Abstract
Pax genes play important roles in Metazoan development. Their evolution has been extensively studied but Lophotrochozoa are usually omitted. We addressed the question of Pax paralog diversity in Lophotrochozoa by a thorough review of available databases. The existence of six Pax families (Pax1/9, Pax2/5/8, Pax3/7, Pax4/6, Paxβ, PoxNeuro) was confirmed and the lophotrochozoan Paxβ subfamily was further characterized. Contrary to the pattern reported in chordates, the Pax2/5/8 family is devoid of homeodomain in Lophotrochozoa. Expression patterns of the three main pax classes (pax2/5/8, pax3/7, pax4/6) during Sepia officinalis development showed that Pax roles taken as ancestral and common in metazoans are modified in S. officinalis, most likely due to either the morphological specificities of cephalopods or to their direct development. Some expected expression patterns were missing (e.g. pax6 in the developing retina), and some expressions in unexpected tissues have been found (e.g. pax2/5/8 in dermal tissue and in gills). This study underlines the diversity and functional plasticity of Pax genes and illustrates the difficulty of using probable gene homology as strict indicator of homology between biological structures.
Introduction
Pax proteins belong to a family of transcription factors playing important roles in development of metazoans, from early specification of cell fate to body patterning through morphogenesis of various tissues and organs [1,2]. The evolution of Pax genes has been extensively studied and the availability of genomes has allowed for the identification of Pax genes in nearly 200 species of chordates in which duplication and subfunctionalization may have occurred several times [3]. Pax genes have also been identified in non-chordates, but no exhaustive study of Pax gene evolution has been conducted regarding these clades. In fact lophotrochozoans, which comprise approximately 30% of all animal species, are usually omitted in studies of Pax gene evolution (see [4] as a recent example).
Pax proteins are characterized by the presence of a DNA-binding domain of 128 amino-acids, referred to as the paired domain (PRD) (review in [5]). Other conserved domains may be present in the sequence of Pax proteins after the PRD: an octapeptide motif (OM) and part or all of a paired-type homeobox DNA-binding domain (HD). A common ancestor (“Ur-pax-gene”) containing the three domains would have led to the “classical” five classes of Pax genes recognized in Ecdysozoa and Chordata (pox-neuro, pax4/6, pax2/5/8, pax1/9 and pax3/7) by gene duplications and subsequent deletions before the divergence of Protostomia and Deuterostomia [6–10]. Moreover, a sixth Pax clade (pax-a/β) has recently been proposed by some authors ([11–13]) and the status of a seventh class (eyg) remains unclear [14]. In addition, further gene or genome duplication events leading to different Pax paralogs, as well as alternative splicing, are known to generate numerous Pax isoforms in chordate (e.g. [15–17]). No basal genome duplication has been demonstrated in Lophotrochozoa and they are thought to use a restricted repertoire of Pax proteins. Nevertheless some Pax isoforms have been characterized. Each of pax6 [18], pax 3/7 [19] and pax β [12] have two isoforms in Helobdella robusta (Annelida). Five isoforms of pax6 resulting from alternative splicing without genome duplication have been characterized in Idiosepius paradoxus (Mollusca Cephalopoda) embryos [20]. A recent article has extensively studied the evolution of pax2/5/8 among molluscs [21] but no extensive search has addressed the question of Pax paralog diversity in Lophotrochozoa. A first objective of this paper is to thoroughly characterize the set of Pax genes in Lophotrochozoa.
The diversity of Pax molecular family is assumed to explain the high functional diversity of Pax proteins [10,22] however there is no unique developmental function for each Pax despite the highly conserved general structure of Pax genes [23,24]. In lophotrochozoan species studied so far (Platyhelminthes [25,26], Annelida [18,19,23,27,28], Mollusca [21,29–34], Brachiopoda [35,36], Nemertea [37]), expression patterns of each Pax gene suggest conserved and consistent roles for pax3/7 in nervous system development, pax2/5/8 in sensory structure formation and pax6 in eye morphogenesis. However, our different on-going studies regarding Sepia officinalis Pax genes during development [33,34,38–40] have called into question the consistency between Pax gene structure, expression and role in cephalopod Pax gene family. Thus, a second objective of this paper is to complete our previous studies on the expression patterns of Pax genes in the cuttlefish Sepia officinalis and to compare these expressions with other Lophotrochozoa. Expression patterns of the three main Pax classes during development show that Pax roles, taken as ancestral (e.g. [21]) and common in metazoans, are modified in S. officinalis. Changes of Pax roles we observed could be linked to an unusual body plan [41], a direct development and numerous morphological novelties which are specific to Cephalopoda, such as muscular and nervous structures related to specific behaviors, locomotion and cognitive abilities (see Fig 1). Comparison of gene expression patterns between metazoans is often used as a footprint for homology in evo-devo studies, disregarding body plan or developmental differences [42]. Therefore, our results remind that such a paradigm should always be used carefully.
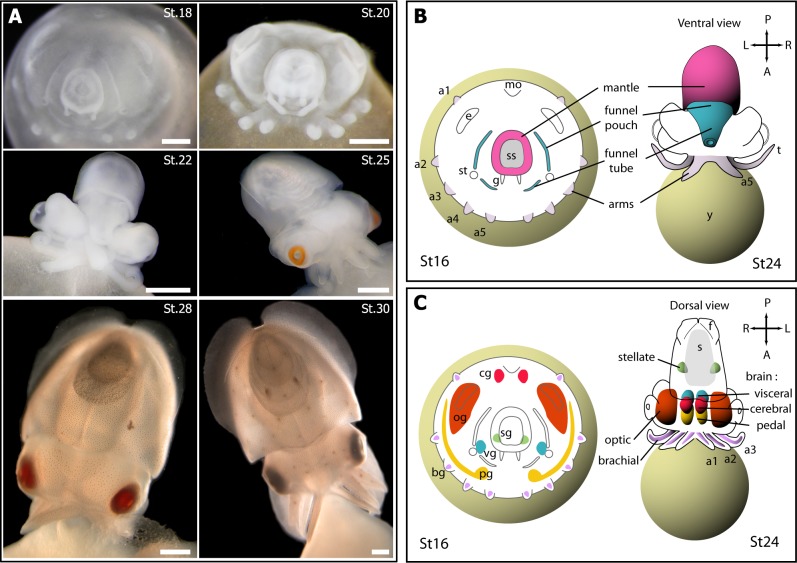
Fig 1. Development of Sepia officinalis and localisation of main nervous structures (after [38], modified).
(A) Organogenesis. St18: Plane phase (Stage 14–18) Stage 18, aboral view; the embryo is “disc-shaped” on the surface of yolk. St20, St22: Extension phase (Stages 19–22). St20: Stage 20, aboral view; the shell sac is closed and the two funnel tube elements grow. St22: Stage 22, ventral view; the funnel tube is formed. St25, St28, St30: Growth phase (Stages 23–30), dorsal view. St25: Stage 25; the shell begins to form, eyes are clearly coloured. St28: Stage 28; skin is clearly coloured. St30: Stage 30, just before hatching. (B) Muscular system: the mantle, the two elements of the funnel (funnel pouches and funnel tube) and arms, essential for locomotion, are coloured. (C) Nervous system: at stage 16, nervous system is composed of sparse ganglia. Cerebroid ganglia (red) will constitute the dorsal supraoesophageal mass of the brain, pedal (yellow) and visceral (blue) ganglia will constitute the anterior/median and posterior (respectively) sub-oesophageal mass of the brain (depicted in stage 24 dorsal view). Optic ganglia (orange) will constitute optic lobes; with the brain, they constitute the central nervous system. Stellate ganglia (green), located symmetrically on the internal side of the mantle and brachial ganglia (one by arm, which develops into a cord inside the arm) belong to the peripheral nervous system. For more information about development see [53]) a1, a2, a3, a4, a5: arms 1 to 5; bg, brachial ganglia; cg, cerebroid ganglia; e: eye; g: gill; mo: mouth; og, optic ganglia; pg, pedal ganglia; s, shell; st, statocyst; sg, stellate ganglia; ss: shell sac; t: tentacle; vg, visceral ganglia; y: yolk. Orientation: A(nterior)–P(osterior), L(eft)–R(ight). Scale bar: 1 mm.
Materials and methods
Sequence analysis
Putative Pax proteins were searched for using protein databases (Uniprot, NCBI protein) and accessible draft genomes (Octopus bimaculoides: metazome website https://metazome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Obimaculoides_er, Pinctada fucata: OIST marinegenomics website http://marinegenomics.oist.jp/pearl/viewer/info?project_id=20 [43]), transcriptome (our EST bank, [44]) or proteome (Mytilus galloprovincialis [45]) to construct a set of sequences restricted to the lophotrochozoan clade. Alignments were done using Jalview2 [46]. Phylogenetic analysis was performed on the “phylogeny.fr” platform [47] accessible at http://www.phylogeny.fr. After alignment, ambiguous regions (i.e. containing gaps and/or poorly aligned) were removed with Gblocks [48] using the following parameters: minimum length of a block after gap cleaning: 10, no gap positions allowed in the final alignment, all segments with contiguous non-conserved positions bigger than 8 rejected, minimum number of sequences for a flank position: 85%. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML (v3.1/3.0 aLRT) program [49]. The default substitution model was selected assuming an estimated proportion of invariant sites (of 0.088) and 4 gamma-distributed rate categories to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data (gamma = 0.716). Reliability for internal branch was assessed using the aLRT test (SH-Like) [50] or using the bootstrapping method (100 bootstrap replicates); all branches with support <50% were collapsed. Graphical representation and edition of the phylogenetic tree were performed with TreeDyn v198.3 [51].
Expression patterns
Sepia officinalis eggs were obtained from captive females (maintained in the biological station of Luc-sur-Mer, France) and maintained at 18°C in oxygenated seawater in the laboratory. The experimental procedures were carried out in strict compliance with the European Communities Council Directive (86/609/EEC) and followed the French legislation requirements (decree 87/848) regarding the care and use of laboratory animals, under the control of the ethics committee of the Muséum National d'Histoire Naturelle (“Comité Cuvier-68”). All efforts were made to minimize animal suffering and to reduce the number of animals used.
