Abstract
Background
Mucinous adenocarcinoma of the salivary gland (MAC) is a lethal cancer with unknown molecular etiology and a high propensity to lymph node metastasis. Mostly due to its orphan status, MAC remains one of the least explored cancers that lacks cell lines and mouse models that could help translational and pre-clinical studies. Surgery with or without radiation remains the only treatment modality but poor overall survival (10-year, 44%) underscores the urgent need for mechanism-based therapies.
Methods
We developed the first patient-derived xenograft (PDX) model for pre-clinical MAC studies and a cell line that produces aggressively growing tumors after subcutaneous injection into nude mice. We performed cytogenetic, exome, and proteomic profiling of MAC to identify driving mutations, therapeutic targets, and pathways involved in aggressive cancers based on TCGA database mining and GEO analysis. Results: We identified in MAC KRAS (G13D) and TP53 (R213X) mutations that have been previously reported as drivers in a variety of highly aggressive cancers. Somatic mutations were also found in KDM6A, KMT2D, and other genes frequently mutated in colorectal and other cancers: FAT1, NBEA, RELN, RLP1B, and ZFHX3. Proteomic analysis of MAC implied epigenetic up-regulation of a genetic program involved in proliferation and cancer stem cell maintenance.
Conclusion
Genomic and proteomic analyses provided the first insight into potential molecular drivers of MAC metastases pointing at common mechanisms of CSC propagation in aggressive cancers. The in vitro/in vivo models that we created should aid in the development and validation of new treatment strategies against MAC.
Keywords: Cancer, Salivary mucinous adenocarcinoma, Metastases, Mutations, KRAS, TP53, KDM6A, KMT2D, Cancer stem cells
Introduction
While mucinous adenocarcinomas occur more frequently in the colon, appendix, breast, lung and ovary, mucinous adenocarcinoma of the salivary gland (MAC) represents only 0.1% of all salivary gland tumors. MAC is a rare cancer: only 73 MAC cases have been registered in the U.S. from 1998 to 2012 [1–3]. The majority of salivary MAC present with cervical nodal metastases (63%) and primary treatment is surgical excision, nodal dissection, with or without post-operative radiation therapy. Despite this aggressive treatment regimen, 10-year survival is only 44% [3]. Because of its orphan status, randomized clinical trials for MAC are not feasible and collection of tumor specimens for clinical research is rare, making salivary MAC one of the least studied cancers with yet unknown molecular drivers. Lack of MAC cell lines or patient-derived xenograft (PDX) models further complicates development of targeted therapies. To catalog molecular defects that may contribute to its MAC aggressive behavior and to identify potential therapeutic targets, we created the first MAC PDX model and cell line from a metastatic lymph node deposit. Cytogenetic analysis, exome sequencing, and shotgun proteomic studies revealed multiple chromosomal and molecular abnormalities that may contribute to aggressive behavior possibly via epigenetically regulated cancer stem cells (CSC).
Materials and methods
Clinical specimen
A high grade tumor with metastases to 3 out of 22 lymph nodes (largest node ~4.8 cm) was removed by parotidectomy and neck dissection in Vanderbilt-Ingram Cancer Center (WGY). A portion of the resected lymphatic metastases was collected after informed consent (IRB#030062, WGY), and diagnosed as a high-grade mucinous adenocarcinoma by a board-certified pathologist (AKE).
Mouse xenograft model
Athymic nude Foxn1 mice (Harlan Laboratories, Indianapolis, IN) were maintained in accordance with the Institutional Animal Care and Use Committee guidelines (Yale IACUC#12-11510). Male mice between 4 and 6 weeks of age were housed on irradiated corncob bedding (Harlan Laboratories) in individually ventilated cages on a 12-h light–dark cycle at 70–74 1F (21–23 1C) and 40–60% humidity. Mice were fed water ad libitum (reverse osmosis, 2 ppm Cl2) and an irradiated standard rodent diet (Teklad 2919) consisting of 18% protein, 5% fat and 4% fiber. To generate patient-derived xenografts (PDX), unprocessed tumor specimens were briefly washed with 70% ethanol, cut into ~30–50 mm3 pieces and implanted subcutaneously in the shoulder area of anesthesized animals using sterile tools and staples for wound closure.
Primary cell culture
Resected tumor specimen was washed with PBS and digested for 30 min at 37 °C in serum-free KGM media (Life Technologies, Carlsbad, CA) that contained trypsin (0.05%) and 1 mg/ml collagenase from Clostridium histolyticum (Sigma-Aldrich, St. Louis, MO). FBS (Life technologies) was then added to the mixture (final conc. 10%) to quench trypsin and cells were pelleted by centrifugation (400g, 5 min) and washed twice with PBS. The cells were then resuspended in KGM with 10% FBS and 1X Antibiotic-Antimitotic (Invitrogen, Carlsbad, CA) and incubated overnight in collagen-coated plates. Attached cells were washed three times with PBS and incubated in antibiotic/antimitotic serum-free KGM media to produce primary culture. After 3 passages in serum-free media, fibroblast-free cells were transferred into KGM media supplemented with serum.
Microsatellite analysis
Using Promega GenePrint 10 STR analysis PCR kit with fluorescent tagging, PCR products were analyzed on Applied Biosystems 3730xL DNA Analyzer at the Yale Keck Facility and data processed using GeneMapper 3.7 (Applied Biosystems) software. The results were then compared to STR databases (http://www.cstl.nist.gov/strbase/) and (http://www.dsmz.de/services/services-human-and-animal-cell-lines/online-str-analysis.html).
