative feedback.
In this sense, E1, under negative feedback, has very little influence over either the flux or the signal molecule X. This implies that the regulated step will be a poor target for manipulating the pathway.
The analysis on the two-step pathway can be extended to larger pathways with negative feedback and the conclusions from the two step study apply equally. For example, consider the four step pathway shown in figure 9. There are standard methods for deriving the control expressions [34,71] which will not be described here, but tools such as Mathematica or open source specialized tools such as SymCA [72] can be used.
In total, there will be four flux control coefficients and 12 concentration control coefficients corresponding to the three species, X1, X2 and X3 and the four enzyme catalysed reactions, E1–E4. The denominator for each control equation is the same and is given by equation (5.7).
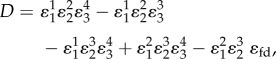 |
5.7 |
where 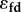 is the feedback elasticity.
is the feedback elasticity. 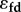 is equivalent to
is equivalent to 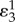 . The symbol
. The symbol 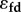 is used to help the reader identify the feedback elasticity more easily. Here we focus on four of the control coefficients shown in table 3.
is used to help the reader identify the feedback elasticity more easily. Here we focus on four of the control coefficients shown in table 3.
Table 3.
Control coefficients and corresponding numerators of control equations. The feedback elasticity is highlighted in underline/red, 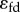 .
.
| control coefficient | numerator |
|---|---|
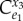 |
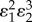 |
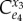 |
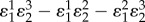 |
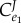 |
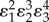 |
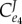 |
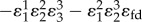 |
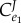 is reduced by the presence of the feedback term
is reduced by the presence of the feedback term 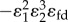 in the denominator. As with the simple two step pathway, increasing the strength of the negative feedback reduces the flux control coefficient for regulated step e1. At the same time, the influence of the last step,
in the denominator. As with the simple two step pathway, increasing the strength of the negative feedback reduces the flux control coefficient for regulated step e1. At the same time, the influence of the last step, 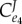 , tends to increase (to a maximum of one, figure 10).
, tends to increase (to a maximum of one, figure 10).
Likewise, the concentration control coefficient, 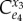 tends to zero as the feedback strength increases. The negative feedback locks X3 into a narrow range. These results illustrate again the partition of the pathway into a supply block between X1 and X3 and a demand block downstream from X3. In general, all steps within the signal loop will have little influence on either the flux or the level of X3 though the actual degree will be influenced by the elasticities inside the loop. From these studies, we conclude the following general statement:
tends to zero as the feedback strength increases. The negative feedback locks X3 into a narrow range. These results illustrate again the partition of the pathway into a supply block between X1 and X3 and a demand block downstream from X3. In general, all steps within the signal loop will have little influence on either the flux or the level of X3 though the actual degree will be influenced by the elasticities inside the loop. From these studies, we conclude the following general statement:
Regulated steps have low flux control coefficients and are therefore not rate-limiting.
What is troubling for many is that the regulated step, 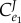 has little in the way of flux control, that is, it is not rate-limiting. This would seem to raise a paradox. On the one hand, the regulated step must be important, and yet this importance is not reflected in the degree of influence the step has on the flux. How do we resolve this?
has little in the way of flux control, that is, it is not rate-limiting. This would seem to raise a paradox. On the one hand, the regulated step must be important, and yet this importance is not reflected in the degree of influence the step has on the flux. How do we resolve this?
5.3. Regulation and negative feedback
Most textbooks and online sites such as Wikipedia refer to phosphofructokinase as the rate-limiting or pacemaker step of glycolysis. There are many reasons why this is considered so. Phosphofructokinase is one of the earliest steps of glycolysis, in vivo it is a non-equilibrium reaction and, most convincing of all, it is regulated by many effectors. What are the effectors there other than to control the glycolytic flux? Many allosteric enzymes such as phosphofructokinase are considered flux controllers by the same reasoning.
From the last section, however, we saw that regulated enzymes in unbranched pathways with end-product inhibition are in fact very poor flux controllers. When determined experimentally the flux control coefficient for phosphofructokinase is found to be invariable small [73–82] and therefore phosphofructokinase is not rate-limiting in many situations. This matches the theoretical expectation even though intuitively it seems suspect.
We therefore have a paradox (sometimes called ‘The PFK paradox’ [83]). Intuition suggests that phosphofructokinase should be controlling glycolytic flux particularly given the multitude of effectors that regulate it. On the other hand, experimental evidence and theory suggest the opposite. The question is how to reconcile these two opposite views?
5.3.1. The phosphofructokinase paradox
We first restate that phosphofructokinase in not rate-limiting when operating in situ. This has been shown experimentally many times [73–82] as well as being consistent with theory. This cannot be easily disputed. And yet the literature, textbooks and online resources still claim that phosphofructokinase is rate-limiting [84]. To reconcile this difference we must introduce a different measure that describes the strength of the regulated step and its ability to throttle flux.
