Abstract
Understanding the processes that generate novel adaptive phenotypes is central to evolutionary biology. We used comparative analyses to reveal the history of tetrodotoxin (TTX) resistance in TTX-bearing salamanders. Resistance to TTX is a critical component of the ability to use TTX defensively but the origin of the TTX-bearing phenotype is unclear. Skeletal muscle of TTX-bearing salamanders (modern newts, family: Salamandridae) is unaffected by TTX at doses far in excess of those that block action potentials in muscle and nerve of other vertebrates. Skeletal muscle of non-TTX-bearing salamandrids is also resistant to TTX but at lower levels. Skeletal muscle TTX resistance in the Salamandridae results from the expression of TTX-resistant variants of the voltage-gated sodium channel NaV 1.4 (SCN4a). We identified four substitutions in the coding region of salSCN4a that are likely responsible for the TTX resistance measured in TTX-bearing salamanders and variation at one of these sites likely explains variation in TTX resistance among other lineages. Our results suggest that exaptation has played a role in the evolution of the TTX-bearing phenotype and provide empirical evidence that complex physiological adaptations can arise through the accumulation of beneficial mutations in the coding region of conserved proteins.
Keywords: Adaptation, Caudata, evolutionary genomics, molecular evolution, Salamandridae
Adaptation plays a central role in driving phenotypic evolution, yet fundamental questions about this process remain unresolved (Fisher 1958; Stapley et al. 2010; Losos et al. 2013; Travisano and Shaw 2013). The process by which novel, complex adaptations arise is still unanswered and key questions about the molecular basis of adaptive evolution remain the focus of considerable attention (Monteiro and Podlaha 2009; Stern and Orgogozo 2009; Nadeau and Jiggins 2010; Barrett and Hoekstra 2011; Rockman 2012; Wray 2013). Specifically, these include questions about the number and phenotypic effect of substitutions associated with adaptive phenotypes (Hoekstra and Coyne 2007; McGregor et al. 2007; Lynch 2010; Orr 2010; Frankel et al. 2011).
Elucidating the dynamics of adaptive evolution is problematic (Rockman 2012; Travisano and Shaw 2013). Many adaptive phenotypes comprised multiple, genetically independent characters. Furthermore, most characters are complex, resulting from the interplay of multiple genes with complicated environmental and epistatic interactions (Weinreich et al. 2006; Stern and Orgogozo 2008; Breen et al. 2012; McCandlish et al. 2013). However, a considerable body of empirical and theoretical work is still focused on efforts to understand the molecular basis of adaptation and resolve these issues (Stapley et al. 2010; Losos et al. 2013; Wray 2013; and see Rockman 2012; Natarajan et al. 2013; Projecto-Garcia et al. 2013 for recent examples).
The comparative method is fundamental to evolutionary biology and studies that elucidate the phenotypic and genotypic history of adaptive traits can provide important insight into the process of adaptation (Garland and Adolph 1994; Jones et al. 2012; Romero et al. 2012; Alfoldi and Lindblad-Toh 2013; Projecto-Garcia et al. 2013). Ultimately these studies can generate information about the specific changes underlying adaptive phenotypes as well as evolutionary history of those changes that, in turn, inform our understanding of the process of adaptive evolution. In cases where adaptive phenotypes are multifactorial, comparative studies of individual component traits can also provide insight into the evolutionary history of the complete phenotype. This is particularly true when the functional and genetic basis of one trait in a complex adaptation is well understood but the genetic or functional basis of other components is not. Here, we document the evolutionary history of tetrodotoxin (TTX) resistance in TTX-bearing salamanders and use that history to provide insight into the evolution of the complete TTX-bearing phenotype.
TTX is a potent small-molecule toxin that serves as a deterrent to predation in an array of taxa that includes fish, frogs, mollusks, flatworms, and salamanders (order: Caudata; Hanifin 2010; Williams 2010). The physiological effect of TTX is simple and well understood. Exposure to submicromolar concentrations of TTX causes paralysis and death by disrupting the initiation and propagation of action potentials (APs) in peripheral nerves and skeletal muscle (Narahashi 1972). In salamanders, the antipredator role of TTX has been well characterized and TTX is the central trait in the coevolution between the TTX-bearing newts of the genus Taricha and garter snake predators (Brodie 1968; Brodie et al. 1974, 2002, 2005; Hanifin et al. 2008; Feldman et al. 2009, 2010).
The first isolation and purification of TTX in salamanders occurred in the genus Taricha (family: Salamandridae; Brown and Mosher 1963; Fuhrman et al. 1963; Kao and Fuhrman 1963; Buchwald et al. 1964). Ongoing work has confirmed its presence in three additional genera: Cynops, Notophthalmus, and Triturus (reviewed in Hanifin 2010). TTX-bearing salamander lineages form a single monophyletic clade known as the modern newts (family: Salamandridae). TTX has not been identified in any other species of salamanders including the sister clade (primitive newts) to the modern newts that diverged from modern newts roughly 75 MYA (Zhang et al. 2008). The modern newts are distributed globally with representative genera in Europe, Asia, and North America. Molecular and morphological evidence suggests that members of this lineage diverged rapidly between about 49 and 69 MYA but have maintained their current forms in the subsequent interval (Zhang et al. 2008). The evolutionary history and distribution of TTX toxicity in this group suggests that the TTX-bearing phenotype arose once in the common ancestor of all modern newts and was subsequently maintained as these lineages diversified and spread. The factors that have contributed to the longevity of these lineages are not clear, but the presence of TTX in skin and other tissues of these otherwise poorly defended animals is likely to have played an important role in their success.
The TTX-bearing phenotype has likely played a critical role in the evolutionary history of modern newts yet the evolutionary pathway that generated this complex phenotype is something of a puzzle (Narahashi 2008; Hanifin 2010; Mebs et al. 2010; Williams 2010). The genes and enzymatic pathways associated with TTX production are unknown. In fact, the ultimate origin of the TTX present in all vertebrates is still a matter of some contention. Some evidence suggests that bacteria produce the TTX present in salamanders, frogs, and even fish, whereas other evidence suggests that salamanders may produce TTX themselves (Hanifin et al. 2002; Cardall et al. 2004; Lehman et al. 2004; Hanifin 2010). Furthermore, the TTX-bearing phenotype is multifactorial and requires both a means to prevent self-intoxication as well as the ability to either synthesize or sequester TTX. TTX specifically binds to the extracellular surface of an integral membrane protein, the voltage-gated Na+ (NaV) channel (Narahashi 1972; Hille 1992; Narahashi 2008). These proteins are present in the excitable tissues (nerve and muscle) of all vertebrates and are directly responsible for the initiation and propagation of APs in nerves and muscles (Hille 1992). TTX selectively blocks the flux of Na+ ions essential for initiating an AP thereby disrupting the function of nerves and muscles (Narahashi 2008). TTX-bearing salamanders neither sequester TTX in specific organs, nor do they possess a means of detoxifying the toxin (Wakely et al. 1966; Mebs et al. 2010). Thus, peripheral nerve and muscle cells as well as associated NaV channels in these salamanders are exposed to high levels of TTX and must be resistant to the action of TTX for the animals to function and survive. There is no evidence that NaV channels play a role in the synthesis (or sequestration) of TTX. As a result, TTX resistance and synthesis of TTX evolved independently in these animals. Ultimately the evolution of the complete TTX-bearing phenotype required adaptations in the NaV channels expressed in nerves and muscles, as well as the enzymatic pathways that allow either the direct synthesis or uptake and sequestration of TTX.
