Abstract
Influences of adenosine 2A receptor (A2AR) activity on the cardiac transcriptome and genesis of endotoxemic myocarditis are unclear. We applied transcriptomic profiling (39 K Affymetrix arrays) to identify A2AR-sensitive molecules, revealed by receptor knockout (KO), in healthy and endotoxemic hearts. Baseline cardiac function was unaltered and only 37 A2AR-sensitive genes modified by A2AR KO (≥1.2-fold change, <5 % FDR); the five most induced are Mtr, Ppbp, Chac1, Ctsk and Cnpy2 and the five most repressed are Hp, Yipf4, Acta1, Cidec and Map3k2. Few canonical paths were impacted, with altered Gnb1, Prkar2b, Pde3b and Map3k2 (among others) implicating modified G protein/cAMP/PKA and cGMP/NOS signalling. Lipopolysaccharide (LPS; 20 mg/kg) challenge for 24 h modified >4100 transcripts in wild-type (WT) myocardium (≥1.5-fold change, FDR < 1 %); the most induced are Lcn2 (+590); Saa3 (+516); Serpina3n (+122); Cxcl9 (+101) and Cxcl1 (+89) and the most repressed are Car3 (−38); Adipoq (−17); Atgrl1/Aplnr (−14); H19 (−11) and Itga8 (−8). Canonical responses centred on inflammation, immunity, cell death and remodelling, with pronounced amplification of toll-like receptor (TLR) and underlying JAK-STAT, NFκB and MAPK pathways, and a ‘cardio-depressant’ profile encompassing suppressed ß-adrenergic, PKA and Ca2+ signalling, electromechanical and mitochondrial function (and major shifts in transcripts impacting function/injury including Lcn2, S100a8/S100a9, Icam1/Vcam and Nox2 induction, and Adipoq, Igf1 and Aplnr repression). Endotoxemic responses were selectively modified by A2AR KO, supporting inflammatory suppression via A2AR sensitive shifts in regulators of NFκB and JAK-STAT signalling (IκBζ, IκBα, STAT1, CDKN1a and RRAS2) without impacting the cardio-depressant gene profile. Data indicate A2ARs exert minor effects in un-stressed myocardium and selectively suppress NFκB and JAK-STAT signalling and cardiac injury without influencing cardiac depression in endotoxemia.
Electronic supplementary material
The online version of this article (doi:10.1007/s11302-016-9536-1) contains supplementary material, which is available to authorized users.
Keywords: Adenosine, Adenosine 2A receptor, Anti-inflammatory, Cytokines, Endotoxemic myocarditis, Gene expression, Inflammation, Microarray, Sepsis
Introduction
Uncontrolled inflammation with sepsis and the systemic inflammatory response syndrome induce multiple organ dysfunction, including cardiac abnormalities key to disease progression and mortality [1]. Unravelling the complex mechanisms governing myocardial injury [1, 2] is not only fundamentally important but also reveals targets for manipulating outcomes. In this regard, adenosine 2A receptors (A2ARs) may fulfil a broadly suppressive role to limit inflammatory injury in multiple tissues [3–6] and enhance myocardial resistance to ischaemic/hypoxic insult, presenting a potentially useful therapeutic target [3, 7]. In heart, this G protein-coupled receptor (GPCR) influences coronary tone and angiogenesis, cardiac contractility, fibroblast growth and fibrosis and may mediate protection via ischaemic pre- and post-conditioning [8–10]. Inflammatory modulation contributes to this latter cardioprotection [9–11], together with the regulation of myocyte kinase signals to limit oxidative stress, mitochondrial dysfunction and cell death [12–14]. However, impacts of the A2AR on integrated myocardial responses to uncontrolled inflammation, and the mechanisms underlying such effects, remain to be elucidated.
While adenosine analogues and A2AR agonists can limit endotoxemic or septic injury in lung [15, 16], liver [17], brain [18] and heart [19–21], the roles of intrinsic A2AR activity are less clear. Receptor deletion fails to modify inflammatory markers/injury in some studies [22], reportedly worsens endotoxemic injury in heart [23] and lung [15, 24] and improves survival in models of polymicrobial sepsis [25, 26]. These divergent outcomes may reflect different cell- and organ-specific effects of A2ARs, for example promoting inflammasome formation and macrophage-dependent injury [27], impairing bacterial clearance [25], while activating cardiac survival signalling and suppressing inflammation [3, 7, 12–14]. Nonetheless, opposing effects of endogenous vs. exogenous (or amplified endogenous) adenosine are evident within cell types, for example inhibiting vs. promoting vascular myocyte NOS activation with inflammation [28]. The chronicity of receptor activation may also be critical to tissue-specific responses; while acute or transient increases in adenosine levels/receptor activity are generally beneficial (improving tissue perfusion, ischemic/hypoxic tolerance and reducing cytokines/inflammation), chronic elevations may be detrimental, exaggerating fibrotic processes via A2BRs in lung [29] or A2A and A2BRs in liver [30, 31], for example. Whether acute vs. chronic A2AR activity induces opposing myocardial outcomes is unknown, though prolonged A2B agonism limits rather than promotes fibrosis in injured myocardium [32], as does prolonged A1 agonism [33].
Additional to questions regarding protective vs. deleterious effects of the A2AR, the evolution of myocardial inflammatory injury and ‘endotoxemic myocarditis’ itself is complex and incompletely defined [1, 2]. Effects in non-cardiac tissues involve TLR/CD14-dependent NFκB and MAPK signalling and interferon/cytokine engagement of the JAK-STAT path [34–36]. These mechanisms likely participate in heart, with evidence NFκB and IκB kinase promotes cardiac dysfunction in sepsis/endotoxemia [37, 38], while JAK-STAT signalling mediates dysfunction and cell death in myocardial ischemia [39, 40]. In vivo observations suggest marked expansion of this signalling in intact myocardium [41, 42], contrasting in vitro evidence that cardiomyocyte NFκB and IκB kinase signalling is only transiently LPS responsive [43]. Importantly, these paths are inhibited by A2ARs in other cells, with exogenous agonism decreasing [44, 45] and A2AR KO increasing NFκB activity [6]. Nonetheless, these paths can also promote myocardial stress-resistance and survival under conditions that include inflammatory cytokine challenge [46–48]. The molecular underpinnings of endotoxemic myocarditis in vivo warrant further investigation, as do the mechanisms countering this dysfunction (including A2AR activity). We thus undertook broad-scale transcriptomic profiling of hearts from WT and A2AR KO mice, at baseline and following endotoxin challenge: shifts in gene expression can shed light on both functions of the A2AR, and pathogenesis and modulation of endotoxemic myocarditis. There are no prior analyses of transcriptome-wide effects of A2AR activity in myocardium, and relatively few of the in situ cardiac response to endotoxemia or sepsis [41, 42, 49].
Materials and methods
Animals
All animal procedures conformed to the Guide for the Care and Use of Laboratory Animals published by the NIH (publication number 85-25, revised 1996). This project (and protocols for care of animals) was approved by the Animal Care and Use Committees of St. Jude Children’s Research Hospital and the University of Tennessee Health Sciences Center. The A2AR KO mice and WT littermates were obtained from a sub-colony of the original lines generated and characterized by Ledent et al. [50]. Analysis of cardiac tissue confirms ablation of A2AR transcript without compensatory shifts in other adenosine receptors (Fig. S1). Mice were bred and maintained on-site at St. Jude Children’s Research Hospital, with standard laboratory food and water available ad libitum. Animal genotype was confirmed by PCR analysis of tail snips. All animals were originated from the same breeding series and were matched for age and weight. Male and female mice were used with equal representation in each experimental group. Both A2AR KO and WT littermate mice were randomly allocated to receive either an intraperitoneal injection of 20 mg/kg LPS isolated from E. coli (Sigma-Aldrich, St. Louis, MO) or an equal volume of sterile saline vehicle. Blood was sampled at 12 and 24 h of LPS challenge to assess shifts in blood cell counts and circulating cytokines/biomarkers, as detailed by us previously [23]. Mice were monitored for the development of symptoms of illness or distress (lethargy, piloerection, hunched posture and/or respiratory distress) during the initial 12 h post-injection and every 4 h thereafter; observation of any combination of symptoms prompted immediate euthanasia. All efforts were made to minimize animal suffering and distress. No analgesic or anaesthetic was administered other than for euthanasia immediately on evidence of illness/distress.
We initially tested responses to 24 h LPS challenge in both young (14 week) and old (46–52 weeks) WT and A2AR KO mice. However, absent overt symptoms of illness/distress, there was nonetheless significant mortality in aged LPS-treated mice, as detailed in the “Results” section and in Supplementary material. Cause of death in older animals was not determined. As a result of mortality, detailed molecular interrogation of cardiac gene expression (n = 6–8 per group), together with analyses of cardiac function in isolated hearts (n = 8–9 per group) and haematological/biomarker assessment (n = 6–9 per group) is necessarily constrained to the young group of mice.
Langendorff perfusion and tissue sampling
After 24 h of LPS challenge, mice were anaesthetised with sodium pentobarbital (50 mg/kg intraperitoneally), a thoracotomy performed and hearts excised into ice-cold Krebs-Henseleit solution for Langendorff perfusion, as detailed previously [23], or sampling of ventricular tissue for RNA preparation. Briefly, for perfusions, the aorta was immediately cannulated and hearts perfused at 80 mmHg with modified Krebs-Henseleit solution containing (in mM): NaCl, 120; NaHCO3, 25; KCl, 4.7; CaCl2, 2.5; MgCl2, 1.2; KH2PO4, 1.2; D-glucose, 15 and EDTA, 0.5. Perfusion fluid was maintained at 37 °C and bubbled with a mix of 95 % O2/5 % CO2 at 37 °C to provide a pH of 7.4. Ventricular function was monitored via a fluid-filled plastic film balloon in the left ventricle, connected to a P23 XL pressure transducer (Viggo-Spectramed, Oxnard, CA, USA). Coronary flow was monitored via a flow-probe in the aortic perfusion line, connected to a T206 flowmeter (Transonic Systems Inc., Ithaca, NY, USA). Functional data were recorded at 1 KHz on MacLab data acquisition system (ADInstruments, Castle Hill, Australia). Statistical comparisons of cardiovascular (Table 1) and blood parameters (Figs. 1 and 2) between groups were made via analysis of variance (ANOVA), with a Newman-Keuls post hoc test for specific comparisons. A P < 0.05 was considered indicative of significance in all tests.
Table 1.
Ex vivo cardiovascular function in perfused for hearts from WT and A2AR KO mice
| Functional parameter | WT (n = 8) | A2AR KO (n = 8) | WT + LPS (n = 9) | A2AR KO + LPS(n = 9) |
|---|---|---|---|---|
| Coronary flow (ml/min/g) | 16.7 ± 0.7 | 15.6 ± 1.2 | 16.9 ± 1.7 | 15.9 ± 0.6 |
| Heart rate (beats/min) | 341 ± 7 | 341 ± 7 | 352 ± 9 | 353 ± 9 |
| Systolic pressure (mmHg) | 113 ± 6 | 118 ± 5 | 80 ± 7* | 86 ± 3* |
| +dP/dt (mmHg/s) | 6039 ± 247 | 6039 ± 163 | 4310 ± 422* | 4489 ± 220* |
| −dP/dt (mmHg/s) | −3664 ± 129 | −3718 ± 132 | 2666 ± 197* | −2846 ± 105* |
| Coronary ‘Supply:Demand’ (ml/min/g/mmHg) | 0.15 ± 0.01 | 0.13 ± 0.02 | 0.21 ± 0.02* | 0.18 ± 0.02* |
All data are means ± S.E.M. Functional parameters were measured after 30 min of normothermic aerobic perfusion. Intrinsic heart rate is recorded immediately prior to pacing. *P < 0.05 vs. corresponding values in Vehicle-treated hearts. Coronary ‘Supply:Demand’ was calculated as the ratio of coronary flow rate (O2 supply) relative to ventricular systolic pressure (reflecting myocardial O2 demand). dP/dt, first derivative of pressure over time. *P < 0.05 vs. untreated. A2AR KO did not independently modify functional parameters
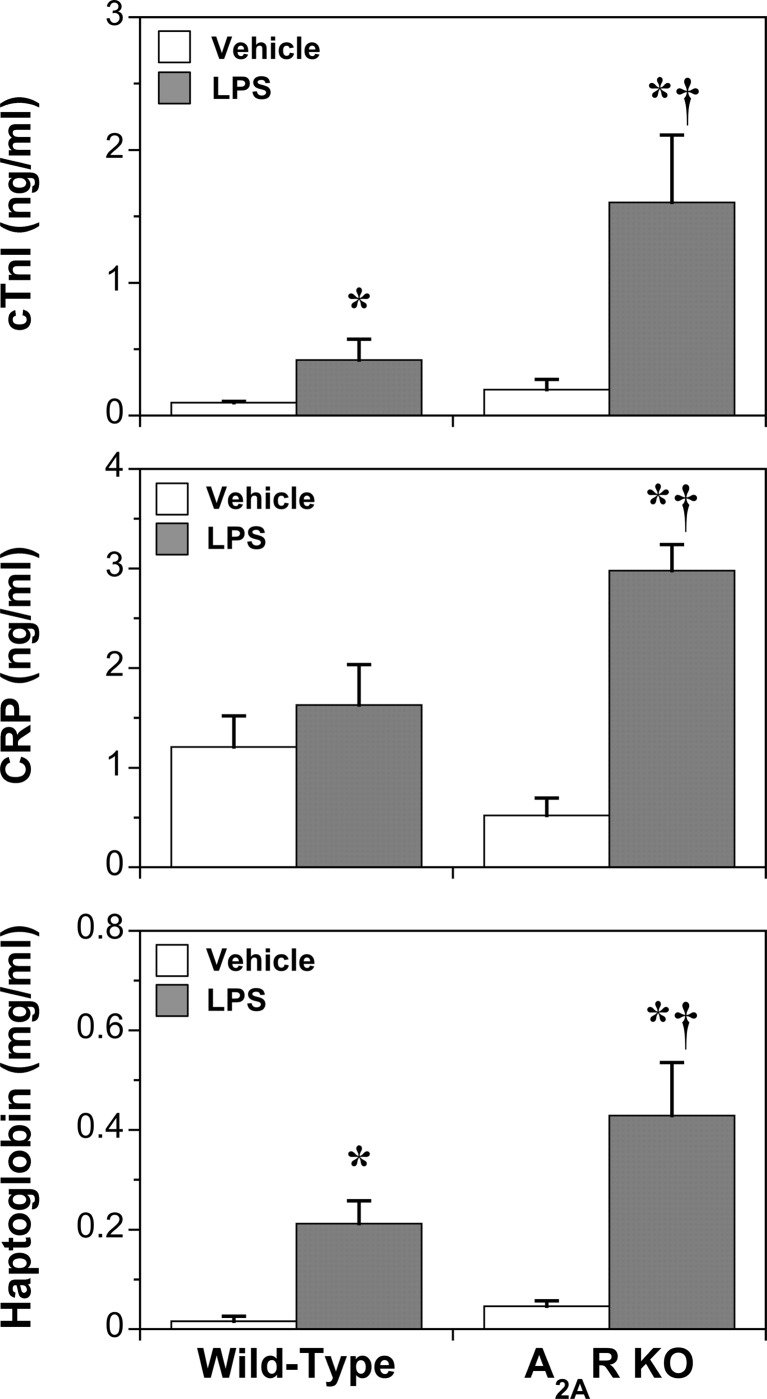
Fig. 1.
