Introduction
The toxicity of misfolded proteins and mitochondrial dysfunction are pivotal factors that promote age-associated functional neuronal decline and neurodegenerative disease1,2. Accordingly, neurons invest considerable cellular resources in chaperones, protein degradation, autophagy, and mitophagy to maintain proteostasis and mitochondrial quality3,4. Complicating the challenges of neuroprotection, misfolded human disease proteins and mitochondria can move into neighboring cells via unclear mechanisms, which may promote pathology spread5,6. Here we document a previously unrecognized capacity of Caenorhabditis elegans adult neurons to extrude large (~4 μm) membrane-surrounded vesicles called “exophers” that can harbor protein aggregates and organelles. Inhibiting chaperone expression, autophagy, or the proteasome, as well as compromising mitochondrial quality, enhances exopher production. Proteotoxically-stressed neurons that extrude exophers subsequently function better than those that do not. The extruded exopher transits through a surrounding tissue where some contents appear degraded, but some non-degradable materials can be subsequently found in remote cells. Our observations suggest that exopher-genesis is a potential “garbage-removal” response to challenged proteostasis and organelle function. We propose that exophers are components of a conserved mechanism that constitutes a fundamental, but formerly unrecognized, branch of neuronal proteostasis and mitochondrial quality control, which, when dysfunctional or diminished with age, might actively contribute to pathogenesis in human neurodegenerative disease and brain aging.
Results
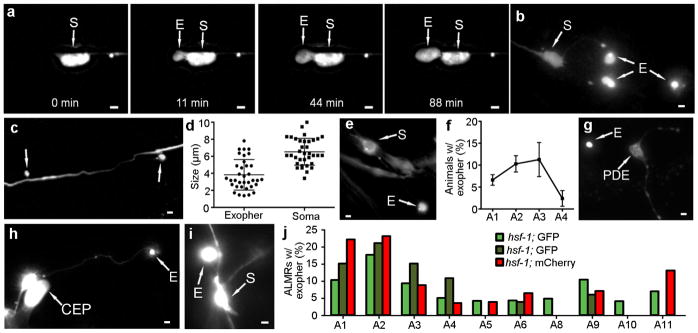
While studying age-associated dendritic restructuring in C. elegans neurons7, we noticed that fluorescent signals originating from neurons sometimes appeared situated outside of the cell in defined vesicle-like structures that we call exophers (Fig. 1a–c, Extended Data Fig. 1a–c, 2g). We first characterized exophers associated with the six gentle touch receptor neurons, for which cell bodies and dendrites are easily visualized. We found that exophers were comparable in size (average 3.8 μM) to neuronal somas (Extended Data Fig. 1d). The size of the vesicles, the morphological stages in their biogenesis (Fig. 1a–c), and the genetic requirements for their production (Extended Data Table 1a), distinguish them from much smaller exosomes (~30–100nm; Extended Data Table 2 compares exophers to characterized extracellular vesicles). Neuronal exophers do not appear to result from classical cell division: a) exophers did not stain with nuclear DNA indicator DAPI (Fig. 1b); b) cell division-inhibiting hydroxyurea8 did not change exopher levels (n > 30 per trial, three trials); and c) RNAi-mediated disruption of cell cycle genes did not change exopher detection (Extended Data Table 1b).
Figure 1. C. elegans touch neurons can extrude cytoplasmic contents.
a. An exopher is generated with a striking concentration of fluorescence segregated to the extrusion. Strain is Is[pmec-4mCh2]. ALM neuron with mCherry-visualized cytoplasm and aggregates. See supplementary Video 1 for corresponding movie: https://youtu.be/HR-G9UphZeA.
b. Exophers do not include nuclear-like levels of DNA. Blue, DAPI; red, cytoplasm. 0/25 exophers but 25/25 soma were DAPI positive. Strain is Is[pmec-4mCh2].
c. An ALMR soma with one attached exopher (right) and one unattached (left). Strain is Is[pmec-4mCh1]; Is[pmec-4GFP].
d. Individual touch neurons differ in their production of detectable exophers. ALMR neurons produce most (~23%) for Is[pmec-18sid-1]; Is[pmec-4mCh3], and PLM neurons fail to generate detectable extrusions (0/>500). 12 trials of RNAi empty vector controls with n > 500 total for each neuron. All animals, adult day 2; E, exophers; S, soma; arrowhead, neuronal process; scale bars, 2 μm; trial means ± s.d. Data in d are mean ± s.d.
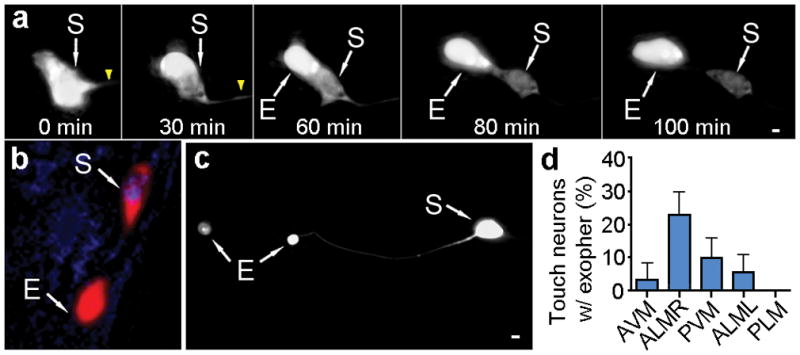
We found that exopher production is not restricted to a specific transgene reporter or line (examples in Fig. 1, Extended Data Fig. 1). Amphid neurons that are dye-filled due to openings to the outside environment9 (Extended Data Fig. 1e, f) can produce exophers, establishing that exophers can form under native/physiological cellular conditions. Exopher production differs dramatically among the six touch receptor neurons, with ALMR producing exophers most frequently (Fig. 1d). Multiple neuronal types can produce exophers, including dopaminergic PDE and CEP neurons (Extended Data Fig. 1g, h), FLP neurons (not shown), sensory ASER neurons (Extended Data Fig. 1i).
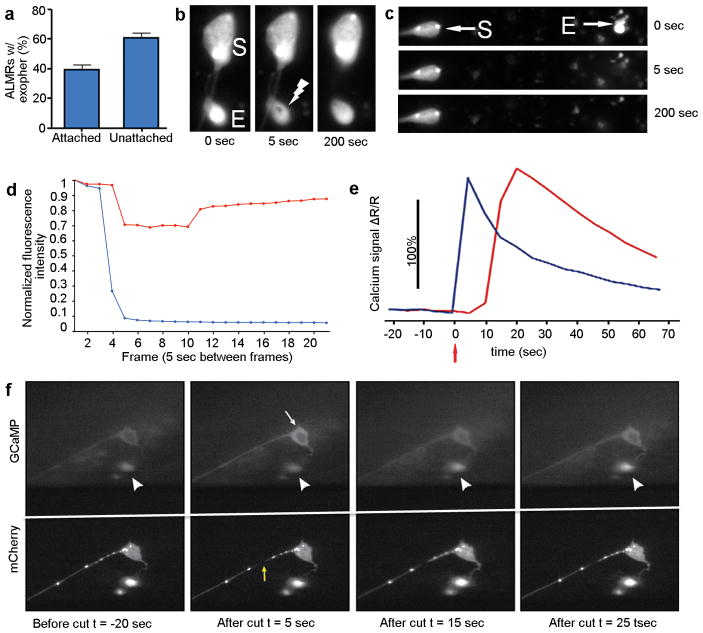
Time-lapse analyses (Supplemental Videos 1–2) revealed that exophers typically arise from the soma by asymmetrically amassing labeled protein to create a balloon-like extrusion via a pinching off event; the exopher compartment then moves outward from the neuronal cell body (extrusion ~15–100 minutes; Fig. 1a, Extended Data Fig. 1a). Plasma membrane reporter Pmec-4PH(plcDelta)::GFP (Extended Data Fig. 2a) and electron microscopy data (Extended Data Fig. 2) confirm that exophers are membrane-bound. Exophers can initially remain connected to the soma by a thin thread-like tube (Fig. 1c) that can allow transfer of tagged proteins and calcium into the attached exopher compartment (Extended Data Fig. 1a, 3, Supplemental Video 2). Exophers ultimately disconnect from the originating neuronal soma (Extended Data Fig. 3).
Why might neurons produce exophers? Time-lapse studies indicated that aggregating mCherry often appeared preferentially concentrated into exophers, and we found that neurons expressing neurotoxic polyglutamine expansion protein huntingtinQ128CFP could also concentrate and extrude this aggregating protein in exophers (Fig. 2a, b). We therefore further queried the relationship of aggregating or toxic protein expression to exopher production. Strains expressing Q128 (toxic with high levels of apparent aggregation10,11) produced significantly more exophers compared to strains that did not express polyQ or expressed huntingtinQ19CFP (non-toxic and low aggregation) (Fig. 2c). Likewise, aggregating mCherry lines exhibited higher average exopher numbers over adult life as compared to lines expressing soluble GFP (example in Fig. 2d). High aggregate load in individual neurons was predictive of increased exopher production on the following day (Fig. 2e). Conversely, mCherry(RNAi) reduced exopher number by ~half in a line producing aggregating mCherry (Fig. 2f). Although our studies cannot determine the relative contribution of aggregate load from protein expression levels, they suggest that proteostatic challenges increase exopher production. Consistent with a potential role for exophers in elimination of potentially harmful neuronal contents, expression of amyloid-forming human Alzheimer’s disease fragment Aβ1–42 in ASER neurons increases exopher numbers (Fig. 2g). Our combined observations on exopher formation, contents, and frequency of detection suggest that exophers preferentially include aggregated, excess, or otherwise neurotoxic proteins for removal.
Figure 2. Touch neurons under proteotoxic stress jettison aggregation-prone proteins into exophers.
