ABSTRACT
Complex media are routinely used to cultivate diverse bacteria. However, this complexity can obscure the factors that govern cell growth. While studying protein acetylation in buffered tryptone broth supplemented with glucose (TB7-glucose), we observed that Escherichia coli did not fully consume glucose prior to stationary phase. However, when we supplemented this medium with magnesium, the glucose was completely consumed during exponential growth, with concomitant increases in cell number and biomass but reduced cell size. Similar results were observed with other sugars and other peptide-based media, including lysogeny broth. Magnesium also limited cell growth for Vibrio fischeri and Bacillus subtilis in TB7-glucose. Finally, magnesium supplementation reduced protein acetylation. Based on these results, we conclude that growth in peptide-based media is magnesium limited. We further conclude that magnesium supplementation can be used to tune protein acetylation without genetic manipulation. These results have the potential to reduce potentially deleterious acetylated isoforms of recombinant proteins without negatively affecting cell growth.
IMPORTANCE Bacteria are often grown in complex media. These media are thought to provide the nutrients necessary to grow bacteria to high cell densities. In this work, we found that peptide-based media containing a sugar are magnesium limited for bacterial growth. In particular, magnesium supplementation is necessary for the bacteria to use the sugar for cell growth. Interestingly, in the absence of magnesium supplementation, the bacteria still consume the sugar. However, rather than use it for cell growth, the bacteria instead use the sugar to acetylate lysines on proteins. As lysine acetylation may alter the activity of proteins, this work demonstrates how lysine acetylation can be tuned through magnesium supplementation. These findings may be useful for recombinant protein production, when acetylated isoforms are to be avoided. They also demonstrate how to increase bacterial growth in complex media.
KEYWORDS: acetylation, complex media, improved cell growth, magnesium, tryptone
INTRODUCTION
Bacteria are routinely grown in complex media containing a rich assortment of nutrients. For example, lysogeny broth (LB) contains tryptone and yeast exact. Together, these components provide a rich mixture of nutrients that supports growth of numerous bacteria. However, the complexity of such media obscures factors governing cell behavior, especially those that limit cell growth.
We routinely grow Escherichia coli aerobically in buffered tryptone broth (TB7) supplemented with 4 g/liter glucose (TB7-glucose) when studying protein acetylation. Under these growth conditions, the cells acetylate their proteins during stationary phase (1–3). This glucose-induced acetylation is due primarily to production of acetyl phosphate (1, 2), a metabolic intermediate of the acetate fermentation pathway (4).
While studying glucose-induced protein acetylation, we found that E. coli continues to consume the glucose in TB7-glucose even after the cultures reach stationary phase. In fact, the majority of the glucose was consumed after the cultures ceased exponential growth. This behavior was contrary to our initial expectations, which were based upon the behavior of E. coli cultures in minimal medium containing glucose as the sole carbon source (5). Under these conditions, the cultures enter stationary phase when the glucose is depleted. We also knew that the concentration of acetyl phosphate peaks during exponential growth in unbuffered TB without glucose supplementation (6) and during growth in MOPS (morpholinepropanesulfonic acid) minimal medium with glucose as the sole carbon source (7). Under both of these growth conditions, acetyl phosphate is undetectable by the time the cultures reach stationary phase. Taken together, these observations suggested that E. coli cells growing in TB7-glucose acetylate their proteins during stationary phase because that is when glucose is converted to acetyl phosphate, the primary acetyl donor in E. coli (1, 2).
In this study, we investigated the growth of E. coli in TB7-glucose. The key finding was that a low magnesium concentration limits growth in TB7-glucose, causing the culture to enter stationary phase before consuming the majority of glucose. However, lack of magnesium did not inhibit glucose consumption or its metabolism to acetate; rather, consumption occurred primarily during stationary phase. When TB7-glucose was supplemented with magnesium, the exponential phase of growth was extended, resulting in complete consumption of glucose prior to stationary phase, increased biomass, and decreased protein acetylation. The same general behavior was observed in other peptide-based media containing glucose, including LB. In fact, LB supplemented with magnesium alone resulted in higher growth yields. These results were not specific to E. coli, as similar behavior was observed for two other bacterial species, Vibrio fischeri and Bacillus subtilis. Collectively, these results further our understanding of bacterial growth in complex media and enable us to control global acetylation levels in E. coli.
RESULTS
A component of TB7 limits growth.
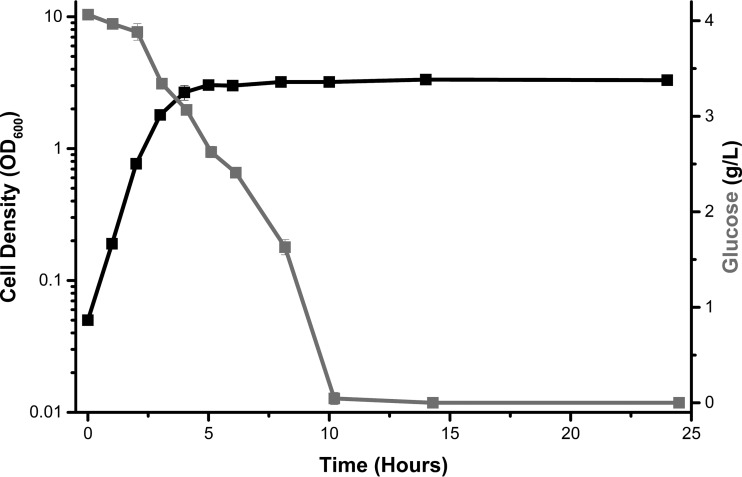
While growing E. coli BW25113 (Table 1) in TB7-glucose, we noticed that the cells continued to consume glucose even after the culture reached stationary phase (Fig. 1). In fact, the majority of glucose was consumed during stationary phase. Because glucose is consumed during exponential growth in minimal media (e.g., M9) with glucose as the sole carbon source (5), we wondered whether tryptone might somehow inhibit glucose consumption by E. coli.
TABLE 1.
