Abstract
Background
Lysophosphatidic acid acyltransferase (LPAAT) encoded by a multigene family is a rate-limiting enzyme in the Kennedy pathway in higher plants. Cotton is the most important natural fiber crop and one of the most important oilseed crops. However, little is known on genes coding for LPAATs involved in oil biosynthesis with regard to its genome organization, diversity, expression, natural genetic variation, and association with fiber development and oil content in cotton.
Results
In this study, a comprehensive genome-wide analysis in four Gossypium species with genome sequences, i.e., tetraploid G. hirsutum- AD1 and G. barbadense- AD2 and its possible ancestral diploids G. raimondii- D5 and G. arboreum- A2, identified 13, 10, 8, and 9 LPAAT genes, respectively, that were divided into four subfamilies. RNA-seq analyses of the LPAAT genes in the widely grown G. hirsutum suggest their differential expression at the transcriptional level in developing cottonseeds and fibers. Although 10 LPAAT genes were co-localised with quantitative trait loci (QTL) for cottonseed oil or protein content within a 25-cM region, only one single strand conformation polymorphic (SSCP) marker developed from a synonymous single nucleotide polymorphism (SNP) of the At-Gh13LPAAT5 gene was significantly correlated with cottonseed oil and protein contents in one of the three field tests. Moreover, transformed yeasts using the At-Gh13LPAAT5 gene with the two sequences for the SNP led to similar results, i.e., a 25–31% increase in palmitic acid and oleic acid, and a 16–29% increase in total triacylglycerol (TAG).
Conclusions
The results in this study demonstrated that the natural variation in the LPAAT genes to improving cottonseed oil content and fiber quality is limited; therefore, traditional cross breeding should not expect much progress in improving cottonseed oil content or fiber quality through a marker-assisted selection for the LPAAT genes. However, enhancing the expression of one of the LPAAT genes such as At-Gh13LPAAT5 can significantly increase the production of total TAG and other fatty acids, providing an incentive for further studies into the use of LPAAT genes to increase cottonseed oil content through biotechnology.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3594-9) contains supplementary material, which is available to authorized users.
Keywords: Gossypium spp, Lysophosphatidic acid acyltransferase (LPAAT), Gene expression patterns, Sequence variation, Seed oil
Background
Cotton (Gossypium spp.) is not only the world's most important fiber crop, but also an irreplaceable oil crop. The cotton fiber provides 85% of the farm gate value of the cotton crop, and the rest is made up by cottonseed, seed meal and seed oil (National Cottonseed Association, www.cottonseed.org). Cottonseed oil represents a complementary product that can be used in foods or as a material for biodiesel production, with quality and price advantages over rapeseed oil and soybean oil. Cottonseed oil, which makes up approximately 16% of the seed weight, is the most valuable product derived from cottonseed [1]. As a result, there has been a stable increase in the demand for cottonseed oil in the global market. At present, there are two main strategies to genetically improve cottonseed oil yields, i.e., traditional breeding to increase the proportion of oil in cottonseed and transgenic approaches to increase the quality and content of cottonseed oil. However, it is currently unknown if an improvement in cottonseed oil production can be achieved through manipulations of genes involved in plant oil biosynthesis.
Plant oils are mainly composed of triacylglycerols (TAGs), the main storage lipids. There are two pathways for TAG biosynthesis in plants [2]. The first pathway is the de novo biosynthesis from glycerol-3-phosphate and acyl-CoA occurs via the Kennedy pathway, which involves three acyltransferases, i.e., glycerol-3-phosphate O-acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAAT), and diacylglycerol acyltransferase (DGAT) [2]. In mammals, LPAATs are identified as part of the 1-acyl-glycerol-3-phosphate O -acyltransferase (AGPAT) family with LPAAT 1 to 5 corresponding to AGPAT 1 to 5, and LPAAT ƞ to AGPAT 7 [3, 4]. DGAT belongs to the membrane-bound O-acyltransferase (MBOAT) family that includes four subfamilies in plants, i.e., MBOAT1, DGAT1, lysophosphatidylcholine acyltransferase (LPCAT) or lysophospholipid acyltransferase (LPLAT), and homologs of Saccharomyces cerevisiae glycerol uptake protein (GUP) [5–7]. GPAT, AGPAT, LPCAT or MBOAT share common features in acyltransferase motifs, but their overall sequences are distinctly different [3]. The second pathway involves the conversion of lipid phosphatidylcholine (PC) to diacylglycerol (DAG), in which, acyl-CoAs are redirected from PC via the activities of phospholipase C, choline phosphotransferase, and phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) [8, 9], or phospholipid:diacylglycerol acyltransferase (PDAT) [10]. An acyl group can be released from PC to generate lysophosphatidylcholine (LPC) by the reverse reaction of acyl-CoA:LPC acyltransferase [11] or by phospholipase A/acyl-CoA synthase [12].
Phosphatidic acids (PAs) are a key intermediate in the biosynthesis of TAGs and the first acylation step catalyzed by GPAT can be partly bypassed by dihydroxyacetone phosphate acyltransferase in some tissues and organisms. Therefore, LPAATs are vital to PA biosynthesis in catalyzing the incorporation of acyl groups into the sn-2 position of the glycerol backbone [13]. In higher plants, two pathways of LPAAT catalysis control the metabolic flow of lysophosphatidate (LPA) into different PAs in diverse tissues. PAs are either dephosphorylated to form TAGs, or are used in the synthesis of phospholipids, which are important components of biological membranes. Therefore, LPAATs are crucial to the biosynthesis of membrane phospholipids and storage lipids in developing seeds. In fact, in higher plants, LPAATs play an essential role in improving the fatty acid component of seeds by reducing the proportion of saturated fatty acids (SFAs).
LPAATs are thought to be among the most stringent acyltransferases in terms of substrate specificity [14–17]. Studies on the acyl specificity of LPAATs in various plant species have shown that they have stronger affinity for 16–18 carbon monounsaturated fatty acids (MUFAs) than for SFAs [18]. There are at least two classes of LPAAT genes (classes A and B) in different plant species [14]. The characteristics of class A microsomal LPAATs, as defined by Frentzen [14], are ubiquitously expressed throughout the plant, and their enzymes show specificity for 18:1-CoA. These are typical features of enzymes that biosynthesize membrane glycerolipids. Class B LPAATs were first cloned and characterized from coconut (Cocos nucifer) [19]. In this subfamily, LPAAT genes are generally expressed in seeds, and they encode enzymes with a strong substrate specificity towards unusual acyls, such as erucic acid (22:1) and lauric acid (12:0), consistent with the seed microsomal activities and oil composition of the above species. However, recent studies have indicated that the characteristics of B-class LPAATs may differ in other species. For example, the B-class LPAATs in castor (Ricinus communi) showed specificity for 18:1-OH [20].
In a recent study, a rapeseed (Brassica napus) LPAAT was shown to be the rate-limiting enzyme catalyzing the conversion of lysophosphatidate (LPA) to PA in the TAG assembly process [21]. The expression of rapeseed LPAAT genes in Arabidopsis enhanced LPAAT enzymatic activities, which redirected more acyl-CoAs from PC and relieved feedback inhibition of TAG biosynthesis. Also, the overexpression of LPAATs increased seed oil content, thus reinforcing the utility of LPAAT genes as valuable biotechnological tools. Consequently, LPAAT homologs have been cloned from poached eggplant (Limnanthes douglasii) [22], A. thaliana [23], rapeseed [24], coconut [19], corn (Zea mays) [25], Indian cress (Tropaeolum majus) [26], Java olives (Sterculia foetida) [27], and castor [20]. The overexpression of a LPAAT gene from the SLCl-1 mutant yeast stain in A. thaliana and B. napus increased the incorporation of long-chain fatty acids into the sn-2 position of TAGs, leading to an increase in seed oil content by 8–48% [28]. However, thus far, no such a transgenic work has been performed using LPAAT genes from cotton.
To date, little is known about the natural variation of the LPAAT genes and their association with seed oil production in higher plants including cotton. Furthermore, none of the genes encoding for these enzymes has been characterized in cotton; therefore, their role in increasing cottonseed oil content is unknown. In addition, since fatty acids are essential for membrane biosynthesis, saturated very-long-chain fatty acids were found to promote fiber cell elongation in cotton [29]. However, the relationship between natural genetic variation in LPAAT genes and fiber development is also currently unknown. Near-isogenic lines (NILs) or backcross inbred lines (BILs) with the same genetic background but differing in oil content, fiber initiation or elongation can provide important genetic stocks to investigate LPAAT genes with relation to fiber development and seed oil content.
The genomic organization of the LPAAT family has been mostly studied in only Arabidopsis and castor, but not in other species including cotton. Upland cotton (G. hirsutum-AD1) with higher yields and wider adaptations produces approximately 95% of the world cotton fibers and seed. Another cultivated tetraploid, i.e., G. barbadense-AD2, accounting for the remaining world cotton production, produces much longer, stronger and finer fibers with higher cottonseed oil content and lower protein content than Upland cotton. Therefore, BILs developed from an interspecific crossing between the two species provide important genetic stocks to address if LPAATs are associated with cottonseed oil content and fiber quality. The genomes of the two cultivated tetraploid cotton species were recently sequenced [30–33]. It is commonly accepted that, like other three wild tetraploid cotton species including G. tomentosum-AD3 , G. mustelinum-AD4, and G. darwinii-AD5, both cultivated tetraploid species originated from a common ancestor of a natural hybrid between an extant diploid A-genome cotton (likely tree cotton G. arboreum-A2 or levant G. herbaceum-A1) and a wild D genome (likely G. raimondii-D5) species [34]. In this study, we identified 40 LPAAT genes in the two allotetraploid species, i.e., G. hirsutum and G. barbadense and their likely ancestral diploid species G. arboreum and G. raimondii whose genomes were sequenced earlier [35–37]. After an in-silico analysis of the phylogeny, genomic organization and gene structure for the LPAAT gene family, we then conducted a detailed analysis of transcription profiles of the LPAAT gene family by analyzing information in two RNA-seq datasets followed by a quantitative real-time RT-PCR (qRT-PCR) analysis. Sequence variations within each gene were further evaluated for development of polymorphic markers and association analysis with cottonseed oil content and fiber quality traits. One of the LPAAT genes with a sequence variation was used in a transgenic yeast study to confirm its effect on TAGs. The study represents one of the most comprehensive genomic approaches in plants that combine an in-silico bioinformatics analysis with transcriptomics at the RNA level, SNP identification and typing at the DNA level and their associations with seed oil and fiber quality.
