Abstract
Many pathogenic Gram-negative bacteria use the type III secretion system (T3SS) to deliver effector proteins into eukaryotic host cells. In Yersinia, the switch to secretion of effector proteins is induced first after intimate contact between the bacterium and its eukaryotic target cell has been established, and the T3SS proteins YscP and YscU play a central role in this process. Here we identify the molecular details of the YscP binding site on YscU by means of nuclear magnetic resonance (NMR) spectroscopy. The binding interface is centered on the C-terminal domain of YscU. Disrupting the YscU-YscP interaction by introducing point mutations at the interaction interface significantly reduced the secretion of effector proteins and HeLa cell cytotoxicity. Interestingly, the binding of YscP to the slowly self-cleaving YscU variant P264A conferred significant protection against autoproteolysis. The YscP-mediated inhibition of YscU autoproteolysis suggests that the cleavage event may act as a timing switch in the regulation of early versus late T3SS substrates. We also show that YscUC binds to the inner rod protein YscI with a dissociation constant (Kd) of 3.8 μm and with 1:1 stoichiometry. The significant similarity among different members of the YscU, YscP, and YscI families suggests that the protein-protein interactions discussed in this study are also relevant for other T3SS-containing Gram-negative bacteria.
Keywords: Gram-negative bacteria, nuclear magnetic resonance (NMR), protein conformation, secretion, type III secretion system (T3SS), YscI, YscP, YscU
Introduction
Yersinia spp. share a common virulence plasmid that encodes a type III secretion system (T3SS)3 supporting the secretion of the Yersinia outer protein (Yop) effectors from the bacterium into eukaryotic host cells (1). After translocation into the host cells, Yop effectors counteract immune defense mechanisms such as phagocytosis or apoptosis and thereby promote the survival and propagation of the extracellular bacteria (2). Many pathogenic and symbiotic Gram-negative bacteria also utilize the T3SS to deliver proteins during the interactions with their hosts (3, 4). The T3SS (also referred to in the literature as the non-flagellar T3SS) evolved from the flagellum, and both systems present a similar architecture (5, 6) featuring a basal body assembly consisting of a multiringed complex that spans the bacterial envelope (5). However, the basal body of the flagellum is connected to an extracellular hook that forms a link with the flagellar filament. In contrast, the T3SS basal body is attached to a needle (in animal pathogens) or a pilus (in plant pathogens) that protrudes from the bacterial surface (5, 7, 8). The basal body and the needle/pilus together form the so-called needle complex that is the hallmark of the T3SS.
It is accepted that the needle complex is assembled in a stepwise manner, and in Yersinia spp., it has been shown that the inner membrane platform (formed by YscR, -S, -T, -U, and -V) is assembled independently of the outer membrane ring-forming proteins (YscC, -J, and -D) (9). After association of the inner and outer rings (mediated by YscJ), the secretion of the so-called early substrates such as the needle subunit YscF begins, which results in elongation of the needle complex. Activation of the T3SS impairs the secretion of the early substrates but triggers the secretion of the Yop effectors (late substrates) (10, 11). This modification of the secretion pattern was first described by Macnab and co-workers (12–14) in the flagellum and is called the substrate specificity switch. It has been shown that YscP plays a critical role in regulating the needle length (15–18). An YscP-null mutant or insertions within the YscP sequence triggered the formation of long needles. in contrast, shorter needles were produced when deletions were introduced in the YscP sequence (19). Furthermore, a minimal needle length is required to support Yop secretion (17). Together, these results suggest that the needle length is tightly regulated and that YscP functions as a molecular ruler (16). Although the ruler model has attracted significant interest, alternative models of needle length control such as the measuring cup model (15) and the molecular clock model (20) have also been proposed, and there is currently no consensus regarding the true nature of the needle length control mechanism. A systematic deletion analysis of YscP in Yersinia enterocolitica led to the identification of domains with specific functions (11, 19), including two distinct secretion signals (residues 1–35 and residues 97–137) and a substrate specificity switch domain located between residues 385 and 500 (11). Homologues of YscP with similar functions are found in the flagella and T3SSs of diverse bacterial species (21, 22).
YscU in Yersinia spp. and FlhB in the flagellum have also been linked to the substrate specificity switch (23). YscU is anchored in the inner membrane via four transmembrane helices and possesses a large C-terminal cytoplasmic domain named YscUC (Fig. 1) (24, 25). YscUC is characterized by a conserved NPTH motif that undergoes an autoproteolytic process (between Asp263 and Pro264) to generate a C-terminal peptide named YscUCC that form a stable complex with the N-terminal part of the cytoplasmic domain named YscUCN. Previous results obtained in our laboratories suggest that the positively charged residues within the linker between YscUC and the membrane domain interact with the membrane lipids to associate YscUC with the inner membrane (26). Accordingly, it has been shown that mutations at Asp263 or Pro264 that block YscU autoproteolysis interfere with both needle formation and Yop secretion (10, 27). Also, we have shown that the dissociation of YscUC is required for Yop effector secretion (28). It was subsequently found that YscUC contains a C-terminal secretion signal domain (29). The deletion of the last 15 residues of YscU triggered an increase in YscF secretion without affecting Yop secretion (29). Thus, like YscP, YscU is involved in the substrate specificity switch. Similar results were reported for FliK and FlhB, which are the flagellar homologues of YscP and YscU, respectively (30, 31). Interestingly, the phenotype observed in a yscP-null mutant was partially suppressed by single amino acid substitutions within YscU (YscUA268F, YscUY287G, and YscUV292T) (10). It was subsequently shown that these suppressor mutants are less stable and exhibit faster dissociation kinetics than the wild-type protein, which enabled Yop secretion to occur in the absence of YscP (10, 28). Direct interactions have been observed between FlhB and FliK (Salmonella enterica serovar Typhimurium) and between the homologous proteins in Pseudomonas aeruginosa, PscU and PscP (residues 31–36), supporting the proposed functional link between YscU and YscP homologues (10, 21, 22).
FIGURE 1.
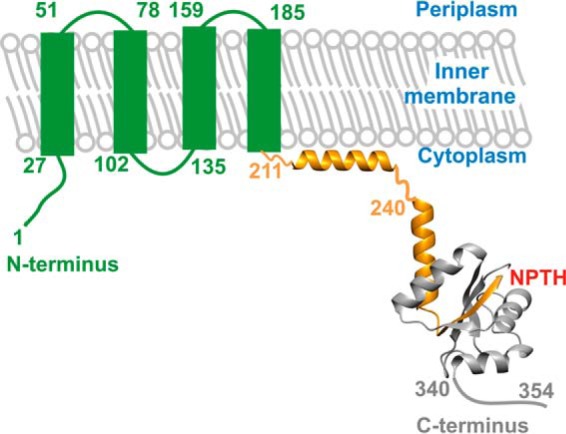
Schematic representation of the YscU domain architecture. YscU features an N-terminal inner membrane-interacting domain (green) consisting of four transmembrane α helices, which were predicted using the TMHMM server. The cytoplasmic domain of YscU, denoted YscUC, has been crystallized with a construct comprising residues 240–339 (Protein Data Bank code 2JLI) and is shown in a schematic representation (25). The NPTH motif is indicated in red, and the polypeptides resulting from autocleavage are colored orange (YscUCN) and gray (YscUCC), respectively. The linker sequence (residues 211–239) connecting the N-terminal domain with YscUC has been shown to interact with the inner membrane (25–27) and is modeled as an α helix.
The homologue of YscI in S. enterica serovar Typhimurium, PrgJ, has been proposed to form the inner rod, a structure that connects the needle to the basal body (20, 32). A study on Yersinia pseudotuberculosis showed that both YscP and YscU regulate YscI export (33). Indeed, YscI secretion by the T3SS is increased in a yscP-null mutant, whereas mutations in YscUC reduced the secretion level of YscI in a ΔyscP strain. Interestingly, it was also recently shown that in Y. enterocolitica YscI is required for the export of YscP-PhoA hybrid into the periplasm (34). Moreover, an interaction between EscI and EscU (homologues of YscI and YscU, respectively) in Escherichia coli has been reported (35).
