Abstract
p300 and CREB-binding protein (CBP), two homologous lysine acetyltransferases in metazoans, have a myriad of cellular functions. They exert their influence mainly through their roles as transcriptional regulators but also via nontranscriptional effects inside and outside of the nucleus on processes such as DNA replication and metabolism. The versatility of p300/CBP as molecular tools has led to their exploitation by viral oncogenes for cellular transformation and by cancer cells to achieve and maintain an oncogenic phenotype. How cancer cells use p300/CBP in their favor varies depending on the cellular context and is evident by the growing list of loss- and gain-of-function genetic alterations in p300 and CBP in solid tumors and hematological malignancies. Here, we discuss the biological functions of p300/CBP and how disruption of these functions by mutations and alterations in expression or subcellular localization contributes to the cancer phenotype.
The lysine acetyltransferases p300 and CBP participate in diverse cellular functions (e.g., transcription, DNA replication, and metabolism). These functions are differentially exploited in cancer, depending on context.
EP300 (hereafter referred to as p300) and its closely related paralog CREB-binding protein (CREBBP, hereafter CBP) are ubiquitously expressed transcriptional coactivators and major lysine acetyltransferases (KATs) in metazoans. They regulate transcription by serving as scaffolds that bridge sequence-specific DNA-binding factors and the basal transcriptional machinery (Chan and La Thangue 2001), and facilitate transcription through acetylation of histones, transcription factors, and autoacetylation (Sterner and Berger 2000; Black et al. 2006; Pugh 2006; Das et al. 2014). p300 and CBP are large proteins with multiple functional domains accommodating diverse protein–protein interactions. This has enabled a large number of disparate transcription factors to use p300/CBP as cofactors in regulating the expression of thousands of genes in essentially all cell types (Chan and La Thangue 2001). The large number of proteins that interact with p300 and CBP underscore the widespread influence of these coactivators on essential cellular functions. p300 and CBP regulate several fundamental biological processes including proliferation, cell cycle, cell differentiation, and the DNA damage response (Shi and Mello 1998; Goodman and Smolik 2000; Grossman 2001; Polesskaya et al. 2001). But the versatility of these proteins has also made it difficult to discern their specific involvements in distinct biological processes and pathophysiological states. Another impediment to understanding the roles of p300 and CBP is the overlapping contribution of these proteins to the same molecular processes such as gene regulation, hence, the commonly used designation “p300/CBP.”
Although p300/CBP have been implicated in cancer development, the specific contributions of each acetyltransferase to the cancer phenotype have been less precisely defined. This is in part attributable to the participation of p300/CBP in diverse and, at times, antagonistic cellular pathways such as tumor-suppressive and pro-oncogenic processes. The challenge is even greater when attempting to understand the consequences of mutations in p300 and CBP, which have been identified in numerous cancer genome studies. To fully understand how genetic or epigenetic alterations of p300/CBP contribute to the cancer phenotype, it is important to determine which cellular pathways are specifically affected by mutations in p300 or CBP. One important consideration is that p300 and CBP, despite significant sequence homology, also perform nonoverlapping cellular functions and can cooperate with distinct binding partners. In this review, we discuss the major functions of p300/CBP in the cell and how cancer cells exploit these functions to their advantage.
p300/CBP ORCHESTRATE THE CELL CYCLE AND REGULATE PROLIFERATION
p300 was initially identified through its physical association with the adenovirus transforming protein E1A and determined to be essential for adenovirus-mediated oncogenic transformation (Whyte et al. 1989; Sawada et al. 1997). Soon after, the E1A–p300 interaction was shown to be critical for the G1–S phase transition in adenovirus-infected cells (Howe et al. 1990). A number of other oncogenic viral proteins (e.g., SV40/polyoma LT, HPV E7) were subsequently shown to also target p300 or CBP to promote cellular transformation (Eckner et al. 1996; Zimmermann et al. 1999; Bernat et al. 2003). The frequent exploitations of p300/CBP as cofactors for viral oncoproteins highlighted the fundamental role of these proteins in regulating cellular proliferation, and raised the possibility that alterations of p300/CBP may also contribute to nonviral mechanisms of tumorigenesis.
E1A was initially reported to inhibit p300/CBP-mediated KAT activity and transcriptional activation, which led to a hypothesis that p300 may normally function as a negative regulator of S phase entry (Arany et al. 1995; Yang et al. 1996; Chakravarti et al. 1999). However, many studies have since revealed a major role for p300/CBP in promoting growth and cell-cycle progression. In fact, on adenoviral infection p300 is recruited to genes with functions in cell cycle and proliferation to promote their full activation and S phase entry in otherwise nondividing cells (Ferrari et al. 2008, 2014).
The functions of p300 and CBP in regulating cell-cycle progression are partly mediated through their influence on transcription by being recruited to gene regulatory regions, such as enhancers and promoters, via sequence-specific DNA-binding transcription factors. Once bound, they facilitate subsequent regulatory events to ultimately direct RNA polymerase II activation. p300 in particular contributes to the formation of the transcription pre-initiation complex, a large multiprotein complex required for expression of genes. p300 does this partly through dynamic association with and dissociation from the transcriptional machinery, which is facilitated by p300 auto-acetylation activity (Black et al. 2006). The pervasive participation of p300 and CBP in transcriptional regulation is evident in their binding to >16,000 genes in human cells (Smith et al. 2004; Ramos et al. 2010). Not all binding events lead to transcriptional activation and a growing body of evidence indicates a gene-repressive role for p300/CBP in certain contexts (Santoso and Kadonaga 2006; Sankar et al. 2008; Ferrari et al. 2014). p300 and CBP also regulate the cell cycle through interactions with or acetylation of proteins involved in cell-cycle progression, such as the DNA replication machinery and histones for the purpose of DNA replication through chromatin.