Bacterial strains
| Strain | Description | Reference or source |
|---|---|---|
| BW25113 | F− λ− Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 rrnB3 rph-1 hsdR514 | 24 |
| MC4100 | F− [araD139]B/r Δ(argF-lac)169 λ− e14− flhD5301 Δ(fruK-yeiR)725(fruA25) relA1 rpsL150(Strr) rbsR22, Δ(fimB-fimE)632(::IS1) deoC1 | 25 |
| MG1655 | F− λ− rph-1 | 26 |
| AJW678 | thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136 lacX74 | 27 |
| AJW5231 | BW25113 Δcrp::kan | 3 |
| AJW5389 | BW25113 Δcra::kan (aka ΔfruR::kan) | 3 |
| BW27422 | BW25113 ΔarcA43 | 28 |
| AJW2573 | BW25113 Δ(ackA pta)::cat | P1: EPB6 × BW25113 |
| AJW3451 | BW25113 ΔackA::FRT | Kan cassette flipped from AJW3443 |
| AJW3443 | BW25113 ΔackA::kan | P1: JW2293 × BW25113 |
| AJW5384 | BW25113 ΔptsG::kan | 3 |
| AJW5441 | BW25113 Δpgi::kan | P1: JW3985 × BW25113 |
| AJW5533 | BW25113 ΔglnA::kan | P1: JW3841 × BW25113 |
| AJW5566 | BW25113 ΔgltA::kan | P1: JW0710 × BW25113 |
| AJW5567 | BW25113 ΔglnD::kan | P1: JW0162 × BW25113 |
| AJW5568 | BW25113 ΔglnE::kan | P1: JW3025 × BW25113 |
| AJW5569 | BW25113 ΔphnP::kan | P1: JW0710 × BW25113 |
| AJW5570 | BW25113 ΔphoQ::kan | P1: JW0710 × BW25113 |
| EPB6 | Δ(ackA pta)::cat | Mark Goulian (University of Pennsylvania) |
| JW2293 | Keio ΔackA::kan | 22 |
| JW2491 | Keio ΔguaA::kan | 22 |
| JW0474 | Keio ΔybaS::kan | 22 |
| JW1517 | Keio ΔyneH::kan | 22 |
| JW2309 | Keio ΔpurF::kan | 22 |
| JW2541 | Keio ΔpurL::kan | 22 |
| JW0660 | Keio ΔasnB::kan | 22 |
| JW0031 | Keio ΔcarB::kan | 22 |
| JW0030 | Keio ΔcarA::kan | 22 |
| JW2005 | Keio ΔhisH::kan | 22 |
| JW1256 | Keio ΔtrpE::kan | 22 |
| JW2408 | Keio ΔptsH::kan | 22 |
| JW2410 | Keio Δcrr::kan | 22 |
| JW2409 | Keio ΔptsI::kan | 22 |
| JW1116 | Keio ΔphoP::kan | 22 |
| JW1115 | Keio ΔphoQ::kan | 22 |
| JW3985 | Keio Δpgi::kan | 22 |
| JW0389 | Keio ΔphoB::kan | 22 |
| JW1328 | Keio Δfnr::kan | 22 |
| JW4099 | Keio ΔaspA::kan | 22 |
| JW0911 | Keio ΔaspC::kan | 22 |
| JW3841 | Keio ΔglnA::kan | 22 |
| JW2537 | Keio ΔglnB::kan | 22 |
| JW0710 | Keio ΔgltA::kan | 22 |
| JW3789 | Keio ΔcorA::kan | 22 |
| JW4201 | Keio ΔmgtA::kan | 22 |
| JW1750 | Keio ΔgdhA::kan | 22 |
| JW3179 | Keio ΔgltB::kan | 22 |
| JW3180 | Keio ΔgltD::kan | 22 |
| JW0440 | Keio ΔglnK::kan | 22 |
| JW3839 | Keio ΔglnG::kan | 22 |
| JW3840 | Keio ΔglnL::kan | 22 |
| JW0162 | Keio ΔglnD::kan | 22 |
| JW3025 | Keio ΔglnE::kan | 22 |
| JW0715 | Keio ΔsucA::kan | 22 |
| PY79 | Bacillus subtilis PY79 | 29 |
| ES114 | Wild-type isolate from Euprymna scolopes | 30 |
FIG 1.
Glucose consumption is incomplete during exponential growth in buffered tryptone broth. Wild-type E. coli cells (strain BW25113) were aerated in tryptone broth buffered to pH 7 (TB7) supplemented with 4 g/liter glucose (TB7-glucose). OD600, optical density at 600 nm.
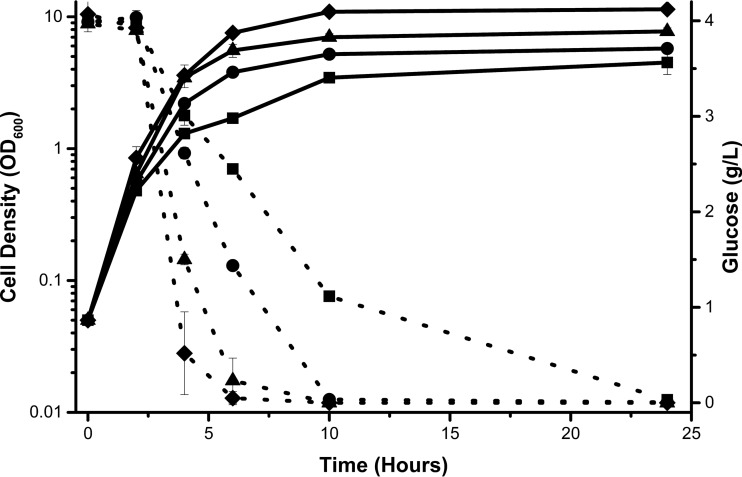
To test whether tryptone inhibits glucose consumption, we grew E. coli in M9 minimal medium supplemented with 4 g/liter glucose and various concentrations of tryptone. Instead of inhibiting glucose consumption as initially hypothesized, tryptone increased glucose consumption in a dose-dependent manner (Fig. 2). Tryptone also increased the growth rate and growth yield (i.e., final cell density). This suggests that tryptone does not inhibit glucose uptake per se but rather that some compound present in M9 minimal medium, but not in TB7, enables better cell growth.