Consider two configurations for the four-step pathway with negative feedback (figure 11).
Figure 11.
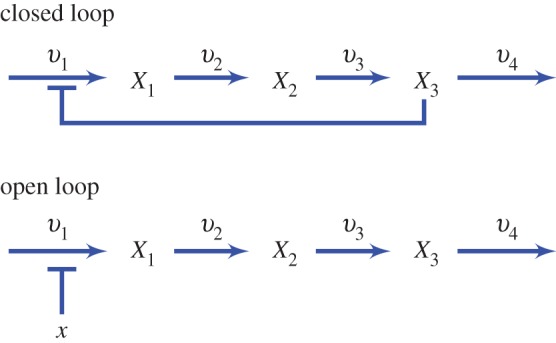
Four-step unbranched pathway shows closed and open loop configurations. (Online version in colour.)
The closed loop configuration has the negative feedback loop connected to v1. The open loop configuration is an equivalent system (or a controlled comparison [85]), but with the negative feedback loop disconnected. Note however that we still have a regulator on v1, except it is now an external regulator rather than an internal one.
For a controlled comparison [85], the two configuration in figure 11 will be equivalent. Given the steady-state concentrations and flux for the closed loop pathway, let us break the negative feedback loop. With the loop broken, the repression on the first step is released. This means that the steady state flux and concentrations will rise. We now make the system equivalent by reducing the concentration of the first enzyme until the steady-state concentrations are back to where they were before the break. This also means that the flux is restored. With the concentrations and flux the same in both configurations, we can safely assume that the elasticities will be the same in each configuration. The only difference is that the feedback elasticity is an external effector in the open loop configuration and means that the feedback elasticity will not appear in the control equations.
We can now ask the question, what are the relative values for the flux control coefficients in step one? We can derive the ratio of the flux control of the regulated step of the open configuration, 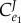 to the closed configuration,
to the closed configuration, 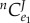 . This can be shown as
. This can be shown as
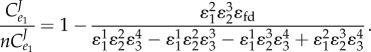 |
The numerator term, 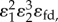 is negative and the denominator positive. The ratio is therefore negative. However, we are subtracting this negative value from 1. This means that overall the expression,
is negative and the denominator positive. The ratio is therefore negative. However, we are subtracting this negative value from 1. This means that overall the expression, 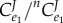 must be
must be  . In other words, the flux control exerted by the open configuration will be greater than the control exerted by the closed configuration. This shows that the regulated step has the ability to throttle the flux. In the closed configuration, this property is hidden and operates silently by actively adjusting the flux so that the end product, X3 is stabilized.
. In other words, the flux control exerted by the open configuration will be greater than the control exerted by the closed configuration. This shows that the regulated step has the ability to throttle the flux. In the closed configuration, this property is hidden and operates silently by actively adjusting the flux so that the end product, X3 is stabilized.
Is there someway to easily measure the regulatory strength of the regulated step and thereby recognize its importance? One clear possibility is to look at the loop gain [65]. As described before, the loop gain is the overall gain around the feedback loop. It was shown previously that the loop gain is the product of the forward gain, A, and the feedback gain, K, that is 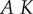 . In terms of elasticities, the loop gain is the product of the feedback elasticity,
. In terms of elasticities, the loop gain is the product of the feedback elasticity, 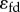 , and the concentration control coefficient,
, and the concentration control coefficient, 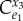 , for the unregulated pathway. In this case, this equals the product of the forward elasticities,
, for the unregulated pathway. In this case, this equals the product of the forward elasticities, 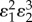 divided by the denominator, D, of the unregulated pathway, equation (5.6)
divided by the denominator, D, of the unregulated pathway, equation (5.6)
| 5.8 |
We can illustrate the use of this measure with an example. Using the four step pathway with negative feedback, the elasticities are set to the following values. All substrate elasticities (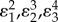 ) are set to 0.5. This corresponds to substrate levels set to the Km of each enzyme. The product inhibition elasticities (
) are set to 0.5. This corresponds to substrate levels set to the Km of each enzyme. The product inhibition elasticities (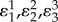 ) are assumed to be small but not negligible and are set to
) are assumed to be small but not negligible and are set to 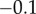 . Lastly, the feedback inhibition elasticity,
. Lastly, the feedback inhibition elasticity, 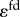 is set to
is set to 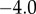 . Given these values, we can compute the flux control coefficient for the regulated step using the expressions in table 3:
. Given these values, we can compute the flux control coefficient for the regulated step using the expressions in table 3:
This tells us that from the perspective of the flux control coefficient, the reaction is not rate-limiting. However, we can compute the loop gain using equation (5.8), this yields
Note that that loop gain is significantly higher than the flux control coefficient. It means that changes to the regulator, X3, will have a significant effect on throttling the pathway. We can compare these calculations to the same pathway but where the negative feedback strength is weak. If we set 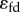 to a small value such as
to a small value such as 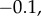 we obtain the following values for the flux control and loop gain:
we obtain the following values for the flux control and loop gain:
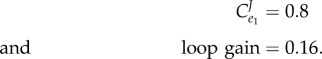 |
The flux control has increased about eightfold and is now rate-limiting with respect to an external perturbation. What is more interesting is that the loop gain has been reduced 40-fold, indicating little or no regulation from the feedback loop. We can access the degree to which a regulated enzyme is actually regulating by looking at the loop gain. The higher the loop gain the stronger the degree of regulation.