Transcriptional Coactivation and the Cell Cycle
One of the earliest cell-based models showing the role of p300/CBP in cell-cycle progression involved depletion of p300 and CBP, through microinjection of an antibody against both proteins, which was found to limit S phase entry (Ait-Si-Ali et al. 2000). This defect was reversed by overexpression of exogenous CBP, indicating a direct function of CBP in promoting cell-cycle progression. p300 and CBP serve as transcriptional coactivators for the E2F transcription factor family, which are central for expression of genes required for G1/S transition (Trouche and Kouzarides 1996; Trouche et al. 1996; Wang et al. 2007). In addition, p300/CBP acetylate the E2F proteins themselves (e.g., E2F1), leading to enhanced DNA-binding and gene activation (Martínez-Balbás et al. 2000; Marzio et al. 2000). The acetyltransferase activity of CBP is regulated in a cell-cycle-dependent manner and peaks at the G1/S boundary possibly as a consequence of cyclin/Cdk-mediated phosphorylation of CBP before initiation of S phase (Ait-Si-Ali et al. 1998). Cell-cycle-dependent transcription of the major histone genes for DNA replication is also dependent on p300/CBP, which are recruited by NPAT, the general histone expression regulator (He et al. 2011). Therefore, the transcriptional coactivator functions of p300/CBP mediate S phase entry through proper expression of DNA replication and cellular growth genes.
Nontranscriptional Effects of p300/CBP on Cell Cycle
p300/CBP may regulate DNA replication through modifying the histones surrounding the DNA replication origins. These two acetyltransferases are responsible for the bulk of histone H3 lysine 18 acetylation (H3K18ac) and H3K27ac, modifications associated with active promoters and enhancers (Horwitz et al. 2008; Jin et al. 2011). H3K18ac is also associated with active DNA replication in certain cell types (Li et al. 2014). p300/CBP may also directly regulate the DNA replication machinery by acetylating two major endonucleases involved in Okazaki fragment processing, FEN1 and Dna2, inhibiting and stimulating their activities, respectively (Hasan et al. 2001; Balakrishnan et al. 2010). This differential regulation is suggested to lead to increased accuracy of DNA replication (Balakrishnan et al. 2010). Complementing these results, pharmacological inhibition of p300 KAT activity prolongs S phase because of reduced replication fork velocity and defects in timing of replication origin firing and synchronization (Prieur et al. 2011). Altogether, p300/CBP regulate various aspects of the DNA replication process, including the choice and timing of origin firing and the assembly of the newly synthesized DNA into chromatin.
p300/CBP function in other phases of the cell cycle as well. Depletion of CBP leads to a delay in mitosis and accumulation of cells in G2/M because of the aberrant activity of the anaphase-promoting complex (APC/C), an E3 ubiquitin ligase required for progression through mitosis (Turnell et al. 2005). Taken together with the above, these findings place p300 and CBP at multiple positions along the cell cycle and emphasize the functions of these proteins in promoting progression through the entire cell cycle (Fig. 1).
Figure 1.
p300/CREB-binding protein (CBP) regulate the cell cycle at multiple points. The diagram summarizes the known functions of p300/CBP at different points along the cell cycle. The E2F family of transcription factors uses p300/CBP as transcriptional coactivators to facilitate expression of E2F target genes that orchestrate the transition from G1 to S. p300 also facilitates S phase progression by acetylating the DNA replication machinery (e.g., FEN1 and Dna2) and the histones surrounding the replication origins (e.g., H3K18). CBP promotes progression through mitosis by regulating the function of the anaphase-promoting complex (APC)/C complex.
Consistent with the critical roles of p300/CBP in cell-cycle regulation, significant growth defects are observed when these proteins are depleted in cells or organisms. Mouse models null for p300 or CBP are embryonic lethal and, although p300-null cells obtained from these embryos are viable, they show reduced proliferation (Yao et al. 1998). This also occurs when p300 is transiently or stably depleted (Yuan et al. 1999; Iyer et al. 2007). Therefore, loss of p300/CBP in most contexts leads to decreased proliferation.
p300 AND CBP IN CANCER
p300/CBP as Classic Tumor Suppressors
Early indications of tumor suppression by p300/CBP came from findings in a rare congenital developmental disorder, Rubinstein–Taybi syndrome (RTS). Germline heterozygous mutations in CBP and less frequently in p300 are observed in RTS and may play a role in the pathogenesis of this disease. RTS patients have an increased incidence of cancer with ∼5% diagnosed with childhood tumors of neural crest origin (Miller and Rubinstein 1995). p300/CBP mutations in RTS are variable and encompass microdeletions, truncating mutations as well as point mutations in different domains (Petrij et al. 1995; Roelfsema and Peters 2007). A number of these genetic lesions reduce acetyltransferase and/or transcriptional activities of p300/CBP implicating the reduction of these functions in the etiology of RTS-associated malignancies (Roelfsema and Peters 2007), a contention that is supported by studies in mice (Tanaka et al. 1997; Rebel et al. 2002; Alarcón et al. 2004).
Investigations of primary tumor samples helped to strengthen the tumor-suppressive functions of p300/CBP in humans. Work by Gayther et al. identified the first cancer-associated inactivating genetic lesions in p300 in breast and colorectal primary tumors and cell lines (Gayther et al. 2000). The majority of cases harbored inactivation or deletion of the second allele of p300. Studies of larger cohorts of solid tumors including colorectal, gastric, ovarian, and hepatocellular carcinomas also detected loss of heterozygosity (LOH) at the p300 or CBP loci at frequencies ranging from 1% to 50% (Bryan et al. 2002; Tillinghast et al. 2003; Koshiishi et al. 2004; Dancy and Cole 2015). A small fraction of p300/CBP LOH events in these studies were accompanied by somatic mutations in the second allele confirming earlier findings. Tumors showing LOH indicate that haploinsufficiency of p300/CBP may be a factor in the pathogenesis of cancer. This is consistent with the idea that a limiting cellular pool of p300/CBP may be a biological determinant of their effects on the cell. In fact there is evidence that these proteins are haploinsufficient because p300/CBP heterozygote null embryos have reduced survival (Yao et al. 1998). Therefore, different molecular pathways have to compete for a limited pool of p300/CBP to regulate their target genes (Kamei et al. 1996; Huang et al. 2007). So the reduced availability of p300/CBP through LOH may contribute to cancer development or progression by altering the equilibrium between the various p300/CBP-dependent pathways. Additional evidence suggesting a tumor-suppressive function for p300/CBP came from oral and cervical carcinoma cell lines. These cell lines, which harbor either a homozygous mutation in p300 or a heterozygous truncation of p300 with inactivation of the normal allele, show reduced proliferation on introduction of a normal copy of p300 (Suganuma et al. 2002).