Results
Genome-wide identification of LPAAT genes and phylogenetic analysis
The whole genome sequence scaffolds from the ancestral diploids G. raimondii (D5), G. arboreum (A2) and their descendant tetraploids G. hirsutum (AD1) and G. barbadense (AD2) were used for a genome-wide search for LPAAT genes in Gossypium. As a result, we identified 40 LPAATs genes in the four genomes, including 8 Gr_LPAATs based on the sequence information of D5 reported by Paterson et al. [35], 9 Ga_LPAATs in the draft A2 genome reported by Li et al. [37], and 13 and 10 LPAATs on the A subgenome (At-Gh (Gb)_LPAATs) or the D subgenome (Dt-Gh (Gb)_LPAATs) in the draft AD1 genome [31] and AD2 genome [33], respectively. The further phylogenetic analysis of putative cotton LPAAT proteins included some previously characterized plant acyltransferases (Fig. 1). Based on their predicted protein sequences, the LPAAT genes were classified into four subfamilies; A-class LPAAT (LPAAT2/3) (11 genes), LPAAT4/5 (12 genes), B-class LPAAT (6 genes), and LPAAT1 (plastidial) (11 genes) (Table 1). Also, some close paralogous relatives of LPAATs were identified in the four Gossypium species.
Fig. 1.
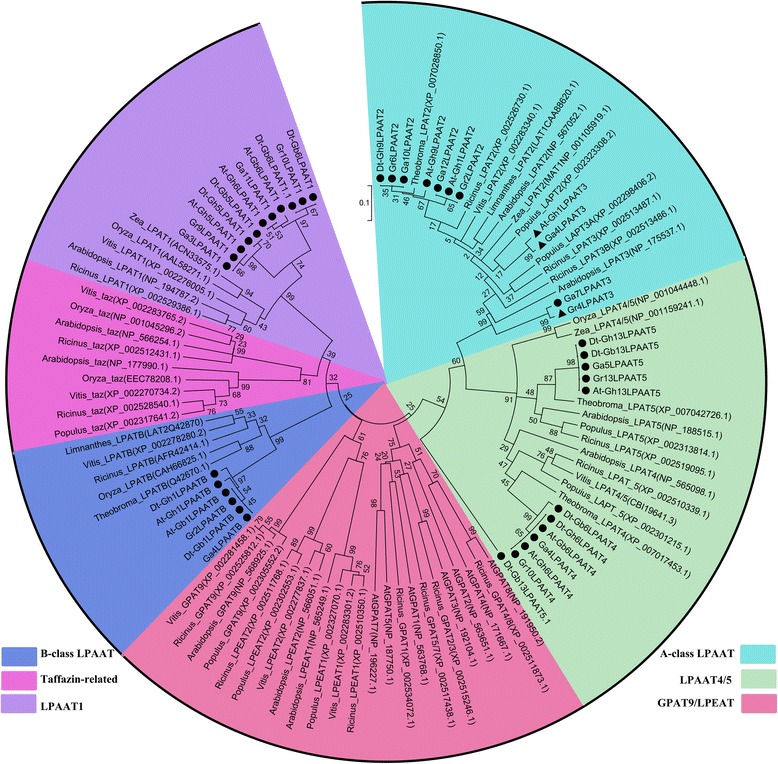
A phylogenetic tree of LPAATs in four Gossypium species. Sequences of other LPAAT genes and LPAAT-related genes were downloaded from NCBI website. ● Gossypium; ▼ three putative isoforms in Gossypium
Table 1.
Characteristics of LPAAT genes and predicted properties of LPAAT proteins in four Gossypium species
| Gene name | Gene identifier | Size (AA) | MW (KD) | pI | Subcellular localization |
|---|---|---|---|---|---|
| A-class LPAAT(LPAAT2/3) (Box I: NHXXDXD; Box III: FVEGTR) | |||||
| Dt-Gh9LPAAT2 | Gh_D09G1169 | 387 | 43.5451 | 9.46 | Endoplasmic reticulum |
| Gr6LPAAT2 | Gorai.006G138600.1 | 387 | 43.501 | 9.53 | Endoplasmic reticulum |
| Ga10LPAAT2 | Cotton_A_07803 | 387 | 43.501 | 9.53 | Endoplasmic reticulum |
| At-Gh9LPAAT2 | Gh_A09G1164 | 387 | 43.492 | 9.63 | Endoplasmic reticulum |
| At-Gh1LPAAT2 | Gh_A01G1672 | 381 | 42.9697 | 9.75 | Endoplasmic reticulum |
| Ga12LPAAT2 | Cotton_A_15334 | 381 | 42.9297 | 9.75 | Endoplasmic reticulum |
| Gr2LPAAT2 | Gorai.002G230000.1 | 381 | 42.9136 | 9.68 | Endoplasmic reticulum |
| At-Gh1LPAAT3 | Gh_A01G1107 | 230 | 25.8612 | 9.65 | Plasma membrane |
| Ga4LPAAT3 | Cotton_A_39902 | 260 | 29.2383 | 8.95 | Plasma membrane |
| Ga7LPAAT3 | Cotton_A_12015 | 370 | 42.0493 | 9.11 | Endoplasmic reticulum |
| Gr4LPAAT3 | Gorai.004G019300.1 | 204 | 22.9039 | 9.4 | Mitochondrial inner membrane |
| LPAAT4/5 LPAAT (Box I: NHXXDXD; Box III: FVEGTR) | |||||
| Dt-Gh6LPAAT4 | Gh_D06G0073 | 377 | 43.6875 | 8.86 | Plasma membrane |
| Gr10LPAAT4 | Gorai.010G012300.1 | 377 | 43.6663 | 8.77 | Plasma membrane |
| At-Gh6LPAAT4 | Gh_A06G0100 | 377 | 43.7905 | 8.77 | Plasma membrane |
| Ga4LPAAT4 | Cotton_A_06880 | 377 | 43.7715 | 8.67 | Plasma membrane |
| Dt-Gb6LPAAT4 | Gbscaffold17837.25.0 | 377 | 43.7625 | 8.77 | Plasma membrane |
| At-Gb6LPAAT4 | Gbscaffold22202.16.0 | 377 | 43.6603 | 8.77 | Plasma membrane |
| Gr13LPAAT5 | Gorai.013G052400.1 | 379 | 44.103 | 8.59 | Plasma membrane |
| At-Gh13LPAAT5 | Gh_A13G0409 | 354 | 41.1643 | 8.9 | Mitochondrial inner membrane |
| Dt-Gh13LPAAT5 | Gh_D13G0458 | 372 | 432.95 | 8.79 | Plasma membrane |
| Ga5LPAAT5 | Cotton_A_17180 | 384 | 44.6355 | 9.06 | Plasma membrane |
| Dt-Gb13LPAAT5.1 | Gbscaffold19187.1.0 | 255 | 29.6291 | 9.14 | Plasma membrane |
| Dt-Gb13LPAAT5 | Gbscaffold19187.1.3 | 372 | 43.332 | 8.9 | Plasma membrane |
| B-class LPAAT (Box I: NHXSXXD; Box III: FPEGT) | |||||
| At-Gh1LPAATB | Gh_A01G0616 | 358 | 40.2893 | 9.69 | Endoplasmic reticulum |
| Dt-Gh1LPAATB | Gh_D01G0632 | 311 | 35.1431 | 9.78 | Mitochondrial inner membrane |
| Ga4LPAATB | Cotton_A_04784 | 288 | 32.8554 | 9.65 | Mitochondrial inner membrane |
| Gr2LPAATB | Gorai.002G088200.1 | 299 | 33.7234 | 9.79 | Mitochondrial inner membrane |
| Dt-Gb1LPAATB | Gbscaffold8424.3.0 | 311 | 35.1891 | 9.75 | plasma membrane |
| At-Gb1LPAATB | Gbscaffold10133.29 | 311 | 35.2282 | 9.77 | plasma membrane |
| LPAAT1 (Plastidial) (Box I: NHQXXXD; Box III: FPEGT) | |||||
| Ga3LPAAT1 | Cotton_A_01233 | 312 | 34.2761 | 9.8 | Microbody (peroxisome) |
| At-Gh5LPAAT1 | Gh_A05G0428 | 358 | 39.6192 | 9.8 | Plasma membrane |
| Gr9LPAAT1 | Gorai.009G056600.1 | 355 | 39.3199 | 9.72 | Plasma membrane |
| Dt-Gh5LPAAT1 | Gh_D05G0546 | 355 | 39.392 | 9.69 | Mitochondrial inner membrane |
| Gr10LPAAT1 | Gorai.010G113400.1 | 335 | 37.306 | 9.81 | Plasma membrane |
| Ga11LPAAT1 | Cotton_A_25757 | 238 | 26.2541 | 9.65 | Plasma membrane |
| Dt-Gb5LPAAT1 | Gbscaffold12734.5.1 | 319 | 35.4066 | 9.59 | Microbody (peroxisome) |
| Dt-Gb6LPAAT1 | Gbscaffold3314.7.0 | 230 | 25.4521 | 9.65 | Endoplasmic reticulum |
| At-Gb6LPAAT1 | Gbscaffold6506.1.0 | 322 | 35.6169 | 9.8 | Plasma membrane |
| Dt-Gb6LPAAT1.1 | Gbscaffold3314.8.0 | 335 | 37.308 | 9.74 | Endoplasmic reticulum |
| At-Gh6LPAAT1 | Gh_A06G0884 | 322 | 35.7141 | 9.85 | Golgi body |
AA amino acid, pI theoretical isoelectric point, MW theoretical molecular weight, KD Kilo Dalton
The LPAAT2/3 subfamily, also known as the A-class LPAAT family, encodes microsomal LPAATs that show a generalized expression pattern. This subfamily includes AtLPAAT2 from Arabidopsis [23]. Within the LPAAT2/3 subfamily, Gr6LPAAT2, Ga10LPAAT2 and two copies of GhLPAAT2 from tetraploid G. hirsutum (Dt-Gh9LPAAT2 and At-Gh9LPAAT2) encoded proteins with a high level of similarity to the cacao TcLPAAT2 protein, and were identified as putative orthologs from different genomes or subgenomes, i.e., homoelogs (Fig. 1). Interestingly, three putative LPAAT2/3 isoforms were identified in Gossypium. These three LPAAT isoforms appeared to have incomplete conserved domains (Additional file 1: Figure S2). Specifically, Ga4LPAAT3 and At-Gh1LPAAT3 had only one LPAAT-characteristic Box III (ΦFPEGTR-G, where Φ is a hydrophobic residue) [38], a motif that is well conserved among proteins with the ΦFVEGTR consensus sequence in the LPAAT2/3 subfamily. Gr4LPAAT3 had Boxes III and IV (ΦPΦΦPΦΦΦ), but an imprecise VLIPRTKG consensus sequence. The sequence around Box I (Φ-NHQS-ΦDΦΦ) was quite similar among the LPAAT2/3 proteins, conforming to the consensus sequence NHXSDIDWL, where X usually indicates an arginine (R) residue [38]. However, this consensus sequence was different (NHVSDSDTΦ) in Gr7LPAAT3. The same held true for sequences around Box II (G-ΦFIDR), where there was a well-conserved E(D)YLFLER motif in most members of the cotton LPAAT2/3 subfamily (Additional file 1: Figure S2), with an exception of Gr7LPAAT3 having a different sequence (ESIFLDR). The consensus sequences of Boxes III and IV were conserved among all of the LPAAT2/3 proteins.