It is worth noting that there are two contributions to the loop gain (5.8), the action of the signal on the regulated step, 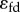 , and the transmission of that signal to cause a change (figure 12),
, and the transmission of that signal to cause a change (figure 12), 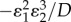 . The effectiveness of the overall regulation is therefore not just a function of the regulated step but of the entire loop. If the transmission elasticities, in this case
. The effectiveness of the overall regulation is therefore not just a function of the regulated step but of the entire loop. If the transmission elasticities, in this case 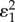 and
and 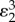 , are small then the loop gain could be significantly reduced. An examination of the elasticity of the regulated step is therefore insufficient to ascertain the effectiveness of the regulation, and it is quite possible that with weak signal transmission, even in the presence of a strongly regulated step, effective regulation could be minimal.
, are small then the loop gain could be significantly reduced. An examination of the elasticity of the regulated step is therefore insufficient to ascertain the effectiveness of the regulation, and it is quite possible that with weak signal transmission, even in the presence of a strongly regulated step, effective regulation could be minimal.
Figure 12.
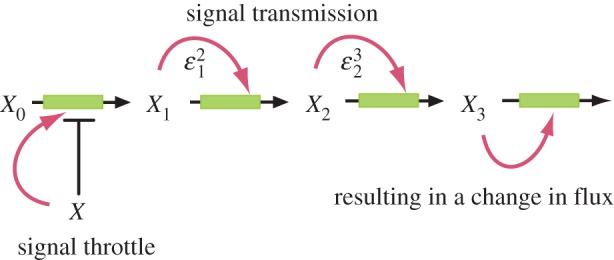
Transmission from signal to flux change. (Online version in colour.)
It would seem that we can resolve the phosphofructokinase paradox. Control is measured by the flux or concentration control coefficients and tells us how much influence an external agent has over the system. How that system reacts is governed by regulatory mechanisms, for example negative feedback where the regulation locks the end product and flux into a narrow range. We can measure the strength of the regulation by looking at the loop gain.
The regulation of glycolysis is more subtle than implied here. Although ATP is a negative regulator of phosphofructokinase, it is also a reactant. This means that increases in ATP levels can activate and inhibit at the same time which would appear to reduce the effectiveness of the ATP feedback. Instead evolution has furnished phosphofructokinase with a more powerful regulator in the form of AMP. There are two interesting aspects related to AMP regulation. The first is that AMP activates phosphofructokinase, the second is that via adenylate kinase, ATP, ADP and AMP are equilibrated via the reaction
The adenylate reaction means that increases in ATP result in decreases in AMP (and vice versa). Therefore, the activation of phosphofructokinase as a result of AMP makes sense in the light of adenylate kinase. When ATP increases, phosphofructokinase activity is reduced owing to a lowering of AMP. Equally interesting is the amplifying effect of adenylate kinase on the levels of AMP. We can see this by examining the sensitivities of AMP to changes in ATP [34]. At any given instant, the total sum of the nucleotide concentrations is fixed, because we can assume that the net synthesis and degradation of nucleotides is small. We also assume that the adenylate kinase reaction that equilibrates the nucleotide pools is fast, so that any changes in one or more of the nucleotide concentrations results in rapid equilibration with the others. Given these assumptions, we can write
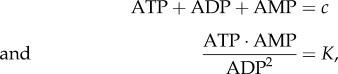 |
where K is the equilibrium constant for the adenylate kinase reaction. Let us now consider the sensitivity of changes in AMP to changes in ATP. Combining the equations and doing implicit differentiation, we obtain
The expression can be scaled by multiplying by ATP and dividing by AMP on both sides and eliminating K by substituting 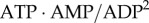 yields
yields
To illustrate the effect of changes in ATP on AMP concentration, we can use the measured concentrations obtained from the literature. For example, Chassagnole et al. [86] reported the following measured concentrations of ATP, ADP and AMP in E. coli K-12 grown under glucose limiting conditions in a stirred-tank bioreactor: ATP, 4.27; ADP, 0.595; and AMP, 0.955 mM. Given these values we can compute the sensitivity to be −3.65. That is a 1% increase in ATP results in a 3.65% decrease in AMP. This shows that AMP can be a more potent indicator of changes in ATP than ATP alone.