Embryos were sampled daily to assemble a complete collection of morphological stages from stage 14 (beginning of organogenesis) according to the developmental table established by Lemaire [52] and revised by Boletzky et al. [53]. The dark pigmented egg capsule and the chorion were removed in seawater. Embryos were maintained on ice in seawater until lethargy. They were then fixed and processed for in situ hybridization (ISH).
RNA extraction, cDNA synthesis and gene cloning of Sof-pax3/7 and Sof-pax6 have been described previously [33,34,54]. For Sof-pax2/5/8, the primers F-5’- ACCTAACCACAGCGTACCGT-3’ (DLTTAYR) and R-5’-GACCATGTTTGCCTGGGAGA-3’ (TMFAWE) were used to obtain a 420 bp fragment. The primers were designed in order to target each gene but without discrimination of their potential splicing variants: in addition to specific regions of pax6, pax3/7 and pax2/5/8 genes, a large part of the conserved Paired Domain was included in all probes (see S1 Primers for primer positions). Procedures for cloning, RNA probes synthesis, whole-mount ISH and sectioning have been described previously [34]. Embryos were observed with a Leica M16 2F binocular stereomicroscope and a Leica DMLB compound microscope. Maximum intensity projections were generated using ImageJ (http://rsbweb.nih.gov/ij/). All images were adjusted for contrast and brightness and assembled into plates using Adobe Photoshop 8 or CS4 (Adobe, San Jose, CA, USA).
Results
Pax alignments and Pax family in Lophotrochozoa
After an extensive search, a set of 216 putative Pax protein sequences restricted to the lophotrochozoan clade was compiled (S1 Table). As expected, the paired domain (PRD) signature of Pax proteins was highly conserved. A first alignment was made by automatic analysis based on the PRD domain but the general alignment of all sequences was manually obtained (S1 Alignment) using the PRD, OM (Octapeptide motif) and HD (Homeodomain) as visual guides.
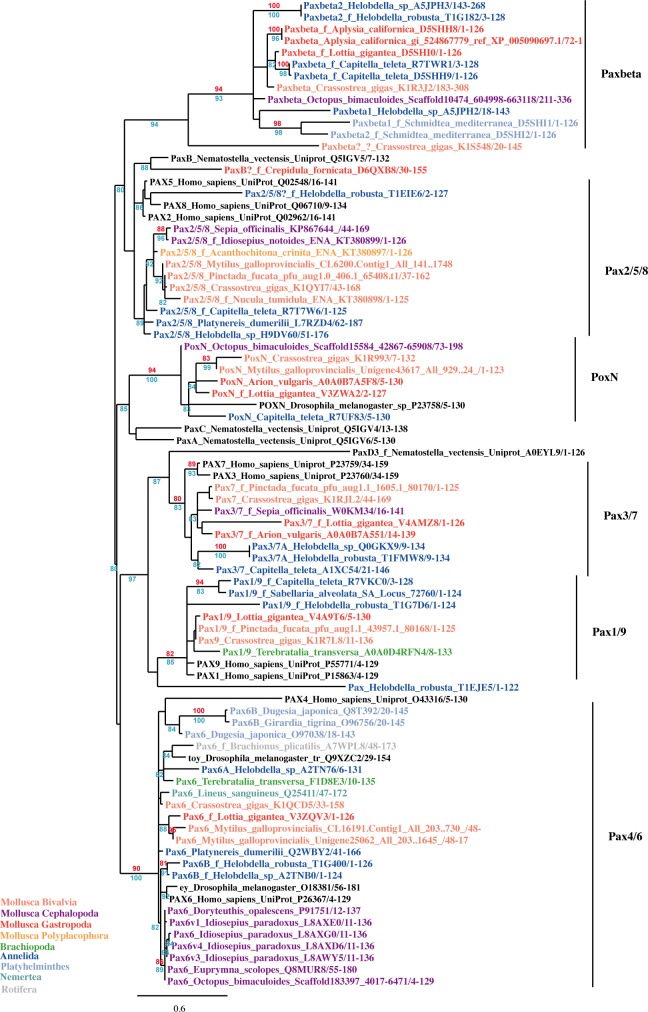
The PRD was used to construct phylogenetic trees of the lophotrochozoan Pax proteins. Human, Drosophila and Nematostella sequences were included to facilitate the identification of the classes of proteins obtained after phylogenetic reconstruction. The sets used in these analyses were obtained by elimination of redundant sequences (e.g. sequences from the same species containing 100% identity in the PRD) and sequences lacking important parts or whole PRD (e.g. 93 partial PRD sequences from various Cephalopoda and all putative eyg sequences). Depending on the stringency of this elimination, the procedure led to sets from 71 to 65 lophotrochozoan PRD sequences, which could be analysed on 108 to 121 positions (respectively). The phylogenetic trees obtained with these different sets were comparable and confirmed the presence of the 6 Pax families in Lophotrochozoa proposed by Schmerer et al. [12] (Fig 2). One representative phylogenetic tree is presented (Fig 3). The following description of each of these families is based on the alignment and supported by the phylogeny. 56 sequences that were previously non- or mis-identified are listed in Table 1.
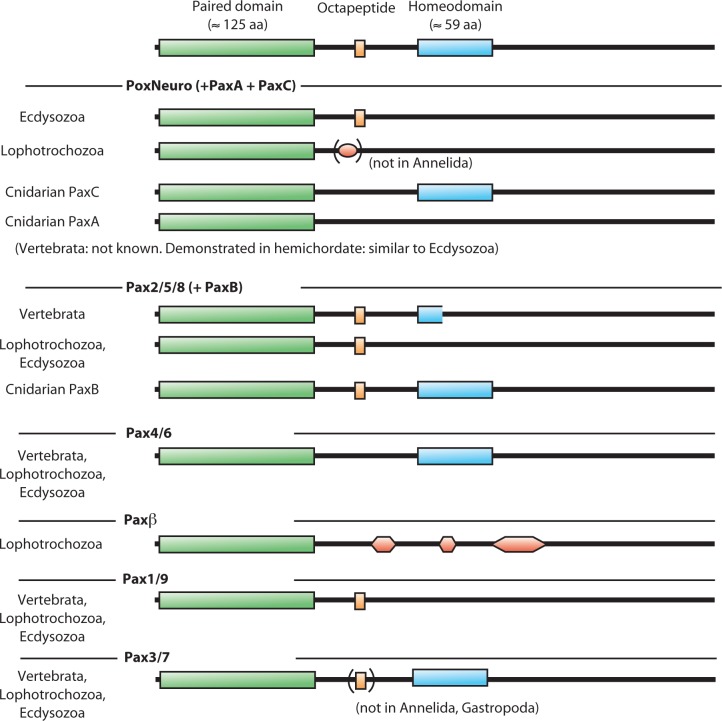
Fig 2. Domain composition of Pax proteins.
Data from [8,9,13] and this work. The known lophotrochozoan sequences present some distinctive features: no OP in PoxNeuro, no HD in Pax2/5/8, presence of Paxβ with characteristic signals.
Fig 3. Unrooted phylogenetic tree of lophotrochozoan Pax proteins.
All branches with support <50% were collapsed. The results of approximate Likelihood-Ratio Test (SH-Like) (blue) and bootstrap (red) are indicated if >80%. The sequences used and their reference numbers are given in S1 Table. The names used in the tree are our proposed identifications of these sequences if different from original annotations (see the differences between submitted name and proposed name in Table 1 and S1 Table).
Table 1. Previously non- or mis-identified lophotrochozoan Pax protein sequences.