Cytogenetics
G-banding and spectral karyotyping (SKY) analyses were performed as previously described [4]. Composite karyotype was generated based on all clonally occurring chromosome abnormalities in 10 analyzed metaphase spreads.
Molecular profiling
Whole exome sequencing was done at the New Generation Sequencing Core at Vanderbilt University Medical Center Core using Illumina Hi Seq 2000 with SureSelect All-Exon Target Enrichment System. Shotgun proteomic analysis was performed by first suspending the samples in LDS sample buffer (Life Technologies), resolving the proteins approximately 1 cm using a 10% Novex pre-cast gel, and then performing in-gel tryptic digestion to recover peptides. These peptides were analyzed via MudPIT (Multidimensional Protein Identification Technology) essentially as previously described [5,6]. Briefly, digested peptides were loaded onto a biphasic pre-column consisting of 4 cm of reversed phase (RP) material followed by 4 cm of strong cation exchange RP material. Once loaded, this column was placed in line with a 20 cm RP analytical column packed into a nanospray emitter tip directly coupled to a linear ion trap mass spectrometer (LTQ). For a total of 11 salt steps, a subset of peptides was eluted from the SCX material onto the RP analytical via a pulse of volatile salt, those peptides separated by an RP gradient, and then ionized directly into the mass spectrometry where both the intact masses (MS) and fragmentation patters (MS/MS) of the peptides were collected. These peptide spectral data were searched against a protein database using Sequest and the resulting identifications collated and filtered using IDPicker and Scaffold (http://www.proteomesoftware.com). Relative protein abundances were evaluated via spectral counting techniques using the Quasitel program for P-value calculations.
Computational gene expression analysis
Expression array studies were performed using Multi-Experiment Matrix software [7,8] available at http://biit.cs.ut.ee/mem/index.cgi.
Results
Generation of a PDX model for metastasizing salivary mucinous adenocarcinoma (MAC)
Patient-derived xenograft models (PDX) produced via implantation of minimally processed tumor tissue into immunodeficient mice are emerging as a novel platform for clinical research that maintains tumor heterogeneity and faithfully recapitulates tumor response to treatment. PDX are also used for studying cancer stem cells (CSC) that have been linked to therapy resistance, metastases, and recurrence [9–11].
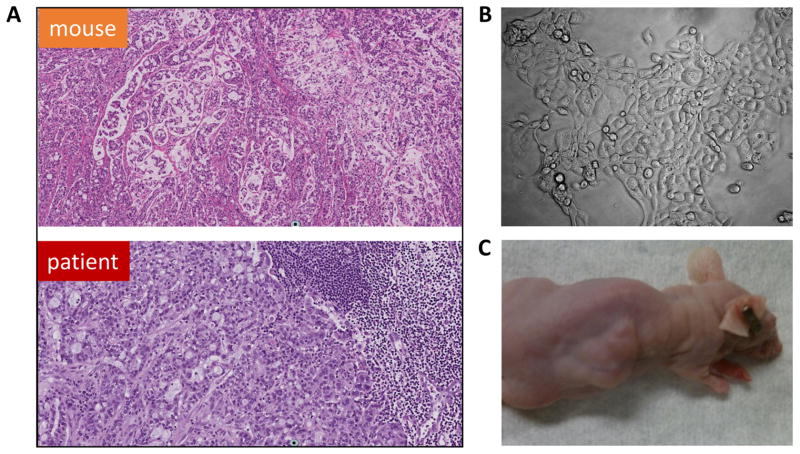
To produce a PDX model, fragments of tumor extracted from a patient’s lymph node were minced and subcutaneously implanted into the flanks of 3 nude mice. Within two months, all animals grew tumors, which rapidly developed lobulated morphology and required frequent (monthly) passages into additional mice due to its aggressive growth. Comparative H&E staining demonstrated that PDX tissue recapitulated the histology of the original tumor (Fig. 1A).
Fig. 1.
PDX and primary cell culture MAC models derived from a lymph node biopsy. (A) H&E staining of the original and grafted tumors (×100); (B) cultured MAC cells (×400); (C) tumor produced in nude mice via s.c. injection of cultured MAC cells.
Establishment of cell culture from MAC tissue
The metastatic MAC specimen was enzymatically digested producing cells that readily adhered and grew well on collagen-coated cell culture plates (Fig. 1B). After several passages, the cells were transferred into uncoated cell culture plates, maintained in DMEM media supplemented with 10% FBS, and passaged >30 times without signs of cell distress, senescence, or death. Short Tandem Repeat (STR) analyses of cultured MAC cells matched a DNA specimen isolated from patient’s blood except for the loss of a Y chromosome marker, AMEL, in cultured cells (Suppl. Table 1). This observation was consistent with the cytogenetic absence of the Y chromosome in cultured cells (see below). Subcutaneous injections of these cells into nude mice at different passages were consistently tumorigenic generating aggressively growing lobulated tumors (Fig. 1C).
