Abstract
c-Myc-interacting zinc finger protein-1 (Miz-1) is a poly-Cys2His2 zinc finger (ZF) transcriptional regulator of many cell cycle genes. A Miz-1 DNA sequence consensus has recently been identified and has also unveiled Miz-1 functions in other cellular processes, underscoring its importance in the cell. Miz-1 contains 13 ZFs, but it is unknown why Miz-1 has so many ZFs and whether they recognize and bind DNA sequences in a typical fashion. Here, we used NMR to deduce the role of Miz-1 ZFs 1–4 in detecting the Miz-1 consensus sequence and preventing nonspecific DNA binding. In the construct containing the first 4 ZFs, we observed that ZFs 3 and 4 form an unusual compact and stable structure that restricts their motions. Disruption of this compact structure by an electrostatically mismatched A86K mutation profoundly affected the DNA binding properties of the WT construct. On the one hand, Miz1–4WT was found to bind the Miz-1 DNA consensus sequence weakly and through ZFs 1–3 only. On the other hand, the four ZFs in the structurally destabilized Miz1–4A86K mutant bound to the DNA consensus with a 30-fold increase in affinity (100 nm). The formation of such a thermodynamically stable but nonspecific complex is expected to slow down the rate of DNA scanning by Miz-1 during the search for its consensus sequence. Interestingly, we found that the motif stabilizing the compact structure between ZFs 3 and 4 is conserved and enriched in other long poly-ZF proteins. As discussed in detail, our findings support a general role of compact inter-ZF structures in minimizing the formation of off-target DNA complexes.
Keywords: fluorescence anisotropy, nuclear magnetic resonance (NMR), protein dynamic, protein-DNA interaction, protein-protein interaction, structural biology, transcription factor, zinc finger
Introduction
Miz-1 (c-Myc-interacting zinc finger protein-1) is an 88-kDa protein that contains a BTB (Broad complex, Tramtrack, and Bric-a-brac)-POZ (poxvirus and zinc finger) (POZ)3 domain at its N terminus followed by 13 ZFs. It was first identified as a direct interactor of the oncogenic protein c-Myc by yeast two-hybrid screening (1). Miz-1 is an activator of cell cycle regulator genes, such as the cyclin-dependent kinase inhibitors p15INK4 (p15), p21CIP1 (p21), and p57KIP2 (p57) (2–5). Miz-1 activates the transcription of those genes by recruiting different co-activators, such as the histone acetyltransferase p300 and the nucleophosmin (3, 6). Moreover, it was shown that in response to TGF-β, Miz-1 forms a complex with the Smad 2/3/4 proteins to activate the expression of p15. This interaction was shown to involve a region located within the first four ZFs of Miz-1 and the MH1 domain of Smad 3 (7). Whereas the role of Miz-1 in cell cycle regulation is well established, recent studies have underlined its implication in other cell cycle-independent processes, such as autophagy, endocytosis, vesicular trafficking, inflammation and DNA repair (8–11). Moreover, some cytoplasmic functions of Miz-1, such as its implication in the regulation of the Wnt pathway, are emerging, revealing the multifunctional nature of this transcription factor (12).
c-Myc can directly bind Miz-1 and repress the expression of p15, p21, and p57, presumably by abolishing the interaction between Miz-1 and its co-activators (2, 3, 6). On the one hand, numerous studies carried out in different cancer cell lines have shown that the transcriptional repression of Miz-1 by c-Myc is relevant in many different stages of the carcinogenesis process (13). Interestingly, on the other hand, another recent study showed that high levels of Miz-1 cause the repression of c-Myc transcriptional transactivation (14). These results have led to the interesting hypothesis that the c-Myc/Miz-1 balance can dictate cell fate by controlling the transcription of gene networks (15).
The specific DNA binding of Miz-1 is mediated by its ZF motifs (16). ZFs are small domains of ∼30 amino acids that share the following consensus sequence: (F/Y)XCX2–5CX3(F/Y)X5LX2HX3–4H. These motifs are among the most abundant DNA-binding domains in eukaryotes and possess a ββα fold that is stabilized by the coordination of a Zn(II) atom by two conserved Cys and two conserved His side chains (17). Classically, the recognition between ZFs and DNA is mediated by specific hydrogen bonds between side chains of residues −1, 2, 3, and 6 (position relative to the beginning of the α-helix) and DNA bases in the Hoogsteen edge (17). Typically, series of ZFs are connected by highly conserved TGEKP sequence linkers and bind in sets of two or more in the major groove of the DNA with each ZF contacting three DNA bases. These linkers are important to allow for consecutive motifs to optimally fit in the major groove of DNA (18, 19). Variations in the consensus sequence can significantly reduce binding affinity (18, 19). Although not infallible, recognition codes and statistical potentials have been derived to predict the DNA sequences bound by ZF tandems from the nature of the residues at positions −1, 2, 3, and 6 of their recognition helix (17, 20, 21). Miz-1-specific DNA binding has been mainly associated with regions of the proximal and core promoters of p15 and p21 (2). Although one of the regions of the p15 promoter bound by Miz-1 contains an initiator element (INR), no other specific or consensus sequences have emerged until recently. Indeed, two groups recently and independently unveiled similar Miz-1 consensus DNA sequences by genome-wide ChIP-sequencing and Bind-n-Seq methods, respectively (8, 16). However, like many other transcription factors, Miz-1 is found to bind elsewhere on the genome, especially when it is stabilized or overexpressed. In fact, in cancer cells, Miz-1 is found to co-localize with c-Myc at many transcription start sites where INR sequences are present. Most interestingly, Miz-1 can inhibit c-Myc transactivation of genes involved in tumorigenic programs (14, 22). Despite such a critical role in the normal and oncogenic biology of the cell, it is not known whether all of the ZFs or only subsets bind and recognize the Miz-1 consensus sequence, the INR element, or other nonspecific DNA sequences.
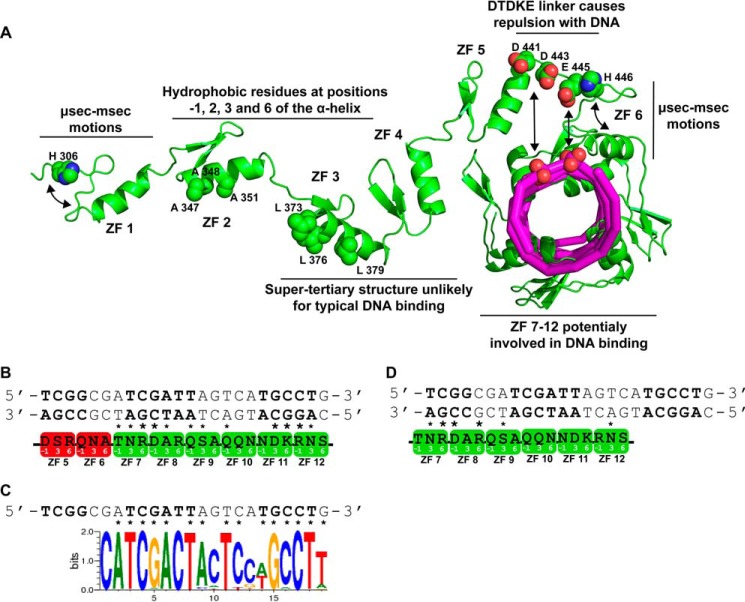
In this context, we have initiated the determination of the solution structure and DNA binding specificity of different constructs of Miz-1 ZF tandems. To date, the structures of Miz-1 ZFs 5–10 have been reported (23, 24). Whereas the ZFs 7–10 possess stable Cys2His2 folds and the usual interdomain dynamics, ZF 6 has been shown to be undergoing conformational exchange on the microsecond to millisecond time scale. Moreover, we showed that the non-canonical DTDKE linker that connects ZFs 5 and 6 is rather unlikely for classical DNA binding because it will cause electrostatic repulsions with the DNA backbone according to the classical ZF recognition mode (24).
Here, we continue with the structural and dynamic characterization as well as the DNA binding properties of Miz-1 with a construct consisting of Miz-1 ZFs 1–4 (Miz1–4). Although the 3D structure of ZFs 2–4 of Miz1–4 present the typical ββα fold, we were not able to determine the structure of the ZF 1 because this motif also undergoes conformational exchange on the microsecond to millisecond time scale. Also, unexpectedly, we discovered that ZFs 3 and 4 make many interdomain contacts, allowing them to form a compact structure that maintains them in a stable orientation unlikely to provide them with typical DNA binding properties.
As can be rationalized from the dynamic and structural characterization of Miz1–4, the construct fails to bind specifically different Miz-1 DNA targets. The motif involved in the stable interdomain interactions between ZFs 3 and 4 was mutated and led to the disruption of the compact structure. This impacted drastically the binding of the Miz1–4 construct to the DNA consensus sequence. Whereas the DNA binding of the WT Miz1–4 construct is mediated by only ZFs 1–3, reintroducing conformational freedom to ZF 4 led to the formation of a complex with all four ZFs bound. Whereas this could suggest a role in the recognition of the consensus by Miz1–4, we show that the actual Miz-1 consensus sequence can be predicted using the identity of the residues in the recognition helices of ZFs 7–12, hence suggesting that recognition of the consensus is accomplished by the C-terminal ZFs. Thus, we instead propose that Miz-1 ZFs 1–3 are involved in scanning DNA and that the compact structure prevents the formation of stable but nonspecific and off-target complexes. In accordance with this view, we find that the motif leading to the formation of the compact structure between ZFs 3 and 4 is conserved and enriched in long poly-ZF (containing 14 ± 6 ZFs).