Effects of A2AR KO and LPS (24 h) on markers of cardiac damage (TnI) and systemic inflammation (CRP) and the acute phase response (haptoglobin). Data represent means ± S.E.M. *P < 0.05 vs. vehicle; †P < 0.05 vs. wild-type
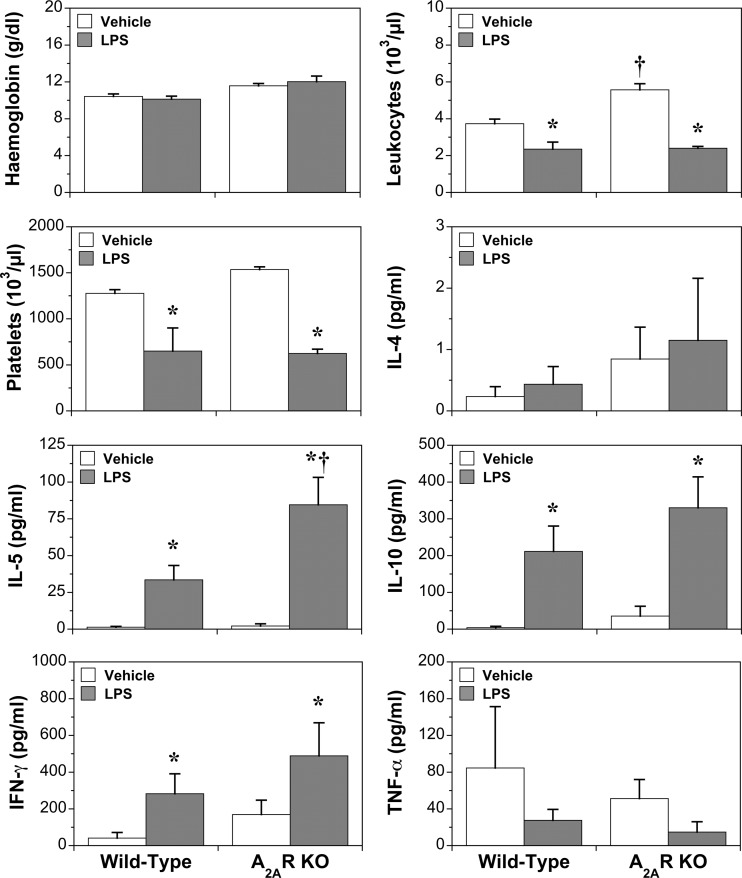
Fig. 2.
Effects of A2AR KO and LPS (24 h) on haematological parameters and cytokines. Data represent means ± S.E.M. *P < 0.05 vs. vehicle; †P < 0.05 vs. wild-type
RNA preparation and microarray hybridisation
Ventricular tissue was isolated, frozen in liquid N2 and stored at −80 °C until analysis. Tissue was subsequently homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA) and total RNA isolated according to manufacturer’s protocol. The RNA was further treated with 50 U of DNase I (Promega, Madison, WI) for 15 min at 37 °C and purified on RNeasy spin columns (Qiagen, Hilden, Germany). Total RNA yield and integrity were determined spectrophotometrically and via capillary electrophoresis on a 2100 BioAnalyzer (Agilent, Palo Alto, CA), respectively.
Microarray experiments were performed at the St. Jude Hartwell Center Core Facility according to manufacturers’ protocols. Briefly, first- and second-strand complementary DNA (cDNA) synthesis reactions were performed using the SuperScript Choice System (Invitrogen, Carlsbad, CA) followed by in vitro transcription using biotin-labelled dNTPs (ENZO Diagnostics, Farmingdale, NY). Complementary RNA (cRNA) samples were fragmented and individually hybridized to GeneChip® Mouse Genome 430 2.0 microarrays (Affymetrix, Santa Clara, CA). Each microarray quantified expression of over 39,000 transcripts, including full-length mRNA sequences and expressed sequence tags (ESTs). Following hybridization, microarrays were washed and stained with streptavidin–phycoerythrin (Molecular Probes, Eugene, OR) before scanning. Image files were converted into probe-set data (*.CEL files) via Affymetrix MAS 5.0 software. Experimental data are accessible through GEO Series accession number GSE44363 at http://www.ncbi.nlm.nih.gov/geo.
Microarray data and statistical analyses
Raw expression data were background corrected, normalized and log2 transformed using the Robust Multichip Average (RMA) method in BioConductor/R [51]. Data were filtered to include only transcripts detected on ≥3 arrays, with 25,646 transcripts passing this quality criterion (i.e. consistently detected in cardiac tissue). Log2 ratio values were generated by subtracting median expression values for WT vehicle-treated animals from each sample per probe before importing into TIGR MeV 4.0 software for statistical analysis [52]. The Significant Analysis of Microarrays (SAM) algorithm was used to correct for multiple comparisons and non-parametrically select differentially expressed genes using a median false discovery rate (FDR) ≤1.0 % [53]. In addition, a two-factor analysis of variance (ANOVA) was performed. To generate a data sets similar in size and false-positive rates, a P value <0.005 was employed. The lists of differentially expressed genes generated via SAM or ANOVA were then compared to identify 8110 shared transcripts for further investigation and validation (Fig. S2). This output was subjected to appropriate pair-wise SAM comparisons, incorporating a 1.5-fold change cut-off [54]. Significant transcripts were annotated using Ingenuity Pathway Analysis (IPA) (v8.7; Ingenuity® Systems, Redwood City, CA, USA), providing insight into biological/molecular themes over-represented in response to A2AR deletion and/or LPS. In brief, for each pathway/process, the fraction of differentially expressed genes was compared with the fraction of total genes in that path (shown in tables as the ‘Ratio’, useful for determining which pathways/processes overlap the most with altered genes in specific datasets). The probability of involvement of the respective number of modified transcripts in a path/process is expressed as a P value or range (values <0.05 considered significant).
Validation of expression changes and Adora expression via RT-qPCR
Two-step RT-qPCR, utilizing SYBR Green I, was employed to confirm differential gene expression for 11 transcripts (primer details provided in Table S1), as previously described [54]. Six additional genes (Actb, Gapdh, Hprt1, Pgk1, Ppia and Ubc) were assessed using GeNorm to determine utility as reference genes [55]. Following GeNorm assessment, Actb was found to be most stable (M = 0.03) and served as the endogenous reference control for all messenger RNAs (mRNAs) assessed via RT-qPCR. Briefly, 1 μg total RNA was used to synthesize cDNA using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. RT-qPCR was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Final reaction volumes (10 μL) included 5 μL iQ SYBR-Green Supermix (Bio-Rad, Hercules, CA), 100 nM of each primer and 4 μL of a 1:20 dilution of cDNA. Optimal qPCR cycling conditions entailed an initial denaturation at 95 °C for 3 min followed by 40 cycles of 95 °C for 15 s/62 °C for 60 s. After the final PCR cycle, reactions underwent melt curve analysis to detect non-specific amplicons. All reactions were performed in triplicate, with each plate containing an equal number of samples from each group, a calibrator control derived from a pool of all cDNA samples and a no-template control. PCR amplification efficiencies (90–110 %) for each primer pair were calculated using a 5-log serial dilution of calibrator sample. PCR data were analysed using CFX Manager v1.6 (Bio-Rad, Hercules, CA). Baseline subtractions and threshold settings above background were applied to all data. The calibrator sample was used to normalize inter-assay variations, with the threshold coefficient of variance for intra- and inter-assay replicates <1 % and <5 %, respectively. Normalized expression (ΔΔCq) was calculated, with mRNAs normalized to Actb levels and the calibrator control then log2-transformed.
Results
Impact of A2AAR deletion on cardiovascular function and inflammatory markers
Deletion of the A2AR was confirmed in cardiac tissue, with no compensatory changes in transcription of the other three sub-types (Fig. S1). Receptor deletion did not modify cardiac or coronary function in healthy or endotoxemic hearts (Table 1). Cardiovascular, blood cell and cytokine responses to LPS in WT and A2AR KO mice have been reported by us previously [23]. Similar outcomes were apparent here for markers of inflammation and injury (Fig. 1), haematological parameters and cytokines (Fig. 2) and cardiac function (Table 1). Data confirm significant inflammatory activation, cellular injury and cardiac dysfunction with LPS, with A2AR deletion amplifying changes in IL-5 and markers of cardiac injury (TnI) and systemic inflammatory stress (haptoglobin, CRP), without altering myocardial dysfunction or circulating levels of IL-4, IL-10, IFNγ or TNFα.
Impact of A2AAR deletion on survival
Interestingly, we acquire preliminary evidence LPS-dependent mortality is age- and sex-dependent and selectively exaggerated by A2AR KO in older males (Fig. S3). Initially, testing responses to LPS challenge in young (14 week) and middle-aged (46–52 week) mice—since age may exaggerate impacts of endotoxemia [56]—we recorded 20 % mortality in older WT mice with A2AR KO specifically reducing survival to <20 % in older males (Fig. S3). No mortality was recorded in young mice challenged with LPS (or any vehicle-treated groups). There is thus a trend to greater mortality with age, with a major survival effect of A2AR activity in older males. While molecular interrogations here are thus limited to young mice (in which LPS was non-lethal), it is possible A2AR activity may be increasingly crucial to survival in older, stress-intolerant males (whereas younger animals and females may possess intrinsically greater resistance to injury/death [57]). This is discussed further in the online Supplementary material.
Cardiac transcriptomic response to A2AR KO
Few transcriptional differences were detected between WT and A2AR KO hearts, with the amplitude of changes modest (up to threefold). Employing an initial 1.5-fold cut-off and 1 % FDR identified only 13 genes (Table 2). To enhance the power of subsequent pathway analysis, these criteria were relaxed to 1.2-fold and 5 %, encompassing 37 altered transcripts (Table 2). There was a little impact of A2AR deletion on inflammatory mediators, with modest induction of a CXC chemokine (Ppbp, +2.1), a regulator of cell migration/adhesion and cytokinesis (Iqgap1, +1.6) and a hemopoietic cytokine (Tslp, +1.6), together with repression of Hp (−2.7) and an Ig adhesion molecule regulating T-lymphocyte development (Mpzl2, −1.7) (Table 2).
Table 2.
Genes modified by A2AR KO in healthy myocardium
| Gene | Gene name | Affymetrix ID | Fold-Change | %FDR |
|---|---|---|---|---|
| UpRegulated | ||||
| Mtr | 5-methyltetrahydrofolate-homocysteine methyltransferase | 1439811_at | 2.83 | 0.00 |
| Ppbp | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 1418480_at | 2.08 | 3.93 |
| Chac1 | ChaC, cation transport regulator homologue 1 (E. coli) | 1451382_at | 1.85 | 0.00 |
| Ctsk | cathepsin K | 1450652_at | 1.84 | 0.00 |
| Cnpy2 | canopy 2 homologue (zebrafish) | 1416507_at | 1.67 | 0.00 |
| Iqgap1 | IQ motif containing GTPase activating protein 1 | 1445724_at | 1.64 | 3.85 |
| Tslp | thymic stromal lymphopoietin | 1450004_at | 1.63 | 3.93 |
| Nav3 | neuron navigator 3 | 1456144_at | 1.61 | 0.00 |
| Cux2 | cut-like homeobox 2 | 1447500_at | 1.60 | 0.00 |
| Slc38a1 | solute carrier family 38, member 1 | 1454764_s_at | 1.47 | 0.00 |
| Nsmce1 | non-SMC element 1 homologue (S. cerevisiae) | 1436121_a_at | 1.42 | 0.00 |
| Gk5 | glycerol kinase 5 (putative) | 1436210_at | 1.38 | 3.93 |
| Rpl22 | ribosomal protein L22 | 1448398_s_at | 1.37 | 0.00 |
| C11orf75 | chromosome 11 open reading frame 75 | 1419403_at | 1.37 | 0.00 |
| Slco5a1 | solute carrier organic anion transporter family, member 5 A1 | 1440874_at | 1.31 | 4.21 |
| Nfatc2 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 1426032_at | 1.28 | 4.70 |
| Downregulated | ||||
| Dgat1 | diacylglycerol O-acyltransferase homologue 1 (mouse) | 1418295_s_at | −1.24 | 2.39 |
| Ticam1 | toll-like receptor adaptor molecule 1 | 1454676_s_at | −1.24 | 3.05 |
| Mrap | melanocortin 2 receptor accessory protein | 1451371_at | −1.31 | 3.05 |
| Ppa1 | pyrophosphatase (inorganic) 1 | 1416939_at | −1.32 | 4.31 |
| Ell2 | elongation factor, RNA polymerase II, 2 | 1450744_at | −1.33 | 2.53 |
| Fmod | fibromodulin | 1456084_x_at | −1.47 | 4.31 |
| Ptger3 | prostaglandin E receptor 3 (subtype EP3) | 1450344_a_at | −1.49 | 2.68 |
| Pkdcc | protein kinase domain containing, cytoplasmic homologue (mouse) | 1454838_s_at | −1.50 | 4.61 |
| Ank3 | ankyrin 3, node of Ranvier (ankyrin G) | 1457288_at | −1.58 | 3.05 |
| Sacs | spastic ataxia of Charlevoix-Saguenay (sacsin) | 1434958_at | −1.61 | 2.39 |
| Pon1 | paraoxonase 1 | 1418190_at | −1.68 | 0.00 |
| Gnb1 | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 1425908_at | −1.72 | 4.54 |
| Mpzl2 | myelin protein zero-like 2 | 1416236_a_at | −1.74 | 0.00 |
| Pde3b | phosphodiesterase 3B, cGMP-inhibited | 1433694_at | −1.74 | 0.00 |
| Prkar2b | protein kinase, cAMP-dependent, regulatory, type II, beta | 1438664_at | −1.78 | 3.05 |
| Slc16a9 | solute carrier family 16, member 9 (monocarboxylic acid transporter 9) | 1429726_at | −1.84 | 0.00 |
| Map3k2 | mitogen-activated protein kinase kinase kinase 2 (MEKK2) | 1438719_at | −2.01 | 0.00 |
| Cidec | cell death-inducing DFFA-like effector c | 1452260_at | −2.08 | 0.00 |
| Acta1 | actin, alpha 1, skeletal muscle | 1427735_a_at | −2.14 | 2.68 |
| Yipf4 | Yip1 domain family, member 4 | 1426417_at | −2.51 | 4.54 |
| Hp | haptoglobin | 1448881_at | −2.70 | 0.00 |
Data shown for transcripts for which myocardial expression levels were modified by A2AR KO by a factor of ≥1.2-fold (FDR <5 %)
Analysis via the IPA suite identified 112 canonical pathways sensitive to A2AR activity in healthy hearts (Table S2), the most highly modified shown in Table 3. The top five included relaxin signalling, cardiac β-adrenergic signalling, cellular effects of sildenafil, PKA signalling and photo-transduction. The influence of A2AR KO on these and other paths can be attributed to repression of Prkar2b (−1.8), Gnb1 (−1.7) and Pde3b (−1.7), collectively impacting G protein-coupled cAMP/PKA dependent signalling (encompassing relaxin, α- and β-adrenergic, NO, Ca2+ and hypertrophic signalling). Additionally, Map3k2 (−2) and Nfat2c (+1.3) span many paths modified by A2AR KO. Modulation of these five transcripts contributes to 48 of the top 50 A2AR-sensitive canonical pathways. Overall, A2AR deletion exerts an inhibitory effect on G protein-coupled, cAMP/PKA, and MEKK2 signalling downstream of this and other GPCRs.
Table 3.
Top 20 canonical pathways modified by A2AAR KO in healthy myocardium
| Canonical pathways | P value | Ratio | Genes |
|---|---|---|---|
| Relaxin signalling | 3.55E-03 | 1.99E-02 | Gnb1, Prkar2b, Pde3b |
| Cardiac β-adrenergic signalling | 3.55E-03 | 2.11E-02 | Gnb1, Prkar2b, Pde3b |
| Cellular effects of sildenafil (Viagra) | 3.98E-03 | 1.99E-02 | Acta1, Prkar2b, Pde3b |
| Protein Kinase A signalling | 6.03E-03 | 1.26E-02 | Gnb1, Prkar2b, Pde3b, Nfatc2 |
| Phototransduction pathway | 6.03E-03 | 3.08E-02 | Gnb1, Prkar2b |
| Germ cell-sertoli cell junction signalling | 6.31E-03 | 1.88E-02 | Iqgap1, Acta1, Map3k2 |
| Calcium signalling | 8.71E-03 | 1.47E-02 | Prkar2b, Nfatc2, Acta1 |
| Glutamate receptor signalling | 9.55E-03 | 2.86E-02 | Gnb1, Slc38a1 |
| Caveolar-mediated endocytosis signalling | 1.41E-02 | 2.35E-02 | Acta1, Map3K2 |
| Nitric oxide signalling in the cardiovascular system | 1.48E-02 | 2.02E-02 | Prkar2b, Pde3b |
| Leptin signalling in obesity | 1.55E-02 | 2.38E-02 | Prkar2b, Pde3b |
| Cardiac hypertrophy signalling | 1.78E-02 | 1.22E-02 | Gnb1, Prkar2b, Map3k2 |
| TR/RXR Activation | 2.09E-02 | 2.02E-02 | Hp, Pde3b |
| SAPK/JNK signalling | 2.14E-02 | 1.98E-02 | Gnb1, Map3k2 |
| Colorectal cancer metastasis signalling | 2.14E-02 | 1.17E-02 | Gnb1, Prkar2b, Ptger3 |
| α-Adrenergic signalling | 2.19E-02 | 1.89E-02 | Gnb1, Prkar2b |
| RANK signalling in osteoclasts | 2.19E-02 | 2.04E-02 | Nfatc2, Map3K2 |
| G Beta Gamma signalling | 2.24E-02 | 1.68E-02 | Gnb1, Prkar2b |
| IL-1 signalling | 2.29E-02 | 1.89E-02 | Gnb1, Prkar2b |
| fMLP signalling in neutrophils | 2.95E-02 | 1.57E-02 | Gnb1, Nfatc2 |
Canonical signalling paths modified by A2AR KO are shown, ranked according to P values determined by a Fisher’s Exact Test. Also shown is a ratio value reflecting the number of molecules in a given path that meet cut-off criteria for differential expression, divided by the total number of molecules in the pathway
Multiple biological functions appear sensitive to A2AR KO (see Table S3), the most significant (Table 4) revolving around cellular development, growth, movement and death, together with humoral immunity and haematological development and intercellular signalling. Toxicological functions included high representation of liver processes (fibrosis, damage, steatosis, hepatitis and inflammation) together with six cardiac functions (Table 4). The latter included cardiac arteriopathy (Ank3, Pon1, Cux2, Gk5, Pde3b, Dgat1 and Nsmce1), infarction (Pon1 and Acta1) and transcripts involved in failure and hypertrophy (Pde3b and Nfat2c).