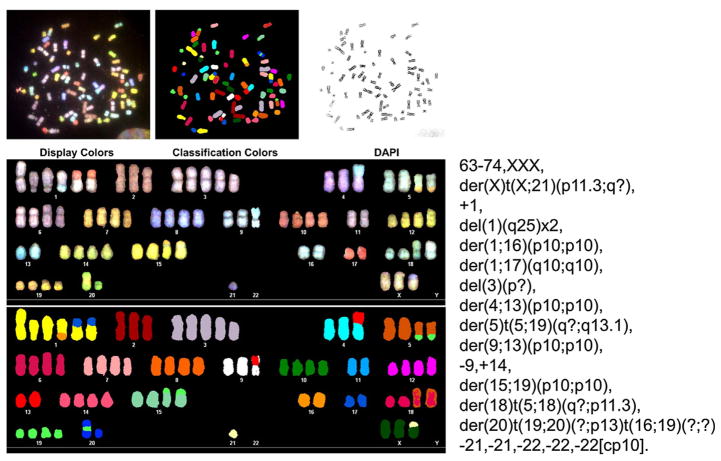
Chromosomal abnormalities detected in MAC cells
Chromosomal painting revealed a highly abnormal nearly triploid karyotype with multiple rearrangements that involved chromosomes X, 1, 4, 5, 9, 13, 15, 16, 17, 18, 19, and 20. Losses of chromosomes 1q25, 3(p?), 9, 21, and 22 were also detected (Fig. 2). The loss of chromosome Y in cultured cells suggested by microsatellite typing (see above) was confirmed karyotypically. Overall, this karyotype was consistent with chromosomal instability observed in highly aggressive cancers.
Fig. 2.
Cytogenetic SKY analysis of cultured MAC cells.
The mutational landscape of MAC
Exome sequencing of cultured MAC cells identified 1170 non-synonymous and 69 stop-gain substitutions as well as 37 insertions and deletions indicative of high genome instability (Suppl. Table 2). Among all mutated genes, 10 potentially driving mutations that have been reported as recurrent or frequent in other cancers were identified (Table 1). Among mutations pivotal for understanding the molecular basis of MAC aggressiveness were the stop-codon-generating (‘‘nonsense”) substitution R213X in TP53 and the ‘‘hot-spot” G13D KRAS mutation. Both Ras and p53 mutations were confirmed by Sanger re-sequencing performed after amplification of message from cultured MAC cells.
Table 1.
Potential driver mutations identified in MAC via pair-end Exome sequencing.
| Gene | Alteration | Alleles in blood/reads | Alleles in tumor cells/reads | Predominant in cancers | Mutation type |
|---|---|---|---|---|---|
| FAT1 | K3714X | 0/0:236, 0:236 | 0/1:101,103:204 | Head and neck squamous, diffuse large B-cell lymphoma | Mostly nonsense/truncating |
| FAT1 | E3653X | 0/0:260,1:262 | 0/1:117,83:200 | ||
| KDM6A | W1258X | 0/0:16,0:16 | 1/1:0,13:13 | Bladder | Mostly nonsense/truncating |
| KRAS | G13D | 0/0:92,0:92 | 0/1:41,32:73 | Pancreatic, colorectal | Recurrent hotspot in colorectal cancers and multiple myeloma |
| KMT2D | S2438X | 0/0:30,0:30 | 0/1:14,18:32 | Bladder | Mostly nonsense/truncating |
| NBEA | R696X | 0/0:12,0:12 | 0/1:3,9:12 | Melanoma, colorectal, etc. | Mosly missense |
| RELN | G2110E | 0/0:57,0:58 | 0/1:39,43:82 | Melanoma, lung, etc. | Mostly missense |
| TP53 | R213X | 0/0:38,0:38 | 1/1:0,19:19 | Colorectal, breast | Recurrent in colorectal cancers |
| LRP1B | W3334X | 0/0:76,0:76 | 0/1:39,34:74 | Melanoma, lung, etc. | Mostly missense |
| ZFHX3 | T3055A | 0/0:113,0:113 | 0/1:48,59:107 | Multiple | Missense and frameshift/truncating |
0 – wild type allele, 1 – mutated allele, 0/0 – wild type homozygous state, 0/1 – heterozygote, 1/1 – mutation in a homozygous or LOH state; 0/1:101,103:204 – a mutation in a heterozygous state as based on 101 wild type and 103 mutation reads, 204 reads in total.
R213X, the most frequent nonsense p53 mutation, is recurrent in esophageal, kidney chromophobe, stomach, colorectal cancers, and melanoma with an average frequency of 3–4% (www.cbiopor-tal.org). Interestingly, aminoglycosides, such as G418, have been shown to induce read-through of the c.637C > T missense (R213X) substitution and expression of full-length p53 [12]. We observed a similar G418 effect on cultured MAC cells: while no wild-type p53 protein was observed in these cells without treatment, incubation with G418 produced full-length p53 (Suppl. Fig. 1). As clinical use of G418 is limited by its toxicity, this finding stimulates development of less toxic read-through-inducing compounds for future MAC therapies.
