Summary
Here, we describe a targeted reverse genetic screen for thermal nociception genes in Drosophila larvae. Using laser capture microdissection and microarray analyses of nociceptive and non-nociceptive neurons we identified 275 nociceptor-enriched genes. We then tested the function of the enriched genes with nociceptor-specific RNAi and thermal nociception assays. Tissue-specific RNAi targeted against 14 genes caused insensitive thermal nociception while targeting of 22 genes caused hypersensitive thermal nociception. Previously uncategorized genes were named for heat resistance (ie. boilerman, fire dancer, oven mitt, trivet, thawb and bunker gear) or heat sensitivity (firelighter, black match, eucalyptus, primacord, jet fuel, detonator, gasoline, smoke alarm, and jetboil). Insensitive nociception phenotypes were often associated with severely reduced branching of nociceptor neurites and hyperbranched dendrites were seen in two of the hypersensitive cases. Many genes that we identified are conserved in mammals.
Introduction
The pomace fly Drosophila melanogaster has been developed as a robust system to study nociception (Babcock et al., 2009; Babcock et al., 2011; Im et al., 2015; Neely et al., 2010; Tracey et al., 2003). Drosophila, with its unparalleled genetic tools, is an excellent model to uncover nociception genes. Drosophila larvae rotate along the long body axis in a corkscrew like fashion in response to noxious stimuli such as heat (>39°C) or harsh mechanical stimuli (Tracey et al., 2003). This highly stereotyped response to harmful stimuli, named nocifensive escape locomotion (NEL) or rolling, serves as a robust behavioral readout of nociception since it is specifically triggered by noxious stimuli and it is clearly distinguishable from normal locomotion and other somatosensory responses.
Several lines of evidence indicate that Class IV multidendritic (md) neurons are polymodal nociceptive sensory neurons responsible for larval thermal and mechanical nociception. The pickpocket and balboa/ppk-26 genes show highly specific expression in these neurons and are required for mechanical nociception (Gorczyca et al., 2014; Guo et al., 2014; Mauthner et al., 2014; Zhong et al., 2010). Similarly, reporter genes for specific dTRPA1 transcripts are specifically expressed in the Class IV cells and dTRPA1 is required for both mechanical and thermal nociception (Zhong et al., 2012). Genetic silencing of Class IV neurons severely impairs thermal and mechanical nociception behavior and optogenetic activation of these neurons is sufficient to evoke NEL (Hwang et al., 2007; Zhong et al., 2012).
Results and Discussion
Laser capture microdissection and microarray analyses identify 275 nociceptor-enriched genes
Genes involved in nociception are often preferentially expressed in nociceptors (Akopian et al., 1996; Caterina et al., 1997; Chen et al., 1995; Dib-Hajj et al., 1998; Mauthner et al., 2014; Nagata et al., 2005; Zhong et al., 2012; Zhong et al., 2010). Thus, to identify Drosophila nociceptor-enriched genes we performed laser capture microdissection to isolate RNAs from nociceptive and non-nociceptive neurons (Mauthner et al., 2014). We then performed microarray analyses on the isolated samples (Mauthner et al., 2014). We compared the gene expression profiles of nociceptive Class IV multidendritic (md) neurons to Class I md neuron profiles (Mauthner et al., 2014) as Class IV md neurons are polymodal nociceptors (their output is both necessary and sufficient for triggering larval nociception behaviors), and Class I md neurons are functionally dispensable for nociception (Hwang et al., 2007). Indeed, as internal validation of these methods, this microarray study successfully detected the enrichment of genes previously thought to be preferentially expressed in Class IV relative to Class I neurons, such as cut, knot, Gr28b, ppk and balboa/ppk26 (Mauthner et al., 2014) (Table S1A).
To further identify nociceptor-enriched genes, we made a side-by-side comparison of the normalized hybridization intensity between Class IV and Class I neurons for all Affymetrix probe sets, and identified 278 probe sets corresponding to 275 genes that showed a greater than two-fold higher expression in Class IV neurons in comparison to Class I neurons (Class IV / Class I > 2; p < 0.05 with Welch t-test) (Figure S1 and Table S1A).