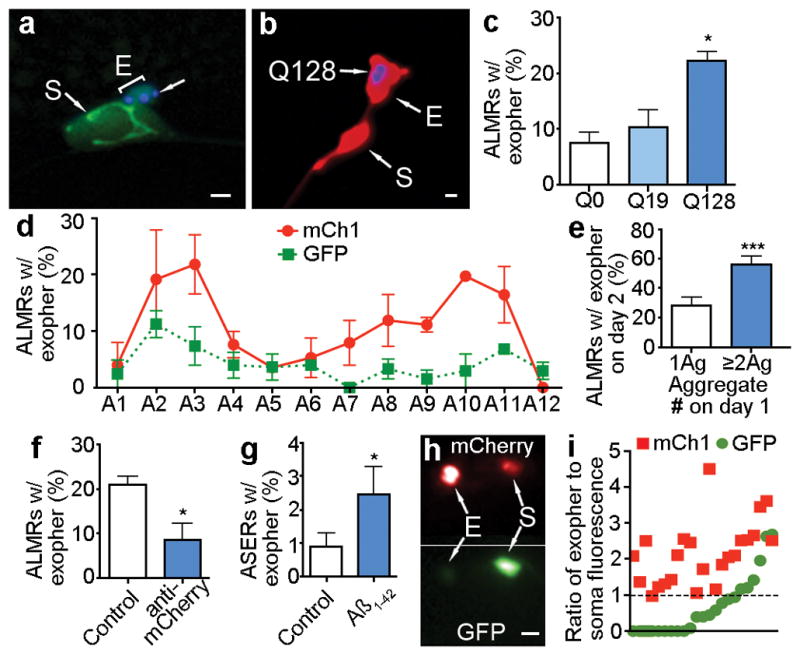
a. CFP-tagged Q128 concentrated into a budding domain. Strain is Is[pmec-4mCh2; pmec-3Q128CFP]. Blue, Q128CFP; green, mitochondria GFP signal.
b. Mature exopher containing Q128CFP aggregates. Strain is Is[pmec-4mCh2; pmec-3Q128CFP]. 5/10 ALM exophers were Q128CFP positive. These five had no detectable Q128CFP in their somas.
c. Neurons expressing Q128CFP produce more exophers than neurons expressing Q19CFP. Is[pmec-4GFP] (Q0), Is[pmec-7YFP; pmec-3Q19CFP] (Q19) and Is[pmec-7YFP; pmec-3Q128CFP] (Q128). 3 trials, n > 100 total for each strain, one-way ANOVA with Tukey’s test, *P < 0.05. PolyQ-expressing strains have similar expression levels10.
d. Touch neuron exophers are detectable in young adults, diminish in abundance in midlife, and increase again in older animals. Longitudinal study of Is[pmec-4GFP] and Is[pmec-4mCh1] (starting n = 75 total per strain), 3 trials, adult days 1–12. Variation between days and between strains was significant (***P < 0.001). A similar early adulthood peak occurs in dye-filled amphid neurons (Extended Data Fig. 1f) and in the hsf-1 mutant (Extended Data Fig. 1j).
e. Multiple early visible aggregates predict later exopher formation. On adult day 1, we segregated animals by number of mCherry aggregates (1 Ag vs ≥ 2 Ag) in the ALMR soma and scored for exophers on adult day 2. 5 trials, n > 130 total per condition, ***P < 0.001. Strain is Is[pmec-4mCh1].
f. Reducing mCherry expression levels using an anti-mCherry RNAi reduces exopher levels. Strain is Is[pmec-18sid-1 pmec-4mCh3]. 3 trials, n > 35 per trial, *P < 0.05.
g. ASER neurons expressing human toxic AD protein fragment exhibit elevated exopher production. Adult day 2 exopher production for 7 trials, n > 800 total for each strain, sesIs2512 [Pgcy-5GFP], and sesIs25 [Pflp-6Aβ1–42; Pgcy-5GFP]. *P < 0.05.
h, i. Aggregation-prone mCherry is preferentially segregated into the exopher compared to non-aggregating GFP, which is relatively more concentrated in the soma. h. mCherry (top) and GFP (bottom) channels from an ALMR exopher and soma pair in a strain co-expressing Is[pmec-4mCh1] and Is[pmec-GFP]. i. Quantification of mCherry and GFP fluorescence ratios for exopher and soma pairs. Each point represents an exopher-to-soma fluorescence ratio for either GFP or mCherry. Each cell has a paired GFP and mCherry ratio, aligned vertically, n = 23 pairs. Mean E/S ratio of mCherry was 2.2, mean E/S ratio of GFP was 0.75, **P < 0.01. All animals, adult day 2; E, exophers; S, soma; scale bars, 2 μm; trial means ± s.e.m.; unpaired t-test (e, f, g), one-way ANOVA (d) and one-way ANOVA with Tukey’s test (c).
To address the hypothesis that aggregation-prone proteins might be selectively extruded in exophers, we constructed a line that expressed both an aggregation-prone mCherry (Is[Pmec4mCh1]) and a non-aggregating GFP (Is[pmec-4GFP]) and compared the red and green fluorescence distribution between exophers and somas (example in Fig. 2h, data in Fig. 2i). In 22/23 exophers, we found higher relative levels of mCherry in the exopher, and higher relative levels of GFP in the soma. That neurons appear to preferentially extrude aggregation-prone mCherry over soluble GFP suggests deleterious materials are identified and sorted for export during exopher-genesis.
To address the hypothesis that proteostatic challenges enhance the exopher production response, we manipulated the in vivo protein-folding milieu. We examined exopher production in an hsf-1(sy441) mutant deficient in core proteostasis transcription factor HSF-1 (and therefore chaperone expression) to record ~6-fold increase in exophers (Fig. 3a). We impaired autophagy by treating animals with a pharmacological inhibitor, Spautin-1 and by RNAi knockdown (lgg-1, atg-7, bec-1, lgg-1/2) in a strain expressing aggregation-prone mCherry and measured a significant increase in exopher incidence (Fig. 3b,c). Impairment of proteasome activity with inhibitor MG132 in strain Is[pmec-4mCh1] also increased exopher production (Fig. 3d). Given that inhibiting multiple facets of proteostasis increases exopher extrusion, we suggest that exophers may constitute a previously undescribed component of the proteostasis network, which may function as a backup or alternative response to rid cells of neurotoxic aggregates/proteins when proteostasis becomes overwhelmed by mounting intracellular proteotoxicity.
Figure 3. Disruption of multiple branches of proteostasis increases exopher formation.
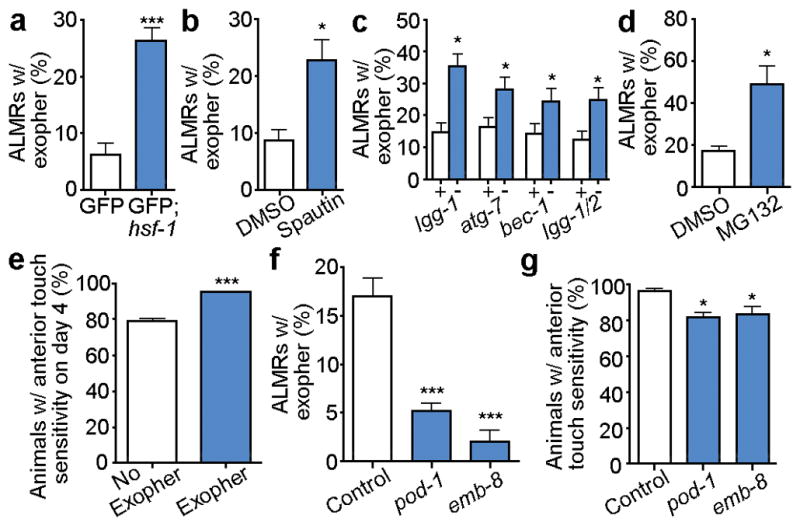
a. Disrupting proteostasis by hsf-1 impairment increases exopher formation. Strains were Is[pmec-4GFP] and Is[pmec-4GFP]; hsf-1(sy441), ***P < 0.001, n > 280 total per strain.
b. Pharmacological inhibition of autophagy by Spautin-1 increases the occurrence of exophers. Strain is Is[pmec-4GFP], n > 80 total per condition, *P< 0.05.
c. RNAi knockdown of autophagy genes lgg-1, atg-7, and bec-1 increases the occurrence of exophers. Strain is Is[pmec-18sid-1 pmec-4mCh3]. White, empty vector control RNAi; blue, RNAi against gene indicated, lgg-1 (5 trials), atg-7 (5 trials), bec-1 (4 trials), lgg-1/2 (5 trials), n > 100 total per condition, *P< 0.05.
d. Pharmacological inhibition of the proteasome by MG132 treatment increases the occurrence of exophers. Strain is Is[pmec-4mCh1], 3 trials, n > 33 per trial, *P < 0.05.
e. Q128-expressing animals with an ALMR exopher on day 2 have better anterior touch sensitivity on adult day 4 compared to animals with no apparent early exopher. We tested sensitized strain Is[pmec-4mCh2; pmec-3Q128CFP], which exhibits accelerated functional decline of touch neurons. 3 trials, n > 100 animals total, ***P < 0.001. Note that the “no exopher” category should include animals that have actually produced exophers, but were not present at the time of sampling, thus differences are likely to be under-estimated in this panel.
f. Knocking down pod-1 or emb-8 by RNAi significantly decreases exopher detection, defining a genetic approach to limiting exopher-genesis. Strain was Is[pmec-18sid-1 pmec-4mCh3]. 4 trials, n > 100 total per condition, ***P< 0.001.
g. Knocking down pod-1 and emb-8 by RNAi is associated with a decrease of anterior touch sensitivity in day 4 adults. Strain was Is[pmec-18sid-1 pmec-4mCh3], an aggregation-prone mCherry line which, like wild type, maintains strong touch sensitivity in young adult life. pod-1 and emb-8 RNAi knockdown had no effect on young animal touch sensitivity from egg-lay RNAi knockdown, but L4 pod-1 and emb-8 knockdown reduced exopher production and decreased touch sensitivity in day 4 adults. 3 trials, n > 130 total per condition, *P < 0.05.
All animals, adult day 2; E, exophers; S, soma; scale bars, 2 μm; trial means ± s.e.m. (a, b, c,), ± s.d. (d, e, f,); unpaired t-test (a, b, c, d, e), one-way ANOVA with Dunnett’s test (f, g).
Exopher production occurs with a striking bimodal distribution over adult life: exophers are most commonly observed on adult days A2-A3, diminish in abundance A4-A8, and then reappear again later in life ~A10-A11 (Fig. 2d; similar young adult pattern with dye-filled amphid neurons, Extended Data Fig. 1f; and with a 1 day earlier onset in an hsf-1 mutant, Extended Data Fig. 1j). The distinctive temporal production profile suggests that conditions permissive for exopher production exist in young adulthood but can then be limited or remain below a threshold until late adulthood. The coincidence of the early peak with a transition in C. elegans young adult proteostasis management12–14 suggests the first wave of exopher-genesis may serve as a normal component of an orchestrated proteostasis reset in young adulthood that involves jettisoning neuronal garbage generated during development; the later adult increase in exopher production may be the consequence of age-associated decline in proteostatic robustness.