TTX has been a critical tool in determining the shape and structure of the pore of NaV channels and there is a considerable body of work describing the interaction between NaV channels and TTX (Narahashi 1972; Catterall 1980b; Terlau et al. 1991; Penzotti et al. 1998; Cestele and Catterall 2000; Choudhary et al. 2003; Catterall et al. 2007; Fozzard and Lipkind 2010; Payandeh et al. 2011; Tikhonov and Zhorov 2011, 2012; Zhorov and Tikhonov 2013). Voltage-gated sodium channels consist of four homologous domains each of which contains a series of six trans-membrane helices that form the voltage sensors and pore of the channel (Catterall 2012). TTX binds to the outer pore of the channel and blocks the influx of Na+ ions (Hille 1992; Fozzard and Lipkind 2010; Tikhonov and Zhorov 2012). Only a small number (≈84) of amino acid residues in the NaV channel regulate TTX binding (Terlau et al. 1991; Payandeh et al. 2011; Tikhonov and Zhorov 2012). The outer pore includes the ion-selectivity filter and is formed by the region that links the fifth and sixth trans-membrane domains from all four domains (Terlau et al. 1991; Payandeh et al. 2011; Tikhonov and Zhorov 2012). This region, also known as the pore loop, consists of two short pore helices (P1 and P2) and a linker region of about four residues that forms the selectivity filter and TTX-binding site (Tikhonov and Zhorov 2012). Within the pore loop, interactions between the pore helices hold the short linker region between them into a structure that forms the selectivity filter and outer pore (Payandeh et al. 2011; Tikhonov and Zhorov 2012). TTX binding can be altered by changes in residues of the pore helices or selectivity filter from any of the four domains (Terlau et al. 1991; Penzotti et al. 1998; Cestele and Catterall 2000; Geffeney et al. 2005; Jost et al. 2008; Tikhonov and Zhorov 2012). Changes in these residues directly disrupt electrostatic interactions between TTX and the outer pore or cause subtle conformational changes in the shape of the outer pore that reduce the binding affinity for TTX (Terlau et al. 1991; Penzotti et al. 1998; Choudhary et al. 2003; Tikhonov and Zhorov 2012). Site-directed mutagenesis experiments have shown that epistatic interactions between mutations in the pore loop and other channel regions do not affect TTX binding (Geffeney et al. 2005). Ultimately this detailed understanding of TTX and its interactions with NaV channel provides clearer picture of genotype–phenotype relationships in regards to TTX resistance in TTX-bearing taxa and has allowed investigators to focus their attention on relatively small regions of the channel when interpreting molecular studies of TTX resistance (Geffeney et al. 2002, 2005; Jost et al. 2008; Feldman et al. 2009, 2010).
The genetic basis and physiology of TTX resistance in other vertebrate lineages has been well characterized but none of these has focused on the evolutionary history of the trait (Geffeney et al. 2005; Jost et al. 2008; Feldman et al. 2010). Genetically based structural variation in the pore loop of the NaV channel expressed in skeletal muscle of vertebrates (NaV 1.4) is associated with adaptive TTX resistance in TTX-bearing puffer fish as well as TTX-resistant snake predators of newts (Geffeney et al. 2005; Jost et al. 2008; Feldman et al. 2009, 2010; Zhorov and Tikhonov 2013). We examined the evolutionary history and genetic basis of TTX resistance in skeletal muscle of TTX-bearing newts by assaying the TTX sensitivity of skeletal muscle fibers from four lineages of salamanders and sequencing of the NaV channel expressed in the skeletal muscle of nine species of salamandrids and an evolutionary out-group, Ambystoma mavortium.
Materials and Methods
ANIMALS
Adult salamanders (n = 2) from each of four species chosen to provide representative taxa of major extant salamandrid lineages (i.e., true salamanders, primitive newts, and modern newts) as well an appropriate out-group taxon (family: Ambystomatidae) were used for electrophysiological recordings to assess phenotype. Specimens of Salamandra salamandra and Tylototriton shanjing were obtained commercially. H. B. Schaeffer (U.C.L.A.) provided A. mavortium, and Taricha granulosa from Benton County, Oregon, were provided by E. D. Brodie, Jr. (Utah State University). Experimental animals were fed a diet of earthworms and crickets prior to their use in physiological and/or molecular biology experiments.
We generated cDNA sequence data for the gene that encoded NaV 1.4 (salSCN4a) from each of the four species used for phenotypic measurements. We also collected sequence data from six additional species: Pleurodeles waltl, Cynops pyrrhogaster, Pachytriton labiatus, Triturus dobrogicus, Notophthalmus viridescens, and Taricha torosa. This sampling strategy allowed us to sample all of the major evolutionary lineages of the Salamandridae and included five of the extant genera of the modern newt clade as well as two of the three extant genera of primitive newts. E. D. Brodie III (University of Virginia) provided specimens of N. viridescens and T. torosa were wild caught by either CTH or WFG in northern Monterey County (CA) under CA scientific collecting permit #8010210–05. All other species used for sequence analysis were obtained commercially. Animal care and experiments were approved under appropriate Institutional Animal Care and Use Committee (IACUC) protocols at Stanford University to WFG (protocol #12300).
ELECTROPHYSIOLOGY
Each of two iliofibularis muscles was dissected from salamanders that had been decapitated and pithed. This muscle has accessible tendon or bone on both ends and is composed largely of parallel fast-twitch fibers (Ashley-Ross and Barker 2002; Walthall and Ashley-Ross 2006). During the dissection, muscles were bathed in a solution containing NaCl 101.5 mM, KCl 2.8 mM, CaCl2 1.75 mM, HEPES 10 mM at pH 7.2. Individual muscles were stretched as far as practical and pinned through a tendon or bone fragment to a Sylgard™-coated dish filled with the same solution. Recordings also employed this solution and were carried out at room temperature (~20°C).
Conventional intracellular recording techniques were used to record action potentials (APs) from individual muscle fibers under a Wild M5A stereomicroscope. Glass microelectrodes filled with 3 M KCl and of 10–20 MΩ resistance were used with an OC-725C amplifier (Warner Instruments, Hampden CT). Signals were digitized at 20 kHz and stored using Digidata 1322A and pClamp9 software (Axon Instruments, Foster City, CA). APs were stimulated using a SD9 stimulator (Grass Instruments, Quincy, MA) through a coaxial, bipolar electrode (SNE-100, Rhodes Medical Instruments, Woodland Hills, CA, USA) applied to a group of muscle fibers.
Recordings were made only from superficial muscle fibers with a resting potential greater than −70 mV. Resting potentials more positive than this were associated with decreased action potential amplitude, presumably due to inactivation of Na+ channels and were not used to control for the effect of time.
EXPERIMENTAL PROCEDURE AND DATA ANALYSIS
Data presented here are based on a minimum of three recordings from a single muscle at a given TTX concentration. At least two (typically three or four) muscle preparations were used for each species. Although individual preparations were exposed to multiple concentrations of TTX, quantification of maximum rate of rise (ROR) in 0.01 μM TTX (Fig. 1B) was carried out using data only from fibers in which this concentration was the first to be applied. After recording control APs (i.e., no TTX) from a minimum of five fibers, each muscle preparation was washed (2×) with a bath solution containing the appropriate TTX concentration. We repeated this 2× wash at 15 min with fresh TTX-containing solution (same concentration). Recordings commenced after an additional 15-min wait. Thirty minutes was thus allowed for the lowest concentration of TTX employed (0.01 μM) to reach a steady-state effect. Empirical tests suggested that this amount of time was adequate.
Figure 1.