Twelve LPAATs were clustered into the LPAAT4/5 subfamily which was a closely related sister group with LPAAT2/3 (Fig. 1), although there were some sequence differences around various LPAAT boxes. For example, there was an aspartic acid (D) residue near Box I in the LPAAT2/3 subfamily, but a glutamic acid (E) residue near Box I in the LPAAT4/5 subfamily. This variation may explain why these sequences were grouped into two subfamilies.
Six Gossypium gene sequences designated as a paralogous LPAAT group clustered together in the B-class LPAAT subfamily, which encodes LPAATs that are usually, but not always, seed-specific and shows substrate specificities for unusual acyl groups [14]. As the sister group to the B-class subfamily the LPAAT1 (plastidial) subfamily contained eleven LPAATs in two paralogous groups from two different branches. The LPAAT1 (plastidial) subfamily includes AtLPAAT1 in Arabidopsis [39] (Fig. 1). A close relationship between B-class LPAATs and plastidial LPAATs was revealed in the phylogenetic analysis, consistent with the results of a previous report [39]. The B-class LPAATs and the plastidial LPAATs shared the same conserved motif (FPEGT) around Box III, except for Ga4LPAATB which lacked Box III (Additional file 2: Figure S3).
In plants and other organisms, LPAAT activities have been detected in the endoplasmic reticulum [23], plasma membrane [39], and mitochondria [40]. Table 1 summarizes their predicted subcellular localizations of the putative LPAAT proteins in cotton, together with information on the predicted length, molecular weight (MW), and isoelectric point (pI). Each LPAAT subfamily was predicted to a specific subcellular location. For example, within the A-class LPAAT (LPAAT2/3) subfamily, 72.7% of the LPAAT proteins were predicted to localize in the endoplasmic reticulum; and LPAAT4/5, B-class LPAATs, and LPAAT1 (plastidial) were predicted to be in the plasma membrane, the mitochondrial inner membrane, and the plasma membrane, respectively (Table 1).
Structure and domain analysis of putative LPAAT genes in sequenced diploid and tetraploid Gossypium genomes
We used the GFF (Generic Feature Format) files of the four Gossypium species to analyze the exon–intron structure of putative LPAAT genes. Figure 2 shows the exon–intron structure of each subfamily. The number and location of introns varied among subfamilies, but there were some common features. In the A-class subfamily, eight deduced genes included 11 exons, but the locations of introns differed. At-Gh1LPAAT3, Ga4LPAAT3 and Gr4LPAAT3 were closely related to LPAAT2, but had only nine, six and five exons, respectively. Within this subfamily, homoelogous LPAATs had the same structure, such as Dt-Gh9LPAAT2, Gr6LPAAT2, At-Gh9LPAAT2 and Ga10LPAAT2. Members of the LPAAT4/5 subfamily had three exons, except for Ga5LPAAT5 and Dt-Gb13LPAAT5 which had four and two exons, respectively. These similarities in gene structure were also observed among members of the B-class and LPAAT1 (plastidial) groups (Fig. 2). The results of the domain analysis indicated the presence of a highly conserved LPLAT_LCLAT1-like domain in the A-class and LPAAT4/5 LPAATs, and an LPLAT_AGPAT-like domain in the B-class and plastidial LPAATs (Additional file 3: Table S3). In conclusion, members belonging to the same subfamilies of the phylogenetic tree had a similar gene structure and conserved motif, consistent with the results of the phylogenetic analysis.
Fig. 2.
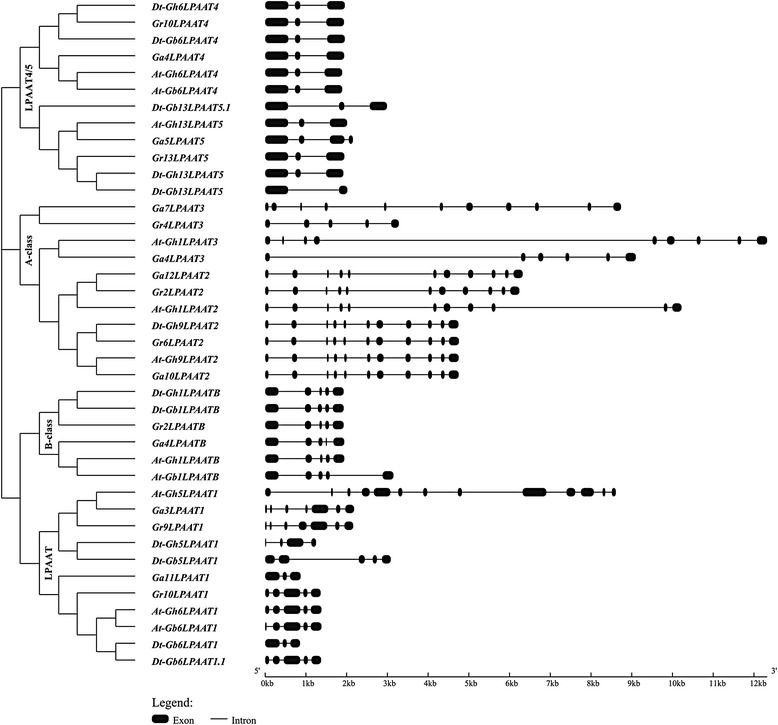
Genetic structures of LPAAT genes in four Gossypium species. Black boxes show exons and lines show introns
Chromosomal distribution of LPAAT genes on Gossypium genomes
After integrating chromosomes of the sequenced cotton genomes excluding G. barbadense as its LPAAT genes were not physically well mapped, we found that the LPAAT family members were distributed unevenly among chromosomes. Among the candidate LPAAT genes, eight GrLPAATs were located in six D5 chromosomes (D5/c2, D5/c4, D5/c6, D5/c9, D5/c10, and D5/c13), and nine GaLPAATs to seven A2 chromosomes (A2/c3, A2/c4, A2/c5, A2/c7, A2/c10, A2/c11, and A2/c12) (Additional file 4: Figure S4). Because there are many syntenic gene blocks between the chromosomes of G. raimondii and G. arboreum, the dotted lines in Additional file 4: Figure S4 link homoelogous LPAATs between A2 and D5 chromosomes, but the chromosomes linked may not be homoelogous between genes from the two subgenomes of the tetraploid species. For G. barbadense, three GbLPAATs matched to the At subgenome, and seven GbLPAATs matched to the Dt subgenome (Additional file 4: Figure S4). For G. hirsutum, five GhLPAATs were on the Dt subgenome, and eight GhLPAATs on the At subgenome (Fig. 3).
Fig. 3.
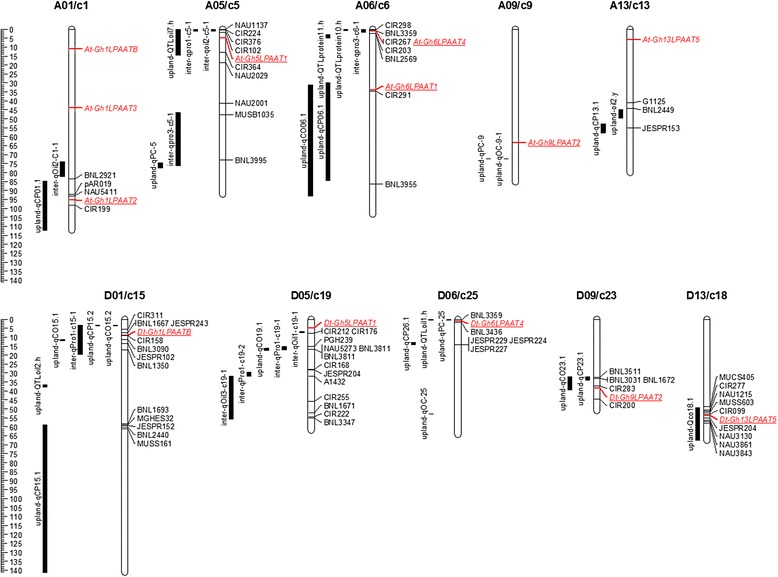
Co-localisation analysis of LPAAT genes with seed oil and protein quantitative trait loci (QTL). Red color shows LPAAT genes and underlines indicate the LPAAT genes co-localisation with seed oil and protein QTL
Analysis of transcript levels of GhLPAAT genes in two RNA-seq databases of tetraploid Upland cotton
To reveal a general pattern of gene expression for the LPAAT genes identified, we analyzed the transcript profiles of LPAAT genes in two RNA-seq datasets: one with transcriptomic information for two Upland BILs, i.e., NMGA-062 vs. NMGA-105 (with G. barbadense germplasm introgression), with differing fiber lengths but similar seed oil content, and the other containing transcriptomic information for Upland Xuzhou 142 vs. its fiberless and fuzzless mutant Xuzhou 142 fl (with a likely G. barbadense origin) [41] which differed in fiber initiation and cottonseed oil content [42]. The transcript abundances of all the 13 putative LPAAT genes were determined at the fiber initiation stage (0 DPA) and the elongation stage (3–10 DPA). Of the 13 genes in the sequenced G. hirsutum genome, two (At-Gh1LPAAT3 and Dt-Gh13LPAAT5) was not expressed. The other 11 genes were transcribed at both stages of fiber development and in all tissues represented in the two RNA-seq datasets. Based on the RNA-seq data of the two BILs, the transcript levels of the A-class LPAAT genes were higher than those of the genes in the other three subfamilies in both genotypes. Interestingly, the transcript levels of Dt-Gh9LPAAT2 and At-Gh9LPAAT2 were similar in maintaining a high expression level at three different stages, but the At-Gh1LPAAT2 had a low level of expression at 0 DPA fibers ovules, with the highest level observed in 10 DPA ovules in both genotypes (Fig. 4a). There was no significant difference in expression levels between the two BIL genotypes differing in fiber length but with similar seed oil content.