Results
Folding of Miz1–4 and Resonance Assignments
Circular dichroism was used to characterize and monitor the folding of the Miz1–4 construct. Folding of ZFs is pH- and zinc-dependent because conserved Cys side chains need to be in their deprotonated form to coordinate the Zn(II) atom. At pH 6.5, the addition of Zn(II) led to a transition from a CD spectrum typical of a random coil to an α-helix-like spectrum with a plateau reached at 4 molar eq of Zn(II), suggesting that under these conditions, the four ZFs of Miz1–4 are well folded (Fig. 1B). Moreover, at 5 eq of Zn(II), increasing the pH from 6.5 to 7.5 did not lead to any change of the Miz1–4 CD spectrum, suggesting that the protein is already optimally folded at pH 6.5. Based on this CD characterization, all NMR experiments were carried out at pH 6.5 and with 5 eq of Zn(II). As anticipated from the CD study, the cross-peaks on the 1H-15N HSQC spectrum of Miz1–4 are well dispersed, indicating the presence of stable tertiary structures (Fig. 1C). The data set recorded for Miz1–4 allowed for the assignment of 96 of 107 (90%) of the 1HN, 96 of 112 (86%) of the 15N, 91 of 112 (81%) of the 13C′, 102 of 112 (91%) of the 13Cα, and 94 of 103 (91%) of the 13Cβ. Interestingly, many cross-peaks of ZF 1 are very broad (weak) or broadened beyond detection, preventing us from assigning 9 of its 23 residues. This observation for ZF 1 suggests that this motif could experience some microsecond to millisecond motions (discussed below). Therefore, excluding the ZF 1, 84 of 85 (99%) of 1HN, 84 of 89 (94%) of 15N, 79 of 89 (89%) of 13C′, 88 of 89 (99%) of 13Cα, and 81 of 82 (99%) of 13Cβ were assigned for backbone atom resonances of residues 24–112. Secondary chemical shift (Δδ(Cα − Cβ)) and DANGLE secondary structure prediction obtained for Miz1–4 are shown in Fig. 1D. The four expected α-helices of Miz1–4 ZFs are predicted by DANGLE (based on φ and ψ angles) and present positive Δδ(Cα − Cβ) values. On the other hand, even if β-strands are not all predicted by DANGLE, mostly negative Δδ(Cα − Cβ) values are observed for these secondary structures. Altogether, CD and NMR data show that the four ZFs of Miz1–4 are folded in the selected conditions.
FIGURE 1.
Folding and secondary structure content of Miz1–4. A, alignment of the primary structures of the 13 ZFs of Miz-1. B, far-UV CD spectra of Miz1–4 at different pH and Zn(II) concentrations, demonstrating that the secondary structure content is optimal at pH 6.5 and 4 eq of Zn(II). C, 1H-15N HSQC of Miz1–4. Many resonances of ZF 1 (residues 4–26) are weak or broadened beyond detection. D, the expected secondary structures for the consensus ZF motifs and the secondary structures determined from the chemical shifts of the backbone atoms and the program DANGLE are shown at the top. Secondary chemical shift values for the Cα and Cβ (Δδ (Cα − Cβ)) along with NOE connectivities are displayed and support the presence of the expected secondary structures for Miz1–4 ZFs.
Miz-1 ZF 1 Undergoes Microsecond to Millisecond Conformational Exchange
Conformational exchange has previously been detected by our group for the ZF 6 of Miz-1 (24). It is worth noting that both ZF 1 and 6 contain a His at position 1 of the motif instead of a conserved Phe or Tyr (Fig. 1A). To verify whether the weak signal of Miz-1 ZF 1 in the 1H-15N HSQC is caused by conformational exchange, we recorded a set of CPMG relaxation dispersion experiments. One can notice that R2,eff values of many ZF 1 amides decrease as the applied refocusing pulse frequency (νCPMG) is increased (Fig. 2A). This observation clearly indicates that ZF 1 undergoes some conformational exchange on the microsecond to millisecond time scale. The program NESSY was used to fit the dispersion curves according to the protocol described by Bieri and Gooley (25). All of the residues presenting a relaxation dispersion were best fit by the Carver-Richard equation, which is valid for all time scales (slow to fast conformational exchange limits) and considering a two-state model (26). Fig. 2B shows all of the residues manifesting relaxation dispersion curves. Interestingly, conformational exchange at the C terminus of the α-helix of ZFs 1, 2, and 4 is also observed. Although such motions at the end of ZF α-helices have been suggested to be frequent for C2H2 ZFs (24), cases of conformational exchange affecting the core of the α-helix and the first β-strand are rather unusual. It is worth mentioning that resonances of nine consecutive amides invisible on the 1H-15N HSQC, located in the second β-strand and in the beginning of the α-helix, do not reappear even at the highest νCPMG used. This suggests the presence of conformational exchange, but on a faster time scale (i.e. toward the microsecond limit not covered by CPMG but in the range of the rotating frame relaxation dispersion experiments (T1ρ)) (27). Such a widespread exchange broadening for ZF 1 supports the notion that a His at position 1 promotes motions that affect the local environment of almost all of the amides of the motif.
FIGURE 2.

Conformational exchange of Miz1–4 probed by 15N relaxation dispersion experiments. A, representative CPMG dispersion curves are shown for some residues of the first ZF 1. The R2,eff values for Lys20, Ile23, Ile25, and His29 decrease as a function of the νCPMG, demonstrating the presence of microsecond to millisecond motions in contrast to Glu29, which shows no dispersion. B, Miz1–4 residues having an Rex contribution are colored in orange, and residues invisible on the 1H-15N HSQC spectrum are shown in red. Error bars, S.D.
Solution Structure of Miz-1 ZF 2 and ZF 3–4
Because of the ZF 1 extensive conformational exchange, we were not able to solve its 3D structure. However, we were able to solve the structures of ZFs 2, 3, and 4 and to characterize their linkers. Tandems of ZFs connected by classical TGEKP linkers are relatively independent from one another but are quasi-ordered in the absence of DNA (28, 29). Indeed, these linkers are moderately flexible, with 15N-{1H} NOE values significantly lower than for the ZF core motif (usually ∼0.5 for the linkers and 0.65 for the motifs). In accordance, no long range NOE involving the TGEKP linkers and consecutive ZFs is usually detected in the absence of DNA (23). However, extensive medium and long range NOEs involving residues from the non-canonical SGEAR linker and ZF 3–4 are observed (Fig. 3). This clearly indicates that they are not independent from one another. On the other hand, no long range NOE was observed for the non-canonical linker between ZFs 2 and 3. For these reasons, we decided to calculate the structure of the ZF 2 alone and ZFs 3 and 4 together.
FIGURE 3.
Stacked bar chart of the NOE used for the calculation of the Miz1–4 structure. Intraresidue, sequential, medium range, and long range NOE are colored in light gray, gray, dark gray, and black, respectively. Note the many long range NOEs involve residues from the linker between ZFs 3 and 4 (linkers are highlighted in light gray).
The NOE restraints, dihedral angles, and hydrogen bonds used for structure calculations of each ZF are summarized in Table 1. The superposition of the 20 lowest energy conformers onto their geometric averages demonstrates that all three ZFs are well defined, with backbone RMSDs of 0.42 ± 0.07, 0.27 ± 0.07, and 0.19 ± 0.05 Å for ZFs 2, 3, and 4, respectively (Fig. 4A). Moreover, their structures reveal that they all present the classical ββα topology. The highest backbone RMSD observed for ZF 2 is most probably due to the presence of a Cys at position 4 of the α-helix (Cys47; Fig. 4B) instead of a conserved Leu in the motif hydrophobic core. Indeed, relatively few long range NOEs were detected for this motif. This can be caused by a less stable tertiary structure and/or by the fact that a Cys side chain has a lesser density of 1H and hence generated less NOE. The 15N-{1H} NOEs recorded for Miz1–4 argue in favor of this last hypothesis and suggest that the amides of ZF 2 are as rigid as those of ZF 3–4 on the picosecond to nanosecond time scale (see below).
TABLE 1.
Structural statistics for Miz1–4 ZF2 and ZF 3–4
| ZF 2 | ZF 3–4 | |
|---|---|---|
| Restraints for final structure calculations | ||
| NOE distance restraints | ||
| Intraresidue (|i − j| = 0) | 246 | 801 |
| Sequential (|i − j| = 1) | 115 | 345 |
| Medium range (1 < |i − j| < 5) | 70 | 290 |
| Long range (|i − j| ≥ 5) | 55 | 291 |
| Ambiguous NOE | 7 | 40 |
| Total NOE distance restraints | 493 | 1767 |
| Hydrogen bonds | 10 × 2 | 21 × 2 |
| Zinc ligands | 4 × 2 | 8 × 2 |
| Dihedral angle restraintsa | ||
| Φ and ψ angles | 48 | 96 |
| χ angles | 3 | 10 |
| Structure statistics (20 structures) | ||
| Number of NOE violations >0.5 Å | 0 | 0 |
| Number of dihedral angle violations >5° | 0 | 0 |
| Root mean square deviations from experimental data | ||
| Average distance restraint violation (Å) | 0.028 ± 0.005 | 0.045 ± 0.001 |
| Average dihedral restraint violation (degrees) | 0.41 ± 0.19 | 0.36 ± 0.10 |
| Root mean square deviation to mean coordinates | ||
| Backbone heavy atoms (Å) | 0.67 ± 0.19 | 0.92 ± 0.36 |
| All heavy atoms (Å) | 1.34 ± 0.28 | 1.30 ± 0.30 |
| For ZF consensus sequencesb | ||
| Backbone heavy atoms (Å) | 0.42 ± 0.07 | 0.27 ± 0.07 (ZF 3); 0.19 ± 0.05 (ZF 4) |
| All heavy atoms (Å) | 1.06 ± 0.15 | 0.90 ± 0.12 (ZF 3); 0.71 ± 0.11 (ZF 4) |
| Ramachandran plot statisticsc (%) | ||
| Residues in most favored regions | 86 | 84.7 |
| Residues in additionally allowed regions | 14 | 14.7 |
| Residues in generously allowed regions | 0 | 0.7 |
| Residues in disallowed regions | 0 | 0 |
a Φ and ψ angles were derived from the program DANGLE.
b ZF consensus sequences comprise residues 32–54 for ZF 2, 60–82 for ZF 3, and 88–110 for ZF 4.
c Ramachandran plot statistics were generated using PROCHECK_NMR.
FIGURE 4.
Solution structures of Miz-1 ZF 2 and ZF 3–4. A, the 20 lowest energy conformers of individual ZFs aligned onto the geometric average structure backbone atoms. B, schematic representations of the lowest energy structures. Residues potentially involved in DNA binding at positions −1, 2, 3, and 6, relative to the beginning of the α-helix, are shown in magenta. Residues involved in Zn(II) coordination are in green, and conserved hydrophobic residues are shown in blue. Zn(II) atoms are displayed as gray balls. C, the 20 lowest energy conformers of ZFs 3 and 4 aligned onto the geometric average structure are shown to the left. A schematic representation of the lowest energy structure is shown to the right. Key residues at the ZF 3–4 interface that stabilize the compact structure are shown as green spheres. Residues normally involved in DNA binding based on classical DNA recognition are shown as magenta sticks. D, attribution of some resonances of Ala86 and Leu96 side chains are shown on the aliphatic region of the 1H-13C HSQC at the top. At the bottom, a strip of the 13C-edited NOESY-HSQC on the Ala Cβ plane (18.5 ppm) shows some NOEs observed between the Ala Hβ and many Leu96 side chain protons that dictate the compact structure of Miz-1 ZF 3–4.