p300/CBP may exert tumor-suppressive effects through promoting the functions of other bona fide tumor suppressors, such as p53, RB1, BRCA1, or through inducing transforming growth factor β (TGF-β)-responsive genes (Nishihara et al. 1998; Pao et al. 2000; Chan et al. 2001; Grossman 2001). The involvement of p300/CBP in p53-mediated functions is extensively studied and occurs at multiple levels. In response to DNA damage, p300/CBP augment p53-dependent transcriptional activation of genes required for cell-cycle arrest and DNA repair (Grossman 2001). In addition, p300 promotes the nuclear accumulation and stability of p53 in response to genotoxic stress. Interestingly, in unstressed conditions and during recovery from DNA damage, p300 is thought to ensure degradation of p53 for resumption of the cell cycle after DNA repair (Grossman et al. 1998; Grossman 2001; Kawai et al. 2001). BRCA1, which is frequently mutated in familial breast and ovarian cancers, plays a role in cell-cycle checkpoint, DNA damage repair, and transcriptional regulation (Monteiro et al. 1996; Wu et al. 2010). The latter role has linked BRCA1 to p300/CBP, which enhance BRCA1-mediated transcriptional activation (Pao et al. 2000; Mullan et al. 2006). Similarly p300/CBP mediate the effects of TGF-β signaling by serving as transcriptional coactivators for Smad3, a downstream effector of this tumor-suppressive pathway (Feng et al. 1998; Derynck et al. 2001).
p300/CBP as Drivers of Cancer Growth
Despite the tumor-suppressive roles of p300/CBP, several lines of evidence suggest that these KATs can also participate in promoting cancer. Although inactivating mutations in p300/CBP are found in certain cancers (Kalkhoven 2004; Pasqualucci et al. 2011), some cancer-linked point mutations are in fact gain-of-function alterations in p300/CBP that could contribute to cancer development (Ringel and Wolberger 2013). In addition to the acetyltransferase domains, important structural features of p300/CBP include three cysteine/histidine-rich zinc-binding domains (CH1-3), a bromodomain, and a recently identified RING (Really Interesting New Gene) domain within the larger CH2 region. The RING domain contacts the active site of the KAT domain blocking substrate binding and decreasing acetyltransferase activity in vitro. Disruption of the RING domain enhances p300 KAT activity (Delvecchio et al. 2013). Mutations in the p300 RING domain are found in malignancies including melanoma, endometrial and colorectal carcinoma (Forbes et al. 2015), as well as in RTS, and may boost p300 KAT activity in these settings (Delvecchio et al. 2013). How increased KAT activity of p300 or CBP promotes malignancy is not clear. In addition to acetylation of H3K18 and H3K27, p300/CBP also mediate the acetylation of histone H3 lysine 56, a modification associated with nucleosome assembly in yeast and DNA replication and repair in mammals (Li et al. 2008; Yuan et al. 2009; Vempati et al. 2010). Increased cellular levels of histone H3K56ac are observed in a number of epithelial tumors and relate to tumor stage and an undifferentiated phenotype (Das et al. 2009). Enhanced KAT activity of p300 or CBP may lead to increased acetylation of H3K56 in certain cancers.
Another mode of p300/CBP acetyltransferase gain-of-function involves translocation events in hematological malignancies such as myelodysplastic syndrome and acute myeloid leukemia. These translocations occur between p300 or CBP and monocytic leukemia zing-finger (MOZ), MOZ-related factor (MORF), or myeloid/lymphoid or mixed-lineage leukemia (MLL) (Kitabayashi et al. 2001; Panagopoulos et al. 2001). The translocation events more commonly generate a fusion protein containing the carboxy-terminal region of CBP with or without its KAT domain. The MOZ/MORF-CBP as well as MOZ-p300 fusion proteins maintain the KAT domains from both parent proteins potentially resulting in highly active lysine acetyltransferases (Yang and Ullah 2007). In essentially all translocation events, the amino-terminal region of CBP is excluded from the fusion protein. The novel functions that are gained by the fusion protein can contribute to the oncogenic nature of these translocations. Mutations in the KAT-inhibitory RING domain of p300 are also detected in myelodysplastic syndrome (Forbes et al. 2015). This further suggests a key role for increased p300/CBP KAT activity in the pathogenesis of these hematological malignancies.
Mutations in p300/CBP Are Nonrandom
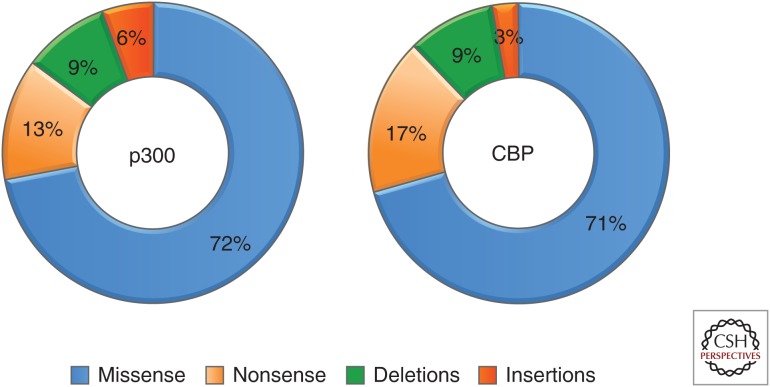
The widespread application of next generation sequencing has revealed an abundance of somatic genetic mutations with frequencies of up to 30% in p300 and CBP in various types of cancer. Although earlier studies of p300/CBP had uncovered gross or partial gene deletions, only a relatively small fraction of all p300/CBP genetic lesions in cancer are of this nature. The majority of alterations are, in fact, missense point mutations (Fig. 2) that occur essentially throughout the p300 and CBP proteins with a higher frequency in the KAT domains, suggesting a selective pressure in cancers for alteration of this activity (Fig. 3A). Certain residues in the KAT domain of p300 and CBP (e.g., D1399 and Y1467 for p300 and Y1450 and Y1503 for CBP) that are known to reduce or abolish the KAT activity when mutated are among the most frequent mutations in cancer (Delvecchio et al. 2013; Forbes et al. 2015). It is unclear, however, whether and how the other frequently mutated residues in this domain are important for the KAT activity. The top four and five most frequent missense mutations in p300 and CBP, respectively, are highlighted in Fig. 3B. These mutations in p300 including those thought to reduce KAT activity are clustered near the site of acetyl-CoA binding (Fig. 3B) (Liu et al. 2008; Maksimoska et al. 2014). The location of the other uncharacterized mutations near this site suggests these may also have an effect on the KAT activity.
Figure 2.
Frequency of the different types of mutations in p300 and CBP in cancer. The majority of genetic lesions to p300 or CBP in cancer are missense mutations followed by nonsense mutations, deletions, and a small number of insertions.