6. Discussion
It is a truism to say that biological cells are complicated [87,88] and yet our approach to dealing with such complex systems has historically been largely qualitative. Part of the problem has been obtaining enough data on cell behaviour to begin to formulate quantitative theories. In the last decade or so, the lack of data has largely disappeared and we have now entered the era of ‘big data’ [89,90] where the emphasis is on identifying patterns, trends and associations without reference to any underlying theory. However, data in the absence of theory are limited as was clearly pointed out by Pigliucci [91]. Control theory and its sister domain dynamical systems theory has much to contribute to providing a framework around which data and new hypotheses can be organized. The synthetic biology community serves as a good example of the interplay between theory, computation and experiment [92,93].
Metabolic engineering, in particular, could benefit considerably from testing and refining the current theories of metabolic regulation and control. As already implied, metabolic engineering is still on the whole a discipline that relies on anecdotal evidence to accomplish specific objectives. A renewed emphasis on how control theory can help engineers could deliver not only more success stories but also a better understanding of how metabolism operates.
Supplementary Material
Acknowledgments
I am indebted to the three reviewers who very carefully read and corrected the manuscript. They efforts significantly improved the final draft.
Endnote
In accordance with standard biochemical notation, an uppercase Roman letter such as X will be used to denote the name of a molecular species, whereas the lower case x will be used to denote the concentration of the species X. This means that the standard practice of using of square brackets to indicate concentration, as in [X], can be avoided to limit syntactic clutter.
Competing interests
I have no competing interests.
Funding
Research reported in this publication was partly supported by NIGMS of the National Institutes of Health under award number R01-GM081070 as well as the National Science Foundation under grants nos. MCB-1158573, MCB-1515280, EF-0827592 and DBI-1355909. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health or National Science Foundation.
References
- 1.Jones R. 2012. Principles of biological regulation: an introduction to feedback systems. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 2.van Heerden JH, Bruggeman FJ, Teusink B. 2015. Multi-tasking of biosynthetic and energetic functions of glycolysis explained by supply and demand logic. BioEssays 37, 34–45. ( 10.1002/bies.201400108) [DOI] [PubMed] [Google Scholar]
- 3.van Heerden JH. et al. 2014. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343, 1245114 ( 10.1126/science.1245114) [DOI] [PubMed] [Google Scholar]
- 4.van Eunen K, Kiewiet JA, Westerhoff HV, Bakker BM. 2012. Testing biochemistry revisited: how in vivo metabolism can be understood from in vitro enzyme kinetics. PLoS Comput. Biol. 8, e1002483 ( 10.1371/journal.pcbi.1002483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teusink B. et al. 2000. Can yeast glycolysis by understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267, 5313–5329. ( 10.1046/j.1432-1327.2000.01527.x) [DOI] [PubMed] [Google Scholar]
- 6.Al-Hassan AY, Hill DR. 1986. Islamic technology; an illustrated history. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 7.Bennett S. 1996. A brief history of automatic control. IEEE Control Syst. Mag. 16, 17–25. ( 10.1109/37.506394) [DOI] [Google Scholar]
- 8.Mayr O. 1975. The origins of feedback control. Cambridge, MA: The MIT Press. [Google Scholar]
- 9.Guest GM, Rapoport S. 1940. Role of diphosphoglycerate and other organic acid-soluble phosphorus compounds of the red blood cells in the electrolyte equilibrium of the blood. In American Association for the Advancement of Science, Symposium, Blood, Heart and Circulation, no. 13, pp. 55–60. Washington, DC: Science Press. [Google Scholar]