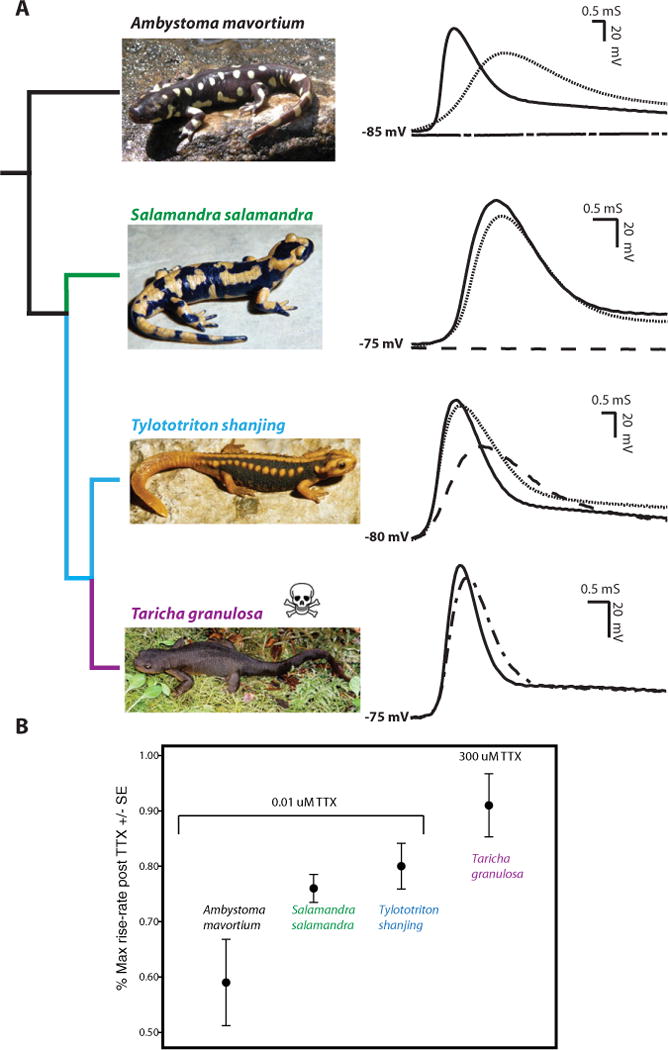
Evolutionary history of TTX resistance in salamanders. The differential resistance to tetrodotoxin as well as the evolutionary relationships of representative species from major salamandrid lineages and an out-group family (Ambystomatidae) are shown. (A) Intracellular action potentials (APs) recorded in skeletal muscle fibers from the indicated species exposed to a range of TTX concentrations. Control APs (no TTX) are shown in solid black lines. APs in the out-group species (Ambystoma mavortium) were completely blocked by 0.025 μM TTX (staggered dashed line), whereas 0.050 μM TTX (dashed line) was required to block APs in the true salamander (S. salamandra), but failed to block APs in the primitive newt (T. shanjing). Conversely, high concentrations of TTX (300 μM; dashed and dotted line) failed to block AP propagation in the TTX-bearing modern newt Taricha granulosa. Also shown are the effects of 0.010 μM TTX (dotted line) on APs from A. mavortium, S. salamandra, and T. shanjing demonstrating a progressive increase in TTX resistance across these species. (B) Comparison of the reductions in maximum rate of rise of the AP in 300 μM TTX for T. granulosa (9% ± 5.7 SE, n = 5) as well as differences (df = 2, F = 3.76, P = 0.06) in 0.010 μM TTX among A. mavortium (41% ± 7.8 SE, n = 5), S. salamandra (24% ± 2.5 SE, n = 3), and T. shanjing (20% ± 4.1 SE, n = 5). Line and font color refer to evolutionary lineages within the Salamandridae (true salamanders, green; primitive newts, blue; and modern newts, purple). Evolutionary relationships are inferred from Zhang et al. (2008) and Zhang and Wake (2009).
We characterized the effect of TTX by determining the reduction in the maximum rate of voltage rise during the AP at a given TTX concentration and by estimating the absolute concentration of TTX required to eliminate the AP. We assumed that maximum rate of voltage change during the rising phase of the AP was proportional to maximum inward Na+ current and the driving force was the same for all recordings for any given muscle. Thus, the relative decrease in maximum ROR is proportional to the decrease in maximum Na+ conductance during the AP, which, in turn, is proportional to the number of Na+ channels that are blocked by TTX. We calculated maximum AP-ROR in the presence and absence of TTX by taking the derivative of the rising phase of the AP, after the recordings were smoothed with box algorithm (smoothing = 5) in IGOR PRO (vs. 6.04, WaveMetrics). Absolute values for the maximum ROR also depend on the effective capacitance of a muscle fiber, and this property may vary somewhat from species to species. Because only relative changes in ROR between different species are compared, this factor is not critical in interpretation of our data.
SEQUENCE DATA
We used a combination of PCR, Reverse transcription PCR (RT-PCR), Rapid Amplification of cDNA Ends (RACE), and cloning to generate full (5735 nt; T. torosa) and partial (≈4450 nt; all other species) sequence of a single α-subunit of the NaV channel expressed in salamander skeletal muscle. Sequences reported here are based on a minimum of 2× coverage, but we generated at least 4× coverage for pore or pore-helix regions for all species. In the case of T. torosa, pore and pore-helix sequences results are based on a minimum of 6× coverage. We isolated and purified total RNA from freshly excised skeletal muscle with the RNeasy Mini Plus Kit and Tissuelyser (Qiagen, Valenica, CA) as directed by the manufacturer’s protocols. Reverse transcription of mRNA to cDNA employed either the Superscript II or Superscript III RT-PCR Kit (Life Technologies, Carlsbad, CA) using degenerate primers for NaV 1.4. We then amplified a series of overlapping pieces of NaV 1.4 to construct a complete fragment of the gene by using a combination of salamander-specific or species-specific degenerate and nondegenerate primers that we designed specifically for salamander NaV 1.4 (Table S1). For T. torosa sequence, we used rapid amplification of cDNA ends (SMART RACE kit, Clontech Laboratories, Mountainview, CA) to generate sequence that included stop and start codons as well as 3 and 5 UTR to ensure sequencing of full length coding sequences. Amplification of RACE cDNA was achieved using nested gene-specific primers based on known T. torosa sequence and primers provided with the kit (Table S2). RACE-based amplicons were either directly sequenced or cloned and amplified as described below. We cleaned amplified products with the QIAquick PCR purification kit (Qiagen). Cleaned PCR products were sequenced on an ABI 3730xi DNA analyzer platform (Sequetech, Mountainview, CA) or in house (Hopkins Marine Station) using cycle-sequencing reactions with Big Dye 3.1 (Carlsbad, CA) followed by isopropanol/ethanol wash and running products on an ABI 3100 capillary electrophoresis. We sequenced all samples in both directions.
For a subset of amplicons (typically pore and pore helix) and species (at least one from each major clade of salamandrids), we used either Zero Blunt or TOPO TA cloning kits (Life Technologies, Carlsbad, CA) to confirm the absence of either allelic variation or expression of multiple NaV isoforms in skeletal muscle. Products were generated from cDNA and amplified as described above. These PCR products were then purified using gel electrophoresis and/or QIAquick PCT purification kit (Qiagen) and cloned using kit protocols. Individual colonies were picked and grown in 1 mL volumes. Plasmid DNA was isolated and purified using Miniprep Kits (Qiagen) and sequenced as described above.
We confirmed the identity of the NaV channel isolated from salamander skeletal muscle by assembling a gene tree of known vertebrate NaV orthologs that included examples of all known mammalian NaV channel orthologs as well as a subset of those identified in fish (Fig. S1). In addition, we included NaV 1.4 orthologs of a lizard, Anolis caralonensis, frog, Xenopus lavaeis, and a snake (Thamnophis sirtalis). Because a single neuronal channel from the TTX-bearing modern newt C. pyrrhogaster is also known to be TTX resistant we included this channel in our analysis.