Analysis of the MAC proteome reveals activation of epigenetically regulated genes involved in proliferation, DNA repair, and CSC maintenance
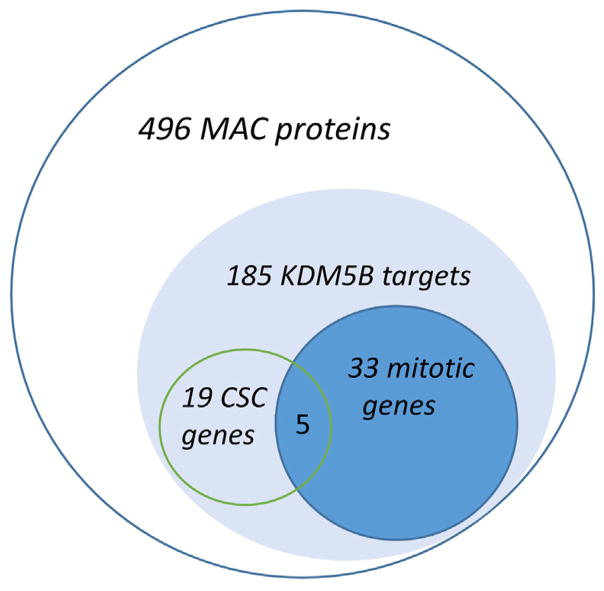
To identify potential MAC drivers at the protein level, cultured MAC cells were compared with benign salivary adenoma and normal salivary gland cells in a ‘‘shotgun” proteomic study. We selected 496 proteins expressed exclusively in MAC (Suppl. Table 3) and assessed their biological activities via gene set enrichment analysis (GSEA) using Enrichr (http://amp.pharm.mssm.edu/Enrichr/). The most remarkable enrichment was found using ChEA and ENCODE databases. Specifically, large and highly significant overlaps were detected with targets of KDM5B, an H3K4-specific methyltransferase (n = 185 or 37%, p = 0) and genes that bear H3K4 methylation marks (n = 158 or 32%, p < 5.6E–17) (Suppl. Table 3 and data not shown), consistent with activation of a large set of genes controlled via H3K4. We also observed significant enrichment with binding sites for MYC, E2F4, MYBL2, CREM, FOXM1, and other transcription factors that have been previously associated with epigenetic regulation and CSC stimulation. To determine possible biological implications of this finding, we performed GSEA study of the list of 185 MAC genes associated with KDM5B against the GO Biological Process database. Remarkably, this study demonstrated that ~21% of these H3K4-dependent genes are involved in mitotic cell cycle regulation (n = 33, p < 4E–18). Other overlaps with robust p values were represented by genes from functionally overlapping fields related to cell division, DNA repair, and chromatin remodeling (E–13 < p < E–4) (Suppl. Table 3). These results suggested that genes regulated via H3K4 methylation may drive MAC proliferation.
Since high tumorigenicity in xenografted MAC is consistent with CSC properties, we then asked if CSC-stimulating genes can be identified among H3K4-dependent MAC genes. Based on our analysis of stem cell datasets collected at http://stemcellnet.sysbi-olab.eu/ and literature mining, we detected among the 185 KDM5B targets 19 well-established regulators of embryonic stem cells (Table 2 and Fig. 4). One of the most interesting of them was an H3K36-specific dimethyltransferase, ASH1L, recently reported to control embryonic and hematopoietic stem cells [13,14]. As significance of ASH1L for solid cancers is not fully appreciated, we interrogated TCGA data collected from 123 cancer genomic studies for DNA copy number variation in the ASH1L gene area. Remarkably, ASH1L was amplified in 34% of neuroendocrine prostate cancers, 13.5% of lung, 17.9% of uterine, 16.6% of liver, and 13.8% of pancreatic cancers (http://www.cbioportal.org/) suggesting that ASH1L is a common oncogene involved in progression of many solid cancers. Two additional CSC-linked factors found in the MAC-185 list were serine/threonine protein kinase BUB1 and abnormal spindle micro-tubule assembly ASPM. Both of them play key roles in CSC proliferation. BUB1 co-operates with BUB1B in phosphorylation of mitotic checkpoint complex members and activating the spindle checkpoint [15], while ASPM regulates spindle organization and rotation [16]. BUB1 and ASPM functions appear to be essential for asymmetrically dividing CSC and thus are appealing as targets for selective CSC elimination [16–18]. Our analysis performed on hundreds of public cancer datasets (https://www.oncomine.org, http://biit.cs.ut.ee/mem) demonstrated that BUB1 and ASPM are overexpressed in a great variety of solid cancers within a highly conserved and substantial gene signature (>500 genes, p < E–70) (data not shown). Remarkably, in addition to BUB1 and ASPM we identified in this gene signature 32 more genes from the same MAC-185 list (highlighted in Suppl. Table 3). Importantly, many of these genes have been individually implicated in cytokinesis, spindle checkpoint, chromosomal instability, and poor prognosis [19–23]. This finding suggests that MAC and other cancers coordinately activate epigenetically regulated genes that promote proliferation.
Table 2.
List of MAC proteins/genes previously linked to cancer stem cells.
| Gene name | Activity | PubMed ID | Inhibitors |
|---|---|---|---|
| ASH1L | H3K36 dimethyltransferase activity involved in hematopoietic SC maintenance | 25866973 | |
| ASPM | Key CSC driver in glioblastoma | 17090670 | |
| AURKB | Maintains tumor-initiating cells in neuroblastoma | 20651058 | AZD1152 |
| BRD4 | Co-operates with Nanog in maintaining ES pluripotency and stimulates CSC | 25146928, 24525235 | BET inhibitors |
| BUB1 | Required for maintenance of breast CSC | 26522589, 25564677 | 2OH-BNPP1 |
| HELLS/LSH | ATP-dependent nucleosome remodeling in ES cells | 25578963 | |
| DNMT1 | Essential for mammary and cancer stem cell maintenance | 25908435 | |
| EED | A polycomb group protein essential for pluripotency | 24457600, 25369470 | Astemizole |
| MCM3 | Protects hematopoietic SC cell from replication stress | 26456157 | |
| KIF11 | Driver of self-renewal in glioblastoma | 26355032 | |
| MYEF2 | Maintains hematopoietic SC | 22801375 | |
| PARP1 | Involved in DNA damage response in glioblastoma stem-like cells | 26282173 | olaparib, etc. |
| NASP | A hub gene involved in ES cell fate determination via histone transport into the nucleus of dividing cells | 18923680 | |
| SMARCAD1 | A part of regulatory network of differentiating ES cells | 20019792 | |
| RNF2 | A polycomb group U3 ubiquitin ligase essential for maintanance of ES cells | 18493325 | PRT 4165 |
| SIN3A | Transcriptional silencing of pluripotency genes | 23217425, 24763403 | Fasoracetam |
| TOP2A | DNA topoisomerase involved in chromosome condensation, chromatid separation, and the relief of torsional stress that occurs during DNA transcription and replication in pluripotent stem cells | 25377277 | PluriSIn#2 |
| WWP2 | An E3 ubiquitin ligase for PTEN involved in embryonic stem fate determination | 21532586 | |
| XAB2 | Transcription-coupled DNA repair and silencing of pluripotency genes | 18832051 |
Fig. 4.