Here we report data regarding the interactions among YscU and the two proteins YscP and YscI. Using NMR spectroscopy, we showed that YscP binds to the C-terminal helix of YscU. The A335N point mutation within the YscP binding interface of YscUC disrupted YscU-YscP interaction without affecting the YscUC structure. The interaction was found to be functionally significant, and a yscU-null mutant expressing the YscUA335N variant did not secrete Yops into the culture supernatant and did not induce cytotoxicity in HeLa cells. Furthermore, unlike a ΔyscP mutant or YscU mutants in the NPTH motif, YscUA335N did not promote secretion of the needle subunit YscF. These results suggest that the YscU-YscP interaction is critical for secretion of both early and late substrates. In addition, we found that binding of YscP to the slowly autocleaving mutant YscUCP264A reduced the rate constant of the autocleavage event in vitro. This finding suggests that the binding of YscP to YscUC can regulate the autocatalytic event, which in turn would affect the ability of YscUC to dissociate and the subsequent secretion of its C-terminal polypeptide (YscUCC). Finally, data from isothermal titration calorimetry and pulldown assays show that YscI interacts with YscUC but not with either of the polypeptides resulting from autoproteolysis and dissociation (YscUCN and YscUCC).
Results
YscU Binds to the Disordered Segment of YscP
Previous genetic analysis showed that yscP deletion abolishes Yop secretion and that the wild-type phenotype can be partially rescued by the A268F, Y287G, and V292T suppressor mutations within YscU (10). Based on these results, it was proposed that an interaction between YscP and YscU was critical for Yop secretion. Here we characterize the interaction between the cytoplasmic domain of YscU (YscUC) and YscP using NMR spectroscopy. A direct interaction between YscP and YscUC homologues in P. aeruginosa (denoted PscP and PscUC, respectively) has been observed previously by NMR (22); it was shown that a conserved N-terminal segment of PscP was responsible for its interaction with PscUC. The domain architecture of PscP has been described as a “ball-and-chain” topology where the “ball”, i.e. the C-terminal domain, is folded and believed to function as a substrate switch, whereas the N-terminal “chain” polypeptide is disordered and flexible in solution (22). A similar architecture has been determined for the YscP homologue FliK from S. enterica serovar Typhimurium (21). A multiple sequence and structural alignment of YscP with FliK, PscP, and other homologues (supplemental Fig. 1) suggests that YscP has a structural topology resembling those described for FliK and PscP.
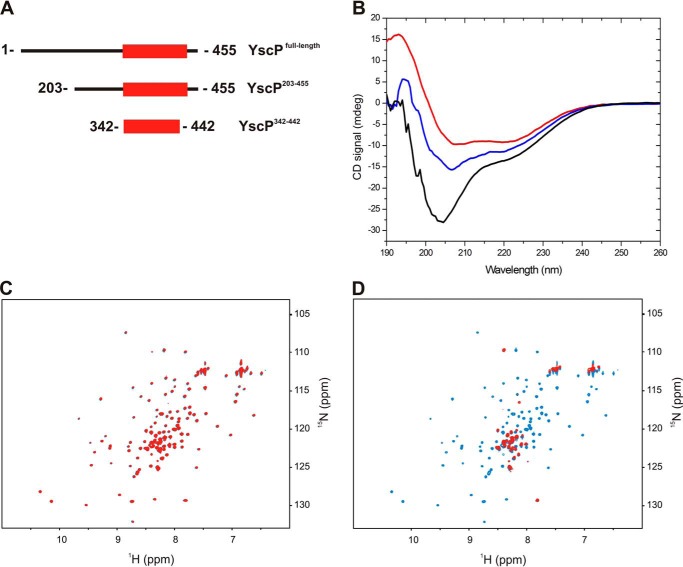
To investigate the domain architecture of YscP, we used three constructs (Fig. 2A) that were designed based on the results of a limited proteolysis experiment of YscP (supplemental Fig. 2) and a multiple sequence alignment (supplemental Fig. 1). These YscP variants were subjected to structural analysis using NMR and circular dichroism (CD) spectroscopy. First, size exclusion chromatography was used to determine the oligomeric state of YscP, YscP(342–442), and YscP(203–455) in solution. Interestingly, although the full-length YscP migrated as a 50-kDa monomer, YscP(342–442) and YscP(203–455) eluted with the column void volume, suggesting that both truncated variants aggregated in solution (supplemental Fig. 3). A CD spectrum of full-length YscP (Fig. 2B) indicated that the protein contained both unstructured and structured regions, and this was confirmed by the observation of bands at 205 and 220 nm in their CD spectra. The structured region of YscP was assigned to the C terminus of the protein by analyzing the CD spectra of the two truncated variants (YscP(203–455) and YscP(342–442)). To confirm that YscP(342–442) contained a structured domain, we acquired a 1H-15N HSQC spectrum of a sample of the protein that was isotopically enriched with 15NH4Cl. This NMR spectrum conclusively demonstrated that YscP(342–442) is a folded domain because it exhibits the large chemical shift dispersion typical of well folded proteins (supplemental Fig. 4). Although folded, the domain displayed tendencies to aggregate. The NMR spectrum was acquired at a concentration of 50 μm, and any increase in concentration resulted in precipitation of the protein. These results together demonstrate that YscP has a ball-and-chain domain architecture like its homologues. Next, we investigated the interaction between YscP and YscUC by observing perturbations in the 1H-15N HSQC spectrum of 15N-isotopically enriched YscUC upon addition of the different YscP constructs. In general, interactions between partners in such cases induce changes in the chemical shifts or the line width of the peaks in the spectrum depending on the molecular masses of the interacting partners. The addition of YscP(342–442) did not trigger any changes in the NMR spectrum of YscUC (Fig. 2C), suggesting that this YscP segment does not harbor the YscUC interaction interface. In contrast, we observed extensive line broadening of the YscUC resonances when YscP(203–455) was added (Fig. 2D), indicating that YscUC becomes integrated into a high molecular weight complex following the addition of YscP(203–455). Based on this analysis, we propose that the YscUC binding segment on YscP is localized between residues Glu203 and Arg341.
FIGURE 2.
YscP domain architecture and its YscUC-interacting segment. A, schematic representation of YscP and its truncated variants that were used in this study. B, far UV-CD spectra of YscP (black), YscP(342–442) (red), and YscP(203–455) (blue). All proteins were used at a concentration of 10 μm in 5 mm sodium phosphate, 30 mm NaCl, 1 mm DTT (pH 7.4) at 20 °C. mdeg, millidegrees. C, overlay of two-dimensional 1H-15N HSQC spectra of free 15N-labeled YscUC (blue) and 15N-labeled YscUC mixed with unlabeled YscP(342–442) (red). D, overlay of two-dimensional 1H-15N HSQC spectra of free 15N-labeled YscUC alone (blue) and mixed with unlabeled YscP(203–455) (red).
YscP Interaction Interface on YscUC
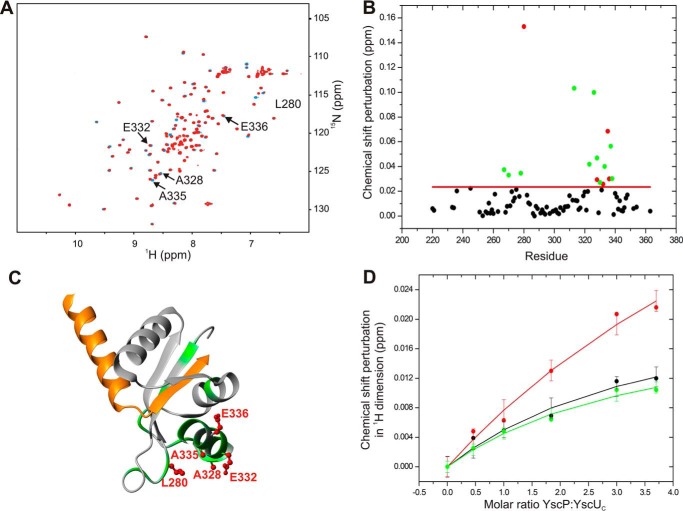
Despite intensive studies on YscP and YscU homologues, there are no high resolution structural data on the interaction interface on YscUC responsible for its interaction with YscP. We identified this interaction interface using NMR spectroscopy by titrating unlabeled YscP to 15N-labeled YscUC. The stepwise addition of unlabeled YscP to 15N-labeled YscUC caused localized perturbations in the chemical shifts of the amide proton and nitrogen resonances of several YscUC residues in the 1H-15N HSQC spectrum of the protein (Fig. 3A). The binding interface was identified by displaying the residues with significant compounded chemical shift perturbations on the YscUC structure (Fig. 3B). As expected for a well defined interaction interface, the YscP-interacting residues on YscUC cluster to form a contiguous surface; the solvent-exposed residues Leu280, Ala328, Glu332, Ala335, and Glu336 seem to form the YscP interaction interface. Interestingly, the interaction interface is centered on the C-terminal α helix of the YscU C-terminal domain (YscUCC) (Fig. 3C). The titration experiment also enabled us to estimate the binding affinity (dissociation constant (Kd)) of the YscP-YscUC interaction by monitoring changes in the chemical shifts of the methyl resonances of YscUC as the concentration of YscP increased (Fig. 3D). These data were then fitted to an analytical function based on an assumed two-state binding reaction. The best fitted binding isotherm gave a Kd value of 430 ± 293 μm. Although the estimated Kd value suggests that the YscU-YscP interaction is relatively weak under our experimental conditions, this interaction may be modulated in vivo by other factors such as membrane anchoring (26) and/or additional protein partners.