Table 4.
Top 20 biological and toxicological functions modified by A2AAR KO in healthy myocardium
| Biological pathway | P values | Genes |
|---|---|---|
| Cellular development | 7.34E-04–4.5E-02 | Mrap, Ctsk, Sacs, Prkar2b, Ptger3, Nfatc2, Tslp, Map3k2 |
| Cellular growth and proliferation | 7.34E-04–4.01E-02 | Mrap, Ppbp, Nfatc2, Tslp |
| Haematological system development and function | 7.34E-04–4.5E-02 | Hp, Ptger3, Ticam1, Ppbp, Nfatc2, Tslp, Map3k2 |
| Humoral immune response | 7.34E-04–4.5E-02 | Nfatc2, Fmod, Tslp |
| Cell-to-cell signalling and interaction | 1.53E-03–4.74E-02 | Ank3, Ptger3, Ticam1, Ppbp, Nfatc2, Iqgap1, Tslp, Map3k2 |
| Amino acid metabolism | 2.55E-03–5.09E-03 | Slc38a1 |
| Carbohydrate metabolism | 2.55E-03–4.72E-02 | Gnb1, Pon1, Ppbp, Ppa1 |
| Cell death | 2.55E-03–4.5E-02 | Ppbp, Nfatc2, Tslp, Map3k2 |
| Cell morphology | 2.55E-03–3.68E-02 | Ank3, Ptger3, Nfatc2, Cnpy2, Iqgap1, Tslp |
| Cellular assembly and organization | 2.55E-03–4.01E-02 | Ank3, Ctsk, Nfatc2, Fmod, Cnpy2, Iqgap1, Acta1 |
| Cellular compromise | 2.55E-03–4.01E-02 | Ank3, Hp, Ctsk, Ptger3, Ppbp |
| Cellular function and maintenance | 2.55E-03–1.52E-02 | Hp, Ppbp, Nfatc2, Tslp, Map3k2 |
| Cellular movement | 2.55E-03–4.58E-02 | Gnb1, Ctsk, Hp, Ticam1, Ppbp, Nfatc2, Iqgap1, Tslp |
| Connective tissue development and function | 2.55E-03–3.02E-02 | Ctsk, Rpl22, Ptger3, Pde3b, Ppbp, Cidec, Nfatc2 |
| Connective tissue disorders | 2.55E-03–5.09E-03 | Nfatc2 |
| Developmental disorder | 2.55E-03–2.55E-03 | Ctsk |
| Genetic disorder | 2.55E-03–4.01E-02 | Ank3, Ctsk, Ptger3, Cidec, Iqgap1, Tslp, Mrap, Gnb1, Pon1, Hp, Prkar2b, Pde3b, Nfatc2, Dgat1, Acta1 |
| Hair and skin development and function | 2.55E-03–5.09E-03 | Ptger3, Dgat1 |
| Immune cell trafficking | 2.55E-03–3.51E-02 | Hp, Ticam1, Ppbp, Nfatc2, Tslp |
| Inflammatory response | 2.55E-03–4.67E-02 | Hp, Ptger3, Ticam1, Ppbp, Nfatc2, Fmod, Tslp |
| Molecular toxicological function | P values | Genes |
| Liver fibrosis | 3.01E-03 | Hp, Tslp |
| Liver damage | 1.43E-02 | Ticam1, Tslp |
| Liver steatosis | 2.45E-02 | Pde3b |
| Cardiac arteriopathy | 3.41E-02 | Ank3, Cux2, Dgat1, Gk5, Nsmce1, Pde3b, Pon1 |
| Cardiac infarction | 3.76E-02 | Acta1, Pon1 |
| Liver hepatitis | 4.41E-02 | Pde3b |
| Hepatocellular carcinoma | 4.43E-02 | Hp, Iqgap1 |
| Liver steatohepatitis | 5.67E-02 | Pde3b |
| Renal proliferation | 7.90E-02 | Tslp |
| Liver inflammation | 9.36E-02 | Hp |
| Pulmonary hypertension | 1.08E-01 | Ptger3 |
| Nephrosis | 1.10E-01 | Pde3b |
| Cardiac congestive cardiac failure | 1.63E-01 | Pde3b |
| Heart failure | 1.63E-01 | Pde3b |
| Renal nephritis | 2.50E-01 | Pde3b |
Enriched biological/toxicological functions of A2AR KO sensitive transcripts are listed according to P values or ranges determined by a Fisher’s Exact Test
Cardiac transcriptomic response to LPS
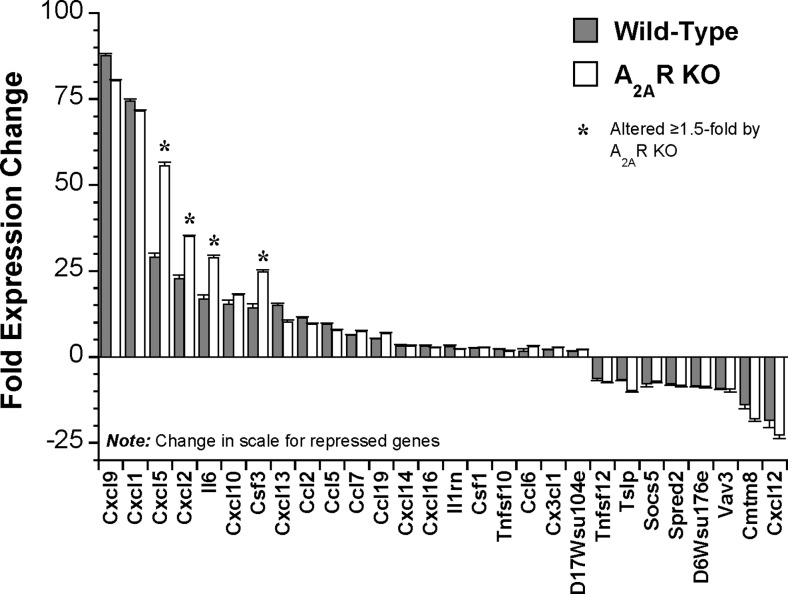
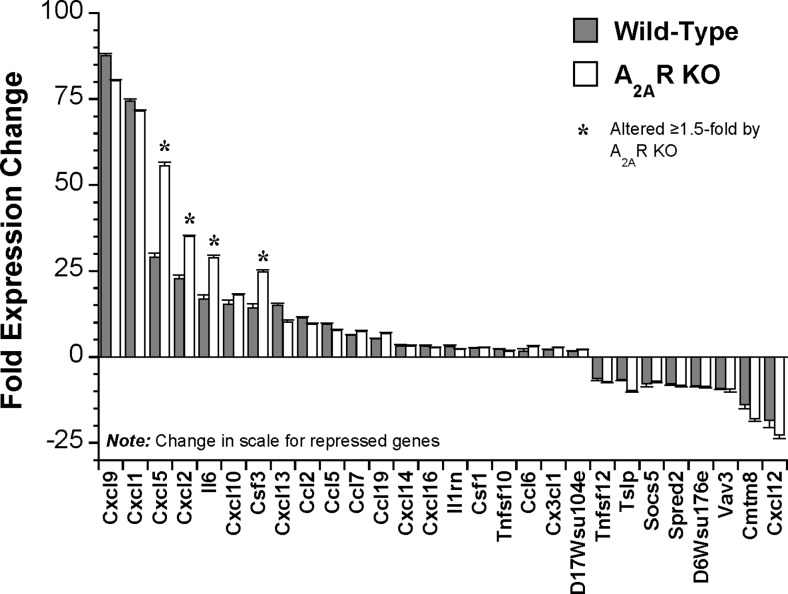
Analysis via the SAM algorithm (1.5-fold threshold, 1 % FDR) identified 4146 transcripts modified after 24 h of LPS challenge (see Table S4 for full details). Figure 3 presents the 25 most induced and repressed transcripts, with responses in A2AR KO hearts shown for comparison. Many of the most highly modified are predictably involved in inflammation/innate immunity, together with tissue development/remodelling. The most LPS-responsive cytokines/chemokines are shown in Fig. 4, with responses in KO hearts highlighted.
Fig. 3.
The 25 most induced and 25 most repressed transcripts in hearts from young (2–3 month) mice challenged with LPS for 24 h. Mice were injected with 20 mg/kg LPS or saline vehicle and left ventricular tissue isolated for analysis 24 h later. Data from both wild-type and A2AR KO hearts are shown for comparison (n = 6–8/group). Data are means ± S.E.M
Fig. 4.
Effects of A2AR KO on the most LPS-responsive cytokines in wild-type hearts. Mice were injected with 20 mg/kg LPS or saline vehicle, and left ventricular tissue isolated for analysis 24 h later. Data from both wild-type and A2AR KO hearts are shown for comparison (n = 6–8/group). Data are means ± S.E.M
Functional classifications, using a twofold threshold to narrow the focus to the most highly modified paths, identify 236 canonical pathways (Table S5 ), with the most significantly modified presented in Table 5. Data confirm a profound inflammatory and immune response, with marked upregulation of TLR/MyD88 and interferon signalling (Fig. 5), the acute phase response (Fig. S4), IRF activation (Fig. S5) and PRR- and RIG-1-like signalling (Figs. S6 and S7). These changes primarily reflect shifts in underpinning NFκB, JAK-STAT, MAPK and PI3K/Akt signalling (Figs. S8–S12). Additionally, LPS upregulated cell death signalling (Figs. S13 and S14) and modified paths involved in cellular differentiation, movement, growth and remodelling (e.g. Figs. S15 and S17). The top LPS-sensitive biological functions are summarized in Table S6, with biologic and cardiovascular toxicological responses fully detailed in Table S7 and S8 in the Supplementary material.
Table 5.
Top 20 canonical pathways modified by LPS (≥2-fold change, <1 % FDR) in WT hearts
| Canonical pathways | P value | Ratio | Genes |
|---|---|---|---|
| Acute phase response signalling | 9.12E-09 | 2.02E-01 | Socs3, Rac2, Hamp, Serping1, Nfkbie, Socs2, Cp, Saa2, Serpina3, Jak2, Il6, Rbp1, Nr3C1, C1r, Hmox1, Shc1, Nfkbia, Cfb, Lbp, Serpine1, Nfkbib, Saa1, C3, Tnfrsf1a, Myd88, C1s, Rac1, Serpinf1, Cebpb, Stat3, Nfkb2, Hp, Rras2, Il1Rn, C4a, C2 |
| Interferon signalling | 1.82E-08 | 4.33E-01 | Ifit3, Oas1, Ptpn2, Mx1, Ifi35, Irf9, Psmb8, Jak2, Tap1, Irf1, Ifitm1, Stat2, Stat1 |
| Activation of IRF by cytosolic pattern recognition receptors | 1.38E-07 | 2.33E-01 | Dhx58, Nfkbie, Zbp1, Irf9, Tbk1, Il6, Nfkb2, Adar, Isg15, Ifih1, Irf7, Nfkbia, Ddx58, Stat2, Nfkbib, Stat1, Ifit2 |
| Hepatic fibrosis/hepatic stellate cell activation | 7.94E-07 | 2.01E-01 | Icam1, Lepr, Myh7b (Includes Eg:668,940), Myh11, Ccl5, Il6, Fas, Pgf, Vegfa, Il1r2, Cxcl3, Igf1, Cyp2e1, Timp1, Lbp, Stat1, Timp2, Egfr, Il4r, Vcam1, Tnfrsf1a, Vegfc, Igfbp5, Nfkb2, Myl7, Csf1, Cd14 |
| Dendritic cell maturation | 9.12E-07 | 1.51E-01 | B2m, Rac2, Icam1, Lepr, Nfkbie, Pik3r5, Cd83, Il6, Jak2, Fcgr2b, Fcgr1a, Nfkbia, Hla-b, Tlr3, Stat1, Nfkbib, Fcgr3a, Hla-c, Myd88, Tnfrsf1a, Relb, Rac1, Nfkb2, Tlr2, Il1rn, Fcer1g, Stat2, Irf8 |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | 2.57E-06 | 1.34E-01 | Socs3, Rac2, Icam1, Mmp3, Nfkbie, Pik3r5, Jak2, Ccl5, Il6, Il17ra, Fcgr1a, Pgf, Myc, C1r, Il1r2, Vegfa, Nfkbia, Traf3Ip2, Wif1, Cfb, Plcb1, Tlr3, Nfkbib, Prkd1, Fcgr3a, Calm1, Adamts4, Sele, Vcam1, Myd88, Tnfrsf1a, C1s, Daam1, Rac1, Vegfc, Stat3, Cebpb, Irak3, Tlr2, Hp, Rras2, Il1Rn, Csf1, Cxcl12, Cebpd, Sost, Wnt5a, Irak2 |
| Retinoic acid mediated apoptosis signalling | 5.37E-06 | 1.94E-01 | Parp10, Art1, Tnfsf10, Parp3, Parp12, Parp9, Tiparp, Irf1, Parp4, Bid, Parp11, Cflar, Parp14 |
| Antigen presentation pathway | 6.61E-06 | 2.56E-01 | B2m, Psmb9, Hla-e, Hla-b, Psmb8, Hla-g, Tap1, Tap2, Tapbp, Hla-c |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 8.32E-06 | 2.12E-01 | Ptx3, Oas1, C3, Oas2, Myd88, Pik3R5, Il6, Nfkb2, Ccl5, Tlr2, Ifih1, Clec7a, Irf7, Syk, Ddx58, Eif2ak2, Tlr3 |
| Death receptor signalling | 1.29E-05 | 2.34E-01 | Hspb3, Tnfrsf1a, Nfkbie, Tnfsf10, Tbk1, Nfkb2, Fas, Daxx, Nfkbia, Bid, Cflar, Nfkbib, Birc3, Birc2, Hspb1 |
| IL-10 signalling | 3.24E-05 | 2.14E-01 | Ccr1, Socs3, Il4r, Nfkbie, Il6, Stat3, Nfkb2, Fcgr2b, Il1r2, Hmox1, Nfkbia, Il1rn, Cd14, Lbp, Nfkbib |
| Role of RIG1-like receptors in antiviral innate immunity | 6.61E-05 | 1.96E-01 | Dhx58, Ifih1, Irf7, Nfkbia, Nfkbie, Ddx58, Tbk1, Nfkb2, Nfkbib, Trim25 |
| IL-6 signalling | 1.00E-04 | 1.94E-01 | Hspb3, Tnfrsf1a, Nfkbie, Il6, Nfkb2, Stat3, Cebpb, Jak2, Il1r2, Shc1, Nfkbia, Rras2, Il1rn, Cd14, Lbp, Nfkbib, Tnfaip6, Hspb1 |
| IL-8 signalling | 1.02E-04 | 1.45E-01 | Rac2, Icam1, Cxcl1, Pik3r5, Pgf, Eif4ebp1, Vegfa, Hmox1, Gng11, Gna13, Nfkbib, Prkd1, Egfr, Vcam1, Rhoc, Rac1, Vegfc, Irak3, Cstb, Myl7, Myl9 (Includes Eg:98,932), Bcl2l1, Itgb2, Rras2, Ccnd2, Itgam, Irak2 |
| Bladder cancer signalling | 1.26E-04 | 1.89E-01 | Fgf16, Mmp3, Fgf9, Mmp14, Mmp15, Vegfc, Pgf, Myc, Vegfa, Rras2, Mmp8, Fgf12, Cdkn1a, Chd1 (Includes Eg:1105), Rps6Ka5, Fgf7, Egfr |
| Pathogenesis of multiple sclerosis | 1.45E-04 | 5.56E-01 | Cxcl10, Ccr1, Cxcl9, Ccl5, Cxcl11 |
| Role of PKR in interferon induction and antiviral response | 1.58E-04 | 2.39E-01 | Nfkbia, Tnfrsf1a, Nfkbie, Bid, Nfkb2, Eif2ak2, Tlr3, Stat1, Nfkbib, Fcgr1a, Irf1 |
| JAK/Stat signalling | 2.00E-04 | 2.19E-01 | Rac2, Socs3, Pias2, Socs2, Rac1, Pik3r5, Stat3, Jak2, Shc1, Rras2, Cdkn1a, Cish, Stat2, Stat1 |
| Complement system | 2.09E-04 | 2.5E-01 | C1R, Cfd, Serping1, C3, C1S, Cfb, C4a, Masp1, C2 |
| Glucocorticoid receptor signalling | 2.14E-04 | 1.25E-01 | Rac2, Icam1, Nfkbie, Pik3R5, Pbx1, Ccl5, Jak2, Il6, Fcgr1a, Nr3c1, Il1R2, Shc1, Cxcl3, Hspa4, Nfkbia, Ar, Ccl13, Cdkn1c, Serpine1, Stat1, Fkbp5, Nfkbib, Sra1, Vcam1, Ccnh, Sele, Rac1, Stat3, Cebpb, Bcl2l1, Rras2, Il1rn, Cdkn1a, Fkbp4, Nr3c2 |
Fig. 5.