The superposition of the 20 lowest energy conformers for ZFs 3 and 4 together shows that they form a well defined compact structure with a backbone RMSD of 0.92 ± 0.36 Å (Fig. 4C). Based on an alignment of 2435 human ZFs realized by Schmidt and Durrett (30), the Ala86, Arg87, and Leu96 have low probabilities to be at their specific position (0, 1.6, and 1.5%, respectively, according to this database). Whereas the Ala86 and the Leu96 participate to the formation of a hydrophobic core between ZFs 3 and 4 with the aliphatic portions of the Lys79 and Lys80 side chains, the Arg87 engages solvent-exposed electrostatic interactions with the conserved Glu85 of the linker (Fig. 4C). Typically, in the DNA-bound state, the conserved Glu forms electrostatic interactions with a conserved basic residue at the position of Lys80. Interestingly, the non-conserved Arg87 appears to compensate for this interaction in the free state. One can also notice that the conserved Lys instead of the Ala86 in the linker would cause electrostatic repulsion with the conserved basic residue at position of Lys80 and destabilize the compact structure. This observation provides an explanation for why this fold is not observed for ZF tandems connected by classical TGEKP linkers. Finally, the non-conserved Ala86, Arg87, and Leu96 stand as the key residues allowing the formation of a cryptic hydrophobic core stabilizing ZFs 3 and 4 in this stable inter-ZF structure.
15N Spin Relaxation Data of Miz1–4 Support the Compact Structure of Miz-1 ZFs 3 and 4
To complement the structure determination of the individual ZF, we recurred to 15N spin relaxation and measured Miz1–4 backbone amide T1, T2, and 15N-{1H} NOEs to characterize the fluctuations in the linkers and the effect of the compact structure on the overall tumbling of Miz1–4 compared with other constructs of four ZFs.
15N-{1H} NOEs give information about local motions of the backbone amides in the picosecond to nanosecond time scale. For a completely rigid amide (S2 = 1), a maximum 15N-{1H} NOE value of 0.87 is expected (at 600 MHz), whereas highly mobile amides (S2 tends toward 0) are expected to have 15N-{1H} NOEs that tend toward zero and negative values. In agreement with the existing literature, most of the residues of ZFs 2–4 have 15N-{1H} NOE values of >0.6 with average values of 0.64 ± 0.12, 0.65 ± 0.09, and 0.66 ± 0.13, respectively (Fig. 5). However, 8 of the 13 residues, for which 15N-{1H} NOE could be recorded for ZF 1, have values <0.6 despite an average of 0.62 ± 0.11. Those results further validate the dynamic nature of the structure of ZF 1 with important fluctuations on both the microsecond to millisecond and the picosecond to nanosecond time scales. The three linkers display average 15N-{1H} NOE values lower than those of ZF core motifs (linker 1, 0.52 ± 0.08; linker 2, 0.35 ± 0.02; linker 3, 0.39 ± 0.23). This suggests that the three linkers undergo more frequent local motions than the four ZFs. Hence, the compact structure adopted by ZFs 3 and 4 does not seem to contribute to rigidify the linker that connects these motifs as compared with what is normally observed for classical linkers in the absence of DNA. That is consistent with the moderately high RMSD observed for ZFs 3 and 4. Those results suggest that residual interdomain motions promote fluctuations of the amide bonds of the linker in that time scale.
FIGURE 5.
Backbone 15N spin relaxation measurements for Miz1–4. Bar plots of {1H}-15N NOE, T1, T2, and T1/T2 values are shown as a function of the Miz1–4 primary structure. Secondary structures expected for the consensus ZF fold are shown at the top and are highlighted in gray on the plots. Error bars, S.D.
The 15N spin relaxation parameters T1, T2, and T1/T2 measured for Miz1–4 are presented in Fig. 5. The average values of T1, T2, and T1/T2 are 541.95 ± 70.82, 99.82 ± 30.67, and 5.75 ± 1.52, respectively. Compared with previous studies of tandems of four ZFs, the average T1/T2 value measured for Miz1–4 is smaller than those of MTF-1 ZFs 1–4 (8.07) and Miz-1 ZFs 5–8 (10.69) (24, 29). For macromolecules, 15N-T1 and T2, respectively, increase and decrease as their effective correlation time (τm) increases. Hence, the smaller T1/T2 ratios observed for Miz1–4 indicate that it tumbles faster than the other two constructs. As described in detail in the supplemental material (rotational diffusion analysis of Miz1–4) and depicted in Fig. 6, this is mainly caused by the compact structure, which orients the N-H vectors of ZF 4 perpendicular to the largest component of Miz1–4 rotational diffusion tensor (Dpar). This leads to a faster effective tumbling for the N-H vectors of this ZF compared with the others. This will result in a decrease in T1 and an increase in T2 for those backbone amides with the net result of decreasing the average T1/T2 ratio (Fig. 5). The details of the simulations presented in Fig. 6 can be found in the supplemental material, and a summary of the simulated parameters is given in Table 2.
FIGURE 6.
15N spin relaxation indicates that Miz1–4 ZFs undergo axially symmetric rotational diffusion in solution and confirms the compact fold of ZFs 3–4. Shown are orientations of ZF 2 (A) and ZF 3 (B) in the axially symmetric diffusional reference frame. Dpar is the rotational diffusion constant parallel to the unique axis (along z) of the diffusion tensor. Dper rotational diffusion constants are perpendicular to the unique axis and aligned along x and y. Both Dper values are equal and smaller than Dpar. The amide bonds used for all analyses are shown as spheres. Experimental (black) and simulated values (red) of the amide bonds used for analysis are shown as a function of the position in the primary structure (C and D), and their α-angle values (E and F) for ZFs 2 and 3, respectively. α-Angles are the angles between the direction of the bond vectors and Dpar. G, ZF 3–4 compact structure aligned in the axially symmetric diffusion tensor (ZF 3 is shown in the same diffusional and molecular frame as in B). Shown are experimental (black) and simulated values (red for completely rigid amides and green for amides with a τe = 1.7 ns and an S2 = 0.75 for ZF 4 amides) as a function of their position in the primary structure (H) and their α-angle values (I). ZF 4 T1/T2 values are best described considering internal motions (τe = 1.7 ns, S2 = 0.75) suggesting a collective wobbling motion of ZF 4 relative to ZF 3. J, the 20 conformers of the final ensemble of ZF 3–4 are aligned for ZF 3 backbone atoms, illustrating the wobbling motions of the ZF 4. The average structure is in magenta. A cone with a semiangle θ of 25°, illustrating the ZF 4 domain motion amplitude considering a S2 of 0.75, is shown. Error bars, S.D.
TABLE 2.
Rotational diffusion analysis of Miz1–4
| ZF analyzed | Isotropic model |
Axially symmetric model |
No. of T1/T2 valuesd | Fe | |||
|---|---|---|---|---|---|---|---|
| Disoa | χ2b | Disoc | Dpar/Dper | χ2 | |||
| 107 s−1 | 107 s−1 | ||||||
| ZF 2 | 2.19 ± 0.02 | 63.15 | 2.28 ± 0.04 | 1.66 ± 0.12 | 5.13 | 9 | 18.4 |
| ZF 3 | 2.19 ± 0.02 | 134.49 | 2.31 ± 0.03 | 1.68 ± 0.08 | 9.31 | 11 | 31.3 |
| ZF 4 | 2.89 ± 0.02 | 8.71 | 2.87 ± 0.05 | 1.29 ± 0.18 | 4.46 | 10 | 1.9 |
| ZF 3–4f | NAg | NA | 2.31 ± 0.03 | 1.68 ± 0.08 | 529.55 | 21 | |
| ZF 3–4h | NA | NA | 2.31 ± 0.03 | 1.68 ± 0.08 | 33.82 | 21 | 125i |
a For the isotropic model, the correlation time (τm) is given by 1/6Diso, where Diso is the isotropic diffusion coefficient.
b χ is an error function, given the sum of the squared difference between the experimental and calculated T1/T2 values divided by the estimated experimental error. See “Experimental Procedures.”
c For the axially symmetric model, the effective correlation time (τm,eff) is given by 1/6Diso, where Diso is equal to (2Dperp + Dpar)/3.
d This column displays the number of T1/T2 values that were used to calculate the rotational diffusion coefficients of the different ZFs.
e Results of the F statistics analysis provided by the R2_R1_Diffusion program.
f Simulations of ZF 3–4 T1/T2 values were realized, keeping ZF 3 in the same molecular and diffusional reference frame as the one obtained from the analysis of ZF 3 alone without considering wobbling motions for the ZF 4 (S2 = 0 and τe = 0 ns).
g NA, not applicable.
h Simulations of ZF 3–4 T1/T2 values were realized, keeping ZF 3 in the same molecular and diffusional reference frame as the one obtained from the analysis of ZF 3 alone considering wobbling motions for the ZF 4 (S2 = 0.75 and τe = 1.7 ns). See “Experimental Procedures.”
i The F parameter was calculated as described by Gagné et al. (63).
The Mutation Ala86 → Lys Destabilizes the Compact Structure Adopted by Miz-1 ZFs 3 and 4
As described above, we hypothesized that in the compact structure fold, the conserved Lys usually present in TGEKP linkers should cause electrostatic repulsions with the conserved basic residue at the position of Miz1–4 Lys80 and destabilize it. Hence, to verify this assertion and to destabilize the compact fold, we mutated the Ala86 to a Lys and prepared the Miz1–4A86K mutant. Interestingly, whereas most of the amide cross-peaks on the mutant 1H-15N HSQC spectrum are at chemical shifts similar to those in the wild type (Fig. 7A), some peaks, mostly residues located in the linker between ZFs 3 and 4 and at the interface of those ZFs, are significantly perturbed (Fig. 7, B and C). This suggests that these amides experience changes in their chemical environment either because of the Ala to Lys substitution or because the Lys side chain prevents the formation of the compact structure as hypothesized. Accordingly, the inspection of a 15N-edited NOESY-HSQC spectrum recorded with the mutant reveals that many NOEs characteristic of the ZF 3–4 compact structure are lost. For instance, we noted that NOEs detected in Miz1–4 between Arg87 HN and Leu97 Hδ*, Phe97 HN, and Thr98 Hα (Fig. 7C) are absent in the spectrum of the mutant.
FIGURE 7.