FIGURE 3.
NMR-based identification of the YscP binding surface on YscUC. A, overlay of the two-dimensional 1H-15N HSQC spectra of free 15N-enriched YscUC (100 μm) before (blue) and after addition of YscP (400 μm) (red). The spectra were acquired in PBS buffer containing 1 mm TCEP at 37 °C. Solvent-exposed residues that exhibit significant chemical shift perturbations upon YscP addition are marked with arrows. B, compounded chemical shift perturbations (ppm) between free 15N-labeled YscUC and 15N-labeled YscUC in complex with unlabeled YscP were calculated using Equation 1 and plotted against the YscUC primary sequence. The threshold value used to define a significant chemical shift perturbation is indicated by the red line, and residues exhibiting significant shifts are represented by green dots except for solvent-accessible amino acid residues with significant chemical shift perturbations (Leu280, Ala328, Glu332, Ala335, and Glu336), which are represented by red dots. C, display of the YscP binding interface on YscUC. The structure of YscUC illustrates the positions of the residues with significant chemical shifts (green) and the solvent-exposed residues with significant chemical shift perturbations (indicated as red ball and sticks). D, determination of the Kd value for the YscP-YscUC interaction obtained by using Equation 2 to fit the changes in the chemical shifts of the methyl groups of YscUC induced by YscP addition. Plots for three peaks in the methyl region are shown in black, red, and green, respectively. The error bars represent S.D.
Mutations at the YscU/YscP Binding Interface Regulate Yop and YscF Secretion
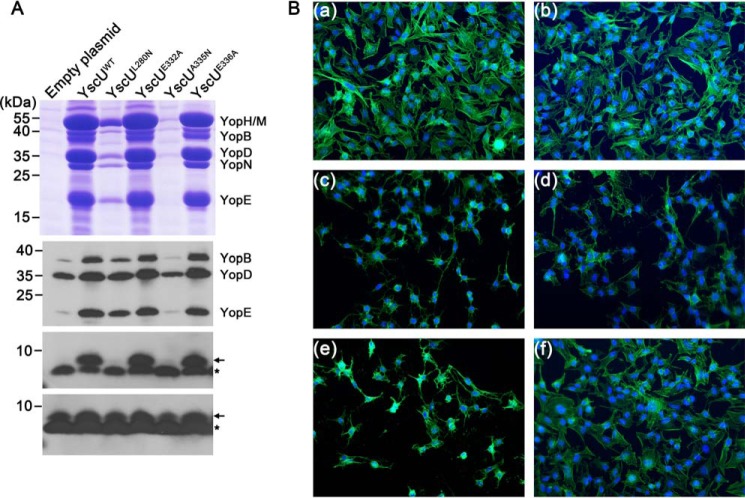
Based on the results presented above, we hypothesized that the interaction of YscU with YscP is mediated by the residues Leu280, Ala328, Glu332, Ala335, and Glu336. To test our hypothesis and determine the physiological relevance of the YscU-YscP interaction, we performed a scanning mutagenesis analysis targeting these residues. The point mutations L280N, A328N, E332A, A335N, and E336A were separately introduced into the yscU coding sequence and cloned into the pBADmycHis-A plasmid. The resulting plasmids were then transformed into the pIB75 (ΔyscU) strain, which carries an in-frame deletion of the yscU gene. The ability of the different YscU variants to complement Yop secretion was assayed by culturing the cells in Ca2+-depleted medium with a temperature shift from 26 to 37 °C. These growing conditions mimic the host cell contact required for activation of secretion via the T3SS and lead to a massive secretion of Yop into the culture supernatant. The empty plasmid and the plasmid encoding YscUWT were used as negative and positive controls, respectively. As expected, Yop secretion was restored by the plasmid encoding YscUWT, and no Yop was detected in the supernatant of the strain carrying the empty plasmid (Fig. 4A). Similar amounts of Yop were detected in the supernatants of the strains expressing YscUWT, YscUE332A, and YscUE336A (Fig. 4A). Thus, mutating residue Glu332 or Glu336 to alanine did not affect the function of YscU or the secretion of Yop via the T3SS. Given that the YscU-YscP interaction is required for Yop secretion, this result suggests that Glu332 and Glu336 may not be important residues for the binding of YscP to YscU. On the other hand, the mutation A335N drastically reduced the secretion of Yop into the supernatant (Fig. 4A, upper panel). The strain expressing YscUL280N exhibited an intermediate phenotype with a decrease of about 80% of the Yop secreted by the wild type (Fig. 4A, upper panel). Lower amounts of intracellular Yop were detected for the strains bearing the empty plasmid or the plasmid expressing YscUA335N (Fig. 4A). The attenuation of Yop secretion represses the expression of the yop genes, reducing the intracellular amount of the corresponding proteins. The ability of these different variants to generate a cytotoxic phenotype in HeLa cells was also assayed. Unlike YscUWT and YscUE332A, no cytotoxicity was generated by YscUA335N (Fig. 4B), and the appearance of the cytotoxicity phenotype was delayed for YscUL280N (Fig. 4B). Thus, both Yop secretion and translocation are affected by the A335N and L280N mutations. Taken together, these results indicate that Ala335 and Leu280 are critical for the interaction of YscU with YscP and Yop secretion via the T3SS. These mutations also impaired the secretion of the needle subunit YscF (Fig. 4A, third and fourth panels from the top). YscF is an early substrate, whereas Yop is a late substrate (34). Previous studies showed that the substrate specificity switch from early to late substrates is regulated by both YscU and YscP (10). The absence of YscF in the supernatant of the strain expressing YscUA335N suggests that this mutation affects the secretion of both early and late substrates.
FIGURE 4.
Secretion profiles and cytotoxicity of Y. pseudotuberculosis expressing various point mutants of YscU. A, secretion profiles of Y. pseudotuberculosis strain YPIII/pIB75 transformed with plasmids encoding different point mutants of YscU. Culture supernatants were precipitated with TCA, separated on a Tris/Tricine gel, and stained with Coomassie R-250 (upper panels). Cell pellets of the corresponding strains were analyzed with anti-Yop antibody (second panel from the top). The amounts of secreted YscF (third panel from the top) and intracellular YscF (bottom panel) were analyzed using an anti-YscF antibody. The position of YscF is indicated by the black arrows, and the black asterisks indicate an unspecific band. B, HeLa cells were infected with bacteria at a multiplicity of infection of 10 for 45 min. After fixation with formaldehyde, the cytoskeleton actin filaments were stained with green phalloidin-Alexa Fluor 488 (Invitrogen) and nucleic acids were stained with DAPI (blue). Uninfected HeLa cells (panel a) exhibited non-cytotoxic morphologies similar to cells infected with strains bearing the empty plasmid (panel b). Cytotoxicity was observed for 100% of HeLa cells infected with the strains expressing YscUWT (panel c) and YscUE332A (panel e). YscUL280N induced delayed cytotoxicity (panel d). YscUA335N generated no cytotoxicity (panel f).