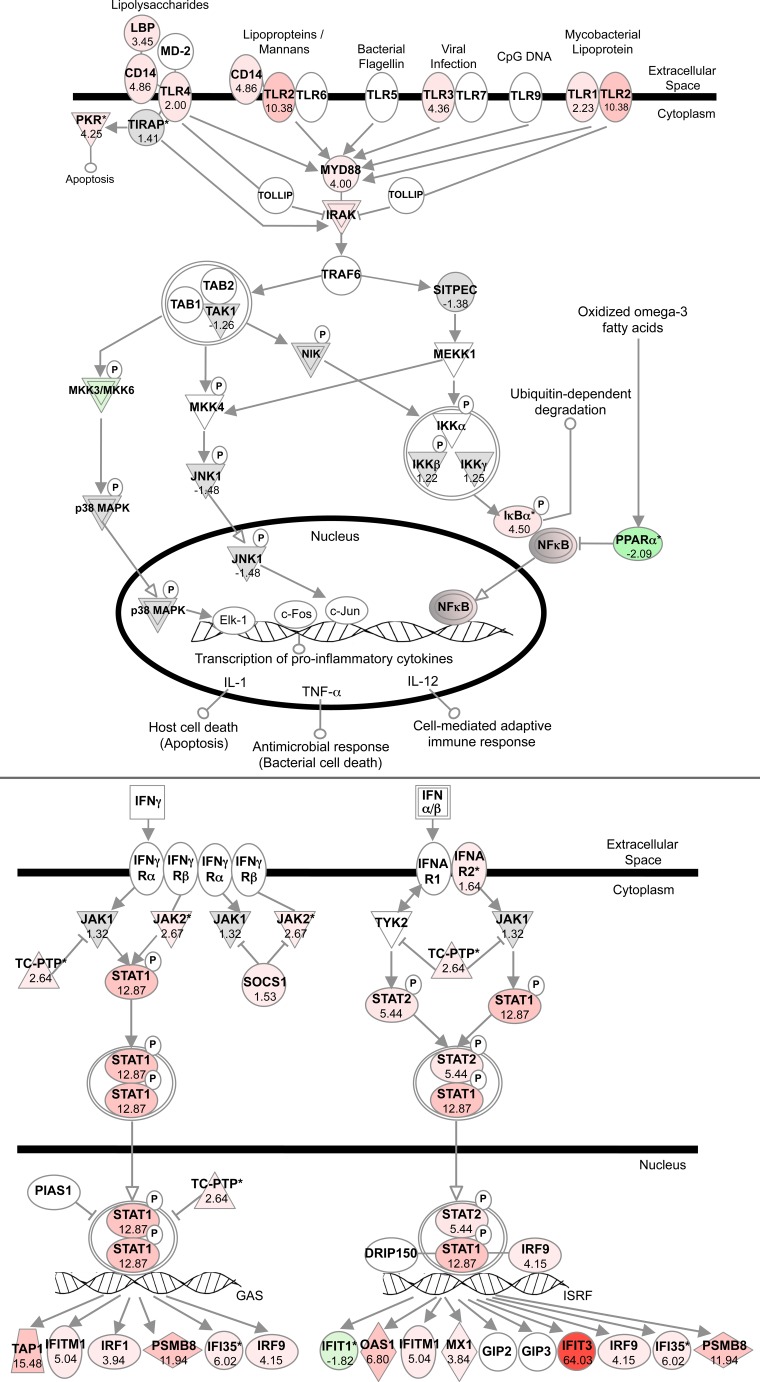
Impact of 24 h LPS challenge on cardiac transcription of the TLR (upper panel) and interferon signalling paths (lower panel) in wild-type mice (n = 6–8/group). Induced transcripts highlighted in red and repressed in green. Shaded transcripts are altered by LPS by less than the threshold value
Unsurprisingly, many highly responsive transcripts were involved in inflammation (Figs. 3 and 4, Table 5): within the most induced are interferon-related genes (Ifit1, Ifit2, Ifit3, Igtp1, Igtp2, Igtp3, Iigp1, Iigp2, Ifi44 and Irf7) and inflammatory/immune modulators (Lcn2, Saa3, Socs3, S100a8, S100a9, Mpa2l, Cxcl1, Cxcl9, Cxcl10, IL6, Ptx3, Serpina3n, Csf3 and Mt2). Several are potentially injurious to the heart: the most highly induced transcript encodes lipocalin-2, which regulates inflammation and matrix degradation, is implicated in heart failure [58] and promotes cardiac apoptosis [59]; CXCL1 is a potent neutrophil chemoattractant; CXCL9 stimulates cytokine production and T-cell proliferation/recruitment; CXCL10 mediates CXCR3+ cell migration and augments inflammation. Conversely, some transcripts encode anti-inflammatory and potentially protective molecules: SERPINA3N (α1-antichymotrypsin) inhibits proteases involved in inflammation; the pentraxin PTX3 is expressed in heart with inflammation and limits cell death; SOCS3 is a negative feedback regulator of IL-6 signalling, implicated in sex-dependent cardiac stress-resistance; metallothionein-2, induced by STAT3-related signalling, mediates cardioprotection and can limit dysfunction in sepsis.
Cardio-depressant molecular profile
Major pathways critical to contractile function were substantially impacted, identifying a ‘cardio-depressed’ molecular profile in hearts of endotoxemic mice (Figs. S18–S21). β-Adrenergic signalling was broadly suppressed (Fig. S18), including reduced transcripts for β1-adrenergic receptor, G protein, adenylate cyclase, PKA and related signalling elements (Ppp1r14c, Gng11, Prkar2b, Pde7a, Ppp1r3c, Ppp1r14a, Pkia and Adcy7), together with induction of Pde4b (AMP-dependent phosphodiesterase). The PKA pathway itself was substantially suppressed (Fig. S19), including reductions in PKA RIα (Prkar1a), PKA RIIα (Prkar2a), PKA RIIβ (Prkar2b), PKAα (Prkaca) and PKA-Cβ (Prkacb), while PKA RIβ (Prkar1b) was modestly induced (+1.5). Transcripts for Gαs and Gαq also declined.
Cellular Ca2+ signalling paths were repressed (Fig. S20), with reductions in key elements including: Camk1d, Ryr3, Casq1, Mef2c, Casq2, Calm1, Hdac9, Myh7b, Tpm2, Myh11, Rcan1, Myl7, Atp2b2, Prkar2b, Tpm3 and Asph. Mitochondrial dysfunction (Fig. S21) is also evidenced by repression of complex I/NADH dehydrogenase components (including Ndufv2 and 3; Ndufs4, 5, 6, 7 and 8; Ndufa3, 4, 5, 6 and 8; Ndufab1, Ndufaf1, Ndufb2, 3, 4, 7, 9, 10 and 11), together with Complex II components (including Sdhc, Sdhd, Uqcrb, Uqcrc1 and Uqcrfs1). Uncoupling proteins Ucp2 and Ucp3 were also induced, while Ucp1 was repressed. Transcript for the major activator of mitochondrial biogenesis (Ppargc1a) was repressed.
Adenosine-related transcripts
There were limited impacts on the adenosinergic system itself, with two- to threefold upregulation of adenosine deaminase, 1.5-fold repression of adenosine kinase, 1.7-fold induction of S-adenosylhomocysteine hydrolase and twofold induction of the ecto-nucleotidase CD39. These changes may modify patterns of cellular adenosine generation vs. uptake and metabolism, potentially altering receptor activation in endotoxemic tissue. A small 1.3-fold rise in Adora2b expression was also detected.
Impact of A2AR KO on transcriptomic responses to endotoxemia
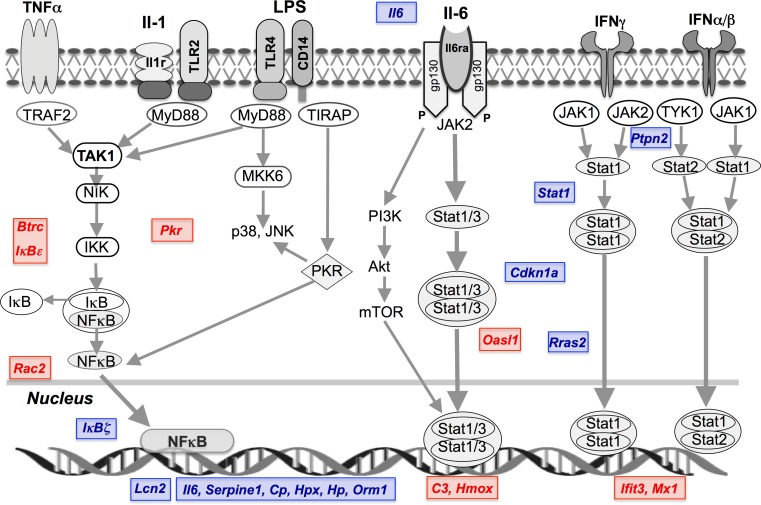
Interestingly, A2AR activity does not broadly suppress the effects of endotoxemia on myocardial gene expression, with 90 % of transcriptomic responses unaltered by receptor KO. For example, Fig. 3 presents response profiles for the 25 most induced or repressed transcripts in WT hearts, with most shown to be insensitive to A2AR KO. Deletion selectively enhanced induction of Lcn2, Igtp, Cxcl5, S100a8, Iigp2, Cxcl2 and Iigp1 and repression of C1qtnf9, Colec11 and Inmt, while reducing induction of Ifit3 and repression of Adipoq, Gpr22, and Scube2, Ifit3 (Fig. 3). These effects support A2AR modulation of inflammatory and interferon-related signalling responses. Nonetheless, the pattern of LPS-dependent cytokine/chemokine change was largely insensitive to A2AR KO, with specific enhancement of Cxcl5, Cxcl2, Il6 and Csf3 induction (Fig. 4). Figure 6 summarizes the select effects of A2AR activity (revealed via KO) on TLR- and interferon-triggered NFκB and JAK-STAT signalling responses to LPS (generally repressing these pathways).
Fig. 6.
Schematic of TLR, IL-1, IL-6 and interferon receptor activation of NFκB and JAK-STAT signalling and transcriptional control. The effects of A2AR KO- on LPS-dependent gene changes are highlighted. These pathways are critical to cellular responses to LPS and orchestration of inflammation. Specific effects of A2AR activity (based on effects of receptor KO) on transcriptional responses to LPS are highlighted (red—increased expression; blue—reduced expression)
To gain further insight into these specific effects of A2AR KO, we identified those LPS-responsive transcripts whose expression changes were increased or reduced ≥1.5-fold by A2AR KO (Table S9). Effects of LPS on 282 genes were augmented ≥1.5-fold following A2AR KO, with a further 9 responsive to LPS specifically in KO and not WT hearts (Table S9). The most highly augmented included 25 transcripts increased ≥twofold by KO. Many A2AR-sensitive transcripts are associated with pro-inflammatory signalling (e.g. Igtp, Lcn2, Cxcl5, S100a8, Iigp2, Cxcl2, Il6, Ifi202b, Hp and Cxcl9), with data supporting inhibitory effects of the A2AR on the responsiveness of canonical pathways that include the acute phase response, glucocorticoid receptor, Erk/MAPK, HIF1α and JAK-STAT signalling (Table 6).
Table 6.