| Accession | Submitted name | Proposed name | species | Classification |
|---|---|---|---|---|
| T1EJE5 | Uncharacterized protein | Pax | Helobdella_robusta | Annelida_Clitellata |
| T1G7D6 | Uncharacterized protein | Pax1/9(f) | Helobdella_robusta | Annelida_Clitellata |
| T1EIE6 | Uncharacterized protein | Pax2/5/8?(f) | Helobdella_robusta | Annelida_Clitellata |
| T1EH18 | Uncharacterized protein | Pax2/5/8(f) | Helobdella_robusta | Annelida_Clitellata |
| T1FMW8 | Uncharacterized protein | Pax3/7A | Helobdella_robusta | Annelida_Clitellata |
| T1G8F8 | Uncharacterized protein | Pax3/7B(f) | Helobdella_robusta | Annelida_Clitellata |
| T1F6U6 | Uncharacterized protein | Pax6A | Helobdella_robusta | Annelida_Clitellata |
| T1G400 | Uncharacterized protein | Pax6B(f) | Helobdella_robusta | Annelida_Clitellata |
| T1EHA5 | Uncharacterized protein | Paxβ1(f) | Helobdella_robusta | Annelida_Clitellata |
| T1G182 | Uncharacterized protein | Paxβ2(f) | Helobdella_robusta | Annelida_Clitellata |
| R7VKC0 | Uncharacterized protein | Pax1/9(f) | Capitella_teleta | Annelida_Polychaeta |
| R7T7W6 | Uncharacterized protein | Pax2/5/8(f) | Capitella_teleta | Annelida_Polychaeta |
| R7TKD0 | Uncharacterized protein | Pax3/7(f) | Capitella_teleta | Annelida_Polychaeta |
| R7TWR1 | Uncharacterized protein | Paxβ(f) | Capitella_teleta | Annelida_Polychaeta |
| R7UF83 | Uncharacterized protein | PoxN | Capitella_teleta | Annelida_Polychaeta |
| K1QWY6 | Paired box protein Pax-6 | eyg | Crassostrea_gigas | Mollusca_Bivalvia |
| K1QYI7 | Paired box protein Pax-2-A | Pax2/5/8 | Crassostrea_gigas | Mollusca_Bivalvia |
| K1R3J2 | Paired box protein Pax-2-A | Paxβ | Crassostrea_gigas | Mollusca_Bivalvia |
| K1S548 | Paired box protein Pax-6 | Paxβ(?) | Crassostrea_gigas | Mollusca_Bivalvia |
| K1R993 | Paired box protein Pax-8 | PoxN | Crassostrea_gigas | Mollusca_Bivalvia |
| Unigene67849_All_[769..2] | Unigene67849_All_[769..2] | Pax1/9(f) | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig1_All_[141..1748] | CL6200.Contig1_All_[141..1748] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig2_All_[141..1640] | CL6200.Contig2_All_[141..1640] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig3_All_[308..1978] | CL6200.Contig3_All_[308..1978] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig4_All_[308..2086] | CL6200.Contig4_All_[308..2086] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig5_All_[308..1885] | CL6200.Contig5_All_[308..1885] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL6200.Contig6_All_[308..1993] | CL6200.Contig6_All_[308..1993] | Pax2/5/8 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| Unigene61312_All_[208..2] | Unigene61312_All_[208..2] | Pax3/7(f) | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL16191.Contig1_All_[203..730] | CL16191.Contig1_All_[203..730] | Pax6 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| Unigene25062_All_[203..1645] | Unigene25062_All_[203..1645] | Pax6 | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| CL16191.Contig2_All_[1..435] | CL16191.Contig2_All_[1..435] | Pax6(f) | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| Unigene43617_All_[929..24] | Unigene43617_All_[929..24] | PoxN | Mytilus_galloprovincialis | Mollusca_Bivalvia |
| Scaffold15272:139788–157863 | Ocbimv22007526m.p | Pax2/5/8 | Octopus_bimaculoides | Mollusca_Cephalopoda |
| Scaffold183397:4017–6471 | Ocbimv22011111m.p | Pax6 | Octopus_bimaculoides | Mollusca_Cephalopoda |
| Scaffold17697:31829–33450 | Ocbimv22010462m.p | Pax9 | Octopus_bimaculoides | Mollusca_Cephalopoda |
| Scaffold10474:604998–663118 | Ocbimv22000807m.p | Paxβ | Octopus_bimaculoides | Mollusca_Cephalopoda |
| Scaffold15584:42867–65908 | Ocbimv22007901m.p | PoxN | Octopus_bimaculoides | Mollusca_Cephalopoda |
| gi|524867779|ref|XP_005090697.1 | PREDICTED: mucin-5AC | Paxβ | Aplysia_californica | Mollusca_Gastropoda |
| A0A0B6YZY1 | Uncharacterized protein (Fragment) GN = ORF42948 | Pax2/5/8?(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B7A551 | Uncharacterized protein (Fragment) GN = ORF96941 | Pax3/7(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B7A6X2 | Uncharacterized protein (Fragment) GN = ORF96938 | Pax3/7(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B7A6Y7 | Uncharacterized protein (Fragment) GN = ORF96935 | Pax3/7(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B6Y326 | Uncharacterized protein (Fragment) GN = ORF11503 | Pax6(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B6YFJ8 | Uncharacterized protein (Fragment) GN = ORF24120 | Pax6(f) | Arion_vulgaris | Mollusca_Gastropoda |
| A0A0B7A5F8 | Uncharacterized protein | PoxN | Arion_vulgaris | Mollusca_Gastropoda |
| gi|908452192|ref|XP_013082659.1 | PREDICTED: mucin-5AC-like | Paxβlike | Biomphalaria_glabrata | Mollusca_Gastropoda |
| A0A0B6VJL1 | Paired box 6 protein | eyg | Lottia_gigantea | Mollusca_Gastropoda |
| V4A9T6 | Uncharacterized protein | Pax1/9 | Lottia_gigantea | Mollusca_Gastropoda |
| V3ZI38 | Uncharacterized protein | Pax2/5/8(f) | Lottia_gigantea | Mollusca_Gastropoda |
| V4AMZ8 | Uncharacterized protein | Pax3/7(f) | Lottia_gigantea | Mollusca_Gastropoda |
| V3ZQV3 | Uncharacterized protein | Pax6(f) | Lottia_gigantea | Mollusca_Gastropoda |
| Contig4275(2463977–2470937) | LgGsHFWreduced.5213 | Paxβ | Lottia_gigantea | Mollusca_Gastropoda |
| V4B0D1 | Uncharacterized protein | Paxβ(f) | Lottia_gigantea | Mollusca_Gastropoda |
| V3ZWA2 | Uncharacterized protein | PoxN(f) | Lottia_gigantea | Mollusca_Gastropoda |
| Q25411 | Pax6-like protein | Pax6 | Lineus_sanguineus | Nemertea_Anopla |
| O96756 | DtPax-6 protein | Pax6B | Girardia_tigrina | Platyhelminthes_Rhabditophora |
Accession: accession number or genome reference. Submitted name: name found in databases. Proposed name: our interpretation (grey background if different from a submitted name); (f) denote a fragment. Sequences are included in S1 Table.
PoxNeuro
To our knowledge, the only PoxNeuro already signalled in molluscs before this study was Pon in Pinctada fucata [55]. Our phylogenetic analysis demonstrated the existence of other PoxNeuro, non- or mis-identified (see Table 1 and S1 Table), in molluscan clades (Gastropoda, Bivalvia and Cephalopoda), as well as in Annelida.
These sequences showed a tribasic signal in the first helix of the PRD and insertions corresponding to the known exon2 / exon3 junction in Drosophila (KPKQVAT) between the N-ter (PAI domain) and C-ter (RED domain) domains of the PRD. The HD was absent. By contrast with PoxNeuro from Ecdysozoa or the hemichordate Saccoglossum kowalevskii ([13,56], and our alignments not shown), no OM was clearly detected in Lophotrochozoa. However, a commonly found [VI]PGLSYP[KR][IL]V motif was present at least in Mollusca after the PRD.
Pax2/5/8
The Pax2/5/8 group was well identified but moderately supported in phylogenetic analysis. Contrary to the structure described in chordate, no partial HD could be identified in any lophotrochozoan Pax2/5/8. By contrast, a clear octapeptide (Y[TS]IX2ILG) was present, followed by a quadribasic/diacid signal already underlined in chordate (KR-rich region of [57]).
The transcriptome of Mytilus galloprovincialis revealed 6 isoforms of Pax2/5/8. The N-ter region flanking the PRD suggested two different groups of sequences, which may reflect the existence of two different genes and alternative splicing. By contrast, the existence of different isoforms of Pax2/5/8 in Annelida remained elusive. The Helobdella sp. complete sequence H9DV60 revealed two N-rich regions, reminiscent of the description of Paxβ by Schmerer et al. [12]. These regions surrounded a clear octapeptide but without the KRKR signal of Pax2/5/8. Nevertheless, this sequence was clearly assigned to the Pax2/5/8 group in accordance with its gene organisation description [28]. Unfortunately, the Helobdella robusta Pax2/5/8 sequence fragment T1EH18, flagged as « complete » but with « non-terminal residues » in Uniprot, does not cover the OM zone. The Helobdella robusta sequence T1EIE6 is quite different (% identity = 72% in the PRD) and appeared linked to different groups, depending on the set analysed.
Pax4/6
The Pax4/6 subfamily was clearly identified. These sequences displayed a canonical structure (PRD, no octapeptide, HD), including the characteristic MDKL pattern between PRD and HD [37].
Two Pax6 isoforms are known in Planaria, one of which (Pax6B) described as specific to this clade [58]. We did not detect any non-planarian homologue to the planarian B isoform which was the only Pax6 devoid of the MDKL motif. Noteworthy, the two planarian isoforms constituted a well-supported clade in phylogenetic analysis using the paired domain, in accordance with the results of Quigley et al. [18].
The transcriptome of Mytilus galloprovincialis contained at least 2 forms of Pax6: a long canonical isoform and a short isoform reduced to the paired zone. The only Pax6 detected in the genome of Octopus bimaculoides was lacking a homeodomain, although complete Pax6 are already known in other cephalopods [20]. The Crassostrea gigas Pax6 showed an insertion in the C-ter part (RED domain) of the PRD, reminiscent of the human Pax6-5a isoform which contains an insertion in the PAI domain thought to modulate the interaction between the Pax transcription factors and their DNA consensus sequence [59–62].
Two sequences previously identified as Pax6 (Lottia gigantea A0A0B6VJL1 and Crassostrea gigas K1QWY6) were in fact identified as eyegone homologues. This gene was also present in Pinctada genome: two sequences have already been signalled, most likely reflecting two allelic copies [55].
Pax beta
A Pax beta subfamily was present in phylogenetic analysis with a good support. This subfamily has been described as lophotrochozoan-specific with two forms known in Annelida (Paxβ1, Paxβ2), corresponding to two genes [12]. Paxβ complete sequences were found in Octopus bimaculatus (Ocbimv22000807m.p), Lottia gigantea (LgGsHFWreduced.5213) and Crassostrea gigas (K1R3J2), the later misidentified as Pax-2-A by automatic annotation. These molluscan sequences did not show the N-rich regions mentioned in Helobdella sequences [12]. Their organization included a PRD followed by a very long and variable region, in which three conserved motifs could be detected: (YDY[NS]LPDRGL), (PLDLS), and (Y[ED][RK]N[LVM]L[LI]FGD[SNQ]E[IVL]EI[MI]SVGKX[KR]W[IV][VI]RNEX[DE]L). None of these patterns was found in databases and their significance remains unknown. Using the longest motif in a Blast search led to the identification of a very short sequence of Arion vulgaris (A0A0B6XZN0), a complete sequence of Aplysia californica (XP_005090697.1, identified as mucin-5AC) and a Biomphalaria glabrata sequence showing all the conserved motifs but lacking the PRD (also identified as mucin-5AC like). Curiously, the corresponding Aplysia mRNA sequence was previously annotated as Pax beta (from automatic annotation) and this sequence was included in our set as a bona fide Paxβ. The identification of the Biomphalaria sequence remains questionable and therefore, we included it in our set as a Paxβ-like.
The Crassostrea gigas sequence K1S548, automatically annotated as Pax6, was grouped with the Paxβ with a good support (aLRT) despite its different PRD sequence (% Identity with K1R3J2 = 58%), a typical Pax6-type PRD N-ter sequence (SGVNQL) and the absence of the three motifs already signalled. This sequence may be the first example of Paxβ duplication outside of the Annelida clade. Interestingly, this sequence contains a quadribasic/quadriacid (K208RKHEDED) signal down to the PRD domain. This is reminiscent of Pax2/5/8 and consistent with proximity between Paxβ and Pax2/5/8 as suggested by Schmerer et al. [12]. However, this proximity was only weakly supported by our phylogenetic analysis.