Figure 3.
The acetyltransferase domains of p300 and CBP are hotspots for mutations in cancer. (A) The number of tumors of different origins with missense mutations along the p300 and CBP proteins domain structures is shown (Forbes et al. 2015). CH1, Cysteine/histidine-rich region 1; BR, bromodomain; PHD, plant homeodomain finger; RING, Really Interesting New Gene finger domain; KAT, lysine acetyltransferase domain; CH3, cysteine/histidine-rich region 3 (also referred to as TAZ2); IBiD, IRF3-binding domain. (B) The residues in the KAT domains of p300 and CBP that are frequently mutated in cancer are indicated. Residues in red are important for the KAT activity. The four most common residues mutated in p300 reside close to the acetyl-CoA-binding site as indicated in the crystal structure (Liu et al. 2008; Maksimoska et al. 2014).
Tables 1 and 2 list the cancers with p300 and CBP mutation frequencies, respectively, of 5% or higher as reported in the COSMIC (Catalogue of Somatic Mutations in Cancer) database (Forbes et al. 2015). Both p300 and CBP are frequently mutated in skin squamous cell carcinoma followed by certain types of lymphomas. Alterations of p300/CBP may therefore be key contributory milestones to the development or progression of these cancers. Alternatively, these specific cellular contexts may provide a more permissive background for accumulation of genetic lesions in p300/CBP with no selective pressure to avoid them. Tables 3 and 4 include lists of cancers with ≤1% frequency of p300 or CBP mutations, respectively. Of interest in these groups are prostate and pancreas carcinomas, which may be under selective pressure to preserve the normal functions of p300/CBP for growth or may alter the functions of p300 or CBP through nongenetic means.
Table 1.
Cancer subtypes with higher frequency of p300 mutations
| Cancer subtype | N | Samples mutated (%) |
|---|---|---|
| Skin squamous cell carcinoma | 82 | 26.8 |
| Marginal zone B-cell lymphoma | 15 | 13.3 |
| Bladder carcinoma | 425 | 9.4 |
| Follicular lymphoma | 68 | 8.8 |
| Lung small-cell carcinoma | 48 | 8.3 |
| Endometrial carcinoma | 387 | 7.2 |
| Esophageal squamous cell carcinoma | 511 | 5.9 |
| Cervical squamous cell carcinoma | 193 | 5.2 |
| Breast carcinoma (estrogen-receptor-positive) | 80 | 5.0 |
Data were obtained from the publicly available COSMIC database (cancer.sanger.ac.uk).
Table 2.
Cancer subtypes with higher frequency of CBP mutations
| Cancer subtype | N | Samples mutated (%) |
|---|---|---|
| Follicular lymphoma | 66 | 33.3 |
| Skin squamous cell carcinoma | 77 | 28.6 |
| Marginal zone B-cell lymphoma | 15 | 13.3 |
| Diffuse large B-cell lymphoma | 242 | 12.0 |
| Salivary gland carcinoma | 63 | 9.5 |
| Bladder carcinoma | 438 | 8.9 |
| Endometrial carcinoma | 337 | 8.0 |
| Lung small-cell carcinoma | 52 | 7.7 |
| Breast carcinoma (estrogen-receptor-positive) | 80 | 7.5 |
Data were obtained from the publicly available COSMIC database (cancer.sanger.ac.uk).
Table 3.
Cancer subtypes with low frequency of p300 mutations
| Cancer subtype | N | Samples mutated (%) |
|---|---|---|
| Acute lymphoblastic leukemia | 438 | 0.9 |
| Clear cell renal cell carcinoma | 692 | 0.9 |
| Hepatocellular carcinoma | 628 | 0.8 |
| Chronic lymphocytic leukemia | 798 | 0.5 |
| Ovarian carcinoma | 693 | 0.3 |
| Prostate adenocarcinoma | 755 | 0.3 |
| Pancreatic carcinoma | 593 | 0.2 |
Data were obtained from the publicly available COSMIC database (cancer.sanger.ac.uk).
Table 4.
Cancer subtypes with low frequency of CBP mutations
| Cancer subtype | N | Samples mutated (%) |
|---|---|---|
| Pancreatic carcinoma | 593 | 1.0 |
| Breast carcinoma (triple-negative) | 121 | 0.8 |
| Clear cell renal cell carcinoma | 692 | 0.6 |
| Chronic lymphocytic leukemia | 798 | 0.5 |
| Prostate adenocarcinoma | 762 | 0.4 |
| Acute myeloid leukemia | 785 | 0.4 |
Data were obtained from the publicly available COSMIC database (cancer.sanger.ac.uk).
Beyond Genetic Alterations
In the absence of genetic defects, mechanisms such as changes in expression or subcellular localization can alter p300/CBP-associated functions in cancer. Analysis of The Cancer Genome Atlas (TCGA) data indicates differences in expression levels of p300 and CBP in multiple types of carcinomas and hematological malignancies. The effects of these changes in mediating the neoplastic phenotype cannot be described under one umbrella as both higher and lower levels of p300/CBP are found. These expression differences in some cases are accompanied by changes in gene copy number as a result of gross gene amplifications or deletions. Several studies of primary tumors of varying origins have also revealed changes in p300/CBP protein levels, which in either direction are prognostic in many cases. A study of 95 prostate cancer lesions revealed that increased p300 protein levels, as compared with adjacent normal tissue, correlate with increased proliferation, tumor volume, and extraprostatic involvement (Debes et al. 2003). A function of p300 in the progression of prostate cancer has also been proposed as it mediates androgen-dependent as well as independent transactivation of the androgen receptor (Debes et al. 2002). Additionally, increased p300 expression correlates with poor survival and aggressive phenotypes in breast, hepatocellular, esophageal, and cutaneous squamous cell carcinoma (Li et al. 2011a,b; Xiao et al. 2011; Chen et al. 2014). Consistently, pharmacological inhibition of p300/CBP KAT activities in a panel of primary melanoma cell lines sensitizes cells to DNA-damaging chemotherapeutic agents (Yan et al. 2013). Higher nuclear CBP protein levels have also been detected in precancerous hyperplastic and dysplastic laryngeal lesions, suggesting overexpression of p300/CBP may contribute to different steps of cancer development and growth (Karamouzis et al. 2002). Conversely, reduced levels of p300/CBP have been detected in certain cancers. Pasqualucci et al. (2011) detected loss of p300 and/or CBP expression in 8% of diffuse large B-cell lymphoma with no genetic lesion in these genes. The significance of these changes has been underscored by the prognostic value of this information. A study of 327 melanoma samples found that decreased nuclear levels of p300 associated with disease progression and poor overall survival (Rotte et al. 2013).