Nociceptor-specific RNAi screens uncover 36 genes required for larval thermal nociception
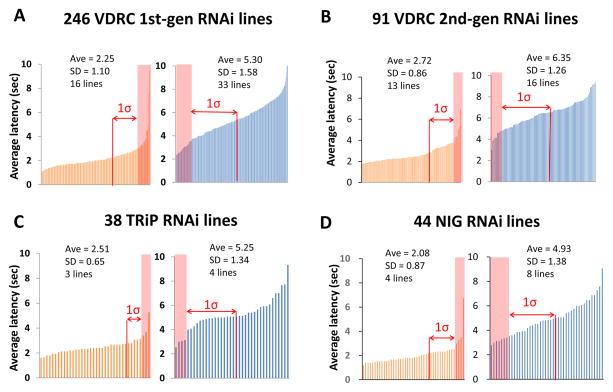
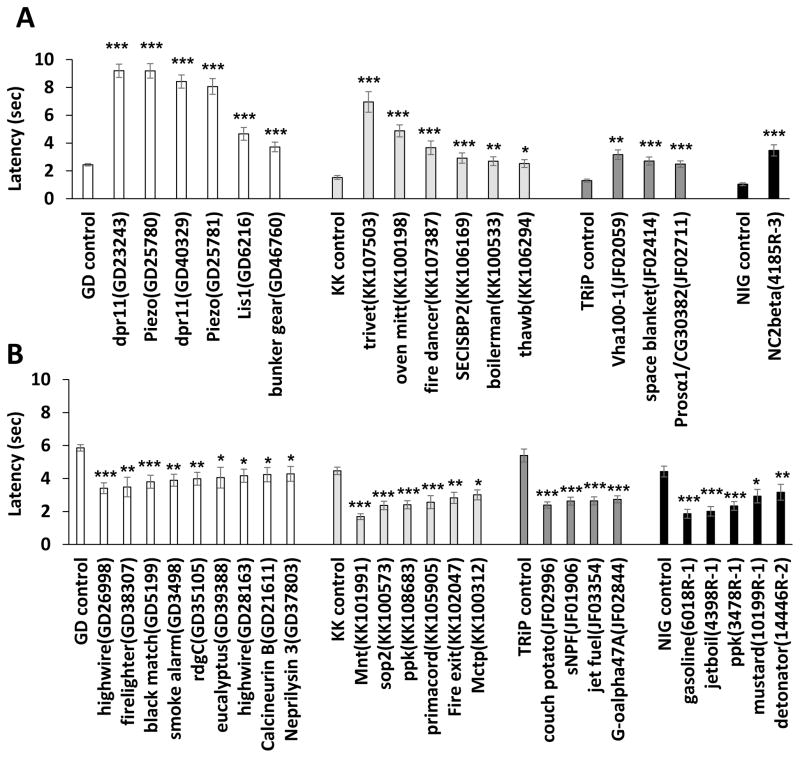
We subsequently tested the function of the nociceptor-enriched genes in thermal nociception responses. In order to test their requirement specifically in nociceptors, we used RNAi to knock down each gene in a tissue-specific pattern using the Class IV specific GAL4 driver ppk1.9-GAL4. UAS-dicer2 was also present in the driver strain in order to enhance RNAi knockdown (Dietzl et al., 2007). A total of 419 UAS-RNAi lines were obtained from the Vienna Drosophila RNAi library (Dietzl et al., 2007), the TRiP RNAi library (Ni et al., 2009), and the National Institute of Genetics RNAi library and were used to knock down 229 of the 275 (83.3%) nociceptor-enriched genes. In control experiments, we found that the baseline nociception responses differed among the genetic backgrounds that were used to generate the different collections of UAS-RNAi strains. Thus, the progeny of our GAL4 driver strain crossed to UAS-RNAi lines from the four different collections (VDRC 1st-generation, VDRC 2nd-generation, TRiP and NIG) were each statistically analyzed in comparison to the relevant genetic controls for parental isogenic background. Progeny of each ppk-GAL4 UAS-dicer-2 × UAS-RNAi cross were tested in an established larval thermal nociception assay (Tracey et al., 2003). In order to identify either insensitive or hypersensitive phenotypes, independent tests were carried out at two different probe temperatures. A 46°C probe was used to screen for insensitive phenotypes (defined as a lengthened latency to respond to the 46°C stimulus) while a 42°C probe was used to assess hypersensitivity (defined as a shortened latency to respond to this stimulus). Average latency to 46°C and 42°C thermal probe stimulation were determined for each RNAi knockdown genotype (Figure 1 and Table S1B–E). We set our initial cut-off line at the +1σ (84.13th percentile) in the insensitivity screen and −1σ (15.87th percentile) in the hypersensitivity screen, and all ppk-GAL4 UAS-dicer-2 × UAS-RNAi pairs that met these cut-offs were subjected to retesting (Figure 1 and Table S1B–E). Only pairs showing significant insensitivity or hypersensitivity in comparison to the appropriate control for genetic background (p < 0.05 with Steel’s test) during the retest were considered as positive hits. Sixteen RNAi lines targeting 14 genes were identified in the insensitivity screen and 24 RNAi lines targeting 22 genes were found in the hypersensitivity screen (Table 1, Figure 2 and S1F–M). We confirmed that these positive RNAi lines did not show the observed nociception phenotypes when crossed to w1118strain (no driver control), suggesting that the phenotypes observed in the screen were dependent on GAL4-driven expression of RNAi (Table S1N and O).
Figure 1. Summary of primary screen results.
Summary of the insensitivity and the hypersensitivity screen with (A) 1st-generation VDRC (GD) lines, (B) 2nd-generation VDRC (KK) lines, (C) TRiP lines, (D) and NIG lines. The left chart in each panel with the orange bars show the results of the insensitivity screen with a 46°C probe. The right chart in each panel, with blue bars, shows the hypersensitivity screen results with a 42°C probe. The average latency and standard deviation of all tested lines and the number of lines which survived the initial cut-off (+1σ for the insensitivity screen and −1σ for the hypersensitivity screen) are indicated with each graph. Shaded areas indicate lines that were selected for retesting. See also Table S1B–E.
Table 1.