MOLECULAR AND PHYLOGENETIC ANALYSIS
All sequence results were edited by eye in Sequencher 4.8 (Gene Codes). Initial sequence alignments were performed with Clustal W2 (European Bioinformatics Institute) followed by hand alignment and cleanup with MacClade 4.08 using translated amino acid sequences as guides (Larkin et al. 2007). We used Bayesian inference with Markov chain Monte Carlo and maximum likelihood (ML) analyses to estimate NaV 1.4 gene trees in salamanders as well as phylogenetic comparisons of our sequences with vertebrate NaV orthologs (see above). In addition, we used the HyPhy package implemented on the Data Monkey web server (University of California at San Diego) to estimate the best-fitting nucleotide substitution model for our dataset using Akaike Information Criterion (AIC) (Pond et al. 2005; Delport et al. 2010).
Bayesian estimates of phylogeny for Figure 3 were generated using the MrBayes software package (version 3.1.2) on the Cyberinfrastructure for Phylogenetic Research (CIPRES) portal teragrid (version 3.1; Miller et al. 2010). We used the GTR + I + G model with Ambystoma as the out-group taxa for 10 million generations with sampling set at every 1000 generation to generate 10,000 pseudoreplicates. We discarded the first 3000 samples and the final majority consensus tree and posterior probabilities are based the remaining 7000 pseudoreplicates. We executed ML analysis in RAxML (version 7.2.7) on the CIPRES portal using a GTR + G model with all parameters set to default states.
Figure 3.
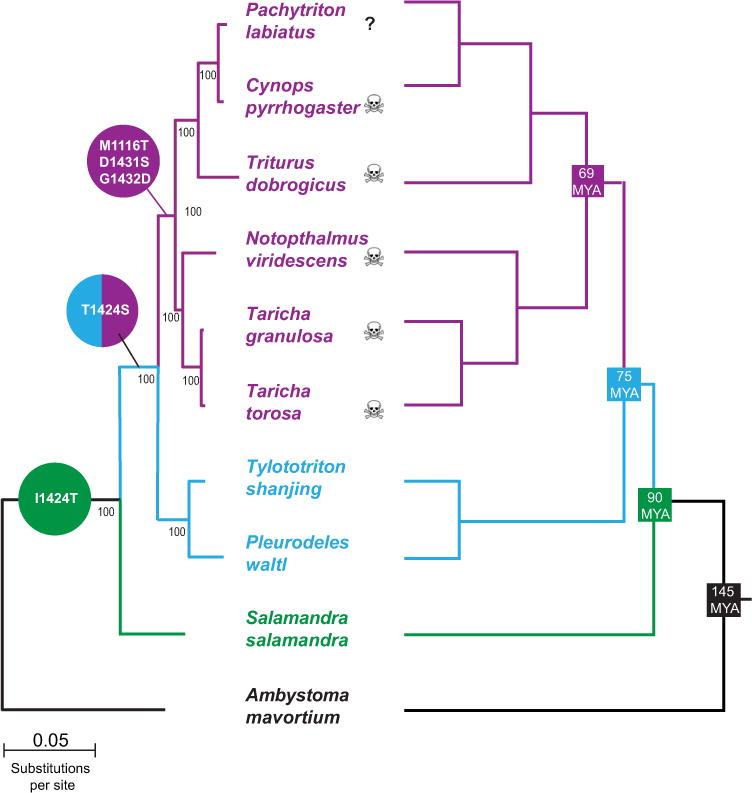
Evolutionary history of adaptation in salamander NaV 1.4. The general congruence of the gene tree for salSCNA4a (salamander NaV 1.4; left) with established evolutionary history (right) of sampled taxa demonstrates the evolutionary history of adaptation to TTX resistance in salamanders. Adaptive amino acid replacements associated with the evolution of TTX resistance are mapped in circles (color coded to salamandrid lineages as Fig. 1). Phylogeny of NaV 1.4 from salamanders is based on 4.6 kb of NaV 1.4 coding sequence. Bootstrap values are shown but topology and nodal support values were also estimated via Bayesian and maximum likelihood tree searches (not shown; Miller et al. 2010). Evolutionary relationships of the Salamandridae and Ambystoma as well as divergence dates (right) are from Zhang and Wake (2009).
Analysis of gene orthology relationships (Fig. S1) was generated using the MrBayes software package (version 3.2.2) on the XSEDE cluster (Miller et al. 2010). We used the GTR + I + G model with no preset out-group with other values set a default for 10 million generations with sampling set at every 1000 generation to generate 10,000 pseudoreplicates. We discarded the first 2500 samples and the final majority consensus tree and posterior probabilities are based the remaining 7500 pseudoreplicates. The final tree contains Bayesian probability support for all nodes.
Protein alignments for Figure S2 were generated using T-Coffee with default parameters (Centre for Genomic Regulation, Barcelona). Final alignments were prepared using BoxShade (version 3.2.1) with default parameters on the ExPASy bioinformatics resource portal.
Results
The peripheral neuromuscular system of the TTX-bearing salamander, T. granulosa, is extremely resistant to TTX (Fig. 1). Submillimolar concentrations of TTX (300 μM) had minimal effect on the AP in T. granulosa (Fig. 1A). In contrast, APs in the non-TTX-bearing out-group species, A. mavortium, were eliminated in submicromolar concentrations of TTX (0.025 μM, Fig. 1A). Results from Taricha are consistent with levels of TTX resistance observed in TTX-resistant garter snakes that eat toxic Taricha as well as puffer fish that also posses high levels of TTX; however, our results suggest that TTX-bearing salamanders are approximately an order of magnitude more resistant to TTX than either of these lineages (Geffeney et al. 2002, 2005; Jost et al. 2008). Results from Ambystoma are also consistent with other TTX-sensitive vertebrate species. In these lineages, nanomolar concentrations of TTX are sufficient to block APs (Hille 1992; Geffeney et al. 2005).
Analysis of the TTX concentrations required to block APs in the extant evolutionarily intermediate lineages of the Salamandridae (true salamanders and primitive newts) demonstrates that all lineages of this clade posses some degree of resistance to TTX (Fig. 1A). However, the degree of TTX resistance in these lineages was variable, with more derived lineages progressively more resistant to TTX (Fig. 1A). In the true salamander, S. salamandra, 0.05 μM TTX was required to block APs (compared to 0.025 μM in Ambystoma; Fig. 1A). In the primitive newt T. shanjing, this same concentration of TTX (0.05 μM) failed to block APs and had a much different qualitative effect on the AP (Fig. 1A, see below).
Skeletal muscle TTX resistance in salamanders originates from decreased TTX sensitivity of the voltage-gated sodium channel expressed in this tissue (Fig. 1B). Because the ROR of an AP is proportional to flux of Na ions through the NaV channels of a given muscle fiber, estimates of AP-ROR provide a measure of the TTX sensitivity of expressed NaV channels (Geffeney et al. 2002, 2005). AP-ROR estimates from salamander muscles also confirmed a progressive pattern of TTX-resistant phenotypes in salamandrid lineages (Fig. 1B). In the TTX-bearing salamander T. granulosa, 300 μM concentrations of TTX generated a minimal, but measurable, reduction in the maximum ROR of the AP (Fig. 1B). In contrast, 0.01 μM TTX produced much larger effects in all three non-TTX-bearing species (Fig. 1B). As with the concentration required to block APs, reduction in the rate of AP rise at this single concentration was graded among non-TTX-bearing species with Ambystoma ROR showing the highest reduction (40%) and the evolutionary intermediate lineages falling sequentially in between Ambystoma and Taricha (Salamandra: 25%; Tylototriton: 19%; Fig. 1B). These results demonstrate that skeletal muscle resistance in these lineages results from the expression of TTX resistant NaV channels and that variability in TTX resistance measured in AP block experiments results from variation in the in TTX sensitivity of expressed NaV channels.