Top canonical responses to LPS modified by A2AAR KO (the 15 most promoted or inhibited by A2AAR KO are shown)
| Canonical Pathways | P value | Ratio | Genes |
|---|---|---|---|
| LPS responses enhanced by A2AR KO (countered by A2AR activity) | |||
| Acute phase response signalling | 7.41E-04 | 5.06E-02 | Hpx, Hp, Rras2, Pik3r1, Cp, Il6, Serpine1, Nr3c1, Orm1 |
| Glucocorticoid receptor signalling | 9.33E-04 | 3.90E-02 | Hspa4, Cxcl3, Rras2, Pik3r1, Cdkn1a, Tgfb2, Il6, Stat1, Serpine1, Hspa2, Nr3c1 |
| Glioma invasiveness signalling | 1.29E-03 | 8.47E-02 | Timp4, Rras2, Pik3r1, Rhoj, Itgb3 |
| Germ cell-sertoli cell junction signalling | 1.45E-03 | 5.00E-02 | Rras2, Tuba8, Pak6, Map3k6, Pak3, Pik3r1, Tgfb2, Rhoj |
| Angiopoietin signalling | 3.16E-03 | 6.58E-02 | Grb14, Rras2, Pak6, Pak3, Pik3r1 |
| ILK signalling | 3.55E-03 | 4.26E-02 | Ppp2r3a, Pik3r1, Vegfc, Atf4, Rhoj, Itgb6, Pgf, Itgb3 |
| Ephrin receptor signalling | 3.98E-03 | 4.04E-02 | Rras2, Pak6, Pak3, Vegfc, Atf4, Epha4, Efnb3, Pgf |
| ERK/MAPK signalling | 4.37E-03 | 4.10E-02 | Rras2, Pak6, Pak3, Ppp2r3a, Ppp1R3c, Pik3r1, Atf4, Stat1 |
| Pancreatic adenocarcinoma signalling | 4.47E-03 | 5.08E-02 | Pik3r1, Cdkn1a, Tgfb2, Vegfc, Stat1, Pgf |
| HER-2 signalling in breast cancer | 5.13E-03 | 6.17E-02 | Rras2, Pik3r1, Cdkn1a, Itgb6, Itgb3 |
| Macrophage NO & ROS production | 7.08E-03 | 3.74E-02 | Map3k6, Ppp2r3a, Ppp1r3c, Pik3r1, Rhoj, Irf8, Stat1 |
| Hepatic fibrosis/stellate cell activation | 1.05E-02 | 4.48E-02 | Cxcl3, Tgfb2, Vegfc, Il6, Stat1, Pgf |
| Circadian rhythm signalling | 1.12E-02 | 8.57E-02 | Per3, Arntl, Atf4 |
| HIF1α signalling | 1.38E-02 | 4.55E-02 | Rras2, Pik3r1, Vegfc, Slc2a4, Pgf |
| JAK/Stat signalling | 1.41E-02 | 6.06E-02 | Rras2, Pik3r1, Cdkn1a, Stat1 |
| LPS responses inhibited by A2AR KO (promoted by A2AR activity) | |||
| PXR/RXR activation | 6.46E-04 | 4.4E-02 | Rac2, Prkar2b, Cyp2b6 |
| Nicotinate and nicotinamide metabolism | 3.98E-03 | 2.94E-02 | Cilp, Art5, Eif2ak2, Bst1 |
| Amyloid processing | 4.57E-03 | 5.08E-02 | Rac2, Prkar2b, Capn10 |
| Glutamate receptor signalling | 5.89E-03 | 4.29E-02 | Glul, Slc38a1, Homer1 |
| AMPK signalling | 9.55E-03 | 2.40E-02 | Rac2, Prkar2b, Adipoq, Eif4ebp1 |
| PRRs in recognition of bacteria & viruses | 9.77E-03 | 3.66E-02 | C3, Ddx58, Eif2ak2 |
| NF-κB signalling | 1.15E-02 | 2.58E-02 | Rac2, Nfkbie, Btrc, Eif2ak2 |
| NF-κB activation by viruses | 1.23E-02 | 3.61E-02 | Rac2, Nfkbie, Eif2ak2 |
| Inhibition of angiogenesis by TSP1 | 1.55E-02 | 5.56E-02 | Rac2, Sdc1 |
| Role of RIG1-like receptors in antiviral innate immunity | 1.66E-02 | 3.92E-02 | Nfkbie, Ddx58 |
| FXR/RXR activation | 1.78E-02 | 2.91E-02 | Rac2, Sdc1 |
| TR/RXR activation | 1.78E-02 | 3.03E-02 | Rac2, Ucp1 |
| PPARα/RXRα activation | 1.86E-02 | 2.22E-02 | Prkar2b, Nfkbie, Adipoq |
| RANK signalling in osteoclasts | 1.95E-02 | 3.06E-02 | Rac2, Nfkbie, Map3k2 |
| Fcγ receptor-mediated phagocytosis in macrophages and monocytes | 2.04E-02 | 2.97E-02 | Hmox1, Rac2, Hck |
Canonical pathways modified by LPS, ranked according to P-values determined by Fisher’s Exact Test. Also shown is the ratio reflecting the number of molecules in a given path that meet cut-off criteria for differential expression, divided by the total number of molecules in the path
Deletion of the A2AR reduced or entirely eliminated 134 gene responses to LPS (Table S9). Responses entirely A2AR-dependent included repression of Sacs, Cidec, Gnb1l, Mtap1b, Msi2, Cdh2, Lpgat1, Tbx3, Slc38a1, Sdc1, Reln, Ext1 and Pvrl3, and induction of Rit1, Ezh1, Eif4ebp1 and Abhd4 (Table S9). The transcripts most sensitive to A2AR KO included Dnmt3a, Qk, Sema5a, Srpk2, Car3, Ucp1, Rsad2 and Dnajb14. Highly LPS-responsive changes counteracted by A2AR KO included: Ifit3, Rsad2, Cxcl13, Ms4a6d, Oasl1, Ms4a4c, Sema5a, Tyki and Hmox1. Intrinsic A2AR activity thus also promotes LPS-dependent shifts in select genes associated with immune/inflammatory processes, including orphan nuclear receptor and PPARα activation, PRR responses to infection, AMPK signalling and NFκB signalling (Table 6).
In terms of biological functions, A2AR activity reduced LPS responsiveness of paths related to cellular movement, growth, signalling and death, immune cell trafficking and haematological and cardiovascular development (Tables 7 and S10). Conversely, A2AR activity appears to augment responses in pathways related to metabolism and transport, together with organ, haematological, cardiovascular and lymphoid development (Table 7), with complete details of pathway responses modified by A2AR KO included in Table S10. Deletion of the A2AR also influenced multiple toxicological functions; of 35 responses countered by A2AR KO, 12 were cardiac-specific categories (Table S11), with reduced Adipoq and Hmox1 responses involved in ~half. Of 46 toxicological function responses enhanced by A2AR KO, a third was cardiac categories. Enhanced responses for Serpine1, Kckn3 and Ppargc1a are involved in a majority of these and are also implicated in cardiac dysfunction.
Table 7.
Functional groupings of genes for which LPS responses were modified by ≥1.5-fold (induction or repression) by A2AR KO
| Functional grouping | P value | Number of genes |
|---|---|---|
| LPS responses enhanced by A2AR KO (countered by A2AR activity) | ||
| Molecular and cellular functions | ||
| Cellular Movement | 1.10E-08–5.10E-03 | 58 |
| Cell-To-Cell Signalling and Interaction | 1.78E-06–5.40E-03 | 46 |
| Cell Death | 1.13E-05–4.87E-03 | 74 |
| Cellular Growth and Proliferation | 1.30E-05–5.40E-03 | 58 |
| Cell Cycle | 2.22E-05–4.86E-03 | 30 |
| Physiological system development and function | ||
| Haematological System Development and Function | 3.17E-07–5.40E-03 | 53 |
| Haematopoiesis | 3.17E-07–4.79E-03 | 36 |
| Tissue Morphology | 3.17E-07–4.79E-03 | 38 |
| Immune Cell Trafficking | 3.90E-07–5.10E-03 | 37 |
| Cardiovascular System Development and Function | 1.54E-06–4.25E-03 | 24 |
| Disease and disorders | ||
| Cancer | 3.71E-08–5.03E-03 | 82 |
| Immunological Disease | 9.17E-08–3.81E-03 | 72 |
| Skeletal and Muscular Disorders | 7.90E-07–3.81E-03 | 77 |
| Inflammatory Response | 1.78E-06–5.05E-03 | 46 |
| Haematological Disease | 2.44E-06–3.68E-03 | 37 |
| LPS responses inhibited by A2AR KO (promoted by A2AR activity) | ||
| Molecular and cellular functions | ||
| Carbohydrate Metabolism | 2.24E-05–2.33E-02 | 6 |
| Molecular Transport | 2.38E-05–2.61E-02 | 24 |
| Small Molecule Biochemistry | 2.38E-05–2.87E-02 | 34 |
| Lipid Metabolism | 4.28E-05–2.87E-02 | 14 |
| Cellular Compromise | 6.16E-05–2.61E-02 | 11 |
| Physiological system development and function | ||
| Cardiovascular System Development and Function | 4.28E-05–2.97E-02 | 5 |
| Organ Development | 4.28E-05–2.33E-02 | 7 |
| Organismal Functions | 9.49E-05–2.61E-02 | 4 |
| Haematological System Development and Function | 2.54E-04–3.11E-02 | 13 |
| Lymphoid Tissue Structure and Development | 2.54E-04–2.61E-02 | 6 |
| Disease and disorders | ||
| Inflammatory Response | 6.16E-05–3.11E-02 | 15 |
| Nutritional Disease | 8.22E-05–6.58E-03 | 10 |
| Genetic Disorder | 1.34E-04–3.00E-02 | 30 |
| Developmental Disorder | 2.54E-04–2.61E-02 | 14 |
| Neurological Disease | 2.54E-04–2.61E-02 | 30 |
Functional groupings of LPS-responsive cardiac transcripts modified by A2AR KO (enhanced or repressed) by a factor of ≥1.5-fold. Functional groups derived from IPA analysis are categorized into molecular and cellular functions, physiological system development and function, and disease and disorders, and ranked according to P value ranges determined by a Fisher’s Exact Test. Total numbers of involved genes are also shown
Importantly, a relatively small set of A2AR responsive genes are implicated in a majority of these A2AR sensitive biological pathways and processes and can be considered A2AR-dependent ‘nodes’ or points of regulatory convergence. Of the top 50 pathway responses augmented by A2AR KO, Pi3kr1 is implicated in 42; Rras2 in 40; Stat1, Vegfc, Pgf and Pak6 each in 12; Atf4 in 11; Cdkn1a in 9 and Rhoj and IL6 each in 8 (Tables 6 and S10). Of the leading 50 canonical responses inhibited by A2AR KO, Rac2 was implicated in 34, Nfkbie in 21, Prkar2b in 10, Hmox1 and Eif2ak2 each in 6 and Eif4ebp1 and Cyp2b6 each in 5 (Tables 6 and S10). These data point to an influence of intrinsic A2AR activity on key signalling responses, including G protein, PKA, STAT1 and NFκB-dependent processes (see Supplementary material for additional discussion). While the absence of A2ARs impacted inflammatory and underlying signalling changes in endotoxemia, paths implicated in endotoxemic cardio-depression (ß-adrenergic and PKA signalling, Ca2+ handling, excitation-contraction coupling and mitochondrial function; Figs. S18–S21) were not among those sensitive to A2AR KO (Table 6), consistent with a lack of effect of A2AR KO on contractile dysfunction itself (Table 1).
PCR analysis of select transcripts
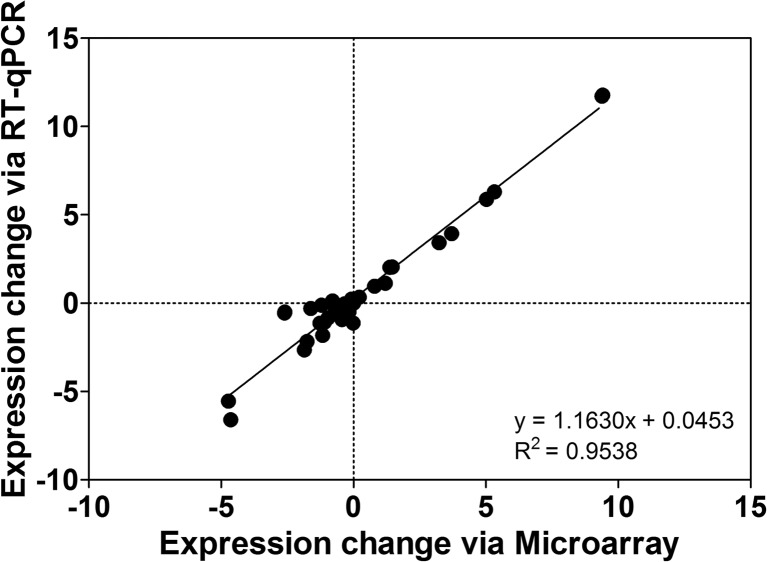
Quantitative RT-PCR analysis supports microarray-determined changes in transcripts selected for differential responsiveness to LPS and A2AR KO (Fig. 7). While expression ratios vary slightly between the two methods, a significant linear relationship was apparent between both measures (with a slope > 1 suggesting greater dynamic range for RT-qPCR measurement).
Fig. 7.
Comparison of expression values for select transcripts assessed via array analysis and quantitative real-time PCR. Expression ratios are relative to values in wild-type (WT) hearts treated with normal saline (NS) and are thus shown for WTs treated with LPS for 24 h (WT LPS) and A2AR KO hearts untreated (KO NS) or treated with LPS (KO LPS). Data are means ± S.E.M, n = 6–8/group
Discussion
The present study characterizes impacts of A2AR deletion on the myocardial transcriptome, markers of inflammation and cardiac function and injury in healthy and endotoxemic mice. Data indicate A2AR activity exerts limited effects in un-stressed myocardium, with transcriptomic changes limited to G protein, cAMP/PKA and cGMP/NO signalling downstream of this and other GPCRs (Tables 2 and 3). However, during endotoxemia, absence of A2ARs exaggerated myocardial injury (and age- and sex-dependent mortality), without substantially modifying patterns of cytokine release, myocardial cytokine/chemokine transcription or contractile depression. The latter is consistent with insensitivity of the ‘cardio-depressant’ profile in endotoxemic hearts to A2AR KO. Rather, data reveal A2AR activity selectively influences transcription of regulators of NFκB and JAK-STAT signalling during endotoxemia, which may limit myocardial inflammation and injury. Additional changes with A2AR KO suggest potential influences on insulin-resistance, hypertrophy/remodelling and vascular control/angiogenesis in endotoxemia.
A2AR activity and the transcriptome in un-stressed myocardium
Modest impacts of A2AR KO in un-stressed hearts (Table 2) are consistent with a largely retaliatory or stress-responsive role for myocardial A2ARs. Indeed, deletion failed to modify cardiac or vascular function, and circulating CRP, haptoglobin and cytokines in healthy animals. Functional annotation of transcripts supports A2AR-dependent shifts in relaxin, adrenergic, Ca2+, PKA, SAPK/JNK and hypertrophic pathways (Tables 3 and S2), involved in cellular growth/movement/death, immune and cell-to-cell signalling, and toxicological processes of fibrosis, cell damage and inflammation. This profile stems from a handful of changes spanning pathways (i.e. Gnb1, Nfat2c, Acta1, Prkar2b, Map3k2 and Pde3b), supporting effects of A2AR activity on G protein and cAMP/PKA signalling downstream of the receptor [5, 8, 60]. Deletion of the A2AR has been shown to reduce cAMP and PKA activation in other cell types [61], consistent with impacts of KO here (Table 3). The altered MEKK2 path is also linked to G protein/Rac-dependent signalling distal to this and other GPCRs. In terms of vasoregulatory functions of the A2AR, shifts in inter-related pathways involved in relaxin signalling, cellular effects of sildenafil and NO signalling (Table 3) support modulation of cGMP/NOS dependent control, while cardiac arteriopathy was identified as a pathologic process sensitive to A2AR KO (Table 4). These rather limited transcriptomic changes in healthy myocardium are consistent with observations in other tissues. For example, Yu et al. [62] found A2AR KO alters a very small sub-set of transcripts in healthy striatum (implicating A2AR sensitive EGR-2 control), with expression changes also modest (a majority ≤twofold). Others report no impact of A2AR KO on myocardial expression of RAC1, ERK1/2, p38-MAPK or JNK [14], though phospho-activation of the latter kinases was impaired, potentially reflecting shifts in cAMP/PKA and MEKK2 signalling.
Transcriptomic profile of endotoxemic myocardium
Myocardial injury and dysfunction are critical determinants of circulatory changes and mortality with uncontrolled inflammation; however, their mechanistic basis is poorly defined. Transcriptomic interrogation can reveal elements of these complex responses, though there are few analyses of myocardial [41, 42, 49] or cardiomyocyte [43, 49] responses to endotoxin/sepsis. Approximately 15 % of the 25,646 transcripts expressed in murine hearts were modified in endotoxemia (Table S4), encompassing a multiplicity of canonical paths and functions (Tables 5, S5–S7). Many are consistent with those highlighted by Wong et al. in more limited analysis of endotoxemic rats [41] and Rudiger et al. in a rat faecal peritonitis model [49]. The myocardial response involves profound upregulation of immune/inflammatory paths (Figs. 3–5 and S4–S7), the most highly modified including acute-phase response, PRR/TLR and interferon/IRF signalling (Table 5). Underlying NFκB, JAK-STAT and MAPK/PI3K paths are up-regulated (Tables 5, S5 and S7; Figs. S8–S12), with NFκB/JAK-STAT mechanistically linking the top five canonical pathway responses. Integrated signalling via NFκB and JAK-STAT paths thus appears central to myocardial endotoxemia, as in other tissues. In other tissues, these paths are also targeted by A2ARs to suppress inflammatory/immune responses [44, 45, 58], with KO augmenting NFκB activation in macrophages, for example [6].
Amplified cardiac TLR and interferon signalling
Since uncontrolled inflammation induces cellular injury/death, pro-inflammatory TLR/CD14 signalling is the subject to negative control. However, this path was transcriptionally amplified in endotoxemic myocardium (Fig. 5), including receptor molecules (Cd14, Tlr1, Tlr2, Tlr3 and Tlr4) and LPS-binding protein (Lbp), transduction molecules (Cr3/Mac1 and Cr4), MyD88, TIRAP and MyD88 targets (Il6 and Il1b), MyD88-independent signal components (Tbk1, Cxcl10, Ccl2 and Ccl5) and downstream JAK-STAT signalling (Fig. S9). Upregulated TLR and MyD88-dependent and independent signalling are consistent with changes in rat sepsis [49], though Tlr4, Cd14 and Tirap induction here was not apparent in the rat model. Sweeney et al. [63] recently reported a novel MyD88-independent path linking TLR2/TLR4 signalling to Ppargc1a, encoding the mitochondrial biogenesis co-regulator PGC-1α. However, despite induction of elements of this path here (including Tlr2, Tlr3, Tlr4 and Irf7), cardiac Ppargc1a was repressed by LPS, a response potentially exaggerating mitochondrial dysfunction and countered by A2AR activity.