Rather than inducing neuronal death or dysfunction, exopher-genesis appears beneficial. First, in hundreds of longitudinal observations, we did not observe neuronal loss after exopher production: exophers are distinct from apoptotic bodies in their biogenesis (Fig. 1a, Extended Data Fig. 1a), and the soma of an exopher-producing neuron retains normal ultrastructural features (Extended Data Fig. 2e). Second, the relative functionality of proteotoxically-stressed neurons that have generated exophers is increased over neurons that did not extrude exophers. In blinded studies in a line expressing Q128CFP, which progressively impairs touch sensation10, we found that midlife touch sensitivity is greater when ALMR had definitely produced an exopher at A2, as compared to age-matched siblings in which ALMR had not produced an exopher (Fig. 3e). Third, we identified pod-1 and emb-8 as polarity genes required in adults for exopher-genesis (Fig. 3f), and found that adult RNAi knockdown impaired midlife touch sensitivity (Fig. 3g). Although we cannot rule out that pod-1 and emb-8 RNAi interventions might generally disrupt adult neuronal function, taken together our data are consistent with a model in which adult neurons that do not make exophers become functionally compromised compared to those neurons that extruded offending contents. Overall, despite a striking expulsion of cellular contents, adult neurons appear to be healthier after throwing out their trash.
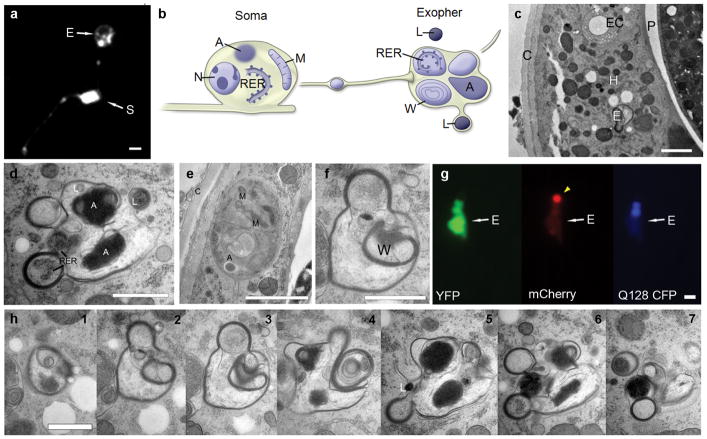
Considering the large volume of exophers, we hypothesized they might include organelles. Indeed, both lysosomes (Extended Data Fig. 4) and mitochondria (Fig. 4a, b; Extended Data Fig. 5) can be extruded in exophers. Mitochondrially-localized GFP reporters revealed mitochondrial inclusion in budding and dissociated exophers, with punctate or filamentous morphology typical of adult mitochondrial networks (Fig. 4a, Extended Data Fig. 5a–c). To address whether impairing mitochondrial quality enhances the production of exophers, we genetically manipulated dct-1/BNIP3 (mitophagy), pink-1/PINK15 and pdr-1/Parkin16 (human Parkinson’s disease homologs implicated in mitochondrial maintenance), and ubl-517(mitochondrial unfolded protein response), (Fig. 4c, d). We conclude that multiple approaches toward genetic impairment of mitochondria can increase exopher-genesis.
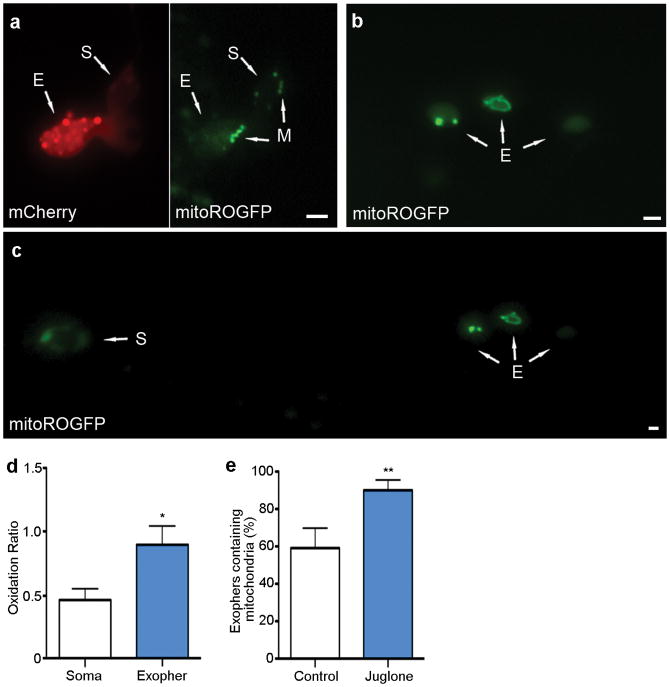
Figure 4. Mitochondria can be extruded in exophers, and mitochondria with higher mitochondrial matrix oxidation might be preferentially extruded.
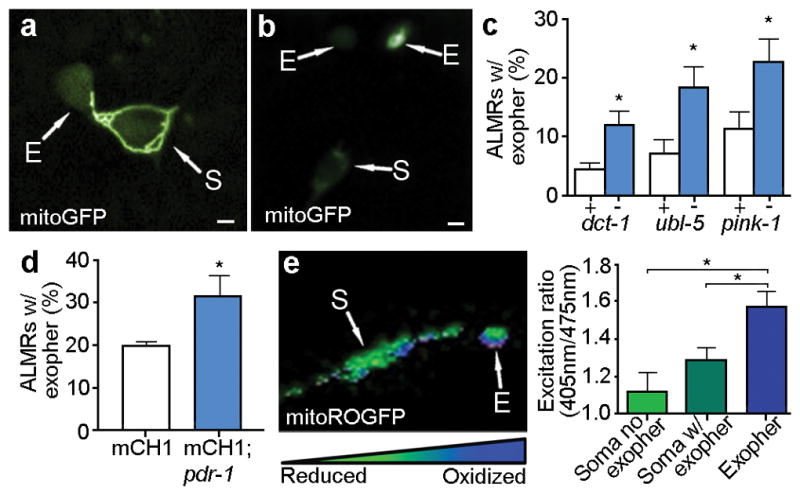
a. Mitochondria in a budding exopher. Mitochondria form a ring around the somatic periphery, typical of young adulthood, with some mitochondria segregated into a putative exopher. Strain is bzIs167[pmec-4mitoGFP].
b. Mitochondrially localized mitoGFP (strain bzIs167[Pmec-4mitoGFP]) can be extruded in exophers; left exopher shown does not include substantial mitochondrial content, but right exopher does. 10/20 exophers scored with this reporter contained mitochondria.
c. RNAi knockdown of mitochondrial health and function genes ubl-5, pink-1, and dct-1 increases the occurrence of exophers. White, empty vector control; blue, RNAi against genes indicated. ubl-5 (3 trials), pink-1(4 trials,) dct-1 (3 trials). n > 80 total per condition, *P < 0.05.
d. Genetic disruption of mitochondrial health and function increases the occurrence of exophers. We compared exopher levels in Is[pmec-4mCh1]; pdr-1(gk448) mutant, a Parkin homolog16 *P < 0.05, n = 30 per trial, 6 trials.
e. Mitochondria segregated into exophers have higher relative oxidation levels than somatic mitochondria, as reported by mitoROGFP. Left, a pseudo-colored image indicating relative emission levels at excitation wavelengths of 405 nm/476 nm (blue, oxidized; green, reduced). Right, redox excitation ratio in exophers vs. soma. n =10 pairs of exophers with mitochondria and originating soma, *P < 0.05, strain is zhsEx17 [Pmec-4mitoLS::ROGFP]. Of note, the soma shown exhibits locally concentrated oxidized mitochondria, indicating that oxidizing conditions are not restricted to exopher. Wild type, unstressed somas have a typical 405 nm/476 nm ratio of 119; cells that form an exopher may experience somewhat elevated levels of oxidation in the soma, overall. All animals, adult day 2; E, exophers; S, soma; scale bars, 2 μm; trial means ± s.e.m. (c, d, e,), unpaired t-test (c, d), one-way ANOVA with Tukey’s test (e).
To address the hypothesis that stressed or damaged mitochondria might be preferentially segregated to exophers, we utilized mitoROGFP, a mitochondrially localized reporter that changes its peak excitation wavelength from ~405 nm (oxidized) to 476 nm (reduced) according to the local oxidative environment18,19. We find a significant increase in the 405 nm (oxidized) / 476 nm (reduced) excitation ratio of mitochondria in exophers as compared to those in somas (Fig. 4e), roughly equivalent to the redox excitation ratio observed in C. elegans neurons subjected to H2O2-induced oxidative stress19. We confirmed higher oxidation scores using MitoTimer, an alternative reporter of mitochondrial matrix oxidation20 (Extended Data Fig. 5d). In addition, touch neurons of juglone-treated21 bzIs166[Pmec-4mCherry]; zhsEx17[Pmec-4mitoLS::ROGFP] animals had significantly higher numbers of mitochondria-including exophers than matched controls (Supplemental Data Fig. 5e). Although compromised mitochondrial health may impair neuronal proteostasis, thus increasing exopher production, our data establish that touch neurons can eject mitochondria via exophers, which raises the intriguing possibility that exopher-genesis may constitute a previously unappreciated removal-based mechanism of mitochondrial homeostasis.
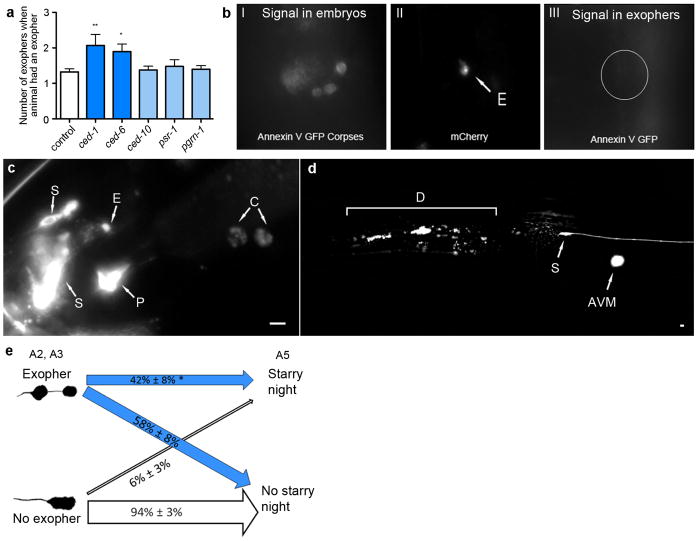
What is the fate of the extruded exopher and its contents? With time, exopher fluorescence intensity diminishes or disappears (persistence times ~1–12 hours), possibly as exopher contents are degraded internally or digested by the neighboring hypodermis that fully surrounds the touch neuron and has degradative capabilities. Consequent to disruption of the C. elegans apoptotic engulfment genes ced-1 (homologue of mammalian CD91, LRP1 and MEGF10, and fly Draper), ced-6 (GULP) and ced-7 (ABC), ALMR neurons are associated with multiple exophers (Fig. 5a; Extended Data Fig. 6a). However, genetic manipulation of a parallel engulfment pathway comprising ced-2, ced-5, ced-10, ced-12, and psr-1, changed neither the frequency of exopher generation nor the detection of multiple exophers. Moreover, we did not detect the apoptotic “eat-me” signal phosphatidylserine (PS) on the exopher surface using a widely expressed PS-binding annexinV::GFP (0/43 exophers; Extended Data Fig. 6b). Our data suggest that hypodermal recognition/degradation of exophers and their contents occurs by mechanisms that are at least in part distinct from classical removal of apoptotic corpses, but involve the CED-1, CED-6, CED-7 proteins. Electron microscopy studies support that the hypodermis may mediate degradation of at least some of exopher contents (Extended Data Fig. 2d–f, h).