GSEA analysis of proteomic data reveals enrichment of 496 MAC-specific proteins with H3K4 methylation-dependent KDM5B targets as well as mitotic and cancer stem cell-associated gene products.
Discussion
Mucinous adenocarcinoma of the salivary gland (MAC) is a highly aggressive cancer that rapidly spreads to lymph nodes. While pre-metastatic growth is relatively slow (>2 years), timely MAC diagnosis is difficult and controversial due to the lack of molecular markers, and 60% of patients are diagnosed at an advanced stage. Without effective therapy, about half of MAC patients die of tumor within 6 years [1]. However, generation of MAC cell lines and mouse models for identification of major molecular drivers has been challenging due to the fact that MAC is an orphan tumor.
In this study, we developed, for the first time, a patient-derived xenograft (PDX) MAC model from a metastatic biopsy and produced robustly growing cell culture. Cultured MAC cells were consistently tumorigenic pointing at the presence in this culture of tumor-initiating cells. We then used cultured MAC cells to perform cytogenetical assessment, molecular profiling, and analyzed molecular traits of this cancer from clinical and therapeutic perspectives.
MAC cells showed a highly abnormal chromosome content consistent with unstable genome and aggressive tumor behavior [24]. Exome sequencing supported this observation revealing >1000 non-synonymous mutations and pointing at potential drivers of aggressive growth and metastases. Specifically, MAC showed hot-spot mutations in prominent cancer drivers TP53 and KRAS, which are among the most frequently mutated genes that cause cancer and promote invasion and metastases [25,26].
The G13D KRAS mutation that we identified in MAC has been reported in 5–19% of colorectal cancers (www.cbioportal.org and [27]) as well as in multiple myeloma (4.4%), stomach (3.5%), uterine (2.9%), and a few other malignancies. Interestingly, both KRAS (G13D) and TP53(R213X) mutations were reported as recurrent alterations in 4 of 4 colorectal cancer studies as well as in stomach (2 of 4 studies), bladder (1 of 6 studies), and uterine (1 of 4 studies) carcinomas. However, only colorectal cancers show cases when both mutations are present (1 patient in each of 4 studies) (www.cbioportal.org). The G12V and G13D as well as other KRAS-activating mutations that lock this oncoprotein in the GTP-bound mode are the best documented ‘‘hot-spot” drivers of aggressive tumors. These mutations activate the PI3K/Akt and Erk1/2 pathways that confer proliferative and survival advantages to cancer cells [28]. The urgent need to develop effective therapies against KRAS-driven cases that constitute about one third of all cancers stimulates research into new treatment approaches that target KRAS and its downstream signaling [29]. Future studies will show if activated KRAS is predominant in MAC.
Inactivation of p53 co-operates with Ras activation in promoting CSC distinguished with resistance to cytotoxic therapies, and propensities to invade, metastasize, and recur [30]. This observation raises interest to next generation inhibitors against KRAS and downstream RAF and MEK1/2 that recently entered clinical trials [31–33]. In addition to KRAS and TP53, we identified in MAC potentially inactivating mutations in FAT1, NBEA, RELN, LRP1B, ZFHX3, KDM6A, and KMT2D (Table 1). Remarkably, all these genes demonstrate high mutation frequencies in a variety of cancers including most aggressive ones, such as pancreatic, colorectal, bladder cancers, and melanoma. According to previously published studies and our analysis performed on TCGA databases, all these genes possess tumor suppressor properties. For designing targeted therapies, it is of great interest therefore to identify downstream effects of these commonly mutated cancer-associated genes.
Glioblastomas, colorectal, and head and neck cancers show mutations in FAT genes, members of the cadherin family involved in Hippo signaling [34,35]. Two nonsense FAT1 mutations that we identified in MAC (Table 1) were similar to the ones recently reported in head and neck squamous carcinoma. In this cancer, FAT1 was mutated in 23% of cases and more than half of these mutations were nonsense/truncating [36]. In colorectal carcinoma, FAT1 mutations were detected in 8% of cases with about one third of them being protein-truncating [37].
According to our exome sequencing data, MAC contained protein-truncating mutations in histone-modifying lysine-specific enzymes KDM6A/UTX (W1258X) and KMT2D/MLL2 (S2438X), two tumor suppressors frequently mutated in various cancers [38–42] (Table 1). Interestingly, mutations in both genes whose products are components of the same H3K4-methylation machinery, have been linked to Kabuki syndrome [43,44]. In cancers, protein-truncating KDM6A and KMT2D mutations are the most common defects of these genes (38 and 24%, respectively [45], www.cbioportal.org).