FIG 2.
Addition of tryptone to M9 increases growth rate and glucose consumption. Wild-type E. coli cells (strain BW25113) were aerated in M9 minimal medium supplemented with 4 g/liter glucose and increasing concentrations of tryptone: 0.1% (squares), 0.2% (circles), 0.5% (triangles), and 1% (diamonds).
Magnesium increases cell growth in TB7 broth.
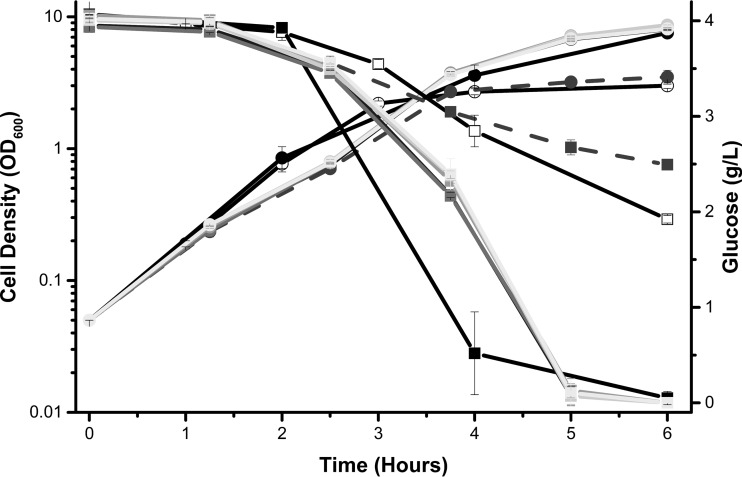
M9 minimal medium contains five compounds not present in TB7 medium: ferric citrate, calcium chloride, magnesium sulfate, thiamine, and ammonia. To determine whether the addition of these compounds would increase cell growth in TB7-glucose, we first grew the cells in TB7-glucose supplemented with four of the five compounds. We expected that if a single compound was responsible for the growth yield increase, then removing it would cause the cells to grow as if they were in TB7-glucose alone. Indeed, we found that when magnesium sulfate was not added to TB7-glucose, the cells grew as if they were in TB7-glucose alone (Fig. 3). Based on these results, we conclude that magnesium is the limiting compound.
FIG 3.
Magnesium is necessary for robust growth in buffered tryptone broth. Cell density (circles) and glucose concentration in the medium (squares) were measured hourly for wild-type E. coli cells (strain BW25113) aerated in TB7 supplemented with 4 g/liter glucose alone (open symbols), with addition of 5 M9 compounds (ferric citrate, calcium chloride, magnesium sulfate, thiamine, and ammonium chloride), or with 4 out of 5 M9 supplements (shades of gray). This was compared to cells of the same strain aerated in M9 supplemented with 4 g/liter glucose and 1% tryptone (black symbols). Only medium supplemented without magnesium sulfate (dashed lines) behaved like TB7-glucose.
While these results demonstrate that magnesium is necessary for robust growth in TB7-glucose, they do not establish that it alone is sufficient. Therefore, we tested cell growth in TB7-glucose supplemented with 1 mM magnesium sulfate. As shown in Fig. S1A in the supplemental material, the addition of magnesium to TB7-glucose increased the growth yield. We further found that the increased growth yield was, in fact, due to magnesium and not to sulfate, as supplementation with magnesium chloride increased the growth yield, whereas potassium sulfate had no effect (Fig. S1A). The magnesium effect was dose dependent, where magnesium sulfate increased the growth yield in a dose-dependent manner up to a concentration of 1 mM (Fig. S1B). We also found that supplementation with magnesium or glucose alone could exert a minor enhancement on the growth yield in TB7, but the growth yield was greatly increased in the presence of both (Fig. S1C). Buffering was not required to achieve an increased growth yield in the presence of both glucose and magnesium; however, magnesium exerted a greater effect when the medium was buffered with either phosphate or MOPS buffer (Fig. S1C).
To determine whether the effect was unique to magnesium, we also tested other divalent cations. No increases in cell growth were observed when TB7-glucose was supplemented with calcium chloride, cobalt chloride, cobalt sulfate, nickel chloride, or manganese sulfate (see Fig. S2A in the supplemental material). We also tested whether magnesium increased the growth yield in TB7 containing seven other carbon sources (lactate, succinate, fructose, lactose, arabinose, pyruvate, and acetate). In all cases, magnesium increased the growth yield in a manner similar to glucose (Fig. S2B). Thus, this magnesium effect is not specific to glucose.
Magnesium increases the growth yield in other peptide-based media.
To determine whether magnesium increases the growth yield in other peptide-based media, we tested growth in phosphate-buffered peptone (P7) and Casamino Acids (CA7), both supplemented with 4 g/liter glucose. The cells grew best in TB7-glucose medium, slightly worse in CA7-glucose medium, and very poorly in P7-glucose medium (see Fig. S3A in the supplemental material). The final cell densities achieved in these media were directly related to the magnesium content per dry weight of these peptide-based media: tryptone, 195 μg/g; Casamino Acids, 48 μg/g; and peptone, 17 μg/g (8). In all three cases, the addition of 1 mM magnesium sulfate increased the growth yield. These results indicate that the lack of magnesium limits the growth yield in these complex, buffered, peptide-based media when carbon is in excess.
We also tested whether the addition of magnesium to the commonly used medium lysogeny broth (LB) could increase the cell yield. The use of unbuffered LB supplemented with both 4 g/liter glucose and 1 mM magnesium resulted in a minor growth yield increase that was comparable to that when 1 mM magnesium alone was added (Fig. S3B). However, buffering LB with either phosphate or MOPS resulted in a greater growth yield when both glucose and magnesium were added, and again magnesium alone resulted in a partial growth yield increase (Fig. S3A and B).
Buffering permits increased growth yield by preventing growth inhibition by the metabolism by-products acetate and ammonium.