Figure 5. Fluorescent mCherry escapes touch neurons and surrounding hypodermis to later concentrate in distant coelomocytes.
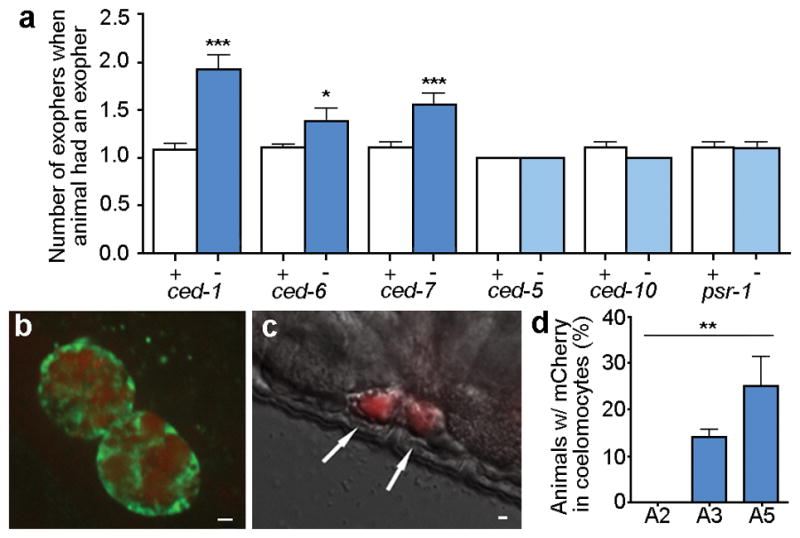
a. ced-1, ced-6, and ced-7 mutations increase the number of ALMR neurons with two or more exophers, and ced-5, ced-10, and psr-1 mutations, acting in a parallel phagocytosis pathway, do not. Strain zdIs5[Pmec-4GFP]), adult day 2, 3 trials, n > 90 total per strain; unpaired t-test, *P < 0.05; ***P < 0.001 (replicating RNAi data in Extended Data Fig. 6a). ced-1, ced-6, and ced-7 mutations do not increase the percentage of ALMRs producing exophers (data not shown), but rather increase multiple exopher detection, suggesting a deficit in persistence rather than generation.
b. In older animals, coelomocytes (green) can concentrate fluorescent proteins that were originally expressed in touch neurons (red). Strain Is[pmec-4mCh1]; pwIs979[Pcup-4GFP::vps-29], adult day 6.
c. mCherry localization in coelomocytes (arrows) can also be visualized with DIC underlay. Strain is Is[pmec-4mCh1], adult day 6.
d. The number of coelomocytes containing mCherry released from touch neurons increases with time. Is[pmec-4mCh1] animals with an exopher on adult day 2 were segregated and scored for red fluorescence in coelomocytes on A2, A3 and A5; n=20 per trial, 3 trials, **P< 0.01, one-way ANOVA with Tukey’s test. Scale bars, 2μm. Data are mean±s.e.m.
The lack of detectable PS “eat me” signal on exophers raised the question as to whether at least some exopher contents might be destined to elude hypodermal degradation. Indeed, fluorescent mCherry protein that was originally expressed specifically in touch neurons, or fluorescent DiI loaded into dye-filling neurons, appeared later in distant scavenger coelomocytes (Fig. 5b–d; Extended Data Fig. 6c). Blocking coelomocyte uptake capacity by cup-4 mutation22 caused fluorescent particles to accumulate outside neurons, possibly within the pseudocoelomic fluid (body cavity; Extended Data Fig. 6d, e). We conclude that some exopher contents transit the hypodermal tissue to be released into the pseudocoelom, from which materials can later be taken up by distant coelomocytes. Exophers can therefore mediate transfer of neuronal materials to remote cells.
Considerable excitement in the neurodegenerative disease field has been generated by the findings that mammalian neurons can extrude conformational disease proteins, including in Alzheimer’s, Parkinson’s and prion disease23. C. elegans exopher production constitutes a newly identified mechanism by which neurons can transfer cellular material (preferentially neurotoxic species) to other cells. Interestingly, in a C. elegans muscle model of prion toxicity, offending prion proteins were transferred among muscle cells and ultimately localized to coelomocytes24. We speculate that the basic mechanism we document here may correspond to a conserved pathway for the transfer of toxic contents out of multiple cell types. In this regard, it may be noteworthy that mammalian aggregated poly-Q expanded huntingtin can transfer between neurons via tunneling nanotubes25–27 that resemble thin connections between C. elegans somas and exophers, and that neuronal polyQ in Drosophila is transferred to glia via a process that requires the CED-1 homolog, DRAPER28.
Recent reports show mitochondria can transfer out of specific cells to contribute positive roles (mesenchymal stem cells via tunneling nanotubes29; astrocytes to neurons in a stroke model30), but our study underscores a generally underappreciated option for mitochondrial quality control: mitochondrial expulsion. The mito-expulsion we report in C. elegans touch neurons has a striking mammalian counterpart: mouse mitochondria originating in retinal ganglion cells can be extruded into neighboring astrocytes for degradation6 (with some intriguingly similar morphology to C. elegans exophers; see Fig. 1e of ref. 6). Although further study will be required to definitively establish the health status and fates of transferred mitochondria in the C. elegans model, it is tempting to speculate that transcellular degradation of mitochondria may be a more broadly utilized mechanism of mitochondrial quality control than currently appreciated, with associated potential importance in neuronal health.
Overall, although further experiments are needed to elucidate the detailed mechanisms at play, and validate the proposed functions of exophers in proteostasis and the removal of damaged organelles, we suggest that exopher production is a previously unrecognized mechanism for clearing out accumulating protein aggregates and dysfunctional organelles that threaten neuronal homeostasis (Extended Data Fig. 7). The analogous process in mammals could promote transfer of misfolded protein and/or dysfunctional mitochondria to neighboring cells, promoting human pathology in neurodegenerative disease if compromised. Mechanistic dissection of this novel facet of proteostasis and mitochondrial homeostasis should thus inform on fundamental mechanisms of neuronal maintenance and suggest novel targets for intervention in neurodegenerative disease.
Methods
Strains and Media
C. elegans strains were cultured at 20 °C with standard methods31. Strains used were SK4005 zdIs5[Pmec-4GFP] (abbreviated in the text as Is[Pmec-4GFP]), ZB4065 bzIs166[Pmec-4mCherry1](abbreviated in the text as Is[Pmec-4mCh1]), ZB4066 bzIs167[Pmec-4mitogfp Pmec-4mCherry2] (abbreviated in the text as Is[Pmec-4mCh2]), ZB4067 bzIs167[Pmec-4mitogfp Pmec-4mCherry4]; igIs1[Pmec-7YFP Pmec-3htt57Q128::cfp lin-15+]10 (abbreviated in the text as Is[mCh2 ; Q128CFP]), sesIs2512[Pgcy-5GFP], sesIs25[Pflp-6 Aβ; Pgcy-5GFP]32, KWN176 rnyIs014[Pmec-4mCherry unc-119(+)], ZB4071 bzIs169[Pmec-18sid-1Psng-1YFP]; bzIs101[Pmec-4mCherry; Punc-119+], ZB4087 bzIs169[Pmec-18sid-1Psng-1YFP]; bzIs101[Pmec-4mCherry; Punc-119+]; hsf-1(sy441), BZ555 egIs1[Pdat-1GFP], ZB4070 bzIs168 [Pmec-7LMP-1::GFP], ZB4509 bzIs166[Pmec-4mCherry]; bzIs168[Pmec-7LMP-1::GFP], ZB4082 cup-4(ok837); bzIs166[pmec-4mCherry], ZB4083 smIS76 [Phsp-16ANV::GFP]33; bzIs166[Pmec-4mCherry], ZB4084 hsf-1(sy441); zdIs5[Pmec-4GFP], ZB4085 hsf-1(sy441); bzIs166 [Pmec-4mCherry], ZB4086 zdIs5[Pmec-4GFP]; bzIs166[Pmec-4mCherry], PTN73 pha-1(e2123); him-5(e1490); zhsEx17[Pmec-4mitoLS::ROGFP], RBW2834 rbw2834Si[Pmec-3::mitotimer::T54, CB-unc-119 + II ttTi5605] in unc-119 (ed3)20, QH3738 ced-1(e1735); zdIs5, QH3737ced-6(n1813); zdIs5, QH4623 ced-5(n1812); zdIs5, QH3768 ced-7(n2690); zdIs5, QH3130 ced-10(n3246); zdIs5, QH3533 psr-1(ok714); zdIs534, ZB4526 bzIs166[Pmec-4mCherry]; pdr-1(gk448), ZB4525 bzIs166[Pmec-4mCherry]; (pwIs979 [Pcup-4GFP::vps-29]cB-unc119), ZB4528 bzIs166[Pmec-4mCherry]; zhsEx17 [Pmec-4mitoLS::ROGFP], ZB4059 bzIs163 [Pmec-4::GCaMP3.0::SL2::mCherry], ZB4524 bzEx242 [Pmec-4::PH(plcDelta)::GFP]35, and wild type N2.
RNAi was administered through feeding animals with RNAi-expressing bacteria with standard methods36 with touch neurons RNAi-enhanced via SID-1 expression37. Exophers are readily visible at 400X total magnification, with high power dissecting microscopes. In general, exophers have the following features: a ~4 μm membrane-bound vesicle extruded from a neuron via a mechanism that temporarily includes a thin filamentous connection to the originating soma, but eventually breaks off. Contents of exophers can include neurotoxic proteins, mitochondria, and lysosomes; exophers are produced by native amphid neurons after dye-filling.