The mutation Ala86 → Lys destabilizes the compact structure adopted by Miz-1 ZFs 3 and 4. A, overlay of Miz1–4 and Miz1–4A86K 1H-15N HSQC spectra. B, the chemical shift displacements (CSD; Δδ = ((δHN)2 + (δN/6.5)2)½) are displayed as a function of the Miz1–4 primary structure. The residues presenting CSD greater than the average + 0.5 of the S.D. values (i.e. 0.06 ppm) are labeled with asterisks, and their amides are shown as spheres on the Miz-1 ZF 3–4 structure in C. The amide at position 86 is labeled in magenta. Red dashed lines represent examples of some NOEs diagnostic of the compact structure involving the Arg87 HN that were lost upon A86K mutation. D, 15N-T1/T2 values of Miz1–4A86K are shown as black bars, whereas the values recorded for the wild type are shown as red dots. E, orientations of ZFs 2, 3, and 4 of Miz1–4A86K in the axially symmetric diffusional reference frame. Error bars, S.D.
To further validate the loss of the compact structure for the mutant, we measured its amide 15N spin T1 and T2 (supplemental Fig. S1). As described in the supplemental material, if it is disrupted, ZF 4 should realign with the other ZF and slow down the overall tumbling of the tandem. Accordingly, the T1 and T2 values of the mutant are systematically higher and lower, respectively, leading to T1/T2 values that are systematically lower with an average of 7.26 ± 2.03. Such a ratio corresponds to an effective τm of 7.9 ns (Fig. 7D), a value larger than wild type Miz1–4 (6.9 ns) and more consistent with those reported for tandems of four ZFs devoid of stable compact structure (e.g. 8.4 ns for MTF-1 ZFs 1–4) (29). Using the T1/T2 values of the same residues as those used with the wild type construct, we characterized the rotational diffusion analysis of ZFs 2–4 of the mutant. The parameters obtained from the analysis are presented in Table 3. As expected, the rotational diffusion of the three ZFs is significantly best simulated using the axially symmetric model (at the 90% confidence interval) with their α-helices preferentially aligned with Dpar (Fig. 7E). Collectively, these results further validate that ZFs 3 and 4 are no longer perpendicular to each other and that the compact structure is not stable in the Miz1–4A86K mutant. Moreover, the Diso values of the three ZFs have significantly decreased in the mutant compared with the wild type, in agreement with a more anisotropic overall rotational diffusion for this protein construct. In fact, the Diso values calculated for Miz1–4A86K ZFs 2, 3, and 4 (1.99, 1.96, and 2.48 × 107 s−1, respectively) are similar to those calculated by Potter et al. (29) for ZFs 2, 3, and 4 of a construct containing the first four ZFs of MTF-1. Altogether, these results clearly show that the mutation of Ala86 to a Lys prevents the formation of the compact structure adopted by Miz-1 ZF 3–4 and validates the role of the non-canonical SGEAR linker in the formation of such a structure.
TABLE 3.
Rotational diffusion analysis of Miz1–4A86K
| ZF analyzed | Isotropic model |
Axially symmetric model |
No. of T1/T2 values | F | |||
|---|---|---|---|---|---|---|---|
| Diso | χ2 | Diso | Dpar/Dper | χ2 | |||
| 107 s−1 | 107 s−1 | ||||||
| ZF 2 | 1.96 ± 0.02 | 41.66 | 1.99 ± 0.03 | 1.58 ± 0.13 | 2.41 | 8 | 21.7 |
| ZF 3 | 2.50 ± 0.04 | 705.11 | 1.96 ± 0.02 | 1.81 ± 0.11 | 4.18 | 11 | 391.7 |
| ZF 4 | 2.44 ± 0.02 | 25.18 | 2.48 ± 0.04 | 1.40 ± 0.18 | 7.53 | 10 | 4.7 |
Role of Miz-1 ZF 3–4 Compact Structure on the DNA Binding
The structure alignment of the ZF 4 of Miz1–4 with the second ZF of Zif268 bound classically to its target DNA is shown in Fig. 8A (31). One can notice that the compact structure adopted by Miz-1 ZF 3–4 is unlikely to favor the classical DNA binding of the four ZFs. Indeed, if Miz-1 ZF 4 was to bind to the major groove of DNA, ZF 3 would reorient ZFs 1 and 2 away from DNA. Conversely, despite the conformational exchange in ZF 1 and the fact that ZFs 2 and 3 mostly bear hydrophobic residues in their recognition helices (Figs. 4A and 8A), if ZFs 1–3 were to bind DNA, they would also orient ZF 4 away from DNA.
FIGURE 8.
Miz1–4 is not involved in the recognition of Miz-1 DNA cognate sequences. A, ZF 4 is aligned with the ZF 2 of Zif268 bound to its DNA target in a classical fashion (PDB code 1AAY). Due to the compact structure, ZFs 1–3 are projected away from DNA, demonstrating that the fold adopted by ZF 3–4 is unlikely for classical DNA binding by these motifs. The DNA backbone is depicted as a magenta ribbon. B, binding curves obtained from fluorescence anisotropy experiments following the addition of Miz1–4 to the fluorescein-dT-labeled DNA. The apparent Kd values were determined as described under “Experimental Procedures” and resulted from two biological replicates and three technical replicates. The binding curve of Miz1–4A86K to the consensus DNA is shown as a dashed line. C, close-up of the 1H-15N HSQC spectra of Miz1–4 before and after the addition of 1 molar eq of the Miz-1 consensus DNA. The amide cross-peaks of the ZFs 1–3 (residues 4–82) disappear upon the addition of DNA, whereas most of the ZF 4 (residues 88–110, boldface labels) cross-peaks are still visible and at similar chemical shifts. The four potential binding site of Miz1–4 (considering ZF 1–3 binding) are depicted. The residues at positions −1, 3, and 6 of the Miz1–4 ZF recognition helices are shown, and predicted contacts are in boldface type in the consensus sequence. Arg residues at position 6 of the ZFs 1 and 6 are displayed in green to emphasize the fact that an Arg at that position generally strongly and specifically binds a guanine. As depicted on the model shown at the right, the ZFs 1–3 of Miz1–4 are most likely simultaneously bound to the different sites, whereas the compact structure adopted by ZF 3–4 prevents the binding of the ZF 4. D, close-up of the 1H-15N HSQC spectra of Miz1–4A86K before and after the addition of 1 molar eq of the Miz-1 consensus DNA. The amide cross-peaks of the ZFs 1–4 (residues 4–110) shift upon the addition of DNA. The two potential binding sites that would allow the Arg at position 6 of ZFs 1 and 4 to contact a guanine are displayed. The mutation destabilizes the compact structure adopted by ZFs 3 and 4, allowing the four ZFs to contact the consensus DNA and form a well defined specific complex. Error bars, S.D.
Fluorescence anisotropy was used to determine the apparent DNA binding affinity of Miz1–4 to the sequences of the p15 promoter identified by Seoane et al. (2) to be engaged by Miz-1 (region −155 to −140 and −2 to +14 that contains an INR element) and to the consensus sequence recently identified by Wolf et al. (8) (supplemental Fig. S2). As shown in Fig. 8B, the Miz1–4 construct binds to all of the sequences tested, including two unrelated and nonspecific sequences, with a similar low apparent affinity in the micromolar range. Reported affinities for series of three ZFs to nonspecific DNA sequences normally lie in the low micromolar range, whereas their specific binding rather lies in the nanomolar to picomolar range (17, 32–34). Hence, our data clearly demonstrate that Miz-1 ZFs 1–4 bind DNA with an affinity and a specificity that are not sufficient to promote specific molecular recognition of the consensus DNA or the p15 core promoter regions by this transcription factor. To identify which ZFs are involved in the weak and nonspecific binding, we recorded 1H-15N HSQC spectra of Miz1–4 in the presence of 1 molar eq of the consensus DNA (Fig. 8C and supplemental Fig. S3). Strikingly, all of the resonances of the ZFs 1, 2, and 3 become invisible upon DNA addition, whereas most of the amide cross-peaks of the ZF 4, even if significantly less intense, are still visible and at similar chemical shifts. This result clearly supports the notion that ZFs 1–3 are more affected by the presence of DNA than ZF 4 and that they are involved in nonspecific and weak interactions. Indeed, a weak binding of DNA by ZFs 1–3 could lead to chemical exchange (and extreme line broadening) on the microsecond to millisecond time scale through sliding and/or hopping mechanisms, as already described for some proteins in complexes with nonspecific DNA (35, 36). However, we ran relaxation dispersion experiments on the complex and were not able to recover any of the cross-peak intensities (data not shown). When a 15N amide is experiencing a conformational exchange (T2 decrease by Rex−1), it leads to line broadening. However, the evolution of the chemical exchange process leading to the line broadening can be refocused during the CPMG pulse train in the course of a relaxation dispersion experiment. The extent of refocusing can be such that the contribution of the Rex−1 can be completely removed at high field strength or high refocusing frequencies with the result that vanishing peak intensities of broadened resonances can be recovered (37). Hence, the fact that we could not recover the cross-peak intensities of ZFs 1–3 argues against the presence of an active conformational exchange process during the CPMG experiments on the microsecond to millisecond time scale as reported by others (40, 41). However, it is possible that a conformational exchange, caused by the scanning of DNA, occurs on a faster time scale than the one covered by the CPMG experiment. Alternately, our results can be explained by the fact that ZFs 1–3 bind at multiple and non-interchanging sites on the consensus sequence (Fig. 8C). Indeed, based on the recognition code proposed by the group of Pabo (17) and on statistical potentials (21), four different sites could be bound by Miz-1 ZFs 1–3 with six or seven predicted contacts considering residues at positions −1, 3, and 6 of the ZF recognition helices (Fig. 8C). It is noteworthy that these four sites would allow the Arg at position 6 of the ZF 1 to contact a guanine, which is generally one of the strongest and more stringent interactions found for specific ZF DNA binding (17). Assuming that all of these sites are bound with comparable affinities by the construct, it is likely that the backbone amides of ZFs 1–3 experience different static chemical shifts in the different complexes. It should be noted that although the apparent binding constant lies in the micromolar range, at the concentration used in the experiments, all of the protein constructs are expected to be bound to DNA. Coupled to the increase of Miz1–4 apparent molecular weight in the complex, this could lead to the disappearance of the cross-peaks. However, because ZF 4 does not contact DNA, it resides in a more chemically isotropic environment independently of where ZFs 1–3 bind on DNA. This can explain the fact that the intensity of its cross-peaks is reduced because of the increase in the molecular weight but not decreased beyond detection by different chemical environments. But, once again, we cannot rule out the contribution of a conformational exchange caused by a sliding of Miz1–4 on DNA on a faster time scale than the one covered by CPMG experiments.