To elucidate the genetic basis of TTX resistance in expressed skeletal muscle NaV channels, we isolated and sequenced approximately 95% (≈4200 nt [1400 amino acids], see Methods) of the single NaV channel expressed in the skeletal muscle of nine species of Salamandridae as well as the out-group taxa (A. mavortium). Alignments with vertebrate NaV channels demonstrate that the channel expressed in skeletal of all salamanders is orthologous to the vertebrate skeletal muscle channel SCN4a or NaV 1.4 (Fig. S1). Although there is evidence that low levels of other NaV channel paralogs are expressed in the skeletal muscle of mammals, amphibians appear to posses a reduced complement of NaV genes relative to amniotes, and paralogs such as NaV 1.7 that have been identified in mammal skeletal muscle do not appear to be present in salamanders (Novak et al. 2006; Zakon et al. 2009, 2011). We saw no evidence that other channel paralogs were expressed in the skeletal muscle of salamanders (see Methods).
Comparisons of the amino acid and nucleotide sequence of salSCNA4a from the non-TTX-bearing, TTX-sensitive out-group species A. mavortium and the highly TTX-resistant, TTX-bearing species T. granulosa show high levels of sequence conservation with 89.4% sequence identity between NaV 1.4 from A. mavortium and T. torosa (Figs. 2, S2). Although we identified multiple variable sites in salSCN4a of T. granulosa relative to A. mavortium, four substitutions (M1116T in Domain III; I1424S, D1431S, and G1432D in Domain IV) in the pore loop of NaV 1.4 (salSCN4a) likely play key roles in the extreme TTX resistance measured in T. granulosa (Figs. 2, S2). The M1116T, I1424S, D1431S, and G1432D substitutions occur in pore helix or selectivity filter-linker residues and substitutions at these sites have either been shown to decrease the TTX sensitivity of NaV channels or are associated with high levels of TTX resistance in other TTX-resistant species (see Discussion). We identified two additional substitutions in the P1 helix of Domain I (S146N and A151S) and one additional substitution in the P2 helix of DIV (N1435I) in Taricha NaV 1.4 (Fig. 2). Although these substitutions occur in the pore loop, there is no evidence that these residues play a role in TTX binding and their distribution among salamandrid lineages does not allow a clear view of their phenotypic contributions (Terlau et al. 1991; Yamagishi et al. 2001; Payandeh et al. 2011).
Figure 2.
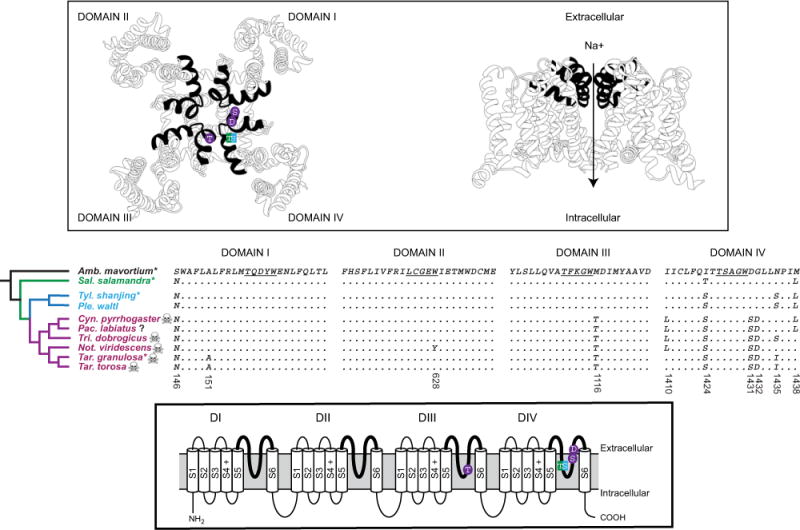
Substitutions in the salamander voltage-gated sodium channel, NaV 1.4. The phylogenetic distribution of amino acid substitutions in the outer pore of salamander NaV 1.4 (middle) is shown with a model of the TTX-binding site/outer pore (in black, above) and the transmembrane organization of NaV 1.4 (below). Candidate substitutions are also shown on the schematic structures of NaV 1.4 (above and below). Sequence alignments of the P1 and P2 helices with the selectivity filter linker (underlined) of all four domains are shown. Four S5–S6 extracellular linker regions (pore loops, bold lines) form the TTX-binding site as well as the part of the pore responsible for selective permeability to Na+ ions. Three substitutions (M116T, D1431S, and G1432D) are coupled with an extreme increase in the TTX resistance of modern newts, and Thr (true salamanders) and Ser (primitive and modern newts) substitutions at I1424 are associated with concomitant increases in TTX resistance of these lineages (Fig. 1). Substitutions and tree topology are color coded as in Fig. 1. Species for which TTX resistance was assayed (∗) and species known to possess TTX (skull) are indicated. The presence of TTX in Pachytriton is unknown (?). The upper model is based on the crystal structure of the bacterial sodium channel and Tikhonov and Zhorov (2012). Phylogentic relationships are inferred from Zhang et al. (2008) and are in agreement with the topology of salSCN4a (Fig. S1). Amino acid positions are based on sequence from Taricha torosa (GenBank numbers, TBD).
The M1116T, I1424S, D1431S, and G1432D substitutions identified in T. granulosa were present in its sister species T. torosa as well as representative species from all extant genera of modern newts (Figs. 2, S2). The S146N present in T. granulosa was present in all species of modern newts, however A151S was only identified in T. torosa and was not present in other modern newts. We identified additional pore-loop substitutions with lineage-specific distributions in modern newts. A W628Y substitution was present in the selectivity filter of Domain II in Notopthalmus viridescens (Fig. 2). Additionally, an I1410L substitution in Domain IV was present in the clade that contains all old world genera of modern newts (Cynops, Pachytriton, and Triturus) as well as the eastern North American genus Notopthalmus (Fig. 2). The N1435I substitution identified in T. granulosa was present in T. torosa but absent in Cynops, Pachytriton, and Notopthlamus, and in Triturus we identified an alternative serine replacement instead of isoleucine at N1435. Finally, a M1438L substitution was present in the two Asian species of modern newts (Cynops and Pachytriton). As with substitutions at A151, S146, and N1435, the phenotypic effect of substitutions observed at I1410 and M1438 in some modern newts cannot be predicted but current models suggest that they have limited effect on the TTX sensitivity of the channel (see Discussion).
Sodium channel (salSCN4a, NaV 1.4) sequence data from one species of true salamander (S. salamandra) and two primitive newts (Pleurodeles watl and T. shanjing) provided insight into the molecular basis of TTX resistance and observed variation in TTX sensitivity of salSCN4a among salamander lineages that are not known to possess TTX (see Methods, Figs. 2, S2). We observed no changes at M1116, D1431, or G1432 in the pore loop of either the primitive newts (P. watl and T. shanjing) or the true salamander (S. salamandra), but the I1424S substitution was present in primitive newts as was an I424T substitution in the true salamander (Fig. 2). The S146N Domain I substitution present in modern newts was present in both primitive newts as well as true salamanders (Fig. 2). No substitutions in either Domain II or III were identified, but an N1435S similar to that seen in Triturus was present in both species of primitive newts as was the M1438L substitution identified in Asian modern newts (Cynops and Pachytriton, Fig. 2).