Age synchronization and RNAi screening
To synchronize animals, L4 stage hermaphrodites were selected and moved to test plates. The day after moving was considered adult day 1, and animals were scored on adult day 2 for the occurrence of exophers. For scoring of exophers, animals were immobilized by adding 100 μL of 10 mM tetramisole to the surface of the plate. Animals were measured on the plate with a Kramer dissecting scope with a 20x objective. The ALMR neuron was scored for the presence of an exopher, which was counted if greater than ¼ the size of the soma, as a threshold against inclusion of smaller species of extracellular vesicles. Exophers were also visible in live animals without anesthetic. RNAi experiments had a negative empty vector control. An mCherry knockdown was used to confirm RNAi had an effect in the neurons of interest. RNAi screens were performed with the strain bzIs169[Pmec-18sid-1 Psng-1YFP]; bzIs101[Pmec-4mCherry; unc-119+]. All genes were independently screened a minimum of three times.
Microscopy Techniques
For imaging, animals were mounted by placing them in a drop of cold, liquid 36% Pluronic F-127 with 1 mM tetramisole solution and pressed between two coverslips. The slides were brought to room temperature, solidifying the Pluronic F-127 gel and immobilizing the animals. Co-localization images were made using iVision software. Images were taken using a Zeiss Imager D1m upright compound microscope with a 40x dry objective. For confocal imaging, animals were immobilized by using 7.5% M9 agarose pads with 2.5 μl PolySciences 0.05 μm polystyrene microspheres. A Zeiss spinning disk confocal upright microscope with 100x oil immersion objective was used for select images to show additional details, including lysosomal imaging and connection imaging.
MitoROGFP Imaging and Quantification
Adult day 2 PTN73 pha-1(e2123); him-5(e1490); zhsEx17[Pmec-4mitoLS::ROGFP] animals were mounted as above on a Leica SP5 II confocal microscope (Leica Microsystems, Exton, PA) with a 63x oil immersion lens. Samples were alternately excited with a 30% power 405 nm UV laser and a 30% power 476 nm visible laser with a sequential line scanning method. Emission was detected by HYD1 photon counting at 508–513 nm. Images were quantified using ImageJ. Images were thresholded to remove background. The 405 nm channel was divided by the 476 nm channel, and ROI measurement was used to quantify mean intensities.
MitoTimer Imaging and Quantification
MitoTimer encodes a dsRed derivative that fluoresces green when reduced (first synthesized), but irreversibly shifts tored fluorescence as it oxidizes 20. Adult day 2 rbw2834Si[Pmec-3::mitotimer::T54, CB-unc-119 + II ttTi5605] in unc-119 (ed3)20 animals were mounted as above on a Zeiss Imager D1m upright compound microscope with a 63x oil immersion lens. Samples were alternately measured under GFP and dsRed channels, keeping light intensity and exposure times constant between images. Images were quantified using ImageJ by selecting the ROI, subtracting the background, measuring red and green intensities, and calculating the red/green ratio.
Fluorescence Quantification
Fluorescence quantification was performed in ImageJ by selecting the ROI, measuring the mean intensity, and subtracting background intensity.
Time-lapse Imaging
Time-lapse imaging was performed with a 100x oil immersion objective with a motorized stage. 15 animals were mounted to a slide using a 7.5% M9 agarose pads with 2.5 μl PolySciences 0.05 μm polystyrene microspheres; coverslip was sealed with a 60:40 mix of Vaseline and paraffin wax. An iVision script was used to image selected locations every 8–15 minutes for 12 hours. Image analysis and video compilation were done manually.
Dye-filling
Animals were washed off a plate into a 1.5 mL centrifuge tube with 1 mL M9 and 10 μL of 1 mM DiI. Animals were allowed to soak at room temperature for 3 hours. Animals were washed with M9 twice before mounting onto slides for imaging.
Longitudinal measurements
50 animals were synchronized at the L4 stage and 25 animals were measured on subsequent adult days, directly from the plate without anesthetics using a Kramer microscope. The animals were transferred to fresh plates every 2 days until adult day 8 to prevent crowding and starvation.
DAPI Staining
DAPI staining was performed after wash-harvesting with PBS and permeabilizing the membrane at −80 °C freezer for 10 minutes. After thawing, the supernatant was removed and animals were re-suspended in 1 mL cold methanol and incubated 5 minutes for fixation. Animals were washed with PBS twice and then stained in a 1 mL DAPI solution (200 ng/mL in PBS) for 30 minutes before mounting for microscopy.
Size Measurement
Exopher and cell size was performed by measuring pixel length with Photoshop and comparing to a calibration scale for each objective used. Width was measured at the widest point.
Drug Assays
MG132 (Sigma-Aldrich C2211) and Spautin-138 (Sigma-Aldrich SML0440) were dissolved in DMSO at 10 mM and 1 mM, respectively, and administered by placing 30 μL of the solution over the bacterial food lawn.
Juglone21 (Sigma-Aldrich 59990) was dissolved to a final concentration of 230 μM in a solution of 0.23% v/v ethanol in M9. Adult day 1 worms were transferred into either a 1 mL tube of the juglone solution or a 1 mL control tube of 0.23% v/v ethanol in M9 for 90 minutes. Animals were washed with M9 buffer, centrifuged, and recovered onto a microscope slide for imaging.
Hydroxyurea (Sigma-Aldrich H8627) was dissolved in distilled water to make a 1 M solution, of which 250 mL was added to a standard seeded NGM plate to reach a working concentration of 25 mM8. Plate was left at room temperature for 6 hours to allow for complete diffusion before transferring adult day 1 animals for measurement 24 hours later on adult day 2.
Touch Sensitivity Assay
To assay for touch sensitivity, animals were stroked with a calibrated force probe on the anterior and posterior halves of the body. Reversal was an indication of a positive response. Animals responding to at least 3 out of 5 touches were considered sensitive. Animals responding to 2 or fewer touches were considered not sensitive.
Aggregate measurements
Q128 aggregates can be visually distinguished in touch neuron somas with a 20x objective11,39. The aggregate exopher prediction experiment was done by separating day 1 adult animals into two populations, those that had one visible aggregate in the ALMR neuron and those that had two or more. The two populations were scored on the next day for exophers extruded from the ALMR neuron.
Electron microscopy
Young adults were screened by light microscopy to identify samples in which the ALM neurons had recently expelled an exopher. These animals were prepared for TEM analysis approximately 3 hours after initial selection by high pressure freezing and freeze substitution (HPF/FS) following a standard protocol for preservation of ultrastructure40. Briefly, after HPF, animals were exposed to 1% osmium tetroxide in acetone with 2% water added, kept at −90 °C for 5–6 days before slowly warming back to room temperature. Samples were rinsed in cold acetone and embedded in plastic resin before curing at high temperature for 1–2 days. Serial thin sections were collected on plastic-coated slot grids, post-stained with uranyl acetate, and examined with a Philips CM10 electron microscope. By looking in transverse sections for landmarks such as the 2nd bulb of the pharynx, it was possible to reach the vicinity of the ALM soma before collecting about 1,500 serial thin transverse sections. Having found the soma, one could then explore the region 30–50 μm posterior to the ALMR for evidence of the exopher.
FRAP Analysis
Synchronized Is[Pmec-4mCh1] adult day 2 animals were immobilized on 7.5% M9 agarose pads with 2.5 μL PolySciences 0.05 μm polystyrene microspheres. Exopher centers were photo-bleached with 7 pulses of the MicroPoint pulsed nitrogen pumped dye laser (neutral density filter at position 9, Lumencor solid state light source) attached to a Zeiss Inverted Axio Observer microscope (100x 1.4 N.A. objective) on an anti-vibration table. 1 frame was recorded every 5 seconds using constant excitation intensity and exposure time with a Qimaging EXi Blue camera. Images were analyzed with ImageJ. Exopher fluorescence intensity was normalized to the intensity of the first data point in each series.
GCaMP studies on axotomized neurons with connected exophers
Adult day 4 bzIs163 [Pmec-4::GCaMP3.0::SL2::mCherry] worms expressing the genetically encoded calcium indicator GCaMP3.0 in the mechanosensory neurons were immobilized with 0.1% tetramisole on 3% agar pads. As described in Gabel et al.41, a Ti:Sapphire laser system was used to perform axotomy (10 KHz pulse rate, 15 nJ/pulse). Axons were cut 20 μm from the soma with five rapid exposures (0.25 seconds) to the laser beam, resulting in vaporization of the axon at the target point. Time-lapse images were taken 20 seconds before cutting and up to a minute following the cut, 1 frame/second. Two individuals with exophers connected to the soma and three individuals with exophers not connected to the soma were analyzed, with only the connected exophers showing any calcium response to axotomy.
Blinding
Blinding was performed by lab members uninvolved in the relevant experiment. Strain and treatment information were recorded in a secret key and replaced with a symbol on the measurement plates. The data were unblinded following completion of the experiment. Animals were allocated to measurement plates randomly.
Statistical Analysis
Sample size was established using G-power software to be able to detect moderate effects with 80% power at P = 0.05 after a replicate for routine measurements. For higher throughput, larger screens were designed to have an 80% power to meet the re-screening cutoff of P = 0.25. Data were considered normal by the Shapiro-Wilk normality test.