Candidate genes
| Insensitive Candidates | ||||||||
|---|---|---|---|---|---|---|---|---|
| CG number | Synonym | New name | RNAi line | IV/I | Latency | Significance | Gene Ontology | Human orthologs |
| CG33202 | dpr11 | GD23243 | 9.83 | 9.20 ± 0.48 | p < 0.001 | Ig family, Membrane protein | ||
| GD40329 | 8.42 ± 0.47 | p < 0.001 | ||||||
| CG18103 | Piezo | GD25780 | 6.12 | 9.19 ± 0.52 | p < 0.001 | mechanically-gated ion channel activity | PIEZO1a, | |
| GD25781 | 8.06 ± 0.56 | p < 0.001 | PIEZO2a,b,c | |||||
| CG8297 | CG8297 | bunker gear (bug) | GD46760 | 2.82 | 3.72 ± 0.34 | p < 0.001 | Thioredoxin-like fold. | TXNDC15 |
| CG8440 | Lis1 | GD6216 | 2.81 | 4.66 ± 0.46 | p < 0.001 | Dynein-binding, Microtubule organization | PAFAH1B1c | |
| CG12681 | CG12681 | boilerman (boil) | KK100533 | 12.48 | 2.70 ± 0.32 | p < 0.01 | Unknown | |
| CG14608 | CG14608 | thawb (thw) | KK106294 | 43.59 | 2.52 ± 0.28 | p < 0.05 | Chitin binding, Chitin metabolic process | |
| CG31976 | CG31976 | oven mitt (ovm) | KK100198 | 15.81 | 4.88 ± 0.43 | p < 0.001 | Unknown | |
| CG34362 | CG12870 | trivet (trv) | KK107503 | 10.06 | 6.96 ± 0.74 | p < 0.001 | Nucleotide binding, Alternative splicing | TIA1b, TIA1L |
| CG7066 | SECIS-binding protein2 | KK106169 | 3.37 | 2.92 ± 0.37 | p < 0.001 | Selenoprotein synthesis | SECISBP2c, SECISBP2L | |
| CG42261 | CG8968 | fire dancer (fid) | KK107387 | 3.06 | 3.66 ± 0.49 | p < 0.001 | Methyltransferase activity | |
| CG18495 | Proteasome α1 subunit | JF02711 | 2.18 | 2.48 ± 0.33 | p < 0.001 | Endopeptidase activity, Proteasome core complex. | PSMA6 | |
| CG1709 | Vha100-1 | JF02059 | 3.24 | 3.17 ± 0.33 | p < 0.01 | V-ATPase, Calmodulin binding, synaptic vesicle exocytosis | ATP6V0A4, ATP6V0A1, ATP6V0A2c | |
| CG8216 | CG8216 | space blanket (spab) | JF02414 | 13.27 | 2.71 ± 0.29 | p < 0.001 | Unknown | |
| CG4185 | NC2 beta | 4185R-3 | 6.994 | 3.47 ± 0.40 | p < 0.001 | histone acetyltransferase activity | Dr1a | |
| Hypersensitive Candidates | ||||||||
|---|---|---|---|---|---|---|---|---|
| CG number | Synonym | New name | RNAi line | IV/I | Latency | Significance | Gene Ontology | Human orthologs |
| CG32592 | highwire | GD26998 | 6.04 | 3.41 ± 0.33 | p < 0.001 | ubiquitin-protein ligase activity | MYCBP2c | |
| GD28163 | 4.17 ± 0.40 | p < 0.05 | ||||||
| CG14946 | CG14946 | firelighter (firl) | GD38307 | 2.627 | 3.48 ± 0.59 | p < 0.01 | Short-chain dehydrogenase/reductase SDR; Glucose/ribitol dehydrogenase; NAD(P)-binding domain. | HSD17B13, SDR16C5, RDH10, HSD17B11c, DHRS3 |
| CG34356 | CG12524 | black match (bma) | GD5199 | 3.85 | 3.80 ± 0.39 | p < 0.001 | ATP binding; protein kinase activity | |
| CG34380 | CG13988 | smoke alarm (smal) | GD3498 | 2.57 | 3.89 ± 0.37 | p < 0.01 | Receptor signaling | DDR1a, DDR2 |
| CG4209 | Calcineurin B | GD21611 | 3.79 | 4.24 ± 0.44 | p < 0.05 | calcium ion binding; cthreonine phosphatase activity | PPP3R1a, PPP3R2, WDR92 | |
| CG6571 | rdgC | GD35105 | 3.74 | 3.98 ± 0.39 | p < 0.01 | Calmodulin binding, phototransduction | PPEF1a,b, PPEF2 | |
| CG12269 | CG12269 | eucalyptus (euc) | GD39388 | 11.57 | 4.05 ± 0.63 | p < 0.05 | sterol binding | |
| CG9565 | Neprilysin 3 | GD37803 | 2.33 | 4.28 ± 0.45 | p < 0.05 | Metalloendopeptidase | ECE1a,c, MMEL1, ECEL1, ECE2a,c, MME, PHEX | |
| CG1079 | fire exit | KK102047 | 14.98 | 2.82 ± 0.33 | p < 0.01 | Unknown | ||
| CG13316 | Mnt | KK101991 | 2.69 | 1.68 ± 0.18 | p < 0.001 | DNA binding, Transcriptional repressor, Negative regulator of cell growth, Phagocytosis | PHF15a, PHF16a, PHF17 | |
| CG15078 | Mctp | KK100312 | 34.97 | 3.00 ± 0.31 | p < 0.05 | calcium ion binding | MCTP1a,c, MCTP2c | |
| CG15704 | CG15704 | primacord (prim) | KK105905 | 4.35 | 2.56 ± 0.39 | p < 0.001 | Unknown | |
| CG3478 | pickpocket1 | KK108683 | 28.03 | 2.41 ± 0.24 | p < 0.001 | acid-sensing ion channel activity | ||
| 3478R-1 | 2.33 ± 0.27 | p < 0.001 | ||||||
| CG8978 | Suppressor of profilin 2 | KK100573 | 15.74 | 2.36 ± 0.26 | p < 0.001 | actin binding; structural constituent of cytoskeleton | ARPC1Ba,b,c | |