Pax1/9
The Pax1/9 subfamily contained a clear octapeptide (H[ST]V[ST][DN][IL]LG) and no HD. In this group, the status of Helobdella robusta T1EJE5, a fragment flagged as « complete » but with « non-terminal residues » in Uniprot, was unclear. This fragment was poorly similar (% identity = 60) to a clear Helobdella robusta Pax1/9 (T1G7D6). Depending of the set of sequences used in the phylogenetic analysis, it was included or not in the Pax1/9 group. Surprisingly, we failed to detect or clone Pax1/9 from S. officinalis whereas bona fide Pax1/9 sequences from Octopus were found in databases (not included in our phylogenetic analysis due to their incomplete PRD). Further exploration is needed to resolve this discrepancy.
Pax3/7
The Pax3/7 subfamily was also well supported, but the structures of these proteins were variable. The PRD and HD were present in all sequences, but the octapeptide was only obvious in cephalopods and in one Bivalvia (Crassostrea gigas). No clear homologue of the octapeptide could be evidenced in Gastropoda, Annelida and in the Bivalvia Pinctada fucata.
Two forms of Pax3/7, Pax3/7A and Pax3/7B, have been described in Helobdella sp. (Austin), corresponding to two different genes [19]. They were also identified in Helobdella robusta but no evidence of Pax3/7B was found in other species including the Annelida Capitella teleta, implying that the duplication suggested by Woodruff et al. [19] was restricted to an Annelida sub-clade.
Three different pax3/7 sequences have been identified in Arion vulgaris, issued from the same contig but with different cDNA sequences. Theses sequences showed the same insertion in PRD, possibly reflecting a particular structure of these genes.
Pax genes expression patterns in Sepia officinalis
We compared the expression patterns of pax6, pax2/5/8 and pax3/7 based on our previous results [33,34,39,40] and new ones obtained during Sepia development. Organogenesis of cephalopods takes place in three main phases (Fig 1A) and is only briefly described here (see [53] for details). First, the animal pole of the embryo is shaped as a disk in which ectoderm and mesendoderm differentiate and organs start delineating: the arms at the periphery and the mantle at the centre of the disc (stage 14 to 18, see Fig 1). In a second phase (stage 19 to 21), the animal pole increases its volume and starts elongating: the arm crown becomes anterior (it surrounds the mouth) and the mantle acquires its definitive posterior position (the anus is located in the mantle cavity). Between both extremities, most of the nervous ganglia concentrate inside the future head and the muscular funnel surrounds the mantle border (Fig 1). In the third phase, the embryo acquires the definitive juvenile shape. Inside the head, nervous ganglia develop as brain lobes and optic lobes. The development of muscular and nervous territories in S. officinalis are summed up in Fig 1B and 1C.
Sof-pax 6 expression
Expression of Sepia officinalis pax6 (Sof-pax6) was observed from early stages (stage 14) and throughout the development. This expression was apparently restricted to structures of ectodermal origin. As organogenesis progresses, Sof-pax6 was strongly expressed in a large area from the optical region to statocystes from stage 15 to 18 (Fig 4A). As in other cephalopods (Loligo opalescens [29]; Euprymna scolopes [32]), pax6 expression in S. officinalis was observed in cerebroid and optic ganglia, from stage 19 (Fig 4A and 4B and [33]). At stage 23, optic lobes (former optic ganglia) were prominent and expressed Sof-pax6, as did the lateral supraoesophageal mass and the sub-pedunculate tissue of the cheek (Fig 4D), a neuro-endocrine tissue coming from the center of optic lobe [63]. Sof-pax6 was not expressed in pedal and visceral ganglia, two ganglia leading to the suboesophageal mass.
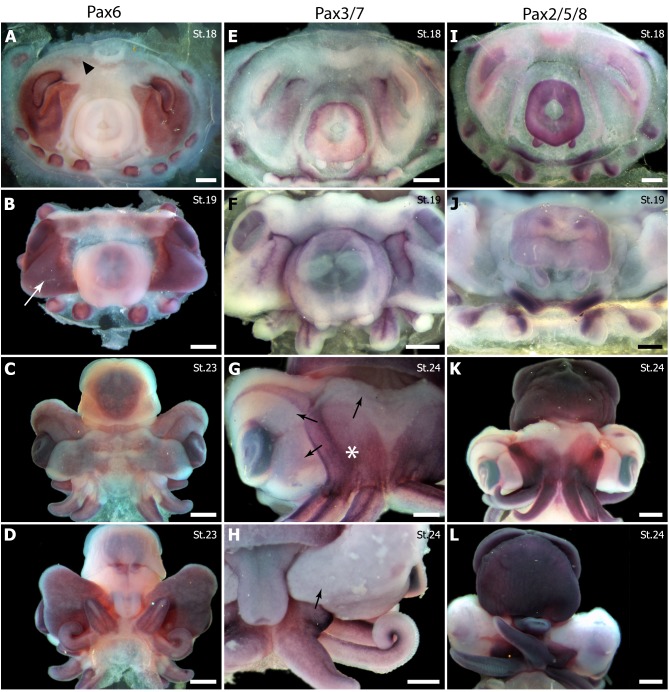
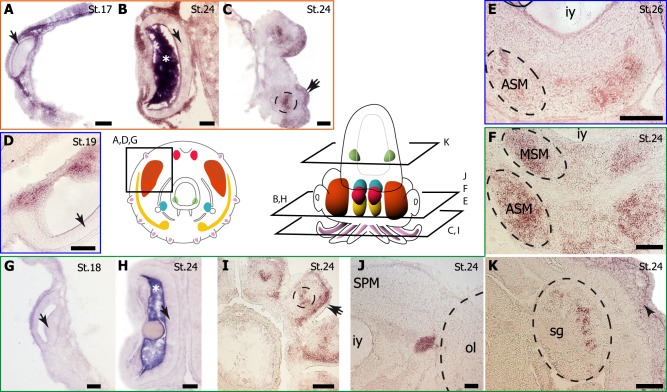
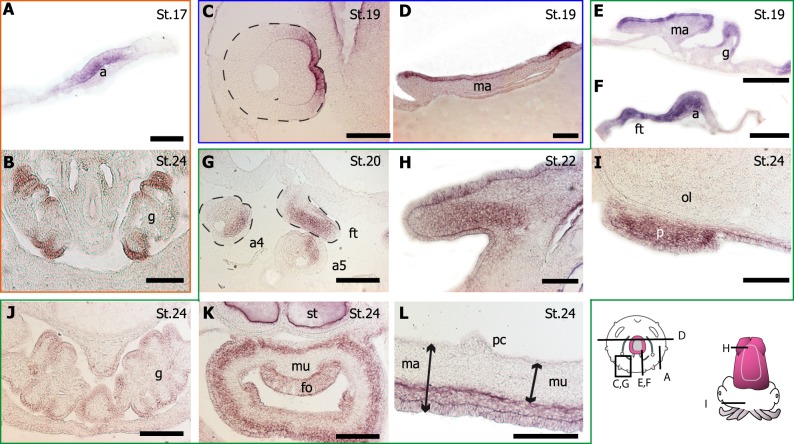
Fig 4. Pax gene expression pattern by in toto ISH during Sepia officinalis development.
(A-D) Pax6. (E-H) Pax3/7. (I-L) Pax2/5/8. Stages are indicated by St#; C,G,K, dorsal view; D,H,L, ventral view. Scale: 1 mm. (A) A large optic area and the five arms are strongly Pax6-positive, a light expression is observed in cerebroid ganglia (arrowhead). (B) All the tissues surrounding the eye express Pax6 including cheek (white arrow). (C/D) The distal part of the arms is Pax6-positive. Note the artefact in the shell area. (E/F) Aboral side of the arms, anterior part of the mantle and funnel tube elements express Pax3/7. (G/H) Dorsal arm pillars (asterisk) and aboral side of the arms are Pax3/7-positive. Black arrows denote the future extension of arm pillars. (I/J) Mantle, gills, funnel tube elements and oral side of the arms are Pax2/5/8-positive. (K/L) Mantle, fins and arms express Pax2/5/8.
Sof-pax6 was expressed from stage 17 in arm epidermis (Fig 4A, Fig 5C, Fig 6A). Later, Sof-pax6 was expressed in the distal part of the arms with a strict and straight limit (Fig 4C). From stage 23, Sof-pax6 was also expressed in intrabrachial nervous cords (Fig 5C). Finally, an expression of Sof-pax6 was observed in gill epithelium at stage 24, where Sof-pax2/5/8 is also detected (Fig 6B and 6J) (see below).
Fig 5. Pax gene expression in eye and the nervous system.
(A-C) Pax6; (D,E) Pax3/7; (F-K) Pax2/5/8. Stages are indicated by St#. Note the artefact in vitreous humour in the eye (asterisk). E, F and J are similar cutting planes, E, being the most anterior. Scale: 150 μm. The retina (arrow) is devoid of Pax6 (A,B), Pax3/7 (D) and Pax2/5/8 (G, H) expression although surrounding tissues express Pax6 and/or Pax3/7. At stage 24, the brachial cord (circle) expresses Pax6 (C) and Pax2/5/8 (I). The arm epidermis also expresses Pax6 but not Pax2/5/8 (double-arrow). The anterior suboesophageal mass expresses Pax3/7 (ASM in E) and Pax2/5/8 (ASM in F). The middle suboesophageal mass expresses Pax2/5/8 (MSM in F). The optic tractus (J) and the stellate ganglia (sg in K) express Pax2/5/8, as the dermal part of the skin (arrowhead). iy: internal yolk sac. ol: optic lobe. SPM: supraoesophageal mass.
Fig 6. Pax gene expression in non neuronal structures.
(A, B) Pax6; (C, D) Pax3/7; (E-L) Pax2/5/8. Stages are indicated by St#. Note the artefact in statocyst (st) in K. Scale 150 μm, except E, F, G: 300 μm. Pax expression in arm (a): Pax6 (A. See also Fig 5C), Pax3/7 (C) and Pax2/5/8 (F, G a4 and a5. See also Fig 5I). Pax expression in mantle (ma): Pax3/7 (D) and Pax2/5/8 (E, L). Pax expression in gill (g): Pax6 (B) and Pax2/5/8 (E, J). Dynamic of Pax2/5/8 expression in funnel tube (ft): compare F, G and K (fo: funnel organ). Pax2/5/8 is also expressed in fin (H), arm pillars (p) (I). ol: optic lobe. mu: muscle. pc: pallial cavity. ss: shell sac.