Because of variable RNAi outcomes in different trials, exopher numbers were always compared to the empty vector control for that particular experiment. Statistics were performed using a two tailed unpaired t-test between the trial means, considering neurons with an exopher as 1 and neurons without an exopher as 0. One-way ANOVA was performed with Dunnett’s test when multiple samples were compared to a single control, and with Tukey’s test when multiple samples were compared to each other. Details of statistics are described in figure legends.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Figure 1. Morphological features of exophers derived from touch neurons.
a. An exopher is generated with evident filling and growth. S indicates the soma of an ALM neuron on adult day 2 with mCherry visualized; E indicates the significant extrusion of a balloon-like exopher, which grows with time. We noted that the size of this exopher increased for more than an hour, with fluorescence intensity increasing specifically in the exopher compartment, possibly via continual delivery of materials to the exopher after the initial formation (see Supplemental Video 2 for striking time-lapse movie corresponding to this image). Strain is Is[pmec-4mCh1].
b. An ALMR soma with multiple exophers. S, soma; E, exopher; strain is Is[pmec-4mCh2], adult day 2; scale bar, 2 μm.
c. A rare instance of an ALM neuron with exophers that appear extruded from the dendrite (arrows). Strain is Is[pmec-4mCh1], adult day 2. Scale bar, 2 μm.
d. Size measurements for somas (squares) and exophers (circles). Data are combined for exophers scored in different backgrounds, n = 35. Values are the basis of the measurements in Extended Data Table 2.
e. Example of a dye-filled exopher derived from an amphid neuron. We dye-filled the amphid neurons, which are open to the environment, in N2 animals and identified exophers in the amphid region, some of which appeared attached. Neuronal exophers are thus not induced solely in response to expression of foreign proteins, but rather can be produced from neurons that express only native proteins. Scale bar, 2 μm. Note that a second example of an exopher labeled by dye-filling is included in Extended Data Fig. 6c.
f. Early adult longitudinal time-course on DiI-soaked wild type N2. Dye-filled chemosensory amphid neurons also produce exophers with a peak early in life in wild type animals. The production of exophers in this study reflects the extrusion of native neuronal contents, as no fluorescent transgene is introduced. Total n > 150, 3 trials; trial means ± s.e.m.; day of adult life indicated on x-axis.
g, h. Dopaminergic PDE and CEP neurons can produce exophers. g. GFP-expressing PDE neuron with two anterior exophers indicated (8/450 had exophers, typical of the low rate observed with GFP reporters in touch neurons); h. CEP neuron with an associated exopher. Transgene is dopaminergic neuron-specific reporter egIs1[pdat-1GFP]; adult day 2; scale bar, 2 μm.
i. ASER neurons can form exophers. Strain is sesIs25 [Pflp-6Aβ; Pgcy-5GFP]32; adult day 2; scale bar, 2 μm.
j. The onset of touch neuron exophers in an hsf-1 mutant background occurs one day earlier than wild type touch neurons, beginning on adult day 1, but follows the general trend of high incidence early in adult life. Longitudinal study with Is[pmec-4GFP]; hsf-1(sy441) (2 green trials, starting n = 25), and Is[pmec-4mCh1]; hsf-1(sy441) (red, starting n = 25), day of adult life indicated on x-axis. We observed a similar temporal pattern for distinct transgenic line Is[pmec-4mCh2]; [pmec-3Q128CFP] (data not shown). A late onset peak might not be apparent due to sickness of hsf-1 mutants later in life. Data are from single longitudinal trials and thus error bars are not included.
Extended Data Figure 2. Electron microscopy images of an extruded exopher.
a. A membrane GFP reporter reveals that exophers contain membrane. Strain is ZB4524 bzEx242 [Pmec-4PH(plcDelta)::GFP]35. PVM neuron. S, soma; E, exopher. Scale bar, 2 μm.
b. General relationship of exopher to ALMR soma. Schematic view from a lateral aspect (anterior to the left). The ALMR soma remains connected to its primary dendrite as represented here, projecting leftward. Several smaller membrane-bound tubes extend away from the soma, containing small items expelled from the ALMR soma, such as the large vesicle shown along the path between the soma and the exopher. The ALMR nucleus (N) remains intact, but pushed to an eccentric position by other cytoplasmic inclusions. The soma contains intact rough endoplasmic reticulum (RER), mitochondria (M), small vesicles (not shown), and larger protein aggregates (A).
The exopher comprises heterogeneous contents. The exterior of the exopher is completely bounded by the hypodermal plasma membrane, so that none of the exopher contents are immediately in contact with the hypodermal cytoplasm. Often, a double membrane is observed in which the exopher is likely to supply the inner bilayer, and the hypodermis is likely to contribute the outer bilayer. Additional, separate membrane-bound items lie peripherally (not shown; but see panel h), whose provenance is uncertain, but may represent breakdown products from an earlier, larger stage of the exopher. Most internal contents of the exopher also have their own membrane boundaries, but some diffuse material (not shown; but see panels c, e) fills the spaces around those membrane-bound objects. Membrane-bound contents include portions of the neuronal cytoplasm holding intact RER, large protein aggregates (A), and complex whorls of membrane (W) that generally seem to enclose empty space. Two lysosomes (L) are shown here, one lying separately, nearby the exopher, and a second one in the process of fusing into the exopher’s outer membrane. Other small lysosomes are seen within the exopher (not shown). A tube is shown extending away (far right) towards the pseudocoelom, which might offer one potential route for elimination of contents that cannot be degraded during hypodermal transit. Cartoon designed by and published with permission of C. Crocker.
Panelsc,d,f,h are TEM views of a single exopher, emitted from the cell body in panel e.
c. The exopher is fully embedded within the hypodermis. Strain is Is[pmec-4mCh1]; Is[pmec-4GFP]. TEM of thin transverse section, seen from anterior aspect (thus left/right are reversed). Shown is an exopher (E) fully embedded within the hypodermis (H). Animal cuticle shown on the left (C). The exopher is ~1.5 μm across, similar in size to the excretory canal (EC). The exopher lies closer to the pseudocoelom (P), while the ALMR neuron soma lies closer to the cuticle (see panel e). Jagged white spaces running vertically through the hypodermis are an artifact where the tissue cracked during processing. Scale bar, 2 μm.
d. Exophers are complex and heterogeneous structures with multiple membranes. The exopher is characterized by many small round protrusions and involutions. The main exopher complex has a complete plasma membrane surrounding it, isolating its contents from the hypodermal cytoplasm. Involuted portions of the exopher can have multiple membrane layers. In this thin section, the exopher displays structures such as protein aggregates (A), rough endoplasmic reticulum (RER), and a possible small lysosome, in addition to loose material floating inside the exopher membrane within a less electron dense fluid matrix. Scale bar, 1 μm.
e. The originating touch neuron soma remains intact. The ALMR neuron still has intact cell and nuclear membranes. Small mitochondria (M) are visible inside the soma, as are protein aggregates (A). Note that the aggregate within the soma is not membrane bound, resembling a mammalian aggresome42. The electron density of the neuronal cytoplasm is darker than that of the surrounding hypodermis, and the mitochondria of the hypodermis are far larger than those of the neuron. Scale bar, 2 μm.
f. The exopher is surrounded by a continuous membrane and contains electron-lucent materials and electron dense membrane whorls (W). The exterior of the exopher is completely bounded by the hypodermal plasma membrane, so that none of the exopher contents are immediately in contact with the hypodermal cytoplasm. The exopher contains complex whorls of membrane (W) that enclose an electron-lucent lumen. Within the exopher membrane, the exopher lumen also is electron-lucent, with some diffuse free-floating material. Scale bar is 1μm.
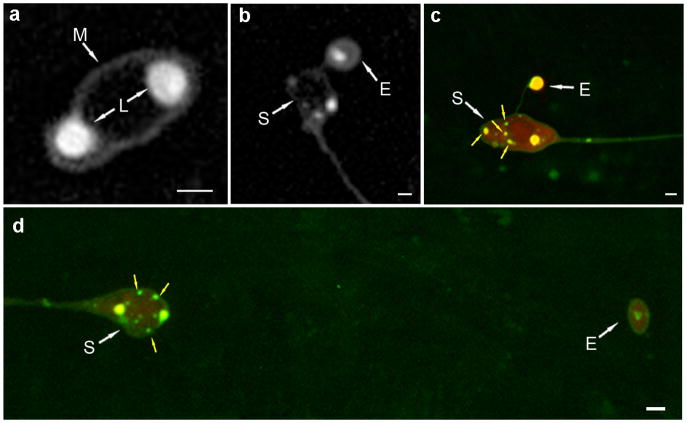
g. Fluorescent images demonstrate exopher features similar to the TEM ultrastructure. Fluorescent microscopy of exophers have irregular shapes and round protrusions, and an irregular distribution of distinct fluorescent signals that resemble the heterogeneous domains we see by TEM. Shown is an exopher in a day 2 animal expressing a soluble YFP, a CFP PolyQ128 fusion, and an aggregation prone mCherry. The mCherry has a bright spot that excludes the other two signals (arrow). The Q128 CFP is localized in the middle portion of the exopher, and the YFP is most strongly localized to the bottom portion of the exopher. The YFP signal also forms a dim ring around the mCherry spot. View is from a lateral aspect, as in panel a. Scale bar, 2 μm.
h. Serial sections of the exopher reveal a complex and heterogeneous structure. Seven views (numbered 1–7) are shown from 50 serial thin sections that traverse the main region of the exopher. As one approaches the edge of the exopher, additional membrane-bound objects can be seen at the fringe of the main body (see panels 6, 7), which likely represent portions that have decayed from the original larger object, and are perhaps more easily phagocytosed by the hypodermis, or shuttled along a tube for release into the pseudocoelom. A small electron dense lysosome (L) lies beside the exopher in panel 5. Scale bar, 1 μm.
Extended Data Figure 3. FRAP and post-axotomy calcium imaging support that exophers that appear connected to the soma can fill with cytoplasmic materials.
a. Both connected and unconnected exophers can be identified at high frequency in the same strain with a 40x objective. Strain is Is[pmec-4mCh1], adult day 2; n = 77 total, 3 trials; trial means ± s.e.m.
b. Example of a connected ALMR exopher recovering after laser-bleaching. Strain is Is[pmec-4mCh1]; Is[pmec-4GFP], adult day 2. 0 seconds is prior to laser treatment, other times are post-treatment.
c. Example of a detached ALMR exopher photo-bleaching and failing to refill. Strain is Is[pmec-4mCh1]; Is[pmec-4GFP], adult day 2.
d. Fluorescence recovery measurements reveal that some connected exophers are able to transport fluorescent material from the cytoplasm to the exopher, while disconnected exophers do not appear to transport fluorescent material. Shown are data for examples in b (red, connected) and c (blue, unconnected) above.
e. Time-lapse measurements of fluorescence intensity of the soma (blue trace) and the exopher (red trace) during the laser axotomy experiment in f show that the injury-induced calcium wave in the soma is followed by a pulse of calcium elevation in the exopher (yellow arrow, laser axotomy at t = 0 seconds). We analyzed two individual neurons with exophers connected to the soma and three individual neurons with what appeared to be non-connected exophers. Only the clearly connected exophers gave a calcium response comparable (~100% signal) to that measured at the cell soma as in this panel. Strain is ZB4059 bzIs163 [Pmec-4GCaMP3.0::SL2::mCherry].
f. The soma calcium wave induced by laser axotomy is followed by a calcium wave to connected exophers. We laser-cut an ALMR neuron that had a connected exopher in a day 2 adult that expressed both mCherry and the calcium sensitive fluorophore, GCaMP3. We made the laser cut 20 μm along the axon (yellow arrow) at time t = 0 while taking simultaneous time-lapse images (1 frame/5 seconds). Selected frames are shown at t = −20 seconds (before laser axotomy), right after laser axotomy at t = 5 seconds (note increased fluorescence in the soma; white arrow), and at t = 15 seconds and 25 seconds post-axotomy (note the later exopher increase in fluorescence; white arrowheads). Signal quantitation is in e above. Supplemental Video 3 shows the calcium wave that travels from soma to exopher.