| CG13968 | short neuropeptide F | JF01906 | 62.4 | 2.63 ± 0.24 | p < 0.001 | hormone activity; neuropeptide hormone activity | ||
| CG31243 | couch potato | JF02996 | 2.57 | 2.39 ± 0.19 | p < 0.001 | mRNA binding; nucleic acid binding; nucleotide binding | RBPMSa, RBPMS2 | |
| CG2204 | G protein o alpha 47A | JF02844 | 2.66 | 2.73 ± 0.22 | p < 0.001 | G-protein beta/gamma-subunit complex binding; G-protein coupled receptor binding; GTPase activity; signal transducer activity | GNAT1, GNAI2a,c, GNAO1a,b,c, GNAI3a, GNAZa, GNAT2, GNAT3 | |
| CG12858 | CG12858 | jet fuel (jef) | JF03354 | 7.1 | 2.64 ± 0.25 | p < 0.001 | transmembrane transport | MFSD6c |
| CG32464 | mustard | 10199R-1 | 7.276 | 2.93 ± 0.41 | p < 0.05 | Peptidoglycan-binding Lysin subgroup, immune response | ||
| CG14446 | CG14446 | detonator (dtn) | 14446R-2 | 4.027 | 3.17 ± 0.48 | p < 0.01 | Unknown | |
| CG4398 | CG4398 | jetboil (jtb) | 4398R-1 | 13.39 | 2.01 ± 0.29 | p < 0.001 | Insect allergen-related | |
| CG6018 | CG6018 | gasoline (gas) | 6018R-1 | 96.18 | 1.86 ± 0.27 | p < 0.001 | carboxylesterase activity, Carboxylesterase, type B | |
Genes enriched in nociceptive lineage neurons compared to proprioeptive lineage neurons in Chiu et al. (2014).
Genes enriched in nociceptors compared to unpurified DRG neurons in Thakur et al. (2014).
Genes enriched in nociceptors compared to cortical neurons in Thakur et al. (2014).
Figure 2. RNAi lines that showed significant insensitivity or hypersensitivity upon retesting with a larger sample size.
The behavioral responses of retested lines in the hypersensitivity and insensitivity screens. Each panel shows average latency on the Y axis and the targeted genes for each genotype are listed along the X axis. (A) ppk-GAL4 dependent insensitive behavioral responses seen with crossing to 1st-generation VDRC (GD) lines, 2nd-generation VDRC (KK) lines, TRiP lines and NIG lines. (B) ppk-GAL4 dependent hypersensitive behavioral responses seen with crossing to VDRC (GD and KK) lines, TRiP lines and NIG lines. Steel’s test was used to statistically compare each genotype to its appropriate control except that Mann-Whitney’s U-test was used to perform the pair-wise comparison of NC2beta versus NIG control (n > 32; * p < 0.05, ** p < 0.01 and *** p < 0.001). Error bars represent S.E.M. See also Table S1F-O, Table S2 and Figure S1.
Neely et al. carried out a genome-wide RNAi screen for nociception genes using adult Drosophila (Neely et al., 2010). Neely and colleagues identified 3 genes of the 14 that we found with insensitive nociception phenotypes (dpr11, lis-1 and vha100-1), and a single gene out of the 22 with hypersensitive phenotypes (retinal degeneration C). As this prior screen relied on thermally-induced paralysis of adult flies as a surrogate for studying larval nociception behavior, it is possible that molecular mechanisms of nociception in adult flies and larval flies may distinct. In addition, Neely et al used broadly expressed knockdown approaches. It is possible that pleiotropy caused by opposing effects in distinct tissues may have masked the effects that we are able to detect with nociceptor specific knockdown. In either case, it is likely that a large number of bona fide nociception signaling genes remain to be discovered using larval nociception assays.
Uncharacterized Genes Identified in the Screens
Our screen identified genes that remain named according to Celera Gene (CG) numbers (Table 1). Knockdown of seven CGs caused insensitivity and nine CGs caused hypersensitivity. Thus, given these loss of function phenotypes we have named each of the heat-insensitivity screen genes (boilerman (boil), fire dancer (fid)), oven mitt (ovm) and trivet (trv), thawb (thw), bunker gear (bug), space blanket (spab)) (Table 1). And we have also named uncharacterized genes that were identified in the hypersensitivity screen (firelighter (firl), black match (bma), eucalyptus (euc), primacord (prim), jet fuel (jef), detonator (dtn) and gasoline (gas)), smoke alarm (smal)), jetboil (jtb)) (Table 1).