Fig. 4.
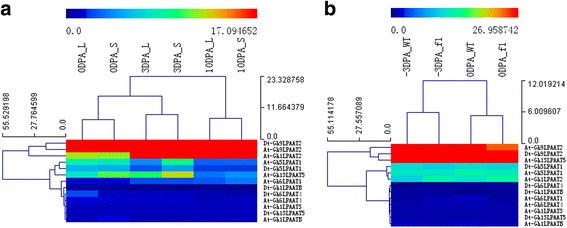
Transcript profiles of LPAAT genes in Gossypium hirsutum. a A heat map showing transcript levels of 13 LPAAT genes in ovules at three stages (0 DPA, 3 DPA, 10 DPA; shown above each lane) in two BILs. b Transcript levels of 13 LPAAT genes in −3 DPA and 0 DPA ovules of Xuzhou 142 and Xuzhou 142 fl. Color scale above dendrogram shows relative transcript levels. "L" and "S" indicate long fiber length line NMGA-062 and short fiber length line NMGA-105, respectively. "WT" and "fl" indicate Xuzhou 142 and its Xuzhou 142 fl mutant, respectively
Similar patterns were observed in the other RNA-seq dataset for Xuzhou 142 and its fiberless and fuzzless mutant Xuzhou 142 fl. The exception was that At-Gh13LPAAT5 showed higher transcript levels in Xuzhou 142 and Xuzhou 142 fl than in the other two BILs due to different genetic backgrounds (Fig. 4b). The results indicate that the expression of LPAAT genes at the transcription level is not involved in the natural variation of fiber length between Upland and G. barbadense and fiber initiation as controlled by the genes in Xuzhou 142 fl.
Quantitative real-time RT-PCR analysis of transcript profiles of eight GhLPAAT genes
Due to a high homology for genes within the same subfamily, we chose eight of the above eleven expressed LPAAT genes in Upland cotton that have homologs with its ancestral diploid species G. raimondii and G. arboreum to design gene-specific primers (Additional file 3: Table S1) for a further evaluation of the transcript levels. The eight genes included two A-class genes, two LPAAT4/5 genes, three plastidial LPAAT genes, and one B-class gene. Using qRT-PCR analyses, we further analyzed the gene transcript levels in roots, stems, leaves, petals, ovules, and fibers of Xuzhou 142 and the Xuzhou 142 fl. We analyzed ovules at different developmental stages, i.e., −3, −1, 0, 1, 3, and 5 DPA (fibers and ovules were not separated), and 10, 15, 20, and 25 DPA (fibers and ovules were separated) as cottonseed oil accumulates rapidly in ovules after 15–20 DPA. The results showed that the eight LPAAT genes were expressed in all the organs and tissues tested throughout the plant. However, seven LPAAT genes (except for Dt-Gh6LPAAT4) showed the highest transcript levels in 10 DPA fibers. At-Gh6LPAAT1, At-Gh1LPAAT2, At-Gh1LPAAT3, At-Gh13LPAAT5 and Dt-Gh1LPAATB were expressed preferentially at the late period of ovule development in Xuzhou 142 fl, especially at 20 DPA. The transcript profiles of the two A-class genes were similar; however, in the LPAAT4/5 subfamily, Dt-Gh6LPAAT4 and At-Gh13LPAAT5 showed an opposite trend in their transcription patterns. At −3 to 5 DPA ovules (Xuzhou 142 and Xuzhou 142 fl) and 10 DPA fibers (Xuzhou 142), Dt-Gh6LPAAT4 showed very low transcript levels, while At-Gh13LPAAT5 was highly expressed (Fig. 5). Moreover, Dt-Gh6LPAAT4 showed very high transcript levels, but At-Gh13LPAAT5 had a low level of expression at 15–25 DPA fibers. In the B-class subfamily, Dt-Gh1LPAATB did not show a seed-specific expression pattern as expected, because it was also expressed in all of the other organs (Additional file 5: Figure S5).
Fig. 5.
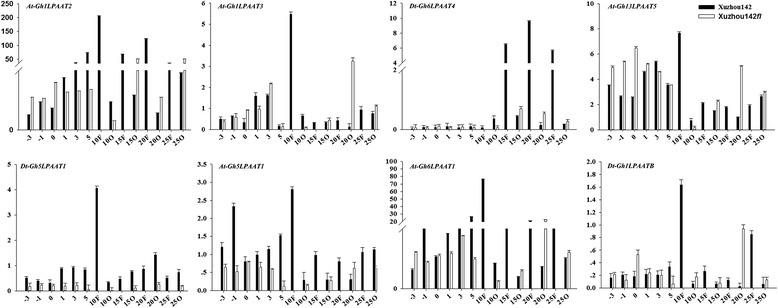
Expression patterns of eight LPAAT genes in Xuzhou 142 and Xuzhou 142 fl. Y-axis, relative expression levels. X-axis, days post anthesis. Fibers and ovules were not separated at −3, −1, 0, 1, 3, and 5 DPA. 10 F, 15 F, 20 F, 25 F DPA and 10O, 15O, 20O, and 25O DPA indicate fibers and ovules, respectively, at different stages. Error bars show standard deviation (S.D.) calculated from three replications
Co-localisation of LPAAT genes with quantitative trait loci (QTL) for seed oil and protein contents
To analyse if any of the GhLPAATs is genetically associated with cottonseed oil content, a co-localisation analysis of GhLPAATs with quantitative trait loci (QTL) for seed oil and protein contents was performed. We first downloaded QTL for these two traits from two types of genetic populations, i.e., intraspecific G. hirsutum and interspecific G. hirsutum × G. barbadense populations [43]. As a result, 10 GhLPAATs on five pairs of homeologous chromosomes were located within a 25 cM region of cottonseed oil or protein QTL, including At-Gh1LPAAT2 on chromosome A01/c1, At-Gh5LPAAT1 on A05/c5, At-Gh6LPAAT4 and At-Gh6LPAAT1 on A06/c6, At-Gh9LPAAT2 on A09/c9, Dt-Gh1LPAATB on D01/c15, Dt-Gh13LPAAT5 on D13/c18, Dt-Gh5LPAAT1 on D05/c19, Dt-Gh9LPAAT2 on D09/c23, and Dt-Gh6LPAAT4 on D06/c25 except for At-Gh13LPAAT5 on A13/c13 (Fig. 3). Interestingly, the respective cottonseed oil or protein QTL co-localised LAPPT genes on six chromosomes belong to three pairs of the homeologous chromosomes, i.e., LPAAT1 on A05 and D05, LPAAT2 on A09 and D09, and LPAAT4 on A06 and D06, while other two pairs of homeologous chromosomes (i.e., A01 vs. D01 and A13 vs. D13) carry cottonseed oil or protein QTL but with different LPAAT genes. These co-localised QTL include five QTL (on A09 and D09, A13 and D13, and D06) from the Upland population only and five (on A01 and D01, A05 and D05, and A06) from both Upland and interspecific populations (Additional file 3: Table S4). However, since a 25-cM chromosomal region may contain several hundred genes [30, 31], the co-localisation of a seed oil QTL with a LPAAT gene may not indicate a causative relationship between the natural variation of the LPAAT gene and seed oil content (see next section).
Sequence variation of LPAAT genes and its association with fiber quality and seed oil and protein contents
Sequence variations in the predicted genes between the sequenced G. hirsutum (TM-1) and G. barbadense (3–79 and Xinhai 21) were further analyzed. The results showed that most of the sequence variations were detected between homeologous genes from the A- and D- subgenomes, while only a few single nucleotide polymorphisms (SNPs) were predicted in homologous genes between the two cultivated species or between 3-79 and Xinhai 21 within G. barbadense (Additional file 6: Figure S6). The results imply that the differences in seed oil and protein contents and fiber quality traits between the two species are unlikely related to the natural sequence variations in the LPAAT genes, which is verified by the following correlation analysis between LPAAT gene markers and the seed and fiber quality traits.
The above RNA-seq datasets were further used to identify SNPs in the LPAAT genes within Upland cotton. In comparison with the sequenced TM-1 genome, we identified 43 SNPs in the cDNA sequences of seven LPAAT genes in the NMGA-062 and NMGA-105 dataset and 63 SNPs in five LPAAT genes in the Xuzhou 142 and Xuzhou 142 fl dataset (Additional file 3: Tables S5 and S6). However, as expected, most of the sequence differences were between TM-1 and the pair of the two BILs in the SG 747 background or the NILs in the Xuzhou 142 background (i.e., BILs vs. TM-1 or Xuzhou 142 WT/fl vs. TM-1). Only eight SNPs between NMGA-062 and NMGA-105 BILs were detected in five LPAAT genes (Additional file 3: Table S5). Between Xuzhou 142 and Xuzhou 142 fl NILs, eight SNPs were identified in four LPAAT genes, confirming the non-near-isogenic status of the two genotypes used in this study, which was recently reported by Ma et al. [41].