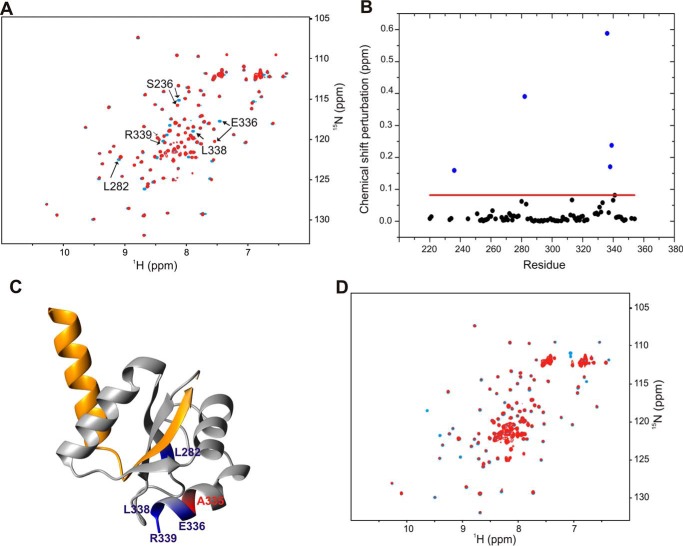
The results of the complementation analysis presented above showed that the residue Ala335 is critical for the function of YscU. The A335N mutation could thus impair either the folding of YscU or its interaction with YscP. To determine whether the A335N mutation affects the structure of YscU, we acquired a 1H-15N HSQC spectrum of YscUCA335N and compared it with that of the wild type. An overlay of the two spectra revealed no significant differences in chemical shifts, and the majority of their cross-peaks have similar intensities and positions (Fig. 5A). The only residues that showed significant chemical shift perturbations are in close proximity to the mutation site, so we can conclude that the mutation does not affect the overall structure of YscUC (Fig. 5, B and C). We then investigated whether YscUCA335N retained the ability to interact with YscP by performing an NMR titration of unlabeled YscP against 15N-labeled YscUCA335N at 37 °C (Fig. 5D) as we had done previously with YscUCWT. A comparison of the spectra from the titrations of YscP against YscUCWT and YscUCA335N showed that the chemical shift perturbations of YscUCA335N were significantly smaller than for YscUCWT (Fig. 5D). Furthermore, a Kd value for the interaction between YscUCA335N and YscP could not be quantified because the chemical shift changes were too small (supplemental Fig. 5). These data clearly indicate that the YscUCA335N mutant retains the structure of the wild-type protein but has lost its YscP binding activity.
FIGURE 5.
Disruption of YscP binding by the A335N replacement in YscUC. A, overlay of two-dimensional 1H-15N HSQC spectra of 15N-labeled YscUCWT (blue) and YscUCA335N (red) at concentrations of 100 μm. The buffer consisted of PBS and 1 mm TCEP at 37 °C. Solvent-exposed residues that exhibit significant chemical shift perturbations upon mutation are indicated with arrows. B, compounded chemical shift perturbations (ppm) between 15N-labeled YscUCWT and 15N-labeled YscUCA335N calculated by Equation 1 and plotted against the YscU residue number. The threshold value used to define significant chemical shift changes is indicated by the red line. The residues with the largest chemical shift perturbations (Ser236, Leu282, Glu336, Leu338, and Arg339) cluster around the mutation site and are shown in blue; position 335 is shown in red on the YscUC crystal structure in C. D, overlay of two-dimensional 1H-15N HSQC spectra of 15N-labeled YscUCA335N at a concentration of 100 μm alone before (blue) and after addition of unlabeled YscP (red) at a molar ratio of 1:4 in PBS.
YscP Protects the Slow Processing Mutant P264A from Autocleavage
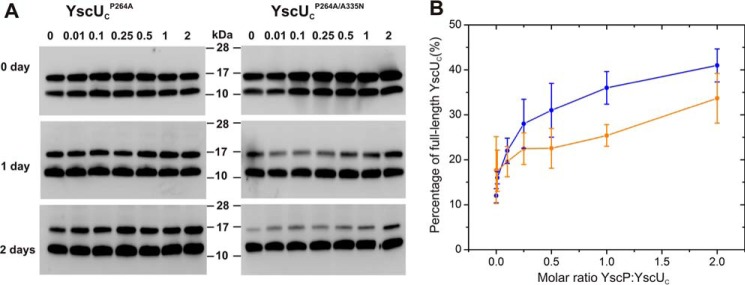
It has been shown previously that the dissociation of YscUC into the two polypeptides YscUCC and YscUCN is required for Yop secretion and that YscUCC is subsequently secreted into the culture supernatant (27, 29). The secretion of YscUCC through the T3SS is facilitated by a C-terminal secretion sequence, and the non-secreting phenotype observed for yscP-null mutants is partially rescued by the secondary suppressor mutants YscUA268F, YscUY287G, and YscUV292T (10, 33). These suppressor mutants display elevated rate constants for the dissociation of YscUC into YscUCN and YscUCC relative to wild-type YscUC (28). To determine whether YscP can affect the dissociation of YscUC, we studied the dissociation kinetics of YscUC in the absence and presence of YscP by NMR spectroscopy as described previously (28). The experiments revealed that YscP does not significantly change the dissociation kinetics of YscUC (supplemental Fig. 6). It is important to note that wild-type YscUC is fully cleaved in these experiments because of the protein's long handling time during purification. To determine whether YscP can modulate the rate of autoproteolysis, we studied the slowly autocleaving mutant P264A (27) whose autoproteolysis proceeds slowly enough to allow purification of partially uncleaved protein. The in vitro cleavage kinetics of this mutant into the two polypeptide fragments YscUCN and YscUCC can be followed by Western blotting with an anti-YscUCC antibody (Fig. 6 and supplemental Fig. 7) at 37 °C at 0, 1, and 2 days after purification. Each sample generated two bands in the immunoblot due to the incomplete autoproteolysis of the YscUCP264A variant; the uppermost band corresponded to uncleaved YscUC, and the lower band corresponded to the cleaved YscUCC polypeptide (Fig. 6). The total protein content for each sample was calculated from the sum of the bands corresponding to YscUC and YscUCC. The relative abundance of the uncleaved full-length protein (i.e. YscUC variant) was then calculated as the percentage of total protein concentration. Following this analysis, it is evident that YscP can reduce the rate of YscUCP264A autoproteolysis in a concentration-dependent manner (Fig. 6). To test whether the effect is specific, we turned to the double mutant YscUCP264A/A335N, which has both slow autoproteolysis and a weakened YscP binding affinity. The YscP dose dependence for the protection against autoproteolysis is significantly weaker for the double mutant in comparison with the YscUCP264A single mutant. This observation suggests that the effect on autoproteolysis mediated by YscP is specific and driven by the interaction of YscP with the identified binding surface of YscUC. Taken together, the data show that YscP can protect the slowly cleaving YscUC mutant P264A from autoproteolysis in vitro.
FIGURE 6.
YscP binding attenuates the rate of YscU autocleavage. A, immunoblotting detection of the degree of autocleavage of YscUCP264A and YscUCP264A/A335N using anti-YscUCC as the primary antibody. Autocleavage was monitored over 2 days at 37 °C with varying concentrations of YscP. The YscUC:YscP ratio was varied from 0 to 2 as indicated at the top of the figure. Each sample shows two bands in the immunoblot: the top band corresponds to uncleaved YscUC, whereas the lower band corresponds to the YscUCC polypeptide. B, the immunoblotting experiments were analyzed by quantifying the proportion of uncleaved YscUCP264A (blue) and YscUCP264A/A335N (orange) in the samples after 2-day incubation. Values shown in the graph are means with error bars representing ± S.D. (the experiment was repeated four times).