Sof-pax3/7 expression
We pushed forward with our previous Sof-pax3/7 expression study on sensory structures in S. officinalis [34]. Sof-pax3/7 is expressed as early as stage 16 in most parts of epidermal tissues. At stage 19, Sof-pax3/7 was expressed in skin epithelium (ectodermal cells) of the mantle and arms (Fig 4E, Fig 6C and 6D). From stage 23/24 to 27, Sof-pax3/7 expression was clearly observed on the arm pillars area that will extend and cover the early cephalic tissue, forming the secondary cornea at the level of the eyes (Fig 4G and 4H).
From stage 22, Sof-pax3/7 was also expressed in the nervous system, first in pedal ganglia and later in the anterior part of the suboesophageal mass (formed by these ganglia), more specifically in pre-brachial and brachial lobes (Fig 5E) involved in arm control [64]. No expression of Sof-pax3/7 was detected in the supraoeophageal part of the brain, in the buccal ganglia or in optic lobes. An expression of Sof-Pax3/7 was observed from stage 16 to 20 in the upper side of optic vesicle corresponding to lentigenic tissues (Fig 5D). This expression disappeared at the same time the lens is forming.
There was no expression of Sof-pax3/7 in mesodermal tissues, such as gills and muscles in the mantle or the funnel. By contrast, Sof-pax3/7 expression was detected in the epidermal funnel organ, a prominent structure producing mucus [65,66], which also expressed So-pax2/5/8 (see below).
Sof-pax2/5/8 expression
In early stages, Sof-pax2/5/8 was detected in mantle, arms, funnel tube territories and gills but not in the funnel pouch territories (Fig 4I–4K). On the head, the covering tissue issued from arm pillars area expressed Sof-pax2/5/8 (Figs 4K and 6I), as observed with Sof-pax3/7.
Expression of Sof-pax2/5/8 in the central nervous structures was only detected during late development (from stage 23), restricted to the ventral side of the brain, in anterior and median suboesophageal masses (Fig 5F). The optic tractus (connecting the optic lobe and the supra oesophageal mass) transiently expressed So-pax2/5/8 at stage 24 (Fig 5J). These expressions stopped by stage 25. In peripheral nervous system, Sof-pax2/5/8 was expressed in the intrabrachial nervous cords from stage 23 (Fig 5I) and in stellate ganglia, at least between stages 21 and 24 (Fig 5K).
In arms, Sof-pax2/5/8 was faintly detected in the epidermis at stage 19 (Fig 6F) but its later expression was clearly mesodermal (Fig 5I, Fig 6F and 6G). Staining intensity was asymmetric (anti oral side), different between the arms (stronger in the most ventral arm 5) and extended along the arms as they grew. A mesodermal Sof-pax2/5/8 expression was also observed in the developing fins (Fig 6H). In the funnel tube a mesodermal signal was also obvious in stage 19–20 (Fig 6F and 6G) and was later restricted to the dermis (Fig 6K). The epithelial funnel organ was clearly stained (Fig 6K). By contrast, Sof-pax2/5/8 expression was epidermal in the mantle at stage 19 (Fig 6E) and later extended to the dermal layers but not to the muscular layer of the mantle (Fig 6L). Finally, an expression in gills was observed from stage 16 to 24 in the epithelium, as for pax6 (Fig 6E and 6J).
Discussion
An expanding repertoire of Pax genes
Our analysis confirms the existence of six Pax subfamilies including the Pax-α/β subfamily already proposed [11–13] and illustrated in our set by a strongly supported Pax-β group which is putatively specific to Lophotrochozoa. The origin of this subfamily is unclear and awaits further analysis. The roles of Paxβ remain largely unknown. Two isoforms Hau-Paxβ1 and Hau-Paxβ2 have been described in the leech Helobdella. Paxβ1 is expressed during early cleavage stages in Helobdella austinensis embryogenesis and is probably implicated in the transition from spiralian to symetrical segmentation during spiralian development [28]. Both paralogs are also expressed at several different stages in segmental mesoderm (Helobdella robusta [12]), with unknown functions. Moreover, under the hypothesis that the Biomphalaria glabrata Paxβ-like is physiologically relevant, the absence of a PD suggests that these proteins may not always act as transcription factors as previously suggested for some Pax splice variants [15].
The Pox neuro subfamily is more common than previously described as it is found in all lophotrochozoan clades (this work) as well as in protostomian clades, hemichordates and echinoderms [11]. Currently, the function of the PoxNeuro protein has only been studied in Drosophila. It is known to play a role in the specification of neuronal identity in central and peripheral nervous system [67,68], in morphogenesis of appendages [69] and in the control of various aspects of male courtship behaviour [70]. This subfamily clearly deserves more attention.
Contrary to the pattern reported in chordate, the Pax2/5/8 subfamily is devoid of HD in all accessible lophotrochozoan sequences; this fact should be taken into account in future proposals for the evolution history of Pax genes (e.g. [11]). Similarly, no HD was detected in Octopus Pax6 sequence which is quite surprising as complete Pax6 are found in other cephalopods. Yoshida et al. [20] have related the numerous splicing isoforms found in Idiosepius paradoxus to the evolution abilities of cephalopods eye, but likewise the HD was clearly modified in three of the five Pax6 Idiosepius isoforms (first alpha-helix absent in Pax6v1 and Pax6v2, alpha 3/4 helix disrupted in Pax6v4 [71]). Analysis of mouse mutants has shown that Pax6 HD plays an important role in eye development [72] but no role in the regulation of neurogenesis in the developing forebrain, although it is partially required together with the PRD for some—but not all—boundary formation in the forebrain [73]. Pax6 HD is also dispensable for pancreas development [74]. The physiological significance of these short forms in Lophotrochozoa is currently unknown.
Finally, our analysis underlined misinterpretations in the databases (see Table 1 and S1 Table). We subscribe to the conclusion of Friedrich {Friedrich:2015fz} about the utility for the community to build a consolidated Pax homolog database.
Pax6 expression: Conserved in the brain and in eye tissues, but not in the retina
As already underlined in [33], Sof-pax6 expression was largely distributed in central nervous system (CNS) areas. The role of pax6 in the development of the CNS appears to be conserved in cephalopods. The expression of Sof-pax6 in the brachial nervous chord suggests that the role of pax6 in neural development is extended to this morphological novelty of cephalopods. Likewise, the restriction of Sof-pax6 expression at the distal tip of growing arms, which is described as a growing/proliferation region in Octopus [75] and Euprymna [76], suggests a role of Sof-pax6 in the growth of the arms.
Pax6 is considered as the master gene in eye development [77,78]. In Drosophila, homologs of Pax6 (Ey and Toy) are atop the retinal determination gene network (RDGN), a group of transcription factors and cofactors controlling eye development and including Pax6, Sine oculis (So), Dashchund and Eyes absent (Eya) (review in [79]). The expression of Pax6 in developing photoreceptors (i.e. retina) has been demonstrated in rhabdomeric [80] as well as in ciliary [35] photoreceptors in numerous metazoans, including Lophotrochozoa. Cephalopods have camerular eyes with rhabdomeric photoreceptors, but it is noteworthy that the role of Pax6 in the development of retina seems far less obvious in cephalopods. In S. officinalis, Sof-Pax6 was expressed from early to late development in cornea and tissues surrounding the eye but never in the retina cells throughout all stages of development (see Fig 5A and 5B). In addition, the probe used in our study covered a common sequence of Sof-pax6 splice variants identified in a recent S. officinalis transcriptomic database (unpublished data), suggesting that none of them are expressed in retina. Likewise, Yoshida et al. [20] have shown that none of the six Idiosepius Pax6 isoforms is expressed in the retina during development, questioning the paradigm of the Pax6-photoreceptor link. No other Pax gene (see below) seems to be involved in the organogenesis of the retina in S. officinalis. By contrast, the role of otx2 in the early development of the retina is probably conserved in S. officinalis as its expression, in a restricted but unidentified set of cells (stem cells? future support cells?), is observed in retina [81].
The widespread expression of Pax6 in developing eyes of metazoans has led to the hypothesis of a monophyletic origin of the structures dedicated to photoreception. The conservation of the genetic control of eye development by Pax6 among all bilaterian animals would then not be due to functional constraints, rather, a consequence of its evolutionary history [78]. These ideas are considered as an oversimplified view by others [61,82], as some examples of eye developing without any Pax6 expression are already known: developing Limulus eyes [83], developing Platynereis adult (but not larval) eyes [84], Hesse organs (eye cups) of amphioxus [85], eye regeneration in planarians [58] as well as formation of the Bolwig organ (larval eyes) in Drosophila [86]. The developing Sepia officinalis eye may be added to this list, at least for the main functional part in photoreception (i.e. retina). Nevertheless, So-Pax6 seems to contribute to the formation of other parts of the visual organ. Studies of Pax expression in basal bilaterian have shown that other Pax classes can master eye formation: PaxB (Pax2/5/8 subfamily) in the cubozoan jellyfish Tripedalia [87], PaxA (PoxN subfamily) in the hydrozoan Cladonema radiatum [88]. The fact that all these « master genes » belong to the Pax family is interpreted by Suga et al. [88] as a confirmation for the hypothesis of a monophyletic origin of photoreceptors. This argument is not validated by our current data regarding Pax gene expression in the retina of S. officinalis as we did not detect any other Pax expression at any stage. However, the hypothesis of a secondary loss of Pax6 role in photoreceptor differentiation with a co-optation of Pax1/9, Pox neuro or Paxβ, cannot be yet discarded.
Pax3/7 expression: In ectodermal tissues but not in muscles
The involvement of the Pax3/7 family in skeletal muscle development [89] is specific to vertebrates (and maybe nematodes [90]). S. officinalis, as other Lophotrochozoa, is not an exception since no expression was detected in any muscular tissues, including systemic heart. The expression of the pax3/7-A paralog is also mesodermal in the leech Helobdella robusta [19], and it is involved in nephridies development and body cavity formation. We did not observe any clear expression of So-pax3/7 in mesodermal tissues.