For an association analysis between the existence of SNPs in the LPAAT genes (detected from a comparison between the two BILs, i.e., NMGA-062 and NMGA-105) and cottonseed oil and protein contents and fiber traits, primers were designed to amplify fragments containing these SNPs using single strand conformation polymorphisms (SSCPs), which were used to screen the BIL population of 146 lines derived from a backcross between Upland SG 747 and G. barbadense Giza 75. As a result, one polymorphic SSCP marker was developed from a pair of primers (Additional file 3: Table S2) designed for At-Gh13LPAAT5, named At-Gh13LPAAT5-342 (Additional file 7: Figure S7). A transition, A/G (or T/C), was located in the 342th nucleotide of At-Gh13LPAAT5 (Additional file 3: Table S6), but the mutation appears to be synonymous with no change in amino acid. To examine the relationship of the SSCP marker with seed oil and protein contents and fiber traits, a correlation analysis was performed in the BIL population of 146 lines tested in 2006, 2008 and 2009. The marker was found to be significantly associated with both seed oil and protein content in the BILs tested in 2008. The correlation between the presence of the SSCP marker and seed oil content was significantly negative (−0.281, P < 0.01) and significantly positive with seed protein content (0.245, P < 0.01) (Table 2). The results indicated that the presence of the SNP allele reduced cottonseed oil and increased protein content. Since seed oil and protein contents are negatively correlated (−0.905, P < 0.01), which was also reported by others [44], the opposite effects of the marker on seed oil and protein contents are expected. Moreover, the SSCP marker had a negative correlation with only one fiber quality trait, i.e., fiber uniformity (−0.176, P < 0.05) (Table 2). However, the above results obtained in 2008 were not confirmed by the results from the same BIL population tested in 2006 and 2009, as no significant correlations were detected between the SNP and seed oil and protein contents or fiber quality (Table 2). Therefore, the inconsistent association between the marker of this LPAAT gene and seed oil and protein contents or fiber quality indicates that the natural variation of this LPAAT gene may not affect seed oil and protein synthesis or fiber development, but more studies are needed. Overall, our results indicated that sequence variations in most if not all of the LPAAT genes are not associated with the natural variation (i.e., QTL) for seed oil content or fiber quality in cotton.
Table 2.
The correlation coefficients between the marker At-Gh13LPAAT5-342 developed from the LPAAT gene and seed oil and protein contents and fiber quality traits in the backcross inbred line population of SG 747 × Giza 75 hybrid tested in 2006, 2008 and 2009
| 2006 | 2008 | 2009 | |
|---|---|---|---|
| Fiber length (mm) | −0.082 | −0.071 | 0.078 |
| Fiber uniformity (%) | −0.063 | −0.176b | 0.02 |
| Micronaire (unit) | −0.003 | −0.089 | 0.091 |
| Fiber strength (cN/tex) | −0.007 | −0.042 | 0.088 |
| Seed oil (%) | −0.122 | −0.281a | 0.119 |
| Seed protein (%) | nt | 0.245a | 0.146 |
a and b indicate correlation at the 0.01 and 0.05 significant levels, respectively. nt not tested
Content of TAG and fatty acids in At-Gh13LPAAT5 transgenic yeast strains
Because At-Gh13LPAAT5 is not located within the QTL region for the cottonseed oil and protein QTL on chromosome A13/c13 (Fig. 3) and the SNP detected in this gene was synonymous with no change in amino acid, the detected correlation between the SNP and cottonseed oil and protein content in the BIL population tested in 2008 should not be causative. To validate the hypothesis that both gene alleles producing polypeptides of the same sequence have the same effect on oil biosynthesis, we compared the effect of the At-Gh13LPAAT5 gene from the Upland, G. hirsutum (as represented by sequences from TM-1 and Xuzhou 142) and G. barbadense (as represented by sequences from G. barbadense 3–79 and G. hirsutum Xuzhou 142 fl) sources on oil synthesis in transgenic yeasts. A full length cDNA with an 1119-bp open reading frame (ORF) from both sources was cloned into the yeast expression vector pPIC3.5 K and transformed into yeasts by LiCl. The GC/MS analysis determined six major fatty acids in the transgenic yeasts (represented by three transgenic yeasts for each source), i.e., palmitic acid (C16:0), hexadecenoic acid (C16:1), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2) and α-linolenic acid (C18:3) (Fig. 6a). The analysis of fatty acid composition revealed that the expression of At-Gh13LPAAT5 in the yeast led to an approximately 25–31% increase in palmitic acid (C16:0) and oleic acid (C18:1) when compared to the non-transgenic control strains (Fig. 6b), and no significant differences were detected between the transgenic yeasts with genes from the Upland and G. barbadense sources. As compared to the empty vector transformants, the total TAGs in the transgenic strains were also increased within the range of 16–29%, and transgenic yeasts with genes from both sources also showed similar results. The qRT-PCR analysis showed that the gene expression pattern was essentially similar to the concentration of TAGs among the three transgenic yeast strains tested (Fig. 6 c, d). These results indicate that expression of the foreign gene At-Gh13LPAAT5 in the yeast can enhance oil content and selectively incorporate fatty acids into TAGs, and the gene alleles from both the Upland and G. barbadense sources has a similar effort.
Fig. 6.
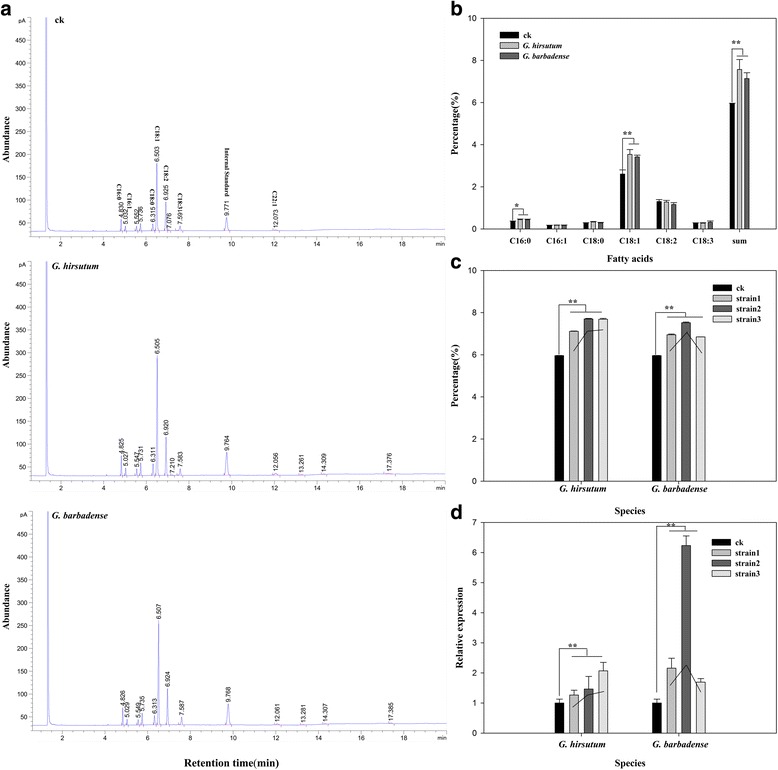
Content of TAG and fatty acids in At-Gh13LPAAT5 transgenic yeast strains. a Gas chromatography/mass spectrometry chromatogram of fatty acids from transgenic yeasts. b The content of fatty acids in transgenic yeasts. c The total TAG content in three transgenic yeast strains from two backgrounds G. hirsutum and G. barbadense, respectively. d Expression patterns of At-Gh13LPAAT5 in three transgenic yeast strains. ** and * indicate correlation at the 0.01 and 0.05 significant levels, respectively
Discussion
The LPAAT gene family in Gossypium
The results of this study revealed details of 40 LPAAT genes in the multigene family in four Gossypium species with sequenced genomes: diploids G. raimondii and G. arboreum, and their descendant tetraploids G. hirsutum and G. barbadense. The translated protein sequences were clustered into four groups (subfamilies), consistent with other previous reports [20]. Consistent with previous studies that whole genome duplication events occurred during the evolution of Gossypium [37], we observed that each GrLPAAT gene or GaLPAAT gene in each of the diploid species corresponded to two GhLPAAT genes in the tetraploid cotton belonging to one homeologous LPAAT group. The eleven putative plastidial LPAATs in cotton showed a high similarity to the previously characterized phospholipid/glycerol acyltransferase family proteins from coconut. Proteins in the A-class LPAAT (LPAAT2/3) group, the B-class LPAAT group, and the plastidial LPAAT group were shown to have LPAAT activity [20]. However, in Arabidopsis, members of the LPAAT4/5 subfamily did not show LPAAT activity in vitro [25].
In this study, At-Gh1LPAATB, Dt-Gh1LPAATB, At-Gb1LPAATB, Dt-Gb1LPAATB, Ga4LPAATB, and Gr2LPAATB, were predicted to be in the B-class LPAAT group in cotton. Previous reports suggested that the B-class proteins are seed-specific isoenzymes, such as LdLPAATB and CnLPAATB isolated from poached eggplant and coconut, respectively [19, 22]. However, recent studies have shown that B-class proteins are not restricted to plant seeds. RcLPAATB (formerly called RcLPATB) identified in castor, was shown to express at similar levels in other tissues/organs of the plant [20]. We obtained a similar result in that there were very low transcript levels of B-class LPAATs in ovules based on our RNA-seq analyses. This is possibly because the RNA-seq data were obtained at the fiber initiation stage, and not the seed oil accumulation stage. The qRT-PCR results showed that Dt-Gh1LPAATB did not show a typical B-class expression pattern, but was expressed throughout the plant.
Expression of LPAAT genes and their natural variations in relation to seed oil content and fiber development
In higher plants, LPAATs catalyze the incorporation of acyl groups into the sn-2 position of LPAs to yield PAs, the key intermediates in the synthesis of membrane phospholipids or TAG storage lipids [11]. Each cotton fiber is a single cell that can be used as a biological model to study cell expansion. During the rapid polarized expansion of the fiber cell, abundant membrane phospholipids are required to synthesize the cytomembrane so that the fiber cell can elongate from 10 to 15 mm at 5–10 DPA to 25–30 mm at 25 DPA. Cottonseed oil accumulates later in ovules after 15–20 DPA. Therefore, in developing cottonseeds, LPAATs maybe play a dual role in that they produce raw materials for the biosynthesis of the membrane as the cell elongates and also catalyze the conversion of LPAs to TAGs. To test this hypothesis, we analyzed LPAATs in Xuzhou 142 and Xuzhou 142 fl, which have a similar genetic background based on the origin of the mutant. Xuzhou 142 fl, the natural fuzzless-lintless mutant from the fibered wild type Xuzhou 142 with both lint and fuzz, not only has few or no fibers, but also has higher seed oil content than that of Xuzhou 142 [42].