- 10.Dische Z. 1976. The discovery of feedback inhibition. Trends Biochem. Sci. 1, N269–N270. ( 10.1016/0968-0004(76)90333-9) [DOI] [Google Scholar]
- 11.Umbarger HE. 1956. Evidence for a negative-feedback mechanism in the biosynthesis of leucine. Science 123, 848 ( 10.1126/science.123.3202.848) [DOI] [PubMed] [Google Scholar]
- 12.Yates RA, Pardee AB. 1956. Control of pyrimidine biosynthesis in Escherichia coli by a feed-back mechanism. J. Biol. Chem. 221, 757–770. [PubMed] [Google Scholar]
- 13.Monod J, Wyman J, Changeux JP. 1965. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118. ( 10.1016/S0022-2836(65)80285-6) [DOI] [PubMed] [Google Scholar]
- 14.Higgins JJ. 1959. A theoretical study of the kinetic properties of sequencial enzyme reactions. Philadelphia, PA: University of Pennsylvania. [Google Scholar]
- 15.Burns JA. et al. 1985. Control analysis of metabolic systems. Trends Biochem. Sci. 10, 16 ( 10.1016/0968-0004(85)90008-8) [DOI] [PubMed] [Google Scholar]
- 16.Higgins J. 1963. Analysis of sequential reactions. Annu. NY Acad. Sci. 108, 305–321. ( 10.1111/j.1749-6632.1963.tb13382.x) [DOI] [PubMed] [Google Scholar]
- 17.Higgins J. 1967. The theory of oscillating reactions. Ind. Eng. Chem. 59, 18–62. ( 10.1021/ie50689a006) [DOI] [Google Scholar]
- 18.Heinrich R, Rapoport TA. 1974. A linear steady-state treatment of enzymatic chains; general properties, control and effector strength. Eur. J. Biochem. 42, 89–95. ( 10.1111/j.1432-1033.1974.tb03318.x) [DOI] [PubMed] [Google Scholar]
- 19.Kacser H, Burns JA. 1973. The control of flux. In Rate control of biological processes (ed. DD Davies). Symp. Soc. Exp. Biol., vol. 27, pp. 65–104. Cambridge University Press. [Google Scholar]
- 20.London WP. 1966. A theoretical study of hepatic glycogen metabolism. J. Biol. Chem. 241, 3008–3022. [PubMed] [Google Scholar]
- 21.Reich J. 1968. Analogue computer analysis of tracer flow patterns through the glycolytic and related pathway in erythrocytes and other intact metabolic systems. Eur. J. Biochem. 6, 395–403. ( 10.1111/j.1432-1033.1968.tb00460.x) [DOI] [PubMed] [Google Scholar]
- 22.Sel'kov EE. 1968. Self-oscillations in glycolysis. 1. A simple kinetic model. Eur. J. Biochem. 4, 79–86. ( 10.1111/j.1432-1033.1968.tb00175.x) [DOI] [PubMed] [Google Scholar]
- 23.Garfinkel D, Garfinkel L, Pring M, Green SB, Chance B. 1970. Computer applications to biochemical kinetics. Annu. Rev. Biochem. 39, 473–498. ( 10.1146/annurev.bi.39.070170.002353) [DOI] [PubMed] [Google Scholar]
- 24.Wright S. 1934. Physiological and evolutionary theories of dominance. Am. Nat. 68, 24–53. ( 10.1086/280521) [DOI] [Google Scholar]
- 25.Savageau MA. 1969. Biochemical systems analysis: II. The steady state solutions for an n-pool system using a power-law approximation. J. Theor. Biol. 25, 370–379. ( 10.1016/S0022-5193(69)80027-5) [DOI] [PubMed] [Google Scholar]
- 26.Savageau MA. 1971. Concepts relating the behaviour of biochemical systems to their underlying molecular properties. Arch. Biochem. Biophys. 145, 612–621. ( 10.1016/S0003-9861(71)80021-8) [DOI] [PubMed] [Google Scholar]
- 27.Savageau MA. 1971. Parameter sensitivity as a criterion for evaluating and comparing the performance of biochemical systems. Nature 229, 542–544. ( 10.1038/229542a0) [DOI] [PubMed] [Google Scholar]
- 28.Savageau MA. 1972. The behaviour of intact biochemical control systems. Curr. Top. Cell Reg. 6, 63–130. ( 10.1016/B978-0-12-152806-5.50010-2) [DOI] [Google Scholar]
- 29.Groen A, Van der Meer R, Westerhoff H, Wanders R, Akerboom T, Tager J. 1982. Control of metabolic fluxes. Metab. Compartment. 9–37. [Google Scholar]
- 30.Groen AK, Wanders RJA, Westerhoff HV, van der Meer R, Tager JM. 1982. Quantification of the contribution of various steps to the control of mitochondrial respiration. J. Biol. Chem. 257, 2754–2757. [PubMed] [Google Scholar]
- 31.Westerhoff HV, Groen AK, Wanders RJ. 1984. Modern theories of metabolic control and their applications. Biosci. Rep. 4, 1–22. ( 10.1007/BF01120819) [DOI] [PubMed] [Google Scholar]