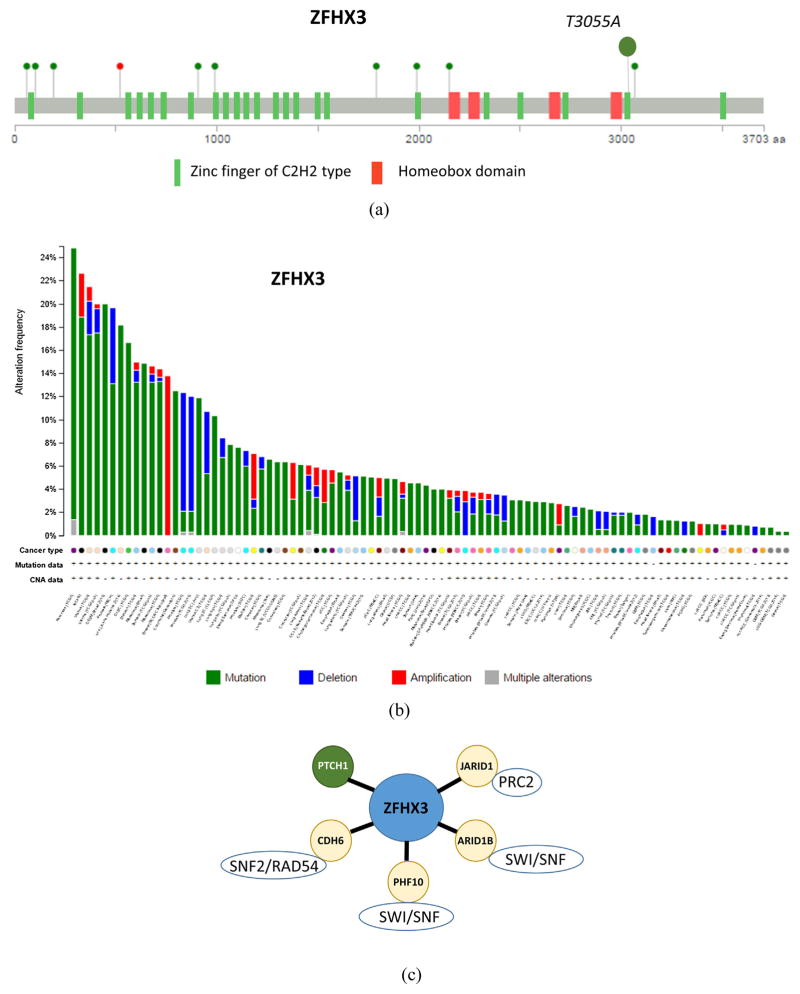
A potentially inactivating mutation in MAC was also identified in ZFHX3/ATBF1, a homeobox transcription factor involved in cell differentiation. This missense mutation, T3055A, was detected in the 22nd of its 23 zinc fingers (Table 1 and Fig. 3A). As ZFHX3 is emerging as a novel tumor suppressor [46], all 123 cancer genomic TCGA datasets were screened for ZFHX3 variations, revealing that this gene is frequently mutated in a broad spectrum of tumors, such as pancreatic (25%), endometrial (17%), prostate (13%), colorectal (12.5%), and stomach (12%) carcinomas, diffuse large B-cell lymphoma (17%), melanoma (20%), small cell lung (12%), and other cancers as well as in 19% of the NCI-60 set of cell lines. Deletions in the vicinity of ZFHX3 were found in ~ 10% of prostate adenocarcinomas and in 5.4% of uterine carcinosarcomas (http://www.cbioportal.org, Fig. 3B). Overall, ZFHX3 was mutated in >15 cancer types at rates that exceed 5%. Intriguingly, our analysis of a publicly available dataset demonstrated that expression of ZFHX3 in colorectal carcinoma correlates with genes that encode chromatin remodeling factors, such as JARID1, ARID1B, CDH6, and PHF10 (Fig. 3C). Taken together with tumor-suppressive activity demonstrated for ZFHX3 [46–48], all these data suggest that ZFHX3 is a novel tumor suppressor involved in epigenetic gene regulation and frequently mutated in various cancers.
Fig. 3.
Non-synonymous substitution in the 22nd zinc finger domain of ZFHX3/ATBF1. (A) Location of non-synonymous T3055A MAC substitution as compared to mutations in colorectal adenocarcinoma patients [37], green circles – missense mutations, red circle – frameshift deletion. (B) Cross-cancer alteration summary for ZFHX3 (123 studies, http://www.cbioportal.org/. (C) ZFHX3 expression in colorectal carcinoma (study E-GEOD-21510) correlates with chromatin remodeling factors (based on rank aggregation analysis with probe 226137_at, http://biit.cs.ut.ee/mem/, p < 10E–3).
Finally, the MAC mutations in NBEA, RELN, and LRP1B that we identified here have been reported in a broad spectrum of cancers that included gastric [49], pancreatic cancers [50], lung adenocarcinomas [51], and other malignancies. Profiling of TCGA datasets revealed NBEA mutations in 18–20% of melanomas and colorectal carcinomas as well as in diffuse large B-cell lymphomas (17%), stomach adenocarcinomas (15%), endometrial carcinomas (15%), and several other cancers. RELN mutations are predominant in melanoma (21–50%) and in lung and stomach adenocarcinomas (16–18%). LRP1B mutations were most frequent in melanomas (39–65%), cutaneous squamous cell carcinomas (48%), small cell lung cancers (45–46%), lung squamous cell carcinomas (39%), and lung adenocarcinomas (32%).