The qRT-PCR results revealed different expression patterns of LPAAT genes, even those in the same subfamily. These findings suggested that LPAAT genes have diverse developmental expressions. Based on their expression patterns, the LPAATs were grouped into two categories. The first category (At-Gh6LPAAT1, At-Gh1LPAAT2, At-Gh1LPAAT3, At-Gh13LPAAT5 and Dt-Gh1LPAATB) showed high expression levels at two significant phases, i.e., during the rapid phase of fiber elongation (10 DPA) in Xuzhou 142 and at the beginning of fatty acid accumulation in ovules of Xuzhou 142 fl (20 DPA). At 10 DPA, fiber cells require membrane phospholipids to form new membranes. It is possible that these genes were preferentially expressed during membrane biosynthesis in cotton. Interestingly, these genes showed a sharp increase in transcript levels in the 20 DPA Xuzhou 142 fl ovules, but slower increases in transcript levels in Xuzhou 142 at 20 DPA ovules and during other stages of Xuzhou 142 fl ovule development. This may be because the fuzzless/lintless mutant Xuzhou 142 fl seed has few or no fibers, and therefore, does not require large amounts of membrane phospholipids for new cytomembranes of fiber development. In that case, the LPAATs would synthesize TAGs for seed oil production, rather than membrane phospholipids for developing fibers. The second group of genes (At-Gh5LPAAT1, At-Gh6LPAAT1, Dt-Gh9LPAAT1 and Dt-Gh6LPAAT4) was expressed preferentially in fiber tissues, especially Dt-Gh6LPAAT4. Their expression levels were very low in ovules of both Xuzhou 142 and Xuzhou 142 fl. This result suggested that these three LPAATs in this group may be specific to membrane biosynthesis, but more studies are needed. However, it should also be pointed out that, the expression magnitudes at the RNA level might not necessarily reflect the expression of these genes at the enzymatic level. In this study, the expression patterns of the LPAAT genes between the two genotypes were overall similar, and no SNPs of LPAATs identified and developed from a comparison between the two lines were located within the two gene regions for the fiberless and fuzzless traits [41]. We therefore conclude that none of the LPAAT genes was directly involved with the fiberless and fuzzless mutation and its higher oil content in Xuzhou 142 fl. Therefore, a segregating population between the two genotypes is unnecessary to address the role of LPAATs in the molecular mechanism of the fiberless and fuzzless trait in this mutant.
To further test if LPAAT genes are related to cotton fiber elongation, a pair of BILs with different fiber length and similar seed oil content was used. Our results indicated that the expression levels of LPAAT genes were similar between the two lines, suggesting that the genetic differences in fiber elongation between the two NILs are not associated with the expression of LPAAT genes. Furthermore, a physical mapping of LPAAT genes with QTL for cottonseed oil content showed that 10 of the 13 LPAAT genes were located within QTL regions (i.e., within 25 cM) for seed oil or protein content. However, to further test if LPAAT genes are genetically related to cottonseed oil production and fiber quality traits, a backcross inbred line population (from which the two BILs were selected) of 146 lines differing in fiber quality traits and seed oil content and fiber quality were used. The results indicated that, of all the potential SNPs identified in the LPAAT genes, only one marker from one gene At-Gh13LPAAT5 showed a significant correlation with cottonseed oil content and fiber uniformity in only one of the three field tests, suggesting that sequence differences in most if not all of the LPAAT genes are not involved in the natural genetic variation for seed oil production and fiber quality traits in cotton.
Furthermore, we designed an experiment to see if the two LPAAT variants of the same gene conferred by a SNP (which differed in a synonymous base change) had a similar effect on oil content and composition in a yeast system. Both variants were supposed to produce the protein with the same sequence and therefore a similar biochemical activity. As expected, the transgenic yeast experiment showed that the total TAG and palmitic acid and oleic acid increased compared to the nontransgenic yeast control, similar to these obtained in other transgenic plant studies [28]. The results further showed that both sources of the At-Gh13LPAAT5 gene had a similar effect, proving the lack of association between the natural variation of the LPAAT gene and fatty acid content and composition.
Conclusions
A total of 40 LPAAT genes were identified and grouped into four distinct subfamilies in four Gossypium species, i.e., i.e., tetraploid G. hirsutum- AD1 and G. barbadense- AD2 and its ancestral diploids G. raimondii- D5 and G. arboreum- A2. The detailed analysis of the sequence variation, QTL co-localisation and content of TAG in transgenic yeasts showed that natural sequence variations in the LPAAT genes are highly limited which are unlikely associated with the natural variations in seed oil and protein contents and fiber quality traits in cotton. However, the cotton LPAATs can increase oil composition and content as demonstrated in the transgenic yeast experiment. The results provide an important lead for further studies to elucidate the involvement of LPAAT genes in the natural variation of cottonseed oil content and a possible strategy in genetic engineering for the improvement of seed oil content and composition in cotton.
Methods
Materials
Xuzhou 142 (wild type), an Upland cotton cultivar and its fuzzless/lintless natural mutant (Xuzhou 142 fl) differing in fiber initiation, and two backcross inbred lines (NMGA-062 with fiber length of 32.58 mm and NMGA-105 with fiber length of 27.06 mm) differing in fiber length, were used for RNA-seq and tissue/organ quantitative real-time RT-PCR analysis. Xuzhou142 fl and its wild type also had significantly different oil contents and fatty acid compositions [42]. Xuzhou 142 and Xuzhou 142 fl were grown at the Institute of Cotton Research (ICR), Chinese Academy of Agricultural Sciences (CAAS), Anyang, Henan province, China. On selected days post anthesis (DPA), ovules were excised from developing bolls, and fibers were scraped from the epidermis of the ovules. Petals were sampled on the day of flowering, and roots, stems and leaves were also collected from seedlings at 2 weeks after germination. All samples were quick-frozen in liquid nitrogen and stored at −80 °C until use. For each time-point or tissue, three biological replications were collected.
Since the other cultivated tetraploid cotton G. barbadense contains higher seed oil content and lower protein content than G. hirsutum, introgression lines between the two species were also used in this study. SG 747 (G. hirsutum) was first crossed with Giza 75 (G. barbadense), and the resulting F1 was backcrossed with SG 747 twice followed by three generations of self-pollination to produce advanced backcross inbred lines (BILs). The two parents and 146 BILs including NMGA-062 and NMGA-105 were grown in two replications using a randomized complete block design in Anyang, Henan in 2006, 2008, and 2009. Crop management practices and boll sampling followed local recommendations [44]. Mature seed after ginning boll samples was determined for oil and protein contents using the near infrared reflectance spectroscopy [44, 45], which were also used in other cotton studies [46, 47]. Another non-destructive measuring techniques, i.e., nuclear magnetic resonance [48] was not used in the current study. Fiber quality traits were measured using High Volume Instrument (HVI) 1000 by ICR, CAAS, Anyang, Henan [49].
To study the gene function of the heterologous gene, At-Gh13LPAAT5 from both Upland cotton and G. barbadense sources, the methylotrophic yeast, Pichia pastoris was used. The P. pastoris strain GS115 and plasmid pPIC3.5 K were kept in our laboratory.
Prediction and cladistic analyses of LPAAT genes
The genome sequences of G. arboreum, G. raimondii, G. hirsutum and G. barbadense were downloaded from CottonGen database (https://www.cottongen.org/home). Putative LPAATs were identified in the PFAM protein family database using HMMER software version 3.0 [50], with the LPAAT domain (PF01553, corresponding to LPLAT_LCLAT1-like and LPLAT_AGPAT-like in NCBI CDD) as the search query [51], with an initial threshold value of E ≤ 10−20. For the cladistics analysis of LPAAT-like proteins, the representative LPAAT protein sequences from other species including the model plant Arabidopsis, and T. cacao, which shared an ancestor with cotton at least 60 million years ago [35], were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/guide/). Amino acid sequences were aligned using Clustal X v. 2.0.11 (http://www.clustal.org/) under the default settings, and were further refined by a visual inspection. The alignment outputs were used to construct cladograms using the Neighbor-Joining (NJ) method, as implemented in MEGA v. 5 (http://www.megasoftware.net/). The bootstrap consensus tree was inferred from 1000 replicates.
In-silico mapping and analysis of LPAAT genes
Mapping of LPAAT genes was performed using MapChart (http://www.earthatlas.mapchart.com/) [52]. Quantitative trait loci (QTL) in this paper were downloaded from CottonQTLdb (http://www.cottonqtldb.org) [43, 53, 54]. The genetic structures of LPAAT genes were generated using the GSDS (Gene Structure Display Server) algorithm (http://gsds.cbi.pku.edu.cn/). All of the putative protein sequences were analyzed using the domain analysis program CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) with the default cut-off parameters. The length, molecular weight, and isoelectric point of each LPAAT protein were calculated using ExPasy (http://web.expasy.org/compute_pi/). The subcellular localization of each protein was analyzed using PSORT Prediction (http://psort.hgc.jp/form.html).
Analysis of LPAAT genes in two RNA-seq datasets
To study the expression of LPAAT genes in ovules and fibers during fiber initiation and elongation stages, the transcriptional profiles of LPAAT genes were analyzed using information from two transcriptome sequencing databases. One database contained transcriptomic information for 0, 3 and 10 DPA ovules for the following two backcross inbred lines (BILs) with significantly different fiber lengths but similar oil and protein contents in seed: NMGA-062 and NMGA-105. The other database contained transcriptomic information for −3 and 0 DPA ovules of the cotton cultivar Xuzhou 142 and its natural fiberless mutant Xuzhou 142 fl. These databases are generated in our own laboratory and can be accessed through NCBI under accession numbers SRP038911, SRP039385, and SRP056184. Transcriptome analyses were conducted with MeV software [55].
RNA isolation and real-time PCR analysis
Based on the LPAAT coding sequences, gene-specific primers for qRT-PCR were designed with Oligo 7 software (Additional file 3: Table S1). Because we attempted to study the differential expression of LPAATs in cotton fiber and seed oil, total RNA was isolated from ovules at the fiber initiation stage (−3, −1 and 0 DPA), ovules and fibers at the early fiber elongation stage (3–10DPA), and ovules and fibers at the seed oil accumulation stage (20–25DPA). Other cotton tissues including roots, stems, leaves, and petals were also sampled and analyzed for a comparison. Each sample possessed 3 biological replicates. The RNA Prep Pure Plant kit (Tiangen, Beijing, China) was used to extract RNA. Then, 0.5 μg purified total RNA was reverse-transcribed into cDNA using the Super Script First-Strand Synthesis System for RT-PCR (PrimeScript, Takara, Dalian, China), following the manufacturer's instructions. Because of high homologies among LPAAT gene sequences within the same subfamily, 8 typical LPAAT genes from four gene subfamilies were chosen. The transcript levels of the 8 LPAAT genes were normalized to the mean value of Histone3 (AF024716) used as an internal control. All qRT-PCR reactions were run on an iCycler iQ5 Fast Real-Time PCR System (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Each 20 μL reaction mixture contained 10 μL SYBR Premix Ex Taq II (2×), 0.4 μL forward and reverse primers (10 μM), 2 μL diluted cDNA, and ddH2O up to 20 μL. The thermal profile used for all PCRs was as follows: 10 min at 95 °C for DNA polymerase activation, 40 cycles of 15 s at 95 °C, 30 s at 58 °C, and 20 s at 72 °C, and then a 5 min elongation step at 72 °C. The default settings were selected for the melting curve analysis. The gene transcript levels were calculated using the 2-△△CT method. Three biological replicates, each with three technical replicates, were evaluated.