We show on Fig. 8D the 1H-15N HSQC of Miz1–4A86K in the presence of 1 eq of the consensus DNA. One can notice that the amide resonances of the four ZFs are present on the spectrum and that most of them are perturbed by the presence of DNA (Fig. 8D and supplemental Fig. S3). This strongly suggests that the disruption of the compact structure allows for the four ZFs to engage DNA to form one well defined and stable complex. To further validate this, we have measured the apparent affinity of the Miz1–4A86K for the consensus DNA (Fig. 8B). Accordingly, we found that the affinity of the mutant for the consensus is increased by 30-fold with an apparent Kd of 0.11 μm. As shown in Fig. 8D, only two positions on the consensus can satisfy the strong and specific Arg-guanine interactions defined by the Arg at position 6 of the α-helices of ZF 1 and 4 (Fig. 8D). However, according to the prediction tools (17, 21), site 1 depicted in Fig. 8D would allow for four additional favorable and observed contacts compared with binding site 2, suggesting that it is probably the site preferentially bound by Miz1–4A86K. Although the structure of the complex would need to be solved to make conclusions about the exact nature of this complex, the results presented here confirm that the compact structure does not allow for the recognition of the consensus and leads to weak DNA binding, whereas the reestablishment of the freedom of ZF 4 leads to much stronger DNA binding.
Miz1–4 Does Not Interact Directly with the MH1 of Smad 3
Based on the model presented in Fig. 8A, we reasoned that Miz1–4 could also be involved in protein-protein interactions. Indeed, the three Ala and Leu residues (Ala45, Ala46, and Ala49 and Leu71, Leu74, and Leu77) present on Miz-1 ZFs 2 and 3 at positions normally involved in DNA binding, could constitute a suitable hydrophobic surface for protein-protein interactions if solvent-exposed. In a series of co-immunoprecipitation experiments, Seoane et al. (7) reported strong evidence for the implication of Miz-1 ZFs 1–4 in the formation of a protein complex containing Miz-1 and Smad 2/3/4 in response to the activation of the TGF-β pathway. Moreover, the authors performed GST pull-down experiments and observed a direct interaction between purified Miz-1 and the Smad 3 MH1 (7). For the sake of completeness in our quest to characterize the structural biology of Miz1–4, we cloned, expressed, and purified the MH1 domain of Smad 3 (residues 1–145) and tried to validate the interaction at the atomic level. The CD spectrum and the thermal denaturation of the Smad 3 MH1 obtained are similar to what have already been published for the folded protein (supplemental Fig. S4A) (38). To detect the interaction between Miz1–4 and the Smad 3 MH1, 1H-15N HSQC spectra of 15N-labeled Miz1–4 were recorded in the absence or in the presence of 1 eq of unlabeled Smad 3 MH1. Surprisingly, no resonance from Miz1–4 was perturbed by the addition of the Smad 3 MH1 domain, suggesting that there is no interaction between the two folded domains (supplemental Fig. S4B). No protein precipitation was observed upon the addition of the MH1 domain, and both proteins were intact and in the right ratio as shown by the SDS-polyacrylamide gel presented in supplemental Fig. S4B. Moreover, no interaction was observed between these proteins by CD and by fluorescence anisotropy (supplemental Fig. S4, C–E). Our results demonstrate that there is no direct interaction between Miz-1 ZFs 1–4 and the Smad 3 MH1 domain. Because the folding of both the MH1 domain of Smad 3 and ZFs 1–4 of Miz-1 depend on the coordination of zinc atoms, we hypothesize that the utilization of EDTA and the absence of a reducing agent in the buffer used by Seoane et al. (7) in their pull-down experiments could have led to the formation of nonspecific intermolecular disulfide bonds between Smad 3 MH1 and Miz-1 in vitro. More investigation will be necessary to identify the protein(s) that bridges Miz-1 to Smad 2/3/4 through its first four ZF motifs.
Toward the Understanding of Miz-1-specific DNA Binding to Its Consensus Sequence
Our results suggest that Miz1–4 is most likely not involved in the formation of the native and specific complex between Miz-1 and its cognate sequence. We can also cast doubt on the possibility that ZFs 5 and 6 participate in Miz-1 DNA binding. Indeed, we have previously shown that the Asp and Glu residues in the DTDKE linker between these ZFs would clash with the DNA phosphates. In addition, as shown here for ZF 1, ZF 6 also undergoes extensive conformational exchange, indicating that its binding-competent conformation is probably not always present (24), and this will further contribute to weaken DNA binding. On the other hand, Miz-1 ZFs 7–10 all present stable canonical ZF structures and no unusual dynamic properties or compact folds that could prevent them from binding DNA classically (23, 24). Indeed, these ZFs are linked by classical or quasi-classical (S/T)GEKP linkers (Fig. 1A). To identify potential Miz-1 ZF series that could recognize Miz-1 target DNA consensus sequence derived by Wolf et al. (8), we have used the code proposed by the group of Pabo (17). It is important to remember that the second conserved region of the sequence identified by Wolf et al. (8) (Fig. 9B) has a high homology with the two consensus sequences recently identified by Barrilleaux et al. (16). Strikingly, the best fit obtained involves ZFs 7–12 with 12 interactions unambiguously predicted by the code. Remarkably, the most conserved regions of the consensus are matched to the strongest and most specific canonical side chain-DNA base interactions (i.e. involving either an Asp with a C or an Arg or a Lys with a G) (Fig. 9B). In addition, we have used the approach proposed by Persikov et al. (21), based on statistical potentials derived from a database of structural and thermodynamic data, to predict the DNA sequence bound by Miz-1 ZFs 7–12 (Fig. 9C). This approach also predicts almost perfectly (one mismatch) the second and third most conserved sequences of the consensus. Also according to the recognition code of Pabo, we found that the least conserved region of the consensus, although almost identical to the second, could alternately be bound by ZFs 7 and 8 (Fig. 9D). Perhaps this degenerated cluster of binding sites is important to favor a high local concentration as a step into the mechanism of specific recognition. It is worth mentioning that two other recognition modes for Miz-1 ZFs along the consensus DNA sequence fit well with nine interactions predicted by the code in both cases (supplemental Fig. S5B). However, because those recognition modes involve ZFs 1–6 and the matches do not correlate well with the conserved regions, we believe that these binding modes, although less stable, could serve in the DNA scanning. In this regard, Zandarashvili et al. (34, 36) recently published studies demonstrating the importance of the presence of a lower affinity DNA binding ZF for optimal target search efficiency by a protein containing three ZFs. Analogously, we propose that ZFs 1–6 serve a similar purpose for the efficient recognition of the consensus by subsets of ZFs 7–12.
FIGURE 9.
Model of Miz-1 recognition of its DNA consensus sequence. A, summary of the dynamic and structural properties of Miz-1 ZFs and their impact on DNA binding. B, the consensus DNA of Miz-1, identified by Wolf et al. (8), is depicted, and the three conserved regions of the consensus are displayed in boldface letters. Residues at positions −1, 3, and 6 of the 12 consecutive Miz-1 ZF recognition helices are shown. Predicted contacts with the consensus are labeled with asterisks. Boldface asterisks identify strong interactions (i.e. Arg and Lys with a G or Asp with a C). ZFs 1–6 are colored in red, and ZFs 7–12 are shown in green. C, sequence logo predicted to be bound by Miz-1 ZFs 7–12 (reverse complement), according to the statistical approach developed by Persikov et al. (21). Note the striking similarity with the consensus sequence. DNA bases predicted to be specifically bound are labeled with asterisks. D, model depicting the recognition mode of Miz-1 ZFs for the binding of the first conserved DNA region of the consensus sequence.
Discussion
Miz-1 is a transcription factor that contains 12 consecutive ZFs. Despite its crucial role in many decisive aspects of cell biology, the precise identity and role of its ZFs in the recognition of cognate DNA sequences remain unknown. In this study, we report the structural and dynamic characterization of ZFs 1–4 of Miz-1 by solution state NMR as well as their DNA binding properties. Our structural and dynamic analysis revealed that ZF 1 undergoes peculiar conformational exchange and revealed the existence of an unusual compact fold involving ZFs 3 and 4. Similar conformational exchange has been reported by us for ZF 6 (24), and those two ZFs represent, to the best of our knowledge, the only reported cases so far. As discussed below, the compact structure we have unveiled has rarely been discussed or seen so far, but it could be conserved in other poly-ZF proteins yet to be studied.
Our results and previous work from our laboratory enable us to propose that the first six ZFs of Miz-1 are involved in DNA scanning in the search for specific DNA binding through ZFs 7–12 (Fig. 9). Indeed, ZFs 1–4 bind to nonspecific DNA with the same marginal affinity as cognate DNA sequences (p15 core promoter (2) and the newly identified consensus (8)). Moreover, the unusual flexibility of the ZF 1 and the compact fold of ZFs 3 and 4 coupled with already published evidence on ZFs 5 and 6 render canonical DNA binding improbable for those ZFs. Interestingly, this region of Miz-1, more specifically ZFs 1–4, has been reported to bind to the MH1 domain of Smad 3 and to play a role in the TGF-β-dependent activation of p15 (7). Unfortunately, using biophysical approaches and the purified constructs, we were unable to validate a direct interaction. More investigations will be necessary to identify the protein(s) that bridges Miz-1 to the Smad proteins.
Of more general interest, although specific DNA recognition by poly-ZFs generally involves series of 3–5 ZFs, the number of ZFs per poly-ZF protein has increased throughout evolution with an average of eight ZFs and an upper limit of around 40 ZFs for humans (17, 30). This observation suggests that these ubiquitous motifs are implicated in biological functions other than DNA binding. Accordingly, there are an increasing number of reported cases of ZFs involved in protein and RNA interactions (39, 40). Some groups have even suggested that the potential of ZFs for protein interactions is likely to be greater than for DNA interaction (41). However, as discussed elsewhere, the structural and dynamic characteristics dictating the functional role of ZFs are poorly understood (40). Tandems of ZFs are generally separated by highly conserved TGEKP sequence linkers that have been shown to be important for optimal DNA binding (18, 19, 42, 43). Accordingly, different groups have proposed that the nature of ZF tandem linkers could enable the prediction of their functions (18, 19, 44). Although many studies have addressed the functional impact of linker variations on DNA binding by ZFs, little is known about the structural impact of such variations. Here we report that the SGEAR linker between Miz-1 ZFs 3 and 4 participates in the formation of a compact structure that maintains those ZFs in an orientation unlikely for typical DNA binding. To assess the recurrence of similar compact structures potentially adopted by ZF tandems among poly-ZF proteins, we searched for other reported cases in the literature and tried to find rules that could allow identification of them.