Interestingly, an increase in cytoplasmic levels of p300 was observed in melanoma and correlated significantly with tumor size and disease progression in early stages (Rotte et al. 2013; Bhandaru et al. 2014). These findings suggest that a shift in the subcellular localization of p300 may be involved in the progression of this cancer and highlight the importance of p300/CBP cytoplasmic functions. However, few investigations addressing cytoplasmic functions of p300/CBP have been conducted. Among these are studies that provide evidence for a p53-directed E4 ligase activity associated with cytoplasmic p300/CBP leading to polyubiquitination and degradation of cytoplasmic p53 (Grossman et al. 2007; Shi et al. 2009). The compartmentalized regulation of p53 by p300/CBP provides an explanation for previously reported opposing effects of these KATs on p53, namely, destabilization of the protein and promoting its nuclear functions. Hence, the physical separation of p300/CBP’s nuclear transcriptional function from the cytosolic E3/E4 ligase activity is an important aspect of p53 regulation by p300/CBP. Extranuclear functions of p53 involve triggering apoptosis through interactions with mitochondrial outer membrane proteins, which contribute to its tumor-suppressive effects (O’Brate and Giannakakou 2003). Inappropriate accumulation of cytosolic p300/CBP can, therefore, suppress p53-mediated apoptosis in response to stress signals.
The Effects of p300/CBP on Chromatin in Cancer
Deregulated chromatin targeting by p300/CBP can have profound effects in cancer. For instance, alterations of H3K27ac, a major in vivo target of p300/CBP at enhancer loci, is observed in numerous cancers indicating disrupted activities of p300/CBP at specific enhancer elements (Akhtar-Zaidi et al. 2012). These “variant enhancer loci” (VELs) as they were termed, correlate with aberrant expression of their putative target genes. The enhancers with inappropriately acquired H3K27ac associate frequently with genes that have known contributions to many phenotypic hallmarks of cancer (Hnisz et al. 2013). Thus, alterations in the distribution of H3K27ac (and likely H3K18ac, the other major target of p300/CBP) and, hence, enhancer activity in cancer can promote tumorigenesis through promoting an oncogenic gene expression program. Possible mechanisms of deregulated chromatin targeting may involve mutant or inappropriately expressed p300/CBP as well as changes in transcription factors that recruit these coactivators to gene regulatory elements. For example, genetic lesions in the acetyllysine-binding bromodomain of p300/CBP can lead to alterations in specificity or strength of chromatin binding and to redistribution of H3K18/27ac across the genome. Such a scenario could also occur through gene amplifications or translocations, giving rise to aberrant formation of enhancer loci in some tumors (Hnisz et al. 2013).
Functions of p300/CBP on chromatin can also be co-opted by viral oncogenes leading to cellular transformation. Adenovirus small E1A (e1a), a splice variant of E1A that is responsible for reprogramming the expression of thousands of host genes, relies on interactions with p300/CBP to coerce normal, primary cell-cycle-arrested fibroblasts into S phase (Howe et al. 1990). The e1a protein causes the recruitment of p300/CBP to and increased H3K18ac (but not H3K27ac) at promoter regions of cell-cycle genes for full transcriptional activation. In parallel, e1a also actively represses cell-type-specific genes by opposing the functions of p300/CBP at promoters and enhancers of these genes as evident by substantial deacetylation of H3K18 and H3K27 at these sites on e1a expression (Ferrari et al. 2010, 2014). Furthermore, e1a represses cellular defense response genes by forming a trimeric complex between e1a, RB1, and, surprisingly, p300 itself, which acetylates RB1 to prevent its normal inactivation by phosphorylation (Ferrari et al. 2014). The repressive RB1-e1a-p300 complex binds to the promoter and gene body regions of defense response genes, and in some cases fully coating entire gene loci and preventing their activation by the host cell. This repression is accompanied by condensation of the local chromatin environment (Ferrari et al. 2014). The overall effect of e1a in the 24 h after entry into a cell is to turn off cell identity and the antiviral cellular defense genes, and to turn on genes that are required for entry into S phase and DNA replication. The bulk of this oncogenic reprogramming depends on interactions of e1a with p300/CBP and RB1. These findings provide a blueprint for understanding how nonviral oncogenesis may also depend on precise exploitations of p300/CBP to achieve similar cellular outcomes.
HISTONE ACETYLATION BALANCE AND ITS IMPLICATIONS FOR CANCER
In addition to targeted recruitment and acetylation of specific genomic loci, histone acetyltransferases function globally throughout the genome in a seemingly nontargeted manner by mechanisms that are not yet clear (Vogelauer et al. 2000). When coupled to the global actions of lysine deacetylases (KDACs), the opposing but continual functions of KATs and KDACs result in fast turnover of histone acetylation (Waterborg 2001), which consumes acetyl coenzyme A and generates acetate anions. In primary tumor tissues, cancer cells show marked differences in the global levels of histone modifications including acetylation, which are prognostic of clinical outcome in many types of solid tumors (Kurdistani 2007). Specifically, lower global level of H3K18ac is associated with cancer-related mortality and/or morbidity in prostate, kidney, lung, pancreatic, and breast cancers (Seligson et al. 2005, 2009; Manuyakorn et al. 2010; Mosashvilli et al. 2010; Kurdistani 2011). Cancer-associated genetic lesions in p300/CBP resulting in reduced KAT activity can certainly lead to the global loss of H3K18/27ac. However, recent work from our laboratory has revealed an unanticipated function for global histone acetylation in regulating intracellular pH (McBrian et al. 2013). We found that in multiple cancer or normal cell lines, the balance of KAT and KDAC activities is shifted toward the latter in response to acidic cellular environment, resulting in histones that are globally and continuously deacetylated. This leads to liberated acetate anions that are in turn used by the membrane-bound monocarboxylate transporters to export protons out of the cell, thus buffering the intracellular pH. Proliferating cells including cancer cells need to maintain an alkaline intracellular pH relative to the outside for cell growth and division (Webb et al. 2011). Because cancer tissues commonly show low pH in vivo, it is possible that enhanced global histone deacetylation serves to maintain a viable intracellular pH in these tumors, providing a growth advantage (Parks et al. 2011). This chromatin response to acidity is an active process resulting in the continuous generation of free acetate molecules through enhanced deacetylation and thus depends on intact or even enhanced histone acetylation. Therefore, the function of KATs in maintaining global histone acetylation is imperative to this pH-regulatory function of chromatin. In this regard, loss of KAT function may in fact reduce fitness in tumors exposed to an acidic environment.