Discussion
The distribution of observed substitutions among salamander lineages coupled with the evolutionary history of salSCN4a provides some insight into the history of skeletal muscle resistance in the Salamandridae (Fig. 3). Because the gene tree for salSCN4a is congruent with the current phylogeny of the Salamandridae, we can use the known species relationships of this clade to infer the likely order in which observed substitutions in salSCN4a occurred (Fig. 3). With the exception of the Ser replacement at I424 single nucleotide changes in the out-group A. mavortium, salSCNA4a could generate three of the primary substitutions (M1116T in Domain III; D1431S and G1432D in Domain IV) that likely contribute to the TTX-resistant phenotype observed in NaV 1.4 of modern newts (Figs. 2, 3). The I1424S replacement observed in both primitive and modern newts required two sequential mutations and the likely order of each of these changes can be observed in the I1424T and subsequent T1424S present in true salamanders and newts (primitive and modern), respectively (Fig. 3). The transition from isoleucine to threonine in true salamanders requires only a single nucleotide change, as does the transition from threonine to serine in primitive and modern newts. This pattern suggests that an ancient mutation (145–90 MYA) resulted in the I1424T replacement in the ancestor of all modern salamandrids (Fig. 3). This mutation was followed by a subsequent mutation at the same position in the ancestor of all newts (primitive and modern) roughly 90–75 MYA that resulted in the T1424S replacement present in these taxa (Fig. 3). These sequential replacements at I1424 were then followed by three mutations (M1116T, D1431S, and G1432D) that occurred over the roughly 6 million years between the emergence of the common ancestor of modern newts and their subsequent divergence roughly 69 MYA (Fig. 3). The order and timing of other observed substitutions is less clear. The Domain I S146N substitution is ubiquitous in the Salamandridae but absent in A. mavortium suggesting that it may also be an ancient replacement in the clade. The other observed substitutions have distributions that prevent a clear interpretation of their evolutionary history.
A longstanding focus in evolution has centered on identifying the location and magnitude of the specific molecular changes underlying adaptive evolution (Dean and Thornton 2007; Dalziel et al. 2009; Storz and Wheat 2010; Natarajan et al. 2013). Although our ability to draw inferences related to these issues is limited, our results suggest that at least some mutations of large effect in the coding region of salSCN4a likely played an important role in the evolutionary history of TTX resistance and the TTX-bearing phenotype in salamanders. We identified clear differences in levels of TTX resistance among salamandrid lineages that are, in turn, correlated with a pattern of substitutions in NaV channel residues known to effect the TTX sensitivity of the channel (Figs. 1 and 2). The single NaV channel (NaV 1.4) expressed in the skeletal muscle of TTX-bearing newts is 30,000-fold more resistant to TTX than NaV 1.4 from TTX-sensitive vertebrates and approximately an order of magnitude more resistant to TTX than any other species of TTX-resistant vertebrate known (Geffeney et al. 2005; Jost et al. 2008). This extreme resistance is coupled with four substitutions in the pore loop of NaV 1.4 (M1116T, I1424S, D1431S, and G1432D) that extensive modeling, empirical manipulations, as well as comparative work from other TTX-resistant taxa have shown to directly affect TTX binding (Terlau et al. 1991; Geffeney et al. 2002, 2005; Jost et al. 2008).
Although direct quantification of each of the individual pore-loop substitutions in our data is not possible, we can draw important inferences about the likely effect of observed substitutions from earlier work on TTX and NaV channels. We identified three candidate substitutions that are found only in TTX-bearing modern newts (M1116T, D1431S, and G1432D) and that occur in pore-loop residues that have been shown to directly disrupt toxin–channel interactions and dramatically affect TTX binding (Terlau et al. 1991; Penzotti et al. 1998; Yamagishi et al. 2001). TTX binds to two conserved rings of negatively charged amino acids in the selectivity filter and P2 helices of the channel. The M1116T, D1431S, and G1432D replacements occur in residues that either form or are adjacent to these rings and are accessible to extracellular toxins (Terlau et al. 1991; Fozzard and Lipkind 2010; Payandeh et al. 2011; Tikhonov and Zhorov 2012). Studies of natural substitutions in other TTX-resistant vertebrates as well as mutagenesis studies of NaV 1.4 demonstrate that the M1116T and charge-neutralizing D1431 replacements, as seen in modern newts, yield, respectively, 15-fold and 300-fold increases in NaV 1.4 resistance to TTX, and extreme TTX resistance of a single population of T. sirtalis corresponds to a Val replacement at the residue equivalent to G1432 in snake NaV 1.4 (Terlau et al. 1991; Geffeney et al. 2005; Jost et al. 2008). Although it is possible that the extreme phenotype observed in Taricha results, in part, from the contributions of other observed pore-loop substitutions (e.g., S146N, A151S in Domain I or replacements at I1410, N1435, or M1438 in Domain IV) or epistatic interactions among pore-loop substitutions, manipulative studies of mammalian NaV 1.4 as well as work with TTX-resistant garter snakes have shown that the large phenotypic effect of the M1116T substitution and substitutions at D1431 are not dependent on the genetic background in which they occur (Geffeney et al. 2005). This evidence suggests that the effect of the M1116T substitution and substitutions at D1431 in salamanders is likely comparable and that the occurrence of these substitutions in the ancestor of all modern newts played a critical role in the evolution of TTX resistance and, ultimately, the full TTX-bearing phenotype.
Ancient sequential replacements at the I1424 residue likely also have direct effects on TTX resistance. The true salamander S. salamandra is measurably more resistant to TTX than our TTX-sensitive out-group species (A. mavortium). Although we identified three substitutions (S146N, M1438, and I1424T) in or near the TTX-binding site of true salamander salSCN4a, changes at the I1424 position are known to affect TTX binding whereas there is no evidence that changes at S146 or M1438 regulate TTX binding (Terlau et al. 1991; Penzotti et al. 1998; Tikhonov and Zhorov 2012). Furthermore, the primitive newts T. shanjing and P. watl are measurably more resistant to TTX than either Ambystoma or Salamandra and posses a novel substitution (Ser) at I1424 but not at S146 or M1438 (Figs. 1, 2). Changes at this position do not appear to directly disrupt toxin–channel interactions but instead generate subtle changes in the shape of the charged rings that help form the TTX-binding site (Payandeh et al. 2011; Tikhonov and Zhorov 2012). In the garter snake T. sirtalis, a substitution at the equivalent residue of snake NaV 1.4 roughly doubles the estimated Kd, the concentration of TTX required to block 50% of available channels, of snake NaV 1.4 (Geffeney et al. 2005). Additionally, direct manipulation of snake NaV 1.4 demonstrated that substitutions at the I1424 position had additive effects when combined with substitutions at D1431 and that these effects are also not dependent on the genetic background in which they occur (Geffeney et al. 2005). These data coupled with our results suggest that ancient sequential changes at the I1424 likely played an important role in the evolution of skeletal muscle TTX resistance in salamanders. Interestingly, the isoleucine to valine substitution at this residue is ubiquitous in TTX-resistant populations of T. sirtalis and phylogenetic reconstructions of the history of TTX resistance in these populations suggest that, like salamanders, changes at this position may have occurred prior to other substitutions that confer higher levels of resistance to TTX (Geffeney et al. 2005).