Inactivation of the histone methyltransferase KDM6A has been linked to stimulation of EMT and cancer stem cell properties [52], and its partner in H3K4 methylation, KMT2D, has been implicated in normal and cancer stem cell regulation [53–56]. Nonsense or insertion/deletion mutations that produce KDM6A or KMT2D truncations are typical for a great variety of cancers: bladder [41,57], head and neck [58], esophageal squamous cell carcinoma [39], diffuse large B-cell lymphoma [56], etc. According to our data, MAC falls into this category of epigenetically deregulated cancers showing truncating mutations in KDM6A/UTX and KMT2D, two components of the H3K4 methyltransferase complex [59]. Validating the role of H3K4 methylation in MAC, our proteomic study reveals that up to ~40% (185/469) of proteins overexpressed in MAC have been previously associated with the activity of KDM5B, an H3K4-specific demethylase. In agreement with their important roles in stemness, about 10% of these H3K4-associated genes (19 of 185) have been previously known as CSC-promoting. Our expression analysis linked another 17% of these genes (32 of 185) to various cancers as elements of a conserved gene signature associated with CSC, aggressive proliferation, and poor prognosis. Of note, several of these CSC-promoting genes activated in MAC are targetable (Table 2).
In conclusion, the major strength of our study is in providing the first insight into molecular drivers of aggressive MAC and pointing at nine potential driver mutations whose therapeutic significance has been explored in a variety of aggressive cancers. At the same time, our comparative analysis highlights common epigenetic mechanisms of CSC maintenance that deserve special attention. Thus, our study provides directions for future MAC studies and mechanism-based therapies that can be tested on patient-derived xenografts. One important limitation of our study, however, is that it is based on a single tumor specimen due to extreme rarity of clinical material. Hence, additional studies on a variety of MAC specimens are required to conclude what chromosomal abnormalities and mutations that we described are representative for MAC.
Supplementary Material
Acknowledgments
Role of the funding source
This study was supported by funds from the Adenoid Cystic Carcinoma Research Foundation and by grants 5R21DE023228 (WGY) and 5R21DE022641 (SVI) from the National Institute of Dental and Craniofacial Research. This work was also supported in part by funds from the Department of Surgery, Yale School of Medicine, by an endowment to the Barry Baker Laboratory for Head and Neck Oncology, by Laura and Isaac Perlmutter Cancer Center Support Grant NIH/NCI P30CA016087, and the National Institutes of Health S10 Grants NIH/ORIP S10OD01058 and S10OD018338.
Abbreviations
- MAC
mucinous adenocarcinoma of the salivary gland
- CSC
cancer stem cells
- PDX
patient-derived xenografts
- GEO
gene expression omnibus
- TCGA
the cancer genome atlas
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.oraloncology.2016.12.011.
Footnotes
Conflict of interest
None declared.
References
- 1.Ide F, Mishima K, Tanaka A, Saito I, Kusama K. Mucinous adenocarcinoma of minor salivary glands: a high-grade malignancy prone to lymph node metastasis. Virchows Arch. 2009;454:55–60. doi: 10.1007/s00428-008-0699-1. [DOI] [PubMed] [Google Scholar]
- 2.Lehmer LM, Ragsdale BD, Crawford RI, Bukachevsky R, Hannah LA. Mucoepidermoid carcinoma of the parotid presenting as periauricular cystic nodules: a series of four cases. J Cutan Pathol. 2012;39:712–7. doi: 10.1111/j.1600-0560.2012.01909.x. [DOI] [PubMed] [Google Scholar]
- 3.Farhood Z, Zhan KY, Lentsch EJ. Mucinous adenocarcinoma of the salivary gland: a review of a rare tumor. Otolaryngol Head Neck Surg. 2016 doi: 10.1177/0194599816630546. [DOI] [PubMed] [Google Scholar]
- 4.Bell D, Zhao YJ, Rao PH, Weber RS, El-Naggar AK. Translocation t(6;14) as the sole chromosomal abnormality in adenoid cystic carcinoma of the base of tongue. Head Neck Pathol. 2007;1:165–8. doi: 10.1007/s12105-007-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A. 2002;99:7900–5. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez MN, Emfinger CH, Overton M, Hill S, Ramaswamy TS, Cappel DA, et al. Obesity and altered glucose metabolism impact HDL composition in CETP transgenic mice: a role for ovarian hormones. J Lipid Res. 2012;53:379–89. doi: 10.1194/jlr.M019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodhia KA, Hadley AM, Haluska P, Scott CL. Prioritizing therapeutic targets using patient-derived xenograft models. Biochim Biophys Acta. 2015;1855:223–34. doi: 10.1016/j.bbcan.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams SA, Anderson WC, Santaguida MT, Dylla SJ. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab Invest. 2013;93:970–82. doi: 10.1038/labinvest.2013.92. [DOI] [PubMed] [Google Scholar]
- 12.Floquet C, Deforges J, Rousset JP, Bidou L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011;39:3350–62. doi: 10.1093/nar/gkq1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J, et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest. 2015;125:2007–20. doi: 10.1172/JCI78124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanellopoulou C, Gilpatrick T, Kilaru G, Burr P, Nguyen CK, Morawski A, et al. Reprogramming of polycomb-mediated gene silencing in embryonic stem cells by the miR-290 family and the methyltransferase Ash1l. Stem Cell Rep. 2015;5:971–8. doi: 10.1016/j.stemcr.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overlack K, Primorac I, Vleugel M, Krenn V, Maffini S, Hoffmann I, et al. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife. 2015;4:e05269. doi: 10.7554/eLife.05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–77. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Hubert CG, Herman J, Corrin P, Toledo CM, Skutt-Kakaria K, et al. Cancer-specific requirement for BUB1B/BUBR1 in human brain tumor isolates and genetically transformed cells. Cancer Discov. 2013;3:198–211. doi: 10.1158/2159-8290.CD-12-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu HH, Zhuang G, Gao WQ. A candidate gastric stem/progenitor cell marker revealed by genome-wide analysis. J Pathol. 2016;238:3–6. doi: 10.1002/path.4601. [DOI] [PubMed] [Google Scholar]