We observed that buffering our media increased the growth yield upon magnesium supplementation to TB-glucose, LB-glucose, or LB alone (Fig. S1C and S3B). Acetate produced from glucose consumption is known to acidify media and inhibit growth of E. coli. Thus, we predicted that the differences between buffered and unbuffered media supplemented with magnesium resulted from inhibitory pH levels. The pH of unbuffered LB-glucose and TB-glucose supplemented with magnesium dropped to about 5, compared to about 7 for the buffered media (Table 2). However, buffering also exerted a positive effect on growth in LB when supplemented with magnesium alone. Ammonium produced from amino acid consumption results in alkalization of the medium. Indeed, unbuffered LB supplemented with magnesium reached pH 8.7, while the buffered medium reached pH 7.5. These results indicate that magnesium is limiting in LB, but the effect is more pronounced when the LB is buffered and contains glucose.
TABLE 2.
Effect of buffering on final cell density and pH
| Supplement | TB |
TB7 |
LB |
LB7 |
||||
|---|---|---|---|---|---|---|---|---|
| ODa | pHa | OD | pH | OD | pH | OD | pH | |
| None | 2.57 ± 0.43 | 8.36 ± 0.17 | 2.55 ± 0.23 | 7.37 ± 0.04 | 4.09 ± 0.80 | 8.73 ± 0.10 | 3.88 ± 0.41 | 7.50 ± 0.03 |
| Magnesium | 3.01 ± 0.42 | 8.40 ± 0.20 | 3.76 ± 0.22 | 7.30 ± 0.01 | 5.71 ± 0.54 | 8.71 ± 0.05 | 6.90 ± 0.64 | 7.35 ± 0.14 |
| Glucose | 3.90 ± 0.45 | 5.41 ± 0.05 | 2.72 ± 0.23 | 7.10 ± 0.01 | 4.76 ± 0.58 | 6.28 ± 0.30 | 4.12 ± 0.68 | 7.16 ± 0.02 |
| Magnesium + glucose | 4.46 ± 0.83 | 4.69 ± 0.03 | 9.10 ± 0.40 | 7.02 ± 0.00 | 5.93 ± 0.79 | 5.06 ± 0.02 | 11.56 ± 0.96 | 7.26 ± 0.07 |
Averages of 9 replicates ± standard deviation.
Magnesium increases cell growth in multiple strains of E. coli and species of bacteria.
The experimental results described above were obtained using E. coli BW25113. To ensure that the magnesium effect was not limited to this particular strain, we tested the growth of the E. coli strains MC4100, MG1655, and AJW678 (Table 1) in TB7-glucose and found that magnesium increased the growth yield for all three strains (see Fig. S4A in the supplemental material). We also tested whether magnesium increased the growth yield of B. subtilis (strain PY79 [Table 1]) and V. fischeri (strain ES114 [Table 1]) grown in TB7-glucose and TB7-glucose supplemented with 2% sodium chloride, respectively. V. fischeri is a marine bacterium that must be grown under higher-salt conditions that mimic its natural seawater environment. For these bacterial species, magnesium increased growth yield, albeit to a lesser extent than for E. coli (Fig. S4B). Even in the absence of glucose, V. fischeri achieved higher biomass with magnesium. Similar results were obtained when these species were, respectively, grown in LB7-glucose and LB7-glucose supplemented with 2% sodium chloride (data not shown). The results indicate that magnesium supplementation increases the growth of diverse bacteria in complex peptide-based media.
Magnesium directs glucose to biomass formation.
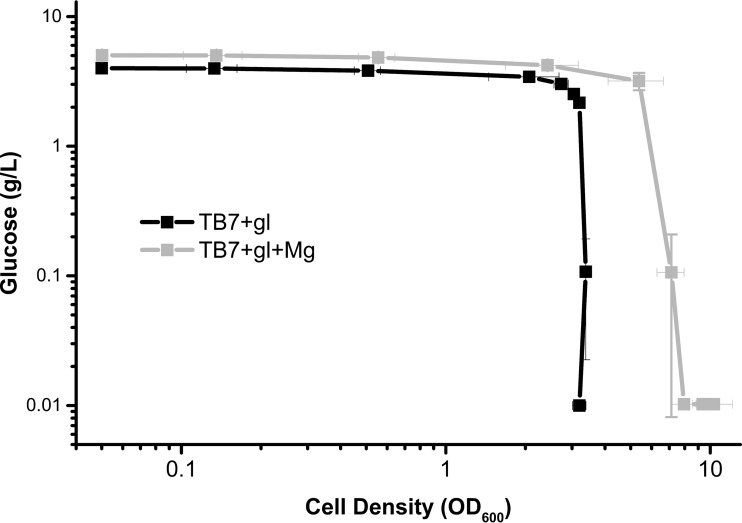
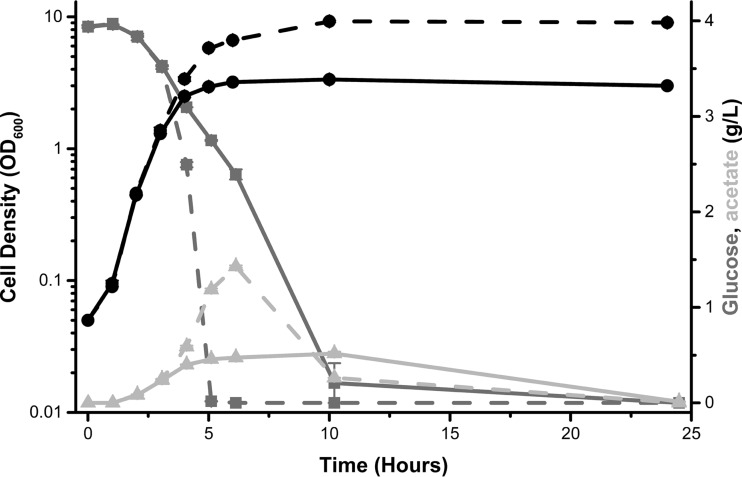
We noted that the addition of magnesium to peptide-based media supplemented with glucose did not alter the growth rate but, rather, extended the duration of exponential growth. Based on this observation, we hypothesized that magnesium depletion causes cell division to cease, thus preventing the remaining carbon from being used for cell biomass. Whereas a TB7-glucose culture supplemented with magnesium increased in optical density (OD) while consuming glucose, a TB7-glucose culture (without added magnesium) did not have increased cell growth even though it consumed glucose during stationary phase (Fig. 4). The cell density of the magnesium-supplemented culture continued to increase, even after glucose was fully consumed. This appears to be due to consumption of excreted acetate (Fig. 5). The magnesium-dependent increase in optical density resulted from both increased cell number and increased biomass (see Fig. S5A and B in the supplemental material). Magnesium-replete cells also were smaller than their magnesium-limited counterparts (Fig. S5B).