In vertebrates, pax3 and pax7 also contribute to the development of the nervous system [62,91,92] and this role seems more largely conserved among Metazoa. Their homologs in Drosophila (Gooseberry and Gooseberry-neuro [7]) are essential segment-polarity genes [93] and are involved in neurogenesis [94]. This role in nervous system development seems conserved in S. officinalis as expression is conserved in brain. Nevertheless this neural expression is late, suggesting that Pax3/7 is not involved in early neurogenesis (the emergence of ganglia), and restricted to the ventral brain in “motor” areas controlling the arms.
Pax3/7 is expressed as longitudinal neuroectodermal bands in two annelids, Platynereis dumerilii [27] and Capitella teleta [23]. In S. officinalis, Sof-pax3/7 is expressed in ectodermal tissues, such as the skin of the arm pillars covering the head. An involvement of Pax3/7 in the development of ectoderm is not surprising. In Drosophila, Pax3/7 (gsb) specifies the epithelial pattern of each segment, including neuroblast specification [95]; in vertebrates, Pax3 and Pax7 regulate the development of neural crests at the origin of epidermal pigmentary cells [92,96]. The exact roles of Pax3/7 in cephalopod skin development, including the differentiation of several types of epidermal sensory cells, remains to be determined.
Pax2/5/8 expression: In brain and mesodermal tissues but not in sensory organs
Pax2/5/8 expression has been characterized from Lophotrochozoa belonging to Annelida [27,28] but its expression during development has only been followed in four molluscan species [21,31].
No clear Pax2/5/8 expression could be observed in S. officinalis statocytes during organogenesis (from stage 16), as also shown by Wollesen et al. [21] in Idiosepius notoides (Cephalopoda) from stage 19. This differs from what is observed in other protostomes. In the developing (until 78hpf) mollusc Haliotis asinina, the statocyst and chemio / mechanosensory cells also express Has-Pax2/5/8 [31] In Drosophila early embryos, Pax2/5/8 is expressed in labial and antennal mechanosensory organs [97]. No expression of Sof-pax2/5/8 has been detected in eyes whereas an expression has been detected in optic area of veliger larvae and in eye area after metamorphosis of Haliotis asinina [30,31] as well as in differentiating Drosophila omatidia [97]. Based on these results, we suggest that the role of Pax 2/5/8 in sensory structures development could have been lost in cephalopod lineage. Nevertheless, Sof-pax2/5/8 transient expression in optic tractus suggests a role for Sof-pax2/5/8 in the development of neuronal circuits for vision. A role of Pax2 in the development of the optic chiasm and in the guidance of axons of the optic nerve has also been shown in vertebrates [98].
Quite unexpectedly, and unlike Sof-Pax3/7, Sof-Pax2/5/8 was expressed in mesodermal structures. Early mesodermal expressions of Sof-Pax2/5/8 in arms and funnel tube are remarkably similar. These observations are in agreement with the hypothesis of a common origin for arms and funnel tube, different from the funnel pouch [99] which does not express Sof-Pax2/5/8. Expression in mesodermal tissues has already been shown in the Annelida Helobdella austinensis where Pax2/5/8 has a role in the symmetric cleavage of the mesodermal proteoblast [28]. Interestingly, Pax2/5/8 is expressed in typical molluscan structures in Haliotis asinina larvae: dorsolateral cells of the foot, right shell muscle and in the pallial chamber [31]. The expression of Sof-Pax2/5/8 in S. officinalis arms and funnel, considered as derived from the molluscan foot, suggests a conserved role in derived structures and morphological novelties.
Pax2/5/8 expression is maintained, at least in muscle, in the adult Haliotis asinina [30]. In S. officinalis, Sof-pax2/5/8 might be involved in early steps of myogenesis in the locomotor structures derived from the foot during evolution, but is clearly not implicated in muscle differentiation. This hypothesis would imply the recruitment of other genes in the formation of muscles, particularly in the mantle. In this context, NK4 is a good candidate [54]. On the other hand, the early expression of Pax2/5/8 in the brain occurs in anterior and middle subœsophageal mass, derived from the pedal ganglia and known to control the arms and the funnel in the adult [63]. The development of S. officinalis is direct but the embryo is able to move and react inside the capsule from stage 24/25, implying a functional connection between muscular and nervous systems from these stages. So-pax2/5/8 could be implicated in the formation of the whole nervous circuitry controlling arms and funnel muscles. In further development, So-pax2/5/8 is expressed in nervous territories involved in motor control: intra-brachial nervous system, subœsophageal masses, stellate ganglia (through which pass the giant fibres innervating muscles). Pax2/5/8 may thus play a major role in the neuro-muscular complex development.
Finally, Pax2/5/8 expression in the gills of S. officinalis underlines an interesting and provocative problem. Expression of Pax2/5/8 in gills has been detected in adult gastropoda [30]. It has also been recorded in urochordate Oikopleura dioica [57], amphioxus [100] as well as in furrows separating branchial arches in Xenopus embryos [101]. This leads to a discussion between a ‘ciliated placode formation’ versus a ‘perforation’ role for Pax2/5/8 (see [57]). The gills in cephalopods emerge from a small cellular blastema; to our knowledge, they are neither considered as homolog to chordate gills nor ciliated. However, on the basis of the common expression of Pax2/5/8 in “gills” from urochordate, cephalochordate, vertebrate and molluscs (cephalopods and gastropods), these structures may be taken as homologous (as defined by [42], see [102]) and should exist in Urbilateria. One possible alternative is that the biological functions governed by Pax2/5/8 in this case are ancestral and used by analogous structures, even under the control of orthologous genes.
Conclusion
The diversity of the Pax family is proposed to explain the high functional diversity of Pax proteins [10,22]. We observed overlapping of different Pax gene expression patterns in S. officinalis, although we cannot yet ensure that these observations correspond to a coexpression at the cellular level or only to a concomitant expression in neighbouring cells. Such overlapping of gene expression patterns has been observed for Platynereis Pax3/7, Pax6 and Pax2/5/8 in the nervous system, where it was interpreted as a “spatial code” defining longitudinal neural progenitor domains [27]. Alternatively, some overlapping areas between paralogs could be regarded as “redundancy islets” allowing further evolution. Functional redundancy between Pax6 and Pax2 has been demonstrated in mouse [103]. Moreover, Pax genes are known to be prone to abundant alternative splicing (e.g. [15,17,101]). The results of Short et al. [24] suggest that the functional network of co-expressed Pax isoforms is more evolutionary constrained than each of the isoforms, because orthologous isoforms, even if conserved during evolution, may individually have very different transcriptional activities in different species. Thus, functional studies are in fact needed to confirm inferences proposed on the basis of simple gene homology.
Supporting information
Location of primers used for probe synthesis. Green: forward primers; pink: reverse primers. The grey blocks correspond to PRD and OM (present only in Pax2/5/8 and Pax3/7 sequences) domains respectively. The HD domain is not presented.
(DOCX)
Accession: accession number or genome reference (DNA/RNA: corresponding sequence). Submitted name: name found in databases. Proposed name: our interpretation (bold type if different from submitted name). (f) denote a fragment. Please note that sequences from Sabellaria alveolata (SA_Locus_72760, SA_Locus_22824, SA_Locus_22463) are unpublished and courtesy of P-J. Lopez and J. Fournier, UMR BOREA (personal communication).
(XLSX)
Alignment of the sequences given in S1_Table.xlsx, in fasta format.
(FAS)
Acknowledgments
We thank J. Henry, C. Zlatyny-Gaudin (BOREA) and the biological station of Luc-sur-Mer (University of Caen-Normandy), S. Henry and the Biological station of Roscoff (UPMC, Sorbonne Universités) for providing Sepia brood. We are grateful to Mitchell Fleming (BOREA, ITN IMPRESS) for his carefully reading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant JC/JC 0043 from Agence Nationale de la Recherche (ANR) and by a “Bourse de la Vocation Marcel Bleustein-Blanchet” to SN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blake JA, Thomas M, Thompson JA, White R, Ziman M. Perplexing Pax: from puzzle to paradigm. Dev Dyn. 2008;237: 2791–2803. 10.1002/dvdy.21711 [DOI] [PubMed] [Google Scholar]
- 2.Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141: 737–751. 10.1242/dev.091785 [DOI] [PubMed] [Google Scholar]
- 3.Paixão-Côrtes VR, Salzano FM, Bortolini MC. Evolutionary history of chordate PAX genes: dynamics of change in a complex gene family. Robinson-Rechavi M, editor. PLoS ONE. 2013;8: e73560 10.1371/journal.pone.0073560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paixão-Côrtes VR, Salzano FM, Bortolini MC. Origins and evolvability of the PAX family. Semin Cell Dev Biol. 2015;44: 64–74. 10.1016/j.semcdb.2015.08.014 [DOI] [PubMed] [Google Scholar]
- 5.Mayran A, Pelletier A, Drouin J. Pax factors in transcription and epigenetic remodelling. Semin Cell Dev Biol. 2015;44: 135–144. 10.1016/j.semcdb.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 6.Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209: 186–197. [DOI] [PubMed] [Google Scholar]
- 7.Breitling R, Gerber JK. Origin of the paired domain. Dev Genes Evol. 2000;210: 644–650. [DOI] [PubMed] [Google Scholar]
- 8.Hoshiyama D, Iwabe N, Miyata T. Evolution of the gene families forming the Pax/Six regulatory network: isolation of genes from primitive animals and molecular phylogenetic analyses. FEBS Lett. 2007;581: 1639–1643. 10.1016/j.febslet.2007.03.027 [DOI] [PubMed] [Google Scholar]
- 9.Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol Dev. 2007;9: 25–38. 10.1111/j.1525-142X.2006.00135.x [DOI] [PubMed] [Google Scholar]
- 10.Underhill DA. PAX proteins and fables of their reconstruction. Crit Rev Eukaryot Gene Expr. 2012;22: 161–177. [DOI] [PubMed] [Google Scholar]
- 11.Hill A, Boll W, Ries C, Warner L, Osswalt M, Hill M, et al. Origin of Pax and Six gene families in sponges: Single PaxB and Six1/2 orthologs in Chalinula loosanoffi. Dev Biol. 2010;343: 106–123. 10.1016/j.ydbio.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 12.Schmerer M, Savage RM, Shankland M. Paxbeta: a novel family of lophotrochozoan Pax genes. Evol Dev. 2009;11: 689–696. 10.1111/j.1525-142X.2009.00376.x [DOI] [PubMed] [Google Scholar]
- 13.Franke FA, Schumann I, Hering L, Mayer G. Phylogenetic analysis and expression patterns of Pax genes in the onychophoran Euperipatoides rowellireveal a novel bilaterian Pax subfamily. Evol Dev. 2015;17: 3–20. 10.1111/ede.12110 [DOI] [PubMed] [Google Scholar]
- 14.Friedrich M. Evo-Devo gene toolkit update: at least seven Pax transcription factor subfamilies in the last common ancestor of bilaterian animals. Evol Dev. 2015;17: 255–257. 10.1111/ede.12137 [DOI] [PubMed] [Google Scholar]
- 15.Short S, Holland LZ. The evolution of alternative splicing in the Pax family: the view from the Basal chordate amphioxus. J Mol Evol. 2008;66: 605–620. 10.1007/s00239-008-9113-5 [DOI] [PubMed] [Google Scholar]