YscU Binds to the Inner Rod Protein YscI at Molar Ratio 1:1
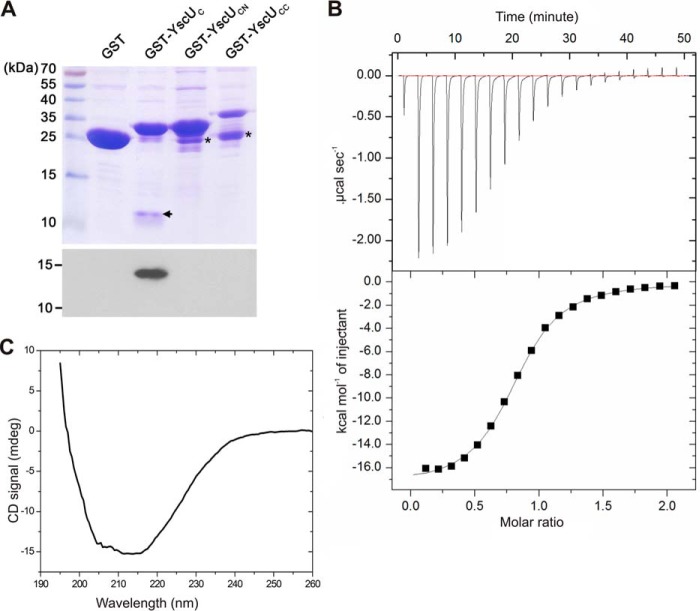
The proposed T3SS structural model suggests that the protein YscI (Yersinia spp.), MxiI (Shigella spp.), or PrgJ (Salmonella spp.) polymerizes into an inner rod to form a conduit for protein secretion through the periplasm (20, 33, 36, 37). YscI is secreted via the T3SS, and it has been shown that YscI secretion is regulated by both YscP and YscU (33). It has also been shown that the inner rod protein EscI, a homologue of YscI in enteropathogenic E. coli interacts with the YscU homologue EscU (35). We therefore investigated whether YscU interacts with YscI and attempted to determine the binding interface (Fig. 7) by performing GST pulldown experiments in which YscUC, YscUCN, and YscUCC were GST-tagged and used as bait to co-purify YscI-His6. YscI-His6 only bound to GST-YscUC; no YscI remained associated to GST alone, GST-YscUCN, or GST-YscUCC (Fig. 7A). This interaction pattern suggests that the interaction of YscI with YscUC is abolished by the dissociation of YscUCC from YscUCN and/or structural changes of YscUCC and YscUCN. To further characterize the binding of YscI to YscU, we analyzed this interaction using isothermal titration calorimetry (ITC) (Fig. 7B). The binding is characterized by a distinct exothermic binding process that can be robustly fitted to a one-site binding model (Fig. 7B) with a Kd of 3.8 μm and a 1:1 stoichiometry.
FIGURE 7.
The inner rod protein YscI interacts with YscUC but not with YscUCN or YscUCC. A, GST pulldown assay using different variants of YscU and polyhistidine-tagged YscI. Top panel, verification of binding of GST, GST-YscUC, GST-YscUCN, and GST-YscUCC to glutathione-Sepharose beads. This was accomplished by applying aliquots of the different proteins to glutathione-Sepharose beads, extensive washing to remove unbound material, and subsequent SDS-PAGE analysis of aliquots of beads bearing the bound proteins. The gels were stained with Coomassie R-250 for detection. Black asterisks indicate degradation products. Bottom panel, GST and GST-tagged YscU variants were incubated with purified YscI. After several washes, resin-associated proteins were eluted and separated on the gel for immunoblotting detection with an anti-His6 antibody. The black arrow indicates the position of YscUCC in the GST-YscUC sample. B, raw data from an ITC experiment involving addition of YscUC to YscI are shown in the upper panel; the lower panel shows the integrated heats obtained after subtracting the heat of dilution. The best fitting binding isotherm is shown as a solid line. The fitted parameters are Kd = 3.8 μm, ΔH0 = −17 kcal mol−1, ΔS0 = −33.8 cal mol−1 degrees−1, and a stoichiometry “n” of 0.8 at 25 °C. C, far-UV CD spectrum of YscI. The experiment was conducted with 10 μm YscI in a 5 mm sodium phosphate buffer (pH 7.4) containing 30 mm NaCl and 1 mm DTT at 25 °C. mdeg, millidegrees.
Previous structural analyses of two YscI homologues, MxiI and PrgJ, using CD and NMR spectroscopy have shown that they are partially folded in solution (36). Although MxiI, PrgJ, and YscI have equivalent functions in the T3SS, their sequence similarity is relatively low (supplemental Fig. 8). The CD spectrum of YscI features distinct CD bands in the region between 205 and 220 nm, which is indicative of significant secondary structure content (Fig. 7C).
We attempted to characterize the binding interface of YscI on YscUC by performing two-dimensional NMR experiments. A titration of unlabeled YscI against 15N-labeled YscUC resulted in massive line broadening of the peaks corresponding to YscUC residues due to aggregation of YscI in solution into complexes that are too large to permit the detection of YscI-bound YscUC resonances. However, a two-dimensional 1H-15N HSQC experiment using 15N-labeled YscUCA335N showed that the binding of YscI was not abolished by the mutation A335N in contrast to that of YscP (supplemental Fig. 9). The interactions of YscP and YscI with YscU thus appear to occur at different binding surfaces.
Discussion
Type III secretion systems are complex nanomachines spanning the cell wall of Gram-negative bacteria. They are constituted by around 25 proteins whose assembly requires that the individual components are added in a specific order (38). Furthermore, the T3SS substrates (Yops in Yersinia) that are destined for the eukaryotic cytosol are not exported before cell contact between the pathogen and the eukaryotic target cell has been established (39). Thus, there exists a secretion hierarchy allowing early T3SS substrates to be secreted before the secretion of the late substrates is allowed. This process was first described for the flagellar T3SS and was named “substrate specificity switching” by Macnab (12). In Yersinia spp., YscP and YscU have been shown to be essential components of the substrate specificity switching mechanism. A yscP mutant assembles elongated needles, but it is unable to secrete Yop effectors (19). Similar phenotypes have also been reported for the homologous proteins in Salmonella and Shigella T3SSs (31, 40). Interestingly mutations in the cytoplasmic domain of yscU can suppress a yscP mutant phenotype, leading to decreased export of the needle protein YscF and increased Yop secretion (10, 27). These genetic results argue strongly for a direct YscP and YscU interaction that is important to enable Yersinia to regulate the substrate switch. YscU and its homologues in other bacteria undergo autocatalytic cleavage at a conserved NPTH motif, generating a C-terminal 10-kDa peptide fragment called YscUCC in Yersinia. YscUCC remains bound to the membrane-anchored part of YscU after cleavage (27). Removal of Ca2+ from the growth medium results in Yop secretion as well as secretion of YscUCC, indicating that YscU is dissociated under these conditions (28). Furthermore, we also showed previously that yscP suppressor mutants mapping to the yscUCC part of the yscU gene destabilized the interaction between the two YscU polypeptides generated by autocleavage, indicating that YscUCC dissociation is an essential step in substrate switching in Yersinia. It has therefore been proposed in line with these findings that the decisive step in substrate switching is the autocatalytic event and that YscP is a key regulator of this process.
Here we used solution state NMR spectroscopy to identify the YscP binding interaction interface on the cytoplasmic domain of YscU (YscUC). The interaction surface is centered on the C-terminal α helix of YscUC, and disruption of the surface by introducing the A335N point mutation dramatically reduced Yop secretion and cytotoxicity on HeLa cells. On the basis of biophysical analyses with circular dichroism and NMR spectroscopy, we show that YscP has a ball-and-chain architecture with an extended N-terminal flexible segment attached to a folded C-terminal domain. A similar architecture has been demonstrated previously for the orthologue PscP from P. aeruginosa (22) and for FliK from S. enterica serovar Typhimurium (21). These results suggest that the ball-and-chain architecture is strongly conserved among the YscP class of bacterial proteins. By performing a deletion analysis of YscP, we showed that the protein interacts with YscU via amino acid residues in its disordered N-terminal segment. This is analogous to the scenario observed in P. aeruginosa where N-terminal residues of PscP interact with the YscU orthologue PscU (22). It has been proposed that the C-terminal folded domain of PscP is located at the growing tip of the needle structure (22) and that the N-terminal flexible segment spans the entire T3SS to make contact with PscU located in the cytoplasm. This model is consistent with the fact that YscP has been identified as a secreted protein in Yersinia (41) and that it is also surface-located. In accordance with these results, YscP has been postulated to function as a “molecular ruler” that defines the correct length of the needle structure (19). An issue that remains to be answered is the nature of the mechanism that enables discrimination between early and late substrates. As discussed, we have shown previously that autoproteolysis of YscU enables secretion of the C-terminal YscU polypeptide (YscUCC) and that its secretion parallels that of the Yops (28). Destabilized YscU mutants that displayed increased rate constants of dissociation also enable secretion of Yops at 30 °C, a temperature at which the wild-type strain effectively displays no Yop secretion. In addition, we previously identified a C-terminal secretion signal in YscUCC (29). Taken together, these findings suggest that YscUCC dissociation and subsequent secretion enabled by autoproteolysis play an important role in regulating Yop secretion. However, a contradictory hypothesis exists whereby the role of autoproteolysis is to shape the three-dimensional structure of YscU and thereby enable the protein to engage in downstream protein-protein interactions (42). This hypothesis is supported by the observation that uncleaved SpaS (the YscU orthologue in Salmonella) could not be detected inside bacteria. In addition, in an elegant experiment, the authors engineered a SpaS variant that was cleaved by an external protease; also with this experiment no regulatory function of SpaS cleavage could be verified. Taken together, these observations suggest that self-cleavage of SpaS has no regulatory function regarding the substrate specificity switch (42). However, it is possible that the time resolution in the experiments did not allow detection of uncleaved SpaS. Hence, in the context of wild-type SpaS, it is currently problematic to test regulatory aspects of self-cleavage. To test whether autoproteolysis could be regulated in Yersinia, we turned to the slowly cleaving YscUC mutant P264A. The YscUCP264A mutant has a half-life for autoproteolysis that enables biochemical experiments aimed at probing the role of YscP for the rate of autoproteolysis and/or dissociation. To test for potential effects on YscUC dissociation kinetics in response to YscP binding, we relied on our established NMR protocol for quantifying this event (28). We could not observe any statistically significant changes in the rate constant of wild-type YscUC dissociation in the absence or presence of YscP. Using the YscUCP264A mutant and a protocol based on Western blotting with an antibody against YscUCC, we showed that the presence of YscP significantly reduced the rate constant of autoproteolysis. This finding opens the possibility that YscU might be maintained in a state that prevents late substrate secretion when bound to YscP and that release of YscP enables YscU self-cleavage, YscUCC dissociation, and activation of late substrate secretion. This inference is consistent with the model of Bergeron et al. (22) where PscP is anchored to PscU on the cytoplasmic side with the N-terminal flexible segment threaded through the needle and the C-terminal folded domain located at the tip of the needle.