FIG 4.
Magnesium diverts carbon from glucose into biomass. Wild-type E. coli cells (strain BW25113) were aerated in TB7-glucose (black line) or TB7-glucose supplemented with MgSO4 (gray line) for 9 h. Optical density and glucose concentration measurements were taken hourly and plotted on a log-log scale.
FIG 5.
Magnesium permits full consumption of glucose prior to stationary phase. Cell density (black lines), glucose concentration (dark gray lines), and acetate concentration (light gray lines) were measured over a 24-h period for wild-type E. coli cells (strain BW25113) grown in TB7-glucose alone (solid lines) or supplemented with 1 mM MgSO4 (dashed lines).
Magnesium concentration influences protein acetylation.
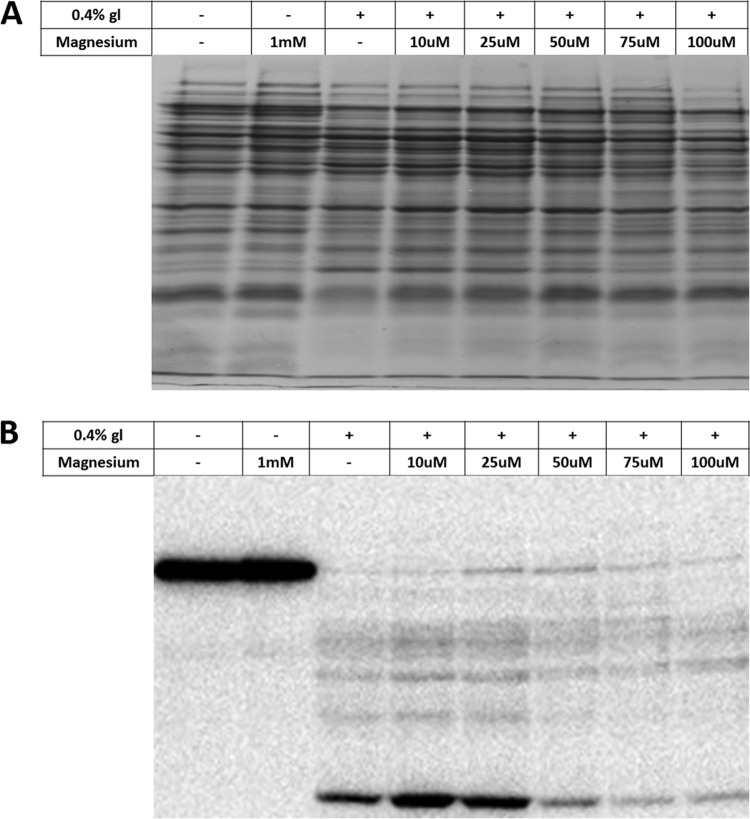
Global acetylation in E. coli is intimately linked to glucose consumption and acetate fermentation through the accumulation of acetyl phosphate, a high-energy acetyl donor (1–3). Therefore, we tested whether magnesium supplementation altered the acetylation profile of E. coli. We specifically compared the acetylation profiles of E. coli grown in TB7 (low acetylation), TB7-glucose (high acetylation), and TB7-glucose supplemented with increasing concentrations of magnesium sulfate from 10 μM to 100 μM (Fig. 6A and B; see Fig. S6 in the supplemental material). When E. coli cells were grown in TB7 alone or with magnesium supplementation, their proteins were weakly acetylated, with the exception of acetyl coenzyme A (acetyl-CoA) synthetase, which is acetylated by the acetyltransferase YfiQ (9). In contrast, when cells were grown in TB7-glucose alone, the proteins were highly acetylated, as reported previously (2, 3). However, magnesium noticeably reduced acetylation when concentrations were greater than 50 μM. As these results demonstrate, the magnesium-dependent biomass increase was met with a reduction in intensity of global acetylation, as determined by antiacetyllysine Western blot analysis. It appears that magnesium limitation promotes acetylation. We conclude that the status of magnesium in peptide-based media determines whether cells put carbon into biomass or into fermentation products, such as acetyl phosphate.
FIG 6.
Magnesium decreases acetylation in a dose-dependent manner. Wild-type E. coli cells (strain BW25113) were aerated in TB7 alone or supplemented with 0.4% glucose and/or the indicated concentrations of MgSO4 for 10 h. (A) Coomassie blue-stained SDS-polyacrylamide gel and loading control for panel B. (B) Antiacetyllysine Western blot of lysates from cells harvested from the indicated conditions at 10 h. The high-molecular-mass band observed in the absence of glucose but significantly diminished in the presence of glucose is acetyl-CoA synthetase, whose gene is repressed by glucose and whose acetylation does not depend upon acetyl phosphate.
Magnesium-dependent cell growth is lost in a ΔglnA mutant.