Genes required for Class IV neuron morphogenesis
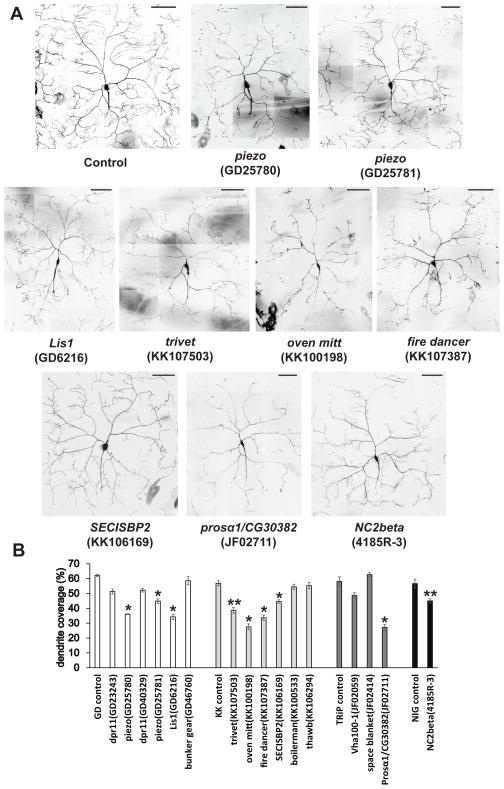
Lis1, which showed an insensitive nociception phenotype in our screen (Figure 2A and Table 1), is a component of a dynein-dependent motor complex that is known to play a role in dendrite and axonal morphogenesis in Class IV neurons (Satoh et al., 2008). Similarly, a reduced dendrite phenotype in another study was associated with insensitive nociception behaviors (Stewart et al., 2012). These observations raised the possibility that nociception phenotypes detected in our screen might also be associated with defects in dendrite morphogenesis.
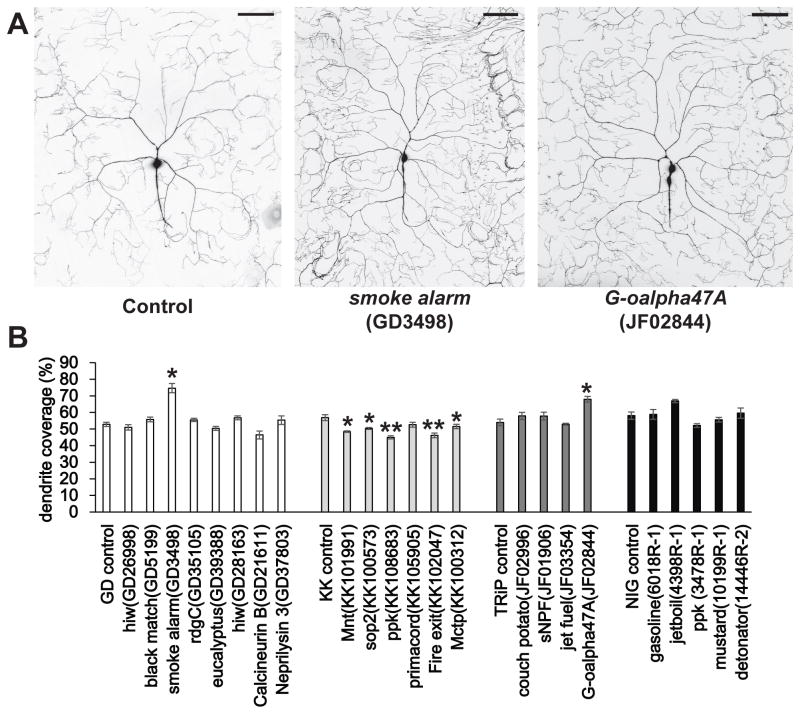
To test for this possibility, we used confocal microscopy to observe and quantify the dendritic coverage of CD8-GFP expressing Class IV neurons in all of the genotypes that showed insensitive or hypersensitive thermal nociception phenotypes (Figure 3 and 4). Consistent with the previous study, Lis1 RNAi showed severe defects in dendrite morphogenesis (Figure 3). Significantly reduced nociceptor dendrites were also found with RNAi lines targeting piezo, oven mitt, trivet, fire dancer, SECISBP2, pros-alpha1 and NC2beta (Figure 3). Among hypersensitive hits, smoke alarm and G-oalpha47A RNAi resulted in significantly increased dendrite phenotypes, which potentially contributes to their hypersensitive nociception phenotypes (Figure 4).
Figure 3. Genes targeted with RNAi that showed a reduced dendrite phenotype.
(A) Representative composite images of the dendritic structure of Class IV ddaC neurons in thermal nociception insensitive RNAi lines that also showed significantly reduced dendritic coverage. Scale bars represent 100 μm. (B) Quantification of dendritic coverage for insensitive RNAi lines. Steel’s test was used for statistical comparisons between each genotype and controls, except that Mann-Whitney’s U-test was used to compare NC2beta and NIG control (n > 4; * p < 0.05 and ** p < 0.01). Error bars represent S.E.M. For representative images of RNAi lines that did not show significantly reduced dendrite, see Figure S2.