It was recently shown by Lloyd and co-workers (33) that the formation of the inner rod by the YscI protein exhibits a critical role in the substrate specificity switch. They also showed that YscU and YscP indirectly control Yop secretion by regulating the export of YscI. These results are in perfect agreement with the view of Marlovits and co-workers (43) who showed that the inner rod was absent in YscP mutants, leading to the conclusion that the inner rod is essential for substrate specificity switching. In addition, it has also been shown that EscI interacts with EscU (35). In line with these observations, we found that YscI interacted with the cytosolic domain (YscUC), but in contrast YscI was unable to interact with either of the two peptides resulting from autoproteolysis. Thus, we conclude that YscI interacts with YscU and that autoproteolysis per se does not affect YscI interaction with the two resulting YscU polypeptides as long as they are interconnected. In addition, we recently showed that induction of the switch to late Yop secretion also resulted in secretion of YscUCC (28). This result shows that YscUCC is dislocated from the remaining part of YscU after induction of the switch (28). One possibility based on these findings is that YscU anchors the inner rod to the T3SS and that induction of the switch results in disassociation of the inner rod followed by Yop secretion. In accordance with this hypothesis and the results presented herein, we further suggest that YscP/YscU binding blocks autoproteolysis, and thus YscUCC dissociation followed by YscI release is inhibited. This mechanism requires that YscUC harbor distinct YscI and YscP binding surfaces, and this feature was established in the current study. In this scenario, YscP could fit very well to serve as the enigmatic T3SS surface sensor because it has also been shown that a domain of YscP is surface-located prior to induction of the switch (41). Thus, YscP can serve as a link between the exterior of the T3SS and proteins regulating the substrate specificity switch, thus fulfilling the basal criteria for a surface sensor. More work is needed in this context, and we now focus our efforts to test this hypothesis.
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in supplemental Table S1. Standard molecular biology methods were used to generate the different plasmid constructs used in the experiments. The PCR primers used in the different clonings are listed in supplemental Table S2. The sequences of all the constructs were verified by sequencing (Eurofins MWG Operon). E. coli strains were grown in Luria-Bertani broth (LB) or on Luria agar plates at 37 °C. Y. pseudotuberculosis strains were grown at 26 °C in LB or on Luria agar plates. Antibiotics were added to the medium for selection according to the resistance markers carried by the plasmid at the following concentrations: kanamycin, 50 μg/ml; carbenicillin, 100 μg/ml; and chloramphenicol, 35 μg/ml.
Yop Secretion Analysis and Immunoblotting Quantification
To induce Yop secretion, Y. pseudotuberculosis strains were first grown at 26 °C for 2 h in Ca2+-depleted LB medium (medium containing 5 mm EGTA and 20 mm MgCl2) and then at 37 °C for 3 h. Cultures were started at an A600 of 0.1. Samples were treated as described previously (28) and separated on a Tris-Tricine polyacrylamide gel. Proteins were either stained with Coomassie R-250 or transferred onto a PVDF membrane for immunoblotting. Anti-Yop, anti-His, and anti-YscF antibodies were diluted at 1:5,000. Horseradish peroxidase-conjugated anti-rabbit antibody was diluted at 1:10,000 (GE Healthcare). Proteins were detected with a chemiluminescence detection kit (GE Healthcare).
To monitor the effect of YscP binding on YscU autocleavage, YscP was mixed with 20 μm YscUC variants at different molar ratios (0, 0.01, 0.1, 0.25, 0.5, 1, and 2) in PBS with 1 mm TCEP and incubated at 37 °C. Samples were taken after 0, 1, and 2 days and diluted to a concentration of 1 μm in the YscUC variant before mixing with Laemmli buffer (Bio-Rad). The proteins in the samples were then separated by SDS-PAGE and transferred to a PVDF membrane (Bio-Rad) for immunoblotting. Anti-YscUCC was diluted at 1:10,000. Goat anti-rabbit IgG (heavy + light)-HRP conjugate antibody was diluted at 1:20,000. Transferred proteins were detected with a chemiluminescence detection kit (Bio-Rad). Densitometric analysis of the bands was performed using the ImageJ software package based on the same immunoblotting samples (44).
HeLa Cell Cytotoxicity Assay
Y. pseudotuberculosis cultures were started at an A600 of 0.1 in LB medium containing 1 mm CaCl2. After 1 h of growth at 26 °C, cultures were shifted to 37 °C for 2 h. HeLa cells were infected for 45 min at a multiplicity of infection of 10. Cytotoxicity assays were performed as described by Rosqvist and co-workers (45) with some modifications. For immunostaining, samples were subsequently fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich). Unspecific binding was suppressed by treatment with 0.1 m glycine in PBS containing 1% bovine serum albumin. Alexa Fluor 488-phalloidin (Life Technologies) and DAPI were used to stain the actin cytoskeleton and nucleic acids of the cells, respectively. Samples were mounted in mounting medium (Dako) and examined with a fluorescence microscope (Nikon Eclipse C1 Plus).
Protein Expression and Purification
Proteins produced in the BL21(DE3)/pLysS strain were grown in Luria broth at 37 °C. When an A600 of 0.6 was reached, protein production was induced by adding 1 mm isopropyl β-d-1-thiogalactopyranoside (IPTG) to the culture medium and allowing the bacteria to grow for a further 4 h. Bacteria were harvested by centrifugation for 30 min at 5,000 × g, and the pellets were frozen at −80 °C. For 15N labeling, bacteria were grown in M9 minimal medium containing 15NH4Cl. After addition of 1 mm IPTG at an A600 of 0.6, 15N-labeled proteins were isolated from bacteria grown overnight at 30 °C and harvested as described above.
GST-tagged variants of YscUC and YscP were purified as described previously (28). Bacteria were resuspended in the lysis buffer (50 mm Tris, 100 mm NaCl, 2 mm DTT (pH 7.4)) and lysed by sonication. The lysates were then centrifuged for 30 min at 30,000 × g at 4 °C. The supernatant was loaded on a GSTrap column (GE Healthcare) pre-equilibrated with the lysis buffer, the column was washed with 10 column volumes of the same buffer, and then the GST-tagged protein was eluted with 50 mm Tris, 100 mm NaCl, 20 mm glutathione (pH 7.4) buffer. Glutathione was eliminated from the eluate using a centrifugal filter device, and the GST tag was cleaved by incubating the resulting samples with 10 units of PreScission protease (GE Healthcare)/mg of protein overnight at 4 °C. The free GST was eliminated by passing the sample through a GSTrap column with the purified proteins remaining in the column flow-through. The purification of YscP was finalized by loading untagged YscP on a HiPrep 26/60 Sephacryl S-100HR gel filtration column (GE Healthcare) after a buffer exchange to PBS containing 2 mm DTT (pH 7.4). YscUC was loaded on a Sepharose SP cation exchange column (Sigma-Aldrich) after a buffer exchange to 10 mm Tris containing 25 mm NaCl (pH 7.4) and eluted using an NaCl gradient from 25 to 800 mm in 10 mm Tris (pH 7.4). Both YscP and YscUC were concentrated in PBS containing 1 mm TCEP (pH 7.4) using centrifugal filter devices.