- 19.Liang PI, Chen WT, Li CF, Li CC, Li WM, Huang CN, et al. Subcellular localisation of anillin is associated with different survival outcomes in upper urinary tract urothelial carcinoma. J Clin Pathol. 2015;68:1026–32. doi: 10.1136/jclinpath-2015-202958. [DOI] [PubMed] [Google Scholar]
- 20.Genga KR, Filho FD, Ferreira FV, de Sousa JC, Studart FS, Magalhaes SM, et al. Proteins of the mitotic checkpoint and spindle are related to chromosomal instability and unfavourable prognosis in patients with myelodysplastic syndrome. J Clin Pathol. 2015;68:381–7. doi: 10.1136/jclinpath-2014-202728. [DOI] [PubMed] [Google Scholar]
- 21.Bie L, Zhao G, Wang YP, Zhang B. Kinesin family member 2C (KIF2C/MCAK) is a novel marker for prognosis in human gliomas. Clin Neurol Neurosurg. 2012;114:356–60. doi: 10.1016/j.clineuro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, et al. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–8. [PubMed] [Google Scholar]
- 23.Janus JR, Laborde RR, Greenberg AJ, Wang VW, Wei W, Trier A, et al. Linking expression of FOXM1, CEP55 and HELLS to tumorigenesis in oropharyngeal squamous cell carcinoma. Laryngoscope. 2011;121:2598–603. doi: 10.1002/lary.22379. [DOI] [PubMed] [Google Scholar]
- 24.Roschke AV, Glebov OK, Lababidi S, Gehlhaus KS, Weinstein JN, Kirsch IR. Chromosomal instability is associated with higher expression of genes implicated in epithelial-mesenchymal transition, cancer invasiveness, and metastasis and with lower expression of genes involved in cell cycle checkpoints, DNA repair, and chromatin maintenance. Neoplasia. 2008;10:1222–30. doi: 10.1593/neo.08682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–67. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Post SM, Lozano G. You can win by losing: p53 mutations in rhabdomyosarcomas. J Pathol. 2010;222:124–8. doi: 10.1002/path.2757. [DOI] [PubMed] [Google Scholar]
- 27.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–62. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 29.Collins MA. PdMM: Kras as a key oncogene and therapeutic target in pancreatic cancer. Front Physiol. 2014:4. doi: 10.3389/fphys.2013.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. P53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–8. doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- 31.Mandal R, Becker S, Strebhardt K. Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene. 2015 doi: 10.1038/onc.2015.329. [DOI] [PubMed] [Google Scholar]
- 32.Luke JJ, Ott PA, Shapiro GI. The biology and clinical development of MEK inhibitors for cancer. Drugs. 2014;74:2111–28. doi: 10.1007/s40265-014-0315-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11:385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed AF, de Bock CE, Lincz LF, Pundavela J, Zouikr I, Sontag E, et al. FAT1 cadherin acts upstream of Hippo signalling through TAZ to regulate neuronal differentiation. Cell Mol Life Sci. 2015;72:4653–69. doi: 10.1007/s00018-015-1955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–61. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125:373–84. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 40.Kantidakis T, Saponaro M, Mitter R, Horswell S, Kranz A, Boeing S, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30:408–20. doi: 10.1101/gad.275453.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickerson ML, Dancik GM, Im KM, Edwards MG, Turan S, Brown J, et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin Cancer Res. 2014;20:4935–48. doi: 10.1158/1078-0432.CCR-14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Laarhoven PM, Neitzel LR, Quintana AM, Geiger EA, Zackai EH, Clouthier DE, et al. Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum Mol Genet. 2015;24:4443–53. doi: 10.1093/hmg/ddv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–68. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim PH, Cha EK, Sfakianos JP, Iyer G, Zabor EC, Scott SN, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun X, Frierson HF, Chen C, Li C, Ran Q, Otto KB, et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407–12. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 47.Sakata N, Kaneko S, Ikeno S, Miura Y, Nakabayashi H, Dong XY, et al. TGF- beta signaling cooperates with AT motif-binding factor-1 for repression of the alpha -fetoprotein promoter. J Signal Transduct. 2014;2014:970346. doi: 10.1155/2014/970346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kataoka H, Miura Y, Joh T, Seno K, Tada T, Tamaoki T, et al. Alpha-fetoprotein producing gastric cancer lacks transcription factor ATBF1. Oncogene. 2001;20:869–73. doi: 10.1038/sj.onc.1204160. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Wu WK, Xing R, Wong SH, Liu Y, Fang X, et al. Distinct subtypes of gastric cancer defined by molecular characterization include novel mutational signatures with prognostic capability. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-15-2443. [DOI] [PubMed] [Google Scholar]
- 50.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–65. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi HJ, Park JH, Park M, Won HY, Joo HS, Lee CH, et al. UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD1 and HDAC1. EMBO Rep. 2015;16:1288–98. doi: 10.15252/embr.201540244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–37. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- 54.Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, et al. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol. 2013;20:1093–7. doi: 10.1038/nsmb.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan X, Liu L, Ding X, Zhou P, Yuan X, Zhou Z, et al. Mll2 controls cardiac lineage differentiation of mouse embryonic stem cells by promoting H3K4me3 deposition at cardiac-specific genes. Stem Cell Rev. 2014;10:643–52. doi: 10.1007/s12015-014-9527-y. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21:1190–8. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27:271–80. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 58.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5:8906–23. doi: 10.18632/oncotarget.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.