Substitutions present in nonpore-loop regions of salSCN4a that are not addressed in detail above are unlikely to play an important role in TTX resistance. Extensive modeling and empirical results have shown these residues do not directly affect TTX binding (Catterall 1980a; Terlau et al. 1991; Penzotti et al. 1998; Cestele and Catterall 2000; Choudhary et al. 2003; Fozzard and Lipkind 2010; Payandeh et al. 2011; Tikhonov and Zhorov 2011, 2012). Additionally, epistatic interactions between nonpore-loop residues and pore-loop residues also do not directly affect TTX binding (Geffeney et al. 2005; Tikhonov and Zhorov 2012). As a result, the distribution of Domains III and IV pore-loop substitutions discussed above likely explains a significant portion of the variation in phenotypes among salamander lineages. The exception to this may be the W628Y identified in N. viridescens. Although current evidence suggests that TTX sensitivity is most strongly regulated by Domains I and IV, this W628Y substitution likely affects TTX binding. Changes at this position have been identified in marine gastropods that are exposed to TTX and the related toxin saxitoxin suggesting that this residue also regulates toxin–channel interactions (Bricelj et al. 2005). We do not have phenotype measures from Notopthalmus but the presence of this substitution warrants further investigation.
One challenge of evolutionary biology is to elucidate how multifactorial, complex phenotypes evolve (Monteiro and Podlaha 2009; Lynch 2010; Wray 2013). Most adaptations integrate multiple, genetically independent characters and result from the complex interplay of multiple genes. In the case of the TTX-bearing phenotype, the ability to use TTX as a defense against predation also requires resistance to TTX itself. The distribution of TTX-resistant phenotypes and associated genotypes among extant lineages of the Salamandridae provides a compelling view of the evolutionary history of the TTX-bearing phenotype as well as insight into this issue.
Our results suggest that some form of exaptation has played a role in the evolution of the full TTX-bearing phenotype. In salamanders, TTX is limited to the modern newts. Six of the seven extant genera of this clade possess TTX but it is not present in other species of the Salamandridae including the sister clade of primitive newts. Modern newts share substitutions that likely confer the extreme TTX resistance observed T. granulosa to all members of the clade and the widespread use of TTX as a de-fensive trait in the modern newts but not in other closely related lineages suggests this extreme resistance is a critical element of the TTX-bearing phenotype. However, we measured moderate levels of skeletal muscle TTX resistance in two lineages (true salamanders and primitive newts) of the Salamandridae that do not posses TTX. Although the role of TTX resistance in these lineages is unclear, the presence of TTX resistance without TTX itself in these basal lineages suggests that TTX resistant in the Salamandridae is ancient and predates the full TTX-bearing phenotype. Whether TTX resistance in basal lineages is indicative of exaptation requires further investigation because the functional role of TTX resistance in non-TTX-bearing lineages or ancestral salamandrid lineages is unclear. Ultimately, our results provide some evidence that mutations in highly conserved protein can play an important role in the evolution of novel complex phenotypes.
Supplementary Material
Table S1. Primers for RT-PCR and PCR cloning.
Table S2. RACE primers (forward).
Figure S1. Phylogenetic tree of voltage-gated sodium channels using Bayesian inference.
Figure S2. Amino acid alignment of partial sequence of NaV 1.4 from 10 species of salamanders.
Acknowledgments
We thank H. B. Schaeffer, E. D. Brodie Jr., and E. D. Brodie III for providing specimens as well as photographs. We thank M. Sotomayor for model parameters for use in construction of Figure 2. B. Williams, E. Brodie Jr., S. Geffeney, and three anonymous reviewers provided helpful comments in improving this manuscript. Z. Lebaric and C. Ellinger provided help with sequencing and laboratory work. Sequence data for 10 species of salamander salSCN4a have been assigned to GenBank under accession numbers (KP118968-KP118977). This work was funded by a R. L. Kirschstein National Research Service Award to CTH.
Footnotes
The authors state that they have no conflict of interest to declare.
DATA ARCHIVING
The doi for our data is 10.5061/dryad.b0vt6.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s website:
LITERATURE CITED
- Alfoldi J, Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23:1063–1068. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley-Ross MA, Barker JU. The effect of fiber-type heterogeneity on optimized work and power output of hindlimb muscles of the salamander Ambystoma tigrinum. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002;188:611–620. doi: 10.1007/s00359-002-0336-4. [DOI] [PubMed] [Google Scholar]
- Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 2012;490:535–538. doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- Bricelj VM, Connell L, Konoki K, MacQuarrie SP, Scheuer T, Catterall WA, Trainer VL. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature. 2005;434:763–767. doi: 10.1038/nature03415. [DOI] [PubMed] [Google Scholar]
- Brodie ED., Jr Investigations on the skin toxin of the adult rough-skinned newt. Taricha granulosa Copeia. 1968;2:307–313. [Google Scholar]
- Brodie ED, Jr, Hensel JL, Jr, Johnson JA. Toxicity of the urodele amphibians Taricha, Notophthalmus, Cynops and Paramesotriton (salamandridae) Copeia. 1974;1:506–511. [Google Scholar]
- Brodie ED, Jr, Ridenhour BJ, Brodie ED., III The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution. 2002;56:2067–2082. doi: 10.1111/j.0014-3820.2002.tb00132.x. [DOI] [PubMed] [Google Scholar]
- Brodie ED, III, Feldman CR, Hanifin CT, Motychak JE, Mulcahy DG, Williams BL, Brodie ED., Jr Parallel arms races between garter snakes and newts involving tetrodotoxin as the phenotypic interface of coevolution. J Chem Ecol. 2005;31:343–356. doi: 10.1007/s10886-005-1345-x. [DOI] [PubMed] [Google Scholar]
- Brown MS, Mosher HS. Tarichatoxin: isolation and purification. Science. 1963;140:295–296. doi: 10.1126/science.140.3564.295. [DOI] [PubMed] [Google Scholar]
- Buchwald HD, Durham L, Fischer HG, Harada R, Mosher HS, Kao CY, Fuhrman FA. Identity of tarichatoxin and tetrodotoxin. Science. 1964;143:474–475. doi: 10.1126/science.143.3605.474. [DOI] [PubMed] [Google Scholar]
- Cardall BL, Brodie ED, Jr, Brodie ED, III, Hanifin CT. Secretion and regeneration of tetrodotoxin in the rough-skin newt (Taricha granulosa) Toxicon. 2004;44:933–938. doi: 10.1016/j.toxicon.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Neurotoxins that act on voltage-sensitive sodium channels in excitable membranes. Annu Rev Pharmacol Toxicol. 1980a;20:15–43. doi: 10.1146/annurev.pa.20.040180.000311. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Pharmacologic properties of voltage-sensitive sodium channels in chick muscle fibers developing in vitro. Dev Biol. 1980b;78:222–230. doi: 10.1016/0012-1606(80)90331-0. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Cestele S, Yarov-Yarovoy V, Yu FH, Konoki K, Scheuer T. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124–141. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Cestele S, Catterall WA. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie. 2000;82:883–892. doi: 10.1016/s0300-9084(00)01174-3. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Yotsu-Yamashita M, Shang L, Yasumoto T, Dudley SC., Jr Interactions of the C-11 hydroxyl of tetrodotoxin with the sodium channel outer vestibule. Biophys J. 2003;84:287–294. doi: 10.1016/S0006-3495(03)74849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel AC, Rogers SM, Schulte PM. Linking genotypes to phenotypes and fitness: how mechanistic biology can inform molecular ecology. Mol Ecol. 2009;18:4997–5017. doi: 10.1111/j.1365-294X.2009.04427.x. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Pond SL Kosakovsky. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Jr, Brodie ED, III, Pfrender ME. The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proc Natl Acad Sci USA. 2009;106:13415–13420. doi: 10.1073/pnas.0901224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Jr, Brodie ED, III, Pfrender ME. Genetic architecture of a feeding adaptation: garter snake (Thamnophis) resistance to tetrodotoxin bearing prey. Proc R Soc B. 2010;277:3317–3325. doi: 10.1098/rspb.2010.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Dover Publications; New York: 1958. [Google Scholar]