LINKING CELLULAR ENERGETICS AND THE EPIGENOME
p300 and CBP target a significant number of nonhistone proteins for acetylation, including cytosolic proteins involved in essential metabolic processes. This can potentially coordinate cytoplasmic and chromatin-related functions of p300/CBP. The involvement of p300 in regulating metabolism via targeting the M2 isoform of pyruvate kinase, PKM2, is one such example (Lv et al. 2013). A majority of cancers express PKM2, which unlike the constitutively active PKM1 isoform, shows lower activity and is allosterically activated by an upstream glycolytic intermediate (Christofk et al. 2008; Wong et al. 2015). The slower enzymatic rate of PKM2 is thought to essentially serve as a road blockage that causes a logjam in upstream glycolytic reactions, forcing glycolysis intermediates into branching pathways, the products of which, such as nucleotide precursors, are required for general cellular biosynthesis. p300 acetylates a lysine residue (K433) unique to PKM2, which abolishes allosteric activation and enhances nuclear localization of this PK isoform. The switch between cytoplasmic metabolic function and nuclear protein kinase activity of PKM2 regulated by p300 occurs in response to mitogens and oncogenic signals and may be involved in tumorigenesis (Lv et al. 2013). In the nucleus PKM2 phosphorylates histone H3 at threonine 11, a modification shown to be required for cell-cycle progression and tumorigenesis (Yang et al. 2012). These findings indicate a role of p300 in mediating the proliferative program in cancer cells through switching a metabolic enzyme to a nuclear kinase to create a chromatin state conducive for cell replication. The regulation of PKM2 localization and activity is just one recent indication of the broader influence of p300/CBP beyond their nuclear functions as transcriptional coactivators.
FUTURE DIRECTIONS AND THERAPEUTIC APPLICATIONS
The central roles of p300/CBP in regulating cell proliferation have spurred efforts to develop specific inhibitors of the enzymatic activities as well as protein–protein interactions of p300/CBP. KAT inhibitors with higher specificity toward p300/CBP show antiproliferative effects in preclinical studies of cancer (Santer et al. 2011; Yang et al. 2013). Small molecules that inhibit p300/CBP interactions with other proteins also show promising clinical use. ICG-001, which specifically inhibits CBP binding to β-catenin, a component of the Wnt signaling pathway, reduces tumorigenic phenotypes and enhances drug sensitivity in both acute lymphoblastic leukemia (ALL) and nasopharyngeal carcinoma (Emami et al. 2004; Gang et al. 2014; Chan et al. 2015). This approach is thought to take advantage of differential co-activator usage by β-catenin. β-catenin may mediate the opposing outcomes of Wnt signaling by using CBP or p300 to either stimulate proliferation or initiate differentiation, respectively (Ma et al. 2005; Teo and Kahn 2010). Interestingly, the effect of ICG-001 is independent of CBP mutational status in ALL (Gang et al. 2014). Most CBP mutations in ALL are found carboxy terminal to the β-catenin binding site where ICG-001 binds (Mullighan et al. 2011). These findings suggest that the preservation of CBP-β-catenin interaction and not the mutations in other regions of CBP may underlie progression of ALL.
CONCLUDING REMARKS
The many functions of p300/CBP can be differentially exploited in cancer depending on the context, cellular identity, and perhaps environmental cues to confer a growth advantage. The paradigm of cancer as an evolutionary system suggests the sequential acquisition of somatic mutations in a fluctuating microenvironment to gain fitness. Considering such a system, the order and nature of other oncogenic events can dictate the selection for or against p300/CBP alterations that are most advantageous for survival and growth, therefore branding these proteins as tumor suppressors or oncogenes.
ACKNOWLEDGMENTS
We thank Michael Carey and Trent Su for valuable input and discussions in preparing this manuscript. N.A. is supported by a Ruth L. Kirschstein National Research Service Award (CA186619-02) and S.K.K. by a National Institutes of Health Grant (CA178415).
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Ait-Si-Ali S, Ramirez S, Barre FX, Dkhissi F, Magnaghi-Jaulin L, Girault JA, Robin P, Knibiehler M, Pritchard LL, Ducommun B, et al. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396: 184–186. [DOI] [PubMed] [Google Scholar]
- Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. 2000. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene 19: 2430–2437. [DOI] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. 2004. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein–Taybi syndrome and its amelioration. Neuron 42: 947–959. [DOI] [PubMed] [Google Scholar]
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374: 81–84. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Stewart J, Polaczek P, Campbell JL, Bambara RA. 2010. Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. J Biol Chem 285: 4398–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat A, Avvakumov N, Mymryk JS, Banks L. 2003. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 22: 7871–7881. [DOI] [PubMed] [Google Scholar]
- Bhandaru M, Ardekani GS, Zhang G, Martinka M, McElwee KJ, Li G, Rotte A. 2014. A combination of p300 and Braf expression in the diagnosis and prognosis of melanoma. BMC Cancer 14: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M. 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell 23: 809–818. [DOI] [PubMed] [Google Scholar]
- Bryan EJ, Jokubaitis VJ, Chamberlain NL, Baxter SW, Dawson E, Choong DYH, Campbell IG. 2002. Mutation analysis of EP300 in colon, breast and ovarian carcinomas. Int J Cancer 102: 137–141. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96: 393–403. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114: 2363–2373. [DOI] [PubMed] [Google Scholar]
- Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol 3: 667–674. [DOI] [PubMed] [Google Scholar]
- Chan KC, Chan LS, Ip JCY, Lo C, Yip TTC, Ngan RKC, Wong RNS, Lo KW, Ng WT, Lee AWM, et al. 2015. Therapeutic targeting of CBP/β-catenin signaling reduces cancer stem-like population and synergistically suppresses growth of EBV-positive nasopharyngeal carcinoma cells with cisplatin. Sci Rep 5: 9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Cai MY, Luo RZ, Tian X, Liao QM, Zhang XY, Han JD. 2014. Overexpression of p300 correlates with poor prognosis in patients with cutaneous squamous cell carcinoma. Br J Dermatol 172: 111–119. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233. [DOI] [PubMed] [Google Scholar]
- Dancy BM, Cole PA. 2015. Protein lysine acetylation by p300/CBP. Chem Rev 115: 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Roy S, Namjoshi S, Malarkey CS, Jones DNM, Kutateladze TG, Churchill ME, Tyler JK. 2014. Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proc Natl Acad Sci 111: E1072–E1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes JD, Schmidt LJ, Huang H, Tindall DJ. 2002. P300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Res 62: 5632–5636. [PubMed] [Google Scholar]
- Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DAL, Tindall DJ. 2003. p300 in prostate cancer proliferation and progression. Cancer Res 63: 7638–7640. [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. 2013. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol 20: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat Genet 29: 117–129. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ludlow JW, Lill NL, Oldread E, Arany Z, Modjtahedi N, DeCaprio JA, Livingston DM, Morgan JA. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol 16: 3454–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo J, Oh SW, Kim HY, et al. 2004. A small molecule inhibitor of β-catenin/CREB-binding protein transcription. Proc Natl Acad Sci 101: 12682–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. 1998. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev 12: 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. 2008. Epigenetic reprogramming by adenovirus e1a. Science 321: 1086–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Berk AJ, Kurdistani SK. 2010. Viral manipulation of the host epigenome for oncogenic transformation. Nat Rev Genet 10: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, et al. 2014. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16: 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang EJ, Hsieh YT, Pham J, Zhao Y, Nguyen C, Huantes S, Park E, Naing K, Klemm L, Swaminathan S, et al. 2014. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene 33: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et al. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat Genet 24: 300–303. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev 14: 1553–1577. [PubMed] [Google Scholar]
- Grossman SR. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem 268: 2773–2778. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell 2: 405–415. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Grossman SR, Deato ME, Tagami H, Nakatani Y, Livingston DM. 2007. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 342: 342–345. [DOI] [PubMed] [Google Scholar]
- Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hübscher U, Hottiger MO. 2001. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell 7: 1221–1231. [DOI] [PubMed] [Google Scholar]
- He H, Yu FX, Sun C, Luo Y. 2011. CBP/p300 and SIRT1 are involved in transcriptional regulation of S phase specific histone genes. PLoS ONE 6: e22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. 2008. Adenovirus small e1a alters global patterns of histone modification. Science 321: 1084–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Mymryk JS, Egan C, Branton PE, Bayley ST. 1990. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci 87: 5883–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Ju TK, Hung MC, Chen CC. 2007. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol Cell 26: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Xian J, Chin SF, Bannister AJ, Daigo Y, Aparicio S, Kouzarides T, Caldas C. 2007. p300 is required for orderly G1/S transition in human cancer cells. Oncogene 26: 21–29. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SYR, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E. 2004. CBP and p300: HATs for different occasions. Biochem Pharmacol 68: 1145–1155. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, et al. 1996. A CBP-integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85: 403–414. [DOI] [PubMed] [Google Scholar]
- Karamouzis MV, Papadas T, Varakis I, Sotiropoulou-Bonikou G, Papavassiliou AG. 2002. Induction of the CBP transcriptional co-activator early during laryngeal carcinogenesis. J Cancer Res Clin Oncol 128: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Nie L, Wiederschain D, Yuan ZM. 2001. Dual role of p300 in the regulation of p53 stability. J Biol Chem 276: 45928–45932. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N, Abe T, Ohki M. 2001. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia 15: 89–94. [DOI] [PubMed] [Google Scholar]
- Koshiishi N, Chong JM, Fukasawa T, Ikeno R, Hayashi Y, Funata N, Nagai H, Miyaki M, Matsumoto Y, Fukayama M. 2004. P300 gene alterations in intestinal and diffuse types of gastric carcinoma. Gastric Cancer 7: 85–90. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK. 2007. Histone modifications as markers of cancer prognosis: A cellular view. Br J Cancer 97: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S. 2011. CRCnetBASE—Histone modifications in cancer biology and prognosis. Prog Drug Res 67: 91–106. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Luo RZ, Chen JW, Cao Y, Lu JB, He JH, Wu QL, Cai MY. 2011a. High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J Transl Med 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang HX, Luo RZ, Zhang Y, Li M, Wang X, Jia WH. 2011b. High expression of p300 has an unfavorable impact on survival in resectable esophageal squamous cell carcinoma. Ann Thorac Surg 91: 1531–1538. [DOI] [PubMed] [Google Scholar]
- Li B, Su T, Ferrari R, Li JY, Kurdistani SK. 2014. A unique epigenetic signature is associated with active DNA replication loci in human embryonic stem cells. Epigenetics 9: 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. 2008. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451: 846–850. [DOI] [PubMed] [Google Scholar]
- Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, et al. 2013. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell 52: 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Nguyen C, Lee KS, Kahn M. 2005. Differential roles for the coactivators CBP and p300 on TCF/β-catenin-mediated survivin gene expression. Oncogene 24: 3619–3631. [DOI] [PubMed] [Google Scholar]
- Maksimoska J, Segura-Peña D, Cole PA, Marmorstein R. 2014. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry 53: 3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, et al. 2010. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: Results from RTOG 9704. J Clin Oncol 28: 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. 2000. Regulation of E2F1 activity by acetylation. EMBO J 19: 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. 2000. E2F family members are differentially regulated by reversible acetylation. J Biol Chem 275: 10887–10892. [DOI] [PubMed] [Google Scholar]
- McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, et al. 2013. Histone acetylation regulates intracellular pH. Mol Cell 49: 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RW, Rubinstein JH. 1995. Tumors in Rubinstein–Taybi syndrome. Am J Med Genet 56: 112–115. [DOI] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H. 1996. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci 93: 13595–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosashvilli D, Kahl P, Mertens C, Holzapfel S, Rogenhofer S, Hauser S, Büttner R, Von Ruecker A, Müller SC, Ellinger J. 2010. Global histone acetylation levels: Prognostic relevance in patients with renal cell carcinoma. Cancer Sci 101: 2664–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Harkin DP. 2006. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25: 5854–5863. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. 2011. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471: 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara A, Hanai JI, Okamoto N, Yanagisawa J, Kato S, Miyazono K, Kawabata M. 1998. Role of p300, a transcriptional coactivator, in signalling of TGF-β. Genes Cells 3: 613–623. [DOI] [PubMed] [Google Scholar]
- O’Brate A, Giannakakou P. 2003. The importance of p53 location: Nuclear or cytoplasmic zip code? Drug Resist Updat 6: 313–322. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Fioretos T, Isaksson M, Samuelsson U, Billström R, Strömbeck B, Mitelman F, Johansson B. 2001. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13). Hum Mol Genet 10: 395–404. [DOI] [PubMed] [Google Scholar]
- Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM. 2000. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci 97: 1020–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks SK, Chiche J, Pouyssegur J. 2011. pH control mechanisms of tumor survival and growth. J Cell Physiol 226: 299–308. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. 1995. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348–351. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Fritsch L, Duquet A, Ait-Si-Ali S, Robin P, Vervisch A, Pritchard LL, Cole P, Harel-Bellan A. 2001. CBP/p300 and muscle differentiation: No HAT, no muscle. EMBO J 20: 6816–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur A, Besnard E, Babled A, Lemaitre JM. 2011. p53 and p16(INK4A) independent induction of senescence by chromatin-dependent alteration of S phase progression. Nat Commun 2: 473. [DOI] [PubMed] [Google Scholar]
- Pugh BF. 2006. HATs off to PIC assembly. Mol Cell 23: 776–777. [DOI] [PubMed] [Google Scholar]
- Ramos YFM, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van Galen M, van Dam H, van Ommen GJB, den Dunnen JT, Zantema A, et al. 2010. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res 38: 5396–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. 2002. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci 99: 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel AE, Wolberger C. 2013. A new RING tossed into an old HAT. Structure 72: 181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema JH, Peters DJM. 2007. Rubinstein–Taybi syndrome: Clinical and molecular overview. Expert Rev Mol Med 9: 1–16. [DOI] [PubMed] [Google Scholar]
- Rotte A, Bhandaru M, Cheng Y, Sjoestroem C, Martinka M, Li G. 2013. Decreased expression of nuclear p300 is associated with disease progression and worse prognosis of melanoma patients. PLoS ONE 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar N, Baluchamy S, Kadeppagari RK, Singhal G, Weitzman S, Thimmapaya B. 2008. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene 27: 5717–5728. [DOI] [PubMed] [Google Scholar]
- Santer FR, Höschele PPS, Oh SJ, Erb HHH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z. 2011. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther 10: 1644–1655. [DOI] [PubMed] [Google Scholar]
- Santoso B, Kadonaga JT. 2006. Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat Struct Mol Biol 13: 131–139. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Ishino M, Miura K, Ohtsuka E, Fujinaga K. 1997. Identification of specific amino acid residues of adenovirus 12 E1A involved in transformation and p300 binding. Virus Genes 15: 161–170. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435: 1262–1266. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. 2009. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 174: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mello C. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev 12: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. 2009. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci 106: 16275–16280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Freebern WJ, Collins I, De Siervi A, Montano I, Haggerty CM, McNutt MC, Butscher WG, Dzekunova I, Petersen DW, et al. 2004. Kinetic profiles of p300 occupancy in vivo predict common features of promoter structure and coactivator recruitment. Proc Natl Acad Sci 101: 11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. 2000. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Kawabata M, Ohshima T, Ikeda MA. 2002. Growth suppression of human carcinoma cells by reintroduction of the p300 coactivator. Proc Natl Acad Sci 99: 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: A partial similarity with Rubinstein–Taybi syndrome. Proc Natl Acad Sci 94: 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JL, Kahn M. 2010. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev 62: 1149–1155. [DOI] [PubMed] [Google Scholar]
- Tillinghast GW, Partee J, Albert P, Kelley JM, Burtow KH, Kelly K. 2003. Analysis of genetic stability at the EP300 and CREBBP loci in a panel of cancer cell lines. Genes Chromosom Cancer 37: 121–131. [DOI] [PubMed] [Google Scholar]
- Trouche D, Kouzarides T. 1996. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci 93: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Cook A, Kouzarides T. 1996. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res 24: 4139–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell AS, Stewart GS, Grand RJA, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. 2005. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature 438: 690–695. [DOI] [PubMed] [Google Scholar]
- Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. 2010. p300–mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem 285: 28553–28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408: 495–498. [DOI] [PubMed] [Google Scholar]
- Wang H, Larris B, Peiris TH, Zhang L, Le Lay J, Gao Y, Greenbaum LE. 2007. C/EBP β activates E2F-regulated genes in vivo via recruitment of the coactivator CREB-binding protein/P300. J Biol Chem 282: 24679–24688. [DOI] [PubMed] [Google Scholar]
- Waterborg JH. 2001. Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry 40: 2599–2605. [DOI] [PubMed] [Google Scholar]
- Webb BA, Chimenti M, Jacobson MP, Barber DL. 2011. Dysregulated pH: A perfect storm for cancer progression. Nat Rev Cancer 11: 671–677. [DOI] [PubMed] [Google Scholar]
- Whyte P, Williamson NM, Harlow E. 1989. Cellular targets for transformation by the adenovirus E1A proteins. Cell 56: 67–75. [DOI] [PubMed] [Google Scholar]
- Wong N, Ojo D, Yan J, Tang D. 2015. PKM2 contributes to cancer metabolism. Cancer Lett 356: 184–191. [DOI] [PubMed] [Google Scholar]
- Wu J, Lu LY, Yu X. 2010. The role of BRCA1 in DNA damage response. Protein Cell 1: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao XS, Cai MY, Chen JW, Guan XY, Kung HF, Zeng YX, Xie D. 2011. High expression of p300 in human breast cancer correlates with tumor recurrence and predicts adverse prognosis. Chinese J Cancer Res 23: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, et al. 2013. Selective inhibition of p300 HAT blocks cell-cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. J Invest Dermatol 133: 2444–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ullah M. 2007. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene 26: 5408–5419. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382: 319–324. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. 2012. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pinello CE, Luo J, Li D, Wang Y, Zhao LY, Jahn SC, Saldanha SA, Planck J, Geary KR, et al. 2013. Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents. Mol Cancer Ther 12: 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93: 361–372. [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, et al. 1999. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem 274: 1883–1886. [DOI] [PubMed] [Google Scholar]
- Yuan J, Pu M, Zhang Z, Lou Z. 2009. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle 8: 1747–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Degenkolbe R, Bernard HU, O’Connor MJ. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol 73: 6209–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]