Interferon signalling transducing inflammatory/TLR responses was also upregulated (Fig. 5), including a majority of gene targets and the path of IRF activation by PRRs (Figs. S5–S7). This entailed induction of membrane (Tlr2, Tlr3 and Tlr4), cytosolic (Ddx58, Ifih1, Eif2ak2 and Oas1) and extracellular (C1q, C3, C3a and Ptx3) receptors, and downstream mediators (Il6, Irf7 and Rantes). These shifts in TLR/CD14, MyD88-dependent and -independent and interferon/IRF signalling are relevant to temporal expansion of molecular changes [41, 42] and injury progression in endotoxemia: CD14 [64] and TLR4 [65, 66] both mediate cardiac dysfunction and injury and MyD88 signalling promotes cardiac hypertrophy [67], inflammation and injury [68, 69]. These profound responses diverge from in vitro data suggesting dampened/transient impacts of LPS on isolated myocyte NFκB and IκB kinase [43, 63].
Endotoxemic cardio-depression
Cardiomyocyte and myocardial function is LPS-sensitive and depressed in endotoxemia [21, 23, 70, 71] (Table 1). This has been linked to abnormalities in myofibrillar Ca2+ sensitivity [70, 71], adrenergic control [71–74] and mitochondrial function [75], together with cell death [23, 76, 77]. Transient changes in preload may also mediate early reversible depression in vivo [78]. Transcriptomic data reveal a cardio-depressant profile entailing suppression of key determinants of contraction, including Ca2+, β-adrenergic and PKA signalling, mitochondrial function and electromechanical coupling (Figs. S18–S21). Importantly, and despite other impacts, A2AR expression did not influence this cardio-depressant profile nor modify contractile depression (Table 1) [23].
Suppression of β-adrenergic and Ca2+ signalling is consistent with desensitization in hearts and myocytes [71–73], and transcriptional changes in a rat model of faecal peritonitis [49]. Despite overlap with the latter response, the β1- rather than β2-adrenoceptor is depressed in endotoxemic mouse heart (Fig. S18), while reductions in AKAP, PP1, PP2A, NCX and L-type Ca2+ channel components observed here were not evident in the peritonitis model. Nonetheless, data collectively implicate β-adrenergic dysfunction as a common component of cardio-depression, consistent with reductions observed in β-adrenoceptor expression [73], tissue noradrenaline and adrenaline [73] and adrenoceptor stimulation of cAMP and Ca2+ fluxes [73]. In terms of approaches to inotropic support, coincident depression of adrenoceptor, PKA and Ca2+ signalling may limit the value of interventions targeting these effectors. Abnormalities within the contractile apparatus itself (repressed α-tropomyosin, troponin-T, α-actin and titin transcription) may also be relevant. Targeting mitochondrial dysfunction may more broadly improve cardiac outcomes: repression of Complex I components (Fig. S21) is consistent with Complex I specific dysfunction and ROS generation in mitochondria from endotoxemic hearts [75, 79].
Additional cardio-depressant changes include altered TLR4 and fibroblast factor signalling, suppression of substrate metabolism and IGF-1 signalling (Fig. S22) and induction of inhibitory S100a8/S100a9, Icam1, Vcam1, Cybb (NOX2), Ptgs2 (COX2) and the TNF-α receptors Tnfr1 and Fas. Marked induction of S100 A8 and S100 A9 (TLR4 activators promoting endotoxemic injury) has been observed in LPS-treated myocytes, involving MyD88 and NFκB signalling [80]. Intriguingly, two adipokines with opposing actions, relevant to cardiac dysfunction, were among the most responsive to LPS. Pro-inflammatory and injurious lipocalin-2 (Lcn2) was the most induced (~600-fold), consistent with changes in rodent and human myocarditis [59], while anti-inflammatory and cardioprotective adiponectin (Adipoq) was the second most repressed. Lipocalin-2 is induced via IκBζ (+13 with LPS) and with heart failure, ischemia and inflammation [81, 82]. Profound induction may promote dysfunction given its inflammatory [83], pro-death [82, 84] and mitochondrial actions [84]. Current data suggest the A2AR may beneficially limit LPS-dependent IκBζ induction to suppress injurious lipocalin-2. Highly repressed adiponectin exerts anti-inflammatory, anti-oxidant and cardioprotective effects [85, 86], thus downregulation is also likely to promote inflammation and sensitize myocardium to oxidative-stress and cell death. Curiously, Adipoq repression was reduced with A2AR KO, suggesting a positive influence of A2AR activity on this response. These and other changes relevant to pathogenesis of cardiac depression are discussed further in the online Supplementary material.
A2AR modulation of the endotoxemic transcriptome
Despite well-established anti-inflammatory effects of acute A2AR agonism, the myocardial transcriptomic response to endotoxemia was largely insensitive to A2AR KO (Figs. 3 and 4; Tables 7 and S9). Rather than a broadly suppressive impact of A2AR activity, only 10 % of the transcriptomic response was modified by KO. This selective outcome is consistent with data for cytokine/chemokine transcription and release—the majority of these responses were also unaltered by A2AR deletion, which specifically modified Il6, Cxcl2, Cxcl5 and Csf3 induction (Fig. 4) and circulating IL-5, haptoglobin and CRP levels (Figs. 1 and 2). These responses are nonetheless relevant to myocardial outcome: IL-6 initiates and expands inflammatory signalling, and CXCL2 and CXCL5 both participate in cardiac inflammation and remodelling. Important IL-6 target genes were highly induced with LPS, including Bcl2l1, Cebpb, Irf1, JunB, Lbp, Socs3 and Timp1. By suppressing IL-6 transcription, potentially through IκBζ induction and STAT1 repression, A2AR activity may limit cardiac IL-6 signalling. Functional annotation of A2AR sensitive transcripts does support modulation of acute phase response, JAK-STAT, MAPK, PRR and NFκB pathways (Tables 7, S6 and S10). Figure 6 summarizes these key changes, highlighting impacts of A2AR KO. Through modifying a small number of key transcript responses (Nfkbie, Nfkbiz, Il6, Lcn2, Stat1, Cdkn1a and Rras2), A2AR activity may limit expansion of injurious inflammation in endotoxemic myocardium. In contrast, the cardio-depressant molecular profile appears insensitive to A2AR KO.
NFκB signalling
The A2AR represses NFκB signalling via multiple mechanisms in non-cardiac cells, including phosphorylation-/SUMOylation-dependent IκB degradation and modulation of the SCF-E3 ubiquitin ligase complex [58]. The absence of A2ARs did modify the NFκB path in endotoxemic myocardium (Fig. 6), amplifying pro-inflammatory Nfkbiz (IκBζ) induction while countering inhibitory Nfkbie (IκBε) induction. Enhanced transcription of IκBζ and its targets Lcn2 and Il6 with A2AR KO suggests the receptor may suppress inflammation by inhibiting IκBζ induction and its transcriptional actions, while promoting IκBε induction to limit nuclear translocation of NFκB (Fig. 6).
In contrast, A2AR KO also reduced induction of molecules promoting NFκB signalling (Fig. 6): Eif2ak2/Pkr inhibits IκBα expression and enhances NFκB signaling and IFN-β expression; Btrc promotes degradation of IκBα and Rac2 promotes cytokine-triggered NFκB signalling and IFN-γ expression. It is unclear what the balance of these changes might be, though repression of the distal transcriptional effector IκBζ is predicted to limit effects of up-stream changes. Failure of A2AR KO to modify changes in genes recently implicated in cardioprotective effects of NFκB signalling [47] (including induction of Ptx3, Plscr1, Sfi1 and Igfbp3 and repression of Car3, Dkk3, ai605517 and Grhl2) suggests receptor activity might selectively modify injurious rather than protective aspects of NFκB signalling.
JAK-STAT signalling
In non-cardiac cells, A2AR agonism also inhibits JAK-STAT signalling, via control of SOCS transcription [58] and ubiquitination/degradation of JAK-phosphorylated STATs [87]. Despite no evidence for A2AR modulation of Socs3 (induced with LPS unaltered by KO), A2AR deletion did modify the JAK-STAT response, exaggerating induction of both Stat1 and the key activator Il6. Since STAT1 is crucial to LPS-induced apoptosis in non-cardiac cells [88], and mediates injury with ischemic insult [39], A2AR-dependent suppression is predicted to be protective. Deletion of A2ARs also enhanced endotoxemic induction of Cdkn1a/p21Cip1/Waf1 (modulator of STAT1-dependent apoptosis and IFN-γ/STAT signalling, linked to poor sepsis outcomes [49]) and the signal transducer Rras2. These data collectively support beneficial A2AR modulation of JAK-STAT activation in endotoxemic myocardium (Fig. 6).
Select impacts on acute phase response vs. TLR and interferon signalling
The acute phase response is triggered by IL-1, IL-6 and TNF-α and transduced via NFκΒ and JAK-STAT signals. Additional to effects of KO on Il6 and JAK-STAT and NFκB paths, A2AR activity appears to counter induction of key gene targets (Serpine1, Il6, Cp, Hpx, Hp and Orm1). Conversely, A2AR activity was also associated with greater induction of target genes C3 and Hmox and reduced induction of the glucocorticoid receptor inhibiting this inflammatory response. While mixed, collective shifts in JAK-STAT/NFκB paths support transcriptional suppression of the acute phase response via A2AR activity. However, while TLR/interferon paths were among the most highly responsive to LPS, A2AR KO did not substantially modify these responses (augmenting induction of MyD88-responsive Irf8 and altering downstream NFκB signalling). Major changes in interferon signalling were also largely un-modified by A2AR KO (Fig. 6), suggesting select impacts of A2AR activity on elements of the acute phase response yet not associated TLR and interferon signalling.
Novel effects of A2AR KO
The impact of A2AR activity is limited primarily to canonical elements of endotoxemia (Tables 6, S10 and S11); however, several of the more A2AR-sensitive transcriptional responses to LPS hint at novel effects of A2AR signalling, including modulation of insulin-sensitivity, hypertrophic growth/remodelling and cardiac rhythmicity, together with influencing angiogenesis and vascular control.
Disruption of insulin-dependent glucose metabolism is an important consequence of endotoxemia, and three of the most A2AR-sensitive transcripts (Ptprf, Glut4/Slc2a4 and Glut12/Slc2a12) govern insulin-dependent glucose uptake (Table S9). Endotoxemic suppression of Glut4 and the secondary insulin-sensitive transporter Glut12 and the induction of Ptprf (encoding protein tyrosine phosphatase, receptor type F; implicated in insulin-resistance) were exaggerated >twofold with A2AR KO (Table S9). Endotoxemic suppression of Glut4 and glucose uptake is reported in skeletal and cardiac muscle [89]. Conversely, increased expression of Glut4 improves glucose uptake and limits myocardial dysfunction in endotoxemia [89]. These responses to A2AR KO suggest regulatory influences of A2AR activity on glucose handling in endotoxemic tissue.
Other highly A2AR-sensitive transcriptional responses suggest A2AR modulation of myocardial remodelling, including shifts in Asb14 and Asb15, Ca3 and Ca4 and Dnmt3a and Srpk2. Endotoxemic repression of transcripts for 2 ankyrin repeat and SOCS box (ASB) proteins, Asb14 and Asb15, was substantially exaggerated with A2AR KO. The ASB family bind and target proteins for degradation and regulate skeletal muscle development, while roles in heart are unclear. The ASB15 protein enhances skeletal muscle protein synthesis [90] and delays differentiation, potentially via modulating MAPK and PI3K/Akt signalling [91]. This control may be important in adaptive response to muscle load/activity. Carbonic anhydrases are also implicated in myocardial hypertrophy, with endotoxemic Car4 induction (the dominant cardiac isoform) exaggerated and Car3 downregulation was inhibited in A2AR KO hearts. The myocardial roles of these enzymes are only beginning to be unravelled, though they may promote hypertrophic responses by facilitating Na+-H+ and Cl-HCO3 exchanger over-activities, while evidence from other cells suggests CAR3 may modify oxidative stress and apoptosis. Similarly, Srpk2 induction was augmented in the absence of A2ARs, encoding a serine/arginine (SR) protein kinase (SRPK) that phosphorylates SR domain-containing proteins within interchromatin granule clusters/nuclear speckles to regulate pre-mRNA splicing. The SRPK2 protein also plays an important role in cell proliferation and apoptosis. Additionally, downregulation of the methyltransferase transcript Dnmt3a, governing growth and function of embryonic myocytes [92], appears to be countered by A2AR activity, which may also limit cardiac fibrosis/remodelling [93]. Collectively, this suite of A2AR responsive changes may limit myocardial hypertrophy and remodelling responses to inflammatory insult. This is consistent with functional groupings sensitive to A2AR KO, including determinants of cell cycle, growth, proliferation and death and cardiovascular development (Table 7) and shifts in canonical hypertrophy pathways (Tables S10 and S11).
Curiously, A2AR activity also limited LPS induction of Hcn1 (Table S9). The HCN proteins mediate the pacemaker (funny) current governing cardiac automaticity/excitability. While HCN4 is the most highly expressed, HCN1 is significantly expressed within the conduction network, contributes to cardiac rhythmicity [94] and is up-regulated in hypertrophy and heart failure. Endotoxemic induction may thus promote arrhythmicity, while A2AR activity appears to suppress this change.
Finally, A2AR KO modified a broader range of vascular control and growth pathways in endotoxemic (vs. healthy) hearts. Vascular dysfunction is a critical to organ damage and mortality and may involve NOS overactivity in some vascular beds. Deletion of the A2AR modified NO signalling together with renin-angiotensin signalling (Table S10), while cardiac arteriopathy was identified as a pathological process sensitive to KO (Table S11). These responses support influences of A2AR signalling on vascular pathology. We have previously shown endotoxemia impairs coronary hyperaemia, a dysfunction mimicked by KO of (and potentially involving) the A2AR [23]. Additionally, there is an apparent coronary ‘over-supply’ relative to myocardial demand in endotoxemic hearts (Table 1), owing to 25–30 % lower contractile function without reductions in coronary perfusion. The absence of coronary coupling to ventricular activity, described by us in this and other models [95, 96], suggests additional coronary dysregulation (though this effect is neither replicated nor modified by A2AR KO). Multiple paths/mediators regulating angiogenesis were also sensitive to A2AR KO in endotoxemic heart, including shifts in VEGF, HIF-1α, angiopoietin, TGF-β, GM-CSF, PDGF and IGF-1 pathways (Table S10) and key regulatory molecules (including exaggerated induction of Il6, Pgf, Tgfb2, Tgfb2r, Angptl4 and Amotl1; exaggerated suppression of Vegfc and reduced suppression of Qk) (Table S9). While the A2B receptor is more broadly implicated in control of angiogenesis, these changes suggest A2AR-dependent processes may influence angiogenesis in the context of uncontrolled inflammation.
RT-qPCR confirmation of microarray-detected responses
Data confirm agreement between transcript expression determined via RT-qPCR and microarray methods (Fig. 7), with PCR further highlighting distinct transcriptional effects of A2AR KO: in some cases, KO eliminates responses to LPS (Acta1); alters baseline yet not LPS-dependent expression (Amid or Dbp); enhances induction (Txnip) or repression (Cidec) in response to vehicle and LPS or reduces gene expression in the vehicle group while negating further changes with LPS (Eif4ebp2 and Slc38a1). Confirming the microarray approach, some of these responses are also functionally relevant. For example, Txnip encodes a stress-responsive protein inhibiting the anti-oxidant/signalling molecule thioredoxin. Reductions in TXNIP inhibit vascular inflammation and TNF-α signalling [97], TXNIP dysregulation promotes inflammatory disease [98] and elevations in TXNIP facilitate apoptosis [99]. Inhibition of Txnip induction by A2AR activity may thus enhance protective thioredoxin functionality.