- Fozzard HA, Lipkind GM. The tetrodotoxin binding site is within the outer vestibule of the sodium channel. Mar Drugs. 2010;8:219–234. doi: 10.3390/md8020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman FA, Kao CY, Mosher HS, Brown MS. Tarichatoxin—a potent neurotoxin from the California newt. Proc West Pharmacol Soc. 1963;6:31–32. [PubMed] [Google Scholar]
- Garland T, Jr, Adolph SC. Why not to do two-species comparative studies: limitations on inferring adaptation. Physiol Zool. 1994;67:797–828. [Google Scholar]
- Geffeney S, Brodie ED, Jr, Ruben PC, Brodie ED., III Mechanisms of adaptation in a predator-prey arms race: TTX-resistant sodium channels. Science. 2002;297:1336–1339. doi: 10.1126/science.1074310. [DOI] [PubMed] [Google Scholar]
- Geffeney SL, Fujimoto E, Brodie ED, 3rd, Brodie ED, Jr, Ruben PC. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature. 2005;434:759–763. doi: 10.1038/nature03444. [DOI] [PubMed] [Google Scholar]
- Hanifin CT. The chemical and evolutionary ecology of tetrodotoxin (TTX) toxicity in terrestrial vertebrates. Mar Drugs. 2010;8:577–593. doi: 10.3390/md8030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanifin CT, Brodie ED, III, Brodie ED., Jr Tetrodotoxin levels of the rough-skin newt, Taricha granulosa, increase in long-term captivity. Toxicon. 2002;40:1149–1153. doi: 10.1016/s0041-0101(02)00115-0. [DOI] [PubMed] [Google Scholar]
- Hanifin CT, Brodie ED, Jr, Brodie ED., III Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 2008;6:e60. doi: 10.1371/journal.pbio.0060060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ionic channels of excitable membranes. Sinauer Associates; Sunderland, MA: 1992. [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost MC, Hillis DM, Lu Y, Kyle JW, Fozzard HA, Zakon HH. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol Biol Evol. 2008;25:1016–1024. doi: 10.1093/molbev/msn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY, Fuhrman FA. Pharmacological studies on tarichatoxin, a potent neurotoxin. J Pharmacol Exp Ther. 1963;140:31–40. [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lehman EM, Brodie ED, Jr, Brodie ED., III No evidence for an endosymbiotic bacterial origin of tetrodotoxin in the newt Taricha granulosa. Toxicon. 2004;44:243–249. doi: 10.1016/j.toxicon.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Losos JB, Arnold SJ, Bejerano G, Brodie ED, III, Hibbett D, Hoekstra HE, Mindell DP, Monteiro A, Moritz C, Orr HA, et al. Evolutionary biology for the 21st century. PLoS Biol. 2013;11:e1001466. doi: 10.1371/journal.pbio.1001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Scaling expectations for the time to establishment of complex adaptations. Proc Natl Acad Sci USA. 2010;107:16577–16582. doi: 10.1073/pnas.1010836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish DM, Rajon E, Shah P, Ding Y, Plotkin JB. The role of epistasis in protein evolution. Nature. 2013;497:E1–E2. doi: 10.1038/nature12219. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Mebs D, Arakawa O, Yotsu-Yamashita M. Tissue distribution of tetrodotoxin in the red-spotted newt Notophthalmus viridescens. Toxicon. 2010;55:1353–1357. doi: 10.1016/j.toxicon.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. IEEE; New York: 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Monteiro A, Podlaha O. Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 2009;7:e37. doi: 10.1371/journal.pbio.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 2010;26:484–492. doi: 10.1016/j.tig.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Narahashi T. Mechanism of action of tetrodotoxin and saxitoxin on excitable membranes. Fed Proc. 1972;31:1124–1132. [PubMed] [Google Scholar]
- Narahashi T. Tetrodotoxin: a brief history. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:147–154. doi: 10.2183/pjab.84.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Inoguchi N, Weber RE, Fago A, Moriyama H, Storz JF. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak AE, Jost MC, Lu Y, Taylor AD, Zakon HH, Ribera AB. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- Orr HA. The population genetics of beneficial mutations. Philos Trans R Soc Lond B Biol Sci. 2010;365:1195–1201. doi: 10.1098/rstb.2009.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC., Jr Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J. 1998;75:2647–2657. doi: 10.1016/S0006-3495(98)77710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Projecto-Garcia J, Natarajan C, Moriyama H, Weber RE, Fago A, Cheviron ZA, Dudley R, McGuire JA, Witt CC, Storz JF. Repeated elevational transitions in hemoglobin function during 110: 20669–20674 the evolution of Andean hummingbirds. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Reger J, Feulner PG, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, Slate J. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;25:705–712. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Heinemann SH, Stuhmer W, Pusch M, Conti F, Imoto K, Numa S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 1991;293:93–96. doi: 10.1016/0014-5793(91)81159-6. [DOI] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Possible roles of exceptionally conserved residues around the selectivity filters of sodium and calcium channels. J Biol Chem. 2011;286:2998–3006. doi: 10.1074/jbc.M110.175406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol Pharmacol. 2012;82:97–104. doi: 10.1124/mol.112.078212. [DOI] [PubMed] [Google Scholar]
- Travisano M, Shaw RG. Lost in the map. Evolution. 2013;67:305–314. doi: 10.1111/j.1558-5646.2012.01802.x. [DOI] [PubMed] [Google Scholar]
- Wakely JF, Fuhrman GJ, Fuhrman FA, Fischer HG, Mosher HS. The occurrence of tetrodotoxin (tarichatoxin) in amphibia and the distribution of the toxin in the organs of newts (Taricha) Toxicon. 1966;3:195–203. doi: 10.1016/0041-0101(66)90021-3. [DOI] [PubMed] [Google Scholar]
- Walthall JC, Ashley-Ross MA. Postcranial myology of the California newt, Taricha torosa. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:46–57. doi: 10.1002/ar.a.20279. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Williams BL. Behavioral and chemical ecology of marine organisms with respect to tetrodotoxin. Mar Drugs. 2010;8:381–398. doi: 10.3390/md8030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. Genomics and the evolution of phenotypic traits. Annu Rev Ecol Evol Syst. 2013;44:51–72. [Google Scholar]
- Yamagishi T, Li RA, Hsu K, Marban E, Tomaselli GF. Molecular architecture of the voltage-dependent Na channel: functional evidence for alpha helices in the pore. J Gen Physiol. 2001;118:171–182. doi: 10.1085/jgp.118.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Jost MC, Zwickl DJ, Lu Y, Hillis DM. Molecular evolution of Na+ channels in teleost fishes. Integr Zool. 2009;4:64–74. doi: 10.1111/j.1749-4877.2008.00136.x. [DOI] [PubMed] [Google Scholar]
- Zakon HH, Jost MC, Lu Y. Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 2011;28:1415–1424. doi: 10.1093/molbev/msq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Papenfuss TJ, Wake MH, Qu L, Wake DB. Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2008;49:586–597. doi: 10.1016/j.ympev.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wake DB. Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2009;53:492–508. doi: 10.1016/j.ympev.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Zhorov BS, Tikhonov DB. Ligand action on sodium, potassium, and calcium channels: role of permeant ions. Trends Pharamacol Sci. 2013;34:154–161. doi: 10.1016/j.tips.2013.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers for RT-PCR and PCR cloning.
Table S2. RACE primers (forward).
Figure S1. Phylogenetic tree of voltage-gated sodium channels using Bayesian inference.
Figure S2. Amino acid alignment of partial sequence of NaV 1.4 from 10 species of salamanders.