- 16.Jiao S, Tan X, Wang Q, Li M, Du SJ. The olive flounder (Paralichthys olivaceus) Pax3 homologues are highly conserved, encode multiple isoforms and show unique expression patterns. Comp Biochem Physiol B, Biochem Mol Biol. 2015;180: 7–15. 10.1016/j.cbpb.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 17.Fabian P, Kozmikova I, Kozmik Z, Pantzartzi CN. Pax2/5/8 and Pax6 alternative splicing events in basal chordates and vertebrates: a focus on paired box domain. Front Genet. 2015;6: 228 10.3389/fgene.2015.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quigley IK, Xie X, Shankland M. Hau-Pax6A expression in the central nervous system of the leech embryo. Dev Genes Evol. 2007;217: 459–468. 10.1007/s00427-007-0156-1 [DOI] [PubMed] [Google Scholar]
- 19.Woodruff JB, Mitchell BJ, Shankland M. Hau-Pax3/7A is an early marker of leech mesoderm involved in segmental morphogenesis, nephridial development, and body cavity formation. Dev Biol. 2007;306: 824–837. 10.1016/j.ydbio.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 20.Yoshida M-A, Yura K, Ogura A. Cephalopod eye evolution was modulated by the acquisition of Pax-6 splicing variants. Sci Rep. 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wollesen T, Rodríguez Monje SV, Todt C, Degnan BM, Wanninger A. Ancestral role of Pax2/5/8 in molluscan brain and multimodal sensory system development. BMC Evol Biol. 2015;15: 231 10.1186/s12862-015-0505-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland LZ, Short S. Alternative splicing in development and function of chordate endocrine systems: a focus on Pax genes. Int Comp Biol. 2010;50: 22–34. [DOI] [PubMed] [Google Scholar]
- 23.Seaver EC, Yamaguchi E, Richards GS, Meyer NP. Expression of the pair-rule gene homologs runt, Pax3/7, even-skipped-1 and even-skipped-2 during larval and juvenile development of the polychaete annelid Capitella teleta does not support a role in segmentation. Evodevo. 2012;3: 8 10.1186/2041-9139-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short S, Kozmik Z, Holland LZ. The function and developmental expression of alternatively spliced isoforms of amphioxus and Xenopus laevis Pax2/5/8 genes: revealing divergence at the invertebrate to vertebrate transition. J Exp Zool. 2012;318: 555–571. [DOI] [PubMed] [Google Scholar]
- 25.Callaerts P, Munoz-Marmol AM, Glardon S, Castillo E, Sun H, Li WH, et al. Isolation and expression of a Pax-6 gene in the regenerating and intact Planarian Dugesia(G)tigrina. Proc Natl Acad Sci USA. 1999;96: 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saló E, Pineda D, Marsal M, Gonzalez J, Gremigni V, Batistoni R. Genetic network of the eye in Platyhelminthes: expression and functional analysis of some players during planarian regeneration. Gene. 2002;287: 67–74. [DOI] [PubMed] [Google Scholar]
- 27.Denes AS, Jékely G, Steinmetz PRH, Raible F, Snyman H, Prud'homme B, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129: 277–288. 10.1016/j.cell.2007.02.040 [DOI] [PubMed] [Google Scholar]
- 28.Schmerer MW, Null RW, Shankland M. Developmental transition to bilaterally symmetric cell divisions is regulated by Pax-mediated transcription in embryos of the leech Helobdella austinensis. Dev Biol. 2013;382: 149–159. 10.1016/j.ydbio.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 29.Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, et al. Squid Pax-6 and eye development. Proc Natl Acad Sci USA. 1997;94: 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien EK, Degnan BM. Expression of POU, Sox, and Pax genes in the brain ganglia of the tropical abalone Haliotis asinina. Mar Biotechnol. 2000;2: 545–557. 10.1007/s101260000039 [DOI] [PubMed] [Google Scholar]
- 31.O'Brien EK, Degnan BM. Expression of Pax258 in the gastropod statocyst: insights into the antiquity of metazoan geosensory organs. Evol Dev. 2003;5: 572–578. [DOI] [PubMed] [Google Scholar]
- 32.Hartmann B, Lee PN, Kang YY, Tomarev S, de Couet HG, Callaerts P. Pax6 in the sepiolid squid Euprymna scolopes: evidence for a role in eye, sensory organ and brain development. Mech Dev. 2003;120: 177–183. [DOI] [PubMed] [Google Scholar]
- 33.Navet S, Andouche A, Baratte S, Bonnaud L. Shh and Pax6 have unconventional expression patterns in embryonic morphogenesis in Sepia officinalis (Cephalopoda). Gene Expr Patterns. 2009;9: 461–467. 10.1016/j.gep.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Buresi A, Croll RP, Tiozzo S, Bonnaud L, Baratte S. Emergence of sensory structures in the developing epidermis in sepia officinalis and other coleoid cephalopods. J Comp Neurol. 2014;522: 3004–3019. 10.1002/cne.23562 [DOI] [PubMed] [Google Scholar]
- 35.Passamaneck YJ, Furchheim N, Hejnol A, Martindale MQ, Lüter C. Ciliary photoreceptors in the cerebral eyes of a protostome larva. Evodevo. 2011;2: 6 10.1186/2041-9139-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Passamaneck YJ, Hejnol A, Martindale MQ. Mesodermal gene expression during the embryonic and larval development of the articulate brachiopod Terebratalia transversa. Evodevo. 2015;6: 10 10.1186/s13227-015-0004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loosli F, Kmita-Cunisse M, Gehring WJ. Isolation of a Pax-6 homolog from the ribbonworm Lineus sanguineus. Proc Natl Acad Sci USA. 1996;93: 2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassaglia Y, Buresi A, Franko D, Andouche A, Baratte S, Bonnaud L. Sepia officinalis: A new biological model for eco-evo-devo studies. J Exp Mar Biol Ecol. 2013;447: 4–13. [Google Scholar]
- 39.Navet S, Baratte S, Bassaglia Y, Andouche A, Buresi A, Bonnaud L. Neurogenesis in Cephalopods:”Eco-Evo-Devo” approach in the cuttlefish Sepia officinalis (Mollusca-Cephalopoda). J Mar Sci Technol. 2014;22: 15–24. [Google Scholar]
- 40.Buresi A, Andouche A, Navet S, Bassaglia Y, Bonnaud-Ponticelli L, Baratte S. Nervous system development in cephalopods: How egg yolk-richness modifies the topology of the mediolateral patterning system. Dev Biol. 2016;415: 143–156. 10.1016/j.ydbio.2016.04.027 [DOI] [PubMed] [Google Scholar]
- 41.Shigeno S, Takenori S, Boletzky S. The origins of cephalopod body plans: a geometrical and developmental basis for the evolution of vertebrate-like organ systems In: Tanabe K, Shigeta Y, Sasaki T, Hirano H, editors. Tokai University Press; Tokyo: Cephalopods-Present and Past; 2010. pp. 23–34. [Google Scholar]
- 42.Arendt D. Genes and homology in nervous system evolution: Comparing gene functions, expression patterns, and cell type molecular fingerprints. Theory Biosci. 2005;124: 185–197. 10.1016/j.thbio.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, et al. Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 2012;19: 117–130. 10.1093/dnares/dss005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassaglia Y, Bekel T, Da Silva C, Poulain J, Andouche A, Navet S, et al. ESTs library from embryonic stages reveals tubulin and reflectin diversity in Sepia officinalis (Mollusca—Cephalopoda). Gene. 2012;498: 203–211. [DOI] [PubMed] [Google Scholar]
- 45.Moreira R, Pereiro P, Canchaya C, Posada D, Figueras A, Novoa B. RNA-Seq in Mytilus galloprovincialis: comparative transcriptomics and expression profiles among different tissues. BMC Genomics. 2015;16: 728 10.1186/s12864-015-1817-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36: W465–9. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17: 540–552. [DOI] [PubMed] [Google Scholar]
- 49.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52: 696–704. [DOI] [PubMed] [Google Scholar]
- 50.Anisimova M, Gascuel O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst Biol. 2006;55: 539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- 51.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7: 439 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemaire J. Table de développement embryonnaire de Sepia officinalis L.(Mollusque Céphalopode). Bull Soc Zool Fr. Bull Soc Zool Fr; 1970;95: 773–782. [Google Scholar]
- 53.Boletzky von S, Andouche A, Bonnaud-Ponticelli L. A developmental table of embryogenesis in Sepia officinalis. Vie et Milieu. 2016;66: 3–9. [Google Scholar]
- 54.Navet S, Bassaglia Y, Baratte S, Martin M, Bonnaud L. Somatic muscle development in Sepia officinalis (cephalopoda—mollusca): a new role for NK4. Dev Dyn. 2008;237: 1944–1951. 10.1002/dvdy.21614 [DOI] [PubMed] [Google Scholar]
- 55.Morino Y, Okada K, Niikura M, Honda M, Satoh N, Wada H. A Genome-Wide Survey of Genes Encoding Transcription Factors in the Japanese Pearl Oyster, Pinctada fucata: I. Homeobox Genes. Zool Sci. 2013;30: 851–857. 10.2108/zsj.30.851 [DOI] [PubMed] [Google Scholar]
- 56.Sun H, Rodin A, Zhou Y, Dickinson DP, Harper DE, Hewett-Emmett D, et al. Evolution of paired domains: isolation and sequencing of jellyfish and hydra Pax genes related to Pax-5 and Pax-6. Proc Natl Acad Sci USA. 1997;94: 5156–5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassham S, Cañestro C, Postlethwait JH. Evolution of developmental roles of Pax2/5/8 paralogs after independent duplication in urochordate and vertebrate lineages. BMC Biol. 2008;6: 35 10.1186/1741-7007-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pineda D, Rossi L, Batistoni R, Salvetti A, Marsal M, Gremigni V, et al. The genetic network of prototypic planarian eye regeneration is Pax6 independent. Development. 2002;129: 1423–1434. [DOI] [PubMed] [Google Scholar]
- 59.Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci USA. 1998;95: 13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16: 6793–6803. 10.1093/emboj/16.22.6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozmik Z. Pax genes in eye development and evolution. Curr Opin Genet Dev. 2005;15: 430–438. 10.1016/j.gde.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 62.Thompson JA, Ziman M. Pax genes during neural development and their potential role in neuroregeneration. Prog Neurobiol. 2011;95: 334–351. 10.1016/j.pneurobio.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 63.Nixon M, Young JZ. The Brains and Lives of Cephalopods. Oxford. UK: Oxford University Press; 2003. [Google Scholar]
- 64.Boycott BB. The functional organization of the brain of the cuttlefish Sepia officinalis. Proc R Soc London B, Biol Sci. 1961;153: 503–534. [Google Scholar]
- 65.Hu MY, Sucré E, Charmantier-Daures M, Charmantier G, Lucassen M, Himmerkus N, et al. Localization of ion-regulatory epithelia in embryos and hatchlings of two cephalopods. Cell Tissue Res. 2010;339: 571–583. 10.1007/s00441-009-0921-8 [DOI] [PubMed] [Google Scholar]
- 66.Derby CD. Cephalopod ink: production, chemistry, functions and applications. Mar Drugs. 2014;12: 2700–2730. 10.3390/md12052700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dambly-Chaudière C, Jamet E, Burri M, Bopp D, Basler K, Hafen E, et al. The paired box gene pox neuro: a determinant of poly-innervated sense organs in Drosophila. Cell. 1992;69: 159–172. [DOI] [PubMed] [Google Scholar]
- 68.Jiang Y, Boll W, Noll M. Pox neuro control of cell lineages that give rise to larval poly-innervated external sensory organs in Drosophila. Dev Biol. 2015;397: 162–174. 10.1016/j.ydbio.2014.10.013 [DOI] [PubMed] [Google Scholar]
- 69.Awasaki T, Kimura K. Multiple function of poxn gene in larval PNS development and in adult appendage formation of Drosophila. Dev Genes Evol. 2001;211: 20–29. [DOI] [PubMed] [Google Scholar]
- 70.Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129: 5667–5681. [DOI] [PubMed] [Google Scholar]
- 71.A comparison with fig 1 of Yoshida et al. (2014) demonstrated a discrepancy between the nomenclature used in their paper and the nomenclature of the sequences in Uniprot. The sequence submitted as « Pax-6 variant form2 » (L8AUT1) is in fact the v3 in the paper, and the sequence deposited as « Pax-6 variant form3 » (L8AWY5) is in fact the v2 in the paper.