To the best of our knowledge, there is only one other reported case of consecutive ZFs separated by a five-residue linker that adopt a compact structure in the absence of DNA. Indeed, the linker between ZFs 5 and 6 of MBP-1 is involved in interfinger residue contacts that maintain them in a conformation different from what is usually observed for classical DNA binding (PDB entry 1BBO, supplemental Fig. S6A) (45). Interestingly, although MBP-1 ZF 5–6 and Miz1–4 ZF 3–4 conformations are quite different, in both cases, the presence of a long side chain amino acid at the position of Miz1–4 Leu96 (Lys40 for MBP-1) instead of a conserved small residue is involved (probability of 74.41% for an Ala, Ser, or Gly at that position according to Schmidt and Durrett (30)). In both cases, this residue participates in the formation of a hydrophobic core at the finger interface with a non-conserved hydrophobic residue in the linker (MBP-1 Val27 and Miz-1 Ala86) (supplemental Fig. S6, B and C) and a residue from the preceding ZF. The observation of the structure of INSM1 ZF 4–5, deposited in the PDB under the code 2D9H (unpublished), shows that these motifs also adopt a compact structure unlikely to be fit for classical DNA binding (supplemental Fig. S6A). Incidentally, these ZFs do not participate in the specific DNA binding of INSM1 (46). Strikingly, the residues of INSM1 involved in hydrophobic interaction that maintain INSM1 ZFs 4 and 5 in a fixed orientation (Leu48, Val56, and Thr65) are at the same position as the one responsible for the compact structure adopted by Miz1–4 ZFs 3–4 (Lys80, Ala86, and Leu96) (supplemental Fig. S6C). However, it should be noted that those ZFs are connected by a shorter linker (4 residues instead of 5) that could also contribute to fix their orientation. Another ZF tandem linked by a non-classical four-residue linker (KKIK) with restrained ZF flexibility was recently identified for ZFAT ZFs 4 and 5 (PDB entry 2RV7) (47). Once again, a hydrophobic residue of the linker (ZFAT Ile63) is involved in hydrophobic interactions that stabilize the orientation of those ZFs. However, in this case, the linker hydrophobic residue does not interact with the residue present at the position of Miz1–4 Leu96 but instead interacts with the ZF 4 Tyr41 (supplemental Fig. S6E). Interestingly, this residue also has a low probability of 0.53% to be at its specific position based on the Schmidt and Durrett alignment (30). This last example suggests that many different scenarios can cause the restriction of ZF orientations. Finally, the inspection of the structure of TFIIIA bound to RNA shows that its ZFs 4 and 5 adopt a non-classical orientation necessary for the specific RNA binding of this poly-ZF (48, 49). Strikingly, once again, hydrophobic interactions between a long side chain residue at the position of Miz1–4 Leu96 (Arg145) and two hydrophobic residues at the position of Miz1–4 Lys80 and Ala86 (TFIIIA Phe127 and Leu133) are observed in this unusual fold (supplemental Fig. S6F). Interestingly, the particular orientation of ZF 4–5 is also observed in the crystal structure of TFIIIA bound to DNA and prevents the ZF 4 from contacting DNA bases (50). In this case, the ZF 4 is acting as a spacer element between the ZFs 1–3 and 5 that mediate the specific DNA recognition (supplemental Fig. S6F). This example of TFIIIA ZF 4–5 illustrates that a better understanding of ZF tandem structural biology could help to predict their functions.
Based on the structural information described above, we ran a PHI-BLAST search against the Swiss-Prot databank to identify ZF tandems that could form compact structures similar to the one observed for Miz-1 ZFs 3 and 4, MBP-1 ZFs 5 and 6, INSM1 ZFs 4 and 5, and TFIIIA ZFs 4 and 5. We found 170 ZF tandems containing a hydrophobic residue at position 3 or 4 of the linker and a non-conserved hydrophobic residue (all except Ala) or a long side chain residue (Arg or Lys) at the position of Miz1–4 Leu96 (the PHI-BLAST pattern used is shown in supplemental Fig. S7). Interestingly, an average of 14 ± 6 ZFs per poly-ZF are observed for the 66 human proteins identified in the BLAST. This suggests an enrichment of unusual tandem fold for poly-ZFs containing more than the average ZFs in human (average of eight ZFs per poly-ZF in humans). In another PHI-BLAST, we identified 37 proteins in the data bank that contain long hydrophobic side chains at the positions of ZFAT Tyr42 and Ile63. It is worth mentioning that the residue numbering used here is the one provided in the different PDB files.
The above observations suggest that compact structures similar to the one observed between Miz-1 ZFs 3 and 4 are probably not uncommon among poly-ZFs and may possess a conserved role. Based on our results, one possible role could be to limit the number of consecutive ZFs binding in a canonical fashion and consequently fine tune the DNA affinity of poly-ZFs proteins, allowing for an optimal DNA scanning speed in the search for specific sequences. Indeed, it can be anticipated that too many ZFs binding in a concerted fashion to nonspecific DNA could lead to the formation of stable nonspecific complexes that slow down or even halt the DNA scanning process. Another role for ZF tandem compact structures could be to prevent DNA binding of ZF subsets so they can serve as platforms to engage other proteins or RNA.
To conclude, the results presented in this study not only contribute to deepen our knowledge of Miz-1 structural biology, but also provide key elements essential to our understanding of Miz-1-specific DNA binding. The structural, dynamic, and functional results presented here for Miz-1 ZFs 1–4 together with previous structural studies provide strong evidence for the implication of Miz-1 ZFs 1–6 in the scanning of DNA and ZFs 7–12 in specific DNA binding. Moreover, the analysis of the unusual compact structure adopted by Miz-1 ZFs 3 and 4 suggests that other ZF tandems could form such structures and contributes to expanding our understanding of these ubiquitous and versatile motifs.
Experimental Procedures
Preparation of the Miz1–4 and Miz1–4A86K Constructs
The cDNA of ZFs 1–4 of Miz-1 (Miz1–4, residues 304–414) was generated by PCR from the complete cDNA of Miz-1 using primers F (5′-CAT ATG GTC ATC CAC AAG TGC GAG GAC TGT GG-3′) and R (5′-GGA TCC CTA GCC GCT GTG CAC CAG CTG GTG-3′). The PCR product was inserted into pDrive (Qiagen), digested by NdeI and BamHI, and inserted in pET-3a (Novagen) by the same restriction sites. The construct was transformed in BL21 star (DE3) competent cells (Invitrogen). Bacteria were grown in M9 medium containing 15NH4Cl and [13C]glucose to an A600 nm of 0.6, were induced for 15 h at 37 °C with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and were harvested by centrifugation. The cell pellet was resuspended in a lysis buffer (700 mm NaCl, 50 mm KH2PO4 (pH 4.5)), frozen at −80 °C for at least 1 h, thawed in hot water, and then sonicated. The lysate was treated with 100 mm DTT and DNase I for 1 h at 37 °C and was centrifuged at 17,500 relative centrifugal force for 30 min. The soluble fraction was diluted 5 times in buffer A (0.12 m citric acid-Na2HPO4 (pH 3), 8 m urea) and purified by FPLC with a HiTrap SP HP column (GE Healthcare). After an extensive wash with buffer B (0.12 m citric acid-Na2HPO4 (pH 3)), Miz1–4 was eluted by a gradient of NaCl. Ultracentrifugation (Amicon Ultra-15, Millipore) was used to concentrate the purified protein in H2O containing 0.1% TFA. The protein was finally purified by HPLC with a Discovery BIO Wide Pore C18 column (Supelco) preconditioned with H2O-0.1% TFA and was eluted by a gradient of acetonitrile. The fractions containing purified Miz1–4 were lyophilized and kept at −20 °C until resuspension in the desired buffer. The mutant Miz1–4A86K was purified according to the same protocol.
CD Spectropolarimetry
CD measurements were realized with a Jasco J-810 spectropolarimeter equipped with a Jasco Peltier-type thermostat. The CD spectra were recorded with a 1-mm path length quartz cell at 20 °C and were averaged over 10 scans with a wavelength step of 0.2 nm. The spectra were smoothed using Spectra Manager (JASCO Corp.). Miz1–4 spectra were recorded at 25 μm in a solution containing 10 mm acetic acid, 10 mm cacodylate, 2 mm TCEP, and the indicated ZnCl2 concentrations. The Smad 3 MH1 spectrum was recorded at 15 μm in a buffer containing 10 mm Tris (pH 8), 50 mm KCl, and 2.5 mm β-mercaptoethanol. Data were converted from CD signal to mean residue ellipticity using the equation, [θ] = (δ·MRW)/(10·c·l), where δ is the ellipticity in degrees, MRW is the mean residue weight, c is the concentration of the sample (g/ml), and l is the path length (cm). Thermal denaturation of Smad 3 MH1 was realized by following the CD signal at 222 nm as a function of the temperature at a rate of 1 °C/min.
NMR Spectroscopy
All NMR experiments were recorded at 25 °C on a Varian (Agilent) Unity INOVA operating at a 1H frequency of 600 MHz equipped with an indirect detection H/C/N room temperature probe with a z axis pulsed field gradient capacity. Samples were prepared at a final concentration of 0.7–1.0 mm Miz1–4 in the NMR buffer (10 mm acetic acid, 10 mm BisTris (pH 6.5), 50 mm KCl, 2 mm TCEP, 5 eq of ZnCl2, and 10% D2O). Miz1–4 backbone and side chain assignments were obtained from standard triple resonance experiments. 2D 1H-15N HSQC and 3D HNCO, HNCACB, and CBCA(CO)NH spectra were used for the 1H, 15N, and 13C assignments of the protein backbone. Side chain 1H and 13C assignments were obtained using 2D 1H-13C HSQC and 3D H(CCCO)NH, (H)CC(CO)NH, and HCCH-TOCSY spectra. The 1H and 13C resonance assignments of the aromatic rings of Phe, Tyr, and His were realized using 3D aliphatic and aromatic 13C-edited NOESY-HSQC and 15N-edited NOESY-HSQC. 3D NOESY spectra were recorded with mixing times of 50 and 150 ms. All of the pulse sequences used were taken from the Biopack repertoire.