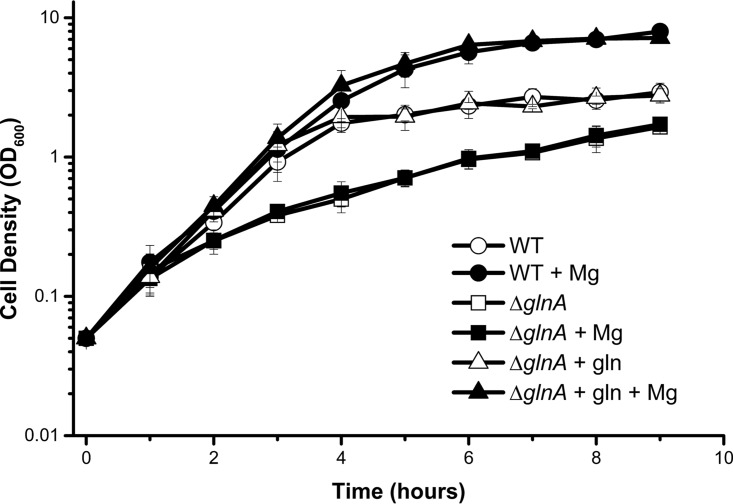
We next tested whether the loss of any genes would eliminate the magnesium effect. If magnesium-dependent cell growth depended simply on magnesium as a nutrient, then only a mutation that reduced maximum cell density would cause the effect to be lost. We tested various mutants with mutations in carbon regulation, amino acid biosynthesis, central metabolism and the tricarboxylic acid (TCA) cycle, magnesium transporters/sensors, and phosphate transporters/regulators. Of the mutants tested, only a ΔglnA mutant lost the magnesium-dependent growth yield increase (Table 3). The glnA gene encodes glutamine synthetase, and thus the ΔglnA mutant is a glutamine auxotroph (10). We hypothesized that these cells could not respond to magnesium because glutamine now was limiting. Indeed, supplementation with glutamine restored the magnesium-dependent biomass increase to ΔglnA mutants (Fig. 7). We further investigated the reason why the ΔglnA mutant does not respond to magnesium by testing mutants with mutations in glutamine-dependent pathways downstream of glnA. We found that three nucleotide biosynthesis mutants, the ΔguaA, ΔpurF, and ΔcarA mutants, also did not respond to magnesium (Table 4). We suspect that the ΔglnA mutant lacks sufficient glutamine for both amino acid and nucleotide biosynthesis and is thus limited for glutamine rather than magnesium in TB7-glucose.
TABLE 3.
Mutants tested for loss of Mg2+-dependent biomass increasea
| Category | Genes |
|---|---|
| Carbon regulators | fnr, crp, cra, arcA |
| Amino acid biosynthesis | aspA, aspC, glnA, gdhA |
| Central metabolism/TCA cycle | gltA, ackA, pta-ackA, pgi, sucA |
| Magnesium transport related | corA, mgtA, phoP |
| GlnA regulators/related | glnB, gltB, glnG, glnL, glnD, glnE, glnK |
| Phosphate related | phoB, phnA, phnC |
Mutants were grown in buffered tryptone broth supplemented with 0.4% glucose alone or with 1 mM MgSO4 for 10 h. Biomass increase was determined by OD600 measurements. Cells were considered limited for magnesium if they had increased biomass with MgSO4 supplementation relative to no supplementation. The ΔglnA mutant, highlighted in bold, did not respond. Growth defects and magnitudes of the effects are not reflected here.
FIG 7.
A glnA mutant does not respond to magnesium unless supplemented with glutamine. Wild-type (WT) E. coli cells (strain BW25113) (circles) or an isogenic glnA mutant (triangles or squares [see below]) were aerated in TB7-glucose alone (open symbols) or with 1 mM MgSO4 (closed symbols). The glnA mutant cells were supplemented with 1.4 g/liter glutamine (triangles) or left untreated (squares).
TABLE 4.
Mutants with mutations related to glnA tested for loss of Mg2+-dependent biomass increasea
| Category | Genes |
|---|---|
| Responded to magnesiumb | ybaS, yneH, asnB, hisH, trpE, crr, ptsG |
| No responsec | guaA, purF, purL, carB, carA, ptsH, ptsI |
Mutants were grown in buffered tryptone broth supplemented with 0.4% glucose alone or with 1 mM MgSO4 for 24 h. Biomass was determined by OD600 measurements.
By 9 h of growth, mutants that had increased biomass with magnesium supplementation were considered responders.
Mutants in bold did not respond even after 24 h; other mutants responded between 9 and 24 h.
DISCUSSION
During growth on buffered, peptide-based media (e.g., TB7) supplemented with a sugar (e.g., glucose), cultures of E. coli entered stationary phase prior to complete consumption of the sugar. The majority of the sugar was consumed after the culture stopped growing. This behavior provides a simple explanation for why protein acetylation occurs primarily during stationary phase, because that is when acetyl phosphate, the major acetyl donor in E. coli (1, 2), is produced as an intermediate of the acetate fermentation pathway (4). When we supplemented TB7-glucose with magnesium, exponential growth was extended, resulting in consumption of glucose prior to entry into stationary phase, increased biomass, and decreased protein acetylation. We observed the same general behavior in other peptide-based media containing glucose and in two other bacterial species, Vibrio fischeri and Bacillus subtilis. Collectively, these results further our understanding of bacterial growth in complex media and enable us to control global acetylation levels in E. coli.
In Gram-negative bacteria such as E. coli, magnesium is involved in stabilization of membrane phospholipids, lipopolysaccharide, polyphosphate compounds like DNA and RNA, and the ribosome (11–13). Magnesium is also required to make ATP biologically active and participates in catalysis of certain enzymatic reactions through either direct or indirect mechanisms (14, 15). Almost half of total magnesium is found associated with nucleoside triphosphates (NTPs) (primarily ATP) (11), approximately one-third is associated with the cell wall (13), and the remaining is bound to various molecules or free in the cytoplasm. Almost a quarter of this cytoplasmic magnesium is thought to stabilize ribosomes (16). As such, magnesium deprivation can result in cessation of translation due to dissociation of ribosomes (16). Under the magnesium-starved conditions found in complex peptide-based media, it would not be surprising for ribosome control to be a mechanism by which cells cease growth and division.
Hiroshi Nikaido proposed that the amount of magnesium in LB is not sufficient to achieve the maximum possible cell growth (17). LB contains between 30 and 200 μM magnesium. In contrast, the magnesium concentration is 528 μM in MOPS medium, a defined minimal medium that avoids the excess of any one ionic species (18). Our data suggest that magnesium is limiting in LB. A minor increase in growth yield was achieved in unbuffered LB supplemented with magnesium, while buffered LB produced a much more pronounced effect. The results were similar for TB, where the addition of magnesium produced a minor effect that was enhanced upon buffering. It is likely that yeast extract provides LB with extra carbon, which allows for increased growth upon magnesium supplementation. However, when both carbon and magnesium were added together to buffered or unbuffered TB or LB, we observed a substantial increase in the cell yield, which then becomes limited due to some other factor(s).