Extended Data Figure 4. Lysosomes can be found in exophers.
We also analyzed GFP-tagged lysosomes in touch neurons (bzIs168 [Pmec-7LMP-1::GFP]) to establish that lysosomes can be extruded in exophers. We observe two types of lysosomal arrangements in exophers: (1) those that have small lysosome-like concentrated fluorescence, with mCherry dispersed (d), and (2) those that are nearly fully loaded with lysosome-like staining in which mCherry is also present throughout (Extended Data Fig. 6c). The inclusion of lysosomes in exophers suggests that some elimination of expelled material might be accomplished via internal degradation. Alternatively, dysfunctional lysosomes might be expelled via exophers. Future studies will be needed to define the role of the lysosome in exophers.
a. Neuron soma featuring typical two large LMP-1::GFP-tagged pericentric lysosome domains, with no smaller ones evident. Strain is ZB4509 Is[pmec-4mCh1]; bzIs168 [Pmec-7LMP-1::GFP], green channel shown. As has previously been observed43, LMP-1::GFP signal clearly marks the plasma membrane (M), but less intensely than the lysosomes (L). Scale bar, 2 μm.
b. LMP-1::GFP reveals lysosome inclusions are frequent and sometimes prominent in exophers. Strain is ZB4070 bzIs168 [Pmec-7LMP-1::GFP]; S, soma; E, exopher. We found that 18/25 (~70%) of exophers scored contained lysosomes in day 2 adults. Note that LMP-1::GFP faintly labels membrane43 and rings the exopher in panels b-d, supporting that the exopher is membrane-bound. Scale bar, 2 μm.
c, d. Co-labeling of aggregating mCherry and lysosome compartments suggests two types of lysosomal organization in exophers. Strain is ZB4509 Is[pmec-4mCh1]; bzIs168 [Pmec-7LMP-1::GFP]. c. Some exophers appear to be filled with LMP-1::GFP and coincident mCherry. d. LMP-1::GFP-tagged lysosomes included in exophers can be small and differentially localized from mCherry. In the absence of stress, neurons typically feature two large intensely-fluorescent pericentric lysosome domains with LMP-1::GFP, with few smaller ones evident (see panel a). Neurons that had an exopher tended to also have additional small mobile lysosomes that we did not observe in cells without an exopher(see panel b, c, d). Additionally, neurons that featured “large lysosome” exophers generally appeared to have fewer of the large perinuclear lysosomes in the soma (example in d). Scale bar, 2 μm.
Extended Data Figure 5. Mitochondria GFP markers exhibit a normal mitochondrial appearance in exophers.
a. Mitochondria in exophers can form a network. Strain Is[Pmec-4mitoLS::ROGFP]; Is[Pmec-4mCh1]). Shown is an exopher (E) budding off from the ALMR soma (S). The exopher contains a disproportionate number of punctate mCherry aggregates; the exopher also includes a GFP signal typical in size for neuronal mitochondria. It is noteworthy that the mitochondria in the exopher exhibit a filamentous structure similar to those in the soma, and the signal does not co-localize with the mCherry signal, but rather may remain within a distinct sub-cellular domain. These two observations are consistent with the mito-GFP label localized to actual mitochondria as opposed to representing mislocalized GFP-labeled protein. Scale bar, 2 μm.
b. Exophers can contain punctate mitochondria, networked mitochondria, or no mitochondria. Strain Is[Pmec-4mitoLS::ROGFP]; Is[Pmec-4mCh1]). Here we show three exophers. In the left exopher, the mitoROGFP signal is localized to two puncta. The middle exopher contains networked mitochondria. The right exopher contains no visible mitoROGFP signal. Scale bar, 2 μm.
c. Zoom out of panel b, to show location of exophers(E) relative to the touch neuron soma (S). Scale bar, 2 μm.
d. MitoTimer reporter reveals a difference in mitochondrial-matrix oxidation environment in exophers vs. somas. MitoTimer encodes a dsRed derivative that fluoresces green when reduced (first synthesized), but irreversibly shifts to red fluorescence as it oxidizes20. We used a single copy pmec-4MitoTimer reporter to measure the relative red/green signal in exopher-soma pairs. Exophers have proportionately more oxidized signal, suggesting “older” mitochondria (with more oxidation of the matrix-localized reporter) are preferentially expelled. n = 7 exophers, scale bar, 2 μm; single exopher means ± s.e.m.; *P < 0.05, paired t-test.
e. Pharmacological disruption of mitochondria leads to higher rates of mitochondrial inclusion in exophers. Juglone exposure leads to an increase in intracellular reactive oxygen species production, most notably superoxide radicals, and can cause mitochondrial membrane depolarization and opening of permeability transition pores, allowing pro-apoptotic Cytochrome C to escape from the mitochondria21. We treated strain Is[Pmec-4mCh1]; zhsEx17 [Pmec-4mitoLS::ROGFP] with 230 μM juglone which increases ROS production.. Exophers from ALMR neurons from animals exposed to juglone (blue, n = 30 total exophers) were significantly more likely to include at least one mitochondrion than exophers from animals in the control (white, n = 22 total exophers). Mitochondrial extrusion increases under conditions of juglone-induced oxidative stress. 3 trials, trial means ± s.e.m.; **P < 0.01, unpaired t-test.
Extended Data Figure 6. RNAi knockdown of ced-1 and ced-6, but not other engulfment machinery, increases occurrence of multiple exophers detected.
a. RNAi knockdown of ced-1 and ced-6 engulfment genes increases the number of ALMR neurons with 2 or more exophers near the touch neurons, supporting conclusions from mutants. RNAi against C. elegans transmembrane receptor ced-1/CD91, ced-6/GULP, GTPase ced-10/RAC1, phosphatidylserine receptor psr-1, and pgrn-1/progranulin-1(RNAi increases apoptotic corpse clearance44). Control is empty vector, strain is ZB4071 bzIs169 [Pmec-18sid-1Psng-1YFP]; bzIs101 [Pmec-4mCherry], at least 3 trials each, n > 30 ALMRs measured per trial; n > 15 cells with exopher per condition graphed; means ± s.e.m., one-way ANOVA with Dunnett’s test, *P < 0.05 and **P < 0.01. Note that ced-1 and ced-6 RNAi do not increase the percentage of ALMRs producing exophers (data not shown), but rather increases the number of animals with multiple exophers detectable at adult day 2. Thus, ced-1 and ced-6 knockdown might affect persistence of expelled materials introduced into the hypodermal compartment.
b. Phosphatidylserine (PS) indicator annexin V::GFP ref. 44 labels apoptotic corpses, but does not label exophers in ZB4083 smIS76 [phsp-16ANV::GFP]; Is[pmec-4mCh1]. Broadly expressed annexinV::GFP binds to PS on developmental apoptotic corpses in embryos (I), but does not bind to exophers (right example compares mCherry-labeled exopher (II) to annexinV::GFP channel for the same image (III), which should visualize bound annexinV::GFP (as occurs for apoptotic corpses in embryos of the same strain (I) if PS is on the exopher surface). Note that in published studies, PS can be recognized on corpses of necrotic touch neurons, showing that touch neurons can produce surface PS, and can be recognized by annexinV-tagging, when inappropriately induced to die33,45. 0/18 fluorescent mCherry-labeled exophers were co-labeled with annexinV::GFP. Knocking down ced-1 by RNAi in the annexinV::GFP line did not enable us to better detect PS on exophers (n = 0/25 additional observations).
c. DiI introduced via amphid neurons can be detected in anterior coelomocytes. In a DiI-soaked N2 animal, an amphid exopher (E) originating from the ASIR soma (S) can be seen proximal to the terminal bulb of the pharynx (P). The anterior coelomocytes ccAR and ccPR (C) also contain DiI. Coelomocytes have no connection to the external environment, suggesting that the DiI must have been uptaken and jettisoned by the chemosensory amphid neurons and subsequently engulfed, analogously to the mCherry detected in Is[pmec-4 mCh1] coelomocytes. This wild type context suggests coelomocytes can scavenge the contents of exophers that are generated under normal physiological conditions, without the added stress of a potentially aggregating product of a transgene. Scale bar 5 μm.
d. In ZB4082 cup-4(ok837); Is[pmec-4mCh1] mutants in which coelomocyte uptake is disrupted, we observe increased incidence of dispersed fluorescence, similar to that shown here (S, ALMR soma; D bracket, “starry night phenotype,” present in 29/200 animals, adult day 4). Similar dispersions are rare in cup-4(+) lines. Scale bar, 2 μm. AVM soma is also visible.
e. Young adult animals that produced an exopher often later exhibit the starry night phenotype, suggesting mCherry material can move through the body. Is[pmec-4mCh1] animals were separated into populations that had an ALMR exopher on either adult day 2 or day 3 (blue arrows), and a population that had no ALMR exopher on either day 2 or 3 (white arrows). Animals were scored again on day 5 for presence of the starry night phenotype. In the exopher-producing group, 42% of animals exhibited a starry night phenotype, while in the non-exopher producing group only 6% of the animals exhibited the starry night phenotype. 3 trials, n = 60 total per group, means ± s.e.m.; *P < 0.05, unpaired t-test. Arrow thickness is weighted according to relative incidence. Note that the “no exopher” category should include animals that have actually produced exophers, but were not present at the time of sampling, thus differences are likely to be under-estimated in this panel.
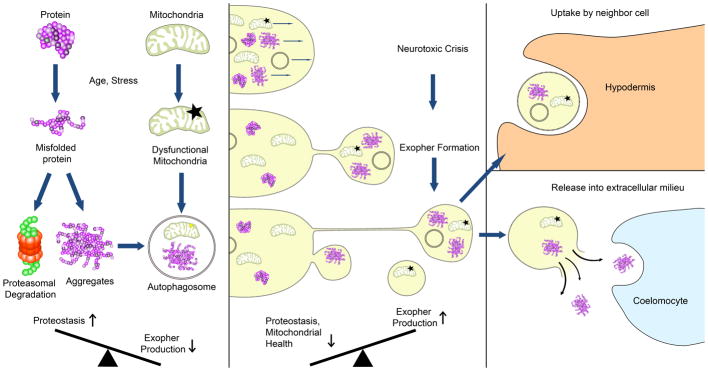
Extended Data Figure 7. Working model for a proposed exopher role in proteostasis.