NMR 1H-15N HSQC experiments run to verify the interaction between Miz1–4 (or Miz1–4A86K) and the Miz-1 consensus DNA have been recorded with a 250 μm concentration of both the protein and the DNA (see Fig. 8B for the DNA sequence) at 25 °C in a solution containing 20 mm BisTris (pH 7), 150 mm KCl, 1.25 mm ZnCl2, 2 mm TCEP, and 10% D2O.
Structure Calculations
All NOEs were assigned manually and converted into distance restraints using CcpNmr Analysis (51). The program DANGLE was used to obtain φ and ψ dihedral angles based on the backbone and 13Cβ chemical shift values (52). Structures were calculated using the program ARIA version 2.2 in conjunction with CNS (53, 54). Calculations for Miz1–4 ZF 2 and ZFs 3 and 4 were first carried out without a zinc atom and zinc coordination restraints. For all three ZFs, the conserved cysteines and histidines were positioned correctly to allow coordination of zinc. In the following calculations, a Zn(II) ion was added, and zinc coordination distance restraints were specified to ARIA (2.3 Å for Zn(II)-Sγ and 2.0 Å for Zn(II)-Nϵ2). The 20 lowest energy conformers of 300 for the final iteration of the calculation were refined in water and submitted to PROCHECK_NMR for initial validation of the structural quality of our models. The final structure ensembles of ZF 2 (residues 30–58) and ZFs 3 and 4 (residues 58–112) were deposited in the PDB under identification codes 2N25 and 2N26, respectively. A full report of the structural quality assessment can be found in the PDB with the access codes. NMR resonance assignments for Miz1–4 were deposited in the BMRB under accession number 25587.
15N Spin Relaxation
15N-T1, T2, and 15N-{1H} NOE experiments were recorded using previously described pulse sequences available in the Biopack repertoire (55). Delays of 0, 10, 30, 90, 120, 150, 250, 350, 450, 550, 650, 800, and 1000 ms and of 0, 10, 30, 50, 70, 90, 110, 130, 150, 170, 190, 210, and 250 ms were used to obtain T1 and T2, respectively. 15N-{1H} NOE measurements were done by comparing 1H-15N HSQC spectra with and without a 6-s proton saturation. 15N backbone CMPG relaxation dispersion profiles were acquired in a constant time (60 ms) and interleaved manner using a modified version of the pulse sequence from the Biopack repertoire based on the work of Palmer et al. (56) and Mulder et al. (57). The field strengths (νcpmg = 1/4τcpmg) used were 28.57, 57.14, 85.71, 114.29, 142.86, 171.43, 200, 228.57, 285.71, 371.43, 428.57, 514.29, 542.86, 571.43, 657.14, 714.29, 771.43, and 800 Hz. Experiments with νcpmg of 28.57, 85.71, 228.57, and 571.43 Hz were repeated twice to estimate the extent of the experimental error. CPMG data (R2,eff) were fitted with the program NESSY according to the protocol described by Bieri and Gooley (25). R2,eff values were calculated using the equation, R2,eff(νcpmg) = 1/T·ln·(I(νcpmg)/I0), where T is the total and constant duration of the CPMG period (60 ms), and I(νcpmg) and I0 are the resonance intensities of the 15N and 1HN correlations in the presence and absence of a refocusing pulse, respectively.
Rotational Diffusion Analysis
The programs pdbinertia and R2R1_diffusion (A.G. Palmer) were used to characterize the rotational diffusion of the different ZFs, as described elsewhere by Tjandra et al. (58). Briefly, for the isotropic case, τm (or 1/6Diso) was determined by calculating T1/T2 and comparing it with the experimental value to optimize with R2_R1_diffusion the following error function,
| (Eq. 1) |
where σ is the experimental uncertainty. As described elsewhere (59), 15N-T1/T2 ratios are independent of S2 (order parameter of the backbone amide vector) and τe (internal correlation time of the backbone amide vector) for rigid amides (high S2 values) and fast internal picosecond motions (τe). For the axially symmetric diffusion case, the molecular reference frames of the PDB files of the geometric averages of ZF 2 (2N25), ZF 3, ZF 4, and ZF 3–4 (2N26) were rotated into their inertia frames with the program pdbinertia. Then a local τm was determined by optimizing the χ2 function. The local τm is a function of cosine angles α (the angle between the amide vectors and the unique axis of the inertia tensor), Dper, and Dpar (the rotational diffusion coefficients perpendicular and parallel to the unique axis of the inertia and diffusion tensors). Hence, during the minimization of the error function, it is in fact Dper, Dpar, and the unique axis of the diffusion tensor that are optimized. An effective correlation time (τm,eff) is also obtained and is given by 1/6Diso, where Diso is equal to (2Dper + Dpar)/3 with an anisotropy or Dratio given by Dpar/Dper. Finally, an F statistics test (F) is run within R2R1_diffusion. Confidence in the isotropic or the axially symmetric diffusion model was evaluated by comparing the F values with the critical F values determined by the R2R1_diffusion program at the 90% confidence interval following 500 Monte Carlo simulations, as described elsewhere (60). A large F value indicates that the improvement in χ2 by using more parameters (axially symmetric versus isotropic) is statistically significant or not obtained by chance. As suggested elsewhere (61), the data (residues) used for the calculations were filtered to exclude residues with T1/T2 ratios higher than the average plus 1.5 S.D. values (considering ZFs individually) and those with the lowest 15N-{1H} NOE. The former residues are likely to undergo conformational exchange and have T2 values artificially lowered (or T1/T2 ratios artificially increased) by Rex−1, and the latter are likely to be unfolded (low order parameter) and undergo concurrent motions in addition to rotational diffusion. Moreover, residues presenting resonance overlap or having T1 or T2 values with >10% uncertainty were removed to use only high quality data for the analysis. A total of 9, 11, 10, and 21 residues were retained for ZF 2, ZF 3, ZF 4, and ZF 3–4, respectively. T1/T2 values of the selected amides presenting uncertainties lower than 5% (generally comprised between 0.5 and 2.5%) were set to 5% in the calculation to account for the inherent deviations of individual 15N chemical shift anisotropy (29). Once the calculations were completed, all of the conformers of the ensemble of structures deposited in the PDB were aligned onto their corresponding average structures in the final diffusional reference frame, and α-angles were extracted for all of the conformers. Using the diffusion parameters estimated, the α-angles and T1/T2 ratios were back-calculated with an in-house written program using the S2-τe spectral density function (with S2 = 1 and τe = 0) (62) with an axially symmetric diffusion (58) for all of the conformers of the ensemble of structures deposited in the PDB to estimate uncertainties on the calculated α and T1/T2 ratios. To simulate the effect of slow nanosecond motions of ZF 4 relative to ZF 3 on its T1/T2 ratios, we used its α values, Dper and Diso, determined as described above and minimized the difference between the calculated and experimental ratio by manually changing S2 and τe (in the nanosecond time scale) with our in-house written program. Note that we neglected the effect of picosecond motions in this simulation.
Fluorescence Anisotropy
Oligonucleotides were purchase from IDT. The different lyophilized fluorescein-dT-labeled oligonucleotides were resuspended in water at 100 μm and were mixed to a 1:1 ratio with the non-labeled complementary strand. The mixtures were then incubated at 95 °C and slowly cooled down to room temperature to form the duplex. The double-stranded probes were diluted to 15 nm in a solution containing 100 mm Tris (pH 7.5), 50 mm KCl, 1 mm TCEP, and 500 μm ZnCl2 and were added to a 1-cm path length quartz cell. Data were recorded on a HITACHI F-2000 spectrofluorimeter mounted in the L-shape configuration with the excitation and emission wavelengths set to 490 and 520 nm, respectively, and both slits set to 10 nm. Miz1–4 protein was gradually added from a stock at 450 μm. An equilibration of 5 min was allowed before the acquisition of each point. The anisotropy was calculated according to the following equation, where r is the anisotropy, I‖ is the fluorescence intensity when the polarizers are paralleli and I⊥ is the fluorescence intensity when they are perpendicular.
| (Eq. 2) |
The dissociation constants were obtained assuming a two-state binding using the following classical equation,
| (Eq. 3) |
where Δrobs is the observed anisotropy change at a particular protein/DNA ratio, Δrmax is the maximum anisotropy change at binding saturation, R is the protein/DNA ratio, and [DNA]0 is the total concentration of the double-stranded oligonucleotide.
Author Contributions
P. L. and M. B. designed the study and wrote the paper. M. B. constructed the vector for the expression of Miz1–4WT and purified the protein. V. R. and M. M. constructed the vector for the expression of Miz1–4A86K and purified the protein. M. B. conducted all of the NMR, CD, and fluorescence experiments and analyzed the data with P. L. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Frank Hänel (Hans-Knöll-Institut für Naturstoff-Forschung e.V, Jena, Germany) for kindly providing the Miz-1 cDNA and Dr. Yves L. Dory (Université de Sherbrooke) for access to the HPLC system and advice on its use. Finally, we thank Prof. Jim Omichinski (Université de Montréal) for help with the refolding protocol of zinc fingers.
This work was supported by the Natural Science and Engineering Research Council (NSERC) of Canada (grant to P. L. and studentships to M. B.) and by the Regroupement Stratégique sur la Fonction, la Structure, et l'Ingénierie des Protéines (PROTEO). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S7.
- POZ
- BTB (Broad complex, Tramtrack, and Bric-a-brac)-POZ (poxvirus and zinc finger)
- CPMG
- Car-Purcell-Meiboom-Gill
- HSQC
- heteronuclear single quantum coherence
- INR
- initiator element
- RMSD
- root mean square deviation
- ZF
- Cys2His2 zinc finger
- PDB
- Protein Data Bank
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane.