LB medium has been reported to be limited for carbon by Sezonov and coworkers (19). In that study, glucose supplementation of cell-free, spent LB medium was shown to support further cell growth upon reinoculation with E. coli. We observed that magnesium supplementation to LB or LB7 was able to permit additional growth, while glucose addition did not. The differences between this study and that of Sezonov and coworkers may be due to possible variation in different preparations of LB. LB can vary between different tryptone and yeast extract preparations from different companies. Thus, our LB medium may have contained a larger amount of carbon, creating a more balanced carbon/magnesium ratio, whereas their LB medium may have been relatively more magnesium rich.
In an attempt to find genes that may mediate the magnesium-dependent growth yield increase, we sought mutants that did not respond to added magnesium and identified the ΔglnA mutant. We determined that the ΔglnA mutant did not respond to magnesium because it is a glutamine auxotroph (10). Glutamine is the scarcest amino acid in tryptone, at 0.1% (8); supplementation of TB7 with glutamine permitted the ΔglnA mutant to respond to magnesium. Glutamine is a required precursor for both protein biosynthesis and nucleotide biosynthesis. Presumably, TB7-glucose does not contain sufficient glutamine for both needs in the ΔglnA mutant, and thus its growth was limited by glutamine rather than by magnesium. Supporting this contention, we found that genes required for nucleotide biosynthesis downstream of glutamine also could not respond to magnesium. Would this have been true for other amino acid auxotrophs? Presumably not, because the other amino acids are present in much higher concentrations than glutamine in tryptone.
The magnesium-dependent biomass increase was not exclusive to E. coli but also was found to occur in Bacillus subtilis, a Gram-positive bacterium, and Vibrio fischeri, another Gram-negative bacterium. Both achieved higher optical density when magnesium was supplemented to TB7-glucose. We predict that magnesium limits growth for multiple bacteria in peptide-based media. Interestingly, V. fischeri achieved increased optical density from magnesium supplementation even without the additional glucose. This suggests the possibility that V. fischeri may be able to utilize amino acids for growth more efficiently than E. coli or B. subtilis or that V. fischeri simply requires more magnesium for growth. O'Shea and coworkers reported that V. fischeri grown in TBS (1% tryptone with 342 mM NaCl) did not benefit from addition of MgSO4 but did benefit from KCl (20). As we utilized TBS7, which is the same medium but buffered with potassium phosphate, our results suggest that V. fischeri may experience potassium limitation, then magnesium limitation, and finally carbon limitation in TBS. This need for high magnesium may reflect the naturally high magnesium concentration (∼50 mM) of seawater in which V. fischeri lives (20).
Finally, we found that magnesium reduces protein acetylation in E. coli. Most acetylation in E. coli is mediated by acetyl phosphate (1, 2), an intermediate in the Pta-AckA acetate fermentation pathway (4), which is induced by glucose (3). Indeed, the vast majority of glucose-induced acetylation depends on acetyl phosphate (3). How does magnesium decrease acetylation? Under magnesium-limiting conditions (TB7-glucose), most glucose is not consumed until stationary phase. Stationary-phase cells do not generate biomass as readily as those cells in exponential phase, resulting in a buildup of acetyl-CoA. To regenerate CoA for further metabolism of glucose, the cell ferments acetate and generates acetyl phosphate (4). Thus, continued metabolism of glucose in a stationary-phase culture with a relatively stable pool of susceptible proteins results in protein acetylation. In contrast, we envision two ways that magnesium could reduce fermentation. First, magnesium supplementation to TB7-glucose allows cells to convert more carbon into biomass as glucose is consumed before the cells reach stationary phase. Diversion of carbon into biomass must reduce the amount of carbon entering acetate fermentation, thus reducing the amount of acetyl phosphate available to acetylate proteins. Second, magnesium supplementation prolongs the exponential phase of growth. During this phase, acetylated proteins are constantly diluted by the synthesis of nascent unmodified proteins and cell growth. In stationary phase, when acetylation would normally accumulate, little carbon remains to be fermented and less acetyl phosphate is available to donate acetyl groups to proteins. It was previously shown that a carbon-to-nitrogen imbalance could increase acetylation (1); in this work, a carbon-to-magnesium imbalance produced the same effect. We propose that any carbon/nutrient imbalance (e.g., a carbon-to-oxygen imbalance) will have the same effect, with the excess carbon being excreted as acetate, resulting in acetylation.
We conclude with the following two observations. First, there is hidden potential in peptide-based media. The addition of excess carbon and magnesium can increase the final cell density anywhere from 2-fold to 17-fold. The addition of magnesium alone could also increase the final cell densities. Buffering the culture is important to ensure that the medium does not become overly acidic or alkaline due to the consumption of glucose and amino acids, respectively. Second, we can now tune the level of acetylation in cultures without using genetic manipulation. Those who use E. coli for recombinant protein production may benefit most from this work. Each acetylated isoform of a recombinant protein has the potential to have different biological implications. To reduce the number of acetylated isoforms, excess carbon should be avoided (21), and the growth medium should be buffered and supplemented with magnesium.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study are listed in Table 1. For growth experiments using E. coli and B. subtilis, the bacteria were aerated at 37°C. For growth experiments using V. fischeri, the bacteria were aerated at 28°C. The Keio collection was used to screen for mutant genes of interest. Derivatives were constructed by moving the appropriate deletions from the Keio collection (22) by generalized transduction with P1kc, as described previously (23). The following media were used for cell growth: LB (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl), M9 minimal medium (11.3 g/liter M9 salts [5×], 0.1 mM CaCl2, 1 mM MgSO4, 2.45 μM ferric citrate, and 0.03 mM thiamine), TB (10 g/liter tryptone and 5 g/liter NaCl), TB7 (10 g/liter tryptone buffered at pH 7.0 with 100 mM potassium phosphate), P7 (10 g/liter peptone buffered at pH 7.0 with 100 mM potassium phosphate), CA7 (10 g/liter Casamino Acids buffered at pH 7.0 with 100 mM potassium phosphate), LB7 (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl buffered at pH 7.0 with 100 mM potassium phosphate), TB7M (10 g/liter tryptone buffered at pH 7.0 with 50 mM MOPS), and LB7M (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl buffered at pH 7.0 with 50 mM MOPS). Sugars were added to the growth media at a concentration of 4 g/liter, unless noted otherwise. In media containing magnesium, MgSO4 was added to a final concentration of 1 mM, unless noted otherwise.