Study limitations
Several study limitations are worth noting. While essential genesis of cardiac injury is studied and understood within the in situ organ, this entails experimental limitations inherent to in vivo studies of organ dysfunction in sepsis/endotoxemia [15, 16, 20, 21, 24, 41, 42, 49]. Organ injury in these settings not only involves intrinsic organ-specific mechanisms but also influenced by extrinsic factors, including shifts in systemic inflammation, haemodynamics and neuroendocrine influences (with relative impacts of these factors difficult to delineate). Although we assess the intrinsic myocardial dysfunction in ex vivo tissue (together with in vivo markers of cardiac damage), we cannot isolate potential influences of A2AR KO on cardiac loading, neurohumoral factors or indeed systemic inflammation. That said, while A2AR KO augmented some inflammatory markers, systemic and cardiac cytokine responses were largely insensitive to KO and the myocardial response was very selectively modified, indicating distinct effects of A2AR activity on cardiac injury processes. We are also unable to detail cell-specific origins of myocardial transcriptional changes. Migrating inflammatory cells could influence the transcriptomic profile for intact myocardium, though, as detailed in the Supplementary material, the expression profile of endotoxemic myocardial tissue is inconsistent with major contamination from such cells. An added limitation relates to the intriguing observation of both age- and sex-dependent mortality effects of LPS and A2ARs (Fig. S3). This unfortunately precluded any detailed interrogation of these outcomes due to poor survival in older KO mice. Future work might focus specifically on responses to LPS/sepsis in clinically relevant aged cohorts, correlating age-dependent transcriptomic and phenotypic outcomes. Finally, while profound transcriptional changes observed in the current profiling study are predicted to translate to functionally relevant protein changes, this exploratory analysis does not directly confirm altered protein expression.
Conclusions
Deletion of the A2AR induces modest effects in healthy myocardium, shifting transcription of G protein coupled cAMP/PKA, MEKK2 and cGMP/NO signalling downstream of this and other GPCRs, without impacting cardiac function. However, the receptor may play a retaliatory role in selectively suppressing inflammatory signalling and injury, yet, not contractile depression, in endotoxemic myocardium. Cardiac endotoxemia itself is characterized by profound upregulation of acute-phase response, PRR/TLR and interferon signalling and cell death and remodelling pathways, together with broad suppression of primary determinants of contractility (Ca2+, ß-adrenergic and PKA signalling; mitochondrial function; electromechanical coupling) and induction of cardio-depressant genes (Lcn2, S100a8/S100a9, Icam1, Vcam1, Nox2, Cox2, Tnfr1 and Fas). This complex transcriptome is selectively A2AR sensitive, with effects of KO implicating modulation of key regulatory molecules (Nfkbie, Nfkbiz, Il6, Lcn2, Stat1, Cdkn1a and Rras2) to suppress myocardial JAK-STAT/NFκB and pro-inflammatory IL-6 and TLR signalling, together with influencing determinants of insulin-sensitivity, hypertrophy/remodelling and vascular function/angiogenesis. In contrast, endotoxemic suppression of β-adrenergic, PKA, Ca2+ signalling, electromechanical and mitochondrial function pathways appears insensitive to A2AR KO, as does contractile dysfunction itself.
Electronic supplementary material
(PDF 5.86 MB)
(PDF 1.52 MB)
Acknowledgments
This work was supported by the National Heart, Lung and Blood Institute grants HL-74001 (RRM), HL-027339 (SJM) and HL-48839 (PAH), an American Heart Association Southeast Affiliate Grant-in-Aid (PAH), a grant from the National Health and Medical Research Council of Australia (JPH) and the American Lebanese Syrian Associated Charities (RRM). We gratefully acknowledge the technical assistance of Kiel J. Headrick and Mary A. Cerniway.
Authors’ contributions
Conceived and designed the experiments: MER, KJA, JPH, RRM, SJM. Performed the experiments: MER, KJA, RRM, BT. Analysed data: KJA, MER, JPH, RRM, BT, LD. Generated and characterized KO mice: CL, PAH. Contributed reagents/materials/analytical tools: RRM, CL, PAH, KJA, JPH. Wrote the paper: KJA, MER, JPH, RRM, LMD, SJM.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Kevin J. Ashton and Melissa E. Reichelt denotes equal first authorship
References
- 1.Merx MW, Weber C. Sepsis and the heart. Circulation. 2007;116:793–802. doi: 10.1161/CIRCULATIONAHA.106.678359. [DOI] [PubMed] [Google Scholar]
- 2.Balija TM, Lowry SF. Lipopolysaccharide and sepsis-associated myocardial dysfunction. Curr Opin Infect Dis. 2011;24:248–253. doi: 10.1097/QCO.0b013e32834536ce. [DOI] [PubMed] [Google Scholar]
- 3.Haskó G, Linden J, Cronstein B, Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sitkovsky MV, Lukashev D, Apasov S, Kojima H, Koshiba M, Caldwell C, et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu Rev Immunol. 2004;22:657–682. doi: 10.1146/annurev.immunol.22.012703.104731. [DOI] [PubMed] [Google Scholar]
- 5.Sitkovsky MV, Ohta A. The ‘danger’ sensors that STOP the immune response: the A2 adenosine receptors? Trends Immunol. 2005;26:299–304. doi: 10.1016/j.it.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Lukashev D, Ohta A, Apasov S, Chen JF, Sitkovsky M. Cutting edge: physiologic attenuation of proinflammatory transcription by the Gs protein-coupled A2A adenosine receptor in vivo. J Immunol. 2004;173:21–24. doi: 10.4049/jimmunol.173.1.21. [DOI] [PubMed] [Google Scholar]
- 7.Ohta A, Sitkovsky M. The adenosinergic immunomodulatory drugs. Curr Opin Pharmacol. 2009;9:501–506. doi: 10.1016/j.coph.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Headrick JP, Peart JN, Reichelt ME, Haseler LJ. Adenosine and its receptors in the heart: regulation, retaliation and adaptation. Biochim Biophys Acta. 2011;1808:1413–1428. doi: 10.1016/j.bbamem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Day YJ, Toufektsian MC, Xu Y, Ramos SI, Marshall MA, et al. Myocardial infarct-sparing effect of adenosine A2A receptor activation is due to its action on CD4+ T lymphocytes. Circulation. 2006;114:2056–2064. doi: 10.1161/CIRCULATIONAHA.106.649244. [DOI] [PubMed] [Google Scholar]
- 10.Rork TH, Wallace KL, Kennedy DP, Marshall MA, Lankford AR, Linden J. Adenosine A2A receptor activation reduces infarct size in the isolated, perfused mouse heart by inhibiting resident cardiac mast cell degranulation. Am J Physiol Heart Circ Physiol. 2008;295:H1825–H1833. doi: 10.1152/ajpheart.495.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glover DK, Riou LM, Ruiz M, Sullivan GW, Linden J, Rieger JM, et al. Reduction of infarct size and postischemic inflammation from ATL-146e, a highly selective adenosine A2A receptor agonist, in reperfused canine myocardium. Am J Physiol Heart Circ Physiol. 2005;288:H1851–H1858. doi: 10.1152/ajpheart.00362.2004. [DOI] [PubMed] [Google Scholar]
- 12.Boucher M, Pesant S, Falcao S, de Montigny C, Schampaert E, Cardinal R, Rousseau G. Post-ischemic cardioprotection by A2A adenosine receptors: dependent of phosphatidylinositol 3-kinase pathway. Cardiovasc Pharmacol. 2004;43:416–422. doi: 10.1097/00005344-200403000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Ribé D, Sawbridge D, Thakur S, Hussey M, Ledent C, Kitchen I, et al. Adenosine A2A receptor signaling regulation of cardiac NADPH oxidase activity. Free Radic Biol Med. 2008;44:1433–1442. doi: 10.1016/j.freeradbiomed.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi J, McIntosh R, Shen X, Lee S, Chanoit G, Criswell H, et al. Adenosine A2A and A2B receptors work in concert to induce a strong protection against reperfusion injury in rat hearts. J Mol Cell Cardiol. 2009;47:684–690. doi: 10.1016/j.yjmcc.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, JL H, Li J, Zhao L, Zhang Y, Zeng YJ, et al. A feedback loop in PPARγ-adenosine A2A receptor signaling inhibits inflammation and attenuates lung damages in a mouse model of LPS-induced acute lung injury. Cell Signal. 2013;25:1913–1923. doi: 10.1016/j.cellsig.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Gonzales JN, Gorshkov B, Varn MN, Zemskova MA, Zemskov EA, Sridhar S, et al. Protective effect of adenosine receptors against lipopolysaccharide-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2014;306:L497–L507. doi: 10.1152/ajplung.00086.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odashima M, Otaka M, Jin M, Komatsu K, Wada I, Matsuhashi T, et al. Selective A2A adenosine agonist ATL-146e attenuates acute lethal liver injury in mice. J Gastroenterol. 2005;40:526–529. doi: 10.1007/s00535-005-1609-9. [DOI] [PubMed] [Google Scholar]
- 18.Rebola N, Simões AP, Canas PM, Tomé AR, Andrade GM, Barry CE, et al. Adenosine A2A receptors control neuroinflammation and consequent hippocampal neuronal dysfunction. J Neurochem. 2011;117:100–111. doi: 10.1111/j.1471-4159.2011.07178.x. [DOI] [PubMed] [Google Scholar]
- 19.Thiel M, Kreimeier U, Holzer K, Moritz S, Peter K, Messmer K. Effects of adenosine on cardiopulmonary functions and oxygen-derived variables during endotoxemia. Crit Care Med. 1998;26:322–337. doi: 10.1097/00003246-199802000-00036. [DOI] [PubMed] [Google Scholar]
- 20.Tofovic SP, Zacharia L, Carcillo JA, Jackson EK. Inhibition of adenosine deaminase attenuates endotoxin-induced release of cytokines in vivo in rats. Shock. 2001;16:196–202. doi: 10.1097/00024382-200116030-00005. [DOI] [PubMed] [Google Scholar]
- 21.Braun-Dullaeus RC, Dietrich S, Schoaff MJ, Sedding DG, Leithaeuser B, Walker G, et al. Protective effect of 3-deazaadenosine in a rat model of lipopolysaccharide-induced myocardial dysfunction. Shock. 2003;19:245–251. doi: 10.1097/00024382-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Reutershan J, Cagnina RE, Chang D, Linden J, Ley K. Therapeutic anti-inflammatory effects of myeloid cell adenosine receptor A2a stimulation in lipopolysaccharide-induced lung injury. J Immunol. 2007;179:1254–1263. doi: 10.4049/jimmunol.179.2.1254. [DOI] [PubMed] [Google Scholar]
- 23.Reichelt ME, Ashton KJ, Tan XL, Mustafa SJ, Ledent C, Delbridge LM, et al. The adenosine A2A receptor—myocardial protectant and coronary target in endotoxemia. Int J Cardiol. 2013;166:672–680. doi: 10.1016/j.ijcard.2011.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Zhao L, He X, Zeng YJ, Dai SS. Sinomenine protects against lipopolysaccharide-induced acute lung injury in mice via adenosine A2A receptor signaling. PLoS One. 2013;8:e59257. doi: 10.1371/journal.pone.0059257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Németh ZH, Csóka B, Wilmanski J, Xu D, Lu Q, Ledent C, et al. Adenosine A2A receptor inactivation increases survival in polymicrobial sepsis. J Immunol. 2006;176:5616–5626. doi: 10.4049/jimmunol.176.9.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belikoff B, Hatfield S, Sitkovsky M, Remick DG. Adenosine negative feedback on A2A adenosine receptors mediates hyporesponsiveness in chronically septic mice. Shock. 2011;35:382–387. doi: 10.1097/SHK.0b013e3182085f12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang X, Ghani A, Malik A, Wilder T, Colegio OR, Flavell RA, et al. Adenosine is required for sustained inflammasome activation via the A2A receptor and the HIF-1α pathway. Nat Commun. 2013;4:2909. doi: 10.1038/ncomms3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassi A, Malorgio F, Tedesco S, Cignarella A, Gaion RM. Upregulation of inducible NO synthase by exogenous adenosine in vascular smooth muscle cells activated by inflammatory stimuli in experimental diabetes. Cardiovasc Diabetol. 2016;15:32. doi: 10.1186/s12933-016-0349-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun CX, Zhong H, Mohsenin A, Morschl E, Chunn JL, Molina JG, et al. Role of A2B adenosine receptor signaling in adenosine-dependent pulmonary inflammation and injury. J Clin Invest. 2006;116:2173–2182. doi: 10.1172/JCI27303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan ES, Montesinos MC, Fernandez P, Desai A, Delano DL, Yee H, et al. Adenosine a(2A) receptors play a role in the pathogenesis of hepatic cirrhosis. Br J Pharmacol. 2006;148:1144–1155. doi: 10.1038/sj.bjp.0706812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoll M, Kim YO, Bebich B, Robson SC, Schuppan D. The selective adenosine 2B receptor antagonist MRS1754 mitigates hepatic collagen deposition during fibrosis progression and induces mild fibrosis regression. Gastroenterology. 2012;142(Suppl 1):S974–S975. doi: 10.1016/S0016-5085(12)63778-0. [DOI] [Google Scholar]
- 32.Wakeno M, Minamino T, Seguchi O, Okazaki H, Tsukamoto O, Okada K, et al. Long-term stimulation of adenosine A2b receptors begun after myocardial infarction prevents cardiac remodeling in rats. Circulation. 2006;114:1923–1932. doi: 10.1161/CIRCULATIONAHA.106.630087. [DOI] [PubMed] [Google Scholar]
- 33.Liao Y, Takashima S, Asano Y, Asakura M, Ogai A, Shintani Y, et al. Activation of adenosine A1 receptor attenuates cardiac hypertrophy and prevents heart failure in murine left ventricular pressure-overload model. Circ Res. 2003;93:759–766. doi: 10.1161/01.RES.0000094744.88220.62. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs AT, Ignarro LJ. Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J Biol Chem. 2001;276:47950–47957. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- 35.Park EJ, Park SY, Joe EH, Jou I. 15d-PGJ2 and rosiglitazone suppress Janus kinase-STAT inflammatory signaling through induction of suppressor of cytokine signaling 1 (SOCS1) and SOCS3 in glia. J Biol Chem. 2003;278:14747–14752. doi: 10.1074/jbc.M210819200. [DOI] [PubMed] [Google Scholar]
- 36.Kim OS, Park EJ, Joe EH, Jou I. JAK-STAT signaling mediates gangliosides-induced inflammatory responses in brain microglial cells. J Biol Chem. 2002;277:40594–40601. doi: 10.1074/jbc.M203885200. [DOI] [PubMed] [Google Scholar]
- 37.Niu J, Wang K, Graham S, Azfer A, Kolattukudy PE. MCP-1-induced protein attenuates endotoxin-induced myocardial dysfunction by suppressing cardiac NF-кB activation via inhibition of IкB kinase activation. J Mol Cell Cardiol. 2011;51:177–186. doi: 10.1016/j.yjmcc.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Coldewey SM, Rogazzo M, Collino M, Patel NS, Thiemermann C. Inhibition of IκB kinase reduces the multiple organ dysfunction caused by sepsis in the mouse. Dis Model Mech. 2013;6:1031–1042. doi: 10.1242/dmm.012435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mascareno E, El-Shafei M, Maulik N, Sato M, Guo Y, Das DK, et al. JAK/STAT signaling is associated with cardiac dysfunction during ischemia and reperfusion. Circulation. 2001;104:325–329. doi: 10.1161/01.CIR.104.3.325. [DOI] [PubMed] [Google Scholar]
- 40.McCormick J, Suleman N, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. STAT1 deficiency in the heart protects against myocardial infarction by enhancing autophagy. J Cell Mol Med. 2012;16:386–393. doi: 10.1111/j.1582-4934.2011.01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong ML, O’Kirwan F, Khan N, Hannestad J, KH W, Elashoff D, et al. Identification, characterization, and gene expression profiling of endotoxin-induced myocarditis. Proc Natl Acad Sci U S A. 2003;100:14241–14246. doi: 10.1073/pnas.2336220100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastronardi CA, Licinio J, Wong ML. Candidate biomarkers for systemic inflammatory response syndrome and inflammation: a pathway for novel translational therapeutics. Neuroimmunomodulation. 2010;17:359–368. doi: 10.1159/000292040. [DOI] [PubMed] [Google Scholar]