Identification of single nucleotide polymorphisms (SNPs) for LPAAT genes and statistical analysis with cottonseed oil and protein content and fiber quality traits
The two RNA-seq datasets were also used to identify SNPs for LPAAT genes. Assembled contigs (unigenes) were scanned for SNPs using SNP detection software SOAPsnp [56]. Putative SNPs were identified to design primers (Additional file 3: Table S2) using Oligo7 for developing single strand conformation polymorphic (SSCP) markers for the BIL population per the method of Lu et al. [57]. The SSCP markers were coded as “1” (i.e., G. hirsutum or G. barbadense allele) for presence and “0” for absence of a SSCP marker and used to perform a simple correlation analysis with the seed oil and protein contents in the BIL population using SPSS software (IBM, New York, USA).
Construction of the At-Gh13LPAAT5 yeast expression vector and yeast transformation
As a SSCP marker developed from the gene At-Gh13LPAAT5 was correlated to cottonseed oil content, the gene was used in this study. First-strand cDNA synthesis was carried out using ReverTra Ace qPCR RT Master Mix (Toyobo, Japan) from Upland TM-1, Xuzhou 142, Xuzhou 142 fl and G. barbadense 3–79 used as a template for PCR to amplify At-Gh13LPAAT5 using the primer pair At-Gh13LPAAT5F/At-Gh13LPAAT5R (5'-ATTATTCGAAGGATCCATGGAAGTTCCAAGTGCGAAA-3'/ 5'-CCGCCCTAGGGAATTCTTAAGCTCCCGACATGAACC-3'), and the PCR products were sequenced in Genewiz for validation of the sequences. Because the sequences from Upland Xuzhou 142 fl (which was likely derived from a natural hybrid between Upland Xuzhou 142 and an unknown G. barbadense based on Ma et al. 2016) [41] and G. barbadense 3–79 were identical which were different from the sequence from Upland TM-1 and Xuzhou 142 (Additional file 8: Figure S1), the cDNA sequences encoding the At-Gh13LPAAT5 from Xuzhou 142 fl (to represent the G. barbadense gene allele) and Xuzhou 142 were separately cloned into the EcoRI site of the expression vector pPIC3.5 K [58, 59]. 50 μL competent cells of Pichia pastoris GS115 was transformed with 10 μg of SacI-linearized pPIC3.5 K that was recombined with At-Gh13LPAAT5 by LiCl according to the Invitrogen manual.
Gas chromatography/mass spectrometry (GC/MS) profiling and statistical analysis
A GC/MS analysis was performed using a gas chromatograph (7890A, Agilent Technologies, USA) equipped with a flame ionization detector (FID) and an HP-FFAP capillary column (30 m × 250 μm × 0.25 μm) by OCRI-CAAS. High purity nitrogen was used as carrier gas. Inlet pressure is 25 psi,and sample volumes of 1 μL were injected with a split ratio of 30:1 using a hot-needle technique. The GC column temperature was programmed from 150 (initial equilibrium time, 1 min) to 230 °C via a ramp of 5 °C/min and maintained at 230 °C for 8 min. The injection temperature was 250 °C, and the test temperature was set to 280 °C [60]. Transgenic yeasts were induced for 72 h and internal standard gas chromatography was performed to determine the fatty acid components and the content of triacylglycerols (TAGs). The t-test was performed using the Microsoft excel.
Acknowledgements
The authors wish to thank New Mexico Agricultural Experiment Station, New Mexico, USA and Xia Wan from Institute of Oilcrops Research, Chinese Academy of Agricultural Sciences (CAAS), Wuhan, China.
Funding
This study was supported by the National Natural Science Foundation of China (31301368), the 863 Project of China (2013AA102601) and the National Key Research and Development Program of China (2016YFD0101400).
Availability of data and materials
The sequence read data from RNA-seq analysis for the 2 BILs, Xuzhou 142 and Xuzhou 142 fl are available in the Sequence Read Archive (SRA) (accession number SRP038911, SRP039385 and SRP056184). The data sets supporting the results of this study are included in the manuscript and its additional files.
Authors’ contribution
SY and JY directed the experiments. WP, HL and XL performed the field cultivation of cotton plants and ovules collection. MW participated in the study. JZ conceived the study and revised the manuscript. JM performed the SSCP experiments. NW performed the experiments, and NW and JZ wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- DGAT
Diacylglycerol acyltransferase
- DPA
Days post-anthesis
- GPAT
Glycerol-3-phosphate O-acyltransferase
- LPA
Lysophosphatidate
- LPAAT
Lysophosphatidic acid acyltransferase
- LPC
Lysophosphatidylcholine
- MUFAs
Monounsaturated fatty acids
- PAs
Phosphatidic acids
- PC
Phosphatidylcholine
- PDAT
Phospholipid:diacylglycerol acyltransferase
- PDCT
Phosphatidylcholine:diacylglycerol cholinephosphotransferase
- SFAs
Saturated fatty acids
- SNPs
Single nucleotide polymorphisms
- SSCP
Single strand conformation polymorphic
- TAGs
Triacylglycerols
- Xuzhou 142 fl
fuzzless/lintless natural mutant of Xuzhou 142
Additional files
An alignment of LPAAT2/3 with LPAAT4/5 by ClustalX2 program. The boxes indicate variation. (TIF 2178 kb)
An alignment of LPAAT1 with B-class LPAAT by ClustalX2 program. (TIF 4401 kb)
Sequences of primers used for studies of GhLPAAT expression. Table S2. Sequences of primers of GhLPAATs to identify single nucleotide polymorphic markers (SNPs) based on single strand conformation polymporphism (SSCP). Table S3. Domain analysis and annotation. Table S4. Oil and protein quantitative trait loci (QTL). Table S5. Identification of putative sequence variation (SNPs) by SOAPsnp in NMGA-062 vs. NMGA-105 database. L indicates SNP only exists in long fiber length line NMGA-062 (fiber length, 32.58 mm) when compared with the sequenced genome sequence of TM-1. S indicates SNP only exists in short fiber length line NMGA-105 (fiber length, 27.06 mm) when compared with the sequenced genome sequence of TM-1. L/S indicates SNP exists in the two lines. Table S6. Identification of putative SNPs by SOAPsnp in Xuzhou 142 vs. Xuzhou 142 fl database. WT indicates SNP only exists in Xuzhou 142 when compared with the sequenced genome sequence of TM-1. fl indicates SNP only exists in Xuzhou 142 fl when compared with the sequenced genome sequence of TM-1. WT/fl indicates SNP exists in the two lines. The polymorphic SSCP marker is shown in red color. (XLSX 26 kb)
Chromosomal distribution of LPAAT genes. Dotted lines link homoelogous LPAATs between A2 and D5 chromosomes. The scale to the left of chromosomes denotes the size of chromosomes in million bases (Mb). (TIF 6248 kb)
Expression patterns of Dt-Gh1LPAATB genes in other organs. Histone3 was used as an internal control. (TIF 46 kb)
Sequence variations in the predicted LPAAT genes between the sequenced G. hirsutum (TM-1) and G. barbadense (3–79 and Xinhai 21). The boxes indicate the variations of the A- and D- subgenomes and the ellipses indicate the SNPs between the two cultivated species; Gb1 and Gb2 indicate 3–79 and Xinhai21 respectively. (PDF 22277 kb)
A polymorphic SSCP marker developed from a LPAAT gene in the backcross inbred population of SG 747 x Giza 75. The arrow indicates the polymorphic marker. (TIF 9674 kb)
An alignment of At-Gh13LPAAT5 in four Gossypium species. The box indicates the variation of At-Gh13LPAAT5 at 342th nucleotide. (TIF 2008 kb)
Contributor Information
Nuohan Wang, Email: wnhan2013@126.com.
Jianjiang Ma, Email: mjj1699@126.com.
Wenfeng Pei, Email: peiwenfeng1988@163.com.
Man Wu, Email: wuman2004@163.com.
Haijing Li, Email: leo812002@163.com.
Xingli Li, Email: 1965075665@qq.com.
Shuxun Yu, Email: yu@cricaas.com.cn.
Jinfa Zhang, Email: jinzhang@nmsu.edu.
Jiwen Yu, Email: yujw666@hotmail.com.