Analytical methods.
Cell growth was measured by optical absorbance at 600 nm. Dry cell weights were determined by vacuum filtration of 4 ml of culture through predried and preweighed nitrocellulose membranes. The membranes were then washed with 4 ml of autoclaved Milli-Q water, allowed to dry at room temperature overnight, and incubated at 80°C for 20 min before being weighed on an analytical balance to observe changes in mass. Membranes were kept in a calcium sulfate desiccator between each step to avoid ambient moisture absorption.
Glucose and acetate concentrations were measured using a Shimadzu high-performance liquid chromatography system equipped with a RID-10A refractive index detector, an Aminex HPX-87H carbohydrate analysis column (Bio-Rad Laboratories, Hercules, CA, USA), and a cation H microguard cartridge (Bio-Rad Laboratories). The column and guard cartridge were kept at 65°C, and 0.5 mM H2SO4 was used as a mobile phase at a constant flow rate of 0.5 ml/min. Prior to analysis, culture samples were first pelleted, and then the supernatant was passed through a 0.22-μm polyethersulfone syringe filter. Peaks were identified and quantified by retention time comparison to authentic standards.
Western blot analysis of protein acetylation.
Pelleted cells were lysed using BugBuster protein extraction reagent (Novagen, Merck Millipore, Billerica, MA, USA). The amount of cell lysate loaded on the gel was normalized to the total protein concentration, as determined by the bicinchoninic acid (BCA) assay (Thermo Scientific Pierce, Waltham, MA, USA). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and normalization was verified by Coomassie staining. Protein acetylation was determined using a rabbit polyclonal antiacetyllysine antibody (Cell Signaling) at a dilution of 1:1,000, as described previously (2, 3). All experiments were performed in triplicate (see Fig. S6 in the supplemental material).
Microscopy and image analysis.
For cell size analysis, 10 μl of culture was mounted onto plain microscope slides (Thermo Fisher Scientific, Waltham, MA, USA) with cover glass. Transmitted-light differential interference contrast (TL-DIC) microscopy images were captured on a Leica AF7000 wide-field fluorescence microscope fitted with a sCMOS PCO.EDGE 5.5 camera, automated stage, and adaptive focus control, and an APO PH3 100×/1.40 oil immersion objective was used (Leica/Nuhsbaum Inc., McHenry, IL).
Supplementary Material
ACKNOWLEDGMENT
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under award number DE-SC0012443.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03034-16.
REFERENCES
- 1.Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, Sorensen DJ, Zemaitaitis B, Gibson BW, Wolfe AJ. 2015. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol 98:847–863. doi: 10.1111/mmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enjalbert B, Letisse F, Portais JC. 2013. Physiological and molecular timing of the glucose to acetate transition in Escherichia coli. Metabolites 3:820–837. doi: 10.3390/metabo3030820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prüss BM, Wolfe AJ. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol 12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 7.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.BD. 2006. BD bionutrients technical manual: advanced bioprocessing, 3rd ed BD, Sparks, MD. [Google Scholar]
- 9.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Backman K, Chen YM, Magasanik B. 1981. Physical and genetic characterization of the glnA–glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A 78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coughlin RT, Tonsager S, McGroarty EJ. 1983. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry 22:2002–2007. doi: 10.1021/bi00277a041. [DOI] [PubMed] [Google Scholar]
- 13.Groisman EA, Kayser J, Soncini FC. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol 179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan JA. 1998. Metal activation of enzymes in nucleic acid. Biochem Chem Rev 98:1067–1088. doi: 10.1021/cr960436q. [DOI] [PubMed] [Google Scholar]
- 15.Sreedhara A, Cowan JA. 2002. Structural and catalytic roles for divalent magnesium in nucleic acid biochemistry. Biometals 15:211–223. doi: 10.1023/A:1016070614042. [DOI] [PubMed] [Google Scholar]
- 16.Pontes MH, Sevostyanova A, Groisman EA. 2015. When too much ATP is bad for protein synthesis. J Mol Biol 427:2586–2594. doi: 10.1016/j.jmb.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H. 2009. The limitations of LB medium. http://schaechter.asmblog.org/schaechter/2009/11/the-limitations-of-lb-medium.html Accessed 28 September 2016.
- 18.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Shea TM, Deloney-Marino CR, Shibata S, Aizawa S, Wolfe AJ, Visick KL. 2005. Magnesium promotes flagellation of Vibrio fischeri. J Bacteriol 187:2058–2065. doi: 10.1128/JB.187.6.2058-2065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szewczak J, Bierczyńska-Krzysik A, Piejko M, Mak P, Stadnik D. 2015. Isolation and characterization of acetylated derivative of recombinant insulin Lispro produced in Escherichia coli. Pharm Res 32:2450–2457. doi: 10.1007/s11095-015-1637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor, Cold Spring Harbor, NY. [Google Scholar]
- 24.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiGiuseppe PA, Silhavy TJ. 2003. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol 185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guyer MS, Reed RE, Steitz T, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spr Harb Symp Quant Biol 45:135–140. [DOI] [PubMed] [Google Scholar]
- 27.Kumari S, Beatty CM, Browning DF, Busby SJ, Simel EJ, Hovel-Miner G, Wolfe AJ. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol 182:4173–4179. doi: 10.1128/JB.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youngman P, Perkins JB, Losick R. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1–9. [DOI] [PubMed] [Google Scholar]
- 30.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.