- 72.Favor J, Peters H, Hermann T, Schmahl W, Chatterjee B, Neuhäuser-Klaus A, et al. Molecular characterization of Pax6(2Neu) through Pax6(10Neu): an extension of the Pax6 allelic series and the identification of two possible hypomorph alleles in the mouse Mus musculus. Genetics. 2001;159: 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haubst N, Berger J, Radjendirane V, Graw J, Favor J, Saunders GF, et al. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development. 2004;131: 6131–6140. 10.1242/dev.01524 [DOI] [PubMed] [Google Scholar]
- 74.Dames P, Puff R, Weise M, Parhofer KG, Göke B, Götz M, et al. Relative roles of the different Pax6 domains for pancreatic alpha cell development. BMC Dev Biol. 2010;10: 39 10.1186/1471-213X-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nödl M-T, Fossati SM, Domingues P, Sánchez FJ, Zullo L. The making of an octopus arm. Evodevo. 2015;6: 19 10.1186/s13227-015-0012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nödl M-T, Kerbl A, Walzl MG, Müller GB, de Couet HG. The cephalopod arm crown: appendage formation and differentiation in the Hawaiian bobtail squid Euprymna scolopes. Front Zool. 2016;13: 44 10.1186/s12983-016-0175-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15: 371–377. [DOI] [PubMed] [Google Scholar]
- 78.Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96: 171–184. 10.1093/jhered/esi027 [DOI] [PubMed] [Google Scholar]
- 79.Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009;1789: 306–314. 10.1016/j.bbagrm.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47: 563–571. [PubMed] [Google Scholar]
- 81.Buresi A, Baratte S, Da Silva C, Bonnaud L. orthodenticle/otx ortholog expression in the anterior brain and eyes of Sepia officinalis (Mollusca, Cephalopoda). Gene Expr Patterns. 2012;12: 109–116. 10.1016/j.gep.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 82.Vopalensky P, Kozmik Z. Eye evolution: common use and independent recruitment of genetic components. Philos Trans R Soc B. 2009;364: 2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blackburn DC, Conley KW, Plachetzki DC, Kempler K, Battelle B-A, Brown NL. Isolation and expression of Pax6 and atonal homologues in the American horseshoe crab, Limulus polyphemus. Dev Dyn. 2008;237: 2209–2219. 10.1002/dvdy.21634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arendt D, Tessmar K, de Campos-Baptista M-IM, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129: 1143–1154. [DOI] [PubMed] [Google Scholar]
- 85.Glardon S, Holland LZ, Gehring WJ, Holland ND. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development. 1998;125: 2701–2710. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki T, Saigo K. Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development. The Company of Biologists Ltd; 2000;127: 1531–1540. [DOI] [PubMed] [Google Scholar]
- 87.Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, et al. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5: 773–785. [DOI] [PubMed] [Google Scholar]
- 88.Suga H, Tschopp P, Graziussi DF, Stierwald M, Schmid V, Gehring WJ. Flexibly deployed Pax genes in eye development at the early evolution of animals demonstrated by studies on a hydrozoan jellyfish. Proc Natl Acad Sci USA. 2010;107: 14263–14268. 10.1073/pnas.1008389107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buckingham M, Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin Cell Dev Biol. 2015;44: 115–125. 10.1016/j.semcdb.2015.09.017 [DOI] [PubMed] [Google Scholar]
- 90.Yi B, Bumbarger D, Sommer RJ. Genetic evidence for pax-3 function in myogenesis in the nematode Pristionchus pacificus. Evol Dev. 2009;11: 669–679. 10.1111/j.1525-142X.2009.00374.x [DOI] [PubMed] [Google Scholar]
- 91.Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90: 169–180. [DOI] [PubMed] [Google Scholar]
- 92.Monsoro-Burq AH. PAX transcription factors in neural crest development. Semin Cell Dev Biol. 2015;44: 87–96. 10.1016/j.semcdb.2015.09.015 [DOI] [PubMed] [Google Scholar]
- 93.Kilchherr F, Baumgartner S, Bopp D, Frei E, Noll M. Isolation of the paired gene of Drosophila and its spatial expression during early embryogenesis. Nature. 1986;321: 493–499. [Google Scholar]
- 94.Colomb S, Joly W, Bonneaud N, Maschat F. A concerted action of Engrailed and Gooseberry-Neuro in neuroblast 6–4 is triggering the formation of embryonic posterior commissure bundles. PLoS ONE. 2008;3: e2197 10.1371/journal.pone.0002197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duman-Scheel M, Li X, Orlov I, Noll M, Patel NH. Genetic separation of the neural and cuticular patterning functions of gooseberry. Development. 1997;124: 2855–2865. [DOI] [PubMed] [Google Scholar]
- 96.Minchin JEN, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317: 508–522. 10.1016/j.ydbio.2008.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Czerny T, Bouchard M, Kozmik Z, Busslinger M. The characterization of novel Pax genes of the sea urchin and Drosophila reveal an ancient evolutionary origin of the Pax2/5/8 subfamily. Mech Dev. 1997;67: 179–192. [DOI] [PubMed] [Google Scholar]
- 98.Thanos S, Püttmann S, Naskar R, Rose K, Langkamp-Flock M, Paulus W. Potential role of Pax-2 in retinal axon navigation through the chick optic nerve stalk and optic chiasm. J Neurobiol. 2004;59: 8–23. 10.1002/neu.20001 [DOI] [PubMed] [Google Scholar]
- 99.Boletzky SV. Recent studies on spawning, embryonic development, and hatching in the Cephalopoda. Adv Mar Biol. 1989;25: 85–115. [Google Scholar]
- 100.Kozmik Z, Holland ND, Kalousova A, Paces J, Schubert M, Holland LZ. Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development. 1999;126: 1295–1304. [DOI] [PubMed] [Google Scholar]
- 101.Heller N, Brändli AW. Xenopus Pax-2 displays multiple splice forms during embryogenesis and pronephric kidney development. Mech Dev. 1997;69: 83–104. [DOI] [PubMed] [Google Scholar]
- 102.p. 191 “If, in two distinct species, orthologous transcription factors are expressed in a sufficiently similar and specific manner, these expression regions are considered homologous, even across phyletic boundaries, and should have been present in the body plan of their last common ancestor.”
- 103.Carbe C, Garg A, Cai Z, Li H, Powers A, Zhang X. An allelic series at the paired box gene 6 (Pax6) locus reveals the functional specificity of Pax genes. J Biol Chem. 2013;288: 12130–12141. 10.1074/jbc.M112.436865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of primers used for probe synthesis. Green: forward primers; pink: reverse primers. The grey blocks correspond to PRD and OM (present only in Pax2/5/8 and Pax3/7 sequences) domains respectively. The HD domain is not presented.
(DOCX)
Accession: accession number or genome reference (DNA/RNA: corresponding sequence). Submitted name: name found in databases. Proposed name: our interpretation (bold type if different from submitted name). (f) denote a fragment. Please note that sequences from Sabellaria alveolata (SA_Locus_72760, SA_Locus_22824, SA_Locus_22463) are unpublished and courtesy of P-J. Lopez and J. Fournier, UMR BOREA (personal communication).
(XLSX)
Alignment of the sequences given in S1_Table.xlsx, in fasta format.
(FAS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.