His6-YscP(203–455) was produced and lysed as described above. The supernatant was then loaded on a HisTrap column (GE Healthcare). The protein was eluted with an imidazole gradient from 10 to 700 mm in PBS. After removal of imidazole using centrifugal filter units, His6-YscP(203–445) was loaded on a 26/60 Sephacryl S-100HR gel filtration column.
His6-maltose-binding protein (MBP)-YscP(342–442) with a double His6-MBP tag was produced overnight at 25 °C by adding 1 mm IPTG when the A600 of the culture reached 0.6. Inclusion bodies were solubilized in 50 mm Tris containing 100 mm NaCl and 8 m urea (pH 7.4). After centrifugation for 30 min at 30,000 × g at 4 °C, the supernatant was dialyzed against the same buffer containing 4 m urea for 2 h at 4 °C. The procedure was then repeated with buffers containing 2 m and then 1 m urea. The final dialysis was carried out overnight against 50 mm Tris containing 100 mm NaCl and 10 mm imidazole after which the refolded protein was loaded onto a HisTrap column and eluted with an imidazole gradient from 10 to 700 mm in the loading buffer. The imidazole concentration was then reduced to 10 mm using centrifugal filter devices, and the His6-MBP tag was removed by overnight incubation at room temperature with 100 units of tobacco etch virus protease/mg of protein. 8 m urea was added to the resulting solution, and the uncleaved His6-MBP-YscP(342–442) was removed by affinity chromatography on a HisTrap column with the YscP(342–442) remaining in the flow-through. The buffer was then exchanged to 6 m urea in PBS, and the protein was purified by size exclusion chromatography using a HiPrep 26/60 Sephacryl S-100HR column.
For YscI-His6 purification, bacteria were resuspended and lysed in 50 mm Tris containing 100 mm NaCl (pH 7.4). The lysates were then centrifuged for 30 min at 30,000 × g at 4 °C. Because YscI-His formed inclusion bodies, the pellet was dissolved in a binding buffer of 50 mm Tris containing 100 mm NaCl and 7 m urea (pH 7.4) and then centrifuged. The resulting supernatant was loaded on a HisTrap column, and urea was eliminated by washing the column with a non-urea buffer of 50 mm Tris containing 100 mm NaCl (pH 7.4). The protein was then eluted with an imidazole gradient from 0 to 1 m in 50 mm Tris containing 100 mm NaCl (pH 7.4). Imidazole was eliminated by dialysis overnight at 4 °C into an appropriate buffer for NMR or isothermal titration calorimetry.
NMR Spectroscopy
15N-Labeled proteins were studied at 100 μm in PBS containing 1 mm TCEP and 10% D2O. In the YscP titration experiments, the quantity of added YscP was chosen to achieve a final concentration of 50, 100, 200, 300, or 400 μm and a YscUC:YscP molar ratio of 0.5, 1, 2, 3, or 4. 1H-15N HSQC spectra were acquired at 37 °C using a Bruker Avance III HD spectrometer at 850 MHz equipped with a 5-mm TCI HCN triple resonance cryoprobe. The NMR spectra were processed with Topspin (Bruker Biospin) or NMRPipe and visualized using ANSIG for Windows (46, 47). Compounded chemical shift perturbations for each backbone 1H-15N signal were computed using the following equation.
| (Eq. 1) |
where ΔδN and ΔδH are the chemical shift perturbations of the amide nitrogen and amide proton of the assigned residue in the free and complexed forms of the protein, respectively. The calculated chemical shift perturbations were plotted against residue numbers to localize the affected surfaces.
In the binding studies, the chemical shifts of the methyl resonances in 1H one-dimensional NMR spectra of YscUC were monitored during YscP addition. The YscP-YscUC interaction occurs under the fast exchange regime on the NMR time scale, so binding constants (Kd) were estimated by fitting the experimental data to Equation 2 based on an assumed 1:1 binding reaction (P + L ↔ PL) where YscP is regarded as “ligand L,” YscUC is regarded as “protein P,” and the YscP-YscUC complex is regarded as “PL”. Δδmax is the perturbation of the chemical shift when the binding site is saturated (48).
| (Eq. 2) |
where P is concentration of protein and x is molar ratio of ligand:protein. The reported Kd is an average value based on the individual Kd values of the analyzed peaks.
Limited Trypsin Proteolysis
Limited trypsin proteolysis was utilized to determine the domain architecture of YscP. A 22 μm solution of the full-length YscP protein in PBS was incubated with sequencing grade trypsin (Promega) at ratios of 1:100, 1:1,000, and 1:10,000, and samples were taken after 1-, 5-, 10-, and 20-min incubation at 4 °C. The peptide content of these samples was separated on a 15% SDS-acrylamide gel and stained with Coomassie R-250. The bands corresponding to stable fragments were then excised and identified by ultraperformance liquid chromatography-mass spectroscopy/mass spectroscopy.
CD
Far-UV CD spectra were recorded in a 0.1-cm quartz cuvette on a Jasco J-810 spectropolarimeter equipped with a Peltier element for temperature control. The spectra were recorded using 10 μm protein solutions in 5 mm sodium phosphate, 30 mm NaCl, 1 mm DTT (pH 7.4) buffer. Spectra were acquired at 20 °C from 260 to 190 nm in continuous scanning mode with 0.5-nm steps, a bandwidth of 2 nm, and a scan speed of 50 nm/min. Five spectra per sample were accumulated and averaged to improve the signal to noise ratio. The appropriate solvent spectrum background was subtracted from each of these spectra.
ITC
ITC experiments were performed in PBS buffer (pH 7.4) at 25 °C using a MicroCal Auto iTC200 instrument (GE Healthcare). A 900 μm YscUC solution in the syringe was titrated against 90 μm YscI in the cell. The experiment involved 20 injections of 2 μl of the titrant protein (YscUC) with 150 s between injections. The resulting raw data were fitted to a 1:1 binding model using the Origin 7.0 software package (GE Healthcare).
Pulldown of GST-tagged YscUC Variants with YscI
Proteins were produced as described above. GST-tagged YscU variants were resuspended in PBS buffer containing 1% Triton X-100, sonicated, and centrifuged for 20 min at 20,000 × g at 4 °C. The resulting supernatants were then incubated with a glutathione-Sepharose 4B slurry (GE Healthcare) for 1 h at 4 °C with gentle rocking. Bound proteins were washed with PBS containing 0.1% Triton X-100 and then used in an interaction assay. The YscI pellet was resuspended in PBS buffer containing 8 m urea, sonicated, and centrifuged for 20 min at 20,000 × g at 4 °C. The supernatant was then incubated with HIS-Select Nickel Affinity Gel (Sigma-Aldrich) for 1 h at 4 °C. Bound YscI was washed with PBS containing 0.1% Triton X-100 and eluted with 0.4 m imidazole in the same buffer. The GST-tagged YscU variants bound to the beads were incubated with 10 μm YscI for 3 h at 4 °C with gentle rocking. The beads were then washed with PBS containing 0.1% Triton X-100, and the proteins that remained bound to the beads were eluted using SDS-PAGE sample buffer and analyzed by immunoblotting.
Author Contributions
O. H., F. H. L., H. W.-W., and M. W.-W. designed experiments, analyzed data, and managed the project. O. H. and P. R. conducted experiments to characterize the domain structure of YscP. O. H. performed experiments to study the binding interface of YscP binding on YscU, YscP protection for the mutant P264A from autocleavage, and interaction of YscU and YscI by ITC. F. H. L. performed Yop and cytotoxicity assays for functional characterization of mutations at the YscU/YscP binding interface in vivo and GST binding assay for YscI to GST-tagged YscUC variants and discussed the project. T. E. performed secretion experiments. The manuscript was written by O. H., M. W.-W., F. H. L., and H. W.-W. All authors discussed the results and commented on the manuscript.