References
- 1. Peukert K., Staller P., Schneider A., Carmichael G., Hänel F., and Eilers M. (1997) An alternative pathway for gene regulation by Myc. EMBO J. 16, 5672–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seoane J., Le H. V., and Massagué J. (2002) Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734 [DOI] [PubMed] [Google Scholar]
- 3. Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massague J., Hänel F., and Eilers M. (2001) Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3, 392–399 [DOI] [PubMed] [Google Scholar]
- 4. Adhikary S., Peukert K., Karsunky H., Beuger V., Lutz W., Elsässer H. P., Möröy T., and Eilers M. (2003) Miz1 is required for early embryonic development during gastrulation. Mol. Cell. Biol. 23, 7648–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Riggelen J., Müller J., Otto T., Beuger V., Yetil A., Choi P. S., Kosan C., Möröy T., Felsher D. W., and Eilers M. (2010) The interaction between Myc and Miz1 is required to antagonize TGFβ-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 24, 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wanzel M., Russ A. C., Kleine-Kohlbrecher D., Colombo E., Pelicci P. G., and Eilers M. (2008) A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat. Cell Biol. 10, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 7. Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., and Massagué J. (2001) TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3, 400–408 [DOI] [PubMed] [Google Scholar]
- 8. Wolf E., Gebhardt A., Kawauchi D., Walz S., von Eyss B., Wagner N., Renninger C., Krohne G., Asan E., Roussel M. F., and Eilers M. (2013) Miz1 is required to maintain autophagic flux. Nat. Commun. 4, 2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Do-Umehara H. C., Chen C., Urich D., Zhou L., Qiu J., Jang S., Zander A., Baker M. A., Eilers M., Sporn P. H., Ridge K. M., Sznajder J. I., Budinger G. R., Mutlu G. M., Lin A., and Liu J. (2013) Suppression of inflammation and acute lung injury by Miz1 via repression of C/EBP-δ. Nat. Immunol. 14, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Rashkovan M., Vadnais C., Ross J., Gigoux M., Suh W. K., Gu W., Kosan C., and Möröy T. (2014) Miz-1 regulates translation of Trp53 via ribosomal protein L22 in cells undergoing V(D)J recombination. Proc. Natl. Acad. Sci. U.S.A. 111, E5411–E5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen P., Wang W., Zhang Y., Yuan Y., and Wu Y. (2016) Decreased MIZ1 expression in severe experimental acute pancreatitis: a rat study. Dig. Dis. Sci. 61, 758–766 [DOI] [PubMed] [Google Scholar]
- 12. Huang Y., Wang P., Chen H., Ding Y., and Chen Y. G. (2015) Myc-interacting zinc-finger protein 1 positively regulates Wnt signalling by protecting Dishevelled from Dapper1-mediated degradation. Biochem. J. 466, 499–509 [DOI] [PubMed] [Google Scholar]
- 13. Wiese K. E., Walz S., von Eyss B., Wolf E., Athineos D., Sansom O., and Eilers M. (2013) The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb. Perspect. Med. 3, a014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peter S., Bultinck J., Myant K., Jaenicke L. A., Walz S., Müller J., Gmachl M., Treu M., Boehmelt G., Ade C. P., Schmitz W., Wiegering A., Otto C., Popov N., Sansom O., et al. (2014) Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol. Med 6, 1525–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schaub F. X., and Cleveland J. L. (2014) Tipping the MYC-MIZ1 balance: targeting the HUWE1 ubiquitin ligase selectively blocks MYC-activated genes. EMBO Mol. Med. 6, 1509–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrilleaux B. L., Burow D., Lockwood S. H., Yu A., Segal D. J., and Knoepfler P. S. (2014) Miz-1 activates gene expression via a novel consensus DNA binding motif. PLoS One 9, e101151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfe S. A., Nekludova L., and Pabo C. O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212 [DOI] [PubMed] [Google Scholar]
- 18. Choo Y., and Klug A. (1993) A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 21, 3341–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryan R. F., and Darby M. K. (1998) The role of zinc finger linkers in p43 and TFIIIA binding to 5S rRNA and DNA. Nucleic Acids Res. 26, 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaplan T., Friedman N., and Margalit H. (2005) Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 1, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Persikov A. V., and Singh M. (2014) De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 42, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walz S., Lorenzin F., Morton J., Wiese K. E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M., Roels F., Wüstefeld T., Fischer M., Teichmann M., Zender L., et al. (2014) Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bédard M., Maltais L., Beaulieu M. E., Bilodeau J., Bernard D., and Lavigne P. (2012) NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10. J. Biomol. NMR 54, 317–323 [DOI] [PubMed] [Google Scholar]
- 24. Bernard D., Bédard M., Bilodeau J., and Lavigne P. (2013) Structural and dynamical characterization of the Miz-1 zinc fingers 5–8 by solution-state NMR. J. Biomol. NMR 57, 103–116 [DOI] [PubMed] [Google Scholar]
- 25. Bieri M., and Gooley P. R. (2011) Automated NMR relaxation dispersion data analysis using NESSY. BMC Bioinformatics 12, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carver J. P., and Richards R. E. (1972) General 2-site solution for chemical exchange produced dependence of t2 upon Carr–Purcell pulse separation. J. Magn. Reson. 6, 89–105 [Google Scholar]
- 27. Kleckner I. R., and Foster M. P. (2011) An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 1814, 942–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brüschweiler R., Liao X., and Wright P. E. (1995) Long-range motional restrictions in a multidomain zinc-finger protein from anisotropic tumbling. Science 268, 886–889 [DOI] [PubMed] [Google Scholar]
- 29. Potter B. M., Feng L. S., Parasuram P., Matskevich V. A., Wilson J. A., Andrews G. K., and Laity J. H. (2005) The six zinc fingers of metal-responsive element binding transcription factor-1 form stable and quasi-ordered structures with relatively small differences in zinc affinities. J. Biol. Chem. 280, 28529–28540 [DOI] [PubMed] [Google Scholar]
- 30. Schmidt D., and Durrett R. (2004) Adaptive evolution drives the diversification of zinc-finger binding domains. Mol. Biol. Evol. 21, 2326–2339 [DOI] [PubMed] [Google Scholar]
- 31. Elrod-Erickson M., Rould M. A., Nekludova L., and Pabo C. O. (1996) Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc finger-DNA interactions. Structure 4, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 32. Greisman H. A., and Pabo C. O. (1997) A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science 275, 657–661 [DOI] [PubMed] [Google Scholar]
- 33. Wolfe S. A., Greisman H. A., Ramm E. I., and Pabo C. O. (1999) Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 285, 1917–1934 [DOI] [PubMed] [Google Scholar]
- 34. Zandarashvili L., Esadze A., Vuzman D., Kemme C. A., Levy Y., and Iwahara J. (2015) Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc. Natl. Acad. Sci. U.S.A. 112, E5142–E5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedman J. I., Majumdar A., and Stivers J. T. (2009) Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 37, 3493–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zandarashvili L., Vuzman D., Esadze A., Takayama Y., Sahu D., Levy Y., and Iwahara J. (2012) Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. U.S.A. 109, E1724–E1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mittermaier A. K., and Kay L. E.. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 34, 601–611 [DOI] [PubMed] [Google Scholar]
- 38. BabuRajendran N., Palasingam P., Narasimhan K., Sun W., Prabhakar S., Jauch R., and Kolatkar P. R. (2010) Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-β effectors. Nucleic Acids Res. 38, 3477–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown R. S. (2005) Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15, 94–98 [DOI] [PubMed] [Google Scholar]
- 40. Brayer K. J., and Segal D. J. (2008) Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131 [DOI] [PubMed] [Google Scholar]
- 41. Brayer K. J., Kulshreshtha S., and Segal D. J. (2008) The protein-binding potential of C2H2 zinc finger domains. Cell Biochem. Biophys. 51, 9–19 [DOI] [PubMed] [Google Scholar]
- 42. Thukral S. K., Morrison M. L., and Young E. T. (1991) Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. Proc. Natl. Acad. Sci. U.S.A. 88, 9188–9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clemens K. R., Zhang P., Liao X., McBryant S. J., Wright P. E., and Gottesfeld J. M. (1994) Relative contributions of the zinc fingers of transcription factor IIIA to the energetics of DNA binding. J. Mol. Biol. 244, 23–35 [DOI] [PubMed] [Google Scholar]
- 44. Laity J. H., Dyson H. J., and Wright P. E. (2000) DNA-induced α-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J. Mol. Biol. 295, 719–727 [DOI] [PubMed] [Google Scholar]
- 45. Omichinski J. G., Clore G. M., Robien M., Sakaguchi K., Appella E., and Gronenborn A. M. (1992) High-resolution solution structure of the double Cys2His2 zinc finger from the human enhancer binding protein MBP-1. Biochemistry 31, 3907–3917 [DOI] [PubMed] [Google Scholar]
- 46. Breslin M. B., Zhu M., Notkins A. L., and Lan M. S. (2002) Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 30, 1038–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tochio N., Umehara T., Nakabayashi K., Yoneyama M., Tsuda K., Shirouzu M., Koshiba S., Watanabe S., Kigawa T., Sasazuki T., Shirasawa S., and Yokoyama S. (2015) Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT. J. Struct. Funct. Genomics 16, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Searles M. A., Lu D., and Klug A. (2003) Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426, 96–100 [DOI] [PubMed] [Google Scholar]
- 49. Lee B. M., Xu J., Clarkson B. K., Martinez-Yamout M. A., Dyson H. J., Case D. A., Gottesfeld J. M., and Wright P. E. (2006) Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 357, 275–291 [DOI] [PubMed] [Google Scholar]
- 50. Nolte R. T., Conlin R. M., Harrison S. C., and Brown R. S. (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl. Acad. Sci. U.S.A. 95, 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 52. Cheung M.-S., Maguire M. L., Stevens T. J., and Broadhurst R. W. (2010) DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202, 223–233 [DOI] [PubMed] [Google Scholar]
- 53. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 54. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., and Nilges M. (2007) ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 55. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., and Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 56. Palmer A. G. 3rd, Kroenke C. D., and Loria J. P. (2001) Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 339, 204–238 [DOI] [PubMed] [Google Scholar]
- 57. Mulder F. A., Skrynnikov N. R., Hon B., Dahlquist F. W., and Kay L. E. (2001) Measurement of slow (micros-ms) time scale dynamics in protein side chains by 15N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 123, 967–975 [DOI] [PubMed] [Google Scholar]
- 58. Tjandra N., Feller S. E., Pastor R. W., and Bax A. (1995) Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc. 117, 12562–12566 [Google Scholar]
- 59. Kay L. E., Torchia D. A., and Bax A. (1989) Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 60. Mandel A. M., Akke M., Palmer A. G. 3rd (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 61. Baber J. L., Szabo A., and Tjandra N. (2001) Analysis of slow interdomain motion of macromolecules using NMR relaxation data. J. Am. Chem. Soc. 123, 3953–3959 [DOI] [PubMed] [Google Scholar]
- 62. Lipari G., and Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104, 4559–4570 [Google Scholar]
- 63. Gagné S. M., Tsuda S., Spyracopoulos L., Kay L. E., and Sykes B. D. (1998) Backbone and methyl dynamics of the regulatory domain of troponin C: anisotropic rotational diffusion and contribution of conformational entropy to calcium affinity. J. Mol. Biol. 278, 667–686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.