- 43.Cuenca J, Goren N, Prieto P, Martín-Sanz P, Boscá L. Selective impairment of nuclear factor-kappaB-dependent gene transcription in adult cardiomyocytes: relevance for the regulation of the inflammatory response in the heart. Am J Pathol. 2007;171:820–828. doi: 10.2353/ajpath.2007.061076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincenzi F, Targa M, Corciulo C, Gessi S, Merighi S, Setti S, et al. The anti-tumor effect of A3 adenosine receptors is potentiated by pulsed electromagnetic fields in cultured neural cancer cells. PLoS One. 2012;7:e39317. doi: 10.1371/journal.pone.0039317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Zheng X, Haugen F, Darè E, Lövdahl C, Schulte G, et al. Adenosine increases LPS-induced nuclear factor kappa B activation in smooth muscle cells via an intracellular mechanism and modulates it via actions on adenosine receptors. Acta Physiol (Oxf) 2014;210:590–599. doi: 10.1111/apha.12176. [DOI] [PubMed] [Google Scholar]
- 46.Boengler K, Hilfiker-Kleiner D, Drexler H, et al. The myocardial JAK/STAT pathway: from protection to failure. Pharmacol Ther. 2008;120:172–185. doi: 10.1016/j.pharmthera.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Wilhide ME, Tranter M, Ren X, Chen J, Sartor MA, Medvedovic M, et al. Identification of a NF-κB cardioprotective gene program: NF-κB regulation of Hsp70.1 contributes to cardioprotection after permanent coronary occlusion. J Mol Cell Cardiol. 2011;51:82–89. doi: 10.1016/j.yjmcc.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergmann MW, Loser P, Dietz R, von Harsdorf R. Effect of NF-kappa B inhibition on TNF-alpha-induced apoptosis and downstream pathways in cardiomyocytes. J Mol Cell Cardiol. 2001;33:1223–1232. doi: 10.1006/jmcc.2001.1385. [DOI] [PubMed] [Google Scholar]
- 49.Rudiger A, Dyson A, Felsmann K, Carré JE, Taylor V, Hughes S, et al. Early functional and transcriptomic changes in the myocardium predict outcome in a long-term rat model of sepsis. Clin Sci (Lond) 2013;124:391–401. doi: 10.1042/CS20120334. [DOI] [PubMed] [Google Scholar]
- 50.Ledent C, Vaugeois JM, Schiffmann SN, Pedrazzini T, El Yacoubi M, Vanderhaeghen JJ, et al. Aggressiveness, hypoalgesia and high blood pressure in mice lacking the adenosine A2a receptor. Nature. 1997;388:674–678. doi: 10.1038/41771. [DOI] [PubMed] [Google Scholar]
- 51.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 53.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Budiono BP, See Hoe LE, Peart JN, Sabapathy S, Ashton KJ, et al. Voluntary running in mice beneficially modulates myocardial ischemic tolerance, signaling kinases and gene expression patterns. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1091–R1100. doi: 10.1152/ajpregu.00406.2011. [DOI] [PubMed] [Google Scholar]
- 55.Everaert BR, Boulet GA, Timmermans JP, Vrints CJ. Importance of suitable reference gene selection for quantitative real-time PCR: special reference to mouse myocardial infarction studies. PLoS One. 2011;6:e23793. doi: 10.1371/journal.pone.0023793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tateda K, Matsumoto T, Miyazaki S, Yamaguchi K. Lipopolysaccharide-induced lethality and cytokine production in aged mice. Infect Immun. 1996;64:769–774. doi: 10.1128/iai.64.3.769-774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Chiazza F, Collino M, Patel NS, Coldewey SM, Thiemermann C. Gender dimorphism of the cardiac dysfunction in murine sepsis: signalling mechanisms and age-dependency. PLoS One. 2014;9:e100631. doi: 10.1371/journal.pone.0100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milne GR, Palmer TM. Anti-inflammatory and immunosuppressive effects of the A2A adenosine receptor. Sci World J. 2011;11:320–339. doi: 10.1100/tsw.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding L, Hanawa H, Ota Y, Hasegawa G, Hao K, Asami F, et al. Lipocalin-2/neutrophil gelatinase-B associated lipocalin is strongly induced in hearts of rats with autoimmune myocarditis and in human myocarditis. Circ J. 2010;74:523–530. doi: 10.1253/circj.CJ-09-0485. [DOI] [PubMed] [Google Scholar]
- 60.Blackburn MR, Vance CO, Morschl E, Wilson CN. Adenosine receptors and inflammation. Handb Exp Pharmacol. 2009;193:215–269. doi: 10.1007/978-3-540-89615-9_8. [DOI] [PubMed] [Google Scholar]
- 61.Nadeem A, Ponnoth DS, Ansari HR, Batchelor TP, Dey RD, Ledent C, Mustafa SJ. A2 A adenosine receptor deficiency leads to impaired tracheal relaxation via NADPH oxidase pathway in allergic mice. J Pharmacol Exp Ther. 2009;330:99–108. doi: 10.1124/jpet.109.151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu L, Haverty PM, Mariani J, Wang Y, Shen HY, Schwarzschild MA, et al. Genetic and pharmacological inactivation of adenosine A2 A receptor reveals an Egr-2-mediated transcriptional regulatory network in the mouse striatum. Physiol Genomics. 2005;23:89–102. doi: 10.1152/physiolgenomics.00068.2005. [DOI] [PubMed] [Google Scholar]
- 63.Sweeney TE, Suliman HB, Hollingsworth JW, Welty-Wolf KE, Piantadosi CA. A toll-like receptor 2 pathway regulates the Ppargc1a/b metabolic co-activators in mice with staphylococcal aureus sepsis. PLoS One. 2011;6:e25249. doi: 10.1371/journal.pone.0025249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barber RC, Maass DL, White DJ, Chang LY, Horton JW. Molecular or pharmacologic inhibition of the CD14 signaling pathway protects against burn-related myocardial inflammation and dysfunction. Shock. 2008;30:705–713. doi: 10.1097/SHK.0b013e31816f6caa. [DOI] [PubMed] [Google Scholar]
- 65.Oyama J, Blais C, Jr, Liu X, Pu M, Kobzik L, Kelly RA, et al. Reduced myocardial ischemia-reperfusion injury in toll-like receptor 4-deficient mice. Circulation. 2004;109:784–789. doi: 10.1161/01.CIR.0000112575.66565.84. [DOI] [PubMed] [Google Scholar]
- 66.Avlas O, Fallach R, Shainberg A, Porat E, Hochhauser E. Toll-like receptor 4 stimulation initiates an inflammatory response that decreases cardiomyocyte contractility. Antioxid Redox Signal. 2011;15:1895–1909. doi: 10.1089/ars.2010.3728. [DOI] [PubMed] [Google Scholar]
- 67.Ha T, Hua F, Li Y, Ma J, Gao X, Kelley J, Zhao A, et al. Blockade of MyD88 attenuates cardiac hypertrophy and decreases cardiac myocyte apoptosis in pressure overload-induced cardiac hypertrophy in vivo. Am J Physiol Heart Circ Physiol. 2006;290:H985–H994. doi: 10.1152/ajpheart.00720.2005. [DOI] [PubMed] [Google Scholar]
- 68.Hua F, Ha T, Ma J, Gao X, Kelley J, Williams DL, et al. Blocking the MyD88-dependent pathway protects the myocardium from ischemia/reperfusion injury in rat hearts. Biochem Biophys Res Commun. 2005;338:1118–1125. doi: 10.1016/j.bbrc.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 69.Feng Y, Zhao H, Xu X, Buys ES, Raher MJ, Bopassa JC, et al. Innate immune adaptor MyD88 mediates neutrophil recruitment and myocardial injury after ischemia-reperfusion in mice. Am J Physiol Heart Circ Physiol. 2008;295:H1311–H1318. doi: 10.1152/ajpheart.00119.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rubin LJ, Keller RS, Parker JL, Adams HR. Contractile dysfunction of ventricular myocytes isolated from endotoxemic Guinea pigs. Shock. 1994;2:113–120. doi: 10.1097/00024382-199408000-00006. [DOI] [PubMed] [Google Scholar]
- 71.Abi-Gerges N, Tavernier B, Mebazaa A, Faivre V, Paqueron X, Payen D, et al. Sequential changes in autonomic regulation of cardiac myocytes after in vivo endotoxin injection in rat. Am J Respir Crit Care Med. 1999;160:1196–1204. doi: 10.1164/ajrccm.160.4.9808149. [DOI] [PubMed] [Google Scholar]
- 72.Bensard DD, Banerjee A, McIntyre RC, Jr, Berens RL, Harken AH. Endotoxin disrupts beta-adrenergic signal transduction in the heart. Arch Surg. 1994;129:198–204. doi: 10.1001/archsurg.1994.01420260094013. [DOI] [PubMed] [Google Scholar]
- 73.Kadoi Y, Saito S, Fujita N, Morita T, Fujita T. Alterations in the myocardial β-adrenergic system during experimental endotoxemia. J Anesth. 1996;10:49–54. doi: 10.1007/BF02482068. [DOI] [PubMed] [Google Scholar]
- 74.Yasuda S, Lew WY. Lipopolysaccharide depresses cardiac contractility and beta-adrenergic contractile response by decreasing myofilament response to Ca2+ in cardiac myocytes. Circ Res. 1997;81:1011–1020. doi: 10.1161/01.RES.81.6.1011. [DOI] [PubMed] [Google Scholar]
- 75.Vanasco V, Magnani ND, Cimolai MC, Valdez LB, Evelson P, Boveris A, Alvarez S. Endotoxemia impairs heart mitochondrial function by decreasing electron transfer, ATP synthesis and ATP content without affecting membrane potential. J Bioenerg Biomembr. 2012;44:243–252. doi: 10.1007/s10863-012-9426-3. [DOI] [PubMed] [Google Scholar]
- 76.McDonald TE, Grinman MN, Carthy CM, Walley KR. Endotoxin infusion in rats induces apoptotic and survival pathways in hearts. Am J Physiol Heart Circ Physiol. 2000;279:H2053–H2061. doi: 10.1152/ajpheart.2000.279.5.H2053. [DOI] [PubMed] [Google Scholar]
- 77.Cai J, Lu S, Yao Z, Deng YP, Zhang LD, JW Y, et al. Glibenclamide attenuates myocardial injury by lipopolysaccharides in streptozotocin-induced diabetic mice. Cardiovasc Diabetol. 2014;13:106. doi: 10.1186/s12933-014-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jianhui L, Rosenblatt-Velin N, Loukili N, Pacher P, Feihl F, Waeber B, Liaudet L. Endotoxin impairs cardiac hemodynamics by affecting loading conditions but not by reducing cardiac inotropism. Am J Physiol Heart Circ Physiol. 2010;299:H492–H501. doi: 10.1152/ajpheart.01135.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanasco V, Cimolai MC, Evelson P, Alvarez S. The oxidative stress and the mitochondrial dysfunction caused by endotoxemia are prevented by alpha-lipoic acid. Free Radic Res. 2008;42:815–823. doi: 10.1080/10715760802438709. [DOI] [PubMed] [Google Scholar]
- 80.Boyd JH, Kan B, Roberts H, Wang Y, Walley KR. S100 A8 and S100 A9 mediate endotoxin-induced cardiomyocyte dysfunction via the receptor for advanced glycation end products. Circ Res. 2008;102:1239–1246. doi: 10.1161/CIRCRESAHA.107.167544. [DOI] [PubMed] [Google Scholar]
- 81.Yndestad A, Landrø L, Ueland T, Dahl CP, Flo TH, Vinge LE, et al. Increased systemic and myocardial expression of neutrophil gelatinase-associated lipocalin in clinical and experimental heart failure. Eur Heart J. 2009;30:1229–1236. doi: 10.1093/eurheartj/ehp088. [DOI] [PubMed] [Google Scholar]
- 82.Xu G, Ahn J, Chang S, Eguchi M, Ogier A, Han S, et al. Lipocalin-2 induces cardiomyocyte apoptosis by increasing intracellular iron accumulation. J Biol Chem. 2012;287:4808–4817. doi: 10.1074/jbc.M111.275719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aigner F, Maier HT, Schwelberger HG, Wallnofer EA, Amberger A, Obrist P, et al. Lipocalin-2 regulates the inflammatory response during ischemia and reperfusion of the transplanted heart. Am J Transplant. 2007;7:779–788. doi: 10.1111/j.1600-6143.2006.01723.x. [DOI] [PubMed] [Google Scholar]
- 84.Yang B, Fan P, Xu A, Lam KS, Berger T, Mak TW, Tse HF, et al. Improved functional recovery to I/R injury in hearts from lipocalin-2 deficiency mice: restoration of mitochondrial function and phospholipids remodeling. Am J Transl Res. 2012;4:60–71. [PMC free article] [PubMed] [Google Scholar]
- 85.Smith CC, Yellon DM. Adipocytokines, cardiovascular pathophysiology and myocardial protection. Pharmacol Ther. 2011;129:206–219. doi: 10.1016/j.pharmthera.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 86.Nanayakkara G, Kariharan T, Wang L, Zhong J, Amin R. The cardioprotective signaling and mechanisms of adiponectin. Am J Cardiovasc Dis. 2012;2:253–266. [PMC free article] [PubMed] [Google Scholar]
- 87.Safhi MM, Rutherford C, Ledent C, Sands WA, Palmer TM. Priming of signal transducer and activator of transcription proteins for cytokine-triggered polyubiquitylation and degradation by the A2A adenosine receptor. Mol Pharmacol. 2010;77:968–978. doi: 10.1124/mol.109.062455. [DOI] [PubMed] [Google Scholar]
- 88.Kristof AS, Marks-Konczalik J, Billings E, Moss J. Stimulation of signal transducer and activator of transcription-1 (STAT1)-dependent gene transcription by lipopolysaccharide and interferon-gamma is regulated by mammalian target of rapamycin. J Biol Chem. 2003;278:33637–33644. doi: 10.1074/jbc.M301053200. [DOI] [PubMed] [Google Scholar]
- 89.Otero YF, Mulligan KX, Barnes TM, Ford EA, Malabanan CM, Zong H, et al. Enhanced glucose transport, but not phosphorylation capacity, ameliorates lipopolysaccharide-induced impairments in insulin-stimulated muscle glucose uptake. Shock. 2016;45:677–685. doi: 10.1097/SHK.0000000000000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McDaneld TG, Hannon K, Moody DE. Ankyrin repeat and SOCS box protein 15 regulates protein synthesis in skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1672–R1682. doi: 10.1152/ajpregu.00239.2005. [DOI] [PubMed] [Google Scholar]
- 91.McDaneld TG, Spurlock DM. Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 alters differentiation of mouse C2C12 myoblasts and phosphorylation of mitogen-activated protein kinase and Akt. J Anim Sci. 2008;86:2897–2902. doi: 10.2527/jas.2008-1076. [DOI] [PubMed] [Google Scholar]
- 92.Fang X, Poulsen RR, Wang-Hu J, Shi O, Calvo NS, Simmons CS, et al. (2016) Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. FASEB J. 2016 Jun 15. [DOI] [PMC free article] [PubMed]
- 93.Tao H, Yang JJ, Chen ZW, SS X, Zhou X, Zhan HY, et al. DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology. 2014;323:42–50. doi: 10.1016/j.tox.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 94.Fenske S, Krause SC, Hassan SI, Becirovic E, Auer F, Bernard R, et al. Sick sinus syndrome in HCN1-deficient mice. Circulation. 2013;128:2585–2594. doi: 10.1161/CIRCULATIONAHA.113.003712. [DOI] [PubMed] [Google Scholar]
- 95.Flood AJ, Willems L, Headrick JP. Coronary function and adenosine receptor-mediated responses in ischemic-reperfused mouse heart. Cardiovasc Res. 2002;55:161–170. doi: 10.1016/S0008-6363(02)00329-2. [DOI] [PubMed] [Google Scholar]
- 96.Reichelt ME, Willems L, Hack BA, Peart JN, Headrick JP. Cardiac and coronary function in the Langendorff-perfused mouse heart model. Exp Physiol. 2009;94:54–70. doi: 10.1113/expphysiol.2008.043554. [DOI] [PubMed] [Google Scholar]
- 97.Yamawaki H, Pan S, Lee RT, Berk BC. Fluid shear stress inhibits vascular inflammation by decreasing thioredoxin-interacting protein in endothelial cells. J Clin Invest. 2005;115:733–738. doi: 10.1172/JCI200523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takahashi Y, Masuda H, Ishii Y, Nishida Y, Kobayashi M, Asai S. Decreased expression of thioredoxin interacting protein mRNA in inflamed colonic mucosa in patients with ulcerative colitis. Oncol Rep. 2007;18:531–535. [PubMed] [Google Scholar]
- 99.Chen CL, Lin CF, Chang WT, Huang WC, Teng CF, Lin YS. Ceramide induces p38 MAPK and JNK activation through a mechanism involving a thioredoxin-interacting protein-mediated pathway. Blood. 2008;111:43. doi: 10.1182/blood-2007-08-106336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 5.86 MB)
(PDF 1.52 MB)