References
- 1.Liu Q, Singh S, Chapman K, Green A. Bridging traditional and molecular genetics in modifying cottonseed oil. Genet Genomics Cotton. 2009;3:353–83. doi: 10.1007/978-0-387-70810-2_15. [DOI] [Google Scholar]
- 2.Bates PD, Browse J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci. 2012;3. [DOI] [PMC free article] [PubMed]
- 3.Yamashita A, Hayashi Y, Matsumoto N, Nemoto-Sasaki Y, Oka S, Tanikawa T, Sugiura T. Glycerophosphate/Acylglycerophosphate acyltransferases. Biology. 2014;3(4):801–30. doi: 10.3390/biology3040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korbes AP, Kulcheski FR, Margis R, Margis-Pinheiro M, Turchetto-Zolet AC. Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family. Mol Phylogenet Evol. 2016;96:55–69. doi: 10.1016/j.ympev.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Arroyo-Caro JM, Chileh T, Alonso DL, Garcia-Maroto F. Molecular characterization of a lysophosphatidylcholine acyltransferase gene belonging to the MBOAT family in Ricinus communis L. Lipids. 2013;48(7):663–74. doi: 10.1007/s11745-013-3797-z. [DOI] [PubMed] [Google Scholar]
- 6.Wang P, Wang Z, Dou Y, Zhang X, Wang M, Tian X. Genome-wide identification and analysis of membrane-bound O-acyltransferase (MBOAT) gene family in plants. Planta. 2013;238(5):907–22. doi: 10.1007/s00425-013-1939-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Gai J, Xu S, Chen Z, Luo Z, Cao Z, Jin L, Li F. Analysis of MBOAT family reveals the diversity of MBOAT1 amplification in Solanaceae. Arch Biol Sci. 2015;67(4):1349–56. doi: 10.2298/ABS150326112W. [DOI] [Google Scholar]
- 8.Hu Z, Ren Z, Lu C. The phosphatidylcholine diacylglycerol cholinephosphotransferase is required for efficient hydroxy fatty acid accumulation in transgenic Arabidopsis. Plant Physiol. 2012;158(4):1944–54. doi: 10.1104/pp.111.192153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, Xin Z, Ren Z, Miquel M. An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci U S A. 2009;106(44):18837–42. doi: 10.1073/pnas.0908848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlqvist A, Ståhl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97(12):6487–92. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stymne S, Stobart AK. Evidence for the reversibility of the acyl-CoA: lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons and rat liver. Biochem J. 1984;223:305–14. doi: 10.1042/bj2230305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen G, Snyder CL, Greer MS, Weselake RJ. Biology and biochemistry of plant phospholipases. Crit Rev Plant Sci. 2011;30(3):239–58. doi: 10.1080/07352689.2011.572033. [DOI] [Google Scholar]
- 13.Okazaki K, Sato N, Tsuji N, Tsuzuki M, Nishida I. The significance of C16 fatty acids in the sn-2 positions of glycerolipids in the photosynthetic growth of Synechocystis sp. PCC6803. Plant Physiol. 2006;141(2):546–56. doi: 10.1104/pp.105.075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frentzen M. Acyltransferases from basic science to modified seed oils. Lipid/Fett. 1998;100(4–5):161–6. doi: 10.1002/(SICI)1521-4133(19985)100:4/5<161::AID-LIPI161>3.0.CO;2-P. [DOI] [Google Scholar]
- 15.Lassner MW, Levering CK, Davies HM, Knutzon DS. Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic rapeseed oil. Plant Physiol. 1995;109(4):1389–94. doi: 10.1104/pp.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent P, Huang AH. Organ-and development-specific acyl coenzyme A lysophosphatidate acyltransferases in palm and meadowfoam. Plant Physiol. 1992;99(4):1711–5. doi: 10.1104/pp.99.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ. Acyltransferase action in the modification of seed oil biosynthesis. N Biotech. 2009;26(1):11–6. doi: 10.1016/j.nbt.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Brown AP, Slabas AR, Denton H. Substrate selectivity of plant and microbial lysophosphatidic acid acyltransferases. Phytochemistry. 2002;61(5):493–501. doi: 10.1016/S0031-9422(02)00290-X. [DOI] [PubMed] [Google Scholar]
- 19.Knutzon DS, Lardizabal KD, Nelsen JS, Bleibaum JL, Davies HM, Metz JG. Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol. 1995;109(3):999–1006. doi: 10.1104/pp.109.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arroyo-Caro JM, Chileh T, Kazachkov M, Zou J, Alonso DL, García-Maroto F. The multigene family of lysophosphatidate acyltransferase (LPAT)-related enzymes in Ricinus communis. Cloning and molecular characterization of two LPAT genes that are expressed in castor seeds. Plant Sci. 2013;199:29–40. doi: 10.1016/j.plantsci.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Maisonneuve S, Bessoule J-J, Lessire R, Delseny M, Roscoe TJ. Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 2010;152(2):670–84. doi: 10.1104/pp.109.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown A, Carnaby S, Brough C, Brazier M, Slabas A. Limnanthes douglasii lysophosphatidic acid acyltransferases: immunological quantification, acyl selectivity and functional replacement of the Escherichia coli plsC gene. Biochem J. 2002;364:795–805. doi: 10.1042/bj20020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HU, Li Y, Huang AH. Ubiquitous and endoplasmic reticulum–located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell. 2005;17(4):1073–89. doi: 10.1105/tpc.104.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgis F, Kader JC, Barret P, Renard M, Robinson D, Robinson C, Delseny M, Roscoe TJ. A plastidial lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol. 1999;120(3):913–22. doi: 10.1104/pp.120.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown AP, Coleman J, Tommey AM, Watson MD, Slabas AR. Isolation and characterisation of a maize cDNA that complements a 1-acyl sn-glycerol-3-phosphate acyltransferase mutant of Escherichia coli and encodes a protein which has similarities to other acyltransferases. Plant Mol Bio. 1994;26(1):211–23. doi: 10.1007/BF00039533. [DOI] [PubMed] [Google Scholar]
- 26.Taylor DC, Francis T, Lozinsky S, Hoffman T, Giblin M, Marillia EF. Cloning and characterization of a constitutive lysophosphatidic acid acyltransferase 2 (LPAT2) gene from Tropaeolum majus L. Open Plant Sci J. 2010;4:7–17. doi: 10.2174/1874294701004010007. [DOI] [Google Scholar]
- 27.Yu XH, Prakash RR, Sweet M, Shanklin J. Coexpressing Escherichia coli cyclopropane synthase with Sterculia foetida lysophosphatidic acid acyltransferase enhances cyclopropane fatty acid accumulation. Plant Physiol. 2014;164(1):455–65. doi: 10.1104/pp.113.230953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou J, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell. 1997;9(6):909–23. doi: 10.1105/tpc.9.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu YX. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007;19(11):3692–704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotech. 2015;33(5):524–30. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotech. 2015;33(5):531–7. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Zhao B, Zheng HJ, Hu Y, Lu G, Yang CQ, Chen JD, Chen JJ, Chen DY, Zhang L, et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep. 2015;5:14139. doi: 10.1038/srep14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:17662. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–86. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- 35.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–7. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 36.Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 37.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–72. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 38.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38(18):5764–71. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 39.Kim HU, Huang AH. Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol. 2004;134(3):1206–16. doi: 10.1104/pp.103.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bursten S, Harris W, Bomsztyk K, Lovett D. Interleukin-1 rapidly stimulates lysophosphatidate acyltransferase and phosphatidate phosphohydrolase activities in human mesangial cells. J Biol Chem. 1991;266(31):20732–43. [PubMed] [Google Scholar]
- 41.Ma Q, Wu M, Pei W, Wang X, Zhai H, Wang W, Li X, Zhang J, Yu J, Yu S. RNA-seq-mediated transcriptome analysis of a fiberless mutant cotton and its possible origin based on SNP markers. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0151994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma J, Wu M, Pei W, Li X, Huang S, Zhang J, Yu J, Yu S. The oil and fatty acid accumulation patterns in developing cottonseeds of Xuzhou 142 and its fiberless and fuzzless mutant. Cotton Sci. 2015;27(2):95–103. [Google Scholar]
- 43.Said JI, Knapka JA, Song M, Zhang J. Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G. hirsutum × G. barbadense populations. Mol Genet Genomics. 2015;290(4):1615–25. doi: 10.1007/s00438-015-1021-y. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Yu S, Gore M, Wu M, Zhai H, Li X, Fan S, Song M, Zhang J. Identification of quantitative trait loci across interspecific F2, F2:3 and testcross populations for agronomic and fiber traits in tetraploid cotton. Euphytica. 2013;191(3):375–89. doi: 10.1007/s10681-013-0875-5. [DOI] [Google Scholar]
- 45.Huang Z, Sha S, Rong Z, Chen J, He Q, Khan DM, Zhu S. Feasibility study of near infrared spectroscopy with variable selection for non-destructive determination of quality parameters in shell-intact cottonseed. Ind Crop Prod. 2013;43:654–60. doi: 10.1016/j.indcrop.2012.08.015. [DOI] [Google Scholar]
- 46.Alfred Q, Liu HY, Xu HM, Li JR, Wu JG, Zhu SJ, Shi CH. Mapping of quantitative trait loci for oil content in cottonseed kernel. J Genet. 2012;91(3):289–95. doi: 10.1007/s12041-012-0184-0. [DOI] [PubMed] [Google Scholar]
- 47.Kohel RJ. Evaluation of near infrared reflectance for oil content of cottonseed. J cotton sci. 1998;2:23–6. [Google Scholar]
- 48.Hinze LL, Horn PJ, Kothari N, Dever JK, Frelichowski J, Chapman KD, Percy RG. Nondestructive measurements of cottonseed nutritional trait diversity in the U.S. National Cotton Germplasm Collection. Crop Sci. 2015;55(2):770–82. doi: 10.2135/cropsci2014.04.0318. [DOI] [Google Scholar]
- 49.Yu J, Zhang K, Li S, Yu S, Zhai H, Wu M, Li X, Fan S, Song M, Yang D. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor Appl Genet. 2013;126(1):275–87. doi: 10.1007/s00122-012-1980-x. [DOI] [PubMed] [Google Scholar]
- 50.Eddy SR. Accelerated profile HMM searches. PLoS Comp Biol. 2011;7(10) doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J. Pfam: the protein families database. Nucl Acids Res. 2013;42(D1):222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 53.Said JI, Song M, Wang H, Lin Z, Zhang X, Fang DD, Zhang J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol Genet Genomics. 2015;290(3):1003–25. doi: 10.1007/s00438-014-0963-9. [DOI] [PubMed] [Google Scholar]
- 54.Said JI, Lin ZX, Zhang XL, Song MZ, Zhang JF. A comprehensive Meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics. 2013;14(1):776. doi: 10.1186/1471-2164-14-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 56.Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19(6):1124–32. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Y, Curtiss J, Percy R, Hughs S, Yu S, Zhang J. DNA polymorphisms of genes involved in fiber development in a selected set of cultivated tetraploid cotton. Crop Sci. 2009;49(5):1695–704. doi: 10.2135/cropsci2008.12.0693. [DOI] [Google Scholar]
- 58.Chen X, Cao Y, Ding Y, Lu W, Li D. Cloning, functional expression and characterization of Aspergillus sulphureus beta-mannanase in Pichia pastoris. J Biotechnol. 2007;128(3):452–61. doi: 10.1016/j.jbiotec.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24(1):45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 60.Gong Y, Guo X, Wan X, Liang Z, Jiang M. Triacylglycerol accumulation and change in fatty acid content of four marine oleaginous microalgae under nutrient limitation and at different culture ages. J Basic Microbiol. 2013;53(1):29–36. doi: 10.1002/jobm.201100487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence read data from RNA-seq analysis for the 2 BILs, Xuzhou 142 and Xuzhou 142 fl are available in the Sequence Read Archive (SRA) (accession number SRP038911, SRP039385 and SRP056184). The data sets supporting the results of this study are included in the manuscript and its additional files.