- 32.Sorribas A, Bartrons R. 1986. Theoretical analysis of the flux control properties of a substrate cycle. Eur. J. Biochem. 158, 107–115. ( 10.1111/j.1432-1033.1986.tb09727.x) [DOI] [PubMed] [Google Scholar]
- 33.Torres N, Mateo F, Melendez-Hevia E, Kacser H. 1986. Kinetics of metabolic pathways. A system in vitro to study the control of flux. Biochem. J. 234, 169–174. ( 10.1042/bj2340169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fell DA, Sauro HM. 1985. Metabolic control analysis: additional relationships between elasticities and control coefficients. Eur. J. Biochem. 148, 555–561. ( 10.1111/j.1432-1033.1985.tb08876.x) [DOI] [PubMed] [Google Scholar]
- 35.Murphy MP, Brand MB. 1987. The control of electron flux through cytochrome oxidase. Biochem. J. 243, 499–505. ( 10.1042/bj2430499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steuer R, Junker BH. 2009. Computational models of metabolism: stability and regulation in metabolic networks. Adv. Chem. Phys. 142, 105 ( 10.1002/9780470475935.ch3) [DOI] [Google Scholar]
- 37.Ingalls BP. 2004. A frequency domain approach to sensitivity analysis of biochemical systems. J. Phys. Chem. B 108, 1143–1152. ( 10.1021/jp036567u) [DOI] [Google Scholar]
- 38.Rao CV, Sauro HM, Arkin AP. 2004. Putting the ‘control’ in metabolic control analysis. In 7th Int. Symp. Dynamics and Control of Process Systems, DYCOPS-7, Cambridge, MA, 5–7 July. [Google Scholar]
- 39.Nelson DL, Lehninger AL, Cox MM. 2008. Lehninger principles of biochemistry. New York, NY: Macmillan. [Google Scholar]
- 40.Sauro HM. 2012. Enzyme kinetics for systems biology, 2nd edn Seattle, WA: Ambrosius Publishing. [Google Scholar]
- 41.Adair GS, With the collaboration of Bock AV, Field H Jr. 1925. The hemoglobin system VI. The oxygen dissociation curve of hemoglobin. J. Biol. Chem. 63, 529–545. [Google Scholar]
- 42.Hofmeyr JH, Cornish-Bowden A. 1997. The reversible Hill equation: how to incorporate cooperative enzymes into metabolic models. Comput. Appl. Biosci. 13, 377–385. ( 10.1093/bioinformatics/13.4.377) [DOI] [PubMed] [Google Scholar]
- 43.Pichersky E. 2005. Is the concept of regulation overused in molecular and cellular biology? Plant Cell 17, 3217–3218. ( 10.1105/tpc.105.038968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauro HM. 1990. Regulatory responses and control analysis: assessment of the relative importance of internal effectors. In Control of Metabolic Processes (eds A Cornish-Bowden, ML Cárdenas). NATO ASI Series, pp. 225–230. New York, NY: Plenum Press. [Google Scholar]
- 45.Hofmeyr JH, Cornish-Bowden A. 1991. Quantitative assessment of regulation in metabolic systems. Eur. J. Biochem. 200, 223–236. ( 10.1111/j.1432-1033.1991.tb21071.x) [DOI] [PubMed] [Google Scholar]
- 46.Hofmeyr JH, Cornish-Bowden A, Rohwer JM. 1993. Taking enzyme kinetics out of control; putting control into regulation. Eur. J. Biochem. 212, 833–837. ( 10.1111/j.1432-1033.1993.tb17725.x) [DOI] [PubMed] [Google Scholar]
- 47.Kahn D, Westerhoff HV. 1993. The regulatory strength: how to be precise about regulation and homeostasis. Acta Biotheor. 41, 85–96. ( 10.1007/BF00712777) [DOI] [PubMed] [Google Scholar]
- 48.Hofmeyr JHS, Cornish-Bowden A. 1996. Co-response analysis: a new experimental strategy for metabolic control analysis. J. Theor. Biol. 182, 371–380. ( 10.1006/jtbi.1996.0176) [DOI] [PubMed] [Google Scholar]
- 49.Rohwer JM, Hofmeyr JHS. 2008. Identifying and characterising regulatory metabolites with generalised supply–demand analysis. J. Theor. Biol. 252, 546–554. ( 10.1016/j.jtbi.2007.10.032) [DOI] [PubMed] [Google Scholar]
- 50.Kacser H, Acerenza L. 1993. A universal method for achieving increases in metabolite production. Eur. J. Biochem. 216, 361–367. ( 10.1111/j.1432-1033.1993.tb18153.x) [DOI] [PubMed] [Google Scholar]
- 51.Small JR, Kacser H. 1993. Responses of metabolic systems to large changes in enzyme activities and effectors 2. The linear treatment of branched pathways and metabolite concentrations. Assessment of the general non-linear case. Eur. J. Biochem. 213, 625–640. ( 10.1111/j.1432-1033.1993.tb17802.x) [DOI] [PubMed] [Google Scholar]