Figure 4. Genes targeted with RNAi that showed a an increased dendrite coverage phenotype.
(A) Representative composite images of the dendritic field of Class IV ddaC neurons in RNAi lines with hypersensitive thermal nociception animals that also showed significantly increased dendritic coverage. Scale bars represent 100 μm. (B) Quantification of dendritic coverage for hypersensitive candidate RNAi lines. Steel’s test was used for statistical comparisons between each genotype and controls, except that Mann-Whitney’s U-test was used to compare NC2beta and NIG control (n > 4; * p < 0.05 and ** p < 0.01). Error bars represent S.E.M. For representative images of RNAi lines that did not show significantly increased dendrite coverage, see Figure S3. The effects seen in the KK collection was not a consequence of unintended Tio expression (See supplemental experimental procedures, Table S2 and Figure S2)
In contrast, some hypersensitive hits actually showed a mild (but statistically significant) reduction in dendrite coverage (Mnt, sop2, ppk, fire exit and Mctp) (Figure 4). Thus, perhaps not surprisingly, the degree of dendrite branching cannot be perfectly correlated with noxious heat sensitivity. An interesting possibility is that targeting these genes with RNAi results in hypersensitivity due to effects in another compartment of the cell such as axons and/or synapses. Or alternatively, hypersensitivity in the dendrites masks a pleiotropic morphologically reduced dendrite phenotype.
highwire, one of hypersensitive candidates (Figure 2B and Table 1), has been shown to be important for dendrite and axonal morphogenesis of Class IV neurons (Wang et al., 2013). Interestingly hiw RNAi did not cause reduced dendritic arbors that have been reported with strong loss of function alleles for hiw. Our detailed analyses of hiw indicate that the hypersensitive nociception phenotype is more sensitive to hiw dosage than are the previously reported dendrite phenotypes (Honjo and Tracey, unpublished observations). As in the case of hiw, RNAi knockdown often results in phenotypes that are less severe than those that would be observed with null alleles.
Indeed, there are other caveats to be considered when using an RNAi screening methodology. The incomplete knockdown effect can also result in false negatives, which are estimated to occur in up to 40% of the UAS-RNAi strains in the major collection of strains used in our screen. Thus, the lack of a phenotype in our screen cannot be used to conclusively infer a lack of function for a particular gene of interest. As well, false positives may occur, presumably due to off target effects. When the UAS-RNAi used in conjunction with UAS-dicer-2 (as in our experiments) the effectiveness of knockdown is enhanced, and off-target effects are seen in approximately 6% of lines (when tested in the very sensitive crystalline lattice of the eye, or in the notum). Applying this estimate to the 36 genes implicated by our screen cautions that two or more of the candidates may represent false positives.
Nociception genes are evolutionarily conserved in mammals
Twenty out of the thirty-six of the genes that are implicated here in nociception have clearly predicted mammalian orthologues (Table 1). Interestingly, published evidence supports roles for some of these orthologues in regulating mammalian nociception. Nociceptor and thermoreceptor-specific knock-out of MYCBP2, a mammalian homologue of highwire, shows prolonged thermal hypersensitivity with formalin-induced hyperalgesia (Holland et al., 2011). Knockdown of highwire caused an intriguingly similar hypersensitivity to heat (Figure 2B and Table 1).
RNAi targeting Neprilysin-3, encoding a neutral endopeptidase, showed hypersensitive nociception (Figure 2B and Table 1). Loss of ECE2, one of six predicted mammalian homologues of Neprilysin-3, has been also implicated in hypersensitive nociception as a knock-out mouse for ECE2 exhibits thermal hypersensitivity and rapid tolerance to morphine induced analgesia (Miller et al., 2011). In addition, knock-out of MME (aka NEP), another homologue of Neprilysin-3, causes thermal hyperalgesia (Fischer et al., 2002). These results thus raise the possibility that inhibitory nociceptive functions of Neprilysin-3 may be conserved between flies and mammals.
To our knowledge, the remaining conserved genes that our studies implicate in nociception remain to be functionally implicated in mammalian pathways. However, it is very intriguing that many of these conserved candidate genes are more highly expressed in nociceptors compared to non-nociceptive neurons or other tissues (Chiu et al., 2014; Goswami et al., 2014; Thakur et al., 2014). The orthologues of the seventeen out of twenty candidate nociception genes that came out from our study have been reported to show significantly enriched expression in nociceptive sensory neurons (Table 1). These genes will thus be particularly promising targets to identify previously uncharacterized molecular pathways involved in nociception.