As neurotoxic events such as protein aggregation or mitochondrial dysfunction occur in the cell, multiple homeostatic mechanisms clear them (left panel). At the young adult transition point to adult proteostasis (heat shock response down, UPR down, proteasome activity up12–14) or when basal levels of damage reach a threshold and overwhelm neuronal proteostasis, aggregates and organelles such as mitochondria and lysosomes are sequestered into a compartment that can be jettisoned from the cell. One possibility is that this compartment might correspond to the aggresome described in mammalian cells42. For touch neurons, extruded exopher contents may be degraded by accompanying lysosomes, digested by the surrounding hypodermis, or may reach the pseudocoelom and be taken up by coelomocytes. The process of exopher-genesis appears to be neuroprotective in young adults, but when dysregulated, might induce toxicity in neighboring tissues. We speculate that exopher contents that cannot be degraded or passed on could remain in the neighboring cell, where they could contribute to dysfunction. Possibly, exopher-genesis may be akin to the process by which protein aggregates and mitochondria become localized to neighboring neurons in humans, promoting disease spread.
Extended Data Table 1. Impact of RNAi knockdown of genes functioning in exosome biogenesis and cell cycle progression on exopher detection.
a, b. Neither ESCRT proteins required for exosome biogenesis, nor cell cycle-related homologs exert major influences on exopher-genesis.
a. RNAi knockdown against ESCRT genes did not significantly affect exopher occurrence. Strain is bzIs169 [Pmec-18sid-1 Psng-1YFP]; bzIs101 [Pmec-4mCherry; unc-119+]; hsf-1(sy441). For each listed gene, P-value is from at least 3 trials, n > 100 total per condition, unpaired t-test. Note that negative results for RNAi screens are not definitive. The data support that the biogenesis of the exopher is distinct from the biogenesis of exosomes (Extended Data Table 2).
b. RNAi knockdown against homologs of cell cycle-related genes did not significantly influence exopher production, with the exception of cki-1. Reducing cki-1 activity by RNAi causes extra larval cell divisions46, so its inhibition of exopher-genesis, although interesting, is not consistent with new cell division underlying exopher production. Note that cdk-4 and cyd-1 are required for cell cycle progression in postembryonic cells. Strain is bzIs169 [Pmec-18sid-1 Psng-1YFP]; bzIs101 [Pmec-4mCherry; unc-119+]. For each listed gene, P-value is from at least 3 trials, n > 100 total per condition, unpaired t-test. FDR statistic for cki-1(RNAi) is q = 0.04, Benjamini-Hochberg FDR calculator.
| a. Exosome Biogenesis | ||
|---|---|---|
|
| ||
| Process Targeted | Gene Name | P-value |
| ESCRT-0 | hgrs-1 | 0.48 |
| stam-1 | 0.21 | |
| ESCRT-1 | tsg-101 | 0.55 |
| vps-28 | 0.67 | |
| vps-37 | 0.22 | |
| ESCRT-2 | vps-22 | 0.50 |
| vps-25 | 0.96 | |
| vps-36 | 0.54 | |
| ESCRT-3 | vps-20 | 0.49 |
| vps-24 | 0.89 | |
| b. Cell Cycle-related | |||
|---|---|---|---|
|
|
|
||
| Gene Name | P-value | Gene Name | P-value |
|
|
|
||
| cki-1 | 0.03* | cit-1.2 | 0.75 |
| ccnk-1 | 0.46 | cki-2 | 0.83 |
| cdk-4 | 0.84 | cya-1 | 0.52 |
| cdk-9 | 0.68 | cya-2 | 0.45 |
| cdk-1 | 0.40 | cyb-2.1 | 0.52 |
| cdk-12 | 0.23 | cyd-1 | 0.95 |
| cdk-5 | 0.72 | cye-1 | 0.95 |
| cdk-8 | 0.79 | cyl-1 | 0.32 |
| cic-1 | 0.95 | dpl-1 | 0.94 |
Extended Data Table 2. Comparison of features of exophers to other characterized extracellular vesicles.
| Exosomes | Microvesicles | Migrasomes | Exophers | |
|---|---|---|---|---|
|
|
||||
| Diameter | 30 nm–100 nm46 | 100 nm–1,000 nm46 | 500 nm–3,000 nm49 | 1,000 nm–7,800 nm |
| Timing of release | Tens of minutes47 | Seconds47 | 40–200 minutes49 | 15–60 minutes |
| Mechanism of Release | Multi-vesicular bodies fuse to the cell membrane46 | Outward budding and scission46 | Expands at tip of retraction fibers49 | Jettisoned from cell body |
| ESCRT Machinery involved | Yes47 | ESCRT3; tsg-10147 | Unknown | No |
| Attachment to releasing cell | No | No | Yes (via retraction fibers)49 | Sometimes (via thin fiber) |
| Actin required | No | No | Yes49 | Yes |
| Vesicular Contents | DNA, RNA, Proteins, Lipids46 | DNA, RNA, Proteins, Lipids46 | Cytosol, Proteins49 | Mitochondria, Lysosomes, Protein Aggregates |
| Phosphatidylserine Distribution | Membrane outer leaflet46 | Membrane outer leaflet48 | Unknown | Not displayed on membrane outer leaflet |
Supplementary Material
Acknowledgments
We thank B. Grant for expert advice, C. Reina for time-lapse microscopy help; N. Kane and J. Kramer for confocal microscopy assistance; and H. Ushakov for construction of some genetic lines. We thank Geoff Perumal and Frank Macaluso for help with HPF fixations, and Chris Crocker for the cartoon in Extended Data Fig. 2b. Federico Sesti and Massimo Hilliard generously supplied C. elegans strains; Alex Mendenhall, Bryan Sands, Roger Brent constructed the MOSCI mitoTimer strain. Research was supported by the National Institutes of Health under award numbers 1R01NS086064 and 1R01AG046358. IM and RG were supported by the National Institute of General Medical Sciences under award number T32 GM008339. KN and DH were supported by NIH OD10943 (to DHH); Core EM facilities (Hall) NICHD P30 HD71593 for the RFK-IDDRC at Albert Einstein College of Medicine. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions I.M., M.L.T., M.L.A., R.J.G. and G.H. conducted and designed experiments along with M.D. I.M. and M.D. wrote the manuscript with input from R.J.G., M.L.A. and M.L.T. C.V.G. and D.T. carried out calcium connection experiments. K.C.N. and D.H.H. carried out electron microscopy. J.A.P. and C.N. supplied the Q128 reagent and manuscript critiques.
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 2.Federico A, et al. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci. 2012;322:254–262. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Gedalya T, Cohen E. Quality control compartments coming of age. Traffic. 2012;13:635–642. doi: 10.1111/j.1600-0854.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee SJ, Desplats P, Sigurdson C, Tsigelny I, Masliah E. Cell-to-cell transmission of non-prion protein aggregates. Nat Rev Neurol. 2010;6:702–706. doi: 10.1038/nrneurol.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis CH, et al. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci U S A. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toth ML, et al. Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J Neurosci. 2012;32:8778–8790. doi: 10.1523/JNEUROSCI.1494-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig AL, Moser SC, Bailly AP, Gartner A. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods in cell biology. 2012;107:321–352. doi: 10.1016/b978-0-12-394620-1.00011-4. [DOI] [PubMed] [Google Scholar]
- 9.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 10.Parker JA, et al. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci U S A. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vayndorf EM, et al. Morphological remodeling of C. elegans neurons during aging is modified by compromised protein homeostasis. NPJ Aging Mech Dis. 2016;2 doi: 10.1038/npjamd.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbadia J, Morimoto RI. Proteostasis and longevity: when does aging really begin? F1000Prime Rep. 2014;6:7. doi: 10.12703/p6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labbadia J, Morimoto RI. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samann J, et al. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. The Journal of biological chemistry. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Springer W, Hoppe T, Schmidt E, Baumeister R. A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum Mol Genet. 2005;14:3407–3423. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]
- 17.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannon MB, Remington SJ. Redox-sensitive green fluorescent protein: probes for dynamic intracellular redox responses. A review. Methods Mol Biol. 2008;476:51–65. doi: 10.1007/978-1-59745-129-1_4. [DOI] [PubMed] [Google Scholar]
- 19.Ghose P, Park EC, Tabakin A, Salazar-Vasquez N, Rongo C. Anoxia-reoxygenation regulates mitochondrial dynamics through the hypoxia response pathway, SKN-1/Nrf, and stomatin-like protein STL-1/SLP-2. PLoS Genet. 2013;9:e1004063. doi: 10.1371/journal.pgen.1004063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laker RC, et al. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. The Journal of biological chemistry. 2014;289:12005–12015. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji YB, Qu ZY, Zou X. Juglone-induced apoptosis in human gastric cancer SGC-7901 cells via the mitochondrial pathway. Experimental and toxicologic pathology : official journal of the Gesellschaft fur Toxikologische Pathologie. 2011;63:69–78. doi: 10.1016/j.etp.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Patton A, et al. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Current biology : CB. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 23.Nath S, et al. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of beta-amyloid. J Neurosci. 2012;32:8767–8777. doi: 10.1523/JNEUROSCI.0615-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussbaum-Krammer CI, Park KW, Li L, Melki R, Morimoto RI. Spreading of a prion domain from cell-to-cell by vesicular transport in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003351. doi: 10.1371/journal.pgen.1003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costanzo M, et al. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- 26.Abounit S, Zurzolo C. Wiring through tunneling nanotubes--from electrical signals to organelle transfer. J Cell Sci. 2012;125:1089–1098. doi: 10.1242/jcs.083279. [DOI] [PubMed] [Google Scholar]
- 27.Gousset K, et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 28.Pearce MM, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun. 2015;6:6768. doi: 10.1038/ncomms7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquier J, et al. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med. 2013;11:94. doi: 10.1186/1479-5876-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayakawa K, et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 2016;535:551–555. doi: 10.1038/nature18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Z, Sesti F. A Caenorhabditis elegans model system for amylopathy study. Journal of visualized experiments : JoVE. 2013:e50435. doi: 10.3791/50435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mapes J, et al. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr Biol. 2012;22:1267–1275. doi: 10.1016/j.cub.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann B, et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–222. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- 35.Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Developmental biology. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 36.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 37.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scerbak C, et al. Insulin signaling in the aging of healthy and proteotoxically stressed mechanosensory neurons. Front Genet. 2014;5:212. doi: 10.3389/fgene.2014.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topalidou I, et al. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22:1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabel CV, Antoine F, Chuang CF, Samuel AD, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- 42.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends in cell biology. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 43.Hermann GJ, et al. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 45.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 46.Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chem Bio Chem. 2014;15:923–928. doi: 10.1002/cbic.201400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 49.Ma L, et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015;25:24–38. doi: 10.1038/cr.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.