- 52.Acerenza L. 2000. On the universality of the universal method. In Technological and medical implications of metabolic control analysis, pp. 33–37. Berlin, Germany: Springer. [Google Scholar]
- 53.Acerenza L. 2003. Metabolic responses: large and small. Comments Theor. Biol. 8, 279–320. ( 10.1080/08948550302447) [DOI] [Google Scholar]
- 54.Acerenza L, Ortega F. 2007. Modular metabolic control analysis of large responses. FEBS J. 274, 188–201. ( 10.1111/j.1742-4658.2006.05575.x) [DOI] [PubMed] [Google Scholar]
- 55.Kacser H, Burns J, Fell D. 1995. The control of flux. Biochem. Soc. Trans. 23, 341–366. ( 10.1042/bst0230341) [DOI] [PubMed] [Google Scholar]
- 56.Fell DA. 1997. Understanding the control of metabolism. London, UK: Portland Press.. [Google Scholar]
- 57.Ingalls BP. 2013. Mathematical modeling in systems biology: an introduction. Cambridge, MA: MIT Press. [Google Scholar]
- 58.Schuster S, Heinrich R. 1992. The definitions of metabolic control analysis revisited. Biosystems 27, 1–15. ( 10.1016/0303-2647(92)90042-W) [DOI] [PubMed] [Google Scholar]
- 59.Heinrich R, Schuster S. 1996. The regulation of cellular systems. New York, NY: Chapman and Hall. [Google Scholar]
- 60.Hofmeyr JHS. 1989. Control-pattern analysis of metabolic pathways. Eur. J. Biochem. 186, 343–354. ( 10.1111/j.1432-1033.1989.tb15215.x) [DOI] [PubMed] [Google Scholar]
- 61.Christensen CD, Hofmeyr JHS, Rohwer JM. 2015. Tracing regulatory routes in metabolism using generalised supply-demand analysis. BMC Syst. Biol. 9, 89 ( 10.1186/s12918-015-0236-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voet D, Voet JGP, Charlotte W. 2013. Fundamentals of biochemistry: life at the molecular level, 4th edn. Hoboken, NJ: Wiley. [Google Scholar]
- 63.d'Azzo JJ, Houpis CD. 1995. Linear control system analysis and design: conventional and modern. New York, NY: McGraw-Hill Higher Education. [Google Scholar]
- 64.Ogata K.Modern control engineering, 5th edn Upper Saddle River, NJ: Lugar; 07458. Prentice Hall; 2009. [Google Scholar]
- 65.Aström KJ, Murray RM. 2010. Feedback systems: an introduction for scientists and engineers. Princeton, NJ: Princeton University Press. [Google Scholar]
- 66.Del Vecchio D, Dy AJ, Qian Y. 2016. Control theory meets synthetic biology. J. R. Soc. Interface 13, 20160380 ( 10.1098/rsif.2016.0380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khammash M. 2016. An engineering viewpoint on biological robustness. BMC Biol. 14, 1 ( 10.1186/s12915-016-0241-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofmeyr JH, Cornish-Bowden A. 2000. Regulating the cellular economy of supply and demand. FEBS Lett. 476, 47–51. ( 10.1016/S0014-5793(00)01668-9) [DOI] [PubMed] [Google Scholar]
- 69.Hofmeyr JHS. 2008. The harmony of the cell: the regulatory design of cellular processes. Essays Biochem. 45, 57–66. ( 10.1042/bse0450057) [DOI] [PubMed] [Google Scholar]
- 70.Hofmeyr JHS, Rohwer JM. 2011. Supply-demand analysis: a framework for exploring the regulatory design of metabolism. Methods Enzymol. 500, 533–554. ( 10.1016/B978-0-12-385118-5.00025-6) [DOI] [PubMed] [Google Scholar]
- 71.Hofmeyr JH. 2001. Metabolic control analysis in a nutshell. In Proc. of the Second Int. Conf. on Systems Biology, pp. 291–300. Pasadena, CA: Caltech. [Google Scholar]
- 72.Rohwer JM, Akhurst TJ, Hofmeyr JHS. 2008. Symbolic control analysis of cellular systems. In Experimental standard conditions of enzyme characterizations. Proc. the 3rd Int. Beilstein Workshop. Beilstein-Institut zur Förderung der Chemischen Wissenschaften, pp. 137–148. [Google Scholar]
- 73.Heinisch J. 1986. Isolation and characterisation of the two structural genes coding for phosphofructokinase in yeast. Mol. Gen. Genet. 202, 75–82. ( 10.1007/BF00330520) [DOI] [PubMed] [Google Scholar]
- 74.Schaaff I, Heinisch J, Zimmermann FK. 1989. Overproduction of glycolytic enzymes in yeast. Yeast 5, 285–290. ( 10.1002/yea.320050408) [DOI] [PubMed] [Google Scholar]