Two ion channel genes from our screen have been implicated in mechanical nociception, ppk and Piezo (Figure 2, Table 1). Since studies of Piezo in Class IV neurons have observed defective phenotypes in mechanical nociception assays but not in thermal nociception assays (Kim et al., 2012) it is surprising that Piezo RNAi causes thermal insensitivity. The apparent discrepancy may be because the two Piezo RNAi strains used in this study specifically target low-abundance exons that were previously annotated as an independent gene, fos28F (Graveley et al., 2011). And in our microarray dataset we found enriched expression for “fos28F” in Class IV neurons but not for piezo. Our interpretation of these microarray data is that a nociceptor specific transcript for piezo exists and it contains sequences from the previously annotated gene fos28F. Both of the RNAi constructs that target the fos28F/piezo exons caused gross abnormalities in dendritic and axonal gross morphology (Figure 3 and data not shown). RNAi lines targeting canonical piezo exons do not cause a similar thermal nociception phenotype (KH unpublished observations). Thus, the nociceptor-specific knockdown of the low-abundance transcriptional variant containing exons from fos28F appears to disrupt the morphology and thermal nociception capacity of Class IV neurons.
It was also unexpected that two independent ppk RNAi strains collected from different libraries showed hypersensitive thermal nociception phenotypes. This contrasts with the severely insensitive mechanical nociception phenotypes that occur with the loss of ppk (Zhong et al., 2010). Our previous studies did not detect thermal hypersensitivity due to the testing with a single higher probe temperature (46 º C). The finding that ppk RNAi causes thermal hypersensitivity highlights the importance of using the 42º C probe temperature in the search for hypersensitive phenotypes. In addition, the results indicate that modality specific phenotypes within the larval nociceptors can be of opposite sign. It is interesting to note that ppk mutants have been found to show a locomotion phenotype in which the animals crawl rapidly in a straight line across the substrate with very infrequent turning (Ainsley et al., 2003). A similar form of locomotion is also seen in a second phase of nociception behavior that immediately follows rolling behavior (NEL) (Ohyama et al., 2013). Thus, it is interesting to speculate that the locomotion phenotype of ppk mutants is actually a consequence of a hypersensitive process in the nociceptors. This in turn may be causing the larvae to continuously manifest the fast crawling phase of nociception escape.
In conclusion, we have carried out a large-scale screen that combines molecular approaches to identify cell type enriched nociceptor RNA with in vivo functional studies of the same identified RNAs in a phenotypic screen. This approach has led to the identification of a set of nociception genes. Many of these genes are evolutionarily conserved and also show enriched expression in mammalian nociceptors, future studies will reveal the physiological importance and molecular mechanisms that depend on these molecules.
Experimental procedures
Laser Capture Microdissection and Microarray analysis
Detailed methods for our Laser Capture Microdissection and Microarray analysis were previously described in Mauthner et al. (Mauthner et al., 2014).
Thermal nociception screen
Three males of each RNAi strain were crossed to six virgin females of ppk1.9-GAL4; UAS-dicer2 strain in a standard molasses cornmeal food vial, and incubated for 5 to 7 days at 25º C prior to harvest of the F1 wandering third instar larvae. Control crosses (a control strain crossed to ppk1.9-GAL4; UAS-dicer2) were performed side-by-side.
Thermal nociception assays were performed as described previously (Hwang et al., 2012; Tracey et al., 2003; Zhong et al., 2012). To detect insensitivity and hypersensitivity phenotype efficiently, each UAS-RNAi × ppk1.9-GAL4; UAS-dicer2 pair was tested by using two different probe temperatures: A custom-made thermal probe heated to 46º C was used to test insensitivity and the probe heated to 42º C was used for hypersensitivity.
Quantifying dendrite coverage of Class IV neurons
Dendritic field coverage was quantified on composite images of maximum intensity projections of confocal micrographs and quantified as described previously (Stewart et al., 2012) with slight modifications. Images of ddaC neurons were overlaid with a grid of 32 × 32 pixel squares (14 × 14 μm), and squares containing dendritic branches were counted to calculate a dendritic field coverage score (i.e. the percentage of squares containing dendritic branches / the total number of squares). Counting dendrite-positive squares was done with a Matlab custom code and then manually curated to eliminate false-positives and false–negatives. One or two neurons from each imaged animal were analyzed.
Statistical analyses
All pair wise comparisons were performed with Mann-Whitney’s U-test. For multiple comparisons, Steel’s test (non-parametric equivalent of Dunnet’s test) was used. Statistical analyses were performed in the R software and Kyplot.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila RNAi Stock Center, TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), and the National Institute of Genetics Fly Stock Center. We are grateful to Dr. Kyriacou for UAS-tio strain. Eashan Kumar and Stephen Jeffirs assisted with crosses. This work was supported by grants from the National Institutes of Health (U24NS51870 to J.H.P.S. and R01GM086458 to W.D.T.) and JSPS KAKENHI (26890025 to K.H.). K.H. was supported by fellowships from the Uehara Memorial Foundation, the Ruth K. Broad Biomedical Research Foundation and the Japan Society for the Promotion of Science.
Footnotes
Author Contributions
KH performed experiments, analyzed data and wrote the manuscript, SEM performed experiments, analyzed data and wrote the manuscript, YW performed LCM experiments, JHPS directed the LCM core for the NIH Microarray Consortium, WDT conceived the project, analyzed data, and wrote the manuscript.