Supplementary Material
Acknowledgments
The NMR experiments were conducted at NMR4life, and we thank the Wallenberg and “Seth M. Kempes Minne” foundations for supporting this infrastructure. We acknowledge Dr. Stefan Frost, the Protein Expertise Platform (Umeå University, Sweden) and the Protein Science Facility (Karolinska Institute, Sweden) for assistance with protein cloning.
This work was supported by Swedish Research Council Grants 2013-5954 (to M. W.-W.) and 2015-02874 (to H. W.-W). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. 1–9 and Tables S1 and S2.
- T3SS
- type III secretion system
- Yop
- Yersinia outer protein
- HSQC
- heteronuclear single quantum coherence
- ITC
- isothermal titration calorimetry
- Kd
- dissociation constant
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TCEP
- tris(2-carboxyethyl)phosphine
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MBP
- maltose-binding protein.
References
- 1. Portnoy D. A., Wolf-Watz H., Bolin I., Beeder A. B., and Falkow S. (1984) Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect. Immun. 43, 108–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosqvist R., Forsberg A., Rimpiläinen M., Bergman T., and Wolf-Watz H. (1990) The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4, 657–667 [DOI] [PubMed] [Google Scholar]
- 3. Hueck C. J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ghosh P. (2004) Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cornelis G. R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 6. Tampakaki A. P., Fadouloglou V. E., Gazi A. D., Panopoulos N. J., and Kokkinidis M. (2004) Conserved features of type III secretion. Cell. Microbiol. 6, 805–816 [DOI] [PubMed] [Google Scholar]
- 7. Yip C. K., Kimbrough T. G., Felise H. B., Vuckovic M., Thomas N. A., Pfuetzner R. A., Frey E. A., Finlay B. B., Miller S. I., and Strynadka N. C. (2005) Structural characterization of the molecular platform for type III secretion system assembly. Nature 435, 702–707 [DOI] [PubMed] [Google Scholar]
- 8. Yip C. K., and Strynadka N. C. (2006) New structural insights into the bacterial type III secretion system. Trends Biochem. Sci. 31, 223–230 [DOI] [PubMed] [Google Scholar]
- 9. Diepold A., Wiesand U., and Cornelis G. R. (2011) The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82, 502–514 [DOI] [PubMed] [Google Scholar]
- 10. Edqvist P. J., Olsson J., Lavander M., Sundberg L., Forsberg A., Wolf-Watz H., and Lloyd S. A. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185, 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agrain C., Callebaut I., Journet L., Sorg I., Paroz C., Mota L. J., and Cornelis G. R. (2005) Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56, 54–67 [DOI] [PubMed] [Google Scholar]
- 12. Macnab R. M. (2003) How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100 [DOI] [PubMed] [Google Scholar]
- 13. Minamino T., and Macnab R. M. (2000) Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182, 4906–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferris H. U., Furukawa Y., Minamino T., Kroetz M. B., Kihara M., Namba K., and Macnab R. M. (2005) FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 280, 41236–41242 [DOI] [PubMed] [Google Scholar]
- 15. Makishima S., Komoriya K., Yamaguchi S., and Aizawa S.-I. (2001) Length of the flagellar hook and the capacity of the type III export apparatus. Science 291, 2411–2413 [DOI] [PubMed] [Google Scholar]
- 16. Journet L., Agrain C., Broz P., and Cornelis G. R. (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 [DOI] [PubMed] [Google Scholar]
- 17. Mota L. J., Journet L., Sorg I., Agrain C., and Cornelis G. R. (2005) Bacterial injectisomes: needle length does matter. Science 307, 1278–1278 [DOI] [PubMed] [Google Scholar]
- 18. Büttner D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agrain C., Sorg I., Paroz C., and Cornelis G. R. (2005) Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 20. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., and Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 [DOI] [PubMed] [Google Scholar]
- 21. Mizuno S., Amida H., Kobayashi N., Aizawa S., and Tate S. (2011) The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus. J. Mol. Biol. 409, 558–573 [DOI] [PubMed] [Google Scholar]
- 22. Bergeron J. R., Fernández L., Wasney G. A., Vuckovic M., Reffuveille F., Hancock R. E., and Strynadka N. C. (2016) The structure of a type 3 secretion system (T3SS) ruler protein suggests a molecular mechanism for needle length sensing. J. Biol. Chem. 291, 1676–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sorg I., Wagner S., Amstutz M., Müller S. A., Broz P., Lussi Y., Engel A., and Cornelis G. R. (2007) YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J. 26, 3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiesand U., Sorg I., Amstutz M., Wagner S., van den Heuvel J., Lührs T., Cornelis G. R., and Heinz D. W. (2009) Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J. Mol. Biol. 385, 854–866 [DOI] [PubMed] [Google Scholar]
- 25. Lountos G. T., Austin B. P., Nallamsetty S., and Waugh D. S. (2009) Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci. 18, 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weise C. F., Login F., H., Ho O., Gröbner G., Wolf-Watz H., and Wolf-Watz M. (2014) Negatively charged lipid membranes promote a disorder-order transition in the Yersinia YscU protein. Biophys. J. 107, 1950–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Björnfot A.-C., Lavander M., Forsberg A., and Wolf-Watz H. (2009) Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J. Bacteriol. 191, 4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frost S., Ho O., Login F. H., Weise C. F., Wolf-Watz H., and Wolf-Watz M. (2012) Autoproteolysis and intramolecular dissociation of Yersinia YscU precedes secretion of its C-terminal polypeptide YscU(CC). PLoS One 7, e49349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Login F. H., and Wolf-Watz H. (2015) YscU/FlhB of Yersinia pseudotuberculosis harbors a C-terminal type III secretion signal. J. Biol. Chem. 290, 26282–26291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kutsukake K., Minamino T., and Yokoseki T. (1994) Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176, 7625–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Williams A. W., Yamaguchi S., Togashi F., Aizawa S. I., Kawagishi I., and Macnab R. M. (1996) Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178, 2960–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sukhan A., Kubori T., and Galán J. E. (2003) Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 185, 3480–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wood S. E., Jin J., and Lloyd S. A. (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190, 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Diepold A., Wiesand U., Amstutz M., and Cornelis G. R. (2012) Assembly of the Yersinia injectisome: the missing pieces. Mol. Microbiol. 85, 878–892 [DOI] [PubMed] [Google Scholar]
- 35. Sal-Man N., Deng W., and Finlay B. B. (2012) EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli. Biochem. J. 442, 119–125 [DOI] [PubMed] [Google Scholar]
- 36. Zhong D., Lefebre M., Kaur K., McDowell M. A., Gdowski C., Jo S., Wang Y., Benedict S. H., Lea S. M., Galan J. E., and De Guzman R. N. (2012) The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J. Biol. Chem. 287, 25303–25311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lefebre M. D., and Galán J. E. (2014) The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 111, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galán J. E., and Wolf-Watz H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 39. Rosqvist R., Magnusson K. E., and Wolf-Watz H. (1994) Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13, 964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Botteaux A., Sani M., Kayath C. A., Boekema E. J., and Allaoui A. (2008) Spa32 interaction with the inner-membrane Spa40 component of the type III secretion system of Shigella flexneri is required for the control of the needle length by a molecular tape measure mechanism. Mol. Microbiol. 70, 1515–1528 [DOI] [PubMed] [Google Scholar]
- 41. Stainier I., Bleves S., Josenhans C., Karmani L., Kerbourch C., Lambermont I., Tötemeyer S., Boyd A., and Cornelis G. R. (2000) YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 37, 1005–1018 [DOI] [PubMed] [Google Scholar]
- 42. Monjarás Feria J. V., Lefebre M. D., Stierhof Y.-D., Galán J. E., and Wagner S. (2015) Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. mBio 6, e01459–01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radics J., Königsmaier L., and Marlovits T. C. (2014) Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol. 21, 82–87 [DOI] [PubMed] [Google Scholar]
- 44. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Akopyan K., Edgren T., Wang-Edgren H., Rosqvist R., Fahlgren A., Wolf-Watz H., and Fallman M. (2011) Translocation of surface-localized effectors in type III secretion. Proc. Natl. Acad. Sci. U.S.A. 108, 1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 47. Helgstrand M., Kraulis P., Allard P., and Härd T. (2000) Ansig for Windows: an interactive computer program for semiautomatic assignment of protein NMR spectra. J. Biomol. NMR 18, 329–336 [DOI] [PubMed] [Google Scholar]
- 48. Williamson M. P. (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.