- 75.Davies SE, Brindle KM. 1992. Effects of overexpression of phosphofructokinase on glycolysis in the yeast Saccharomyces cerevisiae. Biochemistry 31, 4729–4735. ( 10.1021/bi00134a028) [DOI] [PubMed] [Google Scholar]
- 76.Burrell MM, Mooney PJ, Blundy M, Carter D, Wilson F, Green J, Blundy K, ap Rees T. 1994. Genetic manipulation of 6-phosphofructokinase in potato tubers. Planta 194, 95–101. ( 10.1007/BF00201039) [DOI] [Google Scholar]
- 77.Thomas S, Mooney PJ, Burrell MM, Fell DA. 1997. Metabolic control analysis of glycolysis in tuber tissue of potato (Solanum tuberosum): explanation for the low control coefficient of phosphofructokinase over respiratory flux. Biochem. J. 322, 119–127. ( 10.1042/bj3220119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruijter G, Panneman H, Visser J. 1997. Overexpression of phosphofructokinase and pyruvate kinase in citric acid-producing Aspergillus niger. Biochim. Biophys. Acta-Gen. Subj. 1334, 317–326. ( 10.1016/S0304-4165(96)00110-9) [DOI] [PubMed] [Google Scholar]
- 79.Müller S, Zimmermann FK, Boles E. 1997. Mutant studies of phosphofructo-2-kinases do not reveal an essential role of fructose-2, 6-bisphosphate in the regulation of carbon fluxes in yeast cells. Microbiology 143, 3055–3061. ( 10.1099/00221287-143-9-3055) [DOI] [PubMed] [Google Scholar]
- 80.Urbano AM, Gillham H, Groner Y, Brindle KM. 2000. Effects of overexpression of the liver subunit of 6-phosphofructo-1-kinase on the metabolism of a cultured mammalian cell line. Biochem. J. 352, 921–927. ( 10.1042/bj3520921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marín-Hernández A, Rodríguez-Enríquez S, Vital-González PA, Flores-Rodríguez FL, Macías-Silva M, Sosa-Garrocho M, Moreno-Sánchez R. 2006. Determining and understanding the control of glycolysis in fast-growth tumor cells. FEBS J. 273, 1975–1988. ( 10.1111/j.1742-4658.2006.05214.x) [DOI] [PubMed] [Google Scholar]
- 82.Moreno-Sánchez R, Marín-Hernández A, Saavedra E, Pardo JP, Ralph SJ, Rodríguez-Enríquez S. 2014. Who controls the ATP supply in cancer cells? Biochemistry lessons to understand cancer energy metabolism. Int. J. Biochem. Cell Biol. 50, 10–23. ( 10.1016/j.biocel.2014.01.025) [DOI] [PubMed] [Google Scholar]
- 83.Storey KB. 2005. Functional metabolism: regulation and adaptation. London, UK: John Wiley & Sons. [Google Scholar]
- 84.Mor I, Cheung E, Vousden K. 2011. Control of glycolysis through regulation of PFK1: old friends and recent additions. In Cold Spring Harbor symposia on quantitative biology, vol. 76, pp. 211–216. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PubMed] [Google Scholar]
- 85.Savageau MA. 1974. Optimal design of feedback control by inhibition: steady-state considerations. J. Mol. Evol. 4, 139–156. ( 10.1007/BF01732019) [DOI] [PubMed] [Google Scholar]
- 86.Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M. 2002. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 79, 53–73. ( 10.1002/bit.10288) [DOI] [PubMed] [Google Scholar]
- 87.Sauro HM. 2005. The next frontier in cellular networking: systems biology doesn't have to be complicated. The Scientist 19, 20–22. [Google Scholar]
- 88.Colclough RC, Sauro HM. 2005. Cells and chips: it's no contest. The Scientist 19, 10–11.17975652 [Google Scholar]
- 89.Marx V. 2013. Biology: the big challenges of big data. Nature 498, 255–260. ( 10.1038/498255a) [DOI] [PubMed] [Google Scholar]
- 90.May M. 2014. Life science technologies: big biological impacts from big data. Science 344, 1298–1300. ( 10.1126/science.344.6189.1298) [DOI] [Google Scholar]
- 91.Pigliucci M. 2009. The end of theory in science? EMBO Rep. 10, 534–534. ( 10.1038/embor.2009.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mishra D, Rivera PM, Lin A, Del Vecchio D, Weiss R. 2014. A load driver device for engineering modularity in biological networks. Nat. Biotechnol. 32, 1268–1275. ( 10.1038/nbt.3044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsimring LS. 2014. Noise in biology. Rep. Prog. Phys. 77, 026601 ( 10.1088/0034-4885/77/2/026601) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.