Accession numbers:
The ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) accession number for the microarray dataset reported in this study is E-MTAB-3863.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- Babcock DT, Landry C, Galko MJ. Cytokine signaling mediates UV-induced nociceptive sensitization in Drosophila larvae. Curr Biol. 2009;19:799–806. doi: 10.1016/j.cub.2009.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock DT, Shi S, Jo J, Shaw M, Gutstein HB, Galko MJ. Hedgehog Signaling Regulates Nociceptive Sensitization. Curr Biol. 2011 doi: 10.1016/j.cub.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Chen CC, Akopian AN, Sivilotti L, Colquhoun D, Burnstock G, Wood JN. A P2X purinoceptor expressed by a subset of sensory neurons. Nature. 1995;377:428–431. doi: 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife. 2014;3 doi: 10.7554/eLife.04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD, Tyrrell L, Black JA, Waxman SG. NaN, a novel voltage-gated Na channel, is expressed preferentially in peripheral sensory neurons and down-regulated after axotomy. Proc Natl Acad Sci U S A. 1998;95:8963–8968. doi: 10.1073/pnas.95.15.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fischer HS, Zernig G, Hauser KF, Gerard C, Hersh LB, Saria A. Neutral endopeptidase knockout induces hyperalgesia in a model of visceral pain, an effect related to bradykinin and nitric oxide. J Mol Neurosci. 2002;18:129–134. doi: 10.1385/JMN:18:1-2:129. [DOI] [PubMed] [Google Scholar]
- Gorczyca DA, Younger S, Meltzer S, Kim SE, Cheng L, Song W, Lee HY, Jan LY, Jan YN. Identification of Ppk26, a DEG/ENaC Channel Functioning with Ppk1 in a Mutually Dependent Manner to Guide Locomotion Behavior in Drosophila. Cell Reports. 2014;9:1446–1458. doi: 10.1016/j.celrep.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami SC, Mishra SK, Maric D, Kaszas K, Gonnella GL, Clokie SJ, Kominsky HD, Gross JR, Keller JM, Mannes AJ, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain. 2014;15:1338–1359. doi: 10.1016/j.jpain.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Wang Q, Wang Z. The role of PPK26 in Drosophila larval mechanical nociception. Cell Reports. 2014;9:1183–1190. doi: 10.1016/j.celrep.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Holland S, Coste O, Zhang DD, Pierre SC, Geisslinger G, Scholich K. The ubiquitin ligase MYCBP2 regulates transient receptor potential vanilloid receptor 1 (TRPV1) internalization through inhibition of p38 MAPK signaling. J Biol Chem. 2011;286:3671–3680. doi: 10.1074/jbc.M110.154765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Stearns NA, Tracey WD. The Ankyrin Repeat Domain of the TRPA Protein Painless Is Important for Thermal Nociception but Not Mechanical Nociception. PLoS One. 2012;7:e30090. doi: 10.1371/journal.pone.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Takle K, Jo J, Babcock DT, Ma Z, Xiang Y, Galko MJ. Tachykinin acts upstream of autocrine Hedgehog signaling during nociceptive sensitization in Drosophila. eLife. 2015;4:e10735. doi: 10.7554/eLife.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthner SE, Hwang RY, Lewis AH, Xiao Q, Tsubouchi A, Wang Y, Honjo K, Skene JH, Grandl J, Tracey WD., Jr Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr Biol. 2014;24:2920–2925. doi: 10.1016/j.cub.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LK, Hou X, Rodriguiz RM, Gagnidze K, Sweedler JV, Wetsel WC, Devi LA. Mice deficient in endothelin-converting enzyme-2 exhibit abnormal responses to morphine and altered peptide levels in the spinal cord. J Neurochem. 2011;119:1074–1085. doi: 10.1111/j.1471-4159.2011.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Duggan A, Kumar G, Garcia-Anoveros J. Nociceptor and hair cell transducer properties of TRPA1, a channel for pain and hearing. J Neurosci. 2005;25:4052–4061. doi: 10.1523/JNEUROSCI.0013-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. A Genome-wide Drosophila Screen for Heat Nociception Identifies alpha2delta3 as an Evolutionarily Conserved Pain Gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Jovanic T, Denisov G, Dang TC, Hoffmann D, Kerr RA, Zlatic M. High-throughput analysis of stimulus-evoked behaviors in Drosophila larva reveals multiple modality-specific escape strategies. PLoS One. 2013;8:e71706. doi: 10.1371/journal.pone.0071706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- Stewart A, Tsubouchi A, Rolls MM, Tracey WD, Sherwood NT. Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. J Neurosci. 2012;32:11631–11642. doi: 10.1523/JNEUROSCI.0729-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur M, Crow M, Richards N, Davey GI, Levine E, Kelleher JH, Agley CC, Denk F, Harridge SD, McMahon SB. Defining the nociceptor transcriptome. Front Mol Neurosci. 2014;7:87. doi: 10.3389/fnmol.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Kim JH, Bazzi M, Robinson S, Collins CA, Ye B. Bimodal control of dendritic and axonal growth by the dual leucine zipper kinase pathway. PLoS Biol. 2013;11:e1001572. doi: 10.1371/journal.pbio.1001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Bellemer A, Yan H, Honjo K, Robertson J, Hwang RY, Pitt GS, Tracey WD. Thermosensory and non-thermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat sensor domains of a thermoTRP channel. Cell Reports. 2012;1:43–55. doi: 10.1016/j.celrep.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.